Abstract
Tumor mutational burden (TMB) is gaining attention as a biomarker for responses to immune checkpoint inhibitors in cancer patients. In this study, we evaluated the status of TMB in primary and liver metastatic lesions in patients with colorectal cancer (CRC). In addition, the status of TMB in primary and liver metastatic lesions was inferred by radiogenomics on the basis of computed tomography (CT) images. The study population included 24 CRC patients with liver metastases. DNA was extracted from primary and liver metastatic lesions obtained from the patients and TMB values were evaluated by next‐generation sequencing. The TMB value was considered high when it equaled to or exceeded 10/100 Mb. Radiogenomic analysis of TMB was performed by machine learning using CT images and the construction of prediction models. In 7 out of 24 patients (29.2%), the TMB status differed between the primary and liver metastatic lesions. Radiogenomic analysis was performed to predict whether TMB status was high or low. The maximum values for the area under the receiver operating characteristic curve were 0.732 and 0.812 for primary CRC and CRC with liver metastasis, respectively. The sensitivity, specificity, and accuracy of the constructed models for TMB status discordance were 0.857, 0.600, and 0.682, respectively. Our results suggested that accurate inference of the TMB status is possible using radiogenomics. Therefore, radiogenomics could facilitate the diagnosis, treatment, and prognosis of patients with CRC in the clinical setting.
Keywords: colorectal cancer, heterogeneity, metastasis, radiogenomics, tumor mutational burden
This study evaluated the status of tumor mutation burden (TMB) in primary and liver metastatic lesions in patients with colorectal cancer using radiogenomics. In 7 out of 24 patients (29.2%), the TMB status differed between the primary and liver metastatic lesion. Radiogenomic analysis was used to accurately predict whether TMB status was high or low.
Abbreviations
- CRC
colorectal cancer
- CT
computed tomography
- CTCs
circulating tumor cells
- ICI
immune checkpoint inhibitors
- MSI
microsatellite instability
- TMB
tumor mutational burden
- UICC
Union for International Cancer Control
1. INTRODUCTION
Barack Hussein Obama, the 44th President of the United States, announced Precision Medicine Initiative in his State of the Union Address in 2015, which highlighted the importance of precision medicine in cancer therapeutics. Treatment approaches based on data pertaining to gene mutations and amplification have become a common alternative in countries around the world. 1 However, the associated investigation requires time and high costs and not all patients can obtain the benefits. 2 Even if an antitumor drug is developed on the basis of precision medicine, it is often not effective for a majority of patients. 3
The causes of inadequate therapeutic effects are (a) substantial gene mutations or amplification was not detected, (b) antitumor drugs corresponding to the gene mutation is not available, and (c) the patient is intolerant to the drug. Tumor heterogeneity is considered to be a major barrier to personalized medicine. There are 3 types of cancer heterogeneity: (a) genetic heterogeneity in tumors, (b) genetic heterogeneity between primary and metastatic lesions, and (c) genetic changes in the tumor after treatment. 4 , 5 , 6
Radiogenomics is a new field of medical sciences that infers changes such as gene mutations and amplification from general imaging tests such as CT and MRI. 7 The images are considered to be qualitative data, but they are essentially digital data. Therefore, quantifying them using mathematical methods is possible by assuming that the images of the targeted tumors are a matrix of numbers. Numerous specific quantitative values therefore obtained are processed using a variety of analytical methods. This is called an image feature and can be used for various analyses. After quantifying gene mutations and expression levels in the target tumor on the basis of conventional molecular biology methods, the numerical values of the numerous features obtained from the images and the quantified genetic characteristics are integrated to determine underlying predictive and prognostic information. 8 , 9
In this study, we focused on tumor heterogeneity to examine the utility of radiogenomics. TMB indicates the amount of gene mutations that has occurred in the genome of cancer cells. 10 In tumors with high TMB, there is the possibility that inherent DNA repair mechanisms are abnormal and, as a result, gene mutations accumulate. 10 In such tumors, several antigens, recognized by the immune system as foreign, may be produced; therefore, the effect of ICI may be high. 11 , 12 , 13 , 14 In fact, a study has reported that TMB is high in tumors with high microsatellite instability (MSI). These tumors are the targets of ICIs in several carcinomas such as CRC. 15
In this study, we determined the TMB levels of primary and liver metastatic lesions in patients with CRC along with the discordance rate, which is the heterogeneity of TMB values among tumors within the same individual. We also attempted to predict TMB status using radiogenomics for primary and metastatic lesions of CRC.
2. MATERIALS AND METHODS
2.1. Patient characteristics
Patients who were treated at the Chiba Cancer Center between September, 2013 and December, 2020 were recruited for the study. The postoperative clinical course was followed and patients with synchronous or metachronous metastatic liver metastases were included in the study. Radical resection was performed for both the primary and metastatic lesions. Twenty‐four patients were examined retroactively. All patients were pathologically diagnosed with CRC and metastasis. All patients provided written informed consent, and the study was approved by our institutional review board (H29‐006).
2.2. TMB assay
Genomic DNA was extracted from 4 sections (5‐10 μm thick) of paraffin or frozen tissue blocks containing approximately 50‐100 mg of primary or liver metastatic lesion samples using the QIAamp DNA FFPE Tissue Kit (QIAGEN), in accordance with the manufacturer's instructions. The purified DNA was quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Inc). The Oncomine™ Tumor Mutation Load Assay (Thermo Fisher Scientific, Inc), which covers the exon region of 409 cancer‐related genes, was performed to measure the TMB. For each sample, 20 ng of genomic DNA was used for library preparation using the Ion AmpliSeq™ technology. The genomic DNA from the library was quantified using the Ion Library Quantification kit (Thermo Fisher Scientific, Inc), diluted to a final concentration of 8 pM, and amplified by emulsion PCR using the Ion OneTouch ES (Thermo Fisher Scientific, Inc). Target exome sequencing was performed using an Ion Proton sequencer (Thermo Fisher Scientific, Inc) with an Ion PI Chip v3. The sequenced reads were aligned to the human reference genome (hg19) using the Torrent Suite v5.6 software (Thermo Fisher Scientific, Inc) and the resulting BAM file was transferred to the Ion Reporter™ v5.12 and v5.14 (Thermo Fisher Scientific, Inc) for annotation and subsequent TMB calculation. For the detection of TMB values (Mutations / Mb), Oncomine™ Tumor Mutation Load – w3.0 and – w3.1 – DNA – Single Sample workflows (Thermo Fisher Scientific, Inc) were used. A population database (UCSC common single nucleotide polymorphism [SNP]) was used to eliminate germline mutations. The number of non‐synonymous single nucleotide variants and short insertion‐deletion mutations (InDels) with allele frequencies of 10% or greater and with a coverage of 300 or greater was divided by 1.2 Mb (exonic region covered by the panel) to calculate the number of somatic mutations per 1 Mb of patient's genome. A TMB of 10 mut/Mb or more was defined as a high TMB status in both the primary colon and metastatic liver lesions.
2.3. Computed tomography acquisition
All CT scans were performed using a 128‐detector‐row CT system (SOMATOM Definition Flash; Siemens). A contrast agent (Iopamidol, Iopamiron 300; Bayer; 100 mL) was administered through the superficial vein of the upper extremity using a power injector (bodyweight ≥55 kg; 150 mL injected at 4.5 mL/s, bodyweight <55 kg; 100 mL injected at 3.6 mL/s). For primary lesions, CT colonography was performed with the following parameters: tube voltage, 120 kVp; tube current, 210 mAs; pitch, 0.6; and resolution 0.68 × 0.68 × 5 mm. Images of the supine position were acquired 35 s after starting the contrast agent injection. Subsequently, images of the prone position were captured 300 s after the start of the injection. Non‐contrast‐enhanced and enhanced images were acquired for liver metastasis. The following imaging parameters were applied: tube voltage, 120 kVp; tube current, 200 mAs; pitch, 0.6; and resolution 0.68 × 0.68 × 5 mm. After the injection of the contrast agent, imaging slices were taken 70 s after the start of the injection.
The contrast agent could not be injected in 2 patients because of renal failure. Twenty‐two cases were analyzed using CT colonography and contrast‐enhanced CT for liver metastasis, whereas 24 cases were analyzed using non‐contrast‐enhanced CT for liver metastasis.
2.4. Tumor segmentation
A board‐certified diagnostic radiologist and surgeon (15 and 7 y of experience in pancreatic imaging, respectively) delineated the volume of interest in the primary colon and metastatic liver lesions with consensus. Contrast‐enhanced images of the supine and prone positions were used for primary lesions, whereas non–contrast‐enhanced and contrast‐enhanced images were used for liver metastasis. Each phase was segmented individually (Figure 1, 1) Tumor segmentation). In patients with multiple liver metastases, only the lesion whose TMB was evaluated was segmented.
FIGURE 1.
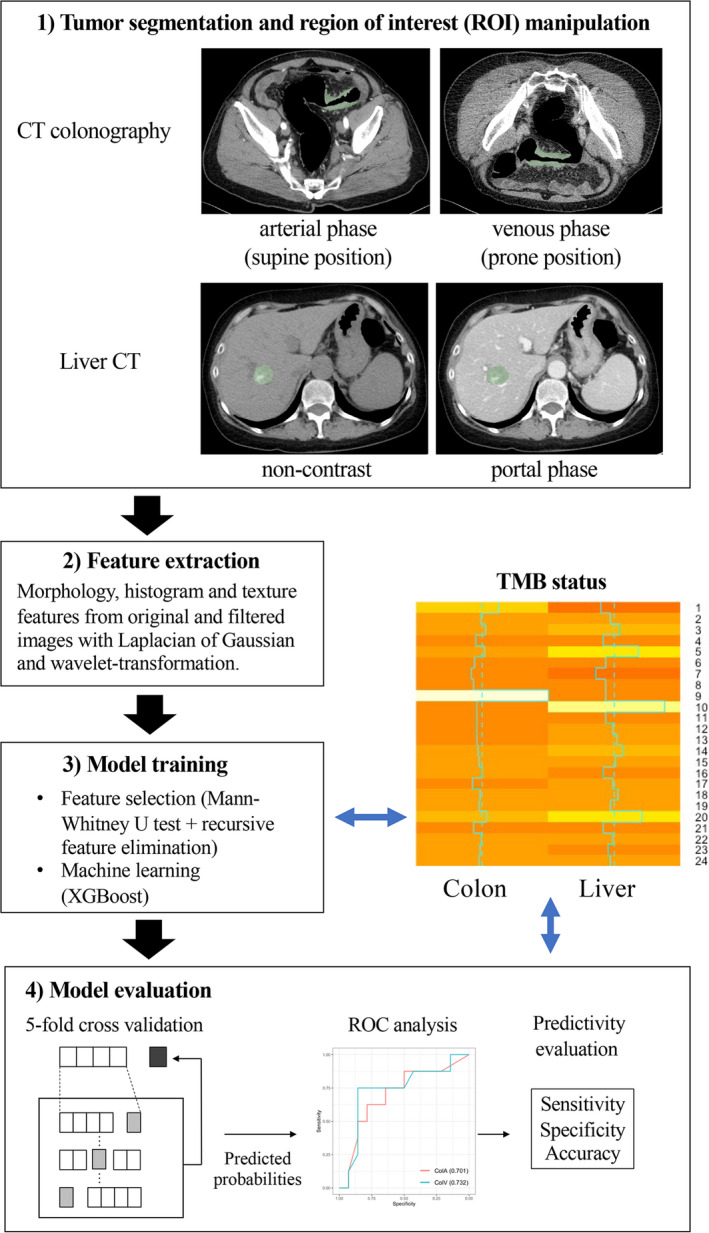
A summary of the steps involved in sample processing
2.5. Imaging feature extraction
Imaging features were extracted using an open‐source Python package, PyRadiomics v2.2.0 (http://www.radiomics.io/pyradiomics.html). 16 Pyradiomics can calculate various quantitative values from images using mathematical methods on the basis of morphological, histogram, and texture analyses. The quantitative values reflect the imaging characteristics of the tumor, such as heterogeneity. The absolute rescaling method (−150 to 500 Hounsfield units) was applied. The bin width was 20 Hounsfield units for making a histogram and gray‐level discretization of the image. Pixel values between the upper and lower limits were resampled into 64 levels and those outside the limits were truncated. The morphological, histogram, and texture features were calculated from the original images. The same types of features were extracted from the Laplacian of Gaussian filtered and wavelet‐transformed images. Finally, in total, 1037 features were extracted from each VOI (Figure 1, 2) Feature extraction).
2.6. Statistical analysis
The significance of the difference between the TMB status (positive/negative) and several clinical and pathologic variables was assessed using the chi‐square (χ2) test, Fisher exact test, or Mann‐Whitney U test. Overall survival (OS) was defined as the period between surgery and the final observation (in days). The 2 groups were defined on the basis of discordance in TMB status between the colon and liver. The discordance was considered positive if the colon had high TMB status and the liver had low TMB status or if the colon had low TMB status and the liver had high TMB status. A survival curve was plotted using the Kaplan‐Meier method, and the log‐rank test was used to assess significant differences between the groups. A P‐value of < .050 was considered significant.
2.7. Machine learning
To avoid collinearity, highly correlated features (Pearson r > .9) were removed. Feature selection consisted of 2 steps to stabilize the predictive power of the model. First, the Mann‐Whitney U test was performed on each imaging feature and only those with significant differences were retained. Second, another feature selection with recursive feature elimination was performed using a random forest function. Finally, 1037 features derived from the early and late phases were put into XGBoost to construct predictive models for TMB. The feature selection and model construction steps were performed using nested cross‐validation. Inner cross‐validation for feature selection and outer cross validations for model construction was 5 fold (Figure 1, 3) Model training).
2.8. Model evaluation
The mean output values of cross‐validation were used for a receiver operating characteristic (ROC) analysis (Figure 1, 4) Model evaluation). To evaluate the survival prediction of machine learning models, cutoff values were defined from the maximum point of the Youden index (ie, sensitivity + specificity − 1). The predictivity of machine learning for TMB status discordance between the colon and liver was calculated at the thresholds computed by the ROC analyses.
All statistical analyses and machine learning were conducted using R version 4.1.0 (R Foundation for Statistical Computing, Vienna, Austria).
3. RESULTS
3.1. Patient background
The observation period lasted from January 2013 to July 2021 with a median of 1063 d (247‐2485 d) from the first surgery of the primary tumor. The median age of the participants was 64.8 y (range, 46‐82 y) and the sex ratio was 12:12. The curative resection was R0 in all cases of primary tumor and liver metastasis. Venous invasion was negative in only 1 case. Lymph node metastasis was observed in 12 patients. As per the classification by Union for International Cancer Control (UICC), 8th edition, T3 was the maximum T factor in 15 cases. As per the TNM classification (UICC, 8th edition), stage IV was the most common in 12 cases because of synchronous liver metastasis. The onset of the right colon was 4 (16.7%), whereas that of the left or rectum was 20 (83.3%). (Table 1).
TABLE 1.
Patient details and clinicopathological features
| CRC with liver metastasis | |
|---|---|
| Number | 24 |
| Gender | |
| Male | 12 (50.0) |
| Female | 12 (50.0) |
| Mean age ± s.d. (y) | 64.8 ± 10.1 |
| Age range (y) | 46‐82 |
| Depth of tumor invasion | |
| T1 | 0 (0.0) |
| T2 | 2 (8.3) |
| T3 | 15 (62.5) |
| T4 | 7 (29.2) |
| Lymph node metastasis | |
| Positive | 12 (50.0) |
| Negative | 12 (50.0) |
| Liver metastasis | |
| Synchronous | 12 (50.0) |
| Metachronous | 12 (50.0) |
| TNM stage (At the first surgery) | |
| I | 1 (4.2) |
| II | 6 (25.0) |
| III | 5 (20.8) |
| IV | 12 (50.0) |
| Vascular invasion | |
| Negative | 1 (4.2) |
| Positive | 23 (95.8) |
| Site of primary tumor | |
| Right | 4 (16.7) |
| Left or rectum | 20 (83.3) |
3.2. TMB status in samples
TMB was evaluated in all 48 samples, which were obtained from 24 patients. TMB was considered high when the mutation frequency was 10 or more per 1 Mb and was considered low when the mutation frequency was less than 10. TMB was found to be high in 8 out of 24 patients with primary lesions and in 5 of 24 patients with liver metastatic lesions (Table 2). The mean TMB for primary lesions was 10.93 and the mean TMB for liver metastatic lesions was 8.14. Therefore, no significant difference was observed in the TMB status of the primary and metastatic lesion samples (P = .296).
TABLE 2.
Tumor mutational burden (mutations/Mb)
| Case no. | Primary tumor | Liver metastasis |
|---|---|---|
| 1 | 23.66 | 3.38 |
| 2 | 10.1 | 6.73 |
| 3 | 13.47 | 10.14 |
| 4 | 5.91 | 4.24 |
| 5 | 12.73 | 16.46 |
| 6 | 5.05 | 5.09 |
| 7 | 2.53 | 1.69 |
| 8 | 4.23 | 5.08 |
| 9 | 61.97 | 5.08 |
| 10 | 6.81 | 25.76 |
| 11 | 6.81 | 5.08 |
| 12 | 6.79 | 7.63 |
| 13 | 6.87 | 9.04 |
| 14 | 7.72 | 11.1 |
| 15 | 8.55 | 8.47 |
| 16 | 11.11 | 4.29 |
| 17 | 3.40 | 7.62 |
| 18 | 9.32 | 9.37 |
| 19 | 8.51 | 6.77 |
| 20 | 14.46 | 17.8 |
| 21 | 4.28 | 4.21 |
| 22 | 10.23 | 7.60 |
| 23 | 9.35 | 5.91 |
| 24 | 8.48 | 6.75 |
| Ave. | 10.93 | 8.14 |
Case numbers in bold indicate cases in which the TMB status differs between the primary lesion and the liver metastatic lesion.
3.3. Heterogeneity of TMB between primary tumor and metastatic liver tumors
In 7 out of 24 patients, the TMB status differed between the primary and liver metastatic lesions (Table 3A, B). In 5 patients, the TMB was high only in the primary lesion. In these cases, the average TMB value in the primary lesion was 23.41 and the average TMB value in the metastatic lesion was 5.42. Therefore, their ratio was 4.93, indicating a significant difference in TMB status. In contrast, in the remaining 2 cases, the TMB was high only in the metastatic lesions. The ratio of these TMB values was 0.48, indicating that the TMB value in the metastatic lesion was half that of the primary lesion. Of the 7 tumor samples, 5 were synchronous and 2 were metachronous tumors. There was a significant difference in OS between positive and negative TMB status discordance in the primary and metastatic lesions (P = .042; Figure 2). In addition, the prognosis curve was drawn separately for the cases with TMB status of primary high/metastasis low and primary low/metastasis high. As a result, the prognosis was poor in the primary low/metastasis high group (Figure S1). There were no differences in the patient details and clinicopathological features between concordant and discordant cases (Table S1).
TABLE 3.
A, Cases with TMB high in the primary lesion. B, Cases with TMB high in the liver metastatic lesion
| A | TMB | Clinicopathological features | ||||||
|---|---|---|---|---|---|---|---|---|
| Case no. | Primary tumor | Liver metastasis | Ratio | Liver metastasis | Stage | Prognosis | Recurrence (after first liver resection) | |
| 1 | 1 | 23.66 | 3.38 | 7.00 | Synchronous | IVA | Dead | + |
| 2 | 2 | 10.1 | 6.73 | 1.50 | Synchronous | IVA | Dead | + |
| 3 | 9 | 61.97 | 5.08 | 12.20 | Metachronous | IIB | Alive | − |
| 4 | 16 | 11.11 | 4.29 | 2.59 | Synchronous | IVA | Alive | − |
| 5 | 22 | 10.23 | 7.60 | 1.35 | Synchronous | IVA | Alive | − |
| ave. | 23.41 | 5.42 | 4.93 | |||||
| B | TMB | Clinicopathological features | ||||||
|---|---|---|---|---|---|---|---|---|
| Case no. | Primary tumor | Liver metastasis | Ratio | Liver metastasis | Stage | Prognosis | Recurrence (after first liver resection) | |
| 1 | 10 | 6.81 | 25.76 | 0.26 | Synchronous | IVA | Alive | + |
| 2 | 14 | 7.72 | 11.1 | 0.70 | Metachronous | IIA | Alive | − |
| ave. | 7.27 | 18.43 | 0.48 | |||||
FIGURE 2.
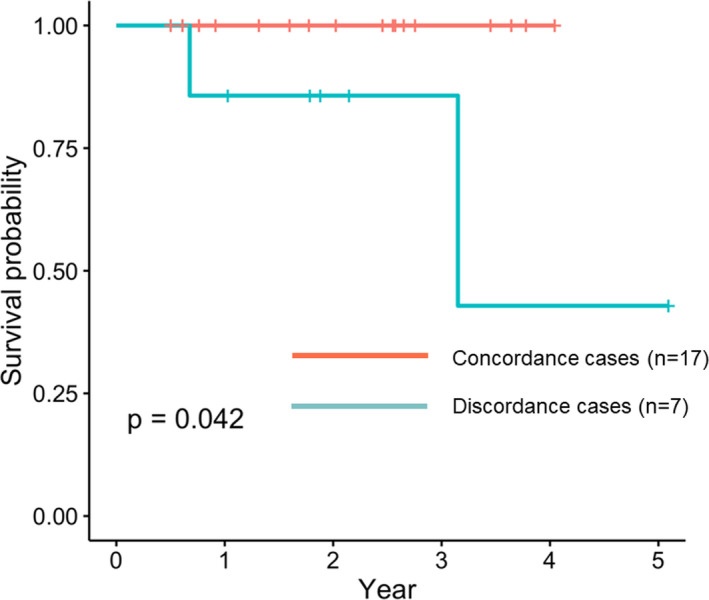
Overall survival curves of patients with positive and negative tumor mutational burden status discordance between primary and metastatic lesions
3.4. Discordance in the gene mutation rate among primary and metastatic lesions
The ranking of genes harboring mutations in the primary and metastatic lesions based on mutation frequencies was performed using the Oncomine™ Tumor Mutation Load Assay (Thermo Fisher Scientific, Inc). Table 4 shows the top 20 mutated genes. We observed that 12 of the top 20 genes were common (Figure 3). However, even in common genes, differences in the mutation frequencies were observed between the primary and liver metastatic lesions. Moreover, when the mutations were analyzed individually, the mismatches in the mutant genes were more evident (data not shown).
TABLE 4.
Frequency of gene mutations in primary and liver metastases
| Ranking | Primary lesion | Liver metastatic lesion | ||
|---|---|---|---|---|
| gene symbol | Mutation frequency (%) | gene symbol | Mutation frequency (%) | |
| 1 | KRAS | 13 (54.2) | KRAS | 11 (45.8) |
| 2 | APC | 11 (45.8) | APC | 10 (41.7) |
| 3 | TAF1L | 10 (41.7) | TP53 | 10 (41.7) |
| 4 | FN1 | 9 (37.5) | ADGRA2 | 4 (16.7) |
| 5 | TP53 | 8 (33.3) | LRP1B | 4 (16.7) |
| 6 | CIC | 6 (25.0) | RNF213 | 4 (16.7) |
| 7 | KMT2D | 4 (16.7) | SMAD4 | 4 (16.7) |
| 8 | ERCC1 | 4 (16.7) | TAF1L | 4 (16.7) |
| 9 | KDR | 4 (16.7) | AMER1 | 3 (12.5) |
| 10 | USP9X | 4 (16.7) | ARID1A | 3 (12.5) |
| 11 | RNF213 | 3 (12.5) | BCL9 | 3 (12.5) |
| 12 | SMAD4 | 3 (12.5) | CREBBP | 3 (12.5) |
| 13 | AKT2 | 3 (12.5) | ERCC1 | 3 (12.5) |
| 14 | LRP1B | 3 (12.5) | FN1 | 3 (12.5) |
| 15 | MYH9 | 3 (12.5) | KDR | 3 (12.5) |
| 16 | SMO | 3 (12.5) | PBX1 | 3 (12.5) |
| 17 | ADGRA2 | 3 (12.5) | PKHD1 | 3 (12.5) |
| 18 | NOTCH1 | 3 (12.5) | SAMD9 | 3 (12.5) |
| 19 | AMER1 | 3 (12.5) | AFF3 | 2 (8.3) |
| 20 | BCYRN1|TAF1 | 3 (12.5) | AKAP9 | 2 (8.3) |
Genes found in both primary and liver metastases are shown in bold.
FIGURE 3.
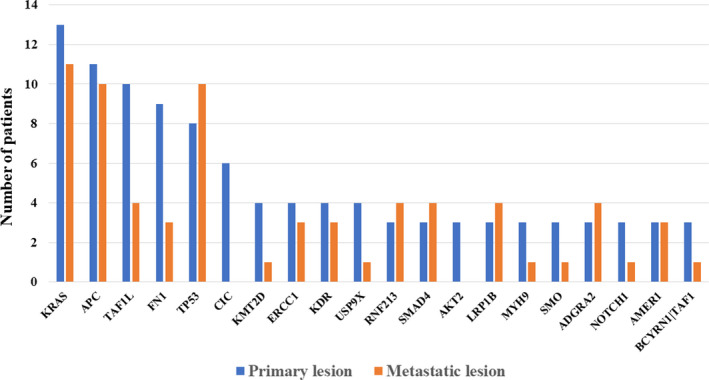
Mutations in genes associated with the primary lesion and metastatic lesion
3.5. Predictivity of machine learning models
The area under the curve (AUC) values for the primary tumor were 0.701 and 0.732 with imaging features of contrast‐enhanced supine and prone images, respectively (Figure 4A). The AUC values for liver metastasis were 0.784 and 0.812 with imaging features of non‐contrast‐enhanced and contrast‐enhanced CT, respectively (Figure 4B). The sensitivities and specificities are listed in Table 5. With the prone images for primary lesions and contrast‐enhanced images for liver metastasis, the sensitivity, specificity, and accuracy for the discordance of TMB status between primary and metastatic lesions were 0.857, 0.600, and 0.682, respectively (Table 6).
FIGURE 4.
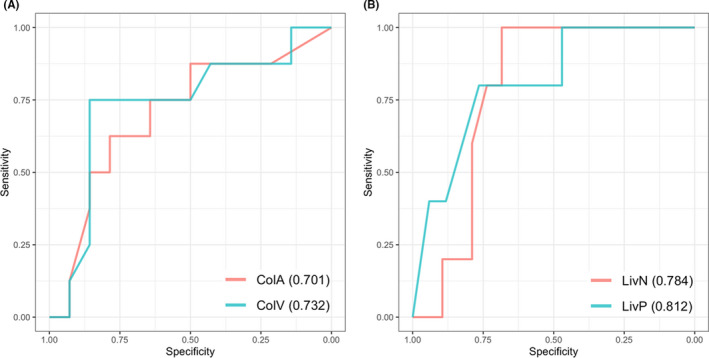
Receiver operating characteristic plots for machine learning prediction for high or low tumor mutational burden in the primary (A) and metastatic (B) lesions. ColA and ColV, computed tomography (CT) of arterial and venous phases for the primary colon lesion. LivN and LivP, CT of non‐contrast enhancement and portal phase for the metastatic liver lesion. Area under the curve is shown in parentheses
TABLE 5.
Predictivities for high TMB status on each CT phase
| Threshold | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|
| CT colonography | ||||
| Arterial phase (supine) | 0.524 | 0.625 | 0.786 | 0.727 |
| Venous phase (prone) | 0.167 | 0.750 | 0.857 | 0.818 |
| Liver CT | ||||
| Non‐contrast | 0.128 | 1.000 | 0.684 | 0.75 |
| Portal phase | 0.082 | 0.800 | 0.765 | 0.773 |
TABLE 6.
Predictivity of machine learning for TMB status discordance
| Machine learning prediction | |||
|---|---|---|---|
| Positive | Negative | ||
| Real | Positive | 6 | 1 |
| Negative | 6 | 9 | |
In this analysis, 22/24 cases were available because contrast‐enhanced CT was not performed in 2 patients with renal failure.
4. DISCUSSION
In this study, we examined differences in the TMB status of the primary and metastatic lesions in the patients with CRC. TMB status is considered to be a predictive biomarker for responses toward ICI treatment. We observed a discordance in TMB status in 7 out of 24 patients (29.2%). We also observed a difference in the frequencies of specific gene mutations, suggestive of tumor heterogeneity in the same individual. In addition, we demonstrated that TMB status can be determined using radiogenomics, omitting the requirement for direct genetic analysis. The TMB assay was developed as an indicator of therapeutic responses, especially as a potential predictive biomarker for immunotherapy. 17 , 18 The Food and Drug Administration (FDA) approved pembrolizumab on June 16, 2020, for the treatment of unresectable or metastatic TMB‐high solid tumors (TMB‐H; ≥10 [mut/Mb]) in adult and pediatric patients as a result of sub‐analysis of the KEYNOTE‐158 trial. 19 , 20
The understanding of the processes involved in tumor heterogeneity is the key to overcoming resistance in cancer treatment. 4 , 21 , 22 Although next‐generation sequencing (NGS) is a well developed technique, sequencing multiple lesions is difficult. This is because biopsy of a large number of metastases samples, particularly that of deep tissues is often technically challenging. It is also inconvenient for patients with advanced‐stage disease. 23 For these reasons, liquid biopsy in combination with molecular profiling has been gaining attention in recent years. A liquid biopsy involves the isolation of CTCs or circulating tumor DNA (ctDNA) from blood samples, on which molecular analysis is performed to obtain the overall tumor profile. Currently, clinically used liquid biopsies typically include plasma‐based ctDNA assays, which use NGS for genomic mutation or copy number determination. 24 , 25 The approval was based on the ENSURE study, a multicenter, open‐label, randomized phase III study to evaluate the efficacy and safety of erlotinib vs gemcitabine plus cisplatin as a first‐line treatment for patients with stage IIIB/IV non‐small cell lung cancer. The extracted plasma tested positive for epidermal growth factor receptor mutations in 76.7% of the tissue‐positive specimens and tested negative in 98.2% of the tissue‐negative specimens. 26 Although liquid biopsy is a promising technique, its utility is limited by tumor heterogeneity. Specifically, the source of ctDNA cannot be tracked in liquid biopsy, which complicates the final analysis. García‐Saenz and colleagues reported that plasma PIK3CA mutation levels correlated with treatment response in a majority of advanced breast cancer patients in their cohort, with a treatment response discrepancy rate of 25% (2/8 patients). The discrepancy was found because of differences in drug susceptibility within metastatic tumors. 27 As mentioned above, tissue and plasma samples provide only limited information about the evolutionary history of tumors.
Therefore, to resolve this problem, we have attempted to collect data pertaining to gene expression and mutation via radiogenomics. Radiogenomics was initially developed for examining brain tumors and breast and lung cancer tissues. 28 , 29 , 30 , 31 It has also been used for gastrointestinal cancer, including that in luminal organs. 7 , 8 , 9 Existing literature on the use of radiogenomics in CRC infers gene mutations from 1 feature of fluorodeoxyglucose‐positron emission tomography (FDG‐PET), CT, and MRI images and does not report the use of true radiogenomics. Chen and colleagues 32 used FDG‐PET to distinguish between KRAS wild‐type and mutant tumors. The results showed that, in multivariate analysis, a threshold level of 40% (TW40%) was considered for the maximal uptake of SUVmax and TW as 2 predictors of KRAS mutations. Yang and co‐workers used contrast‐enhanced CT images of the primary lesion to investigate whether radiation signatures could predict KRAS, NRAS, and BRAF mutations in CRC. The features of 346 images were extracted from the CT images of the primary lesion of the portal vein layer and their correlation with gene mutations was investigated. Image features were extracted using artificial intelligence and the yield of the extracted images was significantly associated with KRAS, NRAS, and BRAF mutations (P < .001). The ROC analysis to predict KRAS, NRAS, and BRAF mutations also showed AUC, sensitivity, and specificity of 0.869, 0.757, and 0.833 in the test cohort and 0.829, 0.686, and 0.857 in the validation cohort, respectively. 33 Lubner and colleagues used a new tool to perform tumor histology analysis of hepatic metastatic lesions on pretreatment contrast‐enhanced CT scans of 77 patients. Non‐uniformity measurements, including entropy, kurtosis, skewness, mean, mean positive pixels (MPP), and SD of the pixel distribution histogram, correspond to fine, medium, and coarse textures, as derived using filter values. Skewness was negatively associated with KRAS mutations (P = .02). 34
None of the above studies, associated with the use of radiogenomics in CRC, focus on TMB determination. However, there is a study on the prediction of microsatellite instability (MSI) using radiogenomics. In this report, contrast‐enhanced CT images using dual‐energy computed tomography (DECT) imaging were used to predict the MSI status of CRC preoperatively. The study included 102 CRC patients, 34 with MSI and 68 with microsatellites. All patients underwent preoperative DECT imaging with either a Revolution CT or Discovery CT 750 HD scanner. Data from the Revolution CT scanner were used to establish a radiogenomics model for predicting MSI status (70% of the samples were randomly selected as a training set and the remaining 30% were used for validation). The radiogenomics model was then tested using data from the Discovery CT 750HD scanner. Nine features were selected to build the radiogenomics model. In the training set, the AUC was 0.961 (accuracy: 0.875; sensitivity: 1.000; specificity: 0.812), whereas in the validation set, the AUC was 0.918 (accuracy, 0.875; sensitivity, 0.875; specificity, 0.857). In the test set, the diagnostic performance was slightly lower, with an AUC of 0.875 (accuracy: 0.788; sensitivity: 0.909; specificity: 0.727). 35
In our results, discordance between the TMB status of the primary lesion and metastatic lesion was observed in 7 out of 24 cases (29.2%) and was considered to be non‐negligible. A discrepancy was observed in the mutation frequency and the presence of mutations between the primary lesion and the hepatic metastatic lesion in the same individual for individual genes. When the TMB status of the primary lesion and the metastatic lesion is different, the gene mutation frequency is high in the TMB‐high lesion group. In this study, when the TMB‐high lesion was used as the standard, the concordance rate of gene mutation with the TMB‐low lesion varied from 37.5% to 87.5%. Therefore, if liver metastases were to be treated on the basis of the TMB status of the primary lesion, treatment in 5 cases may be rendered ineffective. In addition, 2 cases would not have received effective treatment. The diagnostic ability of radiogenomics to detect liver metastasis was appropriate with an AUC value of approximately 0.8 by contrast‐enhanced CT. In our study, the sensitivity, specificity, and accuracy of TMB discordance between primary and metastatic lesions were 0.857, 0.600, and 0.682, respectively. Although these are preliminary results, the discordance rate of the gene profiles between the primary lesion and the metastatic lesion highlights its accuracy. We propose that this radiogenomics approach for TMB status determination is more reliable than genetic analysis of primary lesions. This radiogenomics approach of TMB status determination may be more useful in therapeutic determination than the current TMB status determination by molecular biology techniques using tissues from the primary lesion. Interestingly, patients exhibiting discordance in TMB status showed a worse prognosis than those who did not (Figure 2). None of the patients actually received ICI treatment in this study with the exact cause being unknown. In cases in which the TMB status differs between the primary and metastatic lesions, the gene expression and mutation profiling change and diversity in cancer may increase the malignant tendency of the cancer.
MSI status has been reported to be significantly associated with TMB status in patients with CRC. 30 , 31 When the TMB cutoff value was set to 10 mut/Mb, MSI‐high tumors were diagnosed in 1 out of 5 TMB‐high primary lesions (20%) and 2 out of 3 TMB‐high liver metastatic lesions (66.6%; Table S2). In other words, with a cutoff of 10 mut/Mb, the concordance rate between TMB high and MSI high was 3 out of 8 TMB‐high samples (37.5%). These values were slightly higher, considering that the concordance rate between TMB high and MSI high in CRC patients was 11% to 14% in previous reports. 19 , 20
In addition, Antoniotti and co‐workers reported that 8 out of 11 samples (73%) were MSI high when the TMB‐high cutoff value was set to 17 mut/Mb. 20 In our study, when the TMB cutoff was set to 17 mut/Mb, 1 out of 2 primary lesions of TMB high (50%) and 1 liver metastatic lesion of TMB high were diagnosed with MSI high (100%). As a result, when the cutoff was 17 mut/Mb, the concordance rate between TMB high and MSI high was two‐thirds (66.6%), which was almost the same as the results of the previous report.
In addition, recent studies have reported that many microsatellite stable cases are effective in treating ICI, and a study has reported that these cases might be selected by examining the TMB status. 36
This study has several limitations. The number of participants was small. Moreover, the radiogenomics approach is limited to tumors that can be visualized in images and the results may be inaccurate for small tumors. Further studies involving a higher number of cases with collaborative verification by multiple centers are warranted to obtain more robust results.
5. CONCLUSION
This study showed discordance in TMB status between primary and liver metastatic lesions in patients with CRC. We also showed that accurate inference of the status of TMB using radiogenomics is possible. In conclusion, radiogenomics may be useful in developing effective gene‐based therapies for cancer patients.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Fig S1
Table S1‐2
ACKNOWLEDGMENTS
Not applicable
Hoshino I, Yokota H, Iwatate Y, et al. Prediction of the differences in tumor mutation burden between primary and metastatic lesions by radiogenomics. Cancer Sci.2022;113:229–239. 10.1111/cas.15173
Hajime Yokota contributed equally to this work.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.
REFERENCES
- 1. Ulahannan D, Kovac MB, Mulholland PJ, Cazier JB, Tomlinson I. Technical and implementation issues in using next‐generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109:827‐835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weymann D, Pollard S, Chan B, et al. Clinical and cost outcomes following genomics‐informed treatment for advanced cancers. Cancer Med. 2021;10(15):5131‐5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sunami K, Naito Y, Aimono E, et al. The initial assessment of expert panel performance in core hospitals for cancer genomic medicine in Japan. Int J Clin Oncol. 2021;26:443‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883‐892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology. 2020;158:291‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo C, Lin X, Yin J, et al. Pharmacogenomics signature: a novel strategy on the individual differences in drug response. Cancer Lett. 2018;420:190‐194. [DOI] [PubMed] [Google Scholar]
- 7. Hoshino I, Yokota H. Radiogenomics of gastroenterological cancer: the dawn of personalized medicine with artificial intelligence‐based image analysis. Ann Gastroenterol Surg. 2021;5(4):427‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoshino I, Yokota H, Ishige F, et al. Radiogenomics predicts the expression of microRNA‐1246 in the serum of esophageal cancer patients. Sci Rep. 2020;10:2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwatate Y, Hoshino I, Yokota H, et al. Radiogenomics for predicting p53 status, PD‐L1 expression, and prognosis with machine learning in pancreatic cancer. Br J Cancer. 2020;123:1253‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galuppini F, Dal Pozzo CA, Deckert J, Loupakis F, Fassan M, Baffa R. Tumor mutation burden: from comprehensive mutational screening to the clinic. Cancer Cell Int. 2019;19:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD‐1 inhibition. N Engl J Med. 2017;377:2500‐2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawlor RT, Mattiolo P, Mafficini A, et al. Tumor mutational burden as a potential biomarker for immunotherapy in pancreatic cancer: systematic review and still‐open questions. Cancers. 2021;13:3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valero C, Lee M, Hoen D, et al. Response rates to anti‐PD‐1 immunotherapy in microsatellite‐stable solid tumors with 10 or more mutations per megabase. JAMA Oncol. 2021;7:739‐743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next‐generation sequencing and compared with PD‐L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Can Res. 2017;77:e104‐e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koeppel F, Blanchard S, Jovelet C, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One. 2017;12:e0188174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong X, Sullivan RJ, Kalinich M, et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc Natl Acad Sci USA. 2018;115:2467‐2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marcus L, Fashoyin‐Aje LA, Donoghue M, et al. FDA approval summary: pembrolizumab for the treatment of tumor mutational burden‐high solid tumors. Clin Cancer Res. 2021;27:4685‐4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open‐label, phase 2 KEYNOTE‐158 study. Lancet Oncol. 2020;21:1353‐1365. [DOI] [PubMed] [Google Scholar]
- 21. Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol. 2019;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Z, Boggon TJ, Kobayashi S, et al. Resistance to an irreversible epidermal growth factor receptor (EGFR) inhibitor in EGFR‐mutant lung cancer reveals novel treatment strategies. Cancer Res. 2007;67:10417‐10427. [DOI] [PubMed] [Google Scholar]
- 23. Denis MG, Vallée A, Théoleyre S. EGFR T790M resistance mutation in non small‐cell lung carcinoma. Clin Chim Acta. 2015;444:81‐85. [DOI] [PubMed] [Google Scholar]
- 24. Dearden S, Brown H, Jenkins S, et al. EGFR T790M mutation testing within the osimertinib AURA phase I study. Lung Cancer. 2017;109:9‐13. [DOI] [PubMed] [Google Scholar]
- 25. Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO‐1686). Clin Cancer Res. 2016;22:2386‐2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu YL, Zhou C, Liam CK, et al. First‐line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer: analyses from the phase III, randomized, open‐label, ENSURE study. Ann Oncol. 2015;26:1883‐1889. [DOI] [PubMed] [Google Scholar]
- 27. García‐Saenz JA, Ayllón P, Laig M, et al. Tumor burden monitoring using cell‐free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer. 2017;17:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. Am J Roentgenol. 2012;199:654‐663. [DOI] [PubMed] [Google Scholar]
- 29. Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology. 2014;270:1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nair VS, Gevaert O, Davidzon G, Plevritis SK, West R. NF‐κB protein expression associates with (18)F‐FDG PET tumor uptake in non‐small cell lung cancer: a radiogenomics validation study to understand tumor metabolism. Lung Cancer. 2014;83:189‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gevaert O, Xu J, Hoang CD, et al. Non‐small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology. 2012;264:387‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen SW, Chiang HC, Chen WT, et al. Correlation between PET/CT parameters and KRAS expression in colorectal cancer. Clin Nucl Med. 2014;39:685‐689. [DOI] [PubMed] [Google Scholar]
- 33. Yang L, Dong D, Fang M, et al. Can CT‐based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28:2058‐2067. [DOI] [PubMed] [Google Scholar]
- 34. Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre‐treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331‐2337. [DOI] [PubMed] [Google Scholar]
- 35. Wu J, Zhang Q, Zhao Y, et al. Radiomics analysis of iodine‐based material decomposition images with dual‐energy computed tomography imaging for preoperatively predicting microsatellite instability status in colorectal cancer. Front Oncol. 2019;9:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shitara K, Özgüroğlu M, Bang YJ, et al. Molecular determinants of clinical outcomes with pembrolizumab versus paclitaxel in a randomized, open‐label, phase III trial in patients with gastroesophageal adenocarcinoma. Ann Oncol. 2021;32:1127‐1136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1‐2
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.


