Abstract
Deciphering microbiota functions is crucial to predict ecosystem sustainability in response to global change. High-throughput sequencing at the individual or community level has revolutionized our understanding of microbial ecology, leading to the big data era and improving our ability to link microbial diversity with microbial functions. Recent advances in bioinformatics have been key for developing functional prediction tools based on DNA metabarcoding data and using taxonomic gene information. This cheaper approach in every aspect serves as an alternative to shotgun sequencing. Although these tools are increasingly used by ecologists, an objective evaluation of their modularity, portability, and robustness is lacking. Here, we reviewed 100 scientific papers on functional inference and ecological trait assignment to rank the advantages, specificities, and drawbacks of these tools, using a scientific benchmarking. To date, inference tools have been mainly devoted to bacterial functions, and ecological trait assignment tools, to fungal functions. A major limitation is the lack of reference genomes—compared with the human microbiota—especially for complex ecosystems such as soils. Finally, we explore applied research prospects. These tools are promising and already provide relevant information on ecosystem functioning, but standardized indicators and corresponding repositories are still lacking that would enable them to be used for operational diagnosis.
Keywords: microbiota, metabarcoding, taxonomy, functional inference, ecological traits, soil
Background
Microorganisms are present in all habitats on Earth and are essential for animals, plants, and therefore for the sustainability of human activities [1]. The extraordinary diversity of microbial communities plays an essential role in the various biogeochemical cycles, allows aquatic and terrestrial ecosystems to function properly, and ensures their ability to provide ecological services (e.g., soil structuring, organic matter renewal, nutrient recycling, pollution control, regulation/barrier to pathogens, or even plant productivity) [2–4]. Their fabulous capacity to adapt to different environmental stresses over time is now well known, and the regulation process of their diversity is better and better deciphered. Despite these tremendous improvements in the approaches targeting indigenous microbiotas, our understanding of the link between microbes and their associated functions remains limited [5]. A workshop hosted by the British Ecological Society's Microbial Ecology Special Interest Group (June 2016) recently identified 50 important research questions in microbial ecology. One of the main ones was “What methods can we use to marry microbial diversity with function; how do we link transcriptomics, proteomics and metabolomics?” [6]. This sums up the future challenges facing the scientific community when it comes to improving our understanding of the regulation of the microbiome diversity and functions [7].
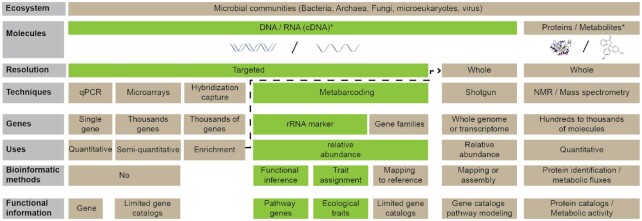
Microbial functions can be characterized from genomic, proteomic, or metabolic data (Fig. 1) [8–10]. Considering genomics, quantitative PCR (qPCR) and microarrays were the first technologies used to describe functional genes or taxa from complex environmental samples [11]. Initially designed to determine the absolute copy number of a single given gene, the latest technical advances can analyze thousands of combinations of samples and targets in parallel [12]. Standardized methods even make it possible to quantify genes of interest (e.g., involved in biogeochemical cycles or pesticide degradation) to estimate soil quality [13]. DNA microarrays were the first high-throughput technologies giving access to gene expression profiles at the individual or community levels [11, 14]. There exist different kinds of microarrays (e.g., PhyloChip, GeoChip, PathoChip, StressChip, CAZyChip). They provide a snapshot of microbial diversity (bacteria, fungi, viruses) and/or of the functional genes present in a given sample (e.g., genes coding for enzymes involved in polysaccharide degradation) [15–18]. Some of these microarrays have become diagnostic tools in many fields, in particular for targeting viruses, bacterial or fungal pathogens, or harmful organisms [19]. More recent and cheaper, various high-throughput sequencing (HTS) alternatives have been developed to explore microbial communities (Fig. 1) [20]. Genome and metagenome sequencing have changed the microbial ecology field: thanks to genome sequencing and meta-omics approaches, gene catalogs can be assessed, and new microorganisms can be discovered [21, 22].
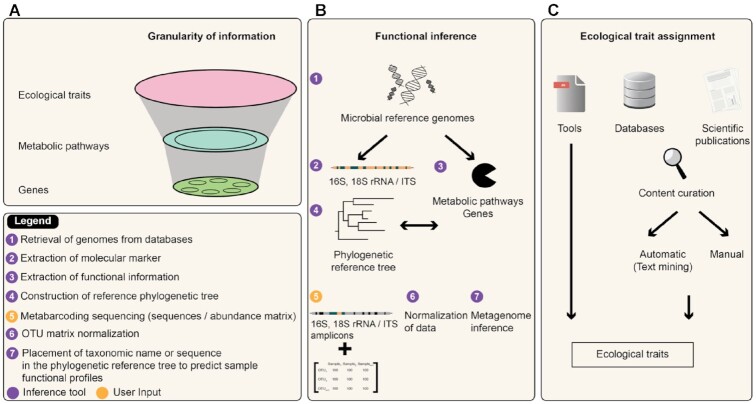
Figure 1:
Schematic diagram of the various strategies available for exploring the functional diversity of the microbiota. Green frames indicate metabarcoding approaches for retrieving putative functions from taxonomic genes by functional inference and ecological trait assignment. cDNA: complementary DNA; NMR: nuclear magnetic resonance; rRNA: ribosomal RNA.
For example, by implementing a metabarcoding approach, microbial ecologists were initially enthusiastic about such huge taxonomic information but quickly pointed out the lack of associated functional information [22]. Taxonomic profiles can indeed change to varying degrees among samples, and predicting to what extent these changes affect the overall functional capacity of the community has remained a technical and scientific challenge to date [6, 23, 24]. Metabarcoding may well be used to directly target functional genes and classify them by taxonomic group, but applications remain limited to a few families [25–29]. In the face of these limitations, 2 solutions have emerged to indirectly obtain functional information from taxonomic profiles, i.e., (i) functional inference, and (ii) ecological trait assignment, using (meta)genome and microbiome big data (Fig. 1). Functional inference predicts the putative functions (e.g., gene catalogs, metabolic pathways) of microbial communities, while ecological trait assignment directly retrieves a trait common to all taxa by linking taxonomic names with a dedicated database. The major difference between these 2 solutions for obtaining functional information is that functional inference retrieves functions even for operational taxonomic units (OTUs) without a taxonomic name thanks to phylogenetic placement of sequences (taxonomic markers) in a reference tree and different evolutionary models.
Many bioinformatic tools have been developed since the first publication about a functional prediction tool using metabarcoding data. To date, only 1 review has addressed functional inference tools; it is focused on aquaculture and on a limited subset of all the tools available to predict functions from 16S ribosomal DNA (rDNA) metabarcoding datasets [30]. Therefore, in the present context where new solutions are proposed regularly to predict putative function profiles, the state of the art needs to be scrutinized more exhaustively to build a scientific and technical benchmark. More precisely, we provide a detailed description of each tool and evaluate their advantages, specificities, and drawbacks by paying special attention to their methods, modularity, portability, and robustness. One of the main objectives of this review is to provide a rationale on the use of the different tools currently available for prokaryote and fungal communities and draw perspectives, with a few suggestions to enhance their usefulness in microbial ecology. Finally, we illustrate the application of these methods with studies focusing on the soil environment. The choice of this particular system is justified by the fact that it is the most diverse and complex one in terms of microbial diversity, ecology, and functional reservoir [4, 31]; therefore, it represents the most challenging environmental matrix for linking diversity and functions. We believe that this work will help scientists working on microbial communities make choices to best take advantage of their high amount of microbial data. This work also shows that although those approaches are promising, they still need improvements to make them operational tools for microbial diagnosis. Repositories using standardized and robust metrics are still lacking when it comes to interpreting the results.
Historical and Recent Increase of Microbial Datasets
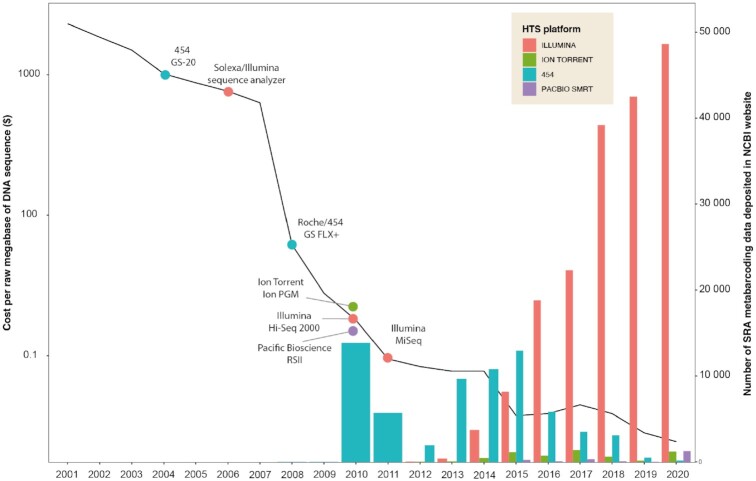
The emergence of HTS in the mid 2000s generated a huge volume of data, leading to a revolution in our way of describing biodiversity. This rise of microbial data can be directly linked to the improvement of HTS technologies, concomitantly with a tremendous decrease in sequencing costs (Fig. 2). This was reflected, with a small time lag, by an increase in the number of sequence read archives (SRAs) linked to metabarcoding data deposited on the NCBI website (Fig. 2).
Figure 2:
Evolution of costs (dollars) per raw megabase of DNA sequence (black line with logarithmic scale), and evolution of the number of SRA metabarcoding data deposited in the NCBI website. The data used to draw this figure are described in Additional File 1, section Figure 2.
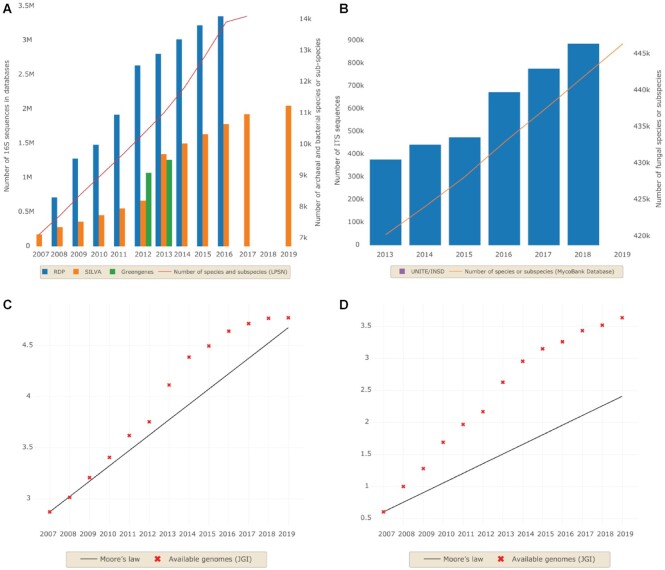
Thanks to the contribution of ecologists, microbiologists, taxonomists, and computer scientists, the databases are continuously enriched and are key to enhance our knowledge about the description and determinism of environmental and human microbiotas [32, 33]. For example, the 16S rDNA sequence data available to analyze bacterial/archaeal diversity were multiplied by 4 and 10 in the RDP and SILVA databases, respectively, between 2007 and 2019 (Fig. 3A). The trend is the same for fungal diversity, with a doubling of internal transcribed spacer (ITS) sequences in the UNITE/INSD database within the past 5 years (Fig. 3B). The 16S rDNA sequences are much more numerous than ITS sequences. However, there were 30 times more fungal species referenced than bacterial ones in 2017 (Fig. 3A and B). The numbers of microbial genomes available, in particular in the Joint Genome Institute (JGI) platform, have increased continuously, and they outpaced Moore's Law mostly from 2013 for bacteria and archaea (Fig. 3C and D).
Figure 3:
Annual cumulative growth of databases in terms of bacterial/archaeal (A) and fungal (B) sequences, and species/subspecies deposited per year. Comparison of the annual cumulative growth of bacterial/archaeal (C) and fungal (D) genomes compared to simulations of Moore's law. The plot is in logarithmic scale. Three databases were compared for 16S rRNA gene sequences: RDP (blue), SILVA (orange), and Greengenes (green). Information is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN [34, 35]) website for bacterial and archaeal species, and on the MycoBank database for fungal species [36, 37]. Information about the bacterial, archaeal, and fungal genomes is based on the Genome OnLine Database (GOLD) [38].
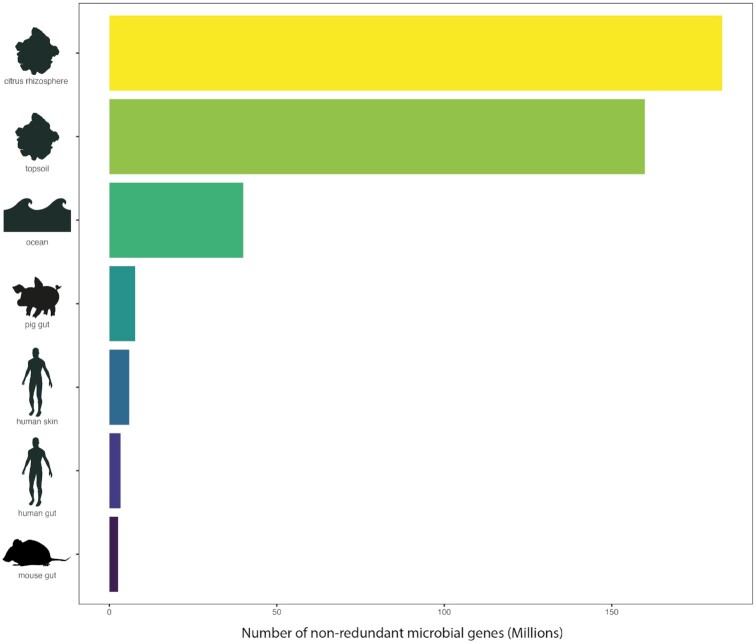
The number of known microbial genes, enzymes, or metabolic pathways available in specialized databases has also considerably increased in the past few years [39–41]. Thousands of functional information files are currently accessible in the KEGG, CAZy, or MetaCyc databases (Table 1). A recent survey predicted the total global estimated bacterial and fungal functions based on KEGG Orthology (KO) to reach 35.5 and 3.2 million, respectively [42]. The authors also indicated that only a tiny fraction of these functions is known today, representing 0.02% and 0.14% for bacteria and fungi, respectively. Although the characterization of gene catalogs using metagenomic approaches was recently criticized [43], the number of non-redundant genes provides an overview of the potential functional reservoir available across various ecosystems [44]. The soil by far seems to harbor the largest pool of functions, followed by the marine, and then animal microbiomes (Fig. 4).
Table 1:
Numbers of organisms, genes, enzymes, and metabolic pathways available in the CAZy, KEGG, and MetaCyc databases
| Database | Organisms | Metabolic pathways | Enzymes/Genes |
|---|---|---|---|
| CAZy | Eukaryotes: 344; Bacteria: 20,421; Archaea: 413 | NA | GH: 171; GT: 114; PL: 41; CE: 19; AA: 16 |
| KEGG | Eukaryotes: 557; Bacteria: 6,317; Archaea: 344 | 547 | KO groups: 24,402 |
| MetaCyc | Total: 3,295 | 2,937 | 13,356 |
When possible, we detailed the number of organisms for the 3 domains of the tree of life. CAZy includes glycoside hydrolases (GH), glycosyl transferases (GT), carbohydrate esterases (CE), polysaccharide lyases (PL), and auxiliary activities (AA). CAZy: Carbohydrate-Active Enzymes; KEGG: Kyoto Encyclopedia of Genes and Genomes; KO: KEGG Orthology; MetaCyc: metabolic pathways and enzymes; NA: not applicable.
Figure 4:
Global microbial gene catalogs from various ecosystems. The references are listed in Additional File 1.
The rapid growth of available genomes is a unique opportunity to predict the putative microbial functions from metabarcoding data by linking taxonomic markers (i.e., rRNA gene amplicons) and their reference genomes or ecological traits. Therefore, the next section is devoted to the different tools and databases dedicated to functional inference and ecological trait assignment for bacterial and fungal communities.
Overview of the Available Tools for Predicting the Potential Functions of the Microbiotas
HTS and the presently increasing collection of functional or ecological traits on a more regular and rigorous basis are promising cues for linking biodiversity and associated functions in the near future [24, 45]. In the literature, the term “function” is used in different ways depending on the study model, the time scale, or even the habitat [46–49]. The notion of function may refer to genes, enzymes, or metabolic pathways but may also represent ecological traits that bring together phenotypic and biochemical notions [50–52].
On the basis of the analysis of 20 papers since 2013, we classified the databases and tools according to the granularity of the results (Fig. 5A), from general information such as ecological traits to more detailed information such as genes or metabolic pathways (Fig. 5). The tools used to obtain fine results, i.e., at the metabolic pathway or gene levels for any taxonomic resolution, are known as functional inference tools (Fig. 5B). On the other hand, we grouped existing tools or databases under the term “ecological trait assignment” when functional information referred to phenotypic or ecological traits and was accessible only for a specific taxonomic rank (Fig. 5C). Indeed, there is a wealth of information often linked to ecological traits in published scientific articles, or of partially formatted metadata (i.e., partial taxonomy or data not linked to the ID of a taxonomic database) [53].
Figure 5:
Diagram of the granularity of the data (A) that can be obtained by functional inference (B) or ecological trait assignment (C).
Tools or methods exist, known under the term “text mining,” to automatically collect data from various sources (e.g., a website, a document in pdf format) through automatic language processing (e.g., natural language processing) [54]. For example, @MInter [55] retrieves information related to microbial interactions from abstracts of articles thanks to a supervised machine learning model. Other tools are based on ontologies; i.e., they use a structured set of terms and concepts from a particular domain by specifying the relationships between these terms and their properties, and thus have a common reference for the use of a common vocabulary. For example, OntoBiotope [56] ontology in the food field retrieves the phenotypes and habitats of microbes from the literature based on the NCBI taxonomy. Another ontology exists, called Ontology of Microbial Phenotype [57]; it brings together a structured set of terms and concepts around microbial phenotypes, and specifies the relationships between these terms and their properties. Tools also based on machine learning such as ProTraits [58] can automatically annotate prokaryotic species on the basis of phenotypic or genomic data from scientific articles or online resources [59].
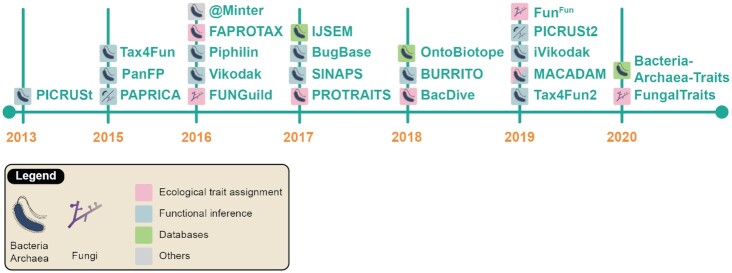
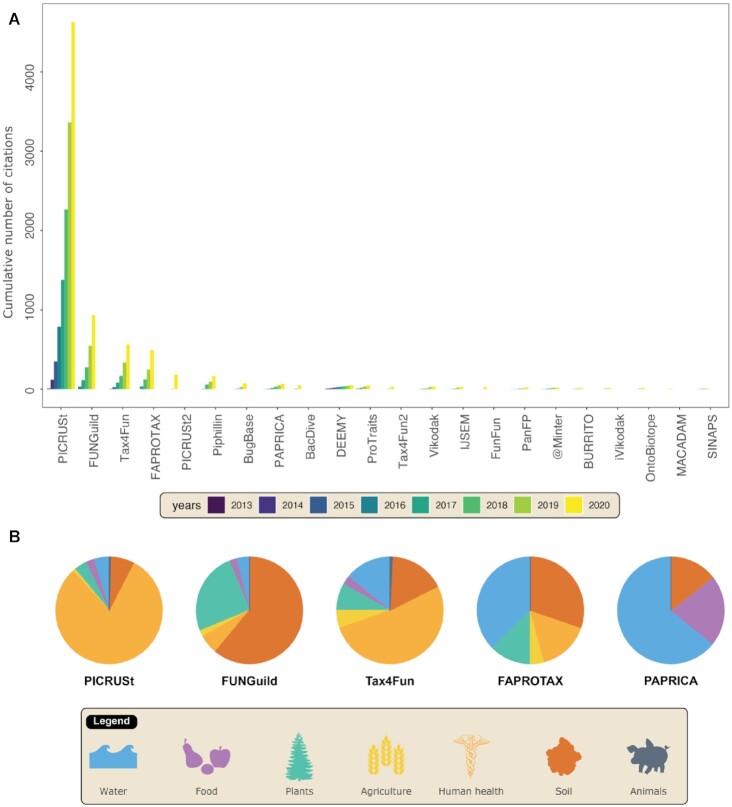
To date, we have recorded ∼20 tools or databases that retrieve functional or ecological data from microbial taxonomic markers, with 2–4 developments per year (Fig. 6 and Table 2). The timeline shows that most of these tools (18 of 23 in total) are only dedicated to bacteria/archaea, 2 are dedicated to bacteria/archaea + fungi, and only 3 are specifically dedicated to fungal organisms. It is important to also underline that most of these tools are devoted to functional inference (13 of 23). The most cited tool is PICRUSt v1 [60], which continued to outrank all others with >4,000 citations in 2020. While FUNGuild [61], Tax4Fun v1 [62], or FAPROTAX [63] are moderately cited, with a few hundred citations, the others are much less so, with only a dozen citations (Fig. 7A). Interestingly, the articles citing functional inference and ecological trait assignment tools fall within the same scope as those for which they were initially developed (Fig 7B.): PICRUSt, FUNGuild, and PAPRICA are mainly cited in articles about human health, the soil, and marine environments, respectively.
Figure 6:
Timeline depicting the historical record of the major tools developed for functional inference or ecological trait assignment. The first version of the DEEMY database dates back to 1996; it was omitted for aesthetic reasons.
Table 2:
List of the functional inference tools, ecological trait assignment tools, and databases
| Tool | Implementation | Targeted genes | Functional prediction | Approaches | Methods | Inputs used | Strengths and Specificities | Limitations |
|---|---|---|---|---|---|---|---|---|
| PanFP | Perl (recently Python) | 16S rRNA | KO, Gene Ontology, Pfam, TIGRFAM | Functional inference | Builds a pangenome | NCBI taxonomy |
|
|
| PAPRICA | Python | 16S/18S rRNA | MetaCyc ontology | Functional inference | Phylogenetic placement | Based on rDNA amplicon sequences |
|
|
| PICRUSt | Python | 16S rRNA | KO, KEGG Pathway, COG, CAZy | Functional inference | ASR (Wagner Parsimony, ACE ML, ACE REML, ACE PIC) | Greengenes taxonomy (18may2012 or v13.5/v13.8) |
|
|
| PICRUSt2 | Python/R | 16S/18S rRNA/ITS | MetaCyc, KO, EC number, COG, Pfam, TIGRFAM | Functional inference | HSP (maximum parsimony, empirical probabilities, subtree averaging, SCP) | Based on rDNA amplicon sequences |
|
|
| Piphillin | Web-based | 16S rRNA | BioCyc, KEGG | Functional inference | Nearest-neighbor matching of 16S rRNA gene amplicons with genomes from reference databases | Based on rDNA amplicon sequences |
|
|
| SINAPS | USEARCH | 16S rRNA | Trait annotation (e.g., energy metabolism, Gram-positive staining, presence of a flagellum) | Functional inference | Word counting | Greengenes, SILVA |
|
|
| Tax4Fun | R package | 16S rRNA | KO | Functional inference | Nearest-neighbor search based on a minimum 16S rRNA sequence similarity | SILVA taxonomy |
|
|
| Tax4Fun2 | R package | 16S rRNA | KO | Functional inference | BLAST | Based on rDNA amplicon sequences |
|
|
| Vikodak | Web-based (not longer available) | 16S rRNA | KEGG pathway, EC number | Functional inference | Microbial co-existence patterns | RDP, SILVA |
|
|
| iVikodak | Web-based | 16S rRNA | KEGG, Pfam, COG, TIGRfam | Functional inference | Microbial co-inhabitance patterns | RDP, Greengenes, SILVA |
|
|
| FUNGuild | Python/Web-based | ITS | Guild type | Trait assignment | Not applicable | Based on UNITE taxonomy (ITS) |
|
|
| FAPROTAX | Python; flat file | 16S rRNA | Ecological functions (e.g., nitrification, denitrification, or fermentation) | Trait assignment, Database | If all type strains of a species at the genus level share the function, FAPROTAX assumes that all uncultured organisms of this genus possess the putative function | SILVA (128, 132) |
|
|
| BacDive | Python and R API, R package | Morphology, physiology (API®-tests), molecular data, and cultivation conditions | Database | Not applicable | NCBI taxonomy |
|
|
|
| BugBase | R/Python | 16S rRNA | KEGG | Functional inference | PICRUSt, custom trait assignment | Greengenes |
|
|
| IJSEM | Flat file with R script for curation | IJSEM | Database | Not applicable | Not applicable |
|
|
|
| ProTraits | Web-based; flat files | Wikipedia, MicrobeWiki, HAMAP proteomes, PubMed abstracts and publications, Bacmap, Genoscope, JGI, KEGG, NCBI, Karyn's Genomes | Database | Not applicable | Not applicable |
|
|
|
| BURRITO | Web-based | 16S rRNA | KO | Functional inference | PICRUSt | Greengenes |
|
|
| MACADAM | Python/web implementation | 16S rRNA | MetaCyc, MicroCyc, FAPROTAX, IJSEM | Functional inference, Trait assignment | Custom methods (provides functional information about upper-rank taxa when organism name is not found) | NCBI taxonomy |
|
|
| FunFun | R package; flat file | Ecological traits | Trait assignment | Not applicable | Based on UNITE taxonomy (ITS) |
|
||
| FungalTraits | Flat files | Guild type, body type, habitat | Trait assignment | Not applicable | Based on UNITE taxonomy (ITS) |
|
|
|
| DEEMY | Web-based | Morphology, anatomy, potential for chemical reactions, or even ecology traits | Database | Not applicable | Not applicable |
|
|
|
| Bacteria-archaea-traits | R package; flat file | 16S rRNA | Traits, phenotypic traits, quantitative genomic traits | Database | Not applicable | NCBI taxonomy, GTDB taxonomy |
|
|
| OntoBiotope | Web-based | Habitats and phenotypes | Database | ToMap (Text to ontology mapping) | NCBI taxonomy |
|
|
|
| @Minter | Python | Microbial interactions | Machine learning | Support-vector machine (SVM)-based classifier | No specific taxonomy, just species level |
|
|
Figure 7:
Annual cumulative number of citations of the major tools (A) and their scope (B). The keywords used for “scope” were retrieved from the titles and abstracts of the articles listed in Additional File 1.
Functional inference
Definition
Functional inference consists of predicting the functional potential of a microbial community from metabarcoding data. The functional potential of a taxon or of a microbial community represents the metabolic capacities based on the presence/absence of genes involved in these pathways. Functional inference methods are based on the assumption that phylogenetic information from marker gene sequences correlates well enough with the genomic content to produce accurate predictions when associated reference genomes are available. In other words, it assumes a significant relationship between (i) the phylogenetic distance between taxonomic markers and (ii) the conservation of the genetic content, referring to vertical gene descent during the evolution of microbial genomes. This is made possible through the relationship between the phylogenetic relatedness of organisms and their gene content [64, 65] (Fig. 5B).
It should be emphasized that the presence of 1 or more genes involved in a function remains “potential” and may not be expressed under environmental conditions. From this point of view, functional inference results may be similar to shotgun metagenomics data; which is often observed in the literature, especially when focusing on a family of genes or a specific biogeochemical cycle [66]. Also, the fact that inferred metagenomes are based only on the reference genomes available in these tools (archaea, bacteria, fungi) means that the lateral gene transfer and gene loss cannot be studied, unlike shotgun metagenomics.
Available tools
PICRUSt.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) v1 [60] is the first tool to have been developed to predict potential functional genes from 16S rRNA metabarcoding and has been the most popular one since it was launched in 2013 (Fig. 5B). PICRUSt v1 needs 3 things: (i) a reference OTU, (ii) a reference genome, and (iii) a reference phylogenetic tree. As regards the reference OTU, the file (in BIOM or tabulated format) is expected to contain a standard OTU abundance table with sequences picked only against the Greengenes taxonomic reference (18 May 2012 or v13.5/v13.8). This tool based on a modified method of ancestral state reconstruction (ASR) deduces functional information for taxa without a match in the reference genomes. The reference genomes are functional proxies that provide a weighting of the functional profiles for the phylogenetically close taxa within a reference phylogenetic tree. The PICRUSt method is divided into 3 main steps that are necessary to obtain relevant information on functional profiles: (i) genome prediction, (ii) metagenome prediction, and (iii) analysis of predictions.
The genome prediction step consists of preparing the trees and checking the quality of the input datasets; then comes the reconstruction of ancestral states in the reference tree (ASR; 4 methodologies are available). Using the output files, the software program predicts traits for leaves of the phylogenetic tree lacking sequenced genomes.
During the metagenome prediction step, normalization of the abundance of each OTU is carried out on the basis of rRNA gene copy numbers (GCNs) to predict the functional category abundances of the metagenome. The user obtains an abundance table for each functional category per sample. The correcting step of the rRNA GCNs allows normalizing to correct the biases towards microorganisms with greater GCNs and improve the estimation of microbial diversity [67]. This step is recommended when the OTUs are phylogenetically closely linked to the genomes [68]. To assess the robustness of the predictions, i.e., to obtain the representativeness of the database towards a community of interest, a nearest sequenced taxon index (NSTI) is generated for each sample. It is calculated using the average of the branches that separate the sequences of interest (OTUs, amplicon sequence variants [ASVs]) in a sample from the reference microbial genome, with a weighting by their relative abundance in the sample. This confidence score is one of the major strengths of this tool. Regarding functional categories, information can be obtained at different levels (genes or metabolic pathways) with more or less detailed descriptions (EC numbers, KEGG pathway [40], cluster of orthologous groups [COG]). Information about all functional categories can also be obtained for each OTU. The last step consists of analyzing the predicted data. This step is essential for interpreting the large number of results generated from a robust statistical analysis.
The major strength of PICRUSt v1 lies in its evolutionary models that infer functions for the complete bacterial community. The portability of this tool with the support of a broad stakeholder community including a forum (Google group) and blogs are advantages that make it a central tool for functional predictions (Table 2). Despite all its benefits, PICRUSt v1 has drawbacks such as focusing only on the 16S rDNA marker and using only Greengenes taxonomy (Table 2). Several specialized tools have emerged to integrate PICRUSt as a sublayer to carry out diagnoses in the medical field [69] or directly in a pipeline [70]. PICRUSt v2 fills the gaps of the first version, with an improvement that allows inference directly based on the sequences and no longer through taxonomy. Another improvement concerns the addition of bacterial but also fungal reference genomes, thus making it possible to infer from 18S rDNA and ITS amplicons [71].
PAPRICA. Pathway Prediction by Phylogenetic Placement (PAPRICA) [72] infers the metabolic potential of prokaryotic and eukaryotic communities from metabarcoding data based on rRNA gene amplicons. It was the first tool that allowed for the functional prediction of 16S and 18S rRNA amplicons. It comes in the form of a pipeline taking the OTU reads as inputs to place them in an rRNA reference tree built from complete genomes. To build this tree, a consensus genome is found for each node in the tree, which then makes it possible to predict metabolic pathways for the sequences of interest without a match in the complete reference genomes. The abundance of metabolic pathways is weighted by rRNA GCNs from known genomes. A strength of this tool is that it also provides an indicator of genomic stability depicting the robustness of the results. However, PAPRICA, like all the tools using a reference phylogenetic tree and sequence placement methods, is dependent on the quality of rRNA resolution, and this represents a drawback when some clades may be affected (Table 2).
Tax4Fun. Tax4Fun [62] is an R [73] package published in 2015 for predicting functional profiles from targeted metagenomic 16S rRNA data. However, the algorithm and statistical efficiency based on a metabolic mixture model in terms of a mixture of pathways was developed in 2013. This R-based architecture is inherently a cross-platform tool, and it may be more accessible for a large number of users with low experience in bioinformatics. This tool uses pre-calculated functional profiles like PICRUSt v1 and taxonomic data formatted from the SILVA database. One of the differences with PICRUSt is the rRNA sequence placement in the reference genomes, which is achieved by a BLAST search (instead of a tree placement approach as for PICRUSt). It is a convenient tool because it provides a confidence score (FTU and FSU) to determine the fraction of OTUs that was not mapped to KEGG organisms or the number of sequences without KEGG Orthology (KO) hits (Table 2). Like PICRUSt v1, it cannot be used for fungal diversity predictions.
Piphillin. Piphillin [74] differs from the PICRUSt or PAPRICA approaches because it does not use a phylogenetic tree or database (16S) but directly maps the OTU sequences on the rRNA of the reference genomes using a nearest-neighbor algorithm. This specificity could avoid faulty sequence placements in the reference phylogenetic tree. It is used online only, which represents both a strength and a weakness: it benefits from computing power (a strength), whose strength depends on the hosting server (e.g., quota management, cluster configuration) (a weakness). A Piphillin sublayer also exists to complete the analysis of the results [75].
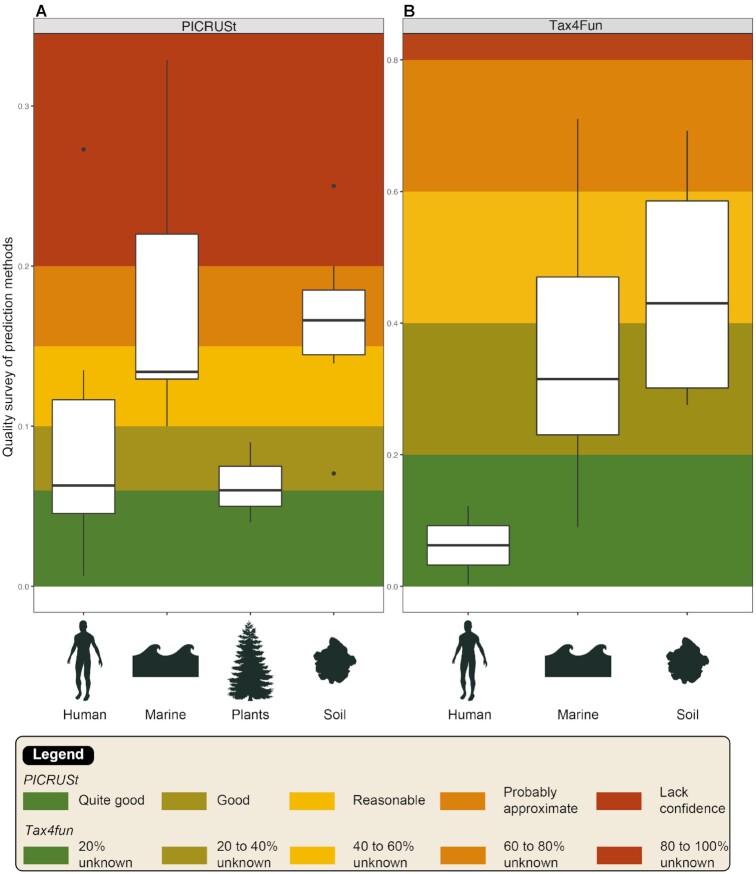
The quality of prediction represents a prerequisite for the application of the above-presented tools to study indigenous microbial communities. It may depend on the tool but also on the type of targeted ecosystem. To test the quality of functional prediction according to the tool and the studied ecosystem, we compiled the NSTI scores for PICRUSt v1 and the FTUs for Tax4Fun from a subsampling of articles that covered a range of ecosystems—human, marine, plant, and soil (Fig. 8). Whatever the tool, the best predictions were obtained for the human microbiotas, and the most approximate ones, for the soil samples. The variability of quality scores across the different soil studies seemed to be lower with PICRUSt than with Tax4Fun. Nevertheless, some soil studies using Tax4fun indicate a high-quality survey, with only ∼30% of OTUs unmapped to a reference. This likely reflects the discrepancy between human reference genome availability and soil microbiota genome availability. In addition, microbial diversity is much more complex in soils than in the human microbiotas. In this case, it is essential that the quality scores from functional inference tools should be taken into account because it is a key to a robust interpretation of the results. Unfortunately, we found few studies indicating these quality scores.
Figure 8:
Overview of the quality of functional prediction based on a subsampling of articles for PICRUSt (A) and Tax4Fun (B) across various ecosystems. For PICRUSt, colors were assigned according NSTI results: <0.06, quite good; 0.06–0.10, good; 0.10–0.15, reasonable but probably approximate; and >0.20, probably unreliable. For Tax4Fun, we split the fraction of OTUs that could not be mapped to KEGG organisms in 5 harmonious groups. References are listed in Additional File 1. The distribution of data are displaying by boxplots and are standardized way of based on a five number summary (minimum, first quartile, median, third quartile, and maximum) and the outliers (shown as black circles).
Ecological trait assignment
Definition
Ecological trait assignment differs from functional inference because it consists of obtaining information on the life strategy, phenotypic, and quantitative genomic traits (e.g., trophic modes, growth strategy) of a taxon from its nomenclature, whatever its taxonomic rank. If the taxon is not present in the database, it will not be possible to know its traits (Fig. 5C). This approach is faster than functional inference for retrieving an item of functional information, but tools dedicated to metabarcoding outputs are lacking, and only a few ecological traits are available (Table 2). The main interest is to get functional information with a possibly not so fine granularity as functional inference does, but obviously more accurate. Ecological traits are indeed often based on results with biochemical experimentations from curated databases or scientific publications. Practically speaking, only the guild will be recovered and for example the fungal sequences identified as belonging to the Serpula genus will be assigned to a wood saprotroph when an ecological trait tool is used; with an inference tool, the abundance of various genes related to polysaccharide degradation will be attributed to all fungal sequences.
Tools
FUNGuild.
FUNGuild [61] is the pioneer and one of the few tools that assigns ecological traits to fungi based on their taxonomy (Table 2). These assignments rely on metabarcoding data. They require providing a contingency table (OTUs or sequence counts per sample) and the link between each OTU and its taxonomy. To carry out the assignment, FUNGuild uses its own curated database, and searches it for the taxon. This database contains several taxonomic levels (e.g., phylum, genus, species). However, the taxonomic name at the genus or species level is necessary to assign traits to the taxa of interest. Trait information is available in 66% of the cases at the genus level, and only in 34% of the cases at the species level [61]. The user obtains a summary table of the different possible ecological traits for each taxon with a robustness indicator and a confidence range (“possible,” “probable,” and “highly probable”).
The strength of this database is that the provided data are based on the literature (primary research), or on reference websites or their own collective research experience if the datum is missing. The authors recommend the use of the UNITE database for taxonomic assignment and therefore the use of the ITS marker, but it can be easily transposed to data based on the 18S rRNA marker. It just requires creating a wrapper to make a link between the taxonomy of the data and FUNGuild to retrieve the traits of interest.
A new database called FunFun [76] is now available. It encompasses 80 fungal ecological traits. In reality, this database is a FUNGuild database overlay with information on genetic, enzymatic, morphological, stoichiometric, life history, and physiological aspects. In addition, the authors mention that FunFun will be updated in terms of taxonomy and associated guilds, which is not necessarily the case with FUNGuild. However, although this database is promising, a lot of information is missing because it integrates literature data for the first time ever, and its improvement relies on the progress of research, as well as the contribution of scientists. This caused an impulse leading to a community of scientists proposing a new database: FungalTraits [77] links information from FUNGuild and FunFun. It is very complete, and offers different levels of life styles. Please note that this database includes species from the fungal kingdom but also fungus-like stramenopiles (e.g., the Oomycota phylum). This may be especially useful because various species are identified as major plant pathogens within Oomycota. For example, the genus Phytophthora gathers several crop pathogens that cause important losses and can represent a risk to global food security [78].
To conclude, the minor drawbacks of FUNGuild, with rare updates or a tool oriented to ITS sequences, have been offset by the new FunFun and FungalTraits databases.
To complete the tools concerning fungal communities, DEEMY [79] is an information system only available online and specialized in ectomycorrhizas [80]. This website references 554 species associated with their respective symbiotic organisms, including 104 genera. To characterize each species, a summary sheet provides taxonomic nomenclature and bibliographical references and photographs, as well as information on morphology, anatomy, potential chemical reactions, or even ecology traits.
FAPROTAX. Functional Annotation of Prokaryotic Taxa (FAPROTAX) [63] is used to assign metabolic functions, ecological traits, or large functional groups relevant to prokaryotes (Table 2). This database was built manually from the scientific literature of the International Journal of Systematic and Evolutionary Microbiology (IJSEM) and Bergey's Manual of Systematic Bacteriology. It contains ∼4,700 unique prokaryotic taxonomies (mostly at the species level) and 90 functional groups. FAPROTAX is based on the implicit assignment of a trait/function to a taxon (whether cultivated or not) if all the cultivated members display this trait/function. Its main limitation is that it is focused on marine prokaryotic organisms, so communities from other biomes can be missing. Another point to be considered is that if the taxa of interest do not have a species name, the tool cannot draw inferences at the upper levels (e.g., genus) to assign an ecological trait.
IJSEM phenotypic database. IJSEM [81] compiles phenotypic and environmental tolerance data about >5,000 bacterial strains. It is an official and unique reference for publishing and validating new strains. These strains cover ∼23 phyla from various habitats (mainly soils). The database appears as a TSV file [82], and available information can be grouped into 5 categories: ancillary data (e.g., article's DOI; taxonomic nomenclature), morphology/phenotype (e.g., Gram stain status; motility), metabolism (e.g., BIOLOG information), environmental preferences (e.g., habitat of isolation; oxygen requirement), and sequence data (e.g., 16S rRNA accession No.).
BacDive. BacDive [83] is one of the largest metadatabases [84] referencing information on bacterial and archaeal diversity (Table 2). The tool links taxonomy and phenotypic information directly, but the database can only be browsed on a website or data can be downloaded from it. However, it provides a complete API to achieve scripts and retrieve the desired information. In the first months of 2020, it offered data on 81,827 bacterial and archaeal strains, including 14,091 type strains, and thereby covered ∼90% of the described species according to their website. This database is interesting because it provides different levels of robust information on taxonomy, morphology, physiology (API®-tests), molecular data, and cultivation conditions. As for physiological data, it provides—for example—the main substrates used for culturing a species and the enzymes present (a link with the EC classification number is available). These data have been more broadly incorporated into a tool (bacteria-archaea-traits) that encompasses numerous traits of bacteria and archaea from 26 sources [51].
To complete this list, a few specialized databases target only 1 or a few traits. For example, Engqvist [85] recently grouped the growth temperatures of 21,498 non-redundant organisms across the whole tree of life. This study showed a strong correlation between the growth temperature of organisms and enzymatic optima, with temperature-dependent increases or decreases of enzymatic functions. This information can be very interesting and complementary to the interpretation of functional inference results, and can be linked—for example—to environmental conditions.
Application of These New Approaches to the Functions of the Soil Microbial Ecosystem
Functional inference
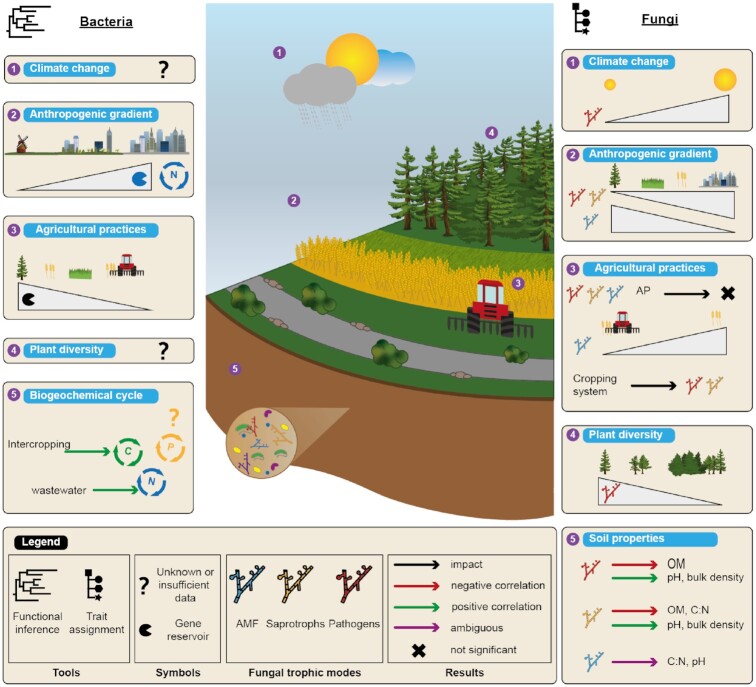
In recent years, meta-omics approaches have been increasingly included in soil monitoring, whether in fundamental research programs or in more operational projects [86]. Most studies (∼60% on the basis of keywords in the titles or abstracts of the publications, see Fig. 7B) have focused on PICRUSt to generate functional predictions from taxonomic data of the soil microbiota. We summarized the most valuable outcomes about soils by grouping them into categories: anthropogenic gradient, agricultural practices, and biogeochemical cycle or soil properties (Fig. 9). For example, a study showed that plant-bacteria interactions in the rhizosphere were mainly related to beneficial cooperation [87] involving the release of root exudates by the plants on the one hand, and hormone production or the ability to break down toxic chemicals by bacteria on the other hand. Another study investigated the stoichiometric regulation of soil carbon cycling by comparing functional predictions by metabarcoding (via PICRUSt) and shotgun sequencing on a wide C:N:P soil gradient in a rice field [66]. A strong correlation was evidenced between the functional predictions from metabarcoding and metagenomics as regards the abundance of some metabolic families involved in the C, N, and P cycles. Still using PICRUSt, another study examined the effects of intercropping by predicting the soil microbial functional profiles. It evidenced that an intercropping system increased the functional potential in terms of carbon fixation pathways and the citrate cycle [88]. Finally, a study focused on the impact of long-term land-use practices (forest, grassland, crops) on soil bacterial communities [89] showed that forest soils harbored the largest reservoir of genes, followed by no-till soils and then grasslands. The plowed soils presented the lowest functional richness.
Figure 9:
Summary diagram of the most relevant microbial soil functions results based on functional inference and ecological trait assignment.
The figure is made up of 2 parts: studies on bacterial communities based on functional inference on the left and studies on fungal communities based on ecological trait assignment on the right. For all studies (climate change, anthropogenic gradient, agricultural practices, plant diversity, or the biogeochemical cycle), if an effect or a correlation was found on the gene reservoir or on microbial communities with a particular ecological trait, a colored arrow indicates the effect and a cross indicates no significant effect. A triangle indicates either a decrease or an increase of the gene reservoir or microbial communities with a particular trait. References are listed in Additional File 1.
Based on Tax4Fun predictions, a study investigated the effect of different irrigation practices with various water qualities (freshwater, treated or untreated wastewater) along with the different land use systems in drylands [90]. The authors compared the potential functional and taxonomic profiles of bacteria. Irrigation with wastewater had an effect on bacterial responses by shaping communities and functional profiles. By bringing more nitrogen, wastewater favored the response of certain genera, in particular Nitrosospira, and increased the relative abundance of the genes involved in nitrification and denitrification.
Among all the functional inference tools available today, 2 of them stand out, i.e., PICRUSt and Tax4Fun. A benchmark study of these tools found no major differences in terms of performance, especially for soil samples [91]. Another benchmark study indicated that these 2 tools provided similar functional profiles but could be complementary for certain gene families found only in one or the other [92]. Moreover, the characterization of the fungal functional potential by PICRUSt2 is too recent for us to have any insights into its robustness concerning soil communities. Compared to trait assignment, the links between diversity and functions still remain tenuous concerning certain biogeochemical cycles or the impact of climate change and plant diversity (Fig. 9).
Ecological trait assignment
The complexity of microbial traits is variable, with simple traits like organic phosphate utilization and more complex ones like methanogenesis [24, 93]. The conservation of prokaryotic traits or core genes varies according to phylogenetic depth [64]. For example, the complex methanogenesis trait seems to be very conserved at the order and family levels, which contrasts with the resistance to specific bacteriophages, which seems to vary at the species level owing to particular point mutations [24]. Below are a few examples of the possible benefits of ecological traits to the analysis of the diversity of soil microbial communities (Fig. 9).
Regarding the assignment of fungal traits, FUNGuild is currently and by far the most implemented tool, if not the only tool implemented by ecologists wishing to supplement their diversity analyses with data on the ecological traits of fungal communities, and mainly in studies on soil fungal communities [94–97]. A study on fungal communities in subtropical forest soils highlighted a negative relationship between the abundance of pathogenic fungi and the phylogenetic diversity of plant communities [98]. Another study showed a positive correlation between soil fungal community dissimilarities (plant pathogens, saprotrophs, and ectomycorrhizas) and plant phylogenetic distances in forest soils [99]. Tropical land uses also affect the functional guild. A massive shift of fungal trophic modes has been shown—notably a decrease in mycorrhizal fungi and an increase in saprophytic and pathogenic fungi—along with increased anthropization levels [100]. Interestingly, several large-scale (national or global) studies have characterized the distribution of trophic types while identifying the environmental parameters that influence them [94, 101–103]. The distribution of these trophic modes seems to vary greatly depending on temperature and precipitation [103]. This supports a recent global study focused on the distribution of pathogens and indicating higher abundance in warm regions [102]. A recent study compared the trophic modes (synonym: life strategies) assigned to the ITS and 18S rDNA molecular markers by FUNGuild [94]. This study indicated that the saprotroph and pathotroph richness levels were directly and negatively correlated with the organic matter content and elevation, and positively correlated with the pH and bulk density. For symbiotroph richness, the relationship differed depending on the molecular marker used: it was positively correlated with the C:N ratio when ITS sequences were used but negatively correlated when 18S rDNA sequences were used. Similarly, the pH was positively correlated on the basis of 18S rDNA data but negatively correlated on the basis of ITS data [94]. These differences may come from the fact that the 2 molecular markers do not cover the same taxonomic range. Therefore, the choice of molecular markers and primers is essential because it affects the global picture obtained by possibly enhancing or decreasing the representation of particular functional groups in the community. For example, arbuscular mycorrhizal fungi are better represented, in particular the Glomeromycota group, when the 18S rDNA marker is used [104, 105]. A study at a smaller scale also showed that saprotroph richness was directly driven by the soil physico-chemical parameters and confirmed the aforementioned results. The authors showed a positive correlation with the pH but a negative one with the C:N ratio [106]. All these studies used the FUNGuild tool dedicated to characterizing fungal community traits.
Regarding the assignment of bacterial traits, various databases exist but few tools have been developed to assign ecological traits from metabarcoding datasets. Only FAPROTAX stands out as a powerful tool for analyzing the functional potential of soil communities [107], although it is dedicated to marine organisms.
Technical and Conceptual Limitations and Biases
The metabarcoding approaches have significant advantages for characterizing indigenous prokaryotic and eukaryotic microbial communities. Standard protocols now exist, from sample preparation to bioinformatic and statistical analyses, and scientists have acquired an important feedback on biases, costs, and efficiency [108–110].
A fundamental limitation of functional inference tools, represented by gene gain and loss, is mainly due to horizontal gene transfer but also gene duplication, gene loss, and de novo gene birth [111–114], which is addressed in the literature and taken into account to some extent in these tools. However, horizontal gene transfer remains difficult to consider accurately for functional prediction, and its influence on microbial communities is hard to estimate. Moreover, the horizontal gene transfer rate varies substantially within the tree of life and according to gene families/pathways [24, 93, 111]. This process is mainly described in prokaryotes but is also found to a lesser extent in eukaryotes, in particular fungi [115]. Microorganisms can gain a function through plasmid transfer, but no information was found in the literature about functional prediction [60]. However, plasmids are extrachromosomal DNA molecules that play a role in the rapid adaptation of microbial communities to environmental changes across all microbiomes [116, 117]. In particular, they are transferred between phylogenetically distant populations for them to acquire genes and beneficial traits for their adaptation (e.g., resistance to antibiotics, biocides, pollutants). This is key for all environments, especially soils, where biotic and abiotic fluctuations are tremendous [118]. The transfer of plasmids is also introduced from phages or viruses into microbial genomes [119].
From a technical point of view, most of the studies on microbial diversity using metabarcoding approaches are based on the sequencing of 1 or more hypervariable regions and remain limited by the size of the amplicon to be sequenced. The most commonly used Illumina sequencing platforms (MiSeq, HiSeq, and NovaSeq) can provide maximum readings of 600 bp (∼550 bp after adapter/tag/primer trimming). Several studies have questioned the most suitable regions for obtaining the best taxonomic resolution [120, 121]; the use of full-length rRNA (∼1,800 bp) seems to be the most appropriate solution [122]. It would significantly enhance phylogenetic resolution for prokaryotic and eukaryotic microorganisms [123] (Fig. 10, second box). Short reads do not allow good enough resolution in taxonomic assignment either (i.e., not down to the species level), although this point is crucial for placing sequences/taxa in the phylogenetic tree to achieve functional inference. With third-generation HTS platforms (e.g., PacBio, Oxford Nanopore), full-length molecular markers can be sequenced, e.g., 16S/18S rRNA genes or the full ITS1 and ITS2 sequences [124, 125]. This will considerably improve taxonomic assignment and make it possible to assign sequences at the species or even the strain level in certain cases [125]. This way, functional inference and ecological trait assignment will be improved. However, if the objective is to obtain the best taxonomic resolution possible, the study of ecological traits at high taxonomic ranks (e.g., the phylum) remains very promising, especially for highly conserved traits [126]. For example, the carbon mineralization rate was positively (e.g., Bacteroidetes) or negatively (e.g., Acidobacteria) correlated with their relative abundance [127].
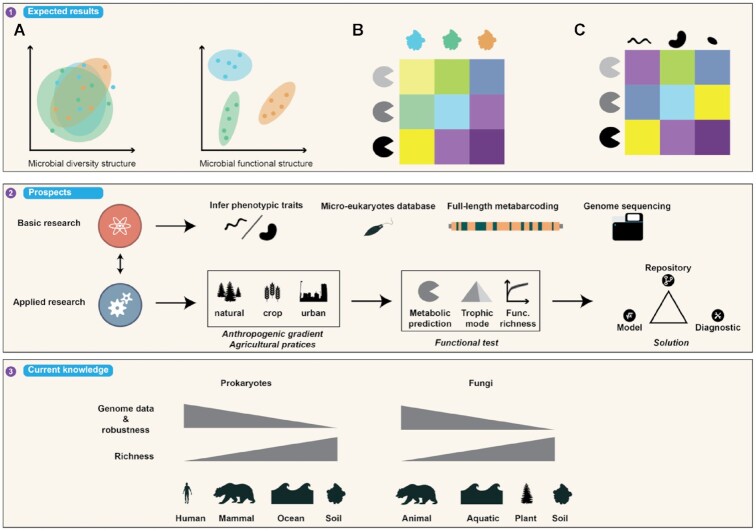
Figure 10:
Summary diagram of the expected results (first box), the functional prediction prospects (second box), and the limits of the microbial genomic data available for different habitats (third box). The first box illustrates a comparative example of data results of community structures and functional structures through a PCA (A). This example illustrates the case when the functional community structure differentiates experimental conditions better than it differentiates the microbial community structure. Illustrative heat maps showing the relative abundance of genes per sample (B) or per OTU (C).
A good practice complementary to the use of full-length amplicon sequencing would be the use of ASVs (also called ZOTUs) to increase the rate of inference with a better sequence placement on the reference tree [71, 128]. Indeed, for those using an OTU clustering approach with a similarity threshold, 1 solution would be to use all the sequences within the OTUs instead of 1 representative sequence for each OTU seed, which could be less accurate. However, this would also increase the analysis time.
Importance of Taxonomy and Genome References: From Accuracy to Resolution
Many tools use taxonomic data to obtain information about microbial functions through a metabarcoding approach. Therefore, it is important to check the bioinformatic strategy used to analyze the amplicon sequences, from the filtering steps to OTU clustering or not (see ASV), including taxonomic assignment.
The use of tools on ecological traits is highly dependent on taxonomic resolution. For example, when using FUNGuild, special attention must also be paid to the fact that a sequence assigned at the genus level may be associated with several trophic types, and that plant-pathogenic fungi are highly host-specific and may be non-pathogenic in the context of the study. For the sequences (or OTUs) without any taxonomic assignment, functions cannot be obtained using tools on ecological traits (Fig. 10, second box). To improve this point, especially for fungal communities, inferences may be drawn on the basis of phylogeny, as done for bacteria, archaea, or macroorganisms [129–133]. One of the avenues to be explored is the use of ASR tools such as PICANTE [34] or CASTOR [36], which infer traits for taxa devoid of ecological data from a phylogenetic tree.
Functional inference tools depend on the reference genomes to establish predictions, so the accuracy of the results can vary among samples. Samples with well-described host-associated communities such as the human microbiome have many reference genomes available and allow good predictive accuracy (Figs 8 and 10 third box). Contrastingly, in more complex and highly biodiverse environments like soils [38], the genomes representing the total taxonomic diversity are much more difficult to obtain. The proportion of cultivable terrestrial strains remains very low (∼25%) compared to the human microbiotas (80%) [134]. Thus, the results estimated for the communities from complex biomes are approximate and debatable.
To improve functional prediction results, it is advisable to provide genomes specific to the habitat of interest [135]. Considerable efforts have to be made to increase the number of habitat-specific reference genomes (animal/human, water, plant, soil), with special attention to the most complex and unknown environments [136]. Tools to routinely update the databases will also need to be developed [137]. This is an ongoing dynamic at the international scale. For example, the annotation of reference genomes in databases is not yet representative of soil microbial diversity [138]. To fill this gap, an effort has been made by creating the Refsoil database [138] (which does not seem to be maintained [139]) or a Refsoil + plasmid database [117].
Discussion and Future Prospects
The possible retrieval of a putative functional potential or ecological traits directly from taxonomic markers and metabarcoding approaches opens new perspectives for our understanding of microbial communities, both from a fundamental and/or an operational point of view (e.g., functional redundancies, diagnostic tool) [69, 140]. This information can be used to (i) understand the main functions potentially expressed in a given environment and identify the possible drivers, (ii) examine the distribution of functions among taxonomic groups, or (iii) supplement the classic diversity metrics used to evaluate the ecological state of environmental matrices (Fig. 10, first box). Beyond providing an overview of the putative functions of an ecosystem, prediction tools could also provide more detailed information than taxonomic markers do for users to significantly distinguish sample groups from each other in certain habitats [122] (Fig. 10A, first box).
A new generation of tools solves the main limitations of the previous generation tools by including improvements in terms of taxonomic marker targeting, methodology, and flexibility.
Future Prospects with Second-Generation Tools
Second-generation tools are currently emerging, e.g., PICRUSt2 [71], Tax4Fun2 [135], or iVikodak [141] (Fig. 6). Indeed, Langille's team of developers bridged the gap for the scientific community working on fungal ecology. PICRUSt2 now includes 18S rDNA and ITS amplicons from the fungal kingdom. Another great improvement is flexibility: the sequence can be used directly, instead of taxonomy based on Greengenes nomenclature. Users are no longer dependent on taxonomy to infer functions; this is a great comfort and provides better robustness of the analyses. However, users should be wary of the results because the number of sequenced fungal genomes currently integrated in the tool is much lower than the number of bacterial genomes. It is recommended to check the quality score (e.g., NSTI) for the robustness of the results and interpretation. However, this limitation can be lifted. For example, the 1000 Fungal Genomes Project [142] is aimed at high-quality sequencing and annotation of fungal genomes so as to build a reference dataset to be used for meta-omics data analysis.
Another downside of these tools is the absence of data support for micro-eukaryotic communities, which are essential to the soil ecosystem. Protists are abundant and diverse, with a large range of functional diversity, and are highly involved in soil food webs and functioning [143, 144]. It would be particularly useful to develop tools dedicated to protists from data on ecological traits available in the literature [145].
Challenges: From Fundamental Research to Diagnosis
Switching from fundamental research to practical applications would be interesting because although operational microbial diversity bioindicators are increasingly emerging, there is a huge gap in the functional information of microbial communities. Even if the number of species can be an indicator of the impact of biotic and abiotic factors [146, 147], the need to characterize the associated functions at the ecosystem level has become obvious to obtain a complete diagnosis with functional information on the soil microbial quality [148, 149].
As regards human health, identifying taxonomic and functional changes to estimate the contributions of taxa associated with a disease is an emerging topic [150], as, e.g., in research into gene markers involved in colorectal or oral cancers [151, 152].
Some interesting examples exist in the biomonitoring and bioassessment of water quality [153, 154], but examples for the soil microbial quality are still scarce. The huge complexity and diversity of the soil microbial community probably still limits such applications to the soil ecosystem, along with a lack of genome references. However, initiatives at the global level are in progress to access soil biodiversity using taxonomic, functional, and environmental data [147, 155]. We can also note that a real dynamic seems to be developing at the international scale to collect, standardize, and disseminate traits through the tree of life via an open science tool called the Open Traits Network (OTN) [92].
To our knowledge, providing robust and operational indicators based on putative functions derived from metabarcoding data is impossible today. The main challenges are to (i) aggregate and summarize the mass of data currently generated, (ii) test the predictions on datasets and compare them with “real” functional measurements, (iii) validate these indicators on datasets under diverse experimental conditions (e.g., land use gradient, agricultural practices) at the local and global scales, and (iv) develop representative repositories to ensure the validity of the diagnosis made from these new tools.
Regarding aggregation and data reduction (item i), a track would be to use a constrained non-negative matrix factorization approach [156], an alternative to the concept of community-aggregated traits [157]. This method has already been used to aggregate functional traits from metagenomes [156]. The authors demonstrated that significant data reduction made it possible to propose simple models to describe a set of complex functions at the scale of an ecosystem (here the potential for fiber degradation in the human intestinal microbiota) while preserving biological data quality [156]. Concerning item ii, it will be interesting, for example, to confront functional predictions with volatile organic compound (VOCs) emissions or microbial respiration rates from soil measurements. Indeed, the very diverse microbial VOCs are secondary metabolites playing various roles, in particular making it possible to carry out more or less long-distance interactions and communication (e.g., growth, motility, antibiotic resistance, expression of stress response genes) [158]. Moreover, to suggest these tools as robust indicators of the soil quality (item iii), it will be essential to use large datasets to determine the best metrics (e.g., functional richness, relative gene abundance, aggregation of traits) and the most sensitive genes or groups of genes depending on the various scientific issues. Once these limitations have been lifted, these tools will provide results of great interest to the scientific community at relatively affordable human, technological, and financial costs. However, maintaining the associated scientific expertise will be essential to support their transfer for operational applications and avoid erroneous interpretations that could potentially have disastrous consequences for soil users and soil policy makers (item iv). For example, interpreting trophic types requires strong expertise, with particular attention to the exploitation of potential pathogenicity information—a highly sensitive task. The responses of the traits vary according to the disturbances applied to the ecosystem [159], and the results must be contextualized to ensure correct interpretation.
Conclusion
The exploration of microbial functional diversity based on taxonomic marker genes in order to improve our knowledge of microbial diversity and functions is just starting. As highlighted in this review, various solutions have emerged over a number of years and are being improved quickly thanks to technological advances. Functional inference results are already robust and representative for some ecosystems with low diversity (specific richness) and with well-characterized genomes such as the human microbiotas. Progress now needs to be made for more complex environments. The upcoming challenge, notably for environmental samples, will be to establish the link between functional predictions on reference datasets and environmental measurements. The new network SoilBON dedicated to monitoring soil biodiversity and functional ecosystems at a global scale, with particular attention to microbial diversity, is a step in this direction [3]. This ambitious framework aims to collect and analyze soil diversity on the basis of soil ecological indicators (i.e., essential biodiversity variables [160]). One purpose of this framework is to inform policy makers and stakeholders so that they can adopt measures to preserve this biodiversity.
Data Availability
Not applicable.
Additional File
Additional file 1
Abbreviations
API: application programming interface; ASV: amplicon sequence variant; BLAST: Basic Local Alignment Search Tool; bp: base pairs; CAT: community-aggregated trait; CAZy: carbohydrate-active enzymes; C, N, and P: carbon, nitrogen, and phosphorus; COG: cluster of orthologous groups; DOI: digital object identifier; EC number: enzyme commission number; FTU: fraction of OTUs; GCN: gene copy number; HTS: high-throughput sequencing; IJSEM: International Journal of Systematic and Evolutionary Microbiology; ITS: internal transcribed spacer; INSD: International Nucleotide Sequence Database; JGI: Joint Genome Institute; KEGG: Kyoto Encyclopedia of Genes and Genomes; KO: KEGG orthology; NCBI: National Center for Biotechnology Information; NSTI: nearest sequenced taxon index; OTN: open traits network; OTU: operational taxonomic unit; qPCR: quantitative PCR; RDP: Ribosomal Database Project; rDNA: ribosomal DNA; rRNA: ribosomal RNA; SRA: Sequence Read Archive; VOC: volatile organic compound; ZOTU: zero-radius OTU.
Competing Interests
The authors declare that they have no competing interests.
Funding
No funding to declare.
Authors' Contributions
C.D. and L.R. conceptualized the manuscript. C.D. drafted the manuscript with contributions from S.T., S.D., A.C., P.-A.M., and L.R. All authors read and approved the final manuscript.
Supplementary Material
Trevor Charles -- 10/31/2021 Reviewed
Axel Künsnter, PhD -- 11/2/2021 Reviewed
ACKNOWLEDGEMENTS
Thanks to Annie Buchwalter for correction and improvement of English language in the manuscript.
Contributor Information
Christophe Djemiel, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
Pierre-Alain Maron, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
Sébastien Terrat, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
Samuel Dequiedt, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
Aurélien Cottin, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
Lionel Ranjard, Agroécologie, AgroSup Dijon, INRAE, Université de Bourgogne, Université de Bourgogne Franche-Comté, F-21000 Dijon, France.
References
- 1. Cavicchioli R, Ripple WJ, Timmis KN, et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat Rev Microbiol. 2019;17:569–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maron PA, Mougel C, Ranjard L. Soil microbial diversity: Methodological strategy, spatial overview and functional interest. C R Biol. 2011;334(5-6):403–11. [DOI] [PubMed] [Google Scholar]
- 3. Guerra CA, Bardgett RD, Caon L, et al. Tracking, targeting, and conserving soil biodiversity: A monitoring and indicator system can inform policy. Science. 2021;371(6526):239–41. [DOI] [PubMed] [Google Scholar]
- 4. Bardgett RD, Van Der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515(7528):505–11. [DOI] [PubMed] [Google Scholar]
- 5. Rivett DW, Bell T. Abundance determines the functional role of bacterial phylotypes in complex communities. Nat Microbiol. 2018;3(7):767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Antwis RE, Griffiths SM, Harrison XA, et al. Fifty important research questions in microbial ecology. FEMS Microbiol Ecol. 2017;93(5):doi: 10.1093/femsec/fix044. [DOI] [PubMed] [Google Scholar]
- 7. Sergaki C, Lagunas B, Lidbury I, et al. Challenges and approaches in microbiome research: From fundamental to applied. Front Plant Sci. 2018;9:doi: 10.3389/fpls.2018.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Starr AE, Deeke SA, Li L, et al. Proteomic and metaproteomic approaches to understand host-microbe interactions. Anal Chem. 2018;90(1):86–109. [DOI] [PubMed] [Google Scholar]
- 9. Aldridge BB, Rhee KY. Microbial metabolomics: Innovation, application, insight. Curr Opin Microbiol. 2014;19:90–6. [DOI] [PubMed] [Google Scholar]
- 10. Knight R, Vrbanac A, Taylor BC, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–22. [DOI] [PubMed] [Google Scholar]
- 11. Porter TM, Hajibabaei M. Scaling up: A guide to high-throughput genomic approaches for biodiversity analysis. Mol Ecol. 2018;27(2):313–38. [DOI] [PubMed] [Google Scholar]
- 12. Mehle N, Dreo T. Quantitative analysis with droplet digital PCR. Methods Mol Biol. 2019;1875:171–86. [DOI] [PubMed] [Google Scholar]
- 13. Thiele-Bruhn S, Schloter M, Wilke BM, et al. Identification of new microbial functional standards for soil quality assessment. Soil. 2020;6(1):17–34. [Google Scholar]
- 14. Sessitsch A, Hackl E, Wenzl P, et al. Diagnostic microbial microarrays in soil ecology. New Phytol. 2006;171(4):719–36. [DOI] [PubMed] [Google Scholar]
- 15. He Z, Gentry TJ, Schadt CW, et al. GeoChip: A comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1(1):67–77. [DOI] [PubMed] [Google Scholar]
- 16. Lee YJ, Van Nostrand JD, Tu Q, et al. The PathoChip, a functional gene array for assessing pathogenic properties of diverse microbial communities. ISME J. 2013;7(10):1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou A, He Z, Qin Y, et al. StressChip as a high-throughput tool for assessing microbial community responses to environmental stresses. Environ Sci Technol. 2013;47(17):9841–9. [DOI] [PubMed] [Google Scholar]
- 18. Abot A, Arnal G, Auer L, et al. CAZyChip: Dynamic assessment of exploration of glycoside hydrolases in microbial ecosystems. BMC Genomics. 2016;17(1):671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tedersoo L, Drenkhan R, Anslan S, et al. High-throughput identification and diagnostics of pathogens and pests: Overview and practical recommendations. Mol Ecol Resour. 2019;19(1):47–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franzosa Ea, Hsu T, Sirota-Madi A, et al. Sequencing and beyond: Integrating molecular “omics” for microbial community profiling. Nat Rev Microbiol. 2015;13(6):360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Segata N, Boernigen D, Tickle TL, et al. Computational meta'omics for microbial community studies. Mol Syst Biol. 2013;9:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frioux C, Singh D, Korcsmaros T, et al. From bag-of-genes to bag-of-genomes: Metabolic modelling of communities in the era of metagenome-assembled genomes. Comput Struct Biotechnol J. 2020;18:1722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fierer N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15(10):579–90. [DOI] [PubMed] [Google Scholar]
- 24. Martiny JBH, Jones SE, Lennon JT, et al. Microbiomes in light of traits: A phylogenetic perspective. Science. 2015;350(6261):doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 25. Penton CR, Johnson TA, Quensen JF, et al. Functional genes to assess nitrogen cycling and aromatic hydrocarbon degradation: Primers and processing matter. Front Microbiol. 2013;4:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hannula SE, van Veen JA. Primer sets developed for functional genes reveal shifts in functionality of fungal community in soils. Front Microbiol. 2016;7:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barbi F, Bragalini C, Vallon L, et al. PCR primers to study the diversity of expressed fungal genes encoding lignocellulolytic enzymes in soils using high-throughput sequencing. PLoS One. 2014;9(12):e116264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fish JA, Chai B, Wang Q, et al. FunGene: The functional gene pipeline and repository. Front Microbiol. 2013;4:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Angel R, Nepel M, Panhölzl C, et al. Evaluation of primers targeting the diazotroph functional gene and development of NifMAP - A bioinformatics pipeline for analyzing nifH amplicon data. Front Microbiol. 2018;9:doi: 10.3389/fmicb.2018.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ortiz-Estrada ÁM, Gollas-Galván T, Martínez-Córdova LR, et al. Predictive functional profiles using metagenomic 16S rRNA data: A novel approach to understanding the microbial ecology of aquaculture systems. Rev Aquac. 2019;11(1):234–45. [Google Scholar]
- 31. Bahram M, Hildebrand F, Forslund SK, et al. Structure and function of the global topsoil microbiome. Nature. 2018;560(7717):233–7. [DOI] [PubMed] [Google Scholar]
- 32. Hahn AS, Konwar KM, Louca S, et al. The information science of microbial ecology. Curr Opin Microbiol. 2016;31:209–16. [DOI] [PubMed] [Google Scholar]
- 33. Farley SS, Dawson A, Goring SJ, et al. Situating ecology as a big-data science: Current advances, challenges, and solutions. Bioscience. 2018;68(8):563–76. [Google Scholar]
- 34. Kembel SW, Cowan PD, Helmus MR, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26(11):1463–4. [DOI] [PubMed] [Google Scholar]
- 35. LPSN - List of Prokaryotic names with Standing in Nomenclature. http://www.bacterio.net. Accessed 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Louca S, Doebeli M. Efficient comparative phylogenetics on large trees. Bioinformatics. 2018;34(6):1053–5. [DOI] [PubMed] [Google Scholar]
- 37. MYCOBANK Database. http://www.mycobank.org. Accessed 2021. [Google Scholar]
- 38. Walters KE, Martiny JBH. Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS One. 2020;15(9):e0233872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caspi R, Billington R, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Starke R, Capek P, Morais D, et al. The total microbiome functions in bacteria and fungi. J Proteomics. 2020;213:103623. [DOI] [PubMed] [Google Scholar]
- 43. Commichaux S, Shah N, Ghurye J, et al. A critical assessment of gene catalogs for metagenomic analysis. Bioinformatics. 2021;37(18):2848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Y, Coventry DR, Gupta V, et al. The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 2020;21(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baldrian P. The known and the unknown in soil microbial ecology. FEMS Microbiol Ecol. 2019;95(2):doi: 10.1093/femsec/fiz005. [DOI] [PubMed] [Google Scholar]
- 46. Blondel J. Guilds or functional groups: Does it matter?. Oikos. 2003;100(2):223–31. [Google Scholar]
- 47. Mlambo MC. Not all traits are “functional”: Insights from taxonomy and biodiversity-ecosystem functioning research. Biodivers Conserv. 2014;23(3):781–90. [Google Scholar]
- 48. Volaire F, Gleason SM, Delzon S. What do you mean “functional” in ecology? Patterns versus processes. Ecol Evol. 2020;10(21):11875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Escalas A, Hale L, Voordeckers JW, et al. Microbial functional diversity: From concepts to applications. Ecol Evol. 2019;9(20):12000–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malik AA, Martiny JBH, Brodie EL, et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J. 2020;14:doi: 10.1038/s41396-019-0510-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Madin JS, Nielsen DA, Brbic M, et al. A synthesis of bacterial and archaeal phenotypic trait data. Sci Data. 2020;7(1):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lajoie G, Kembel SW. Making the most of trait-based approaches for microbial ecology. Trends Microbiol. 2019;27(10):814–23. [DOI] [PubMed] [Google Scholar]
- 53. Reimer LC, Söhngen C, Vetcininova A, et al. Mobilization and integration of bacterial phenotypic data—Enabling next generation biodiversity analysis through the Bac Dive metadatabase. J Biotechnol. 2017;261:187–93. [DOI] [PubMed] [Google Scholar]
- 54. Endara L, Cui H, Burleigh JG. Extraction of phenotypic traits from taxonomic descriptions for the tree of life using natural language processing. Appl Plant Sci. 2018;6(3):e1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim KMK, Li C, Chng KR, et al. @MInter: Automated text-mining of microbial interactions. Bioinformatics. 2016;32(19):2981–7. [DOI] [PubMed] [Google Scholar]
- 56. Chaix E, Deléger L, Bossy R, et al. Text mining tools for extracting information about microbial biodiversity in food. Food Microbiol. 2019;81:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chibucos MC, Zweifel AE, Herrera JC, et al. An ontology for microbial phenotypes. BMC Microbiol. 2014;14(1):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brbić M, Piškorec M, Vidulin V, et al. Phenotype inference from text and genomic data. In: Altun Y, et al., ed. Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2017. Cham: Springer; 2017:373–7. [Google Scholar]
- 59. Protraits. http://protraits.irb.hr. Accessed 2021. [Google Scholar]
- 60. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nguyen NH, Song Z, Bates ST, et al. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–8. [Google Scholar]
- 62. Aßhauer KP, Wemheuer B, Daniel R, et al. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–7. [DOI] [PubMed] [Google Scholar]
- 64. Segata N, Huttenhower C. Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies. PLoS One. 2011;6(9):e24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21(1):108–10. [DOI] [PubMed] [Google Scholar]
- 66. Hartman WH, Ye R, Horwath WR, et al. A genomic perspective on stoichiometric regulation of soil carbon cycling. ISME J. 2017;11(12):2652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kembel SW, Wu M, Eisen JA, et al. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol. 2012;8(10):e1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Louca S, Doebeli M, Parfrey LW. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome. 2018;6(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Woloszynek S, Mell JC, Zhao Z, et al. Exploring thematic structure and predicted functionality of 16S rRNA amplicon data. PLoS One. 2019;14(12):e0219235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Douglas GM, Maffei VJ, Zaneveld JR, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bowman JS, Ducklow HW. Microbial communities can be described by metabolic structure: A general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic Peninsula. PLoS One. 2015;10(8):doi: 10.1371/journal.pone.0135868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria; 2020. https://www.r-project.org/. [Google Scholar]
- 74. Iwai S, Weinmaier T, Schmidt BL, et al. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11(11):e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mitchell K, Ronas J, Dao C, et al. PUMAA: A platform for accessible microbiome analysis in the undergraduate classroom. Front Microbiol. 2020;11:584699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zanne AE, Abarenkov K, Afkhami ME, et al. Fungal functional ecology: Bringing a trait-based approach to plant-associated fungi. Biol Rev. 2020;95(2):409–33. [DOI] [PubMed] [Google Scholar]
- 77. Põlme S, Abarenkov K, Henrik Nilsson R, et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020;105(1):1–16. [Google Scholar]
- 78. Fones HN, Bebber DP, Chaloner TM, et al. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat Food. 2020;1(6):332–42. [DOI] [PubMed] [Google Scholar]
- 79. Agerer R, Rambold G. DEEMY—An information system for characterization and determination of ectomycorrhizae. Mycorrhiza. 1997;7(2):113–6. [Google Scholar]
- 80. DEEMY An Information System for Characterization and DEtermination of EctoMYcorrhizae. http://www.deemy.de. Accessed 2021. [Google Scholar]
- 81. Barberán A, Caceres Velazquez H, Jones S, et al. Hiding in plain sight: Mining bacterial species records for phenotypic trait information. mSphere. 2017;2(4):doi: 10.1128/mSphere.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Barberan Albert. International Journal of Systematic and Evolutionary Microbiology (IJSEM) phenotypic database. figshare. Dataset. (2006). 10.6084/m9.figshare.4272392.v3. [DOI]
- 83. Reimer LC, Vetcininova A, Carbasse JS, et al. Bac Dive in 2019: Bacterial phenotypic data for high-throughput biodiversity analysis. Nucleic Acids Res. 2019;47(D1):D631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. BacDive: Explore Bacterial Diversity. https://bacdive.dsmz.de. Accessed 2021. [Google Scholar]
- 85. Engqvist MKM. Correlating enzyme annotations with a large set of microbial growth temperatures reveals metabolic adaptations to growth at diverse temperatures. BMC Microbiol. 2018;18(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nkongolo KK, Narendrula-Kotha R. Advances in monitoring soil microbial community dynamic and function. J Appl Genet. 2020;61(2):249–63. [DOI] [PubMed] [Google Scholar]
- 87. Jin T, Wang Y, Huang Y, et al. Taxonomic structure and functional association of foxtail millet root microbiome. Gigascience. 2017;6(10):doi: 10.1093/gigascience/gix089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lian T, Mu Y, Jin J, et al. Impact of intercropping on the coupling between soil microbial community structure, activity, and nutrient-use efficiencies. PeerJ. 2019;7:e6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sengupta A, Hariharan J, Grewal PS. et al. Bacterial community dissimilarity in soils is driven by long-term land-use practices. Agrosyst Geosci Environ. 2020;3(1):doi: 10.1002/agg2.20031. [DOI] [Google Scholar]
- 90. Lüneberg K, Schneider D, Siebe C, et al. Drylands soil bacterial community is affected by land use change and different irrigation practices in the Mezquital Valley, Mexico. Sci Rep. 2018;8(1):1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun S, Jones RB, Fodor AA. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome. 2020;8(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Koo H, Hakim JA, Morrow CD, et al. Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee, Antarctica. J Microbiol Methods. 2017;140:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7(4):830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. George PBL, Creer S, Griffiths RI, et al. Primer and database choice affect fungal functional but not biological diversity findings in a national soil survey. Front Environ Sci. 2019;7:doi: 10.3389/fenvs.2019.00173. [DOI] [Google Scholar]
- 95. Yang T, Adams JM, Shi Y, et al. Soil fungal diversity in natural grasslands of the Tibetan Plateau: Associations with plant diversity and productivity. New Phytol. 2017;215(2):756–65. [DOI] [PubMed] [Google Scholar]
- 96. Makiola A, Dickie IA, Holdaway RJ, et al. Land use is a determinant of plant pathogen alpha- but not beta-diversity. Mol Ecol. 2019;28(16):3786–98. [DOI] [PubMed] [Google Scholar]
- 97. Buscardo E, Souza RC, Meir P, et al. Effects of natural and experimental drought on soil fungi and biogeochemistry in an Amazon rain forest. Commun Earth Environ. 2021;2(1):55. [Google Scholar]
- 98. Liang M, Liu X, Parker IM, et al. Soil microbes drive phylogenetic diversity-productivity relationships in a subtropical forest. Sci Adv. 2019;5(10):eaax5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yang T, Tedersoo L, Soltis PS, et al. Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. ISME J. 2019;13(3):686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Brinkmann N, Schneider D, Sahner J, et al. Intensive tropical land use massively shifts soil fungal communities. Sci Rep. 2019;9(1):3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Egidi E, Delgado-Baquerizo M, Plett JM, et al. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun. 2019;10(1):2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Delgado-Baquerizo M, Guerra CA, Cano-Díaz C, et al. The proportion of soil-borne pathogens increases with warming at the global scale. Nat Clim Chang. 2020;10(6):550–4. [Google Scholar]
- 103. Větrovský T, Kohout P, Kopecký M, et al. A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat Commun. 2019;10(1):5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Öpik M, Davison J, Moora M, et al. DNA-based detection and identification of Glomeromycota: The virtual taxonomy of environmental sequences. Botany. 2014;92(2):135–47. [Google Scholar]
- 105. Berruti A, Desirò A, Visentin S, et al. ITS fungal barcoding primers versus 18S AMF-specific primers reveal similar AMF-based diversity patterns in roots and soils of three mountain vineyards. Environ Microbiol Rep. 2017;9(5):658–67. [DOI] [PubMed] [Google Scholar]
- 106. Anthony MA, Frey SD, Stinson KA. Fungal community homogenization, shift in dominant trophic guild, and appearance of novel taxa with biotic invasion. Ecosphere. 2017;8(9):e01951. [Google Scholar]
- 107. Sansupa C, Wahdan SFM, Hossen S, et al. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria?. Appl Sci. 2021;11(2):688. [Google Scholar]
- 108. Nilsson RH, Anslan S, Bahram M, et al. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat Rev Microbiol. 2019;17(2):95–109. [DOI] [PubMed] [Google Scholar]
- 109. Comeau AM, Douglas GM, Langille MGI. Microbiome Helper: A custom and streamlined workflow for microbiome research. mSystems. 2017;2(1):doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Piper AM, Batovska J, Cogan NOI, et al. Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. Gigascience. 2019;8(8):doi: 10.1093/gigascience/giz092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3(9):679–87. [DOI] [PubMed] [Google Scholar]
- 112. Douglas GM, Langille MGI. Current and promising approaches to identify horizontal gene transfer events in metagenomes. Genome Biol Evol. 2019;11(10):2750–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Seiler E, Trappe K, Renard BY. Where did you come from, where did you go: Refining metagenomic analysis tools for horizontal gene transfer characterisation. PLoS Comput Biol. 2019;15(7):e1007208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. van Dijk B, Hogeweg P, Doekes HM, et al. Slightly beneficial genes are retained by bacteria evolving DNA uptake despite selfish elements. Elife. 2020;9:e56801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79(2):243–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Smalla K, Jechalke S, Top EM. Plasmid detection, characterization, and ecology. Microbiol Spectr. 2015;3(1):doi: 10.1128/microbiolspec.PLAS-0038-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dunivin TK, Choi J, Howe A, et al. RefSoil+: A reference database for genes and traits of soil plasmids. mSystems. 2019;4(1):doi: 10.1128/mSystems.00349-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol. 2011;2:doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Brito IL. Examining horizontal gene transfer in microbial communities. Nat Rev Microbiol. 2021;19(7):442–53. [DOI] [PubMed] [Google Scholar]
- 120. Banos S, Lentendu G, Kopf A, et al. A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol. 2018;18(1):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bukin YS, Galachyants YP, Morozov IV, et al. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci Data. 2019;6(1):190007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Xu Z, Malmer D, Langille MGI, et al. Which is more important for classifying microbial communities: Who's there or what they can do?. ISME J. 2014;8(12):2357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Karst SM, Dueholm MS, McIlroy SJ, et al. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol. 2018;36(2):190–5. [DOI] [PubMed] [Google Scholar]
- 124. Tedersoo L, Anslan S. Towards PacBio-based pan-eukaryote metabarcoding using full-length ITS sequences. Environ Microbiol Rep. 2019;11(5):659–68. [DOI] [PubMed] [Google Scholar]
- 125. Johnson JS, Spakowicz DJ, Hong B-Y, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Philippot L, Andersson SGE, Battin TJ, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8(7):523–9. [DOI] [PubMed] [Google Scholar]
- 127. Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88(6):1354–64. [DOI] [PubMed] [Google Scholar]
- 128. Callahan BJ, Wong J, Heiner C, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47(18):e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Feldbauer R, Schulz F, Horn M, et al. Prediction of microbial phenotypes based on comparative genomics. BMC Bioinformatics. 2015;16:doi: 10.1186/1471-2105-16-S14-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Weimann A, Mooren K, Frank J, et al. From genomes to phenotypes: Traitar, the microbial trait analyzer. mSystems. 2016;1(6):043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Goberna M, Verdú M. Predicting microbial traits with phylogenies. ISME J. 2016;10(4):959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Levatić J, Brbić M, Perdih TS, et al. Phenotype prediction with semi-supervised classification trees. In: Appice A, Loglisci C, Manco Geds. New Frontiers in Mining Complex Patterns. NFMCP 2017. Cham: Springer; 2018:doi: 10.1007/978-3-319-78680-3_10. [DOI] [Google Scholar]
- 133. Zanne AE, Powell JR, Flores-Moreno H, et al. Finding fungal ecological strategies: Is recycling an option?. Fungal Ecol. 2020;46:100902. [Google Scholar]
- 134. Martiny AC. High proportions of bacteria are culturable across major biomes. ISME J. 2019;13:2125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wemheuer F, Taylor JA, Daniel R, et al. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiome. 2020;15(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Cheifet B. Where is genomics going next?. Genome Biol. 2019;20(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Piro VC, Dadi TH, Seiler E, et al. ganon: Precise metagenomics classification against large and up-to-date sets of reference sequences. Bioinformatics. 2020;36(Supplement_1):i12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Choi J, Yang F, Stepanauskas R, et al. Strategies to improve reference databases for soil microbiomes. ISME J. 2017;11(4):829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. RefSoil: a repository provide the list of reference soil. https://github.com/germs-lab/ref_soil. Accessed 2021. [Google Scholar]
- 140. Louca S, Jacques SMS, Pires APF, et al. High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol. 2017;1(1):15. [DOI] [PubMed] [Google Scholar]
- 141. Nagpal S, Haque MM, Singh R, et al. iVikodak—A platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front Microbiol. 2019;9:doi: 10.3389/fmicb.2018.03336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Grigoriev IV, Nikitin R, Haridas S, et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(D1):D699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bonkowski M, Dumack K, Fiore-Donno AM. The protists in soil—A token of untold eukaryotic diversity. In: van Elsas JD, Trevors JT, Rosado AS et al., et al., eds. Modern Soil Microbiology, third ed. Boca Raton: CRC; 2019:125–40. [Google Scholar]
- 144. Xiong W, Jousset A, Guo S, et al. Soil protist communities form a dynamic hub in the soil microbiome. ISME J. 2018;12(2):634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Fiore-Donno AM, Richter-Heitmann T, Degrune F, et al. Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate grassland. Front Microbiol. 2019;10:doi: 10.3389/fmicb.2019.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Delgado-Baquerizo M, Trivedi P, Trivedi C, et al. Microbial richness and composition independently drive soil multifunctionality. Funct Ecol. 2017;31(12):2330–43. [Google Scholar]
- 147. Terrat S, Horrigue W, Dequietd S, et al. Mapping and predictive variations of soil bacterial richness across France. PLoS One. 2017;12:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Schloter M, Nannipieri P, Sørensen SJ, et al. Microbial indicators for soil quality. Biol Fertil Soils. 2018;54:1–10. [Google Scholar]
- 149. Hariharan J, Sengupta A, Grewal P, et al. Functional predictions of microbial communities in soil as affected by long-term tillage practices. Agric Environ Lett. 2017;2(1):170031. [Google Scholar]
- 150. Manor O, Borenstein E. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe. 2017;21(2):254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang Q, Zhao H, Wu D, et al. A comprehensive analysis of the microbiota composition and gene expression in colorectal cancer. BMC Microbiol. 2020;20(1):308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhang L, Liu Y, Zheng HJ, et al. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2020;9:doi: 10.3389/fcimb.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ji P, Rhoads WJ, Edwards MA, et al. Impact of water heater temperature setting and water use frequency on the building plumbing microbiome. ISME J. 2017;11(6):1318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Maguvu TE, Bezuidenhout CC, Kritzinger R, et al. Combining physicochemical properties and microbiome data to evaluate the water quality of South African drinking water production plants. PLoS One. 2020;15(8):e0237335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ramirez KS, Döring M, Eisenhauer N, et al. Toward a global platform for linking soil biodiversity data. Front Ecol Evol. 2015;3:doi: 10.3389/fevo.2015.00091. [DOI] [Google Scholar]
- 156. Raguideau S, Plancade S, Pons N, et al. Inferring aggregated functional traits from metagenomic data using constrained non-negative matrix factorization: Application to fiber degradation in the human gut microbiota. PLoS Comput Biol. 2016;12(12):e1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Fierer N, Barberán A, Laughlin DC. Seeing the forest for the genes: Using metagenomics to infer the aggregated traits of microbial communities. Front Microbiol. 2014;5:doi: 10.3389/fmicb.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Weisskopf L, Schulz S, Garbeva P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat Rev Microbiol. 2021;19(6):391–404. [DOI] [PubMed] [Google Scholar]
- 159. Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105(Suppl 1):11512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Navarro LM, Fernández N, Guerra C, et al. Monitoring biodiversity change through effective global coordination. Curr Opin Environ Sustain. 2017;29:158–69. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Barberan Albert. International Journal of Systematic and Evolutionary Microbiology (IJSEM) phenotypic database. figshare. Dataset. (2006). 10.6084/m9.figshare.4272392.v3. [DOI]
Supplementary Materials
Trevor Charles -- 10/31/2021 Reviewed
Axel Künsnter, PhD -- 11/2/2021 Reviewed
Data Availability Statement
Not applicable.