Summary
Necrotrophic fungus Rhizoctonia solani Kühn (R. solani) causes serious diseases in many crops worldwide, including rice and maize sheath blight (ShB). Crop resistance to the fungus is a quantitative trait and resistance mechanism remains largely unknown, severely hindering the progress on developing resistant varieties. In this study, we found that resistant variety YSBR1 has apparently stronger ability to suppress the expansion of R. solani than susceptible Lemont in both field and growth chamber conditions. Comparison of transcriptomic profiles shows that the photosynthetic system including chlorophyll biosynthesis is highly suppressed by R. solani in Lemont but weakly in YSBR1. YSBR1 shows higher chlorophyll content than that of Lemont, and inducing chlorophyll degradation by dark treatment significantly reduces its resistance. Furthermore, three rice mutants and one maize mutant that carry impaired chlorophyll biosynthesis all display enhanced susceptibility to R. solani. Overexpression of OsNYC3, a chlorophyll degradation gene apparently induced expression by R. solani infection, significantly enhanced ShB susceptibility in a high‐yield ShB‐susceptible variety ‘9522’. However, silencing its transcription apparently improves ShB resistance without compromising agronomic traits or yield in field tests. Interestingly, altering chlorophyll content does not affect rice resistance to blight and blast diseases, caused by biotrophic and hemi‐biotrophic pathogens, respectively. Our study reveals that chlorophyll plays an important role in ShB resistance and suppressing chlorophyll degradation induced by R. solani infection apparently improves rice ShB resistance. This discovery provides a novel target for developing resistant crop to necrotrophic fungus R. solani.
Keywords: sheath blight, chlorophyll, resistant breeding, transcriptomics, rice (Oryza sativa), maize (Zea mays)
Introduction
Rhizoctonia solani Kühn (R. solani) is a necrotrophic fungus that infects a very wide range of crops, including staple crops rice, wheat and maize (Molla et al., 2020). In rice, R. solani (AG1 IA) causes the sheath blight (ShB) disease, a serious worldwide rice disease posing an increasing threat due to the lack of resistant germplasms and intensive cultivation practices that favour disease development (Jia et al., 2009; Molla et al., 2020). R. solani causes lesions on both leaves and sheaths, and ultimately affects grain filling and yield in rice, resulting in yield loss up to 45% (Singh et al., 2019).
Comparatively, genetic improvement of rice varieties with ShB resistance is the most sustainable and effective strategy to control this disease (Helliwell and Yang, 2013). Li et al. (2019) recently identified ZmFBL4 through a genome‐wide association study that regulates maize ShB resistance by strengthening lignin biosynthesis, and confirmed that this mechanism also exists in rice against R. solani. Currently, around 50 quantitative trait loci (QTLs) have been identified that confer only partial resistance to R. solani in rice (Molla et al., 2020). However, due to the absence of an accurate, high‐throughput phenotyping assay and ShB‐resistant germplasms, only few of these QTLs have been fine‐mapped and none are characterized so far, which has severely hindered the progress in breeding for rice ShB resistance (Jia et al., 2013; Singh et al., 2019; Zuo et al., 2013).
Over the past decades, studies using transgenic techniques to manipulate potential defence‐responsive genes have expanded our understanding of rice‐ R. solani interaction (Molla et al., 2020). Such defence‐responsive genes mainly belong to the SA or ET‐JA phytohormone signalling components, pathogenesis‐related (PR) proteins, antimicrobial peptides/compounds, secondary messengers and transcription factors (TFs). Besides these universal defence‐responsive genes in host‐pathogen interaction systems, Qiao et al. (2020) recently reported a rice siRNA (siR109944) and its target (OsFBL55) that affect rice ShB resistance via regulating auxin homeostasis. In addition, Gao et al. (2018) found that R. solani facilitates its nutrient acquisition by inducing expression of OsSWEET11, a sugar transporter. As expected, suppression of OsSWEET11 enhanced rice ShB resistance and did not produce evident yield penalty (Gao et al., 2018). Most recently, activation of SWEET14‐mediated signalling by tissue‐specific expression of DOF11 was found to increase rice ShB resistance (Kim et al., 2020). These results demonstrate that rice components, known as susceptible genes, are hijacked by R. solani to induce host susceptibility, and provide new insights into both rice‐ R. solani interaction mechanisms and strategies for developing ShB‐resistant varieties.
Chlorophyll, as a key compound in chloroplasts, is mostly assembled with apoproteins to form light‐harvesting complexes for light energy harvesting and conversion to chemical energy (Liu et al., 2004). The accumulation of adequate amounts of chlorophyll is therefore vital for plants to establish photosynthetically active chloroplasts (Jarvis and Lopez‐Juez, 2013). The chlorophyll metabolism pathways in rice have been well characterized with the isolation of chlorophyll biosynthesis (DVR, YGL8, CAO1, etc.) and catabolism (NYC3, SGR, etc.) genes (Peng et al., 2019; Yamatani et al., 2013; Yang et al., 2016). Besides its basic function in photosynthesis, some studies have demonstrated that chlorophyll also affects the defence response in plants (Kariola et al., 2005; Mach et al., 2001), because porphyrin compounds, the precursors and breakdown products of chlorophyll, are extremely phototoxic and lead to reactive oxygen species (ROS) overproduction and cell death or hypersensitive responses (Tanaka and Tanaka, 2007). Therefore, the biosynthesis and degradation of chlorophyll are highly compartmentalized and regulated. The absence of ferredoxin‐dependent glutamate synthase1 (Fd‐GOGAT1), which participates in chlorophyll biosynthesis, leads to accumulation of excessive ROS levels, conferring broad‐spectrum bacterial blight resistance in rice (Chen et al., 2016). The chlorophyll‐deficient, Mg‐chelatase H subunit (ChlH)‐ and phytoene desaturase (PDS)‐silenced leaves accelerate hemibiotrophs Zymoseptoria tritici‐induced cell death in wheat (Lee et al., 2015). The Cucumis sativus mutant version of SGR, which is involved in chlorophyll degradation, displays a broad‐spectrum resistance against oomyceteous downy mildew, bacterial angular leaf spot and fungal anthracnose pathogens (Wang et al., 2018). ROS production and cell death are generally effective defence responses against infection by biotrophic pathogens, which feed on host live tissues. On the contrary, host cell death is in general beneficial to necrotrophic pathogens, which uptake nutrition from dead cells (Pitsili et al., 2019). Thus, the signals associated with cell death could be employed by necrotrophic pathogens to facilitate infection (Faris et al., 2010; Haugrud et al., 2019). However, available lines of evidence and our understanding are very limited.
Here, we report our new findings that chlorophyll plays an important role in regulating rice ShB resistance. We employed the resistant rice variety YSBR1 and the highly susceptible Lemont for inoculation with R. solani under natural environment conditions, and compared their transcriptomic profiles. We found that the two varieties showed a significant difference in their photosynthetic system, especially on chlorophyll metabolism in response to R. solani infection: highly suppressed in Lemont but almost no change in YSBR1. We found that YSBR1 had higher chlorophyll content than that of Lemont, and inducing chlorophyll degradation by dark treatment significantly reduces its resistance. Using rice and maize chlorophyll‐deficient mutants, we further confirmed that chlorophyll played an important role in affecting both rice and maize resistance to ShB. Subsequently, we found that OsNYC3, a chlorophyll degradation gene (Morita et al., 2009), was apparently induced expression by R. solani in a high yield ShB‐susceptible variety ‘9522’, widely planted in Jiangsu province, China. Notably, silencing OsNYC3 significantly improves rice ShB resistance with almost no inferior effects on important agronomic traits or yield of ‘9522’ in field tests. However, the OsNYC3 transgenic lines did not show difference on resistance to biotrophic pathogen Xanthomonas oryzae pv. oryzae (Xoo) and hemi‐biotrophic pathogen Magnaporthe oryzae (M. oryzae), which cause blight and blast diseases, respectively. This discovery provides a novel target for improving crop resistance to R. solani.
Results
YSBR1 shows excellent resistance to sheath blight
In order to find potentially novel components that contribute to rice resistance to R. solani, we selected rice variety YSBR1, identified previously for carrying high resistance to R. solani (Zuo et al., 2009), and compared it to a highly susceptible variety Lemont. We first confirmed that YSBR1 is clearly more resistant to R. solani than Lemont, showing significantly lower disease scores (scores 2.86 ± 0.86 vs 8.28 ± 1.05), lesion lengths and lesion areas in both field and growth chamber tests (Figure 1a,b). We then observed R. solani infection and development on the inner sheaths, and found no clear difference on hyphae behaviour between the two varieties at 24 h after inoculation (HAI) (Figure 1c). However, at 60–72 HAI, the infection hyphae and cushions on Lemont were intact, plump and tightly tangled and closely entrenched into the inner surface of host sheaths, whereas YSBR1 contained sparse, shrivelled and chapped infection hyphae and cushions (Figure 1c). This is consistent with the data that both lesion lengths and areas showed no visible difference between the two varieties at 1 day post inoculation (DPI); while the disease lesions on YSBR1 were developed apparently slowly compared with Lemont after 2 DPI (Figure 1b). Together, these data indicate that YSBR1 is a reliable resistance variety to ShB and is able to suppress R. solani expansion after its initial infection.
Figure 1.
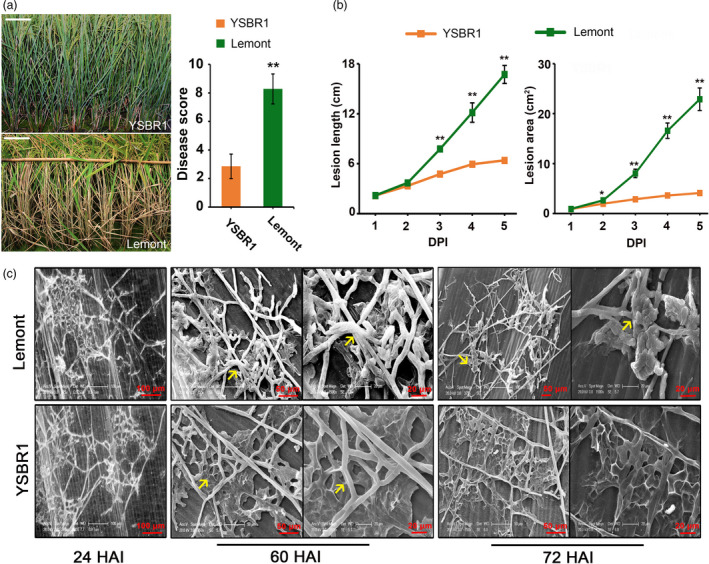
YSBR1 shows high resistance to sheath blight (ShB) with a strong ability to suppress Rhizoctonia solani (R. solani) infection. (a) Evaluation of YSBR1 and Lemont in resistance to ShB disease in a field. Artificial inoculation was performed at booting stage for both varieties and ShB disease scores were rated at 30 days after heading. Photographs were taken at 30 days after heading. Values are means±s.d. Scale bar, 10 cm. (b) Lesion lengths and areas of adult plants were measured at different days post inoculation (DPI) in the growth chamber. Values are means ± SD. (c) Scanning electron microscopy (SEM) of R. solani hyphal behaviour on the surface of leaf sheaths of Lemont and YSBR1 at 24, 60 and 72 h after inoculation (HAI). 8‐week‐old plants grown in natural conditions were inoculated. The inoculated sheaths were collected at the indicated time points. Infection cushions are labelled with yellow arrows. All data are presented as mean ± SD, *, P < 0.05, **, P < 0.01 using Student’s t‐tests.
Transcriptomic analysis reveals that the known defence signals were activated in both YSBR1 and Lemont with the infection of R. solani
To investigate the differential response to R. solani between YSBR1 and Lemont, we performed transcript analyses of the two varieties inoculated and un‐inoculated with R. solani. Two time‐points (10 and 20 HAI) were selected for the transcriptomic analysis using the Affymetrix Rice Genome Array. We detected over 22 000 genes expressed in leaf sheath among the 46 000 unigene models in the microarray chips. We found an excellent consistency among the three biological samples reflected by the correlation coefficient ranging from 0.9702 to 0.9954 (Table S1). Clear groupings of the 22 000 genes into rice varieties, treatments and time points support this conclusion (Figure S1a). Of these genes, we found 2632 significantly differentially expressed genes (DEGs) from the two varieties (2429 in Lemont and 714 in YSBR1) in response to R. solani infection (Figure 2a, Table S2). We randomly tested 10 DEGs using reverse transcription‐quantitative PCR (RT‐qPCR) and found a good correlation (R 2 = 0.7947) with the microarray data (Figure S1b). Noticeably, the circadian cycle caused a stronger effect on transcriptomic reprogramming than pathogen infection, because much more DEGs were found from circadian cycle (Figure 2a). Hierarchical clustering using the 2632 DEGs allowed us to conclude that the time‐point factor has the primary effect, then the variety factor on R. solani ‐induced transcriptomic reprogramming, because the 10 and 20 HAI time‐point factor separated these DEGs into two primary clusters, then the variety factor further separated the DEGs within the clusters (Figure 2b).
Figure 2.
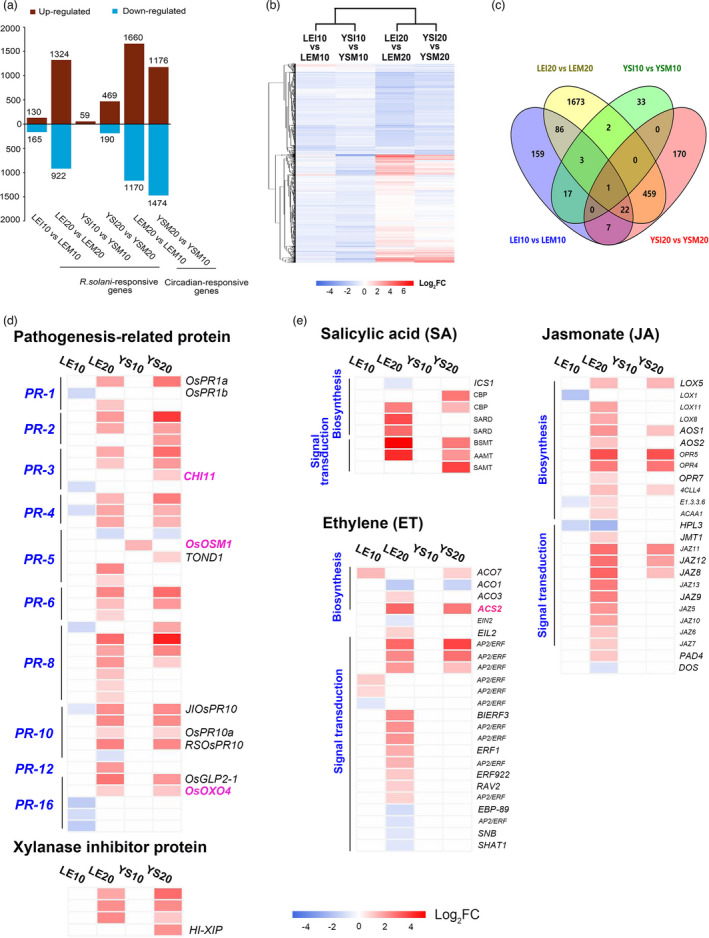
Resistant responses were activated by Rhizoctonia solani (R. solani) invasion as revealed by transcriptomics. (a) Numbers of R. solani‐ and circadian‐responsive genes detected in each comparison. (b) Hierarchical clustering analysis of 2632 differentially expressed genes (DEGs) in response to R. solani. DEGs were identified using |Log2‐Fold change (FC)| ≥0.75 and q‐value <0.1 as the cut‐offs. (c) Four‐way Venn diagram showing the number of common and unique R. solani‐responsive genes among four comparisons. (d) Comparison of YSBR1 and Lemont DEGs involved in pathogenesis‐related proteins (PRs) and xylanase inhibitor proteins (XIPs). (d) Comparison of YSBR1 and Lemont DEGs involved in pathogenesis‐related proteins (PRs) and xylanase inhibitor proteins (XIPs). (e) R. solani infection affects the hormone signal in both varieties. The DAVID (https://david.ncifcrf.gov/) and KEGG (https://www.kegg.jp/kegg/tool/) websites were used for gene annotation The scale bars display the Log2‐FC, shown in red when >0.75, in blue when <−0.75, and in white when −0.75 ~ 0.75.
The two varieties shared 393 up‐regulated and 113 down‐regulated DEGs and each contained a certain number of specific DEGs (1918 in Lemont and 203 in YSBR1) (Figure 2c, Table S2). Functional category indicated that susceptible Lemont had apparently much more defence signalling related DEGs than that in YSBR1, and almost all defence signalling‐related DEGs from YSBR1 were included in Lemont DEGs with similar expression pattern (Table S3). However, the most downstream pathogenesis‐related genes (PRs) and xylanase inhibitor protein‐encoding genes (XIPs) were induced more strongly in YSBR1 than in Lemont (Figure 2d, Table S3). Some of these PR genes, like OsOXO4, CHI11 and OSM1, have been validated as having significant effects on enhancing ShB resistance under transgenic overexpression conditions (Molla et al., 2020). The expression levels of OsOXO4 significantly increased in both varieties, but CHI11 and OSM1 levels only increased in YSBR1 (Figure 2d). For phytohormone signals, we found most DEGs, associated with the biosynthesis and signalling of ET, JA and SA, were up‐regulated and 17 of them were detected in both Lemont and YSBR1 (Figure 2e, Table S3); in contrast, most cytokinins (CKs) and auxin‐related DEGs were down‐regulated and mainly found in Lemont (Table S3). This is consistent with previous reports showing that ET, JA and SA signals were involved in rice defence response to R. solani (Molla et al., 2020). For example, ET biosynthesis gene OsACS2 significantly increased expression in both inoculated varieties (Figure 2e) and was demonstrated to enhance ShB resistance by increasing endogenous ET levels in rice (Helliwell et al., 2013). In addition, many DEGs encoding receptor‐like kinases or proteins (RLKs/RLPs), MAP kinases, TFs and heat shock factors/proteins (HSFs/HSPs) that are widely involved in plant defence against pathogens were activated in both varieties (Table S3). Therefore, based on these data, we conclude that the classic defence responses to R. solani are activated in both varieties although YSBR1 has much less DEGs compared with Lemont.
Photosynthesis and antioxidant systems are clearly suppressed by R. solani in Lemont but not in YSBR1
Necrotrophs, such as R. solani, produce various phytotoxic metabolites or peptides to promote host cell death for their benefit (Foley et al., 2016). Chlorophyll loss and disintegration of photosynthetic apparatus are marker events in a cell death/senescence process (Woo et al., 2019). Gene ontology (GO) analysis performed for R. solani‐responsive DEGs showed that the photosynthetic system was clearly suppressed in Lemont but remained little change in YSBR1 (Figure S2). Chlorophyll metabolism was significantly disturbed in R. solani‐infected Lemont reflected by reducing the expression of chlorophyll biosynthesis genes, such as YGL8, OsDVR and OsCAO1, and elevating the expression of chlorophyll degradation genes, including SGR and NYC3 (Figure 3a, Table S4). Moreover, we found that the down‐regulation of photosynthesis, chloroplast development and light‐responsive genes were mostly Lemont‐specific upon R. solani infection (Figure S3, Table S4). This suggests that chlorophyll is probably involved in rice response to R. solani infection.
Figure 3.
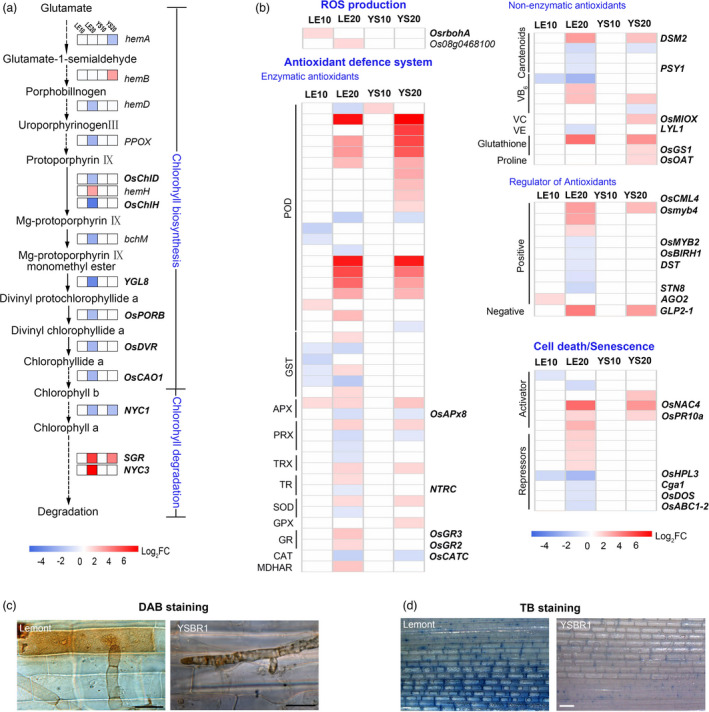
Rhizoctonia solani triggered susceptible responses through the repression of chlorophyll and ROS metabolism. (a,b) Comparison of YSBR1 and Lemont DEGs involved in chlorophyll (a) and ROS metabolism (b). The scale bars display the Log2‐FC, shown in red when >0.75, in blue when <−0.75, and in white when −0.75 ~ 0.75. (c,d) Detection of hydrogen peroxide (H2O2) accumulation by 3,3′‐diaminobenzidine (DAB) staining (c) and cell death by trypan blue (TB) (d) in infected epidermal cells of leaf sheath at 20 HAI. Plants grown in natural conditions were inoculated at tilling stage. Scale bar, 20 μm in (c); 1 mm in (d).
Previous studies have demonstrated that chlorophyll reduction may accompany accumulation of ROS and reduction of host antioxidant capacity (Mur et al., 2010; Yang et al., 2016). We found that DEGs, like RbohA encoding respiratory burst oxidase homologs and Os08g0468100 encoding nitrate reductase, required for ROS generation, were Lemont‐specific (Figure 3b, Table S4). The antioxidant defence system, including enzymatic and non‐enzymatic antioxidants, mainly functions to counter the accumulation of ROS for reducing its damage on cells (Pitsili et al., 2019). Comparatively, although the antioxidant defence system was activated by the pathogen in both varieties, it clearly showed a much stronger response in YSBR1 than in Lemont as revealed by the fold changes and numbers of POD encoding DEGs (Figure 3b, Table S4). In addition, Lemont showed evidently more down‐regulated DEGs (26 vs 55, Down‐regulated vs Total) than YSBR1 (7 vs 34) (Figure 3b and Table S4). Among them, down‐regulated genes encoding four glutathione S‐transferases (GSTs), two peroxidases and one vitamin B6 (VB6) biosynthetic enzyme involved in early response to R. solani in Lemont at 10 HAI. This implies that R. solani may effectively suppress the antioxidant defence system in Lemont but not in YSBR1, and ultimately block ROS scavenge in Lemont. Excessive ROS may lead to cell death and benefit necrotrophic infection (Pitsili et al., 2019). For validating the differences between the two varieties in ROS accumulation and cell death, we further conducted DAB and TB staining, and confirmed that YSBR1 had lower levels of ROS and cell death than Lemont upon R. solani infection (Figure 3b–d, Table S4).
Taken together, these data suggest that the obvious differences in chlorophyll biosynthesis/degradation and ROS production/scavenging between the two varieties are probably one of the key resistance mechanisms of YSBR1 to ShB.
Abolishing chlorophyll biosynthesis increases rice susceptibility to R. solani
Compared with Lemont, YSBR1 displayed more green and had significantly higher chlorophyll content in both leaf and leaf sheath at either later tillering stage or heading stage (Figure S4a). Ultrastructure observation revealed that the two varieties had no difference on chloroplast structure (Figure S4b). These implied that the high chlorophyll content of YSBR1 was not caused by the genes involved in chloroplast development but probably by the genes associated with chlorophyll metabolism. Furthermore, to empirically test the influence of chlorophyll on YSBR1 resistance to ShB, we treated YSBR1 plants with darkness to suppress chlorophyll biosynthesis and accelerate its degradation, then inoculated them with R. solani. The dark treatment significantly reduced the levels of chlorophyll biosynthesis genes (CHLM, CAO1) while up‐regulated chlorophyll degradation genes (SGR1, NYC3) (Figure 4a). We found that the dark‐treated YSBR1 plants developed lesions 1.4–1.6‐fold longer than non‐treated plants at both 3 and 7 DPI (Figure 4b). More significantly, when tested in a detached leaf assay, dark‐treated leaves developed over fivefold larger lesion areas than non‐treated leaves at 6 DPI (Figure 4c). Notably, the dark‐treated YSBR1 has reached 60%–65% of the susceptibility level of non‐treated Lemont (Figure 4b,c). These data suggest that chlorophyll degradation signals play an important role in maintaining the high ShB resistance of YSBR1.
Figure 4.
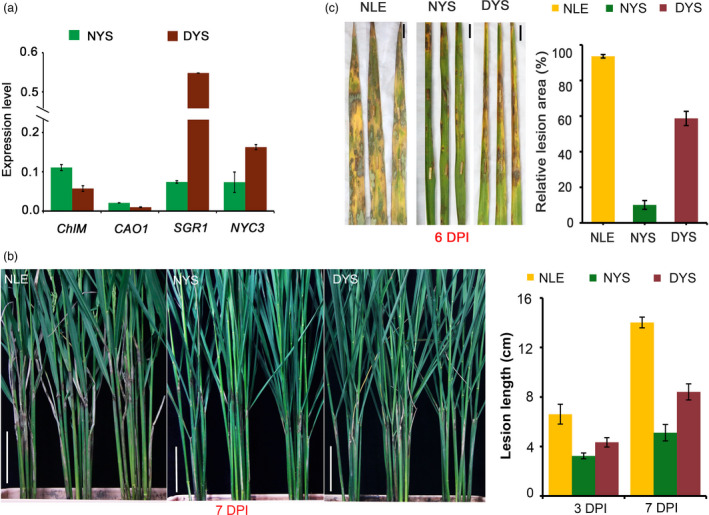
Dark treatment reduces YSBR1 resistance to Rhizoctonia solani (R. solani). (a) Changes in transcript levels of chlorophyll biosynthesis (CHLM, CAO1) and degradation (SGR1, NYC3) genes in YSBR1 untreated (NYS) and treated (DYS) with darkness. All qPCR data were normalized to ubiquitin (Ubq) and were presented as mean ± SD. The experiment was repeated three times with similar trends among each other. (b,c) The whole plants (b) and the detached leaves (c) were used for inoculation, respectively. DYS: dark‐treated YSBR1; NYS: normal YSBR1; NLE: normal Lemont. Disease symptoms were investigated at 6 DPI. Scale bar, 10 cm in (b); 1 cm in (c). Values are means ± SD.
To further assess the role of chlorophyll in host resistance to R. solani, we employed three chlorophyll‐biosynthesis defective rice mutants, dvr, yg18 and cao1 (Figure S5a–c). Two of the mutants carry a single‐nucleotide substitution and one has a 5‐bp deletion in the corresponding gene causing a significant reduction in chlorophyll content (Figure S5a–c). When inoculated with R. solani, we found that all 3 mutants developed lesion areas and lesion lengths significantly larger than their corresponding controls: up to twofold lesion areas at 3 DPI in detached leaf assay and around twofold lesion lengths at 8 DPI in whole plant inoculation assay (Figure 5a). Staining of infected tissues with DAB and TB showed that these mutants accumulated more ROS upon R. solani infection, resulting in greater cell death (Figure 5b). To test if maize defective in chlorophyll biosynthesis also responds similarly to R. solani infection, we inoculated a maize yellow‐green leaf mutant, ygl‐1, defective in chlorophyll biosynthesis (Figure S5d) (Guan et al., 2016), with R. solani isolated from both rice and maize. As expected, the ygl‐1 mutant displayed lesion areas approximately twofold as large as wild type control when inoculated with the rice R. solani isolate and approximately 3.5‐fold as the control when inoculated with the maize R. solani isolate at 5 DPI (Figure 5c). These data demonstrate that chlorophyll is essential against R. solani infection in both rice and maize.
Figure 5.
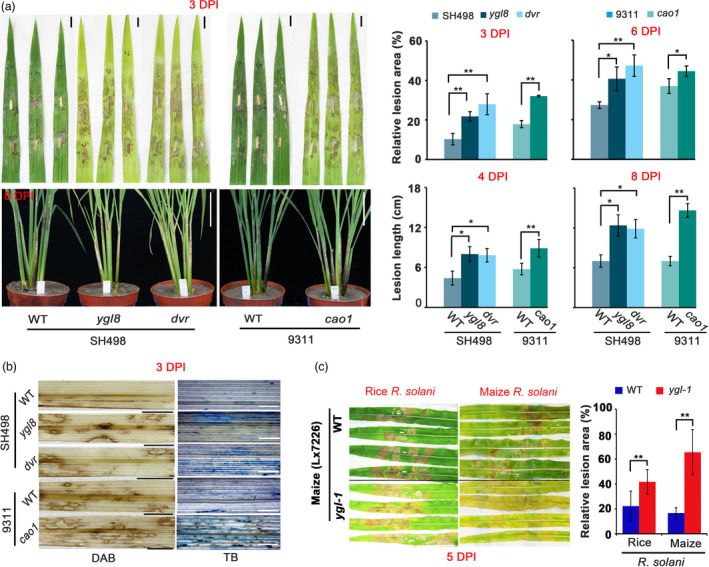
Rice and maize mutants with defects in chlorophyll biosynthesis were impaired in resistance to Rhizoctonia solani (R. solani). (a) Comparison of three rice mutants (ygl8, dvr and cao1) with abolished chlorophyll biosynthesis with wild type on disease symptoms post R. solani inoculation in detached leaf (upper) and whole plant (bottom). Pictures of inoculated leaves were taken at 3 and 6 DPI, which were used to calculate the lesion areas (upper), scale bar, 1 cm; pictures of inoculated whole plants were taken at 8 DPI, and the lesion lengths were measured at 4 and 8 DPI (bottom), scale bar, 10 cm. (b) DAB and TB staining of the three mutants and wild type at 3 DPI. Scale bar, 1 cm in left; 100 μm in right. (c) Comparison of a maize chlorophyll biosynthesis mutant ygl‐1 with wild type on disease symptoms post R. solani inoculation. Scale bar, 10 cm. All data are presented as mean ± SD, *, P < 0.05, **, P < 0.01 using Student’s t‐tests.
Suppression of chlorophyll degradation by down‐regulation of OsNYC3 gene results in enhanced resistance to R. solani with almost no detrimental effects
In our transcriptomic data, the OsNYC3 (Non‐Yellow Coloring 3) gene, encoding a plastid‐localized α/β hydrolase‐fold family protein involved in chlorophyll degradation (Morita et al., 2009), was significantly induced in Lemont but not in YSBR1 (Figure 3a). We then employed qPCR to test its expression in susceptible rice variety 9522, a popular super rice (high‐yield) variety in Jiangsu, China (Figure 6a). We found that the OsNYC3 transcription level, after an initial reduction at 10 HAI in both 9522 and YSBR1, was up‐regulated over sevenfold within 96 HAI in 9522, but was down‐regulated in most time‐points in YSBR1, indicating that R. solani may use OsNYC3 to degrade host chlorophyll (Figure 6a). To further assess the role of OsNYC3, we generated two OsNYC3 overexpression (NYC3‐OX) and two RNA interference (NYC3‐RI) transgenic lines in variety 9522. OsNYC3 RNA levels were confirmed significantly higher in NYC3‐OX and lower in NYC3‐RI lines compared to the wild type (Figure S6a). Consistently, the NYC3‐OX lines displayed a distinct yellow‐green phenotype and the NYC3‐RI lines showed a slightly dark‐green phenotype (Figure S6b). Using detached leaf inoculation assay, we found that the NYC3‐OX lines displayed significantly larger (20%–40% larger) lesion areas than the wild type (WT) control whereas the NYC3‐RI lines displayed significantly smaller (40%–50% smaller) lesion areas (Figure 6b). Inoculation of whole plants gave similar results as detached leaf assay (Figure 6c). DAB and TB staining showed that the NYC3‐OX lines produced significantly more H2O2 and cell death than WT while the NYC3‐RI lines produced less upon R. solani infection (Figure 6d), indicating that chlorophyll probably functions to scavenge ROS, limiting cell death. Together, these data demonstrate that R. solani induces rice chlorophyll degradation to facilitate its infection, and suppression of this process improves rice ShB resistance.
Figure 6.
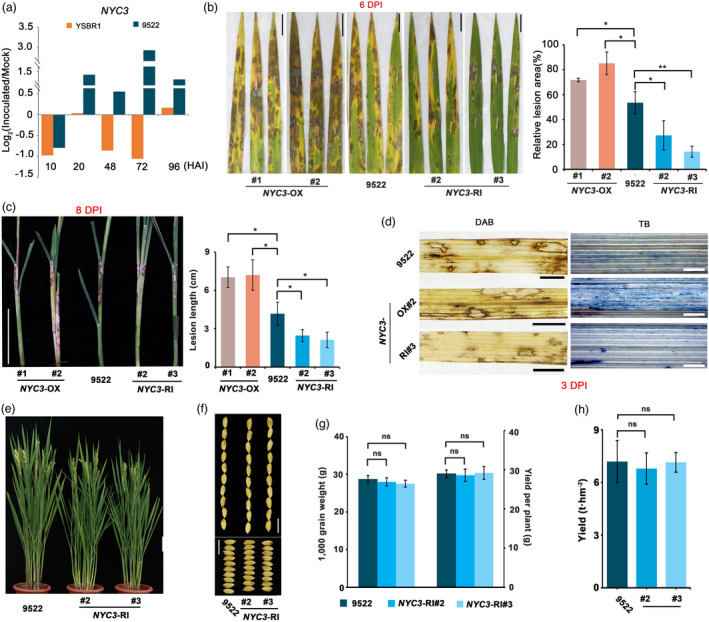
The effect of NYC3, a regulator of chlorophyll degradation, on rice ShB resistance and yield‐related traits. (a) NYC3 is more significantly induced by Rhizoctonia solani (R. solani) in variety 9522 than in YSBR1. Data are shown as means ± SD. The experiment was repeated three times with similar trends among each other. (b,c) NYC3‐RI and NYC3‐OX alter rice resistance to R. solani in detached leaf (b) and whole plant (c) inoculations. All transgenic lines are in the 9522 variety. Images were taken and data collected at 6 and 8 DPI, respectively. Scale bar, 2 cm in (b); 5 cm in (c). (d) DAB and TB staining for H2O2 accumulation and cell death of NYC3‐RI, NYC3‐OX and wild type 9522 at 3 DPI. Scale bar, 1 cm in left, 100 μm in right. (e–h) Effects of NYC3 silencing on the yield‐related agronomic traits. Pictures displayed the morphology of whole plant (e), grain length and width (f). (g) shown the 1000 grain weight and yield per plant, (h) shown the yield test. Scale bar, 10 cm in (e); 1 cm in (f). All data are presented as mean ± SEM; *P < 0.05; **P < 0.01; ns, not significant using Student’s t‐tests.
Because the enhanced resistance to R. solani achieved by down‐regulation of OsNYC3 in NYC3‐RI plants carries a potential value in application, we further examined these transgenic lines for their major agronomic traits in the field. We found that, except for plant height, the two NYC3‐RI lines shared almost the same agronomic traits, such as tiller number, seed setting rate, panicle length, grain length and width, 1000‐grain weight, with WT 9522 (Figure 6e–g, Figure S7). Most importantly, we found that these NYC3‐RI lines did not affect grain yield, giving similar yields as wild type 9522 in field tests (Figure 6h). For plant height, both NYC3‐RI lines are moderately shorter than the WT by approximately 4.0 cm (Figure S7a). Shorter rice plant height is often a preferred breeding target because it offers certain advantages, such as reduction in lodging problems. Thus, the shorter plant height trait of NYC3‐RI plants may be a desirable one while all other traits remain essentially unchanged. This indicates that OsNYC3 would be a new high‐potential target for engineering ShB‐resistant varieties.
Altering chlorophyll content not affects rice resistance to biotrophic and hemi‐biotrophic pathogens
In order to test if chlorophyll is essential for rice resistance to other pathogens, we employed Xoo and M. oryzae to inoculate two rice chlorophyll‐deficient mutants (ygl1 and dvr) and OsNYC3‐related transgenic lines. Biotrophic pathogen Xoo and hemi‐biotrophic pathogen M. oryzae are the causal agent of seriously global rice diseases, blight and blast, respectively. We found that all 6 M. oryzae isolates from Jiangsu province, China, could not infect WT control SH498 and two chlorophyll‐deficient mutants in its genetic background (Figure S8a). Three of them could infect OsNYC3 transgenic lines and its WT control 9522, but no difference on lesion size was observed among NYC3OX#1, NYC3RI#3 and the control 9522 (Figure S8a). With regard to Xoo, two strains were used and both could infect all plants tested, while no difference was found between all pairwise comparisons (Figure S8b). Together, these data suggest that altering chlorophyll content does not affect rice resistance to M. oryzae and Xoo.
Discussion
Comparing each time point with its mock control eliminates circadian genes and explains the discrepancy to previous transcriptomic data
Previously, Zhang et al. (2017) identified 4802 DEGs responsive to R. solani infection using Teqing and Lemont, and Yuan et al. (2018) reported 7624 DEGs responsive to R. solani infection. In contrast, we identified only 2632 DEGs responsive to R. solani infection, including 2429 in Lemont and 714 in YSBR1 (Table S2). Clearly the number of DEGs identified in this report is much less than previous reports. This discrepancy may be explained by our design which included more controls than previous reports as well as conducting inoculations all under natural conditions. Specifically, we included mock controls at both 10 and 20 HAI, whereas previous reports compared different R. solani‐infected time points all to time zero (Yuan et al., 2018; Zhang et al., 2017). Many of the DEGs identified in previous reports are probably not specifically induced by R. solani, but simply due to circadian rhythm. This could be validated by comparing the transcriptomic profiles of the two time points in either Lemont or YSBR1 without infection treatment. Comparing the two time points produced more than 2500 DEGs for each variety, demonstrating that circadian rhythm has a great effect on gene expression (Figure 2a). Also, our study used only leaf sheath tissues near the infected sites instead of nearly whole leaves or shoot tissues of infected plants previously reported (Karmakar et al., 2019; Suharti et al., 2016; Zhang et al., 2017). In addition, we grew and inoculated the plants all under natural conditions (Figure S9a), which was different from the previous studies that used a greenhouse or growth chamber (Ghosh et al., 2017; Karmakar et al., 2019; Prathi et al., 2018). Because of these designs, we obtained high‐quality transcriptomic data, reflected by high correlation coefficients among biological samples and RT‐qPCR validating data (Figure S1, Table S1).
Besides the above strict designs, the resistant YSBR1 used in this study should be the other reason leading to this small number of total DEGs, which produced much less DEGs than that in susceptible Lemont. At 20 HAI, most inoculation sites displayed yellowing and water‐soaked lesions in both varieties (Figure S9d), and the samples with similar diseased symptoms at this time point were collected for transcriptomic analysis. Therefore, we could exclude that much less DEGs of YSBR1 than that of Lemont at 20 HAI were caused by apparently different disease processes due to inconsistent inoculation amount or activities of inoculum. With respect to 10 HAI, however, due to no visible symptoms, we employed microscopy to observe the behaviour of infection hyphae on both varieties (Figure S9f), and which allowed us to choose the samples with similar infection behaviour of R. solani for transcriptomic analysis. Therefore, we could also exclude that the less DEGs of YSBR1 than that of Lemont at 10 HAI was caused by inconsistent inoculation amount or activities of inoculum. In addition, based on the similar growth status of six randomly selected inoculum on PDA plates (Figure S9c) and the data of cluster analysis and correlation coefficients using DEGs (Figure S1a, Table S1), we could further infer that inocula used in this study contained an almost same amount and/or activities of R. solani among each other. Together, we believe that the much less DEGs in YSBR1 than that in Lemont was due to an unknown resistant mechanism of YSBR1 to R. solani.
Chlorophyll reduction regulated by R. solani invasion probably represents an important susceptible response of host to this necrotrophic fungus
ET, JA and SA constitute the backbone of the plant defence signalling system triggering expression of various defence genes (Pieterse et al., 2009), which have been widely found to regulate resistance against R. solani (Karmakar et al., 2017; Molla et al., 2020). Here, we found that all the three signalling systems were significantly activated in both YSBR1 and Lemont after inoculation, although SA signalling was generally considered a defence response against biotrophic pathogens (Figure 2e). In addition, many defence‐related DEGs encoding RLK/RLPs, calcium signalling‐related proteins, GTP‐binding proteins, MAP kinases, HSFs, HSPs, WRKY TFs and NAC TFs were detected in response to R. solani in both varieties (Table S3), which provides a valuable information for future study. PR proteins, the most downstream component of plant defence functioning to directly battle against pathogens (Helliwell and Yang, 2013). We found that, nine out of 17 groups of PR proteins displayed remarkably elevated levels in both YSBR1 and Lemont after inoculation, but the levels in YSBR1 were significantly higher than those in Lemont (Figure 2d). A similar observation was noted for xylanase inhibitor proteins (XIPs) (Figure 2d), which mainly function to inhibit pathogen xylanases for reducing their ability to degrade cell walls (Zhan et al., 2017). All these together allowed us to infer that YSBR1 has a slightly stronger resistance response than Lemont.
Besides the known defence‐related DEGs, in the comparison of variety‐specific DEGs, we observed a striking difference that many DEGs associated with the functions of photosystems and ROS scavenging capacity were significantly suppressed in Lemont but not or only slightly suppressed in YSBR1 (Table S4). Inhibition of chlorophyll biosynthesis and acceleration of chlorophyll degradation will reduce plant photosynthesis, promoting ROS production and cell death or senescence (Ishikawa, 2005). Several previous studies have also found that photosynthesis is apparently affected by the invasion of R. solani as well as the destruction of chloroplast (Ghosh et al., 2017). Considering the fact that cell death is generally required for pathogenesis of necrotrophic pathogens, we infer that suppressing these processes will favour pathogen infection through accelerating cell death, but disfavour host resistance, which could be designated as the susceptible response of the host to the pathogen. In this study, we noted that many chlorophyll biosynthetic genes were greatly suppressed and chlorophyll catabolic genes elevated in Lemont, but only slightly affected in YSBR1 (Figure 3a). Through dark treatment, we found that suppression of chlorophyll biosynthesis and activation of chlorophyll degradation could significantly reduce YSBR1 resistance (Figure 4). Subsequently, we employed three chlorophyll biosynthesis‐deficient mutants (OsCAO1, OsDVR and OsYGL8) and found they all displayed higher susceptibility to R. solani than the wild type (Figure 5a). We further demonstrated similar results in a maize chlorophyll biosynthesis‐deficient mutant (ygl‐1) (Figure 5c). Specific defects in the biosynthesis of chlorophyll have been shown to reduce the ability of ROS scavenging (Yang et al., 2016). We found that compared with WT after inoculation, the three chlorophyll‐deficient mutants indeed accumulated more ROS, which should be the major factor resulting in cell death (Figure 5b). In addition, the combination of omics and histochemical staining results suggest that R. solani infection clearly suppresses ROS scavenging capacity in Lemont but not in YSBR1 (Figure 3b–d). This means that YSBR1 is able to withstand ROS due to a stronger ROS‐detoxifying or scavenging capacity. Together, we hypothesize that chlorophyll is required for maintaining the high resistance of YSBR1 to ShB, which is probably associated with ROS scavenging ability.
From the side of pathogen, we could infer that R. solani manipulates chlorophyll biosynthesis and degradation, and ROS scavenging processes to induce ROS burst and cell death for its invasion. As a result, after the initial infection, we think that R. solani could not easily induce cell death through manipulating chlorophyll‐related signals in YSBR1, and which affected its acquiring nutrients from dead cells and further infection. This could account for that the sparse, shrivelled and chapped infection hyphae on YSBR1 at 60‐72 HAI but not on susceptible Lemont (Figure 1c). The lesion length and areas were very slowly developed on YSBR1 after 2 DPI, which is consistent with this abnormal hyphal morphology (Figure 1c). Besides this possibility, other factors, like the accumulation of secondary metabolites and the thickness of cell wall, may also contribute to resisting R. solani invasion, and lastly results in this kind of abnormal hyphal morphology, which is worthy of being further investigated. In addition, whether this kind of hyphae behaviour related to ShB resistance still needs further validation using many more varieties as well as the detailed observation of infection structure.
Preventing R. solani‐induced chlorophyll reduction provides a practical tool for breeding ShB‐resistant varieties
Manipulating resistant response genes has been widely reported to improve rice ShB resistance, but few susceptible responsive genes were investigated for immunity against R. solani (Molla et al., 2020). Here, we have demonstrated that chlorophyll reduction is probably an important susceptible responsive of rice and maize during the infection of R. solani, and maintaining the chlorophyll content may play a universal role in protecting crops against necrotrophic pathogens like R. solani. Interestingly, we found that altering chlorophyll content did not affect rice resistance to biotrophic pathogen Xoo and hemi‐biotrophic pathogen M. oryzae, implying its specific role in crop resistance to necrotrophic pathogens. We guess this is probably due to that the immune responses in these mutants or transgenic lines are not constitutively activated, which is not like most lesion mimic mutants with constitutively activated immunity and chlorophyll‐deficient phenotype. In total, this discovery provides a novel target or strategy for developing ShB‐resistant varieties through manipulation of chlorophyll degradation‐related genes. We found that R. solani infection significantly induces expression of the OsNYC3 gene in susceptible varieties Lemont and 9522 but not or only slightly changed in YSBR1 (Figures 3a and 6a). We then developed transgenic overexpression and RNAi lines of OsNYC3 in the background of 9522 (Figure S6). The OsNYC3 overexpression lines significantly enhanced ShB susceptibility and cell death after R. solani infection, while the RNAi lines evidently increased ShB resistance and reduced cell death (Figure 6b–d). More interestingly, we found that, except for a slight reduction of plant height, the NYC3‐RI lines are not significantly different from wild type plants on important breeding traits in field tests (Figure 6e–h, Figure S7). This means that OsNYC3 is an excellent target for rice breeding against ShB resistance, considering that overexpression or down‐regulation of many genes generally carries detrimental effects on rice development, especially on economically important traits (Xue et al., 2016). Consequently, our findings provide not only a new target for developing ShB‐resistant varieties but also new insights into the interaction mechanism between crop and the necrotrophic fungus R. solani.
Methods
Plant materials
Rice variety YSBR1, developed by pedigree breeding from the progeny of a japonica/indica hybrid, displayed a high resistance level to rice ShB disease in both field and growth chamber (Zuo et al., 2009). Rice variety Lemont, a japonica rice cultivar from Louisiana, USA, was highly susceptible to ShB disease.
Both ygl8 and dvr plants were natural mutants selected from an indica variety Shuihui498 (SH498). The cao1 mutant was described by Yang et al. (2014). The maize mutant ygl‐1 was described by Guan et al. (2016).
For the generation of the OsNYC3‐RI construct, the partial exon 1 and 2 of OsNYC3 containing was amplified from japonica rice Nipponbare cDNA using primers RI‐F (5′‐AAAGGATCCACAATAGCAAGGCACCG‐3′) and RI‐R (5′‐AAAACTAGTA GCGAGGAGATGTAGCAG‐3′). The target fragment was then cloned into the p1022 vector and transferred into the plant binary vector p1301UbiNOS expressed under the control of the maize ubiquitin promoter. To generate the OsNYC3‐OX construct, the coding region of OsNYC3 was amplified from Nipponbare cDNA using OX‐F (5′‐AAAACTAGTATGGAAGTGGTTTCTT CCAGCCACTC‐3′) and OX‐R (5′‐AAAGAGCTCTTATCTAGATATTACCCA TGTGTTGGA‐3′) inserted into the p1301UbiNOS vector. All the constructs were transformed into japonica variety 9522 via Agrobacterium tumefaciens‐mediated transformation.
Culture of fungal isolate
R. solani AGI‐1A isolate RH‐9 isolated from rice (Zuo et al., 2013), with moderate pathogenicity, was used to inoculate rice and maize. R. solani strain YWK196 isolated from maize was used to inoculate maize (Li et al., 2019). The isolates were cultured on potato dextrose agar (PDA) medium at 26 °C for 3 days. Mycelial discs (c. 0.7 cm diameter) were transformed into potato dextrose broth (PDB) medium containing autoclaved, truncated thin toothpicks with a length of c. 1.0 cm at 26 °C until the mycelium twined around the toothpicks. The toothpicks with mycelia were used as the inoculum (Figure S9b). We randomly re‐cultured six inocula in fresh PDA plates and found that their growing status was very similar at 24 and 48 HAI respectively (Figure S9c), which indicates that the amount of activities of the inocula used in this study was very close.
Sampling for microarray analysis
Rice plants were sown in pots (12 cm diameter, 20 cm high) and all grew in a pool with water under natural conditions (Figure S9a). Pesticides and fungicides were sprayed at 10 days before inoculation for ensuring plants were healthy without disease or pests. Plants at similar developmental progress, like the same tiller number and height, were selected for inoculation. A toothpick inoculum colonized with R. solani was placed at the third leaf sheath from the top, as described in detail by Zuo et al. (2013). Toothpicks without pathogens were used to inoculate plants as mock controls. Considering the relatively high humidity in the evening, the inoculation was conducted at 8 p.m. and kept under natural conditions.
The leaf sheath tissues at 1.0 cm away from the inoculation site with a total length of around 4.0 cm were harvested at 10 and 20 HAI for subsequent transcriptomic analysis. At 20 HAI, most inoculation sites displayed yellowing and water‐soaked lesions (Figure S9d), while no visible symptoms could be observed at 10 HAI. Therefore, to ensure the 10 HAI samples receive successful inoculation, we collected and stained its corresponding inoculation site samples by lacto‐phenol cotton blue (Figure S9e, f), and checked by microscopy for the formation of infection cushion, a specialized infection structure containing distinct hyphal aggregates (Figure S9f). Three biological replicates were conducted at 10 and 20 HAI for transcriptomic analysis, and the remaining inoculated plants were kept for confirmation of their differences in ShB resistance (Figure S9g–i).
Transcriptome profiling and statistical analysis
Transcriptome profiling was performed according to the standard protocol of the Affymetrix Rice Genome Array (CapitalBio). Differentially expressed genes were selected using | Log2 Fold change | ≥0.75 and q‐value < 0.1 as the cut‐offs and analysed with DAVID (https://david.ncifcrf.gov), GO (http://geneontology.org) and KEGG (http://www.kegg.jp/kegg) for significant functional enrichment (Huang et al., 2009; Kanehisa et al., 2019; Mi et al., 2019). Heat maps were generated with R package to show gene expression levels.
ShB fungus inoculation and disease scoring
All plants were grown in plots with sterilized soil in greenhouse till adult developmental stages (around 8 weeks), and then were transferred to growth chamber with a relative humidity of 45%–60%, a photoperiod of 13 h (30 °C) : 11 h (26 °C), light : dark for 2 days before inoculation. Five tillers in each plant were inoculated by inserting the toothpick inoculums into the third leaf sheath from the top. Lesion lengths and areas were measured with a ruler at the designed times. Disease index per variety, calculated by the average of lesion lengths or areas with five replicates per line with three plants each replicate, was used to represent the resistance level of the variety.
Field inoculation and disease scoring method used to evaluate ShB resistance at adult stage was as described by Zuo et al. (2013). The disease severity was rated approximately 30 days after heading. Disease index per variety, calculated by the average disease score of 30 plants from three replications, was used to represent the resistant degree.
Detached leaf inoculation method as described by Jia et al. (2013) was also conducted on rice and maize lines. The average of relative lesion areas of 10 leaves per line was used to represent the resistance level of the variety. The experiment was repeated three times. Disease index expressed as the relative lesion area is expressed as the ratio of Lesion area: Leaf area calculated from digital photographs using the measuring tools of Adobe Photoshop CS6 (Basu et al., 2016).
Histological observation of rice in response to R. solani
Scanning electron microscope (XL‐30E SEM, Philips, Amsterdam, the Netherlands) was used for observation of the hyphal behaviour; samples were prepared as described by Sun et al. (2019). The lacto‐phenol cotton blue staining and preparation of inoculated sheaths at 10 HAI were performed as described by Tiwari et al. (2017), and light microscope (Leica DMLS, 20×‐100×) was used for observation of the hyphal development. 3,3′‐Diaminobenzidine (DAB) staining of rice mutants, transgenic lines and WT leaves was conducted using a previously described protocol (Yang et al., 2017). In all these experiments, the toothpick inoculum described above was used. Particularly, in DAB staining assay for YSBR1 and Lemont, we adopted mycelial suspension as the inoculum, which was obtained by resuspending the smashed mycelia with ddH2O. When inoculation, the mycelial suspension was injected into the leaf sheath. This kind of inoculation may lead to a much slower penetration and colonization of the tissue and more dispersedly tiny infection sites on leaf sheath compared to the toothpick inoculum covered by more intact mycelium. It allowed us to more easily compare the difference between the two varieties in response to R. solani at more early stage. After inoculation, the observation was performed as the description in De Vleesschauwer et al. (2006), and the images were taken with a light microscope. Cell death was visualized using trypan blue (TB) staining according to the protocol of Dietrich et al. (1994). At least five plants were stained at the same time for each genotype. All pictures were taken by the camera (Canon).
RNA extraction and quantitative RT‐PCR (qRT‐PCR)
Total RNA was extracted from liquid‐nitrogen‐frozen sheath samples using the TRizol reagent (Invitrogen, Carlsbad, CA). First‐strand cDNA was synthesized from 2 μg of purified total RNA following the manufacturer's protocol (Primescript™ First‐Strand cDNA Synthesis kit; Takara, Dalian, China). RT‐qPCR was performed on CFX96TM Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA) using SYBR Premix ExTaq Ⅱ kit (Takara) with gene‐specific primers (Table S5). The rice ubiquitin (UBQ) transcript was used as an internal control to quantify relative transcript levels.
Evaluation of agronomic traits in field
WT (9522) and OsNYC3‐RI lines were grown in fields at Yangzhou (Jiangsu) with three replicates/plots per line. Each plot occupied 30 m2 with 5 m in width and 6 m in length. The distances between two plants and between rows were 13.3 and 25 cm, respectively. The middle 10 plants in the central row of each plot were selected for measuring agronomic traits at the maturing stage. Various parameters of agronomic traits such as plant height, tiller number, panicle length, grain width and length, seed‐setting rate, 1000 grain weight and yield per plant. The seed‐setting rate was calculated as: (Total number of filled grains per main panicle/total number of spikelets per main panicle) × 100. Plot yields were determined when the seeds were harvested, and then normalized to yield per 10 000 m2 (hm2).
Transmission electron microscopy
Leaves (second from the top) of 8‐week old YSBR1 and Lemont were prepared for transmission electron microscopy. The leaf segments were fixed in a 2.5% glutaraldehyde solution and then in 1% osmic acid solution. Samples were prepared as described previously (Qiu et al., 2018), and were observed using a JEOL JEM‐1010 electron microscope (Tokyo, Japan).
Chlorophyll measurements
For pigment extraction, leaf tissues of 3‐week‐old ygl8, dvr and cao1 mutants and wild‐type (SH498; 93‐11) grown in the greenhouse were homogenized and resuspended in ice‐cold 80% acetone in dim green light. Residual plant debris were removed by centrifugation. Supernatants were analysed with a visible‐light spectrophotometer and chlorophyll contents were determined using the equation of Lichtenthaler (1987). Total chlorophyll was normalized to leaf fresh weight.
Dark treatment
YSBR1 and Lemont were grown in a growth chamber (13 h : 11 h, light : dark). Then, at the tillering stage, one‐half of the plants were treated with darkness for 3 days to suppress chlorophyll biosynthesis and induce chlorophyll degradation, and the other half of plants without darkness treatment were used as control. The experiment was repeated three times with 12 plants per replication for both dark treatment and control. For detached leaf inoculation assay, the second leaves from the top of YSBR1 and Lemont were harvested in the field. All leaves were placed on wet filter papers, and half of them were treated with darkness for 3 days. The experiment was performed three replications with 10 leaves in each replication for both dark treatment and control. After treatment, all plants or leaves were inoculated with R. solani using the method described above.
Magnaporthe oryzae inoculation
M. oryzae isolates, JD19‐1‐1, YN670, WJA‐2, ZJB‐4, GYC‐3 and HYE‐5, were used for inoculation. All isolates were cultured on complete medium (CM) for 2 weeks in dark at 28°C, and then exposed to fluorescent lights for 1 week at room temperature for sporulation. Spore concentration was adjusted to 5 × 105 spores/mL with a haemocytometer before spores were applied by punch inoculation (Park et al., 2012). Lesions were photographed at 7 DPI. Disease index per variety, the average lesion lengths of three replicates per line with 10 leaves per replicate, was used to represent the resistance level of the variety.
Xoo inoculation
To evaluate the resistance of rice bacterial blight disease, Xoo strain PX099 and PX099a were grown in NA media. Rice plants at the booting stage were inoculated using the leaf‐clipping method (Sun et al., 2004). Disease phenotype was photographed at 15 DPI. Disease index per variety, the average lesion lengths of three replicates per line with 10 leaves per replicate, was used to represent the resistance level of the variety.
Conflicts of interest
The authors have declared no conflict of interest.
Author contributions
S.Z. and W.C. conceived the project. W.C. analysed the data, performed most of the experiments and drafted the manuscript with other co‐authors. H.Z. performed R. solani inoculation, phenotypic scorings and histochemical staining of genetic materials; Y.Z. constructed the NYC3‐transgenic lines; J.Z. performed the effect of dark‐treatment on ShB resistance of YSBR1, and provided R language support; L.Y. performed the scanning electron microscopy; De Vleesschauwer performed histochemical staining. H.G generated ygl‐1 maize mutant; X.Q and S.L. performed blast and bacterial blight inoculation; G.W., W.S., Z.L. and X.S. provided field management and phenotypic scoring; J.G conducted yield test of NYC3‐transgenic lines; M.G. generated ygl8, dvr, and cao1 rice mutants; X.C., Z.F., Z.C., Y.Z., X.P., W.L., G.L. C.Y, K.H. and Q.L. supervised aspects of the project. S.Z. designed experiments, interpreted data and revised the manuscript.
Supporting information
Figure S1 Quality control of microarray data.
Figure S2 Functional categories of unique genes in each variety.
Figure S3 Photosynthesis‐related genes were responsive to R. solani.
Figure S4 Difference between YSBR1 and Lemont exists in chlorophyll content, rather than chloroplast structure.
Figure S5 Phenotypic characterization of green‐yellow leaf mutants in rice and maize.
Figure S6 The validation of NYC3 transgenic plants.
Figure S7 Effects of NYC3 silencing on other yield‐related agronomic traits.
Figure S8 The correlation of chlorophyll with rice resistance to Magnaporthe oryzae (M. oryzae) and Xanthomonas oryzae pv. oryzae (Xoo).
Figure S9 Experimental design and execution for the multi‐omics analyses.
Table S1 The correlation coefficient between biological replicates of each detected group.
Table S2 List of differentially expressed probes of YSBR1 and Lemont under R. solani infection.
Table S3 List of differentially expressed genes (DEGs) involving with general defence response.
Table S4 List of DEGs involving a photosynthetic and antioxidant defence system.
Table S5 qRT‐PCR primers used in the present study.
Acknowledgements
The authors are grateful to Prof. Zhaohui Chu (Shandong Agricultural University, China) for providing R. solani strain YWK196 isolated from maize. This research was supported by the grants from the National Key Research and Development Program of China (2016YFD0100601), the Natural Science Foundation of China (31672013), Fok Ying Tung Education Foundation (151026), China Postdoctoral Science Foundation (2015M581871), Jiangsu Key Research and Development Program for modern agriculture (BE2019339) and a Program Development of Jiangsu Higher Education Institutions (PAPD), respectively.
Cao, W. , Zhang, H. , Zhou, Y. , Zhao, J. , Lu, S. , Wang, X. , Chen, X. , Yuan, L. , Guan, H. , Wang, G. , Shen, W. , De Vleesschauwer, D. , Li, Z. , Shi, X. , Gu, J. , Guo, M. , Feng, Z. , Chen, Z. , Zhang, Y. , Pan, X. , Liu, W. , Liang, G. , Yan, C. , Hu, K. , Liu, Q. and Zuo, S. (2022) Suppressing chlorophyll degradation by silencing OsNYC3 improves rice resistance to Rhizoctonia solani, the causal agent of sheath blight. Plant Biotechnol. J., 10.1111/pbi.13715
References
- Basu, A. , Chowdhury, S. , Chaudhuri, T.R. and Kundu, S. (2016) Differential behaviour of sheath blight pathogen Rhizoctonia solani in tolerant and susceptible rice varieties before and during infection. Plant. Pathol. 65, 1333–1346. [Google Scholar]
- Chen, H. , Li, C. , Liu, L. , Zhao, J. , Cheng, X. , Jiang, G. and Zhai, W. (2016) The Fd‐GOGAT1 mutant gene lc7 confers resistance to Xanthomonas oryzae pv. oryzae in rice. Sci Rep. 6, 26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer, D. , Cornelis, P. and Hofte, M. (2006) Redox‐active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol. Plant Microbe Interact. 19, 1406–1419. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A. , Delaney, T.P. , Uknes, S.J. , Ward, E.R. , Ryals, J.A. and Dangl, J.L. (1994) Arabidopsis mutants simulating disease resistance response. Cell, 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Faris, J.D. , Zhang, Z. , Lu, H. , Lu, S. , Reddy, L. , Cloutier, S. , Fellers, J.P. et al. (2010) A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proc. Natl Acad. Sci. USA, 107, 13544–13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, R.C. , Kidd, B.N. , Hane, J.K. , Anderson, J.P. and Singh, K.B. (2016) Reactive oxygen species play a role in the infection of the necrotrophic fungi, Rhizoctonia solani in wheat. PLoS ONE, 11, e0152548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Zhang, C. , Han, X. , Wang, Z.Y. , Ma, L. , Yuan, P. , Wu, J.N. et al. (2018) Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 19, 2149–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S. , Kanwar, P. and Jha, G. (2017) Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani . Sci. Rep. 7, 41610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H. , Xu, X. , He, C. , Liu, C. , Liu, Q. , Dong, R. , Liu, T. et al. (2016) Fine mapping and candidate gene analysis of the leaf‐color gene ygl‐1 in maize. PLoS ONE, 11, e0153962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugrud, A.R.P. , Zhang, Z. , Richards, J.K. , Friesen, T.L. and Faris, J.D. (2019) Genetics of variable disease expression conferred by inverse gene‐for‐gene interactions in the wheat‐Parastagonospora nodorum pathosystem. Plant Physiol. 180, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, E.E. , Wang, Q. and Yang, Y. (2013) Transgenic rice with inducible ethylene production exhibits broad‐spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani . Plant Biotechnol. J. 11, 33–42. [DOI] [PubMed] [Google Scholar]
- Helliwell, E.E. and Yang, Y. (2013) Molecular strategies to improve rice disease resistance. Methods Mol. Biol. 956, 285–309. [DOI] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. and Lempicki, R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ishikawa, A. (2005) Tetrapyrrole metabolism is involved in lesion formation, cell death, in the Arabidopsis lesion initiation 1 mutant. Biosci. Biotechnol. Biochem. 69, 1929–1934. [DOI] [PubMed] [Google Scholar]
- Jarvis, P. and Lopez‐Juez, E. (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Jia, Y. , Liu, G. , Costanzo, S. , Lee, S. and Dai, Y. (2009) Current progress on genetic interactions of rice with rice blast and sheath blight fungi. Front Agric China, 3, 231–239. [Google Scholar]
- Jia, Y. , Liu, G. , Park, D.S. and Yang, Y. (2013) Inoculation and scoring methods for rice sheath blight disease. Methods Mol. Biol. 956, 257–268. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Sato, Y. , Furumichi, M. , Morishima, K. and Tanabe, M. (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariola, T. , Brader, G. , Li, J. and Palva, E.T. (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell, 17, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar, S. , Datta, K. , Molla, K.A. , Gayen, D. , Das, K. , Sarkar, S.N. and Datta, S.K. (2019) Proteo‐metabolomic investigation of transgenic rice unravels metabolic alterations and accumulation of novel proteins potentially involved in defence against Rhizoctonia solani . Sci. Rep. 9, 10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar, S. , Molla, K.A. , Das, K. , Sarkar, S.N. , Datta, S.K. and Datta, K. (2017) Dual gene expression cassette is superior than single gene cassette for enhancing sheath blight tolerance in transgenic rice. Sci. Rep. 7, 7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P. , Xue, C.Y. , Song, H.D. , Gao, Y. , Feng, L. , Li, Y. and Xuan, Y.H. (2020) Tissue‐specific activation of DOF11 promotes rice resistance to sheath blight disease and increases grain weight via activation of SWEET14. Plant Biotechnol J. 9, 409–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.S. , Devonshire, B.J. , Hammond‐Kosack, K.E. , Rudd, J.J. and Kanyuka, K. (2015) Deregulation of plant cell death through disruption of chloroplast functionality affects asexual sporulation of Zymoseptoria tritici on wheat. Mol. Plant Microbe Interact. 28, 590–604. [DOI] [PubMed] [Google Scholar]
- Li, N. , Lin, B. , Wang, H. , Li, X. , Yang, F. , Ding, X. , Yan, J. et al. (2019) Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. [Google Scholar]
- Liu, Z. , Yan, H. , Wang, K. , Kuang, T. , Zhang, J. , Gui, L. , An, X. et al. (2004) Crystal structure of spinach major light‐harvesting complex at 2.72 A resolution. Nature, 428, 287–292. [DOI] [PubMed] [Google Scholar]
- Mach, J.M. , Castillo, A.R. , Hoogstraten, R. and Greenberg, J.T. (2001) The Arabidopsis‐accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl Acad. Sci. USA, 98, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , Ebert, D. , Huang, X. and Thomas, P.D. (2019) PANTHER version 14: more genomes, a new PANTHER GO‐slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla, K.A. , Karmakar, S. , Molla, J. , Bajaj, P. , Varshney, R.K. , Datta, S.K. and Datta, K. (2020) Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol J. 18, 895–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, R. , Sato, Y. , Masuda, Y. , Nishimura, M. and Kusaba, M. (2009) Defect in non‐yellow coloring 3, an alpha/beta hydrolase‐fold family protein, causes a stay‐green phenotype during leaf senescence in rice. Plant J. 59, 940–952. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Aubry, S. , Mondhe, M. , Kingston‐Smith, A. , Gallagher, J. , Timms‐Taravella, E. , James, C. et al. (2010) Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis . New Phytol. 188, 161–174. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.Y. , Liao, L.L. , Liu, S. , Nie, M.M. , Li, J. , Zhang, L.D. , Ma, J.F. et al. (2019) Magnesium deficiency triggers SGR‐mediated chlorophyll degradation for magnesium remobilization. Plant Physiol. 18, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pitsili, E. , Phukan, U.J. and Coll, N.S. (2019) Cell death in plant immunity. Cold Spring Harb. Perspect. Biol. 12, a036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathi, N.B. , Palit, P. , Madhu, P. , M, R. , Laha, G.S. , Balachandran, S.M. , Madhav, M.S. et al. (2018) Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani . Plant Physiol. Biochem. 130, 258–266. [DOI] [PubMed] [Google Scholar]
- Qiao, L. , Zheng, L. , Sheng, C. , Zhao, H. , Jin, H. and Niu, D. (2020) Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J. 102, 948–964. [DOI] [PubMed] [Google Scholar]
- Qiu, Z. , Chen, D. , He, L. , Zhang, S. , Yang, Z. , Zhang, Y. , Wang, Z. et al. (2018) The rice white green leaf 2 gene causes defects in chloroplast development and affects the plastid ribosomal protein S9. Rice (N Y), 11, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Mazumdar, P. , Harikrishna, J.A. and Babu, S. (2019) Sheath blight of rice: a review and identification of priorities for future research. Planta, 250, 1387–1407. [DOI] [PubMed] [Google Scholar]
- Suharti, W.S. , Nose, A. and Zheng, S.‐H. (2016) Metabolite profiling of sheath blight disease resistance in rice: in the case of positive ion mode analysis by CE/TOF‐MS. Plant Product. Sci. 19, 279–290. [Google Scholar]
- Sun, W. , Xu, X.H. , Li, Y. , Xie, L. , He, Y. , Li, W. , Lu, X. et al. (2019) OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol. 226, 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Cao, Y. , Yang, Z. , Xu, C. , Li, X. , Wang, S. and Zhang, Q. (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase‐like protein. Plant J. 37, 517–527. [DOI] [PubMed] [Google Scholar]
- Tanaka, R. and Tanaka, A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. [DOI] [PubMed] [Google Scholar]
- Tiwari, I.M. , Jesuraj, A. , Kamboj, R. , Devanna, B.N. , Botella, J.R. and Sharma, T.R. (2017) Host delivered RNAi, an efficient approach to increase rice resistance to sheath blight pathogen (Rhizoctonia solani). Sci. Rep. 7, 7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Tan, J. , Wu, Z. , VandenLangenberg, K. , Wehner, T.C. , Wen, C. , Zheng, X. et al. (2018) STAYGREEN, STAY HEALTHY: a loss‐of‐susceptibility mutation in the STAYGREEN gene provides durable, broad‐spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 221, 415–430. [DOI] [PubMed] [Google Scholar]
- Woo, H.R. , Kim, H.J. , Lim, P.O. and Nam, H.G. (2019) Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. [DOI] [PubMed] [Google Scholar]
- Xue, X. , Cao, Z. , Zhang, X. , Wang, Y. , Zhang, Y. , Chen, Z. , Pan, X. et al. (2016) Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis. 100, 1634–1642. [DOI] [PubMed] [Google Scholar]
- Yamatani, H. , Sato, Y. , Masuda, Y. , Kato, Y. , Morita, R. , Fukunaga, K. , Nagamura, Y. et al. (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll – protein complexes during leaf senescence. Plant J. 74, 652–662. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Li, W. , Cao, J. , Meng, F. , Yu, Y. , Huang, J. , Jiang, L. et al. (2017) Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. [DOI] [PubMed] [Google Scholar]
- Yang, H.L. , Liu, M. , Guo, M. , Li, R.D. , Zhang, H.G. and Yan, C.J. (2014) Genetic analysis and position cloning of a yellow green leaf 10 (ygl10) Gene, responsible for leaf color in rice. Chinese J. Rice Sci. 28, 41–48. [Google Scholar]
- Yang, Y. , Xu, J. , Huang, L. , Leng, Y. , Dai, L. , Rao, Y. , Chen, L. et al. (2016) PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 67, 1297–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z.J. , Zhang, Y. , Xu, G.J. , Bi, D.L. , Qu, H.Y. , Zou, X.W. , Gao, X.Q. et al. (2018) Comparative transcriptome analysis of Rhizoctonia solani‐resistant and ‐susceptible rice cultivars reveals the importance of pathogen recognition and active immune responses in host resistance. J. Plant Biol. 61, 143–158. [Google Scholar]
- Zhan, Y. , Sun, X. , Rong, G. , Hou, C. , Huang, Y. , Jiang, D. and Weng, X. (2017) Identification of two transcription factors activating the expression of OsXIP in rice defence response. BMC Biotechnol. 17, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Chen, L. , Fu, C. , Wang, L. , Liu, H. , Cheng, Y. , Li, S. et al. (2017) Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front. Plant Sci. 8, 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, S. , Wang, Z. , Chen, X. , Gu, F. , Zhang, Y. , Chen, Z. and Pan, X. (2009) Evaluation of resistance of a novel rice germplasm YSBR1 to sheath blight. Acta Agronomica Sin. 35, 608–614. [Google Scholar]
- Zuo, S. , Yin, Y. , Pan, C. , Chen, Z. , Zhang, Y. , Gu, S. , Zhu, L. et al. (2013) Fine mapping of qSB‐11(LE), the QTL that confers partial resistance to rice sheath blight. Theor. Appl. Genet. 126, 1257–1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Quality control of microarray data.
Figure S2 Functional categories of unique genes in each variety.
Figure S3 Photosynthesis‐related genes were responsive to R. solani.
Figure S4 Difference between YSBR1 and Lemont exists in chlorophyll content, rather than chloroplast structure.
Figure S5 Phenotypic characterization of green‐yellow leaf mutants in rice and maize.
Figure S6 The validation of NYC3 transgenic plants.
Figure S7 Effects of NYC3 silencing on other yield‐related agronomic traits.
Figure S8 The correlation of chlorophyll with rice resistance to Magnaporthe oryzae (M. oryzae) and Xanthomonas oryzae pv. oryzae (Xoo).
Figure S9 Experimental design and execution for the multi‐omics analyses.
Table S1 The correlation coefficient between biological replicates of each detected group.
Table S2 List of differentially expressed probes of YSBR1 and Lemont under R. solani infection.
Table S3 List of differentially expressed genes (DEGs) involving with general defence response.
Table S4 List of DEGs involving a photosynthetic and antioxidant defence system.
Table S5 qRT‐PCR primers used in the present study.


