ABSTRACT
Background
Healthy eating is associated with lower risks of disease and mortality, but the mechanisms underlying these associations are unclear. Age is strongly related to health outcomes, and biological age can be estimated using the blood methylome.
Objectives
To determine whether healthy eating patterns are associated with methylation-based measures of biological age.
Methods
Among women in the Sister Study, we calculated scores on 4 recommendation-based healthy eating indexes [Dietary Approaches to Stop Hypertension diet, Healthy Eating Index–2015, Alternative Healthy Eating Index (aHEI-2010), and the Alternative Mediterranean diet] using a validated 110-item Block FFQ completed at enrollment. Genome-wide DNA methylation data were generated using the HumanMethylation450 BeadChip on whole blood samples collected at enrollment from a case-cohort sample of 2694 women and were used to calculate 4 measures of epigenetic age acceleration (Hannum AgeAccel, Horvath AgeAccel, PhenoAgeAccel, and GrimAgeAccel). Linear regression models, adjusted for covariates and cohort sampling weights, were used to examine cross-sectional associations between eating patterns and measures of biological age.
Results
All 4 healthy eating indexes had inverse associations with epigenetic age acceleration, most notably with PhenoAgeAccel and GrimAgeAccel. Of these, the strongest associations were for aHEI-2010 [per 1-SD increase in diet quality, PhenoAgeAccel β = −0.5 y (95% CI: −0.8 to −0.2 y) and GrimAgeAccel β = −0.4 y (95% CI: −0.6 to −0.3 y)]. Although effect modification was not observed for most lifestyle factors, in analyses stratified by physical activity, the benefits of a healthy diet on epigenetic age acceleration were more pronounced among women who did not meet physical activity guidelines (reporting <2.5 h/wk of exercise).
Conclusions
Higher diet quality is inversely associated with methylation-based measures of biological age. Improving diet could have the most benefits in lowering biological age among women with lower levels of physical activity. This trial was registered at clinicaltrials.gov as NCT00047970.
Keywords: diet quality, healthy eating, epigenetic clocks, DNA methylation, biological age
See corresponding editorial on page 6 and article on page 163.
Introduction
Better diet quality is associated with decreased risks of cardiometabolic disease, cancer, and death (1–8). The biological mechanisms that link diet quality and disease risks are complex and may involve epigenetic modifications (9). This hypothesis is supported by studies showing that DNA methylation at individual cytosine-phosphate-guanine (CpG) sites in candidate gene regions can mediate the relationship between diet quality and the prevalence of metabolic diseases and cancer (10, 11). Although studies of individual CpGs can provide a useful means of exploring biological mechanisms, other epigenetic-based metrics that combine large sets of CpGs and are designed to estimate biological age, also known as epigenetic clocks, may provide a novel view into the pathways that link diet and disease.
Several epigenetic clocks have been developed; some were designed as predictors of chronological age, whereas others were designed as predictors of mortality. Perhaps the most utilized of the age predictors are the Hannum (12) and Horvath (13) epigenetic clocks, which were developed using DNA methylation from blood (Hannum) or dozens of tissues (Horvath). In contrast, the CpGs used in the PhenoAge clock (14) were selected to predict PhenoAge, a score designed using a combination of clinical measures as a predictor of all-cause mortality. Finally, the GrimAge clock (15) was developed using panels of CpGs, each correlated with concentrations of different circulating proteins or other measures, which were combined with chronological age and sex to produce a predictor of age-related disease and mortality. All of these clocks produce epigenetic age estimates that are highly correlated with a person's chronological age. People whose epigenetic age is older than their chronological age (termed “positive age acceleration”) are hypothesized to be biologically older and have an elevated risk of disease, whereas those with younger epigenetic ages relative to their chronological age are hypothesized to be biologically younger and have a decreased risk. Positive age acceleration metrics from the various epigenetic clocks are reported to be associated with a variety of disease-associated environmental exposures and unhealthy lifestyle factors, as well as increased age-specific disease incidence and mortality (16–25).
In prior studies, associations between diet and age acceleration metrics have predominately focused on individual dietary components, such as food items or specific nutrients (14, 15, 26, 27). For example, fish and poultry intakes have been reported to be inversely associated with age acceleration metrics (26), whereas red meat intake has shown positive associations (14, 15). A limitation of these studies is that by focusing on individual food components, the broader beneficial effects of an improved diet may be missed. We hypothesize that better diet quality, as determined by different recommendation-based guidelines, will be associated with lower age acceleration, with stronger associations for the epigenetic clocks designed as predictors of mortality. Here, we use a population of women to examine how different dietary indexes are related to age acceleration as determined by various epigenetic clocks.
Methods
Study population
The Sister Study is an ongoing, longitudinal cohort of 50,884 women from the United States recruited between 2003 and 2009 designed to identify novel environmental and biological risk factors for breast cancer (28). Eligible women were between ages 35 and 74 and had a biological sister previously diagnosed with breast cancer but were themselves free of breast cancer. At enrollment, women completed a computer-assisted telephone interview that included information on lifestyle factors and demographics. A questionnaire on dietary factors was self-completed and retrieved during a home visit where written informed consent, anthropomorphic measurements, and whole blood samples were collected (28). Information about obtaining Sister Study data can be found at https://sisterstudy.niehs.nih.gov/English/researchers.htm. The study was approved by the Institutional Review Boards of the National Institute of Environmental Health Sciences and the Copernicus Group.
Dietary assessment and healthy eating index calculation
Dietary data were collected using a modified version of the validated 110-item 1998 Block FFQ (29), which has been shown to be valid and reliable among a population of women with similar characteristics (30). The questionnaire was structured to measure average food consumption in the prior 12 mo, which was calculated by multiplying the frequency of consumption (9 possible frequencies ranging from “never” to “every day”) by the specified quantity (3 or 4 choices per each food item or group of similar food items). Based on information obtained via the FFQ, food groups were created using the Food Patterns Equivalents Database, and nutrient consumption was estimated using the Food and Nutrient Database for Dietary Studies, which the USDA developed for US women (31).
The FFQ data were then used to calculate scores on 4 recommendation-based dietary indexes: the Dietary Approaches to Stop Hypertension (DASH) diet operationalized by Fung et al. (32), the Healthy Eating Index (HEI) 2015 (33), the Alternative Healthy Eating Index (aHEI) 2010 (34), and the Alternative Mediterranean (aMed) diet developed by Fung et al. (35). The DASH diet includes various foods and nutrients known to be protective against hypertension (36). For the DASH diet components of fruits, vegetables, whole grains, nuts and legumes, and low-fat dairy, those in the lowest quintile of intake were assigned 1 point and an additional point was assigned for each increasing quintile. For red and processed meat, sugar-sweetened beverages, and sodium, those in the highest quintile of intake were assigned 1 point and an additional point was assigned for each decreasing quintile. The DASH diet component scores were summed, with a potential range between 8 and 40. The HEI-2015 is a summary score for adherence to the USDA 2015–2020 Dietary Guidelines for Americans (33). It is based on intakes of total fruits, whole fruits, total vegetables, greens and beans, total protein foods, seafood and plant proteins, whole grains, dairy, fatty acids, refined grains, sodium, added sugars, and saturated fats. Each component received a score, with higher scores assigned for more favorable intakes. The first 6 items carried a maximum score of 5 points each, whereas intakes of the other items carried a maximum score of 10 points each. The total HEI-2015 score had a potential range between 0 and 100. The aHEI-2010 incorporates additional evidence on diet and health to better predict chronic disease (34). This score is based on intakes of 11 foods and nutrients. Higher scores were assigned for higher intakes of vegetables, fruits, whole grains, nuts and legumes, PUFAs, and omega-3 fatty acids, whereas lower scores were assigned for higher intakes of sugar-sweetened beverages, red and processed meats, trans fatty acids, and sodium; moderate intake was rewarded for alcohol. Each component received a score from 0 (least favorable) to 10 (most favorable), with partial scores that were proportional to intake. Component scores were summed for a total aHEI-2010 score with a potential range between 0 to 110. Finally, the Mediterranean diet incorporates foods found to be protective against heart disease and other chronic diseases (37, 38). The aMed diet is based on 9 components. For vegetables, fruits, legumes, nuts, whole grains, fish, and the MUFA to SFA ratio, intake above the median was assigned 1 point; for red and processed meats, intake below the median was assigned 1 point; and for alcohol, moderate intake was assigned 1 point. The aMed diet component scores were summed for a potential range between 0 to 9 points.
DNA methylation processing and age acceleration calculation
In July 2014, to determine whether leukocyte DNA methylation profiles are markers of breast cancer risk, a case-cohort subsample of 2878 self-identified non-Hispanic White women was selected for genome-wide DNA methylation analysis. To limit confounding by ancestry, only self-identified non-Hispanic White women were eligible for selection. Overall, 1336 women were randomly selected (approximately 3%) from the eligible women enrolled in the Sister Study. Another 1542 women were also chosen because they were diagnosed with incident breast cancer between enrollment and the end of February 2014 (39).
DNA methylation assessment procedures have been previously described (40). DNA methylation preprocessing was completed using the ENmixR package (Xu, Niu, Taylor; Bioconductor, https://www.bioconductor.org/packages/release/bioc/html/ENmix.html), which included: reducing background noise using the ENmix method (41); correcting fluorescent dye bias using the regression on logarithm of internal control probes method (42); and quantile normalization to make overall array fluorescence intensity distributions comparable between arrays and to reduce Infinium I and II probe design-type bias using the regression on correlated probes method (43). Of the 2878 genomic DNA samples assayed on Illumina's Infinium HumanMethylation450 BeadChip, 102 were excluded because they failed quality control checks (39). Of these samples, 91 had a mean bisulfate intensity less than 4000 or had greater than 5% of probes with low-quality methylation values (detection P > 0.000001, <3 beads, or values outside 3 times the IQR), 4 were outliers for their methylation beta value distributions, 1 had missing phenotype data, and 6 were from women whose date of breast cancer diagnosis preceded blood collection. For the remaining 2776 samples, 4 epigenetic clocks (Hannum, Horvath, PhenoAge, and GrimAge) were obtained using an online calculator (https://dnamage.genetics.ucla.edu/home).
Age acceleration, as a general concept, is defined as the difference between the estimated epigenetic age and observed, chronological age (44). An age acceleration value is quantified for an individual as the residual from a linear regression model using the entire sample population, where epigenetic age, based on a single epigenetic clock, is treated as the dependent variable, and observed, chronological age is treated as the independent variable. For each of the 4 epigenetic clocks (Hannum, Horvath, PhenoAge, and GrimAge), separate regression models were performed to generate their 4 corresponding metrics of age acceleration for the study population (i.e., Hannum AgeAccel, Horvath AgeAccel, PhenoAgeAccel, and GrimAgeAccel (12–15).
Statistical analysis
In all analyses, to account for the case-cohort sampling scheme, we applied inverse probability of selection weights, thereby standardizing the DNA methylation subsample to approximate the entire population of self-identified non-Hispanic White women enrolled in the Sister Study. We describe the sample population's characteristics using survey-weighted means and SDs or survey-weighted proportions. We examined the dietary indexes' distributions using weighted histograms and we calculated correlations with chronological age separately for the 4 epigenetic clocks and 4 age acceleration metrics using weighted Pearson correlation coefficients. Weighted correlations were also examined among the dietary indexes and the age acceleration metrics.
We used separate weighted linear regression models to investigate associations between the 4 dietary indexes (independent variables: DASH diet, HEI-2015, aHEI-2010, aMed diet) and the 4 age acceleration metrics (dependent variables: Hannum AgeAccel, Horvath AgeAccel, PhenoAgeAccel, GrimAgeAccel). All models were adjusted for educational attainment (high school degree/equivalency or less, some college or bachelor's degree, or advanced degree), number of live births, physical activity (metabolic equivalent tasks/wk), menopause status (premenopausal, postmenopausal), smoking status (never, former, current), and total caloric intake. Models testing associations with the DASH diet and HEI-2015 were further adjusted for recent alcohol use (average drinks per wk). In all regression analyses, the dietary indexes were transformed to have a mean of 0 and an SD of 1; thus, all association estimates reported are based on a 1-SD increase in diet quality. To examine whether women with the highest diet qualities drove associations, we substituted a 4-level, categorical variable for the continuous dietary quality indexes described above; significant linear trends were determined by treating the dietary index quartiles as ordinal, using the significance cut point of a P value ≤0.05. In supplemental analyses, we additionally adjusted for the BMI and waist-to-hip ratio by including these continuous variables in the models mentioned above (i.e., treating the dietary indexes as continuous or, when assessing linear trends in the model, treating the dietary index as a 4-level, ordinal variable). We examined effect modification by lifestyle factors using cross-product terms in the models, treating the dietary indexes as continuous, with a significant interaction declared at a P value ≤0.05. We examined associations in stratified analyses by physical activity [below recommended CDC guidelines (<2.5 h per wk) compared with meeting recommended guidelines (≥2.5 h/wk)] (45), BMI (<30 compared with ≥30 kg/m2), educational attainment (high school or less compared with college or more), menopause status (pre- compared with postmenopause), and smoking status (ever compared with never) in the models treating the dietary indexes as continuous. Among the 2776 women with available DNA methylation data, we excluded women with extreme age acceleration values (greater than 4 SDs from the mean; n = 3), incomplete FFQ data (n = 38), missing dietary indices (n = 11), or missing covariate information (n = 30; Figure 1). Our final analytic set included 2694 women. All analyses were conducted using Stata version 16 (Stata Corp).
FIGURE 1.
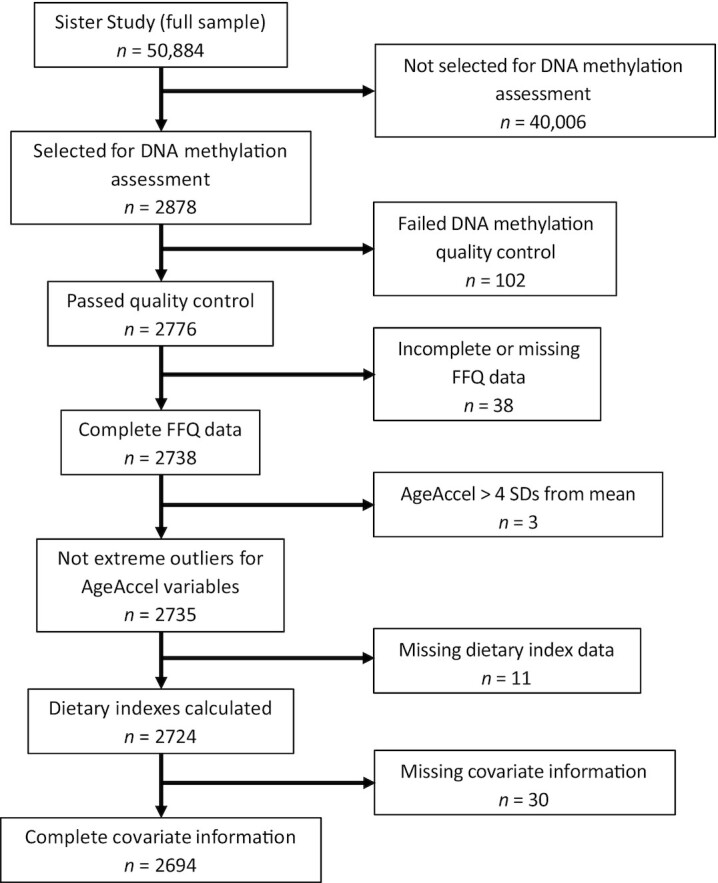
Flow chart for the study population. Description of the inclusion criteria for the subset of Sister Study participants included in this study.
Results
By design, all women selected into the Sister Study's DNA methylation subsample self-identified as non-Hispanic White. Overall, the women had a mean age of 56 y (± 9 y), and a majority attended at least some college, were never smokers, and were postmenopausal at enrollment (Table 1). The weighted mean (± SD) dietary index scores were 24 ± 5 for the DASH diet, 72 ± 9 for HEI-2015, 61 ± 11 for the aHEI-2010, and 4.2 ± 2 for the aMed diet (Table 1; Supplementary Figure 1). In general, the dietary indexes were positively correlated with each other; the DASH diet and HEI-2015 scores had the strongest correlation (weighted ρ = 0.78) and the HEI-2015 and aHEI-2010 scores had the weakest (weighted ρ = 0.59; Supplementary Figure 2). The epigenetic age estimates from the 4 clocks were all positively correlated with chronological age, with the GrimAge clock showing the strongest correlation (weighted ρ = 0.92; Supplementary Figure 3). As expected, the age acceleration estimates were not meaningfully correlated with chronological age (Supplementary Figure 4) but were positively correlated with each other (Supplementary Figure 5). The strongest correlation was for Horvath AgeAccel and PhenoAgeAccel (weighted ρ = 0.50) and the weakest was for Horvath AgeAccel and GrimAgeAccel (weighted ρ = 0.11; Supplementary Figure 5).
TABLE 1.
Weighted participant characteristics for women selected into the Sister Study methylation subsample (n = 2694)
| DASH diet, mean score ± SD | 24 ± 5 |
| Healthy Eating Index–2015, mean score ± SD | 72 ± 9 |
| Alternative Healthy Eating Index–2010, mean score ± SD | 61 ± 11 |
| Alternative Mediterranean diet, mean score ± SD | 4.2 ± 2 |
| Total calories, mean kcals ± SD | 1630 ± 559 |
| Age, mean y ± SD | 56 ± 9 |
| BMI, mean kg/m2 ± SD | 27 ± 6 |
| Waist-to-hip ratio, mean ratio ± SD | 0.8 ± 0.1 |
| Parity, mean live births ± SD | 2.0 ± 1 |
| Physical activity, mean METs/wk ± SD | 52 ± 32 |
| Physical activity, mean h/wk ± SD | 2.9 ± 3 |
| Alcohol consumption, mean drinks/wk S± D | 2.9 ± 4 |
| Educational attainment, % | |
| High school diploma/GED or less | 16.5 |
| Some college/Bachelor's degree | 59.7 |
| Advanced degree | 12.8 |
| Smoking status, % | |
| Never | 52.7 |
| Former | 40.0 |
| Current | 7.3 |
| Menopause status, % | |
| Premenopausal | 32.6 |
| Postmenopausal | 67.4 |
Abbreviations: DASH, Dietary Approaches to Stop Hypertension; GED, General Educational Development; MET, metabolic equivalent task.
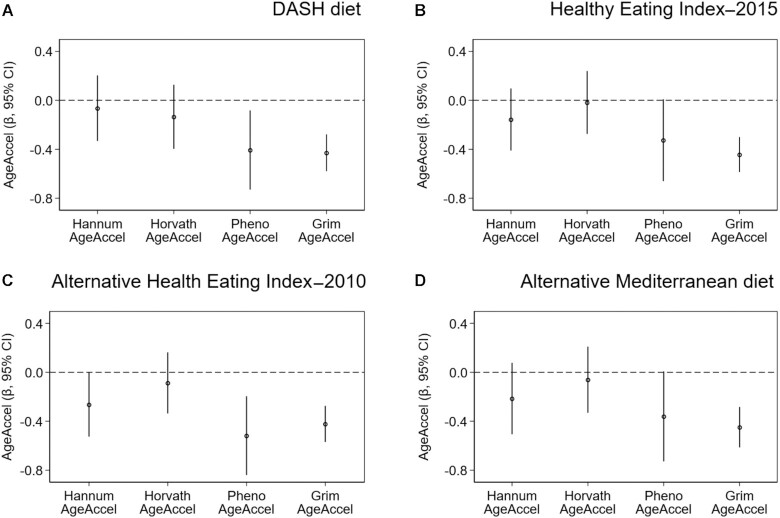
In weighted linear regression models adjusted for confounders, all dietary indexes were inversely associated with the PhenoAgeAccel and GrimAgeAccel metrics (Figure 2). The strongest associations were observed for the aHEI-2010 [per 1-SD increase in diet quality; PhenoAgeAccel β = −0.5 (95% CI: −0.8 to −0.2; P = 0.002) and GrimAgeAccel β = −0.4 (95% CI: −0.6 to −0.3; P = 2.3 × 10–8)]. A higher aHEI-2010 score was also inversely associated with Hannum AgeAccel (per 1-SD increase; β = −0.3; 95% CI: −0.4 to −0.0; P = 0.05). In analyses of the dietary index quartiles, the aHEI-2010 showed significant trends with Hannum AgeAccel, PhenoAgeAccel, and GrimAgeAccel, whereas the HEI-2015 and the DASH and aMed diets showed significant trends only with GrimAgeAccel (Table 2). Although further adjustment for the BMI and waist-to-hip ratio attenuated associations, the aHEI-2010 remained inversely associated with Hannum AgeAccel, PhenoAgeAccel, and GrimAgeAccel and the HEI-2015, DASH, and aMed diets remained inversely associated with GrimAgeAccel (Supplementary Table 1).
FIGURE 2.
Associations between recommendation-based diets and the 4 measures of epigenetic age acceleration. Plots display the β-coefficients and 95% CIs from adjusted linear regression models, which represent the adjusted mean difference for the 4 individual AgeAccel metrics per 1-SD increase in diet quality, for the (A) DASH diet, (B) Healthy Eating Index–2015, (C) Alternative Health Eating Index–2010, and (D) Alternative Mediterranean diet (n = 2694). Abbreviation: DASH, Dietary Approaches to Stop Hypertension.
TABLE 2.
Association estimates (and 95% CIs) for relationships between recommendation-based diet quartiles and epigenetic age acceleration metrics (n = 2694)
| Dietary index score quartiles | P linear-trend | ||||
|---|---|---|---|---|---|
| DASH diet | 10–211 | 22–25 | 26–28 | 29–37 | |
| Hannum AgeAccel | 0 | −0.6 (−1.2, 0.1) | −0.4 (−1.1, 0.3) | −0.1 (−0.9, 0.7) | 0.82 |
| Horvath AgeAccel | 0 | −0.1 (−0.8, 0.5) | −0.4 (−1.1, 0.3) | −0.4 (−1.1, 0.4) | 0.20 |
| PhenoAgeAccel | 0 | −1.3 (−2.1, −0.4) | −1.2 (−2.0, −0.3) | −0.8 (−1.8, 0.1) | 0.10 |
| GrimAgeAccel | 0 | −0.8 (−1.2, −0.4) | −0.8 (−1.2, −0.4) | −1.1 (−1.6, −0.7) | 1.6 × 10–6 |
| HEI-2015 | 40–661 | 67–73 | 74–79 | 80–95 | |
| Hannum AgeAccel | 0 | −0.3 (−1.0, 0.4) | −0.2 (−0.9, 0.5) | −0.4 (−1.1, 0.3) | 0.34 |
| Horvath AgeAccel | 0 | −0.1 (−0.8, 0.5) | 0.1 (−0.6, 0.8) | 0.1 (−0.6, 0.8) | 0.71 |
| PhenoAgeAccel | 0 | −1.1 (−2.0, −0.2) | −1.0 (−1.9, −0.1) | −0.8 (−1.8, 0.1) | 0.09 |
| GrimAgeAccel | 0 | −0.5 (−0.9, −0.1) | −0.7 (−1.2, −0.3) | −0.9 (−1.3, −0.6) | 1.3 × 10–6 |
| aHEI-2010 | 27–521 | 53–60 | 61–69 | 69–100 | |
| Hannum AgeAccel | 0 | −0.5 (−1.2, 0.3) | −0.3 (−1.0, 0.4) | −0.9 (−1.6, −0.2) | 0.02 |
| Horvath AgeAccel | 0 | −0.2 (−0.9, 0.4) | −0.4 (−1.0, 0.3) | −0.2 (−0.9, 0.5) | 0.48 |
| PhenoAgeAccel | 0 | −1.2 (−2.1, −0.4) | −0.9 (−1.8, −0.1) | −1.5 (−2.3, −0.6) | 0.004 |
| GrimAgeAccel | 0 | −0.2 (−0.6, 0.2) | −0.6 (−1.0, −0.2) | −1.0 (−1.4, −0.5) | 1.6 × 10–6 |
| aMed diet | 0–31 | 4–4 | 5–6 | 7–9 | |
| Hannum AgeAccel | 0 | 0.3 (−0.4, 0.9) | −0.1 (−0.7, 0.5) | −0.9 (−1.8, −0.0) | 0.11 |
| Horvath AgeAccel | 0 | 0.2 (−0.5, 0.9) | −0.1 (−0.7, 0.6) | −0.0 (−0.8, 0.8) | 0.90 |
| PhenoAgeAccel | 0 | 0.2 (−0.7, 1.1) | −0.3 (−1.1, 0.5) | −0.9 (−2.0, 0.2) | 0.12 |
| GrimAgeAccel | 0 | −0.1 (−0.5, 0.3) | −0.7 (−1.0, −0.3) | −1.1 (−1.6, −0.6) | 1.8 × 10–6 |
Abbreviations: aHEI, Alternative Healthy Eating Index; aMed, Alternative Mediterranean; DASH, Dietary Approaches to Stop Hypertension; HEI, Healthy Eating Index; MET, metabolic equivalent task.
Represents the referent category against which the other quartiles are compared. Estimates were obtained using separate linear regression models, treating the individual AgeAccel metrics as the dependent variables and the diet index score quartiles (treating the lowest as the referent category) as the independent variables. Models were adjusted for educational attainment (high school, college, advanced degree), parity (live births), physical activity (METs/wk), smoking status (current/former/never), menopause status, and caloric intake. Models for DASH and the HEI were additionally adjusted for alcohol intake (drinks/wk).
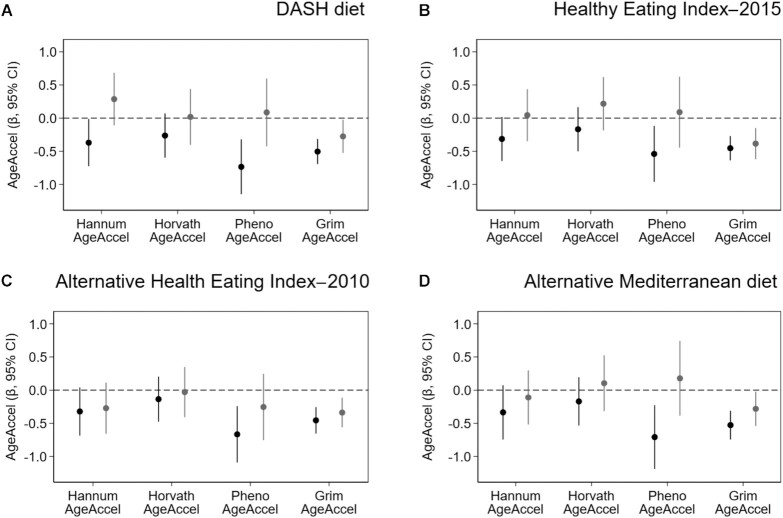
Stratification by common lifestyle factors revealed that the inverse associations of PhenoAgeAccel with the DASH diet and aMed diet were stronger among women who did not meet physical activity recommendations [DASH diet, <2.5 h/wk β = −0.7 (95% CI: −1.1 to −0.3), ≥ 2.5 h/wk β = 0.1 (95% CI: −0.4 to 0.6; P-interaction = 0.04); aMed diet, <2.5 h/wk β = −0.7 (95% CI: −1.1 to −0.2), ≥2.5 h/wk β = 0.2 (95% CI: −0.4 to 0.7; P-interaction = 0.04); Figure 3]. Although a similar pattern was observed for PhenoAgeAccel associations with HEI-2015, the interaction was not statistically significant [<2.5 h/wk β = −0.5 (95% CI: −1.0 to −0.1), ≥ 2.5 h/wk β = 0.1 (95% CI: −0.4 to 0.6); P-interaction = 0.16]. BMI did not modify associations between diet quality and the Horvath AgeAccel, PhenoAgeAccel, or GrimAgeAccel metrics (Supplementary Figure 6). However, the BMI did appear to modify associations between the HEI-2015 index and Hannum AgeAccel [<30 kg/m2 β = 0.1 (95% CI: −0.2 to 0.4 kg/m2), ≥30 kg/m2 β = −0.5 (95% CI: −0.9 to −0.1 kg/m2); P-interaction = 0.03; Supplementary Figure 6]. Diet quality associations with Hannum AgeAccel also varied by smoking status, such that the inverse associations with the aHEI-2010 and aMed were stronger among never smokers [aHEI-2010, never smokers β = −0.5 (95% CI: −0.9 to −0.2), ever smokers β = 0.0 (95% CI: −0.4 to 0.4; P-interaction = 0.01); aMed, never smokers β = −0.4 (95% CI: −0.8 to −0.0), ever smokers β = 0.1 (95% CI: −0.4 to 0.5; P-interaction = 0.02); Supplementary Figure 7]. Age acceleration associations with the dietary indexes did not vary by educational attainment (Supplementary Figure 8) or menopause status (Supplementary Figure 9).
FIGURE 3.
Associations between recommendation-based diets and the 4 measures of epigenetic age acceleration, stratified by physical activity level. Plots display the β-coefficients and 95% CIs from adjusted linear regression models, which represent the adjusted mean difference for the 4 individual AgeAccel metrics per 1-SD increase in diet quality, for the (A) DASH diet, (B) Healthy Eating Index–2015, (C) Alternative Health Eating Index–2010, and (D) Alternative Mediterranean diet among women who did not meet physical activity guidelines (<2.5 h/wk, black lines; n = 1541) and for women who did (≥2.5 h/wk, gray lines; n = 1153). Significant statistical interactions determined by cross-product terms were observed for physical activity on the relationships between PhenoAgeAccel and the DASH diet (P-interaction = 0.04) and the aMed diet (P-interaction = 0.04). Abbreviation: DASH, Dietary Approaches to Stop Hypertension.
Discussion
We found that a higher diet quality was only weakly associated with the 2 epigenetic clocks designed as predictors of chronological age but has strong inverse associations with the 2 epigenetic clocks designed as predictors of mortality. In a stratified analysis, physical activity appeared to modify the relationships of the DASH diet and aMed diet with PhenoAgeAccel, such that inverse associations between diet quality and age acceleration were only observed among women who did not meet physical activity guidelines. Overall, our study shows that diet quality is related to the subset of epigenetic clocks designed to reflect mortality risks, and suggests that improving diet quality may have the most benefits in lowering the biological age for women with lower levels of physical activity.
Previous investigations into the relationship between diet and methylation-based measures of biological age have primarily focused on individual food items and nutrients (14, 15, 26, 27). The most consistent association has been for red meat intake, which appears to be positively associated with the Horvath AgeAccel, PhenoAgeAccel, and GrimAgeAccel metrics (14, 15, 26, 27). Among women enrolled in the Women's Health Initiative, both fruit and vegetable intakes have been reported to be negatively correlated with Hannum AgeAccel and GrimAgeAccel (15, 26); however, the association with vegetable intake was not replicated in a subsequent Australian cohort study (27). Despite the inconsistent findings for vegetable intake, consistent negative correlations have been reported between circulating carotenoid concentrations and all 4 age acceleration metrics (14, 15, 26).
Our study focused on established healthy eating dietary indexes (32–35), which may provide more integrated assessments of food and nutrient intakes (46). Consistent with earlier null reports from the Melbourne Collaborative Cohort Study and the European Project on Nutrition in Elderly People clinical trial (27, 47), we found little evidence that the dietary indexes were associated with either the Hannum or Horvath AgeAccel metrics, which are based on the epigenetic clocks developed solely as predictors of chronological age. Conversely, we found robust associations between all 4 dietary indexes and the PhenoAgeAccel and GrimAgeAccel metrics, which are the epigenetic clocks developed as predictors of mortality. Prior studies have reported that PhenoAgeAccel and GrimAgeAccel are associated with other lifestyle factors, including body composition and physical activity (18), and such factors might interact with diet. We found some evidence for an interaction, with inverse associations between diet quality and PhenoAge Accel most apparent in women with lower levels of physical activity. These findings introduce the hypothesis that the health benefits of physical activity and diet quality may operate on the same epigenetic pathways that are captured by the PhenoAge clock.
Our study is not without limitations. First, our study is cross-sectional, with both diet and age acceleration assessed at enrollment. While the beneficial health effects of a good diet are well known, there is also evidence that poor health is associated with lower adherence to healthy eating (48–50). Another limitation is that our sample population only included self-identified non-Hispanic White women. This design limits potential confounding by ancestry or sex, but associations among other races/ethnicities or men cannot be examined. Although our use of an FFQ to assess diet has the potential for exposure misclassification, misclassification of healthy eating by FFQ tends to be more common among racial/ethnic minorities than Whites (51). Given that our sample population is fairly homogenous, we believe any exposure misclassification would most likely be nondifferential, potentially biasing results towards the null. Finally, although we investigated several recommendation-based healthy eating patterns, the diet indexes were strongly correlated, making it difficult to determine whether some specific dietary patterns are more beneficial than others.
In summary, we found that healthy eating patterns are associated with some methylation-based measures of biological age. The benefits of healthy eating appear to accrue particularly in women reporting lower levels of physical activity. These findings support the hypothesis that a higher diet quality may slow aging and lower disease and mortality risks. Prospective studies with DNA methylation and diet quality measurements at multiple time points can help clarify any remaining questions about directionality, and interventional studies can address the reversibility of biological age effects.
Supplementary Material
Acknowledgments
We thank Drs Kristen Moore and Nicole Niehoff for providing an internal review of the manuscript.
The authors’ responsibilities were as follows—DPS: is Principal Investigator of the Sister Study; DPS, JAT: designed the methylation subsample; JKK, Y-MMP: conceptualized the research question; JAK: managed the data; JKK: performed the analysis; JKK, JAT: drafted the original version of the manuscript. DPS, Y-MMP, JAK: provided significant intellectual contributions; and all authors read and approved the final manuscript. The authors report no conflicts of interest.
Notes
This work was supported by the NIH's Intramural Research Program, National Institute of Environmental Health Sciences (Z01-ES044005, Z01-ES049033, Z01-ES049032).
JKK and Y-MMP are co-first authors. DPS and JAT are co-senior authors.
Supplemental Figures 1–9 and Supplemental Table 1 are available from the “Supplementary data” link in the online posting of the article at https://academic.oup.com/ajcn/.
Abbreviations used: aHEI, Alternative Health Eating Index; aMed, Alternative Mediterranean; CpG, cytosine-phosphate-guanine; DASH, Dietary Approaches to Stop Hypertension; HEI, Healthy Eating Index.
Contributor Information
Jacob K Kresovich, Epidemiology Branch, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC, USA.
Yong-Moon Mark Park, Epidemiology Branch, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC, USA; Department of Epidemiology, Fay W. Boozman College of Public Health, University of Arkansas for Medical Sciences, Little Rock, AR, USA.
Jean A Keller, Westat, Durham, NC, USA.
Dale P Sandler, Epidemiology Branch, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC, USA.
Jack A Taylor, Epidemiology Branch, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC, USA; Epigenetic and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, NIH, Research Triangle Park, NC, USA.
Data Availability
Data described in the manuscript, code book, and analytic code will be made available upon request.
References
- 1. Reedy J, Krebs-Smith SM, Miller PE, Liese AD, Kahle LL, Park Y, Subar AF. Higher diet quality is associated with decreased risk of all-cause, cardiovascular disease, and cancer mortality among older adults. J Nutr. 2014;144(6):881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park YM, Choi MK, Lee SS, Shivappa N, Han K, Steck SE, Hebert JR, Merchant AT, Sandler DP. Dietary inflammatory potential and risk of mortality in metabolically healthy and unhealthy phenotypes among overweight and obese adults. Clin Nutr. 2019;38(2):682–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park YM, Steck SE, Fung TT, Zhang J, Hazlett LJ, Han K, Merchant AT. Mediterranean diet and mortality risk in metabolically healthy obese and metabolically unhealthy obese phenotypes. Int J Obes. 2016;40(10):1541–9. [DOI] [PubMed] [Google Scholar]
- 4. Park YM, Steck SE, Fung TT, Zhang J, Hazlett LJ, Han K, Lee SH, Kwon HS, Merchant AT. Mediterranean diet, Dietary Approaches to Stop Hypertension (DASH) style diet, and metabolic health in U.S. adults. Clin Nutr. 2017;36(5):1301–9. [DOI] [PubMed] [Google Scholar]
- 5. Siervo M, Lara J, Chowdhury S, Ashor A, Oggioni C, Mathers JC. Effects of the Dietary Approach to Stop Hypertension (DASH) diet on cardiovascular risk factors: A systematic review and meta-analysis. Br J Nutr. 2015;113(1):1–15. [DOI] [PubMed] [Google Scholar]
- 6. Schwingshackl L, Hoffmann G. Adherence to Mediterranean diet and risk of cancer: A systematic review and meta-analysis of observational studies. Int J Cancer. 2014;135(8):1884–97. [DOI] [PubMed] [Google Scholar]
- 7. Petimar J, Park YM, Smith-Warner SA, Fung TT, Sandler DP. Dietary index scores and invasive breast cancer risk among women with a family history of breast cancer. Am J Clin Nutr. 2019;109(5):1393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park YM, Steck SE, Fung TT, Merchant AT, Elizabeth Hodgson M, Keller JA, Sandler DP. Higher diet-dependent acid load is associated with risk of breast cancer: Findings from the sister study. Int J Cancer. 2019;144(8):1834–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeisel SH. Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr. 2009;89(5):1488S–93S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lai CQ, Parnell LD, Smith CE, Guo T, Sayols-Baixeras S, Aslibekyan S, Tiwari HK, Irvin MR, Bender C, Fei D et al. Carbohydrate and fat intake associated with risk of metabolic diseases through epigenetics of CPT1A. Am J Clin Nutr. 2020;112(5):1200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fasanelli F, Giraudo MT, Vineis P, Fiano V, Fiorito G, Grasso C, Polidoro S, Trevisan M, Grioni S, Krogh V et al. DNA methylation, colon cancer and Mediterranean diet: Results from the EPIC-Italy cohort. Epigenetics. 2019;14(10):977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, Taylor JA, Sandler DP. Air pollution, particulate matter composition and methylation-based biologic age. Environ Int. 2019;132:105071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White AJ, Kresovich JK, Xu Z, Sandler DP, Taylor JA. Shift work, DNA methylation and epigenetic age. Int J Epidemiol. 2019;48(5):1536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kresovich JK, Garval EL, Martinez Lopez AM, Xu Z, Niehoff NM, White AJ, Sandler DP, Taylor JA. Body composition and physical activity associations with multiple measures of epigenetic age acceleration. Am J Epidemiol. 2021;190(6):984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kresovich JK, Harmon QE, Xu Z, Nichols HB, Sandler DP, Taylor JA. Reproduction, DNA methylation and biological age. Hum Reprod. 2019;34(10):1965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kresovich JK, Xu Z, O'Brien KM, Weinberg CR, Sandler DP, Taylor JA. Methylation-based biological age and breast cancer risk. J Natl Cancer Inst. 2019;111(10):1051–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kresovich JK, Xu Z, O'Brien KM, Weinberg CR, Sandler DP, Taylor JA. Epigenetic mortality predictors and incidence of breast cancer. Aging. 2019;11(24):11975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian birth cohort 1936. Int J Epidemiol. 2015;44(4):1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kresovich JK, Martinez Lopez AM, Garval EL, Xu Z, White AJ, Sandler DP, Taylor JA. Alcohol consumption and methylation-based measures of biological age. J Gerontol A Biol Sci Med Sci. 2021. doi:10.1093/gerona/glab149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dugue PA, Bassett JK, Joo JE, Baglietto L, Jung CH, Wong EM, Fiorito G, Schmidt D, Makalic E, Li S et al. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am J Epidemiol. 2018;187(3):529–38. [DOI] [PubMed] [Google Scholar]
- 28. Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D'Aloisio AA, Suarez LM, Kleeberger CA, Shore DL, DeRoo LA, Taylor JA et al. The Sister study cohort: Baseline methods and participant characteristics. Environ Health Perspect. 2017;125(12):127003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Block G, Hartman AM, Dresser CM, Carroll MD, Gannon J, Gardner L. A data-based approach to diet questionnaire design and testing. Am J Epidemiol. 1986;124(3):453–69. [DOI] [PubMed] [Google Scholar]
- 30. Boucher B, Cotterchio M, Kreiger N, Nadalin V, Block T, Block G. Validity and reliability of the Block98 food-frequency questionnaire in a sample of Canadian women. Public Health Nutr. 2006;9(1):84–93. [DOI] [PubMed] [Google Scholar]
- 31. Bowman SA, Clemens JC, Friday JE, Thoerig RC, Moshfegh AJ. Food patterns equivalents database 2011–12: Methodology and user guide [Internet]. Beltsville, MD: Food Surveys Research Group, Beltsville Human Nutrition Research Center, Agricultural Research Service, USDA; 2014. Available from: www.ars.usda.gov/Services/docs.htm?docid=23871. [Google Scholar]
- 32. Fung TT, Chiuve SE, McCullough ML, Rexrode KM, Logroscino G, Hu FB. Adherence to a DASH-style diet and risk of coronary heart disease and stroke in women. Arch Intern Med. 2008;168(7):713–20. [DOI] [PubMed] [Google Scholar]
- 33. Krebs-Smith SM, Pannucci TE, Subar AF, Kirkpatrick SI, Lerman JL, Tooze JA, Wilson MM, Reedy J. Update of the Healthy Eating Index: HEI-2015. J Acad Nutr Diet. 2018;118(9):1591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, Stampfer MJ, Willett WC. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142(6):1009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fung TT, Rexrode KM, Mantzoros CS, Manson JE, Willett WC, Hu FB. Mediterranean diet and incidence of and mortality from coronary heart disease and stroke in women. Circulation. 2009;119(8):1093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sacks FM, Obarzanek E, Windhauser MM, Svetkey LP, Vollmer WM, McCullough M, Karanja N, Lin PH, Steele P, Proschan MA et al. Rationale and design of the Dietary Approaches to Stop Hypertension trial (DASH). A multicenter controlled-feeding study of dietary patterns to lower blood pressure. Ann Epidemiol. 1995;5(2):108–18. [DOI] [PubMed] [Google Scholar]
- 37. Estruch R, Ros E, Salas-Salvado J, Covas MI, Corella D, Aros F, Gomez-Gracia E, Ruiz-Gutierrez V, Fiol M, Lapetra J et al. Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med. 2013;368(14):1279–90. [DOI] [PubMed] [Google Scholar]
- 38. Sotos-Prieto M, Bhupathiraju SN, Mattei J, Fung TT, Li Y, Pan A, Willett WC, Rimm EB, Hu FB. Changes in diet quality scores and risk of cardiovascular disease among US men and women. Circulation. 2015;132(23):2212–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kresovich JK, O'Brien KM, Xu Z, Weinberg CR, Sandler DP, Taylor JA. Prediagnostic immune cell profiles and breast cancer. JAMA Network Open. 2020;3(1):e1919536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Brien KM, Sandler DP, Xu Z, Kinyamu HK, Taylor JA, Weinberg CR. Vitamin D, DNA methylation, and breast cancer. Breast Cancer Res. 2018;20(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu Z, Niu L, Li L, Taylor JA. ENmix: A novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3):e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu Z, Langie SA, De Boever P, Taylor JA, Niu L. RELIC: A novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics. 2017;18(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niu L, Xu Z, Taylor JA. RCP: A novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics. 2016;32(17):2659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging. 2016;8(9):1844–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piercy KL, Troiano RP, Ballard RM, Carlson SA, Fulton JE, Galuska DA, George SM, Olson RD. The physical activity guidelines for Americans. JAMA. 2018;320(19):2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu FB. Dietary pattern analysis: A new direction in nutritional epidemiology. Curr Opin Lipidol. 2002;13(1):3–9. [DOI] [PubMed] [Google Scholar]
- 47. Gensous N, Garagnani P, Santoro A, Giuliani C, Ostan R, Fabbri C, Milazzo M, Gentilini D, di Blasio AM, Pietruszka B et al. One-year Mediterranean diet promotes epigenetic rejuvenation with country- and sex-specific effects: A pilot study from the NU-AGE project. Geroscience. 2020;42(2):687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu EA, Toledo E, Diez-Espino J, Estruch R, Corella D, Salas-Salvado J, Vinyoles E, Gomez-Gracia E, Aros F, Fiol M et al. Lifestyles and risk factors associated with adherence to the Mediterranean diet: A baseline assessment of the PREDIMED trial. PLoS One. 2013;8(4):e60166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leung AWY, Chan RSM, Sea MMM, Woo J. An overview of factors associated with adherence to lifestyle modification programs for weight management in adults. Int J Environ Res Public Health. 2017;14(8):922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patino-Alonso MC, Recio-Rodriguez JI, Belio JF, Colominas-Garrido R, Lema-Bartolome J, Arranz AG, Agudo-Conde C, Gomez-Marcos MA, Garcia-Ortiz L, Group E . Factors associated with adherence to the Mediterranean diet in the adult population. J Acad Nutr Diet. 2014;114(4):583–9. [DOI] [PubMed] [Google Scholar]
- 51. Olendzki B, Procter-Gray E, Magee MF, Youssef G, Kane K, Churchill L, Ockene J, Li W. Racial differences in misclassification of healthy eating based on food frequency questionnaire and 24-hour dietary recalls. J Nutr Health Aging. 2017;21(7):787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript, code book, and analytic code will be made available upon request.