Abstract
OBJECTIVE
Gestational diabetes mellitus (GDM) is associated with an increased risk of obesity and insulin resistance in offspring later in life, which might be explained by epigenetic changes in response to maternal hyperglycemic exposure.
RESEARCH DESIGN AND METHODS
We explored the association between GDM exposure and maternal blood and newborn cord blood methylation in 536 mother-offspring pairs from the prospective FinnGeDi cohort using Illumina MethylationEPIC 850K BeadChip arrays. We assessed two hypotheses. First, we tested for shared maternal and offspring epigenetic effects resulting from GDM exposure. Second, we tested whether GDM exposure and maternal methylation had an epigenetic effect on the offspring.
RESULTS
We did not find any epigenetic marks (differentially methylated CpG probes) with shared and consistent effects between mothers and offspring. After including maternal methylation in the model, we identified a single significant (false discovery rate 1.38 × 10−2) CpG at the cg22790973 probe (TFCP2) associated with GDM. We identified seven additional FDR-significant interactions of maternal methylation and GDM status, with the strongest association at the same cg22790973 probe (TFCP2), as well as cg03456133, cg24440941 (H3C6), cg20002843 (LOC127841), cg19107264, and cg11493553 located within the UBE3C gene and cg17065901 in FAM13A, both susceptibility genes for type 2 diabetes and BMI, and cg23355087 within the DLGAP2 gene, known to be involved in insulin resistance during pregnancy.
CONCLUSIONS
Our study reveals the potential complexity of the epigenetic transmission between mothers with GDM and their offspring, likely determined by not only GDM exposure but also other factors indicated by maternal epigenetic status, such as maternal metabolic history.
Introduction
Gestational diabetes mellitus (GDM) is a condition in which hyperglycemia develops during pregnancy. GDM is one of the most common metabolic complications, affecting 8–25% of pregnancies, and prevalence rates show considerable variation among countries depending on the test strategies used (1). Observational studies support evidence that GDM is associated with an increased future offspring risk of obesity and insulin resistance, key risk factors in type 2 diabetes and other cardiometabolic diseases in adulthood (2–4), suggesting that hyperglycemia exposure in utero has a direct adverse impact on offspring. There is also evidence for a genetic basis of GDM, with a history of diabetes in either parent increasing the risk of GDM at least twofold (5) and identified shared genetic risk variants that confer risk of both GDM and type 2 diabetes (6). Epigenetic mechanisms likely contribute to GDM, because evidence suggests that risks factors arise from prenatal exposure to a diabetic intrauterine environment (5,7). Targeted and candidate gene approaches have found several differentially methylated positions (DMPs) in leukocytes associated with offspring GDM exposure (8). However, recent technological advances have allowed for a more systematic and genome-wide approach to the identification of novel methylation loci. Indeed, studies have identified several DMPs associated with GDM (9–11). The most recent epigenome-wide association study (EWAS) was a meta-analysis of cord blood DNA methylation from six birth cohorts, from nine countries, including a total of 3,677 newborn offspring, of whom 317 were exposed to GDM. Despite the relatively large meta-analysis sample size, the authors did not observe any individual cord blood CpG probes associated with GDM exposure using the false discovery rate (FDR) for testing significance (12). Here, we used the FinnGeDi (Finnish Gestational Diabetes) study cohort of 298 mother-offspring pairs with GDM and 238 without, all with maternal blood and cord blood samples (13), to investigate whether epigenetic changes occur in response to GDM exposure during pregnancy, both in mothers and their offspring. Specifically, we tested two hypotheses: whether exposure to GDM is associated with 1) epigenetic changes shared by mother-offspring pairs and 2) specific epigenetic changes in the offspring.
Research Design and Methods
Cohort
We designed a case-control study as part of the FinnGeDi prospective multicenter case-control cohort, including a total of 1,072 participants (536 mother-offspring pairs), of whom 298 (55.6%) had GDM during pregnancy. As described in detail (13), women with GDM were recruited from delivery units when arriving to give birth, and the next woman without GDM was recruited as a control. Maternal venous blood and offspring cord blood samples were collected during delivery. Exclusion criteria were multifetal births, prepregnancy diabetes, and smoking during pregnancy. GDM was diagnosed following the Finnish Current Care Guideline (14), which is modified from the criteria of the International Association of the Diabetes and Pregnancy Study Groups. The Finnish guideline recommends the oral glucose tolerance test, with an oral dose of 75 g glucose and the following cutoff values: fasting glucose, ≥5.3 mmol/L (95 mg/dL); 1-h glucose, ≥10 mmol/L (180 mg/dL); and 2-h glucose, ≥8.6 mmol/L (155 mg/dL). Mothers without GDM and with normal oral glucose tolerance test results after the 24th week of gestation were eligible controls. Mothers in each group were carefully group matched by age and prepregnancy BMI to select an equal number of women in each stratum. However, there were insufficient eligible control mothers in the older age and BMI categories (Supplementary Table 1). Therefore, a total of 238 women were selected as controls. A summary of the cohort characteristics is shown in Table 1. The samples were collected and approved following the ethics committee of Oulu University Hospital (reference 33/2008; Oulu, Finland).
Table 1.
Cohort characteristics of mother-offspring pairs used in this study
| Characteristic | Control (n = 238) | GDM (n = 298) | P * |
|---|---|---|---|
| Age of the mother, years | 31.5 (5.2) | 32.5 (5.3) | 0.042 |
| BMI of the mother, kg/m2 | 25.6 (4.8) | 27.9 (6.1) | <0.001 |
| Gestation, weeks | 40.32 (1.19) | 39.60 (1.33) | <0.001 |
| Sex of the offspring | 0.3 | ||
| Male | 109 (46) | 152 (51) | |
| Female | 129 (54) | 146 (49) | |
| Birth weight, g | 3,703 (473) | 3,705 (474) | >0.9 |
| Birth weight SD score | 0.15 (1.00) | 0.38 (1.08) | 0.011 |
| Primipara (yes) | 119 (50) | 127 (43) | 0.10 |
| Previous deliveries | 1.28 (2.29) | 1.46 (2.27) | 0.4 |
| Mode of delivery (vaginal) | 198 (83) | 245 (82) | 0.8 |
| Large for gestational age ( >90th percentile)† | 24 (10) | 50 (17) | 0.032 |
| Small for gestational age ( >90th percentile)‡ | 15 (6.3) | 20 (6.7) | >0.9 |
| Maternal weight gain, kg | 15.1 (5.6) | 11.9 (5.6) | <0.001 |
| Unknown | 4 | 20 | |
| Participant’s mother had GDM in same pregnancy (yes) | 5 (2.5) | 9 (3.5) | 0.6 |
| Unknown | 37 | 44 | |
| Participant’s mother had GDM in any pregnancy (yes) | 9 (100) | 23 (100) | >0.9 |
| Unknown | 229 | 275 | |
| Mother’s father had diabetes (yes) | 30 (100) | 48 (100) | >0.9 |
| Unknown | 208 | 250 | |
| Mother’s mother had diabetes (yes) | 13 (100) | 50 (100) | >0.9 |
| Unknown | 225 | 248 | |
| Maternal socioeconomic status | 0.4 | ||
| 1 (highest) | 53 (28) | 65 (26) | |
| 2 | 73 (39) | 112 (45) | |
| 3 | 13 (7.0) | 22 (8.8) | |
| 4 | 47 (25) | 50 (20) | |
| Unknown | 52 | 49 | |
| Maternal education | 0.027 | ||
| 1 (basic) | 2 (0.9) | 11 (3.9) | |
| 2 (secondary) | 81 (37) | 127 (45) | |
| 3 (lower-level tertiary) | 71 (33) | 77 (28) | |
| 4 (upper-level tertiary) | 64 (29) | 65 (23) | |
| Unknown | 20 | 18 |
Data presented as mean (SD) or n (%).
Welch two-sample Student t test or Fisher exact test.
> +2 SD (40).
Infinium MethylationEPIC BeadChip
DNA was extracted from whole blood for the mothers and from cord blood for the offspring using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Bilsulfite conversion of genomic DNA was performed using the EZ-96 DNA Methylation Kit (Zymo Research) following manufacturer protocols, and bisulfite-converted DNA was subjected to a genome-wide DNA methylation analysis performed using the Illumina Infinium MethylationEPIC 850K BeadChip array (San Diego, CA), which covers a total of 853,307 CpG sites. All samples were randomized across the chips and analyzed on the same machine by the same technician to reduce batch effects. After single-base extension and staining, the BeadChips were imaged with the Illumina iScan. Raw fluorescence intensities of the scanned images were extracted with the GenomeStudio Methylation module (Illumina). The fluorescence intensity ratio was used to calculate the β-value, which corresponds to the methylation score for each analyzed site according to the following equation: β-value = IM/(IU + IM + 100), where IM is the intensity of the methylated allele and IU the intensity of the unmethylated allele. DNA methylation β-values range from 0 (completely unmethylated) to 1 (completely methylated). All samples had high bisulfite conversion efficiency (signal intensity >4,000) and were included for further analysis based on GenomeStudio quality control. Quality control was performed using R software (version 4.0.0) (15). The DNA methylation IDAT files were imported using the R package minfi for preprocessing and quality control (16).
The following probes were excluded from further analysis: probes with a detection P value ≥0.01 for at least one sample, cross-hybridizing probes (17), probes with a bead count less than three in at least 5% of the samples, non-CpG probes, and probes that lie near single-nucleotide polymorphisms (18). Probes on chromosomes X and Y were used for sex estimation and then excluded from downstream analyses. Quality control identified one offspring sex-discordant sample, which was excluded from further analysis. Samples with <99% probes with a detection P value <0.01 were excluded, and one offspring sample was excluded with a call rate <99%. Probe design biases and batch effects were normalized using R packages ENmix (19) and sva (20), respectively. After quality control, 1,070 samples (534 offspring and 536 mothers) and 724,671 CpG probes were available for further downstream analysis.
Additionally, because blood and cord blood samples are expected to include a variety of cell types, which might have a potential confounding effect on DNA methylation, cell composition was estimated based on whole blood (for mothers) and cord blood (for neonates) reference panels: the Bioconductor R EPIC (21) and FlowSorted.CordBloodCombined.450k (22).
Statistical Analyses
P values were corrected for multiple testing using the FDR method from Benjamini-Hochberg (23). For all analyses, the β-value has been transformed into an M value (24), where M value = log2(β/1 − β). The relationship between β and M values follows a logistic curve with, for example, β-values of 0.2, 0.5, and 0.8 corresponding to M values of −2, 0, and 2, respectively.
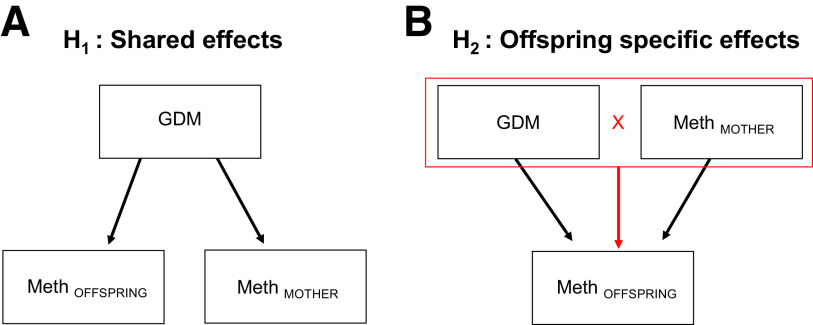
Three sets of analyses were performed to test the two study hypotheses described in Fig. 1. The first hypothesis was that both offspring and mothers might share the same epigenetic effects as a result of shared GDM exposure. The second hypothesis tested for potential epigenetic effects that are specific or unique to the offspring as a result of GDM exposure. Equations for the implemented models are described in Supplementary Table 2. The three analyses implemented were as follows:
Figure 1.
The investigated study hypotheses. A: Hypothesis 1: assess whether there are epigenetic effects that are shared by both offspring and their mothers as a result of shared exposure to GDM (offspring and maternal methylation regressed upon GDM status; see Research Design and Methods). B: Hypothesis 2: test if there are epigenetic effects as a result of exposure to GDM that are specific to the offspring (offspring DNA methylation regressed upon GDM status, maternal methylation for the same probe, and interaction between GDM and maternal methylation status; see Research Design and Methods).
1. EWAS for Mothers and Offspring
Two separate EWAS were conducted to identify epigenetic changes associated with GDM exposure in both mothers and offspring, separately, using the R package limma (25). Linear regression was performed using CpG methylation levels to identify differentially methylated positions in offspring and mothers with GDM exposure compared with nonexposed controls. Offspring methylation probes were adjusted using the following covariates: sex, birth weight, and gestational age at delivery (Supplementary Fig. 1). For the mothers, we adjusted methylation probes for age, gestational weight gain, and prepregnancy BMI. Cell composition was adjusted for cord blood cell composition for the offspring (Supplementary Fig. 1) and blood cell composition for the mothers (Supplementary Fig. 2).
2. Shared Epigenetic Associations
A linear mixed model (pooled data model) was created to identify potentially shared epigenetic changes between the maternal and offspring methylomes. This was performed using the combined methylome probe data for the mothers and the offspring, accounting for the correlation between each mother and her offspring, via the R package lme4 (26). This test was performed to identify potential shared causal paths for the mother and offspring in response to in utero exposure to GDM that could explain a shared effect (Fig. 1A). In the first step, linear regression was performed on the methylomes of the offspring and mothers using the same covariates described above. Cell composition was performed for offspring (cord blood) and mothers (blood), separately. Residuals from the first step were then used in a mixed linear model as the traits of interest and regressed upon the explanatory variables of GDM, offspring or maternal status, and interaction between GDM and offspring or maternal status.
3. Epigenetic Changes Specific to Offspring
Linear regression was performed to identify offspring-specific effects in response to GDM exposure and maternal environment (i.e., methylation levels). This test was performed to identify whether methylation changes of the offspring are determined not only by GDM status but also by maternal methylation levels (Fig. 1B). To do this, the offspring methylome was linearly regressed on GDM exposure status, with the methylome of the mother (preadjusted for age, BMI, and blood cell composition) included as a covariate. In addition, an interaction term between GDM exposure and maternal methylation status at the same probe was included, in addition to sex, birth weight, gestational week, and cord blood cell composition. To control for outlying genomic data points, offspring-mother pairs were excluded from this model, following Tukey method, if the M value for either the offspring or the mother was lower than three times the interquartile range below the first quartile or higher than three times the interquartile range above the third quartile.
Bioinformatic Analyses
The Genotype-Tissue Expression Portal (https://gtexportal.org/home/) was used to identify the expression patterns of genes in 44 tissues. The expression level was plotted using the median transcripts per kilobase million.
Data and Resource Availability
The MethylationEPIC 850K array data will be made available upon request to E.Ka., P.F., or T.A. The code to perform the analyses in this manuscript is available at https://github.com/umr1283/EpxGDM (https://doi.org/10.5281/zenodo.4709136).
Results
To investigate epigenetic changes that occur in response to maternal GDM exposure, we performed a whole-methylome analysis in a total of 534 mother-offspring pairs from the FinnGeDi study. We addressed two questions: 1) whether there were epigenetic changes in response to GDM exposure that were shared between mother and offspring, using a paired mother-offspring study design, and 2) whether any individual DMPs associated with GDM exposure were specific to the offspring. We used two approaches to address the first question (models 1 and 2, described above), both of which yielded the same results. We did not observe any FDR-significant DMPs associated with GDM exposure in the EWAS for the mothers (n = 536) or offspring (n = 534), separately (Supplementary Tables 3 and 4), consistent with recent findings in newborn GDM (12). Using the paired mother-offspring study design, following two separate EWAS for mother and offspring, we implemented a generalized linear mixed model for the combined methylation data to formally test for DMPs resulting from GDM exposure shared by mothers and their offspring. We found no strong evidence (FDR <0.05) to support shared epigenetic changes associated with GDM exposure (Supplementary Fig. 3).
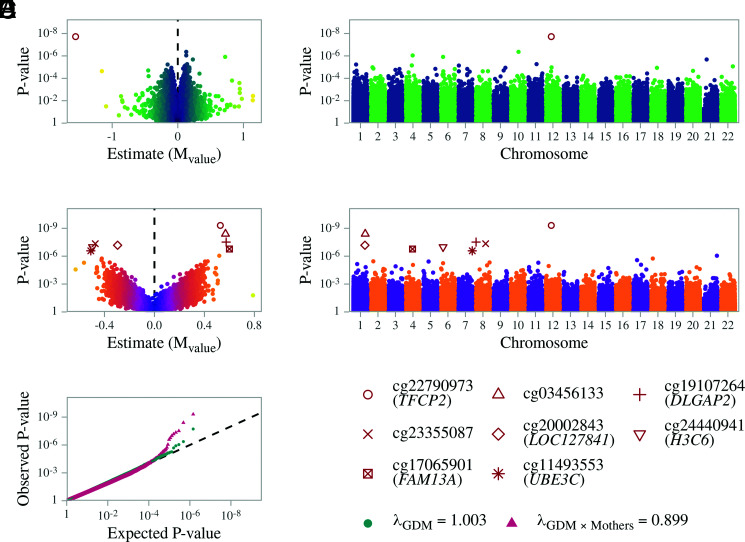
Second, we used the same paired design in linear regression to assess whether both maternal methylation status and GDM exposure had an effect on offspring methylation. Using this model, we observed a hypomethylation at the cg22790973 probe (average methylation 1.03%; M value estimate −1.56 ± 0.27; FDR 1.38 × 10−2) associated with GDM exposure (Fig. 2A and 2B). This probe is located at the TSS1500 (1,500 base pairs upstream of the transcription start site) of the ubiquitously expressed transcription factor CP2 (TFCP2) gene (Supplementary Fig. 4). This CpG probe is also located in a CpG island (chr12:51566680–51567072), a region in the genome with high CpG density that has been previously shown to regulate gene expression (27).
Figure 2.
Summary results for offspring EWAS associations. Linear model between offspring methylation and exposure to maternal GDM, including the methylation of mothers in the model, and interaction between GDM exposure and maternal methylation, adjusted for offspring sex, gestational week, birth weight, and cell composition. A: Volcano plot for offspring probe differential methylation by GDM exposure; cg22790973 has an estimate of −1.56 (FDR 1.38 × 10−2), equivalent to a β-value of 1.03%. B: Manhattan plot for the GDM exposure main effect, showing the genome-wide results for all the CpGs. C: Volcano plot of the GDM exposure interaction effect. D: Manhattan plot of the GDM exposure interaction effect, showing the genome-wide results for all the CpGs. E: Probability-probability plot of the GDM exposure main effect (green) and interaction term (red) on methylation of offspring, with the black line indicating the expected distribution.
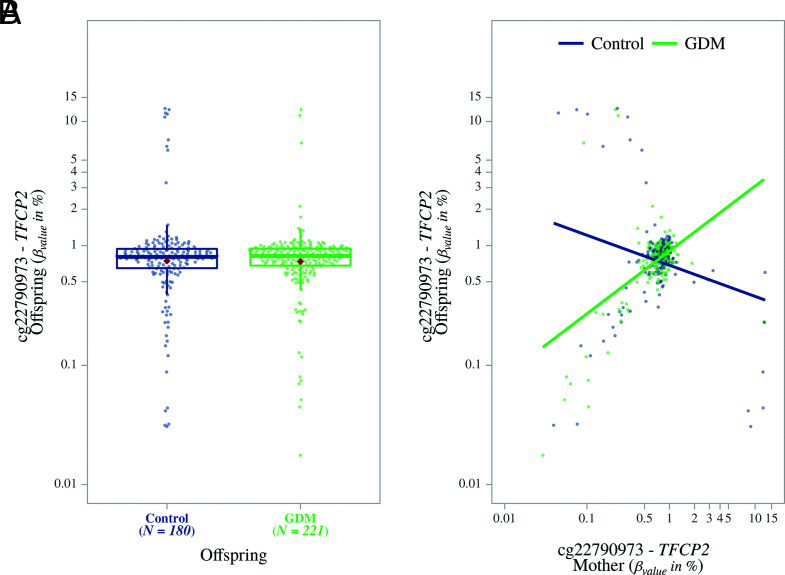
To test for potential offspring-specific effects, we also fitted an interaction term in our model between GDM exposure and maternal methylation (i.e., the impact of GDM exposure in the context of maternal methylation status) (Supplementary Table 5 and Fig. 2C–E). We observed that the same locus (TFCP2 cg22790973) showed the highest FDR-significant association between offspring and maternal methylation (average methylation 1.03%; M value estimate 0.52 ± 0.08; FDR 3.59 × 10−4). We observed that as maternal GDM methylation increased at this locus, so did the GDM-exposed offspring methylation, compared with a decrease in nonexposed GDM offspring (Fig. 3A and 3B).
Figure 3.
Differential methylation observed for offspring at probe cg22790973 (TFCP2). A: Box plot showing the methylation differences between offspring exposed to maternal GDM compared with nonexposed controls. B: Scatterplot showing increased methylation at this probe for offspring exposed to maternal GDM along with increased maternal methylation at the same probe. For nonexposed offspring, methylation decreased with increased maternal methylation at this probe.
We observed seven additional DMPs that reached FDR significance for the interaction between maternal GDM status and methylation of the mother (Supplementary Fig. 5 and Supplementary Table 5). At the cg03456133 probe, located in an intergenic region, we found an increase in methylation of the GDM-exposed offspring as maternal methylation increased, compared with a decrease in nonexposed controls (average methylation 96.48%; M value estimate 0.56 ± 0.09; FDR 1.46 × 10−3). We also found a similar trend at the cg19107264 probe (average methylation 96.64%; M value estimate 0.57 ± 0.10; FDR 7.37 × 10−3), located within the gene body of DLGAP2, encoding disks large–associated protein 2, which is a gene most strongly expressed in the brain (Supplementary Fig. 6). We also found a decrease in methylation at the cg23355087 probe, located in an intergenic region, in GDM offspring as methylation of the mother with GDM increased, compared with nonexposed controls (average methylation 95.66%; M value estimate −0.47 ± 0.08; FDR 8.24 × 10−3). A similar trend was found at the cg20002843 probe, located within the LOC127841 gene body (average methylation 47.37%; M value estimate −0.29 ± 0.05; FDR 9.68 × 10−3) and at the cg24440941 probe (average methylation 1.48%; M value estimate −0.49 ± 0.09; FDR 1.24 × 10−2), located in the TSS1500 of H3C6 (H3 clustered histone 6) (Supplementary Fig. 7). At the cg17065901 probe, located within the FAM13A gene body, we found an increase in methylation as methylation of the mother with GDM increased, compared with non–GDM-exposed offspring (average methylation 98.25%; M value estimate 0.60 ± 0.11; FDR 1.76 × 10−2); the FAM13A gene is highly expressed in adipose tissue (Supplementary Fig. 8). Lastly, for the cg11493553 probe located within the ubiquitously expressed UBE3C gene, we found a decrease in methlylation in the GDM-exposed offspring as maternal methylation increased, compared with the nonexposed controls (average methylation 97.07%; M value estimate −0.50 ± 0.09; FDR 2.41 × 10−2; Supplementary Fig. 9).
Conclusions
We present a comprehensive and in-depth study investigating epigenetic associations in response to GDM exposure in mother-offspring pairs using the prospective FinnGeDi cohort. Our data did not support robust epigenetic associations for women and offspring exposed to GDM during pregnancy. However, in terms of epigenetic associations that are specific to offspring, our study has identified a potentially novel perspective in maternal transmission, which includes not only GDM but also maternal methylation as the exposure that could have an effect on offspring methylation.
Our study suggests that although there is no strong direct association between GDM exposure and the methylome of offspring, the context of maternal environment (i.e., maternal methylome) may contribute to a multiplicative causal effect. Because we included maternal methylation in our model, we were able to account for the possible modifying effect of maternal methylation. This is illustrated with the observed significant association of methylation at the TFCP2 gene with offspring exposure to GDM. TFCP2 is ubiquitously expressed, and elucidating the function of this gene in various tissues has been a challenge. However, some studies have identified roles of TFCP2 in reproduction and embryonic development (28), which suggests pleiotropic effects of this transcription factor later in adulthood. We found that this CpG also had an interaction association between maternal methylation and GDM status. Although methylation was lower in offspring exposed to maternal GDM, we found that methylation at TFCP2 in offspring increased as maternal methylation increased. Interestingly, the reverse was observed for nonexposed controls. This suggests that the higher the maternal GDM methylation at this locus, the smaller the effect on the GDM-exposed offspring. It is important to note that for each locus, extreme methylation values within the offspring and mothers could lead to bias, mainly in the interaction term, and were thus excluded. To avoid this bias, >20% of the sample pairs were excluded for the TFCP2 cg22790973 probe, based on the Tukey method.
Altogether, our data suggest that the maternal gestational environment (i.e., GDM and methylation status) has an impact on the offspring. This is demonstrated in seven additional DMPs, where the interaction between the mother’s methylation and GDM status was FDR significant: within DLGAP2, H3C6, FAM13A, LOC127841, UBE3C, and two loci within intergenic regions. These CpGs were only FDR significant for the interaction association and not GDM alone.
Interestingly, a few of these genes have previously been identified to have a role in diabetes, namely DLGAP2 and FAM13A. For both CpGs, we found a similar methylation trend as in TFCP2 (i.e., the methylation of the offspring was increased as a function of the methylation status of the GDM mother, compared with the nonexposed offspring). A recent study found that methylation changes in the DLGAP2 gene were associated with maternal insulin sensitivity in pregnancy (29). The authors found that hypermethylation in the CpG was associated with a decrease in the Matsuda index, which is a measure of insulin sensitivity, suggesting that maternal methylation may mediate an effect on insulin sensitivity in future offspring risk.
Another study found that DLGAP2 is involved in determining β-cell fate through AMP-activated protein kinase signaling (30), which has a role in maintaining insulin sensitivity and glucose homeostasis (31). Moreover, a recent study showed that FAM13A represses hepatic AMP-activated protein kinase activity, thereby inducing insulin resistance in mice (32). Single-nucleotide polymorphisms in FAM13A, in addition to UBE3C, another gene identified in our analysis, have previously been found to be associated with type 2 diabetes (33,34), BMI, and lipid traits (35,36), highlighting a possible role of their methylation in the future risk of obesity and type 2 diabetes in offspring. Interestingly, two separate meta-analyses in neonates identified methylation changes in both FAM13A and DLGAP2 as being associated with environmental changes during pregnancy (i.e., prenatal air pollution and smoking, respectively) (37,38).
We did not identify any individual CpGs as being directly associated with GDM exposure in our EWAS stratified for both offspring and mothers. This was consistent with our shared (i.e., pooled) approach of mother-offspring pairs, which increased the sample size (because both mothers and offspring were included in our model) and thus increased the statistical power. Therefore, our study strongly suggests that for this relatively homogenous population, there is no strong association with GDM exposure alone, irrespective of maternal environmental factors.
This study exhibits several strengths. To our knowledge, this is the largest, single-cohort study investigating GDM epigenetic associations. Moreover, with the carefully selected mother-offspring pairs, we were able to investigate the effect of maternal methylation on offspring in response to maternal GDM exposure. In addition, the individuals in our cohort were from a relatively homogeneous population, and all were nonsmokers, thereby bypassing any confounding associations resulting from ethnicity or any obvious confounding health risk factors. Because most published studies and cohorts have solely investigated offspring methylation, this study indeed warrants further investigation and provides a basis for follow-up studies.
In addition to strengths, this study has some limitations, which are important to consider. Given the absence of a comparable study design (i.e., sample size and paired mother-offspring cohort), our study does not include a replication cohort, and therefore, the results should be interpreted with caution. Although our study is the largest, single-population GDM epigenetic study, it is possible that we still did not have the power to detect an association with GDM exposure alone, which might be too small to detect at birth. It is important to note that our two-step approach, in addition to the different adjustments for the tissues in our study (i.e., mother’s blood and offspring cord blood), may have resulted in a loss of power in favor of reduced bias. It is also important to note that Finland has very high standards for GDM care, and maternal glucose levels are likely to be well controlled. As a result, any shared effects of GDM exposure on mother and offspring methylomes might be modest, despite the relatively large sample size of our study. Although our study design matched individuals by age and prepregnancy BMI, our cohort had insufficient numbers of controls with higher BMI. However, these individuals were included, because they were representative of patients in Finnish clinics.
The clinical and biological effects of these loci need to be further investigated in target tissues to explore the biological and clinical relevance of these findings. Previous evidence demonstrates that changes in DNA methylation in the blood, although well associated with tissue-specific changes, can be less intense. Indeed, in 2016, a study reported that methylation changes in the pancreatic islet were associated with age, implicating genes that have important roles in the pancreatic islet and insulin secretion (39). A majority of these associations were also observed, to a lesser extent, in blood. So although we observed small methylation differences in cord blood, it could be speculated that this change could also occur with more substantial effects in target tissues. Therefore, in our study, it is indeed possible that the effects in target genes are more pronounced and their role needs to be further elucidated, particularly for the very small effect size observed for TFCP2, which is a ubiquitous gene and may have multiple roles in different tissues.
The biological significance of the loci we identified as interacting, suggesting moderation by maternal methylation, is open to interpretation. There are precedents to our study that support plausible biological significance. For instance, one study included a similar two-hit hypothesis where the level of an individual’s MTHFR enzyme activity was determined by an interaction between DNA methylation status at the MTHFR gene and folate levels. The authors observed that individuals with compromised enzyme activity had the same methylation levels if they were folate sufficient (and vice versa). Therefore, only individuals with both alterations resulted in depletion of genome-wide methylation levels.
To our knowledge, to date this is the largest GDM methylation study involving a single cohort with access to phenotypic and methylation data from both mothers and offspring. The current study complements previous EWAS by incorporating the exposure and/or possible modulating effects of maternal DNA methylation status and found that there is no strong direct association between GDM exposure and the methylome of offspring; however, the context of the maternal environment may contribute to a moderating effect. Our study reveals the potential complexity of the epigenetic transmission between mothers with GDM and their offspring, likely determined by not only GDM exposure but also other factors indicated by maternal epigenetic status, which are involved in establishing epigenetic signatures in offspring.
Article Information
Funding. This study was sponsored by the French National Research Agency (Agence Nationale de la Recherche), ANR-16-CE17-0017-01; Fondation Francophone pour la Recherche sur le Diabète (FFRD), which is sponsored by Fédération Française des Diabètiques (FFD), Abbott, AstraZeneca, Eli Lilly, Merck Sharp & Dohme (MSD) and Novo Nordisk. This work was also supported by the Agence Nationale de la Recherche (ANR) grants European Genomic Institute for Diabetes (E.G.I.D), ANR-10-LABX-0046, a French State fund managed by ANR under the frame program Investissements d′Avenir I-SITE ULNE / ANR-16-IDEX-0004 ULNE and from the National Center for Precision Diabetic Medicine – PreciDIAB, which is jointly supported by the French National Agency for Research (ANR-18-IBHU-0001), by the European Union (FEDER), by the Hauts-de-France Regional Council and by the European Metropolis of Lille (MEL). The co-authors were supported by PREcisE; LongITools; and Medical Research Council UK (grants MR/M013138/1 and MRC/BBSRC MR/S03658X/1 to Joint Programming Initiative–A Healthy Diet for a Healthy Life). The FinnGeDi study was supported by the Academy of Finland, Signe and Ane Gyllenberg Foundation, Sigrid Jusélius Foundation, Foundation for Pediatric Research, Finnish Diabetes Research Foundation, and Novo Nordisk Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.C. verified the underlying data, performed the statistical analyses, and generated the associated figures and tables. M.C. and A.K. wrote the manuscript. M.C., P.F., and T.A. designed the study. E.Ke., S.M., M.V., and E.Ka. provided clinical data. S.H., A.B., F.D., E.T., M.V., M.-R.J., S.S., E.Ka., P.F., and T.A. contributed data. S.L. performed the wet laboratory experiments to generate the MethylationEPIC array data. All authors critically reviewed and edited the manuscript. T.A. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
M.C. and A.K. contributed equally to this work.
This article contains supplementary material online at https://doi.org/10.2337/figshare.14488899.
References
- 1. Guariguata L, Linnenkamp U, Beagley J, Whiting DR, Cho NH. Global estimates of the prevalence of hyperglycaemia in pregnancy. Diabetes Res Clin Pract 2014;103:176–185 [DOI] [PubMed] [Google Scholar]
- 2. Boerschmann H, Pflüger M, Henneberger L, Ziegler A-G, Hummel S. Prevalence and predictors of overweight and insulin resistance in offspring of mothers with gestational diabetes mellitus. Diabetes Care 2010;33:1845–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nolan CJ, Damm P, Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet 2011;378:169–181 [DOI] [PubMed] [Google Scholar]
- 4. Vääräsmäki M, Pouta A, Elliot P, et al. Adolescent manifestations of metabolic syndrome among children born to women with gestational diabetes in a general-population birth cohort. Am J Epidemiol 2009;169:1209–1215 [DOI] [PubMed] [Google Scholar]
- 5. Williams MA, Qiu C, Dempsey JC, Luthy DA. Familial aggregation of type 2 diabetes and chronic hypertension in women with gestational diabetes mellitus. J Reprod Med 2003;48:955–962 [PubMed] [Google Scholar]
- 6. Kwak SH, Kim S-H, Cho YM, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes 2012;61:531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruchat S-M, Houde A-A, Voisin G, et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 2013;8:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Hajj N, Pliushch G, Schneider E, et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes 2013;62:1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hjort L, Martino D, Grunnet LG, et al. Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight 2018;3:e122572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haertle L, El Hajj N, Dittrich M, et al. Epigenetic signatures of gestational diabetes mellitus on cord blood methylation. Clin Epigenetics 2017;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias S, Adam S, Rheeder P, Louw J, Pheiffer C. Altered genome-wide DNA methylation in peripheral blood of South African women with gestational diabetes mellitus. Int J Mol Sci 2019;20:5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Howe CG, Cox B, Fore R, et al. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the Pregnancy and Childhood Epigenetics Consortium. Diabetes Care 2020;43:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keikkala E, Mustaniemi S, Koivunen S, et al. Cohort profile: the Finnish Gestational Diabetes (FinnGeDi) study. Int J Epidemiol 2020;49:762–763g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Working Group established by the Finnish Medical Society Duodecim, the Medical Advisory Board of the Finnish Diabetes Association, and the Finnish Gynecological Association . Current Care Guideline. Gestational Diabetes. Helsinki, Finnish Medical Society Duodecim, 2008 [Google Scholar]
- 15. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2020. Available from https://www.R-project.org/ [Google Scholar]
- 16. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nordlund J, Bäcklin CL, Wahlberg P, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol 2013;14:r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou W, Laird PW, Shen H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 2017;45:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res 2016;44:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salas LA, Koestler DC. FlowSorted.Blood.EPIC: Illumina EPIC data on immunomagnetic sorted peripheral adult blood cells. Available from https://github.com/immunomethylomics/FlowSorted.Blood.EPIC
- 22. Salas LA, Gervin K, Jones MC. FlowSorted.CordBloodCombined.450k: Illumina 450k/EPIC data on FACS and MACS umbilical blood cells. Available from https://github.com/immunomethylomics/FlowSorted.CordBloodCo mbined.450k
- 23. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 24. Du P, Zhang X, Huang C-C, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48 [Google Scholar]
- 27. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taracha A, Kotarba G, Wilanowski T. Neglected functions of TFCP2/TFCP2L1/UBP1 transcription factors may offer valuable insights into their mechanisms of action. Int J Mol Sci 2018;19:2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hivert M-F, Cardenas A, Allard C, et al. Interplay of placental DNA methylation and maternal insulin sensitivity in pregnancy. Diabetes 2020;69:484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kone M, Pullen TJ, Sun G, et al. LKB1 and AMPK differentially regulate pancreatic β-cell identity. FASEB J 2014;28:4972–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coughlan KA, Valentine RJ, Ruderman NB, Saha AK. AMPK activation: a therapeutic target for type 2 diabetes? Diabetes Metab Syndr Obes 2014;7:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin X, Liou Y-H, Li Y, et al. FAM13A represses AMPK activity and regulates hepatic glucose and lipid metabolism. iScience 2020;23:100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scott RA, Scott LJ, Mägi R, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017;66:2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji Y, Yiorkas AM, Frau F, et al. Genome-wide and abdominal MRI data provide evidence that a genetically determined favorable adiposity phenotype is characterized by lower ectopic liver fat and lower risk of type 2 diabetes, heart disease, and hypertension. Diabetes 2019;68:207–219 [DOI] [PubMed] [Google Scholar]
- 36. Shungin D, Winkler TW, Croteau-Chonka DC, et al.; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium . New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015;518:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gruzieva O, Xu C-J, Yousefi P, et al. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect 2019;127:57012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 2016;98:680–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bacos K, Gillberg L, Volkov P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun 2016;7:11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sankilampi U, Hannila M-L, Saari A, Gissler M, Dunkel L. New population-based references for birth weight, length, and head circumference in singletons and twins from 23 to 43 gestation weeks. Ann Med 2013;45:446–454 [DOI] [PubMed] [Google Scholar]