Abstract
Helicobacter pylori is one of the most frequent human pathogens and a leading etiological agent of various gastric diseases. As stringent response, coordinated by a SpoT protein, seems to be crucial for the survivability of H. pylori, the main goal of this article was to use in silico computational studies to find phytochemical compounds capable of binding to the active site of SpoT from H. pylori and confirm the ability of the most active candidates to interfere with the virulence of this bacterium through in vitro experiments. From 791 natural substances submitted for the virtual screening procedure, 10 were chosen and followed for further in vitro examinations. Among these, dioscin showed the most interesting parameters (the lowest MIC, the highest anti-biofilm activity in static conditions, and a relatively low stimulation of morphological transition into coccoids). Therefore, in the last part, we extended the research with a number of further experiments and observed the ability of dioscin to significantly reduce the formation of H. pylori biofilm under Bioflux-generated flow conditions and its capacity for additive enhancement of the antibacterial activity of all three commonly used antibiotics (clarithromycin, metronidazole, and levofloxacin). Based on these results, we suggest that dioscin may be an interesting candidate for new therapies targeting H. pylori survivability and virulence.
Keywords: Helicobacter pylori, alternative therapy, phytotherapy, virulence, biofilm, coccoid forms, dioscin, molecular docking, Bioflux
1. Introduction
The rapid spread of drug-resistant bacteria is now occurring all over the world, posing a threat to the forms and availability of currently known treatments [1,2,3]. This situation has worsened over time and now, according to many scientists and clinicians, medicine is gradually entering the post-antibiotic era [4,5]. The universality of antibiotic usage in various sectors of human life generates a selective pressure on human microflora and pathogens colonizing the human body [6]. Keeping in mind that the digestive system is the largest human reservoir of bacteria and is constantly subjected to the selection pressure to diverse antimicrobials, monitoring resistance and improving methods aimed at combating pathogens persistently colonizing this system is of paramount importance [7,8]. In this context, Helicobacter pylori seems to be one of the most important threats [9,10].
H. pylori is one of the most frequent human pathogens and a leading etiological agent of various gastric diseases, including gastritis, peptic ulcers, and gastric cancers (adenocarcinomas and mucosa-associated lymphoid tissue lymphomas) [11]. The success of H. pylori as a gastric pathogen is associated with its ability to survive in harsh conditions generated by the stomach [12]. Complex physiological processes related to virulence, such as biofilm formation [13,14,15] and morphological transformation [16,17,18], although described for first time for H. pylori a long time ago, just recently began to be shown as crucial to the survival of this microorganism and the relationship to therapeutic failures. This is reflected not only in the increasing amount of research conducted on both biofilms and coccoid forms of H. pylori, but also in recommendations encouraging efforts to intensify the search for methods effective in interfering with these mechanisms [13,16,19,20].
In the past few years, the relevance of computational chemistry tools as partners to experimental investigations has been emphasized. Among several application fields, computer aided drug design is the most promising and intensively utilized. These in silico studies focus on searching for possible drug candidates and have become a strategy of choice in examining and developing novel compounds with the desired activity due to their satisfying results [21]. Not only does this approach significantly reduce the time required to conduct studies by screening the entire library in a short period of time and thus discarding inactive compounds at the outset, but it also provides opportunities to discover novel pharmaceuticals that may be used in the treatment of certain diseases such as cancers [22] or neurodegenerative disorders [23]. This is especially true nowadays, as the methodology is being coupled with artificial intelligence, which improves its accuracy and, therefore, leads to new discoveries [24].
Plant substances are well known for their beneficial, health-promoting properties [25]. Studies show that phytochemicals have a multidirectional mechanism of action in their antibacterial activity: disruption of cytoplasmic membrane function and structure (including the efflux system), interaction with the membrane proteins (ATPases and others), interruption of DNA/RNA synthesis and function, destabilization of the proton motive force and induction of ion leakage, prevention of enzyme synthesis, coagulation of cytoplasmic constituents, and interruption of microbial communication [26]. Some publications highlight the potential of natural products to influence the survivability of H. pylori, either by reducing resistance to specific antibiotics [27] or by postponing biofilm development [28].
It seems that stringent response may be an excellent target in novel therapies fighting pathogenic microorganisms [29]. The stringent response is a phenomenon related to microbial reprogramming during which ribosomal synthesis is downregulated, while stress-associated genes are upregulated. This process is controlled by the intracellular concentrations of alarmon (guanosine tetra- and pentaphosphate [(p)ppGpp]), a molecule existing in most bacteria that is synthesized and degraded by RelA and SpoT, respectively. In the case of H. pylori, however, the stringent response is coordinated only by a one bifunctional SpoT protein regulating the level of this messenger substance [30,31]. Although the amount of data on SpoT of H. pylori is still very limited, studies by some research teams have observed that activity of SpoT is associated with stress and defense responses in H. pylori, and thus, interfering with its protective activity might be a promising way of fighting this pathogen [32,33,34].
The main goal of our research was to use in silico computational studies to find phytochemical compounds capable of binding to the active site of SpoT from H. pylori and to confirm the ability of the most active candidates to interfere with the virulence of this bacterium through in vitro experiments.
2. Results and Discussion
2.1. Virtual Screening
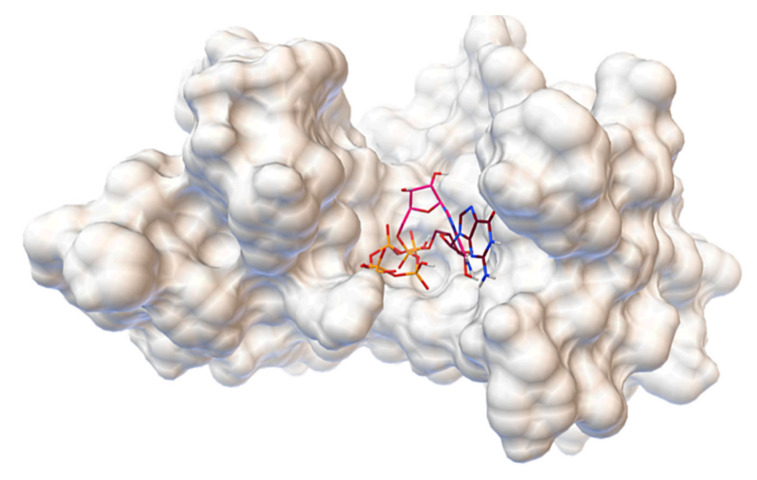
We were able to evaluate binding sites and discover interactions that are likely to be relevant for possible competitive inhibitors by docking natural ligands, ATP and GTP, to the constructed structure of SpoT protein third domain. The ligands are located in the common cavity in the 3D visualization of the aforementioned process, which is shown in Figure 1. It should be noted that, despite the fact that the docking box was defined for the whole domain, the ligands appear only in the presented region, although in various conformations. Furthermore, with the exception of ATP’s ribose, the structures are in close proximity to one another, suggesting the potential for Pi transfer from ATP to GTP, which corresponds to diphosphokinase activity.
Figure 1.
The 3D structure of SpoT domain exhibiting kinase activity with ATP and GTP bound to the common cavity.
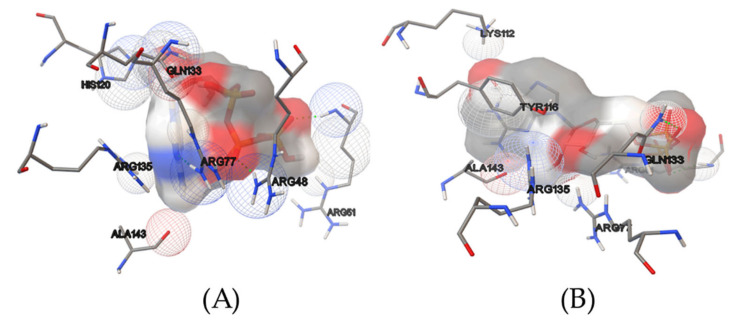
Figure 2 displays amino acids that interact with ligands and thereby contribute to their binding. Both natural ligands are surrounded by ARG77, ARG51, GLN133, ARG135, and ALA143 amino acids. Hydrogen bonds are formed with ATP via ARG51, ARG77, and ARG48, and with GTP via ARG51 and GLN133. Furthermore, a slew of van der Waals interactions can be seen, which are known to maintain ligands within the pocket.
Figure 2.
Amino acids involved in the binding of ATP (A) and GTP (B). Van der Waals interactions are depicted as wireframed spheres, with red representing oxygen, blue representing nitrogen, and gray representing carbon origin. The surfaces of the ligands are colored in the same way.
To choose the compounds to be submitted for the experimental part of the research, the following approach was adopted. To assert the substance’s competitive nature, the preliminary screening results were filtered out of compounds having binding free energies of less than −7.6 kcal/mol, which corresponds to that of GTP (ATP has a binding free energy of −6.4 kcal/mol). They are listed in Table 1, and the whole list is available in Table S1. Furthermore, inhibition constants (Ki) were calculated, as described in the Section 3. This preliminary screening yielded 105 of the most promising candidates, from which 10 hits were selected based on the following criteria: (1) comparably low binding free energies, (2) amino acid composition interacting similarly to those previously found for natural ligands, (3) visible structural differences, and (4) negligible toxicity to humans. A toxicity profile was established only for selected compounds with favorable parameters (e.g., comparably low binding free energies and/or the ability to interact with a high amount of amino acids in the target site of SpoT in a way similar to those found for natural ligands). The literature was searched for the toxicity data of these compounds (Table S2).
Table 1.
Virtual screening hits, their binding free energies, and theoretical inhibition constants (at T = 310.15 K).
| Compound | Class | ΔG [kcal/mol] | Ki [μM] |
|---|---|---|---|
| Dioscin | Saponine glycoside | −9.8 | 0.122 |
| Zeaxanthin | Carotenoid | −8.9 | 0.527 |
| Glycyrrhizic acid | Saponine glycoside | −8.8 | 0.620 |
| Ginsenoside Rg3 | Saponine glycoside | −8.6 | 0.858 |
| Robinin | Flavone glycoside | −8.6 | 0.858 |
| Amentoflavone | Biflavonoid | −8.5 | 1.009 |
| Hypericin | Naphthodianthrone | −8.5 | 1.009 |
| Procyanidin A2 | Proanthocyanidin | −8.5 | 1.009 |
| Procyanidin C1 | Proanthocyanidin | −8.2 | 1.642 |
| Aescin | Saponine glycoside | −8.2 | 1.642 |
The inhibition constants were obtained with the Autodock Vina software as described in the Section 3.1.3.
2.2. MICs
In the first step, we decided to determine the antimicrobial activity of 10 previously selected compounds by determining their minimal inhibitory concentration (MIC) values (Table 2). We noticed that dioscin (MIC = 64 µg/mL) was the most active one, followed by amentoflavone and hypericin (both with MIC = 128 µg/mL), and aescin (MIC = 256 µg/mL). The remaining six compounds showed no antibacterial activity against H. pylori J99 in the concentration range used in this study (MIC > 256 µg/mL). Despite numerous studies on the anti-inflammatory [35,36,37] and antitumor [38,39,40,41] activity of dioscin or diosgenin (an aglycone form of dioscin), the data on their antimicrobial action are very scarce. Among the few articles available, three concerning fungi [42,43,44] and one indicating activity against anaerobic bacteria [45] could be found. For this reason, we believe that the results obtained by our team bring the aspect of novelty in the context of using dioscin in anti-bacterial therapy.
Table 2.
Minimal inhibitory concentration (MIC) values of 10 selected compounds of natural origin against H. pylori J99.
| Tested Compounds | MIC [µg/mL] |
|---|---|
| Dioscin | 64 |
| Hypericin | 128 |
| Amentoflavone | 128 |
| Aescin | 256 |
| Glycyrrhetinic acid | >256 |
| Ginsenoside Rg3 | >256 |
| Procyanidin A2 | >256 |
| Procyanidin C1 | >256 |
| Robinin | >256 |
| Zeaxanthin | >256 |
2.3. Anti-Biofilm Activity
To obtain a more complete picture of the activity of the tested compounds against H. pylori, we extended our screening to include the determination of anti-biofilm activity and the effect on bacterial morphology. Both biofilm formation [13,14,15] and transformation into coccoid morphotype [16,17,18] are phenomena related to the tolerance of H. pylori to antimicrobial substances. Therefore, we assumed that ideally the tested compounds should be able to reduce the amount of biofilm without or with minimal stimulation of the spiral-to-coccoid transition (coccoid forms arise often intensively during exposure to antibiotics or environmental conditions unfavorable to bacterial physiology [16,17,18]).
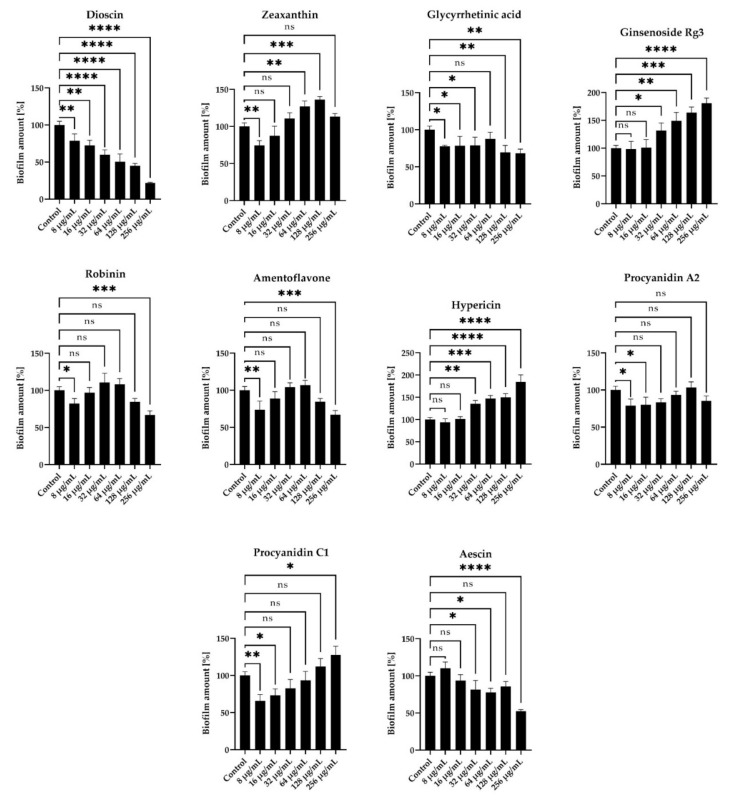
We observed that dioscin was characterized by the most intense inhibitory activity against biofilm formation under static conditions, i.e., MICs reduced this process by about half (up to 50.2 ± 10.6%) (Figure 3). Importantly, at these concentrations, we detected a low level of coccoid H. pylori forms (approx. 20%) (Figure S1). Other compounds at MIC values for which we noticed an ability to decrease the amount of H. pylori biofilm include aescin (decrease to 52.3 ± 2.2%) or amentoflavone (decrease to 84.5 ± 4.4%) (Figure 3); however, for both of them, the number of coccoid forms was unsatisfactorily high (40–60%) (Figure S1).
Figure 3.
Anti-biofilm activity of 10 selected compounds of natural origin against H. pylori J99. The amount of biofilm under stationary conditions was determined using the crystal violet staining method. The study was performed with triplicate biological replications (n = 3). The p-value represented by ns, *, **, ***, and **** is equal to > 0.05, < 0.05, < 0.01, < 0.001, and < 0.0001, respectively.
In line with our earlier deduction, which indicated only a few publications describing the antimicrobial activity of dioscin, in this case also, it was difficult for us to refer to other data in the literature. Despite this, in the original article by Yang et al. (2018) [43], the ability of dioscin to interfere with a number of virulence factors of fungi from the Candida genus has been proved. This mechanism covered not only their biofilm formation, but also morphological transformation (transition to filamentous forms) and secretion of lytic exoenzymes. This seemed to confirm our primary assumptions about the possibility of using dioscin in anti-virulence therapy against H. pylori.
Surprisingly, for some substances—hypericin, ginsenoside Rg3, procyanidin C1, and zeaxanthin—we noticed a stimulating effect on the amount of H. pylori biofilm. For example, for hypericin at a concentration equal to 2 × MIC (256 µg/mL) the amount of biofilm increased to 184.4 ± 16% compared to the control (Figure 3). In our opinion, these observations carry great research and clinical implications. They indicate that the primary selection of antimicrobials based only on the MIC measurement may be insufficient or even confusing (hypericin, next to amentoflavone, as the second most active antimicrobial substance in this study) and emphasize the need to determine additional antimicrobial parameters [46]. Moreover, the above observations throw a shadow on the widespread hypothesis that microorganisms are unable to generate resistance or activate defense mechanisms against plant extracts or plant-derived compounds, in agreement with others [47,48,49]. On the other hand, it has been shown by several research groups that the activity of phytochemicals may be modulated by a concentration-dependent formation of dimers/trimers/polymers and aggregation-related changes in the antimicrobial action of these molecules [50,51,52].
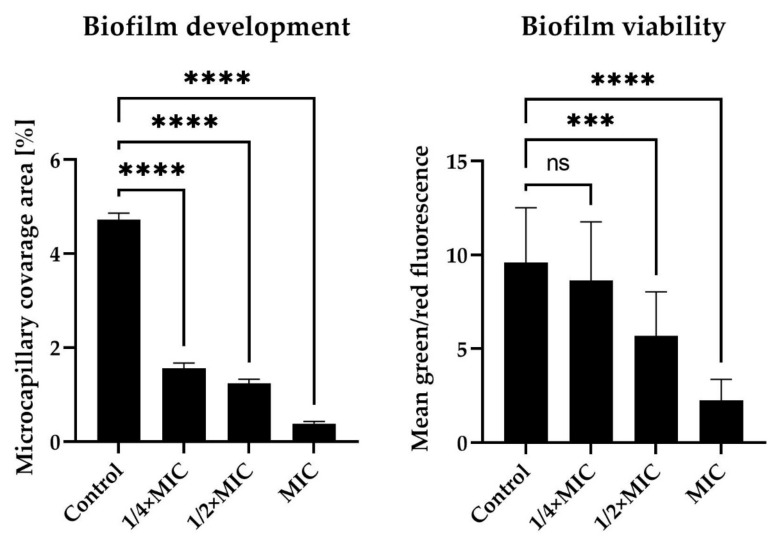
Based on the most favorable parameters obtained for dioscin (the lowest MIC, the highest anti-biofilm activity in static conditions, and a relatively low stimulation of morphological transition into coccoids), we decided to extend the panel of studies on dioscin with the verification of its anti-biofilm activity against H. pylori J99 under flow conditions. For this purpose, we used an automatic system, namely Bioflux 1000, generating the flow of culture medium and enabling observation of microbes in real time [53]. Although this equipment has previously been used to determine the antibacterial and/or anti-biofilm activity of antibiotics and newly tested compounds against different bacterial genera [54,55,56], our team is the first to apply it to H. pylori (previously [57] and hereby). Analysis of time-lapse microscopy recordings of bacteria exposed to dioscin allowed us to confirm the concentration-dependent anti-biofilm effect of this compound (Figure 4, Figure 5 and Figure S2). After a 24-h incubation of H. pylori under the above flow conditions, we noticed a significant reduction in the amount of biofilm formed (4.72 ± 0.14% vs. 0.38 ± 0.05% of the capillary coverage in the control and MIC-exposed samples, respectively) (Figure 4 and Figure 5). Moreover, we observed a decrease in the green/red fluorescence ratio of dioscin-treated bacterial cells (9.59 ± 1.17 vs. 2.26 ± 0.11 for the control and MIC-treated samples, respectively) (Figure 4 and Figure 5).
Figure 4.
Representative photos obtained after 24 h of experiments determining anti-biofilm activity of dioscin under flow conditions against H. pylori J99. The flow conditions were generated using the Bioflux 1000 system. The MIC, ½ × MIC, and ¼ × MIC had values of 64 µg/mL, 32 µg/mL, and 16 µg/mL, respectively. Control samples were bacteria not exposed to any concentration of dioscin. The pictures on the right show cells stained with the LIVE/DEAD kit (green and red fluorescence showing live and dead cells, respectively) and viewed under a fluorescence microscope, while the pictures on the left show cells under a light microscope. Scale bars show 10 µm.
Figure 5.
Data quantifying anti-biofilm activity of dioscin against H. pylori J99 after 24-h incubation under flow conditions. The flow conditions were generated using the Bioflux 1000 system. The MIC, ½ × MIC, and ¼ × MIC had values of 64 µg/mL, 32 µg/mL, and 16 µg/mL, respectively. Control samples were bacteria not exposed to any concentration of dioscin. Viability was defined as the ratio of green to red fluorescence of bacterial cells stained with the LIVE/DEAD kit, while the development of biofilm was verified by determining the degree of capillary coverage (both parameters were determined using the Bioflux Montage software). The study was performed with triplicate biological replications (n = 3). The p-value represented by ns, ***, and **** is equal to > 0.05, < 0.001, and < 0.0001, respectively.
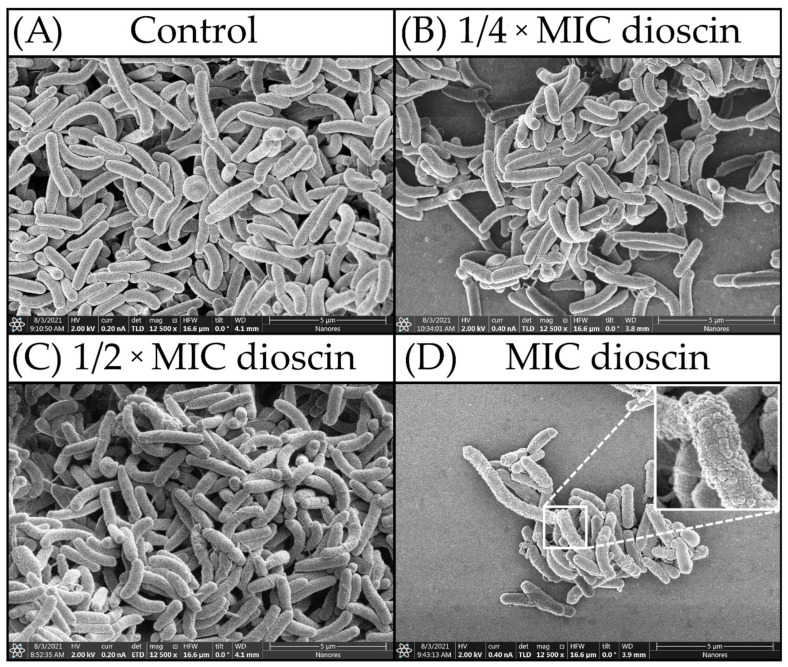
2.4. Morphostructural Analysis of Bacterial Cells
To confirm our assumptions about the lack of degradation of H. pylori cells exposed to concentrations of ¼ × MIC—MIC of dioscin, we performed morphostructural analysis of bacterial cells using scanning electron microscopy (SEM). The results obtained from the micrographs were unequivocal (Figure 6), and in all cases the bacterial cells were unchanged in size. H. pylori cells treated with dioscin at ¼ × MIC, ½ × MIC, and MIC concentrations showed normal morphology (Figure 6B–D), where they presented typically spiral/rod forms and non-numerous coccoids (both morphological forms were with intact cell surface). Similarly, images of the untreated control cells showed undeformed structure of their cell membrane (Figure 6A). It is noteworthy that the cells treated with a MIC value of dioscin displayed several apparent, distinguished signs showcasing a specific defense mechanism of H. pylori expressed in secretion of membrane vesicles (Figure 6D).
Figure 6.
SEM micrographs of H. pylori cells: (A) untreated cells (control); (B) cells treated with ¼ × MIC of dioscin; (C) cells treated with a ½ × MIC of dioscin; (D) cells treated with a MIC of dioscin.
Membrane vesicles are nanometric structures that are secreted by both Gram-positive and Gram-negative bacteria [58,59,60]. Historically, they have been considered as evidence of microbial cell membrane instability and lysis. Nowadays, however, it has been proved that the increase in secretion of these structures is associated with adaptation to unfavorable environmental conditions, as they may promote cell–cell interactions and biofilm development [61]. For diverse groups of bacteria, an intensification in the production of membrane vesicles has been observed in response to exposure of bacteria to different classes of antibiotics and other antimicrobial compounds [62]. For H. pylori, also, it has been shown that treatment with antibiotics (including clarithromycin and levofloxacin) increases the amount of membrane vesicles with a positive effect on their survival in these conditions [63]. On this basis, we suspect that at MIC levels, dioscin may also induce a similar mechanism in H. pylori cells. However, this type of research is beyond the main scope of this research article, and the hypothesis we presented will require verification in the future.
Summarizing this part of the analysis, we did not observe any significant structural changes in cells treated with dioscin regardless of the applied concentrations. This led us to believe that the observed anti-biofilm activity of dioscin did not depend directly on the destruction of H. pylori cells and that a different mechanism was responsible for the death of the bacteria.
2.5. Checkerboard Assay
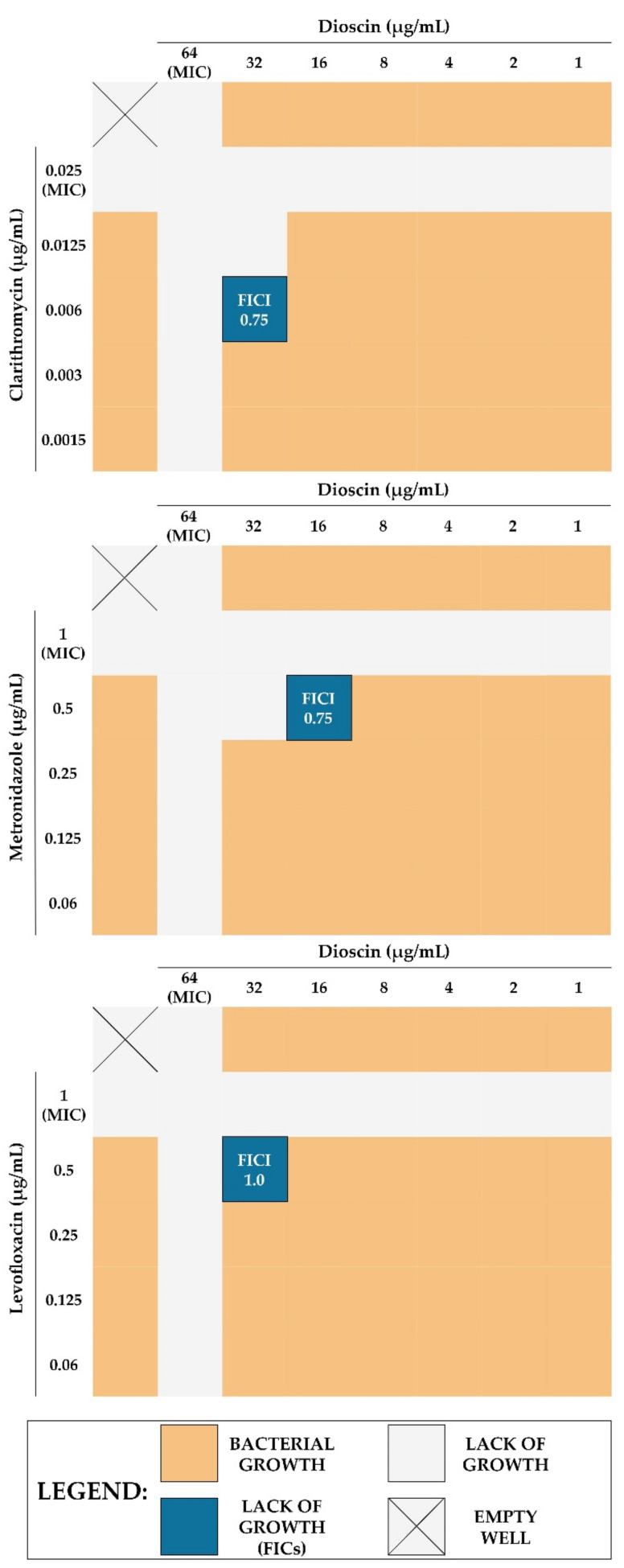
Next, we decided to confirm our proof-of-concept for the usefulness of dioscin in H. pylori therapy. In this context, using the checkerboard method, we determined the types of interactions between dioscin and three antibiotics classically used in H. pylori treatment (clarithromycin [CLR], metronidazole [MTZ], and levofloxacin [LEV] [64]). For all of them, the existence of additive interactions was demonstrated (FICI = 0.75 for both CLR and MTZ, and FICI = 1.0 for LEV), allowing us to reduce the concentrations of the tested components by 2–4 times (Table 3 and Figure 7). At this point, it is worth mentioning that this effect could be potentially stronger in relation to antibiotic-resistant strains of H. pylori; however, the necessity to use a genomically characterized reference strain in these studies forced us to choose an antibiotic-sensitive strain. As we noticed positive interactions in anti-H. pylori activity for all tested antibiotics when combined with dioscin, we suspected that the mechanism of sensitizing bacterial cells to these antibiotics could be non-specific and related rather to the anti-virulent activity of dioscin, i.e., reduction of autoaggregation and biofilm formation or production of antibiotic-tolerant coccoid forms.
Table 3.
Numerical values showing interactions between dioscin and the three classically used antibiotics (clarithromycin [CLR], metronidazole [MTZ], and levofloxacin [LEV]) in antibacterial activity against H. pylori J99. Interactions were determined using the checkerboard assay.
| Tested Combination |
MIC [µg/mL] | FICI (Outcome) |
|||||
|---|---|---|---|---|---|---|---|
| Dioscin | Antibiotic | ||||||
| Alone | Combination | Fold Change | Alone | Combination | Fold Change | ||
| Dioscin + CLR |
64 | 32 | 2 | 0.025 | 0.006 | 4 | 0.75 (additive) |
| Dioscin + MTZ | 64 | 16 | 4 | 1 | 0.5 | 2 | 0.75 (additive) |
| Dioscin + LEV |
64 | 32 | 2 | 1 | 0.5 | 2 | 1.0 (additive) |
The fractional inhibitory concentration index (FICI) values of ≤ 0.5, > 0.5 to ≤ 1, and > 1 to ≤ 4 were expressed as synergistic, additive, and neutral, respectively.
Figure 7.
Graphics showing the interactions between dioscin and the three classically used antibiotics (clarithromycin [CLR], metronidazole [MTZ], and levofloxacin [LEV]) in antibacterial activity against H. pylori J99. Interactions were determined using the checkerboard assay. FICI values of ≤ 0.5, > 0.5 to ≤ 1, and > 1 to ≤ 4 were expressed as synergistic, additive, and neutral, respectively.
2.6. Concluding Computational Studies
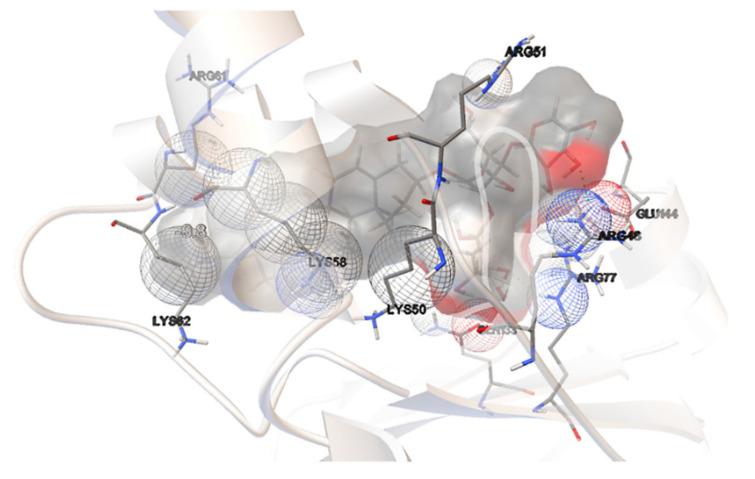
Among the compounds tested, dioscin was shown to be a very promising candidate for reducing H. pylori biofilm formation and sensitizing to certain antibiotics. This substance was shown to be the most promising during molecular docking experiments since it had one of the lowest binding free energies of −9.8 kcal/mol. Only madecassoside was discovered to have a lower value (−10.6 kcal/mol) and shall be considered for further studies as a potent SpoT inhibitor.
Figure 8 illustrates our hit’s interactions with the binding pocket, using coloring schemes adequate to those discussed earlier. The hit and natural ligands shared four amino acids: ARG77, ARG61, GLN133, and ARG48. Although just two hydrogen bonds, interacting with dioscin saccharides, have been discovered in this binding mechanism, they arise from the amino groups of ARG48 and ARG77, just as they do for ATP and GTP, suggesting their relevance for the binding mode. Instead, we observed a number of van der Waals interactions, which underlined that the binding mechanism is strongly hydrophobic.
Figure 8.
Dioscin interactions with the binding pocket of a SpoT protein of H. pylori.
2.7. Limitations and Future Perspectives
In the last part of this article, we would like to highlight some limitations of our research. The results of molecular docking on the inhibition of SpoT from H. pylori by selected phytochemical compounds have been confirmed in experiments with indirect methods. The inclusion of these methods was related to the supervisory function of SpoT in the survivability of H. pylori, i.e., biofilm formation, morphological transformation, and antibiotic resistance. The capacity of dioscin to interfere with the above processes was demonstrated by reducing the amount of biofilm under stationary conditions (a crystal violet-based staining) and flow conditions (a microfluidic Bioflux system), low stimulation of coccoid formation (light and electron microscopy), and enhancement of the antibiotics’ activity (checkerboard assays). To directly confirm the ability of dioscin to inhibit SpoT of H. pylori, we should finally extend our research to the purified protein and isothermal titration calorimetry assays, which we intend to do in the future. In our opinion, this article may be an important voice in raising the scientific community’s awareness to the potential of SpoT in promoting H. pylori survival. The first reports on SpoT of H. pylori were published relatively long ago—2006 [30,31], but the continuation of research on this issue was averted for over a decade, and just in recent years has slowly reappeared [32,33,34]. We hope that the results presented in the current article, although preliminary, may help in the search for alternative therapies to combat the virulence of this pathogen.
3. Materials and Methods
3.1. Computational Analysis
3.1.1. Bioinformatic Protocol
Due to the lack of the crystal structure of SpoT protein for any H. pylori strain at the time of writing this paper, a homology modeling pathway had to be undertaken. In the first step, an ATCC Genome Portal entry corresponding to the J99/ATCC 700824 strain [65] used within this study was thoroughly searched for the sequence encoding the SpoT protein. Since none of the identified product was classified as such, the UniProt database [66] was explored for the known sequence responsible for the aforementioned activity. The found entry (O25466), corresponding to the HP_0775 gene in H. pylori (strain ATCC 700392/26695) [67], was submitted for BLAST procedure [68], using BLOSUM62 matrix [69], against the genome of J99, yielding a sequence described by the satisfying results of 98% identity, 99% positivity, and 0% gaps.
It was trimmed out and used as an input for ThreaDomEx [70] to establish domains. With the cutoff of 0.56, five were identified, and each was submitted for I-TASSER [71,72,73,74], known for its continuous success in CASP experiments [75], to predict their structure and the activity resulting from it. Based on the anticipated GO:Scores, we decided to pick the first model of the third domain for further studies as the only one possessing a positive and satisfying C-Score (1.24), and the only one expressing kinase activity (GO:0016301).
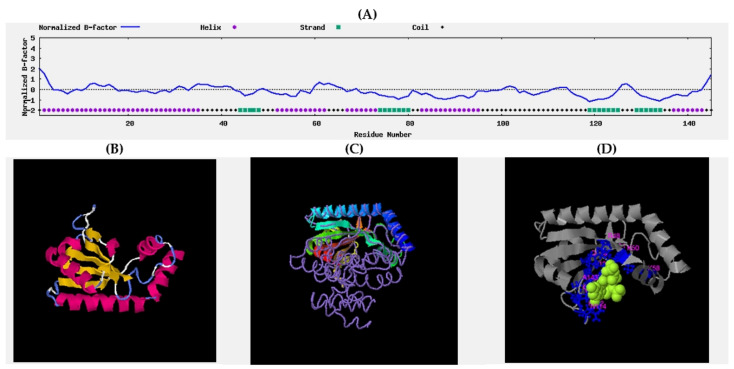
The predicted secondary structure of the protein was satisfying, both with a confidence score average of 7.5 and a predicted normalized B-factor (Figure 9) [76,77] mostly below 0, which indicated that most of the residues are relatively stable in the structure. Three entries were found in the top 10 threading templates: 1vj7A (6 times), 5iqrA (2 times), and 5kpvA (2 times), and their corresponding Z-scores of 4.43, 4.09, and 3.32 indicated very good alignment between sequences. Importantly, 1vj7A represents a bifunctional catalytic fragment of RelSeq, the RelA/SpoT homolog from Streptococcus equisimilis. Four final tertiary structures were predicted, but only the first one represented a satisfying C-score parameter of 1.24, whereas the range may vary from −5 to 2, thus pointing to its satisfyingly correct global topology; corresponding to a TM-score equaling 0.88 ± 0.07 and a corresponding RMSD = 2.3 + 1.8 Å.
Figure 9.
Supporting graphs: (A) B-factor plot; (B) homology model; (C) structure superposition of modeled protein (cartoon) and best hit—1vj7A (coil); (D) predicted ligand binding site.
Apart from the structural features, function was also established. Although the highest C-score was only 0.40, the corresponding ligand was found to be guanosine 5′-(tetrahydrogen triphosphate) 3′-(trihydrogen diphosphate). This corresponds with the established EC numbers that represent activities of GTP diphosphokinase and guanosine-3′,5′-bis(diphosphate) 3′-diphosphatase and a GO:Score of 0016301 as one expressing kinase activity (C-score of all of these was 0.450).
3.1.2. Molecular Dynamics
The obtained domain was submitted for MD simulation using Gromacs 2019.4. [78]. Its topology was prepared with CHARMM36m forcefield [79]. The domain was centered in a rectangular box, with 10.0 Å distance from the edge, and surrounded by TIP3 water molecules. Next, the system was neutralized with Na+ and Cl- ions placed randomly using the Monte Carlo algorithm [80]. The temperature of the system was set to 310.15 K (37 °C) to resemble that in the stomach.
Energy minimization was performed using the steepest descend algorithm. The minimized system was then submitted for NVT ensemble equilibration, which was carried out at 125 ps and 310.15 K temperature using a Nose-Hoover thermostat [81,82]. The production was performed in 1 ns of NPT ensemble, with the temperature controlled as previously, and pressure maintained at 1 bar with Parrinello-Rahman pressure coupling [83].
At each step, electrostatic interactions were subjected to the Particle Mesh Ewald method [84] with a cut-off value of 1.2 Å; the same cut-off value was set for Van der Waals interactions. The LINCS algorithm was used to convert bonds with hydrogen atoms to constraints [85].
3.1.3. Virtual Screening
Due to the unknown active site, it was necessary to evaluate where the natural ligands (ATP and GTP) bind [86,87]. Firstly, a conformational search procedure was utilized in a molecular dynamic framework implemented in Gabedit [88], and three conformers of each of these molecules were obtained. These were submitted for optimization and frequency calculations in Gaussian quantum chemistry software package (revision D.01) [89], using density functional theory method B3LYP in 6-311++G(d,p) basis set, accompanied with GD3 dispersion correction [90] and SMD water solvent [91]. No imaginary frequency was present. The lowest one for each was chosen for the molecular docking procedure performed with Autodock Vina [92]. The water molecules were removed from the MD output and Gasteiger chargers added. Grid box size was set to cover the entire domain, and the docking procedure was started.
Having detected the eventual binding site, the grid box was moved and resized to cover it. Afterwards, an in-house library consisting of 791 natural substances belonging to the collection of phytochemicals from the Department of Pharmacognosy and Herbal Medicines, Wroclaw Medical University, Poland was submitted for a virtual screening procedure, generating a list of the most probable inhibitors.
The inhibition constant (Ki) was obtained from the binding free energy of the compound (ΔG) using the equation:
| Ki = exp(ΔG/RT) |
where, R is the gaseous constant (1.987 cal K−1 mol−1) and T is the temperature (310.15 K).
3.2. Laboratory Analysis
3.2.1. Storage and Revival of Bacteria
The research was conducted using the reference H. pylori J99 (ATCC 700824) strain, obtained by courtesy of Dr. Anna Pawilk from Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences. Bacteria were stored in Tryptic Soy Broth (TSB; Oxoid, Dardilly, France) supplemented with 15% glycerol at −70 °C [93,94]. After thawing, the tested bacterial strain was cultured on Columbia agar (Difco, Lublin, Poland) with 7% horse blood. The bacteria were grown for 3 to 5 days at 37 °C in a microaerophilic atmosphere (Genbox microaer kits, BioMerieux, Marcy-l’Étoile, France), and eventually sub-cultured again in the same conditions if needed.
3.2.2. Antibacterial Activity Determination
The designation of antibacterial activity of 10 selected, plant-originated compounds (dioscin, aescin, glycyrrhetinic acid, ginsenoside Rg3, procyanidin A2 and C1, hypericin, robinin, amentoflavone, and zeaxanthin; all from Sigma-Aldrich, St. Louis, MO, USA) against H. pylori J99 was made by checking their minimum inhibitory concentrations (MICs) [93,94]. To obtain this, bacteria in a density of 107 CFU/mL were cultured in 12-well titration plates (Bionovo, Legnica, Poland) filled with 1 mL of Brain Heart Infusion broth (BHI; Oxoid, Dardilly, France) with 5% fetal calf serum (FCS; Gibco, Paisley, Scotland, UK) and a concentration gradient of the tested compounds (2–256 µg/mL). The titration plates were then followed for 3 days of microaerophilic incubation at 37 °C and 100 rpm shaking (MaxQ 6000, ThermoFisher, Waltham, MA, USA). Then, bacterial turbidity in each well of the plates was determined visually, and the lowest concentration with the lack of growth was considered as the MIC.
3.2.3. Impact on the Morphology and/or Morphostructure
The screening determination of the effect of high concentrations (64–256 µg/mL) of all 10 tested compounds on the H. pylori morphology was performed using light microscopy combined with a Gram-staining [95]. For each sample, bacterial suspension in a volume of 50 μL was dropped onto a coverslip and stained by Gram’s method. From each of the obtained preparations, 100 cells from three to five different view pools were counted and classified as spiral/rod-shaped or coccoid forms. Observations were made under the Olympus BX50 light microscope (Olympus Optical, Tokyo, Japan), using a 100× oil immersion objective with numerical aperture of 1.3.
In order to more accurately define the impact of dioscin on H. pylori J99 cells, bacteria were exposed to different concentrations of the compound and their morphostructure was visualized by scanning electron microscopy [95]. For this purpose, each well of the 12-well titration plates was filled with 1 mL BHI broth with 5% fetal calf serum containing 16 µg/mL, 32 µg/mL, and 64 µg/mL of dioscin (representing ¼ × MIC, ½ × MIC and MIC, respectively) and inoculated with H. pylori (107 CFU/mL). Such plates were then followed for a 72-h incubation period at 37 °C under microaerophilic conditions. Untreated bacteria were a positive control. After incubation, each bacterial culture was centrifugated, and the pellets were fixed with 4% glutaraldehyde and post-fixed in 2% OsO4. Fixed samples were dehydrated in ethanol-acetone series and, air-dried, and a silver coating was sputtered on the bacteria surface, providing optimal conductivity. SEM microphotographs were obtained by a DualBeam SEM/Xe-PFIB FEI Helios G4 PFIB CXe microscope (Thermo Fisher Scientific, Eindhoven, Netherlands).
3.2.4. Anti-Biofilm Activity Determination
Activity in Static Conditions
To assess the anti-biofilm activity of all 10 tested compounds against H. pylori J99, the crystal violet staining method was applied [96]. Bacteria in a final density of 107 CFU/mL were suspended in 1 mL of BHI broth with 5% fetal calf serum and a gradient concentration (2–256 µg/mL) of the tested compounds, and were cultured for 3 days under microaerophilic atmosphere, 37 °C, and 50 rpm shaking. Then, the entire bacterial suspension was removed from each well, and the plates were dried and stained with a 0.1% crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA) for 15–30 min. The dye was removed, and the wells were then rinsed twice with a phosphate buffer solution (PBS; Sigma-Aldrich, St. Louis, MO, USA) and flushed with 1 mL of 96% ethanol (Stanlab, Lublin, Poland). Two hundred microliters of crystal violet-stained alcohol was withdrawn from each well, transferred to a 96-well microtiter plate (Bionovo, Legnica, Poland), and measured for absorbance at OD590 with an Asys UVM 340 microplate reader (Biochrom Ltd., Cambridge, UK). The negative control consisted of wells without bacteria (the absorbance of these samples was used each time to subtract background from the tested samples).
Activity in the Flow Conditions
Confirmation of the anti-biofilm activity of dioscin against H. pylori J99 was performed under flow conditions created by the Bioflux 1000 system (Fluxion, San Francisco, CA, USA) [57]. To generate these conditions, 48-well low-shear plates composed of inlet and outlet wells connected by a capillary (Fluxion, San Francisco, CA, USA) were used. In the first stage, inlet wells were filled with 900 µL of BHI broth with 5% fetal calf serum and the desired concentration of dioscin (16 µg/mL, 32 µg/mL, and 64 µg/mL, representing ¼ × MIC, ½ × MIC, and MIC, respectively). Wells without dioscin served as a positive control. In the next stage, the above-mentioned medium was flowed through capillaries with a pressure of 10 dyne/cm2 for 10 sec, allowing components of the medium to cover the capillaries’ surfaces. Then, 100 µL of bacterial suspension (108 CFU/mL) was placed into inlet wells to obtain the final bacterial density of 107 CFU/mL in each well. A 1-day flow of the medium at 37 °C and microaerophilic atmosphere (Pecon Incubator XL S1, Carl Zeiss, Jena, Germany) was then directed from the input to the output wells with a pressure of 0.1 dyne/cm2. Following this, the flow of medium was interrupted, and the remaining medium from inlet wells was taken. The emptied inlet wells were filled with 100 µL of dye solution made from a LIVE/DEAD kit (Thermo Fisher, Waltham, MA, USA). The flow of 0.1 dyne/cm2 was turned on again to provide a 1-h staining of bacterial microcolonies and biofilms. After this, photos from capillaries were obtained with an inverted Axio Observer 7 microscope (GmbH, Carl Zeiss, Jena, Germany). Both the degree of capillary overgrowth and the ratio of green/red fluorescence (the ratio of live/dead bacteria) was determined using the Bioflux Montage software (Fluxion, San Francisco, CA, USA).
3.2.5. Checkerboard Assays
The determination of interactions between dioscin and the most commonly used antibiotics (clarithromycin, metronidazole, and levofloxacin; all from Sigma-Aldrich, St. Louis, MO, USA) was achieved with checkerboard assays [96]. All experiments were carried out in four combined 12-well plates, creating a 48-well panel. Each well of these plates was loaded with 1 mL of BHI broth with 5% fetal calf serum, a bacterial suspension of 107 CFU/mL, and a mixture of both tested substances (dioscin and one of the above-mentioned antibiotics). The concentration gradients of antibiotics (MIC to 1/16 × MIC) and dioscin (MIC to 1/64 × MIC) were generated in the vertical and horizontal direction, respectively. Plates with such content were followed for a 3-day period of microaerophilic culture at 37 °C with 100 rpm shaking. The interactions between dioscin and each of the antibiotics were checked by calculating a fractional inhibitory concentration index (FICI). The FICIs with values of ≤ 0.5, > 0.5 to ≤ 1, and > 1 to ≤ 4 were expressed as synergistic, additive, and neutral, respectively [27].
3.2.6. Statistical Analysis
Calculations were performed using the GraphPad Prism version 9 software (GraphPad Co., San Diego, CA, USA). The normality of distribution was assessed by means of the Shapiro–Wilk test. As all values were normally distributed, the one-way ANOVA test was further applied. The results of statistical analyses were considered significant if they produced p-values < 0.05.
4. Conclusions
Using in silico studies and in vitro laboratory experiments, we proved that dioscin has a number of favorable therapeutic parameters against H. pylori, including antimicrobial activity, ability to interfere with biofilm formation under stationary and flow conditions, and additive enhancement of antibacterial capacity of all three commonly used antibiotics (clarithromycin, metronidazole, and levofloxacin). Based on these results, we suggest that dioscin may be an interesting candidate for new therapies targeting H. pylori survivability and virulence.
Acknowledgments
Molecular dynamics and virtual screening were carried out with the computational computer of the Department of Pharmacognosy and Herbal Medicines, Wroclaw Medical University, Poland. Quantum chemistry studies were performed in Poznan Supercomputing and Networking Center (grant number 467), and SEM images were obtained in cooperation with the nanotechnology laboratory Nanores Sp. z o. o. Sp. K in Wroclaw, Poland.
Supplementary Materials
Table S1: List of phytochemicals tested by virtual screening with their binding free energies required to attach to the active site of the SpoT protein of H. pylori; Table S2. Results of literature search showing a low or acceptable toxicity profile of phytochemicals selected for testing in the current study [97,98,99,100,101,102,103,104,105,106,107,108]; Figure S1: Representative photos presenting bacterial morphology observed after a 72-h exposure of H. pylori J99 to the tested compounds; Figure S2: Representative photos obtained during time-lapse microscopy recordings from experiments determining anti-biofilm activity of dioscin under flow conditions against H. pylori J99.
Author Contributions
Conceptualization, P.K., M.S., E.D. and Z.S.; methodology, P.K. and M.S.; software, M.S.; validation, P.K. and E.D.; formal analysis, P.K. and M.S.; investigation, P.K., M.S., E.D. and R.A.; writing—original draft preparation, P.K. and M.S.; writing—review and editing, P.K., M.S., E.D., Z.S. and R.A.; visualization, P.K. and M.S.; supervision, P.K., M.S., E.D. and Z.S.; funding acquisition, P.K. and E.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Wroclaw Medical University, grant numbers SUB.A130.19.021 and SUB.A130.21.031. The Bioflux system was funded the National Centre for Research and Development (NCBiR) No. IA/SP/453975/2020.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds dioscin, aescin, glycyrrhetinic acid, ginsenoside Rg3, procyanidin A2 and C1, hypericin, robinin, amentoflavone, and zeaxanthin are available from Zbigniew Sroka (Department of Pharmacognosy and Herbal Medicines, Wroclaw Medical University. Poland).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ventola C.L. The antibiotic Resistance Crisis: Causes and Threats. PTJ. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 2.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wall S. Prevention of Antibiotic Resistance—An Epidemiological Scoping Review to Identify Research Categories and Knowledge Gaps. Glob. Health Action. 2019;12:1756191. doi: 10.1080/16549716.2020.1756191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin R. Imagining the Postantibiotic Future: The Visual Culture of a Global Health Threat. Med. Humanit. 2020 doi: 10.1136/medhum-2020-011884. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C.H., Hsieh Y.H., Powers Z.M., Kao C.Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020;21:1061. doi: 10.3390/ijms21031061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serwecińska L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water. 2020;12:3313. doi: 10.3390/w12123313. [DOI] [Google Scholar]
- 7.Schjørring S., Krogfelt K. Assessment of Bacterial Antibiotic Resistance Transfer in the Gut. Int. J. Microbiol. 2011;2011:312956. doi: 10.1155/2011/312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subirats J., Domingues A., Topp E. Does Dietary Consumption of Antibiotics by Humans Promote Antibiotic Resistance in the Gut Microbiome? J. Food Prot. 2019;82:1636–1642. doi: 10.4315/0362-028X.JFP-19-158. [DOI] [PubMed] [Google Scholar]
- 9.Contreras-Omaña R., Escorcia-Saucedo A., Velarde-Ruiz Velasco J. Prevalence and Impact of Antimicrobial Resistance in Gastrointestinal Infections: A Review. Rev. Gastroenterol. Mex. 2021;86:265–275. doi: 10.1016/j.rgmxen.2021.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 11.Haley K.P., Gaddy J.A. Helicobacter pylori: Genomic Insight into the Host-Pathogen Interaction. Int. J. Genomics. 2015;2015:386905. doi: 10.1155/2015/386905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagoonee S., Pellicano R. Helicobacter pylori: Molecular Basis for Colonization and Survival in Gastric Environment and Resistance to Antibiotics. A Short Review. Infect. Dis. 2019;51:399–408. doi: 10.1080/23744235.2019.1588472. [DOI] [PubMed] [Google Scholar]
- 13.Krzyżek P., Grande R., Migdał P., Paluch E., Gościniak G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens. 2020;9:1062. doi: 10.3390/pathogens9121062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hathroubi S., Servetas S.L., Windham I., Merrell D.S., Ottemann K.M. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018;82:e00001-18. doi: 10.1128/MMBR.00001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonezawa H., Osaki T., Kamiya S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. Biomed Res. Int. 2015;2015:914791. doi: 10.1155/2015/914791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krzyżek P., Grande R. Transformation of Helicobacter pylori into Coccoid Forms as a Challenge for Research Determining Activity of Antimicrobial Substances. Pathogens. 2020;9:184. doi: 10.3390/pathogens9030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krzyżek P., Gościniak G. Morphology of Helicobacter pylori as a Result of Peptidoglycan and Cytoskeleton Rearrangements. Prz. Gastroenterol. 2018;13:182–195. doi: 10.5114/pg.2018.78284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reshetnyak V.I., Reshetnyak T.M. Significance of Dormant Forms of Helicobacter pylori in Ulcerogenesis. World J. Gastroenterol. 2017;23:4867–4878. doi: 10.3748/wjg.v23.i27.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cammarota G., Sanguinetti M., Gallo A., Posteraro B. Review article: Biofilm Formation by Helicobacter pylori as a Target for Eradication of Resistant Infection. Aliment. Pharmacol. Ther. 2012;36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 20.Gladyshev N., Taame M., Kravtsov V. Clinical and Laboratory Importance of Detecting Helicobacter pylori Coccoid Forms for the Selection of Treatment. Prz. Gastroenterol. 2020;15:294–300. doi: 10.5114/pg.2020.101557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talele T., Khedkar S., Rigby A. Successful Applications of Computer Aided Drug Discovery: Moving Drugs from Concept to the Clinic. Curr. Top. Med. Chem. 2010;10:127–141. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- 22.Cui W., Aouidate A., Wang S., Yu Q., Li Y., Yuan S. Discovering Anti-Cancer Drugs via Computational Methods. Front. Pharmacol. 2020;11:733. doi: 10.3389/fphar.2020.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrella C., Farioli-Vecchioli S., Cisale G.Y., Strimpakos G., Borg J.J., Ceccanti M., Fiore M., Monteleone G., Nisticò R. A Healthy Gut for a Healthy Brain: Preclinical, Clinical and Regulatory Aspects. Curr. Neuropharmacol. 2020;19:610–628. doi: 10.2174/1570159X18666200730111528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Wang Y., Byrne R., Schneider G., Yang S. Concepts of Artificial Intelligence for Computer-Assisted Drug Discovery. Chem. Rev. 2019;119:10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 25.Rasouli H., Farzaei M.H., Khodarahmi R. Polyphenols and Their Benefits: A Review. Int. J. Food Prop. 2017;20:1700–1741. doi: 10.1080/10942912.2017.1354017. [DOI] [Google Scholar]
- 26.Gorlenko C.L., Kiselev H.Y., Budanova E.V., Zamyatnin A.A., Ikryannikova L.N. Plant Secondary Metabolites in the Battle of Drugs and Drug-Resistant Bacteria: New Heroes or Worse Clones of Antibiotics? Antibiotics. 2020;9:170. doi: 10.3390/antibiotics9040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krzyżek P., Paluch E., Gościniak G. Synergistic Therapies as a Promising Option for the Treatment of Antibiotic-Resistant Helicobacter pylori. Antibiotics. 2020;9:658. doi: 10.3390/antibiotics9100658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonifácio B.V., dos Santos Ramos M.A., da Silva P.B., Bauab T.M. Antimicrobial Activity of Natural Products against Helicobacter pylori: A Review. Ann. Clin. Microbiol. Antimicrob. 2014;13:54. doi: 10.1186/s12941-014-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinchen W., Zegarra V., Bange G. (p)ppGpp: Magic Modulators of Bacterial Physiology and Metabolism. Front. Microbiol. 2020;11:2072. doi: 10.3389/fmicb.2020.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells D.H., Gaynor E.C. Helicobacter pylori Initiates the Stringent Response upon Nutrient and pH Downshift. J. Bacteriol. 2006;188:3729. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouery K., Rader B.A., Gaynor E.C., Guillemin K. The Stringent Response is Required for Helicobacter pylori Survival of Stationary Phase, Exposure to Acid, and Aerobic Shock. J. Bacteriol. 2006;188:5494–5500. doi: 10.1128/JB.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng X., Li W., Chen Z., Gao S., Hong W., Ge X., Hou G., Hu Z., Zhou Y., Zeng B., et al. The Bifunctional Enzyme SpoT Is Involved in the Clarithromycin Tolerance of Helicobacter pylori by Upregulating the Transporters HP0939, HP1017, HP0497, and HP0471. Antimicrob. Agents Chemother. 2017;61:e02011-16. doi: 10.1128/AAC.02011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge X., Cai Y., Chen Z., Gao S., Geng X., Li Y., Li Y., Jia J., Sun Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP) Antimicrob. Agents Chemother. 2018;62:e00957-18. doi: 10.1128/AAC.00957-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poursina F., Fagri J., Mirzaei N., Safaei H.G. Overexpression of spoT Gene in Coccoid Forms of Clinical Helicobacter pylori Isolates. Folia Microbiol. 2018;63:459–465. doi: 10.1007/s12223-017-0557-0. [DOI] [PubMed] [Google Scholar]
- 35.Yao H., Hu C., Yin L., Tao X., Xu L., Qi Y., Han X., Xu Y., Zhao Y., Wang C., et al. Dioscin Reduces Lipopolysaccharide-Induced Inflammatory Liver Injury via Regulating TLR4/MyD88 Signal Pathway. Int. Immunopharmacol. 2016;36:132–141. doi: 10.1016/j.intimp.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Zhu H., Yang X. Dioscin Exhibits Anti-inflammatory Effects in IL-1β-Stimulated Human Osteoarthritis Chondrocytes by Activating LXRα. Immunopharmacol. Immunotoxicol. 2020;42:340–345. doi: 10.1080/08923973.2020.1775248. [DOI] [PubMed] [Google Scholar]
- 37.Wu S., Xu H., Peng J., Wang C., Jin Y., Liu K., Sun H., Qin J. Potent Anti-inflammatory Effect of Dioscin Mediated by Suppression of TNF-α-Induced VCAM-1, ICAM-1 and EL Expression via the NF-κB Pathway. Biochimie. 2015;110:62–72. doi: 10.1016/j.biochi.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Ma T., Wang R., Zou X. Dioscin Inhibits Gastric Tumor Growth through Regulating the Expression Level of lncRNA HOTAIR. BMC Complement. Altern. Med. 2016;16:383. doi: 10.1186/s12906-016-1360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P., Wang C., Liu C. Antitumor Effects of Dioscin in A431 Cells via Adjusting ATM/p53-Mediated Cell Apoptosis, DNA Damage and Migration. Oncol. Lett. 2021;21:59. doi: 10.3892/ol.2020.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He S., Yang J., Hong S., Huang H., Zhu Q., Ye L., Li T., Zhang X., Wei Y., Gao Y. Dioscin Promotes Prostate Cancer Cell Apoptosis and Inhibits Cell Invasion by Increasing SHP1 Phosphorylation and Suppressing the Subsequent MAPK Signaling Pathway. Front. Pharmacol. 2020;11:1099. doi: 10.3389/fphar.2020.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin H., Zhang M.-J., An R.-F., Zhou J., Liu W., Morris-Natschke S.L., Cheng Y.-Y., Lee K.-H., Huang X.-F. Diosgenin Derivatives as Potential Antitumor Agents: Synthesis, Cytotoxicity, and Mechanism of Action. J. Nat. Prod. 2020;84:616–629. doi: 10.1021/acs.jnatprod.0c00698. [DOI] [PubMed] [Google Scholar]
- 42.Barros Cota B., Batista Carneiro de Oliveira D., Carla Borges T., Cristina Catto A., Valverde Serafim C., Rogelis Aquiles Rodrigues A., Kohlhoff M., Leomar Zani C., Assunção Andrade A. Antifungal Activity of Extracts and Purified Saponins from the Rhizomes of Chamaecostus cuspidatus against Candida and Trichophyton Species. J. Appl. Microbiol. 2021;130:61–75. doi: 10.1111/jam.14783. [DOI] [PubMed] [Google Scholar]
- 43.Yang L., Liu X., Zhong L., Sui Y., Quan G., Huang Y., Wang F., Ma T. Dioscin Inhibits Virulence Factors of Candida albicans. Biomed Res. Int. 2018;2018:4651726. doi: 10.1155/2018/4651726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho J., Choi H., Lee J., Kim M.S., Sohn H.Y., Lee D.G. The Antifungal Activity and Membrane-Disruptive Action of Dioscin Extracted from Dioscorea nipponica. Biochim. Biophys. Acta Biomembr. 2013;1828:1153–1158. doi: 10.1016/j.bbamem.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Cong S., Tong Q., Peng Q., Shen T., Zhu X., Xu Y., Qi S. In Vitro Anti-bacterial Activity of Diosgenin on Porphyromonas gingivalis and Prevotella intermedia. Mol. Med. Rep. 2020;22:5392–5398. doi: 10.3892/mmr.2020.11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalska-Krochmal B., Dudek-Wicher R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens. 2021;10:165. doi: 10.3390/pathogens10020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samoilova Z., Muzyka N., Lepekhina E., Oktyabrsky O., Smirnova G. Medicinal Plant Extracts Can Variously Modify Biofilm Formation in Escherichia coli. Antonie Van Leeuwenhoek. 2014;105:709–722. doi: 10.1007/s10482-014-0126-3. [DOI] [PubMed] [Google Scholar]
- 48.Slobodníková L., Fialová S., Hupková H., Grančai D. Rosmarinic Acid Interaction with Planktonic and Biofilm Staphylococcus aureus. Nat. Prod. Commun. 2013;8:1747–1750. doi: 10.1177/1934578X1300801223. [DOI] [PubMed] [Google Scholar]
- 49.Vacheva A., Mustafa B., Staneva J., Marhova M., Kostadinova S., Todorova M., Ivanova R., Stoitsova S. Effects of Extracts from Medicinal Plants on Biofilm Formation by Escherichia coli Urinary Tract Isolates. Biotechnol. Biotechnol. Equip. 2014;25:92–97. doi: 10.5504/BBEQ.2011.0111. [DOI] [Google Scholar]
- 50.Luganini A., Terlizzi M.E., Catucci G., Gilardi G., Maffei M.E., Gribaudo G. The Cranberry Extract Oximacro® Exerts In Vitro Virucidal Activity against Influenza Virus by Interfering with Hemagglutinin. Front. Microbiol. 2018;9:1826. doi: 10.3389/fmicb.2018.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D., Deng J., Joshi S., Liu P., Zhang C., Yu Y., Zhang R., Fan D., Yang H., D’Souza D.H. Monomeric Catechin and Dimeric Procyanidin B2 against Human Norovirus Surrogates and their Physicochemical Interactions. Food Microbiol. 2018;76:353. doi: 10.1016/j.fm.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattio L.M., Dallavalle S., Musso L., Filardi R., Franzetti L., Pellegrino L., D’Incecco P., Mora D., Pinto A., Arioli S. Antimicrobial Activity of Resveratrol-Derived Monomers and Dimers against Foodborne Pathogens. Sci. Rep. 2019;9:19525. doi: 10.1038/s41598-019-55975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian S., Huiszoon R.C., Chu S., Bentley W.E., Ghodssi R. Microsystems for Biofilm Characterization and Sensing—A Review. Biofilm. 2020;2:100015. doi: 10.1016/j.bioflm.2019.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao R., Tong Z., Lin Y., Xue Y., Wang W., Kuang R., Wang P., Tian Y., Ni L. Antimicrobial and Antibiofilm Activity of Pleurocidin against Cariogenic Microorganisms. Peptides. 2011;32:1748–1754. doi: 10.1016/j.peptides.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Díez-Aguilar M., Morosini M.I., Köksal E., Oliver A., Ekkelenkamp M., Cantón R. Use of Calgary and Microfluidic BioFlux Systems To Test the Activity of Fosfomycin and Tobramycin Alone and in Combination against Cystic Fibrosis Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2017;62:e01650-17. doi: 10.1128/AAC.01650-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J., Xu C., Fang R., Cao J., Zhang X., Zhao Y., Dong G., Sun Y., Zhou T. Resistance and Heteroresistance to Colistin in Pseudomonas aeruginosa Isolates from Wenzhou, China. Antimicrob. Agents Chemother. 2019;63:e00556-19. doi: 10.1128/AAC.00556-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krzyżek P., Junka A., Słupski W., Dołowacka-Jóźwiak A., Płachno B.J., Sobiecka A., Matkowski A., Chodaczek G., Płusa T., Gościniak G., et al. Antibiofilm and Antimicrobial-Enhancing Activity of Chelidonium majus and Corydalis cheilanthifolia Extracts against Multidrug-Resistant Helicobacter pylori. Pathogens. 2021;10:1033. doi: 10.3390/pathogens10081033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwechheimer C., Kuehn M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015;13:619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jan A.T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front. Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y., Defourny K.A.Y., Smid E.J., Abee T. Gram-positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease. Front. Microbiol. 2018;9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W., Chanda W., Zhong M. The Relationship between Biofilm and Outer Membrane Vesicles: A Novel Therapy Overview. FEMS Microbiol. Lett. 2015;362:fnv117. doi: 10.1093/femsle/fnv117. [DOI] [PubMed] [Google Scholar]
- 62.Mozaheb N., Mingeot-Leclercq M.-P. Membrane Vesicle Production as a Bacterial Defense Against Stress. Front. Microbiol. 2020;11:600221. doi: 10.3389/fmicb.2020.600221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray B.O., Dawson R.A., Alsharaf L.M., Winter J.A. Protective Effects of Helicobacter pylori Membrane Vesicles against Stress and Antimicrobial Agents. Microbiology. 2020;166:751–758. doi: 10.1099/mic.0.000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malfertheiner P., Megraud F., O’Morain C.A., Gisbert J.P., Kuipers E.J., Axon A.T., Bazzoli F., Gasbarrini A., Atherton J., Graham D.Y., et al. Management of Helicobacter pylori Infection—The Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 65.ATCC Helicobacter pylori (Marshall et al.) Goodwin et al. [(accessed on 21 October 2021)]. Available online: https://www.atcc.org/products/700824.
- 66.Consortium T.U., Bateman A., Martin M.-J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.HP_0775—Penta-Phosphate Guanosine-3′-pyrophosphohydrolase (SpoT)—Helicobacter pylori (strain ATCC 700392/26695)—_0775 Gene & Protein. [(accessed on 21 October 2021)]. Available online: https://www.uniprot.org/uniprot/O25466.
- 68.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 69.Henikoff S., Henikoff J.G. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Wang J., Li R., Shi Q., Xue Z., Zhang Y. ThreaDomEx: A Unified Platform for Predicting Continuous and Discontinuous Protein Domains by Multiple-Threading and Segment Assembly. Nucleic Acids Res. 2017;45:W400–W407. doi: 10.1093/nar/gkx410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roy A., Kucukural A., Zhang Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J., Zhang Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moult J., Pedersen J.T., Judson R., Fidelis K. A Large-Scale Experiment to Assess Protein Structure Prediction Methods. Proteins Struct. Funct. Bioinform. 1995;23:2–4. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 76.Johnson T.W., Gallego R.A., Brooun A., Gehlhaar D., McTigue M. Reviving B-Factors: Retrospective Normalized B-Factor Analysis of c-ros Oncogene 1 Receptor Tyrosine Kinase and Anaplastic Lymphoma Kinase L1196M with Crizotinib and Lorlatinib. ACS Med. Chem. Lett. 2018;9:878–883. doi: 10.1021/acsmedchemlett.8b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J., Wang Y., Zhang Y. ResQ: An Approach to Unified Estimation of B-Factor and Residue-Specific Error in Protein Structure Prediction. J. Mol. Biol. 2016;428:693–701. doi: 10.1016/j.jmb.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Páll S., Abraham M.J., Kutzner C., Hess B., Lindahl E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. Lect. Notes Comput. Sci. 2014;8759:3–27. [Google Scholar]
- 79.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kroese D.P., Brereton T., Taimre T., Botev Z.I. Why the Monte Carlo Method Is So Important Today. Wiley Interdiscip. Rev. Comput. Stat. 2014;6:386–392. doi: 10.1002/wics.1314. [DOI] [Google Scholar]
- 81.Nosé S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 2006;52:255–268. doi: 10.1080/00268978400101201. [DOI] [Google Scholar]
- 82.Hoover W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A. 1985;31:1695. doi: 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- 83.Parrinello M., Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1998;52:7182. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 84.Darden T., York D., Pedersen L. Particle Mesh Ewald: An N⋅log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1998;98:10089. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 85.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 86.Nel A.E., Mädler L., Velegol D., Xia T., Hoek E.M.V., Somasundaran P., Klaessig F., Castranova V., Thompson M. Understanding Biophysicochemical Interactions at the Nano–Bio Interface. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 87.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. Conformational Antagonism between Opposing Active Sites in a Bifunctional RelA/SpoT Homolog Modulates (p)ppGpp Metabolism during the Stringent Response. Cell. 2004;117:57–68. doi: 10.1016/S0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 88.Allouche A.-R. Gabedit—A Graphical User Interface for Computational Chemistry Softwares. J. Comput. Chem. 2011;32:174–182. doi: 10.1002/jcc.21600. [DOI] [PubMed] [Google Scholar]
- 89.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., et al. Gaussian 09, Revision A.02. [(accessed on 21 October 2021)]. Available online: https://gaussian.com/g09citation/
- 90.Grimme S., Antony J., Ehrlich S., Krieg H. A Consistent and Accurate ab initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 91.Marenich A.V., Cramer C.J., Truhlar D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B. 2009;113:6378–6396. doi: 10.1021/jp810292n. [DOI] [PubMed] [Google Scholar]
- 92.Trott O., Olson A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. In Vitro Activity of 3-Bromopyruvate, an Anti-Cancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers. 2019;11:229. doi: 10.3390/cancers11020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. In Vitro Activity of Sertraline, an Antidepressant, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Pathogens. 2019;8:228. doi: 10.3390/pathogens8040228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krzyżek P., Biernat M.M., Gościniak G. Intensive Formation of Coccoid Forms as a Feature Strongly Associated with Highly Pathogenic Helicobacter pylori Strains. Folia Microbiol. 2019;64:273–281. doi: 10.1007/s12223-018-0665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krzyżek P., Migdał P., Paluch E., Karwańska M., Wieliczko A., Gościniak G. Myricetin as an Antivirulence Compound Interfering with a Morphological Transformation into Coccoid Forms and Potentiating Activity of Antibiotics against Helicobacter pylori. Int. J. Mol. Sci. 2021;22:2695. doi: 10.3390/ijms22052695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L., Ren S., Xu F., Ma Z., Liu X., Wang L. Recent Advances in the Pharmacological Activities of Dioscin. Biomed Res. Int. 2019;2019:5763602. doi: 10.1155/2019/5763602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kang K.B., Ryu J., Cho Y., Choi S.Z., Son M., Sung S.H. Combined Application of UHPLC-QTOF/MS, HPLC-ELSD and 1H–NMR Spectroscopy for Quality Assessment of DA-9801, A Standardised Dioscorea Extract. Phytochem. Anal. 2017;28:185–194. doi: 10.1002/pca.2659. [DOI] [PubMed] [Google Scholar]
- 99.Karioti A., Bilia A.R. Hypericins as Potential Leads for New Therapeutics. Int. J. Mol. Sci. 2010;11:562–594. doi: 10.3390/ijms11020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schempp C.M., Winghofer B., Langheinrich M., Schöpf E., Simon J.C. Hypericin Levels in Human Serum and Interstitial Skin Blister Fluid after Oral Single-Dose and Steady-State Administration of Hypericum perforatum Extract (St. John’s Wort) Skin Pharmacol. Physiol. 1999;12:299–304. doi: 10.1159/000066256. [DOI] [PubMed] [Google Scholar]
- 101.Rizk Y.S., Santos-Pereira S., Gervazoni L., de Hardoim D.J., de Cardoso F.O., de Souza C.D.S.F., Pelajo-Machado M., Carollo C.A., de Arruda C.C.P., Almeida-Amaral E.E., et al. Amentoflavone as an Ally in the Treatment of Cutaneous Leishmaniasis: Analysis of Its Antioxidant/Prooxidant Mechanisms. Front. Cell. Infect. Microbiol. 2021;11:615814. doi: 10.3389/fcimb.2021.615814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suryavanshi S.V., Kulkarni Y.A. Toxicity of Escin-Triterpene Saponins from Aesculus. Toxicol. Environ. Chem. 2021 doi: 10.1080/02772248.2021.1996577. ahead of print. [DOI] [Google Scholar]
- 103.Cosmetic Ingredient Review Expert Panel Final Report on the Safety Assessment of Glycyrrhetinic Acid, Potassium Glycyrrhetinate, Disodium Succinoyl Glycyrrhetinate, Glyceryl Glycyrrhetinate, Glycyrrhetinyl Stearate, Stearyl Glycyrrhetinate, Glycyrrhizic Acid, Ammonium Glycyrrhizate, Dipotassium Glycyrrhizate, Disodium Glycyrrhizate, Trisodium Glycyrrhizate, Methyl Glycyrrhizate, and Potassium Glycyrrhizinate. Int. J. Toxicol. 2007;26((Suppl. 2)):79–112. doi: 10.1080/10915810701351228. [DOI] [PubMed] [Google Scholar]
- 104.Pan L., Zhang T., Sun H., Liu G. Ginsenoside Rg3 (Shenyi Capsule) Combined with Chemotherapy for Digestive System Cancer in China: A Meta-Analysis and Systematic Review. Evid. Based. Complement. Alternat. Med. 2019;2019:2417418. doi: 10.1155/2019/2417418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rauf A., Imran M., Abu-Izneid T., Lahtisham-Ul-Haq, Patel S., Pan X., Naz S., Sanches Silva A., Saeed F., Rasul Suleria H.A. Proanthocyanidins: A Comprehensive Review. Biomed. Pharmacother. 2019;116:108999. doi: 10.1016/j.biopha.2019.108999. [DOI] [PubMed] [Google Scholar]
- 106.Tsiklauri L., Švík K., Chrastina M., Poništ S., Dráfi F., Slovák L., Alania M., Kemertelidze E., Bauerova K. Bioflavonoid Robinin from Astragalus falcatus Lam. Mildly Improves the Effect of Metothrexate in Rats with Adjuvant Arthritis. Nutrients. 2021;13:1268. doi: 10.3390/nu13041268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Edwards J.A. Zeaxanthin: Review of Toxicological Data and Acceptable Daily Intake. J. Ophthalmol. 2016;2016:3690140. doi: 10.1155/2016/3690140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murillo A.G., Hu S., Fernandez M.L. Zeaxanthin: Metabolism, Properties, and Antioxidant Protection of Eyes, Heart, Liver, and Skin. Antioxidants. 2019;8:390. doi: 10.3390/antiox8090390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.