Abstract
Ferroptosis is a novel form of regulated cell death characterized by accumulated lipid reactive oxygen species (ROS) and inactivation of glutathione peroxidase 4 (GPX4). The present study aimed to investigate the role of microRNA (miRNA/miR)-15a in ferroptosis of prostate cancer cells. Bioinformatics analysis was performed to predict the potential interaction between miR-15a and the 3′-untranslated region (UTR) of GPX4 mRNA. The prostate cancer cell line, LNCAP was transfected with miR-15a mimics or small interfering (si)-GPX4. Reverse transcription-quantitative PCR and western blot analyses were performed to detect the mRNA and protein expression levels of GPX4, respectively. Biotin-RNA pull-down and dual-luciferase reporter assays were performed to verify the interaction between miR-15a and GPX4 mRNA. The Cell Counting Kit-8 assay was performed to assess cell proliferation, while lactate dehydrogenase (LDH) and intracellular ferrous iron levels were detected via ELISA. Lipid ROS and mitochondrial membrane potential (MMP) were assessed via flow cytometry and staining with C11-BIODIPY probes or JC-1. Furthermore, lipid peroxidation was identified by measuring malondialdehyde (MDA) levels. The results demonstrated that transfection with miR-15a mimics decreased GPX4 protein expression. Bioinformatics analysis revealed potential binding sites between miR-15a and the 3′-UTR region of GPX4, and RNA pull-down and the dual-luciferase reporter assays further confirmed the interaction between miR-15a and GPX4 mRNA. Both transfection with miR-15a mimics and si-GPX4 suppressed cell proliferation, elevated LDH release, accumulated intracellular ferrous iron and ROS, disrupted MMP and increased MDA levels. Taken together, the results of the present study suggest miR-15a induces ferroptosis by regulating GPX4 in prostate cancer cells, which provides evidence for investigating the therapeutic strategies of prostate cancer.
Keywords: miR-15a, GPX4, prostate cancer, ferroptosis
Introduction
Prostate cancer is a malignancy that arises from the lateral posterior lobes or glands of the prostate, which seriously threatens the health of those affected (1). Over the past decades, the therapeutic strategies for prostate cancer have made significant progression, particularly androgen-deprivation therapy (2). However, the acquired resistance to medicines targeting androgen receptor has proved a major challenge in effectively treating prostate cancer. Thus, it is important to identify novel therapeutic targets for prostate cancer (2).
Ferroptosis is a newly established non-apoptotic form of regulated cell death caused by failed regulation of glutathione-dependent lipid-peroxide-scavenging (3). Ferroptosis is biochemically characterized by generation of intracellular ferrous iron (Fe2+), elevated lipid peroxidation and accumulated lethal reactive oxygen species (ROS) (3). Ferroptosis plays an important role in regulating different types of cancer, including triple-negative breast cancer, renal cancer and gastric cancer (3). For example, cysteine dioxygenase 1 suppresses gastric cancer progression by inducing ferroptosis of gastric cancer cells (4). In addition, tumor suppressor BRCA1-associated protein 1 inhibits the development of renal cancer by inducing ferroptosis via upregulation of Solute carrier family 7 member 11 (5). However, the role and regulatory mechanisms of ferroptosis in prostate cancer remain unclear.
Several enzymes play critical roles during ferroptosis, of which glutathione peroxidase 4 (GPX4) has been well studied (6–10). GPX4 is an antioxidant enzyme that downregulates lipid hydroperoxides and protects cells from damage of accumulated ROS, and it is considered a suppressor of ferroptosis (7). Increasing evidence suggest that GPX4 plays an important role in tumors (6,8). A recent study revealed that RSL3 induces cell death in colorectal cancer cells by triggering ferroptosis via the inactivation of GPX4 (9). In addition, GPX4 contributes to the vulnerability of ferroptosis in clear-cell carcinomas (CCCs), and thus is a therapeutic target in CCCs (10). However, the regulatory mechanisms of GPX4 in prostate cancer remain unknown.
Various genes are involved in tumorigenesis. For example, mutations of acetyl-CoA carboxylase alpha and DEP domain containing MTOR interacting protein are associated with lung cancer (11). MicroRNAs (miRNAs/miRs) are a principal member of non-coding RNAs, defined as highly conserved short RNAs, 21–25 nucleotides in length (12). In the past decades, the functions of miRNAs have been widely studied in various diseases and pathological processes, particularly in cancers (13–15). However, only a few miRNAs, including miR-9, miR-137 and miR-522 have been identified to be associated with ferroptosis (16–19). miR-15a is a widely studied miRNA in cancers, including prostate cancer (20,21). A bioinformatics study revealed that miR-15a expression was notably downregulated in blood samples of patients with prostate cancer compared with healthy subjects, suggesting that circulating miR-15a may be a potential diagnostic biomarker for prostate cancer (22). Jin et al (23) and Bonci et al (24) reported that miR-15a functions as a tumor suppressor to inhibit the proliferation and invasion of prostate cancer cells by downregulating Wnt/β-catenin and TGF-β signaling.
The present study aimed to investigate the function of miR-15a on ferroptotic cell death of prostate cancer cells via regulation of GPX4 to determine whether miR-15a and GPX4 may be used as potential therapeutic targets for prostate cancer.
Materials and methods
Cell culture
The human prostate cancer cell line, LNCAP was purchased from the American Type Culture Collection and maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin, at 37°C with 5% CO2.
Target prediction
Targetscan (http://www.targetscan.org) was used to predict the binding sites between miRNA-15a and the 3′-untranslated region (UTR) of GPX4 mRNA.
Cell transfection
miR-15a mimics, miR-15a inhibitor (anti-miR-15a) and negative control (NC), small interfering (si)-GPX4 and si-NC were designed and purchased from Guangzhou RiboBio Co., Ltd. The following sequences were used: MiR-15a mimics, 5′-UAGCAGCACAUAAUGGUUUGUG-3′; miR-15a inhibitor, 5′-CACAAACCAUUAUGUGCUGCUA-3′; mimic NC, 5′-UUUGUACUACACAAAAGUACUG-3′; inhibitor NC, 5′-UCUACUCUUUCUAGGAGGUUGUGA-3′; si-GPX4 forward, 5′-GUGGAUGAAGAUCCAACCCdTdT-3′ and reverse, 3′-GGGUUGGAUCUUCAUCCACdTdT-5′; si-NC forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 3′-ACGUGACACGUUCGGAGAATT-5′. LNCAP cells were seeded into 6-well plates at a density of 1×105 cells/well and incubated overnight to achieve monolayer confluence. Following starvation with serum-free medium for 12 h, cells were transfected with miR-15a mimics or si-GPX4 at 50 nmol/l using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions and cultured at 37°C for 48 h, after which subsequent experiments were performed.
Reverse transcription-quantitative (RT-qPCR)
Total RNA was extracted from LNCAP cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Total RNA was reverse transcribed into cDNA using the cDNA synthesis kit (Takara Bio, Inc.). The reverse transcribed reaction was performed as 42°C for 60 min and 85°C for 5 min. qPCR was subsequently performed using the SYBR Green Super Mix kit (BD Biosciences) and the thermocycling conditions were as follows: 40 Cycles of 95°C for 30 sec, 95°C for 15 sec and 60°C for 15 sec. The following primer sequences were used for qPCR: U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; miR-15a forward, 5′-ACACTCCAGCTGGGTAGCAGCACATAATGGTTTG-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; GAPDH forward, 5′-AAGAAGGTGGTGAAGCAGGC-3′ and reverse, 5′-TCCACCACCCAGTTGCTGTA-3′; and GXP4 forward, 5′-GCCGGGACCATGTGCGCGTC-3′ and reverse, 5′-CAGGATCCGCAAACCACACTC-3′. Small endogenous nucleolar U6 snRNA and GAPDH were used as internal controls for normalization of miR-15a and GXP4 mRNA expression levels, respectively. Relative expression levels were calculated using the 2−ΔΔCq method (17).
Western blotting
Following transfection, LNCAP cells were lysed in 1× ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology), centrifuged at 13,000 × g at 4°C for 10 min, and the deposit was discarded. Protein concentration was determined using the BCA kit (Takara Biotechnology, Inc.). A total of 35 ug protein was determined via 10% SDS-PAGE, transferred onto PVDF membranes and blocked with 5% non-fat milk dissolved in 1×TBST for 1 h at room temperature. The membranes were incubated with primary antibodies against GPX4 (1:1,000; cat. no. ab252833; Abcam) and GAPDH (1:1,000; cat. no. ab8245; Abcam) overnight at 4°C. Following the primary incubation, membranes were incubated with corresponding secondary antibodies including goat anti-rabbit IgG H&L (HRP; 1:20,000; cat. no. ab6721; Abcam) and rabbit anti-mouse IgG H&L (HRP; 1:20,000; cat. no. ab6728; Abcam) for 2 h at room temperature. Protein bands were visualized using the ECL kit (Invitrogen; Thermo Fisher Scientific, Inc.) and captured using the Gel imaging system (BD Biosciences). All antibodies were purchased from Abcam and diluted according to the manufacturer's instructions.
Biotin RNA pull-down assay
Biotin-labelled wild-type (WT) miR-15a (Bio-miR-15a-WT), biotin-labelled mutated (MUT) miR-15a (Bio-miR-15a-MUT) and biotin-labelled NC (Bio-NC) were purchased from Guangzhou RiboBio Co., Ltd. The sequences were as follows: Bio-miR-15a-WT, 5′-Bio-UAGCAGCACAUAAUGGUUUGUG-3′; Bio-miR-15a-MUT, 5′-Bio-UGAUGAUGCAUAAUGGUUUGUG-3′; and Bio-NC, 5′-Bio-UUUGUACUACACAAAAGUACUG-3′. Next, RNA pull-down was performed by Pierce™ Magnetic RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. Briefly, LNCAP cells were transfected with Bio-miR-15a-WT, Bio-miR-15a-MUT or the NC. Following transfection for 48 h at 37°C, cells were lysed and whole cell extractions were collected by centrifuging at 13,000 × g at 4°C for 10 min. Subsequently, 100 µl lysates were incubated with Streptavidin Magnetic Beads (MedChemExpress) overnight at 4°C according to manufacturer's protocol. The biotinylated nucleic acids coated beads were subsequently diluted using magnetic to separate the co-precipitated RNA. The samples were purified, reverse transcribed into cDNA and detected via RT-qPCR analysis.
Dual-luciferase reporter assay
The sequence of GPX4 mRNA (NM_002085.5) was obtained from GenBank of The National Center for Biotechnology Information. The WT complete sequence and MUT binding sites of miR-15a complete sequence of GPX4 3′-UTR were subcloned into the psiCHECK™−2 vector (Promega Corporation) and named as GPX4-UTR WT and GPX4-UTR MUT, respectively. LNCAP cells were transfected with GPX4-UTR WT or GPX4-UTR MUT and miR-15a mimics, anti-miR-15a or mi-NC using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Cells were lysed and the luciferase activities were measured using the dual-luciferase reporter assay kit (Promega Corporation) 48 h post-transfection, which was normalized by comparing with Renilla luciferase activity.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay (Abcam) was performed to assess the proliferation of LNCAP cells. Cells were digested and seeded into 96-well plates at a density of 1×103 cells/well and transfected with mi-NC, miR-15a mimics, si-NC, siGPX4 or treated with normal DMEM (Gibco; Thermo Fisher Scientific, Inc.) as the control, respectively. CCK-8 solution (10 µl) was added to each well and further incubated for 2 h. Absorbance was measured at a wavelength of 450 nm using an absorbance reader (Multiscan MK3; Thermo Fisher Scientific, Inc.). The inhibitory ratio of each group was calculated as follows: (1-average absorbance value ÷ average absorbance value of control group) ×100%.
Lactate dehydrogenase (LDH) detection
LDH is considered a cell damage biomarker (4). First, 1×105 cells were seeded per well of the 6-well plates and transfected with mi-NC, miR-15a mimics, si-NC, siGPX4 and siGPX4. The levels of LDH were detected using the L-Lactate Dehydrogenase (Crude Enzyme) kit (cat. no. P3532-100 ml) purchased from Beyotime Institute of Biotechnology according to the manufacturer's instructions.
Iron assay
LNCAP cells were transfected with mi-NC, miR-15a mimics, si-NC and siGPX4 for 48 h at 37°C and then the intracellular ferrous iron (Fe2+) level was determined using the Iron Assay kit (Abcam), according to the manufacturer's instructions. Cells were collected and washed twice in PBS, and subsequently homogenized with buffer in the kit. The supernatant was collected and incubated with iron reducer at room temperature for 30 min. Next, the supernatant was mixed with iron probe and incubated at room temperature for 1 h. Absorbance was measured at a wavelength of 593 nm using a Multiscan MK3 absorbance reader (Thermo Fisher Scientific, Inc.).
ROS activity assay
Cells were seeded into 6-well plates and transfected with mi-NC, miR-15a mimics, si-NC and siGPX4. Following the culture at 37°C for 48 h, cells were stained with fresh medium containing C11-BODIPY dye (Beyotime Institute of Biotechnology) at 37°C for 20 min in culture incubator. Next, cells were collected, washed and suspended in PBS. The cell suspension was detected using a FACSC Calibur (BD Biosciences).
Evaluation of the mitochondrial membrane potential (MMP)
LNCAP cells were transfected with mi-NC, miR-15a mimics, si-NC and siGPX4 at 37°C for 48 h, and their MMP was subsequently analyzed via staining using the JC-1 Assay kit (Beyotime Institute of Biotechnology). Results were detected using the FACSC Calibur (BD Biosciences), according to the manufacturer's instructions.
Lipid peroxidation detection
LNCAP cells were lysed with 1X ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology), centrifuged at 13,000 × g at 4°C for 10 min and the deposit was discarded. The supernatant was used to detect lipid peroxidation using the Lipid Peroxidation MDA Assay kit (Beyotime Institute of Biotechnology), according to the manufacturer's instructions.
Statistical analysis
Data are presented as the mean ± SD. Analysis of statistical differences were performed using SPSS 20 software (IBM Corp.) Unpaired Student's t-test was used to compare differences between two groups, while one-way ANOVA followed by Tukey's post hoc test was used to compare differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference. Each experiment was performed three times.
Results
miR-15a affects GPX4 expression in prostate cancer cells
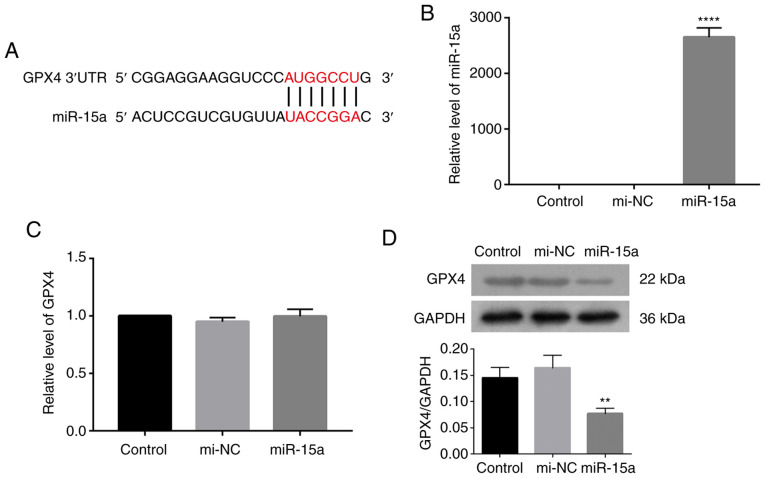
GPX4 plays a critical role in ferroptosis, whereby its suppression is considered a characteristic of ferroptosis (4–7). To determine the role of miR-15a in prostate cancer, Targetscan was used to predict the potential targets of miR-15a. The results revealed a potential binding between miR-15a and GPX4 (Fig. 1A). The present study aimed to investigate the direct regulation of miR-15a to GPX4 by transfecting LNCAP cells with miR-15a mimics. Transfection efficiency of miR-15a mimics was assessed via RT-qPCR analysis. Results indicated that transfection of miR-15a mimics significantly upregulated miR-15a level in LNCAP cells compared with that of LNCAP cells transfected with mimic controls (Fig. 1B). As presented in Fig. 1C and D, transfection with miR-15a mimics decreased GPX4 protein expression, while little change was observed at the mRNA level. Taken together, these results suggest that miR-15a may regulate GPX4 expression at a translational level.
Figure 1.
miR-15a affects GPX4 expression in prostate cancer cells. (A) Targetscan was used to predict the potential binding between miR-15a and GPX4. (B) RT-qPCR analysis was performed to assess transfection efficiency of miR-15a mimics and mi-NC in LNCAP cells. (C) RT-qPCR and (D) western blot analyses were performed to detect GPX4 mRNA and protein expression levels following transfection with miR-15a mimics, respectively. The experiments were performed in triplicate. **P<0.01 vs. control group; ****P<0.0001 vs. control group. mir, microRNA; GPX4, glutathione peroxidase 4; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; UTR, untranslated region.
Validation of direct interaction between miR-15a and GPX4
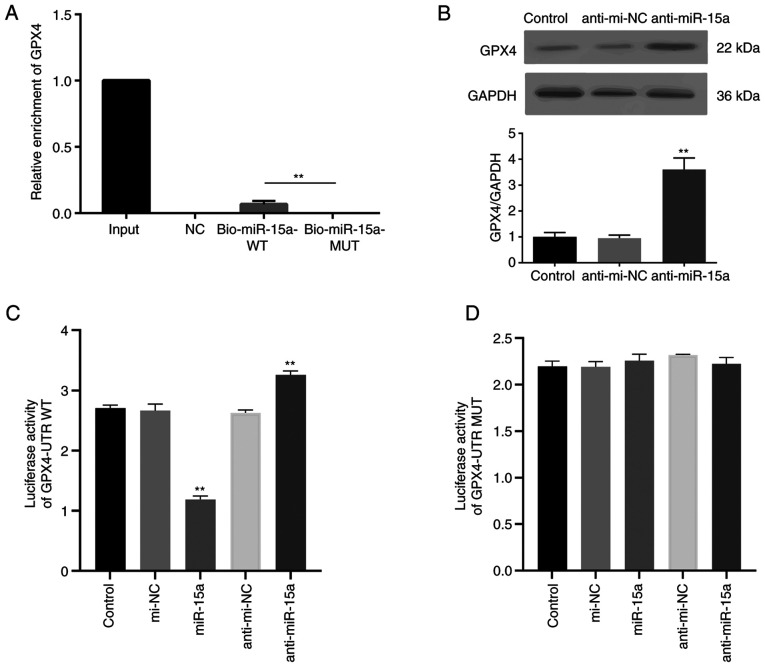
The direct interaction between miR-15a and GPX4 was assessed. Bio-miR-15a-WT, Bio-miR-15a-MUT and Bio-NC were transfected into LNCAP cells, and the enrichment of GPPX4 was assessed. As presented in Fig. 2A, GPX4 mRNA was detected in the WT miR-15a group but not the MUT group. To further identify the regulatory effect of miR-15a on GPX4, miR-15a inhibitor (anti-miR-15a) were used. Usually, miRNA inhibitors do not decrease miRNA expression (18,20). The results of the present study demonstrated that transfection with miR-15a inhibitor did not decrease miR-15a expression (Fig. S1). MiRNA inhibitors can competitively bind with miRNA to suppress the association between miRNA and target mRNA, and reverse the inhibitory effect of miRNA on the translation of the target mRNA (18,20). As presented in Fig. 2B, transfection with miR-15a inhibitor notably increased GPX4 protein expression. The dual-luciferase reporter assay was performed with transfection of plasmids containing GPX4-UTR WT or GPX4-UTR MUT miR-15a binding sequences of GPX4 mRNA 3′-UTR. The luciferase activity of the GPX4-UTR WT group decreased following co-transfection with miR-15a mimics, the effects of which were reversed following transfection with anti-miR-15a (Fig. 2C). However, the luciferase activity of the GPX4-UTR MUT group did not change following transfection (Fig. 2D). Collectively, these results highlight the direct regulatory role of miR-15a on GPX4.
Figure 2.
miR-15a directly interacts with the 3′-UTR of GPX4. (A) The biotin RNA pull-down assay was performed to assess the enrichment of GPX4 following transfection with Bio-miR-15a-WT, Bio-miR-15a-MUT or Bio-NC. (B) Western blot analysis was performed to detect GPX4 protein expression following transfection with anti-miR-15a inhibitor. Luciferase activity of reporter plasmids containing (C) GPX4-UTR WT or (D) GPX4-UTR MUT miR-15a binding sequences of GPX4 3′-UTR, co-transfected with miR-15a mimics, anti-miR-15a inhibitor and the respective NCs. The experiments were performed three times. **P<0.01 vs. control or Bio-miR-15a-WT group. miR, microRNA; UTR, untranslated region; GPX4, glutathione peroxidase 4; WT, wild-type; MUT, mutant; NC, negative control.
miR-15a inhibits prostate cancer cell proliferation and causes cell death via GPX4
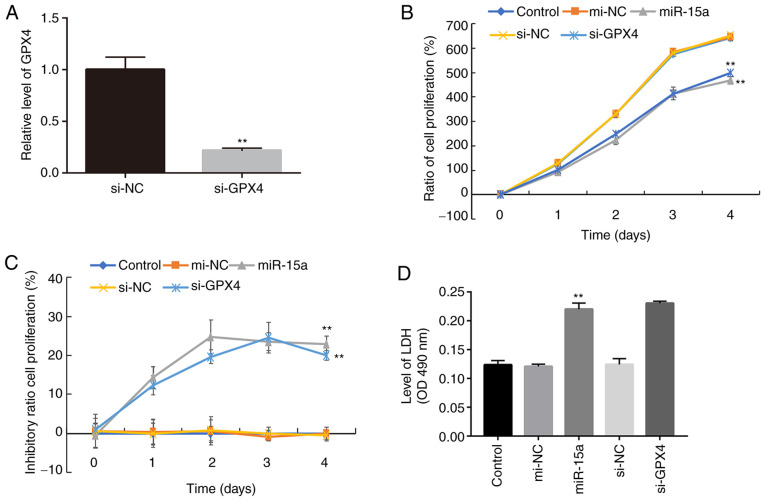
To further elucidate the role of miR-15a and GPX4 in prostate cancer cells, cell proliferation was assessed following transfection with miR-15a mimics or si-GPX4. The results demonstrated that transfection with si-GPX4 significantly decreased GPX4 mRNA expression in LNCAP cells (Fig. 3A). In addition, the results of the CCK-8 assay demonstrated a significant decline in the proliferation of cells transfected with miR-15a mimics or si-GPX4 compared with the control groups (Fig. 3B). Similar inhibitory effects were observed in the miR-15 mimics and si-GPX4 groups (Fig. 3C), further confirming that miR-15a functions by regulating GPX4.
Figure 3.
miR-15a inhibits the proliferation of prostate cancer cells by regulating GPX4. (A) Reverse transcription-quantitative PCR analysis was performed to detect GPX4 mRNA expression in LNCAP cells transfected with or without si-GPX4. LNCAP cells were transfected with miR-15a mimics, si-GPX4 or NCs and the (B) Cell Counting Kit-8 assay was performed to assess cell proliferation and the (C) inhibitory ratio. (D) LDH levels in the supernatant of cultured cells were measured. The experiments were performed in triplicate. **P<0.01 vs. control or si-NC group. miR, microRNA; GPX4, glutathione peroxidase 4; si, small interfering; NC, negative control; LDH, lactate dehydrogenase; OD, optical density.
Ferroptosis is a novel type of necrosis, characterized by destruction of the cell membrane structure and the release of LDH (4). Thus, the present study detected LDH levels in the culture medium. As presented in Fig. 3D, transfection with miR-15a mimics and si-GPX4 significantly increased LDH levels, which is associated with elevated cell death (4).
miR-15a induces prostate cancer cell ferroptosis via GPX4
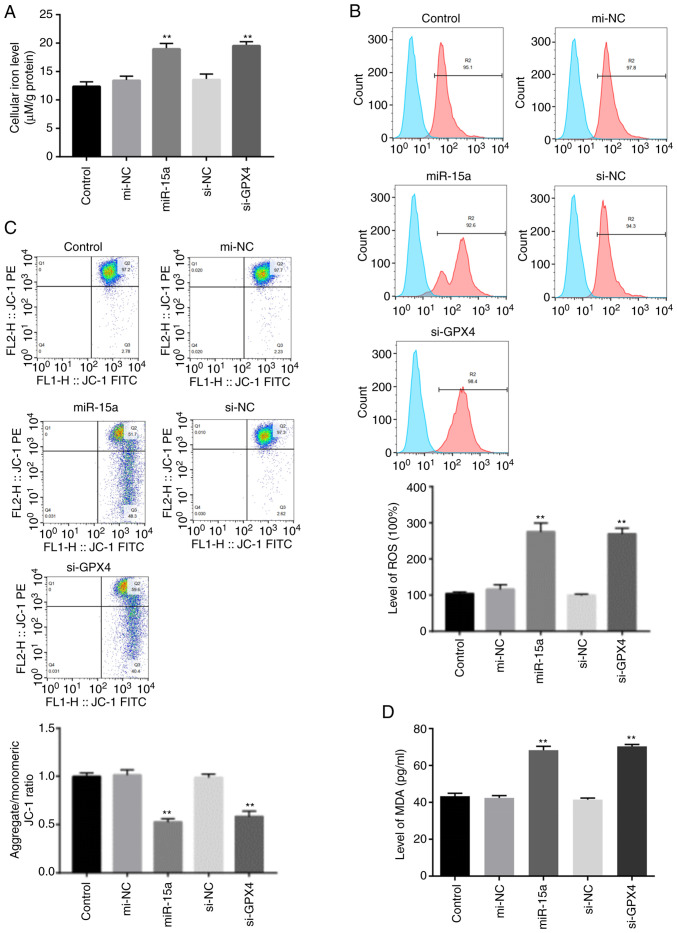
Ferroptosis is characterized by elevated lipid peroxidation, disrupted mitochondrial membrane density and accumulation of intracellular iron (3,4,9). Thus, the present study assessed the accumulation of redox active metal Fe2+, ROS, integrity of mitochondrial membrane and lipid peroxidation. The levels of intracellular Fe+ and lipid ROS accumulation notably increased in LNCAP cells transfected with miR-15a mimics or si-GPX4 (Fig. 4A and B). In addition, the present study detected the integrity of mitochondrial membrane via flow cytometric analysis using JC-1 staining. The decreased membrane potential following transfection with miR-15a mimics and si-GPX4 indicated disrupted membrane structure (Fig. 4C). Furthermore, transfection with both miR-15a mimics and si-GPX4 significantly increased the levels of MDA (Fig. D), which is a marker of lipid peroxidation (9). Taken together, these results suggest that miR-15a triggers ferroptosis via GPX4 in prostate cancer cells.
Figure 4.
miR-15a induces prostate cancer cell ferroptosis via GPX4. (A) Intracellular ferrous iron levels were determined via the iron assay in LNCAP cells transfected with miR-15a mimics, si-GPX4 or NCs. (B) Lipid ROS levels were measured via flow cytometric analysis using C11-BODIPY in LNCAP cells transfected with miR-15a mimics, si-GPX4 or NCs. (C) MMP was detected via JC-1 staining and flow cytometric analysis in LNCAP cells transfected with miR-15a mimics, si-GPX4 or NCs. (D) MDA levels were measured in LNCAP cells transfected with miR-15a mimics, si-GPX4 or NCs. **P<0.01 vs. control group. miR, microRNA; GPX4, glutathione peroxidase 4; si, small interfering; NC, negative control; ROS, reactive oxygen species; MMP, mitochondrial membrane potential; MDA, malondialdehyde.
Discussion
Recent studies have reported several novel strategies for diagnosis and therapies of cancers, among which, serum miRNAs are promising targets for cancer research and treatment (25–27). miRNAs manly function by interacting with the 3′-UTR of target mRNAs to induce degradation of mRNAs or suppress their translation, leading to the alteration of regulatory factors in cellular physiological processes, including cell proliferation, differentiation, autophagy and apoptosis (28). Several studies have revealed the function of miRNAs in prostate cancer (29,30). For example, one analysis of miRNAs in prostate lesions identified global loss of miRNA expression during progression of prostate cancer (31). In addition, downregulated miR-15a expression has been observed in patients with prostate cancer compared with healthy subjects (32). Several studies have implicated that miR-15a may inhibit the proliferation and metastasis of prostate cancer cells (23,33).
In the present study, bioinformatics analysis revealed the potential interaction between miR-15a and the 3′-UTR of GPX4 mRNA, suggesting that miR-15a may regulate GPX4 expression. In addition, transfection with miR-15a mimics altered GPX4 protein expression, but not GPX4 mRNA expression, suggesting that miR-15a regulates the translation of GPX4. miRNA pull-down and the dual-luciferase reporter assays were also performed. The enrichment of GPX4 and the enhanced luciferase activity demonstrated the direct binding of miR-15a to the 3-™UTR of GPX4 mRNA.
A previous study reported that apoptosis of prostate cancer cells is involved in the development of prostate cancer (34). However, the role of ferroptosis in prostate cancer progression remains unclear. Ferroptosis is a newly discovered form of ferrous iron-associated cell death (3,4), first described in 2012 by Dixon et al (35). Within the past decade, several studies have revealed the regulatory function of ferroptosis in different types of cancer (36,37). As the key suppressor for ferroptosis, GPX4 interacts with glutathione to decrease peroxidase reaction within biological membranes, thereby suppressing the accumulation of lipid ROS and activation of ferroptosis (38). In the present study, the suppressed cell proliferation and elevated levels of released cell death biomarker LDH (4) confirmed the induced cell death following transfection with miR-15a mimics and si-GPX4. The unique biochemical characteristics of ferroptosis consist of accumulated ferrous iron and lipid ROS (10,36). The destructed cell membrane structure induced by ferroptosis can results in extracellular release of LDH (4). Here, to confirm that the suppressed cell proliferation was indeed caused by ferroptosis, lipid ROS accumulation was measured via flow cytometric analysis using C11-BIODIPY probes, the MMP via JC-1 staining, as well as the cellular iron level. In accordance with the aforementioned features of ferroptosis, transfection with miR-15a mimics and si-GPX4 simultaneously increased ROS and Fe2+ generation, and disrupted the structure of the mitochondrial membrane.
The present study is not without limitations. First, only one prostate cancer cell line was assessed. Thus, prospective studies will focus on assessing androgen receptor negative prostate cancer cell lines. In addition, it would be useful to assess normal cell lines to determine whether the protocol is selective.
In conclusion, the results of the present study demonstrated that miR-15a induced ferroptosis of prostate cancer cells by targeting GPX4. These findings may provide novel insights and targets for the treatment of prostate cancer.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- miRNA
microRNA
- GPX4
glutathione peroxidase 4
- siRNA
small interfering RNA
- ROS
reactive oxygen species
- MMP
mitochondrial membrane potential
- LDH
lactate dehydrogenase
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant nos. 81772631 and 81974362), the Shenzhen Science and Technology Innovation Committee (grant nos. JCYJ20190814121001751, JCYJ201908143000229 and JCYJ20190814110203636), the Shenzhen Key Medical Discipline Construction Fund (grant no. SZXK013) and the Natural Science Foundation of Guangdong Province (grant No. 2018A030313530).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XP and PX initiated the project, designed the experiments and analyzed the data. PX made the figures and drafted the initial manuscript. YW performed the experiments and revised the manuscript. ZD and ZT performed the experiments. XP and PX confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Omar MI, Roobol MJ, Ribal MJ, Abbott T, Agapow PM, Araujo S, Asiimwe A, Auffray C, Balaur I, Beyer K, et al. Introducing PIONEER: A project to harness big data in prostate cancer research. Nat Rev Urol. 2020;17:351–362. doi: 10.1038/s41585-020-0355-3. [DOI] [PubMed] [Google Scholar]
- 2.Hu C, Xia H, Bai S, Zhao J, Edwards H, Li X, Yang Y, Lyu J, Wang G, Zhan Y, et al. CUDC-907, a novel dual PI3K and HDAC inhibitor, in prostate cancer: Antitumour activity and molecular mechanism of action. J Cell Mol Med. 2020;24:7239–7253. doi: 10.1111/jcmm.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Hao S, Yu J, He W, Huang Q, Zhao Y, Liang B, Zhang S, Wen Z, Dong S, Rao J, et al. Cysteine dioxygenase 1 mediates erastin-induced ferroptosis in human gastric cancer cells. Neoplasia. 2017;19:1022–1032. doi: 10.1016/j.neo.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, Sirohi K, Li X, Wei Y, Lee H, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20:1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seibt TM, Proneth B, Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med. 2019;133:144–152. doi: 10.1016/j.freeradbiomed.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Louandre C, Marcq I, Bouhlal H, Lachaier E, Godin C, Saidak Z, François C, Chatelain D, Debuysscher V, Barbare JC, et al. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015;356:971–977. doi: 10.1016/j.canlet.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Sui X, Zhang R, Liu S, Duan T, Zhai L, Zhang M, Han X, Xiang Y, Huang X, Lin H, Xie T. RSL3 drives ferroptosis through GPX4 Inactivation and ROS production in colorectal cancer. Front Pharmacol. 2018;9:1371. doi: 10.3389/fphar.2018.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dančík V, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10:1617. doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldassarri M, Fallerini C, Cetta F, Ghisalberti M, Bellan C, Furini S, Spiga O, Crispino S, Gotti G, Ariani F, et al. Omic approach in non-smoker female with lung squamous cell carcinoma pinpoints to germline susceptibility and personalized medicine. Cancer Res Treat. 2018;50:356–365. doi: 10.4143/crt.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karbasforooshan H, Roohbakhsh A, Karimi G. SIRT1 and microRNAs: The role in breast, lung and prostate cancers. Exp Cell Res. 2018;367:1–6. doi: 10.1016/j.yexcr.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 15.Guelfi G, Cochetti G, Stefanetti V, Zampini D, Diverio S, Boni A, Mearini E. Next Generation Sequencing of urine exfoliated cells: An approach of prostate cancer microRNAs research. Sci Rep. 2018;8:7111. doi: 10.1038/s41598-018-24236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, Wu L, Zhang P, Luo M, Du J, Gao T, O'Connell D, Wang G, Wang H, Yang Y. miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol Carcinog. 2018;57:1566–1576. doi: 10.1002/mc.22878. [DOI] [PubMed] [Google Scholar]
- 17.Luo M, Wu L, Zhang K, Wang H, Zhang T, Gutierrez L, O'Connell D, Zhang P, Li Y, Gao T, et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ. 2018;25:1457–1472. doi: 10.1038/s41418-017-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita K, Fukumoto M, Itoh K, Kuwahara Y, Igarashi K, Nagasawa T, Suzuki M, Kurimasa A, Sato T. MiR-7-5p is a key factor that controls radioresistance via intracellular Fe2+ content in clinically relevant radioresistant cells. Biochem Biophys Res Commun. 2019;518:712–718. doi: 10.1016/j.bbrc.2019.08.117. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, Zhang Q, Lin D, Ge S, Bai M, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Yang Y, Ren L, Yang J, Wang B, Xing T, Chen H, Chen M. miR-15a-3p suppresses prostate cancer cell proliferation and invasion by targeting SLC39A7 via downregulating Wnt/β-catenin signaling pathway. Cancer Biother Radiopharm. 2019;34:472–479. doi: 10.1089/cbr.2018.2722. [DOI] [PubMed] [Google Scholar]
- 21.Jia X, Liu H, Xu C, Han S, Shen Y, Miao X, Hu X, Lin Z, Qian L, Wang Z, Gong W. MiR-15a/16-1 deficiency induces IL-10-producing CD19+ TIM-1+ cells in tumor microenvironment. J Cell Mol Med. 2019;23:1343–1353. doi: 10.1111/jcmm.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Kafaji G, Said HM, Alam MA, Al Naieb ZT. Blood-based microRNAs as diagnostic biomarkers to discriminate localized prostate cancer from benign prostatic hyperplasia and allow cancer-risk stratification. Oncol Lett. 2018;16:1357–1365. doi: 10.3892/ol.2018.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin W, Chen F, Wang K, Song Y, Fei X, Wu B. miR-15a/miR-16 cluster inhibits invasion of prostate cancer cells by suppressing TGF-β signaling pathway. Biomed Pharmacother. 2018;104:637–644. doi: 10.1016/j.biopha.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 24.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 25.Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S, Shimizu C, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qadir MI, Faheem A. miRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. doi: 10.1615/CritRevEukaryotGeneExpr.2017019494. [DOI] [PubMed] [Google Scholar]
- 27.Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda T, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319. doi: 10.1038/s41467-018-06434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omidkhoda N, Wallace Hayes A, Reiter RJ, Karimi G. The role of MicroRNAs on endoplasmic reticulum stress in myocardial ischemia and cardiac hypertrophy. Pharmacol Res. 2019;150:104516. doi: 10.1016/j.phrs.2019.104516. [DOI] [PubMed] [Google Scholar]
- 29.Jiang J, Jia P, Zhao Z, Shen B. Key regulators in prostate cancer identified by co-expression module analysis. BMC Genomics. 2014;15:1015. doi: 10.1186/1471-2164-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart M, Nolte E, Wach S, Szczyrba J, Taubert H, Rau TT, Hartmann A, Grässer FA, Wullich B. Comparative microRNA profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol Cancer Res. 2014;12:250–263. doi: 10.1158/1541-7786.MCR-13-0230. [DOI] [PubMed] [Google Scholar]
- 31.Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sañudo A, Camara-Lopes LH, Srougi M. MicroRNA expression profiles in the progression of prostate cancer-from high-grade prostate intraepithelial neoplasia to metastasis. Urol Oncol. 2013;31:796–801. doi: 10.1016/j.urolonc.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Zidan HE, Abdul-Maksoud RS, Elsayed WSH, Desoky EAM. Diagnostic and prognostic value of serum miR-15a and miR-16-1 expression among egyptian patients with prostate cancer. IUBMB Life. 2018;70:437–444. doi: 10.1002/iub.1733. [DOI] [PubMed] [Google Scholar]
- 33.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 34.Antognelli C, Mezzasoma L, Fettucciari K, Mearini E, Talesa VN. Role of glyoxalase I in the proliferation and apoptosis control of human LNCaP and PC3 prostate cancer cells. Prostate. 2013;73:121–132. doi: 10.1002/pros.22547. [DOI] [PubMed] [Google Scholar]
- 35.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M, Gai C, Li Z, Ding D, Zheng J, Zhang W, Lv S, Li W. Targeted exosome-encapsulated erastin induced ferroptosis in triple negative breast cancer cells. Cancer Sci. 2019;110:3173–3182. doi: 10.1111/cas.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin D, Kim EH, Lee J, Roh JL. Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer. Free Radic Biol Med. 2018;129:454–462. doi: 10.1016/j.freeradbiomed.2018.10.426. [DOI] [PubMed] [Google Scholar]
- 38.Bebber CM, Müller F, Prieto Clemente L, Weber J, von Karstedt S. Ferroptosis in cancer cell biology. Cancers (Basel) 2020;12:164. doi: 10.3390/cancers12010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.