Abstract
The Hengduan Mountains Region (HMR) is the largest “evolutionary frontier” of the northern temperate zone, and the origin and maintenance of species in this area is a research hotspot. Exploring species-specific responses to historical and contemporary environmental changes will improve our understanding of the role of this region in maintaining biodiversity. In this study, mitochondrial and microsatellite diversities were used to assess the contributions of paleogeological events, Pleistocene climatic oscillations, and contemporary landscape characteristics to the rapid intraspecific diversification of Liangshantriton taliangensis, a vulnerable amphibian species endemic to several sky-island mountains in the southeastern HMR. Divergence date estimations suggested that the East Asian monsoon, local uplifting events (Xigeda Formation strata), and Early-Middle Pleistocene transition (EMPT) promoted rapid divergence of L. taliangensis during the Pleistocene, yielding eight mitochondrial lineages and six nuclear genetic lineages. Moreover, population genetic structures were mainly fixed through isolation by resistance. Multiple in situ refugia were identified by ecological niche models and high genetic diversity, which played crucial roles in the persistence and divergence of L. taliangensis during glacial-interglacial cycles. Dramatic climatic fluctuations further promoted recurrent isolation and admixing of populations in scattered glacial refugia. The apparent mitonuclear discordance was likely the result of introgression by secondary contact and/or female-biased dispersal. Postglacial expansion generated two major secondary contact zones (Ganluo (GL) and Chuhongjue (CHJ)). Identification of conservation management units and dispersal corridors offers important recommendations for the conservation of this species.
Keywords: Liangshantriton taliangensis, Phylogeography, Landscape genetics, Mitonuclear discordance
INTRODUCTION
Pleistocene climatic oscillations and Meso-Cenozoic intracontinental orogenesis are considered essential drivers of the current geographical distribution and genetic divergence of organisms (Favre et al., 2015; Gillespie & Roderick, 2014; Rana et al., 2021). Topographical changes, such as mountain and river systems caused by orogenesis, are commonly regarded as geographical barriers that promote biodiversity, especially in species with limited dispersal abilities (Rana et al., 2021; Sánchez-Montes et al., 2018). Mountainous regions often become hotspots of species diversity and lineage divergence (Kozak & Wiens, 2010; Noroozi et al., 2018), with the uplift of the Qinghai-Xizang (Tibet) Plateau (QTP) receiving considerable attention (He et al., 2021). The barrier effect caused by this uplift has been documented for various fauna and flora taxa from the QTP and adjacent areas (Huang et al., 2017; Wang et al., 2017; Ye et al., 2019). However, the geological history of these regions is highly complex, and many details on the timing and patterns of geological uplift remain controversial (Harrison et al., 1992; Li & Fang, 1999; Renner, 2016; Spicer et al., 2021; Su et al., 2019). Thus, interpretation of the effect of QTP uplift on species formation and lineage divergence needs to focus on differences in biota from the QTP and surrounding regions (Xing & Ree, 2017). This is particularly important for relatively young species that developed from the Miocene onward. Uplift of the QTP during the Miocene further contributed to the establishment of the South Asian and East Asian monsoon climate (An et al., 2001; Liu & Dong, 2013). Furthermore, QTP uplift shaped the warm and humid environment, dominated by tropical and subtropical vegetation on the southeastern edge, especially the Hengduan Mountains Region (HMR). Many unoccupied ecological niches formed by new habitats in the region promoted the evolutionary radiation of organisms and enhanced their diversification potential (Favre et al., 2015; Wang et al., 2015, 2018; Xing & Ree, 2017).
Climatic fluctuations in the Quaternary caused repeated and extreme environmental changes, which profoundly affected the distribution, dispersal, and adaptation of organisms (Davis & Shaw, 2001; Hewitt, 2000). Different distributional and physiological characteristics allowed species to respond to glaciation differently (Huang et al., 2017; Qu et al., 2010; Yu et al., 2013). Alpine species adapted to temperate and warm habitats retreated southward to low-altitude refugia during glacial periods, thus avoiding glacial cover and the cold-dry climate, and expanded and recolonized the northern and plateau areas when the climate again warmed (Gutiérrez-Rodríguez et al., 2017; Qiu et al., 2011). Cold-tolerant species or species with distribution areas in climatically stable zones often survivedin situ instead of migrating to refugia (Sánchez-Montes et al., 2019; Wang et al., 2017). Microrefugia play an important role in the rapid divergence and persistence of lineages in mountainous regions with severe climatic fluctuations (He et al., 2021; Rana et al., 2021; Vasconcellos et al., 2019). Furthermore, shifts in species range caused by glacial-interglacial cycles provide recurrent opportunities for expansion and admixture of lineages, thus facilitating the occurrence of mitonuclear discordance (Dufresnes et al., 2020; Phuong et al., 2017).
The HMR is a transition region between the eastern circum-Pacific belt and the western ancient Mediterranean belt and is a constitutional part of the QTP. The area (~ N23°–33° and E94°–103°) is characterized by a series of parallel high mountains and great rivers running north to south. The terrain becomes gentle from the northwest to southeast, but with sharp altitudinal differences. The HMR, Yunnan-Guizhou Plateau, and Bashan Mountains are known as “the southwest China sky-island complex” (He & Jiang, 2014). Dispersal ability and habitat preference may affect the response patterns of species to landscape features (Robertson et al., 2018). The mountains and rivers of the sky-island complex constitute a potential barrier for gene flow and thus may promote rapid species diversification (Atlas & Fu, 2019; Deng et al., 2020; Pan et al., 2020) or may provide dispersal corridors for other species (Qu et al., 2011; Zhang et al., 2010). The southeastern fringe of the HMR consists of large ice-free areas, especially at low altitudes, which are considered as glacial refugia for a variety of plants and animals (Xie et al., 2019; Zhan et al., 2011). The HMR has a unique geographical history (Xing & Ree, 2017). Notably, while the area likely uplifted during the late Miocene to Pliocene (Favre et al., 2015), recent fossil evidence indicates that part of the HMR may have uplifted earlier, i.e., during the early Oligocene (Su et al., 2019). Evolutionary radiation of sky-island species can often be traced to the Pleistocene or earlier, especially for endemic species (He & Jiang, 2014; Yu et al., 2013). The HMR plays an important role in maintaining biodiversity and is considered the largest “evolutionary frontier” of the northern temperate zone (Boufford, 2014; López‐Pujol et al., 2011) and one of the most vulnerable biodiversity hotspots due to climate change and human disturbance (Ding et al., 2020; Myers et al., 2000). The inter-relationships between specific sky-island species responses and topography as well as climatic fluctuations in the HMR need to be further explored (He & Jiang, 2014). This is particularly important for amphibians with generally weak dispersal ability and more apparent genetic structure.
The Taliang knobby newt, Liangshantriton taliangensis Liu, 1950 belongs to Salamandridae Goldfuss, 1820 (Fei et al., 2012). The species is endemic to several sky-island mountains in the southeastern HMR and is distributed at altitudes from 1 390 to 3 000 m a.s.l. (Fei et al., 2012), where it usually inhabits ponds and wetlands with high humidity and heavy vegetation. Its distribution area is in the transition zone between the first and second steps of China’s terrain, containing a diverse topography and belonging to the branches of the Daxueshan Mountains. The L. taliangensis population continues to decline and is threatened by habitat degradation and excessive collection. Consequently, it has been classified as vulnerable by the International Union for Conservation of Nature red list (IUCN SSC Amphibian Specialist Group, 2020) and the red list of China’s vertebrates (Jiang et al., 2016). Research on the origin and lineage divergence of L. taliangensis not only improves our understanding of the interaction mechanism between historical and contemporary environmental factors and sky-island species divergence in the southwest mountainous area in China (especially the HMR), but also provides a reference for the conservation of L. taliangensis.
In this study, both mitochondrial and microsatellite markers were used to assess the rapid lineage divergence and genetic structure of L. taliangensis populations in southeastern HMR. The ecological niche model (ENM) and landscape genetics (isolation-by-resistance (IBR) and isolation-by-environment (IBE)) were used to evaluate the role of regional uplift events, Quaternary climatic oscillations, and landscape factors on intraspecific diversification. Based on the study objectives, we: (a) explored the phylogenetic relationship and genetic structure of L. taliangensis; (b) evaluated the historical and contemporary landscape and climatic factors that contributed to the rapid divergence of L. taliangensis; (c) identified the responses of L. taliangensis to Pleistocene glaciation; and (d) suggested conservation implications for L. taliangensis.
MATERIALS AND METHODS
Sampling, DNA extraction, and amplification
Tissue samples (426) of L. taliangensis were collected from 16 sites throughout the mountain ranges of the southeastern HMR in China, including the Gonggar (GG), Xiaoxiangling (XXL), and Liangshan mountains (LS), which include Xiaoliangshan (XLS) and Daliangshan (DLS) (Figure 1; Supplementary Table S1). All procedures followed the guidelines established by the Animal Care and Use Committee of the Chengdu Institute of Biology, Chinese Academy of Sciences (Permit No.: 2017-AR-JJP-03). Three mitochondrial genes were amplified, including NADH dehydrogenase subunit 2 (ND2), cytochrome b (cyt b), and cytochrome c oxidase subunit I (COI). Multiple primer pairs were used for each gene amplification (Supplementary Table S2). Nine species from four clades of Tylototriton sensu lato (s.l.) were used as outgroups to root the trees (Wang et al., 2018). Among these species, the DNA of Tylototriton pseudoverrucosus Hou, Gu, Zhang, Zeng, and Lu, 2012 was extracted from a specimen (Specimen No.: CIB100802), while all others were obtained from the GenBank database (Supplementary Table S3). Eleven high polymorphism microsatellite markers (DLY32,DLY39, DLY42,DLY74, DLY76, DLY96, DLY124, DLY131, DLY134, DLY148, and DLY153) were developed and used to characterize the genotypes of each individual (Chen et al., 2019; Shu, 2020). Further details are presented in the Supplementary Methods.
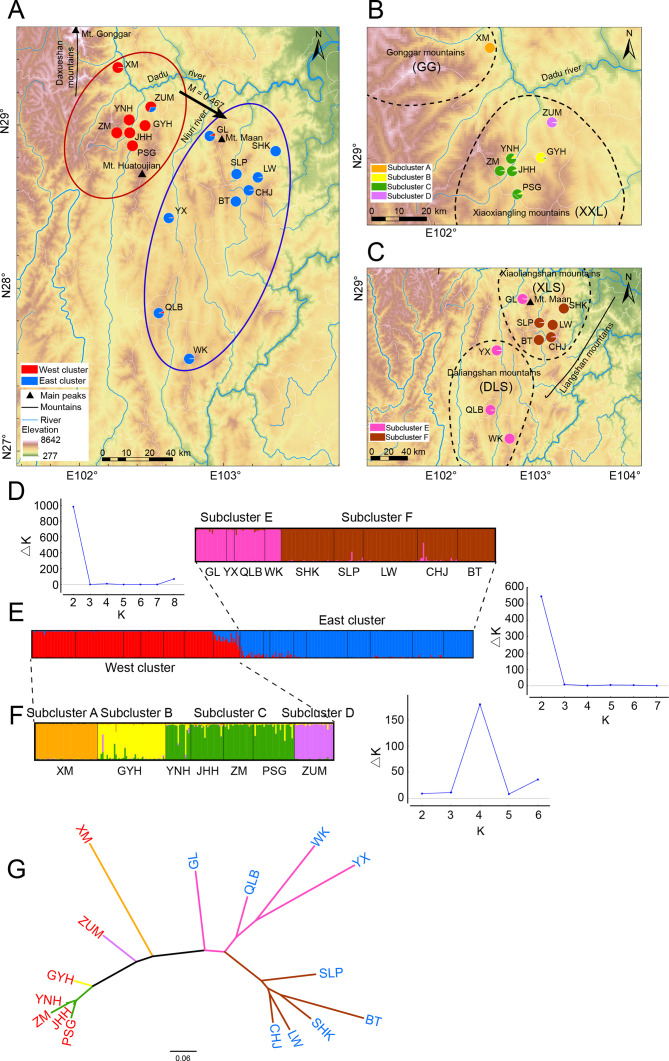
Figure 1.
Results of the Bayesian clustering analysis across Liangshantriton taliangensis distribution range
Black dashed line represents four main distribution areas of L. taliangensis (Gonggar (GG), Xiaoxiangling (XXL), Xiaoliangshan (XLS), and Daliangshan mountains (DLS)). A: Geographic distribution of two main genetic clusters, as identified by microsatellite data. West cluster (red) contains populations of XXL and GG mountains (XXL-GG); East cluster (blue) contains populations of Liangshan Mountains (LS), including DLS and XLS. Black arrows indicate gene flow. B: Geographic distribution of four subclusters of West cluster. Subcluster A (orange) contains populations of GG mountains; subclusters B (yellow), C (green), and D (purple) belong to XXL mountains. C: Geographic distribution of two subclusters in East cluster. Subcluster E (magenta) contains DLS and GL populations; subcluster F (brown) contains populations of XLS mountains except for GL. Populations are represented by pie charts, which also represent assignment probabilities between clusters or subclusters. Population codes, sampling information, and number of analyzed individuals are shown in Supplementary Table S1. D: Plot on left shows functional relationship between ΔK and K; K value corresponding to ΔKmax represents optimal grouping; right plot shows membership assignment of East cluster for K=2. E: Left plot shows membership assignment of all populations for K=2; right plot shows maximum value of ΔK corresponding to K=2. F: Left plot shows membership assignment of West cluster for K=4; right plot represents maximum value of ΔK corresponding to K=4. Black lines separate populations. G: Neighbor-joining tree of L. taliangensis populations based on DA distances. Branch color represents subclusters (A–F) results of STRUCTURE, population code color represents two main clusters.
Mitochondrial DNA (mtDNA) analysis
DNA sequences were edited by MEGA v6.06 (Tamura et al., 2013) and aligned using SeaView v4.7 (Gouy et al., 2010). Each protein-coding DNA sequence could be translated into amino acid sequences and no nucleotide gaps were found. Lasergene v7.1 (DNASTAR, USA) was used to combine gene fragments (cyt b+ND2+COI) for further phylogenetic analysis.
DNASP v5.1 (Librado & Rozas, 2009) was used to generate the haplotype file, and to calculate haplotypic diversity (Hd) and nucleotide diversity (π). Geographic patterns of Hd and π were visualized using the inverse distance weighted (IDW) method in ArcGIS v10.3 (ESRI, USA). Haplotype networks were generated with the TCS network module (Clement et al., 2000) in PopART v1.7 (Leigh & Bryant, 2015). PartitionFinder v2.1.1 (Lanfear et al., 2017) was used to identify the best partition scheme of mitochondrial concatenated genes (cytb+ND2+COI) and to explore the best evolutionary model for each partition based on Bayesian Information Criterion (BIC) and the greedy algorithm. Bayesian inference (BI) analyses were conducted in MrBayes v.3.2.2 (Ronquist et al., 2012) with four Markov chain Monte Carlo (MCMC) chains of 20 000 000 generations, with samples taken every 1 000 generations and the first 25% discarded as burn-in. Maximum-likelihood (ML) analyses were performed in RAxML v7.0.4 (Silvestro & Michalak, 2012) with 1 000 bootstrap replicates for the evaluation of branch confidences. Optimal trees were rooted and visualized using Figtree v1.4.2 (Rambaut & Drummond, 2012).
The two-step method (Hedges & Kumar, 2004) was used to estimate divergence time based on mitochondrial concatenated genes in BEAST v1.8.2 (Drummond & Rambaut, 2007). For the first step, four Salamandridae fossil records were used as primary calibration points (C1–C4, GenBank Nos. in Supplementary Table S4) (Wang et al., 2018) to establish the divergence date of the most recent common ancestor (MRCA) of both L. taliangensis and T. pseudoverrucosus; in the second step, the above divergence date was used for secondary calibration, which yielded the divergence times of the internal clades of L. taliangensis. The dataset resulting from the second step in BEAST was used for ancestral area reconstruction in RASP v3.2 (Yu et al., 2015) with the statistical dispersal-vicariance analysis (S-DIVA) model. Four distributional areas were delimited according to the mountains across the distribution range of L. taliangensis: i.e., DLS, XLS, XXL, and GG. The max areas at each node were set to two.
Neutrality tests (Tajima’s D (Tajima, 1989), Fu’s FS (Fu & Li, 1993)), and mismatch distribution analysis (via sum of squares differences (SSD) and Harpending’s raggedness index (Hrag) (Harpending, 1994; Rogers & Harpending, 1992)) were implemented for all populations and two clades using Arlequin v3.0 (Excoffier & Lischer, 2010). These tests were employed to estimate whether the species has experienced historical population expansion. Under the sudden demographic expansion model, the goodness-of-fit of the observed mismatch distribution was tested via 1 000 bootstrap replicates. The mismatch distribution curve was drawn by DnaSP v5.0 (Librado & Rozas, 2009). Bayesian Skyline Plots (BSPs) were drawn in BEAST v1.8.2 to analyze effective population size fluctuations of all populations and two clades through evolutionary time. Results were displayed using TRACER v1.7 (Rambaut et al., 2018).
Further details on mtDNA analysis are shown in the Supplementary Methods.
Microsatellite analysis
Null alleles and allele dropout of microsatellite data were assessed by MICROCHECKER v2.2.3 (Van Oosterhout et al., 2004). Significant deviations from both the Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium between all pairs of microsatellite loci were identified by Genepop v4.2 (Rousset, 2008). Significance values were Bonferroni corrected (Rice, 1989).
Total number of alleles (TA), number of effective alleles (Ne), observed heterozygosity (Ho), unbiased expected heterozygosity (uHe), and inbreeding coefficients (FIS) for each population were calculated by GenAlEx v6.5 (Peakall & Smouse, 2012). Allelic richness (Ar) and private allelic richness (pAr) were calculated in ADZE v1.0 (Szpiech et al., 2008) and were corrected for the effects of different sample sizes of each population by the rarefaction approach. Geographic patterns of genetic diversity (uHe and Ar) were visualized by the inverse distance weighted (IDW) method in ArcGIS v10.3.
Population structures were investigated using STRUCTURE v2.3.1 (Pritchard et al., 2000) with Bayesian clustering based on microsatellite loci with a burn-in of 100 000 iterations, followed by 1 000 000 MCMC iterations. Number of genetic clusters (K) was tested from 1 to 8 with 10 replicates for each K value. The criterion for optimal K was the highest value of delta K (ΔK) (Evanno et al., 2005), which was calculated in STRUCTURE HARVEST (Earl & vonHoldt, 2012). The outputs of optimal K with 10 replicates were averaged and calculated using CLUMPP v1.1 (Jakobsson & Rosenberg, 2007). The diagram of the best genetic clusters was drawn by DISTRUCT v1.1 (Rosenberg, 2004). According to the population structure, hierarchical analysis of molecular variance (AMOVA) was performed both in whole populations and two main genetic clusters using Arlequin v3.5.1 with 1 000 permutations. Principal coordinate analysis (PCoA) was conducted by GenAlEx v6.5, and the results were visualized in R v3.6.1 using the software package “ggpolt2”. Furthermore, Nei’s genetic distance (DA; Nei et al., 1983) of the microsatellite data was also calculated and a neighbor-joining (NJ) tree was constructed in POPTREE2 (Takezaki & Tamura, 2014) with 1 000 bootstraps.
Heterozygosity excess, which presents evidence of a population bottleneck, was estimated by the sign-test and Wilcoxon sign-rank test using the stepwise mutation model (SMM) and two-phase model (TPM) (proportion of SMM in TPM=95%, and variance of TPM=12). BOTTLENECK v1.2.02 was used with 1 000 replicates (Piry et al., 1999). To estimate the direction of migration and gene flow between major genetic clusters, mutation-scaled migration rates (M) and effective population sizes (θ) of each cluster were calculated using the Bayesian coalescent approach in MIGRATE-N v3.7.2 (Beerli & Palczewski, 2010). Differences in the inbreeding coefficient (FIS), mean corrected assignment index (mAIC), and variance of corrected assignment index (vAIC) were compared between sexes in the whole population and two genetic clusters (Goudet et al., 2002). For this, Fstat v2.9.4 (Goudet, 1995) was used with 1 000 permutations to estimate sex-biased dispersal. FIS andvAIC of the dispersing sex are higher than the philopatric sex, while mAIC is lower for the dispersing sex (Goudet et al., 2002).
Additional details on microsatellite analysis are shown in the Supplementary Methods.
Ecological niche modeling
To infer the potential distribution range of L. taliangensis under historical and current climatic conditions and to estimate whether populations have experienced contraction during periods of climatic fluctuations, ENM was applied (Elith et al., 2011). This approach combines current distribution GPS information of the species and environmental variable modeling to project historical distributions.
Current distribution information was obtained from field investigations, as well as records from the Global Biodiversity Information Facility (https://www.gbif.org). Only one GPS site within each pixel (30 arc-seconds, 1 km×1 km) was retained. Moreover, 19 predictor climate variables in four periods were downloaded from WorldClim (https://www.worldclim.org). According to the distribution records, the current distribution region and surrounding 200 m buffer area (N25–31°, E100–105°) were selected as the range of interest. ENM was conducted in MaxEnt v3.4.1 (Phillips et al., 2017) using five bioclimatic variables and maximum entropy method based on optimal parameters. The area under the receiver operating characteristic curve (AUC) (Swets, 1988) was used as the evaluation index of the overall fit of the model (Araújo & Guisan, 2006). The MaxEnt output was converted into the binary classification of either suitable (i.e., 1) or unsuitable (i.e., 0) based on the maximum training sensitivity plus specificity (MTSS) threshold (Liu et al., 2016). To identify the climatic stability area, the distribution prediction rasters of four periods were overlapped by ArcGIS and a raster with classification values ranging from 0 (i.e., not present in any of the four time periods) to 4 (i.e., present during all four periods) was generated.
For more detailed information, please refer to the Supplementary Methods.
Landscape genetics
To assess whether the population structure and patterns of gene flow are related to geographical barriers or environmental differences, IBR (which uses resistance distance instead of Euclidean geographical distance) and IBE were examined. These are often used to study the effects of landscape characteristics on genetic structuring (Atlas & Fu, 2019; Myers et al., 2019; Vasconcellos et al., 2019; Waraniak et al., 2019).
To alleviate the greater subjectivity associated with expert experiences (Myers et al., 2019; Spear et al., 2010), the resistance distance for the IBR model was calculated in CIRCUITSCAPE v4.0 (McRae, 2006; McRae et al., 2016) based on the resistance surfaces derived from the ENM result rasters. The environmental dissimilarity matrix, used for IBE analysis, was calculated from 20 environmental parameters. The pairwise genetic distance matrix (FST) of the microsatellite data was calculated in Arlequin v3.5.1. All above distance matrices were standardized by R v3.6.1 using the “scale” function. The variance inflation factor (VIF) was calculated before regression analysis to avoid multicollinearity of independent variables. The multiple matrix regression with randomization (MMRR) function was employed to evaluate the significance of effects of multiple independent factors (environmental dissimilarity and resistance distance in this study) on genetic divergence (Wang, 2013), conducted in R v3.6.1 with 10 000 permutations. The backward-stepwise method was used to select the optimal model. In addition, significance of the rejected variables was also tested.
Further details on resistance surface construction and environmental dissimilarity matrix calculation are presented in the Supplementary Methods.
Dispersal route analysis
To evaluate the influence of landscape features on the connectivity of dispersal routes of L. taliangensis, least-cost corridors (LCCs) and least-cost paths (LCPs) were calculated using SDMTOOLBOX v1.1c (Brown, 2014) in ArcGIS v10.3. The resistance raster used in IBR analysis was used as an analysis raster and sampling site coordinates were entered in decimal degrees. The LCPs were weighted by resistance values and classified into three categories using the “percentage of LCP” method (cutoff values: 1%, 2%, and 5%); finally, an LCC dispersal network was created.
RESULTS
Mitochondrial phylogeography
After manual sequence correction, alignment, and trimming of ends, concatenated gene sequences with a total length of 3 255 bp were obtained with three gene fragments (i.e., ND2 (945 bp), cyt b (762 bp), and COΙ (1 548 bp)) from 407 samples. No gaps were found, and all sequences could be correctly translated into amino acid sequences based on vertebrate genetic codes. The sequence data of three mtDNA haplotypes were deposited in GenBank under accession Nos.: cyt b (MZ368891–MZ368957), ND2 (MZ368958–MZ369024), and COI (MZ369025–MZ369091).
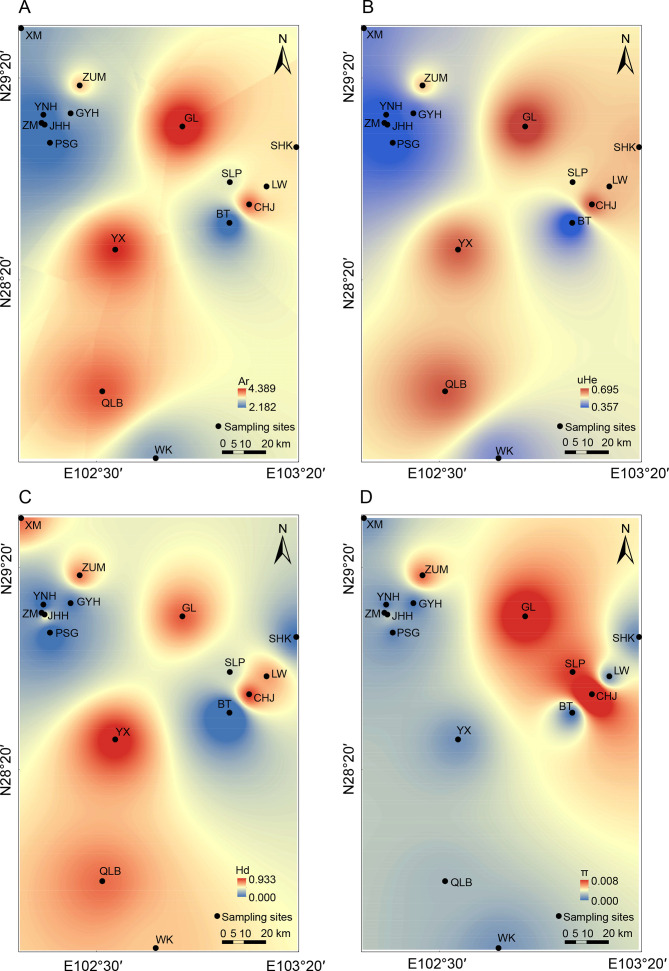
For the combined gene sequence, 156 mutation sites were found, and 67 haplotypes were defined (i.e., H1–H67), including 35 private haplotypes belonging to six populations (Supplementary Table S5). The haplotype network showed that adjacent clades or subclades were separated by at least seven mutational steps. The non-single star-shaped haplotype network suggested that the population has experienced multiple local expansions (Supplementary Figure S1). Populations Yuexi (YX) and Chuhongjue (CHJ) exhibited the highest population haplotype diversity and highest nucleotide diversity, respectively (Supplementary Table S6). The North clade had a much higher Hd and π than the South clade (Figure 2; Supplementary Table S7).
Figure 2.
Spatial interpolation of genetic diversity parameters among populations of Liangshantriton taliangensis
A: Allelic richness (Ar); B: Unbiased expected heterozygosity (uHe); C: Haplotype diversity (Hd); D: Nucleotide diversity (π). Warmer colors indicate higher genetic diversity. Black dots represent sampling sites. Further information on population codes is given in Supplementary Table S1.
The best substitution models (i.e., K80+I for the 1st COI and cyt b codon partition, HKY+I for the 2nd COI, cyt b and ND2 codon partition, GTR+G for the 3rd COI, cyt b and ND2 codon partition, HKY+G for 1st ND2 codon partition) were identified using PartitionFinder v2.1.1. Both BI and ML phylogenetic trees clearly divided L. taliangensis into two monophyletic lineages (i.e., North and South clades, BI/ML: 100%/100) (Supplementary Figure S1). Both clades had mostly allopatric distributions, with the South clade concentrated in the DLS, and the North clade concentrated in the XLS and XXL-GG (Supplementary Figure S1). The North clade could also be divided into seven lineages (subclades B–H) , although the relationships between specific subclades could not be completely resolved (Supplementary Figure S1). Subclades B, G, and H were from XXL-GG and Subclades C–E were from XLS. The Xinmin (XM) population from GG was isolated as Subclade H. Subclade F consisted of four populations (i.e., Jinghuahu (JHH), Pusagang (PSG), Zima (ZM), and GL) from XXL and XLS (Supplementary Figure S1 and Table S5).
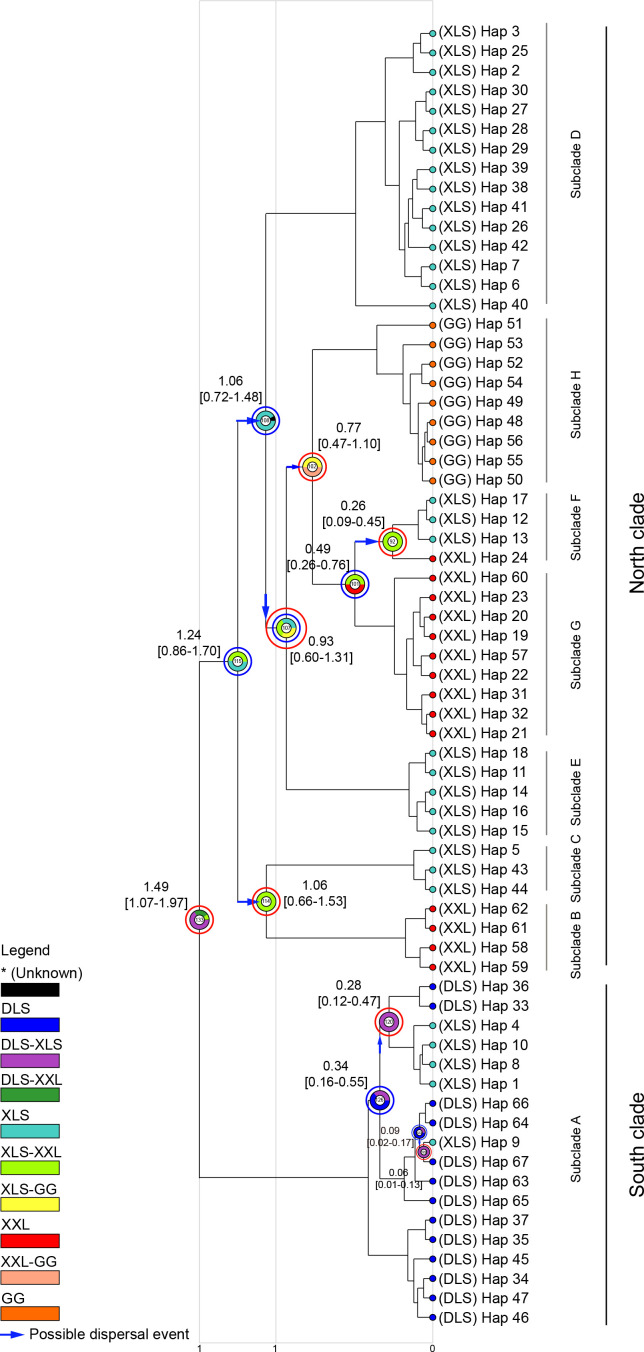
Divergence time results indicated that L. taliangensis diverged from its sister species T. pseudoverrucosus during the early Pleistocene period, approximately 2.44 million years ago (Ma) (95% highest posterior density (HPD): 1.65–3.28 Ma) (Supplementary Figure S2). The most recent common ancestor (MRCA) of L. taliangensis was estimated at 1.49 Ma (95% HPD: 1.07–1.97 Ma) and the first vicariance event occurred simultaneously (Figure 3). The seven subclades formed before the Guxiang Glaciation (0.3–0.13 Ma) and underwent four dispersal and three vicariance events. The Zhuma (ZUM) and GL populations were the first colony for XXL and transit point between XLS and XXL, respectively. In addition, two dispersal events (both from DLS to XLS) and two vicariance events occurred within the South clade during the Guxiang glacial-interglacial and last glacial-interglacial cycles (Figure 3). The LS was the most likely original distribution area ofL. taliangensis (node 133), with a probability of 66.1% (Figure 3), with the DLS geographically closest to the T. pseudoverrucosus outgroup. The BSP for all populations and the North clade suggested that the species experienced a strong decrease in effective population size after the Last Interglacial (LIG), followed by a very recent increase after the Last Glacial Maximum (LGM) (Figure 4A, C). However, the BSP for the South clade confirmed general stabilization during the LGM and accelerated expansion after the LGM (Figure 4B). Neutrality and mismatch distribution analyses basically supported these results (Supplementary Figure S3 and Table S7).
Figure 3.
Ancestor distribution areas of Liangshantriton taliangensis inferred from S-DIVA analyses based on mitochondrial data
Sectorial annular represents proportions of ancestral range and different ancestral ranges are denoted by different colors. Blue arrows represent direction of dispersal event, blue circles represent dispersal events, and red circles represent vicariance events. Number in sectorial annular is node ID. Divergent times (Ma; based on mitochondrial concatenated genes) are annotated beside main nodes. Numbers in square brackets represent 95% HPD of divergence time. Clades and subclades are named on the right.
Figure 4.
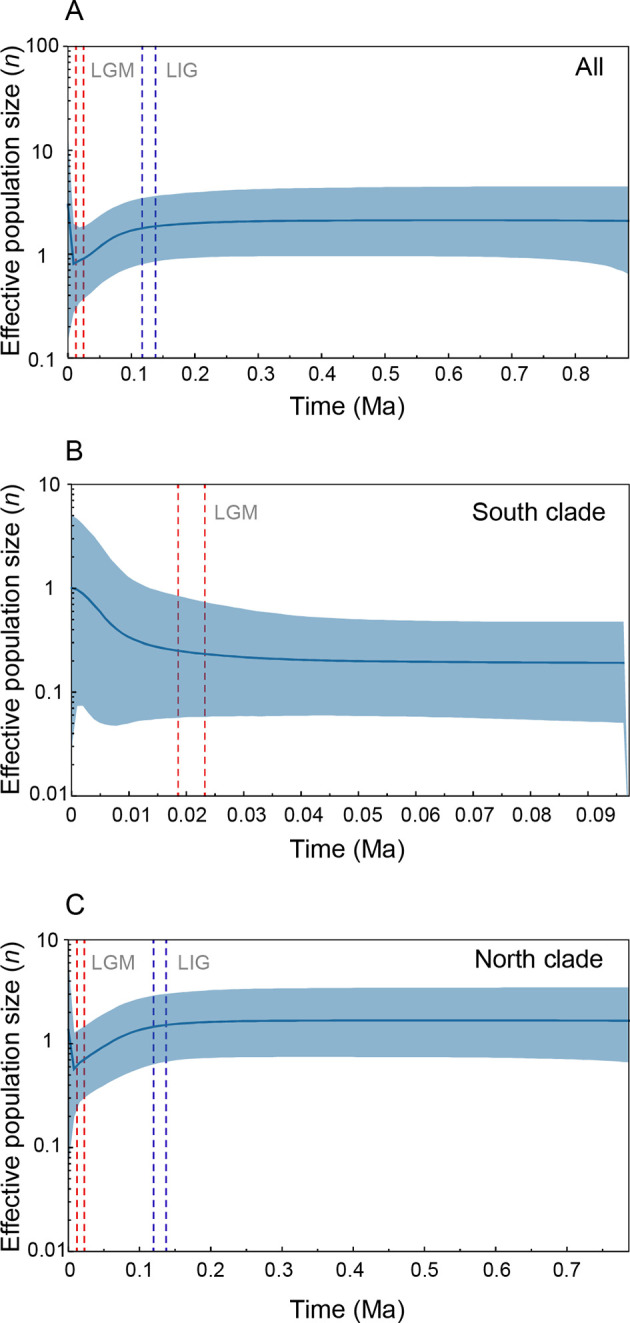
Bayesian skyline plots (BSP) showing variation in effective population sizes through time
A–C: For all populations (A), South clade (B), and North clade (C) of Liangshantriton taliangensis based on mitochondrial concatenated gene sequences. Blue area represents 95% confidence interval of effective population size. Ranges within red dotted lines represent LGM and ranges within blue dotted lines represent LIG.
Genetic structure and dispersal pattern based on microsatellite data
No significant linkage disequilibrium was detected at any loci after Bonferroni correction. Except for homozygous loci in the population, which could not be evaluated for HWE, two loci showed significant departures from HWE in one to six populations following Bonferroni correction. DLY32 indicated significant departure from HWE in one population (i.e., Sanhekou (SHK)), while DLY96 showed significant departure in five populations (i.e., XM, ZUM, Gongyihai (GYH), ZM, and PSG). However, this may be the result of the population genetic structure. In addition, deviations from HWE were not consistent across populations, with most loci in the population conforming to HWE. The microsatellite genetic diversity of each population was estimated (Supplementary Table S8). Ho (mean=0.488) was lower than uHe (mean=0.506), indicating that the population suffered from heterozygosity deficiency, and the mean value was 0.308 for Ar (2.182–4.389). The higher genetic diversity was mostly concentrated in LS, together with the ZUM population in XXL (Figure 2). The positive FIS values (mean FIS=0.022>0) suggested that all populations had an inbreeding tendency. The results of population genetic distance (FST) showed that there was significant genetic divergence (P<0.001), and the average genetic distance was 0.342 (0.033 (JHH, PSG)–0.598 (ZM, Bingtu (BT))) (Supplementary Table S9).
Structural analysis showed that ΔK reached the highest peak at K=2, indicating that all individuals could be divided into two genetic clusters (Figure 1A, E). The red cluster was defined as the West cluster (XXL-GG) and the blue cluster was defined as the East cluster (LS) (Figure 1A). Relatively high admixture was detected where the genetic clusters were in contact, particularly in the ZUM population (assignment proportion: 68.7% from XXL-GG and 31.3% from LS). Furthermore, four subclusters were revealed in the West cluster (Figure 1B, F) and K=2 was the optimal grouping for the East cluster (Figure 1C, D). Genetic diversity of the East cluster was higher than that of the West cluster (Supplementary Table S8). Similarly, PCoA suggested that all populations could be divided into two genetic clusters (i.e., East and West cluster), which did not overlap on the PCoA1 axis (Supplementary Figure S4). The NJ tree (inferred from DA distances) also identified two clusters and six subclusters (Figure 1G). Subclusters C and F showed the highest genetic divergence (Figure 1G). AMOVA showed that genetic variation was slightly lower between two clusters than among populations within clusters (Supplementary Table S10). Within the West cluster, genetic variation was much higher among groups than among populations within groups. However, within the East cluster, genetic variation was lower among groups than among populations within groups and no significant inbreeding was found (Supplementary Table S10). The TPM model, which is suitable for microsatellite data (Di Rienzo et al., 1994), indicated that L. taliangensis was not subjected to a bottleneck effect (Supplementary Table S11).
The migration patterns among genetic clusters were from XXL-GG to LS, with the highest Bezier approximation score of −71 223.03 lmL (Figure 1; Supplementary Table S12). Under this model, the mutation-scaled population size of XXL-GG (ΘXXL-GG=5.97, 95% CI: 2.27–9.47) was almost two times higher than that of LS (ΘLS=2.99, 95% CI: 0.00–6.40) and had a very low M value (i.e., immigration rate/mutation rate) (MX-L=0.467<1) for one-way gene flow. These results indicated thatL. taliangensis experienced very limited unidirectional gene flow from XXL-GG to LS.
FIS,mAIC, and vAIC were compared between females and males over the whole population. Results showed that FIS was significantly higher in males than in females, indicating that males were the dispersing sex, while vAIC was significantly higher for females than for males, indicating that females were the dispersing sex. mAIC showed no significant differences between the sexes. Thus, dispersal sex was not fixed within the whole population (Table 1). We checked the two main clusters, respectively. Only mAIC showed a significant difference, with FIS and vAIC showing non-significant sex differences in the two clusters. For the West cluster, the mAIC values were significantly lower in males than in females, suggesting male-biased dispersal; for the East cluster, the mAIC values were significantly lower in females than in males, indicating that females were the dominant dispersal sex (Table 1).
Table 1. Differences in inbreeding coefficient (FIS), mean of corrected assignment index (mAIC), and variance of corrected assignment index (vAIC) between males and females of L. taliangensis from total range, West cluster, and East cluster .
| N | F IS | mAI C | vAI C | |
| N: Sample size. *: 0.01<P<0.05. | ||||
| ALL | ||||
| Females | 83 | –0.023 | 0.536 | 82.169 |
| Males | 324 | 0.043 | –0.137 | 50.659 |
| P | 0.014* | 0.533 | 0.028* | |
| West cluster (XXL-GG) | ||||
| Females | 49 | –0.014 | 1.645 | 38.690 |
| Males | 152 | 0.072 | –0.530 | 57.506 |
| P | 0.061 | 0.046* | 0.120 | |
| East cluster (LS) | ||||
| Females | 34 | –0.023 | –3.949 | 86.440 |
| Males | 172 | 0.024 | 0.781 | 54.285 |
| P | 0.247 | 0.017* | 0.133 | |
Changes in distribution area during glaciations
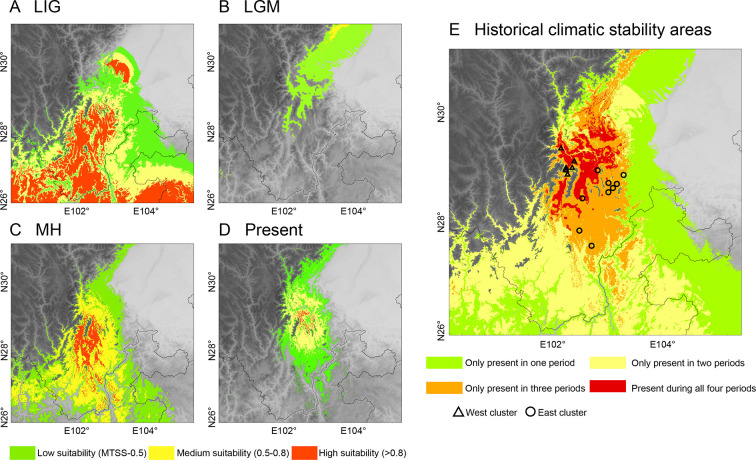
Linear and quadratic features and a regularization multiplier of 0.5 (LQ 0.5) were optimal model parameters, and the very high AUC values (0.963±0.018) indicated that this model had high predictive power. The MTSS threshold was 0.2064. The ENM predicted wider areas of suitable L. taliangensis habitat in the tropical and subtropical regions of the southeastern HMR during the LIG than during the LGM, mid-Holocene (MH), or the present (Figure 5). Based on the distribution predictions from the three general circulation models (GCMs), the potentially suitable distribution areas ofL. taliangensis shrank during the dry and cold LGM (Figure 5B; Supplementary Figure S5). The predicted distribution range during the MH was similar to the current range but much wider (Figure 5C, D; Supplementary Figure S6). Overlap of the predictions in four periods showed that the climatic stability zone was mainly distributed in the current distribution range (Figure 5E). Isothermality (Bio03), minimum temperature of coldest month (Bio06), annual precipitation (Bio12), and precipitation of driest month (Bio14) were high in contribution and permutation importance. The cumulative contribution rate was 97.59% (Supplementary Table S13). The most critical climatic predictor in the model was minimum temperature of coldest month (Bio06), with a contribution rate of 36.93% (Supplementary Table S13).
Figure 5.
Species distribution range of Liangshantriton taliangensis under four different periods and overlapping map yielding historical climatic stability areas
A: Last Interglacial (LIG). B: Last Glacial Maximum (LGM). Projection of LGM was the average of three general circulation models (GCMs) (CCSM4, MIROC-ESM, and MPI-ESM-P) (see Supplementary Figure S5). C: mid-Holocene (MH). Projection of MH was the average of four GCMs (CCSM4, MIROC-ESM, MPI-ESM-P, and BCC-CSM1-1) (see Supplementary Figure S6). D: Present. Based on MTSS, which represents maximum training sensitivity plus specificity threshold, projection distribution area was divided into areas with low, medium, and high suitability, denoted in green, yellow, and red, respectively. E: Showing historical climatic stability areas, from presence in only one period to presence during all four periods. Two major genetic clusters are represented by hollow triangles and circles.
Critical landscape factors affecting population structure
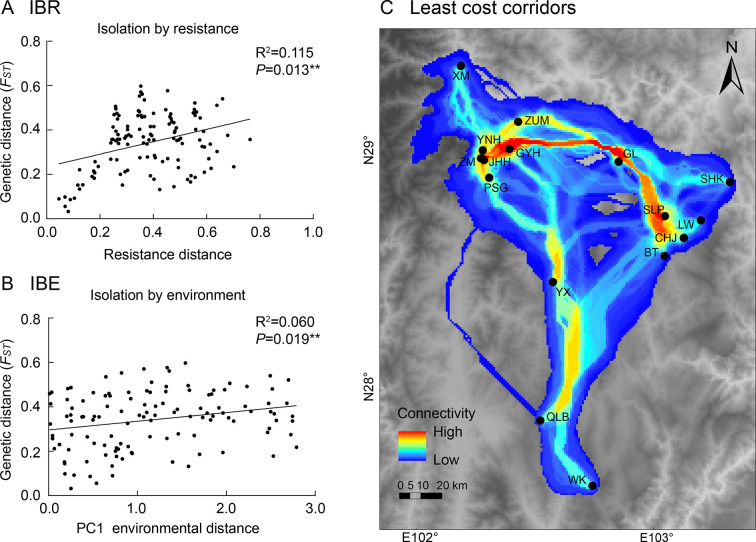
The ENM used for IBR analysis showed good performance in predicting the potential distribution of L. taliangensis (AUC=0.936) and MTSS was 0.2301 (Supplementary Figure S7). The VIF did not show significant multicollinearity between resistance distance and environmental dissimilarity (√VIF<2). A multiple matrix regression model combining resistance distance and environmental dissimilarity was generated via MMRR. The model indicated that genetic distance (FST) of all populations (Supplementary Table S9) was significantly positively correlated with resistance distance (R2=0.115, P=0.013) and all environmental variables were excluded from the final model (Figure 6A; Supplementary Table S14). However, the removed factors were used for repeat MMRR analysis, which showed that only PC1 (temperature-related variables, Supplementary Table S15) was significantly, though weakly, positively correlated with genetic distance (R2=0.060, P=0.019) (Figure 6B; Supplementary Table S14). For the West cluster, only resistance distance (IBR) strongly explained genetic divergence (R2=0.938, P=0.001) (Supplementary Figure S8 and Table S14). No landscape characteristics were associated with genetic divergence in the East cluster (Supplementary Table S14). Evaluations of variable contribution and permutation importance identified Bio14 (precipitation of driest month) and altitude as the most important factors affecting species distribution and dispersal, while normalized difference vegetation index (NDVI) and Bio12 (annual precipitation) were the next most significant variables (Supplementary Table S16).
Figure 6.
Multiple matrix regression with randomization (MMRR) plots for isolation-by-resistance (IBR) and isolation-by-environment (IBE) as well as construction of least cost corridors (LCCs) forLiangshantriton taliangensis based on resistance raster
A: Correlations between genetic distance (FST) and resistance distance. B: Correlations between genetic distance (FST) and PC1 environmental distance. C: LCCs among 16 sampled populations of L. taliangensis. Warmer colors indicate higher connectivity and lower dispersal cost. Sampling sites are represented by black dots.
Dispersal route analyses
According to the LCC map, neighboring populations had better connectivity than distantly separated populations (Figure 6C). There were few corridors with low resistance between DLS and both XLS and XXL-GG (Figure 6C). The dispersal corridor between the XM population and other populations showed high geographic and environmental resistance, implying that individuals from the XM population and other populations were not easy dispersal each others. Of note, a narrow dispersal corridor with low resistance through the GL and Niuri River (tributary of the Dadu River) between XXL and XLS indicated the possibility of gene flow between populations of both mountains.
DISCUSSION
Regional uplift and Early-Middle Pleistocene transition (EMPT) drove L. taliangensis formation and divergence
The causes of species evolution and diversification are diverse and complex, and generally include both biotic and abiotic factors or a combination of both, e.g., climate change, geographic barriers, and inter- and intra-specific interactions (Bouchenak-Khelladi et al., 2015; Doebeli & Dieckmann, 2003). In this study, we discovered that L. taliangensis originated from the LS mountains, separated from its sister species (T. pseudoverrucosus) during the Pleistocene (~2.44 Ma) (Supplementary Figure S2), and dispersed to higher latitudes. This is mostly consistent with the divergence times and radiation direction of Tylototriton s.l. (Wang et al., 2018), although the divergence time estimated by secondary calibration should be interpreted with caution (Hedges & Kumar, 2004; McCormack et al., 2011).
The QTP uprising promoted the formation of the East Asian monsoon, creating a warm, moist, and vegetation-rich habitat on the southeastern edge of the HMR, which was suitable for newts (Wang et al., 2018). Climatic shifts and increased niche vacancies drove the radiation of ancestral populations of L. taliangensis from low latitudes to high latitudes. The South and North clades separated ~1.49 Ma (Figure 3), with the uplift of the southeastern HMR. The first vicariance event was detected during this period. Although details on the timing and pattern of uplift remain controversial, consensus has been achieved that the uplift of the HMR happened recently and rapidly (Xing & Ree, 2017). Speciation and divergence of amphibians are presumed to be the result of ecological isolation caused by uneven uplift of the QTP (Hofmann et al., 2017; Wu et al., 2020). About 2.5–1.2 Ma, the Xigeda Formation strata on the southeastern margin of the QTP (widely distributed between the Dadu River, Anning River, and Jinsha River valley in southwestern Sichuan) gradually began to uplift and complete the accretionary orogenic process (Jiang & Wu, 1998). Consequently, the formation of species and divergence of L. taliangensis clades may be related to the East Asian monsoon climate and regional uplift events (e.g., Xigeda Formation strata). The LS mountains (DLS and XLS) showed the highest probability as the ancestral area for L. taliangensis. The existence of multiple ancestral areas suggests that the divergence patterns of the main clades may be consistent with the fragmentation model, where isolation and fragmentation occurred over a wide range of ancestral distributions.
The North clade of L. taliangensis underwent phylogenetic divergence around the EMPT, although the phylogenetic relationships among subclades remain unclear. The EMPT (~1.4–0.4 Ma, Head & Gibbard, 2015) was characterized by a gradual increase in the amplitude of climatic oscillations and in global ice volume, with a shift from 41 thousand year (ka) glacial cycles to 100 ka cycles since ~1.2 Ma (Clark et al., 2006; Head & Gibbard, 2015). From ~1.25 Ma onwards, the East Asian summer monsoon during the interglacial periods intensified (Sun et al., 2010), which further promoted the dispersal of L. taliangensis from XLS to XXL-GG. At ~0.9 Ma, a dispersal and vicariance event of L. taliangensis was recorded, simultaneously. This period, known as the “0.9 Ma event”, was the beginning of the first large-scale expansion of continental ice sheets and was interrupted by the weak interglacial period (Marine Isotope Stage (MIS) 23 ) (Clark et al., 2006; Head & Gibbard, 2015). A total of three dispersal and two vicariance events occurred during the early phase of the EMPT, with divergence of the XLS population from the XXL-GG population. The Naynayxungla Glaciation (0.78–0.5 Ma) had a profound influence on the formation of the internal lineage of the XXL-GG, and one dispersal and one vicariance event occurred during this period. The XM populations diverged into a monophyletic lineage at ~0.77 Ma. This time point, called the Matuyama-Brunhes boundary (773 ka), is the primary guide for the Lower-Middle Pleistocene Subseries boundary (Head & Gibbard, 2015). During this period, the global temperature decreased significantly, ice sheets expanded, and glacial cycles were dominated by 100 ka periodicity (Clark et al., 1999, 2006; Head & Gibbard, 2015). Furthermore, the QTP had a glacier coverage of close to 500 000 km2 (Zheng et al., 2002). The Shaluli Mountain region and Daocheng Kuzhaori, which are relatively close geographically to the current distribution area of L. taliangensis, were the main areas and more strongly influenced by QTP glaciation (Wang et al., 2006; Xu et al., 2005). A dispersal event from XXL to GL occurred ~0.49 Ma during the great interglacial period, which was related to the strong summer monsoon throughout the Northern Hemisphere (Figure 3). The overall dispersal direction was from low to high latitudes, and ZUM was identified as the first colony of XXL (Figure 3). Subclade divergence mainly supported a stepping-stone colonization mode, with dispersal from one mountain to another, followed by vicariance. GL was an important stepping-stone between XLS and XXL (Figure 3).
Frequent vicariance and dispersal of populations due to Pleistocene climatic fluctuations may be a common cause of lineage divergence in many rapidly diverging species (Wang et al., 2017, 2018; Ye et al., 2019), including L. taliangensis. The formation and differentiation of L. taliangensis also provide further biological reference for the timing controversy associated with orogenesis.
Multiple glacial refugia patterns respond to LGM
Refugia tend to maintain moderate temperature and precipitation levels, and are thus commonly identified as climatically stable regions, which enabled species to survive during the LGM (Tang et al., 2018; Tzedakis et al., 2002). Different from Europe and North America, there was no unified ice sheet covering the QTP in China and many ice-free low-altitude areas could be found on its southeastern margin (Owen et al., 2008). The southeastern HMR was a large glacial refugium for endemic species and its complex topographic features allowed for lineage maintenance in “refugia within refugia” (Xie et al., 2019; Yu et al., 2013). In this study, the current distribution area of L. taliangensis represents such a climatically stable zone, mainly located in the Dadu River valley and the LS and XXL intersection area (Figure 5E). Liangshantriton taliangensis showed multiple glacial refugia patterns in response to the LGM, which is common in species with weak dispersal capacity in southwest China (Liu et al., 2015; Wang et al., 2017). The single refugium hypothesis suggests that genetic diversity decreases from glacial refugia to surrounding areas, and refugia usually harbor private haplotypes and alleles (Hewitt, 2000). The following glacial refugia forL. taliangensis were identified (Figures 2, 5E; Supplementary Tables S6, S8): one in GG (XM), two potential major glacial refugia in XXL (ZUM and JHH), two in DLS (YX and Qiliba (QLB)), and two main glacial refugia in XLS (GL and CHJ). The response patterns of population dynamics to Pleistocene climatic fluctuations vary among species and distribution ranges (Lu et al., 2012; Qu et al., 2010). Here, two major lineages (i.e., South and North clades) displayed diverse population dynamics (Figure 4). The South clade population was located at relatively low latitudes and on medium-high altitude mountains below the snow line. Therefore, L. taliangensis in this region was probably rarely affected by glaciation. The GG mountains are considered as the glaciation center of the HMR, where mountains above the snow line became a source of cold, the influence of which extends from north to south (Li et al., 1991). The North clade (i.e., GG, XXL, and XLS) population was distributed at higher latitudes and adjacent to the GG mountains. Thus, L. taliangensis in this range was subjected to glacial influences.
Mitonuclear discordance
Amphibians, especially frogs, commonly show mitonuclear discordance; however, it is rare in salamanders (Chan & Levin, 2005; Bryson et al., 2014; Toews & Brelsford, 2012). In general, mitonuclear discordance results from female-biased dispersal, incomplete lineage sorting, hybridization/introgression in cases of secondary contact, and selective sweep of mtDNA haplotypes (Garrick et al., 2019; Lucati et al., 2020; Toews & Brelsford, 2012). In this study, female-biased dispersal was found in the East cluster (Table 1), and introgression by secondary contact was identified in several regions and at different times (Figure 3). Thus, we speculated that these two factors may be the main reasons for the observed mitonuclear discordance.
Inferred from mtDNA, L. taliangensis could be divided into two well-supported genetic lineages, i.e., the South and North clades (separated at ~N28.6°). In contrast, two main genetic clusters of L. taliangensis were recognized by the microsatellite data, i.e., West and East clusters, which exhibited a more explicit biogeographic pattern and were separated by the XXL mountains (Figure 1), similar to the sympatric red pandas (Ailurus fulgens F. Cuvier, 1825) (Hu et al., 2011). Although contrasting divergence patterns, associated with the main genetic structures in the mitochondrial and nuclear genotypic data, were detected in the current study, fine-scale structures within these main genetic clusters showed strong concordant patterns.
Within the East cluster, two subclusters (i.e., E and F) were recognized. Subcluster E mainly contained populations from DLS (YX, QLB, and Wuke (WK)), and partly corresponded to the mitochondrial South clade at the base. Ancestral range reconstruction analysis identified this as a plausible independent biogeographic area. Therefore, if populations from DLS were to be accepted as an independent genetic cluster, the BT population and a few individuals from the CHJ population, with mitochondrial haplotypes nested within the South clade, may be the result of introgression due to secondary contact between the South and North clades. Two secondary contacts may have occurred between the South and North clades based on ancestral range reconstruction analysis. Combined with haplotype information (Supplementary Table S5), the first introgression may have occurred ~0.34 Ma, with dispersal from the QLB population (DLS) to the BT and CHJ populations (XLS); the second introgression was a dispersal from the YX population (DLS) to the CHJ population (XLS), which occurred during the LIG (Figure 3). Sex-biased dispersal suggested that the LS population was dominated by female dispersal, indicating that females drove asymmetrical mtDNA introgression (Table 1). The CHJ population may represent an important secondary contact zone between DLS and XLS.
It is worth noting that neither mitochondrial nor microsatellite data showed significant gene flow between GL population and other populations in the XLS (Figure 1; Supplementary Figure S1). Although dispersal route analysis revealed a narrow low-resistance dispersal corridor between GL and both SLP and CHJ populations, and weak nuclear gene mixing between them was detected by microsatellites, the limited barrier effect of Ma'an Mountains (altitude: 4288 m a.s.l.) cannot be ignored. Mitochondrial data suggested secondary contact between the GL and XXL populations at ~0.49 Ma and isolation from the XXL population during the Guxiang Glaciation (0.3–0.13 Ma), with mitochondria of the GL population partially replaced by that of the XXL population (Figure 3). The good landscape connectivity between the GL and XXL populations (especially GYH and JHH) and migration analysis both suggested the possibility of limited gene flow from XXL to XLS (Figures 1, 6C). GL may represent an important secondary contact zone between XXL and XLS. The GL population belonged to the same nuclear genotypic cluster as populations from DLS. However, this pattern contrasted with the mitochondrial phylogeographic patterns and dispersal route analysis, which indicated high dispersal resistance between the DLS and GL populations and a low potential for gene flow (Figure 6C). These findings suggest that the distinct genetic differences of the GL population from other populations may be diminished by size homoplasy of microsatellites or strong selection pressure on sequence-based loci, which often occur in populations with large effective population size (Epperson, 2005). However, further studies are needed to confirm this speculation using more robust genetic and morphological data.
All populations from XLS (except for GL) belonged to subcluster F, which was mainly associated with the two mitochondrial subclades (C and D). This divergence in the mitochondrial clades occurred during the Pleistocene, as tracers of Quaternary glacial events, indicating that repeated expansions and contractions of the population ranges was accompanied by lineage sorting. However, the homogenization of nuclear genotypic data and mitochondrial introgressions between subclades C and D may reflect recent gene flow, resulting from population expansion after the Quaternary. Within the XXL-GG, two subclusters corresponded to two mitochondrial subclades. Apparent mitonuclear concordances were reflected by nuclear subcluster A and mitochondrial subclade H (XM population) as well as nuclear subcluster D and mitochondrial subclade B (from the ZUM population). Discordance in the distribution of nuclear genotypes with mitochondrial clades mainly manifested in the BT, CHJ, GL, and GYH populations. The GYH population showed a distinct nuclear genotype but was characterized by mitochondrial subclade G haplotypes. The geographical approximation between GYH and other populations of subclade G and the high landscape connectivity in this region suggested frequent gene flow (Figure 6C); thus, mitochondrial capture from neighboring populations may explain the above (Hausdorf et al., 2011).
Dramatic climatic fluctuations and scattered glacial refugia can promote the occurrence of frequent population expansion and isolation, which may be an important cause of mitonuclear discordance. These effects provide opportunities for gene admixture and fusion, and traces of asymmetric gene flow at distribution boundaries are identifiable for longer in mitochondrial than in nuclear genes (Currat et al., 2008; Dufresnes et al., 2020; Excoffier et al., 2009). Here, GL and CHJ in XLS were important glacial refugia and major secondary contact zones for L. taliangensis, they also distributed at the geographic edge of the intersection of two different genetic clades/clusters.
Resistance distance plays a dominant role in population structure
Resistance distance was a main reason for the population genetic structure of L. taliangensis (Figure 6A; Supplementary Table S14), suggesting that strong gene flow may have occurred in adjacent regions and regions with high geographic connectivity. The barrier effect of geographic distance is ubiquitous among organisms, especially in amphibians due to their low dispersal ability (Bani et al., 2015; He et al., 2021; Waraniak et al., 2019). In addition, the effect of temperature difference was weak but significant when environmental dissimilarity was analyzed alone (Figure 6B; Supplementary Table S14). The West cluster, with its finer population structure and large genetic variation, was strongly influenced by geographic resistance (Supplementary Figure S8 and Table S14), while the genetic structure of the East cluster was not significantly influenced by landscape features (Supplementary Table S14).
Precipitation of the driest month and altitude were primary factors influencing gene flow between populations, while NDVI and annual precipitation were secondary factors (Supplementary Table S16). Precipitation is crucial for the survival and dispersal of species (especially during the dry season), not only for amphibians but also for reptiles, even though they are perfectly adapted to terrestrial life (Myers et al., 2019). Heavy precipitation contributes to the establishment of temporary connections between adjacent water systems, which facilitate salamander dispersal (Wu et al., 2013). A 70% reduction in forest connectivity can diminish dispersal and further reduce gene flow among amphibians (Campos et al., 2020; Curtis & Taylor 2004; Gibbs, 1998). Salamanders have a particularly high demand for vegetation, not only as terrestrial shelters (Belasen et al., 2013; Otto et al., 2014) but also as microenvironments created by the interaction between precipitation and vegetation (Escoriza & Hassine, 2014). The barrier effect on gene flow, imposed by elevational differences, has been demonstrated in many amphibians, and is particularly important for high-altitude and pond-breeding species (Atlas & Fu, 2019; He et al., 2021; Sánchez-Montes et al., 2018). The north-south running XXL mountains, with a highest peak of 4 791 m a.s.l., may have driven divergence among the East and West genetic clusters ofL. taliangensis. However, these mountains were not absolute barriers (Sánchez-Montes et al., 2018) and limited gene flow could still be detected between the two genetic clusters (Figures 1, 6C). The elevational range of XXL-GG mountains is about 780–5 793 m a.s.l., consisting of steep mountains that are high in the northwest and low in the southeast. Such topographic features may have fragmented the distribution of the West cluster populations. However, the LS mountains consist of the northern XLS with steep topography and altitude range of ~500–4 500 m a.s.l. as well as the southern DLS with a gentle topography and altitude range of about 2 000–3 500 m a.s.l. (Yao et al., 2017). No landscape features that critically affected the genetic structure of the East cluster populations were found. Whether the relationship between population genetic differences and landscape features in the East cluster was masked by variations in the resistance effects of different topographies on gene flow, the size homoplasy of microsatellites, and strong selection pressure on sequence-based loci still need to be further clarified. The population differentiation ofL. taliangensis showed weak correlation with temperature (IBE), especially during the dry and cold seasons, which probably reflects differential adaptation and natural selection to the local climate (Foll & Gaggiotti, 2006; Zhang et al., 2016).
Conservation implications
Research on genetic diversity and population structure provides an important reference and foundation for the development of conservation measures (Ouborg, 2010).
Based on mitochondrial and microsatellite data, the overall genetic diversity of L. taliangensis was high and did not show a bottleneck event (Supplementary Tables S6, S8, S11). None of the sampled L. taliangensis populations currently suffer from genetic deterioration. Although evolutionarily significant units (ESUs) could not be clearly defined (according to Moritz’s strict criteria for ESUs, which require significant differentiation in both mitochondrial and nuclear genes (Moritz, 1994)), two major genetic clusters (i.e., West and East clusters) could be separated as two management units. As critical regions for the persistence and evolution of biodiversity, glacial refugia have conservation priority. Former glacial refugia, including XM, ZUM, JHH, GL, CHJ, YX, and QLB, harbor rich genetic diversity. GL and CHJ are crucial secondary contact zones, and their protection should be prioritized. In addition, attention should be paid to maintaining interpopulation connectivity, especially between XLS and XXL (Figure 6C). Although precipitation, altitude, and vegetation affect the construction of natural corridors (Supplementary Table S16), human population expansion, agricultural development, deforestation, and related problems may put these corridors at risk of disappearing, especially in XXL (Hu et al., 2011). Therefore, field surveys should be conducted for the establishment of alternative corridors while maintaining those that exist.
CONCLUSIONS
Based on multiple genetic markers (i.e., mitochondrial concatenated genes (cyt b+ND2+COI) and 11 microsatellite loci), the genetic divergence and driving factors ofL. taliangensis, a vulnerable amphibian species endemic in the southeastern HMR, were analyzed. Regional (Xigeda Formation strata) uplift events, the East Asian monsoon, glacier development during the EMPT, multiple in situ refugia patterns during glacial-interglacial cycles, and geographic resistance (precipitation, altitude, and vegetational cover) were identified as the main driving forces for the rapid divergence and population genetic structure formation of this species. Furthermore, secondary contact and/or female-biased dispersal were found to be the main reasons for the observed mitonuclear discordance. The two main microsatellite markers confirmed genetic clusters should be considered as independent management units. Seven major glacial refugia as well as crucial natural corridors between XXL and XLS should also be protected as a priority. This study improves our understanding of the contribution of historical and contemporary environmental factors to the genetic divergence of species in the HMR and provides valuable knowledge for the conservation and genetic management of L. taliangensis.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
F.X., J.P.J., C.L., and Z.B.S. designed the study. X.X.S, Y.M.H., M.Y.C., and G.C.S. collected and processed samples. X.X.S., Y.M.H., G.C.S., B.W., and X.Q.L. analyzed the data. X.X.S. and Y.M.H. interpreted the data and wrote the manuscript. F.X. and J.P.J. improved the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to the Liziping National Nature Reserve for field assistance, Ying-Hao Wu and Kan Zhang for assistance with sample collection, and Jin-Long Liu for help with data analysis.
Funding Statement
This study was supported by the National Key Research and Development Programs of China (2017YFC0505202), Construction of Basic Conditions Platform of Sichuan Science and Technology Department (2019JDPT0020), and Species Conservation Project of Liziping National Nature Reserve
Contributor Information
Jian-Ping Jiang, Email: Jiangjp@cib.ac.cn.
Feng Xie, Email: xiefeng@cib.ac.cn.
References
- 1.An ZS, Kutzbach JE, Prell WL, Porter SC Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature. 2001;411(6833):62–66. doi: 10.1038/35075035. [DOI] [PubMed] [Google Scholar]
- 2.Araújo MB, Guisan A Five (or so) challenges for species distribution modelling. Journal of Biogeography. 2006;33(10):1677–1688. doi: 10.1111/j.1365-2699.2006.01584.x. [DOI] [Google Scholar]
- 3.Atlas JE, Fu J Isolation by resistance analysis reveals major barrier effect imposed by the Tsinling Mountains on the Chinese wood frog. Journal of Zoology. 2019;309(1):69–75. doi: 10.1111/jzo.12702. [DOI] [Google Scholar]
- 4.Bani L, Pisa G, Luppi M, Spilotros G, Fabbri E, Randi E, et al Ecological connectivity assessment in a strongly structured fire salamander (Salamandra salamandra) population . Ecology and Evolution. 2015;5(16):3472–3485. doi: 10.1002/ece3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beerli P, Palczewski M Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185(1):313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belasen A, Burkett E, Injaian A, Li K, Allen D, Perfecto I Effect of sub-canopy on habitat selection in the Blue-spotted Salamander (Ambystoma laterale-jeffersonianum unisexual complex) . Copeia. 2013;2013(2):254–261. doi: 10.1643/CE-12-051. [DOI] [Google Scholar]
- 7.Bouchenak-Khelladi Y, Onstein RE, Xing YW, Schwery O, Linder HP On the complexity of triggering evolutionary radiations. New Phytologist. 2015;207(2):313–326. doi: 10.1111/nph.13331. [DOI] [PubMed] [Google Scholar]
- 8.Boufford DE Biodiversity hotspot: Chinaʼs Hengduan Mountains. Arnoldia. 2014;72(1):24–35. [Google Scholar]
- 9.Brown JL SDMtoolbox: a python‐based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods in Ecology and Evolution. 2014;5(7):694–700. doi: 10.1111/2041-210X.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryson Jr RW, Smith BT, Nieto-Montes de Oca A, García-Vázquez UO, Riddle BR The role of mitochondrial introgression in illuminating the evolutionary history of Nearctic treefrogs. Zoological Journal of the Linnean Society. 2014;172(1):103–116. doi: 10.1111/zoj.12169. [DOI] [Google Scholar]
- 11.Campos FS, Lourenço-de-Moraes R, Ruas DS, Mira-Mendes CV, Franch M, Llorente GA, et al Searching for networks: ecological connectivity for amphibians under climate change. Environmental Management. 2020;65(1):46–61. doi: 10.1007/s00267-019-01240-0. [DOI] [PubMed] [Google Scholar]
- 12.Chan KMA, Levin SA Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution. 2005;59(4):720–729. doi: 10.1111/j.0014-3820.2005.tb01748.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen DQ, Dong ZJ, Fang M, Fu JJ, Jia XY, Jiang BJ, et al Microsatellite records for volume 11, issue 1. Conservation Genetics Resources. 2019;11(1):109–112. doi: 10.1007/s12686-019-01085-z. [DOI] [Google Scholar]
- 14.Clark PU, Alley RB, Pollard D Northern Hemisphere ice-sheet influences on global climate change. Science. 1999;286(5442):1104–1111. doi: 10.1126/science.286.5442.1104. [DOI] [Google Scholar]
- 15.Clark PU, Archer D, Pollard D, Blum JD, Rial JA, Brovkin V, et al The middle Pleistocene transition: characteristics, mechanisms, and implications for long-term changes in atmospheric pCO2. Quaternary Science Reviews. 2006;25(23-24):3150–3184. doi: 10.1016/j.quascirev.2006.07.008. [DOI] [Google Scholar]
- 16.Clement M, Posada D, Crandall KA TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 17.Currat M, Ruedi M, Petit RJ, Excoffier L The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62(8):1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 18.Curtis JMR, Taylor EB The genetic structure of coastal giant salamanders (Dicamptodon tenebrosus) in a managed forest . Biological Conservation. 2004;115(1):45–54. doi: 10.1016/S0006-3207(03)00092-2. [DOI] [Google Scholar]
- 19.Davis MB, Shaw RG Range shifts and adaptive responses to Quaternary climate change. Science. 2001;292(5517):673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- 20.Deng JY, Fu RH, Compton SG, Liu M, Wang Q, Yuan C, et al Sky islands as foci for divergence of fig trees and their pollinators in Southwest China. Molecular Ecology. 2020;29(4):762–782. doi: 10.1111/mec.15353. [DOI] [PubMed] [Google Scholar]
- 21.Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB Mutational processes of simple-sequence repeat loci in human populations. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(8):3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding WN, Ree RH, Spicer RA, Xing YW Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science. 2020;369(6503):578–581. doi: 10.1126/science.abb4484. [DOI] [PubMed] [Google Scholar]
- 23.Doebeli M, Dieckmann U Speciation along environmental gradients. Nature. 2003;421(6920):259–264. doi: 10.1038/nature01274. [DOI] [PubMed] [Google Scholar]
- 24.Drummond AJ, Rambaut A BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7(1):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dufresnes C, Nicieza AG, Litvinchuk SN, Rodrigues N, Jeffries DL, Vences M, et al Are glacial refugia hotspots of speciation and cytonuclear discordances? Answers from the genomic phylogeography of Spanish common frogs. Molecular Ecology. 2020;29(5):986–1000. doi: 10.1111/mec.15368. [DOI] [PubMed] [Google Scholar]
- 26.Earl DA. vonHoldt BM STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 27.Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ A statistical explanation of MaxEnt for ecologists. Diversity and Distributions. 2011;17(1):43–57. doi: 10.1111/j.1472-4642.2010.00725.x. [DOI] [Google Scholar]
- 28.Epperson BK Mutation at high rates reduces spatial structure within populations. Molecular Ecology. 2005;14(3):703–710. doi: 10.1111/j.1365-294X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 29.Escoriza D, Hassine JB Microclimatic variation in multiple Salamandra algira populations along an altitudinal gradient: phenology and reproductive strategies . Acta Herpetologica. 2014;9(1):33–41. [Google Scholar]
- 30.Evanno G, Regnaut S, Goudet J Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 31.Excoffier L, Foll M, Petit RJ Genetic consequences of range expansions. Annual Review of Ecology, Evolution, and Systematics. 2009;40:481–501. doi: 10.1146/annurev.ecolsys.39.110707.173414. [DOI] [Google Scholar]
- 32.Excoffier L, Lischer HEL Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 33.Favre A, Päckert M, Pauls SU, Jähnig SC, Uhl D, Michalak I, et al The role of the uplift of the Qinghai‐Tibetan Plateau for the evolution of Tibetan biotas. Biological Reviews. 2015;90(1):236–253. doi: 10.1111/brv.12107. [DOI] [PubMed] [Google Scholar]
- 34.Fei L, Ye CY, Jiang JP. 2012. Colored Atlas of Chinese Amphibians and Their Distributions. Chengdu: Sichuan Publishing House of Science and Technology, 1–620. (in Chinese)
- 35.Foll M, Gaggiotti O Identifying the environmental factors that determine the genetic structure of populations. Genetics. 2006;174(2):875–891. doi: 10.1534/genetics.106.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu YX, Li WH Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrick RC, Banusiewicz JD, Burgess S, Hyseni C, Symula RE Extending phylogeography to account for lineage fusion. Journal of Biogeography. 2019;46(2):268–278. doi: 10.1111/jbi.13503. [DOI] [Google Scholar]
- 38.Gibbs JP Amphibian movements in response to forest edges, roads, and streambeds in Southern New England. The Journal of Wildlife Management. 1998;62(2):584–589. doi: 10.2307/3802333. [DOI] [Google Scholar]
- 39.Gillespie RG, Roderick GK Geology and climate drive diversification. Nature. 2014;509(7500):297–298. doi: 10.1038/509297a. [DOI] [PubMed] [Google Scholar]
- 40.Goudet J FSTAT (version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86(6):485–486. doi: 10.1093/oxfordjournals.jhered.a111627. [DOI] [Google Scholar]
- 41.Goudet J, Perrin N, Waser P Tests for sex‐biased dispersal using bi‐parentally inherited genetic markers. Molecular Ecology. 2002;11(6):1103–1114. doi: 10.1046/j.1365-294X.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- 42.Gouy M, Guindon S, Gascuel O SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 43.Gutiérrez-Rodríguez J, Barbosa AM, Martínez-Solano Í Integrative inference of population history in the Ibero-Maghrebian endemic Pleurodeles waltl (Salamandridae) . Molecular Phylogenetics and Evolution. 2017;112:122–137. doi: 10.1016/j.ympev.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 44.Harpending HC Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology. 1994;66(4):591–600. [PubMed] [Google Scholar]
- 45.Harrison TM, Copeland P, Kidd WSF, Yin A Raising Tibet. Science. 1992;255(5052):1663–1670. doi: 10.1126/science.255.5052.1663. [DOI] [PubMed] [Google Scholar]
- 46.Hausdorf B, Wilkens H, Strecker U Population genetic patterns revealed by microsatellite data challenge the mitochondrial DNA based taxonomy of Astyanax in Mexico (Characidae, Teleostei) . Molecular Phylogenetics and Evolution. 2011;60(1):89–97. doi: 10.1016/j.ympev.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 47.He JK, Lin SL, Ding CC, Yu JH, Jiang HS Geological and climatic histories likely shaped the origins of terrestrial vertebrates endemic to the Tibetan Plateau. Global Ecology and Biogeography. 2021;30(5):1116–1128. doi: 10.1111/geb.13286. [DOI] [Google Scholar]
- 48.He K, Jiang XL Sky islands of southwest China. I: an overview of phylogeographic patterns. Chinese Science Bulletin. 2014;59(7):585–597. doi: 10.1007/s11434-013-0089-1. [DOI] [Google Scholar]
- 49.Head MJ, Gibbard PL Early-Middle Pleistocene transitions: linking terrestrial and marine realms. Quaternary International. 2015;389:7–46. doi: 10.1016/j.quaint.2015.09.042. [DOI] [Google Scholar]
- 50.Hedges SB, Kumar S Precision of molecular time estimates. Trends in Genetics. 2004;20(5):242–247. doi: 10.1016/j.tig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Hewitt G The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann S, Stöck M, Zheng YC, Ficetola FG, Li JT, Scheidt U, et al Molecular phylogenies indicate a Paleo-Tibetan origin of Himalayan lazy toads (Scutiger) . Scientific Reports. 2017;7(1):3308. doi: 10.1038/s41598-017-03395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu YB, Guo Y, Qi DW, Zhan XJ, Wu H, Bruford MW, et al Genetic structuring and recent demographic history of red pandas (Ailurus fulgens) inferred from microsatellite and mitochondrial DNA . Molecular Ecology. 2011;20(13):2662–2675. doi: 10.1111/j.1365-294X.2011.05126.x. [DOI] [PubMed] [Google Scholar]
- 54.Huang ZS, Yu FL, Gong HS, Song YL, Zeng ZG, Zhang Q Phylogeographical structure and demographic expansion in the endemic alpine stream salamander (Hynobiidae: Batrachuperus) of the Qinling Mountains . Scientific Reports. 2017;7(1):1871. doi: 10.1038/s41598-017-01799-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.IUCN SSC Amphibian Specialist Group. 2020. Tylototriton taliangensis. The IUCN Red List of Threatened Species 2020: e. T59486A63871228. https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T59486A63871228.en. (Accessed 15 Feb. 2020)
- 56.Jakobsson M, Rosenberg NA CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23(14):1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 57.Jiang FC, Wu XH Late Cenozoic tectonic movement in geomorphologic boundary belt of southeastern Qinghai-Xizang Plateau. Journal of Chengdu University of Technology. 1998;25(2):162–168. [Google Scholar]
- 58.Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, et al Red list of China’s vertebrates. Biodiversity Science. 2016;24(5):500–551. doi: 10.17520/biods.2016076. [DOI] [Google Scholar]
- 59.Kozak KH, Wiens JJ Niche conservatism drives elevational diversity patterns in Appalachian salamanders. The American Naturalist. 2010;176(1):40–54. doi: 10.1086/653031. [DOI] [PubMed] [Google Scholar]
- 60.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution. 2017;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 61.Leigh JW, Bryant D POPART: full‐feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6(9):1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 62.Li JJ, Fang XM Uplift of the Tibetan Plateau and environmental changes. Chinese Science Bulletin. 1999;44(23):2117–2124. doi: 10.1007/BF03182692. [DOI] [Google Scholar]
- 63.Li ZW, Chen ZR, Wang ML Classification and correlation of the quaternary glacial epoch in the Hengduan (Transverse) Mountains. Geological Review. 1991;37(2):125–132. [Google Scholar]
- 64.Librado P, Rozas J DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 65.Liu CR, Newell G, White M On the selection of thresholds for predicting species occurrence with presence‐only data. Ecology and Evolution. 2016;6(1):337–348. doi: 10.1002/ece3.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Wang CM, Fu DL, Hu XJ, Xie XM, Liu PF, et al Phylogeography of Nanorana parkeri (Anura: Ranidae) and multiple refugia on the Tibetan Plateau revealed by mitochondrial and nuclear DNA . Scientific Reports. 2015;5(1):9857. doi: 10.1038/srep09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu XD, Dong BW Influence of the Tibetan Plateau uplift on the Asian monsoon-arid environment evolution. Chinese Science Bulletin. 2013;58(34):4277–4291. doi: 10.1007/s11434-013-5987-8. [DOI] [Google Scholar]
- 68.López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography. 2011;38(7):1267–1280. doi: 10.1111/j.1365-2699.2011.02504.x. [DOI] [Google Scholar]
- 69.Lu B, Zheng YC, Murphy RW, Zeng XM Coalescence patterns of endemic Tibetan species of stream salamanders (Hynobiidae: Batrachuperus) . Molecular Ecology. 2012;21(13):3308–3324. doi: 10.1111/j.1365-294X.2012.05606.x. [DOI] [PubMed] [Google Scholar]
- 70.Lucati F, Poignet M, Miró A, Trochet A, Aubret F, Barthe L, et al Multiple glacial refugia and contemporary dispersal shape the genetic structure of an endemic amphibian from the Pyrenees. Molecular Ecology. 2020;29(15):2904–2921. doi: 10.1111/mec.15521. [DOI] [PubMed] [Google Scholar]
- 71.McCormack JE, Heled J, Delaney KS, Peterson AT, Knowles LL Calibrating divergence times on species trees versus gene trees: implications for speciation history of Aphelocoma jays . Evolution. 2011;65(1):184–202. doi: 10.1111/j.1558-5646.2010.01097.x. [DOI] [PubMed] [Google Scholar]
- 72.McRae B, Shah V, Edelman A. 2016. Circuitscape: modeling landscape connectivity to promote conservation and human health. Fort Collins: The Nature Conservancy.
- 73.McRae BH Isolation by resistance. Evolution. 2006;60(8):1551–1561. doi: 10.1111/j.0014-3820.2006.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 74.Moritz C Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology & Evolution. 1994;9(10):373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 75.Myers EA, Xue AT, Gehara M, Cox CL, Davis Rabosky AR, Lemos‐Espinal J, et al Environmental heterogeneity and not vicariant biogeographic barriers generate community‐wide population structure in desert‐adapted snakes. Molecular Ecology. 2019;28(20):4535–4548. doi: 10.1111/mec.15182. [DOI] [PubMed] [Google Scholar]
- 76.Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 77.Nei M, Tajima F, Tateno Y Accuracy of estimated phylogenetic trees from molecular data: II. Gene frequency data. Journal of Molecular Evolution. 1983;19(2):153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- 78.Noroozi J, Talebi A, Doostmohammadi M, Rumpf SB, Linder HP, Schneeweiss GM Hotspots within a global biodiversity hotspot-areas of endemism are associated with high mountain ranges. Scientific Reports. 2018;8(1):10345. doi: 10.1038/s41598-018-28504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otto CRV, Roloff GJ, Thames RE Comparing population patterns to processes: abundance and survival of a forest salamander following habitat degradation. PLoS One. 2014;9(4):e93859. doi: 10.1371/journal.pone.0093859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ouborg NJ Integrating population genetics and conservation biology in the era of genomics. Biology Letters. 2010;6(1):3–6. doi: 10.1098/rsbl.2009.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Owen LA, Caffee MW, Finkel RC, Seong YB Quaternary glaciation of the Himalayan-Tibetan orogen. Journal of Quaternary Science. 2008;23(6−7):513–531. [Google Scholar]
- 82.Pan T, Wang H, Yan P, Zhang CL, Zhou WL, Wu XB, et al The impact of stream landscape on genetic structure and dispersal patterns in stream salamander (Pachyhynobius shangchengensis) . Asian Herpetological Research. 2020;11(3):205–218. [Google Scholar]
- 83.Peakall R, Smouse PE GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28(19):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Phillips SJ, Dudík M, Schapire RE. 2017(2021-11-26). Maxent software for modeling species niches and distributions (Version 3.4. 1). http://biodiversityinformatics.amnh.org/open_source/maxent/.
- 85.Phuong MA, Bi K, Moritz C Range instability leads to cytonuclear discordance in a morphologically cryptic ground squirrel species complex. Molecular Ecology. 2017;26(18):4743–4755. doi: 10.1111/mec.14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piry S, Luikart G, Cornuet JM Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity. 1999;90(4):502–503. doi: 10.1093/jhered/90.4.502. [DOI] [Google Scholar]
- 87.Pritchard JK, Stephens M, Donnelly P Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qiu YX, Fu CX, Comes HP Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution. 2011;59(1):225–244. doi: 10.1016/j.ympev.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 89.Qu Y, Lei F, Zhang R, Lu X Comparative phylogeography of five avian species: implications for Pleistocene evolutionary history in the Qinghai‐Tibetan plateau. Molecular Ecology. 2010;19(2):338–351. doi: 10.1111/j.1365-294X.2009.04445.x. [DOI] [PubMed] [Google Scholar]
- 90.Qu YH, Luo X, Zhang RY, Song G, Zou FS, Lei FM Lineage diversification and historical demography of a montane bird Garrulax elliotii-implications for the Pleistocene evolutionary history of the eastern Himalayas . BMC Evolutionary Biology. 2011;11(1):147. doi: 10.1186/1471-2148-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rambaut A, Drummond A. 2012. FigTree: Tree Figure Drawing Tool, v1.4. 0. Institute of Evolutionary Biology, University of Edinburgh.
- 92.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology. 2018;67(5):901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rana SK, Luo D, Rana HK, O'Neill AR, Sun H Geoclimatic factors influence the population genetic connectivity of Incarvillea arguta (Bignoniaceae) in the Himalaya-Hengduan Mountains biodiversity hotspot . Journal of Systematics and Evolution. 2021;59(1):151–168. doi: 10.1111/jse.12521. [DOI] [Google Scholar]
- 94.Renner SS Available data point to a 4‐km‐high Tibetan Plateau by 40 Ma, but 100 molecular‐clock papers have linked supposed recent uplift to young node ages. Journal of Biogeography. 2016;43(8):1479–1487. doi: 10.1111/jbi.12755. [DOI] [Google Scholar]
- 95.Rice WR Analyzing tables of statistical tests. Evolution. 1989;43(1):223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 96.Robertson JM, Murphy MA, Pearl CA, Adams MJ, Páez‐Vacas MI, Haig SM, et al Regional variation in drivers of connectivity for two frog species (Rana pretiosa and R. luteiventris) from the U. S. Pacific Northwest . Molecular Ecology. 2018;27(16):3242–3256. doi: 10.1111/mec.14798. [DOI] [PubMed] [Google Scholar]
- 97.Rogers AR, Harpending H Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9(3):552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 98.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosenberg NA DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4(1):137–138. [Google Scholar]
- 100.Rousset F GENEPOP’ 007: a complete re‐implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources. 2008;8(1):103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 101.Sánchez-Montes G, Recuero E, Barbosa AM, Martínez‐Solano Í Complementing the Pleistocene biogeography of European amphibians: Testimony from a southern Atlantic species. Journal of Biogeography. 2019;46(3):568–583. doi: 10.1111/jbi.13515. [DOI] [Google Scholar]
- 102.Sánchez-Montes G, Wang JL, Ariño AH, Martínez‐Solano Í Mountains as barriers to gene flow in amphibians: quantifying the differential effect of a major mountain ridge on the genetic structure of four sympatric species with different life history traits. Journal of Biogeography. 2018;45(2):318–331. doi: 10.1111/jbi.13132. [DOI] [Google Scholar]
- 103.Shu XX, 2020. Conservation Genetics and Habitat Selection of Taliang Knobby Newt (Liangshantriton taliangensis). Sichuan University, Chengdu. (in Chinese)
- 104.Silvestro D, Michalak I raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution. 2012;12(4):335–337. [Google Scholar]
- 105.Spear SF, Balkenhol N, Fortin MJ, McRae BH, Scribner K Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Molecular Ecology. 2010;19(17):3576–3591. doi: 10.1111/j.1365-294X.2010.04657.x. [DOI] [PubMed] [Google Scholar]
- 106.Spicer RA, Su T, Valdes PJ, Farnsworth A, Wu FX, Shi GL, et al Why ‘the uplift of the Tibetan Plateau’ is a myth. National Science Review. 2021;8(1):nwaa091. doi: 10.1093/nsr/nwaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Su T, Spicer RA, Li SH, Xu H, Huang J, Sherlock S, et al Uplift, climate and biotic changes at the Eocene-Oligocene transition in South-eastern Tibet. National Science Review. 2019;6(3):495–504. doi: 10.1093/nsr/nwy062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun YB, An ZS, Clemens SC, Bloemendal J, Vandenberghe J Seven million years of wind and precipitation variability on the Chinese Loess Plateau. Earth and Planetary Science Letters. 2010;297(3−4):525–535. doi: 10.1016/j.epsl.2010.07.004. [DOI] [Google Scholar]
- 109.Swets JA Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 110.Szpiech ZA, Jakobsson M, Rosenberg NA ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24(21):2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tajima F Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takezaki N, Nei M, Tamura K POPTREEW: web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Molecular Biology and Evolution. 2014;31(6):1622–1624. doi: 10.1093/molbev/msu093. [DOI] [PubMed] [Google Scholar]
- 113.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang CQ, Matsui T, Ohashi H, Dong YF, Momohara A, Herrando-Moraira S, et al Author Correction: identifying long-term stable refugia for relict plant species in East Asia. Nature Communications. 2018;9(1):5241. doi: 10.1038/s41467-018-07727-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Toews DPL, Brelsford A The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology. 2012;21(16):3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 116.Tzedakis PC, Lawson IT, Frogley MR, Hewitt GM, Preece RC Buffered tree population changes in a Quaternary refugium: evolutionary implications. Science. 2002;297(5589):2044–2047. doi: 10.1126/science.1073083. [DOI] [PubMed] [Google Scholar]
- 117.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P MICRO‐CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4(3):535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 118.Vasconcellos MM, Colli GR, Weber JN, Ortiz EM, Rodrigues MT, Cannatella DC Isolation by instability: Historical climate change shapes population structure and genomic divergence of treefrogs in the Neotropical Cerrado savanna. Molecular Ecology. 2019;28(7):1748–1764. doi: 10.1111/mec.15045. [DOI] [PubMed] [Google Scholar]
- 119.Wang B, Nishikawa K, Matsui M, Nguyen TQ, Xie F, Li C, et al Phylogenetic surveys on the newt genus Tylototriton sensu lato (Salamandridae, Caudata) reveal cryptic diversity and novel diversification promoted by historical climatic shifts . PeerJ. 2018;6:e4384. doi: 10.7717/peerj.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang B, Xie F, Li JN, Wang G, Li C, Jiang JP Phylogeographic investigation and ecological niche modelling of the endemic frog species Nanorana pleskei revealed multiple refugia in the eastern Tibetan Plateau . PeerJ. 2017;5:e3770. doi: 10.7717/peerj.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang IJ Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution. 2013;67(12):3403–3411. doi: 10.1111/evo.12134. [DOI] [PubMed] [Google Scholar]
- 122.Wang J, Raisbeck G, Xu XB, Yiou F, Bai SB In situ cosmogenic 10Be dating of the Quaternary glaciations in the southern Shaluli Mountain on the southeastern Tibetan Plateau . Science in China Series D:Earth Sciences. 2006;49(12):1291–1298. doi: 10.1007/s11430-006-2026-5. [DOI] [Google Scholar]
- 123.Wang ZW, Chen ST, Nie ZL, Zhang JW, Zhou Z, Deng T, et al Climatic factors drive population divergence and demography: insights based on the phylogeography of a riparian plant species endemic to the Hengduan Mountains and adjacent regions. PLoS One. 2015;10(12):e0145014. doi: 10.1371/journal.pone.0145014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Waraniak JM, Fisher JDL, Purcell K, Mushet DM, Stockwell CA Landscape genetics reveal broad and fine‐scale population structure due to landscape features and climate history in the northern leopard frog (Rana pipiens) in North Dakota . Ecology and Evolution. 2019;9(3):1041–1060. doi: 10.1002/ece3.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu YH, Yan F, Stuart BL, Prendini E, Suwannapoom C, Dahn HA, et al A combined approach of mitochondrial DNA and anchored nuclear phylogenomics sheds light on unrecognized diversity, phylogeny, and historical biogeography of the torrent frogs, genus Amolops (Anura: Ranidae) . Molecular Phylogenetics and Evolution. 2020;148:106789. doi: 10.1016/j.ympev.2020.106789. [DOI] [PubMed] [Google Scholar]
- 126.Wu YK, Wang YZ, Jiang K, Hanken J Significance of pre-Quaternary climate change for montane species diversity: insights from Asian salamanders (Salamandridae: Pachytriton) . Molecular Phylogenetics and Evolution. 2013;66(1):380–390. doi: 10.1016/j.ympev.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 127.Xie C, Xie DF, Zhong Y, Guo XL, Liu Q, Zhou SD, et al The effect of Hengduan Mountains Region (HMR) uplift to environmental changes in the HMR and its eastern adjacent area: tracing the evolutionary history of Allium section Sikkimensia (Amaryllidaceae) . Molecular Phylogenetics and Evolution. 2019;130:380–396. doi: 10.1016/j.ympev.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 128.Xing YW, Ree RH Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(17):E3444–E3451. doi: 10.1073/pnas.1616063114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu LB, Zhou SZ, Wang J Pleistocene glaciations in the Shaluli Shan and the influences of southwest monsoon on the glaciations during the last glacial period. Quaternary Sciences. 2005;25(5):620–629. [Google Scholar]
- 130.Yao Q, Song J, Cheng J, Yang W, Xu C, Zhao J Integral uplift and anticlockwise rotation of the Daliangshan sub-block. Chinese Journal of Geology. 2017;52(2):328–342. [Google Scholar]
- 131.Ye XY, Ma PF, Yang GQ, Guo C, Zhang YX, Chen YM, et al Rapid diversification of alpine bamboos associated with the uplift of the Hengduan Mountains. Journal of Biogeography. 2019;46(12):2678–2689. doi: 10.1111/jbi.13723. [DOI] [Google Scholar]
- 132.Yu GH, Zhang MW, Rao DQ, Yang JX Effect of pleistocene climatic oscillations on the phylogeography and demography of red knobby newt (Tylototriton shanjing) from Southwestern China . PLoS One. 2013;8(2):e56066. doi: 10.1371/journal.pone.0056066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu Y, Harris AJ, Blair C, He XJ RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 134.Zhan XJ, Zheng YF, Wei FW, Bruford MW, Jia CX Molecular evidence for Pleistocene refugia at the eastern edge of the Tibetan Plateau. Molecular Ecology. 2011;20(14):3014–3026. doi: 10.1111/j.1365-294X.2011.05144.x. [DOI] [PubMed] [Google Scholar]
- 135.Zhang DR, Chen MY, Murphy RW, Che J, Pang JF, HU JS, et al Genealogy and palaeodrainage basins in Yunnan Province: phylogeography of the Yunnan spiny frog, Nanorana yunnanensis (Dicroglossidae) . Molecular Ecology. 2010;19(16):3406–3420. doi: 10.1111/j.1365-294X.2010.04747.x. [DOI] [PubMed] [Google Scholar]
- 136.Zhang YH, Wang IJ, Comes HP, Peng H, Qiu YX Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae) . Scientific Reports. 2016;6(1):24041. doi: 10.1038/srep24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng BX, Xu QQ, Shen YP. 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai–Tibetan Plateau: review and speculation. Quaternary International, 97–98: 93–101.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.