Abstract
Aims
Vertebrate heart development requires the complex morphogenesis of a linear tube to form the mature organ, a process essential for correct cardiac form and function, requiring coordination of embryonic laterality, cardiac growth, and regionalized cellular changes. While previous studies have demonstrated broad requirements for extracellular matrix (ECM) components in cardiac morphogenesis, we hypothesized that ECM regionalization may fine tune cardiac shape during heart development.
Methods and results
Using live in vivo light sheet imaging of zebrafish embryos, we describe a left-sided expansion of the ECM between the myocardium and endocardium prior to the onset of heart looping and chamber ballooning. Analysis using an ECM sensor revealed the cardiac ECM is further regionalized along the atrioventricular axis. Spatial transcriptomic analysis of gene expression in the heart tube identified candidate genes that may drive ECM expansion. This approach identified regionalized expression of hapln1a, encoding an ECM cross-linking protein. Validation of transcriptomic data by in situ hybridization confirmed regionalized hapln1a expression in the heart, with highest levels of expression in the future atrium and on the left side of the tube, overlapping with the observed ECM expansion. Analysis of CRISPR-Cas9-generated hapln1a mutants revealed a reduction in atrial size and reduced chamber ballooning. Loss-of-function analysis demonstrated that ECM expansion is dependent upon Hapln1a, together supporting a role for Hapln1a in regionalized ECM modulation and cardiac morphogenesis. Analysis of hapln1a expression in zebrafish mutants with randomized or absent embryonic left–right asymmetry revealed that laterality cues position hapln1a-expressing cells asymmetrically in the left side of the heart tube.
Conclusion
We identify a regionalized ECM expansion in the heart tube which promotes correct heart development, and propose a novel model whereby embryonic laterality cues orient the axis of ECM asymmetry in the heart, suggesting these two pathways interact to promote robust cardiac morphogenesis.
Keywords: Heart morphogenesis, Extracellular matrix, Laterality, Zebrafish, Heart development
Graphical Abstract
1. Introduction
Congenital heart defects are the most common human birth abnormality, with an incidence of approximately 1% of live births.1 These structural malformations arise due to abnormal morphogenesis and maturation of the heart during embryonic development. A key stage in cardiac development occurs when the heart transitions from a linear tube to an asymmetric organ, a process including initial looping morphogenesis of the tube and subsequent ballooning of the cardiac chambers. Correct cardiac morphogenesis is vital for ensuring normal blood flow through the heart, proper chamber and vessel alignment, valve formation and septation, and is therefore a tightly controlled process requiring intricate coordination of heart-extrinsic signalling cues, cardiac growth, and tissue-intrinsic changes in cell shape.2
The requirement for embryonic left–right signalling pathways in promoting directionality of heart morphogenesis is well established, with asymmetric Nodal signalling playing a key role in driving rightward looping of the linear heart tube in multiple organisms.3–6 However, while embryos with defective asymmetric Nodal signalling display disrupted directionality of heart looping, the heart still undergoes looping morphogenesis.5–7 This indicates that while extrinsic asymmetric cues provide directional information to the heart, regionalized intrinsic signals may help to promote morphogenesis. How the interplay of both extrinsic and intrinsic regionalized signalling and cell behaviours ensures the coordination of directionality and morphogenesis required to orient and shape the heart remains unknown.
The developing heart tube is composed of two tissue layers: an outer tube of myocardium surrounding an inner layer of specialized endothelial cells (endocardium). These two layers are separated by an extracellular matrix (ECM), termed the cardiac jelly. The ECM consists of collagens, glycosaminoglycans, and glycoproteins and plays a pivotal role in providing mechanical cues and modulating extracellular signalling in the heart during cardiac development.8 Classic embryological experiments demonstrated that the cardiac jelly is important for heart morphogenesis9 while more recent studies have begun to identify specific ECM constituents with distinct roles in heart development.10–16 Hyaluronic acid (HA) is a glycosaminoglycan with conserved roles in heart tube formation, cardiac morphogenesis and atrioventricular valve development,10,17,18 suggesting multiple requirements for HA at various stages during cardiac development. While broad disruption of the cardiac ECM has profound effects on heart morphology,17,19 it is likely that the ECM plays distinct functions in regulating regionalized morphogenesis of the heart tube.
In this study, we demonstrate that the cardiac ECM of the zebrafish heart tube exhibits regionalized expansion prior to onset of heart tube morphogenesis, with a thicker ECM in both the left side and future atrium of the heart tube. Loss-of-function analyses demonstrate that this ECM expansion is dependent upon the ECM cross-linking protein Hyaluronan and Proteoglycan Link Protein 1a (Hapln1a), and that Hapln1a promotes heart morphogenesis. Finally, we show that while asymmetric hapln1a expression is independent of laterality cues, the axis of hapln1a asymmetry in the heart is dictated by embryonic laterality, supporting a new model where embryonic left–right asymmetry tightly defines the orientation of ECM asymmetry in the heart tube, and together these pathways fine tune asymmetric cardiac morphogenesis.
2. Methods
2.1 Zebrafish maintenance
Adult zebrafish were maintained according to standard laboratory conditions. The following lines were used: AB, Tg(myl7:eGFP),20 Tg(myl7:lifeActGFP),21 Tg(fli1a:AC-TagRFP),sh511 22 spawt30973 7, Tg(lft2BAC:Gal4FF); Tg(UAS;RFP), pkd2hu2173, hapln1aΔ187 (allele designation hapln1ash611), hapln1aΔ241 (allele designation hapln1ash580). Embryos older than 24hpf were treated with 0.2 mM 1-phenyl-2-thiourea (PTU) in E3 medium to inhibit melanin production. All animals were euthanized by immersion in overdose of Tricaine methanesulfonate (1.33 g/L). Animal work was approved by the local Animal Welfare and Ethical Review Body (AWERB) at the University of Sheffield, conducted in accordance with UK Home Office Regulations under PPLs 70/8588 and PA1C7120E, and in line with the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
2.2 Generation of hapln1a mutants
CRISPR guide RNAs (gRNAs) were designed to target the putative promoter region of hapln1a (GRCz11: ENSDART00000122966.4, g1: 5′-TCGTCTCTCTCTAAGGGGAGGGG-3′) and downstream of the translation start site (g2: 5′-GATGATTGCTCTGTTTTCTGTGG-3′). Sequence-specific CRISPR RNAs (crRNA, Merck) were resuspended in MilliQ water to 21.4 μM, and injected with equimolar tracrRNA (Merck) together with Cas9 protein (NEB M0386T) into 1-cell stage embryos in a volume of 1nl. CRISPR-Cas9-injected embryos were raised to adulthood and individuals transmitting germline promoter deletions were identified by PCR using the following primers: forward 5′-ACATTTTGCATGCCCTCGAA-3′; reverse 5′-TGCATCCTGGACCTTCATTCA-3′. Promoter deletions were identified by Sanger sequencing. F0 founders transmitting a desirable mutation were established as stable lines at F2. Two hapln1a promoter deletion alleles were recovered: hapln1aΔ187 and hapln1aΔ241.
2.3 mRNA in situ hybridization
Embryos were fixed overnight in 4% PFA, and mRNA in situ hybridizations carried out as previously described.7 Fluorescent in situ hybridizations were performed using the TSA kit (Perkin-Elmer).23 Primers used to generate new mRNA in situ probe constructs and information on published probes are detailed in Supplementary material online, Methods. Riboprobes were transcribed from linearized template in the presence of DIG-11-UTP or Fluorescein-11-UTP (Roche).
2.4 Immunohistochemistry
Embryos were fixed overnight in 4% PFA with 4% sucrose at 4°C. For Hapln1a immunostaining samples were stored overnight in MeOH at −20°C. After rehydration if applicable, embryos were blocked for 1 h at room temperature in 0.2% PBS-Triton-X (PBS-Tx) with 10% goat serum. Embryos were incubated overnight at 4°C with primary and secondary antibodies diluted in PBS-Tx/10% goat serum with 1% DMSO. The following commercially available primary antibodies were used: αGFP (1:1000 Aves lab), αCT3 (1:100, Developmental Studies Hybridoma Bank), αCdh524 (1:100). A rabbit polyclonal antibody targeting amino acids 117–134 (DGMNDMTLEVDLEVQGKD) of zebrafish Hapln1a was designed and produced by Proteintech. Test bleeds were used to determine cross-reactivity with Hapln1a by comparing protein localization at 26hpf with mRNA in situ hybridization. Subsequently, affinity-purified Hapln1a antibody was used at 1:100. Fluorophore-conjugated secondary antibodies (Jackson labs) were used at 1:200.
2.5 Tomo-seq
Hearts were dissected from Tg(myl7: eGFP) zebrafish embryos at 26hpf and placed into OCT cryofreezing medium (Sakura Finetek). Blue Affy-gel beads (BioRad) were placed at each end of the heart tube to aid visualization during sectioning, and the hearts were rapidly frozen and stored at −80°C. Hearts were sectioned using a cryostat at 9 nm resolution. RNA extraction, aRNA synthesis, library preparation, sequencing, and data analysis were performed as previously described.25,26
2.6 Morpholino-mediated knockdown and hapln1a overexpression construct generation and analysis
A morpholino was designed to target the translational start site of hapln1a (5′-AGAGCAAT[CAT]CTTCACGTTTGTTA-3′, brackets denote hapln1a ATG reverse complement). Morpholinos blocking tp53,27 (Zfin tp53 MO-4) and has228 (Zfin has2 MO-1) are previously described. All morpholinos were supplied by GeneTools and diluted to a 1 mM stock. Working concentrations were as follows: hapln1a 500 nM or 250 nM, has2 250 nM, combinatorial has2/hapln1a 250 nM each, tp53 250 nM. has2 and hapln1a morpholinos were co-injected together with tp53 morpholino. Embryos were injected with 1 nL of morpholino solution.
The hapln1a coding sequence was amplified using the following primers containing AttB sequences for Gateway cloning and a Kozak sequence (underlined): forward: 5′ggggacaagtttgtacaaaaaagcaggctTCGCCGCCACCATGATTGCTCTGTTTTCTGT 3′; reverse: 5′GGGGACCACTTTGTACAAGAAAGCTGGGTTTTACTGCTGGGCTTTGTAGCAATA-3′. The resulting PCR product was ligated into the pDONR221 middle entry Gateway vector, generating a pMEhapln1aCDS vector. Full-length hapln1a was subsequently recombined with a p5E myl7 promoter sequence,29 and a p3E polyA sequence into the pDestTol2pA3 destination vector30 to generate the pDestmyl7: hapln1a construct. Gateway cloning was performed using the Tol2kit via standard protocols.30 60 pg of pDestmyl7: hapln1a was co-injected with 25 pg of tol2 mRNA into the cell of 1-cell stage embryos. Analysis of hapln1a overexpression and cardiac morphogenesis was performed using double in situ hybridization to assess hapln1a and myl7 expression.
2.7 RNA injections
ssNcan-GFP mRNA was synthesized from the ssNcan-GFP plasmid as previously described.31 Embryos were injected with 100 pg of mRNA in 1 nL volume at the 1-cell stage and screened for GFP at 24hpf.
2.8 Imaging and image quantification
Live zebrafish embryos were imaged on a ZEISS Lightsheet Z.1 microscope. To assess cardiac morphology at 50hpf and 72hpf embryos were anesthetized by immersion in 8.4% Tricaine (Merck 10521) before mounting in 1% low melting point agarose in E3 with 8.4% Tricaine. To stop the heart the imaging chamber was filled with E3 with 8.4% Tricaine and the temperature maintained at 10°C. All samples were imaged using a 20× lens and 1.0 zoom at 0.47–0.65 µm z-step size, with sufficient z slices to capture the entire heart. Dual side lasers with dual side fusion and pivot scan were used for sample illumination.
Embryos injected with ssNcan-GFP mRNA were fixed overnight in 4% PFA with 4% sucrose, and the GFP signal amplified by immunohistochemistry. Dissected embryos were imaged using a Zeiss Airyscan microscope, z stacks were obtained with a step size of 1 µm.
Detailed image quantification methodology is included in Supplementary material online.
3. Results
3.1 The cardiac ECM is asymmetrically expanded at early stages of heart looping morphogenesis
During cardiac development the myocardial and endocardial layers of the heart are separated by the cardiac ECM. We hypothesized there may be regional differences in the ECM of the zebrafish heart tube which could promote local changes in tissue shape to drive cardiac morphogenesis. To examine regional ECM thickness in the heart tube, we used live in vivo light-sheet microscopy to image quadruple transgenic zebrafish embryos at 26 h post-fertilization (hpf).
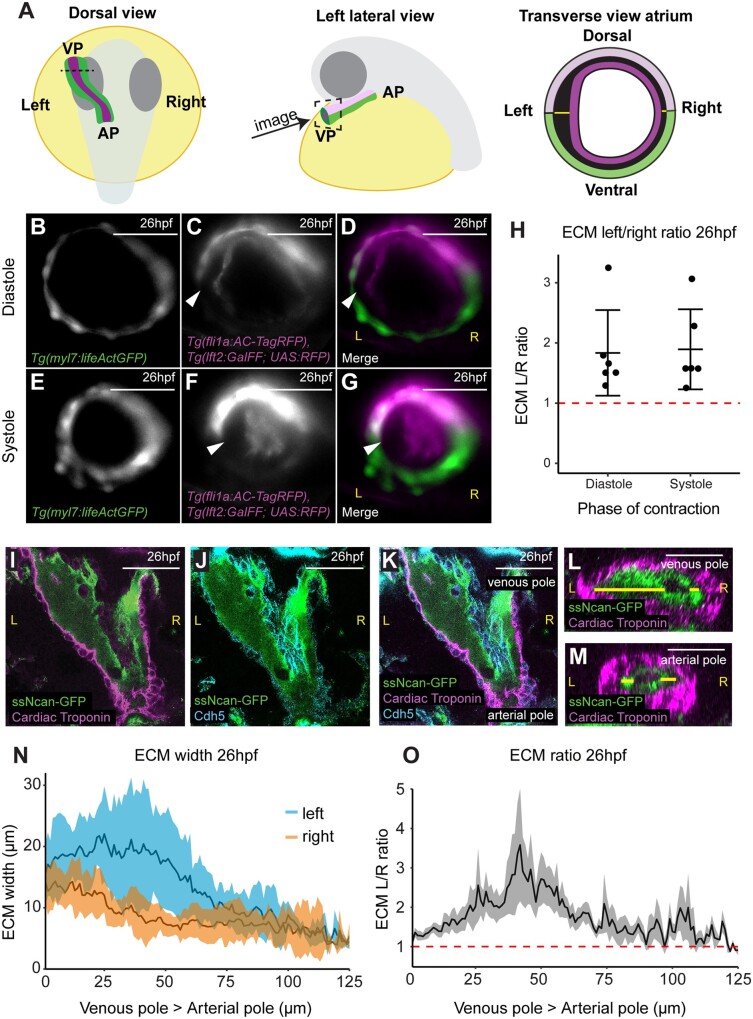
Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP); Tg(lft2BAC:Gal4FF); Tg(UAS:RFP) zebrafish express actin-tagged GFP in the myocardium21 and actin-localized RFP in the endothelium including the endocardium,22 allowing visualization of the two tissue layers in the heart tube. The Tg(lft2BAC:Gal4FF); Tg(UAS:RFP) double transgenic drives RFP in lefty2-expressing cells, comprising the dorsal myocardium of the heart tube at 26hpf (Figure 1A and Supplementary material online, Figure S1).18,32 This combination of transgenes allowed imaging of optical cross-sections through the heart tube at 26hpf, just before the onset of looping morphogenesis, and enabled dorsal–ventral axis orientation of the heart tube (Figure 1A–G and Supplementary material online, Movie S1). We consistently observed an asymmetry in the extracellular space between the myocardial and endocardial layers of the heart in the future atrium, with an apparent thickening of the ECM on the left side of the tube which is maintained throughout the cardiac cycle (Figure 1D and G). We quantified the extracellular space between the two tissue layers and calculated the ECM left: right ratio (left ECM thickness divided by right ECM thickness, where >1 indicates a left sided expansion). Using this method, we detected a reproducible expansion of the ECM in the left side of the heart tube (Figure 1H) which is maintained in the atrium at 50hpf (Supplementary material online, Figure S2).
Figure 1.
The hyaluronan-rich ECM is asymmetric during early zebrafish heart development. (A) Schematic depicting the developmental stage and orientation of zebrafish embryos used in live imaging experiments. Optical transverse sections of the heart tube are imaged at the position of the dotted line/dotted square. Green, myocardium; magenta, endocardium; light pink, dorsal myocardium. (B–G) Light-sheet optical cross-sections through the heart tube of a 26hpf Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP); Tg(lft2BAC:Gal4FF); Tg(UAS:RFP) transgenic embryo during diastole (B–D) and systole (E–G) at the level of the dotted line in (A). The myocardium is marked in green (B, D, E, and G), and the dorsal myocardium and endocardium are marked in magenta (C, D, F, and G). The extracellular space between myocardium and endocardium is expanded on the left side of the heart tube (white arrowhead). Scale bar = 50 μm. (H) Quantification of left–right ECM ratio in heart tubes, >1 (red dotted line) denotes left-sided expansion. Mean ± SD are plotted, n = 6. (I–K) Single confocal z-planes longitudinally through the heart at 26hpf of embryos injected with ssNcan-GFP (green), counterstained with cardiac troponin (magenta, I, K) and VE-Cadherin (cyan J, K). (L and M) Transverse optical reslice through the heart tube the venous pole (L) or arterial pole (M). ECM width is measured using the ssNcan-GFP signal (yellow line) on left and right sides of the tube. (N) Quantification of ECM width on the left (blue) and right (orange) sides of the heart tube from venous pole to arterial pole at 26hpf. Mean ± SD are plotted, n = 7. (O) Left–right ECM ratio in the heart tube from venous pole to arterial pole, where >1 (red dotted line) indicates a left-sided expansion. Mean ± SEM are plotted, n = 7. The mean L/R ratio across the heart is 1.667, and analysis using a one-sample t-test shows this significantly differs from 1, P < 0.0001 L, left; R, right; VP, venous pole; AP, arterial pole. Scale bar = 50μm.
Due to technical limitations in imaging deeper cardiac tissue with sufficient resolution at 26hpf, we could only image the superficially located venous pole/atrium of the heart tube in live embryos. Therefore, to determine whether ECM left–right asymmetry is restricted to the venous pole or is maintained along the atrioventricular axis of the heart, we performed fixed tissue imaging. Previous studies have demonstrated that hyaluronic acid (HA) is present in the cardiac jelly during vertebrate heart development.17,33,34 To visualize the HA-rich ECM, wild-type embryos were injected with the HA sensor ssNcan-GFP31 at the 1-cell stage, fixed at 26hpf, and the GFP signal detected by immunohistochemistry before imaging the entire heart tube as a z-stack using confocal microscopy (Figure 1I–K). Optical reslicing of z-stacks generated cross-sections of the heart tube from the venous pole to the arterial pole, allowing us to quantify the width of the ssNcan-GFP-positive ECM on left and right sides of the tube along the entire pole-to-pole length of the heart (Figure 1L–N). We confirmed that the ECM is thicker on the left side of the heart tube compared to the right, however this asymmetry is more profound at the venous pole/future atrium than at the arterial pole/future ventricle (Figure 1N and O). Furthermore, the cardiac ECM is thicker in the future atrium when compared to the ventricle (Figure 1L–N). Together these data demonstrate that the heart tube exhibits a regionally expanded ECM prior to onset of looping morphogenesis.
3.2 hapln1a exhibits regionalized cardiac expression prior to heart tube formation and looping morphogenesis
The asymmetric expansion of the cardiac ECM could be due to regionalized synthesis of ECM components. However, we did not observe any clear asymmetry in levels of HA deposition in the cardiac ECM in either live embryos injected with the ssNcan-GFP sensor (Supplementary material online, Figure S3) or in fixed hearts (Figure 1I–K). We also did not find left–right asymmetry in the heart tube in the expression of hyaluronan synthase 2 (has2, the major HA producing enzyme), chondroitin sulfate synthase 1 (chsy1), or the ECM proteoglycans versican (vcana/b), aggrecan (acana/b), all of which have previously been implicated in heart development11,17–19,35,36 (Supplementary material online, Figure S3), suggesting that regionalized synthesis of these proteins is not responsible for ECM asymmetry. We therefore hypothesized that a protein required for HA modification or cross-linking may be regionally expressed in the heart tube.
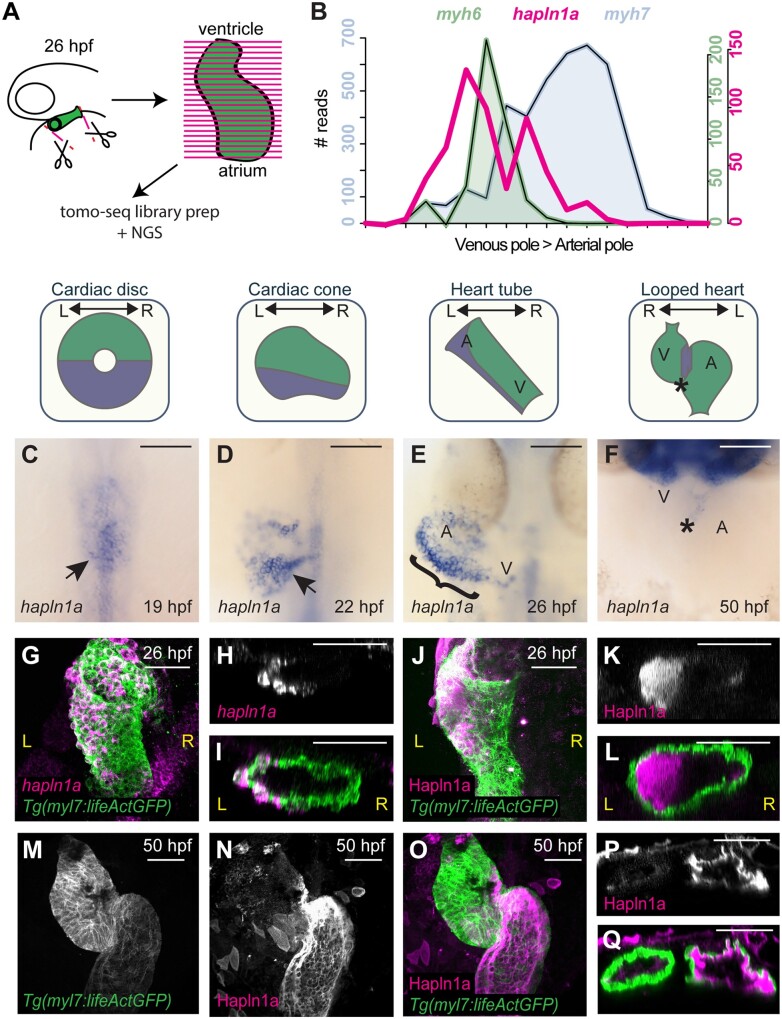
To identify candidate genes which modulate cardiac ECM expansion, we took a genome-wide unbiased approach to identify genes expressed in the heart tube at 26hpf, prior to the onset of looping morphogenesis. Since we observed the strongest left-sided ECM expansion in the putative atrium, as well as a generally more expanded ECM at the venous pole of the heart compared to the arterial pole, we used the previously described Tomo-seq technique to generate a regionalized map of gene expression from pole-to-pole in the heart tube25,26 (Figure 2A). We sectioned two individual hearts along the atrioventricular axis, identifying 6787 and 8916 expressed genes (Supplementary material online, Tables S2–S5), of which approximately half were expressed in more than one section. By identifying which sections express the atrial marker myh6 (myosin, heavy chain 6, cardiac muscle, alpha), we defined a subset of sections with atrial identity. We subsequently filtered genes that were up-regulated in atrial sections compared to ventricular sections in both hearts and examined this list for genes implicated in ECM modification.
Figure 2.
hapln1a is regionally expressed in the heart tube and secreted asymmetrically into the cardiac jelly. (A) Schematic representation of Tomo-seq pipeline. GFP-expressing hearts are manually excised from embryos at 26hpf, and frozen in tissue freezing medium. Heart tubes are sectioned along the atrioventricular axis. RNA extracted from individual slices is labelled with a slice-specific molecular barcode during reverse transcription before generating sequencing libraries. (B) Example Tomo-seq traces from a single 26hpf heart tube, with individual slices from venous pole to arterial pole along the x-axis and normalized read number along the y-axis. Read numbers for atrial marker myh6 (green) and ventricular marker myh7 (blue) allows identification of chamber position within the dataset. hapln1a expression (magenta) is up-regulated in atrial sections. (C–F) mRNA in situ hybridization analysis of hapln1a expression in the heart between 19hpf and 50hpf. At cardiac disc stage, hapln1a is up-regulated in the posterior (arrow C), which is maintained as the heart forms the cardiac cone prior to tube formation (arrow D), with lower expression in the anterior cone. Once the heart cone has extended to form the tube, the previously posterior hapln1a expression is positioned on the left side of the tube (bracket, E), and expressed at higher levels in the atrium (A) than the ventricle (V). By 50hpf hapln1a expression in the heart is restricted to low levels in the atrioventricular canal (AVC, asterisk, F). Schematics above in situ panels indicate heart morphology at each stage, and hapln1a expression domain within the heart (blue) V, Ventricle; A, Atrium. Scale bar = 100 μm. (G–I) Fluorescent in situ hybridization analysis of hapln1a (magenta) in Tg(myl7:lifeActGFP) transgenic embryos shows hapln1a is expressed in myocardial cells at 26hpf. (J–L) Fluorescent immunostaining of Hapln1a (magenta) in Tg(myl7:lifeActGFP) transgenic embryos demonstrating the protein is secreted into the extracellular space predominantly on the left side of the heart tube (magenta) at 26hpf. (G and J) Dorsal views. (H and I, K and L) Transverse views. (M–Q) Fluorescent immunostaining of Hapln1a (magenta) in Tg(myl7:lifeActGFP) transgenic embryos at 50hpf revealing Hapln1a is maintained in the cardiac ECM as looping progresses. (M–O) Ventral views. (P and Q) Transverse views. L, left; R, right, scale bar = 50 μm
Using this approach, we identified hyaluronan and proteoglycan link protein 1a (hapln1a, formerly crtl1) as a candidate to drive regionalized ECM expansion (Figure 2B). The Hapln family of proteins are secreted into the ECM where they cross-link HA to proteoglycans,37 suggesting Hapln1a may modify the cardiac ECM environment. mRNA in situ hybridization analysis revealed that hapln1a is expressed in the posterior of the heart disc and cardiac cone prior to formation of the linear heart tube (Figure 2C and D). At 26hpf, hapln1a expression is up-regulated on the left side of the heart tube with elevated levels of expression in the future atrium compared to the future ventricle, recapitulating the regionalized ECM expansion in the heart (compare Figures 2E and 1K). This dynamic hapln1a expression is in line with recent studies demonstrating that the posterior compartment of the cardiac disc is re-positioned to the left side of the heart tube.38 By 50hpf hapln1a expression is restricted to low levels in the atrioventricular canal (Figure 2F). Fluorescent in situ hybridization demonstrates hapln1a is expressed in the myocardium (Figure 2G–I), while analysis of Hapln1a protein localization confirms it is deposited in the ECM (Figure 2J–L). Despite the absence of hapln1a expression in the heart at 50hpf (Figure 2F), Hapln1a protein is maintained in the ECM at 50hpf (Figure 2M–Q), suggesting that the ECM environment established during early stages prior to heart tube formation is maintained during heart development and may be important for subsequent cardiac morphogenesis.
3.3 Hapln1a is required for heart morphogenesis and promotes ECM expansion
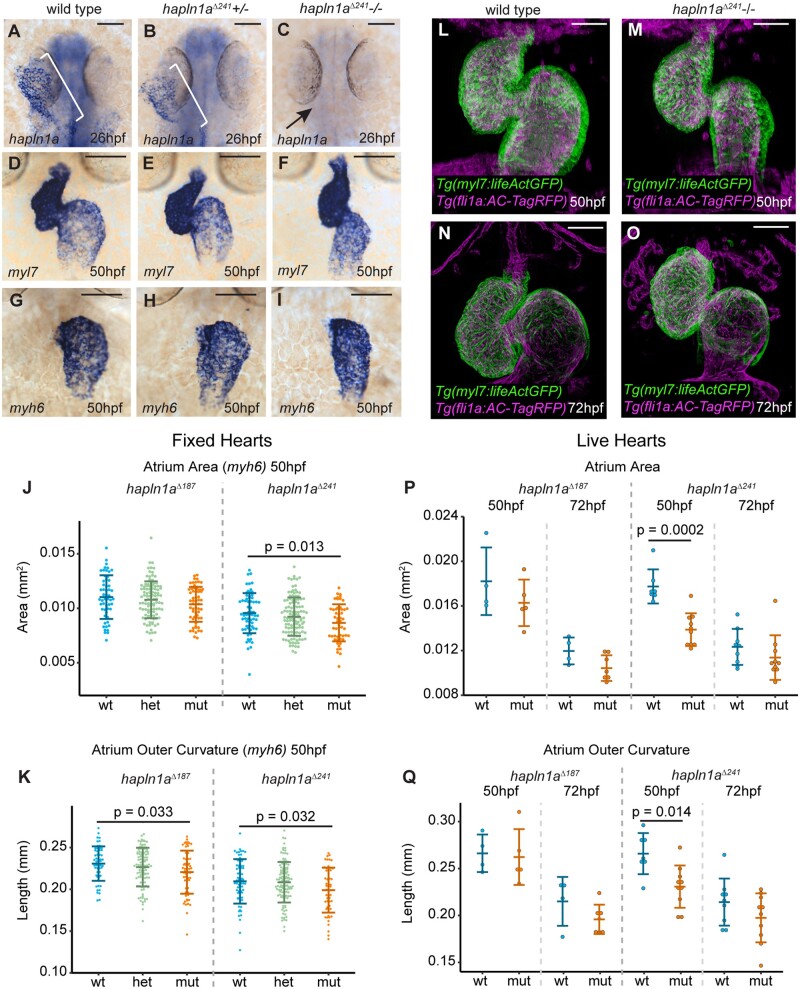
To determine whether Hapln1a is required for cardiac morphogenesis, we used CRISPR-Cas9-mediated genome editing to generate hapln1a mutants by deleting the putative hapln1a promoter therefore abolishing hapln1a expression (Supplementary material online, Figure S4). We recovered two alleles; a 187-bp deletion (hapln1aΔ187) and a 241-bp deletion (hapln1aΔ241) and established both as stable lines at F2. Both alleles remove the initiating ATG and upstream sequence. To confirm both deletions removed the hapln1a promoter and abrogated transcription, hapln1a expression was analyzed at 26hpf in F3 mutant embryos. Homozygous hapln1a promoter mutants of either allele exhibit a complete loss of hapln1a expression at 26hpf compared to wild-type embryos (Figure 3A–C and Supplementary material online, Figure S4), demonstrating successful deletion of the hapln1a promoter. While analysis of heart development in hapln1a Δ241 mutants at 50hpf by in situ hybridization of the pan-cardiac marker myl7 (myosin light chain 7) did not reveal striking abnormalities in cardiac morphogenesis (Figure 3D–F), we did observe mild heart malformations and an apparent reduction in atrial size. To investigate this further, we quantified heart size and chamber morphology at 50hpf by in situ hybridization analysis of myl7, the atrial marker myh6 and the ventricular marker myh7l (myosin heavy chain 7-like)39 (Figure 3 and Supplementary material online, Figures S4 and S5). We observed a reduction in atrial size in hapln1aΔ241 mutants and a reduction in the length of the outer curvature of the atrium in both hapln1aΔ241 and hapln1aΔ187 mutants, suggesting a defect in atrial growth (Figure 3J and K). We performed similar analyses by generating z-projections of live light-sheet images of Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP) transgenic wild-type sibling and hapln1a mutant hearts at 50hpf and 72hpf, allowing us to assess ongoing heart morphogenesis without chamber morphology being altered during fixation and processing (Figure 3L–O and Supplementary material online, Figure S6). We confirmed a reduction in atrial size and ballooning of the atrial outer curvature in live hapln1aΔ241 mutants at 50hpf (Figure 3P and Q), as well as reductions in atrial perimeter and elongation at 72hpf in hapln1aΔ187 mutants (Supplementary material online, Figure S6), with similar trends in reduction in all atrial parameters across alleles. At 72hpf hapln1a mutants also display abnormally positioned atria compared to wild-type siblings, with reduced compaction of the heart, and mispositioning of the atrium and ventricle relative to each other (Supplementary material online, Figure S6). While expressivity of phenotype appears variable between alleles, both exhibit significant changes in morphological parameters indicative of defective atrial growth. Supporting a requirement in atrial growth specifically after heart tube formation, we did not observe significant defects in heart size or tube position at 26hpf, or in looping morphology, or ventricular growth at 50hpf in hapln1a mutants (Supplementary material online, Figure S5). Both hapln1a mutant alleles are adult viable and we did not observe more profound defects in maternal-zygotic hapln1a mutants (data not shown). Together this demonstrates that similar to mouse,15 hapln1a is required for atrial morphogenesis in zebrafish.
Figure 3.
Hapln1a promotes atrial growth. (A–C) mRNA in situ hybridization analysis of hapln1a expression at 26hpf in embryos from an incross of hapln1aΔ241 heterozygous carriers. Wild-type and heterozygous siblings express hapln1a in the heart (bracket A, B, respectively), whereas hapln1a is absent in homozygous mutants (arrow C). (D–I) mRNA in situ hybridization expression analysis at 50hpf of myl7 (D–F) and myh6 (G–I) in wild-type siblings (D and G), hapln1aΔ241 heterozygous siblings (E and H) or hapln1aΔ241 homozygous mutant embryos (F and I). Scale bar = 50 μm. (J and K) Quantification of atrial area (J), and atrial outer curvature (K) in ISH-processed sibling embryos (wt/het) and hapln1aΔ241 or hapln1aΔ187 mutants (mut) at 50hpf. Atrial area is significantly reduced in hapln1aΔ241 mutants compared to wild-type siblings (P = 0.013), and atrial outer curvature is significantly reduced in both hapln1aΔ187 and hapln1aΔ241 mutants (P = 0.033 and P = 0.032). In both J and K, n = 54 hapln1aΔ187 wt; 104 hapln1aΔ187 het; 60 hapln1aΔ187 mut, 66 hapln1aΔ241 wt; 116 hapln1aΔ241 het; 53 hapln1aΔ241 mut. (L–O) Maximum intensity projections of light-sheet z-stacks of live 50hpf (L and M) and 72hpf (N and O) Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP) transgenic wild-type (L and N), and hapln1aΔ241 mutant embryos (M and O). Scale bar = 50μm. (P and Q) Quantification of atrial area (P), and atrial outer curvature (Q) in live light-sheet z-projections from wild-type sibling embryos and hapln1aΔ241 or hapln1aΔ187 mutants at 50hpf and 72hpf. Atrial area and atrial outer curvature are significantly reduced in hapln1aΔ241 mutants (mut) compared to wild-type siblings (wt/het) at 50hpf (P = 0.0002, and P = 0.014). In both (P) and (Q), n = 4 hapln1aΔ187 50hpf wt; 5 hapln1aΔ187 50hpf mut, 7 hapln1aΔ241 50hpf wt; 10 hapln1aΔ241 50hpf mut; 4 hapln1aΔ187 72hpf wt; 7 hapln1aΔ187 72hpf mut, 9 hapln1aΔ241 72hpf wt; 10 hapln1aΔ241 72hpf mut. Comparative statistics carried out using a Kruskal–Wallis test with multiple comparisons.
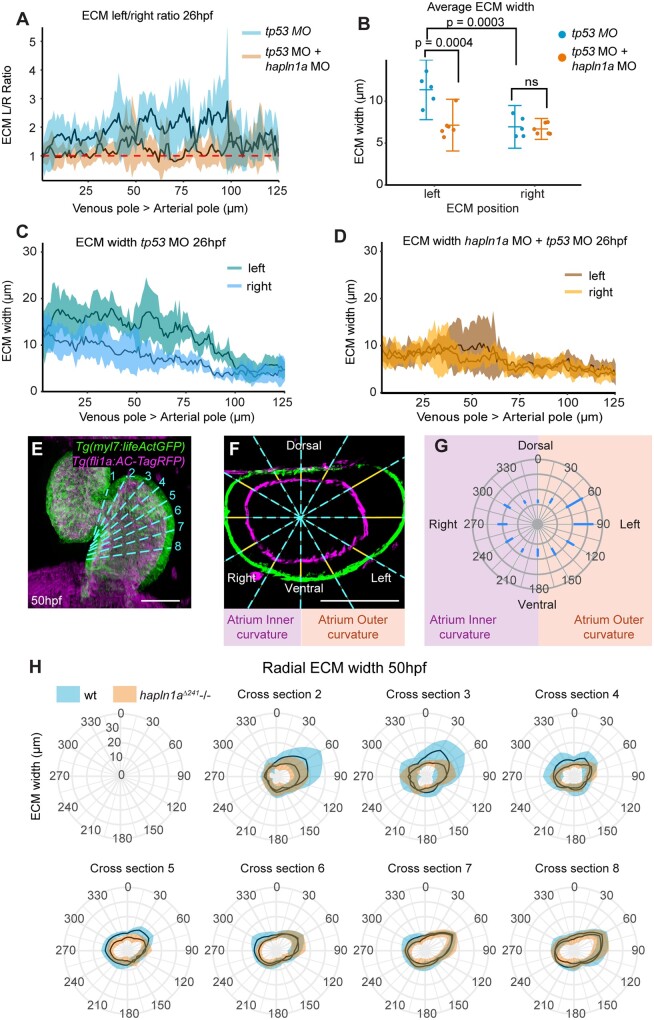
Hapln1 functions as an ECM binding protein and its localization recapitulates the regionalized ECM expansion in the heart tube, therefore, we hypothesized that Hapln1a promotes cardiac morphogenesis by driving regionalized ECM expansion in the heart. Since both hapln1a promoter deletion alleles carry the Tg(myl7:lifeActGFP) transgene, this prevented analysis of ECM width throughout the heart tube of hapln1a mutants using the ssNcan-GFP HA sensor. We instead injected a morpholino (MO) against hapln1a into zebrafish embryos at the 1-cell stage together with a tp53 MO control and the ssNcan-GFP HA sensor and assessed ECM expansion in the heart tube at 26hpf. Analysis of Hapln1a protein levels in hapln1a morphants confirms successful blocking of Hapln1a translation in the morphants (Supplementary material online, Figure S7). Control embryos injected with tp53 MO demonstrate the regionalized ECM expansion previously observed, with a left-sided expansion of the ECM (Figure 4A and B), and a higher level of ECM expansion in the atrium vs. the ventricle (Figure 4C), although overall ECM width was slightly reduced compared to uninjected embryos (compare Figures 4C and 1N). Embryos injected with hapln1a MO + tp53 MO did not exhibit either atrial or left-sided ECM expansion (Figure 4A, B, and D), suggesting that Hapln1a drives regionalized ECM expansion in the heart tube. To confirm a role for Hapln1a in ECM regionalization, we analyzed ECM size in a series of optical cross-sections from live light-sheet images of wild-type and hapln1a mutant Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP) transgenic embryos, taking sections from the atrioventricular canal through the atrium towards the venous pole at 50hpf and 72hpf (Figure 4E–H and Supplementary material online, Figure S8). Analysis of radial ECM width in the atrium of hapln1a mutants revealed a reduction in ECM specifically in the outer curvature of the atrium proximal to the AVC (Figure 4H and Supplementary material online, Figure S8, cross-sections 2–4), where ECM expansion is most profound in wild-type hearts and defects in atrial morphology occur in hapln1a mutants (Figure 3K and Q). Together this supports a role for Hapln1a in regionally regulating cardiac ECM size to promote normal cardiac morphogenesis.
Figure 4.
Hapln1a drives regionalized ECM expansion. (A) Quantification of ECM left/right width along the longitudinal axis of the heart at 26hpf in embryos injected with either tp53 MO (blue, n = 5) or hapln1a MO + tp53 MO (orange, n = 6). Mean ± SD are plotted. (B) Average ECM width on the left or right side of the heart tube in embryos injected with tp53 MO (blue, n = 5) or hapln1a MO + tp53 MO (orange, n = 6). tp53-injected controls display an expanded ECM on the left side of the heart tube compared to the right, (P = 0.0003, 2-way ANOVA) whereas embryos injected with hapln1a MO + tp53 MO have a reduced left ECM size when compared to tp53 MO injected (P = 0.0004), resulting in loss of left-sided ECM expansion. Mean ± SD are plotted. (C and D) Quantification of ECM width on the left and right sides of the heart tube from venous pole to arterial pole at 26hpf in embryos injected with tp53 MO (C, n = 5) or hapln1a MO + tp53 MO (D, n = 6). Mean ± SD are plotted. The cardiac ECM in tp53 morphants exhibits atrial and left side expansion, whereas the ECM in hapln1a morphants is more uniform in width from atrium to ventricle and is not expanded on the left side. Mean ± SD are plotted. (E) Maximum intensity projection of light-sheet z-stacks of 50hpf Tg(myl7:lifeActGFP); Tg(fli1a:AC-TagRFP) transgenic embryo. Dashed cyan line indicates position of optical cross-sections. (F) Example orthogonal view through the atrium of wild-type embryo in (E). Blue dashed lines indicate radial positions for measuring ECM thickness (yellow lines). Scale bar = 50μm. (G) Schematic of radial plot corresponding to radial ECM positions in (F). (H) Quantification of ECM width in atrial cross-sections at defined angular positions along the longitudinal axis of the atrium from AVC (cross-section 2) towards the venous pole (cross-section 8) at 50hpf in wild-type (blue) or hapln1aΔ241 mutants (orange). Asymmetric ECM width is reduced in the outer curvature close to the AVC (cross-sections 2–4) in hapln1aΔ241 mutants compared to wild-type siblings. Mean ± SD are plotted, radial-axis is consistent between plots, n ≥ 4 at each location.
3.4 Hapln1a and HA interact to drive heart morphogenesis
Hapln1a is a member of a family of ECM binding proteins which cross-link HA with proteoglycans.37 Since hapln1a is transiently expressed during early heart development at cardiac disc and early tube stage (Figure 2), this suggests that the cardiac ECM driving continued morphogenesis of the heart is established at early stages of heart development and requires the interaction of Hapln1a with HA. To interrogate the temporal requirements for HA in heart looping, we applied the HA synthesis inhibitor 4-methylumbelliferone (4-MU40,41) to embryos prior to the onset of heart tube formation at 18hpf, and either washed the drug off at 22hpf, or left the embryos to develop to 48hpf, when we assessed heart morphology. Inhibiting HA synthesis from cardiac disc stage (18hpf) until 48hpf often arrested heart development mid-way during tube formation (Supplementary material online, Figure S9), a more profound phenotype than that observed in has2 zebrafish morphants or Has2 mouse mutants.17,18 However, inhibition of HA synthesis during the short time window between cardiac disc (18hpf) and cardiac cone (22hpf) stage, prior to tube formation, resulted in normal tube formation but a specific disruption to subsequent cardiac looping morphogenesis (Supplementary material online, Figure S9). This supports the hypothesis that HA synthesized prior to formation of the heart tube is required for ongoing morphogenesis of the heart.
Having demonstrated a requirement for HA synthesis in heart morphogenesis during early cardiac development when hapln1a expression is initiated, we wanted to confirm the interaction of hapln1a and HA in heart looping morphogenesis. Injection of sub-phenotypic doses of morpholinos targeting either has2 or hapln1a did not result in significant defects in cardiac morphology at 48hpf (Supplementary material online, Figure S9). However, co-injection of both has2 and hapln1a morpholinos results in profound defects in heart development at 48hpf (Supplementary material online, Figure S9), including a reduction in heart looping ratio, and abnormal atrial morphology. This is a more severe phenotype than that observed by either injection of hapln1a MO + tp53 MO, has2 MO + tp53 MO, 4MU treatment or deletion of the hapln1a promoter, suggesting that while timely HA synthesis drives heart morphogenesis subsequent to tube formation, hapln1a is an important regional modulator of this process.
3.5 Hapln1a misexpression results in abnormal heart morphogenesis
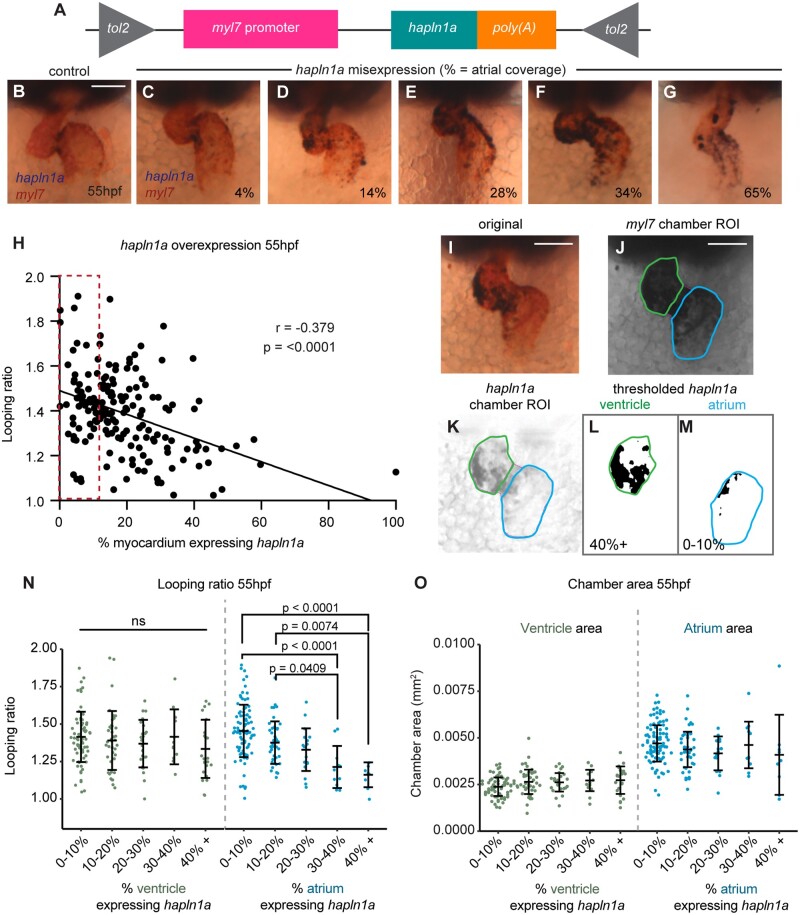
While analysis of hapln1a mutants demonstrates a requirement for Hapln1a in heart development (Figure 3 and Supplementary material online, Figure S6), we wished to investigate whether the regionalization of hapln1a expression is important for cardiac morphogenesis. We generated a DNA construct in which the full-length hapln1a coding sequence is driven by the pan-myocardial myl7 promoter, flanked by Tol2 transposon sites to allow integration into the genome (Figure 5A). We co-injected myl7: hapln1a DNA with tol2 transposase mRNA at the 1-cell stage and analyzed both myl7 and hapln1a expression at 55hpf, allowing us to visualize heart morphology alongside assessing the extent of hapln1a misexpression (Figure 5B–G). We analyzed heart morphology by quantifying looping ratio and assessed this as a function of percentage coverage of hapln1a expression in the whole heart (Figure 5H). Increasing the domain of hapln1a expression in the heart results in a reduction in looping morphogenesis (Figure 5H), suggesting that regionalized expression of hapln1a in the heart is important for cardiac morphogenesis. Since hapln1a expression and ECM asymmetry is greater in the atrium than the ventricle, we hypothesized that hapln1a misexpression in each chamber may impact differently on heart morphogenesis. We quantified hapln1a misexpression in each chamber by calculating the percentage of the chamber which expresses hapln1a (Figure 5I–M), and found that while misexpression of hapln1a in the ventricle did not impact upon heart morphogenesis, misexpression of hapln1a in the atrium decreased looping ratio (Figure 5N). Conversely, we did not find significant changes in chamber area upon hapln1a overexpression (Figure 5O). Together with our observations of abnormal heart morphology in hapln1a mutants this suggests that spatially restricted hapln1a expression in the atrium drives cardiac morphogenesis.
Figure 5.
Regionalized hapln1a expression in the atrium promotes heart morphogenesis. (A) Schematic of DNA construct used to misexpress hapln1a specifically in cardiomyocytes. (B–G) Example images of heart morphology (myl7, red) upon different levels of hapln1a misexpression (blue) at 55hpf, related to the quantification method depicted in (I–M). Percentage indicates hapln1a coverage of the atrium. (H) Scatter plot depicting looping ratio as a function of percentage of the heart covered by hapln1a expression together with linear regression of the data (n = 174). Spearman’s correlation coefficient (r) deviates significantly from zero (P < 0.0001) demonstrating that increased coverage of hapln1a in the myocardium results in reduced heart looping morphogenesis. Red dashed box indicates embryos with wild-type levels of hapln1a expression. (I–M) Quantification approach to analyze pan-cardiac or chamber-specific hapln1a misexpression at 55hpf. The number of hapln1a-positive pixels within each chamber is measured alongside the total chamber area, quantifying the percentage of the chamber expressing hapln1a. (N–O) Analysis of looping ratio (N) and chamber area (O) as a function of the level of hapln1a expression in each chamber of the heart. Embryos are categorized depending on the percentage of the chamber expressing hapln1a. Misexpression of hapln1a in the ventricle does not affect looping ratio, whereas misexpression of hapln1a in the atrium significantly reduces looping ratio at ≥30% coverage (N). hapln1a misexpression in either chamber does not appear to alter overall heart size (O). ns = not significant. In both (N) and (O), atrium misexpression categories: n = 92 (0–10%); 43 (10–20%); 17 (20–30%); 12 (30–40%); 9 (40+%); ventricle misexpression categories: n = 73 (0–10%); 41 (10–20%); 25 (20–30%); 12 (30–40%); 23 (40+%). Comparative statistics carried out using a Kruskal–Wallis test with multiple comparisons.
3.6 Embryonic left–right asymmetry orients the axis of ECM asymmetry in the heart tube
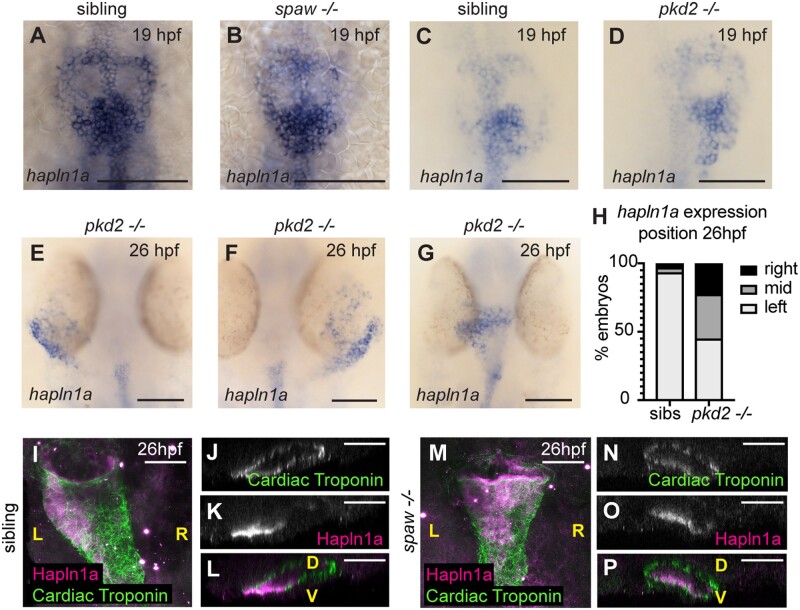
Finally, since hapln1a is asymmetrically expressed on the left side of the heart tube, and is required for heart morphogenesis, we hypothesized that it may contribute to a previously-described tissue-intrinsic mechanism of heart looping morphogenesis7 and thus is expressed independent of embryonic left–right asymmetry cues. Embryos with mutations in pkd2 (polycystic kidney disease 2), which is required for Kupffer’s Vesicle function,42 exhibit defects in left–right asymmetry including a disruption to normal leftward displacement of the heart tube,42 while spaw mutants lack asymmetric Nodal expression prior to asymmetric organ morphogenesis resulting in midline positioning of the heart tube.7 We hypothesized that induction of hapln1a expression occurs independent of embryonic laterality cues, but that asymmetric positioning of hapln1a-expressing cells in the heart tube may be tightly linked to the direction of heart tube position, and therefore dictated by embryonic left–right asymmetry. We analyzed hapln1a expression in an incross of pkd2hu2173 and spaw heterozygotes and found that consistent with our hypothesis hapln1a is always expressed in the posterior of the heart disc in both pkd2hu2173 mutants and spaw mutants at 19hpf (Figure 6A–D). Importantly, at 26hpf, we observed that positioning of hapln1a-expressing cells is dependent upon cardiac position—in pkd2hu2173 mutants where the heart is positioned to the right, hapln1a is up-regulated on the right side of the tube, whereas if the heart remains midline, hapln1a does not exhibit a clear left–right asymmetry in up-regulation (Figure 6E–H). Similarly, analysis of Hapln1a deposition in spaw mutants at 26hpf reveals Hapln1a is no longer positioned on the left side of the heart tube, but instead is secreted into the cardiac ECM on the ventral face of the heart (Figure 6I–P). These data support a model where laterality cues do not initiate hapln1a expression but are required for its subsequent position in the heart tube.
Figure 6.
Posterior up-regulation of hapln1a in the cardiac disc is independent of left–right asymmetry. (A–G) mRNA in situ hybridization analysis of hapln1a expression in an incross of spaw (A and B) or pkd2hu2173 (C–G) heterozygous carriers. At 19hpf hapln1a is expressed in the posterior cardiac disc of both spaw mutant embryos (B, n = 17/19), and pkd2 mutant embryos (D, n = 13/13), similar to sibling embryos. At 26hpf pkd2 mutant hearts that have jogged to the left exhibit left side up-regulation of hapln1a (E, n = 18), pkd2 mutant hearts on the right have right side up-regulation of hapln1a (F, n = 9), and pkd2 mutant hearts that remain at the midline have no clear left–right asymmetry in expression (G, n = 13). (H) Quantification of position of hapln1a expression in sibling and pkd2hu2173 mutant embryos at 26hpf, n = 138 pkd2hu2173 siblings, 40 pkd2hu2173 mutants. (I–P) Fluorescent immunostaining of Hapln1a (magenta) and cardiac troponin (green) at 26hpf in wild-type siblings (I–L) or spaw mutant embryos (M–P). Wild-type siblings exhibit left-sided deposition of Hapln1a in the heart tube (I–L, n = 6), whereas spaw mutant embryos exhibit ventral localization of Hapln1a (M–P, n = 6). (I and M) Dorsal views. (J–L and N–P) Optical transverse sections. Scale bar = 50μm. L, left; R, right; D, dorsal; V, ventral.
Together, this supports a model where initiation of hapln1a expression in the posterior cardiac disc is independent of laterality cues, but the subsequent cell movements which occur during heart tube formation reposition this population of cells to the left side of the heart, dictating the axis of ECM asymmetry in the heart tube.
4. Discussion
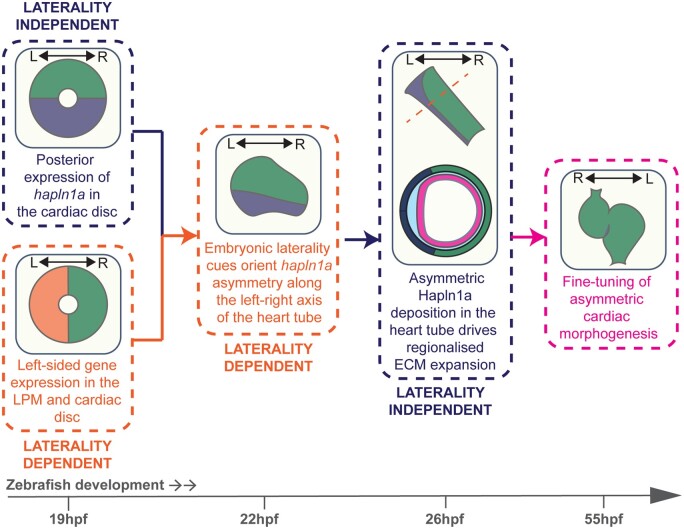
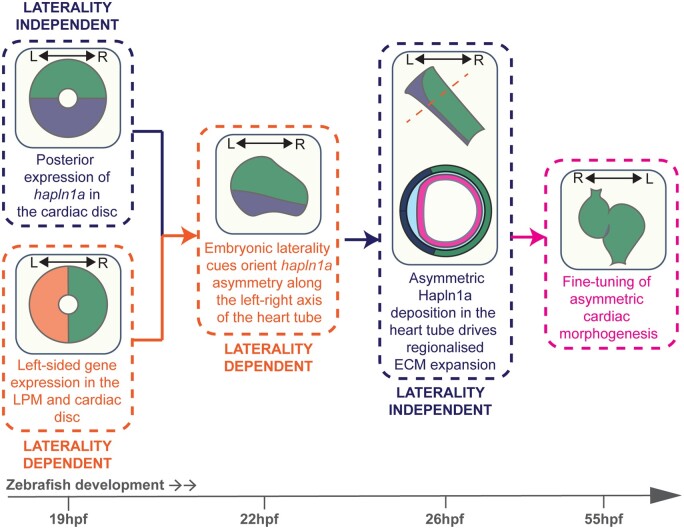
The process of forming the complex heart from a simple linear tube requires careful coordination of spatially restricted extrinsic signals and highly regionalized changes in cell shape and tissue growth to ensure correct shaping of the developing tissue. Our data show that the zebrafish heart tube exhibits regionalized ECM expansion that is dependent upon localized expression of the ECM binding protein hapln1a and this promotes cardiac morphogenesis. Our finding that while the onset of hapln1a in the posterior cardiac disc is independent of embryonic left–right asymmetry, laterality cues are required to orient asymmetric Hapln1a deposition along the left–right axis of the heart tube, allows us to propose a new model in which heart-extrinsic embryonic asymmetry orients tissue-intrinsic cardiac ECM asymmetry, ensuring that directionality and growth of the heart are tightly coordinated to fine tune cardiac morphogenesis (Figure 7).
Figure 7.
Left–right asymmetry orients ECM asymmetry in the heart tube to fine tune cardiac morphogenesis. Model depicting the interaction between embryonic laterality and ECM regionalization in cardiac morphogenesis. LPM, lateral plate mesoderm; L, left; R, right.
Interestingly, the only other gene thus far identified specifically in the posterior cardiac disc/left heart tube in zebrafish is meis2b.38 meis2b mutants exhibit defects in atrial morphology at juvenile and adult stages, supporting our conclusion that early anterior–posterior asymmetry in the heart disc/left–right asymmetry in the heart tube are important for continual cardiac morphogenesis. However, contrary to our study which reveals a reduced atrial size in hapln1a mutants, meis2b mutant adult zebrafish exhibit an enlarged atrium,38 suggesting these two genes play opposing roles in atrial morphogenesis. Investigating hapln1a expression in meis2b mutants may uncover any interactions between these two genes in heart development. Similarly, hapln1a mutants are adult viable, and comparing atrial phenotypes in juveniles and adults will better define the phenotypic relationship between these two genes, as well as the impact of abnormal embryonic morphology on adult heart form and function. Furthermore, recent studies have shown that cross-talk between the myocardium and endocardium modulates atrial growth,43 and our study suggests that differential ECM composition and/or degradation may help regionally fine tune this process to dictate chamber morphology.
The mechanism by which regionalized ECM composition modulates cardiac morphogenesis remains unclear. A major role of the ECM in tissue morphogenesis is to provide structural or biomechanical cues to neighbouring tissues. Alternatively, Hapln1a-mediated cross-linking may modulate regional stiffness of the cardiac ECM, and differential matrix stiffness has been shown to regulate cardiomyocyte form and function.44–47 Importantly, we do not see defects in heart rate in hapln1a mutants (Supplementary material online, Figure S5), suggesting this regionalized ECM is not regulating cardiac function. In addition to provision of mechanical cues to the surrounding cells, the ECM also modulates diffusion and availability of extracellular signalling molecules.48 It is therefore tempting to speculate that the specific ECM environment allows precise regionalized cellular responses to pan-cardiac or chamber specific signalling pathways.
Hapln proteins cross-link HA to proteoglycans. HA and Versican have previously been implicated in heart development, however global loss of these genes throughout the heart result in profound morphological defects.17,19 We suggest that regionalized modification of the ECM by Hapln1a allows the developing heart to generate different regional responses to globally deposited ECM components, resulting in spatial fine-tuning of the specific morphological rearrangements required for complex tissue shaping. While regional ECM cross-linking may change the biomechanical properties of the ECM by stabilizing these components in specific regions of the heart tube, HA and proteoglycan cleavage products can also act as signalling molecules.49 Our finding that hapln1a and has2 work synergistically to promote heart morphogenesis suggests that Hapln1a could promote regional atrial growth by regulating HA signalling in the heart.
Importantly, while our data show that ECM regionalization appears to be the result of asymmetric hapln1a expression and deposition, other factors may support and maintain ECM regionalization during development. Although we do not observe asymmetric has2 expression, we cannot rule out asymmetries in deposition which current analytical methods are not sensitive enough to detect. Similarly, we cannot discard the possibility that asymmetric activity of additional ECM modifiers or hyaluronidases enhance ECM regionalization through localized degradation, and there is some evidence that hyaluronidases themselves are regionally localized in the heart tube.50 This suggests multiple regulatory mechanisms may interact to regionalize HA/ECM activity in the developing heart. Comparative quantitative analysis of cardiac morphology in single and combinatorial knockout models will help define how ECM components and modifiers interact to contribute to heart morphogenesis. Our relatively simplistic analysis of 3D chamber morphology suggests that 2D analyses do not capture all aspects of cardiac morphology during development, and thus defining the links between spatiotemporal ECM dynamics and cardiac morphology requires better definition of changes in ECM composition and morphological cardiac parameters in 3D.
Additionally, Hapln1 mouse mutants exhibit decreased protein levels of the proteoglycan Versican,15 suggesting that Hapln1-mediated HA-Versican cross-linking prevents degradation of one or both of these components. Further supporting an interaction between Hapln1a, Versican, and HA in heart morphogenesis, both mice and medaka lacking Versican exhibit severe cardiac malformations.19,36 Versican proteins can be subject to cleavage by ADAMTS proteases,51 depending on isoform and domain structure, and reduction in ADAMTS protease activity results in reduced Versican cleavage and cardiac abnormalities.52,53 We have shown that of the two zebrafish versican paralogs, only vcana expression overlaps the hapln1a expression domain (Supplementary material online, Figure S3). Zebrafish Vcana appears to be a small V3 or V4-like isoform which is not predicted to undergo cleavage (Uniprot ID A0A2R8Q3K1). This suggests that in zebrafish Hapln1a may not stabilize Versican in the ECM, and alternatively Hapln1a cross-linking may promote regional degradation of HA in the heart tube.
Hapln1 mutant mouse embryos also exhibit relatively mild structural cardiac malformations consistent with abnormal early cardiac morphogenesis.15 However, while that study describes Hapln1 expression in the valve leaflets it does not address a potentially conserved role for transiently asymmetric Hapln1 expression during earlier heart development. Zebrafish have two hapln1 paralogs, each with a distinct expression profile in the heart during development, with hapln1b expressed primarily in the endocardium (data not shown). Thus, zebrafish provide an opportunity to define tissue-specific requirements for Hapln1 function in either the myocardium or endocardium during cardiac morphogenesis.
Together this study identifies a novel functional role for ECM regionalization in the developing heart, mediated by the HA cross-linking protein Hapln1a, and provides a new model in which ECM regionalization acts together with embryonic laterality cues to drive cardiac morphogenesis.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
C.J.D. and E.S.N. conceived the study and designed the experiments. C.J.D., J.S.-P., F.H., E.J.G.P., and E.S.N. carried out experimental work. A.M.S., R.N.W., and T.J.C. shared the Tg(fli1a: AcTagRFP) transgenic line prior to publication. F.T. and J.B. generated the Tg(lft2BAC: Gal4FF) transgenic line, and F.J.v.E. characterized the pkdhu2173 allele. J.B. provided financial support for the Tomo-seq experiments. E.S.N. wrote the manuscript with input from C.J.D., J.S.-P., E.J.G.P., F.T., and T.J.C.
Supplementary Material
Acknowledgements
Additional imaging work was performed at the Wolfson Light Microscopy Facility, using Airyscan and Nikon A1 microscopes. We thank Kelly Smith for the ssNcan-GFP construct, Markus Affolter for the VE-Cadherin antibody, and Aylin Metzner for help characterizing the pkd2hu2173 allele. We also thank Tanya Whitfield, David Strutt, and Simon Johnston for helpful comments on the manuscript.
Conflict of interest: none declared.
Funding
This work was supported by the British Heart Foundation (FS/16/37/32347 to E.S.N.; IG/15/1/31328) and the Academy of Medical Sciences (Springboard Award to E.S.N.). J.B. and F.T. acknowledge support from the Netherlands Cardiovascular Research Initiative: An initiative with support of the Dutch Heart foundation, CVON2014-18 CONCOR-GENES.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Translational Perspective
This study reveals that the cardiac ECM exhibits regional specialization required for heart morphogenesis, and sheds light on how embryonic left–right asymmetry acts in concert with ECM regionalization to fine tune heart shape. This work can help us understand the origins of congenital heart defects, and in particular the nature of morphological heart abnormalities in patients with heterotaxia-associated heart malformations. Furthermore, recent studies suggest the ECM is a key regulator of regenerative potential in the heart, thus defining how distinct ECM composition impacts upon heart form and function has implications for developing regenerative therapies in the future.
References
- 1. Linde D V D, Konings EEM, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJM, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 2011;58:2241–2247. [DOI] [PubMed] [Google Scholar]
- 2. Desgrange A, Garrec J-FL, Meilhac SM. Left-right asymmetry in heart development and disease: forming the right loop. Development 2018;145:dev162776. [DOI] [PubMed] [Google Scholar]
- 3. Levin M, Johnson RL, Sterna CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 1995;82:803–814. [DOI] [PubMed] [Google Scholar]
- 4. Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 2003;130:2303–2316. [DOI] [PubMed] [Google Scholar]
- 5. Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev 2002;16:2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toyoizumi R, Ogasawara T, Takeuchi S, Mogi K. Xenopus nodal related-1 is indispensable only for left-right axis determination. Int J Dev Biol 2005;49:923–938. [DOI] [PubMed] [Google Scholar]
- 7. Noël ES, Verhoeven M, Lagendijk AK, Tessadori F, Smith K, Choorapoikayil S, Hertog J D, Bakkers J. A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat Commun 2013;4:2754. [DOI] [PubMed] [Google Scholar]
- 8. Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol 2010;341:126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barry A. The functional significance of the cardiac jelly in the tubular heart of the chick embryo. Anat Rec 1948;102:289–298. [DOI] [PubMed] [Google Scholar]
- 10. Chowdhury B, Xiang B, Liu M, Hemming R, Dolinsky VW, Triggs-Raine B. Hyaluronidase 2 deficiency causes increased mesenchymal cells, congenital heart defects, and heart failure. Circ Cardiovasc Genet 2017;10:e001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rambeau P, Faure E, Théron A, Avierinos J-F, Jopling C, Zaffran S, Faucherre A. Reduced aggrecan expression affects cardiac outflow tract development in zebrafish and is associated with bicuspid aortic valve disease in humans. Int J Cardiol 2017;249:340–343. [DOI] [PubMed] [Google Scholar]
- 12. Strate I, Tessadori F, Bakkers J. Glypican4 promotes cardiac specification and differentiation by attenuating canonical Wnt and Bmp signaling. Development 2015;142:1767–1776. [DOI] [PubMed] [Google Scholar]
- 13. Mittal A, Pulina M, Hou S-Y, Astrof S. Fibronectin and integrin alpha 5 play essential roles in the development of the cardiac neural crest. Mech Dev 2010;127:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tao G, Levay AK, Peacock JD, Huk DJ, Both SN, Purcell NH, Pinto JR, Galantowicz ML, Koch M, Lucchesi PA, Birk DE, Lincoln J. Collagen XIV is important for growth and structural integrity of the myocardium. J Mol Cell Cardiol 2012;53:626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wirrig EE, Snarr BS, Chintalapudi MR, O’Neal JL, Phelps AL, Barth JL, Fresco VM, Kern CB, Mjaatvedt CH, Toole BP, Hoffman S, Trusk TC, Argraves WS, Wessels A. Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev Biol 2007;310:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trinh LA, Stainier DYR. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell 2004;6:371–382. [DOI] [PubMed] [Google Scholar]
- 17. Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest 2000;106:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith KA, Chocron S, S von der H, E de P, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M, Bakkers J. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell 2008;14:287–297. [DOI] [PubMed] [Google Scholar]
- 19. Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, Markwald RR. TheCspg2Gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev Biol 1998;202:56–66. [DOI] [PubMed] [Google Scholar]
- 20. Huang C, Tu C, Hsiao C, Hsieh F, Tsai H. Germ‐line transmission of a myocardium‐specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn 2003;228:30–40. [DOI] [PubMed] [Google Scholar]
- 21. Reischauer S, Arnaout R, Ramadass R, Stainier DYR. Actin binding GFP allows 4D in vivo imaging of myofilament dynamics in the zebrafish heart and the identification of Erbb2 signaling as a remodeling factor of myofibril architecture. Circ Res 2014;115:845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Savage AM, Kurusamy S, Chen Y, Jiang Z, Chhabria K, MacDonald RB, Kim HR, Wilson HL, van Eeden FJM, Armesilla AL, Chico TJA, Wilkinson RN. tmem33 is essential for VEGF-mediated endothelial calcium oscillations and angiogenesis. Nat Commun 2019;10:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Welten MCM, Haan S. D, Boogert N, van den Noordermeer JN, Lamers GEM, Spaink HP, Meijer AH, Verbeek FJ. ZebraFISH: fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish 2006;3:465–476. [DOI] [PubMed] [Google Scholar]
- 24. Blum Y, Belting H-G, Ellertsdottir E, Herwig L, Lüders F, Affolter M. Complex cell rearrangements during intersegmental vessel sprouting and vessel fusion in the zebrafish embryo. Dev Biol 2008;316:312–322. [DOI] [PubMed] [Google Scholar]
- 25. Burkhard SB, Bakkers J. Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. Elife 2018;7:e31515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Junker JP, Noël ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, Oudenaarden A. V. Genome-wide RNA tomography in the zebrafish embryo. Cell 2014;159:662–675. [DOI] [PubMed] [Google Scholar]
- 27. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet 2007;3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bakkers J, Kramer C, Pothof J, Quaedvlieg NEM, Spaink HP, Hammerschmidt M. Has2 is required upstream of Rac1 to govern dorsal migration of lateral cells during zebrafish gastrulation. Development 2004;131:525–537. [DOI] [PubMed] [Google Scholar]
- 29. Veerkamp J, Rudolph F, Cseresnyes Z, Priller F, Otten C, Renz M, Schaefer L, Abdelilah-Seyfried S. Unilateral dampening of Bmp activity by nodal generates cardiac left-right asymmetry. Dev Cell 2013;24:660–667. [DOI] [PubMed] [Google Scholar]
- 30. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C. The Tol2kit: a multisite gateway‐based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007;236:3088–3099. [DOI] [PubMed] [Google Scholar]
- 31. Angelis JED, Lagendijk AK, Chen H, Tromp A, Bower NI, Tunny KA, Brooks AJ, Bakkers J, Francois M, Yap AS, Simons C, Wicking C, Hogan BM, Smith KA. Tmem2 regulates embryonic vegf signaling by controlling hyaluronic acid turnover. Dev Cell 2017;40:123–136. [DOI] [PubMed] [Google Scholar]
- 32. Baker K, Holtzman NG, Burdine RD. Direct and indirect roles for Nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart. Proc Natl Acad Sci U S A 2008;105:13924–13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grassini DR, Lagendijk AK, Angelis JED, Silva JD, Jeanes A, Zettler N, Bower NI, Hogan BM, Smith KA. Nppa and Nppb act redundantly during zebrafish cardiac development to confine AVC marker expression and reduce cardiac jelly volume. Development 2018;145:dev.160739. [DOI] [PubMed] [Google Scholar]
- 34. Lagendijk AK, Goumans MJ, Burkhard SB, Bakkers J. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ Res 2011;109:649–657. [DOI] [PubMed] [Google Scholar]
- 35. Peal DS, Burns CG, Macrae CA, Milan D. Chondroitin sulfate expression is required for cardiac atrioventricular canal formation. Dev Dyn 2009;238:3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mittal N, Yoon SH, Enomoto H, Hiroshi M, Shimizu A, Kawakami A, Fujita M, Watanabe H, Fukuda K, Makino S. Versican is crucial for the initiation of cardiovascular lumen development in medaka. Sci Rep 2019;9:9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spicer AP, Joo A, Bowling RA. A hyaluronan binding link protein gene family whose members are physically linked adjacent to chrondroitin sulfate proteoglycan core protein genes The Missing Links. J Biol Chem 2003;278:21083–21091. [DOI] [PubMed] [Google Scholar]
- 38. Guerra A, Germano RFV, Stone O, Arnaout R, Guenther S, Ahuja S, Uribe V, Vanhollebeke B, Stainier DYR, Reischauer S. Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish. Elife 2018;7:e32833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yelon D, Horne SA, Stainier DYR. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol 1999;214:23–37. [DOI] [PubMed] [Google Scholar]
- 40. Nakamura T, Funahashi M, Takagaki K, Munakata H, Tanaka K, Saito Y, Endo M. Effect of 4-methylumbelliferone on cell-free synthesis of hyaluronic acid. Biochem Mol Biol Int 1997;43:263–268. [DOI] [PubMed] [Google Scholar]
- 41. Ouyang X, Panetta NJ, Talbott MD, Payumo AY, Halluin C, Longaker MT, Chen JK. Hyaluronic acid synthesis is required for zebrafish tail fin regeneration. PLoS One 2017;12:e0171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schottenfeld J, Sullivan-Brown J, Burdine RD. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development 2007;134:1605–1615. [DOI] [PubMed] [Google Scholar]
- 43. Bornhorst D, Xia P, Nakajima H, Dingare C, Herzog W, Lecaudey V, Mochizuki N, Heisenberg C-P, Yelon D, Abdelilah-Seyfried S. Biomechanical signaling within the developing zebrafish heart attunes endocardial growth to myocardial chamber dimensions. Nat Commun 2019;10:4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wan W, Bjorkman KK, Choi ES, Panepento AL, Anseth KS, Leinwand LA. Cardiac myocytes respond differentially and synergistically to matrix stiffness and topography. bioRxiv 2019;682930. [Google Scholar]
- 45. Bhana B, Iyer RK, Chen WLK, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol Bioeng 2010;105:1148–1160. [DOI] [PubMed] [Google Scholar]
- 46. Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res 2002;91:877–887. [DOI] [PubMed] [Google Scholar]
- 47. Majkut S, Idema T, Swift J, Krieger C, Liu A, Discher DE. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol 2013;23:2434–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Müller P, Schier AF. Extracellular movement of signaling molecules. Dev Cell 2011;21:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol 2015;42:11–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith KA, Lagendijk AK, Courtney AD, Chen H, Paterson S, Hogan BM, Wicking C, Bakkers J. Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development 2011;138:4193–4198. [DOI] [PubMed] [Google Scholar]
- 51. Nandadasa S, Foulcer S, Apte SS. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol 2014;35:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kern CB, Wessels A, McGarity J, Dixon LJ, Alston E, Argraves WS, Geeting D, Nelson CM, Menick DR, Apte SS. Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol 2010;29:304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim KH, Nakaoka Y, Augustin HG, Koh GY. Myocardial angiopoietin-1 controls atrial chamber morphogenesis by spatiotemporal degradation of cardiac jelly. Cell Rep 2018;23:2455–2466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.