Abstract
Recently, a novel coronavirus, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has raised global concerns, being the etiological agent of the current pandemic infectious coronavirus disease 2019 (COVID-19). Specific prophylactic treatments like vaccines, have been authorized for use by regulatory bodies in multiple countries, however there is an urgent need to identify new, safe, and targeted therapeutics as post-exposure therapy for COVID-19. Among a plethora of potential pharmacological targets, the angiotensin-converting enzyme 2 (ACE2) membrane receptor, which plays a crucial role in viral entry, is representing an attractive intervention opportunity for SARS-CoV-2 antiviral discovery process.
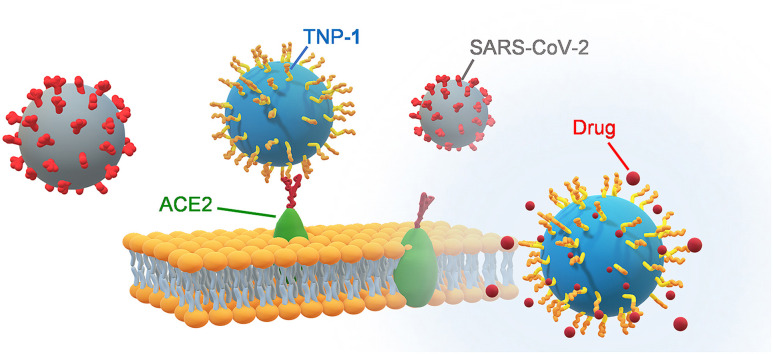
In this scenario, we envisioned that binding to ACE2 by multivalent attachment of ligands to nanocarriers incorporating antiviral therapeutics, it would increase receptor avidity and impart specificity to these nanovectors for host cells, particularly in the pulmonary tract, which is the primary entry route for SARS-CoV-2.
Herein, we report the design and development of novel polymeric nanoparticles (NP), densely grafted with various ligands to selectively bind to ACE2, as innovative nanovectors for targeted drug delivery. We first evaluated the impact of these biocompatible targeted NP (TNP) on ligand binding toward ACE2 and measured their competition ability vs a model of spike protein (Lipo-S1). Next, we tested the effectiveness of the most performing nanoprotopype, TNP-1, loaded with a model anti-SARS-CoV-2 drug such as remdesivir (RDV), on antiviral activity against SARS-CoV-2 infected Vero E6 cells. The RDV-TNP-1 exhibited a significantly improved antiviral effect compared to RDV at the same concentration. Interestingly, unloaded TNP (TNP-1E) also exhibited a basal antiviral activity, potentially due to a direct competitive mechanism with viral particles for the ACE2 binding site. We also measured the anti-exopeptidase activity of TNP-1E against ACE2 protein. Collectively, these insights warrant in-depth preclinical development for our nanoprototypes, for example as potential inhalable drug carriers, with the perspective of a clinical translation.
Keywords: SARS-CoV-2, ACE2, Antiviral activity, Targeted nanoparticles, Targeted drug delivery
Graphical abstract
Abbreviations used
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- COVID-19
coronavirus disease 19
- ACE2
human angiotensin converting enzyme 2
- TMRPSS
Transmembrane Serine Proteases (TACE/ADAM-17) Tumor necrosis factor-alpha-converting enzyme
- RAS
renin-angiotensin- system
- Ang II
angiotensine II
- Ang (1–7)
angiotensin-(1–7)
- AT1R
angiotensine II type 1 receptor
- NP
nanoparticles
- TNP
targeted nanoparticles
- PLGA
poly-(D,L-lactide-co-glycolide)
- PLGA-PEG
poly(D,L-lactic-co-glycolic acid)-block-poly(ethylene glycol)
- PCL
poly(epsilon-caprolactone)
- RDV
remdesivir
- Boc2O
di-tert-butyl dicarbonate
- DIPEA
dihysopropylethylamine
- TEA
triethylamine
- CH2Cl2
dichloromethane
- CHCl3
chloroform
- DCE
dichloroethane
- THF
tetrahydrofuran
- NaB(OAc)3H
sodium triacethoxyborohydride
- DMF
dimethylformamide
- DMSO
dimethysulphoxyde
- EDC
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide
- h
hours
- NMR
nuclear magnetic resonance
- NOESY
Nuclear Overhauser Effect Spectroscopy
- COSY
1H–1H Correlation Spectroscopy (COSY)
- HSQC
Heteronuclear Single Quantum Correlation
- MS
mass spectrometry
- HRMS
high-resolution mass spectrometry
- HPLC
high performance liquid chromatography
- Rho
rhodamine
- C6
Coumarin-6
- RFU
relative fluorescence units
- Lipo-S1
Rho-loaded liposome-derived SP
- ELISA
enzyme-linked immunosorbent assay
- CPE
cytopatic effect
- MOI
multeplicity od infection
- DMEM
Dulbecco's modified eagle medium
- FBS
Fetal Bovine Serum
- qRT-PCR
quantitative real-time polymerase chain reaction
- DAPI
4',6-diamidino-2-phenylindole
1. Introduction
Novel beta-coronavirus SARS-CoV-2 is the pathogenic agent responsible for coronavirus disease 2019 (COVID-19) and for the current global pandemic [[1], [2], [3], [4]]. This emergency highlights the urgent need to expand and focus current research tools on “neglected infectious diseases” to prevent future potential epidemics [[5], [6], [7], [8]]. Impressive efforts have been done at elucidating the viral structure, life cycle and to identify the potential targets in SARS-CoV-2 [[9], [10], [11], [12], [13], [14]]. Although some promising drug candidates are expected soon [15], to date not novel approved antiviral pharmaceutical drugs exist. Only one drug, the RNA-dependent RNA polymerase (RdRp) inhibitor remdesivir (RDV [15,16], brand name Veklury, developed by Gilead Sciences), with broad-spectrum activity against RNA viruses, has been issued by the U.S. Food and Drug Administration (FDA) for emergency use authorization (EUA) as a treatment for hospitalized patients with severe COVID-19 symptoms [[17], [18], [19], [20], [21]]. Recent data showed that RDV can enhance clinical outcomes [22], but its efficacy is limited by its poor physicochemical and pharmacokinetic properties that also prevent the feasibility of alternative routes such oral and intramuscular administration [23]. For this reason, the pulmonary delivery route by the inhalation may maximize direct delivery of RDV to the target site. This would avoid gastrointestinal proteolysis and hepatic first-pass metabolism, boosting local antiviral activity in the lungs, and limiting systemic off-target effects, thus improving its therapeutic profile [24,25] as well as that of other antiviral agents currently involved in the development process. For example, since systemic adverse effects such as hepatotoxicity preclude escalation of the Remdesivir dose to more than 200 mg/day, (e.g. 100–200 mg of RDV is the current intravenous dose regimen in humans to achieve effective levels of drug against SARS-CoV-2), other therapeutic strategies which combine tools and different administration routes are needed [[25], [26]].
Drug loaded polymer nanoparticles (NP) are the most recent advanced technology of drug formulations towards pulmonary delivery of various therapeutics due to their abilities to overcome drug resistance and to improve pharmacokinetics and biodistribution profiles of the administered drugs, thereby maximizing direct delivery and retention at the sites of infection while minimizing systemic exposure, which might provide pharmacological benefits [[27], [28], [29], [30]]. In the face of drug development, a promising anti-SARS-CoV-2 prototype must reach its site of action (e.g., lung cells and tissues) in sufficient quantity to block the virus replication and limit toxicological issues.
In this work, we have developed a novel nanotechnology platform by engineered NP as a powerful strategy to selectively deliver payload antiviral therapeutics to cellular targets primarily involved in the route of SARS-CoV-2 infection process, with the potential to improve treatment of COVID-19 to overcome intrinsic limitations of drugs in the context of a clinical translation [[31], [32], [33]].
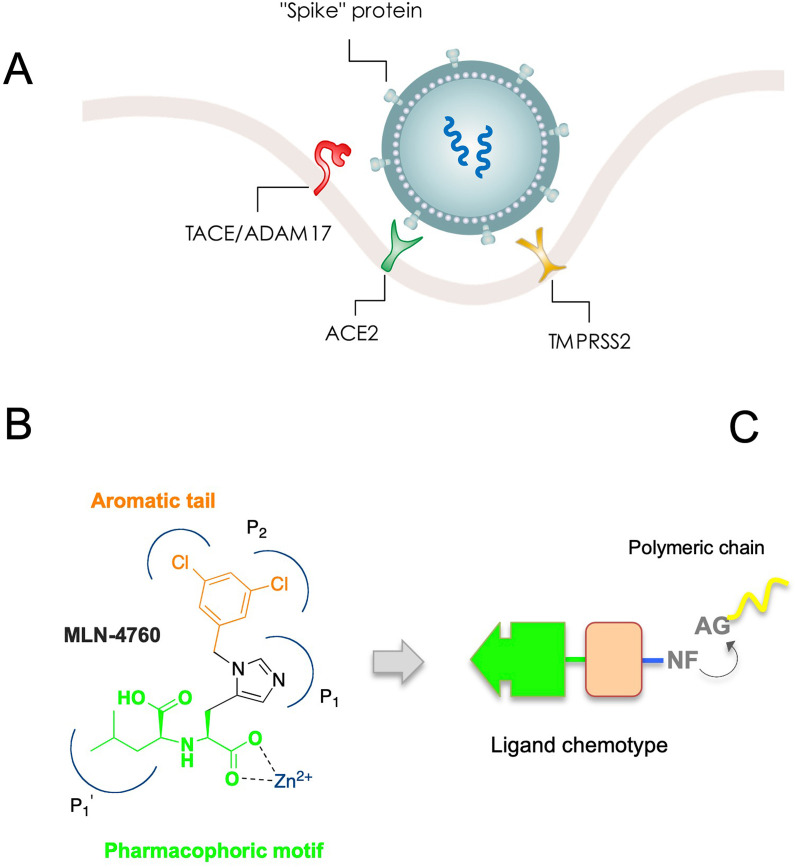
Among the potential pharmacological targets in SARS-CoV-2, the angiotensin-converting enzyme 2 (ACE2) membrane receptor is still relatively unexplored [34,35]. SARS-CoV-2 enters cells through the interaction of its surface “spike” protein (SP) receptor binding domain (RBD) with the host receptor ACE2 (Chart 1 A ) [[36], [37], [38]], in which high expression and cell surface localization are particularly observed in lung alveolar epithelial cells, but also present in the heart, kidney, and in other different cells and tissue [34]. Subsequent activation/proteolytic cleavage of the SP into S1 and S2 domains by the host cell membrane-associated serine proteases such as transmembrane protease serine 2 (TMPRSS2) [39], allows definitive cell entry by endocytosis into the host cells, where the viral nucleocapsid with its genome payload is released into the cytoplasm (Chart 1A). We envisioned that the biological process pursued by the virus to recognize and infect the human host cells could be reproduced and translated into a therapeutic strategy.
Chart 1.
(A) Simplified model of SARS-CoV-2 entry into the host cell, with potential pharmacological targets. Binding of the spike protein (SP) to the cellular ACE2 receptor followed by enzymatic activation/cleavage of the SP into S1 and S2 domains by cell membrane-associated serine proteases such as TMPRSS2 results in membrane attachment and subsequent receptor-mediated internalization, which triggers an increased TACE/ADAM17-mediated ACE2 shedding. (B) Structure of the ACE2 inhibitor MLN-4760 and pharmacophoric features. (C) Chemotype of targeting ligands and conjugation with co-polymers for nanoformulation.
This enzyme-related carboxypeptidase (i.e. ACE2) has high homology with the other ACE isoform, with approximately 40% sequence identity between ACE2 and ACE, and 60% similarity in the zinc binding on the catalytic domains [[40], [41], [42]]. It plays a key role in balancing the physiological levels of vasopressor octapeptide angiotensin II (Ang II), a component of the classical renin-angiotensin system (RAS), and the angiotensin (1–7) (Ang (1–7) [43]. In particular, by catalyzing the conversion of Ang II to Ang (1–7), ACE2 constitutes a regulator pathway of cardiovascular homeostasis [43]. Specifically, Ang (1–7) acts on Mas receptors producing cardiovascular protective functions and counter-regulating the detrimental effects of ACE/Ang II axis [44,45]. Thus, the physiophatological role of ACE2 in the homeostatic regulation of cardiovascular system, as well as the vascular complications associated with COVID-19 disease, should be taken into consideration in studying its potential as a pharmacological target. For example, the implication of antihypertensive drugs targeting the cognate ACE, as well as Ang II-type 1 receptor (AT1R) blockers, to modulate the expression/function of ACE2, in order to putatively interfere with SARS-CoV-2 infection, have been disclosed recently [46]. The interaction of SP of SARS-CoV with ACE2 receptors can affect the RAS, inducing a cellular “protective” ACE2 shedding feedback response that initially attenuates viral entry [47]. Again, tumor necrosis factor-alpha-converting enzyme (TACE/ADAM17)-mediated ACE2 shedding, in combination with ACE2 enzymatic activity, have been shown to correlate positively with viral infection and related disease complications [[48], [49], [50]]. Collectively, these and other reports [[51], [52], [53]] indicate that, to date, it is not clear what is the downstream effect of the ACE2, and no clinical and consistent studies were reported for ACE2 inhibition, particularly in the interface with SARS-CoV-2 infection. However, the possibility to target ACE2 as an antiviral strategy appears to be attractive, and ACE2 targeted SARS-CoV-2 SP blockers hold greater promise. A significant example is represented by the lipoglycopeptide antibiotic dalbavancin, which directly binds to ACE2 with high affinity, thereby blocking the SARS-CoV-2 SP/ACE2 interaction, effectively inhibiting SARS-CoV-2 replication in vitro, and preventing viral infection and histopathological injuries in in vivo models [54]. Although other authors argue that it can be conceivable that targeting ACE2 provides an effective means for COVID-19 prevention and treatment [55], this approach objectively requires careful consideration of the pros and cons of the availability of specific ACE2 modulators/targeting in the therapeutical armamentarium, as well as the clinical contexts in which they are likely to be effective.
Among various small molecules capable of targeting ACE2 [13], to date, one of the most effective compounds is the His-Leu mimetic MLN-4760 (Chart 1B) [56], which is reported to inhibit the catalytic activity of human ACE2 with an IC50 of 440 pM [56] (or low nanomolar concentration in other assays) [57], and it has excellent selectivity versus related carboxypeptidase enzymes [43,44]. Structurally, the S,S diastereoisomer demonstrated the highest potency, compared to the other isomers [56,57]. However, no antiviral SARS-CoV-2 data for MLN-4760 are available.
We thus hypothesized that ACE2 could be used as a key structure for targeted drug delivery by polymeric nanoparticles (NP), to improve the efficient transport of bioactive molecules/drugs and their specific and strong release in the ACE2 expressing cells, the primary target by SARS-CoV-2, which are expected to be infected by the virus. In fact, in our previous studies [58,59], inspired by Paul Ehrlich's concept of “magic bullet” [60], we shifted the targeting paradigm in nanomedicine to enable enhanced selectivity of nanosystems for cells and tissues by a triple (tissue, cellular, molecular) targeting approach [31,61,62].
On this idea, we recognised that a ligand capable of targeting ACE2 can be used to engineer nanosystems to obtain targeted NP (TNP). A suitable ligand chemotype should contain crucial features such as an appropriate pharmacophoric motif involved in chelation with the zinc metal ion on the active site, and a tail bearing a nucleophilic function (NF) to link with activated group (AG) of polymers used for nanoformulation (Chart 1C).
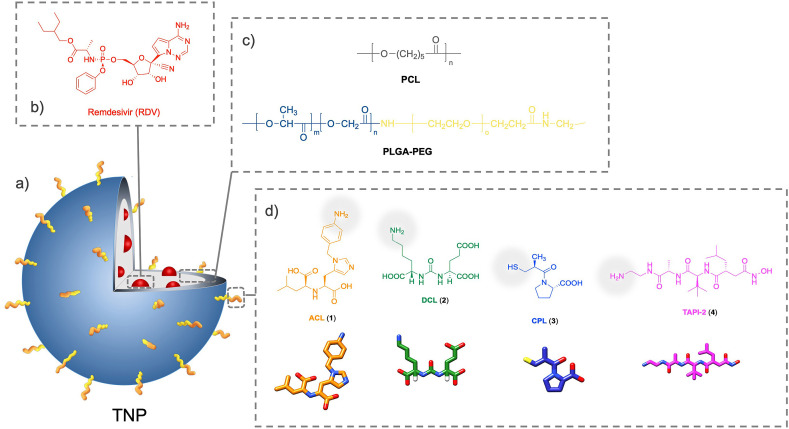
Herein, we report the design and development of novel biocompatible TNP covered by small organic molecules as targeting ligands in their polymeric shell surface, to selectively bind to ACE2 highly expressed in a model cell line (Fig. 1). We used pseudo-tri-block-copolymer PLGA-PEG-ligands as well as the parent di-block-copolymer PLGA-PEG-COOH to generate viral size comparable NP, which were characterized for their technological parameters, and evaluated for their biological profile by ACE2 binding affinity, competition assays (TNP vs SP model), ACE2 inhibition, antiviral activity for the most performing nanosystem loaded (and nonloaded) with RDV, and uptake and internalization into host cells.
Fig. 1.
Schematic representation of the design and development of (a) targeted nanoparticles (TNP). (b) Encapsulated model drug (remdesivir, RDV). To perform binding/upkate studies fluorescent dye (C6 or Rho) is loaded rather than the drug. (c) The PCL and PLGA-PEG blended polymeric matrix. (d) The targeting ligands (ACL, 1; DCL, 2; CPL, 3, and TACE-2, 4). The conjugation terminus site (amino or thiol functionality) for each ligand is also indicated (pale grey).
2. Results and discussion
2.1. Design of targeted nanoparticles
We designed the NP to target the ACE2 enzyme for site-directed antiviral drug delivery to cells potentially and primarily infected by SARS-CoV-2, with the aim of optimizing and validating their binding affinity against ACE2, and the antiviral impact of the most effective prototype. As a first step, we incorporated a fluorescent dye (Coumarin-6, C6) into polymeric NP conjugated to targeting ligands (Fig. 1), which allowed easy tracking.
According to the aforementioned plan (Chart 1C), we structurally modified the ACE2 inhibitor MLN-4760 to generate the (S)-2-(((S)-2-(1-(4-aminobenzyl)-1H-imidazole-5-yl)-1-carboxyethyl)amino)-4-methylpentanoic acid (ACL, 1), on which the 3,5-dichlorobenzyl moiety of MLN-4760 was replaced by a nucleophilic 4-amino-benzyl funtionality. To ascertain the set of putative ACE2 ligands, we also selected two other structures: a) the pseudomimetic dipeptide N-[N-[(S)-1,3-dicarboxypropyl]carbamoyl]-(S)-lysine (DCL, 2) [59,63,64], previously used as a prostate-specific membrane antigen (PSMA) targeting agent (PSMA is a glutamate carboxypeptidase which is highly expressed in prostate cancer cells and in nonprostatic tumor neovasculature) [65], and b) the ACE1 inhibitor captopril (CPL, 3), which binds to the ACE active site [66]. Finally, since TACE/ADAM17 system take part in viral shedding [50], we additionally tested a specific TACE inhibitor, i.e. the pseudo-peptide N-(R)-(2-(hydroxyaminocarbonyl)methyl)-4-methylpentanoyl-L-t-butyl-glycine-L-alanine 2-aminoethyl amide (TAPI-2, 4) [13,67], as a ligand to bind to this putative additional target cell membrane (Fig. 1), which was also used as a negative control for ACE2 binding.
Based on the structural X-ray information of the ACE2 binding site with the inhibitor bound MLN-4760 [68] (for details see Experimental Section) and the abovementioned considerations, a comparative docking simulation study on ligands 1, 2, and 3, and on their respective polymeric constructs (PEG-ACL, PEG-DCL, and PEG-CPL, Fig. S1A) simulating the NP’ terminus anchors (an amide bond for 1 and 2, and the maleimide conjugation for 3), was performed.
First, free 1, 2, and 3, were docked to the ACE2 active site, and crucial protein-ligand interactions were analyzed (Fig. 2A, C, E). In parallel, the corresponding interactions of alkyl-conjugated-ligands (PEG-ACL, PEG-DCL, and PEG-CPL), were simulated (Fig. 2B, D, F), and the binding modes of these model structures were compared.
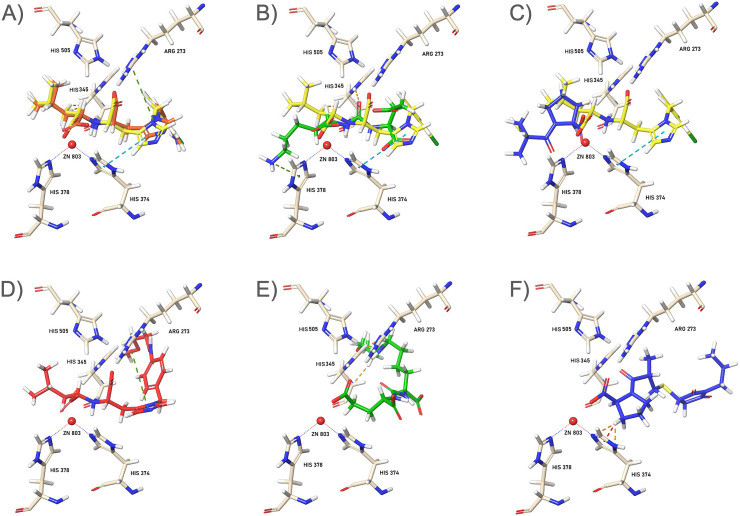
Fig. 2.
3D interaction images of the ligands 1–3 (top) and their PEGylated models (bottom) in the ACE2 active site. The inhibitor MLN-4760 (yellow) is used for comparison. (A) Binding modes of ACL (1, orange), (B) DCL (2, green), and (C) CPL (3, blue), superimposed with MLN-4760 onto the ACE2 receptor. (D, E, and F) Respective alkylated-ligands (Fig. S1A): PEG-ACL (orange), PEG-DCL (green), and PEG-CPL (blue), simulating the polymeric termini of NP within the ACE2 tunnel. Zinc ion cofactor is represented as red sphere.
Analysis of the docking results showed an overlapping alignment for both 1 (1 -S,S) and MLN-4760 within the active site (Fig. 2A), and specific binding interactions of the inhibitors with ACE2 were in agreement with that of MLN-4760, as previoulsy reported [68]. Both ligands showed a consistent interaction with respect to the dicarboxylate moiety: the active site Zn2+ metal ion is well coordinated by the proline-based carboxylate oxygens, where the hystidine-derived carboxylate established strong hydrogen lengths with the His 345.
DCL (2) also revealed a good energy binding score but with an opposite disposition within the active site: the aminoalkyl side chain exends along the deeply buried active site (Fig. 2B), while the carboxylate groups directed towards the zinc ion. Again, CPL (3) was well accomodated within the site where the proline carboxylate establishes interations with the metal ion cofactor (Fig. 2C). However, due to the small molecular size, the ligand 3 showed the highest binding energies compared to the other ligands (ACE2 binding affinities were −11.268, −10.402, and −6.278 kcal/mol, for 1-S,S, 2, and 3, respectively) as well as to the parent MLN-4760 (ACE2 binding energy: −10.193 kcal/mol).
With regards to the polyether-modified ligands (PEG-ACL, PEG-DCL, and PEG-CPL, Fig. 2D, E, and F, respectively), their alyphatic chains protrude outside the tunnel, while the pharmacophoric motifs (for all compounds) pointed toward the active site metal ion. The comparative docking study showed that the insertion of a PEG linker chain might have had a significant impact on ligand binding to ACE2, establishing a most favourable orientation of ligands for binding (especially for 2, which aligned similarly with the other models). Although all polymer-conjugated ligands shared a slightly lower ACE2 binding affinity (in terms of absolute energy values) with respect their respective free ligand counterparts, they maintain a comparable profile. We also evaluated the S,R-1 diastereosomer, which did show a slightly reduced affinity (−9.431 kcal/mol) than 1 -S,S, as indicated in the previous inhibition studies [56,57].
Following the same strategy, the compound 4 and its polymeric model (PEG-TACE-2, Fig. S1A) were docked to TACE receptor [69], and the effect of polymer conjugation was evaluated (Fig. S1B). Both the ligand 4 and its polymeric-based model mutually matched within the pocket, and a similar orientation pattern and disposition within the TACE catalytic site was observed. These evalutations support that the polymeric conjugated inhibitor can be consequently introduced onto the NP surface.
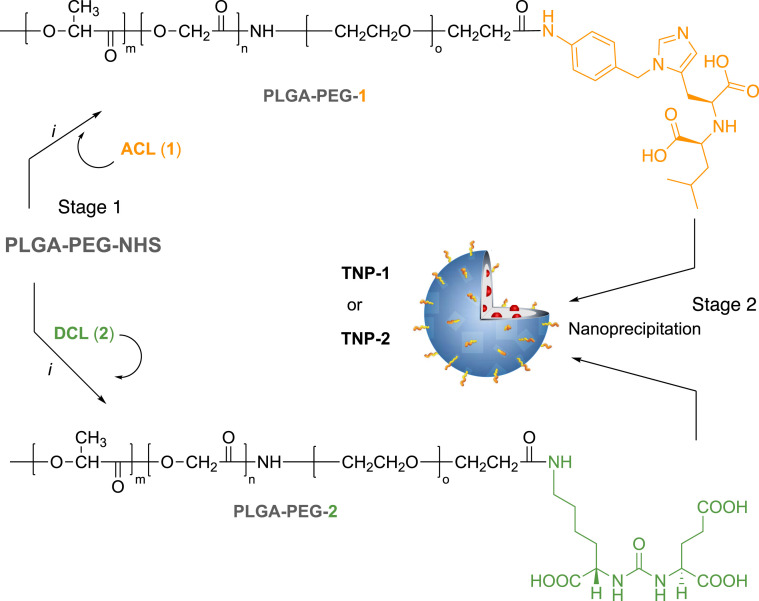
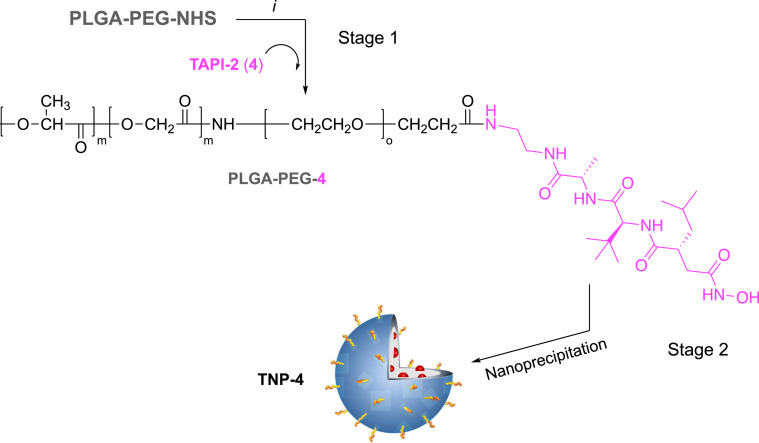
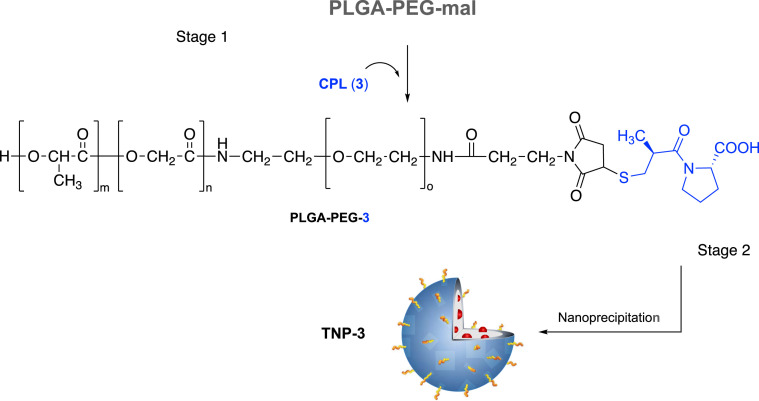
Ligands 1, 2, and 4 contain an amine functional group which allows for amide bond formation with carboxyl terminal groups of polymer monomer. To obtain a better control of ligand-polymer functionalization, these ligands were conjugated to their respective polymeric systems prior to nanoformulation. We used poly(D,L-lactide-coglycolide)poly(ethylene glycol) carboxylic acid (PLGA-PEG-COOH) as a biocompatible/biodegradable polymer for the preparation of NPs. PEGylation it is known to confer immune shielding properties to nanosystems, acting as a spacer between the targeting ligand and the NP surface. This starting carboxylate-functionalized di-block-copolymer was activated to PLGA-PEG-NHS for the preparation of the tri-block-co-polymers (i.e., PLGA-PEG-1, PLGA-PEG-2, and PLGA-PEG-4) to obtain the TNP (Scheme 1, Scheme 3 ). Else, the thiol containing CPL (3) was reacted with maleimide terminal groups of PEG in the PLGA-PEG-maleimide (PLGA-PEG-mal) to obtain PLGA-PEG-3 (Scheme 2 ).
Scheme 1.
Synthesis of copolymers PLGA-PEG-1 and PLGA-PEG-2 (Stage 1) and nanoformulation (Stage 2). Reagents and conditions: (i) ACL (1, 10 equiv.) or DCL (2, 4 equiv.), DIPEA, DMF, N2, r. t. for 24 h.
Scheme 3.
Synthesis of copolymer PLGA-PEG-4 (Stage 1) and nanoformulation (Stage 2). Reagents and conditions: (i) TAPI-2 (4, 4 equiv.), DIPEA, DMF, N2, r. t. for 24 h.
Scheme 2.
Synthesis of copolymer PLGA-PEG-3 (Stage 1) and nanoformulation (Stage 2). Reagents and conditions: (i) CPL (3, 1.5 equiv.), ACN:DMF (1:1), N2, r. t. for 24 h.
2.2. Synthesis of pseudo-tri-block-copolymers
We prepared four sets of prefunctionalized biopolymer, the pseudo-tri-block-copolymers PLGA-PEG-1, PLGA-PEG-2, PLGA-PEG-3, and PLGA-PEG-4 (Scheme 1, Scheme 2, Scheme 3, Stage 1).
Conjugation of targeting agents 1, 2, and 4 to functional PEG termini was performed with the use of electrophilic NHS esters of PEG carboxylic acids, i.e. PLGA-PEG-NHS, in DMF in the presence of DIPEA, via standard carbodiimide/NHS-mediated chemistry (Scheme 1, Scheme 3). For conjugation, increased equivalent ratio of PLGA-PEG-NHS and 1 (from 1:4 to 1:10) improved the reaction efficiency. Preparation of activated block copolymer PLGA-PEG-NHS was carried out using a previous procedure (Scheme S1) [58,64].
Characterization of copolymers was performed by 1H NMR, where the resonance shifts of the characteristic PLGA-PEG backbone were detected (Fig. S2, S3, and S5). Specifically, overlapping signals at 1.46–1.56 ppm are assigned to the lactide methyl repeat units of PLGA, for all copolymers. The multiplets at 5.16–5.29 and 4.82–4.96 ppm correspond to the lactide methine (CH) and to the glycolide protons (CH2) of PLGA. Singlet signals centered at 3.51 ppm confirmed the sequence of ethoxy repeat units of PEG. 1H NMR spectra recorded for PLGA-PEG-1/2/4 also exhibited a series of multiple peaks attributable to the pattern signals of each free ligand (Fig. S2, S3, and S5, and Experimental Section), which supported the successful conjugation of 1, 2, and 4 to PLGA-PEG copolymer.
Conjugation of the targeting agents 3 to carboxylate PEG terminal group was performed using the electrophilic maleimide functionality of PEG activated carboxylic acids (Scheme S2). Reaction between the terminal –SH group of CPL (3) with PLGA-PEG capped maleimide resulted in the formation of PLGA-PEG-3 copolymer, which was synthesized by reacting the activated PLGA-PEG-mal with 3 in DMSO (Scheme 2, Stage 1). The intermediate PLGA-PEG-mal was prepared using a two-step reaction by conjugating bifunctional PEG, NH2-PEG-mal, to activated PLGA-NHS polymer, following a procedure previously described by our group (Scheme S1, and Supporting Information) [59]. The structure of PLGA-PEG-3 was also confirmed by 1H NMR spectroscopy, which revealed resonance shifts at 1.44–1.50, 3.51, 4.83–4.97, and 5.17–5.27 ppm, characteristics of PLGA-PEG backbone (Fig. S4). After conjugation, we observed a clear reduction of the maleimide methine free proton at 6.70 ppm of the PLGA-PEG-mal. Simultaneously, the appearance of a series of partially overlapped multiple peaks, especially in the aliphatic zone, attributable to the 3 signal pattern (Fig. S4 and Experimental Section for details) were detected.
2.3. Synthesis of ligands 1 and 2
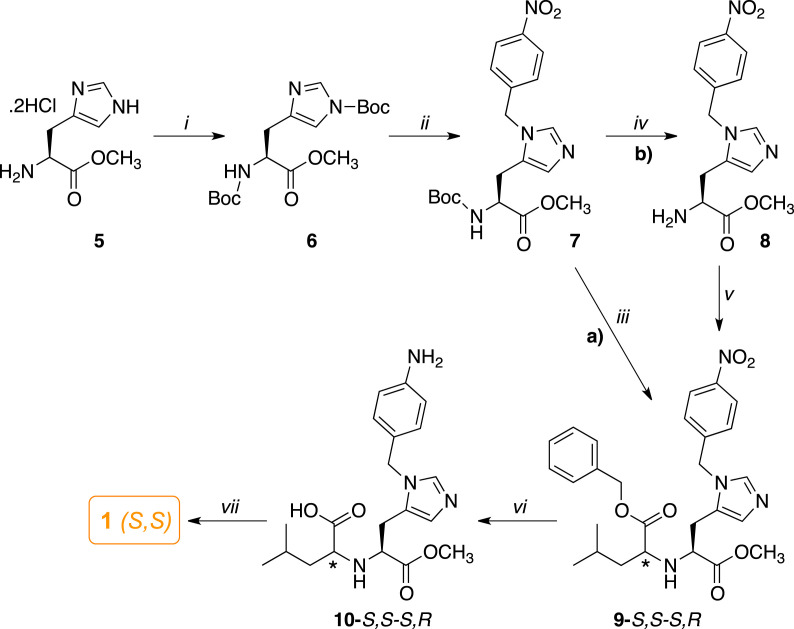
Compound 1 was synthesized by revisiting the same methodology reported for the synthesis of MLN-4760 (Scheme 4 ) [56].
Scheme 4.
Synthetic route for the preparation of 1. Reagents and conditions: (i) Boc2O, TEA, MeOH, r. t., 24 h; (ii) 4-NO2-benzyl alcohol, CH2Cl2, DIPEA, trifluoromethane sulfonic anhydride, −78 °C, N2, 24 h; a) (iii) 4 N HCl in dioxane, r. t., 2 h, next DCE, keto-ester (benzyl 4-methyl-2-oxo-pentanoate (11), r. t. 1 h, then, NaB(OAc)3H, r. t., 24 h; b) (iv) 4 N HCl in dioxane, r. t., 2 h, 1 N NaOH, CH2Cl2; (v) DCE, 11, r. t., 1 h, then, NaB(OAc)3H, acetic acid, r. t., 24 h; (vi) (i) HCOONH4, 10% Pd–C, r. t., 1.5 h; (vii) 1 N NaOH, EtOH, r. t., 12 h.
In order to regioselectively provide the N-3 imidazole substitution, the readily available (S)-histidine methyl ester (5) was fully protected with Boc2O in methanolic solution and TEA to afford the di-Boc-derivative 6. Alkylation of the imidazole nitrogen was conducted by treating 6 with the triflate of a 4-nitro benzyl alcohol in dichloromethane solution and DIPEA at −78 °C, which provided the N-benzyl-histidine derivative 7. This intermediate was converted to diester 9 as a mixture of diastereoisomers by a tandem reaction (Scheme 4, method a), where 7 was first treated with 4 N HCl in dioxane to remove the Boc protecting group, then the di-hydrochloride salt was coupled with the benzyl 4-methyl-2-oxo-pentanoate (11, Scheme S3) in the presence of NaB(OAc)3H [70]. Alternatively, 9 was obtained by a Boc deprotection to give the neutral aminoester 8, which was isolated (Scheme 4, method b), and then submitted to the same reductive amination with the α-keto ester, in the presence of acetic acid, to give 9 as a mixture of S,S-R,S diastereoisomers in comparable overall yields. Reduction of the nitro group in position 4 of the benzyl moiety with ammonium formate and Pd/C, followed by chromatographic purification, furnished the amino acid diastereoisomers 10. Hydrolysis and purification of 10 afforded the desired ligand 1, isolated in S,S configuration, which was fully characterized by NMR, MS, HPLC, and elemental analyses (see in Experimental Section and Fig. S6-S11). To support the S,S stereochemistry, the structure of 1 diastereoisomer was determined by 2D-NOESY (Fig. S7). We realized that preferable conformations of this diastereoisomer led the position of the methyl pentanoic acid moiety in the same side of the imidazole ring, thus NOE correlation between the imidazole proton in the position 4 of the ring and the isopropyl group could be detectable.
On the other hand, the ligand DCL (2) was prepared following a procedure previously reported by us and by others [58,64], with slight modifications (Scheme S4, and Figs. S11 and S12).
2.4. Nanoformulation and characterization of nontargeted and targeted NP
To evaluate the binding affinity, the competition against the lentivirus-spike and cellular uptake of the NP, five batches of C6-encapsulated NP were successfully prepared using a modified nanoprecipitation method as previously described (Schemes 1, 2 and 3, stage 2) [58,64,71]. We used a blend of hydrophobic polymer PCL and amphiphilic block PLGA-PEG copolymers (PLGA-PEG-COOH, PLGA-PEG-1, PLGA-PEG-2, PLGA-PEG-3, and PLGA-PEG-4) to prepare nontargeted and targeted NP. During the nanoprecipitation in water, the polymers self-assemble to form NP, in which the hydrophobic PLGA blocks interact with the PCL into a core, minimizing their exposure to aqueous surroundings [72]. Simultaneously, the hydrophilic termini of the polymers (i.e. the carboxylate surfaces and PEG-1-4 flexible moieties) extend from the shell to stabilize the core by preventing degradation and aggregation. As mentioned above, PEG should impart “stealth” properties to the nanoconstructs, and the PEG methylene chain enables high-affinity binding of ligands to ACE2. Moreover, the spacer portion of the polymer would remain outside the tunnel as long as the pharmacophoric motif of 1, 2, 3, or 4 is anchored to the protein active site [73].
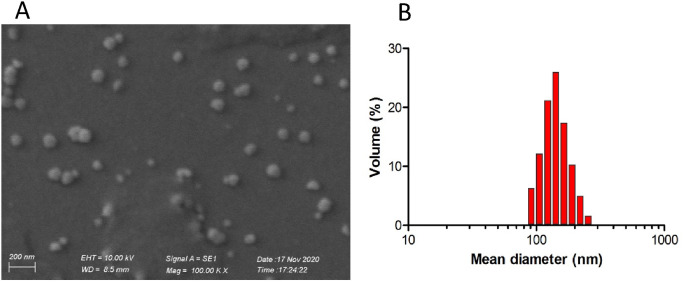
All prepared NP batches showed well dispersed particles characterized by similar morphological properties, with a smooth surface and spherical shape (see below in Fig. 5a as example). As summarized in Table 1 , the mean diameter of the NP ranged from 103 nm to 130 nm (analogous to a viral particle size), without significant differences within batches prepared with different copolymer conjugates. Only NP formulated with ACL (1) conjugated polymer exhibited significantly lower particle size (102.97 ± 7.23 nm) compared with nontargeted batches.
Fig. 5.
SEM image of RDV-loaded TNP-1 (A) and its hydrodynamic size distribution (B), chosen as examples. The scale bar is 200 nm.
Table 1.
Average Diameter, Polydispersity Index (PDI), percentage of Encapsulation Efficiency (EE %), Loading Capacity (LC %), Yield of Production (YP %), and ζ potential (mV) of dye-loaded (C6) formulated NP.
| Batch | Mean diameter (nm) | PDI | EE (%) | LC (%) | YP (%) | ζ potential (mV) |
|---|---|---|---|---|---|---|
| C6-NP-0 | 130.21 ± 5.84 | 0.125 ± 0.01 | 76.42 ± 3.16 | 0.038 ± 0.002 | 72.93 ± 4.67 | −25.2 ± 3.9 |
| C6-TNP-1 | 102.97 ± 7.23∗ | 0.140 ± 0.03 | 73.86 ± 2.19 | 0.037 ± 0.001 | 65.44 ± 2.39 | −28.8 ± 4.2 |
| C6-TNP-2 | 118.87 ± 9.47 | 0.113 ± 0.05 | 73.41 ± 0.91 | 0.037 ± 0.001 | 68.67 ± 4.77 | −27.8 ± 5.4 |
| C6-TNP-3 | 111.29 ± 7.47 | 0.107 ± 0.02 | 73.55 ± 3.37 | 0.038 ± 0.002 | 70.67 ± 1.67 | −24.5 ± 6.1 |
| C6-TNP-4 | 119.32 ± 8.75 | 0.126 ± 0.04 | 74.93 ± 3.38 | 0.037 ± 0.002 | 74.20 ± 3.52 | −21.1 ± 5.7 |
Each value is the mean ± SD of triplicate determinations. ∗Significantly different from C6-NP-0 at p < 0.05.
In all cases, the NP dispersions were characterized by low PDI values (below 0.2) (Table 1), indicating a narrow and unimodal distribution, as a typical behavior of monodispersed systems (see below in Fig. 5b as example) [74]. We detected a negativeζpotential for all bathches of C6-TNP-0/1/2/3, due to the presence of carboxylate groups on the PLGA-PEG termini. Also, C6-TNP-4 showed a negative ζ potential, which is coherent with their surface hydroxamic funtionalities. With regard to the dye entrapment efficiencies, the obtained values resulted in about 75%, with no significant differences between the different NP batches, suggesting a good affinity between the polymeric blend and the dye loaded molecules. Moreover, yield of production ranging from 65 to 74% were obtained for all preparations over multiple experiments.
2.5. Evaluation of ACE2 binding by NP
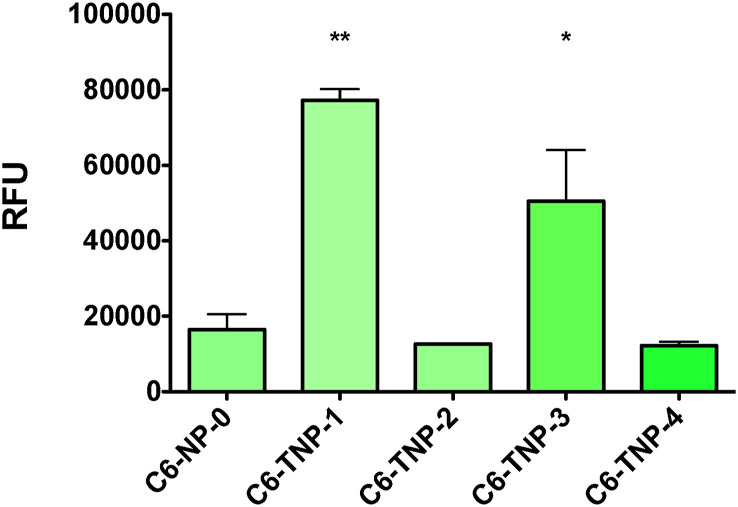
The ability of the TNP to specifically bind to ACE2 protein, in comparison with the nontargeted NP, was assessed by interaction analysis using direct ELISA experiments. Affinity evaluation and determination of binding specificity were conducted by using fluorescent C6-NP against hACE2 protein. NP-ACE2 binding affinity was evaluated by C6-fluorescence measurement and expressed as relative fluorescence units (RFU). We observed that the TNP-1, containing the ACL ligand, was the strongest nanoprotype in binding to ACE2 (Fig. 3).
Fig. 3.
Elisa-based comparative binding analysis of C6-loaded targeted and non targeted NP (C6-TNP-1-4, and C6-NP-0) with ACE2. Histograms represent the best fitting of the association processes. Affinity results are expressed as difference in response of relative fluorescent units (RFU). Data shown are representative of three independent experiments. (∗) and (∗∗) significantly (∗∗p < 0.01 and ∗p < 0.05 vs. C6-NP-0, n = 4) by one-way ANOVA and post-hoc Dunnett's test.
A consistent affinity was also provided by the TNP-3, which was about 35% less effective than TNP-1. Conversely, the TNP-2 (carrying DCL) only showed a minimal residual interaction with ACE2. Finally, both the nontargeted NP (NP-0) and the TNP-4 did not exhibite any appreciable binding to the protein, as expected.
2.6. Lipo-S1 protein vs NP competitive assays
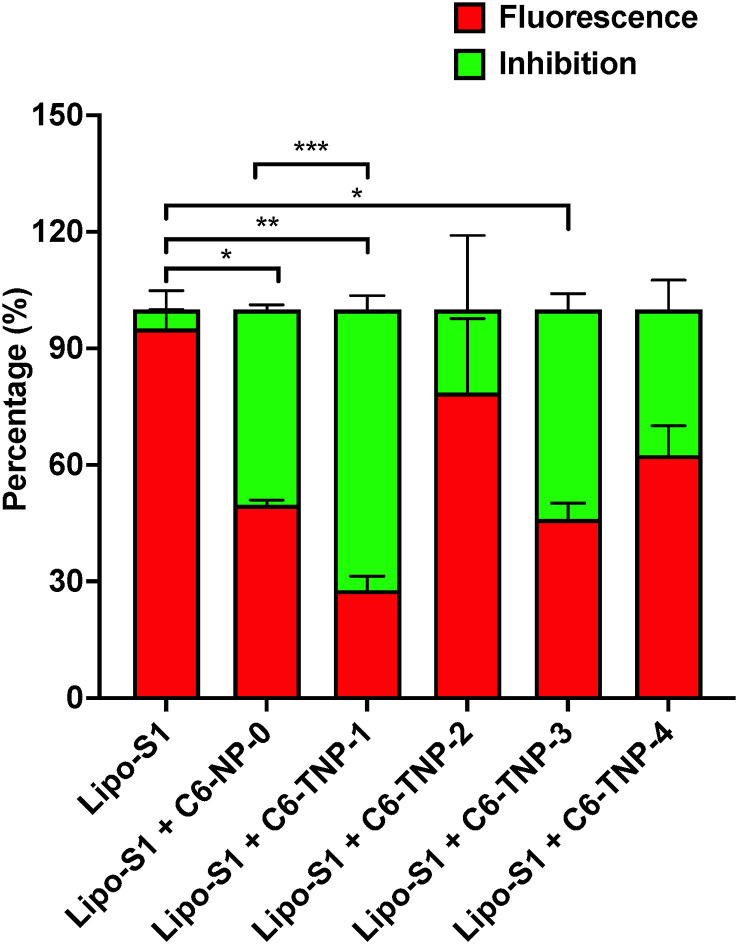
To evaluate whether the ACE2 targeting ability by NP is capable to provide a physical and topological blockage of SARS-CoV-2 binding with host cells, a competition experiment was carried out using rhodamine (Rho)-loaded liposome-derived SP (Lipo-S1) vs C6-loaded NP (in 1:1 ratio) in the ternary mixture with ACE2, which were assessed through hACE2 ELISA assay. The Lipo-S1 (measuring about 100 nm in size) were prepared according with the procedure we recently developed [75]. We observed that the C6 fluorescent TNP-1 strongly inhibited the SP model in binding to ACE2, with ∼75% and ∼25% for TNP-1 and Lipo-S1, respectively (Fig. 4).
Fig. 4.
A) Rho-incorporated Lipo-S1 vs C6-NP in 1:1 ratio in the ELISA ACE2 experiments. Fluorescent red signal (Ex 544 nm, Em 668 nm) was read for Rho and indicates the binding of Lipo-S1. Conversely, the competition/inhibition by C6-NP (C6-TNP-1-4 and C6-NP-0) was detected from the green fluorescence (Ex 488 nm, Em 510 nm). (∗) and (∗∗) significantly (∗∗p < 0.01 and ∗p < 0.05 vs Lipo-S1, n = 4) by one-way ANOVA and post-hoc Dunnett's test. Statistical analysis was also performed among the candidates, C6TNP-1 showed statistical significance against C6-NP-0 (∗∗∗p < 0.001 vs C6-NP-0 one-way ANOVA and post-hoc Dunnett's test).
As observed in the results of the binding affinity experiments (Fig. 3), TNP-3 was also able to significantly compete (for ∼50%) with Lipo-S1, whilst the TNP-2 (as well as TNP-4) did not show any evident binding interference. Finally, a certain quota of competition observed by NP-0 in these experimental conditions could warrant further mechanistic elucidation. These results demonstrated an excellent affinity of TNP-1 toward ACE2 receptor, thus supporting that this specific interaction would allow for selective binding toward host infected cells by the respective drug-loaded NP. TNP-1 resulted the most performing targeting nanosystems in binding with ACE2, and it was thus selected to determine its ability to inhibit the SARS-CoV-2 replication in cell-based assays. TNP-1 loaded with RDV was developed to obtain a model of drug-encapsulated TNP, together with the corresponding unloaded NP (TNP-1E). Additionally, other two sets of Rho-TNP-1 and Rho-NP-0 for further uptake/internalization experiments (Table 2 ) were prepared.
Table 2.
Average Diameter, Polydispersity Index (PDI), percentage of Encapsulation Efficiency (EE %), RDV Loading Capacity (LC %), and Yield of Production (YP %) of RDV-loaded and unloaded NP (RDV-TNP-1, and TNP-1E, respectively), and Rho-loaded NP (Rho-NP-0 and Rho-NP-1). Data are mean ± SD, n = 3.
| Batch | Mean diameter (nm) | PDI | EE (%) | LC (%) | YP (%) |
|---|---|---|---|---|---|
| RDV-TNP-1 | 101.27 ± 5.39 | 0.105 ± 0.03 | 51.35 ± 1.37 | 1.03 ± 0.03 | 73.17 ± 2.02 |
| TNP-1E∗ | 111.29 ± 7.47 | 0.104 ± 0.03 | – | – | 69.67 ± 1.15 |
| Rho-NP-0 | 130.12 ± 7.28 | 0.112 ± 0.02 | 34.89 ± 5.43 | 0.017 ± 0.003 | 63.47 ± 4.39 |
| Rho-TNP-1 | 129.68 ± 5.87 | 0.136 ± 0.04 | 29.19 ± 0.75 | 0.015 ± 0.001 | 61.13 ± 1.23 |
Each value is the mean ± SD of triplicate determinations. ∗E indicates “empty” NP.
The results indicated that TNP-1 batches displayed similar particle mean diameter (101 nm and 111 nm for RDV-TNP-1 and TNP-1E, respectively), independently of the drug loading, and exhibit a unimodal distribution with a PDI values of 0.105 (Table 2 and Fig. 5). Also, TNP-1 exhibited a good loading efficiency of RDV as confirmed from the obtained EE% and LC% values (51% and 1.03%, respectively).
2.7. Antiviral activity of TNP-1
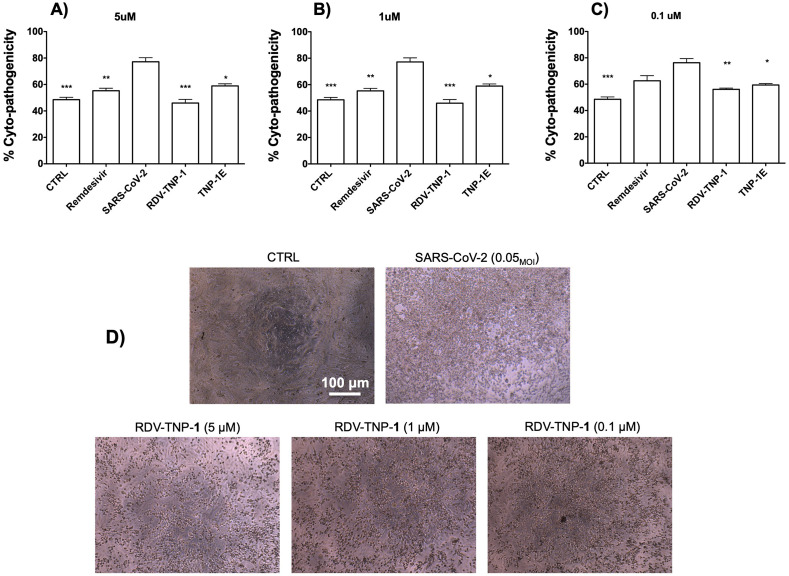
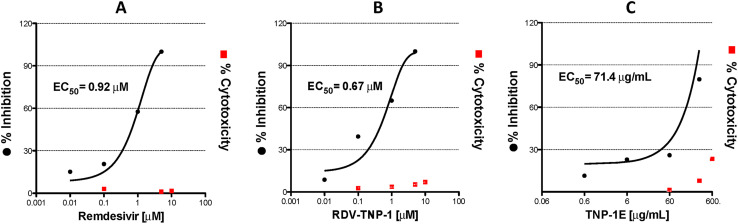
To confirm whether the effectiveness of TNP-1 in binding to ACE2 could be exploited against the SARS-CoV-2 virus, a series of cell-based experiments were conducted. First, we evaluated the impact of TNP-1 loaded with RDV (chosen as a model drug) on the cytopathic effect (CPE) induced by SARS-CoV-2 infection in Vero E6 cells, as a relatively highly ACE2 expression model. Cytopathogenic assays were assessed as a preliminary antiviral evaluation, also to explore cell infection damage [76,77]. Cells were infected with SARS-CoV-2 isolate USA-WA1/2020 at a multiplicity of infection (MOI) of 0.05, together with different concentrations of the RDV loaded into NP (in the sequence 5, 1, 0.1 μM). Medium and SARS-CoV-2 were used as negative and positive controls, respectively, and free RDV was used as control drug for comparison. Efficacies were evaluated by quantification and visualization of virus nucleoprotein expression, and they were expressed as percentage of absorbance by viable SARS-CoV-2 viral cells, evaluated via AlamarBlue (AB) assay. Next, we tested the antiviral activity of the RDV-TNP-1 and the unloaded (empty) analogue (TNP-1E). Results in Fig. 6A-C showed that RDV-TNP-1 blocked virus infection at low-micromolar concentration. TNP-1 demonstrated a significantly improved cytoprotective effect compared to RDV at the same concentration. Specifically, we observed that RDV-TNP-1 significantly reduced the SARS-CoV-2-induced CPE in Vero E6 cells at 5 μM, when compared to SARS-CoV-2 treated cells. Similar results were obtained using a 5-fold decreased concentration (i. e. 1 μM, at 72 h post-infection), while a lesser effect in CPE inhibition was observed at 0.1 μM (Fig. 6A-C).
Fig. 6.
Protection against SARS-CoV-2 induced cytopatogenic effect by TNP-1 treatment. (A-C) Cells were evaluated at different concentrations (5, 1, 0.1 μM) of RDV-loaded and nonloaded NP, using SARS-CoV-2 and RDV as controls. Empty (E) NP were tested at the same NP w/v concentration (μg/mL) of the parent RDV-loaded nanosample (300, 60, 6 μg/mL, for TNP-1E, which correspond to 5, 1, 0.1 μM, for RDV-TNP-1, respectively). Vero E6 cells were treated with samples and infected with SARS-CoV-2 at 0.05MOI. At 72 h post infection, inhibition of SARS-CoV-2-induced CPE was determined as survival of infected or not infected cells, and measured by using AB assay. The results were evaluated setting the uninfected control cells as 100% viability and the remaining values represented as a relative value. Data are mean ± SD, n = 6 replicates. (D) Representative microscopic images showing inhibition of virus-induced CPE in Vero E6 cells. From left to right: not infected Vero E6 cell monolayer (complete absence of CPE) as a control (CTRL), SARS-CoV-2 infected cells at MOI of 0.05, after 72 h post infection, and SARS-CoV-2 infected Vero E6 after RDV-TNP-1 exposure at different concentrations (5, 1, 0.1 μM). (∗), (∗∗) and (∗∗∗) significantly (∗∗∗p < 0.001, ∗∗p < 0.01 and ∗p < 0.05vs. C6-NP-0, n = 4) by one-way ANOVA and post-hoc Dunnett's test.
Furthermore, empty NP, i.e. TNP-1E, exhibited a significant cytoprotective profile (calculated by comparing the w/v concentration with those of RDV-TNP-1), likely by competing with the viral particles in binding to the ACE2, which would contribute to the antiviral potency of nanosystems.
Images showing protection against virus-induced CPE in Vero E6 cells were evaluated at 72 h post-infection, and at the same time the culture media was collected from the wells, for viral titer measurement (Figs. 6D and S13). Ranging from the CTRL, with a complete absence of CPE, and the SARS-CoV-2 infected cells with a consistent production of CPE, we observed a dose-dependent reduction of CPE production by the TNP-1 (as well as by RDV used as positive control), thus supporting the cytoprotective property of our prototypes.
To further substantiate the cytoprotective and antiviral activity results, we evaluated the ability of TNP-1 to inhibit SARS-CoV-2 in vitro by quantitative real-time polymerase chain reaction (qRT-PCR) assays, using 4-fold concentration scale ranging between 5 and 0.01 μM (Fig. 7A-C). The samples were added at the same time point of SARS-CoV-2 multiplicity of infection (MOI = 0.05). We confirmed that RDV-TNP-1 exhibited a good inhibitory activity (half-maximun viral infection, EC50 = 0.67 μM), which was greater than RDV (EC50 = 0.92 μM) in the same assays (Fig. 7A-C), and no cytoxicity was observed in similarly treated uninfected cultures across the dose range tested.
Fig. 7.
Antiviral activities of TNP-1. Vero E6 cells were infected with SARS-CoV-2 at 0.05MOI, and treated at different concentrations (5, 1, 0.1, 0.01 μM) of RDV-TNP-1 (B) using RDV (A) as control. TNP-1E (C) were tested at the same NP w/v concentration (μg/mL) of the parent RDV-loaded nanosample (300, 60, 6, 0.6 μg/mL, for TNP-1E, which correspond to 5, 1, 0.1, and 0.01 μM, for RDV-TNP-1, respectively). At 72 h post infection, the viral yield in the cell supernatant was then quantified by RT-PCR. Cytotoxicity of these drugs to Vero E6 cells was measured in parallel by AlamarBlue assays. The left and right Y-axis of the graphs represent mean % inhibition (EC50s) of virus yield and cytotoxicity of the drugs, respectively. The experiments were done in triplicates and are represented as median of the values.
We also confirmed that unloaded NP were able to significantly reduce the SARS-CoV-2 replication at all concentrations tested, potentially due to the high affinity of the TNP-1 for the ACE2 receptor, which would prevent the viral entry. In our opinion, the targeting approach should not necessarily provide a more than two-fold difference in activity with respect to free drug, but should impart a site-specific drug delivery towards biological targets, which would allow to use a significant low dose of drug, to improve the PK of antiviral drugs and, most importantly, to prevent off-target distribution of a drug in the body. Moreover, RDV-TNP-1 resulted in about 2-fold highly active than the respective unloaded TNP-1E (by comparing their w/v concentrations). The basal activity demonstrated by TNP-1E would in part explain the increased antiviral activity observed for TNP-1. This general behaviour can be attributed to targeting ability of TNP-1 to ACE2, which promotes an enhanced binding between NP and the cells, thus resulting in increased adhesion in the membrane, and subsequent cell uptake through receptor-mediated endocytosis. Retrospective analysis on antiviral results allows to speculate that increased relative antiviral potency demonstrated by TNP-1, and the possibility to selectively deliver drugs to host cells, might be better highlighted in in vivo assays. In fact, while in the wells a drug diffuses through the cell membrane, the TNP would act by delivering the payload drug into the cells by a receptor (ACE2)-mediated endocytosis, thus preventing/reducing potential side effects.
2.8. ACE2 inhibition of TNP-1
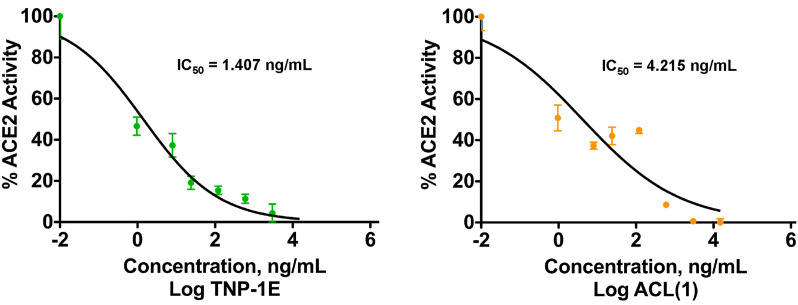
To evaluate the putative ACE2 inhibition by TNP-1, we measured the effect of TNP-1E, compared with that of ligand 1, on the exopeptidase activity of recombinant human (rh)ACE2 employing a fluorometric assay (BPS Bioscience). We found that TNP-1E and 1 inhibited rhACE2 activity in the same concentration range (to compare both chemical entities the IC50s were determined and expressed as ng/mL), with TNP-1E being 3-fold more effective than the ligand 1 (IC50 = 1.407 and 4.215 ng/mL, for TNP-1E and 1, respectively, Fig. 8).
Fig. 8.
Inhibition of ACE2 exopeptidase activity by TNP-1E and ligand 1. IC50s were determined and expressed as ng/mL. Data are expressed as the mean ± SD of at least triplicate determinations.
1 proved to be more potent in inhibiting ACE2 activity than MLN-4760 (IC50 = 12.72 ng/mL, Fig. S14), which was predicted by the sligthly higher binding energies observed in the preliminary docking calculations. The behaviour of TNP-1 can be explained by the concept of ligand multivalency, the effect capable to enhance affinity and selectivity of NP to the biological target [[78], [79], [80]]. Specifically, multiple ligands coated NP (as our targeted nanosystems) consist of a combination of several molecular entities resulting in simultaneous and reversible local molecular recognition interactions, which are amplified over simple monovalent ligands. This strategy allows high avidity for the target receptor by the NP, which supports the targeting ability of TNP-1, but also induces inhibition of the ACE2 function. Although the interaction with the ACE2 receptor from any protein or particle could temporarily affect the RAS, the predicted short time for the antiviral treatment should not generate any significant interference or potential compensatory shift in the Ang II/Ang (1–7)/Mas receptor axis. Again, it should be taken into account that a single NP cannot be structurally compared with a small organic molecule ligand inhibitor such 1 or MLN-4760. To simplify this concept, we can assume that the inhibitory effect due to a single NP can be pursued by the same amount in weight of inhibitors, which should have a more consistent distribution and localization in the same district. Moreover, since SARS-CoV-2 prevalently infects (ACE2 enriched) epithelial cells in the respiratory system, our TNP-1 are suggested for inhalation route delivery, in order to limit the interference within ACE2-related pathways in cardiovascular system and kidney. Inhaled antiviral drugs can be localized to the target organ/tissues, especially if site-specifically targeted, which can allow for a lower dose compared to the systemic delivery. This would lead to reduced and less severe adverse effects concentration-dependent [29,30,81]. However, although a low amount of TNP could statistically evade the respiratory tract, we should consider that polymeric NP are biocompatible/biodegradable materials, and that only a fewer amount of drug can be adsorbed in the systemic circulation with respect to the other administration routes. Inhibiting intracellular targets of the SARS-CoV-2 by targeted drug delivery, also preventing SARS-CoV-2 internalization by competing for the ACE2, may be effective in disrupting key protein/protein interactions involved in this pathogenic event [82]. Therefore, the benefit of our methodology can be higher than presumed side effects, which should be carefully evaluated in the next step of the study.
2.9. Cellular uptake
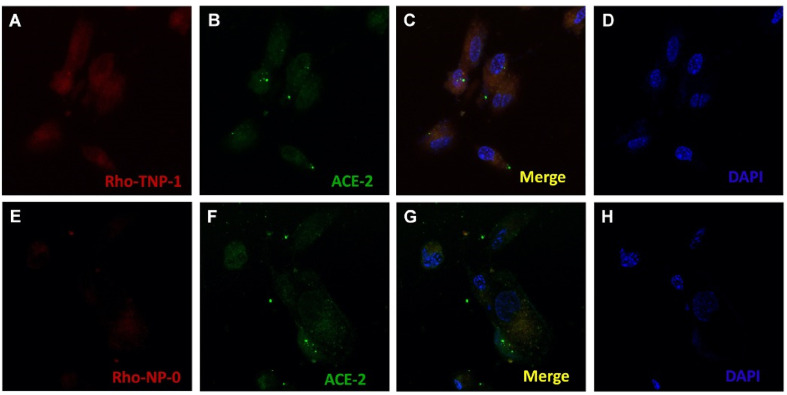
The correlation between antiviral activity of the nanosystems with cellular uptake was explored by uptake/internalization of Rho-loaded NP in Vero E6 cells, which were visualized by confocal fluorescence microscopy. Fig. 9 shows images (scale bar 20 μm) of cellular uptake efficiency between targeted and nontargeted NP (Rho-TNP-1 and Rho-NP-0, respectively).
Fig. 9.
Uptake assays for NP. Panels show live cell confocal fluorescence microscopy images of Vero E6 cells after exposure to targeted (TNP-1) and nontargeted (NP-0) Rho-loaded NP. Panels A and E: Rho-TNP-1 and Rho-NP-0, respectively. Panels B and F: visualization of ACE2 cell receptors (in green) overexposed by lenti-hACE2. Panels C and G: merge images of Vero E6 cells incubated for 3 h with Rho-TNP-1 and Rho-NP-0, respectively. Panels D and H: blue fluorescence of the nuclei stained with DAPI dye. Cellular binding/uptake visualized by overlaying images (scale bar 20 nm).
After 3 h incubation, the Rho-TNP-1 showed higher cellular binding and uptake than the Rho-NP-0, as visible by a strong fluorescence in cell membrane and in the cytoplasm (Fig. 9C), which further confirmed their effective interaction with the targeted cells. This approach would support that TNP-1 will be internalized in the cell membrane, potentially via endocytosis. This putative mechanism can be triggered by targeting the ability of TNP-1 to ACE2, which results in cell uptake in stepwise manner. Importantly, antiviral results also confirm the effectiveness of the TNP-1, thus confirming an efficient endosomal escape of payload RDV, after disaggregation of the polymeric matrix in the cytosol. The selectivity of the endosomal mechanism by TNP-1, in terms of membrane diffusion/permeation cytosolic uptake by free drug, might have major impact in in vivo assays. These results indicate that the TNP-1 prototypes were able to effectively address active compounds to ACE-2 expressing cells, potentially infected/infectable by SARS-CoV-2, building the basics for further optimization.
3. Conclusions
When the COVID-19 pandemic started, based on the structural and functional information available for SARS-CoV-2, we sought to develop engineered targeted nanosystems for ACE2 site-directed antiviral drug delivery which could represent a suitable and powerful approach to improve the antiviral profile of anti-SARS-CoV-2 therapeutics.
In this study, we developed drug-encapsulated NP targeting to ACE2 enzyme receptors, with TNP-1 demonstrated to be the most effective prototype. We highlighted that the TNP-1 efficiently binds to ACE2 receptors compared to the nontargeted ones, confirming similar behaviour when tested in both binding assays and in competition experiments against a model of SP. This is strongly associated with the antiviral potency, with increased anti-SARS-CoV-2 activity for RDV-TNP-1 compared with the free RDV in cell-based assays, without affecting cell viability. This corroborates our rationale in which the primary role of targeting agents is to increase cellular uptake of RDV-loaded NP and their distibution into ACE2-expressing cells, crucially involved in infection such as epithelial cells of the mucosa oral cavity and lung. We further observed a direct antiviral effect from unloaded NP, likely due to a competitive interference with spike attachment for the ACE2 membrane receptors. These insights might be useful to develop suitable therapeutic tools for treating SARS-CoV-2 infection, through different administration routes, such as inhalation/aerosol systems [[24], [25], [26]]. Furthermore, administering RVD-TNPs by inhalation may be a reasonable approach to achieve facilitate distribution at the site of infection, and to reduce systemic toxicity and adverse drug reactions. This can potentially be achieved in combination with its intravenous drug administration [25], as in the current protocols or to be evaluated in different and/or modulated COVID-19 drug regimens. Herein, our NP show promising results and they might be optimised for an inhaled delivery, so that RDV can be distributed in a optimal concentrations in the primary site of infection.
In summary, our findings provide key proof-of-principle showing that targeted delivery systems may improve the pharmacological anti-SARS-CoV-2 therapy, leading the way for novel and effective treatment in COVID-19 patients. Further work is in progress to evalute their therapeutic potential against COVID-19.
4. Experimental section
4.1. Molecular modelling
Molecular modeling was performed on a personal Macbook installed with IOS operating system, and on a Dell workstation (Intel(R) core i5-5200U CPU, 2.20 GHz, 8 GB, installed with Windows 10 operating system. Discovery Studio Biovia 2020 (Dassault Systèmes, Vélizy-Villacoublay, France) was employed to prepare the protein, and to visualize and modify receptor and ligand structures. Detailed analysis of the interactions was carried out using the Accelerys Discovery Studio Visualizer. All the ligands 1–4 were constructed and modelled, and energy minimized in neutral form by standard molecular mechanics methods. Operatively, the atomic charges were assigned using the Gasteiger-Marsili procedure. For 1–3 (docking with ACE2) the crystal structure of ACE2 in complex with the inhibitor MLN-4760 was retrieved from the RCSB Protein Data Bank (PDB ID: 1r4l; resolution: 3.0 Å) [68]. For 4 (docking with TACE), the crystal structure of TACE in complex with the inhibitor TAPI-2 was retrieved from the RCSB Protein Data Bank (PDB ID: 2ddf; resolution: 1.70 Å) [69]. The protein targets were refined using AutoDock 4.25, using a well-established cross docking protocol to identify the conformations that have significant binding affinity with ACE2 and TACE [[83], [84], [85], [86]]. Briefly, water molecules were removed while hydrogen atoms were added using the ADT module implemented in AutoDock. The charges were adjusted using Gasteiger charge module for proteins implemented in AutoDock. Docking was performed for both complexes using the empirical free energy function and the Lamarckian protocol [87]. Mass-centered grid maps were generated with 40 grid points for every direction and with 0.375 Å spacing by the AutoGrid program for the catalytic domains of ACE2 and TACE. The results were analyzed for the presence of hydrogen bonding, hydrophobic and p-p interactions between hits and the active site of the protein. The common interactions in both complexes were scrutinized. All the figures were rendered with PyMOL Molecular Graphics System.
4.2. Chemistry
Poly(D,L-lactide-co-glycolide) carboxylic acid end group, (PLGA-COOH, PLGA-A, Purasorb Polymer PDLG 5002A, Mw ∼17,000) (lactide/glycolide ratio of 50:50, viscosity range: 0.20 dL/g in CHCl3) was kindly provided by Corbion Purac (Gorinchem, The Netherlands). Poly(epsilon-caprolactone) (PCL, Mw ∼80,000), polyvinyl alcohol (PVA, Mw Mw 31,000–50,000), Coumarin-6, Rhodamine, MLN-4760 were purchased from Merck/Sigma-Aldrich (Steinheim, Germany). The heterobifunctional PEG polymer with a terminal amine and carboxylic acid functional group, NH2-PEG-COOH (Mw = 3400), and with a terminal amine and maleimide functionality NH2-PEG-mal (TFA salt, Mw = 3500), were purchased from JenKem Technology USA. All solvents and other chemicals (used for the preparation of 1 and 2) were purchased from Sigma-Aldrich or Carlo Erba (analytical grade and were used without further purification). Captopril (CPL, 3) was purchased from DBA Italia s.r.l. (MedChemExpress). The pseudo-peptide TAPI-2 (4) was purchased from DC Chemicals Limited, Shanghai, China. Melting points (mp) were determined using an Electrothermal melting point or a Köfler apparatus and are uncorrected. Nuclear magnetic resonance (1H NMR, 13C NMR, NOESY, COSY, HSQC) spectra were determined in CDCl3, or DMSO‑d 6 were recorded on 400 MHz Bruker Avance III. Chemical shifts are reported in parts per million (ppm) downfield from tetramethylsilane (TMS), used as an internal standard. Splitting patterns are designated as follows: s, singlet; d, doublet; t, triplet; q, quadruplet; m, multiplet; brs, broad singlet; dd, double doublet. The assignment of exchangeable protons (OH and NH) was confirmed by the addition of D2O. HRMS spectra, ESI-TOF were obtained on Thermo Scientific™ Q Exactive Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer, with API-HESI source. Samples were introduced as 0.1 mg/L solutions in MS grade water added with 0.1% acetic acid with a 5.0 μL/min flow and the following source parameters for positive and negative polarities. Analytical thin-layer chromatography (TLC) was carried out on Merck silica gel F-254 plates. Flash chromatography purifications were performed on Merck Silica gel 60 (230–400 mesh ASTM) as a stationary phase. Analytical HPLC was carried out on an Agilent 1260 Infinity II Quaternary pump HPLC apparatus with a Poroshell 120 EC-C18 reversed column (4.6 × 100 mM, 4 μm) at a flow rate of 0.5 mL/min (injection volume 20 μL) and detected at 254 nm. Elemental analyses for 1 and 2 were performed on a PerkinElmer 2400 spectrometer, and were within ±0.4% of the theoretical values, thus confirming ≥95% purity.
4.2.1. Synthesis of PLGA-PEG-1, PLGA-PEG-2, and PLGA-PEG-4 copolymers
The PLGA–PEG-1, PLGA–PEG-2 or PLGA-PEG-4 were obtained by adding a solution of ligand (1, mg 37.44, 10 equiv; 2, mg 21.0, 4 equiv.; 4, mg 16.62, 4 equiv.) in DMF (1.0 mL), and DIPEA (140 equiv. for 1, or 55 equiv. for 2 and 4) to a solution of PLGA–PEG-NHS (220 mg, 0.010 mmol for 1, 2, or 4) in dimethylformamide (DMF, 3.0 mL). The reaction mixture was magnetically stirred at room temperature for 24 h, under nitrogen atmosphere. Then, diethyl ether was added to the solution, and the product that formed was submitted to centrifugation (650 r.p.m.) to obtain a amorphous beige solid. The crude product was washed several times (3) with cold diethyl ether and methanol, with cold diethyl ether (3 times), then dried under vacuum. A white solid was obtained for each copolymer, which was dried with nitrogen, and then characterized by 1H NMR. The polymers were stored at −20 °C before use.
PLGA-PEG-1: (yield, 79%). 1H NMR (DMSO‑d 6): δ 5.28–5.16 (m, 1H, -OC-CH(CH3)O-, PLGA), 4.96–4.82 (m, 2H, -OC-CH 2O-, PLGA), 3.51 (brs, 2H, -CH 2CH 2O-, PEG), 1.50–1.44 (brs, 3H, -OC-CH(CH 3)O-, PLGA). Signals for conjugated 1: δ 7.60 (s), 6.88 (d), 6.67 (s), 6.52 (d), 3.11–3.01 (m), 2.88–2.69 (m), 1.75–1.66 (m), 1.40–1.25 (m), 0.89-0.84 (m). PLGA-PEG-2: (yield, 92%). 1H NMR (DMSO‑d 6): δ 5.25–5.16 (m, 1H, -OC-CH(CH3)O-, PLGA), 4.91–4.83 (m, 2H, -OC-CH 2O-, PLGA), 3.51 (brs, 2H, -CH 2CH 2O-, PEG), 1.58–1.55 (brs, 3H, -OC-CH(CH 3)O-, PLGA). Signals for conjugated 2: δ 4.13–4.05 (m), 2.80–2.73 (m), 2.32–2.21 (m), 2.00–1.89 (m), 1.75–1.60 (m), 1.40–1.24 (m). PLGA-PEG-4: (yield, 71%). 1H NMR (DMSO‑d 6): δ 5.29–5.16 (m, 1H, -OC-CH(CH3)O-, PLGA), 4.92–4.82 (m, 2H, -OC-CH 2O-, PLGA), 3.51 (brs, 2H, -CH 2CH 2O-, PEG), 1.48–1.44 (brs, 3H, -OC-CH(CH 3)O-, PLGA). Signals for conjugated 4: δ 8.18 (s), 7.96 (s), 4.20–4.15 (m), 4.00 (s), 3.10–3.03 (m), 3.95–3.90 (m), 2.45–2.40 (m), 2.24-2-20 (m), 1.43-1-29 (m), 1.12–1.07 (m), 0.98–0.85 (m).
4.2.2. Synthesis of PLGA-PEG-3
PLGA-PEG-mal (0.200 g, 0.00846 mmol) and 3 (2.76 mg, 1.5 equiv.) were dissolved in 4 mL of 1:1 solution of acetonitrile and DMF, and stirred for 24 h under nitrogen atmosphere. The desired copolymer was obtained by precipitation with cold diethyl ether, centrifugation (650 r.p.m.) to obtain a white solid, which was washed several times (3) with cold diethyl ether, then dried under vacuum, and used for NP preparation without further treatment (yield, 84%). 1H NMR (DMSO‑d 6): δ 5.27–5.17 (m, 1H, -OC-CH(CH3)O-, PLGA), 4.97–4.83 (m, 2H, -OC-CH 2O-, PLGA), 3.51 (brs, 2H, -CH 2CH 2O-, PEG), 1.50–1.44 (brs, 3H, -OC-CH(CH 3)O-, PLGA). Signals for conjugated 3: δ 4.55–4.51 (m), 3.72–3.61 (m), 2.90–2.68 (m), 2.51–2.39 (m), 1.98–1.82 (m), 1.43-1-29 (m), 1.12–1.07 (m).
4.2.3. Synthesis of (S)-tert-butyl 4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1H-imidazole-1-carboxylate (6) [58]
Di-tert-butyl dicarbonate (Boc2O, 27.04 g, 123.92 mmol, 2 eq.) in MeOH (15 mL) was added dropwise to a solution of (S)-histidine methyl ester dihydrochloride 5 (15 g, 61.96 mmol, 1 equiv) in MeOH (150 mL, 0.4 M) and triethylamine (17.18 mL, 123.92 mmol, 2 equiv), and the reaction was stirred at r. t. for 24 h. Next, the solvent was evaporated and the residue was dissolved in CH2Cl2 and water, and then extracted three times with CH2Cl2. The organic phase was washed with brine, dried over Na2SO4, filtered and concentrated to provide a yellow oil, which was stored at -4-8 °C. An amorphous beige solid appeared, which was triturated with hexane to afford the di-Boc protected imidazole 6 as white solid (yield, 95%). 1H NMR (CDCl3): δ 7.99 (s, 1H), 7.14 (s, 1H), 5.70–5.68 (m, 1H), 4.58–4.56 (m, 1H), 3.64 (s, 3H), 3.05–3.04 (m, 2H), 1.61 (s, 9H), 1.44 (s, 9H). MS (ESI): m/z [M+H]+ 370.
4.2.4. Synthesis of (S)-methyl 2-((tert-butoxycarbonyl)amino)-3-(1-(4-nitrobenzyl)-1H-imidazole-5-yl)propanoate (7)
A solution of 4-nitro-benzyl alcohol (2.28 g, 14.89 mmol, 1.1 equiv) in diisopropylethylamine (DIPEA) (2.60 mL, 14.89 mmol, 1.1 equiv) and CH2Cl2 (10 mL, 1 M) was added slowly to a cooled solution (−78 °C) of trifluoromethane sulfonic anhydride (2.50 mL, 14.89 mmol, 1.1 equiv) in CH2Cl2 (50 mL, 0.3 M) under nitrogen. After 30 min, a solution of the di-Boc protected histidine 6 (5.0 g, 13.53 mmol, 1 equiv) in CH2Cl2 (1 M) was added slowly to the triflate solution. The reaction mixture warmed to room temperature overnight. After 20 h, the reaction mixture was concentrated completely, dissolved in MeOH (70 mL) and heated for 1 h. The reaction mixture was concentrated and diluted with CH2Cl2. The organic phase was washed with saturated NaHCO3 solution (3 times) and brine (1 time), then dried over Na2SO4, filtered and concentrated. The resulting oil was purified by column chromotagraphy (gradient 1–10% MeOH in CH2Cl2) to give the N-3 alkylated imidazole derivative 7 (yield 73%): 1H NMR (CDCl3): δ 8.22–8.17 (d, 2H), 8.16 (s, 1H), 7.55–7.52 (m, 1H), 7.47 (d, 1H), 7.19 (dd, 2H), 6.88 (d, 1H), 5.30–5.15 (bm, 3H), 4.48–4.44 (m, 1H), 3.74 (s, 3H), 2.99–2.91 (m, 2H), 1.42 (s, 9H). MS (ESI): m/z [M+H]+ 405.5.
4.2.5. Synthesis of benzyl (R/S)-2-(((S)-1-methoxy-3-(1-(4-nitrobenzyl)-1H-imidazole-5-yl)-1-oxopropan-2-yl)amino)-4-methylpentanoate (9)
To remove the Boc protecting group, the intermediate 7 (g. 3.0, 7.42 mmol) was treated with 4 N HCl in dioxane (43 mL, 23 equiv.) for 2 h. A solid corresponding to the resulting di-HCl salt appears, and then the reaction was concentrated completely. Trituration of the solid with EtOAc provided a white solid, which was suspended in dichloroethane (DCE, 70 mL). Method a) The keto-ester 16 (3.27 g. 14.84 mmol, 2 equiv) was added to this suspension, and the mixture was stirred for 1 h. Then, NaB(OAc)3H (4.71 g, 22.25 mmol, 3 equiv) was added slowly, and the reaction was magnetically stirred at room temperature for 24 h. The reaction was then quenched by addition of a saturated NaHCO3 solution (until pH ∼8–9), and the mixture was stirred for 1 h. Next, the aqueous phase was separated and extracted with EtOAc (2 times). After drying over Na2SO4, the organic layers were concentrated to give a yellow oil which was purified by flash chromatography (EtOAc/hexane = 5/5; EtOAc; EtOAc/MeOH = 9/1) to provide the diester as about 2:1 mixture of diastereomers (yield 59%). 1H NMR (CDCl3): δ 8.22–8.17 (m), 7.60 (s), 7.55 (s), 7.37–7.31 (m), 7.22–7.20 (d), 7.16–7.13 (m), 6.95.6.93 (d), 6.88 (s), 5.29 (d), 5.09 (d), 3.68-3-64 (m), 3.74 (s), 3.66 (s), 3.19 (t), 2.80–2.73 (m), 1.70–1.53 (m), 1.49–1.32 (m), 0.88–0.78 (m). MS (ESI): m/z [M+H]+ 509.2404. Method b) According to previous procedure, the resulting di-HCl salt obtained was neutralized with 1 N NaOH to obtain a pale beige oil, which has been purified by flash cromatography to give the aminoester 8 as green oil. (S)-methyl 2-amino-3-(1-(4-nitrobenzyl)-1H-imidazole-5-yl)propanoate (8). 1H NMR (CDCl3): δ 8.21 (d, 2H), 7.56 (s, 1H), 7.21 (d, 2H), 6.87 (s, 1H), 6.18 (brs, 1H), 5.35–5.22 (q, 2H), 4.69–4.64 (m, 1H), 3.73 (s, 3H), 3.02–2.90 (m, 2H). MS (ESI): m/z [M+H]+ 305.4. The compound 8 (1.10 g, 3.61 mmol) has been treated with NaB(OAc)3H, but with the addition of 1 equiv. of acetic acid to the reaction mixture, and following the abovementioned procedure, to obtain 9 in comparable overall yield.
4.2.6. Synthesis of (R/S)-2-(((S)-3-(1-(4-aminobenzyl)-1H-imidazole-5-yl)-1-methoxy-1-oxopropan-2-yl)amino)-4-methylpentanoic acid (10-S,S-R,S)
A suspension of the diester 9 (0.45 g, 0.885 mmol) and 10% Pd–C (0.19 g) in dry methanol (5 mL) was magnetically stirred for 5 min at room temperature, then ammonium formate (0.25 g, 3.98 mmol, 4.5 equiv.) was added in a single portion. The mixture was stirred for 1.5 h, and the catalyst was removed by filtration through a Celite pad, and washed with dry methanol. The filtrate was evaporated, and the resulting residue was purified by flash cromatography (CH2Cl2/MeOH = 9:1) to give the diastereoisomers 10 as brown oil (Yield: 65%). 1H NMR (CDCl3): δ: δ 7.74 (s), 7.65 (s), 7.18 (s), 6.95 (s), 6.92–6.88 (d), 6.65-6-61 (d), 5.01–4.99 (d), 3.88–3.71 (brs), 3.73 (s), 3.51 (s), 3.50–3.45 (m), 3.15–3.19 (m), 2.94–2.76 (m), 1.82–1.39 (m), 0.94–0.83 (m). MS (ESI): m/z [M+H]+ 389.22.
4.2.7. (S)-2-(((S)-2-(1-(4-aminobenzyl)-1H-imidazol-5-yl)-1-carboxyethyl)amino)-4-methylpentanoic acid (1)
The compound 10 (0.200 g., 0.5148 mmol) was treated with 1N NaOH (3.1 mL, 3.089 mmol, 6 equiv.) in EtOH (1.5 mL) for 12 hours, then neutralized and concentrated to provide a yellow solid. The crude product was purified by flash cromatography (gradient elution CH2Cl2/MeOH = 8:2 to CH2Cl2/MeOH = 6:4), then treated with EtOH (to remove traces of inorganic salt) to give 1 as a pale yellow powder (Yield: 55%). S,S diastereomer: 1H-NMR (DMSO-d6): δ 7.62 (s, 1H), 6.85 (d, 2H), 6.80 (s, 1H), 6.51 (d, 2H), 4.95 (d, 2H), 3.70-3.10 (brs, 4H), 3.21-3.19 (m, 1H), 2.92-2.77 (m, 3H), 1.76-1.72 (m, 1H), 1.41-1.23 (m, 2H), 0.87-0.83 (m, 6H). 1H-NMR (D2O): δ 8.67 (d, 1H), 7.44 (d, 1H), 7.26 (t, 2H), 7.15 (t, 2H), 5.36 (s, 2H), 3.56-3.40 (m, 2H), 3.22-3.21 (d, 2H), 1.87-1.51 (m, 3H), 0.88-0.85 (m, 6H). 13C-NMR (DMSO-d6): δ 173.9, 171.3, 148.2, 137.3, 128.2, 126.7, 123.5, 121.0, 113.8, 69.7, 59.2, 48.5, 47.4, 26.8, 24.2, 22.6, 22.0. MS (ESI): m/z [M+H]+ 375.20; [M+Na]+ 397.18. Anal. (C19H26N4O4) C, H, N. HPLC Analysis. The mobile phases were mixtures of A = acetonitrile + 0.1% trifluoroacetic acid and B = H2O + 0.1% trifluoroacetic acid and are indicated as the ratio A/B. ACL (1), HPLC (A/B = 60:40), Rt 1.72 min (100%). QD 394-Me, HPLC (A/B = 60:40), Rt 1.156 min (95.11%).
4.3. Preparation of NP
NP were prepared using a solvent displacement method previously reported with slight modifications [50,51,56,63]. Briefly, an acetonitrile solution containing PCL and PLGA-PEG-COOH or PLGA-PEG-ligand conjugated (mass ratio of 1.5:1), and the fluorescent dye (C6 or Rho) (0.05% w/w), or RDV (2%, w/w), was added dropwise to a Pluronic F-127 aqueous solution (0.1% w/w) with magnetic stirring giving a final polymer concentration of 7.0 mg/mL. Later the dispersion was kept for magnetic stirring at room temperature to evaporate the organic solvent, then centrifuged and washed to remove the non-encapsulated compound. Drug-free NPs were produced in a similar manner and used as comparison. The obtained NP suspension was used immediately for assay or lyophilized for storage at −50 °C.
4.3.1. Characterization of NP
4.3.1.1. Morphology, size, and ζ potential
Morphological examination of NP was performed by environmental scanning electron microscopy (ESEM) (Zeiss LS10, Germany). The samples were placed on aluminum stub, then analyzed at 10 kV acceleration voltage after gold sputtering (Sputter Coater Edwards S150A), under an argon atmosphere. Mean diameter and polydispersity index (PDI) of NPs were measured by using photon correlation spectroscopy (Zetasizer Nano ZS, Malvern Instruments, U.K.) at 25 °C and a scattering angle of 90° after dilution of samples with Milli-Q water. Each sample was measured in triplicate. The ζ potential values of the NP were detected with a Zeta Plus analyzer (Brookhaven, USA) at 25 °C. Each sample of NP was diluted with distilled water (1.0 mg/2.0 mL) and sonicated before measurement. All data were obtained with the average of three measurements.
4.3.2. Loading capacity, Encapsulation efficiency, and yields of production
To determine the fluorescent probe loading, a weighed amount of dye loaded NP was dissolved in acetonitrile. The solution was filtered through a 0.2 μm syringe filter and analyzed spectrophotometrically (LLG uniSPEC 2 Uv–Vis spectrophotometer, BDL Czech Republic sro, Turnov, Czech Republic) at 449 nm for 6C and 548 nm for Rho, respectively. The amount of 6C and Rho were calculated by referring to the calibration curves in acetonitrile in the range of 0.5–5 μg/mL (y = 0.2411x - 0.0396, R2 = 0.9999 for 6C and y = 0.2733x - 0.0019, R2 = 1 for Rho, respectively). The amount of the encapsulated RDV was determined by dissolving a weighed amount of dried loaded NP in acetonitrile and measured using a modified HPLC method. Chromatographic analysis was performed on a 1260 Infinity II (Agilent Technologies, U.S.) liquid chromatography system equipped with a diode array detector of the same series, using an Infinity Lab Poroshell 120, EC-C18 column (4 μm, 4.6 × 100 mm) (Agilent Technologies, U.S.). The elution was carried out in gradient mode using a binary solvent mixture composed of water acidified with 0.1% TFA (solvent A) and acetonitrile with 0.1% TFA (solvent B) under the following conditions: from 99% to 98% A, 0–1 min; 98%–2% A, 1–15 min; 2–99% A, 15–25 min. The injection volume was 20 μL, the flow rate was 0.75 mL/min, the wavelength for UV detection was 254 nm, and the retention time was 11 min. The calibration curve was found to be linear in the range of 10–200 μg/mL (y = 40.262x + 83.739; R2 = 0.9999).
The yields of production were expressed as the weight percentage of the final product after drying, regarding the initial total amount of solid materials used for the preparation.
The loading capacity (LC%), the entrapment efficiency (EE%), and yield of NP (YP%) were calculated by the following equations, respectively:
LC% = (Weight of Dye or RDV in NP/Weight of NP) × 100.
EE% = (Actual Dye or RDV content/Theoretical Dye or RDV content) × 100.
YP% = (Weight of NP recovered/Weight of polymer and Dye or RDV fed initially) × 100.
4.3.3. Statistical analysis
The data of NP characterization were processed by GraphPad Prism software (version 6, GraphPad software. Inc., San Diego, CA, USA) and subjected to one-way analysis of variance. Individual differences were evaluated using a nonparametric post hoc test (Tukey's test) and considered statistically significant at p < 0.05.
4.4. SARS-CoV-2 infection studies in Biosafety Level 3 laboratory
SARS-CoV-2, isolate USA-WA1/2020, was acquired from the Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID). SARS-CoV-2 was passaged once in Vero E6 cells (ATCC) and viral stocks were aliquoted and stored at −80 °C. Virus titer was determined by plaque assay using Vero E6 Cells. 0.05MOI of SARS-CoV-2 was used for the experiments. Studies involving live SARS-CoV-2 virus were approved by the University of California, Los Angeles Institutional Biosafety Committee (IBC), and were performed in compliance with approved Biosafety Level 3 (BSL3) standard operating procedures. The UCLA BSL-3 laboratory was designed in compliance with the guidelines recommended by the Biosafety in Microbiological and Biomedical Laboratories (BMBL), the U.S. Department of Health and Human Services, the Los Angeles Department of Public Health (LADPH) and the Center for Disease Control and Prevention (CDC).
4.5. Construction of custom-designed liposome spike S1
Construction of nano-liposomes (diameter = about 100 nm) containing S1 spike proteins (Lipo-S1), biotinylated to an external lipid bilayer, was performed as previously reported [75]. Briefly, Rho-containing liposomes (Encapsula Nanoscience) prepared with Immunosome-Biotinyl Cap were coupled to SARS-CoV-2 S1 protein, His, Avitag (ACROBiosystems; No. S1N–C82E8), in a 1:2 ratio and excess S1 protein, dialyzed out from the liposome preparation. Control liposome preparations were prepared identically in the absence of SARS-CoV-2 S1 protein. Binding of S-protein liposomes to ACE2 was verified by modified ELISA and in ACE2-transfected human embryonic kidney 293 cells. Liposome preparations were diluted to 1:500 before use, added to cultures or perfusion culture media for 24 h. Additional details are available in ref. 75.
4.6. NP-ACE2 binding and competition assay
C6-incorporated NP affinity bond with ACE2 was assessed through direct Elisa ACE2 (Abcam). NP were incubated at 4 °C overnight, then the plate was washed 5 times to eliminate the unbound NP, and binding between the ACE and NP was evaluated through fluorescence emission using a CLARIOstar® microplate reader (BMG Labtech). The C6 fluorescence signal was read at λex 488 nm λem 510 nm. Elisa ACE2 was also used to assess the competition between Lipo-S1 and NP, according with a published procedure [75]. Briefly, Rho-incorporated Lipo-S1 vs 6C-loaded NP were mixed in the ratio of 1:1 (0.98 μg/mL for each sample, considering the same amount of dye-loaded into NP) and incubated overnight at 4 °C in the ELISA ACE2. Next, the Elisa plate was washed 5 times according to manufacture and binding between the hACE2 and Lipo-S1 or NP was evaluated through fluorescence emission using a CLARIOstar® microplate reader (BMG Labtech). Fluorescent signal was read for Rho (λex 570 nm; λem ∼610 nm) and 6C (λex 488 nm - λem 510 nm).
4.7. AlamarBlue assays
AlamarBlue (AB) assay was used both to elucidate the inhibitory effect on CPE induction caused by SARS-CoV-2 infection and to assess the NP cytotoxicity. This assay was carried out according to manufacturer's instructions. Briefly, Vero E6 cells (infected and non infected) were incubated with Dulbecco's Modified Eagle Medium (DMEM) without FBS or supplements, and AB solution (5% [v/v] solution of AB dye). Following 3 h incubation, AB absorbance was quantified at 600 nm using a Fisher AccuSkan FC microplate reader. Six technical replicates per experiment at concentration of 5-1-0.1 μM were carried out (n = 3) for cytopatic effect evaluation of SARS-CoV-2 in the presence of NP, while scale dose 10–0.01 μM were used to test NP cytoxicity. Finally, for each plate the reading was also done in triplicate (the median values obtained from 3 different wells was calculated) in order include the technical variability due to the efficiency of AB assay, sensitivity of the plate reader or simply related to the sample preparation. The relative cell viability rate (%) was calculated by comparing infected and non-infected wells [(mean fluorescence of treated-SARS-CoV-2 infected wells/mean fluorescence of treated non-infected wells) x 100]. Positive (infected, non-treated with NP, and infected treated drug) and negative (non-infected, non-treated, non-treated exposed to propylene glycol medium with cells) control wells were also included. Each assay was carried out in triplicate. To score SARS-CoV-2 induced CPE, microscopy was performed in cells infected with virus, alone and after NP treatment.
4.8. Viral RNA extraction and quantification of SARS-CoV-2 viral RNA genome copy number by quantitative real-time (qRT-PCR)
One hundred microliter cell culture supernatant was harvested for viral RNA extraction using Quick-RNATM Microprep Kit (R1050) according to the manufacturer's instructions. RNA was eluted in 10 μL RNase-free water. Reverse transcription was assessed with a 5x All-In-One RT Mastermix (abm), and viral RNA was quantified by qRT-PCR on BioRad Real-time PCR system (BioRad) with TB BrightGreen Premix Taq II (abm). cDNA was synthesized in 20 μL reaction and 2 μL cDNA was used as template for quantitative PCR. The SARS-CoV-2 N gene was amplified using RBD forward 5′-CAATGGTTTAACAGGCACAGG-3′ and RBD-Reverse 5′-CTCAAGTGTCTGTGGATCACG-3. PCR amplification was performed as follows: 95 °C for 5 min followed by 40 cycles consisting of 95 °C for 15 s, 54 °C for 15 s, 72 °C for 30 s. The dose-response curves were plotted form viral RNA copies versus the drug concentrations by using GraphPad Prism 6 software.
4.9. ACE2 inhibition assays
The ACE2 inhibitor screening assay kit (BPS Bioscience, San Diego, USA) was purchased to determine the exopeptidase activity of ACE2 and, consequently, its putative inhibition by TNP-1E and ligand 1, using MLN-4760 as control inhibitor. The assays were performed according to the manufacture's protocol (catalog #79923). Briefly, enzyme (ACE2) stocks were prepared from the supplied kit; 20 μL of enzyme solution (0.5 ng/μL in ACE2 buffer) were added to each wells designated for the assay (considering positive “test inhibitors” and negative “blank” control). The samples (TNP-1E, 1, and MLN-4760, used as control inhibiton) were serially diluted in 1% DMSO as follows: 15, 3, 0.6, 0.12, 0.0024, 0.0048, 0.00096 μg/mL). Next, 5 μL of sample inhibitor solutions were added to respective designated wells, and the reactions were incubated at room temperature for 5 min. All experiments were performed in triplicates. After incubation, 25 μL ACE2 fluorogenic substrate were added to the wells, and reactions were incubated for 1 h at room temperature. The fluorescence intensities (RFU) of the samples (λ excitation = 555 nm, λ emission = 585 nm) were detected using a Beckman Coulter DTX880 multimode microplate plate reader. The background hydrolysis was subtracted by the data, which were fitted to a variable slope equation using GraphPad prism software 8.4.3. The IC50 values were calculated based on the positive controls and set to 100%.
4.10. Lentivirus production in HEK293T cells
Expression clone DNA (625 ng) was mixed with packaging plasmids pVSV-G and pCMV delta R8.2 at the ratio of 8:1 by mass. Viafect transfection reagent (Promega) was incubated with OPTIMEM (Gibco) (1: 12.5, v/v) for 20–30 min at room temperature, prior to adding to the DNA mix. HEK293T cells were plated until 70–90% confluence prior to transfection. After 16 h, the medium was replaced with fresh medium. After 48 h, the virus was harvested and filtered in 0.45 μm in a 15 mL tube. Lenti-X concentrator (Takara) was mixed with the medium containing the virus in the ratio of 1:3 media:Lenti-X. The mix was left overnight at 4 °C following a 45-min centrifugation at 1500g. The supernatant was removed by aspiration and the pellet was resuspended in 400 μL of 1XPBS (Gibco).
4.11. Construction of pLentivirus-hACE2-GFP vectors
The mature polypeptide of human ACE2 (GenBank NM_021804.3) was cloned in to the XbaI-BamHI site of pLenti-CMV-MCS-GFP-SV-puro (Addgene #73582). IRES linker was previously inserted into pcDNA3-sACE2v4-sfGFP via BamHI restriction enzyme site. The pcDNA3-sACE2v4-IRES-sfGFP insert was removed by restriction enzymes using NheI- XhoI. Next, the NheI-XhoI sites were converted into XbaI-BamHI by PCR (Primers: Forward-TAGCCTAGAGCCACCATGTCAAGCT, Reverse-CACCTGATCCCATTTGTAG AGCTCATCCATGCCATG) to be compatible with pLenti-CMV-MCS-SV40-PURO vector. Amplicon size was evaluated through gel electrophoresis in agarose 1.2%. DNA band was excised from the gel and purified using gel clean up reaction (Macherey Nagel). DNA insert and linear destination vector were mixed at a 1:1 ratio respectively. T4 was used for the ligase reaction and incubated overnight at 16 °C. Successful ligation of the inserted vector was verified through gel electrophoresis in agarose 2%. The DNA band was excised from the gel and purified using gel clean up reaction (Macherey Nagel).
4.12. Fluorescence microscopy
Cells were seeded in clear bottom black plates and allowed to adhere overnight, and then treated with indicated samples for 3 h at 37 °C. Next, cells were washed with 1 x PBS to remove excess NP (Rho-TNP-1 or Rho-NP-0) and dye. Cells were then imaged using BD Pathway 435 High-Content Bioimager (BD Biosciences) using 20x objective.
Author contributions
The work was performed through contributions of all authors. All authors have given approval to the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully thank Dr. Maria Orecchioni for her professional and unvaluable assistance with NMR experiments, Dr. Pankaj Kumar Singh for his great help in docking calculations, and Dr. Barbara J. Dillon at UCLA COVID-19 Basic, Translational, and Clinical Science Team for the use of Biosafety Level 3 Laboratory and critical support during this work. MS is grateful to the Biovia Dassault Systèmes SE Team for the opportunity to evaluate a trial of the latest version of software Biovia Discovery Studio, released in the context of a SARS-CoV-2 related research. T. H. thanks the National Institute of Heath (HL118650, HL129727, HL111437, HL149808, BX004356) and the American Heart Association COVID-19 Rapid Response Award (HA20203858) for support on biological work. We also thank Prof. Eugenio Garribba, Dr. Federico Pisanu, Prof. Mauro Carcelli, Prof. Dominga Rogolino for their support with mass analyses, Mr. Salvatore Marceddu for SEM analyses, Mr. Antonello Unali for his administrative assistance, Dr. Paolo D'Aquila for his kind support, and Dr. Nicolino Pala and Dr. Alessandro Arca for their friendly and fruitful discussion during the pandemic period.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114121.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Naming the Coronavirus Disease (COVID-19) and the Virus that Causes it.https://www.who.int/emergencies/diseases/novelcoronavirus-2019/technical-guidance. Virological.org Novel 2019 Coronavirus Genome. [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respirator disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osman E.E.A., Toogood P.L., Neamati N. COVID-19: living through another pandemic. ACS Infect. Dis. 2020;6:1548–1552. doi: 10.1021/acsinfecdis.0c00224. [DOI] [PubMed] [Google Scholar]
- 6.Preparing for the next pandemic. Nat. Med. 2021;27:357. doi: 10.1038/s41591-021-01291-z. [DOI] [PubMed] [Google Scholar]
- 7.Adamson C.S., Chibale K., Goss R.J.M., Jaspars M., Newman D.J., Dorrington R.A. Antiviral drug discovery: preparing for the next pandemic. Chem. Soc. Rev. 2021;50:3647–3655. doi: 10.1039/d0cs01118e. [DOI] [PubMed] [Google Scholar]
- 8.Meganck R.M., Baric R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021;27:401–410. doi: 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- 9.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.Á., Urquiza J., Ramírez D., Alonso C., Campillo N.E., Martinez A. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 12.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses, Drug Discov. Today Off. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugin K., Woodcock J. Trends in COVID-19 therapeutic clinical trials. Nat. Rev. Drug Discov. 2021;20:254–255. doi: 10.1038/d41573-021-00037-3. [DOI] [PubMed] [Google Scholar]
- 15.Ledford H. COVID antiviral pill: what scientists still want to know. Nature. 2021;599:358–359. doi: 10.1038/d41586-021-03074-5. [DOI] [PubMed] [Google Scholar]
- 16.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolin R., Hirsch M.S. Remdesivir - an important first step. N. Engl. J. Med. 2020;383:1886–1887. doi: 10.1056/NEJMe2018715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remdesivir emergency Use authorization. Gilead Sci., Inc. 2020 https://www.gilead.com/-/media/files/pdfs/remdesivir/euafda-authorization-letter_01may2020-old.pdf?la=en&hash= 2AE25800C7C9612D2F67F28D3F3B9921 [Google Scholar]
- 19.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai C.C., Chen C.H., Wang C.Y., Chen K.H., Wang Y.H., Hsueh P.R. Clinical efficacy and safety of remdesivir in patients with COVID-19: a systematic review and network meta-analysis of randomized controlled trials. J. Antimicrob. Chemother. 2021;76:1962–1968. doi: 10.1093/jac/dkab093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottlieb R.L., Vaca C.E., Paredes R., Mera J., Webb B.J., Perez G., Oguchi G., Ryan P., Nielsen B.U., Brown M., Hidalgo A., Sachdeva Y., Mittal S., Osyiemi O., Skarbinski J., Juneja K., Hyland R.H., Osinusi A., Chen S., Gamus G., Abdelghany M., Davies S., Behenna-Renton N., Duff F., Marty F.M., Katz M.J., Ginde A.A., A, Brown S., M., Schiffer J.T., Hill J.A. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2116846. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deb S., Reeves A.A., Hopefl R., Bejusca R. ADME and pharmacokinetic properties of remdesivir: its drug interaction potential. Pharmaceuticals. 2021;14:655. doi: 10.3390/ph14070655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahakijpijarn S., Moon C., Koleng J.J., Christensen D.J., Williams R.O., III Development of remdesivir as a dry powder for inhalation by thin film freezing. Pharmaceutics. 2020;12:1002. doi: 10.3390/pharmaceutics12111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun D. Remdesivir for treatment of COVID-19: combination of pulmonary and IV administration may offer additional benefit. AAPS J. 2020;22:77. doi: 10.1208/s12248-020-00459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vartak R., Patil S.M., Saraswat A., Patki M., Kunda N.K., Patel K. Aerosolized nanoliposomal carrier of remdesivir: an effective alternative for COVID-19 treatment in vitro. Nanomedicine. 2021;16:1187–1202. doi: 10.2217/nnm-2020-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour H.M., Rhee Y.S., Wu X. Nanomedicine in pulmonary delivery. Int. J. Nanomed. 2009;4:299–319. doi: 10.2147/ijn.s4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer R., Hsia C.C., Nguyen K.T. Nano-therapeutics for the lung: state-of-the-art and future perspectives. Curr. Pharmaceut. Des. 2015;21:5233–5244. doi: 10.2174/1381612821666150923095742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim Y.H., Tiemann K.M., Hunstad D.A., Elsabahy M., Wooley K.L. Polymeric nanoparticles in development for treatment of pulmonary infectious diseases. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2016;8:842–871. doi: 10.1002/wnan.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praphawatvet T., Peters J.I., 3rd Williams R.O. Inhaled nanoparticles-An updated review. Int. J. Pharm. 2020;587:119671. doi: 10.1016/j.ijpharm.2020.119671. [DOI] [PubMed] [Google Scholar]
- 31.Sanna V., Sechi M. Therapeutic potential of targeted nanoparticles and perspective on nanotherapies. ACS Med. Chem. Lett. 2020;11:1069–1073. doi: 10.1021/acsmedchemlett.0c00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min Y., Caster J.M., Eblan M.J., Wang A.Z. Clinical translation of nanomedicine. Chem. Rev. 2015;115:11147–11190. doi: 10.1021/acs.chemrev.5b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z., Ukidve A., Kim J., Mitragotri S. Targeting strategies for tissue-specific drug delivery. Cell. 2020;181:151–167. doi: 10.1016/j.cell.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 41.Guy J.L., Jackson R.M., Acharya K.R., Sturrock E.D., Hooper N.M., Turner A.J. Angiotensin-converting enzyme-2 (ACE2): comparative modeling of the active site, specificity requirements, and chloride dependence. Biochemistry. 2003;42:13185–13192. doi: 10.1021/bi035268s. [DOI] [PubMed] [Google Scholar]
- 42.Lubbe L., Cozier G.E., Oosthuizen D., Acharya K.R., Sturrock E.D. ACE2 and ACE: structure-based insights into mechanism, regulation and receptor recognition by SARS-CoV. Clin. Sci. 2020;134:2851–2871. doi: 10.1042/CS20200899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang F., Yang J., Zhang Y., Dong M., Wang S., Zhang O., Liu F.F., Zhang K., Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1-7: novel therapeutic targets. Nat. Rev. Cardiol. 2014;11:413–426. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos R.A., Ferreira A.J., Verano-Braga T., Bader M. Angiotensin-converting enzyme 2, angiotensin-(1-7) and Mas: new players of the renin-angiotensin system. J. Endocrinol. 2013;216:R1–R17. doi: 10.1530/JOE-12-0341. [DOI] [PubMed] [Google Scholar]
- 45.Iwai M., Horiuchi M. Devil and angel in the renin-angiotensin system: ACE-angiotensin II-AT1 receptor axis vs. ACE2-angiotensin-(1–7)-Mas receptor axis. Hypertens. Res. 2009;32:533–536. doi: 10.1038/hr.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saponaro F., Rutigliano G., Sestito S., Bandini L., Storti B., Bizzarri R., Zucchi R. ACE2 in the era of SARS-CoV-2: controversies and novel perspectives. Front. Mol. Biosci. 2020;7:588618. doi: 10.3389/fmolb.2020.588618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamai L. The Yin and Yang of ACE/ACE2 pathways: the rationale for the use of renin-angiotensin system inhibitors in COVID-19 patients. Cells. 2020;9:1704. doi: 10.3390/cells9071704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I., Hooper N.M., Turner A.J. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J. Biol. Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α-production and facilitates viral entry. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zipeto D., Palmeira J.d.F., Argañaraz G.A., Argañaraz E.R. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front. Immunol. 2020;11:576745. doi: 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/Angiotensin-(1-7)/MAS axis of the renin-angiotensin system: focus on angiotensin-(1-7) Physiol. Rev. 2018;98:505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J., Sun P.D. High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. J. Biol. Chem. 2020;295:18579–18588. doi: 10.1074/jbc.RA120.015303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G., Yang M.L., Duan Z.L., Liu F.L., Jin L., Long C.B., Zhang M., Tang X.P., Xu L., Li Y.C., Kamau P.M., Yang L., Liu H.Q., Xu J.W., Chen J.K., Zheng Y.T., Peng X.Z., Lai R. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia H., Neptune E., Cui H. Targeting ACE2 for COVID-19 therapy: opportunities and challenges. Am. J. Respir. Cell Mol. Biol. 2021;64:416–425. doi: 10.1165/rcmb.2020-0322PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dales N.A., Gould A.E., Brown J.A., Calderwood E.F., Guan B., Minor C.A., Gavin J.M., Hales P., Kaushik V.K., Stewart M., Tummino P.J., Vickers C.S., Ocain T.D., Patane M.A. Substrate based design of the first class of angiotensin-converting enzyme-related carboxypeptidase (ACE2) inhibitors. J. Am. Chem. Soc. 2002;124:11852–11853. doi: 10.1021/ja0277226. [DOI] [PubMed] [Google Scholar]
- 57.Joshi S., Balasubramanian N., Vasam G., Jarajapu Y.P.R. Angiotensin converting enzyme versus angiotensin converting enzyme-2 selectivity of MLN-4760 and DX600 in human and murine bone marrow-derived cells. Eur. J. Pharmacol. 2016;774:25–33. doi: 10.1016/j.ejphar.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanna V., Pintus G.P., Roggio A.M., Punzoni S., Posadino A.M., Arca A., Marceddu S., Bandiera P., Uzzau S., Sechi M. Targeted biocompatible nanoparticles for the delivery of (-)-epigallocatechin 3-gallate to prostate cancer cells. J. Med. Chem. 2011;54:1321–1332. doi: 10.1021/jm1013715. [DOI] [PubMed] [Google Scholar]
- 59.Sanna V., Nurra S., Pala N., Marceddu S., Pathania D., Neamati N., Sechi M. Targeted nanoparticles for the delivery of novel bioactive molecules to pancreatic cancer cells. J. Med. Chem. 2016;59:5209–5220. doi: 10.1021/acs.jmedchem.5b01571. [DOI] [PubMed] [Google Scholar]
- 60.Strebhardt K., Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nat. Rev. Drug Discov. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 61.Ahmad A., Khan F., Mishra R.K., Khan R. Precision cancer nanotherapy: evolving role of multifunctional nanoparticles for cancer active targeting. J. Med. Chem. 2019;62:10475–10496. doi: 10.1021/acs.jmedchem.9b00511. [DOI] [PubMed] [Google Scholar]
- 62.Shi J., Xiao Z., Kamaly N., Farokhzad O.C. Self-assembled targeted nanoparticles: evolution of technologies and bench to bedside translation. Acc. Chem. Res. 2011;44:1123–1134. doi: 10.1021/ar200054n. [DOI] [PubMed] [Google Scholar]
- 63.Maresca K.P., Hillier S.M., Femia F.J., Keith D., Barone C., Joyal J.L., Zimmerman C.N., Kozikowski A.P., Barrett J.A., Eckelman W.C., Babich J.W. A series of halogenated heterodimeric inhibitors of prostate specific membrane antigen (PSMA) as radiolabeled probes for targeting prostate cancer. J. Med. Chem. 2009;52:347–357. doi: 10.1021/jm800994j. [DOI] [PubMed] [Google Scholar]
- 64.Sanna V., Singh C.K., Jashari R., Adhami V.M., Chamcheu J.C., Islam R., Sechi M., Mukhtar H., Siddiqui I.A. Targeted nanoparticles encapsulating (-)-epigallocatechin-3-gallate for prostate cancer prevention and therapy. Sci. Rep. 2017;7:41573. doi: 10.1038/srep41573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schülke N., Varlamova O.A., Donovan G.P., Ma D., Gardner J.P., Morrissey D.M., Arrigale R.R., Zhan C., Chodera A.J., Surowitz K.G., Maddon P.J., Heston W.D., Olson W.C. The homodimer of prostate-specific membrane antigen is a functional target for cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12590–12595. doi: 10.1073/pnas.1735443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Natesh R., Schwager S.L.U., Evans H.R., Sturrock E.D., Ravi Acharya K. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry. 2004;43:8718–8724. doi: 10.1021/bi049480n. [DOI] [PubMed] [Google Scholar]
- 67.Haga S., Nagata N., Okamura T., Yamamoto N., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antivir. Res. 2010;85:551–555. doi: 10.1016/j.antiviral.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ingram R.N., Orth P., Strickland C.L., Le H.V., Madison V., Beyer B.M. Stabilization of the autoproteolysis of TNF-alpha converting enzyme (TACE) results in a novel crystal form suitable for structure-based drug design studies. Protein Eng. Des. Sel. 2006;19:155–161. doi: 10.1093/protein/gzj014. [DOI] [PubMed] [Google Scholar]
- 70.Abdel-Magid A.F., Carson K.G., Harris B.D., Maryanoff C.A., Shah R.D. Reductive amination of aldehydes and ketones with sodium triacetoxyborohydride. studies on direct and indirect reductive amination procedures (1) J. Org. Chem. 1996;61:3849–3862. doi: 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- 71.Sanna V., Roggio A.M., Posadino A.M., Cossu A., Marceddu S., Mariani A., Alzari V., Uzzau S., Pintus G., Sechi M. Novel docetaxel-loaded nanoparticles based on poly(lactide-cocaprolactone) and poly(lactide-co-glycolide-co-caprolactone) for prostate cancer treatment: formulation, characterization and cytotoxicity studies. Nanoscale Res. Lett. 2011;6:260. doi: 10.1186/1556-276X-6-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu F., Langer R., Farokhzad O.C. Formulation/preparation of functionalized nanoparticles for in vivo targeted drug delivery. Methods Mol. Biol. 2009;544:589–598. doi: 10.1007/978-1-59745-483-4_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otsuka H., Nagasaki Y., Kataoka K. PEGylated nanoparticles for biological and pharmaceutical applications. Adv. Drug Deliv. Rev. 2003;55:403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 74.Bhattacharjee S. DLS and zeta potential. What they are and what they are not? J. Contr. Release. 2016;235:337–351. doi: 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 75.Kaneko N., Satta S., Komuro Y., Muthukrishnan S.D., Kakarla V., Guo L., An J., Elahi F., Kornblum H.I., Liebeskind D.S., Hsiai T., Hinman J.D. Flow mediated susceptibility and molecular response of cerebral endothelia to SARS-CoV-2 infection. Stroke. 2021;52:260–270. doi: 10.1161/STROKEAHA.120.032764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mo C., Yamagata R., Pan A., Reddy J., Hazari N., Duke G. Development of a high-throughput Alamar blue assay for the determination of influenza virus infectious dose, serum antivirus neutralization titer and virus ca/ts phenotype. J. Virol. Methods. 2008;150:63–69. doi: 10.1016/j.jviromet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 77.De Meyer S., Bojkova D., Cinatl J., Van Damme E., Buyck C., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mammen M., Choi S.K., Whitesides G.M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 79.Mulder A., Huskens J., Reinhoudt D.N. Multivalency in supramolecular chemistry and nanofabrication. Org. Biomol. Chem. 2004;2:3409–3424. doi: 10.1039/b413971b. [DOI] [PubMed] [Google Scholar]
- 80.Badjić J.D., Nelson A., Cantrill S.J., Turnbull W.B., Stoddart J.F. Multivalency and cooperativity in supramolecular chemistry. Acc. Chem. Res. 2005;38:723–732. doi: 10.1021/ar040223k. [DOI] [PubMed] [Google Scholar]
- 81.Toots M., Yoon J.J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G., Shean R.C., Bluemling G.R., Kolykhalov A.A., Greninger A.L., Natchus M.G., Painter G.R., Plemper R.K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tammam S.N., Safy S.E., Ramadan S., Arjune S., Krakor E., Mathur S. Repurpose but also (nano)-reformulate! the potential role of nanomedicine in the battle against SARS-CoV2. J. Contr. Release. 2021;337:258–284. doi: 10.1016/j.jconrel.2021.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boraei A.T.A., Singh P.K., Sechi M., Satta S. Discovery of novel functionalized 1,2,4-triazoles as PARP-1 inhibitors in breast cancer: design, synthesis and antitumor activity evaluation. Eur. J. Med. Chem. 2019;182:111621. doi: 10.1016/j.ejmech.2019.111621. [DOI] [PubMed] [Google Scholar]
- 85.Singh P.K., Silakari O. Molecular dynamics and pharmacophore modeling studies of different subtype (ALK and EGFR (T790M)) inhibitors in NSCLC. SAR QSAR. Environ. Res. 2017;28:221–233. doi: 10.1080/1062936X.2017.1300189. [DOI] [PubMed] [Google Scholar]
- 86.Pala N., Stevaert A., Dallocchio R., Dessì A., Rogolino D., Carcelli M., Sanna V., Sechi M., Naesens L. Virtual screening and biological validation of novel influenza virus PA endonuclease inhibitors. ACS Med. Chem. Lett. 2015;6:866–871. doi: 10.1021/acsmedchemlett.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.