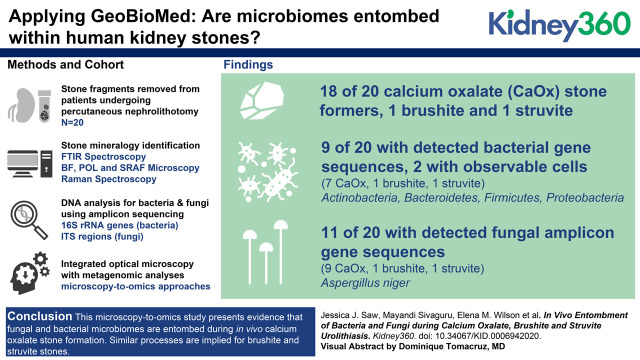
Visual Abstract
Keywords: nephrolithiasis, bacteria, basic science, fungi entombment, geomicrobiology, kidney stone mineralogy, microbiome, nephrolithiasis, Raman spectroscopy, super-resolution autofluorescence (SRAF) microscopy, urolithiasis
Abstract
Background
Human kidney stones form via repeated events of mineral precipitation, partial dissolution, and reprecipitation, which are directly analogous to similar processes in other natural and manmade environments, where resident microbiomes strongly influence biomineralization. High-resolution microscopy and high-fidelity metagenomic (microscopy-to-omics) analyses, applicable to all forms of biomineralization, have been applied to assemble definitive evidence of in vivo microbiome entombment during urolithiasis.
Methods
Stone fragments were collected from a randomly chosen cohort of 20 patients using standard percutaneous nephrolithotomy (PCNL). Fourier transform infrared (FTIR) spectroscopy indicated that 18 of these patients were calcium oxalate (CaOx) stone formers, whereas one patient formed each formed brushite and struvite stones. This apportionment is consistent with global stone mineralogy distributions. Stone fragments from seven of these 20 patients (five CaOx, one brushite, and one struvite) were thin sectioned and analyzed using brightfield (BF), polarization (POL), confocal, super-resolution autofluorescence (SRAF), and Raman techniques. DNA from remaining fragments, grouped according to each of the 20 patients, were analyzed with amplicon sequencing of 16S rRNA gene sequences (V1–V3, V3–V5) and internal transcribed spacer (ITS1, ITS2) regions.
Results
Bulk-entombed DNA was sequenced from stone fragments in 11 of the 18 patients who formed CaOx stones, and the patients who formed brushite and struvite stones. These analyses confirmed the presence of an entombed low-diversity community of bacteria and fungi, including Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Aspergillus niger. Bacterial cells approximately 1 μm in diameter were also optically observed to be entombed and well preserved in amorphous hydroxyapatite spherules and fans of needle-like crystals of brushite and struvite.
Conclusions
These results indicate a microbiome is entombed during in vivo CaOx stone formation. Similar processes are implied for brushite and struvite stones. This evidence lays the groundwork for future in vitro and in vivo experimentation to determine how the microbiome may actively and/or passively influence kidney stone biomineralization.
Introduction
Calcium-rich human kidney stones composed of the minerals calcium oxalate (CaOx, CaC2O4), calcium phosphate (hydroxyapatite Ca10[PO4]6[OH]2), and brushite (CaHPO4) are the most common products globally of urolithiasis in industrialized nations (1). Integrated approaches from geology, biology, and medicine to study universal biomineralization in humans, animals, and plants, called GeoBioMed, has recently shown these kidney stones are formed via repeated events of precipitation, partial dissolution, and reprecipitation (2,3). Each of these biomineralization steps is promoted and/or inhibited by a variety of physical, chemical, and biologic processes and mechanisms (4). Importantly, the deposition of minerals that form human kidney stones (5) is directly comparable to biomineralization processes in other natural and manmade environments (6), all of which are strongly influenced by microorganisms (collectively called the microbiome) (7). For example, in hot-spring calcium carbonate (CaCO3) travertine deposits, bacteria control mineral growth rate, mineralogy, and crystalline structure (6). Additionally, fungi excrete a number of organic acids (e.g., citric, oxalic, and formic acids, among many others) that drive limestone dissolution and reprecipitation in both natural and manmade settings (8). Similar types of microbe-urine-mineral interactions are thought to influence human kidney stone formation (5). For instance, previous microbial culturing and 16S rRNA gene sequencing from human kidney stones have detected both bacteria and fungi (9–12). As another example, oxalate-metabolizing bacterial networks in the gut microbiome have also been identified as a contributing factor to hyperoxaluria and the simultaneous formation of CaOx kidney stones (13). Furthermore, struvite stones have been shown to be influenced by the metabolic activity of urease-producing bacteria (14).
Given this background, this study has integrated optical microscopy with metagenomic analyses (herein called microscopy-to-omics) to collect definitive direct evidence of in vivo microbiome diversity, entombment, and preservation during human kidney stone formation. This project was undertaken to analyze stone fragments collected from a randomly chosen cohort of 20 patients using standard percutaneous nephrolithotomy. This experimental design requires that analytical results from all 20 patients be presented, which included 18 CaOx stone formers, one CaHPO4 stone former, and one struvite stone former. The relatively large number of patients who formed CaOx stones has permitted conclusive interpretations to be drawn regarding in vivo microbiome diversity, entombment, and preservation. In contrast, the small number of patients who form CaHPO4 and struvite stones prevents similarly conclusive characterizations. However, their microscopy-to-omics analyses are nonetheless extremely valuable, and serve as a useful pilot study comparison with previously published microbiome analyses of CaOx, CaHPO4, and struvite kidney stone fragments. Similar applications of microscopy-to-omics analyses, aided by comparisons with biomineralization in other natural and manmade environments, can be used to design future in vitro and in vivo experimentation dedicated to finding new therapeutic interventions for the prevention and treatment of urolithiasis.
Materials and Methods
The methods used in this study are briefly summarized here and presented in detail in the Supplemental Materials. This includes a flow chart of the systematic analyses applied in this study (Figure 1). Kidney stone fragments were collected from a cohort of 20 randomly chosen patients at the Mayo Clinic (Supplemental Table 1). All patients received antibiotics for a minimum of 7 days before surgery. Medical history, standard serum labs, medication intake, and comorbid conditions were assessed. In addition, 24-hour urine supersaturation profiles were obtained from all patients after surgery (Supplemental Table 2). To collect enough entombed microbial DNA, multiple stone fragments were grouped from each patient, washed in deionized water, air dried, and analyzed for bulk mineralogic composition using Fourier Transform Infrared Spectroscopy at the Mayo Clinic Metals Laboratories (Supplemental Table 3). Subsets from seven of the patient-specific fragment groups (five CaOx, one CaHPO4, one struvite; Figure 1) were three-dimensionally oriented, impregnated with epoxy, and made into 25 µm thick, doubly polished, uncovered thin section by Wagner Petrographic (Lindon, Utah) (3). Thin section microscopy analyses were carried out in the Microscopy Core of the Carl R. Woese Institute for Genomic Biology on a Zeiss Axio Zoom.V16, a Zeiss Axio Observer Widefield System, a Zeiss LSM 880 Laser Scanning Microscope with Airyscan Superresolution, and a WITec Alpha 300RAS Raman System.
Figure 1.
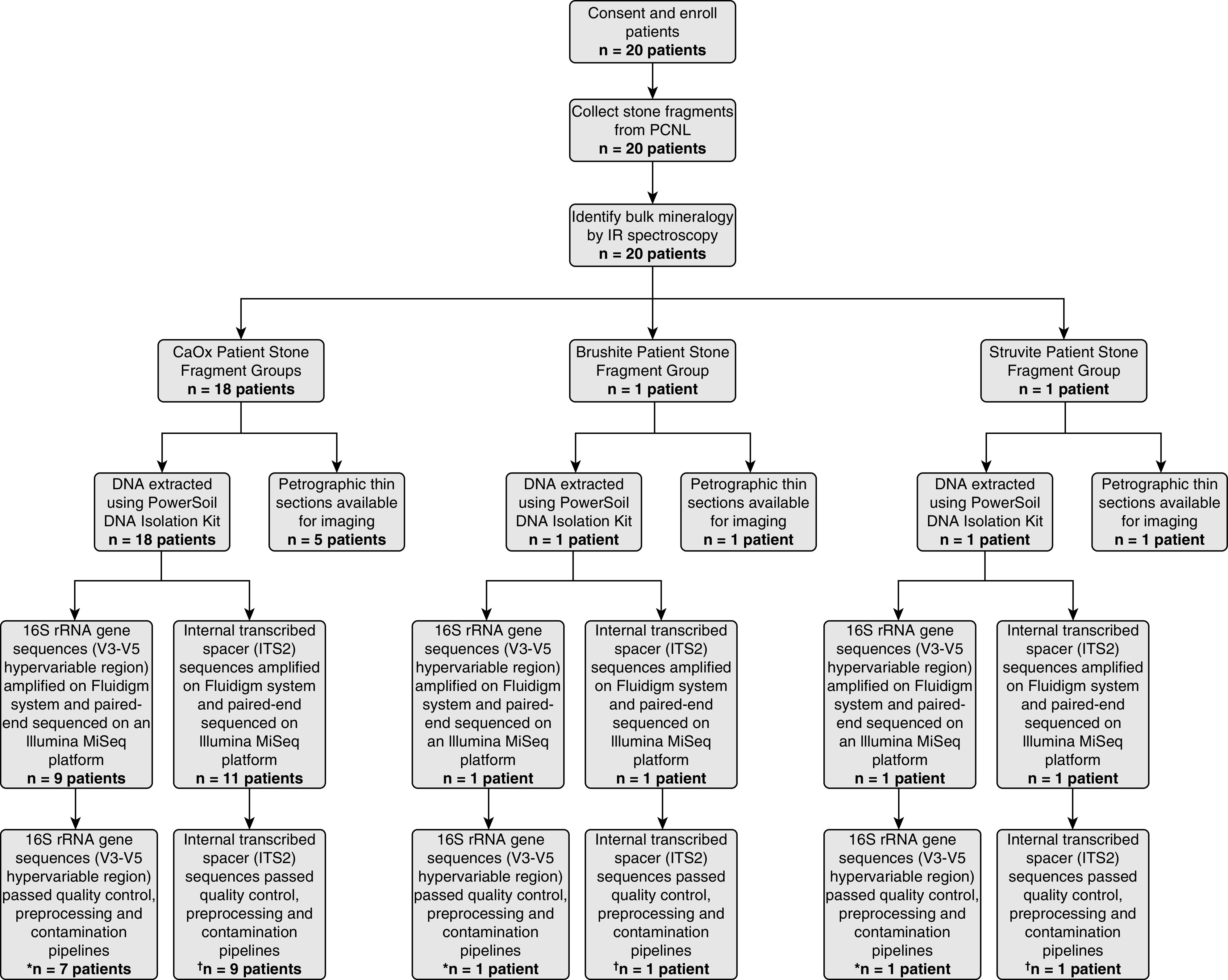
Study flow. A total of 20 patients consented and enrolled in the study. Mineralogies analyzed in this study include calcium oxalate (CaOx) (n=18 patients), struvite (n=1 patient), and brushite (n=1 patient). After filtering reads through pipelines for quality, preprocessing, and contamination, *16S rRNA gene sequences (V3–V5 hypervariable region) were detected in nine out of 20 patients (45%), whereas internal transcribed spacer †internal transcribed spacer 2 (ITS2) sequences were detected in 11 out of 20 patients (55%). PCNL, percutaneous nephrolithotomy; IR, infrared.
Another subset of each of the 20 patient-specific kidney stone fragment groups were collected for metagenomic analyses by flash freezing in the operating room immediately after standard percutaneous nephrolithotomy collection via placement in a −80°C Taylor-Wharton CX Series dry shipper dewar (Borehamwood, UK). The dewar was shipped overnight to the Carl R. Woese Institute for Genomic Biology, where the fragment cohorts were stored at −80°C until analyzed. Thawed fragment groups were powdered with a sterilized mortar and pestle under a sterile laminar flow hood. Molecular sequencing and bioinformatic analysis of the V1–V3 and V3–V5 hypervariable regions of bacterial 16S rRNA gene sequences, human host nonribosomal DNA fragments, and fungal internal transcribed spacer (ITS) regions (ITS1 and ITS2) were conducted in the Roy J. Carver Biotechnology Center. The 16S rRNA gene sequences and ITS regions were analyzed on a Fluidigm system. Paired-end sequencing was completed on an Illumina MiSeq platform. Amplicon sequence variants (ASVs) in control samples were identified as contaminants and removed. Phylogenetic diversity analysis and statistical analyses were completed using the Phyloseq v1.22.3 (15) and R programs (16). Statistical correlations were assessed using Wilcoxon signed-rank and Fisher’s exact test.
Ethics Approval and Consent to Participate
This basic medical research study was reviewed and approved by the Institutional Review Board (09–002083) at the Mayo Clinic. Written informed consent was obtained from all participants and are on file with the Mayo Clinic in Rochester, Minnesota.
Availability of Data and Materials
The raw metagenomic sequencing data and other raw images and finalized images can be retrieved from the following link.
Kidney stone fragment thin sections and all data curation is available from Dr. Mayandi Sivaguru.
Results
Entombment of Bacterial, Fungal, and Human Host Amplicon Sequences
The V1–V3 and V3–V5 hypervariable regions of bacterial 16S rRNA gene sequences and human host nonribosomal DNA fragments were detected in nine out of 20 patient-specific kidney stone fragment groups (seven CaOx, one CaHPO4, one struvite; Figures 1 and 2A, Supplemental Figures 1 and 2). From these, a total of 214 unique ASVs were identified (Figure 2A, Supplemental Table 4). Host human nonribosomal DNA sequences were dominant in four of the seven CaOx fragment groups (Figure 3A, Supplemental Figure 1). In the CaHPO4 stone fragment group, 99.5% of the total sequence reads were classified as host human nonribosomal DNA sequences (Figure 3C). Conversely, no host human nonribosomal DNA was detected in the struvite fragment group (Figure 3E). The 40 unique ASVs identified in the struvite fragment group were slightly more diverse than those detected in the CaOx and CaHPO4 fragment groups (Shannon Index 0.84, Simpson Index 0.35). The struvite fragment group bacterial community composition was dominated by Staphylococcus (80% of reads, Figure 3E) and included Porphyromonas (7%), Abiotrophia (6%), and Haemophilus (2%). Fungal amplicon sequences were detected in 11 out of 20 fragment groups (nine CaOx, one CaHPO4; one struvite; Figures 1 and 2B, Supplemental Figure 3). ASVs attained from all stone fragment groups were predominantly Aspergillus niger, with the remaining 36 ASVs affiliated with Aspergillus, Basidiomycota, and Agaricomycetes. In addition, Aspergillus nomius, Aspergillus costaricensis, Candida albicans, Candida dubliniensis, and Dothideaceae (family-level) constituted <1% of the total community (Figure 3, B, D, and F; Supplemental Figures 2 and 4).
Figure 2.
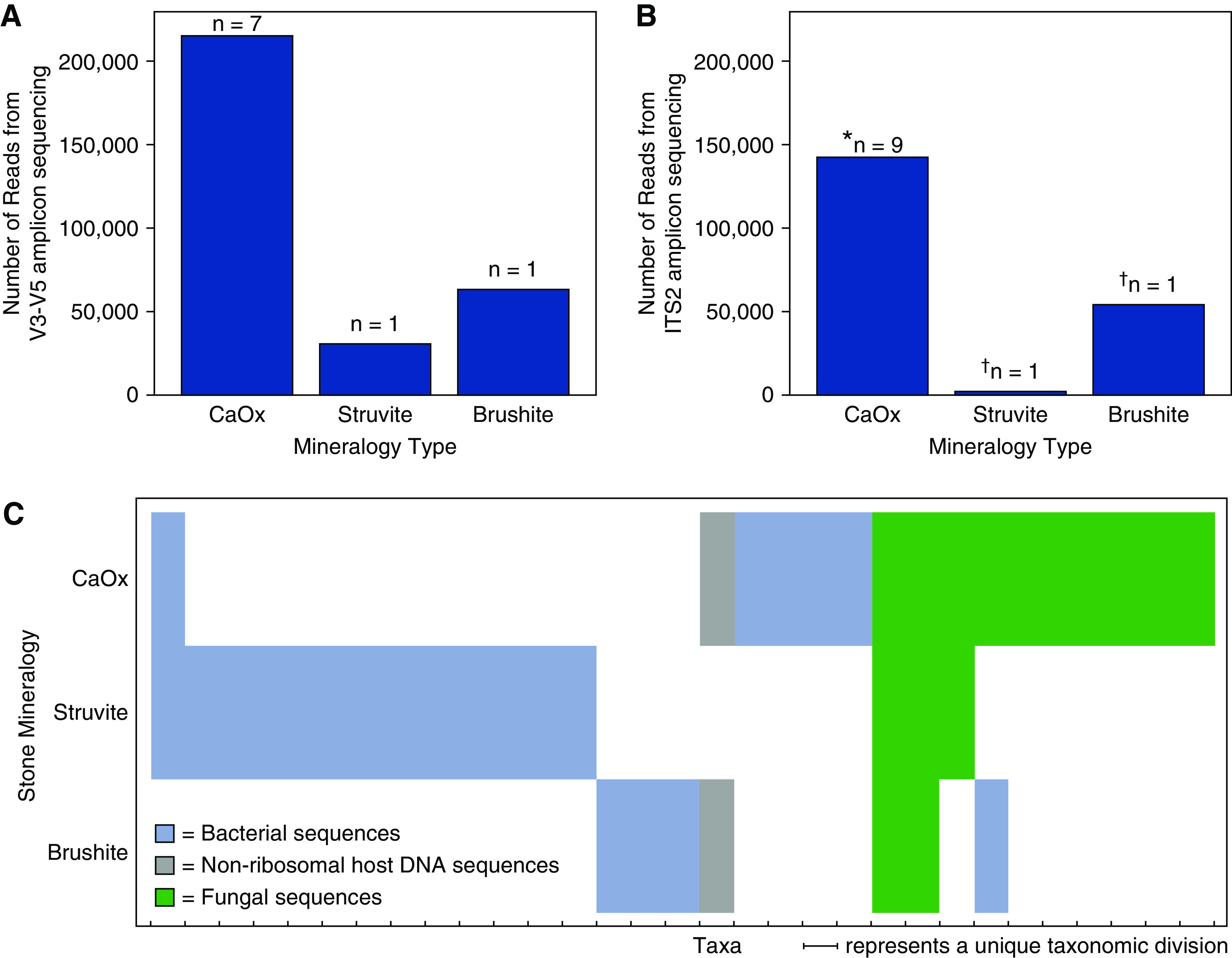
Total number of reads and taxa grouped by kidney stone mineralogical type. Number of amplicon sequence variants (ASVs) detected within calcium oxalate (CaOx), brushite, and struvite kidney stones, respectively, targeting the (A) V3–V5 hypervariable region of the 16S rDNA gene and (B) internal transcribed spacer 2 (ITS2) hypervariable region of the fungal rDNA gene. The number of reads for each individual patient stone fragment group are presented in Supplemental Figure 1. The asterisk (*) represents six of the stone fragment groups presented in (A), and an additional three stone fragment groups included in calculation. The dagger (†) represents the same stone fragment groups presented in (A). (C) Binary heatmap indicating the presence or absence of unique taxonomical divisions identified from ASVs (blue = bacteria; gray = nonribosomal host human DNA; green = fungi). The n values correspond to individual patient stone fragment groups.
Figure 3.
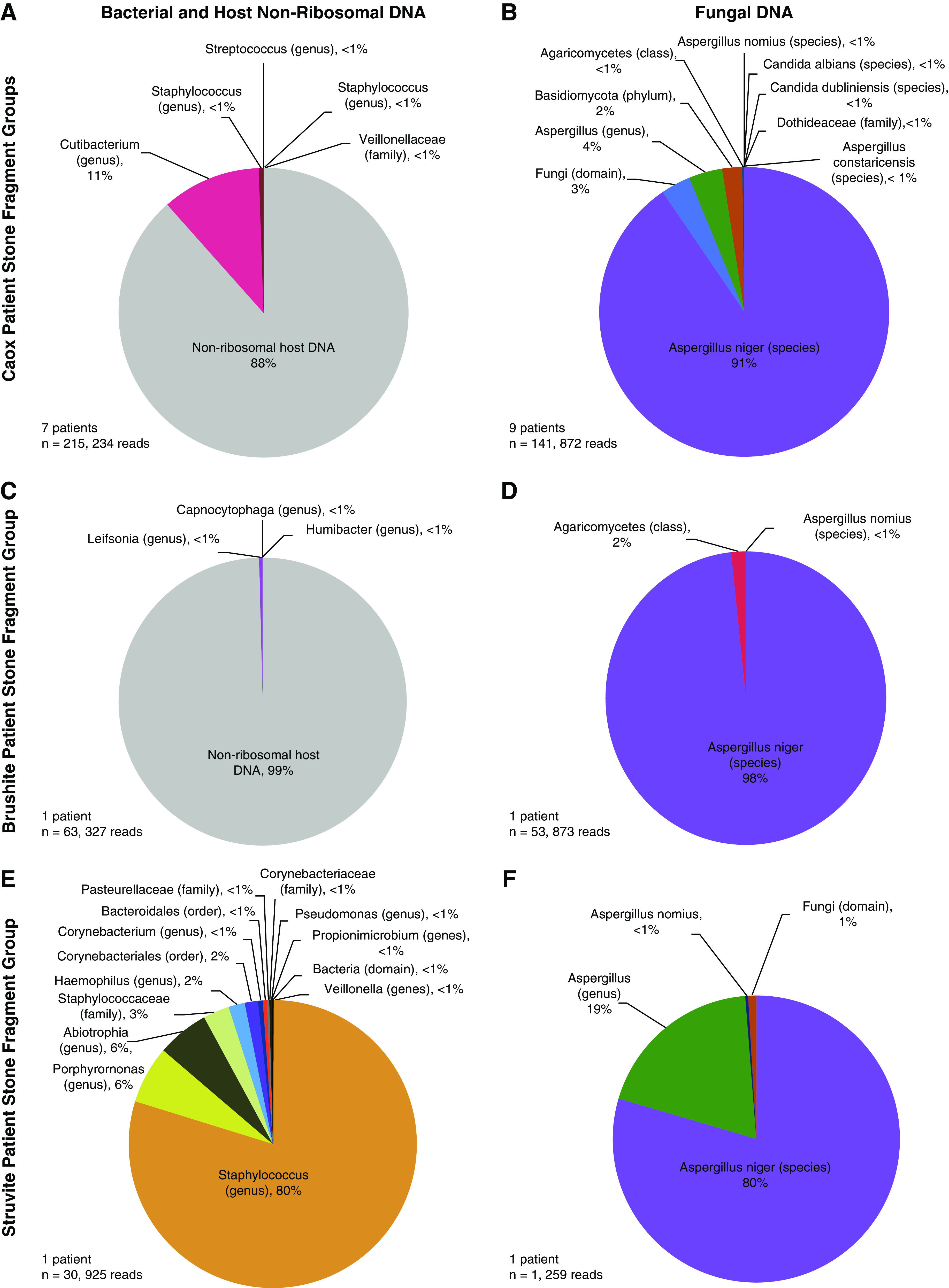
Phylogenetic diversity of three mineralogical types of human kidney stones. Pie charts represent the community diversity of bacterial 16S rRNA gene sequences, nonribosomal human host (A, C, and E), and internal transcribed spacer (ITS) fungal sequences (B, D, and F) from calcium oxalate (CaOx), brushite, and struvite patient-specific kidney stone fragment groups, respectively. The community diversity of bacterial and fungal sequences for each individual patient kidney stone fragment group is presented in Supplemental Figure 1.
Microbiome Partitioning between Stone Mineralogy Types
The 18 patients that formed CaOx kidney stone fragment groups permit firm conclusions to be drawn with respect to in vivo microbiome diversity, entombment, and preservation. However, the single CaHPO4 and struvite fragment groups permit only pilot study–level initial comparison with the CaOx fragment groups. The CaOx, CaHPO4, and struvite fragment groups each contain significantly different entombed bacterial communities (Figure 2C). Bacteria in the CaHPO4 fragment group share only one taxa (Staphylococcus) with the CaOx and struvite fragment groups. Additionally, host nonribosomal DNA was identified in the CaOx and CaHPO4 fragment groups, but not in the struvite fragment group (Figure 2C). Fungal communities exhibited more overlap among the three CaOx, CaHPO4, and struvite fragment group mineralogy types (Figure 2C). A. niger dominated the fungal communities of all three fragment mineralogy types, whereas A. nomius constituted <1% of these fungal communities. CaOx and struvite fragment groups shared Aspergillus, whereas the CaOx and CaHPO4 fragment groups shared Agaricomycetes.
Correlation of Fungal Microbiome with Urinary Calcium and Oxalate Excretion
The 11 patient-specific kidney stone fragment groups containing fungal sequences do not exhibit statistically significant correlations with higher patient urine calcium excretion at the α=0.05 level (Figure 4A; P=0.07). However, mean calcium concentrations in 24-hour urine analyses are higher in the presence of fungal sequences (“absent” mean = 180.3 mg/24 hour; “present” mean = 308.8 mg/24 hour; Figure 4A). Similarly, there were no statistically significant correlations observed between the presence and absence of fungal sequences and urine oxalate concentrations (Figure 4B; P=0.15). However, patients with fungal sequences in their stone fragment groups exhibited lower mean urine oxalate concentrations (20.2 mg/24 hour) than patients without fungal sequences (33.0 mg/24 hour). Patient-specific fragment groups with fungal sequences also exhibited statistically insignificant (P=0.33) increases in average 24-hour urine CaOx saturation indices (1.6 Delta Gibbs Free Energy/24 hour without; 2.0 Delta Gibbs Free Energy/24 hour with). Additionally, there was no statistically significant correlation between the presence or absence of fungal sequences and patient 24-hour urine citrate levels (P=0.4). Finally, the presence or absence of apatite in each patient-specific fragment group correlates with the presence of fungal sequences (P=0.02; odds ratio, 0.05; 95% confidence interval, 0 to 0.65).
Figure 4.
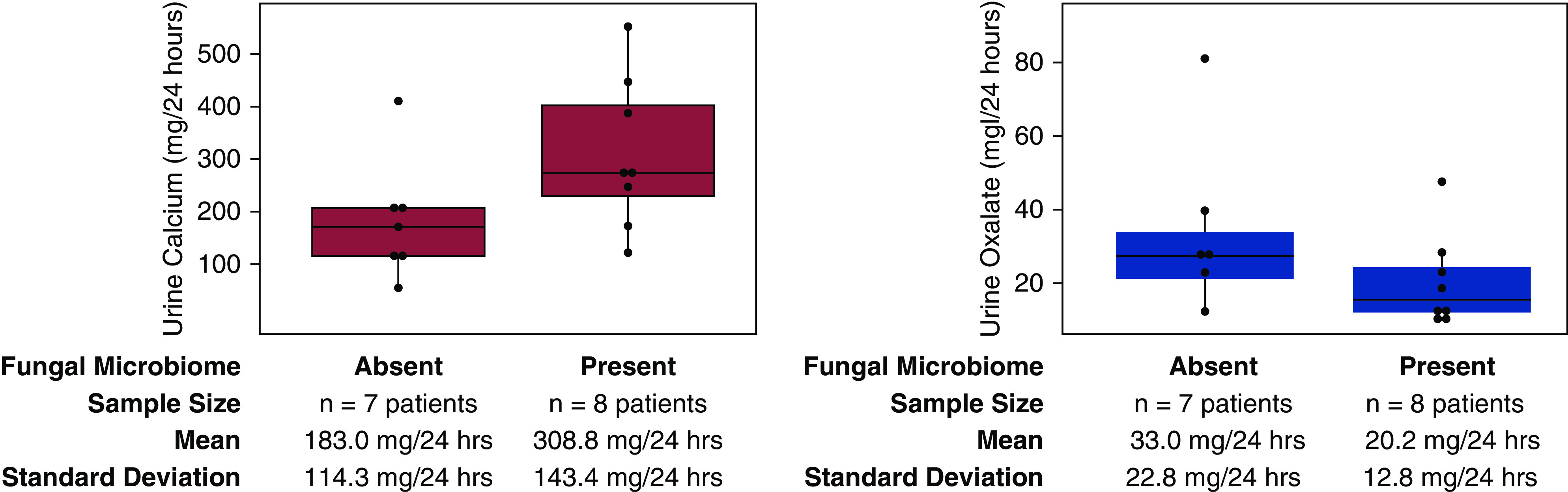
Patient metadata analysis. Wilcoxon signed-rank tests that the detection of fungal sequences is not significantly correlated with elevated urine calcium excretion at the α=0.05 level (P=0.07). No statistically significant correlation was observed between the presence or absence of fungal sequences and urine oxalate excretion. Mean and SD for each sample group are depicted in the text below the graphs. Purple and blue boxes are box plots for urine calcium and oxalate levels, respectively. The box plot summary statistics are as follows: lower boundary of box = first quartile, line in center of box = second quartile (median), upper boundary of box = third quartile. Dots represents individual patient data points. A dot unattached to the vertical lines represents outlier points, which were included when calculating the interquartile range.
Microbiome Entombment within Amorphous and Crystalline Minerals
Microscopy was completed on five CaOx, one CaHPO4, and one struvite patient-specific kidney stone fragment. Whereas bacteria and fungi were detected in the metagenomic analyses for the CaOx stone fragments (Figure 3), neither entombed bacterial cells or fungal hyphae borings were observed with microscopy. It is important to note that metagenomic analyses result from a significantly higher volume coverage of each of the patient-specific stone fragment groups, and are therefore expected to yield more evidence of an entombed microbiome than the small 25 μm thick cross-sectional area investigated by thin section microscopy analyses. Conversely, CaHPO4 and struvite stone fragments contained both bacterial and fungal metagenomic sequences (Figure 3). Although they clearly exhibited entombed bacterial cells within their amorphous spherulitic hydroxyapatite (Figures 5–8, Supplemental Figure 4), no fungal hyphae were observed. In the struvite stone fragment, concentrically layered amorphous hydroxyapatite spherules and radiating needle-like (acicular) struvite crystals contain entombed coccoid- and rod-shaped bacteria approximately 1 μm in diameter (Figures 5 and 6). Similarly, the CaHPO4 fragment contained entombed bacteria within concentrically layered amorphous hydroxyapatite spherules and radiating acicular CaHPO4 crystals (Figures 6, E and F and 8). In addition, Raman spectroscopy has identified entombed organic biomolecules, including lipids and proteins, and confirmed that amorphous spherules are comprised of hydroxyapatite (Figure 7).
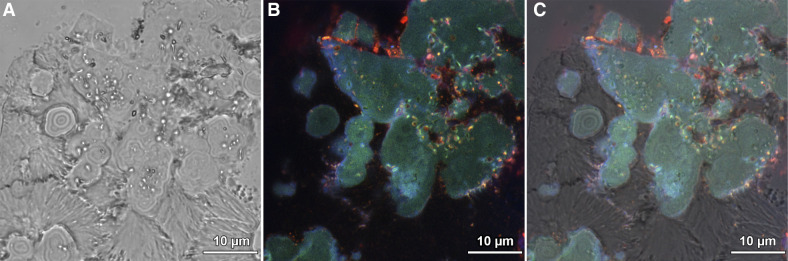
Figure 5.
Microscopy evidence of microorganism entombment within human kidney stones. Mineralogical identifications determined with a combination of bulk stone Fourier Transform Infrared (FTIR) analyses and determination of individual crystal morphologies (2) and Raman spectroscopy (Figure 7) in thin sections. (A) transmitted light photomultiplier (TPMT), (B) super-resolution autofluorescence (SRAF), and (C) TPMT overlaid on SRAF image from a struvite kidney stone documenting entombed bright orange autofluorescent coccoidal and rod-shaped bacterial cells. See also Supplemental Figure 5 for contextualization of the occurrence of both coccoidal and rod-shaped bacteria in surrounding regions of the thin section.
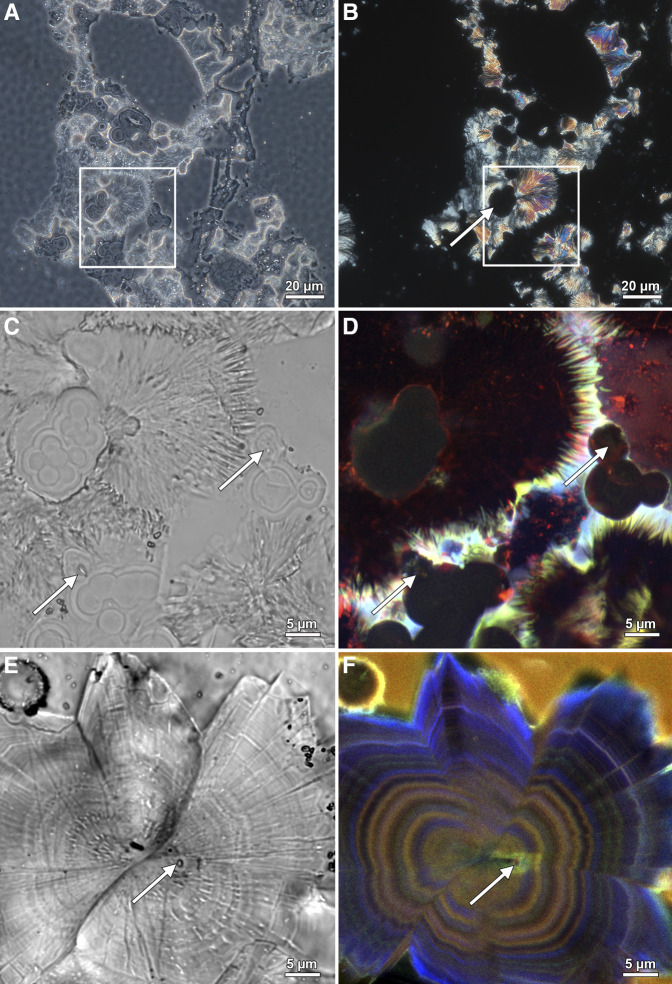
Figure 6.
Evidence for bacteria entombed within struvite and brushite kidney stones. Mineralogical identifications determined with a combination of bulk stone Fourier Transform Infrared (FTIR) analyses and determination of individual crystal morphologies (2) and Raman spectroscopy (Figure 7) in thin sections. (A) Color brightfield (BF) image of a 25 µm thin section prepared from a struvite stone. (B) The same field of view as in a indicating that concentrically layered spherulitic hydroxyapatite exhibits extinction under polarized light (POL) and are therefore amorphous (non-crystalline). Conversely, the radiating needle-like (acicular) crystals of struvite are strongly birefringent. (C) Color BF image of enlargement box shown in (A). White arrows in (C and D) indicate cross-sections at various oblique angles of entombed coccoidal and rod-shaped bacteria. (D) Super-resolution autofluorescence (SRAF) image of the same field of view shown in (C). (E) BF image of polymorphic twinning of radiating acicular brushite crystals. This is an approximately 600 nm thick slice in reflection mode, therefore not all BF objects are not in focus. White arrows in (E and F) indicate cross-sections at various oblique angles of entombed coccoidal and rod-shaped bacteria. (F) SRAF image of the same field of view shown in (E). Regions of blurred (fuzzy) concentric zonations represent regions of mimetic dissolution and replacement (2,4).
Figure 7.
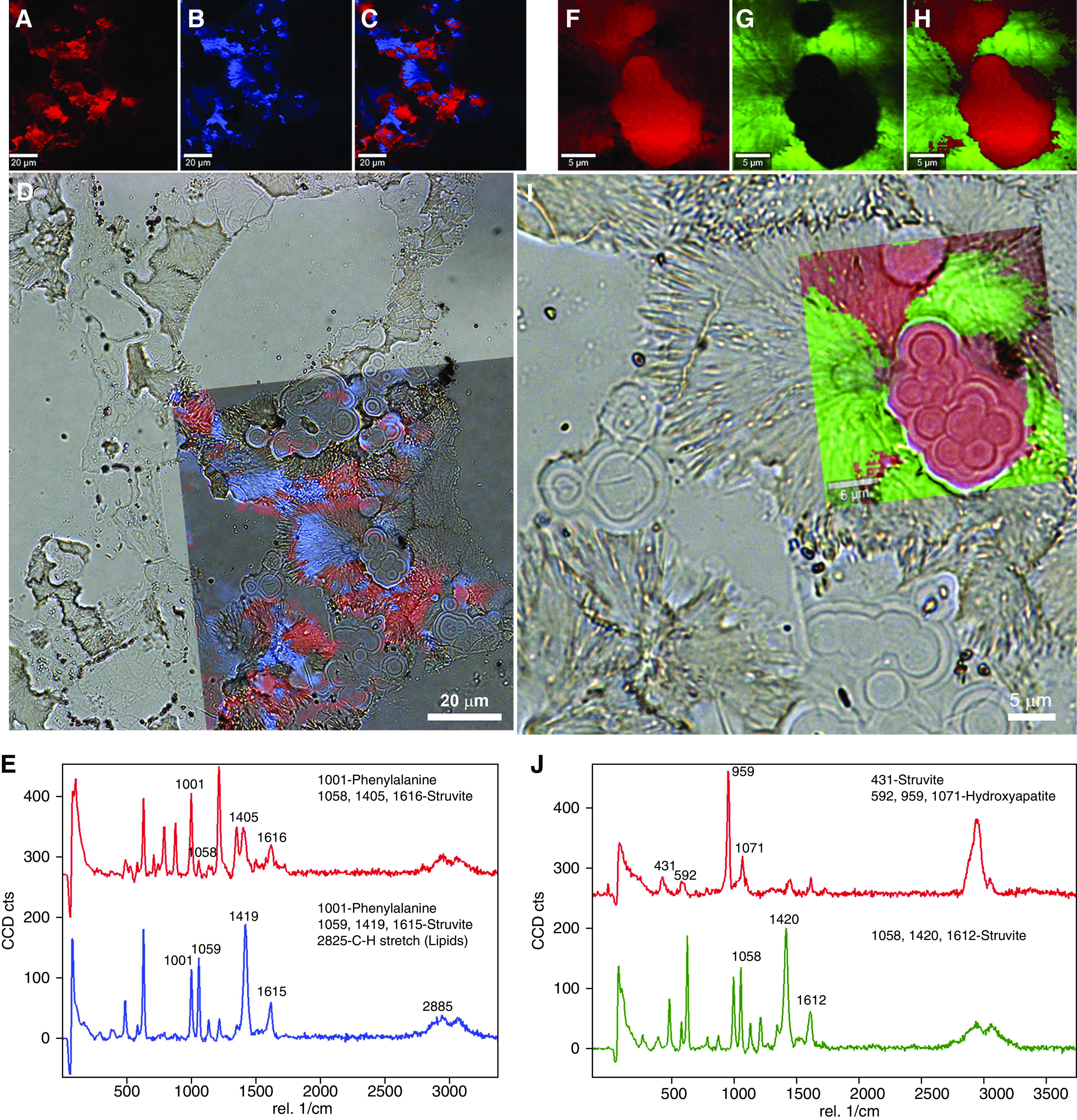
Raman spectroscopy evidence for entombed bacteria within a struvite (NH4MgPO40.6H2O) human kidney stone. (A–C) Mineral component 1 (A, pseudo-colored red), mineral component 2 (B, pseudo-colored blue), and the corresponding merged images extracted from Raman spectra (C). (D) Transparent overlay of image (C) on a lower magnification color brightfield (BF) image illustrating optical microscopy correlated with Raman spectroscopy. (E) Raman spectra for mineral components 1 (A) and 2 (B), with legends highlighting chemical components for identified peaks on the basis of Takasaki (Supplemental Reference 11) and Balan et al. (Supplemental Reference 12). (F–J) Enlargement of box in (D) similar to Figure 5C but with high magnification Raman scan pseudo-colored red for hydroxyapatite Ca10(PO4)6(OH2) and green for struvite. Note the 959 peak (italicized in J) is the “high” peak for hydroxyapatite among the other peaks. Also note the similarity between struvite peaks in (E) (larger field of view) and (J) (smaller field of view).
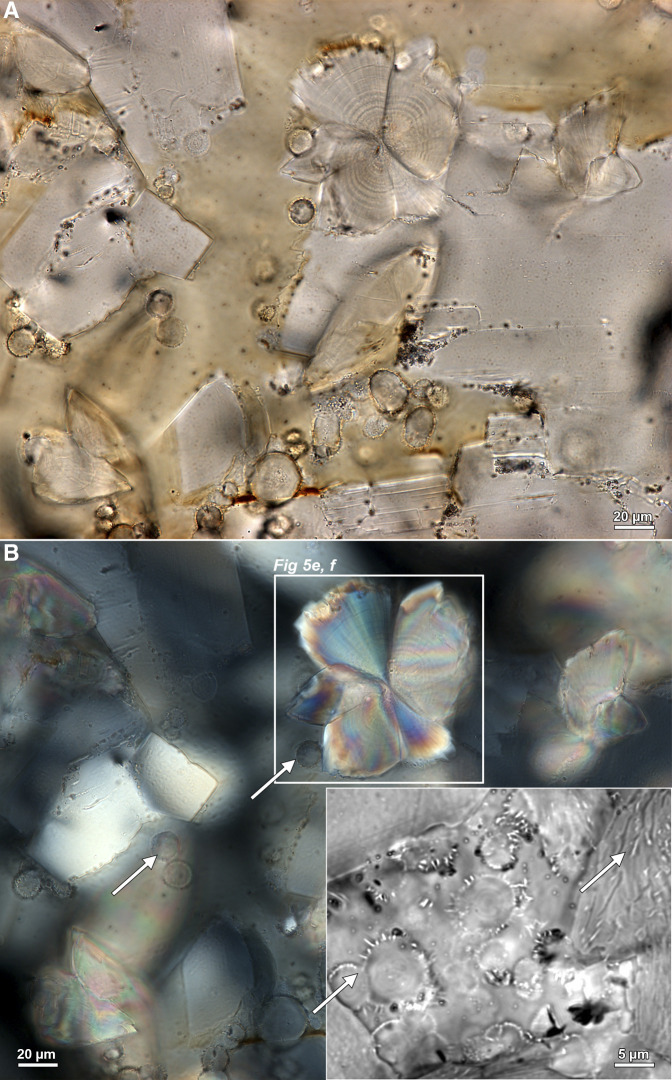
Figure 8.
Microscopy evidence for entombed bacteria in brushite human kidney stones. (A) Color brightfield (BF) image of a 25 µm thin section prepared from a brushite stone. (B) Same field of view as in a indicating that concentrically layered spherules exhibit extinction under polarized light (POL) and are therefore amorphous (noncrystalline; white arrows). Conversely, the radiating acicular crystals of brushite (white box in B, see also Figure 5, E and F) are strongly birefringent. Inset in lower right in (B) is a transmitted light photomultiplier (TPMT) image of spherules with entombed coccoidal and rod-shaped bacteria throughout each crystal (white arrows). White arrows in (B) indicate cross-sections at various oblique angles of entombed coccoidal and rod-shaped bacteria.
Discussion
Phylogenetic Diversity of the Entombed In Vivo Microbiome
The microscopy-to-omics analyses assembled in this study provide important microscopy and genomic evidence of entombment of a low-diversity community of microorganisms during in vivo CaOx stone formation (Figures 2 and 3). Optical and genomic evidence in this pilot study analyses of the CaHPO4 and struvite patient-specific kidney stone fragments show similar processes are implied for CaHPO4 and struvite stones (Figures 2–8). The human microbiome, in combination with both genetic and environmental factors, has been implicated as playing an important role in urolithiasis (5,17–19). This is supported by several previous studies using both culturing and sequencing techniques that have detected a microbiome within CaOx stones (10–12), CaHPO4 stones (12), struvite stones (14), and urine (12,20).
Staphylococcus was detected in both this study (Figure 3A) and previous analyses of CaOx stone fragments (10–12). However, no overlap was observed in bacterial diversity between this pilot study (Figure 3C) and previous studies of CaHPO4 stone microbiology (12). Conversely, Pseudomonas and Staphylococcus were found in common between this pilot study (Figure 3E) and previous struvite stone analyses (14). Pseudomonas, along with other microorganisms associated with urinary tract infections (e.g., Proteus, Klebsiella, and yeast), produce ammonia and increase urine alkalinity (pH>7), which ultimately form struvite infection stones (14,21). In addition, this study identified fungal sequences affiliated with Aspergillus and Candida preserved within CaOx, CaHPO4, and struvite stone fragment groups (Figure 3, B, D, and F). This is the first amplicon gene sequencing identification of fungi in these three mineralogies, and is consistent with previous culturing studies that documented Candida growth from powdered CaOx stone fragments (9).
It has also recently been observed that patients who are idiopathic stone formers have developed an imbalance in the normal healthy composition of the bacterial communities inhabiting the gut (dysbiosis) (17,19,22). These dysbiosis studies have focused on gut microbiome networks that include the oxalate-degrading anaerobic bacterium Oxalabacter formigenes, whose activity is strongly controlled by changes in patient diet (19,23). The metabolic activity of Oxalabacter formingenes causes oxalate degradation, which decreases oxalate concentration in the serum to make intratubular CaOx precipitation less likely (24,25). Alternatively, enzymatic reactions might effectively degrade oxalate without the presence of Oxalabacter formingenes, which would similarly serve to decrease enteric hyperoxaluria (26–28). In addition to CaOx stones, hypercalciuria and elevated urine pH in CaHPO4 stones may also result from altered metabolic activity of the gut microbiome (12,29).
Antibiotics, probiotics, and microbial transplants further affect Oxalobacter formigenes colonization (17). Hyperoxaluria and CaOx urine supersaturation have also been shown to be attenuated by probiotic treatment (5,12,30). These studies have led to the hypothesis that the gut microbiome (27) acts as a metabolic modulator that directly influences CaOx kidney stone formation, creating a connection within the human body called the gut-kidney axis (19,31).
Biomineralization in Human Kidneys and Natural and Manmade Environments
Microbial communities play a fundamental role in the precipitation, dissolution, and recrystallization of phosphate, carbonate, and silicate biomineralization in natural and manmade environments around the world (e.g., coral reefs, hot springs, deep subsurfaces; Figure 9) (2). In these settings, cell wall surfaces, extracellular polymeric substances, and other organic molecules produced and influenced by microbial metabolism can act as direct surface controls on the rate and composition of crystalline mineral growth (6). In situ kinetic experiments in Yellowstone National Park carbonate hot springs have demonstrated the physical cell presence and biomolecules produced by living microorganisms can more than double mineral precipitation rates compared with when they are absent (32). These direct catalytic influences (protein catalysis) can now be linked to microbial gene targets identified by future microscopy-to-omics analyses in kidney stones. Additionally, microbes also cause dissolution across a wide range of rock types, most commonly via secretion of metabolic byproducts such as organic acids and production of biofilms. These processes, which make significant contributions to biogeochemical cycles in everything from natural rock formation to building stone degradation, have previously been well documented in both field and controlled laboratory conditions (33). Overall, microbially mediated mineral precipitation and dissolution is ubiquitous around the world (6).
Figure 9.
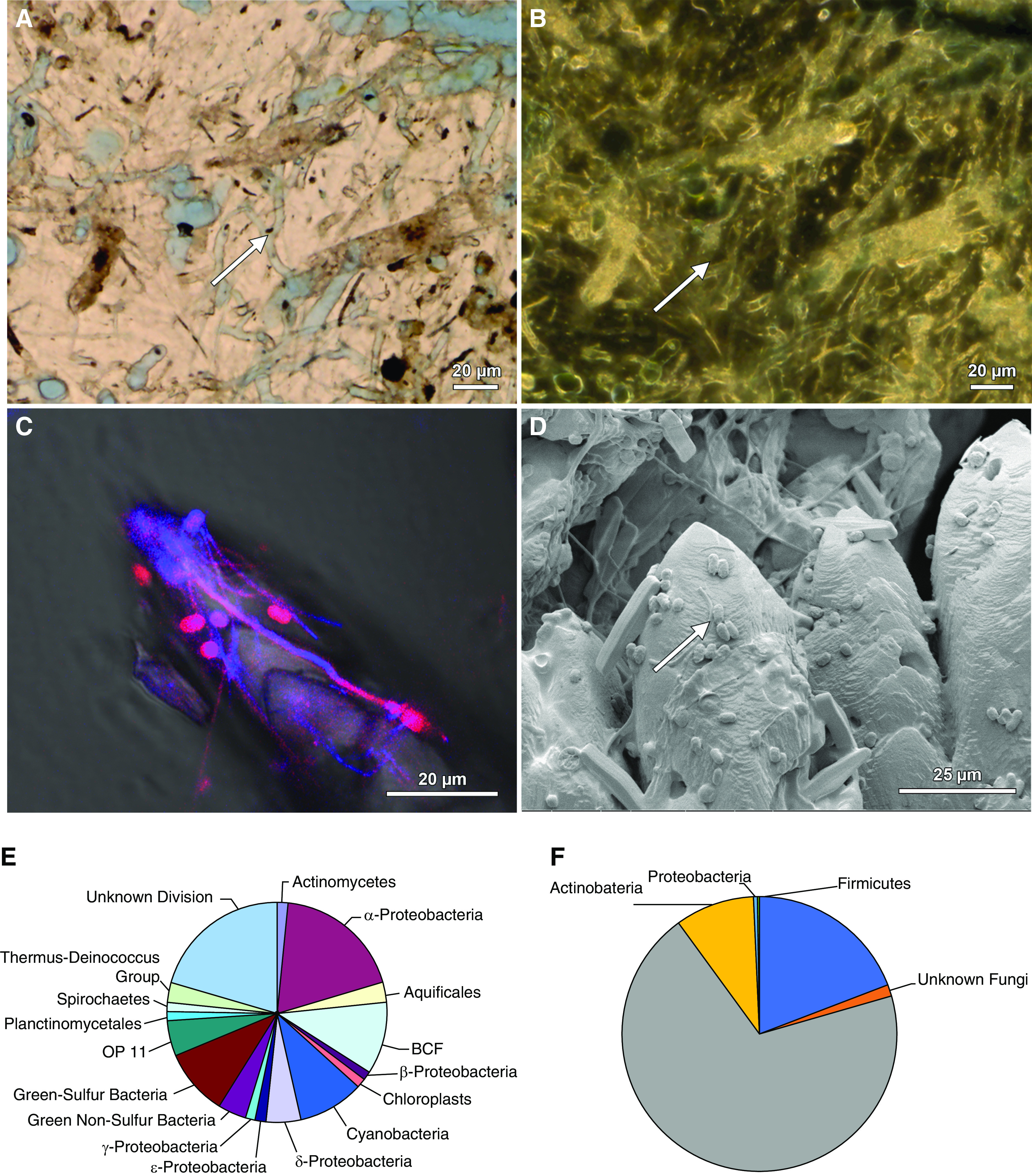
Microscopy-to-omics evidence for microbiome entombment during biomineralization in natural environments. Images and figures modified from previous publications (6,7). (A) A skeleton of the scleractinian coral Orbicella annularis from the leeward reef tract of Curaçao exhibits extensive fungal hyphae and borings (white arrow) in brightfield (BF). (B) Phase contrast (PC) image of the same field of view shown in (A). (C) Rapid growth and accretion of CaCO3 travertine at Mammoth Hot Springs in Yellowstone National Park is coated by and entombs coccoidal, rod, and filaments of the bacterium Sulfurihydrogenibium yellowstonense. A merged blue and red super-resolution autofluorescence (SRAF) image (violet to red color) overlaid on a BF image. (D) Environmental scanning electron microscope (ESEM) of the same sample shown in (C). (E) Phylogenetic diversity pie chart of the microbiome associated the deposition of hot spring travertine at Mammoth Hot Springs. (F) Phylogenetic diversity pie chart of the microbiome entombed in all calcium oxalate (CaOx) kidney stones analyzed in this study.
Initially, microbial populations are directly influenced by the environmental conditions in which they live (34). However, microbial populations can also rise to control the surrounding physical, chemical, and biologic environment (6). Similar types of dynamic feedback may occur during human kidney stone formation. This includes interactions among kidney physiology, microbiome community structure, hydraulic properties, biogeochemical composition of the urine (e.g., chemistry, temperature, pH, saturation, flow rate, nutrient substrates), and the biomolecule composition and reactivity of internal renal surfaces (e.g., tissues, stones, biofilms) (3). Acting in concert, these key factors of the renal environment will affect numerous aspects of kidney stone formation, including stone crystalline architecture, mineralogy, rate of crystallization, and timing and extent dissolution. As a result, microbiome-driven biomineralization and dissolution processes within the human kidney likely play a significant role in kidney stone formation. These insights can now be utilized to generate new testable hypotheses for mechanisms of the microbially mediated diagenetic phase transitions that may be intimately involved in kidney stone formation.
Microbial communities can play vitally important roles in the bulk concentration of ions (e.g., changes in fluid urine chemistry and kidney stone mineralogy and crystallography) that may in turn modify nucleation, crystallization, aggregation, and dissolution during the overall history of stone growth (2,3). An example of this is struvite stone formation, which has already been shown to be due to the effects of urine alkalinity generated by the metabolic activity of urease-producing bacteria (14). The similarities between kidney stones and rocks in the natural environment, as revealed in this study and previous studies, can now be used to develop new hypotheses regarding their formation. For example, the partitioning of fungal and bacterial communities between CaOx, CaHPO4, and struvite stone types is analogous to that found in many other natural rock formations, such as hot spring travertine (6) and marine coral skeletons (Figure 9) (35). As another example, kidney stones contain entombed organic matrices (proteins, lipids, and glycosaminoglycans) that are derived from the host human kidney, the microbiome, and urine (36–38). Organic matrices entombed within mineral deposits are observed in many other common examples of biomineralization, such as in human bones, coral reefs, and pearls (39–42). These biomolecules will play a similarly crucial role in both promotion and inhibition of crystal growth and dissolution during kidney stone formation (4,43). In the natural environment, fungal species such as A. niger ubiquitously mediate diagenetic phase transitions (44,45).These microscopy-to-omics results pave the way for future experimentation, in which these hypotheses can be systematically tested using highly controlled microfluidic devices (4,46). For instance, microfluidic testbeds can be used to quantitatively track the intermediate steps of kidney stone formation in real time under high-resolution microscopy, as in recently published reports (2,3,46). This would allow specific mechanisms and processes controlling kidney stone growth and dissolution to be tested in the context of microbiome community, phylogenetic diversity, functional activity, and biochemistry. Examples include determination of the extent to which Tamm-Horsfall and other proteins serve to promote or inhibit stone growth, while also evaluating its role in protecting the urinary tract and other human organs from fungal infections (4,11,20,25). Other experiments might include tracking stone growth rate, crystalline structure, and mineralogy while changing the microbiome to reflect actual urinary microbiome community diversity, structure, and metabolism (11,20,25). This could include mimicking the dysbiosis observed in urine from the renal calyx and bladder of calcium-based male stone formers (11,20,25). The effect of all types of microbial imbalances on kidney stone growth could therefore be tested, including those resulting from changes in the gut and urinary microbiome due to diet, lifestyle, and frequent antibiotic use (22,47).
Disclosures
A. Krambeck reports consultancy agreements with Boston Scientific, Lumenis, Sonomotion, and Virtuoso; receiving research funding from Boston Scientific, Lumenis; receiving honoraria from Boston Scientific, Lumenis, Sonomotion, and Virtuoso; reports having patents and inventions b7h1 and Survivin as a marker for Renal Cell Carcinoma; and reports being a scientific advisor or member of Boston Scientific, Sonomotion, and Virtuoso. B. Fouke reports receiving research funding from Dornier MedTech. D. Lange reports having consultancy agreements with AdvaTec, BD/Bard, Boston Scientific, Cook Medical, and Kisolite; having an ownership interest in Kisolite Corp; reports receiving research funding from AdvaTec, BD/Bard, Boston Scientific, and Cook Medical; and reports scientific advisor or membership of Kisolite Corp. J. Lieske reports having consultancy agreements with Alnylam, Allena, American Board of Internal Medicine, Dicerna, Orfan, OxThera, Retrophin, and Siemens; reports receiving research funding from Allena, Alnylam, Dicerna, OxThera, Retrophin, and Siemens; reports receiving honoraria from Alnylam, Allena, American Board of Internal Medicine, Dicerna, Retrophin, Novobiome, Orfan, OxThera, and Synlogic; scientific advisor or membership of American Board of Internal Medicine, Hyperoxaluria Foundation, Kidney International, and Oxalosis. M. Rivera reports consultancy agreements with Boston Scientific, Cook Medical and Lumenis. M. Romero reports scientific advisor or membership of Kidney360 - Associate Editor, American Journal of Physiology-Renal Physiology, Hyperoxaluria Foundation, Oxalosis, National Institute of Diabetes and Digestive and Kidney Diseases study sections, ad hoc. N. Chia reports receiving research funding from Archer Daniels Midland. T. Large reports having consultancy agreements with Boston Scientific and Lumenis. Y. Dong reports being a scientific advisor or member of Frontiers in Microbiology. All remaining authors have nothing to disclose. All remaining authors have nothing to disclose.
Funding
This research was supported by the Mayo Clinic and University of Illinois Strategic Alliance for Technology-Based Healthcare, Mayo Clinic Center for Individualized Medicine and Mayo Clinic O’Brien Urology Research Center (DK100227), Mayo Nephrology/Urology Summer Undergraduate Research Fellowship (DK101405), and the National Aeronautics and Space Administration Astrobiology Institute (Cooperative Agreement NNA13AA91A).
Acknowledgments
This study is a collaboration between University of Illinois Urbana-Champaign and the Mayo Clinic under the Center for Individualized Medicine and the Mayo Clinic & Illinois Alliance for Technology-Based Healthcare. The authors acknowledge the invaluable contributions of the staff of the High-Performance Computing Bioinformatics core within the Roy J. Carver Biotechnology Center at the University of Illinois Urbana-Champaign. The authors also sincerely thank Dr. James Williams, Dr. Patricio Jeraldo, and Dr. Andy Schwaderer for their support in sample collection, bioinformatic processing, and discussions. The authors also thank Angela Waits, Clinical Research Coordinator at the Mayo Clinic in Rochester, Minnesota, for assisting in sample collection and obtaining patient consent.
Author Contributions
Annette C. Merkel was responsible for the data curation, formal analysis, methodology, and validation. Dr. Amy E. Krambeck was responsible for the data curation, methodology, and resources. Dr. Ananda S. Bhattacharjee was responsible for the data curation, formal analysis, methodology, and software. Dr. Bruce W. Fouke was responsible for the funding acquisition, project administration, and supervision. Chris J. Fields was responsible for the data curation, formal analysis, investigation, and supervision. Dr. Dirk Lange was responsible for the data curation, methodology, resources, and writing review and editing. Elena M. Wilson was responsible for the data curation, formal analysis, investigation, and writing review and editing. Dr. Jessica J. Saw was responsible for the data curation, formal analysis, investigation, methodology, supervision, validation, visualization, writing the original draft, and writing review and editing. Dr. John C. Lieske was responsible for the project administration, resources, and supervision. Dr. Joseph R. Weber and Dr. Melissa A. Cregger were responsible for the data curation, formal analysis, and methodology. Dr. Marcelino E. Rivera, Dr. Nicholas Chia, and Dr. Timothy Large were responsible for the data curation, methodology, and resources. Dr. Michael F. Romero was responsible for the methodology, resources, and supervision. Dr. Mayandi Sivaguru was responsible for the data curation, formal analysis, investigation, methodology, supervision, validation, visualization, writing the original draft, and writing review and editing. Dr. Robert A. Sanford was responsible for the data curation, formal analysis, and investigation. Dr. William J. Bruce was responsible for the data curation, formal analysis, investigation, and methodology. Dr. Yiran Dong was responsible for the data collection, curation, formal analysis, investigation, and validation. All authors read and approved the final manuscript.
Footnotes
J.J.S. and M.S. are joint first authors.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0006942020/-/DCSupplemental.
Download Supplemental Methods, PDF file, 2 MB (1.8MB, pdf)
Relative Abundance for Individual CaOx Stones. Download Supplemental Figure 1, PDF file, 2 MB (1.8MB, pdf)
Comparison of 16S rRNA bacterial amplicon gene sequencing (V1-V3 versus V3-V5 hypervariable region). Download Supplemental Figure 2, PDF file, 2 MB (1.8MB, pdf)
Comparison of ITS region fungal amplicon gene sequencing (ITS1 versus ITS2 region). Download Supplemental Figure 3, PDF file, 2 MB (1.8MB, pdf)
Overlay of SRAF image transparency on a larger field of view brightfield (BF) image. Download Supplemental Figure 4, PDF file, 2 MB (1.8MB, pdf)
Demographic and clinical characteristics of patients. Download Supplemental Table 1, PDF file, 2 MB (1.8MB, pdf)
24-hour Urine Supersaturation Profile. Download Supplemental Table 2, PDF file, 2 MB (1.8MB, pdf)
Full IR Spectroscopy Results. Download Supplemental Table 3, PDF file, 2 MB (1.8MB, pdf)
Read Summary for 16S rRNA (V1-V3 and V3-V5 hypervariable region) and ITS (ITS1 and ITS2 regions) Amplicon Gene Sequencing. Download Supplemental Table 4, PDF file, 2 MB (1.8MB, pdf)
Download Supplemental References, PDF file, 2 MB (1.8MB, pdf)
References
- 1.GBD Chronic Kidney Disease Collaboration: Global, regional, and national burden of chronic kidney disease, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 395: 709–733, 2020. 10.1016/S0140-6736(20)30045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivaguru M, Lieske JC, Krambeck AE, Fouke BW: GeoBioMed sheds new light on human kidney stone crystallization and dissolution. Nat Rev Urol 17: 1–2, 2020. 10.1038/s41585-019-0256-5 [DOI] [PubMed] [Google Scholar]
- 3.Sivaguru M, Saw JJ, Williams JC Jr, Lieske JC, Krambeck AE, Romero MF, Chia N, Schwaderer AL, Alcalde RE, Bruce WJ, Wildman DE, Fried GA, Werth CJ, Reeder RJ, Yau PM, Sanford RA, Fouke BW: Geobiology reveals how human kidney stones dissolve in vivo. Sci Rep 8: 13731, 2018. 10.1038/s41598-018-31890-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basavaraj DR, Biyani CS, Browning AJ, Cartledge JJ: The role of urinary kidney stone inhibitors and promoters in the pathogenesis of calcium containing renal stones. EAU-EBU Update Ser 5: 126–136, 2007. 10.1016/j.eeus.2007.03.002 [DOI] [Google Scholar]
- 5.Schwaderer AL, Wolfe AJ: The association between bacteria and urinary stones. Ann Transl Med 5: 32, 2017. 10.21037/atm.2016.11.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouke BW: Hot-spring systems geobiology: Abiotic and biotic influences on travertine formation at Mammoth Hot Springs, Yellowstone National Park, USA. Sedimentology 58: 170–219, 2011. 10.1111/j.1365-3091.2010.01209.x [DOI] [Google Scholar]
- 7.Dong Y, Sanford RA, Inskeep WP, Srivastava V, Bulone V, Fields CJ, Yau PM, Sivaguru M, Ahrén D, Fouke KW, Weber J, Werth CR, Cann IK, Keating KM, Khetani RS, Hernandez AG, Wright C, Band M, Imai BS, Fried GA, Fouke BW: Physiology, metabolism, and fossilization of hot-spring filamentous microbial mats. Astrobiology 19: 1442–1458, 2019. 10.1089/ast.2018.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warscheid T, Braams J: Biodeterioration of stone: A review. Int Biodeterior Biodegradation 46: 343–368, 2000. 10.1016/S0964-8305(00)00109-8 [DOI] [Google Scholar]
- 9.de Cógáin MR, Lieske JC, Vrtiska TJ, Tosh PK, Krambeck AE: Secondarily infected nonstruvite urolithiasis: A prospective evaluation. Urology 84: 1295–1300, 2014. 10.1016/j.urology.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr-Beare E, Saxena V, Hilt EE, Thomas-White K, Schober M, Li B, Becknell B, Hains DS, Wolfe AJ, Schwaderer AL: The interaction between enterobacteriaceae and calcium oxalate deposits. PLoS One 10: e0139575, 2015. 10.1371/journal.pone.0139575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose E, Zampini AM, Nguyen AH, Monga M, Miller AW: MP24-05 the urine and stone microbiome in kidney stone patients. J Urol 199[4S]: e291, 2018. 10.1016/j.juro.2018.02.758 [DOI] [Google Scholar]
- 12.Dornbier RA, Bajic P, Van Kuiken M, Jardaneh A, Lin H, Gao X, Knudsen B, Dong Q, Wolfe AJ, Schwaderer AL: The microbiome of calcium-based urinary stones. Urolithiasis 48: 191–199, 2020. 10.1007/s00240-019-01146-w [DOI] [PubMed] [Google Scholar]
- 13.Miller AW, Choy D, Penniston KL, Lange D: Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int 96: 180–188, 2019. 10.1016/j.kint.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannigan R, Choy WH, Chew B, Lange D: Renal struvite stones--pathogenesis, microbiology, and management strategies. Nat Rev Urol 11: 333–341, 2014. 10.1038/nrurol.2014.99 [DOI] [PubMed] [Google Scholar]
- 15.McMurdie PJ, Holmes S: phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217, 2013. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Core Team: R: A Language and Environment for Statistical Computing, Vienna, Austria, R Foundation for Statistical Computing, 2013 [Google Scholar]
- 17.Mehta M, Goldfarb DS, Nazzal L: The role of the microbiome in kidney stone formation. Int J Surg 36: 607–612, 2016. 10.1016/j.ijsu.2016.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaux CA, Raoult D: The microbiological memory, an epigenetic regulator governing the balance between good health and metabolic disorders. Front Microbiol 9: 1379, 2018. 10.3389/fmicb.2018.01379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ticinesi A, Nouvenne A, Chiussi G, Castaldo G, Guerra A, Meschi T: Calcium oxalate nephrolithiasis and gut microbiota: Not just a gut-kidney axis. A nutritional perspective. Nutrients 12: 548, 2020. 10.3390/nu12020548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zampini A, Nguyen AH, Rose E, Monga M, Miller AW: Defining dysbiosis in patients with urolithiasis. Sci Rep 9: 5425, 2019. 10.1038/s41598-019-41977-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prywer J, Torzewska A, Płociński T: Unique surface and internal structure of struvite crystals formed by Proteus mirabilis. Urol Res 40: 699–707, 2012. 10.1007/s00240-012-0501-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tasian G, Miller A, Lange D: Antibiotics and kidney stones: Perturbation of the gut-kidney axis. Am J Kidney Dis 74: 724–726, 2019. 10.1053/j.ajkd.2019.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison MJ, Dawson KA, Mayberry WR, Foss JG: Oxalobacter formigenes gen. nov., sp. nov.: Oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol 141: 1–7, 1985. 10.1007/BF00446731 [DOI] [PubMed] [Google Scholar]
- 24.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, Cave DR: Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 19: 1197–1203, 2008. 10.1681/ASN.2007101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J, Huang JS, Huang XJ, Peng JM, Yu Z, Yuan YQ, Xiao KF, Guo JN: Profiling the urinary microbiome in men with calcium-based kidney stones. BMC Microbiol 20: 41, 2020. 10.1186/s12866-020-01734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peck AB, Canales BK, Nguyen CQ: Oxalate-degrading microorganisms or oxalate-degrading enzymes: Which is the future therapy for enzymatic dissolution of calcium-oxalate uroliths in recurrent stone disease? Urolithiasis 44: 45–50, 2016. 10.1007/s00240-015-0845-6 [DOI] [PubMed] [Google Scholar]
- 27.Lange D: Editorial Comment on: Calcium Oxalate Urolithiasis: A Case of Missing Microbes? by Batagello et al. J Endourol 32: 1006, 2018. 10.1089/end.2018.0606 [DOI] [PubMed] [Google Scholar]
- 28.Lieske JC, Goldfarb DS, De Simone C, Regnier C: Use of a probiotic to decrease enteric hyperoxaluria. Kidney Int 68: 1244–1249, 2005. 10.1111/j.1523-1755.2005.00520.x [DOI] [PubMed] [Google Scholar]
- 29.Siener R, Netzer L, Hesse A: Determinants of brushite stone formation: A case-control study. PLoS One 8: e78996, 2013. 10.1371/journal.pone.0078996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golechha S, Solanki A: Bacteriology and chemical composition of renal calculi accompanying urinary tract infection. Indian J Urol 17: 111–117, 2001 [Google Scholar]
- 31.Pluznick JL: The gut microbiota in kidney disease. Science 369: 1426–1427, 2020. 10.1126/science.abd8344 [DOI] [PubMed] [Google Scholar]
- 32.Kandianis MT, Fouke BW, Johnson RW, Veysey J, Inskeep WP: Microbial biomass: A catalyst for CaCO3 precipitation in advection-dominated transport regimes. Geol Soc Am Bull 120: 442–450, 2008. 10.1130/B26188.1 [DOI] [Google Scholar]
- 33.Seiffert F, Bandow N, Kalbe U, Milke R, Gorbushina AA: Laboratory tools to quantify biogenic dissolution of rocks and minerals: A model rock biofilm growing in percolation columns. Front Earth Sci 4: 31, 2016. 10.3389/feart.2016.00031 [DOI] [Google Scholar]
- 34.Madigan MT, Bender KS, Buckley DH, Sattley WM, Stahl DA: Brock Biology of Microorganisms, 15th Ed., New York, Pearson, 2017 [Google Scholar]
- 35.Sivaguru M, Fouke KW, Todorov L, Kingsford MJ, Fouke KE, Trop JM, Fouke BW: Correction factors for δ18 O-derived global sea surface temperature reconstructions from diagenetically altered intervals of coral skeletal density banding. Front Mar Sci 6, 2019. 10.3389/fmars.2019.00306 [DOI] [Google Scholar]
- 36.Khan SR, Atmani F, Glenton P, Hou Z, Talham DR, Khurshid M: Lipids and membranes in the organic matrix of urinary calcific crystals and stones. Calcif Tissue Int 59: 357–365, 1996. 10.1007/s002239900140 [DOI] [PubMed] [Google Scholar]
- 37.Verkoelen CF: Crystal retention in renal stone disease: A crucial role for the glycosaminoglycan hyaluronan? J Am Soc Nephrol 17: 1673–1687, 2006. 10.1681/ASN.2006010088 [DOI] [PubMed] [Google Scholar]
- 38.Canales BK, Anderson L, Higgins L, Ensrud-Bowlin K, Roberts KP, Wu B, Kim IW, Monga M: Proteome of human calcium kidney stones. Urology 76: 1017.e13–1017.e20, 2010. 10.1016/j.urology.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 39.Skinner CWH: In: The Scientific Basis of Orthopaedics, edited by Albright J. A., Brand R., Norwalk, CT, Appleton and Lange, 1987, pp 198–212 [Google Scholar]
- 40.Liu X, Li J, Xiang L, Sun J, Zheng G, Zhang G, Wang H, Xie L, Zhang R: The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc Biol Sci 279: 1000–1007, 2012. 10.1098/rspb.2011.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Addadi L, Weiner S: Biomineralization: Mineral formation by organisms. Phys Scr 89: 098003, 2014. 10.1088/0031-8949/89/9/098003 [DOI] [Google Scholar]
- 42.Pernice M, Raina JB, Rädecker N, Cárdenas A, Pogoreutz C, Voolstra CR: Down to the bone: The role of overlooked endolithic microbiomes in reef coral health. ISME J 14: 325–334, 2020. 10.1038/s41396-019-0548-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wesson JA, Worcester EM, Wiessner JH, Mandel NS, Kleinman JG: Control of calcium oxalate crystal structure and cell adherence by urinary macromolecules. Kidney Int 53: 952–957, 1998. 10.1111/j.1523-1755.1998.00839.x [DOI] [PubMed] [Google Scholar]
- 44.Gadd GM, Bahri-Esfahani J, Li Q, Rhee YJ, Wei Z, Fomina M, Liang X: Oxalate production by fungi: Significance in geomycology, biodeterioration and bioremediation. Fungal Biol Rev 28: 36–55, 2014. 10.1016/j.fbr.2014.05.001 [DOI] [Google Scholar]
- 45.Sturm VE, Frank-Kamenetskaya O, Vlasov D, Zelenskaya M, Sazanova K, Rusakov A, Kniep R: Crystallization of calcium oxalate hydrates by interaction of calcite marble with fungus Aspergillus niger. Am Mineral 100: 2559–2565, 2015. 10.2138/am-2015-5104 [DOI] [Google Scholar]
- 46.Kuliasha CA, Rodriguez D, Lovett A, Gower LB: In situ flow cell platform for examining calcium oxalate and calcium phosphate crystallization on films of basement membrane extract in the presence of urinary ‘inhibitors’. CrystEngComm 22: 1448–1458, 2020. 10.1039/C9CE01587F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al KF: Characterizing the role of the microbiome in kidney stone disease. Doctor of Philosophy Thesis, University of Western Ontario, 2020. Available at: https://ir.lib.uwo.ca/etd/7122 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Download Supplemental Methods, PDF file, 2 MB (1.8MB, pdf)
Relative Abundance for Individual CaOx Stones. Download Supplemental Figure 1, PDF file, 2 MB (1.8MB, pdf)
Comparison of 16S rRNA bacterial amplicon gene sequencing (V1-V3 versus V3-V5 hypervariable region). Download Supplemental Figure 2, PDF file, 2 MB (1.8MB, pdf)
Comparison of ITS region fungal amplicon gene sequencing (ITS1 versus ITS2 region). Download Supplemental Figure 3, PDF file, 2 MB (1.8MB, pdf)
Overlay of SRAF image transparency on a larger field of view brightfield (BF) image. Download Supplemental Figure 4, PDF file, 2 MB (1.8MB, pdf)
Demographic and clinical characteristics of patients. Download Supplemental Table 1, PDF file, 2 MB (1.8MB, pdf)
24-hour Urine Supersaturation Profile. Download Supplemental Table 2, PDF file, 2 MB (1.8MB, pdf)
Full IR Spectroscopy Results. Download Supplemental Table 3, PDF file, 2 MB (1.8MB, pdf)
Read Summary for 16S rRNA (V1-V3 and V3-V5 hypervariable region) and ITS (ITS1 and ITS2 regions) Amplicon Gene Sequencing. Download Supplemental Table 4, PDF file, 2 MB (1.8MB, pdf)
Download Supplemental References, PDF file, 2 MB (1.8MB, pdf)
Data Availability Statement
The raw metagenomic sequencing data and other raw images and finalized images can be retrieved from the following link.
Kidney stone fragment thin sections and all data curation is available from Dr. Mayandi Sivaguru.