ABSTRACT
Cytophaga hutchinsonii is a Gram-negative bacterium belonging to the phylum Bacteroidetes. It digests crystalline cellulose with an unknown mechanism and possesses a type IX secretion system (T9SS) that can recognize the C-terminal domain (CTD) of the cargo protein as a signal. In this study, the functions of the CTD in the secretion and localization of T9SS substrates in C. hutchinsonii were studied by fusing the green fluorescent protein (GFP) with the CTD from CHU_2708. The CTD is necessary for the secretion of GFP by C. hutchinsonii T9SS. The GFP-CTDCHU_2708 fusion protein was found to be glycosylated in the periplasm, with a molecular mass about 5 kDa higher than that predicted from its sequence. The glycosylated protein was sensitive to peptide-N-glycosidase F, which can hydrolyze N-linked oligosaccharides. Analyses of mutants obtained by site-directed mutagenesis of asparagine residues in the N-X-S/T motif of CTDCHU_2708 suggested that N-glycosylation occurred on the CTD. CTD N-glycosylation is important for the secretion and localization of GFP-CTD recombinant proteins in C. hutchinsonii. Glycosyltransferase-encoding gene chu_3842, a homologous gene of Campylobacter jejuni pglA, was found to participate in the N-glycosylation of C. hutchinsonii. Deletion of chu_3842 affected cell motility, cellulose degradation, and cell resistance to some chemicals. Our study provided evidence that the CTD as the signal of T9SS was N-glycosylated in the periplasm of C. hutchinsonii.
IMPORTANCE The bacterial N-glycosylation system has previously been found only in several species of Proteobacteria and Campylobacterota, and the role of N-linked glycans in bacteria is still not fully understood. C. hutchinsonii has a unique cell contact cellulose degradation mechanism, and many cell surface proteins, including cellulases, are secreted by the T9SS. In this study, we found that C. hutchinsonii, a member of the phylum Bacteroidetes, has an N-glycosylation system. Glycosyltransferase CHU_3842 was found to participate in the N-glycosylation of C. hutchinsonii proteins and had effects on cell resistance to some chemicals, cell motility, and cellulose degradation. Moreover, N-glycosylation occurs on the CTD translocation signal of T9SS. The glycosylation of the CTD appears to play an important role in affecting T9SS substrate transportation and localization. This study enriched our understanding of the widespread existence and multiple biological roles of N-glycosylation in bacteria.
KEYWORDS: Cytophaga hutchinsonii, T9SS, glycosylation, CTD, protein secretion
INTRODUCTION
Protein secretion systems are crucial for bacteria to grow and survive, including getting nutrients and responding to various extracellular milieux (1). The recently discovered type IX secretion system (T9SS) is exclusively widespread in the Bacteroidetes phylum (2). It involves the secretion of abundant proteins that play important roles in the survival of bacteria, such as pathogenicity related to soluble or cell-associated peptidases in Porphyromonas gingivalis, motility required for cell surface adhesins in Flavobacterium johnsoniae, and obtaining nutrition from the environment through other hydrolytic enzymes (3). It is a common feature that most cargo proteins of the T9SS have an N-terminal signal peptide (SP) and a conserved C-terminal domain (CTD) (4). The N-terminal signal peptide allows substrates to be transported across the plasma membrane by the Sec system. The CTD, the signal recognized by the T9SS, assists cargo proteins across the outer membrane (5). The majority of T9SS CTDs are divided into two protein domain families, the family TIGR04183 (type A CTDs) and the family TIGR04131 (type B CTDs) (6, 7). Many studies have focused on type A CTDs of proteinaceous virulence factors of P. gingivalis (8, 9). Most CTDs are removed by PorU sortase during or after secretion in P. gingivalis (10). After CTDs detach from the cargo proteins, some cargo proteins are anchored to the cell surface by anionic lipopolysaccharide (A-LPS) modification, while other T9SS substrates are released into the extracellular space in soluble form (11). Type A CTDs of P. gingivalis contain approximately 80 amino acids and have five conserved sequential motifs (2, 4). A recent study has shown that the two-terminal-β-strand structure of the type A CTD of RgpB in P. gingivalis may contain the signal recognized by the T9SS (5). Type B CTDs are very different in sequence from type A CTDs, and there have been relatively few studies on them (9). The C-terminal region (218 amino acids) of the type B CTD of gliding motility adhesin SprB is capable of targeting proteins to be secreted by the T9SS in F. johnsoniae (12).
Cytophaga hutchinsonii is an aerobic Gram-negative cellulolytic bacterium and widely distributed in the soil. Different from free cellulases or cellulosome strategies, its mechanism of cellulose hydrolysis is unique and mysterious (13). It relies heavily on cell surface-localized cellulases and other factors to digest crystalline cellulose (14, 15). C. hutchinsonii as a member of the Bacteroidetes has all homologous genes encoding T9SS component proteins (16). Deletion of T9SS component protein PorU or SprP causes defects in the secretion of CTD proteins, cellulose degradation, and gliding motility in C. hutchinsonii (17, 18). Recently, we found that the T9SS components GldN, SprA, and SprT of C. hutchinsonii also take part in ion assimilation (19, 20). Bioinformatic analysis showed that C. hutchinsonii has the largest amount of CTD proteins in the phylum Bacteroidetes (4). There are at least 147 CTD proteins processed by the T9SS, of which 118 contain type A CTDs, including carbohydrate binding proteins, cellulases, and other hypothetical proteins (18). CHU_3220, with a CHU_C domain closely related to type B CTDs, is secreted to the outer membrane by the T9SS and participates in utilization of the cellulose crystalline region (21). However, the features and functions of CTDs of T9SS substrates in C. hutchinsonii have rarely been reported. A previous study found that the cell envelope fraction of C. hutchinsonii has weak reactivity with the A-LPS antibody MAb-1B5 of P. gingivalis (4). How these T9SS substrates attach to the outer membrane of C. hutchinsonii is worthy of further study.
CHU_2708, with a typical type A CTD, is the only reported T9SS cargo protein with a clear CTD cleavage site in C. hutchinsonii (4). In this study, the CTD of CHU_2708 was linked to the C terminus of green fluorescent protein (GFP) to study its function in the secretion of T9SS cargo proteins. We found that CTDCHU_2708 in the fusion protein was glycosylated in the periplasm of C. hutchinsonii and that glycosylation affects the secretion and localization of the fusion protein. This study provides insights on the role of glycosylation of type A CTDs in C. hutchinsonii T9SS substrates, which may help to explain the recognition mechanism of T9SS in the phylum Bacteroidetes.
RESULTS
The CTD in the GFP-CTDCHU_2708 is necessary for its secretion by the T9SS in C. hutchinsonii.
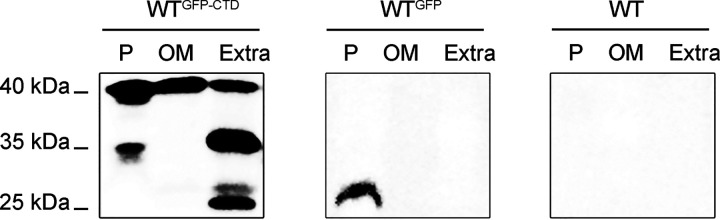
It is a common feature that N-terminal signal peptides assist T9SS cargo proteins across the cytoplasmic membrane by the Sec system, and CTDs target T9SS substrates to the outer membrane in the phylum Bacteroidetes (22). Though C. hutchinsonii has the largest number of CTD-containing proteins among members with the novel secretion system (2), the hypothetical protein CHU_2708 is the only reported T9SS substrate with a defined CTD cleavage site. The typical type A CTD of CHU_2708 contains 88 amino acid residues from the C terminus (4). Moreover, a previous study reported that the length of the CTD cleavage region affects the modification and anchorage of T9SS cargo proteins on the cell surface (23). In this study, the N-terminal signal peptide and C-terminal 110 amino acids from CHU_2708 were fused to the N and C termini of the green fluorescent protein (GFP), respectively, and the recombinant protein (signal peptide-GFP-CTDCHU_2708) was expressed in C. hutchinsonii to explore the function of the CTD. GFP antibody was used to detect the secretion and localization of the GFP recombinant protein. Western blotting results showed that GFP-related proteins were found in the outer membrane and extracellular fraction of the wild-type (WT) strain containing promoterCHU_2708-signal peptideCHU_2708-GFP-CTDCHU_2708 (WTGFP-CTD). However, when GFP only with the signal peptide was expressed in C. hutchinsonii, GFP (25 kDa) was detected only in the periplasm of the WT strain containing promoterCHU_2708-signal peptideCHU_2708-GFP (WTGFP) (Fig. 1). These results showed that the CTD of CHU_2708 facilitated the translocation of GFP-related proteins across the outer membrane. Protein bands with a molecular size as predicted by the GFP-CTDCHU_2708 polypeptide sequence of 35 kDa were found in the periplasm and culture medium. They were designated GFP-CTDCHU_2708 fusion proteins. We previously found that PorU sortase of C. hutchinsonii is involved in removing CTDs of T9SS cargo proteins (17). Both the GFP-CTDCHU_2708 fusion protein and GFP were detected in the culture fluid of the WTGFP-CTD strain, revealing that the CTD of CHU_2708 was removed after secretion. It was unexpected to find that a GFP-related protein of about 40 kDa occurred in the periplasm of the WTGFP-CTD strain, which is about 5 kDa bigger than the GFP-CTDCHU_2708 fusion protein. Moreover, protein bands of approximately 40 kDa also appeared in the outer membrane and extracellular fraction of the WTGFP-CTD strain, suggesting that recombinant GFP might be in a modified form after secretion. So, we designated the protein (40 kDa) on the outer membrane modified GFP.
FIG 1.
The CTD in GFP-CTDCHU_2708 is necessary for its secretion by the T9SS in C. hutchinsonii. The periplasmic (P), outer membrane (OM), and extracellular (Extra) proteins of the different C. hutchinsonii expression strains were identified by Western blotting with anti-GFP antibody. The wild type (WT) was used as the control. The amount of protein loaded in each lane came from cells of equal culture volumes.
The GFP-CTDCHU_2708 fusion protein may be N-glycosylated in the periplasm.
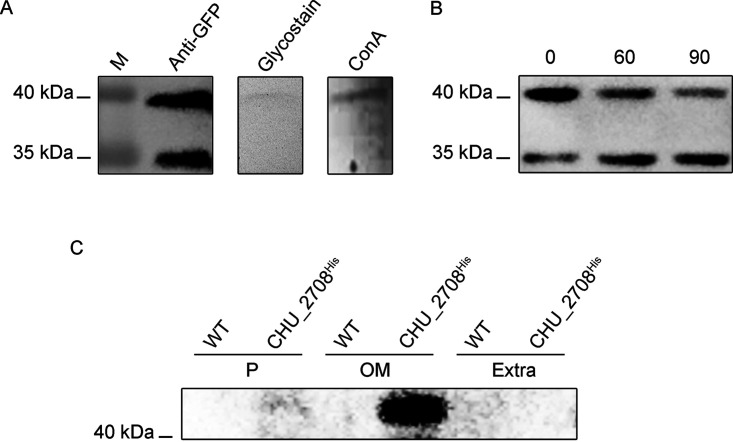
Previous studies reported that T9SS cargo proteins were extensively modified by A-LPS for anchoring after outer membrane translocation (2). It is surprising that the protein around 40 kDa was also detected in the periplasm of the WTGFP-CTD strain. In addition, this unique protein could not be detected in the cytoplasm of the WTGFP-CTD strain (Fig. S1). Pierce anti-GFP magnetic agarose beads were used to purify the GFP-related proteins from the periplasmic fraction of the WTGFP-CTD strain. Then the purified proteins were analyzed by liquid chromatography-tandem mass spectrometry. The results showed that both the proteins of 40 kDa and 35 kDa contained peptide fragments derived only from GFP and CTDCHU_2708. Glycosylation is one of the most prevalent modification forms in protein processing (24). In order to determine whether the protein of 40 kDa in the periplasmic space of the WTGFP-CTD strain was glycosylated, the purified GFP-related proteins were assayed by periodate-Schiff staining. As shown in Fig. 2A, the protein of 40 kDa had a positive staining reaction, while the protein of 35 kDa was not stained. We also used the lectin affinity approach for further verification, and the results showed that only the protein of 40 kDa reacted with concanavalin A (ConA). These results proved that the GFP-CTDCHU_2708 fusion protein was glycosylated to 40 kDa in the periplasm of the WTGFP-CTD strain. Then the periplasmic space glycoprotein (40 kDa) was designated glycosylated GFP. Peptide-N-glycosidase F (PNGase F) is the most effective enzyme to remove almost all N-linked oligosaccharides from asparagine residues of glycoproteins (25). For further study, the periplasmic proteins of the WTGFP-CTD strain were subjected to PNGase F treatment. Western blot results showed that the glycosylated GFP gradually decreased when being treated with PNGase F, while the content of the GFP-CTDCHU_2708 fusion protein (35 kDa) increased over time (Fig. 2B). Glycosylated GFP is susceptible to PNGase F enzymatic deglycosylation. The results not only confirmed that it is a glycoprotein but also suggested that it may be N-glycosylated.
FIG 2.
Glycosylation of the GFP-related proteins in the C. hutchinsonii WTGFP-CTD strain. (A) The purified proteins from the periplasmic fraction of WTGFP-CTD were separated by SDS-PAGE, detected with the anti-GFP antibody, stained with periodate-Schiff, or probed with ConA lectin. Lane M, molecular mass marker. (B) Periplasmic proteins of the WTGFP-CTD separated by SDS-PAGE after treatment with PNGase F for 0 min, 60 min, and 90 min and then detected with anti-GFP antibody. (C) The periplasmic, outer membrane, and extracellular proteins of the WT and CHU_2708His strains were Western blotted with the anti-His tag.
To determine the mature form of T9SS substrate CHU_2708 in C. hutchinsonii, a His tag was added after its signal peptide. Different cellular fraction lysates were analyzed by Western blotting using antibody to His tag. CHU_2708 could only be detected in the outer membrane fraction, and its molecular weight was higher than the predicted 33 kDa, indicating that the mature CHU_2708 was modified on the outer membrane (Fig. 2C). The modified CHU_2708 could not react with PNGase F, suggesting that the modification form of T9SS substrate on the cell surface is different from that of the periplasm.
Potential N-glycosylation sites of CTDCHU_2708.
As shown in Fig. 1, glycosylation occurred only when GFP was fused to CTDCHU_2708, so glycosylation may occur on the CTD region. Glycans are usually attached to asparagine residues in the N-X-S/T amino acid motif (X ≠ P) in the N-glycosylation system (26). Three asparagine residues (N273, N276, and N296) of CTDCHU_2708 are potential N-linked glycosylation sites conforming to the conservative N-X-S/T motif (Fig. 3A). Site-directed mutagenesis was then used to individually mutate asparagine to glutamine. The mutation of N276 in the NVT motif resulted in the apparent disappearance of the glycosylated GFP and GFP-CTDCHU_2708 fusion protein in the periplasm (Fig. 3B). None of the GFP-related proteins could be detected on the outer membrane or the extracellular fraction of the N276Q mutant strain (Fig. 3C and D). When N296 in the NGS motif was mutated, compared with the WTGFP-CTD strain, the glycosylated GFP in the periplasmic space was reduced (Fig. 3B). There was no modified GFP on the outer membrane of the N296Q strain and less GFP-CTDCHU_2708 fusion protein and GFP in the extracellular milieu (Fig. 3C and D). The changing of N273 in the NNS motif to glutamine had no significant effect on the glycosylated GFP in the periplasm of the N273Q strain (Fig. S2A). These results showed that the site N296 of CTDCHU_2708 had an effect on the glycosylated GFP. The destruction of site N296 of CTDCHU_2708 led to defective secretion and localization of GFP-related proteins, suggesting that the glycosylation of CTDCHU_2708 may play roles in substrate recognition and transportation of T9SS. Moreover, site N276 may also affect the stability of periplasmic space GFP-related proteins in C. hutchinsonii, because the mutation of N276 caused disappearance of all GFP-related proteins in the periplasm of N276Q. Proteins of the HtrA family are effective in removing misfolded or damaged proteins in the periplasm (27). We previously reported that CHU_0052 (DegQ) is involved in periplasmic protein quality control, and the high expression level of degQ reflects the protein folding pressure in the periplasmic space (20, 28). We found that the expression level of degQ in the N276Q mutant strain increased 3 times compared with that in the WTGFP-CTD strain when the unique asparagine site N276 of CTDCHU_2708 was mutated (Fig. 3E). The result supported that the site N276 of CTDCHU_2708 may contribute to the stability of GFP-related proteins in the periplasm.
FIG 3.
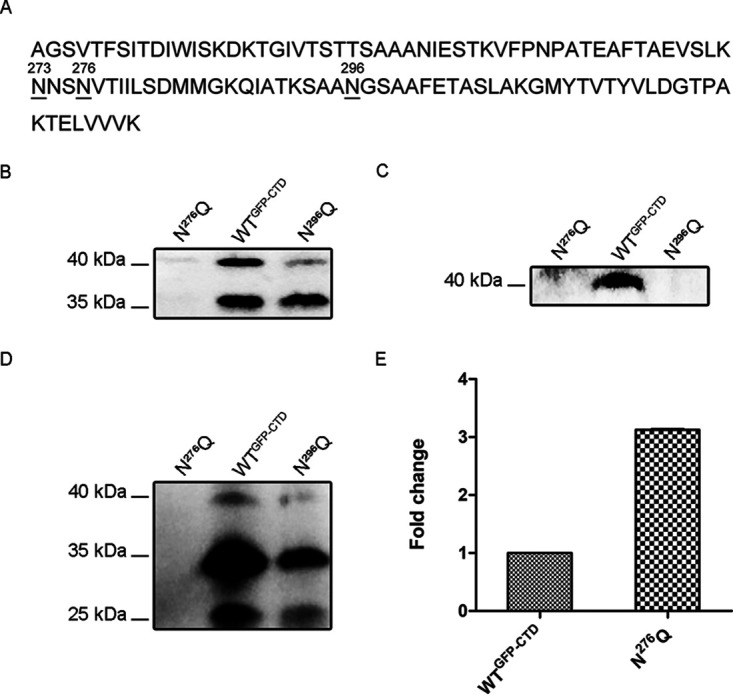
Site-directed mutagenesis of CTDCHU_2708 and transcription level of C. hutchinsonii degQ in the mutant. (A) Amino acid sequence of the CTD of CHU_2708. The residues underlined were mutated to glutamine. The asparagine residues within the conserved N-X-S/T amino acid motif were numbered. (B to D) The periplasmic protein (B), outer membrane proteins (C), and extracellular proteins (D) of the WTGFP-CTD, N276Q, and N296Q strains were detected with anti-GFP antibody. (E) Transcription level of degQ in the WTGFP-CTD and N276Q mutant strains. The mean values and standard deviations from three biological replicates are shown.
CHU_3842 participated in the N-glycosylation system of C. hutchinsonii proteins.
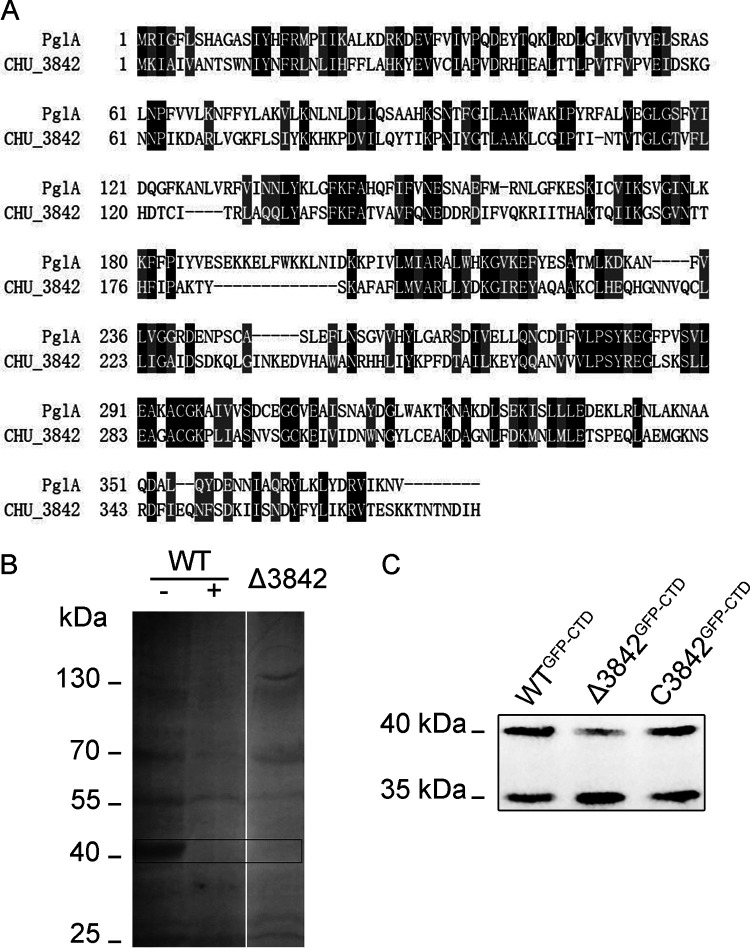
The mutation of the two asparagine residues in the N-X-S/T amino acid motif of CTDCHU_2708 led to the disappearance or decrease of glycosylation of the GFP-CTDCHU_2708 fusion protein. These phenomena hint that C. hutchinsonii has an N-glycosylation system. The gastrointestinal pathogen Campylobacter jejuni was the first bacterium demonstrated to have a well-characterized N-linked glycosylation pathway (29). A 16-kb pgl (protein glycosylation) gene cluster encodes enzymes that are required for N-glycan synthesis and protein linkage in C. jejuni (30). There are 5 glycosyltransferases involved in the N-glycosylation system of C. jejuni, PglA, PglC, PglH, PglI, and PglJ (31). Bioinformatics analysis showed that PglH and the key oligosaccharyltransferase PglB are not found in C. hutchinsonii. Moreover, the amino acid sequence identity of PglI and PglJ between C. hutchinsonii and C. jejuni is very low. In addition, the homologous genes of C. jejuni pglA and pglC were predicted to be chu_3842 and chu_1211 in C. hutchinsonii. However, the deletion of C. hutchinsonii CHU_1211, which has a 43% amino acid sequence identity with C. jejuni PglC, had no significant effect on N-glycosylated proteins (data not shown). PglA is responsible for adding N-acetylgalactosamine (GalNAc) residues on Und-P-diNAcBac in C. jejuni (29). There is a 28% amino acid sequence identity between C. jejuni PglA and CHU_3842 (Fig. 4A). In addition, glycosyltransferase CHU_3842 and C. jejuni PglA have the same conserved GT4_CapM-like functional domain.
FIG 4.
CHU_3842 is involved in the N-glycosylation of C. hutchinsonii proteins. (A) Sequence alignment of PglA from C. jejuni and CHU_3842. Black and gray shading shows identical and similar amino acid residues, respectively. The lines show the gaps between the two sequences. (B) The periplasmic glycoproteins of the Δ3842 mutant and WT. ConA lectin was used to enrich the periplasmic proteins of the WT and Δ3842 strains, treated with PNGase F (+) or not (-), then separated by SDS-PAGE, and probed with ConA lectin. (C) The periplasmic fraction proteins of the WTGFP-CTD, Δ3842GFP-CTD, and C3842GFP-CTD strains were detected with anti-GFP antibody.
To explore the role of CHU_3842 in the glycosylation system of C. hutchinsonii, the linear DNA double-crossover method was used to knock it out. The periplasmic glycoproteins of the wild type and the Δ3842 mutant were enriched by ConA lectin affinity chromatography. As lectin Western blotting showed, there were many glycoproteins in the periplasmic space of the wild type, especially the glycoproteins around 40 kDa, and they disappeared after PNGase F treatment (Fig. 4B), indicating that these proteins may be N-glycosylated. The periplasmic glycoproteins of the Δ3842 mutant were much less than that of the wild type. No glycoproteins at 40 kDa were found in the Δ3842 mutant. When the recombinant GFP (GFP-CTDCHU_2708) was expressed in the Δ3842 mutant, compared with the WTGFP-CTD strain, the glycosylated GFP (40 kDa) in the periplasm of the Δ3842GFP-CTD mutant was decreased. After supplementing the function of CHU_3842, glycosylated GFP returned to normal in the periplasm of the C3842GFP-CTD strain (Fig. 4C). All these results suggested that glycosyltransferase CHU_3842 is engaged in the N-glycosylation process of C. hutchinsonii.
Phenotypic properties of the Δ3842 mutant.
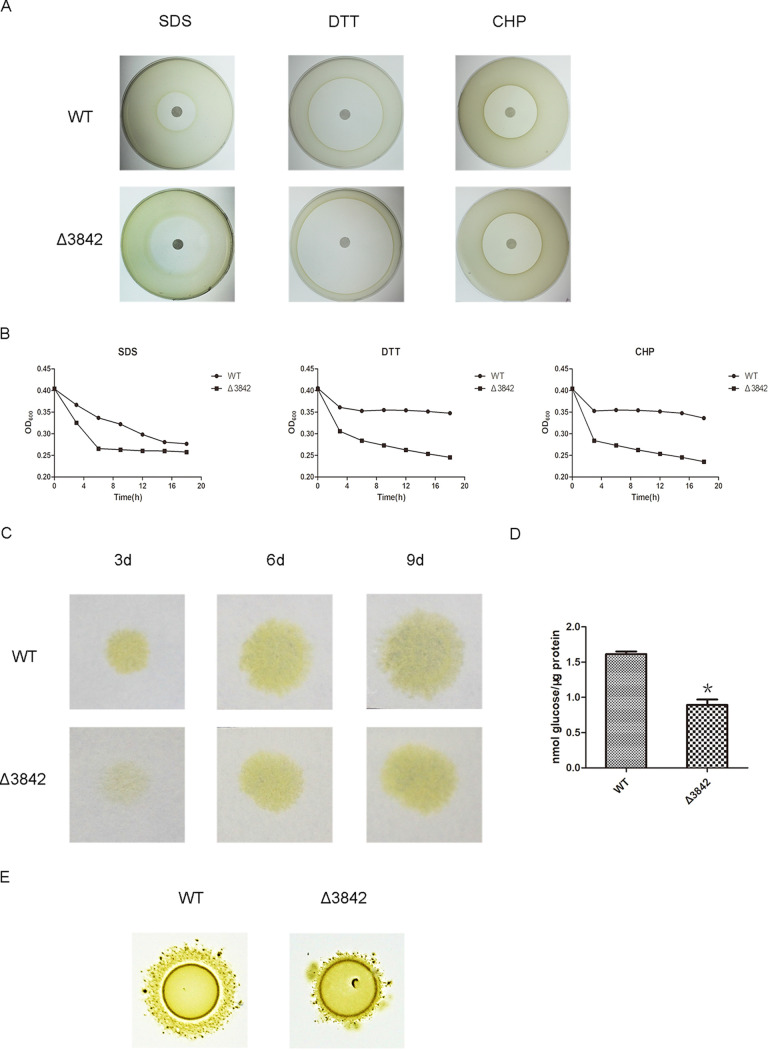
N-Glycosylation was shown to be associated with stress resistance and motility in bacteria (32). The disk diffusion susceptibility test found that the cells of the Δ3842 mutant had bigger inhibition zone diameters than the wild type on sodium dodecyl sulfate, cumene hydroperoxide, and dithiothreitol, showing that the Δ3842 mutant was more sensitive to these agents than the wild type (Fig. 5A). Quantitative cell stress resistance to chemical analysis by absorbance changes found that the cell density of the Δ3842 mutant was more quickly reduced than that of the wild type in the early stages of detection (Fig. 5B). The results showed that the cell resistance to chemicals was affected by CHU_3842.
FIG 5.
CHU_3842 has effects on cell resistance, cellulose degradation, and cell motility. (A) Disk diffusion susceptibility assay of the Δ3842 and WT strains to sodium dodecyl sulfate (SDS), cumene hydroperoxide (CHP), and dithiothreitol (DTT). (B) Resistance of the Δ3842 and WT strains to the agents. (C) Filter paper degradation ability of the Δ3842 and WT strains. (D) Cell surface cellulase activity analysis of the Δ3842 and WT strains. The standard deviations from three biological replicates are shown. *, P < 0.05. (E) Colony spreading ability of the Δ3842 and WT strains.
Previous reports showed that some cellulases containing type A CTDs are secreted and anchored to the outer membrane by the C. hutchinsonii T9SS (4, 18). We detected the capability for cellulose utilization after the deletion of CHU_3842. The growth rate of the Δ3842 mutant was lower than that of the wild type on the Whatman filter paper (Fig. 5C). Further assay of the cellulase activity, as in Fig. 5D, showed that the cell surface cellulase activity of Δ3842 was decreased in contrast to that of the wild type. These results suggested that CHU_3842 had effects on the cellulose degradation of C. hutchinsonii. Cells of the Δ3842 mutant formed smaller spreading colonies in contrast to the wild-type cells on soft agar, suggesting that the motility of cells of the Δ3842 mutant was deficient (Fig. 5E). These results indicated that the deletion of CHU_3842 caused the loss of N-glycosylation of some proteins, leading to pleiotropic defects on cell resistance, cell mobility, and cellulose degradation in C. hutchinsonii.
DISCUSSION
Protein glycosylation is the most common and complex posttranslational modification, which not only enlarges an organism’s proteome but also affects many vital processes (26). For a long time, the protein glycosylation system was thought to be restricted to eukaryotes. It was not until the discovery of glycoproteins in Halobacterium and Clostridium that it was proved that protein glycosylation systems exist in all forms of life (33, 34). Several types of protein glycosylation systems have been identified in bacteria, including N-, O-, C-, and S-linked protein glycosylation systems (26). The enteropathogen C. jejuni was the first bacterium reported to have a general N-linked protein glycosylation (pgl) system (35). Further comparative genomic analysis showed that the conserved pgl locus is widespread in Campylobacter species (36). Other atypical bacterial N-glycosylation systems have also been uncovered and characterized (37–39). Many studies reported that bacterial N-glycosylation systems are limited within the species of Proteobacteria and Campylobacterota (formerly Epsilonproteobacteria) (40). Our study provides the evidence that C. hutchinsonii, a member of the phylum Bacteroidetes, also has an N-glycosylation system. The N-glycosylation system of C. hutchinsonii has some similarities to that of C. jejuni. First, glycosyltransferase PglA was shown to engage in N-glycosylation of the C. jejuni periplasmic lipoprotein AcrA (41). Bioinformatic analysis revealed that chu_3842 is the homologous gene of pglA, and the deletion of CHU_3842 led to the disappearance of the periplasmic N-linked glycoproteins in C. hutchinsonii (Fig. 4). Second, the N-glycosylation of the GFP-CTD recombinant protein occurred in the periplasmic space of C. hutchinsonii, just as most N-glycosylated proteins in C. jejuni. Third, proteins are decorated with a heptasaccharide glycan in the N-glycosylation system of C. jejuni. Glycosylated GFP was 5 kDa higher than the theoretical molecular weight, indicating that there might also be complex glycans transferred to proteins of C. hutchinsonii, which is different from the monosaccharide or disaccharide modified protein in the atypical N-glycosylation system. However, there are also some differences between the N-glycosylation systems of C. hutchinsonii and C. jejuni. Our study showed that the glycosylated GFP is sensitive to PNGase F, while the N-linked glycoprotein of C. jejuni contains unique linking sugar bacillosamine, which makes it resistant to PNGase F (42). We also mutated T278 in the NVT motif of CTDCHU_2708 to alanine, and no glycosylated GFP was found in the periplasm of the mutant strain (Fig. S2B). The possible N-glycosylation sites N276 and N296 of CTDCHU_2708 are located in the N-X-S/T (X ≠ P) sequence, which is consistent with the motif recognized by the eukaryotic N-glycosylation system but different from the D/E-X-N-X-S/T sequon of C. jejuni. The characteristics of the N-glycosylation system in C. hutchinsonii need to be further studied.
The O-glycosylation system is extensively distributed in the phylum Bacteroidetes (43). Flagellins, pilins, and S-layer proteins are common O-glycosylated polymeric proteins (44). Plenty of studies have focused on the A-LPS glycosylation of T9SS substrates on the cell surface in P. gingivalis (45). Previous studies showed that the CTD signal is essential for the secretion, A-LPS modification, and attachment of T9SS substrates to the cell surface. In addition, the T9SS CTD signal was not the site of modification (3). C. hutchinsonii has the largest number of T9SS substrates in the phylum Bacteroidetes. There are many CTD proteins with type A CTD processing by the C. hutchinsonii T9SS (18). However, studies that focused on the features and functions of C. hutchinsonii CTDs are very limited. Our study reports that CTDCHU_2708 as the signal for the T9SS is N-glycosylated in the periplasm before being transported across the outer membrane. A previous study reported that the extensive mutation of potential glycosylation sites in the CTD of RgpB did not yield any carbohydrate modification sites and had no effect on the secretion or cell surface A-LPS modification of the virulence factor RgpB in P. gingivalis (46). No GFP-related proteins were detected in the periplasm when GFP fused with CTD of HBP35 was expressed in P. gingivalis (8). Moreover, attachment of the CTD of F. johnsoniae RemA to the superfolder GFP had no obvious modification (9). Therefore, whether there is N-glycosylation of CTDs in other Bacteroidetes species requires further exploration.
The contributions of N-linked glycan to proteins are usually associated with protein folding, stability, intracellular trafficking, subcellular targeting, and other properties in eukaryotic cells (47). But many of the roles assumed by protein N-glycosylation in eukaryotes are not applicable to Bacteria (48). A previous study reported that one N-glycosylation site of the T4SS component VirB10 is important for the stability of the T4SS in C. jejuni (49). A recent study showed that N-linked glycans take part in enhancing the thermostability of the multidrug efflux pump CmeABC in C. jejuni (50), but the role of N-linked glycans in bacteria is still not fully understood. Our study found that the N-glycosylation of the T9SS recognition signal CTDCHU_2708 plays an important role in the secretion and localization of the recombinant cargo protein. It was reported that glycosylation sites are often located in flexible parts of folded proteins in the bacterial system (51). Our study showed that mutation of N296 in CTDCHU_2708 to glutamine resulted in a significant reduction of the glycosylated protein in the periplasmic space of N296Q (Fig. 3B). In the predicted three-dimensional structure of CTDCHU_2708, the N296 site is in the amorphous region of a typical sandwich-like fold of the immunoglobulin superfamily domain (Fig. S3). The mutation of N296 might affect the conformation of the CTD, leading to changes in the accessibility of glycosyltransferase, thereby reducing the glycosylation rate of the GFP-CTDCHU_2708 fusion protein. The decreased secretion of GFP-related proteins in the N296Q mutant also indicated that glycosylation of CTDCHU_2708 is important for GFP-CTDCHU_2708 recombinant protein secretion. No modified GFP was detected on the outer membrane of the N296Q mutant strain, indicating that the N-glycosylation of the CTD may be linked with the anchoring of T9SS substrates to the cell surface. Glycans of proteins play a role as labels in binding and recognition by other proteins in eukaryotes (52). Our results suggested that glycosylation of the CTD might be involved in the recognition of cargo proteins by the T9SS. Moreover, the mutation of N276 of CTDCHU_2708 to glutamine caused not only the loss of the glycosylated GFP but also the disappearance of the GFP-CTDCHU_2708 fusion protein in the periplasm (Fig. 3B). A similar phenomenon occurred when N276 was converted to alanine (Fig. S2C). The weak band of GFP-CTDCHU_2708 fusion protein in the periplasm of the N276A was detected. Increasing the amount of proteins detected by Western blotting, a weak band of GFP-CTDCHU_2708 fusion protein could also be found in the periplasm of the N276Q mutant strain. These results showed that site N276 was also very important for the stabilization of GFP recombinant proteins. Proteases of the HtrA family play a central role in the degradation of aberrant proteins (53). Previously, we found that CHU_0052 is DegQ of C. hutchinsonii, which participates in the degradation or refolding of periplasmic misfolded proteins (28). The mutation of site N276 in CTDCHU_2708 caused a significant increase of the transcription level of degQ, suggesting that there is high folding stress in the periplasmic space of the N276Q strain. Whether the instability of GFP-CTDCHU_2708 recombinant protein was caused by direct change of N276 or by lack of glycosylation of N276 deserves further study.
Our study not only expanded the scope of the N-glycosylation system in bacteria, but also enriched the function of N-glycosylation in bacterial proteins. The N-glycosylation of CTD is important for the secretion and localization of the C. hutchinsonii T9SS cargo protein. Moreover, the N-glycosylation of C. hutchinsonii affects stress resistance, cell motility, and cellulose degradation. The wide range of phenotypic characteristics of C. hutchinsonii N-glycosylation also enriched our understanding of the bacterial N-glycosylation system. Our further study will focus on identifying more glycoproteins and analyzing the glycostructure of them in C. hutchinsonii.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general growth conditions.
Bacterial strains and plasmids are listed in Table 1. C. hutchinsonii strain ATCC 33406 and mutants were cultured in PY6 medium at 30°C with shaking at 160 rpm (17). Unless otherwise specified, solid medium with 1% agar was used. Filter paper-covered solid Stanier medium (14) was used to detect cell cellulose degradation. The soft-agar medium was made up of PY2 medium (54). The Escherichia coli DH5α strain was grown in Luria-Bertani medium at 37°C. Antibiotic concentrations were as follows: ampicillin, 100 μg/mL; erythromycin, 30 μg/mL; cefoxitin, 15 μg/mL; and chloramphenicol, 15 μg/mL. The primers are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21 | Strain with vector pUT-miniTn5-gfp-tet used for GFP gene cloning | TaKaRa |
| DH5α | Strain used for plasmid replication | TaKaRa |
| C. hutchinsonii strains | ||
| ATCC 33406 | Wild type | ATCC |
| WTGFP strain | WT containing promoterCHU_2708-signal peptideCHU_2708-GFP | This study |
| WTGFP-CTD strain | WT containing promoterCHU_2708-signal peptideCHU_2708-GFP-CTDCHU_2708 | This study |
| CHU_2708His strain | WT with His-tagged CHU_2708 | This study |
| N273Q mutant | Site-directed mutation of N273 of WTGFP-CTD | This study |
| N276Q mutant | Site-directed mutation of N276 of WTGFP-CTD | This study |
| T278A mutant | Site-directed mutation of T278 of WTGFP-CTD | This study |
| N276A mutant | Site-directed mutation of N276 of WTGFP-CTD | This study |
| N296Q mutant | Site-directed mutation of N296 of WTGFP-CTD | This study |
| C3842GFP-CTD strain | C3842 containing promoterCHU_2708-signal peptideCHU_2708-GFP-CTDCHU_2708 | This study |
| Δ3842 strain | chu_3842 deleted | This study |
| C3842 strain | Complementation of Δ3842 | This study |
| Plasmids | ||
| pTSK | Gene deletion template plasmid with ermF flanked by two FRT sites; Apr Emr | 17 |
| pTSK3328 | Plasmid for deletion of chu_3328; Apr Emr | 20 |
| pCFXSK3328 | Plasmid for deletion of chu_3328; Apr Cfxr | 59 |
| pCHF | Plasmid for complementation of Δ 3842; Apr Cmr | 17 |
Apr, ampicillin resistance; Emr, erythromycin resistance; Cfxr, cefoxitin resistance; Cmr, chloramphenicol resistance.
TABLE 2.
Sequences of primers used in this study
| Primer | Sequencea |
|---|---|
| 3328H1F | TTAGCATGCTCTGTTGAGCAGGTTCTACTGGG |
| 3328H2R | ATAGGATCCTCTATAATTGGCTGACCGACACG |
| P2708F | TGTTGTCGACCAAATCGCAGCGAAGAAATAAT |
| PS2708R | CTTCTCCTTTGCTTACCATTGAATTCCAAAGTACAA |
| GFPF | ATTGTACTTTGGAATTCAATGGTAAGCAAAGGAGAAG |
| GFPR | GGGGGAGCTCTTATTTGTATAGTTCATCCATGCCA |
| GFPR2708 | AAAGTTACTGATCCAGCTTTGTATAGTTCATCCAT |
| CTDF2708 | ATGGATGAACTATACAAAGCTGGATCAGTAACTTT |
| CTDR2708 | CCCCGAGCTCTTACTTAACAACTACTAATTCAGTTTTAG |
| 2708H1F | AGCTTCTAGAATCATGGTTCTGGAGTTCTCAAAAC |
| HISR | TTGGTGATGGTGATGGTGATGTGCGCTAGCAGTCATTACA |
| HISF | GCACATCACCATCACCATCACCAAAATCAAATTGTACTTT |
| SHISF | CATCACCATCACCATCACCAAAATCAAATTGTACTTTGGAA |
| 2708H1R | CCCCGAGCTCCTTAACAACTACTAATTCAGTTTTA |
| 2708H2F | GCGAGGGTACCTTAACCGCTCTATAATAGTATTAA |
| 2708H2R | ACCAGGATCCTAAAGAGTAATCATGGTACGAAGGT |
| EMF | CAAGTTGTCGGTTGTGATT |
| 2708UF | GAGTAGGGATGGATATTTATTGCCCGTTAT |
| 2708UR | AGGAATCGGTACAGACTTTCATAGCGGT |
| 273NQR | CGTTTGAATTTTGTTTCAAAGAAACTTCAGCTGTGAAA |
| 273NQF | CTTTCACAGCTGAAGTTTCTTTGAAACAAAATTCAAAC |
| 276NQR | TGATTGTTACTTGTGAATTGTTTTTCAAAGAAACTTCA |
| 276NQF | TGAAGTTTCTTTGAAAAACAATTCACAAGTAACAATCA |
| 276NAR | GATTGTTACAGCTGAATTGTTTTTCAAAGAAACTTCAG |
| 276NAF | CTGAAGTTTCTTTGAAAAACAATTCAGCTGTAACAATC |
| 278TAR | TCAGATAAGATGATAGCTACGTTTGAATTGTTTTTCAAA |
| 278TAF | TTTGAAAAACAATTCAAACGTAGCTATCATCTTATCTGA |
| 296NQR | AGCAGAACCTTGAGCAGCAGATTTAGTAGCAATTTGCT |
| 296NQF | AGCAAATTGCTACTAAATCTGCTGCTCAAGGTTCTGCT |
| Q0052F | GGTGCGTATCGGTGAGTGG |
| Q0052R | TATGCTCCCGTAGGACTTG |
| 3842H1F | GAAAGAGCTCAAGCAACAGATGACAACTGGATGAA |
| 3842H1R | GGTGGTCGACATATTGGGCTTGATGGTATACTGCA |
| 3842H2F | TGAGCCATGGGGCAATAACGTACAGTGTTTACTCA |
| 3842H2R | AATGGGATCCATAAGGTTCTTGCCTGCTGACTGGA |
| 3842UF | ACATGCCAAAACACAGATTATTAAAGGCTC |
| 3842UR | GAAGTGTCTGCTCAAAGTAAATCGAATC |
| C3842F | ACGGGAGCTCCATAGACATAAATCAATGCGGCGGC |
| C3842R | CGCGGTCGACTCAGTGAATGTCATTTGTATTCGT |
| YF | GTTCTCTCCTGAAATACGCT |
| YR1 | AACCCATTGTCAAACGTGTT |
| YR2 | GCAACAAAAAATCACTATGAAACGGCTACT |
Restriction sites are underlined.
Generation of expression strains.
The basic expression fragment contained the promoter and N-terminal signal peptide (SP) of CHU_2708 followed by GFP (promoterCHU_2708-SPCHU_2708-GFP). On this basis, CTDCHU_2708 with different operations was linked to the C terminus of GFP (promoterCHU_2708-SPCHU_2708-GFP-CTD). The fragment spanning the promoter and N-terminal signal peptide of CHU_2708 was obtained with primers P2708F (engineered SalΙ site) and PS2708R. The primers GFPF and GFPR (engineered SacΙ site) were used to amplify a 720-bp GFP region with a stop codon from the vector pUT-miniTn5-gfp-tet (Table 1). Using primers P2708F and GFPR, a fragment containing the promoter, signal peptide, and GFP was obtained by overlapping PCR. The fragment was ligated to pTSK3328 with SalI and SacI sites, producing pTSK3328-PSGFP. Then primers 3328H1F and 3328H2R were used to amplify the expression cassette. The PCR product was transformed into the competent cells of C. hutchinsonii by electroporation. The pseudogene chu_3328 of C. hutchinsonii was replaced with the expression cassette by the linear DNA double-crossover method (55). Transformants were grown on PY6 solid medium with erythromycin. The PCR products from two sets of primers (YF/YR1 and YF/YR2) were sequenced to screen the correct expression strains. Primers GFPF and GFPR2708 were used to amplify GFP without a stop codon. A fragment of the CTD was amplified with primers CTDF2708 and CTDR2708 (engineered SacI site). By overlap PCR, the complete fragment containing GFP and all necessary elements for T9SS secretion was obtained. Then the fragment was ligated to pTSK3328 or pCFXSK3328. The expression cassette was expressed in the C. hutchinsonii wild type or mutant strains for further study. The linear DNA double-crossover deletion method and overlap PCR were used to construct C. hutchinsonii with His-tagged CHU_2708.
Site-directed mutagenesis of CTDCHU_2708.
The amino acids of CTDCHU_2708 were individually mutated to glutamine or alanine by overlap PCR. Primer CTDF2708 and mutagenic primers with the desired mutation points (273NQR, 276NQR, 276NAR, 278TAR, and 296NQR) were used to amplify the first half fragment of the CTD. The second half fragment of the CTD was obtained by primers with the mutation points (273NQF, 276NQF, 276NAF, 278TAF, and 296NQF) and CTDR2708. Then the CTD fragments containing mutation sites were obtained by overlap PCR. The complete fragment containing the promoter, N-terminal signal peptide, and site-directed mutation CTD was digested and ligated into plasmid pTSK3328. The obtained expression cassettes were expressed in C. hutchinsonii. The strains were designated the N273Q, N276Q, N276A, T278A, and N296Q mutants, respectively.
Cell fractionation procedures.
The extracellular and outer membrane proteins were extracted according to the method of Zhao et al. (55). The mid-exponential-phase cells of C. hutchinsonii were harvested by centrifugation (5,000 × g, 4°C, 10 min). Cells in the supernatants were further removed with a 0.22-μm-pore-size polyvinylidene difluoride (PVDF) filter. Then the collected supernatants were precipitated with 10% (wt/vol) trichloroacetic acid for at least 1 h to obtain extracellular proteins. The washed cell pellets were resuspended in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (containing 0. 5 M NaCl) with shaking at 150 rpm (4°C, 30 min). The supernatant without cell debris was ultracentrifuged (100,000 × g, 4°C, 30 min), and the outer membrane proteins were in the sediment. The periplasmic space proteins were obtained as described by Soares et al. (56). The collected cells were incubated in sucrose buffer (20% sucrose, 1 mM EDTA, and 0. 3 mM Tris [pH 8. 0]) at 4°C for 15 min. Then the cells were pelleted (5,000 × g, 4°C, 10 min) and then incubated in double-distilled water (4°C, 15 min). The cell pellet was removed by ultrafiltration (25,000 × g, 4°C, 10 min), and the resulting supernatant was the periplasmic proteins.
Western blot analysis.
The Western blot operation was performed according to the method of Wang et al. (17). Equal biomasses of extracellular, outer membrane, and periplasmic proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then proteins on the gel were transferred onto a 0.45-μm-pore-size PVDF membrane by semidry electrophoresis. The obtained PVDF membrane was blocked in TBS (10 mM Tris, 150 mM NaCl [pH 7.4]) containing 5% nonfat powdered milk overnight. The anti-GFP rabbit monoclonal antibody was supplemented at a dilution of 1:5,000. After 4 h of incubation, the PVDF membrane was washed 5 times with TBST (TBS containing 0.1% Tween 20) for 8 min each time. The PVDF membrane was probed with anti-rabbit IgG polyclonal secondary antibody at a 1:10,000 dilution for 2 h. After washing the membrane with TBST, development was carried out as per Sparkjade ECL super (Shandong Sparkjade Biotechnology Co., Ltd.) instructions. For the detection of His-tagged proteins, anti-His mouse monoclonal antibody (MAb) was used at a 1:6,000 dilution.
Purification of GFP-related proteins.
The GFP-related proteins in the periplasmic space of the C. hutchinsonii strain were purified by anti-GFP tag nanobody-conjugated agarose beads (Engibody, Shanghai, China) following the manufacturer’s instructions.
Protein preparation for liquid chromatography-tandem mass spectrometry analysis.
The proteins in the SDS-PAGE gel were reduced with dithiothreitol (DTT) and then alkylated with iodoacetamide. Trypsin was used to further digest the proteins. The resulting product was acidified with trifluoroacetic acid (TFA) and then analyzed by liquid chromatography-tandem mass spectrometry (57).
Detection of glycosylation.
The purified GFP-related proteins on the SDS-PAGE gel were stained by periodate-Schiff chemicals (58). The purified proteins were also transferred to a PVDF membrane by semidry electrophoresis and then probed with ConA lectin per manufacturer’s instructions.
Deglycosylation of protein.
Proteins were processed using a deglycosylation kit (Rhinozyme, Suzhou, China) per the manufacturer’s instructions. The kit utilizes peptide-N-glycosidase F, which is specially designed for N-glycosylation. Then processed proteins were separated, blotted, and probed with anti-GFP rabbit MAb to detect the deglycosylation of GFP-related proteins.
Real-time quantitative PCR analysis.
The cells of the WTGFP-CTD and N276Q strains were harvested when the optical density at 600 nm (OD600) was 0.7. Then the total RNA was extracted referring to the method by Guan et al. (59). Quantitative PCR was performed with the SYBR green Premix Pro Taq HS qPCR kit (AG, Hunan, China) on a LightCycler 480 system. The relative quantitation/comparative threshold cycle (ΔΔCT) method was used to analyze data from three biological repeats of C. hutchinsonii (60). The 16S rRNA gene served as an endogenous control.
Construction of the chu_3842 deletion mutant.
The 3842H1 fragment, of about 2.5 kbp, containing the region upstream of chu_3842 and the first 291 bp of chu_3842 was amplified using primers 3842H1F and 3842H1R. 3842H1 was ligated to the pTSK plasmid through the SacΙ and SalΙ sites, generating pTSK-3842H1. The 3842H2 fragment, of approximately 2.5 kbp, was amplified using primers 3842H2F and 3842H2R. The fragment included the last 478 bp of chu_3842 and the region downstream of chu_3842. 3842H2 was cloned into pTSK-3842H1 through the NcoΙ and BamHΙ sites, producing pTSK-3842H1H2. Other procedures were as previously described. The chu_3842 deletion mutant was complemented using the complementation plasmid pCH. The fragment including chu_3842 and its promoter was obtained with primers C3842F and C3842R. Then it was ligated into pCHF by the SacΙ and SalΙ sites, generating pCHF-3842. Plasmid pCHF-3842 was electroporated into the Δ3842 mutant. Chloramphenicol was used to select the transformants.
Phenotypic assay of the Δ3842 mutant.
The periplasmic proteins of C. hutchinsonii strains were enriched using ConA lectin affinity chromatography per the manufacturer’s instructions. The disk diffusion test of susceptibility of cells to chemicals was performed as described by Bai et al. (61). The agents were sodium dodecyl sulfate (10%), dithiothreitol (2 mM), and cumene hydroperoxide (2 mM). For the quantitative detection of susceptibility, cells of the WT and Δ3842 mutant in the same growth phase were treated with different chemicals, and the absorbance at 600 nm was monitored. The filter paper degradation and colony spreading assay were conducted as previously described by Guan et al. (59). Cellulase activity was detected as described by Wang et al. (21). Significance of the data was calculated using Student’s t test. The protein concentration was quantified as described by Bradford (62). The data are the averages and standard deviations from three independent experiments.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31770080) and National Key Research and Development Program of China (2021YFC2100500).
We sincerely thank Mark J. McBride (University of Wisconsin—Milwaukee, Milwaukee, WI) for providing Cytophaga hutchinsonii ATCC 33406. We thank Edward C. Mignot, Shandong University, for linguistic advice.
Footnotes
Supplemental material is available online only.
Contributor Information
Xuemei Lu, Email: luxuemei@sdu.edu.cn.
Maia Kivisaar, University of Tartu.
REFERENCES
- 1.Abby SS, Cury J, Guglielmini J, Neron B, Touchon M, Rocha EP. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasica AM, Ksiazek M, Madej M, Potempa J. 2017. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol 7:215. 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veith PD, Glew MD, Gorasia DG, Reynolds EC. 2017. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 4.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res 12:4449–4461. 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 5.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thogersen IB, Gao J, Kwan AH, Trewhella J, Dubin G, Gomis-Ruth FX, Nguyen KA, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6:23123. 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 8.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni SS, Zhu Y, Brendel CJ, McBride MJ. 2017. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J Bacteriol 199:e00884-16. 10.1128/JB.00884-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni SS, Johnston JJ, Zhu Y, Hying ZT, McBride MJ. 2019. The carboxy-terminal region of Flavobacterium johnsoniae SprB facilitates its secretion by the type IX secretion system and propulsion by the gliding motility machinery. J Bacteriol 201:e00218-19. 10.1128/JB.00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, McBride MJ. 2017. The unusual cellulose utilization system of the aerobic soil bacterium Cytophaga hutchinsonii. Appl Microbiol Biotechnol 101:7113–7127. 10.1007/s00253-017-8467-2. [DOI] [PubMed] [Google Scholar]
- 14.Stanier RY. 1942. The Cytophaga group: a contribution to the biology of Myxobacteria. Bacteriol Rev 6:143–196. 10.1128/br.6.3.143-196.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie G, Bruce DC, Challacombe JF, Chertkov O, Detter JC, Gilna P, Han CS, Lucas S, Misra M, Myers GL, Richardson P, Tapia R, Thayer N, Thompson LS, Brettin TS, Henrissat B, Wilson DB, McBride MJ. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol 73:3536–3546. 10.1128/AEM.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci USA 107:276–281. 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Wang Z, Cao J, Guan Z, Lu X. 2014. FLP-FRT-based method to obtain unmarked deletions of CHU_3237 (porU) and large genomic fragments of Cytophaga hutchinsonii. Appl Environ Microbiol 80:6037–6045. 10.1128/AEM.01785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. 10.1007/s00253-013-5355-2. [DOI] [PubMed] [Google Scholar]
- 19.Gao L, Tan Y, Zhang W, Qi Q, Lu X. 2021. Cytophaga hutchinsonii SprA and SprT are essential components of the type IX secretion system required for Ca2+ acquisition, cellulose degradation, and cell motility. Front Microbiol 12:195. 10.3389/fmicb.2021.628555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L, Guan Z, Gao P, Zhang W, Qi Q, Lu X. 2020. Cytophaga hutchinsonii gldN, encoding a core component of the type IX secretion system, is essential for ion assimilation, cellulose degradation, and cell motility. Appl Environ Microbiol 86:e00242-20. 10.1128/AEM.00242-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Zhao D, Bai X, Zhang W, Lu X. 2017. Identification and characterization of a large protein essential for degradation of the crystalline region of cellulose by Cytophaga hutchinsonii. Appl Environ Microbiol 83:e02270-16. 10.1128/AEM.02270-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorasia DG, Veith PD, Reynolds EC. 2020. The type IX secretion system: advances in structure, function and organisation. Microorganisms 8:1173. 10.3390/microorganisms8081173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou XY, Gao JL, Hunter N, Potempa J, Nguyen KA. 2013. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol Microbiol 89:903–917. 10.1111/mmi.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473:4–8. 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 25.Zeituni AE, McCaig W, Scisci E, Thanassi DG, Cutler CW. 2010. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J Bacteriol 192:4103–4110. 10.1128/JB.00275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichler J. 2019. Protein glycosylation. Curr Biol 29:R229–R231. 10.1016/j.cub.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Kim DY, Kim KK. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol 38:266–274. 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zhao D, Gao L, Zhang W, Lu X. 2020. Cage-like polyhedrons of DegQ from Cytophaga hutchinsonii show stable proteolytic activity and strong chaperone activity. Biochem Eng J 159:107585. 10.1016/j.bej.2020.107585. [DOI] [Google Scholar]
- 29.Cain JA, Dale AL, Sumer-Bayraktar Z, Solis N, Cordwell SJ. 2020. Identifying the targets and functions of N-linked protein glycosylation in Campylobacter jejuni. Mol Omics 16:287–304. 10.1039/d0mo00032a. [DOI] [PubMed] [Google Scholar]
- 30.Lu Q, Li S, Shao F. 2015. Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol 23:630–641. 10.1016/j.tim.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Szymanski CM, Logan SM, Linton D, Wren BW. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol 11:233–238. 10.1016/s0966-842x(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 32.Cain JA, Dale AL, Niewold P, Klare WP, Man L, White MY, Scott NE, Cordwell SJ. 2019. Proteomics reveals multiple phenotypes associated with N-linked glycosylation in Campylobacter jejuni. Mol Cell Proteomics 18:715–734. 10.1074/mcp.RA118.001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleytr UB. 1975. Heterologous reattachment of regular arrays of glycoproteins on bacterial surfaces. Nature 257:400–402. 10.1038/257400a0. [DOI] [PubMed] [Google Scholar]
- 34.Mescher MF, Strominger JL. 1976. Purification and characterization of a prokaryotic glycoprotein from cell-envelope of Halobacterium salinarium. J Biol Chem 251:2005–2014. 10.1016/S0021-9258(17)33647-5. [DOI] [PubMed] [Google Scholar]
- 35.Szymanski CM, Yao RJ, Ewing CP, Trust TJ, Guerry P. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol 32:1022–1030. 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 36.Nothaft H, Scott NE, Vinogradov E, Liu X, Hu R, Beadle B, Fodor C, Miller WG, Li J, Cordwell SJ, Szymanski CM. 2020. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics 19:913. 10.1074/mcp.AAC120.002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Qaidi S, Chen K, Halim A, Siukstaite L, Rueter C, Hurtado-Guerrero R, Clausen H, Hardwidge PR. 2017. NleB/SseK effectors from Citrobacter rodentium, Escherichia coli, and Salmonella enterica display distinct differences in host substrate specificity. J Biol Chem 292:11423–11430. 10.1074/jbc.M117.790675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassak J, Keilhauer EC, Fürst M, Wuichet K, Gödeke J, Starosta AL, Chen J-M, Søgaard-Andersen L, Rohr J, Wilson DN, Häussler S, Mann M, Jung K. 2015. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat Chem Biol 11:299. 10.1038/nchembio0415-299d. [DOI] [PubMed] [Google Scholar]
- 39.Choi K-J, Grass S, Paek S, St Geme JW, III, Yeo H-J. 2010. The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS One 5:e15888. 10.1371/journal.pone.0015888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nothaft H, Szymanski CM. 2013. Bacterial protein N-glycosylation: new perspectives and applications. J Biol Chem 288:6912–6920. 10.1074/jbc.R112.417857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nothaft H, Liu X, McNally DJ, Szymanski CM. 2010. N-Linked protein glycosylation in a bacterial system. Methods Mol Biol 600:227–243. 10.1007/978-1-60761-454-8_16. [DOI] [PubMed] [Google Scholar]
- 42.Scott NE, Bogema DR, Connolly AM, Falconer L, Djordjevic SP, Cordwell SJ. 2009. Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J Proteome Res 8:4654–4664. 10.1021/pr900544x. [DOI] [PubMed] [Google Scholar]
- 43.Coyne MJ, Fletcher CM, Chatzidaki-Livanis M, Posch G, Schaffer C, Comstock LE. 2013. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol Microbiol 88:772–783. 10.1111/mmi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 8:765–778. 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 45.Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. 2013. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol 89:14–28. 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 46.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J Bacteriol 193:132–142. 10.1128/JB.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73:1019–1049. 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 48.Eichler J, Koomey M. 2017. Sweet new roles for protein glycosylation in prokaryotes. Trends Microbiol 25:662–672. 10.1016/j.tim.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JC, Szymanski C, Guerry P. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni. J Bacteriol 186:6508–6514. 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abouelhadid S, Raynes J, Bui T, Cuccui J, Wren BW. 2020. Characterization of posttranslationally modified multidrug efflux pumps reveals an unexpected link between glycosylation and antimicrobial resistance. mBio 11:e02604-20. 10.1128/mBio.02604-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowarik M, Numao S, Feldman MF, Schulz BL, Callewaert N, Kiermaier E, Catrein I, Aebi M. 2006. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314:1148–1150. 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- 52.Haltiwanger RS, Lowe JB. 2004. Role of glycosylation in development. Annu Rev Biochem 73:491–537. 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 53.Clausen T, Southan C, Ehrmann M. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10:443–455. 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 54.Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA 94:12139–12144. 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao D, Wang Y, Wang S, Zhang W, Qi Q, Lu X. 2020. A disulfide oxidoreductase (CHU_1165) is essential for cellulose degradation by affecting outer membrane proteins in Cytophaga hutchinsonii. Appl Environ Microbiol 86:e02789-19. 10.1128/AEM.02789-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soares CR, Gomide FI, Ueda EK, Bartolini P. 2003. Periplasmic expression of human growth hormone via plasmid vectors containing the lambdaPL promoter: use of HPLC for product quantification. Protein Eng 16:1131–1138. 10.1093/protein/gzg114. [DOI] [PubMed] [Google Scholar]
- 57.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O’Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 58.McGuckin WF, McKenzie BF. 1958. An improved periodic acid fuchsin sulfite staining method for evaluation of glycoproteins. Clin Chem 4:476–483. 10.1093/clinchem/4.6.476. [DOI] [PubMed] [Google Scholar]
- 59.Guan Z, Wang Y, Gao L, Zhang W, Lu X. 2018. Effects of the histone-like protein HU on cellulose degradation and biofilm formation of Cytophaga hutchinsonii. Appl Microbiol Biotechnol 102:6593–6611. 10.1007/s00253-018-9071-9. [DOI] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Bai X, Zhu S, Wang X, Zhang W, Liu C, Lu X. 2017. Identification of a fabZ gene essential for flexirubin synthesis in Cytophaga hutchinsonii. FEMS Microbiol Lett 364:fnx197. 10.1093/femsle/fnx197. [DOI] [PubMed] [Google Scholar]
- 62.Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3. Download AEM.01606-21-s0001.pdf, PDF file, 0.4 MB (362.3KB, pdf)