Abstract
It is well established that the gut microbiota plays an important role in host health and is perturbed by several factors including antibiotics. Antibiotic‐induced changes in microbial composition can have a negative impact on host health including reduced microbial diversity, changes in functional attributes of the microbiota, formation, and selection of antibiotic‐resistant strains making hosts more susceptible to infection with pathogens such as Clostridioides difficile. Antibiotic resistance is a global crisis and the increased use of antibiotics over time warrants investigation into its effects on microbiota and health. In this review, we discuss the adverse effects of antibiotics on the gut microbiota and thus host health, and suggest alternative approaches to antibiotic use.
Keywords: antibiotic resistance, gut bacteria, microbiome
Antibiotics can have several negative impacts on host health; both direct and indirect effects. The gut microbiota plays a crucial role in host health regulation. Effects of antibiotics on the host through the gut microbiome are immense and can affect various functions including immune regulation, metabolic activities, and thus overall health.
1. INTRODUCTION
Since their discovery, antibiotics have revolutionized the treatment of infectious diseases on a global scale. They are recognized as one of the contributing factors to increased life expectancy in the 20th century owing to the decline in infectious disease mortality (Adedeji, 2016). However, their overuse and misuse in human and veterinary medicine and animal husbandry have resulted in the current global antibiotic resistance crisis (Llor & Bjerrum, 2014; Vidovic & Vidovic, 2020) which is exacerbated by the slow rate of new drug development (Simpkin et al., 2017). Despite this, antibiotics are still widely prescribed in disease treatment and studies have reported increased consumption of antibiotics in certain countries in the past number of years (Adriaenssens et al., 2011; Klein et al., 2018).
More recently, scientists have uncovered the detrimental impact of broad‐spectrum antibiotics on the gut microbiota. Home to bacteria, archaea, microeukaryotes, and viruses, the gut microbiota plays a fundamental role in human health. It prevents pathogen colonization, regulates gut immunity, provides essential nutrients and bioactive metabolites, and is involved in energy homeostasis (Mills et al., 2019). In infants, the gut microbiota is acquired during birth and thereafter plays an essential role in the development of infant gut immunity. Evidence to date strongly suggests that balanced microbiota composition and rich species diversity are essential to its optimal functioning (Heiman & Greenway, 2016), which can be compromised in disease states (Mosca et al., 2016). Likewise, reduced diversity and imbalanced microbiota composition in the infant's gut are associated with intestinal illnesses and a predisposition to certain diseases later in life (Milani et al., 2017; Volkova et al., 2021).
Broad‐spectrum antibiotics reduce gut microbiota diversity (Dubourg et al., 2014), and as well as killing the pathogen of concern can eradicate beneficial microbes (Blaser, 2011), with deleterious consequences for the host. Despite this, in Western countries, up to 35% of women are exposed to an antibiotic during pregnancy and delivery, and antibiotics comprise 80% of the drugs a woman is exposed to during pregnancy (Kuperman & Koren, 2016; Stokholm et al., 2013). Mothers are frequently prescribed intrapartum antibiotics prophylactically to prevent and treat infections (Verani et al., 2010).
Clostridioides difficile (formerly known as Clostridium difficile) infection is an example of a disease brought about directly through antibiotic disruption of the gut microbiota (Theriot et al., 2014). Illness ranges from mild diarrhea to death (Guh & Kutty, 2018). Antibiotic eradication of beneficial bacteria in the gut enables C. difficile to flourish (Rea et al., 2011). A recent study also concluded that oral antibiotic use is associated with an increased risk of colon cancer (S. Zhang & Chen, 2019). These are just some of the examples of how antibiotic therapy can compromise health.
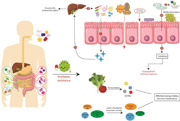
This review thus focuses on the negative impacts of antibiotics on human health from pregnancy through to adulthood, most of which are microbiota‐dependent, although we also provide evidence of nonmicrobiota‐associated negative impacts. We discuss the changes to microbiota composition and functionality and the consequences for host health (Figure 1). We look at the impact of antibiotics at the single bacterial cell level, and how antibiotic use and misuse result in antibiotic resistance development. Further, we consider alternative approaches to antibiotic therapy and discuss therapeutics that can be used to maintain and improve host health and minimize the effects of antibiotics when used.
Figure 1.
The negative impacts that can occur on host health due to overuse and misuse of antibiotics
2. INTRODUCTION TO MICROBIOTA COMPOSITION FROM INFANCY TO ADULTHOOD
It was previously believed that infants are protected in the mother's womb which is a sterile environment, but studies have now demonstrated that amniotic fluid samples, placenta from mothers, and meconium samples from infants contain bacterial DNA suggesting the early exposure of infants to bacteria (Aagaard et al., 2014; de Goffau et al., 2019; Moles et al., 2013; Stinson et al., 2019). However, this is much debated (Perez‐Muñoz et al., 2017) due to issues with contamination and varying interpretations, thus we emphasize on microbiota development from infancy to adults in this review.
The gut microbiota in the early stages of life becomes more diverse until it reaches a stable adult‐like composition by 2–4 years of age. Following birth, the gut is colonized by facultative anaerobes due to the partially aerobic or microaerophilic environment. These then generate the appropriate atmosphere for the development of anaerobes by consuming the available oxygen. Thereafter, followed by early exposure to food (breast milk or infant formula), this composition changes, and facultative anaerobes such as Bifidobacterium, Bacteroides, and Clostridium dominate (Voreades et al., 2014). An initial decrease in Proteobacteria and Enterobacteriaceae accompanied by increases in Bacteroidetes and bifidobacteria have been reported in many studies (Bäckhed et al., 2015; Bokulich et al., 2017). The establishment of the adult‐like microbiota occurs at 2–4 years of age, which is represented by the high relative abundance of Bacteroidetes and Firmicutes (Fouhy et al., 2019).
It is largely accepted that the mother is the most important source of the gut microbiota for infants (Asnicar et al., 2017; Ferretti et al., 2018). The establishment of the healthy infant gut microbiota and its subsequent development is a continuous process that is influenced by several factors. Mode of delivery is one of the first factors that influence the infant gut microbiota, with vaginally delivered infants having microbiota that is more diverse and similar to their mothers' vaginal microbiota while cesarean section‐born infants are deprived of this exposure and thus have gut microbiota similar to their mothers' skin and the hospital environment. These differences are significant and studies have demonstrated an increased association of Propionibacterium, Corynebacterium, Staphylococcus, C. difficile, and Streptococcus and lower abundances of bifidobacteria and Bacteroides with cesarean‐born neonates, whereas Lactobacillus, Prevotella, and Sneathia spp. are associated with vaginally delivered neonates (Azad et al., 2013; Dominguez‐Bello et al., 2010; Penders et al., 2006). Feeding habit is another crucial factor affecting the infant's gut microbiota composition. Because of the presence of oligosaccharides in human milk (human milk oligosaccharides) that are largely used by bifidobacteria, breastfed infants show higher levels of bifidobacteria compared to formula‐fed infants, and the proportions remain high even postweaning (Bezirtzoglou et al., 2011; Fallani et al., 2011). Bacteroides, Streptococcus, and Lactobacillus have also been reported in breastfed infants (Harmsen et al., 2000). Formula‐fed infants present higher abundances of Escherichia coli, C. difficile, the Bacteroides fragilis group, and lactobacilli than their breastfed counterparts (Penders et al., 2006). Gestational age is another factor that affects the gut microbiota composition with preterm infants showing lower diversity, higher abundance of Proteobacteria and reduced levels of obligate anaerobes such as Bifidobacterium, Bacteroides, and Atopobium compared to full‐term infants (Arboleya et al., 2012; C. J. Hill et al., 2017; Moles et al., 2013). Another factor is antibiotic administration, as shown in Figure 2, and is discussed below.
Figure 2.
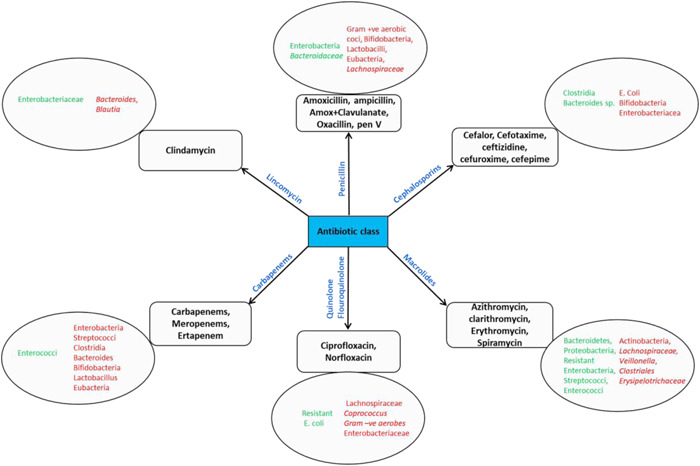
Diagrammatic representation of the effect of various antibiotics on human gut bacteria. Green (left) denotes increasing levels, while red (right) indicates decreasing levels
The gut microbial composition changes throughout pregnancy and a healthy pregnancy is characterized by an increase in the bacterial load and profound alterations in the composition of gut microbiota (Nuriel‐Ohayon et al., 2016). During the first to the third trimester of pregnancy, major changes such as an overall increase in Proteobacteria and Actinobacteria and reduced richness have been reported owing to the major physiological changes (Koren et al., 2012). Also, the vaginal microbiota during pregnancy has been reported to change—represented by the high bacterial load with high Lactobacillus abundance, low richness, and diversity compared to the vaginal microbiota of nonpregnant females (Aagaard et al., 2012; Freitas et al., 2017).
The gut microbiota of healthy adults is predominantly composed of the phyla Firmicutes and Bacteroidetes, representing the majority, followed by Actinobacteria, Proteobacteria, and Verrucomicrobia (Arumugam et al., 2011). Indeed, the gut microbiome can be perturbed by short‐term use or even low‐doses of antibiotics that can have long‐term effects on health (Jernberg et al., 2010), this cautions against the misuse and overuse of antibiotics, particularly in pregnant women and young children.
3. ANTIBIOTIC TYPES COMMONLY ADMINISTERED
The use of antibiotic therapy during pregnancy and lactation varies depending on the underlying condition, country, and medical guidelines, but some of the most commonly prescribed antibiotics during pregnancy are β‐lactam antibacterials (Petersen et al., 2010). Some of the other antibiotic classes prescribed include sulfonamides/trimethoprim and macrolides/lincosamides/streptogramins (de Jonge et al., 2014). Common infections for which antibiotics are prescribed during pregnancy include urinary tract infections, respiratory tract infections, skin or ear infections, bacterial vaginosis, and fever of unknown origin (Heikkila, 1993; Petersen et al., 2010). Antibiotics are frequently administered to mothers during labor to prevent Group B Streptococcus transmission, to reduce and prevent infections in the endometrium, and to prevent wound infections (Braye et al., 2019); though the WHO advises against the prophylactic use of antibiotics post uncomplicated delivery. This is of concern as infant antibiotic exposure through intrapartum antibiotic prophylaxis (IAP) has been shown to alter infant gut microbial diversity (Tapiainen et al., 2019). Further, antibiotics are commonly prescribed to newborns, owing to their high susceptibility to infections and lowered immunity, particularly in premature infants (Clark, 2006; Vergnano et al., 2005). The most commonly used antibiotics for infants include amoxicillin, co‐amoxiclav, benzylpenicillin, cephalosporins, gentamycin, vancomycin, clindamycin, and azithromycin. These antibiotics are indicated in respiratory and ear infections, bronchitis, pharyngitis, and high temperature (CDC, 2017). Table 1 summarizes the commonly used antibiotics and their indication for use.
Table 1.
Summary of the various antibiotics used commonly, their mode of action, and indications when used
| Antibiotic | Class of antibiotic | Broad/narrow spectrum | Mode of action | Target pathogen | Reference |
|---|---|---|---|---|---|
| Amoxicillin, ampicillin | β‐Lactam | Broad‐spectrum | Bactericidal | Rhinosinusitis, respiratory and genitourinary tract infections, septicemia | Akhavan et al. (2020); Peechakara and Gupta (2021) |
| Cephalosporin | β‐Lactam | Broad‐spectrum | Bactericidal | Urinary and respiratory tract infection, Gram‐negative bacteria | Bui and Preuss (2021) |
| Azithromycin/erythromycin | Macrolides | Broad‐spectrum | Bactericidal | Sinusitis, pneumonia, respiratory tract, and skin infection, urogenital and chlamydial infection | Pitsouni et al. (2007); Patel and Hashmi (2021) |
| Metronidazole | Nitroimidazole | Broad‐spectrum | Bactericidal | Protozoal infections, C. difficile infections, Gram‐negative bacterial infections | Rineh et al. (2014); Weir and Le (2020) |
| Gentamycin | Aminoglycoside | Broad‐spectrum | Bactericidal | Urinary tract infection, Gram‐negative bacterial infections | Habak and Griggs, (2020); Chaves and Tadi (2021) |
4. IMPACT OF ANTIBIOTICS ON GUT MICROBIAL COMPOSITION
4.1. Impact of antibiotics during pregnancy and lactation
Perinatal and peripartum antibiotic use can impact gut microbial colonization and the resistome profile in infants (Wong et al., 2020; Zhou et al., 2020). To understand the potential impact of antibiotic administration on offspring during pregnancy, scientists examined the temporal impact of cefoperazone, on both maternal and offspring microbiota when administered during the peripartum period in an interleukin 10 (IL‐10)‐deficient murine model of colitis (Miyoshi et al., 2017). Offspring from cefoperazone‐exposed dams developed altered gut microbial communities into adulthood and had increased susceptibility to spontaneous and chemically induced colitis (Miyoshi et al., 2017). Similar results were demonstrated by Schulfer et al. (2018), who inoculated germ‐free pregnant mice with an antibiotic‐altered microbial community. They observed that the altered microbial community was transmitted to the IL‐10‐deficient offspring which resulted in the development of markedly increased colitis (Schulfer et al., 2018). Another study demonstrated that uptake of antibiotics during pregnancy can lead to alterations in the vaginal microbial composition before birth (Stokholm et al., 2014); this can impact the microbial composition infants receive at birth (Dobbler et al., 2019). Maternal antibiotic uptake during pregnancy has been reported to be associated with altered microbial composition depending on the antibiotic type (Azad et al., 2016; Coker et al., 2020). It is also associated with increased risk of asthma and allergy in the infant (Stokholm et al., 2014) although there is some controversy around this (Kim et al., 2019), as well as with functional impairment in development and cognition (Kenyon et al., 2008), obesity (Mueller et al., 2015), immunological alterations and development of diabetes (Tormo‐Badia et al., 2014) in offspring.
Many studies have demonstrated that IAP‐exposed infants in the first weeks of life have lower proportions of Actinobacteria and Bacteroidetes (Aloisio et al., 2016; Nogacka et al., 2017), high oral Proteobacteria levels (Gomez‐Arango et al., 2017), and lower levels of bifidobacteria (Mazzola et al., 2016). At 3 months, they show under‐representation of Bacteroides, Parabacteroides, and higher Enterococcus and Clostridium (Azad et al., 2016), as well as a higher abundance of Enterobacteriaceae (Mazzola et al., 2016), as compared to nonantibiotic exposed infants. Similar results were reported in a recent study including 22 newborns which demonstrated that maternal intrapartum antibiotics can affect the infant oral microbiota with phyla Actinobacteria, Bacteroidetes, and Proteobacteria being more abundant in infants of antibiotic‐treated mothers (Li et al., 2019). Another study in 28 preterm infants (with fecal samples collected on Day 7 and 14 from birth) demonstrated similar results with decreases in Bacteroidetes and Bifidobacterium in prenatal antibiotic‐exposed infants and suggested that the altered microbiota resembled the resistant bacteria in the neonatal intensive care unit during the same period (Zou et al., 2018). Many of these effects are similar to those observed following postnatal antibiotic administration (Tapiainen et al., 2019). Ampicillin use as an intrapartum prophylactic drug in mothers against Group B Streptococcus is also demonstrated to reduce the levels of bifidobacteria in infants (Aloisio et al., 2014). Similar effects of perinatal antibiotic exposure have been observed in preterm infants (n = 40); marked by increased levels of Enterobacteriaceae species and reduced levels of Bacteroidaceae (Arboleya et al., 2016).
Maternal antibiotic administration during lactation also influences the milk microbiota (Hermansson et al., 2019; Soto et al., 2014), which can in turn influence the infant gut microbial composition.
4.2. Impact of antibiotic administration directly to infants on the infant gut microbiota
Premature infants are very often treated with antibiotics owing to their health conditions, and antibiotics are one of the most commonly prescribed drugs in the NICU (Clark, 2006). Many of the prophylactic antibiotics are broad‐spectrum and thus affect a huge proportion of the gut bacterial community, leading to many alterations in the early establishment pattern. Studies have reported that very preterm infants who received prolonged antibiotic treatment had less diverse bacterial populations and reduced species richness in their gut and more antibiotic resistance genes (ARGs; Gasparrini et al., 2019; Gibson et al., 2016). Both short‐term and long‐term exposure of preterm infants to antibiotics can alter their gastrointestinal microbiota. These changes include decreases in bifidobacteria and Bacteroidetes relative abundance, and an increase in Enterococcus abundance (Gasparrini et al., 2019; Zou et al., 2018; Zwittink et al., 2018). Extended antibiotic treatment in premature infants can result in an increased risk of developing late‐onset sepsis (primarily caused by Group B Streptococcus), necrotizing enterocolitis (NEC), and overall mortality (Esaiassen et al., 2017; Kuppala et al., 2011).
Amoxicillin treatment in infants (n = 31) for 7 days was shown to completely eradicate Bifidobacterium adolescentis species which was accompanied by decreased diversity of the bifidobacteria population (Mangin et al., 2010). In a cohort of 18 infants, we have previously found that the use of ampicillin and gentamicin in early life resulted in higher levels of Proteobacteria and lower proportions of Bifidobacterium and Lactobacillus 4 and 8 weeks after the treatment (Fouhy et al., 2012). A study reporting on the effects of administering the broad‐spectrum antibiotic, cefalexin, in 26 infants, in the first 4 days of life revealed that the gut microbiota of antibiotic‐treated infants showed less diversity than the control antibiotic‐free infants (Tanaka et al., 2009). The antibiotic also arrested the growth of some bacterial groups such as bifidobacteria and resulted in unusual colonization of Enterococcus in the first week. Further to this, a high Enterobacteriaceae population was observed 1 month following antibiotic treatment as compared to the control antibiotic‐free infants (Tanaka et al., 2009). Antibiotics change the dominant members of the bacterial community which could have profound effects on immune development, metabolism, and growth of the infant (Cox et al., 2014; Nobel et al., 2015).
Similar results of reduced gut microbiota diversity have been observed in children (n = 39, monthly sampling in the first 3 years of life) who have undergone antibiotic therapy. The authors also reported the presence of ARGs on mobile genetic elements long after cessation of treatment (Yassour et al., 2016). Antibiotic use during infancy and childhood has been associated with the altered microbial composition and metabolic functions (Korpela et al.,2016), higher risk of asthma and allergy development (Ni et al., 2019; Yamamoto‐Hanada et al., 2017), and obesity (L. C. Bailey et al., 2014) in later life.
4.3. Impact of antibiotics on the gut and oral microbiota in adults
To investigate the long term effects of antibiotics on the healthy state microbial composition, the antibiotics amoxicillin (n = 6; Cochetière et al., 2005), ciprofloxacin (n = 3; Les Dethlefsen et al., 2008), and cefprozil (n = 24; Raymond et al., 2016) were administered to healthy individuals. These studies reported changes in microbial composition persisting for up to 12 weeks after treatment had ended with the incomplete restoration of microbial composition and emergence of antibiotic‐resistant strains. In one study, the authors examined the distal gut microbiota of three individuals over 10 months post‐administration of the antibiotic ciprofloxacin. They reported that the effect of ciprofloxacin on the gut microbiota was profound and rapid with a decrease in richness and diversity of microbiota accompanied by shifts in levels of Bacteroidetes, Lachnospiraceae, and the Ruminococcaceae. By 1 week after the end of each course, communities began to return to their initial state, but the return was incomplete and variable from the initial stage (Dethlefsen & Relman, 2011). Many studies have investigated the long‐term impact on gut microbiota following a course of antibiotics. A short term course of clindamycin (7 days) resulted in significant disturbances in the bacterial community such as a sharp decline in Bacteroides (Jernberg et al., 2007; Löfmark et al., 2006) and enterococcal colonies (Lindgren et al., 2009) that remained for up to 2 years post‐treatment and was accompanied by increased levels of ARGs and strains (Jernberg et al., 2007; Lindgren et al., 2009; Löfmark et al., 2006). Another study (n = 10) that analyzed adult fecal samples from 11 days to 12 months post antibiotic treatment demonstrated that the use of ciprofloxacin for 10 days resulted in lower abundances of Bifidobacterium but did not affect Lactobacillus and Bacteroides levels, but clindamycin treatment for the same number of days caused Lactobacillus and Bifidobacterium to decrease and Bifidobacterium did not normalize until 1‐year post‐treatment, showing that different bacterial groups require different times for normalization post‐treatment (Rashid et al., 2015).
Pérez‐Cobas and Gosalbes et al. (2013) also reported that the type of antibiotic, whether static or cidal, had varying effects on the gut microbiota which was reflected at the functional level. For instance, in a study of four subjects, a bacteriostatic drug resulted in the flourishing of Gram‐negative bacteria which was related to an increase in the number of genes involved in lipopolysaccharide (LPS) synthesis, while the bactericidal drug was associated with an increase in Gram‐positive bacteria which was accompanied by an over‐representation of genes involved in endospore formation (Pérez‐Cobas & Artacho et al., 2013). Antibiotic administration for the eradication of Helicobacter pylori demonstrated that the antibiotics could affect the indigenous microbiota and lead to the development of resistant strains which can persist for years after treatment (Jakobsson et al., 2010; Sjölund et al., 2003).
Furthermore many antibiotics are used for dentistry procedures routinely. These antibiotics can increase the number of resistant strains present orally, can increase the minimum inhibitory concentrations, and can also eliminate the nonpathogenic strains (Harrison et al., 1985; Ready et al., 2004), which can lead to systemic infections and inflammation.
5. CONSEQUENCES OF ANTIBIOTIC‐INDUCED MICROBIOTA CHANGES FOR HEALTH AND DISEASE
5.1. In adults
Due to the role of the microbiota in host metabolism and physiology, many studies postulate that microbial imbalances can be related to obesity (Riley et al., 2013; Scott et al., 2016), diabetes (Boursi et al., 2015; Mikkelsen et al., 2015), and asthma (Arrieta et al., 2015; Kozyrskyj et al., 2007). Blaser and Falkow (2009) have suggested a link between the “missing microbes” and modern conditions such as obesity and juvenile diabetes. These multifactorial conditions can be controlled by identifying the controllable factors such as the microbiota component and dietary habits, thus preventing them from occurring if possible.
Studies have reported a link between antibiotic usage and obesity (Del Fiol et al., 2018). Some studies suggest that an increased ratio of Firmicutes to Bacteroidetes rather than specific levels is associated with obesity (Kasai et al., 2015), though results are conflicting (Duncan et al., 2008; Schwiertz et al., 2010). While the microbial component of obesity is debated, studies have reported a common change at functional microbial levels. Indeed, obese individuals have higher short‐chain fatty acid (SCFA) content compared to lean individuals (Schwiertz et al., 2010; Turnbaugh et al., 2006). Furthermore, obesity is associated with metabolic alterations related to glucose homeostasis and insulin resistance and linked to the development of diabetes (Cani et al., 2012). In a study in 96 humans (48 each antibiotic group and controls), researchers reported significant and persistent weight gain after an episode of infectious endocarditis in patients who had been treated with vancomycin and gentamycin (Thuny et al., 2010).
An association between antibiotic‐induced changes in microbial colonization and type 1 diabetes in male mice was reported (Candon et al., 2015). A combination of broad‐spectrum antibiotics or vancomycin alone was given to neonatal nonobese diabetic mice that spontaneously developed autoimmune type 1 diabetes. The microbiota was significantly altered with an increase in Escherichia and Lactobacillus species and a decrease of the Clostridiales order compared to controls. A major reduction of IL‐17‐producing cells was also observed in the lamina propria of the ileum and the colon of vancomycin‐treated mice (Candon et al., 2015), which can affect host defense mechanisms. Some studies in human populations also suggest a link between repeated use of broad‐spectrum antibiotics and diabetes (Boursi et al., 2015; Mikkelsen et al., 2015), while some suggest a protective and preventative role of antibiotics and diet in diabetes development in diabetes‐prone animals partly due to lowering of specific antigenic load or development of tolerogenic APCs (Brugman et al., 2006; Hu et al., 2015).
Associations between altered microbial composition and type 2 diabetes are more established, with decreased levels of butyrate‐producing bacteria reported in type 2 diabetic patients (Gurung et al., 2020). X. Zhang et al. (2013) studied 121 subjects with normal glucose tolerance, prediabetes, and newly diagnosed diabetes and reported that there is modulation of the gut microbial composition at the prediabetes stage which can act as a marker for the development of diabetes state.
Antibiotics can lead to antibiotic‐associated diarrhea (AAD) and studies have demonstrated that clindamycin can result in alteration of the microbial community which can promote the colonization of potential pathogens such as C. difficile which can lead to diarrhea and colitis (Buffie et al., 2012; McDonald, 2017). Another study in a mouse model reported that antibiotic treatment resulted in decreased alpha and beta diversity, which potentially caused a decrease in levels of serotonin, tryptophan hydrolase, and secondary bile acids which can further affect gut motility and metabolism (Ge et al., 2017).
5.2. During pregnancy and infancy
Extrinsic factors such as antibiotics can alter the diversity of the maternal microbiota that can affect the infant's gut microbiota diversity, immunity, and disease development in later life, both directly and indirectly (Azad et al., 2016; Nyangahu et al., 2018; Tapiainen et al., 2019; Tormo‐Badia et al., 2014). According to the hygiene hypothesis, if the host is not exposed to a diverse range of microbiota early in childhood or in the developing stages, immune‐related disorders may develop such as asthma and allergic sensitization. Antibiotics can have a similar effect when administered during infancy. Preterm infants are often exposed to antibiotics which results in an altered microbial composition (Clark, 2006; Gibson et al., 2016; Zou et al., 2018), predisposing them to probable infections such as NEC, and invasive fungal infections (Esaiassen et al., 2017). Animal studies have reported that administration of low dose or subtherapeutic concentrations of antibiotics in early life can disturb the microbial composition, affecting the expression of genes involved in immunity and carbohydrate metabolism, and can alter metabolic homeostasis predisposing the host to adiposity later in life (Cho et al., 2012; Cox et al., 2014; Schulfer et al., 2019). Obesity has been widely linked with altered microbial colonization during early life owing to the role of the gut microbiota in dietary metabolism. Studies have demonstrated that early‐life antibiotic exposure has potential links to increases in body mass index, overweight and central adiposity, and this can be gender‐specific affecting males more than females (Azad et al., 2014; Murphy et al., 2014). Both murine models (Cho et al., 2012; Cox et al., 2014) and human studies have reported that antibiotic exposure in the first few months of life is associated with increases in body mass index and risk of overweight (L. C. Bailey et al., 2014; Scott et al., 2016; Trasande et al., 2013) and asthma (Kozyrskyj et al., 2007; Risnes et al., 2011) in childhood. Similarly, studies have reported that antibiotic administration in early life can be associated with a heightened risk of asthma, allergy, and atopic dermatitis, and IBD (Johnson et al., 2005; Kronman et al., 2012; Ni et al., 2019; Yamamoto‐Hanada et al., 2017). NEC and AAD have also been associated with prolonged or prophylactic antibiotic uptake in early life (Alexander et al., 2011; Kuppala et al., 2011; Michael Cotten et al., 2009).
5.3. Changes in immune response
The immune system is trained to fight pathogens during infancy, and this is the time when microbial colonization takes place. Any disturbance to microbial colonization has been shown to affect immune maturation due to this co‐developmental process. Studies in germ‐free mice have confirmed that the absence of microbes in the gut results in both physiological and immunological changes to the gut environment. These changes include alterations in mucus thickness and composition (Szentkuti et al., 1990), reduced gastric motility (Abrams & Bishop, 1967), improper development and functioning of intestinal cells and immune cells (Cahenzli et al., 2013; Vaishnava et al., 2008) and improper development of the immune system (Bauer et al., 1963; Macpherson & Uhr, 2004). Antibiotic treatment has been shown to reduce the thickness of the colonic mucus layer thus increasing the risk of pathogen invasion and intestinal inflammation in 8–10‐week‐old mice (Wlodarska et al., 2011). Another study in mice reported that antibiotic‐induced alterations in the microbiota shift the TH1/TH2 balance toward TH2‐dominant immunity—which leads to atopy development, accompanied by a reduced number of lymphocytes (Oyama et al., 2001). Altered microbial composition and altered gene maturation profile such as downregulation of genes coding for MHC class 1b and class II proteins and products of Paneth cells such as defensins were seen post clamoxyl treatment in neonatal rats, which could affect mucosal barrier development (Schumann et al., 2005).
A study demonstrated that certain molecules produced by bacteria in the gut are involved in immune system maturation. Indeed, germ‐free mice colonized with B. fragilis producing a bacteria polysaccharide resulted in correcting T‐cell‐deficiencies and improving T(H)1/T(H)2 imbalances along with promoting lymphoid organogenesis; this was not observed in the case of the nonpolysaccharide‐producing mutant B. fragilis (Mazmanian et al., 2005). Studies have reported that the secretion of antimicrobial peptides by intestinal epithelial cells is regulated by the microbiota in the microenvironment. Germ‐free mice colonized with conventional or human microbiota or specific probiotic species or LPS demonstrated increased production of antimicrobial peptides like REGIII‐γ, boosting the innate immune response (Brandl et al., 2008; Cash et al., 2006; Natividad et al., 2013).
A study in mice demonstrated that prenatal antibiotics not only altered the pattern of microbiota colonization in infant mice but also negatively affected the activity of CD8+ T lymphocytes towards viral infections affecting their immune responses (Gonzalez‐Perez et al., 2016). Further, they also observed that infant mice were more susceptible to infection when born in stricter hygienic facilities (Gonzalez‐Perez et al., 2016). Similar results were reported post‐antibiotic treatment in another murine model with low bacterial diversity alongside reduced cytokine production by CD4+ T lymphocytes and reduced production of interferon‐γ (D. A. Hill et al., 2010). In a study involving influenza patients, the authors reported that subjects with low titers of infection and treated with antibiotics had low immune responses accompanied with microbiota loss, low immunoglobulin (Ig) G1, IgA, and secondary bile acid levels against the infection (Hagan et al., 2019).
These studies demonstrate the complex relationship between the microbiota and the host immune response, and the impact of antibiotics on this interaction which needs to be further studied. It can also impact the effectiveness of vaccines used postantibiotic treatment.
6. INFLUENCE OF ANTIBIOTIC‐INDUCED CHANGES ON MICROBIOTA FUNCTIONALITY AND BACTERIAL BEHAVIOR AT THE SINGLE‐CELL LEVEL
6.1. Changes in metabolites
The gut microbiota is responsible for the production of many essential metabolites including SCFAs and amino acids (Mills et al., 2019). Studies have reported that commensal‐produced butyrate and propionate have anti‐inflammatory roles, promoting the generation and differentiation of regulatory T cells (Arpaia et al., 2013; Furusawa et al., 2013), with roles in energy metabolism (den Besten et al., 2013; De Vadder et al., 2014). By impacting the composition of the microbial community, antibiotics also alter microbiota functionality and thus the metabolites produced (Ferrer et al., 2017).
For example, metabolomics profiles were analyzed in antibiotic‐treated piglets fed a corn‐soy basal diet with or without in‐feed antibiotics from postnatal days 7–42. The antibiotic‐treated group had higher concentrations of metabolites associated with amino acid metabolism, decreasing the concentration of amino acids. A reduction in SCFA production was also reported as levels of butyrate and propionate were decreased (Mu et al., 2017). Another study in piglets (Days 7–21) demonstrated that fecal microbiota transplant (FMT) and antibiotic (amoxicillin) treatment both resulted in lowering of fatty acid oxidative catabolism and amino acid biosynthesis, though antibiotics had a more significant effect (Wan et al., 2019).
Multiple studies performed in mouse models have elucidated the effects of antibiotic treatment on host metabolic functions. A study in antibiotic (streptomycin)‐treated mice reported that antibiotic administration can affect pathways of hormone synthesis such as steroids and eicosanoids, along with altering the pathways involved in sugar, bile acids, and amino acid metabolism, thus suggesting the role of the microbiota in these pathways (Antunes et al., 2011). Sun et al. (2019) reported that mice treated with antibiotics (enrofloxacin, vancomycin, and polymixin B sulfate) showed upregulated gene expression of various cytokines in the colon, with significant metabolic shifts in valine, leucine, and isoleucine biosynthesis pathways. These alterations correlated to changes in microbial composition. One study reported that clindamycin treatment in mice resulted in significant changes in metabolite composition (30% of the compounds analyzed); the restoration of which was associated with recovery of the altered microbiota (Jump et al., 2014). Zhao et al. (2013) reported that antibiotic treatment altered the products of bacterial metabolism. This included decreased levels of SCFAs, amino acids (correlated with the abundance of Prevotella, Alistipes, and Barnesiella), and increased precursors like bile acids and oligosaccharides (associated with high levels of facultative anaerobic bacteria, Enterococcus faecalis, Enterococcus faecium, and Mycoplasma; Zhao et al., 2013). Microbial depletion in mice due to antibiotic uptake decreased baseline serum glucose levels, improved insulin sensitivity (Zarrinpar et al., 2018), altered systemic glucose metabolism along with changes in expressions of the genes in the liver and ileum involved in glucose and bile acid metabolism (Rodrigues et al., 2017). This was reported to be accompanied by reduced levels of SCFAs and secondary bile acid pools (Zarrinpar et al., 2018). Similar results were reported following vancomycin treatment (Vrieze et al., 2014). This can lead to impairment of barrier function (Kelly et al., 2015); act as a causative factor in the development of ulcerative colitis (Machiels et al., 2013), and Salmonella infection (Gillis et al., 2018).
Studies have also reported that antibiotic uptake can result in changes in protein expression, energy metabolism in the microbiota, with a slight increase following antibiotic therapy, which may be as a coping mechanism to antibiotic stress but decreased at later stages and post‐antibiotic usage (Pérez‐Cobas & Artacho et al., 2013). Another study demonstrated that antibiotics have a sex‐dependent effect on host metabolism. They reported that vancomycin and ciprofloxacin–metronidazole treatment resulted in significant reductions of Firmicutes and SCFAs in female mice, which was only observed after vancomycin treatment in males. They also reported that both antibiotic exposures significantly decreased the levels of alanine, branched‐chain amino acids (leucine, isoleucine, and valine), and aromatic amino acids in colonic contents of female mice but not in male mice (Gao et al., 2019).
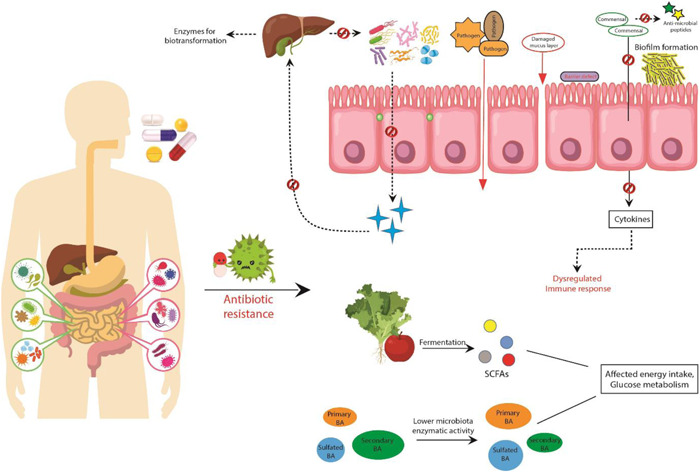
6.2. Accumulation of metabolites/xenobiotics
Xenobiotics (including antibiotics, heavy metals, and environmental chemicals) have an impact on gut microbial composition. The effect here is cyclical in that the microbiota is necessary for xenobiotic biotransformation (Figure 3). The metabolism of xenobiotics before it reaches its target organ site is largely dependent on the microbiota. The gut microbiota can affect the xenobiotic half‐life in the host, the extent to which they reach the target receptor, and may also influence the host's capacity to metabolize xenobiotics (Koppel et al., 2017). Both in vivo and in vitro studies have shown that the gut microbiota is involved in the biotransformation of xenobiotics (Lu et al., 2013). Studies using germ‐free and conventional rodents reported that the absence of gut microbiota‐affected gene expression of many liver enzymes including active androstane receptor and host detoxifying enzymes such as glutathione peroxidases, sulfotransferase, epoxide hydrolases, and N‐acetyltransferases (Björkholm et al., 2009; Meinl et al., 2009). However, the absence of microbes necessary for metabolizing particular compounds can result in their accumulation in the host which may lead to toxicity. Ciprofloxacin administration to SPF and germ‐free mice revealed decreases in hepatic Cyp3a11 expression, this was associated with the gut microbiota. The lower lithocholic acid (LCA) levels due to decreases in LCA‐producing bacteria post antibiotic administration could be responsible for decreases in the expression of Cyp3a11. Similar effects in humans can result in less clearance of multiple CYP3A4 (human analog of Cyp3a11)‐dependent medications (Lynch & Price, 2007).
Figure 3.
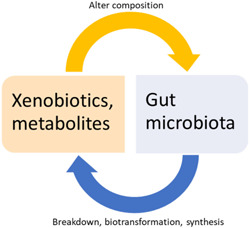
Figure demonstrating cyclical relation between gut microbiota, xenobiotics, and metabolites
Another study speculated that increased levels of hepatic lipid accumulation and TG levels in mice were due to antibiotic treatment and the altered gut microbial composition (Jin et al., 2016). Studies have shown that antibiotic treatment in mice increases the levels of sialic acid and succinate which increase the susceptibility of the host to Salmonella and C. difficile infections (Ferreyra et al., 2014; K. M. Ng et al., 2013). In the same manner, an antibiotic‐altered microbial composition can result in deficiencies of certain metabolites or vitamins that are solely produced by bacteria. For example, one study reported that antibiotic‐induced changes in the gut microbiota of mice resulted in shifts in copper (Cu) metabolism. This can have consequences for immunity and the intestinal barrier due to the role of Cu in these functions (Miller et al., 2019).
Furthermore, cross‐feeding is a significant feature of the gut microbiota. For example, B. adolescentis produces lactate and acetate by utilizing fructo‐oligosaccharides and starch. Butyrate‐producing anaerobes cannot utilize fructo‐oligosaccharides and starch but rely on lactate and acetate as growth substrates. Therefore, B. adolescentis indirectly facilitate the proliferation and expansion of butyrate‐producing species through cross‐feeding. This type of dependency is also seen in many other bacterial groups (Heinken & Thiele, 2015; Rowland et al., 2018). This codependency can be disturbed by antibiotic use leading to increased accumulation or deficiency of some metabolites/compounds. For example, a study reported a decrease in abundance of Gram‐negative bacteria in the gut by vancomycin which is a Gram positive‐targeting antibiotic (Ubeda et al., 2010). This can be due to the interdependency of bacteria in their community.
6.3. Changes in bacterial signaling pattern
Antibiotics can alter the transcription of several major functional genes such as those encoding transport proteins, genes involved in the metabolism of carbohydrates, and protein synthesis (Goh et al., 2002; J. T. Lin et al., 2005). A study demonstrated induced expression of virulence‐associated genes in Pseudomonas aeruginosa leading to higher secretion of rhamnolipids and phenazines on exposure to subinhibitory concentrations of antibiotics (Shen et al., 2008). Many studies have reported that aminoglycosides (Hoffman et al., 2005), β‐lactams (Kaplan et al., 2012; K. M. Ng et al., 2014), vancomycin, and oxacillin (Mirani & Jamil, 2010) can induce biofilm formation even at sublethal concentrations. These biofilms then act as reservoirs of antibiotic resistance. This confers additional resistance to bacteria against several antibiotics and host defense, which can make treatment difficult in humans and can cause several issues such as blockage of pipes/equipment in healthcare settings and food industries (Muhammad et al., 2020).
Bacteria interact with their host using pattern recognition receptors (PRR) via signaling through the production of bile acids, SCFAs, fatty acids, amino acids, LPS, lipoteichoic acid, flagellin, CpG DNA, and peptidoglycan. These signaling molecules either serve as a source of energy for other cells, regulate or modulate the function of immune cells such as monocytes, macrophages, T cells via G‐protein coupled receptor and nuclear receptor family, free fatty acid receptors (Brestoff & Artis, 2013). Antibiotic use results in a reduction of these bacteria and hence the PRRs such as TLR signaling and downstream regulation of innate defenses (Willing et al., 2011). A study reported that an antibiotic‐mediated decrease in butyrate‐producing bacteria resulted in reduced epithelial signaling through the intracellular butyrate sensor peroxisome proliferator‐activated receptor γ (Byndloss et al., 2017). Similarly, antibiotic disruption of the commensal microbiota in newborn mice increases their susceptibility to pneumonia, due to interrupted migration of IL‐22 producing lymphoid cells (IL‐22 + ILC3; Gray et al., 2017). This effect could be reversed by the transfer of commensal microbiota to mice at birth (Gray et al., 2017). Another study reported the importance of commensals in protecting against colon injury and maintaining intestinal homeostasis (Rakoff‐Nahoum et al., 2004). In this case, commensals induced release of protective factors via TLRs; these factors were not released in mice treated with antibiotics lacking the commensal bacteria (Rakoff‐Nahoum et al., 2004). Antibiotics can thus impact complex host‐microbial interactions due to changes in microbial community composition.
6.4. ARG reservoir
ARGs are now reported to be found in the environment including oceans and freshwater bodies (Hatosy & Martiny, 2015), soil (Cycoń et al., 2019), glaciers (Segawa et al., 2013), the food chain, and also within humans. Apart from bacteria, viruses have also been reported to be carriers of ARGs (Debroas & Siguret, 2019). Some of this spread of ARGs is historical such as in some untouched/uncontaminated environments (Van Goethem, 2018) but much of it is because of the wide use of antibiotics by humans.
For instance, apart from their use in treating infections in humans, antibiotics have been widely used as growth promoters for weight gain in animals (Butaye et al., 2003) and to treat and control infections (Ding et al., 2014; Mcmanus et al., 2002). They have also been used in aquaculture for similar reasons (Cabello, 2006; Lulijwa et al., 2019). Some of these antibiotics are the same as, or are structurally similar to the ones used to treat human infections such as erythromycin, gentamycin, enrofloxacin, neomycin, streptomycin (Marshall & Levy, 2011). The development of antibiotic‐resistant bacteria in agriculture and aquaculture is a serious concern as such bacteria can enter humans through the food chain promoting cross‐resistance and also reducing the susceptibility of infectious bacteria to antibiotic treatment.
Humans are reservoirs of ARGs (J. K. Bailey et al., 2010; Clemente et al., 2015; Sommer et al., 2010) and the gut has been pinpointed as the epicenter of antibiotic resistance (Carlet, 2012). Intriguingly, some studies have reported the presence of ARGs in humans from remote communities who have had very limited exposure to antibiotic therapy. They reported high levels of acquired resistance to antibiotics such as tetracycline, ampicillin, trimethoprim/sulfamethoxazole, streptomycin, and chloramphenicol (Bartoloni et al., 2009). Similarly, ARGs in healthy humans have been reported despite the absence of antibiotic use (Bengtsson‐Palme et al., 2015; Sommer et al., 2009; de Vries et al., 2011). Studies in healthy infants and children who have never been exposed to antibiotics report the presence of genes that confer resistance to β‐lactams, fluoroquinolones, tetracycline, macrolide, sulfonamide, or multiple drug classes. The major ARG carriers were found to be Enterococcus spp., Staphylococcus spp., Klebsiella spp., Streptococcus spp., and Escherichia/Shigella spp. (Casaburi et al., 2019; Karami et al., 2006; Moore et al., 2013; L. Zhang et al., 2011).
In the gut, bacteria can horizontally and vertically transmit genes to other related or unrelated bacteria due to their proximity via mobile genetic elements (Table 2). ARGs in the infant gut microbiota can be derived from that of their mothers, as AR bacteria can be transmitted from mother to infant through breastfeeding (Parnanen et al., 2018). The presence of ARGs in humans is a global crisis because it makes the treatment of infections more difficult, costly, and inefficient.
Table 2.
Various mobile genetic elements that can be used for the transfer of ARGs
| Mobile elements and transfer of resistance genes | ||
|---|---|---|
| Vehicle element | Description | Some antibiotic resistance genes transported |
| Plasmids; Rozwandowicz et al. (2018) | Extrachromosomal material | Colistin resistance, extended‐spectrum β‐lactams, β‐lactams, aminoglycoside, quinolone, sulfonamides, tetracycline, chloramphenicol, trimethoprim |
| Integrons; Partridge et al. (2009) | Genetic elements with site‐specific recombination system | Sulfonamide resistance (sulI), aminoglycosides, β‐lactams, quinolones, chloramphenicol, and trimethoprim |
| Transposons; Lupski (1987) | Mobile elements that need integration into chromosome or plasmid | β‐Lactams, macrolides, aminoglycoside, chloramphenicol, tetracycline |
7. NONMICROBIOTA‐ASSOCIATED EFFECTS OF ANTIBIOTICS
7.1. In pregnancy
Associations between antibiotic usage during pregnancy with neonatal and congenital abnormalities have been reported. For example, studies have reported an increased risk of developing cerebral palsy, epilepsy, cardiac and genital malformations in infants born to mothers treated with macrolides during pregnancy and which are more harmful when consumed during the first trimester (Källén & Danielsson, 2014; Kenyon et al., 2008; Meeraus et al., 2015). Similarly, amoxicillin use during the first trimester was linked to cleft lip and cleft palate development, and though reported in only a small number of cases, it exemplifies the adverse effects of antibiotic use during pregnancy (Lin et al., 2012). Birth defects such as microphthalmia, hypoplastic left heart syndrome, atrial septal defects, and cleft lip with cleft palate have also been associated with the use of sulfonamides and nitrofurantoin during the first trimester of pregnancy (Crider et al., 2009). Similarly, trimethoprim–sulfonamide use during pregnancy is associated with a higher risk of cardiovascular malformations (Czeizel et al., 2001). The evidence in all cases is mixed (Muanda & Sheehy, 2017), and is usually linked with consumption during the early months of pregnancy. This could be due to the antibacterial impacting the neonate during the organogenesis and early development stages in utero. Though the examples cited represent potential associations and not causal relationships, a reappraisal of antibiotic usage during pregnancy is warranted.
7.2. General effects
Studies have reported that antibiotics can exert direct toxic effects on host tissues such as mitochondrial damage, suppressed ribosomal gene expression (Morgun et al., 2015), and oxidative tissue damage in mammalian cells (Kalghatgi et al., 2013). Though controversial, some studies suggest that antibiotic use can be related to increased risk of breast cancer (Tamim et al., 2007; Velicer et al., 2004) and increased risk of miscarriage (Fan et al., 2019). Sometimes, antibiotics can worsen the condition they are meant to treat. The bactericidal action of many β‐lactam antibiotics has been reported to increase toxin production such as Shiga toxin which is released from entero‐hemorrhagic E. coli, predisposing the host to a higher risk of a hemolytic uremic syndrome (Kimmitt et al., 1999; Wong, 2000). Antibiotics can also affect host metabolism directly without microbes as a mediator while making the targeted pathogen less susceptible to the antibiotic. These changes in host metabolites are mostly local to the site of infection and include high levels of AMP which decrease the efficacy of antibiotics and also increase phagocytic activity. This was accompanied by impaired immune function because of the inhibitory effect of antibiotics on the respiratory activity of immune cells (Yang et al., 2017).
8. ANTIBIOTIC ALTERNATIVES AND USE OF PROBIOTICS FOR RESTORING THE MICROBIAL COMMUNITY AND BETTERMENT OF HEALTH
It is now well established that antibiotic use results in changes in microbial composition, the consequences of which can be detrimental for the host. Certain approaches can be used along with or post antibiotic therapy to restore the microbial composition faster (Figure 4). Probiotics are widely used for this purpose and have been shown to increase the abundance of beneficial microbes, stabilize the microbial community and thus alleviate the effects of antibiotics (Ki Cha et al., 2012; Korpela et al., 2018). Probiotics exert their effects by promoting antimicrobial peptide production, producing bacteriocins, suppressing the growth of non‐commensals via competing for nutrients and receptors on the intestinal mucosa, enhancing barrier function in the gut, and modulating immunity (Bron et al., 2011; Cazorla et al., 2018; Collado et al., 2007; O'Shea et al., 2012; Xue et al., 2017), but the use of probiotics may not lead to complete restoration of the gut microbiota.
Figure 4.
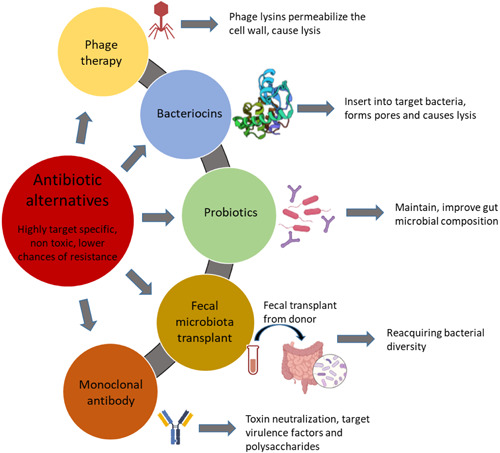
Various alternatives to antibiotics that can be used alone or in some cases in combination with antibiotic treatment
FMT can be more beneficial for regaining microbial balance in the gut (Suez et al., 2018). FMT has been widely used therapeutically for rebalancing the microbiota of C. difficile‐infected patients; restoring their microbial and metabolic activity (Weingarden et al., 2014). FMT can also utilize donor fecal material from the patient itself before antibiotic therapy, known as autologous FMT. Many factors such as efficacy, cost, and suitability make FMT an attractive option but detailed studies are needed to optimize the process and understand other probable therapeutic applications beyond gut disorders (Allegretti et al., 2019; Ramai et al., 2019).
One of the major issues of antibiotic use is the development of bacterial resistance. Alternatives such as bacteriophage (phage) therapy and bacteriocins are now being investigated as substitutes for antibiotics or complementary therapies with antibiotics to overcome the issue of resistance. Phage therapy was first described and used in the early 1900s, but due to the introduction of antibiotics, phage therapy was dismissed in western medicine (D. M. Lin et al., 2017), although it is still practiced in certain regions of the world including Georgia, Russia, and Poland (C. Hill et al., 2018). With the growing antibiotic resistance crisis, phage therapy is now being revisited. The use of phage therapy to reduce biofilm and treat lung infections has been studied in mouse models and it has been demonstrated that phage therapy successfully treats respiratory P. aeruginosa infection (F. Cao et al., 2015; Fong et al., 2017; Waters et al., 2017). A report based on using personalized phage therapy for treating a patient with multidrug resistant A. baumannii reported clearance of infection and success of the treatment (Schooley et al., 2017). Furthermore, case reports of patients with bacterial prostatitis and septicemia and acute kidney injury reported pathogen eradication and decrease in clinical symptoms post phage therapy (Jennes et al., 2017; Letkiewicz et al., 2009). Another study reported that 67 of 96 patients' wounds and ulcers healed post phage therapy; healing was associated with a reduction in pathogens (Markoishvili et al., 2002). The use of engineered bacteriophages to treat drug‐resistant Mycobacterium abscessus was reported to show clinical improvement in a patient with cystic fibrosis (Dedrick et al., 2019). Another promising approach is the use of phage lytic proteins as antimicrobial compounds (Mondal et al., 2020), making phages a strong antibacterial contender of antibiotics.
Bacteriocins represent another category of potential antibiotic alternative. Bacteriocins are ribosomally produced antibacterial peptides produced by bacteria which themselves are immune to the killing peptide due to specific immunity mechanisms. To date, bacteriocins have been mainly used in the food industry as food safety and preservative agents (Silva et al., 2018). Bacteriocins have shown promising results as antimicrobials in animal studies. For example, mouse model studies have reported the successful use of pyocin to treat P. aeruginosa lung infections with high efficacy and without any adverse effects (McCaughey et al., 2016; Merrikin & Terry, 1972). Another study in mice reported that administration of nisin‐ and pediocin‐producing Lactococcus lactis and P. acidilactici strains helped reduce intestinal vancomycin‐resistant enterococci colonization (Millette et al., 2008). Bacteriocins have been successfully used to treat and prevent bovine mastitis with comparable efficacy to antibiotics (L. T. Cao et al., 2007; Crispie et al., 2004; Kitching et al., 2019). Furthermore, nisin has proven to be effective in treating mastitis caused by Staphylococcus in eight lactating females (Fernández et al., 2008).
Another effective way of addressing the problem of antibiotic resistance is with the use of monoclonal antibodies as alternatives or in conjunction with antibiotics. Monoclonal antibodies bypass the complications of toxicity, resistance development, and early clearance by the immune system which is seen in the case of antibiotics. The use of monoclonal antibodies for treating bacterial infections is emerging in the past few years, before which monoclonal antibodies were mostly used for treating cancer, autoimmune diseases, or viral infections (Zurawski & McLendon, 2020). A recent study in rabbits demonstrated the success of the use of monoclonal antibody obiltoxaximab against anthrax protective antigen. The authors reported that the use of obiltoxaximab improved the survival of rabbits that received a lethal dose of B. anthracis spores (Henning et al., 2018). In a recent clinical trial of 2655 participants, authors reported that the use of bezlotoxumab (monoclonal antibody against C. difficile toxin) for treating C. difficile infection resulted in a lower recurrence of infection (Wilcox et al., 2017). In another recent study, Watson et al. (2021) generated a monoclonal antibody from B cells of a patient to be used against Mycobacterium tuberculosis infection in mice. Monoclonal antibodies are costlier than producing antibiotics but have many benefits and more studies in this direction will help can help transform medicine.
One of the major advantages of bacteriocins, phages and their endolysins, and monoclonal antibodies is that they can be highly target‐specific thus rendering minimal if any, collateral damage to the microbiota.
9. CONCLUSION
This review has summarized the importance of the gut microbiota in host metabolism and immune functions such as immunity development, colonization resistance, cell signaling, and with the help of advanced omics technologies, the complex interactions between host and microbiota are now becoming clear. Antibiotics disrupt the microbial balance and hence the networking within the bacterial community, and that with the host. The resulting resistant bacteria make clinical treatment difficult. Due to this complex link between the host and microbiota, the current usage of antibiotics requires careful stewardship, with an emphasis on the application of antibiotic alternatives, while limiting collateral damage. To this end, we need to design, develop, and translate new antibiotic alternatives from bench to bedside, in addition to methodologies that are efficient in conserving and restoring the microbial community after antibiotic‐associated perturbations.
CONFLICT OF INTERESTS
None declared.
ETHICS STATEMENT
None required.
AUTHOR CONTRIBUTIONS
Dhrati Patangia: Conceptualization (lead); writing – original draft (lead), writing – review and editing (equal), Paul Ross: Supervision (equal); conceptualization (supporting); writing – review and editing (equal), Catherine Stanton: Supervision (equal); conceptualization (supporting); writing – review and editing (equal), Tony Ryan: Supervision (equal); conceptualization (supporting); writing – review and editing (equal), Eugene Dempsey: Supervision (equal); conceptualization (supporting); writing – review and editing (equal).
ACKNOWLEDGMENTS
The authors were funded in part by Science Foundation Ireland and APC Microbiome Ireland.
Patangia, D. V. , Anthony Ryan, C. , Dempsey, E. , Paul Ross, R. , & Stanton, C. (2022). Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen, 11, e1260. 10.1002/mbo3.1260
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- Aagaard, K. , Ma, J. , Antony, K. M. , Ganu, R. , Petrosino, J. , & Versalovic, J. (2014). The placenta harbors a unique microbiome. Science Translational Medicine, 6(237), 237ra65. 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard, K. , Riehle, K. , Ma, J. , Segata, N. , Mistretta, T. A. , Coarfa, C. , Raza, S. , Rosenbaum, S. , Van den Veyver, I. , Milosavljevic, A. , Gevers, D. , Huttenhower, C. , Petrosino, J. , & Versalovic, J. (2012). ‘A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy’. PLoS One, 7(6), e36466. 10.1371/journal.pone.0036466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams, G. D. , & Bishop, J. E. (1967). Effect of the normal microbial flora on gastrointestinal motility. Proceedings of the Society for Experimental Biology and Medicine, 126(1), 301–304. 10.3181/00379727-126-32430 [DOI] [PubMed] [Google Scholar]
- Adedeji, W. A. (2016). The treasure called antibiotics. Annals of Ibadan Postgraduate Medicine, 14(2), 56–57. [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens, N. , Coenen, S. , Versporten, A. , Muller, A. , Minalu, G. , Faes, C. , Vankerckhoven, V. , Aerts, M. , Hens, N. , Molenberghs, G. , & Goossens, H. , ESAC Project Group . (2011). European surveillance of antimicrobial consumption (ESAC): Outpatient antibiotic use in Europe (1997‐2009). Journal of Antimicrobial Chemotherapy, 66(Suppl. 6), 3–12. 10.1093/jac/dkr453 [DOI] [PubMed] [Google Scholar]
- Akhavan, B. J. , Khanna, N. R. , & Vijhani, P. (2020). Amoxicillin, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Alexander, V. , Northrup, V. , & Bizzarro, M. (2011). Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. Journal of Pediatrics, 159(3), 392–397. 10.1016/j.jpeds.2011.02.035.Antibiotic [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti, J. R. , Mullish, B. H. , Kelly, C. , & Fischer, M. (2019). The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. The Lancet, 394(10196), 420–431. 10.1016/S0140-6736(19)31266-8 [DOI] [PubMed] [Google Scholar]
- Aloisio, I. , Mazzola, G. , Corvaglia, L. T. , Tonti, G. , Faldella, G. , Biavati, B. , & Di Gioia, D. (2014). Influence of intrapartum antibiotic prophylaxis against group B Streptococcus on the early newborn gut composition and evaluation of the anti‐Streptococcus activity of Bifidobacterium strains. Applied Microbiology and Biotechnology, 98(13), 6051–6060. 10.1007/s00253-014-5712-9 [DOI] [PubMed] [Google Scholar]
- Aloisio, I. , Quagliariello, A. , De Fanti, S. , Luiselli, D. , De Filippo, C. , Albanese, D. , Corvaglia, L. T. , Faldella, G. , & Di Gioia, D. (2016). Evaluation of the effects of intrapartum antibiotic prophylaxis on newborn intestinal microbiota using a sequencing approach targeted to multi hypervariable 16S rDNA regions. Applied Microbiology and Biotechnology, 100(12), 5537–5546. 10.1007/s00253-016-7410-2 [DOI] [PubMed] [Google Scholar]
- Antunes, L. C. , Han, J. , Ferreira, R. B. , Lolić, P. , Borchers, C. H. , & Finlay, B. B. (2011). Effect of antibiotic treatment on the intestinal metabolome. Antimicrobial Agents and Chemotherapy, 55(4), 1494–1503. 10.1128/AAC.01664-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya, S. , Binetti, A. , Salazar, N. , Fernández, N. , Solís, G. , Hernández‐Barranco, A. , Margolles, A. , los Reyes‐Gavilán, C. G. , & Gueimonde, M. (2012). Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiology Ecology, 79(3), 763–772. 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- Arboleya, S. , Sánchez, B. , Solís, G. , Fernández, N. , Suárez, M. , Hernández‐Barranco, A. M. , Milani, C. , Margolles, A. , de Los Reyes‐Gavilán, C. G. , Ventura, M. , & Gueimonde, M. (2016). Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: A functional inference study. International Journal of Molecular Sciences, 17(5), 649. 10.3390/ijms17050649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia, N. , Campbell, C. , Fan, X. , Dikiy, S. , van der Veeken, J. , de Roos, P. , Liu, H. , Cross, J. R. , Pfeffer, K. , Coffer, P. J. , & Rudensky, A. Y. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature, 504(7480), 451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta, M.‐C. , Stiemsma, L. T. , Dimitriu, P. A. , Thorson, L. , Russell, S. , Yurist‐Doutsch, S. , Kuzeljevic, B. , Gold, M. J. , Britton, H. M. , Lefebvre, D. L. , Subbarao, P. , Mandhane, P. , Becker, A. , McNagny, K. M. , Sears, M. R. , Kollmann, T. , The CHILD Study Investigators , Mohn, W. W. , Turvey, S. E. , & Finlay, B. B. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine, 7(307), 307ra152. 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- Arumugam, M. , Raes, J. , Pelletier, E. , Le Paslier, D. , Yamada, T. , Mende, D. R. , Fernandes, G. R. , Tap, J. , Bruls, T. , Batto, J. M. , Bertalan, M. , Borruel, N. , Casellas, F. , Fernandez, L. , Gautier, L. , Hansen, T. , Hattori, M. , Hayashi, T. , Kleerebezem, M. , … Bork, P. (2011). Enterotypes of the human gut microbiome. Nature, 473(7346), 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnicar, F. , Manara, S. , Zolfo, M. , Truong, D. T. , Scholz, M. , Armanini, F. , Ferretti, P. , Gorfer, V. , Pedrotti, A. , Tett, A. , & Segata, N. (2017). Studying vertical microbiome transmission from mothers to infants by strain‐level metagenomic profiling. mSystems, 2(1), e0016416. 10.1128/mSystems.00164-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, M. B. , Bridgman, S. L. , Becker, A. B. , & Kozyrskyj, A. L. (2014). Infant antibiotic exposure and the development of childhood overweight and central adiposity. International Journal of Obesity, 38(10), 1290–1298. 10.1038/ijo.2014.119 [DOI] [PubMed] [Google Scholar]
- Azad, M. B. , Konya, T. , Maughan, H. , Guttman, D. S. , Field, C. J. , Chari, R. S. , Sears, M. R. , Becker, A. B. , Scott, J. A. , & Kozyrskyj, A. L. (2013). Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Canadian Medical Association Journal, 185(5), 385–394. 10.1503/cmaj.121189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, M. B. , Konya, T. , Persaud, R. R. , Guttman, D. S. , Chari, R. S. , Field, C. J. , Sears, M. R. , Mandhane, P. J. , Turvey, S. E. , Subbarao, P. , Becker, A. B. , Scott, J. A. , & Kozyrskyj, A. L. , The CHILD Study Investigators . (2016). Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG: An International Journal of Obstetrics and Gynaecology, 123(6), 983–993. 10.1111/1471-0528.13601 [DOI] [PubMed] [Google Scholar]
- Bäckhed, F. , Roswall, J. , Peng, Y. , Feng, Q. , Jia, H. , Kovatcheva‐Datchary, P. , Li, Y. , Xia, Y. , Xie, H. , Zhong, H. , Khan, M. T. , Zhang, J. , Li, J. , Xiao, L. , Al‐Aama, J. , Zhang, D. , Lee, Y. S. , Kotowska, D. , Colding, C. , … Wang, J. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host and Microbe, 17(5), 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Bailey, J. K. , Pinyon, J. L. , Anantham, S. , & Hall, R. M. (2010). Commensal Escherichia coli of healthy humans: A reservoir for antibiotic‐resistance determinants. Journal of Medical Microbiology, 59(11), 1331–1339. 10.1099/jmm.0.022475-0 [DOI] [PubMed] [Google Scholar]
- Bailey, L. C. , Forrest, C. B. , Zhang, P. , Richards, T. M. , Livshits, A. , & DeRusso, P. A. (2014). Association of antibiotics in infancy with early childhood obesity. JAMA Pediatrics, 168(11), 1063–1069. 10.1001/jamapediatrics.2014.1539 [DOI] [PubMed] [Google Scholar]
- Bartoloni, A. , Pallecchi, L. , Rodríguez, H. , Fernandez, C. , Mantella, A. , Bartalesi, F. , Strohmeyer, M. , Kristiansson, C. , Gotuzzo, E. , Paradisi, F. , & Rossolini, G. M. (2009). Antibiotic resistance in a very remote Amazonas community. International Journal of Antimicrobial Agents, 33(2), 125–129. 10.1016/j.ijantimicag.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Bauer, H. , Horowitz, R. E. , Levenson, S. M. , & Popper, H. (1963). The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. The American Journal of Pathology, 42(4), 471–483. [PMC free article] [PubMed] [Google Scholar]
- Boursi, B. , Mamtani, R. , Haynes, K. , & Yang, Y.‐X. (2015). The effect of past antibiotic exposure on diabetes risk. European Journal of Endocrinology, 172(6), 639–648. 10.1530/eje-14-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson‐Palme, J. , Angelin, M. , Huss, M. , Kjellqvist, S. , Kristiansson, E. , Palmgren, H. , Larsson, D. G. J. , & Johansson, A. (2015). The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59(10), 6551–6560. 10.1128/AAC.00933-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten, G. , Lange, K. , Havinga, R. , van Dijk, T. H. , Gerding, A. , van Eunen, K. , Müller, M. , Groen, A. K. , Hooiveld, G. J. , Bakker, B. M. , & Reijngoud, D. J. (2013). Gut‐derived short‐chain fatty acids are vividly assimilated into host carbohydrates and lipids. American Journal of Physiology ‐ Gastrointestinal and Liver Physiology, 305(12), 900–910. 10.1152/ajpgi.00265.2013 [DOI] [PubMed] [Google Scholar]
- Bezirtzoglou, E. , Tsiotsias, A. , & Welling, G. W. (2011). Anaerobe microbiota profile in feces of breast‐ and formula‐fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe, 17(6), 478–482. 10.1016/j.anaerobe.2011.03.009 [DOI] [PubMed] [Google Scholar]
- Björkholm, B. , Bok, C. M. , Lundin, A. , Rafter, J. , Hibberd, M. L. , & Pettersson, S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One, 4(9), e6958. 10.1371/journal.pone.0006958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser, M. (2011). Stop the killing of beneficial bacteria. Nature, 476(7361), 393–394. 10.1038/476393a [DOI] [PubMed] [Google Scholar]
- Blaser, M. , & Falkow, S. (2009). What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology, 7, 887–894. 10.1038/nrmicro2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N. A. , Chung, J. , Battaglia, T. , Henderson, N. , Jay, M. , Li, H. , Lieber, A. D. , Wu, F. , Perez‐Perez, G. I. , Chen, Y. , Schweizer, W. , Zheng, X. , Contreras, M. , Dominguez‐Bello, M. G. , & Blaser, M. J. (2017). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Science Translational Medicine, 8(343), 1–14. 10.1126/scitranslmed.aad7121.Antibiotics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl, K. , Plitas, G. , Mihu, C. N. , Ubeda, C. , Jia, T. , Fleisher, M. , Schnabl, B. , DeMatteo, R. P. , & Pamer, E. G. (2008). Vancomycin‐resistant enterococci exploit antibiotic‐induced innate immune deficits. Nature, 455(7214), 804–807. 10.1038/nature07250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braye, K. , Foureur, M. , de Waal, K. , Jones, M. , Putt, E. , & Ferguson, J. (2019). Group B streptococcal screening, intrapartum antibiotic prophylaxis, and neonatal early‐ onset infection rates in an Australian local health district: 2006‐2016. PLoS One, 14(4), e0214295. 10.1371/journal.pone.0214295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff, J. , & Artis, D. (2013). Population genetic tools to dissect innate immunity in humans. Nature Immunology, 14(7), 676–684. 10.1038/ni.2640.Commensal [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron, P. A. , Van Baarlen, P. , & Kleerebezem, M. (2011). Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nature Reviews Microbiology, 10(1), 66–78. 10.1038/nrmicro2690 [DOI] [PubMed] [Google Scholar]
- Brugman, S. , Klatter, F. A. , Visser, J. T. , Wildeboer‐Veloo, A. C. , Harmsen, H. J. , Rozing, J. , & Bos, N. A. (2006). Antibiotic treatment partially protects against type 1 diabetes in the bio‐breeding diabetes‐prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia, 49(9), 2105–2108. 10.1007/s00125-006-0334-0 [DOI] [PubMed] [Google Scholar]
- Buffie, C. G. , Jarchum, I. , Equinda, M. , Lipuma, L. , Gobourne, A. , Viale, A. , Ubeda, C. , Xavier, J. , & Pamer, E. G. (2012). Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile‐induced colitis. Infection and Immunity, 80(1), 62–73. 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, T. , & Preuss, C. V. (2021). Cephalosporins, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Butaye, P. , Devriese, L. A. , & Haesebrouck, F. (2003). Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on Gram-positive bacteria. Clinical Microbiology Reviews, 16(2), 175–188. 10.1128/CMR.16.2.175-188.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss, M. X. , Olsan, E. E. , Rivera‐Chávez, F. , Tiffany, C. R. , Cevallos, S. A. , Lokken, K. L. , Torres, T. P. , Byndloss, A. J. , Faber, F. , Gao, Y. , Litvak, Y. , Lopez, C. A. , Xu, G. , Napoli, E. , Giulivi, C. , Tsolis, R. M. , Revzin, A. , Lebrilla, C. B. , & Bäumler, A. J. (2017). Microbiota‐activated PPAR‐γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science, 357(6351), 570–575. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello, F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environmental Microbiology, 8(7), 1137–1144. 10.1111/j.1462-2920.2006.01054.x [DOI] [PubMed] [Google Scholar]
- Cahenzli, J. , Köller, Y. , Wyss, M. , Geuking, M. B. , & McCoy, K. D. (2013). Intestinal microbial diversity during early‐life colonization shapes long‐term IgE levels. Cell Host and Microbe, 14(5), 559–570. 10.1016/j.chom.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candon, S. , Perez‐Arroyo, A. , Marquet, C. , Valette, F. , Foray, A. P. , Pelletier, B. , Milani, C. , Ventura, M. , Bach, J. F. , & Chatenoud, L. (2015). Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin‐dependent diabetes. PLoS One, 10(5), 1–16. 10.1371/journal.pone.0125448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani, P. D. , Osto, M. , Geurts, L. , & Everard, A. (2012). Involvement of gut microbiota in the development of low‐grade inflammation and type 2 diabetes associated with obesity. Gut Microbes, 3(4), 279–288. 10.4161/gmic.19625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, F. , Wang, X. , Wang, L. , Li, Z. , Che, J. , Wang, L. , Li, X. , Cao, Z. , Zhang, J. , Jin, L. , & Xu, Y. (2015). Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. BioMed Research International, 2015, 2015–2019. 10.1155/2015/752930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L. T. , Wu, J. Q. , Xie, F. , Hu, S. H. , & Mo, Y. (2007). Efficacy of nisin in treatment of clinical mastitis in lactating dairy cows. Journal of Dairy Science, 90(8), 3980–3985. 10.3168/jds.2007-0153 [DOI] [PubMed] [Google Scholar]
- Carlet, J. (2012). The gut is the epicentre of antibiotic resistance. Antimicrobial Resistance and Infection Control, 1, 1–7. 10.1186/2047-2994-1-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaburi, G. , Duar, R. M. , Vance, D. P. , Mitchell, R. , Contreras, L. , Frese, S. A. , Smilowitz, J. T. , & Underwood, M. A. (2019). Early‐life gut microbiome modulation reduces the abundance of antibiotic‐resistant bacteria. Antimicrobial Resistance & Infection Control, 8(1), 131. 10.1186/s13756-019-0583-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash, H. L. , Whitham, C. V. , Behrendt, C. L. , & Hooper, L. V. (2006). ‘Symbiotic bacteria direct expression of an intestinal bactericidal lectin’. Science, 313(5790), 1126–1130. 10.1126/science.1127119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla, S. I. , Maldonado‐Galdeano, C. , Weill, R. , De Paula, J. , & Perdigón, G. (2018). ‘Oral administration of probiotics increases Paneth cells and intestinal antimicrobial activity’. Frontiers in Microbiology, 9, 1–14. 10.3389/fmicb.2018.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centre for Disease Control (CDC) . (2017). Pediatric treatment recommendations. https://www.cdc.gov/antibiotic-use/community/for-hcp/outpatient-hcp/pediatric-treatment-rec.html
- Chaves, B. J. , & Tadi, P. (2021). Gentamicin, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Cho, I. , Yamanishi, S. , Cox, L. , Methé, B. A. , Zavadil, J. , Li, K. , Gao, Z. , Mahana, D. , Raju, K. , Teitler, I. , Li, H. , Alekseyenko, A. V. , & Blaser, M. J. (2012). ‘Antibiotics in early life alter the murine colonic microbiome and adiposity’. Nature, 488(7413), 621–626. 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. H. (2006). Reported medication use in the neonatal intensive care unit: Data from a large national data set. Pediatrics, 117(6), 1979–1987. 10.1542/peds.2005-1707 [DOI] [PubMed] [Google Scholar]
- Clemente, J. C. , Pehrsson, E. C. , Blaser, M. J. , Sandhu, K. , Gao, Z. , Wang, B. , Magris, M. , Hidalgo, G. , Contreras, M. , Noya‐Alarcón, Ó. , Lander, O. , McDonald, J. , Cox, M. , Walter, J. , Oh, P. L. , Ruiz, J. F. , Rodriguez, S. , Shen, N. , Song, S. J. , … Dominguez‐Bello, M. G. (2015). ‘The microbiome of uncontacted Amerindians’. Science Advances, 1(3), 3047. 10.1126/sciadv.1500183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker, M. O. , Hoen, A. G. , Dade, E. , Lundgren, S. , Li, Z. , Wong, A. D. , Zens, M. S. , Palys, T. J. , Morrison, H. G. , Sogin, M. L. , Baker, E. R. , Karagas, M. R. , & Madan, J. C. (2020). ‘Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: A prospective cohort study’. BJOG: An International Journal of Obstetrics & Gynaecology, 127(2), 217–227. 10.1111/1471-0528.15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado, M. C. , Meriluoto, J. , & Salminen, S. (2007). ‘Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus’. Letters in Applied Microbiology, 45(4), 454–460. 10.1111/j.1472-765X.2007.02212.x [DOI] [PubMed] [Google Scholar]
- Cotten, C. M. , Taylor, S. , Stoll, B. , Goldberg, R. N. , Hansen, N. I. , Sánchez, P. J. , Ambalavanan, N. , & Benjamin, D. K. (2009). Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics, 123(1), 58–66. 10.1542/peds.2007-3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, L. M. , Yamanishi, S. , Sohn, J. , Alekseyenko, A. V. , Leung, J. M. , Cho, I. , Kim, S. G. , Li, H. , Gao, Z. , Mahana, D. , Zárate Rodriguez, J. G. , Rogers, A. B. , Robine, N. , Loke, P. , & Blaser, M. J. (2014). ‘Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences’. Cell, 158(4), 705–721. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider, K. S. , Cleves, M. A. , Reefhuis, J. , Berry, R. J. , Hobbs, C. A. , & Hu, D. J. (2009). Antibacterial medication use during pregnancy and risk of birth defects. Archives of Pediatrics and Adolescent Medicine, 163(11), 978–985. 10.1001/archpediatrics.2009.188 [DOI] [PubMed] [Google Scholar]
- Crispie, F. , Flynn, J. , Ross, R. P. , Hill, C. , & Meaney, W. J. (2004). ‘Bismuth‐based intramammary teat seal in combination with the bacteriocin lacticin 3147 Crispie Flynn’. Irish Veterinary Journal, 57(11), 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cycoń, M. , Mrozik, A. , & Piotrowska‐Seget, Z. (2019). ‘Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity’. Frontiers in Microbiology, 10, 2677. 10.3389/fmicb.2019.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel, A. E. , Rockenbauer, M. , Sørensen, H. T. , & Olsen, J. (2001). The teratogenic risk of trimethoprim‐sulfonamides: A population based case‐control study. Reproductive Toxicology, 15, 637–646. 10.1016/S0890-6238(01)00178-2 [DOI] [PubMed] [Google Scholar]
- De Vadder, F. , Kovatcheva‐Datchary, P. , Goncalves, D. , Vinera, J. , Zitoun, C. , Duchampt, A. , Bäckhed, F. , & Mithieux, G. (2014). Microbiota‐generated metabolites promote metabolic benefits via gut‐brain neural circuits. Cell, 156(1–2), 84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Debroas, D. , & Siguret, C. (2019). Viruses as key reservoirs of antibiotic resistance genes in the environment. ISME Journal, 13(11), 2856–2867. 10.1038/s41396-019-0478-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick, R. M. , Guerrero‐Bustamante, C. A. , Garlena, R. A. , Russell, D. A. , Ford, K. , Harris, K. , Gilmour, K. C. , Soothill, J. , Jacobs‐Sera, D. , Schooley, R. T. , Hatfull, G. F. , & Spencer, H. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug‐resistant Mycobacterium abscessus . Nature Medicine, 25(5), 730–733. 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Fiol, F. S. , Balcão, V. M. , Barberato‐Fillho, S. , Lopes, L. C. , & Bergamaschi, C. C. (2018). Obesity: A new adverse effect of antibiotics? Frontiers in Pharmacology, 9, 1–10. 10.3389/fphar.2018.01408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen, L. , & Relman, D. A. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America, 108(Suppl_1), 4554–4561. 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, G. C. , Radl, V. , Schloter‐Hai, B. , Jechalke, S. , Heuer, H. , Smalla, K. , & Schloter, M. (2014). Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS One, 9(3), e92958. 10.1371/journal.pone.0092958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbler, P. , Mai, V. , Procianoy, R. S. , Silveira, R. C. , Corso, A. L. , & Roesch, L. (2019). The vaginal microbial communities of healthy expectant Brazilian mothers and its correlation with the newborn's gut colonization. World Journal of Microbiology and Biotechnology, 35(10), 1–14. 10.1007/s11274-019-2737-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez‐Bello, M. G. , Costello, E. K. , Contreras, M. , Magris, M. , Hidalgo, G. , Fierer, N. , Knight, R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences, 107(26), 11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg, G. , Lagier, J. C. , Robert, C. , Armougom, F. , Hugon, P. , Metidji, S. , Dione, N. , Dangui, N. P. M. , Pfleiderer, A. , Abrahao, J. , Musso, D. , Papazian, L. , Brouqui, P. , Bibi, F. , Yasir, M. , Vialettes, B. , & Raoult, D. (2014). Culturomics and pyrosequencing evidence of the reduction in gut microbiota diversity in patients with broad‐spectrum antibiotics. International Journal of Antimicrobial Agents, 44(2), 117–124. 10.1016/j.ijantimicag.2014.04.020 [DOI] [PubMed] [Google Scholar]
- Duncan, S. H. , Lobley, G. E. , Holtrop, G. , Ince, J. , Johnstone, A. M. , Louis, P. , & Flint, H. J. (2008). Human colonic microbiota associated with diet, obesity and weight loss. International Journal of Obesity, 32(11), 1720–1724. 10.1038/ijo.2008.155 [DOI] [PubMed] [Google Scholar]
- Esaiassen, E. , Fjalstad, J. W. , Juvet, L. K. , van den Anker, J. N. , & Klingenberg, C. (2017). Antibiotic exposure in neonates and early adverse outcomes: A systematic review and meta‐analysis. Journal of Antimicrobial Chemotherapy, 72(7), 1858–1870. 10.1093/jac/dkx088 [DOI] [PubMed] [Google Scholar]
- Fallani, M. , Amarri, S. , Uusijarvi, A. , Adam, R. , Khanna, S. , Aguilera, M. , Gil, A. , Vieites, J. M. , Norin, E. , Young, D. , Scott, J. A. , Doré, J. , & Edwards, C. A. , The Infabio Team . (2011). Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology, 157(5), 1385–1392. 10.1099/mic.0.042143-0 [DOI] [PubMed] [Google Scholar]
- Fan, H. , Li, L. , Wijlaars, L. , & Gilbert, R. E. (2019). Associations between use of macrolide antibiotics during pregnancy and adverse child outcomes: A systematic review and meta‐analysis. PLOS ONE, 14(2), e0212212. 10.1371/journal.pone.0212212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, L. , Delgado, S. , Herrero, H. , Maldonado, A. , & Rodríguez, J. M. (2008). The Bacteriocin Nisin, an Effective Agent for the Treatment of Staphylococcal Mastitis During Lactation. Journal of Human Lactation, 24(3), 311–316. 10.1177/0890334408317435 [DOI] [PubMed] [Google Scholar]
- Ferrer, M. , Méndez‐García, C. , Rojo, D. , Barbas, C. , & Moya, A. (2017). Antibiotic use and microbiome function. Biochemical Pharmacology, 134, 114–126. 10.1016/j.bcp.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Ferretti, P. , Pasolli, E. , Tett, A. , Asnicar, F. , Gorfer, V. , Fedi, S. , Armanini, F. , Truong, D. T. , Manara, S. , Zolfo, M. , Beghini, F. , Bertorelli, R. , De Sanctis, V. , Bariletti, I. , Canto, R. , Clementi, R. , Cologna, M. , Crifò, T. , Cusumano, G. , … Segata, N. (2018). Mother‐to‐infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host and Microbe, 24(1), 133–145. 10.1016/j.chom.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra, J. A. , Wu, K. J. , Hryckowian, A. J. , Bouley, D. M. , Weimer, B. C. , & Sonnenburg, J. L. (2014). Gut microbiota‐produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host & Microbe, 16(6), 770–777. 10.1016/j.chom.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, S. A. , Drilling, A. , Morales, S. , Cornet, M. E. , Woodworth, B. A. , Fokkens, W. J. , Psaltis, A. J. , Vreugde, S. , & Wormald, P. J. (2017). Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Frontiers in Cellular and Infection Microbiology, 7, 1–11. 10.3389/fcimb.2017.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy, F. , Guinane, C. M. , Hussey, S. , Wall, R. , Ryan, C. A. , Dempsey, E. M. , Murphy, B. , Ross, R. P. , Fitzgerald, G. F. , Stanton, C. , & Cotter, P. D. (2012). High‐throughput sequencing reveals the incomplete, short‐term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrobial Agents and Chemotherapy, 56(11), 5811–5820. 10.1128/AAC.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy, F. , Watkins, C. , Hill, C. J. , O'Shea, C. A. , Nagle, B. , Dempsey, E. M. , O'Toole, P. W. , Ross, R. P. , Ryan, C. A. , & Stanton, C. (2019). Perinatal factors affect the gut microbiota up to four years after birth. Nature Communications, 10(1), 1–10. 10.1038/s41467-019-09252-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas, A. C. , Chaban, B. , Bocking, A. , Rocco, M. , Yang, S. , Hill, J. E. , & Money, D. M. , The VOGUE Research Group . (2017). The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non‐pregnant women. Scientific Reports, 7(1), 1–16. 10.1038/s41598-017-07790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa, Y. , Obata, Y. , Fukuda, S. , Endo, T. A. , Nakato, G. , Takahashi, D. , Nakanishi, Y. , Uetake, C. , Kato, K. , Kato, T. , Takahashi, M. , Fukuda, N. N. , Murakami, S. , Miyauchi, E. , Hino, S. , Atarashi, K. , Onawa, S. , Fujimura, Y. , Lockett, T. , … Ohno, H. (2013). Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature, 504(7480), 446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Gao, H. , Shu, Q. , Chen, J. , Fan, K. , Xu, P. , Zhou, Q. , Li, C. , & Zheng, H. (2019). Antibiotic exposure has sex‐dependent effects on the gut microbiota and metabolism of short‐chain fatty acids and amino acids in mice. mSystems, 4(4), 1–16. 10.1128/msystems.00048-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparrini, A. J. , Wang, B. , Sun, X. , Kennedy, E. A. , Hernandez‐Leyva, A. , Ndao, I. M. , Tarr, P. I. , Warner, B. B. , & Dantas, G. (2019). Persistent metagenomic signatures of early‐life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nature Microbiology, 4(12), 2285–2297. 10.1038/s41564-019-0550-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. , Ding, C. , Zhao, W. , Xu, L. , Tian, H. , Gong, J. , Zhu, M. , Li, J. , & Li, N. (2017). Antibiotics‐induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. Journal of Translational Medicine, 15(1), 1–9. 10.1186/s12967-016-1105-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, M. K. , Wang, B. , Ahmadi, S. , Burnham, C. A. D. , Tarr, P. I. , Warner, B. B. , & Dantas, G. (2016). Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nature Microbiology, 301903, 1–25. 10.1038/nmicrobiol.2016.24.Developmental [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis, C. C. , Hughes, E. R. , Spiga, L. , Winter, M. G. , Zhu, W. , Furtado de Carvalho, T. , Chanin, R. B. , Behrendt, C. L. , Hooper, L. V. , Santos, R. L. , & Winter, S. E. (2018). Dysbiosis‐associated change in host metabolism generates lactate to support Salmonella growth. Cell Host and Microbe, 23(1), 54–64. 10.1016/j.chom.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem, M. W. , Pierneef, R. , Bezuidt, O. K. I. , Van De Peer, Y. , Cowan, D. A. , & Makhalanyane, T. P. (2018). A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome, 6(1). 10.1186/s40168-018-0424-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau, M. C. , Lager, S. , Sovio, U. , Gaccioli, F. , Cook, E. , Peacock, S. J. , Parkhill, J. , Charnock‐Jones, D. S. , & Smith, G. (2019). Human placenta has no microbiome but can contain potential pathogens. Nature, 572(7769), 329–334. 10.1038/s41586-019-1451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, E. B. , Yim, G. , Tsui, W. , McClure, J. , Surette, M. G. , & Davies, J. (2002). Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proceedings of the National Academy of Sciences of the United States of America, 99(26), 17025–17030. 10.1073/pnas.252607699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Arango, L. F. , Barrett, H. L. , McIntyre, H. D. , Callaway, L. K. , Morrison, M. , & Dekker Nitert, M. (2017). Antibiotic treatment at delivery shapes the initial oral microbiome in neonates. Scientific Reports, 7, 1–10. 10.1038/srep43481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Perez, G. , Hicks, A. L. , Tekieli, T. M. , Radens, C. M. , Williams, B. L. , & Lamousé‐Smith, E. S. (2016). Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. The Journal of Immunology, 196(9), 3768–3779. 10.4049/jimmunol.1502322 [DOI] [PubMed] [Google Scholar]
- Gray, J. , Oehrle, K. , Worthen, G. , Alenghat, T. , Whitsett, J. , & Deshmukh, H. (2017). Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Science Translational Medicine, 9(376), 1–14. 10.1126/scitranslmed.aaf9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guh, A. Y. , & Kutty, P. K. (2018). Clostridioides difficile infection. Annals of Internal Medicine, 169(7), ITC49–ITC64. 10.7326/AITC201810020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, M. , Li, Z. , You, H. , Rodrigues, R. , Jump, D. B. , Morgun, A. , & Shulzhenko, N. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine, 51, 102590. 10.1016/j.ebiom.2019.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habak, P. J. , & Griggs, R. P., Jr. (2021). Urinary tract infection in pregnancy, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Hagan, T. , Cortese, M. , Rouphael, N. , Boudreau, C. , Linde, C. , Maddur, M. S. , Das, J. , Wang, H. , Guthmiller, J. , Zheng, N. Y. , Huang, M. , Uphadhyay, A. A. , Gardinassi, L. , Petitdemange, C. , McCullough, M. P. , Johnson, S. J. , Gill, K. , Cervasi, B. , Zou, J. , … Pulendran, B. (2019). Antibiotics‐driven gut microbiome perturbation alters immunity to vaccines in humans. Cell, 178(6), 1313–1328. 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen, H. J. , Wildeboer‐Veloo, A. C. , Raangs, G. C. , Wagendorp, A. A. , Klijn, N. , Bindels, J. G. , & Welling, G. W. (2000). ‘Analysis of intestinal flora development in breast‐fed and formula‐fed infants by using molecular identification and detection methods’. Journal of Pediatric Gastroenterology and Nutrition, 30(1), 61–67. http://journals.lww.com/jpgn/Fulltext/2000/01000/Analysis_of_Intestinal_Flora_Development_in.19.aspx [DOI] [PubMed] [Google Scholar]
- Harrison, G. A. , Rubin, M. P. , Davies, R. M. , & Speller, D. C. (1985). ‘Resistance in oral streptococci after repetition of a single‐dose amoxycillin prophylactic regimen’. Journal of Antimicrobial Chemotherapy, 15(4), 501–503. 10.1093/jac/15.4.501 [DOI] [PubMed] [Google Scholar]
- Hatosy, S. M. , & Martiny, A. C. (2015). The ocean as a global reservoir of antibiotic resistance genes. Applied and Environmental Microbiology, 81(21), 7593–7599. 10.1128/AEM.00736-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkila, A. M. (1993). Antibiotics in pregnancy—a prospective cohort study on the policy of antibiotic prescription. Annals of Internal Medicine, 25(5), 467–471. 10.3109/07853899309147314 [DOI] [PubMed] [Google Scholar]
- Heiman, M. L. , & Greenway, F. L. (2016). A healthy gastrointestinal microbiome is dependent on dietary diversity. Molecular Metabolism, 5(5), 317–320. 10.1016/j.molmet.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinken, A. , & Thiele, I. (2015). Anoxic conditions promote species‐specific mutualism between gut microbes in silico. Applied and Environmental Microbiology, 81(12), 4049–4061. 10.1128/AEM.00101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning, L. N. , Carpenter, S. , Stark, G. V. , & Serbina, N. V. (2018). Development of protective immunity in New Zealand White rabbits challenged with Bacillus anthracis spores and treated with antibiotics and obiltoxaximab, a monoclonal antibody against protective antigen. Antimicrobial Agents and Chemotherapy, 62(2), e01590‐17. 10.1128/AAC.01590-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson, H. , Kumar, H. , Collado, M. C. , Salminen, S. , Isolauri, E. , & Rautava, S. (2019). Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Frontiers in Nutrition, 6, 475. 10.3389/fnut.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C. , Mills, S. , & Ross, R. P. (2018). Phages & antibiotic resistance: Are the most abundant entities on earth ready for a comeback? Future Microbiology, 13(6), 711–726. 10.2217/fmb-2017-0261 [DOI] [PubMed] [Google Scholar]
- Hill, C. J. , Lynch, D. B. , Murphy, K. , Ulaszewska, M. , Jeffery, I. B. , O'Shea, C. A. , Watkins, C. , Dempsey, E. , Mattivi, F. , Tuohy, K. , Ross, R. P. , Ryan, C. A. , O’ Toole, P. W. , & Stanton, C. (2017). Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome, 5, 1–18. 10.1186/s40168-016-0213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D. A. , Hoffmann, C. , Abt, M. C. , Du, Y. , Kobuley, D. , Kirn, T. J. , Bushman, F. D. , & Artis, D. (2010). Metagenomic analyses reveal antibiotic‐induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunology, 3(2), 148–158. 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. R. , D'Argenio, D. A. , MacCoss, M. J. , Zhang, Z. , Jones, R. A. , & Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature, 436(7054), 1171–1175. 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Peng, J. , Tai, N. , Hu, C. , Zhang, X. , Wong, F. S. , & Wen, L. (2015). Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. The Journal of Immunology, 195(9), 4176–4184. 10.4049/jimmunol.1500884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson, H. E. , Jernberg, C. , Andersson, A. F. , Sjölund‐Karlsson, M. , Jansson, J. K. , & Engstrand, L. (2010). Short‐term antibiotic treatment has differing long‐term impacts on the human throat and gut microbiome. PLoS One, 5(3), e9836. 10.1371/journal.pone.0009836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennes, S. , Merabishvili, M. , Soentjens, P. , Pang, K. W. , Rose, T. , Keersebilck, E. , Soete, O. , François, P. M. , Teodorescu, S. , Verween, G. , Verbeken, G. , De Vos, D. , & Pirnay, J. P. (2017). Use of bacteriophages in the treatment of colistin‐only‐sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury‐a case report. Critical Care, 21(1), 2016–2018. 10.1186/s13054-017-1709-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg, C. , Löfmark, S. , Edlund, C. , & Jansson, J. K. (2007). Long‐term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME Journal, 1(1), 56–66. 10.1038/ismej.2007.3 [DOI] [PubMed] [Google Scholar]
- Jernberg, C. , Löfmark, S. , Edlund, C. , & Jansson, J. K. (2010). Long‐term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology, 156(11), 3216–3223. 10.1099/mic.0.040618-0 [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Wu, Y. , Zeng, Z. , Jin, C. , Wu, S. , Wang, Y. , & Fu, Z. (2016). From the cover: Exposure to oral antibiotics induces gut microbiota dysbiosis associated with lipid metabolism dysfunction and low‐grade inflammation in mice. Toxicological Sciences, 154(1), 140–152. 10.1093/toxsci/kfw150 [DOI] [PubMed] [Google Scholar]
- Johnson, C. C. , Ownby, D. R. , Alford, S. H. , Havstad, S. L. , Williams, L. K. , Zoratti, E. M. , Peterson, E. L. , & Joseph, C. L. (2005). Antibiotic exposure in early infancy and risk for childhood atopy. Journal of Allergy and Clinical Immunology, 115, 1218–1224. 10.1016/j.jaci.2005.04.020 [DOI] [PubMed] [Google Scholar]
- de Jonge, L. , Bos, H. J. , van Langen, I. M. , de Jong‐van den Berg, L. T. , & Bakker, M. K. (2014). Antibiotics prescribed before, during and after pregnancy in the Netherlands: a drug utilization study. Pharmacoepidemiology and Drug Safety, 23, 60–68. 10.1002/pds.3492 [DOI] [PubMed] [Google Scholar]
- Jump, R. L. , Polinkovsky, A. , Hurless, K. , Sitzlar, B. , Eckart, K. , Tomas, M. , Deshpande, A. , Nerandzic, M. M. , & Donskey, C. J. (2014). Metabolomics analysis identifies intestinal microbiota‐derived biomarkers of colonization resistance in clindamycin‐treated mice. PLoS One, 9(7), e101267. 10.1371/journal.pone.0101267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalghatgi, S. , Spina, C. S. , Costello, J. C. , Liesa, M. , Morones‐Ramirez, J. R. , Slomovic, S. , Molina, A. , Shirihai, O. S. , & Collins, J. J. (2013). Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Science Translational Medicine, 5(192), 192ra85. 10.1126/scitranslmed.3006055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källén, B. , & Danielsson, B. R. (2014). Fetal safety of erythromycin. An update of Swedish data. European Journal of Clinical Pharmacology, 70(3), 355–360. 10.1007/s00228-013-1624-3 [DOI] [PubMed] [Google Scholar]
- Kaplan, J. B. , Izano, E. A. , Gopal, P. , Karwacki, M. T. , Kim, S. , Bose, J. L. , Bayles, K. W. , & Horswill, A. R. (2012). Low levels of β‐Lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio, 3(4), 2–9. 10.1128/mBio.00198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami, N. , Nowrouzian, F. , Adlerberth, I. , & Wold, A. E. (2006). Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrobial Agents and Chemotherapy, 50(1), 156–161. 10.1128/AAC.50.1.156-161.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, C. , Sugimoto, K. , Moritani, I. , Tanaka, J. , Oya, Y. , Inoue, H. , Tameda, M. , Shiraki, K. , Ito, M. , Takei, Y. , & Takase, K. (2015). Comparison of the gut microbiota composition between obese and non‐obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next‐generation sequencing. BMC Gastroenterology, 15(100), 1–10. 10.1186/s12876-015-0330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C. J. , Zheng, L. , Campbell, E. L. , Saeedi, B. , Scholz, C. C. , Bayless, A. J. , Wilson, K. E. , Glover, L. E. , Kominsky, D. J. , Magnuson, A. , Weir, T. L. , Ehrentraut, S. F. , Pickel, C. , Kuhn, K. A. , Lanis, J. M. , Nguyen, V. , Taylor, C. T. , & Colgan, S. P. (2015). Crosstalk between microbiota‐derived short‐chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host & Microbe, 17(5), 662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, S. , Pike, K. , Jones, D. R. , Brocklehurst, P. , Marlow, N. , Salt, A. , & Taylor, D. J. (2008). Childhood outcomes after prescription of antibiotics to pregnant women with spontaneous preterm labour: 7‐year follow‐up of the ORACLE II trial. The Lancet, 372(9646), 1319–1327. 10.1016/S0140-6736(08)61203-9 [DOI] [PubMed] [Google Scholar]
- Ki Cha, B. , Mun Jung, S. , Hwan Choi, C. , Song, I. D. , Woong Lee, H. , Joon Kim, H. , Hyuk, J. , Kyung Chang, S. , Kim, K. , Chung, W. S. , & Seo, J. G. (2012). ‘The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea‐dominant irritable bowel syndrome: A randomized, double‐blind, placebo‐controlled trial’. Journal of Clinical Gastroenterology, 46(3), 220–227. 10.1097/MCG.0b013e31823712b1 [DOI] [PubMed] [Google Scholar]
- Kim, H. , Sitarik, A. R. , Woodcroft, K. , Johnson, C. C. , & Zoratti, E. (2019). Birth mode, breastfeeding, pet exposure, and antibiotic use: Associations with the gut microbiome and sensitization in children. Current Allergy and Asthma Reports, 19(4), 435. 10.1007/s11882-019-0851-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmitt, P. T. , Harwood, C. R. , & Barer, M. R. (1999). Induction of type 2 Shiga toxin synthesis in Escherichia coli 0157 by 4‐quinolones. Lancet, 353(9164), 1588–1589. 10.1016/S0140-6736(99)00621-2 [DOI] [PubMed] [Google Scholar]
- Kitching, M. , Mathur, H. , Flynn, J. , Byrne, N. , Dillon, P. , Sayers, R. , Rea, M. C. , Hill, C. , & Ross, R. P. (2019). A live bio‐therapeutic for mastitis, containing Lactococcus lactis DPC3147 with comparable efficacy to antibiotic treatment. Frontiers in Microbiology, 10, 1–11. 10.3389/fmicb.2019.02220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, E. Y. , Van Boeckel, T. P. , Martinez, E. M. , Pant, S. , Gandra, S. , Levin, S. A. , Goossens, H. , & Laxminarayan, R. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proceedings of the National Academy of Sciences of the United States of America, 115(15), E3463–E3470. 10.1073/pnas.1717295115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel, N. , Rekdal, V. M. , & Balskus, E. P. (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science, 356(6344), 1246–1257. 10.1126/science.aag2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, O. , Goodrich, J. K. , Cullender, T. C. , Spor, A. , Laitinen, K. , Bäckhed, H. K. , Gonzalez, A. , Werner, J. J. , Angenent, L. T. , Knight, R. , Bäckhed, F. , Isolauri, E. , Salminen, S. , & Ley, R. E. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell, 150(3), 470–480. 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela, K. , Salonen, A. , Virta, L. J. , Kekkonen, R. A. , Forslund, K. , Bork, P. , & de Vos, W. M. (2016). Intestinal microbiome is related to lifetime antibiotic use in Finnish pre‐school children. Nature Communications, 7, 1–8. 10.1038/ncomms10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela, K. , Salonen, A. , Vepsäläinen, O. , Suomalainen, M. , Kolmeder, C. , Varjosalo, M. , Miettinen, S. , Kukkonen, K. , Savilahti, E. , Kuitunen, M. , & de Vos, W. M. (2018). Probiotic supplementation restores normal microbiota composition and function in antibiotic‐treated and in caesarean‐born infants. Microbiome, 6(1), 1–11. 10.1186/s40168-018-0567-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyrskyj, A. L. , Ernst, P. , & Becker, A. B. (2007). Increased risk of childhood asthma from antibiotic use in early life. Chest, 131(6), 1753–1759. 10.1378/chest.06-3008 [DOI] [PubMed] [Google Scholar]
- Kronman, M. P. , Zaoutis, T. E. , Haynes, K. , Feng, R. , & Coffin, S. E. (2012). Antibiotic exposure and IBD development among children: A population‐based cohort study. Pediatrics, 130(4), e794–e803. 10.1542/peds.2011-3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman, A. A. , & Koren, O. (2016). Antibiotic use during pregnancy: How bad is it?’. BMC Medicine, 14(1), 1–7. 10.1186/s12916-016-0636-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppala, V. S. , Meinzen‐Derr, J. , Morrow, A. L. , & Schibler, K. R. (2011). Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. The Journal of Pediatrics, 159(5), 720–725. 10.1016/j.jpeds.2011.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Les Dethlefsen, S. H. , Sogin, M. L. , & Relman, D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biology, 6(11), e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letkiewicz, S. , Miedzybrodzki, R. , Fortuna, W. , Weber‐Dabrowska, B. , & Górski, A. (2009). Eradication of Enterococcus faecalis by phage therapy in chronic bacterial prostatitis—case report. Folia Microbiologica, 54(5), 457–461. 10.1007/s12223-009-0064-z [DOI] [PubMed] [Google Scholar]
- Li, H. , Xiao, B. , Zhang, Y. , Xiao, S. , Luo, J. , & Huang, W. (2019). Impact of maternal intrapartum antibiotics on the initial oral microbiome of neonates. Pediatrics & Neonatology, 60(6), 654–661. 10.1016/j.pedneo.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Lin, D. M. , Koskella, B. , & Lin, H. C. (2017). Phage therapy: An alternative to antibiotics in the age of multi‐drug resistance. World Journal of Gastrointestinal Pharmacology and Therapeutics, 8(3), 162–173. 10.4292/wjgpt.v8.i3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. T. , Connelly, M. B. , Amolo, C. , Otani, S. , & Yaver, D. S. (2005). Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrobial Agents and Chemotherapy, 49(5), 1915–1926. 10.1128/AAC.49.5.1915-1926.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K. J. , Mitchell, A. A. , Yau, W.‐P. , Louik, C , & Hernández‐Díaz, S. (2012). Maternal exposure to amoxicillin and the risk of oral clefts. Epidemiology, 23(5), 699–705. 10.1097/ede.0b013e318258cb05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, M. , Lofmark, S. , Edlund, C. , Huovinen, P. , & Jalava, J. (2009). Prolonged impact of a one‐week course of clindamycin on Enterococcus spp. in human normal microbiota. Scandinavian Journal of Infectious Diseases, 41(3), 215–219. 10.1080/00365540802651897 [DOI] [PubMed] [Google Scholar]
- Llor, C. , & Bjerrum, L. (2014). Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Therapeutic Advances in Drug Safety, 5(6), 229–241. 10.1177/2042098614554919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfmark, S. , Jernberg, C. , Jansson, J. K. , & Edlund, C. (2006). Clindamycin‐induced enrichment and long‐term persistence of resistant Bacteroides spp. and resistance genes. Journal of Antimicrobial Chemotherapy, 58(6), 1160–1167. 10.1093/jac/dkl420 [DOI] [PubMed] [Google Scholar]
- Lu, K. , Cable, P. H. , Abo, R. P. , Ru, H. , Graffam, M. E. , Schlieper, K. A. , Parry, N. M. , Levine, S. , Bodnar, W. M. , Wishnok, J. S. , Styblo, M. , Swenberg, J. A. , Fox, J. G. , & Tannenbaum, S. R. (2013). Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chemical Research in Toxicology, 26(12), 1893–1903. 10.1021/tx4002868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulijwa, R. , Rupia, E. J. , & Alfaro, A. C. (2019). Antibiotic use in aquaculture, policies and regulation, health and environmental risks: A review of the top 15 major producers. Reviews in Aquaculture, 12(2), 640–663. 10.1111/raq.12344 [DOI] [Google Scholar]
- Lupski, J. R. (1987). Molecular mechanisms for transposition of drug‐resistance genes and other movable genetic elements. Reviews of Infectious Diseases, 9(2), 357–368. 10.1093/clinids/9.2.357 [DOI] [PubMed] [Google Scholar]
- Lynch, T. , & Price, A. (2007). The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. American Family Physician, 76(3), 391–396. [PubMed] [Google Scholar]
- Machiels, K. , Joossens, M. , Sabino, J. , De Preter, V. , Arijs, I. , Eeckhaut, V. , Ballet, V. , Claes, K. , Van Immerseel, F. , Verbeke, K. , Ferrante, M. , Verhaegen, J. , Rutgeerts, P. , & Vermeire, S. (2013). A decrease of the butyrate‐producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut, 63, 1–9. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- Macpherson, A. J. , & Uhr, T. (2004). Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science, 303(1662), 1662–1665. 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- Mangin, I. , Suau, A. , Gotteland, M. , Brunser, O. , & Pochart, P. (2010). Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe, 16(4), 433–438. 10.1016/j.anaerobe.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Markoishvili, K. , Tsitlanadze, G. , Katsarava, R. , Glenn, J. , & Sulakvelidze, A. (2002). A novel sustained‐release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. International Journal of Dermatology, 41, 453–458. 10.1046/j.1365-4362.2002.01451.x [DOI] [PubMed] [Google Scholar]
- Marshall, B. M. , & Levy, S. B. (2011). Food animals and antimicrobials: Impacts on human health. Clinical Microbiology Reviews, 24(4), 718–733. 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian, S. K. , Liu, C. H. , Tzianabos, A. O. , & Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell, 122(1), 107–118. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Mazzola, G. , Murphy, K. , Ross, R. P. , Di Gioia, D. , Biavati, B. , Corvaglia, L. T. , Faldella, G. , & Stanton, C. (2016). Early gut microbiota perturbations following intrapartum antibiotic prophylaxis to prevent group B streptococcal disease. PLoS One, 11(6), e0157527. 10.1371/journal.pone.0157527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey, L. C. , Ritchie, N. D. , Douce, G. R. , Evans, T. J. , & Walker, D. (2016). Efficacy of species‐specific protein antibiotics in a murine model of acute Pseudomonas aeruginosa lung infection. Scientific Reports, 6, 1–8. 10.1038/srep30201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, L. C. (2017). Effects of short‐ and long‐course antibiotics on the lower intestinal microbiome as they relate to traveller's diarrhea. Journal of Travel Medicine, 24(1), S35–S38. 10.1093/jtm/taw084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmanus, P. S. , Stockwell, V. O. , Sundin, G. W. , & Jones, A. L. (2002). Antibiotic use in plant agriculture. Annual Review of Phytopathology, 40, 443–465. [DOI] [PubMed] [Google Scholar]
- Meeraus, W. H. , Petersen, I. , & Gilbert, R. (2015). Association between antibiotic prescribing in pregnancy and cerebral palsy or epilepsy in children born at term: A cohort study using the health improvement network. PLoS One, 10(3), e0122034. 10.1371/journal.pone.0122034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinl, W. , Sczesny, S. , Brigelius‐Flohé, R. , Blaut, M. , & Glatt, H. (2009). Impact of gut microbiota on intestinal and hepatic levels of phase 2 xenobiotic‐metabolizing enzymes in the rat. Drug Metabolism and Disposition, 37(6), 1179–1186. 10.1124/dmd.108.025916 [DOI] [PubMed] [Google Scholar]
- Merrikin, D. J. , & Terry, C. S. (1972). Use of pyocin 78‐C2 in the treatment of Pseudomonas aeruginosa infection in mice. Applied Microbiology, 23(1), 164–165. 10.1128/aem.23.1.164-165.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, K. H. , Frost, M. , Bahl, M. I. , Licht, T. R. , Jensen, U. S. , Rosenberg, J. , Pedersen, O. , Hansen, T. , Rehfeld, J. F. , Holst, J. J. , Vilsbøll, T. , & Knop, F. K. (2015). Effect of antibiotics on gut microbiota, gut hormones and glucose metabolism. PLOS ONE, 10(11), e0142352. 10.1371/journal.pone.0142352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, K. H. , Knop, F. K. , Frost, M. , Hallas, J. , & Pottegård, A. (2015). Use of antibiotics and risk of type 2 diabetes: A population‐based case‐control study. Journal of Clinical Endocrinology and Metabolism, 100(10), 3633–3640. 10.1210/jc.2015-2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Duranti, S. , Bottacini, F. , Casey, E. , Turroni, F. , Mahony, J. , Belzer, C. , Palacio, S. D. , Montes, S. A. , Mancabelli, L. , Lugli, G. A. , Rodriguez, J. M. , Bode, L. , de Vos, W. , Gueimonde, M. , Margolles, A. , van Sinderen, D. , & Ventura, M. (2017). The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiology and Molecular Biology Reviews, 81(4), 1–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. A. , Vicentini, F. A. , Hirota, S. A. , Sharkey, K. A. , & Wieser, M. E. (2019). Antibiotic treatment affects the expression levels of copper transporters and the isotopic composition of copper in the colon of mice. Proceedings of the National Academy of Sciences of the United States of America, 116(13), 5955–5960. 10.1073/pnas.1814047116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millette, M. , Cornut, G. , Dupont, C. , Shareck, F. , Archambault, D. , & Lacroix, M. (2008). Capacity of human nisin‐ and pediocin‐producing lactic acid bacteria to reduce intestinal colonization by vancomycin‐resistant enterococci. Applied and Environmental Microbiology, 74(7), 1997–2003. 10.1128/AEM.02150-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, S. , Stanton, C. , Lane, J. , Smith, G. , & Ross, R. (2019). ‘Precision nutrition and the microbiome, part I: Current state of the science’. Nutrients, 11(4), 1–45. 10.3390/nu11040923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirani, Z. A. , & Jamil, N. (2010). Effect of sub‐lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus . Journal of Basic Microbiology, 51(2), 191–195. 10.1002/jobm.201000221 [DOI] [PubMed] [Google Scholar]
- Miyoshi, J. , Bobe, A. M. , Miyoshi, S. , Huang, Y. , Hubert, N. , Delmont, T. O. , Eren, A. M. , Leone, V. , & Chang, E. B. (2017). Peripartum antibiotics promote gut dysbiosis, loss of immune tolerance, and inflammatory bowel disease in genetically prone offspring. Cell Reports, 20(2), 491–504. 10.1016/j.celrep.2017.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles, L. , Gómez, M. , Heilig, H. , Bustos, G. , Fuentes, S. , de Vos, W. , Fernández, L. , Rodríguez, J. M. , & Jiménez, E. (2013). Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One, 8(6), e66986. 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal, S. I. , Draper, L. A. , Ross, R. P. , & Hill, C. (2020). Bacteriophage endolysins as a potential weapon to combat Clostridioides difficile infection. Gut Microbes, 12(1), 1–15. 10.1080/19490976.2020.1813533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A. M. , Patel, S. , Forsberg, K. J. , Wang, B. , Bentley, G. , Razia, Y. , Qin, X. , Tarr, P. I. , & Dantas, G. (2013). Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS ONE, 8(11), e78822. 10.1371/journal.pone.0078822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgun, A. , Dzutsev, A. , Dong, X. , Greer, R. L. , Sexton, D. J. , Ravel, J. , Schuster, M. , Hsiao, W. , Matzinger, P. , & Shulzhenko, N. (2015). Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut, 64(11), 1732–1743. 10.1136/gutjnl-2014-308820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca, A. , Leclerc, M. , & Hugot, J. P. (2016). Gut microbiota diversity and human diseases: Should we reintroduce key predators in our ecosystem? Frontiers in Microbiology, 7(MAR), 1–12. 10.3389/fmicb.2016.00455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, C. , Yang, Y. , Yu, K. , Yu, M. , Zhang, C. , Su, Y. , & Zhu, W. (2017). Alteration of metabolomic markers of amino‐acid metabolism in piglets with in‐feed antibiotics. Amino Acids, 49(4), 771–781. 10.1007/s00726-017-2379-4 [DOI] [PubMed] [Google Scholar]
- Muanda, F. T. , & Sheehy, O. (2017). Use of antibiotics during pregnancy and the risk of major congenital malformations: A population based cohort study. Pharmacoepidemiology, 83, 2557–2571. 10.1111/bcp.13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, N. T. , Whyatt, R. , Hoepner, L. , Oberfield, S. , Dominguez‐Bello, M. G. , Widen, E. M. , Hassoun, A. , Perera, F. , & Rundle, A. (2015). Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. International Journal of Obesity, 39(4), 665–670. 10.1038/ijo.2014.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad, M. H. , Idris, A. L. , Fan, X. , Guo, Y. , Yu, Y. , Jin, X. , Qiu, J. , Guan, X. , & Huang, T. (2020). Beyond risk: Bacterial biofilms and their regulating approaches. Frontiers in Microbiology, 11, 1–20. 10.3389/fmicb.2020.00928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R. , Stewart, A. W. , Braithwaite, I. , Beasley, R. , Hancox, R. J. , Mitchell, E. A. , & ISAAC Phase Three Study, G. (2014). Antibiotic treatment during infancy and increased body mass index in boys: An international cross‐sectional study. International Journal of Obesity, 38(8), 1115–1119. 10.1038/ijo.2013.218 [DOI] [PubMed] [Google Scholar]
- Natividad, J. M. , Hayes, C. L. , Motta, J. P. , Jury, J. , Galipeau, H. J. , Philip, V. , Garcia‐Rodenas, C. L. , Kiyama, H. , Bercik, P. , & Verdu, E. F. (2013). Differential induction of antimicrobial REGIII by the intestinal microbiota and Bifidobacterium breve NCC2950. Applied and Environmental Microbiology, 79(24), 7745–7754. 10.1128/AEM.02470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K. M. , Ferreyra, J. A. , Higginbottom, S. K. , Lynch, J. B. , Kashyap, P. C. , Gopinath, S. , Naidu, N. , Choudhury, B. , Weimer, B. C. , Monack, D. M. , & Sonnenburg, J. L. (2013). Microbiota‐liberated host sugars facilitate post‐antibiotic expansion of enteric pathogens. Nature, 502(7469), 96–99. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M. , Epstein, S. B. , Callahan, M. T. , Piotrowski, B. O. , Simon, G. L. , Roberts, A. D. , Keiser, J. F. , & Kaplan, J. B. (2014). Induction of MRSA biofilm by low‐dose β‐lactam antibiotics: Specificity, prevalence and dose‐response effects. Dose‐Response, 12(1), 152–161. 10.2203/dose-response.13-021.Kaplan [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J. , Friedman, H. , Boyd, B. C. , McGurn, A. , Babinski, P. , Markossian, T. , & Dugas, L. R. (2019). Early antibiotic exposure and development of asthma and allergic rhinitis in childhood. BMC Pediatrics, 19(1), 1–8. 10.1186/s12887-019-1594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel, Y. R. , Cox, L. M. , Kirigin, F. F. , Bokulich, N. A. , Yamanishi, S. , Teitler, I. , Chung, J. , Sohn, J. , Barber, C. M. , Goldfarb, D. S. , Raju, K. , Abubucker, S. , Zhou, Y. , Ruiz, V. E. , Li, H. , Mitreva, M. , Alekseyenko, A. V. , Weinstock, G. M. , Sodergren, E. , & Blaser, M. J. (2015). Metabolic and metagenomic outcomes from early‐life pulsed antibiotic treatment. Nature Communications, 6, 1–15. 10.1038/ncomms8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogacka, A. , Salazar, N. , Suárez, M. , Milani, C. , Arboleya, S. , Solís, G. , Fernández, N. , Alaez, L. , Hernández‐Barranco, A. M. , de Los Reyes‐Gavilán, C. G. , Ventura, M. , & Gueimonde, M. (2017). Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full‐term neonates. Microbiome, 5(1), 1–10. 10.1186/s40168-017-0313-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriel‐Ohayon, M. , Neuman, H. , & Koren, O. (2016). Microbial changes during pregnancy, birth, and infancy. Frontiers in Microbiology, 7, 1–13. 10.3389/fmicb.2016.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyangahu, D. D. , Lennard, K. S. , Brown, B. P. , Darby, M. G. , Wendoh, J. M. , Havyarimana, E. , Smith, P. , Butcher, J. , Stintzi, A. , Mulder, N. , Horsnell, W. , & Jaspan, H. B. (2018). Disruption of maternal gut microbiota during gestation alters offspring microbiota and immunity. Microbiome, 6(1), 1–10. 10.1186/s40168-018-0511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, E. F. , Cotter, P. D. , Stanton, C. , Ross, R. P. , & Hill, C. (2012). Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: Bacteriocins and conjugated linoleic acid. International Journal of Food Microbiology, 152(3), 189–205. 10.1016/j.ijfoodmicro.2011.05.025 [DOI] [PubMed] [Google Scholar]
- Oyama, N. , Sudo, N. , Sogawa, H. , & Kubo, C. (2001). Antibiotic use during infancy promotes a shift in the TH1/TH2 balance toward TH2‐dominant immunity in mice. Journal of Allergy and Clinical Immunology, 107(1), 153–159. 10.1067/mai.2001.111142 [DOI] [PubMed] [Google Scholar]
- Pärnänen, K. , Karkman, A. , Hultman, J. , Lyra, C. , Bengtsson‐Palme, J. , Larsson, D. , Rautava, S. , Isolauri, E. , Salminen, S. , Kumar, H. , Satokari, R. , & Virta, M. (2018). Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nature Communications, 9(1), 1–11. 10.1038/s41467-018-06393-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge, S. R. , Tsafnat, G. , Coiera, E. , & Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons: Review article. FEMS Microbiology Reviews, 33(4), 757–784. 10.1111/j.1574-6976.2009.00175.x [DOI] [PubMed] [Google Scholar]
- Patel, P. H. , & Hashmi, M. F. (2021). Macrolides, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Peechakara, B. V. , & Gupta, M. (2021). Ampicillin/sulbactam, StatPearls [Internet]. StatPearls Publishing. [Google Scholar]
- Penders, J. , Thijs, C. , Vink, C. , Stelma, F. F. , Snijders, B. , Kummeling, I. , van den Brandt, P. A. , & Stobberingh, E. E. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics, 118(2), 511–521. 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- Pérez‐Cobas, A. E. , Artacho, A. , Knecht, H. , Ferrús, M. L. , Friedrichs, A. , Ott, S. J. , Moya, A. , Latorre, A. , & Gosalbes, M. J. (2013). Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One, 8(11), e80201. 10.1371/journal.pone.0080201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cobas, A. E. , Gosalbes, M. J. , Friedrichs, A. , Knecht, H. , Artacho, A. , Eismann, K. , Otto, W. , Rojo, D. , Bargiela, R. , von Bergen, M. , Neulinger, S. C. , Däumer, C. , Heinsen, F. A. , Latorre, A. , Barbas, C. , Seifert, J. , dos Santos, V. M. , Ott, S. J. , Ferrer, M. , & Moya, A. (2013). Gut microbiota disturbance during antibiotic therapy: A multi‐omic approach. Gut, 62(11), 1591–1601. 10.1136/gutjnl-2012-303184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Muñoz, M. E. , Arrieta, M. C. , Ramer‐Tait, A. E. , & Walter, J. (2017). A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome, 5(1), 1–19. 10.1186/s40168-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, I. , Gilbert, R. , Evans, S. , Ridolfi, A. , & Nazareth, I. (2010). Oral antibiotic prescribing during pregnancy in primary care: UK population‐based study. Journal of Antimicrobial Chemotherapy, 65, 2238–2246. 10.1093/jac/dkq307 [DOI] [PubMed] [Google Scholar]
- Pitsouni, E. , Iavazzo, C. , Athanasiou, S. , & Falagas, M. E. (2007). Single‐dose azithromycin versus erythromycin or amoxicillin for Chlamydia trachomatis infection during pregnancy: a meta‐analysis of randomised controlled trials. International Journal of Antimicrobial Agents, 30(3), 213–221. 10.1016/j.ijantimicag.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Rakoff‐Nahoum, S. , Paglino, J. , Eslami‐Varzaneh, F. , Edberg, S. , & Medzhitov, R. (2004). Recognition of commensal microflora by toll‐like receptors is required for intestinal homeostasis. Cell, 118, 229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Ramai, D. (2019). Fecal microbiota transplantation: Donor relation, fresh or frozen, delivery methods, cost‐effectiveness. Annals of Gastroenterology, 32(1), 30–38. 10.20524/aog.2018.0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, M. U. , Zaura, E. , Buijs, M. J. , Keijser, B. J. , Crielaard, W. , Nord, C. E. , & Weintraub, A. (2015). Determining the long‐term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clinical Infectious Diseases, 60(Suppl 2), S77–S84. 10.1093/cid/civ137 [DOI] [PubMed] [Google Scholar]
- Raymond, F. , Ouameur, A. A. , Déraspe, M. , Iqbal, N. , Gingras, H. , Dridi, B. , Leprohon, P. , Plante, P. L. , Giroux, R. , Bérubé, È. , Frenette, J. , Boudreau, D. K. , Simard, J. L. , Chabot, I. , Domingo, M. C. , Trottier, S. , Boissinot, M. , Huletsky, A. , Roy, P. H. , … Corbeil, J. (2016). The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME Journal, 10(3), 707–720. 10.1038/ismej.2015.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, M. C. , Dobson, A. , O'Sullivan, O. , Crispie, F. , Fouhy, F. , Cotter, P. D. , Shanahan, F. , Kiely, B. , Hill, C. , & Ross, R. P. (2011). Effect of broad‐ and narrow‐spectrum antimicrobials on Clostridium dif fi cile and microbial diversity in a model of the distal colon. Proceedings of the National Academy of Sciences of the United States of America, 108, 4639–4644. 10.1073/pnas.1001224107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready, D. , Lancaster, H. , Qureshi, F. , Bedi, R. , Mullany, P. , & Wilson, M. (2004). Effect of amoxicillin use on oral microbiota in young children. Antimicrobial Agents and Chemotherapy, 48(8), 2883–2887. 10.1128/AAC.48.8.2883-2887.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, L. W. , Raphael, E. , & Faerstein, E. (2013). Obesity in the United States—dysbiosis from exposure to low-dose antibiotics? Frontiers in Public Health, 1, 1–8. 10.3389/fpubh.2013.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rineh, A. , Kelso, M. J. , Vatansever, F. , Tegos, G. P. , & Hamblin, M. R. (2014). Clostridium difficile infection: Molecular pathogenesis and novel therapeutics. Expert Review of Anti‐Infective Therapy, 12(1), 131–150. 10.1586/14787210.2014.866515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risnes, K. R. , Belanger, K. , Murk, W. , & Bracken, M. B. (2011). Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. American Journal of Epidemiology, 173(3), 310–318. 10.1093/aje/kwq400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, R. R. , Greer, R. L. , Dong, X. , DSouza, K. N. , Gurung, M. , Wu, J. Y. , Morgun, A. , & Shulzhenko, N. (2017). Antibiotic‐induced alterations in gut microbiota are associated with changes in glucose metabolism in healthy mice. Frontiers in Microbiology, 8, 1–14. 10.3389/fmicb.2017.02306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, I. , Gibson, G. , Heinken, A. , Scott, K. , Swann, J. , Thiele, I. , & Tuohy, K. (2018). Gut microbiota functions: Metabolism of nutrients and other food components. European Journal of Nutrition, 57(1), 1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozwandowicz, M. , Brouwer, M. , Fischer, J. , Wagenaar, J. A. , Gonzalez‐Zorn, B. , Guerra, B. , Mevius, D. J. , & Hordijk, J. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy, 73(5), 1121–1137. 10.1093/jac/dkx488 [DOI] [PubMed] [Google Scholar]
- Schooley, R. T. , Biswas, B. , Gill, J. J. , Hernandez‐Morales, A. , Lancaster, J. , Lessor, L. , Barr, J. J. , Reed, S. L. , Rohwer, F. , Benler, S. , Segall, A. M. , Taplitz, R. , Smith, D. M. , Kerr, K. , Kumaraswamy, M. , Nizet, V. , Lin, L. , McCauley, M. D. , Strathdee, S. A. , … Hamilton, T. (2017). Development and use of personalized bacteriophage‐based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrobial Agents and Chemotherapy, 61(10), 1–15. 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer, A. F. , Battaglia, T. , Alvarez, Y. , Bijnens, L. , Ruiz, V. E. , Ho, M. , Robinson, S. , Ward, T. , Cox, L. M. , Rogers, A. B. , Knights, D. , Sartor, R. B. , & Blaser, M. J. (2018). Intergenerational transfer of antibiotic‐perturbed microbiota enhances colitis in susceptible mice. Nature Microbiology, 3(2), 234–242. 10.1038/s41564-017-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer, A. F. , Schluter, J. , Zhang, Y. , Brown, Q. , Pathmasiri, W. , McRitchie, S. , Sumner, S. , Li, H. , Xavier, J. B. , & Blaser, M. J. (2019). The impact of early‐life sub‐therapeutic antibiotic treatment (STAT) on excessive weight is robust despite transfer of intestinal microbes. ISME Journal, 13(5), 1280–1292. 10.1038/s41396-019-0349-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, A. , Nutten, S. , Donnicola, D. , Comelli, E. M. , Mansourian, R. , Cherbut, C. , Corthesy‐Theulaz, I. , & Garcia‐Rodenas, C. (2005). Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiological Genomics, 23(2), 235–245. 10.1152/physiolgenomics.00057.2005 [DOI] [PubMed] [Google Scholar]
- Schwiertz, A. , Taras, D. , Schäfer, K. , Beijer, S. , Bos, N. A. , Donus, C. , & Hardt, P. D. (2010). Microbiota and SCFA in lean and overweight healthy subjects’. Obesity, 18(1), 190–195. 10.1038/oby.2009.167 [DOI] [PubMed] [Google Scholar]
- Scott, F. I. , Horton, D. B. , Mamtani, R. , Haynes, K. , Goldberg, D. S. , Lee, D. Y. , & Lewis, J. D. (2016). Administration of antibiotics to children before age 2 years increases risk for childhood obesity. Gastroenterology, 151(1), 120–129. 10.1053/j.gastro.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa, T. , Takeuchi, N. , Rivera, A. , Yamada, A. , Yoshimura, Y. , Barcaza, G. , Shinbori, K. , Motoyama, H. , Kohshima, S. , & Ushida, K. (2013). Distribution of antibiotic resistance genes in glacier environments. Environmental Microbiology Reports, 5(1), 127–134. 10.1111/1758-2229.12011 [DOI] [PubMed] [Google Scholar]
- Shen, L. , Shi, Y. , Zhang, D. , Wei, J. , Surette, M. G. , & Duan, K. (2008). Modulation of secreted virulence factor genes by subinhibitory concentrations of antibiotics in Pseudomonas aeruginosa. The Journal of Microbiology, 46(4), 441–447. 10.1007/s12275-008-0054-x [DOI] [PubMed] [Google Scholar]
- Silva, C. C. G. , Silva, S. P. M. , & Ribeiro, S. C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Frontiers in Microbiology, 9, 169. 10.3389/fmicb.2018.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpkin, V. L. , Renwick, M. J. , Kelly, R. , & Mossialos, E. (2017). Incentivising innovation in antibiotic drug discovery and development: Progress, challenges and next steps. Journal of Antibiotics, 70(12), 1087–1096. 10.1038/ja.2017.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund, M. , Wreiber, K. , Andersson, D. I. , Blaser, M. J. , & Engstrand, L. (2003). Long‐term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori . Annals of Internal Medicine, 139(6), 483–487+I42. 10.7326/0003-4819-139-6-200309160-00011 [DOI] [PubMed] [Google Scholar]
- Sommer, M. O. A. , Church, G. M. , & Dantas, G. (2010). The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence, 1(4), 299–303. 10.4161/viru.1.4.12010 [DOI] [PubMed] [Google Scholar]
- Sommer, M. O. A. , Dantas, G. , & Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science, 325(5944), 1128–1131. 10.1126/science.1176950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto, A. , Martín, V. , Jiménez, E. , Mader, I. , Rodríguez, J. M. , & Fernández, L. (2014). Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. Journal of Pediatric Gastroenterology and Nutrition, 59(1), 78–88. 10.1097/MPG.0000000000000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson, L. F. , Boyce, M. C. , Payne, M. S. , & Keelan, J. A. (2019). The not‐so‐sterile womb: Evidence that the human fetus is exposed to bacteria prior to birth. Frontiers in immunology, 10, 1–15. 10.3389/fmicb.2019.01124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm, J. , Schjørring, S. , Pedersen, L. , Bischoff, A. L. , Følsgaard, N. , Carson, C. G. , Chawes, B. L. K. , Bønnelykke, K. , Mølgaard, A. , Krogfelt, K. A. , & Bisgaard, H. (2013). Prevalence and predictors of antibiotic administration during pregnancy and birth. PLoS One, 8(12), e82932. 10.1371/journal.pone.0082932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm, J. , Sevelsted, A. , Bønnelykke, K. , & Bisgaard, H. (2014). Maternal propensity for infections and risk of childhood asthma: A registry‐based cohort study. The Lancet Respiratory Medicine, 2(8), 631–637. 10.1016/S2213-2600(14)70152-3 [DOI] [PubMed] [Google Scholar]
- Suez, J. , Zmora, N. , Zilberman‐Schapira, G. , Mor, U. , Dori‐Bachash, M. , Bashiardes, S. , Zur, M. , Regev‐Lehavi, D. , Ben‐Zeev Brik, R. , Federici, S. , Horn, M. , Cohen, Y. , Moor, A. E. , Zeevi, D. , Korem, T. , Kotler, E. , Harmelin, A. , Itzkovitz, S. , Maharshak, N. , … Elinav, E. (2018). Post‐antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell, 174(6), 1406–1423. 10.1016/j.cell.2018.08.047 [DOI] [PubMed] [Google Scholar]
- Sun, L. , Zhang, X. , Zhang, Y. , Zheng, K. , Xiang, Q. , Chen, N. , Chen, Z. , Zhang, N. , Zhu, J. , & He, Q. (2019). Antibiotic‐induced disruption of gut microbiota alters local metabolomes and immune responses. Frontiers in Cellular and Infection Microbiology, 9, 1–13. 10.3389/fcimb.2019.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentkuti, L. , Riedesel, H. , Enss, M. L. , Gaertner, K. , & Von Engelhardt, W. (1990). Pre‐epithelial mucus layer in the colon of conventional and germ‐free rats. The Histochemical Journal, 22(9), 491–497. 10.1007/BF01007234 [DOI] [PubMed] [Google Scholar]
- Tamim, H. M. , Hanley, J. A. , & Hajeer, A. H. (2007). Risk of breast cancer in relation to antibiotic use y. Pharmacoepidemiology and Drug Safety, 17, 144–150. 10.1002/pds.1512 [DOI] [PubMed] [Google Scholar]
- Tanaka, S. , Kobayashi, T. , Songjinda, P. , Tateyama, A. , Tsubouchi, M. , Kiyohara, C. , Shirakawa, T. , Sonomoto, K. , & Nakayama, J. (2009). Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunology and Medical Microbiology, 56(1), 80–87. 10.1111/j.1574-695X.2009.00553.x [DOI] [PubMed] [Google Scholar]
- Tapiainen, T. , Koivusaari, P. , Brinkac, L. , Lorenzi, H. A. , Salo, J. , Renko, M. , Pruikkonen, H. , Pokka, T. , Li, W. , Nelson, K. , Pirttilä, A. M. , & Tejesvi, M. V. (2019). Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Scientific Reports, 9(1), 10635. 10.1038/s41598-019-46964-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot, C. M. , Bowman, A. A. , & Young, B. (2014). Antibiotic‐induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature Communications, 5(3114), 1–16. 10.1128/mSphere.00045-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuny, F. , Richet, H. , Casalta, J.‐P. , Angelakis, E. , Habib, G. , & Raoult, D. (2010). Vancomycin treatment of infective endocarditis is linked with recently acquired obesity. PLoS ONE, 5, e90742. 10.1371/journal.pone.0009074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo‐Badia, N. , Håkansson, Å. , Vasudevan, K. , Molin, G. , Ahrné, S. , & Cilio, C. M. (2014). Antibiotic treatment of pregnant non‐obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scandinavian Journal of Immunology, 80(4), 250–260. 10.1111/sji.12205 [DOI] [PubMed] [Google Scholar]
- Trasande, L. , Blustein, J. , Liu, M. , Corwin, E. , Cox, L. M. , & Blaser, M. J. (2013). Infant antibiotic exposures and early‐life body mass. International Journal of Obesity, 37(1), 16–23. 10.1038/ijo.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh, P. J. , Ley, R. E. , Mahowald, M. A. , Magrini, V. , Mardis, E. R. , & Gordon, J. I. (2006). An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Ubeda, C. , Taur, Y. , Jenq, R. R. , Equinda, M. J. , Son, T. , Samstein, M. , Viale, A. , Socci, N. D. , van den Brink, M. R. , Kamboj, M. , & Pamer, E. G. (2010). Vancomycin‐resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of Clinical Investigation, 120(12), 4332–4341. 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava, S. , Behrendt, C. L. , Ismail, A. S. , Eckmann, L. , & Hooper, L. V. (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host‐microbial interface. Proceedings of the National Academy of Sciences of United States of America, 105(52), 20858–20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer, C. M. , Heckbert, S. R. , Lampe, J. W. , Potter, J. D. , Robertson, C. A. , & Taplin, S. H. (2004). Antibiotic use in relation to the risk of breast cancer. Journal of the American Medical Association, 291(7), 827–835. 10.1001/jama.291.7.827 [DOI] [PubMed] [Google Scholar]
- Verani, J. R. , McGee, L. , & Schrag, S. J. (2010). Morbidity and mortality weekly report prevention of perinatal group B streptococcal disease. Revised guidleines from CDC, 2010. Morbidity and Mortality Weekly Report, 59(RR‐10), 1–36. [PubMed] [Google Scholar]
- Vergnano, S. , Sharland, M. , Kazembe, P. , Mwansambo, C. , & T. H., P. (2005). Neonatal sepsis: An international perspective. Archives of Disease in Childhood, 90, 220–224. 10.1136/adc.2002.022863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic, N. , & Vidovic, S. (2020). Antimicrobial resistance and food animals: Influence of livestock environment on the emergence and dissemination of antimicrobial resistance. Antibiotics, 9(2), 52. 10.3390/antibiotics9020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova, A. , Ruggles, K. , Schulfer, A. , Gao, Z. , Ginsberg, S. D. , & Blaser, M. J. (2021). Effects of early‐life penicillin exposure on the gut microbiome and frontal cortex and amygdala gene expression. iScience, 24(7), 102797. 10.1016/j.isci.2021.102797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voreades, N. , Kozil, A. , & Weir, T. L. (2014). Diet and the development of the human intestinal microbiome. Frontiers in Microbiology, 5, 1–9. 10.3389/fmicb.2014.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, L. E. , Vallès, Y. , Agersø, Y. , Vaishampayan, P. A. , García‐Montaner, A. , Kuehl, J. V. , Christensen, H. , Barlow, M. , & Francino, M. P. (2011). The gut as reservoir of antibiotic resistance: Microbial diversity of tetracycline resistance in mother and infant. PLoS One, 6(6), e21644. 10.1371/journal.pone.0021644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze, A. , Out, C. , Fuentes, S. , Jonker, L. , Reuling, I. , Kootte, R. S. , van Nood, E. , Holleman, F. , Knaapen, M. , Romijn, J. A. , Soeters, M. R. , Blaak, E. E. , Dallinga‐Thie, G. M. , Reijnders, D. , Ackermans, M. T. , Serlie, M. J. , Knop, F. K. , Holst, J. J. , van der Ley, C. , … Nieuwdorp, M. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. Journal of Hepatology, 60(4), 824–831. 10.1016/j.jhep.2013.11.034 [DOI] [PubMed] [Google Scholar]
- Wan, J.‐J. , Lin, C. H. , Ren, E. D. , Su, Y. , & Zhu, W. Y. (2019). Effects of early intervention with maternal fecal bacteria and antibiotics on liver metabolome and transcription in neonatal pigs. Frontiers in Physiology, 10, 1–12. 10.3389/fphys.2019.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, E. M. , Neill, D. R. , Kaman, B. , Sahota, J. S. , Clokie, M. , Winstanley, C. , & Kadioglu, A. (2017). Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa . Thorax, 72(7), 666–667. 10.1136/thoraxjnl-2016-209265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, A. , Li, H. , Ma, B. , Weiss, R. , Bendayan, D. , Abramovitz, L. , Ben‐Shalom, N. , Mor, M. , Pinko, E. , Bar Oz, M. , Wang, Z. , Du, F. , Lu, Y. , Rybniker, J. , Dahan, R. , Huang, H. , Barkan, D. , Xiang, Y. , Javid, B. , & Freund, N. T. (2021). Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis. Nature Communications, 12, 602. 10.1038/s41467-021-20930-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden, A. R. , Chen, C. , Bobr, A. , Yao, D. , Lu, Y. , Nelson, V. M. , Sadowsky, M. J. , & Khoruts, A. (2014). Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. American Journal of Physiology ‐ Gastrointestinal and Liver Physiology, 306(4), 310–319. 10.1152/ajpgi.00282.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, C. B. , & Le, J. K. (2021). Metronidazole, StatPearls [Internet]. StatPearls Publishing. [PubMed] [Google Scholar]
- Wilcox, M. H. , Gerding, D. N. , Poxton, I. R. , Kelly, C. , Nathan, R. , Birch, T. , Cornely, O. A. , Rahav, G. , Bouza, E. , Lee, C. , Jenkin, G. , Jensen, W. , Kim, Y. S. , Yoshida, J. , Gabryelski, L. , Pedley, A. , Eves, K. , Tipping, R. , Guris, D. , … MODIFY I and MODIFY II Investigators . (2017). Bezlotoxumab for prevention of recurrent Clostridium difficile infection. New England Journal of Medicine, 376, 305–317. [DOI] [PubMed] [Google Scholar]
- Willing, B. P. , Russell, S. L. , & Finlay, B. B. (2011). Shifting the balance: Antibiotic effects on host‐microbiota mutualism. Nature Reviews Microbiology, 9(4), 233–243. 10.1038/nrmicro2536 [DOI] [PubMed] [Google Scholar]
- Wlodarska, M. , Willing, B. , Keeney, K. M. , Menendez, A. , Bergstrom, K. S. , Gill, N. , Russell, S. L. , Vallance, B. A. , & Finlay, B. B. (2011). Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium‐induced colitis. Infection and Immunity, 79(4), 1536–1545. 10.1128/IAI.01104-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, W. S. W. , Sabu, P. , Deopujari, V. , Levy, S. , Shah, A. A. , Clemency, N. , Provenzano, M. , Saadoon, R. , Munagala, A. , Baker, R. , Baveja, R. , Mueller, N. T. , Dominguez‐Bello, M. G. , Huddleston, K. , Niederhuber, J. E. , & Hourigan, S. K. (2020). Prenatal and peripartum exposure to antibiotics and cesarean section delivery are associated with differences in diversity and composition of the infant meconium microbiome. Microorganisms, 8(2), 179. 10.3390/microorganisms8020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongng (2000). The risk of hemolytic‐uremic syndrome after antibiotic treatment of Escherichia coli O157:h7 infections. New England Journal of Medicine, 342(26), 1930–1936. 10.1056/NEJM200006293422601.THE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L. , He, J. , Gao, N. , Lu, X. , Li, M. , Wu, X. , Liu, Z. , Jin, Y. , Liu, J. , Xu, J. , & Geng, Y. (2017). Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Scientific Reports, 7, 1–13. 10.1038/srep45176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto‐Hanada, K. , Yang, L. , Narita, M. , Saito, H. , & Ohya, Y. (2017). Influence of antibiotic use in early childhood on asthma and allergic diseases at age 5. Annals of Allergy, Asthma and Immunology, 119(1), 54–58. 10.1016/j.anai.2017.05.013 [DOI] [PubMed] [Google Scholar]
- Yang, J. H. , Bhargava, P. , McCloskey, D. , Mao, N. , Palsson, B. O. , & Collins, J. J. (2017). Antibiotic‐induced changes to the host metabolic environment inhibit drug efficacy and alter immune function. Cell Host and Microbe, 22(6), 757–765. 10.1016/j.chom.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour, M. , Vatanen, T. , Siljander, H. , Hämäläinen, A. M. , Härkönen, T. , Ryhänen, S. J. , Franzosa, E. A. , Vlamakis, H. , Huttenhower, C. , Gevers, D. , Lander, E. S. , Knip, M. , DIABIMMUNE Study Group , & Xavier, R. J. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatments on strain‐level diversity and stability. Science Translational Medicine, 8(343), 343ra81. 10.1126/scitranslmed.aad0917.Natural [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar, A. , Chaix, A. , Xu, Z. Z. , Chang, M. W. , Marotz, C. A. , Saghatelian, A. , Knight, R. , & Panda, S. (2018). Antibiotic‐induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nature Communications, 9(1), 1469. 10.1038/s41467-018-05336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Kinkelaar, D. , Huang, Y. , Li, Y. , Li, X. , & Wang, H. H. (2011). Acquired antibiotic resistance: Are we born with it? Applied and Environmental Microbiology, 77(20), 7134–7141. 10.1128/AEM.05087-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , & Chen, D. C. (2019). Facing a new challenge: The adverse effects of antibiotics on gut microbiota and host immunity. Chinese Medical Journal, 132(10), 1135–1138. 10.1097/CM9.0000000000000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Shen, D. , Fang, Z. , Jie, Z. , Qiu, X. , Zhang, C. , Chen, Y. , & Ji, L. (2013). Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One, 8(8), e71108. 10.1371/journal.pone.0071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wu, J. , Li, J. V. , Zhou, N. Y. , Tang, H. , & Wang, Y. (2013). Gut microbiota composition modifies fecal metabolic profiles in mice. Journal of Proteome Research, 12(6), 2987–2999. 10.1021/pr400263n [DOI] [PubMed] [Google Scholar]
- Zhou, P. , Zhou, Y. , Liu, B. , Jin, Z. , Zhuang, X. , Dai, W. , Yang, Z. , Feng, X. , Zhou, Q. , Liu, Y. , Xu, X. , & Zhang, L. (2020). Perinatal antibiotic exposure affects the transmission between maternal and neonatal microbiota and is associated with early‐onset sepsis. mSphere, 5(1), e00984–19. 10.1128/mSphere.00984-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatkovic, N. , Aleksić, G. , & Gašić, K. (2005). ‘Resilience of the dominant human fecal microbiota upon short‐course antibiotic challenge’. Journal of Clinical Microbiology, 43(11), 5588–5592. 10.1128/JCM.43.11.5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Z. H. , Liu, D. , Li, H. D. , Zhu, D. P. , He, Y. , Hou, T. , & Yu, J. L. (2018). Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Annals of Clinical Microbiology and Antimicrobials, 17(1), 1–11. 10.1186/s12941-018-0264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski, D. V. , & McLendon, M. K. (2020). Monoclonal antibodies as an antibacterial approach against bacterial pathogens. Antibiotics, 9(4), 155. 10.3390/antibiotics9040155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwittink, R. D. , Renes, I. B. , van Lingen, R. A. , van Zoeren‐Grobben, D. , Konstanti, P. , Norbruis, O. F. , Martin, R. , Groot Jebbink, L. , Knol, J. , & Belzer, C. (2018). Association between duration of intravenous antibiotic administration and early‐life microbiota development in late‐preterm infants. European Journal of Clinical Microbiology & Infectious Diseases, 37, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


