Tumor tissue collection and processing under physioxia allow highly relevant detection of signaling networks and drug sensitivity.
Abstract
Preclinical studies of primary cancer cells are typically done after tumors are removed from patients or animals at ambient atmospheric oxygen (O2, ~21%). However, O2 concentrations in organs are in the ~3 to 10% range, with most tumors in a hypoxic or 1 to 2% O2 environment in vivo. Although effects of O2 tension on tumor cell characteristics in vitro have been studied, these studies are done only after tumors are first collected and processed in ambient air. Similarly, sensitivity of primary cancer cells to anticancer agents is routinely examined at ambient O2. Here, we demonstrate that tumors collected, processed, and propagated at physiologic O2 compared to ambient air display distinct differences in key signaling networks including LGR5/WNT, YAP, and NRF2/KEAP1, nuclear reactive oxygen species, alternative splicing, and sensitivity to targeted therapies. Therefore, evaluating cancer cells under physioxia could more closely recapitulate their physiopathologic status in the in vivo microenvironment.
INTRODUCTION
The past two decades witnessed substantial advances in development of targeted therapies against pathways altered in cancer cells (1). These agents act against specific cellular dependencies, most often pathways that have undergone oncogenic dysregulation. These advances led to significant improvements in outcome in select groups of patients. Examples include patients with lung cancer with specific mutations in epidermal growth factor receptor (EGFR) who benefit from EGFR-targeted therapies (1). Similar advances have been achieved with immune checkpoint inhibitor therapies (2). While these two are major success stories, many more targeted therapies were less effective in clinic despite promising preclinical data. The best example is PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha)-targeted therapies. PIK3CA, which is mutated in ~30% of all cancer (the second most common mutation after TP53), is part of the major growth factor–activated signaling networks involved in cell proliferation, survival, and drug resistance. Many drugs targeting PIK3CA failed in clinic, and responses observed with a few tumors did not correlate with PIK3CA mutation status (3). Various mechanisms of resistance to these targeted therapies have been proposed. A convergence-based framework for cancer drug resistance was recently proposed to aid in discovery of new classes of drugs that can overcome resistance (4). Three major mechanisms of resistance described include pathway reactivation, pathway bypass, and pathway indifference. Pathway reactivation involves alterations in drug target as well as upstream, parallel, and downstream effectors of the signaling network targeted by the drug. For example, insulin feedback loop induced by phosphatidylinositol 3-kinase (PI3K) inhibitors reactivates the PI3K-mTOR (mammalian target of rapamycin) signaling axis, thus compromising the effectiveness of PI3K inhibitors. In cases of pathway bypass, the cancer cell finds alternative ways to reengage more downstream oncogenic output. Pathway bypass is often observed in PI3K inhibitor therapies where PIM (proviral integration site for moloney murine leukemia virus) kinases converge onto mTOR pathway (3). Pathway indifference is a situation in which cancer cells acquire an alternative cell state such as mesenchymal state, which permits cancer cells to escape the effects of targeted therapies. Cancer stem cells (CSCs) are typically resistant to many therapies including targeted therapies, and CSC phenotype often overlaps with mesenchymal state (5). Thus, cancers enriched for CSCs may use pathway indifference to become resistant to drugs. In addition, a clear understanding of multifactorial targeted therapy resistance mechanisms is needed to develop effective durable responses to treatment.
Most preclinical studies of cancer models including CSC characterization are done at ambient atmospheric oxygen (O2) of ~21%, while physiologic O2 concentrations vary between organs, usually ranging from 3 to 10% (6). O2 concentrations at 5, 2, 1, and 0.4% are defined as physioxia, physiologic hypoxia, pathologic hypoxia, and radiobiological hypoxia, respectively (6). While median O2 concentration in the normal breast is 6.8%, it is 1.3% in breast tumors. This range in O2 concentrations has been described for organs such as brain, liver, head and neck, lung, pancreas, and prostate and for tumors originating in these organs (6). While the effect of O2 concentration on tumor cell characteristics has been studied for a number of years through comparative analyses of cells grown under hypoxia (1%), an O2 concentration required for classic hypoxia-inducible factor (HIF) pathway activation, and ambient air, it was not recognized until recently that exposure of cells to ambient air for even minutes significantly alters the biology and enhances the differentiation of stem cells, effects noted with stem cells in young and old (7, 8). Our group demonstrated that hematopoietic stem cells (HSCs) collected and processed at ambient air undergo extensive differentiation and demonstrate lower engrafting capacity compared to HSCs collected and processed at physioxia (collection at 3% O2 and growth under 5% O2) (8). Exposure of HSCs to ambient air caused extra-physiologic oxygen shock/stress (EPHOSS), which enhanced differentiation of HSCs to hematopoietic progenitor cells.
The goal of this study was to determine how collection and processing of tumor tissues under physioxia and ambient air affect CSC characteristics, cellular signaling networks, and sensitivity to therapies. We used two transgenic mammary tumor models and ascites fluids of patients experiencing ovarian and other cancers that metastasize into the peritoneal cavity as model systems. Colon tissues from mice collected and processed at physioxia and ambient air were also analyzed as controls. In every case, tissue was harvested at physioxia, minced, split into two, and then processed at physioxia and ambient air to ensure that comparison is between two fractions of the same tissue but differ only with respect to immediate and subsequent O2 exposure. We demonstrate distinct tumor biology in cells collected and processed under physioxia compared to those collected at 3% O2 and then processed in ambient air and propose that characterizing tumors at physioxia is critical to identify physiologically/pathologically relevant signaling networks and effective responsiveness to drugs that correctly recognize their in vivo status.
RESULTS
Tumor cells display distinct CSC marker profiles under physioxia collection/processing compared to processing under ambient air
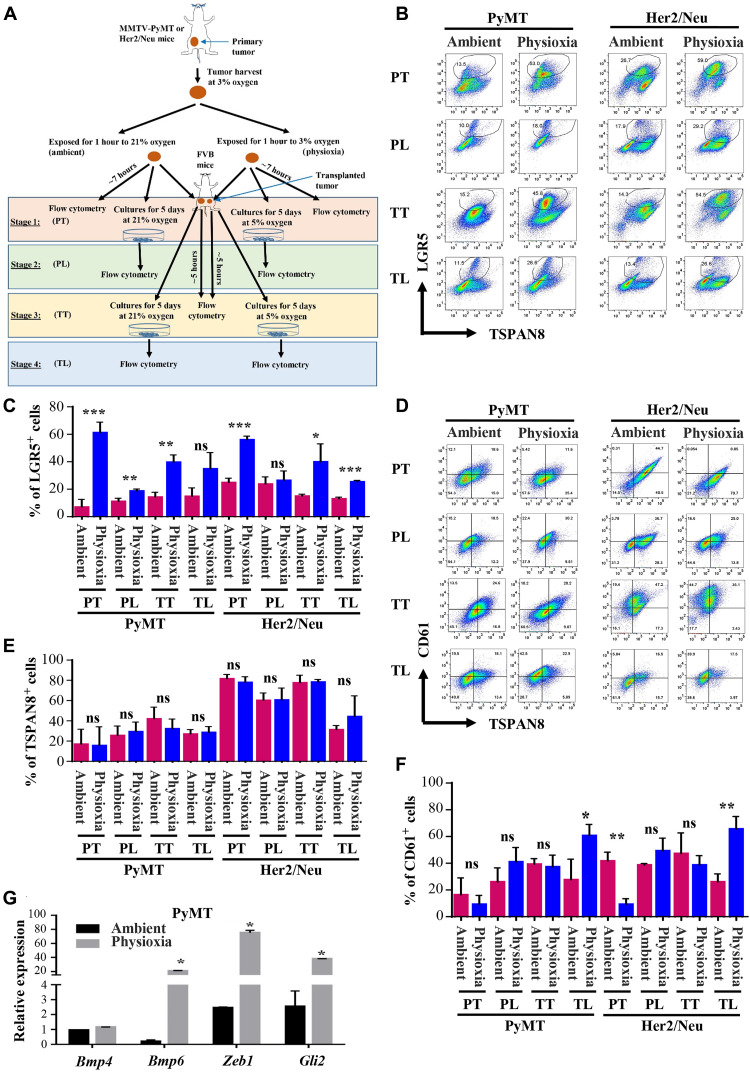
To determine whether continued exposure of tumor cells under physioxia during their collection and processing versus ambient air influences CSC marker profiles, we harvested mammary tumors from MMTV-PyMT (mouse mammary tumor virus-polyomavirus middle tumor-antigen) and MMTV-Her2/Neu+ (mouse mammary tumor virus-human epidermal growth factor receptor 2/Neu+) mice under 3% O2, minced, and divided the tumors into two fractions. Gene expression patterns in mammary tumors of MMTV-PyMT and MMTV-Her2/Neu+ transgenic mice represent Luminal B and HER2–amplified breast cancer subtypes, respectively (9). One fraction was kept at 3% O2, and the other fraction was placed at ambient air for 1 hour. We had previously demonstrated using mouse bone marrow cells that results were similar in terms of collecting cells at ambient air or placing them in ambient air for 1 hour after collecting at 3% O2 (8). After 1 hour of exposure, samples were processed at 3% O2 and ambient air, and cells were either processed immediately for CSC marker profiles, reinjected into animals, or cultured at 5% O2 versus 21% O2. Cells collected, processed, and grown at 3 to 5% O2 are referred to as physioxia and those at 21% as ambient air from here onward. We selected a 1-hour time point because it typically takes at least 1 hour for tumor tissue from surgery to go to pathology and then to research laboratories for functional studies. Samples were processed, stained, fixed, and subjected to flow cytometry for cell surface marker profiling. The cells derived from tumors were characterized in four stages as indicated in Fig. 1A. We selected LGR5 (leucine-rich repeat-containing G-protein coupled receptor 5) and TSPAN8 (tetraspanin 8) as stem cell markers of mammary tumor cells for the following reasons. LGR5-expressing cells are sufficient for postnatal mammary gland organogenesis, and LGR5 is a marker of fetal mammary stem cells (10). In addition, LGR5 is a functional biomarker for CSCs that imparts proliferative and self-renewal capacity to stem cells and has the potential to produce reactive oxygen species (ROS) (11). TSPAN8 is a marker of CSCs and activates sonic Hedgehog signaling (12).
Fig. 1. Effects of EPHOSS on CSC-associated cell surface marker levels and stemness-associated gene expression.
(A) Schematic view of the experimental design. Stage 1 (PT, primary tumor) involved processing of tumors under physioxia and ambient air, flow cytometry characterization of cells, cell propagation, and reimplantation of cells into female FVB/N mice. Stage 2 (PL, primary line) involved flow cytometry characterization of cultured cells of stage 1. Stage 3 (TT, Transplanted Tumor) involved recharacterization of tumors obtained from cell implantation of stage 1. Stage 4 (TL, transplanted line) involved flow cytometry characterization of cells grown from stage 3 tumor. PT, Primary Tumor; PL, Primary Line; TT, Transplanted Tumor; TL, Transplanted Line. (B) Representative flow cytometry profile of tumor cells stained for antibodies against LGR5 and TSPAN8. Antibodies against lineage markers CD31-PE (phycoerythrin)/Cy7, CD45-PE/Cy7, and CD140a-PE/Cy7 were used to label endothelial cells, hematopoietic cells, and fibroblasts, respectively, and only lineage-negative cells were included in the analysis. (C) Quantitation of LGR5+ cells. Differences in LGR5+ cells between ambient air and physioxia are significant [n = 3 to 6, one-way analysis of variance (ANOVA)]. (D) CD61/TSPAN8 staining patterns of tumor cells (n = 3 to 6, one-way ANOVA). (E) Quantitation of TSPAN8+ cells. (F) Quantitation of CD61+ cells. (G) Tumor cells collected and processed at physioxia express higher levels of stemness-associated genes compared to ambient air (n = 3, one-way ANOVA, P = 0.0004). *P < 0.05, **P < 0.01, and ***P < 0.001 by ANOVA. ns, not significant.
The number of LGR5+ cells was significantly higher in both models under most of the experimental stages when the tumor cells were collected and maintained at physioxia (Fig. 1, B and C). Culturing cells for 5 days reduced the number of LGR5+ cells under both O2 levels (Fig. 1, B and C). We did not observe significant changes in levels of TSPAN8 (Fig. 1, D and E). O2 levels had an effect on the number of CD61+ cells, which correspond to luminal progenitor cells that differentiate into mature luminal cells under appropriate growth conditions (13). We noted lower levels of CD61+ cells under physioxia compared to ambient air in the case of tumor cells evaluated directly, particularly Her2/Neu+ tumor cells, while culturing increased the levels of CD61+ cells under physioxia compared to ambient air (Fig. 1, D and F). We did not observe an effect of O2 levels on CD49f/epithelial cell adhesion molecule (EpCAM) staining patterns in both models except in cases of cultured Her2/Neu tumor cells (fig. S1, A to C). Cells grown under physioxia showed a modest increase in CD49f+/EpCAM+ luminal progenitor cells. Thus, O2 tension during collection and processing of tumor tissues affects cell surface levels of markers that are used to estimate and isolate CSCs in tumors. Note that staining patterns of isotype controls for flow cytometry, which were used for gating and to determine the quadrants, are shown in fig. S1 (D to F).
To determine whether the effects of transient exposure of tumor cells to physioxia versus ambient air on CSC marker profiles are permanent or reversible upon regrowth of tumor cells in vivo, we processed and retransplanted the tumor cells derived from tumors of stage 3 (stage 3, TT). The number of LGR5+ cells was substantially higher in both models in tumors derived from retransplanted cells under physioxia compared to ambient air (fig. S2A). Similarly, TSPAN8+/CD61+ double-positive cells were higher under physioxia compared to ambient air in the case of PyMT tumor cell–derived tumors (fig. S2B). As with the first set of analysis, CD49f/EpCAM staining patterns were similar under physioxia and ambient air (fig. S2C), suggesting that physioxia affects the status of select CSC markers in repeated cycles of transplantation.
To independently verify enrichment of cells with stemness-associated gene expression at physioxia compared to ambient air, we measured expression levels of Bmp4, Bmp6, Zeb1, and Gli2. Bmp4 and Bmp6 expression is reported to be higher in LGR5+/TSPAN8hi cells compared to LGR5−/TSPAN8hi, LGR5+/TSPAN8−, and LGR5−/TSPAN8− cells (10), whereas Zeb1 expression is linked to stemness and aggressive claudin-low tumor growth phenotypes (14). Gli2 is part of the major stemness-associated hedgehog signaling pathway (15). Bmp6, Zeb1, and Gli2 levels were higher in cells collected/processed under physioxia compared to ambient air (Fig. 1G).
EPHOSS influences levels of additional cancer cell surface markers, including biomarkers of immunotherapy response
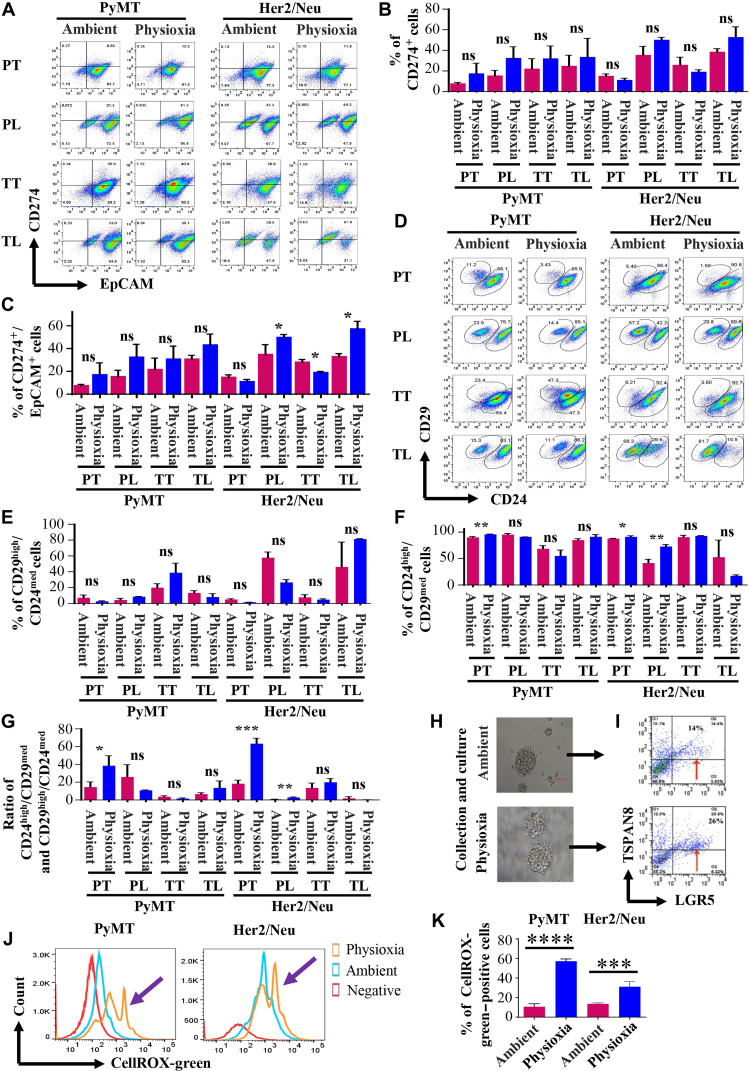
CD274/PD-L1 (programmed death-ligand 1) expression is used as a predictive biomarker of immunotherapy (16). We determined the possibility that EPHOSS affected CD274 levels, which would potentially affect the measurement of PD-L1 levels in clinical samples. Exposure to ambient air O2 during culturing, but not tissue harvest/processing, influenced CD274 levels in cancer cells in an oncogene-dependent manner (Fig. 2, A to C). For example, tumor cells cultured from Her2/Neu, but not PyMT, mice contained higher levels of EpCAM+/CD274+ cells at physioxia compared to ambient air (Fig. 2, A to C). The mechanisms responsible for oncogene-specific effects on CD274 expression under physioxia are unknown but could involve an interplay between Her2 oncogene–induced signals and transcription factors such as HIF-1, signal transducer and activator of transcription 3, nuclear factor κB (NF-κB), and AP-1 (activator protein 1) that regulate PD-L1 expression under physioxia (17).
Fig. 2. Cancer cells in physioxia and ambient air display different levels of additional cell surface markers including CD274.
(A) CD274/EpCAM staining pattern of cancer cells under physioxia and ambient air (n = 3 to 6, one-way ANOVA). (B) Quantitation of CD274+ cells. (C) Culturing under physioxia increased CD274+/EpCAM+ Her2/Neu but not PyMT cancer cells. (D) CD24/CD29 staining patterns (n = 3 to 6, one-way ANOVA). (E) O2 tension had no effect on CD29high/CD24medium basal cells. (F) Physioxia increased the levels of CD24high/CD29medium luminal cells. (G) O2 tension affected the ratio between CD24high/CD29medium luminal and CD29hi/CD24medium basal cells. (H and I) Mammospheres derived from PyMT tumor cells under physioxia contained higher levels of LGR5+ cells compared to mammospheres under ambient air. (J) Cells processed under physioxia display higher levels of nuclear ROS (n = 4, one-way ANOVA). (K) Quantitative differences in nuclear ROS in cells collected and processed under physioxia and cells processed under ambient air. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by ANOVA.
CD29hi/CD24+ basal cells in the mouse mammary gland display highest repopulating frequency in mammary gland reconstitution assays and are suggested to contain a population of quiescent stem cells (18). CD29lo/CD24+/hi cells are enriched for luminal epithelial cells. First, we observed tumor-specific differences in a subpopulation of cells. Tumors contained a distinct small population of CD29hi/CD24medium cells (Fig. 2, D to G), although their numbers did not differ between physioxia and ambient air. Culturing of cells substantially increased their number in both models and in both O2 levels, suggesting that disruption of the environment triggers proliferation of this population of cells. A ratio between CD29lo/CD24+/hi luminal and CD24medium/CD29hi basal cells was higher under physioxia compared to that in ambient air (Fig. 2G). Thus, O2 tension during tissue processing and culturing affects measurements of mammary cell hierarchy.
Because plating cells under two-dimensional conditions reduced the levels of LGR5+ cells, which are known to have CSC activity (19), we plated cells under mammosphere growth conditions, which favor self-renewal of CSC-like cells. Tumor cells formed mammospheres under both physioxia and ambient air (Fig. 2H). However, mammospheres from tumors collected, processed, and grown under physioxia contained higher numbers of LGR5+/TSPAN8medium cells compared to mammospheres from tumors processed and grown under ambient air (Fig. 2I, arrow).
Cancer cells under physioxia maintain higher levels of nuclear ROS without changes in cytoplasmic and mitochondrial ROS
ROSs, generated as by-products of O2 metabolism, play a critical role in orchestrating intracellular signaling pathways in a concentration-dependent manner (20). While high levels of ROS are deleterious, intermediate levels cause loss of quiescence and promote senescence. Low basal levels promote balanced self-renewal and differentiation, whereas extremely low levels cause impaired proliferation, differentiation, and self-renewal (20). Stemness-associated LGR5 increases ROS through the Nox1 pathway (11). ROS is also compartmentalized in cells, and effects of ROS on cellular functions are compartment dependent (21). We used membrane-permeant redox-sensitive fluorescent probes to measure ROS in different compartments of cells. While CELLROX Green dye measures ROS in the nucleus and mitochondria, CELLROX Orange dye measures cytoplasmic ROS, and MitoSOX measures mitochondrial ROS. We observed a significantly higher level of nuclear ROS under physioxia compared to ambient air (Fig. 2, J and K). To determine whether there is a correlation between the observed nuclear ROS and LGR5 or TSPAN8 positivity, we performed concurrent staining of the tumor cells collected and processed under physioxia and ambient air with the CELLROX Green and LGR5 and TSPAN8 markers. LGR5+ cells were enriched in the CellROX-green+ population both under physioxia and ambient air. This enrichment was significantly higher under physioxia compared to ambient air (fig. S3, A and B). In addition, the LGR5+ cells in the CellROX-green−/low were also significantly higher under physioxia than ambient air (fig. S3, A and B). We did not observe significant changes in levels of TSPAN8, except for the Her2/Neu cells, which exhibited a reduced level of TSPAN8+ cells in the CellROX-green+ population under physioxia than ambient air (fig. S3, C and D). These findings suggest that the LGR5+ cells generate ROS, which probably influences their homeostasis, quiescence, and differentiation properties. However, relative levels of cytoplasmic and mitochondrial ROS were similar under both O2 concentrations (fig. S3, E and F). Therefore, EPHOSS has significant effects on levels of ROS, predominantly in the nucleus, which can affect stem/differentiation characteristics of cancer cells and activities of nuclear ROS–sensitive transcription factors. In this respect, activity of several transcription factors, including those that regulate stemness-associated genes and CD274 (AP-1, NF-κB, p53, and glucocorticoid receptor), is sensitive to nuclear ROS (21).
Effects of EPHOSS on tumor incidence and characteristics of transplanted tumor cells
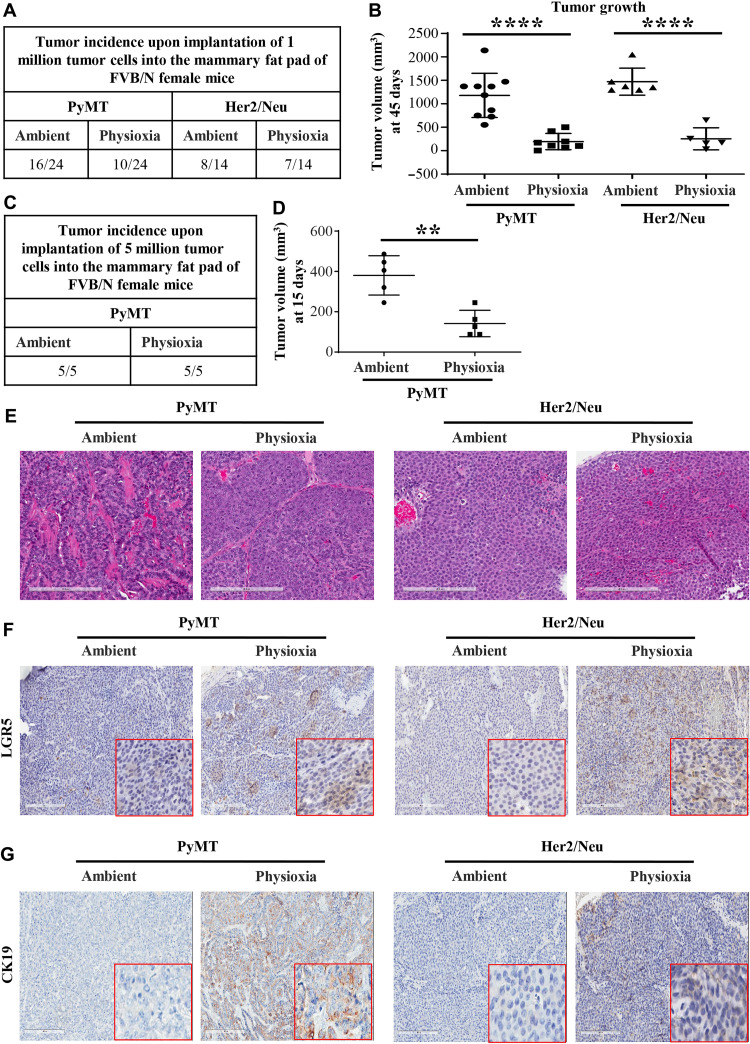
Implantation of human tumor cells at limiting dilution in immunocompromised mice is a standard assay to measure CSC activity in vivo (5). However, in assays that involved mouse tumor to mouse transplantation under immunocompetent conditions, CSC activity is not routinely observed (22). Because mouse cancer cells collected and processed under physioxia showed CSC marker profiles distinct from that of cells collected and processed under ambient air (Fig. 1), we used limiting dilution assays to determine whether tumor cells collected and processed at physioxia versus ambient air show distinct differences in tumorigenicity under syngeneic growth conditions. Note that the tumor cells used here were derived from the same mammary tumor. Tumor was collected under physioxia, and a fraction was processed at physioxia or ambient air and reimplanted into syngeneic FVB/N female mice without expanding cells in culture. While no tumors developed when 10,000, 50,000, and 100,000 cells collected under physioxia were implanted, 100,000 cells collected under ambient air developed tumors at low frequency (three of five animals). When 1 million cells were injected, cells collected under both conditions grew out as tumors (Fig. 3A). We consistently noted higher growth rates of tumors originating from tumor cells processed at ambient air (Fig. 3B). With 5 million cells in injection, tumor incidence was 100% under both conditions (Fig. 3C). However, tumors originating from tumor cells processed at ambient air initially displayed higher growth rates (Fig. 3D). We note that these differences in growth patterns persisted only for a short time when 5 million cells were implanted, as tumor developed from tumor cells collected and processed at physioxia reached a similar growth rate as ambient air–processed tumor cells when tumors were allowed to grow for more than 40 days (see below). Hematoxylin and eosin (H&E) staining of resulting tumors did not show major differences in histotypes of tumors with most of the tumors being adenocarcinomas or adenosquamous carcinomas (Fig. 3E and fig. S4A). Immunohistochemistry analysis of tumors displayed a higher level of LGR5 in tumors that developed from cells collected and processed under physioxia compared to tumors developed from cells processed under ambient air (Fig. 3F), which is consistent with the flow cytometry results. TSPAN8 positivity and EpCAM staining intensity varied between samples and did not show any consistent patterns (fig. S4, B and C). Tumors developed from cells collected and processed under physioxia retained luminal characteristics as these tumors expressed higher levels of luminal keratin CK19 (cytokeratin 19) (Fig. 3G) and were negative for basal keratin CK14 (fig. S4D). Thus, exposure of tumor tissues to ambient air during collection/processing influences growth characteristics and specific marker levels in tumors that develop in reimplantation studies.
Fig. 3. In vivo growth properties of tumor cells processed under physioxia versus ambient air.
(A) Tumor incidence by 1 million reimplanted tumor cells processed at physioxia and ambient air. (B) Tumor volume at 45 days after implantation of 1 million PyMT or Her2/Neu cells. Few outliers were removed for statistical analysis. (C) Tumor incidence by 5 million reimplanted PyMT tumor cells processed at physioxia and ambient air. (D) Tumor volume at 15 days after implantation of 5 million PyMT cells. (E) H&E staining patterns of tumors (n = 7). (F) Representative LGR5 immunohistochemistry (n = 7). (G) Representative CK19 immunohistochemistry (n = 7). ****P < 0.0001 by ANOVA.
EPHOSS induces changes in the levels of key signaling molecules
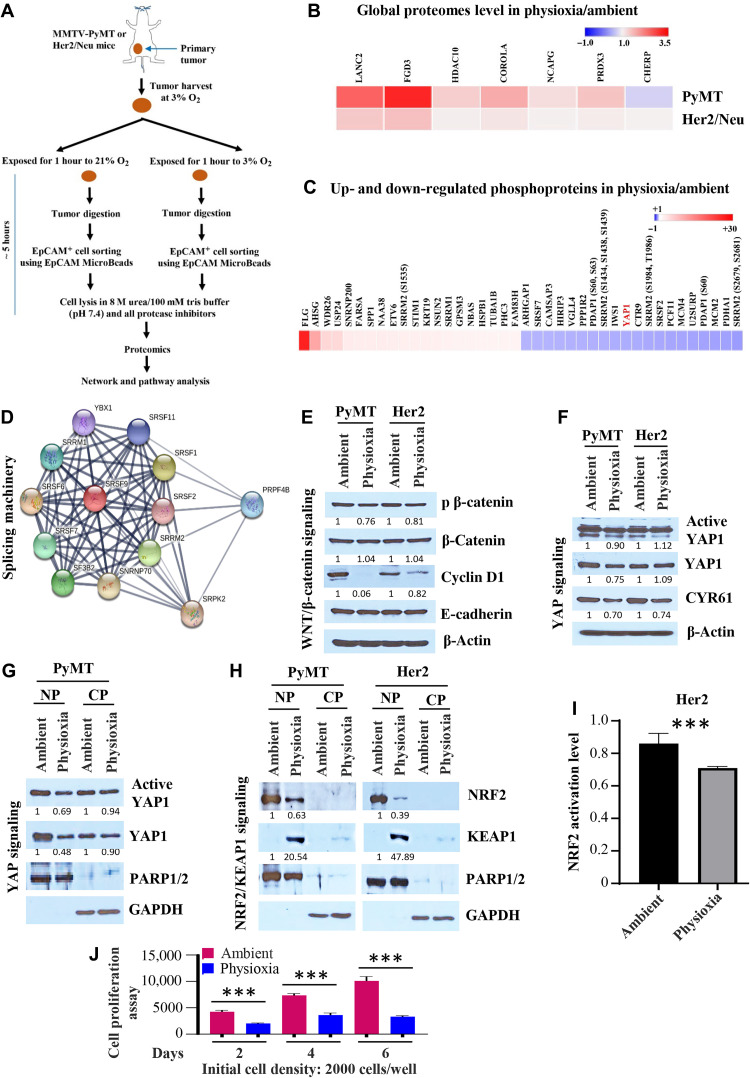
To gain insight into the effects of EPHOSS on cellular proteome and signaling networks, we collected tumors under physioxia as above and processed tumors under physioxia or ambient air. EpCAM+ epithelial cells from processed tumors were isolated under respective processing conditions using EpCAM antibody–conjugated magnetic beads to enrich for cancer cells, and cells were lysed using urea-containing buffer. Lysates were subjected to global proteomic and phosphoproteomic analysis, and proteomes unique to physioxia were identified (Fig. 4A). The entire process from tumor harvest to cell lysis took ~5 hours. Phosphoproteomics was restricted to PyMT models, as tumors in Her2 model are relatively small, and it was difficult to obtain a large quantity of proteins needed for phosphoproteomics. In the global proteome analysis, there were differences in the levels of 105 proteins in PyMT and 25 proteins in Her2 tumor cells processed under physioxia compared to those in tumor cells processed under ambient air (table S1). The top seven proteins elevated under physioxia compared to ambient air in both models are FGD3, HDAC10, CORO1a, NCAPG, PRDX3, LANC2, and CHERP (Fig. 4B). PRDX3 is a thioredoxin-dependent peroxide reductase that functions in the cytoplasm, catalyzes the reduction of H2O2, and protects cells against oxidative stress (23). Phosphoproteome analysis showed abundance of 382 phosphopeptides under physioxia and abundance of 368 phosphopeptides under ambient air (table S2). Figure 4C shows differentially expressed phosphoproteins, and specific residues affected are listed in table S2. We used STRING (search tool for the retrieval of interacting genes/proteins) interaction network to determine whether differentially expressed phosphoproteins are linked to specific signaling networks. We found six signaling clusters in phosphopeptides enriched under physioxia (fig. S5A) and seven under ambient air (fig. S5B). Phosphoproteins uniquely elevated under physioxia or ambient air are involved in RNA splicing machinery, and the splicing network includes PRPF4B, SRSF1, SRSF2, SRPK2, SRRM2, SRSF11, SNRNP70, SF3B2, SRSF9, SRSF6, SRSF7, SRRM1, and YBX1 proteins (Fig. 4D). Therefore, EPHOSS induced by ambient air affects key signaling networks in cancer cells.
Fig. 4. Impact of O2 tension during tumor collection and processing on signaling networks.
(A) Schematic view of experimental design. (B) Heatmap shows elevated protein levels under physioxia compared to ambient air. (C) Heatmap shows the top 40 differentially expressed phosphoproteins in EpCAM+ PyMT tumor cells under physioxia compared to ambient air. (D) STRING network analysis revealed that phosphoproteins described in (C) are part of RNA splicing machinery. (E) Wnt signaling network showed lower activity under physioxia compared to ambient air (n = 3). (F) Levels of total YAP1 protein and its downstream target CYR61 were lower in EpCAM+ PyMT tumor cells under physioxia compared to ambient air (n = 3). (G) Nuclear and cytoplasmic extracts were analyzed for the levels of active and total YAP1. Poly(adenosine diphosphate–ribose) polymerase 1/2 (PARP1/2) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as loading controls for nuclear (NP) and cytoplasmic (CP) extracts, respectively. (H) EpCAM+ tumor cells processed under ambient air contained higher levels of NRF2 but lower levels of its degrader, KEAP1. PARP1/2 and GAPDH were used as loading controls for nuclear and cytoplasmic extracts, respectively (n = 4). (I) Nuclear extracts of PyMT tumor cells processed under ambient air displayed higher levels of NRF2 activity compared to physioxia (n = 4). (J) PyMT tumor cells processed and grown under ambient air displayed a higher proliferation rate compared to physioxia (n = 6). ***P < 0.001 by ANOVA.
EPHOSS targets Yes-associated protein 1 and nuclear factor erythroid 2–related factor 2/Kelch-like ECH-associated protein 1 signaling networks
We then performed a detailed analysis of different signaling networks based on the noted preceding data and observed differential expression levels of Yes-associated protein 1 (YAP1), nuclear factor erythroid 2–related factor 2 (NRF2), and Wnt/β-catenin signaling. LGR5, which showed EPHOSS-dependent changes in cell surface levels, is associated with Wnt/β-catenin signaling network (19, 24). However, LGR5 needs to be activated by its ligand R-spondins 1-3 to stimulate the Wnt/β-catenin pathway, which plays a major role in mammary luminal cell differentiation (19, 24). Phosphoproteome showed lower levels of phosphorylation of YAP1 within amino acids 147 to 166 under physioxia compared to ambient air (table S2), with the decreased phosphorylation occurring on either residue S148 or S149 (fig. S5C). These residues are analogous to residues S163 and S164 within amino acids 162 to 181 of the human YAP1 protein (fig. S5D). YAP1, the Hippo pathway effector, displays context-dependent oncogenic and tumor suppressor activity in several cancer types, although its role in breast cancer remains controversial (25). Furthermore, our previous study has shown that ambient air exposure induces EPHOSS, which triggers the activation of cellular antioxidant defense mechanism involving the NRF2/KEAP1 (Kelch-like ECH-associated protein 1) signaling axis to maintain cellular redox homeostasis (8).
Although we observed elevated levels of LGR5 on the surface of cancer cells collected and processed under physioxia compared to cancer cells processed under ambient air (Fig. 1), LGR5 appears to remain in an inactive state as levels of phosphorylated (activated, pS675) β-catenin were lower in EpCAM+ PyMT tumor cells collected and processed under physioxia compared to that collected and processed under ambient air (Fig. 4E). Consequently, the level of cyclin D1, a downstream target of β-catenin (26), was lower in cells under physioxia compared to ambient air. There were no differences in E-cadherin levels, suggesting that exposure to physioxia and ambient air affects specific signaling molecules.
We examined levels of active YAP1 protein and total YAP1 protein in lysates collected under physioxia or ambient air. In the PyMT, but not in the Her2 model, levels of both active and total YAP1 proteins were lower under physioxia compared to ambient air (Fig. 4F). In addition, because phosphorylation controls the nuclear translocation of YAP1, we evaluated the levels of active (unphosphorylated) and total YAP1 proteins in the nuclear extract of PyMT tumor cells. We observed a higher expression of active and total YAP1 proteins in the nuclear extract of PyMT tumor cells under ambient air compared to physioxia. However, the expression levels of active and total YAP1 proteins in the cytoplasmic extracts showed no clear distinction in both O2 tensions (Fig. 4G). It is likely that exposure to ambient air activates the YAP1 pathway as the levels of its well-characterized downstream target CYR61 (cysteine-rich angiogenic inducer 61) were higher in lysates prepared from cells processed in ambient air compared to those collected/processed under physioxia (Fig. 4F).
Phosphorylation of YAP1 at specific residues has inhibitory effects through its degradation or stimulatory effects through transcription activation (27). For example, phosphorylation at S127 causes YAP1 degradation, whereas phosphorylation at Tyr357 stabilizes the protein (27). S164 is a known phosphorylation site in YAP1, but the physiologic function of this phosphorylation is unknown (27). Because the phosphopeptide spanning amino acids 147 to 166 of YAP1 (peptides 162 to 181 in human YAP1) was lower under physioxia compared to ambient air (fig. S5, C and D), we proceeded to evaluate the possible impact of the mutation of serine residues to nonphosphorylatable alanine residue in the YAP1 protein. To approach this, we cotransfected human embryonic kidney (HEK) 293T cells grown in physioxia and ambient air with 8xGTIIC-luciferase reporter construct containing a YAP1-responsive promoter either with or without wild-type YAP1 and S127A, S163A, S164A, or S163/164A mutant YAP1 and measured the luciferase activity. We had to use 293T cells instead of primary tumor cells for these studies because of the relatively poor transfection efficiency of primary cells. Protein expression of wild-type and mutant YAP1 constructs was confirmed in transfected cells under both ambient air and physioxia (fig. S5E). Consistent with the above results of lower endogenous active YAP1 level and its downstream target protein CYR61 under physioxia compared to ambient air, YAP1 activity, as measured through a reporter gene, was significantly lower under physioxia compared to ambient air (fig. S5F). However, exogenously overexpressed wild-type YAP1 and mutants displayed similar properties under both physioxia and ambient air (fig. S5G). For example, under both conditions, S163A mutant displayed lower activity than wild-type YAP1, whereas S164A mutant displayed higher activity. Thus, phosphorylation at S163 likely increases YAP1 activity, whereas phosphorylation at S164 reduces YAP1 activity. Differences between physioxia and ambient air are likely at the level of phosphorylation of these residues with consequent effects on YAP1 activity.
Previous studies have demonstrated NRF2 activation under stress, and we had demonstrated that ambient air exposure induces EPHOSS (7, 8). Consistent with activation of antioxidant pathway in response to EPHOSS, we found abundant nuclear levels of NRF2 in cells processed under ambient air compared to cells collected and processed under physioxia (Fig. 4H). KEAP1 levels showed an expected opposite pattern as cells prepared under physioxia contained higher levels of KEAP1. Note that collection and processing of cells in ambient air are stress inducers (8). O2 tension–dependent differences in NRF2/KEAP1 were observed in both models assessed, suggesting that these changes occur independent of oncogenes. Consistent with changes in protein levels, NRF2 activity was higher under ambient air compared to physioxia because of EPHOSS (Fig. 4I).
Signaling pathway alterations, particularly cyclin D1 level differences, based on O2 tension, suggested that cells under physioxia and ambient air proliferate at different rates. To test this possibility, we prepared cells from fresh tumors as above, sorted EpCAM+ cancer cells under physioxia or ambient air, and measured cell proliferation rates using CellTiter-Glo cell viability assay. Cells propagated under ambient air grew at much faster rates than cells grown under physioxia (Fig. 4J and fig. S5, H and I). Thus, ambient air exposure accelerates cell cycle machinery, which is not likely in vivo under physioxia.
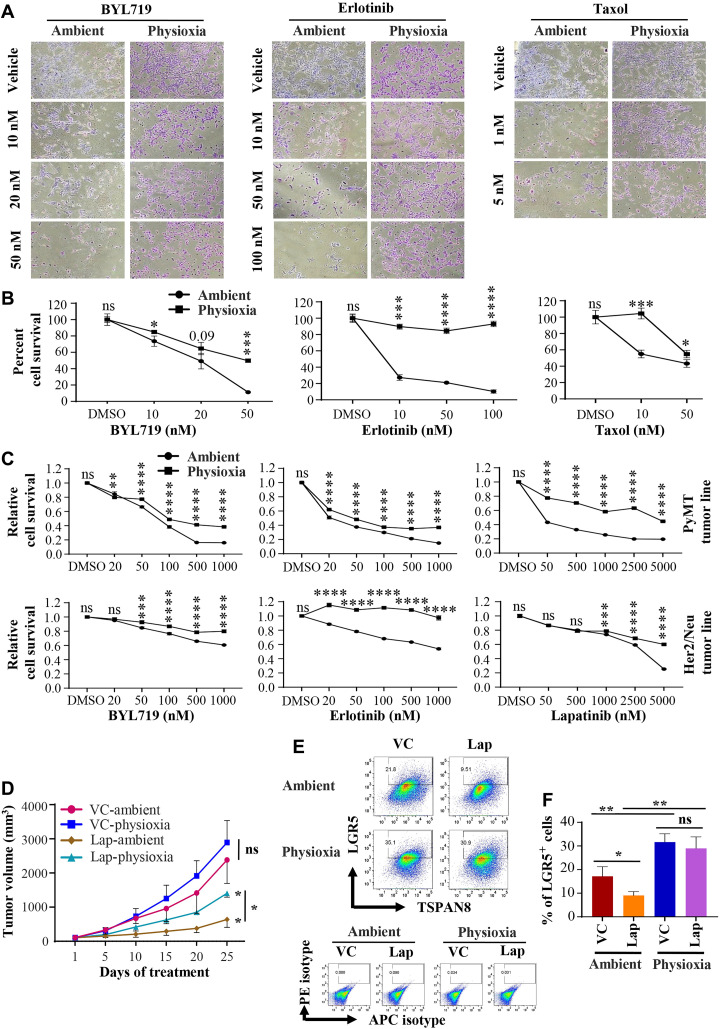
Tumor cells grown under physioxia compared to ambient air are more resistant to targeted therapies
Targeted drugs and chemotherapy are generally more effective on highly proliferating cells, and quiescent cells as well as cells with CSC-like properties tend to evade the effects of these drugs (4). Because proliferation rates and stemness-associated marker expression showed O2 tension–dependent differences, we examined whether cells collected and propagated under physioxia and propagated under ambient air show distinct differences in sensitivity to targeted therapies and to the chemotherapeutic agent Taxol. We assessed the sensitivity of tumor cells grown in ambient and physioxia to the PIK3CAα-specific inhibitor BYL719 (alpelisib), which has recently been approved as a treatment for breast cancer (28), the EGFR inhibitor erlotinib, and the dual EGFR and HER2/ERBB2/Neu inhibitor lapatinib. We selected these drugs because PI3K and EGFR/ERBB2 signaling pathways are major signaling pathways in both PyMT and Her2/Neu tumor cells (29, 30). PyMT cells cultured in physioxia were significantly more resistant to BYL719, erlotinib, lapatinib, and Taxol than cells cultured at ambient air (Fig. 5, A to C). Sensitivity of Her2/Neu tumor cells to BYL719, erlotinib, and lapatinib also varied on the basis of O2 levels, as those cells collected and propagated under physioxia showed significant resistance to these drugs (Fig. 5C).
Fig. 5. Tumor cells prepared and propagated under physioxia displayed lower sensitivity to targeted therapies and Taxol compared to tumor cells processed and propagated under ambient air.
Note that tumor cells for both conditions were derived from the same tumor. (A) Phase-contrast images of PyMT tumor–derived cells with and without drug treatment (n = 3, one-way ANOVA). (B) Quantitative measurement of cell survival data from (A). DMSO, dimethyl sulfoxide. (C) Cell proliferation rate at variable concentrations of drugs was measured using bromodeoxyuridine incorporation enzyme-linked immunosorbent assay (n = 6, one-way ANOVA). Cancer cells used in this experimental series and in (A) were derived from a different PyMT+ and Her2/Neu+ mice. (D) Mice with PyMT tumor xenograft developed from tumor cells collected and processed under ambient air and physioxia were administered daily with lapatinib (Lap) (100 mg/kg of body weight) or vehicle control (VC) via oral gavage for 25 days (n = 10, Student’s t test). Tumor growth was monitored every 5 days for 25 days. Treatment was initiated only after tumors reached similar size in both groups. (E) LGR5/TSPAN8 staining pattern of tumor cells from PyMT tumor xenografts from (D) (n = 3, one-way ANOVA). APC, allophycocyanin. (F) Quantitation of LGR5+ cells. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by ANOVA and Student’s t test.
To expand our in vitro observations, we treated xenografts of PyMT cells collected and processed both in physioxia and ambient air with lapatinib (100 mg/kg per day) or vehicle control for 25 days. Treatments were started when tumors were well established and reached around 100 to 150 mm3 in both groups. As seen in the in vitro studies, we observed that the tumors that developed from PyMT cells collected and processed under physioxia were more resistant to lapatinib compared to those in ambient air, further substantiating the in vitro data (Fig. 5D). To determine whether lapatinib targeted a different subpopulation of cells in tumors developed from cells collected/processed under physioxia compared to ambient air, we harvested and processed untreated tumors and residual tumors from treated mice under respective O2 tensions and stained cells for stemness-associated cell surface markers. LGR5+ cell population was significantly higher under physioxia in the vehicle control groups as previously observed. Notably, a significant decline in LGR5+ cells was observed following treatment in the ambient air group, whereas no significant difference was observed in the LGR5+ cell populations under the physioxia group (Fig. 5, E and F). Thus, these data suggest resistance of LGR5+ cells to lapatinib under physioxia compared to ambient air. However, there was no significant difference in the levels of TSPAN8- and CD61-expressing cells in both treated and vehicle control groups both under physioxia and ambient air (fig. S5J), indicating specificity in lapatinib targets.
Effects of EPHOSS on cells from colon
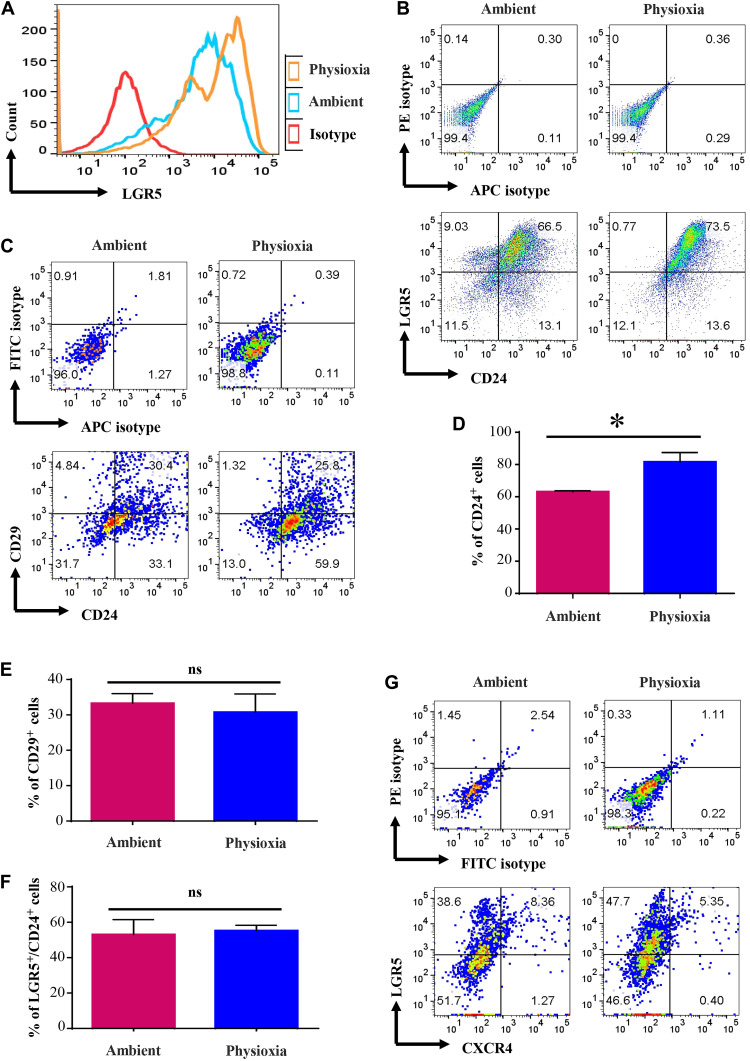
LGR5 is a major signaling molecule in colonic epithelium, and LGR5+/CD24+ population in colon is considered to be enriched for stem cells (31). In addition, LGR5 is used as a marker of CSC in colon cancer (32). Because we observed an effect of EPHOSS on LGR5+ cells in mammary tumors, we investigated whether EPHOSS has a similar effect on mouse colonic epithelial cells. Although numbers of LGR5+ cells did not differ in cells collected at the two different O2 levels, the intensity of LGR5 staining was stronger if cells were collected and processed at physioxia (Fig. 6, A and B). In addition, the number of CD24+ cells was substantially higher under physioxia compared to ambient air (Fig. 6, C and D). We did not observe significant changes in levels of CD29+ or CXCR4+ cells (Fig. 6, E to G). These results indicate that effects of EPHOSS are not restricted to mammary tissues, and EPHOSS during sample processing affects the characterization of colon CSCs.
Fig. 6. Cells derived from mouse colon collected and processed under physioxia display distinct marker profile compared to that of cells in colon processed under ambient air.
Mouse colon was collected under physioxia, washed, and minced. A fraction was transferred to ambient air for processing, whereas the other fraction was processed under physioxia (n = 3, one-way ANOVA). (A) Intensity of LGR5 staining. (B) LGR5 and CD24 staining patterns of colonic cells under physioxia and ambient air. Only lineage-negative cells were gated for the analysis. (C) CD24/CD29 staining patterns of colonic cells. FITC, fluorescein isothiocyanate. (D) Percentage of CD24+ cells under physioxia and ambient air. CD24+ cells were higher under physioxia. (E) CD29 positivity did not differ between physioxia and ambient air. (F) Number of LGR5+ cells did not differ between physioxia and ambient air, although staining intensity was higher under physioxia. (G) Physioxia did not have an influence on CXCR4 positivity. *P < 0.05 by ANOVA.
Effects of EPHOSS on human metastatic cancer cells
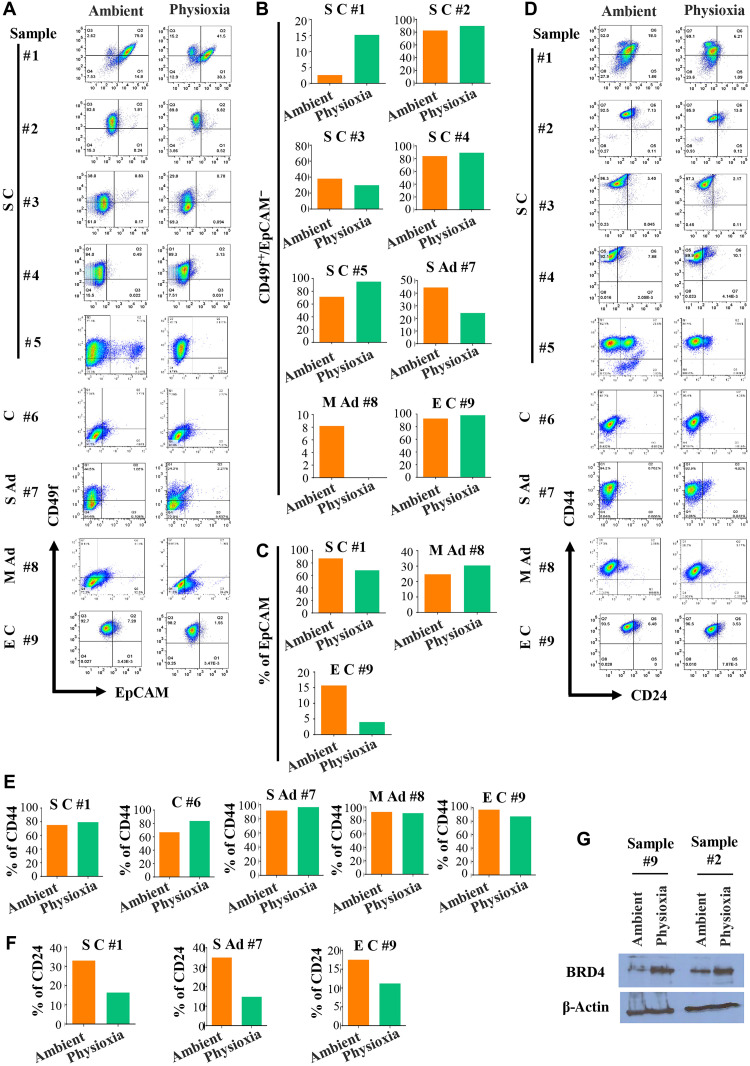
All the studies described above were performed using mouse tissues. To ensure that the above observations are relevant to human cancer, we devised a strategy to collect ascites fluids from patients with cancer, particularly ovarian cancer, under physioxia and ambient air. A uniquely designed syringe/needle (see Materials and Methods for syringe/needle information) calibrated to physioxia was used to drain ascites fluids, and another batch was drained using a regular syringe/needle at ambient air. Samples were transferred immediately to their respective O2 tension work area, and the samples were processed. Fractions of samples in three cases were washed extensively in phosphate-buffered saline (PBS) under appropriate O2 levels and lysed in RNA lysis buffer for RNA sequencing (RNA-seq). Because of the presence of a significant number of noncancer cells in the fluids, direct flow cytometry analysis of cancer cells for CSC markers was not possible. Therefore, we cultured cells for 5 days using our recently developed primary cell culturing technique not requiring feeder layer cells to enrich for adherent cells and then measured cell surface markers (33). We used ascites fluids from patients with serous ovarian carcinoma (S C), carcinoma (C), serous adenocarcinoma (S Ad), mucinous adenocarcinoma (M Ad), and endometrioid carcinoma (E C) for analysis. A representative staining pattern of cells grown from various ascites fluids is shown in Fig. 7 and fig. S6. Because of variabilities in organs from which the tumors originated and mutation patterns in tumor cells, there were sample-specific variations in the effects of physioxia and ambient air on collected and processed cells. In addition, because culturing affects cell surface marker profiles, as evident from data presented in Fig. 1, the differences in cell surface marker profiles between physioxia and ambient air were modest. For example, CD49f+/EpCAM− cells were generally higher under physioxia compared to ambient air in cases of five S C samples examined (Fig. 7, A and B). EPHOSS affected EpCAM and CD24 levels in a tumor type–specific manner (Fig. 7, A and C to F). Ovarian CSC marker CD133 expression was modest, which was unaffected by O2 tension (fig. S6A). In one cultured S C sample, which contained distinct CD274±/EpCAM+ cells, O2 tension affected CD274 positivity (fig. S6, B and C). These results are consistent with data presented in Fig. 2B on CD274 levels in cultured Her2/Neu tumor cells under physioxia. In both cases, physioxia favored CD274 expression. A potential CSC marker CD271, associated with metastatic capacity and chemoresistance (34), displayed a modest increase in levels in serous carcinoma and mucinous adenocarcinoma under physioxia, while decreased levels were observed in carcinoma and serous adenocarcinoma samples collected and processed under physioxia compared to ambient air (fig. S6, D and E). Staining patterns of isotype controls for flow cytometry are shown in fig. S6 (F and G).
Fig. 7. Cells obtained from ascites fluids of patients with cancers such as serous ovarian cancer show O2 tension–dependent variability in cell surface marker profiles.
Cells for flow cytometry analysis were cultured for 5 days, and only lineage-negative (CD31-PE, CD45-PE, and CD140b-PE negative) cells were gated for the analysis. (A) CD49f/EpCAM staining patterns (n = 9). (B) The number of CD49f+/EpCAM− cells varied on the basis of O2 tension and tumor type. (C) EpCAM positivity also showed tumor-specific variability under physioxia compared to ambient air. (D) CD24/CD44 staining patterns of cells from ascites fluid under physioxia and ambient air (n = 9). (E) Percentage of CD44+ cells in each sample under physioxia and ambient air. (F) Percentage of CD24+ cells under physioxia and ambient air. CD24+ cells were lower under physioxia compared to ambient air. (G) Cells from ascites fluids collected under physioxia contained elevated levels of BRD4 protein compared to cells collected under ambient air.
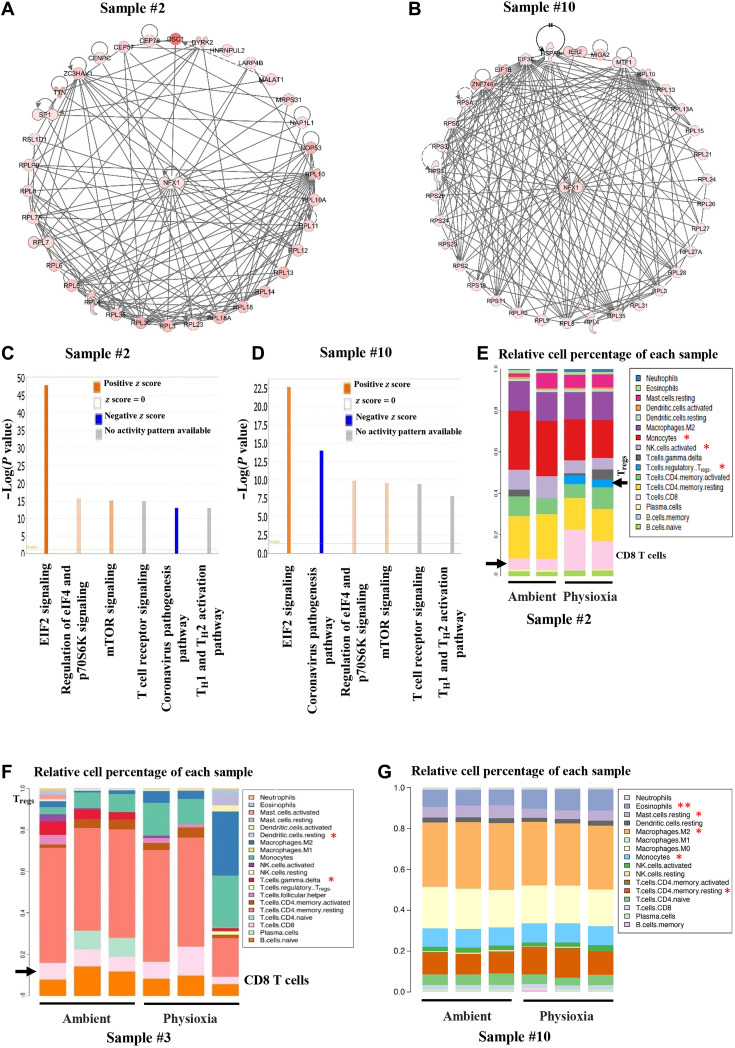
To determine EPHOSS-induced signaling pathway alterations in human cancer cells, we performed RNA-seq of cells from ascites fluids of three human patients with metastatic ovarian cancer (samples #2, #3, and #10; table S3). Unlike flow cytometry studies, RNA-seq was done directly after washing cells extensively in PBS and lysing in RNA lysis buffer. In each of these samples, EPHOSS affected the levels of >3000 genes (P value cutoff of <0.01; table S3). A heatmap of genes that showed differential levels (P < 0.01 and >2 fold change) in sample #2 (serous carcinoma of ovarian origin) and sample #10 (clear cell carcinoma of ovarian origin) collected and processed under physioxia compared to ambient air is shown in fig. S7A. Expression levels of CCR4 and CCR7, chemokine receptors involved in ovarian cancer progression or immune response (35, 36), and transforming growth factor–β pathway modulator SMAD7 (37) were different in samples collected and processed under physioxia compared to samples collected and processed under ambient air. Several epigenetic regulators including histone methyltransferases, histone demethylases, and the bromodomain protein BRD4 showed O2 tension–dependent variability in levels. This is biologically relevant as a few histone demethylases, particularly KDM5A and KDM6B, have recently been shown to have O2-sensing functions (38, 39). Transcripts of BRD4 were differentially expressed in two sets of samples. We confirmed differential levels of BRD4 in cells grown from two samples under physioxia and ambient air (Fig. 7G). Ingenuity pathway analysis of differentially expressed genes showed specific effects of O2 tension on a transcription factor network involving the transcription factor NFX1 (Fig. 8, A and B). Although not studied extensively in human, NFX1-regulated genes control H2O2 levels and ROS in Arabidopsis (40). We also identified several biological pathway differences in cells collected and processed under physioxia compared to ambient air. The top six common pathways in the two samples were EIF2 signaling, regulation of eIF4 and p70S6K signaling, mTOR signaling, T cell receptor signaling, and T helper cell 1 (TH1) and TH2 activation pathways (Fig. 8, C and D). Thus, EPHOSS likely targets protein synthesis machinery, and results obtained by collection and processing of cells in ambient air likely do not reflect these pathways in vivo, thereby providing an inaccurate measure.
Fig. 8. RNA-seq analysis of cells from ascites fluid collected and processed under physioxia shows distinct gene expression patterns compared to that of cells collected and processed under ambient air.
(A and B) Ingenuity pathway analysis of differentially expressed genes demonstrated enrichment of NFX1 signaling network under physioxia compared to ambient air. (C and D) Major signaling network alterations under physioxia compared to ambient air in cells from ascites fluids. (E to G) Differences in immune cell composition in cells collected and processed under physioxia and ambient air were determined using the CIBERSORT bioinformatics tool. Data are from duplicate (sample #2) or triplicate (samples #3 and #10) RNA-seq analysis. Asterisks denote significant differences (P < 0.05). Tregs, regulatory T cells; NK, natural killer.
We subjected RNA-seq data for the CIBERSORT tool to estimate the immune cell composition of the ovarian cancer ascites fluids processed in physioxia and ambient air. O2 tension affects a specific population of immune cells, particularly T cell subpopulations (Fig. 8, E to G).
Because phosphoproteomics and global proteomics revealed the specific effects of O2 tension on splicing machinery, we analyzed RNA-seq data for splice variants uniquely present in samples processed under physioxia compared to ambient air. As shown in fig. S7B and table S4, multiple splicing events were uniquely observed under physioxia compared to ambient air. Ingenuity pathway analysis of alternatively spliced genes showed their role in cMyc-mediated apoptosis (fig. S7C). Collectively, these results show the distinct biology of cancer cells collected and processed under physioxia compared to that processed under ambient air.
DISCUSSION
Limited reproducibility of preclinical data and translatability of preclinical data into clinical studies are two major concerns in the biomedical research field, which are potentially responsible for the limited success of clinical trials. As highlighted in a relatively recent review, current drug screening strategies are suboptimal and are facing a reproducibility crisis (41). For example, two large-scale pharmacogenomic high-throughput screening studies, the Cancer Cell Line Encyclopedia (1036 cell lines, 24 drugs) and Cancer Genome Project (727 cell lines and 138 drugs), evaluated sensitivity and gene expression profiles in drug-treated cancer cell lines. While there was significant concordance in drug-induced changes in gene expression, drug sensitivities were inconsistent between the two studies (41). Our results identify another factor that likely contributes to a lack of correlation in drug sensitivity observed in in vitro studies and spontaneously developing tumors in vivo. Our results reveal that even short-term exposure of tumor cells to ambient air is sufficient to trigger signaling changes that alter their biology and responsiveness. Proteomic analyses of cells collected and processed under physioxia and ambient air revealed differential signaling networks. We were able to demonstrate differential expression patterns of relevant signaling networks, including YAP1, NRF2/KEAP1, and WNT/β-catenin signaling. Notably, phosphoproteomic analysis showed a decreased phosphorylation of YAP1 on residue S148 or S149, which are analogous to S163 and S164 residues in human YAP1. S164 has been shown to be phosphorylated by large tumor suppressor (LATS), but the physiologic function of this phosphorylation is yet to be fully understood (42). While mutation of other LATS phosphorylation sites S127 and S381 to alanine increased the oncogenic activity of YAP1, mutation of S164 did not affect oncogenic activity. Therefore, additional studies are required to decipher the effects of O2 on YAP1 phosphorylation at S163 or S164, particularly under physiologic O2 levels. In this regard, we observed that substitution of the S164 residue to alanine resulted into an increase in YAP1 activity, whereas S163 substitution to alanine caused reduced activity, as shown via a luciferase reporter assay. Thus, further studies are needed to elucidate the effects of differential phosphorylation of YAP1 under physioxia compared to ambient air and kinases that phosphorylate these residues in O2 tension–dependent manner. Complexities in YAP signaling in cancer are just beginning to emerge, as a recent study defined anticancer activity of YAP in neural/neuroendocrine cancers and procancer activities in adenocarcinomas and small cell lung cancers with intact retinoblastoma proteins (43). However, these studies were performed under ambient air. Whether this binary function of YAP1 is reproduced under physioxia needs further investigation.
Cells processed and maintained under physioxia compared to ambient air displayed distinct differences in levels of stemness-associated signaling molecules, antioxidant response signals, and cell cycle–associated signals. It is likely that all of these signaling pathway differences are interconnected to allow cells under physioxia to maintain higher levels of quiescence, a characteristic most often observed in cells with higher stem cell activity. We observed elevated levels of BRD4 in human cancer cells collected and processed at physioxia, which helps to maintain CSC phenotype, protects against DNA damage, and confers resistance to targeted therapies (44). In addition, BRD4 signaling generates ROS (45), which is consistent with our observation of elevated nuclear ROS in cells under physioxia compared to ambient air.
One of the major consequential effects of exposing cancer cells to ambient air is on cell proliferation. We observed elevated levels of cyclin D1 in cells processed under ambient air, and these cells also proliferated at a faster rate than cells propagated under physioxia. The effects of transient exposure to ambient air on cell proliferation appear to be a long-term change, as cells exposed to ambient air established tumors rapidly in vivo compared to tumors that develop from tumor cells collected and processed under physioxia (Fig. 3A). Mechanisms associated with O2 tension–dependent changes in cyclin D1 levels need to be determined.
NRF2 levels in cells are typically kept low under basal conditions because of targeting of newly synthesized NRF2 for degradation through KEAP1 (46). Stress signals promote NRF2 stabilization and NRF2-dependent expression of antioxidant and stress response genes. We observed that processing of tumors under ambient air, but not physioxia itself, triggered NRF2 stabilization and loss of KEAP1. EPHOSS due to exposure to ambient air, coupled with stress of tissue processing under ambient air, likely activates an antioxidant response in cancer cells. We speculate that this elevation in NRF2 activity under ambient air may have contributed to the observed reduction in nuclear ROS in ambient air, relative to the cells under physioxia. NRF2 has recently been demonstrated to be required for stem cell activity of lung cancer cells, as measured by spheroid formation assay under ambient air (47). In breast cancer, two distinct types of CSCs have been described. One is highly proliferative epithelial-like CSCs (E-BCSCs) and the other, mesenchymal CSCs (M-BCSCs). E-BCSCs have higher NRF2 activity compared to M-BCSCs (48). Our results show elevated NRF2 activity in cancer cells exposed to ambient air, suggesting that many of the previously described CSC characteristics, which were mostly measured in cells collected and processed under ambient air, are influenced by O2 tension and may not truly reflect cancer cell characteristics in vivo. Furthermore, our findings provide a likely explanation as to why it appears that cancer cells constitutively express NRF2 with an impairment of KEAP1 activity. The NRF2/KEAP1 signaling pathway is one of the most extensively studied signaling pathways in the field of free radical and cancer biology. Future studies of NRF2 signaling pathway may need to consider O2 tension in experimental settings. We emphasize that none of the signaling pathway differences noted between physioxia and ambient air is due to classic hypoxia signaling pathway, as we did not observe induction of HIF-1 under physioxia growth conditions. In this respect, several O2-sensing mechanisms independent of HIF-1 signaling that include histone demethylases, E3 ubiquitin ligases such as FBXL5, and reliance on iron-sulfur cluster biosynthesis are beginning to be elucidated (38, 39, 49, 50).
How differences in O2 tension during collection and processing of tissues for a relatively short duration (2 to 5 hours, depending on assays) affect levels of various cell surface markers or cell composition, as seen with immune cells, is unknown. A possibility is that select cell types undergo rapid apoptosis or differentiation based on O2 tension. Changes previously noted for HSCs to progenitor cell fate decision under ambient air collection and processing were due to increased differentiation of HSCs rather than HSC apoptosis (8). Another possibility is that certain cell surface molecules are internalized and degraded under specific O2 levels. However, we do not have any evidence to support this latter possibility, as processing of tissues in media containing clathrin or proteasome inhibitors did not alter cell surface marker profiles.
Results presented here have practical implications in studies relevant to stem cell and cancer biology as well as drug discovery. Notably, our findings suggest a possible impact of differential O2 tensions on biomarkers of immunotherapy response. This is of clinical importance, considering the fact that the utilization of the PD-L1 biomarker as an indicator for a probable response to immune checkpoint therapy is rife with inconsistencies. For example, patients positive for PD-L1 expression do not necessarily respond to therapy. In addition, some patients who tested negative for PD-L1 have benefitted from immune checkpoint therapy (51). In line with this, a study that was carried out to evaluate PD-L1 as a predictive biomarker for immunotherapy showed PD-L1 to be predictive in only 28.9% of cases (52). It can be assumed that most of the analyses of patient samples for PD-L1 expression are typically done in ambient air. Therefore, our data suggest that the expression of PD-L1 is probably confounded by the effects of increased O2 tension and a differential effect of oncogenes (such as Her2) on the expression level, potentially through their influence on transcription factors that regulate PD-L1 expression in an O2-dependent fashion. Although it may be less logistical, but not impossible practically, to collect human tumor tissues without exposing to ambient air, minimizing exposure to ambient air before fixing tissue for pathology purpose or transfer to chambers with physiologic air should help to overcome the effects of ambient air on cell signaling. However, even here, short-term (minutes) exposure to ambient air could compromise results. Alternatively, we have found that collecting and processing mouse bone marrow and human cord blood cells at 4°C, in part, mimic the effects of physioxia collection and processing of cells (53).
In this study, we were able to demonstrate that cancer cells collected and processed under physioxia were resistant to chemotherapy and targeted therapy, both in in vitro and tumor xenograft studies. Therefore, with respect to drug discovery, drugs that prove to be effective in in vitro studies under ambient air may need to be first tested on cells derived from spontaneous mouse tumor models under physioxia condition, as we have done here before starting large-scale in vivo studies. Furthermore, drugs that work in xenograft models, which typically use cells grown under ambient air, may need to be tested in transgenic or knockout models in which tumors develop spontaneously under physioxia before considering clinical studies. Alternatively, the cells can be collected and processed at 3% O2 and injected into recipient mice under conditions that do not allow the cells to be exposed to ambient air, as we have done with mouse bone marrow and human cord blood (7, 8). These proposed steps may likely enhance clinical translation of preclinical studies for more efficacious treatment results.
MATERIALS AND METHODS
Study design
We hypothesized that 1 hour of exposure to ambient atmospheric O2 during tissue collection results in phenotypic alteration of several cell surface markers that are used for characterizing CSCs in murine mammary tumor and human ovarian cancer. We confirmed that this phenomenon not only occurs during tissue collection and handling but also is sustained when the cells from murine mammary tumors are cultured or transplanted into FVB mice. We used tumors from MMTV-PyMT and MMTV-Her2/Neu+ mouse models of breast cancer, where we first collected the solid tumors in an oxygen-controlled chamber at 3% O2, minced them, and divided them into two fractions. One fraction was kept at 3% O2, and the other was kept at ambient air for 1 hour. After 1 hour of exposure, tissues were dissociated and processed for the experiments. We extended studies to human system by collecting and processing ascites fluids of patients with cancer using a uniquely designed syringe/needle calibrated to 3% O2. Another batch of ascites fluid was collected using a regular syringe/needle at ambient air. Samples were transferred immediately to their respective O2 tension work area. We used a randomized study design to determine the cell signaling and effects of drugs on cancer cells under physioxia and ambient air.
Study approval
Collection of tissues for research purpose was approved by the Institutional Review Board of Indiana University and conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects (protocol #1106005767). All patients provided written informed consent before ascites fluid samples were used. The Indiana University Animal Care and Use Committee approved the use of animals in this study, and all procedures were performed as per the National Institutes of Health guidelines (protocol #18035).
Tissue harvesting, creating, and propagating primary tumor cells
We acclimatized all the required materials at their respective O2 conditions, i.e., 3% O2 and 21% O2 for 24 hours before experiments. Solid tumors were harvested from MMTV-PyMT and MMTV-Her2/Neu+ under 3% O2 and minced, and the tumors were divided into two fractions. One fraction was kept at 3% O2, and the other fraction was kept at ambient air (21% O2) for 1 hour. After 1 hour of exposure, tissues were dissociated using a 10% collagenase/hyaluronidase mixture (#07919, STEMCELL Technologies) in culture media for 90 min at 37°C (54). Dissociated cells were filtered through a sterile 70-μm filter, washed in media, and centrifuged for 5 min at 1000 rpm. Samples were processed at 3% O2 and ambient air, and cells were either processed immediately for CSC marker profiles, reinjected into animals, or cultured at 5% O2 and 21% O2 using Dulbecco’s modified Eagle’s medium (10-013-CV, Corning), minimum essential medium nonessential amino acids (25-025-CI, Corning), 10% fetal bovine serum, and 1% penicillin and streptomycin.
Ascites fluids were collected using a uniquely designed syringe/needle calibrated to 3% O2 (Dual Function Blue Cap with Male and Female End, B2000B, Braun), and another batch was collected using a regular syringe/needle at ambient air. Samples were transferred immediately to their respective O2 tension work area. Ascites fluids were centrifuged at 1000 rpm for 5 min to collect cell pellet and washed extensively in PBS under appropriate O2 levels, and cells were either processed immediately for RNA-seq or cultured at 5% O2 and 21% O2 on 804G conditioned media–precoated plates using a primary cell culture medium described previously (33). Medium was changed the next day to remove cellular debris and floating cells. The cells were cultured for 5 days and analyzed for the cell surface marker profiles.
Flow cytometry analysis and CellROX and MitoSOX assays
Red blood cells were lysed/removed by red blood cell lysis solution (130-094-183, Miltenyi Biotec) according to the manufacturer’s instructions. Deoxyribonuclease I (DNase I; D4513, Sigma-Aldrich) was used to clear DNA released by dead cells. Flow cytometry was performed as described previously (33). Briefly, cells were either processed directly or collected from cultured cells by trypsinization and stained using specific antibodies. The antibodies used against mouse cells were CD31-PE/Cyanine7 (A14715) from Molecular Probes; CD45-PE/Cyanine7 (25-0451-82), EpCAM-APC (allophycocyanin) (17-5791-82), and CD29-FITC (fluorescein isothiocyanate) (11-0291-82) from Invitrogen; CD140a-PE/Cyanine7 (323508) from BioLegend; LGR5-PE (phycoerythrin) (FAB8240P), TSPAN8-APC (FAB6524A), and CD49f-PE (FAB13501P) from R&D Systems; and CD61-FITC (561911), CD274-PE (558091), CD24-APC (562349), and CXCR4-FITC (551967) from BD Pharmingen; isotype control antibodies used included PE/Cyanine7 (400522) and APC (400612) from BioLegend and PE (554689) and FITC (553971) from BD Pharmingen. The antibodies used against human cells were CD31-PE (130-092-653), CD45-PE (130-080-201), EpCAM-VioBlue (130-113-266), and CD133-APC (130-090-826) from Miltenyi Biotec; CD140b-PE (558821), CD44-APC (559942), CD24-FITC (555427), and CD274-APC (563741) from BD Pharmingen; CD49f-APC (FAB13501A) from R&D Systems; and CD271-APC (345108) from BioLegend; isotype control antibodies used included PE (555749), APC (555576), and FITC (555573) from BD Pharmingen and VioBlue (130-094-670) from Miltenyi Biotec. Stained cells were acquired using a BD LSR II flow cytometer, and data were analyzed using FlowJo software.
Tumor cells were processed and stained with CELLROX Green (C10444, Life Technologies), CELLROX Orange (C10443, Life Technologies), and MitoSOX red (M36008, Molecular Probes) reagents separately at a 5 μM concentration for the detection of nuclear, cytoplasmic, and mitochondrial ROS, respectively, under 3% O2 and ambient air according to the manufacturer’s instructions.
RNA isolation and quantitative reverse transcription polymerase chain reaction
Total RNA was isolated using the RNeasy Kit (74106, Qiagen), and 2 μg of RNA was used to synthesize complementary DNA (cDNA) with the iScript cDNA Synthesis Kit (1708891, Bio-Rad) according to the manufacturer’s instructions. Quantitative reverse transcription polymerase chain reaction (PCR) was performed using TaqMan universal PCR mix (4324018, Thermo Fisher Scientific) and predesigned TaqMan assay primers with best coverage from Applied Biosystems. The following assays were used in our study: ACTB (Hs01060665_g1), Bmp4 (Mm00432087_m1), Bmp6 (Mm01332882_m1), Zeb1 (Mm00495564_m1), and Gli2 (Mm01293117_m1). Applied Biosystems StepOnePlus real-time PCR system was used for PCR and fluorescence detection, and the StepOne software was used for analysis.
Mammosphere and cell proliferation assays
For mammosphere assay, single-cell suspensions were cultured at 5% O2 and 21% O2 to form mammospheres in an ultralow-attachment six-well plate at a density of 5000 cells per well in MammoCult basal medium supplemented with MammoCult proliferation supplement (#05620), heparin (#07980) from STEMCELL Technologies Inc., hydrocortisone (#H0888, Sigma-Aldrich), and penicillin/streptomycin (#30-002-CI, Corning) as described previously (55). Phase-contrast images were captured, and mammospheres were trypsinized and processed for staining at day 5. For cell proliferation assay, cells were seeded at the indicated density in 96-well plates and grown for 2, 4, and 6 days. At the end of time points, proliferation rates were determined using CellTiter-Glo luminescent cell viability assay (G7571, Promega) according to the manufacturer’s instructions.
In vivo tumorigenicity and immunohistochemistry
Tumor cells (10,000, 50,000, 100,000, 1 × 106, or 5 × 106, total 100-μl volume) processed at 3% O2 and 21% O2 were implanted into the fifth inguinal mammary fat pad of each 6- to 8-week-old syngeneic FVB/N female mice without expanding in culture. Tumor growth was measured weekly, and tumor volume was calculated as described previously (55). Tumor tissues were harvested and divided into two fractions at their respective 3% O2 or ambient air. One fraction was collected for immunohistochemistry, and the other fraction was dissociated using a 10% collagenase/hyaluronidase enzyme mixture and processed at their respective 3% O2 or ambient air, and cells were either processed immediately for CSC marker profiles, reinjected into animals, or cultured at 5% O2 and 21% O2.
H&E, LGR5, TSPAN8, EpCAM, CK19, and CK14 immunostaining was performed at the CLIA (clinical laboratory improvement amendments)-certified Indiana University Health Pathology Laboratory, and the whole-slide digital imaging system of Aperio (ScanScope CS) was used for imaging according to a protocol described previously (55). The following antibodies were used: LGR5 (ab75732, Abcam), TSPAN8 (PA5-75311, Invitrogen), EpCAM (MA5-29698, Invitrogen), CK19 (IR 615, Dako), and CK14 (LL002, ab7800, Abcam).
Immunomagnetic separation of EpCAM+ tumor cells
Tumor tissues were dissociated using a mouse tumor dissociation kit (130-096-730, Miltenyi Biotec) and gentleMACS dissociator (Miltenyi Biotec) according to the manufacturer’s instructions and processed into single cells under 3% O2 and ambient air. Red blood cells were lysed/removed by red blood cell lysis solution, and DNase I was used to remove DNA released by dead cells. In addition, dead cells were removed via magnetic separation using the MACS dead cell removal kit (130-090-101, Miltenyi Biotec) following the manufacturer’s instructions. Before enrichment of EpCAM+ tumor cells, hematopoietic cells were removed via magnetic separation using mouse CD45 MicroBeads (130-052-301, Miltenyi Biotec) following the manufacturer’s instructions. EpCAM+ tumor cells were separated using mouse CD326 (EpCAM) MicroBeads (130-105-958, Miltenyi Biotec) following the manufacturer’s instructions.
Sample preparation, global proteome, phosphoproteome, and data analysis
Sample preparation, mass spectrometry analysis, bioinformatics, and data evaluation were performed in the Proteomics Core Facility at Indiana University School of Medicine, Indianapolis, USA, as described in the previous report (56) and vendor-provided protocols. Briefly, EpCAM+ tumor cells processed at respective O2 tensions were lysed in 8 M urea and 50 mM tris-HCl (pH 8.5) solution. Samples were sonicated in a Bioruptor sonication system from Diagende Inc. (30-s/30-s on/off cycles for 15 min, 4°C). Following centrifugation at 12,000g for 15 min, protein concentrations were determined using a Bradford protein assay (5000002, Bio-Rad). Protein samples in equal amounts (20 μg) were reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride and alkylated with 10 mM chloroacetaminde. Samples were diluted with 100 mM tris-HCl to a final urea concentration of 2 M and digested overnight with trypsin/Lys-C mix mass spectrometry (1:100 protease:substrate ratio; V5072, Promega). For phosphopeptide analysis, 360 μg of protein from EpCAM+ PyMT tumor cells under 3% O2 and ambient air was prepared as described above.
Peptide purification and labeling
Peptides were desalted on a 50-mg Sep-Pak Vac (Waters Corporation) using a vacuum manifold. After elution from the column in 70% acetonitrile (ACN) and 0.1% formic acid (FA), peptides were dried by speed vacuum and resuspended in 24 μl of 50 mM triethylammonium bicarbonate. Peptide concentration was measured using the Pierce Quantitative Colorimetric Peptide Assay Kit (23275, Thermo Fisher Scientific) to ensure that an equal amount of each sample was labeled. Samples were then tandem mass tag (TMT)–labeled with 0.2 and 2 mg of reagent for global and phosphoproteomic studies, respectively, for 2 hours at room temperature (global: TMT lot UD278759A: PyMT ambient-1 126, PyMT physioxia-1 127C, PyMT ambient-2 128C, PyMT physioxia-2 129C, Her2 ambient-1 127N, Her2 ambient-2 128N, Her2 physioxia-1 129N, and Her2 physioxia-2 130N; phospho: TMTpro lot UL297970: PyMT ambient 126N and PyMT physioxia 128N). Labeling reactions were quenched with hydroxylamine at room temperature for 15 min. Labeled peptides were then mixed by global and phospho sets, respectively, and dried by speed vacuum.
Phosphopeptide enrichment
Phosphopeptides were enriched by applying the larger samples prepared above to High-Select TiO2 Phosphopeptide Enrichment tips (A32993, Thermo Fisher Scientific). Tips were washed and eluted using vendor-described protocols.
High-pH basic fractionation
The peptide mixtures (global and phosphopeptide-enriched) were resuspended in 0.1% trifluoroacetic acid and fractionated on Pierce High-pH reversed-phase peptide fractionation spin columns using vendor methodology (84868, Thermo Fisher Scientific). Eight global fractions and 4 phospho fractions were then dried by speed vacuum and resuspended in 24 μl of 0.1% FA.
Nano–liquid chromatography–tandem mass spectrometry analysis
Nano–liquid chromatography (LC)–tandem mass spectrometry analyses were performed on an EASY-nLC HPLC system coupled to an Exploris 480 mass spectrometer (Thermo Fisher Scientific). For global samples, one-third of each fraction was loaded onto a reversed-phase PepMap RSLC C18 column with an Easy-Spray tip at 400 nl/min (ES802A, Thermo Fisher Scientific; 2 μm, 100 Å, 75 μm by 25 cm). Peptides were eluted from 4 to 35% B over 160 min, 35 to 50% B for 14 min, and dropping from 50 to 10% B over the final 1 min (mobile phases A: 0.1% FA and water; B: 0.1% FA and 80% ACN). Mass spectrometer settings include a capillary temperature of 300°C, and ion spray voltage was kept at 1.8 kV. The mass spectrometer method was operated in positive-ion mode with a 4-s cycle time data-dependent acquisition with advanced peak determination and Easy-IC on (internal calibrant). Precursor scans [mass/charge ratio (m/z), 400 to 1750] were done with an Orbitrap resolution of 120,000, 40% RF (radio frequency) lens, auto maximum inject time (IT) and standard automatic gain control (AGC) target, including charges of 2 to 6 for fragmentation with 45-s dynamic exclusion. Higher-energy collisional dissociation (HCD) MS2 scans were performed at 50k Orbitrap resolution, fixed collision energy of 36%, 200% normalized AGC target, and 50-ms maximum IT.
For phosphopeptide samples, one-half of each fraction was analyzed with the same LC conditions as above on a Lumos Orbitrap mass spectrometer (Thermo Fisher Scientific). Mass spectrometer settings include a capillary temperature of 300°C, and ion spray voltage was kept at 1.9 kV. The mass spectrometer method was operated in positive-ion mode with a 4-s cycle time data-dependent acquisition with advanced peak determination and Easy-IC on (internal calibrant). Precursor scans (m/z, 400 to 1750) were done with an Orbitrap resolution of 120,000, 30% RF lens, 50-ms maximum IT, and standard AGC target, including charges of 2 to 6 for fragmentation with 45-s dynamic exclusion. HCD MS2 scans were performed at 50k Orbitrap resolution, fixed collision energy of 34%, 100% normalized AGC target, and 90-ms maximum IT.
Proteomic data analysis
Resulting RAW files were analyzed in Proteome Discover 2.4 (Thermo Fisher Scientific) with FASTA databases including Swiss-Prot UniProt Mus musculus sequences plus common contaminants. Quantification methods used isotopic impurity levels available from Thermo Fisher Scientific. SEQUEST HT searches were conducted with a maximum number of two missed cleavages, a precursor mass tolerance of 10 parts per million, and a fragment mass tolerance of 0.02 Da. Static modifications used for the search were (i) carbamidomethylation on cysteine (C) residues and (ii) TMT sixplex (global or TMTpro for phosphor) label on lysine (K) residues and the N termini of peptides. Dynamic modifications used for the search were oxidation of methionine and acetylation of N termini. Percolator false discovery rate (FDR) was set to a strict setting of 0.01 and a relaxed setting of 0.05. Values from both unique and razor peptides were used for quantification. In the consensus workflow, peptides were normalized by total peptide amount with no scaling. Resulting grouped abundance values for each sample type and abundance ratio values were exported to Microsoft Excel and are available as supplementary material files. Proteome data have been deposited with ProteomeXchange Consortium, project accession: PXD023133.
RNA-seq and data analysis
The concentration and quality of total RNA samples were first assessed using Agilent 2100 Bioanalyzer. An RNA integrity number of five or higher was required to pass the quality control. Five hundred nanograms of RNA per sample was used to prepare a dual-indexed strand-specific cDNA library using the TruSeq Stranded mRNA Library Prep Kit (Illumina). The resulting libraries were assessed for their quantity and size distribution using Qubit and Agilent 2100 Bioanalyzer. Pooled libraries (200 pM) were used per flowcell for clustering amplification on cBot using the HiSeq 3000/4000 PE Cluster Kit and sequenced with 2 × 75–base pair paired-end configuration on HiSeq 4000 (Illumina) using the HiSeq 3000/4000 PE SBS Kit. A Phred quality score (Q score) was used to measure the quality of sequencing. More than 90% of the sequencing reads reached Q30 (99.9% base call accuracy).
The sequencing data were first assessed using FastQC (Babraham Bioinformatics) for quality control. All sequenced libraries were mapped to the human genome (UCSC hg19) using STAR RNA-seq aligner with the following parameter: “--outSAMmapqUnique 60.” The read distribution across the genome was assessed using bamutils (from ngsutils) (57). Uniquely mapped sequencing reads were assigned to hg19 refGene genes using featureCounts (from subread) with the following parameters: “-s 2 –p –Q 10.” Genes with a read count per million (CPM) of >0.5 in more than three of the samples were kept. The data were normalized using the TMM (trimmed mean of M values) method. Differential expression analysis was performed using edgeR R package (57). FDR was computed from P values using the Benjamini-Hochberg procedure. The multidimensional scaling (MDS) plot was drawn using the plotMDS function in edgeR to visualize the differences between the expression profiles of different samples in two dimensions.
Sequence alignment and gene counts
The sequencing data were first assessed using FastQC (v.0.11.5, Babraham Bioinformatics, Cambridge, UK) for quality control. All sequenced libraries were mapped to the human genome (UCSC hg38) using STAR RNA-seq aligner (v.2.5) with the following parameter: “--outSAMmapqUnique 60.” The read distribution across the genome was assessed using bamutils (from ngsutils v.0.5.9) (57). Uniquely mapped sequencing reads were assigned to hg38 refGene genes using featureCounts (subread v.1.5.1) with the following parameters: “-s 2 –p –Q 10.” Each patient was analyzed independently, and genes with a read CPM of <0.5 in more than the number of sample replicates in one group were removed from the comparisons. The data were normalized using the TMM method. MDS analysis was done with limma (v.3.38.3). Differential expression analysis was performed using edgeR (v.3.12.1) (57). FDR was computed from P values using the Benjamini-Hochberg procedure.
Splicing analysis
Splicing analysis was performed using rMATS (v.4.1.0). Fastq files were again aligned with STAR RNA-seq aligner (v.2.5) to the human genome (UCSC hg38) but with parameters optimized for detection of reads across splice junctions according to the default rMATS settings. Specifically, the options used were “--chimSegmentMin 2 --outFilterMismatchNmax 3 --alignEndsType EndToEnd --outSAMstrandField intronMotif --alignSJDBoverhangMin 6 --alignIntronMax 299999.”
Bioinformatic analysis and CIBERSORT analysis
Proteomic and transcriptomic data were sorted on the basis of a P value cutoff of 0.01 and analyzed using the QIAGEN Ingenuity Pathway Analysis software. A core analysis was carried out to identify relevant canonical pathways and gene networks. Proteomic data were also analyzed with the STRING database for the identification of potential protein-protein interactions (58). The proteomic heatmaps were generated using the Morpheus web-based tool from Broad Institute to show differences in protein levels. The CIBERSORT tool was used to estimate the immune cell composition in RNA-seq data of ovarian cancer ascites fluids processed in physioxia and ambient air (59).
Nuclear/cytoplasmic protein extraction and NRF2 transcription factor activation assay
Tumor tissues were dissociated and processed into single cells under 3% O2 and ambient air, and red blood cells were lysed/removed using red cell lysis buffer. Nuclear and cytoplasmic proteins were extracted from tumor cells under respective O2 conditions using NE-PER nuclear and cytoplasmic extraction reagents (78833, Thermo Fisher Scientific) following the manufacturer’s instructions. The NRF2 activation assay was carried out with the NRF2 transcription factor assay kit (colorimetric, ab207223, Abcam) using 6.5 μg of nuclear proteins as per the manufacturer’s instructions.
Antibodies and Western blotting
Primary antibodies used included rabbit anti–phospho-β-catenin (#4176), rabbit anti-cyclin D1 (#55506), rabbit anti-YAP1 (#4912), rabbit anti-CYR61(#39382), rabbit anti-KEAP1 (#4678), mouse anti–E-cadherin (#5296), rabbit anti-BRD4 (#13440), and rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (#5174) from Cell Signaling Technology; rabbit anti-active YAP1 (ab205270) and rabbit anti-NRF2 (ab137550) from Abcam; mouse anti–β-catenin (610154) from BD Transduction Laboratories; rabbit anti–poly(adenosine diphosphate–ribose) polymerase 1/2 (sc-7150) from Santa Cruz Biotechnology; and mouse anti–β-actin (A5441, Sigma-Aldrich). Anti-mouse (#7076) and anti-rabbit (#7074) horseradish peroxidase–linked secondary antibodies were purchased from Cell Signaling Technology. Tumor tissues were dissociated/processed into single cells at 3% O2 and ambient air, and we lysed/removed the red blood cells. Cell lysates were prepared in radioimmunoassay buffer and analyzed by Western blotting as described previously (55).
Transfections and luciferase assays
HEK293T cells (5 × 105) grown in physioxia and ambient air were cotransfected with 2 μg of 8xGTIIC-luciferase reporter plasmid (#34615, Addgene) containing YAP1-responsive promoter and either pcDNA4/HisMaxB-YAP1 wild type (#18978, Addgene) or pcDNA4/HisMaxB-YAP1-S127A (#18988, Addgene)/pcDNA4/HisMaxB-YAP1-S163A (#18989, Addgene)/pcDNA4/HisMaxB-YAP1-S164A (#18990, Addgene)/pCMV-Flag-YAP-S163/164A (#33089, Addgene) mutant plasmids (42) in 60-mm dishes using Lipofectamine 3000 (L3000-015, Invitrogen Life Technologies) according to the manufacturer’s instructions. Renilla luciferase (0.25 μg) was added to each transfection as a control. Luciferase activity was measured using a Dual-Luciferase Reporter assay system (E1910, Promega) 48 hours after transfection according to the manufacturer’s instructions.
In vitro and in vivo drug sensitivity assays
Cells (3 × 105) collected and processed at 3% O2 and processed at ambient air were seeded in a six-well plate under respective O2 levels and treated with indicated concentrations of drugs BYL719 (S2814, Selleckchem), erlotinib (S7786, Selleckchem), Taxol (paclitaxel, T1912, Sigma-Aldrich), and lapatinib (S2111, Selleckchem) for 72 hours. Colonies were stained with crystal violet (C0775, Sigma-Aldrich) according to the manufacturer’s instructions and imaged under a microscope. For cell proliferation assay, 5000 cells were seeded in 96-well plates under 3% O2 and ambient air followed by treatment with indicated concentrations and drugs for 72 hours. At the end of treatments, bromodeoxyuridine (BrdU) incorporation enzyme-linked immunosorbent assay was performed using a BrdU cell proliferation assay kit (QIA58, Calbiochem/Millipore) according to the manufacturer’s instructions.
PyMT tumor cells (5 × 106, total 100-μl volume) collected and processed at 3% O2 and 21% O2 were implanted into the fifth inguinal mammary fat pad of each 6- to 8-week-old syngeneic FVB/N female mouse (total of 40 mice, 10 for each group). The growth of xenograft tumors was measured thrice a week using calipers before the initiation of treatment, and tumor volume in cubic millimeters was calculated as described above. When tumors reached an average size of 100 to 150 mm3, mice were randomized, and treatment was started. Lapatinib (100 mg/kg of body weight) or vehicle control was administered daily via oral gavage in polyethylene glycol 300 (S6704, Selleckchem), Tween 80 (S6702, Selleckchem), and water as per the manufacturer’s instructions for 25 days, and tumor growth during treatment was monitored every 5 days. Data are presented as mean volumes for each group, and error bars represent the SD of mean. Upon termination of treatment, mice were euthanized, and tumor tissues were harvested, processed, and stained for cell surface marker analysis under respective O2 tensions.
Statistical analysis
The experiments were performed in three or more biological replicates. Statistical analyses were conducted using the Prism software program (version 6.0). Data were analyzed using one-way analysis of variance (ANOVA). P values below 0.05 were considered statistically significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by ANOVA.
Acknowledgments
We thank members of the IUSCC flow cytometry core, Medical Genomics, animal facility, and tissue procurement cores at the NCI-designated IU Simon Comprehensive Cancer Center for their services (P30-CA082709). Acquisition of the IUSM Proteomics core instrumentation used for this project, the Orbitrap Fusion Lumos, was provided by the Indiana University Precision Health Initiative.
Funding: Falk Medical Trust Catalyst and Transformative Awards (to H.N. and H.E.B.) and an R35 HL139599 Outstanding Investigator Award (to H.E.B.) funded this study. Work in the IUSM Proteomics Core was supported, in part, by the Indiana Clinical and Translational Sciences Institute, which is funded by award number UL1TR002529 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Author contributions: Conception and design: B.K., H.E.B., and H.N. Development of methodology: B.K., A.K.A., M.P., M.L.C., P.B.-N., and H.N. Acquisition of data: B.K., A.K.A., M.P., M.L.C., R.W., P.B.-N., M.A., E.S., D.C., Y.L., J.M.S., A.B.C., C.M., C.J.T., G.S., E.H.D., A.B.W., A.L.M., H.E.B., and H.N. Analysis and interpretation of data: B.K., A.K.A., M.P., P.B.-N., A.B.C., G.S., M.L.C., H.E.B., and H.N. Administrative, technical, or material support: P.B.-N., H.E.B., and H.N. Writing, review, and/or revision of the manuscript: B.K., A.K.A, M.L.C., H.E.B., and H.N. Study supervision: H.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw mass spectrometry proteomic datasets of mouse tumor reported in this paper are available at ProteomeXchange with identifier PXD023133. Lists of differentially expressed genes and splicing events under physioxia compared to ambient air are presented in tables S3 and S4. The raw RNA-seq data can be provided by H.N. pending scientific review and a completed material transfer agreement. Requests for the unique reagents should be submitted to H.N.
Supplementary Materials
This PDF file includes:
Legends for tables S1 to S4
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.de Bono J. S., Ashworth A., Translating cancer research into targeted therapeutics. Nature 467, 543–549 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Kalbasi A., Ribas A., Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20, 25–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le X., Antony R., Razavi P., Treacy D. J., Luo F., Ghandi M., Castel P., Scaltriti M., Baselga J., Garraway L. A., Systematic functional characterization of resistance to PI3K inhibition in breast cancer. Cancer Discov. 6, 1134–1147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konieczkowski D. J., Johannessen C. M., Garraway L. A., A convergence-based framework for cancer drug resistance. Cancer Cell 33, 801–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clevers H., The cancer stem cell: Premises, promises and challenges. Nat. Med. 17, 313–319 (2011). [DOI] [PubMed] [Google Scholar]
- 6.McKeown S. R., Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 87, 20130676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capitano M. L., Mohamad S. F., Cooper S., Guo B., Huang X., Gunawan A. M., Sampson C., Ropa J., Srour E. F., Orschell C. M., Broxmeyer H. E., Mitigating oxygen stress enhances aged mouse hematopoietic stem cell numbers and function. J. Clin. Invest. 131, e140177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantel C. R., O’Leary H. A., Chitteti B. R., Huang X., Cooper S., Hangoc G., Brustovetsky N., Srour E. F., Lee M. R., Messina-Graham S., Haas D. M., Falah N., Kapur R., Pelus L. M., Bardeesy N., Fitamant J., Ivan M., Kim K. S., Broxmeyer H. E., Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell 161, 1553–1565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfefferle A. D., Herschkowitz J. I., Usary J., Harrell J. C., Spike B. T., Adams J. R., Torres-Arzayus M. I., Brown M., Egan S. E., Wahl G. M., Rosen J. M., Perou C. M., Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 14, R125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu N. Y., Rios A. C., Pal B., Law C. W., Jamieson P., Liu R., Vaillant F., Jackling F., Liu K. H., Smyth G. K., Lindeman G. J., Ritchie M. E., Visvader J. E., Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 19, 164–176 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Ohata H., Shiokawa D., Obata Y., Sato A., Sakai H., Fukami M., Hara W., Taniguchi H., Ono M., Nakagama H., Okamoto K., NOX1-dependent mTORC1 activation via S100A9 oxidation in cancer stem-like cells leads to colon cancer progression. Cell Rep. 28, 1282–1295.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Zhu R., Gires O., Zhu L., Liu J., Li J., Yang H., Ju G., Huang J., Ge W., Chen Y., Lu Z., Wang H., TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat. Commun. 10, 2863 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asselin-Labat M. L., Sutherland K. D., Barker H., Thomas R., Shackleton M., Forrest N. C., Hartley L., Robb L., Grosveld F. G., van der Wees J., Lindeman G. J., Visvader J. E., Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Morel A. P., Hinkal G. W., Thomas C., Fauvet F., Courtois-Cox S., Wierinckx A., Devouassoux-Shisheboran M., Treilleux I., Tissier A., Gras B., Pourchet J., Puisieux I., Browne G. J., Spicer D. B., Lachuer J., Ansieau S., Puisieux A., EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLOS Genet. 8, e1002723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh Y., Katoh M., Hedgehog target genes: Mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 9, 873–886 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z. Q., Zhao J. J., Pan Q. Z., Chen C. L., Liu Y., Tang Y., Zhu Q., Weng D. S., Xia J. C., PD-L1 expression is a predictive biomarker for CIK cell-based immunotherapy in postoperative patients with breast cancer. J. Immunother. Cancer 7, 228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerdes I., Matikas A., Bergh J., Rassidakis G. Z., Foukakis T., Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: Biology and clinical correlations. Oncogene 37, 4639–4661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.dos Santos C. O., Rebbeck C., Rozhkova E., Valentine A., Samuels A., Kadiri L. R., Osten P., Harris E. Y., Uren P. J., Smith A. D., Hannon G. J., Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc. Natl. Acad. Sci. U.S.A. 110, 7123–7130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L., Tang H., Kong Y., Xie X., Chen J., Song C., Liu X., Ye F., Li N., Wang N., Xie X., LGR5 promotes breast cancer progression and maintains stem-like cells through activation of Wnt/β-catenin signaling. Stem Cells 33, 2913–2924 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Tan D. Q., Suda T., Reactive oxygen species and mitochondrial homeostasis as regulators of stem cell fate and function. Antioxid. Redox Signal. 29, 149–168 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Go Y. M., Jones D. P., Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780, 1273–1290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly P. N., Dakic A., Adams J. M., Nutt S. L., Strasser A., Tumor growth need not be driven by rare cancer stem cells. Science 317, 337 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ismail T., Kim Y., Lee H., Lee D. S., Lee H. S., Interplay between mitochondrial peroxiredoxins and ROS in cancer development and progression. Int. J. Mol. Sci. 20, 4407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oskarsson T., Acharyya S., Zhang X. H., Vanharanta S., Tavazoie S. F., Morris P. G., Downey R. J., Manova-Todorova K., Brogi E., Massagué J., Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 17, 867–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamar J. M., Stern P., Liu H., Schindler J. W., Jiang Z. G., Hynes R. O., The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl. Acad. Sci. U.S.A. 109, E2441–E2450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecarpentier Y., Schussler O., Hebert J. L., Vallee A., Multiple targets of the canonical WNT/β-catenin signaling in cancers. Front. Oncol. 9, 1248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Li Y., Luo J., Hou N., Molecular mechanism of hippo-YAP1/TAZ pathway in heart development, disease, and regeneration. Front. Physiol. 11, 389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayan P., Osgood C. L., Singh H., Chiu H. J., Ricks T. K., Chow E. C. Y., Qiu J., Song P., Yu J., Namuswe F., Guiterrez-Lugo M., Hou S., Pierce W. F., Goldberg K. B., Tang S., Amiri-Kordestani L., Theoret M. R., Pazdur R., Beaver J. A., FDA approval summary: Fam-trastuzumab deruxtecan-Nxki for the treatment of unresectable or metastatic HER2-positive breast cancer. Clin. Cancer Res. 27, 4478–4485 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pylayeva Y., Gillen K. M., Gerald W., Beggs H. E., Reichardt L. F., Giancotti F. G., Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J. Clin. Invest. 119, 252–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorpe L. M., Spangle J. M., Ohlson C. E., Cheng H., Roberts T. M., Cantley L. C., Zhao J. J., PI3K-p110α mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85α. Proc. Natl. Acad. Sci. U.S.A. 114, 7095–7100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King J. B., von Furstenberg R. J., Smith B. J., McNaughton K. K., Galanko J. A., Henning S. J., CD24 can be used to isolate Lgr5+ putative colonic epithelial stem cells in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G443–G452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merlos-Suarez A., Barriga F. M., Jung P., Iglesias M., Cespedes M. V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Munoz P., Clevers H., Sancho E., Mangues R., Batlle E., The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Prasad M., Kumar B., Bhat-Nakshatri P., Anjanappa M., Sandusky G., Miller K. D., Storniolo A. M., Nakshatri H., Dual TGFβ/BMP pathway inhibition enables expansion and characterization of multiple epithelial cell types of the normal and cancerous breast. Mol. Cancer Res. 17, 1556–1570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lathia J. D., Li M., Sinyuk M., Alvarado A. G., Flavahan W. A., Stoltz K., Rosager A. M., Hale J., Hitomi M., Gallagher J., Wu Q., Martin J., Vidal J. G., Nakano I., Dahlrot R. H., Hansen S., McLendon R. E., Sloan A. E., Bao S., Hjelmeland A. B., Carson C. T., Naik U. P., Kristensen B., Rich J. N., High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep. 6, 117–129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng S., Guo J., Yang Q., Yang X., Crk-like adapter protein regulates CCL19/CCR7-mediated epithelial-to-mesenchymal transition via ERK signaling pathway in epithelial ovarian carcinomas. Med. Oncol. 32, 47 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Chang D. K., Peterson E., Sun J., Goudie C., Drapkin R. I., Liu J. F., Matulonis U., Zhu Q., Marasco W. A., Anti-CCR4 monoclonal antibody enhances antitumor immunity by modulating tumor-infiltrating Tregs in an ovarian cancer xenograft humanized mouse model. Onco. Targets. Ther. 5, e1090075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q., Inhibitory SMADs: Potential regulators of ovarian function. Biol. Reprod. 92, 50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batie M., Frost J., Frost M., Wilson J. W., Schofield P., Rocha S., Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty A. A., Laukka T., Myllykoski M., Ringel A. E., Booker M. A., Tolstorukov M. Y., Meng Y. J., Meier S. R., Jennings R. B., Creech A. L., Herbert Z. T., McBrayer S. K., Olenchock B. A., Jaffe J. D., Haigis M. C., Beroukhim R., Signoretti S., Koivunen P., Kaelin W. G. Jr., Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisso J., Schröder F., Schippers J. H., Müssig C., NFXL2 modifies cuticle properties in Arabidopsis. Plant Signal. Behav. 7, 551–555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatzis C., Bedard P. L., Birkbak N. J., Beck A. H., Aerts H. J., Stem D. F., Shi L., Clarke R., Quackenbush J., Haibe-Kains B., Enhancing reproducibility in cancer drug screening: How do we move forward? Cancer Res. 74, 4016–4023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L., A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson J. D., Huang K., Pacal M., McCurdy S. R., Lu S., Aubry A., Yu T., Wadosky K. M., Zhang L., Wang T., Gregorieff A., Ahmad M., Dimaras H., Langille E., Cole S. P. C., Monnier P. P., Lok B. H., Tsao M. S., Akeno N., Schramek D., Wikenheiser-Brokamp K. A., Knudsen E. S., Witkiewicz A. K., Wrana J. L., Goodrich D. W., Bremner R., Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell 39, 1115–1134.e12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doroshow D. B., Eder J. P., LoRusso P. M., BET inhibitors: A novel epigenetic approach. Ann. Oncol. 28, 1776–1787 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Zhu W., Wu R. D., Lv Y. G., Liu Y. M., Huang H., Xu J. Q., BRD4 blockage alleviates pathological cardiac hypertrophy through the suppression of fibrosis and inflammation via reducing ROS generation. Biomed. Pharmacother. 121, 109368 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Tebay L. E., Robertson H., Durant S. T., Vitale S. R., Penning T. M., Dinkova-Kostova A. T., Hayes J. D., Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 88, 108–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi N., Cho P., Selfors L. M., Kuiken H. J., Kaul R., Fujiwara T., Harris I. S., Zhang T., Gygi S. P., Brugge J. S., 3D culture models with CRISPR screens reveal hyperactive NRF2 as a prerequisite for spheroid formation via regulation of proliferation and ferroptosis. Mol. Cell 80, 828–844.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo M., Wicha M. S., Targeting cancer stem cell redox metabolism to enhance therapy responses. Semin. Radiat. Oncol. 29, 42–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ast T., Mootha V. K., Oxygen and mammalian cell culture: Are we repeating the experiment of Dr. Ox? Nat. Metab. 1, 858–860 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Alvarez S. W., Sviderskiy V. O., Terzi E. M., Papagiannakopoulos T., Moreira A. L., Adams S., Sabatini D. M., Birsoy K., Possemato R., NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 551, 639–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribas A., Hu-Lieskovan S., What does PD-L1 positive or negative mean? J. Exp. Med. 213, 2835–2840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis A. A., Patel V. G., The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 7, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broxmeyer H. E., Cooper S., Capitano M. L., Enhanced collection of phenotypic and engrafting human cord blood hematopoietic stem cells at 4°C. Stem Cells 38, 1326–1331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar B., Prasad M. S., Bhat-Nakshatri P., Anjanappa M., Kalra M., Marino N., Storniolo A. M. V., Rao X., Liu S., Wan J., Liu Y., Nakshatri H., Normal breast-derived epithelial cells with luminal and intrinsic subtype-enriched gene expression document inter-individual differences in their differentiation cascade. Cancer Res. 78, 5107–5123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar B., Bhat-Nakshatri P., Maguire C., Jacobsen M., Temm C. J., Sandusky G., Nakshatri H., Bidirectional regulatory cross-talk between cell context and genomic aberrations shapes breast tumorigenesis. Mol. Cancer Res. 19, 1802–1817 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peck Justice S. A., Barron M. P., Qi G. D., Wijeratne H. R. S., Victorino J. F., Simpson E. R., Vilseck J. Z., Wijeratne A. B., Mosley A. L., Mutant thermal proteome profiling for characterization of missense protein variants and their associated phenotypes within the proteome. J. Biol. Chem. 295, 16219–16238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhat-Nakshatri P., Kumar B., Simpson E., Ludwig K. K., Cox M. L., Gao H., Liu Y., Nakshatri H., Breast cancer cell detection and characterization from breast milk-derived cells. Cancer Res. 80, 4828–4839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., Mering C. V., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newman A. M., Liu C. L., Green M. R., Gentles A. J., Feng W., Xu Y., Hoang C. D., Diehn M., Alizadeh A. A., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for tables S1 to S4
Figs. S1 to S7
Tables S1 to S4