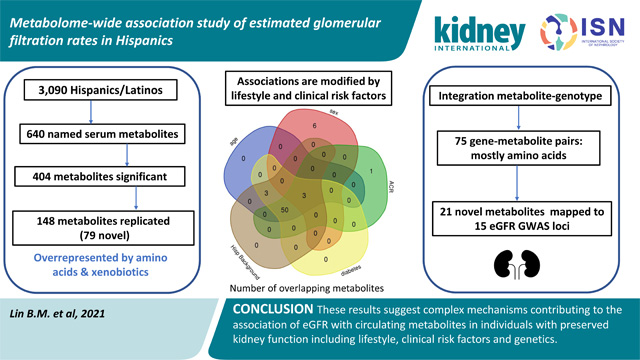
Abstract
Circulating metabolites are by-products of endogenous metabolism or exogenous sources and may inform disease states. Our study aimed to identify the source of variability in the association of metabolites with estimated glomerular filtration rate (eGFR) in Hispanics/Latinos with low chronic kidney disease prevalence and tested the association of 640 metabolites in 3,906 participants of the Hispanic Community Health Study/Study of Latinos. Metabolites were quantified in fasting serum through non-targeted mass spectrometry analysis. eGFR was regressed on inverse normally transformed metabolites in models accounting for study design and covariates. To identify the source of variation on eGFR associations, we tested the interaction of metabolites with lifestyle and clinical risk factors with results integrated with genotypes to identify metabolite genetic regulation. The mean age was 46 years, 43% were men, 22% were current smokers, 47% had a Caribbean Hispanic background, 19% had diabetes and the mean cohort eGFR was 96.4 ml/min/1.73 m2. We identified 404 eGFR-metabolite associations (False Discovery Rate under 0.05). Of these, 69 were previously reported, and 79 were novel associations with eGFR replicated in one or more published studies. There were significant interactions with lifestyle and clinical risk factors, with larger differences in eGFR-metabolite associations within strata of age, urine albumin to creatinine ratio, diabetes and Hispanic/Latino background. Several newly identified metabolites were genetically regulated, and variants were located at genomic regions previously associated with eGFR. Thus, our results suggest complex mechanisms contribute to the association of eGFR with metabolites and provide new insights into these associations.
Keywords: metabolomics, eGFR, risk factors, genetics
Graphical Abstract
Introduction
Circulating metabolites are end products of biological processes from endogenous metabolism (e.g. blood lipids, amino acids, peptides) or exogenous sources (e.g. xenobiotics), which can be altered in disease states such as diabetes and chronic kidney disease (CKD).1 The kidney has important roles in maintaining circulating metabolites levels through glomerular filtration and/or tubular secretion or reabsorption. Studies have shown increased levels of circulating metabolites in CKD2–4 and association of metabolites with estimated glomerular filtration rate (eGFR).5–10 Additional research has shown that some metabolites are extracted or released in the blood as part of intrarenal metabolism such as for renal gluconeogenesis and ammoniagenesis.8, 11 Therefore, kidney function is only one of the mechanisms accounting for circulating metabolites levels.
Studies have shown that circulating metabolites are influenced by lifestyle, the microbiome and genetics.12, 13 These relationships were recently examined in a study that integrated clinical, lifestyle, gut microbiome and genetic data to identify sources of correlation between metabolites based on these features.14 Clinical data (age, sex, anthropometrics, cardiometabolic factors) were better predictors of circulating lipids, amino acids and peptides, whereas the gut microbiome better predicted xenobiotics and unidentified compounds. Circulating metabolites have been shown to vary within clinical disease status such as among individuals with and without diabetes, although studies have included some individuals with CKD.15 Genetic studies have identified genetic regulated metabolites or metabolomic quantitative trait loci (mQTL), including a recent study done in Hispanics/Latinos.16 However, the association of eGFR with circulating metabolites and their interaction with demographic and clinical factors have not been systematically studied, specifically in Hispanics/Latinos, who have a high burden of cardiometabolic conditions.
The main goal of this study is to identify the eGFR-associated metabolites in a population-based cohort of Hispanics/Latinos with low prevalence of CKD. We were also interested in estimating if associations vary within strata of known risk factors including demographics, lifestyle, clinical disease, and Hispanic/Latino background, as these factors are usually not accounted in metabolomic studies. We used data on approximately 4,000 Hispanic Community Health Study/Study of Latinos (HCHS/SOL) participants who were randomly selected for serum metabolomic profiling. Hispanic/Latino participants self-identified by country of origin were grouped as Caribbean background (Cuban, Puerto Rican, Dominican), and Mainland background (Mexican, Central American, and South American). Hispanic/Latino background correlates with proportion of Amerindian and African ancestry in HCHS/SOL, with individuals of Mainland backgrounds having higher Amerindian ancestry and individuals of Caribbean backgrounds having higher African ancestry.17 Given availability of genetic data in HCHS/SOL, we integrated the metabolite-genotype data (metabolomic quantitative trait loci, mQTL) and eGFR-genotype data (genome-wide association study, GWAS) to our findings to identify loci and genes associated with our newly identified eGFR-associated metabolites.16, 18 These analyses provide insights into underlying metabolic pathways related to identified metabolites.
Methods
HCHS/SOL study design and participants
The HCHS/SOL is a longitudinal study of 16,415 Hispanics/Latinos (aged 18–74 years) recruited from households in predefined census-block groups from four US field centers (Chicago, Miami, the Bronx, and San Diego) between 2008 and 201119. Participants self-reported country of origin (Hispanic/Latino background) with Central America (n = 1,730), Cuba (n = 2,348), Dominican Republic (n = 1,460), Mexico (n = 6,471), Puerto Rico (n = 2,728), and South America (n = 1,068). A baseline clinical examination included clinical, behavioral, and sociodemographic assessments, and collection of fasting blood and spot urine samples. The study was approved by the institutional review boards at each field center and the coordinating center, and all subjects provided written informed consent. This study used a subsample of 3,972 participants randomly selected for serum metabolomic profiling, of which 3,906 had complete data on covariates at the same visit that the metabolites were measured (visit 1)(Model 1). There were 17 individuals missing a variable in models adjusted for body mass index (BMI)(Model 2), and within strata of current smoking or diabetes (n=3,889)(Model 3). None of the participants were on renal replacement therapy or had a kidney transplant.
Kidney traits and categories of covariates
The main trait was eGFR, which was estimated using the serum creatinine-based Chronic Kidney Disease Epidemiology Collaboration equation, and a secondary trait was eGFRcys based on cystatin C.20 Serum creatinine was measured using a creatinase enzymatic method traceable to an isotope dilution mass spectrometry (IDMS) reference.21 Serum cystatin C was measured using a turbidimetric method on the Roche Modular P Chemistry Analyzer (Gentian AS, Moss, Norway). Urine albumin (mg/dl) and creatinine (g/dl) were measured using an immunoturbidometric and a creatinase enzymatic method, respectively.
The following categories of covariates/risk factors were used in stratified analyses: age (50 years or older vs <50 years), sex (male vs female), urine albumin to creatinine (ACR, ≥ 30 mg/g vs <30 mg/g), diabetes (defined by a fasting glucose >126 mg/dl or use of diabetes medications), current smoking assessed using a questionnaire (vs past/never smoker) and Hispanic/Latino background. Hispanic/Latino background included self-identified country of origin with additional clustering based on genetic similarity within subgroups using principal components estimated from the genome-wide genotypes (HumanOmni2.5-8v1-1) as previously described.17 Participants were grouped into Caribbean (Puerto Rico, Dominican Republic, Cuba) and Mainland (Central America, South America, Mexico) Hispanic/Latino background.
Metabolite quantification and quality control
Serum metabolites (n=1,136, n=782 named and n=354 unnamed metabolites) were quantified in fasting serum through non-targeted mass spectrometry (MS) analysis using ultra-performance liquid chromatography (UPLC)-MS/MS (DiscoveryHD4™ platform, Metabolon Inc, NC) as previously described.22 Identification and classification of metabolites use a comparison of the ion features in the experimental samples to a reference library of chemical standard entries (e.g., molecular weight (m/z), preferred adducts, in-source fragments and associated MS spectra) and known chemical entities. Peaks were quantified using area-under-the-curve. Raw area counts for each metabolite in each sample were normalized to correct for variation resulting from instrument inter-day tuning differences. To avoid batch effects, samples were randomly allocated across the platform. Duplicate samples (replicas) were used to determine endogenous variability, with representative relative standard deviation of 10% across all biochemicals.
Statistical analyses
Continuous data were presented as the mean ± standard deviation (SD) or median with interquartile range, and categorical variables as number and percentage. Metabolites were inverse normally transformed to approximate a normal distribution. We excluded unknown metabolites and metabolites with 25% or more missing data (amino acids, n=13; carbohydrates, n=2; cofactor a, n=5; lipid, n=23; nucleotide, n=2; peptide, n=3; xenobiotics, n=97). For metabolites with less than 25% missing data, missing values were imputed with the observed minimum value of the metabolite in the sample. This imputation approach optimized the mean squared error in a validation study comparing imputation approaches. We tested pairwise correlation of metabolites using Spearman correlation to identify higher correlated (r>0.70) metabolites. The correlation matrix was displayed using a heatmap. Pathways are defined according to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway metabolism sub-pathways for Homo sapiens (http://www.genome.jp/kegg/pathway.html).
We first examined the main association of eGFR with each metabolite in the overall sample, while adjusting for age, sex, field center and Hispanic/Latino background (Model 1). Additional adjustments for body mass index (BMI, Model 2) did not change results so Model 1 results are reported. We additionally adjusted for diabetes and current smoking in Model 3. We also performed association analyses within pre-specified categories of age, sex, Hispanic/Latino background, diabetes, current smoking and ACR. In a secondary analysis, we tested the association of significantly identified metabolites with eGFRcys. All analyses were performed using a R script and a survey package to account for study design, including strata and sampling weights, given the HCHS/SOL oversampling of subgroups as previously described19. P-values were adjusted for multiple testing using a false discovery rate (FDR) of 0.05, given discovery setting and correlation among some metabolites likely due to their biochemical interrelation as part of interconnected enzymatic reactions and pathways.23 Raw p-values are also shown in supplementary tables.
We replicated associations using publicly available summary statistics of studies of eGFR and metabolites from three published studies that used Metabolon platform like our study.5, 7, 9 This includes studies of Europeans (KORA, n=2,726 discovery sample), a multi-ethnic study of African Americans and European Americans (Bogalusa Heart Study, n=1,202 discovery sample) and a study of African Americans (Atherosclerosis Risk in Communities, n=1,921 discovery sample). We considered replication a p-value <1.2 × 10−4 (Bonferroni correction for 404 metabolites carried forward for replication in these datasets) and a concordant direction of effect in the associations comparing discovery and replication. Some replication studies have used an early version of the Metabolon platform which had limited number of metabolites (n=204 to 493)5, 9 compared to the most recent platform used in this study. Therefore, some metabolites were not available for replication.
We performed several sensitivity analyses. We examined the interaction by obesity using a definition based on a BMI of 30 kg/m2 or more, and tested the associations after removing individuals with an eGFR <60 ml/min/1.73 m2 (n=126, 3% total).
Integration of metabolite-eGFR findings with genotypes. We extracted metabolite-single nucleotide polymorphism (SNP) and locus data from a recently published metabolome-wide association study performed in the HCHS/SOL16 for our newly identified metabolites. We looked up the SNP mQTLs for our newly identified metabolites and the SNPs identified in GWAS of eGFR using data from our metabolome-wide association study16, the Genome Catalog for variants reported in studies of European ancestry, East Asians and trans-ethnic meta-analyses, and our recent GWAS of eGFR in the HCHS/SOL.18
Disclosure statement
The authors have nothing to disclose.
Results
The characteristics of 3,906 participants with complete data on metabolites, eGFR and covariates are shown in Table 1. The mean age was 46 years, 43% were men, 22% were current smokers and 19% had diabetes. Mean BMI was 30 kg/m2, mean eGFR 96.4 ml/min/1.73 m2, and 47% had a Caribbean Hispanic background. Approximately 3% (126 individuals) had CKD based on an eGFR <60 ml/min/1.73 m2, and most of these individuals had mild CKD (Stage G3a). Of 1,136 metabolites available, we excluded those with 25% or more missing values and unnamed metabolites, for a total of 640 metabolites (n=164 amino acids, n=20 carbohydrates, n=20 cofactors, n=10 energy-related, n=292 lipids, n=32 nucleosides, n=32 peptides, n=70 xenobiotics) used in analyses.
Table 1.
Descriptive characteristics of 3,906 HCHS/SOL participants at visit 1.
| Characteristics | HCHS/SOL participants |
|---|---|
| Mean age (SD), years | 45.9 (13.8) |
| Age ≥50 years (vs < 50 years) | 1563 (40.0) |
| Male N (%) | 1670 (42.8) |
| Mean body mass index (SD) kg/m2 | 29.8 (6.1) |
| Current smoker N (%) | 847 (21.7) |
| Caribbean (vs Mainland) Hispanic background N (%) | 1830 (46.9) |
| Diabetes N (%) | 736 (18.8) |
| Mean eGFR (SD), ml/min/1.73 m2 | 96.4 (19.0) |
| eGFR stage G1 | 2595 (66.4) |
| eGFR stage G2 | 1185 (30.3) |
| eGFR stage G3a | 89 (2.9) |
| eGFR stage G3b | 22 (0.6) |
| eGFR stage G4 or G5 | 15 (0.4) |
| Median ACR (interquartiles), mg/g | 6.49 (4.55, 12.12) |
| ACR stage A1 | 3350 (85.8) |
| ACR stage A2 | 319 (8.2) |
| ACR stage A3 | 67 (4.4) |
SD, standard deviation; N, number
N=170 with missing ACR; diabetes defined by a fasting glucose >126 mg/dl or use of medications.
eGFR associated metabolites
We identified 404 metabolites significantly associated with eGFR (FDR <0.05) in models adjusted for age, sex, recruitment center and Hispanic/Latino background (Table S1, Figure 1. When additionally adjusting for diabetes and current smoking, 18 metabolites were no longer significant (Table S1). As expected, creatinine was the most significantly associated metabolite with eGFR and had a larger effect size than other metabolites (Figure S1). Amino acids (p<0.001) and xenobiotics (p=0.02) were more often eGFR associated metabolites, and lipids (p<0.001) less commonly associated with eGFR (Figure 1). Metabolites with lowest p-values were creatinine, 1-methylhistidine, N2,N2-dimethylguanosine, N-acetylcarnosine, N-formylmethionine, N-acetylalanine and N-acetylthreonine, all inversely associated with eGFR (Table S1). Among the significantly associated metabolites, 69 metabolites have been previously described for association with eGFR or CKD including indoxyl sulfate (p=7.7 × 10−20), p-cresol sulfate (p=3.9 × 10−10), hippurate (p=1.3 × 10−4), phenyl acetyl glutamine (p=1.1 × 10−12) and serine (p=4.0 × 10−9) (Table S1). Of significant metabolites newly identified in this study, 79 replicated in one or more published studies. The remaining 256 newly identified metabolites were not available for replication in published studies. Overall, 148 metabolites were previously associated with eGFR or newly associated with replication. Among them, 23 metabolites were highly correlated with at least one metabolite (Figure 1 and Figure 2, Table S2).
Figure 1.
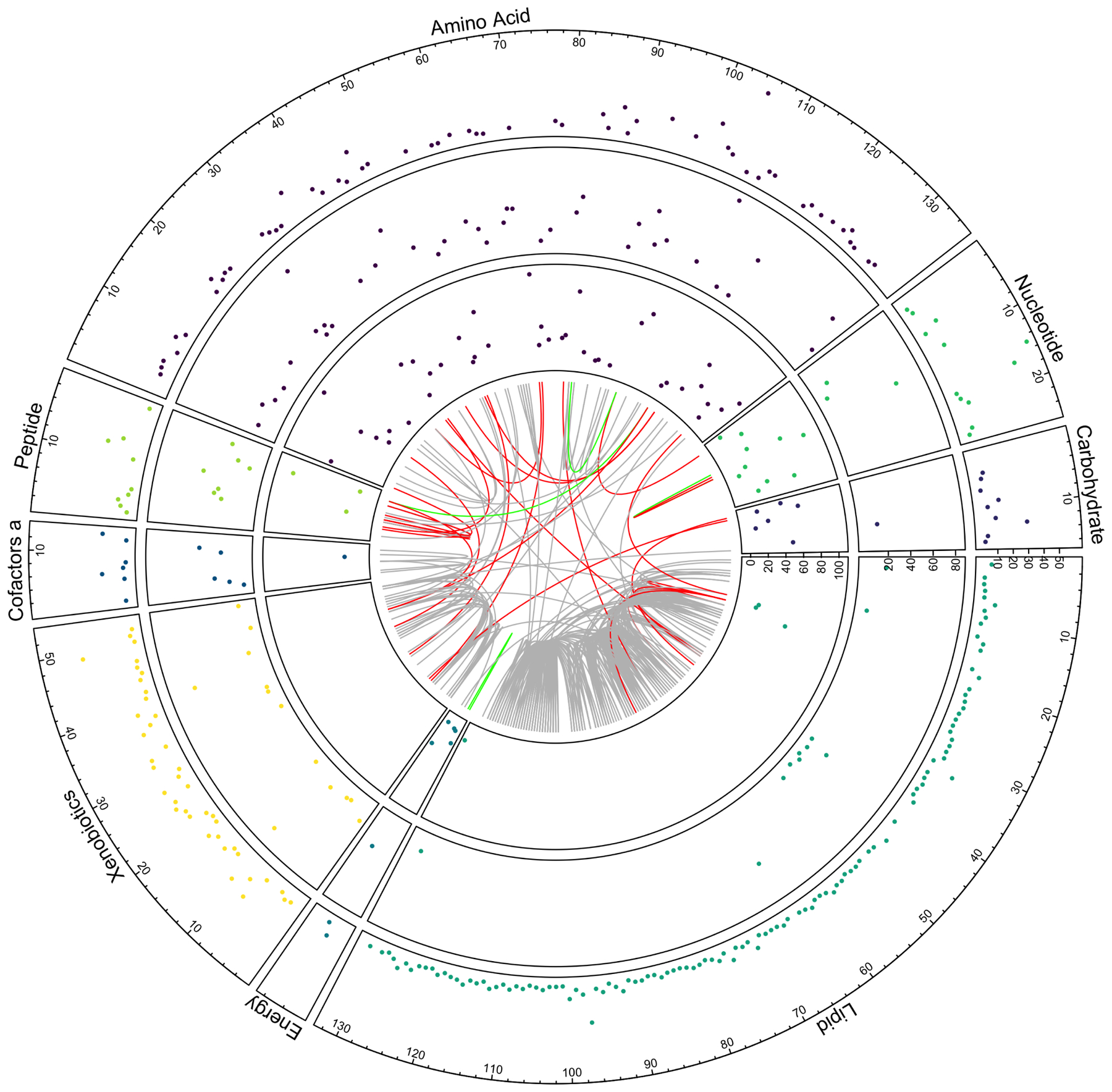
Circus plot showing metabolites significantly associated with eGFR (FDR<0.05) clustered based on super pathways, with novel (outer circle), novel replicated (middle circle) and previously published associated metabolites (inner circle). Each dot is a metabolite p-value, shown as −10 log(p-value). The internal chord diagram represents the pairwise correlations among metabolites for Pearson correlation (r) >0.7 with links related to gray=two newly associated metabolite, red=previously published with newly associated metabolites, green=two previously published metabolites. Note the p-values are lower for previously published metabolites compared to newly identified metabolites.
Figure 2.
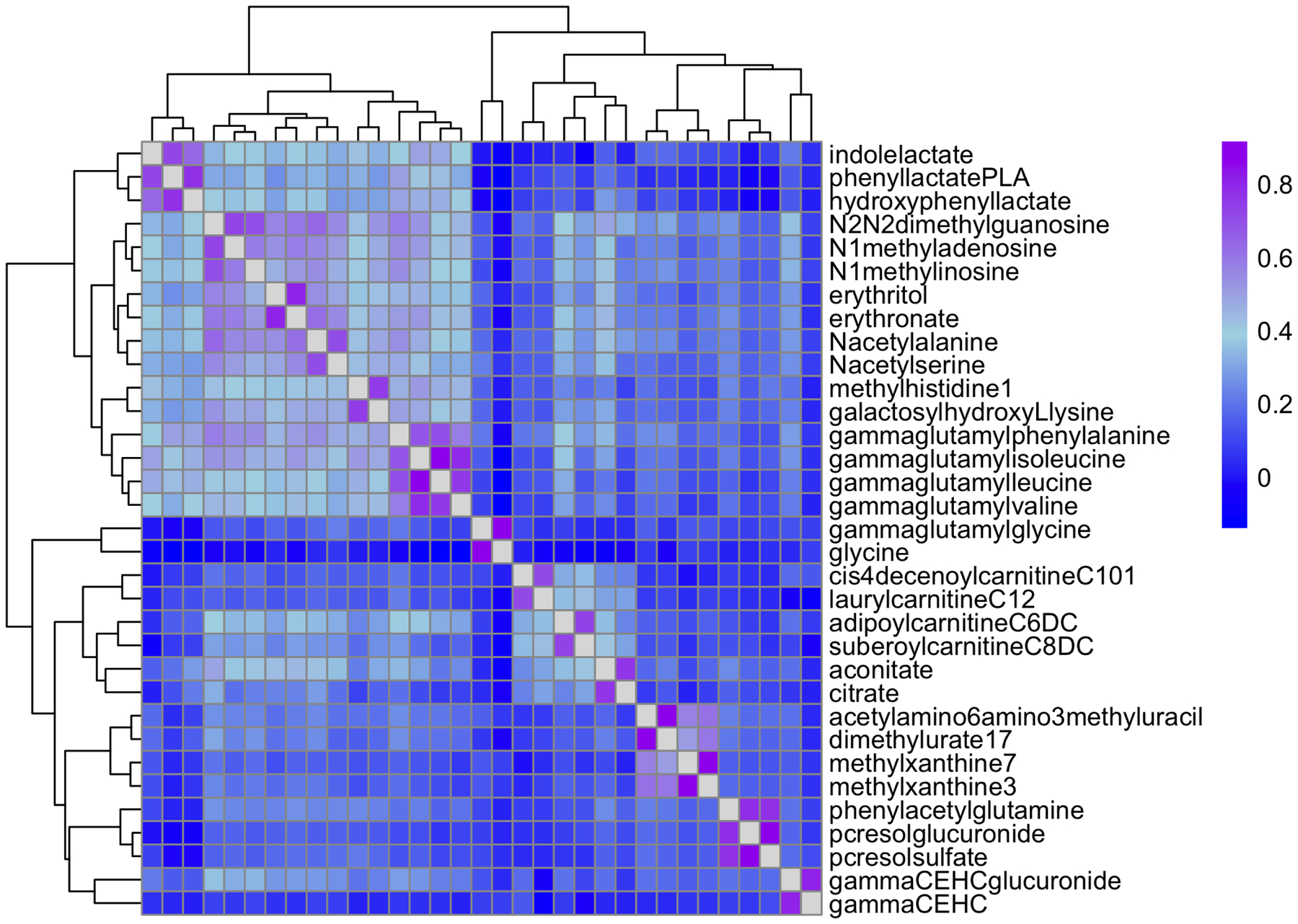
Spearman correlation for metabolites with maximum correlation >0.7 among 148 replicated metabolites (n=69 known and n=79 newly identified).
In a sensitivity analysis using cystatin C-based eGFR equation (eGFRcys), 320 of 404 associated metabolites remained significant including 143 of the 148 metabolites that replicated. Isobutyrylglycine, N-acetylglycine (aminoacids), hippurate, stachydrine, and 4-methylcatechol sulfate (xenobiotics) were not associated with eGFRcys. These metabolites had larger effect estimates in analyses using eGFR compared to eGFRcys (Table S3). Creatinine metabolite was significantly associated with eGFRcys but the effect estimates were smaller compared to models using eGFR based on serum creatinine (beta −0.023 vs −0.042, eGFRcys vs eGFR, respectively) (Table S3, Figure S2). Same was observed for N-acetyl-1-methylhistidine.
In sensitivity analysis excluding individuals with an eGFR <60 ml/min/1.73 m2 (n=126), nine metabolites were no longer significantly associated with eGFR (N-acetylleucine, gamma-CEHC, hexanoylglutamine, 3-hydroxybutyrylcarnitine, 3-hydroxysebacate, 4-methylcatechol sulfate, methylsuccinate, stachydrine, succinate). Two of them overlap with non-significant findings based on eGFRcys (stachydrine, 4-methylcatechol sulfate).
Metabolites associated with eGFR in categories of risk factors
We next examined if the association of metabolites (known and novel replicated, n=148) varied within categories of age, sex, current smoking, diabetes, ACR, BMI and Hispanic/Latino background (Tables S4–S9). Sixty-three metabolites showed a significant interaction within one or more demographic/clinical characteristics. Age, ACR, diabetes and Hispanic/Latino background had the largest number of metabolites showing interactions, and we did not observe any interactions within the current smoker strata (Figure 3) or BMI strata. The estimates varied in the magnitude of effect across strata, with larger effects among older vs younger age, diabetic vs non-diabetics, those with an ACR ≥30 mg/g vs an ACR <30 mg/g, and those with a Mainland Hispanic/Latino background compared to Caribbeans (Figure S3. A–E). None of the metabolites showed differences in the direction of effects across any strata.
Figure 3.
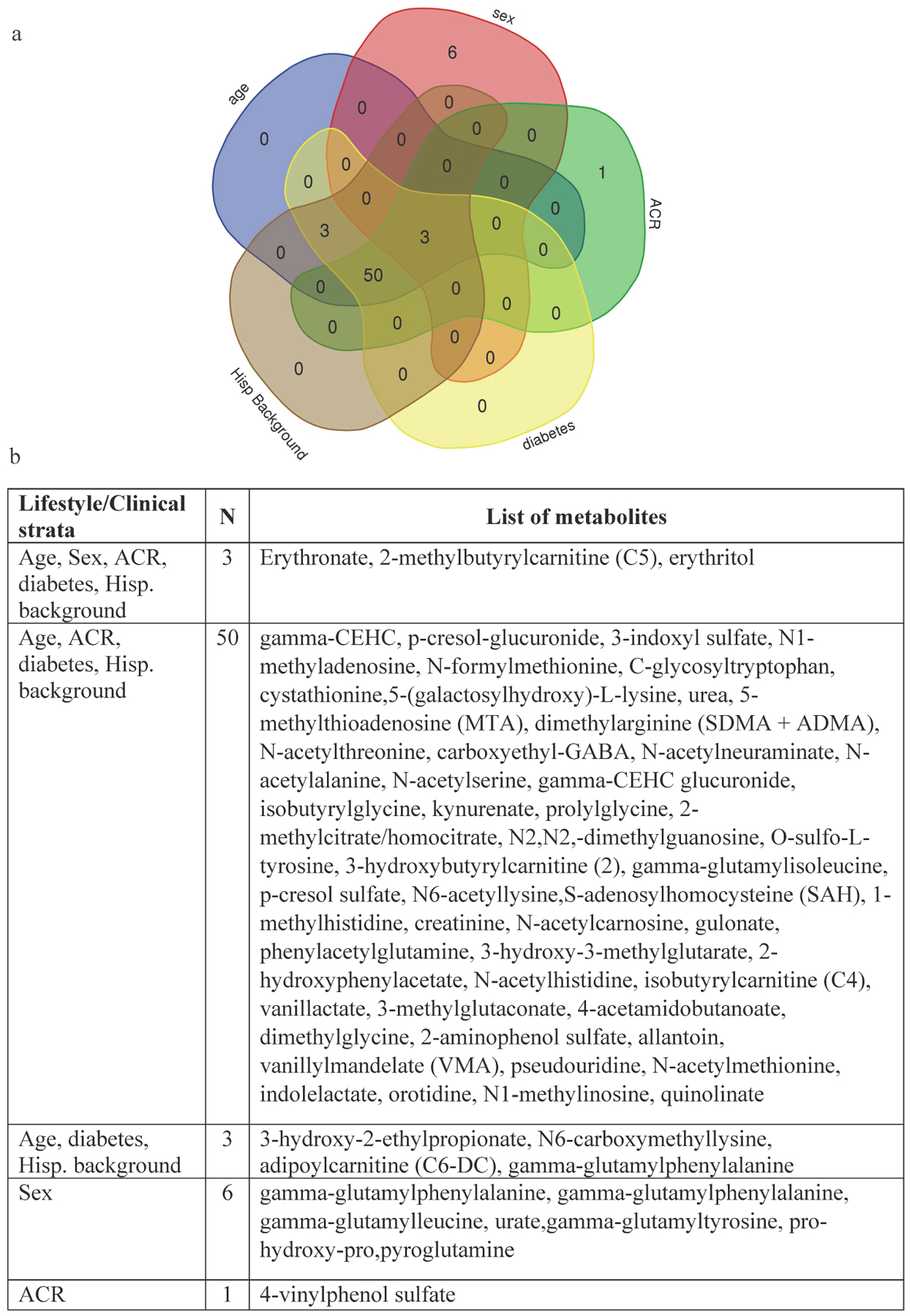
Metabolites showing significant interaction by age, sex, diabetes, ACR and Hispanic/Latino background (n=63). a. Venn diagram showing the number of metabolites identified within and across strata of lifestyle/clinical risk factors and b. List of metabolites identified for one or more lifestyle/clinical risk factors.
Integration of metabolites with genotypes.
To further uncover relationships among eGFR-associated metabolites, we integrated the data with information from mQTLs from a recent genome-wide association study of metabolites performed in HCHS/SOL. Among our 79 newly identified eGFR-associated metabolites, there were 94 SNP-metabolite genome-wide associated pairs (Table 2, Table S10), with 44 metabolites associated with one or more genomic regions within 54 loci (Table S11), for a total of 75 gene/loci-metabolite pairs (Table S12). Most of these metabolites were amino acids (Table S13). Among these genomic regions, 15 metabolite loci have been previously mapped to eGFR GWAS regions (Table S11), including ALMS1/NAT8 (N-acetylhistidine, N-acetylphenylalanine, 2-hydroxyoctanoate, N-acetyl-1-methylhistidine, N-delta-acetylornithine, methionine sulfone), CPS1 (3-methylglutaconate, 3-methylglutarylcarnitine, gamma-glutamylglycine, N-acethylglycine isovalerylglycine, isovalerylglycine) and SLC6A13 (3-aminoisobutyrate, imidazole lactate). Although these genomic regions have been identified in GWAS of eGFR, the metabolites listed have not been previously associated with eGFR (Table 2). The associations at the CPS1 gene were driven by a coding missense variant rs1047891. Among loci not previously identified in GWAS of eGFR were ACADS/SPPL3 (ethylmalonate, isobutyrylglycine, methylsuccinate), GOT2 (imidazole lactate, phenyllactate) and GCH1 (3-methoxytyramine sulfate, dopamine 3-O-sulfate).
Table 2.
Gene-metabolite pairs of newly identified eGFR-associated metabolites at previously reported loci from GWAS of eGFR
| eGFR associated gene/loci | Newly identified eGFR-associated metabolites in this study |
|---|---|
| ABCC1 | cys-gly, oxidized |
| ACSM2A | indoleacetylglutamine |
| ALMS1, NAT8 | N-acetylhistidine, N-acetylphenylalanine, 2-hydroxyoctanoate, N-acetyl-1-methylhistidine, methionine sulfone, N-delta-acetylornithine |
| BC032469, SLC6A19 | methionine sulfone |
| CPS1 | N-acetylglycine, 3-methylglutaconate, 3-methylglutarylcarnitine(2), gamma-glutamylglycine, isobutyrylglycine, isovalerylglycine |
| DMGDH | dimethylglycine |
| DPEP1 | cys-gly-,oxidized |
| FBXL20 | methionine sulfone |
| IGF2R | acisoga |
| LHPP | dihydroorotate |
| PLA2G15 | N6,N6,N6-trimethyllysine |
| SLC22A1 | Acisoga, suberoylcarnitine (C8-DC) |
| SLC25A45 | N6,N6,N6-trimethyllysine |
| SLC6A13 | 3-aminoisobutyrate, imidazole lactate |
| TPRKB | N-acetyl-1-methylhistidine |
Discussion
This study identified and replicated 69 previously reported (including some not validated in the original study) and 79 newly eGFR-associated metabolites in a cohort of Hispanics/Latinos with a low prevalence of CKD (3%), with most of the CKD participants having stage G3a CKD. These findings are not surprising given the role of kidney in filtration, tubular secretion and metabolism of several metabolites. Some metabolites such as indoxyl sulfate, p-cresol sulfate, hippurate and phenyl acetyl glutamine are cleared by renal tubular secretion and have been previously shown to accumulate in CKD.2 Among significant findings, eGFR more often was associated with amino acid and xenobiotic metabolites than lipids, likely due to a key role of the kidney in amino acid metabolism (proximal tubular reabsorption, degradation, synthesis, modification), in addition to detoxification of exogenous compounds.24
Amino acids have important physiological roles in kidneys. For example, some amino acids are substrates for renal gluconeogenesis, and glutamine metabolism in proximal tubule cells is a source of ammonia, an important component in net acid excretion in the kidney.8, 11, 25 The kidney is also a major source for the synthesis of serine, which is involved in methyl group transfer of intermediate substrates in cells.26, 27 Studies examining arterial to renal vein metabolite differences in individuals within normal range of eGFR have shown that the kidney extracts citrulline and releases alanine, leucine, tyrosine, and arginine.11 Rhee et al showed that polar metabolites including amino acids were decreased in the renal vein relative to the aorta, but lipid metabolites were less often changed.8 In our study, eGFR was inversely associated with citrulline, threonine, glutamine and positively associated with serine and lysine. Only citrulline has been previously associated with eGFR in metabolome-wide studies.9 Complex and diverse mechanisms are involved in the regulation of circulating metabolites including kidney metabolism, glomerular filtration and tubular transport, and these factors need to be considered when interpreting associations. A challenge would be to disentangle these relationships while accounting for additional factors such as diet and the microbiome, which were not examined in our study.
This study uncovered substantial interactions in associations of eGFR with metabolites based on lifestyle and clinical risk factors. A larger number of metabolites showed differences in effect estimates within the strata of age, diabetes, ACR and Hispanic/Latino background. Most of the metabolites showing interactions overlap among three or more lifestyle/clinical risk factors. These metabolites were predominantly amino acids and included compounds such as dimethylarginine (SDMA + ADMA), which are inhibitors of nitric oxide, and associate with endothelial dysfunction and disease states28. A small number of metabolites were gender- (i.e., urate, previously described) or ACR-specific interactions. These results support the need to account for lifestyle and clinical diseases in metabolomic studies. In summary, these findings have implications for using serum metabolites as biomarkers of glomerular filtration rate or as predictors for CKD in individuals with preserved renal function.
The integration of our findings with genome-wide association studies has provided insights into the genetic regulation of some metabolites related to eGFR. Some metabolites identified in this study are associated with genetic variants or genes previously described in GWAS of eGFR (Table 2). The missense variant rs1047891 at CPS1 was associated with six newly eGFR-associated metabolites (N-acetylglycine, 3-methylglutaconate, 2-methylglutarylcarnitine, gamma-glutamylglycine, isobutyrylglycine, isovalerylglycine) in addition to the previously reported association with glycine9, 29. This SNP was also the leading variant associated with eGFR in our recent trans-ethnic GWAS meta-analyses of eGFR.30 CPS1 encodes the carbamoyl phosphate synthetase 1, a mitochondrial matrix enzyme that catalyzes the rate-limiting step in the urea cycle, which converts toxic ammonia to urea primarily in liver, although extrahepatic synthesis of urea from arginine or citrulline can occur in kidney and gut.31 Two of our newly identified metabolites (imidazole lactate, phenyllactate) were associated to the GOT2 gene which encodes the mitochondrial glutamic-oxaloacetic transaminase enzyme involved in the urea and tricarboxylic acid cycles. Several other metabolites involved in the urea cycle were significantly associated with eGFR in our data, including citrulline, homocitrulline, N2,N5-diacetylornithine, trans-4-hydroxyproline, N-delta-acetylornithine, urea and argininate (Table S1). The urea cycle generates creatine, which is converted to creatinine in muscles. Our analyses using eGFRcys showed consistent association of these metabolites except for isobutyrylglycine, suggesting that associations may be unrelated to creatinine metabolism.
Additional findings from the gene-metabolite pairs are the associations with cys-gly, oxidized, a metabolite derived from glutathione metabolism. Glutathione mediates detoxification of xenobiotic and endogenous compounds, and has antioxidant functions32. The multi-drug resistance ABCC1 gene encoded protein is an organic anion transporter that mediates efflux of glutathione and glucuronide conjugates from cells33, and the DPEP1 encoded kidney membrane enzyme is involved in the renal metabolism of glutathione in addition to regulate leukotriene activity34.
Among interesting findings, the effect estimate (beta) for the association of eGFR with creatinine metabolite was twice larger in analyses using serum creatinine-based eGFR compared to the cystatin C-based equation, though p-values were similarly low. The association with N-acetyl-1-methylhistidine was also larger for eGFR than eGFRcys, and for five additional metabolites which were associated with eGFR but not eGFRcys, suggesting an overestimation of association effects for some metabolites when using eGFR based on serum creatinine. Among the associated metabolites with eGFR, a subset of metabolites was highly correlated. This includes p-cresol sulfate and p-cresol glucoronide (r=0.91), byproducts of p-cresol which is generated from tyrosine metabolism by anaerobic gut bacteria, and phenylacetylglutamine, derived from the metabolism of phenylalanine by gut microbes and associated with cardiovascular disease.35
This study is the largest study of circulating metabolites associated with eGFR in a population-based study of Hispanics/Latinos. We identified and replicated 148 metabolites including 79 novel associations with eGFR and showed important interaction for some metabolites with lifestyle and clinical risk factors in individuals with preserved kidney function. Our participants had low prevalence of diabetes and were on average obese, although BMI was not an important confounder or modifier in associations. Some of our identified metabolites showed shared genetic regulation based on mQTLs, and shared genetic loci with GWAS of eGFR. This study is limited to untargeted metabolites and a single visit data. Several metabolites identified were not available in replication studies (n=258). They should be examined in future studies using more current metabolomic platforms. Our results suggest complex mechanisms contributing to the variation of circulating metabolites associated with eGFR including lifestyle, clinical risk factors and genetic regulation.
Supplementary Material
Table S1. Association of 404 metabolites with eGFR (FDR<0.05) and replication findings in three published studies (*replication based on p<1.2E-04 and consistent directtion of effects between discovery and replication samples)
Table S2. Pairwise Pearson correlation of 148 associated metabolites that replicated
Table S3. Association of metabolites with eGFRcys for the 404 significantly associated metabolites associated with eGFR based on serum creatinine (in red, metabolites significantly associated with eGFR based on serum creatinine but not eGFRcys)
Table S4. Association of 148 metabolites with eGFR in strata of age (young=<50 years, old: 50 years or older) and interaction p-values
Table S5. Association of 148 metabolites with eGFR in strata of gender and interaction p-values
Table S6. Association of 148 metabolites with eGFR in strata of current smoking and interaction p-values
Table S7. Association of 148 metabolites with eGFR in strata of ACR (<30 mg/g vs 30 mg/g or more) and interaction p-values
Table S8. Association of 148 metabolites with eGFR in strata of diabetes and interaction p-values
Table S9. Association of 148 metabolites with eGFR in strata of Hispanic background (Carib=Caribbean; Main=Mainland) and interaction p-values
Table S10. SNP-metabolite pairs for newly associated metabolites with eGFR (n=79 metabolites queried)
Table S11. Gene/loci associated with newly identified metabolites for eGFR
Table S12. Gene-metabolite pairs identified for newly associated metabolites with eGFR
Table S13. Newly associated metabolites with eGFR and number of genes/loci in mQTL data
Figure S1. Volcano plot showing the association of metabolites with eGFR in models adjusted for age, gender, center and Hispanic Background (n=640 metabolites).
Figure S2. Comparison of betas of associations with metabolites when using eGFR based on serum creatinine and eGFR based on cystatin C. In grey, metabolites not significant in models using eGFRcys.
Figure S3. Comparison of betas across strata of lifestyle and clinical risk factors. For each metabolite results, see Tables S4–S9
Acknowlegdement
The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at Washington University was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) grant DK063491. Support for metabolomics data was graciously provided by the JLH Foundation (Houston, Texas). This study is supported by the following grants: National Institutes of Health DK117445, MD012765, HL140385 and HL123677 to N. Franceschini.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newgard CB. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab 2017; 25: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sirich TL, Funk BA, Plummer NS, et al. Prominent accumulation in hemodialysis patients of solutes normally cleared by tubular secretion. J Am Soc Nephrol 2014; 25: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubin RF, Rhee EP. Proteomics and Metabolomics in Kidney Disease, including Insights into Etiology, Treatment, and Prevention. Clin J Am Soc Nephrol 2020; 15: 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwan B, Fuhrer T, Zhang J, et al. Metabolomic Markers of Kidney Function Decline in Patients With Diabetes: Evidence From the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis 2020; 76: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekula P, Goek ON, Quaye L, et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J Am Soc Nephrol 2016; 27: 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goek ON, Doring A, Gieger C, et al. Serum metabolite concentrations and decreased GFR in the general population. Am J Kidney Dis 2012; 60: 197–206. [DOI] [PubMed] [Google Scholar]
- 7.Nierenberg JL, He J, Li C, et al. Novel associations between blood metabolites and kidney function among Bogalusa Heart Study and Multi-Ethnic Study of Atherosclerosis participants. Metabolomics 2019; 15: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee EP, Clish CB, Ghorbani A, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 2013; 24: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu B, Zheng Y, Nettleton JA, et al. Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 2014; 9: 1410–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Titan SM, Venturini G, Padilha K, et al. Metabolites related to eGFR: Evaluation of candidate molecules for GFR estimation using untargeted metabolomics. Clin Chim Acta 2019; 489: 242–248. [DOI] [PubMed] [Google Scholar]
- 11.Tizianello A, De Ferrari G, Garibotto G, et al. Renal metabolism of amino acids and ammonia in subjects with normal renal function and in patients with chronic renal insufficiency. J Clin Invest 1980; 65: 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet 2014; 46: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu T, Holzapfel C, Dong X, et al. Effects of smoking and smoking cessation on human serum metabolite profile: results from the KORA cohort study. BMC Med 2013; 11: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar N, Korem T, Weissbrod O, et al. A reference map of potential determinants for the human serum metabolome. Nature 2020; 588: 135–140. [DOI] [PubMed] [Google Scholar]
- 15.Barrios C, Zierer J, Wurtz P, et al. Circulating metabolic biomarkers of renal function in diabetic and non-diabetic populations. Sci Rep 2018; 8: 15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feofanova EV, Chen H, Dai Y, et al. A Genome-wide Association Study Discovers 46 Loci of the Human Metabolome in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2020; 107: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conomos MP, Laurie CA, Stilp AM, et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2016; 98: 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian H, Kowalski MH, Kramer HJ, et al. Genome-Wide Association of Kidney Traits in Hispanics/Latinos Using Dense Imputed Whole-Genome Sequencing Data: The Hispanic Community Health Study/Study of Latinos. Circ Genom Precis Med 2020; 13: e002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorlie PD, Aviles-Santa LM, Wassertheil-Smoller S, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 2010; 20: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inker LA, Schmid CH, Tighiouart H, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012; 367: 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thyagarajan B, Howard AG, Durazo-Arvizu R, et al. Analytical and biological variability in biomarker measurement in the Hispanic Community Health Study/Study of Latinos. Clin Chim Acta 2016; 463: 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans A, Bridgewater B, Liu Q, et al. : High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. 2014. Preprint avaialble at: https://www.hilarispublisher.com/open-access/high-resolution-mass-spectrometry-improves-data-quantity-and-qualityas-compared-to-unit-mass-resolution-mass-spectrometry-in-hight-2153-0769-1000132.pdf.
- 23.Vinaixa M, Samino S, Saez I, et al. A Guideline to Univariate Statistical Analysis for LC/MS-Based Untargeted Metabolomics-Derived Data. Metabolites 2012; 2: 775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young GA. Amino acids and the kidney. Amino Acids 1991; 1: 183–192. [DOI] [PubMed] [Google Scholar]
- 25.Weiner ID, Verlander JW. Renal ammonia metabolism and transport. Compr Physiol 2013; 3: 201–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosnan JT, Hall B. Renal serine production in vivo: effects of dietary manipulation of serine status. Can J Physiol Pharmacol 1989; 67: 1058–1061. [DOI] [PubMed] [Google Scholar]
- 27.Lowry M, Hall DE, Hall MS, et al. Renal metabolism of amino acids in vivo: studies on serine and glycine fluxes. Am J Physiol 1987; 252: F304–309. [DOI] [PubMed] [Google Scholar]
- 28.Tain YL, Hsu CN. Toxic Dimethylarginines: Asymmetric Dimethylarginine (ADMA) and Symmetric Dimethylarginine (SDMA). Toxins (Basel) 2017; 9. [Google Scholar]
- 29.Suhre K, Shin SY, Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011; 477: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AP, Le TH, Wu H, et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun 2019; 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Poll MC, Soeters PB, Deutz NE, et al. Renal metabolism of amino acids: its role in interorgan amino acid exchange. Am J Clin Nutr 2004; 79: 185–197. [DOI] [PubMed] [Google Scholar]
- 32.Lu SC. Glutathione synthesis. Biochim Biophys Acta 2013; 1830: 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole SP. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J Biol Chem 2014; 289: 30880–30888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa H, Inazawa J, Inoue K, et al. Assignment of the human renal dipeptidase gene (DPEP1) to band q24 of chromosome 16. Cytogenet Cell Genet 1992; 59: 258–260. [DOI] [PubMed] [Google Scholar]
- 35.Witkowski M, Weeks TL, Hazen SL. Gut Microbiota and Cardiovascular Disease. Circ Res 2020; 127: 553–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association of 404 metabolites with eGFR (FDR<0.05) and replication findings in three published studies (*replication based on p<1.2E-04 and consistent directtion of effects between discovery and replication samples)
Table S2. Pairwise Pearson correlation of 148 associated metabolites that replicated
Table S3. Association of metabolites with eGFRcys for the 404 significantly associated metabolites associated with eGFR based on serum creatinine (in red, metabolites significantly associated with eGFR based on serum creatinine but not eGFRcys)
Table S4. Association of 148 metabolites with eGFR in strata of age (young=<50 years, old: 50 years or older) and interaction p-values
Table S5. Association of 148 metabolites with eGFR in strata of gender and interaction p-values
Table S6. Association of 148 metabolites with eGFR in strata of current smoking and interaction p-values
Table S7. Association of 148 metabolites with eGFR in strata of ACR (<30 mg/g vs 30 mg/g or more) and interaction p-values
Table S8. Association of 148 metabolites with eGFR in strata of diabetes and interaction p-values
Table S9. Association of 148 metabolites with eGFR in strata of Hispanic background (Carib=Caribbean; Main=Mainland) and interaction p-values
Table S10. SNP-metabolite pairs for newly associated metabolites with eGFR (n=79 metabolites queried)
Table S11. Gene/loci associated with newly identified metabolites for eGFR
Table S12. Gene-metabolite pairs identified for newly associated metabolites with eGFR
Table S13. Newly associated metabolites with eGFR and number of genes/loci in mQTL data
Figure S1. Volcano plot showing the association of metabolites with eGFR in models adjusted for age, gender, center and Hispanic Background (n=640 metabolites).
Figure S2. Comparison of betas of associations with metabolites when using eGFR based on serum creatinine and eGFR based on cystatin C. In grey, metabolites not significant in models using eGFRcys.
Figure S3. Comparison of betas across strata of lifestyle and clinical risk factors. For each metabolite results, see Tables S4–S9


