Abstract
Comparative population genomics is an ascendant field using genomic comparisons between species to draw inferences about forces regulating genetic variation. Comparative phylogeography, by contrast, focuses on the shared lineage histories of species codistributed geographically and is decidedly organismal in perspective. Comparative phylogeography is approximately 35 years old, and, by some metrics, is showing signs of reduced growth. Here, we contrast the goals and methods of comparative population genomics and comparative phylogeography and argue that comparative phylogeography offers an important perspective on evolutionary history that succeeds in integrating genomics with landscape evolution in ways that complement the suprageographic perspective of comparative population genomics. Focusing primarily on terrestrial vertebrates, we review the history of comparative phylogeography, its milestones and ongoing conceptual innovations, its increasingly global focus, and its status as a bridge between landscape genomics and the process of speciation. We also argue that, as a science with a strong “sense of place,” comparative phylogeography offers abundant “place-based” educational opportunities with its focus on geography and natural history, as well as opportunities for collaboration with local communities and indigenous peoples. Although comparative phylogeography does not yet require whole-genome sequencing for many of its goals, we conclude that it nonetheless plays an important role in grounding our interpretation of genetic variation in the fundamentals of geography and Earth history.
Keywords: Gott Earth projection, whole-genome sequencing, landscape genomics, place-based education, indigenous knowledge
Significance
Comparative phylogeography shares many concepts and techniques with comparative population genomics, yet differs in its emphasis on geography, codistribution of species, and genetic structure within species. We review the history of comparative phylogeography and its global expansion in the last 12 years and suggest that it complements comparative population genomics in its ability to engage students, indigenous groups and the public via its focus on geography, prominent landscape features, and Earth history.
Introduction
Comparative population genomics is an emerging field that leverages variation in genomes of multiple species to understand the origins of genomic diversity and natural selection and to make inferences about patterns of divergence between closely related species. By comparing genomic variation across multiple species, comparative population genomics aims to discover common forces acting on homologous or divergent regions of the genome. Comparative population genomics approaches have been used with great success since the 1970s, but with the rise of whole-genome sequencing (WGS) in recent years, the approach has gained renewed vigor and detail. In its most recent incarnation, the emphasis in comparative population genomics is less on the details of geography of each species, and more on sampling each species sufficiently so as to capture major patterns of variation and identify the demographic and genetic forces that have shaped them. Appropriate for its goals, the focus of modern comparative genomics is often on genetics, rather than geography.
Here we argue that comparative phylogeography—in many ways a more mature discipline than comparative population genomics, but one arguably with less ongoing conceptual innovation—stands as an important counterpoint to and partner of comparative population genomics. In so far as comparative population genomics suffers from a lack of a rigorous geographic perspective, and often seeks to marginalize over geography (see Comparative Phylogeography as Place-Based Evolutionary Biology section), comparative phylogeography brings an explicit geographic perspective that has not only revealed details of species history, but can complement the goals of comparative population genomics. Comparative phylogeography seeks to understand biotic history by finding landscape features—mountains, rivers, transition zones—that create vicariant breaks in genetic variation across a suite of species. A major emphasis in comparative phylogeography is to find drivers of genomic splits shared across suites of codistributed species, and thereby find landscape features that ultimately explain patterns of species occurrence, community composition, and genetic diversity (table 1). A related discipline, landscape genomics, usually focuses on a smaller spatial scale and emphasizes genomic adaptation to landscape and climate features (Manel et al. 2003; Manel and Holderegger 2013; Rissler 2016). The focus of comparative phylogeography is usually on organismal histories, rather than on the diversity of evolutionary forces shaping the genome (table 1). To understand the relationships between comparative phylogeography and comparative population genomics, we first ask whether comparative phylogeography is a viable field of inquiry and on scientific growth trajectory. We briefly review the historical origins and recent milestones of comparative phylogeography from our perspective as empiricists working on terrestrial vertebrates. We then update our understanding of the global distribution of phylogeographic studies and argue that, as a “place-based” discipline, comparative phylogeography has great appeal for local and regional scientific communities and public education around the world in ways that are challenging for comparative population genomics. We conclude that, although it has yet to fully embrace the genomics revolution, comparative phylogeography is still a vibrant discipline that stands to make important contributions to evolutionary biology and in particular can leverage its “sense of place” to help include diverse communities in the research enterprise around the globe.
Table 1.
Conceptual Relationships between the Fields of Comparative Population Genomics, Landscape Genomics, and Comparative Phylogeography
| Concept/Parameter | Comparative Population Genomics | Landscape Genomics | Comparative Phylogeography |
|---|---|---|---|
| Comparative perspective | Growing | Nascent | Mature |
| Emphasis on space | No | Yes | Yes |
| Geographic scale | Random mating population | Region | Biome |
| Temporal scale | Arbitrary | Recent | Deep |
| Focus on: | |||
| selection versus neutrality | Both | Both | Neutrality |
| recombination | Yes | Not yet considered | Not yet considered |
| geography versus environment | Nuisance parameters | Environment | Both |
| Future use of whole-genome sequencing | Yes | Likely | Unlikely |
| Growth out of museum collections community | No | No | Partial |
Is Comparative Phylogeography Dead?
A Brief History of Comparative Phylogeography
Early in its evolution, phylogeography adopted a comparative perspective, seeking to find common patterns across codistributed species. Although the first phylogeographic studies were conducted on single species (Avise et al. 1979), very quickly the field adopted a comparative approach, motivated to identify patterns of intraspecific genetic variation shared across species and so point to common historical processes (Bermingham and Avise 1986; Avise et al. 1987). At the same time, using morphology and other cladistic characters, the field of vicariance biogeography had begun to decipher shared geographic patterns of speciation among closely related and codistributed taxa with phylogenetic trees (Cracraft 1982, 1986). Although the problems addressed in early comparative phylogeographic studies by definition adopted molecular tools, they were often inspired by morphological studies of vicariance biogeography and cladistic analyses, often on the same organismal lineages (e.g., Cracraft 1986; Edwards 1993; Dolman and Joseph 2012; Cracraft 1991; Nyari and Joseph 2013). Indeed, phylogeography was envisaged then as the bridge between population genetics and vicariance-centered phylogenetic systematics, a perspective that remains true today but which has also been expanded in modern parlance into a bridge between the divergent scales of landscape genetics and processes of speciation.
A literal focus on gene trees in space defined phylogeography from the outset and distinguished it from prior analyses of allele frequency variation, as revealed by blood types Edwards and Cavalli-Sforza 1963; Cavalli-Sforza and Edwards 1967; Bodmer 2015) or multilocus allozymes (Allendorf 2017). Using allozyme electrophoresis, researchers had accumulated hundreds of studies documenting the geographic pattern of genetic variation within species. Allozyme electrophoresis proved to be a useful tool to quantify levels of variation in protein-coding regions whose alleles could be differentiated by electric charge. Studies of allozyme variation were relatively inexpensive and were usually couched in terms of quantifying geographic variation or investigating spatial variation in selection and clines, and eventually developed into quantitative tests of modes of speciation. Many studies employing allozymes, as well as more recent studies employing microsatellites or single-nucleotide polymorphisms (SNPs), focused on the geographic distribution of alleles and models of isolation by distance that provided a sensitive framework for interpreting patterns of geographic variation (Slatkin 1985; Novembre and Slatkin 2009; François et al. 2010). There was also considerable interest in the ecological correlates and life history predictors of genetic variation—how the ecology and behavior of organisms modulated levels of genetic variation. Although phylogenetic trees were routinely made from allozyme data, the emphasis was on variation in allele frequencies: For example, the commonly used statistic for allele frequency differentiation, average Fst, was largely used to estimate levels of gene flow among populations; only occasionally was Fst used to attempt to detect natural selection, primarily because the number of loci routinely surveyed in allozyme studies (anywhere from ∼20 to 60 loci) was too small to detect Fst outliers.
Restriction enzyme analysis was introduced to population genetics in the mid-1970s and fundamentally changed the way empiricists viewed genetic variation; whereas in the allozyme era, alleles were distinguished categorically, as similar or different to one another. By contrast, restriction fragment length polymorphisms (RFLPs) allowed researchers to quantify the degree of difference between alleles. Perhaps more fundamentally, RFLPs permitted the phylogenetic analysis of alleles, first deployed on a large scale with animal mitochondrial DNA (mtDNA). The first intraspecific “gene tree” was published by Avise et al. (1979). This rather understated publication ushered in a dramatically new view of genetic variation, one in which genealogies of alleles, within and between species, rather than allele frequencies, were the mode of description for genetic variation. Because of its genetic simplicity, rapid evolution, and abundance in animal cells, mtDNA provided easy access to genetic variation and geographic patterns in animals. In time, massive gene trees consisting of hundreds of alleles, allowed fine dissection of geographic patterns within species, such as the human species (Cann et al. 1987). A plethora of statistics of genetic variation, such as nucleotide diversity (π) and Fst incorporating divergence among haplotypes, arose in the context of RFLP variation (Nei and Li 1979; Allendorf 2017); many are still in use today.
Milestones and Recent Innovations in Comparative Phylogeography
The introduction of the polymerase chain reaction to phylogeography in the early 1980s, to both nuclear and mitochondrial genes, finally allowed direct visualization at the nucleotide level to the DNA changes underlying RFLPs; although phylogeographers still struggled then with the apparently low levels of variation detectable at nuclear genes and the challenge of recombination within loci, we suggest that the modern form of phylogeography, in which sequence changes at multiple loci are analyzed in allele-frequency and phylogenetic frameworks, was established by the early 1990s. With the establishment of phylogeography as a discipline, it was only a matter of time before researchers endeavored to compare phylogeographic histories of multiple species. Often phylogeographic comparisons between species were conducted in the context of codistributed species that may have responded similarly to the same physical or environmental barriers on the landscape. Indeed, codistribution of species under study is a central tenet of comparative phylogeography and is often included in its definition (Gutiérrez-García and Vázquez-Domínguez 2011). In many ways, comparative phylogeography refocused attention to the geographic context of genetic variation in ways that the earlier focus on single species did not, or could only do so haphazardly. By analyzing the genetic diversity of codistributed species at key transition zones around the globe, comparative phylogeography allowed the identification of zones of phylogenetic divergence and promoted the idea that landscape features, such as mountains, rivers, and marine archipelagoes, could serve as powerful generators of biodiversity and genetic divergence across multiple lineages. A special issue of Molecular Ecology in 1998 focused on comparative phylogeography. Bermingham and Moritz (1998) celebrated 10 years of achievement since Avise’s seminal 1987 review, but they bemoaned the overreliance on the single locus provided by animal mtDNA.
The 2000s brought continued expansion of frameworks and methods in comparative phylogeography. Beheregaray (2008) noted spectacular growth of phylogeography and comparative phylogeography in the ensuing decade, with concomitant diversification in molecular markers used, by then routinely including nuclear SNPs and microsatellites (Garrick et al. 2015). However, Beheregaray also noted a gross paucity of studies from the Southern Hemisphere and warned of making general conclusions of process without more equal representation of the hemispheres. Hickerson et al. (2010) documented further expansion of the molecular marker toolkit and pointed toward the integration of ecological niche models, community assembly perspectives, and model-based inference of demographic histories into comparative phylogeography. The incorporation of techniques such as approximate Bayesian computation (ABC) and coalescent simulations flowed naturally from the increased distinction by practitioners between gene histories and population histories, and the focus, appropriately, on the population history as the end-goal of phylogeography. Hierarchical approximate Bayesian computation represents an important statistical approach specifically tailored to comparative phylogeography and the detection of concerted demographic responses to environmental events. Increasingly, estimates of demographic history are integrated with paleospecies distribution models (Carstens and Richards 2007; Alvarado‐Serrano and Knowles 2014). Additional recent innovations in comparative phylogeography include the use of machine learning to identify comparative phylogeographic trends across clades (Espíndola et al. 2016; Carstens et al. 2018; Barrow et al. 2020; Fonseca et al. 2021). Finally, the use of phenotypic traits to guide phylogeographic sampling and erect hypotheses of intraspecific differentiation and interspecific codiversification have been proposed (Papadopoulou and Knowles 2016; Sullivan et al. 2019). Many of these innovations were celebrated in a 2016 symposium on comparative phylogeography organized by John Avise, Brian Bowen, and Francisco Ayala for the National Academy of Sciences (Avise et al. 2016). These tools have yet to be widely applied but suggest multiple promising avenues for future research and help promote a predictive, rather than descriptive, framework for comparative phylogeography.
As the number and scale of comparative phylogeographic studies increased, the complexity and heterogeneity of species histories across individual barriers became increasingly evident. Indeed, most studies of multiple taxa across individual barriers find heterogeneity in the timing of vicariant splits and population dynamics across the barrier (for a few of many examples, see Schneider et al. 1998; Barber and Klicka 2010; Naka and Brumfield 2018; Thom et al. 2020; Provost et al. 2021). Another facet of comparative phylogeography that has expanded in recent years is the ubiquity of dispersal as a mechanism for obscuring common vicariant patterns. Indeed, dispersal and population expansion from Pleistocene refugia have been common characteristics across many multispecies communities investigated, particularly in temperate regions and the marine realm (Hewitt 1996, 1999; Bernatchez and Wilson 1998; Floeter et al. 2007; Burney and Brumfield 2009; Lohman et al. 2011; Bagley and Johnson 2014; Smith et al. 2014). The extent to which lineages depart from a strict vicariant patterns is in many cases linked to morphological and life history traits related to dispersal and vagility (Paz et al. 2015; Papadopoulou and Knowles 2016; Puritz et al. 2017; Schiebelhut and Dawson 2018; Sullivan et al. 2019; Ribeiro et al. 2020). Along with historical niche modeling, the intergration of trait-based ecology into comparative phylogeography has had a profound influence on the trajectory of the field.
Coalescent Theory and the Death of Classical Phylogeography
Coalescent theory, focusing on statistical properties of gene trees and introduced to a wide audience by the mid-1980s, was an important addition to the phylogeographers toolkit. Strikingly for empiricists, an authoritative history of the origin of coalescent theory does not mention Avise, Allan Wilson, or any of the empiricists working in phylogeography during the transition to coalescent theory, which was driven by more theoretical considerations (Kingman 2000). Theoreticians quickly adopting coalescent theory, such as Tajima (1983), provide a much more direct link with empirical data sets, and the production and visualization of gene trees, particularly large ones of the human and other iconic species (Cann et al. 1987), was an important driver of coalescent theory’s early development. The early connection between phylogeography and coalescent theory—for example, to estimate population size trajectories (Rienzo and Wilson 1991; Slatkin and Hudson 1991) or population migration rates (Slatkin and Maddison 1989)—was an exciting time, even for single-locus mtDNA or cpDNA studies, which generated new insights, many of which have proved remarkably resilient. The development of spatially realistic coalescent theory remains a vibrant area of population genetics today (Bradburd and Ralph 2019).
As coalescent theory developed to address the burgeoning multilocus data from next-generation sequencing approaches, it is reasonable to ask whether comparative phylogeography—in the sense of interpreting gene trees in space—has had its day. As in early empirical surveys, the first applications of coalescent theory in phylogeography viewed gene trees as fixed entities, requiring researchers to estimate them first and infer parameters second (Slatkin and Maddison 1989). However, a key transition point in the field was the shift away from “gene tree” thinking and toward population and lineage thinking: the realization that gene trees themselves are nuisance parameters from the perspective of estimating population genetic parameters (Edwards and Beerli 2000; Hey and Machado 2003; Rosenberg and Nordborg 2002), just as they are considered now for estimation of phylogenetic histories of species (Edwards 2009). New software for simulating coalescent histories within diverging lineages helped reinforce this view among empiricists (Beerli and Felsenstein 1999; Excoffier et al. 2000). Around the time of Hickerson et al.’s review, disagreements arose as to how phylogeography should incorporate the variation and stochasticity found in gene trees as the number of markers expanded. Lacey Knowles coined the term “statistical phylogeography” to acknowledge the inherent noisiness of gene trees and to encourage the use of summary statistics and ensemble analyses going forward (Knowles 2009). Others, like Alan Templeton, had promoted methods, such as nested clade analysis (NCA), that focused on more literal interpretation of branching patterns in gene trees as signals of population inferences (Templeton 1998). Coalescent simulations suggested that NCA overinterpreted signals in gene trees, resulting in many false positives (Panchal and Beaumont 2007), and arguments ensued as to whether the signals utilized by NCA were valuable, or whether likelihood or approximate Bayesian methods that factored in uncertainty in the gene trees were the way forward (Beaumont et al. 2010; Templeton 2010). Likelihood and simulation methods appear to have won that argument (Petit 2008)—while at the same time ushering in a suite of new analytical approaches that moved the field away from the gene trees that marked its founding.
The solution arising over the last two decades is to estimate population divergence histories from extensive multilocus data and coalescent models, often using simulations and ABC methods to compare different hypotheses (Knowles 2009), themselves generated from spatial modeling of species or habitats through past climates (Hugall et al. 2002; Carnaval et al. 2009). Many such methods avoid gene tree estimation entirely, for example, by using SNP data and the multidimensional site frequency spectrum (mdSFS) directly (Xue and Hickerson 2015). In turn, hierarchical methods using the mdSFS across species were developed to test for common histories of vicariance or population expansion without interrogating gene trees directly (Overcast et al. 2017; Xue and Hickerson 2017, 2020). Indeed, the software msBayes and its updates and relatives (Oaks 2014, 2019; Robinson et al. 2014; Bunnefeld et al. 2018) are arguably the only software focused specifically on comparative phylogeography, as opposed to single-species phylogeography.
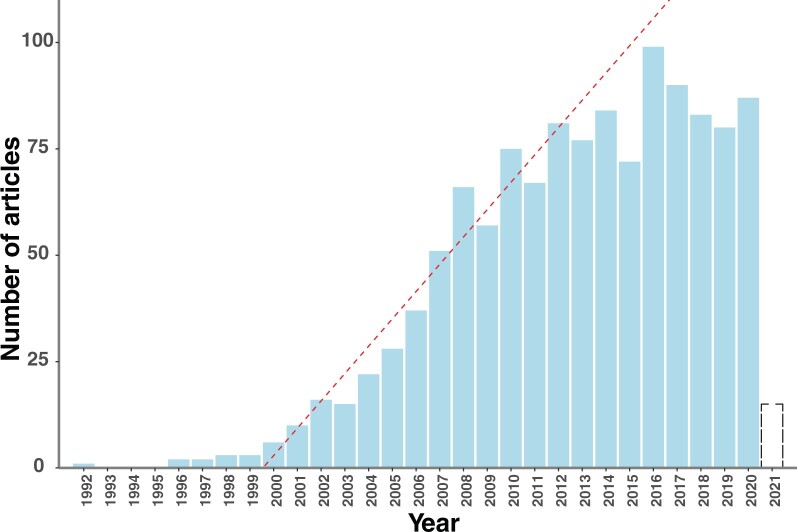
Against this backdrop of innovation, however, comparative phylogeography as a field seems no longer to be growing, as judged by numbers of publications (fig. 1). The number of publications related to comparative phylogeography seems to have plateaued since about 2010, no longer growing linearly or even exponentially as many new fields do (supplementary tables S1 and S2, Supplementary Material online). The transformation of the tools of comparative phylogeography—from the data-analysis perspective of empiricists—may help explain these trends. For example, some might argue that the eclipse of gene trees from phylogeography, as well as the leveling-off of publications per year (fig. 1), is sufficient grounds to proclaim the field dead, or at least superseded by modern, gene-tree-free approaches. So, is comparative phylogeography dead, or transformed? We assert the latter. Although the focus on gene trees in space, across multiple codistributed taxa will likely be replaced by SNP- or haplotype-based approaches, the core goals of comparative phylogeography remain in the population genomics era. The major change in perspective, we suggest, is the increased focus on the expectations of coalescence histories across space and taxa, rather than the estimated gene trees themselves. Empirical estimation of single-locus gene trees may now more profitably serve the valuable role of generating hypotheses and sampling designs for more intensive, multitaxon, genome-wide SNP-based approaches. Although there are now many examples of discordance between organelle and nuclear gene histories, often due to hybridization, selection, or nonrandom lineage extinction, rapid phylogeographic surveys of a region via estimated mitochondrial or chloroplast gene trees still prove very useful and have particular value in relatively underexplored regions, including much of the biodiverse tropics.
Fig. 1.
Number of publications per year with some variant of “comparative phylogeography” in the title, as referenced in the InCite database (accessed March 28, 2021). Dashed red line indicates a line of constant increase in numbers of publications from 1999 to 2008 (slope, increase of 6.42 publications per year). Dashed bar for 2021 indicates that a partial year is represented. Data are from the 1229 articles in supplementary tables S1 and S2, Supplementary Material online.
At the same time, it is unclear whether comparative phylogeography will, or needs to, embrace WGS as a methodological standard. Even with dropping sequencing costs, WGS is unlikely to be adopted widely by diverse researchers around the globe. Moreover, WGS is likely overkill and is not required to answer the core questions of the field (table 1); genome subsampling approaches such as ddRadseq, or capture methods, will likely carry the field forward for the foreseeable future (Oswald et al. 2017; Alter et al. 2017; Thom et al. 2020; DiBattista et al. 2020; de Medeiros and Farrell 2020). Conceivably, the greater access to WGS methods will cause the conceptual boundaries of comparative phylogeography to expand, incorporating more questions traditionally in the domain of population genetics (Edwards et al. 2015). By contrast, landscape genomics may well embrace WGS (table 1), given its focus on adaptation of organisms to environmental gradients and effects of habitat heterogeneity on genetic diversity, as well as connections with life history, dispersal, and population connectivity (Gagnaire 2020). On the other hand, large meta-analyses of genetic diversity across many species across the globe often require restricting attention to commonly employed markers, such as mtDNA (“comparative mitogenomics”), with concomitant uncertainty about what genome-wide processes might look like (Miraldo et al. 2016; Manel et al. 2020; Millette et al. 2020; Theodoridis et al. 2020).
The importance of museum specimens in phylogeographic work in general has been widely discussed, and the value of specimens linked to the genomic resources forming the basis of phylogeographic studies—as means of replication and verification, further research, or as direct sources of historical DNA—is increasingly appreciated (Besnard et al. 2016; Yao et al. 2017; Schmitt et al. 2018; Nakahama 2021). However, the relationship of the three fields outlined in table 1 to the museum collections community is uneven. To the extent that comparative phylogeography emerged in part from the systematics community—and conceptual antecedents like vicariance biogeography suggest that this is at least partly true—the field also emerged from the museum community and its values. But its prime exponent, John Avise, was not a museum scientist and, in truth, archiving of specimens for phylogeographic research is highly patchy and inconsistent. Poor and inconsistent archiving of museum specimens for genomics work is well documented (Buckner et al. 2021), and museum specimens in landscape genomics are unusual, especially given that field’s orientation toward ecology. Thus, whether comparative phylogeography in the future will continue to benefit from the extra value provided by museum specimens is unclear.
Comparative Phylogeography: A Global Perspective
Is Comparative Phylogeography Becoming More Global?
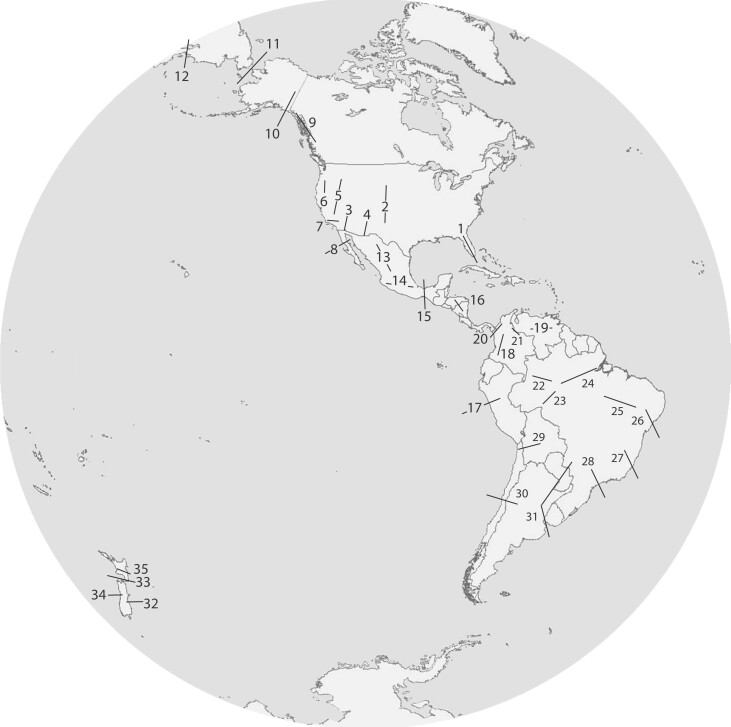
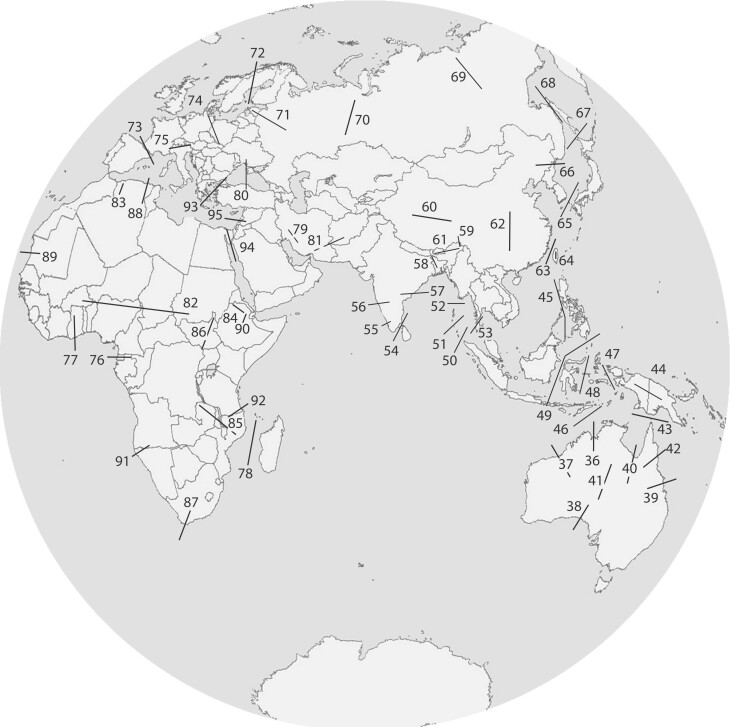
Figures 2 and 3 and supplementary table S4, Supplementary Material online, present summaries of some of the major sites of comparative phylogeographic investigation today in the New and Old Worlds, respectively, built on earlier work, identified, or restudied since the first investigations of the codistributed faunas around the Gulf of Mexico and Atlantic Ocean in the mid-1980s (Bermingham and Avise 1986). Building on the overview of phylogeographic breaks presented by Riddle (2016), and focusing primarily on terrestrial vertebrates, we document 93 major sites of divergence across the globe, many in the tropics. It seems likely that the number of geomorphological barriers facilitating divergence in the tropics is still underestimated, and we present only some of the known physical barriers in the tropics, especially around the barrier-rich regions of the northern Andes and Amazon basin in South America (Naka et al. 2012; Cuervo 2013; Naka and Brumfield 2018). Tropical Southeast Asia is likely another region whose barrier numbers have been underestimated, although the full extent of bias against the tropics versus temperate regions has not been studied.
Fig.2.
Major sites of comparative phylogeographic exploration in the New World, on a map with the new Gott et al. (2021) projection, which minimizes 2D distortion and orientation of landmasses less so than any previous projection. Numbers correspond to listings in supplementary table S4, Supplementary Material online.
Fig. 3.
Major phylogeographic breaks in the Old World, with projection details as in figure 2. Numbers correspond to listing of breaks in supplementary table S4, Supplementary Material online.
For figures 2 and 3, we have used a recently proposed Earth projection that provides technical and ethical improvements upon classical projections used in comparative phylogeography. The particular global projection used in figures 2 and 3 was proposed only in February 2021 and represents the best 2D projection yet as measured by six distortion metrics, including isotropy, area, flexion, skewness, distances, and boundary cuts (Gott et al. 2021). It is well known that many projections of the globe induce biases in how researchers and the public perceive the world; this awareness of implicit biases in data visualization is beginning to influence many areas of science, including modern genomics (Narayan et al. 2021). For example, the Mercator projection, which causes the impression that many high-latitude countries are larger in area than those at the equator, is known to cause biases in perception of the global order (Haemer 1949; Castex 1993). Riddle (2016) appears to have used the Gall–Peter projection, first presented in 1855 but promoted widely in the 1970s. Although the Gall–Peter projection captures a more “equal-area” perspective than the Mercator, it still grossly distorts the shapes of landmasses at the poles and the equator (Vujakovic 1989). The Gott et al. (2021) projection is a radical and elegant departure from previous projections and requires two discs, rather than a single flat image, to represent the entire globe. We suggest that the Gott et al. (2021) projection presents the most scientifically and socially equitable context yet proposed for depicting geography; we encourage its more widespread use, in comparative phylogeography and other fields.
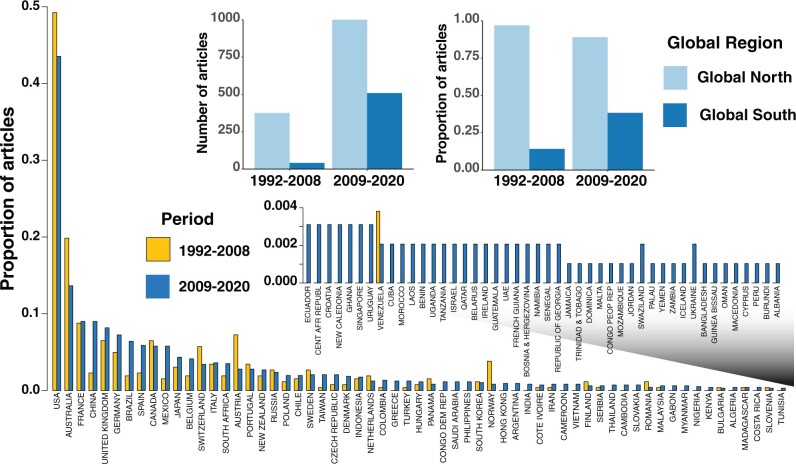
In an important review, Beheregaray (2008) pointed out that comparative phylogeography was underrepresented by what today we call the Global South, and encouraged international collaborations as one way of empowering countries in the developing world. We update his investigation in figure 4, comparing geographical trends in publications focusing on comparative phylogeography before and since his review (supplementary table S3, Supplementary Material online). Assigning one paper contribution to each country that is represented in the author line, we find that, although there still appears to be a strong bias toward publications coming from the Global North, since 2008 there has been a noted increase in publications coming from the Global South. Although, on average since 2008, the proportional contribution of the Global North to the literature on comparative phylogeography has increased by 39.1%, the contribution of Global South countries has increased by 97.5% (supplementary fig. S1, Supplementary Material online). The difference in the distribution of contributions by country between the two time periods is significant (two-sided Kolmogorov–Smirnov test, D = 0.59813, P < 2.2e−16). By tabulating country-of-origin of authors of papers on comparative phylogeography, our analysis focuses on where the work is getting done, and not directly on the geographic focus of the work. Still, it is likely that most authors of papers in comparative phylogeography are focusing on systems close to home, with the result that more regions of the globe are being studied as the geographical diversity of authorlines expands. For example, since 2008, and even before that date, there has been a steady accumulation of studies in the regions like the Atlantic Forest and Amazon of South America, many of them driven by researchers in Brazil, whose contribution to the literature in comparative phylogeography has grown since 2008 by 140% (Raposo do Amaral et al. 2013; Thom et al. 2020). The contribution of China, considered a member of the Global South, to the comparative phylogeography literature since 2008 has grown by 236%. Although only three African countries (Madagascar, Côte d'Ivoire, and South Africa) contributed to the pre-2008 literature on comparative phylogeography, 21 countries in Africa published at least one paper in comparative phylogeography after 2008, with an average of 4.85 publications per country in the latter period. There has been growth in Africa as a focus of comparative phylogeography in general (Hydeman et al. 2017; Beddek et al. 2018; van Asch et al. 2019; Helmstetter et al. 2020), although the extent to which in-country researchers participate in this increase is variable (Lorenzen et al. 2012; Freilich et al. 2016). Although we record only five comparative phylogeography publications from two countries (Indonesia and Malaysia) in SE Asia before 2008, in the later period ten SE Asian countries (not including Singapore, a Global North country) produced a total of 73 publications, with greater than 7 coming from authors in each of Vietnam, India, Thailand, Cambodia, Philippines, and Indonesia (fig. 4). Very few phylogeographic studies have focused on the rich biodiversity of India (Reddy 2014); the few studies since 2008 have revealed exciting phylogeographic patterns in birds (Robin et al. 2010, 2015), mammals (Karanth et al. 2010), amphibians (Meegaskumbura et al. 2002; Bossuyt et al. 2004; Vijayakumar et al. 2016), and reptiles (Agarwal and Ramakrishnan 2017), with several leading to descriptions of new taxa, particularly in the Western Ghats. We hope these trends continue and that new regulations promoted by the Convention on Biological Diversity, including the Nagoya Protocol, not only empower researchers from the Global South but also encourage international collaboration, data sharing and open access, especially for fields like comparative phylogeography, which are generally not driven with an eye for profit.
Fig. 4.
Global trends in geographic locations of authors of publications in comparative phylogeography in two time periods, 1992–2008 and 2009–present. (A) Proportion of publications with authors from countries listed on the x-axis to total publication output for two time periods. (B) Shifts in number of publications and proportion of publication by authors in countries listed as Global North or Global South (country designations from https://meta.wikimedia.org/wiki/List_of_countries_by_regional_classification). Data on which these figures are based are in supplementary table S3, Supplementary Material online. We recognize that the terms “Global North” and “Global South” may offend some readers and for that we apologize.
One reason for the plateau in publication number observed in figure 4 is that comparative phylogeographic studies, by necessity, are more challenging and more expensive to complete than single-species studies. Garrick et al. (2015) noted that the number of loci in phylogeographic studies increased over time only in single-species studies, which consistently employed larger numbers of loci than comparative phylogeographic studies. The embrace of genome-wide methods in comparative phylogeography will be most pronounced in wealthier countries (Edwards et al. 2016; Afonso Silva et al. 2017; Oswald et al. 2017), even as the Global South increases its participation in the field. It is unclear whether comparative phylogeography will ever fully embrace WGS (except perhaps in meta-analyses of single-species studies), or whether it requires it to achieve its goals (table 1).
Comparative Phylogeography: A Bridge between Landscape Genetics and Speciation
The focus on geography and potential isolating barriers on the landscape puts comparative phylogeography in a privileged position to link the micro- and macroevolutionary ends of the speciation continuum (Harvey et al. 2019) (fig. 5). Speciation research is highly heterogeneous in its focus. At one extreme, there are the many empirical studies and mathematical theories focusing on the genetics of reproductive isolating mechanisms and the role of pre- and postzygotic isolating mechanisms. On the other extreme, there are numerous studies focusing on the geography and demography of the speciation process, with phylogeographic approaches, bottlenecks, and introgression having prominent roles. Moreover, a large body of literature focuses on ecological factors and organismal traits influencing the rate of speciation across higher level clades (Rosenblum et al. 2012). Comparative phylogeography stands to bridge these extremes by grounding empirical studies in the demography of speciation and quantifying the contribution of the landscape and environmental change to the likelihood of speciation (Singhal and Bi 2017; Potter et al. 2018; Edwards et al. 2019; Harvey et al. 2019). Recent genomic studies of speciation have repeatedly shown the prevalence of ongoing gene flow as species diverge, producing in many cases sister species that differ phenotypically but genomically only at a few places along the genome. Prominent examples of this outcome are found in the butterfly genus Heliconius (Martin et al. 2013) and many bird species (Toews et al. 2016). The prevalence of gene flow during the speciation process has highlighted the important role of incompatibilities and premating isolating mechanisms in keeping gene pools functionally distinct at few or many loci across the genome. Comparative phylogeography can aid such studies by estimating, for example, how long recent species have been in sympatry or what landscape features may have facilitated the achievement of sympatry. Although these new speciation models have made processes like sympatric speciation and ecological speciation due to local adaptation more plausible, they have not seriously undermined the long-appreciated role of geographic isolation in many speciation scenarios. Geographic isolation, in turn, is a major cause of phylogeographic concordance across lineages that is a core focus of comparative phylogeography.
Fig. 5.
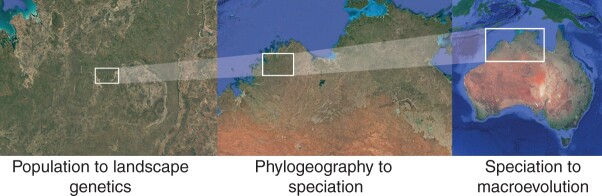
Depiction of how different approaches in comparative genetics scale geographically. In this example, focused on low dispersal vertebrates northern Australia, population to landscape studies might be done in the north Kimberly at scales of 10–200 km, phylogeographic to speciation studies at the scale of 100s—1000 km (Kimberley to Top End), and speciation to macroevolution at continental scale. Images are from Google Earth; see Potter et al. (2018) as an example.
Comparative phylogeographic studies, particularly when conducted with genome-wide methods, can also help determine the prevalence of genome-wide genetic differentiation of interacting and diverging populations, and thereby help us gauge how often speciation is accompanied by limited genomic divergence. Many studies in comparative phylogeography are conducted with a primary focus on demographic process and genetic diversity, rather than the genetics of speciation per se. Nonetheless, as we accumulate studies in comparative phylogeography, we obtain a valuable catalog—unbiased by a focus on unusual and genetically unique speciation scenarios—of the environmental, demographic, and genomic context in which speciation takes place (Moritz et al. 2009). Harvey et al. (2019) suggested that the metapopulation framework, with its emphasis on gene flow, population divergence, and local extinction, will be a useful addition to models and studies aiming to understand the speciation process. Comparative phylogeography, with its clear connections to the metapopulation concept, can help estimate the population sizes, rates of gene flow, and divergence times that accompany suites of speciation events. Such demographic factors have been argued as a key ingredient to ground modern speciation studies in the messy realities of real populations in nature (Harvey et al. 2019). Moreover, by accumulating repeated instances of divergence across specific geographic barriers, comparative phylogeography can help determine whether different lineages have responded to similar geographic contexts and landscape barriers uniformly or variably (Peñalba et al. 2019).
Finally, comparative phylogeography can help gauge the frequency of population extinction and “ephemeral speciation,” which have been suggested to play an important role in unifying the disparate estimates of speciation rates across studies (Rosenblum et al. 2012). Although population extinction and the presence of ghost populations is intrinsically challenging to detect (Slatkin 2005), a variety of phylogeographic tools now permit researchers to postulate and test for the existence of now extinct species and populations and their contribution to extant genetic diversity. Such approaches can leverage direct interrogation of the genetics of extinct populations via ancient DNA or can infer such populations by investigating anomalies in the genetic landscape of extant populations. Recent studies of extinct hominin species, such as Neanderthals, Denisovans, and other groups provide compelling examples of the former approach. Direct interrogation of ancient DNA and integration with extant samples is becoming more prevalent in nonhuman species, particularly in Africa and the Arctic (Shapiro et al. 2004; Soubrier et al. 2016; Palkopoulou et al. 2018; de Manuel et al. 2020). A few studies have leveraged enough samples from multiple ancient codistributed species to warrant status as a comparative phylogeographic study (Lorenzen et al. 2011). Notably, Lorenzen et al. (2011) found that responses of different large mammals to climate change in the Late Quarternary were idiosyncratic, reflecting the frequent finding from studies of extant populations that different species respond to similar barriers in different ways and at different times. Other lines of evidence, such as evidence for deep mtDNA lineages having been overridden by nuclear DNA gene flow from larger, expanding populations, as well as theory(Alcala and Vuilleumier 2014), have been combined with distributional modeling to infer extinction of populations by genetic swamping (Singhal and Moritz 2012).
Comparative Phylogeography as Place-Based Evolutionary Biology
Marginalizing over Geography in Comparative Population Genomics
Given its roots back to the 1980s and 1990s, it is reasonable to suggest that comparative phylogeography predates comparative population genomics, especially when we consider that population genomics as a field did not really emerge until genome-wide analyses even for single species became routine in the late 2000s. One can additionally argue that—perhaps because of its relative novelty—comparative population genomics is more trendy, and certainly more ascendant, than comparative phylogeography. Still, as reviewed earlier, there have been important conceptual and methodological innovations coming from comparative phylogeography that reinforce its relevance. However, comparative phylogeography continues to focus on the core questions of codiversification that have dominated it since its inception. Comparative population genomics, on the other hand, already seems to be asking questions that hitherto had not been asked before by single-species studies, as this special issue of GBE amply shows. Thus, it is helpful to clarify the relationships between comparative phylogeography and comparative population genetics as a means of understanding what each has to offer the other.
A major strand of comparative population genomics seeks to explain the levels of genetic diversity observed across species. We now know that multiple ecological, demographic, and phylogenetic factors can influence levels of genetic diversity, a deceptively simple summary of genetic variation that is influenced most directly by effective population size, local recombination rate, and mutation rate. This simplicity remains, but researchers now seek multiple upstream factors influencing these two primary determinants (Ellegren and Galtier 2016). The frequency with which the equilibrium level of diversity in a species is realized is now thought to be relatively rare, because pervasive linked selection tends to reduce the realized diversity compared with that expected without linked selection, especially in species with large effective population size (Corbett-Detig et al. 2015). Although the field is far from settled, some of the important suggested factors include life history traits, such as where the species lands on the r to K spectrum, including its mating system and average offspring number (Romiguier et al. 2014). Recent population genetic surveys across multiple species in a clade—studies in butterflies (Mackintosh et al. 2019) and pinnipeds (Peart et al. 2020) are recent examples—while not downplaying the role of geographic context in modulating genetic diversity, tend to marginalize over it, striving instead for a generality that is suprageographic. Geographic and demographic history necessarily plays an important role in modulating genetic diversity; but one of the main goals of comparative population genetics is to develop predictions for genetic diversity that transcend geography.
“Space is the Place” in Comparative Phylogeography
In contrast to the goals for comparative population genetics, for comparative phylogeography, “space is the place” (Battey et al. 2020). In seeking to understand how environmental variation across space and time, for a given region, has shaped the evolution of genetic diversity and opportunity for speciation, comparative phylogeography is place-based evolutionary biology. The emphasis on multiple taxa across a common region contrasts with the comparative population genomics, where often geography is explicitly ignored or factored out as a nuisance parameter, in order to make generalizations that transcend geography (Leroy et al. 2021) (table 1).
Comparative phylogeography shares with comparative population genomics a desire for generality, but that generality often comes in the form of discovering common geographic boundaries influencing the location and timing of lineage splits. A given comparative phylogeographic study is unabashedly regional—its focus is on how the specific geological and ecological history of a region may have impacted multiple organismal lineages. We suggest, however, that this regionality does not imply a more narrow focus than a typical study in comparative population genomics. Instead, although the primary focus of comparative population genomics is the genome and the factors modulating its variation, comparative phylogeography is primarily concerned with the history of organismal lineages (table 1). Secondarily, comparative population genomics is concerned with organisms in the context of their environmental histories to the extent that the environment shapes patterns in the genome; by contrast, for comparative phylogeography, genetic interactions and natural selection on the genome are secondary. As phylogeography entered the genomic era, its purview necessarily expanded to include topics traditionally the domain of population genetics, such as selection on the genome, genomic islands of differentiation, linkage disequilibrium, and selective sweeps (Edwards et al. 2015). However, at its core, phylogeography maintains its emphasis on organismal history and its link to regional environmental and landscape history.
Comparative Phylogeography, Place-Based Education, and Mutual Understanding
With its strong emphasis on geography and landscape features, and its focus on specific biomes and regions, comparative phylogeography is appealing as a means to facilitate place-based education. Conducted inclusively, comparative phylogeography can also serve as a forum for mutual understanding between scientists and local communities and indigenous groups around the world. Place-based education is education using examples from student’s local communities and environments and has emerged as an important vehicle for conveying the immediacy and relevance of scientific principles while at the same time inculcating principles of sustainability (Smith 2002, 2007). Although place-based education has usually been envisioned at the level of very local communities—much smaller in scale than the typical comparative phylogeographic study (fig. 5)—the importance of geography to place-based education has also been acknowledged, both as an object of study but also as a means of analysis and connecting different scales of inquiry (Shimeld 2012; Preston 2015). The study of phylogeography is intrinsically place-based and can help convey important scientific principles to students, particularly when paired with tangible experiences like use of museum specimens (Cook et al. 2014). By drawing attention to details of specific regions of the globe, comparative phylogeography can serve as a powerful place-based context in which students can learn geography and the landscape features that have influenced local plants and animals. Another strength of comparative phylogeography, and phylogeography in general, is its integration of organismal biology and geography. This integration and relevance to organisms found in local communities can be a powerful force in engaging students in evolutionary biology.
Comparative phylogeography can also help mediate dialogs with local and indigenous communities in ways that comparative population genomics alone would find challenging. Indigenous knowledge systems are recognized as powerful sources for place-based education and learning communities (Davidson-Hunt and O’Flaherty 2007), and we suggest comparative phylogeography has great potential for dialogue and study codesign with indigenous peoples. With its frequent references to prominent landscape features and pathways for movement of organisms, comparative phylogeography provides compelling examples not only of scientific principles with clear connections to indigenous knowledge but also of the potential for collaboration between the scientific community and indigenous communities (Moritz et al. 2013; Colwell 2016). Indeed, many comparative phylogeographic studies have been conducted in the spirit of collaboration with in-country scientists and indigenous peoples. Modern genomics and museum science, fields allied with comparative phylogeography, have both had exploitative relationships with indigenous peoples in the past. However, the record of collaboration is improving, and there is an increasing legal and moral expectation of consultative processes and a desire for mutual learning and understanding (Colwell 2016; Card et al. 2021). Recent work, such as the sequencing of the genome of the Tuatara (Sphenodon punctatus) of New Zealand, offers compelling high-profile examples of this inclusive approach to evolutionary biology (Gemmell et al. 2020).
Conclusion
Comparative phylogeography, with roots going back to the 1980s, offers an instructive counterpoint to the relatively new field of comparative population genomics. Although comparative population genomics, in its most extreme forms, eschews the details of geography in an effort to focus on selective forces acting on the genome, comparative phylogeography embraces geography and strives to link landscape history with codiversification of organismal lineages. Although the output of papers in comparative phylogeography has plateaued in recent years, we suggest that comparative phylogeography as a discipline is still vibrant and innovative, having been transformed from its roots in gene tree analysis by fields like coalescent theory and tools like simulation, trait-based hypotheses, and machine learning. The geographic breadth of comparative phylogeographic studies has increased in the last decade, with a greater contribution to the literature by countries in the Global South than in the previous two decades. Comparative phylogeography offers a compelling bridge between landscape genomics at the microgeographic level and the speciation process. Finally, with its emphasis on geography and landscape features, comparative phylogeography—more so, we suggest, than comparative population genomics—offers great opportunities for promoting place-based education and engaging local and indigenous communities. We hope this review will encourage continued dialogue and cross-fertilization between comparative population genomics and comparative phylogeography, because both fields stand to be enriched by the other.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Tim Sackton and Russ Corbett-Detig for inviting us to write this review. We thank Gustavo Bravo, Archita Sharma, Joe Cook, and Naman Goyal for helping with summarizing phylogeographic barriers in figures 2 and 3 and the references to the barriers (supplementary table S2, Supplementary Material online). We thank Elizabeth Vieira for expertly tabulating the citations for figures 1 and 4 and Supplementary Material online. We thank Brett Riddle for helpful comments and encouragement, John Wakeley for helpful discussion, and Monty Slatkin and two anonymous reviewers for helpful comments on the manuscript. Richard Gott and Robert Vanderbei provided access to Earth images with the Gott et al. (2021) projection (figs. 2 and 3).
S.V.E. acknowledges funding from the National Science Foundation Dimensions of Biodiversity Program (DEB - 1831560) and the National Institutes of Health (1R01HG011485-01). V.V.R. acknowledges funding from the SERB Early Career Grant (ECR/2016/001065), Department of Science & Technology, Government of India. C.M. thanks the Australian Research Council for support via a Laureate Fellowship, Discover and Linkage Projects.
Literature Cited
- Afonso Silva AC, et al. 2017. Tropical specialist vs. climate generalist: diversification and demographic history of sister species of Carlia skinks from northwestern Australia. Mol Ecol. 26(15):4045–4058. [DOI] [PubMed] [Google Scholar]
- Agarwal I, Ramakrishnan U.. 2017. A phylogeny of open-habitat lizards (Squamata: Lacertidae: Ophisops) supports the antiquity of Indian grassy biomes. J Biogeogr. 44(9):2021–2032. [Google Scholar]
- Alcala N, Vuilleumier S.. 2014. Turnover and accumulation of genetic diversity across large time-scale cycles of isolation and connection of populations. Proc Biol Sci. 281(1794):20141369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW. 2017. Genetics and the conservation of natural populations: allozymes to genomes. Mol Ecol. 26(2):420–430. [DOI] [PubMed] [Google Scholar]
- Alter SE, Munshi-South J, Stiassny MLJ.. 2017. Genomewide SNP data reveal cryptic phylogeographic structure and microallopatric divergence in a rapids-adapted clade of cichlids from the Congo River. Mol Ecol. 26(5):1401–1419. [DOI] [PubMed] [Google Scholar]
- Alvarado-Serrano DF, Knowles LL.. 2014. Ecological niche models in phylogeographic studies: applications, advances and precautions. Mol Ecol Resour. 14(2):233–248. [DOI] [PubMed] [Google Scholar]
- Amaral FR, Cabanne GS, Michelangeli FA, Miyaki CY. Forthcoming 2021. Exploring the evolution of a hyperdiverse Neotropical biome, the Atlantic Forest. Mol Phyl Evol. Available from: https://www.sciencedirect.com/journal/molecular-phylogenetics-and-evolution/special-issue/10GHQ3HF5V6. [Google Scholar]
- Avise JC, Bowen BW, Ayala FJ.. 2016. In the light of evolution X: comparative phylogeography. Proc Natl Acad Sci U S A. 113(29):7957–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Lansman RA, Shade RO.. 1979. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics 92(1):279–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, et al. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 18(1):489–522. [Google Scholar]
- Bagley JC, Johnson JB.. 2014. Phylogeography and biogeography of the lower Central American Neotropics: diversification between two continents and between two seas. Biol Rev Camb Philos Soc. 89(4):767–790. [DOI] [PubMed] [Google Scholar]
- Barber BR, Klicka J.. 2010. Two pulses of diversification across the Isthmus of Tehuantepec in a montane Mexican bird fauna. Proc Biol Sci. 277(1694):2675–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow LN, Fonseca EM, da Thompson CEP, Carstens BC.. 2020. Predicting amphibian intraspecific diversity with machine learning: challenges and prospects for integrating traits, geography, and genetic data. Mol Ecol Resour. 1–14. doi: 10.1111/1755-0998.13303. [DOI] [PubMed] [Google Scholar]
- Battey CJ, Ralph PL, Kern AD.. 2020. Space is the place: effects of continuous spatial structure on analysis of population genetic data. Genetics 215(1):193–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, et al. 2010. In defence of model-based inference in phylogeography. Mol Ecol. 19(3):436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddek M, et al. 2018. Comparative phylogeography of amphibians and reptiles in Algeria suggests common causes for the east-west phylogeographic breaks in the Maghreb. PLoS One 13(8):e0201218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J.. 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152(2):763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheregaray LB. 2008. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol Ecol. 17(17):3754–3774. [DOI] [PubMed] [Google Scholar]
- Bermingham E, Avise JC.. 1986. Molecular zoogeography of freshwater fishes in the southeastern United States. Genetics 113(4):939–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham E, Moritz C.. 1998. Comparative phylogeography: concepts and applications. Mol Ecol. 7(4):367–369. [Google Scholar]
- Bernatchez L, Wilson CC.. 1998. Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol. 7(4):431–452. [Google Scholar]
- Besnard G, et al. 2016. Valuing museum specimens: high-throughput DNA sequencing on historical collections of New Guinea crowned pigeons (Goura). Biol J Linn Soc. 117(1):71–82. [Google Scholar]
- Bodmer W. 2015. Genetic characterization of human populations: from ABO to a genetic map of the British people. Genetics 199(2):267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt F, et al. 2004. Local endemism within the Western Ghats-sri Lanka biodiversity hotspot. Science 306(5695):479–481. [DOI] [PubMed] [Google Scholar]
- Bradburd GS, Ralph PL.. 2019. Spatial population genetics: it’s about time. Annu Rev Ecol Evol Syst. 50(1):427–449. [Google Scholar]
- Buckner JC, Sanders RC, Faircloth BC, Chakrabarty P.. 2021. The critical importance of vouchers in genomics. Elife 10:e68264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnefeld L, Hearn J, Stone GN, Lohse K.. 2018. Whole-genome data reveal the complex history of a diverse ecological community. Proc Natl Acad Sci U S A. 115(28):E6507–E6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney CW, Brumfield RT.. 2009. Ecology predicts levels of genetic differentiation in neotropical birds. Am Nat. 174(3):358–368. [DOI] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC.. 1987. Mitochondrial DNA and human evolution. Nature 325(6099):31–36. [DOI] [PubMed] [Google Scholar]
- Card D, Shapiro B, Giribet G, Moritz C, Edwards SV. Forthcoming 2021. Museum genomics. Annu Rev Genet. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C.. 2009. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323(5915):785–789. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Morales AE, Field K, Pelletier TA.. 2018. A global analysis of bats using automated comparative phylogeography uncovers a surprising impact of Pleistocene glaciation. J Biogeogr. 45(8):1795–1805. [Google Scholar]
- Carstens BC, Richards CL.. 2007. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution 61(6):1439–1454. [DOI] [PubMed] [Google Scholar]
- Castex GM. 1993. Frames of reference: the effects of ethnocentric map projections on professional practice. Soc Work. 38:685–693. [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF.. 1967. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Colwell C. 2016. Collaborative archaeologies and descendant communities. Annu Rev Anthropol. 45(1):113–127. [Google Scholar]
- Cook JA, et al. 2014. Natural history collections as emerging resources for innovative education. Bioscience 64(8):725–734. [Google Scholar]
- Corbett-Detig RB, Hartl DL, Sackton TB.. 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13(4):e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracraft J. 1982. Geographic differentiation, cladistics, and vicariance biogeography: reconstructing the tempo and mode of evolution. Am Zool. 22(2):411–424. [Google Scholar]
- Cracraft J. 1986. Origin and evolution of continental biotas: speciation and historical congruence within the Australian avifauna. Evolution 40(5):977–996. [DOI] [PubMed] [Google Scholar]
- Cracraft J. 1991. Patterns of diversification within continental biotas: hierarchical congruence among areas of endemism of Australian vertebrates. Aust Syst Bot. 4:211–227. [Google Scholar]
- Cuervo A. 2013. Evolutionary Assembly of the Neotropical Montane Avifauna. LSU Doctoral Dissertations. Available from: https://digitalcommons.lsu.edu/gradschool_dissertations/275. 131 pp.
- Davidson-Hunt IJ, O’Flaherty RM.. 2007. Researchers, indigenous peoples, and place-based learning communities. Soc Nat Resour. 20(4):291–305. [Google Scholar]
- de Medeiros BAS, Farrell BD. 2020. Evaluating insect-host interactions as a driver of species divergence in palm flower weevils. Commun Biol. 3(1):749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, et al. 2020. The evolutionary history of extinct and living lions. Proc Natl Acad Sci U S A. 117(20):10927–10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista JD, et al. 2020. Population genomic response to geographic gradients by widespread and endemic fishes of the Arabian Peninsula. Ecol Evol. 10(10):4314–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolman G, Joseph L.. 2012. A species assemblage approach to comparative phylogeography of birds in southern Australia. Ecol Evol. 2(2):354–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AWF, Cavalli-Sforza LL.. 1963. The reconstruction of evolution. Ann Hum Genet. 27:104–105. [Google Scholar]
- Edwards SV. 1993. Long-distance gene flow in a cooperative breeder detected in genealogies of mitochondrial DNA sequences. Proc R Soc Lond B Biol Sci. 252:177–185. [DOI] [PubMed] [Google Scholar]
- Edwards SV. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63(1):1–19. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Beerli P.. 2000. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54(6):1839–1854. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Hopkins R, Mallet J.. 2019. Speciation. In: The theory of evolution. Chicago: University of Chicago Press. p. 296–318. Available from: https://press.uchicago.edu/ucp/books/book/chicago/T/bo45713136.html (accessed April 2, 2021). [Google Scholar]
- Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C.. 2016. Reticulation, divergence, and the phylogeography-phylogenetics continuum. Proc Natl Acad Sci U S A. 113(29):8025–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Shultz AJ, Campbell-Staton SC.. 2015. Next-generation sequencing and the expanding domain of phylogeography. J Vertebr Biol. 64(3):187–206. [Google Scholar]
- Ellegren H, Galtier N.. 2016. Determinants of genetic diversity. Nat Rev Genet. 17(7):422–433. [DOI] [PubMed] [Google Scholar]
- Espíndola A, et al. 2016. Identifying cryptic diversity with predictive phylogeography. Proc R Soc B Biol Sci. 283(1841):20161529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Novembre J, Schneider S.. 2000. SIMCOAL: a general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J Hered. 91(6):506–509. [DOI] [PubMed] [Google Scholar]
- Floeter SR, et al. 2007. Atlantic reef fish biogeography and evolution. J Biogeogr. 35(1):22–47. [Google Scholar]
- Fonseca EM, da Colli GR, Werneck FP, Carstens BC.. 2021. Phylogeographic model selection using convolutional neural networks. Mol Ecol Resour. doi: 10.1111/1755-0998.13427. [DOI] [PubMed]
- François O, et al. 2010. Principal component analysis under population genetic models of range expansion and admixture. Mol Biol Evol. 27(6):1257–1268. [DOI] [PubMed] [Google Scholar]
- Freilich X, et al. ; Evolutionary Genetics - Class of 2013. 2016. Comparative phylogeography of Ethiopian anurans: impact of the Great Rift Valley and Pleistocene climate change. BMC Evol Biol. 16(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire P-A. 2020. Comparative genomics approach to evolutionary process connectivity. Evol Appl. 13(6):1320–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick RC, et al. 2015. The evolution of phylogeographic data sets. Mol Ecol. 24(6):1164–1171. [DOI] [PubMed] [Google Scholar]
- Gemmell NJ, et al. ; Ngatiwai Trust Board. 2020. The tuatara genome reveals ancient features of amniote evolution. Nature 584(7821):403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JR, Goldberg DM, Vanderbei RJ.. 2021. Flat Maps that improve on the Winkel Tripel. ArXiv210208176 Astro-Ph. Available from: http://arxiv.org/abs/2102.08176 (accessed April 12, 2021).
- Gutiérrez-García TA, Vázquez-Domínguez E.. 2011. Comparative phylogeography: designing studies while surviving the process. BioScience 61(11):857–868. [Google Scholar]
- Haemer KW. 1949. Area bias in map presentation. Am Stat. 3(2):19. [Google Scholar]
- Harvey MG, Singhal S, Rabosky DL.. 2019. Beyond reproductive isolation: demographic controls on the speciation process. Annu Rev Ecol Evol Syst. 50(1):75–95. [Google Scholar]
- Helmstetter AJ, Béthune K, Kamdem NG, Sonké B, Couvreur TLP.. 2020. Individualistic evolutionary responses of Central African rain forest plants to Pleistocene climatic fluctuations. Proc Natl Acad Sci U S A. 117(51):32509–32518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 58(3):247–276. [Google Scholar]
- Hewitt GM. 1999. Post-glacial re-colonization of European biota. Biol J Linn Soc. 68(1–2):87–112. [Google Scholar]
- Hey J, Machado CA.. 2003. The study of structured populations—new hope for a difficult and divided science. Nat Rev Genet. 4(7):535–543. [DOI] [PubMed] [Google Scholar]
- Hickerson MJ, et al. 2010. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol Phylogenet Evol. 54:291–301. [DOI] [PubMed] [Google Scholar]
- Hugall A, Moritz C, Moussalli A, Stanisic J.. 2002. Reconciling paleodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875). Proc Natl Acad Sci U S A. 99(9):6112–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydeman ME, et al. 2017. Prevalence and genetic diversity of Batrachochytrium dendrobatidis in Central African island and continental amphibian communities. Ecol Evol. 7(19):7729–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanth KK, Nichols JD, Karanth KU, Hines JE, Christensen NL.. 2010. The shrinking ark: patterns of large mammal extinctions in India. Proc Biol Sci. 277(1690):1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman JFC. 2000. Origins of the Coalescent: 1974–1982. Genetics 156(4):1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LL. 2009. Statistical phylogeography. Annu Rev Ecol Evol Syst. 40(1):593–612. [Google Scholar]
- Leroy T, et al. 2021. Island songbirds as windows into evolution in small populations. Curr Biol. 31(6):1303–1310.e4. [DOI] [PubMed] [Google Scholar]
- Lohman DJ, et al. 2011. Biogeography of the Indo-Australian Archipelago. Annu Rev Ecol Evol Syst. 42(1):205–226. [Google Scholar]
- Lorenzen ED, Heller R, Siegismund HR.. 2012. Comparative phylogeography of African savannah ungulates. Mol Ecol. 21(15):3656–3670. [DOI] [PubMed] [Google Scholar]
- Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479(7373):359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh A, et al. 2019. The determinants of genetic diversity in butterflies. Nat Commun. 10(1):3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel S, Holderegger R.. 2013. Ten years of landscape genetics. Trends Ecol Evol. 28(10):614–621. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P.. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 18(4):189–197. [Google Scholar]
- Manel S, et al. 2020. Global determinants of freshwater and marine fish genetic diversity. Nat Commun. 11(1):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, et al. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meegaskumbura M, et al. 2002. Sri Lanka: an amphibian hot spot. Science 298(5592):379. [DOI] [PubMed] [Google Scholar]
- Millette KL, et al. 2020. No consistent effects of humans on animal genetic diversity worldwide. Ecol Lett. 23(1):55–67. [DOI] [PubMed] [Google Scholar]
- Miraldo A, et al. 2016. An Anthropocene map of genetic diversity. Science 353(6307):1532–1535. [DOI] [PubMed] [Google Scholar]
- Moritz C, Ens EJ, Potter S, Catullo RA.. 2013. The Australian monsoonal tropics: an opportunity to protect unique biodiversity and secure benefits for Aboriginal communities. Pac Conserv Biol. 19(4):343–355. [Google Scholar]
- Moritz C, et al. 2009. Identification and dynamics of a cryptic suture zone in tropical rainforest. Proc Biol Sci. 276(1660):1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka LN, Bechtoldt CL, Henriques LMP, Brumfield RT.. 2012. The role of physical barriers in the location of avian suture zones in the Guiana Shield, Northern Amazonia. Am Nat. 179(4):E115–E132. [DOI] [PubMed] [Google Scholar]
- Naka LN, Brumfield RT.. 2018. The dual role of Amazonian rivers in the generation and maintenance of avian diversity. Sci Adv. 4(8):eaar8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama N. 2021. Museum specimens: an overlooked and valuable material for conservation genetics. Ecol Res. 36(1):13–23. [Google Scholar]
- Narayan A, Berger B, Cho H.. 2021. Assessing single-cell transcriptomic variability through density-preserving data visualization. Nat Biotechnol. 39(6):765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH.. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Slatkin M.. 2009. Likelihood-based inference in isolation-by-distance models using the spatial distribution of low-frequency alleles. Evolution 63(11):2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyari AS, Joseph L. 2013. Comparative phylogeography of Australo-Papuan mangrove-restricted and mangrove-associated avifaunas. Biol J Linn Soc Lond. 109(3):574–598. [Google Scholar]
- Oaks JR. 2014. An improved approximate-Bayesian model-choice method for estimating shared evolutionary history. BMC Evol Biol. 14:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks JR. 2019. Full Bayesian comparative phylogeography from genomic data. Syst Biol. 68(3):371–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald JA, Overcast I, Mauck WM, Andersen MJ, Smith BT.. 2017. Isolation with asymmetric gene flow during the nonsynchronous divergence of dry forest birds. Mol Ecol. 26(5):1386–1400. [DOI] [PubMed] [Google Scholar]
- Overcast I, Bagley JC, Hickerson MJ.. 2017. Strategies for improving approximate Bayesian computation tests for synchronous diversification. BMC Evol Biol. 17(1):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkopoulou E, et al. 2018. A comprehensive genomic history of extinct and living elephants. Proc Natl Acad Sci U S A. 115(11):E2566–E2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal M, Beaumont MA.. 2007. The automation and evaluation of nested clade phylogeographic analysis. Evolution 61(6):1466–1480. [DOI] [PubMed] [Google Scholar]
- Papadopoulou A, Knowles LL.. 2016. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc Natl Acad Sci U S A. 113(29):8018–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz A, Ibanez R, Lips KR, Crawford AJ.. 2015. Testing the role of ecology and life history in structuring genetic variation across a landscape: a trait-based phylogeographic approach. Mol Ecol. 24(14):3723–3737. [DOI] [PubMed] [Google Scholar]
- Peart CR, et al. 2020. Determinants of genetic variation across eco-evolutionary scales in pinnipeds. Nat Ecol Evol. 4(8):1095–1104. [DOI] [PubMed] [Google Scholar]
- Peñalba JV, Joseph L, Moritz C.. 2019. Current geography masks dynamic history of gene flow during speciation in northern Australian birds. Mol Ecol. 28(3):630–643. [DOI] [PubMed] [Google Scholar]
- Petit RJ. 2008. The coup de grâce for the nested clade phylogeographic analysis? Mol Ecol. 17(2):516–518. [DOI] [PubMed] [Google Scholar]
- Potter S, et al. 2018. Pleistocene climatic changes drive diversification across a tropical savanna. Mol Ecol. 27(2):520–532. [DOI] [PubMed] [Google Scholar]
- Preston L. 2015. The place of place-based education in the Australian Primary Geography Curriculum. Geogr Educ. 28:41–49. [Google Scholar]
- Provost KL, Myers EA, Smith BT.. 2021. Community phylogeographic patterns reveal how a barrier filters and structures taxa in North American warm deserts. J Biogeogr. 48(6):1267–1283. [Google Scholar]
- Puritz JB, et al. 2017. Life-history predicts past and present population connectivity in two sympatric sea stars. Ecol Evol. 7(11):3916–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo do Amaral F, Albers PK, Edwards SV, Miyaki CY.. 2013. Multilocus tests of Pleistocene refugia and ancient divergence in a pair of Atlantic Forest antbirds (Myrmeciza). Mol Ecol. 22(15):3996–4013. [DOI] [PubMed] [Google Scholar]
- Reddy S. 2014. What’s missing from avian global diversification analyses? Mol Phylogenet Evol. 77:159–165. [DOI] [PubMed] [Google Scholar]
- Ribeiro TD, Batalha H, Silveira LF, Miyaki CY, Maldonado-Coelho M.. 2020. Life history and ecology might explain incongruent population structure in two co-distributed montane bird species of the Atlantic Forest. Mol Phylogenet Evol. 153:106925. [DOI] [PubMed] [Google Scholar]
- Riddle BR. 2016. Comparative phylogeography clarifies the complexity and problems of continental distribution that drove A. R. Wallace to favor islands. Proc Natl Acad Sci U S A. 113(29):7970–7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienzo AD, Wilson AC.. 1991. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 88(5):1597–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissler LJ. 2016. Union of phylogeography and landscape genetics. Proc Natl Acad Sci U S A. 113(29):8079–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin VV, Gupta P, Thatte P, Ramakrishnan U.. 2015. Islands within islands: two montane palaeo-endemic birds impacted by recent anthropogenic fragmentation. Mol Ecol. 24(14):3572–3584. [DOI] [PubMed] [Google Scholar]
- Robin VV, Sinha A, Ramakrishnan U.. 2010. Ancient geographical gaps and paleo-climate shape the phylogeography of an endemic bird in the sky islands of Southern India. PLoS One 5(10):e13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JD, Bunnefeld L, Hearn J, Stone GN, Hickerson MJ.. 2014. ABC inference of multi-population divergence with admixture from unphased population genomic data. Mol Ecol. 23(18):4458–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, et al. 2014. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515(7526):261–263. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Nordborg M.. 2002. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet. 3(5):380–390. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, et al. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol Biol. 39(2):255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebelhut LM, Dawson MN.. 2018. Correlates of population genetic differentiation in marine and terrestrial environments. J Biogeogr. 45(11):2427–2441. [Google Scholar]
- Schmitt CJ, Cook JA, Zamudio KR, Edwards SV.. 2018. Museum specimens of terrestrial vertebrates are sensitive indicators of environmental change in the Anthropocene. Philos Trans R Soc Lond B Biol Sci. 374(1763):20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CJ, Cunningham M, Moritz C.. 1998. Comparative phylogeography and the history of endemic vertebrates in the Wet Tropics rainforests of Australia. Mol Ecol. 7(4):487–498. [Google Scholar]
- Shapiro B, et al. 2004. Rise and fall of the Beringian steppe bison. Science 306(5701):1561–1565. [DOI] [PubMed] [Google Scholar]
- Shimeld J. 2012. (Dis)Entangling place. Geogr Educ. 25: 39–42. [Google Scholar]
- Singhal S, Bi K.. 2017. History cleans up messes: the impact of time in driving divergence and introgression in a tropical suture zone. Evolution 71(7):1888–1899. [DOI] [PubMed] [Google Scholar]
- Singhal S, Moritz C.. 2012. Testing hypotheses for genealogical discordance in a rainforest lizard. Mol Ecol. 21(20):5059–5072. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 1985. Rare alleles as indicators of gene flow. Evolution 39(1):53–65. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 2005. Seeing ghosts: the effect of unsampled populations on migration rates estimated for sampled populations. Mol Ecol. 14(1):67–73. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR.. 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129(2):555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP.. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123(3):603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BT, et al. 2014. The drivers of tropical speciation. Nature 515(7527):406–409. [DOI] [PubMed] [Google Scholar]
- Smith GA. 2002. Place-based education: learning to be where we are. Phi Delta Kappan. 83(8):584–594. [Google Scholar]
- Smith GA. 2007. Place-based education in the global age: local diversity. 1st ed. New York: Routledge. [Google Scholar]
- Soubrier J, et al. 2016. Early cave art and ancient DNA record the origin of European bison. Nat Commun. 7:13158. doi: 10.1038/ncomms13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, et al. 2019. Integrating life history traits into predictive phylogeography. Mol Ecol. 28(8):2062–2073. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1983. Evolutionary relationships of DNA sequences in finite populations. Genetics 105(2):437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. 1998. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol. 7(4):381–397. [DOI] [PubMed] [Google Scholar]
- Templeton AR. 2010. Coalescent-based, maximum likelihood inference in phylogeography. Mol Ecol. 19(3):431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis S, et al. 2020. Evolutionary history and past climate change shape the distribution of genetic diversity in terrestrial mammals. Nat Commun. 11(1):2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom G, et al. 2020. Climatic dynamics and topography control genetic variation in Atlantic Forest montane birds. Mol Phylogenet Evol. 148:106812. [DOI] [PubMed] [Google Scholar]
- Toews DPL, et al. 2016. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr Biol. 26(17):2313–2318. [DOI] [PubMed] [Google Scholar]
- van Asch B, et al. 2019. Phylogeography, genetic diversity, and population structure of Nile crocodile populations at the fringes of the southern African distribution. PLoS One 14(12):e0226505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar SP, Menezes RC, Jayarajan A, Shanker K.. 2016. Glaciations, gradients, and geography: multiple drivers of diversification of bush frogs in the Western Ghats Escarpment. Proc R Soc B Biol Sci. 283(1836):20161011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujakovic P. 1989. Mapping for world development. Geography 74:97–105. [Google Scholar]
- Xue AT, Hickerson MJ.. 2015. The aggregate site frequency spectrum for comparative population genomic inference. Mol Ecol. 24(24):6223–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue AT, Hickerson MJ.. 2017. multi-dice: r package for comparative population genomic inference under hierarchical co-demographic models of independent single-population size changes. Mol Ecol Resour. 17(6):e212–e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue AT, Hickerson MJ.. 2020. Comparative phylogeographic inference with genome-wide data from aggregated population pairs. Evolution 74(5):808–830. [DOI] [PubMed] [Google Scholar]
- Yao L, Li H, Martin RD, Moreau CS, Malhi RS.. 2017. Tracing the phylogeographic history of Southeast Asian long-tailed macaques through mitogenomes of museum specimens. Mol Phylogenet Evol. 116:227–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.