Abstract
Background:
Epigenetic age acceleration has been studied as a promising biomarker of age-related conditions, including cognitive aging. This pilot study aims to explore potential cognitive aging-related biomarkers by investigating the relationship of epigenetic age acceleration and cognitive function and by examining the epigenetic age acceleration differences between successful cognitive aging (SCA) and normal cognitive aging (NCA) among Korean community-dwelling older adults (CDOAs).
Methods:
We used data and blood samples of Korean CDOAs from the Korean Frailty and Aging Cohort Study. The participants were classified into two groups, SCA (above the 50th percentile in all domains of cognitive function) and NCA. The genome-wide DNA methylation profiling array using Illumina Infinium MethylationEPIC BeadChip was used to calculate the following: the DNA methylation age, universal epigenetic age acceleration, intrinsic epigenetic age acceleration (IEAA), and extrinsic epigenetic age acceleration (EEAA). We also used Pearson correlation analysis and independent t-tests to analyze the data.
Results:
Universal age acceleration correlated with the Frontal Assessment Battery test results (r = −0.42, p = 0.025); the EEAA correlated with the Word List Recognition test results (r = −0.41, p = 0.027). There was a significant difference between SCA and NCA groups in IEAA (p = 0.041, Cohen’s d = 0.82) and EEAA (p = 0.042, Cohen’s d = 0.78).
Conclusions:
Epigenetic age acceleration can be used as a biomarker for early detection of cognitive decline in Korean community-dwelling older adults. Large longitudinal studies are warranted.
Keywords: cognitive function, successful aging, biomarkers, DNA methylation, epigenomics
Aging populations are rapidly increasing worldwide. During the aging process, many physiological changes occur, including age-related cognitive decline. A decline in cognitive function has been associated with the normal aging process, but it has been used as one of the important indicators of neurological conditions such as Alzheimer’s diseases and other dementias (Smith, 2016). Age-related cognitive decline or normal cognitive aging (NCA) is a preclinical stage of deterioration of cognitive function related to advancing age due to changes in structure and brain function, that are influenced by several factors including lifestyle and environmental factors (Harada et al., 2013; T. A. Salthouse, 2019). While some older adults with NCA develop dementia (Harada et al., 2013), some older adults maintain their cognitive function as they age (successful cognitive aging [SCA]). Expanding our understanding of the underlying biological mechanism to explain these differences in cognitive decline patterns (NCA vs SCA) may help us develop interventions and/or treatments that can delay or prevent these progressive cognitive declines.
Increasing evidence highlights the potential involvement of the epigenetic clock as an underlying mechanism of these age-related changes, including cognitive decline. The epigenetic clock is a mathematical algorithm that uses values assigned to specific CpGs in the genome to calculate the biological age of a person (Horvath, 2013). The epigenetic clock that used the DNA methylation-based age estimator was found to be a better predictor of sleep difficulties, cognitive impairment, or mortality among patients with chronic conditions such as cancer and Alzheimer’s disease (Carroll et al., 2017; Dugué et al., 2018; Levine, 2015, 2018; Lu et al., 2017; Marioni et al., 2015). The epigenetic age acceleration means the DNA methylation age is faster than the chronological age (Horvath & Raj, 2018). Three studies explored the associations of epigenetic age acceleration and cognitive performance. The epigenetic age acceleration was positively correlated with neuropathological measurements (diffuse, neurite, and amyloid-beta plaques) and negatively correlated with the proportion of neurons relative to glial cells (Levine et al., 2015, 2018; Lu et al., 2017). However, the majority of the participants in that study were white individuals with Alzheimer’s disease. It is unknown if these relationship patterns are the same in NCA people with other ethnic backgrounds.
Our study aims to 1) investigate the associations of epigenetic age acceleration with specific domains of cognitive function, and 2) examine the differences in epigenetic age acceleration between SCA and NCA in the Korean community-dwelling older adults (CDOAs), hoping to explore potential biomarkers of cognitive aging.
Materials and Methods
Participants
This cross-sectional pilot study included 29 participants from the Korean Frailty and Aging Cohort Study (KFACS), a longitudinal, multi-center cohort study of Korean older adults that aimed to identify the causes and risk factors of frailty in that population (Won et al., 2020). Starting in 2016, the parent study (KFACS) recruited 3,000 community-dwelling older adults 70–84 years of age from 10 urban and rural medical facilities starting. The participants’ physical, mental, and psychosocial status, health conditions, and routine laboratory tests were collected every 2 years.
The participants in our study were selected based on available neuropsychological test scores which were measured by the Korean version of the modified Consortium to Establish a Registry for Alzheimer’s Disease Assessment Battery (CERAD-K). Using the CERAD-K domains (global cognition, memory, attention, and executive function) score, 14 participants scored above the 50th percentile mark (SCA group) compared to general Korean older adults CERAD-K scores (Negash et al., 2011) (Supplement Table 1). A total of 15 age- and gender-matched participants who scored below the 50th percentile of CERAD-K (NCA group) were selected as a comparison group.
Measures
Cognitive functions
Comprehensive cognitive function was measured by the CERAD-K, a standardized assessment for the evaluation of patients with Alzheimer’s disease (J. H. Lee et al., 2002). It consists of scores from four different domains: (1) Mini-Mental State Exam (MMSE-KC), (2) Word List Memory/Recall/ Recognition, (3) Digit Span Forward/Backward, and (4) Frontal Assessment Battery (FAB).
Domain 1: The MMSE-KC domain is a screening tool to evaluate the global cognitive function in older adults (D. Y. Lee et al., 2004). The MMSE-KC has good internal consistency (Cronbach’s α: 0.92) and validity (J. H. Lee et al., 2002; K. Lee et al., 2009).
Domain 2: The Word List Memory/Recall/Recognition domain assesses the imputation of new information, recalls the information, and distinguishes the inputted information (D. Y. Lee et al., 2004). Participants were asked to complete three memory tasks: (1) recall a series of 10 words presented thrice in a different order within 90 seconds; (2) recall any words from the previous task in 90 seconds, and (3) discriminate words from the previous 10 words from the word list memory task from 10 new words.
Domain 3: The Digit Span Forward and Backward domain assessed attention and working memory (Choi et al., 2014). The participants were asked to repeat a numerical sequence of two to nine digits in the exact order of how they were presented, then repeat those numbers in backward order.
Domain 4: The FAB domain is a test for executive function (T. H. Kim et al., 2010), and consists of six tasks to evaluate conceptualization, mental flexibility, motor programming, sensitivity to interference, inhibitory control, and environmental autonomy. The total score is 18. A detailed description of how this domain was measured is included in Supplement Table 1.
DNA methylation microarray and data processing
The DNA methylation microarray was conducted using Illumina Infinium MethylationEPIC BeadChip kits (Illumina, Inc., San Diego, CA) by Macrogen, Inc. (Seoul, Republic of Korea). The 500 ng of genomic DNA from blood was extracted and bisulfite sodium was administered to deaminate cytosine into uracil using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). The 5-methylcytosine (methylated cytosine) was unchanged to bisulfite conversion. The bisulfite-converted DNA was hybridized to the BeadChip. After washing the unhybridized DNA, each of the CpG probe sites was calculated at the methylation level. DNA methylation data at CpG was represented by fluorescent signals from the M (methylated) and U (unmethylated) alleles. The background intensity was subtracted as a set of negative control at the CpG sites. The ratio of fluorescent signals was calculated from the two alleles. The β-value reflected the methylation level range from 0 to 1 at each CpG site. The data were exported and analyzed using Illumina GenomeStudio v2011.1 (Methylation Module v1.9.0) and R statistical software version 3.0.2 (The R Foundation for Statistical Computing, Vienna, Austria).
Epigenetic age acceleration
DNA methylation age prediction and epigenetic age acceleration were performed using the online DNA Methylation Age Calculator (https://dnamage.genetics.ucla.edu/home). Horvath’s clock is the first multi-tissue age estimator validated by 8,000 microarray samples and consisting of selected 353 CpGs, 193 of CpGs are positively correlated with age, and 160 CpGs are negatively correlated (Horvath, 2013). The estimator was based on data from Illumina Infinium Human Methylation 27 BeadChip and Illumina Infinium Human Methylation 450K BeadChip arrays. Despite the missing 17 CpGs in the Illumina Infinium MethylationEPIC BeadChip array, the estimated DNA methylation age was unaffected by the missing CpGs (Dhingra et al., 2019).
We measured three types of epigenetic age acceleration: universal age acceleration, intrinsic epigenetic age acceleration (IEAA), and extrinsic epigenetic age acceleration (EEAA), and the DNA methylation age. The universal age acceleration, denoted as “AgeAccelerationResidual” in the calculator, is defined as residuals from regressing DNA methylation age based on chronological age (R language: residuals (lm(DNAmAge − Age). Positive values suggested that the DNA methylation age was faster than the chronological age, whereas negative values suggested that the DNA methylation age was slower than the chronological age. The major feature of blood-based estimates is that the DNA methylation age can be changed by blood cell counts (Horvath & Raj, 2018). The proportions of naive or senescent cytotoxic (CD8) T-cells have changed with advancing age. For example, sleep disturbance was associated with a decrease of CD8 T-cells and an increase of EEAA (Carroll et al., 2017). The IEAA is independent of estimated blood cell type proportion by Horvath’s method; EEAA is affected by the blood cell estimates by Hannum’s method and might represent immunosenescence (Chen et al., 2016). The Horvath’s clock is not likely to affect the composition of the blood cells (Horvath & Raj, 2018); however, both IEAA and EEAA were measured in this study to measure accurate epigenetic age acceleration.
Ethical Statements
The original study was approved by the institutional review board (IRB) at the Kyung Hee Medical Center, Seoul, Republic of Korea (KHUH 2015-12-103). This study was exempt from review by the IRB (KHSIRB-20-129).
Power Analysis
The sample size for this secondary analysis was calculated using G*Power 3.1.9.5. A previous study investigated the “Mean epigenetic delta age” between two groups (Maintainer group vs Decliner group) and used an independent t-test to report an effect size of (Cohen’s d 0.67; Degerman et al., 2017). Using that study, we estimated that the enrollment of 70 participants can achieve an α of 0.05 and a power of 0.8. The purpose of this pilot project, however, was to provide preliminary evidence of the relationship of epigenetic age acceleration and cognitive function and inform power analyses for future full-scale studies. Hetzog (2008) suggested that a pilot study should have a sample size of at least 10% of the full study sample size to achieve a reasonable effect size. For this pilot study, we enrolled 29 participants, 41.4% of the estimated full study sample size.
Statistical analysis
Parametric data were denoted as mean ± standard deviation, while non-parametric data were indicated as median (interquartile range, IQR), and categorical variables were expressed in numbers and percentages. A p-value of ≤0.05 was considered statistically significant. Scatter plots were generated to explore the relationships between chronological age and DNA methylation age. We calculated the Pearson correlation coefficient to illustrate the relationships between epigenetic age acceleration and cognitive function. The differences in demographic, general characteristics, DNA methylation age, and epigenetic age accelerations between the SCA and NCA groups were tested using χ2 test, independent t-tests, and Mann-Whitey U tests. All statistical analyses were conducted using SPSS software 25.0 (IBM Corp., Armonk, NY, USA); graphical works were produced by R statistical software version 3.6.1 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Demographic and general characteristics of the participants
Twenty-nine participants were included in this pilot analysis. The mean chronological age of the participants was 75.7 ± 4.3 years, with 16 (55.2%) females. The average duration of education was 8 years (IQR: 6.0–12.0). Nearly 70% of the participants were married and living with others; most were residing in an urban setting. The demographic and general characteristics of these participants are presented in Table 1.
Table 1.
Demographic, General Characteristics, Neuropsychological Testing, and Epigenetic Age Acceleration Between the NCA and SCA Groups.
| Variables | Categories |
Total
(n = 29) |
NCA
(n = 15) |
SCA
(n = 14) |
p | Cohen’s d |
|---|---|---|---|---|---|---|
| Age (year) | 75.7 ± 4.3 | 75.1 ± 4.9 | 76.4 ± 3.6 | 0.430 | ||
| Sex | 0.867 | |||||
| Male | 13 (44.8%) | 9 (60.0%) | 7 (50.0%) | |||
| Female | 16 (55.2%) | 6 (40.0%) | 7 (50.0%) | |||
| Body mass index (kg/m2) | 25.2 ± 3.2 | 25.7 ± 4.0 | 24.6 ± 2.2 | 0.332 | ||
| Smoking | 0.885 | |||||
| Never | 18 (62.1%) | 10 (66.7%) | 8 (57.1%) | |||
| Ever | 11 (37.9%) | 5 (33.3%) | 6 (42.9%) | |||
| Alcohol consumption | 1.000 | |||||
| Never | 10 (34.5%) | 5 (33.3%) | 5 (35.7%) | |||
| Ever | 19 (65.5%) | 10 (66.7%) | 9 (64.3%) | |||
|
Physical activity (day)
|
Vigorous† | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.334 | |
| Moderate† | 3.0 (0.0–6.0) | 3.0 (0.0–6.0) | 3.5 (0.0–7.0) | 0.639 | ||
| Light† | 7.0 (6.5–7.0) | 7.0 (7.0) | 7.0 (6.0–7.0) | 0.684 | ||
| Education (year)† | 8.0 (6.0–12.0) | 7.0 (6.0–12.0) | 12.0 (6.0–13.0) | 0.317 | ||
| Marital status | 0.798 | |||||
| Married | 19 (65.5%) | 9 (60.0%) | 10 (71.4%) | |||
| Divorced & widowed | 10 (34.5%) | 6 (40.0%) | 4 (28.6%) | |||
| Living status | 1.000 | |||||
| Alone | 8 (27.6%) | 4 (26.7%) | 4 (28.6%) | |||
| With others | 21 (72.4%) | 11 (73.3%) | 10 (71.4%) | |||
| Living area | 0.445 | |||||
| Urban | 22 (75.9%) | 10 (66.7%) | 12 (85.7%) | |||
| Rural | 7 (24.1%) | 5 (33.3%) | 2 (14.3%) | |||
| Neuropsychological Test | MMSE-KC† | 26.0 (25.0–27.0) | 29.0 (28.0–29.0) | <0.001 | ||
| Word List Memory | 16.7 ± 2.5 | 21.7 ± 2.3 | <0.001 | |||
| Word List Recall† | 5.0 (4.0–7.0) | 8.0 (7.0–9.0) | <0.001 | |||
| Word List Recognition† | 9.0 (8.0–10.0) | 10.0 (10.0) | 0.017 | |||
| Digit Span Forward† | 6.0 (4.0–7.0) | 7.0 (6.0–8.0) | 0.009 | |||
| Digit Span Backward | 3.6 ± 0.8 | 5.6 ± 1.2 | <0.001 | |||
| Frontal Assessment Battery† | 14.0 (11.0-15.0) | 16.0 (16.0–17.0) | <0.001 | |||
| Epigenetic age acceleration | DNA methylation age (year) | 71.1 ± 7.5 | 68.8 ± 4.9 | 0.340 | ||
| Universal Age Acceleration | 2.0 ± 5.5 | −1.5 ± 3.4 | 0.054 | 0.77 | ||
| IEAA | 2.0 ± 5.4 | −1.6 ± 3.1 | 0.041 | 0.82 | ||
| EEAA | 1.8 ± 4.2 | −1.3 ± 3.7 | 0.042 | 0.78 |
Note. †Mann-Whitney U test. NCA = normal cognitive aging; SCA = successful cognitive aging; MMSE-KC = Mini-Mental State exam in the Korean version; IEAA = intrinsic epigenetic age acceleration; EEAA = extrinsic epigenetic age acceleration.
Associations between cognitive function and epigenetic age acceleration
Table 2 shows the associations between three measures of epigenetic age acceleration and neuropsychological tests. The universal age acceleration showed significant correlation with IEAA (r = 0.96, p < 0.001) and EEAA (r = 0.41, p = 0.028). The universal age acceleration was significantly correlated with FAB (r = −0.42, p = 0.025); EEAA was significantly correlated with Word List Recognition (r = −0.41, p = 0.027).
Table 2.
Correlation Between the Neuropsychological Tests and Epigenetic Age Acceleration.
| Variables | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. Universal Age Acceleration | 1 | .95** | .41* | −.29 | −.36 | −.27 | −.10 | .01 | −.07 | −.42* |
| 2. IEAA | 1 | .30 | −.26 | −.35 | −.33 | −.05 | .09 | −.04 | −.34 | |
| 3. EEAA | 1 | −.23 | −.26 | −.28 | −.41* | −.31 | −.12 | −.20 | ||
| 4. MMSE-KC | 1 | .56** | .64** | .42* | .39* | .62** | .73** | |||
| 5. Word List Memory | 1 | .81** | .50** | .31 | .54** | .65** | ||||
| 6. Word List Recall | 1 | .54** | .28 | .44* | .56** | |||||
| 7. Word List Recognition | 1 | .54** | .38* | .63** | ||||||
| 8. Digit Span Forward | 1 | .65** | .60** | |||||||
| 9. Digit Span Backward | 1 | .55** | ||||||||
| 10. FAB | 1 |
Note. MMSE-KC = Mini-Mental State exam in the Korean version; IEAA = intrinsic epigenetic age acceleration; EEAA = extrinsic epigenetic age acceleration; FAB = frontal assessment battery.
*p < 0.05. **p < 0.01.
Epigenetic age acceleration differences between SCA and NCA groups
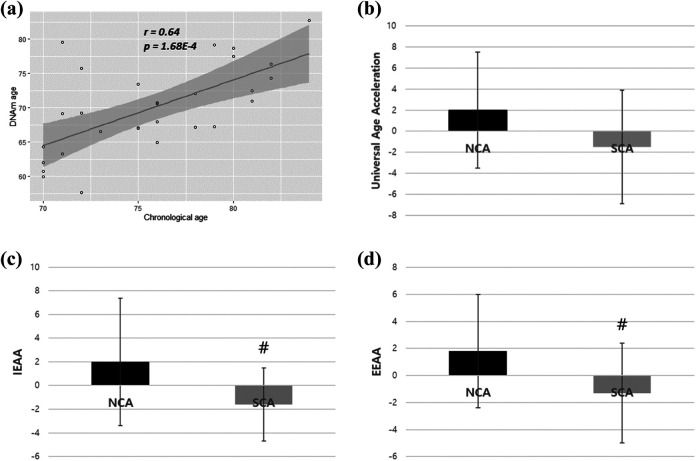
There was no significant difference between the two groups in age, sex, body mass index, health behaviors, and sociodemographic characteristics (Table 1). The epigenetic age acceleration result used to determine the methylation age, as predicted by Horvath’s clock, was highly correlated with chronological age (r = 0.64). While there were no significant differences in biological (DNA methylation) age between the two groups, participants in the SCA group had higher chronological vs. biological age (7.6 years’ difference) compared to those in the NCA group (4 years’ difference; Table 1). Moreover, we found there were significant differences of IEAA (p = 0.041, Cohen’s d = 0.82), EEAA (p = 0.042, Cohen’s d = 0.78), and universal age acceleration (p = 0.054, Cohen’s d = 0.77) between the NCA and SCA groups (Table 1 and Figure 1).
Figure 1.
Plots of epigenetic age acceleration analysis. (a) Scatter plot of the relationship between chronological age and DNA methylation age. (b) Bar graph of universal epigenetic age acceleration comparison between SCA and NCA. (c) Bar graph of IEAA compare son between SCA and NCA. (d) Bar graph of EEAA comparison between SCA and NCA. NCA = normal cognitive aging; SCA = successful cognitive aging; IEAA = intrinsic epigenetic age acceleration; EEAA = extrinsic epigenetic age acceleration. # p < .05 versus NCA group.
Discussion
By 2050, approximately 3 million Korean older adults are estimated to be diagnosed with dementia (K. Kim et al., 2012). The condition not only impacts an individual’s quality of life but also imposes a huge burden on families and their communities (M. D. Kim et al., 2009). The prevention of dementia by detecting the early stages of cognitive decline has become more critical because of the lack of effective pharmacologic treatments (Iqbal et al., 2014). Our findings suggest that the epigenetic clock can be used as a biomarker for cognitive decline in the Korean CDOAs. In this study, we demonstrated that the SCA group showed delayed epigenetic age acceleration despite no significant difference in demographic characteristics. To the best of our knowledge, this study is the first to explore the associations of epigenetic age acceleration and NCA in older adults who live in community-based dwellings in Asia.
Personal factors (chronological age, sex, education), health-related behaviors, history of depression, and sociodemographic characteristics are known risk factors for age-related conditions including dementia and cognitive decline (Baumgart et al., 2015; Legdeur et al., 2018; Lipnicki et al., 2013; Petersen et al., 2010). Our study, however, did not find significant differences in these risk factors between the NCA and SCA groups. This result may be due to the small sample size. Evidence suggests that a history of depression is one of the influential factors of cognitive decline. Our study was not able to provide depression scores because it was not collected in the parent study (KFACS). Future studies need to investigate the influence of depression in the epigenetic ages of NCA and SCA individuals.
Our study showed significant associations between universal age acceleration with executive function, as well as extrinsic epigenetic age acceleration (EEAA) with recognition memory function. Both executive and recognition memory functions are known to decrease in the aging process (Harada et al., 2013; T. Salthouse, 2012). In a previous study, EEAA, which is a measure of immunosenescence as well as biological age, was associated with cognitive decline in healthy men living in urban areas (Beydoun et al., 2020). Our study supports the idea that universal age acceleration and EEAA can be used to detect specific age-associated cognitive decline.
We also found that the NCA group had significantly higher IEAA and EEAA levels than the SCA group (Cohen’s d = 0.78 and 0.82). This suggests that people with good cognitive health (SCA group) have slower epigenetic age acceleration than people with normal age-related cognitive decline (NCA group). This is consistent with studies where people with cognitive decline have faster epigenetic age acceleration than the people with good cognitive health (Beydoun et al., 2020; Degerman et al., 2017; Levine et al., 2015). Studies have suggested that the cumulative effects of personal factors, health-related behaviors, and sociodemographic factors influence a phenotypic change through modifications at the DNA methylation level (Alegría-Torres et al., 2011; Martin & Fry, 2018), which can lead to mild cognitive impairment and dementia (Alegría-Torres et al., 2011). Our results highlight the promising role of epigenetic age acceleration as a potential biomarker of age-related cognitive decline.
Our study provides initial evidence that blood-based epigenetic biomarkers can be used as a potential biological age estimator to detect early-stage cognitive dysfunction in Korean CDOAs. The assessment of epigenetic age blood-based biomarkers is less invasive than the testing of cerebrospinal fluid amyloid-beta and tau protein levels, which are used in standard practice. The results of this pilot study should be interpreted with caution because of the small sample size and the cross-sectional nature of the study.
Conclusions
We found that epigenetic age acceleration can be used as a potential biomarker for cognitive decline in Korean CDOAs. These findings provide critical preliminary evidence to consider the role of epigenetic age acceleration in assessing cognitive health in aging. Because of the pilot nature of this cross-sectional study, longitudinal studies using larger sample sizes, comparisons with different ethnicities, and using DNAs from various tissues, are needed to validate these findings.
Supplemental Material
Supplemental Material, sj-pdf-1-brn-10.1177_1099800420983896 for Accelerated Epigenetic Age in Normal Cognitive Aging of Korean Community-Dwelling Older Adults by Jongmin Park, Chang Won Won, Leorey N. Saligan, Youn-Jung Kim, Yoonju Kim and Nada Lukkahatai in Biological Research For Nursing
Footnotes
Authors’ Note: Jongmin Park and Chang Won Won are co-first authors.
Author Contributions: Park, J contributed to conception and design contributed to acquisition, analysis, and interpretation; drafted manuscript; critically revised manuscript; and gave final approval agrees to be accountable for all aspects of work ensuring integrity and accuracy. Won, CW contributed to design, contributed to acquisition, critically revised manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Saligan, LN contributed to interpretation, critically revised manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Kim, Y-J contributed to conception, critically revised manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Kim, Y contributed to acquisition and analysis, critically revised manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Lukkahatai, N contributed to analysis and interpretation, drafted manuscript, critically revised manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no conflicts of interest with respect to the authorship and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by a grant from the Korea Health Technology R&D Project through the Korean Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI15C3153) and National Research Foundation of Korea funded by the Ministry of Education (grant number: 2019R1C1C1005519).
ORCID iDs: Jongmin Park  https://orcid.org/0000-0001-6176-5670
https://orcid.org/0000-0001-6176-5670
Yoonju Kim  https://orcid.org/0000-0003-4147-0490
https://orcid.org/0000-0003-4147-0490
Supplemental Material: Supplemental material for this article is available online.
References
- Alegría-Torres J. A., Baccarelli A., Bollati V. (2011). Epigenetics and lifestyle. Epigenomics, 3(3), 267–277. 10.1212/wnl.0000000000008756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M., Snyder H. M., Carrillo M. C., Fazio S., Kim H., Johns H. (2015). Summary of the evidence on modifiable risk factors for cognitive decline and dementia: A population-based perspective. Alzheimer’s & Dementia, 11(6), 718–726. 10.1016/j.jalz.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Beydoun M. A., Shaked D., Tajuddin S. M., Weiss J., Evans M. K., Zonderman A. B. (2020). Accelerated epigenetic age and cognitive decline among urban-dwelling adults. Neurology, 94(6), e613–e625. 10.1212/wnl.0000000000008756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. E., Irwin M. R., Levine M., Seeman T. E., Absher D., Assimes T., Horvath S. (2017). Epigenetic aging and immune senescence in women with insomnia symptoms: Findings from the women’s health initiative study. Biological Psychiatry, 81(2), 136–144. 10.1016/j.biopsych.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. H. Marioni R. E. Colicino E. Peters M. J. Ward-Caviness C. K. Tsai P. C. Roetker N. S. Just A. C. Demerath E. W. Guan W. Bressler J. Fornage M. Studenski S. Vandiver A. R. Moore A. Z. Tanaka T. Kiel D. P. Liang L. Vokonas P.…Horvath S. (2016). DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging, 8(9), 1844–1865. 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. J., Lee D. Y., Seo E. H., Jo M. K., Sohn B. K., Choe Y. M., Byun M. S., Kim J. W., Kim S. G., Yoon J. C., Jhoo J. H., Kim K. W., Woo J. I. (2014). A normative study of the digit span in an educationally diverse elderly population. Psychiatry Investigation, 11(1), 39–43. 10.4306/pi.2014.11.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degerman S., Josefsson M., Adolfsson A. N., Wennstedt S., Landfors M., Haider Z., Pudas S., Hultdin M., Nyberg L., Adolfsson R. (2017). Maintained memory in aging is associated with young epigenetic age. Neurobiology of Aging, 55, 167–171. 10.1016/j.neurobiolaging.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Dhingra R., Kwee L. C., Diaz-Sanchez D., Devlin R. B., Cascio W., Hauser E. R., Gregory S., Shah S., Kraus W. E., Olden K., Olden K. (2019). Evaluating DNA methylation age on the Illumina methylationepic bead chip. PLoS One, 14(4), e0207834. 10.1371/journal.pone.0207834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugué P., Bassett J. K., Joo J. E., Jung C., Ming Wong E., Moreno-Betancur M., Schmidt D., Makalic E., Li S., Severi G., Hodge A. M., Buchanan D. D., English D. R., Hopper J. L., Southey M. C., Giles G. G., Severi G. (2018). DNA methylation-based biological aging and cancer risk and survival: Pooled analysis of seven prospective studies. International Journal of Cancer, 142(8), 1611–1619. 10.1002/ijc.31189 [DOI] [PubMed] [Google Scholar]
- Harada C. N., Natelson Love M. C., Triebel K. L. (2013). Normal cognitive aging. Clinics in Geriatric Medicine, 29(4), 737–752. 10.1016/j.cger.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M. A. (2008). Considerations in determining sample size for pilot studies. Research in Nursing & Health, 31(2), 180–191. 10.1002/nur.20247 [DOI] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10), 3156. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S., Raj K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics, 19(6), 371. 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- Iqbal K., Liu F., Gong C. (2014). Alzheimer disease therapeutics: Focus on the disease and not just plaques and tangles. Biochemical Pharmacology, 88(4), 631–639. 10.1016/j.bcp.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Gwak K. P., Kim B. J., Kim S. Y., Kim S. K., Kim J. L., Kim T. H., Moon S. W., Park J. H., Bae J. N. (2012). 2012 National study on the prevalence of dementia in Korean elders. Seoul National University Bundang Hospital. [Google Scholar]
- Kim M. D., Hong S. C., Lee C. I., Kim S. Y., Kang I. O., Lee S. Y. (2009). Caregiver burden among caregivers of Koreans with dementia. Gerontology, 55(1), 106–113. 10.1159/000176300 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Huh Y., Choe J. Y., Jeong J. W., Park J. H., Lee S. B, Lee J. J., Jhoo J. H., Lee D. Y., Woo J. I., Kim K. W. (2010). Korean version of frontal assessment battery: Psychometric properties and normative data. Dementia and Geriatric Cognitive Disorders, 29(4), 363–370. 10.1159/000297523 [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Lee K. U., Lee J. H., Kim K. W., Jhoo J. H., Kim S. Y., Yoon J. C., Woo S. I., Ha J., Woo J. I. (2004). A normative study of the CERAD neuropsychological assessment battery in the Korean elderly. Journal of the International Neuropsychological Society, 10(1), 72–81. 10.1017/s1355617704101094 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Lee K. U., Lee D. Y., Kim K. W., Jhoo J. H., Kim J. H., Lee K. H., Kim S. Y., Han S. H., Woo J. I. (2002). Development of the Korean version of the consortium to establish a registry for Alzheimer’s disease assessment packet (CERAD-K) clinical and neuropsychological assessment batteries. The Journals of Gerontology Series B: Psychological Sciences and Social Sciences, 57(1), P47–P53. 10.1093/geronb/57.1.p47 [DOI] [PubMed] [Google Scholar]
- Lee K., Cheong H., Oh B., Hong C. (2009). Comparison of the validity of screening tests for dementia and mild cognitive impairment of the elderly in a community: K-MMSE, MMSE-K, MMSE-KC, and K-HDS. Journal of Korean Neuropsychiatric Association, 48(2), 61–69. [Google Scholar]
- Legdeur N., Heymans M., Comijs H., Huisman M., Maier A., Visser P. (2018). Age dependency of risk factors for cognitive decline. BMC Geriatrics, 18(1), 187. 10.1186/s12877-018-0876-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. E., Lu A. T., Bennett D. A., Horvath S. (2015). Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging, 7(12), 1198–1211. 10.18632/aging.100864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. E., Lu A. T., Quach A., Chen B. H., Assimes T. L., Bandinelli S., Hou L., Baccarelli A. A., Stewart J. D., Li Y., Whitsel E. A., Wilson J. G., Reiner A. P., Aviv A., Lohman K., Liu Y., Ferrucci L., Horvath S. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging, 10(4), 573–591. https://doi:10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipnicki D. M., Sachdev P. S., Crawford J., Reppermund S., Kochan N. A., Trollor J. N., Draper B., Slavin M. J., Kang K., Mather K. A., Lux O., Lux O. (2013). Risk factors for late-life cognitive decline and variation with age and sex in the Sydney memory and ageing study. PLoS One, 8(6), e65841. 10.1371/journal.pone.0065841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. T., Hannon E., Levine M. E., Crimmins E. M., Lunnon K., Mill J., Geschwind D. H., Horvath S. (2017). Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nature Communications, 8, 15353. 10.1038/ncomms15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni R. E., Shah S., McRae A. F., Ritchie S. J., Muniz-Terrera G., Harris S. E., Gibson J., Redmond P., Cox S. R., Pattie A., Corley J., Taylor A., Murphy L., Starr J. M., Horvath S., Visscher P. M., Wray N. R., Pattie A. (2015). The epigenetic clock is correlated with physical and cognitive fitness in the Lothian birth cohort 1936. International Journal of Epidemiology, 44(4), 1388–1396. 10.1093/ije/dyu277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E. M., Fry R. C. (2018). Environmental influences on the epigenome: Exposure-associated DNA methylation in human populations. Annual Review of Public Health, 39, 309–333. 10.1146/annurev-publhealth-040617-014629 [DOI] [PubMed] [Google Scholar]
- Negash S., Smith G. E., Pankratz S., Aakre J., Geda Y. E., Roberts R. O., Knopman D. S., Boeve B. F., Ivnik R. J., Petersen R. C. (2011). Successful aging: Definitions and prediction of longevity and conversion to mild cognitive impairment. The American Journal of Geriatric Psychiatry, 19(6), 581–588. 10.1097/JGP.0b013e3181f17ec9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R. C., Roberts R. O., Knopman D. S., Geda Y. E., Cha R. H., Pankratz V. S., Boeve B. F., Tangalos E. G., Ivnik R. J., Rocca W. A. (2010). Prevalence of mild cognitive impairment is higher in men: The Mayo Clinic Study of Aging. Neurology, 75(10), 889–897. 10.1212/WNL.0b013e3181f11d85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse T. (2012). Consequences of age-related cognitive declines. Annual Review of Psychology, 63, 201–226. 10.1146/annurev-psych-120710-100328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse T. A. (2019). Trajectories of normal cognitive aging. Psychology and Aging, 34(1), 17. 10.1037/pag0000288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E. (2016). Healthy cognitive aging and dementia prevention. American Psychologist, 71(4), 268. 10.1037/a0040250 [DOI] [PubMed] [Google Scholar]
- Won C. W., Lee S., Kim J., Chon D., Kim S., Kim C., Kim M. K., Cho B., Choi K. M., Roh E., Jang H. C., Son S. J., Lee J. H., Park Y. S., Lee S. G., Kim B. J., Kim B. J., Kim H. J., Choi J., Ga H., Lee K. J., Lee Y., Roh E. (2020). Korean frailty and aging cohort study (KFACS): Cohort profile. BMJ Open, 10(4), e035573. 10.1136/bmjopen-2019-035573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-brn-10.1177_1099800420983896 for Accelerated Epigenetic Age in Normal Cognitive Aging of Korean Community-Dwelling Older Adults by Jongmin Park, Chang Won Won, Leorey N. Saligan, Youn-Jung Kim, Yoonju Kim and Nada Lukkahatai in Biological Research For Nursing