Abstract
Lack of rapid and comprehensive microbiological diagnosis in patients with community acquired pneumonia (CAP) hampers appropriate antimicrobial therapy. This study evaluates the real-world performance of the BioFire FilmArray Pneumonia panel plus (FAP plus) and explores the feasibility of evaluation in a randomised controlled trial. Patients presenting to hospital with suspected CAP were recruited in a prospective feasibility study. An induced sputum or an endotracheal aspirate was obtained from all participants. The FAP plus turnaround time (TAT) and microbiological yield were compared with standard diagnostic methods (SDs). 96/104 (92%) enrolled patients had a respiratory tract infection (RTI); 72 CAP and 24 other RTIs. Median TAT was shorter for the FAP plus, compared with in-house PCR (2.6 vs 24.1 h, p < 0.001) and sputum cultures (2.6 vs 57.5 h, p < 0.001). The total microbiological yield by the FAP plus was higher compared to SDs (91% (162/179) vs 55% (99/179), p < 0.0001). Haemophilus influenzae, Streptococcus pneumoniae and influenza A virus were the most frequent pathogens. In conclusion, molecular panel testing in adults with CAP was associated with a significant reduction in time to actionable results and increased microbiological yield. The impact on antibiotic use and patient outcome should be assessed in randomised controlled trials.
Subject terms: Bacterial infection, Viral infection, Respiratory tract diseases, Clinical microbiology, Infectious-disease diagnostics
Introduction
Lower respiratory tract infections (LRTIs), including community acquired pneumonia (CAP), are one of the leading causes of death and years of life lost globally1,2. The microbiological etiology of CAP is often difficult to ascertain, due to difficulties in obtaining representative respiratory specimens and insufficient methods for microbial detection3–5. Empiric antimicrobial therapy is often initiated in the primary care setting, further hampering diagnostic attempts based on conventional bacterial culture in hospitalised patients. In Norway, data on the etiology of CAP in hospitalised patients is limited to a few studies, which do not include a comprehensive assessment of viral and bacterial respiratory pathogens6,7.
Polymerase chain reaction (PCR) based assays targeting respiratory viruses and atypical bacteria have widened our understanding of CAP etiology, and CAP is no longer considered to be exclusively of bacterial origin3,8–13. Improved detection of viruses could potentially reduce unnecessary use of antibiotics, although coinfections can be difficult to rule out owing to the lack of comparable methods for the detection of bacterial pathogens. Recently, rapid syndromic PCR panels for viruses and bacteria involved in CAP have been developed. Several studies have evaluated the diagnostic performance of these panels, concluding that they are highly accurate and detect pathogens in a higher proportion compared with standard diagnostic methods (SDs)14–19. However, reports on the incorporation of syndromic tests in clinical practice are scarce, and there is limited evidence of the clinical impact of these tests.
Our primary objective was to explore the feasibility of introducing the BioFire FilmArray Pneumonia panel plus (FAP plus) (bioMérieux S.A., Marcy-l’Etoile, France) in the diagnostic workup of patients admitted with suspected CAP, with a view to inform the design of a subsequent randomised controlled trial. We hypothesized that syndromic PCR-based testing of lower respiratory tract samples is achievable in acute settings. The secondary objectives were to investigate the real-world turnaround time (TAT) and microbiological yield of the FAP plus compared to SDs at our hospital.
Methods
Patients and study design
This was a prospectively recruited cohort study conducted at Haukeland University Hospital, a tertiary care referral centre in Bergen, Norway between December 2nd 2019, and February 17th 2020. In addition to conventional microbiological diagnostics, samples from the lower respiratory tract were systematically analysed with a commercial rapid syndromic PCR panel, the FAP plus. The study was conducted as a feasibility study to inform the design of a larger randomised controlled trial evaluating the clinical impact of the FAP plus assay on antibiotic use and outcome (NCT04660084). Patients were eligible for inclusion if they were ≥ 18 years, presenting to the emergency department (ED) with a suspicion of CAP (evaluated by investigating physicians and/or study nurses) and fulfilling at least two of the following criteria: new or worsening cough; new or worsening expectoration of sputum; new or worsening dyspnoea; haemoptysis; pleuritic chest pain; radiological evidence of pneumonia; abnormalities on chest auscultation and/or percussion; fever (≥ 38.0 °C). Exclusion criteria were cystic fibrosis, severe bronchiectasis (defined as patients in need of regular follow-up and treatment by a pulmonologist due to bronchiectasis), hospitalisation within the last 14 days prior to admission, a palliative approach (defined as life expectancy below two weeks documented by a treating physician; either by preexisting estimates in the electronic journal, or estimations made at admission), or if the patient was not willing or able to provide a lower respiratory tract sample (by either sputum induction or endotracheal aspiration).
Data collection and sampling
Patients were enrolled on weekdays between 08:00 a.m. and 09:00 p.m. Most patients were enrolled in the ED shortly after admission. Investigating physicians and/or study nurses screened all electronic triage documents. Patients with respiratory complaints and/or a suspected infection of any type were then evaluated for eligibility, according to the inclusion criteria. To compensate for the restricted study operating hours, some cases were included at the wards up to a maximum of 24 h after admission. These patients were identified through retrospective screening of electronic triage documents and medical records. Relevant baseline information was collected by study nurses or investigating physicians through a structured interview. Symptoms and findings upon clinical examinations were recorded. Data pertaining to treatment and results from laboratory tests and medical imaging were obtained from electronic medical records and charts. Data were registered in an electronic case report form (eCRF) from VieDoc (Viedoc Technologies, Uppsala, Sweden).
The final diagnosis (as per the four different categories enumerated below) was determined retrospectively, following patient discharge, by the use of pre-specified diagnostic criteria in consensus meetings among investigating physicians. The diagnostic criteria thus differed from the inclusion criteria, which were designed to be used as a screening tool based on the available information at admission in the ED. The complete definitions are provided in the Supplementary material (S2). Patients were categorized into four different categories: (1) Confirmed CAP (with radiological confirmation); (2) Clinical CAP (without radiological confirmation); (3) Other respiratory tract infections (RTIs), i.e., bronchitis, acute infectious exacerbation in patients with chronic obstructive pulmonary disease (COPD), acute infectious exacerbation of asthma, and upper RTIs; or (4) Other diagnosis, i.e., CAP suspected at admission but RTI later disproved. To avoid observer bias an independent agreement between the treating physicians and the study investigators was desired. If any disagreement, an additional study investigator would arbitrate.
Microbiological sampling and methods
At inclusion, a lower respiratory tract sample for the FAP plus and standard culture was obtained from all patients. Depending on clinical symptoms, vital signs and medical history, sputum induction with either nebulized isotonic (0.9%) or hypertonic (5.8%) saline was attempted. Patients with known obstructive lung disease and patients with hypoxemia or signs of airway obstruction upon physical examination, were additionally treated with a bronchodilator (salbutamol and/or ipratropium bromide) prior to sampling. If sputum induction was unsuccessful, endotracheal aspiration was performed. The detailed procedures are provided in the Supplementary material (S3).
The FAP plus is an automated multiplex PCR test for the detection of 27 bacteria and viruses, as well as seven genetic markers of antibiotic resistance, validated for lower respiratory tract samples (Supplementary material, S1). The hands-on time is around two minutes and the total analysis time about 1 h20. Typical bacterial detections are reported in a semi-quantitative manner and categorized as negative if ≤ 103.5 copies/ml. Above this level, results are reported as positive and semi-quantitatively specified as 104, 105, 106 or ≥ 107 copies/ml21.
The SDs included culture of respiratory tract samples and blood according to current guidelines (adapted from22). Blood culture isolates and relevant respiratory isolates were identified with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF MS) using the Bruker's microflex LT instrument, MBT Compass software ver. 4.1 and Compass Library DB-8468 (Bruker Daltonics, Massachusetts, U.S.). Nasopharyngeal and/or oropharyngeal swabs were examined by an in-house real-time PCR test to detect respiratory viruses and atypical bacteria (influenza A and B, human parainfluenza viruses 1–3, respiratory syncytial virus, human metapneumovirus, rhinovirus, Bordetella pertussis, Bordetella parapertussis, Mycoplasma pneumoniae and Chlamydia pneumoniae). SDs also included rapid diagnostic tests; the pneumococcal urine antigen test (Quidel Corporation, San Diego, U.S.) and a point of care (POC) test for influenza virus A and B (ID NOW, Illinois, U.S.). Any additional tests requested by the treating physician were also noted and counted as part of SDs.
Gram staining was utilized to evaluate the representativeness of all sputum samples (adapted from22). Samples containing ≥ 10 squamous epithelial cells per field in at least 10 fields with 10 × enlargement were considered non-representative. However, this criterion was disregarded if a significant amount of both leukocytes (≥ 10 times the amount of squamous epithelial cells per field of view) and a morphologically uniform microbe (> 5 microbes per field of view with 100× enlargement) were present. All samples were analysed by the FAP plus and cultured on agar-plates, irrespective of the representativeness. Abundant growth of plausible respiratory pathogens was reported regardless of the representativeness of the sputum sample. Non-abundant growth was only reported in samples considered representative.
The clinical relevance of all microbiological findings, in terms of categorization as a relevant pathogen for the current RTI or not, was established retrospectively using pre-specified criteria (Supplementary material, S4).
Statistical analysis
Descriptive statistics for continuous variables are reported as median with interquartile range (IQR). Turn-around time for microbiological methods are compared with Student’s paired t-test on logarithm-transformed times. For paired categorical samples, McNemar’s test with risk differences and 95% confidence intervals (CIs) are used. A two tailed p-value ≤ 0.05 was considered statistically significant for all analyses. The statistics were performed using IBM SPSS Statistics (version 26.0; Armonk, NY, U.S.), the statistical environment R (version 3.6.3; Vienna, Austria) and the GraphPad QuickCalcs Web site: http://www.graphpad.com/quickcalcs/McNemar1.cfm (last accessed 18. March 2021).
Ethics
The study was approved by the Regional Committee for Medical and Health Research Ethics in South-Eastern Norway (REK ID: 31935) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants or from their legal guardian/close relative at the time of recruitment.
Results
Patient characteristics
A total of 104 patients with suspected CAP were enrolled (Fig. 1). Eight patients were subsequently diagnosed with an alternative diagnosis (non-infectious exacerbation of COPD (n = 2), hearth failure (n = 2), unspecified non-infectious dyspnoea (n = 2), urinary tract infection (n = 1), aortic graft infection (n = 1)), and were excluded from further analyses. Baseline characteristics for the final cohort of 96 patients with an RTI are shown in Table 1. The majority of patients (69/96 (72%)) were recruited in the ED, with a median (IQR) time of 31 (18–77) minutes from hospital admission to enrolment. For the 27 (28%) patients recruited in the wards, the median (IQR) time to enrolment was 17 (15–21) hours. Overall, 61 (64%) patients had a radiologically confirmed CAP, 11 (11%) patients had CAP without radiological confirmation, and 24 (25%) patients had other RTIs (acute bronchitis (n = 10), acute infectious exacerbation of COPD (n = 8), acute infectious exacerbation of asthma (n = 4) and upper RTI (n = 2)). In the following sections, radiological confirmed CAP and clinically diagnosed CAP without radiological confirmation are considered as a single entity (n = 72).
Figure 1.
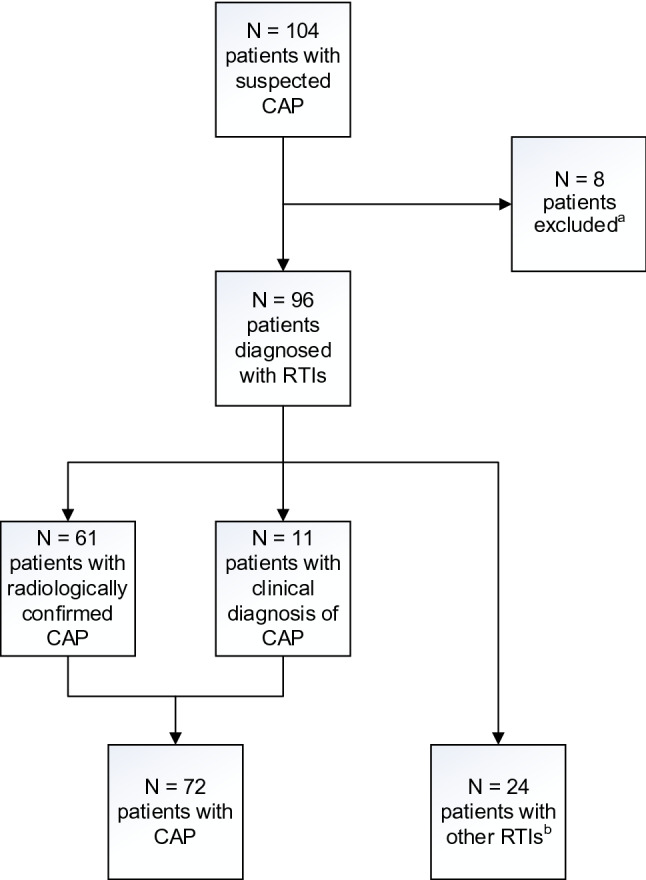
Study flowchart. CAP, community acquired pneumonia; RTI, respiratory tract infection; COPD, chronic obstructive pulmonary disease. aEight patients were excluded due to other diagnoses: non-infectious exacerbation of COPD (n = 2); heart failure (n = 2); unspecified non-infectious dyspnea (n = 2); urinary tract infection (n = 1); aortic graft infection (n = 1). bAcute bronchitis (n = 10), acute infectious exacerbation of COPD (n = 8), acute infectious exacerbation of asthma (n = 4) and upper RTI (n = 2).
Table 1.
Characteristics of the study cohort of n = 96 patients with community acquired respiratory tract infections.
| CAP (n = 72) |
Other RTIs (n = 24) |
|
|---|---|---|
| A: Baseline characteristics | ||
| Demography | ||
| Age | 74 (61–81) | 65 (51–75) |
| Male | 35 (49) | 9 (38) |
| Comorbidity | ||
| Cardiovascular disease | 38 (53) | 10 (42) |
| Diabetes mellitus | 5 (7) | 6 (25) |
| Asthma/COPD | 26 (36) | 12 (50) |
| Kidney disease | 12 (17) | 3 (13 ) |
| Previous smoker | 37 (51) | 6 (25) |
| Current smoker | 10 (14) | 10 (42) |
| Vaccine statusa | ||
| Influenza virus | 42 (58) | 16 (67) |
| Pneumococcal | 17 (24) | 6 (25) |
| B: Severity and outcome | ||
| Biochemistryb | ||
| WBC count | 13.7 (9.3–16.6) | 9.5 (7.1–10.9) |
| CRP level | 175 (112–250) | 57 (34–82) |
| Severity scorec | ||
| CURB-65 | 1.0 (1.0–2.0) | – |
| PSId | 93 (71–111) | – |
| Outcome | ||
| Length of stay (days) | 3.6 (2.2–5.2) | 1.9 (0.4–3.2) |
| HDU or ICU admission | 6 (8.3) | 4 (16.7) |
| Case fatality rate | ||
| In-hospital | 1 (1) | 0 (0) |
| 30 days | 1 (1) | 0 (0) |
| 60 days | 4 (6) | 0 (0) |
Data shown as count (%) or median (IQR).
CAP, community acquired pneumonia; RTI, respiratory tract infection; COPD, chronic obstructive pulmonary disease; WBC, white blood cells; CRP, C-reactive protein; CURB-65, confusion, urea, respiratory rate, blood pressure, age ≥ 65 years; PSI, pneumonia severity index; HDU, high dependency unit; ICU, intensive care unit; IQR, interquartile range.
aVaccinated for influenza virus within the last year; for Streptococcus pneumoniae within the last 5 years.
bHighest value during hospital stay.
cOnly validated for CAP patients.
dMissing in five patients with CAP.
Respiratory tract specimens were obtained in all RTI-patients, mainly by sputum induction (91/96 (95%)), the remaining by endotracheal aspiration (5/96 (5%)). By Gram-staining, 56 of 96 (58%) respiratory tract samples were classified as not representative of the lower respiratory tract. All samples were investigated using the FAP plus rapid PCR panel in addition to standard culture-based methods. Blood cultures were performed in 95 (99%), in-house PCR testing in 87 (91%), the POC influenza test in 76 (79%), and a pneumococcal urine antigen test in 62 (65%) of the patients.
Time to results
The TAT, i.e., time from sampling to a reported test result, varied considerably by method (Table 2). Median TAT for the FAP plus was strikingly shorter, compared with in-house PCR (2.6 vs 24.1 h, median difference of 21.0 h (IQR 16.3–24.7), p < 0.001) and sputum cultures (both negative and positive combined) (2.6 vs 57.5 h, median difference of 48.6 h (IQR 24.0–91.5), p < 0.001). Further, results from sputum cultures were not reported until antimicrobial susceptibility testing (AST) was completed, implying an even larger difference in TAT when the FAP plus was compared with solely positive sputum cultures (2.6 vs 92.6 h, median difference of 89.1 h (IQR 47.4–130.2), p < 0.001).
Table 2.
Turnaround time for the BioFire FilmArray Pneumonia panel plus versus standard microbiological methods.
| Diagnostic method | Patients (n) | Turnaround time | Time differencea | p-valuea |
|---|---|---|---|---|
| FAP plus | 96 | 2.6 (2.2–3.4) | N.A | N.A |
| In-house PCR | 87 | 24.1 (19.6–27.8) | 21.0 (16.3–24.7) | < 0.001 |
| Sputum culture | 96 | 57.5 (26.9–94.4) | 48.6 (24.0–91.5) | < 0.001 |
| Pneumococcal antigen | 62 | 1.3 (0.9–1.8) | − 1.2 (− 2.0 to (− 0.5)) | < 0.001 |
| POC influenzab | 76 | –b | – | – |
Comparison of the turnaround time for different microbiological diagnostic methods used in this study. Numbers presented are median hours with IQR if not otherwise specified. P-values are calculated with Student’s paired t-test on logarithm-transformed times.
FAP plus, Biomérieux BioFire FilmArray Pneumonia panel plus; PCR, polymerase chain reaction; POC, point of care; N.A., not applicable; IQR, inter-quartile range.
aCompared to the FAP plus.
bTurnaround time was not recorded for the POC Influenza test (ID NOW). It was directly available in the ED, with an analysis time of approximately 15 min.
Microbiological findings
Eighty-nine out of the 96 (93%) RTI-patients had at least one positive microbiological test, yielding a total of 179 microbes. A considerable proportion of the microbes, 64 of 179 (36%), were deemed to be of uncertain relevance. The remaining 115 (64%) microbes were deemed clinically relevant pathogens and were recovered from 83 (86%) patients, including 61 of the 72 (85%) CAP patients. Among the total CAP patients, 24 (33%) had pure bacterial detections, 23 (32%) bacterial and viral co-detections, 13 (18%) pure viral detections, 11 (15%) had no detections, while one (1%) was diagnosed with a Pneumocystis jirovecii infection. Haemophilus influenzae (32% (23/72)), Streptococcus pneumoniae (28% (20/72)) and influenza A virus (22% (16/72)) were the most frequently detected pathogens. More than one relevant microbe was detected in 35% (25/72) of CAP patients. Among the 24 patients with other RTIs, a viral etiology was detected in 92% (22/24) of patients, most frequently influenza A virus (54% (13/24)).
Comparison of the FAP plus to SDs
Except for three cases of human metapneumovirus, all microbes found by the in-house PCR test were also detected by the FAP plus. A relevant detection of Staphylococcus aureus by blood culture, two positive S. pneumoniae urinary antigen tests and a case of P. jirovecii were solely made by SDs. The complete microbiological findings by the FAP plus versus SDs are listed in Tables 3 and 4, stratified by evaluation of relevance and diagnostic category. The total microbiological yield was significantly higher by use of the FAP plus compared to SDs (91% (162/179) vs 55% (99/179), difference of 35% (95%CI 25–45%, p < 0.0001)). Using the FAP plus affected both the proportion of relevant, assumed causative detections (94% (108/115) vs 69% (79/115), difference of 25% (95%CI 14–36%, p < 0.0001)), and the proportion of detections with uncertain relevance (84% (54/64) vs 31% (20/64), difference of 53% (95%CI 33–73%, p < 0.0001)). In CAP patients included at the wards (n = 23), where 21 (91%) had received antibiotic treatment before the collecting of respiratory specimens, the FAP plus detected 92% (12/13) of relevant bacterial pathogens compared to 31% (4/13) by SDs (difference of 62% (95%CI 20–103%, p = 0.0269)). For the CAP patients enrolled in the ED (n = 49), the FAP plus detected 95% (37/39) of relevant bacterial pathogens compared to 64% (25/39) by SDs (difference of 31% (95%CI 11–51%, p = 0.006)).
Table 3.
Microbiological findings by use of the BioFire FilmArray Pneumonia panel plus versus standard microbiological methods in 72 patients with CAP.
| Microbes | Community acquired pneumonia (CAP) | |||||
|---|---|---|---|---|---|---|
| Total detections* | Detections deemed a relevant** | |||||
| All methods combined | FAP plus | Standard methods | All methods combined | FAP plus | Standard methods | |
| Viruses | 38 | 35 (92) | 28 (74) | 38 | 35 (92) | 28 (74) |
| Influenza A virus | 16 | 16 (100) | 13 (81) | 16 | 16 (100) | 13 (81) |
| Human metapneumovirus | 13 | 10 (77) | 10 (77) | 13 | 10 (77) | 10 (77) |
| Respiratory syncytial virus | 3 | 3 (100) | 3 (100) | 3 | 3 (100) | 3 (100) |
| Coronavirus | 3 | 3 (100) | – | 3 | 3 (100) | – |
| Parainfluenza virus | 2 | 2 (100) | 2 (100) | 2 | 2 (100) | 2 (100) |
| Rhino-/enterovirus | 1 | 1 (100) | 0 (0) | 1 | 1 (100) | 0 (0) |
| Bacteria | 91 | 81 (89) | 41 (45) | 52 | 49 (94) | 30 (58) |
| H. influenzae | 25 | 25 (100) | 14 (56) | 23 | 23 (100) | 13 (57) |
| S. pneumoniae | 20 | 18 (90) | 11 (55) b | 20 | 18 (90) | 11 (55) b |
| M. catarrhalis | 8 | 8 (100) | 2 (25) | 2 | 2 (100) | 2 (100) |
| E. coli | 6 | 5 (83) | 2 (33) | 0 | 0 (–) | 0 (–) |
| S. aureus | 5 | 4 (80) | 2 (40) | 2 | 1 (50) | 1 (50) c |
| S. agalactiae | 5 | 5 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| S. epidermidis | 5 | – | 5 (100) | 0 | – | 0 (–) |
| K. pneumoniae | 3 | 3 (100) | 1 (33) | 1 | 1 (100) | 0 (0) |
| S. marcescens | 3 | 3 (100) | 1 (33) | 1 | 1 (100) | 1 (100) |
| P. aeruginosa | 3 | 2 (67) | 1 (33) | 0 | 0 (–) | 0 (–) |
| Proteus spp. | 2 | 2 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| K. oxytoca | 1 | 1 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| A. calcoaceticus–A. baumanii complex | 1 | 1 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| E. cloacae complex | 1 | 1 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| M. pneumoniae | 2 | 2 (100) | 1 (50) | 2 | 2 (100) | 1 (50) |
| L. pneumophila | 1 | 1 (100) | 1 (100)d | 1 | 1 (100) | 1 (100)d |
| Other detections | 4 | – | 4 (100) | 1 | – | 1 (100) |
| C. albicans | 3 | – | 3 (100) | 0 | – | 0 (–) |
| P. jirovecii | 1 | – | 1 (100) | 1 | – | 1 (100) |
| Total | 133 | 116 (87) | 73 (55) | 91 | 84 (92) | 59 (65) |
Microbiological findings provided by the syndromic PCR panel (FAP plus) compared to SDs in CAP patients (n = 72). Detections deemed as clinically relevant a pathogens are further specified. *Data are shown as number of detections with percentage of the respective microbe’s total detections (All methods combined) in brackets. **Data are shown as number of relevant detections with percentage of the respective microbe’s total relevant detections (All methods combined) in brackets. Statistically significant differences (McNemar’s test, p < 0.05) between the FAP plus and SDs, are marked with bold fonts.
CAP, community acquired pneumonia; FAP plus, Biomérieux BioFire FilmArray Pneumonia panel plus; SDs, standard diagnostic methods; –, not applicable; FIA, fluorescent immunoassay.
aThe clinical relevance of all detected microbes was evaluated retrospectively using pre-specified criteria (Supplementary material, S4).
bSputum culture detected five patients with S. pneumoniae. A pneumococcal antigen detection test in urine (Sofia S. Pneumoniae FIA, Quidel) was positive in ten patients, of which five were unique findings, whereas one was in combination with a positive blood culture.
cDetected in blood culture only.
dLegionella pneumophila antigen detection test in urine (Sofia Legionella FIA, Quidel).
Table 4.
Microbiological findings by use of the BioFire FilmArray Pneumonia panel plus versus standard microbiological methods in 24 patients with other respiratory tract infections.
| Microbes | Other respiratory tract infections (other RTIs) | |||||
|---|---|---|---|---|---|---|
| Total detections* | Detections deemeda relevant** | |||||
| All methods combined | FAP plus | Standard methods | All methods combined | FAP plus | Standard methods | |
| Viruses | 24 | 24 (100) | 20 (83) | 24 | 24 (100) | 20 (83) |
| Influenza A virus | 13 | 13 (100) | 13 (100) | 13 | 13 (100) | 13 (100) |
| Human metapneumovirus | 3 | 3 (100) | 2 (67) | 3 | 3 (100) | 2 (67) |
| Respiratory syncytial virus | 4 | 4 (100) | 3 (75) | 4 | 4 (100) | 3 (75) |
| Coronavirus | 2 | 2 (100) | – | 2 | 2 (100) | – |
| Parainfluenza virus | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| Rhino-/enterovirus | 2 | 2 (100) | 2 (100) | 2 | 2 (100) | 2 (100) |
| Bacteria | 22 | 22 (100) | 6 (27) | 0 | 0 (–) | 0 (–) |
| H. influenzae | 10 | 10 (100) | 4 (40) | 0 | 0 (–) | 0 (–) |
| S. pneumoniae | 4 | 4 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| M. catarrhalis | 3 | 3 (100) | 1 (33) | 0 | 0 (–) | 0 (–) |
| E. coli | 2 | 2 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| S. aureus | 2 | 2 (100) | 1 (50) | 0 | 0 (–) | 0 (–) |
| S. agalactiae | 1 | 1 (100) | 0 (0) | 0 | 0 (–) | 0 (–) |
| S. epidermidis | 0 | – | 0 (–) | 0 | – | 0 (–) |
| K. pneumoniae | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| S. marcescens | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| P. aeruginosa | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| Proteus spp. | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| K. oxytoca | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| A. calcoaceticus–A. baumanii complex | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| E. cloacae complex | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| M. pneumoniae | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| L. pneumophila | 0 | 0 (–) | 0 (–) | 0 | 0 (–) | 0 (–) |
| Other detections | 0 | – | 0 (–) | 0 | – | 0 (–) |
| C. albicans | 0 | – | 0 (–) | 0 | – | 0 (–) |
| P. jirovecii | 0 | – | 0 (–) | 0 | – | 0 (–) |
| Total | 46 | 46 (100) | 26 (57) | 24 | 24 (100) | 20 (83) |
Microbiological findings provided by the syndromic PCR panel (FAP plus) compared to SDs in patients with other RTIs (n = 24). Detections deemed as clinically relevant a pathogens are further specified. *Data are shown as number of detections with percentage of the respective microbe’s total detections (All methods combined) in brackets. **Data are shown as number of relevant detections with percentage of the respective microbe’s total relevant detections (All methods combined) in brackets. Statistically significant differences (McNemar’s test, p < 0.05) between the FAP plus and SDs, are marked with bold fonts.
RTI, respiratory tract infection; FAP plus, Biomérieux BioFire FilmArray Pneumonia panel plus; SDs, standard diagnostic methods; –, not applicable; FIA, fluorescent immunoassay.
aThe clinical relevance of all detected microbes was evaluated retrospectively using pre-specified criteria (Supplementary material, S4).
Discussion
This prospective study is one of the first to evaluate the real-world performance and time to results of a rapid syndromic PCR panel in patients presenting to hospital with suspected CAP. We demonstrated that the routine collection of lower respiratory tract specimens (induced sputum or endotracheal aspirates) in the ED was feasible and found a large improvement in both time to microbiological results and the microbiological yield by use of FAP plus compared to SDs. Study enrolment and collection of specimens were in general performed before medical imaging and laboratory results were available. Our patient population is therefore confined to suspected rather than confirmed CAP patients, with the rationale to maximize the potential impact on initial diagnostics and treatment decisions, and reflect actual clinical practice.
During the last decade, research on the etiology of CAP has often included PCR-based methods, although usually limited to a restricted selection of bacterial targets or specimens from the upper respiratory tract3,6,23,24. One of the most comprehensive studies is a British investigation of sputum from 323 patients with CAP8. Like our results, a pathogen was identified in 87% of patients representing a substantial improvement compared to standard diagnostic testing (30–40%)3,7. However, the British study analysed frozen samples retrospectively with resource-consuming in-house PCR procedures. A short analysis time and ease of use are major advantages of automated commercial PCR panels. When embedded in routine clinical practice, we observed a real-world median TAT of 2.6 h for the FAP plus, considerably faster than the standard in-house PCR test and sputum culture (Table 2). A direct consequence of a rapid TAT is the provision of near real-time information to treating physicians. This study thus demonstrates a promising potential for informed initial decisions on antimicrobial treatment and isolation. Still, further exploration is needed, preferably in a larger randomised controlled trial, before implementation as a routine test can be considered.
The most frequent pathogens in our CAP cohort were H. influenzae and S. pneumoniae, a finding similar to that in other studies6,8,17,24–27. In Norway, monotherapy with benzylpenicillin, 1.2 g every six hours, is the current empirical treatment recommendation for mild and moderate hospitalised CAP patients. The high prevalence of H. influenzae is consistent with previous PCR-based studies and needs further exploration, moreover, EUCAST has stated that there is still insufficient data for H. influenzae to set clinical breakpoints for benzylpenicillin28. Viral pathogens were also frequently detected in our CAP cohort, in agreement with other studies influenced by winter enrolment6,23. The growing recognition of an important role for viruses in CAP is thus supported by our work3,9–13.
Samples from the lower respiratory tract are often difficult to obtain in a clinically meaningful time frame. Previously reported collection rates of sputum range from 30 to 60%3,6,13,23 and this represents a major barrier to introducing rapid molecular diagnostics for CAP. Induced sputum was obtained from the majority of included patients (95%) in this study, underscoring that successful collection of this material is feasible, well tolerated and achievable in an ED setting. By traditional microscopic criteria, 42% of specimens were considered representative, a rate not different from other reports3,6,23. These microscopic criteria have been developed as a pragmatic quality check of samples in traditional culture-based diagnostics. Modern PCR methods can detect and quantify potentially pathogenic bacteria from complex background microbial populations. In this context, the value of pre-analytic microscopy remains uncertain29. Detections of viruses and atypical bacteria in RTI-patients are generally considered pathogenic3,10,12,30–32. Some data supports that detection of higher quantities of bacterial pathogens are rare among adults without RTI10,13,31,32; but this is disputed, especially in patients with chronic obstructive pulmonary disease33,34. Intended to aid in the differentiation between colonising bacteria and causative agents, the FAP plus provides semi-quantitative information on copy numbers for the bacterial pathobionts. Like others, we chose not to consider semi-quantitative values in our study, because of limited evidence from prior use, variable impact on antimicrobial treatment, and the possible influence of different comorbidities on patterns of colonisation and susceptibility to infections18. The correlation between semi-quantitative bacterial PCR levels and clinical relevance requires further exploration in larger settings.
Recent studies reporting on the performance of FAP plus also show a considerable increase in bacterial detections compared to SDs14–19. However, important limitations include heterogeneous patient populations (random combinations of community-, hospital-, and/or ventilator acquired pneumonia, as well as other RTIs), various sample materials (sputum, tracheal aspirates or broncho-alveolar lavage) and inconsistent testing by SDs14–17,19. Clinical data are often scarce, and incompletely accounted for. Therefore, the relevance of many of the detected microbes is uncertain. Evaluation of the clinical relevance of all microbiological results is thus a major strength of this study. Only one other recent FAP plus study in CAP patients has made an attempt to determine potential colonisers18. Unlike our study, their algorithm included an evaluation of procalcitonin levels to determine relevance and category of infection. The utility of procalcitonin to distinguish viral from bacterial infections is, however, uncertain35. Anyhow, determining relevance is difficult, and although not being causative per se, microbes of uncertain relevance could still contribute to an ongoing infectious process. Information about these microbes could therefore be valuable in the case of antibiotic treatment failure.
The pragmatic nature of this study i.e., embedded in routine clinical care hospital practice, the prospective enrolment of patients with community acquired RTIs, application of a stringent case definition of CAP and the analysis of rigorously collected, homogeneous lower respiratory tract samples are major strengths of our work. The study has limitations; the inclusion of a limited number of CAP patients and a short recruitment window during the winter months, suggesting that the findings should be confirmed in further studies. Moreover, patients that were unable to provide a sample from the lower respiratory tract were excluded; resulting in the most severe cases of CAP not being represented, which in turn could explain the low in-hospital mortality rate observed in this study (1%) compared to similar settings36.
In conclusion, the use of a rapid syndromic PCR panel for respiratory pathogens was associated with a dramatically reduced time to actionable results and increased detection of clinically relevant pathogens, compared to SDs. A stepwise algorithm for sampling of respiratory specimens with induced sputum and tracheal aspiration was feasible in routine clinical practice and led to obtaining a lower respiratory tract sample in the majority of patients. This suggests that syndromic PCR-based testing may be feasible in acute settings and holds potential to provide clinically actionable results in near real-time. The clinical impact of rapid syndromic pneumonia panels on antibiotic use and patient outcome should therefore be urgently assessed in randomised controlled trials.
Supplementary Information
Acknowledgements
We gratefully acknowledge the CAPNOR study group, the study nurses and the staff at the emergency department, and staff at the Department of Microbiology. Last, we thank all the enrolled patients.
Author contributions
H.M.S.G. with advice from C.R., D.F.J., T.W.C., C.H.V.W., Ø.K., and E.U. conceptualized and designed the study. D.M., S.T.K., S.S., and R.B. coordinated patient recruitment and follow-up. M.E. supervised the microbiology studies. S.S., and S.T.K., wrote the first draft of the manuscript which subsequently underwent revisions with contribution from all co-authors. D.M. proposed the section on “Determining the clinical relevance of microbiological detections” detailed in the supplementary material. S.S. and S.T.K. performed the data analysis and generated tables and figures, with advice from Ø.K., M.E., T.W.C. and C.R. C.R. reviewed the statistical analysis. H.M.S.G. and S.T.K. had primary responsibility for the final content of the manuscript. All authors have approved and contributed to the final manuscript.
Funding
This work was supported by the Trond Mohn Foundation (RESPNOR; BFS2019TMT06), The Research Council of Norway (NORCAP; 288718), The University of Bergen and Haukeland University Hospital.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
C. H. van Werkhoven has received non-financial and financial research support from BioMérieux. Tristan W. Clark has received consulting fees, honoraria for lectures and manuscript writing/educational events, support for attending meetings and other services from BioMérieux and BioFire LLC. The other authors have nothing to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: D. Markussen and M. Ebbesen.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
H. M. S. Grewal, Email: harleen.grewal@uib.no
The CAPNOR Study Group:
R. Bjørneklett, T. W. Clark, M. Ebbesen, D. Faurholt-Jepsen, H. M. S. Grewal, L. Heggelund, S. T. Knoop, Ø. Kommedal, D. Markussen, P. Ravn, C. Ritz, S. Serigstad, E. Ulvestad, and C. H. van Werkhoven
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-03741-7.
References
- 1.World Health Organization: The top 10 causes of death, https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (Accessed Mar 2021).
- 2.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain S, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N. Engl. J. Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett JG. Diagnostic tests for agents of community-acquired pneumonia. Clin. Infect. Dis. 2011;52(Suppl 4):S296–304. doi: 10.1093/cid/cir045. [DOI] [PubMed] [Google Scholar]
- 5.Musher DM, et al. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: Results of a one-year study. J. Infect. 2013;67:11–18. doi: 10.1016/j.jinf.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holter JC, et al. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: A 3-year prospective study in Norway. BMC Infect. Dis. 2015;15:64. doi: 10.1186/s12879-015-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roysted W, et al. Aetiology and risk factors of community-acquired pneumonia in hospitalized patients in Norway. Clin. Respir. J. 2016;10:756–764. doi: 10.1111/crj.12283. [DOI] [PubMed] [Google Scholar]
- 8.Gadsby NJ, et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin. Infect. Dis. 2016;62:817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karhu J, Ala-Kokko TI, Vuorinen T, Ohtonen P, Syrjala H. Lower respiratory tract virus findings in mechanically ventilated patients with severe community-acquired pneumonia. Clin. Infect. Dis. 2014;59:62–70. doi: 10.1093/cid/ciu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Self WH, et al. Respiratory viral detection in children and adults: Comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016;213:584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennings LC, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 13.Clark TW, et al. Adults hospitalised with acute respiratory illness rarely have detectable bacteria in the absence of COPD or pneumonia; viral infection predominates in a large prospective UK sample. J. Infect. 2014;69:507–515. doi: 10.1016/j.jinf.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webber DM, Wallace MA, Burnham CA, Anderson NW. Evaluation of the BioFire FilmArray pneumonia panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J. Clin. Microbiol. 2020;58:e00343-20. doi: 10.1128/JCM.00343-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchan BW, et al. Practical comparison of the BioFire FilmArray pneumonia panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J. Clin. Microbiol. 2020;58:e00135-20. doi: 10.1128/JCM.00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rand KH, et al. Performance of a semi-quantitative multiplex bacterial PCR panel compared with standard microbiological laboratory results: 396 patients studied with the BioFire® pneumonia panel. Open Forum Infect. Dis. 2020 doi: 10.1093/ofid/ofaa560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edin A, Eilers H, Allard A. Evaluation of the Biofire Filmarray Pneumonia panel plus for lower respiratory tract infections. Infect. Dis. 2020 doi: 10.1080/23744235.2020.1755053. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert DN, et al. Enhanced detection of community-acquired pneumonia pathogens with the BioFire(R) pneumonia FilmArray(R) panel. Diagn. Microbiol. Infect. Dis. 2021;99:115246. doi: 10.1016/j.diagmicrobio.2020.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gastli N, et al. Multicentric evaluation of BioFire FilmArray Pneumonia Panel for rapid bacteriological documentation of pneumonia. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Biomérieux. BioFire® FilmArray Pneumonia plus, https://www.biomerieux-diagnostics.com/biofire-filmarray-pneumonia-panel (Accessed Apr 2021).
- 21.Biomérieux. FilmArray® Pneumonia Panel plus, Instructions for use, https://www.biofiredx.com/support/documents/ (2019) (Accessed Jan 2021).
- 22.Leber, A. M. Clinical Microbiology Procedures Handbook, Vol. 1‐3, 4th ed (2016).
- 23.Bjarnason A, et al. Incidence, etiology, and outcomes of community-acquired pneumonia: A population-based study. Open Forum Infect. Dis. 2018;5:ofy010. doi: 10.1093/ofid/ofy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stralin K, Olcen P, Tornqvist E, Holmberg H. Definite, probable, and possible bacterial aetiologies of community-acquired pneumonia at different CRB-65 scores. Scand. J. Infect. Dis. 2010;42:426–434. doi: 10.3109/00365540903552353. [DOI] [PubMed] [Google Scholar]
- 25.Fally M, et al. The increasing importance of Haemophilus influenzae in community-acquired pneumonia: Results from a Danish cohort study. Infect. Dis. 2020 doi: 10.1080/23744235.2020.1846776. [DOI] [PubMed] [Google Scholar]
- 26.Woodhead M. Community-acquired pneumonia in Europe: Causative pathogens and resistance patterns. Eur. Respir. J. Suppl. 2002;36:20s–27s. doi: 10.1183/09031936.02.00702002. [DOI] [PubMed] [Google Scholar]
- 27.Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. Etiology of community-acquired pneumonia: Increased microbiological yield with new diagnostic methods. Clin. Infect. Dis. 2010;50:202–209. doi: 10.1086/648678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, Version 11.0, https://www.eucast.org/ (2021).
- 29.Saukkoriipi A, Palmu AA, Jokinen J. Culture of all sputum samples irrespective of quality adds value to the diagnosis of pneumococcal community-acquired pneumonia in the elderly. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:1249–1254. doi: 10.1007/s10096-019-03536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman D, et al. Respiratory viruses in adults with community-acquired pneumonia. Chest. 2010;138:811–816. doi: 10.1378/chest.09-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stralin K, Tornqvist E, Kaltoft MS, Olcen P, Holmberg H. Etiologic diagnosis of adult bacterial pneumonia by culture and PCR applied to respiratory tract samples. J. Clin. Microbiol. 2006;44:643–645. doi: 10.1128/JCM.44.2.643-645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunnarsson RK, Holm SE, Soderstrom M. The prevalence of potential pathogenic bacteria in nasopharyngeal samples from healthy children and adults. Scand. J. Prim. Health Care. 1998;16:13–17. doi: 10.1080/028134398750003340. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs DM, Ochs-Balcom HM, Zhao J, Murphy TF, Sethi S. Lower Airway Bacterial Colonization Patterns and Species-Specific Interactions in Chronic Obstructive Pulmonary Disease. J. Clin. Microbiol. 2018;56:e00330-18. doi: 10.1128/JCM.00330-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouquet J, et al. Microbial burden and viral exacerbations in a longitudinal multicenter COPD cohort. Respir. Res. 2020;21:77. doi: 10.1186/s12931-020-01340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamat IS, Ramachandran V, Eswaran H, Guffey D, Musher DM. Procalcitonin to distinguish viral from bacterial pneumonia: A systematic review and meta-analysis. Clin. Infect. Dis. 2020;70:538–542. doi: 10.1093/cid/ciz545. [DOI] [PubMed] [Google Scholar]
- 36.Fally M, et al. Improved treatment of community-acquired pneumonia through tailored interventions: Results from a controlled, multicentre quality improvement project. PLoS ONE. 2020;15:e0234308. doi: 10.1371/journal.pone.0234308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


