Abstract
Microbial organisms have diverse populations, where using a single linear reference sequence in comparative studies introduces reference-bias in downstream analyses, and leads to a failure to account for variability in the population. Recently, pan-genome graphs have emerged as an alternative to the traditional linear reference with many successful applications and a rapid increase in the number of methods available in the literature. Despite this enthusiasm, there has been no attempt at exploring these graph construction methods in depth, demonstrating their practical use. In this study, we aim to develop a general guide to help researchers who may want to incorporate pan-genomes in their analyses of microbial organisms. We evaluated the state-of-the art pan-genome construction tools to model a collection of 70 Acinetobacter baumannii strains. Our results suggest that all tools produced pan-genome graphs conforming to our expectations based on previous literature, and that their approach to homologue detection is likely to be the most influential in determining the final size and complexity of the pan-genome. The graphs overlapped most in the core pan-genome content while the cloud genes varied significantly among tools. We propose an alternative approach for pan-genome construction by combining two of the tools, Panaroo and Ptolemy, to further exploit them in downstream analyses, and demonstrate the effectiveness of our pipeline for structural variant calling in beta-lactam resistance genes in the same set of A. baumannii isolates, identifying various transposon structures for carbapenem resistance in chromosome, as well as plasmids. We identify a novel plasmid structure in two multidrug-resistant clinical isolates that had previously been studied, and which could be important for their resistance phenotypes.
Keywords: Acinetobacter baumannii, bacterial pangenomics, pan-genome graphs, string graphs, comparative genomics, plasmids
Data Summary
A dataset of 70 Acinetobacter baumannii strains has been curated from a published dataset used in a comparative study of adaptation in niche environments by removing the oldest assemblies of low quality [1]. This particular dataset was selected as the use-case for evaluating pan-genomes because (i) it comprises only full-length genome assemblies, (ii) it includes strains isolated from different environments and thus is diverse, and (iii) the original study provides a common ground on which a baseline evaluation can be performed to compare the results of different tools. Sequence and annotation data have been obtained from the NCBI RefSeq database [2]; the accession numbers of assemblies used in this work are listed in Table S1 (available in the online version of this paper).
Impact Statement.
Use of a single linear reference in comparative genomics introduces reference-bias, especially in diverse populations such as microbial organisms. As a solution, pan-genome graphs have found many successful applications, and now we have several methods available in the literature. However, there is a lack of comparative studies in the field and the sheer number of choices can be overwhelming. To address this gap, we present an explorative study to introduce the average user to pan-genome graphs and guide them in using pan-genomes to study microbial organisms. We evaluated the state-of-the art pan-genome construction tools to model Acinetobacter baumannii populations. While each tool produced valid graphs in line with previous work, there were significant differences in the cloud genes. Next, we demonstrate their use in a pipeline we have developed to call structural variants; we detected transposons carrying beta-lactam resistance genes, and identified a novel plasmid structure associated with multidrug resistance. The novelty of our study is two-fold: first, it is among the rare work in the field to provide insight into the current state-of-the-art in pan-genome tools for microbial organisms, and second, it shows that we can combine two of these tools in a pipeline to call structural variants successfully.
Introduction
As the amount of DNA sequence data available has increased dramatically, the conventional, reference-based approach in bioinformatics is being re-examined. Relying on a single linear reference sequence in comparative genomic studies can lead to reference-bias in downstream analyses, and to failure to account for population variance which may be valuable [3].
Pan-genome graphs have been proposed as an alternative to a linear reference to model a collection of DNA sequences [4], representing genes shared across multiple genomes in a compact structure, and hence, they have found many applications in many tasks such as genome alignment, read mapping and variant calling [5–8]. Several methods have been developed in the literature to construct gene-based pan-genome graphs, and we can summarize these methods in roughly three steps: (i) identifying homologue genes based on all-vs-all pairwise alignments [9–11] and clustering [12, 13], (ii) paralogue splitting and (iii) linking families to preserve the genomic order. For paralogue splitting, two different approaches stand out: tree-based ones that make use of the phylogeny in gene families and synteny-based ones in which the neighbourhood of each gene family guides the paralogue splitting process. The final step may vary depending on the output of the tool and, in some algorithms, it may be absent unless a final graph is produced.
Currently, there has been no attempt at bringing these graph construction methods to a common ground, assessing both their weaknesses and strengths independent of their computational performance. In this study, we evaluate the state-of-the-art pan-genome construction tools to propose general guidelines and rules-of-thumb to help with researchers who may want to incorporate pan-genomes in their analyses, particularly in those of microbial organisms. The aim is to explore what questions each tool might be useful in answering, and in what ways we can make use of these answers to gain valuable biological insight. We performed a comparative study on a collection of 70 Acinetobacter baumannii strains of different isolation sources that has previously been published [1]. A. baumannii , a multi-drug-resistant bacteria classified as an ESKAPE pathogen, is among the leading causes of nosocomial infections, and thus plays a vital role in understanding antibiotic resistance [14]. Studies have established genes associated with several traits including virulence, pathogenicity and adaptation to its niche, probably acquired through horizontal transfer in large clusters via plasmids [15]. A. baumannii , as a population, has a diverse gene repertoire, and exhibits large, structural rearrangements; hence it has the prominent characteristics of bacterial genomes and presents as a good example use-case for application of pan-genome graphs in bacterial species. Given its typical average genome size and plasmid content for bacteria, it should not pose any additional challenges to the algorithms which would interfere with the comparison. In this work, first, we verify that our results confirm the original analyses, and are in parallel with previous studies on A. baumannii . Next, we propose to combine two of the pan-genome construction tools we have evaluated, Panaroo and Ptolemy, to further exploit them in downstream analyses; the effectiveness of this approach is demonstrated by calling structural variants in A. baumannii species to gain more insights in the data set. We analysed different structures of transposons carrying the bla OXA-23 carbapenemase gene in the set of A. baumannii strains. In addition, we explore A. baumannii plasmids, and locate novel structures that might be involved in transferring multiple antimicrobial resistance genes.
Methods
In this section, we first describe our approach for comparing state-of-the-art pan-genome tools. The aim in the first part of this study is to evaluate existing tools in both qualitative and quantitative terms, and to provide an overview of the current field. In the second part, two of these tools, Panaroo and Ptolemy, are used in conjunction for calling structural variants. The final pan-genome graph serves as a compact model of a set of genomes, utilizing Panaroo’s error correction mechanisms while it also retains the sequence continuity in each genome with the guidance of Ptolemy’s indexing and anchoring.
Data preprocessing
The A. baumannii dataset was curated from a previous study by Yakkala et al. which analysed niche-specific adaptations of A. baumannii [1]. We removed the oldest, low-quality assemblies to retain 70 in total. Nucleotide sequences and annotations were downloaded from the NCBI RefSeq database (assembly accessions are listed in Table S1) [2]. The genomes were not re-annotated in our study since all the assemblies had been annotated by NCBI’s prokaryotic annotation pipeline, and thus they had gone through the same process. NCBI annotations were corrected and the corresponding nucleotide sequences were appended to the GFF input files using the python script ‘convert_refseq_to_prokka_gff.py’ provided by Panaroo (see Supplementary Text) [16].
Tools
In this study, we have compared the tools Roary (v3.13.0), Ptolemy (v1.0), PPanGGOLiN (v1.0.13), PIRATE (v1.0.3) and Panaroo (v1.1.2) [16–20]. The set of tools are by no means comprehensive; however, they are diverse enough in their methodology and at the same time sufficiently similar in their purpose to make comparison meaningful. In addition, the pan-genome representation is consistent across these tools; the pan-genome is a graph in which the nodes are formed by at least one gene (a node may contain multiple genes forming an orthologous cluster) and the edges indicate sequence continuity between two nodes. All the tools were run in their default settings according to instructions provided by their authors. Tools which allow for some options without the need for parameter tuning were also run with these options. A full list of commands and arguments used in this study can be found in the Supplementary Text.
Qualitative and quantitative assessment
In the first part, we compared different tools qualitatively in their usage first in terms of software availability input/output file formats and compatibility with existing downstream analyses. Input is usually sequences with their annotations in FASTA and GFF files, or GenBank and GFF3 files that contain both the nucleotide sequences and annotations in a single file. If a tool provides sequence annotation as well, then FASTA sequences alone can be used. Since these tools are often run within a pipeline, once a pan-genome is obtained, it might be used for aligning reads and whole genomes, calling structural variants or performing genome-wide association studies (GWAS). To establish compatibility, tools produce outputs in commonly used file formats such as DOT, GML, GEXF or GFA for graphs, NEWICK for phylogenic trees and tab-separated text files for the remaining types of outputs. In addition to these, we attempt to compare the core algorithms of the tools by breaking down pan-genome construction into multiple steps in (i) detection of homologue genes, as there are different methods (blast, DIAMOND, CD-HIT, minimap2) to determine sequence similarity; (ii) paralogue identification (and splitting) to differentiate paralogues from orthologues and find repeats in a genome, which can be achieved using the local context of genes (synteny), phylogenetic information or graph-based approaches; (iii) type of final output, a directed/undirected graph if a graph is produced, or gene clusters; and (iv) additional functions the tools provide for correction of annotations, assembly errors, or pre-/post-processing for variant calling, converting file formats, etc [9–11, 21].
For quantitative comparison, the numbers of nodes, edges and connected components were used as metrics to assess the graph size. Pan-genome content was measured based on the average number of genomes per node, and the soft-core thresholding approach, which is implemented frequently in the literature to classify gene clusters [22]: core genome is observed in over 99 %, soft core in 95–99 %, shell in 15–95 % and the cloud genome is observed in less than 15 % of the strains in the dataset. Unique genes are defined as the singleton nodes on the graph; they are present in only a single strain. Finally, we established a pairwise comparison in core pan-genome content using the Jaccard index: , where A and B are two different sets of core genes identified by different tools.
Replication of Yakkala et al.
Pan-genome graphs were also used to replicate the following findings of the previous study from which the dataset had been obtained [1]:
Identify genes related to different carbon catabolism, and iron acquisition in environmental A. baumannii strain isolated from soil, DS002.
Find antimicrobial resistance genes associated with biofilm formation, efflux pumps and beta-lactamases in the clinical strains.
We processed pan-genome graphs constructed by all five of the tools to identify the nodes which contain genes from only the environmental A. baumannii strain DS002; these nodes represent the unique genome content of DS002. Next, we extracted all the genes contained in these nodes to analyse gene enrichment. In our replication study, gene enrichment analyses were performed using the python package GOATOOLS (v0.9.9) [23]. Gene ontology (GO) hierarchy in OBO format was retrieved from the Gene Ontology Website [24], and all the annotated ORFs in our dataset were mapped to GO terms using the ID mapping tool on UniProt [25]. Gene enrichment was performed with correction for false discovery rate (fdr option in GOAtools), and GOATOOLS was also used for plotting GO subgraphs in python. The in-house python script used to perform the analysis (see ‘runGOE.py’) is provided in the Supplementary Text.
Combining methods
Individual steps from two of the tools in our comparative study, Panaroo and Ptolemy, were combined. Briefly, Panaroo uses CD-HIT with a high threshold for sequence identity to obtain gene clusters. These clusters are then collapsed according to synteny information, which is also used to find missing genes and correct for possible errors in assembly and annotation [16]. The error correction in Panaroo can disrupt sequence continuity by breaking up genomes. For that reason, we used Ptolemy to index genomes and connect the nodes to retain the sequence continuity so that each genome can be traversed as a path on the graph.
Panaroo was first run with default parameters in relaxed option (‘mode –relaxed’) in order to get an initial estimate of the pan-genome that consists of gene families as nodes in the graph output file ‘final_graph.gml’. Next, all sequences were indexed with Ptolemy (‘extract’), and Panaroo’s gene families were reformatted to match Ptolemy’s indexing and conform to the format of the syntenic anchor input file in python (see the script ‘createSA.py’ in Supplementary Text). Gene families were then used as input to the canonical quiver construction step in Ptolemy’s algorithm (‘canonical-quiver’). The final output is a directed graph stored in a GFA file. The Supplementary Text provides the full list of commands, as well as the in-house scripts used to perform the analysis.
Results and discussion
Qualitative and quantitative comparison
To evaluate different tools for pan-genome construction, we selected a number of tools from the literature, Table 1 provides a qualitative overview as described in the methods section for the set of five tools we applied to our A. baumannii dataset. The most prominent feature among the tools is their compatibility with other software; they accept inputs in standard formats for sequence and annotation data, and produce graph outputs in formats compatible with common graph visualization software. Since all tools construct gene-based pan-genomes, sequences should be annotated with predicted ORFs beforehand, with the exception of PPanGGoLin which can run Prodigal internally for annotation. The tools differ most in their choice of sequence similarity, while usually the synteny or phylogeny (tree-based) information is used for paralogue detection with the exception of Ptolemy, which opts for a repeat graph.
Table 1.
Summary overview of qualitative features of pan-genome tools implemented in this study
|
Method |
Software |
Input |
Graph output |
Pan-genome |
Sequence homology |
Paralogue identification |
|---|---|---|---|---|---|---|
|
Roary (v3.13.0) |
Conda package |
GFF3 |
DOT |
Directed graph |
blast |
Synteny |
|
Ptolemy (v1.0) |
Java executable |
FASTA+GFF |
GFA |
Directed graph |
minimap2 |
Graph-based |
|
PPanGGoLin (v1.0.13) |
Conda package |
GBK or FASTA |
GEXF |
Undirected graph |
MMseq2 |
Synteny |
|
PIRATE (v1.0.3) |
Conda package |
GFF3 |
GFA |
Directed graph |
blast (/DIAMOND) |
Synteny |
|
Panaroo (v1.1.2) |
Conda package |
GFF3 |
GML |
Directed graph |
CD-HIT |
Synteny |
Depending on the aim of pan-genome analysis, some tools could be preferred for the outputs they generate in addition to a graph, although our quantitative comparison on our A. baumannii dataset is limited to the graphs and we did not investigate these additional features in our use-case. Both Panaroo and Ptolemy have modules to identify structural rearrangements, while PIRATE, Panaroo and Roary can perform core gene alignment, which can be useful for downstream phylogenetic studies. Similarly, the binary gene presence/absence outputs from PPanGGoLin, PIRATE, Panaroo and Roary can also be used to make a quick and dirty tree or run pan-genome association studies.
Another feature of these tools is that they can be packaged with auxiliary scripts for pre-/post-processing, which can save user time. For instance, Panaroo and Roary both include scripts to perform quality control on the input data before generating a pan-genome graph. Moreover, Roary, Panaroo and PIRATE provide scripts for querying the pan-genome. All tools, except for Ptolemy, have built-in modules to plot pan-genome statistics in various ways. For visualizing the pan-genome graph, we found Ptolemy and PIRATE to be the most straightforward since the GFA outputs can be used directly in Bandage [26]. However, depending on the use-case, Panaroo might be preferred for its GML output, which can be visualized more extensively using Cytoscape and combining additional metadata with the graph [27]. Finally, we note that among these projects, Roary is the only one that is no longer maintained actively. However, the PPanGGoLin, PIRATE and Panaroo packages have all been extended since completion of our study.
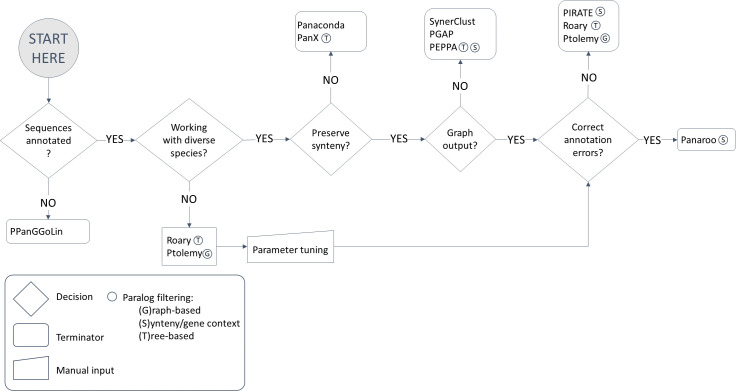
The flowchart in Fig. 1 also includes tools not implemented in our study, to provide a guide to help users choose among the state-of-the-art pan-genome tools. For some applications, the flowchart in Fig. 1 can lead to multiple tools to choose from. In that case, one can differentiate the tools according to the (i) required inputs or (ii) additional outputs they produce and whether they could benefit from these in the downstream analysis. For instance, Panaconda and PanX both provide visualization of the results [27, 28]. Panaconda’s GEXF graph can be viewed using JS visualizer, while panX, having an accompanying web-based interactive application, has more extensive options to visualize the outputs. PanX also provides several statistics on genes (count, length, distribution, etc.), and a phylogenetic tree, all of which can be manipulated and adjusted through its graphical interface. Note that it is not possible to perform the analysis for one’s own dataset using the web interface alone. Moreover, Panaconda requires input in PATRIC’s feature tab format, in comparison to GBK format as in PanX, which might be less convenient to prepare depending on the data available [28].
Fig. 1.
Flowchart for users to choose among the current state-of-the-art pan-genome construction tools in the literature, accompanying the analyses in this study.
If one ends up having to decide between SynerClust, PGAP and PEPPA, the input requirements could be considered. PEPPA has the fewest requirements, with only annotations (GFF) and the nucleotide sequence; for PGAP, protein sequences and more extensive gene annotations, including their functional descriptions and COG classifications, must be available [29–31]. For SynerClust, a phylogenetic tree of the genomes needs to be available along with gene annotations (GFF). In terms of the outputs, these three tools are not significantly different except for PGAP, which also performs species evolution and gene function enrichment analysis, and could eliminate the need to run additional tools downstream. Note that, in general, for tools that require annotations beyond to the most basic ORF predictions, the performance of the upstream annotation step may become even more important, possibly more than the choice of the pan-genome construction tool itself.
While all tools used on the A. baumanni dataset for quantitative comparison were run in default settings, we note that all tools considered in Fig. 1 are flexible enough to allow for parameter tuning to tailor for different use-cases. The subset of tools implemented in our study are mostly suited for use in microbial organisms out-of-the-box. It is estimated that the A. baumannii pan-genome contains 20 000 genes [32, 33], although some studies report fewer than or approximately 10 000 genes [1, 34]. We presume this number should vary depending on the diversity among the strains in a particular dataset, in addition to the method of choice. A. baumannii can colonize a variety of ecological niches; its extreme adaptability is driven by a flexible genome that allows for the acquisition of new genes, and these niche-specific genes can inflate the pan-genome. Except for two strains (SDF and DS002), all genomes in our dataset were isolated from clinical sources, and thus there is little variation in their habitat, and it is likely that the smaller estimate of the pan-genome size is more applicable in our example. Regardless, with an average genome size of 3 Mbp, around 3000 coding sequences, and an estimated pan-genome of 10 000 genes, we found A. baumannii to be a middle-of-the-road species among other bacteria [35]. We also note that the A. baumannii pan-genome has been classified as open, and that our findings may not generalize to closed pan-genomes [36].
We observe that all tools produced graphs within the range of previous studies (Table 2). In terms of graph size and complexity, there is little variation between PPanGGoLin, PIRATE and Panaroo, possibly due to their similar algorithms. Roary and Ptolemy, on the other hand, stand out with the largest graphs, which indicates a more stringent threshold for sequence similarity. When the synteny window size was increased in Ptolemy, the number of nodes varied by as much as 15 % (~3000 nodes, see Table S2). PPanGGoLin and Panaroo also have the option to run in different modes (-defrag in PPanGGoLin, and -relaxed in Panaroo) that might allow for some adjustment without parameter tuning (Table 2); graph size decreased by 17 % (~2000 nodes) in the former (PPanGGoLin with -defrag mode) and increased by less than 5 % (~400 nodes) in the latter (Panaroo in -relaxed mode). Costa et al. also report significant changes in bacterial pan-genomes when the sequence identity thresholds are altered [35]. We recommend that users try different values and settings for the parameters in these tools to improve their results. External annotations, such as known orthologous clusters from the COG database, protein families from the Pfam database, or KEGG pathways can be used to check for the integrity of nodes in the pan-genome graph and possibly guide the parameter tuning, which is beyond the scope of this work.
Table 2.
Quantitative comparison of pan-genome graphs in terms of size and complexity, and the pan-genome content, all run in default configurations, except for the three that also include a different mode
|
Method |
Roary |
Ptolemy |
PPanGGoLin |
PIRATE |
Panaroo |
||
|---|---|---|---|---|---|---|---|
|
Settings |
Default |
Default |
Default |
Defrag |
blast |
Relaxed |
Strict |
|
No. of nodes |
13 928 |
20 140 |
11 329 |
9318 |
7871 |
10 776 |
10 336 |
|
No. of edges |
21 032 |
31 946 |
17 751 |
14 989 |
11 331 |
16 270 |
14 709 |
|
No. of connected components |
10 |
34 |
7 |
6 |
416 |
13 |
13 |
|
Mean sequence length (bp) |
803.7 |
870.5 |
815.5 |
849.8 |
850 |
824.6 |
832.5 |
|
Average no. of genomes per node |
18.7 |
11.7 |
22.4 |
27.1 |
31.6 |
24.4 |
25 |
|
Core genes |
1996 (14.3 %) |
1623 (8.1 %) |
2025 (17.9 %) |
2223 (23.8 %) |
2126 (27 %) |
1910 (17.7 %) |
2353 (22.8 %) |
|
Soft core genes |
429 (3.1 %) |
624 (3.1 %) |
509 (4.5 %) |
389 (4.2 %) |
447 (5.7 %) |
783 (7.3 %) |
322 (3.1 %) |
|
Shell genes |
1912 (13.7 %) |
1367 (6.8 %) |
1624 (14.3 %) |
1526 (16.4 %) |
1516 (19.3 %) |
1655 (15.4 %) |
1609 (15.6 %) |
|
Cloud |
9591 (68.9 %) |
16 526 (82.1 %) |
7171 (63.3 %) |
5180 (55.6 %) |
3782 (48 %) |
6419 (59.6 %) |
6052 (58.5 %) |
|
Unique |
5276 (37.9 %) |
13 963 (69.3%) |
4271 (37.7 %) |
2522 (27.1 %) |
1629 (20.7 %) |
3299 (30.6 %) |
2968 (28.7 %) |
We also report pan-genome content with the soft-core approach as outlined in the Methods section, in terms of both the number of nodes (or clusters) and the percentage with the respect to the entire pan-genome. Previous studies estimate the core content of the A. baumannii pan-genome to be in the range 1500–2500 [37]; in a recent analysis of 2112 A . baumannii strains, Mangas et al. identified 2221 core genes while the entire pan-genome comprised 19 000 genes in total [32]. Yakkala et al. also found a total of 7683 genes in the pan-genome, 1344 of which are core genes and 1695 are unique (present in only one genome) [1]. In our analyses, we found the variance in the core gene size to be much smaller compared to the entire pan-genome. While the difference in core genome size ranges from 130 to 503 genes, all tools predict the core content to be within the established range from previous studies. We also computed the pairwise Jaccard index for the core genome content, and observed that it varies from 0.65 to 0.91 (Table 3). However, when core and soft core genes are considered together, the difference in the number of genes is much smaller.
Table 3.
Pairwise similarity of core genome content
|
Roary |
Ptolemy |
PPanGGoLin |
PIRATE |
Panaroo |
|
|---|---|---|---|---|---|
|
Roary |
0.68 |
0.87 |
0.87 |
0.84 |
|
|
Ptolemy |
0.68 |
0.74 |
0.71 |
0.65 |
|
|
PPanGGoLin |
0.87 |
0.74 |
0.91 |
0.85 |
|
|
PIRATE |
0.87 |
0.71 |
0.91 |
0.88 |
|
|
Panaroo |
0.84 |
0.65 |
0.85 |
0.88 |
We observed the largest differences among tools in the cloud genes, as Ptolemy and Roary both stand out with the largest set of cloud genes (Table 2). Both tools have a relatively pared-down approach with fewer steps to identify homologous genes, which could possibly lead to a more stringent algorithm that produces clusters with fewer number of genes on average, and an inflated pan-genome. This suggests that the homologue detection step is likely to have the largest influence on the cloud gene content, although it should be further investigated by changing parameter settings, which is beyond the scope of this work. For applications in diverse species, or in cases where the strain speciation is of primarye interest, cloud gene content could become more important since the cloud content reflects how an organism has evolved and diversified to adapt to different conditions and environments. In that case, we presume the cloud gene content to play the most important role in deciding which tool to use.
Replication of Yakkala et al.
Following the preliminary assessment of pan-genome graphs, we attempted to replicate the major conclusions drawn in a previous study performed on the same A. baumannii dataset [1]. Yakkala et al. had: (i) first identified genes related to the survival mechanism of A. baumannii in diverse environments, and observed that the non-clinical isolate DS002 carried genes which take part in detoxifying aromatic compounds to generate energy. An absence of genes with these functions in clinical isolates suggests the environmental strain had developed an adaptation mechanism in order to survive in soil that is often polluted with phenol-based insecticides. Strain DS002 also showed differences in iron acquisition mechanisms. (ii) Second, the authors reported the absence of genes involved in biofilm formation and efflux pumps, as well as modification of aminoglycoside molecules in non-clinical isolates.
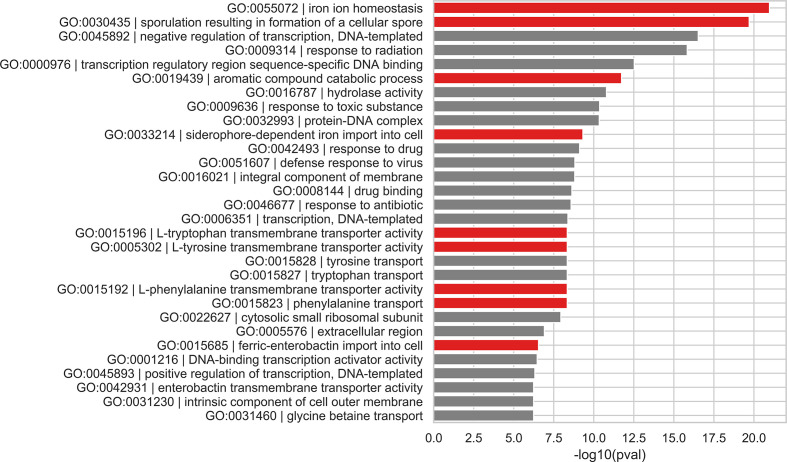
In order to replicate these findings, nodes containing genes from only the soil isolate were identified in the pan-genome output, and a gene enrichment analysis was performed against the entire pan-genome. Note that for this part, only the results from the Ptolemy graph are reported here. Compared to the other tools, we found Ptolemy to have the largest set of significant terms, but when the most significant terms in common were considered, the resulting sets would overlap more than 50 % (column 1 in Table S3, proportion of common GO terms). The remaining raw gene enrichment test results can be found in the Tables S4 and S5. Fig. 2 shows a bar chart of the most significant GO terms associated with these nodes; GO terms related to unique carbon catabolism and iron acquisition are highlighted in red. Conforming with Yakkala et al., the unique gene content of the soil isolate dataset is preserved in the pan-genome graph as well.
Fig. 2.
Gene enrichment analysis identifying the unique carbon catabolome and iron acquisition mechanisms in the soil isolate, DS002; bar plot of GO terms significantly enriched in DS002. GO IDs and term names are displayed on the y-axis, and the x-axis shows the −log10(P-value) of each GO term. GO terms associated with specialized mechanisms for carbon catalysis and iron acquisition are in red.
Next, we looked into GO terms significantly enriched in all the clinical isolates compared to the environmental strain DS002 isolated from soil, and a subgraph of GO hierarchy was extracted for the GO terms significantly enriched in clinical isolates; Fig. S1 shows this subgraph detailing the portion including terms associated with aminoglycoside-modifying enzymes and efflux pumps. A. baumannii strains are also known to have developed mechanisms for biofilm formation as a fundamental strategy for survival that contribute to their antibiotic resistance. GO terms associated with biofilm formation are identified as significant in clinical isolates; these terms are drawn in Fig. S2 in the context of GO hierarchy as well.
Combining methods
To further demonstrate the power of pan-genome graphs we created a hybrid downstream application for calling structural variants in pan-genome graphs. Following the results in the first part of this study, we attempted to combine individual steps from Panaroo and Ptolemy to obtain a pan-genome that we assert is more suited for this task than using either tool on its own. We report the results from variant calling first in the context of genes involved in carbapenem and amikacin resistance and then for validating and identifying possible novel plasmid structures in our A. baumannii dataset.
In the preliminary exploratory part of this study, we found that Panaroo produced average-sized pan-genomes, which suggests it achieves a good balance between over- and under-clustering for our particular use-case. However, we also observed that the error correction step in Panaroo could disrupt the continuity in certain chromosomes, thereby making it difficult to place them within the context of individual genomes. It is more challenging to analyse structural differences without sufficient contextual information. To circumvent this, we attempted to re-introduce sequence continuity by making use of the indexing and path construction steps in Ptolemy. While Panaroo’s error correction was shown to be highly useful for handling fragmented assemblies as well as annotation errors in the original study, our dataset consists of only chromosome-level complete assemblies and thus such errors should be negligible and we presume there will not be any major benefits from correcting the assemblies.
Unlike other tools, Ptolemy was developed modularly to the extent that each module can essentially be used independently, provided that the inputs are in the correct format. Hence, it is relatively straightforward to use Ptolemy to (i) index all the genomes, so we can keep track of the order and place of each gene within a genome, and (ii) construct a graph using indexed genes as a guide to connect genomes broken down into separate islands of gene clusters. The resulting graph contains the same set of nodes as those produced by Panaroo initially, but with an increased number of edges to establish sequence continuity. Thus, we obtain a pan-genome graph that preserves whole genomes at a coarse level, and can easily be queried for structural variant calling at small distances.
Structural variation in transposons carrying the bla OXA-23 carbapenemase gene
Using the combined method, we explored the structural variation in β-lactamase-carrying transposons in A. baumannii . Beta-lactam resistance in A. baumannii is mainly driven by class D β-lactamase enzymes (also called oxacillinases or OXAs). The bla OXA-23 gene encoding OXAs is readily carried on transposons and thus frequently observed in clinical isolates, both on the chromosome and on plasmids. It is hypothesized that bla OXA-23 (red arrows in Fig. 3) was mobilized with the help of insertion sequence (IS) ISAb1 (green arrows in Fig. 3) [38]. ISAba1 acts as a promotor, and only in the presence of this sequence is the level of gene expression enough to lead to significant imipenem, meropenem and doripenem resistance [39, 40]. Hence, it is also important to investigate the context of the gene in the A. baumannii genome and the mechanisms through which it is mobilized among the strains [41].
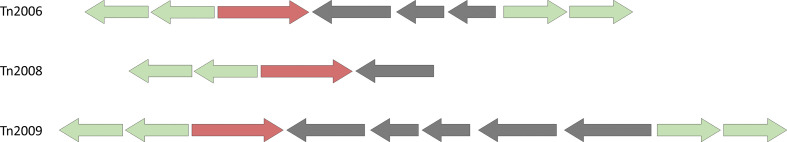
Fig. 3.
Three different structures of A. baumannii transposons in which the bla OXA-23 gene is present: ISAba1 is shown with green and bla OXA-23 with red arrows; all arrows are placed according to the direction of the ORFs.
So far, bla OXA-23 has been observed in five contexts in the literature, Tn2006, Tn2007, Tn2008, Tn2008B and Tn2009. Among these, the A. baumannii dataset contains strains that harbour three: Tn2006, Tn2008 and Tn2009 as reported in the literature (Fig. 3).
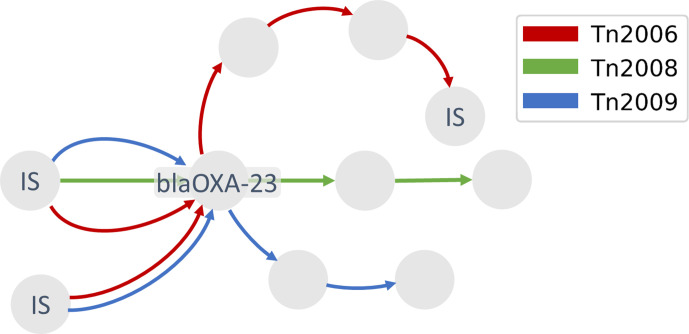
It is possible to locate the bla OXA-23 gene in our pan-genome, and extract the local neighbourhood around this gene. This allows us to visually assess different contexts. Strain 15A5, for instance, has two copies of the β-lactamase gene in a Tn2006 context in its chromosome, and Fig. 4 shows one of these copies (node labelled bla OXA-23 in Fig. 4) and their surrounding structure. Edges are coloured according to which path they belong to among these three different structures.
Fig. 4.
Local context of bla OXA-23 in A. baumannii strains extracted from the pan-genome graph reveals three different structures of transposons on which the gene is located. Edge colours indicate which structure each path belongs to: red for Tn2006, green for Tn2008 and blue for Tn2009. Only the nodes containing IS and bla OXA-23 are labelled; the unlabeled nodes represent ORFs.
Two ISAb1 sequences are located upstream of bla OXA-23 in reverse direction, and the forward ISs downstream are placed after two proteins of unknown function. Both copies of the upstream IS could be identified for Tn2006 and Tn2009 (two IS nodes left of the bla OXA-23 node in Fig. 4, connected with blue and red edges), whereas the downstream IS was detected only for Tn2006. In contrast, the Tn2008-carrying strains AbPK1 and CBA7 are both lacking this second instance of IS, and we did not observe it in the pan-genome graph either. According to the literature, strains BJAB0868 and BJAB07104 carry the Tn2009 transposon, but it was not possible to extract this transposon in its entirety due to the presence of four additional proteins of unknown function, since they increase the length of this syntenic region, thereby making it more difficult to capture it.
Exploring different plasmid structures
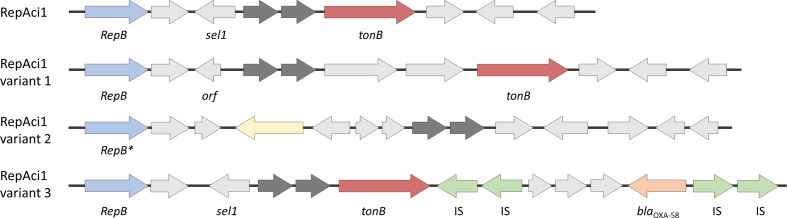
In addition to the resistance islands located in the chromosome, antimicrobial resistance genes related to carbapenems and amikacins are also frequently observed in plasmids. Conjugative plasmids play a crucial role in the spread of antibiotic resistance since they facilitate in transferring resistance genes by carrying transposons on which they are contained [42]. RepAci1 and RepAci2 types of plasmids are characterized by the replication proteins (RepB) and the dif modules they contain, and there is only little variance in the DNA sequence (sequence identity over 99.9 %), and hence can be represented by the RepAci1 plasmid pA1-1 (accession number CP010782.1) contained in an early strain A1 (also present in our A. baumannii dataset). While pA1-1 does not carry resistance genes, they can be found inserted in downstream of plasmid pA1-1 on transposons [43]. Blackwell et al. identified several RepAci1 and RepAci2 plasmids and their variants across different strains in [42]. In this section, we analysed the context of the pA1-1 plasmid in the pan-genome graph to find four different variants of this plasmid, three of which had been studied by Blackwell et al. (variants 1–3 in Fig. 5) and a novel one carrying resistance genes related to multiple drugs in our collection of strains.
Fig. 5.
RepAci1 plasmid structure and its three variants observed in our A. baumannii dataset: variant 1 in p2ABAYE, variant 2 in pD36-3 and variant 3 in pABa3207a; note that variant numbers are assigned arbitrarily in this study to help follow the results.
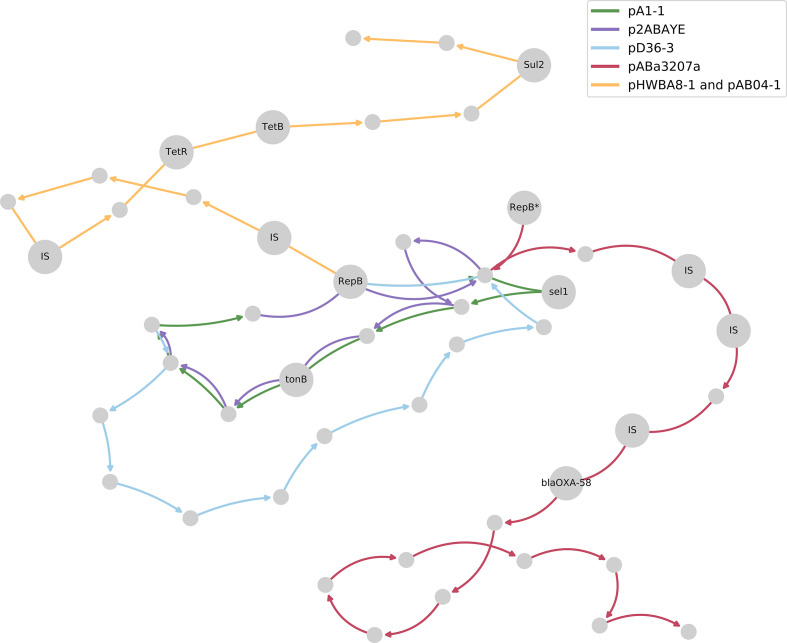
The pA1-1 plasmid comprises the green path in Fig. 6; this path is shared across all plasmids in the tonB domain but not in sel1. Plasmids p2ABAYE and pD36-3 in strains AYE and D36 are both classified as RepAci1, and they share common paths with pA1-1 but diverge where the Sel1 protein is replaced with a different dif module (variant 1 and variant 2 in Fig. 5, respectively), also reported by Blackwell et al. [42]. The pABa3207a plasmid from strain 3207 carries the carbapenemase gene bla OXA-58, which had been introduced by repeating IS elements upstream (variant 3 in Fig. 5). It is suggested that the RepB protein carried by pABa3207a had been mistaken for a variant of RepB, and while it also appears as a separate node in the graph (labelled RepB* protein in Figs 5 and 6), it remains within close proximity due to shared genomic context with other RepAci1 plasmids.
Fig. 6.
Variants of the pA1-1 plasmid sequence across A. baumannii strains; unique plasmid structures are differentiated by their edge colours (see key), and all the significant nodes mentioned in the text are labelled with the corresponding product name.
We also observed a novel variant of a RepAci1 plasmid, pHWBA8-1 and pAB04-1 in strains HWBA8 and Ab04-mff, respectively (yellow path in Fig. 6, not shown in Fig. 5) which were neither reported by Blackwell et al. nor studied yet to our knowledge. These plasmids would be interesting to study as they had been isolated from multidrug-resistant strains and they carry the tetracyline resistance genes TetB and TetR, as well as the Sul1 gene, which has been linked to sulfonamide resistance.
Conclusions
In this study, we have evaluated the state-of-the-art different pan-genome construction methods to understand the ways in which they can be the most useful to incorporate into existing pipelines and gain insight. We curated a list of tools diverse enough to describe the current literature in pan-genome construction, while still similar in their algorithms so that a meaningful comparison could be made. We provide a flowchart to guide users to select the tool most suited for their application, and we replicate a previous study analysing various survival mechanisms of A. baumannii .
Our results on A. baumannii suggest that while all the tools produced pan-genome graphs in line with previous work on the same species, they differed significantly in cloud genes. In addition, we found that graph size is likely to be influenced the most by the homologue detection step in the algorithm, and that it can be vary considerably when the parameter settings are changed. Thus, if one desires to go one step forward, and use these tools in more specialized downstream analyses, one must consider parameter tuning or moulding the algorithms available to suit one's own specific purpose. We recommend that users utilize external databases of known annotations to validate their results for the species they are working on.
Finally, we have provided an example case of structural variant calling in the same A. baumannii dataset by combining two of the tools in order to explore (i) the context of the bla OXA-23 carbapenemase gene carried on Acinetobacter transposons and (ii) different structures of RepAci1 plasmids in A. baumannii that play a significant role in transmission of antimicrobial resistance genes. Interestingly, we have also identified a novel variant of the RepAci1 plasmid in two clinical strains, carrying resistance genes associated with more than one resistance phenotype, and that would play an important role in understanding the mechanisms of multidrug resistance in A. baumannii . We assert the added benefit of combining different tools strategically instead of using any of the tools on their own. Akin to ensemble modelling in the field of machine learning, mixing and matching different methods might be a viable option to consider for constructing pan-genome graphs.
While A. baumannii is a good representative of bacterial organisms, our findings are limited to the particular use-case and thus may not be generalizable to species on the more extreme ends, such as Escherichia coli , which is reported to be have a higher genome plasticity as well as a larger average genome size (~5 Mbp long), or Campylobacter jejuni , for which the core genome forms a substantial part of the whole pan-genome although on average its genome is much smaller than that of A. baumannii (~1.5 Mbp long, with 40 % core genome content). In addition, since the A. baumannii pan-genome has been classified as open, our findings may not generalize well to bacterial species with closed pan-genomes. We presume such extreme cases would be the most to benefit from parameter tuning.
Supplementary Data
Funding information
This work received no specific grant from any funding agency.
Author contributions
T.A. and A.U., conceptualized the study; A.U., performed investigation and wrote the original draft; T.A., supervised work and reviewed and edited the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: GO, gene ontology; IS, insertion sequence; NCBI, National Center for Biotechnology Information; OBO, Open Biological and Biomedical Ontology; OXA, oxacillinase.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and two supplementary figures are available with the online version of this article.
References
- 1.Yakkala H, Samantarrai D, Gribskov M, Siddavattam D. Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLOS ONE. 2019;14:e0218204. doi: 10.1371/journal.pone.0218204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Center for Biotechnology Information (NCBI) Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1988. https://www.ncbi.nlm.nih.gov/
- 3.Yang X, Lee W-P, Ye K, Lee C. One reference genome is not enough. Genome Biol. 2019;20:104. doi: 10.1186/s13059-019-1717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marschall T, Marz M, Abeel T, Dijkstra L, Dutilh BE, et al. Computational pan-genomics: status, promises and challenges. Brief Bioinformatics. 2018;19:118–135. doi: 10.1093/bib/bbw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dilthey A, Cox C, Iqbal Z, Nelson MR, McVean G. Improved genome inference in the MHC using a population reference graph. Nat Genet. 2015;47:682–688. doi: 10.1038/ng.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrison E, Sirén J, Novak AM, Hickey G, Eizenga JM, et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol. 2018;36:875–879. doi: 10.1038/nbt.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biederstedt E, Oliver JC, Hansen NF, Jajoo A, Dunn N, et al. NovoGraph: Human genome graph construction from multiple long-read de novo assemblies. F1000Res. 2018;7:1391. doi: 10.12688/f1000research.15895.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Feng X, Chu C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 2020;21:265. doi: 10.1186/s13059-020-02168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 11.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright AJ. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinegger M, Söding J. Clustering huge protein sequence sets in linear time. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-04964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013;41:11–19.:S0924-8579(12)00373-1. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Salto IP, Torres Tejerizo G, Wibberg D, Pühler A, Schlüter A, et al. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-26180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, et al. Producing polished prokaryotic pangenomes with the panaroo pipeline. bioRxiv. 2020:2020.01.28.922989. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar AN, Abeel T. Approximate, simultaneous comparison of microbial genome architectures via syntenic anchoring of quiver representations. Bioinformatics. 2018;34:i732–42. doi: 10.1093/bioinformatics/bty614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautreau G, Bazin A, Gachet M, Planel R, Burlot L, et al. PPanGGOLiN: Depicting microbial diversity via a Partitioned Pangenome Graph. bioRxiv. 2019:836239. doi: 10.1371/journal.pcbi.1007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience. 2019;8:598391. doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klopfenstein D, Zhang L, Pedersen BS, Ramírez F, Vesztrocy AW, et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci Rep. 2018;8:1–17. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carbon S, Mungall C. Gene ontology data archive. Dataset on Zenodo. 2018 [Google Scholar]
- 25.The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–9. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgescu CH, Manson AL, Griggs AD, Desjardins CA, Pironti A, et al. SynerClust: a highly scalable, synteny-aware orthologue clustering tool. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Wu J, Yang J, Sun S, Xiao J, et al. PGAP: Pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Charlesworth J, Achtman M. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 2020;30:1667–1679. doi: 10.1101/gr.260828.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangas EL, Rubio A, Álvarez-Marín R, Labrador-Herrera G, Pachón J, et al. Pangenome of Acinetobacter baumannii uncovers two groups of genomes, one of them with genes involved in CRISPR/Cas defence systems associated with the absence of plasmids and exclusive genes for biofilm formation. Microb Genom. 2019;5:e000309. doi: 10.1099/mgen.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galac MR, Snesrud E, Lebreton F, Stam J, Julius M, et al. A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob Agents Chemother (Bethesda) 2020;64 doi: 10.1128/AAC.00840-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii . Genome Biol. 2015;16:143. doi: 10.1186/s13059-015-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa SS, Guimarães LC, Silva A, Soares SC, Baraúna RA. First Steps in the Analysis of Prokaryotic Pan-Genomes. Bioinform Biol Insights. 2020;14:117793222093806. doi: 10.1177/1177932220938064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan AP, Sutton G, DePew J, Krishnakumar R, Choi Y, et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii . Genome Biol. 2015;16:143. doi: 10.1186/s13059-015-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 38.Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob Agents Chemother (Bethesda) 2008;52:1252–1256. doi: 10.1128/AAC.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal H, Jacobson RK, Garny S, Bamford CM, Elisha BG. Extended -10 promoter in ISAba-1 upstream of blaOXA-23 from Acinetobacter baumannii [3] Antimicrob Agents Chemother (Bethesda) 2007;51:3040–3041. doi: 10.1128/AAC.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Héritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii . Antimicrob Agents Chemother (Bethesda) 2005;49:3198–3202. doi: 10.1128/AAC.49.8.3198-3202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nigro SJ, Hall RM. Structure and context of acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother. 2016;71:1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 42.Blackwell GA, Hall RM. Mobilisation of a small Acinetobacter plasmid carrying an oriT transfer origin by conjugative RepAci6 plasmids. Plasmid. 2019;103:36–44. doi: 10.1016/j.plasmid.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Gao J, Zhang H, Ying C. Spread of the blaOXA-23-containing Tn2008 in carbapenem-resistant Acinetobacter baumannii isolates Grouped in CC92 from China. Front Microbiol. 2017;8:163. doi: 10.3389/fmicb.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.