Abstract
Large numbers of expression quantitative trait loci (eQTLs) have recently been identified in humans, and many of these regulatory variants have large allele frequency differences between populations. Here, we conducted genome-wide scans of selection to identify adaptive eQTLs (i.e., eQTLs with large population branch statistics). We then tested if tissue pleiotropy affects whether eQTLs are more or less likely to be adaptive and identified tissues that have been key targets of positive selection during the last 100,000 years. Top adaptive eQTL outliers include rs1043809, rs66899053, and rs2814778 (a SNP that is associated with malaria resistance). We found that effect sizes of eQTLs were negatively correlated with population branch statistics and that adaptive eQTLs affect two-thirds as many tissues as do non-adaptive eQTLs. Because the tissue breadth of an eQTL can be viewed as a measure of pleiotropy, these results imply that pleiotropy inhibits adaptation. The proportion of eQTLs that are adaptive varies by tissue, and we found that eQTLs that regulate expression in testis, thyroid, blood, or sun-exposed skin are enriched for signatures of positive selection. By contrast, eQTLs that regulate expression in the cerebrum or female-specific tissues have a relative lack of adaptive outliers. Scans of selections also reveal that many adaptive eQTLs are closely linked to disease-associated loci. Taken together, our results indicate that eQTLs have played an important role in recent human evolution.
Keywords: adaptation, eQTLs, gene expression, pleiotropy, natural selection, population genetics
Introduction
Regulatory mutations and changes in gene expression can lead to functional differences in anatomy, physiology, and behavior that are evolutionarily important.1, 2, 3, 4, 5, 6 Polymorphic sites that influence gene expression are known as expression quantitative trait loci (eQTLs), and many of these sites are relevant to human health and disease.7, 8, 9 Although identifying specific nucleotides that cause differences in gene expression can be challenging,10 many eQTLs have been identified in model and non-model organisms.11, 12, 13 In recent years, hundreds of thousands of human eQTLs have been cataloged in the Genotype-Tissue Expression (GTEx) project and RegulomeDB databases.14, 15, 16, 17 Many of these eQTLs act in a tissue-specific manner, and by studying adaptive eQTLs, it is possible to identify the tissues that have been the primary targets of recent human evolution. Although eQTL effect sizes and directions of effect tend to be conserved among human populations,18 array and sequence data reveal that gene expression patterns vary across populations19,20 (though some of these differences may be due to technical artifacts21). Many eQTLs have divergent allele frequencies across populations, and local adaptation may underlie these differences.22,23 Hereditary disease risks have evolved in the recent past,24,25 and many of these changes are likely due to positive selection acting on regulatory DNA.
Adaptation is a fundamental concern of evolutionary biology, and recent years have seen a contentious debate about whether adaptation tends to proceed via non-synonymous changes in coding regions (amino acid changes) or due to changes in gene regulation.26, 27, 28 Of particular relevance is the fact that less than only 1.5% of the human genome is coding,29 and many scans of positive selection have implicated intergenic regions of the human genome.30,31 Regardless of the proportion of the human genome that is functional,32,33 there are multiple reasons why some changes in gene expression may be beneficial. Unlike non-synonymous changes that affect protein sequences across the body, eQTLs can modify gene expression in a tissue-specific manner, and gene expression can also be optimized for a given environment.34 Although many eQTLs are likely to be evolving neutrally, there is a growing body of empirical evidence that regulatory DNA is an important target of selection in humans.35, 36, 37, 38
Many methods of detecting positively selected alleles exist, including population branch statistics (PBS).39, 40, 41 These within-species scans of selection use genetic distances between multiple populations to identify outlier loci that have undergone accelerated evolution along one branch of a population-level phylogenetic tree. Scans of selection that examine population differentiation, such as PBS, are well-suited for detecting selection that has occurred on a continental scale during the last 100,000 years.42 Because PBS scores do not rely on extended haplotype heterozygosity, they are robust to whether adaptive alleles are due to new mutations or standing genetic variation. PBS scores are also able to detect partial sweeps.
Evolutionary theory informs our understanding about which types of eQTLs are expected to be adaptive. As formulated by Fisher and Orr, the geometric model of adaptation posits that mutations of a small effect are more likely to be positively selected than mutations of a large effect when populations are close to a fitness optimum.43, 44, 45, 46 Because of this, we predict that adaptive eQTLs are unlikely to involve large changes in gene expression. Similarly, pleiotropy (including tissue breadth) can inhibit adaptation,47 which leads to the prediction that most adaptive eQTLs will affect a small number of tissues. Scans of selection in human genomes have revealed that immunity genes tend to be fast-evolving.48,49 There is also evidence from Drosophila that testis-expressed genes evolve quickly,50 and reproductive genes have experienced elevated rates of evolution in many vertebrate lineages.51 Because of this, eQTLs that affect fast-evolving tissues are expected be enriched for adaptive PBS outliers. Despite these predictions, multiple knowledge gaps exist. The extent to which eQTL effect sizes and tissue breadth constrain human adaptation has yet to be tested empirically. It is also unknown whether tissues that have been targets of recent human adaptation are the same tissues that experienced accelerated evolution over deeper timescales. Importantly, affordable sequencing has ushered in an era of population genomics, and thousands of tissue-specific eQTLs have recently been identified.16 For the first time, a comprehensive understanding of adaptive eQTLs in human populations is possible.
Here, we combine continental allele frequencies from the 1000 Genomes Project (1KGP) with eQTL data from the GTEx project and RegulomeDB to identify adaptive eQTLs in human populations. We focus on five questions: (1) Which eQTLs exhibit signatures of local adaptation? (2) Are pleiotropic eQTLs less likely to be positively selected? (3) Does the effect size of an eQTL affect whether it is adaptive? (4) Which tissues tend to be targets of local adaptation? (5) To what extent do adaptive eQTLs overlap with genome-wide association study (GWAS) results?
Material and methods
Population genomic data and eQTL datasets
Allele frequencies at 80,701,406 autosomal SNPs were obtained from phase 3 of the 1KGP.52 Continental super-populations from the 1KGP were used: Africa (African Caribbean in Barbados [ACB], African Ancestry in Southwest USA [ASW], Esan in Nigeria [ESN], Gambian in Western Division – Mandinka [GWD], Luhya in Webuye, Kenya [LWK], Mende in Sierra Leone [MSL], and Yoruba in Ibadan, Nigeria [YRI]), Europe (Utah residents with Northern and Western European ancestry [CEU], Finnish in Finland [FIN], Iberian populations in Spain [IBS], British from England and Scotland [GBR], and Toscani in Italy [TSI]), and East Asia (Chinese Dai in Xishuangbanna, China [CDX], Han Chinese in Beijing, China [CHB], Han Chinese South [CHS], Kinh in Ho Chi Minh City, Vietnam [KHV]. and Japanese in Tokyo, Japan [JPT]). Sample sizes varied for each continent population: 661 individuals of African descent, 503 individuals of European descent, and 504 individuals of East Asian descent. Biallelic SNPs from phase 3 of the 1KGP (ascertained via whole-genome sequencing) were merged with rs identifiers from the Illumina Omni 2.5M array, RegulomeDB, and GTEx project. RegulomeDB scores of 1a, 1b, 1c, 1d, 1e, or 1f indicate that a SNP is a RegulomeDB eQTL.17 For V7 GTEx eQTLs, we required sample sizes of at least 70 individuals per tissue, yielding 48 tissues. To correct for multiple statistical tests, GTEx eQTLs were required to have a p value ≤ 10−9 for at least one tissue. Allele frequency and eQTL data were merged using the dplyr package in R.53 Genomic positions described here are from the GRCh37/hg19 assembly.
Genetic distances and scans of selection
Weir and Cockerham's FST was calculated for each pairwise combination of populations (Africa-Europe [AFR-EUR], Africa-East Asia [AFR-EAS], and Europe-East Asia [EUR-EAS]).54,55 This method of calculating genetic distances corrects for small sample sizes. Six types of SNPs were analyzed: variants from the 1KGP (ascertained via whole-genome sequencing), variants on the Illumina Omni 2.5M array, V7 GTEx eQTLs, RegulomeDB eQTLs, and simulated neutral loci that were generated via SLiM.56 For each type of SNP, empirical cumulative distribution functions and mean values of FST were found for each population pair. To identify adaptive SNPs, PBS40,41 were then calculated for V7 GTEx eQTLs using the following equations:
| (Equation 1A) |
| (Equation 1B) |
| (Equation 1C) |
Undefined and negative values of Weir and Cockerham's FST were treated as zero for PBS calculations. Genome-wide distributions of PBS scores were calculated for each branch (Africa, Europe, and East Asia). In total, this yielded 1,154,731 PBS scores for V7 GTEx eQTLs. Negative PBS scores were treated as zero. We classified eQTLs as adaptive outliers if they had PBS scores above the 99th percentile of all eQTLs. Previous studies have indicated that this is a reasonable PBS threshold.57, 58, 59 To correct for the effects of linkage disequilibrium (LD), we selected the eQTL with the top PBS score in each 100 kb genomic window. To increase rigor, analyses were repeated using a cutoff of the top 0.1% eQTL PBS scores (also LD pruned).
Integrated haplotype score (iHS) statistics
iHSs quantify selection acting on new mutations (i.e., they can be used to identify targets of hard selective sweeps).60 Here, we used precomputed iHS statistics from hapbin61,62 to examine whether PBS outliers exhibit additional signatures of selection. Representative populations from the 1KGP were chosen for each continent (YRI: Africa, CEU: Europe, and CHB: East Asia). For each population, iHS statistics were available for autosomal loci with minor allele frequencies (MAF) >0.05. To enable comparisons between different types of variants, raw iHSs were converted to population-specific percentiles. Distributions of iHS statistics for adaptive outliers were compared with non-outlier eQTLs using Wilcoxon rank-sum tests (these comparisons did not involve LD pruning of either type of eQTL).
SLiM simulations
Computer simulations were used to generate a null dataset of neutrally evolving loci in Africa, Europe, and East Asia. Using v.3.2.1 of SLiM,56 we simulated the Gravel model of human demography.63 This model includes a Eurasian bottleneck followed by gene flow between each continental population. The recipe for the Gravel model (Section 5.4 of the SLiM 3 Manual) was modified to include larger chromosome sizes, recent explosive population growth,64 and outputs analogous to 1KGP data (661 AFR, 503 EUR, and 504 EAS individuals). This code was run 22 times to generate a set of virtual autosomes, and PLINK 2 was used to obtain allele frequencies from simulated vcf files.65 As noted above, PBS scores were calculated for a total of 47,643 simulated loci. The SLiM code is available at https://github.com/LachanceLab/adaptive_eQTLs.
Pleiotropy and tissue breadth
Highly pleiotropic eQTLs modify the expression of a large number of tissues. We generated tissue breadth scores for each eQTL by counting the number of tissues affected by each eQTL. These scores range between 1 and 48 for V7 GTEx eQTLs. To identify whether adaptive eQTLs affect a different number of tissues than non-adaptive eQTLs, empirical cumulative distribution functions and mean values of tissue breadth scores were calculated for non-adaptive eQTLs and adaptive PBS outliers. Derived allele frequencies in Africa, Europe, and East Asia were used as proxies of allele age for comparisons of testis-specific eQTLs with multi-tissue eQTLs that affect gene expression in the testis.66 Comparisons of derived allele frequency distributions used Wilcoxon rank-sum tests.
Effect size comparisons
We tested whether eQTLs with large PBS scores yield large differences in gene expression. Here, we focused on LD-pruned eQTLs that only affect a single tissue. Effect sizes for each eQTL and tissue combination were quantified by taking the absolute value of normalized effect size (NES) under a fixed effect model, i.e., |βFE|. The “ggscatter” function in the ggpubr R package was used for local regression (loess) fitting and to quantify how PBS scores and |βFE| values are correlated. Note that loess fitting allows non-linear relationships to be identified. This analysis was repeated for three different continental populations (African, European, and East Asian) and for all 48 tissues in the V7 GTEx dataset. Wilcoxon rank-sum tests were used to determine whether single-tissue eQTLs with PBS scores in the top 1% had effect sizes that differ from eQTLs with PBS scores in the bottom 99%.
Tissue-specific adaptation
For this analysis, we required adaptive outliers to have PBS scores above the 99th percentile of all eQTLs. After LD pruning, the proportion of eQTLs that are adaptive was found by taking the geometric mean of the number of LD-pruned PBS outliers divided by the number of eQTLs for each tissue:
| (Equation 2) |
k refers to the total number of tissues. We then found the expected number of LD-pruned adaptive PBS outliers for each tissue:
| (Equation 3) |
Enrichment ratio statistics were calculated for each tissue by comparing the observed number of adaptive PBS outliers to the expected number of adaptive PBS outliers:
| (Equation 4) |
The geometric mean used in Equation 2 ensures that the sum of all tissue-specific enrichment ratios generated using Equation 4 is zero. Positive enrichment ratios indicate tissues with a relative excess of adaptive eQTLs, and negative enrichment ratios indicate tissues with a relative lack of adaptive eQTLs. Enrichment ratio statistics were calculated for 48 V7 GTEx tissues. 95% confidence intervals for enrichment ratios were found by using the Agresti-Coull approach to find the lower and upper bounds for tissue-specific outlier proportions.67 We also adjusted for sample size as a covariate using the following equation:
| (Equation 5) |
ntissue refers to the number of individuals sampled per tissue. Tissue-specific enrichment ratios were linearly regressed against sample size, yielding an intercept (m) of 0.0012 and a slope (b) of −0.2517.
Overlap with GWAS loci
We assessed the overlap between adaptive eQTLs and GWAS loci by downloading data from the EBI-NHGRI GWAS Catalog.68,69 For this analysis, LD-pruned V7 GTEx eQTLs with PBS scores in the top 1% were considered to be adaptive. Distances between adaptive eQTLs and GWAS loci were found using the “closest” function in BEDTools.70
Results
Scans of selection identify adaptive eQTLs
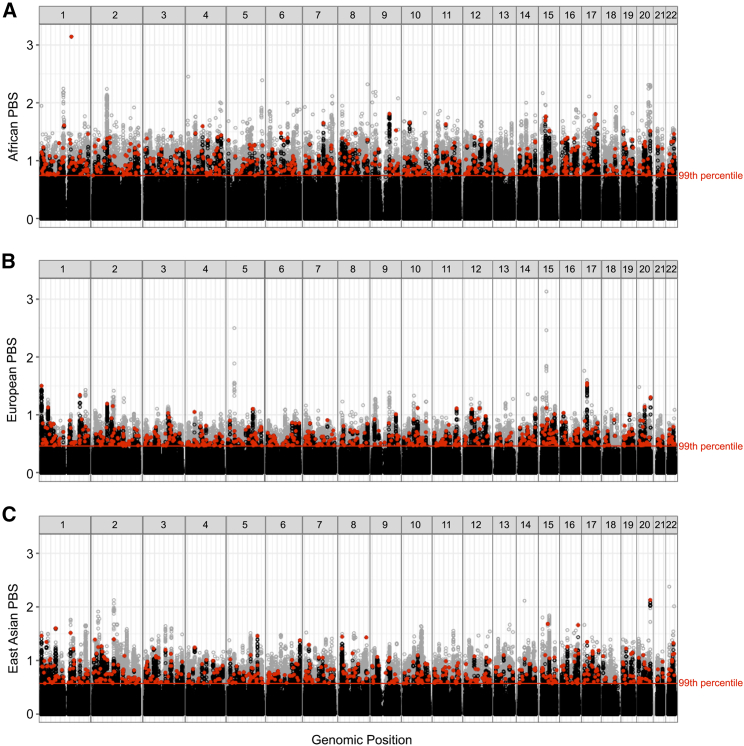
Many presently known eQTLs have large allele frequency differences between populations, and some of these eQTLs are targets of local adaptation. We calculated PBS scores for individuals from Africa, Europe, and East Asia at every biallelic SNP in the 1KGP (Figure 1). We then identified the top 1% of all V7 GTEx eQTLs with respect to PBS for each population. LD pruning yielded 614 eQTLs that are adaptive outliers for Africa, 561 eQTLs that are adaptive outliers for Europe, and 524 eQTLs that are adaptive outliers for East Asia. Different numbers of LD-pruned adaptive outliers were observed for each branch because African genomes have smaller haplotype blocks than do European and Asian genomes.52 In Figure 1, LD-pruned adaptive eQTLs are represented by filled red circles, other eQTLs are represented by open black circles, and 1KGP SNPs that are not eQTLs are represented by open gray circles. Scans of selection reveal that the strongest PBS signal in many adaptive regions of the genome is an eQTL (visualized as red-tipped peaks in the Manhattan plots of Figure 1). Overall, we find that GTEx eQTLs are 2.53 times as likely than SNPs from the 1KGP to have signatures of positive selection, i.e., PBS scores above the red lines in Figure 1 (p < 0.0001, chi-square test of independence).
Figure 1.
Genome-wide scans of selection identify adaptive eQTLs in human populations
(A–C) Manhattan plot of population branch statistics (PBS) versus genomic position for each population: (A) Africa, (B) Europe, and (C) East Asia. All variants in the 1KGP, including V7 GTEx eQTLs, are shown here. LD-pruned adaptive eQTLs are represented by filled red circles, other eQTLs are represented by open black circles, and 1KGP SNPs that are not eQTLs are represented by open gray circles.
Validation of adaptive PBS outliers
Additional lines of evidence reinforce the claim that LD-pruned eQTLs with high PBS scores are positively selected loci, including iHS statistic comparisons between eQTLs and non-regulatory DNA as well as computer simulations of neutrally evolving loci. PBS outliers are more likely to be found in genomic regions with extended haplotype homozygosity than are other variants from the 1KGP. Examining non-LD-pruned eQTLs, we find that these differences are statistically significant for Africa, Europe, and East Asia (p < 2.2 × 10−16, Wilcoxon rank-sum tests). Computer simulations also reveal that eQTLs with PBS scores above the outlier cutoffs (red lines) in Figure 1 are unlikely to be neutral. Using SLiM and a modified version of the Gravel model of human demography, we simulated neutrally evolving loci in Africa, East Asia, and Europe. Most simulated neutral loci and non-regulatory SNPs from the 1KGP have small FST statistics (Figure S1). Overall, we find that 99.8% of all simulated loci have PBS scores that are below the adaptive outlier cutoffs shown in Figure 1. Similarly, 99.6% of non-regulatory variants from the 1KGP have PBS scores that are below the cutoffs used in this paper. Collectively, these findings provide additional support that PBS outliers are due to adaptive evolution, especially in light of recent claims about the pervasiveness of positive selection.71
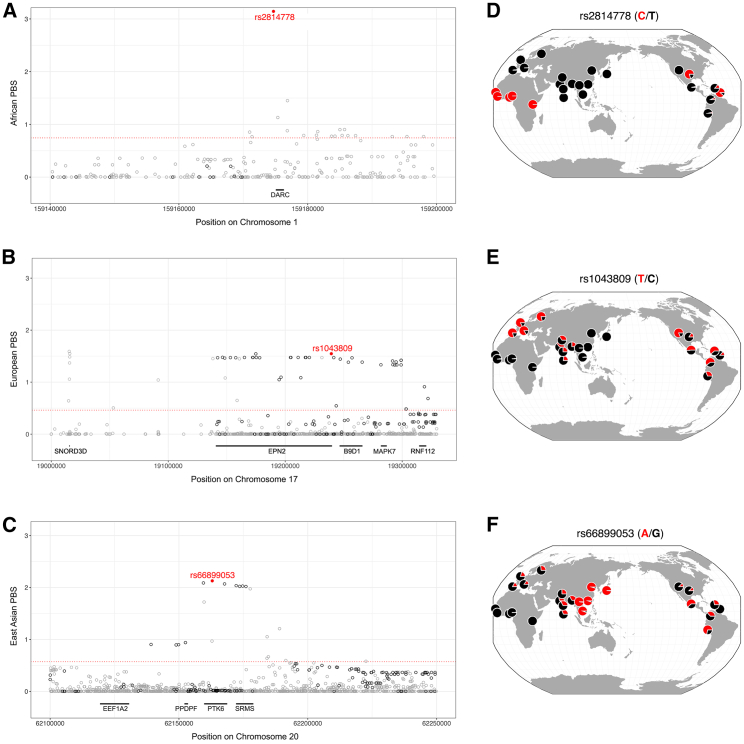
eQTLs with the highest PBS scores
Here, we highlight the strongest signatures of adaptation for each population (Figure 2). rs2814778 has the highest PBS score for the African branch. This C/T SNP has a striking geographic pattern: African frequencies of the C allele are >96% and non-African allele frequencies of the C allele are <1%. rs2814778 affects the gene expression of DARC, also known as ACKR1. rs2814778 is in the promoter of the DARC gene, and the C allele at this regulatory locus confers a null phenotype.72 DARC encodes the Duffy blood group antigen, which is known to be adaptive with respect to Plasmodium vivax and malaria.73,74 rs2814778 is also predictive of neutrophil counts in African Americans.75 Despite a lack of local recombination hotspots, rs2814778 has negligible amounts of LD with nearby SNPs. This hints that selection acting on the Duffy blood group may have acted on standing genetic variation.73 rs2814778 only modifies expression in whole blood, and this tissue-specificity and lack of pleiotropy may contribute strong signatures of positive selection at this eQTL.
Figure 2.
Adaptive eQTLs with strong signatures of positive selection
(A–C) Genomic regions flanking eQTLs with the highest PBS score for each continental population. LD-pruned adaptive eQTLs are represented by filled red circles, other eQTLs are represented by open black circles, and 1KGP SNPs are represented by open gray circles. hg19 coordinates are shown. Population-specific 99th percentiles of PBS scores are represented by dashed red lines.
(D–F) Allele frequencies for 26 populations from the 1KGP (modified from the Geography of Genetics Variants Browser105).
rs1043809 is the eQTL with the highest PBS score for the European branch. This C/T SNP is near the EPN2, B9D1, and RNF112 genes at 17p11. At present, the reason why this genomic region was positively selected in Europe is unknown. Many eQTLs that are closely linked to rs1043809 have similar PBS statistics (visualized as a plateau of points in Figure 2B), which suggests the existence of an adaptive haplotype rather than of a single SNP.
rs66899053 is the eQTL with the highest PBS score for the East Asian branch. This A/G SNP modifies expression of the EEF1A2, PPDPF, PTK6, and SRMS genes, and it is found in an adaptive haplotype at 20q13. Scans of selection have previously implicated this genomic region with respect to Helicobacter pylori infection and gastric cancer.76 Consistent with this cause, rs66899053 affects gene expression in the stomach and in many other tissues. Intriguingly, rs66899053 is found in a genomic region that has previously been shown to contain adaptively introgressed Neanderthal alleles in non-African populations.77,78 rs66899053 is also 427 kb away from HAR1, a genomic region that has undergone accelerated evolution in humans following the split between human and chimpanzee lineages.79
Pleiotropy and the tissue breadth of eQTLs
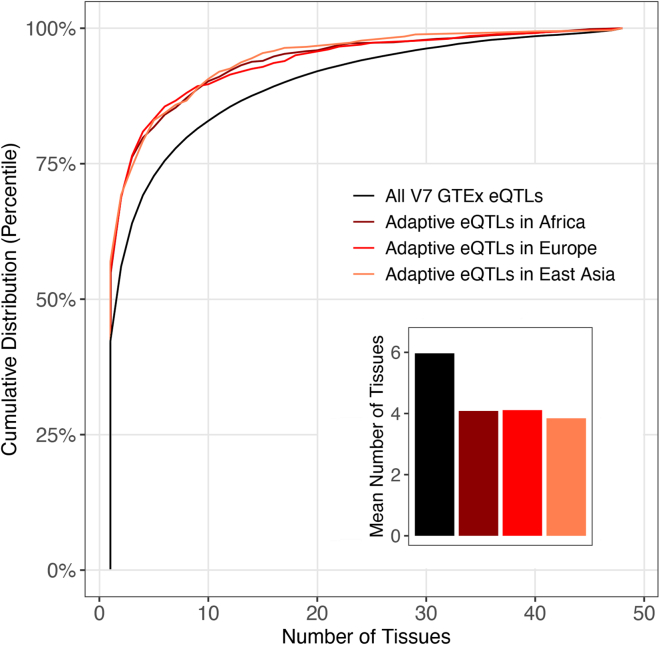
We compared the number of tissues affected by adaptive and non-adaptive eQTLs to infer whether tissue breadth constrains adaptive evolution. Each of the V7 GTEx eQTLs analyzed here modifies expression in up to 48 tissues. Overall, adaptive eQTL outliers affect fewer tissues than non-adaptive eQTLs (Figure 3). This pattern occurs regardless of whether eQTLs have high PBS scores in Africa, Europe, or East Asia. Non-adaptive eQTLs affect a mean number of 5.97 tissues. By contrast, adaptive eQTL outliers affect a mean number of 4.08, 4.11, and 3.84 tissues (African, European, and East Asian outliers, respectively). These differences between adaptive outliers and other eQTLs are statistically significant (p < 2.2 × 10−16 for all comparisons, Wilcoxon rank-sum tests). Furthermore, 55.3% of all LD-pruned adaptive outliers affect a single tissue. Similar patterns arise if a more stringent PBS score cutoff is used to identify positive selected eQTLs (Figure S2). Because tissue breadth (the number of different tissues that each eQTL affects) can be viewed as a form of pleiotropy, our results indicate that pleiotropy appears to inhibit adaptation.
Figure 3.
Highly pleiotropic eQTLs are less likely to be adaptive
Cumulative distributions and mean number of tissues are shown for adaptive and non-adaptive eQTLs. Here, adaptive outliers are LD-pruned eQTLs that have PBS scores in the top 1% of all GTEx eQTLs. In general, adaptive eQTLs modify gene expression in fewer tissues than do non-adaptive eQTLs. For each population, differences in the number of tissues affected by adaptive outliers and the overall set of GTEx eQTLs are statistically significant (p < 2.2 10−16 for all comparisons, Wilcoxon rank-sum tests).
Many new genes are known to have testis-specific expression patterns,80 which suggests that tissue breadth may be related to allele age. We find that testis-specific eQTLs tend to have lower derived allele frequencies than do eQTLs that affect multiple tissues (African p = 9.334 × 10−16, European p < 2.2 × 10−16, East Asian p = 6.446 × 10−10, Wilcoxon rank-sum tests). As derived allele frequencies can be used as a proxy of allele age,66 this indicates that testis-specific eQTLs are younger, on average, than are eQTLs that affect multiple tissues.
Effect sizes of single tissue eQTLs
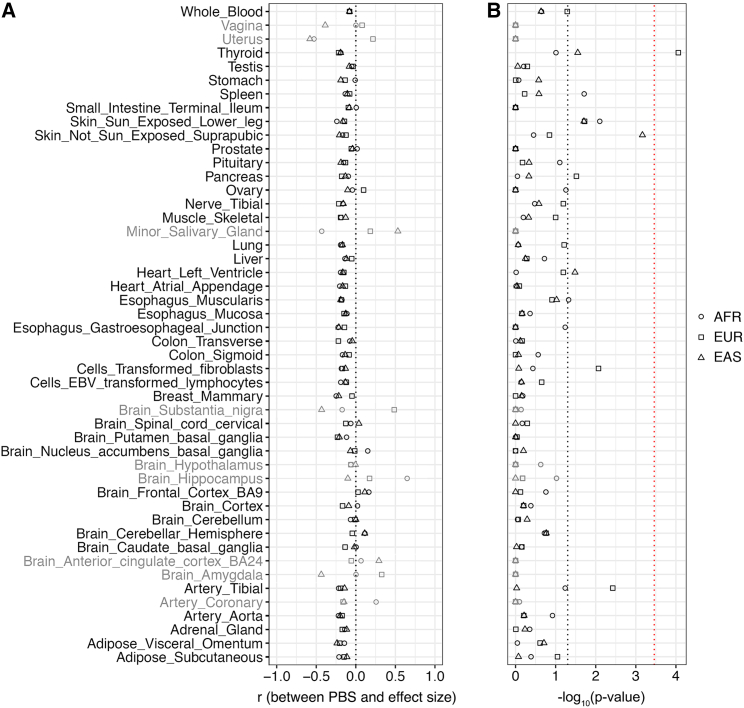
For each combination of tissue and population, we tested whether adaptive PBS outliers have different expression effect sizes than do other eQTLs. Consistent with the Fisher-Orr geometric model of adaptation, most tissues yield a weak negative correlation between PBS scores and effect sizes (Figure 4A; r ≈ −0.2, local regression). In other words, single-tissue eQTLs with large PBS scores tend to have a slightly smaller effect on gene expression than do single tissue eQTLs with small PBS scores. However, due to the relatively small number of single-tissue eQTLs, most comparisons yield p values that fail to meet a Bonferroni correction cutoff (Figure 4B; Wilcoxon rank-sum tests). The one exception to this pattern involves thyroid eQTLs in Europe (p = 8.72 × 10−5, Wilcoxon rank-sum test between outlier and non-outlier eQTLs). In general, our results indicate that there are small differences in the effect sizes of adaptive eQTLs compared with in other eQTLs.
Figure 4.
Most adaptive eQTLs do not have a large effect on gene expression
(A) Population-specific correlations between PBS and effect size for each tissue.
(B) Results of Wilcoxon rank-sum tests comparing the effect sizes of adaptive outlier and non-outlier eQTLs. Uncorrected (p < 0.05) and Bonferroni corrected p value cutoffs are indicated by dotted black and red lines, respectively. Gray shading indicates tissues that have fewer than 200 LD-pruned single-tissue eQTLs.
Tissue-specificity of adaptive eQTLs
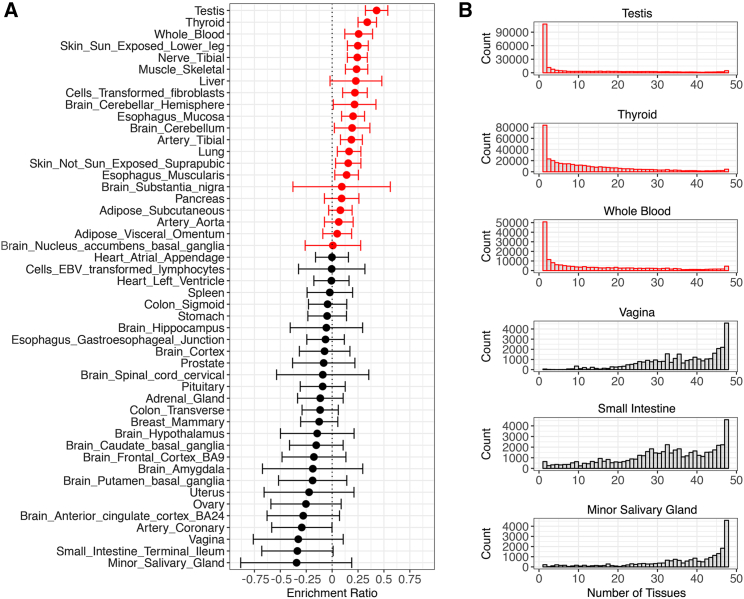
The proportion of eQTLs that are adaptive varies by tissue. Here, we used enrichment ratio scores to compare the observed and expected counts of adaptive PBS outliers for each tissue (i.e., we compared the relative proportions of tissue-specific eQTLs with PBS scores above or below each outlier threshold). Positive enrichment ratio scores indicate tissues that have an excess of adaptive eQTLs compared with null expectations, and negative enrichment ratio scores indicate tissues that have a relative lack of adaptive eQTLs compared with null expectations. Because enrichment ratio scores use a natural log scale, a difference of one unit translates to a 2.718-fold difference in the relative proportion of eQTLs that are adaptive outliers.
Focusing on individual tissues, testis eQTLs are the most likely to have high PBS scores, followed by eQTLs that modify gene expression in the thyroid, whole blood, and sun-exposed skin (Figure 5A). Our results suggest that recent human adaptation may have been driven by sexual selection, metabolism, pathogens, and local environmental conditions. We note that adipose, pancreas, and liver are moderately enriched for adaptive eQTLs—indicating that diet has also had an evolutionary impact. By contrast, we find that eQTLs that affect expression in the prostate, ovary, uterus, or vagina have a relative lack of adaptive outliers. We also find that eQTLs that affect expression in the cerebellum are more likely to be adaptive than are eQTLs that affect expression in the cerebrum.
Figure 5.
Tissue enrichment of adaptive eQTLs
Positive enrichment ratio scores indicate a relative excess of adaptive eQTLs for a particular tissue (red), and negative enrichment ratio scores indicate a relative lack of adaptive eQTLs for a particular tissue (black).
(A) Tissues are ranked by enrichment ratio score. Here, adaptive outliers are LD-pruned eQTLs that have PBS scores in the top 1% of all GTEx eQTLs. 95% confidence intervals for each enrichment ratio are shown.
(B) Tissues with high enrichment ratios tend to have eQTLs that affect less than 10 tissues, and tissues with low enrichment ratios tend to have eQTLs that affect more than 40 tissues.
Pleiotropy contributes to whether tissues are enriched for adaptive outliers. eQTLs that modify expression in tissues with positive enrichment ratio scores tend to be eQTLs that affect a small number of additional tissues. For example, testis eQTLs that are adaptive tend to affect only a small number of additional tissues, if at all (Figures S3, S4, and S5). By contrast, eQTLs that modify expression in tissues with negative enrichment ratio scores tend to be eQTLs that affect many additional tissues (Figure 5B).
Some tissues have large sample sizes in the GTEx database, including the thyroid, whole blood, sun-exposed skin, and skeletal muscle (Figure S6). To correct for sample size as a covariate, we linearly regressed tissue-specific enrichment ratios against sample size, yielding a set of adjusted enrichment ratios. Adjusted enrichment ratios measure how much a given tissue is above or below the regression line in Figure S7, i.e., they are the residuals. Many tissues that have positive enrichment ratios also have positive adjusted enrichment ratios, including testis, thyroid, liver, transformed fibroblasts, and the cerebellum. These findings indicate that tissue-specific differences in the proportions of adaptive outliers are not simply due to differences in sample sizes.
Overlap with GWAS results
Noting that colocalization of eQTLs and GWAS signals has previously been used to prioritize target genes that are associated with complex traits,81, 82, 83, 84 we tested the extent to which adaptive outliers overlap with loci that are associated with complex traits and disease susceptibility. A total of 29 adaptive eQTLs are exact matches with GWAS loci from the EBI-NHGRI Catalog. Traits associated with these adaptive eQTLs include white blood cell count, schizophrenia risk, BMI, and eye color. An important consideration is that GWAS loci tag genomic regions that are associated with complex traits, i.e. they are sentinel SNPs.85 Causal SNPs are often 10–100 kb distant from sentinel SNPs,86 and many eQTLs are closely linked to GWAS loci. Indeed, the median distance of V7 GTEx eQTLs to GWAS loci is 26.4 kb. Adaptive PBS outliers have a similar median distance to GWAS loci (26.9 kb). By contrast, random SNPs from the 1KGP have a median distance to GWAS loci of 30.2 kb. These differences are statistically significant: p < 2.2 × 10−16 for all eQTLs compared with 1KGP SNPs, and p = 7.6 × 10−4 for adaptive PBS outliers compared with 1KGP SNPs (Wilcoxon rank-sum tests). Although close proximity to GWAS loci need not imply that regulatory variants are causal, physical linkage can have implications for health inequities.87 This is because local adaptation results in large allele frequency differences between populations for not only the direct targets of selection but also for the linked loci.88 The combination of positive selection acting on eQTLs and genetic hitchhiking may contribute to population-level differences in functionally important traits, including hereditary disease risks.
Discussion
We found that most adaptive alleles do not have large effect sizes and that tissue pleiotropy appears to inhibit adaptation.89 eQTLs that modify expression in a small number of tissues are more likely to have large allele frequency differences between populations than are eQTLs that affect a large number of tissues. These results are consistent with the Fisher-Orr model, which posits that large phenotypic changes are less likely to be adaptive than are small changes.43, 44, 45, 46
Multiple tissues that are enriched for adaptive outliers are involved in resistance to pathogen pressure (e.g. blood, esophageal mucosa, and the skin), and there is prior evidence that the immune response is an important selective pressure.90 We also found that eQTLs that modify gene expression in the cerebellum are more likely to be adaptive than are eQTLs that modify gene expression in other brain tissues. Furthermore, eQTLs that affect male-specific tissues (testis and prostate) are more likely to be adaptive than are eQTLs that affect female-specific tissues (uterus, ovary, and vagina). One contributing factor is that eQTLs that regulate expression in female tissues tend to be more pleiotropic, i.e., they affect a larger number of additional tissues. An additional contributing factor is that variability in reproductive success is greater for males than for females (Bateman's principle).91 This suggests that the accelerated evolution of testis eQTLs may be driven by increased sexual selection acting on males. There is also prior evidence that spermatogenesis has been a key target of positive selection in human evolution.92 An additional consideration is that young mammalian duplicate genes often have divergent expression patterns, particularly with respect to the testis.93,94 The evolution of tissue specificity is an important mechanism of neofunctionalization and subfunctionalization, and multiple adaptive eQTLs (e.g., rs796497, rs11668109, and rs4630823) modify the expression of recently duplicated genes. Overall, our results indicate that eQTLs are important targets of adaptation. Indeed, there is increasing evidence of selection-driven expression differences between human populations.95
We focused on evolution that occurred in the last 100,000 years; different eQTLs and tissues may have been adaptive over longer timescales. That said, our findings mirror results from comparative transcriptomics of humans and chimpanzees: recent genetic changes are more likely to affect expression in testes than in the brain.96 Here, we focused on detecting signatures of positive selection. We note that gene expression is a trait that can also be under stabilizing97 or purifying98,99 selection. Furthermore, enrichment ratio statistics are not the only way to identify tissues that are key targets of adaptation. Some tissues may have played an important role in human evolution simply because they have more eQTLs than other tissues, i.e., they have a larger mutational target size. With this in mind, we note that a large number V7 GTEx eQTLs modify gene expression in thyroid tissue, tibial nerves, and sun-exposed skin (Figure S8). Finally, large PBS scores may be due to genetic hitchhiking and selection acting on closely linked loci. Nevertheless, we note that the PBS outliers identified in this paper constitute the set of eQTLs with the largest allele frequency differences between continental populations.
We note that eQTLs have largely been ascertained in individuals that have European ancestry. This ascertainment bias causes known eQTLs to be enriched for common alleles in Europe.100 Despite this bias, adaptive eQTLs in Africa, Europe, and East Asia yielded similar patterns of pleiotropy and tissue specificity. An additional consideration is that rare alleles are underrepresented in the set of known eQTLs. This occurs because the statistical power to detect an eQTL is maximized when SNPs have large MAFs.101 As sample sizes increase in the future, additional rare eQTLs will be able to be discovered.102 However, these rare eQTLs are unlikely to have high FST statistics and PBS scores.54
In conclusion, many eQTLs have been positively selected, and these adaptive eQTLs reveal important details about the recent evolution of our species. Going forward, our evolutionary understanding of eQTLs will grow as future studies examine other timescales,103 additional types of natural selection (including stabilizing and purifying selection),104 eQTLs affecting newly duplicated genes, and archaic introgression of regulatory DNA.38
Data and code availability
SLiM code is available at https://github.com/LachanceLab/adaptive_eQTLs.
Acknowledgments
We thank Greg Gibson, Michelle Kim, Urko Martinez-Marigorta, Corinne Simonti, Biao Zeng, and Mimi Brown for helpful comments and suggestions. This work was supported by startup funds from Georgia Institute of Technology and an NIGMS MIRA grant to J.L. (R35GM133727). M.H.Q. was also supported by an NIH training grant (T32GM105490).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100083.
Supplemental information
References
- 1.Wittkopp P.J., Haerum B.K., Clark A.G. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 2.Fraser H.B. Gene expression drives local adaptation in humans. Genome Res. 2013;23:1089–1096. doi: 10.1101/gr.152710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferea T.L., Botstein D., Brown P.O., Rosenzweig R.F. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci U S A. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babbitt C.C., Haygood R., Nielsen W.J., Wray G.A. Gene expression and adaptive noncoding changes during human evolution. BMC Genomics. 2017;18:435. doi: 10.1186/s12864-017-3831-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King M.-C., Wilson A.C. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 6.Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 7.Marigorta U.M., Denson L.A., Hyams J.S., Mondal K., Prince J., Walters T.D., Griffiths A., Noe J.D., Crandall W.V., Rosh J.R., et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn's disease. Nat Genet. 2017;49:1517–1521. doi: 10.1038/ng.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 9.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng B., Lloyd-Jones L.R., Holloway A., Marigorta U.M., Metspalu A., Montgomery G.W., Esko T., Brigham K.L., Quyyumi A.A., Idaghdour Y., et al. Constraints on eQTL fine mapping in the presence of multisite local regulation of gene expression. G3 (Bethesda) 2017;7:2533–2544. doi: 10.1534/g3.117.043752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia K., Shabalin A.A., Huang S., Madar V., Zhou Y.H., Wang W., Zou F., Sun W., Sullivan P.F., Wright F.A. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lappalainen T., Sammeth M., Friedlander M.R., t Hoen P.A., Monlong J., Rivas M.A., Gonzalez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keen J.C., Moore H.M. The genotype-tissue expression (GTEx) project: linking clinical data with molecular analysis to advance personalized medicine. J Pers Med. 2015;5:22–29. doi: 10.3390/jpm5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M., et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Wang I.X., Li Y., Bruzel A., Richards A.L., Toung J.M., Cheung V.G. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey J.D., Madeoy J., Strout J.L., Wurfel M., Ronald J., Akey J.M. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell J.K., Gilad Y., Pritchard J.K. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park L. Evidence of recent intricate adaptation in human populations. PLoS One. 2016;11:e0165870. doi: 10.1371/journal.pone.0165870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudaravalli S., Veyrieras J.B., Stranger B.E., Dermitzakis E.T., Pritchard J.K. Gene expression levels are a target of recent natural selection in the human genome. Mol Biol Evol. 2009;26:649–658. doi: 10.1093/molbev/msn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berens A.J., Cooper T.L., Lachance J. The genomic health of ancient hominins. Hum Biol. 2017;89:7–19. doi: 10.13110/humanbiology.89.1.01. [DOI] [PubMed] [Google Scholar]
- 25.Sanz J., Randolph H.E., Barreiro L.B. Genetic and evolutionary determinants of human population variation in immune responses. Curr Opin Genet Dev. 2018;53:28–35. doi: 10.1016/j.gde.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Hoekstra H.E., Coyne J.A. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 27.Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 28.Jones F.C., Grabherr M.G., Chan Y.F., Russell P., Mauceli E., Johnson J., Swofford R., Pirun M., Zody M.C., White S., et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vernot B., Stergachis A.B., Maurano M.T., Vierstra J., Neph S., Thurman R.E., Stamatoyannopoulos J.A., Akey J.M. Personal and population genomics of human regulatory variation. Genome Res. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachance J., Vernot B., Elbers C.C., Ferwerda B., Froment A., Bodo J.M., Lema G., Fu W., Nyambo T.B., Rebbeck T.R., et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman S.R., Andersen K.G., Shlyakhter I., Tabrizi S., Winnicki S., Yen A., Park D.J., Griesemer D., Karlsson E.K., Wong S.H., et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graur D. An upper limit on the functional fraction of the human genome. Genome Biol Evol. 2017;9:1880–1885. doi: 10.1093/gbe/evx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Maury L., Marguerat S., Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 35.Johnson K.E., Voight B.F. Patterns of shared signatures of recent positive selection across human populations. Nat Ecol Evol. 2018;2:713–720. doi: 10.1038/s41559-018-0478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Robinson-Rechavi M. Robust inference of positive selection on regulatory sequences in the human brain. Sci Adv. 2020;6 doi: 10.1126/sciadv.abc9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh D., Yi S.V. Enhancer pleiotropy, gene expression, and the architecture of human enhancer-gene interactions. Mol Biol Evol. 2021 doi: 10.1093/molbev/msab085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telis N., Aguilar R., Harris K. Selection against archaic hominin genetic variation in regulatory regions. Nat Ecol Evol. 2020;4:1558–1566. doi: 10.1038/s41559-020-01284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lachance J., Tishkoff S.A. Population genomics of human adaptation. Annu Rev Ecol Evol Syst. 2013;44:123–143. doi: 10.1146/annurev-ecolsys-110512-135833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S., et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shriver M.D., Kennedy G.C., Parra E.J., Lawson H.A., Sonpar V., Huang J., Akey J.M., Jones K.W. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peyregne S., Boyle M.J., Dannemann M., Prufer K. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 2017;27:1563–1572. doi: 10.1101/gr.219493.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenaillon O. The utility of Fisher's geometric model in evolutionary genetics. Annu Rev Ecol Evol Syst. 2014;45:179–201. doi: 10.1146/annurev-ecolsys-120213-091846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr H.A. Adaptation and the cost of complexity. Evolution. 2000;54:13–20. doi: 10.1111/j.0014-3820.2000.tb00002.x. [DOI] [PubMed] [Google Scholar]
- 45.Fisher R.A. The Clarendon Press; 1930. The Genetical Theory of Natural Selection. [Google Scholar]
- 46.Dittmar E.L., Oakley C.G., Conner J.K., Gould B.A., Schemske D.W. Factors influencing the effect size distribution of adaptive substitutions. Proc Biol Sci. 2016;283 doi: 10.1098/rspb.2015.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otto S.P. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc Biol Sci. 2004;271:705–714. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barreiro L.B., Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 49.Fumagalli M., Sironi M., Pozzoli U., Ferrer-Admetlla A., Pattini L., Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assis R., Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 2013;110:17409–17414. doi: 10.1073/pnas.1313759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grassa C.J., Kulathinal R.J. Elevated evolutionary rates among functionally diverged reproductive genes across deep vertebrate lineages. Int J Evol Biol. 2011;2011:274975. doi: 10.4061/2011/274975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham H., Francois R. 2017. Dplyr: A Grammar of Data Manipulation. https://CRAN.R-project.org/package=dplyr. [Google Scholar]
- 54.Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 55.Weir B.S. Sinauer Associates; 1996. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. [Google Scholar]
- 56.Haller B.C., Messer P.W. SLiM 3: forward genetic simulations beyond the wright-Fisher model. Mol Biol Evol. 2019;36:632–637. doi: 10.1093/molbev/msy228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ayub Q., Mezzavilla M., Pagani L., Haber M., Mohyuddin A., Khaliq S., Mehdi S.Q., Tyler-Smith C. The Kalash genetic isolate: ancient divergence, drift, and selection. Am J Hum Genet. 2015;96:775–783. doi: 10.1016/j.ajhg.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yelmen B., Marnetto D., Molinaro L., Flores R., Mondal M., Pagani L. Improving selection detection with population branch statistic on admixed populations. Genome Biol Evol. 2021;13 doi: 10.1093/gbe/evab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harlemon M., Ajayi O., Kachambwa P., Kim M.S., Simonti C.N., Quiver M.H., Petersen D.C., Mittal A., Fernandez P.W., Hsing A.W., et al. A custom Genotyping array reveals population-level heterogeneity for the genetic risks of prostate cancer and other cancers in Africa. Cancer Res. 2020;80:2956–2966. doi: 10.1158/0008-5472.CAN-19-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maclean C.A., Chue Hong N.P., Prendergast J.G. Hapbin: an efficient program for performing haplotype-based scans for positive selection in large genomic datasets. Mol Biol Evol. 2015;32:3027–3029. doi: 10.1093/molbev/msv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prendergast J.G., Maclean C.A., Chue Hong N.P. University of Edinburgh. Roslin Institute; 2015. Hapbin: An Efficient Program for Performing Haplotype Based Scans for Positive Selection in Large Genomic Datasets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gravel S., Henn B.M., Gutenkunst R.N., Indap A.R., Marth G.T., Clark A.G., Yu F., Gibbs R.A., Genomes P., Bustamante C.D. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keinan A., Clark A.G. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slatkin M., Rannala B. Estimating allele age. Annu Rev Genomics Hum Genet. 2000;1:225–249. doi: 10.1146/annurev.genom.1.1.225. [DOI] [PubMed] [Google Scholar]
- 67.Agresti A., Coull B.A. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52:119–126. [Google Scholar]
- 68.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burdett T., Hall P., Hastings E., Hindorff L., Junkins H., Klemm A., et al. 2018. https://www.ebi.ac.uk/gwas/
- 70.Quinlan A.R. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kern A.D., Hahn M.W. The neutral theory in light of natural selection. Mol Biol Evol. 2018;35:1366–1371. doi: 10.1093/molbev/msy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tournamille C., Colin Y., Cartron J.P., Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 73.McManus K.F., Taravella A.M., Henn B.M., Bustamante C.D., Sikora M., Cornejo O.E. Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLoS Genet. 2017;13:e1006560. doi: 10.1371/journal.pgen.1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwiatkowski D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reich D., Nalls M.A., Kao W.H., Akylbekova E.L., Tandon A., Patterson N., Mullikin J., Hsueh W.C., Cheng C.Y., Coresh J., et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jha P., Lu D., Yuan Y., Xu S. Signature of positive selection of PTK6 gene in East Asian populations: a cross talk for Helicobacter pylori invasion and gastric cancer endemicity. Mol Genet Genomics. 2015;290:1741–1752. doi: 10.1007/s00438-015-1032-8. [DOI] [PubMed] [Google Scholar]
- 77.Racimo F., Marnetto D., Huerta-Sanchez E. Signatures of archaic adaptive introgression in present-day human populations. Mol Biol Evol. 2017;34:296–317. doi: 10.1093/molbev/msw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sankararaman S., Mallick S., Dannemann M., Prufer K., Kelso J., Paabo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pollard K.S., Salama S.R., King B., Kern A.D., Dreszer T., Katzman S., Siepel A., Pedersen J.S., Bejerano G., Baertsch R., et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hormozdiari F., van de Bunt M., Segre A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL signals detects target genes. Am J Hum Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stefánsson, K., and Gulcher, J.R. On sequence variants that influence the risk of common diseases. In Handbook of Human Molecular Evolution, D.N. Cooper, and H. Kehrer-Sawatzki, eds. (Wiley), pp. 1635-1639.
- 83.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 84.Umans B.D., Battle A., Gilad Y. Where are the disease-associated eQTLs? Trends Genet. 2021;37:109–124. doi: 10.1016/j.tig.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A., et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lachance J., Berens A.J., Matthew E.B.H., Teng A.K., Tishkoff S.A., Rebbeck T.R. Genetic hitchhiking and population bottlenecks contribute to prostate cancer disparities in men of African descent. Cancer Res. 2018;78:2432–2443. doi: 10.1158/0008-5472.CAN-17-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith J.M., Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 89.Wagner G.P., Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011;12:204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 90.Nédélec Y., Sanz J., Baharian G., Szpiech Z.A., Pacis A., Dumaine A., Grenier J.C., Freiman A., Sams A.J., Hebert S., et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 91.Bateman A.J. Intra-sexual selection in Drosophila. Heredity (Edinb) 1948;2:349–368. doi: 10.1038/hdy.1948.21. [DOI] [PubMed] [Google Scholar]
- 92.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 93.Assis R., Bachtrog D. Rapid divergence and diversification of mammalian duplicate gene functions. BMC Evol Biol. 2015;15:138. doi: 10.1186/s12862-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guschanski K., Warnefors M., Kaessmann H. The evolution of duplicate gene expression in mammalian organs. Genome Res. 2017;27:1461–1474. doi: 10.1101/gr.215566.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang X., Assis R. Population-specific genetic and expression differentiation in Europeans. Genome Biol Evol. 2020;12:358–369. doi: 10.1093/gbe/evaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Paabo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 97.Gibson G., Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 98.Popadin K.Y., Gutierrez-Arcelus M., Lappalainen T., Buil A., Steinberg J., Nikolaev S.I., Lukowski S.W., Bazykin G.A., Seplyarskiy V.B., Ioannidis P., et al. Gene age predicts the strength of purifying selection acting on gene expression variation in humans. Am J Hum Genet. 2014;95:660–674. doi: 10.1016/j.ajhg.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hernandez R.D., Uricchio L.H., Hartman K., Ye C., Dahl A., Zaitlen N. Ultrarare variants drive substantial cis heritability of human gene expression. Nat Genet. 2019;51:1349–1355. doi: 10.1038/s41588-019-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lachance J., Tishkoff S.A. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun W. A statistical framework for eQTL mapping using RNA-seq data. Biometrics. 2012;68:1–11. doi: 10.1111/j.1541-0420.2011.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fraser H.B. Genome-wide approaches to the study of adaptive gene expression evolution: systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. Bioessays. 2011;33:469–477. doi: 10.1002/bies.201000094. [DOI] [PubMed] [Google Scholar]
- 103.Fair B.J., Blake L.E., Sarkar A., Pavlovic B.J., Cuevas C., Gilad Y. Gene expression variability in human and chimpanzee populations share common determinants. Elife. 2020;9 doi: 10.7554/eLife.59929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hill M.S., Vande Zande P., Wittkopp P.J. Molecular and evolutionary processes generating variation in gene expression. Nat Rev Genet. 2021;22:203–215. doi: 10.1038/s41576-020-00304-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marcus J.H., Novembre J. Visualizing the geography of genetic variants. Bioinformatics. 2017;33:594–595. doi: 10.1093/bioinformatics/btw643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SLiM code is available at https://github.com/LachanceLab/adaptive_eQTLs.