Abstract
Vascular calcification (VC), or calcium deposition inside the blood vessels, is common in patients with atherosclerosis, cardiovascular disease, and chronic kidney disease. Although several treatments are available to reduce calcification, the incidence of VC continues to rise. Recently, there have been several reports describing the regulation of circular RNAs (circRNAs) in various diseases. However, the role of circRNAs in VC has not yet been fully explored. Here, we investigated the function of circSmoc1-2, one of the circRNAs generated from the Smoc1 gene, which is downregulated in response to VC. CircSmoc1-2 is localized primarily to the cytoplasm and is resistant to exonuclease digestion. Inhibition of circSmoc1-2 worsens VC, while overexpression of circSmoc1-2 reduces VC, suggesting that circSmoc1-2 can prevent calcification. We went on to investigate the mechanism of circSmoc1-2 as a microRNA sponge and noted that miR-874-3p, the predicted target of circSmoc1-2, promotes VC, while overexpression of circSmoc1-2 reduces VC by suppressing miR-874-3p. Additionally, we identified the potential mRNA target of miR-874-3p as Adam19. In conclusion, we revealed that the circSmoc1-2/miR-874-3p/Adam19 axis regulates VC, suggesting that circSmoc1-2 may be a novel therapeutic target in the treatment of VC.
Keywords: Circular RNA, circSmoc1-2, miRNA-874-3p, Adam19, vascular calcification, vascular smooth muscle cells
Graphical abstract
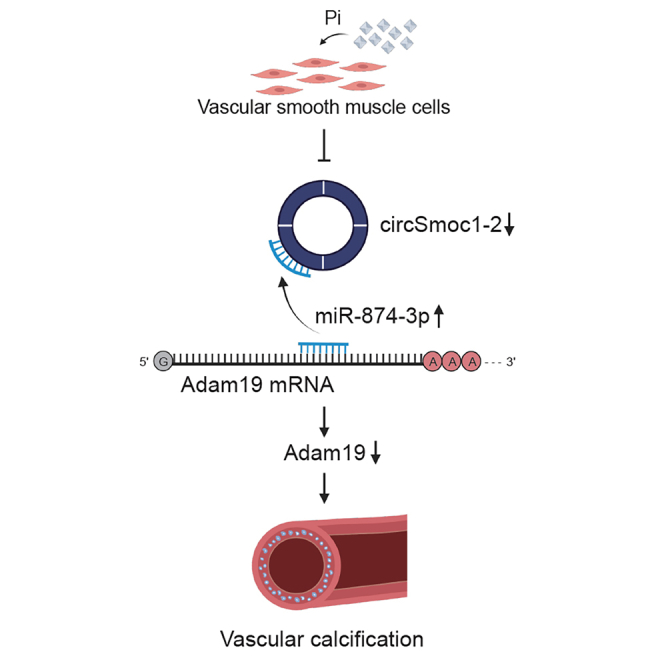
Vascular calcification, or calcium deposition inside the blood vessels, is common in patients with atherosclerosis, cardiovascular disease, and chronic kidney disease. In this study, the authors demonstrate that the circSmoc1-2/miR-874-3p/Adam19 axis regulates vascular calcification, suggesting the circSmoc1-2 may be a novel therapeutic target in the treatment of vascular calcification.
Introduction
Vascular calcification (VC) is the accumulation of calcium phosphate inside blood vessels. VC is known to be an independent risk factor of cardiovascular diseases (CVDs)1 and results in vessel stiffness, which can ultimately lead to cardiac hypertrophy and ischemia.2 Although several therapies used in patients with osteoporosis and chronic kidney disease (CKD) have been shown to be effective in the control of VC,3 the prevalence of VC continues to increase.4, 5, 6 Thus, it is necessary to investigate novel therapeutic targets for the treatment of patients with VC.
Recently, non-coding RNAs (ncRNAs) including microRNAs (miRNAs), long non-coding RNAs (lncRNA), and circular RNAs (circRNAs) have been reported to regulate diverse diseases.7, 8, 9, 10 Most ncRNAs have low protein-coding potential, but they play important roles in various biological processes.7 However, although coding RNAs have been extensively evaluated, relatively little is known about the roles of ncRNAs. miRNAs are small ncRNAs with an average length of 23 nt that bind to the 3′ untranslated region of messenger RNAs (mRNAs), inhibiting their translation and upregulating their degradation.7 LncRNAs are ncRNAs of more than 200 nt in length with a very diverse pool of functions, most of which are determined by their cellular localization. LncRNAs localized to the nucleus may directly regulate transcription, while lncRNAs localized in the cytoplasm can act as miRNA sponges.11 CircRNAs are produced primarily by back-splicing when the 3′ end of the downstream exon binds to the 5′ end of the upstream exon, generating ncRNAs with a circular structure. Because circRNAs do not contain 5′ or 3′ ends, they are not easily digested by the exonucleases known to degrade linear RNAs.12 CircRNAs can also act as miRNA sponges and transcriptional or post-transcriptional regulators.13, 14, 15 Some circRNAs may interact with proteins15 or be involved in translation.16,17 Although various mechanisms of action have been reported for circRNAs, the molecular mechanism underlying their role as miRNA sponges is best understood,18 as this seems to be the major function of cytoplasmic circRNAs.19
Our research group has focused on evaluating the various regulatory functions of ncRNAs in VC. We previously reported several studies describing a number of ncRNAs involved in VC.20 ncRNAs can function as both positive and negative regulators of VC.21 We recently reported that miR-134-5p induces VC by suppressing histone deacetylase 5,22 and miR-27a-3p targets ATF3 to inhibit VC in vascular smooth muscle cells (VSMCs).23 In addition, we found that the lncRNA Lrrc75a-as1 works as a negative mediator of VC and inhibits osteoblast-related factors.24 Moreover, we revealed that circSamd4a has an anti-calcification effect and acts as a miRNA sponge, regulating Camsap2 and Flna.25 During this study of characterization of circRNAs in VC, we noticed that circSmoc1-2 was significantly downregulated following the induction of VC.25 As the role of circSmoc1-2 has not been reported in any other studies, we decided to investigate the function and mechanism of this novel circRNA.
Results
Characterization of circSmoc1-2 in vascular smooth muscle cells
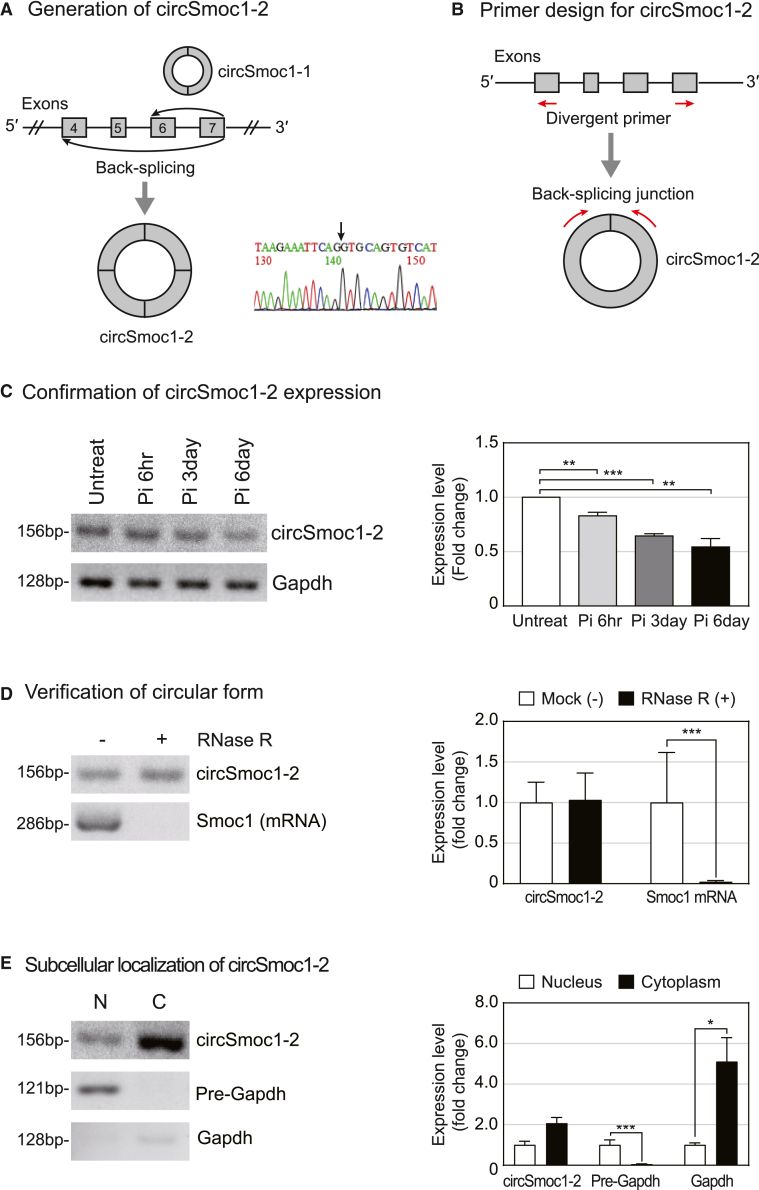
We previously identified two VC-associated circRNAs generated from the Smoc1 gene, circSmoc1-1 and circSmoc1-2.25 In the present study, we went on to evaluate the role of circSmoc1-2 as one of the circRNAs known to be downregulated in response to VC.25 CircSmoc1-2 comprises four exons, namely, exons 4–7, of the Smoc1 gene. The back-splicing junction for circSmoc1-2 was confirmed using Sanger sequencing, which is indicated by an arrow in Figure 1A. To confirm the differential expression of circSmoc1-2 under different conditions, we designed a set of divergent primers targeting the exons where back-splicing occurs (Figure 1B). CircSmoc1-2 was downregulated after the induction of VC (Figure 1C), as previously shown in RNA sequencing (RNA-seq) data.25 To verify the circular structure, total RNA was treated with RNase R exonuclease. CircSmoc1-2 was shown to be resistant to RNase R digestion, while the Smoc1 mRNA transcript was completely degraded (Figure 1D). We went on to investigate the subcellular localization of circSmoc1-2 and found that circSmoc1-2 resides mainly in the cytoplasm (Figure 1E).
Figure 1.
Characterization of circSmoc1-2 in rat vascular smooth muscle cells (VSMCs)
(A) CircRNAs generated from Smoc1. (B) Construction of circSmoc1-2 primers around the splicing junction. Divergent primers were designed to detect circSmoc1-2 expression. (C) Verification of circSmoc1-2 expression following VC induction by RT-PCR (n = 3). Expression of circSmoc1-2 was normalized against Gapdh. Primary rat VSMCs were treated with 2 mM Pi for 6 h, 3 days, or 6 days. (D) Confirmation of circSmoc1-2 structure using RNase R digestion (n = 3). Linear RNA is degraded by RNase R, while circRNA is not. (E) Subcellular localization of circSmoc1-2 (n = 3). Pre-Gapdh and Gapdh were used as controls for the nuclear and cytoplasmic fractions, respectively. Data represent mean ± SEM. Statistical significance was determined using Student’s t test (∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤0.001).
CircSmoc1-2 inhibits VC
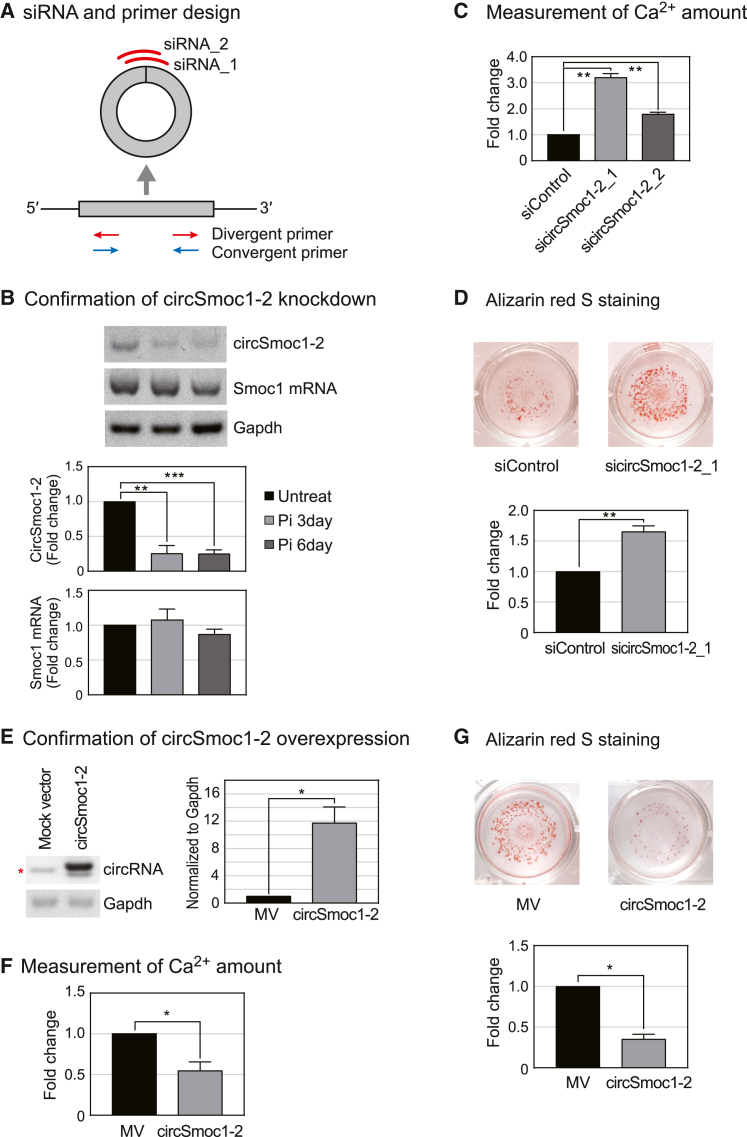
To conduct the functional study of circRNA, circSmoc1-2 was silenced using small interfering RNAs (siRNAs) (Figure 2A). As circSmoc1-2 is found predominately in the cytoplasm (Figure 1E), where RNA interference-mediated gene silencing is effective, we designed two siRNAs to suppress circSmoc1-2 expression by binding to its splicing junction (Figure 2A). CircSmoc1-2 expression was markedly inhibited by these siRNAs, but there was no significant effect on Smoc1 mRNA expression (Figure 2B). Expression of circSmoc1-2 was confirmed using the divergent primers, while Smoc1 mRNA expression was verified using the convergent primers (Figure 2A). These siRNAs were then applied in the calcium assay, which revealed that inhibition of circSmoc1-2 increased calcium deposition (Figure 2C). The effect of silencing circSmoc1-2 was further confirmed using Alizarin red S (ARS) staining, which showed that calcium deposition increased in response to decreased circSmoc1-2 expression (Figure 2D). Given the results of calcium assay on the circSmoc1-2 inhibition, we went on to evaluate the effect of circSmoc1-2 overexpression. The circSmoc1-2 overexpression vector was created using a Laccase2 MCS Exon vector26 backbone, which circularizes any inserted sequence. Transfection with this vector clearly upregulated circSmoc1-2 expression compared with the control (Figure 2E). Overexpression of circSmoc1-2 was confirmed using Sanger sequencing. The effects of this overexpression were then evaluated using the calcium assays and ARS staining assays. These assays revealed that the overexpression of circSmoc1-2 significantly reduces calcium deposition (Figures 2F and 2G). Taken together these results suggest that regulation of circSmoc1-2 affects VC.
Figure 2.
Inhibitory role of circSmoc1-2 in vascular calcification (VC)
(A) Construction of the siRNA and PCR primers. Two sets of siRNAs were designed against the splicing junction of circSmoc1-2. Divergent primers were then used to verify the expression of circSmoc1-2, while convergent primers were used to confirm the expression of Smoc1 mRNA. (B) Verification of circSmoc1-2 by knockdown using semi-quantitative RT-PCR (n = 3). CircSmoc1-2 and Smoc1 expression were both normalized against Gapdh. (C) Calcium concentration (mg Ca/mg protein) following circSmoc1-2 knockdown (n = 3). A10 cells were then treated with 2 mM inorganic phosphate (Pi) and cultured for an additional 3 days following transfection. (D) Alizarin red S (ARS) staining (mM) following circSmoc1-2 knockdown (n = 3). Representative images of ARS staining in each group are shown. The sicircSmoc1-2_1 was used to inhibit circSmoc1-2, and the cells were then treated with 4 mM Pi for an additional 3 days after transfection. (E) Validation of circSmoc1-2 overexpression (n = 3). The red asterisk denotes the endogenous circRNA band. Mock vector (MV) indicates the addition of the Laccase2 vector without an insert, while circSmoc1-2 refers to the circSmoc1-2 overexpression vector that includes a circSmoc1-2 sequence within the Laccase2 vector. (F) Calcium concentrations (mg Ca/mg protein) after circSmoc1-2 overexpression (n = 3). Cells were treated with 2 mM Pi for an additional 3 days following transfection. (G) ARS staining (mM) after circSmoc1-2 overexpression (n = 3). Transfected cells were also treated with 4 mM Pi and cultured for an additional 3 days prior to evaluation. Data represent mean ± SEM. Statistical significance was determined using Student’s t test (p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤0.001).
CircSmoc1-2 acts as a sponge for miR-874-3p
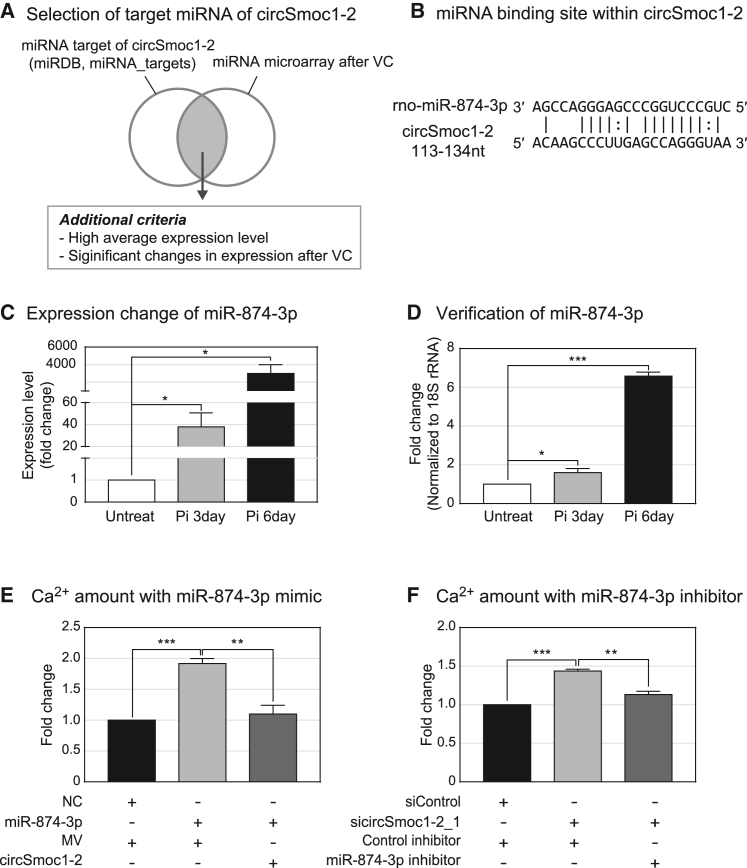
As circSmoc1-2 is localized to the cytoplasm, we hypothesized that circSmoc1-2 may function as miRNA sponges. We used bioinformatics databases, miRDB,27 and miRNA_targets28 to produce a list of potential miRNA partners for circSmoc1-2. Then we narrowed this list by evaluating the expression of these miRNAs in a microRNA microarray assay and identified those miRNAs exhibiting a negative correlation with circSmoc1-2 and significant differential expression in response to VC induction (see Materials and methods for more details) (Figure 3A). This analysis allowed us to identify miR-874-3p, which had a highly complement sequence in circSmoc1-2 at its seed region (Figure 3B). miR-874-3p was significantly upregulated following the induction of VC according to miRNA microarray (GEO: GSE130486)23 (Figure 3C), and this expression profile was confirmed using quantitative real-time polymerase chain reaction (PCR) (Figure 3D). Furthermore, the regulatory relationship between circSmoc1-2 and miR-874-3p was evaluated using a calcium assay. The addition of a miR-874-3p mimic promoted calcium deposition, while circSmoc1-2 overexpression in the presence of the miR-874-3p mimic reduced calcium accumulation (Figures 3E and S1A). In addition, the inhibition of circSmoc1-2 induced calcium deposition, while the addition of a miR-874-3p inhibitor with a circSmoc1-2 siRNA reduced calcium deposition (Figure 3F). These results suggest that circSmoc1-2 functions as a miR-874-3p sponge during VC.
Figure 3.
circSmoc1-2 acts as a microRNA (miRNA) sponge
(A) Schematic describing the identification of miRNA targets for circSmoc1-2. (B) miR-874-3p recognition sites within circSmoc1-2. (C) Expression of miR-874-3p as evaluated by miRNA microarray. Rat VSMC samples were collected at days 3 or 6 after Pi treatment, and miRNA microarrays were performed using these samples (GEO: GSE130486). (D) Verification of miR-874-3p expression using quantitative real-time PCR (n = 4). (E) Calcium concentrations (mg Ca/mg protein) after introducing miR-874-3p in A10 cells (n = 4). (F) Calcium concentration (mg Ca/mg protein) after adding an miR-874-3p inhibitor in A10 cells (n = 3). Cells were treated with 4 mM inorganic phosphate (Pi) and cultured for 2 days following each transfection. Data represent mean ± SEM. Statistical significance was determined using Student’s t test (∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤0.001).
The circSmoc1-2/miR-874-3p/Adam19 axis regulates VC
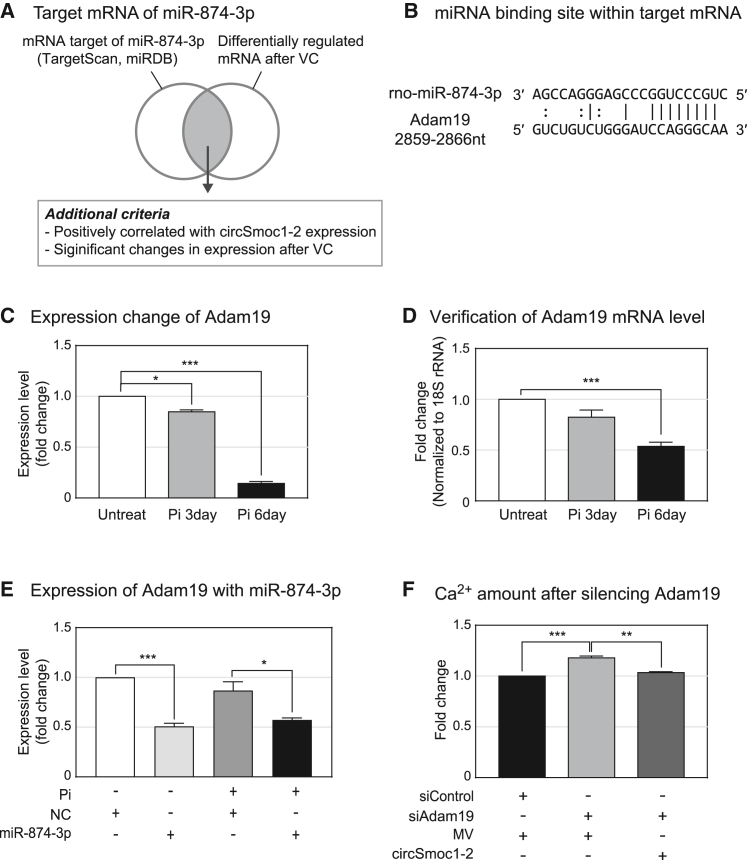
As we were able to establish that circSmoc1-2 functions as a miR-874-3p sponge, we went on to identify the mRNA target of miR-874-3p. We first produced a list of potential mRNA targets using bioinformatics databases, namely, miRDB27 and TargetScan,28 and then narrowed this list by evaluating RNA-seq data from the previous experiment.25 This allowed us to narrow the list to mRNAs that negatively correlate with miR-874-3p expression and undergo a significant change following VC (see Materials and methods for more details) (Figure 4A). This analysis revealed several potential transcripts, but only Adam19 (a disintegrin and metalloproteinase domain 19) fulfilled all of the above selection criteria and included a miR-874-3p binding sequence (Figure 4B). RNA-seq data show that Adam19 is significantly downregulated following the induction of VC (Figure 4C), and this expression pattern was confirmed using quantitative real-time PCR (Figure 4D). Next, we investigated whether miR-874-3p directly targets Adam19 in VSMCs using a quantitative real-time PCR and found that the addition of miR-874-3p significantly inhibited Adam19 expression (Figure 4E). In addition, the role of Adam19 in VC was examined using the calcium assay. Calcium deposition increased following the suppression of Adam19, while the overexpression of circSmoc1-2 reduced this effect (Figures 4F and S1B). Moreover, we also verified the protein level of Adam19, SMA (smooth muscle actin), Col1a1 (collagen type I alpha 1), and Alpl (alkaline phosphatase, tissue-nonspecific isozyme), which further supported results of mRNA level and calcium assay. The calcification marker Alpl was upregulated along with Col1a1 after the suppression of circSmoc1-2 in VSMCs with VC. On the contrary, the protein level of Adam19 and SMA were reduced after inhibition of circSmoc1-2 (Figure S2). The regulatory axis of circSmoc1-2/miR-874-3p/Adam19 was further validated in two in vivo VC models such as the CVD model with VC (Figure S1C) and vitamin D3-administered mice model (Figure 5). Although the expression of circSmoc1-2 and Adam19 was reduced, expression of miR-874-3p was upregulated in these in vivo VC models (Figures 5 and S1C). Taken together these data reveal the circSmoc1-2/miR-874-3p/Adam19 axis, the novel regulatory access that contributes to VC (Figure 6).
Figure 4.
miR-874-3p targets Adam19 during vascular calcification (VC)
(A) Schematic describing the identification of mRNA targets for miR-874-3p. (B) miR-874-3p/Adam19 binding sites. (C) The expression level of Adam19 in RNA-seq. (D) Verification of Adam19 expression using qualitative real-time PCR (n = 4). (E) Expression of Adam19 after adding a miR-873-3p mimic with or without Pi treatment. Cells were treated with 4 mM inorganic phosphate (Pi) for 2 days post-transfection. (F) Calcium concentration (mg Ca/mg protein) after Adam19 inhibition (n = 4). Cells were treated with 4 mM Pi and then cultured for an additional 2 days post-transfection. Data represent mean ± SEM. Statistical significance was determined using Student’s t test (∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤0.001).
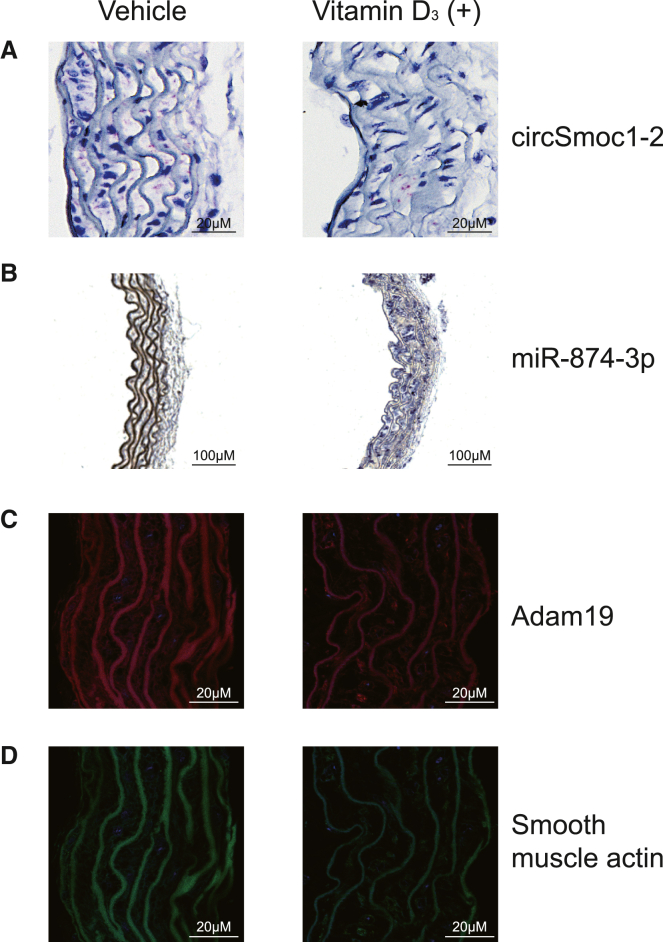
Figure 5.
Histological examination of circSmoc1-2, miR-874-3p, and Adam19 in mouse aorta with vitamin D3-induced vascular calcification (VC)
(A and B) Expression of (A) circSmoc1-2 and (B) miR-874-3p in mouse aortas with or without vitamin D3 treatment for 6 days detected by in situ hybridization. circSmoc1-2 expression was identified using circSmoc1-2 probe specifically targeting the splicing junction of circSmoc1-2. Each red dot represents the single circSmoc1-2 transcript. Expression of circSmoc1-2 was downregulated in vitamin D3-treated mouse aorta. miR-874-3p expression was detected using 5′-digoxigenin (DIG) and 3′-DIG-labeled miR-874-3p probe and shown as dark blue. miR-874-3p signals were upregulated in vitamin D3-treated mouse aorta. (C and D) Expression of (C) Adam19 and (D) smooth muscle actin in mouse aortas with or without vitamin D3 treatment for 6 days detected by fluorescent immunohistochemistry. Expression of Adam19 (red) and smooth muscle actin (green) was detected using anti-Adam19 antibody and anti-smooth muscle actin antibody, respectively. Expression of Adam19 and smooth muscle actin was downregulated in vitamin D3-treated mouse aorta.
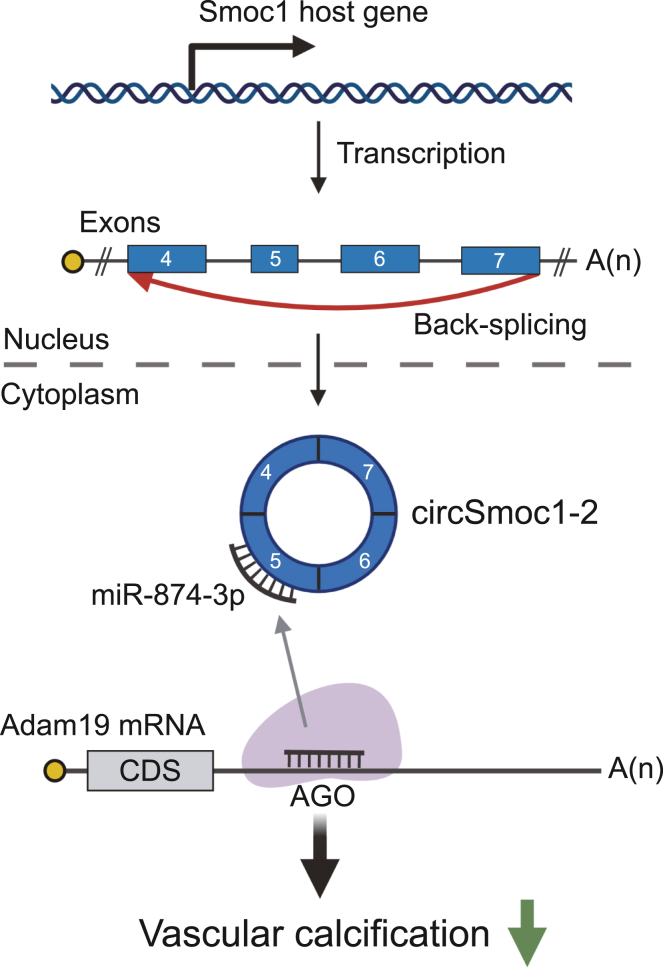
Figure 6.
Schematic representation of the circSmoc1-2/miR-874-3p/Adam19 regulatory axis in vascular calcification (VC)
CircSmoc1-2 is a circRNA generated from the Smoc1 locus via back-splicing. CircSmoc1-2 functions as an inhibitor of VC and acts as a miR-874-3p sponge regulating Adam19.
Discussion
Although circRNAs have recently been evaluated in several diseases, there have been no previous reports describing the role of circSmoc1-2. This is the first study to evaluate the function of circSmoc1-2 in VC, and our key finding is that circSmoc1-2 acts as a negative regulator of VC by functioning as a miR-874-3p sponge and indirectly regulating Adam19 (Figure 5).
CircSmoc1-2, a circRNA produced from the Smoc1 gene (Figure 1A), is known to be downregulated in response to VC (Figure 1C). It is RNase R resistant and localizes primarily to the cytoplasm (Figures 1D and 1E), demonstrating the general characteristics of most circRNAs.29 Upregulation of circSmoc1-2 decreases calcium deposition (Figures 2F and 2G), suggesting that circSmoc1-2 plays an anti-calcification role in VC. As VC is commonly found in patients with CKD or CVD, it is important to understand the underlying mechanisms regulating its severity and onset, as this may reveal a new therapeutic target. Because circSmoc1-2 regulates VC, it could be a novel target in the treatment of VC. Moreover, circSmoc1-2 is conserved in the human, mouse, and rat genomes (Figure S3A), and its expression in human coronary artery smooth muscle cells (HCASMCs) has been verified (Figure S3B). Thus, circSmoc1-2 could be studied in other animals and humans. Importantly, the function of the circSmoc1-2/miR-874-3p/Adam19 axis needs to be investigated in other cell types within vasculature and adverse effects of circSmoc1-2 should be tested before considering its clinical use as a diagnostic biomarker or therapeutic agent. In addition, circSmoc1-2 was found to be expressed in other cell types in liver, lung, and brain30,31 (Table S1). Therefore, the role of circSmoc1-2 in other organs can be further explored.
Because cytoplasmic circRNAs have been reported to function as miRNA sponges,13,32,33 we decided to investigate the potential action of circSmoc1-2 as a miRNA suppressor. We predicted that miR-874-3p is a likely target of circSmoc1-2, as its expression increased following the induction of VC, demonstrating an inverse relationship with circSmoc1-2 expression. Their interaction was further supported by the presence of a complementary binding site in both RNA transcripts (Figure 3B), and circSmoc1-2 sequestration of miR-874-3p was confirmed using the VC model (Figures 3E and 3F). A miR-874-3p mimic induced VC, while circSmoc1-2 inhibited VC (Figure 3E). Likewise, inhibition of circSmoc1-2 promoted VC, while the addition of a miR-874-3p inhibitor suppressed VC (Figure 3F). Previously, miR-874-3p has been studied in various types of cancer,34, 35, 36 stroke,37 and spinal degenerative disease.38 However, the role of miR-874-3p in VC has not been established and this is the first study to show that miR-874-3p promotes VC.
We then went on to identify the target mRNA of miR-874-3p using bioinformatics databases and RNA-seq data25 (Figure 4A). Adam19, which demonstrated a similar expression pattern to circSmoc1-2 and included miR-874-3p binding sites, was predicted as a target of miR-874-3p (Figures 4B–4D). Overexpression of miR-874-3p inhibited Adam19, verifying that miR-874-3p inhibits Adam19 expression in VC (Figure 4E). Finally, the relationship between circSmoc1-2 and Adam19 was evaluated using the calcium assay. Suppression of Adam19 increased VC, while overexpression of circSmoc1-2 reduced VC (Figure 4F). Thus, we identified that the circSmoc1-2/miR-874-3p/Adam19 axis is involved in VC pathogenesis (Figure 5). Previously, Adam19 has been studied in diverse diseases, such as congenital heart disease39,40 and cancer.41,42 Interestingly, miR-874-3p was reported to target Adam19 in nasopharyngeal carcinoma.43 However, there have been no reports linking Adam19 to VC, suggesting that this is the first study to identify the value of Adam19 in VC pathogenesis.
Although our evaluations revealed that circSmoc1-2 acts as a miRNA sponge in VC, there may be other roles for this circRNA that should be evaluated in the future. As very few circRNAs have been reported to encode any kind of protein,16 the protein-coding potential of circSmoc1-2 was assessed using a bioinformatics database, circDb,44 and was found to be low (Table S2); therefore, it is unlikely that circSmoc1-2 translates into any protein products. Our previous study also demonstrated that circRNA and its host gene, Smoc1, exhibit a positive correlation in their expression.25 Therefore, further studies should be performed to assess circSmoc1-2 as a transcriptional regulator. In addition, several circRNAs were reported to interact with proteins or act as protein sponges.15 Thus, the potential of circSmoc1-2 interacting with proteins can be investigated.
ncRNAs have emerged as novel therapeutic targets for various diseases. They can regulate cellular pathways and modulate diverse diseases. Accumulating evidence suggests that RNA therapeutics can act as novel therapies in previously untreatable diseases.45 Here, we revealed that circSmoc1-2 inhibits VC and functions as a miR-874-3p sponge modulating Adam19 expression. As there is no approved therapy for the treatment of VC, researchers should consider evaluating circSmoc1-2 as a novel therapeutic target for combating VC.
Materials and methods
Cell culture
VSMCs were isolated from 6-week-old male Sprague-Dawley rats and grown in 100 mm dishes (Eppendorf) with Dulbecco’s modified eagle’s medium (DMEM) (LM001-05; Welgene) and 10% Hyclone fetal bovine serum (FBS) (GE Healthcare Life Sciences) up to passages 2–7. The rat VSMC cell line, A10 (CRL-1476; ATCC), was cultured in DMEM (LM001-05) with 10% FBS (Welgene). Human coronary artery smooth muscle cells (SMCs) were grown in medium 231 and SMC growth or differentiation supplement and were used at passages 4–8. The animal experiment was conducted following the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 8023, revised 1978). All animal experiment protocols were reviewed and approved by the Chonnam National University Medical School Research Institutional Animal Care and Use Committee.
Cellular models of VC and RNA isolation
The in vitro induction of VC was completed as previously described.25 Primary rat VSMCs were treated with 2 mM inorganic phosphate (Pi) for 6 h, 3 days, or 6 days to induce VC. Total RNA from each of these four time points was then extracted using the TRIzol reagent (Thermo Fisher Scientific) as described in the manufacturer’s instruction, and residual DNA was removed following DNase I (Takara) treatment.
RNase R treatment
We confirmed the circular structure of circSmoc1-2 using RNase R and 10× RNase R buffer (Epicentre). Although experimental samples were treated with RNase R, control samples were treated only with 10× RNase R buffer. The RNase R treatment was conducted at 37°C for 20 min and then inactivated at 95°C for 3 min.
Cell fractionation
Cell fractionation was completed using the protocol described in our previous publication.25 In brief, A10 cells were washed twice with phosphate-buffered saline (PBS) and then removed from their 100 mm dish. The cells were pelleted and then resuspended in cool lysis buffer (20 mM HEPES, pH 7.4, 250 mM sucrose, 1 mM EDTA, 1 mM DTT) and then placed on ice for 25 min before adding 10% NP which was allowed to react for another 2 min on ice. These samples were then centrifuged at 2,000 × g and 4°C for 20 min, the cytoplasmic fraction was collected by centrifugation at 18,000 × g and 4°C for 20 min. The cell pellets containing the nuclear fraction were mixed with K100 buffer D (100 mM KCl, 20 mM Tris at pH 8.0, 0.2 mM EDTA) and centrifuged again at 10,000 rpm for 5 min at 4°C.
cDNA synthesis and PCR
Total RNA from A10 cells, HCASMCs, and mouse aortas from ApoE knockout (KO) mice was converted to cDNA and applied in the reverse transcriptase polymerase chain reaction (RT-PCR). Each RNA sample was mixed with 1 μL random hexamer (Thermo Fisher Scientific) and 1 μL RevertAid reverse transcriptase to produce cDNA and then used as a template in the semi-quantitative PCR to assess the expression levels of circRNA or mRNA. For miRNA cDNA synthesis, TaqMan Advanced miRNA assay (Applied Biosystems) or miScript II RT Kit (Qiagen) was used. Expression levels of circRNAs were evaluated using ImageJ (http://imagej.nih.gov/ij/)46 and normalized against Gapdh. Quantitative real-time PCR was used to evaluate the relative mRNA expression of various targets and was completed using a Power Sybr Green Master Mix (Applied Biosystems) or QuantiTect SYBR Green PCR kit (Qiagen) in a Rotor-Gene Q (Qiagen). The expression levels for the quantitative real-time PCR evaluations were normalized against 18S rRNA (4332641; Applied Biosystems), β-actin, or U6 (MS00033740; Qiagen) using the 2−ΔCt method. Expression of miR-874-3p was detected using miR-874-3p primer purchased from Applied Biosystems or Qiagen. The primer sequences are summarized in Table S3.
circSmoc1-2 cloning
The full circSmoc1-2 sequence was cloned into a pcDNA3.1(+) Laccase2 MCS exon vector,26 a gift from Jeremy Wilusz (Addgene plasmid #69893), to facilitate circSmoc1-2 overexpression. The primer sequences used to amplify circSmoc1-2 are provided in Table S3. After cloning, the sequence of the circSmoc1-2 overexpression vector was verified using Sanger sequencing.
Design of siRNAs, miRNA mimics, and miRNA inhibitors
The small interfering RNAs against circSmoc1-2 were produced using two different tools, the siDesign center from Horizon Discovery (https://horizondiscovery.com/design-center/) and the i-Score designer (https://www.med.nagoya-u.ac.jp/neurogenetics/i_Score/i_score.html).47 The siRNAs against circSmoc1-2 were designed to target the splicing junction, and siRNA against Adam19 mRNA for humans was constructed to base pair with exons within Adam19. These designed siRNAs were synthesized by Bioneer. The predesigned Adam19 siRNA for rat species, negative control siRNA, miR-874-3p mimic, and control miRNA were purchased from Bioneer. In addition, both the miR-874-3p inhibitor and negative miRNA inhibitor control were purchased from Qiagen.
Transfection
A total of 3.5 × 105 A10 cells were seeded in 60 mm dishes and used to evaluate circSmoc1-2 overexpression. Following seeding, 2.5 μg of Laccase2 vector, with or without the circSmoc1-2 insert, was transfected into the A10 cells using Lipofectamine 3000 (Invitrogen). Additionally, 2 × 105 A10 cells were seeded for suppression of cirSmoc1-2 in 60 mm dishes. After overnight incubation, these cells were transfected with 30 nM of each siRNA construct using Lipofectamine 3000. Negative control siRNAs were purchased from Bioneer, and the siRNA sequences are provided in Table S3. Furthermore, A10 cells were seeded in 60 mm dishes and transfected with 10 nM miR-874-3p mimic or control miRNA using Lipofectamine RNAiMAX after the cell reached 80% confluence to assess the expression change of Adam19 after 4 mM Pi treatment for 2 days.
Calcium assay
Calcium deposition was evaluated in A10 cells and HCASMCs grown in 24-well plates. When cells reached about 40% confluence, these cells were transfected with 0.25 μg of the Laccase2 vector encoding circSmoc1-2 or 30 nM circSmoc1-2 siRNAs to evaluate the effects of circSmoc1-2 overexpression or inhibition, respectively. To assess the effect of miR-874-3p in VC, A10 cells and HCASMCs were grown in 24-well plates and then co-transfected with 30 nM miR-874-3p mimic and 0.25 μg of the Laccase2 vector with circSmoc1-2. This was also validated by co-transfecting cells with 10 nM miR-874-3p inhibitor and 10 nM circSmoc1-2 siRNA in A10 cells. Additionally, to evaluate the role of Adam19 mRNA in VC, A10 cells and HCASMCs were grown in 24-well plates and then co-transfected with 30 nM Adam19 siRNA and 0.25 μg of the Laccase2 vector expressing circSmoc1-2.
Following transfection, these cells were treated with 2–5 mM Pi for 1–3 days to induce VC. After the cells were washed with 1× phosphate-buffered saline, cells were decalcified using 0.6 mol/L HCl incubated at 4°C for 1 day. These cells were then evaluated using a QuantiChrom calcium assay kit (BioAssay Systems), and the calcium concentration was normalized against the protein concentration for each experiment.
ARS staining and quantification
We verified the effects of both circSmoc1-2 knockdown and overexpression using A10 cells cultured in 24-well plates. Cells with 40% confluence were transfected with either 30 nM siRNA or 0.25 μg overexpression vector. After 1 day, cells were treated with 4 mM Pi for 3 days. The cells were washed in PBS, fixed in 10% formalin for 1 h, washed three times with deionized water, and air-dried. These samples were then treated with 40 mM ARS solution (pH 4.2) (Sigma-Aldrich) and kept at 20°C–25°C. Then the ARS solution was removed, and samples were washed using PBS and deionized water. ARS staining was then quantified by dissolving the ARS precipitate in 10% cetylpyridinium chloride (Sigma-Aldrich) and evaluating the absorbance of this solution at 562 nm using a microplate spectrophotometer (BioTek).
miRNA predictions
Bioinformatics databases, including miRDB27 and miRNA_targets,28 were used to identify the miRNA targets of circSmoc1-2. The expression of these miRNAs was retrieved from bioinformatics databases cataloging miRNA microarray from rat VSMCs with Pi treatment for 3 or 6 days23 (GEO: GSE130486). These target miRNA sequences were further filtered using the following criteria: average expression level ≥ 100, 4-fold change on day 6 of Pi treatment, and miRNA seed region similarities. This evaluation identified miR-874-3p as the most likely target for cirSmoc1-2.
mRNA prediction
mRNA targets of miR-874-3p were identified using several bioinformatics databases, including miRDB27 and TargetScan.48 The expression values for each of the potential target mRNAs were determined using the RNA-seq data from rat VSMCs following Pi treatment for 3 or 6 days.25 These target mRNA candidates were then further validated using the following criteria: average expression level ≥ 1,000, 4-fold change in expression on day 6 of Pi treatment, and a high degree of sequence similarity to the miRNA seed region. This selection process revealed that Adam19 shared a positive correlation with circSmoc1-2 and included a binding sequence for miR-874-3p, making this the obvious choice for our evaluations.
Western blot
Either negative control siRNA or circSmoc1-2 siRNA was transfected into A10 cells. On the next day, 2 mM Pi was added to cells for 3 days. Cells were treated with RIPA buffer (Translab) and were collected by scrapper. After protein quantification using the Pierce BCA Assay kit (Thermo Fisher Scientific), 13 μg proteins were loaded into 8%–10% SDS-polyacrylamide gel and separated. Then proteins were transferred to polyvinylidene difluoride membrane (Millipore) after methanol activation. After blocking with 5% skim milk (BD Biosciences) and washing with 1× TBS-T (Tris-buffered saline, 0.1% Tween 20) three times, membranes were incubated overnight at 4°C with primary antibodies. Primary antibodies were anti-Alpl antibody (11187-1-Ap; Proteintech), anti-Adam19 antibody (NBP1-69364; Novus Biologicals), anti-SMA antibody (sc-53015; Santa Cruz Biotechnology), anti-β tubulin antibody (sc-9935; Santa Cruz Biotechnology) with a 1:1,000 dilution, and anti-collagen type I (ab34710, Abcam) with a 1:4,000 dilution. An anti-SMA antibody was used to detect α-SMA expression. Then membranes were washed three times with 1× TBS-T and were incubated with secondary antibodies at room temperature (RT) for 1 h. Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat immunoglobulin G antibody (Jackson Immunoresearch). Finally, the membranes were treated with Immobilon Western Chemiluminescent HRP substrate (Millipore) and were detected using Fusion Solo (Vilber). For analysis of western blot image, ImageJ (https://imagej.nih.gov/ij/) was used, and β-tubulin was selected to normalize expression levels.
Animal model of vascular calcification
Vitamin D3 was administered in mice to induce VC as described previously.23 Vitamin D3 in 70 μL 100% ethanol was added to 500 μL Cremophor (Alkamuls EL-620; Sigma-Aldrich) and incubated for 15 min at RT. Then the solution was transferred into 6.2 mL sterilized water including 250 mg dextrose and incubated for 15 min at RT. Finally, vitamin D3 (150 μL/25 g, 5 × 105 IU/kg/day) was administered subcutaneously for 3 days in 6- to 7-week-old C57BL/6 male mice, and mice were kept for 6 days. Age- and gender-matched vehicle mice received the solution prepared in the same manner without the inclusion of vitamin D3 for the same period. After mice were sacrificed using intraperitoneal injection of 2,2,2-tribromoethanol (T48402; Sigma-Aldrich) at a dose of 240 mg/kg, the aorta was isolated and fixed in neutral buffered formalin at RT for 1 day and embedded in paraffin. Arterial sections (5 μm) of vitamin D3-administered mice were used for in situ hybridization and immunohistochemistry.
To induce atherosclerotic vascular calcification, 8-week-old ApoE knockout male mice were fed a high-fat and calcium-supplemented diet for 8 weeks. Simultaneously, age- and gender-matched ApoE knockout mice, vehicle group, were supplied with a normal chow diet for the same period. Then total RNA was isolated from the mice aorta using TRIzol reagent.
In situ hybridization
The expression of circSmoc1-2 was detected in arterial sections vehicle or vitamin D3-administered mice using BaseScope Detection Reagent Kit v2-Red (323900; Advanced Cell Diagnostics [ACD]) as indicated in the manufacturer’s protocol with slight modification. The circSmoc1-2 probe targeting splicing junctions of circSmoc1-2 was designed and synthesized from ACD. Briefly, slides were deparaffinized by xylene and ethanol and incubated with RNAscope hydrogen peroxide for 10 min at RT. After washing with deionized water three times, slides were immersed in RNAscope 1× target retrieval reagents at 98°C for 15 min. Then slides were incubated with RNAscope protease IV at 40°C for 30 min in a HybEz oven and were hybridized with the circSmoc1-2 probe at 40°C overnight in a HybEz oven. Then signal of circSmoc1-2 was amplified with AMP 1-8 reagents. Finally, to detect the circSmoc1-2 chromogenic signal, slides were incubated with Basescope Fast RED for 10 min and counterstained with Gill’s hematoxylin and 0.02% ammonia water. After mounting the slides with VectaMount (Vector Laboratories), the images were visualized on the following day using an Axio scan.Z1 scanner (Carl Zeiss).
Expression of miR-874-3p was assessed in arterial sections of vehicle or vitamin D3-treated mice using miR-874-3p miRCURY LNA miRNA detection probe (YD00616271-BEG; Qiagen) as described in the manufacturer’s protocol with few modifications. Briefly, slides were deparaffinized using xylene and ethanol and incubated with Proteinase K for 10 min at 37°C. Then slides were washed with PBS-T two times and incubated with a hybridization mixture containing 5′-digoxigenin (DIG) and 3′-DIG-labeled 40 nM LNA miR-874-3p probe and miRNA ISH buffer at 55°C for overnight. After stringent washes with saline sodium citrate buffer, slides were incubated with blocking solution for 15 min at RT in a humidified chamber. Then slides were incubated with an anti-DIG antibody (1:800) for 1 h at RT. Then slides were washed with PBS-T three times and incubated with AP substrate at 30°C overnight. After counterstaining with Nuclear Fast Red for 1 min, slides were mounted and observed on the next day using an Axio scan.Z1 scanner (Carl Zeiss).
Immunohistochemistry
Protein expression in aortic sections of vitamin D3-administered mice was assessed using fluorescent immunohistochemistry as previously described23 using anti-Adam19 antibody (NBP1-69364; Novus Biologicals) and anti-smooth muscle actin antibody (sc-53015; Santa Cruz Biotechnology) with a 1:100 dilution. The photographs of aortic sections were visualized using a Zeiss LSM 710 confocal microscope (Carl Zeiss).
Statistical analysis
Data are shown as mean ± standard error of the mean (SEM). Statistical significance was determined using Student’s t test, and differences with p values ≤ 0.05 were considered statistically significant.
Acknowledgments
We would like to take this opportunity to convey our gratitude to all of our colleagues from the Departments of Pharmacology and Biochemistry at Chonnam National University Medical School who made significant contributions to this research. This study was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning (NRF-2021R1A2B5B02001501, NRF-2018R1A2B3001503, NRF-2019R1A4A1028534, NRF-2021R1C1C2009289). The funders played no role in designing this study, collecting or analyzing the data, or preparing or publishing the manuscript. The graphical abstract and Figure 6 was created using BioRender.com
Author contributions
J.R., H.K., and Y.-K.K. designed the study. J.R., N.C., D.-H.K., S.S., Y.-H.L., G.Y., and J.H.K. conducted the experiments. J.R. collected the data. J.R., H.K., Y.-K.K. analyzed and interpreted the data. J.R. wrote the first draft. J.R., H.S.K., I.-K.L., Y.A., W.J.P., H.K., and Y.-K.K. revised the manuscript. J.R., H.K., and Y.-K.K. acquired funding.
Declaration of interests
There authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.031.
Contributor Information
Hyun Kook, Email: kookhyun@jnu.ac.kr.
Young-Kook Kim, Email: ykk@jnu.ac.kr.
Supplemental information
References
- 1.Sage A.P., Tintut Y., Demer L.L. Regulatory mechanisms in vascular calcification. Nat. Rev. Cardiol. 2010;7:528–536. doi: 10.1038/nrcardio.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizobuchi M., Towler D., Slatopolsky E. Vascular calcification: the killer of patients with chronic kidney disease. J. Am. Soc. Nephrol. 2009;20:1453–1464. doi: 10.1681/ASN.2008070692. [DOI] [PubMed] [Google Scholar]
- 3.Wu M., Rementer C., Giachelli C.M. Vascular calcification: an update on mechanisms and challenges in treatment. Calcified Tissue Int. 2013;93:365–373. doi: 10.1007/s00223-013-9712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus M.A., Kalra P.A., Hunter J., Menoyo J., Stankus N. The prevalence of vascular calcification in patients with end-stage renal disease on hemodialysis: a cross-sectional observational study. Ther. Adv. Chronic Dis. 2015;6:84–96. doi: 10.1177/2040622315578654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu Q., Zhao H., Wu B., Tsai S., Wu J., Zhang M., Lu L., Qiao J., Men C., Zuo L., et al. Study on the prevalence of vascular calcification in different types of arteries and influencing factors in maintenance peritoneal dialysis patients. Blood Purif. 2019;47:8–16. doi: 10.1159/000496218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X.-R., Zhang J.-J., Xu X.-X., Wu Y.-G. Prevalence of coronary artery calcification and its association with mortality, cardiovascular events in patients with chronic kidney disease: a systematic review and meta-analysis. Ren. Fail. 2019;41:244–256. doi: 10.1080/0886022X.2019.1595646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Boeckel J.N., Jae N., Heumuller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 9.Gruner H., Cortes-Lopez M., Cooper D.A., Bauer M., Miura P. CircRNA accumulation in the aging mouse brain. Scientific Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim Y.H., Ryu J., Kook H., Kim Y.K. Identification of long noncoding RNAs involved in differentiation and survival of vascular smooth muscle cells. Mol. Ther. Nucleic Acids. 2020;22:209–221. doi: 10.1016/j.omtn.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 12.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Peng L., Yuan X.Q., Li G.C. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol. Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 15.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can Be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X., Cai Y., Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int. J. Mol. Sci. 2019;20:3926. doi: 10.3390/ijms20163926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Q., Yang Z., Jia R., Ge S. The novel roles of circRNAs in human cancer. Mol. Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu J., Ahn Y., Kook H., Kim Y.K. The roles of non-coding RNAs in vascular calcification and opportunities as therapeutic targets. Pharmacol. Ther. 2021;218:107675. doi: 10.1016/j.pharmthera.2020.107675. [DOI] [PubMed] [Google Scholar]
- 21.Kwon D.H., Ryu J., Kim Y.K., Kook H. Roles of histone acetylation modifiers and other epigenetic regulators in vascular calcification. Int. J. Mol. Sci. 2020;21:3246. doi: 10.3390/ijms21093246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe N., Shin S., Joung H., Ryu J., Kim Y.K., Ahn Y., Kook H., Kwon D.H. The microRNA miR-134-5p induces calcium deposition by inhibiting histone deacetylase 5 in vascular smooth muscle cells. J. Cell Mol. Med. 2020;24:10542–10550. doi: 10.1111/jcmm.15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choe N., Kwon D.H., Ryu J., Shin S., Cho H.J., Joung H., Eom G.H., Ahn Y., Park W.J., Nam K.I., et al. miR-27a-3p targets ATF3 to reduce calcium deposition in vascular smooth muscle cells. Mol. Ther. Nucleic Acids. 2020;22:627–639. doi: 10.1016/j.omtn.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong G., Kwon D.H., Shin S., Choe N., Ryu J., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcification. Sci. Rep. 2019;9:5848. doi: 10.1038/s41598-019-42283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu J., Kwon D.H., Choe N., Shin S., Jeong G., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Characterization of circular RNAs in vascular smooth muscle cells with vascular calcification. Mol. Ther. Nucleic Acids. 2020;19:31–41. doi: 10.1016/j.omtn.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar A., Wong A.K.L., Tizard M.L., Moore R.J., Lefèvre C. miRNA_Targets: a database for miRNA target predictions in coding and non-coding regions of mRNAs. Genomics. 2012;100:352–356. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu L., Wang J., Khanabdali R., Kalionis B., Tai X., Xia S. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14:1715–1721. doi: 10.1080/15476286.2017.1367886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Cheng D., Wang J., Dong Z., Li X. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int. 2021;21:11. doi: 10.1186/s12935-020-01703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia B., Lin M., Dong W., Chen H., Li B., Zhang X., Hou Y., Lou G. Upregulation of miR-874-3p and miR-874-5p inhibits epithelial ovarian cancer malignancy via SIK2. J. Biochem. Mol. Toxicol. 2018;32 doi: 10.1002/jbt.22168. [DOI] [PubMed] [Google Scholar]
- 35.Liu W.G., Zhuo L., Lu Y., Wang L., Ji Y.X., Guo Q. miR-874-3p inhibits cell migration through targeting RGS4 in osteosarcoma. J. Gene Med. 2020;22 doi: 10.1002/jgm.3213. [DOI] [PubMed] [Google Scholar]
- 36.Yuan R.B., Zhang S.H., He Y., Zhang X.Y., Zhang Y.B. MiR-874-3p is an independent prognostic factor and functions as an anti-oncomir in esophageal squamous cell carcinoma via targeting STAT3. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7265–7273. doi: 10.26355/eurrev_201811_16261. [DOI] [PubMed] [Google Scholar]
- 37.Xie K., Cai Y., Yang P., Du F., Wu K. Upregulating microRNA-874-3p inhibits CXCL12 expression to promote angiogenesis and suppress inflammatory response in ischemic stroke. Am. J. Physiol. Cell Physiol. 2020;319:C579–C588. doi: 10.1152/ajpcell.00001.2020. [DOI] [PubMed] [Google Scholar]
- 38.Song Q., Zhang F., Wang K., Chen Z., Li Q., Liu Z., Shen H. MiR-874-3p plays a protective role in intervertebral disc degeneration by suppressing MMP2 and MMP3. Eur. J. Pharmacol. 2021;895:173891. doi: 10.1016/j.ejphar.2021.173891. [DOI] [PubMed] [Google Scholar]
- 39.Arai H.N., Sato F., Yamamoto T., Woltjen K., Kiyonari H., Yoshimoto Y., Shukunami C., Akiyama H., Kist R., Sehara-Fujisawa A. Metalloprotease-dependent attenuation of BMP signaling restricts cardiac neural crest cell fate. Cell Rep. 2019;29:603–616.e5. doi: 10.1016/j.celrep.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H.M., Weskamp G., Chesneau V., Sahin U., Vortkamp A., Horiuchi K., Chiusaroli R., Hahn R., Wilkes D., Fisher P., et al. Essential role for ADAM19 in cardiovascular morphogenesis. Mol. Cell Biol. 2004;24:96–104. doi: 10.1128/MCB.24.1.96-104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoyne G., Rudnicka C., Sang Q.X., Roycik M., Howarth S., Leedman P., Schlaich M., Candy P., Matthews V. Genetic and cellular studies highlight that A Disintegrin and Metalloproteinase 19 is a protective biomarker in human prostate cancer. BMC Cancer. 2016;16:151. doi: 10.1186/s12885-016-2178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Lian Y.M., Ge C.Y. MiR-145 changes sensitivity of non-small cell lung cancer to gefitinib through targeting ADAM19. Eur. Rev. Med. Pharmacol. Sci. 2019;23:5831–5839. doi: 10.26355/eurrev_201907_18323. [DOI] [PubMed] [Google Scholar]
- 43.Feng X., Xue H., Guo S., Chen Y., Zhang X., Tang X. MiR-874-3p suppresses cell proliferation and invasion by targeting ADAM19 in nasopharyngeal carcinoma. Panminerva Med. 2019;63:238–239. doi: 10.23736/S0031-0808.19.03682-6. [DOI] [PubMed] [Google Scholar]
- 44.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Scientific Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y.K. RNA therapy: current status and future potential. Chonnam Med. J. 2020;56:87–93. doi: 10.4068/cmj.2020.56.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichihara M., Murakumo Y., Masuda A., Matsuura T., Asai N., Jijiwa M., Ishida M., Shinmi J., Yatsuya H., Qiao S., et al. Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res. 2007;35:e123. doi: 10.1093/nar/gkm699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.