ABSTRACT
Background
Probiotic lozenges have been developed to harvest the benefits of probiotics for oral health, but their long-term consumption may encourage the transfer of resistance genes from probiotics to commensals, and eventually to disease-causing bacteria.
Aim
To screen commercial probiotic lozenges for resistance to antibiotics, characterize the resistance determinants, and examine their transferability in vitro.
Results
Probiotics of all lozenges were resistant to glycopeptide, sulfonamide, and penicillin antibiotics, while some were resistant to aminoglycosides and cephalosporins. High minimum inhibitory concentrations (MICs) were detected for streptomycin (>128 µg/mL) and chloramphenicol (> 512 µg/mL) for all probiotics but only one was resistant to piperacillin (MIC = 32 µg/mL). PCR analysis detected erythromycin (erm(T), ermB or mefA) and fluoroquinolone (parC or gyr(A)) resistance genes in some lozenges although there were no resistant phenotypes. The dfrD, cat-TC, vatE, aadE, vanX, and aph(3”)-III or ant(2”)-I genes conferring resistance to trimethoprim, chloramphenicol, quinupristin/dalfopristin, vancomycin, and streptomycin, respectively, were detected in resistant probiotics. The rifampicin resistance gene rpoB was also present. We found no conjugal transfer of streptomycin resistance genes in our co-incubation experiments.
Conclusion
Our study represents the first antibiotic resistance profiling of probiotics from oral lozenges, thus highlighting the health risk especially in the prevailing threat of drug resistance globally.
KEYWORDS: Probiotics, antibiotic resistance, probiotic supplements, oral probiotics, probiotic lozenges, dental probiotics, lactobacillus, horizontal gene transfer
Introduction
Oral lozenges containing a formulation of probiotic bacteria have been recently developed to harvest the benefits of probiotics for oral health. It is long known that probiotics can confer various health benefits such as improving the host immune system [1,2], preventing cancer and virus-inflammatory lung damage [3,4], decreasing cholesterol and preventing cardiovascular diseases [5], improving blood glucose and lipid profiles [6,7], preventing diabetes [8], and enhancing cognitive function and mental health [9]. As the health claims of probiotics continue to emerge, the number of foods containing probiotics also increases [10,11]. This is exemplified by the overwhelmingly popular probiotic health or dietary supplements targeted for improving intestinal health with beneficial claims ranging from regulating the gut microbiota [12], and reducing lactose intolerance [13], increasing bioavailability of nutrients [14], preventing gastrointestinal infections [15,16], and treating gastroenteritis and antibiotic-associated diarrhea [17]. In recent years, probiotics have also been proposed as alternative to or as adjuvant for antibiotic treatments [18–22]. For instance, evidence in vitro showed that lactobacilli exhibit anti-carbapenem resistant enterococci while evidence in mouse models showed that Lactobacillus paracasei CNCM I-3689 reduces the drug resistant enterococci amount in the feces [23,24]. Moreover, recent clinical trials also showed that the colonization of multi-drug resistant pathogens in the human gut, can be counteracted by treatment with a mixture of probiotics during antibiotic therapies [25,26].
The benefits of probiotics in oral health have been consistently reported for the treatment of caries [27,28], periodontal disease [29–32], fungal infection [33], and halitosis [34,35], through synergistic mechanisms that include the inhibition of bacteria commonly associated with oral diseases such as Streptococcus mutans, Porphyromonas gingivalis, and Candida albicans by antimicrobial compounds, enhancement of local or systemic immune responses, and out-competing disease-causing bacteria for adhesion [36]. Collectively, probiotics lead to direct antagonistic effects against pathogens and/or reduction of inflammation and tissue destruction [37,38]. Intestinal swallowable probiotic supplements in the form of capsules or tablets do not remain in the oral cavity long enough for probiotics to be retained in the mouth. As such, probiotic oral lozenges were developed to deliver probiotics directly to the mouth and enable them to adhere, form biofilms, and colonize oral cavity surfaces [39,40].
Much like probiotics for gut health, probiotic strains for oral health are predominantly Lactobacillus and Streptococcus strains, which are ‘Generally Recognized as Safe (GRAS)’ according to the European Food Safety Authority (EFSA) [41]. They were also granted the ‘Qualified Presumption of Safety (QPS)’ status by the U.S. Food and Drug Administration (FDA) [42] on the condition that they do not harbor known genes conferring resistance to clinically and veterinary important drugs [43]. However, recent studies have reported that probiotics in food supplements are resistant to multiple antibiotics [44,45]. This is in addition to the numerous reports of drug resistance in probiotics from other foods [46–50]. Since probiotic supplements contain much higher bacteria per serving compared to other foods, any adverse health effects would therefore be more pronounced [51,52]. One such health concern is the risk of transmitting resistance determinants. The long-term consumption of oral probiotic lozenges may encourage the transfer of resistance genes from probiotics to over 700 bacterial species in the oral cavity [53]. Over time, the oral microbiota may act as a reservoir for antibiotic resistance genes which could then be transferred to bacteria commonly associated with oral diseases such as S. mutans and P. gingivalis [54,55], thus rendering antibiotic treatments ineffective.
This health concern has been raised for probiotic foods harboring antibiotic resistance genes [46,51,52,56–68], and the same applies to the oral cavity because much like the gut, it contains a complex and rich diversity of microbiota [69]. Moreover, the nutrient-rich environment, suitable and stable temperature of around 37°C, a stable pH range of 6.5–7.0 considered ideal for most bacteria, as well as the moist and large surface areas in the oral cavity, further encourage the trafficking of resistance determinants [70]. While drug resistance of gut probiotic supplements has been reported, the resistance profiles of oral probiotic lozenges have not been examined. Thus, this study aims to analyze the resistance profiles of probiotics from oral lozenges and the transferability of resistance determinants.
Materials and methods
Probiotic lozenges, probiotic drinks, and antibiotics
Six popular brands of probiotic oral lozenges were purchased. Only probiotic supplements in the form of lozenges intended for dental applications or oral health were selected. Those in the form of capsules or tablets intended for gut health were eliminated from our selection. Products that contain heterogenous populations of Lactobacillus spp. were prioritized. Other selection factors such as the overall reputation of the manufacturers, and the product ratings and reviews, were also considered. The probiotic oral lozenges are subsequently designated as A, B, C, D, E, F, G, and H, the product information such as the bacteria strains and amounts, and the country of manufacture are listed in Table 1. Commercially available Lactobacillus containing probiotic drinks such as probiotic milk, yogurt, and juice were purchased from local food markets in China.
Table 1.
Oral probiotic lozenges product information
| Product label | Probiotic bacteria | Bacteria amount | Country of manufacture |
|---|---|---|---|
| A |
Lactobacillus paracasei Lactobacillus reuteri Lactobacillus sakei Lactobacillus salivarius |
1–3 billion CFU per lozenge |
USA |
| B |
Streptococcus salivarius M18 Lactobacillus plantarum (heat treated) |
1 billion CFU per lozenge |
USA |
| C |
Lactobacillus reuteri DSM 17938 Lactobacillus reuteri ATCC PTA 5289 |
0.2 billion CFU per lozenge |
Belgium |
| D |
Lactobacillus acidophilus HA-122 Lactobacillus casei HA-108 Bifidobacterium bifidum HA-132 Lactobacillus rhamnosus HA-111 Lactobacillus salivarius HA-188 |
1–2 billion CFU per lozenge |
USA |
| E |
Lactobacillus brevis CECT 7480 Lactobacillus plantarum CECT 7481 Pediococcus acidilactici CECT 8633 |
1–5 billion CFU per lozenge |
USA |
| F |
Lactobacillus acidophilus Lactobacillus reuteri Lactobacillus paracasei Lactobacillus salivarius Streptococcus thermophilus Streptococcus salivarius K12 Streptococcus salivarius M18 |
2–6 billion CFU per lozenge |
Canada |
| G |
Lactobacillus paracasei Lactobacillus reuteri |
1–3 billion CFU per lozenge |
USA |
| H | Lactobacillus reuteri DSM 17938 | 0.2 billion CFU per 5 drops |
Belgium |
Powdered or crystallized antibiotics: chloramphenicol, doxycycline, erythromycin, piperacillin, and streptomycin, were purchased from Sigma, USA. The antibiotics were dissolved in sterile Milli-Q water or ethanol to 10 mg/mL stock concentrations and stored at −20°C. Prior to the broth microdilution experiments, antibiotic stocks were diluted to 1 mg/mL and 0.1 mg/mL working concentrations as required.
Bacteria recovery and enumeration
o recover probiotic bacteria, oral probiotic lozenges were ground and dissolved in phosphate buffered saline (PBS) and immediately spread on De Man, Rogosa and Sharpe (MRS) agar which is selective for growing lactobacilli. The dissolved samples were cultured overnight in MRS broth at 37°C in an orbital shaker at 250 rpm for the enrichment of probiotic bacteria. To culture S. mutans, Streptococcus gordonii, Streptococcus sanguinis, and Enterococcus faecalis, the Brain Heart Infusion (BHI) medium was used. Glycerol stocks (50%, v/v) were prepared as required and stored at −80°C.
he drop plate method was used to enumerate probiotic bacteria. One hundred milligrams of probiotic lozenges were dissolved in 1 mL PBS and a series of dilutions from 10−1 to 10−7 were prepared. Five microliters of each diluted sample were dropped onto MRS agar using aseptic methods and incubated for 48 h at 37°C. MRS plates with the appropriate dilutions that produce discernable single colonies of bacteria, were photographed, and enumerated using ImageJ. The enumerated viable bacteria were compared with the claims of the manufacturers on the product label or information sheet (Table 1). Only contributions from Lactobacillus strains were considered. Bacteria enumeration was conducted in biological triplicates with each containing nine droplets of the dissolved samples.
Identification of probiotic isolates by 16S rRNA sequencing
The bacterial genomic DNA was extracted with lysozyme treatment following a Gram-positive bacteria lysis protocol using the QIAprep Miniprep Kit (Qiagen, MD) according to the manufacturer’s instructions. The concentration and purity of extracted DNA were determined on a NanoDropOne spectrophotometer (ND-ONE-W, ThermoFischer, WI) before being sent to Genewiz, Inc. (Suzhou, China) for PCR detection and sequencing of the 16S rRNA using the universal primers 27 F: 50-AGAGTTTGATCCTGGCTCAG-30, and 1492 R: 50-GGTTACCTTGTTACGACTT-30. Resulting sequences of approximately 1,500 bp were compared to the NCBI GenBank database using the blastn tool [71,72].
Antibiotic susceptibility test
The disc diffusion method was used to screen for antibiotic susceptibility against a wide range of antibiotics. Commercial antibiotic discs of different classes were purchased from HiMedia, India. Probiotic lozenges dissolved in PBS were incubated overnight in MRS broth at 37°C and adjusted to 7 × 106 CFU/mL (OD600 = 0.6) prior to the disc diffusion and broth microdilution assays. In the diffusion test, one hundred microliters of bacteria culture were spread evenly onto MRS agar before carefully placing the antibiotic ring on the bacteria lawn. One antibiotic ring contained 12 antibiotic discs and a total of 4 different antibiotic rings: Dodeca G-I-Plus (DE002), Dodeca G-II-Plus (DE009), Dodeca G-III-Plus (DE018), and Dodeca G-IV-Plus (DE023), containing a total of 31 unique antibiotics with different mode of actions, were used. After 48 h of incubation at 37°C, the plates were photographed and the diameter of inhibition (clear) zones forming around the antibiotic discs were measured by ImageJ. Antibiotic susceptibility tests were conducted at least twice, and each antibiotic was tested at least four times on the same sample. Bacteria are determined to be resistant to an antibiotic if the inhibition zones had diameters less than 2 × the diameter of the antibiotic disc i.e. <12 mm, while they were determined to be partially resistant if the inhibition zones had diameters between 12 and 15 mm. The minimum inhibitory concentrations (MICs) were determined for the representative antibiotics: chloramphenicol, doxycycline, erythromycin, piperacillin, and streptomycin, representing different classes of antibiotics, on 96-well plates using the broth dilution method. The MICs were determined from the dose response curves and compared to the cut-off values for resistance as determined in the guidelines provided for Lactobacillus spp. by the European Food Safety Authority (EFSA) [43]. In this study, a conservative approach was adopted in assigning resistance to probiotics. For the disc diffusion assay, the quality control data in the manufacturer’s technical guide which followed the performance standards for antimicrobial disk susceptibility tests of the Clinical and Laboratory Standards Institute (CLSI) [73], suggested that inhibition zones with diameters less than three times the diameters of the antibiotic discs (<18 mm) were considered resistant for most antibiotics when tested on representative Gram positive (Staphylococcus aureus ATCC 259230) and Gram negative (Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853) bacteria but, we assigned resistance only if the inhibition zones were less than two times the diameter of the antibiotic discs (<12 mm). Likewise, for the broth microdilution assays, resistance was only assigned if the MIC values exceeded the thresholds of all Lactobacillus spp. listed by the EFSA [43].
Detection of antibiotic resistance genes by PCR
Genomic DNA from the probiotics of oral lozenges were extracted using the QIAprep Miniprep Kit (Qiagen, MD). The concentration and purity of extracted DNA was determined on a NanoDropOne spectrophotometer (ND-ONE-W, ThermoFischer, WI). Gene-specific primers for known antibiotic resistance genes, such as gentamicin, streptomycin, kanamycin, neomycin, tetracycline, erythromycin, clindamycin, chloramphenicol, ampicillin, vancomycin, quinupristin/dalfopristin, linezolid, trimethoprim, rifampicin, ciprofloxacin, and others, were purchased from Sangon Biotech, Shanghai, China. The respective annealing temperatures and amplicon sizes were determined from the literature (Table 2). The amplification program was as follows: initial denaturation step at 94°C for 5 min; 30 cycles of: 94°C for 45 s, annealing temperature for 45 s and 72°C for 45 s; and a final extension of 10 min at 72°C. The amplicons were analyzed on 1% (w/v) agarose gel to confirm the DNA fragment size.
Table 2.
Primer sequences and PCR conditions for the detection of antibiotic resistance genes (ARG)
| Antibiotic | ARG | Primers 5ʹ-3ʹ (Forward, top; Reverse, bottom) |
Annealing T. (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| Gentamicin | aac(6ʹ)-aph(2”) | CCAAGAGCAATAAGGGCATA | 60 | 220 | [128] |
| CACTATCATAACCACTACCG | |||||
| aac(6ʹ)Ieaph(2”)Ia | CAGAGCCTTGGGAAGATGAAG | 58 | 348 | [74] | |
| CCTCGTGTAATTCATGTTCTGGC | |||||
| Streptomycin | strA | CTTGGTGATAACGGCAATTC | 55 | 548 | [129] |
| CCAATCGCAGATAGAAGGC | |||||
| strB | ATCGTCAAGGGATTGAAACC | 56 | 509 | [129] | |
| GGATCGTAGAACATATTGGC | |||||
| aadA | ATCCTTCGGCGCGATTTTG | 56 | 282 | [129] | |
| GCAGCGCAATGACATTCTTG | |||||
| aadE | ATGGAATTATTCCCACCTGA | 50 | 565 | [129] | |
| TCAAAACCCCTATTAAAGCC | |||||
| ant(6) | ACTGGCTTAATCAATTTGGG | 53 | 597 | [124] | |
| GCCTTTCCGCCACCTCACCG | |||||
| Kanamycin | aph(3”)-III | GCCGATGTGGATTGCGAAAA | 52 | 292 | [129] |
| GCTTGATCCCCAGTAAGTCA | |||||
| ant(2”)-I | GGGCGCGTCATGGAGGAGTT | 67 | 329 | [129] | |
| TATCGCGACCTGAAAGCGGC | |||||
| Neomycin | aph(3”)-I | AACGTCTTGCTCGAGGCCGCG | 68 | 670 | [129] |
| GGCAAGATCCTGGTATCGGTCTGCG | |||||
| aph(3”)-III | GCCGATGTGGATTGCGAAAA | 52 | 292 | [129] | |
| GCTTGATCCCCAGTAAGTCA | |||||
| Tetracycline | tet(M) | GGTGAACATCATAGACACGC | 55 | 401 | [75] |
| CTTGTTCGAGTTCCAATGC | |||||
| tet(K) | TCGATAGGAACAGCAGTA | 55 | 169 | [135] | |
| CAGCAGATCCTACTCCTT | |||||
| tet(W) | GAGAGCCTGCTATATGCCAGC | 64 | 168 | [76] | |
| GGGCGTATCCACAATGTTAAC | |||||
| tet(L) | GTMGTTGCGCGCTATATTCC | 55 | 696 | [76] | |
| GTGAAMGRWAGCCCACCTAA | |||||
| tet(O) | AATGAAGATTCCGACAATTT | 55 | 781 | [135] | |
| CTCATGCGTTGTAGTATTCCA | |||||
| tet(S) | ATCAAGATATTAAGGAC | 55 | 573 | [135] | |
| TTCTCTATGTGGTAATC | |||||
| Erythromycin | erm(A) | AAGCGGTAAACCCCTCTGAG | 55 | 441 | [77] |
| TCAAAGCCTGTCGGAATTGG | |||||
| erm(B) | GAAAAGGTACTCAACCAAATA | 54 | 639 | [135] | |
| AGTAACGGTACTTAAATTGTTTAC | |||||
| erm(B)-1 | CATTTAACGACGAAACTGGC | 54 | 405 | [75] | |
| GGAACATCTGTGGTATGGCG | |||||
| erm(C) | TCAAAACATAATATAGATAAA | 50 | 642 | [135] | |
| GCTAATATTGTTTAAATCGTCAAT | |||||
| erm(T) | TATTATTGAGATTGGTTCAGGG | 55 | 395 | [78] | |
| GGATGAAAGTATTCTCTAGGGATTT | |||||
| mefA | AGTATCATTAATCACTAGTGC | 55 | 348 | [79] | |
| TTCTTCTGGTACTAAAAGTGG | |||||
| Clindamycin | lnu(A) | GGTGGCTGGGGGGTAGATGTATTAACTGG | 55 | 323 | [76] |
| GCTTCTTTTGAAATACATGGTATTTTTCGATC | |||||
| lnu(B) | CCTACCTATTGTTTGTGGAA | 54 | 925 | [80] | |
| ATAACGTTACTCTCCTATTTC | |||||
| Chloramphenicol | catA | GGATATGAAATTTATCCCTC | 50 | 486 | [80] |
| CAATCATCTACCCTATGAAT | |||||
| cat | TTAGGTTATTGGGATAAGTTA | 48 | 300 | [81] | |
| GCATGRTAACCATCACAWAC | |||||
| cat-TC | CAATAGCGACGGAGAGTTAGG | 55 | 384 | [82] | |
| AATCCTGCATGATAACCATCAC | |||||
| Ampicillin | blaZ | ACTTCAACACCTGCTGCTTTC | 58 | 240 | [135] |
| TAGGTTCAGATTGGCCCTTAG | |||||
| bla | CATARTTCCGATAATASMGCC | 51 | 297 | [81] | |
| CGTSTTTAACTAAGTATSGY | |||||
| mecA | GGGATCATAGCGTCATTATTC | 58 | 1429 | [135] | |
| AGTTCTGCAGTACCGGATTTGC | |||||
| Vancomycin | vanA | ATGAATAGAATAAAAGTTGCAATAC | 62 | 1028 | [83] |
| CCCCTTTAACGCTAATACGAT | |||||
| vanB | CCCGAATTTCAAATGATTGAAAA | 59 | 457 | [83] | |
| CGCCATCCTCCTGCAAAA | |||||
| vanC | GCTGAAATATGAAGTAATGACCA | 58 | 832 | [83] | |
| CGGCATGGTGTTGATTTCGTT | |||||
| vanE | TGTGGTATCGGAGCTGCAG | 52 | 513 | [76] | |
| GTCGATTCTCGCTAATCC | |||||
| vanX | TCGCGGTAGTCCCACCATTCGTT | 55 | 454 | [80] | |
| AAATCATCGTTGACCTGCGTTAT | |||||
| Quinupristin/ Dalfopristin |
vatC | GAAATGGTTGGGAGAAGCATACC CAGCAATCGCGCCCGTTTG |
64 | 392 | [76] |
| vatE | CTATACCTGACGCAAATGC | 52 | 490 | [76] | |
| GGTTCAAATCTTGGTCCG | |||||
| Linezolid | cfr | TGAAGTATAAAGCAGGTTGGGAGTCA | 55 | 746 | [84] |
| ACCATATAATTGACCACAAGCAGC | |||||
| Trimethoprim | dfrA | CTTTTCTACGCACTAAATGTAAG | 50 | 474 | [80] |
| CATTATCAATAATTGTCGCTCAC | |||||
| dfrD | GGAAGGGCTTTACCTGACAGAAG | 50 | 175 | [80] | |
| CGACATAAGGCAAGAACATAACATA | |||||
| Rifampicin | rpoB | TAACCGTGGTGCTTGGCTDGAATWYGAAAC | 59 | 1100 | [85] |
| ATCAAACCAATGTTAGGNCCTTCWGGDGTTTC | |||||
| Ciprofloxacin | gyrA | GAYTATGCWATGTCAGTTATTGT | 45 | 286 | [129] |
| GGAATRTTRGAYGTCATACCAAC | |||||
| parC | TATTCYAAATAYATCATTCARGA | 50 | 286 | [81] | |
| GCYTCNGTATAACGCATMGCCG | |||||
| int | GCGTGATTGTATCTCACT | 50 | 1028 | [86] | |
| GACGCTCCTGTTGCTTCT | |||||
| Macrolide antibiotics | msrA/B | GCAAATGGTGTAGGTAAGACAACT | 52 | 399 | [87] |
| ATCATGTGATGTAAACAAAAT | |||||
| msrA | GGCACAATAAGAGTGTTTAAAGG | 40 | 939 | [88] | |
| AAGTTATATCATGAATAGATTGTCCTGTT | |||||
| msrC | AAGGAATCCTTCTCTCTCCG | 55 | 343 | [89] | |
| GTAAACAAAATCGTTCCCG | |||||
| Tn554 | AAGCGGTAAACCCCTCTGAG | 55 | 440 | [77] | |
| TCAAAGCCTGTCGGAATTGG |
Conjugative transfer of resistance genes from probiotics to bacteria implicated in diseases
To examine the transmissibility of resistance genes from probiotics of oral lozenges to bacteria commonly associated with diseases during co-incubation in vitro, the liquid culture and filter mating technique were employed. Representative probiotics from one oral lozenge which was determined to be resistant to streptomycin, were mixed with the recipients i.e. S. mutans, S. gordonii, S. sanguinis, or E. faecalis that were susceptible to streptomycin at a probiotic-to-recipient ratio of 1:1 and 10:1, respectively. The mixed cultures were incubated for 24 h at 37°C and then spread on MRS agar which is selective for lactobacilli with and without 100 µg/mL of streptomycin, and on Mannitol Salt Agar (MSA) which is selective for Streptococcus strains and E. faecalis with and without 100 µg/mL of streptomycin, respectively. As controls, monocultures of probiotics and recipients were grown and spread on plates alongside the co-cultures.
Another 2 mL of bacteria mixtures were filtered through a sterile nitrocellulose MCE membrane filter MF-Millipore (2.5 cm diameter, 0.45 µm pore size, Merck Millipore, Ireland) using a Millipore pump with a negative pressure of −50 kPa. The membrane filters were then carefully placed onto BHI agar, in which both donor and recipient are culturable. As controls, 2 mL of pure probiotics and recipient cultures were also passed through the membrane filters and grown on BHI agar plates respectively. After 72 h of incubation at 37°C, the membrane filters were placed in 1 mL PBS in a sterile microcentrifuge tube and vortexed to free the bacteria. Another 1 mL PBS was used to wash the plates and the washings were placed in the sterile tube. Serial dilutions were made and 50 µL of each diluted sample were spread onto selective agar plates.
The liquid culture and filter-mating experiments were repeated at least three times in duplicates. The colonies on the selective agar were observed after 2 days of culture. If there was a transfer of resistance gene from the streptomycin resistant probiotic to the recipient, the colonies of the recipient bacteria would be detected on the streptomycin containing MSA agar.
Results and discussion
Recovery and enumeration of probiotics from oral lozenges
Probiotic bacteria, predominantly Lactobacillus spp. from eight commercial probiotic lozenges labelled A, B, C, D, E, F, G, and H (Table 1), were recovered on MRS agar, and enumerated using the drop plate method. Since MRS is selective for Lactobacillus, only contributions from these strains of bacteria were taken into consideration in our analysis. Except for product B, all probiotic lozenges contained live bacteria. Product B contained heat-killed probiotic bacteria, and thus were unable to be recovered in the laboratory. Although previous studies have shown that heat-inactivated probiotics could still inhibit oral bacteria such as S. mutans, P. gingivalis, Fusobacterium nucleatum, and Aggregatibacter actinomycetemcomitans, it was suggested that long-term protection against these presumed pathogens would require active probiotics [90]. Based on the boxplot in Figure 1, all products except product A contained live bacteria that were about 0.2–10% fewer than that claimed by the manufacturers. Product A contained live bacteria that was comparable to the claims of the manufacturer. It must be noted that the oral lozenges dissolved in PBS were immediately spread onto MRS agar to avoid exposure to conditions that might affect bacteria viability in vitro.
Figure 1.
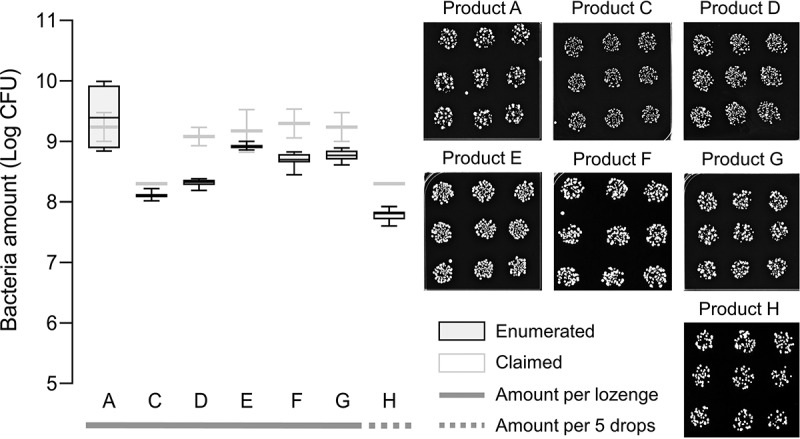
Enumeration of probiotic bacteria from oral lozenges.
Left panel shows a box and whisker plot of enumerated bacteria amounts for probiotics recovered on agar plates (dark boxes) from commercially available oral lozenges labelled A, B, C, D, E, F, G, and H, respectively, compared to the amounts claimed by the manufacturers (light boxes). Product B contained heat-killed probiotic bacteria and were thus not viable. Oral lozenges were dissolved in PBS and immediately dropped onto MRS agar plates and incubated for 24–48 h at 37°C. Agar plates were then photographed for processing on ImageJ. Representative images of recovered probiotic bacteria at the appropriate dilutions that gave rise to single bacteria colonies, that are shown on the right panel. All experiments were done in biological duplicates where each contained nine drops of the respective PBS-dissolved oral lozenge. Note: Only Lactobacillus strains were recovered and considered in our enumeration.
Underestimation of bacteria amounts in probiotic food products is not uncommon as previous studies have also reported discrepancies in bacteria amounts for yogurt and fermented milk [91–94], and importantly also for intestinal probiotic supplements [44,45] and other commercially available oral lozenges [95]. However, the enumerated amounts are above the threshold of 106 colony-forming units (CFU) per serving regarded to be sufficient to confer health benefits [96], or the recommended daily consumption of 109–1010 CFU [97]. Other studies have also reported poor tolerance of probiotic bacteria to stomach pH and bile salts [98–102]. While encapsulation technologies and other additives have to some extent improved the viability of probiotics transiting through the gastrointestinal tract [103–106], their stability under oral conditions such as antimicrobial proteins in the saliva and the inhibitory effects exerted by the native oral microbiota, remain uncertain [107–109]. Since probiotic lozenges are designed to dissolve gradually in the mouth, it is therefore critical that probiotic bacteria tolerate those conditions long enough for them to adhere on the surfaces of, and colonize, the oral cavity [39,40].
Screening for antibiotic resistance in probiotics from oral lozenges
The antibiotic susceptibility of Lactobacillus probiotic strains from oral lozenges A, C, D, E, F, and G, was analyzed by the disc diffusion method. Resistance to more than 30 antibiotics representing different modes of actions i.e. acting on the cell wall, ribosomal subunits 30S and 50S, or DNA, were examined. The antibiotic disc had a diameter of 6 mm. Probiotics were determined to be resistant to an antibiotic if the inhibition zones had diameters less than 2 × the diameter of the antibiotic disc i.e. <12 mm, while they were determined to be partially resistant if the inhibition zones had diameters between 12 and 15 mm. Antibiotic resistance profiles of probiotics of all products are summarized as a heatmap in Figure 2, and the corresponding bar graphs of their antibiograms are shown in Supplemental Figure S1.
Figure 2.
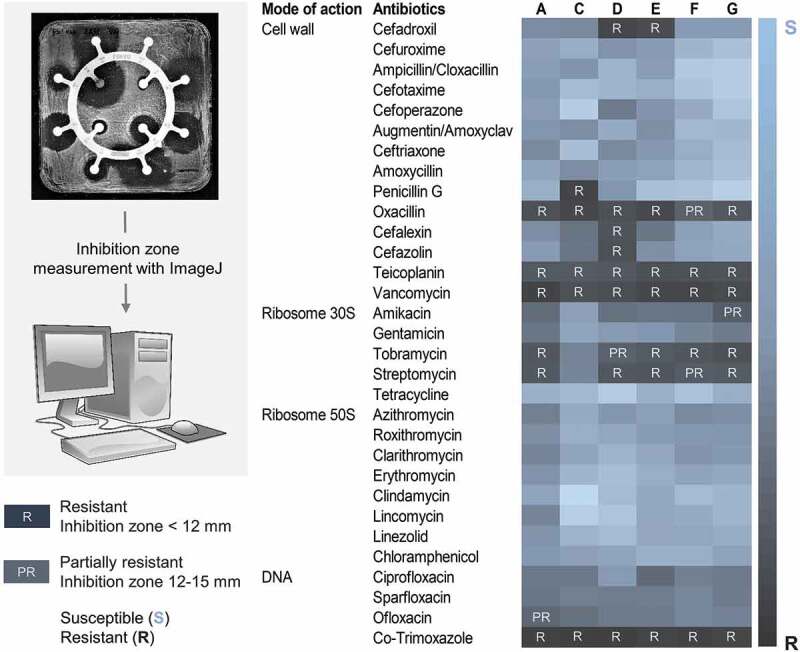
Antibiotic resistance profiles of probiotics from oral lozenges.
I noticed that all figure legends have been omitted from the figures here and in the PDF version of the proof. Is this deliberate? If not, please make sure that they are included for every figure which I belief would be more informative and increase clarity for the readers. I have resend the manuscript word document again by email.
Left panel illustrates the workflow of the antibiotic susceptibility test by disc diffusion. PBS-dissolved oral lozenges were spread onto MRS agar in square petri dishes and a ring of antibiotic discs was carefully placed onto the bacteria lawn. Agar plates were incubated for 24–48 h at 37°C and then photographed for processing on ImageJ. The right panel shows the antibiotics used in this study and their modes of action, as well as a heatmap of the resistant profiles of Lactobacillus probiotic bacteria from oral lozenges labelled A, C, D, E, F, and G, respectively. The diameter of the antibiotic disc was 6 mm. Probiotics were determined to be resistant to an antibiotic if the inhibition zones had diameters less than 2 × the diameter of the antibiotic disc i.e. <12 mm (R), while they were determined to be partially resistant if the inhibition zones had diameters between 12 and 15 mm (PR). Bar graphs of all antibiograms were shown in Supplemental Figure 1. All experiments were done in biological duplicates. Note: Antibiotic resistance profiles belong only to Lactobacillus strains of the respective oral lozenge.
Probiotics from all oral lozenges were resistant to vancomycin, teicoplanin, and co-trimoxazole (Figure 2). Resistance to vancomycin and teicoplanin was expected as Lactobacillus harbors chromosomally encoded D-Ala-D-lactate in the peptidoglycan instead of the D-Ala-D-Ala dipeptide, which prevents the binding of these antibiotics [110]. Since it is an intrinsic property of Lactobacillus which is not transferable, it is therefore not a clinical concern even though vancomycin is used intravenously and orally to treat various bacterial infections, including Clostridium difficile and methicillin-resistant S. aureus (MRSA) [111]. However, previous studies in mice colonized with human microbiota have demonstrated the conjugative transfer of vanA gene clusters among E. faecium strains, and importantly also, from Enterococcus faecium strains to L. acidophilus [112,113]. This has raised clinical concerns especially on the treatment of Lactobacillus bacteremia in patients with existing conditions such as ulcerative colitis [114–116]. More recently, vanA plasmids transmission between different Enterococcus spp. was reported to be prevalent in hospital settings [117].
Co-trimoxazole, which is a combination of sulfamethoxazole and trimethoprim, is commonly used to treat a broad range of bacterial infections including those caused by MRSA [118]. Since it acts on the biosynthetic pathway of the vitamin folic acid that is lacking in most lactobacilli, resistance to co-trimoxazole is considered intrinsic [119]. However, it was speculated that the high exposure to co-trimoxazole especially in developing countries [120] could lead to a high resistance of lactobacilli especially given the fact that mobile resistance determinants of trimethoprim have been identified and are becoming increasingly prevalent [121–123].
Except for product C, resistance to streptomycin and tobramycin was detected in probiotics from all lozenges (Figure 2). Consistently, our broth microdilution assays also showed MIC for streptomycin of 128 µg/mL or higher for probiotics from all lozenges except for product C, which had a MIC of 8 µg/mL (Figure 3). Aminoglycoside antibiotics bind irreversibly to the bacterial 30S ribosomal subunit and interfere with the initiation complex of mRNA and the 30S subunit during protein synthesis. High resistance to streptomycin has also been reported in Lactobacillus from human samples and commercial probiotic foods including starter cultures, curd, yoghurt, and dairy products [56,124–127]. Since lactobacilli lack the cytochrome-mediated electron transport that enables the uptake of aminoglycoside drugs, their resistance is thought to be intrinsic [46,128]. However, genes encoding for aminoglycoside-modifying enzymes which are localized on transposons or plasmids, such as acetyltransferases, nucleotidyltransferases and phosphotransferases, have been identified in lactobacilli [128–133]. Moreover, there is high frequency of mutation affording high resistance to streptomycin and kanamycin in lactobacilli which has compounded the impact of resistance gene acquisition along the food chain as aminoglycoside resistance genes such as nucleotidyltransferase ant(6) from lactobacilli are highly identical to those from pathogens and commensals in animals [46,48,128].
Figure 3.
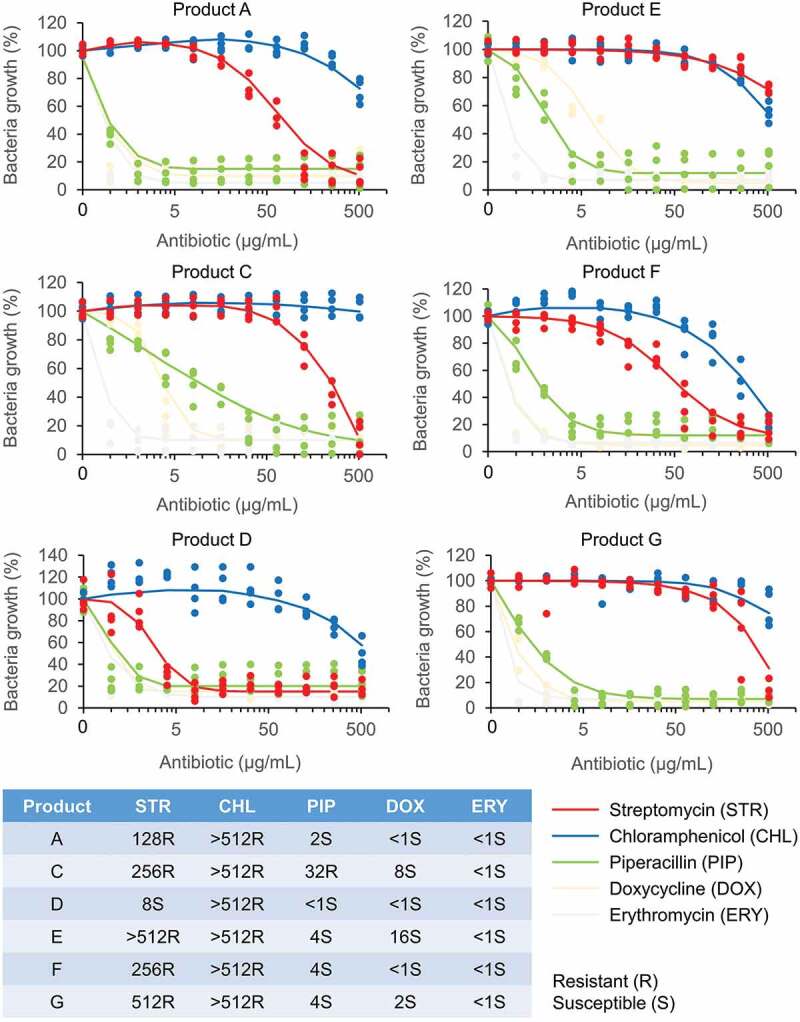
Dose–response curves and minimum inhibitory concentration (MIC) values of probiotics of oral lozenges.
Lactobacillus probiotic strains from oral lozenges labelled A, C, D, E, F, and G, were respectively grown on MRS broth containing a gradient of antibiotics (0–512 µg/mL) in quadruplicates on 96-well plates. The plates were incubated for 24 h at 37°C with shaking, and the absorbance at OD600 was measured. Bacteria growth of wells containing antibiotics were expressed as percentage (%) of the no-antibiotic wells. Representative antibiotics of different classes: penicillins (piperacillin), aminoglycosides (streptomycin), macrolides (erythromycin), tetracyclines (doxycycline) and chloramphenicol (chloramphenicol), were used in this study. The MIC values of the respective antibiotics were determined from the dose-response curves and summarized in the table at the bottom panel. Resistance to the respective antibiotics was determined based on the microbiological breakpoints for Lactobacillus strains as determined by the European Food Safety Authority (EFSA) [43]. According to EFSA, the highest cut-off values for Lactobacillus strains are 64 µg/mL for streptomycin, 8 µg/mL for chloramphenicol, 4 µg/mL for ampicillin (used as reference for piperacillin), 32 µg/mL for tetracycline (used as reference for doxycycline), and 1 µg/mL for erythromycin. Note: Antibiotic resistance profiles belong only to Lactobacillus strains of the respective oral lozenge.
Probiotics of all lozenges were resistant to oxacillin which is a beta-lactam antibiotic that binds to penicillin-binding proteins at the cell wall but only probiotics of lozenge C showed resistance to penicillin G (Figure 2), which belongs to the same antibiotic class as oxacillin. Although usually sensitive to penicillin, Lactobacillus resistant to oxacillin has been reported in human milk, curd, and raw milk cheeses [126,134]. They could be conferred by the plasmid-encoded resistance genes blaZ and mecA found in lactobacilli from raw and processed pork and chicken meat products, as well as staphylococci from humans and animals [135–138]. Beta-lactamases encoded by blaZ or the alternative penicillin binding protein, PBP2a encoded by mecA, could also confer protection against other beta-lactam antibiotics such as cephalosporins: cefadroxil, cefalexin, and cefazolin, which were ineffective against probiotics of oral lozenge D and E (Figure 2). Lactobacillus resistance to cephalosporins have been reported in curd, fermented table olives, commercial dairy products and fermented plant materials, turkeys, and human milk samples [126,139–141]. The resistance could be conferred by plasmid-born genes such as the extended-spectrum β-lactamases blaTEM and blaSHV, which have been shown through whole-genome sequencing, to be disseminated in E. coli from farm animals and humans [142]. Intermediate resistance to ofloxacin and amikacin was also detected in probiotics from products A and G, respectively (Figure 2). In the case of ofloxacin, mutations in the GyrA or ParC genes have been associated with quinolone resistance in Lactobacillus [143].
In agreement with the disc diffusion experiments, the broth microdilution assays showed that probiotics from all lozenges were susceptible to erythromycin with MICs <1 µg/mL (Figure 3), although erythromycin resistance is commonly reported in lactobacilli from starter cultures, dairy, plant and poultry products, fermented foods, swine meat, and human samples. Their corresponding resistance genes such as erm(B), erm(C), mefA, and lnuA, have also been characterized [125,129,130,135,139,140,144]. Doxycycline which has a mode of action similar to tetracycline, was effective against probiotics from all products but has higher MICs for resistant lactobacilli also commonly reported in probiotics from products C, E, and G, respectively (Figure 3). Consistently, no resistance to tetracycline was detected in the disc diffusion experiments (Figure 2). Tetracycline starter cultures, dairy, fermented sausages, plant and poultry products, swine meat, and human samples and their corresponding resistance genes tet(W), tet(L), tet(K), tet(S), and tet(M) have also been characterized [56,124,125,129,130,135,140,144,145]. Piperacillin, which has a mode of action similar to penicillin, was effective against probiotics of all products except for oral lozenge C which had a MIC of 32 µg/mL (Figure 3). Consistently, in the disc diffusion experiments, probiotics of product C also showed resistance to penicillin G (Figure 2). In contrast to the disc diffusion experiments where all probiotics were susceptible to chloramphenicol (Figure 2), the broth microdilution studies showed high MICs (>512 µg/mL) for probiotics of all products (Figure 3). The chloramphenicol acetyltransferase cat-TC gene was previously reported in Lactobacillus from dairy products such as raw milk, cream, yogurt, cheese, and human samples such as mouth, feces, and vagina [144]. Since the cat genes are plasmid-borne [146], they could be transmitted to other lactobacilli more effectively in broth cultures than on agar plates.
Detection of antibiotic resistance genes by PCR
We attempted to characterize the antibiotic resistance genes by PCR using gene-specific primers (Table 2). The trimethoprim resistance gene dfrD which is a plasmid-encoded dihydrofolate reductase, was detected in probiotics from lozenges A, C, D, and E (Table 3). It could account for their resistant phenotypes and pose a risk of intra- and inter-species acquisitions (Figure 2) [147].
Table 3.
PCR detection of antibiotic resistance genes in probiotic lozenges
| Product | Lactobacillus strain | Antibiotic resistance genes |
|---|---|---|
| A |
Lactobacillus paracasei Lactobacillus reuteri Lactobacillus sakei Lactobacillus salivarius |
erm(T), rpoB, gyr(A), parC, dfrD |
| C |
Lactobacillus reuteri DSM 17938 Lactobacillus reuteri ATCC PTA 5289 |
erm(T), rpoB, gyr(A), parC, dfrD |
| D |
Lactobacillus acidophilus HA-122 Lactobacillus casei HA-108 Lactobacillus rhamnosus HA-111 Lactobacillus salivarius HA-188 |
aph(3”)-III, erm(B), mefA, cat-TC, rpoB, gyr(A), parC, dfrD |
| E |
Lactobacillus brevis CECT 7480 Lactobacillus plantarum CECT 7481 |
gyr(A), dfrD |
| F |
Lactobacillus acidophilus Lactobacillus reuteri Lactobacillus paracasei Lactobacillus salivarius |
ant(2”)-I, erm(B), vanX, vatE |
| G |
Lactobacillus paracasei Lactobacillus reuteri |
aadE, aph(3”)-III, cat-TC, vanX, rpoB, gyr(A) |
Note: Detected resistance genes were reflective of Lactobacillus in the oral lozenge and not specific to individual strains.
Although the erythromycin resistant phenotype was not observed in the disc-diffusion and broth microdilution studies, the plasmid-encoded erythromycin resistance gene erm(T) was detected in probiotics of lozenge A and C while erm(B) was detected in probiotics from products D and F, respectively. Additionally, the macrolide resistance gene mefA that encodes for efflux channels, was also detected in probiotics from product D (Table 3). The plasmid-encoded chloramphenicol acetyltransferase cat-TC gene was detected in probiotics from lozenge D and G, which could account for the resistant phenotype observed in the broth microdilution studies (Table 3) (Figure 3).
The aminoglycoside resistance gene aadE detected in probiotics from oral lozenge G could account for the resistant phenotype. The aminoglycoside 3ʹ-phosphotransferase aph(3”)-III and aminoglycoside-2″-O-nucleotidyltransferase ant(2”)-I genes which are known to confer resistance to kanamycin, were detected in probiotics of the respective products D, G, and F (Table 3). They could confer cross-protection to streptomycin which has a similar mode of action.
The DNA topoisomerase and DNA gyrase genes parC and/or gyr(A) were detected in probiotics of oral lozenge A, C, D, and E, but there were no resistant phenotypes to fluoroquinolones: ciprofloxacin and ofloxacin, except for probiotics of oral lozenge A which were only partially resistant to ofloxacin (Table 3) (Figure 2). On the other hand, while resistance to oxacillin was detected in probiotics of all lozenges, and also to penicillin G in the case of product C (Figure 2), the corresponding transmissible genes blaZ and mecA were not detected. The RNA polymerase B subunit rpoB gene that confers resistance to rifampicin, was detected in probiotics from products A, C, D and G. Oral lozenges F and G harbored the vancomycin resistance gene vanX while only product F harbored the quinupristin/dalfopristin resistance gene vatE (Table 3).
The resistance determinants detected in our studies were mostly consistent with those of previous studies on lactobacilli from various foods, animals, and human sources [48,56,124,125,130,135,140,141,148]. One notable difference was the promiscuity of tetracycline resistance genes tet(W), tet(M), tet(S), tet(O), tet(Q), tet(36), tet(Z), tet(W/O), tet(O/W/32/O/W/O), tet(K), and tet(L) in lactobacilli from poultry and meat products, starter cultures, fermented foods, and the human intestine [125,135,149–151], which were not detected in this study. Notably, previous studies conducted on lactobacilli from intestinal probiotic supplements reported resistance to a broad range of antibiotics including teicoplanin, vancomycin, amikacin, tobramycin, streptomycin, cephalexin, all of which, were also reported in this study. Also consistent with our data, no resistance to erythromycin, clindamycin, tetracycline, and ampicillin was reported from the previous studies [42,45].
Single strain analysis of antibiotic resistance
To examine the contribution of individual strains to the resistance profile of the probiotic lozenges, we selected as representatives, single bacteria colonies from products C and D, extracted their genomic DNA, and resolved their identities by 16S rRNA sequencing. All the four strains L. acidophilus, L. casei, L. salivarius and L. rhamnosus listed on the label of product D, were represented in our 16S rRNA sequencing analysis showing >98% identities to known sequences deposited in the NCBI GenBank [71]. From the disc diffusion assay, apart from the expected intrinsic resistance of Lactobacillus, we observed that none of the single isolates from product D was resistant to cephalosporin antibiotics such as cefadroxil, cefalexin, and cefazolin although resistance to these drugs was detected in the heterogenous populations of bacteria of product D (Figure 4). This may be due to the absence of plasmids carrying cephalosporin resistance genes in the pure isolates tested. Oxacillin resistance was also not found in the single strains, and this could be attributed to the absence of the mecA gene known to confer resistance to oxacillin although it was detected in the heterogenous populations of product D. It is conceivable that plasmids carrying the resistance determinants are present only in a fraction of bacterial cells in product D, thus affording them resistance to the same antibiotics in the mixed populations. All other resistance genes detected in product D, were present in the single isolates. As expected, the cat-TC gene conferring resistance to chloramphenicol was present in all single isolates of product D while the rpoB gene responsible for rifampicin resistance was detected in three out of four isolates. The GyrA or ParC genes associated with quinolone resistance in Lactobacillus, were detected in L. casei, L. salivarius and L. rhamnosus, respectively, while the erythromycin resistance gene erm(B), was detected only in L. casei. Although only L. rhamnosus was phenotypically partially resistant to streptomycin, the streptomycin resistance gene aph(3”)-III was only detected in L. acidophilus. On the contrary, amikacin resistance which was not detected in the mixed populations of product D, was found to be present in three of the four single isolates of product D. L. reuteri of product C was resistant to co-trimoxazole, teicoplanin, vancomycin, penicillin, and oxacillin, which is consistent with the antibiogram of product C, although it also showed additional resistance to cefazolin. In the broth microdilution assay, the MICs for all tested antibiotics except for streptomycin, were generally lower for the single strains than for the mixed populations of product D. Similarly, L. reuteri from product C also gave lower MICs than that of product C although it was the only Lactobacillus strain present in this product. Except for erm(T), all the resistance genes parC, rpoB, gyr(A), and dfrD, detected in product C, were present in the L. reuteri isolate (Figure 4). While single strain resistance profiles enable comparisons with threshold values determined by the European Food Safety Authority (EFSA) [43] antibiograms of the oral lozenges as a whole, is more reflective of the actual diet where heterogenous populations of probiotics are normally consumed. Furthermore, a conservative approach was adopted in this study where resistance was only assumed if the MIC values exceeded the thresholds of all Lactobacillus spp. listed by the EFSA.
Figure 4.
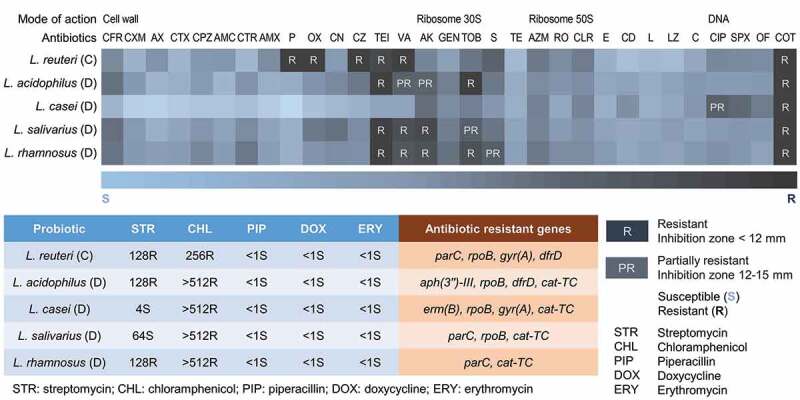
Antibiograms of single strains isolated from probiotic lozenges.
Top panel shows the antibiotics used in this study and their mode of actions, as well as a heatmap of the resistance profiles of Lactobacillus from products C and D. The diameter of the antibiotic disc was 6 mm. Probiotics were determined to be resistant to an antibiotic if the inhibition zones had diameters less than 2 × the diameter of the antibiotic disc i.e. <12 mm (R), while they were determined to be partially resistant if the inhibition zones had diameters between 12 and 15 mm (PR). All experiments were done in biological duplicates. The bottom panel shows the MIC values of the corresponding isolates. Lactobacillus strains were grown on MRS broth containing a gradient of antibiotics (0–512 µg/mL) in quadroduplicates on 96-well plates. The plates were incubated for 24 h at 37°C with shaking, and the absorbance at OD600 was measured. Bacteria growth of wells containing antibiotics were expressed as percentage (%) of the no-antibiotic wells. Representative antibiotics of different classes: penicillins (piperacillin), aminoglycosides (streptomycin), macrolides (erythromycin), tetracyclines (doxycycline) and chloramphenicol (chloramphenicol) were used. Resistance to the respective antibiotics was determined based on the microbiological breakpoints for Lactobacillus strains as determined by the European Food Safety Authority (EFSA) [43]. According to EFSA, the highest cut-off values for Lactobacillus strains are 64 µg/mL for streptomycin, 8 µg/mL for chloramphenicol, 4 µg/mL for ampicillin (used as reference for piperacillin), 32 µg/mL for tetracycline (used as reference for doxycycline), and 1 µg/mL for erythromycin. Abbreviation of antibiotics: AZM: Azithromycin; AK: Amikacin; GEN: Gentamicin; CIP: Ciprofloxacin; CFR: Cefadroxil; CXM: Cefuroxime; RO: Roxithromycin; AX: Ampicillin/Cloxacillin; CTX: Cefotaxime; CPZ: Cefoperazone; CLR: Clarithromycin; SPX: Sparfloxacin; AMC: Augmentin/Amoxyclav; CTR: Ceftriaxone; AMX: Amoxycillin; E: Erythromycin; CD: Clindamycin; OF: Ofloxacin; L: Lincomycin; TOB: Tobramycin; P: Penicillin G; OX: Oxacillin; LZ: Linezolid; COT: Co-Trimoxazole; VA: Vancomycin; TE: Tetracycline; C: Chloramphenicol; CN: Cefalexin; CZ: Cefazolin; S: Streptomycin; TEI: Teicoplanin.
Despite some variations, the single isolates generally exhibited lower resistance to antibiotics compared to the mixed populations in the oral lozenges as determined through our disc diffusion, broth microdilution and molecular characterization studies. Our data implied synergistic effect or cooperativity operating in the heterogenous populations of probiotic lozenges including mechanisms such as horizontal gene transfer that is strengthened through surface adherence and biofilm formations and extracellular DNA, thus affording resistance to a broader range of antibiotics [60,152]. More recently, it has been shown that even without antibiotic pressure, horizontal gene transfer helps establish low frequency of resistance genes to potentiate adaptation of bacteria to future environmental changes such as when antibiotics are present [153]. As such, a comparative metagenomics analysis conducted in conditions as close as possible to the oral cavity, would be required to observe the change in the gene pool with and without antibiotics. Since it is conceivable that plasmids carrying the resistance determinants are present only in a fraction of bacterial cells, the long-term consumption of heterogenous populations of probiotics in the form of health supplements such as oral lozenges, could exacerbate the spread of antimicrobial resistance in the oral cavity.
Conjugative transfer of resistance genes from probiotics to bacteria implicated in diseases
The capacity of conjugal transfer of streptomycin resistance genes from probiotics represented by product G to S. mutans, S. sanguinis, S. gordonii, and E. faecalis, were examined. An illustration of the liquid culture co-incubation and filter mating conjugative transfer workflow and representative images of transconjugant selections, are shown in Figure 5. Since aadE and aph(3”)-III genes that confer resistance to streptomycin were detected in probiotics of product G, they were selected as the donor in both liquid co-culture incubation and filter mating conjugative transfer experiments. Previously, co-transfer of plasmid-encoded aminoglycoside and macrolide resistance genes erm(B)-Tn5405-like element and aac(6ʹ)-Ie-aph(2”)-Ia was detected in vitro and in the gut of mice [154]. Notably, the transfer of the erythromycin resistance plasmid pAM81 between S. gordonii and E. faecalis has been observed ex vivo using prepared root canals of sterilized teeth [155]. However, we detected no transconjugants on the streptomycin agar plates (Figure 5). Thus, our results indicated that the antibiotic resistance genes were not transferred between the donor and the recipient strains in the current experimental settings.
Figure 5.
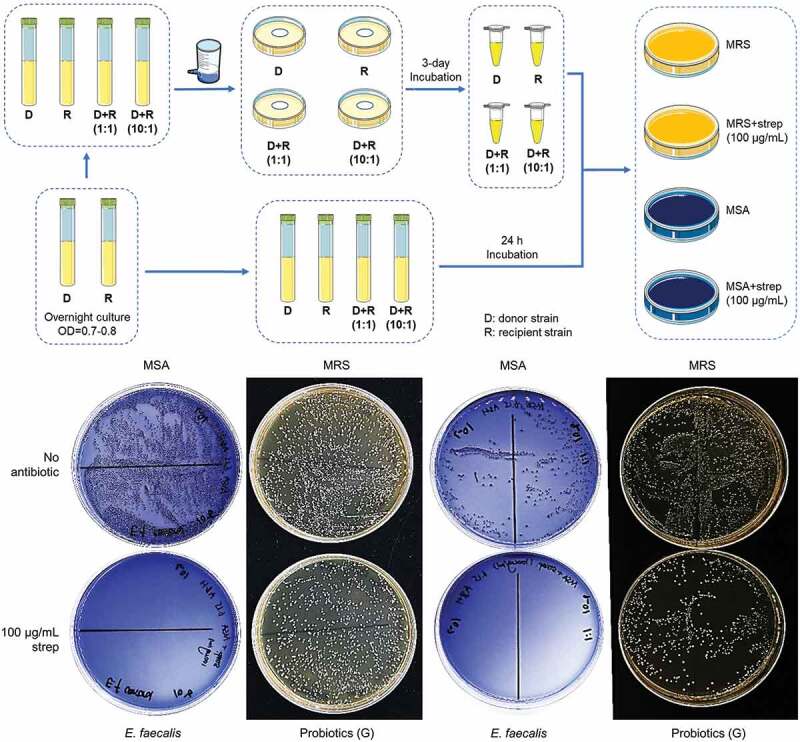
Transmission of resistance genes by conjugative transfer.
An illustration of the liquid culture co-incubation and filter mating conjugative transfer workflow is shown in the top panel. Probiotics donor from oral lozenges which are resistant to one antibiotic, and recipient which is susceptible to the same antibiotic, were incubated at 1:1 or 10:1 probiotics-to-recipient ratio in BHI broth for 24 h at 37°C with shaking. The liquid co-cultures were then spread on MSA and MRS agar plates with and without antibiotics, to detect conjugative transfer of resistance determinants from probiotics to the recipient. Lateral transfer of resistance genes was also examined by filter mating where probiotics resistant to one antibiotic and recipient susceptible to the same antibiotic, were localized on a 0.45 µm filter at 1:1 and 10:1 probiotics-to-pathogen ratio. The filter was placed on BHI agar and incubated for 24 h at 37°C. Bacteria were released from the filter in PBS and immediately spread onto MSA and MRS agar plates with and without antibiotics. The presence of recipient bacteria colonies on MSA agar plates containing antibiotics, would indicate transfer of resistance genes from probiotics. Representative pictures of agar plates from the filter mating experiment are shown in the bottom panel. Strep represents streptomycin; D, donor probiotic from product G; R, recipient S. mutans, S. gordonii, S. sanguinis, or E. faecalis. Note: MRS is selective for Lactobacillus strains which were unable to grow on MSA agar.
It was previously shown that the erythromycin resistance plasmid pLFE1 in L. plantarum isolated from raw milk cheese could be transferred to another Lactobacillus and to the pathogens Listeria innocua, Listeria monocytogenes, and E. faecalis, through filter-mating experiments [156]. Likewise, the tetracycline resistance gene tet(M) located on the Tn916 transposon in L. paracasei could also be transferred to E. faecalis [150]. Moreover, conjugal transfer of erythromycin and tetracycline resistance genes from Lactobacillus to pathogens in the animal gut, in vitro, and during food fermentation, were also detected [149]. Yet, there are also studies that detected no conjugal transfer of resistance genes from lactobacilli isolated from fermented milk and sausages to E. faecalis and S. aureus [145,147]. L. fermentum strains isolated from human feces and commercial dairy products were also unable to transfer their tetracycline and erythromycin resistance genes to pathogens such as Staphylococcus and Listeria strains by filter mating [140], while rifampicin and fusidic acid resistant lactobacilli from human origins also failed to transfer their resistance determinants to other lactobacilli, and to the pathogens E. faecium and E. faecalis, respectively [125]. Moreover, the transfer of plasmid encoded pediocin PA-1 like bacteriocin from L. plantarum to E. faecalis examined in vitro by filter mating as well as in situ using a soymilk model, were also not detected [157].
As conditions in vitro are not representative of the complexity and dynamics of the oral cavity and gut, actual rates of resistance gene transfer were thought to be underestimated [158]. Factors that may affect resistance gene transmission include the presence of commensal and pathogenic bacteria populations in healthy, antibiotic treated, and different genetic backgrounds of subjects, chemical and physical parameters such as bile salts, temperature, oxygen levels, and the requirement of biofilm formation [158–163]. As such, resistance gene trafficking must be examined in vivo taking into consideration various health, genetics, and nutritional factors. Notably, a recent metagenomics profiling of mice and human gastrointestinal tracts reported that probiotics worsen the resistome expansion caused by a prior course of antibiotics, as evidenced by an elevated number of strains carrying antibiotic resistance genes, thus directly linking probiotics with the alteration of antibiotic resistance gene reservoir in the human gut [164].
Comparative analysis of probiotic lozenges antibiograms with probiotic drinks
We also assessed the antibiotic resistance profiles of commonly available probiotic drinks such as probiotic milk, yogurt, and juice. Like many probiotic drinks, the probiotic drinks examined in this study contained only one type of Lactobacillus. From our enumeration studies of the recovered probiotic strains, the probiotic drinks contained approximately two to three orders of magnitude fewer bacteria compared to the oral lozenges per weight. Their antibiograms showed mostly intrinsic resistance such as resistance to vancomycin, teicoplanin and co-trimoxazole. However, resistance to tobramycin, streptomycin, and ciprofloxacin, were detected in one or more probiotic drinks. They also generally had lower MICs than the probiotic lozenges (Supplemental Figure S2). The high amounts of heterogenous populations of probiotic bacteria in supplements such as oral lozenges have been previously thought to encourage the spread of antimicrobial resistance [45,54], thus their long-term consumption may pose a higher risk to human health.
Conclusion
In conclusion, we report that probiotics from oral lozenges are resistant to multiple antibiotics belonging to glycopeptides, aminoglycosides, penicillins, and/or cephalosporins. The resistant probiotics display high MICs for streptomycin (>128 µg/mL), chloramphenicol (>512 µg/mL), and piperacillin (32 µg/mL), but are susceptible to doxycycline and erythromycin. Our PCR analysis detected genes conferring resistance to erythromycin, chloramphenicol, fluoroquinolone, aminoglycosides, vancomycin, rifampicin, and quinupristin/dalfopristin, in the probiotic lozenges. Additionally, our analysis of single strains isolated from probiotic lozenges and of probiotic drinks showed generally lower resistance to antibiotics compared to the mixed populations in the oral lozenges. Although we detected no conjugal transfer of antibiotic resistance genes in vitro, the presence of plasmid-encoded resistance genes in probiotics of oral lozenges, highlighted the potential of probiotics to acquire resistance genes during food processing or along the food chain from farm to fork [51,57–60,64,66,165–168]. This notion is further strengthened by the fact that the healthy oral microbiome resistome revealed recently through whole-genome sequencing and real-time quantitative PCR microarray, contain highly prevalent genes conferring resistance to macrolides, lincosamides, streptogramins, and tetracyclines [55].
Taken together, our study represents the first antibiotic resistance profiling of probiotics from oral lozenges which serves not only to inform consumers and medical practitioners on the potential health risk, but also to encourage a more comprehensive study on the mechanisms underlying the transferability of resistance genes, especially in probiotics that do not carry the resistance determinants. Considering the global threat of drug resistance [169] and concomitant with the rising trend of dietary or health supplements [10], our data indicate a potential threat to human health.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Eric Yang, the Vice Chancellor for Academic Affairs of the Wenzhou-Kean University and Associate Provost of the Kean University, and the laboratory center of the Wenzhou-Kean University, for providing logistic and administrative support to this research.
Funding Statement
This research received funding from Wenzhou-Kean University under the Student Partnering with Faculty (SpF) research program (WKU201920009 & SpF2021002) awarded to A.W.; and Natural Science Foundation of Zhejiang Province (LQ20H140002) and Wenzhou Science and Technology bureau (Y20190100) awarded to Y.W.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
YW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Roles/Writing – original draft, Writing – review and editing; JD: Data curation, Formal analysis, Investigation, Methodology; Roles/Writing – original draft; JW: Data curation, Formal analysis, Investigation, Methodology; WC: Data curation, Formal analysis, Investigation, Methodology; WZ: Data curation, Formal analysis, Investigation, Methodology; QT: Data curation, Formal analysis, Investigation, Methodology; YH: Data curation, Formal analysis, Investigation, Methodology; XZ: Methodology, Formal analysis, Project administration, Resources, Supervision, Validation; HY: Methodology, Formal analysis, Project administration, Resources, Supervision, Validation; XT: Methodology, Formal analysis, Project administration, Resources, Supervision, Validation, Writing – review and editing; RH: Funding acquisition, Project administration, Resources, Supervision, Validation; AW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Roles/Writing - original draft, Writing – review and editing.
Data availability statement
The original contributions presented in the study are included in the article or Supplemental Information, further inquiries can be directed to the corresponding authors.
Ethics statement
There are no animal or human experiments involved in this study
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Perdigón G, Fuller R, Raya R.. Lactic acid bacteria and their effect on the immune system. Curr Issues Intest Microbiol. 2001;2(1):27–20. [PubMed] [Google Scholar]
- [2].Kaur IP, Chopra K, Saini A. Probiotics: potential pharmaceutical applications. Eur J Pharm Sci. 2002;15(1):1–9. [DOI] [PubMed] [Google Scholar]
- [3].Lakritz JR, Poutahidis T, Levkovich T, et al. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int J Cancer. 2014;135(3):529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zelaya H, Tsukida K, Chiba E, et al. Immunobiotic Lactobacilli reduce viral-associated pulmonary damage through the modulation of inflammation-coagulation interactions. Int Immunopharmacol. 2014;19(1):161–173. [DOI] [PubMed] [Google Scholar]
- [5].Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, et al. Effect of probiotic yogurt containing Lactobacillus acidophilus and Bifidobacterium lactis on lipid profile in individuals with type 2 diabetes mellitus. J Dairy Sci. 2011;94(7):3288–3294. [DOI] [PubMed] [Google Scholar]
- [6].He J, Zhang F, Han Y. Effect of probiotics on lipid profiles and blood pressure in patients with type 2 diabetes: a meta-analysis of RCTs. Medicine (Baltimore). 2017;96(51):e9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nikbakht E, Khalesi S, Singh I, et al. Effect of probiotics and synbiotics on blood glucose: a systematic review and meta-analysis of controlled trials. Eur J Nutr. 2016;57(1):95–106. [DOI] [PubMed] [Google Scholar]
- [8].Sun J, Buys NJ. Glucose- and glycaemic factor-lowering effects of probiotics on diabetes: a meta-analysis of randomised placebo-controlled trials. Br J Nutr. 2016;115(7):1167–1177. [DOI] [PubMed] [Google Scholar]
- [9].Foster JA, Lyte M, Meyer E, et al. Gut microbiota and brain function: an evolving field in neuroscience. Int J Neuropsychopharmacol. 2016;19(5):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cunningham M, Azcarate-Peril MA, Barnard A, et al. Shaping the future of probiotics and prebiotics. Trends Microbiol. 2021;29(8):667–685. [DOI] [PubMed] [Google Scholar]
- [11].Stanton C, Gardiner G, Meehan H, et al. Market potential for probiotics. Am J Clin Nutr. 2001;73(2):476S–483S. [DOI] [PubMed] [Google Scholar]
- [12].Thomas LV, Ockhuizen T, Suzuki K. Exploring the influence of the gut microbiota and probiotics on health: a symposium report. Br J Nutr. 2014;112(Suppl. 1):S1–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Savaiano DA, Ritter AJ, Klaenhammer TR, et al. Improving lactose digestion and symptoms of lactose intolerance with a novel galacto-oligosaccharide (RP-G28): a randomized, double-blind clinical trial. Nutr J. 2013;12(1):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scholz-Ahrens KE, Ade P, Marten B, et al. Prebiotics, probiotics, and synbiotics affect mineral absorption, bone mineral content, and bone structure. J Nutr. 2007;137(3 Suppl. 2):838S–846S. [DOI] [PubMed] [Google Scholar]
- [15].Parvez S, Malik KA, Ah Kang S, et al. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100(6):1171–1185. [DOI] [PubMed] [Google Scholar]
- [16].Ringel Y, Quigley EMM, Lin HC. Using probiotics in gastrointestinal disorders. Am J Gastroenterol Suppl. 2012;1(1):34–40. [Google Scholar]
- [17].Guarino A, Guandalini S, Lo Vecchio A. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol. 2015;49(Suppl 1):S37–S45. [DOI] [PubMed] [Google Scholar]
- [18].Silva DG, Sardi JCO, Pitangui NS, et al. Probiotics as an alternative antimicrobial therapy: current reality and future directions. J Funct Foods. 2020;73:104080. [Google Scholar]
- [19].Ji J, Yang H. Using probiotics as supplementation for Helicobacter pylori antibiotic therapy. Int J Mol Sci. 2020;21(3):1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352(6285):535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nguyen T, Brody H, Radaic A, et al. Probiotics for periodontal health-Current molecular findings. Periodontol 2000. 2021;87(1):254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rodrigues JZS, Passos MR, Silva de Macêdo Neres N, et al. Antimicrobial activity of Lactobacillus fermentum TcUESC01 against Streptococcus mutans UA159. Microb Pathog. 2020;142:104063. [DOI] [PubMed] [Google Scholar]
- [23].Chen CC, Lai CC, Huang HL, et al. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front Microbiol. 2019;10:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Crouzet L, Derrien M, Cherbuy C, et al. Lactobacillus paracasei CNCM I-3689 reduces vancomycin-resistant Enterococcus persistence and promotes Bacteroidetes resilience in the gut following antibiotic challenge. Sci Rep. 2018;8(1):5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ljungquist O, Kampmann C, Resman F, et al. Probiotics for intestinal decolonization of ESBL-producing Enterobacteriaceae: a randomized, placebo-controlled clinical trial. Clin Microbiol Infect. 2020;26(4):456–462. [DOI] [PubMed] [Google Scholar]
- [26].Wieërs G, Verbelen V, Van Den Driessche M, et al. Do probiotics during in-hospital antibiotic treatment prevent colonization of gut microbiota with multi-drug-resistant bacteria? A randomized placebo-controlled trial comparing Saccharomyces to a mixture of Lactobacillus, Bifidobacterium, and Saccharomyces. Front Public Health. 2021;8:578089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Srivastava S, Saha S, Kumari M, et al. Effect of probiotic curd on salivary pH and Streptococcus mutans: a double blind parallel randomized controlled trial. J Clin Diagn Res. 2016;10(2):ZC13–ZC16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nishihara T, Suzuki N, Yoneda M, et al. Effects of Lactobacillus salivarius-containing tablets on caries risk factors: a randomized open-label clinical trial. BMC Oral Health. 2014;14(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Routier A, Blaizot A, Agossa K, et al. What do we know about the mechanisms of action of probiotics on factors involved in the pathogenesis of periodontitis? A scoping review of in vitro studies. Arch Oral Biol. 2021;129:105196. [DOI] [PubMed] [Google Scholar]
- [30].Alshareef A, Attia A, Almalki M, et al. Effectiveness of probiotic lozenges in periodontal management of chronic periodontitis patients: clinical and immunological study. Eur J Dent. 2020;14(2):281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Khalaf H, Nakka SS, Sandén C, et al. Antibacterial effects of Lactobacillus and bacteriocin PLNC8 αβ on the periodontal pathogen Porphyromonas gingivalis. BMC Microbiol. 2016;16(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Matsubara VH, Bandara HM, Ishikawa KH, et al. The role of probiotic bacteria in managing periodontal disease: a systematic review. Expert Rev Anti Infect Ther. 2016;14(7):643–655. [DOI] [PubMed] [Google Scholar]
- [33].Matsubara VH, Wang Y, Bandara HM, et al. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl Microbiol Biotechnol. 2016;100(14):6415–6426. [DOI] [PubMed] [Google Scholar]
- [34].Lee SH, Baek DH. Effects of Streptococcus thermophilus on volatile sulfur compounds produced by Porphyromonas gingivalis. Arch Oral Biol. 2014;59(11):1205–1210. [DOI] [PubMed] [Google Scholar]
- [35].Bustamante M, Oomah BD, Mosi-Roa Y, et al. Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob Proteins. 2020;12(2):325–334. [DOI] [PubMed] [Google Scholar]
- [36].Mahasneh SA, Mahasneh AM. Probiotics: a promising role in dental health. Dent J. 2017;5(4):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Haukioja A. Probiotics and oral health. Eur J Dent. 2010;4(3):348–355. [PMC free article] [PubMed] [Google Scholar]
- [38].Chugh P, Dutt R, Sharma A, et al. A critical appraisal of the effects of probiotics on oral health. J Funct Foods. 2020;70:103985. [Google Scholar]
- [39].Lin CW, Chen YT, Ho HH, et al. Lozenges with probiotic strains enhance oral immune response and health. Oral Dis. 2021. 10.1111/odi.13854. [DOI] [PubMed] [Google Scholar]
- [40].Chua JCL, Hale JDF, Silcock P, et al. Bacterial survival and adhesion for formulating new oral probiotic foods. Crit Rev Food Sci Nutr. 2020;60(17):2926–2937. [DOI] [PubMed] [Google Scholar]
- [41].FDA . Microorganisms & microbial-derived ingredients used in food (partial list). 2018. Available online: https://www.fda.gov/food/generally-recognized-safe-gras/microorganisms-microbial-derived-ingredients-used-food-partial-list (accessed on 2021 Aug 4).
- [42].Koutsoumanis K, Allende A, Alvarez-Ordóñez A, EFSA Panel on Biological Hazards (BIOHAZ) . Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: suitability of taxonomic units notified to EFSA until march 2020. EFSA J. 2020;18(7):e06174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rychen G, Aquilina G, Azimonti G. EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16(3):e05206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wong A, Ngu DYS, Dan LA, et al. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr J. 2015;14(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wang Y, Jiang Y, Deng Y, et al. Probiotic Supplements: hope or Hype? Front Microbiol. 2020;11:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gueimonde M, Sánchez B, G de Los Reyes-gavilán C, et al. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tóth AG, Csabai I, Maróti G, et al. A glimpse of antimicrobial resistance gene diversity in kefir and yoghurt. Sci Rep. 2020;10(1):22458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Campedelli I, Mathur H, Salvetti E, et al. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol. 2018;85(1):e01738–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mathur S, Singh R. Antibiotic resistance in food lactic acid bacteria–a review. Int J Food Microbiol. 2005;105(3):281–295. [DOI] [PubMed] [Google Scholar]
- [50].Abriouel H, Casado Muñoz MDC, Lavilla Lerma L, et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res Int. 2015;78:465–481. [DOI] [PubMed] [Google Scholar]
- [51].Kothari D, Patel S, Kim SK. Probiotic supplements might not be universally-effective and safe: a review. Biomed Pharmacother. 2019;111:537–547. [DOI] [PubMed] [Google Scholar]
- [52].Zheng M, Zhang R, Tian X, et al. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front Microbiol. 2017;8:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kilian M, Chapple I, Hannig M, et al. The oral microbiome – an update for oral healthcare professionals. Br Dent J. 2016;221(10):657–666. [DOI] [PubMed] [Google Scholar]
- [54].Gendron R, Grenier D, Maheu-Robert L. The oral cavity as a reservoir of bacterial pathogens for focal infections. Microbes Infect. 2000;2(8):897–906. [DOI] [PubMed] [Google Scholar]
- [55].Caselli E, Fabbri C, D’Accolti M, et al. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 2020;20(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sirichoat A, Flórez AB, Vázquez L, et al. Antibiotic susceptibility profiles of lactic acid bacteria from the human vagina and genetic basis of acquired resistances. Int J Mol Sci. 2020;21(7):2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Das DJ, Shankar A, Johnson JB, et al. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition. 2020;69:110567. [DOI] [PubMed] [Google Scholar]
- [58].Li T, Teng D, Mao R, et al. A critical review of antibiotic resistance in probiotic bacteria. Food Res Int. 2020;136:109571. [DOI] [PubMed] [Google Scholar]
- [59].Jose NM, Bunt CR, Hussain MA. Implications of antibiotic resistance in probiotics. Food Rev Int. 2015;31(1):52–62. [Google Scholar]
- [60].Roberts AP, Kreth J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Broaders E, Gahan CG, Marchesi JR. Mobile genetic elements of the human gastrointestinal tract: potential for spread of antibiotic resistance genes. Gut Microbes. 2013;4(4):271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Penders J, Stobberingh E, Savelkoul P, et al. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol. 2013;4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rolain JM. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol. 2013;4:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci. 2015;370(1670):20140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Imperial IC, Ibana JA. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front Microbiol. 2016;7:1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Duranti S, Lugli GA, Mancabelli L, et al. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl Environ Microbiol. 2017;83(3):e2894–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lerner A, Shoenfeld Y, Matthias T. Probiotics: if it does not help it does not do any harm. really? Microorganisms. 2019;7(4):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chon JW, Seo KH, Bae D, et al. Status and prospect of lactic acid bacteria with antibiotic resistance. J Dairy Sci Biotechnol. 2020;38(2):70–88. [Google Scholar]
- [69].Lerner A, Matthias T, Aminov R. Potential effects of horizontal gene exchange in the human gut. Front Immunol. 2017;8:1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Humphries R, Bobenchik AM, Hindler JA, et al. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol. 2021;59(12):e0021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bujnakova D, Strakova E. Safety, probiotic and technological properties of Lactobacilli isolated from unpasteurised ovine and caprine cheeses. Ann Microbiol. 2017;67(12):813–826. [Google Scholar]
- [75].Gad GF, Abdel-Hamid AM, Farag ZS. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz J Microbiol. 2014;45(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kastner S, Perreten V, Bleuler H, et al. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst Appl Microbiol. 2006;29(2):145–155. [DOI] [PubMed] [Google Scholar]
- [77].Jensen LB, Frimodt-Moller N, Aarestrup FM. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett. 1999;170(1):151–158. [DOI] [PubMed] [Google Scholar]
- [78].Tannock GW, Luchansky JB, Miller L, et al. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31(1):60–71. [DOI] [PubMed] [Google Scholar]
- [79].Giovanetti E, Brenciani A, Lupidi R, et al. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob Agents Chemother. 2003;47(9):2844–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu C, Zhang ZY, Dong K, et al. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed Environ Sci. 2009;22(5):401–412. [DOI] [PubMed] [Google Scholar]
- [81].Hummel AS, Hertel C, Holzapfel WH, et al. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl Environ Microbiol. 2007;73(3):730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Egervärn M, Roos S, Lindmark H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J Appl Microbiol. 2009;107(5):1658–1668. [DOI] [PubMed] [Google Scholar]
- [83].Miele A, Bandera M, Goldstein BP. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39(8):1772–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Morales G, Picazo JJ, Baos E, et al. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis. 2010;50(6):821–825. [DOI] [PubMed] [Google Scholar]
- [85].Shevtsov AB, Kushugulova AR, Kojakhmetov SS, et al. Detection of Lactobacillus species using a gene fragment of the RNA polymerase beta subunit rpoB. Moscow Univ Biol Sci Bull. 2011;66(1):22–27. [Google Scholar]
- [86].Doherty N, Trzcinski K, Pickerill P, et al. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44(11):2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sutcliffe J, Grebe T, Tait-Kamradt A, et al. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ojo KK, Striplin MJ, Ulep CC, et al. Staphylococcus efflux msr(A) gene characterized in Streptococcus, Enterococcus, Corynebacterium, and Pseudomonas isolates. Antimicrob Agents Chemother. 2006;50(3):1089–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Werner G, Hildebrandt B, Witte W. The newly described msrC gene is not equally distributed among all isolates of Enterococcus faecium. Antimicrob Agents Chemother. 2001;45(12):3672–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen YT, Hsieh PS, Ho HH, et al. Antibacterial activity of viable and heat-killed probiotic strains against oral pathogens. Lett Appl Microbiol. 2020;70(4):310–317. [DOI] [PubMed] [Google Scholar]
- [91].Lin WH, Hwang CF, Chen LW, et al. Viable counts, characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 2006;23(1):74–81. [DOI] [PubMed] [Google Scholar]
- [92].Carr JP, Ibrahim SA. Viability of bifidobacteria in commercial yogurt products in North Carolina. Milchwissenschaft. 2005;60:414–416. [Google Scholar]
- [93].Al-Otaibi MM. Evaluation of some probiotic fermented milk products from Al-Ahsa markets, Saudi Arabia. Am J Food Technol. 2009;4(1):1–8 [Google Scholar]
- [94].Davis C. Enumeration of probiotic strains: review of culture-dependent and alternative techniques to quantify viable bacteria. J Microbiol Methods. 2014;103:9–17. [DOI] [PubMed] [Google Scholar]
- [95].Banas JA, Popp ET. Recovery of viable bacteria from probiotic products that target oral health. Probiotics Antimicrob Proteins. 2013;5(3):227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shah NP. Probiotic bacteria: selective enumeration and survival in dairy foods. J Dairy Sci. 2000;83(4):894–907. [DOI] [PubMed] [Google Scholar]
- [97].Rupa P, Mine Y. Recent advances in the role of probiotics in human inflammation and gut health. J Agric Food Chem. 2012;60(34):8249–8256. [DOI] [PubMed] [Google Scholar]
- [98].Nagyzbekkyzy E, Abitayeva G, Anuarbekova S, et al. Investigation of acid and bile 2tolerance, antimicrobial activity and antibiotic resistance of Lactobacillus strains isolated from kazakh dairy foods. Asian J Appl Sci. 2016;9(4):143–158. [Google Scholar]
- [99].Hassanzadazar H, Ehsani A, Mardani K, et al. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. Vet Res Forum. 2012;3(3):181–185. [PMC free article] [PubMed] [Google Scholar]
- [100].Liong MT, Shah NP. Acid and bile tolerance and cholesterol removal ability of Lactobacilli strains. J Dairy Sci. 2005;88(1):55–66. [DOI] [PubMed] [Google Scholar]
- [101].Succi M, Tremonte P, Reale A, et al. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol Lett. 2005;244(1):129–137. [DOI] [PubMed] [Google Scholar]
- [102].Sahadeva RPK, Leong SF, Chua KH, et al. Survival of commercial probiotic strains to pH and bile. Int Food Res J. 2011;18:1515–1522. [Google Scholar]
- [103].Wang RM, Li N, Zheng K, et al. Enhancing acid tolerance of the probiotic bacterium Lactobacillus acidophilus NCFM with trehalose. FEMS Microbiol Lett. 2018;365(19):fny217. [DOI] [PubMed] [Google Scholar]
- [104].Gbassi GK, Vandamme T. Probiotic encapsulation technology: from microencapsulation to release into the gut. Pharmaceutics. 2012;4(1):149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Rodrigues FJ, Cedran MF, Bicas JL, et al. Encapsulated probiotic cells: relevant techniques, natural sources as encapsulating materials and food applications - A narrative review. Food Res Int. 2020;137:109682. [DOI] [PubMed] [Google Scholar]
- [106].Yao M, Xie J, Du H, et al. Progress in microencapsulation of probiotics: a review. Compr Rev Food Sci Food Saf. 2020;19(2):857–874. [DOI] [PubMed] [Google Scholar]
- [107].Lynge Pedersen AM, Belstrøm D. The role of natural salivary defences in maintaining a healthy oral microbiota. J Dent. 2019;80(Suppl 1):S3–S12. [DOI] [PubMed] [Google Scholar]
- [108].Haukioja A, Yli-Knuuttila H, Loimaranta V, et al. Oral adhesion and survival of probiotic and other lactobacilli and bifidobacteria in vitro. Oral Microbiol Immunol. 2006;21(5):326–332. [DOI] [PubMed] [Google Scholar]
- [109].Wade WG. Resilience of the oral microbiome. Periodontol 2000. 2021;86(1):113–122. [DOI] [PubMed] [Google Scholar]
- [110].Delcour J, Ferain T, Deghorain M, et al. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76(1/4):159–184. [PubMed] [Google Scholar]
- [111].Bruniera FR, Ferreira FM, Saviolli LR, et al. The use of vancomycin with its therapeutic and adverse effects: a review. Eur Rev Med Pharmacol Sci. 2015;19(4):694–700. [PubMed] [Google Scholar]
- [112].Mater DD, Langella P, Corthier G, et al. Evidence of vancomycin resistance gene 6transfer between enterococci of human origin in the gut of mice harbouring human microbiota. J Antimicrob Chemother. 2005;56(5):975–978. [DOI] [PubMed] [Google Scholar]
- [113].Mater DD, Langella P, Corthier G, et al. A probiotic Lactobacillus strain can acquire vancomycin resistance during digestive transit in mice. J Mol Microbiol Biotechnol. 2008;14(1–3):123–127. [DOI] [PubMed] [Google Scholar]
- [114].Meini S, Laureano R, Fani L, et al. Breakthrough Lactobacillus rhamnosus GG bacteremia associated with probiotic use in an adult patient with severe active ulcerative colitis: case report and review of the literature. Infection. 2015;43(6):777–781. [DOI] [PubMed] [Google Scholar]
- [115].Vahabnezhad E, Mochon AB, Wozniak LJ, et al. Lactobacillus bacteremia associated with probiotic use in a pediatric patient with ulcerative colitis. J Clin Gastroenterol. 2013;47(5):437–439. [DOI] [PubMed] [Google Scholar]
- [116].Sadowska-Krawczenko I, Paprzycka M, Korbal P, et al. Lactobacillus rhamnosus GG suspected infection in a newborn with intrauterine growth restriction. Benef Microbes. 2014;5(4):397–402. [DOI] [PubMed] [Google Scholar]
- [117].Fujiya Y, Harada T, Sugawara Y, et al. Transmission dynamics of a linear vanA-plasmid during a nosocomial multiclonal outbreak of vancomycin-resistant enterococci in a non-endemic area, Japan. Sci Rep. 2021;11(1):14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Goldberg E, Bishara J. Contemporary unconventional clinical use of co-trimoxazole. Clin Microbiol Infect. 2012;18(1):8–17. [DOI] [PubMed] [Google Scholar]
- [119].Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3(1):118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Eliopoulos GM, Huovinen P. Resistance to Trimethoprim-Sulfamethoxazole. Clin Infect Dis. 2001;32(11):1608–1614. [DOI] [PubMed] [Google Scholar]
- [121].Ambrose SJ, Hall RM. Novel trimethoprim resistance gene, dfrA35, in IncC plasmids from Australia. J Antimicrob Chemother. 2019;74(7):1863–1866. [DOI] [PubMed] [Google Scholar]
- [122].Lee JC, Oh JY, Cho JW, et al. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J Antimicrob Chemother. 2001;47(5):599–604. [DOI] [PubMed] [Google Scholar]
- [123].Miranda A, Ávila B, Díaz P, et al. Emergence of Plasmid-Borne dfrA14 Trimethoprim Resistance Gene in Shigella sonnei. Front Cell Infect Microbiol. 2016;6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhou N, Zhang JX, Fan MT, et al. Antibiotic resistance of lactic acid bacteria isolated from Chinese yogurts. J Dairy Sci. 2012;95(9):4775–4783. [DOI] [PubMed] [Google Scholar]
- [125].Klare I, Konstabel C, Werner G, et al. Antimicrobial susceptibilities of lactobacillus, pediococcus and lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother. 2007;59(5):900–912. [DOI] [PubMed] [Google Scholar]
- [126].Sharma C, Gulati S, Thakur N, et al. Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. 3 Biotech. 2017;7(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sharma P, Tomar SK, Goswami P, et al. Antibiotic resistance among commercially available probiotics. Food Res Int. 2014;57:176–195. [Google Scholar]
- [128].Rojo-Bezares B, Sáenz Y, Poeta P, et al. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int J Food Microbiol. 2006;111(3):234–240. [DOI] [PubMed] [Google Scholar]
- [129].Ouoba LI, Lei V, Jensen LB. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of African and European origin to antimicrobials: determination and transferability of the resistance genes to other bacteria. Int J Food Microbiol. 2008;121(2):217–224. [DOI] [PubMed] [Google Scholar]
- [130].Dec M, Urban-Chmiel R, Stępień-Pyśniak D, et al. Assessment of antibiotic susceptibility in lactobacillus isolates from chickens. Gut Pathog. 2017;9(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5(6):234–240. [DOI] [PubMed] [Google Scholar]
- [132].Sanz-García F, Anoz-Carbonell E, Pérez-Herrán E, et al. Mycobacterial aminoglycoside acetyltransferases: a little of drug resistance, and a lot of other roles. Front Microbiol. 2019;10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13(6):151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Rodríguez-Alonso P, Fernández-Otero C, Centeno JA, et al. Antibiotic resistance in lactic acid bacteria and Micrococcaceae/Staphylococcaceae isolates from artisanal raw milk cheeses, and potential implications on cheese making. J Food Sci. 2009;74(6):M284–M293. [DOI] [PubMed] [Google Scholar]
- [135].Aquilanti L, Garofalo C, Osimani A, et al. Isolation and molecular characterization of antibiotic-resistant lactic acid bacteria from poultry and swine meat products. J Food Prot. 2007;70(3):557–565. [DOI] [PubMed] [Google Scholar]
- [136].Okiki PA, Eromosele ES, Ade-Ojo P, et al. Occurrence of mecA and blaZ genes in methicillin-resistant Staphylococcus aureus associated with vaginitis among pregnant women in Ado-Ekiti, Nigeria. New Microbes New Infect. 2020;38:100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Garofalo C, Vignaroli C, Zandri G, et al. Direct detection of antibiotic resistance genes in specimens of chicken and pork meat. Int J Food Microbiol. 2007;113(1):75–83. [DOI] [PubMed] [Google Scholar]
- [138].Chen MM, Boardman WS, Smith I, et al. Characterisation of β-lactam resistance mediated by blaZ in staphylococci recovered from captive and free-ranging wallabies. J Glob Antimicrob Resist. 2015;3(3):184–189. [DOI] [PubMed] [Google Scholar]
- [139].Muñoz Mdel C C, Benomar N, Lerma LL, et al. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented aloreña table olives throughout fermentation process. Int J Food Microbiol. 2014;172:110–118. [DOI] [PubMed] [Google Scholar]
- [140].Anisimova EA, Yarullina DR. Antibiotic resistance of LACTOBACILLUS strains. Curr Microbiol. 2019;76(12):1407–1416. [DOI] [PubMed] [Google Scholar]
- [141].Dec M, Nowaczek A, Stępień-Pyśniak D, et al. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018;18(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].de Been M, Lanza VF, de Toro M, et al. Dissemination of cephalosporin resistance genes between escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014;10(12):e1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Li S, Li Z, Wei W, et al. Association of mutation patterns in GyrA and ParC genes with quinolone resistance levels in lactic acid bacteria. J Antibiot. 2015;68(2):81–87. [DOI] [PubMed] [Google Scholar]
- [144].Çataloluk O, Gogebakan B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol Lett. 2004;236(1):7–12. [DOI] [PubMed] [Google Scholar]
- [145].Gevers D, Huys G, Swings J. In vitro conjugal transfer of tetracycline resistance from lactobacillus isolates to other gram-positive bacteria. FEMS Microbiol Lett. 2003;225(1):125–130. [DOI] [PubMed] [Google Scholar]
- [146].Schwarz S, Kehrenberg C, Doublet B, et al. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28(5):519–542. [DOI] [PubMed] [Google Scholar]
- [147].Guo H, Pan L, Li L, et al. Characterization of antibiotic resistance genes from lactobacillus isolated from traditional dairy products. J Food Sci. 2017;82(3):724–730. [DOI] [PubMed] [Google Scholar]
- [148].Shazali N, Foo HL, Loh TC, et al. Prevalence of antibiotic resistance in lactic acid bacteria isolated from the faeces of broiler chicken in Malaysia. Gut Pathog. 2014;6(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Thumu SC, Halami PM. Presence of erythromycin and tetracycline resistance genes in lactic acid bacteria from fermented foods of Indian origin. Antonie Van Leeuwenhoek. 2012;102(4):541–551. [DOI] [PubMed] [Google Scholar]
- [150].Devirgiliis C, Coppola D, Barile S, et al. Characterization of the Tn916 conjugative transposon in a food-borne strain of lactobacillus paracasei. Appl Environ Microbiol. 2009;75(12):3866–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ammor MS, Gueimonde M, Danielsen M, et al. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in lactobacillus sakei Rits 9. Appl Environ Microbiol. 2008;74(5):1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Madsen JS, Burmølle M, Hansen LH, et al. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65(2):183–195. [DOI] [PubMed] [Google Scholar]
- [153].Woods LC, Gorrell RJ, Taylor F, et al. Horizontal gene transfer potentiates adaptation by reducing selective constraints on the spread of genetic variation. Proc Natl Acad Sci U S A. 2020;117(43):26868–26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lester CH, Frimodt-Moller N, Hammerum AM. Conjugal transfer of aminoglycoside and macrolide resistance between Enterococcus faecium isolates in the intestine of streptomycin-treated mice. FEMS Microbiol Lett. 2004;235(2):385–391. [DOI] [PubMed] [Google Scholar]
- [155].Sedgley CM, Lee EH, Martin MJ, et al. Antibiotic resistance gene transfer between Streptococcus gordonii and Enterococcus faecalis in root canals of teeth ex vivo. J Endod. 2008;34(5):570–574. [DOI] [PubMed] [Google Scholar]
- [156].Feld L, Schjørring S, Hammer K, et al. Selective pressure affects transfer and establishment of a lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J Antimicrob Chemother. 2008;61(4):845–852. [DOI] [PubMed] [Google Scholar]
- [157].Devi SM, Halami PM. Conjugal transfer of bacteriocin plasmids from different genera of lactic acid bacteria into Enterococcus faecalis JH2-2. Ann Microbiol. 2013;63(4):1611–1617. [Google Scholar]
- [158].Neil K, Allard N, Rodrigue S. Molecular mechanisms influencing bacterial conjugation in the intestinal Microbiota. Front Microbiol. 2021;12:673260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].García-Quintanilla M, Ramos-Morales F, Casadesús J. Conjugal transfer of the salmonella enterica virulence plasmid in the mouse intestine. J Bacteriol. 2008;190(6):1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Toomey N, Monaghan A, Fanning S, et al. Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog Dis. 2009;6(8):925–933. [DOI] [PubMed] [Google Scholar]
- [161].Aviv G, Rahav G, Gal-Mor O. Horizontal transfer of the salmonella enterica serovar infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio. 2016;7(5):e1395–e1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Licht TR, Christensen BB, Krogfelt KA, et al. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology (Reading). 1999;145(9):2615–2622. [DOI] [PubMed] [Google Scholar]
- [163].Ott LC, Stromberg ZR, Redweik GAJ, et al. Mouse genetic background affects transfer of an antibiotic resistance plasmid in the gastrointestinal tract. mSphere. 2020;5(1):e847–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Montassier E, Valdés-Mas R, Batard E, et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol. 2021;6(8):1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hu Y, Yang X, Lu N, et al. The abundance of antibiotic resistance genes in human guts has correlation to the consumption of antibiotics in animal. Gut Microbes. 2014;5(2):245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Caniça M, Manageiro V, Abriouel H, et al. Antibiotic resistance in foodborne bacteria. Trends Food Sci Technol. 2019;84:41–44. [Google Scholar]
- [167].Hudson JA, Frewer LJ, Jones G, et al. The agri-food chain and antimicrobial resistance: a review. Trends Food Sci Technol. 2017;69(Part A):131–147. [Google Scholar]
- [168].Salvetti E, O’Toole PW. When regulation challenges innovation: the case of the genus Lactobacillus. Trends Food Sci Technol. 2017;66:187–194. [Google Scholar]
- [169].de Kraker MEA, Stewardson AJ, Harbarth S, et al. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13(11):e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article or Supplemental Information, further inquiries can be directed to the corresponding authors.


