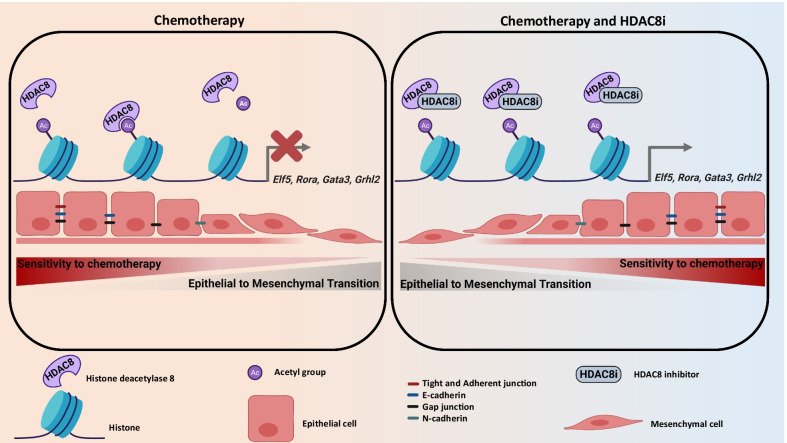
Abstract
Background
Basal-like breast cancer (BLBC) is one of the most aggressive malignant diseases in women with an increased metastatic behavior and poor prognosis compared to other molecular subtypes of breast cancer. Resistance to chemotherapy is the main cause of treatment failure in BLBC. Therefore, novel therapeutic strategies counteracting the gain of aggressiveness underlying therapy resistance are urgently needed. The epithelial-to-mesenchymal transition (EMT) has been established as one central process stimulating cancer cell migratory capacity but also acquisition of chemotherapy-resistant properties. In this study, we aimed to uncover epigenetic factors involved in the EMT-transcriptional program occurring in BLBC cells surviving conventional chemotherapy.
Results
Using whole transcriptome data from a murine mammary carcinoma cell line (pG-2), we identified upregulation of Hdac4, 7 and 8 in tumor cells surviving conventional chemotherapy. Subsequent analyses of human BLBC patient datasets and cell lines established HDAC8 as the most promising factor sustaining tumor cell viability. ChIP-sequencing data analysis identified a pronounced loss of H3K27ac at regulatory regions of master transcription factors (TFs) of epithelial phenotype like Gata3, Elf5, Rora and Grhl2 upon chemotherapy. Interestingly, impairment of HDAC8 activity reverted epithelial-TFs levels. Furthermore, loss of HDAC8 activity sensitized tumor cells to chemotherapeutic treatments, even at low doses.
Conclusion
The current study reveals a previously unknown transcriptional repressive function of HDAC8 exerted on a panel of transcription factors involved in the maintenance of epithelial cell phenotype, thereby supporting BLBC cell survival to conventional chemotherapy. Our data establish HDAC8 as an attractive therapeutically targetable epigenetic factor to increase the efficiency of chemotherapeutics.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-022-01228-4.
Keywords: HDAC8, MET, EMT, BLBC, TNBC, Chemotherapy, Epigenetics, H3K27ac, Epithelial transcription factors
Background
Triple-negative breast cancer (TNBC) is considered one of the most aggressive breast cancer (BC) subtypes. TNBC accounts for 10–15% of all BC and tends to be more common in younger women (under 40 years old) with an African-American origin bearing a BRCA1 mutation [1]. TNBCs distinguish themselves from other histological BC subtypes by the absence of estrogen receptor (ER), progesterone receptor (PR) expression and human epidermal growth factor 2 (HER2) gene amplification, as assessed by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH), respectively [2]. This characteristic makes the group of TNBC lesions largely overlapping with the basal-like (BLBC) and to a much lower extend with normal-like molecular subtypes [1, 3], thereby rendering this type of malignancies insensitive to hormone and anti-HER2-targeted therapies. Consequently, therapeutic options for TNBC are relatively scarce, mostly limited to surgical removal of the lesion eventually combined with radiotherapy and/or chemotherapy. Here, anthracycline, cyclophosphamide, taxanes or platinum salts are most commonly selected and often used in combination [3, 4]. Unfortunately, the efficiency of such therapies frequently declines because of adaptations of the tumor cells and rapid acquisition of chemoresistant traits. Patients with relapses or residual disease have a high probability to develop metastatic outgrowth and subsequently succumb to their disease [2].
Epithelial–mesenchymal transition (EMT) and the reverse mesenchymal-to-epithelial transition (MET) are both indispensable for various processes in multicellular organisms, like during embryogenesis, wound repair, placentation or inflammation [5–7]. However, increasing evidence established a strong implication of the EMT also in the phenomenon of therapy resistance, cancer cell dissemination and distant organ colonization [8–10]. Interestingly, deep involvement of epigenetic mechanisms in the cellular plasticity of tumor cells is nowadays well established [11]. Due to the reversible nature of these pathological alterations, targeting epigenetic factors controlling EMT and MET processes represent an interesting strategy to revert aggressive chemotherapy-resistant metastatic phenotypes [12]. Histone deacetylases (HDACs) are epigenetic effectors categorized in four classes based on their structure, enzymatic function and subcellular localization. HDACs enact a major gene silencing role to regulate multiple gene expression programs involved in, among others, inflammation, cell proliferation, cancer stemness and EMT processes via de-acetylating histones [13].
The WAP-T mammary carcinoma mouse model was designed to investigate processes involved in the progression and metastasis of basal-like malignancies (22–27). We previously reported that WAP-T mammary carcinomas cells treated with conventional cytotoxic therapy develop strategies to survive the treatment in vivo. Interestingly, we identified in the surviving tumor cells pronounced mesenchymal and stem cells specific features that are characteristic for more aggressive phenotypes. These cells showed an increased tendency to disseminate to distant organs (28). To get a better understanding of the mechanisms allowing dynamic gene expression changes responsible for the EMT-driven refractory response to chemotherapy, we designed a combined high-throughput mRNA- and ChIP-sequencing (mRNA-seq and ChIP-seq) approach and analyzed epigenetic changes occurring in a murine mammary carcinoma cell line (pG-2) surviving conventional cytotoxic chemotherapy. We identified three upregulated HDACs, Hdac4, Hdac7 and Hdac8. Interestingly, BLBC patients with high HDAC8 expression demonstrated the poorest prognosis. Our results showed that HDAC8 promotes EMT program and therapy resistance by inhibiting the expression of gatekeeper transcription factors of the epithelial phenotype (Elf5, Gata3, Rora and Grhl2). Accordingly, the combination of conventional chemotherapies with HDAC8 silencing or HDAC8-inhibition considerably potentiated the therapeutic effectiveness. In summary, we identified HDAC8 as a promising targetable epigenetic regulator to enhance the effectiveness of BLBC treatments.
Results
Chemotherapy induces EMT in murine and human BLBCs
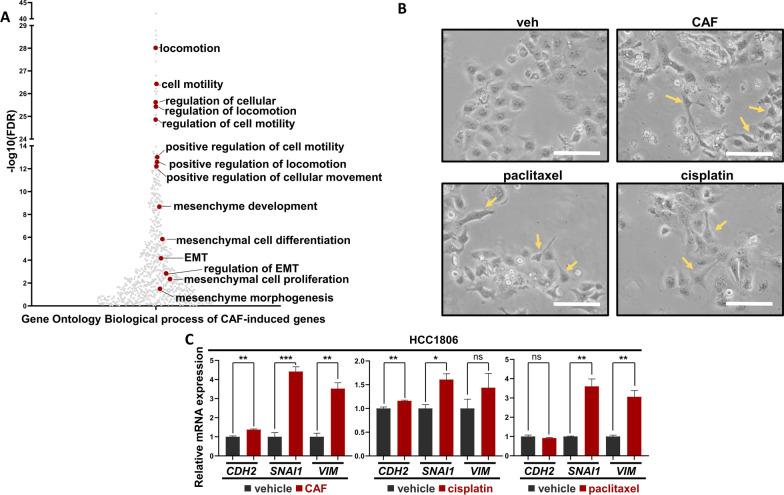
In the past, we and others have shown that BLBC cells activate the EMT transcriptional program to survive conventional chemotherapy [14–16]. In a recent study, we leveraged the WAP-T mammary carcinoma mouse model system (parental G-2 cells; pG-2) to study the mechanisms underlying resistance to combined cytotoxic chemotherapy (cyclophosphamide, adriamycin, 5-fluorouracil; short CAF) and confirmed the enrichment of EMT-associated features along with the disease progression (Fig. 1A), [17, 18]. Hence, we sought to validate the activation of the EMT transcriptional program in a human BLBC cell line (HCC1806) upon different conventional chemotherapy treatments (CAF, cisplatin and paclitaxel). Indeed, we observed changes of cell morphology typically observed during the acquisition of mesenchymal traits in treated cells (Fig. 1B). Furthermore, an increase of typical EMT markers CDH2, SNAI1 and VIM expression was observed in all treated cells, confirming the activation of the EMT-transcriptional program upon chemotherapy (Fig. 1C).
Fig. 1.
Chemotherapy induces EMT in murine and human BLBC cells. A Scatter plot showing EMT signatures, using the online pathway enrichment analysis tool gProfiler, on pG-2 cells upon CAF treatment for 48 h. B Brightfield pictures of HCC1806 cells showing a mesenchymal phenotype upon different chemotherapies (CAF, cisplatin and paclitaxel) for 48 h. The yellow arrows indicate cells with pronounced mesenchymal morphology. White scale bars = 50 µm, C Quantitative real-time PCR (qRT-PCR) on EMT-related genes in vehicle- and CAF-, cisplatin- or paclitaxel-treated (for 48 h) HCC1806 cells. All experiments were performed in biological triplicate. ns = not significant, *p-val < 0.05, **p-val < 0.01, ***p-val < 0.005. C Student's t test. Error bars are standard error of the mean (SEM)
BLBC cells surviving chemotherapy upregulates HDAC4, HDAC7 and HDAC8
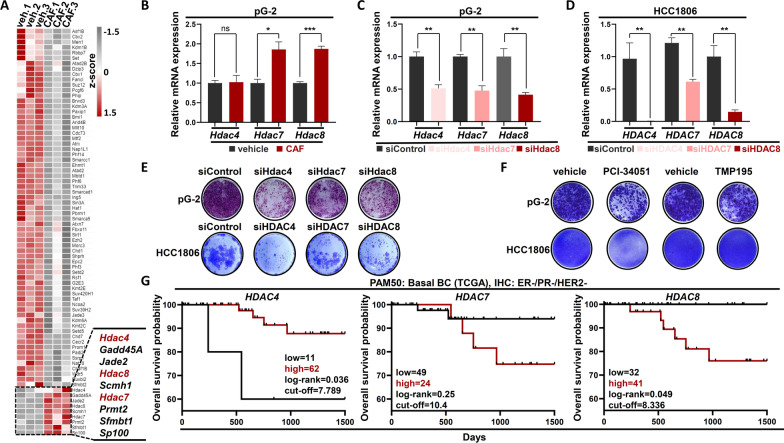
Previous studies of our group and others identified that epigenetic mechanisms are frequently involved in the EMT transcriptomic program induction of cancer cells [18–22]. To better understand factors that could be responsible for the transcriptional changes observed in BLBC cells upon chemotherapy treatment and to identify potentially druggable upregulated epigenetic effectors, we re-analyzed the mRNA-seq data of our previous study [18]. We identified 76 differentially regulated epigenetic factors (log2FC ≥ l0.7l, p-adj < 0.05), of which only 9 were significantly upregulated (Fig. 2A). A closer look at these factors identified a group of three histone deacetylases (Hdac4, Hdac7, Hdac8) with reported implications in cancer patient's prognosis and with a wide range of available specific inhibitory compounds [23, 24]. qRT-PCR confirmed the increased expression of Hdac7, Hdac8 upon CAF treatment in pG-2 cells (Fig. 2B). To study the role of Hdac4, Hdac7 and Hdac8 on the viability of murine pG-2 and human HCC1806 cells, we performed siRNA-mediated knockdown for every single gene (Fig. 2C, D). Interestingly, all siRNA treatments were detrimental for pG-2 cells, siHdac4 and siHdac8 showing the strongest phenotype impairment (Fig. 2E and Additional file 1: Fig. S1A). We observed a similar strong impairment of HCC1806 cell proliferation upon HDAC4 and HDAC8 loss (Fig. 2E and Additional file 1: Fig. S1B). To next test whether the enzymatic activities of these HDACs play a role in tumor cell fitness, we treated pG-2 and HCC1806 cells with specific inhibitors for HDAC8 (PCI-34051) and HDAC4/7 (TMP195) and subsequently assessed tumor cell proliferation. Interestingly, loss of HDAC4 and 7 activity impaired pG-2 cells (Fig. 2F, Additional file 1: Fig. S1C and S1F) and was not able to inhibit HCC1806 growth (Fig. 2F and Additional file 1: Fig. S1D). In contrast, PCI-34051 treatment robustly impaired the proliferation of both cell lines (Fig. 2F, Additional file 1: Fig. S1C-E). To test whether the combination of both HDAC8 and HDAC4/7 could increase the sensitivity of the cells to the treatment, we simultaneously treated pG-2 cells with increasing doses of both inhibitors. Here, we could not identify any improved sensitization with the combination therapy. On the contrary, the anti-proliferative effects of the PCI-34051 inhibitor (5–10 μΜ) in pG-2 cells were attenuated with increasing doses of TMP195 (Additional file 1: Fig. S1G). To next estimate if the identified HDAC4, HDAC7 and HDAC8 play a potential role in BLBC patient outcome, we analyzed data from the Breast Cancer Adenocarcinoma (BRCA) dataset from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and the Kaplan–Meier Plotter database (www.kmplot.com). In line with our results in vitro, HDAC8 was the only member of the three selected HDACs whose expression significantly correlated with a poor BLBC patient prognosis (Fig. 2G and Additional file 1: Fig. S1H). Taken together, our results suggest an important role of HDAC8 in murine and human BLBC aggressiveness.
Fig. 2.
HDAC4, HDAC7 and HDAC8 are upregulated in CAF-treated pG-2 and in CAF-resistant clones. A Heatmap of differentially regulated epigenetic factors in pG-2 cells upon CAF treatment (48 h) (basemean ≥ 15, p-adj < 0.05, log2FC ≥|0.7|). B qRT-PCR of Hdac4, Hdac7, Hdac8 in vehicle- and CAF-treated (48 h) pG-2 cells. C, D qRT-PCR validating the efficiency of HDAC4, HDAC7 and HDAC8 knockdowns in pG-2 (C) and HCC1806 (D), respectively. E Proliferation assay of pG-2 and HCC1806 cells upon HDAC4, HDAC7 and HDAC8 silencing, assessed by crystal violet staining. F Proliferation assay of pG-2 and HCC1806 cells upon HDAC8 inhibition (5 μM and 20 μM PCI34051, respectively) or HDAC4/HDAC7 inhibition (4 μM and 2 μM TMP195, respectively), assessed by crystal violet staining. G Kaplan–Meier plots showing the overall survival probability of HDAC4-, HDAC7- and HDAC8-expressing BLBC patients (expression and survival data are from the TCGA-BRCA database). All experiments were performed in triplicate. ns = not significant, *p-val < 0.05, ***p-val < 0.005. Statistical test: B Student's t test, error bars: standard error of the mean (SEM); G Log-rank test
HDAC8 supports the induction of EMT by suppressing the MET pathway in BLBC cells surviving chemotherapy
HDAC8 is a well-studied deacetylase exerting a function on numerous histone and non-histone substrates involved in various cellular homeostatic mechanisms. Structural Maintenance of Chromosomes 3 (SMC3), a subunit of the cohesin complex, represents one of the best characterized target for deacetylation by HDAC8 [24]. Therefore, we reasoned that HDAC8 could enact its pro-survival function by deacetylating SMC3, thereby facilitating the progression of the cell cycle as proposed by Dasgupta et al. [25]. Surprisingly, chemotherapy treatment of pG-2 cells did not led to a reduction of total acetylated SMC3 (ac-SMC3), as assessed by western blot (Additional file 1: Fig. S1I). Furthermore, PCI-34051 treatment at concentrations showing tumor cell phenotype impairment (5–10 μΜ) did not result in a further increase of ac-SMC3 neither in basal growth conditions nor upon chemotherapy (Additional file 1: Fig. S1I). Therefore, we concluded that HDAC8 function in BLBC cell fitness is rather independent of SMC3.
Since our previous results showed that HDAC8 possesses tumor-supportive properties in BLBC cells, we posited that these effects may result from its ability to positively influence the EMT transcriptional program in chemotherapy surviving cells. To test this hypothesis, we assessed the expression of well-known EMT-drivers (Vim, Cdh2, Wnt5a, Zeb1) in pG-2 cells upon CAF treatment and/or Hdac8 knockdown via qRT-PCR (Fig. 3A and Additional file 1: Fig. S2A). As expected, chemotherapy treatment induced their expression. However, loss of HDAC8 strongly reverted the induction of these genes upon chemotherapy treatment. Under basal growth conditions, only the expression of Wnt5a was affected by HDAC8 knockdown. The EMT process relies on one hand on the activation of EMT-TFs and effectors, but also on the repression of epithelial genes capable of inhibiting or reversing this process. Through its enzymatic activity, HDAC8 removes acetyl groups among others from histones, leading to gene inactivation in the deacetylated regions [26]. Therefore, we hypothesized that HDAC8 may support the induction of the EMT-transcriptional program by repressing genes driving epithelial phenotype (MET genes). To test this assumption, we examined whether genes commonly downregulated during EMT were also downregulated in chemotherapy-treated cells. Indeed, Gene Set Enrichment Analyses (GSEA) supported this assumption (Fig. 3B). Furthermore, by analyzing our ChIP-seq data from pG-2 cells surviving 48-h CAF treatment, we observed that the majority of strongly downregulated genes (log2FC ≤ − 1.5) upon chemotherapy showed a pronounced loss of H3K27ac in their TSS-proximal region (Fig. 3C). To assess whether HDAC8 is involved in the repression of these genes, we determined all H3K27ac occupied regions and investigated the changes of H3K27ac occupancy in pG-2 cells upon CAF treatment via Differential Binding (DiffBind) analysis. Pathway enrichment analysis of TSS-proximal regions losing H3K27ac using the Genomic Regions Enrichment of Annotations Tool (GREAT) showed a strong enrichment for tight junction and cell-to-cell adhesion signatures. This result supports that genes characteristic for epithelial cell phenotype maintenance markedly lose H3K27ac in their regulatory region upon CAF treatment (Additional file 1: Fig. S2B). A closer analysis of the DiffBind results identified a great number of EMT genes with increased H3K27ac (n = 101 EMT genes) and several MET genes (n = 23 MET genes) with a loss of H3K27ac at their TSS-proximal region (Fig. 3D). We decided to focus on the same strongly downregulated gene cohort (Fig. 3C) with concomitant loss of TSS-proximal H3K27ac enrichment upon CAF treatment (n = 94 genes, Fig. 3E) to perform a pathway enrichment analysis on these genes using the online tool ‘’Enrichr’’ (https://maayanlab.cloud/Enrichr/). Strikingly, this particular group of genes was strongly enriched with epithelial differentiation signatures (Fig. 3E, F). Tight junctions were the most prominently lost class of factors in the different MET signatures upon chemotherapy treatment. To validate the regulation of the identified gene signature, we selected two tight junction genes (Cldn3, Cldn8) and confirmed the loss of TSS-proximal H3K27ac and their decreased gene expression in chemotherapy-treated pG-2 cells by ChIP-qPCR and qRT-PCR, respectively (Fig. 3G, Additional file 1: Fig. S2C).
Fig. 3.
HDAC8 supports the induction of EMT by suppressing key-MET TFs in BLBC cells surviving cytotoxic therapy. A Assessment of Vim, Wnt5a, Cdh2, Zeb1 mRNA expression levels by qRT-PCR in vehicle- or CAF-treated (48 h) pG-2 cells, with or without Hdac8 silencing. B GSEA profile showing an enrichment of the "HOLLERN_EMT_BREAST_TUMOR_DN" signature (MSigDB: C2 curated gene sets) in pG-2 cells at basal state (veh) compared to CAF-treated. NES: Normalized Enrichment Score. C Aggregate profile showing changes of H3K27ac at TSS regions of strongly downregulated genes in vehicle- and CAF-treated pG-2 cells (normalized counts basemean ≥ 15, p-adj < 0.05, log2FC ≤ -− 1.5). D Differential binding analysis (DiffBind) depicting changes of H3K27ac occupancy between vehicle- and 48 h CAF-treated pG-2 cells. Genomic regions showing significant changes of H3K27ac occupancy at EMT- or MET-associated genes (MSigDB C2, see method section) were labeled with light red and dark red dots, respectively. E Venn diagram showing genes simultaneously downregulated (normalized counts basemean > 15, p-adj < 0.05, log2FC < − 1.5) and loosing H3K27ac at TSS regions (Diffbind parameters: conc > 3, p-adj < 0.05, FC < -0.5) upon CAF treatment in pG-2 cells. F Pathway enrichment analysis showing that genes with loss of expression and H3K27ac enrich for MET- and cell differentiation-associated signatures (labeled in red). Analysis performed with the online tool. G qRT-PCR and ChIP-RT-PCR of epithelial-specific markers (Cldn8, Cldn3) in vehicle- and CAF-treated pG-2 cells. H Venn diagram showing 10 transcription factors (TFs) with simultaneous gene expression (normalized counts basemean > 15, p-adj < 0.05, log2FC < -− 0.7) and H3K27ac occupancy loss (Diffbind parameters: conc > 3, p-adj < 0.05, FC < − 0.5) upon CAF treatment in pG-2 cells. The murine transcription factor list was retrieved from the Animal TFDB3.0 TF database. I qRT-PCR assessing changes of MET-TFs Elf5, Gata3, Rora, Grhl2 upon pG-2 CAF treatment for 48 h, w/ or w/o Hdac8 silencing. Statistical test: A, I one-way ANOVA, G Student's t test. *p-val < 0.05, **p-val < 0.01, ***p-val < 0.005. Error bars are standard error of the mean (SEM)
Similar to EMT, the MET-driven cell differentiation is a well-documented transcriptional program involving MET-committed transcription factors (MET-TFs) in various biological contexts [27–29]. Hence, we sought to identify MET-TFs potentially under control of HDAC8 repression, therefore showing decreased promoter-proximal H3K27ac occupancy with a concomitant gene expression loss upon CAF treatment. We identified 10 downregulated TFs showing a prominent decrease of TSS-proximal H3K27ac occupancy upon CAF treatment, some of those with a well-established functional role in epithelial cell differentiation [29–36] (Fig. 3H, Additional file 1: Fig. S2D–E). To next assess the potential repressive function of HDAC8 on these genes, we performed a knockdown and a PCI-34051-mediated inhibition of HDAC8 in pG-2 cells with or without CAF treatment. We selected four MET-TF genes and confirmed their decreased expression upon chemotherapy alone, as expected from our mRNA-seq results (Fig. 3I). Strikingly, both HDAC8 knockdown or inhibition partially rescued the expression levels of the four MET-TFs (Elf5, Gata3, Rora and Grhl2) tested here (Fig. 3I and Additional file 1: Fig. S2H), but also rescued the expression of epithelial-specific markers (Cdh1, Cldn3, Cldn8) (Additional file 1: Fig. S2F and S2G). Of note, inhibition of HDAC4 and HDAC7 by TMP195 alone or in combination with PCI-34051 could not restore the expression of MET-TFs and epithelial markers in chemotherapy-treated in p-G2 cells to the same extent as PCI-34051 alone. Concordantly, mesenchymal markers vimentin (Vim) and Zeb1 were efficiently suppressed only in PCI34051-treated cells upon chemotherapy and not by single or co-treatment with TMP195 (Additional file 1: Fig. S2I). Finally, to further exclude SMC3’s potential involvement in the regulation of this MET transcriptional signature, we re-analyzed a publically available microarray gene expression dataset (GSE38252) and confirmed that MCF7 BC-cells do not regulate the expression of any of these genes upon SMC3 knockdown (Additional file 1: Fig. S2J). Therefore, our findings strongly support a transcriptional repressive function of HDAC8 exerted on the MET program.
High HDAC8 expression correlates with repression of epithelial differentiation program in BLBC patients
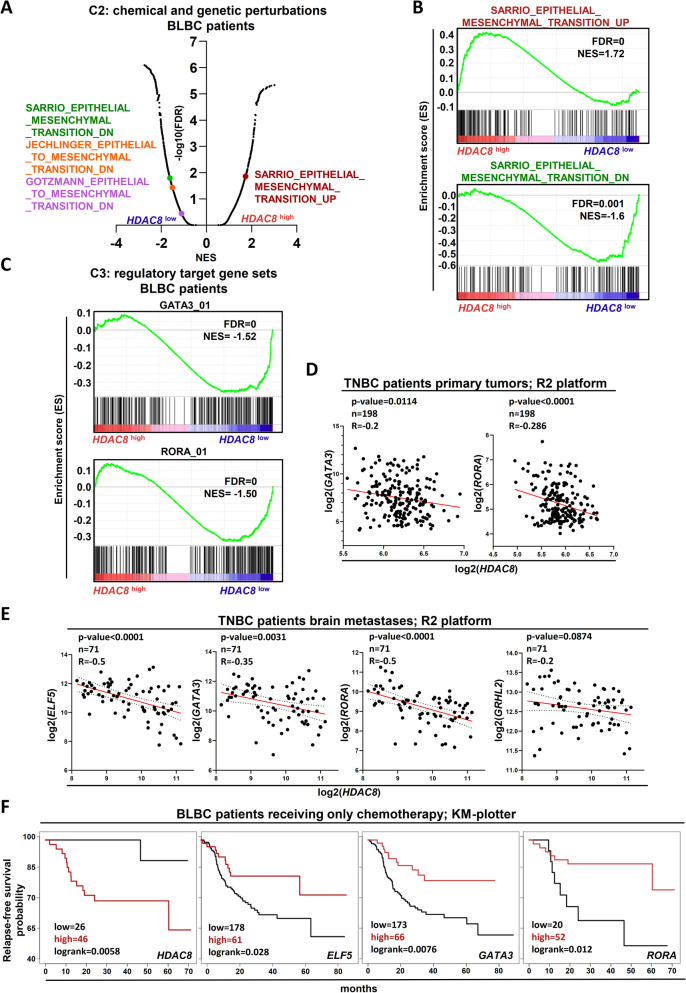
To corroborate our results with the clinical situation of BLBC patients, we analyzed the gene expression profiles of low and high HDAC8-expressing lesions from the BRCA-TCGA dataset. As suggested by our previous in vitro investigations, we observed a significant enrichment of EMT gene expression signatures in HDAC8high patients, whereas HDAC8low lesions-enriched gene sets generally downregulated upon activation of the EMT program (Fig. 4A, B). Further analyses of this patient collective on the C3 library (MSigDB) identified that HDAC8low patients enrich for genes with at least one binding element for GATA3 or RORA in their regulatory region (Fig. 4C). These last results point at a possible higher activity of MET-TFs in HDAC8low BLBC-lesions. We reasoned that if HDAC8 represses MET-TFs also in BLBC-patients, a negative correlation between HDAC8- and MET-TFs should be detectable. Therefore, we retrieved expression data from the R2-platform. Here, HDAC8 expression negatively correlated with the expression of GATA3 and RORA in primary TNBC tumors (Fig. 4D). Accordingly, further analyses on brain metastases of TNBC patients confirmed a significant negative correlation of HDAC8 with ELF5, GATA3, RORA and a mild but insignificant negative correlation with GHRL2 (Fig. 4E). Finally, we asked whether these particular MET-TFs have a prognostic value for BLBC patients' outcomes. Interestingly, whereas HDAC8high-patients had the highest probability of relapse, ELF5high-, GATA3high- and RORAhigh-patients displayed the lowest relapse rate (Fig. 4F). Together, these data strongly indicate a tumor-promoting role of HDAC8 through a repressive function on the MET program in BLBC patients.
Fig. 4.
High HDAC8 expression correlates with repression of the epithelial differentiation program in BLBC patients. A Plot depicting all C2 curated gene sets (MSigDB) enriched in GSEA analyses of HDAC8high- and HDAC8low-expressing BLBC patients. B Representative gene set enrichment profiles of the EMT and MET transcription programs in HDAC8low- and HDAC8high-BLBC patients. NES: Normalized Enrichment Score. C Gene set enrichment profiles of the MET-TFs GATA3 and RORA whose gene targets are particularly expressed in HDAC8low-BLBC patients. D–E Scatter plot showing the anti-correlation of the MET-TFs with HDAC8 in primary tumors (D) and brain metastases (E) of TNBC patients. Data were retrieved from the R2 platform. F Kaplan–Meier plots showing the relapse-free survival probability of HDAC8-, ELF5-, GATA3- and RORA-expressing BLBC patients receiving only chemotherapy as a treatment option (Log-rank test). Data were retrieved from the KM plotter. A–C
Source of patient gene expression data: TCGA. Source of gene set enrichment profiles: MSigDB
HDAC8 silencing or inhibition sensitizes BLBC cells to chemotherapy treatment
Altogether, the upregulation of Hdac8 in CAF-surviving pG-2 cells, its support of the EMT program as well as its implication in patient relapse-free survival in BLBC patients strongly suggested that elevated HDAC8 levels might confer to the tumor cells chemotherapy-resistant features. To test this hypothesis, we first assessed the response of BLBC patients to cytotoxic therapies according to their HDAC8 levels using the ROC plotter platform (Fig. 5A). In accordance with our previous results, patients showing poor responsiveness to the chemotherapy harbored higher HDAC8 expression. In line with these data, BLBC patients with better response to cytotoxic therapies displayed significantly higher ELF5, GATA3 and RORA expression levels (Additional file 1: Fig. S3A–C). To experimentally assess the impact of HDAC8 knockdown or inhibition in BLBC cells upon CAF treatment, we performed in vitro proliferation assays. Interestingly, loss of HDAC8 in pG-2 cells potentiated the effect of the chemotherapy on tumor cell proliferation (Fig. 5B). The increased chemotherapy efficiency was particularly visible in one resistant variant of the pG-2 cells (rG-2) [18], where siHdac8 alone had no significant effect on the cell growth (Fig. 5C). To further support the important role of HDAC8 in BLBC cell survival to chemotherapy, we simultaneously treated the murine and human cell lines with PCI-34051 and CAF. In line with the knockdown results, HDAC8 inhibition sensitized the tumor cells to the cytotoxic treatment in all cases (Fig. 5D, F). Notably, this combinatory treatment had a remarkable long-term effect on pG-2 and rG-2 cell proliferation. Finally, to translate these functional assay findings to the human situation, we investigated the effects of HDAC8 inhibition in combination with CAF chemotherapy in HCC1806 cells. Strikingly, at PCI-34051 concentrations not showing tumor cell impairment as a monotherapy, we observed a robust proliferative defect compared to the control counterparts (Fig. 5F). Collectively, our findings identified HDAC8 as a druggable factor supporting tumor cell survival to conventional chemotherapies by promoting the EMT transcriptional program in BLBC lesions.
Fig. 5.
HDAC8 silencing or inhibition sensitizes BLBC cells to chemotherapy treatment. A ROC analysis from publically available TNBC data demonstrating that patients with poor response to chemotherapy harbor high expression levels of HDAC8. Box plots: Mann–Whitney test. B, C Proliferation assay of CAF-treated pG-2 (B) and rG-2 (C) cells upon Hdac8 silencing. Representative crystal violet pictures are at the right panel of the bar charts. D Proliferation assay of pG-2 treated with low CAF doses (156.25 ng/ml cyclophosphamide, 7.8 ng/ml doxorubicin, 156.25 ng/ml 5-FU) for 48 h, w/ or w/o HDAC8 inhibition (5 μM PCI34051). E rG-2 cells treated with CAF for 48 h, w/ or w/o HDAC8 (5 μM PCI34051). Representative crystal violet pictures are at the left and right panel of proliferation kinetic graphs for pG-2 and rG-2, respectively. F Proliferation assay of HCC1806 treated with low CAF doses (156.25 ng/ml cyclophosphamide, 7.8 ng/ml doxorubicin, 156.25 ng/ml 5-FU) for 48 h, w/ or w/o HDAC8 inhibition (10 μM PCI34051). All experiments were performed in biological triplicates. ns = no difference, *p-val < 0.05, ***p-val < 0.005, ****p-val < 0.001. Statistical test: Student's t test. Error bars are standard error of the mean (SEM)
Discussion
EMT is a process naturally occurring in normal cells under physiologic conditions but is also frequently implicated in the gain of cancer cell aggressiveness, metastatic behavior and therapy resistance [8]. Intensive research efforts in the past decades allowed us to increase our understanding of the cascade of events, the role played by central EMT-TFs during the acquisition of mesenchymal features and their consequence for tumor cell survival to chemotherapy [12, 37]. However, to date, druggable factors regulating the EMT transcriptional program induction and their maintenance are scarce and insufficiently studied. In this context, epigenetic regulators represent attractive factors for the development of novel therapeutic approaches, as their enzymatic activity is per definition reversible and does not imply a modification of the genetic code [38]. We recently leveraged the WAP-T mammary carcinoma mouse model to get insights into transcriptional mechanisms underlying the survival of BLBC cells to a CAF chemotherapy [14]. We thereby demonstrated that the loss of PRC2 repressive activity can drive aggressive BLBC tumor cell phenotypes by promoting NFATc1 expression [18]. In the present study, we analyzed RNA-seq and ChIP-seq data from the same model and identified HDAC8 as an upregulated epigenetic factor supporting BLBC cell chemotherapy resistance by repressing the epithelial differentiation program, and with a potential as a drug target. Our results align with increasing number of studies reporting a relationship between high HDAC8 expression levels and parameters characteristic for aggressive BC [39–41]. In 2016, HDAC8 was shown to promote cell cycle progression by deacetylating the SMC3 cohesin subunit in MCF7 cells [25]. Our present investigations in BLBC malignancies identified an alternative tumor-promoting function by HDAC8 supporting the activation of EMT transcriptional program. This HDAC8-dependent mechanism is rather likely to happen in an SMC3 independent manner, as no changes of SMC3 acetylation levels were observed in tumor cells sensitized by HDAC8i treatment. Interestingly, several groups described an association between HDAC8 activity and EMT-driving signaling. Tang and colleagues reported in 2020 that HDAC8 stimulates the TGF-beta signaling by cooperating with the SMAD3/4 complex to deacetylate SIRT7 promoter region, thereby promoting BC cell migration [42]. In two consecutive studies, An et al. established a similar implication of HDAC8 controlling BC cell dissemination through the YAP signaling and via the AKT/GSK-3β/SNAI1 axis [43, 44]. Our results complement this knowledge and establish that HDAC8 can support the induction of EMT by actively repressing the expression of MET-TFs like GATA3, EFL5, RORA and GHRL2. Of note, An et al. reported a mechanism by HDAC8 enacting a stabilization of the EMT-TF SNAI1 at the protein level via phosphorylation through the AKT/GSK-3β axis [43]. SNAI1 expression levels remained here unchanged. In contrast, we observed that loss of HDAC8 in pG-2 cells led to increased Snai1 expression levels, both under basal culture conditions and chemotherapy treatment (data not shown). Although changes in SNAI1 protein levels were not assessed in the frame of this study, it seems reasonable to hypothesize that the loss of HDAC8 activity should dampen the increased SNAI1 gene expression, and that parallel independent epigenetic mechanisms by HDAC8 repress the epithelial transcriptional program in a cooperative way. Importantly, the activity of HDAC8 was necessary for chemotherapy-resistant features of BLBC cells. This function is likely enacted through the repression of TFs driving the epithelial differentiation program. For example, loss of GATA3 was associated with tumor progression and poor patient outcomes in BC and urothelial carcinoma [45, 46]. In an elegant study, Singh et al. established the ELF5 transcription factor as a tumor suppressor gene inhibiting TNBC progression and metastasis by promoting IFN-γ signaling and immunosurveillance [30]. Similarly, RORA expression was shown to possess tumor-suppressive functions and to inhibit BC tumor invasion [34]. In line, GHRL2 has emerged as an inhibitor of the EMT expression program in multiple cancer entities [35, 37, 47, 48]. Furthermore, GRHL2 expression in human mammary epithelial cells was shown to induce a metabolic switch responsible for reduction of stem cell properties and anchorage independent cell survival. We and others established in the past a strong involvement of HDACs Class I in the EMT transcriptional program activation of several cancer and fibrotic diseases [19, 49, 50]. In this context, Class I HDAC inhibitors (HDACi) showed a general good capacity to revert aggressive tumor cell phenotypes to more sensitive epithelial ones. Further investigations by Choi et al. on renal fibrosis demonstrated that Class I HDACi can revert the TGFβ1-induced repression of E-cadherin (CDH1), a major gatekeeper of the epithelial phenotype [50]. Comparing treatments with different small molecule inhibitors and siRNA-mediated knockdowns, the authors identified HDAC8 as a key factor controlling TGFβ1-mediated E-cadherin repression. Interestingly, HDAC8 enzymatic activity was not found to be necessary here. Our investigations in murine BLBC cells align with these observations, as HDAC8 knockdown in pG-2 cells rescued Cdh1 expression upon chemotherapy. Surprisingly, inhibition of HDAC8 by PCI-34051 also led to a pronounced rescue of Cdh1 levels, pointing at a possible context-dependent necessity of its catalytic activity to enact this repressive function.
In the past decades, growing research efforts led to the development of several pan-HDACi with very promising efficiency and FDA approval for the treatment of hematological malignancies like T-cell lymphoma and multiple myeloma [51, 52]. However, despite good results in experimental models in vitro and in vivo, the applicability of pan-HDACi compounds for the treatment of patients with solid tumors was quite deceiving, mostly because of a moderate bioavailability and relatively high toxicity at pharmacological doses, leading to severe side effects [52, 53]. Therefore, compounds targeting specific members of the HDAC family have been proposed to have a more specific anti-tumor activity, being at the same time better tolerated by patients [54]. Based on our and other studies, specific inhibitors of HDAC8 represent very attractive anti-cancer drugs. PCI-34051, used in the present work, shows a very high specificity for HDAC8 with a reported IC50 of 10 nM [55]. Its efficiency as a combination therapy with BRAF inhibitor was recently validated in an in vivo mouse model for melanoma [56]. However, the applicability of this compound as an in vivo drug has been questioned, as PCI-34051 half-life in rat brains was estimated to be 15 min only [57]. In 2015, the group of Ina Oehme successfully utilized PCI-48012, a more stable variant of PCI-34051, for the treatment of two neuroblastoma xenograft models. Strikingly, the authors observed an anti-tumor activity comparable to pan-HDACi coming along with a much lower degree of toxicity. Three years later, the same group successfully demonstrated an increased efficiency of co-treatments with AKLi and a specific HDAC8i (compound 20a) in zebrafish xenografts of neuroblastoma, thereby emphasizing the attractiveness of HDAC8i-based combined therapies.
Together, our results suggest that BLBC patients with low expression of MET-TFs may benefit from therapies combining conventional chemotherapy with HDAC8-specific inhibition.
Conclusions
Our results establish HDAC8 as a transcriptional repressor of the MET process and as a factor targetable through epigenetic therapies with a strong potential to increase the efficiency of conventional chemotherapies.
Methods
Cell culture
The murine mammary carcinoma pG-2 cell line that was created in a previous study [17] as well their resistant variant (rG-2) [Mieczkowska et al. 2021, in press], was cultured in DMEM GlutaMAX™ (Invitrogen). The triple-negative breast cancer HCC1806 human cell line was purchased from the ATCC (American Type Culture Collection, USA) and was cultured in RPMI 1640 (Gibco) medium. All media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S), and all cell lines were maintained at 37 °C, 5% CO2.
Chemotherapy treatment
CAF chemotherapy was acquired through the pharmacy of the Medical School of Göttingen at following concentrations: cyclophosphamide [20 mg/ml], doxorubicin [2 mg/ml] and 5-fluorouracil (5-FU) [50 mg/ml). In all experiments, pG-2 and rG-2 were treated with a concentration of 312.5 ng/ml cyclophosphamide, 15.6 ng/ml doxorubicin and 312.5 ng/ml 5-FU, except if differently stated in the figure legend. HCC1806 was treated with 625 ng/ml cyclophosphamide, 31.2 ng/ml doxorubicin and 625 ng/ml 5-FU, except if differently stated in the figure legend, or 2.5 nM paclitaxel or 2 µM Cisplatin. The different chemotherapies were applied for 48 h on the cells.
siRNA transfection
Cells were reverse transfected with siRNA using Lipofectamine® RNAiMAX, according to the manufacturer’s instruction. siGENOME siRNAs (Dharmacon) were acquired at Horizon Discovery Ltd. and are listed in Additional file 1: Table S5. siRNAs: Non-targeting control #5 [D-001206–13], murine Hdac4 [M-043626–01], murine Hdac7: [M-040703–01], murine Hdac8 [M-058613–01], human HDAC7 [D-009330–02, D-009330–04, D-009330–05, D-009330–06], human HDAC8 [D-003500–01, D-003500–02, D-003500–03, D-003500–06]. Smart pools of 4 different siRNAs were either commercially acquired or manually mixed at an equimolar concentration of each 5 µM.
Inhibitor treatment
Three cycles of 48-h treatment with PCI34051 (HDAC8 inhibitor, Cayman chemical, cat. no 10444) or TMP 195 (HDAC4 and HDAC7 inhibitor, Cayman chemical, cat. no 23242) were performed. 24 h after seeding, the cells were treated with one first cycle of inhibition. Chemotherapy treatment was performed together with the second cycle of inhibition.
Cell proliferation assay
7000–10,000 cells per well were seeded in 24-well plates, depending on treatment and cell line. In case of reverse transfection with siRNA, cells were treated with chemotherapy 48 h post-transfection. The confluence was measured using a Celigo® Cell Cytometer device (Nexcelom Bioscience) and an Incucyte® Live-Cell Analysis System (Sartorius). At the last day of the experiment, cells were fixed with 100% methanol for 10 min and stained using crystal violet for 20 min. The confluency of scanned pictures was analyzed using the ImageJ software. Graphs were designed using GraphPad Prism (v8.0.1).
RNA extraction
mRNA extraction was performed on cells cultured in 6-well plates. After removing the culture media, cells were washed twice with PBS and lysed by adding 500 µl of Qiazol®. After an incubating of 10 min at room temperature, cells were resuspended and transferred into RNase-free tubes. According to the manufacturer’s manual, 100 µl chloroform was added, vortexed for 15 s and centrifuged (12,000 g, 15 min, 4 °C). The upper aqueous phase was transferred to a new tube and 200 µl of chloroform was added, vortexed and centrifuged as before. The aqueous phase was transferred to a new tube, and mRNAs were precipitated by adding an equal amount of isopropanol (RNA grade) and incubating at − 80 °C for at least one hour. mRNAs were then pelleted (20 min/12000 g/4 °C) and washed with cold 70% ethanol (10 min,12000 g,4 °C). After removal of ethanol, the pellets were allowed to air-dry for 5 to 10 min and diluted in 30 μl of RNase-free water. RNA quantification was performed using a DS-11 + spectrophotometer (Denovix).
cDNA synthesis
The manufacturer’s instructions in the First Strand cDNA Synthesis Kit were followed (M-MLuV, NEB). In short, 1 µg of RNA was diluted to a total volume of 10 µl with RNase-free water and mixed with 2 µl of random 9mer primers (60 µM) and 1 µl of dNTPs (10 mM). The mixture was incubated for 5 min at 65 °C, after which 2 µl of 10 × M-MuLV Buffer, 0.2 µl RNase Inhibitor (40 U/μl), 1 µl M-MuLV Reverse Transcriptase (200 U/μl) and 6.8 µl DEPC water were added and incubated at 25 °C for 5 min and then at 42 °C for 60 min. This was followed by the inactivation of the enzyme at 65 °C for 20 min. The reaction mix was diluted to a total volume of 200 µl and stored at -20 °C.
Quantitative real-time polymerase chain reaction (qRT-PCR)
For qRT-PCR, 5 ng cDNA was utilized per reaction in 25 µl total volume (SYBR-green). Samples were run with the following steps: denaturation (95 °C for 2 min), 40 cycles (for gene expression) or 46 cycles (for ChIP-RT-PCR) amplification (95 °C for 15 s followed by 60 °C for 30 s). A melting curve analysis was subsequently generated (60 °C to 95 °C, 0.5 °C per sec). Samples were quantified by using the method of standard curve. The samples were normalized to the Rplp0 or RPLP0 housekeeping genes. Graphs were designed using GraphPad Prism (v8.0.1).
Western blotting
Proteins were extracted from six-well plates with 500 µl ice-cold RIPA buffer (10 mM Tris–HCl pH 8, 1 mM EDTA, 1% v/v Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl) supplemented with protease inhibitors (1 mM Pefabloc, 1 ng/µl aprotinin/leupeptin, 10 mM BGP, 1 mM NEM). Protein samples were then sonicated three times for five cycles (30 s ON/30 s OFF) in a Bioruptor Pico (Diagenode, Belgium). Lysates were then mixed with Lämmli buffer (6 × , 375 mM Tris–HCl, 10% SDS, 30% glycerol, 0.02% bromophenol blue, 9.3% DTT) and cooked for 5 min at 95 °C. Same amounts of protein per sample were then separated using 10–12% SDS polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Immobilon, Millipore, USA). Membranes were blocked with 5% skimmed milk in TBS-T and incubated overnight at 4 °C with specific primary antibodies diluted in the same blocking solution. Next, membranes were washed with TBS-T, incubated 1 h with secondary antibodies (Dianova GmbH, Germany) diluted in blocking buffer, washed again with TBS-T and finally developed using HRP substrate (Cyanogen WESTAR) in a ChemoStar imaging system (INTAS science imaging, Germany). Antibody list in Additional file 1: Table S6.
RNA sequencing analysis
RNA-seq raw data (Fastq files) were processed in the GWDG Galaxy environment (https://galaxy.gwdg.de) provided by the “Gesellschaft für wissenschaftliche Datenverarbeitung mbH Göttingen” (GWDG). After quality check using FastQC (v0.72) [58], sequencing data were trimmed (FASTQ Trimmer tool, v0.0.1), aligned to the murine reference genome (mm9) with the TopHat tool (v2.1.1) [59]. Next, reads were assigned to their respective genomic features using htseq-count (v0.1.9) [60]. Finally, a differential gene expression analysis was performed using DESeq2 (v2.11.40.5) [61]. Analyses of gene signature enrichment were performed using the Gene Set Enrichment Analysis (GSEA, v4.1.0) tool (http://www.broadinstitute.org/gsea/downloads.jsp) and online pathway enrichment analysis tool Enrichr (http://amp.pharm.mssm.edu/Enrichr/) as well as gProfiler (https://biit.cs.ut.ee/gprofiler/gost). Heatmap representations of gene expression were generated with the online Morpheus tool (https://software.broadinstitute.org/morpheus/). RNA-seq raw data are deposited at ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-9547.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation for H3K27me3 and H3K27ac was performed 48 h after chemotherapy treatment, as described previously [62]. Briefly, pG-2 cells were cultured in 15 cm plates. Protein–DNA complexes were crosslinked with 1% formaldehyde (Sigma), and nuclear fraction was extracted and sonicated with a Bioruptor pico (Diagenode). After controlling the size of the DNA fragments and a pre-cleaning step, the same amounts of samples were incubated with 1 µg anti-H3K27me3 or anti-H3K27ac antibody overnight at 4 °C and immunoprecipitated with protein A-sepharose. Finally, DNA–protein complexes were reverse-crosslinked, DNA fragments were purified by phenol–chloroform extraction, and concentration was determined using Qubit fluorimeter (Invitrogen).
Chromatin immunoprecipitation-sequencing analysis
ChIP-seq data were processed and analyzed in the Galaxy environment (https://galaxy.gwdg.de/). After a quality check (FastQC, v0.72), reads were aligned to the mouse reference genome (mm9) using Bowtie2 [58, 63]. H3K27ac peaks were identified with the MACS2 tool (v 2.1.1.20160309.0), and Differential Binding Analysis as well volcano plots were performed with Diffbind (Bioconductor, v3.6.3) [64]. The deep tools suite was used for the generation of normalized coverage files (bamcoverage, v3.2.0.0.0) [65]. To visualize occupied regions, region scoring matrix was computed (computeMatrix, v3.2.0.0.0) and profiles plots or heatmaps were generated [plotProfile (v3.2.0.0.0) and plotHeatmap (v3.2.0.0.1)] [65]. Histone modification occupancy at specific genomic regions was visualized with the integrative genome Viewer (IGV, v2.8.0, http://software.broadinstitute.org/software/igv/). Pathway enrichment analysis of differentially regulated genomic regions was performed using the Genomic Regions of Annotations Tool (GREAT, http://great.stanford.edu/public/html/). ChIP-seq raw data are deposited at ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-9584.
List of EMT and MET genes in Fig. 3D: EMT genes were extracted from MSigDB C2: HOLLERN_EMT_BREAST_TUMOR_UP, VERHAAK_GLIOBLASTOMA_MESENCHYMAL and JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP; MET genes were extracted from MSigDB C2: HOLLERN_EMT_BREAST_TUMOR_DN). The murine transcription factor list was retrieved from the Animal TFDB3.0 TF database (source: http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/).
Publically available data
Overall survival data of BLBC patients from The Cancer Genome Atlas (TCGA) program (https://portal.gdc.cancer.gov/) were retrieved from the xenabrowser web-based tool (http://xena.ucsc.edu/) using the following patient selection criteria: PAM50 subtype (Nature, 2012): basal-like, ER, PR, HER2 status (Nature, 2012): negative. The Cutoff Finder online tool (https://molpathoheidelberg.shinyapps.io/CutoffFinder_v1/) was used to determine the best cutoffs and the corrected p-values [66].
Relapse-free survival data of BLBC patients the online KM plotter tool (https://kmplot.com) were analyzed in the dedicated web-based platform [67]. Used datasets: 223345_at (HDAC8), 220624_s_at (ELF5), 209602_s_at (GATA3) and 240951_at (RORA). Used patient selection criteria: PAM50: basal-like, endocrine therapy excluded, only neoadjuvant therapy. Overall survival data of BLBC patients were acquired from the same online tool. Used datasets: 228813_at (HDAC4), 217937_s_at (HDAC7), 223345_at (HDAC8). Used patient selection criteria: PAM50: basal-like, endocrine therapy excluded, any chemotherapy.
Finally, gene expression data of HDAC8 and the MET-TFs for Fig. 4D, E were retrieved from the online database tool “R2 Genomic Analysis and Visualization Platform” (R2.amc.nl). Graphs were designed using GraphPad Prism (v8.0.1).
Receiver operating characteristic (ROC) patient data were retrieved from the ROC plotter (source: https://rocplot.org/site/treatment) using the relapse-free survival (RFS; at 5 years) and pathological complete response (pCR) data. RFS: 223345_at (HDAC8), 209604_s_at (GATA3). pCR: 220625_s_at (ELF5), 1562682_at* (RORA). Patient selection criteria: TNBC patients, receiving any chemotherapy.
Gene expression microarray data of siControl- and siSMC3-treated MCF7 cells were retrieved from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE38252 [68], re-analyzed with the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) and visualized in a volcano plot using R (v 4.1.1).
Supplementary Information
Additional file 1: Figure S1. HDAC8 supports the induction of EMT by suppressing the MET pathway in TNBC cells surviving Chemo. Table S1. Concentration of CAF dilutions. Table S2. List of ChIP-qPCR primers. Table S3. List of RT-qPCR mouse primers. Table S4. List of RT-qPCR human primers, excel file with the list of epigenetic regulators. Table S5. List of siRNAs.
Acknowledgements
The authors would like to thank Prof. Steven A. Johnsen’s group for giving access to laboratory space, equipment and material, as well as for the constructive discussions. We furthermore thank the group of Prof. Matthias Dobbelstein for sharing with their cell- and flow-cytometer devices. We also would like to thank Prof. Wolfgang Sippl for sharing with us several HDAC8i compounds that we, unfortunately, could not utilize in the frame of this study. Finally, we thank Sonja Blume for the technical help during the revision of this work.
Abbreviations
- BLBC
Basal-like breast cancer
- TNBC
Triple-negative breast cancer
- EMT
Epithelial–mesenchymal transition
- MET
Mesenchymal–epithelial transition
- HDAC8
Histone deacetylation 8
- TF
Transcription factor
- H3K27ac
Acetylation of lysine 27 on histone H3
- CAF
Cyclophosphamide, adriamycin, 5-fluorouracil
Authors' contributions
GPP helped in experimentation and analyses, bioinformatics analyses, figure conception, manuscript writing and edition; IKM was involved in ChIP-seq experimentation and analyses; EP contributed to cell culture experiments, bioinformatics analyses, figure conception, manuscript edition and revision; GES helped in mRNA-seq approach; SF contributed to cell culture experiments, gene expression analyses, figure conception; JG helped in conception and manuscript edition; FW was involved in project conception and supervision, manuscript edition, revision and finalization. All authors have read and agreed to the published version of the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was funded through a grant from the “Erich und Gertrud Roggenbuck-Stiftung zur Förderung der Krebsforschung” entitled “Unravelling epigenetic mechanisms of CAF-chemotherapy resistance in mammary carcinoma” to FW.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
All patient data analyzed and presented in this study were retrieved from publically available platforms with appropriate ethical approvals. All data were accessed in an anonymized form.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Garyfallia Pantelaiou-Prokaki and Iga Mieczkowska contributed equally to this work
References
- 1.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Nunnery SE, Mayer IA, Balko JM. Triple-negative breast cancer: breast tumors with an identity crisis. Cancer J. 2021;27:2–7. doi: 10.1097/PPO.0000000000000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borri F, Granaglia A. Pathology of triple negative breast cancer. Semin Cancer Biol. 2021;72:136–145. doi: 10.1016/j.semcancer.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Garutti M, Pelizzari G, Bartoletti M, Malfatti MC, Gerratana L, Tell G, et al. Platinum salts in patients with breast cancer: a focus on predictive factors. Int J Mol Sci. 2019;20:3390. doi: 10.3390/ijms20143390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abaurrea A, Araujo AM, Caffarel MM. The role of the il-6 cytokine family in epithelial–mesenchymal plasticity in cancer progression. Int J Mol Sci. 2021;22:8334. doi: 10.3390/ijms22158334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto MA, Huang RYYJ, Jackson RAA, Thiery JPP. Emt: 2016. Cell Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Williams ED, Gao D, Redfern A, Thompson EW. Controversies around epithelial–mesenchymal plasticity in cancer metastasis. Nat Rev Cancer. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye X, Weinberg RA. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gooding AJ, Schiemann WP. Epithelial-mesenchymal transition programs and cancer stem cell phenotypes: mediators of breast cancer therapy resistance. Mol Cancer Res. 2020;18:1257–1270. doi: 10.1158/1541-7786.MCR-20-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bill R, Christofori G. The relevance of EMT in breast cancer metastasis: correlation or causality? FEBS Lett. 2015;589:1577–1587. doi: 10.1016/j.febslet.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong B, Qiu Z, Wu Y. Tackle epithelial-mesenchymal transition with epigenetic drugs in cancer. Front Pharmacol. 2020;11:596239. doi: 10.3389/fphar.2020.596239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanaei M, Kavoosi F. Histone deacetylases and histone deacetylase inhibitors: molecular mechanisms of action in various cancers. Adv Biomed Res. 2019;8:63. doi: 10.4103/abr.abr_142_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jannasch K, Wegwitz F, Lenfert E, Maenz C, Deppert W, Alves F. Chemotherapy of WAP-T mouse mammary carcinomas aggravates tumor phenotype and enhances tumor cell dissemination. Int J Cancer. 2015;137:25–36. doi: 10.1002/ijc.29369. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Zhang L, He X, Zhang P, Sun C, Xu X, et al. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun. 2018;502:160–165. doi: 10.1016/j.bbrc.2018.05.139. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Strietz J, Bleilevens A, Stickeler E, Maurer J. Chemotherapeutic stress influences epithelial–mesenchymal transition and stemness in cancer stem cells of triple-negative breast cancer. Int J Mol Sci. 2020;21:404. doi: 10.3390/ijms21020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegwitz F, Kluth MA, Mänz C, Otto B, Gruner K, Heinlein C, et al. Tumorigenic WAP-T mouse mammary carcinoma cells: A model for a self-reproducing homeostatic cancer cell system. PLoS ONE. 2010;5:e12103. doi: 10.1371/journal.pone.0012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mieczkowska IK, Pantelaiou-Prokaki G, Prokakis E, Schmidt GE, Müller-Kirschbaum LC, Werner M, et al. Decreased PRC2 activity supports the survival of basal-like breast cancer cells to cytotoxic treatments. Cell Death Dis. 2021;12:1118. doi: 10.1038/s41419-021-04407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra VK, Wegwitz F, Kosinsky RL, Sen M, Baumgartner R, Wulff T, et al. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4-and MYC-dependent manner. Nucleic Acids Res. 2017;45:6334–6349. doi: 10.1093/nar/gkx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galle E, Thienpont B, Cappuyns S, Venken T, Busschaert P, Van Haele M, et al. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin Epigenetics. 2020;12:27. doi: 10.1186/s13148-020-0821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Dai M, Zheng Y, Wu J, Yu B, Zhang H, et al. Epigenetic suppression of E-cadherin expression by Snail2 during the metastasis of colorectal cancer. Clin Epigenetics. 2018;10:154. doi: 10.1186/s13148-018-0592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asfaha Y, Schrenk C, Alves Avelar LA, Hamacher A, Pflieger M, Kassack MU, et al. Recent advances in class IIa histone deacetylases research. Bioorgan Med Chem. 2019;27:115087. doi: 10.1016/j.bmc.2019.115087. [DOI] [PubMed] [Google Scholar]
- 24.Chakrabarti A, Oehme I, Witt O, Oliveira G, Sippl W, Romier C, et al. HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol Sci. 2015;36:481–492. doi: 10.1016/j.tips.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Dasgupta T, Antony J, Braithwaite AW, Horsfield JA. HDAC8 inhibition blocks SMC3 deacetylation and delays cell cycle progression without affecting cohesin-dependent transcription in MCF7 cancer cells. J Biol Chem. 2016;291:12761–12770. doi: 10.1074/jbc.M115.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha SD, Reid C, Meshkibaf S, Kim SO. Inhibition of interleukin 1 β (IL-1 β) expression by anthrax Lethal Toxin (LeTx) is reversed by histone deacetylase 8 (HDAC8) inhibition in murine macrophages. J Biol Chem. 2016;291:8745–8755. doi: 10.1074/jbc.M115.695809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogachek MV, De Andrade JP, Weigel RJ. Regulation of epithelial-mesenchymal transition through sumoylation of transcription factors. Cancer Res. 2015;75:11–15. doi: 10.1158/0008-5472.CAN-14-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boivin FJ, Schmidt-Ott KM. Transcriptional mechanisms coordinating tight junction assembly during epithelial differentiation. Ann N Y Acad Sci. 2017;1397:80–99. doi: 10.1111/nyas.13367. [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukačišin M, Romano RA, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh S, Kumar S, Srivastava RK, Nandi A, Thacker G, Murali H, et al. Loss of ELF5–FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-γ signalling. Nat Cell Biol. 2020;22:591–602. doi: 10.1038/s41556-020-0495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu W, Huang W, Yang Y, Qiu R, Zeng Y, Hou Y, et al. GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. Cell Death Dis. 2019;10:1–16. doi: 10.1038/s41419-019-2062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahi P, Wang CY, Lawson DA, Slorach EM, Lu A, Yu Y, et al. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc Natl Acad Sci U S A. 2017;114:3169–3174. doi: 10.1073/pnas.1701690114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Xu R. RORα, a potential tumor suppressor and therapeutic target of breast cancer. Int J Mol Sci. 2012;13(12):15755–15766. doi: 10.3390/ijms131215755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong G, Wang C, Evers BM, Zhou BP, Xu R. RORα suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012;72:1728–1739. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cieply B, Riley P, IV, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, et al. Suppression of the epithelial-mesenchymal transition by grainyhead-like-2. Cancer Res. 2012;72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mooney SM, Talebian V, Jolly MK, Jia D, Gromala M, Levine H, et al. The GRHL2/ZEB feedback loop—a key axis in the regulation of EMT in breast cancer. J Cell Biochem. 2017;118:2559–2570. doi: 10.1002/jcb.25974. [DOI] [PubMed] [Google Scholar]
- 37.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khaled N, Bidet Y. New insights into the implication of epigenetic alterations in the EMT of triple negative breast cancer. Cancers (Basel) 2019;11:559. doi: 10.3390/cancers11040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menbari MN, Rahimi K, Ahmadi A, Elyasi A, Darvishi N, Hosseini V, et al. MiR-216b-5p inhibits cell proliferation in human breast cancer by down-regulating HDAC8 expression. Life Sci. 2019;237:116945. doi: 10.1016/j.lfs.2019.116945. [DOI] [PubMed] [Google Scholar]
- 40.Menbari MN, Rahimi K, Ahmadi A, Mohammadi-Yeganeh S, Elyasi A, Darvishi N, et al. Association of hdac8 expression with pathological findings in triple negative and non-triple negative breast cancer: Implications for diagnosis. Iran Biomed J. 2020;24:283–289. doi: 10.29252/ibj.24.5.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahmani G, Sameri S, Abbasi N, Abdi M, Najafi R. The clinical significance of histone deacetylase-8 in human breast cancer. Pathol Res Pract. 2021;220:153396. doi: 10.1016/j.prp.2021.153396. [DOI] [PubMed] [Google Scholar]
- 42.Tang X, Li G, Su F, Cai Y, Shi L, Meng Y, et al. HDAC8 cooperates with SMAD3/4 complex to suppress SIRT7 and promote cell survival and migration. Nucleic Acids Res. 2020;48:2912–2923. doi: 10.1093/nar/gkaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An P, Chen F, Li Z, Ling Y, Peng Y, Zhang H, et al. HDAC8 promotes the dissemination of breast cancer cells via AKT/GSK-3β/Snail signals. Oncogene. 2020;39:4956–4969. doi: 10.1038/s41388-020-1337-x. [DOI] [PubMed] [Google Scholar]
- 44.An P, Li J, Lu L, Wu Y, Ling Y, Du J, et al. Histone deacetylase 8 triggers the migration of triple negative breast cancer cells via regulation of YAP signals. Eur J Pharmacol. 2019;845:16–23. doi: 10.1016/j.ejphar.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 45.Inoue S, Mizushima T, Fujita K, Meliti A, Ide H, Yamaguchi S, et al. GATA3 immunohistochemistry in urothelial carcinoma of the upper urinary tract as a urothelial marker and a prognosticator. Hum Pathol. 2017;64:83–90. doi: 10.1016/j.humpath.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Hisamatsu Y, Tokunaga E, Yamashita N, Akiyoshi S, Okada S, Nakashima Y, et al. Impact of GATA-3 and FOXA1 expression in patients with hormone receptor-positive/HER2-negative breast cancer. Breast Cancer. 2015;22:520–528. doi: 10.1007/s12282-013-0515-x. [DOI] [PubMed] [Google Scholar]
- 47.Riethdorf S, Frey S, Santjer S, Stoupiec M, Otto B, Riethdorf L, et al. Diverse expression patterns of the EMT suppressor grainyhead-like 2 (GRHL2) in normal and tumour tissues. Int J Cancer. 2016;138:949–963. doi: 10.1002/ijc.29841. [DOI] [PubMed] [Google Scholar]
- 48.Xiang J, Fu X, Ran W, Wang Z. Grhl2 reduces invasion and migration through inhibition of TGFβ-induced EMT in gastric cancer. Oncogenesis. 2017;6:e284. doi: 10.1038/oncsis.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meidhof S, Brabletz S, Lehmann W, Preca B, Mock K, Ruh M, et al. ZEB 1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7:831–847. doi: 10.15252/emmm.201404396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi SY, Kee HJ, Kurz T, Hansen FK, Ryu Y, Kim GR, et al. Class I HDACs specifically regulate E-cadherin expression in human renal epithelial cells. J Cell Mol Med. 2016;20:2289–2298. doi: 10.1111/jcmm.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suraweera A, O’Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front Oncol. 2018;8:92. doi: 10.3389/fonc.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho TCS, Chan AHY, Ganesan A. Thirty years of HDAC inhibitors: 2020 insight and hindsight. J Med Chem. 2020;63:12460–12484. doi: 10.1021/acs.jmedchem.0c00830. [DOI] [PubMed] [Google Scholar]
- 53.McClure JJ, Li X, Chou CJ. Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv Cancer Res. 2018;138:183–211. doi: 10.1016/bs.acr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Chakrabarti A, Melesina J, Kolbinger FR, Oehme I, Senger J, Witt O, et al. Targeting histone deacetylase 8 as a therapeutic approach to cancer and neurodegenerative diseases. Future Med Chem. 2016;8:1609–1634. doi: 10.4155/fmc-2016-0117. [DOI] [PubMed] [Google Scholar]
- 55.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 56.Emmons MF, Faião-Flores F, Sharma R, Thapa R, Messina JL, Becker JC, et al. HDAC8 regulates a stress response pathway in melanoma to mediate escape from BRAF inhibitor therapy. Cancer Res. 2019;79:2947–2961. doi: 10.1158/0008-5472.CAN-19-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joshi EM, Need A, Schaus J, Chen Z, Benesh D, Mitch C, et al. Efficiency gains in tracer identification for nuclear imaging: can in vivo LC-MS/MS evaluation of small molecules screen for successful PET tracers? ACS Chem Neurosci. 2014;5:1154–1163. doi: 10.1021/cn500073j. [DOI] [PubMed] [Google Scholar]
- 58.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, et al. Manipulation of FASTQ data with galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamdan FH, Johnsen SA. DeltaNp63-dependent super enhancers define molecular identity in pancreatic cancer by an interconnected transcription factor network. Proc Natl Acad Sci U S A. 2018;115:E12343–E12352. doi: 10.1073/pnas.1812915116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff finder: a comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 68.Prenzel T, Kramer F, Bedi U, Nagarajan S, Beissbarth T, Johnsen SA. Cohesin is required for expression of the estrogen receptor-alpha (ESR1) gene. Epigenetics and chromatin. Epigenetics Chromatin. 2012;5:13. doi: 10.1186/1756-8935-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. HDAC8 supports the induction of EMT by suppressing the MET pathway in TNBC cells surviving Chemo. Table S1. Concentration of CAF dilutions. Table S2. List of ChIP-qPCR primers. Table S3. List of RT-qPCR mouse primers. Table S4. List of RT-qPCR human primers, excel file with the list of epigenetic regulators. Table S5. List of siRNAs.
Data Availability Statement
Not applicable.