ABSTRACT
Glucuronan lyases (EC 4.2.2.14) catalyze depolymerization of linear β-(1,4)-polyglucuronic acid (glucuronan). Only a few glucuronan lyases have been characterized until now, most of them originating from bacteria. Here we report the discovery, recombinant production, and functional characterization of the full complement of six glucuronan specific polysaccharide lyases in the necrotic mycoparasite Trichoderma parareesei. The enzymes belong to four different polysaccharide lyase families and have different reaction optima and glucuronan degradation profiles. Four of them showed endo-lytic action and two, TpPL8A and TpPL38A, displayed exo-lytic action. Nuclear magnetic resonance revealed that the monomeric end product from TpPL8A and TpPL38A underwent spontaneous rearrangements to tautomeric forms. Proteomic analysis of the secretomes from T. parareesei growing on pure glucuronan and lyophilized A. bisporus fruiting bodies, respectively, showed secretion of five of the glucuronan lyases and high-performance anion-exchange chromatography with pulsed amperometric detection analysis confirmed the presence of glucuronic acid in the A. bisporus fruiting bodies. By systematic genome annotation of more than 100 fungal genomes and subsequent phylogenetic analysis of the putative glucuronan lyases, we show that glucuronan lyases occur in several ecological and taxonomic groups in the fungal kingdom. Our findings suggest that a diverse repertoire of glucuronan lyases is a common trait among Hypocreales species with mycoparasitic and entomopathogenic lifestyles.
IMPORTANCE This paper reports the discovery of a set of six complementary glucuronan lyase enzymes in the mycoparasite Trichoderma parareseei. Apart from the novelty of the discovery of these enzymes in T. parareesei, the key importance of the study is the finding that the majority of these lyases are induced when T. parareesei is inoculated on Basidiomycete cell walls that contain glucuronan. The study also reveals putative glucuronan lyase encoding genes in a wealth of other fungi that furthermore points at fungal cell wall glucuronan being a target C-source for many types of fungi. In a technical context, the findings may lead to controlled production of glucuronan oligomers for advanced pharmaceutical applications and pave the way for development of new fungal biocontrol agents.
KEYWORDS: Trichoderma, enzyme discovery, polysaccharide lyases, glucuronan lyases
INTRODUCTION
Trichoderma parareesei is a clonal sympatric agamospecies to the well-known cellulase producer and industrial workhorse T. reesei. Phylogenetic analysis of conserved genes shows that T. parareesei genetically resembles the ancient ancestor of both T. reesei and T. parareesei. Although many Trichoderma species are regarded as generalistic cosmopolitans (1), the majority of specialized ecological behaviors observed in this species is rooted in their fundamental nature as mycoparasites (1). T. parareesei is thus known to exert aggressive mycoparasitism and antagonistic action against various fungi, including phytopathogens such as Fusarium oxysporum (2), Rhizoctonia solani (2), and Botrytis cinerea (3, 4).
Depolymerizing carbohydrate-active enzymes (CAZymes) are necessary for all fungi to restructure and recycle their own cell wall polysaccharides. Although necrotrophic mycoparasites often utilize the same classes of enzymes to attack their prey (both living and dead prey), genome comparisons of Trichoderma and Hypomyces species indicate that the glycoside hydrolase families harboring enzymes presumed involved in mycoparasitic attack are highly expanded, suggesting a strong evolutionary niche adaptation (5, 6).
To our knowledge, only few polysaccharide lyases have been mentioned in genome comparisons or reported in relation to mycoparasitic traits like the pectin lyase PEL12 in Clonostachys rosea, which was concluded not to be relevant for mycoparasitic abilities of the fungus (7).
Polysaccharide lyases (EC 4.2.2.-) (PLs) catalyze cleavage of uronic acid polysaccharides by β-elimination, producing Δ4,5 unsaturated bonds at the nonreducing end of one of the reaction products (8). In the enzyme database of CAZymes (www.cazy.org/) (9), glucuronan lyases (EC 4.2.2.14) are presently distributed in six PL families (PL5, PL7, PL14, PL20, PL31, PL38) but only represented by seven experimentally characterized sequences, distributed among these families. Most notably the PL20 family was created on the basis of a characterized Trichoderma reesei lyase (10–12).
Glucuronan (β-1,4-polyglucuronic acid) is a linear homopolymer consisting of glucuronic acid residues linked by β-1,4-glycosidic bonds. To date, glucuronan has only been identified in a few organisms, notably in the cell wall of green algae (13), in the cell wall and extracellular matrix of certain Mucor species (14, 15), and as an exopolysaccharide of a mutated strain of the bacterium Rhizobium meliloti (16).
RESULTS
Genomic annotation.
Genome-wide annotation of CAZymes in the T. parareesei genome revealed six genes encoding polysaccharide lyases belonging to the families PL7, 8, 20 and 38. All enzymes contained a predicted signal peptide and a single catalytic domain. The six genes were located on different genomic contigs in the T. parareesei genome. Functional prediction using peptide clustering (CUPP) suggested that TpPL20A, B and TpPL38A are glucuronan specific lyases (EC 4.2.2.14) (Table 1). The two PL7 sequences did not yield a functional prediction but were assigned to the PL7 subfamily 4 both by the dbCAN hidden Markov model (HMM) and by CUPP (Table 1). PL7_4 contains all the members of terrestrial fungal origin assigned to the PL7 family and includes one functionally characterized glucuronan lyase from the actinobacterium Catenulispora acidiphila (17).
TABLE 1.
Predicted features of the polysaccharide lyases in T. parareesei
| Genbank accession no. | Designationa | Native size (aa) | Family predictionb (dbCAN) | Domain (aa) | Family predictionb (CUPP) | EC prediction (CUPP) | Secretion signal (aa) | Predicted size of recombinant protein (kDa) |
|---|---|---|---|---|---|---|---|---|
| OTA00986.1 | TpPL7A | 252 | PL7_4 | 31-249 | PL7_4 | – | 1-20 | 26 |
| OTA01262.1 | TpPL7B | 248 | PL7_4 | 29-248 | PL7_4 | – | 1-18 | 25 |
| OTA05836.1 | TpPL8A | 785 | PL8_4 | 387-632 | nd. | – | 1-19 | 85 |
| OTA06285.1 | TpPL38A | 422 | PL38 | 82-358 | PL38 | 4.2.2.14 | 1-27 | 46 |
| OSZ99920.1 | TpPL20A | 258 | PL20 | 23-257 | PL20 | 4.2.2.14 | 1-19 | 29 |
| OTA07247.1 | TpPL20B | 258 | PL20 | 24-257 | PL20 | 4.2.2.14 | 1-20 | 29 |
This study.
nd., not determined; −, no EC number prediction; _4 denotes predicted subfamily.
One enzyme was assigned to PL8_4 (Table 1). The PL8 family mainly contains bacterial endo- and exo-lyases acting on glycosaminoglycans, and one fungal enzyme from Paradendryphiella salina recently characterized as an exo-mannuronate lyase (18). Until now, glucuronan specificity has not been reported for other PL8 family members.
Recombinant enzyme production.
The sequences of the six lyases were codon optimized and successfully expressed in Pichia pastoris. EndoH treatment revealed the recombinant enzymes, except TpPL20A and B, to be N-glycosylated. The molecular weights of the deglycosylated proteins were in accord with their predicted sizes (Table 1, Fig. S1 in the supplemental material).
Biochemical characterization.
The purified recombinant enzymes had no activity on alginate, chondroitin or hyaluronan, but all six lyases showed activity on glucuronan. The glucuronan had a molar weight of approximately 12 kDa corresponding to an average degree of polymerization (DP) of 68 (Fig. S2). The six lyases exhibited optimal activity on glucuronan at pH 4–7 and 40–50°C (Table 2). All enzymes except TpPL20B had >35% activity between 25–55°C. TpPL20B had more than 40% increased activity between 25 and 50°C (Fig. S3). None of the lyases required NaCl to work, but except for TpPL7B and TpPL8A they all showed enhanced activity in the presence of 100–200 mM NaCl, with TpPL38A having a 6-fold increased activity at 200 mM NaCl (Table 2). For TpPL8A an inhibitory effect occurred already at 100 mM (Fig. S3). Except for TpPL38A, all enzymes showed decreased activity at salt addition levels above 300 mM NaCl. Salts and ions can affect the stability, solubility, and surface charge of the enzymes but may also shield the negative charge on the substrate (19). TpPL20A and B required Ca2+ for activity with optimal levels of 6 mM and 4 mM at the protein concentration tested. This indicates that Ca2+ is necessary for the function or stability of the enzymes in agreement with a previous structural study of a PL20 lyase (12). TpPL8A activity was unaffected, while TpPL7A, TpPL7B, and TpPL38A were inhibited by Ni2+ and Zn2+ but, except for TpPL7B, unaffected by Ca2+, Mg2+ and Mn2+ (Table 2).
TABLE 2.
Kinetic parameters and biochemical characterization of the T. parareesei glucuronan lyasesa
| Parameter | TpPL7A | TpPL7B | TpPL8A | TpPL38A | TpPL20A | TpPL20B |
|---|---|---|---|---|---|---|
| Kinetic parameters | ||||||
| kcat (s−1) | 2276.5 ± 20.1 | 23.8 ± 3.1 | 102.8 ± 2.2 | 50.4 ± 0.4 | 0.3 ± 0.01 | 5.6 ± 0.2 |
| Km (mM) | 3.7 ± 0.2 | 3.8 ± 1.1 | 14.1 ± 0.6 | 12.1 ± 0.3 | 1.5 ± 0.1 | 1.0 ± 0.2 |
| kcat / Km (s−1 mM−1) | 615.9 | 6.3 | 7.3 | 4.2 | 0.2 | 5.5 |
| Optimal conditions | ||||||
| pH optimum | 7 | 5 | 4 | 6 | 6 | 6 |
| Temp. optimum (°C) | 40 | 50 | 45 | 50 | 45 | 40 |
| Salt (0–500 mM NaCl) | 129% (100 mM) | 100% (0 mM) | 100% (0 mM) | 668% (200 mM) | 181% (150 mM) | 170% (200 mM) |
| Divalent cations (2 mM) | ||||||
| Ca2+ | 99% | 83% | 108% | 97% | 100% (6 mM)b | 100% (4 mM)b |
| Mg2+ | 102% | 85% | 101% | 100% | 0% | 0% |
| Mn2+ | 96% | 77% | 103% | 94% | 0% | 0% |
| Ni2+ | 13% | 67% | 101% | 70% | 0% | 0% |
| Zn2+ | 18% | 41% | 110% | 51% | 0% | 0% |
aActivity level achieved at optimal NaCl addition level, with the optimal level given in mM in parentheses. ± indicates standard deviation from triplicate experiments.
bOptimal concentration determined for the activator cation.
Kinetic parameters.
The molar extinction coefficient for the Δ4,5 unsaturated bond required for calculating product formation was determined experimentally to 6368 ± 317 cm−1 M−1 (Table S1 in the supplemental material). This value is in good agreement with the coefficient value of 6150 cm−1 M−1 used for alginate lyases (18, 20). All six lyases exhibited classical Michaelis-Menten kinetics (Fig. S4). TpPL7A had highest turnover number (kcat) of 2277 s−1 and consequently a high specificity constant (kcat/Km) of ∼616 (s−1 mM−1). The kcat of TpPL7A was approximately 22-fold higher than that of the second-fastest enzyme, TpPL8A, which had kcat of 103 s−1 (Table 2). TpPL20A had the lowest activity of them all (Table 2). Except for the extraordinarily fast TpPL7A the kinetic parameters were comparable (within a 10-fold range) to fungal pectin lyases (21), fungal alginate lyases (18), and bacterial chondroitin lyases (22).
Product profiles.
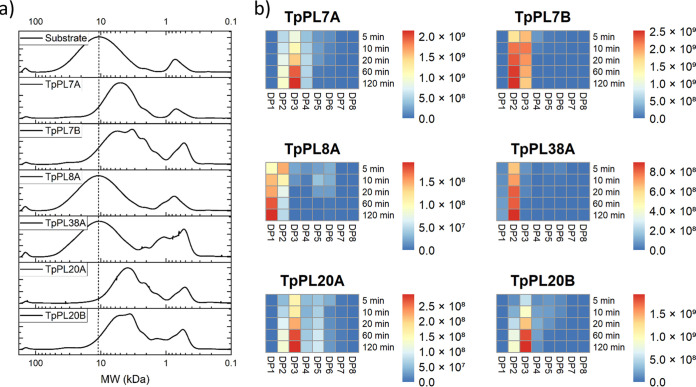
Size exclusion patterns of the enzymatic reaction products revealed a major shift in size of the large polymeric part (12 kDa) of the substrate after reacting with TpPL7A, TpPL7B, TpPL20A and TpPL20B, indicating endo-action. In contrast, no significant shift in the molecular weight of the main polymer fraction, but increases in DP1 and DP2, were observed in the TpPL8A and TpPL38A reactions, indicating exo-action (Fig. 1a). LC-ESI-MS analyses revealed accumulation of products ranging from DP2 to DP6 for the four endo-acting lyases, with DP3 being the most abundant oligosaccharide for TpPL7A, TpPL20A and TpPL20B and DP2 being most abundant for TpPL7B (Fig. 1b). TpPL8A and TpPL38A only accumulated DP2 and DP1, which is consistent with their proposed exo-action.
FIG 1.
Product profiles of each of the six recombinant glucuronan lyases from T. parareesei during reaction on glucuronan. a) High performance size exclusion chromatography patterns of 2h enzyme reactions on glucuronan. b) Heat maps of averaged relative intensities derived from LC-ESI-MS analysis of oligoglucuronan formation during time course experiments.
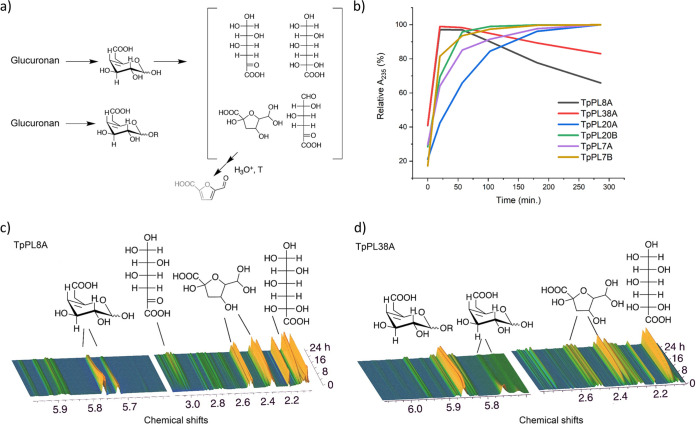
The action-mode of TpPL8A and TpPL38A were further examined by in-situ near magnetic resonance (NMR) analysis (Fig. 2). The 1H signals near 5.75 ppm represent the olefinic hydrogen signals of ΔGlcA, which in the absence of 1H signals for free saturated GlcA affirm that TpPL8A and TpPL38A cleave glucuronan from the reducing end (Fig. 2c and d). The NMR spectroscopy data also validated that TpPL8A liberates monomeric ΔGlcA, while TpPL38A liberates labile monomeric ΔGlcA in addition to short stable oligoglucuronan. The combined results show that the entire complement of glucuronan lyases in T. parareesei catalyze the complete degradation of glucuronan to dimers and monomers under various conditions.
FIG 2.
Nonenzymatic conversion of unsaturated glucuronic acid monomers. a) Proposed reaction scheme. b) Degradation kinetics of the six glucuronan lyases. A235 was measured continuously at 40°C and pH 5 with 8 mM glucuronan, 100 mM NaCl, 2 mM CaCl and under otherwise standard assay parameters. c) 1H NMR spectrum of 24h time series for TpPL8A reaction at pH 4 and 30°C. d) 1H NMR spectrum of 24h time series for TpPL38A reaction at pH 6 and 30°C.
Nonenzymatic conversion of unsaturated glucuronic acid monomers.
As shown for alginate lyases (23, 24) and a bacterial PL38 glucuronan lyase (25), unsaturated monosaccharide products, including α-keto-glucuronic acid, may convert to saturated compounds nonenzymatically (Fig. 2a). Indeed, the reactions of TpPL8A and TpPL38A resulted in a detectable decrease of the absorbance after approximately 1 h, signifying the gradual loss of the formed C4–C5 double bond on the monomer (Fig. 2b). Time-resolved in situ 1H NMR for the TpPL8A and TpPL38A reactions for 24 h (Fig. 2b and c) allowed identification of the main products of monomer rearrangements to products without olefinic double bonds (Fig. S5 in the supplemental material): First, the cyclic ΔGlcA dominates, then, nonenzymatic ring opening of ΔGlcA leads to the formation of an aldo-hydrate that retains a keto group at position 5 and a ketoaldo-hydrate. The aldo-hydrate then undergoes intermolecular cyclization leading to the formation of a 5-membered hemi-ketal (Fig. 2a). The nonenzymatic conversion of ΔGlcA was faster at pH 6 (TpPL38A) than at pH 4 (TpPL8A) (Fig. 2c and d). TpPL38A was found to form both monomeric and oligomeric product at comparable amounts, consistent with signal loss of the 4,5 double bond that is intermediate between rearrangements upon TpPL8A catalyzed monomer formation and the formation of stable nonmonomeric producs from TpPL7 and TpPL20 enzymes (Fig. 2b).
Proteomic analysis of T. parareseei secretomes.
To examine how ascomycete and basidiomycete cell walls and glucuronan might affect the induction and secretion of glucuronan lyases and other CAZymes in T. parareesei, we analyzed the extracellular proteome of the fungus after 24 h of fermentation on liquid media containing i) fruiting body cell walls from Agaricus bisporus (fungal-cell-wall-medium-basidiomycete, FCWM-B), ii) mycelial cell wall from Botrytis cinerea (ascomycete, FCWM-A), iii) pure glucuronan, and iv) glucose (Glc) (as negative control). The ability of T. parareesei to use the substrates as carbon sources was confirmed prior to the experiment (Fig. S6 in the supplemental material).
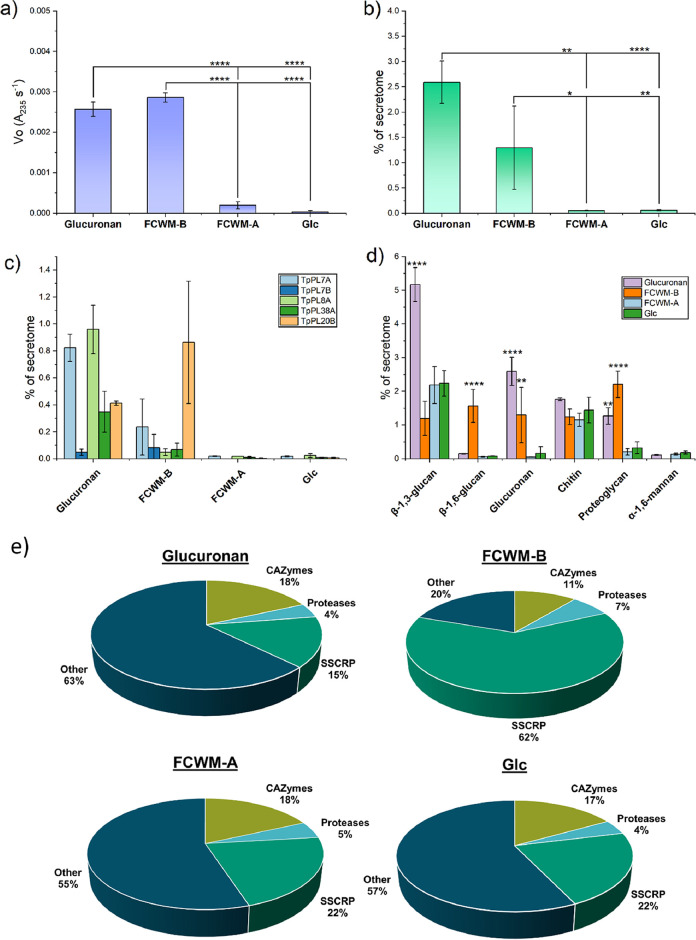
The glucuronan lyase activity of the fungal supernatants was only observed in the glucuronan and FCWM-B fermentations (Fig. 3a). The total protein content of the glucuronan fermentation was 5-fold higher than the glucose control (Fig. S7). This could indicate that the glucuronan used in this study does not act as a catabolic repressor as opposed to pure glucose. Proteomic analysis of the supernatants confirmed the presence of glucuronan lyases in the supernatants from the glucuronan and FCWM-B fermentations (Fig. 3b–d).
FIG 3.
Secreted proteome analysis from T. parareesei after 24 h of growth on four different carbon sources. FCWM-B = Fungal Cell Wall Medium-Basidiomycete (A. bisporus), FCWM-A = fungal cell wall medium-ascomycete (B. cinerea), Glc = Glucose. The intensity based abundances of each detected protein were normalized to the sum of each replicate. Error bars represent standard deviation of triplicate experiments. Asterisks represents P values from one-way ANOVA post hoc Tukey comparison of means. * = P ≤ 0.05, ** = P ≤ 0.01, **** = P ≤ 0.0001. a) Glucuronan assays of the fermentation supernatants under standard assay conditions. b) Summed abundances of the detected glucuronan lyases, described as percentage of the secretome for each substrate. c) Abundances of each individual glucuronan lyases. d) Abundances of CAZymes categorized by substrate specificity (Table S2). The six most abundant categories and with relevance for fungal cell wall degradation are shown. P values were calculated against the glucose fermentation only. e) Pie charts representing the averaged overall distribution of abundances in the secreted proteomes. SSCRP = Small Secreted Cysteine-Rich Proteins.
Of the six glucuronan lyases, only TpPL20A was not detected in any of the supernatants and TpPL7B was only detected in small amounts on glucuronan and FCWM-B (Fig. 3c). Significant differences of ratios between CAZymes related to fungal cell wall carbohydrates were observed between glucuronan, FCWM-B and Glc, but not between Glc and FCWM-A. Significant abundances of glucuronan lyases, β-1,3-glucanases, β-1,6-glucanases and proteoglycan active hydrolases were observed on glucuronan and FCWM-B (Fig. 3d). No significant differences in relative abundance of GH18 chitinases were observed between the fermentations, which agrees with the role of chitin as a structural and recalcitrant polysaccharide and therefore not a target for depolymerization in the early onset of enzymatic attack. Glucuronan has been reported resistant to acid hydrolysis (15, 16), but A. bisporus cell walls were found to contain approximately 2.7% glucuronic acid of all detected sugars. None was detected in B. cinerea and only trace amounts in T. parareesei (Fig. S8 in the supplemental material).
Observations of the distribution of all the detected proteins in the secreted proteome showed similar profiles regarding CAZymes and proteases across the fermentations. Interestingly, the relative amount of small secreted cysteine-rich proteins (SSCRPs) dominated in the FCWM-B supernatant (Fig. 3e). SSCRPs in fungi are proposed to play roles in virulence, blocking proteases and protecting their chitin from foreign chitinases (26).
Polysaccharide lyases in the fungal kingdom.
To further investigate and map the occurrence of genes encoding proteins from the same and similar PL families to the ones found in T. parareesei, we carefully selected 107 publicly available fungal genomes from the NCBI database, covering the major taxonomic groups and ecological niches in the fungal kingdom, and annotated their full repertoires of polysaccharide lyases using HMMs (Fig. S9 in the supplemental material). The annotated sequences were used for phylogenetic analysis in combination with CAZy reported sequences and for a comparison of PL genes across the fungal kingdom.
Nonredundant (<90% similarity) subsets of the catalytic domains predicted by dbCAN HMM models, were used to generate maximum likelihood phylogenetic trees of the families containing at least one member characterized as a glucuronan lyase (Fig. 4).
FIG 4.
Maximum likelihood phylogenetic trees of protein sequences from the four PL families: a) PL7 family, b) PL20 family, c) PL38 family, and d) PL8 family. CUPP clusters are shown by the colored strips preceding the species names. All sequences were subjected to redundancy, fragmentation and alignment checks. All six lyases from T. parareesei were similar (90–100%) to T. reesei sequences (orthologous). For that reason the T. reesei sequences were omitted during the redundancy check. Characterized members are highlighted by symbols at the node ending and subfamily annotations are highlighted by the colored ellipsis next to the species names. Branch numbers indicate bootstrap values above 50.
TpPL7A and TpPL7B both located centrally in the fungal part of the subfamily 4 clade under two different subclades, suggesting a general glucuronan substrate specificity for this entire subfamily (Fig. 4a). Because of the strong connection to alginate, sequences from terrestrial fungi assigned to this family are often annotated as alginate lyases, which in the light of our present results warrants a re-evaluation. Different from the PL7 family, the clade formation in the PL20 phylogenetic tree was seemingly governed by taxonomy, with fungi in the Hypocreales order including TpPL20A and B, separated from bacteria and other ascomycetes (Fig. 4b). The PL20 family was created on the basis of a T. reesei glucuronan lyase TrGL (accession BAG80639.1) (11), which is 100% identical to TpPL20B. Similarly TpPL20A is identical to an uncharacterized T. reesei protein (XP_006969120.1).
Because no subfamily classification has been assigned to this family in the CAZy database, we performed peptide-based clustering by CUPP (27, 28). The clustering produced a single cluster (data not shown), indicating that the overall sequence similarity of members of the PL20 family is highly conserved suggesting that the family is most likely monospecific to glucuronan.
Sequences from the recently created PL38 family (created 2020) (29) were also subjected to peptide clustering, which revealed numerous clusters that generally followed the seemingly taxonomically driven clade divergence of the tree. The largest cluster comprised the ascomycete sequences including TpPL38A and an adjacent clade of Actinobacteria, but not the nearby Proteobacteria clade containing the characterized Brevundimonas lyase (29) (Fig. 4c).
The pattern of clade divergence and cluster groups mirrored the large sequence variety within the family, which in this case is not necessarily indicative of a wide substrate specificity. The sequences listed in the CAZy database already include a variety of bacterial and fungal sequences, the latter group comprising solely Basidio- and Mycomycota sequences.
The discovery of TpPL38A expands the PL38 family to include a fungally derived member, while the identification of TpPL8A expands the PL8 family to include a glucuronan lyase. As for family PL7, the clade divergence of the PL8 tree was governed strongly by substrate specificity. This correlates with the subfamily annotations potentially classifying subfamily 4 in the fungal clade as glucuronan and alginate specific. The latter specificity originates from a PL8 mannuronate specific lyase from the marine ascomycete Paradendryphiella salina (18) (Fig. 4d).
Genomewide comparisons of predicted fungal polysaccharide lyase genes.
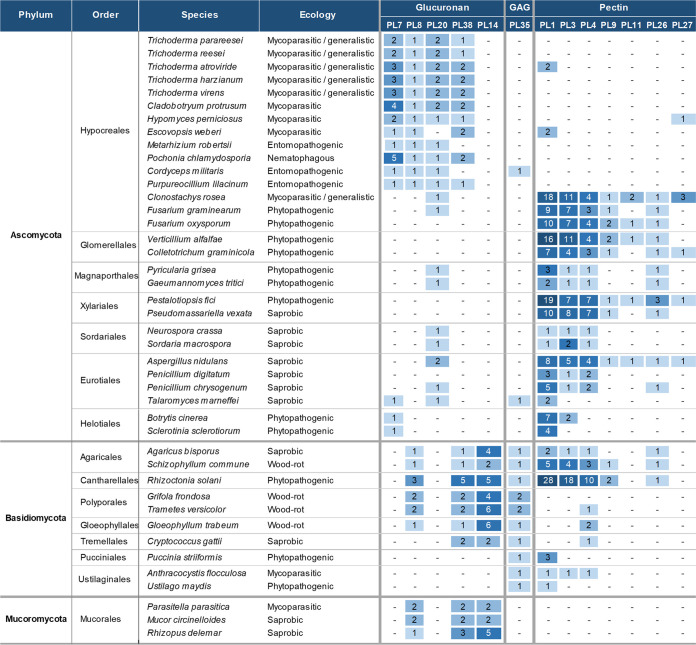
Based on the enzyme characterizations and phylogenetic analyses of the PL7, 8, 20, and 38 families, it is likely that the putative lyases assigned to these families from the terrestrial ascomycetes share substrate specificity toward glucuronan. These PL families were found highly prevalent in Trichoderma, Hypomyces, and Cladobotryum species (Fig. 5, Fig. S9 in the supplemental material) that exhibit necrotrophic mycoparasitic traits toward a wide range of fungi, but most notably toward basidiocarp producing saprobic basidiomycetes (30–32). A somewhat similar pattern was found in the entomopathogenic and nematophagous members of Hypocreales (Fig. 5, Fig.S9).
FIG 5.
Genome annotation of PL genes in a broad selection of fungi, covering major ecological niches and taxonomic groups. This is a representative subset of a larger table (Fig. S9). The putative substrate categories, glucuronan, GlucosAminoGlycans (GAG) and pectin are inferred hypothetically by substrate specificities of experimentally characterized members of the CAZy families and phylogenetic analysis of the protein sequences.
In contrast, except for one PL20 gene Clonostachys rosea and Anthracocystis flocculosa (previously Pseudozyma flocculosa) had no putative glucuronan lyases annotated. Both fungi have reported mycoparasitic traits strictly toward ascomycetes (33, 34). Another distinct pattern of genes belonging to the families 8, 14, and 38 was identified in wood-rotting basidiomycetes and members of the Mucoromycota phyla (Fig. 5).
With very few exceptions, families PL7 and PL20 seem exclusive to ascomycetes, whereas PL14 seem exclusive to basidiomycetes and mucoromycetes. To our knowledge, no fungal PL14 family enzymes have been characterized, but a glucuronan specific PL14 from a Chlorovirus strain is known (9). A phylogenetic analysis of this family revealed the putative PL14 sequences from Basidio- and Mycomycota to form two distinct clades closest to the root of the tree. The Chlorovirus clade containing the characterized glucuronan lyase was placed between the two clades suggesting a possible shared substrate specificity (Fig. S10).
DISCUSSION
The very distinct pattern of glucuronan lyase genes in all the analyzed Trichoderma spp. varied only slightly in copy numbers within the PL7 and PL38 families, but not in the family types. This indicates a sustained genetic adaptation and poses interesting questions about the origin of the substrate which, given the cosmopolitan and generalistic nature of Trichoderma spp. indicates it as a normal occurrence or even stable carbon source in the microenvironments. Furthermore, the active secretion of glucuronan lyases by T. parareesei in the presence of pure glucuronan and basidiomycete cell wall material, as well as the detection of glucuronic acid in said cell walls and the absence of both observations regarding ascomycete cell walls, suggests that glucuronan-like polysaccharides are present in the A. bisporus fruiting bodies. The secretion of the majority of the glucuronan lyases may be part of a true catabolic response of T. parareesei, which may be connected to the fungus mycoparasitic behavior. However, further in-depth studies using gene regulation analysis and reverse genetics would be required to properly elucidate this subject.
The patterns of polysaccharide lyases with a putative similar specificity in basidiomycetes is interesting and if considering the hypothesis that they produce glucuronan themselves and they would probably need depolymerizing enzymes for restructuring and recycling the polysaccharide through growth cycles as observed with chitinases and β-1,3/1,6-glucanases (35). Furthermore, similar patterns of PL8, 38, and 14 families were found in members of the Mucoromycota phylum, which contains the only fungal species reported to produce glucuronan in in the cell wall and as an exopolysaccharide (14, 15). These observations suggest that a diverse repertoire of glucuronan lyases is not strictly a Hypocrealian or common mycoparasitic trait but may also be connected to certain ecological niches and opportunistic behaviors. Indeed, the only fungal genomes harboring very few or no putative glucuronan lyase genes across the phyla were fungi classified as phytopathogenic. Instead, high numbers of pectin lyases were prevalent in these species, suggesting that glucuronan is not found in plants. However, considering the vast amounts of putative bacterial glucuronan lyases already classified in the CAZy database, it is tempting to speculate that glucuronan occurs much more widely in ecological niches connected to soil and the rhizosphere than what is currently recognized.
Given the simple structural nature of their substrate, the whole complement of glucuronan lyases in T. parareesei appears to constitute an enzymatic machinery that facilitates complete degradation of glucuronan. The discovery of TpPL8A and TpPL38A as novel exo-acting enzymes lead to a complete degradation of the polymer to easily absorbable dimers and monomers. It is likely that these monomers then further restructure before entering the d‐glucuronate catabolic pathway similar to the one found in aspergilli (36).
Oligosaccharides, such as oligoglucuronans, have numerous potentially attractive applications in various industries, such as drug carriers, solvents, stabilizers, binders and swelling agents (37). They also have proven potential in bioprotection applications, eliciting strong defense-related responses in agricultural plants like tomatoes (38) and apples against blue and gray mold (39). Also biological activities on mammalian cells and immune-stimulating effects on human blood using oligoglucuronans have been observed (40).
In conclusion, a set of six glucuronan lyases belonging to 4 different PL families (PL7, PL8, PL20, and PL38) was discovered in T. parareesei. The complement of these glucuronan lyases in T. parareesei represents sufficient diversity to constitute an enzymatic machinery that facilitates complete glucuronan degradation. The high catalytic rate of TpPL7A makes this enzyme an ideal candidate for the elucidation of the structure-function relationship along with the exo-lytic TpPL8A and TpPL38A. The observations from the genome comparisons opens questions and invites further studies aimed at the underlying complexity of ecological traits of certain groups of fungi, lyase function, mycoparasitism, ecological adaptation, and the presence or absence of glucuronan-like polysaccharides in fungal cell walls. The discovery of fungal glucuronan lyases may also have practical implications in bioprotection applications, as oligoglucuronans have shown promising potential as bioagents (38, 39). The six glucuronan lyases were effectively produced in P. pastoris, indicating a good comparability for recombinant production in foreign hosts. We therefore anticipate that the enzyme machinery described herein harbors promising options for industrial applications and constitute a good foundation for further characterization studies of glucuronan lyases in the bacterial and fungal kingdom.
MATERIALS AND METHODS
Materials.
Trichoderma parareesei CBS125925 (4) isolated from soil in South America was obtained from CBS-KNAW culture collection (www.westerdijkinstitute.nl/collections/). Botrytis cinerea IBT42711 was obtained from the IBT Fungal Culture Collection (Technical University of Denmark, Kongens Lyngby, Denmark). Organically grown mature A. bisporus fruiting bodies were procured commercially (Salling Group, Brabrand, Denmark). Sodium alginate, chondroitin AC, chondroitin B and hyaluronic acid, were purchased from Sigma (Sigma-Aldrich, St. Louis, MO, USA). Polymannuronic acid and polyguluronic acid were purchased from Carbosynth (Carbosynth, Compton, UK).
Sequence analysis and phylogeny.
Predicted protein sequences from all genomes in this study (Table S3 in the supplemental material) including that of T. parareesei CBS125925 (GCA_001050175.1) were downloaded from NCBI (www.ncbi.nlm.nih.gov/) (41). In the few cases where predicted protein sequences were not available for the chosen genomes, they were predicted with the WebAUGUSTUS server (https://bioinf.uni-greifswald.de/webaugustus) (42) using pretrained parameters from Fusarium graminearum. Putative catalytic domains of CAZymes were predicted by HMMER v. 3.3.1 (43) using the most recent (30 July 2020) HMMs from the dbCAN server (http://bcb.unl.edu/dbCAN2) (44). The results underwent strict post analysis cutoff values by applying a coverage threshold of 0.35 and an E-value threshold of 1 × 10−5. When meeting the necessary thresholds the results included inferred subfamilies. Subfamily classifications in CAZy were used as positive controls. The Interproscan server (www.ebi.ac.uk/interpro/) (45) was also used as a tool for secondary conformational domain prediction along with prediction of proteins other than CAZymes. Signal peptide predictions were performed using the Phobius webserver (https://phobius.sbc.su.se/) (46). CUPP (27) was used for protein clustering under default parameters. Broad substrate categories of CAZymes were inferred manually by cross-referencing listed functions in www.cazy.org (9) and the BRENDA database (www.brenda-enzymes.org) (47).
Phylogenic analysis.
Isolated catalytic domains from sequences listed in CAZy and HMM annotated sequences from the fungal genomes were screened for redundancy using CD-HIT (48) (<90% similarity) under default parameters. The sequences were aligned with Mafft (49) and manually inspected in CLC main workbench. The alignments were used to perform maximum likelihood phylogeny with RaxML blackbox (50) using LeGascuel substitution matrix (51) and otherwise default parameters at the CIPRESS server (www.phylo.org/) (52). RaxML stopped the rapid bootstrap search after meeting the MRE-based Bootstopping criterion.
Preparation of glucuronan.
Glucuronan (β-1,4-polyglucuronic acid) was prepared by TEMPO-mediated oxidation of commercial rayon (Krestoffer, Søborg, Denmark) as previously described (53) using TEMPO (2,2,6,6-tetramethylpipedine-1-oxyl radical; Sigma-Aldrich), sodium bromide (Sigma-Aldrich) and 11% sodium hypochlorite solution (Merck, Darmstadt, Germany). The reaction time was adjusted to 30 min and reactions were spun down for 10 min to remove potential residual un-oxidized cellulose prior to quenching by ethanol 96% ethanol (Merck) and washed twice in 70% acetone (Sigma-Aldrich).
Cloning, expression and purification of glucuronan lyases.
The six mature genes of TpPL7A (GenBank accession no. OTA00986.1), TpPL7B (OTA01262.1), TpPL8A (OTA05836.1), TpPL20A (OSZ99920.1), TpPL20B (OTA07247.1) and TpPL38A (OTA06285.1) excluding their predicted signal peptides were codon optimized for P. pastoris expression (GenScript, Piscataway, NJ, USA) and cloned into the pPICZaA vector (Invitrogen, Carlsbad, CA, USA) using EcoRI and SalI restriction sites. Plasmids were linearized by PmeI (ThermoFisher, Waltham, MA, USA) and transformed into P. pastoris X-33. Production and purification of all recombinant proteins were performed as previously described (54). Based on the sequences of the recombinant enzymes the theoretical molar extinction coefficient and sizes were calculated using ProtParam (http://web.expasy.org/protparam) (55). Protein concentrations were determined by A280 using the obtained molar extinction coefficients. Deglycosylation of the purified recombinant enzymes was performed by adding EndoH (New England Biolabs, Ipswich, USA) to the purified glycoproteins and incubating at 25°C for 3 h. The purity of the recombinant proteins and the effect of the EndoH treatment were checked on 4–12% gradient SDS-PAGE gels (Fig. S1).
Standard assay conditions.
Polysaccharide lyase activity was determined by monitoring the formation of 4,5-unsaturated bonds at the nonreducing end by monitoring absorbance at A235 (235 nm) for 10 min using a Multiskan GO spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Unless otherwise stated, reactions were carried out in triplicate at 30°C in 20 mM UB4 buffer (56) pH 6 with 1.5 g liter−1 of glucuronan in 96-well quartz plates (Corning, New York, USA). Final enzyme concentrations; TpPL7A (0.00015 μM), TpPL7B (0.0029 μM), TpPL8A (0.0018 μM), TpPL38A (0.0066 μM), TpPL20A (0.2804 μM) and TpPL20B (0.0524 μM).
Biochemical characterization.
The substrate specificities were assessed for each enzyme under standard assay conditions at pH 4, 6 and 8 using glucuronan, alginate, polymannuronic acid, polyguluronic acid, chondroitin AC, chondroitin B, and hyaluronic acid as substrates. Once the specific substrates for all recombinant enzymes were established, the pH optimum was determined for each of them using 20 mM UB4 buffer ranging from pH 2 to 8. The optimum temperature was determined at the optimum pH for each enzyme within a range of 25–55°C. The effect of NaCl (0–500 mM) and divalent ions (2 mM final concentration of CaCl2, ZnCl2, MgCl2, NiCl2, and MnCl2), respectively on enzyme activity were investigated under optimum pH, temperature. Prior to the reactions with cations the enzymes were incubated with 2 mM EDTA for 10 min and then dialyzed twice against the activity buffer. In case of TpPL20A and B, concentrations of CaCl2 ranging from 0 to 7 mM were tested to identify the optimum under standard assay conditions.
Molar extinction coefficient of the Δ4,5 bond.
The molar extinction coefficient of the C4-C5 double bond (Δ4,5 bond) formed from the cleavage by glucuronan lyases was determined using the Beer-Lambert law, by measuring the absorbance at 235 nm and the reducing ends generated during the reaction of TpPL7B under optimum conditions. The reactions were carried out at three different substrate concentrations (0.8, 1 and 1.5 g liter−1) at 40°C, 20 mM UB4 buffer pH 5 for 5 min. The reducing ends resulting from glucuronan degradation by TpPL7B were quantified by the para-hydroxybenzoic acid hydrazide (PAHBAH) assay (57, 58) using glucuronic acid (Sigma-Aldrich) as a standard.
Kinetic parameters.
For each enzyme, initial velocities were quantified on glucuronan with concentrations varying from 0.1 to 10 g liter−1 at optimum conditions specific to each enzyme. The average initial velocities quantified in milli-absorbance units (mAU) at A235 per second were converted to mM of product generated via measuring the amount of Δ4,5 bonds formed per second using the experimentally confirmed extinction coefficient of 6368 M−1 cm−1. Kinetic parameters were determined for each enzyme by plotting initial velocities against substrate concentrations. Km and kcat were obtained by fitting the Michaelis-Menten equation v0 = Vmax/(1+(Km/[S0]) using Origin 2019 software (OriginLab Corporation, Northampton, MA, USA) where v0 is the initial velocity, [S0] the initial substrate concentration, Vmax the maximum rate, and Km the Michaelis constant.
High-performance size exclusion chromatography (HP-SEC).
HP-SEC of 120 min enzyme reactions under optimum conditions using 10 g liter−1 of glucuronan was performed using a TSKgel G3000PW column and refractive index detection as previously described (21). A polynomial relationship between logarithmic molecular weight and retention time was established using pullulan standards (Sigma-Aldrich) as calibration references.
Liquid chromatography-mass spectrometry.
Duplicate reactions were prepared under optimal assay conditions for each recombinant enzyme. For pH under 7, 20 mM sodium acetate buffers were used, for pH 7 an ammonium chloride buffer was chosen. Samples were taken at different reaction times (5, 10, 20, 60, and 120 min). The reactions were stopped by heating the samples at 95°C for 5 min. Final sample preparation and liquid chromatography-mass spectrometry analysis was carried out as previously described (18).
NMR.
NMR samples (500 μl, 95% H2O/5% D2O) were prepared with 100 mM NaCl and 5 mM acetate buffer to study degradation of β-glucuronan (20 mg) by TpPL8A (pH 4.0) and TpPL38A (pH 6.0), respectively. All NMR spectra were recorded at 30°C (reactions at 40°C were deemed too fast for identification of the various intermediates and products) on an 800 MHz Bruker Avance III NMR spectrometer equipped with an 18.7 T Oxford Magnet and a Bruker TCI CryoProbe. The reaction was followed in situ over time by a sequence of one-dimensional 1H NMR experiments using excitation sculpting for water suppression. This time-series of one-dimensional 1H NMR was implemented as a pseudo-2D experiment, which sampled the NMR signal (with spectral width of 14 ppm) during an acquisition time of 1.46 s and employed an inter-scan relaxation delay of 5 s. Up to 512 time points were recorded for a reaction time of up to 29 h. A series of two-dimensional assignment spectra was acquired to identify the residual reaction intermediates and products on the postreaction material sample. 1H-1H TOCSY NMR spectra were acquired sampling the FID for 128 and 32 ms by acquiring 1,024 × 256 complex data points in the direct and indirect dimension, respectively, and using an excitation sculpting scheme for water suppression. 1H-1H COSY spectra were recorded by sampling the FID for 160 and 40 ms by acquiring 1,024 × 256 complex data points in the direct and indirect dimension, respectively, and employing presaturation of water signal suppression. 1H-13C HSQC spectra were recorded by sampling the FID for 128 and 8 ms by acquiring 1,024 × 200 complex data points, and 1H-13C HMBC spectra (with a spectral width of 220 ppm around an offset of 110 ppm in the 13C dimension) were recorded by sampling the FID for 197 and 4.5 ms by acquiring 2,048 × 200 complex data points. All NMR spectra were processed with ample zero filling in all spectral dimensions in Bruker Topspin 3.5 pl7 software and integrated in the same software.
Culture conditions of fungal fermentations.
All actively growing fungi were maintained on potato dextrose agar at room temperature. B. cinerea cell walls were prepared by inoculating 200 ml of potato dextrose medium with spore suspension and grown in an orbital shaker at 160 rpm, at 25°C for 7 days. The culture was spun down, washed twice in sterile water, lyophilized and powdered. Matured A. bisporus fruiting bodies (stem and cap) were washed, lyophilized and powdered. Both of the fungal cell wall preparations were added to the base medium at a final concentration of 2% wt/vol before autoclaved at 121°C for 20 min. The glucuronan and glucose were filter sterilized before added to the base medium at a final concentration of 0.1% wt/vol. The base medium contained 0.1% (wt/vol) tryptone (Bacto, NSW, Australia), 0.03% (wt/vol) urea, 0.2% (wt/vol) KH2PO4, 0.14% (wt/vol) (NH4)2SO4, 0.03% (wt/vol) MgSO4·7H2O, 0.06% Tween 20 and 2% (vol/vol) trace elements solution containing 0.025% (wt/vol) FeSO4, 0.0085% (wt/vol) MnSO4, 0.007% (wt/vol) ZnSO4 and 0.01% (wt/vol) CaCl2. T. parareesei spore suspension was added at a final concentration of 105 spores/ml to 250 ml baffled flask containing 100 ml of Yeast Peptone, Dextrose (YPD) medium, sealed with AeraSeal film and grown for 4 days, at 120 rpm, at 28°C. The produced mycelium was then spun down at 4400 rpm for 5 min, washed in 50 ml of sterile water and then transferred to the base medium containing the four different carbon sources each in triplicates. These cultures were grown for 24 h, at 120 rpm and 28°C before being spun down and the supernatant removed for assaying and proteomic analysis. All fermentations were routinely checked for contamination by microscopy.
Total protein content was assessed by Pierce BCA assay (Thermo Scientific, MA, USA) using bovine serum albumin as standard. Glucuronan activity was assessed under standard assay conditions with 100 mM NaCl and 4 mM CaCl2.
Global proteomic analysis of fungal secretomes.
The total protein content in 10 ml of the fungal supernatants was precipitated by adding 40 ml of ice cold acetone (Sigma) and stored overnight at −20°C. The protein pellet was centrifuged at 10,000 g for 10 min and residual acetone was evaporated at room temperature. The protein pellet was resolubilized in 6 M Guanidinium hydrochloride and then diluted in digestion buffer, consisting of 10% Acetonitrile (Sigma) and 50 mM HEPES buffer pH 8.5 (Sigma). Ten micrograms of protein from each sample was reduced, alkylated, in-solution digested with LysC (Wako, Osaka, Japan) and trypsin (Sigma) and desalted on C18 filters (CDS Empore, Oxford, USA) before being concentrated in an Eppendorf Speedvac and resolubilized in a solution containing 2% acetonitrile, 1% trifluoroacetic acid and iRT peptides (Biognosys, Schlieren, Schweiz). One microgram from each sample were loaded onto a 2 cm C18 trap column (ThermoFisher), connected in-line to a 15 cm C18 reverse-phase analytical column (Thermo EasySpray) using 100% Buffer A (0.1% Formic acid in water) at 750 bar in a Thermo EasyLC 1200 HPLC system (ThermoFisher). Peptides were eluted over a 140 min gradient ranging from 10% to 60% of 80% acetonitrile, 0.1% formic acid at 250 nL/min, and a Q-Exactive instrument (Thermo Scientific) was run in a DD-MS2 top 10 method. Full MS spectra were collected at a resolution of 70,000, with an automatic gain control target of 3 × 106 or maximum injection time of 20 ms and a scan range of 300 to 1750 m/z. The MS2 spectra were obtained at a resolution of 17,500, with an automatic gain control target value of 1 × 106 or maximum injection time of 60 ms, a normalized collision energy of 25, and an intensity threshold of 1.7 × 104. Dynamic exclusion was set to 60 s, and ions with a charge state <2 or unknown were excluded. The raw files were analyzed using Proteome Discoverer 2.4. Label-free quantitation (LFQ) was enabled in the processing and consensus steps, and spectra were matched against the predicted proteins from the T. parareesei CBS125925 genome (GCA_001050175.1). Carbamidomethylation of cysteines was defined as fixed modification and oxidation of methionines, deamidation and N-terminal acetylation as variable modifications. The remaining settings were kept on default, including a maximum peptide and protein false discovery rate of 1%. The LFQ intensities for each protein hit present in at least two of the biological replicates, were converted to a percentage of the sum of all LFQ values in the given replicate. Significant differences of abundance were estimated by performing one-way ANOVA post hoc Tukey comparison of means in Origin 2019. Mass spectrometry analysis was performed at the DTU Proteomics Core, Technical University of Denmark.
Carbohydrate monomer composition of fungal cell walls.
Glucuronan and lyophilized fungal cell walls were subjected to a two-step sulfuric acid hydrolysis as described in the NREL method (59). Monosaccharides were quantified by high performance anion-exchange chromatography with pulsed amperometric detection using a Dionex ICS-5000 system (DionexCorp, Sunnyvale, CA, USA) equipped with a CarboPac PA1 analytical column (250 × 4 mm) and a CarboPac PA1 guard column (250 × 4 mm) as described previously (60) with elution at a flow rate of 1 ml/min with water for 38 min, a wash with 500 mM NaOH for 15 min and equilibration with water for 22 min. Detection was done by postcolumn addition of 500 mM NaOH at 0.2 ml/min (60). Standards of fucose, arabinose, rhamnose, galactose, glucose, xylose, mannose, galacturonic acid, and glucuronic acid were used for quantification.
Data availability.
All data supporting the findings of this study are available within the paper (and its supplemental material). All relevant data generated during this study or analyzed in this published article (and its supplemental material) are available from the corresponding author upon reasonable request. All sequences used are available in the NCBI database under the accession numbers listed in this study.
ACKNOWLEDGMENT
B.P. conceived the study. B.P. conducted the majority of the experiments assisted by M.V., L.M., J.H., S.M. and C.W. Data analysis was performed by B.P. with contributions by C.W. and A.M. regarding data interpretation, and A.M. and C.W. also supervised the work. The paper was written by B.P., C.W., and A.M. All authors reviewed the manuscript.
This work was funded by the Department of Biotechnology and Biomedicine (DTU Bio-engineering), Enzyme Discovery & Engineering Program, Technical University of Denmark.
All authors declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Casper Wilkens, Email: cwil@dtu.dk.
Anne S. Meyer, Email: asme@dtu.dk.
Irina S. Druzhinina, Nanjing Agricultural University
REFERENCES
- 1.Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. 2011. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol 9:749–759. 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 2.Naher L, Syawani N, Amieza N, Kamarudin AB, Karim SMR. 2019. Trichoderma species diversity in rhizosphere soils and potential antagonism with Fusarium oxysporum. Biosci J 35:13–26. 10.14393/BJ-v35n1a2019-41605. [DOI] [Google Scholar]
- 3.Druzhinina IS, Komoń-Zelazowska M, Atanasova L, Seidl V, Kubicek CP. 2010. Evolution and ecophysiology of the industrial producer Hypocrea jecorina (anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PLoS One 5:e9191. 10.1371/journal.pone.0009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanasova L, Jaklitsch WM, Komoń-Zelazowska M, Kubicek CP, Druzhinina IS. 2010. Clonal species Trichoderma parareesei sp. nov. likely resembles the ancestor of the cellulase producer Hypocrea jecorina/T. reesei. Appl Environ Microbiol 76:7259–7267. 10.1128/AEM.01184-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson M, Atanasova L, Jensen DF, Zeilinger S. 2017. Necrotrophic mycoparasites and their genomes. Microbiol Spectr 5. 10.1128/microbiolspec.FUNK-0016-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Döhren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gómez-Rodríguez EY, Gruber S, Han C, Henrissat B, Hermosa R, Hernández-Oñate M, Karaffa L, Kosti I, Le Crom S, Lindquist E, Lucas S, Lübeck M, Lübeck PS, Margeot A, Metz B, Misra M, Nevalainen H, Omann M, Packer N, Perrone G, Uresti-Rivera EE, Salamov A, Schmoll M, Seiboth B, et al. 2011. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol 12:R40. 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atanasova L, Dubey M, Grujić M, Gudmundsson M, Lorenz C, Sandgren M, Kubicek CP, Jensen DF, Karlsson M. 2018. Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol 18:178. 10.1186/s12866-018-1310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yip VL, Withers SG. 2006. Breakdown of oligosaccharides by the process of elimination. Curr Opin Chem Biol 10:147–155. 10.1016/j.cbpa.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delattre C, Michaud P, Keller C, Elboutachfaiti R, Beven L, Courtois B, Courtois J. 2006. Purification and characterization of a novel glucuronan lyase from Trichoderma sp. GL2. Appl Microbiol Biotechnol 70:437–443. 10.1007/s00253-005-0077-8. [DOI] [PubMed] [Google Scholar]
- 11.Konno N, Igarashi K, Habu N, Samejima M, Isogai A. 2009. Cloning of the Trichoderma reesei cDNA encoding a glucuronan lyase belonging to a novel polysaccharide lyase family. Appl Environ Microbiol 75:101–107. 10.1128/AEM.01749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konno N, Ishida T, Igarashi K, Fushinobu S, Habu N, Samejima M, Isogai A. 2009. Crystal structure of polysaccharide lyase family 20 endo-β-1,4-glucuronan lyase from the filamentous fungus Trichoderma reesei. FEBS Lett 583:1323–1326. 10.1016/j.febslet.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 13.Redouan E, Cedric D, Emmanuel P, Mohamed EG, Bernard C, Philippe M, Cherkaoui EM, Josiane C. 2009. Improved isolation of glucuronan from algae and the production of glucuronic acid oligosaccharides using a glucuronan lyase. Carbohydr Res 344:1670–1675. 10.1016/j.carres.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 14.Dow JM, Darnall DW, Villa VD. 1983. Two distinct classes of polyuronide from the cell walls of a dimorphic fungus, Mucor rouxii. J Bacteriol 155:1088–1093. 10.1128/jb.155.3.1088-1093.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ruiter GA, Josso SL, Colquhoun IJ, Voragen AGJ, Rombouts FM. 1992. Isolation and characterization of β(1–4)-d-glucuronans from extracellular polysaccharides of moulds belonging to Mucorales. Carb Polym 18:1–7. 10.1016/0144-8617(92)90181-O. [DOI] [Google Scholar]
- 16.Heyraud A, Courtois J, Dantas L, Colin-Morel P, Courtois B. 1993. Structural characterization and rheological properties of an extracellular glucuronan produced by a Rhizobium meliloti M5N1 mutant strain. Carbohydr Res 240:71–78. 10.1016/0008-6215(93)84172-3. [DOI] [PubMed] [Google Scholar]
- 17.Helbert W, Poulet L, Drouillard S, Mathieu S, Loiodice M, Couturier M, Lombard V, Terrapon N, Turchetto J, Vincentelli R, Henrissat B. 2019. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci USA 116:6063–6068. 10.1073/pnas.1815791116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilgaard B, Vuillemin M, Holck J, Wilkens C, Meyer AS. 2021. Specificities and synergistic actions of novel PL8 and PL7 alginate lyases from the marine fungus Paradendryphiella salina. J Fungi 7:80. 10.3390/jof7020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hof DJ, Versteeg EMM, van de Lest CHA, Daamen WF, van Kuppevelt TH. 2019. A versatile salt-based method to immobilize glycosaminoglycans and create growth factor gradients. Glycoconj J 36:227–236. 10.1007/s10719-019-09872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsubara Y, Iwasaki K, Muramatsu T. 1998. Action of poly (α-L-guluronate) lyase from Corynebacterium sp. ALY-1 strain on saturated oligoguluronates. Biosci Biotechnol Biochem 62:1055–1060. 10.1271/bbb.62.1055. [DOI] [PubMed] [Google Scholar]
- 21.Zeuner B, Thomsen TB, Stringer MA, Krogh KBRM, Meyer AS, Holck J. 2020. Comparative characterization of Aspergillus pectin lyases by discriminative substrate degradation profiling. Front Bioeng Biotechnol 8:873. 10.3389/fbioe.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaya D, Hahn B-S, Park NY, Sim J-S, Kim YS, Cygler M. 2008. Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926. Biochemistry 47:6650–6661. 10.1021/bi800353g. [DOI] [PubMed] [Google Scholar]
- 23.Hobbs JK, Lee SM, Robb M, Hof F, Barr C, Abe KT, Hehemann J-H, McLean R, Abbott DW, Boraston AB. 2016. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc Natl Acad Sci USA 113:6188–6193. 10.1073/pnas.1524214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Aarstad OA, Li J, McKee LS, Sætrom GI, Vyas A, Srivastava V, Aachmann FL, Bulone V, Hsieh YS. 2018. Preparation of 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH) and guluronic acid rich alginate using a unique exo-alginate lyase from Thalassotalea crassostreae. J Agric Food Chem 66:1435–1443. 10.1021/acs.jafc.7b05751. [DOI] [PubMed] [Google Scholar]
- 25.Konno N, Habu N, Maeda I, Azuma N, Isogai A. 2006. Cellouronate (β-1,4-linked polyglucuronate) lyase from Brevundimonas sp. SH203: Purification and characterization. Carb Polym 64:589–596. 10.1016/j.carbpol.2005.11.015. [DOI] [Google Scholar]
- 26.Stergiopoulos I, de Wit PJGM. 2009. Fungal effector proteins. Annu Rev Phytopathol 47:233–263. 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 27.Barrett K, Lange L. 2019. Peptide-based functional annotation of carbohydrate-active enzymes by conserved unique peptide patterns (CUPP). Biotechnol Biofuels 12:102. 10.1186/s13068-019-1436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busk PK, Pilgaard B, Lezyk MJ, Meyer AS, Lange L. 2017. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics 18:1–9. 10.1186/s12859-017-1625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi M, Konno N, Suzuki T, Fujii Y, Kodama Y, Isogai A, Habu N. 2020. A bacterial endo-β-1,4-glucuronan lyase, CUL-I from Brevundimonas sp. SH203, belonging to a novel polysaccharide lyase family. Protein Expr Purif 166:105502. 10.1016/j.pep.2019.105502. [DOI] [PubMed] [Google Scholar]
- 30.Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O. 2002. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94:146–170. 10.2307/3761854. [DOI] [PubMed] [Google Scholar]
- 31.Gray DJ, Morgan-Jones G. 1981. Host-parasite relationships of Agaricus brunnescens and a number of mycoparasitic hyphomycetes. Mycopathologia 75:55–59. 10.1007/BF00439068. [DOI] [Google Scholar]
- 32.Kubicek CP, Steindorff AS, Chenthamara K, Manganiello G, Henrissat B, Zhang J, Cai F, Kopchinskiy AG, Kubicek EM, Kuo A, Baroncelli R, Sarrocco S, Noronha EF, Vannacci G, Shen Q, Grigoriev IV, Druzhinina IS. 2019. Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20:485. 10.1186/s12864-019-5680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson M, Durling MB, Choi J, Kosawang C, Lackner G, Tzelepis GD, Nygren K, Dubey MK, Kamou N, Levasseur A, Zapparata A, Wang J, Amby DB, Jensen B, Sarrocco S, Panteris E, Lagopodi AL, Pöggeler S, Vannacci G, Collinge DB, Hoffmeister D, Henrissat B, Lee Y-H, Jensen DF. 2015. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol Evol 7:465–480. 10.1093/gbe/evu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammami W, Castro CQ, Rémus-Borel W, Labbé C, Bélanger RR. 2011. Ecological Basis of the interaction between Pseudozyma flocculosa and powdery mildew fungi. Appl Environ Microbiol 77:926–933. 10.1128/AEM.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latgé J-P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66:279–290. 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuivanen J, Richard P. 2018. NADPH-dependent 5-keto-D-gluconate reductase is a part of the fungal pathway for D-glucuronate catabolism. FEBS Lett 592:71–77. 10.1002/1873-3468.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerkenaar A, Besemer AC. 1991. Polyglucuronic acid, process for preparing polyglucuronic acid and use of polyglucuronic acid. European patent WO1991004988A1. https://patents.google.com/patent/WO1991004988A1/en.
- 38.Caillot S, Rat S, Tavernier M-L, Michaud P, Kovensky J, Wadouachi A, Clément C, Baillieul F, Petit E. 2012. Native and sulfated oligoglucuronans as elicitors of defence-related responses inducing protection against Botrytis cinerea of Vitis vinifera. Carb Polym 87:1728–1736. 10.1016/j.carbpol.2011.09.084. [DOI] [Google Scholar]
- 39.Abouraïcha EF, El Alaoui-Talibi Z, Tadlaoui-Ouafi A, El Boutachfaiti R, Petit E, Douira A, Courtois B, Courtois J, El Modafar C. 2017. Glucuronan and oligoglucuronans isolated from green algae activate natural defense responses in apple fruit and reduce postharvest blue and gray mold decay. J Appl Phycol 29:471–480. 10.1007/s10811-016-0926-0. [DOI] [Google Scholar]
- 40.Elboutachfaiti R, Delattre C, Petit E, Michaud P. 2011. Polyglucuronic acids: Structures, functions and degrading enzymes. Carb Polym 84:1–13. 10.1016/j.carbpol.2010.10.063. [DOI] [Google Scholar]
- 41.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res 44:D67–D72. 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoff KJ, Stanke M. 2013. WebAUGUSTUS—a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res 41:W123–W128. 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction: Tthe Phobius web server. Nucleic Acids Res 35:W429–W432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeske L, Placzek S, Schomburg I, Chang A, Schomburg D. 2019. BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res 47:D542–D549. 10.1093/nar/gky1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 49.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 51.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 52.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In 2010 Gateway Computing Environments Workshop. IEEE. 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- 53.Isogai A, Kato Y. 1998. Preparation of polyuronic acid from cellulose by TEMPO-mediated oxidation. Cellulose 5:153–164. 10.1023/A:1009208603673. [DOI] [Google Scholar]
- 54.Pilgaard B, Wilkens C, Herbst F-A, Vuillemin M, Rhein-Knudsen N, Meyer AS, Lange L. 2019. Proteomic enzyme analysis of the marine fungus Paradendryphiella salina reveals alginate lyase as a minimal adaptation strategy for brown algae degradation. Sci Rep 9:1–13. 10.1038/s41598-019-48823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy Server, p 571–607. In Walker JM (ed), The Proteomics Protocols Handbook. Humana Press. [Google Scholar]
- 56.Brooke D, Movahed N, Bothner B. 2015. Universal buffers for use in biochemistry and biophysical experiments. Biophysics 2:336–342. 10.3934/biophy.2015.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lever M. 1977. Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal Biochem 81:21–27. 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
- 58.Thomassen LV, Vigsnæs LK, Licht TR, Mikkelsen JD, Meyer AS. 2011. Maximal release of highly bifidogenic soluble dietary fibers from industrial potato pulp by minimal enzymatic treatment. Appl Microbiol Biotechnol 90:873–884. 10.1007/s00253-011-3092-y. [DOI] [PubMed] [Google Scholar]
- 59.Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker DP. 2012. Determination of structural carbohydrates and lignin in biomass: Laboratory Analytical Procedure (Technical Report TP-510–42618). NREL, Golden, CO. [Google Scholar]
- 60.Zeuner B, Muschiol J, Holck J, Lezyk M, Gedde MR, Jers C, Mikkelsen JD, Meyer AS. 2018. Substrate specificity and transfucosylation activity of GH29 α-L-fucosidases for enzymatic production of human milk oligosaccharides. N Biotechnol 41:34–45. 10.1016/j.nbt.2017.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S10, Tables S1 to S3. Download AEM.01819-21-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
All data supporting the findings of this study are available within the paper (and its supplemental material). All relevant data generated during this study or analyzed in this published article (and its supplemental material) are available from the corresponding author upon reasonable request. All sequences used are available in the NCBI database under the accession numbers listed in this study.