Abstract
Steroid receptors (SRs) are members of the nuclear hormonal receptor family, many of which are transcription factors regulated by ligand binding. SRs regulate various human physiological functions essential for maintenance of vital biological pathways, including development, reproduction, and metabolic homeostasis. In addition, aberrant expression of SRs or dysregulation of their signaling has been observed in a wide variety of pathologies. SR activity is tightly and finely controlled by post-translational modifications (PTMs) targeting the receptors and/or their coregulators. Whereas major attention has been focused on phosphorylation, growing evidence shows that methylation is also an important regulator of SRs. Interestingly, the protein methyltransferases depositing methyl marks are involved in many functions, from development to adult life. They have also been associated with pathologies such as inflammation, as well as cardiovascular and neuronal disorders, and cancer. This article provides an overview of SR methylation/demethylation events, along with their functional effects and biological consequences. An in-depth understanding of the landscape of these methylation events could provide new information on SR regulation in physiology, as well as promising perspectives for the development of new therapeutic strategies, illustrated by the specific inhibitors of protein methyltransferases that are currently available.
Keywords: ERα, PR, AR, GR, protein arginine methyltransferases, lysine methyltransferases, protein demethylases, coregulators, steroid receptors, methylation
Graphical Abstract
Graphical Abstract.
ESSENTIAL POINTS
Steroid hormones and their receptors play many critical roles in human physiology and pathology through specific gene regulation and modulation of cytoplasmic signaling pathways.
In addition to other types of post-translational modifications, methylation of lysine and arginine is recently shown to regulate steroid receptor activity.
Methylation of histones and nonhistone proteins also participate indirectly in the activities of steroid receptors.
Protein methyltransferases and demethylases play important roles in physiology and their dysregulation contributes to human pathologies.
Increased understanding of the biology of steroid receptor methylation, along with specific methylation enzyme inhibitors, will result in new potential therapeutic options.
Introduction
Steroid hormones play critical roles in various target tissues regulating body homeostasis. Their ability to easily transit through cell membranes aroused interest of scientists for their therapeutic potential. Glucocorticoid (GC) was the first type of steroid to be used in the clinic. The physician Philip Hench successfully treated rheumatoid arthritis symptoms with cortisone, a discovery for which the Nobel Prize in Medicine was awarded in 1950 (1). Thereafter, a plethora of experiments documented the influence of steroid hormones on many biological processes throughout the lifespan. The female hormone estrogen is well known for its effects on mammary gland and reproductive tract development. Progesterone, the other female hormone, plays a vital role during pregnancy. In males, the androgen hormone is important for sexual development and reproductive function. The mediators of these hormones remained elusive until the early 1960s, when radiolabeled lipophilic ligands were developed by Jensen and Jacobson (2). Their innovative experiments led to the identification of “radioligand-binding proteins,” now known as steroid receptors (SRs). Interestingly, these receptors were able to migrate from the cytoplasm to the nucleus, implying that ligand-bound receptors could influence transcription. This hypothesis was confirmed by Ashburner, who showed that the addition of the ecdysteroid hormone was responsible for the activation of a specific subset of genes in drosophila salivary glands (3). This “Ashburner hormonal model” was the foundation for our current knowledge on the regulation of transcription by these receptors and is still, nowadays, unexpectedly relevant.
Since then, numerous clinical therapies have been developed based on this hormone–receptor association. Among them, mifepristone targets the progesterone receptor (PR) to treat endometriosis (4); dexamethasone acts on the glucocorticoid receptor (GR) to induce anti-inflammatory and immunosuppressive effects (5), and is for instance currently used in the treatment of SARS-Cov2 viral infection (6, 7); tamoxifen blocks the estrogen receptor (ERα), thus inhibiting estrogen binding and consequently reducing the progression of hormone-dependent breast cancers (BCs) (8); antiandrogens targeting the androgen receptor (AR) are frequently associated with chemical or rarely used surgical castration for improving the overall survival of prostate cancer (PC) patients (9).
The nuclear receptor (NR) superfamily consists of evolutionarily related DNA-binding transcription factors (TFs), encoded by 48 genes in humans. Within this superfamily, a subgroup of 5 ligand-regulated receptors, the SRs, has been extensively exploited for the development of selective modulators and raised promising expectations for novel treatment strategies to address diverse medical conditions (10, 11). We will herein focus on SRs for which methylation events have been well documented, namely ERα, PR, AR, and GR.
Methylation has emerged as a major post-translational modification (PTM) of proteins, with wide-ranging consequences on their activity, in modulating their properties, such as subcellular localization, stability, or interactions with partners. Most protein methyltransferases preferentially methylate arginine and lysine residues. Based on the residue targeted, these enzymes are classified as lysine methyltransferases (KMTs) or protein arginine methyltransferases (PRMTs). Protein methylation was initially identified on histone proteins, a process deeply involved in local chromatin remodeling and then in gene regulation. However, a growing number of studies have reported methylation events on nonhistone proteins, revealing that methylation is involved in a large variety of biological effects. Currently, hundreds of methylated substrates have been identified, in various cellular processes including RNA metabolism, DNA repair, or gene transcription. Indeed, SR-dependent transcription is strongly influenced by histone methylation (12, 13), as well as by the methylation of SRs themselves and their associated coregulators. Consistent with other PTMs (ie, phosphorylation, acetylation, etc.), protein methylation is a dynamic and reversible process controlled by the activity of protein demethylases, which are able to remove methyl marks.
It is particularly relevant to note that KMTs and PRMTs are frequently overexpressed in cancer cells compared with normal cells, and that their upregulation is often associated with poor prognosis. In line with this, specific inhibitors targeting the methyltransferase activities have recently been developed to assess the involvement of this PTM in tumorigenesis, making these enzymes interesting targets to modulate SR properties (14, 15).
The aim of this review is to summarize current and emerging knowledge about the influence of protein methylation in the biology of SRs. As these receptors and their signaling are involved in numerous pathologies, targeting protein methyltransferase activities could provide promising new therapeutic tools.
Steroid Receptors
Structure
Phylogenetic studies in eukaryotic organisms have placed the emergence of SRs a long time before the divergence between vertebrates and invertebrates (16). This universality has been a powerful tool for scientists, allowing them to study and to decrypt the regulation of crucial hormone receptor–dependent physiological processes, such as development, cell growth, reproduction, and homeostasis, in simple organisms (compared with humans). Since the 1980s, many NR cDNAs have been cloned and characterized, including GR (16), ERα (17), PR (18), and AR (19).
The cloning of the first SR genes (ie, GR and ERα) was a major breakthrough in the field, highlighting a surprising homology between them. This observation provided the first chemical evidence that distinct hormones bind structurally related receptors. Indeed, SRs share a common organization with 5 functional domains (A/B, C, D, E, F), with varying degrees of structural homology (20, 21) (Fig. 1A). The central DNA binding domain (DBD or C domain) is the most conserved region and is crucial for the association between the receptor and specific hormone-response elements (HREs) (22, 23). The ligand binding domain (LBD) is the second domain with considerable structural homology among NRs (24) (Fig. 1A). It is essential for ligand binding, but its role is much broader. The LBD structure allows the LBD itself to undergo conformational changes leading to the recruitment of a SR partner for dimerization (25), as well as coregulators (26) through a ligand-dependent transactivation domain (also called AF-2 for activation function-2) (27). Between these 2 highly conserved regions, a less-conserved interdomain linker is present in all members of the superfamily (28) (Fig. 1A), called the hinge region (or D domain). This domain supports the DBD in the transcriptional activity of SRs (29, 30) by ensuring their nuclear localization with a nuclear localization signal, and by cooperating with the LBD to mediate SR dimerization (31) and its interactions with certain coregulators (32). At the N-terminal end, each member exhibits an aminoterminal domain (NTD or A/B domain), highly variable in size, amino acid composition, and PTMs (33) (Fig. 1B). This A/B domain notably includes a ligand-independent transactivation domain (or AF-1), which can act independently, although optimal receptor activity generally requires a synergistic cooperation between AF-1 and the ligand-dependent transactivation domain AF-2 (34). At the other extremity, the F region is highly variable and not present in all members of the superfamily. Although little is known about it, mutations or complete deletions can strongly alter the transcriptional activity of the receptor, ligand binding, as well as interactions with coregulators (35).
Figure 1.
Common structural organization of steroid receptors. (A) Schematic representation of steroid receptors (SRs) structure. The amino-terminal domain (NTD) is variable in size and composition and contains a ligand-independent transactivation domain (AF-1). The DNA binding domain (DBD) is the most conserved region, in which 2 zinc fingers maintain the core of the domain and bind to DNA. A less-conserved hinge region is present between the DBD and the ligand binding domain (LBD) and contains a nuclear localization signal (NLS). The ligand associates with the receptor through the LBD, which also contains a ligand-dependent transactivation domain (AF-2). The functions associated with the F domain are still not clearly understood. (B) The members of the steroid receptor (ie, ERα, PR, GR, and AR) subgroup share a deeply conserved structure of functional domains with some specificities. The main biological roles of these SRs, and the associated pathological disorders when SR signaling are dysregulated, are pointed out on the right.
Biological and Pathological Roles
SRs are ligand-inducible TFs playing crucial roles in the regulation of diverse and essential aspects of mammalian physiology, by controlling the expression of genetic programs to achieve synchronized and accurate functional responses (36–38). Responding to endocrine hormones, they trigger adaptive signals and serve as a potent regulatory interface between the cellular and organismal environment and the genome. However, they also drive pathological states when their signaling pathways become dysregulated. This feature has led to the development of numerous treatments to control endocrine-associated diseases, including neurological disorders, chronic inflammatory diseases, or cancers (39).
ERα acts in concert with PR to regulate the physiological development of female reproductive tissues and mammary glands. Variable concentrations of ovarian progesterone and estrogen hormones regulate decisive phases of human female life (puberty, pregnancy, and menopause). ERα knockout in pubescent mice totally impairs mammary gland development (40), whereas in mature women, ERα is a crucial modulator of breast cell proliferation and survival, strongly influencing their risk of developing BC. In bone tissue, ERα modulators such as tamoxifen maintain bone mineral density in postmenopausal women and prevent osteoporosis. A set of studies has also reported that ERα signaling is deeply involved in metabolic homeostasis and metabolic disorders, such as diabetes, dyslipidemia, or obesity, and influences numerous biological systems, including the immune and cardiovascular systems, due to the presence of ERα in all these sites (41).
Progesterone is the “hormone of pregnancy” in postpubescent women, as it is essential for the establishment and maintenance of pregnancy (42). Hormone-associated PRs target genes in multiple organs or systems to prepare the body for pregnancy, including the endometrial epithelium, the venous walls or in mammary glands (43). Another notable role for PR is the modulation of immune functions, with deep and distinct consequences in autoimmune diseases, including rheumatoid arthritis and systemic lupus erythematosus (44, 45). Recent studies have also highlighted the pleiotropic protective effects in brain cells exerted by progestin-liganded PR, making PR a key mediator of neuroprotection after stroke, in both sexes (46, 47).
The effects of GCs, including the natural human GC cortisol, are mediated by intracellular GR, which is expressed throughout the body, but with a substantial heterogeneity in GC sensitivity and biological responses across tissues. Once released into the bloodstream, GCs affect all of the major body systems, including cardiovascular, musculoskeletal, immune, nervous, and reproductive systems (48). GCs exert anti-inflammatory and immunosuppressive effects, and have thus been widely used in the clinic for treating autoimmune diseases, inflammatory diseases, and hematological cancers (49). Moreover, their ability to trigger apoptosis of immature B and T lymphocytes (50), has been exploited by clinicians to treat cancers, including leukemia and lymphoma.
The most remarkable role of AR is the maintenance of male reproductive tissues and spermatogenesis, as demonstrated by the complete ablation of the male reproductive tract (seminal vesicles, vas deferens, epididymis, and prostate; small inguinal testes with arrested spermatogenesis) in male AR-knock out (KO) mice (51). Not surprisingly, androgen-associated AR is strongly involved in several aspects of PC, the second most common solid cancer in men (52). Beyond the reproductive activity, AR signaling controls several homeostatic processes, such as bone growth, and glucose and lipid metabolism. It seems that the receptor acts as a negative regulator of adipocyte development in adult males, resulting in late-onset obesity when knocked out (53). Similarly to other SRs, the field of action of AR is vast, and a decisive role for androgen-activated receptors in neurodegenerative processes, such as spinal bulbar muscular atrophy, has been depicted (54, 55). Interestingly, in vivo studies highlighted underestimated and essential roles for AR and androgen signaling in female reproduction, including ovarian and breast physiology, such that dysregulation leads to dysfunctions and cancers (51, 56–58). In contrast, a recent work showed that AR acts as a tumor suppressor in ERα-positive BC (59).
Signaling Pathways
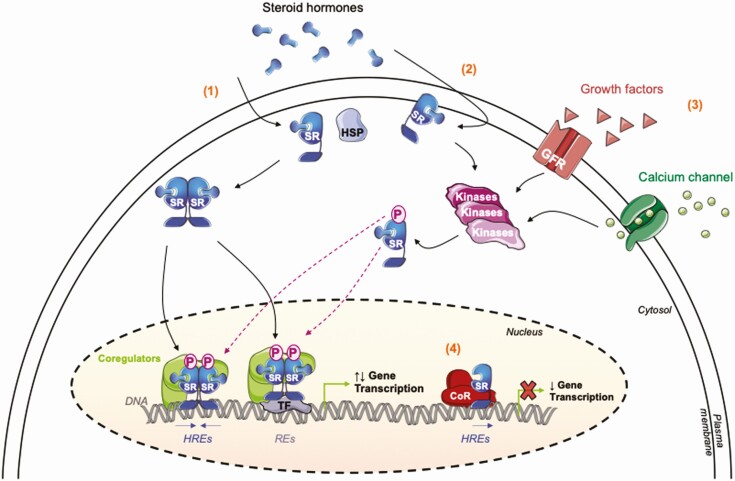
The binding of steroid hormones to their receptors triggers a conformational change within the LBD and functions as a critical step, shifting the receptor function from an inactive to an active state (60, 61). Steroid-bound receptors generally dimerize as homodimers and translocate to the nucleus (Fig. 2). They target specific HREs, generally palindromic, which fully or partially resemble 2 consensus half-sites (5′-AGGACA-3′ for GR, or 5′-AGGTCA-3′ for ERα), arranged as inverted repeats and typically separated by 3 nucleotides (20). The ligand-dependent conformational change also primes the receptor for the recruitment of coregulators and chromatin-modifying complexes, 2 major components of NR signaling (27, 62, 63). Indeed, ligand binding ultimately turns the receptor into a potent transcriptional regulator, which then assembles huge multiprotein complexes, also called coregulator complexes, to activate or repress the expression of target genes (Fig. 2). Alternatively, ligand-stimulated SRs can regulate gene expression by an indirect binding on chromatin, whereby they associate with other DNA-bound TFs, such as AP-1 or SP-1 (64–66) (Fig. 2).
Figure 2.
Steroid receptor signaling pathways. The steroid hormone enters into the cell by passive diffusion through the plasma membrane and binds with high affinity to its specific receptor. (1) Classical steroid hormone nuclear signaling. The ligand–receptor complex undergoes conformational changes triggering its dissociation from the chaperone heat-shock protein (HSP), receptor dimerization, and its translocation into the nuclear compartment. Inside the nucleus, the ligand-associated SR binds to specific DNA sequences that serve as enhancer or silencer elements, recruits coregulators and enzyme-modifying chromatin complexes to locally perturb the chromatin organization and regulate assembly or disassembly of an active transcription complex. SR-dependent multiprotein complexes target selective hormone-response elements (HREs) on target promoters, or indirectly interact with chromatin through transcription factors (TFs) on their response elements (REs). This could affect the level of growth factor receptors (GFRs), calcium signaling actors, or cellular proliferation effectors, among many other cellular pathways regulated by SR target genes. (2) Nongenomic signaling. The steroid ligand binds to SRs located at the plasma membrane or in the cytoplasm and triggers rapid post-translational modifications, often dependent on activating kinase cascades (MAPK, PI3K, Src), that in turn result in the transcriptional activation of the receptor. Conversely, SR genomic effects can regulate rapid nongenomic events, highlighting a potent crosstalk dependent on ligand-bound SRs. (3) Nonclassical steroid hormone nuclear signaling. Apart from the binding of the specific steroid hormone, SRs can also be indirectly activated by growth factors, leading the recruitment and the activity of cytoplasmic phosphorylation cascades, the same involved in the classical signaling, namely MAPK and PI3K/Akt kinases. (4) Unliganded-receptor nuclear signaling. More recent data revealed that unliganded forms of SRs play critical roles on chromatin and deeply take part in gene repression of a subset of target genes after recruitment of corepressors (CoR).
In addition to these agonist-induced transcriptional effects, most SRs are able to act on chromatin in a ligand-independent fashion. This “unliganded form” (ie, receptor without ligand), first demonstrated for the nonsteroid thyroid hormone receptors and later for PR, GR, and ERα, maintains gene silencing prior to hormonal activation (67–69) (Fig. 2). In spite of solid evidence demonstrating that ligand-independent SRs resort to a passive repression, either by competition for binding sites with coactivators or through the formation of inactive heterodimers with liganded homodimers (70, 71), recent reports suggest that inhibition of gene transcription is brought about by a more active mechanism. Advances in high-resolution and comparative techniques, such as cistrome analysis, clearly showed that unliganded receptors can bind their own targets, resulting in a proper and optimal expression of singular genetic programs (growth, apoptosis, development, etc.) (72). They act as active repressors and target the basal transcription machinery. How these unliganded forms are addressed to the nucleus rather than to the membrane/cytosol is still unknown, but some mechanistic studies have revealed that the presence of specific marks on chromatin (ie, histone methylation marks) most of the time associated with collaborating epigenetic silencers definitely could play a role in the targeting and the binding of native receptors to chromatin (72). Interaction with the ligand reverses the epigenetic landscape, consistent with the hypothesis that liganded SRs undergo a conformational change that enables the loading of coactivators needed for chromatin remodeling and gene activation. Further investigations are required to fully understand this regulation, certainly dependent on kinase-inducing regulatory phosphorylation and growth factor signaling, similarly to ligand-induced SRs (73, 74). Nevertheless, it is not surprising that SRs can exert strong and decisive effects independently of ligands, as they evolved from an ancestral SR unable to bind ligands (75).
In addition to these nuclear effects, SRs exhibit nongenomic regulatory properties (76) (Fig. 2). Generally, too rapid to be dependent on gene transcription and protein synthesis, and insensitive to mRNA or protein synthesis inhibitors, they involve SRs at the cell surface and/or in the cytoplasm. In human mammary cancer cell lines for instance, the addition of hormones (ie, estradiol and progesterone) leads to the formation of cytoplasmic multiprotein complexes in which SRs coexist with protein kinases and adaptor proteins. The steroid ligand, as well as some decisive growth factors acting through their receptors, like epidermal growth factor (77), activate a membrane-associated receptor and downstream-associated pathways, such as the tyrosine kinase/p21ras/mitogen-activated protein kinase, Src, and PI3K/Akt (phosphoinositide 3-kinase/protein kinase B) (78–80). Importantly, the final and overall biological effects of the hormone in the target tissue correspond to the convergence of both cytoplasmic and nuclear effects. Indeed, cytoplasmic kinase-dependent phosphorylation of SRs converts them into a transcriptionally active form, suggesting a convergence between classic genomic and rapid, nongenomic signaling pathways (81) (Fig. 2). While many studies have revealed a strong impact of the nongenomic effects on SR nuclear signaling, recent data illustrated how, inversely, hormone-mediated gene activation could affect nongenomic responses. The regulation of calcium homeostasis, playing a crucial role in tumorigenic progression, is a highly relevant illustration. SRs broadly regulate the transcription of many genes encoding components of the calcium pathway (calcium channels, calcium receptors, etc.), which once translated into proteins and activated, propagate nongenomic signals into the cell, contributing to cancer progression and survival (82, 83).
Mechanisms of Regulation
Many different signals can influence the adaptive responses mediated by SRs in cells, and, among them, PTMs trigger subtle but potent adjustments in SR signaling. Indeed, PTMs introduce structural constraints into functional domains that, in turn, alter their properties.
For a long time, phosphorylation was the main modification identified to regulate SR functions. More recently, several other PTMs, such as acetylation, methylation, palmitoylation, ubiquitylation, and SUMOylation, that mostly target alkaline residues (ie, lysine and arginine) have been described (84, 85). They are involved in every step of the receptor signaling pathways and contribute to the overall process of transcription. For instance, receptor stability, hormone binding and sensitivity, subcellular localization, and interactions with partners or DNA are affected by PTMs (86). Most of the time, these modifications are both dynamic and reversible, dependent on enzymes that deposit the marks and enzymes that remove them. This tightly controlled dynamic is crucial for an optimal regulation of SR activity. Concerning transcriptional regulation, receptor-modifying enzymes, like methyltransferases or deacetylases, act as influential SR coregulators, allowing a tight regulation of gene expression by directly modifying receptors (87). Moreover, their interactions with SRs can be direct or indirect, being recruited to enhancers or promoters by upstream coregulators (88, 89). They also remodel chromatin structure by modifying histones, and they promote (coactivator) or inhibit (corepressor) the recruitment and activation of RNA polymerase II, depending on the specific genes and cellular environment (89-92). Depending on which genes need to be activated (or repressed), specific coregulators are preferentially required for hormonal regulation of selected physiological responses that are dependent on a given TF (89). Since the first SR (ie, GR) was described, over 300 coregulators have been discovered (93). Different combinations of these coregulators are required for hormonal regulation of different target genes of the same SR in a given cell type, enabling the independent regulation of different subsets of the SR target genes to control different physiological response pathways (89).
Protein Methyltransferases/Demethylases
Protein methylation is a common covalent PTM that consists in the addition of methyl groups from a donor, S-adenoslyl-L-methionine (AdoMet, or SAM) to specific substrates (94). These substrates are preferentially methylated on either a lysine residue, and thus are modified by enzymes, KMTs, or an arginine residue, by PRMTs.
This PTM is a complex phenomenon, as lysine residues can be mono-, di-, or tri-methylated and arginine residues can be mono- or dimethylated, symmetrically or asymmetrically (95) (Fig. 3). The addition of methyl groups on both residues hardly affects the positive charge of the amino acids but deeply influences the bulkiness and hydrophobicity of the targeted protein. As such, protein methylation mostly functions as a potent signal for the recruitment of effector partners, called readers (because they distinguish the modified from the unmodified form of the target protein and translate that into a function) (96). As for other PTMs, methylation is a reversible process, involving enzymes able to remove methyl marks on lysines (lysine demethylases or KDMs) or on arginines (arginine demethylases) (97, 98). Both protein methyltransferases and protein demethylases are strong actors in transcription, as they influence the histone methylation landscape and consequently the opening or compaction of chromatin (99). However, growing evidence highlights that methylation of nonhistone proteins plays important roles in cells and sharply affects crucial homeostatic cellular processes. Inevitably, dysregulation of these processes can lead to diverse pathologies, including cancer.
Figure 3.
Process of protein methylation. Lysine residues are methylated by lysine methyltransferases (KMTs, green arrow) to generate, mono- (Kme1), di- (Kme2), or tri-methyllysines (Kme3). (A) KMTs use the methyl donor S-Adenosylmethionine (AdoMet) to add methyl (-CH3) groups on targets and produce S-adenosylhomocysteine (AdoHcy) in addition to methyllysines. This process is highly dynamic and can be reversed by lysine demethylases (KDMs, red arrow). (B) Arginine methylation is catalyzed by a family of 9 PRMTs, divided into 3 subgroups (type I, II, or III, green arrows). All use the methyl donor AdoMet to add methyl (-CH3) groups on targets and produce AdoHcy in addition to methylarginines. PRMTs that promote monomethylation (MMA), symmetric dimethylation (sDMA), or asymmetric dimethylation (aDMA) lead to the production of monomethylarginine, asymmetric dimethylarginine, or symmetric dimethylarginine respectively. JMJD6 is currently the only enzyme identified with an arginine demethylase activity (red arrow). PRMTs on which we focus in this article are highlighted in bold.
KMTs, or Lysine (K) Methyltransferases
The first KMT was uncovered on a bacterial flagellar protein in 1959 (100). Numerous other enzymes were discovered since then, making this family of proteins 1 of the largest class of epigenetic enzymes (95). KMTs catalyze the transfer of 1 or more methyl group(s) from the AdoMet donor to the ε-nitrogen of a lysine residue, generating mono-, di-, or trimethyllysine (Kme1, Kme2, and Kme3, respectively) (Fig. 3A) (101). KMTs methylate a wide variety of substrates: it has been predicted that human cells contain more than 1000 proteins with 1 form of methyllysine (102). To date, 2 KMT families have been described: the SET KMT family, containing the majority of KMTs (103), and the seven β-strand methyltransferases (7βS) or class I family (104).
The SET KMT family is characterized by the presence of the evolutionarily conserved Su(var)3-9n Enhancer-of-zeste and trithorax (SET) domain. This is the catalytic core of the enzymes, flanked by the pre- and post-SET domains, each displaying a defined substrate and product specificity (105). In their structure, SET KMTs also express the peptide binding and the AdoMet binding pockets (95). The SET KMT family is classified into subfamilies, based on the sequence of the pre- and post-SET domains.
The 7βS family differs from the SET KMTs family by the catalytic domain. It is composed of 4 motifs, designated I, post-I, -II, and -III. Motifs I and post-I play an important role in the interaction with the methyl donor AdoMet. However, many KMTs in this family do not possess all of the motifs but may harbor different ones. Although this family of KMTs is smaller than the SET family, a proteomic study predicted that the human genome encodes more than 100 7βS KMTs (106).
KMT substrates are involved in various cellular pathways including transcription, cell proliferation, DNA damage repair, inflammation and immune response (Table 1) (107). They were first described as histone KMTs, because numerous KMTs methylate lysine residues in histone N-terminal tails (169). However, recent technical advances in mass spectrometry (MS)–based proteomics have highlighted that numerous nonhistone proteins are modified by lysine methylation, revealing that KMT substrates extend far beyond histones (170).
Table 1.
Lysine methylated substrates
| KMT | Substrate | Site | Effect of lysine methylation | References |
|---|---|---|---|---|
| GLP | HIF-1α | K674me1/2 | Represses HIF-1α transcriptional activity | (108) |
| p53 | K373me2 | Negatively regulates p53 activity | (109) | |
| ATF7IP | K16me3 | Stimulates formation of ATF7IP / MPP8 complex | (110) | |
| DNMT3A | K47me2 | Induces the formation of Dnmt3a–MPP8–GLP/G9a inactive complex | (111) | |
| LIG1 | K126me2/3 | Induces LIG1-mediated recruitment of UHRF1 to replication foci | (112) | |
| G9a | CDYL1 | K135 | Negatively regulates its binding to chromatin | (113) |
| C/EBPβ | K39 | Represses C/EBPβ transactivation | (114) | |
| HIF-1α | K674me1/2 | Represses HIF-1α transcriptional activity | (108) | |
| MEF2D | K267 | Inhibits its chromatin recruitment and transcriptional activity | (115) | |
| MTA1 | K532 | Positively regulates its corepressor activity in NuRD complex | (116) | |
| MyoD | K104 | Inhibits MyoD transcriptional activity | (117) | |
| Pontin | K265, K267, K268, K274, K281, K285 | Enhances p300 recruitment and increases HIF1 transcriptional activity | (118) | |
| Reptin | K67me1 | Negatively regulates transcription of hypoxia genes | (119) | |
| RUNX3 | K129me1/2, K171me1/2 | Suppresses its transcriptional activity | (120) | |
| p53 | K373me2 | Negatively regulates p53 activity | (109) | |
| PLK1 | K209me1 | Supports DNA damage repair | (121) | |
| ATF7IP | K16me3 | Stimulates formation of ATF7IP/MPP8 complex | (110) | |
| DNMT3A | K47me2 | Induces the formation of Dnmt3a–MPP8–GLP/G9a inactive complex | (111) | |
| FOXO1 | K273me1/2 | Decreases FOXO1 stability | (122) | |
| G9a | K165me2/3 | Induces G9a interaction with HP1γ | (123) | |
| K239me3 | (124) | |||
| LIG1 | K126me2/3 | Induces recruitment of UHRF1 to replication foci | (112) | |
| SMYD2 | EZH2 | K307me1/2 | Represses transcription | (125) |
| GFI1 | K8 | Promotes GFI1-mediated transcriptional repression though LSD1 recruitment | (126) | |
| p53 | K370me1 | Negatively regulates p53 activity | (127) | |
| pRb | K860me1 | Regulates RB Binding to the Transcriptional Repressor L3MBTL1 | (128) | |
| K810me1 | Promotes E2F transcriptional activity | (129) | ||
| PARP1 | K528me1 | Enhances PARP1 activity in response to DNA damage | (130) | |
| β-catenin | K133me1 | Activates Wnt signaling | (131) | |
| MAPKAPK3 | K355me1 | Activates MAPKAPK3 | (132) | |
| PTEN | K313me2 | Activation of the phosphatidylinositol 3-kinase-AKT pathway | (133) | |
| HSP90AB1 | K531, K574me1 | Enhances its polymerization and the chaperone complex formation | (134) | |
| SET7/9 | FoxO3 | K271me1 | Decreases FoxO3 protein stability and increasing transcriptional activity | (135) |
| FXR | K206 | Supports the transactivation of FXR target genes | (136) | |
| HIV Tat | K51me1 | Activates HIV transcription | (137) | |
| K71me1 | (138) | |||
| LIN28A | K135me1 | Modifies transcription of c-myc target genes | (139) | |
| PGC-1α | K779 | Allows transcription of PGC-1α target genes | (140) | |
| pRb | K873 | Supports Rb-dependent transcriptional repression | (141) | |
| RelA | K314me1, K315me1 | Negatively regulates NF-κB transcriptional activation | (142) | |
| K37me1 | Stabilizes the DNA-RelA complexes and induces the transcription of a subset of NF-κB-regulated genes | (143) | ||
| RORα2 | K87 | Enhances its target gene transcription | (144) | |
| YY1 | K173me1, K411me1 | Positively regulates YY1 DNA-binding activity | (145) | |
| YY2 | K247me1 | Positively regulates YY2 DNA-binding activity | (146) | |
| p53 | K372me1 | Stabilizes p53 chromatin-bound fraction | (147) | |
| PARP1 | K508 | Stimulates ARTD1 mediated ADP-ribosylation | (148) | |
| SIRT1 | K333, K235, K236, K238 | Enhances p53 acetylation in response to DNA damage | (149) | |
| SUV39H1 | K105me1, K123me1 | Negatively regulates it activity in response to DNA damage | (150) | |
| UHRF1 | K385me1 | Enhances the formation of UHRF1–PARP1 complex at DNA damage site | (151) | |
| ATG16L1 | K151me1 | Inhibits autophagy by impairing the formation of the ATG12–ATG5- ATG16L1 complex | (152) | |
| β-catenin | K180me1 | Decreases β-catenin stability | (153) | |
| DNMT1 | K142me1 | Facilitates DNMT1 ubiquitin-dependent degradation | (154) | |
| E2F1 | K185me1 | Promotes E2F1 ubiquitin-dependent degradation | (155) | |
| eL42 | K53me1, K80me1, K100me1 | Enhances translation | (156) | |
| HIF-1α | K32 | Enhances HIF-1α stability | (157) | |
| IFITM3 | K88me1 | Reduces IFITM3 antiviral activity | (158) | |
| MYPT1 | K442me1 | Increases MYPT1 stability | (159) | |
| PLK1 | K191me2 | Promotes dynamic kinetochore-microtubule attachments | (160) | |
| RIOK1 | K411me1 | Promotes ubiquitin-dependent degradation of RIOK1 | (161) | |
| Rpl29 | K5me2 | Facilitates Rpl29 nuclear localization | (162) | |
| Sam68 | K208 | Positively regulates Sam68 protein stability | (163) | |
| Smad7 | K70me1 | Induces Smad7 ubiquitination and proteasomal degradation | (164) | |
| Sox2 | K119me1 | Induces Sox2 ubiquitination and proteasomal degradation | (165) | |
| STAT3 | K140me2 | Promotes STAT3 binding to SOCS3 promoter | (166) | |
| TAF10 | K189me1 | Increases TAF10 interaction with RNA polymerase II | (167) | |
| Yap | K494me1 | Promotes Yap cytoplasmic sequestration by the Hippo pathway | (168) |
Abbreviations: K, lysine; Kme1, monomethyllysine; Kme2, dimethyllysine; Kme3, trimethyllysine.
In this review, we will focus on the enzymes involved in SR signaling, namely G9a, GLP (G9a-like protein), SMYD2, SET7/9 (SETD7), and DOT1L (DOT1 like histone KMT).
G9a/GLP
G9a and GLP are SET KMTs belonging to the Suv39h family (171). They cooperatively play a predominant role during early embryonic development of mice, as G9a or GLP knockout induced growth retardation and early lethality (171, 172). G9a and GLP were first described as histone methyltransferases, performing Kme1 and Kme2 at lysine 9 of histone H3 (H3K9) (171). Once methylated, H3K9me1/2 becomes a docking site for effectors, especially the heterochromatin proteins HP1α, HP1β, and HP1γ that strongly influence gene silencing (173). Since then, nonhistone substrates have been identified including trimethylation of substrates such as G9a itself (124) or ATF7IP (110). Most of their substrates are involved in DNA damage response, cell cycle regulation, cell proliferation, and chromatin modulation, but also in skeletal muscle differentiation (Table 1). Although the role of G9a in the initiation and progression of cancer is well known (174), it also appears to be implicated in other diseases, such as Alzheimer’s disease (175). These enzymes preferentially methylate a nonexclusive motif such as ARKS/T (124).
SMYD2
SMYD2 is a protein methyltransferase that is capable of performing Kme1 and Kme2 (Table 1) (125). Initially described to methylate histone H3 at lysine 36 (H3K36) (176), it seems that this KMT can also methylate diverse nonhistone proteins (Table 1) (177). For instance, SMYD2 was reported to monomethylate p53 at K370 and pRb at K860 (127, 128), to inhibit their activities. Large-scale analysis revealed a low level of specificity for SMYD2 towards its substrates, as only the LF-K motif has been identified (178).
SMYD2-deficient zebrafish show malformation of both the atrium and ventricle, and a reduced heart rate and cardiac function (179). Mechanistically, in the cytoplasm of cardiomyocytes, SMYD2 monomethylates the heat shock protein 90 (HSP90) at K616 and interacts with the sarcomeric I-band region at the titin N2A domain via its N-terminal and extreme C-terminal regions to influence cardiac contraction (180). Since aberrant SMYD2 expression and its dysfunction are often closely related to cardiovascular diseases and cancer, SMYD2 is a promising candidate for the treatment of these pathologies.
SET7
SET7/9 is a SET KMT that performs mono- and dimethylation (127). It was first identified to monomethylate H3K4, leading to the recruitment of RNA polymerase II and thus maintains the transcription of target genes and the structure of active or potentially active euchromatin (181). As other KMTs, SET7/9 also has nonhistone substrates involved in DNA damage response, cell cycle regulation, cell proliferation, chromatin modulation and cell differentiation (Table 1). SET7/9 preferentially methylates proteins containing the K/R-S/T/A sequence (105). Interestingly, SET7/9 knockout mice are viable and develop normally (182). SET7/9 seems to have different effects on carcinogenesis depending on the cancer type (183). SET7/9 also appears to play a role in diabetes and atherosclerosis by activating inflammatory genes (184).
DOT1L
DOT1L is a 7βS KMT that performs mono-, di-, and trimethylation. For a long time, its unique substrate was H3K79, the methylation of which contributes to activating the transcription of genes involved in DNA damage response and cell cycle regulation (185), but also in immune response (186, 187). In addition, DOT1L is involved in neointimal hyperplasia development, as it is upregulated in the rat injured artery wall (188). Contrary to the other KMTs presented before, DOT1L has no identified nonhistone substrate, aside from AR which we will described later in this review (189). Interestingly, the loss of DOT1L in mice induces developmental problems (ie, growth retardation, cardiac dilatation) and death in utero (190). Furthermore, DOT1L seems to be implicated in diseases such as obesity and cancer. Indeed, inhibition of DOT1L increases adiposity, making it a potential therapeutic target for obesity (191), and DOT1L mislocalization in leukemia promotes H3K79 methylation and activation of the leukemic transcriptional program (185).
KMTs targeting
In view of these data, it has become evident that members of the KMT family constitute attractive new therapeutic targets. BIX-01294, the first G9a inhibitor, was identified in 2009, and numerous inhibitors targeting G9a have since been produced (192). Targeting this enzyme gave rise to promising results in bladder cancer, especially with the selective inhibitor CM-272 that leads to cancer regression in vivo (193). Similar results were obtained in non–small cell lung cancer, where the selective inhibitor UNC0638 prevents tumor growth in vitro and in vivo (194).
The first SMYD2 inhibitor was identified in 2011, and the same year the SMYD2 crystal structure was reported (195). Since then, increasingly potent and selective SMYD2 inhibitors have been produced (196). Inhibition of SMYD2 was shown to increase the sensitivity of ovarian cancer cells to the PARP inhibitor olaparib (197) and to sensitize non–small cell lung cancer cells to the anticancerous agent cisplatin (198).
For SET7/9 inhibitors, cyproheptadine was demonstrated to selectively inhibit SET7/9 activity (199), and many derivatives of this inhibitor have since been studied (200).
Of the KMTs presented in this review, only the DOT1L inhibitor pinometostat is currently undergoing clinical trial according to the clinical trials website (www.clinicltrials.gov). To date, pinometostat is registered in 2 phase Ib/II clinical trials for different hematological malignancies (NCT03724084 and NCT03701295).
KDMs, or Lysine (K) DeMethylases
The discovery of KMTs quickly raised the question of the existence of lysine demethylating proteins. The first KDM was identified in the early 2000s, and numerous other KDMs have since been discovered (201). They are classified into 2 groups based on their structure and the type of lysine demethylation they perform (Fig. 3A).
The first group, called KDM1, includes KDM1A (LSD1) and KDM1B (LSD2). LSD1 contains a flavin adenine dinucleotide–dependent amine oxidase domain and performs demethylation of Kme1 and Kme2 only (202, 203) (Fig. 3A). The second group is larger and includes JmjC domain-containing histone demethylases (JHDMs). The demethylase activity of the JmjC domains requires Fe2+, 2-oxoglutarate and oxygen. JHDMs are capable of demethylating Kme1, Kme2, and Kme3 through hydroxylation (204) (Fig. 3A). JHDMs have been classified into subgroups (KDM2-7), according to their JmjC domain sequence and domain architecture. Indeed, KDMs contain DNA and histone binding domains, such as zinc fingers, Tudor domains, and PH domains. For example, KDM4A, KDM4B, and KDM4C from the KDM4 subgroup have 2 PH domains and 2 Tudor domains in addition to their JmjC and JmjN domains, but no zinc fingers; and KDM4D only has JmjC and JmjN domains (205).
In this section, we will focus on LSD1 and KDM4 family members, which are currently the main KDMs involved in SR regulation.
LSD1
Similarly to KMTs, the first KDM substrates described were initially histones, and nonhistone substrates were later identified. The first identified substrate for LSD1 was H3K4. LSD1-mediated demethylation of H3K4me1/2 induces a repression of target gene transcription by interacting with TFs harboring a SNAG domain, a N-terminal highly conserved repressive domain (eg, SNAIL1/2 and GFI1/B) (206). In addition, LSD1 demethylates nonhistone proteins, among which p53, DNMT1, and HSP90, as well as ERα, which we will further explore later in this review (207). Nevertheless, LSD1 is also a coactivator of several SRs. The relief of repressive histone marks, such as the demethylation of H3K9me1/2, triggers chromatin and DNA conformational changes that are essential for promoting AR and ERα-dependent transcription (208, 209).
KDM4
Enzymes from the KDM4 subgroup can activate or repress transcription depending on the targeted lysine residue. KDM4A was first identified as a H3K9me2/3 and H3K36me2/3 demethylase (210). By demethylating H3K36me3, KDM4A antagonizes HP1γ and allows cell cycle progression (211). Like LSD1, nonhistone substrates of KDM4A have been identified. Indeed, KDM4A was recently identified in a complex with SCFFbxo22 and methylated p53. Indeed, it was reported that KDM4A-mediated p53 demethylation is necessary for the destabilization of methylated p53 induced by SCFFbxo22 (212).
KDMs targeting
Some KDMs are very attractive therapeutic targets. For instance, LSD1 is dysregulated in many cancer types including small cell lung cancer and acute myeloid leukemia (213). Since the discovery of LSD1, many potent and selective inhibitors have been identified, including GSK2879552, which displays an antitumor activity in small cell lung cancer xenograft mouse models (214). Currently, the clinical trials website (www.clinicltrials.gov), references 5 LSD1 inhibitors undergoing clinical trials mostly in cancer patients (215). Among them, the GSK2879552 clinical trial in myelodysplastic syndrome patients completed phase II in 2019, but the outcome does not advocate for completing such trials (NCT02929498). In addition, the dual LSD1/MAO-B inhibitor ORY-2001 efficiently rescues memory and behavioral alterations in mouse models of Alzheimer’s disease. Of note, the observed effects were essentially attributed to the inhibition of LSD1 (216). Interestingly, ORY-2001 is currently undergoing a phase II clinical trial in patients with Alzheimer’s disease (NCT03867253).
KDMs from the KDM4 subgroup are dysregulated in several diseases such as cancers, cardiovascular diseases, and mental retardation (217). Despite the therapeutic potential of the KDM4 subgroup, there are few potent and selective inhibitors to date (218).
PRMTs, or Protein Arginine (R) Methyltransferases
Arginine methylation was first described in the 1970s (219–222), though the first PRMT was only identified in 1996 (223). Similarly to KMTs, PRMTs are structurally defined as S-adenosylmethionine (AdoMet)-dependent methyltransferases. PRMT enzymes belong to the class I methyltransferases, which is characterized by a 7-stranded β-sheet structure. They also harbor additional conserved sequences, including the motifs I, post-I, and the Thr-His-Trp (THW) loop that forms the AdoMet binding pocket (224). PRMTs transfer 1 or 2 methyl group(s) from the AdoMet methyl donor to the guanidine nitrogen atoms of the targeted arginine, resulting in S-adenosylhomocysteine (AdoHcy) and methylarginine production (Fig. 3B). Three main forms of methylated arginines exist in eukaryotes: monomethylarginine (MMA), ω-NG,NG-asymmetric dimethylarginine (aDMA) in which 2 methyl groups are added to the same guanidine nitrogen, and ω-NG,N’ G-symmetric dimethylarginine (sDMA) where 1 methyl group is attached to each guanidine nitrogen (225) (Fig. 3B). So far, 9 PRMTs have been characterized as active and structurally conventional arginine methyltransferases in human cells (226). They are classified into 3 fundamental subgroups, based on the type of methylation they catalyze. Type I PRMTs (PRMT1, PRMT2, PRMT3, CARM1 for Coactivator Associated Arginine Methyltransferase 1 [PRMT4], PRMT6, and PRMT8) produce MMA and aDMA, while type II enzymes (PRMT5 and PRMT9) deposit MMA and sDMA marks. Type III contains only PRMT7 and is responsible for MMA only (227) (Fig. 3B).
Here, we will focus on PRMT1, CARM1, PRMT5 and PRMT6 which are currently the 4 main PRMTs involved in SR regulation.
PRMTs are involved in many essential cellular processes and KO of most of them causes embryonic lethality. PRMT1 and PRMT5 KO are lethal (ie, induce embryonic or post-natal death) (228). Moreover, CARM1-KO mice die at birth and display a reduced size (229). In contrast, PRMT6-KO mice are viable (230). Tissue ablation of PRMTs has contributed to determining their involvement in metabolic, immune, muscular, and neurodegenerative disorders and cancers (228, 231). The different PRMTs regulate a wide variety of important cellular processes (eg, DNA repair, transcriptional regulation, immune system response, RNA processing and signal transduction) (232) by methylating a growing number of substrates (Table 2). It is well known that proteins harboring glycine and arginine-rich motifs (GARs) are often targets for PRMTs (346). However, even if it is the case for PRMT1, 5, and 6, CARM1 prefers to methylate its substrates within a PGM (proline, glycine, methionine) motif (347).
Table 2.
Arginine methylated substrates
| PRMT | Substrate | Site | Effect of arginine methylation | Reference |
|---|---|---|---|---|
| PRMT1 | BRCA1 | 504–802 | Facilitates its binding to promoters | (233) |
| C/EBPα | R35, R156, R165 | Dissociates from SWI/SNF Mediator complex | (234) | |
| c-Myc | R299, R346 | Activates its transcriptional activity by promoting its binding to p300 | (235) | |
| EZH2 | R342 | Suppresses EZH2 target transcription | (236) | |
| FOXO1 | R248, R250 | Blocks FOXO1 phosphorylation by Akt | (237) | |
| FOXP3 | R48, R51 | Enhances its transcriptional activity | (238) | |
| GLI1 | R597 | Enhances its transcriptional activity | (239) | |
| MyoD | R121 | Activates its transcriptional activity by promoting its DNA-binding | (240) | |
| Nrf2 | R437 | Enhances its transcriptional activity | (241) | |
| RACO-1 | R98, R109 | Enhances its binding to c-jun, activates AP1 transcription | (242) | |
| RelA | R30 | Inhibits its binding to DNA | (243) | |
| RIP40 | R240, R650, R948 | Decreases its corepressor function | (244) | |
| RunX1 | R206, R210 | Abrogates Sin3a binding, promoting its transcriptional activity | (245) | |
| STAT1 | R31 | Dissociates from PIAS1 and enhances IFNα/β induced transcription | (246) | |
| TAF15 | R203 | Enhances TAF15-depend transcription | (247) | |
| TLS | R216, R218, R242, R394 | Enhances transcription of surviving | (248) | |
| TOP3B | R833, R835 | Involved in interaction with TDRD3, promoting its topoisomerase activity | (249) | |
| Twist 1 | R34 | Represses import into nucleus and E-cadherin expression | (250) | |
| 53BP1 | 1319–1480 | Localizes to DNA breaks | (251) | |
| APE1 | R301 | Protects mitochondrial DNA from oxidative damage | (252) | |
| DNA polβ | R137me1 | Inhibits its interaction with PCNA, enhances base excision repair | (253) | |
| E2F1 | R109 | Induces PARP cleavage in response to DNA damage | (254) | |
| hnRNPK | R296, R299 | Inactivates caspase 3 after DNA damage | (255) | |
| hnRNPUL1 | R584, R618, R620, R645, R656 | Stimulates its recruitment to DNA damage | (256) | |
| MRE11 | GAR domain | Enhances its exonuclease activity | (251) | |
| ASK1 | R78, R80 | Negatively regulates ASK1 signaling | (257) | |
| Axin | R378 | Increases Axin stability and inhibits Wnt signaling | (258) | |
| CaMKII | R9, R275 | Suppresses cardiac CaMKII hyperactivation | (259) | |
| EGFR | R198, R200 | Promotes EGFR activation | (260) | |
| p38 MAPK | R49, R149 | Enhances p38α activation | (261) | |
| Smad4 | R272 | Activates wnt signaling | (262) | |
| Smad6 | R74, R81 | Activates BMP signaling | (263) | |
| R74, R81 | Inhibits NFkB signaling | (264) | ||
| Smad7 | R57, R67 | Enhances TGF-β signaling | (265) | |
| TSC2 | R1457, R1459 | Regulates mTORC1 activity | (266) | |
| BAD | R94, R96 | Inhibits its association with 14-3-3 | (267) | |
| CDK4 | R55, R73, R82, R163 | Destabilizes CDK4-Cyclin-D3 complex and inhibits cell cycle progression | (268) | |
| CNBP | R25me1/2a, R27me1/2a | Decreases its RNA binding | (269) | |
| cTnI | R146me1/2a, R148me1/2a | Inhibits cardiomyocytes hypertrophy | (270) | |
| EIF4G1 | R689me1, R698me1 | Contributes to its stability and facilitates translation initiation complex assembly | (271) | |
| EZH2 | R342me2a | Enhances its stability | (236) | |
| G3BP1 | R435 me1/2a, R447 me1/2a | Prevents stress granule formation | (272) | |
| hnRNP A1 | R214, R226, R223, R240 | Enhances its RNA binding | (273) | |
| HSP70 | R416, R447 | Protects PDAC cells from apoptosis | (274) | |
| INCENP | R887 | Facilitates interaction with AURKB (maintains chromosomal alignment) | (275) | |
| KCNQ | R333, R345, R353, R435 | Facilitates its ion channel activity by PIP2 interaction | (276) | |
| MYCN | R65 | Stabilizes MYCN protein | (277) | |
| RBM15 | R578 | Facilitates its degradation by CNOT4 (RNA splicing) | (278) | |
| rps3 | R64, R65, R67 | Targets rps3 into ribosomes (translation) | (279) | |
| RunX1 | R233, R237 | Resists to apoptosis under stress condition | (280) | |
| TRAF6 | R88, R125 | Inhibits its ubiquitin ligase activity | (281) | |
| CARM1 | BAF155 | R1064 | Regulates transcription related to c-Myc pathway | (282) |
| C/EBPβ | R3 | Dissociates from SWI/SNF mediator complex | (283) | |
| CARM1 | R551 | Promotes its effect on transcription and mRNA splicing | (284) | |
| HSP70 | R469me1 | Activate RA-induced RARβ2 transcription | (285) | |
| LSD1 | R838 | Stabilizes LSD1, enhancing E-cadherin and decreasing vimentin transcription | (286) | |
| Pax7 | R161 | Activates Myf5 transcription via MLL1/2 complex | (287) | |
| Pontin | R333, R339 | Activates Foxo3-induced autophagy gene expression | (288) | |
| PRMT5 | R505 | Enhances its enzymatic activity, decreasing γ-globin gene transcription | (289) | |
| RNA pol II | R1810 | Activates the transcription of small nuclear RNAs | (290) | |
| RUNX1 | R223 | Induces the repressor complex formation | (291) | |
| SOX2 | R113 | Enhances Sox2-mediated transactivation by self-association | (292) | |
| p300 | R754 | Promotes BRCA1 recruitment to p21 promoter during DNA damage | (293) | |
| GAPDH | R234 | Inhibits glycolysis by repressing its activity | (294) | |
| HuD | R236, R248 | Decreases p21 stability | (295) | |
| HuR | R217 | Stabilizes mRNAs | (296) | |
| MDH1 | R248me1/2a | Inhibits Gln metabolism | (297) | |
| PKM2 | R445, R447 | Enhances its pyruvate kinase activity | (298) | |
| RPIA | R42 | Enhances its enzymatic activity (pentose phosphate pathway) | (299) | |
| PRMT5 | Actin | R256me1 | Either activates or represses transcription | (300) |
| BCL6 | R305 | Facilitates its transcriptional repressive activity | (301) | |
| E2F1 | R111, R113 | Inhibits its transcriptional activity | (302) | |
| GATA4 | R317 | Inhibits its transcriptional activity | (303) | |
| HOXA9 | R140 | Promotes transcription of E-selectin | (304) | |
| RelA | R30 | Enhances NFKB transcriptional activity | (305) | |
| R30me1, R35 | (306) | |||
| R174 | (307) | |||
| RNA pol II | R1810 | Controls termination of transcription | (308) | |
| SHP | R57 | Facilitates its transcriptional repressive activity | (309) | |
| SPT5 | ND | Releases SPT5 from Bscl2 promoter (lipid metabolism) | (310) | |
| SREBP1a | R321 | Enhances SREBP1transcriptional activity | (311) | |
| 53BP1 | GAR motif (both ADMA and SDMA) | Enhances DNA repair process | (312) | |
| FEN1 | R192 | Facilitates DNA repair by binding to PCNA | (313) | |
| p53 | R333me1, R335, R337 | stimulates p53-dependent G1 arrest in response to DNA damage | (314) | |
| RAD9a | R172, R174, R175 | Regulates cell cycle checkpoints | (315) | |
| RUVBL1 | R205 | Removes 53BP1 from DNA breaks then enhances HR-mediated DSB repair | (316) | |
| TDP1 | R361, R586 | Stimulates TDP1/XRCC1 recruitment to DNA breaks | (317) | |
| ASK1 | R89 | Inhibits H2O2-induced ASK1 activation | (318) | |
| BRAF | R671 | Inhibits ERK activation (EGFR signaling) | (319) | |
| CRAF | R563 | |||
| DUSP14 | R17me1/me2s R38, R45me1 | Promotes its ubiquitination, inhibiting TCR signaling | (320) | |
| EGFR | R1175me1 | Inhibits EGF-induced ERK pathway | (271) | |
| YBX1 | R205 | Activates NF-κB signaling | (321) | |
| G3BP1 | R460 | Prevents stress granule assembly | (272) | |
| GLI1 | R990, R1018 | Stabilizes GLI1 protein | (322) | |
| GM 130 | R18, R23 | Regulates GA ribbons, maintaining Golgi architecture | (323) | |
| hnRNP A1 | R218, R225 | Enhances interaction with IREs RNA to promote translation | (324) | |
| Facilitates HIV-1 IRES-mediated translation | (325) | |||
| HSP90A | R345, R368 | Suppresses the cell apoptosis | (326) | |
| KLF-4 | R374, R376, R377 | Inhibits its ubiquitination, maintaining genome stability | (327) | |
| LSH | R309 | Decreases stem-like properties | (328) | |
| PDCD4 | R110 | Inhibits its tumor suppressive activity | (329) | |
| RPS10 | R158, 160 | Facilitates its assembly into ribosome | (330) | |
| ZNF326 | R175 | Regulates alternative splicing | (331) | |
| PRMT6 | FOXO3 | R188, R249 | Activates transcriptional activity | (332) |
| HIV-1 Tat | R52, R53 | Inhibits Tat transcriptional activation | (333) | |
| HIV-1 nucleocapsid | R10, R32 | Inhibits reverse transcription | (334) | |
| RFX5 | R466, R468 | Down-regulates transcription | (335) | |
| TOP3B | R833, R835 | Promotes transcription | (249) | |
| DNA pol β | R83, R152 | Promotes Polβ activity in DNA strand break repair | (336) | |
| CRAF | R100 | Diminishes MEK/ERK signaling | (337) | |
| PTEN | R159 | Inhibits PI3K–AKT signaling | (338) | |
| BAG5 | R15, R24 | Represses cell autophagy | (339) | |
| GPS2 | R323 | Prevents GPS2 degradation | (340) | |
| HIV-1 Rev | R38 | Inhibits viral RNA export to the cytoplasm | (341) | |
| p21 | R156me1/me2a | Enhances cytoplasmic localization of p21 | (132) | |
| p16 | R22, R131, R138 | Weakens p16-mediated apoptosis | (342) | |
| PRMT6 | R35 | Stabilizes PRMT6 protein level | (343) | |
| RCC1 | R214 | Induces its association with chromatin and activation of RAN | (344) | |
| SIRT7 | R388me1/me2a | Inhibits its deacetylase activity (mitochondria biogenesis) | (345) |
When the type of methylation is not specified it is Rme2a for PRMT1, CARM1, and PRMT6, and Rme2s for PRMT5.
PRMT1
PRMT1 was initially shown to catalyze the methylation of H4R3, an epigenetic active mark (348). To date, many nonhistones substrates have also been identified (Table 2). For instance, BReast CAncer 1 (BRCA1) methylation by PRMT1 affects its tumor suppressive capacity in BC cells and samples (233). More recently, PRMT1 was shown to dimethylate the KMT EZH2 (236). This event inhibited its ubiquitylation and consequently increased the stability of the protein, which further impaired expression of EZH2 target genes, contributing to a sustained and aggressive phenotype of BC cells (epithelial mesenchymal transition, invasion, and metastasis).
CARM1
CARM1-dependent methylation of various substrates notably contributes to tumorigenesis (347). For instance, methylation of the core subunit BAF155 of the chromatin complex SWI/SNF promotes proliferation, migration and metastasis of BC cells in vivo. Moreover, BAF155 methylation was associated with poor survival of BC patients (282). CARM1 methylation of the lysine demethylase LSD1 stabilizes the protein, activating vimentin transcription through histone demethylation, which triggers invasion and metastasis of BC (286).
PRMT5
PRMT5 was shown to methylate, among other substrates, the HOXA9 protein, a TF that plays a crucial role in hematopoietic stem cell expansion and is commonly dysregulated in acute leukemia (304). This modification is involved in epithelial mesenchymal transition activation, due to induced expression of proinflammatory endothelial–leukocyte adhesion molecules, such as E-selectin. As such, PRMT5 seems to be a critical actor in the induction of the proinflammatory function of HOXA9, which is important in the pathobiology of inflammation and cardiovascular inflammatory diseases. PRMT5 also controls carcinogenesis by methylating the TF E2F1. This PTM increases the stability of the target, promoting cell cycle progression and cell growth (302). Despite its oncogenic role, new reports highlighted that PRMT5 is also involved outside of the cancer field. For instance, PRMT5-induced methylation of SREBP1 (311) and SPT5 (310) participate in regulating lipid metabolism.
PRMT6
Histone H3R2 was thought to be the major histone target site of PRMT6 in cells, and PRMT6 was widely considered to be a transcriptional repressor (349, 350). However, in several cases, PRMT6 was reported to act as a transcriptional coactivator, by depositing the H3R17me2a mark, similarly to CARM1 (351). PRMT6 also methylates nonhistone proteins regulating transcription, DNA repair and cell signaling (Table 2).
PRMTs targeting
The majority of PRMTs and their variants have been shown to be overexpressed in cancer compared with normal tissues (352). PRMT1 overexpression has been reported in breast, prostate, lung, colon, and bladder cancer, as well as in leukemia. Similarly, overexpression of CARM1 and PRMT5 has been observed in PC, colon cancer, and BC, whereas PRMT6 has been shown to be overexpressed in bladder, lung, and BC (15). Moreover, their expression is often associated with poor prognosis (15). In view of these findings, it has become increasingly evident that members of the PRMT family constitute new targets for treating pathologies, including cancer (353-355). The first PRMT inhibitor, AMI-1, was discovered in 2004 by Bedford et al. (356). Many potent and selective inhibitors have since been produced (357). Although none of the inhibitors selectively inhibit PRMT1, several inhibitors specific for other Type I PRMTs are available. TP-064 (358) and GSK3359088 (359) targeting CARM1 are effective in inhibiting tumor growth for multiple myeloma. EPZ2020411 selectively inhibits PRMT6, but this inhibitor is under preclinical development (360). Some inhibitors are currently undergoing clinical trials in patients with different types of cancers. On the clinical trials website (www.clinicltrials.gov), 6 clinical trials for agents that target PRMTs are referenced (1 for type I enzymes, and 5 for PRMT5). Most of the inhibitors in clinical trials are in phase I, assessing their safety, pharmacokinetics, pharmacodynamics, and clinical activity. GSK3368715 is the only type I PRMT inhibitor in clinical trials (361). It was shown to inhibit in vitro tumor cell growth in lymphoma, acute myeloid leukemia, and numerous solid tumor cell lines.
JNJ64619178 is a PRMT5 inhibitor that provokes tumor growth inhibition and regression in patient-derived xenografts. In addition, PF-06939999, PRT811, and PRT543 are PRMT5 inhibitors with antiproliferative and antineoplastic activities in cancer cell lines. GSK3326595 is the only inhibitor currently in a phase II clinical trial. Recently, this inhibitor was shown to circumvent palbociclib (CDK4/CDK6 inhibitor) resistance in melanoma (362).
Arginine (R) Demethylases
JMJD6
With regards to arginine methylation, only 1 demethylase has so far been identified, and this role was initially a matter of controversy (363). Indeed, in 2007, JMJD6 (Jumonji domain-containing protein 6) was described as a JmJC-containing iron-and 2 oxoglutarate-dependent dioxygenase, able to remove dimethyl groups from H3R2 and H4R3 (98) (Fig. 3B). However, at that time, Webby et al did not confirm these results and reported that arginine-rich (RS) domains of U2AF65 and LUC7-like2 synthesized with dimethylated arginine residues could be Jmjd6 substrates for hydroxylation (364). JMJD6 also catalyzes the hydroxylation of lysine residues of histones H2A and H2B (365).
Despite this multifaceted role for JMJD6, more recent studies confirmed that this enzyme demethylates arginines of some nonhistone substrates. Among them, ERα (366), RNA helicase A (367), the TF PAX3 (368), HSP70 (285), the Ras-GTPase activating SH3-domain-binding-protein 1 (G3BP1) (369) and the ubiquitin ligase TRAF6 (281). It is now broadly accepted that JMJD6 acts as a dual demethylase and lysyl hydroxylase, able to modify proteins on both arginine and lysine residues. Interestingly, JMJD6 is upregulated in a large spectrum of cancers and its enzymatic activities have been associated with tumorigenic roles, making JMJD6 a promising novel therapeutic target (370). However, so far only 1 inhibitor has been developed and its effect on JMJD6 demethylase activity has not been investigated, although it displayed promising antiproliferative effects on ovarian cancer cells (371).
Of note, the fact that JMJD6 is unable to demethylate H3R8, H3R17, H3R26, or H2A sites (98) suggests that other arginine demethylases may play a role in the dynamic regulation of arginine methylation. Indeed, other works demonstrated that some KDMs are also able to remove methyl marks from arginine residues. KDM3A, KDM4A, KDM5, and KDM6B display arginine demethylase activity in vitro on histones and on certain nonhistone peptides (372). JMJD1B (or KDM3B) was recently reported to demethylate H4R3me2s and its intermediate H4R3me1, during the development of hematopoietic stem cells (373). However, its very narrow specificity strongly suggests that other unknown enzymes may display arginine demethylase properties. In addition to true demethylation, arginine methylation levels are further modulated by the conversion of arginine into citrulline by protein arginine deiminase (374).
Methylation/Demethylation of Steroid Receptors: Biological Implications
Estrogen Receptor α, or ERα
Lysine methylation/demethylation
K302.
In 2008, Vertino’s team identified the first lysine methylation of a SR (375). They found that SET7/9 catalyzes ERα methylation on lysine 302 (K302), located in the hinge region, to promote ERα transactivation. Moreover, they linked K302 methylation with the stabilization of ERα protein levels (Fig. 4A and Table 3). Indeed, estrogen-induced ubiquitylation of ERα and its subsequent degradation by the proteasome is an important step in the transcriptional activity of the receptor (390, 391). As such, SET7/9 was recognized as a potent modulator of ERα-dependent gene expression. Interestingly, K302 is located in a PTM “hot-spot,” where acetylation (K299, K302 and K303), ubiquitylation (K302), and phosphorylation (S305) were previously reported (85). Similarly to what happens on histone tails, the presence of other PTMs in the vicinity of K302 could affect the methylation event. Consistently, previous research reported the existence of connections between acetylation and phosphorylation, which markedly regulates the phenotype of cells. ERα nonacetylated K303 variant (K303R) was detected in primary ductal hyperplasia and aggressive BC (392, 393). K303R promotes phosphorylation on the nearby serine 305 (S305) and promotes high transcriptional activity, even with low estrogen levels. Therefore, the crosstalk between K302 methylation and K303 acetylation can contribute to invasive breast tumors, highlighting that PTMs deeply influence SR signaling in normal and malignant contexts.
Figure 4.
Biological consequences of SR methylation. All the methylation events targeting the steroid receptors on arginine (R) and lysine (K) residues and reported at this time are represented for (A) ERα, (B) PR, (C) AR, and (D) GR. When identified, the protein methyltransferases involved are noted in black and the demethylases in brown. The methylation events leading to repressive functions are represented in red and the activating functions are in green. For ERα, we enlarged the hinge domain as it is the main region modified by methylation. When decrypted and reported, the biological consequences of the methylation event on the physiology/pathology have been indicated (in green for activating functions, red for repressive functions and blue when no effect). NTD, N-terminal domain; DBD, DNA-binding domain; h, hinge; LBD, ligand binding domain; NLS, nuclear localization signal; NES, nuclear export signal; BC, breast cancer; PC, prostate cancer.
Table 3.
Lysine and arginine methylation of steroid receptors
| Steroid receptor methylation by lysine methyltransferases | ||||
|---|---|---|---|---|
| Receptor | Enzyme | Residue | Biological effect | References |
| ERα | SET7/9 | K302me1 | Promotes transcriptional activity by protein stabilization | (375) |
| SMYD2 | K266me1 | Represses transcriptional activity | (376) | |
| G9a | K235me2 | Promotes transcriptional activity | (377) | |
| PR | ND | K464me1 | Decreases ligand sensitivity | (378) |
| K481me1 | Represses AF1 activity | (379) | ||
| AR | SET7/9 | K632me1 | Promotes its transcriptional activity | (380) |
| K630me1 | (381) | |||
| AR | DOT1L | K349 | Activates its transcriptional activity | (189) |
| Steroid receptor methylation by protein arginine methyltransferases | ||||
| ERα | PRMT1 | R260me2a | Participates in E2 non genomic signaling | (382, 383) |
| Participates in IGF-1 signaling | (384) | |||
| Participates in vascular protective effects | (385) | |||
| PR | ND | R492me1 | Decreases transcriptional efficiency | (379) |
| PRMT1 | R637me2a | Regulates stability and transcriptional activity | (386) | |
| AR | PRMT5 | R761me1/2s | Represses genes involved in differentiation | (387) |
| PRMT6 | R210me2a, R212me2a, R787me2a, R789me2a | Activates its transcriptional activity in SBMA, by inhibiting phosphorylation by Akt | (388) | |
| GR | PRMT5 | Rme2s | ND | (389) |
K, lysine; R, arginine; Kme1, monomethyllysine; Kme2, dimethyllysine; Rme1, monomethyarginine; Rme2a, asymmetric dimethyarginine; Rme2s, symmetric dimethylarginine; ND, nondetermined; IGF-1, insulin-like growth factor; SBMA, spinal and bulbar muscular atrophy. ND, not determined
K266.
Later, K266 was shown to be methylated by SMYD2 (376). In the absence of estrogen, this modification impairs ERα binding to chromatin to prevent gene activation. Upon estrogen stimulation, K266 methylation is diminished, enabling p300-induced acetylation on this lysine residue to activate ERα transcriptional activity (Fig. 4A and Table 3). Moreover, cells with ERα-K266R mutation have a higher capacity to proliferate than wild-type (WT) cells under estrogen-depleted growth conditions, likely indicating that SMYD2-mediated K266 methylation blocks the estradiol-induced (E2) cellular response. As LSD1 has been shown to remove methyl marks on SMYD2 substrates, including p53 and HSP90 (394, 395), the authors investigated whether it could participate in the regulation of ERα K266 methylation. They showed that LSD1 is able to remove SMYD2-K266 methylation, allowing ERα acetylation on the same residue by p300, activating its transcriptional activity (Fig. 4A). More recently, HSP90 and its cochaperone p23 have been shown to bind SMYD2, inducing an increase in its ability to methylate ERα (396). We can thus speculate that the well-known association between ERα and HSP90 in the cytoplasm before hormonal stimulation could involve SMYD2 in order to maintain ERα in an inactive state in the absence of estrogen.
K235.
The latest described ERα methylation on lysine is catalyzed by the KMT G9a, which dimethylates the receptor at K235, in its DBD (Fig. 4A and Table 3). This modification functions as an activator for the expression of some estrogen target genes, activating the growth of ERα-positive BC cells (377). K235 dimethylation is a recognition site for the Tudor domain of PHF20, a member of the MOF histone acetyltransferase complex, which catalyzes the acetylation of histone H4K16, as well as nonhistone proteins involved in transcription. The association of ERα with PHF20 through its K235 methylation site then recruits the MOF complex to deposit acetylation to H4K16 of E2 target genes, supporting access of ERα to chromatin and improving its transcriptional activity. As previously reported for other methylation events, in the vicinity of K235, S236 was reported to be phosphorylated by protein kinase A. Unlike K235 methylation, S236 phosphorylation is an obstacle for ERα dimerization and its recruitment to DNA. Therefore, these 2 adjacent modifications compete to regulate the transactivation activity of ERα. Aside from K235, K303 in the hinge domain was also revealed to be methylated by G9a in vitro; however, this has not been confirmed in vivo.
Taken together, it seems that ERα activity is regulated by 3 different lysine methylations carried out by 3 different enzymes, in which K302 and K235 methylation strengthens ERα activity, whereas K266 methylation functions as a repressor. Importantly, these 3 ERα residues are located in the same “hot-spot,” which undergoes intensive posttranslational modifications, and it is thus fundamental to study the importance of context-dependent effects of methylation events to better understand their interplay with other modification marks within the context of ERα signaling.
Arginine methylation/demethylation
R260.
Our team was the first to identify an arginine methylation event for a SR. Indeed, we showed that PRMT1 dimethylates ERα on arginine 260 (R260), located at the junction between the DBD and the hinge domain (Fig. 4A and Table 3). This modification is a crucial event for estrogen nongenomic signaling. Mechanistically, estrogen triggers a rapid and transient ERα methylation, which is required for its interaction with the kinases Src and PI3K. The formation of this complex is a prerequisite for activating the downstream Akt pathway (382). In addition, we demonstrated that insulin-like growth factor (IGF-1) also triggers ERα methylation via PRMT1, an important event for IGF-1 signaling in BC, highlighting that targeting PRMT1 activity could be a good strategy to concomitantly impact estrogen and IGF-1 pathways (384).
Together, these results emphasize the importance of PRMT1 as a regulator of both estrogen and IGF-1 signaling, highlighting PRMT1 as a promising target for treating ERα-positive BC patients.
As ERα methylation is a transient event, we investigated whether a demethylase could be involved in the regulation of ERα methylation. We showed that, upon estrogen stimulation, JMJD6 is integrated into the hallmark nongenomic complex metER/Src/PI3K, where it demethylates ERα, causing dissociation of the complex and termination of downstream signaling (366). Moreover, JMJD6 expression is associated with poor prognosis in BC (397), but its enzymatic activities have not yet been fully associated with BC.
Most of the studies on SR methylation have been conducted in cancer cell models; however, metERα expression has been evaluated in human breast tissues concomitantly with ERα/Src and ERα/PI3K complexes, hallmarks of ERα nongenomic signaling. Their expression has been detected at low levels in human breast tissues and high levels in a subset of breast tumors. Interestingly, their high expression is associated with BC aggressiveness (383). More recently, we showed that ERα nongenomic signaling is increased in BC resistant to tamoxifen treatment (398). Later, using a mouse model harboring ERα mutated R264A (equivalent to R260 in human), the role of metERα in physiology was investigated. It was shown that although this arginine is not required for the physiological regulation of the skeleton (399) nor for fertility (385), this residue is involved in ERα activity, such as the rapid dilatation of mesenteric arteries and the endothelial repair of carotid (385).
Altogether, these findings highlight the importance of arginine methylation in ERα nongenomic pathways. Probably because R260 is not conserved among NR, this modification has never been involved in nongenomic signaling triggered by other members of the family.
Progesterone Receptor, or PR
Lysine methylation
K464, K481.
PR is largely post-translationally modified, especially by phosphorylation on serine and threonine residues (400). Methylation on lysine residues has also been reported, in the NTD close to the DBD. Chung et al. (378) reported that K464 methylation is essential in PR ligand–independent and –dependent transcriptional activities (Fig. 4B and Table 3). Using MS, they showed that both PR is endogenously monomethylated in T47D BC cells (378). Interestingly, K464 is located in the AF-1 hormone-independent transactivation domain, previously narrowed down to a 91 amino acid sequence preceding the DBD in PRs (Fig. 4B) (401). Site-directed mutagenesis on PR revealed that nonmethylable mutants display a higher ligand-independent activation, in particular perceptible by an increase in PR phosphorylation at S400, a basally phosphorylated site (402). Importantly, K464 mutations have a significant effect on ligand-induced activity of PR, implying that K464 methylation impedes the transcriptional activity of PR.
A more recent study identified K481me1 acting in cooperation with K464 in the ligand-induced transcriptional activity of the receptor (379) (Fig. 4B). Although the KMT is not identified yet, this study is the first to argue in favor of the importance of methylation in regulating PR signaling. These studies suggest that methylation of lysine residues of the AF-1 domain disrupts PR transcriptional activity, supporting the notion that methylation is a modulator of PR signaling in BC cells.
Arginine methylation
R492.
Within the same study conducted by Woo et al., R492me1 was shown to synergize with the 2 preceding lysine methylation events (K464 and K481), in the transcriptional activity of PR (379) (Fig. 4B and Table 3). When residues were substituted to neutral polar glutamine (K464Q/K481Q/R492Q), the defective triple-methylation mutant exhibited a strong increase in transcriptional activity in response to progestin, compared with WT PR, suggesting that positive charge due to methylation could act as a brake for transcription efficiency. Moreover, data showed that these key residues provide not only the interaction interface with major PR coregulators, like SRC-1, but also with the AF-2-containing LBD, described as acting with AF-1 to bring PR towards a full transcriptional activation.
R637.
Our team recently demonstrated a functional crosstalk between arginine methylation, PR transcriptional activity, and progestin-induced proliferation in BC cells (386). We showed that PRMT1 is an important modulator of the transcriptional activity of PR, at least in part through methylation in the hinge region (Fig. 4B and Table 3). Under progestin treatment, PRMT1 dimethylates PR in the nucleus, at R637, within a RGG methylation consensus motif (346, 403). This methylation event modulates PR oncogenic properties, as cells expressing a nonmethylable mutant (R637K) displayed a retarded cell growth and a reduced expression of a subset of genes that promote proliferation and survival of BC cells. R637 methylation facilitates PR degradation, which in turn constitutes a critical stimulatory switch that accelerates the recycling of PR from pre-initiation complexes, which is required for active hormone-dependent transcription (390, 404, 405). Moreover, BC patients with PR-positive tumors expressing high level of PRMT1 show a worse survival than patients with low PRMT1, suggesting that targeting PRMT1 could constitute a new therapeutic option.
Interestingly, SUMOylation of PR on K388 has also been shown to regulate PR stability by competing with ubiquitination. Indeed, the E3 ubiquitin ligase CUEDC2 binding attenuates sumoylation SUMOylation of K388, while promoting ubiquitination (406). As crosstalk between different PTMs have largely been described (85), we can hypothesize that the integration of these different signals tightly regulates PR stability and transcriptional activity.
Taken together, methylation is an indisputable mechanism involved in the regulation of the transcriptional activity of PR, sometimes acting as a repressor or as an activator, once again highlighting the importance of context-dependent effects of this PTM in cells.
Androgen Receptor, or AR
Lysine methylation
K630, 632.
In 2011, 2 different teams reported that AR activity can be regulated by SET7/9. The first 1 reported that AR interacts with the methyltransferase, which in turn monomethylates the K632 residue within a KLKK motif, similarly to the sequence methylated in the hinge domain of ERα (407) (Fig. 4C and Table 3). This methylation induced upon ligand binding is necessary for enhancing AR transcriptional activity. As SET7/9 functions as a proproliferative and antiapoptotic factor, highly expressed in PC, it could constitute a potential target to treat these tumor (407). In parallel, a study from another research group demonstrated that K630, and not K632, is methylated by SET7/9, and globally linked with the same functions (381). The discrepancy between these 2 works has so far not been resolved. K632 modification better matches the consensus site of SET7/9, but the identification was performed with a small peptide. MS analyses on the full protein or specific antibodies are needed to clarify this point. Nonetheless, these 2 methylation sites belong to a rich acetylation target domain. Indeed, K630, 632, and 633 residues are acetylated by p300, p/CAF (408), and TIP60 (409), suggesting, similarly to ERα, a crosstalk between acetylation and methylation.
K349.
Lysine methylation is also involved in the regulation of AR transcriptional activity, through the binding of 2 long noncoding RNAs, namely PRNCR1 and PCGEM1, in PC cells. The KMT DOT1L was required for AR binding to PCGEM1. Indeed, DOT1L methylates AR on K349, located in the N-terminal region, a critical step for the recruitment of PCGEM1 to AR (189) (Fig. 4C and Table 3). Moreover, DOT1L is overexpressed in PC and is associated with poor clinical outcome, and this KMT selectively regulates the tumorigenicity of AR-positive PC cells in vitro and in vivo, making a promising therapeutic target (410). However, the results about AR methylation on K349 are matter of debate since they were refuted by Chinnaiyan’s team (411).
Arginine methylation
R210, R212, R787, R789.
The first link between PRMT6 and AR was unveiled by Sun et al. (412), showing a direct PRMT6/AR interaction in Cos7 cells overexpressing AR. Furthermore, PRMT6 was able to methylate AR, although specific methylation sites were not explored. More recently, this interaction was confirmed with the AR mutant containing polyglutamine stretch in its NTD and implicated in the X-linked transmitted spinal and bulbar muscular atrophy (388). For this interaction, the required regions were the AF-2 domain of AR, the catalytic domain of PRMT6 as wells as a LXXLL motif present in PRMT6. PRMT6 proved to methylate AR and, to a higher extent, polyglutamine-expanded AR. The AR arginine methylation sites were identified in both the NTD and the LBD (R210/212 and R787/789) (Fig. 4C and Table 3), within RXRXXS motifs involved in Akt-mediated AR phosphorylation. Overall, AR transactivation by PRMT6-mediated methylation was regulated by phosphorylation through a mutually exclusive interaction, making PRMT6 a modifier of polyglutamine-expanded AR neurotoxicity. A model is suggested in which arginine methylation of polyglutamine-expanded AR by PRMT6 at the Akt consensus site motif enhances function and toxicity leading to neurodegeneration, whereas phosphorylation by Akt prevents binding to testosterone, thereby protecting neurons from degeneration (388).
Exemplified in this disease model, the PRMT6/AR interaction is likely to play relevant roles in other diseases, such as PC, where PRMT6 is overexpressed in comparison with normal prostate tissue (413, 414) or even physiological conditions. Thus, overexpression of PRMT6 has been observed in the testes of AR-KO mice (51, 415). Results from this study suggest that downregulation of PRMT6 by AR (binding to an androgen response element present in PRMT6 promoter) is necessary for AR-controlled spermatogenesis (415).
R761.
PRMT5 has been shown to methylate AR on R761 in the LBD (Fig. 4C and Table 3). This modification was described in PC cases expressing the TMPRSS2:ERG fusion gene. Mechanistically, the TF ERG recruits PRMT5 to AR target genes involved in prostate differentiation, where the enzyme methylates the receptor (387). This event attenuates AR recruitment to these sites and subsequent transcription from the associated gene promoter. Consistently, R761 methylation supports proliferative functions of the oncoprotein ERG. Thus, R761 methylation could serve as a potent biomarker for AR-dependent proliferation in TMPRSS2:ERG-positive PC, and inhibiting PRMT5 activity could be beneficial for treating this tumor subtype.
Altogether, it appears that AR and its variants are highly methylated on both lysine and arginine residues, participating actively in its full transcriptional activity, even though the interplay between these modifications is still unknown.
Glucocorticoid Receptor, or GR
GR was not known to undergo methylation until recently. Indeed, using pan methyl antibodies recognizing only dimethylated arginines, our team found that GR is methylated by PRMT5 within the nucleus of the ERα-positive BC cell line MCF-7 (Fig. 4D and Table 3) (389). Although the methylated arginine residue(s) and the role of this modification in GR activity is still under investigation, our finding is highly promising as it suggests a new molecular regulation of GR activity involving PRMT5.
Indirect Role of Methylation/Demethylation in Steroid Receptor Transcriptional Activity
Additional indirect methylation events modulate the transcriptional activity of SRs. On the one hand, SR coregulators are also methylated. These methylation events regulate, among other processes, molecular interactions, stability, and subcellular localization of SRs, in order to tightly modulate their transcriptional activity. Specific coregulators are recruited on the promoter/enhancer regions of their target genes to locally remodel the chromatin structure and to orchestrate the assembly or disassembly of an active transcription complex. On the other hand, lysine or arginine methyltransferases, or demethylases, modify residues in histone tails and thereby participate in SR-dependent target gene expression. In this section, we selected key examples of these indirect methylation events to illustrate their impact on SR transcriptional activity.
Lysine Methylation/Demethylation
G9a/GLP methylation and GR regulation
G9a and its paralogue GLP are interesting illustrations of KMTs involved in SR transcriptional activity through more indirect mechanisms than those previously described. First identified as KMTs methylating the repressive mark H3K9, these enzymes also act as GR coactivators under certain circumstances (90). The self-methylation of G9a/GLP on K185 and K205, respectively, provide a binding site for HP1γ, which facilitates the recruitment of RNA polymerase II, activating the transcription of a subset of GR target genes in specific cell contexts (Fig. 5A). In contrast, phosphorylation of the adjacent threonine (T186 for G9a and T206 for GLP) by Aurora kinase B (AURKB) prevents binding to HP1γ and reduces coactivator functions of G9a and GLP (416), resulting in distinct biological effects in a tissue-specific manner. For instance, the coactivating activity of G9a/GLP regulates migration of the lung cancer cell line A549 (416), and GC-induced cell death in B-acute lymphoblastic leukemia (417) (Fig. 5A). Moreover, a panel of KDM inhibitors identified the JmjC KDM family as demethylases for G9a and GLP. JmjC inhibitors increased G9a methylation, expression of G9a-HP1γ-dependent GR target genes and GC-induced cell death in B-acute lymphoblastic leukemia cell lines (418). In vitro demethylation assays unveiled the KDM4 family as demethylases for G9a and GLP (Fig. 5A). This study was a proof of concept that a methylation/phosphorylation event could influence a coregulator, functioning as a coactivator or corepressor, tightly regulating the transcriptional activity of a SR.
Figure 5.
Indirect methylation events regulating SR signaling. Here, we highlight 2 examples, in (A) GR and in (B) ERα, of indirect methylation events (ie, not directly on SRs), regulating the transcriptional activity of these 2 receptors. This concerns the methylation of histone tails on chromatin and/or the methylation of coregulators. When identified, the targeted lysines (K) or arginines (R) and the methyltransferases are noted in black and the demethylases in brown. The methylation events leading to repressive functions are represented with red lines and the activating functions with green arrows. Me, methylation; GRE, GR response elements; ERE, estrogen response elements; H3, histone H3; H4, histone H4; CoA, coactivators; Dex, dexamethasone; E2, estrogens; BC, breast cancer.
LSD1 regulation of histone marks
Among the well-known coregulators that influence SR signaling by altering protein methylation status, LSD1 was the first demethylase described to regulate histone methylation in a hormone-dependent context. Indeed, the recruitment of LSD1 and subsequent H3K4me1/2 demethylation induces repression of androgen-dependent AR target gene expression (including AR itself) (419). In addition to its well-established corepressor functions (203, 420), LSD1 acts as a coactivator for several TFs, including AR and ERα (208, 421-424). This effect was not only observed in PC cells but also in kidney cancer cells, which are usually not considered to be androgen-sensitive. In this study, using pargyline, an inhibitor of LSD1, investigators were able to block demethylation of H3K9 and subsequently AR-dependent transcriptional activation (425). In another recent study, LSD1 demethylated the pioneer TF FOXA1, thereby facilitating its DNA-binding, notably in androgen response element and estrogen response element, where it is a well-known active partner in AR- and ER-mediated transactivation (426). Lastly, LSD1 acts as a coactivator of both AR and its main splice variant, AR-V7, which has been implicated in resistance to androgen deprivation therapy in castration-resistant PC (427). Overall, through enhanced transactivation of AR (and possibly AR variants), LSD1 may favor proliferation and invasiveness of PC cells and impair apoptosis under androgen deprivation therapy (425, 428).
Interestingly, phosphorylation of H3T6 by protein kinase C beta I prevents LSD1 from demethylating H3K4me1/2 and induces a switch in substrate specificity from H3K4me1/2 to H3K9me1/2, orienting LSD1 towards a transcriptional coactivator role (208, 429). Again, this set of experiments clearly underlines that protein methylation can interfere with other PTMs and induce a switch in the functions of SR coregulators, resulting in a fine-tuning of gene expression.
Arginine Methylation/Demethylation
Similarly to lysine residues, methylation on arginine residues of histone tails and coregulators has been reported to indirectly regulate SR transcriptional activities.
Methylation of histone marks by PRMT1 and CARM1
Twenty years ago, numerous studies demonstrated the strong impact of PRMT1 and CARM1 as coregulators that modify histone tails in order to modulate SR-dependent transcription. Historically, CARM1 was characterized as a methyltransferase owing to its capacity to methylate the N-terminal tail of H3 in vitro (430). This finding was then confirmed in vivo on hormone-regulated promoters. Indeed, the transcriptional activities of ERα, GR, and AR are known to be regulated by CARM1-induced methylation of both R17 and R26 of histone H3 (431-433) (Fig. 5A). As CARM1, PRMT1 also acts as a coactivator of SR through histone methylation (12, 13, 434). H4R3 is methylated by PRMT1 and plays a critical role in transcriptional activation, making PRMT1 a potent coactivator of AR transcriptional activity (13). Interestingly, as it was described for lysine methylation on histone tails, cross-talks between arginine methylation and other PTMs, namely lysine acetylation, were reported to be involved in the regulation of SR-dependent transcription. For instance, stimulation by estrogens results in the acetylation of H3K18 by the acetyltransferase CREB binding protein (CBP). This event stimulates CARM1 recruitment to the H3 tail, which methylates H3R17, leading to the implementation of estrogen-regulated gene expression (435). More recently, PRMT5 associated with pICln was described as an epigenetic activator of AR transcription in castration-resistant PC cells, suggesting that targeting its enzymatic activity could represent a novel therapeutic approach (436).
Methylation of coregulators by CARM1
Aside from direct histone methylation, some transcriptional coregulators are also methylated on arginine residues, making the arginine methyltransferases potent transcriptional modulators, such as CARM1 for ERα- and AR-dependent transcription (437). For example, CARM1 dimethylates CBP on R742 in vivo, playing a role in estrogen-induced gene activation (438) (Fig. 5B). P300 is methylated by CARM1 on R2142, localized in its C-terminal GRIP1 binding domain, inhibiting p300 binding to the coregulator glucocorticoid receptor interacting protein 1 (GRIP1/SRC2), a key coregulator of SRs in vitro and in vivo (439). It was later shown that CARM1-dependent CBP methylation on R742, 768, and 2151 stimulates its histone acetyltransferase activity by increasing its autoacetylation (440). Interestingly, using antibodies specific for the individual methylation sites, they showed that methylation of CBP is required for its recruitment by ERα to specific target genes (Fig. 5B). Using genome-wide analyses, different patterns of binding to ERα target genes were observed for the various methylated CBP species (440). These data suggest that CARM1-dependent CBP methylation induces the expression of specific target genes, diversifying the ERα transcriptional program.
CARM1 was also shown to methylate SR coactivator proteins of the SRC/p160 family, including SRC-3 (441, 442). SRC proteins serve as primary coregulators to recruit secondary coactivators, such as p300/CBP or CARM1, to the promoters of specific estrogen-dependent target genes (443). SRC-3 methylation on R1171 takes place in its C-terminal domain containing its binding sites with p300/CBP and CARM1 (441, 442). Consequently, SRC-3 methylation induced by estrogens triggers its dissociation from CBP and CARM1, a decrease in its stability and a subsequent decrease in ERα-mediated transcription (Fig. 5B). The authors suggested that this methylation event serves as a molecular switch for disassembly of the SRC-3 transcriptional coactivator complex (441).
More recently, 2 new substrates for CARM1 were identified: BAF155 in the SWI/SNF chromatin remodeling complex and MED12, a component of the mediator complex that facilitates RNA polymerase II recruitment (282). CARM1-methylation of MED12 regulates its binding to the chromatin in order to regulate p21 gene expression, increasing BC cell sensitivity to chemotherapy in vitro and in vivo (282, 444) (Fig. 5B). An additional study confirmed that MED12 is methylated by CARM1 in a cluster of arginine residues located in its C-terminal domain, regulating MED12 recruitment to ERα enhancers (445) (Fig. 5B). These methylated residues are recognized by the coactivator protein TDRD3 to activate ERα target genes. Interestingly, a high-resolution MS analysis of CARM1 substrates identified a list of coregulators involved in ERα transcription (called CARM1 methylome), such as acetyltransferases (P300, P400), KMTs (KMT2C and KMT2D) and components of the SWI/SNF, NuRD, and mediator complexes (232). Of note, JMJD6 regulates also the activation of ERα enhancers by participating in the interaction between MED12 and CARM1 (446) (Fig. 5B).
Aside from CARM1, PRMT1 also dimethylates coregulators to modulate SR signaling. For example, PRMT1 dimethylates arginines on the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), a coactivator of many SRs, including ERα (447). PRMT1 and its catalytic activity enhances PGC-1α coactivator activity on ERα reporter genes (Fig. 5B). Endogenously, PRMT1 regulates the expression of PGC-1α target genes that are important for mitochondrial biogenesis (447).
Altogether, these results highlight that methylation of coregulators and histones is heavily involved in the fine regulation of the transcriptional activity of SRs, making these enzymes promising targets to modulate SR functions.
Outlook
There is increasing evidence that SRs are tightly regulated by protein interactions and PTMs targeting the receptors themselves, but also histone tails and coregulatory proteins. Among these PTMs, methylation has assumed an increasingly important role, and given the crosstalk between PTMs, we can extrapolate that the effects extend beyond current knowledge.
Even if ERα appears to be the most post-translationally modified SR (probably because it is the most studied), it is likely that other receptors are also methylated on residues that remain to be identified. This review focused on 4 SRs, but we can easily imagine that other NRs are modified by methylation. It has already been shown for the orphan receptor HNF4, the transcriptional activity of which is regulated by PRMT1 in 2 ways (448): it methylates HNF4-DBD, enhancing its binding to chromatin, and methylates H4R3. In addition, RARα is also methylated on lysine residues, impacting its transcriptional activity (449). Although the thyroid hormone receptor has so far not been shown to be methylated, PRMT1 can regulate its transcriptional activity through H4R3 methylation, contributing to T3-induced metamorphosis of Xenopus laevis (450). LSD1 is also suspected to be a functionally important cofactor for the mineralocorticoid (MR) and MR-related disease, such as high blood pressure (451).
In this review, we focused on some methyltransferases though other enzymes may also regulate SR function via methylation. For instance, PRMT2 regulates the transcriptional activities of ERα (452) and AR (453), likely through its enzymatic activity. In addition, PRMT6 enhances ERα ligand–dependent and –independent activity (412), even if the mechanism involved has not been clearly identified. Most studies have been performed in cancer cell lines, but as the effects of methylation impact key functions of SRs, such as ligand sensitivity and DNA binding, it is likely that methylation is also important for other physiological roles of SRs. For instance, ERα methylation on R260 has been identified firstly in BC, but the role of this methylation has recently been linked in vivo to vascular functions (385). Unfortunately, not all methylation events have been as extensively deciphered, likely due to the difficulty in detecting these modifications in vivo (lack of specific antibodies, lack of mouse models). More investigation is required to identify more thoroughly each methylation event for SRs themselves but also for their coregulators using MS analyses. This may provide a description of the specific pattern of methylation in each tissue and its dysregulation in specific pathologies.
Since SR activities are often linked with cancer, and methylases/demethylases also play a key role in tumorigenesis, future perspectives in cancer treatment could include targeting these enzymes, but their use could be extended to other pathologies where SRs, KMTs, and PRMTs are involved. Compared with other small molecule inhibitors, such as HDAC or RTK, developing small inhibitors of protein methyltransferases is a rising challenge for cutting-edge, drug-designing industrial enterprises.
Acknowledgments
We thank V. Vlaemink-Guillem for her advice and B. Manship for proofreading the manuscript. The illustrations were created by using Servier Medical Art.
Financial Support: M.L.R, L.M., H.T.P, L.E., and C.P.’s laboratory is funded with grants from “La Ligue contre le Cancer,” the “Fondation ARC Cancer,” and the “Fondation de France.” L.M. was supported by a fellowship from “Fondation ARC Cancer.” L.E. was supported by a fellowship from “La Ligue contre le Cancer.” H.T.P. was supported by Ambassade de France au Vietnam. M.R.S. acknowledges support from the United States National Institutes of Health (Grants DK043093) and the Margaret E. Early Medical Research Trust.
Glossary
Abbreviations
- AR
androgen receptor
- BC
breast cancer
- ER
estrogen receptor
- GAR
glycine and arginine-rich motif
- GC
glucocorticoid
- GR
glucocorticoid receptor
- IGF
insulin-like growth factor
- JHDM
JmjC domain-containing histone demethylase
- KMT
lysine methyltransferase
- LBD
ligand binding domain
- MS
mass spectrometry
- NLS
nuclear localization signal
- NR
nuclear receptor
- NTD
aminoterminal domain
- PC
prostate cancer
- PR
progesterone receptor
- PRMT
protein arginine methyltransferase
- PTM
post-translational modification
- SR
steroid receptor
- TF
transcription factor
Additional Information
Disclosures: The authors declare that they have no conflict of interest.
References
- 1. Baschant U, Lane NE, Tuckermann J. The multiple facets of glucocorticoid action in rheumatoid arthritis. Nat Rev Rheumatol. 2012;8(11):645-655. [DOI] [PubMed] [Google Scholar]
- 2. Jensen EV, Jacobson HI, Walf AA, Frye CA. Estrogen action: a historic perspective on the implications of considering alternative approaches. Physiol Behav. 2010;99(2):151-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39(1):141-157. [DOI] [PubMed] [Google Scholar]
- 4. Islam MS, Afrin S, Jones SI, Segars J. Selective progesterone receptor modulators-mechanisms and therapeutic utility. Endocr Rev. 2020;41(5):643-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nicolaides NC, Galata Z, Kino T, Chrousos GP, Charmandari E. The human glucocorticoid receptor: molecular basis of biologic function. Steroids. 2010;75(1):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Selvaraj V, Dapaah-Afriyie K, Finn A, Flanigan TP. Short-term dexamethasone in Sars-CoV-2 patients. R I Med J (2013). 2020;103(6):39-43. [PubMed] [Google Scholar]
- 7. Xiang Z, Liu J, Shi D, et al. Glucocorticoids improve severe or critical COVID-19 by activating ACE2 and reducing IL-6 levels. Int J Biol Sci. 2020;16(13):2382-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Traboulsi T, El Ezzy M, Gleason JL, Mader S. Antiestrogens: structure-activity relationships and use in breast cancer treatment. J Mol Endocrinol. 2017;58(1):R15-R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chi KN, Agarwal N, Bjartell A, et al. ; TITAN Investigators . Apalutamide for metastatic, castration-sensitive prostate cancer. N Engl J Med. 2019;381(1):13-24. [DOI] [PubMed] [Google Scholar]
- 10. Chambon P. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol. 2005;19(6):1418-1428. [DOI] [PubMed] [Google Scholar]
- 11. Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17(10):554-556. [DOI] [PubMed] [Google Scholar]
- 12. Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276(2):1089-1098. [DOI] [PubMed] [Google Scholar]
- 13. Wang H, Huang ZQ, Xia L, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853-857. [DOI] [PubMed] [Google Scholar]
- 14. Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26(10):880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poulard C, Corbo L, Le Romancer M. Protein arginine methylation/demethylation and cancer. Oncotarget. 2016;7(41):67532-67550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escriva H, Safi R, Hänni C, et al. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci U S A. 1997;94(13):6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Green S, Walter P, Kumar V, et al. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320(6058):134-139. [DOI] [PubMed] [Google Scholar]
- 18. Conneely OM, Sullivan WP, Toft DO, et al. Molecular cloning of the chicken progesterone receptor. Science. 1986;233(4765):767-770. [DOI] [PubMed] [Google Scholar]
- 19. Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240(4850):327-330. [DOI] [PubMed] [Google Scholar]
- 20. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson-Rechavi M, Escriva Garcia H, Laudet V. The nuclear receptor superfamily. J Cell Sci. 2003;116(Pt 4):585-586. [DOI] [PubMed] [Google Scholar]
- 22. Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352(6335):497-505. [DOI] [PubMed] [Google Scholar]
- 23. Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75(3):567-578. [DOI] [PubMed] [Google Scholar]
- 24. Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10(3):384-391. [DOI] [PubMed] [Google Scholar]
- 25. Fawell SE, Lees JA, White R, Parker MG. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60(6):953-962. [DOI] [PubMed] [Google Scholar]
- 26. Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402(6757):93-96. [DOI] [PubMed] [Google Scholar]
- 27. Renaud JP, Rochel N, Ruff M, et al. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378(6558):681-689. [DOI] [PubMed] [Google Scholar]
- 28. Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11-26. [DOI] [PubMed] [Google Scholar]
- 29. Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67(9):4514-4523. [DOI] [PubMed] [Google Scholar]
- 30. Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005;19(11):2671-2684. [DOI] [PubMed] [Google Scholar]
- 31. Tetel MJ, Jung S, Carbajo P, Ladtkow T, Skafar DF, Edwards DP. Hinge and amino-terminal sequences contribute to solution dimerization of human progesterone receptor. Mol Endocrinol. 1997;11(8):1114-1128. [DOI] [PubMed] [Google Scholar]
- 32. Zou JX, Guo L, Revenko AS, et al. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69(8):3339-3346. [DOI] [PubMed] [Google Scholar]
- 33. Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391(Pt 3):449-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benecke A, Chambon P, Gronemeyer H. Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 2000;1(2):151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patel SR, Skafar DF. Modulation of nuclear receptor activity by the F domain. Mol Cell Endocrinol. 2015;418(Pt 3):298-305. [DOI] [PubMed] [Google Scholar]
- 36. Beato M. Gene regulation by steroid hormones. Cell. 1989;56(3):335-344. [DOI] [PubMed] [Google Scholar]
- 37. Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3(11):950-964. [DOI] [PubMed] [Google Scholar]
- 38. Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265-266:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ko YJ, Balk SP. Targeting steroid hormone receptor pathways in the treatment of hormone dependent cancers. Curr Pharm Biotechnol. 2004;5(5):459-470. [DOI] [PubMed] [Google Scholar]
- 40. Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2(4):323-334. [DOI] [PubMed] [Google Scholar]
- 41. Xu B, Lovre D, Mauvais-Jarvis F. Effect of selective estrogen receptor modulators on metabolic homeostasis. Biochimie. 2016;124:92-97. [DOI] [PubMed] [Google Scholar]
- 42. DeMayo FJ, Lydon JP. 90 YEARS OF PROGESTERONE: new insights into progesterone receptor signaling in the endometrium required for embryo implantation. J Mol Endocrinol. 2020;65(1):T1-T14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science. 2000;289(5485):1751-1754. [DOI] [PubMed] [Google Scholar]
- 44. Hughes GC. Progesterone and autoimmune disease. Autoimmun Rev. 2012;11(6-7):A502-A514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martocchia A, Stefanelli M, Cola S, Falaschi P. Sex steroids in autoimmune diseases. Curr Top Med Chem. 2011;11(13):1668-1683. [DOI] [PubMed] [Google Scholar]
- 46. Fréchou M, Zhu X, Liere P, et al. Dose-dependent and long-term cerebroprotective effects of intranasal delivery of progesterone after ischemic stroke in male mice. Neuropharmacology. 2020;170:108038. [DOI] [PubMed] [Google Scholar]
- 47. Zhu X, Fréchou M, Liere P, et al. A role of endogenous progesterone in stroke cerebroprotection revealed by the neural-specific deletion of its intracellular receptors. J Neurosci. 2017;37(45):10998-11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Timmermans S, Souffriau J, Libert C. A general introduction to glucocorticoid biology. Front Immunol. 2019;10:1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids–new mechanisms for old drugs. N Engl J Med. 2005;353(16):1711-1723. [DOI] [PubMed] [Google Scholar]
- 50. Pufall MA. Glucocorticoids and cancer. Adv Exp Med Biol. 2015;872:315-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matsumoto T, Shiina H, Kawano H, Sato T, Kato S. Androgen receptor functions in male and female physiology. J Steroid Biochem Mol Biol. 2008;109(3-5):236-241. [DOI] [PubMed] [Google Scholar]
- 52. Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140(3):223-238. [DOI] [PubMed] [Google Scholar]
- 53. Fan W, Yanase T, Nomura M, et al. Androgen receptor null male mice develop late-onset obesity caused by decreased energy expenditure and lipolytic activity but show normal insulin sensitivity with high adiponectin secretion. Diabetes. 2005;54(4):1000-1008. [DOI] [PubMed] [Google Scholar]
- 54. Arnold FJ, Pluciennik A, Merry DE. Impaired nuclear export of polyglutamine-expanded androgen receptor in spinal and bulbar muscular atrophy. Sci Rep. 2019;9(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cicardi ME, Cristofani R, Crippa V, et al. Autophagic and proteasomal mediated removal of mutant androgen receptor in muscle models of spinal and bulbar muscular atrophy. Front Endocrinol (Lausanne). 2019;10:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giovannelli P, Di Donato M, Galasso G, Di Zazzo E, Bilancio A, Migliaccio A. The androgen receptor in breast cancer. Front Endocrinol (Lausanne). 2018;9:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mizushima T, Miyamoto H. The role of androgen receptor signaling in ovarian cancer. Cells. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shiina H, Matsumoto T, Sato T, et al. Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci U S A. 2006;103(1):224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hickey TE, Selth LA, Chia KM, et al. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat Med. 2021;27(2):310-320. [DOI] [PubMed] [Google Scholar]
- 60. Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21(10):381-388. [DOI] [PubMed] [Google Scholar]
- 61. Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29(6):317-324. [DOI] [PubMed] [Google Scholar]
- 62. Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10(10):1167-1177. [DOI] [PubMed] [Google Scholar]
- 63. Wurtz JM, Bourguet W, Renaud JP, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3(2):206. [DOI] [PubMed] [Google Scholar]
- 64. Goldhar AS, Duan R, Ginsburg E, Vonderhaar BK. Progesterone induces expression of the prolactin receptor gene through cooperative action of Sp1 and C/EBP. Mol Cell Endocrinol. 2011;335(2):148-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schüle R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242(4884):1418-1420. [DOI] [PubMed] [Google Scholar]
- 66. Wang C, Mayer JA, Mazumdar A, et al. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25(9):1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fondell JD, Roy AL, Roeder RG. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7(7B):1400-1410. [DOI] [PubMed] [Google Scholar]
- 68. Ritter HD, Mueller CR. Expression microarray identifies the unliganded glucocorticoid receptor as a regulator of gene expression in mammary epithelial cells. BMC Cancer. 2014;14:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vicent GP, Nacht AS, Zaurin R, et al. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013;27(10):1179-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Forman BM, Casanova J, Raaka BM, Ghysdael J, Samuels HH. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6(3):429-442. [DOI] [PubMed] [Google Scholar]
- 71. Glass CK, Holloway JM, Devary OV, Rosenfeld MG. The thyroid hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid hormone and estrogen response elements. Cell. 1988;54(3):313-323. [DOI] [PubMed] [Google Scholar]
- 72. Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Denner LA, Weigel NL, Maxwell BL, Schrader WT, O’Malley BW. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science. 1990;250(4988):1740-1743. [DOI] [PubMed] [Google Scholar]
- 74. Kato S, Endoh H, Masuhiro Y, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491-1494. [DOI] [PubMed] [Google Scholar]
- 75. Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301(5640):1714-1717. [DOI] [PubMed] [Google Scholar]
- 76. Revelli A, Massobrio M, Tesarik J. Nongenomic actions of steroid hormones in reproductive tissues. Endocr Rev. 1998;19(1):3-17. [DOI] [PubMed] [Google Scholar]
- 77. Migliaccio A, Castoria G, Giovannelli P, Auricchio F. Cross talk between epidermal growth factor (EGF) receptor and extra nuclear steroid receptors in cell lines. Mol Cell Endocrinol. 2010;327(1-2):19-24. [DOI] [PubMed] [Google Scholar]
- 78. Björnström L, Sjöberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833-842. [DOI] [PubMed] [Google Scholar]
- 79. Hagan CR, Daniel AR, Dressing GE, Lange CA. Role of phosphorylation in progesterone receptor signaling and specificity. Mol Cell Endocrinol. 2012;357(1-2):43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vlaeminck-Guillem V, Gillet G, Rimokh R. SRC: marker or actor in prostate cancer aggressiveness. Front Oncol. 2014;4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wilkenfeld SR, Lin C, Frigo DE. Communication between genomic and non-genomic signaling events coordinate steroid hormone actions. Steroids. 2018;133:2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Asuthkar S, Velpula KK, Elustondo PA, Demirkhanyan L, Zakharian E. TRPM8 channel as a novel molecular target in androgen-regulated prostate cancer cells. Oncotarget. 2015;6(19):17221-17236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang L, Barritt GJ. Evidence that TRPM8 is an androgen-dependent Ca2+ channel required for the survival of prostate cancer cells. Cancer Res. 2004;64(22):8365-8373. [DOI] [PubMed] [Google Scholar]
- 84. La Rosa P, Pesiri V, Leclercq G, Marino M, Acconcia F. Palmitoylation regulates 17β-estradiol-induced estrogen receptor-α degradation and transcriptional activity. Mol Endocrinol. 2012;26(5):762-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Le Romancer M, Poulard C, Cohen P, Sentis S, Renoir JM, Corbo L. Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocr Rev. 2011;32(5):597-622. [DOI] [PubMed] [Google Scholar]
- 86. Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60(9):520-528. [DOI] [PubMed] [Google Scholar]
- 87. Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011;30(23):4764-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Farcas AM, Nagarajan S, Cosulich S, Carroll JS. Genome-wide estrogen receptor activity in breast cancer. Endocrinology. 2021;162(2):bqaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stallcup MR, Poulard C. Gene-specific actions of transcriptional coregulators facilitate physiological plasticity: evidence for a physiological coregulator code. Trends Biochem Sci. 2020;45(6):497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bittencourt D, Wu DY, Jeong KW, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2012;109(48):19673-19678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20(5):1855-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99(26):16701-16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lonard DM, O’malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27(5):691-700. [DOI] [PubMed] [Google Scholar]
- 94. Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29(18):3784-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Boriack-Sjodin PA, Swinger KK. Protein methyltransferases: a distinct, diverse, and dynamic family of enzymes. Biochemistry. 2016;55(11):1557-1569. [DOI] [PubMed] [Google Scholar]
- 96. Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Arifuzzaman S, Khatun MR, Khatun R. Emerging of lysine demethylases (KDMs): from pathophysiological insights to novel therapeutic opportunities. Biomed Pharmacother. 2020;129:110392. [DOI] [PubMed] [Google Scholar]
- 98. Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444-447. [DOI] [PubMed] [Google Scholar]
- 99. Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26(2):147-170. [DOI] [PubMed] [Google Scholar]
- 100. Ambler RP, Rees MW. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56-57. [DOI] [PubMed] [Google Scholar]
- 101. Wu Z, Connolly J, Biggar KK. Beyond histones - the expanding roles of protein lysine methylation. FEBS J. 2017;284(17):2732-2744. [DOI] [PubMed] [Google Scholar]
- 102. Cao XJ, Garcia BA. Global proteomics analysis of protein lysine methylation. Curr Protoc Protein Sci. 2016;86:24.8.1-24.8.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6(8):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Falnes PØ, Jakobsson ME, Davydova E, Ho A, Małecki J. Protein lysine methylation by seven-β-strand methyltransferases. Biochem J. 2016;473(14):1995-2009. [DOI] [PubMed] [Google Scholar]
- 105. Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr Opin Struct Biol. 2006;16(6):753-760. [DOI] [PubMed] [Google Scholar]
- 106. Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol Cell Proteomics. 2011;10(1):M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Levy D. Lysine methylation signaling of non-histone proteins in the nucleus. Cell Mol Life Sci. 2019;76(15):2873-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bao L, Chen Y, Lai HT, et al. Methylation of hypoxia-inducible factor (HIF)-1α by G9a/GLP inhibits HIF-1 transcriptional activity and cell migration. Nucleic Acids Res. 2018;46(13):6576-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Huang J, Dorsey J, Chuikov S, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285(13):9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tsusaka T, Kikuchi M, Shimazu T, et al. Tri-methylation of ATF7IP by G9a/GLP recruits the chromodomain protein MPP8. Epigenetics Chromatin. 2018;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chang Y, Sun L, Kokura K, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun. 2011;2:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ferry L, Fournier A, Tsusaka T, et al. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell. 2017;67(4):550-565.e5. [DOI] [PubMed] [Google Scholar]
- 113. Rathert P, Dhayalan A, Murakami M, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4(6):344-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Pless O, Kowenz-Leutz E, Knoblich M, et al. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J Biol Chem. 2008;283(39):26357-26363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Choi J, Jang H, Kim H, et al. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic Acids Res. 2014;42(1):224-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nair SS, Li DQ, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013;49(4):704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ling BM, Bharathy N, Chung TK, et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2012;109(3):841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lee JS, Kim Y, Bhin J, et al. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci U S A. 2011;108(33):13510-13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lee JS, Kim Y, Kim IS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39(1):71-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lee SH, Hyeon DY, Yoon SH, et al. RUNX3 methylation drives hypoxia-induced cell proliferation and antiapoptosis in early tumorigenesis. Cell Death Differ. 2021;28(4):1251-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Li W, Tang J, Terry RN, et al. Long-acting reversible contraception by effervescent microneedle patch. Sci Adv. 2019;5(11):eaaw8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chae YC, Kim JY, Park JW, et al. FOXO1 degradation via G9a-mediated methylation promotes cell proliferation in colon cancer. Nucleic Acids Res. 2019;47(4):1692-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sampath SC, Marazzi I, Yap KL, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27(4):596-608. [DOI] [PubMed] [Google Scholar]
- 124. Chin HG, Estève PO, Pradhan M, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35(21):7313-7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zeng Y, Qiu R, Yang Y, et al. Regulation of EZH2 by SMYD2-mediated lysine methylation is implicated in tumorigenesis. Cell Rep. 2019;29(6):1482-1498.e4. [DOI] [PubMed] [Google Scholar]
- 126. Velinder M, Singer J, Bareyan D, et al. GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. Biochem J. 2017;474(17):2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629-632. [DOI] [PubMed] [Google Scholar]
- 128. Saddic LA, West LE, Aslanian A, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285(48):37733-37740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cho HS, Hayami S, Toyokawa G, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14(6):476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Piao L, Kang D, Suzuki T, et al. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia. 2014;16(3):257-264, 264.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Deng X, Hamamoto R, Vougiouklakis T, et al. Critical roles of SMYD2-mediated β-catenin methylation for nuclear translocation and activation of Wnt signaling. Oncotarget. 2017;8(34):55837-55847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Reynoird N, Mazur PK, Stellfeld T, et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev. 2016;30(7):772-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Nakakido M, Deng Z, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. Dysregulation of AKT pathway by SMYD2-mediated lysine methylation on PTEN. Neoplasia. 2015;17(4):367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hamamoto R, Toyokawa G, Nakakido M, Ueda K, Nakamura Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 2014;351(1):126-133. [DOI] [PubMed] [Google Scholar]
- 135. Calnan DR, Webb AE, White JL, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY). 2012;4(7):462-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Balasubramaniyan N, Ananthanarayanan M, Suchy FJ. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G937-G947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pagans S, Kauder SE, Kaehlcke K, et al. The cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe. 2010;7(3):234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ali I, Ramage H, Boehm D, et al. The HIV-1 tat protein is monomethylated at lysine 71 by the lysine methyltransferase KMT7. J Biol Chem. 2016;291(31):16240-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kim SK, Lee H, Han K, et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell. 2014;15(6):735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Aguilo F, Li S, Balasubramaniyan N, et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 2016;14(3):479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29(16):2357-2367. [DOI] [PubMed] [Google Scholar]
- 142. Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A. 2009;106(45):18972-18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Song H, Chu JW, Park SC, et al. Isoform-specific lysine methylation of RORalpha2 by SETD7 is required for association of the TIP60 coactivator complex in prostate cancer progression. Int J Mol Sci. 2020;21(5):1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhang WJ, Wu XN, Shi TT, et al. Regulation of transcription factor Yin Yang 1 by SET7/9-mediated lysine methylation. Sci Rep. 2016;6:21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wu XN, Shi TT, He YH, et al. Methylation of transcription factor YY2 regulates its transcriptional activity and cell proliferation. Cell Discov. 2017;3:17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chuikov S, Kurash JK, Wilson JR, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432(7015):353-360. [DOI] [PubMed] [Google Scholar]
- 148. Kassner I, Andersson A, Fey M, Tomas M, Ferrando-May E, Hottiger MO. SET7/9-dependent methylation of ARTD1 at K508 stimulates poly-ADP-ribose formation after oxidative stress. Open Biol. 2013;3(10):120173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Liu X, Wang D, Zhao Y, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc Natl Acad Sci U S A. 2011;108(5):1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wang D, Zhou J, Liu X, et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci U S A. 2013;110(14):5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hahm JY, Kim JY, Park JW, et al. Methylation of UHRF1 by SET7 is essential for DNA double-strand break repair. Nucleic Acids Res. 2019;47(1):184-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Song H, Feng X, Zhang M, et al. Crosstalk between lysine methylation and phosphorylation of ATG16L1 dictates the apoptosis of hypoxia/reoxygenation-induced cardiomyocytes. Autophagy. 2018;14(5):825-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Shen C, Wang D, Liu X, et al. SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. FASEB J. 2015;29(10):4313-4323. [DOI] [PubMed] [Google Scholar]
- 154. Estève PO, Chin HG, Benner J, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A. 2009;106(13):5076-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39(1):152-160. [DOI] [PubMed] [Google Scholar]
- 156. Mahesh A, Khan MIK, Govindaraju G, et al. SET7/9 interacts and methylates the ribosomal protein, eL42 and regulates protein synthesis. Biochim Biophys Acta Mol Cell Res. 2020;1867(2):118611. [DOI] [PubMed] [Google Scholar]
- 157. Kim Y, Nam HJ, Lee J, et al. Methylation-dependent regulation of HIF-1α stability restricts retinal and tumour angiogenesis. Nat Commun. 2016;7:10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Shan Z, Han Q, Nie J, et al. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J Biol Chem. 2013;288(49):35093-35103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Cho HS, Suzuki T, Dohmae N, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71(3):655-660. [DOI] [PubMed] [Google Scholar]
- 160. Yu R, Wu H, Ismail H, et al. Methylation of PLK1 by SET7/9 ensures accurate kinetochore-microtubule dynamics. J Mol Cell Biol. 2020;12(6):462-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Hong X, Huang H, Qiu X, et al. Targeting posttranslational modifications of RIOK1 inhibits the progression of colorectal and gastric cancers. Elife. 2018;7:e29511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162. Hamidi T, Singh AK, Veland N, et al. Identification of Rpl29 as a major substrate of the lysine methyltransferase Set7/9. J Biol Chem. 2018;293(33):12770-12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Vasileva E, Shuvalov O, Petukhov A, et al. KMT Set7/9 is a new regulator of Sam68 STAR-protein. Biochem Biophys Res Commun. 2020;525(4):1018-1024. [DOI] [PubMed] [Google Scholar]
- 164. Elkouris M, Kontaki H, Stavropoulos A, et al. SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 2016;15(12):2733-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Fang L, Zhang L, Wei W, et al. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell. 2014;55(4):537-551. [DOI] [PubMed] [Google Scholar]
- 166. Yang J, Huang J, Dasgupta M, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107(50):21499-21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14(2):175-182. [DOI] [PubMed] [Google Scholar]
- 168. Oudhoff MJ, Freeman SA, Couzens AL, et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev Cell. 2013;26(2):188-194. [DOI] [PubMed] [Google Scholar]
- 169. Murray K. The occurrence of Epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10-15. [DOI] [PubMed] [Google Scholar]
- 170. Lukinović V, Casanova AG, Roth GS, Chuffart F, Reynoird N. Lysine methyltransferases signaling: histones are just the tip of the iceberg. Curr Protein Pept Sci. 2020;21(7):655-674. [DOI] [PubMed] [Google Scholar]
- 171. Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19(7):815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27(20):2681-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol. 2015;6:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Griñán-Ferré C, Marsal-García L, Bellver-Sanchis A, et al. Pharmacological inhibition of G9a/GLP restores cognition and reduces oxidative stress, neuroinflammation and β-Amyloid plaques in an early-onset Alzheimer’s disease mouse model. Aging (Albany NY). 2019;11(23):11591-11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Brown MA, Sims RJ 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yi X, Jiang XJ, Fang ZM. Histone methyltransferase SMYD2: ubiquitous regulator of disease. Clin Epigenetics. 2019;11(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Weirich S, Schuhmacher MK, Kudithipudi S, Lungu C, Ferguson AD, Jeltsch A. Analysis of the substrate specificity of the SMYD2 protein lysine methyltransferase and discovery of novel non-histone substrates. Chembiochem. 2020;21(1-2):256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Sesé B, Barrero MJ, Fabregat MC, Sander V, Izpisua Belmonte JC. SMYD2 is induced during cell differentiation and participates in early development. Int J Dev Biol. 2013;57(5):357-364. [DOI] [PubMed] [Google Scholar]
- 180. Voelkel T, Andresen C, Unger A, Just S, Rottbauer W, Linke WA. Lysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function. Biochim Biophys Acta. 2013;1833(4):812-822. [DOI] [PubMed] [Google Scholar]
- 181. Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58(1):185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kurash JK, Lei H, Shen Q, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29(3):392-400. [DOI] [PubMed] [Google Scholar]
- 183. Gu Y, Wang Y, Wang X, Gao L, Yu W, Dong WF. Opposite effects of SET7/9 on apoptosis of human acute myeloid leukemia cells and lung cancer cells. J Cancer. 2017;8(11):2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Li Y, Reddy MA, Miao F, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283(39):26771-26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wood K, Tellier M, Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Kealy L, Di Pietro A, Hailes L, et al. The histone methyltransferase DOT1L is essential for humoral immune responses. Cell Rep. 2020;33(11):108504. [DOI] [PubMed] [Google Scholar]
- 187. Kwesi-Maliepaard EM, Aslam MA, Alemdehy MF, et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc Natl Acad Sci U S A. 2020;117(34):20706-20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Huang Y, Urabe G, Zhang M, et al. Nullifying epigenetic writer DOT1L attenuates neointimal hyperplasia. Atherosclerosis. 2020;308:22-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Jones B, Su H, Bhat A, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4(9):e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Yi D, Nguyen HP, Dinh J, et al. Dot1l interacts with Zc3h10 to activate Ucp1 and other thermogenic genes. Elife. 2020;9:e59990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Charles MRC, Dhayalan A, Hsieh HP, Coumar MS. Insights for the design of protein lysine methyltransferase G9a inhibitors. Future Med Chem. 2019;11(9):993-1014. [DOI] [PubMed] [Google Scholar]
- 193. Segovia C, San José-Enériz E, Munera-Maravilla E, et al. Inhibition of a G9a/DNMT network triggers immune-mediated bladder cancer regression. Nat Med. 2019;25(7):1073-1081. [DOI] [PubMed] [Google Scholar]
- 194. Zhang K, Wang J, Yang L, et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1α and APC2 gene expression in non-small cell lung cancer. Mol Cancer. 2018;17(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ferguson AD, Larsen NA, Howard T, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19(9):1262-1273. [DOI] [PubMed] [Google Scholar]
- 196. Fabini E, Manoni E, Ferroni C, Rio AD, Bartolini M. Small-molecule inhibitors of lysine methyltransferases SMYD2 and SMYD3: current trends. Future Med Chem. 2019;11(8):901-921. [DOI] [PubMed] [Google Scholar]
- 197. Kukita A, Sone K, Oda K, et al. Histone methyltransferase SMYD2 selective inhibitor LLY-507 in combination with poly ADP ribose polymerase inhibitor has therapeutic potential against high-grade serous ovarian carcinomas. Biochem Biophys Res Commun. 2019;513(2):340-346. [DOI] [PubMed] [Google Scholar]
- 198. Shang L, Wei M. Inhibition of SMYD2 sensitized cisplatin to resistant cells in NSCLC through activating p53 pathway. Front Oncol. 2019;9:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Takemoto Y, Ito A, Niwa H, et al. Identification of cyproheptadine as an inhibitor of SET domain containing lysine methyltransferase 7/9 (Set7/9) that regulates estrogen-dependent transcription. J Med Chem. 2016;59(8):3650-3660. [DOI] [PubMed] [Google Scholar]
- 200. Hirano T, Fujiwara T, Niwa H, et al. Development of novel inhibitors for histone methyltransferase SET7/9 based on cyproheptadine. Chemmedchem. 2018;13(15):1530-1540. [DOI] [PubMed] [Google Scholar]
- 201. Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49(4):e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Karytinos A, Forneris F, Profumo A, et al. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284(26):17775-17782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941-953. [DOI] [PubMed] [Google Scholar]
- 204. Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811-816. [DOI] [PubMed] [Google Scholar]
- 205. Accari SL, Fisher PR. Emerging roles of JmjC domain-containing proteins. Int Rev Cell Mol Biol. 2015;319:165-220. [DOI] [PubMed] [Google Scholar]
- 206. Lin Y, Wu Y, Li J, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Majello B, Gorini F, Sacca CD, Amente S. Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers (Basel). 2019;11(3):324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436-439. [DOI] [PubMed] [Google Scholar]
- 209. Perillo B, Ombra MN, Bertoni A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202-206. [DOI] [PubMed] [Google Scholar]
- 210. Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467-481. [DOI] [PubMed] [Google Scholar]
- 211. Black JC, Allen A, Van Rechem C, et al. Conserved antagonism between JMJD2A/KDM4A and HP1γ during cell cycle progression. Mol Cell. 2010;40(5):736-748. [DOI] [PubMed] [Google Scholar]
- 212. Johmura Y, Sun J, Kitagawa K, et al. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat Commun. 2016;7:10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Karakaidos P, Verigos J, Magklara A LSD1/KDM1A, a gate-keeper of cancer stemness and a promising therapeutic target. Cancers (Basel). 2019;11(12):1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell. 2015;28(1):57-69. [DOI] [PubMed] [Google Scholar]
- 215. Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol. 2019;12(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Maes T, Mascaró C, Rotllant D, et al. Modulation of KDM1A with vafidemstat rescues memory deficit and behavioral alterations. PLoS One. 2020;15(5):e0233468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Young LC, Hendzel MJ. The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Biochem Cell Biol. 2013;91(6):369-377. [DOI] [PubMed] [Google Scholar]
- 218. Lee DH, Kim GW, Jeon YH, Yoo J, Lee SW, Kwon SH. Advances in histone demethylase KDM4 as cancer therapeutic targets. FASEB J. 2020;34(3):3461-3484. [DOI] [PubMed] [Google Scholar]
- 219. Baldwin GS, Carnegie PR. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171(3971):579-581. [DOI] [PubMed] [Google Scholar]
- 220. Brostoff S, Eylar EH. Localization of methylated arginine in the A1 protein from myelin. Proc Natl Acad Sci U S A. 1971;68(4):765-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Kakimoto Y, Akazawa S. Isolation and identification of N-G,N-G- and N-G,N’-G-dimethyl-arginine, N-epsilon-mono-, di-, and trimethyllysine, and glucosylgalactosyl- and galactosyl-delta-hydroxylysine from human urine. J Biol Chem. 1970;245(21):5751-5758. [PubMed] [Google Scholar]
- 222. Paik WK, Kim S. Omega-N-methylarginine in protein. J Biol Chem. 1970;245(1):88-92. [PubMed] [Google Scholar]
- 223. Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271(25):15034-15044. [DOI] [PubMed] [Google Scholar]
- 224. Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120(Pt 24):4243-4246. [DOI] [PubMed] [Google Scholar]
- 225. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113(1):50-87. [DOI] [PubMed] [Google Scholar]
- 227. Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J Biol Chem. 2012;287(11):7859-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8-24. [DOI] [PubMed] [Google Scholar]
- 229. Yadav N, Lee J, Kim J, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100(11):6464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012;40(19):9513-9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Baldwin RM, Morettin A, Côté J. Role of PRMTs in cancer: could minor isoforms be leaving a mark? World J Biol Chem. 2014;5(2):115-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Peng C, Wong CC. The story of protein arginine methylation: characterization, regulation, and function. Expert Rev Proteomics. 2017;14(2):157-170. [DOI] [PubMed] [Google Scholar]
- 233. Guendel I, Carpio L, Pedati C, et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS One. 2010;5(6):e11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Liu LM, Sun WZ, Fan XZ, Xu YL, Cheng MB, Zhang Y. Methylation of C/EBPα by PRMT1 inhibits its tumor-suppressive function in breast cancer. Cancer Res. 2019;79(11):2865-2877. [DOI] [PubMed] [Google Scholar]
- 235. Tikhanovich I, Zhao J, Bridges B, Kumer S, Roberts B, Weinman SA. Arginine methylation regulates c-Myc-dependent transcription by altering promoter recruitment of the acetyltransferase p300. J Biol Chem. 2017;292(32):13333-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Li Z, Wang D, Lu J, et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020;27(12):3226-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Yamagata K, Daitoku H, Takahashi Y, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32(2):221-231. [DOI] [PubMed] [Google Scholar]
- 238. Kagoya Y, Saijo H, Matsunaga Y, et al. Arginine methylation of FOXP3 is crucial for the suppressive function of regulatory T cells. J Autoimmun. 2019;97:10-21. [DOI] [PubMed] [Google Scholar]
- 239. Wang Y, Sun C, Jiang J, et al. GLI1 expression is an important prognostic factor that contributes to the poor prognosis of rhabdomyosarcoma. Histol Histopathol. 2016;31(3):329-337. [DOI] [PubMed] [Google Scholar]
- 240. Liu Q, Zhang XL, Cheng MB, Zhang Y. PRMT1 activates myogenin transcription via MyoD arginine methylation at R121. Biochim Biophys Acta Gene Regul Mech. 2019;1862(10):194442. [DOI] [PubMed] [Google Scholar]
- 241. Liu X, Li H, Liu L, et al. Methylation of arginine by PRMT1 regulates Nrf2 transcriptional activity during the antioxidative response. Biochim Biophys Acta. 2016;1863(8):2093-2103. [DOI] [PubMed] [Google Scholar]
- 242. Davies CC, Chakraborty A, Diefenbacher ME, Skehel M, Behrens A. Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation. EMBO J. 2013;32(11):1556-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Reintjes A, Fuchs JE, Kremser L, et al. Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFα/NF-κB response. Proc Natl Acad Sci U S A. 2016;113(16):4326-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Mostaqul Huq MD, Gupta P, Tsai NP, White R, Parker MG, Wei LN. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25(21):5094-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Mowen KA, Tang J, Zhu W, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104(5):731-741. [DOI] [PubMed] [Google Scholar]
- 247. Jobert L, Argentini M, Tora L. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp Cell Res. 2009;315(7):1273-1286. [DOI] [PubMed] [Google Scholar]
- 248. Du K, Arai S, Kawamura T, Matsushita A, Kurokawa R. TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem Biophys Res Commun. 2011;404(4):991-996. [DOI] [PubMed] [Google Scholar]
- 249. Huang L, Wang Z, Narayanan N, Yang Y. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 2018;46(6):3061-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, et al. PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J Biol Chem. 2015;290(21):13479-13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Boisvert FM, Déry U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19(6):671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Zhang Y, Zhang Q, Li L, et al. Arginine methylation of APE1 promotes its mitochondrial translocation to protect cells from oxidative damage. Free Radic Biol Med. 2020;158:60-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. El-Andaloussi N, Valovka T, Toueille M, et al. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007;21(1):26-34. [DOI] [PubMed] [Google Scholar]
- 254. Zheng S, Moehlenbrink J, Lu YC, et al. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol Cell. 2013;52(1):37-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Yang JH, Chiou YY, Fu SL, et al. Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation. Nucleic Acids Res. 2014;42(15):9908-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Gurunathan G, Yu Z, Coulombe Y, Masson JY, Richard S. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci Rep. 2015;5:10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Cho JH, Lee MK, Yoon KW, Lee J, Cho SG, Choi EJ. Arginine methylation-dependent regulation of ASK1 signaling by PRMT1. Cell Death Differ. 2012;19(5):859-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Cha B, Kim W, Kim YK, et al. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 2011;30(20):2379-2389. [DOI] [PubMed] [Google Scholar]
- 259. Pyun JH, Kim HJ, Jeong MH, et al. Cardiac specific PRMT1 ablation causes heart failure through CaMKII dysregulation. Nat Commun. 2018;9(1):5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Liao HW, Hsu JM, Xia W, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125(12):4529-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Liu MY, Hua WK, Chen CJ, Lin WJ. The MKK-dependent phosphorylation of p38alpha is augmented by arginine methylation on Arg49/Arg149 during erythroid differentiation. Int J Mol Sci. 2020;21(10):3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Albrecht LV, Ploper D, Tejeda-Muñoz N, De Robertis EM. Arginine methylation is required for canonical Wnt signaling and endolysosomal trafficking. Proc Natl Acad Sci U S A. 2018;115(23):E5317-E5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Xu J, Wang AH, Oses-Prieto J, et al. Arginine methylation initiates BMP-induced smad signaling. Mol Cell. 2013;51(1):5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Zhang T, Wu J, Ungvijanpunya N, et al. Smad6 methylation represses NFκB activation and periodontal inflammation. J Dent Res. 2018;97(7):810-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Katsuno Y, Qin J, Oses-Prieto J, et al. Arginine methylation of SMAD7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J Biol Chem. 2018;293(34):13059-13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Gen S, Matsumoto Y, Kobayashi KI, Suzuki T, Inoue J, Yamamoto Y. Stability of tuberous sclerosis complex 2 is controlled by methylation at R1457 and R1459. Sci Rep. 2020;10(1):21160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci U S A. 2011;108(15):6085-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Dolezal E, Infantino S, Drepper F, et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat Immunol. 2017;18(8):911-920. [DOI] [PubMed] [Google Scholar]
- 269. Wei HM, Hu HH, Chang GY, et al. Arginine methylation of the cellular nucleic acid binding protein does not affect its subcellular localization but impedes RNA binding. FEBS Lett. 2014;588(9):1542-1548. [DOI] [PubMed] [Google Scholar]
- 270. Onwuli DO, Samuel SF, Sfyri P, et al. The inhibitory subunit of cardiac troponin (cTnI) is modified by arginine methylation in the human heart. Int J Cardiol. 2019;282:76-80. [DOI] [PubMed] [Google Scholar]
- 271. Hsu JH, Hubbell-Engler B, Adelmant G, et al. PRMT1-mediated translation regulation is a crucial vulnerability of cancer. Cancer Res. 2017;77(17):4613-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem. 2016;291(43):22671-22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Wall ML, Lewis SM. Methylarginines within the RGG-Motif region of hnRNP A1 affect its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J Mol Biol. 2017;429(2):295-307. [DOI] [PubMed] [Google Scholar]
- 274. Wang L, Jia Z, Xie D, et al. Methylation of HSP70 orchestrates its binding to and stabilization of BCL2 mRNA and renders pancreatic cancer cells resistant to therapeutics. Cancer Res. 2020;80(20):4500-4513. [DOI] [PubMed] [Google Scholar]
- 275. Deng X, Von Keudell G, Suzuki T, et al. PRMT1 promotes mitosis of cancer cells through arginine methylation of INCENP. Oncotarget. 2015;6(34):35173-35182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Kim HJ, Jeong MH, Kim KR, et al. Protein arginine methylation facilitates KCNQ channel-PIP2 interaction leading to seizure suppression. Elife. 2016;5:e17159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Eberhardt A, Hansen JN, Koster J, et al. Protein arginine methyltransferase 1 is a novel regulator of MYCN in neuroblastoma. Oncotarget. 2016;7(39):63629-63639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Zhang L, Tran NT, Su H, et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. 2015;4:e07938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Shin HS, Jang CY, Kim HD, Kim TS, Kim S, Kim J. Arginine methylation of ribosomal protein S3 affects ribosome assembly. Biochem Biophys Res Commun. 2009;385(2):273-278. [DOI] [PubMed] [Google Scholar]
- 280. Matsumura T, Nakamura-Ishizu A, Muddineni SSNA, et al. Hematopoietic stem cells acquire survival advantage by loss of RUNX1 methylation identified in familial leukemia. Blood. 2020;136(17):1919-1932. [DOI] [PubMed] [Google Scholar]
- 281. Tikhanovich I, Kuravi S, Artigues A, et al. Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates toll-like receptor signaling. J Biol Chem. 2015;290(36):22236-22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Wang L, Zhao Z, Meyer MB, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25(1):21-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29(6):1105-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Kuhn P, Chumanov R, Wang Y, Ge Y, Burgess RR, Xu W. Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Res. 2011;39(7):2717-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Gao WW, Xiao RQ, Peng BL, et al. Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation. Proc Natl Acad Sci U S A. 2015;112(26):E3327-E3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Liu J, Feng J, Li L, et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 2020;21(2):e48597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11(3):333-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Yu YS, Shin HR, Kim D, et al. Pontin arginine methylation by CARM1 is crucial for epigenetic regulation of autophagy. Nat Commun. 2020;11(1):6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Nie M, Wang Y, Guo C, et al. CARM1-mediated methylation of protein arginine methyltransferase 5 represses human γ-globin gene expression in erythroleukemia cells. J Biol Chem. 2018;293(45):17454-17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Sims RJ 3rd, Rojas LA, Beck DB, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Vu LP, Perna F, Wang L, et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 2013;5(6):1625-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Zhao HY, Zhang YJ, Dai H, Zhang Y, Shen YF. CARM1 mediates modulation of Sox2. PLoS One. 2011;6(10):e27026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Lee YH, Bedford MT, Stallcup MR. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 2011;25(2):176-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Zhong XY, Yuan XM, Xu YY, et al. CARM1 methylates GAPDH to regulate glucose metabolism and is suppressed in liver cancer. Cell Rep. 2018;24(12):3207-3223. [DOI] [PubMed] [Google Scholar]
- 295. Fujiwara T, Mori Y, Chu DL, et al. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol. 2006;26(6):2273-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Li H, Park S, Kilburn B, et al. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277(47):44623-44630. [DOI] [PubMed] [Google Scholar]
- 297. Wang YP, Zhou W, Wang J, et al. Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol Cell. 2016;64(4):673-687. [DOI] [PubMed] [Google Scholar]
- 298. Abeywardana T, Oh M, Jiang L, et al. CARM1 suppresses de novo serine synthesis by promoting PKM2 activity. J Biol Chem. 2018;293(39):15290-15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Guo J, Zhang Q, Su Y, et al. Arginine methylation of ribose-5-phosphate isomerase A senses glucose to promote human colorectal cancer cell survival. Sci China Life Sci. 2020;63(9):1394-1405. [DOI] [PubMed] [Google Scholar]
- 300. Kumar A, Zhong Y, Albrecht A, et al. Actin R256 mono-methylation is a conserved post-translational modification involved in transcription. Cell Rep. 2020;32(13):108172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Lu X, Fernando TM, Lossos C, et al. PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood. 2018;132(19):2026-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Cho EC, Zheng S, Munro S, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31(7):1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Chen M, Yi B, Sun J. Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J Biol Chem. 2014;289(35):24325-24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Bandyopadhyay S, Harris DP, Adams GN, et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Wei H, Wang B, Miyagi M, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc Natl Acad Sci U S A. 2013;110(33):13516-13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Harris DP, Bandyopadhyay S, Maxwell TJ, Willard B, DiCorleto PE. Tumor necrosis factor (TNF)-α induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-κB p65 methylation. J Biol Chem. 2014;289(22):15328-15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Harris DP, Chandrasekharan UM, Bandyopadhyay S, Willard B, DiCorleto PE. PRMT5-mediated methylation of NF-κB p65 at Arg174 is required for endothelial CXCL11 gene induction in response to TNF-α and IFN-γ costimulation. PLoS One. 2016;11(2):e0148905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Zhao DY, Gish G, Braunschweig U, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529(7584):48-53. [DOI] [PubMed] [Google Scholar]
- 309. Kanamaluru D, Xiao Z, Fang S, et al. Arginine methylation by PRMT5 at a naturally occurring mutation site is critical for liver metabolic regulation by small heterodimer partner. Mol Cell Biol. 2011;31(7):1540-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Jia Z, Yue F, Chen X, et al. Protein arginine methyltransferase PRMT5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues. Adv Sci (Weinh). 2020;7(23):2002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Liu L, Zhao X, Zhao L, et al. Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 2016;76(5):1260-1272. [DOI] [PubMed] [Google Scholar]
- 312. Hwang JW, Kim SN, Myung N, et al. PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation. Commun Biol. 2020;3(1):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Guo Z, Zheng L, Xu H, et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol. 2010;6(10):766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Jansson M, Durant ST, Cho EC, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431-1439. [DOI] [PubMed] [Google Scholar]
- 315. He W, Ma X, Yang X, Zhao Y, Qiu J, Hang H. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 2011;39(11):4719-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Clarke TL, Sanchez-Bailon MP, Chiang K, et al. PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol Cell. 2017;65(5):900-916.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Rehman I, Basu SM, Das SK, et al. PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res. 2018;46(11):5601-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Chen M, Qu X, Zhang Z, et al. Cross-talk between Arg methylation and Ser phosphorylation modulates apoptosis signal-regulating kinase 1 activation in endothelial cells. Mol Biol Cell. 2016;27(8):1358-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Andreu-Pérez P, Esteve-Puig R, de Torre-Minguela C, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011;4(190):ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Yang CY, Chiu LL, Chang CC, Chuang HC, Tan TH. Induction of DUSP14 ubiquitination by PRMT5-mediated arginine methylation. FASEB J. 2018;fj201800244RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Hartley AV, Wang B, Mundade R, et al. PRMT5-mediated methylation of YBX1 regulates NF-κB activity in colorectal cancer. Sci Rep. 2020;10(1):15934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Abe Y, Suzuki Y, Kawamura K, Tanaka N. MEP50/PRMT5-mediated methylation activates GLI1 in Hedgehog signalling through inhibition of ubiquitination by the ITCH/NUMB complex. Commun Biol. 2019;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Zhou Z, Sun X, Zou Z, et al. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010;20(9):1023-1033. [DOI] [PubMed] [Google Scholar]
- 324. Gao G, Dhar S, Bedford MT. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 2017;45(8):4359-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Barrera A, Ramos H, Vera-Otarola J, et al. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020;48(18):10479-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Ichikawa T, Shanab O, Nakahata S, et al. Novel PRMT5-mediated arginine methylations of HSP90A are essential for maintenance of HSP90A function in NDRG2low ATL and various cancer cells. Biochim Biophys Acta Mol Cell Res. 2020;1867(2):118615. [DOI] [PubMed] [Google Scholar]
- 327. Hu D, Gur M, Zhou Z, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Liu N, Yang R, Shi Y, et al. The cross-talk between methylation and phosphorylation in lymphoid-specific helicase drives cancer stem-like properties. Signal Transduct Target Ther. 2020;5(1):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011;71(16):5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem. 2010;285(17):12695-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Rengasamy M, Zhang F, Vashisht A, et al. The PRMT5/WDR77 complex regulates alternative splicing through ZNF326 in breast cancer. Nucleic Acids Res. 2017;45(19):11106-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Choi S, Jeong HJ, Kim H, et al. Skeletal muscle-specific Prmt1 deletion causes muscle atrophy via deregulation of the PRMT6-FOXO3 axis. Autophagy. 2019;15(6):1069-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Xie B, Invernizzi CF, Richard S, Wainberg MA. Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J Virol. 2007;81(8):4226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Invernizzi CF, Xie B, Frankel FA, et al. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. Aids. 2007;21(7):795-805. [DOI] [PubMed] [Google Scholar]
- 335. Stavride P, Arampatzi P, Papamatheakis J. Differential regulation of MHCII genes by PRMT6, via an AT-hook motif of RFX5. Mol Immunol. 2013;56(4):390-398. [DOI] [PubMed] [Google Scholar]
- 336. El-Andaloussi N, Valovka T, Toueille M, et al. Arginine methylation regulates DNA polymerase beta. Mol Cell. 2006;22(1):51-62. [DOI] [PubMed] [Google Scholar]
- 337. Chan LH, Zhou L, Ng KY, et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep. 2018;25(3):690-701.e8. [DOI] [PubMed] [Google Scholar]
- 338. Feng J, Dang Y, Zhang W, et al. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc Natl Acad Sci U S A. 2019;116(14):6868-6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Che N, Ng KY, Wong TL, et al. PRMT6 deficiency induces autophagy in hostile microenvironments of hepatocellular carcinoma tumors by regulating BAG5-associated HSC70 stability. Cancer Lett. 2021;501:247-262. [DOI] [PubMed] [Google Scholar]
- 340. Huang J, Cardamone MD, Johnson HE, et al. Exchange factor TBL1 and arginine methyltransferase PRMT6 cooperate in protecting G protein pathway suppressor 2 (GPS2) from proteasomal degradation. J Biol Chem. 2015;290(31):19044-19054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Invernizzi CF, Xie B, Richard S, Wainberg MA. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology. 2006;3:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Wang X, Huang Y, Zhao J, Zhang Y, Lu J, Huang B. Suppression of PRMT6-mediated arginine methylation of p16 protein potentiates its ability to arrest A549 cell proliferation. Int J Biochem Cell Biol. 2012;44(12):2333-2341. [DOI] [PubMed] [Google Scholar]
- 343. Singhroy DN, Mesplède T, Sabbah A, Quashie PK, Falgueyret JP, Wainberg MA. Automethylation of protein arginine methyltransferase 6 (PRMT6) regulates its stability and its anti-HIV-1 activity. Retrovirology. 2013;10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Huang T, Yang Y, Song X, et al. PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol Cell. 2021;81(6):1276-1291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Yan WW, Liang YL, Zhang QX, et al. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018;19(12):e46377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Najbauer J, Johnson BA, Young AL, Aswad DW. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J Biol Chem. 1993;268(14):10501-10509. [PubMed] [Google Scholar]
- 347. Suresh S, Huard S, Dubois T. CARM1/PRMT4: making its mark beyond its function as a transcriptional coactivator. Trends Cell Biol. 2021;31(5):402-417. [DOI] [PubMed] [Google Scholar]
- 348. Stallcup MR, Chen D, Koh SS, et al. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem Soc Trans. 2000;28(4):415-418. [PubMed] [Google Scholar]
- 349. Guccione E, Bassi C, Casadio F, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449(7164):933-937. [DOI] [PubMed] [Google Scholar]
- 350. Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20(10):642-657. [DOI] [PubMed] [Google Scholar]
- 351. Cheng D, Gao G, Di Lorenzo A, et al. Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6. J Biol Chem. 2020;295(50):17060-17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Baldwin RM, Morettin A, Paris G, Goulet I, Côté J. Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells. Cell Cycle. 2012;11(24):4597-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Hamamoto R, Nakamura Y. Dysregulation of protein methyltransferases in human cancer: An emerging target class for anticancer therapy. Cancer Sci. 2016;107(4):377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Xiao W, Chen X, Liu L, Shu Y, Zhang M, Zhong Y. Role of protein arginine methyltransferase 5 in human cancers. Biomed Pharmacother. 2019;114:108790. [DOI] [PubMed] [Google Scholar]
- 355. Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37-50. [DOI] [PubMed] [Google Scholar]
- 356. Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279(23):23892-23899. [DOI] [PubMed] [Google Scholar]
- 357. Kaniskan HÜ, Jin J. Recent progress in developing selective inhibitors of protein methyltransferases. Curr Opin Chem Biol. 2017;39:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Nakayama K, Szewczyk MM, Dela Sena C, et al. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9(26):18480-18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Drew AE, Moradei O, Jacques SL, et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7(1):17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Mitchell LH, Drew AE, Ribich SA, et al. Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med Chem Lett. 2015;6(6):655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Fedoriw A, Rajapurkar SR, O’Brien S, et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell. 2019;36(1):100-114.e25. [DOI] [PubMed] [Google Scholar]
- 362. AbuHammad S, Cullinane C, Martin C, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci U S A. 2019;116(36):17990-18000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Zhang J, Jing L, Li M, He L, Guo Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review). Mol Med Rep. 2019;19(5):3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Webby CJ, Wolf A, Gromak N, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90-93. [DOI] [PubMed] [Google Scholar]
- 365. Unoki M, Masuda A, Dohmae N, et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J Biol Chem. 2013;288(9):6053-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Poulard C, Rambaud J, Hussein N, Corbo L, Le Romancer M. JMJD6 regulates ERα methylation on arginine. PLoS One. 2014;9(2):e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Lawrence P, Conderino JS, Rieder E. Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: role of Jumonji C-domain containing protein 6 in RHA demethylation. Virology. 2014;452-453:1-11. [DOI] [PubMed] [Google Scholar]
- 368. Wu TF, Yao YL, Lai IL, Lai CC, Lin PL, Yang WM. Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with Waardenburg syndrome. J Biol Chem. 2015;290(33):20556-20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Tsai WC, Reineke LC, Jain A, Jung SY, Lloyd RE. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J Biol Chem. 2017;292(46):18886-18896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Yang J, Chen S, Yang Y, et al. Jumonji domain-containing protein 6 protein and its role in cancer. Cell Prolif. 2020;53(2):e12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. Zheng H, Tie Y, Fang Z, et al. Jumonji domain-containing 6 (JMJD6) identified as a potential therapeutic target in ovarian cancer. Signal Transduct Target Ther. 2019;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Walport LJ, Hopkinson RJ, Chowdhury R, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun. 2016;7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Li S, Ali S, Duan X, et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 2018;23(2):389-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279-283. [DOI] [PubMed] [Google Scholar]
- 375. Subramanian K, Jia D, Kapoor-Vazirani P, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30(3):336-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Zhang X, Tanaka K, Yan J, et al. Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci U S A. 2013;110(43):17284-17289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Zhang X, Peng D, Xi Y, et al. G9a-mediated methylation of ERα links the PHF20/MOF histone acetyltransferase complex to hormonal gene expression. Nat Commun. 2016;7:10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Chung HH, Sze SK, Woo AR, et al. Lysine methylation of progesterone receptor at activation function 1 regulates both ligand-independent activity and ligand sensitivity of the receptor. J Biol Chem. 2014;289(9):5704-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Woo ARE, Sze SK, Chung HH, Lin VC. Delineation of critical amino acids in activation function 1 of progesterone receptor for recruitment of transcription coregulators. Biochim Biophys Acta Gene Regul Mech. 2019;1862(4):522-533. [DOI] [PubMed] [Google Scholar]
- 380. Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33(1):13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol. 2011;25(3):433-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Le Romancer M, Treilleux I, Leconte N, et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31(2):212-221. [DOI] [PubMed] [Google Scholar]
- 383. Poulard C, Treilleux I, Lavergne E, et al. Activation of rapid oestrogen signalling in aggressive human breast cancers. EMBO Mol Med. 2012;4(11):1200-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Choucair A, Pham TH, Omarjee S, et al. The arginine methyltransferase PRMT1 regulates IGF-1 signaling in breast cancer. Oncogene. 2019;38(21):4015-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Adlanmerini M, Fébrissy C, Zahreddine R, et al. Mutation of Arginine 264 on ERα (Estrogen Receptor Alpha) selectively abrogates the rapid signaling of estradiol in the endothelium without altering fertility. Arterioscler Thromb Vasc Biol. 2020;40(9):2143-2158. [DOI] [PubMed] [Google Scholar]
- 386. Malbeteau L, Poulard C, Languilaire C, et al. PRMT1 is critical for the transcriptional activity and the stability of the progesterone receptor. Iscience. 2020;23(6):101236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Mounir Z, Korn JM, Westerling T, et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the Androgen Receptor. Elife. 2016;5:e13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Scaramuzzino C, Casci I, Parodi S, et al. Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron. 2015;85(1):88-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Poulard C, Jacquemetton J, Pham TH, Le Romancer M. Using proximity ligation assay to detect protein arginine methylation. Methods. 2020;175:66-71. [DOI] [PubMed] [Google Scholar]
- 390. Métivier R, Penot G, Hübner MR, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751-763. [DOI] [PubMed] [Google Scholar]
- 391. Reid G, Hübner MR, Métivier R, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11(3):695-707. [DOI] [PubMed] [Google Scholar]
- 392. Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64(24):9199-9208. [DOI] [PubMed] [Google Scholar]
- 393. Fuqua SA, Wiltschke C, Zhang QX, et al. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60(15):4026-4029. [PubMed] [Google Scholar]
- 394. Abu-Farha M, Lanouette S, Elisma F, et al. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3(5):301-308. [DOI] [PubMed] [Google Scholar]
- 395. Huang J, Sengupta R, Espejo AB, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105-108. [DOI] [PubMed] [Google Scholar]
- 396. Obermann WMJ. A motif in HSP90 and P23 that links molecular chaperones to efficient estrogen receptor α methylation by the lysine methyltransferase SMYD2. J Biol Chem. 2018;293(42):16479-16487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Poulard C, Rambaud J, Lavergne E, et al. Role of JMJD6 in breast tumourigenesis. PLoS One. 2015;10(5):e0126181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Poulard C, Jacquemetton J, Tredan O, et al. Oestrogen non-genomic signalling is activated in tamoxifen-resistant breast cancer. Int J Mol Sci. 2019;20(11):2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Gustafsson KL, Farman HH, Nilsson KH, et al. Arginine site 264 in murine estrogen receptor-α is dispensable for the regulation of the skeleton. Am J Physiol Endocrinol Metab. 2021;320(1):E160-E168. [DOI] [PubMed] [Google Scholar]
- 400. Abdel-Hafiz HA, Horwitz KB. Post-translational modifications of the progesterone receptors. J Steroid Biochem Mol Biol. 2014;140:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Meyer ME, Quirin-Stricker C, Lerouge T, Bocquel MT, Gronemeyer H. A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J Biol Chem. 1992;267(15):10882-10887. [PubMed] [Google Scholar]
- 402. Pierson-Mullany LK, Lange CA. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol Cell Biol. 2004;24(24):10542-10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65-131. [DOI] [PubMed] [Google Scholar]
- 404. Dennis AP, Lonard DM, Nawaz Z, O’Malley BW. Inhibition of the 26S proteasome blocks progesterone receptor-dependent transcription through failed recruitment of RNA polymerase II. J Steroid Biochem Mol Biol. 2005;94(4):337-346. [DOI] [PubMed] [Google Scholar]
- 405. Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97(3):1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Zhang PJ, Zhao J, Li HY, et al. CUE domain containing 2 regulates degradation of progesterone receptor by ubiquitin-proteasome. EMBO J. 2007;26(7):1831-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Gaughan L, Stockley J, Wang N, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011;39(4):1266-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Fu M, Wang C, Reutens AT, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853-20860. [DOI] [PubMed] [Google Scholar]
- 409. Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J Biol Chem. 2002;277(29):25904-25913. [DOI] [PubMed] [Google Scholar]
- 410. Vatapalli R, Sagar V, Rodriguez Y, et al. Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer. Nat Commun. 2020;11(1):4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Prensner JR, Sahu A, Iyer MK, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5(6):1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Sun Y, Chung HH, Woo AR, Lin VC. Protein arginine methyltransferase 6 enhances ligand-dependent and -independent activity of estrogen receptor α via distinct mechanisms. Biochim Biophys Acta. 2014;1843(9):2067-2078. [DOI] [PubMed] [Google Scholar]
- 413. Almeida-Rios D, Graça I, Vieira FQ, et al. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget. 2016;7(33):53018-53028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Vieira FQ, Costa-Pinheiro P, Ramalho-Carvalho J, et al. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr Relat Cancer. 2014;21(1):51-61. [DOI] [PubMed] [Google Scholar]
- 415. Luo M, Li Y, Guo H, et al. Protein arginine methyltransferase 6 involved in germ cell viability during spermatogenesis and down-regulated by the androgen receptor. Int J Mol Sci. 2015;16(12):29467-29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Poulard C, Bittencourt D, Wu DY, Hu Y, Gerke DS, Stallcup MR. A post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP. EMBO Rep. 2017;18(8):1442-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Poulard C, Kim HN, Fang M, et al. Relapse-associated AURKB blunts the glucocorticoid sensitivity of B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2019;116(8):3052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Poulard C, Baulu E, Lee BH, Pufall MA, Stallcup MR. Increasing G9a automethylation sensitizes B acute lymphoblastic leukemia cells to glucocorticoid-induced death. Cell Death Dis. 2018;9(10):1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20(4):457-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857-864. [DOI] [PubMed] [Google Scholar]
- 421. Cai C, He HH, Gao S, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9(5):1618-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Garcia-Bassets I, Kwon YS, Telese F, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128(3):505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423. Wang J, Scully K, Zhu X, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446(7138):882-887. [DOI] [PubMed] [Google Scholar]
- 424. Wissmann M, Yin N, Müller JM, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9(3):347-353. [DOI] [PubMed] [Google Scholar]
- 425. Lee KH, Kim BC, Jeong SH, et al. Histone demethylase LSD1 regulates kidney cancer progression by modulating androgen receptor activity. Int J Mol Sci. 2020;21(17):6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Gao S, Chen S, Han D, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet. 2020;52(10):1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Regufe da Mota S, Bailey S, Strivens RA, et al. LSD1 inhibition attenuates androgen receptor V7 splice variant activation in castration resistant prostate cancer models. Cancer Cell Int. 2018;18:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Wang M, Liu X, Chen Z, Zhang L, Weng X. Downregulation of lysine-specific demethylase 1 enhances the sensitivity of hormone-sensitive prostate cancer cells to androgen deprivation therapy. Oncol Lett. 2021;21(2):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Metzger E, Imhof A, Patel D, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464(7289):792-796. [DOI] [PubMed] [Google Scholar]
- 430. Chen D, Ma H, Hong H, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174-2177. [DOI] [PubMed] [Google Scholar]
- 431. Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3(1):39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Ma H, Baumann CT, Li H, et al. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol. 2001;11(24):1981-1985. [DOI] [PubMed] [Google Scholar]
- 433. Schurter BT, Koh SS, Chen D, et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40(19):5747-5756. [DOI] [PubMed] [Google Scholar]
- 434. Strahl BD, Briggs SD, Brame CJ, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11(12):996-1000. [DOI] [PubMed] [Google Scholar]
- 435. Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12(24):2090-2097. [DOI] [PubMed] [Google Scholar]
- 436. Beketova E, Fang S, Owens JL, et al. Protein arginine methyltransferase 5 promotes pICln-dependent androgen receptor transcription in castration-resistant prostate cancer. Cancer Res. 2020;80(22):4904-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Majumder S, Liu Y, Ford OH 3rd, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66(12):1292-1301. [DOI] [PubMed] [Google Scholar]
- 438. Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21(20):5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102(10):3611-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Ceschin DG, Walia M, Wenk SS, et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25(11):1132-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26(21):7846-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Naeem H, Cheng D, Zhao Q, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27(1):120-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108(4):465-474. [DOI] [PubMed] [Google Scholar]
- 444. Wang L, Zeng H, Wang Q, et al. MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci Adv. 2015;1(9):e1500463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Peng BL, Li WJ, Ding JC, et al. A hypermethylation strategy utilized by enhancer-bound CARM1 to promote estrogen receptor α-dependent transcriptional activation and breast carcinogenesis. Theranostics. 2020;10(8):3451-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446. Gao WW, Xiao RQ, Zhang WJ, et al. JMJD6 licenses ERα-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 co-activator complex. Mol Cell. 2018;70(2):340-357.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19(12):1466-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Barrero MJ, Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol Cell. 2006;24(2):233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Huq MD, Ha SG, Wei LN. Modulation of retinoic acid receptor alpha activity by lysine methylation in the DNA binding domain. J Proteome Res. 2008;7(10):4538-4545. [DOI] [PubMed] [Google Scholar]
- 450. Matsuda H, Paul BD, Choi CY, Hasebe T, Shi YB. Novel functions of protein arginine methyltransferase 1 in thyroid hormone receptor-mediated transcription and in the regulation of metamorphic rate in Xenopus laevis. Mol Cell Biol. 2009;29(3):745-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451. Baudrand R, Pojoga LH, Romero JR, Williams GH. Aldosterone’s mechanism of action: roles of lysine-specific demethylase 1, caveolin and striatin. Curr Opin Nephrol Hypertens. 2014;23(1):32-37. [DOI] [PubMed] [Google Scholar]
- 452. Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277(32):28624-28630. [DOI] [PubMed] [Google Scholar]
- 453. Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107(1-2):1-14. [DOI] [PubMed] [Google Scholar]