Summary
Parent-of-origin (PoO) effects refer to the differential phenotypic impacts of genetic variants dependent on their parental inheritance due to imprinting. While PoO effects can influence complex traits, they may be poorly captured by models that do not differentiate the parental origin of the variant. The aim of this study was to conduct a genome-wide screen for PoO effects on a broad range of clinical traits derived from electronic health records (EHR) in the DiscovEHR study enriched with familial relationships. Using pairwise kinship estimates from genetic data and demographic data, we identified 22,051 offspring among 134,049 individuals in the DiscovEHR study. PoO of ~9 million variants was assigned in the offspring by comparing offspring and parental genotypes and haplotypes. We then performed genome-wide PoO association analyses across 154 quantitative and 611 binary traits extracted from EHR. Of the 732 significant PoO associations identified (p < 5 × 10−8), we attempted to replicate 274 PoO associations in the UK Biobank study with 5,015 offspring and replicated 9 PoO associations (p < 0.05). In summary, our study implements a bioinformatic and statistical approach to examine PoO effects genome-wide in a large population study enriched with familial relationships and systematically characterizes PoO effects on hundreds of clinical traits derived from EHR. Our results suggest that, while the statistical power to detect PoO effects remains modest yet, accurately modeling PoO effects has the potential to find new associations that may have been missed by the standard additive model, further enhancing the mechanistic understanding of genetic influence on complex traits.
Keywords: parent-of-origin effect, imprinting, familial relationship, GWAS, electronic health record
Genetic variants can have differential phenotypic effects based on parent-of-origin (PoO) due to imprinting. PoO of variants can be inferred by comparing parental and offspring genetic data. Genome-wide PoO-aware association testing reveals PoO effects across numerous clinical traits. PoO statistical models can identify novel associations undetected by standard additive models.
Introduction
Genomic imprinting, the non-equal expression of the two parental alleles, can lead to parent-of-origin (PoO)-specific effects of genetic variants on traits. For example, when a gene is silenced on the paternally inherited DNA strand and only expressed from the maternally inherited DNA strand, functional variants of the gene that are maternally inherited may have observable phenotypic impacts, while those that are paternally inherited may not, leading to maternal-specific effects. Imprinting has been characterized in approximately 1% of the genome and can influence complex traits in a PoO-specific manner1,2 and contribute to their heritability.
There have been studies that aimed to find PoO effects of genetic variants on traits. Many studies focused on associations identified using standard additive models in or near imprinted genes and then examined whether the additive signals are capturing PoO effects.2, 3, 4, 5, 6, 7 Other studies tested for PoO effects genome-wide to identify novel signals for specific traits, including type 2 diabetes (T2D), height, and autism spectrum disorder.2,8,9 Recently, a study performed genome-wide PoO association analyses for 21 common quantitative traits in a Hutterite population isolate.10 These studies have confirmed the presence of PoO effects in multiple loci, including imprinted KCNQ1, KLF14, IGF2, DLK1, and GNAS loci,2, 3, 4, 5, 6, 7, 8,11 and observed diverse patterns of PoO effects, including simple uniparental effects and differential effects between the parental alleles.
Extending previous efforts, we performed a genome-wide screen for PoO effects over a broad spectrum of clinical traits in the DiscovEHR study consisting of 134,049 individuals of European ancestry. The DiscovEHR study is enriched with parent-offspring relationships,12 allowing the assignment of PoO of genetic variants in as many as 22,051 individuals. We employed statistical models that take into account the PoO of the variants to identify PoO-specific associations with clinical traits derived from EHR. The systematic characterization of PoO effects across the genome and across hundreds of traits in a large population study can provide a valuable baseline for future studies of PoO effects.
Subjects and methods
Study participants
The DiscovEHR study is a collaborative project between the Regeneron Genetics Center (RGC) and the Geisinger health system with participants enrolled in Geisinger’s MyCode Community Health Initiative.13 All participants consented to provide genetic data and clinical data from their electronic health records (EHR) for broad research use, including genetic analyses. The study was approved by the Institutional Review Board (IRB) at Geisinger. DNA samples from 143,575 individuals were genotyped at the RGC using either OmniExpress or Global Screening Array (GSA). 1,601 samples were excluded based on quality control criteria, including gender mismatch, low call rate, discordancy compared to exomes, or suspected duplication. Since family-based imputation methods may have only incrementally improved performance for variants passing association test quality control described below, we used population-based imputation to reduce computational burden. The array genotypes were imputed on the Michigan imputation server using Haplotype Reference Consortium (HRC) data as reference.14 hg19 coordinates were converted to hg38 coordinates using Picard LiftoverVCF. Only individuals of European ancestry (n = 134,049) as estimated by a previously described method13 and variants with imputation score ≥ 0.3 were used for subsequent analysis.
The UK Biobank study is a prospective cohort study consisting of 500,000 individuals from the United Kingdom.15 All participants consented to the use of their genetic and medical information for research purpose. The study was approved by the North West Centre for Research Ethics Committee. As described previously, DNA samples were genotyped using UK BiLEVE and UK Biobank Axiom arrays, and the array genotypes were pre-phased by SHAPEIT3 and imputed by IMPUTE4 using the HRC panel as reference.15 Since the publicly available imputed sequence of the UK Biobank study did not retain phase information, the array genotypes of offspring and parents were imputed on the Michigan imputation server to obtain phased haplotypes, which were used for PoO assignment as described in the following sections.
Kinship estimation and identification of offspring
Genome-wide identity-by-descent (IBD) proportions were estimated in all pairs of individuals using PLINK v.1.916 and array genotype data filtered by quality control metrics (individual missingness < 0.1, variant missingness < 0.1, Hardy-Weinberg equilibrium p > 1 × 10−15), minor allele frequency (MAF) (≥0.05), and linkage disequilibrium (LD) pruning (r2 < 0.2). Pairs of individuals who have genome-wide probability of sharing one allele IBD (IBD1) proportions > 0.8 were inferred as parent-offspring relationships.17 Age and sex information was used to identify offspring, father, and mother in these relationships.
PoO assignment of genetic variants in offspring
Imputed variants were filtered based on the quality control criteria (individual missingness < 0.1, variant missingness < 0.1, Hardy-Weinberg equilibrium p > 1 × 10−15), minor allele count (MAC) threshold (≥10 among the offspring), and LD pruning (r2 < 0.2) using PLINK, resulting in 9,085,657 variants for PoO assignment. For offspring with a heterozygous genotype at a given variant site, the parental origin of the minor allele was determined using two methods: Mendelian and haplotype-based methods, as described in detail in the Results section (Figure 1C). The haplotype method was not performed when the number of polymorphic nucleotides observed in either parental or offspring haplotype within the 1 Mb window of the index variant was less than 5.
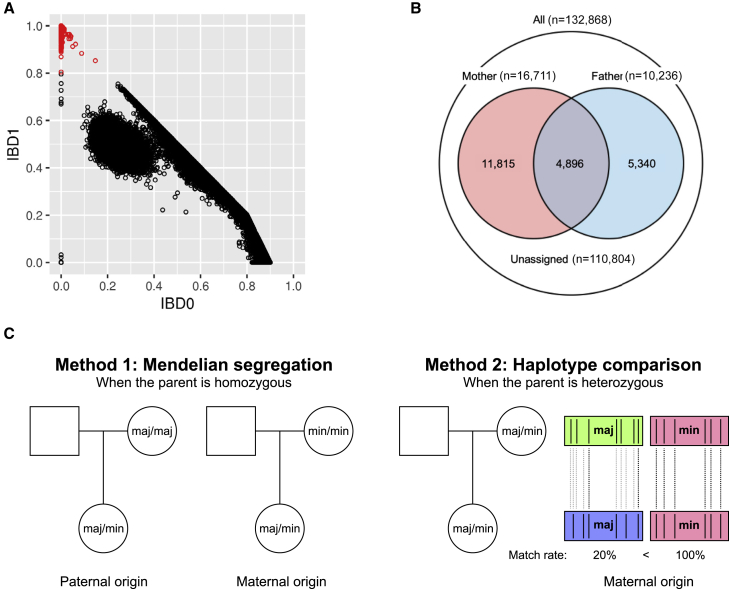
Figure 1.
Identification of parent-offspring relationships and PoO assignment in DiscovEHR study
(A) Genome-wide identity-by-descent (IBD) was estimated from genetic data between every pair of individuals in the DiscovEHR study. Pairs with genome-wide probability of sharing one allele IBD (IBD1) > 0.8 (in red) were inferred to be in parent-offspring relationships.
(B) In each parent-offspring relationship, offspring, father, and mother were inferred based on age and sex information. The number of offspring with one parent or both parents in the study is indicated in the corresponding area of the Venn diagram.
(C) Parent-of-origin (PoO) of variants was assigned among offspring with at least one parent available. For each heterozygous genotype, PoO of the minor allele was assigned using two methods. When at least one available parental genotype is homozygous, PoO was determined based on Mendelian segregation (left). When the available parental genotype(s) is/are heterozygous, PoO was estimated by comparing the haplotypes around the variant between offspring and each available parent (right). See text for detailed methods.
PoO association tests
Quantitative and binary traits were derived from EHR. Quantitative traits were normalized using rank inverse normal transformation prior to testing. As the DiscovEHR study is enriched with familial relationships,12 the associations were tested using mixed models as implemented in BOLT11 for quantitative traits and SAIGE18,19 for binary traits to control type 1 error that may result from relatedness in the samples. Age, age2, sex, age-by-sex interaction, indicator variable for genotyping array, and 10 principal components of ancestry calculated from array genotypes were included as covariates in the model. The associations were tested under additive and PoO models as described in detail in the result section. Traits were not included in downstream analyses or interpretation if there was no estimated heritability or if the genomic inflation factor was greater than 1.5 under additive or PoO models. To control multiplicity, variants were not tested if there were fewer than 5 heterozygous individuals of paternal inheritance or 5 heterozygous individuals of maternal inheritance with observed traits, assuming these variants have minimal power to reject the null hypothesis. Note that since neither the traits derived from EHR nor the additive and various PoO models are independent, a strict Bonferroni correction based on the product of the number of variants, traits, and models tested will result in an overly conservative threshold. For simplicity, we have corrected for multiple testing using an experiment-wise approach based on the number of variants, unless otherwise indicated.
Power simulations
The power estimates to detect PoO effects were simulated under the assumption that the true effects exhibit uniparental, polar dominance, or bipolar dominance patterns. The following parameters were used: total sample size = 134,049, number of offspring = 22,051 (as in the DiscovEHR study), alpha = 5 × 10−8, and ranges of MAFs and effect sizes. The genotypic counts for a given MAF were derived assuming Hardy-Weinberg equilibrium. The phenotypes were simulated from a normal distribution with the assumed difference in means per genotype and standard deviation of 1. For the additive model, half of the heterozygous genotypic counts were simulated from paternal distribution, while the other half were simulated from maternal distribution. The association between simulated genotypes and phenotypes was then tested under additive, parental, and differential models. The process was repeated 10,000 times and the proportion of simulations yielding p values more significant than alpha was used to estimate power.
Results
Identification of offspring in DiscovEHR study
To find parent-offspring relationships among the 134,049 participants of European ancestry in the DiscovEHR study, we estimated the genome-wide IBD in all pairs of individuals using array genotype data. There were 28,562 pairs with genome-wide IBD1 greater than 0.8, which were inferred as parent-offspring relationships (Figure 1A). Then, we used age and sex information to assign father, mother, and offspring in these relationships. As a result, we found a total of 22,051 offspring, of which 4,896 had both parents, 11,815 had mothers, and 5,340 had fathers present in the study (Figure 1B).
PoO assignment of genetic variants in offspring
We assigned the PoO of a total of 9,085,657 imputed variants that passed quality control and had MAC ≥ 10 among offspring and were LD pruned (r2 < 0.2). At each variant site, we assigned the PoO of the minor allele in the heterozygous offspring using two methods (Figure 1C). (1) Mendelian method: when at least one available parental genotype was homozygous for either major or minor allele, PoO was determined based on Mendelian segregation. For example, if the genotype of the mother was homozygous major allele, then the PoO of the minor allele was inferred as paternal, while if the genotype of the mother was homozygous minor allele, then the PoO of the minor allele was inferred as maternal. (2) Haplotype method: since the Mendelian method is uninformative when the available parental genotype(s) is/are heterozygous, we also estimated PoO by comparing offspring and parental haplotypes within a 1 Mb window that carry the same allele (major or minor) of the variant. The haplotype comparison was performed between offspring and each available parent. The percent match rate, or percent of identical by state polymorphic nucleotides, was calculated as below.
The allele that is carried on the offspring-parent haplotype pair that has a greater match rate was inferred as the allele that is inherited from the parent. We assessed the accuracy of the haplotype method for each variant by first applying it to the heterozygous offspring for whom PoO could be determined by the Mendelian method. As a quality control measure, when the PoO estimated by the haplotype method was <80% concordant with the PoO determined by the Mendelian method, only the PoO determined by the Mendelian method was included in follow-up analysis. Using this approach, we assigned the PoO of 9,085,586 and 8,801,949 variants (corresponding to 4,178,033,204 and 2,958,665,691 heterozygous genotypes) by the Mendelian and haplotype methods, respectively, leading to the overall PoO assignment rate of 99.2% of all heterozygous genotypes among the offspring. Among the 8,801,949 variants for which the haplotype method was used to estimate PoO, the overall concordance between the haplotype and Mendelian methods was 98.5%.
Statistical models to find PoO-specific associations
To identify PoO-specific effects, we tested associations under parental and differential models as described below. Parental models include paternal and maternal models that contrast trait values in the heterozygous offspring of the paternally and maternally inherited minor allele, respectively, with those in the homozygous individuals of the major allele. Heterozygous individuals with opposite or unknown inheritance and homozygous individuals of the minor alleles are excluded in parental models. Paternal- and maternal-specific associations were defined as being associated under one parental model and not the other. We also evaluated a differential model, which tests for a difference in trait values between paternally and maternally inherited alleles, contrasting heterozygous offspring of paternal and maternal inheritance. For variant-trait pairs with significant PoO associations, we also included associations under the additive model among all available individuals for comparison. While the parental models may offer greater power to detect PoO effects due to the larger sample size, only the differential model directly measures differences between paternal and maternal alleles.
Testing PoO specificity of additive associations within imprinted regions
Functional variants that affect imprinted genes with monoallelic expression may be most likely to exert PoO-specific effects on traits. Therefore, we examined whether variants with additive associations within the known imprinted regions have detectable PoO-specific associations. We first tested 137,304 variants within a ±500 kb window from 69 known or suggested imprinted genes2,20 (Table S1) under the additive model across 173 quantitative traits derived from EHR (Table S2) using the BOLT linear mixed model.18 These regions constitute approximately 1.3% of the genome. Six traits failed to produce results, due to the absence of estimated heritability. Among the remaining 167 traits, we found 667 additive associations that were statistically significant (p < 3.6 × 10−7, after correction for 137,304 variants). We tested these additive associations under paternal, maternal, and differential models and found 12 significant PoO-specific associations (p < 7.5 × 10−5, after correction for 667 variant-trait associations): 4 were paternal-specific and 8 were maternal-specific associations, among which 4 also had differential associations (Table 1).
Table 1.
PoO specificity among the additive associations identified within imprinted regions
| Imprinted region | Variant | Nearest gene | Variant effect | Trait | MAF | Num.all | Num.offspring | Beta.add | pval.add | Beta.pat | pval.pat | Beta.mat | pval.mat | Beta.diff | pval.diff |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GRB10 | 7:50234619:T:C | C7orf72 | Intergenic | % monocytes | 0.055 | 102,797 | 17,544 | −0.09 | 9.6 × 10−27 | −0.13a | 1.9 × 10−5a | −0.04 | 0.15 | 0.10 | 0.028 |
| 7:50541672:T:C | FIGNL1DDC | intronic intronic | % monocytes | 0.034 | 102,797 | 17,544 | 0.07 | 4.9 × 10−10 | 0.15a | 7.9 × 10−5a | 0.04 | 0.32 | −0.12 | 0.021 | |
| 7:50359449:C:T | IKZF1 | Intronic | % monocytes | 0.023 | 102,797 | 17,544 | −0.12 | 3.2 × 10−19 | −0.19a | 7.9 × 10−5a | −0.08 | 0.084 | 0.14 | 0.029 | |
| IGF2R, SLC22A2, SLC22A3 | 6:160270606:T:G | SLC22A2 | Intronic | EGFR | 0.133 | 116,211 | 19,467 | 0.03 | 2.4 × 10−9 | 0.06a | 2.3 × 10−5a | 0.01 | 0.46 | −0.05 | 0.021 |
| 6:160743692:C:T | PLG | intronic | cholesterol | 0.031 | 93,077 | 14,619 | 0.07 | 1.1 × 10−7 | 0.07 | 0.14 | 0.20a | 1.5 × 10−5a | 0.13 | 0.052 | |
| CPA4, MEST, MESTIT1, COPG2IT1, COPG2, KLF14 | 7:130738173:T:C | KLF14 | upstream | HDL-C | 0.481 | 93,383 | 14,642 | 0.04 | 1.2 × 10−23 | 0.03 | 0.064 | 0.13a | 2.1 × 10−16a | 0.10a | 3.1 × 10−6a |
| upstream | TC/HDL-C ratio | 0.481 | 93,381 | 14,629 | −0.04 | 1.4 × 10−17 | −0.003 | 0.84 | −0.11a | 2.2 × 10−10a | −0.10a | 8.1 × 10−6a | |||
| upstream | triglyceride | 0.481 | 93,365 | 14,625 | −0.04 | 5.1 × 10−17 | −0.01 | 0.56 | −0.10a | 1.4 × 10−9a | −0.09 | 1.3 × 10−4 | |||
| H19, IGF2, IGF2-AS, INS, ASCL2, TRPM5, KCNQ1, KCNQ1OT1, KCNQ1DN, CDKN1C, SLC22A18AS, SLC22A18, PHLDA2, OSBPL5 | 11:2875083:G:A | CDKN1C | intergenic | bilirubin | 0.087 | 108,535 | 17,913 | 0.05 | 7.7 × 10−13 | 0.05 | 0.054 | 0.10a | 3.5 × 10−5a | 0.05 | 0.17 |
| 11:3017489:T:A | CARS | intronic | bilirubin | 0.200 | 108,535 | 17,913 | 0.03 | 8.2 × 10−9 | 0.03 | 0.073 | 0.07a | 5.3 × 10−5a | 0.03 | 0.16 | |
| DLK1, MEG3 | 14:100704203:T:C | DLK1 | upstream | platelets | 0.329 | 115,308 | 19,552 | −0.04 | 3.2 × 10−20 | 0.02 | 0.12 | −0.10a | 2.3 × 10−11a | −0.12a | 7.8 × 10−10a |
| GNAS, GNAS-AS1 | 20:58872268:G:A | GNAS | intronic | TSH | 0.012 | 101,160 | 17,401 | 0.10 | 1.7 × 10−7 | −0.04 | 0.54 | 0.40a | 2.4 × 10−9a | 0.44a | 3.0 × 10−6a |
Among the 667 associations within the known imprinted regions (p < 3.6 × 10−7) identified under the additive model across 167 quantitative traits, 12 were PoO specific (p < 7.5 × 10−5). Variant is denoted as chromosome:position:reference allele:alternate allele on GRCh38 genome build. Num.all, number of all individuals with given traits; Num.offspring, number of offspring with given traits; Beta.add and pval.add, beta coefficient and p values under additive model; Beta.pat and pval.pat, beta coefficient and p values under paternal model; Beta.mat and pval.mat, beta coefficient and p values under maternal model; Beta.diff and pval.diff, beta coefficient (modeled on maternal allele compared to the paternal allele) and p values under differential model.
Indicates significant PoO-specific associations.
Several of these associations were in LD with previously identified PoO associations, serving as positive controls. For example, the 7:130738173:T:C variant with maternal-specific association with high-density lipoprotein cholesterol (HDL-C) levels, total cholesterol (TC)/HDL-C ratios, and triglyceride levels is near the maternally expressed KLF14 (MIM: 609393). The variant is in LD (r2 = 0.97) with rs4731702, which was previously found to have maternal-specific associations with KLF14 gene expression and type 2 diabetes risk in Icelandic and American Indian populations.2,5 While additive associations of KLF14 locus with HDL-C and triglyceride levels have been reported, our results suggest a previously uncharacterized maternal-specific pattern. Another example is the maternal-specific association of the 14:100704203:T:C variant with platelet counts near the paternally expressed DLK1 (MIM: 176290), which replicates a previously reported association of rs7141210 (r2 = 0.81) in the Icelandic population.11 This study showed that rs7141210 is associated with a maternal-specific DNA methylation pattern, suggesting that the maternally inherited allele may impair the silencing of maternal gene expression. A third example is the maternal-specific association of the 20:58872268:G:A variant with thyroid-stimulating hormone (TSH) levels near the maternally expressed GNAS (MIM: 139320), which confirms a previously reported association of rs139242164 (r2 = 0.62) in the Icelandic population.11 Genetic variations in the GNAS gene are a well-established cause of pseudo-hypoparathyroidism (PHP) that is characterized by end-organ resistance to parathyroid hormone and high levels of circulating TSH.21
Genome-wide PoO association analyses for 154 quantitative traits
To identify PoO-specific effects beyond the known imprinted regions, we performed genome-wide PoO association analyses for quantitative traits under parental and differential models using the BOLT linear mixed model.18 Among the 167 traits with non-zero estimated heritability, 13 traits with high genomic inflation (>1.5) under PoO models were omitted, yielding results for 154 traits (Figure S1). We found a total of 732 PoO associations (p < 5 × 10−8) for 725 unique variants, including 341 paternal-specific, 344 maternal-specific, and 49 differential associations (Tables S3–S5). Notably, only 19 of these associations were within known imprinted regions. We attempted to replicate these associations in the UK Biobank study consisting of 462,453 individuals of European ancestry, including 5,015 offspring with at least one parent. Of the 725 variants and 83 traits with associations in the DiscovEHR study, 721 variants and 37 traits could be unambiguously mapped in the UK Biobank study, allowing the examination of 274 associations for replication. We replicated 9 PoO associations at the nominal significance threshold (p < 0.05) across 6 traits: 4 were paternal-specific and 5 were maternal-specific associations, among which one also had differential association (Table 2). The strongest PoO association found was the aforementioned association of the variant near DLK1 with platelet counts. When the 9 significant PoO associations were examined under the additive model for comparison, 5 had stronger associations under the parental or differential model than the additive model and 4 had no association under the additive model (p values > 0.05). Quantile-quantile plots for the 6 traits under each statistical model are provided in Figure S2.
Table 2.
PoO-specific associations for quantitative traits identified in the DiscovEHR study (p < 5 × 10−8) that are replicated in the UK Biobank study (p < 0.05)
| Variant | Nearest gene | Variant effect | Trait | Study | MAF | Num.all | Num.offspring | Beta.add | pval.add | Beta.pat | pval.pat | Beta.mat | pval.mat | Beta.diff | pval.diff |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22:17116671:G:A | IL17RACECR6 | downstream 3′ UTR | % monocytes | DiscovEHR | 0.004 | 102,797 | 17,544 | −0.42 | 1.6 × 10−39 | −0.67∗ | 1.1 × 10−8∗ | −0.17 | 0.16 | 0.48 | 4.8 × 10−3 |
| UK Biobank | 0.007 | 448,877 | 4,846 | −0.45 | 2.2 × 10−307 | −0.41∗ | 4.7 × 10−3∗ | −0.62 | 0.011 | −0.22 | 0.45 | ||||
| 2:233925128:A:G | TRPM8 | upstream | total bilirubin | DiscovEHR | 0.003 | 108,535 | 17,913 | 0.35 | 4.9 × 10−29 | 0.60∗ | 2.8 × 10−8∗ | 0.11 | 0.30 | −0.45 | 5.9 × 10−3 |
| UK Biobank | 0.005 | 440,287 | 4,768 | 0.28 | 9.4 × 10−146 | 0.39∗ | 0.012∗ | 0.11 | 0.62 | −0.28 | 0.30 | ||||
| 1:161478451:C:G | FCGR2A | intergenic | protein | DiscovEHR | 0.197 | 108,144 | 17,857 | −0.04 | 1.1 × 10−17 | −0.10∗ | 2.9 × 10−8∗ | −0.03 | 0.096 | 0.07 | 4.3 × 10−3 |
| UK Biobank | 0.172 | 405,404 | 4,354 | −0.04 | 5.0 × 10−58 | −0.10∗ | 3.5 × 10−3∗ | −0.06 | 0.20 | 0.04 | 0.50 | ||||
| 17:46812337:C:T | WNT3 | intronic | red blood cells | DiscovEHR | 0.439 | 115,524 | 19,569 | 0.004 | 0.23 | 0.08∗ | 3.2 × 10−9∗ | 0.02 | 0.25 | −0.06 | 2.2 × 10−4 |
| UK Biobank | 0.444 | 449,656 | 4,855 | 0.02 | 9.6 × 10−23 | 0.07∗ | 3.4 × 10−3∗ | 0.04 | 0.075 | −0.03 | 0.43 | ||||
| 1:115200874:T:G | NGF | intergenic | DiscovEHR | 0.476 | 115,524 | 19,569 | 0.01 | 0.019 | 0.02 | 0.13 | 0.09a | 3.0 × 10−11a | 0.07 | 1.7 × 10−5 | |
| UK Biobank | 0.473 | 449,656 | 4,855 | 0.001 | 0.67 | −0.0002 | 0.99 | 0.05a | 0.019a | 0.05 | 0.094 | ||||
| 6:170235280:G:C | DLL1 | intergenic | DiscovEHR | 0.482 | 115,524 | 19,569 | 0.001 | 0.76 | 0.02 | 0.16 | 0.08a | 3.1 × 10−8a | 0.06 | 5.0 × 10−4 | |
| UK Biobank | 0.489 | 449,656 | 4,855 | 0.003 | 0.04 | 0.04 | 0.081 | 0.05a | 0.028a | 0.01 | 0.73 | ||||
| 20:55475672:A:G | CBLN4 | intergenic | DiscovEHR | 0.384 | 115,524 | 19,569 | 0.003 | 0.36 | 0.01 | 0.31 | 0.08a | 4.8 × 10−8a | 0.06 | 7.4 × 10−4 | |
| UK Biobank | 0.376 | 449,656 | 4,855 | 0.002 | 0.19 | 0.06 | 0.014 | 0.06a | 9.1 × 10−3a | 0.002 | 0.94 | ||||
| 18:66392331:A:T | CDH19 | intergenic | HDL cholesterol | DiscovEHR | 0.004 | 93,383 | 14,642 | −0.04 | 0.25 | 0.15 | 0.21 | 0.72a | 1.7 × 10−8a | 0.49 | 9.1 × 10−3 |
| UK Biobank | 0.003 | 405,671 | 4,361 | −0.01 | 0.65 | 0.12 | 0.40 | 0.46a | 0.047a | 0.33 | 0.22 | ||||
| 14:100704203:T:C | DLK1 | intergenic | platelets | DiscovEHR | 0.329 | 115,308 | 19,552 | −0.04 | 3.2 × 10−20 | 0.02 | 0.12 | −0.10a | 2.3 × 10−11a | −0.12a | 7.8 × 10−10a |
| UK Biobank | 0.335 | 449,652 | 4,855 | −0.04 | 1.9 × 10−95 | −0.001 | 0.96 | −0.10a | 4.5 × 10−4a | −0.10a | 9.5 × 10−3a |
Nine of the PoO-specific associations (p < 5 × 10−8) identified from the genome-wide screen for 154 traits in the DiscovEHR study were replicated in the UK Biobank study (p < 0.05). Variant is denoted as chromosome:position:reference allele:alternate allele on GRCh38 genome build. Num.all, number of all individuals with given traits; Num.offspring, number of offspring with given traits; Beta.add and pval.add, beta coefficient and p values under additive model; Beta.pat and pval.pat, beta coefficient and p values under paternal model; Beta.mat and pval.mat, beta coefficient and p values under maternal model; Beta.diff and pval.diff, beta coefficient (modeled on maternal allele compared to the paternal allele) and p values under differential model.
Indicates significant PoO associations.
Genome-wide PoO association analyses for 611 binary traits
We also conducted genome-wide PoO association analyses for binary traits based on ICD10 3-digit codes using SAIGE linear mixed model.19 To mitigate the computational burden required for testing association for binary traits, we first tested the differential model as a screen for PoO effects genome-wide and subsequently examined significant differential associations under additive and parental models. Of the 980 traits with at least 20 cases and 20 controls among the offspring, 612 traits were estimated to have non-zero heritability. All traits except one had genomic inflation factors below 1.5 (Figure S1), yielding results for 611 traits. We found 27 significant differential associations (p < 5 × 10−8) (Table S6), including two variants near the imprinted IGF2 (MIM: 147470) locus that were associated with ICD E11 code for T2D. One of the associated variants, 11:1680825:A:T, is in LD (r2 = 0.84) with rs2334499, which was previously identified for PoO-specific association with T2D in the Icelandic population.2 Consistent with the previous report, the paternal allele was associated with increased risk (odds ratio [OR] = 1.13, p = 0.01), while the maternal allele was associated with decreased risk (OR = 0.73, p = 4.0 × 10−10), with significant differential association (p = 8.5 × 10−10). The other variant, 11:1690902:G:A, was in low LD measured by r2 (0.17) with the 11:1680825:A:T variant, but had high D′ (0.98), indicating LD. Interestingly, while it was the maternal allele of 11:1680825:A:T that was associated with reduced risk, it was the paternal allele of 11:1690902:G:A that was associated with reduced risk. We attempted to replicate the 27 differential associations in the UK Biobank study, but none reached nominal statistical significance (p < 0.05), likely due to limited power based on low numbers of cases and offspring in the UK Biobank study.
Power simulations for PoO effects
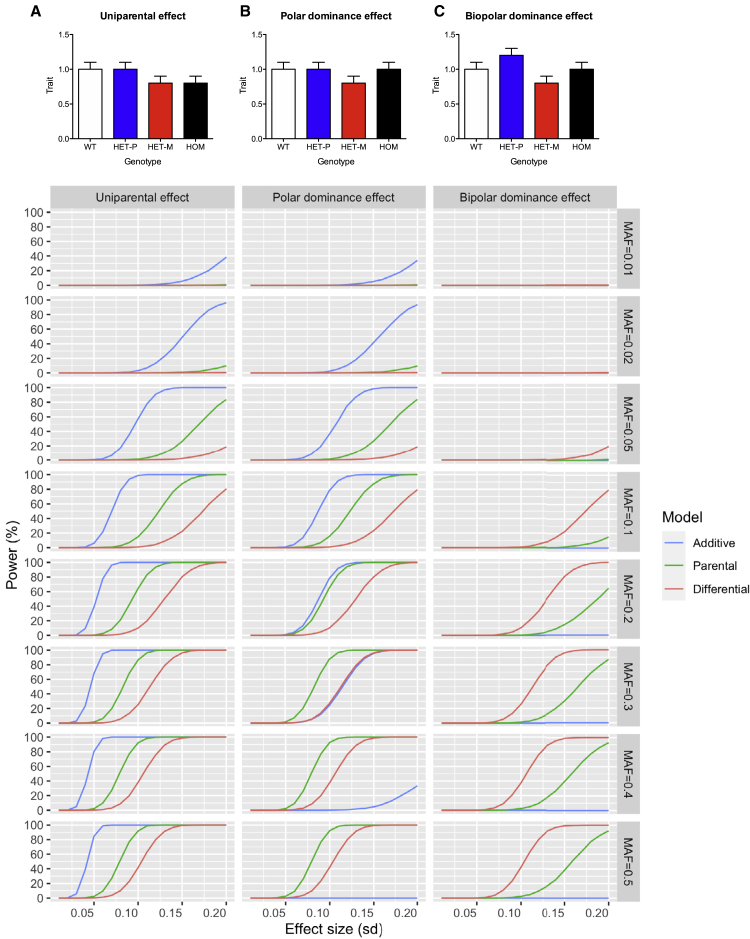
Imprinting can give rise to diverse patterns of PoO effects of genetic variants: uniparental, polar dominance, and bipolar dominance.1 We simulated the power to detect different types of PoO effects under additive, parental, and differential models across ranges of effect sizes and MAFs with fixed parameters matching the DiscovEHR PoO analysis: total sample size, number of offspring, and alpha (type 1 error). When a variant affects an imprinted gene with monoallelic expression from one parental DNA strand, it can have a uniparental (either maternal or paternal) effect. Uniparental effects impact phenotypes in the homozygous and heterozygous individuals who inherited the variant from a specific parent but not in the heterozygous individuals who inherited it from the other parent (Figure 2A). Our simulations showed that the additive model has the greatest power for uniparental effects, even though it does not take into account the phenotypic difference between heterozygous individuals with paternal and maternal inheritance. This is due to the fact that the additive model has the greatest sample size, since it includes all heterozygous and homozygous individuals, while parental models only include homozygous major individuals and heterozygous offspring with PoO from a specific parent, and the differential model only includes heterozygous offspring with PoO assignment and excludes all homozygous individuals. The simulated result is consistent with our observation in the DiscovEHR study that most of the PoO associations near the known imprinted genes with monoallelic expression, likely arising from uniparental effects, had stronger p values under the additive model than the parental or differential model (Table 1). Alternatively, when a variant exerts phenotypic impacts only in the heterozygous individuals who inherited the variant from a specific parent and not in the homozygous individuals or heterozygous individuals who inherited it from the other parent, the variant has a polar dominance effect (Figure 2B). Simulated polar dominance effects have the greatest power in additive or parental models, depending on the MAF of the variant. Specifically, the power of the additive model to capture polar dominance effects increases as the MAF increases up to ~0.2, but beyond ~0.2, the power diminishes because increasing number of homozygous individuals with no phenotypic alteration reduces effect estimates under the additive model. Instead, the parental model has greater power than the additive model when MAF is greater than ~0.2. This may explain some of the observed paternal- and maternal-specific associations in Tables 2, S3, and S4 that have stronger associations under parental models than the additive model. For example, a common intergenic variant near NGF (1:115200874:T:G, MAF = 0.48) was significantly associated with red blood cell counts under the maternal model (beta = 0.09, p = 3.0 × 10−11) but only had weak association under the additive model (beta = 0.01, p = 0.019) (Table 2). Lastly, a bipolar dominance effect occurs when a variant has diverging phenotypic impacts between individuals of paternal and maternal inheritance without any impacts on homozygous individuals (Figure 2C). For bipolar dominance effects, the differential model has the greatest power even though it has the smallest sample size, with just heterozygous offspring with PoO assignment, because the effect estimates are largest when contrasting heterozygous offspring with paternal and maternal inheritance. On the contrary, the additive model has very limited power, even though it has the largest sample size, because the effects of paternal and maternal alleles will cancel each other, and there are no phenotypic impacts in homozygous individuals. This may explain some of the observed differential associations in Tables S5 and S6 that have stronger associations under the differential model than under the additive or parental models. For example, the association of a 3′ UTR variant of ST8SIA5 (18:46674932:C:A) with total cholesterol to HDL-C ratio was strongest under the differential model (p = 1.4 × 10−9) and weaker under the parental models with opposite effect directions (beta = 0.85, p = 1.7 × 10−6 under the paternal model and beta = −0.60, p = 8.8 × 10−5 under the maternal model) but had no association under the additive model (beta = 0.02, p = 0.71) (Table S5).
Figure 2.
Simulation of power to detect PoO effects under different statistical models
Power to detect PoO effects under additive, parental, and differential models was simulated across ranges of minor allele frequencies (MAFs) and effect sizes assuming diverse patterns of PoO effects that could result from imprinting: (A) uniparental, (B) polar dominance, and (C) bipolar dominance effect. Bar plots at the top are illustrative examples of the various patterns of PoO effects that can result from imprinting. The horizontal axis displays a range of simulated effect sizes. The left vertical axes display the % power to detect the effect across a range of MAFs ordered on the right vertical. See text for detailed methods.
Discussion
Variants that affect imprinted genes can have PoO effects on traits and contribute to their variance; however, these effects may not be well captured by standard additive models that do not model parental origin. To address this, studies have employed approaches to assign parental origin of genetic variants based on known pedigrees and employed statistical models to specifically test for PoO effects, leading to the discovery of PoO-specific associations for various traits.2,8, 9, 10 The current study extends previous efforts by assigning the parental origin of genetic variants based on parent-offspring relationships inferred from genetic and demographic data in the absence of known pedigree and employing a high-throughput approach to detect PoO effects across hundreds of traits extracted from EHR in a large clinically ascertained study.
From the genome-wide screen for PoO effects in the DiscovEHR study, we identified 732 PoO associations across 154 quantitative traits and 27 PoO associations across 611 binary traits. Many of the associations that we found in the DiscovEHR study could not be replicated in the UK Biobank study primarily due to the relatively modest number of offspring (5,015), as well as lack of matching quantitative traits, and limited number of cases for binary clinical outcomes. This is evident by the observation that even well-established PoO associations with strong p values in the DiscovEHR study (i.e., KLF14 locus for lipids,2 GNAS locus for TSH,11 and IGF2 locus for type 2 diabetes2) (Tables S3–S6), did not reach nominal significance for replication (p < 0.05) in the UK Biobank study. Therefore, we anticipate that some of the associations that failed to replicate in the UK Biobank study may replicate in future studies that are better powered. Power for detecting PoO effects can be enhanced by employing study populations enriched for familial relationships and approaches that enable PoO assignment in a larger number of individuals. For example, approaches to leverage second-degree relatives for PoO assignment, such as in a study of the Icelandic population with extended pedigree information,2 may further improve power. This may be possible in the absence of known pedigree structures by identifying second-degree relationships based on kinship estimates from genetic data, determining ancestors and descendants based on age, and assigning the parental side of the ancestors based on additional information such as mitochondrial DNA or Y chromosome.
Interestingly, of nine associations that we discovered and replicated, only one association at the DLK1 locus resides within the known imprinted regions. This raises the possibility that PoO effects may be more common and widespread than were previously thought. While around 100 genes in humans are known to be imprinted based on the currently available evidence,20,22 high-throughput approaches applied to multiple tissue types in various developmental stages may reveal tissue- and time-specific imprinting effects on a larger number of genes. In line with this, recent studies that examined genome-wide PoO-specific DNA methylation patterns suggested that the DNA methylation pattern associated with imprinting is widespread and extends beyond the known imprinted regions.11,23 In addition to imprinting, parental genetic effects where the genotypes of the parents directly affect the phenotype of the offspring can lead to apparent PoO effects.24, 25, 26 As we gain more evidence that PoO effects influence multiple complex traits, it would be important to estimate the extent to which PoO effects explain the variance of those traits.
Imprinting can give rise to diverse patterns of PoO effects of genetic variants on traits,1 which in turn influence the power to detect these effects under different statistical models. Based on our power simulations, the additive model has better power to detect associations resulting from uniparental effects than the parental model in the given parameter space. Nonetheless, parental models provide more accurate effect size estimates for the causative parental allele because the additive model would underestimate them by not differentiating the causative and non-causative parental alleles. In addition, parental and differential models can be better powered for detecting associations resulting from polar or bipolar dominance effects than the additive model. This suggests that accurately accounting for the PoO effects of genetic variants can increase the statistical power and identify novel association signals that are not captured under the additive model. Furthermore, knowing the PoO-specific nature of genetic associations and effects can help accurately assess the risk conferred by the variants in the carriers and understand the molecular mechanism behind the genetic associations.
Many of the strongest PoO associations that we identified from the genome- and phenotype-wide scan for PoO effects in the DiscovEHR study have stronger associations under the additive model and are suspected to result from uniparental effects (based on their proximity to known imprinted genes with monoallelic expression). This observation might give the impression that a large portion of the associations with underlying PoO effects could have been discovered with the additive model alone at lower computational burden. However, we note that PoO models have smaller sample sizes than the additive model in the current study and that there are many PoO associations that have stronger associations under PoO models than the additive model, potentially driven by either polar or bipolar dominance effects, which could be missed if tested only under the additive model. In addition, without regular follow-up evaluation of the associations found under the additive model with PoO models, there may be a substantial missed opportunity to discriminate between additive and PoO effects. While it is not possible to extrapolate a universal cost-benefit from our current study due to limited power and the small number of true positives, we anticipate that future studies with greater statistical power and better mechanistic understanding of the PoO effects could help address this issue with more confidence.
In summary, we report a bioinformatic and statistical approach to screen for PoO effects of genetic variants and its application in the DiscovEHR study enriched with familial relationships and phenotypic data derived from ERH. The current study provides a valuable reference point for future studies aimed to find PoO effects and suggests the need for approaches that can increase statistical power to detect PoO effects, methods to assess the contribution of PoO effects to genetic heritability of complex traits, and efforts to delineate the mechanisms behind the observed PoO associations by incorporating epigenetic and transcriptomic resources and experimental models.
Acknowledgments
We thank the DiscovEHR participants and all research staff and teams at the Regeneron Genetics Center and Geisinger who contributed to the current study. We also thank the participants, researchers, and organizers of the UK Biobank study. This work has been conducted using UK Biobank application 26041. We thank the Regeneron postdoctoral program for their support and guidance for H.K. The study is funded by the Regeneron Genetics Center and Regeneron Pharmaceuticals.
Declaration of interests
H.K., B.Y., J.S., A.M., C.G., A.R.S., and C.V.V.H. are current or former employees and/or stockholders of Regeneron Genetics Center. H.K. is a current employee of Pfizer, but work was conducted at Regeneron Genetics Center. C.V.V.H. is a current employee of the National Autonomous University of Mexico, but work was largely conducted at Regeneron Genetics Center.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100039.
Contributor Information
Hye In Kim, Email: hyein.kim267@gmail.com.
Cristopher V. Van Hout, Email: cvanhout@liigh.unam.mx.
Data and code availability
Summary statistics of all significant results are provided in the article. The data that support the reported findings and the codes used for the analyses are available from authors upon reasonable request.
Web resources
Michigan Imputation Server, http://imputationserver.sph.umich.edu/index.html
OMIM, https://www.omim.org
PLINK, www.cog-genomics.org/plink
Supplemental information
References
- 1.Lawson H.A., Cheverud J.M., Wolf J.B. Genomic imprinting and parent-of-origin effects on complex traits. Nat. Rev. Genet. 2013;14:609–617. doi: 10.1038/nrg3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K.T., Jonasdottir A., et al. DIAGRAM Consortium Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace C., Smyth D.J., Maisuria-Armer M., Walker N.M., Todd J.A., Clayton D.G. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Small K.S., Hedman A.K., Grundberg E., Nica A.C., Thorleifsson G., Kong A., Thorsteindottir U., Shin S.Y., Richards H.B., Soranzo N., et al. GIANT Consortium. MAGIC Investigators. DIAGRAM Consortium. MuTHER Consortium Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson R.L., Guo T., Muller Y.L., Fleming J., Knowler W.C., Kobes S., Bogardus C., Baier L.J. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62:2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry J.R., Day F., Elks C.E., Sulem P., Thompson D.J., Ferreira T., He C., Chasman D.I., Esko T., Thorleifsson G., et al. Australian Ovarian Cancer Study. GENICA Network. kConFab. LifeLines Cohort Study. InterAct Consortium. Early Growth Genetics (EGG) Consortium Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoledziewska M., Sidore C., Chiang C.W.K., Sanna S., Mulas A., Steri M., Busonero F., Marcus J.H., Marongiu M., Maschio A., et al. UK10K consortium. Understanding Society Scientific Group Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 2015;47:1352–1356. doi: 10.1038/ng.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benonisdottir S., Oddsson A., Helgason A., Kristjansson R.P., Sveinbjornsson G., Oskarsdottir A., Thorleifsson G., Davidsson O.B., Arnadottir G.A., Sulem G., et al. Epigenetic and genetic components of height regulation. Nat. Commun. 2016;7:13490. doi: 10.1038/ncomms13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly S., Anney R., Gallagher L., Heron E.A. A genome-wide investigation into parent-of-origin effects in autism spectrum disorder identifies previously associated genes including SHANK3. Eur. J. Hum. Genet. 2017;25:234–239. doi: 10.1038/ejhg.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mozaffari S.V., DeCara J.M., Shah S.J., Sidore C., Fiorillo E., Cucca F., Lang R.M., Nicolae D.L., Ober C. Parent-of-origin effects on quantitative phenotypes in a large Hutterite pedigree. Commun. Biol. 2019;2:28. doi: 10.1038/s42003-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zink F., Magnusdottir D.N., Magnusson O.T., Walker N.J., Morris T.J., Sigurdsson A., Halldorsson G.H., Gudjonsson S.A., Melsted P., Ingimundardottir H., et al. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 2018;50:1542–1552. doi: 10.1038/s41588-018-0232-7. [DOI] [PubMed] [Google Scholar]
- 12.Staples J., Maxwell E.K., Gosalia N., Gonzaga-Jauregui C., Snyder C., Hawes A., Penn J., Ulloa R., Bai X., Lopez A.E., et al. Profiling and Leveraging Relatedness in a Precision Medicine Cohort of 92,455 Exomes. Am. J. Hum. Genet. 2018;102:874–889. doi: 10.1016/j.ajhg.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C., et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staples J., Qiao D., Cho M.H., Silverman E.K., Nickerson D.A., Below J.E., University of Washington Center for Mendelian Genomics PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B., et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morison I.M., Ramsay J.P., Spencer H.G. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv. Exp. Med. Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- 22.Monk D., Mackay D.J.G., Eggermann T., Maher E.R., Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019;20:235–248. doi: 10.1038/s41576-018-0092-0. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y., Amador C., Xia C., Marioni R., Sproul D., Walker R.M., Morris S.W., Bretherick A., Canela-Xandri O., Boutin T.S., et al. Parent of origin genetic effects on methylation in humans are common and influence complex trait variation. Nat. Commun. 2019;10:1383. doi: 10.1038/s41467-019-09301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf J.B., Wade M.J. What are maternal effects (and what are they not)? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:1107–1115. doi: 10.1098/rstb.2008.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gleason G., Liu B., Bruening S., Zupan B., Auerbach A., Mark W., Oh J.E., Gal-Toth J., Lee F., Toth M. The serotonin1A receptor gene as a genetic and prenatal maternal environmental factor in anxiety. Proc. Natl. Acad. Sci. USA. 2010;107:7592–7597. doi: 10.1073/pnas.0914805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong A., Thorleifsson G., Frigge M.L., Vilhjalmsson B.J., Young A.I., Thorgeirsson T.E., Benonisdottir S., Oddsson A., Halldorsson B.V., Masson G., et al. The nature of nurture: Effects of parental genotypes. Science. 2018;359:424–428. doi: 10.1126/science.aan6877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics of all significant results are provided in the article. The data that support the reported findings and the codes used for the analyses are available from authors upon reasonable request.