ABSTRACT
Distinct Burkholderia strains were isolated from soil samples collected in tropical northern Australia (Northern Territory and the Torres Strait Islands, Queensland). Phylogenetic analysis of 16S rRNA and whole genome sequences revealed these strains were distinct from previously described Burkholderia species and assigned them to two novel clades within the B. pseudomallei complex (Bpc). Because average nucleotide identity and digital DNA-DNA hybridization calculations are consistent with these clades representing distinct species, we propose the names Burkholderia mayonis sp. nov. and Burkholderia savannae sp. nov. Strains assigned to B. mayonis sp. nov. include type strain BDU6T (=TSD-80; LMG 29941; ASM152374v2) and BDU8. Strains assigned to B. savannae sp. nov. include type strain MSMB266T (=TSD-82; LMG 29940; ASM152444v2), MSMB852, BDU18, and BDU19. Comparative genomics revealed unique coding regions for both putative species, including clusters of orthologous genes associated with phage. Type strains of both B. mayonis sp. nov. and B. savannae sp. nov. yielded biochemical profiles distinct from each other and from other species in the Bpc, and profiles also varied among strains within B. mayonis sp. nov. and B. savannae sp. nov. Matrix-assisted laser desorption ionization time-of-flight (MLST) analysis revealed a B. savannae sp. nov. cluster separate from other species, whereas B. mayonis sp. nov. strains did not form a distinct cluster. Neither B. mayonis sp. nov. nor B. savannae sp. nov. caused mortality in mice when delivered via the subcutaneous route. The addition of B. mayonis sp. nov. and B. savannae sp. nov. results in a total of eight species currently within the Bpc.
IMPORTANCE Burkholderia species can be important sources of novel natural products, and new species are of interest to diverse scientific disciplines. Although many Burkholderia species are saprophytic, Burkholderia pseudomallei is the causative agent of the disease melioidosis. Understanding the genomics and virulence of the closest relatives to B. pseudomallei, i.e., the other species within the B. pseudomallei complex (Bpc), is important for identifying robust diagnostic targets specific to B. pseudomallei and for understanding the evolution of virulence in B. pseudomallei. Two proposed novel species, B. mayonis sp. nov. and B. savannae sp. nov., were isolated from soil samples collected from multiple locations in northern Australia. The two proposed species belong to the Bpc but are phylogenetically distinct from all other members of this complex. The addition of B. mayonis sp. nov. and B. savannae sp. nov. results in a total of eight species within this significant complex of bacteria that are available for future studies.
KEYWORDS: Burkholderia mayonis sp. nov., Burkholderia savannae sp. nov., Burkholderia pseudomallei complex, BDU6T, MSMB266T, Burkholderia, novel species
INTRODUCTION
The genus Burkholderia was recently divided into Burkholderia sensu stricto, Paraburkholderia, Caballeronia, Robbsia, and Pararobbsia. Together, these taxonomic groups comprise over 100 described species (https://www.bacterio.net/) that can have pathogenic, mutualistic, and/or commensal relationships with plants, animals, and/or humans (1–3). This division resulted in Burkholderia sensu stricto containing most of the opportunistic pathogens which belonged to one of two species groups: the Burkholderia pseudomallei complex (Bpc) and the Burkholderia cepacia complex (Bcc). New species are regularly described in Burkholderia sensu stricto (4–9) and the majority of species within it are naturally found in the environment, primarily in soil and water (10).
Members of the Bpc exhibit diverse niche adaptation. B. pseudomallei has adapted to opportunistic pathogenicity, B. mallei to obligate pathogenicity, and B. thailandensis (11), B. oklahomensis (12), B. humptydooensis (5), and B. singularis (6) to environmental saprophytism with (except for B. humptydooensis) occasional pathogenicity. B. pseudomallei is the causative agent of the serious human disease melioidosis and is commonly isolated from soil and water in endemic areas (13). B. mallei is a clone within B. pseudomallei that has undergone host-adapted reductive niche specialization toward obligate pathogenicity in the form of the disease glanders (14). Given these niche differences, the ongoing study of the Bpc can provide insights into the evolutionary mechanisms driving bacterial virulence and niche adaptation. Moreover, the classification of pathogenic members of the Bpc (B. pseudomallei and B. mallei) as U.S. Tier 1 Select Agents due to their potential to be aerosolized and used as biowarfare agents (13, 15), and the suggestion that global melioidosis cases may be severely underestimated (16), means that closely related species are of great interest, due to their potential for cross-reactivity in diagnostic/detection technologies used across defense, health, and environmental applications. In addition, novel Burkholderia species are of significant interest to multiple scientific fields because previously described members of this genus, including members of the Bpc, have been shown to be important sources of new natural products (17, 18).
In this study, we propose the addition of two additional members to the Bpc: B. mayonis sp. nov. and B. savannae sp. nov. We used a polyphasic approach, including bioinformatic and biochemical analyses, to confirm that they are distinct species and to investigate their unique coding region sequences, as well as those they share with other members of the Bpc, to better understand diversification and evolution within this group.
RESULTS AND DISCUSSION
Bacterial growth and characteristics.
Growth of both type strains, BDU6T (B. mayonis sp. nov.) and MSMB266T (B. savannae sp. nov.), was observed on all media types tested in plate format (Ashdown’s, Columbia Blood, MacConkey, and Luria-Bertani) after 24 h when incubated at 25°C and 37°C, with optimal growth for both strains observed at 37°C on all media types after at least 48 h of incubation. Incubation at 25°C for at least 48 h resulted in optimal growth only on Columbia blood agar, and on all other media types after at least 72 h of incubation. Limited to no growth was observed at 42°C for all strains on the four media types. Colony morphology varied depending on media type (Fig. S2 and S3). Unless otherwise noted, Luria-Bertani agar was the medium used during various analyses, and strains were stored long-term in cryovials containing Luria-Bertani broth with 20% glycerol at −80°C.
Biochemical differentiation of the type strain of B. mayonis sp. nov. (BDU6T) from its closest genetic near neighbor, B. oklahomensis (Fig. 1), was observed in the inability of B. mayonis sp. nov. to hydrolyze esculin and assimilate arabinose. Biochemical differentiation of the type strain of B. savannae sp. nov. (MSMB266T) from B. oklahomensis was observed in the inability of B. savannae sp. nov. to hydrolyze esculin and assimilate both arabinose and maltose. Type strains of all three of these species were positive for arginine, adipate, caprate, citrate, gelatin, gluconate, glucose, malate, mannitol, mannose, nitrate, N-acetylglucosamine, and phenylacetate. All three type strains were negative for glucose (acidification), tryptophan, urea, and 4-nitrophenyl-β-d-galactopyranoside (PNPG) (Table 1).
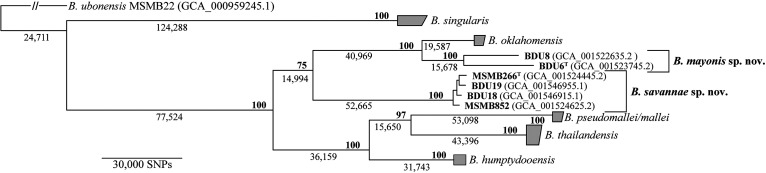
FIG 1.
Core genome phylogeny of 66 strains (Table S3) in the B. pseudomallei complex, including two B. mayonis sp. nov. strains and four B. savannae sp. nov. strains. This maximum-likelihood phylogeny was created using core genome SNPs shared by all strains and rooted on B. ubonensis strain MSMB22 as an outgroup. Bold numbers at nodes indicate bootstrap support values and non-bolded numbers indicate the number of core SNPs defining that node. Collapsed nodes are shown in gray. The type strains are reflected with a T superscript in the strain name.
TABLE 1.
Differential phenotypic characteristics of strains of B. mayonis sp. nov. and B. savannae sp. nov., as well as representative strains from closely related species within the B. pseudomallei complexa
| Biochemical reaction | Characteristic (compound present in medium or assimilated by strain) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bpb K96243 |
Bt E264 |
Bo C6786 |
Bm BDU6T |
Bm BDU8 |
Bs MSMB266T |
Bs MSMB852 |
Bs BDU18 |
Bs BDU19 |
|
| Nitrate | +c | + | + | + | + | + | + | - | + |
| Tryptophan | + | - | - | - | - | - | - | - | - |
| Arginine | + | - | + | + | + | + | + | + | + |
| Esculin | - | + | + | - | + | - | - | - | + |
| Gelatin | + | + | + | + | + | + | - | + | + |
| Arabinose assimilation | - | + | + | - | - | - | + | - | - |
| Maltose assimilation | - | + | + | + | - | - | + | - | - |
aSpecies: Bp, Burkholderia pseudomallei; Bt, B. thailandensis; Bo, B. oklahomensis; Bm, B. mayonis sp. nov.; Bs, B. savannae sp. nov. All strains were positive for the assimilation of adipate, caprate, citrate, gluconate, glucose, malate, mannitol, mannose, N-acetylglucosamine, and phenylacetate; and all strains were negative for glucose (acidification), urea, and PNPG (these data not shown).
bData obtained from a previous study (34).
c+, positive reaction; -, negative reaction.
Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) of the two B. mayonis sp. nov. and four B. savannae sp. nov. strains revealed that they cluster with other members of the Bpc and with B. ubonensis. Within the MALDI-TOF MS cluster which contained the Bpc species and B. ubonensis, the four Burkholderia savannae sp. nov. strains formed a cluster separate from other species, whereas Burkholderia mayonis sp. nov. strains did not form a distinct cluster (Fig. S4).
Antimicrobial susceptibility screening.
All six B. mayonis sp. nov. and B. savannae sp. nov. strains were susceptible in vitro to amoxicillin/clavulanate, ceftazidime, doxycycline, imipenem, and trimethoprim-sulfamethoxazole based on CLSI breakpoints for B. pseudomallei (M45) (19). All of these strains were susceptible in vitro to meropenem and were susceptible or intermediate to chloramphenicol, with the exception of BDU6T, which displayed resistance based on the CLSI breakpoints for B. cepacia complex (M100) (20). All MICs are reported in Table 2, including those for other antimicrobials for which no breakpoints are established.
TABLE 2.
Summary of MICs of antimicrobials determined in duplicate by the microdilution method for B. mayonis sp. nov. (Bm) and B. savannae sp. nov. (Bs)
| Antimicrobial substance | MIC (mg/liter) |
||||||
|---|---|---|---|---|---|---|---|
| Resistance breakpoint (mg/liter) if available | Bm BDU6T |
Bm BDU8 |
Bs MSMB266T |
Bs MSMB852 |
Bs BDU18 |
Bs BDU19 |
|
| Amoxicillin/clavulanic acida | ≥32/16 (40) | 8/4 | ≤4/2 | 8/4 | 8/4 | 8/4 | 8/4 |
| Azithromycin | >64 | >64 | >64 | >64 | >64 | >64 | |
| Carbenicillin | 128 | 64 | 64 | 32 | 64 | 64 | |
| Ceftazidime | ≥32 (40) | ≤4 | ≤4 | ≤4 | ≤4 | ≤4 | ≤4 |
| Ceftazidime-avibactamb | 4/4 | 1/4 | ≤0.5/4 | ≤0.5/4 | ≤0.5/4 | ≤0.5/4 | |
| Chloramphenicol | ≥32 (41) | 32 | 16 | 8 | 16 | 8 | 16 |
| Ciprofloxacin | 2 | ≤0.5 | ≤0.5 | ≤0.5 | 1 | 1 | |
| Doripenem | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| Doxycycline | ≥16 (40) | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Gentamicin | 32 | >64 | 32 | 32 | 64 | 64 | |
| Imipenem | ≥16 (40) | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Kanamycin | 16 | 32 | 16 | 16 | 16 | 32 | |
| Meropenem | ≥16 (41) | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Piperacillin | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 | |
| Piperacillin-tazobactam | ≤8/4 | ≤8/4 | ≤8/4 | ≤8/4 | ≤8/4 | ≤8/4 | |
| Polymyxin B | 512 | >2048 | 512 | >2048 | >2048 | >2048 | |
| Sulfamethoxazole | >512 | 256 | >512 | >512 | >512 | >512 | |
| Tigecycline | 1 | 0.5 | 2 | 1 | 1 | 1 | |
| Trimethoprim | 4 | 4 | 2 | ≤1 | 4 | 2 | |
| Trimethoprim/sulfamethoxazole | ≥4/76 (40) | 2/38 | ≤1/19 | ≤1/19 | ≤1/19 | ≤1/19 | ≤1/19 |
For amoxicillin-clavulanic acid, clavulanic acid was maintained at 4 μg/ml in all wells.
For ceftazidime-avibactam, avibactam was maintained at 4 μg/ml in all wells.
Virulence screening.
Although none of the examined B. pseudomallei virulence genes were conserved in any of the B. mayonis sp. nov. or B. savannae sp. nov. genomes, there was a homolog to the type VI secretion system in the B. savannae sp. nov. genome (Table S1). B. mayonis sp. nov. strain BDU6T, B. savannae sp. nov. strain MSMB266T, and B. thailandensis strain E264T did not cause mortality in any mice at any of the doses when delivered via the subcutaneous route, nor did any mice show outward signs of illness. In comparison, subcutaneous infections of fully virulent B. pseudomallei results in 50% mortality within 10 days at a dose of 103 CFU (21). It remains unknown if delivery via the inhalation route might increase the pathogenicity of these species; B. thailandensis E264T can cause high mortality in mice at doses of 104 to 106 CFU when delivered as an aerosol (22–24). The lack of mortality in mice suggests that B. mayonis sp. nov. and B. savannae sp. nov. are likely environmental saprophytes, similar to most other members of the Bpc.
Genetic and genomic comparative analysis.
The 16S rRNA phylogeny revealed two novel clades for B. mayonis sp. nov. and B. savannae sp. nov. that were distinct from each other and from the other closely related Burkholderia species in the Bpc (Fig. S5). As in B. pseudomallei, B. thailandensis, B. humptydooensis, B. oklahomensis, and B. singularis (6), four rRNA operons were present in all examined B. mayonis sp. nov. and B. savannae sp. nov. strains, with the exception of B. savannae sp. nov. strain BDU19, which has six rRNA operons. B. mayonis sp. nov. strains BDU6T and BDU8, and B. savannae sp. nov. MSMB266T, each had two unique versions among the four copies of 16S rRNA, whereas the four copies within B. savannae sp. nov. strains MSMB852 and BDU18, and the six copies within BDU19, were all identical (Fig. S5). A pairwise similarity matrix shows the percent identity and number of single nucleotide polymorphisms (SNPs) between each of the unique16 rRNA sequences (Table S2). Briefly, within B. mayonis sp. nov. and B. savannae sp. nov., the percent identity of the 16S rRNA sequences ranged from 99.1 to 99.9% (1 to 13 SNPs) and from 99.7 to 100% (0 to 4 SNPs), respectively (Table S2). The species most closely related to B. mayonis sp. nov. in the 16S rRNA phylogeny (Fig. S5) was B. thailandensis (strain E264), with a percent identity ranging from 99.1 to 99.9% (12 to 14 SNPs; Table S2) depending on the B. mayonis sp. nov. strain. The species most closely related to B. savannae sp. nov. in the 16S rRNA phylogeny was B. mayonis sp. nov., with a percent identity ranging from 98.6 to 99.0% (16 to 21 SNPs; Table S2) depending on the strain (Fig. S5).
Each strain in this study was assigned a distinct sequence type (ST) using the B. pseudomallei complex MLST system (Table 3), demonstrating the significant genetic diversity found within both species. This is especially notable considering that four of the strains (B. mayonis sp. nov., BDU6T and BDU8; B. savannae sp. nov., BDU18 and BDU19) were collected from the same soil sample. Although BDU18 and BDU19 appear closely related on the core genome phylogeny (Fig. 1), there are 4,962 SNPs separating these two isolates.
TABLE 3.
Whole genome sequence, sequence type (ST), and epidemiology data for B. pseudomallei complex species, including B. mayonis sp. nov. and B. savannae sp. novg
| Species and strain | GC content (%) | Genome size (Mb) | No. of CDSd | NCBI assembly accession no. | STf | Region of isolation or country | yr | Originating lab |
|---|---|---|---|---|---|---|---|---|
| B. mayonis sp. nov. BDU6T | 66.25 | 6.6a | 5,672 | ASM152374v2e | 1003 | Badu Island, QLD | 2011 | James Cook University |
| B. mayonis sp. nov. BDU8 | 66.47 | 7.4a | 6,368 | ASM152263v2e | 962 | Badu Island, QLD | 2011 | James Cook University |
| B. savannae sp. nov. MSMB266T | 67.05 | 7.4a,c | 6,408 | ASM152444v2e | 646 | Acacia Hills, NT | 2006 | Menzies School of Health and Research |
| B. savannae sp. nov. MSMB852 | 67.32 | 7.1a,c | 6,024 | ASM152462v2e | 1773 | Robin Falls, NT | 2010 | Menzies School of Health and Research |
| B. savannae sp. nov. BDU18 | 67.25 | 7.2b | 6,056 | ASM154691v1 | 963 | Badu Island, QLD | 2011 | James Cook University |
| B. savannae sp. nov. BDU19 | 67.49 | 6.9b | 5,785 | ASM154695v1 | 964 | Badu Island, QLD | 2011 | James Cook University |
| B. singularis MSMB175 | 64.80 | 5.7 | 4,715 | ASM171887v1e | n/a | Australia | 2004 | Menzies School of Health and Research |
| B. humptydooensis MSMB43 | 67.14 | 7.3c | 6,324 | ASM151374v1e | 318 | Australia | 1995 | Menzies School of Health and Research |
| B. thailandensis E264 | 67.60 | 6.7 | 5,652 | ASM1236v1e | 80 | Thailand | 1994 | n/a (external genome) |
| B. oklahomensis C6786 | 66.90 | 7.1 | 6,097 | ASM17037v1 | 81 | United States | 1973 | n/a (external genome) |
| B. pseudomallei K96243 | 68.05 | 7.2 | 5,948 | ASM1154v1e | 10 | Thailand | 1998 | n/a (external genome) |
| B. mallei ATCC 23344 | 68.50 | 5.8 | 5,006 | ASM1170v1 | 40 | Burma | 1944 | n/a (external genome) |
PacBio sequencing from this study resulting in a complete genome.
PacBio sequencing from this study resulting in four contigs.
One plasmid present.
CDS = coding DNA sequences.
Complete genome assembly available from NCBI.
Based on the B. pseudomallei MLST (https://pubmlst.org).
Two chromosomes are present for all genomes, and some also include a single plasmid. All B. mayonis sp. nov. and B. savannae sp. nov. strains originated from Australia, and all were isolated from soil. n/a, not applicable.
Finished assemblies were completed for both B. mayonis sp. nov. strains (BDU6T and BDU8) and for two of the four B. savannae sp. nov. strains (MSMB266T and MSMB852) using PacBio sequencing. The assemblies for B. mayonis sp. nov. strains BDU6T and BDU8 consist of two contigs, corresponding to the two chromosomes typical of Burkholderia spp.: chromosomes 1 and 2 of BDU6T are 3,838,800 bp and 2,752,114 bp, respectively, whereas chromosomes 1 and 2 of BDU8 are 4,439,942 bp and 2,917,588 bp, respectively. The assemblies for B. savannae sp. nov. strains MSMB266T and MSMB852 consist of three contigs each, corresponding to two chromosomes and one plasmid each: chromosome 1, chromosome 2, and the plasmid of MSMB266T (pMSMB0266) are 4,228,278 bp, 2,824,254 bp, and 375,023 bp, respectively, whereas chromosome 1, chromosome 2, and the plasmid of MSMB852 (pMSMB0852) are 4077888 bp, 2934072 bp, and 69213 bp, respectively. The PacBio assemblies for the other two B. savannae sp. nov. strains, BDU18 and BDU19, consist of four contigs each, with contig sizes of 4,097,543 bp, 249,544 bp, 66,284 bp, and 2,746,170 bp for BDU18, and 2,833,644 bp; and sizes of 2,161,131 bp, 1,648,896 bp, and 215,161 bp for BDU19 (Table 3).
The core genome phylogeny revealed the phylogenetic positions of B. mayonis sp. nov. and B. savannae sp. nov. in relation to each other and to other species in the Bpc (Fig. 1). B. savannae sp. nov. forms a distinct clade that is separate from all other species in the Bpc. Although B. mayonis sp. nov. is most closely related to B. oklahomensis, it also forms a distinct and separate clade with >35,000 core genome SNPs separating it from B. oklahomensis.
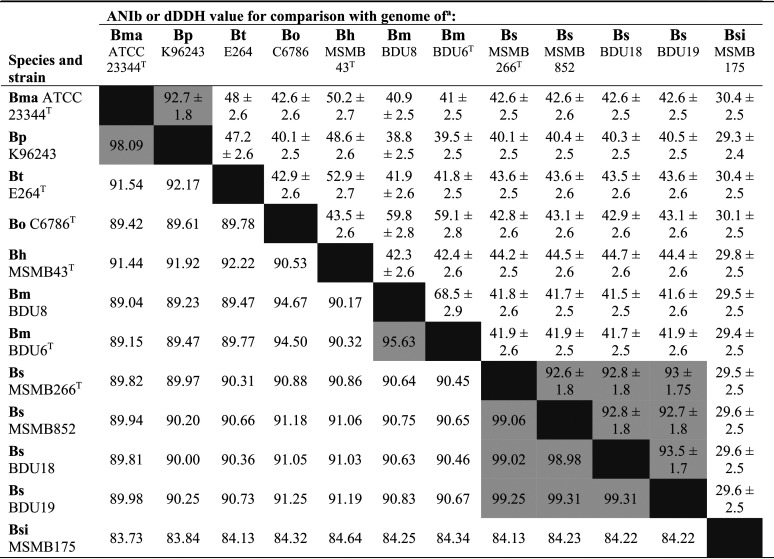
The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values calculated among the B. mayonis sp. nov. and B. savannae sp. nov. strains, and between them and strains from other species in the Bpc, supports our proposal that the B. mayonis sp. nov. and B. savannae sp. nov. strains belong to their corresponding species and that B. mayonis sp. nov. and B. savannae sp. nov. are distinct from all other Bpc species. Although the two B. mayonis sp. nov. strains have a dDDH value of 68.5 ± 2.9, which is slightly below the similarity threshold defining members of the same species, the ANI value (95.63%) supports the proposal that these two strains belong to the same species. The amount of genetic diversity observed between these two B. mayonis sp. nov. strains is quite intriguing, especially given that both strains were collected from not only the same location but also from the same soil sample. Isolating additional B. mayonis sp. nov. strains from soil collected in other locations will shed important new insights on overall levels of genetic diversity within this novel species. The ANI and dDDH values for the four B. savannae sp. nov. strains (ANI: 98.98% to 99.31%, dDDH: 92.6 ± 1.8 to 93.5 ± 1.7; Table 4) clearly suggest that these strains are members of the same species. Collectively, ANI values above 95% and/or dDDH values above 70 indicate that each set of strains belongs to its corresponding single species, including the proposed B. mayonis sp. nov. type strains BDU6T and the proposed B. savannae sp. nov. type strain MSMB266T. As expected, ANI values between B. pseudomallei and its host-adapted clone, B. mallei, were >95%, as previously shown (4, 14). However, the remaining ANI values <95% and dDDH values <70% indicate separate species for B. mayonis sp. nov., B. savannae sp. nov., and for the other Bpc species, with ANI values ranging from 83.73% to 94.67% and dDDH values ranging from 29.3 ± 2.4 to 59.8 ± 2.8 (Table 4). This confirms that the B. mayonis sp. nov. strains comprise a distinct species from B. oklahomensis and the other species in the Bpc, as do the B. savannae sp. nov. strains.
TABLE 4.
ANI and dDDH values for whole-genome sequence similaritiesb
Average nucleotide identity (ANIb) are shown in the bottom left half of the matrix (below the line of identity, i.e., the line formed by blank cells for comparison of strains with themselves); digital DNA-DNA hybridization (dDDH) (with confidence intervals) are shown in the top right half of the matrix. Values in shaded boxes represent values above the similarity threshold that defines members of the same species.
Assemblies used for analyses are listed in Table 3. Species are as follows: Bma, B. mallei; Bp, Burkholderia pseudomallei; Bt, B. thailandensis; Bo, B. oklahomensis; Bh, B. humptydooensis; Bm, B. mayonis sp. nov.; Bs, B. savannae sp. nov.; Bsi, B. singularis
The sizes of the pan-genomes in B. mayonis sp. nov. and B. savannae sp. nov. were 7,460 and 7,804 coding DNA sequences (CDSs), respectively, with core-genome sizes of 4,448 and 5,435 CDSs, respectively. There were 223 CDSs within B. mayonis sp. nov. and 159 CDSs within B. savannae sp. nov. that shared no close homolog to those within all other examined public Burkholderia genome assemblies (n = 3,269). An analysis based on clusters of orthologous genes (COGs) identified the broad functional categories of some of these unique genes (Fig. 2), although the majority of CDSs could not be classified or else their function was unknown. Many unique CDSs in both B. mayonis sp. nov. and B. savannae sp. nov. were identified in clusters. For example, a number of unique coding regions in a contiguous cluster were associated with phage (B. mayonis sp. nov., in strain BDU8 WS71_RS21930 to WS71_RS22315; B. savannae sp. nov., in strain BDU18 WS72_RS13230 to WS72_RS13570), suggesting that these regions are mobile genetic elements associated with phage integration into the chromosome. Although other phages have been associated with virulence in Burkholderia (25), the function of these particular phages is not known and could be the focus of future study.
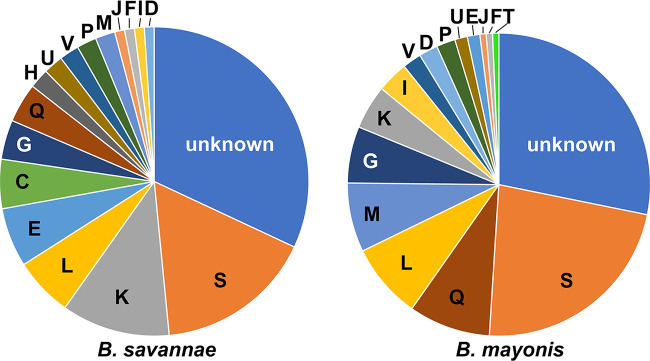
FIG 2.
Cluster of orthologous genes (COG) classifications (n = 18) of unique coding DNA sequences (CDSs) in B. savannae sp. nov. strains (n = 97 unique CDSs) and B. mayonis sp. nov. strains (n = 149 unique CDSs), including some unique CDSs that have no homolog, 31 in B. savannae sp. nov. and 42 in B. mayonis sp. nov., which are assigned to the “unknown” category. The COG categories are as follows with the number of unique CDSs for B. savannae sp. nov. and B. mayonis sp. nov., respectively, listed after each COG category: (C) energy production and conversion (5; 0), (D) cell cycle control and mitosis (1; 3), (E) amino acid metabolism and transport (6; 2), (F) nucleotide metabolisms and transport (1; 1), (G) carbohydrate metabolism and transport (4; 9), (H) coenzyme metabolism (2; 0), (I) lipid metabolism (1; 5), (J) translation (1; 1), (K) transcription (11; 7), (L) replication, recombination and repair (6; 12), (M) cell wall/membrane/envelop biogenesis (2; 11), (P) inorganic ion transport and metabolism (2; 3), (Q) secondary structure (4; 13), (S) function unknown (16; 34), (T) signal transduction (0; 1), (U) intracellular trafficking and secretion (2; 2), (V) defense mechanisms (2; 3). All classifications were performed with the eggNOG-mapper.
The ability to distinguish between B. mayonis sp. nov. or B. savannae sp. nov. and other commonly isolated species of the Bpc, such as B. pseudomallei and B. thailandensis, in environmental and (less likely) clinical samples is important. Obviously, this could be achieved via whole genome sequencing of isolates, but often this is not possible, particularly in developing areas of the world. Different colony morphologies on Ashdown’s agar should provide a clear distinction between these two novel species and B. pseudomallei and B. thailandensis; however, there could be morphological differences within species based on differences among strains, across geographic locations, and among different laboratories. Fortunately, distinguishing B. mayonis sp. nov. or B. savannae sp. nov. from other Burkholderia spp. can be achieved with biochemical tests. B. mayonis sp. nov. and B. savannae sp. nov. can be distinguished from B. pseudomallei with tryptophan and from B. thailandensis with arginine. Of course, the most definitive way to distinguish among any of the Bpc species would be to use whole genome sequencing (4) or species-specific PCR assays, if available.
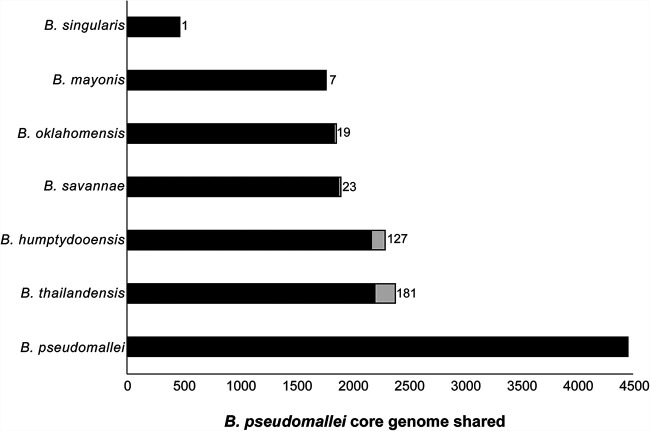
There are several reasons why members of the Bpc, including B. mayonis sp. nov. and B. savannae sp. nov., are of interest to the wider scientific community. The Bpc includes the U.S. Tier 1 Select Agents B. pseudomallei and B. mallei. Previously, we demonstrated the importance of including near-neighbor genomes when designing sensitive and specific diagnostics for B. pseudomallei (4, 26). B. mayonis sp. nov. and B. savannae sp. nov. share 7 and 23 CDSs, respectively, with the B. pseudomallei core genome that are not shared by other species in the Bpc (Fig. 3). Thus, the addition of genomes from these novel species further constrains CDSs in the B. pseudomallei core genome that can be used as diagnostic targets for that species. As such, the B. mayonis sp. nov. and B. savannae sp. nov. whole genome sequences provided here should be utilized when designing DNA-based assays specific for B. pseudomallei. Members of the Bpc, and Burkholderia species in general, also can be sources of novel natural products (17, 18). Indeed, Burkholderia species have been demonstrated to be useful for bioremediation (27, 28), biocontrol (29), and as potential sources of novel antibiotics (30). The detailed genomic data generated in this study, and the deposition of the type strains in public strain collections, will hopefully facilitate detailed bioprospecting studies of B. mayonis sp. nov. and B. savannae sp. nov.
FIG 3.
Overlap of the B. pseudomallei core genome (n = 4,452 CDSs) with pan-genomes from other species in the B. pseudomallei complex (Bpc). The included B. pseudomallei CDSs have a blast score ratio (BSR) value of >0.8 in at least one genome from the near-neighbor species. Gray regions for each bar represent CDSs that are uniquely covered by at least one genome from that species; the number at the end of the bar corresponds to these CDSs. Black bars represent B. pseudomallei core CDSs found in the indicated species and other species in the Bpc.
Description of Burkholderia mayonis sp. nov.
Burkholderia mayonis sp. nov. (ma.yo′nis. N.L. gen. n. mayonis, pertaining to Mark Mayo, an experienced and highly respected Burkholderia scientist in Australia whose family is linked culturally to Badu Island, an island located in the Torres Strait archipelago of Queensland, Australia, where the first group of members of this species was isolated). Mark Mayo was present on Badu Island when the strain was collected, and he serves as a mentor for local indigenous and non-indigenous scientists in northern Australia and elsewhere.
The organism is Gram-negative, rod-shaped, and non-spore forming. Growth is observed at 25°C and 37°C within 24 h on Ashdown’s selective agar, Columbia blood agar, MacConkey agar, and Luria-Bertani agar. Optimal growth at 37°C for 48 to 72 h and at 25°C for 72 to 96 h aerobically. No hemolysis on Columbia blood agar.
Assimilation (API 20NE) was found for arginine, adipate, caprate, citrate, gluconate, glucose, malate, mannitol, mannose, nitrate, N-acetylglucosamine, and phenylacetate, whereas it is negative for arabinose, glucose (acidification), urea, 4-nitrophenyl-β-d-galactopyranoside (PNPG), and tryptophan. Gelatin is hydrolyzed. Assimilation of maltose and esculin hydrolysis is strain-dependent (Table 1).
The organism is positive (API ZYM) for acidic phosphatase, alkaline phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, and naphthol-AS-BI-phosphohydrolase. Enzymes absent on API ZYM are trypsin, α-chymotrypsin, α- and β-galactosidase, β-glucuronidase, α- and β-glucosidase, α-mannosidase, and α-frucosidase, with inconsistent results for cystin arylamidase, N-acetyl-β-glucosaminidase, and valine arylamidase. This species is aerobic, oxidase-positive, and catalase-negative with no immediate bubbling.
B. mayonis sp. nov. strains are resistant to gentamicin and polymyxin B, and have resistance or immediate resistance to chloramphenicol, but are susceptible to amoxicillin-clavulanic acid, ceftazidime, doxycycline, imipenem, meropenem, and trimethoprim-sulfamethoxazole.
The type strain is BDU6T, which has been deposited to the American Type Culture Collection as TSD-80 and to the Belgian Coordinated Collections of Microorganisms as LMG 29941.
Description of Burkholderia savannae sp. nov.
Burkholderia savannae sp. nov. (sa.van′nae. N.L. gen. n. savannae, of a savanna pertaining to grassy plains with scattered trees in tropical regions with distinct wet and dry seasons where the first group of members of this species was isolated).
The organism is Gram-negative, rod-shaped, and non-spore forming. Growth is observed at 25°C and 37°C within 24 h on Ashdown’s selective agar, Columbia blood agar, MacConkey agar, and Luria-Bertani agar. Optimal growth at 37°C for 48 to 72 h aerobically. No hemolysis on Columbia blood agar. Colony morphology varied between strains.
Assimilation (API 20NE) was found for arginine, adipate, caprate, citrate, gluconate, glucose, malate, mannitol, mannose, N-acetylglucosamine, and phenylacetate, whereas it is negative for glucose (acidification), urea, 4-nitrophenyl-β-d-galactopyranoside (PNPG), and tryptophan. Hydrolysis of gelatin and esculin and the assimilation of arabinose, maltose, and nitrate are strain-dependent (Table 1).
The organism is positive (API ZYM) for acidic phosphatase, alkaline phosphatase, cystin arylamidase, esterase, esterase lipase, lipase, leucine arylamidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Enzymes absent on API ZYM are trypsin, α-chymotrypsin, α- and β-galactosidase, β-glucuronidase, α- and β-glucosidase, α-mannosidase, and α-frucosidase, with inconsistent results for N-acetyl-β-glucosaminidase. This species is aerobic, oxidase positive, and catalase-negative with no immediate bubbling.
B. savannae sp. nov. strains are resistant to gentamicin and polymyxin B, but are susceptible to amoxicillin-clavulanic acid, ceftazidime, doxycycline, imipenem, meropenem, and trimethoprim-sulfamethoxazole; immediate resistance or susceptibility to chloramphenicol is strain-dependent.
The type strain is MSMB266T, which was deposited to the American Type Culture Collection as TSD-82 and to the Belgian Coordinated Collections of Microorganisms as LMG 29940.
MATERIALS AND METHODS
Strain isolation.
The two B. mayonis sp. nov. strains (BDU6T, BDU8) and the four B. savannae sp. nov. strains (MSMB266T, MSMB852, BDU18, BDU19) were all isolated from soil collected in tropical northern Australia (Table 3; Fig. S1). A subset of the strains (BDU6T, BDU8, BDU18, BDU19) was collected by James Cook University from a single soil sample collected at a depth of approximately 30 cm on Badu Island, in the Torres Strait Islands (Queensland, Australia) in late October 2011, near the end of the dry season. The soil sample was moist, sandy, and collected less than a meter from stagnant water within an exposed root system of trees. Strains MSMB266T and MSMB852 were collected by investigators from the Menzies School of Health Research from two different locations in the tropical “Top End” of the Northern Territory, Australia in 2006 and 2010, respectively. The BDU strains were recovered using a two-stage culture technique (31), and the MSMB strains were cultured from soil using standard Burkholderia culturing techniques (32); all strains were presumptively identified as Burkholderia based upon colony morphology but confirmed to not be B. pseudomallei via PCR (33). The proposed novel species, B. mayonis sp. nov. and B. savannae sp. nov., were previously reported as putative species 2 and putative species 3, respectively, by Sahl et al. (4) based upon a whole genome analysis.
Bacterial growth and characteristics.
All strains were cultivated at temperatures of 25°C, 37°C, and 42°C for 24, 48, 72, and 96 h on Ashdown’s selective agar, Columbia blood agar, MacConkey agar, and Luria-Bertani agar. Biochemical data were obtained for the two strains of B. mayonis sp. nov. (BDU6T and BDU8) and the four strains of B. savannae sp. nov. (MSMB266T, MSMB852, BDU18, BDU19) using the API 20NE and API ZYM (bioMérieux) systems according to the manufacturer’s instructions. These data were compared to data generated for B. thailandensis strain E264T and B. oklahomensis strain C6786, as well as previous data generated for B. pseudomallei strain K96243 (34). MALDI-TOF MS analysis was also performed for all B. mayonis sp. nov. and B. savannae sp. nov. strains listed above (see the text in the supplemental material for a detailed description of the methods).
Antimicrobial susceptibility screening.
The MIC was determined using the broth microdilution method in biological duplicate using 96-well microtiter custom Micronaut-S plates (Merlin, Bornheim-Hersel, Germany) following manufacturer instructions. In total, 20 antimicrobials were tested with a 2-fold serial dilution at the following concentrations: amoxicillin-clavulanic acid (4/2-128/64 mg/liter), azithromycin (4 to 64 mg/liter), carbenicillin (4 to 512 mg/liter), ceftazidime (4 to 128 mg/liter), ceftazidime-avibactam (0.5/4-256/4 mg/liter), chloramphenicol (4 to 128 mg/liter), ciprofloxacin (0.5 to 16 mg/liter), doripenem (0.5 to 16 mg/liter), doxycycline (1 to 32 mg/liter), gentamicin (2 to 64 mg/liter), imipenem (1 to 32 mg/liter), kanamycin (8 to 256 mg/liter), meropenem (1 to 64 mg/liter), piperacillin (8 to 256 mg/liter), piperacillin-tazobactam (8-256/4 mg/liter), polymyxin B (1 to 2048 mg/liter), sulfamethoxazole (1 to 512 mg/liter), tigecycline (0.25 to 32 mg/liter), trimethoprim (1 to 32 mg/liter), and trimethoprim-sulfamethoxazole (1/19-16/304 mg/liter). Two broth and growth controls containing no antimicrobials were included on each plate, and each strain was screened twice using biological duplicates on separate days. Briefly, for each strain, individual colonies were mixed in 3 ml of sterile saline solution (0.85% NaCl) to achieve a 0.5 McFarland Standard. The suspension (0.2 ml) was added to 20 ml of cation-adjusted Mueller-Hinton II broth (catalog number B12322; Fisher Scientific). Following this, 100 μl was added into each well for a particular strain, excluding the growth control wells. Plates were incubated at 37°C for 20 h and then measured using an accuSkan FC plate spectrophotometer (Fisher Scientific) at a wavelength of 620 nm.
Virulence gene screening.
Peptide sequences for genes associated with bimA (BPSS1492), the type III secretion system (BPSS1390-BPSS1410), and the type VI secretion system 5 (BPSS0091-BPSS0117) were screened against all B. mayonis sp. nov. and B. savannae sp. nov. genomes (Table 3), using LS-BSR v1.2.3 (35) in conjunction with tblastn v2.9.0 (36). The blast score ratio (BSR) (37) was calculated for each gene across each genome assembly.
Virulence testing in mouse models.
The pathogenic potential of B. mayonis sp. nov. and B. savannae sp. nov. was investigated in silico by looking for the presence of three key virulence factors in B. pseudomallei: the type 5 secretion system autotransporter (BimA), the type 3 secretion system (Bsa), and the type 6 secretion system 5 (Hcp-1). B. mayonis sp. nov. strain BDU6T and B. savannae sp. nov. strain MSMB266T were investigated in a BALB/c mouse model using methods previously reported (5); B. thailandensis strain E264T also was included as a comparison. Briefly, live culture was cultivated to the logarithmic phase (OD620 ∼ 1.0) in Luria-Bertani (LB) broth as previously described (22). Sterile 1× phosphate-buffered saline was used to wash cells twice before making dilutions for injecting mice. Viability counts of the final inocula were made on LB agar plates. BALB/c mice 6 to 8 weeks old were utilized, in treatment groups of 5 mice per cage; food and water were provided ad libitum. All mice in a single cage received the same infectious dose (B. mayonis sp. nov.: 3.82 × 104, 105, or 106 CFU; B. savannae sp. nov.: 0.92 × 104, 105, or 106 CFU; B. thailandensis: 3.4 × 104, 105, or 106 CFU) via a single subcutaneous injection in the scruff of the neck. Mice were monitored daily for health status. All mice were euthanized on day 21 postinjection. This work was conducted under approved protocols from the Northern Arizona University’s Institutional Animal Care and Use Committee (Protocol 14-011) and the US Department of Defense’s Animal Care and Use Review Office (HDTRA1-12-C-0066_Wagner).
16S rRNA gene analysis.
16S rRNA genes were extracted from genome assemblies for the two B. mayonis sp. nov. strains (BDU6T, BDU8) and the four B. savannae sp. nov. strains (MSMB266T, MSMB852, BDU18, BDU19) as previously described (11). We investigated the number of 16S rRNA operons present in the B. mayonis sp. nov. and B. savannae sp. nov. genomes using the publicly available rapid rRNA prediction tool barrnap v0.9 (https://github.com/tseemann/barrnap). A maximum likelihood phylogeny was inferred with IQ-TREE v2.0.3 (38), using the HKY+F+I substitution model (39) with 16S rRNA sequences, and was rooted with B. ubonensis. The number of pairwise SNPs between unique 16S rRNA gene copies was calculated with snp-dists v0.7.0 (https://github.com/tseemann/snp-dists).
Genome assembly and core genome phylogeny.
Genomes for the two B. mayonis sp. nov. (BDU6T, BDU8) and four B. savannae sp. nov. (MSMB266T, MSMB852, BDU18, BDU19) strains were previously sequenced on the PacBio platform (4). To construct the core genome phylogeny, assemblies were aligned against the genome of B. pseudomallei strain K96243 (GCA_000011545.1) (40) using NUCmer (41). The reference K96243 genome was also aligned against itself using NUCmer to identify duplicated regions, which were masked from subsequent analyses; these methods were wrapped by NASP v1.1.2 (42). A maximum-likelihood phylogeny was inferred from an alignment of 434,216 SNPs with IQ-TREE v1.6.10, using the TVM+F+ASC+R3 substitution model and 1,000 bootstrap replicates.
Multilocus sequence typing (MLST).
Genes for the seven MSLT loci in the B. pseudomallei pubMLST typing scheme (14) were extracted in silico from the genomes of the two B. mayonis sp. nov. strains (BDU6T, BDU8) and the four B. savannae sp. nov. strains (MSMB266T, MSMB852, BDU18, BDU19) using blastn v2.5.0 (36). The seven genes in this MLST typing scheme are ace, gltB, gmhD, lepA, lipA, narK, and ndh. As of 21 June 2021, a total of 1,934 sequence types (STs) had been identified in B. pseudomallei and in closely related species by MLST (http://pubmlst.org).
Average nucleotide identity values and digital DNA-DNA hybridization.
Average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) were calculated using complete genome assemblies for B. mayonis sp. nov. strains BDU6T and BDU8, complete genome assemblies for B. savannae sp. nov strains MSMB266T and MSMB852, and genome assemblies with four contigs for B. savannae sp. nov. strains BDU18 and BDU19 (NCBI accession numbers listed in Table 3). These assemblies were compared to genome assemblies (using complete genome assemblies when available) of the following Bpc strains: B. humptydooensis MSMB43T, B. mallei ATCC 23344T, B. oklahomensis C6786T, B. pseudomallei K96243, B. singularis MSMB175, and B. thailandensis E264T (NCBI accession numbers listed in Table 3).
For ANI, all assemblies were uploaded to JSpecies WS and analyzed using the ANIb algorithm (43); the authors of JSpecies suggested that ANI values <95% suggest separate species. The digital DNA-DNA hybridization (dDDH) values were produced by the genome-to-genome distance calculator (GGDC), which correlates with values obtained by conventional DDH and also provides a confidence-interval estimation (44). In brief, with this approach, two strains are considered as belonging to different species if the DNA-DNA relatedness between them is less than 70%. The dDDH values were calculated using formula 2 in the GGDC, which summed the identities found in high-scoring segment pairs (HSP) and then divided the sums by the overall HSP length (44).
Comparative genomics.
To better understand the composition of the genomes of the putative new species, annotated locus tags for each genome were obtained from GenBank. For both putative species, combined locus tags were dereplicated with cd-hit v4.8.1 (45) at an ID of 0.8, and the pan genome for each species was defined by the total number of cluster representatives. Unique locus tags were screened with LS-BSR v1.2.2 (35) against a set of 3,273 Burkholderia genome assemblies downloaded with the NCBI-genome-download tool (https://github.com/kblin/ncbi-genome-download). Any locus with a blast score ratio value (37) of <0.4 in all non-target genomes was identified to be unique to that species. The functional profile of each unique region was identified with eggNOG mapper v2.0.1 (46), and regions suspected to contain phage sequence were further classified using PHAST (47). The core genome for each putative species was distinguished by identifying coding regions with a BSR value of ≥0.8 across all target genomes.
To understand the overlap of the B. pseudomallei core genome with other species in the Bpc, including B. mayonis sp. nov. and B. savannae sp. nov., a set of 1,744 B. pseudomallei genomes were annotated with Prokka v1.14.6 (48) and the pan-genome was calculated with Panaroo v1.2.3 (49). The amount of overlap was determined for a coding region if it had a BSR value of ≥0.8 in any genome from another species in the Bpc.
Data availability.
The PacBio whole genome sequence NCBI accession numbers for BDU6T are CP013386.1 for chromosome 1 and CP013387.1 for chromosome 2; the accession numbers for MSMB266T are CP013417.1 for chromosome 1, CP013418.1 for chromosome 2, and CP013419.1 for pMSMB0266. The PacBio whole-genome assembly NCBI accession numbers for all strains are listed in Table 3.
ACKNOWLEDGMENTS
We declare no conflicts of interest. This work was funded by the DOD Defense Threat Reduction Agency (DTRA; HDTRA1-12-C-0066 and HDTRA1-17-1-0051), the Australian National Health and Medical Research Council, and the Australian Research Council. We are grateful to Aharon Oren for assistance with nomenclature.
Footnotes
Supplemental material is available online only.
Contributor Information
David M. Wagner, Email: Dave.Wagner@nau.edu.
Jeremy D. Semrau, University of Michigan-Ann Arbor
REFERENCES
- 1.Eberl L, Vandamme P. 2016. Members of the genus Burkholderia: good and bad guys. F1000Res 5:1007. doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beukes CW, Palmer M, Manyaka P, Chan WY, Avontuur JR, van Zyl E, Huntemann M, Clum A, Pillay M, Palaniappan K, Varghese N, Mikhailova N, Stamatis D, Reddy TBK, Daum C, Shapiro N, Markowitz V, Ivanova N, Kyrpides N, Woyke T, Blom J, Whitman WB, Venter SN, Steenkamp ET. 2017. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front Microbiol 8:1154. doi: 10.3389/fmicb.2017.01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin QH, Lv YY, Gao ZH, Qiu LH. 2020. Pararobbsia silviterrae gen. nov., sp. nov., isolated from forest soil and reclassification of Burkholderia alpina as Pararobbsia alpina comb. nov. Int J Syst Evol Microbiol 70:1412–1420. doi: 10.1099/ijsem.0.003932. [DOI] [PubMed] [Google Scholar]
- 4.Sahl JW, Vazquez AJ, Hall CM, Busch JD, Tuanyok A, Mayo M, Schupp JM, Lummis M, Pearson T, Shippy K, Colman RE, Allender CJ, Theobald V, Sarovich DS, Price EP, Hutcheson A, Korlach J, LiPuma JJ, Ladner J, Lovett S, Koroleva G, Palacios G, Limmathurotsakul D, Wuthiekanun V, Wongsuwan G, Currie BJ, Keim P, Wagner DM. 2016. The effects of signal erosion and core genome reduction on the identification of diagnostic markers. mBio 7:e00846–16. doi: 10.1128/mBio.00846-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuanyok A, Mayo M, Scholz H, Hall CM, Allender CJ, Kaestli M, Ginther J, Spring-Pearson S, Bollig MC, Stone JK, Settles EW, Busch JD, Sidak-Loftis L, Sahl JW, Thomas A, Kreutzer L, Georgi E, Gee JE, Bowen RA, Ladner JT, Lovett S, Koroleva G, Palacios G, Wagner DM, Currie BJ, Keim P. 2017. Burkholderia humptydooensis sp nov., a new species related to Burkholderia thailandensis and the fifth member of the Burkholderia pseudomallei complex. Appl Environ Microbiol 83:e02802–16. doi: 10.1128/AEM.02802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandamme P, Peeters C, De Smet B, Price EP, Sarovich DS, Henry DA, Hird TJ, Zlosnik JEA, Mayo M, Warner J, Baker A, Currie BJ, Carlier A. 2017. Comparative genomics of Burkholderia singularis sp nov., a low G plus C content, free-living bacterium that defies taxonomic dissection of the genus Burkholderia. Front Microbiol 8:1679. doi: 10.3389/fmicb.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martina P, Leguizamon M, Prieto CI, Sousa SA, Montanaro P, Draghi WO, Stammler M, Bettiol M, de Carvalho C, Palau J, Figoli C, Alvarez F, Benetti S, Lejona S, Vescina C, Ferreras J, Lasch P, Lagares A, Zorreguieta A, Leitao JH, Yantorno OM, Bosch A. 2018. Burkholderia puraquae sp. nov., a novel species of the Burkholderia cepacia complex isolated from hospital settings and agricultural soils. Int J Syst Evol Microbiol 68:14–20. doi: 10.1099/ijsem.0.002293. [DOI] [PubMed] [Google Scholar]
- 8.Ong KS, Aw YK, Lee LH, Yule CM, Cheow YL, Lee SM. 2016. Burkholderia paludis sp. nov., an antibiotic-siderophore producing novel Burkholderia cepacia complex species, isolated from Malaysian tropical peat swamp soil. Front Microbiol 7:2046. doi: 10.3389/fmicb.2016.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeshita K, Tamaki H, Ohbayashi T, Meng XY, Sone T, Mitani Y, Peeters C, Kikuchi Y, Vandamme P. 2018. Burkholderia insecticola sp. nov., a gut symbiotic bacterium of the bean bug Riptortus pedestris. Int J Syst Evol Microbiol 68:2370–2374. doi: 10.1099/ijsem.0.002848. [DOI] [PubMed] [Google Scholar]
- 10.Compant S, Nowak J, Coenye T, Clement C, Barka EA. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev 32:607–626. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 11.Brett PJ, Deshazer D, Woods DE. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect 118:137–148. doi: 10.1017/s095026889600739x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass MB, Steigerwalt AG, Jordan JG, Wilkins PP, Gee JE. 2006. Burkholderia oklahomensis sp nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int J Syst Evol Microbiol 56:2171–2176. doi: 10.1099/ijs.0.63991-0. [DOI] [PubMed] [Google Scholar]
- 13.Currie BJ. 2015. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med 36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 14.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol 41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limmathurotsakul D, Golding N, Dance DA, Messina JP, Pigott DM, Moyes CL, Rolim DB, Bertherat E, Day NP, Peacock SJ, Hay SI. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol 1. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 17.Kunakom S, Eustaquio AS. 2019. Burkholderia as a source of natural products. J Nat Prod 82:2018–2037. doi: 10.1021/acs.jnatprod.8b01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XY, Cheng YQ. 2014. Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol 41:275–284. doi: 10.1007/s10295-013-1376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. M45, 3rd edition. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing. M100, 30th edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 21.Barnes JL, Ketheesan N. 2005. Route of infection in melioidosis. Emerg Infect Dis 11:638–639. doi: 10.3201/eid1104.041051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morici LA, Heang J, Tate T, Didier PJ, Roy CJ. 2010. Differential susceptibility of inbred mouse strains to Burkholderia thailandensis aerosol infection. Microb Pathog 48:9–17. doi: 10.1016/j.micpath.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West TE, Frevert CW, Liggitt HD, Skerrett SJ. 2008. Inhalation of Burkholderia thailandensis results in lethal necrotizing pneumonia in mice: a surrogate model for pneumonic melioidosis. Trans R Soc Trop Med Hyg 102(Suppl 1):S119–S126. doi: 10.1016/S0035-9203(08)70028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiersinga WJ, de Vos AF, de Beer R, Wieland CW, Roelofs JJ, Woods DE, van der Poll T. 2008. Inflammation patterns induced by different Burkholderia species in mice. Cell Microbiol 10:81–87. doi: 10.1111/j.1462-5822.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- 25.Summer EJ, Gill JJ, Upton C, Gonzalez CF, Young R. 2007. Role of phages in the pathogenesis of Burkholderia, or ‘Where are the toxin genes in Burkholderia phages?’ Curr Opin Microbiol 10:410–417. doi: 10.1016/j.mib.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson CHD, Wagner DM, Keim P, Sahl JW. 2018. Developing inclusivity and exclusivity panels for testing diagnostic and detection tools targeting Burkholderia pseudomallei, the causative agent of melioidosis. J Aoac Int 101:1920–1926. doi: 10.5740/jaoacint.18-0014. [DOI] [PubMed] [Google Scholar]
- 27.Afzal M, Khan S, Iqbal S, Mirza MS, Khan QM. 2013. Inoculation method affects colonization and activity of Burkholderia phytofirmans PsJN during phytoremediation of diesel-contaminated soil. Int Biodeterior Biodegradation 85:331–336. doi: 10.1016/j.ibiod.2013.08.022. [DOI] [Google Scholar]
- 28.Yang ZH, Zhang Z, Chai LY, Wang Y, Liu Y, Xiao RY. 2016. Bioleaching remediation of heavy metal-contaminated soils using Burkholderia sp. Z-90. J Hazard Mater 301:145–152. doi: 10.1016/j.jhazmat.2015.08.047. [DOI] [PubMed] [Google Scholar]
- 29.Sarli DA, Sanchez LA, Delgado OD. 2021. Burkholderia gladioli MB39 an Antarctic strain as a Biocontrol Agent. Curr Microbiol 78:2332–2344. doi: 10.1007/s00284-021-02492-y. [DOI] [PubMed] [Google Scholar]
- 30.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. 2016. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl Microbiol Biotechnol 100:5215–5229. doi: 10.1007/s00253-016-7520-x. [DOI] [PubMed] [Google Scholar]
- 31.Baker A, Tahani D, Gardiner C, Bristow KL, Greenhill AR, Warner J. 2011. Groundwater seeps facilitate exposure to Burkholderia pseudomallei. Appl Environ Microbiol 77:7243–7246. doi: 10.1128/AEM.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, Wagner DM, Tuanyok A, Wertheim H, Yoke Cheng T, Mukhopadhyay C, Puthucheary S, Day NP, Steinmetz I, Currie BJ, Peacock SJ. 2013. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis 7:e2105. doi: 10.1371/journal.pntd.0002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol 44:85–90. doi: 10.1128/JCM.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuthiekanun V, Smith MD, Dance DA, Walsh AL, Pitt TL, White NJ. 1996. Biochemical characteristics of clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol 45:408–412. doi: 10.1099/00222615-45-6-408. [DOI] [PubMed] [Google Scholar]
- 35.Sahl JW, Caporaso JG, Rasko DA, Keim P. 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. Peerj 2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasko DA, Myers GSA, Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. Bmc Bioinformatics 6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, Hepp CM, Smith DE, Roe C, Engelthaler DM, Wagner DM, Keim P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter M, Rossello-Mora R, Glockner FO, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Liang YJ, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD, Parkhill J. 2020. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol 21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Fig. S1 to S5, Tables S1 to S3. Download AEM.01583-21-s0001.pdf, PDF file, 4.5 MB (4.5MB, pdf)
Data Availability Statement
The PacBio whole genome sequence NCBI accession numbers for BDU6T are CP013386.1 for chromosome 1 and CP013387.1 for chromosome 2; the accession numbers for MSMB266T are CP013417.1 for chromosome 1, CP013418.1 for chromosome 2, and CP013419.1 for pMSMB0266. The PacBio whole-genome assembly NCBI accession numbers for all strains are listed in Table 3.