Abstract
Structure-based virtual screening is a key, routine computational method in computer-aided drug design. Such screening can be used to identify potentially highly active compounds, to speed up the progress of novel drug design. Molecular docking-based virtual screening can help find active compounds from large ligand databases by identifying the binding affinities between receptors and ligands. In this study, we analyzed the challenges of virtual screening, with the aim of identifying highly active compounds faster and more easily than is generally possible. We discuss the accuracy and speed of molecular docking software and the strategy of high-throughput molecular docking calculation, and we focus on current challenges and our solutions to these challenges of ultra-large-scale virtual screening. The development of Web services helps lower the barrier to drug virtual screening. We introduced some related web sites for docking and virtual screening, focusing on the development of pre- and post-processing interactive visualization and large-scale computing.
Keywords: Molecular docking, Virtual screening, Supercomputing, Ultra-large-scale computing
Introduction
Disease has posed an immense burden to human civilizations throughout history. At present, COVID-19 has spread around the world, with the cumulative number of confirmed cases surpassing 166 million worldwide, and the cumulative number of deaths exceeding 3.45 million as of May 25, 2021 (WHO 2021) The development of drugs to treat disease is a long and arduous process. However, scientists recognized early on that the rational application of computer-aided drug design methods can help to improve the efficiency the development of new drugs (Vartikatomar et al. 2019). Currently, new drug development generally follows the process shown in Fig. 1. Recent studies have shown that it takes more than 10 years to develop a new drug, with an investment of about $US2–3 billion (Dhasmana et al. 2019). There has been considerable research into the development of new technologies aimed at increasing the success rate of new drug development and reducing the expenses incurred. The long timeline and high financial cost of new drug development is mainly due to the iterative optimization of lead compounds and the failure of late-stage trials of drug candidates. A lead compound is a compound with a desirable biological activity and chemical structure. The quality of a lead compound directly affects the quality of the drug candidate and is the key to the success of novel drug design. In the process of lead compound discovery and optimization, a very important research tool is virtual screening technology, which can screen out a small number of active molecules from a large number of candidate small molecules, narrowing the scope for subsequent experimental assays, to improve the hit rate.
Fig. 1.
Process of drug discovery and development
Virtual screening in drug discovery can traditionally be divided into two main categories: ligand-based and receptor-based. The former requires a set of known active ligands for 3D quantitative structure activity relationship analysis (3D-QSAR), and is commonly used for targeting receptors and optimizing lead compounds (Srinivasarao et al. 2017). The latter requires the 3D structure of the receptor and ligand molecules to be known. These structures are used to perform molecular docking calculations, to predict the binding site of the molecules by calculating their binding energies in different conformations. Receptor-based virtual screening is often used in the lead compound discovery step. With the development of technologies for protein structural science, including cryo-electron microscopy and X-ray crystallography, more and more proteins are being resolved in three dimensions. It is also possible to predict the three-dimensional structure of proteins through homology modeling. Receptor-based virtual screening is becoming increasingly important. Molecular docking-based virtual screening is more demanding than ligand-based virtual screening in terms of computational resources. In addition to protein–ligand docking, molecular docking also covers protein–protein docking, protein–peptide docking and other large-molecule-related docking problems using biomolecules such as DNA. The use of molecular docking software for virtual screening has dominated the literature on drug development research since 2000 (Wikipedia 2021), and therefore, in this study we focused on protein–ligand docking-based virtual screening. High-throughput virtual screening (HTVS) can be used to find promising compounds from a large molecule library by evaluating their binding modes and affinity to a receptor of interest (Dhasmana et al. 2019). A convenient and easy-to-use web platform is also important for general users, especially those who are not familiar with Linux operations. In this paper, we describe the development of HTVS, especially a new approach to ultra-HTVS that makes full use of molecular docking methods for HTVS in high-performance computing environments.
Molecular docking-based virtual screening in drug design
Molecular docking-based virtual screening techniques are essential for screening out promising drug precursors from the vast amount of structural data available. Improving the hit rate of a virtual screening depends on the algorithm used by the molecular docking software and on the scale of the virtual screening. The ability to quickly screen highly active compounds from a large library of small molecules is the major goal of virtual screening in drug design. In the following sections, the development of molecular docking software and HTVS are described.
Development of molecular docking software
Molecular docking is the process of identifying binding sites between two or more molecules using geometric and energy matching. The use of this approach can help in the prediction of the binding conformation and binding mode or orientation between a receptor and a ligand and is especially valuable for studying changes of substrate conformation during the formation of complexes. Molecular docking is the basis for determining the mechanism of binding of drugs to targets and is very important for receptor-based virtual screening.
The possibility of a receptor and ligand binding to each other and the strength of the binding depend on the change in free energy that occurs during the binding process.
| 1 |
where Ki is the binding constant. The enthalpy effect (Hbinding) during molecular docking includes the interaction energy between the ligand and the receptor, and the interaction energy includes electrostatic, van der Waals, and hydrogen bonding interactions.
| 2 |
The enthalpy effect includes the interaction energy of the ligand with the solvent and the solvent–protein interaction energy. Entropic effects (Sbinding) include entropic changes due to molecular rotation and translation, conformational changes, hydrophobic interactions, and vibrations. Most molecular docking methods ignore the entropic effects and only consider the ligand–receptor interaction energy in the enthalpy effect.
Molecular docking methods can be broadly classified into three categories at different levels of simplification: rigid docking, flexible docking and semi-flexible docking (Prieto-Martínez et al. 2018). Rigid docking means that neither the conformation of the receptor nor that of the ligand changes during the docking process. Rigid docking is often used to examine large systems, such as protein–protein interactions. For example, Juan et al. presented an efficient pseudo-Brownian rigid-body docking procedure and tested it on 24 protein–protein docking examples (Juan et al. 2002). Flexible docking, in which the ligand and target structures are free to change during the docking process, is generally used to examine the recognition between molecules, as it needs more computational power (Bonvin 2006). MedusaDock is one such flexible docking method, which models the flexibility of both the ligand and the receptor simultaneously, with sets of discrete rotamers (Wang 2019). In semi-flexible docking, the conformation of the target or the ligand is allowed to change within a certain range during the docking process, usually with the ligand being allowed to be flexible while the receptor structure remains rigid. Docking methods such as those developed by Morris et al. are commonly used for virtual screening calculations in drug discovery, as ligand molecules are relatively small, and the impact of examining conformational changes can be well balanced with the overall computational power required (Morris et al. 2009).
The number of software applications for molecular docking exceeds 100 as of 2019, not including various accelerated docking software versions (Pagadala 2017; Wikipedia 2021). Table 1 lists 10 commonly used molecular docking applications, of which the first five are free and the last five are commercial software.
Table 1.
Some commonly used docking software
| Software | Algorithm features | Home page |
|---|---|---|
| AutoDock (Morris et al. 1998; Morris et al. 2009) | Lamarckian Genetic Algorithm and empirical binding free energy function | http://autodock.scripps.edu/ |
| AutoDock Vina (Olson 2010) | Iterated local search global optimizer, with sophisticated gradient optimization method in its local optimization procedure. The derivation of its scoring function combines certain advantages of knowledge-based potentials and empirical scoring functions | http://vina.scripps.edu/ |
| rDock (Sergio et al. 2014) | Evolved from RiboDock, rDock uses a combination of stochastic and deterministic search techniques to generate low-energy ligand pose. rDock includes fast intermolecular scoring functions (vdW, polar, desolvation) and implements several pseudo-energy scoring functions that are added to the total scoring function under optimization and a restricted search protocol | http://rdock.sourceforge.net |
| Dock 6 (William et al. 2015) | Anchor-and-grow search algorithm is a breadth-first method for small-molecule conformational sampling. It utilizes a footprint similarity scoring function | http://dock.compbio.ucsf.edu/ |
| LeDock (Zhao et al. 2013; Zhao et al. 2011) | Simulated annealing and genetic algorithm optimization. The scoring function, based on AutoDock 4 scoring function, calculates hydrogen bonding penalty associated with ligand binding to improve binding | http://lephar.com/ |
| Glide (Friesner et al. 2004) | Complete systematic search of the conformational, orientational, and positional space of the docked ligand. Its scoring function, named as Emodel, combines empirically based ChemScore function, force-field-based terms from the Coulomb and vdW interaction energies between the ligand and the receptor, and the solvation model | http://www.schrodinger.com/ |
| Gold (Jones et al. 1997) | Genetic algorithm to explore the full range of ligand conformational flexibility with partial flexibility of the protein. Its scoring function comprised terms for hydrogen bonding, pairwise dispersion potentials, and molecular mechanics terms | http://www.ccdc.cam.ac.uk/ |
| FlexX (Rarey et al. 1996) | Fragment growth method to find the best conformation and empirical scoring function to compute the binding affinity | https://www.biosolveit.de/products/ |
| Surflex (Jain et al. 2003) | Employs a “protomol” that can be automatically generated or user defined to generate putative poses of molecules or molecular fragments. The scoring function, based on Hammerhead scoring function, uses an updated and re-parameterized empirical scoring function | http://www.tripos.com/ |
| LigandFit (Krammer et al. 2005; Venkatachalam et al. 2003) | Shape-directed docking methodology. Ligand conformations are generated by a Monte Carlo conformational search for generating ligand poses consistent with the active site shape. Its scoring function is called LigScore, which consist of three distinct terms that describe the van der Waals interaction, the polar attraction, and the desolvation penalty | https://www.3dsbiovia.com/ |
The core of molecular docking software has two aspects: a conformational search algorithm and a scoring function (Inbal Halperin 2002; Yadava. 2018). The conformational search algorithm helps find the optimal binding site for the receptor and ligand, while the scoring function is used to evaluate the strength of the binding between the docked molecules. The performance of a conformational search step followed by a scoring step is called a run, and molecular docking software typically performs multiple runs to overcome randomness, before outputting the final binding conformation, binding energy, and the ranking of multiple runs.
Common conformational search algorithms include stochastic search methods, simulation methods, and systematic search methods. Stochastic search methods include simulated annealing, Monte Carlo, genetic algorithms (GAs), and Tabu Search. Simulation methods perform structural search by means of molecular dynamics or energy minimization. Systematic search methods include fragment growth methods (Inbal Halperin 2002; Yadava 2018).
There are four common scoring functions: force field-based, empirical, knowledge-based, and machine learning. Of these, force field-based scoring functions use force fields, a collection of equations and associated constants to evaluate van der Waals interactions and electrostatic interactions between and within docked molecules. Methods such as molecular mechanics energies combined with the Poisson–Boltzmann or generalized Born and surface area continuum solvation methods are also commonly used to evaluate the desolvation energy between ligands and receptors (Pu et al. 2017). Empirically based scoring functions are calculated based on the type of interactions counted for the docked molecules, such as hydrophobic interactions, hydrophilic interactions, number of hydrogen bonds, and number of rotatable bonds. The coefficients of the scoring function are fitted by means of multiple linear regression. Knowledge-based scoring functions originate from the statistical mechanics analysis of liquids, also known as the potential of mean force scoring function. The fitness is calculated by solving for a statistical potential function of protein–ligand pairs. The method uses a set of protein–ligand complex structures from the PDB database (PDB bank 2021; Berman et al. 2003) as a training set, or “knowledge base”, in which the atoms of proteins and ligands are classified into a number of simplicial types according to their molecular environment, and the distance-dependent potential of each possible pair is derived based on the frequency of occurrence of the atomic pair. Machine learning-based scoring functions use a variety of descriptors, such as electrostatic interactions, hydrogen bonding or aromatic stacking, surface or shape properties, ligand molecular weight, and rotatable bonds, to build a machine learning model, which is then used by the machine learning algorithm, a branch of artificial intelligence focused on the use of data and algorithms to imitate the way that human learns, to derive a non-linear energy function for the docking score.
Although there are differences in conformational search algorithms and scoring functions in different molecular docking software, research has indicated that the differences in conformational ranking and scoring results between free software and commercial software are not significant, and there is no one docking software that is superior to the others in all respects (Wang et al. 2016). In a virtual screening context, consensus docking can improve the reliability of docking by using more than one docking program to predict the binding pose (Houston et al. 2013).
The speed of individual molecular docking calculation depends on the conformational search algorithm used by the molecular docking software, the implementation of the program, and some parameter settings. For example, AutoDock has used the Lamarckian Genetic Algorithm since version 3.0. This algorithm combines a genetic algorithm for global search and a local search for energy optimization and is more efficient than traditional GAs and simulated annealing algorithms. For program implementation, MPI, threaded, or GPU acceleration can be used. For example, Dock 6 supports MPI parallelism, Autodock Vina supports multi-threaded parallelism, and AutoDock GPU (Diogo et al. 2019) implements acceleration on the GPU. In terms of parameter settings, by limiting the search space to, for example, rigid docking or semi-flexible docking, the search freedom is greatly reduced compared to that in full-flexible docking, speeding up the search. rxDock (Sergio et al. 2014) uses a stepwise scoring setting, for example, starting with five runs of docking for all ligands, an additional 10 runs for ligands with a score of less than a specified binding energy, and a total of 50 runs for ligands with a score of less than another lower specific energy. In this way, the accuracy of the molecular docking is improved, and the computational time consumption is reduced by a factor of approximately 7.5.
Development of HTVS
The application of molecular docking to HTVS involves a receptor performing molecular docking with multiple different ligands over a period of time. The results are ranked according to the docking score to identify highly active compounds. There are several approaches to handling HTVS.
-
i.
Dividing the docking task into multiple files or folders and sequentially calling the molecular docking software for calculation
When using AutoDock Vina for HTVS, a certain number of small ligand molecules can be cut and stored in different file directories in advance, and the software can be invoked in each file directory to perform molecular docking operations. The binding energy of the molecules in the directory are calculated sequentially, and the output is provided. When there are sufficient computational resources, different file directories can be executed independently to optimize the computation time. rDock and rxDock software, on the other hand, use “sdsplit” to divide a sdf file containing multiple ligand files on demand. A sdf file containing 1000 molecules may be divided into 10 sdf files, each containing 100 molecules. Each sdf file can be used to initiate a molecular docking operation independently on local computing nodes or be scheduled to another compute node via a queue scheduling system.
-
ii.
Performing HTVS using docking software
iDock, which is based on the AutoDock Vina software, has improved IO usage by using thread pools to manage calculation tasks for user-specified molecules in the task folder, enabling receptor files and corresponding grid point calculation files to be reused. They also adjust docking scoring thresholds dynamically and use step-by-step scoring settings. The iDock software is 7.5 times faster than AutoDock Vina.
Vina MPI uses an MPI wrapper to enable the simultaneous launching of thousands of parallel AutoDock Vina executables and has been run on the Oak Ridge Leadership Computing Facility Jaguar and Titan supercomputers. For molecules in user-specified task folders, it uses MPI wrappers to execute docking operations on 85,672 cores on the OLCF Jaguar and Titan supercomputers for four protein molecules and 392,656 ligand molecules (98,164 ligands per receptor dock). A total of 14,278 Audodock Vina program calls, with each Audodock Vina program using six computation threads, were set up, and the total computing time was 900 s.
-
iii.
Building parallel workflows with tools such as GNU Parallel
GNU Parallel is a command line tool with the main command “xargs”, which can capture the output of specific commands to concatenate the execution of different commands. GNU Parallel executes jobs in parallel mode, depending on the number of CPU threads assigned by the user. This tool enables complete and powerful utilization of CPU resources. For example, the POAP (Samdani et al. 2018) workflow software uses GNU Parallel to connect the OpenBabel and AutoDock/AutoDock Vina packages to process ligand preparation, receptor preparation, docking tasks, and result processing via a shell command line.
The above-mentioned approaches can only perform HTVS calculations for approximately 106–107 ligand molecules. Strategies that can handle ultra-HTVS calculations are seriously needed.
As well as molecular docking-based HTVS, artificial intelligence (AI) techniques are also used for exploring the binding possibilities of targets and ligands. For example, the MolAICal (Bai et al. 2020) software uses a generative adversarial network approach to train ligand fragment libraries for specific receptors, constructs ligand molecules using a fragment growth method, and determines the optimal conformation and binding energy in conjunction with molecular docking software. The DEEPScreen (Rifaioglu et al. 2020) software was trained individually for 704 targets using a deep convolutional neural network approach from the SMILES of 2D molecules. Each receptor and at least 100 active ligands from the ChEMBL (Gaulton et al. 2017) compound library was used for training, and specific predictive models were obtained for each target. Because of the poor interpretability of AI methods, and the limited accuracy of prediction results for molecules with large structural differences from the training molecules, molecular docking-based HTVS rather than AI methods remain the dominant choice in drug design.
It is important to increase the number of small molecules that can be used for docking. Research shows that the absolute value of binding energies for the top 50 compounds increases with the size of the ligand library for HTVS (Gorgull et al. 2020). This observation indicates that the screening of large ligand libraries can identify more effective active compounds than can be achieved with a small library. The higher the binding affinity of the ligand, the lower the drug dose required. Off-target effects are therefore reduced, and compounds with favorable pharmacokinetics and low cytotoxicity can be identified.
According to chemists’ estimates (Kirkpatrick et al. 2004), the number of stable small molecule compounds meeting Lipinski’s Rule of Five (Benet et al. 2016) (i.e., a molecule with a molecular mass less than 500 Da, no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, and an octanol–water partition coefficient log P not greater than 5) is around 1060, with the number of active molecules smaller than 30 atoms ranging from 1020 to 1024. With our increasing understanding of the human genome, and the development of structural biology, a large number of proteins have been identified as potential drug targets. With the ongoing development of technology, researchers can generate large numbers of small molecule compounds through methods such as fragmentation combination libraries and deep learning (Erlanson et al. 2012; Zhavoronkov et al. 2019).
The application of computationally powerful supercomputers in drug design has brought new opportunities for the discovery of drug precursor structures, enabling a dramatic increase in both the speed and success rate of drug design. After years of development and iteration, supercomputers have now entered the era of exascale computing. Exascale computing can provide the capability to tackle challenges in scientific discovery and national security at levels of complexity and performance that previously were out of reach (ECP 2021). The United States, Japan, and Spain have developed E-class computing programs, and China also plans to deploy and implement large-scale heterogeneous E-class supercomputing systems. Theoretically, supercomputers could be used to perform ultra-HTVS in a shorter period of time than is currently possible. However, some challenges will be faced in the process of using supercomputers for ultra-HTVS. Supercomputers are good at handling large files, but the problem of storing and processing massive amounts of small molecule files is challenging. The best way in which to effectively use heterogeneous accelerators to enhance computational efficiency has not been established. It is unclear how to ensure load balancing of computational nodes in various situations without causing communication pressure on the system. There are significant challenges posed by the large number of IO operations on supercomputing file systems. Ultra-high-throughput jobs must be effectively managed, especially in the event of errors. Finally, it not clear whether a supercomputing approach can be compatible with various clusters and queue scheduling systems for fast deployment and use.
Research into solving the above challenging problems to improve the efficiency of ultra-HTVS on supercomputers is ongoing. For example, the mD3DOCKxb software used by Peng et al. on Tianhe-2 involved a novel scalable parallel algorithm that can obtain over 80% acceleration on 8000 nodes, using CPU + MIC co-computing to screen 42 million old drug compounds in one day (Peng et al. 2017). Christoph Gorgull et al. published a hyperscale drug discovery platform with a VirtualFlow for Virtual Screening module, developed using Shell scripts, with task distribution in the form of file lists, using 8000 CPU cores and screening about 1.3 billion compounds in 4 weeks (Gorgull et al. 2020). The above two research teams have gone some way to solving the challenges encountered in ultra-large-scale virtual screening.
Ultra-large-scale virtual screening platform using aweVS
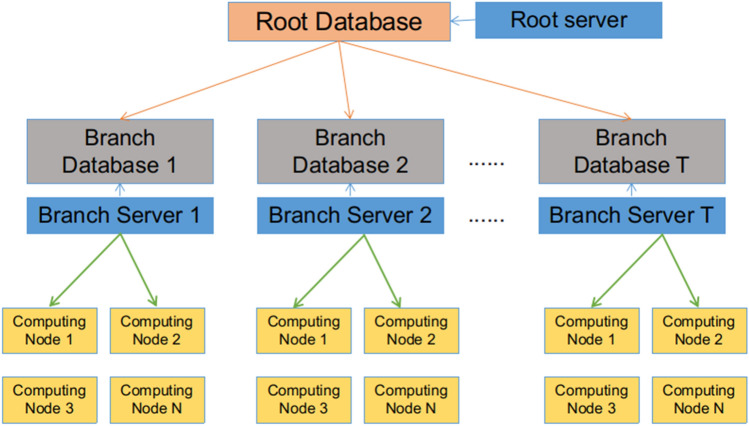
In order to address the challenges encountered in ultra-HTVS on supercomputers, we designed a package named aweVS, which utilizes multi-layer databases to dynamically distribute the large number of docking tasks (Fig. 2). aweVS can integrate all of the docking tasks in the virtual screening process, ideally scaling linearly with the number of CPUs or GPUs, efficiently handle billions of ligands, minimize the input and output loads, and run robustly. It can use different docking software, including heterogeneous acceleration software, and different hardware platforms, including various types of heterogeneous GPU platforms, to perform ultra-HTVS. aweVS is user-friendly and easy to use for non-computational scientists.
Fig. 2.
The design architecture of aweVS
aweVS uses a combination of multiple tiers of databases to manage and distribute docking jobs. At the beginning of the program, the “Root Server” gets the first-level tasks from the “Root Database” and then disassembles and stores the tasks in the “Branch Databases”. After that, each “Branch Server” obtains tasks from each “Branch Database” and distribute them to the foreman of each compute node for execution. In each computing node, the foreman calls multiple workers to complete the tasks distributed on this node, including data acquisition, molecular docking calculation, and post-processing analysis of this batch of tasks. During task execution, the tasks on each computing node are fully loaded, achieving a balanced load. After the tasks on the computing node are completed, the foreman on the computing node fetches new computing tasks from the “Branch Database” until the tasks are completed, and no new tasks exist in the “Root Database” (Fig. 2).
The effective processing of massive files is important for stability of the file system of super computers. We compressed the massive files in multiple layers, using multiple databases. the “Root Database” corresponds to the top-level ligand directory, which includes several top-level compressed files. The secondary compressed file contains the molecules to be docked and corresponds to the “Branch Database”. At the time of calculation, the foreman on the computing node fetches a secondary tar-archive, copies it to the local temporary file systems of that compute node, and then decompresses it to perform the computation. After the calculation is completed, the task is rank ordered, and the top 20% of the results are stored in the database, while all the output files are deleted. After the TOP N molecules are selected after HTVS, re-docking should be done to get all needed results. This approach saves storage space and reduces the IO pressure on the file system caused by data migration.
Network communication between the nodes is required to fetch tasks from the database or to save results to the database. At the beginning of the computation, nodes concurrently communicate with the higher-level database, a situation that can cause excessive instantaneous communication loads. For example, if 500 computing nodes fetch tasks from one “Branch Database”, there are 500 instantaneous communications for the node of the “Branch Database”. We included a 100 ms communication delay to reduce this instantaneous communication. The computation process is performed entirely locally in the computation node, and after the task has been computed it communicates with the database node to store the results and obtain the next task. The computing node communicates with the database node every 30 min to update the job time, and only the energy and ligand names are retained and stored in the database at the end of the task, significantly reducing the amount of data transferred across the network.
aweVS automatically scans nodes and quickly restarts database services during computation, provides effective fault-tolerance management for hardware errors and problems caused by software computation, and ensures that tasks execute automatically to completion. The program shields the user from the complexity of the processing details, and provides the flexibility to adjust the inputs and running parameters using configuration files.
We used aweVS to call Autodock GPU software to complete a virtual screening of over 1.6 billion compounds in less than a day, using 26,000 GPU cards (the computing performance is between NVIDIA Tesla P100 and V100) on a domestic supercomputer. In the future, we expect to be able to use this ultra-large-scale virtual screening platform based on supercomputers to perform larger and faster virtual screening, providing powerful computational simulations for drug candidate discovery.
Web services for molecular docking-based HTVS
Molecular docking calculations not only confirm whether a receptor and ligand can bind but also determine the binding conformation and binding strength. The virtual screening of a large number of ligand molecules based on molecular docking allows the identification of some of the most strongly bound ligand molecules by ranking their binding strength, which reduces the scope and cost for subsequent calculations and experimental activity measurements.
As mentioned above, HTVS plays a significant role in the drug development process. During the research process, several HTVS may need to be performed for different binding sites of a target, a process that can be repetitive. HTVS calculation usually involves the following steps: obtaining the protein structure, performing structural modifications and format conversion, defining the binding pocket and size, modifying the ligand structure and format conversion, setting the docking parameters, using suitable molecular docking software to perform molecular docking calculations for the receptor and the ligand library, and post-processing the results. Post-processing may involve scoring, ranking scores, extracting the molecular structure and energy, and visualizing the binding complex. Some of the pre- and post-processing steps mentioned above, such as structure modification, format conversion, and identification of binding pockets, can be assisted by auxiliary tools such as PyMOL (Schrodinger 2015) which provides parameter setup and structure visualization for AutoDock Vina. Other steps of HTVS must be done via the Linux command line. This process is difficult for biologists or pharmacologists who are not familiar with Linux. Pharmacologists, who focus on experimental or clinical research, are often not familiar with the steps of virtual screening, and learning different tools or software can be difficult.
With the development of the Internet, Web services for HTVS have gradually emerged. A Web service can be used to predict the molecular interactions that may occur between a target protein and a small molecule. They can effectively assist drug development as they lower the barriers to use, and increase the efficiency of HTVS. There are several websites that provide Web-based protein–ligand molecular docking services, most of which run standalone software on the server side, and provide an interface and computational resources via the Web.
Web services generally fall into two categories. The first one is docking of target proteins to individual small molecules, such as the SwissDock platform run by the Swiss Institute of Bioinformatics (based on the software EADock DSS software developed by the same research institute) and the Achilles Blind Docking server run by the Bioinformatics and High-Performance Computing Research Group at the University of Vucón in Spain. Another type of service provides screening of target proteins against specific small molecule databases. As shown in Table 2, DOCK Blaste r (Irwin et al. 2009), based on the DOCK3 software, can use PocketPickker (Coleman et al. 2010) to predict binding sites and the ZINC database (Irwin et al. 2005) for HTVS. Drug Discovery@TACC (Tacc 2021), based on AutoDock Vina and the ZINC database, requires user-defined binding sites. istar is based on idock and ZINC databases, requires user-defined binding sites, and supports a WebGL-based visual interface for previewing results. FINDSITE (Zhou et al. 2013) is based on FINDSITEcomb and supports ZINC, KEGG Compound (Kanehisa et al. 2000), and BindingDB databases. iScreen (Tsai et al. 2011) uses PLANTS as the molecular docking engine, and the ligand database is the TCM@Taiwan database, supporting ab initio ligand design with the LEA3D tool.
Table 2.
Web services for HTVS
| Name | Feature | Developer/maintainer | Website |
|---|---|---|---|
| SwissDock (Grosdidier et al. 2011) | Based on the docking software EADock DSS | Swiss Institute of Bioinformatics | http://www.swissdock.ch |
| Achilles Blind Docking server | Blinding docking server, can use Cloud resources, provide top clusters result preview | Catholic University of Murcia, South East Spain | http://bio-hpc.eu/software/blind-docking-server |
| DOCK Blaster (Irwin et al. 2009) | Based on DOCK software and ZINC database. It provides self-assessment, which estimates the anticipated reliability of the automated screening results using pose fidelity and enrichment | University of California, San Francisco | http://blaster.docking.org |
| Drug Discovery@TACC | Can access to Autodock Vina running on the Lonestar 5 supercomputer at TACC. Ligand libraries were extracted from the ZINC database | Texas Advanced Computing Center, The University of Texas at Austin | https://drugdiscovery.tacc.utexas.edu/#/ |
| Istar (Li et al. 2014) | Based on idock and ZINC database. The results provide binding affinity predicted by RF-Score, putative hydrogen bonds, and supplier information for easy purchase | Chinese University of Hong Kong | http://istar.cse.cuhk.edu.hk/idock/ |
| FINDSITE (Zhou et al. 2013) | Based on FINDSITEcomb. Compound library includes ZINC8, KEGG Compound and BindingDB database | Georgia Institute of Technology | https://sites.gatech.edu/cssb/findsite-comb/ |
| iScreen (Tsai et al. 2011) | Based on PLANTS docking program and traditional Chinese medicine database | China Medical University, Taiwan | http://iscreen.cmu.edu.tw/ |
The above Web services, which implement protein–ligand molecular docking services for specific molecular docking software and databases, are less involved in interactive pre- and post-processing and efficient use of supercomputing resources.
Based on our ultra-HTVS software aweVS and the China National Grid (CNGrid), which provides unified high-performance computing services based on multiple heterogeneous HPCs, we developed a Web server that provides easy access to researchers who wish to perform HTVS but who do not have the necessary computer resources and/or computational biology background. It is a one-stop service platform, on which users can pre- and post-process targets and ligands visually, and submit jobs to HPCs in CNGrid without tedious software installation. The site is currently based on the AutoDock Vina software, ZINC and Enamine REAL databases (Enamine 2021; Irwin et al. 2005), and also supports user-defined ligand uploads for docking.
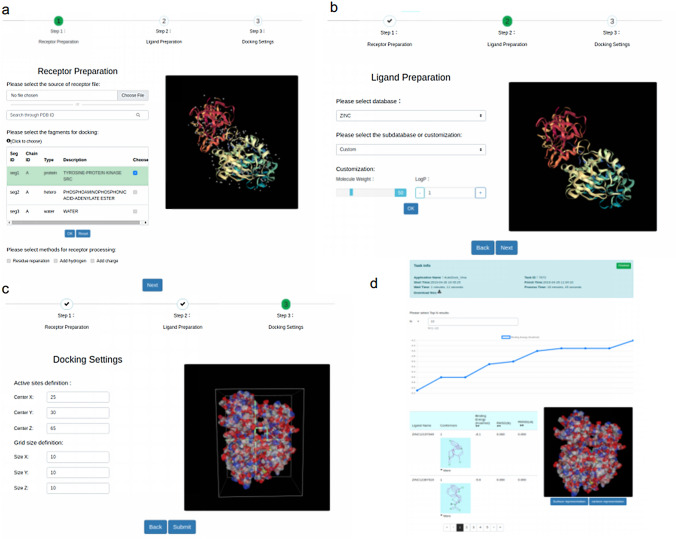
Our Web server uses a front- and back-end separated Web development model, with the Vue.js framework on the front-end for display and Node.js on the back-end to handle the various requests sent from the front-end. MongoDB databases (https://www.mongodb.com) are used to manage the ligands database and calculation results, and virtual screening tasks are submitted to the HPCs through a RESTful API provided by CNGrid. For the pre-processing part of the docking calculation, we designed a guided input file and parameter preparation step based on the virtual screening process. The three steps include receptor file preparation (Fig. 3a), ligand file preparation (Fig. 3b), and docking parameter settings (Fig. 3c). The site uses the NGL viewer plugin to provide real-time visualization of targets and binding pockets (Alexander et al. 2016; Hildebrand et al. 2015). In the receptor file preparation step, the user can upload a PDB file or fill the PDB ID to fetch the initial receptor structure. The receptor file preparation step also includes modifications to the PDB structure, such as hydrogen and charge addition, residue complementation, and other operations, which are assisted by tools such as AutoDock Tools on the back end of the platform. In the ligand preparation phase, the ligand structures can be selected from the ZINC database, or compressed files uploaded in pdbqt format by the user. In the docking parameter setting stage, the position of the crystallized ligand with the protein is automatically identified as a reference pocket and the size of ligand for pocket size, which is shown in the right side of the web page, to enable the user to select the proper docking parameters. The user can specify the parameter TOP N, which is the results of the N top active molecules to be returned after virtual screening.
Fig. 3.
Screenshots of our web site. a Receptor preparation page. b Ligand preparation page. c Parameter setup page. d Results analysis page
The back-end of the Web server is connected with HPCs by a RESTful API from CNGrid. The HTVS calculation is completed by aweVS and Autodock Vina. The user can view the job status in real time and kill the job if necessary. When the job is done, the post-processing program is executed to get the TOP N active molecules. The user can analyze the binding energies and different conformations intuitively and effectively on the website (Fig. 3d).
Conclusions
In this research, we discussed several aspects that affect the accuracy, efficiency, and ease-of-use of molecular docking-based virtual drug screening. With respect to the accuracy of the binding affinity calculation, the difference between commercial and free software is not significant. Consensus scoring can be done by multiple applications to improve the calculation accuracy. The docking score of the top active compounds improves with the size of the ligand library during screening. Efficient screening of chemical spaces on a large scale should make full use of the power of supercomputers. The development of high-throughput concurrent programs to address the challenge of virtual screening at an ultra-large scale is required to obtain virtual screening results in a limited time frame. In this study, we introduced a ultra-large-scale virtual screening platform named aweVS, which can integrate all of the docking tasks in the virtual screening process, ideally scaling linearly with the number of CPUs or GPUs, efficiently handle billions of ligands and run robustly.
In addition, web services for molecular docking-based HTVS is an effective way to reduce the barriers to HPC accessibility and software usage and improve productivity. Based on aweVS, we developed a web site, which is designed based on the characteristics and steps of molecular docking calculations, and it can combine calculations and data processing to provide an intuitive, efficient, and easy-to-use virtual screening service for drug discovery.
In the future, some features, such as adaptive determination of the number of ‘Branch Server’ according to different type of super computers, are required to be implemented in aweVS. Also, more docking software and databases should be added to the web site, and consensus docking should be considered.
Acknowledgements
We thank the support from Ministry of Science and Technology of China (NO. 2016YFB0201700), Guangdong Provincial Key Laboratory of Biocomputing (2016B030301007) and China National Grid (CNGrid)
Biographies
Baohua Zhang
born in 1985, Ph.D. candidate. Her research interests include computer aided drug design and high-performance computing application.
Hui Li
born in 1996, M.S. candidate, His research interests include computer aided drug design.
Kunqian Yu
born in 1975, Ph.D, professor. Ph.D. supervisor. His research interests include molecule modeling and drug design.
Zhong Jin
born in 1974, Ph.D, professor, Ph.D. supervisor. His research interests include high performance computing.
Funding
Guangdong Provincial Key Laboratory of Prevention and Control for Severe Clinical Animal Diseases, 2016B030301007, Baohua Zhang, Ministry of Science and Technology of China, 2016YFB0201700, Baohua Zhang.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Kunqian Yu, Email: yukunqian@simm.ac.cn.
Zhong Jin, Email: zjin@sccas.cn.
References
- Allen WJ, Sudipto Mukherjee TEB, Brozell SR, Moustakas DT, Therese Lang P, Case DA, Kuntz ID, Rizzo RC. DOCK 6: impact of new features and current docking performance. J. Comput. Chem. 2015;36(15):1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Tan S, Xu T, Liu H, Huang J, Yao X. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank, P: www.wwpdb.org. (2021)
- Benet LZ, Ursu O, Oprea TI. BDDCS, the Rule of 5 and drugability. Adv. Drug. Deliv. Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Bonvin AMJJ. Flexible protein-protein docking. Curr. Opin. Struct. Biol. 2006;16(2):194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Chunlan P, Yan G, Shi J, Li R. Assessing the performance of docking scoring function, FEP, MM-GBSA, and QM/MM-GBSA approaches on a series of PLK1 inhibitors. Med. Chem. Commun. 2017;7:1452–1458. doi: 10.1039/c7md00184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RG, Sharp KA. Protein pockets: inventory, shape, and comparison. J. Chem. Inf. Mod. 2010;50:589–603. doi: 10.1021/ci900397t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasmana A, Raza S, Jahan R, Lohani M, Arif MJ. High-throughput virtual screening (htvs) of natural compounds and exploration of their biomolecular mechanisms: an in silico approach. Cambridge: Academic Press; 2019. [Google Scholar]
- ECP. https://www.exascaleproject.org/. (2021)
- Enamine. https://enamine.net/hit-finding/compound-collections/real-database. (2021)
- Erlanson DA. Introduction to fragment-based drug discovery. Top Curr. Chem. 2012;317:1–32. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- Fernández-Recio J, Totrov M, Abagyan R. Soft protein–protein docking in internal coordinates. Protein Sci. 2002;11(2):280–291. doi: 10.1110/ps.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Hersey A, Nowotka M, Bento AP, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrián-Uhalte E, Davies M, Dedman N, Karlsson A, Magariños MP, Overington JP, Papadatos G, Smit I, Leach AR. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(1):945–954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgull C, Boeszoermenyi A, Wang ZF, Fischer PD, Coote PW, Padmanabha Das KM, Arthanari H. An open-source drug discovery platform enables ultra-large virtual screens. Nature. 2020;580:663–668. doi: 10.1038/s41586-020-2117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47(4):409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- Hildebrand ARP. NGL Viewer: a web application for molecular visualization. Nucl Acids Res. 2015;43:576–579. doi: 10.1093/nar/gkv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DR, Walkinshaw MD. Consensus docking: improving the reliability of docking in a virtual screening context. J. Chem. Inf. Model. 2013;53(2):384–390. doi: 10.1021/ci300399w. [DOI] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45(1):177–182. doi: 10.1021/ci049714+”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52(18):5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- Jones G, Willet P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000 doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick P, Ellis C. Chemical space. Nature. 2004;432:823–823. doi: 10.1038/432823a. [DOI] [Google Scholar]
- Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M. LigScore: a novel scoring function for predicting binding affinities. J Mol Graph Modell. 2005;23:395–407. doi: 10.1016/j.jmgm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Li H, Leung KS, Ballester PJ, Wong MH. istar: a web platform for large-scale protein-ligand docking. PLoS ONE. 2014;9(1):e85678. doi: 10.1371/journal.pone.0085678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson OTAJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W.H. https://covid19.who.int/?gclid=EAIaIQobChMI0dH68s2X6gIVrNSzCh306wCSEAAYASAAEgIuqfD_BwE. (2021)
- Pagadala NS, Syed K, Tuszynski F. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S., Zhang, X., Yang, S., Su, W., Zhang, Z., Dong, D., Li, K.-C: mD3DOCKxb: an ultra-scalable CPU-MIC coordinated virtual screening framework. In: 17th IEEE/ACM international symposium on cluster, cloud and grid computing (CCGRID), 671–676. (2017). doi: 10.1109/CCGRID.2017.131
- Prieto-Martínez FD, Arciniega M, Medina-Franco JL. Molecular docking: current advances and challenges. TIP Revista Especializada En Ciencias Químico-Biológicas. 2018;21:65–87. doi: 10.22201/fesz.23958723e.2018.0.143. [DOI] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261(3):470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- Rifaioglu AS, Nalbat E, Atalay V, Martin MJ, Cetin-Atalay R, Doğan T. DEEPScreen: high performance drug–target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem. Sci. 2020;11(9):2351–2557. doi: 10.1039/C9SC03414E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A.S., Valasatava, Y., Duarte, J. M., Prlić, A., Rose, P.W.: Web-based molecular graphics for large complexes. In: ACM Proceedings of the 21st international conference on Web3D technology, 185–186. (2016)
- Ruiz-Carmona S, Alvarez-Garcia D, Foloppe N, Garmendia-Doval AB, Juhos S, Schmidtke P, Morley SD. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014;10(4):1003571. doi: 10.1371/journal.pcbi.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samdani A, Vetrivel U. POAP: A GNU parallel based multithreaded pipeline of Open Babel and AutoDock suite for boosted high throughput virtual screening. Comput. Biol. Chem. 2018;74:39–48. doi: 10.1016/j.compbiolchem.2018.02.012. [DOI] [PubMed] [Google Scholar]
- Santos-Martins, D., Solis-Vasquez, L., Koch, A., Forli, S: Accelerating AutoDock4 with GPUs and gradient-based local search. ChemRxiv (2019) [DOI] [PMC free article] [PubMed]
- Schrodinger, LLC: The PyMOL molecular graphics system, Version 1.8. (2015).
- Srinivasarao M, Low PS. Ligand-targeted drug delivery. Chem. Rev. 2017;117(19):12133–12164. doi: 10.1021/acs.chemrev.7b00013. [DOI] [PubMed] [Google Scholar]
- TACC. https://drugdiscovery.tacc.utexas.edu/#. (2021).
- Tsai TY, Chang K-W, Chen CY-C. iScreen: world's first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. J. Comput. Aided Mol. Des. 2011;25(6):525–531. doi: 10.1007/s10822-011-9438-9. [DOI] [PubMed] [Google Scholar]
- Vartika T, Mazumder M, Chandra R, Yang J, Sakharkar KM. Small molecule drug design. Amsterdam: Elsevier; 2019. [Google Scholar]
- Venkatachalam CM, Oldfield T, Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Modell. 2003;21:289–307. doi: 10.1016/S1093-3263(02)00164-X. [DOI] [PubMed] [Google Scholar]
- Wang J, Dokholyan NV. MedusaDock 2.0: efficient and accurate protein-ligand docking with constraints. J. Chem. Inf. Model. 2019;59:2509–2515. doi: 10.1021/acs.jcim.8b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sun H, Yao X, Li D, Xu L, Li Y, Hou T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016;18(18):12964–12975. doi: 10.1039/C6CP01555G. [DOI] [PubMed] [Google Scholar]
- Wikipedia: https://en.wikipedia.org/wiki/List_of_protein-ligand_docking_software. (2021)
- Yadava U. Search algorithms and scoring methods in protein-ligand docking. Endocrinol. Metab. Int. J. 2018;6(6):359–367. [Google Scholar]
- Zhao H, Caflisch A. Discovery of ZAP70 inhibitors by high-throughput docking into a conformation of its kinase domain generated by molecular dynamics. Bioorg. Med. Chem. Lett. 2013;23(20):5721–5726. doi: 10.1016/j.bmcl.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Zhao H, Huang D. Hydrogen bonding penalty upon ligand binding. PLoS ONE. 2011;6(6):e19923. doi: 10.1371/journal.pone.0019923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A, Ivanenkov YA, Aliper A, Veselov MS, Aladinskiy VA, Aladinskaya AV, Aspuru-Guzik A. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- Zhou H, Skolnick J. FINDSITEcomb: a threading/structure-based, proteomic-scale virtual ligand screening approach. J. Chem. Inf. Model. 2013;53(1):230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]