Expression of miR-3607 in embryonic mammalian cerebral cortex was lost in rodents, limiting progenitor cell proliferation.
Abstract
The evolutionary expansion and folding of the mammalian cerebral cortex resulted from amplification of progenitor cells during embryonic development. This process was reversed in the rodent lineage after splitting from primates, leading to smaller and smooth brains. Genetic mechanisms underlying this secondary loss in rodent evolution remain unknown. We show that microRNA miR-3607 is expressed embryonically in the large cortex of primates and ferret, distant from the primate-rodent lineage, but not in mouse. Experimental expression of miR-3607 in embryonic mouse cortex led to increased Wnt/β-catenin signaling, amplification of radial glia cells (RGCs), and expansion of the ventricular zone (VZ), via blocking the β-catenin inhibitor APC (adenomatous polyposis coli). Accordingly, loss of endogenous miR-3607 in ferret reduced RGC proliferation, while overexpression in human cerebral organoids promoted VZ expansion. Our results identify a gene selected for secondary loss during mammalian evolution to limit RGC amplification and, potentially, cortex size in rodents.
INTRODUCTION
The mammalian cerebral cortex went through a remarkable expansion in size and folding during evolution from its stem ancestor (1), a process recapitulated during embryonic development (2). Understanding the cellular and molecular mechanisms of cerebral cortex expansion in mammalian evolution is a major challenge. At the cellular level, this seems to result from the increased pool size of neural stem and progenitor cells and their proliferative capacity (3, 4). At the molecular level, multiple genes have been identified that emerged specifically in the recent human lineage and promote neural stem cell proliferation and brain growth (5–7). More generally, this was achieved by regulating the levels or patterns of expression of highly conserved genes and signaling pathways (6, 8). In some clades such as New World monkeys and, particularly, rodents, the general trend in evolution toward brain expansion and folding was reversed at some point: Brains evolved, becoming smaller and smoother than those of their ancestors (9). This may have resulted, in part, from the secondary loss of developmental features key for brain size and folding, stemming from gene expression regulation (2, 4). Unfortunately, our understanding of the mechanisms that regulate gene expression and signaling pathway activity across mammalian phylogeny, particularly related to brain evolution, remains limited (10).
Neural stem and progenitor cells in the developing mammalian cerebral cortex are organized in germinal zones. The ventricular zone (VZ) is the inner (apical) layer of the embryonic cortex and is essentially composed of apical radial glia cells (aRGCs), the primary type of cortical progenitor cell. Following mitotic division, aRGCs produce either more aRGCs or basal progenitors, namely, intermediate progenitor cells (IPCs) and basal RGCs (bRGCs). Basal progenitors coalesce into a secondary germinal layer, the subventricular zone (SVZ), and produce the majority of cortical excitatory neurons (11). In species with large brains such as carnivores and primates, the abundance and proliferation of basal progenitor cells increases massively during development, particularly bRGCs, and the SVZ becomes subdivided into inner SVZ (ISVZ) and outer SVZ (OSVZ) (3, 12–16). bRGCs contribute critically to increase neurogenesis and to promote cerebral cortex growth and folding in these species (17). At early stages of cortical development, the abundance of aRGCs increases by self-amplifying divisions, but as neurogenesis starts, aRGCs gradually switch to producing the other cell types. Given that aRGCs are the origin of excitatory cortical cell lineages, the duration of the amplificative period and the rate of aRGC proliferation will determine the pool size of aRGCs, thus the number of ontogenetic cell lineages and, ultimately, cortex size (18). At the onset of neurogenesis, aRGC amplification and the size of VZ are markedly different between species with a large cortex (i.e., human, macaque, and ferret) and a small cortex (i.e., mouse) (3, 12).
Primate- and human-specific genes promote cortical progenitor cell amplification and neurogenesis, including protein-coding genes (5, 7, 19) and noncoding microRNAs (miRNAs) that target cell cycle proteins (20, 21). Whereas these genes may contribute to the emergence of primate-specific features, they do not explain the more general cases of cortex expansion along mammalian evolution or the secondary reduction in cortex size, as in rodents (i.e., human or ferret versus mouse). In contrast, transcriptomic analyses of cortical germinal zones in various paradigms have identified significant differences in expression levels of highly conserved genes: (i) between mouse and macaque or human embryos (20, 22), (ii) between aRGCs that self-amplify and those that self-consume to form bRGCs (23), and (iii) related to the differential expansion of the cerebral cortex between folds and fissures in ferret (24). The identification of several pre-miRNAs in these comparisons suggests that they may be important genetic regulators of cortical size across mammals (25).
Here, we focused on miR-3607-5p (SNORD138), a small noncoding RNA expressed as pre–miR-3607 in the germinal layers of the developing ferret cerebral cortex (23, 26). We found that mature miR-3607-5p (MIR3607 from hereon) is expressed in cortical germinal layers of the developing human, macaque, and ferret embryo but not mouse, and then took advantage of this to study how the development of the mouse cerebral cortex might be affected by MIR3607 expression. At mid-neurogenesis, MIR3607 initially increased cell cycle reentry of aRGCs, linked to increased expression of Pax6 (Paired box 6), but later, it increased neurogenesis, and neurons born prematurely displayed migration and axon guidance defects. We found that MIR3607 promotes transcriptomic signatures of progenitor cell proliferation and stemness by acting as a positive regulator of the canonical Wnt/β-catenin pathway, via directly targeting adenomatous polyposis coli (APC), a key repressor of β-catenin function. In agreement with this, MIR3607 activated β-catenin signaling in cortical progenitors of early mouse embryos, leading to excess aRGC amplification and the formation of proliferative rosettes, which was rescued by coexpression of APC. Conversely, loss of function of endogenous MIR3607 in ferret caused a loss of aRGC polarity, delamination, and impaired proliferation, which was phenocopied by APC overexpression alone. Last, expression of MIR3607 in human cerebral organoids led to expansion of the VZ, in agreement with aRGC amplification. Our results identify MIR3607 as a key regulator of Wnt/β-catenin signaling in aRGCs underlying the evolutionary expansion of the mammalian cerebral cortex and that the secondary loss of MIR3607 expression during rodent evolution led to a marked reduction in cortical size.
RESULTS
Mature MIR3607 is expressed in the embryonic cerebral cortex of human, macaque, and ferret but not mouse
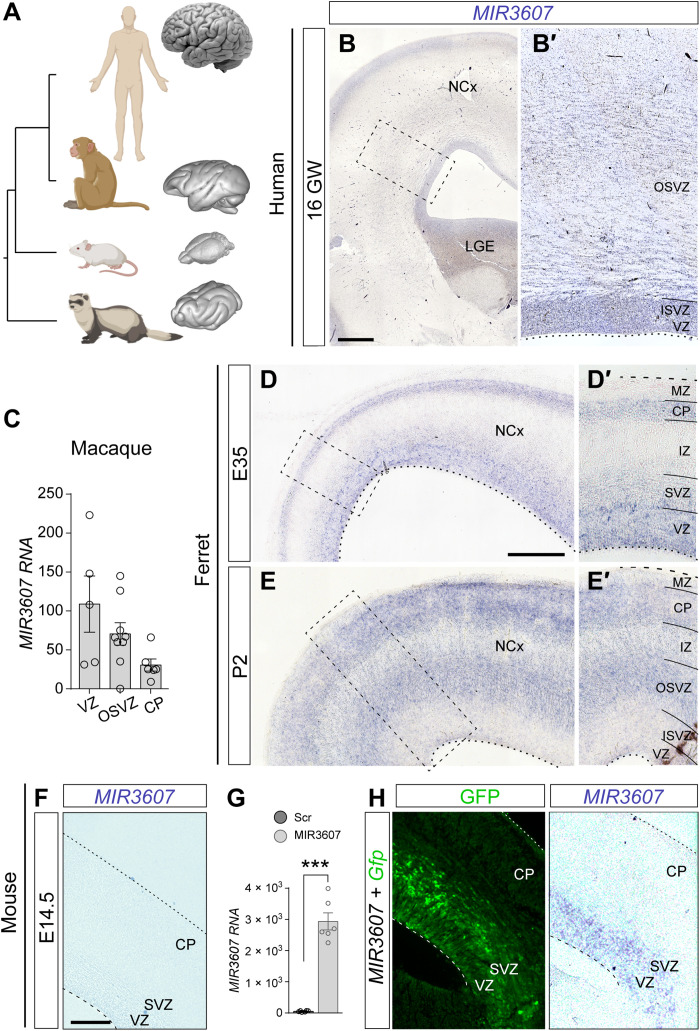
Our previous transcriptomic analyses revealed that pre–miR-3607 is expressed in all germinal layers of the ferret cerebral cortex, which is large and folded (Fig. 1A) (23, 26). This includes an early period of cortical development critical for the formation of the OSVZ by massive seeding of bRGCs (23) and a late period preceding the marked expansion and folding of the cerebral cortex (26). In contrast, pre–miR-3607 has never been reported to be expressed in the developing mouse cortex, small and smooth (Fig. 1A). These observations prompted the idea that MIR3607 might be relevant for the expansion and folding of the mammalian cerebral cortex. Unfortunately, expression of pre-miRNAs does not always parallel miRNA activity, as they must be enzymatically processed and cleaved to the shorter, functionally mature miRNAs. To gain insights into the potential function of MIR3607 in cerebral cortex expansion and folding, we began elucidating the expression level and pattern of MIR3607 in the developing human brain. In situ hybridization (ISH) stains on the embryonic human cortex at 16 gestational weeks (GW; circa the peak of neurogenesis) revealed the highest expression levels of MIR3607 in the VZ and ISVZ and second in the cortical plate (CP), where cortical neurons finish radial migration and begin differentiating their dendritic and axonal arbors (Fig. 1B). Expression was also detected in the OSVZ but at lower levels compared to the other germinal zones (Fig. 1B′). We next extended our analyses to nonhuman primates by mining public sequencing datasets (20). We found that MIR3607 is expressed in the developing cerebral cortex of macaque monkey embryos at similar developmental stages [embryonic day 79 (E79) to E82], at particularly high levels in VZ and OSVZ, and less in CP, similar to human embryos (Fig. 1C).
Fig. 1. Mature MIR3607 is expressed in cortical germinal layers of human, macaque, and ferret but not mouse embryos.
(A) Phylogenic relationship between human, macaque, mouse, and ferret and external appearance of their brains (not at scale). (B and B′) Expression pattern of MIR3607 in the human brain at 16 GW. Area boxed in (B) is shown in (B′). MIR3607 expression is high in the three germinal zones: VZ, ISVZ, and OSVZ. (C) Expression levels of MIR3607 in germinal layers of the embryonic macaque cerebral cortex at E79 to E82. Data are from (33). Circles indicate values for individual replicas. (D to E′) Sagittal sections of the ferret cortex at the indicated ages showing the pattern of MIR3607 expression. Areas boxed in (D) and (E) are shown in (D′) and (E′). (F) Pattern of MIR3607 expression in the cerebral cortex of an E15.5 mouse embryo, with null endogenous expression. (G) MIR3607 expression levels [quantitative polymerase chain reaction (qPCR), arbitrary units] in human embryonic kidney 293 cells transfected with psil-Scr and psil-MIR3607. Histograms indicate means ± SEM; circles indicate values for individual replicas. t test, ***P = 9.76 × 10–7. (H) Patterns of green fluorescent protein (GFP) and MIR3607 expression in the cerebral cortex of an E15.5 mouse embryo electroporated at E14.5 with Gfp plus MIR3607-encoding plasmids. Note the high expression levels in VZ and SVZ. Scale bars, 500 μm (B), 200 μm (D and E), and 100 μm (F and H).
Next, we analyzed MIR3607 expression in the developing ferret, phylogenetically distant from primates but also with a large and folded cortex (Fig. 1A). At E35, expression was high in VZ, low in SVZ, and undetectable in the intermediate zone (IZ) (Fig. 1, D and D′). At this stage, there is no distinction between ISVZ and OSVZ, so this pattern was reminiscent of the difference between VZ and OSVZ in human embryos at 16 GW. However, at later stages [postnatal day 2 (P2)], when ISVZ and OSVZ are clearly distinct in ferret, expression was reduced to background levels in VZ and ISVZ, being high in OSVZ and intermediate in IZ (Fig. 1, E and E′). In addition, and similar to primates, MIR3607 was also expressed in CP at both developmental stages. In contrast to human, macaque, and ferret, ISH stains in the embryonic mouse cortex revealed a complete absence of MIR3607 expression, both at early stages (E12.5) and at late stages (E14.5; Fig. 1F). A search on public datasets further revealed that MIR3607 is also not expressed in stem cells of the embryonic rat striatum (27). This was remarkable because rodents are phylogenetically closer to primates than carnivores, such as ferret (Fig. 1A), so together, these results were consistent with the notion that expression of MIR3607 in the developing cerebral cortex could be linked to its expansion and folding. Extending this argument, the ancestor of carnivores, primates, and rodents may have expressed MIR3607 in the cortical germinal layers at embryonic stages and, at some point after the split between primates and rodents, this expression may have been lost or silenced in the mouse lineage. Hence, a secondary loss of MIR3607 expression in the developing cortex during rodent evolution may have been relevant to limit cortical growth and suppress its folding. To test this idea, we took advantage of the absence of MIR3607 expression in mouse to investigate its role in cortical development and its potential relevance in the limited growth of the mouse cerebral cortex as compared to ferret and human.
miRNAs usually have a high degree of conservation across animal species (28). For MIR3607-5p, sequence conservation compared to human ranges from 100% in hominids to 92% in the house mouse (table S1). Nucleotide changes mostly occur in the 10th and 7th base of the loop sequence (in 29 and 19 species, respectively, of 29 species with mismatches) and secondarily in the 4th base of the seed sequence (in 18 of 29 species; always G to A changes; table S1). Outside primates, the mature MIR3607-5p sequence is 100% conserved in only 4 of the 26 species compared, which belong to three far-related orders: guinea pig (Rodentia), ferret (Carnivora), and wild boar and horse (Artiodactyla). In mouse, the seed sequence is fully conserved, but the first base of the mature MIR3607-5p is changed (G to A), and four nucleotides are changed or missing in the loop sequence (table S1). The notable similarity of the mature MIR3607 in human, macaque, ferret, and mouse is suggestive of a preserved functionality for this miRNA in the mouse brain. Therefore, we reasoned that reexpression of MIR3607 in the developing brain of mouse embryos may shed light on its role during cortical development. We confirmed expression of mature MIR3607 from our artificial DNA vector both in a human cell line by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and in the cerebral cortex of electroporated mouse embryos by ISH (Fig. 1, G and H).
MIR3607 drives amplification of Pax6+ progenitor cells
The expression patterns of MIR3607 in the developing cortex of human, macaque, and ferret supported the notion that it may be involved in multiple steps of cortical development, including VZ progenitor cell proliferation, OSVZ formation and amplification, neurogenesis, radial migration through IZ, and neuronal differentiation in CP (Fig. 1). We first studied its potential involvement in progenitor cell proliferation and neurogenesis. Taking advantage of the absence of MIR3607 in mouse, we expressed its mature form alongside a reporter plasmid in apical progenitor cells of the E14.5 mouse cortex by in utero electroporation. One day after electroporation (E15.5), the majority of green fluorescent protein–positive (GFP+) cells in control, Scrambled (Scr)–electroporated embryos populated the VZ, with only half as many found in SVZ and very few in IZ (Fig. 2, A and B). This distribution of GFP+ cells was similar in embryos expressing MIR3607, except for a clear tendency to having fewer cells in IZ and SVZ while more in VZ (Fig. 2, A and B). This suggested that the immediate early effect of MIR3607 expression might be to reduce neurogenesis and promote aRGC amplification. In agreement with this notion, bromodeoxyuridine (BrdU) labeling analyses revealed a 36% increase in BrdU-incorporating, cycling progenitor cells in E15.5 embryos expressing MIR3607 compared to control littermates and a 65% reduction in cell cycle exit between E14.5 and E15.5 (Fig. 2, C and D). This confirmed that the earliest effects immediately upon MIR3607 expression are reduced neurogenesis and marked amplification of progenitor cells.
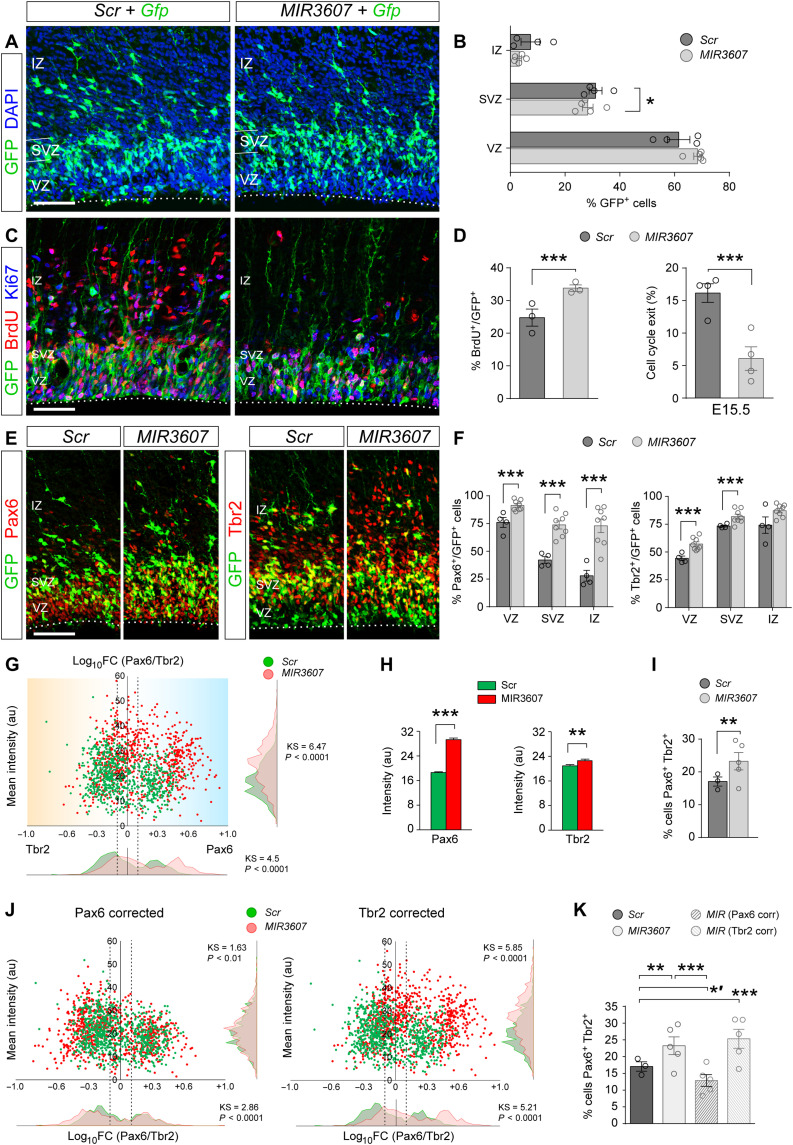
Fig. 2. Amplification of Pax6+ progenitor cells immediately upon MIR3607 expression.
(A to F) Parietal cortex of E15.5 mouse embryos electroporated at E14.5 with Gfp plus Scr- or MIR3607-encoding plasmids (A, C, and E), following a single injection of BrdU 4 hours before (C), and quantifications of laminar distribution (B), BrdU+ labeling and cell cycle exit in any layer (D), and abundance of Pax6+ and Tbr2+ cells. (G) Ratio Pax6/Tbr2 expression level over mean intensity [arbitrary units (au)] in individual GFP+ cells as in (C) and frequency distribution for each parameter. Dashed vertical lines delimit Pax6/Tbr2 coexpression (−0.1 < log10FC < +0.1). N = 738 cells, three embryos for Scr; N = 790 cells, five embryos for MIR3607. (H and I) Average expression intensity of Pax6 and Tbr2 among individual cells (H) and proportion of cells coexpressing both factors (I). (J) Scatterplots and frequency distribution plots after Pax6 or Tbr2 values of each MIR3607 cell were corrected to the average Scr value (H). (K) Proportion of cells coexpressing Pax6 and Tbr2, as defined in (I). Open circles in plots indicate values for individual embryos. Mean ± SEM; n = 3 to 8 embryos per group. Chi-square test (B, D, F, I, and K), t test (H), Kolmogorov-Smirnov test (G and J); *P < 0.05, *′P < 0.02, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. DAPI, 4′,6-diamidino-2-phenylindole.
The vast majority of mouse cortical progenitor cells in VZ are aRGCs, which characteristically express the transcription factor Pax6, whereas most progenitors in SVZ are IPCs, recognized by expression of the transcription factor Tbr2 (T-box brain protein 2) (29). To determine whether the amplification of progenitor cells immediately upon MIR3607 expression regarded aRGCs or IPCs, we next analyzed the expression of Pax6 and Tbr2 in E15.5 embryos. In control embryos, a majority (76%) of GFP+ cells in VZ were positive for Pax6, as expected (Fig. 2, E and F). Pax6 was also expressed at detectable levels by 42% of SVZ and 28% of IZ cells. Tbr2 was detected mostly in SVZ and IZ cells (74%) and, to a lesser extent, in VZ (44%; Fig. 2F). In embryos expressing MIR3607, the proportion of Pax6+ cells increased in all three layers, most markedly in SVZ and IZ (1.7- and 2.6-fold, respectively; Fig. 2F). Tbr2+ cell abundance also increased in VZ and SVZ upon expression of MIR3607, but much less than Pax6+ cells (Fig. 2F). While increased Pax6+ cells in VZ was consistent with greater abundance of aRGCs, it was puzzling to find such high frequency in SVZ and IZ because these layers are normally populated by IPCs and newborn neurons, negative for Pax6 (29). The high frequencies of Pax6+ and Tbr2+ cells in MIR3607 embryos were only compatible with a significant increase in the coexpression of these two proteins, which is one of the defining features of OSVZ progenitors in ferret and primates (12, 30). Analyses of staining intensity in individual cells demonstrated an average 60% increase in Pax6 protein levels upon MIR3607 expression, with only an 8% increase in Tbr2 levels (Fig. 2, G and H). This selective change resulted in a significant increase in the abundance of cells coexpressing Pax6 and Tbr2 (Fig. 2, G and I), largely attributable to basal cells in SVZ (fig. S1). In silico analyses revealed that the observed increase in Pax6 levels, but not Tbr2, was sufficient to explain the differences between control and MIR3607-expressing embryos in Pax6-Tbr2 coexpression (Fig. 2, J and K). Together, the above results indicated that MIR3607 expression in mouse cortex at mid-embryonic stages increases expression of Pax6 protein, resulting in high Pax6/Tbr2 coexpression, particularly in basal progenitors, and promotes progenitor cell self-amplification, both landmark features of the developing large and folded brains of ferret and macaque (12, 13, 30).
Changes in neurogenesis, neuron migration, and axon growth following MIR3607 expression
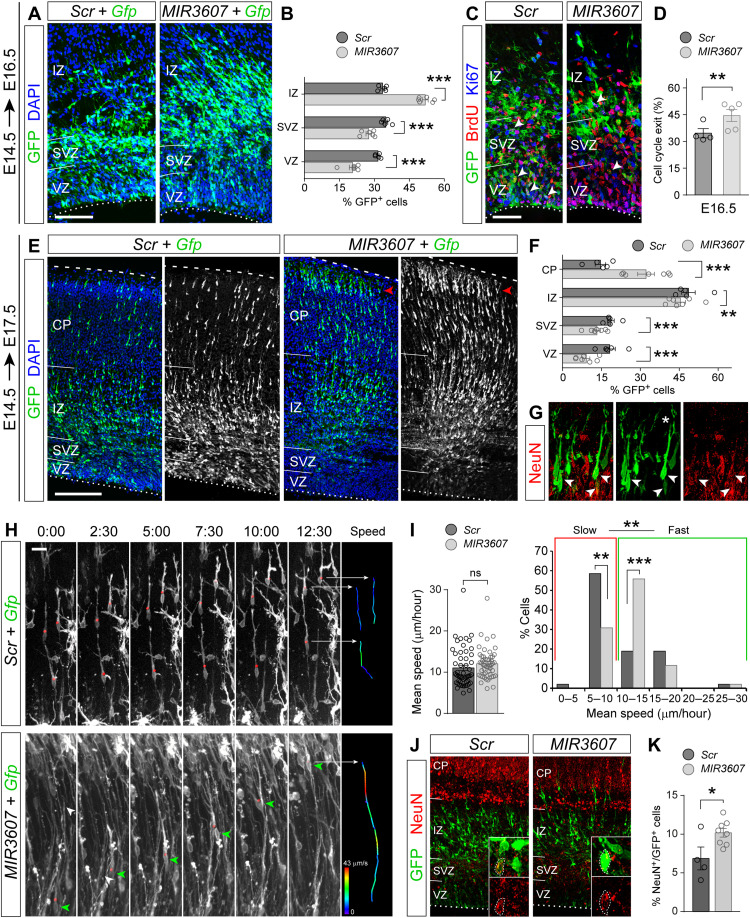
In contrast to our observations at E15.5 (1 day after electroporation), by E16.5, we observed a 21 to 35% reduction of GFP+ cells in VZ and SVZ of MIR3607-expressing embryos, bound to a 54% increase in IZ, as compared to control embryos (Fig. 3, A and B). During normal development, once neurons are born in VZ or SVZ, they spend some time within the SVZ undergoing short-distance multipolar migration, and only after that, they acquire bipolarity, enter the IZ, and undergo fast radial migration to the CP (31, 32). Hence, the increase in IZ cells suggested that MIR3607 expression augmented the relative abundance of radially migrating neurons. We reasoned that this could be originated by a greater abundance of neurons due to increased neurogenesis. Analysis of cell cycle exit revealed that E16.5 embryos expressing MIR3607 since E14.5 had a 29% higher rate compared to controls (Fig. 3, C and D), demonstrating more self-consuming, neurogenic divisions of progenitor cells at this stage.
Fig. 3. Alterations in neurogenesis and neuron migration upon expression of MIR3607.
(A to D) Parietal cortex of E16.5 mouse embryos electroporated at E14.5 with the indicated plasmids (A and C), followed by a single pulse of BrdU at E15.5 (C), and quantification of laminar distribution of GFP+ cells (B) and cell cycle exit (D). N = 4 embryos Scr; N = 5 embryos MIR3607. (E to G) Distribution of GFP+ cells in E17.5 embryos electroporated at E14.5 with the indicated plasmids (E), quantification (F), and coexpression of NeuN in CP (G) (arrowheads; the asterisk indicates immature apical dendrite). (H and I) Selected videomicroscopy frames showing neurons migrating through IZ (H) and quantification of mean migration speed (I). Time elapsed since first frame is indicated in hour:min. Instant speed of indicated cells (red dots, green arrowheads) during the period shown is color-coded in (H). N = 53 cells Scr; N = 52 cells MIR3607. (J and K) NeuN expression in E16.5 embryos electroporated at E14.5 (J) and frequency of GFP+ cell labeling (K). Insets are high magnifications illustrating GFP+ cells positive for NeuN (dotted circles). N = 4 embryos Scr; N = 8 embryos MIR3607. Circles in plots indicate values for individual embryos. Mean ± SEM; chi-square test (B, D, F, I, and K), t test (I); ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001. Scale bars, 100 μm (A, C, and J), 200 μm (E), and 10 μm (H).
Next, we reasoned that if MIR3607 expression increased the ratio of neurogenic over self-renewing divisions, too many neurons would be generated too early. This would, in turn, result in an increase of radially migrating neurons in IZ at subsequent stages, as we observed at E16.5 (Fig. 3A), as well as the subsequent arrival of these neurons to the CP ahead of control-electroporated neurons. In agreement with this notion, E17.5 embryos expressing MIR3607 since E14.5 showed lower proportions of GFP+ cells in VZ, SVZ, and IZ, and a twofold increase of GFP+ cells in CP (Fig. 3, E and F). In contrast to cells in control embryos, MIR3607-expressing cells already started to settle and accumulate at the top of CP (Fig. 3E), where neurons end radial migration and start differentiating. The neuronal identity of CP cells was confirmed by anti-NeuN stains (Fig. 3G). These results seemed to indicate that the primary effect of MIR3607 expression was to promote neurogenesis, which secondarily translated into the premature radial migration and settling of neurons in the CP without directly affecting the movement of neurons. To directly visualize migrating neurons and ascertain whether MIR3607 has any effect on their movement, we performed videomicroscopy experiments (Fig. 3H). We found that the overall speed of radial neuron migration was statistically similar in control and MIR3607-expressing embryos. However, a detailed analysis revealed that in MIR3607 embryos, the proportion of fast-migrating neurons (>10 μm/hour on average) was much greater than in control embryos (Fig. 3, H and I, and movies S1 and S2). This demonstrated that expression of MIR3607 accelerates the radial migration of cortical neurons. Last, we analyzed the proportion of GFP+ cells expressing the neuronal marker NeuN at E16.5 to find that it was higher in the cortex of MIR3607 embryos (Fig. 3, J and K), further confirming that MIR3607 increased neurogenesis first and then accelerated neuron migration.
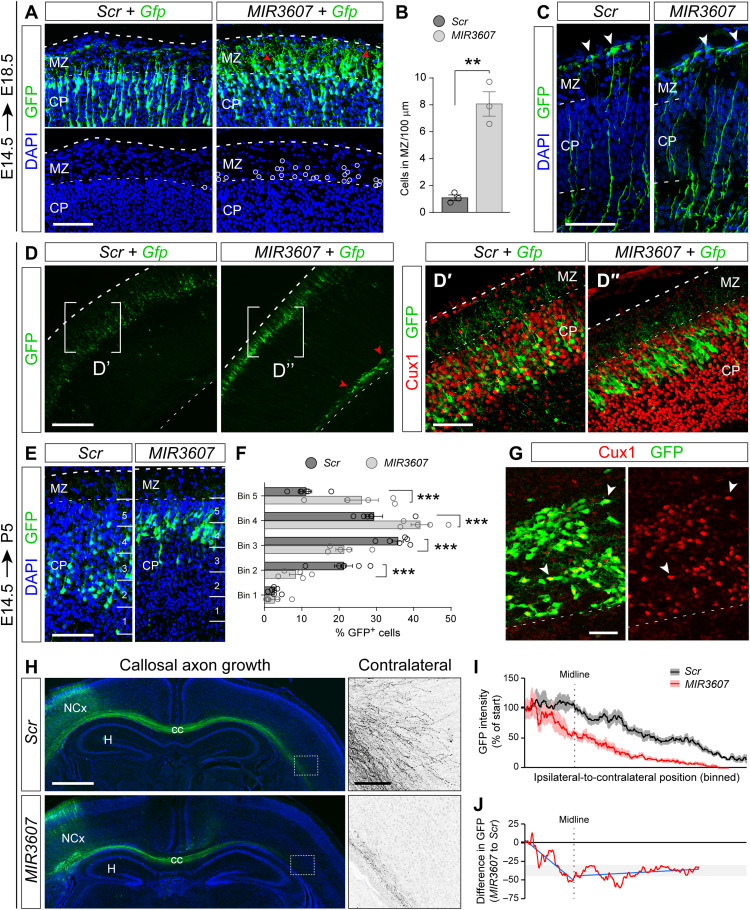
Analyses yet 1 day later (E18.5 embryos electroporated at E14.5) revealed that, in MIR3607 embryos, many GFP+ cells actually migrated past their arrival zone at the CP-MZ (marginal zone) border, invading ectopically the MZ (Fig. 4, A and B), which indicated a defective termination of radial migration. Radial migration of cortical neurons largely depends on their interaction with the basal processes of RGCs, such that their perturbation leads to defective migration. Analysis of the detailed morphology of RGCs revealed no overt abnormalities in cell polarity or in their basal process upon MIR3607 expression (Fig. 4C), suggesting that defects in neuronal migration were cell autonomous. Defects in migration of MIR3607-expressing neurons were confirmed at P5 after cortical lamination is complete (Fig. 4D). GFP+ neurons in MIR3607-electroporated embryos were found in the CP (prospective layer 2/3 at this stage) and expressed the layer 2/3 marker Cux1 (Cut like homeobox 1), exactly like in control embryos, consistent with them maintaining a normal laminar fate (Fig. 4, D′ and D″). However, in MIR3607 embryos, they occupied the top of CP, whereas in control-electroporated embryos, they mostly occupied central positions within the CP (Fig. 4, E and F). Considering that expression of MIR3607 induced premature neurogenesis, according to the inside-out gradient of cortex development, this should have resulted in neurons accumulating in deeper positions within the CP, not more superficial. However, our result was consistent with the overmigration defect observed previously at E18.5. In addition to defects in the CP, we also observed that many MIR3607 cells accumulated in the cortical white matter, forming subcortical ectopias (Fig. 4D). Marker analysis showed that the vast majority of these ectopic cells expressed Cux1 (Fig. 4G), demonstrating both their neuronal identity and conservation of the normal fate for upper cortical layers. Together, these observations demonstrated that MIR3607 expression altered radial neuron migration in the developing mouse cerebral cortex.
Fig. 4. MIR3607 expression causes long-term defects in neuron position and axon growth.
(A and B) GFP+ cells in CP and MZ of E18.5 embryos electroporated at E14.5 (A) and quantification (B). Circles and arrowheads indicate ectopic cells. (C) Details of the basal process of aRGCs in E15.5 embryos electroporated at E14.5. Arrowheads point at basal end feet. (D to G) Parietal cortex of P5 mouse pups electroporated at E14.5 and stained as indicated and distribution of GFP+ cells within CP (E and F). (D′ and D″) Magnifications of areas indicated in (D). (G) Magnification of the white matter in a MIR3607 embryo. All GFP+ cells in CP (normotopic) and white matter (ectopic, arrowheads) were positive for Cux1 (red). (H) Coronal sections through the parietal cortex of P5 pups electroporated at E14.5 revealing GFP+ callosal axons. Boxes indicate areas shown in high magnification (Contralateral). (I and J) Quantification of GFP intensity along the ipsi-to-contralateral track of callosal axons, relative to intensity at the start site (I), and difference between conditions (J). Dashed line indicates location of the midline. Blue is for trend lines before and after midline crossing. Data in (I) are means ± SEM; n = 9 to 10 pups per group. Circles in plots indicate values for individual embryos. Histograms indicate means ± SEM; n = 3 to 7 embryos per group; t test (B), chi-square test (E); **P < 0.01 and ***P < 0.001. Scale bars, 100 μm (A, C, D′, D″, E, and inset in H), 40 μm (G), 300 μm (D), and 1 mm (H).
Given the effects of MIR3607 on neurogenesis and neuron migration, we next enquired whether axonal growth was also affected. We electroporated E14.5 embryos to target progenitor cells producing layer 2/3 neurons and then analyzed their growing axons across the corpus callosum (CC) at P5. In both control and MIR3607-expressing embryos, GFP+ axons crossed the telencephalic midline at the level of the CC and extended along the white matter of the contralateral hemisphere (Fig. 4H). However, the density of GFP+ callosal axons extending along the white matter was lower in MIR3607 embryos than in controls. The deficit in callosal axons increased as these approached the midline, while after midline crossing, it remained largely similar to controls (Fig. 4, I and J). In the contralateral cortex, we further observed the invasion of axons from the white matter toward the CP in control embryos, which was virtually absent in MIR3607 embryos (Fig. 4H).
Together, our results demonstrated that overexpression of MIR3607 at intermediate stages of cortical development has effects at multiple levels. It has an immediate early effect of inducing progenitor cell amplification, and then, it induces premature neurogenesis. It accelerates radial neuron migration and alters its termination, leading to mild defects in lamination and formation of white matter ectopias. Last, it delays growth of cortical callosal axons, both in their initial navigation toward the midline and in their subsequent invasion of the contralateral gray matter.
MIR3607 activates signaling pathways driving progenitor proliferation
To elucidate the mechanism of action of MIR3607 that leads to the immediate early amplification of cortical progenitor cells reported above, we next investigated the impact of MIR3607 expression at the transcriptomic level. We electroporated in utero the cerebral cortex of E14.5 embryos with plasmids encoding MIR3607 or Scr plus Gfp, and 24 hours later, RNA sequencing (RNA-seq) was performed on fluorescence-activated cell sorting (FACS)–purified GFP+ cells (Fig. 5A and fig. S2). We identified 173 genes differentially expressed (DEGs) in cortical progenitors upon MIR3607 expression [false discovery rate (FDR) < 0.01; Fig. 5B and table S2]. A majority of DEGs were down-regulated (58%), as expected from the action of a miRNA (Fig. 5B). Of the 76 genes in the mouse genome computationally predicted to be direct targets of MIR3607, 63 were expressed in our samples and 9 of them were DEGs (Fig. 5C). The great majority of those DEGs were down-regulated (8 of 9), again as predicted from the action of a miRNA: Dnm3, Opcml, Pde4d, Tmem169, Cnr1, Bsn, Apc, and Rnf38 (Fig. 5C). Several functional enrichment analyses were performed to capture biological information on DEGs. Gene Ontology (GO) analysis highlighted the Wnt signaling pathway and axon development as having the highest enrichment (Fig. 5D). Similarly, functional grouping of gene networks highlighted Wnt signaling pathway, neuroblast proliferation, regulation of neural precursor cell proliferation, and L1CAM (L1 cell adhesion molecule) interactions (Fig. 5E). L1CAM interactions are relevant for axon development, so these results were consistent with the observed deficient growth of callosal axons in P5 mice expressing MIR3607 (Fig. 4G). Functional annotation clustering analysis highlighted again, as top ranked, a cluster topped by the terms Wnt signaling pathway, lateral plasma membrane, and signaling pathways regulating pluripotency of stem cells (Fig. 5F). Together, these analyses revealed a prominent role of MIR3607 in the regulation of Wnt/β-catenin signaling pathway, proliferative activity, axon development, and lateral plasma membrane (Fig. 5, D to F, and table S2). All these biological functions closely matched the developmental processes that we found altered in our above phenotypic analyses of the developing cortex upon expression of MIR3607 at E14.5. MIR3607 expression in cortical progenitor cells led to decreased expression of Apc, a key negative regulator of the canonical Wnt/β-catenin signaling pathway, and, concomitantly, to increased levels of Fgfr3, Fzd8, Ctnn1a, and Ctnn1b transcription (Fig. 5, B, C, and E to H). Consistent with the GO analysis, gene set enrichment analyses (GSEAs) further confirmed a strong modulation by MIR3607 of genes regulating Wnt/β-catenin signaling, the apical junctional complex, and cell division in cortical progenitors (Fig. 5, I to K; fig. S3; and table S2). In summary, our transcriptomic analyses revealed that MIR3607 expression causes marked changes in the expression levels of genes participating in biological functions and signaling pathways that are key for the amplification, polarity, and delamination of cortical progenitor cells.
Fig. 5. MIR3607 promotes cell proliferation and activates Wnt signaling.
(A) Schema of experimental design. Cells expressing high GFP were FACS-sorted, and their pooled RNA was analyzed by RNA-seq. (B) Fold change of gene expression in MIR3607-electroporated versus Scr-electroporated cells, over the gene’s average expression level. DEGs (Adj. P < 0.01) are in red (up-regulated, 73 genes) and blue (down-regulated, 100 genes). Top three DEGs among MIR3607-predicted targets are named. (C) Fold change of 63 predicted MIR3607 targets detected; DEGs are named (*Adj. P < 0.1 and **Adj. P < 0.01). (D) Functional enrichment analysis on DEGs, showing most significant enriched GO terms. Ontology: Biological process. (E) Functionally grouped network based on functional enrichment of DEGs. Size of nodes indicates statistical significance of terms (Bonferroni step down corrected P values); color indicates percentage of DEGs. (F) Heatmap of DEGs highlighting genes associated to the top-ranked annotation cluster from functional annotation clustering analysis. Top three terms and their statistical significance (FDR) are indicated. ES, enrichment score. (G and H) Visualization of normalized coverage tracks from RNA-seq data for whole transcript (left, middle) and expression levels (right) for Apc (G) and Ctnn1b (H) in Scr- and MIR3607-expressing cells. TPM, transcripts per million; n = 3 replicates per condition; Student two-sample t test; *P < 0.05. (I to K) Enrichment plots from GSEA for MSigDB Hallmark Apical Junction (P = 0.01; Adj. P = 0.021), WNT β-catenin Signaling (P = 0.002; Adj. P = 0.006), and GO term Cell Division GO:0051301 (P = 0.002; Adj. P = 0.008).
Amplification of aRGCs and disruption of VZ in the early cortex by MIR3607
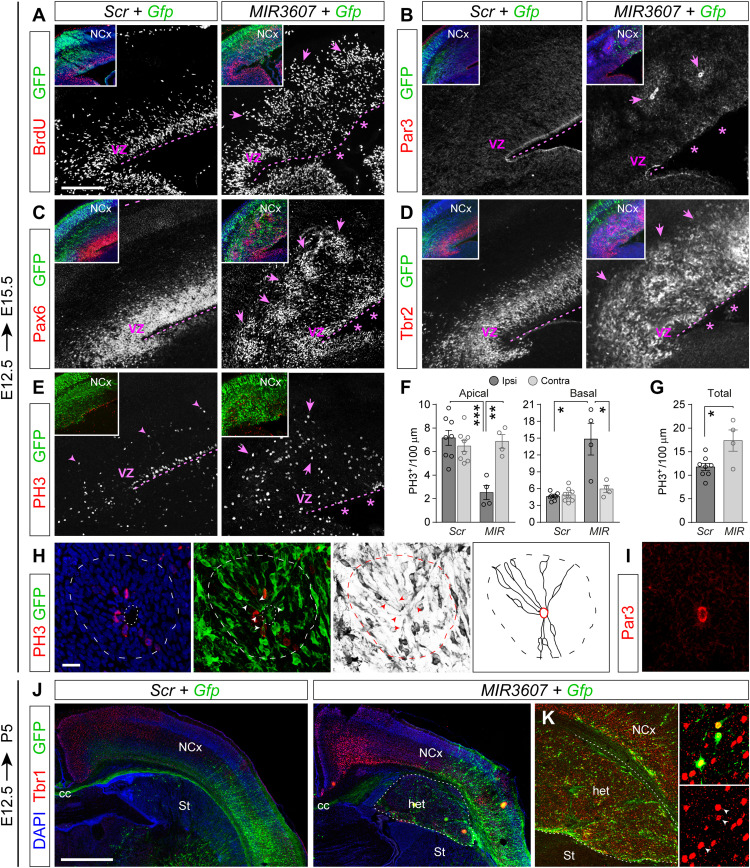
Progenitor cell amplification is singularly important at early stages of cortical development, before and at the onset of neurogenesis, so our above results prompted us to investigate the effects of expressing MIR3607 in the early embryonic mouse cerebral cortex. To this end, we electroporated MIR3607-encoding plasmids into the cortical primordium of mouse embryos at E12.5. To investigate the immediate early effects, we analyzed the cortex of electroporated embryos at E13.5, which revealed severe alterations in the organization of progenitor cells (Fig. 6). Single-pulse BrdU labeling experiments revealed that cells in S-phase were perfectly aligned in the basal aspect of the VZ in control-electroporated and non-electroporated cortices (Fig. 6A and fig. S4A). In contrast, expression of MIR3607 caused the spreading of BrdU-incorporating cells over the entire VZ thickness (Fig. 6A and fig. S4A). BrdU+ cells were frequently arranged in circles, resembling rosettes. This disorganization was paralleled by a severe disruption of the apical adherens junction belt, as identified by Par3 stains (Fig. 6B). In the area of cortex expressing MIR3607, Par3 was completely absent from the apical surface, instead forming multiple small closed domains within the cortical parenchyma, as if forming the lumen of presumptive rosettes (Fig. 6B). The typical band of PH3+ mitotic cells in the apical side of the normal VZ was also not recognizable in MIR3607 embryos, which instead formed clusters of abventricular mitoses within the VZ parenchyma (Fig. 6C). The distribution of Pax6+, Tbr2+, and NeuN+ cells was similarly disrupted, in agreement with a highly disorganized VZ (Fig. 6, D to F). This disorganization was consistent with those cells being constituent parts of rosettes: the core containing a mass of mitotic Pax6+ cells around Par3+ apical junction circles and this being surrounded by basal Tbr2+ progenitor cells and, lastly, NeuN+ neurons (Fig. 6, B, and D to F). Rosettes formed in 13 of 16 MIR3607 embryos (81%) and affected the entire parietal electroporated area, including rostral to caudal regions of the cerebral cortex. This phenotype was accompanied by abundant apoptosis, in both VZ and CP (fig. S4B), as well as by a 30% increase in NeuN-expressing GFP+ cells, indicative of augmented neurogenesis (Fig. 6, F and G). The major disruption of the apical surface and the formation of rosettes following MIR3607 expression was never observed in embryos electroporated at E14.5 (Fig. 2), further supporting the particular relevance of this miRNA in progenitor cell amplification at early stages of cortical development.
Fig. 6. MIR3607 disrupts the VZ with formation of rosettes.
(A to M) Parietal cortex of E13.5 mouse embryos electroporated at E12.5 with the indicated plasmids, stained as indicated, and quantification of NeuN coexpression (G). Circles in plot indicate values for individual embryos (n = 3 embryos per group). In (A) to (F), areas with disrupted apical surface (arrowheads), rosettes of progenitor cells (arrows), and areas with normal organization (asterisks) are indicated. Images in (H) to (M) show that the parallel disposition of aRGCs (green) and alignment of Par3 (red) at the ventricular surface (H and I) was severely disrupted upon expression of MIR3607, with the loss of apical surface integrity (J) (dash-dotted lines), folding of the apical surface (K), and basal formation of apical junction circles (J, L, and M) (arrows). Arrowheads indicate the apical process (I, K, and L) and basal process (M) of aRGCs. (N and O) Clonal analysis of aRGCs in E13.5 embryos electroporated and infected with GFP-encoding retroviruses at E12.5 (N) and quantification of cell type abundance per clone (O). Arrowheads indicate the apical process of aRGCs. Each clone shown contains two cells: one neuron plus one aRGC in Scr and two aRGCs in MIR3607. N = 74 clones, 10 embryos Scr; N = 144 clones, 12 embryos MIR3607. Histograms indicate means ± SEM; chi-square test; *′P = 0.08, *P = 0.013, and ***P < 0.001. Scale bar, 100 μm (A to E), 75 μm (F, H, and J), 10 μm (I), and 30 μm (K to M).
To reveal cellular changes related to the disorganization of the early mouse cortex upon MIR3607 overexpression, we examined the detailed morphology of electroporated aRGCs. In control embryos, GFP+ aRGCs had the typical highly polarized morphology, with an apical process anchored to the Par3+ apical junction belt by a thick end foot (Fig. 6, H and I). In MIR3607-expressing embryos, aRGCs retained their apico-basal polarity, with the apical process anchored to the Par3+ apical junction belt by a thick end foot (Fig. 6J). However, the perfectly parallel arrangement of aRGCs in control embryos was markedly modified in MIR3607 embryos, where these cells acquired a cartwheel conformation around the Par3+ circles in the cortical parenchyma (Fig. 6, J to M). These results further supported the interpretation that expression of MIR3607 in the early embryonic mouse cortex caused the formation of proliferative rosettes by a combination of enhancing progenitor cell proliferation, maintenance of aRGC polarity, and impairing integrity of the apical junction belt. However, proliferative symmetric divisions of aRGCs are favored by their apical anchoring (11), so the impaired apical junction belt and increased neurogenesis observed above seemed contrary to this interpretation. To clarify this conundrum, we performed a clonal cell lineage analysis of aRGCs following MIR3607 overexpression, which revealed a 60% increase in aRGC abundance per clone on average, compared to control embryos (as revealed by cell morphology and Pax6 expression; Fig. 6, N and O). This confirmed the enhanced proliferation and self-renewal of aRGCs upon MIR3607 overexpression, even in the context of a more modest increase in neurogenesis and the overall loss of integrity of the apical junction belt. We also found a remarkable doubling of multipolar cells (potential IPCs) expressing Pax6 (Fig. 6O), which was consistent with our above observations of increased Pax6 expression in SVZ (containing IPCs) and increased coexpression with Tbr2 (marker of IPCs). Together, these results supported the notion that MIR3607 restrains the transition of aRGCs to IPCs, possibly as part of the mechanism favoring overall aRGC amplification.
The general disorganization of germinal layers in MIR3607-electroporated embryos persisted at later stages (E15.5), becoming even more marked. Cycling BrdU+ progenitor cells failed to remain within the basal side of VZ, typical of control embryos, but were widespread from the apical VZ surface to IZ (Fig. 7A). Stains against Par3 demonstrated a persistent absence of the apical adherens junction belt in VZ of MIR3607-expressing embryos, and their continued presence as small circular structures at basal positions within the cortical parenchyma (Fig. 7B). Pax6 stains showed that in MIR3607-expressing embryos, aRGCs no longer formed a compact VZ as in controls but spread from there through the IZ forming basal clusters of cells, coincident with the location of Par3+ circles (Fig. 7, B and C). This massive disorganization of the VZ also affected Tbr2+ IPCs, which were distributed ectopically in MIR3607-expressing embryos, extending from the apical VZ surface through SVZ and IZ, where they formed distinct circular clusters (Fig. 7D). Expression of MIR3607 also led to a 65% loss of PH3+ apical mitoses alongside a threefold increase in basal mitoses, which no longer aligned in a distinct SVZ but spread basally through the IZ, occasionally forming small clusters (Fig. 7, E and F). The combined abundance of apical and basal mitoses was 48% greater in MIR3607-expressing cortices than in controls (Fig. 7G), demonstrating a persistent significantly increased progenitor cell proliferation. Together, the cortex of E15.5 MIR3607 embryos displayed proliferative rosettes similar to those observed in E13.5 embryos: Par3+ lumen, apical layer of Pax6+ cells, and surrounding basal layer of Tbr2+ cells. Detailed examination of GFP+ cells forming the core of rosettes also confirmed their aRGC morphology, with distinct apical and basal processes radially aligned, and mitosis at the apical surface, limited by a Par3+ adherens junction belt (Fig. 7, H and I).
Fig. 7. MIR3607 overexpression drives expansion of aRGCs and formation of rosettes leading to subcortical heterotopia.
(A to D) Parietal cortex of mouse embryos electroporated at E12.5 with the indicated plasmids, analyzed at E15.5 and stained with the indicated markers. Asterisks indicate disrupted organization of VZ and Par3+ adherens junction belt. Arrows indicate rosettes. Insets are low magnifications of the same areas showing DAPI (blue) and GFP. (E to G) Distribution and quantification of mitotic cells in E15.5 embryos electroporated at E12.5 with the indicated plasmids. Histograms indicate means ± SEM for the electroporated (Ipsi) and non-electroporated, contralateral hemisphere (Contra). Circles in plots indicate values for individual embryos. Decreased apical mitoses (asterisks) and increased basal mitoses (arrows) are indicated. N = 4 to 8 sections from two to four embryos per group; t test; *P < 0.05, **P < 0.01, and ***P < 0.001. (H and I) Details of a rosette (dashed line) stained as indicated. Dotted line indicates inner lumen; arrowheads indicate apical processes of aRGCs, Par3 reveals the luminal surface (I). Schematic drawing shows line reconstructions of GFP+ aRGCs within the rosette, with apical processes radiating from the inner lumen (red). (J and K) Cortex of P5 mouse pups electroporated at E12.5 with the indicated plasmids and stained as indicated. Tbr1+ neurons (red) are abundant in the heterotopia (het) of MIR3607-expressing brains (K) between the normal neocortex (NCx) and striatum (St), including many GFP+ cells (arrowheads in insets). Scale bars, 200 μm (A to E), 10 μm (H), and 1 mm (I).
We lastly investigated the long-term consequences of MIR3607 expression and the highly disorganized cortical development. Examination of P5 mouse pups electroporated at E12.5 revealed the occasional formation of subcortical heterotopia underneath the electroporation site (Fig. 7J). The size of this heterotopia varied between animals and was never observed in Scr-electroporated control mice. Heterotopias contained both GFP+ and GFP− cells, many of which were Tbr1+, a marker of deep layer cortical neurons, including a high proportion of GFP+ cells (Fig. 7K). These heterotopias were also composed of a large number of GFP− cells, indicating that the mechanism underlying this phenotype had a significant non–cell-autonomous component.
In summary, expression of MIR3607 in the developing cerebral cortex of young mouse embryos caused the amplification of aRGCs and destabilization of the apical junction belt, leading to the massive delamination of aRGCs. However, the apico-basal polarity of aRGCs was strictly maintained, favoring the formation of intrinsically structured proliferative rosettes accompanied by IPCs. These effects were fully consistent with our previous functional analyses of transcriptomic changes in these embryos, overall highlighting effects on neural progenitor cell proliferation, cell division, lateral plasma membrane, and apical junction (Fig. 5).
MIR3607 promotes aRGC amplification and rosettes by derepression of β-catenin signaling
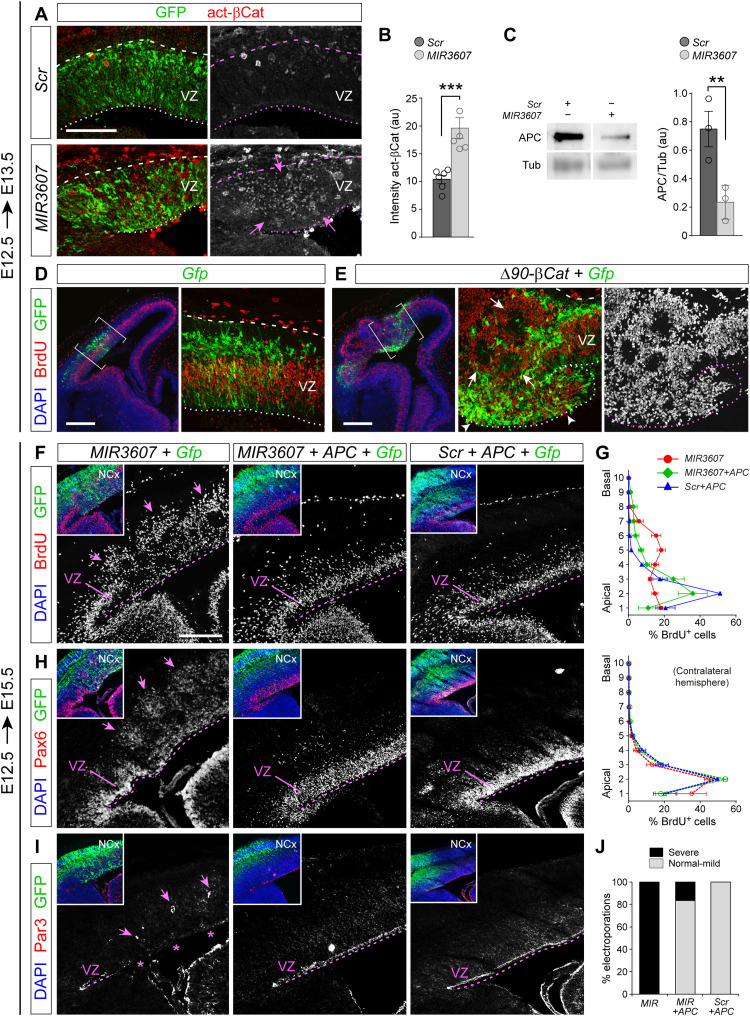
The effects of expressing MIR3607 in the embryonic mouse cerebral cortex, causing a massive amplification of cortical progenitors and disturbance of VZ integrity, resembled the effects produced by overactivation of the Wnt/β-catenin signaling pathway (33–35). Accordingly, our transcriptional profiling experiments revealed the strong and preferential activation of this pathway at the transcriptional level upon MIR3607 expression (Fig. 5). We confirmed these transcriptomic results at the protein level by immunostains against activated β-catenin, which revealed a twofold increase in the VZ of MIR3607-expressing mouse embryos 24 hours after electroporation (Fig. 8, A and B). This further supported the idea that the effects of MIR3607 on cortical progenitor amplification and rosette formation might be caused by the overactivation of β-catenin signaling. To investigate this possibility, we electroporated E12.5 embryos with a constitutively active form of β-catenin (Δ90-β-Cat). One day after electroporation (E13.5), the organization of the cerebral cortex and its VZ were severely disrupted, with multiple proliferative rosettes and massive disorganization of cycling progenitor cells (Fig. 8, D and E), phenocopying the effects of MIR3607 expression but at an even greater level of disruption. This further supported the notion that the alterations caused by MIR3607 were mediated by the overactivation of β-catenin signaling.
Fig. 8. MIR3607 promotes aRGC amplification and VZ overgrowth by derepressing β-catenin signaling.
(A and B) Parietal cortex of E13.5 mouse embryos electroporated at E12.5 with the indicated plasmids and stained against activated β-catenin, and quantification of signal intensity (arbitrary units). Dashed lines indicate pial surface; dotted lines indicate ventricular surface. Arrows indicate high abundance of β-catenin. (C) Western blots for APC and tubulin (loading control) in Mpf cells transfected with the indicated plasmids and densitometry quantifications of APC relative to tubulin. (D and E) Parietal cortex of E13.5 embryos electroporated at E12.5 with the indicated plasmids. Brackets indicate areas shown in the detailed images. Constitutively, active β-catenin severely disrupted the integrity of the VZ (arrowheads), with massive amplification and delamination of cycling (BrdU+) progenitor cells and formation of rosettes (arrows). (F to I) Parietal cortex of E15.5 embryos electroporated at E12.5 with the indicated plasmid combinations and stained as indicated (F, H, and I) and binned distribution of BrdU+ cells across the cortical thickness (G). Inset images show triple stains of the same area as shown for each individual marker (white). In (G), the top graph is the data from electroporated, ipsilateral hemispheres; the bottom graph is from non-electroporated, contralateral hemispheres. Arrows indicate rosettes; asterisks indicate the absence of the Par3+ adherens junction belt. (J) Frequency of embryos with germinal layer disturbance. Circles in plots indicate values for individual embryos. Data are means ± SEM; t test; **P < 0.01 and ***P < 0.001. Scale bars, 100 μm (A), 300 μm (D and E), and 200 μm (F, H, and I).
A major repressor of Wnt/β-catenin signaling is APC, a key component of a protein complex that phosphorylates β-catenin, driving it for proteasome degradation. MIR3607-5p has been shown to target and bind the 3′ untranslated region (3′UTR) of human APC, blocking its expression (36). Our bioinformatics analyses using TargetScanHuman confirmed the presence in the 3′UTR of human APC of two consensus target sequences of MIR3607-5p (table S3). We further found that one or both of these sequences, and their position within the 3′UTR of APC, are highly conserved across vertebrate phylogeny, from primates to bony fishes, suggesting a strong evolutionary selection (table S3). Consistent with these observations, our above transcriptomic analyses in mouse cortical progenitor cells showed a significant decrease of APC mRNA levels upon MIR3607 expression (Fig. 5, F and G). Western blot analyses demonstrated that MIR3607 expression reduces APC expression markedly also at the protein level (Fig. 8C).
The above results suggested that MIR3607 may activate β-catenin signaling indirectly, via repressing APC expression. If this was the case, the deleterious effect of MIR3607 expression on the embryonic cerebral cortex should be rescued by additionally expressing APC. We tested this possibility by expressing MIR3607 together with the coding sequence of APC without 3′UTR, hence resistant to MIR3607. Expression of MIR3607 alone caused a very severe disorganization of germinal zones in the developing cerebral cortex, as shown above (Fig. 8, F to I). Cycling progenitor cells identified by BrdU incorporation were distributed through the thickness of the VZ and spread basally to the IZ forming conspicuous rosettes (Fig. 8, F and G). This was accompanied by the severe disruption of the Par3+ adherens junction belt and the delamination of clusters of Pax6+ cells from VZ to SVZ and IZ, where they constituted proliferative rosettes (Fig. 8, H and I). Coelectroporation of MIR3607 with APC completely rescued all these defects in a majority of embryos, whereas expression of APC with a control, scrambled miRNA sequence, had null effect on the normal organization of cortical germinal layers (Fig. 8, F to J). APC expression also rescued the neurogenesis defects that we had observed previously when overexpressing MIR3607 at E14.5 (Fig. 3, C and D, and fig. S5), demonstrating that this mechanism remains during development. Together, our results demonstrated that expression of MIR3607 in the embryonic mouse cerebral cortex reduces the levels of APC, which leads to an abnormal accumulation of active β-catenin and the overactivation of the canonical Wnt pathway. This caused the overproliferation of aRGCs and expansion of the VZ that, bound to a partial loss of apical junction integrity, led to the formation of proliferative rosettes.
MIR3607 preserves aRGC polarity and promotes VZ expansion in higher mammals
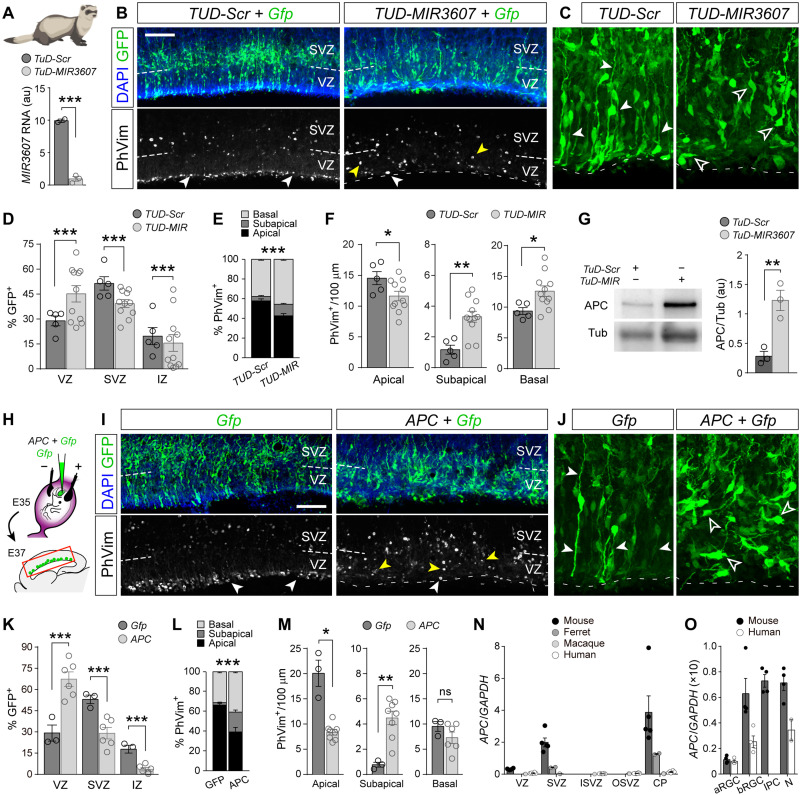
After characterizing the effects of exogenous expression of MIR3607 in the developing small and smooth mouse cortex, where normally it is not expressed, we turned to investigate the function of endogenous MIR3607 in ferret, with a folded and much larger cerebral cortex. To this aim, we generated loss-of-function constructs (Tough-Decoy inhibitors, TUD), antisense sequences that bind miRNAs with perfect complementarity and block them from binding to their endogenous targets, driving miRNAs for degradation (Fig. 9A) (37). We performed loss-of-function experiments by in utero electroporation of TUD-MIR3607 in the VZ of E35 ferret embryos, a stage when the endogenous expression levels of MIR3607 are high (Fig. 1D). Two days later (E37), electroporated (GFP+) cells were mislocated compared to control embryos, with an accumulation of cells in VZ and deficit in SVZ and IZ (Fig. 9, B to D). The loss of MIR3607 also disrupted the location of mitotic cells, with a significant decrease in apical mitoses and a several-fold increase in subapical mitoses (Fig. 9, B, E, and F). Analysis of the detailed morphology of aRGCs revealed a marked loss of polarity in TUD-MIR3607 embryos, where basal and apical processes were not recognizable, including the apical end foot (Fig. 9C). The loss of aRGC polarity likely impaired interkinetic nuclear migration, leading to the mislocalization of apical mitoses to subapical positions. These results were consistent with our findings in mouse where MIR3607 promotes β-catenin abundance, as this is a structural part of apical adherens junctions and loss of β-catenin impairs the integrity of aRGCs and VZ (38).
Fig. 9. MIR3607 is required for β-catenin–dependent aRGC amplification in ferret cortex.
(A) MIR3607 expression levels (qPCR, arbitrary units) in ferret Mpf cells transfected with the indicated plasmids; t test. (B to F) Parietal cortex of ferret embryos electroporated at E35 with the indicated plasmids, analyzed at E37 and stained as indicated (B and C), and quantifications of distribution of GFP+ cells and PhVim+ mitoses (D to F). In (B), white arrowheads indicate apical mitoses, and yellow arrowheads indicate subapical mitoses. In (C), solid arrowheads indicate apical or basal process, and open arrowheads indicate aRGCs without apical-basal polarity. N = 5 to 11 sections, 2 to 4 embryos per group. In (D) and (E), chi-square test; in (F), t test. (G) Western blots for APC and tubulin of Mpf cells transfected with the indicated plasmids and densitometry quantifications; t test. (H to M) Schema of experimental design (H), parietal cortex of ferret embryos electroporated at E35 with the indicated plasmids, analyzed at E37 and stained as indicated (I and J), and quantifications of distribution of GFP+ cells and PhVim+ mitoses (K to M). Arrowheads are used to indicate as in (B) and (C). N = 3 to 6 sections, one to three embryos per group. In (K) and (L), chi-square test; in (M), t test. (N and O) Normalized expression of APC mRNA in the layers and cell types of the developing cerebral cortex of the indicated species. Data are from (8, 36, 69, 70). Human, 13 to 16 wpc (weeks post conception); macaque, E76 to E92; ferret, E35; mouse, E14.5. Histograms show means ± SEM; circles indicate values for individual replicas; *P < 0.05, **P < 0.01, and ***P < 0.001. Scale bars, 100 μm (B and I).
Next, we investigated the molecular mechanism of action of MIR3607 in ferret. Our above results in mouse demonstrated that MIR3607 activates β-catenin signaling indirectly by targeting its repressor APC and driving it for degradation. The target sequence of MIR3607 in the 3′UTR of APC is conserved across vertebrates, from primates to bony fishes, including ferret (table S3), suggesting that its molecular mechanism of action in cerebral cortex development may also be conserved. To determine whether this is the case, we first performed loss-of-function experiments by expressing TUD-MIR3607 in the ferret cell line Mpf. We found that the levels of endogenous APC protein increased several-fold compared to control TUD-Scr cells (Fig. 9G), confirming APC as a conserved target of MIR3607 in ferret, and that endogenous levels of MIR3607 are sufficient to significantly down-regulate APC expression. Together with our previous findings, this result prompted us to test whether direct overexpression of APC might elicit changes on aRGCs and cortical development similar to those observed upon loss of MIR3607. Our prediction was that if APC is down-regulated by MIR3607, overexpressing APC would have even stronger effects. We electroporated the developing cortex of E35 ferret embryos with APC-encoding plasmids and analyzed the effects at E37, as above (Fig. 9H). Similar to the loss of MIR3607 function, APC overexpression caused the loss of polarity in aRGCs, together with a high accumulation of GFP+ cells in VZ and a concomitant reduction of cells in SVZ and IZ (Fig. 9, I to K). This was also accompanied by a very significant reduction in progenitor cell proliferation (30.4 ± 3.4 PhVim+/100 μm, Gfp; 19.9 ± 1.5 PhVim+/100 μm, APC; P = 0.01, t test), which was specifically related to the loss of apical mitoses (Fig. 9, I, L, and M). We also observed a fivefold increase in subapical mitoses, likely delaminated from the apical surface but not compensating for their massive loss. These effects of APC overexpression phenocopied MIR3607 loss of function but at a much greater level, consistent with our prediction.
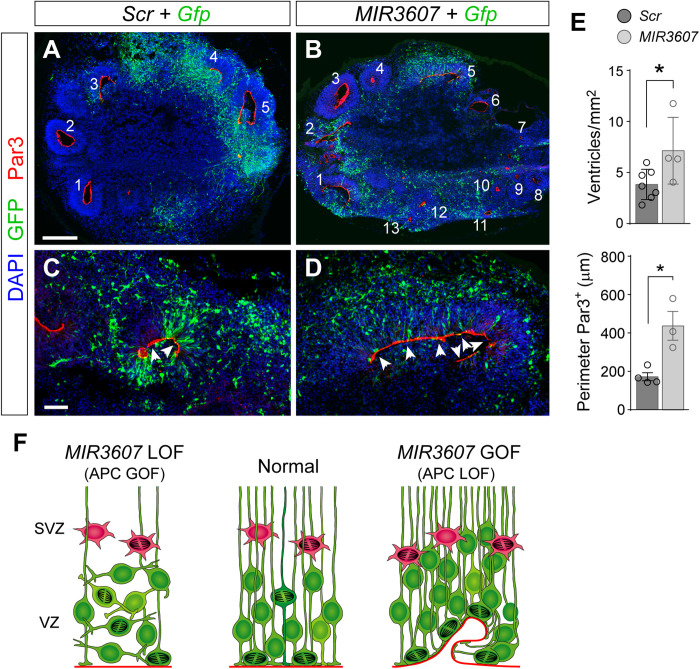
Our functional results in the developing ferret and mouse cortex, together with the expression levels of MIR3607 in mouse, ferret, macaque, and human (Fig. 1), and the conservation of MIR3607 targets in APC across phylogeny (table S3), supported the notion that MIR3607 expression in the developing cortex was selected during evolution to block APC levels and enhance β-catenin signaling, favoring amplification of aRGCs and cortical expansion. Consistent with this idea, analysis of public gene expression datasets revealed that APC mRNA levels are much higher in the developing cortex of mouse than in ferret, macaque, or human, at equivalent developmental stages, both at the level of tissue and single cells (Fig. 9, N and O). More specifically, APC mRNA is expressed at higher levels in mouse than human in all cortical progenitor cell types except aRGCs, where relative levels are similar (Fig. 9O). Thus, to investigate whether the function of MIR3607 in cortical development, via APC repression, is conserved in humans, we overexpressed MIR3607 in human cerebral organoids generated from hiPSCs (human induced Pluripotent Stem Cells). The cortical identity of our organoids was validated by the expression of PAX6 and TBR1 and the overall absence of the subcortical marker proteins NKX2.1 (NK2 homeobox 1) and GSX2 (GS homeobox 1) (fig. S6). Compared to controls, the ventricle of MIR3607-expressing organoids was highly fragmented, showing a greater number of individual ventricles decorated with a Par3+ apical adherens junction belt, reminiscent of delaminated rosettes in mouse (Fig. 10, A to E). The Par3+ apical surface of electroporated ventricles was, on average, much longer in MIR3607 organoids than in controls (Fig. 10, C to E), consistent with VZ expansion by amplification of aRGCs and maintenance of their apico-basal polarity, as we previously found in mouse and ferret.
Fig. 10. MIR3607 drives aRGC amplification in human cortex.
(A to E) Cross sections through human cerebral organoids electroporated at 39 days in culture and stained 7 days later as indicated (A to D) and quantifications (E). Individual ventricles are numbered in panoramic views, and details show the Par3+ apical surface (arrowheads). Histograms represent density of ventricles per surface area of organoid and average length of Par3+ apical surface of individual electroporated ventricles per organoid. All ventricles from each organoid were measured. Histograms show means ± SEM; circles indicate values for individual replicas; n = 3 to 7 organoids per group; 116 to 133 ventricles (density), 8 to 12 ventricles (Par3); t test; *P < 0.05. LOF, loss-of-function; GOF, gain-of-function. Scale bars, 200 μm (A and B) and 50 μm (C and D). (F) Schematic summary of results in this study.
DISCUSSION
Our results demonstrate that MIR3607 is a critical regulator of cortical development by promoting the amplification of aRGCs and maintaining their polarity. We show that MIR3607 directly represses APC expression, which, in turn, activates β-catenin signaling, a major pathway regulating cortical development and expansion (33). Our experimental manipulations in the developing cortex of mouse and ferret embryos in vivo, and in human cerebral organoids in vitro, show that an excess of MIR3607 leads to the sustained maintenance of polarity and amplification of cortical aRGCs, with overgrowth of the VZ. In contrast, a loss of MIR3607, or excess of APC, leads to the loss of aRGC polarity, impaired proliferation, and delamination (Fig. 10F). Because aRGC amplification and VZ growth set the basis for increasing cortical surface area, our findings support the notion that cortical expansion and folding during mammalian evolution may have been boosted by high expression levels of MIR3607 during embryonic development, and hence, the secondary loss of this expression contributed to the subsequent evolution of the small and smooth murine cortex.
Gene regulation and secondary reduction of cortex size during evolution
Cortex expansion and folding during evolution was very prominent in humans and great apes, but it took place in all major mammalian clades. In some clades, this process was reversed during subsequent evolution, particularly in extant species with a small and smooth cortex, like small world monkeys such as marmoset, and rodents like mice, thereby undergoing a secondary reduction of both brain size and folding (9). This secondary reduction during evolution was the result of changes recapitulated during embryonic development. For example, in species with a large and folded cerebral cortex such as human, macaque, or ferret, embryonic development involves a large abundance of highly proliferative neural stem and progenitor cells, such as aRGCs in VZ and bRGCs forming the OSVZ, key for cortex folding (12–16, 30). In species with a smaller and smooth cortex, such as mouse, embryonic cortical development involves much fewer progenitor cells, forming small and simple VZ and SVZ (29, 39). This provides a unique opportunity to test whether developmental features specific to large-brained species are important for cortical expansion by introducing them into small-brained species such as mouse.
An increasing number of transcriptomic studies have identified protein-coding genes expressed in germinal layers of the developing cerebral cortex that are either specific to large-brained species or expressed at different levels according to brain size or progenitor cell type (5–8, 19). Functional testing of some of these genes demonstrates their relevance in the acquisition of features typical of large brains, for example, driving expansion of the cortical progenitor cell pool when expressed in the developing mouse cortex (5, 7, 19, 40, 41). The number of human-specific genes, newly emerged during recent evolution, is relatively very small (19), whereas the number of conserved genes that are expressed in the developing cortex at different levels across phylogeny is much larger (6, 22). Hence, a key question now emerging is how the expression levels of such highly conserved gene networks became regulated so differently during evolution. Despite the power of miRNAs to modulate the expression of protein-coding genes, they have received unexpectedly limited attention in the context of brain evolution and expansion (20, 21). Here, we have investigated this largely unexplored possibility focusing on MIR3607, a candidate miRNA attractive for three main reasons: first, it has a highly conserved sequence across mammals with very different brain sizes, including human, ferret, and mouse (table S1). Second, it is computationally predicted to target a large set of genes functionally related to early neural development, including progenitor proliferation and neurogenesis, and third, it is expressed in the developing cerebral cortex of large-brained species, such as human, macaque, and ferret, but not of the smooth and small-brained mouse. This poses the intriguing possibility that MIR3607 expression in cortical progenitor cells may have been lost systematically in mammalian species undergoing secondary simplification of their cerebral cortex, including some of the small New World monkeys, such as marmoset, which deserves further investigation.
The absence of MIR3607 expression in the embryonic mouse cortex, in contrast to human, macaque, and ferret, raises the key question of how this is regulated. The region of the human genome encoding MIR3607 contains several candidate cis-regulatory elements (CREs), which display functional signatures of promoter, proximal enhancer, and distal enhancer (fig. S7A). Sequence comparison of these regions across the genomes of gyrencephalic and lissencephalic species identifies particular subregions that are highly conserved, especially within candidate promoter elements. Several other subregions are exclusively conserved in gyrencephalic species but not lissencephalic, and at least one subregion is found and conserved mostly in lissencephalic species (fig. S7B). All these brain type–linked subregions within candidate functional CREs contain highly conserved binding site motifs for identified transcription factors, most of which are well-known regulators of embryonic brain development (i.e., INSM1 (insulinoma 1), TEAD1/3/4 (TEA domain transcription factor), SP8 (Sp8 transcription factor), SOX8 (SRY-box transcription factor 8), and POU4F1 (POU class 4 homeobox 1) ) (42–44). Hence, differences in regulatory sequences may control the expression of MIR3607 in the developing cortex of different species, which prompts one to speculate that these sequences may have been under selection pressure during mammalian evolution. Positive selection of CRE sequences conserved in gyrencephalic species may have favored the expression of MIR3607 in their embryonic cerebral cortex, while negative selection may have underlined the secondary loss of this expression, possibly followed by the reduction of cortex size and folding in lissencephalic species. A second mechanism potentially regulating the presence of MIR3607 in embryonic human but not mouse is related to its specific sequence, which differs in 5 nucleotides. Four of these nucleotides belong to the loop region of the pre-MIR3607, three of which are absent in mouse (table S1). This change is predicted to significantly affect the size and secondary structure of the hairpin (fig. S8A), key for processing of pre-miRs into mature miRs by Dicer, thus potentially modifying the efficiency of this processing and limiting the availability of functional MIR3607.
Our results in mouse demonstrate that expression of MIR3607 in the embryonic cerebral cortex is in itself sufficient to strongly promote the proliferation and increase the pool of aRGCs while maintaining their polarity, hence expanding the VZ. These are features greatly enhanced in the developing cerebral cortex of human, macaque, and ferret, linked to the early lateral expansion of the VZ and, later, to the abundant formation of bRGCs (12–16), which contribute critically to the large size of these cortices (13, 25). Overexpression of MIR3607 in cerebral human organoids also expanded the VZ (Fig. 10E). We have found that expansion of aRGCs and VZ in mouse following overexpression of MIR3607 leads to the formation of proliferative rosettes. These are possibly an epiphenomenon unrelated to natural cortical expansion or folding, resulting from the combination of high aRGC proliferation and maintained polarity, with unstable apical junctions, which are the consequence of overactivation of β-catenin signaling in mouse cortex, as rosettes also form upon this manipulation. Whereas overexpression of MIR3607 in mouse VZ caused progenitor cell expansion, loss of endogenous MIR3607 in ferret decreased aRGC polarity and proliferation, promoting neurogenesis (subapical and basal mitoses). Altered or lost cell polarity in aRGCs may directly relate to increased neurogenesis in several ways, for example, by the loss of contact with the lateral ventricle. This represents a loss of signaling from many growth factors and other molecules in the cerebrospinal fluid that promote progenitor cell amplification [i.e., Shh and fibroblast growth factor (FGF)], as well as impaired Notch signaling with neighboring cells, both scenarios leading to the alternative fate: neurogenesis. While cortical expansion requires increased neuron production, premature neurogenesis depletes the pool of progenitor cells, causing the opposite effect: cortical reduction. In summary, our results indicate that expression of MIR3607 in the developing cortex may have been secondarily lost during rodent evolution as a mechanism contributing to reduce cerebral cortex size.
MIR3607 is a major regulator of canonical Wnt/β-catenin signaling
The strong effect of MIR3607 expression on aRGC amplification in the early embryonic cortex is much stronger than late stages, yet our results demonstrate that at both stages, the complex phenotype observed is dominated by the overactivation of the canonical Wnt/β-catenin pathway (Figs. 5 and 8). The formation of rosettes upon expressing MIR3607 in early mouse embryos was phenocopied by the acute expression of constitutively active β-catenin, and reminiscent of defects in transgenic mice expressing active β-catenin in the cortical primordium (33). Furthermore, our transcriptomic analyses demonstrate that MIR3607 promotes canonical Wnt/β-catenin signaling at multiple levels by targeting and modifying mRNA levels of several major regulators of this pathway, including up-regulation of β-Catenin itself and down-regulation of the pathway’s key repressor APC. Previous studies showed that β-catenin is key to maintain the population of aRGCs and suggested that down-regulation of β-catenin signaling may be critical to facilitate the transition to IPCs (45). Our findings demonstrate that MIR3607 has a central role in regulating this process: overexpression of MIR3607 in mouse led to excessive amplification of aRGCs and defective transition to IPCs, to the point of disrupting VZ structure and forming rosettes, whereas down-regulation in ferret led to the loss of aRGC polarity and delamination (Fig. 10F). Overexpression of APC rescued the defects of MIR3607 overexpression in mouse, both at early and late stages, and phenocopied the defects of MIR3607 loss of function in ferret, demonstrating the direct implication of Wnt/β-catenin signaling and its functional link to MIR3607. Moreover, the defects observed in ferret upon loss of endogenous MIR3607 are reminiscent to those previously found in mouse under low β-catenin levels, which severely altered the organization of the neuroepithelium, including translocation of apical mitoses to subapical positions, loss of adherent junctions, decreased proliferation, and premature disassembly of the radial fiber scaffold (38).
In the embryonic cerebral cortex, β-catenin has two very distinct functions: structural component of apical adhesions between aRGCs and transcriptional regulator of progenitor cell proliferation and amplification. In mouse embryos, overexpression of MIR3607 caused the formation of rosettes from early stages. In this scenario, high MIR3607 blocks APC expression and, hence, derepresses β-catenin signaling, causing the overamplification of aRGCs. Increased β-catenin signaling seems to also affect its structural function, as the polarity of RGCs and their apical adhesion are strictly maintained despite the overproliferation, together leading to the formation of rosettes. In contrast, the loss of function of MIR3607 in ferret embryos derepresses (increases) APC levels and, hence, represses β-catenin. In this second scenario, the decrease of β-catenin signaling seems to primarily affect its structural function, destabilizing the apical junctions of aRGCs and causing their individual delamination and loss of polarity but not the formation of rosettes.
In contrast to the strong effect of MIR3607 expression on aRGC amplification in the early embryonic cortex, the largest effect of late-onset MIR3607 expression was on postmitotic neurons, altering their migration, laminar position, and growth of their axons toward the CC. This was consistent with part of the transcriptomic phenotype caused by MIR3607 expression, with significant changes related to mTORC1 (mammalian target of rapamycin complex 1) signaling, axonal development, and L1CAM interactions (46). While the origin of differences in phenotypes between MIR3607 expression at early and mid-developmental stages remains to be identified, existing evidence suggests that the lower impact on aRGC amplification at later stages may be related to a dominant effect of the default genetic program, driving aRGCs to undergo self-consuming neurogenic divisions. Endogenous levels of β-catenin activity are normally down-regulated in the embryonic cerebral cortex from E12.5 through E16.5 (47), which suggests that Wnt/β-catenin activity gradually becomes less relevant in progenitor cells as cortical development progresses. Expression of APC remains similarly high through development (48), suggesting that the late endogenous down-regulation of β-catenin activity is not related to increased APC. Therefore, lowering APC levels by MIR3607 may be more impactful at early than late stages, having progressively less influence on progenitor cell amplification, consistent with our results of MIR3607 overexpression at different stages. Thus, our study confirms the critical role of APC and Wnt signaling in progenitor cells during cerebral cortex development (48, 49) and demonstrates that MIR3607 is a fundamental negative regulator of APC and activator of Wnt/β-catenin signaling in this process. This is in agreement with previous studies showing the regulatory interplay between MIR3607 and APC to control the proliferative activity of lung cancer cells (36).
It has been previously shown that the 3′UTR of human APC contains a sequence computationally predicted to be directly bound by MIR3607 and that this represses APC transcription (36). We have extended these observations showing that this target sequence in APC is highly conserved across amniotes, supporting a strong phylogenic conservation of this regulatory mechanism. Coding genes are usually targeted by a large number and variety of miRNAs, suggesting functional redundancy on gene regulation (28, 50). Accordingly, the loss of MIR3607 expression during rodent evolution might have been compensated by expression of other targeting miRNAs, thus being functionally innocuous. In agreement with this notion, computational analyses of the APC locus in human, macaque, rat, and mouse predict that it is targeted by a very large number and variety of miRNAs (fig. S8B). Although most miRNAs are unique to each species, a majority are shared only between primates (gyrencephalic) or rodents (lissencephalic), suggestive of potentially having some conserved relevance for cortical expansion and/or folding. However, our rescue experiments show that overexpression of APC completely rescues the cell cycle reentry and rosette phenotypes caused by MIR3607 overexpression in mouse. This indicates not only that the primary cause of these phenotypes is the loss of APC but also that if alternative APC-targeting miRNAs are expressed in the embryonic mouse cortex to compensate for the absence of endogenous MIR3607, overexpression of APC is counterbalancing their action. Accordingly, overexpression of APC in a control scenario (without MIR3607) would overload the endogenous APC-targeting miRNAs and cause some phenotype, but this is not the case. In contrast, excess of APC is noxious in ferret (with endogenous MIR3607), as is the loss of MIR3607. Hence, expression of APC in mouse below some threshold level is deleterious for cortical development, but expression above some threshold causes no major defect, so it is unlikely that APC-targeting miRNAs alternative to MIR3607 have a relevant function on mouse cortical development.
In summary, we have identified MIR3607 as a major regulator of Wnt/β-catenin signaling, a fundamental pathway with key functions in the embryonic development of the cerebral cortex (33, 48, 49). Our findings are consistent with recent discoveries on the key importance of miRNAs in early cortical development, regulating neural stem cell amplification and germinal layer homeostasis (51). From the evolutionary point of view, our results suggest that a loss of expression of MIR3607 in cerebral cortex development may have been a key factor for the secondary reduction of brain size during rodent evolution.
MATERIALS AND METHODS
Animals
Timed-pregnant sable ferrets (Mustela putorius furo) were obtained from Euroferret (Denmark) and maintained on a 16-hour light/8-hour dark cycle at the Animal Facilities of the Universidad Miguel Hernández. Wild-type mice in ICR (Institute of Cancer Research) background were obtained from a breeding colony at the Animal Facility of the Instituto de Neurociencias. The day a vaginal plug was detected was considered E0.5. Animals were treated according to Spanish (RD 53/2013) and European regulations, and experimental protocols were approved by the CSIC (Consejo Superior de Investigaciones Científicas) Ethics Committee.
DNA constructs
For gain-of-function experiments, we used pCAG-GFP expressing GFP under the CAG promoter, mixed with psil-pre-MIR3607 encoding pre-MIR3607-5p under the U6 promoter, or with psil-pre-miRScr under U6 as control. Oligos encoding pre-MIR3607 (forward, 5′ GATCCACTGATTTCCTTCATGTCAAGCTTCAAGAGAGCATGTGATGAAGCAAATCAGTTTTTTTGGAAA 3′; reverse, 5′ AGCTTTCCAAAAAAACTGATTTGCTTCATCACATGCTCTCTTGAAGCTTGACATGAAGGAAATCAGTG 3′) were designed with Bam HI–Hind III sites and cloned into pSilencer2.1-U6 puro (Thermo Fisher Scientific, catalog no. AM5762). pSilencer puro negative control plasmid provided with the kit was used as psil-pre-miRScr, encoding a small interfering RNA sequence not found in the mouse, human, or rat genome databases. For loss-of-function experiments, we constructed Tough-Decoy plasmids against MIR3607 or a scrambled sequence as control. These were designed as in (52) and cloned into pSilencer2.1-U6 puro (Ambion) following the same protocol as above. Other plasmids used were pCMV-GFP, pCAG-mGFP (loxp-EGFP-farnesylated) mixed with pCAG-Cre, pCMV-Neo-Bam APC (Addgene, #16507), and pCAG-Δ90βCateninGFP (Addgene, #26645) encoding stabilized β-catenin. Plasmid DNA was purified with a NucleoBond Xtra Midi kit (Cultech, 22740410.50) and resuspended in nuclease-free water (Sigma-Aldrich).
Validation of MIR3607 expression
Human embryonic kidney 293T cells were transfected with 2 μg of psil-pre-miRScr or psil-pre-MIR3607 plus 2 μg of pCAG-GFP using Lipofectamine and harvested 2 days later, and RNA was isolated using mirVana miRNA isolation kit (catalog no. AM1560). Quantitative RT-PCR was carried out using TaqMan microRNA assays. All kits and reagents, including primers and probes for MIR3607 (assay #463448) and U6 small nucleolar RNA control (assay #001973), were from Thermo Fisher Scientific (catalog no. 4427975).
In utero electroporation
In utero electroporation of mouse and ferret embryos was performed as described elsewhere (8, 23). Briefly, timed-pregnant females were deeply anesthetized with isoflurane, the abdominal cavity was open, and the uterine horns were exposed. DNA solution (1 μl) was injected into the lateral ventricle using pulled glass micropipettes, and square electric pulses (mouse: 35 V for E12.5 and 45 V for E14.5; ferret: 75 V; 50 ms on to 950 ms off, five pulses) were applied with an electric stimulator (Cuy21EDIT, Bex C. Ltd.) using round electrodes (CUY650P5, CUY650P7, Nepa Gene). Plasmid concentrations for gain-of-function experiments were as follows: Gfp (0.7 μg/μl), MIR-Scr (1 μg/μl), and MIR3607 (1 μg/μl); for rescue: MIR-Scr (0.75 μg/μl), MIR3607 (0.75 μg/μl), APC (0.75 μg/μl), and Gfp (0.5 μg/μl); and for loss of function: TUD-Scr (1 μg/μl), TUD-MIR3607 (1 μg/μl), and Gfp (0.75 μg/μl). Uterine horns were placed back into the abdominal cavity, suture was closed, and the pregnant female was allowed to fully recover on a heating pad before returning to the home cage.
Retroviral stocks preparation and concentration
High-titer murine Moloney leukemia virus–based retroviruses encoding GFP under the CAG promoter were prepared by transient transfection (together with CMV-vsvg and CMV-gp plasmids) of human embryonic kidney 293T cells as a package cell line and concentrated by ultracentrifugation, and viral titer was estimated by clonal infection (53). Viral solutions were injected using pulled glass micropipettes.
Single progenitor clonal analysis
ICR control pregnant females carrying E12.5 embryos were deeply anesthetized with isoflurane, and individual embryos were injected with 1 μl of Gfp-encoding retroviruses into the telencephalic ventricle immediately after electroporation of Scr or MIR3607 + Gfp encoding plasmids in the same telencephalic ventricle. After 24 hours of survival, embryos were sacrificed, their heads fixed in 4% paraformaldehyde (PFA), cryostat-sectioned at 40 μm, and processed for immunohistochemistry as described above.
hiPSC culture
Human iPSCs [American Type Culture Collection (ATCC)] were cultured at 37°C, 5% CO2, and ambient oxygen level on Geltrex-coated plates in mTeSR Plus medium (STEMCELL Technologies, 05825) with daily medium change. For passaging, iPSC colonies were incubated with StemPro Accutase Cell Dissociation Reagent (A1110501, Life Technologies) diluted 1:4 in phosphate-buffered saline (PBS) for 4 min. Pieces of colonies were washed off with Dulbecco’s Modified Eagle’s Medium (DMEM)/F12, centrifuged for 5 min at 300g, and resuspended in mTeSR Plus supplemented with 10 μM Rock inhibitor Y-27632(2HCl) for the first day.
Generation of human cerebral organoids
Cerebral organoids were generated as previously described (51, 54). Briefly, mycoplasma-free iPSCs from the parental cell line HMGU1 obtained from male foreskin fibroblasts (ATCC, CRL-2522) were dissociated into single cells using StemPro Accutase Cell Dissociation Reagent (A1110501, Life Technologies) and plated in the concentration of 9000 single iPSCs per well into low attachment 96-well tissue culture plates in hES medium [DMEM/F12GlutaMAX supplemented with 20% KnockOut Serum Replacement, 3% embryonic stem–grade fetal bovine serum (FBS), 1% non-essential amino acids, 0.1 mM 2-mercaptoethanol, basic fibroblast growth factor (bFGF; 4 ng/ml), and 50 μM Rock inhibitor Y27632] for 6 days to form embryoid bodies (EBs). Rock inhibitor Y27632 and bFGF were removed on the fourth day. On day 6, EBs were transferred into low attachment 24-well plates in NIM medium [DMEM/F12GlutaMAX supplemented with 1:100 N2 supplement, 1% non-essential amino acids and heparin (5 μg/ml)] and cultured for an additional 6 days. On day 12, EBs were embedded in Matrigel drops, and then, they were transferred to 10-cm tissue culture plates in NDM minus A medium [DMEM/F12GlutaMAX and Neurobasal in ratio 1:1 supplemented with 1:100 N2 supplement 1:100 B27 without vitamin A, 0.5% non-essential amino acids, insulin (2.5 μg/ml), 1:100 penicillin-streptomycin (P/S), and 50 μM 2-mercaptoethanol] to form organoids. Four days after Matrigel embedding, cerebral organoids were maintained in constant agitation using an orbital shaker and cultured until electroporation in NDM plus A medium [DMEM/F12GlutaMAX and Neurobasal in ratio 1:1 supplemented with 1:100 N2 supplement 1:100 B27 with vitamin A, 0.5% non-essential amino acids, insulin (2.5 μg/ml), 1:100 P/S, and 50 μM 2-mercaptoethanol]. During the whole period of cerebral organoid generation, cells were kept at 37°C, 5% CO2, and ambient oxygen level with medium changes every other day. After transferring the cerebral organoids to the shaker, medium was changed twice per week.
Electroporation of human cerebral organoids
Cerebral organoids were kept in antibiotics-free conditions before electroporation. Electroporations were performed in 39-day-old cerebral organoids and fixed 7 days after electroporation. During the electroporation, cerebral organoids were placed in an electroporation chamber (Harvard Apparatus, Holliston, MA, USA) under a stereoscope and using a glass microcapillary 1 to 2 μl of plasmid DNAs was injected together with Fast Green (0.1%, Sigma-Aldrich) into different ventricles of the organoids. Plasmid DNA concentrations were as follows: GFP (0.7 μg/μl), miRScr (1 μg/μl), and MIR3607 (1 μg/μl). Cerebral organoids were subsequently electroporated with five pulses applied at 80 V for 50 ms each at intervals of 500 ms (ECM830, Harvard Apparatus). Following electroporation, cerebral organoids were kept for an additional 24 hours in antibiotics-free media, and then changed into the normal media until fixation. Cerebral organoids were fixed using 4% PFA for 1 hour at 4°C, cryopreserved with 30% sucrose, and stored at −20°C. For immunofluorescence, 20-μm cryosections were prepared.
BrdU labeling experiments
BrdU (Sigma-Aldrich) solution (10 mg/ml in 0.9% NaCl) was administered intraperitoneally at 50 mg/kg body weight. To identify progenitor cells in S phase, a single dose of BrdU was injected 30 min before fixation. To measure cell cycle exit, embryos electroporated with Scr or MIR3607 + Gfp at E14.5 were injected with a single dose of BrdU (50 mg/kg body weight) 24 hours before sacrifice and analysis (E15.5 or E16.5). For embryos analyzed at E15.5, BrdU was injected 4 hours before electroporation. All GFP+BrdU+ and GFP+BrdU+Ki67+ cells were counted in the electroporated area across the entire thickness of all layers containing GFP cells (VZ + SVZ + IZ). Four to five embryos were analyzed per condition, and for each embryo, counting was done in two different sections. Cell cycle exit rate was calculated as the proportion of GFP+BrdU+ cells that were negative for Ki67 (GFP+BrdU+Ki67−/GFP+BrdU+). Results were analyzed for statistical difference using chi-square test.
Tissue processing
Embryonic brains were immersion-fixed in phosphate-buffered 4% PFA at 4°C. Postnatal pups were perfused transcardially with PFA, and the heads were postfixed overnight at 4°C. After fixation, brains were washed with PBS and cryoprotected overnight with 30% sucrose. For ISH, brains were treated under ribonuclease-free conditions and cryoprotected in 30% sucrose and 2% PFA. Brains were finally cryosectioned at 20 to 30 μm.
Immunohistochemistry
Brain sections were incubated with primary antibodies overnight, followed by appropriate fluorescently conjugated secondary antibodies and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, D9542). Primary antibodies used were against BrdU (1:500; Abcam, ab6326), GFP (1:1000; Aves Lab, GFP-1020), Ki67 (1:300; Abcam, ab15580), phosphohistone H3 (1:1000; Upstate, 06-570), Tbr1 (1:500; Abcam, ab31940), Tbr2 (1:500; Millipore, ab31940), Par3 (1:500; Millipore, MABF28), Pax6 (1:500; Millipore, AB2237), anti-NeuN (1:1000; Millipore, ABN78), Cux1 (1:500; Santa Cruz Biotechnology), and activated β-catenin (1:500; Merck, 05-665). Secondary antibodies were from Vector Lab [biotinylated anti-rat immunoglobulin G (IgG; 1:200; BA-9400) and biotinylated anti-rabbit IgG (1:200; BA-1000)], from Jackson ImmunoResearch [Cy3 Fab fragment anti-rabbit IgG (1:200; 711-167-003), Alexa Fluor 488 anti-chicken IgY (1:200; 703-545-155), and Cy5 streptavidin (1:200; 016-170-084)], and from Invitrogen [Alexa Fluor 555 anti-rabbit IgG (1:200; A-31572)].
Western blotting and quantification
For Western blot assays, ferret Mpf cells (ATCC, CRL-1656) were transfected using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 hours, cells were harvested and washed with cold PBS, and lysate was prepared with pH 7.4 sterile cold lysis buffer (20 mM Hepes, 150 mM KCl, 1 mM EGTA, 1 mM EDTA, 0,1 mM dithiothreitol, 40 mM NaF, 1 mM Na3VO4, 1% Triton X-100, and protease inhibitor). The amount of protein in each sample was then quantified as follows. The soluble fraction was collected from the lysate after centrifugation at 15,000 rpm for 15 min. The protein concentration was measured using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; reference: #23227). Samples were heat-treated at 95°C for 5 min in 1× Laemmli Sample Buffer. Total protein (25 μg) per well and per sample condition was run on an 8% SDS–polyacrylamide gel electrophoresis gel for 2 hours at 120 mV in 1× Running Buffer (1× tris-glycine and 0.1% SDS). The wet transfer to a 0.45-μm nitrocellulose membrane (GE Healthcare Life Science; reference: 10600002) was carried out overnight at 4°C, 30 mV in cold transfer buffer (1× tris-glycine, 1% methanol, and 0.01% SDS). After washing and fixing with Ponceau S, the membrane was blocked in 5% milk TBS-T (tris-buiffered saline with 0.1% Tween 20 detergent) buffer in shaker for 1 hour at room temperature. Antibodies were diluted on blocking solution at a concentration of 1:1000 anti-APC rabbit IgG (Cell Signaling Technology; reference: 2504S), 1:1000 anti-cMyc mouse IgG (Santa Cruz Biotechnology; reference: sc40), 1:5000 goat peroxidase anti-rabbit IgG (Thermo Fisher Scientific; reference: 31462), and 1:2000 goat peroxidase anti-mouse IgG (Thermo Fisher Scientific; reference: 31444). Primary antibody incubation was carried out overnight at 4°C in a shaker and incubated with secondary antibody for 2 hours at room temperature. To detect the labeling, Chemiluminescent HRP substrate (Millipore; reference: WBKLS0100) was applied, and the membrane was exposed in an AI680 Bioimager. The quantification was normalized to the tubulin amount using anti-Tub mouse primary antibody 1:1000 (Sigma-Aldrich; reference: T5168) and 1:2000 goat peroxidase anti-mouse IgG (Thermo Fisher Scientific; reference: 31444). Mean intensity was measured by ImageJ Fiji and statistically analyzed by unpaired Student’s t test.
miRNA in situ hybridization
miRNA ISH was performed as described previously (55). Cryostat, cryotome, or deparaffinized paraffin sections were washed briefly with PBS, permeabilized with radioimmunoprecipitation assay buffer (150 mM NaCl, Triton X-100 1%, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and 50 mM tris), prehybridized at 54°C for 1 hour with hybridization solution [50% formamide (Ambion), 5× SSC (Sigma-Aldrich), 5× Denhardt’s (from a 50× stock; Sigma-Aldrich), yeast RNA (250 μg/ml), and salmon sperm DNA (500 μg/ml)], and hybridized overnight at 54°C (30°C below RNA Rm) with miRCURY LNA microRNA ISH Detection Probe (5′-3′-ACTGATTTGCTTCATCACATGC/3Dig_N/; product #612280-350, Exiqon) in hybridization solution. Sections were washed 2, at 1 hour per wash, with prewarmed posthybridization solution (50% formamide, 2× SSC, and 0.1% Tween), washed briefly with MABT buffer [1× maleic acid–NaCl (pH 7.5) and 0.1% Tween], blocked with blocking solution (10% bovine serum in MABT buffer) for 1 hour at room temperature, and incubated with alkaline phosphatase coupled anti-digoxigenin Fab fragments (1:2000; Roche) in blocking solution overnight at 4°C. After brief washes with MABT and NTMT [100 mM tris (pH 9.5), 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween], sections were incubated in nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) solution {3.4 μg/ml from NBT stock and 3.5 μl/ml from BCIP stock in NTMT buffer [NBT stock (100 mg/ml) in 70% dimethylformamide and BCIP stock (50 mg/ml) in 100% dimethylformamide; Roche]}. Sections were lastly dehydrated and coverslipped.
FACS sorting of electroporated brains
Electroporated mouse embryos with similar rostro-caudal and lateromedial location of GFP fluorescence in the neocortex were selected, and the GFP+ cortical area was dissected out in ice-cold Hanks’ balanced salt solution (HBSS; Thermo Fisher Scientific). Dissected tissue was dissociated with trypsin diluted in HBSS in the presence of deoxyribonuclease at 37°C for 8 min. HBSS supplemented with 10% FBS and 1% P/S was then added, and tissue was gently triturated. The cell suspension was centrifuged, and cells in pellet were resuspended in 500 μl of medium, filtered, and FACS-sorted (FACSAria II, BD). Cells with high GFP intensity were collected in Neurobasal medium supplemented with 10% FBS and 1% P/S, and RNA was extracted using Arcturus PicoPure RNA isolation kit (Thermo Fisher Scientific, catalog no. KIT0202) according to the manufacturer’s protocol. RNA integrity was analyzed using Bioanalyzer (Agilent 2100), and samples with RIN values above 9.5 were selected for RNA-seq. For each biological replica, cells from two embryos were pooled together, and three independent replicas per condition were sequenced. Libraries were prepared with the SMART-Seq v4 Library Prep Kit and sequenced on Illumina HiSeq 2500 sequencer using 50–base pair (bp) single reads.
RNA-seq and differential expression analysis
Quality control of sequenced reads was performed using FastQC v0.11.7 (Babraham Institute). Sequencing reads were aligned using HISAT2 v2.1.0 to GRCm38/mm10 mouse genome index masked with the mouse Single Nucleotide Polymorphism Database build 142 (56). Integrative Genomics Viewer (57) was used to visualize aligned reads and normalized coverage tracks [RPM (reads per million)]. Reads overlapping annotated genes (Ensemble GRCm38.93) were counted using HTSeq v0.11.1. DEGs were detected using DESeq2 v1.18.1 package in R language. Genes with adjusted P < 0.01 were considered significantly differentially expressed. For some comparisons, mRNA abundance of RNA-seq data was obtained using transcript per million normalization. RNA-seq data reported in this study are accessible through the Gene Expression Omnibus (GEO) database with the GEO series accession number GSE135321.
Gene set enrichment and pathway analyses
Identification of enriched biological functions and processes in DEGs was performed using clusterProfiler (58) package in R. Overrepresentation test (59) was performed using the enrichGO function of clusterProfiler with the following parameters: gene = DEGs, OrgDb = org.Mm.eg.db, ont = BP, pAdjustMethod = BH, pvalueCutoff = 0.05, and qvalueCutoff = 0.1. Functional annotation analysis of DEGs was performed using DAVID v6.8 with the following annotations: three functional categories (COG_ONTOLOGY, UP_KEYWORDS, and UP_SEQ_FEATURE), three gene ontologies (GOTERM_BP_DIRECT, GOTERM_CC_DIRECT, and GOTERM_MF_DIRECT), two pathways (BIOCARTA and KEGG_PATHWAY), and three protein domains (INTERPRO, PIR_SUPERFAMILY, and SMART). Visualization of interrelations of terms and functional groups in biological networks was performed using ClueGO v2.5.0 plug-in of Cytoscape v3.6.0 with the following parameters: gene list = DEGs; ontologies = GO_MolecularFunction-EBI-QuickGO-GOA_22.03.2018_00h00, GO_BiologicalProcess-EBI-QuickGO-GOA_22.03.2018_00h00, WikiPathways_10.01.2019, KEGG_10.01.2019, REACTOME_Reactions_10.01.2019, and REACTOME_Pathways_10.01.2019; Statistical Test Used = Enrichment/Depletion (Two-sided hypergeometric test); Correction Method Used = Bonferroni step down; Min GO Level = 3; Max GO Level = 8; Number of Genes = 3; Min Percentage = 4.0; Combine Clusters With ‘Or’ = true; Percentage for a Cluster to be Significant = 60.0; GO Fusion = true; GO Group = true; Kappa Score Threshold = 0.4; Over View Term = SmallestPValue; Group By Kappa Statistics = true; Initial Group Size = 1; and Sharing Group Percentage = 50.0. GSEA (60) was performed using GSEAPreranked tool and GSEA function on clusterProfiler package in R, on 21,822 unique features (genes) that were preranked on the log2 of the fold change from DESeq2 analysis. The following parameters were used for GSEA: exponent = 0 (scoring_scheme = classic), nPerm = 1000, minGSSize = 15, and maxGSSize = 500. GSEA was performed using MSigDB gene sets Hallmark (h.all.v6.2.symbols.gmt) and GO (c5.all.v6.2.symbols.gmt).
MIR3607 target and structure prediction
miRNA targets were predicted using available online tools: TargetScanHuman 7.2, miRDB, TargetScanMouse Custom (versions 4 and 5.2), and miRDB custom prediction for identification of mouse target genes. Predictions of the secondary structures of human and mouse pre-MIR3607 were obtained using MXfold2 (61).
Collection of genomic sequences and analysis for presence of transcription factor binding motifs
Sequences from 5 kbp upstream to 1 kbp downstream of the MIR3607 transcription start site were collected using UCSC Table Browser (62) from 18 different species using the option xenoRefGene: human (Homo sapiens), chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), pig (Sus domesticus), cow (Bos taurus), sheep (Ovis aries), ferret (Mustela furo), cat (Felis silvestris), marmoset (Callithrix jacchus), tarsier (Tarsius tarsier), mouse (Mus musculus), rat (Rattus norvegicus), Chinese hamster (Cricetulus barabensis), guinea pig (Cavia porcellus), rabbit (Oryctolagus cuniculus), and pika (Ochotona princeps). Collected sequences were sorted into two distinct groups: gyrencephalic and lissencephalic species. Sequences were then aligned using Clustal Omega (63) and visualized using Jalview (64), where sequences were colored according to their percent identity. CRE and gene sequences were identified via UCSC Genome Browser, using known H. sapiens hg38 assembly from NCBI RefSeq Genes (65, 66), and SCREEN by ENCODE for CREs (67). Within the CRE regions selected by SCREEN, two types of areas were selected manually by sequence homology. Areas presumably conserved between gyrencephalic and lissencephalic species and areas presumably conserved only in one of the groups were subjected to motif discovery using MEME suite (68) with default parameters. Collected motifs were scanned against motif in sequence databases using TOMTOM, a part of MEME Suite, using the default options except alignment, which was selected by Euclidean distance.
Videomicroscopy
For live imaging of radially migrating cells, embryos were electroporated in utero at E14.5 with Scr- or MIR3607-encoding plasmids (1 μg/μl) together with pCAG-mGFP (loxp-EGFP-farnesylated; 0,8 μg/μl) and pCAG-Cre (0.1 μg/μl). Three days after electroporation, the brains were dissected out and vibratome-sliced at 300 μm in ice-cold DMEM/F12 (Sigma-Aldrich) supplemented with P/S (100 U/ml), glucose (0.7 g/liter), and sodium bicarbonate (0.3 g/liter), and bubbled with carbogen (5% carbon dioxide + 95% oxygen). Slices were embedded in collagen matrix (Nitta gelatin) on a filter membrane (Millipore) and cultured in DMEM/F12 (Sigma), 5% FBS, 5% horse serum, N2 (1:100; Invitrogen), B27 (1:50; Invitrogen), P/S (100 U/ml), glucose (0.7 g/liter), and sodium bicarbonate (0.3 g/liter) (8). Imaging was performed on an Andor Dragonfly spinning disk inverted confocal microscope under a 25× lens in a 5% CO2/37°C atmosphere. Stacks of images separated 3 μm were captured every 30 min during at least 24 hours.
Image analysis, quantification, and statistics
Images were acquired using a florescence microscope (Zeiss Axio Imager Z2) with Apotome.2 and coupled to two different digital cameras (AxioCam MRm and AxioCam ICc) or an inverted confocal microscope (Olympus FluoView FV1000). All images were analyzed with ImageJ (Fiji). Colocalization studies were performed on single-plane confocal images from 40× Z stacks. Cell distribution analyses were performed using Neurolucida and NeuroExplorer software (MBF Bioscience). All quantifications in embryos were performed at the same rostral-caudal and lateromedial levels on at least three different subjects from at least two independent litters. ISH images were acquired using Zeiss Axio Imager Z2. Brightness and contrast of the images shown in figures were homogeneously adjusted for clarity using Adobe Photoshop. Analysis and quantification of videomicroscopy images were performed using Imaris software. Quantification of Par3+ ventricular surface in cerebral organoids was performed in electroporated ventricles, comparing MIR3607 with Scr electroporation as control. Non-electroporated ventricles were not used as internal controls because of the high non–cell-autonomous effect of MIR3607 on rosette formation observed in our embryo electroporations. Statistical analyses were carried out in Microsoft Excel or GraphPad Software using analysis of variance (ANOVA) with post hoc Bonferroni correction (equal variances) or the Welch test with post hoc Games-Howell (different variances), Kolmogorov-Smirnov test, chi-square test, or independent samples t test, where appropriate and upon normality testing. Significance was set at P = 0.05. In the analyses of Pax6 and Tbr2 coexpression, the influence of each protein’s increased abundance over their increased coexpression in MIR3607-expressing embryos was tested by mathematical correction. This was performed by dividing the Pax6 intensity value of each cell by the average increase in Pax6 intensity between MIR3607 and Scr embryos. The same method was used for Tbr2.
Acknowledgments
We thank G. Exposito, V. Villar, and A. Caler for excellent technical assistance with imaging and FACS; A. Espinós and J. Brotons-Mas for help with statistics; and M. Drukker for hiPSCs. We also thank members of our laboratory for insightful discussions and critical reading of the manuscript.
Funding: This work was supported by Santiago Grisolía predoctoral fellowship (K.C.), Generalitat Valenciana I+D+i programs grant APOSTD/2019/059 (A.C.), Fundación Tatiana Pérez de Guzmán el Bueno predoctoral fellowship (A.P.-C.), Agencia Estatal de Investigación SVP-2014-068671 (A.V.), Spanish State Research Agency FPI contract (R.S.), Spanish State Research Agency grant RYC-2015-18056 (J.P.L.-A.), Spanish State Research Agency grant RTI2018-102260-B-100 (J.P.L.-A.), Spanish State Research Agency grant SAF2015-69168-R (V.B.), Spanish State Research Agency grant PGC2018-102172-B-I00 (V.B.), Spanish State Research Agency “Severo Ochoa” Programme for Centers of Excellence in R&D grant SEV-2017-0723 (V.B.), and European Research Council grant 309633 (V.B.).
Author contributions: Conceptualization: K.C. and V.B. Experimentation: K.C., A.V., A.P.-C., Y.N., A.C., E.L., and U.T. Analysis: K.C., A.V., Á.M.-G., A.P.-C., Y.N., A.C., R.S., and V.B. Visualization: V.B. Supervision: J.P.L.-A. and V.B. Writing—original draft: V.B. Writing—review and editing: A.V., A.P.-C., U.T., J.P.L.-A., and V.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data reported in this study are accessible through the GEO database with the GEO series accession number GSE135321. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S3
Legends for movies S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
Movies S1 and S2
REFERENCES AND NOTES
- 1.O’Leary M. A., Bloch J. I., Flynn J. J., Gaudin T. J., Giallombardo A., Giannini N. P., Goldberg S. L., Kraatz B. P., Luo Z. X., Meng J., Ni X., Novacek M. J., Perini F. A., Randall Z. S., Rougier G. W., Sargis E. J., Silcox M. T., Simmons N. B., Spaulding M., Velazco P. M., Weksler M., Wible J. R., Cirranello A. L., The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Borrell V., Reillo I., Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 72, 955–971 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Dehay C., Kennedy H., Cell-cycle control and cortical development. Nat. Rev. Neurosci. 8, 438–450 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Fernandez V., Llinares-Benadero C., Borrell V., Fernández V., Llinares-Benadero C., Borrell V., Cerebral cortex expansion and folding: What have we learned? EMBO J. 35, 1021–1044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiddes I. T., Lodewijk G. A., Mooring M., Bosworth C. M., Ewing A. D., Mantalas G. L., Novak A. M., van den Bout A., Bishara A., Rosenkrantz J. L., Lorig-Roach R., Field A. R., Haeussler M., Russo L., Bhaduri A., Nowakowski T. J., Pollen A. A., Dougherty M. L., Nuttle X., Addor M.-C., Zwolinski S., Katzman S., Kriegstein A., Eichler E. E., Salama S. R., Jacobs F. M. J., Haussler D., Human-specific NOTCH2NL genes affect Notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florio M., Borrell V., Huttner W. B., Human-specific genomic signatures of neocortical expansion. Curr. Opin. Neurobiol. 42, 33–44 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Suzuki I. K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F., Detours V., Vanderhaeghen P., Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cárdenas A., Villalba A., de Juan Romero C., Picó E., Kyrousi C., Tzika A. C., Tessier-Lavigne M., Ma L., Drukker M., Cappello S., Borrell V., Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell 174, 590–606.e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelava I., Lewitus E., Huttner W. B., The secondary loss of gyrencephaly as an example of evolutionary phenotypical reversal. Front. Neuroanat. 7, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Villalba, M. Götz, V. Borrell, in Current Topics in Developmental Biology (Academic Press Inc., 2021), vol. 142, pp. 1–66. [DOI] [PubMed] [Google Scholar]
- 11.Taverna E., Götz M., Huttner W. B., The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Reillo I., Borrell V. V., Germinal zones in the developing cerebral cortex of ferret: Ontogeny, cell cycle kinetics, and diversity of progenitors. Cereb. Cortex 22, 2039–2054 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Reillo I., De Juan Romero C., García-Cabezas M. Á., Borrell V., A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hansen D. V., Lui J. H., Parker P. R. L. L., Kriegstein A. R., Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Fietz S. A., Kelava I., Vogt J., Wilsch-Brauninger M., Stenzel D., Fish J. L., Corbeil D., Riehn A., Distler W., Nitsch R., Huttner W. B., Wilsch-Bräuninger M., Stenzel D., Fish J. L., Corbeil D., Riehn A., Distler W., Nitsch R., Huttner W. B., OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Smart I. H. M., Dehay C., Giroud P., Berland M., Kennedy H., Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrell V., Götz M., Role of radial glial cells in cerebral cortex folding. Curr. Opin. Neurobiol. 27, 39–46 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Rakic P., A small step for the cell, a giant leap for mankind: A hypothesis of neocortical expansion during evolution. Trends Neurosci. 18, 383–388 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Florio M., Heide M., Pinson A., Brandl H., Albert M., Winkler S., Wimberger P., Huttner W. B., Hiller M., Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 7, e32332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arcila M. L., Betizeau M., Cambronne X. A., Guzman E., Doerflinger N., Bouhallier F., Zhou H., Wu B., Rani N., Bassett D. S., Borello U., Huissoud C., Goodman R. H., Dehay C., Kosik K. S., Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns. Neuron 81, 1255–1262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowakowski T. J., Rani N., Golkaram M., Zhou H. R., Alvarado B., Huch K., West J. A., Leyrat A., Pollen A. A., Kriegstein A. R., Petzold L. R., Kosik K. S., Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 21, 1784–1792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fietz S. A., Lachmann R., Brandl H., Kircher M., Samusik N., Schröder R., Lakshmanaperumal N., Henry I., Vogt J., Riehn A., Distler W., Nitsch R., Enard W., Pääbo S., Huttner W. B., Schroder R., Lakshmanaperumal N., Henry I., Vogt J., Riehn A., Distler W., Nitsch R., Enard W., Paabo S., Huttner W. B., Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. U.S.A. 109, 11836–11841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Martínez M. Á., De Juan Romero C., Fernández V., Cárdenas A., Götz M., Borrell V., A restricted period for formation of outer subventricular zone defined by Cdh1 and Trnp1 levels. Nat. Commun. 7, 11812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Juan Romero C., Borrell V., Genetic maps and patterns of cerebral cortex folding. Curr. Opin. Cell Biol. 49, 31–37 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Llinares-Benadero C., Borrell V., Deconstructing cortical folding: Genetic, cellular and mechanical determinants. Nat. Rev. Neurosci. 20, 161–176 (2019). [DOI] [PubMed] [Google Scholar]
- 26.De Juan Romero C., Bruder C., Tomasello U., Sanz-Anquela J. M., Borrell V. V., Sanz-Anquela J. M., Borrell V. V., Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 34, 1859–1874 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pati S., Supeno N. E., Muthuraju S., Abdul Hadi R., Ghani A. R. I., Idris F. M., Maletic-Savatic M., Abdullah J. M., Jaafar H., MicroRNA profiling reveals unique miRNA signatures in IGF-1 treated embryonic striatal stem cell fate decisions in striatal neurogenesis in vitro. Biomed. Res. Int. 2014, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebert L. F. R., MacRae I. J., Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Englund C., Fink A., Lau C., Pham D., Daza R. A. M., Bulfone A., Kowalczyk T., Hevner R. F., Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247–251 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betizeau M., Cortay V., Patti D., Pfister S., Gautier E., Bellemin-Ménard A., Afanassieff M., Huissoud C., Douglas R. J., Kennedy H., Dehay C., Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80, 442–457 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Martínez M. Á., Ciceri G., Espinós A., Fernández V., Marín O., Borrell V., Extensive branching of radially-migrating neurons in the mammalian cerebral cortex. J. Comp. Neurol. 527, 1558–1576 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Tabata H., Nakajima K., Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 23, 9996–10001 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chenn A., Walsh C. A., Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Herrera A., Saade M., Menendez A., Marti E., Pons S., Sustained Wnt/β-catenin signalling causes neuroepithelial aberrations through the accumulation of aPKC at the apical pole. Nat. Commun. 5, 4168 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Woodhead G. J., Mutch C. A., Olson E. C., Chenn A., Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J. Neurosci. 26, 12620–12630 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., Gu Q., Sun Z., Sheng B., Qi C., Liu B., Fu T., Liu C., Zhang Y., Upregulation of miR-3607 promotes lung adenocarcinoma proliferation by suppressing APC expression. Biomed. Pharmacother. 95, 497–503 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Yoo J., Hajjar R. J., Jeong D., Generation of efficient miRNA inhibitors using tough decoy constructs. Methods Mol. Biol. 1521, 41–53 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Machon O., van den Bout C. J., Backman M., Kemler R., Krauss S., Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience 122, 129–143 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Tsai J.-W. W., LaMonica B., Kriegstein A. R., A new subtype of progenitor cell in the mouse embryonic neocortex. Nat. Neurosci. 14, 555–561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F. K., Peters J., Guhr E., Klemroth S., Prufer K., Kelso J., Naumann R., Nusslein I., Dahl A., Lachmann R., Paabo S., Huttner W. B., Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Boyd J. L., Skove S. L., Rouanet J. P., Pilaz L. J., Bepler T., Gordân R., Wray G. A., Silver D. L., Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol. 25, 772–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farkas L. M., Haffner C., Giger T., Khaitovich P., Nowick K., Birchmeier C., Pääbo S., Huttner W. B., Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Mukhtar T., Breda J., Grison A., Karimaddini Z., Grobecker P., Iber D., Beisel C., van Nimwegen E., Taylor V., Tead transcription factors differentially regulate cortical development. Sci. Rep. 10, 4625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borello U., Berarducci B., Delahaye E., Price D. J., Dehay C., SP8 transcriptional regulation of Cyclin D1 during mouse early corticogenesis. Front. Neurosci. 12, 119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrobel C. N., Mutch C. A., Swaminathan S., Taketo M. M., Chenn A., Persistent expression of stabilized β-catenin delays maturation of radial glial cells into intermediate progenitors. Dev. Biol. 309, 285–297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid R. S., Maness P. F., L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 18, 245–250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mutch C. A., Funatsu N., Monuki E. S., Chenn A., β-Catenin signaling levels in progenitors influence the laminar cell fates of projection neurons. J. Neurosci. 29, 13710–13719 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokota Y., Kim W. Y., Chen Y., Wang X., Stanco A., Komuro Y., Snider W., Anton E. S., The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron 61, 42–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa N., Li J., Yabuno-Nakagawa K., Eom T. Y., Cowles M., Mapp T., Taylor R., Anton E. S., APC sets the Wnt tone necessary for cerebral cortical progenitor development. Genes Dev. 31, 1679–1692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prieto-Colomina A., Fernández V., Chinnappa K., Borrell V., MiRNAs in early brain development and pediatric cancer. Bioessays 43, e2100073 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Fernández V., Martínez-Martínez M. Á., Prieto-Colomina A., Cárdenas A., Soler R., Dori M., Tomasello U., Nomura Y., López-Atalaya J. P., Calegari F., Borrell V., Repression of Irs2 by let-7 miRNAs is essential for homeostasis of the telencephalic neuroepithelium. EMBO J. 39, e105479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haraguchi T., Ozaki Y., Iba H., Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37, e43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borrell V., Cárdenas A., Ciceri G., Galcerán J., Flames N., Pla R., Nóbrega-Pereira S., García-Frigola C., Peregrín S., Zhao Z., Ma L., Tessier-Lavigne M., Marín O., Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76, 338–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lancaster M. A., Knoblich J. A., Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Pietri Tonelli D., Pulvers J. N., Haffner C., Murchison E. P., Hannon G. J., Huttner W. B., miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherry S. T., Ward M., Sirotkin K., dbSNP—Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 9, 677–679 (1999). [PubMed] [Google Scholar]
- 57.Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., Cherry J. M., Sherlock G., GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato K., Akiyama M., Sakakibara Y., RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 12, 941 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W., Haussler D., Kent W. J., The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madeira F., Park Y., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A., Potter S., Finn R., Lopez R., The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C. M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V. S., Kodali V. K., Li W., Maglott D., Masterson P., McGarvey K. M., Murphy M. R., O’Neill K., Pujar S., Rangwala S. H., Rausch D., Riddick L. D., Schoch C., Shkeda A., Storz S. S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R. E., Vatsan A. R., Wallin C., Webb D., Wu W., Landrum M. J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T. D., Pruitt K. D., Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarro Gonzalez J., Zweig A. S., Speir M. L., Schmelter D., Rosenbloom K. R., Raney B. J., Powell C. C., Nassar L. R., Maulding N. D., Lee C. M., Lee B. T., Hinrichs A. S., Fyfe A. C., Fernandes J. D., Diekhans M., Clawson H., Casper J., Benet-Pagès A., Barber G. P., Haussler D., Kuhn R. M., Haeussler M., Kent W. J., The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 49, D1046–D1057 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abascal F., Acosta R., Addleman N. J., Adrian J., Afzal V., Aken B., Akiyama J. A., Al Jammal O., Amrhein H., Anderson S. M., Andrews G. R., Antoshechkin I., Ardlie K. G., Armstrong J., Astley M., Banerjee B., Barkal A. A., Barnes I. H. A., Barozzi I., Barrell D., Barson G., Bates D., Baymuradov U. K., Bazile C., Beer M. A., Beik S., Bender M. A., Bennett R., Bouvrette L. P. B., Bernstein B. E., Berry A., Bhaskar A., Bignell A., Blue S. M., Bodine D. M., Boix C., Boley N., Borrman T., Borsari B., Boyle A. P., Brandsmeier L. A., Breschi A., Bresnick E. H., Brooks J. A., Buckley M., Burge C. B., Byron R., Cahill E., Cai L., Cao L., Carty M., Castanon R. G., Castillo A., Chaib H., Chan E. T., Chee D. R., Chee S., Chen H., Chen H., Chen J. Y., Chen S., Cherry J. M., Chhetri S. B., Choudhary J. S., Chrast J., Chung D., Clarke D., Cody N. A. L., Coppola C. J., Coursen J., D’Ippolito A. M., Dalton S., Danyko C., Davidson C., Davila-Velderrain J., Davis C. A., Dekker J., Deran A., DeSalvo G., Despacio-Reyes G., Dewey C. N., Dickel D. E., Diegel M., Diekhans M., Dileep V., Ding B., Djebali S., Dobin A., Dominguez D., Donaldson S., Drenkow J., Dreszer T. R., Drier Y., Duff M. O., Dunn D., Eastman C., Ecker J. R., Edwards M. D., El-Ali N., Elhajjajy S. I., Elkins K., Emili A., Epstein C. B., Evans R. C., Ezkurdia I., Fan K., Farnham P. J., Farrell N. P., Feingold E. A., Ferreira A. M., Fisher-Aylor K., Fitzgerald S., Flicek P., Foo C. S., Fortier K., Frankish A., Freese P., Fu S., Fu X. D., Fu Y., Fukuda-Yuzawa Y., Fulciniti M., Funnell A. P. W., Gabdank I., Galeev T., Gao M., Giron C. G., Garvin T. H., Gelboin-Burkhart C. A., Georgolopoulos G., Gerstein M. B., Giardine B. M., Gifford D. K., Gilbert D. M., Gilchrist D. A., Gillespie S., Gingeras T. R., Gong P., Gonzalez A., Gonzalez J. M., Good P., Goren A., Gorkin D. U., Graveley B. R., Gray M., Greenblatt J. F., Griffiths E., Groudine M. T., Grubert F., Gu M., Guigó R., Guo H., Guo Y., Guo Y., Gursoy G., Gutierrez-Arcelus M., Halow J., Hardison R. C., Hardy M., Hariharan M., Harmanci A., Harrington A., Harrow J. L., Hashimoto T. B., Hasz R. D., Hatan M., Haugen E., Hayes J. E., He P., He Y., Heidari N., Hendrickson D., Heuston E. F., Hilton J. A., Hitz B. C., Hochman A., Holgren C., Hou L., Hou S., Hsiao Y. H. E., Hsu S., Huang H., Hubbard T. J., Huey J., Hughes T. R., Hunt T., Ibarrientos S., Issner R., Iwata M., Izuogu O., Jaakkola T., Jameel N., Jansen C., Jiang L., Jiang P., Johnson A., Johnson R., Jungreis I., Kadaba M., Kasowski M., Kasparian M., Kato M., Kaul R., Kawli T., Kay M., Keen J. C., Keles S., Keller C. A., Kelley D., Kellis M., Kheradpour P., Kim D. S., Kirilusha A., Klein R. J., Knoechel B., Kuan S., Kulik M. J., Kumar S., Kundaje A., Kutyavin T., Lagarde J., Lajoie B. R., Lambert N. J., Lazar J., Lee A. Y., Lee D., Lee E., Lee J. W., Lee K., Leslie C. S., Levy S., Li B., Li H., Li N., Li X., Li Y. I., Li Y., Li Y., Li Y., Lian J., Libbrecht M. W., Lin S., Lin Y., Liu D., Liu J., Liu P., Liu T., Liu X. S., Liu Y., Liu X. S., Long M., Lou S., Loveland J., Lu A., Lu Y., Lécuyer E., Ma L., Mackiewicz M., Mannion B. J., Mannstadt M., Manthravadi D., Marinov G. K., Martin F. J., Mattei E., McCue K., McEown M., McVicker G., Meadows S. K., Meissner A., Mendenhall E. M., Messer C. L., Meuleman W., Meyer C., Miller S., Milton M. G., Mishra T., Moore D. E., Moore H. M., Moore J. E., Moore S. H., Moran J., Mortazavi A., Mudge J. M., Munshi N., Murad R., Myers R. M., Nandakumar V., Nandi P., Narasimha A. M., Narayanan A. K., Naughton H., Navarro F. C. P., Navas P., Nazarovs J., Nelson J., Neph S., Neri F. J., Nery J. R., Nesmith A. R., Newberry J. S., Newberry K. M., Ngo V., Nguyen R., Nguyen T. B., Nguyen T., Nishida A., Noble W. S., Novak C. S., Novoa E. M., Nuñez B., O’Donnell C. W., Olson S., Onate K. C., Otterman E., Ozadam H., Pagan M., Palden T., Pan X., Park Y., Partridge E. C., Paten B., Pauli-Behn F., Pazin M. J., Pei B., Pennacchio L. A., Perez A. R., Perry E. H., Pervouchine D. D., Phalke N. N., Pham Q., Phanstiel D. H., Plajzer-Frick I., Pratt G. A., Pratt H. E., Preissl S., Pritchard J. K., Pritykin Y., Purcaro M. J., Qin Q., Quinones-Valdez G., Rabano I., Radovani E., Raj A., Rajagopal N., Ram O., Ramirez L., Ramirez R. N., Rausch D., Raychaudhuri S., Raymond J., Razavi R., Reddy T. E., Reimonn T. M., Ren B., Reymond A., Reynolds A., Rhie S. K., Rinn J., Rivera M., Rivera-Mulia J. C., Roberts B. S., Rodriguez J. M., Rozowsky J., Ryan R., Rynes E., Salins D. N., Sandstrom R., Sasaki T., Sathe S., Savic D., Scavelli A., Scheiman J., Schlaffner C., Schloss J. A., Schmitges F. W., See L. H., Sethi A., Setty M., Shafer A., Shan S., Sharon E., Shen Q., Shen Y., Sherwood R. I., Shi M., Shin S., Shoresh N., Siebenthall K., Sisu C., Slifer T., Sloan C. A., Smith A., Snetkova V., Snyder M. P., Spacek D. V., Srinivasan S., Srivas R., Stamatoyannopoulos G., Stamatoyannopoulos J. A., Stanton R., Steffan D., Stehling-Sun S., Strattan J. S., Su A., Sundararaman B., Suner M. M., Syed T., Szynkarek M., Tanaka F. Y., Tenen D., Teng M., Thomas J. A., Toffey D., Tress M. L., Trout D. E., Trynka G., Tsuji J., Upchurch S. A., Ursu O., Uszczynska-Ratajczak B., Uziel M. C., Valencia A., Van Biber B., van der Velde A. G., Van Nostrand E. L., Vaydylevich Y., Vazquez J., Victorsen A., Vielmetter J., Vierstra J., Visel A., Vlasova A., Vockley C. M., Volpi S., Vong S., Wang H., Wang M., Wang Q., Wang R., Wang T., Wang W., Wang X., Wang Y., Watson N. K., Wei X., Wei Z., Weisser H., Weissman S. M., Welch R., Welikson R. E., Weng Z., Westra H. J., Whitaker J. W., White C., White K. P., Wildberg A., Williams B. A., Wine D., Witt H. N., Wold B., Wolf M., Wright J., Xiao R., Xiao X., Xu J., Yan K. K., Yan Y., Yang H., Yang X., Yang Y. W., Yardimci G. G., Yee B. A., Yeo G. W., Young T., Yu T., Yue F., Zaleski C., Zang C., Zeng H., Zeng W., Zerbino D. R., Zhai J., Zhan L., Zhan Y., Zhang B., Zhang J., Zhang J., Zhang K., Zhang L., Zhang P., Zhang Q., Zhang X. O., Zhang Y., Zhang Z., Zhao Y., Zheng Y., Zhong G., Zhou X. Q., Zhu Y., Zimmerman J., Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey T. L., Johnson J., Grant C. E., Noble W. S., The MEME Suite. Nucleic Acids Res. 43, W39–W49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.L. Shi, X. Luo, J. Jiang, Y. Chen, C. Liu, T. Hu, M. Li, Q. Lin, Y. Li, J. Huang, H. Wang, Y. Niu, Y. Shi, M. Styner, J. Wang, Y. Lu, X. Sun, H. Yu, W. Ji, B. Su, Transgenic rhesus monkeys carrying the human MCPH1 gene copies show human-like neoteny of brain development. Natl. Sci. Rev. 6, 480–493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. B. Johnson, X. Sun, A. Kodani, R. Borges-Monroy, K. M. Girskis, S. C. Ryu, P. P. Wang, K. Patel, D. M. Gonzalez, Y. M. Woo, Z. Yan, B. Liang, R. S. Smith, M. Chatterjee, D. Coman, X. Papademetris, L. H. Staib, F. Hyder, J. B. Mandeville, P. E. Grant, K. Im, H. Kwak, J. F. Engelhardt, C. A. Walsh, B. I. Bae, Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature. 556, 370–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bailey T., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S3
Legends for movies S1 to S3
References
Tables S1 to S3
Movies S1 and S2