Abstract
Identification of new genetic markers may improve the prediction of colorectal cancer prognosis. Our objective was to examine genome-wide associations of germline genetic variants with disease-specific survival in an analysis of 16,964 cases of colorectal cancer. We analyzed genotype and colorectal cancer-specific survival data from a consortium of 15 studies. Approximately 7.5 million SNPs were examined under the log-additive model using Cox proportional hazards models, adjusting for clinical factors and principal components. Additionally, we ran secondary analyses stratifying by tumor site and disease stage. We used a genome-wide p-value threshold of 5 × 10–8 to assess statistical significance. No variants were statistically significantly associated with disease-specific survival in the full case analysis or in the stage-stratified analyses. Three SNPs were statistically significantly associated with disease-specific survival for cases with tumors located in the distal colon (rs698022, HR = 1.48, CI 1.30–1.69, p = 8.47 × 10–9) and the proximal colon (rs189655236, HR = 2.14, 95% CI 1.65–2.77, p = 9.19 × 10–9 and rs144717887, HR = 2.01, 95% CI 1.57–2.58, p = 3.14 × 10–8), whereas no associations were detected for rectal tumors. Findings from this large genome-wide association study highlight the potential for anatomical-site-stratified genome-wide studies to identify germline genetic risk variants associated with colorectal cancer-specific survival. Larger sample sizes and further replication efforts are needed to more fully interpret these findings.
Subject terms: Genetic association study, Cancer genetics, Colorectal cancer
Introduction
The global incidence of colorectal cancer (CRC) has been increasing while the mortality rate has been decreasing1,2. Advances in scientific knowledge, treatment modalities, and medical screening programs are considered among the major factors contributing to improved survival from this disease1,3,4. In the USA, Canada, Australia, and Europe, 5-year survival is around 65%5–8.
Currently, CRC prognostication relies primarily on clinicopathological features with a primary focus on tumor characteristics, such as stage. Several additional factors, in relation to both the tumor (e.g. KRAS and BRAF mutations) and the individual, have been associated with survival times and clinical outcomes9–18. Germline genetic variants are commonly investigated as candidate prognostic markers; they are abundant in the human genome, are polymorphic among patients, are thought to remain unchanged over time, and may biologically modify disease characteristics and risk of progression or clinical outcomes19–21. These characteristics of germline genetic variants, therefore, make them attractive for cancer research studies.
There is now a large body of research on genetic variants in relation to CRC incidence, yet no variants have been confidently associated with CRC survival or used in clinical practice. Many approaches have been attempted in survival outcomes studies, such as candidate SNP, gene, or pathway analyses22,23, including the examination of associations with patient outcomes for variants identified in susceptibility studies. Compared with other study designs, genome-wide association studies (GWAS) offer a comprehensive, agnostic approach. Some CRC survival GWAS have been performed24–27, which have identified a small number of genetic variants at the genome-wide significance level25,26. While these studies have advanced the knowledge of the genetic basis of CRC survival, they have also been limited by relatively small number of cases, restricting the ability to identify modest associations or low frequency risk variants, which may only be apparent when large case cohorts are examined.
In this study, we evaluated germline genetic loci associated with CRC-specific survival using data from 16,964 CRC participants included in an international consortium comprising 15 epidemiologic and clinical studies. As a secondary goal, we evaluated stage- and tumor site-specific associations between genetic loci and CRC survival.
Results
Participant demographic and clinical characteristics are provided in Table 1. Median follow-up time after diagnosis was 13.8 years. Overall, 6,033 (36%) CRC cases died during follow up, of which 4,010 deaths (66%) were attributed to CRC. As expected, participants with stage 4 tumors at diagnosis were more likely to die from CRC (of those who died, 51% were stage 4 compared with 6% stage 1). Participants were ~ 50% female with a median age of 67 years (range 20–94 years).
Table 1.
Demographics and tumor characteristics of 16,964 colorectal cancer patients.
| Overall (n = 16,964) | Died of CRC (n = 4,010) | Did not die of CRC (n = 12,954) | ||||
|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | |
| Age at diagnosis (median (range)) | 67 | (20–94) | 67 | (24–94) | 67 | (20–94) |
| Age category | ||||||
| < 50 | 984 | (5.8) | 263 | (6.6) | 721 | (5.6) |
| 50–60 | 2,751 | (16.2) | 705 | (17.6) | 2,046 | (15.8) |
| 60–70 | 6,520 | (38.4) | 1,435 | (35.8) | 5,085 | (39.3) |
| > 70 | 6,709 | (39.5) | 1,607 | (40.1) | 5,102 | (39.4) |
| All-cause deaths | 6,033 | (35.6) | 4,010 | (100.0) | 2,023 | (15.6) |
| CRC survival, years (median (IQR)) | 5.5 | (3.40–9.77) | – | – | – | – |
| Overall survival, years (median (IQR)) | 6.1 | (3.87–11.24) | – | – | – | – |
| Male Sex | 8,528 | (50.3) | 2,045 | (51.0) | 6,483 | (50.0) |
| Stage | ||||||
| Stage 1 or local | 3,338 | (19.7) | 157 | (3.9) | 3,181 | (24.6) |
| Stage 2/3 or regional | 6,420 | (37.8) | 1,209 | (30.1) | 5,211 | (40.2) |
| Stage 4 or distant | 1,847 | (10.9) | 1,448 | (36.1) | 399 | (3.1) |
| Missing | 5,359 | (31.6) | 1,196 | (29.8) | 4,163 | (32.1) |
| Tumor location | ||||||
| Proximal | 6,214 | (36.6) | 1,433 | (35.7) | 4,781 | (36.9) |
| Distal | 4,881 | (28.8) | 978 | (24.4) | 3,903 | (30.1) |
| Rectal | 4,749 | (28.0) | 1,045 | (26.1) | 3,704 | (28.6) |
| Missing | 1,120 | (6.6) | 544 | (13.8) | 566 | (4.4) |
| Study^ | ||||||
| CCFR | 2,446 | (14.4) | 538 | (13.4) | 1,908 | (14.7) |
| CPSII | 819 | (4.8) | 186 | (4.6) | 633 | (4.9) |
| DACHS | 2,659 | (15.7) | 537 | (13.4) | 2,122 | (16.4) |
| DALS | 1,098 | (6.5) | 210 | (5.2) | 888 | (6.9) |
| EDRN | 191 | (1.1) | 14 | (0.3) | 177 | (1.4) |
| EPIC | 1,821 | (10.7) | 471 | (11.7) | 1,350 | (10.4) |
| HPFS | 348 | (2.1) | 85 | (2.1) | 263 | (2.0) |
| MCCS | 750 | (4.4) | 194 | (4.8) | 556 | (4.3) |
| N9741 | 426 | (2.5) | 366 | (9.1) | 60 | (0.5) |
| NHS | 591 | (3.5) | 161 | (4.0) | 430 | (3.3) |
| PHS | 323 | (1.9) | 130 | (3.2) | 193 | (1.5) |
| PLCO | 972 | (5.7) | 174 | (4.3) | 798 | (6.2) |
| UKB | 2,919 | (17.2) | 581 | (14.5) | 2,338 | (18.0) |
| VITAL | 270 | (1.6) | 67 | (1.7) | 203 | (1.6) |
| WHI | 1,331 | (7.8) | 296 | (7.4) | 1,035 | (8.0) |
^CRC Colorectal Cancer, CCFR Colon Cancer Family Registry, CPSII Cancer Prevention Study II, DACHS Darmkrebs: Chancen der Verhütung durch Screening Study, DALS Diet, Activity and Lifestyle Study, EDRN Early Detection Research Network, EPIC European Prospective Investigation into Cancer, HPFS Health Professionals Follow-up Study, IQR interquartile range, MCCS Melbourne Collaborative Cohort Study, NHS Nurses’ Health Study, PHS Physicians' Health Study, PLCO Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial, UKB UK Biobank, VITAL Vitamins and Lifestyle, WHI Women’s Health Initiative.
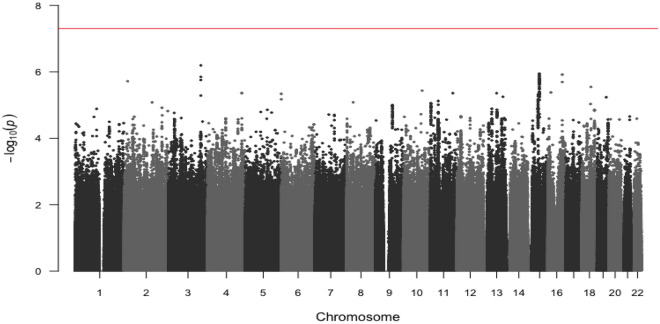
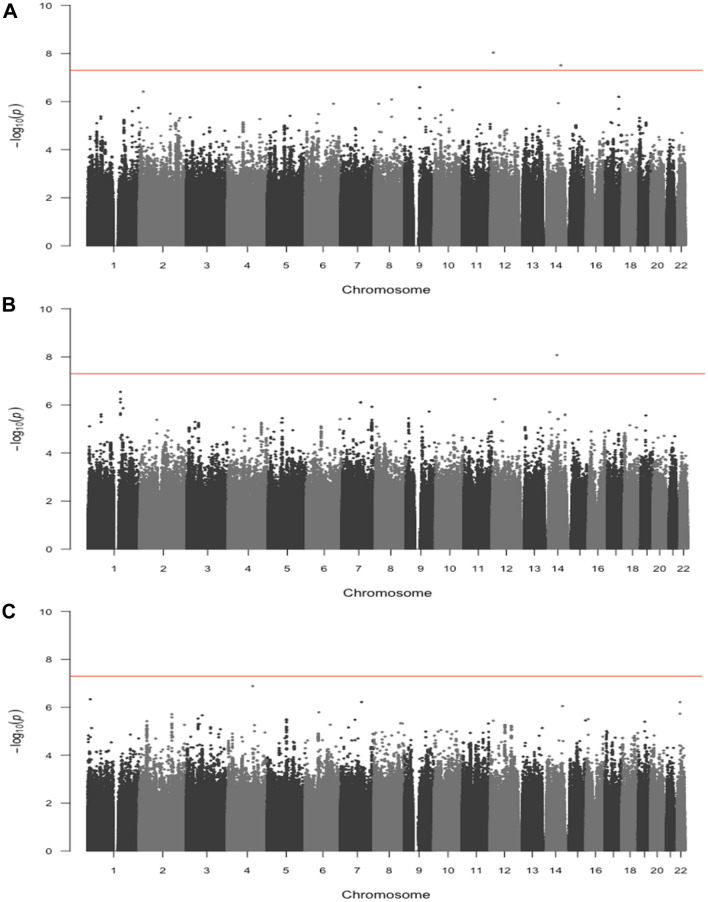
No substantial systemic inflation was identified from quantile-quantile (QQ) plots (Supplementary Fig. 1). No variants reached genome-wide significance for the primary GWAS (Fig. 1) or for the stage-stratified analysis (Supplementary Fig. 2, Supplementary Table 3). Two variants were statistically significant at a genome-wide P-value threshold among proximal colon tumors and one among distal colon tumors (Table 2, Fig. 2). No variants reached genome-wide significance among rectal tumors. The significant variants identified among proximal colon tumors were located on chromosomes 12 (rs189655236, hazard ratio [HR] = 2.14, 95% confidence interval [CI]: 1.65–2.77, p = 9.19 × 10–9) and 14 (rs144717887, HR = 2.01, 95% CI: 1.57–2.58, p = 3.14 × 10–8). Both variants were low frequency (MAF 1.4% and 1.5%, respectively) and neither were in linkage disequilibrium (LD; defined as R2 > 0.6) with nearby variants. The rs189655236 variant was located within the intronic region of BORCS5 and rs144717887 was located in an intergenic region. The variant significantly associated with CRC survival among distal colon tumors was located in an intergenic region on chromosome 14 (rs698022, HR = 1.48, CI 1.30–1.69, p = 8.47 × 10–9), was common with a minor allele frequency (MAF) of 11%, and was not in LD (R2 < 0.6) with nearby variants. The two chromosome 14 variants identified in proximal and distal colon analyses were not in linkage disequilibrium with each other. None of the statistically significant variants were predicted to have regulatory effects (ranking scores = 4 or 5) as reported in the RegulomeDB database.
Figure 1.
Manhattan plot of -log10 p-values by genomic position for the genome-wide analysis of colorectal cancer survival in 16,964 cases. The red line indicates genome-wide significance threshold (p = 5 × 10–8).
Table 2.
Variants associated with colorectal cancer survival at P < 5 × 10–8, stratified by tumor site.
| Chromosome | Variant rsID | Alleles (risk/alternative) | RAF | RegulomeDB rank | HRa | 95% CI | P-value | Imputation quality (info score) |
|---|---|---|---|---|---|---|---|---|
| Proximal colon | ||||||||
| 12 | rs189655236 | C/T | 0.014 | 5 | 2.14 | (1.65, 2.77) | 9.19 × 10–09 | 0.85 |
| 14 | rs144717887 | G/A | 0.015 | 5 | 2.01 | (1.57, 2.58) | 3.14 × 10–08 | 0.93 |
| Distal colon | ||||||||
| 14 | rs698022 | C/T | 0.111 | 4 | 1.48 | (1.30, 1.69) | 8.47 × 10–09 | 0.93 |
Proximal colon tumor-specific analyses included 6,214 cases and distal colon tumor-specific analyses included 4,881 cases.
RAF Risk Allele Frequency, HR Hazard Ratio, CI Confidence Interval. aAdjusted for age at diagnosis, sex, genotyping batch/study, and the first five principal components of genetic ancestry.
Figure 2.
Manhattan plots of -log10 p-values by genomic position for the genome-wide analysis of colorectal cancer survival stratified by tumor site. (A) proximal colon tumor-specific in 6,214 cases, (B) distal colon tumor-specific in 4,881 cases, (C) rectal tumor-specific in 4,749 cases. The red line indicates genome-wide significance threshold (p = 5 × 10–8).
Discussion
In this analysis of common genetic variants in a sizeable study population, we did not identify any SNPs associated with CRC prognosis at the genome-wide significance level. We also found no SNPs associated with survival for specific tumor stages. However, our results suggest that there may be variants that predict CRC-survival for distal and proximal colon cancer cases.
The distal and proximal regions of the colon differ biologically and in terms of tumor incidence rates28,29. In addition, research shows that tumors located in these anatomical subsites display differences in molecular alterations involved in tumorigenesis, and are characterized by different disease progression and prognosis30–32. The identification of different sets of variants with survival for cases with distal and proximal colon tumors in this study is, therefore, not surprising. Using eQTLGen, rs189655236 was predicted to be in cis-eQTL with DUSP16, which has been associated with chemotherapy resistance in colorectal cancer33. However, none of the identified SNPs were predicted to have putative regulatory functions using RegulomeDB, and according to the dbSNP database, they are within intronic or intergenic sequences34. Also, the two significant variants for proximal colon tumors are very low frequency. These considerations lead us to interpret our findings with caution pending further investigation.
This study has several strengths. By leveraging data from 15 population-based and clinical studies, we were able to confidently examine with good statistical power genetic associations with CRC survival. Covariates were well characterized with detailed information on epidemiologic and clinical factors which allowed us to conduct subgroup analysis by stage at diagnosis and tumor anatomical location. In addition, we had a relatively long follow-up period and cause of deaths were uniformly ascertained. We also used an agnostic discovery-based approach to identify variants associated with CRC survival.
Our study also has some limitations. Although we had a large enough sample size to identify significant SNPs in CRC cases with European ancestry, we were unable to evaluate other ancestry groups. Additionally, we were unable to evaluate other tumor markers that might be associated with survival in our population. Another limitation inherent to the GWAS approach is the high likelihood of false-negative findings due to the stringent P-value threshold for genome-wide significance. This threshold is set to account for multiple testing and is designed to reduce the number of false-positive findings; however, a consequence of this stringency is that some important SNP-survival associations may have been missed. Finally, no replication analysis or functional follow-up was conducted.
In summary, in this largest yet GWAS for CRC specific survival, our analyses indicate that genetic variants in the form of SNPs are unlikely to explain variable risk of death from colorectal cancer in people of European ancestry. However, a few SNPs were identified that may be prognostic markers for distal or proximal colon cancers and these should be further examined in other populations, including cases from other ancestry groups.
Methods
Study population
Analyses utilized data from the International Survival Analysis in Colorectal Cancer Consortium (ISACC), a compilation of participants with incident, invasive CRC obtained from clinical trials, case–control, and cohort studies from around the world. Study participants included people of European genetic ancestry diagnosed with invasive CRC and with available genotyping and CRC-specific survival data (as described in the Supplementary Methods). The following 15 ISACC studies were included: the Cancer Prevention Study-II (CPS-II)35, the German Darmkrebs: Chancen der Verhutung durch Screening Study (DACHS)36, the Diet Activity and Lifestyle Study (DALS)37, the Early Detection Research Network (EDRN)38, the European Prospective Investigation into Cancer (EPIC)39, the Health Professionals Follow-up Study (HPFS)40, the Melbourne Collaborative Cohort Study (MCCS)41, the Nurses’ Health Study (NHS)42,43, the N9741 clinical trial44, the Physician’s Health Study (PHS)45,46, the Prostate, Lung, Colorectal, and Ovarian Study (PLCO)47,48, the UK Biobank (UKB)49, the VITamins And Lifestyle Study (VITAL)50, the Women’s Health Initiative (WHI)51,52, and four Colon Cancer Family Registry (CCFR) sites53,54: Seattle, Ontario, Australia, and the Mayo Clinic. Study-specific details are described in the Supplementary Tables 1 and 2.
Ethical considerations
Study protocols were approved by the Institutional Review Board or Independent Ethics Committee overseeing the respective clinical sites. Participants provided informed consent for genetic testing and research participation. The study protocol has been approved by Fred Hutchinson Cancer Research Center Institutional Review Board. All methods were performed in accordance with the relevant guidelines and regulations.
Ascertainment of CRC-specific survival
Protocols for survival outcomes assessment in this study population have been described previously35–38,43–45,49,50,53,55–61. Briefly, studies ascertained vital status via linkage to the National Death Index, state cancer registries, state death records, or population registers with cause of death verified by death certificates (CPSII, DACHS, DALS, EPIC, MCCS, UKB, VITAL), or via active follow-up (CCFR, HPFS, NHS, PHS, PLCO, WHI, N9741) with dates and cause of death confirmed via regional mortality databases, review of death certificates and/or medical records by trained adjudicators. In all studies, cases alive at the most recent study follow-up or data linkage were censored on that date. In VITAL, individuals who moved outside of Washington State were censored at their date of move. CRC-specific survival was calculated as days from diagnosis to CRC-related death or end of follow-up. Individuals who died from causes other than CRC were censored at the time of death.
Tumor stage and location classification
Tumor stage was obtained from pathology and registry reports at the time of diagnosis. The Surveillance, Epidemiology, and End Results (SEER) summary stage categorizations of localized, regional, and distant were used, also incorporating extent of disease information when available. Additionally, the American Joint Committee on Cancer (AJCC) TNM classification of malignant tumors (TNM) categorizations were utilized to assign values I through IV.
Tumor location was obtained from registry and pathology reports. Location was grouped based on ICD-9 codes as follows: (1) “Proximal” (153.0/Hepatic flexure, 153.1/Transverse colon, 153.4/Cecum, 153.6/Ascending colon), (2) “Distal” (153.2/Descending colon, 152.3/Sigmoid colon, 153.7/Splenic flexure), 3) “Rectal” (154.0/Rectosigmoid junction, 154.1/Rectum).
Genotype data
Genotyping methods have been reported previously62–66. Briefly, genomic DNA was extracted from blood or buccal samples using conventional methods, and samples were genotyped using the platforms listed in Supplementary Table 1. Each genotyping platform dataset underwent standard quality control analyses, including exclusion of samples and SNPs with low call rates (< 97% and < 98%, respectively), exclusion of variants departing from Hardy–Weinberg Equilibrium (p < 1 × 10–4), exclusion of individual with discrepant reported and genotyped sex based on X chromosome heterozygosity, and exclusion of duplicates and individuals that were second-degree or more closely related based on identity by descent (IBD) calculations. Additionally, we inferred genetic ancestry using principal components analysis and excluded individuals of non-European ancestry from analyses due to small sample sizes. Participants with a value within one standard deviation of the median for the first and second eigenvectors were categorized as European genetic ancestry and included in the analysis (Supplementary Methods). A total of 16,964 individuals passed quality control filtering. Only variants passing quality control analyses and with missing call rates ≤ 2% were used for imputation.
Phasing and imputation were performed on each pooled set of studies with the same or similar genotyping platforms. Autosomal variants were phased using SHAPEIT2 and imputed to the Haplotype Reference Consortium panel release 1.1 (~ 39 million variants) using the University of Michigan Imputation Server67–69. Genotype probabilities were converted to allelic dosages. Evaluation was restricted to variants with MAF ≥ 1% and imputation accuracy R2 > 0.3. A total of 7,829,749 genetic variants were included in the analyses. All imputed and cleaned individual-level genotype data were pooled for survival analyses.
We used PLINK (v1.9) for principal components analysis on pruned sets of autosomal variants obtained by removing regions with extensive long-range linkage disequilibrium. The first five principal components were used as coviariates to account for population substructure in analysis.
Statistical analysis
We used Cox proportional hazards regression to estimate HRs and 95% CIs for associations of each genetic variant with CRC-specific survival. A log-additive model was used, relating variant genotype dosage to CRC-specific survival. All models were adjusted for age at diagnosis, sex, a categorical variable encompassing genotyping platform and study, and five principal components to account for population substructure. The proportional hazards assumptions for age and sex were evaluated by testing for a non-zero slope of the scaled Schoenfeld residuals on ranked failure time70. The tests for both age and sex were statistically significant (p < 0.05), suggesting the proportionality assumption may not hold. For age, the non-proportionality was resolved by including both a continuous and categorical variable (dichotomized at median age; ≤ 57 years versus > 57 years). We stratified our analysis by sex.
As a secondary goal, we evaluated the association of genetic loci and CRC survival stratified by tumor stage at diagnosis and anatomical location. Tumor stage strata included regional (stages 2 and 3) and distant metastatic (stage 4) disease; local (stage 1) disease was not evaluated due to a low percentage of deaths among this group. Tumor anatomical location was grouped as “proximal colon” (ICD-9-CM 153.0/Hepatic flexure, 153.1/Transverse colon, 153.4/Cecum, 153.6/Ascending colon), “distal colon” (153.2/Descending colon, 152.3/Sigmoid colon, 153.7/Splenic flexure), or “rectum” (154.0/Rectosigmoid junction, 154.1/Rectum).
We evaluated QQ plots of log-transformed p-values and calculated genomic control coefficients to assess for possible systemic inflation. We produced Manhattan plots and specified a genome-wide statistical significance level of p ≤ 5 × 10–8. We performed statistical analyses using R version 3.5.2.
In silico analyses
The NCI ‘LDassoc’ web tool (https://ldlink.nci.nih.gov/) was used to evaluate LD (defined as R2 > 0.6) in 1000 Genomes Phase 3 ‘EUR’ population) for SNPs of interest71. The putative functional effects of variants were inferred based on information in the RegulomeDB database72. This database ranks variants ranging from 1–7 such that lower ranks represent variants with greater predicted regulatory impact (https://regulomedb.org/regulome-help/). For example, eQTLs (expression quantitative trait loci) have a rank of 1; variants locate in a transcription factor binding motif and DNAse peak have a rank of 4; and variants that locate in a transcription factor binding motif or a DNAse peak have a rank of 5. We additionally assessed cis-eQTLs using the eQTLGen Consortium database (https://www.eqtlgen.org/). 73.
Supplementary Information
Acknowledgements
CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program. CCFR: The Colon CFR graciously thanks the generous contributions of their study participants, dedication of study staff, and the financial support from the U.S. National Cancer Institute, without which this important registry would not exist. The authors would like to thank the study participants and staff of the Seattle Colon Cancer Family Registry and the Hormones and Colon Cancer study (CORE Studies). DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Utz Benscheid, Muhabbet Celik and Ursula Eilber for excellent technical assistance. EDRN: We acknowledge all the following contributors to the development of the resource: University of Pittsburgh School of Medicine, Department of Gastroenterology, Hepatology and Nutrition: Lynda Dzubinski; University of Pittsburgh School of Medicine, Department of Pathology: Michelle Bisceglia; and University of Pittsburgh School of Medicine, Department of Biomedical Informatics. EPIC: EPIC-Umeå (NSHDS) investigators thank the Biobank Research Unit at Umeå University, the Västerbotten Intervention Programme, the Northern Sweden MONICA study and Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (VR 2017-00650). Harvard cohorts (HPFS, NHS, PHS): The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the HPFS, NHS and PHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. PLCO: The authors thank the PLCO Cancer Screening Trial screening center investigators and the staff from Information Management Services Inc and Westat Inc. Most importantly, we thank the study participants for their contributions that made this study possible. WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: https://www-whi-org.s3.us-west-2.amazonaws.com/wp-content/uploads/WHI-Investigator-Long-List.pdf.
Author contributions
Design of work or data acquisition: J.L., S.S., T.H., D.B., P.C., S.G., G.G., M.G., M.H., M.J., S.O., S.P., M.S., B.V.G., J.C.F., R.P., R.N., L.Q., A.C., U.P., P.A.N. Analysis or interpretation of data: J.L., S.S., T.H., B.B., Y.H., L.N., Y.L., R.S., W.S., X.H., P.A.N. Drafting manuscript: J.L., S.S., T.H., Y.H., P.A.N.
Funding
Fred Hutch core grant: This research was funded in part through the NIH/NCI Cancer Center 14 Support Grant P30 CA015704. ISACC: National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (R01 CA176272). CPS-II: The American Cancer Society funds the creation, maintenance, and updating of the Cancer Prevention Study-II (CPS-II) cohort. This study was conducted with Institutional Review Board approval. DACHS: This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1 and BR1704/17-1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany, and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B). DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery). EDRN: This work is funded and supported by the NCI, EDRN Grant (U01 CA 84968-06). EPIC: The coordination of EPIC is financially supported by the European Commission (DGSANCO) and the International Agency for Research on Cancer. The national cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRCItaly and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPICOxford) (United Kingdom). Harvard cohorts (HPFS, NHS, PHS): HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993 and R35 CA197735), NHS by the National Institutes of Health (R01 CA137178, P01 CA087969, UM1 CA186107, R01 CA151993 and R35 CA197735) and PHS by the National Institutes of Health (R01 CA042182). MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 509348, 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database. PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH. UK Biobank: This research has been conducted using the UK Biobank Resource under Application Number 8614. VITAL: National Institutes of Health (K05 CA154337). WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology, and End Results (SEER) Program and the following U.S. state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The CCFR Set-1 (Illumina 1 M/1 M-Duo) and Set-2 (Illumina Omni1-Quad) scans were supported by NIH awards U01 CA122839 and R01 CA143247 (to GC). The CCFR Set-3 (Affymetrix Axiom CORECT Set array) was supported by NIH award U19 CA148107 and R01 CA81488 (to SBG). The CCFR Set-4 (Illumina OncoArray 600 K SNP array) was supported by NIH award U19 CA148107 (to SBG) and by the Center for Inherited Disease Research (CIDR), which is funded by the NIH to the Johns Hopkins University, contract number HHSN268201200008I. Additional support for the SFCCR was provided through NCI/NIH awards U01/U24 CA074794 and R01 CA076366 (to PAN). The content of this manuscript does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government, any cancer registry, or the CCFR.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-03945-x.
References
- 1.The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study The lancet. Gastroenterol. Hepatol. 2017;4(913–933):2019. doi: 10.1016/s2468-1253(19)30345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Edwards BK, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–573. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boland GM, et al. Association between adherence to National Comprehensive Cancer Network treatment guidelines and improved survival in patients with colon cancer. Cancer. 2013;119:1593–1601. doi: 10.1002/cncr.27935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Public Health Agency of, C., Statistics, C., Canadian Cancer, S. & Provincial/Territorial Cancer, R. Release notice—Canadian Cancer Statistics 2019. Health Promot Chronic Dis. Prev. Can.39, 255–255, doi:10.24095/hpcdp.39.8/9.04 (2019).
- 7.Brouwer NPM, et al. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int. J. Cancer. 2018;143:2758–2766. doi: 10.1002/ijc.31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karuppannan S, Kelty E, Sodhi-Berry N, Ee HC, Preen DB. Trends in incidence, mortality rates, and survival of colorectal cancer in Western Australia from 1990 to 2014: A retrospective whole-population longitudinal study. Int. J. Colorectal Dis. 2020;35:1719–1727. doi: 10.1007/s00384-020-03644-5. [DOI] [PubMed] [Google Scholar]
- 9.Al-Husseini MJ, et al. Outcome disparities in colorectal cancer: A SEER-based comparative analysis of racial subgroups. Int. J. Colorectal Dis. 2019;34:285–292. doi: 10.1007/s00384-018-3195-3. [DOI] [PubMed] [Google Scholar]
- 10.Compton CC. Pathologic prognostic factors in the recurrence of rectal cancer. Clin. Colorectal Cancer. 2002;2:149–160. doi: 10.3816/CCC.2002.n.020. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y, et al. The long-term survival characteristics of a cohort of colorectal cancer patients and baseline variables associated with survival outcomes with or without time-varying effects. BMC Med. 2019;17:150. doi: 10.1186/s12916-019-1379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berian JR, Benson AB, 3rd, Nelson H. Young age and aggressive treatment in colon cancer. JAMA. 2015;314:613–614. doi: 10.1001/jama.2015.9379. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, et al. Gender differences in colorectal cancer survival: A meta-analysis. Int. J. Cancer. 2017;141:1942–1949. doi: 10.1002/ijc.30827. [DOI] [PubMed] [Google Scholar]
- 14.SafaeeArdekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: A systematic review and meta-analysis. PLoS ONE. 2012;7:e47054. doi: 10.1371/journal.pone.0047054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton CC, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch. Pathol. Lab. Med. 2000;124:979–994. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 16.Tsilimigras DI, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018;27:280–288. doi: 10.1016/j.suronc.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Phipps AI, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol. Biomarkers Prev. 2012;21:1792–1798. doi: 10.1158/1055-9965.EPI-12-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur. J. Cancer. 2005;41:2060–2070. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Genomes Project C et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zienolddiny S, Skaug V. Single nucleotide polymorphisms as susceptibility, prognostic, and therapeutic markers of nonsmall cell lung cancer. Lung Cancer. 2012;3:1–14. doi: 10.2147/lctt.S13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutras A, Kotoula V, Fountzilas G. Prognostic and predictive role of vascular endothelial growth factor polymorphisms in breast cancer. Pharmacogenomics. 2015;16:79–94. doi: 10.2217/pgs.14.148. [DOI] [PubMed] [Google Scholar]
- 22.HajaMohideen AM, et al. Examining the polymorphisms in the hypoxia pathway genes in relation to outcome in colorectal cancer. PLoS ONE. 2014;9:e113513. doi: 10.1371/journal.pone.0113513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang MY, et al. Multiple genetic polymorphisms in the prediction of clinical outcome of metastatic colorectal cancer patients treated with first-line FOLFOX-4 chemotherapy. Pharmacogenet. Genom. 2011;21:18–25. doi: 10.1097/FPC.0b013e3283415124. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, et al. A genome wide association study on Newfoundland colorectal cancer patients' survival outcomes. Biomarker Res. 2015;3:6. doi: 10.1186/s40364-015-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penney KL, et al. Genetic variant associated with survival of patients with stage II–III colon cancer. Clin. Gastroenterol. Hepatol. 2019 doi: 10.1016/j.cgh.2019.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phipps AI, et al. Common genetic variation and survival after colorectal cancer diagnosis: A genome-wide analysis. Carcinogenesis. 2016;37:87–95. doi: 10.1093/carcin/bgv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penney ME, Parfrey PS, Savas S, Yilmaz YE. A genome-wide association study identifies single nucleotide polymorphisms associated with time-to-metastasis in colorectal cancer. BMC Cancer. 2019;19:133. doi: 10.1186/s12885-019-5346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi M, et al. Colorectal cancer: A tale of two sides or a continuum? Gut. 2012;61:794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi M, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slattery ML, et al. A comparison of colon and rectal somatic DNA alterations. Dis. Colon. Rectum. 2009;52:1304–1311. doi: 10.1007/DCR.0b013e3181a0e5df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, et al. Characteristics of differently located colorectal cancers support proximal and distal classification: A population-based study of 57,847 patients. PLoS ONE. 2016;11:e0167540. doi: 10.1371/journal.pone.0167540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low HB, et al. DUSP16 promotes cancer chemoresistance through regulation of mitochondria-mediated cell death. Nat. Commun. 2021;12:2284. doi: 10.1038/s41467-021-22638-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. doi: 10.1101/gr.9.8.677. [DOI] [PubMed] [Google Scholar]
- 35.Calle EE, et al. The American Cancer Society Cancer Prevention Study II nutrition cohort: Rationale, study design, and baseline characteristics. Cancer. 2002;94:2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 36.Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M. Protection from colorectal cancer after colonoscopy: A population-based, case-control study. Ann. Intern. Med. 2011;154:22–30. doi: 10.7326/0003-4819-154-1-201101040-00004. [DOI] [PubMed] [Google Scholar]
- 37.Slattery ML, et al. Energy balance and colon cancer–beyond physical activity. Cancer Res. 1997;57:75–80. [PubMed] [Google Scholar]
- 38.Amin W, Singh H, Dzubinski LA, Schoen RE, Parwani AV. Design and utilization of the colorectal and pancreatic neoplasm virtual biorepository: An early detection research network initiative. J. Pathol. Inform. 2010;1:22. doi: 10.4103/2153-3539.70831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riboli E, Kaaks R. The EPIC project: Rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int. J. Epidemiol. 1997;26:S6–14. doi: 10.1093/ije/26.suppl_1.s6. [DOI] [PubMed] [Google Scholar]
- 40.Rimm EB, et al. Validity of self-reported waist and hip circumferences in men and women. Epidemiology. 1990;1:466–473. doi: 10.1097/00001648-199011000-00009. [DOI] [PubMed] [Google Scholar]
- 41.Giles GG, English DR. The Melbourne collaborative cohort study. IARC Sci. Publ. 2002;156:69–70. [PubMed] [Google Scholar]
- 42.Belanger CF, Hennekens CH, Rosner B, Speizer FE. The nurses' health study. Am. J. Nurs. 1978;78:1039–1040. [PubMed] [Google Scholar]
- 43.Colditz GA, Manson JE, Hankinson SE. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J. Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 44.Goldberg RM, et al. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J. Clin. Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 45.Christen WG, Gaziano JM, Hennekens CH. Design of Physicians' Health Study II–a randomized trial of beta-carotene, vitamins E and C, and multivitamins, in prevention of cancer, cardiovascular disease, and eye disease, and review of results of completed trials. Ann. Epidemiol. 2000;10:125–134. doi: 10.1016/s1047-2797(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 46.Steering Committee of the Physicians' Health Study Research Final report on the aspirin component of the ongoing Physicians' Health Study. N. Engl. J. Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 47.Prorok PC, et al. Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin. Trials. 2000;21:273S–309S. doi: 10.1016/S0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 48.Gohagan JK, Prorok PC, Hayes RB, Kramer BS. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin. Trials. 2000;21:251S–272S. doi: 10.1016/S0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 49.Sudlow C, et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White E, et al. VITamins And Lifestyle cohort study: Study design and characteristics of supplement users. Am. J. Epidemiol. 2004;159:83–93. doi: 10.1093/aje/kwh010. [DOI] [PubMed] [Google Scholar]
- 51.Group T.W. Design of the Women's Health Initiative clinical trial and observational study. Control Clin. Trials. 1998;19:61–109. doi: 10.1016/S0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 52.Hays J, et al. The Women's Health Initiative recruitment methods and results. Ann. Epidemiol. 2003;13:S18–77. doi: 10.1016/s1047-2797(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 53.Newcomb PA, et al. Colon cancer family registry: An international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.Epi-07-0648. [DOI] [PubMed] [Google Scholar]
- 54.Jenkins MA, et al. Cohort profile: The colon cancer family registry cohort (CCFRC) Int. J. Epidemiol. 2018;47:387–388i. doi: 10.1093/ije/dyy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riboli E, et al. European prospective investigation into cancer and nutrition (EPIC): Study populations and data collection. Public Health Nutr. 2002;5:1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 56.Milne RL, et al. Cohort profile: The Melbourne collaborative cohort study (Health 2020) Int. J. Epidemiol. 2017;46:1757–1757i. doi: 10.1093/ije/dyx085. [DOI] [PubMed] [Google Scholar]
- 57.Miller, A. B., Yurgalevitch, S., Weissfeld, J. L., Prostate, L. C. & Ovarian Cancer Screening Trial Project, T Death review process in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin. Trials. 2000;21:400–406. doi: 10.1016/S0197-2456(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 58.Curb JD, et al. Outcomes ascertainment and adjudication methods in the Women's Health Initiative. Ann. Epidemiol. 2003;13:S122–128. doi: 10.1016/S1047-2797(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 59.Final report on the aspirin component of the ongoing Physicians' Health Study. N. Engl J Med321, 129–135, doi:10.1056/nejm198907203210301 (1989). [DOI] [PubMed]
- 60.Rimm EB, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 61.Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters U, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–807. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phipps AI, et al. Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology. 2012;143:51–54. doi: 10.1053/j.gastro.2012.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLeod HL, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: Results from North American gastrointestinal intergroup trial N9741. J. Clin. Oncol. 2010;28:3227–3233. doi: 10.1200/jco.2009.21.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huyghe JR, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 2013;93:687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schoenfeld D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–241. doi: 10.2307/2335876. [DOI] [Google Scholar]
- 71.Genomes Project C et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vosa U, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.