Significance
Photosystem II (PSII) is a photo-oxidoreductase that harnesses light energy to use water to make fuel. Water oxidation occurs at a metal cluster in the active site called the oxygen-evolving complex (OEC). Understanding PSII function has provided design principles for synthetic solar fuel catalysts; however, the details of water oxidation are obscured by the multiple states through which the mechanism proceeds, differences between species, and lability of the OEC. To better understand PSII function, we solved its structure from Synechocystis sp. PCC 6803. We observe significant differences compared with PSII from thermophilic cyanobacteria that highlight the need for reexamination of previous data using this structure for interpretation. The structure also provides a platform for studies of site-directed mutations of PSII.
Keywords: photosynthesis, photosystem II, water oxidation, oxygen-evolving complex, PsbQ
Abstract
Photosystem II (PSII) enables global-scale, light-driven water oxidation. Genetic manipulation of PSII from the mesophilic cyanobacterium Synechocystis sp. PCC 6803 has provided insights into the mechanism of water oxidation; however, the lack of a high-resolution structure of oxygen-evolving PSII from this organism has limited the interpretation of biophysical data to models based on structures of thermophilic cyanobacterial PSII. Here, we report the cryo-electron microscopy structure of PSII from Synechocystis sp. PCC 6803 at 1.93-Å resolution. A number of differences are observed relative to thermophilic PSII structures, including the following: the extrinsic subunit PsbQ is maintained, the C terminus of the D1 subunit is flexible, some waters near the active site are partially occupied, and differences in the PsbV subunit block the Large (O1) water channel. These features strongly influence the structural picture of PSII, especially as it pertains to the mechanism of water oxidation.
Photosystem II (PSII) is a multisubunit membrane protein complex found in oxygenic phototrophs. It is the only global-scale catalyst for solar fuel production (1). Thus, understanding its enzymatic function is relevant to a variety of fields including synthetic photocatalysis, crop optimization, biofuel production, and evolutionary biology. PSII catalyzes redox processes that result in the transfer of electrons from water on the “donor side” (lumenal side) to lower potential plastoquinone on the “acceptor side” (stromal side). The active site of PSII contains an inorganic metallocofactor, a Mn4CaO5 cluster termed the oxygen-evolving complex (OEC) that is coordinated primarily by one of the core subunits, D1. The mechanism of water oxidation at the OEC proceeds through a series of five intermediate storage states, or S-states, S0 through S4, referred to as the Kok cycle, which have been the subject of extensive study (2–4). Various water channels extend away from the OEC toward the lumen and are involved in substrate water delivery, proton transfer, and oxygen release (2–5). In cyanobacteria, the extrinsic subunits bound to the lumenal side, PsbO, PsbQ, PsbU, and PsbV, play different roles in stabilizing the OEC and protecting it from reductants that can disrupt water oxidation, and they also contribute to water channel formation (6, 7). In plants and algae, analogous but evolutionarily distinct extrinsic subunits are bound. Despite many years of interdisciplinary efforts aimed at understanding the details of water oxidation, many fundamental aspects remain unclear due to the highly complicated nature of the reaction mechanism.
The ability to perform site-directed mutagenesis has contributed greatly to our understanding of PSII, especially the mechanism of water oxidation. A genetic system was first reported in the late 1980s in a mesophilic cyanobacterium, Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) (8), which is a convenient model organism to study due to its ability to grow in the absence of PSII when supplemented with glucose. Given the ease of genetic transformation (9, 10), many site-directed mutated PSII complexes have been investigated (11–13). PSII point mutants have been used in biophysical experiments such as Fourier transform infrared spectroscopy (FTIR) (14, 15), electron paramagnetic resonance spectroscopy (16), mass spectrometry (17), and more to assess the role of individual amino acids in PSII function. Equally as informative have been studies of PSII using structural approaches such as X-ray crystallography (4) and more recently cryo-electron microscopy (cryo-EM) (18), although no structures have yet been solved of PSII with point mutations.
All active cyanobacterial PSII structures reported to date have been solved from the thermophiles Thermosynechococcus vulcanus and Thermosynechococcus elongatus, which are nearly identical in sequence and structure. Typically, it is assumed that the structure and function of PSII are highly conserved between mesophilic and thermophilic cyanobacteria. As a result, biophysical data obtained from PSII isolated from mesophilic cyanobacteria are commonly interpreted using molecular structures of PSII obtained from thermophilic cyanobacteria. This approach may be problematic because membrane proteins from mesophilic and thermophilic organisms are generally known to exhibit differences in molecular interactions (19), and there are obvious sequence differences between PSII subunits from mesophilic and thermophilic cyanobacteria that imply structural and functional variation (SI Appendix, Fig. S1 and Table S1). Thus, a glaring roadblock in the study of PSII is the lack of a molecular structure from the easily transformable mesophilic cyanobacterium, Synechocystis 6803, that is the source of so much biochemical and biophysical data.
Here, we present the cryo-EM structure of PSII from Synechocystis 6803 at 1.93-Å resolution. This structure is compared with previously solved structures of PSII, especially a recent cryo-EM structure from T. vulcanus (20). The extrinsic subunit PsbQ that is missing in other cyanobacterial PSII structures is bound at a site nearly identical to that of PsbQ in higher plants and algae. The OEC exhibits differences in its coordination by the C terminus of the D1 subunit and variable positions of some nearby amino acids and waters are observed. The Large water channel (also known as the O1 channel) present in PSII from thermophilic cyanobacteria (21) is blocked in the Synechocystis 6803 PSII structure, owing to differences in the PsbV subunit. To assess radiation damage to the sample, a low-dose Synechocystis 6803 PSII cryo-EM structure was determined at a resolution of 2.01 Å, which shows negligible differences compared with the full-dose structure. The high-resolution structure presented here provides a view of fully assembled PSII from a mesophilic cyanobacterium. This provides a model from which past and future biochemical and biophysical data can be more accurately interpreted and it establishes a method to solve molecular structures of PSII with single amino acid substitutions that will address questions regarding PSII activity.
Results
Cryo-EM and Overall Structure.
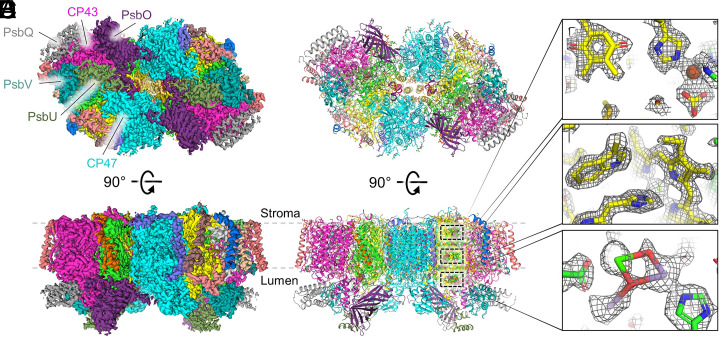
Highly active PSII core complexes were isolated from Synechocystis 6803 and used for cryo-EM as described in Materials and Methods. Data processing and statistics are reported in SI Appendix, Figs. S2–S4 and Table S2 and led to an electrostatic potential (ESP) map at 1.93-Å global resolution (Fig. 1). The structure of PSII from Synechocystis 6803 is of the dimeric complex, with each monomer containing 21 subunits: the core D1 and D2 subunits, the core antenna CP47 and CP43 subunits, 13 peripheral transmembrane subunits (PsbE, PsbF, PsbH, PsbI, PsbJ, PsbK, PsbL, PsbM, Psb30, PsbT, PsbY, PsbX, and PsbZ), and four lumenal extrinsic subunits (PsbO, PsbQ, PsbU, and PsbV). In each PSII monomer, these subunits coordinate one OEC, one nonheme Fe, one bicarbonate, two hemes, two Cl ions, two plastoquinones, two pheophytins, three Ca ions in addition to the Ca ion in the OEC, 10 carotenoids, 24 lipids, 31 n-dodecyl β-D-maltoside (β-DM) molecules, 35 chlorophyll (Chl) a molecules, and 618 water molecules.
Fig. 1.
Cryo-EM structure of PSII from Synechocystis 6803 at 1.93-Å resolution. (A) The unsharpened map (4σ) where ESP map regions assigned to subunits are colored individually. Top shows a lumenal view in which the lumenal domains of CP47 and CP43 and the extrinsic subunits of one monomer are labeled. Bottom shows a membrane plane view. (B) The structural model derived from the cryo-EM map in the same views as A. In the membrane plane view, three regions are boxed which correspond to panels C–E from top to bottom. (C) Map and model of QA and the nonheme Fe coordinated by D2-His214 and bicarbonate (13σ). (D) Map and model of the axial ligation of PB in P680 by D2-His197 and the nearby D2-Trp191 (13σ). (E) Map and model of the OEC and nearby D1-Asp170 and D1-His332 residues (12σ). In C–E, the sharpened map is shown.
Comparing the Synechocystis 6803 and T. vulcanus PSII cryo-EM structures, the cofactors are conserved other than one carotenoid, BCR101 (SI Appendix, Fig. S5). BCR101 was recently suggested to play an important role in stabilizing the dimeric interface in PSII from thermophilic cyanobacteria (22), which our data do not support for PSII from Synechocystis 6803. Cα superpositions of the subunits show that most maintain relatively similar structures (SI Appendix, Table S3), except some peripheral subunits, especially the transmembrane subunits Psb30, PsbT, PsbX, PsbY, and PsbZ, and the extrinsic subunits PsbO, PsbU, and PsbV. A general view of subunit similarity that includes the sequence and structure from Synechocystis 6803 PSII compared with T. vulcanus PSII can be gleaned by plotting the ratio of sequence identity to the RMSD of their Cα superposition (SI Appendix, Fig. S6). The results are consistent with the Cα superpositions alone in which a cluster of subunits exhibit especially low similarity: PsbO, Psb30, PsbT, PsbU, PsbV, PsbX, PsbY, and PsbZ. In all cases, these subunits are the most peripheral from the core D1 and D2 subunits. The poor conservation of PsbO, PsbU, and PsbV is somewhat surprising, because one might expect that water and proton channels, which are partially formed by these extrinsic subunits and are important for water oxidation, would be conserved among PSII structures. The poor conservation of extrinsic subunits is also exemplified by the unique Ca-binding sites on the lumenal surface of Synechocystis 6803 PSII compared with T. vulcanus PSII (SI Appendix, Text S1 and Fig. S7).
Binding, Conservation, and Role of Cyanobacterial PsbQ.
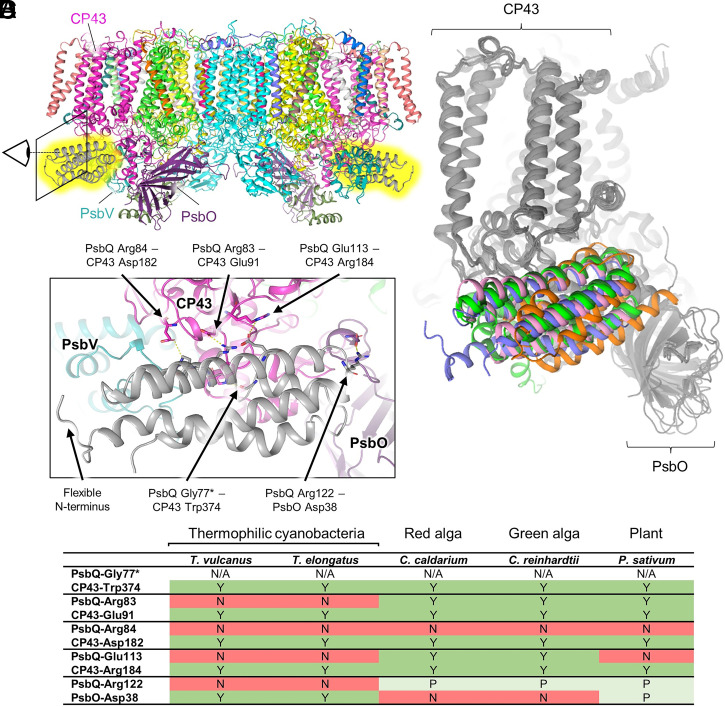
Fig. 2 shows the binding of PsbQ to the Synechocystis 6803 PSII core. PsbQ is a membrane-tethered, 4-helix bundle extrinsic subunit of PSII (23) first identified in PSII preparations from Synechocystis 6803 (24) and has been shown to enhance PSII oxygen evolution (25–27). It is thought to be similar to PsbQ and PsbQ′ from higher plants and algae and also similar to Psb27, a transiently bound subunit involved in PSII assembly (23), all of which are also 4-helix bundle subunits (28). X-ray diffraction (XRD) structures of PsbQ isolated from Synechocystis 6803 and T. elongatus have been reported (29, 30) but not in complex with PSII. The PsbQ subunit is bound to the Synechocystis 6803 PSII structure on the lumenal domain of CP43 between PsbO and PsbV (Fig. 2A), a site analogous to where PsbQ and PsbQ′ are found in PSII structures from plants and algae and to where Psb27 is bound in other cyanobacterial PSII structures (Fig. 2B). This is surprising due to low sequence identity (SI Appendix, Table S4) and somewhat poor Cα superposition RMSDs (SI Appendix, Table S5) between PsbQ homologs. Mass spectrometry–based cross-linking experiments previously suggested that in cyanobacteria, PsbQ binds closer to the center of the PSII dimer, associated with PsbO and CP47 (31), which is inconsistent with our observations. However, the authors of recent cyanobacterial PSII structures containing Psb27 bound between PsbO and PsbV suggested that PsbQ also binds between PsbO and PsbV (32, 33), which is consistent with our observations. Further discussion regarding PsbQ can be found in SI Appendix, Text S2. Most notably, PsbQ appears to be partially occupied in the cryo-EM map (SI Appendix, Fig. S8). Docking simulations suggest that its binding is primarily driven by specific electrostatic interactions (SI Appendix, Table S6 and Figs. S9 and S10) that are not conserved in thermophilic cyanobacteria (Fig. 2), possibly explaining the absence of PsbQ in XRD and cryo-EM structures of T. vulcanus and T. elongatus PSII. We also see no structural evidence that the binding of PsbQ to the PSII core induces a significant conformational change in the protein complex. Thus, it is possible that PsbQ provides stability to the cyanobacterial PSII complex by increasing the number of molecular interactions needing to be broken before all the extrinsic subunits dissociate, leaving the OEC exposed to external reductants.
Fig. 2.
Interactions of PsbQ with the cyanobacterial PSII core and conservation in PsbQ homologs. (A) Membrane plane view of Synechocystis 6803 PSII highlighting in yellow the location of PsbQ on each monomer. The eye and box show the view in C. (B) Superposition of the D1, D2, CP43, CP47, and PsbO subunits shown in gray and the PsbQ homologs and Psb27 subunits shown in color. PsbQ from Synechocystis 6803 is blue (mesophilic cyanobacterium), PsbQ from Pisum sativum is dark green (plant, PDB 5XNL) (34), PsbQ from Chlamydomonas reinhardtii is bright green (green algae, PDB 6KAC) (35), PsbQ′ from Cyanidium caldarium is pink (red algae, PDB 4YUU) (36), and Psb27 from T. vulcanus is orange (thermophilic cyanobacterium, PDB 7CZL) (32). (C) View near PsbQ (gray) and its interactions with CP43 (magenta) and PsbO (purple). Important interacting residues and the N terminus are labeled. (D) Amino acid pairs that interact between PsbQ and CP47 or PsbO are listed. Whether the corresponding amino acid in the Synechocystis 6803 sequence is conserved in other organisms is provided, in which Y = full conservation (green), P = partial conservation in which charge is maintained (light green), and N = no conservation (red). The asterisk (*) denotes the residue in which the backbone carbonyl oxygen atom is involved in the interaction, therefore the conservation of the sidechain is labeled "N/A" (not applicable).
Unique Characteristics near the OEC.
The positions of the OEC metal ions in the Synechocystis 6803 PSII structure are similar to other PSII structures, but some of its ligands exhibit variable positions. The metal-bridging oxygen atoms in the OEC are also modeled in similar positions; however, the spatial resolution of negatively charged oxygen atom ligands adjacent to the positively charged metal ions in the OEC is generally lower than that of uncharged atoms away from the OEC, rendering the model positions of metal-bridging oxygen atoms in the OEC relatively unreliable.
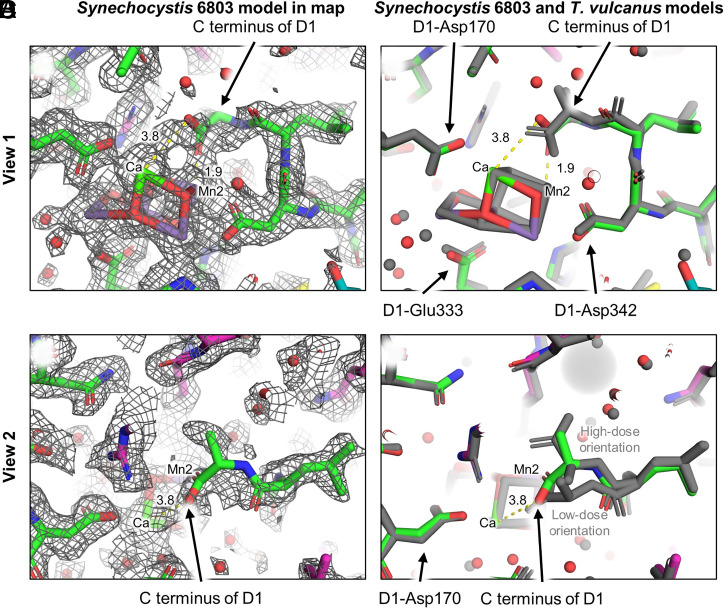
In other PSII structures, the C terminus of D1, D1-Ala344, has a bridging bidentate coordination to the OEC in which one of the carboxylate oxygen atoms ligates Mn2 and the other ligates Ca. In the Synechocystis 6803 PSII structure, however, the map corresponding to the C-terminal Ala344 is poorly resolved (Fig. 3), causing its orientation to be ambiguous, and is therefore modeled with low confidence. The map ambiguity is most likely a consequence of the C terminus being flexible, found in different orientations within individual particles used to create the ensemble cryo-EM map. It is unclear whether this flexibility is an intrinsic characteristic of Synechocystis 6803 PSII or whether it is a result of isolation procedures or experimental conditions as discussed recently (37). Kato et al. recently suggested that electron radiation damage induces structural changes in the orientation of the D1 C terminus in thermophilic PSII (20). Our modeled position of the D1 C terminus in the Synechocystis 6803 PSII structure is found between the two orientations modeled by Kato et al. associated with high-dose and low-dose orientations (Fig. 3D). Thus, the flexibility observed for the D1 C terminus in the Synechocystis 6803 PSII structure may at least partially be due to a fraction of radiation-damaged OECs in the ensemble data used for three-dimensional reconstruction of the ESP map.
Fig. 3.
ESP map and structure comparison near the D1 C terminus. (A) View 1 of the Synechocystis 6803 PSII structure within the map showing ambiguity in the C terminus orientation. (B) View 1 showing the superposition of the Synechocystis 6803 PSII (colored) and T. vulcanus PSII (gray) cryo-EM structures. (C) View 2 of the Synechocystis 6803 PSII structure within the map showing ambiguity in the C terminus orientation. (D) View 2 showing the superposition of the Synechocystis 6803 PSII (colored) and T. vulcanus PSII cryo-EM structures (gray). Note that in the high-dose PSII cryo-EM structure from T. vulcanus, two orientations of the C terminus were modeled that were assigned as high-dose and low-dose orientations (20), which are labeled. Maps are shown at 7σ. Indicated distances are in units of angstrom.
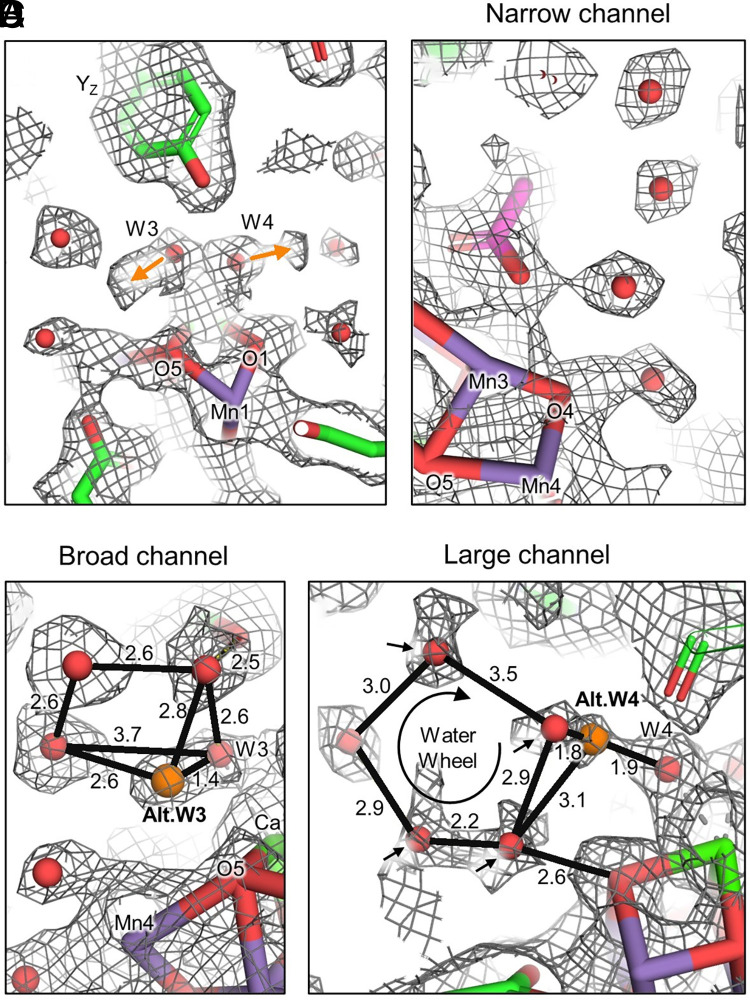
In all PSII structures, the water ligands W3 and W4 ligate the Ca ion of the OEC and are involved in nearby H-bonding with water clusters and YZ. W3, W4, and the nearby water clusters are maintained in the Synechocystis 6803 PSII structure, but some of their associated ESP lack spherical symmetry and/or have low signal intensity when compared with other high-confidence waters in the structure, suggesting alternate positions and decreased occupancy, respectively (Fig. 4). The primary position of W3 ligates the Ca ion of the OEC, as is typically observed, and is within H-bonding distance of YZ, a feature that is also found in the cryo-EM structures of T. vulcanus PSII (20) but not in XRD structures of PSII (SI Appendix, Fig. S11). W3 is also within H-bonding distance of an adjacent water in the Broad channel (also known as the Cl1 channel) as is also typically observed; however, the directionality of the map asymmetry corresponding to W3 suggests an alternate position where the ligation to the Ca ion is lost (it is moved out of H-bonding distance from YZ) and W3 moves toward some Broad channel waters, H-bonding with a second water molecule (Fig. 4C). Similarly, the map asymmetry for W4 suggests either coordination to the Ca, and within H-bonding distance to YZ, or possible alternate positions out of H-bonding distance with YZ and at or near a water in the Large channel that is incorporated in a “water wheel,” a cluster of five water molecules near O1 previously suggested to be involved in substrate water delivery (38) (Fig. 4D). Furthermore, the waters closest to the OEC in the water wheel appear to have decreased signal intensity implying relatively low occupancy compared with waters in the Narrow channel (also known as the O4 channel, Fig. 4B), suggesting high mobility. We note that these water mobility observations are probably not a result of radiation damage, because they were not observed in either the full-dose or low-dose cryo-EM structures of T. vulcanus PSII (20).
Fig. 4.
Alternate positions of W3 and W4 and low-occupancy waters near the OEC in Synechocystis 6803 PSII. (A) Alternate positions of W3 and W4. Both of these waters exhibit primary positions as ligands to the Ca ion of the OEC, but alternate positions are observed toward each of two water clusters (orange arrows). (B) First four waters of the Narrow water channel that exhibit relatively high occupancy and spherical distribution of ESP, suggesting low mobility. (C) The primary position of W3 coordinated to the Ca of the OEC and H-bonded to a nearby water in the Broad channel. The alternate position moves W3 out of a bonding distance with the Ca, and the H-bond is replaced with an H-bond to another Broad channel water. (D) The primary position of W4 coordinated to the Ca of the OEC and H-bonded to low-occupancy waters (black arrows) nearby in the Large channel’s water wheel. Indicated distances are in units of angstrom.
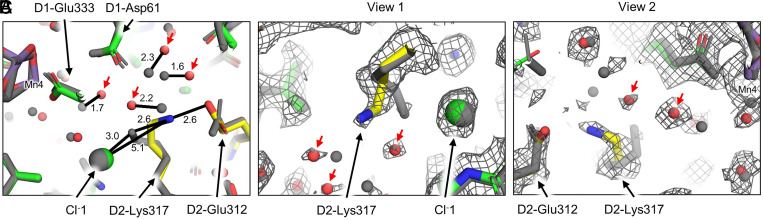
In addition to the ambiguous position of the D1 C terminus, variable positions of W3 and W4, and low-occupancy waters in the water wheel, the Synechocystis 6803 PSII structure also exhibits differences compared with other cyanobacterial PSII structures near D2-Lys317 (Fig. 5). Canonically, D2-Lys317 interacts with Cl−1 that is coupled to the OEC and influences S-state transitions (39, 40). D2-Lys317 and Cl−1 are thought to be involved in a proton-transfer pathway from the OEC to the lumen (39, 40), the latter shielding formation of a salt bridge between D2-Lys317 and D1-Asp61 (40). The position of Cl−1 is the same in the Synechocystis 6803 and T. vulcanus PSII structures, but the position of the D2-Lys317 sidechain in the former is 5.1 Å away from Cl−1 compared with 3.0 Å in the latter (Fig. 5 and SI Appendix, Fig. S12). This results in the D2-Lys317 sidechain of Synechocystis 6803 being positioned within H-bonding distance of the D2-Glu312 sidechain at 2.6 Å. Four nearby water molecules that form the beginning of the Broad channel are also shifted ∼1.5 to 2.5 Å that may be involved in proton egress. We note that D2-Lys317 in the Synechocystis 6803 PSII cryo-EM map appears to exhibit a possible alternate conformation in a site closer to Cl−1, more similar to the D2-Lys317 position observed in most other PSII structures (SI Appendix, Fig. S12). The primary and possible alternate positions are consistent with FTIR results showing that Cl−1, D2-Lys317, D2-Glu312, and other residues are involved in a proton egress pathway (41), which is an important aspect of the water-oxidation mechanism. The primary D2-Lys317 position is reminiscent of the XRD structure of herbicide-treated T. elongatus PSII in which a putative alternate Cl−1-binding site was detected near Cl−1 (42), supporting a two-site Cl−1-binding model (43). Although no alternate Cl−1-binding site is identified in the Synechocystis 6803 PSII cryo-EM structure, we cannot exclude a low-occupancy Cl− species at a site where water is presently modeled.
Fig. 5.
Structural differences near D1-Lys317. (A) Superposition of the Synechocystis 6803 (colored, PDB 7N8O) and T. vulcanus (gray, PDB 7D1T) PSII cryo-EM structures. Indicated distances are shown between the two structures in units of angstrom. (B) View 1 from a perspective near PsbO additionally showing the full-dose cryo-EM map. (C) View 2 from a perspective near D1-Asp61 additionally showing the full-dose cryo-EM map. In all panels, waters in the Synechocystis 6803 PSII structure that are in significantly different positions compared with waters in the T. vulcanus structure are denoted with red arrows. Maps are shown at 5σ. A magnified view near D2-Lys317 is shown in SI Appendix, Fig. S12 that highlights a possible alternate position of its sidechain.
Blockage of the Large Water Channel.
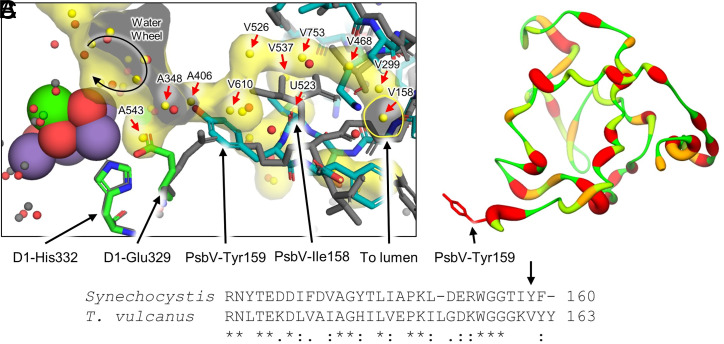
The water channels in PSII are important for substrate water delivery and proton release from the OEC (44, 45). Poor conservation of the extrinsic PSII subunits between Synechocystis 6803 and T. vulcanus (SI Appendix, Fig. S6) suggests possible differences in the water channels that may have important functional implications. We examined the water channels as defined previously (46), comparing the PSII structures from Synechocystis 6803 and T. vulcanus. Within ∼15 Å of the OEC, waters in the Broad and Narrow channels are relatively well conserved; however, the Large channel is disrupted due to differences in the PsbV subunit (Fig. 6). In T. vulcanus PSII, the Large channel begins with the water wheel and extends away from the OEC into PsbV and into the lumen. Though the water wheel waters are maintained in the Synechocystis 6803 PSII structure, 11 of the waters that create the main channel toward the lumen within ∼15 Å of the OEC are not present in the Synechocystis 6803 PSII model (red arrows in Fig. 6A), which in the deposited Protein Data Bank coordinates of T. vulcanus PSII (PDB 3ARC) are waters A543, A348, A406, V610, V526, V537, U512, V753, V468, V299, and V158 (these waters are named by their associated “chain” followed by their “residue number” in the PDB file). Those waters closest to the OEC, A543, A348, and A406 are adjacent to the water wheel. The primary blockage is due to the nonconserved PsbV-Tyr159 of Synechocystis 6803 that extends toward the OEC, filling the position of A406 and causing a shift in the D1-Glu329 sidechain toward the OEC that in turn fills the position of A543 (Fig. 6A). It is possible that A348, which fills the position between PsbV-Tyr159 and D1-Glu329, is present but at very low occupancy. The other eight waters further away from the water wheel, V610, V526, V537, U512, V753, V468, V299, and V158, could also be shifted to different regions and exhibit high mobility, but the presence of PsbV-Ile158 occupies the positions of V537 and U523. Regardless, the blockage by PsbV-Tyr159 cuts off significant portions of the Large channel to the lumen.
Fig. 6.
Comparison of the positions of waters in the Large channel and differences associated with PsbV. (A) Superposition of the T. vulcanus PSII structure (gray sticks, PDB 3ARC) with its Large channel and associated waters colored in yellow and the Synechocystis 6803 PSII structure in colors (waters are colored red). Due to nonconserved C-terminal residues in PsbV, PsbV-Tyr159 of Synechocystis 6803 blocks the network of waters found in T. vulcanus, which also causes a shift in the orientation of D1-Glu329 in Synechocystis 6803. The water wheel, previously suggested to be important for substrate water delivery, is shown as a circular arrow. Red arrows denote those water positions that are not conserved in the Synechocystis 6803 PSII structure. (B) Sequence similarity of PsbV comparing Synechocystis 6803 with T. vulcanus. High to low identity is shown as thin and green to thick and red ribbons, respectively. The stick model of PsbV-Tyr159 is labeled. (C) Partial sequence alignment of PsbV from Synechocystis 6803 and T. vulcanus. The PsbV-Tyr159 designated in B is shown in the sequence alignment with a black arrow. The overall sequence identity between the two full PsbV sequences is 41.25%. Sequence alignments and identity were generated with Clustal Omega (47), and conservation identifiers are shown below the alignment.
Electron Radiation Damage.
To test whether radiation damage influenced the molecular structure of Synechocystis 6803 PSII, the micrograph movie stacks were truncated using a small fraction of the frames. This low-dose data set produced a cryo-EM map at 2.01-Å resolution with the sample having experienced a cumulative electron radiation dose of 4.37 e− A−2, using the same particle set as was used for the full-dose data set (SI Appendix, Fig. S2). No major differences were observed between the full- and low-dose maps near the OEC, nor in any other region of the maps, which received cumulative doses of 40.8 and 4.37 e− A−2, respectively. Kato et al. recently compared “high” and “low” dose cryo-EM structures of PSII from T. vulcanus at 83 and 3.3 e− A−2, respectively (20). Their study suggested that the D1 C terminus that typically ligates the OEC is found in variable orientations in the high-dose structure (20) unlike the low-dose structure that exhibited a larger population of a single orientation ligating the OEC. They additionally observed a breakage of a conserved disulfide bond in PsbO at high dose. Both our full-dose and low-dose maps exhibit ambiguous orientations of the D1 C terminus (SI Appendix, Fig. S13) and mostly broken PsbO disulfide bonds (SI Appendix, Fig. S14), but at least in the range from 40.8 to 4.37 e− A−2, we see no structural dependence upon radiation dose as discussed below in the Discussion.
Because the oxidation states of the Mn ions are expected to be either III or IV, following a high–oxidation state model of the OEC, we also assessed possible radiation damage by calculating the Mulliken spin densities using single-point density functional theory (DFT; Materials and Methods). This approach allows for a calculated assignment of oxidation state to each Mn ion using the molecular structure as an input model. These results suggested that the four Mn ions are in oxidation states II or III regardless of input spin parameters, which supports the hypothesis that the OEC is more reduced in the cryo-EM structure than would be expected for a high–oxidation state model of the OEC (SI Appendix, Text S3 and Table S7); however, it is important to note that these calculations are highly dependent on the atomic coordinates of the OEC, the O atoms of which are modeled with low confidence and probably exist in multiple states due to OEC heterogeneity.
Discussion
The structure of PSII from Synechocystis 6803 contains unique characteristics that further our understanding of PSII. From the basis of thermophilic cyanobacterial PSII structures, the Large channel has been proposed to play roles in oxygen release (48), substrate water delivery (48–50), and proton transfer (51, 52). Molecular dynamics simulations also based on T. vulcanus PSII structures suggest that the Large channel is more hydrophobic than the Broad and Narrow channels (48, 53), and other computational analyses determined that the Large channel is less likely to transfer protons than the Broad and Narrow channels (5). However, the Large channel is blocked in the Synechocystis 6803 PSII structure. Although the water wheel is near the blockage of the Large channel observed in the Synechocystis 6803 PSII structure, it is important to consider that waters near the OEC are highly interconnected (5); therefore, the mobility observed in the water wheel (Fig. 4) does not necessarily require the involvement of the Large channel. The Large channel’s blockage in Synechocystis 6803 PSII may suggest that it is never involved in PSII activity and perhaps is not a channel at all. Rather, the Narrow and Broad channels may be the only ones important for function, hence their being conserved in all structures. This is consistent with FTIR studies showing that mutation of D1-Glu329 to Ala in Synechocystis 6803 does not perturb the OEC structure or waters involved in the catalytic cycle (54), although the mutation does modify the overall H-bonding network around the OEC; the sidechain of D1-Glu329, a second-shell amino acid associated with the Large channel, is in a different position in the Synechocystis 6803 PSII structure compared with other PSII structures. Another possibility is that the channel-blocking PsbV residues are involved in a gating mechanism of the Large channel. Indeed, a gating mechanism of waters to the OEC has been suggested previously for PSII but from an analysis of T. vulcanus PSII (55, 56). The proposed mechanism involved residues near the water wheel in the Large channel (SI Appendix, Fig. S15) and was substantiated by similarities in substrate gating of acetylcholinesterase (57). In part, it involved conformational changes in aromatic residues. It may be that the PsbV-Tyr159 sidechain undergoes a conformational change in the gating of the Large channel. Future molecular dynamics simulations may allow for more insight into this possibility. If a gating mechanism does exist in Synechocystis 6803 PSII involving PsbV, it is presently unclear whether similar mechanisms are observed in other cyanobacteria because of low sequence identity between PsbV from different species (SI Appendix, Table S8). In any case, the differences observed between PSII structures from mesophilic and thermophilic cyanobacteria suggest differences in proton egress and/or substrate water delivery between organisms. However, the light-saturated rate of oxygen evolution by PSII appears comparable between mesophilic and thermophilic cyanobacteria, ∼5,500 μmol of O2 (mg of Chl)−1 h−1 [refer to Materials and Methods and Sugiura et al. (58) for oxygen evolution rates of PSII core complexes isolated from Synechocystis 6803 and T. elongatus, respectively]. This similarity in rates suggests identical mechanisms of O-O bond formation between species, which is consistent with various spectroscopic studies concluding that the OEC structures are nearly identical between mesophilic cyanobacteria, thermophilic cyanobacteria, and spinach (59–66).
Structural features in the map near the OEC are especially challenging to interpret due to the dynamic nature of the structure and challenges in modeling OEC atoms. Based on the asymmetric ESP and low occupancy of some structural elements, it seems likely that the cryo-EM map is a superposition of multiple states due to contributions of damaged OECs and/or S-state mixing. One contribution to heterogeneity may be that current cryo-EM sample preparation procedures require plunge-freezing in low white light, probably resulting in a mixture of S-states. Future endeavors to plunge freeze PSII samples in complete darkness or plunge-freezing immediately following a saturating light pulse may allow for a large fraction of the sample to be poised in a single S-state. Heterogeneity due to OEC damage may also arise from sample preparation procedures. The alternate positions of W3 and W4 and the uncertain position of the D1 C terminus may indicate that a fraction of the OECs have lost Ca. The waters bound to the Ca were calculated to have high energy barriers for dissociation (67), so the alternate positions seem unlikely if the Ca is present at full occupancy. Some fraction of centers lacking Ca may be present naturally in vivo prior to protein isolation, or Ca depletion could have been induced due to the sample preparation procedure. Indeed, Ca is known to be easily lost from the OEC while otherwise maintaining the general structure of the active site (68).
It seems likely that both the full- and low-dose cryo-EM structures of Synechocystis 6803 PSII represent reduced states of the OEC due to electron radiation exposure. First, flexibility in the D1 C terminus and the absence of a disulfide bond in PsbO due to radiation damage would be consistent with the analysis from Kato et al. that showed the same characteristics in their full-dose structure (20). The low-dose cryo-EM structure of PSII from T. vulcanus did not exhibit these characteristics, but the low-dose cryo-EM structure of PSII from Synechocystis 6803 does, despite their similar radiation doses, and this may suggest that PSII from mesophilic cyanobacteria is more prone to radiation damage than PSII from thermophilic cyanobacteria. If the D1 C terminus of PSII is damaged due to radiation exposure, its modeling is especially challenging, because the nature of the damage and chemical identity of the C terminus would be unknown, and damaged protein is unlikely to obey the stereochemical principles of undamaged protein. Second, radiation-induced OEC reduction is supported by our calculation of Mulliken spin densities. In the generally favored high–oxidation state model of the OEC, the Mn ions are always found in oxidation states ≥III. Instead, our DFT calculation based on the model coordinates suggest the Mn oxidation states are either II or III (SI Appendix, Text S3 and Table S7). Third, X-ray radiation is thought to result in elongation of the mean Mn–Mn distance in the OEC (69–72), and the mean Mn–Mn distances of the high-resolution cryo-EM PSII structures from both Synechocystis 6803 and T. vulcanus are larger than those of two representative low-dose X-ray crystal structures (SI Appendix, Table S9). It is important to note, however, that placement of the OEC atoms during modeling is a nontrivial task, and thus, comparison of the OEC atom distance differences should be considered with great caution. This is due to differences in experiment type, data resolution, and even modeling procedures between experimentalists. For example, in the latter case, Kato et al.’s modeling of the OEC relies to a small extent upon previously determined model restraints (20), whereas our OEC atoms are modeled based solely on the ESP maps. Both of these approaches are reasonable but may produce significantly different results. Development of better methods to standardize the placement of OEC atoms in cryo-EM maps may help to improve the precision of these distance comparisons.
It is also important to note that our full-dose map and the previously determined “high dose” map from Kato et al. (20) contain the sum of all frames collected (i.e., contributions from low to high radiation); therefore, neither our “full dose,” nor the “high dose” structure from Kato et al., are produced using only the latter part of the movie stacks that have received the highest radiation doses. Thus, a more in-depth study of the time course of structural changes during electron radiation exposure is needed to provide a better understanding of electron radiation–induced damage in PSII, together with procedures that minimize heterogeneity due to other causes.
In summary, the high-resolution cryo-EM structure of PSII from the mesophilic cyanobacterium Synechocystis 6803 provides insight into important features of PSII function including the PsbQ subunit and a blockage of the Large water channel. Alternate positions of structural elements near the OEC are challenging to interpret due to structural heterogeneity that may arise from a variety of causes. The method for solving the cryo-EM structure reported here can be used to solve future cryo-EM structures of Synechocystis 6803 PSII with point mutations to better understand the roles of individual amino acids, but it is important to develop strategies that stabilize individual states and/or deconvolute contributions from multiple states and damaged centers in the cryo-EM maps.
Materials and Methods
PSII Purification and Activity.
Wild-type cells of Synechocystis 6803 containing a single psbA gene (psbA2) and containing a hexahistidine-tag fused to the C terminus of CP47 (73) were propagated as described previously (40, 74). Oxygen-evolving PSII core complexes were purified with Ni-NTA superflow affinity resin (Qiagen, Inc., Valencia) at 4 °C under dim green light as described previously (74). Purified PSII core complexes (in 1.2 M betaine, 10% [vol/vol] glycerol, 50 mM MES-NaOH [pH 6.0], 20 mM CaCl2, 5 mM MgCl2, 50 mM histidine, 1 mM EDTA, and 0.03% [wt/vol] β-DM) were concentrated via ultrafiltration to 1 mg of Chl/mL, aliquoted, frozen in liquid N2, and stored at −80 °C. To prepare samples for cryo-EM analysis, aliquots (50 μg of Chl a) were exchanged into cryo-EM buffer (0.5 M betaine, 50 mM MES-NaOH [pH 6.8], 20 mM CaCl2, 5 mM MgCl2, 0.02% [wt/vol] β-DM) by passage through Bio-Rad Micro Bio-Spin 6 centrifugal gel filtration columns (Bio-Rad Laboratories, Hercules) at 50 × g and then concentrated to ∼2 mg of Chl/mL with Amicon Ultra 0.5-mL 100-kDa centrifugal filter devices (EMD Millipore, Billerica). Samples were maintained at ∼4 °C until being applied to cryo-EM grids. After being exchanged into cryo-EM buffer, samples exhibited light-saturated O2 evolution activities of 5,300 ± 200 µmol O2 (mg of Chl)−1/hr−1 as measured (75) with a Clark-type oxygen electrode.
Cryo-EM Grid Preparation.
A holey-carbon C-flat 2/1 Cu 300-mesh EM grid (Electron Microscopy Sciences) was glow discharged for 30 s at 25 mA. The PSII sample was kept in the dark until 3 µL was quickly applied to the grid in an FEI Vitrobot (Thermo Fisher Scientific) at 4 °C and 100% humidity in low fluorescent light. The grid was blotted immediately for 3 s, plunged into liquid ethane, and stored in liquid nitrogen for data collection. In total, the PSII sample was exposed to low fluorescent light for <15 s before it was plunged into the liquid ethane.
Cryo-EM Data Collection.
The frozen grid was imaged on a Titan Krios G2 transmission electron microscope (Thermo Fisher Scientific/FEI) operated at 300 kV equipped with a Gatan K3 direct electron detector in superresolution mode. The defocus varied from −1.2 to −2.0 μm, and the nominal magnification was 105,000×, corresponding to a superresolution pixel size of 0.416 Å. The dose rate was 25.2 e− physical pixel−1/s−1. The GIF setting was a slit size of 20 eV. The total exposure time was 1.12 s per exposure with a total dose of 40.8 e− (Å) −2. SerialEM was used to collect 12,237 micrograph movies with 28 images per stack.
Cryo-EM Data Processing.
A flowchart for data processing is shown in SI Appendix, Fig. S2. Data processing was performed using Relion 3.1 (76). To construct the full-dose map, micrograph movies using all 28 frames were corrected, aligned, and dose weighted using MotionCor2 (77). Ctffind (78) was used to estimate the contrast transfer function (CTF). An initial set of ∼2,000 particles was selected manually, and their two-dimensional classification was used for autopicking templates. Autopicking and manual removal of obviously incorrectly picked particles resulted in a selection of 1,896,906 particles. The Initial Model function was used to create an ab initio model, which was subsequently used as a low-resolution reference for two rounds of three-dimensional classification that resulted in 208,901 particles. Rounds of three-dimensional refinement, CTF refinement, and Bayesian Polishing were used that resulted in a three-dimensional reconstruction at 2.1-Å resolution. Particles with the metadata value rlnCtfMaxResolution less than or equal to 5.0 Å were removed from the particle set, resulting in a new particle set of 202,844 particles. These were used in further rounds of CTF refinement and Bayesian Polishing, yielding a resolution of 1.96 Å. Micelle subtraction led to a final map at 1.93-Å resolution.
To generate the low-dose map, micrographs were remotion corrected using only the first three frames. The final particle set was used in rounds of CTF refinement and Bayesian Polishing, leading to a map at 2.01-Å resolution. Micelle subtraction did not impact the final resolution but, for consistency of map processing workflow between the full- and low-dose structures, was performed regardless. Map resolutions were calculated using the Gold-standard Fourier Shell Correlation of 0.143. Local resolution was calculated using Relion 3.1 (76) (SI Appendix, Fig. S3).
Model Building.
Except for the PsbY (chain R) and PsbQ (chain Q) subunits, the initial model of Synechocystis 6803 PSII was created by extracting the coordinates of individual subunits from the T. vulcanus PSII XRD structure (PDB 3WU2), which were used as structural templates with the corresponding Synechocystis 6803 sequences to create Synechocystis 6803 subunit homology models using SwissModel (79). For PsbY, the same strategy was used, but the structural template was taken from the XRD structure of PSII containing PsbY from T. elongatus (PDB 6W1O). The XRD structure of PsbQ from Synechocystis 6803 (PDB 3LS0) was used as its initial model. These components were fit into the ESP map using UCSF Chimera (80). Manual fitting and editing were performed in Coot (81), and automated refinement was performed using real_space_refine (82) in Phenix (83). Metal ions in the OEC were placed by visual inspection and then fit into the map using Chimera’s “fit in map” function (80) individually, then the coordinates for each were used for the final positions in the model. Oxygen atoms in the OEC were placed by visual inspection.
Molecular Docking.
PsbQ PIPER free energies were calculated using the ClusPro server (84). The ClusPro server requires two models for a docking calculation, that of the “ligand” and the “receptor.” For Synechocystis 6803 input modes, PsbQ was separated from the structure reported here to be used as the “ligand,” and the rest of the PSII model (i.e., the model with PsbQ deleted) was used as the “receptor.” For T. vulcanus input models, the “ligand” was a PsbQ homology model created using SwissModel (79) with the Synechocystis 6803 PsbQ model as its template and the “receptor” was the cryo-EM structure of T. vulcanus PSII that lacks PsbQ (PDB 7D1T) (20). For both docking calculations, the simulation was restrained to maintain >1 Å but <4 Å between the CP43-Trp and PsbQ-Gly that is conserved in all PsbQ homologs (SI Appendix, Figs. S9 and S10).
DFT Calculations of Mulliken Spin Densities for Mn Ions.
We performed single-point DFT calculations (72, 85, 86) using the structure of the OEC from the Synechocystis 6803 PSII structure with Gaussian 16 (87). Hydrogen atoms were added with Maestro (88). The OEC, 10 amino acids (D1-D61, D1-D170, D1-D189, D1-H332, D1-E333, D1-H337, D1-D342, D1-A344, CP43-E341, and CP43-R357), and 10 water molecules surrounding the OEC were included in the system. The calculations were carried out at the B3LYP level of theory (89, 90). The LanL2DZ basis set (91, 92) was used for Mn and Ca atoms and 6–31G(d) (93) was used for all C, H, N, and O atoms.
Supplementary Material
Acknowledgments
This work was supported by Department of Energy, Office of Basic Energy Sciences, and Division of Chemical Sciences Grant DE-FG02-05ER15646 to G.W.B. (cryo-EM analyses), Grant DE-SC0005291 to R.J.D. (mutant construction, PSII purification, and FTIR studies), and Grant DE-SC0001423 to M.R.G. and V.S.B. (computational modeling). Research reported in this publication was also supported by the National Institute of General Medical Sciences of the NIH under Award K99GM140174 to C.J.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We acknowledge the Science Hill Cryo-EM Laboratory and Yale Cryo-EM Resource for sample screening and high-resolution data collection, respectively. We also acknowledge the Office of the Dean at the Yale School of Medicine and the Office of the Provost at Yale University for funding of the Yale Cryo-EM Resource. We acknowledge Dr. Shenping Wu for her assistance in data collection on the Titan Krios at the Yale Cryo-EM Resource laboratory. We thank Professor Charles Sindelar at Yale University for helpful discussions regarding radiation damage.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
1Retired.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116765118/-/DCSupplemental.
Data Availability
Cryo-EM structures have been deposited in the Protein Data Bank (https://www.rcsb.org) and Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/). For the full-dose structure, the accession codes are 7N8O and EMD-24239. For the low-dose structure, the accession codes are 7RCV and EMD-24407.
References
- 1.Barber J., Engine of life and big bang of evolution: A personal perspective. Photosynth. Res. 80, 137–155 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Vinyard D. J., Ananyev G. M., Dismukes G. C., Photosystem II: The reaction center of oxygenic photosynthesis. Annu. Rev. Biochem. 82, 577–606 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Cox N., Pantazis D. A., Lubitz W., Current understanding of the mechanism of water oxidation in photosystem II and its relation to XFEL data. Annu. Rev. Biochem. 89, 795–820 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Shen J.-R., The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Kaur D., et al. , Proton exit pathways surrounding the oxygen evolving complex of photosystem II. Biochim. Biophys. Acta Bioenerg. 1862, 148446 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., Eaton-Rye J. J., The extrinsic proteins of photosystem II. Biochim. Biophys. Acta 1817, 121–142 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Roose J. L., Frankel L. K., Mummadisetti M. P., Bricker T. M., The extrinsic proteins of photosystem II: Update. Planta 243, 889–908 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Williams J. G. K., “Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803” in Methods in Enzymology, Packer L., Glazer A. N., Eds. (Academic Press, 1988), pp. 766–778. [Google Scholar]
- 9.Kufryk G. I., Sachet M., Schmetterer G., Vermaas W. F. J., Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: Optimization of efficiency. FEMS Microbiol. Lett. 206, 215–219 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Ikeuchi M., Tabata S., Synechocystis sp. PCC 6803 - A useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 70, 73–83 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Vermaas W., Carpenter S., Bunch C., “Specific mutagenesis as a tool for the analysis of structure/function relationships in photosystem II” in Molecular Biology and Bioenergetics, Singhal G. S., et al., Eds. (Springer Berlin Heidelberg, 1989), pp. 21–35. [Google Scholar]
- 12.Vermass W. F. J., Rutherford A. W., Hansson Ö., Site-directed mutagenesis in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: Donor D is a tyrosine residue in the D2 protein. Proc. Natl. Acad. Sci. U.S.A. 85, 8477–8481 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debus R. J., Barry B. A., Babcock G. T., McIntosh L., Site-directed mutagenesis identifies a tyrosine radical involved in the photosynthetic oxygen-evolving system. Proc. Natl. Acad. Sci. U.S.A. 85, 427–430 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debus R. J., FTIR studies of metal ligands, networks of hydrogen bonds, and water molecules near the active site Mn4CaO5 cluster in photosystem II. Biochim. Biophys. Acta 1847, 19–34 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Noguchi T., Fourier transform infrared difference and time-resolved infrared detection of the electron and proton transfer dynamics in photosynthetic water oxidation. Biochim. Biophys. Acta 1847, 35–45 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Lubitz W., Chrysina M., Cox N., Water oxidation in photosystem II. Photosynth. Res. 142, 105–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisz D. A., Gross M. L., Pakrasi H. B., The use of advanced mass spectrometry to dissect the life-cycle of photosystem II. Front Plant Sci 7, 617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müh F., Zouni A., Structural basis of light-harvesting in the photosystem II core complex. Protein Sci. 29, 1090–1119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meruelo A. D., Han S. K., Kim S., Bowie J. U., Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci. 21, 1746–1753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K., et al. , High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams. Commun. Biol. 4, 382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt L., Vinyard D. J., Khan S., Brudvig G. W., Oxygen-evolving complex of photosystem II: An analysis of second-shell residues and hydrogen-bonding networks. Curr. Opin. Chem. Biol. 25, 152–158 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Yu H., et al. , Cryo-EM structure of monomeric photosystem II at 2.78 Å resolution reveals factors important for the formation of dimer. Biochim. Biophys. Acta Bioenerg. 1862, 148471 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Fagerlund R. D., Eaton-Rye J. J., The lipoproteins of cyanobacterial photosystem II. J. Photochem. Photobiol. B 104, 191–203 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Kashino Y., et al. , Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Roose J. L., Kashino Y., Pakrasi H. B., The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summerfield T. C., Shand J. A., Bentley F. K., Eaton-Rye J. J., PsbQ (Sll1638) in Synechocystis sp. PCC 6803 is required for photosystem II activity in specific mutants and in nutrient-limiting conditions. Biochemistry 44, 805–815 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Kashino Y., Inoue-Kashino N., Roose J. L., Pakrasi H. B., Absence of the PsbQ protein results in destabilization of the PsbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. J. Biol. Chem. 281, 20834–20841 (2006). [DOI] [PubMed] [Google Scholar]
- 28.C. J. Gisriel, G. W. Brudvig, Comparison of PsbQ and Psb27 in photosystem II provides insight into their roles. Photosynth. Res. In Press, DOI: 10.1007/s11120-021-00888-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson S. A., Fagerlund R. D., Wilbanks S. M., Eaton-Rye J. J., Crystal structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: Implications for binding and function in cyanobacterial photosystem II. Biochemistry 49, 2765–2767 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Michoux F., et al. , Crystal structure of CyanoQ from the thermophilic cyanobacterium Thermosynechococcus elongatus and detection in isolated photosystem II complexes. Photosynth. Res. 122, 57–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., et al. , MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 4638–4643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang G., et al. , Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus. Proc. Natl. Acad. Sci. U.S.A. 118, e2018053118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabret J., et al. , Structural insights into photosystem II assembly. Nat. Plants 7, 524–538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su X., et al. , Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357, 815–820 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Sheng X., et al. , Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 5, 1320–1330 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Ago H., et al. , Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. Wanget al., . Heterogeneous composition of oxygen-evolving complexes in crystal structures of dark-adapted photosystem II. Biochemistry 60, 3374–3384 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Ibrahim M., et al. , Untangling the sequence of events during the S2 → S3 transition in photosystem II and implications for the water oxidation mechanism. Proc. Natl. Acad. Sci. U.S.A. 117, 12624–12635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki H., et al. , Functional roles of D2-Lys317 and the interacting chloride ion in the water oxidation reaction of photosystem II as revealed by Fourier transform infrared analysis. Biochemistry 52, 4748–4757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokhrel R., Service R. J., Debus R. J., Brudvig G. W., Mutation of lysine 317 in the D2 subunit of photosystem II alters chloride binding and proton transport. Biochemistry 52, 4758–4773 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Service R. J., Hillier W., Debus R. J., Evidence from FTIR difference spectroscopy of an extensive network of hydrogen bonds near the oxygen-evolving Mn(4)Ca cluster of photosystem II involving D1-Glu65, D2-Glu312, and D1-Glu329. Biochemistry 49, 6655–6669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broser M., et al. , Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn. J. Biol. Chem. 286, 15964–15972 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Vliet P., Rutherford A. W., Properties of the chloride-depleted oxygen-evolving complex of photosystem II studied by electron paramagnetic resonance. Biochemistry 35, 1829–1839 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Linke K., Ho F. M., Water in photosystem II: Structural, functional and mechanistic considerations. Biochim. Biophys. Acta 1837, 14–32 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Guskov A., et al. , Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Sakashita N., Watanabe H. C., Ikeda T., Saito K., Ishikita H., Origins of water molecules in the photosystem II crystal structure. Biochemistry 56, 3049–3057 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 1–6. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassiliev S., Zaraiskaya T., Bruce D., Exploring the energetics of water permeation in photosystem II by multiple steered molecular dynamics simulations. Biochim. Biophys. Acta 1817, 1671–1678 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Kim C. J., Debus R. J., Evidence from FTIR difference spectroscopy that a substrate H2O molecule for O2 formation in photosystem II is provided by the Ca ion of the catalytic Mn4CaO5 cluster. Biochemistry 56, 2558–2570 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Ugur I., Rutherford A. W., Kaila V. R. I., Redox-coupled substrate water reorganization in the active site of photosystem II - The role of calcium in substrate water delivery. Biochim. Biophys. Acta 1857, 740–748 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Bondar A.-N., Dau H., Extended protein/water H-bond networks in photosynthetic water oxidation. Biochim. Biophys. Acta 1817, 1177–1190 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Chrysina M., de Mendonça Silva J. C., Zahariou G., Pantazis D. A., Ioannidis N., Proton translocation via tautomerization of Asn298 during the S2–S3 state transition in the oxygen-evolving complex of photosystem II. J. Phys. Chem. B 123, 3068–3078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho F. M., Styring S., Access channels and methanol binding site to the CaMn4 cluster in photosystem II based on solvent accessibility simulations, with implications for substrate water access. Biochim. Biophys. Acta 1777, 140–153 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Kim C. J., Debus R. J., Roles of D1-Glu189 and D1-Glu329 in O2 formation by the water-splitting Mn4Ca cluster in photosystem II. Biochemistry 59, 3902–3917 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Zhou H.-X., McCammon J. A., The gates of ion channels and enzymes. Trends Biochem. Sci. 35, 179–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho F. M., Uncovering channels in photosystem II by computer modelling: Current progress, future prospects, and lessons from analogous systems. Photosynth. Res. 98, 503–522 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Zhou H.-X., Wlodek S. T., McCammon J. A., Conformation gating as a mechanism for enzyme specificity. Proc. Natl. Acad. Sci. U.S.A. 95, 9280–9283 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugiura M., Boussac A., Noguchi T., Rappaport F., Influence of histidine-198 of the D1 subunit on the properties of the primary electron donor, P680, of photosystem II in Thermosynechococcus elongatus. Biochim. Biophys. Acta 1777, 331–342 (2008). [DOI] [PubMed] [Google Scholar]
- 59.McDermott A. E., et al. , Characterization of the manganese O2-evolving complex and the iron-quinone acceptor complex in photosystem II from a thermophilic cyanobacterium by electron paramagnetic resonance and X-ray absorption spectroscopy. Biochemistry 27, 4021–4031 (1988). [DOI] [PubMed] [Google Scholar]
- 60.Boussac A., Sugiura M., Inoue Y., Rutherford A. W., EPR study of the oxygen evolving complex in His-tagged photosystem II from the cyanobacterium Synechococcus elongatus. Biochemistry 39, 13788–13799 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Yamanari T., Kimura Y., Mizusawa N., Ishii A., Ono T.-A., Mid- to low-frequency Fourier transform infrared spectra of S-state cycle for photosynthetic water oxidation in Synechocystis sp. PCC 6803. Biochemistry 43, 7479–7490 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Suzuki H., Taguchi Y., Sugiura M., Boussac A., Noguchi T., Structural perturbation of the carboxylate ligands to the manganese cluster upon Ca2+/Sr2+ exchange in the S-state cycle of photosynthetic oxygen evolution as studied by flash-induced FTIR difference spectroscopy. Biochemistry 45, 13454–13464 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Su J.-H., et al. , The electronic structures of the S(2) states of the oxygen-evolving complexes of photosystem II in plants and cyanobacteria in the presence and absence of methanol. Biochim. Biophys. Acta 1807, 829–840 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Cox N., et al. , Effect of Ca2+/Sr2+ substitution on the electronic structure of the oxygen-evolving complex of photosystem II: A combined multifrequency EPR, 55Mn-ENDOR, and DFT study of the S2 state. J. Am. Chem. Soc. 133, 3635–3648 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Milikisiyants S., et al. , The structure and activation of substrate water molecules in the S2 state of photosystem II studied by hyperfine sublevel correlation spectroscopy. Energy Environ. Sci. 5, 7747–7756 (2012). [Google Scholar]
- 66.Glöckner C., et al. , Structural changes of the oxygen-evolving complex in photosystem II during the catalytic cycle. J. Biol. Chem. 288, 22607–22620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.E. M. Sproviero, K. Shinopoulos, J. A. Gascón, J. P. McEvoy, G. W. Brudvig, V. S. Batista, QM/MM computational studies of substrate water binding to the oxygen-evolving centre of photosystem II. Philos. Trans. R Soc. Lond. B Biol. Sci. 363, 1149–1156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lohmiller T., Shelby M. L., Long X., Yachandra V. K., Yano J., Removal of Ca2+ from the oxygen-evolving complex in photosystem II has minimal effect on the Mn4O5 core structure: A polarized Mn X-ray absorption spectroscopy study. J. Phys. Chem. B 119, 13742–13754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grundmeier A., Dau H., Structural models of the manganese complex of photosystem II and mechanistic implications. Biochim. Biophys. Acta 1817, 88–105 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Yano J., et al. , X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. U.S.A. 102, 12047–12052 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luber S., et al. , S1-state model of the O2-evolving complex of photosystem II. Biochemistry 50, 6308–6311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pal R., et al. , S0-State model of the oxygen-evolving complex of photosystem II. Biochemistry 52, 7703–7706 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Debus R. J., et al. , Does histidine 332 of the D1 polypeptide ligate the manganese cluster in photosystem II? An electron spin echo envelope modulation study. Biochemistry 40, 3690–3699 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Debus R. J., Evidence from FTIR difference spectroscopy that D1-Asp61 influences the water reactions of the oxygen-evolving Mn4CaO5 cluster of photosystem II. Biochemistry 53, 2941–2955 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Hays A.-M. A., Vassiliev I. R., Golbeck J. H., Debus R. J., Role of D1-His190 in proton-coupled electron transfer reactions in photosystem II: A chemical complementation study. Biochemistry 37, 11352–11365 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guex N., Peitsch M. C., Schwede T., Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30, S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Pettersen E. F., et al. , UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kozakov D., et al. , The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Askerka M., Wang J., Brudvig G. W., Batista V. S., Structural changes in the oxygen-evolving complex of photosystem II induced by the S1 to S2 transition: A combined XRD and QM/MM study. Biochemistry 53, 6860–6862 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghosh I., et al. , D1-S169A substitution of photosystem II reveals a novel S2-state structure. Biochim. Biophys. Acta Bioenerg. 1861, 148301 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Frisch M. J., et al. , Gaussian 16 (Gaussian, Inc., Wallingford, CT, 2016). [Google Scholar]
- 88.Release S., 2021-2: Maestro (Schrödinger, LLC, New York, NY, 2021). [Google Scholar]
- 89.Becke A. D., Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A Gen. Phys. 38, 3098–3100 (1988). [DOI] [PubMed] [Google Scholar]
- 90.Becke A. D., Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993). [Google Scholar]
- 91.Hay P. J., Wadt W. R., Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J. Chem. Phys. 82, 299–310 (1985). [Google Scholar]
- 92.da Silva Filho D. A., et al. , Hole-vibronic coupling in oligothiophenes: Impact of backbone torsional flexibility on relaxation energies. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 365, 1435–1452 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Hariharan P. C., Pople J. A., The influence of polarization functions on molecular orbital hydrogenation energies. Theor. Chim. Acta 28, 213–222 (1973). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM structures have been deposited in the Protein Data Bank (https://www.rcsb.org) and Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/). For the full-dose structure, the accession codes are 7N8O and EMD-24239. For the low-dose structure, the accession codes are 7RCV and EMD-24407.