Abstract
Anopheles coluzzii is one of the primary vectors of human malaria in sub-Saharan Africa. Recently, it has spread into the main cities of Central Africa threatening vector control programs. The adaptation of An. coluzzii to urban environments partly results from an increased tolerance to organic pollution and insecticides. Some of the molecular mechanisms for ecological adaptation are known, but the role of transposable elements (TEs) in the adaptive processes of this species has not been studied yet. As a first step toward assessing the role of TEs in rapid urban adaptation, we sequenced using long reads six An. coluzzii genomes from natural breeding sites in two major Central Africa cities. We de novo annotated TEs in these genomes and in an additional high-quality An. coluzzii genome, and we identified 64 new TE families. TEs were nonrandomly distributed throughout the genome with significant differences in the number of insertions of several superfamilies across the studied genomes. We identified seven putatively active families with insertions near genes with functions related to vectorial capacity, and several TEs that may provide promoter and transcription factor binding sites to insecticide resistance and immune-related genes. Overall, the analysis of multiple high-quality genomes allowed us to generate the most comprehensive TE annotation in this species to date and identify several TE insertions that could potentially impact both genome architecture and the regulation of functionally relevant genes. These results provide a basis for future studies of the impact of TEs on the biology of An. coluzzii.
The deadly success of the malaria mosquito Anopheles coluzzii is rooted in its extraordinary ecological plasticity, inhabiting virtually every habitat in West and Central Africa where it spreads the human malaria parasite (Fontaine et al. 2015; Tene Fossog et al. 2015). Noteworthy, the larvae of An. coluzzii exploit more disturbed and anthropogenic sites than its sister species An. gambiae. An. coluzzii shows a higher tolerance to salinity and organic pollution and, as a consequence, is the predominant species in coastal and urban areas (Tene Fossog et al. 2013, 2015; Kengne et al. 2019; Longo-Pendy et al. 2021). However, this mosquito not only has a greater resilience to ion-rich aquatic environments, but it has also become resistant to DDT and pyrethroid insecticides used for vector control (Wiebe et al. 2017; Fouet et al. 2018; Vontas et al. 2018). The adaptive flexibility of An. coluzzii is also exemplified by its rapid competence to expand its range of peak biting times to avoid insecticide-treated bed nets (Perugini et al. 2020). This extraordinary adaptive capacity makes this malaria vector a threat for malaria control. Thus, elucidating the natural genetic variants underlying the ecological and physiological responses to fluctuating environments in this species is key for its control.
At the molecular level, several genetic mechanisms have been related back to the adaptive capacity of An. coluzzii. The most prominent and historically studied are chromosomal inversions (Coluzzi et al. 2002; Ayala et al. 2017). An. coluzzii shows a large number of polymorphic inversions (Costantini et al. 2009; Simard et al. 2009), many of them associated with environmental adaptation through environmental clines and/or correlation with specific climatic variables (Coluzzi et al. 1979; Fouet et al. 2012; Ayala et al. 2017, 2019). Other types of rearrangements, such as gene duplications, have been involved in insecticide resistance. For example, the acetylcholinesterase (ACE1) gene has been duplicated, maintaining at least a sensitive and a resistance copy, to counteract the fitness cost of the resistant phenotype (Labbé et al. 2007; Assogba et al. 2015; Weetman et al. 2018). Moreover, a recent genome-wide analysis showed that genes containing copy number variants were enriched for insecticide functions (Lucas et al. 2019). However, although several of the candidate genes responsible for the adaptive capacity of An. coluzzii have been identified, including detoxification and immune-related genes, our knowledge of the genetic variants underlying differences in these genes lags behind (Tene Fossog et al. 2013; Mitri et al. 2015; Kamdem et al. 2017; King et al. 2017). In particular, very little is known about natural variation in transposable element (TE) insertions in An. coluzzii.
TEs are key players in multiple adaptive processes across species owing to their capacity to generate a wide variety of mutations (Casacuberta and González 2013; Schrader and Schmitz 2019). TEs can disperse across the genome regulatory sequences such as promoters, enhancers, and repressive elements thus affecting nearby gene expression (Chuong et al. 2017; Ullastres et al. 2021). Additionally, they can also act as substrates for ectopic recombination leading to chromosomal rearrangements (Mathiopoulos et al. 1998; Gray 2000; Reis et al. 2018). However, TEs are often ignored when analyzing functional variants in genomes. Because they are repetitive sequences, TEs are difficult to annotate, and reads derived from TEs are often discarded in genome-wide analyses (Goerner-Potvin and Bourque 2018). Long-read sequencing techniques are needed to get a comprehensive view of TE variation in genomes, because they allow to annotate TE insertions in the genome rather than merely inferring their position (Logsdon et al. 2020; Shahid and Slotkin 2020).
Although TE insertions have been annotated genome-wide in several anopheline species including An. coluzzii, most studies to date have characterized the TE repertoire in a single genome for each species (for review, see Vargas-Chávez and González 2020). To capture the full extent of TE natural variation and the potential consequences of TE insertions, it is necessary to evaluate multiple genomes to comprehensively assess diversity within a species (Yang et al. 2019; Bayer et al. 2020; Weissensteiner et al. 2020). This becomes especially relevant when attempting to identify recent TE insertions and their effect in the genome structure and genome function, given that they might be restricted to local populations. So far, our knowledge of An. coluzzii genome variation attributed to TE insertions is limited to a few well-characterized families that have been found to vary across genomes (Quesneville et al. 2006; Boulesteix et al. 2007; Esnault et al. 2008; Santolamazza et al. 2008b; Salgueiro et al. 2013).
In this work, we sequenced (using long-read technologies) and assembled the genomes of An. coluzzii larvae collected in six natural breeding sites in two major cities in Central Africa: Douala (Cameroon) and Libreville (Gabon). Our work aims at generating a comprehensive TE annotation that could be used to identify insertions that can potentially impact both genome architecture and gene regulation in An. coluzzii.
Results
Six new whole-genome assemblies of An. coluzzii from two major cities in Central Africa
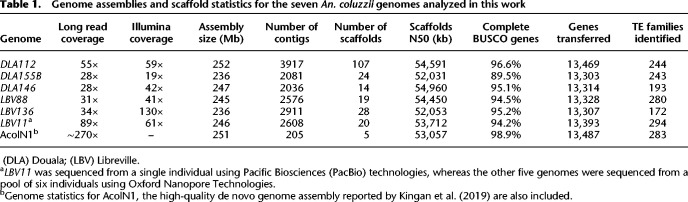
To explore the TE diversity in An. coluzzii, we used long-read sequencing to generate whole-genome assemblies of larvae collected from six natural urban breeding sites: three from Douala (Cameroon), and three from Libreville (Gabon) (Table 1; Fig. 1A; Supplemental Table S1A). Reference-guided scaffolding was performed for the six assemblies and for the available An. coluzzii AcolN1 genome (Kingan et al. 2019). Although the seven genomes analyzed varied in sequencing coverage and read length, these differences only had an effect on contig N50 but did not affect other assembly and scaffold metrics (Supplemental Table S1B). Although the number of scaffolds varied from 5 to 107 (median = 20), the scaffolds’ N50 was similar across the seven genomes (Table 1). The BUSCO percentages of complete genes ranged from 94.2% to 96.6% except for the DLA155B sample, which had a lower completeness value (89.5%) (Table 1; Simão et al. 2015). These completeness values were similar to those from the AcolN1 genome assembly (98.9%) (Table 1; Supplemental Table S1). Thus, the analyzed genomes are overall comparable in terms of scaffold contiguity and completeness (Table 1; Supplemental Table S1).
Table 1.
Genome assemblies and scaffold statistics for the seven An. coluzzii genomes analyzed in this work
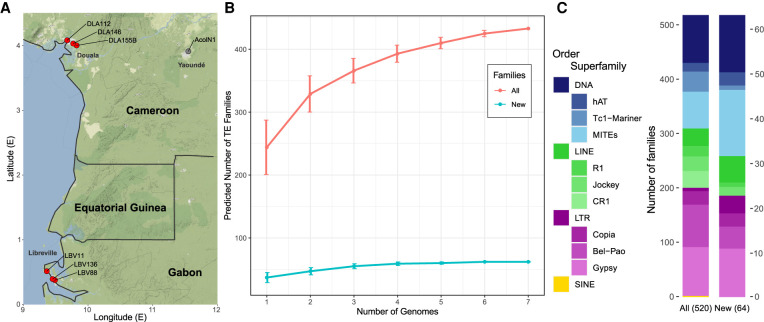
Figure 1.
Transposable elements in three urban populations of An. coluzzii. (A) Geographic location of the six breeding sites analyzed, three in Douala (DLA) and three in Libreville (LBV) (in red), and of the place of origin of the Ngousso colony (in gray) that was used to generate the AcolN1 genome. (B) Number of TE families identified when using a single genome or when using all possible combinations of more than one genome. The red line shows the total number of TE families, and the blue line shows the number of newly described families. Note that on average 76% of all the TE copies where already identified when analyzing a single genome (Supplemental Fig. S1). (C) Classification of all TE families and newly described families in An. coluzzii. The three most abundant superfamilies from each order are shown.
Sixty-four new anopheline TE families discovered in An. coluzzii
To identify the TE families present in each of the genomes, we used the TEdenovo pipeline from the REPET package (Flutre et al. 2011). After several rounds of manual curation, we identified between 172 and 294 TE families per genome (Table 1; Supplemental Table S2; Platt et al. 2016). We discovered that differences in sequencing and assembly metrics correlated with the number of families identified in only one of the genomes (Supplemental Table S1B). Thus, although using a single reference would have only allowed the identification of a median of 244 TE families, clustering the TE libraries from an increasing number of genomes allowed the identification of 435 well-supported TE families (Fig. 1B; Methods). Sixty-four of these families are described here for the first time (Fig. 1B; see below).
To annotate the individual copies in each one of the genomes analyzed, besides the 435 families identified by REPET, we also added to our library 85 TE families from the TEfam database that were also present in the seven An. coluzzii genomes (Methods). Although the majority of these families were indeed identified by REPET, we initially discarded them during manual curation (Supplemental Table S3; Methods). The final total of 520 families were classified into 23 superfamilies and then further grouped into four orders (Fig. 1C). The 82 families initially discovered in all the genomes had a higher copy number (Wilcoxon test, P-value < 0.001) and were more abundant in euchromatin (two proportion Z-test P-value < 0.001) compared with the 353 families only discovered in some of the genomes.
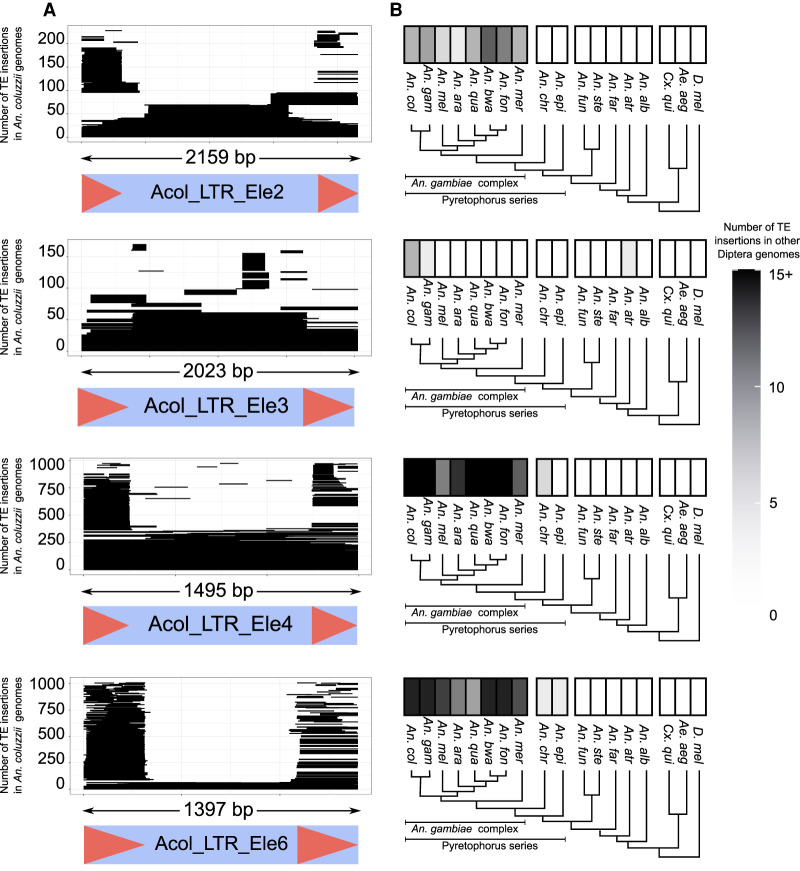
To further characterize the novel families, we estimated their average number of insertions in the seven An. coluzzii genomes (Fig. 2; Supplemental Fig. S2; Supplemental Table S4). Copies from all 64 new families were found in all seven An. coluzzii genomes, further suggesting that these are bona fide families. Although the majority of families contain full-length copies in at least one of the seven genomes analyzed, truncated copies were the most abundant (Fig. 2A; Supplemental Table S3). We identified a median of 72 insertions (ranging from 16 to 1445) per family and genome (Fig. 2A; Supplemental Fig. S2), with new families having lower copy numbers compared with previously described families (Wilcoxon test, P-value = 0.0214). No differences were found in the number of genomes containing new and previously described families (Wilcoxon test, P-value = 0.9381). Two of the four TRIM elements identified (Acol_LTR_Ele 4 and Acol_LTR_Ele 6) are among the most abundant new families, with more than 150 insertions (Fig. 2A). Indeed, TRIM elements have not been previously described in anopheline genomes and are still underexplored in insect genomes in general (Marsano et al. 2012; Zhou and Cahan 2012; Elsik et al. 2014). In plants, some TRIM elements have the capacity to restructure genomes by acting as target sites for retrotransposon insertions, alter host gene structure, and transduce host genes (Witte et al. 2001; Gao et al. 2016). Although we found TRIM elements in An. coluzzii genomes to be underrepresented in gene bodies (χ2 test P-value > 0.05), they were overrepresented in nested insertions (χ2 test P-value < 0.05).
Figure 2.
Structure, abundance, and phylogenetic distribution of novel TE families. The four newly identified TRIM families are shown; for the remaining 60 novel families see Supplemental Figure S2. (A) The structure of each new family is displayed: the light blue box represents the full extension of the TE, and the red arrows represent LTRs. All insertions for each TE family were identified and are shown as a coverage plot in which each line represents a copy in the genome. The large number of stacked horizontal lines in the extremes of the plot represent an abundance of solo LTRs. (B) Phylogenetic distribution of the TE family insertions in 15 members of the Anopheles genus, including the eight members of the An. gambiae complex, two more distantly related mosquito species (Culex quinquefasciatus and Aedes aegypti), and Drosophila melanogaster. The number of insertions with >80% identity and spanning at least 80% of the consensus in each species is shown using a black and white gradient: species with no insertions are shown in white, and species with 15 or more insertions are shown in black.
Finally, the 64 new families were unevenly distributed among the members of the Anopheles genus (Fig. 2B; Supplemental Fig. S2; Supplemental Table S4). Ten families were exclusively found in members of the Pyretophorus series, suggesting that these elements emerged after the split of this series from the Cellia subgenus. Moreover, 13 families were also found in at least one of the other three non-anopheline species (Supplemental Fig. S2C). The distribution of these 13 families was patchy, with some of them present only in distantly related species whereas others were present in members of the Anopheles genus or in members of the Pyretophorus series, suggesting that some of them might have been acquired through horizontal transfer events (de Melo and Wallau 2020).
TEs are nonrandomly distributed throughout the genome
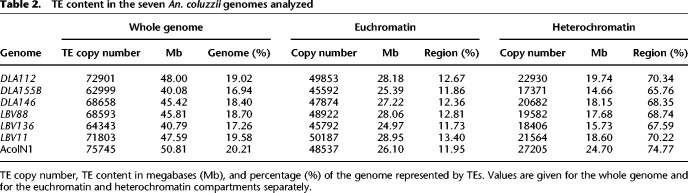
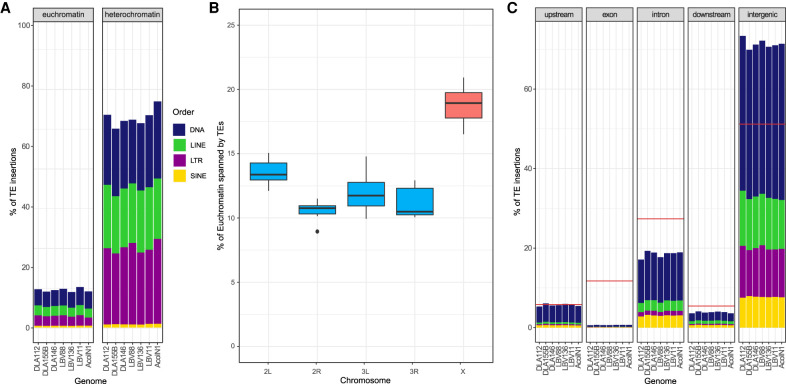
The percentage of the genome represented by TEs across the seven genomes varied between 16.94% and 20.21% (Table 2). However, differences across genomes in assembly and scaffolding statistics did not explain the variation in TE content (Supplemental Table S1B). We found a positive correlation between TE content and genome size as has been previously described in Anopheles and other species (Pearson's r = 0.90, significance = 0.007) (Supplemental Fig. S3; Sessegolo et al. 2016; de Melo and Wallau 2020). The percentage of TEs in euchromatin (11.73%–13.40%) was much lower than the percentage of TEs in heterochromatin (65.76%–74.77%; χ2 test P-value < 0.05) (Table 2; Fig. 3A), making heterochromatin a more variable compartment (Sharma et al. 2020).
Table 2.
TE content in the seven An. coluzzii genomes analyzed
Figure 3.
TE insertion distributions throughout the genomes. (A) Percentage of euchromatin and heterochromatin occupied by TEs in each of the seven analyzed genomes. Each order is shown in a different color. (B) Box plots of the percentage of the euchromatin of each chromosome covered by TEs. Autosomes are shown in blue, and the X Chromosome is in red. (C) Percentage of TE insertions in each genome that fall in a specific genomic region. A red line is used to display the expected percentage that should be covered by TEs taking into consideration the size of the genomic region. Each order is shown in a different color as in A. Significant differences were found across orders and superfamilies (Supplemental Table S5).
Because we are mostly interested in the potential functional impact of TE insertions, for the rest of this work we focused on the TE insertions present in euchromatin. We first assessed whether the seven analyzed genomes differed in TE content at the order and superfamily levels, and we found this to be the case (χ2 test P-value = 1.07 × 10−21 and P-value = 1.69 × 10−14, respectively). The largest differences were found in the LTR order: LTRs were more abundant in the DLA112 and LBV88 genomes and less abundant in AcolN1 (Supplemental Fig. S4A). At the superfamily level, we found that the largest differences were in the Gypsy superfamily, which belongs to the LTR order. Indeed, most of the differences in TE content between the evaluated genomes appear to be in retrotransposon families (Supplemental Fig. S4B). Superfamily abundance did not clearly reflect the geographical population structure (Supplemental Fig. S4B,C).
When comparing the TE content in autosomes versus the X Chromosome, as expected, the X Chromosome had a larger fraction of its euchromatin spanned by TEs (Fig. 3B; Xia et al. 2010). Finally, to evaluate the distribution of TE insertions regarding genes, we divided the genome in five regions: 1 kb upstream, exon, intron, 1 kb downstream, and intergenic (Ruiz et al. 2021). The number of insertions in intergenic regions was higher than expected by chance, whereas the number of insertions in exons and introns was lower (χ2 test P-value < 0.001) (Fig. 3C; Supplemental Table S5). Although the upstream regions were neither enriched nor depleted for TE insertions, the downstream regions had a smaller amount of TEs than expected by chance, consistent with downstream regions being more commonly found in a closed chromatin state (Ruiz et al. 2021).
Overall, TEs were not randomly distributed in the genome, because they were more abundant in heterochromatic than in euchromatic regions, more abundant in the X Chromosome euchromatin than in autosomes, and more abundant in euchromatic intergenic regions than in gene bodies or gene flanking regions.
MITE insertions are present in several inversion breakpoints
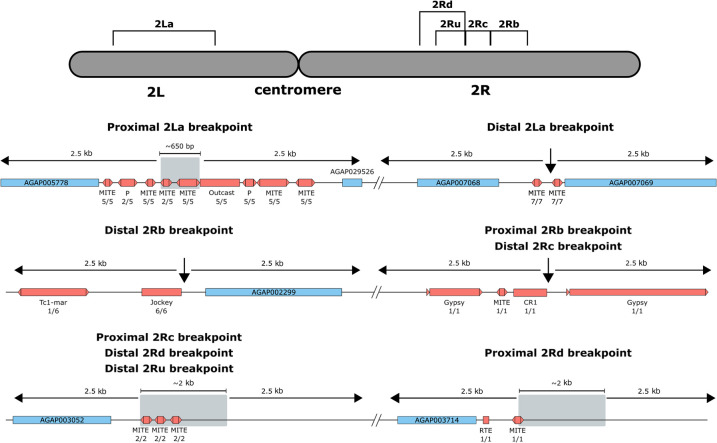
TEs have been found in close proximity to the breakpoints of the 2La in An. gambiae and An. melas, and to the breakpoints of the 2Rb inversions in An. gambiae and An. coluzzii (Sharakhov et al. 2006; Lobo et al. 2010). We thus explored the TE content in the breakpoints of these two inversions and in three other common polymorphic inversions in An. coluzzii: 2Rc, 2Rd, and in the distal breakpoint of the 2Ru (Corbett-Detig et al. 2019). The analysis of these breakpoint regions suggested that the analyzed genomes have the standard conformation for all five inversions (Methods; Supplemental Table S6). We identified several TEs nearby the proximal and the distal breakpoints of 2La and 2Rb, in agreement with previous studies (Fig. 4; Mathiopoulos et al. 1998; Sharakhov et al. 2006; Lobo et al. 2010). For the standard 2La proximal breakpoint, Sharakhov et al. (2006) identified several DNA transposons and a SINE insertion. We also identified a cluster of MITE insertions, which are DNA transposons, and we additionally identified an Outcast (LINE) element (Fig. 4). Regarding the standard 2La distal breakpoint, we observed two MITEs similar to one of the insertions in the proximal breakpoint, in agreement with the findings by Sharakhov et al. (2006) (Fig. 4). We also observed similar TE content in the 2Rb breakpoints as the one described by Lobo et al. (2010): tandem repeats flanking the inversion in the standard and inverted forms, and TEs in the internal sequences of both breakpoints (Fig. 4). Finally, for the 2Rd inversion, we identified MITEs near both breakpoints.
Figure 4.
TE insertions near known inversion breakpoints. Diagram of Chromosome 2 with the analyzed inversions. For each inversion both breakpoints, proximal (closer to the centromere) and distal (farther from the centromere), plus 2.5 kb on each side are shown. When the position of a breakpoint was not identified at the single base pair level, the interval where the breakpoint is predicted to be is shown in a gray box. Genes are shown as blue boxes, and TEs are shown as red boxes. Below each TE, the family and the number of genomes where the insertion was found/the number of genomes where the breakpoint region was identified are given. Breakpoints are shared among some of the inversions.
Six of the seven potential active families are LTR insertions
To identify potentially active TE families, we first estimated their relative age by analyzing the TE landscapes (Smit et al. 2013–2015; Diesel et al. 2019). We observed an “L” shape landscape in all genomes which is indicative of a recent TE burst (Supplemental Fig. S5; Fonseca et al. 2019). This “L” shape landscape, dominated by retrotransposons, had previously been described for the sister species An. gambiae (Diesel et al. 2019; Petersen et al. 2019), where numerous Gypsy LTR retrotransposons (up to 75%) might currently be active (Tubío et al. 2005, 2011). Indeed, piRNAs predominantly target LTR retrotransposons in An. gambiae (George et al. 2015). We further investigated the families in the peak of the landscape, and we identified eight families with more than two identical full-length fragments and with more than half of their copies identical to the consensus (Supplemental Table S7A). Additionally, we assessed the potential ability of our candidates to actively transpose by identifying their intact open read frames (ORFs), LTRs (in the case of LTR retrotransposons), and target site duplications (TSDs) and determined that seven of these families are potentially fully capable of transposing. For the LTR families, we further evaluated the identity between LTRs of the same copy. Mean identities ranged from 97.38% to 99.44% across the six LTR families, with 17.64%–40.35% of the copies having identical LTRs (Supplemental Table S7B). These results further suggest that these families might be responsible for the recent retrotransposon burst in An. coluzzii (Supplemental Table S7A). LTR elements accounted for most of the differences in TE content across genomes (Supplemental Fig. S4).
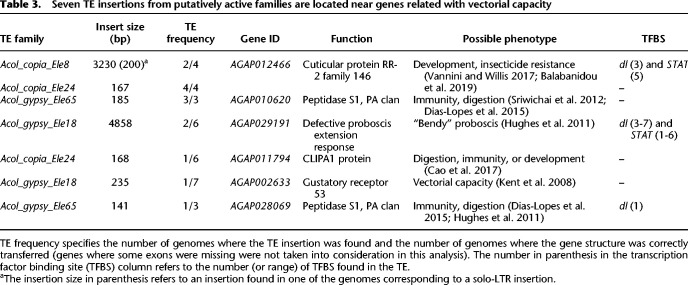
As a first step toward assessing the potential functional consequences of the TE insertions from these seven putatively active families, we focused on insertions that occurred in gene bodies, and 1 kb upstream or downstream from a gene. We identified 66 genes with insertions from these families, with four genes containing up to two insertions in the same gene region (Supplemental Fig. S6; Supplemental Table S8). Six of the genes have functions related to vectorial capacity: insecticide resistance, immunity, and biting ability (Table 3). We checked whether the TE insertions nearby these genes contained binding sites for transcription factors or promoter motifs (Supplemental Table S9). We focused on identifying binding sites for three transcription factors that are known to be involved in response to xenobiotics (cap'n' collar: cnc) and in immune response and development (dorsal: dl; signal transducer and activator of transcription: STAT) given the availability of matrix profiles from D. melanogaster (Osta et al. 2004; Ingham et al. 2017). We identified binding sites for dl, STAT, or both in three insertions; with the Acol_gypsy_Ele18 and the Acol_copia_Ele8 insertions having more than three binding sites for the same transcription factors, suggesting that they might be functional (Table 3; Xie et al. 2010). Additionally, the genes that contained these TE insertions also contained binding sites for the same transcription factors, which suggest that they already played a role in their regulation. We also identified a putative promoter sequence in the Acol_copia_Ele24 insertion found upstream of the CLIPA1 protease encoded by AGAP011794 that could also potentially lead to changes in the regulation of this gene (Supplemental Table S9C).
Table 3.
Seven TE insertions from putatively active families are located near genes related with vectorial capacity
TE insertions could influence the regulation of genes involved in insecticide resistance
The usage of pyrethroids, carbamates, and DDT as vector control mechanisms has led to the rapid dispersion of insecticide resistance alleles in natural populations (Dabiré et al. 2014; Silva et al. 2014; Cheung et al. 2018; Elanga-Ndille et al. 2019; Fadel et al. 2019). Among the best characterized resistance point mutations are L1014F (kdr-west), L1014S (kdr-east), N1575Y in the voltage gated sodium channel para (also known as VGSC), and G119S in the acetylcholinesterase ACE1 gene (Santolamazza et al. 2008a; Jones et al. 2012; Essandoh et al. 2013). We first investigated whether the seven genomes analyzed in this work contained these resistance alleles. We found the kdr-west mutation in the six genomes from Douala and Libreville but not in the AcolN1 genome (Kingan et al. 2019), whereas previous estimates in Douala reported a 68.2% frequency of the kdr resistant allele (Antonio-Nkondjio et al. 2011). None of the other mutations were identified in our data set; however, a previously undescribed nonsynonymous substitution (L1688M) in the fourth domain of para was identified in the aforementioned six genomes. Whether this replacement also increases insecticide resistance is yet to be assessed.
TEs have been hypothesized to play a relevant role specifically in response to insecticides (Wilson 1993; ffrench-Constant et al. 2006; Rostant et al. 2012), and a few individual insertions affecting insecticide tolerance in anopheline mosquitoes have already been described (Weedall et al. 2020). Thus, we searched for TE insertions in the neighborhood of insecticide-related genes that could potentially lead to differences in their regulation. We focused on eight well-known insecticide resistance genes: ACE1, CYP6P3, GSTD1-6, para, Rdl, CYP6M2, CYP6Z1, and GSTE2. We also considered differentially expressed genes in response to insecticides in An. gambiae (Supplemental Fig. S7; Supplemental Table S10; Tene Fossog et al. 2013; Main et al. 2018; Adolfi et al. 2019; Bamou et al. 2019). We found that 21 of the 43 genes analyzed contained at least one TE insertion, which is similar to the number of genes containing insertions genome-wide (χ2 test P-value = 0.6285). Overall, insertions were enriched in intronic regions (χ2 test P-value < 0.05), although 10 were located in the 1-kb gene upstream region, one in the 3′ UTR, and five in the first intron, and thus are more likely to have a functional impact. Moreover, the majority (44/59) of TE insertions located nearby insecticide-related genes were absent or present at low frequencies (<10%) in two rural populations from the Ag1000G project (Anopheles gambiae 1000 Genomes Consortium 2020), suggesting that they might have increased in frequency in urban populations (Supplemental Table S11).
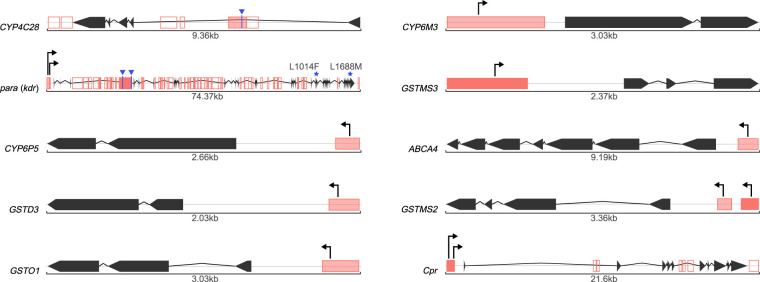
To further explore if TEs could influence the regulation of insecticide resistance genes, we focused on polymorphic (present in two or more genomes) and fixed (present in all seven genomes analyzed) insertions located in gene bodies or 1 kb upstream or downstream from the gene. We searched for cnc binding sites; for those insertions located in gene upstream regions, we also looked for promoter motifs (Supplemental Table S9). We identified 15 insertions in 10 genes containing either cnc binding sites or promoter sequences (Fig. 5). Three insertions contained cnc bindings sites: one located in CYP4C28 and two in para, although the genes did not contain binding sites for this transcription factor (Fig. 5). The other 12 insertions contained promoter motifs. In some cases, such as the Acol_m2bp_Ele10 MITE insertion in ABCA4 or the tSINE insertion in GSTMS2, although the same TE insertion was found in six and seven genomes, respectively, the promoter motifs were found only in four and one genome, respectively (Fig. 5; Supplemental Table S9). We analyzed the consensus sequence of these two families and found that although the Acol_m2bp_Ele10 had the promoter motif, the tSINE did not, suggesting that some of the Acol_m2bp_Ele10 elements lost the promoter motifs, whereas the tSINE copies acquired them.
Figure 5.
TE insertions in the neighborhood of genes involved in insecticide resistance. Gene structures are shown in black with arrows representing the exons. TE insertions are depicted as red boxes. When containing a TFBS for cnc or a promoter they are filled in red, otherwise they are empty. The red color is darker on fixed TEs and lighter on polymorphic TEs. Promoters are shown as arrows, and cnc binding sites are shown in blue. Resistance mutations are shown for para (kdr).
Immune-related genes could also be potentially affected by TEs
Mosquitoes breeding in urban and polluted aquatic environments overexpress immune-related genes, suggesting that immune response is relevant for urban adaptation (Cassone et al. 2014). To assess the potential role of TEs in immune response, we searched for TE insertions in genes putatively involved in immunity according to ImmunoDB (Supplemental Table S12; Waterhouse et al. 2007). We identified TE insertions in 148 of the 281 genes analyzed, similar to the number of genes containing insertions genome-wide (χ2 test P-value = 0.7788). Eleven of the 148 genes containing TE insertions are differentially expressed in response to a Plasmodium invasion (Supplemental Table S12). These 11 genes participate in several pathways of the immune response including the small regulatory RNA pathway, pathogen recognition, the nitric oxide response, and ookinete melanization (Osta et al. 2004; Volz et al. 2006; Oliveira et al. 2012; Dennison et al. 2015). The majority of insertions were located in the 1-kb gene flanking regions (χ2 test P-value < 0.05), with 56 of them located in the first intron and 112 in the 5′ upstream gene region. More than half (262/438) of the TE insertions located nearby immune-related genes in urban populations were absent or present at low frequencies (<10%) in two rural populations (Supplemental Table S11).
Finally, we further explored polymorphic and fixed insertions to identify binding sites for dl and STAT and promoter motifs. We found that 19 TEs contained binding sites for dl, 21 TEs contained binding sites for STAT, and 12 TEs contained binding sites both for dl and STAT (Supplemental Table S12). Additionally, we identified 81 insertions, in the upstream region of 56 genes, which carried putative promoter sequences. Five of the TEs located nearby Plasmodium responsive genes added TFBS and promoter sequences, thus suggesting that these TE insertions can potentially influence the response to this pathogen (Supplemental Table S12; Ruiz et al. 2019).
Discussion
In this study, we de novo annotated transposable element (TE) insertions in seven genomes of An. coluzzii, six of them sequenced here. A comprehensive genome-wide TE annotation was possible because we used long-read technologies to perform the genome sequencing and assembly. Long reads allow identifying TE insertions with high confidence given that the entire TE insertion sequence can be spanned by a single read (Logsdon et al. 2020; Shahid and Slotkin 2020). Although the genome-wide TE repertoire has been studied in other anopheline species, particularly in An. gambiae, to our knowledge there are no other studies that have explored TE variation in multiple genomes from a single species (Holt et al. 2002; Fernández-Medina et al. 2012; Marinotti et al. 2013; Neafsey et al. 2015; Diesel et al. 2019; de Melo and Wallau 2020). As reported in other species, we observed that increasing the number of available genomes analyzed allowed us to increase the number of identified TE families from a median of 244 (172–294) to 435 (Fig. 1B; Hufford et al. 2021). Moreover, having the full sequences of seven genomes also allowed us to discover 64 new TE families, including four TRIM families previously undescribed in anopheline genomes. This might be relevant because TRIM elements have been shown to be important players in genome evolution in other species (Witte et al. 2001; Gao et al. 2016). The wide range of families identified across genomes was not directly related to the quality of the genome assembly taking into consideration the more generally used quality parameters such as read length, number of contigs, and contig N50 (Ou et al. 2020). This suggests that there are possibly other characteristics of each genome that affected the identification of high-quality TE families, such as biases in the location of the TE insertions, given that TE families are challenging to identify in regions with low complexity or with numerous nested TEs. Nonetheless, the identification of TE families is dependent on the methodology used to perform TE annotations; therefore, other annotation strategies could lead to the discovery of still undescribed families (Vargas-Chávez and González 2020).
The availability of several genome assemblies also allowed us to determine that the majority of the intraspecies differences in the TE content were in heterochromatic regions. Although we cannot discard that these differences are at least partly explained by differences in the quality of the genome assemblies, it is known that the heterochromatin compartment is highly variable even among members of the same species (Jagannathan et al. 2017; Sharma et al. 2020). Additionally, there were also significant differences in the TE content in euchromatic regions, as has been previously observed in several organisms including Drosophila (Kofler et al. 2015; Rech et al. 2019), mammals (Rishishwar et al. 2015; Diehl et al. 2020), maize (Haberer et al. 2020), and Arabidopsis (Quadrana et al. 2016). TE insertions were not randomly distributed throughout the genome and instead were consistently enriched in intergenic regions, most likely owing to purifying selection, as suggested in the wild grass Brachypodium distachyon (Stritt et al. 2020). We also analyzed the TE content in the breakpoints of five common polymorphic inversions, three of them analyzed here for the first time. We found TE insertions in all but one of the inversion breakpoints, with MITE elements being the most common TE family (Fig. 4).
As a first step toward identifying the potential role of TEs in rapid adaptation to urban habitats (Johnson and Munshi-South 2017), we focused on insertions from recently active families located near genes that are relevant for the vectorial capacity of An. coluzzii (Table 3). Because adaptation can also happen from standing variation, in the case of insecticide resistance genes, which have been shown to be shaped by TE insertions in several organisms, and immune-related genes, we analyzed all insertions independent of age (Mateo et al. 2014; Salces-Ortiz et al. 2020; Weedall et al. 2020). Although the role of nonsynonymous substitutions and copy number variation in resistance to insecticides commonly used in urban environments has been studied, the potential role of TEs has not yet been comprehensively assessed in An. coluzzii or any other anopheline species (Kamgang et al. 2018; Bamou et al. 2019; Lucas et al. 2019; Grau-Bové et al. 2020; Anopheles gambiae 1000 Genomes Consortium 2020). We identified several insertions that were polymorphic or fixed nearby functionally relevant genes (Table 3; Fig. 5; Supplemental Table S12). Some of the identified candidate insertions contained binding sites for transcription factors related to the function of the nearby genes, and promoter regions. Besides adding regulatory regions, TEs can also affect the regulation of nearby genes by affecting gene splicing and generating long noncoding RNAs among many other molecular mechanisms (Sundaram et al. 2014; Chuong et al. 2017; Jiang and Upton 2019; Villanueva-Cañas et al. 2019; Sundaram and Wysocka 2020). Thus, it is possible that the candidate TE insertions identified, which lack binding sites and promoters, could be affecting nearby genes through other molecular mechanisms. Our results are a first approximation to the potential role of TEs in An. coluzzii adaptation to the challenging environment that urban ecosystems entail. Establishing a direct link between the TEs and the traits involved in urban adaptation will require sampling a larger number of individuals and characterizing the phenotypes associated with the insertions. A better understanding of the biology of An. coluzzii and its ability to rapidly adapt to urban environments should further facilitate the development of novel strategies to combat malaria. Better management strategies can be implemented if we understand and are able to predict changes in the frequency of genetic variants relevant for the vectorial capacity of this species.
Methods
Sample collection and DNA isolation
We sampled An. coluzzii larvae in two cities of Central Africa: Libreville (Gabon) in January 2016 and Douala (Cameroon) in April 2018 (Supplemental Table S1). We collected immature third and fourth stage larvae of Anopheles from water bodies using the standard dipping method (Service 1993). All the samples were PCR tested to differentiate An. coluzzii larvae from An. gambiae larvae before library preparation.
For Pacific Biosciences (PacBio) sequencing, DNA from a single An. coluzzii larva from the LBV11 site was extracted using the MagAttract HMW DNA extraction kit (Qiagen) following the manufacturer's instructions. For Nanopore sequencing, DNA from six larvae from each of the five breeding sites was extracted either with the QiaAMP UCP DNA kit (Qiagen) or MagAttract HMW DNA extraction kit (Qiagen). For Illumina sequencing, DNA from an additional larva from each of the six different breeding sites was extracted following the same extraction protocol as for Nanopore sequencing (for further details, see Supplemental Methods).
Library preparation and sequencing
Quality control of the DNA sample for PacBio sequencing (Qubit, NanoDrop, and Fragment analyzer) was performed at the Center for Genomic Research facility of the University of Liverpool before library preparation. The library was prepared by shearing DNA to obtain fragments of approximately 30 kb and sequenced on two SMRT cells using Sequel SMRT cell, 3.0 chemistry. Nanopore libraries were constructed using the Native Barcoding Expansion 1–12 (PCR-free) and the Ligation Sequencing Kit following the manufacturer's instructions. A minimum of 400 ng of DNA from each larva was used to start with the library workflow. For each breeding site, six larvae were barcoded, and equal amounts of each barcoded sample were pooled before sequencing. The samples from the same breeding site were run in a single R9.4 flow cell in a 48-h run, except for sample DLA112, which was run in two flow cells. The DNA concentration was assessed during the whole procedure to ensure enough DNA was available for sequencing.
The quality control of the samples, library preparation, and Illumina sequencing was performed at the Center for Genomic Research facility of the University of Liverpool. Low input libraries were prepared with the NEBNext Ultra II FS DNA library kit (300-bp inserts) on the Mosquito platform, using a 1/10 reduced volume protocol. Paired-end sequencing was performed on the Illumina NovaSeq platform using S2 chemistry (2 × 150 bp).
Genome assemblies
The PacBio sequenced genome was assembled using Canu version 1.8 (Koren et al. 2017). The Nanopore genomes were assembled using Canu version 1.8 followed by a round of polishing using Racon version 1.3.3 (Vaser et al. 2017), followed by nanopolish version 0.11.1 (Loman et al. 2015) and Pilon version 1.23-0 (Walker et al. 2014) with the fix parameter set on “bases.” Although we cannot discard that using Illumina data from an additional individual could introduce novel variants, BUSCO values increased after the polishing step. Allelic variants were identified and removed using Purge Haplotigs version 1.0.4 (Roach et al. 2018) with the “-l 15 -m 100 -h 195” parameters. Finally, BlobTools version 1.1.1 (Laetsch and Blaxter 2017) was used to remove contamination from all six genome assemblies taking into consideration fragment sizes, their taxonomic assignation, and the coverage using the Illumina reads (for further details, see Supplemental Methods).
As a proxy of the completeness, the BUSCO values for the six newly assembled genomes plus the AcolN1 genome were obtained using BUSCO version 3.0.2 (Simão et al. 2015) with the diptera_odb9 set as reference. Finally, the contigs for all seven assemblies were scaffolded with RaGOO v1.1 (Alonge et al. 2019) using the chromosome level An. gambiae AgamP4 assembly with default parameters.
Gene annotation transfer
The GFF for the genome annotation for AgamP4 was transferred into the newly assembled genomes using Liftoff (Shumate and Salzberg 2021) with default parameters. The annotation was manually inspected using UGENE version 35 (Okonechnikov et al. 2012) and whenever needed the annotation was accordingly corrected. Ninety-six percent of the AgamP4 genes were correctly transferred.
Construction of the curated TE library and de novo TE annotation
We ran the TEdenovo pipeline (Flutre et al. 2011) independently on each of the seven genomes with default parameters. The obtained consensus in each genome were manually curated (for further details, see Supplemental Methods). To ensure that we identified as much of the TE diversity as possible, we also annotated our genomes with the mosquito libraries present in the TEfam database (tefam.biochem.vt.edu). The consensus from the TEfam library identified in our genomes were added to the REPET library, and all the consensuses were clustered using CD-HIT version 4.8.1 (Fu et al. 2012) with the -c and -s parameters set to 0.8. The sequences belonging to the same cluster were used to perform a multiple sequence alignment and the consensuses were obtained.
The consensuses were classified using PASTEC (Hoede et al. 2014) with default parameters. Next, their bidirectional best hits were calculated using BLAST (Camacho et al. 2009) against the TEfam (tefam.biochem.vt.edu), AnoTExcel (Fernández-Medina et al. 2011), and Repbase (Bao et al. 2015) databases (for further details, see Supplemental Methods). These classified consensuses were used to reannotate the assembled genomes with the TEannot pipeline using default parameters, and we discarded copies whose length overlapped >80% with satellite annotations (Quesneville et al. 2005).
Transfer of TE annotations to the AcolN1 reference genome
We transferred the euchromatic TE annotations from the six genomes we sequenced to the AcolN1 genome. Briefly, we built a GFF file composed by the coordinates of two 500-bp-long “anchors” adjacent to each TE. We transferred these features considering each pair of anchors as exons from a single gene using the Liftoff tool (Shumate and Salzberg 2021). We conserved only transfers in which both anchors were transferred to the AcolN1 genome. Next, following the same strategy we transferred these regions from the AcolN1 genome to the other six genomes. This step allowed the identification of TEs that were present in these genomes but that had not been initially annotated by REPET. When both anchors were separated by less than 10 bp, we considered the TE to be absent; when the anchors were found more than 10 bp away, the TE was considered to be present; finally, when any of the anchors was not transferred the TE was not transferred either. Overall, we transferred 53,893 TEs. A manual inspection of 98 of these insertions (686 TE calls) lead to the annotation of six insertions that were initially missed and the removal of 16 TEs that were incorrectly annotated. Thus, both the false positive (2.3%) and the false negative rate (0.87%) of our annotations were low (for further details, see Supplemental Methods).
Identification of newly described families in other species
To determine the presence of the newly described TE families in other species, RepeatMasker version open-4.0.9 (Smit et al. 2013–2015) was run on 18 dipteran genomes with default parameters and using the 64 newly described families as the library (for the list of the species, see Supplemental Methods).
Identification of heterochromatin
The coordinates for the pericentric heterochromatin, compact intercalary heterochromatin, and diffuse intercalary heterochromatin in An. gambiae AgamP3 were obtained from a previous work (Sharakhova et al. 2010). The An. gambiae AgamP3 genome assembly was mapped against the seven An. coluzzii genome assemblies using progressiveMauve (Darling et al. 2010), and the corresponding coordinates on each of the assemblies were retrieved (Supplemental Table S1C). To confirm that the heterochromatin coordinates were accurately transferred to each genome, we plotted the TE abundance throughout the whole genome and, as expected, we observed a sharp decrease in TE density near the heterochromatin–euchromatin boundaries (Supplemental Figure S8).
Transfer of known inversion breakpoints
The coordinates for the breakpoints of inversions 2La, 2Rb, 2Rc, and 2Rd, and the distal 2Ru breakpoint were obtained from Corbett-Detig et al. (2019). Fifty-kilobase regions flanking each side of the insertion were obtained and mapped using minimap2 (Li 2018) against the scaffolded genome assemblies to transfer the breakpoints. To validate the breakpoints, we analyzed long reads spanning the breakpoints using the Integrative Genomics Viewer (IGV) version 2.4.19 (Robinson et al. 2011).
Detection of putatively active TE families
To identify potentially active TE families, we identified families with more than two identical full-length copies in at least six of the seven annotated genomes. We determined the fraction of identical copies of these families by identifying all their insertions in the genome and calculating the sequence identity of all their bases against the consensus by performing a nucleotide BLAST. Given that long reads have a higher rate of sequencing errors that could affect the age estimation (although we used Illumina reads for polishing), we also used dnaPipeTE (Goubert et al. 2015) to estimate the relative age of the TE families using the raw Illumina reads for the six genomes that we sequenced. We compared the TE landscape obtained using dnaPipeTE with that obtained using the BLAST procedure, using a Kolmogorov–Smirnov test corrected for multiple testing using the Benjamini–Hochberg procedure (Supplemental Table S13). Because we observed few significant differences, we continued using the landscape data obtained using the BLAST procedure. We identified the families where the majority of the bases of their insertions were on the peak of identical sequences in the TE landscape (>50% of the bases with >99% base identity) in more than five of the seven genomes we analyzed. Finally, we assessed the ability to actively transpose strong candidates by identifying their intact ORFs, LTRs (in the case of LTR retrotransposons), and target site duplication (TSD), and estimated the percentage identity between the two LTR of each TE copy.
Classification of TEs by their genomic location
To determine the location of TEs we used the findOverlaps function from the GenomicAlignments R package (Lawrence et al. 2013) using default parameters. Both the TE and the gene annotation were converted to GenomicRanges objects ignoring strand information in the case of TEs.
Insecticide resistance and immune-related genes
A list with a total of 43 relevant insecticide resistance genes was generated taking several works into consideration (Supplemental Table S10; Tene Fossog et al. 2013; Main et al. 2018; Adolfi et al. 2019; Bamou et al. 2019). To determine the position of the L to M nonsynonymous substitution that we observed in AGAP004707 (para) we used the position from the CAM12801.1 reference sequence.
The full list of 414 immune-related genes from An. gambiae was downloaded from ImmunoDB (Waterhouse et al. 2007). We focused on the 281 most reliable genes filtering by the STATUS field and conserving only those with A or B scores (A refers to genes confirmed with high confidence and expert-refined cDNA supplied, and B refers to genes confirmed with high confidence, no refinement required).
TFBS and promoter identification
The matrices for dl (MA0022.1), cnc::maf-S (MA0530.1), and Stat92E (MA0532.1) were downloaded from JASPAR (https://jaspar.genereg.net/) (Sandelin et al. 2004). The sequences for the TEs of interest were obtained using getSeq from the Biostrings R package. The TFBS in the sequences were identified using the web version of FIMO (Grant et al. 2011) from the MEME SUITE (Bailey et al. 2009) with default parameters. The ElemeNT online tool was used to identify promoter motifs (Sloutskin et al. 2015).
Insertion frequency estimation in rural populations
We compared the frequencies of the insertions where the presence/absence status was unambiguously determined in at least four of the seven genomes analyzed with the frequencies of these insertions in two rural populations from the Ag1000G project: Bana in Burkina Faso and Tiassalé in Ivory Coast (Anopheles gambiae 1000 Genomes Consortium 2020). We used the PoPoolationTE2 v-1.10.03 pipeline (Kofler et al. 2016) to compute the TE insertion frequencies in these two rural populations. Briefly, we used the AcolN1 reference genome and the newly generated TE library for An. coluzzii as reference and mapped the Illumina paired-end data from all samples. Next PoPoolationTE2 detected signatures of TE presence/absence and estimated their frequencies in every sample (for additional information, see Supplemental Material).
Data access
The sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA676011. TE and gene annotations in each of the seven genomes analyzed are provided as GFF files available at https://digital.csic.es/handle/10261/224416 and as Supplemental Files 1–14, respectively. The TE library and the transferred annotations across the seven genomes are also available at https://digital.csic.es/handle/10261/224416 and as Supplemental Files 15, 16, respectively. The TE consensus sequences have also been deposited at Dfam release 3.6 (Storer et al. 2021).
Supplementary Material
Acknowledgments
We thank members of the González laboratory for comments on the manuscript. We thank the Ecology of Vectorial Systems team at the Centre Interdisciplinaire de Recherches Médicales de Franceville (CIRMF) (Franceville, Gabon) for their support in field collections. We thank Jean Pierre Agbor and Serge Donfanck for their commitment in larvae collections in Douala (Cameroon). This study was supported by grants from the Ministerio de Economía, Industria y Competitividad, Gobierno de España (MINECO/AEI/FEDER, EU) (BFU2017-82937-P) and grant PID2020-115874GB-I00 funded by Ministerio de Ciencia e Innovación/AEI 10.13039/501100011033 awarded to J.G. D.A. was supported by an Agence Nationale de la Recherche grant (ANR-18-CE35-0002-01—WILDING). N.M.L.P. was funded by Agence universitaire de la Francophonie (AUF) and CIRMF scholarships.
Author contributions: D.A. and J.G. conceived and designed the experiments. N.M.L.P., S.E.N., and L.A. generated data. C.V.-C., D.A., and J.G. performed the data analysis. C.V.-C. and J.G. wrote and revised the manuscript with input from all authors. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275761.121.
Competing interest statement
The authors declare no competing interests.
References
- Adolfi A, Poulton B, Anthousi A, Macilwee S, Ranson H, Lycett GJ. 2019. Functional genetic validation of key genes conferring insecticide resistance in the major African malaria vector, Anopheles gambiae. Proc Natl Acad Sci 116: 25764–25772. 10.1073/pnas.1914633116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20: 224. 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anopheles gambiae 1000 Genomes Consortium. 2020. Genome variation and population structure among 1,142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. Genome Res 30: 1533–1546. 10.1101/gr.262790.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Fossog BT, Ndo C, Djantio BM, Togouet SZ, Awono-Ambene P, Costantini C, Wondji CS, Ranson H. 2011. Anopheles gambiae distribution and insecticide resistance in the cities of Douala and Yaoundé (Cameroon): influence of urban agriculture and pollution. Malar J 10: 154. 10.1186/1475-2875-10-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assogba BS, Djogbénou LS, Milesi P, Berthomieu A, Perez J, Ayala D, Chandre F, Makoutodé M, Labbé P, Weill M. 2015. An ace-1 gene duplication resorbs the fitness cost associated with resistance in Anopheles gambiae, the main malaria mosquito. Sci Rep 5: 14529. 10.1038/srep14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Acevedo P, Pombi M, Dia I, Boccolini D, Costantini C, Simard F, Fontenille D. 2017. Chromosome inversions and ecological plasticity in the main African malaria mosquitoes. Evolution 71: 686–701. 10.1111/evo.13176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Zhang S, Chateau M, Fouet C, Morlais I, Costantini C, Hahn MW, Besansky NJ. 2019. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol Ecol 28: 1333–1342. 10.1111/mec.14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202-8. 10.1093/nar/gkp091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanidou V, Kefi M, Aivaliotis M, Koidou V, Girotti JR, Mijailovsky SJ, Juárez MP, Papadogiorgaki E, Chalepakis G, Kampouraki A, et al. 2019. Mosquitoes cloak their legs to resist insecticides. Proc Biol Sci 286: 20191091. 10.1098/rspb.2019.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamou R, Sonhafouo-Chiana N, Mavridis K, Tchuinkam T, Wondji CS, Vontas J, Antonio-Nkondjio C. 2019. Status of insecticide resistance and its mechanisms in Anopheles gambiae and Anopheles coluzzii populations from forest settings in south Cameroon. Genes (Basel) 10: 741. 10.3390/genes10100741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 4–9. 10.1186/s13100-015-0035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D. 2020. Plant pan-genomes are the new reference. Nat Plants 6: 914–920. 10.1038/s41477-020-0733-0 [DOI] [PubMed] [Google Scholar]
- Boulesteix M, Simard F, Antonio-Nkondjio C, Awono-Ambene HP, Fontenille D, Biémont C. 2007. Insertion polymorphism of transposable elements and population structure of Anopheles gambiae M and S molecular forms in Cameroon. Mol Ecol 16: 441–452. 10.1111/j.1365-294X.2006.03150.x [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Gulati M, Jiang H. 2017. Serine protease-related proteins in the malaria mosquito, Anopheles gambiae. Insect Biochem Mol Biol 88: 48–62. 10.1016/j.ibmb.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol 22: 1503–1517. 10.1111/mec.12170 [DOI] [PubMed] [Google Scholar]
- Cassone BJ, Kamdem C, Cheng C, Tan JC, Hahn MW, Costantini C, Besansky NJ. 2014. Gene expression divergence between malaria vector sibling species Anopheles gambiae and An. coluzzii from rural and urban Yaoundé Cameroon. Mol Ecol 23: 2242–2259. 10.1111/mec.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Mahmood A, Kalathur R, Liu L, Carlier PR. 2018. Structure of the G119S mutant acetylcholinesterase of the malaria vector Anopheles gambiae reveals basis of insecticide resistance. Structure 26: 130–136.e2. 10.1016/j.str.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg 73: 483–497. 10.1016/0035-9203(79)90036-1 [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. 10.1126/science.1077769 [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Said I, Calzetta M, Genetti M, McBroome J, Maurer NW, Petrarca V, della Torre A, Besansky NJ. 2019. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics 213: 1495–1511. 10.1534/genetics.119.302385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo WM, Pombi M, Some CY, Bassole IHN, Ose K, Fotsing JM, Sagnon NF, Fontenille D, et al. 2009. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol 9: 16. 10.1186/1472-6785-9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiré RK, Namountougou M, Diabaté A, Soma DD, Bado J, Toé HK, Bass C, Combary P. 2014. Distribution and frequency of kdr mutations within Anopheles gambiae s.l. populations and first report of the ace.1 G119S mutation in Anopheles arabiensis from Burkina Faso (West Africa). PLoS One 9: e101484. 10.1371/journal.pone.0101484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo ES, Wallau GdL. 2020. Mosquito genomes are frequently invaded by transposable elements through horizontal transfer. PLoS Genet 16: e1008946. 10.1371/journal.pgen.1008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison NJ, BenMarzouk-Hidalgo OJ, Dimopoulos G. 2015. MicroRNA-regulation of Anopheles gambiae immunity to Plasmodium falciparum infection and midgut microbiota. Dev Comp Immunol 49: 170–178. 10.1016/j.dci.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Lopes G, Borges-Veloso A, Saboia-Vahia L, Domont GB, Britto C, Cuervo P, De Jesus JB. 2015. Expression of active trypsin-like serine peptidases in the midgut of sugar-feeding female Anopheles aquasalis. Parasit Vectors 8: 296. 10.1186/s13071-015-0908-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AG, Ouyang N, Boyle AP. 2020. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat Commun 11: 1796. 10.1038/s41467-020-15520-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesel JF, Ortiz MF, Marinotti O, Vasconcelos ATR, Loreto ELS. 2019. A re-annotation of the Anopheles darlingi mobilome. Genet Mol Biol 42: 125–131. 10.1590/1678-4685-gmb-2017-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanga-Ndille E, Nouage L, Ndo C, Binyang A, Assatse T, Nguiffo-Nguete D, Djonabaye D, Irwing H, Tene-Fossog B, Wondji CS. 2019. The G119S acetylcholinesterase (Ace-1) target site mutation confers carbamate resistance in the major malaria vector Anopheles gambiae from Cameroon: a challenge for the coming IRS implementation. Genes (Basel) 10: 790. 10.3390/genes10100790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. 2014. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15: 86. 10.1186/1471-2164-15-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Boulesteix M, Duchemin JB, Koffi AA, Chandre F, Dabiré R, Robert V, Simard F, Tripet F, Donnelly MJ, et al. 2008. High genetic differentiation between the M and S molecular forms of Anopheles gambiae in Africa. PLoS One 3: e1968. 10.1371/journal.pone.0001968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essandoh J, Yawson AE, Weetman D. 2013. Acetylcholinesterase (Ace-1) target site mutation 119S is strongly diagnostic of carbamate and organophosphate resistance in Anopheles gambiae s.s. and Anopheles coluzzii across southern Ghana. Malar J 12: 404. 10.1186/1475-2875-12-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel AN, Ibrahim SS, Tchouakui M, Terence E, Wondji MJ, Tchoupo M, Wanji S, Wondji CS. 2019. A combination of metabolic resistance and high frequency of the 1014F kdr mutation is driving pyrethroid resistance in Anopheles coluzzii population from Guinea savanna of Cameroon. Parasit Vectors 12: 263. 10.1186/s13071-019-3523-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Medina RD, Struchiner CJ, Ribeiro JMC. 2011. Novel transposable elements from Anopheles gambiae. BMC Genomics 12: 260. 10.1186/1471-2164-12-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Medina RD, Ribeiro JMC, Carareto CMA, Velasque L, Struchiner CJ. 2012. Losing identity: structural diversity of transposable elements belonging to different classes in the genome of Anopheles gambiae. BMC Genomics 13: 272. 10.1186/1471-2164-13-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ffrench-Constant R, Daborn P, Feyereisen R. 2006. Resistance and the jumping gene. Bioessays 28: 6–8. 10.1002/bies.20354 [DOI] [PubMed] [Google Scholar]
- Flutre T, Duprat E, Feuillet C, Quesneville H. 2011. Considering transposable element diversification in de novo annotation approaches. PLoS One 6: e16526. 10.1371/journal.pone.0016526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca PM, Moura RD, Wallau GL, Loreto ELS. 2019. The mobilome of Drosophila incompta, a flower-breeding species: comparison of transposable element landscapes among generalist and specialist flies. Chromosome Res 27: 203–219. 10.1007/s10577-019-09609-x [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347: 1258524. 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C, Gray E, Besansky NJ, Costantini C. 2012. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One 7: e34841. 10.1371/journal.pone.0034841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C, Atkinson P, Kamdem C. 2018. Human interventions: driving forces of mosquito evolution. Trends Parasitol 34: 127–139. 10.1016/j.pt.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li Y, Kim KD, Abernathy B, Jackson SA. 2016. Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol 17: 7. 10.1186/s13059-015-0867-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P, Jensen S, Pogorelcnik R, Lee J, Xing Y, Brasset E, Vaury C, Sharakhov IV. 2015. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin 8: 50. 10.1186/s13072-015-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerner-Potvin P, Bourque G. 2018. Computational tools to unmask transposable elements. Nat Rev Genet 19: 688–704. 10.1038/s41576-018-0050-x [DOI] [PubMed] [Google Scholar]
- Goubert C, Modolo L, Vieira C, ValienteMoro C, Mavingui P, Boulesteix M. 2015. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol Evol 7: 1192–1205. 10.1093/gbe/evv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Bové X, Tomlinson S, O'Reilly AO, Harding NJ, Miles A, Kwiatkowski D, Donnelly MJ, Weetman D, The Anopheles gambiae 1000 Genomes Consortium. 2020. Evolution of the insecticide target Rdl in African Anopheles is driven by interspecific and interkaryotypic introgression. Mol Biol Evol 37: 2900–2917. 10.1093/molbev/msaa128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YH. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet 16: 461–468. 10.1016/S0168-9525(00)02104-1 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, et al. 2020. European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet 52: 950–957. 10.1038/s41588-020-0671-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoede C, Arnoux S, Moisset M, Chaumier T, Inizan O, Jamilloux V, Quesneville H. 2014. PASTEC: an automatic transposable element classification tool. PLoS One 9: e91929. 10.1371/journal.pone.0091929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JC, Wides R, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, Ricci WA, Guo T, Olson A, Qiu Y, et al. 2021. De novo assembly of 26 diverse maize genomes. Science 373: 655–662. 10.1126/science.abg5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Ren X, Ramirez JL, Sakamoto JM, Bailey JA, Jedlicka AE, Rasgon JL. 2011. Wolbachia infections in Anopheles gambiae cells: transcriptomic characterization of a novel host-symbiont interaction. PLoS Pathog 7: e1001296. 10.1371/journal.ppat.1001296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham VA, Pignatelli P, Moore JD, Wagstaff S, Ranson H. 2017. The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics 18: 669. 10.1186/s12864-017-4086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Warsinger-Pepe N, Watase GJ, Yamashita YM. 2017. Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 (Bethesda) 7: 693–704. 10.1534/g3.116.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Upton KR. 2019. Human transposons are an abundant supply of transcription factor binding sites and promoter activities in breast cancer cell lines. Mob DNA 10: 16. 10.1186/s13100-019-0158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MTJ, Munshi-South J. 2017. Evolution of life in urban environments. Science 358: eaam8327. 10.1126/science.aam8327 [DOI] [PubMed] [Google Scholar]
- Jones CM, Liyanapathirana M, Agossa FR, Weetman D, Ranson H, Donnelly MJ, Wilding CS. 2012. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci 109: 6614–6619. 10.1073/pnas.1201475109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C, Fouet C, Gamez S, White BJ. 2017. Pollutants and insecticides drive local adaptation in African malaria mosquitoes. Mol Biol Evol 34: 1261–1275. 10.1093/molbev/msx087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamgang B, Tchapga W, Ngoagouni C, Sangbakembi-Ngounou C, Wondji M, Riveron JM, Wondji CS. 2018. Exploring insecticide resistance mechanisms in three major malaria vectors from Bangui in Central African Republic. Pathog Glob Health 112: 349–359. 10.1080/20477724.2018.1541160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengne P, Charmantier G, Blondeau-Bidet E, Costantini C, Ayala D. 2019. Tolerance of disease-vector mosquitoes to brackish water and their osmoregulatory ability. Ecosphere 10: e02783. 10.1002/ecs2.2783 [DOI] [Google Scholar]
- Kent LB, Walden KKO, Robertson HM. 2008. The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem Senses 33: 79–93. 10.1093/chemse/bjm067 [DOI] [PubMed] [Google Scholar]
- King SA, Onayifeke B, Akorli J, Sibomana I, Chabi J, Manful-Gwira T, Dadzie S, Suzuki T, Wilson MD, Boakye DA, et al. 2017. The role of detoxification enzymes in the adaptation of the major malaria vector Anopheles gambiae (Giles; Diptera: Culicidae) to polluted water. J Med Entomol 54: 1674–1683. 10.1093/jme/tjx164 [DOI] [PubMed] [Google Scholar]
- Kingan SB, Heaton H, Cudini J, Lambert CC, Baybayan P, Galvin BD, Durbin R, Korlach J, Lawniczak MKN. 2019. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes (Basel) 10: 62. 10.3390/genes10010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schlötterer C. 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet 11: e1005406. 10.1371/journal.pgen.1005406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Gómez-Sánchez D, Schlötterer C. 2016. PoPoolationTE2: comparative population genomics of transposable elements using Pool-Seq. Mol Biol Evol 33: 2759–2764. 10.1093/molbev/msw137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé P, Berthomieu A, Berticat C, Alout H, Raymond M, Lenormand T, Weill M. 2007. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol 24: 1056–1067. 10.1093/molbev/msm025 [DOI] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. 2017. BlobTools: interrogation of genome assemblies. F1000Res 6: 1287. 10.12688/f1000research.12232.1 [DOI] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NF, Sangaré DM, Regier AA, Reidenbach KR, Bretz DA, Sharakhova MV, Emrich SJ, Traore SF, Costantini C, Besansky NJ, et al. 2010. Breakpoint structure of the Anopheles gambiae 2Rb chromosomal inversion. Malar J 9: 293. 10.1186/1475-2875-9-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE. 2020. Long-read human genome sequencing and its applications. Nat Rev Genet 21: 597–614. 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12: 733–735. 10.1038/nmeth.3444 [DOI] [PubMed] [Google Scholar]
- Longo-Pendy NM, Tene-Fossog B, Tawedi RE, Akone-Ella O, Toty C, Rahola N, Braun JJ, Berthet N, Kengne P, Costantini C, et al. 2021. Ecological plasticity to ions concentration determines genetic response and dominance of Anopheles coluzzii larvae in urban coastal habitats of Central Africa. Sci Rep 11: 15781. 10.1038/s41598-021-94258-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas ER, Miles A, Harding NJ, Clarkson CS, Lawniczak MKN, Kwiatkowski DP, Weetman D, Donnelly MJ. 2019. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res 29: 1250–1261. 10.1101/gr.245795.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Everitt A, Cornel AJ, Hormozdiari F, Lanzaro GC. 2018. Genetic variation associated with increased insecticide resistance in the malaria mosquito, Anopheles coluzzii. Parasit Vectors 11: 225. 10.1186/s13071-018-2817-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Cerqueira GC, De Almeida LGP, Ferro MIT, Da Silva Loreto EL, Zaha A, Teixeira SMR, Wespiser AR, E Silva AA, Schlindwein AD, et al. 2013. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res 41: 7387–7400. 10.1093/nar/gkt484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsano RM, Leronni D, D'Addabbo P, Viggiano L, Tarasco E, Caizzi R. 2012. Mosquitoes LTR retrotransposons: a deeper view into the genomic sequence of Culex quinquefasciatus. PLoS One 7: e30770. 10.1371/journal.pone.0030770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo L, Ullastres A, González J. 2014. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet 10: e1004560. 10.1371/journal.pgen.1004560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiopoulos KD, Della Torre A, Predazzi V, Petrarca V, Coluzzi M. 1998. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc Natl Acad Sci 95: 12444–12449. 10.1073/pnas.95.21.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri C, Markianos K, Guelbeogo WM, Bischoff E, Gneme A, Eiglmeier K, Holm I, Sagnon NF, Vernick KD, Riehle MM. 2015. The kdr-bearing haplotype and susceptibility to Plasmodium falciparum in Anopheles gambiae: genetic correlation and functional testing. Malar J 14: 391. 10.1186/s12936-015-0924-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arcà B, Arensburger P, Artemov G, et al. 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347: 6217. 10.1126/science.1258522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, UGENE team. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- Oliveira GdA, Lieberman J, Barillas-Mury C. 2012. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335: 856–859. 10.1126/science.1209678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osta MA, Christophides GK, Vlachou D, Kafatos FC. 2004. Innate immunity in the malaria vector Anopheles gambiae: comparative and functional genomics. J Exp Biol 207: 2551–2563. 10.1242/jeb.01066 [DOI] [PubMed] [Google Scholar]
- Ou S, Liu J, Chougule KM, Fungtammasan A, Seetharam AS, Stein JC, Llaca V, Manchanda N, Gilbert AM, Wei S, et al. 2020. Effect of sequence depth and length in long-read assembly of the maize inbred NC358. Nat Commun 11: 2288. 10.1038/s41467-020-16037-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perugini E, Guelbeogo WM, Calzetta M, Manzi S, Virgillito C, Caputo B, Pichler V, Ranson H, Sagnon NF, Della Torre A, et al. 2020. Behavioural plasticity of Anopheles coluzzii and Anopheles arabiensis undermines LLIN community protective effect in a Sudanese-savannah village in Burkina Faso. Parasit Vectors 13: 277. 10.1186/s13071-020-04142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Armisén D, Gibbs RA, Hering L, Khila A, Mayer G, Richards S, Niehuis O, Misof B. 2019. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol Biol 19: 11. 10.1186/s12862-018-1324-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN 2nd, Blanco-Berdugo L, Ray DA. 2016. Accurate transposable element annotation is vital when analyzing new genome assemblies. Genome Biol Evol 8: 403–410. 10.1093/gbe/evw009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Bortolini Silveira A, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5: e15716. 10.7554/eLife.15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, Anxolabehere D. 2005. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol 1: e22. 10.1371/journal.pcbi.0010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesneville H, Nouaud D, Anxolabéhère D. 2006. P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC Genomics 7: 214. 10.1186/1471-2164-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech GE, Bogaerts-Márquez M, Barrón MG, Merenciano M, Villanueva-Cañas JL, Horváth V, Fiston-Lavier AS, Luyten I, Venkataram S, Quesneville H, et al. 2019. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genet 15: e1007900. 10.1371/journal.pgen.1007900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M, Vieira CP, Lata R, Posnien N, Vieira J. 2018. Origin and consequences of chromosomal inversions in the virilis group of Drosophila. Genome Biol Evol 10: 3152–3166. 10.1093/gbe/evy239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishishwar L, Tellez Villa CE, Jordan IK. 2015. Transposable element polymorphisms recapitulate human evolution. Mob DNA 6: 21. 10.1186/s13100-015-0052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR. 2018. Purge haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19: 460. 10.1186/s12859-018-2485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostant WG, Wedell N, Hosken DJ. 2012. Chapter 2: Transposable elements and insecticide resistance. In Advances in Genetics (ed. Goodwin SF, et al. ), pp. 169–201. Academic Press, San Diego. [DOI] [PubMed] [Google Scholar]
- Ruiz JL, Yerbanga RS, Lefèvre T, Ouedraogo JB, Corces VG, Gómez-Díaz E. 2019. Chromatin changes in Anopheles gambiae induced by Plasmodium falciparum infection. Epigenetics Chromatin 12: 5. 10.1186/s13072-018-0250-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JL, Ranford-Cartwright LC, Gómez-Díaz E. 2021. The regulatory genome of the malaria vector Anopheles gambiae: integrating chromatin accessibility and gene expression. NAR Genomics Bioinforma 3: lqaa113. 10.1093/nargab/lqaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salces-Ortiz J, Vargas-Chavez C, Guio L, Rech GE, González J. 2020. Transposable elements contribute to the genomic response to insecticides in Drosophila melanogaster. Philos Trans R Soc B: Biol Sc 375: 20190341. 10.1098/rstb.2019.0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgueiro P, Moreno M, Simard F, O'Brochta D, Pinto J. 2013. New insights into the population structure of Anopheles gambiae s.s. in the Gulf of Guinea islands revealed by Herves transposable elements. PLoS One 8: e62964. 10.1371/journal.pone.0062964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94. 10.1093/nar/gkh012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Calzetta M, Etang J, Barrese E, Dia I, Caccone A, Donnelly MJ, Petrarca V, Simard F, Pinto J, et al. 2008a. Distribution of knock-down resistance mutations in Anopheles gambiae molecular forms in west and west-central Africa. Malar J 7: 74. 10.1186/1475-2875-7-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, Della Torre A. 2008b. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J 7: 163. 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Schmitz J. 2019. The impact of transposable elements in adaptive evolution. Mol Ecol 28: 1537–1549. 10.1111/mec.14794 [DOI] [PubMed] [Google Scholar]
- Service MW. 1993. Sampling the larval population. In Mosquito ecology (ed. Service MW), pp. 75–209. Springer, Dordrecht. [Google Scholar]
- Sessegolo C, Burlet N, Haudry A. 2016. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol Lett 12: 20160407. 10.1098/rsbl.2016.0407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Slotkin RK. 2020. The current revolution in transposable element biology enabled by long reads. Curr Opin Plant Biol 54: 49–56. 10.1016/j.pbi.2019.12.012 [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo NF, Santolamazza F, Della Torre A, Simard F, Collins FH, Besansky NJ. 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci 103: 6258–6262. 10.1073/pnas.0509683103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, George P, Brusentsova IV, Leman SC, Bailey JA, Smith CD, Sharakhov IV. 2010. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics 11: 459. 10.1186/1471-2164-11-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Kinney NA, Timoshevskiy VA, Sharakhova MV, Sharakhov IV. 2020. Structural variation of the X chromosome heterochromatin in the Anopheles gambiae complex. Genes (Basel) 11: 327. 10.3390/genes11030327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Salzberg SL. 2021. Liftoff: accurate mapping of gene annotations. Bioinformatics 37: 1639–1643. 10.1093/bioinformatics/btaa1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva APB, Santos JMM, Martins AJ. 2014. Mutations in the voltage-gated sodium channel gene of anophelines and their association with resistance to pyrethroids—a review. Parasit Vectors 7: 450. 10.1186/1756-3305-7-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, Pombi M, Etouna J, Ose K, Fotsing JM, Fontenille D, Besansky NJ, Costantini C. 2009. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol 9: 17. 10.1186/1472-6785-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloutskin A, Danino YM, Orenstein Y, Zehavi Y, Doniger T, Shamir R, Juven-Gershon T. 2015. ElemeNT: a computational tool for detecting core promoter elements. Transcription 6: 41–50. 10.1080/21541264.2015.1067286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Sriwichai P, Rongsiryam Y, Jariyapan N, Sattabongkot J, Apiwathnasorn C, Nacapunchai D, Paskewitz S. 2012. Cloning of a trypsin-like serine protease and expression patterns during Plasmodium falciparum invasion in the mosquito, Anopheles dirus (Peyton and Harrison). Arch Insect Biochem Physiol 80: 151–165. 10.1002/arch.21034 [DOI] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA 12: 2. 10.1186/s13100-020-00230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Wyler M, Gimmi EL, Pippel M, Roulin AC. 2020. Diversity, dynamics and effects of long terminal repeat retrotransposons in the model grass Brachypodium distachyon. New Phytologist 227: 1736–1748. 10.1111/nph.16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Phil Trans R Soc B: Biol Sci 375: 20190347. 10.1098/rstb.2019.0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Poupardin R, Costantini C, Awono-Ambene P, Wondji CS, Ranson H, and Antonio-Nkondjio C. 2013. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS One 8: e61408. 10.1371/journal.pone.0061408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Ayala D, Acevedo P, Kengne P, Ngomo Abeso Mebuy I, Makanga B, Magnus J, Awono-Ambene P, Njiokou F, Pombi M, et al. 2015. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol Appl 8: 326–345. 10.1111/eva.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubío JMC, Naveira H, Costas J. 2005. Structural and evolutionary analyses of the Ty3/gypsy group of LTR retrotransposons in the genome of Anopheles gambiae. Mol Biol Evol 22: 29–39. 10.1093/molbev/msh251 [DOI] [PubMed] [Google Scholar]
- Tubío JMC, Tojo M, Bassaganyas L, Escaramis G, Sharakhov IV, Sharakhova MV, Tornador C, Unger MF, Naveira H, Costas J, et al. 2011. Evolutionary dynamics of the Ty3/Gypsy LTR retrotransposons in the genome of Anopheles gambiae. PLoS One 6: e16328. 10.1371/journal.pone.0016328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullastres A, Merenciano M, González J. 2021. Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol 22: 265. 10.1186/s13059-021-02471-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Willis JH. 2017. Localization of RR-1 and RR-2 cuticular proteins within the cuticle of Anopheles gambiae. Arthropod Struct Dev 46: 13–29. 10.1016/j.asd.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Chávez C, González J. 2020. Transposable elements in Anopheles species: refining annotation strategies towards population-level analysis. In Population genomics: insects (ed. Dupuis ORaJ) Springer, Cham, Switzerland. [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27: 737–746. 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Cañas JL, Horvath V, Aguilera L, González J. 2019. Diverse families of transposable elements affect the transcriptional regulation of stress-response genes in Drosophila melanogaster. Nucleic Acids Res 47: 6842–6857. 10.1093/nar/gkz490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. 2006. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol 8: 1392–1405. 10.1111/j.1462-5822.2006.00718.x [DOI] [PubMed] [Google Scholar]
- Vontas J, Grigoraki L, Morgan J, Tsakireli D, Fuseini G, Segura L, de Carvalho N, Nguema J, Weetman R, Slotman D, et al. 2018. Rapid selection of a pyrethroid metabolic enzyme CYP9K1 by operational malaria control activities. Proc Natl Acad Sci 115: 4619–4624. 10.1073/pnas.1719663115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, ξ Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316: 1738–1743. 10.1126/science.1139862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Riveron JM, Hearn J, Irving H, Kamdem C, Fouet C, White BJ, Wondji CS. 2020. An Africa-wide genomic evolution of insecticide resistance in the malaria vector Anopheles funestus involves selective sweeps, copy number variations, gene conversion and transposons. PLoS Genet 16: e1008822. 10.1371/journal.pgen.1008822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Djogbenou LS, Lucas E. 2018. Copy number variation (CNV) and insecticide resistance in mosquitoes: evolving knowledge or an evolving problem? Curr Opin Insect Sci 27: 82–88. 10.1016/j.cois.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissensteiner MH, Bunikis I, Catalán A, Francoijs KJ, Knief U, Heim W, Peona V, Pophaly SD, Sedlazeck FJ, Suh A, et al. 2020. Discovery and population genomics of structural variation in a songbird genus. Nat Commun 11: 3403. 10.1038/s41467-020-17195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe A, Longbottom J, Gleave K, Shearer FM, Sinka ME, Massey NC, Cameron E, Bhatt S, Gething PW, Hemingway J, et al. 2017. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar J 16: 85. 10.1186/s12936-017-1734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TG. 1993. Transposable elements as initiators of insecticide resistance. J Econ Entomol 86: 645–651. 10.1093/jee/86.3.645 [DOI] [PubMed] [Google Scholar]
- Witte CP, Le QH, Bureau T, Kumar A. 2001. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Nat Acad Sci 98: 13778–13783. 10.1073/pnas.241341898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia A, Sharakhova MV, Leman SC, Tu Z, Bailey JA, Smith CD, Sharakhov IV. 2010. Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS One 5: e10592. 10.1371/journal.pone.0010592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. 2010. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 20: 804–815. 10.1101/gr.100594.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee WP, Ye K, Lee C. 2019. One reference genome is not enough. Genome Biol 20: 104. 10.1186/s13059-019-1624-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cahan SH. 2012. A novel family of terminal-repeat retrotransposon in miniature (TRIM) in the genome of the red harvester ant, Pogonomyrmex barbatus. PLoS One 7: e53401. 10.1371/journal.pone.0053401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.