Abstract
Sleep deprivation can generate inflammatory responses in the central nervous system. In turn, this inflammation increases sleep drive, leading to a rebound in sleep duration. Microglia, the innate immune cells found exclusively in the CNS, have previously been found to release inflammatory signals and exhibit altered characteristics in response to sleep deprivation. Together, this suggests that microglia may be partially responsible for the brain's response to sleep deprivation through their inflammatory activity. In this study, we ablated microglia from the mouse brain and assessed resulting sleep, circadian, and sleep deprivation phenotypes. We find that microglia are dispensable for both homeostatic sleep and circadian function and the sleep rebound response to sleep deprivation. However, we uncover a phenomenon by which microglia appear to be essential for the protection of fear-conditioning memories formed during the recovery sleep period following a period of sleep deprivation. This phenomenon occurs potentially through the upregulation of synaptic-homeostasis related genes to protect nascent dendritic spines that may be otherwise removed or downscaled during recovery sleep. These findings further expand the list of known functions for microglia in synaptic modulation.
Keywords: Microglia, Sleep, Sleep deprivation, Memory
Highlights
-
•
Microglia are important for memory maintenance during a sleep deprivation period following a learning event.
-
•
Microglia are dispensable for most features of circadian regulation and the sleep deprivation rebound response.
-
•
Sleep deprivation activates a synaptic regulatory response in microglia.
1. Introduction
Over the last decade, microglia have been increasingly implicated in the homeostatic processes of the Central Nervous System (CNS) (Tremblay et al., 2011). In addition to their canonical role as active immune defense cells responding to primary infections, they have also demonstrated roles in the response to traumatic brain injury (TBI) (Hirbec et al., 2017), ischemia (Wake et al., 2009), and neurodegenerative conditions (Galloway et al., 2019; Kang et al., 2018). Research has uncovered bidirectional relationships between microglia and neurons (Eyo and Wu, 2013) that suggest the former play an important function in homeostatic neuromodulatory processes, especially during sleep (Wisor et al., 2011a; Choudhury et al., 2020).
Previously, microglia have been shown to be associated with sleep processes primarily through their inflammatory functions (Ingiosi et al., 2013). Inhibitors of microglia cytokine production, such as minocycline (Wadhwa et al., 2017a, 2017b), have been found to decrease non-Rapid Eye Movement (NREM) sleep and Delta power- a key metric of sleep drive- following sleep deprivation (SD) (Wisor et al., 2011b). CNS Administration of broadly pro-inflammatory agents such as TNFα, known to be involved with increased reactivity of many cell types including microglia (Karrer et al., 2015; Ji et al., 2013), increases both subsequent sleep and delta power in mice (Rockstrom et al., 2018; KRUEGER et al., 2011). These phenomena are strikingly similar to the rebound sleep response observed following a period of extended wakefulness, and indeed, SD increases the concentrations of pro-inflammatory molecules in the brain (KRUEGER et al., 2011).
Correspondingly, microglia also appear to be ‘reactive’ after periods of SD; a phenotype typified by various degrees of altered morphology, increased motility, phagocytotic behavior, and inflammatory cytokine release (Ingiosi et al., 2013; Deurveilher et al., 2021; Tuan and Lee, 2019; Bellesi et al., 2017; Hsu et al., 2003). However, the exact characteristics of microglia reactivity during SD are unclear. Accounting for varied definitions of microglia reactivity, it nonetheless seems that sleep loss modifies microglial morphology in a manner dependent on the duration and means of deprivation (Bellesi et al., 2017; Hsu et al., 2003). Together, these data imply a causal relationship between microglia inflammatory activity and subsequent sleep drive (Ingiosi et al., 2013; Nadjar et al., 2017).
In addition to their emerging role in sleep biology, microglia have been increasingly implicated in mammalian memory and maintenance of neuronal homeostasis at baseline, processes that are in turn linked to sleep (Eyo and Wu, 2013). Microglia localize to synapse junction sites and similarly to astrocytes appear to play a regulatory role at their interface with these synapses (Webb and Orton, 2019). For a number of years, it has been known that microglia phagocytose damaged synapses in response to brain injury (Galloway et al., 2019) and that their removal can lead to alleviation of cognitive losses in pathogenic conditions (Rice et al., 2015; Janova et al., 2017; Acharya et al., 2016; Krukowski et al., 2018). In the last decade, it has been consistently demonstrated that microglia mediate phagocytosis of healthy adult synapses in homeostatic conditions (e.g. during forgetting) (Tremblay et al., 2011; Wang et al., 2020). There is discussion over whether microglia may exhibit similar behaviors during sleep or sleep deprivation (Choudhury et al., 2020; Bellesi et al., 2017; Nadjar et al., 2017). For example, there is evidence from anesthetized mice that suggests that in the sleeping brain, microglia are more ‘alert’, surveilling their environment and performing roles in neuronal plasticity (Liu et al., 2019; Stowell et al., 2019), but also that they may be increasingly reactive during periods of SD and contribute to cognitive deterioration (Nadjar et al., 2017). However, evidence remains sparse and experimental paradigms varied, so it remains unknown the degree to which microglia play a role in the functions of sleep or sleep deprivation recovery.
Here, we describe the first set of findings suggesting that during a sleep period following SD, microglia maintain homeostasis in the synaptic microenvironment to preserve the strong, memory-associated synapses formed during or before SD. Microglia do not play a primarily inflammatory role in response to the sleep deprivation insult, and are dispensable for the SD recovery response, suggesting that previous research into anti-inflammatory’ effects on sleep are likely mediated through other pathways and cell types. Further, they do not appear to be necessary for phagocytosis of synapses during sleep.
2. Material and methods
2.1. Lead contact and materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ying-Hui Fu (Ying-Hui.Fu@ucsf.edu). All reagents, cell lines and mouse models used in this manuscript are available upon request.
2.2. Animal source
P2RY12CreER mice (Jackson Labs, 034727) and Rpl22HA RiboTag mice (Jackson Labs, 029977) were kindly acquired from Thomas Arnold. Double heterozygous mice were used for TrapSeq experiments. All mice were on C57BL/6J background, and used between 14 and 20 weeks of age. Animals were singly housed on a light-dark (LD) 12:12 cycle and given ad libitum access to food and water. When possible, littermates were used for studies comparing wild type (WT) and transgenic mice. Male and female mice were used in comparable numbers for all experiments unless otherwise noted. In all cases where both sexes were used, no significant differences were found relative to variables and conditions of interest.
All experimental protocols were approved by the University of California, San Francisco Institutional Animal Care and Use Committee following the NIH's Guide for the Care and Use of Laboratory Animals.
2.3. Ablation of microglia
Mice were fed PLX5622 chow containing Colony-Stimulating Factor 1 Receptor (CSF1R) inhibitor (1200 mg/kg) or a supplier-designed control diet for at least 20 days prior to all ablation experiments. Ablation was confirmed by Iba1+ immunostaining prior to all experimentation, finding 95%+ whole brain removal of microglia in each test, confirming previous findings (Elmore et al., 2014a; Zhan et al., 2019). For most trials, each condition was tested in parallel with ablated and non-ablated mice undergoing testing simultaneously. For wheel running assays, mice were recorded on control chow, then swapped to ablation chow for 19 or 20 days before an additional recording period.
2.4. Tissue extraction, sectioning, and immunohistochemistry
All mice were anesthetized using a sublethal (0.25–0.5 mg/kg) dose of Avertin (Tribromoethanol) before transcardial perfusion with Phosphate Buffered Serum (PBS) and a 4% paraformaldehyde (PFA) in Saline solution. Brains were extracted and postfixed in the same 4% PFA solution for 24–30 h before subsequently being transferred to a 30% sucrose solution for another 24–48 h, until the brain had saturated. Brains were then flash frozen in OCT Tissue Freezing medium at −80C. Only male mice were used for immunohistochemistry, following validation of similar phenotypes in both male and female mice in the present study, including fear conditioning, electroencephalography (EEG), wheel running, and Sholl's analysis.
Brains were coronally sectioned (Bregma −3.5 to −4.5 mm) at 25 μM thickness using a Leica CM1850 cryostat at −22C, washed in 1X Phosphate Buffered Saline (PBS) 3 × 30 min, then blocked with 2% Bovine Serum Albumin (BSA) blocking solution with 5% Normal Donkey Serum (Jax 017-000-121) in 1X Tris-Buffered Saline, 0.1% Tween (TBST) for 2 h. Labeling was performed on floating sections using relevant antibodies at 4C overnight, then washed 3 × 30 min in PBS before addition of relevant fluorescent secondaries in BSA+TBST before additional washing. Slices were then placed on SuperFrost slides with Glycerol DAPI mounting solution (EMS #17989-61), or Eukitt Quick-hardening Mounting Medium (Sigma 03989) for sections requiring blue staining that were not DAPI. Primary antibodies and concentrations used were anti-Iba1+ (1:300, Novus Bio NB100-1028 or Wako 019-19741), anti-PSD95 (1:500, Santa Cruz Biotechnology sc-32290) anti-C1q (1:500, abcam ab71940) and anti-GFAP (1:2000, Millipore #AB5804). Secondary antibodies used were Alexa Fluor 405-conjugated donkey anti-rabbit (abcam ab175651), Alexafluor 488-conjugated donkey anti-goat (Fisher, A-11055), AlexaFluor 594-conjugated donkey anti-rat (thermo A21209), all at 1:1000 concentration.
2.5. Western blotting
To Western blot, NuPage 4–12% Bis-Tris Gels (NP0323BOX) were submerged in 1X running buffer in XCell SureLock Mini-Cell containers. Sample concentrations were standardized using a Bradford protein assay, then mixed correspondingly with diluted 10x NUPAGE Reducing agent and diluted 1x LDS Sample Buffer, for a total of 15 μL per sample. Gel was run at 200V until dye reached bottom of gel. PVDF transfer membranes (Thermo Scientific #88518) were prepared by soaking in methanol then 1x transfer buffer each for 30 s. Gels were sandwhiched next to the transfer membrane in between filter and wool, then submerged in a transfer tank in 1x transfer buffer cooled to 4C and surrounded by ice. Transfer reaction was run for approximately 2 h at 300 mA. Following transfer, membranes were removed, rewashed in methanol then distilled water for 30 s each, then prepared for antibody staining. For staining, membranes were blocked in 5% milk in TBST for 15–30 min, then placed in fresh 5% milk+ TBST with desired antibody concentrations (see below) at 4C with gentle shaking for overnight. The following day, membranes were washed 3 times in 1xTBST for 5 min, then placed in another 5% milk + TBST solution with secondary antibodies conjugated to HRP for 2 h. Finally, membranes were placed in Luminol + peroxide development mix with water (1:4 or 1:10 ratio depending on strength of signal) and then imaged using a film and X-ray machine.
Primary antibodies used were anti-GluR1 (1:1500, abcam ab31232), GluR1-831P (1:1500, Millipore #04-823), BDNF (1:600, Novus #NB100-98682), TNFa (1:600, abcam #ab183218), secondary was HRP-conjugated anti-rabbit (1:10,000, CST #7074).
2.6. Surgery for electroencephalography (EEG)
Animals were anesthetized with 2% isoflurane and placed in a stereotaxic head frame on a heating pad. Opthalmic ointment was applied to the eyes to prevent drying. A midline incision was made down the scalp. Four guide holes in the skull were made using a 23-gauge surgical needle placed epidurally over the frontal cortical area (1 mm anterior to bregma and 1 mm lateral to the midline) and over the parietal area (3 mm posterior to bregma and 2.5 mm lateral to midline). One ground screw and three screws with leads were placed into the skull through the holes. The screws with leads were then soldered onto a six-pin connector EEG/EMG headset (Pinnacle Technologies). For EMG recordings, an EMG lead from the headset was placed into the neck muscle. The headset was then covered with black dental cement to form a solid cap atop the mouse's head. The incision was then closed with Vetbond (3M, Santa Cruz Biotech), and animals were given a subcutaneous injection of marcaine (0.05 mg/kg) before recovery on a heating pad. Mice underwent surgery at 12 weeks of age, and behavioral experiments were conducted 3 weeks after surgery to allow for sufficient recovery.
2.7. EEG recording and scoring
EEG recording was performed as previously described (Xing et al., 2019; Shi et al., 2019). In brief, for EEG/EMG recording, 16 week old mice were singly housed and habituated to the recording cable for 7 days in LD 12:12 at 100 Lux. Tethered 3-channel mouse preamplifiers (Pinnacle Technologies, #8202) were attached to the headset of the mice. The signals were relayed through commutators that allowed the animal to move freely. Data were acquired through the Sirenia software package (Pinnacle Technologies).
All EEG analysis was done using Python scripts. Sleep was hand scored in 10 s epochs by researchers blinded to genotype with the assistance of spectral analysis using fast Fourier transformation. In general, wakefulness was defined as desynchronized low-amplitude EEG and heightened tonic electromyograph (EMG) activity with phasic bursts. NREM sleep was defined by a synchronized, high-amplitude, low-frequency (0.5–4 Hz) EEG signal and substantially reduced EMG activity compared with wakefulness. REM sleep was defined as having a pronounced, synchronized theta rhythm (4–9 Hz) with minimal EMG activity.
To examine sleep-wake behavior under baseline conditions, EEG/EMG signals were recorded and analyzed for the entire two consecutive days from the onset of the light phase. Sleep (NREM and REM sleep) time and power spectrum were averaged data from two consecutive days. For SD, mice were sleep deprived for 6 h from the onset of the light phase by gentle handling, involving addition bedding or paper towels to promote wakefulness and light touching when the mice exhibited signs of sleepiness. Food and water were freely available. EEG/EMG signals were recorded and analyzed during and for 24 h after SD.
For spectral analysis, EEG artefacts- epochs with signals extending beyond the measurement range- were excluded. All power spectra were binned at 0.1 Hz frequencies. Absolute EEG power spectra were calculated by the average power across all epochs in that state and time period (Mang and Franken, 2012). Individual differences were normalized (for normalized power) by expressing each frequency bin as a percentage of total EEG power recorded (0.1–25 Hz) over a 24-h period for each mouse. As various behavioral states tend to have different EEG power—which could affect total power depending on each individual's relative time spent in each state—the relative contribution to relative total power of each state was then weighted by the respective time spent in that state (Vassalli and Franken, 2017). The time course of delta power in NREM sleep was computed as previously described, using 54 h of continuous recording (Mang and Franken, 2012; Maret et al., 2005). Change of NREM sleep delta power across the light-dark cycle was determined by the delta band (1.0–4.0 Hz) of NREM sleep and normalized to the average NREM sleep delta power during ZT9–12 of the baseline recording day (Mang and Franken, 2012; Franken et al., 2006). In the dark phase, especially the early dark phase, NREM sleep is absent in some time points. As a result, delta power is not presented for every hour. Intervals were chosen so that every mouse had equal duration of NREM sleep for each time point shown in the figure, to allow better comparison between conditions. The 12 h light period was split into 12 intervals of equal number of NREM epochs, and the dark phase 6 intervals of equal numbers of NREM epochs. See previous publications for further details (Mang and Franken, 2012; Maret et al., 2005).
2.8. Sleep deprivation
All SD for EEG recordings was performed by a form of gentle handling. In short; mice were transferred to new cages initially and provided new bedding, which provoked wakefulness for 2–3 h. Subsequently, when the mice appeared sleepy and began to rest their heads or remain immobile, they were gently touched until they moved (Colavito et al., 2013). For experiments requiring brain tissue, prior to perfusion, mice were placed in a container with a rod spinning at ∼3 rpm to keep them awake for 6 h. Both such forms of SD have been shown to induce minimal stress in mice (Graves et al., 2003; Ramesh et al., 2009; Nollet et al., 2020). The rotating bar method was unable to be used for EEG-connected mice, due to tangling of the wires. For immunostaining of microglia, mice were returned to their cages with food and water before sacrifice 18 h later.
2.9. Contextual fear conditioning
12–14 week old mice were handled gently for 2 min/day for five consecutive days before each experiment and were placed in the experiment room 30 min before initiation at ZT = 0 before each test for acclimatization. On the day of training, mice were allowed to explore the enclosed chamber for 3 min before two 2-s 0.6 mA foot shocks were applied with a 2-min interval. Animals were left in the chamber for additional 30 s after the final shock and returned to their cage. Each cage was wiped down with ethanol between mice. Baseline data was collected by assessing freezing time before either shock, as no learning had occurred. Following training, mice were sleep deprived by rotating bar until ZT 6. On the probe day (subsequent day to testing), the same paradigm was conducted with no foot shock, and automated tracking software (AnyMaze) was used to record freezing times for 5 min.
2.10. Microscopy and sholl analysis
Microglia for Sholl analysis and other IHC assays were imaged using a 40x objective (Plan-Neofluar lens, NA 0.9) on a Zeiss Axioscope Image Z.2 with a motorized stage with Apotome, using a Hamamatsu camera and ZEN Blue software. For each condition, n = 3 or 4 mice were used. 10–20 microglia were randomly identified and selected for intactness on 2–3 coronal slices (bregma −3.5 to −4.5 mm) for Iba1+ staining in dorsal hippocampus, for a total of 75–100 microglia measured. Sholl analysis was performed using a FIJI software plugin (Ferreira et al., 2014) which measures cell branching and outgrowth through sampling at evenly spaced intervals (0.1 μm) from a designated soma center point. Sholl measurements from each microglia was averaged across the relevant condition and compared using first a two-way ANOVA between conditions, followed by a post-hoc test identifying differences in each 0.1 μm bin.
10–20 slices from n = 4 mice were selected and measured via IHC for C1q, PSD95, and GFAP stains. 14 or 15 week old mice were used. For C1q and PSD95, puncta larger than 0.8 μm3 were excluded. Colocalization was established using FIJI's JaCOP (Bolte and Cordelières, 2006) program's object based analysis with standardized parameters for all images.
2.11. Translating ribosome affinity purification (TRAP)
Following two weeks of chow-based tamoxifen administration (400 mg/kg) and health and weight assessments, Rpl22 x P2RY12-CreER, 15 week old mice were anesthetized using a sublethal (0.25–0.5 mg/kg) dose of Avertin (Tribromoethanol) before transcardial perfusion with PBS and subsequent decapitation. Brains were extracted and manually homogenized in 2% w/v supplemented homogenization buffer (10% NP-40, 1M KCl, 1.5M Tris, 1M MgCl2 supplemented with protease inhibitors, Heparin 100 mg/mL, cycloheximide, RNAsin, DTT 1M). Samples were then centrifuged at 4C for 10’ at 10,000g, then the supernatant gently mixed with HA antibody on a cold room spinner for 4 h 200 μL per sample of Pierce Protein A/G Magnetic Beads (cat #8802) were equilibrated during this time to the homogenization buffer. After the 4 h incubation, the beads in homogenization buffer were added to the sample for overnight incubation at 4C. The following day, samples were washed multiple times with a high salt buffer at 4C. Finally, lysis buffer was added to the beads and vigorously vortexed to dislodge the antibody-ribosome complexes from the beads. These samples were run through a Qiagen RNeasy RNA extraction kit to isolate RNA from ribosomes before being sent for sequencing. Two biological replicates were pooled into each technical replicate to ensure enough sample for sequencing, leading to 4 mice for each condition.
2.12. RNA sequencing and analysis
Extracted RNA was stored in ddH20 at −80C and sent to Novogene for RNAsequencing for the ribotagged sequencing. Raw data was returned and analyzed as follows. FASTQ files were extracted, mapped, and counted using salmon. The resulting count files were run through DEseq2 to determine differential expression between conditions. Downstream visualizations and analyses, including KEGG and GO term calling, IPAGE binning, and volcano plots were performed using Python and R software with GOseq, clusterProfiler and EnhancedVolcano (EnhancedVolcano: publicat) visualization packages. RNAseq data found at GEO accession number GSE174231.
2.13. Wheel running behavior
To assess daily activity and period length, 12–15 control and ablated mice (14 weeks old) were placed in individually housed cages with running wheels. Locomotor activity was determined by electronically counting revolutions of the running wheel in 6 min intervals using Clocklab software (Colbourn Instruments). To determine period, the duration between daily onsets of locomotor activity was calculated. For each animal, period was averaged over 15+ days and values were evaluated statistically using Student's t-test. A different cohort of 10–12 animals were tested for the 24 h dark condition, were placed in constant darkness at 14 weeks of age and allowed to acclimatize for 10 days before testing began. Animals on control diet were subsequently swapped to ablation diet and measured again 2 weeks later.
2.14. Golgi staining and dendritic measurement
All Golgi stains were performed using the FD Rapid Golgistain Kit (#PK401) according to manufacturer's recommendations. In brief, brains were placed in a mercuric chloride solution (Solution A) which was replaced after 24 h. After impregnation with occasional gentle stirrings every 48 h for 2 weeks, the brains were transferred to a second solution (Solution B) for a further 72 h. Brains were then frozen, coronally sliced at 120 μm thickness (bregma −3.5 to −4.5 mm), and washed, then stained before mounting on slides. CA3 pyramidal cell secondary apical dendrites were identified that had been impregnated by the golgi staining process. 10 μm stretches as close to the soma as possible that had consistent and clear staining were chosen. A total of 300–400 spines were measured from 30 to 60 neuron segments across 4 animals. Stained brain slices were imaged were imaged using a 100x objective (Plan-Neofluar lens, NA 1.3) on a Zeiss Axioscope Image Z.2 with a motorized stage with Apotome, using a Hamamatsu camera and ZEN Blue software. Analysis of images was performed using the RECONSTRUCT program (Fiala, 2005) (Fiala, 2005) which provided unbiased, automated analysis and classification of spine morphology along designated randomly selected 10 μM stretches of CA3 dendrites (Fiala, 2005).
2.15. Quantification and statistical analysis
Information on all statistical analysis, and n values can be found in the figure legends corresponding to the respective data. For all studies comparing two conditions unless otherwise indicated, statistical analysis was performed using unpaired two-tailed student's t-test. For EEG and Sholl's analysis, we performed a two way ANOVA to verify a significant difference between conditions, then followed up with post-hoc Tukey's tests with Bonferroni multiple comparisons corrections at each equivalent bin for EEG analysis and Holm-Sidak test for Sholl's analysis. All data expressed as mean ± SD unless otherwise noted. Significance was defined with a cutoff of p = 0.05. All statistical tests were performed using Graphpad's PRISM software, including verification of normality of data using Anderson-Darling.
3. Results
3.1. Ramification of microglia increases during SD
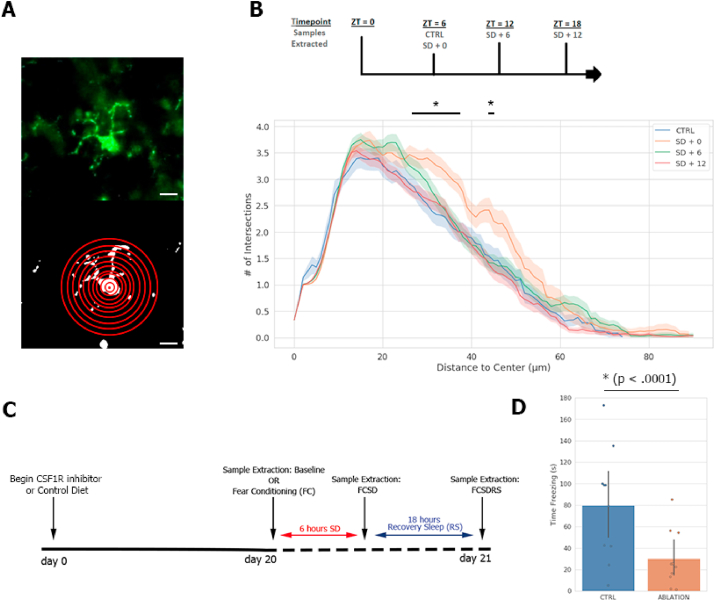
We first set out to observe changes in microglial morphology after SD. Based on Iba1+ immunofluorescent staining and Sholl's analysis we found that microglia ramifications in the hippocampus increased during a period of 6 h of SD by rotating bar; a phenotype that was lost quickly (within 6 h) after recovery sleep (Fig. 1A and B).
Fig. 1.
Microglia prevent severe memory defects caused by sleep deprivation and subsequent recovery sleep (A), Representative microglia stained with Iba1 (left) and example schematic of Sholl's analysis on thresholded image (right). Scale bar = 10 μm. (B), top; timeline schematic of the ZT at which each sample for Sholl's analysis was taken, bottom; Sholl's analysis results measuring extent of hippocampal microglia ramifications at various time points after SD. Microglia in 25 μm slices fully visible in the CA3 were selected at random and stained for Iba1. n = 3 or 4 mice per condition. Statistical analysis performed using two way ANOVA for main effect between the CTRL and each other condition. Post-hoc Holm-Sidak tests with multiple comparisons corrections were then performed for each 0.1 μm bin between the two groups found to be statistically different; CTRL and SD + 0 (* = p < 0.01). A significant difference was found between 3.1 and 3.8 μM and at 4.5 μm. (C), Schematic of experimental paradigm for samples collected in this study. Contextual fear conditioning (FC) was performed at ZT = 0, and mice were kept awake during this time, followed by SD for the remainder of the 6 h before ZT = 6 (SD, ZT = 6), then 18 h of recovery sleep (RS, ZT = 0). Schematic also portrays when samples for other experiments requiring tissue were collected in separate mouse cohorts: at baseline without FC, after FCSD, and after FCSDRS. (D), Total time freezing in seconds out of 300 s in testing chamber during FC probe trial 24 h after training and subsequent 6 h SD (n = 10 mice each condition). Statistical analysis performed using an unpaired parametric two-tailed student's t-test. Data expressed as mean ± SD. Related Fig. S1 demonstrates results obtained immediately after SD and without SD.
3.2. Microglia prevent memory defects caused by sleep deprivation and subsequent recovery sleep
Previous research has shown in a variety of learning paradigms and models that the absence of microglia does not appear to significantly affect memory in adult mice under normal conditions (Elmore et al., 2014a; Han et al., 2017a). However, the role of microglia in memory consolidation in the SD state has not been explored. As SD reliably impairs memory consolidation following contextual fear conditioning (FC) (Graves et al., 2003), and microglia are a primary mediator of neuroinflammation which may impair memory, we reasoned that the removal of microglia may alleviate the negative consequences of SD on memory.
Microglia were efficiently ablated using the CSF1R drug PLX5622, consistently shown to reduce overall microglia populations in the CNS by 90%+, for at least 20 days (Elmore et al., 2014b), hereby referred to as the ‘ablated’ condition. We first confirmed microglial ablation in our mice (Fig. S1A). After 20 days, ablated and control mice were trained in an FC apparatus, a hippocampal-dependent memory assay. They were then sleep deprived until ZT6, for a total of 6 h, following completion of the training, using a low-stress rotating bar deprivation tank (Fear conditioning + sleep deprivation, FCSD). Mice were then tested the following morning after an 18 h period of recovery sleep (RS, time point called FCSDRS) (Fig. 1C). Separate cohorts of mice were sacrificed at each of these three conditions- baseline, FCSD, and FCSDRS for experiments requiring tissue.
Interestingly, the microglia-ablated animals demonstrated far poorer memory, as measured by freezing time, of the fear stimulus (CTRL, 54.97s ± 10.76 s; ABL, 24.97s ± 4.65 s; Fig. 1D) when probed the following day compared to the training day (Fig. S1B) and in comparison to testing without sleep deprivation (Fig. S1C). Additionally, this effect appeared to be sleep dependent, as testing immediately following the period of SD did not reveal any differences between conditions (Fig. S1D). These results suggest that microglia activity is not important for the prevention of SD-induced memory deficits, and instead are important for memory consolidation following SD and subsequent recovery sleep.
3.3. Microglia are dispensable for core sleep and circadian features
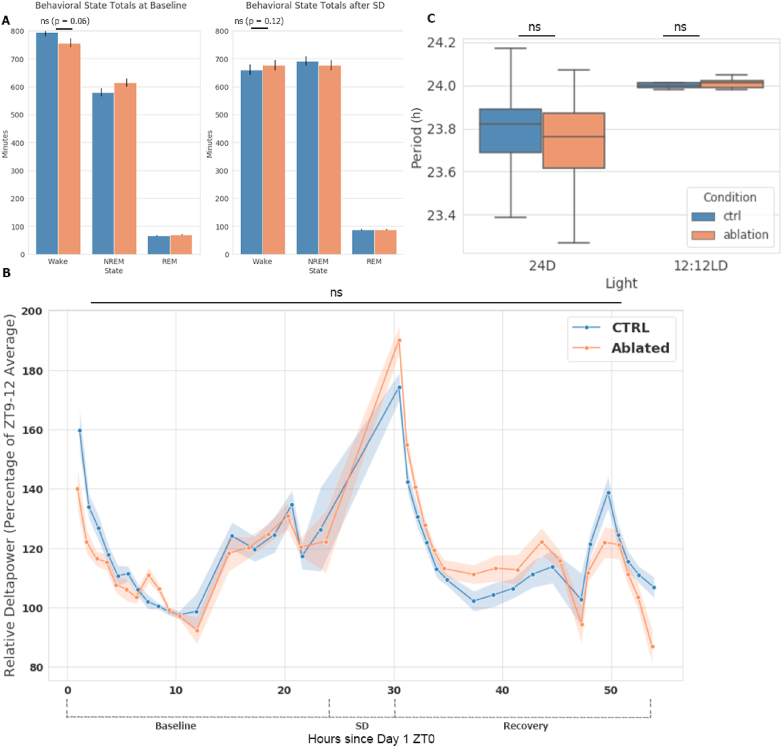
We next verified if there were any major features of sleep disrupted in the absence of microglia, as poor or diminished recovery sleep may compromise memory consolidation. Further, if inflammatory signaling driven by microglia partially mediates the sleep rebound response, we would expect a reduction in sleep rebound following acute SD which may contribute to our observed effects on memory. Control and microglia-ablated mice were recorded by electroencephalograph (EEG) for 24 h under normal conditions and after a period of 6 h of SD by gentle handling. We found sleep architecture to be largely unaffected; mice with ablated microglia trended toward slightly more sleep at baseline (∼25 min) but showed no reduction in post-SD sleep rebound (∼90–120 min) (Fig. 2A). Relative delta power, a commonly used marker for sleep drive comprising electrophysiological power between 1 and 4 Hz, was largely unaffected by microglia ablation over the baseline 24 h period nor after a period of SD (Fig. 2B). Correspondingly, we found no change in hippocampal TNFα (known to affect sleep rebound (Rockstrom et al., 2018) after FCSD) in microglia-ablated mice (Fig. S2). In-depth processing of the EEG data revealed no further significant differences in relative power; however, an increase in absolute delta power signal in ablated mice was observed at baseline in the 1.0–4.0 Hz frequency range, and SD from 0.9 to 3.3 Hz, with a trend towards increased power along the entire frequency spectrum measured (Fig. S3A).
Fig. 2.
Microglia are dispensable for core sleep and circadian features (A), Minutes spent in each behavioral state measured by EEG. Measurement of 24 h periods were split into 10 s epochs for classification then summed for the graphic. Left; baseline, right; the 24 h following 6 h of SD. n = 12-16 mice. Data expressed as mean ± SD. Statistical analysis performed using an unpaired two-tailed student's t-test. (B), Relative Delta Power (1–4 Hz Frequency range) normalized to average Delta Power at ZT9-12, the lowest point in a 24 h period. Figure covers 2+ days of continuous recording in a 12:12 light/dark setting. The first 24 h are baseline measurements, followed by 6 h of SD (ZT 24-30) and 24 h of recovery period. Only the preceding 24 h before SD were used for this calculation, to allow for the use of 54 h of continuous recording data. See methods for details on calculation and selection of x-axis data points. Data expressed as mean ± SD n = 12-16 mice. Statistical analysis was performed using two way ANOVA between the two conditions across the entire 54 h period, with post-hoc Bonferroni multiple comparisons tests with corrections at each equivalent time point. Two-way ANOVA found a statistically main effect significant difference (p = 0.0015) between the two time-courses, but no differences after Tukey's test with multiple testing corrections at any individual ZT bin. Related Fig. S3A illustrates the entire power-frequency spectrum. Fig. S3D displays wake time for each condition by hour. (C), Period length measured by wheel running in 24 h dark and 12:12 light:dark conditions. An automated system tracked number of wheel revolutions, and period length was defined as the time between the onset of wheel running activity of subsequent circadian days. n = 8-10 mice per condition, 3 weeks of data averaged for each mouse. Data expressed as mean ± SD. Statistical analysis performed using unpaired two-tailed student's t-test. Related Figs. S3B and C display additional features of circadian periodicity.
Sleep timing may also affect cognitive responses, and microglial regulation of synaptic strength has been connected to their circadian functions (Hayashi et al., 2013). We therefore measured circadian behavior by free wheel running, a well-established method of determining endogenous circadian period (LeGates and Altimus, 2010). Contrary to the results of Hayashi et al. (2013), who removed a core microglia clock gene (CatS) and observed circadian alterations in sleep times and activity, we found period length was not significantly altered in either normal 12:12 Light:Dark conditions or in a 24 h Dark condition, which unveils endogenous circadian period through the removal of zeitgebers (Fig. 2C). There were no differences in other metrics investigating activity, including bout length on the wheel (Figs. S3B and C), though we did observe a small decrease in active time in ablated mice, similar to that observed at baseline by EEG. For both EEG and circadian phenotypes, male and female mice were analyzed separately and revealed no significant discrepancies between sexes (data not shown). Together, these experiments suggest that microglia are not essential for periodicity, normal sleep wake patterning, or the SD recovery response.
3.4. Microglia participate in synaptic modulation during sleep deprivation
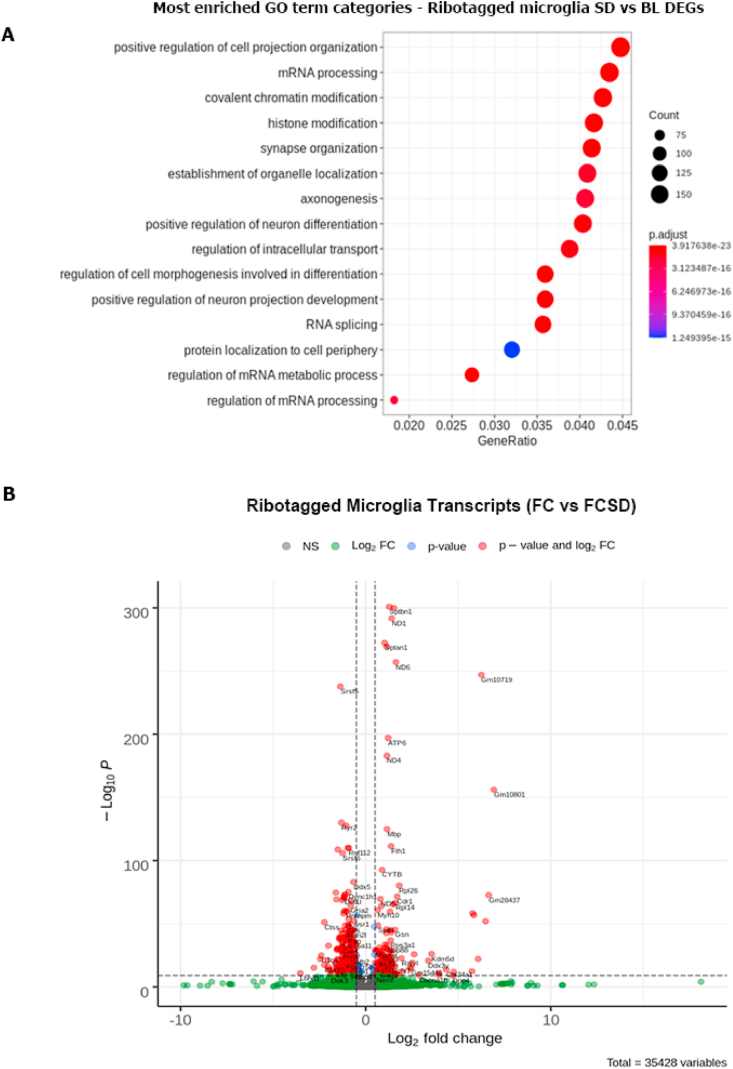
We next sought to characterize transcriptional changes underlying microglia responses to FCSD. We performed RNA-seq on tagged hippocampal ribosomes from double transgenic P2RY12-CreER/Rpl22 mouse (McKinsey et al., 2020) microglia at baseline (6 h after an FC learning event) and after FC with a subsequent 6 h of SD. We selected P2RY12 mice following evidence suggesting it is more specific to microglia than other canonically used genetic markers (McKinsey et al., 2020), such as Cx3cr1, which may label other peripheral or perivascular macrophages. With FCSD, 4102 genes of the more than 34,000 tested were differentially expressed. Unexpectedly, Gene Ontology (GO) term analysis of these differentially expressed genes (DEGs) revealed little change in immunomodulatory terms (Fig. 3A). Instead, analysis found substantial enrichment in DEGs for terms associated with transcriptional regulation, synaptic homeostasis, and intracellular trafficking (Fig. 3B, Table S1). For example, multiple genes related to glutamate signaling, glutamate receptors, and trafficking were dysregulated; Gria2 was one of the most downregulated genes in our dataset, along with reduced expression of Grid2, Grin2b, Gls and Sptbn2. Many specific splicing factor subunits also changed in expression, including Sf3a1, Sf3b1, and Sf1. These observed changes are consistent with the well-characterized role of microglia in synaptic modulation. The large amount of differentially expression microglia genes in the SD context warrants future investigation into the role of the cell type in this context.
Fig. 3.
Sleep deprivation modifies the microglial transcriptome to activate synaptic-associated genes (A), RNA-seq analysis comparing mice at ZT = 6 following fear conditioning with and without SD. Mice were either sacrificed 6 h after FC 6 h of fear conditioning, at ZT = 6, with or without SD from ZT = 0 to ZT = 6. GO term analysis of a submitted list of ∼4000 differentially expressed genes to Python's clusterProfiler package which utilizes PANTHER's GO system. This analysis indicates relative enrichment for terms compared to a random sampling; the color of each circle represents the result of a relative enrichment analysis within each category compared to a random sample of genes, GeneRatio represents total proportion of sample, and size of circle indicates absolute number of genes in category. n = 4 for each condition. For detailed analysis on RNAseq processing, see methods. (B), Volcano plot of all genes identified in sample and analyzed for significance by DEseq2 created using EnhancedVolcano package. Cutoffs for coloring are at a -log10 value of 10 and at a 2 fold expression change. A selection of the top 50 most significantly differentially expressed genes were selected for labeling on the chart. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
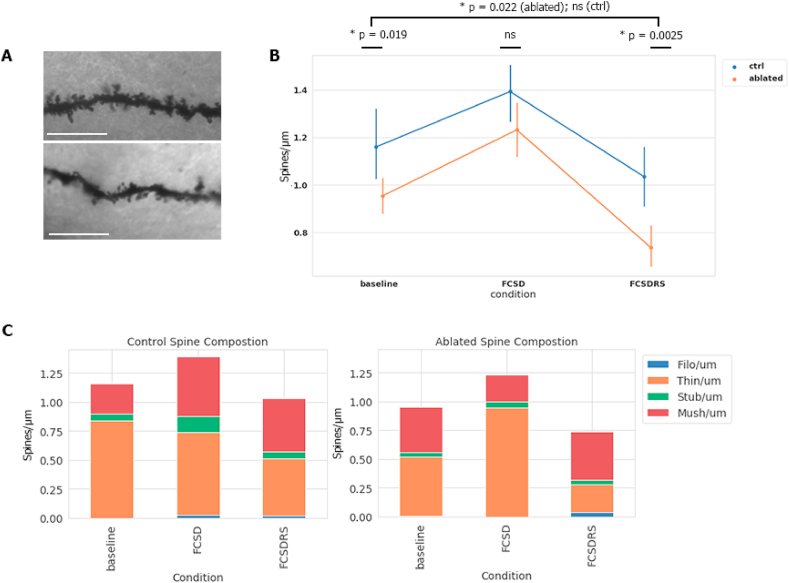
Recent research has found microglia to play a function in the pruning and phagocytosis of synapses during development, forgetting, and other contexts requiring synaptic removal (Ji et al., 2013; Wang et al., 2020; Tremblay et al., 2010). To test whether these functions may be compromised in our ablated FCSD context, we assessed the density and morphology of spines in the dorsal hippocampus by using Golgi staining and dendrite measurements to determine the potential effects associated with the absence of microglia (Fig. 4A). We took samples at three time points- baseline (BL), in mice trained on the FC apparatus and sleep deprived for 6 h (FCSD), and mice which underwent FCSD and allowed 18 h of recovery sleep (FCSDRS). The sample were taken from the dorsal CA3 region, previously associated with the acquisition of contextual memory in FC. (Hunsaker et al., 2009; Wen et al., 2015). At all time points, microglia-ablated mice exhibited fewer total dendritic spines in the CA3 region of the dorsal hippocampus; however, the greatest discrepancy was found after recovery sleep, where spine number decreased drastically, past baseline conditions (Fig. 4B and C), whereas in control mice spine numbers returned to baseline levels after FCSDRS. Morphological analysis indicated that this was primarily due to a significant decrease in thin spines in microglia-ablated mice (Table S2).
Fig. 4.
Microglial ablation leads to altered spine density dynamics (A), Representative image of Golgi staining in FCSD condition for control (top) and ablated (bottom). Only in-focus 10 μm stretches were measured. Scale bar = 10 μm. (B), Total spines with error values for each condition. Baseline = no intervention, ZT = 0; FCSD = fear conditioning plus 6 h of SD, ZT = 6; FCSDRS = fear conditioning plus 6 h of SD and 18 h subsequent recovery sleep, where samples were taken at ZT = 0. Different cohorts of mice used for each condition. Golgi stained 120 μm brain slices were selected and viewed at 100x magnification. Random visible 10 μm stretches of CA3 dendrite were chosen and imaged for downstream analysis. All comparisons between conditions and microglia status are statistically significant except for those shown as not-significant (ctrl BL vs FCSD; p = 0.010. ctrl FCSD vs FCSDRS; p < 0.001. ctrl BL vs ctrl FCSDRS; ns. abl BL vs FCSD; p = 0.0015. abl FCSD vs abl FCSDRS; p < 0.001. abl BL vs FCSDRS; p = 0.022). Statistical analysis performed using an unpaired parametric two-tailed student's T test between relevant conditions. (C), Spine density of each of spine type as categorized by the Reconstruct software, showing the same population of spines as (B) split by type. After the Reconstruct program was provided stretches of spines, it quantified and categorized the relative composition of each morphological spine type. Categories ‘thin’ and ‘long thin’ spines were combined for this analysis. Left; control spines in each condition, right; spines in microglia ablated mice. Data expressed as mean. n = 3 or 4 mice with 30-60 dendritic stretches. Related Fig. S4 displays this data in FC and SD contexts individually, and Table S2 provides statistical testing results between numerous spine type and context comparisons.
Additionally, the composition of spines was significantly different in the FCSD condition in microglia-ablated mice compared to control; there was a significantly increased proportion of thin compared to mushroom spines (Fig. 4B and C, Table S2). In comparison, in neither an FC condition; where mice were trained and sacrificed without SD at ZT = 6, nor a condition where mice were subjected to SD without FC, did control or microglia ablated mice exhibited significant differences in spine density. However, we did observe an increase in mushroom spines in the FCSD control mice compared to both SD and FC individually. This suggests that the effect of FCSD on spine density is unique to this context, or potentially a cumulative result of small, nonsignificant differences in the FC and SD conditions alone (Fig. S4, Table S2). This difference was not observed in the ablated mice. Mushroom spines are associated with long-term memory consolidation and thin spines are generally considered ‘immature’ (Bourne and Harris, 2007), suggesting that following a learning event during extended wakefulness, ablation of microglia led to difficulty stabilizing thin spines into mature, mushroom synapses, as our data demonstrates. Subsequently, in the recovery sleep period, these immature synapses are lost (Fig. 4C), potentially mediating the memory differences observed. Another metric of synapse maturation and long term potentiation of memory; phosphorylated GluR1-831 ratio (Lee et al., 2003), was similarly compromised in the hippocampus of ablated mice (Fig. S5A), despite no significant difference between total GluR1 between ablated and control mice in each condition (Fig. S5B). However, one proxy metric of total mature synapse number- PSD95- found no significant differences between conditions (Figs. S5C and E). Since BDNF concentration has been implicated in microglia's modulatory functions at the synapse (Ferrini and De Koninck, 2013; Ulmann et al., 2008) and in delta power (Faraguna et al. 2008), we examined BDNF levels in the hippocampus of these mice, but found it was unchanged by microglia ablation (Fig. S2).
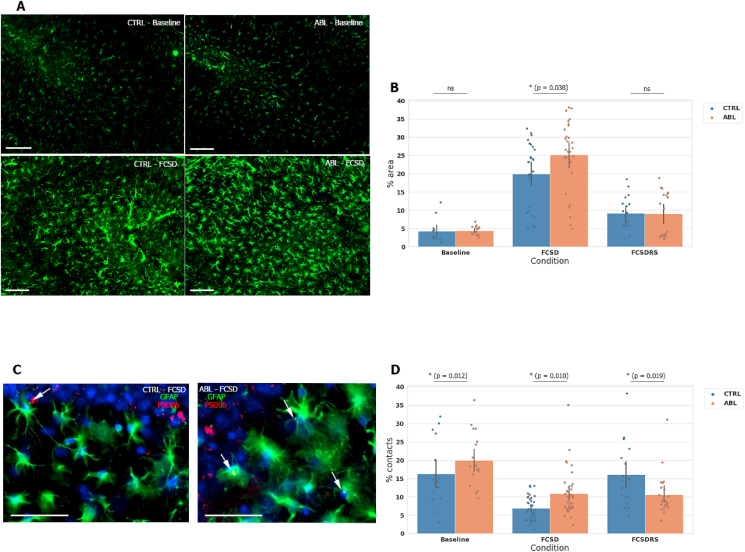
3.5. Microglia restrain astrocytic reactivity after FCSD
Finally, we assessed how astrocytes, which comprise the third component of the ‘tripartite synapse,’ (Schafer et al., 2013) behave with microglia depleted in FCSD conditions. We found astrocytic coverage as measured by GFAP in the dorsal hippocampus was increased following FCSD in both control and ablated conditions, but was increased significantly further in the ablated mice (Fig. 5A and B). We also found that the proportion of postsynaptic elements as measured by PSD95 staining that colocalized with astrocyte ramifications in the DG-CA3 transition region greater in ablated mice than in mice with microglia (Fig. 5C and D), and- similarly to the phenomenon observed with spine density- such colocalization failed to return to baseline levels after RS only in ablated mice. Together, these data suggest that microglia are important for the stabilization of synapses following FCSD and subsequent recovery sleep, possibly through restraining astrocytic phagocytic activity.
Fig. 5.
Microglia restrain astrocytic activity after FCSD (A), Representative hippocampal GFAP staining for astrocytes in 14–20 week old mice at baseline (top) and after FCSD (bottom) with microglia (left) and with microglia ablated (right) to illustrate increase in reactivity following FCSD. Scale bar = 50 μM. Images were passed through a standardized threshold. (B), Quantification of (A); the percent area of a 500 × 500 μm region of CA3 hippocampus covered by GFAP astrocytic staining. n = 10-15 slices per mouse, 4 mice per condition. Statistical analysis performed using nonparametric Mann-Whitney test between relevant conditions. (C), Representative CA3 PSD95 (red) + GFAP (green) double stain with DAPI (blue). Images illustrating the difference between control (left) and ablated (right) conditions after FCSD. Arrows indicate colocalization of astrocytes and PSD95 marks after standardized thresholding, as defined by the JaCOP FIJI program. Different cohorts of mice were used for each condition. Scale bar = 20 μm. (D), Quantification of (C). Percent of PSD95 puncta in the DG to CA3 transition region contacted by astrocytic ramifications. GFAP and PSD95 were imaged in different fluorescent channels, passed through a standardized threshold, and overlaps measured by FIJI's colocalization plugin. Data expressed as mean ± SD. n = 4 mice per condition. Statistical analysis performed using an unpaired parametric two-tailed student's T test between relevant conditions. Related Fig. S5 displays further measured features of synaptic maturity and astrocytic reactivity. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion and conclusions
Microglia have been studied far more extensively in the past two decades than ever before, in part due to the emergence of a variety of techniques capable of specific targeting of this cell type. The array of roles microglia play in the brain has thus expanded significantly beyond their canonical immunological functions (Han et al., 2017b), they are now known to prune (Paolicelli et al., 2011) and remodel synaptic connections (Torres et al., 2016; De Luca et al., 2020), mediate neurogenesis (Ji et al., 2013), and more generally promote homeostasis of neuronal connectivity in the brain (Eyo and Wu, 2013; Liu et al., 2019; Han et al., 2017a). Despite this expansion of knowledge, theories around the role microglia play in sleep and sleep deprivation have largely remained centered around their inflammatory functions (Ingiosi et al., 2013; KRUEGER et al., 2011; Tuan and Lee, 2019; Krueger et al., 2010; Wisor et al., 2011c; Wisor et al., 2011d).
Inflammation in the brain is associated with increased sleep drive (Imeri and Opp, 2009). Microglia release cytokines- inflammatory molecules- in response to stressors, and they have been proposed as a major agent in the sleep rebound process following inflammatory insult (Nadjar et al., 2017). One such intervention with a consistent sleep rebound is SD. SD following a learning event elicits a marked reduction in hippocampal-dependent memory the following day (Graves et al., 2003; McCoy and Strecker, 2011; Sweatt and Hawkins, 2016). Administration of anti-inflammatory drugs- for example, minocycline- have been found to alleviate some of these cognitive deficits seen following SD as well as the sleep rebound response itself (Wadhwa et al., 2017a; Wisor et al., 2011b).
Our data here indicate that microglia have a specific, previously undescribed role in maintaining synaptic homeostasis during recovery sleep following short-term acute SD. Sleep helps consolidate memories formed during preceding waking periods; with microglia depleted, a mouse's ability to stabilize memories formed after a period of SD is greatly compromised.
We first demonstrate that microglia exhibit a greater degree of ramification during SD, possibly as a mechanism through which to directly contact and stabilize more synaptic microenvironments during the stressful process of SD. The effects of such increased contact may not manifest as a memory difference until after recovery sleep when more synaptic remodeling occurs; this is explored later in this discussion. SD results in a number of changes to neuronal expression and physiology which may potentially mediate this behavior, signaling microglia to buffer against its negative consequences, though further research is necessary to confirm this theory. Our finding contradicts some previous studies which found an amoeboid, deramified morphology after SD. (Wadhwa et al., 2017a; Hsu et al., 2003) This discrepancy may be due to the varying durations and techniques for SD used- we sleep deprived mice for 6 h after the beginning of the sleep (light) phase using noninvasive methods, whereas other researchers have employed arguably more stressful methods and sleep deprived for as many as 5 consecutive days (Hsu et al., 2003; Fenzl et al., 2007). Other studies employing a shorter period of SD found similar or more modest modifications to microglial morphology than we document here (Tuan and Lee, 2019; Bellesi et al., 2017). Long duration SD or chronic sleep restriction techniques are less likely to reflect ‘homeostatic’ responses of the brain to SD and may reflect other microglial behaviors observed during severe insult or chronic perturbation of the neuronal environment. Longer SD paradigms may also overwhelm the stabilizing response we observed here during short periods of SD. Alternatively, some researchers have suggested the Iba1+ staining used to identify microglia morphology in this study can also label peripheral macrophages which may infiltrate the brain, though there is evidence to suggest the effect of this cell type on overall Iba1+ staining in a sleep deprived context is minimal (Hall et al., 2020). Further research is necessary to validate the findings, given the complex picture painted by our and previous research.
Using thorough EEG, (the gold standard for mouse sleep-wake assessment), we also found that microglia are dispensable for most other features of homeostatic sleep and SD recovery. Namely, microglia-ablated mice exhibit normal baseline sleep dynamics and a robust SD rebound response, as well as circadian periodicity. Within these experiments, we document an interesting phenomenon whereby absolute EEG power was broadly increased; significantly so in frequencies with high power (e.g. the Delta frequency band), and exhibiting a trend towards it at all other frequencies. The effect was largely lost when power was normalized to total power across all states and frequencies, indicating a broad increase. Recent research has hinted at such a characteristic before, as elimination of microglia broadly increases some correlates of neural circuit activity in the cortex (Torres et al., 2016; Schafer et al., 2012). Further research is necessary to understand the functional consequences of this change.
Microglia-specific transcriptome sequencing revealed a suite of upregulated differentially expressed genes upon FCSD enriched for processes associated with neuronal modulation, synaptic plasticity, and transcriptional regulation and translation, further confirming the novel role we have described. Many of these genes have previously been identified in SD of various lengths (Vecsey et al., 2012; Hor et al., 2019; Gaine et al., 2021), and this is first study to our knowledge specifically attributing some of these genes modified in SD to microglia (Hirbec et al., 2017). Microglia, according to this finding, may primarily perform a role during sleep deprivation in stabilizing the neuronal microenvironment, rather than mediating more deleterious inflammatory or other immune-relate processes. These effects may then manifest as morphological differences in neurons in the subsequent sleep period. The observed differences in expression patterns were supported by our morphological analyses which found a reduction in CA3 dendritic spine density in mice with ablated microglia and fewer mature synapses, especially after a subsequent period of recovery sleep (Fig. 4B). More specifically, the processes enriched in the transcript data may be reflected by the different spine type compositions found between microglia-ablated and control mice (Fig. 4C). From baseline to FCSD, control mice exhibited increases in mushroom spine density both proportionally and in absolute density, whereas mice with microglia depleted exhibited a reduction in both. Thin spine density increased after FCSD in this condition. This may signify that microglia-derived signals- the synapse-related terms identified in our RNA-seq- are important for maturation of thin spines into mushroom spines during a learning event that occurs before SD. The lack of these signals may drive the failure to mature, seen as an increase in thin spines which are subsequently lost during the recovery sleep period.
The molecular mechanism for the microglial role in protecting memories and maturing synapses after sleep deprivation remains unknown. We were unable to identify changes with microglial ablation after FCSD in BNDF, a common neurotrophic factor previously associated with microglia activity (Ferrini and De Koninck, 2013; Ulmann et al., 2008). We did, however, observe an increase in astrocytic association with PSD95-positive synapses by GFAP following microglial ablation during baseline and FCSD, despite an overall decrease in spine density. After FCSDRS, astrocytic contacts to PSD95 puncta increased to approximately baseline levels in control mice, but remained unchanged in microglia-ablated mice at lower levels (Figs. 4B and 5D). Astrocytes are the third member of the ‘tripartite synapse’ model and have been shown to modulate sleep responses (Bellesi et al., 2017; Halassa et al., 2009) and synaptic homeostasis (Havekes et al., 2012). Microglia participate in extensive crosstalk with astrocytes in a concerted manner to regulate the synapse microenvironment (Schafer et al., 2013; Pascual et al., 2012). Astrocytes and microglia may both be essential for proper regulation of the synaptic microenvironment after a learning event during SD. Our results therefore suggest that microglia are important for the stability of synaptic maturation following FCSDRS possibly through the restraint of astrocytic phagocytic activity, though this theory requires further validation, as colocalization is only one indicator of phagocytosis. In the absence of microglia, astrocytes could be ‘overactive’ and negatively affect maturation of thin spines during recovery sleep, leading to the observed drastic decrease in overall spine density and proportion of thin spines after FCSDRS and resulting in fewer mature spine-astrocyte points of contact. Alternatively, other mechanisms through which microglia have been found to modulate synapses could play a role in the SD context, such as through the process of remodeling by trogocytosis (Weinhard et al., 2018). Overall, astrocytes and microglia may counterbalance each other to maintain memory and cognitive function during acute SD; we found that in the absence of microglia, astrocytes appear more reactive, perhaps in compensation.
The Synaptic Homeostasis hypothesis (SHY) postulates that synapse density and strength increase not only with learning but with wakefulness (Tononi and Cirelli, 2014a). During sleep, synapses are downscaled proportionally to the strength they gained during this wake, consolidating memories (Vyazovskiy et al., 2008; Tononi and Cirelli, 2014b). Our spine density analyses confirmed an increase following FCSD and a subsequent decrease to baseline levels following recovery sleep in control mice. Initially, due to our observations of microglia-dependent memory formation during the sleep recovery period following SD, we hypothesized that microglia may be responsible for synaptic downscaling during sleep. Indeed, previous research has found they may behave in this manner (Choudhury et al., 2020), suggesting that without microglia we may expect a decreased reduction in synaptic strength after recovery sleep, which may interfere with memory consolidation. However, we observed a sharp loss in spine density following FCSDRS in mice with microglia depleted, primarily in immature thin spines- those most likely formed during the recent learning period (Bourne and Harris, 2007; Pchitskaya and Bezprozvanny, 2020; Berry and Nedivi, 2017). C1q, a phagocytic marker for microglia (Fonseca et al., 2017), was also largely unaffected by ablation (Figs. S5D and F) in another region of the hippocampus, suggesting that microglia are not necessary for sleep-dependent downscaling and may instead perform a protective role, aiding the maturation of spines during a period of SD or identifying those important to maintain during the remodeling that occurs during sleep after an SD period. Altogether, this indicates that the SHY alone cannot explain our findings. Additionally, according to our data, astrocytes may be more important for the synaptic downscaling and sleep rebound functions during sleep, functions which may be tempered by the presence of microglia, a theory reflected by the exacerbated loss of spines in mice with ablated microglia. This hypothesis is supported by the findings of Bellesi et al. (2017), which found astrocytes were the primary drivers of phagocytosis during short-term, acute sleep loss (Bellesi et al., 2017).
Further, recent research in the modulation of astrocytic signaling, primarily through Ca2+ GPCRs, was found to influence sleep duration and depth (Vaidyanathan et al., 2021; Bojarskaite et al., 2020). To gain a more complete picture of the relative synaptic modulatory roles of astrocytes and microglia in the SD context, future research should examine the colocalization of astrocytes with presynaptic markers such as VGLUT or synapsin, as well as the direct relationship between spines and C1q in identical brain regions, or preferably, the same brain slices. Another possibility is that circadian timing or sleep duration interacts with the ablation condition to play a role in spine type, as we observed a distribution of mushroom spines more similar to control conditions following FC (6 h of recovery sleep at ZT6) than after FCSDRS (18 h recovery sleep, ZT0).
Consistency in interventions is an issue across the SD literature. Differences in response between even 6 and 24 h of SD may be large, and caution should be taken in broadly interpreting data discussing the effects of SD. Data finding an increase during SD of canonical microglial markers of inflammation and reactive phenotypes has promoted the idea that microglia are responsible for driving the deleterious side effects of SD or other insults (Nadjar et al., 2017). However, studies differ in duration of SD, method of modifying microglia behavior, and brain region investigated. For example; some research has found that downregulation of microglial activity leads to increased spine density, counter to our findings (Choudhury et al., 2020). These studies employed anti-inflammatory treatments which may have also affected astrocyte activity, or perhaps reduced certain crosstalk pathways between microglia and astrocytes. Similar conclusions can be made for the research finding that anti-inflammatory drugs reduced the increases in delta power and sleep following SD. These studies also primarily used anti-inflammatory treatments (Wisor et al., 2011b; Aono et al., 2017), and may not reflect the role of microglia, but rather astrocytes. If the inflammatory function of microglia was a main player in delta power increases following SD, removal of this cell type would be expected to attenuate such a phenomenon. Indeed, evidence is building that astrocytes are important for sleep dynamics (Vaidyanathan et al., 2021). However, in homeostatic conditions, it appears that microglia acts to protect the brain from the negative consequences of SD, not drive it, though validation of specific gene players in this process needs to be validated. Perhaps in certain situations, like severe neurodegeneration or long-term sleep restriction, microglia may lose this function. Increased microglia reactivity appears to be associated with deleterious outcomes in such highly disruptive states (Rice et al., 2015; Liu et al., 2021; Spangenberg et al., 2019; Ni et al., 2019; Frank-Cannon et al., 2009); however, this and other current research suggests that their activity in such contexts may not be wholly representative of their function in all contexts, especially in the healthy mammalian brain (Hirbec et al., 2017). The phenomenon we observed may be quite context specific, as previous similar experiments using more severe inflammatory insults yielded differing results (Acharya et al., 2016).
The normal function of microglia was once a mystery, and they were broadly studied mostly in contexts of deleterious activity in the diseased brain. Only recently has the diversity of their roles begun to come to light and their importance realized. Our research here furthers our understanding of the complexity and nuance with which microglia support the synaptic microenvironment, as they appear to have an essential role in maintaining memories formed before or during SD.
4.1. Limitations of the study
There are a number of limitations of this study pertaining to breadth of biological variables measured. Firstly, the PSD95 and C1q stains would provide more reliable information from which to form synapse-related conclusions if they were performed on identical regions of the hippocampus as the golgi staining measuring spine density. For the astrocyte experiments, GFAP may not label all astrocytes equally, and may not label finer processes which could be also contacting synapses. Additionally, we only labeled for PSD95, a postsynaptic marker, to identify synapse density. Synapsin, VGLUT, or other markers of pre- or post-synaptic densities would provide a fuller picture. To more fully establish the process at play during synapse-astrocyte colocalization, lysosomal markers are additionally necessary. Future research should measure astrocytic lysosomal-mediated phagocytosis in the sleep deprivation context.
Sleep deprivation for most mice was conducted by rotating bar, found to be effective at causing sleep deprivation. However, SD for the EEG experiment was by necessity done through gentle handling, as the rotating bar is incompatibly with the leads used in EEG recording- the mice would get tangled and injured. Potential differences in a rotating bar versus gentle handling could lead to confounds.
Further, we looked at spine differences in the CA3 hippocampal region specifically, as it has been associated with contextual fear learning. However, the CA1 and DG regions have also been implicated in this form of memory, and would be additionally useful to investigate. Measuring the CA3 provides only one useful, but crude, approximation of spine density in relevant brain regions.
Finally, though we tested for sex differences by using male and female mice in some experiments, this was not possible in all paradigms, namely our immunostaining which had small numbers of mice used. We established no differences in the assays we did use both sexes in, but some may still exist in those we did not, especially as microglia behavior and characteristics appear to have a sex-dependent component (Villa et al., 2018), and small differences in physiology may not necessarily manifest as larger-scale behavioral or EEG effects.
Declaration of interests
The authors declare no competing interests for this study.
CRediT authorship contribution statement
Nicholas W. Gentry: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. Thomas McMahon: Methodology, Resources. Maya Yamazaki: Methodology, Resources. John Webb: Methodology. Thomas D. Arnold: Resources, Conceptualization. Susanna Rosi: Resources, Conceptualization. Louis J. Ptáček: Funding acquisition, Supervision, Writing – review & editing. Ying-Hui Fu: Funding acquisition, Supervision, Writing – review & editing.
Acknowledgements
We thank all members in the Ptáček and Fu labs for helpful discussions. This work was supported by NIH grants NS117929, NS072360 and NS104782 (L.J.P. and Y-H.F.), R01 AG 56770 (S.R.), and by the William Bowes Neurogenetics Fund (L.J.P. and Y.H.F.). The generation of mouse models was also supported by NIH P30 DK063720 to the Diabetes Center at UCSF.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbscr.2021.100073.
Contributor Information
Louis J. Ptáček, Email: ljp@ucsf.edu.
Ying-Hui Fu, Email: Ying-Hui.Fu@ucsf.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Acharya M.M., Green K.N., Allen B.D., Najafi A.R., Syage A., Minasyan H., Le M.T., Kawashita T., Giedzinski E., Parihar V.K., et al. Elimination of microglia improves cognitive function following cranial irradiation. Sci. Rep. 2016;6:31545. doi: 10.1038/srep31545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono H., Choudhury M.E., Higaki H., Miyanishi K., Kigami Y., Fujita K., Akiyama J.I., Takahashi H., Yano H., Kubo M., et al. Microglia may compensate for dopaminergic neuron loss in experimental Parkinsonism through selective elimination of glutamatergic synapses from the subthalamic nucleus. Glia. 2017;65:1833–1847. doi: 10.1002/glia.23199. [DOI] [PubMed] [Google Scholar]
- Bellesi M., Vivo L. De, Chini M., Gilli F., Tononi G., Cirelli C. Sleep loss promotes astrocytic phagocytosis and microglial activation in mouse cerebral cortex. J. Neurosci. 2017;37:5263–5273. doi: 10.1523/JNEUROSCI.3981-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry K.P., Nedivi E. Spine dynamics: are they all the same? Neuron. 2017;96:43–55. doi: 10.1016/j.neuron.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarskaite L., Bjørnstad D.M., Pettersen K.H., Cunen C., Hermansen G.H., Åbjørsbråten K.S., Chambers A.R., Sprengel R., Vervaeke K., Tang W., et al. Astrocytic Ca2+ signaling is reduced during sleep and is involved in the regulation of slow wave sleep. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-17062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Bourne J., Harris K.M. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Choudhury M.E., Miyanishi K., Takeda H., Islam A., Matsuoka N., Kubo M., Matsumoto S., Kunieda T., Nomoto M., Yano H., et al. Phagocytic elimination of synapses by microglia during sleep. Glia. 2020;68:44–59. doi: 10.1002/glia.23698. [DOI] [PubMed] [Google Scholar]
- Colavito V., Fabene P.F., Grassi-Zucconi G., Pifferi F., Lamberty Y., Bentivoglio M., Bertini G. Experimental sleep deprivation as a tool to test memory deficits in rodents. Front. Syst. Neurosci. 2013;7:106. doi: 10.3389/fnsys.2013.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca S.N., Soch A., Sominsky L., Nguyen T.X., Bosakhar A., Spencer S.J. Glial remodeling enhances short-term memory performance in Wistar rats. J. Neuroinflammation. 2020;17:52. doi: 10.1186/s12974-020-1729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R.P., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., et al. Colony-Stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore M.R.P., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.bioconductor.org/packages/release/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling.
- Eyo U.B., Wu L.J. Bidirectional microglia-neuron communication in the healthy brain. Neural Plast. 2013 doi: 10.1155/2013/456857. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenzl T., Romanowski C.P.N., Flachskamm C., Honsberg K., Boll E., Hoehne A., Kimura M. Fully automated sleep deprivation in mice as a tool in sleep research. J. Neurosci. Methods. 2007;166:229–235. doi: 10.1016/j.jneumeth.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ferreira T.A., Blackman A.V., Oyrer J., Jayabal S., Chung A.J., Watt A.J., Sjöström P.J., Van Meyel D.J. Neuronal morphometry directly from bitmap images. Nat. Methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrini F., De Koninck Y. Microglia control neuronal network excitability via BDNF signalling. Neural Plast. 2013 doi: 10.1155/2013/429815. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C. Reconstruct: a free editor for serial section microscopy. J. Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Fonseca M.I., Chu S.-H., Hernandez M.X., Fang M.J., Modarresi L., Selvan P., MacGregor G.R., Tenner A.J. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J. Neuroinflammation. 2017;141 14:1–15. doi: 10.1186/s12974-017-0814-9. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Cannon T.C., Alto L.T., McAlpine F.E., Tansey M.G., Andersen P., Marklund S., Koistinaho J., Mai T., Mou S., Pye Q., et al. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol. Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P., Dudley C.A., Estill S.J., Barakat M., Thomason R., O'Hara B.F., McKnight S.L. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7118–7123. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaine M.E., Bahl E., Chatterjee S., Michaelson J.J., Abel T., Lyons L.C. Altered hippocampal transcriptome dynamics following sleep deprivation. Mol. Brain. 2021;141(14):1–17. doi: 10.1186/s13041-021-00835-1. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway D.A., Phillips A.E.M., Owen D.R.J., Moore C.S. Phagocytosis in the brain: homeostasis and disease. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L.A., Heller E.A., Pack A.I., Abel T. Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn. Mem. 2003;10:168–176. doi: 10.1101/lm.48803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa M.M., Florian C., Fellin T., Munoz J.R., Lee S.-Y., Abel T., Haydon P.G., Frank M.G. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61:213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S., Deurveilher S., Robertson G.S., Semba K. Homeostatic state of microglia in a rat model of chronic sleep restriction. Sleep. 2020;43 doi: 10.1093/sleep/zsaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Harris R.A., Zhang X.M. An updated assessment of microglia depletion: current concepts and future directions. Mol. Brain. 2017;10:25. doi: 10.1186/s13041-017-0307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Harris R.A., Zhang X.M. An updated assessment of microglia depletion: current concepts and future directions. Mol. Brain. 2017;10 doi: 10.1186/s13041-017-0307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havekes R., Vecsey C.G., Abel T. The impact of sleep deprivation on neuronal and glial signaling pathways important for memory and synaptic plasticity. Cell. Signal. 2012;24:1251–1260. doi: 10.1016/j.cellsig.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Koyanagi S., Kusunose N., Okada R., Wu Z., Tozaki-Saitoh H., Ukai K., Kohsaka S., Inoue K., Ohdo S., et al. The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Sci. Rep. 2013;3:2744. doi: 10.1038/srep02744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H.E., Noristani H.N., Perrin F.E. Microglia responses in acute and chronic neurological diseases: what microglia-specific transcriptomic studies taught (and did not teach) Us. Front. Aging Neurosci. 2017;9 doi: 10.3389/fnagi.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor C.N., Yeung J., Jan M., Emmenegger Y., Hubbard J., Xenarios I., Naef F., Franken P. Sleep–wake-driven and circadian contributions to daily rhythms in gene expression and chromatin accessibility in the murine cortex. Proc. Natl. Acad. Sci. Unit. States Am. 2019 doi: 10.1073/pnas.1910590116. 201910590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.C., Lee Y.S., Chang C.N., Chuang H.L., Ling E.A., Lan C.T. Sleep deprivation inhibits expression of NADPH-d and NOS while activating microglia and astroglia in the rat hippocampus. Cells Tissues Organs. 2003;173:242–254. doi: 10.1159/000070380. [DOI] [PubMed] [Google Scholar]
- Hunsaker M.R., Tran G.T., Kesner R.P. A behavioral analysis of the role of CA3 and CA1 subcortical efferents during classical fear conditioning. Behav. Neurosci. 2009;123:624–630. doi: 10.1037/a0015455. [DOI] [PubMed] [Google Scholar]
- Imeri L., Opp M.R. How (and why) the immune system makes us sleep. Nat. Rev. Neurosci. 2009;10:199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingiosi A.M., Opp M.R., Krueger J.M. Sleep and immune function: glial contributions and consequences of aging. Curr. Opin. Neurobiol. 2013;23:806–811. doi: 10.1016/j.conb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janova H., Arinrad S., Balmuth E., Mitjans M., Hertel J., Habes M., Bittner R.A., Pan H., Goebbels S., Begemann M., et al. Microglia ablation alleviates myelin-associated catatonic signs in mice. J. Clin. Invest. 2017;128:734–745. doi: 10.1172/JCI97032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji K., Miyauchi J., Tsirka S.E. Microglia: an active player in the regulation of synaptic activity. Neural Plast. 2013 doi: 10.1155/2013/627325. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.S., Ebbert M.T.W., Baker K.E., Cook C., Wang X., Sens J.P., Kocher J.-P., Petrucelli L., Fryer J.D. Microglial translational profiling reveals a convergent APOE pathway from aging, amyloid, and tau. J. Exp. Med. 2018;215:2235–2245. doi: 10.1084/jem.20180653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger J.M., Taishi P., De A., Davis C.J., Winters B.D., Clinton J., Szentirmai E., Zielinski M.R. ATP and the purine type 2 X7 receptor affect sleep. J. Appl. Physiol. 2010;109:1318–1327. doi: 10.1152/japplphysiol.00586.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger J.M., Majde J.A., Rector D.M. Cytokines in immune function and sleep regulation. Handb. Clin. Neurol. 2011;98:229. doi: 10.1016/B978-0-444-52006-7.00015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukowski K., Feng X., Paladini M.S., Chou A., Sacramento K., Grue K., Riparip L.-K., Jones T., Campbell-Beachler M., Nelson G., et al. Temporary microglia-depletion after cosmic radiation modifies phagocytic activity and prevents cognitive deficits. Sci. Rep. 2018;8:7857. doi: 10.1038/s41598-018-26039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Takamiya K., Han J.S., Man H., Kim C.H., Rumbaugh G., Yu S., Ding L., He C., Petralia R.S., et al. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- LeGates T.A., Altimus C.M. Measuring circadian and acute light responses in mice using wheel running activity. JoVE. 2010;48 doi: 10.3791/2463. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Y.U., Ying Y., Li Y., Eyo U.B., Chen T., Zheng J., Umpierre A.D., Zhu J., Bosco D.B., Dong H., et al. Neuronal network activity controls microglial process surveillance in awake mice via norepinephrine signaling. Nat. Neurosci. 2019;22:1771–1781. doi: 10.1038/s41593-019-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-J., Spangenberg E.E., Tang B., Holmes T.C., Green K.N., Xu X. Microglia elimination increases neural circuit connectivity and activity in adult mouse cortex. J. Neurosci. 2021;41:1274–1287. doi: 10.1523/JNEUROSCI.2140-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer M., Lopez M.A., Meier D., Mikhail C., et al. Cytokine-induced sleep: neurons respond to TNF with production of chemokines and increased expression of Homer1a in vitro. Brain Behav. Immun. 2015;47:186–192. doi: 10.1016/j.bbi.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Mang G.M., Franken P. Current Protocols in Mouse Biology. John Wiley & Sons, Inc.); 2012. Sleep and EEG phenotyping in mice; pp. 55–74. [DOI] [PubMed] [Google Scholar]
- Maret S., Franken P., Dauvilliers Y., Ghyselinck N.B., Chambon P., Tafti M. Retinoic acid signaling affects cortical synchrony during sleep. Science. 2005;310:111–113. doi: 10.1126/science.1117623. 80-. [DOI] [PubMed] [Google Scholar]
- McCoy J.G., Strecker R.E. The cognitive cost of sleep lost. Neurobiol. Learn. Mem. 2011;96:564–582. doi: 10.1016/j.nlm.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey G.L., Lizama C.O., Keown-Lang A.E., Niu A., Santander N., Larpthaveesarp A., Chee E., Gonzalez F.F., Arnold T.D. A new genetic strategy for targeting microglia in development and disease. Elife. 2020;9:1–34. doi: 10.7554/eLife.54590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjar A., Wigren H.-K.M., Tremblay M.-E. Roles of microglial phagocytosis and inflammatory mediators in the pathophysiology of sleep disorders. Front. Cell. Neurosci. 2017;11:250. doi: 10.3389/fncel.2017.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Wu Z., Meng J., Saito T., Saido T.C., Qing H., Nakanishi H. An impaired intrinsic microglial clock system induces neuroinflammatory alterations in the early stage of amyloid precursor protein knock-in mouse brain. J. Neuroinflammation. 2019;16:173. doi: 10.1186/s12974-019-1562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet M., Wisden W., Franks N.P. Sleep deprivation and stress: a reciprocal relationship. Interface Focus. 2020;10 doi: 10.1098/rsfs.2019.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L., et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. 80. [DOI] [PubMed] [Google Scholar]
- Pascual O., Achour S. Ben, Rostaing P., Triller A., Bessis A. Microglia activation triggers astrocyte-mediated modulation of excitatory neurotransmission. Proc. Natl. Acad. Sci. U. S. A. 2012;109 doi: 10.1073/pnas.1111098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchitskaya E., Bezprozvanny I. Dendritic spines shape analysis—classification or clusterization? Perspective. Front. Synaptic Neurosci. 2020;12:31. doi: 10.3389/fnsyn.2020.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V., Kaushal navita, Gozal D. Sleep fragmentation differentially modifies EEG delta power during slow wave sleep in socially isolated and paired mice. Sleep Sci. 2009;2:64–75. [Google Scholar]
- Rice R.A., Spangenberg E.E., Yamate-Morgan H., Lee R.J., Arora R.P.S., Hernandez M.X., Tenner A.J., West B.L., Green K.N. Elimination of microglia improves functional outcomes following extensive neuronal loss in the Hippocampus. J. Neurosci. 2015;35:9977–9989. doi: 10.1523/JNEUROSCI.0336-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockstrom M.D., Chen L., Taishi P., Nguyen J.T., Gibbons C.M., Veasey S.C., Krueger J.M. Tumor necrosis factor alpha in sleep regulation. Sleep Med. Rev. 2018;40:69–78. doi: 10.1016/j.smrv.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurveilher S., Golovin T., Hall S., Semba K. Microglia dynamics in sleep/wake states and in response to sleep loss. Neurochem. Int. 2021;143 doi: 10.1016/j.neuint.2020.104944. [DOI] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Stevens B. The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia. 2013;61:24–36. doi: 10.1002/glia.22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Xing L., Wu D., Bhattacharyya B.J., Jones C.R., McMahon T., Chong S.Y.C., Chen J.A., Coppola G., Geschwind D., et al. A rare mutation of β1-adrenergic receptor affects sleep/wake behaviors. Neuron. 2019;103:1044–1055. doi: 10.1016/j.neuron.2019.07.026. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg E., Severson P.L., Hohsfield L.A., Crapser J., Zhang J., Burton E.A., Zhang Y., Spevak W., Lin J., Phan N.Y., et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model. Nat. Commun. 2019;10:1–21. doi: 10.1038/s41467-019-11674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell R.D., Sipe G.O., Dawes R.P., Batchelor H.N., Lordy K.A., Whitelaw B.S., Stoessel M.B., Bidlack J.M., Brown E., Sur M., et al. Noradrenergic signaling in the wakeful state inhibits microglial surveillance and synaptic plasticity in the mouse visual cortex. Nat. Neurosci. 2019;22:1782–1792. doi: 10.1038/s41593-019-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt J.D., Hawkins K.E. The molecular neurobiology of the sleep-deprived, fuzzy brain. Sci. Signal. 2016;9:fs7. doi: 10.1126/scisignal.aaf6196. [DOI] [PubMed] [Google Scholar]
- Tononi G., Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron. 2014;81:12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G., Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron. 2014;81:12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres L., Danver J., Ji K., Miyauchi J.T., Chen D., Anderson M.E., West B.L., Robinson J.K., Tsirka S.E. Dynamic microglial modulation of spatial learning and social behavior. Brain Behav. Immun. 2016;55:6–16. doi: 10.1016/j.bbi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.Ě., Lowery R.L., Majewska A.K. Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 2010;8:1000527. doi: 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.È., Stevens B., Sierra A., Wake H., Bessis A., Nimmerjahn A. The role of microglia in the healthy brain. J. Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan L.H., Lee L.J. Microglia-mediated synaptic pruning is impaired in sleep-deprived adolescent mice. Neurobiol. Dis. 2019;130:104517. doi: 10.1016/j.nbd.2019.104517. [DOI] [PubMed] [Google Scholar]
- Faraguna U., Vyazovskiy V.V., Nelson A.B., et al. A causal role for brain-derived neurotrophic factor in the homeostatic regulation of sleep. J. Neurosci. 2008;28:4088–4095. doi: 10.1523/JNEUROSCI.5510-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmann L., Hatcher J.P., Hughes J.P., Chaumont S., Green P.J., Conquet F., Buell G.N., Reeve A.J., Chessell I.P., Rassendren F. Up-regulation of P2X4 receptors in spinal microglia after peripheral nerve injury mediates BDNF release and neuropathic pain. J. Neurosci. 2008;28:11263–11268. doi: 10.1523/JNEUROSCI.2308-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan T.V., Collard M., Yokoyama S., Reitman M.E., Poskanzer K.E. Cortical astrocytes independently regulate sleep depth and duration via separate GPCR pathways. Elife. 2021;10 doi: 10.7554/eLife.63329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A., Franken P. Hypocretin (orexin) is critical in sustaining theta/gamma-rich waking behaviors that drive sleep need. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E5464–E5473. doi: 10.1073/pnas.1700983114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey C.G., Peixoto L., Choi J.H.K., Wimmer M., Jaganath D., Hernandez P.J., Blackwell J., Meda K., Park A.J., Hannenhalli S., et al. Genomic analysis of sleep deprivation reveals translational regulation in the hippocampus. Physiol. Genom. 2012;44:981–991. doi: 10.1152/physiolgenomics.00084.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A., Gelosa P., Castiglioni L., Cimino M., Rizzi N., Pepe G., Lolli F., Marcello E., Sironi L., Vegeto E., et al. Sex-specific features of microglia from adult mice. Cell Rep. 2018;23:3501. doi: 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy V.V., Cirelli C., Pfister-Genskow M., Faraguna U., Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci. 2008;11:200–208. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- Wadhwa M., Prabhakar A., Ray K., Roy K., Kumari P., Jha P.K., Kishore K., Kumar S., Panjwani U. Inhibiting the microglia activation improves the spatial memory and adult neurogenesis in rat hippocampus during 48h of sleep deprivation. J. Neuroinflammation. 2017;14 doi: 10.1186/s12974-017-0998-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wadhwa M., Kumari P., Chauhan G., Roy K., Alam S., Kishore K., Ray K., Panjwani U. Sleep deprivation induces spatial memory impairment by altered hippocampus neuroinflammatory responses and glial cells activation in rats. J. Neuroimmunol. 2017;312:38–48. doi: 10.1016/j.jneuroim.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Wake H., Moorhouse A.J., Jinno S., Kohsaka S., Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yue H., Hu Z., Shen Y., Ma J., Li J., Wang X.-D., Wang L., Sun B., Shi P., et al. Microglia mediate forgetting via complement-dependent synaptic elimination. Science. 2020;367:688–694. doi: 10.1126/science.aaz2288. [DOI] [PubMed] [Google Scholar]
- Webb S.D., Orton L.D. bioRxiv; 2019. Iba1+ Microglia Colocalize with Synaptophysin+ and Homer1+ Puncta and Reveal Two Novel Sub-types of GABAergic Neuron in Guinea Pig Inferior Colliculus; p. 606509. [Google Scholar]
- Weinhard L., Di Bartolomei G., Bolasco G., Machado P., Schieber N.L., Neniskyte U., Exiga M., Vadisiute A., Raggioli A., Schertel A., et al. Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-03566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J.L., Xue L., Wang R.H., Chen Z.X., Shi Y.W., Zhao H. Involvement of the dopaminergic system in the consolidation of fear conditioning in hippocampal CA3 subregion. Behav. Brain Res. 2015;278:527–534. doi: 10.1016/j.bbr.2014.10.049. [DOI] [PubMed] [Google Scholar]
- Wisor J.P., Schmidt M.A., Clegern W.C. Evidence for neuroinflammatory and microglial changes in the cerebral response to sleep loss. Sleep. 2011;34:261–272. doi: 10.1093/sleep/34.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor J.P., Schmidt M.A., Clegern W.C. Evidence for neuroinflammatory and microglial changes in the cerebral response to sleep loss. Sleep. 2011;34:261–272. doi: 10.1093/sleep/34.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor J.P., Clegern W.C., Schmidt M.A. Toll-like receptor 4 is a regulator of monocyte and electroencephalographic responses to sleep loss. Sleep. 2011;34:1335–1345. doi: 10.5665/SLEEP.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor J.P., Schmidt M.A., Clegern W.C. Evidence for neuroinflammatory and microglial changes in the cerebral response to sleep loss. Sleep. 2011;34:261–272. doi: 10.1093/sleep/34.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Shi G., Mostovoy Y., Gentry N.W., Fan Z., McMahon T.B., Kwok P.-Y., Jones C.R., Ptáček L.J., Fu Y.-H. Mutant neuropeptide S receptor reduces sleep duration with preserved memory consolidation. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Krabbe G., Du F., Jones I., Reichert M.C., Telpoukhovskaia M., Kodama L., Wang C., Cho S., Sayed F., et al. Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.