Abstract
Organisms acquire and use information from their environment to guide their behaviour. However, it is unclear whether this information quantitatively limits their behavioural performance. Here, we relate information to the ability of Escherichia coli to navigate up chemical gradients, the behaviour known as chemotaxis. First, we derive a theoretical limit on the speed with which cells climb gradients, given the rate at which they acquire information. Next, we measure cells’ gradient-climbing speeds and the rate of information acquisition by their chemotaxis signaling pathway. We find that E. coli make behavioural decisions with much less than the one bit required to determine whether they are swimming up-gradient. Some of this information is irrelevant to gradient climbing, and some is lost in communication to behaviour. Despite these limitations, E. coli climb gradients at speeds within a factor of two of the theoretical bound. Thus, information can limit the performance of an organism, and sensory-motor pathways may have evolved to efficiently use information acquired from the environment.
Organisms’ survival depends on their ability to perform behavioral tasks. These tasks require that the organism measure signals in its environment and respond appropriately. Information theory is a natural language for quantifying the fidelity of measurements and responses, but it is unclear how an abstract quantity like information might limit an organism’s performance at real-world tasks. Past studies have used information theory to understand the maximum amount of information biological systems can acquire and transmit about environmental signals1–6 and have shown that they can approach biophysical limits6–9. But high information transfer is not sufficient for high performance because not all of the information contained in the signal is relevant (i.e. contributes to performing the task), and not all of it is appropriately acted on10. What limits does information place on performance, and how efficiently do organisms use the information they acquire relative to these limits?
We address these questions using one of the best-understood behaviors in biology: bacterial chemotaxis. The bacterium Escherichia coli alternates between “runs,” which propel the cell forward, and “tumbles,” which randomly reorient its swimming direction11 (Fig. 1). E. coli continuously sense the concentration c(t) of chemoattractant they encounter using transmembrane receptors. Relative changes in concentration12 , which we define to be the “signal”, induce changes in activity a(t) of receptor-associated CheA kinases. Thus, CheA kinase activity encodes E. coli’s estimate of the signal. CheA activity then modulates transitions in the cell’s swimming behavior m(t) between run and tumble states via a signal transduction pathway13,14. If the attractant concentration is increasing (s(t) > 0), the cell runs for longer on average, thereby biasing its random motion up the gradient11. However, noise in sensing and signal transduction corrupt the signal15–18. Since the goal of chemotaxis is to climb chemical gradients, we quantify performance as the cell’s drift velocity vd up a static gradient. E. coli chemotaxis has been studied extensively, but how much information a cell acquires about chemical signals19–22 and its relationship to chemotactic performance are unknown.
Figure 1. Information sets an upper limit on chemotaxis performance.
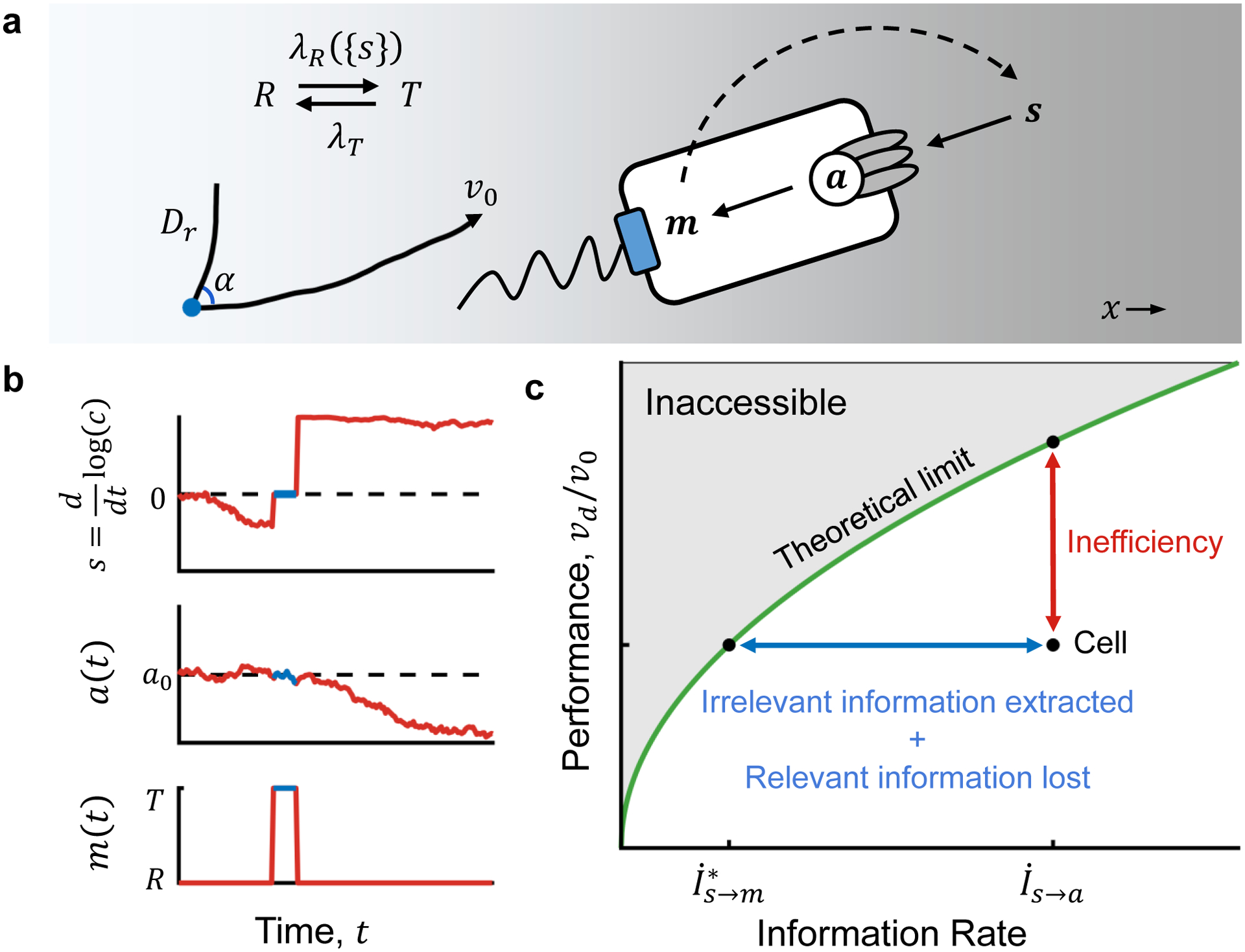
A) In our model, a cell navigates chemical gradients by sensing relative changes in attractant concentration c over time using receptors, which influence the activity a of receptor-associated kinases. a encodes E. coli’s estimate of the signal, i.e. whether it’s going up or down the gradient. a then modulates stochastic transitions between behavioral states m: a run state R, in which the cell swims with constant speed v0 and rotational diffusion Dr; and a tumble state T, in which it reorients randomly with directional persistence a (see Supplementary Fig. S1). Responses to past signals {s} that are transduced from a to m are described by changes in the cell’s transition rate λR({s}) from run to tumble states. The cell’s motion creates the signal that it experiences (dashed arrow). B) Signals the cell experience s(t) (top) are encoded in noisy kinase activity a(t) (middle), which influences stochastic behavioral state transitions m(t) (bottom). The flow of information from s(t) to a(t) to m(t) implies that . C) Given an information rate , there is an upper limit on chemotaxis performance (green line), defined as the cell’s up-gradient drift speed vd relative to v0 (Eqn. 1). The cell’s performance is ultimately set by how much relevant information contained in is communicated to its behavior, quantified by . Thus, real cells cannot reach the limit because of two factors (quantified by the length of the blue line): some of the information they encode in CheA kinase activity is irrelevant for climbing the gradient, and some of the relevant information in a is lost during transfer to the motors. These factors result in reduced performance (quantified by the length of the red line) and reduced efficiency of information usage.
To use information efficiently, a cell must only encode information in CheA that is relevant to gradient-climbing and act on that information appropriately at the motors to guide behavior. An inefficient cell may acquire abundant information about the signal, but it is either irrelevant to chemotaxis or goes unused. To determine how efficiently E. coli use information to navigate, we first must derive the maximum gradient-climbing speed a cell can achieve given the amount of information available in CheA. Second, we need to quantify how close to this theoretical bound E. coli cells operate by measuring both the amount of information available in CheA and cells’ performance with that information (Fig. 1C). In doing so, we will quantify information transfer on two different length scales: the information about signals available in kinase activity, and the information used to perform behaviors.
To do this, we must define a measure of information acquisition. Bacteria continuously encode chemical signals they experience. Thus, unlike most studies in biological systems1–3,5,6, we cannot use the one-shot mutual information23 between signal s(t) and kinase activity a(t) as a measure of information transfer because it is blind to past signals the cell has experienced. Instead, we need to quantify information transfer between signal and kinase trajectories, in this case using a steady state information rate24. Furthermore, signals are generated by the cell’s own motion in the gradient25. This makes a natural extension, the mutual information rate ṀI(s; a) between s(t) and a(t), also unsuitable because kinase activity and signal are correlated even for cells that don’t respond to the signal (Fig. 1AB). We address these challenges by decomposing the mutual information rate into the sum of two directed information terms (SI Section 1; see also26,27): the transfer entropy21,28 rates from kinase activity to signal and from signal to kinase activity , or . The first term quantifies the feedback of kinase activity onto signal via the behavior. The second term measures the influence of the signal on kinase activity, in bits/s, and must be nonzero for the cell to climb the gradient.
Next, we need to derive the theoretical limit on gradient-climbing performance vd imposed by information . The drift speed depends on how the cell’s motors use information about past signals that is provided by CheA. While information at the motors cannot exceed the information in kinase activity 29, at best, all information in kinase activity is preserved at the motors, in which case . Therefore, to derive the bound, we sought to determine how , and thus , bounds performance vd.
To do this, we constructed a mathematical model of run-and-tumble navigation (Fig. 1A; SI Section 2). During runs, cells swim with a constant speed v0 and lose direction with rotational diffusion coefficient Dr. During tumbles, they randomly reorient with directional persistence α. In the absence of a gradient, cells switch from run to tumble and vice versa with rates λR0 and λT. The fraction of time the cell spends running is . In shallow gradients, like those we consider here, the tumble rate λR({s}) depends approximately linearly on the history of signals experienced {s}30–33.
Using the model above, we derived how vd and depend on the behavioral response λR({s}). While any response to signal implies information transfer, it does not imply high drift speed. To find the limit, we derived the maximum drift speed vd possible given an information rate by optimizing over responses λR({s}). This revealed that the optimal behavioral response only depends on the current rate of change of log-concentration, s(t) (SI Section 4). Thus, information about the current signal is relevant to gradient-climbing (SI Section 5). Bacteria can’t implement the optimal behavioral response because they must make comparisons of concentrations over a finite time to infer s(t)15. Nevertheless, the performance achieved by any behavioral response is bounded by (Fig. 1C):
| (1) |
where we have used that , and . This expression makes rigorous the intuition that information transfer sets a limit on chemotaxis performance15,20,22,24,34,35. Of the information contained in , the amount that is both relevant and transferred to behavior, , ultimately determines performance: (SI Section 5). Thus, the green line in Fig. 1C directly maps a cell’s performance to . Furthermore, the length of the blue line in Fig. 1C measures the amount of irrelevant information encoded in CheA plus the amount of relevant information lost in communication to the motors. These factors lower the cell’s performance relative to the theoretical limit by an amount quantified by the length of the red line. With this, we define the efficiency of information usage as the cell’s actual drift speed relative to the maximum possible with the information available in CheA, .
We next set out to measure how efficiently E. coli use information to navigate, η. First, we quantified the bound by measuring the rotational diffusion coefficient Dr and behavioral parameters θ from trajectories of swimming E. coli (Supplementary Figs. S1, S2). Individual cells in a clonal population exhibit nongenetic differences in behavioral parameters36–39, which in E. coli are highly correlated with Prun36,38. From this data, we find f(θ) = 0.531 ± 0.005 for the median Prun (± one standard error throughout; Supplementary Table S1). This quantifies the minimum amount of information needed to climb a gradient. Surprisingly, the bound predicts that run-and-tumble navigation is theoretically possible with very small information rates: a hundredth of a bit per second is sufficient to climb gradients at ~6% of the run speed. This is far less than the 1 bit per run (~1 bit/s) required to distinguish whether concentration is currently increasing or decreasing before every tumble decision24.
To compare E. coli to the theoretical limit, we measured the rate at which cells encode information about signals in their CheA kinase activity, , during chemotaxis. Directly measuring kinase activity in individual swimming cells is infeasible. Instead, we quantified the mutual information rate24,40 between signal and kinase activity, ṀI(s; a), in immobilized cells by measuring their kinase response and noise properties and by separately measuring the signal statistics they would experience during navigation (Fig. 2A). In these immobilized cells, since there is no feedback of kinase activity onto the signal. quantified this way in immobilized cells differs from in a swimming cell because it does not take into account higher-order correlations between s and a. However, these correlations contribute terms that are multiplied by higher powers of the gradient steepness g = d log(c)/dx, and thus can be neglected in shallow gradients (SI Section 8).
Figure 2. Measuring the rate of information transfer from signal to intracellular kinase.
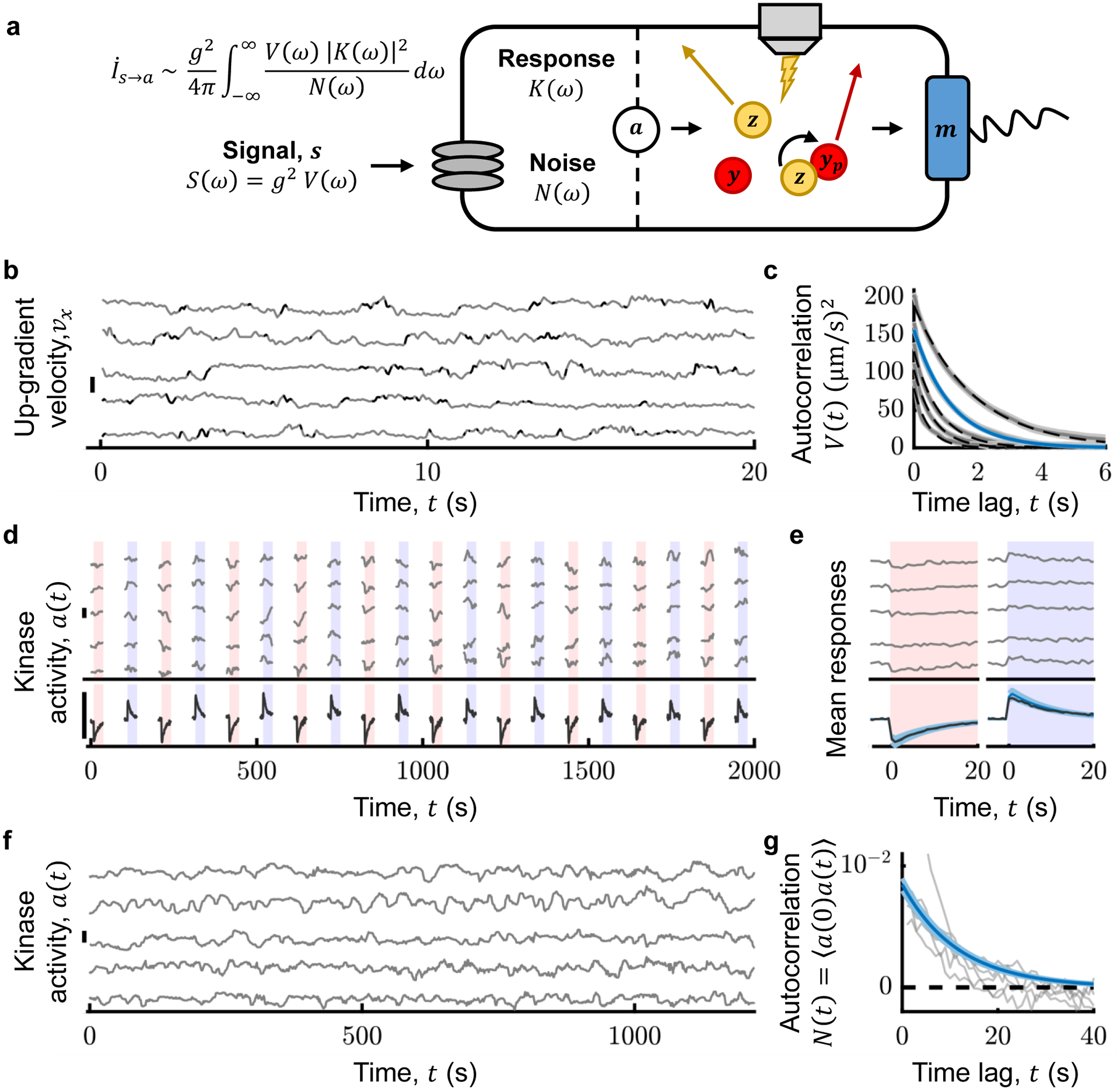
A) Information rate from signal s to kinase activity a depends on the signal power spectrum S(ω), the kinase frequency response K(ω), and the kinase noise power spectrum N(ω). The signal is s(t) = g vx(t), where g is the gradient steepness and vx is the cell’s up-gradient velocity. Thus, S(ω) = g2 V(ω), where V(ω) is the power spectrum of vx. Kinase activity a was quantified from the FRET between the substrate of the kinase, CheY-mRFP, and its phosphatase, CheZ-mYFP (Methods; SI Sections 10–15; Supplementary Figs. S3–S6). All experiments were performed in a background of 100 μM MeAsp. B) (Gray) Individual cells’ vx(t). (Black: tumbles; scale bar 40 μm/s.) C) (Gray) Average autocorrelation of vx, V(t), for Prun = 0.93, 0.89, 0.84, 0.79, 0.74 (top to bottom; throughout, shading is ± one standard error; black dashed lines are fits to V(t) = av exp(−λtot |t|)). (Blue) Best fit to the median bin, Prun~0.89 (Supplementary Fig. S2; SI Section 21). D) Immobilized cells were delivered 10 μM steps up (red shading) and down (blue shading). (Top, gray) a(t) for five cells (here and in (F), smoothed with 10th order median filter, and scale bars represent Δa = 0.3; Methods; SI Section 16; Supplementary Fig. S5). (Bottom, black) Population average a(t) (n = 442 cells). E) Single-cell average (top, gray) and population-average (bottom, black) response functions K(t) to positive and negative stimuli. (Blue line) K(t) = G exp(−t/τ2)(1 − exp(−t/τ1)) H(t), where H(t) is the Heaviside step function and G, τ1, and τ2 are the median parameters extracted from fits to single-cell responses. F,G) (Gray) Kinase activity (F) and corresponding autocorrelations (G) in single cells (Methods; SI Section 17; Supplementary Fig. S6). (Blue, G) , where σn and τn are the median parameters extracted from fits to single-cell traces (n = 262 cells). V(ω), K(ω), and N(ω) shown in Supplementary Fig. S7.
The signal statistics are characterized by their power spectrum S(ω). It is often difficult to know the natural signal statistics an organism experiences7,41. But during bacterial chemotaxis in static gradients, the signal is generated from the cell’s own motion. Thus, the signal power spectrum is S(ω) = g2 V(ω), where V(ω) is the power spectrum of the cell’s up-gradient velocity vx(t). Furthermore, in shallow gradients, the statistics of vx(t) are nearly identical to those in the absence of a gradient (SI Sections 8,9). To quantify the autocorrelation function of vx, V(t), and thus V(ω), we tracked individual swimming cells in a 100 μM background of the attractant α-methyl-aspartate (MeAsp) (Fig. 2BC; Supplementary Fig. S2; Methods; SI Sections 18–21). For each value of Prun, we fit the measured V(t) with a decaying exponential, , giving av = 157.1 ± 0.5 (μm/s)2 and λtot = 0.862 ± 0.005 s−1 for the median phenotype (~104 s total trajectory time, 7s average duration; Supplementary Table S1).
Next, we measured the response and noise properties of CheA kinase activity using Förster resonance energy transfer (FRET) between the kinase’s substrate CheY and the phosphatase CheZ inside single cells17,18,42 (Fig. 2A). In the linear regime, the kinase response is characterized by its frequency-response function, K(ω), or its response K(t) to an impulse of signal . To measure K(t), we used a microfluidic device that can rapidly switch (in ~100 ms) the concentration of attractant delivered to hundreds of immobilized cells42 (Supplementary Figs. S3–S4). To ensure cells were in the log-sensing regime12,43, we first adapted them to a background of 100 μM MeAsp. We then delivered 10 positive and 10 negative 10% step changes of MeAsp concentration (impulses of s) (Fig. 2D; Methods), which were small enough to be in the linear-response regime44,45 (Supplementary Fig. S5). Cell responses exhibited a stereotypical shape (Fig. 2E) described by a phenomenological model , with gain G, rise time τ1, and adaptation time τ2, and H(t) is the Heaviside step function. We fit this model to each cell’s responses to the positive and negative stimuli simultaneously (SI Section 16) and then determined the population-median parameter values (n = 442 cells) (Supplementary Table S1; Supplementary Fig. S5): G = 1.73 ± 0.03, τ1 = 0.22 ± 0.01 s, and τ2 = 9.9 ± 0.3 s. This value of τ1 includes the stimulus switching time and CheY/CheZ binding kinetics, making it longer than the kinase response time, which was previously measured to be τ1 ~ 1/60 s46,47. After verifying that our results are insensitive to the value of τ1 (SI Section 9; Supplementary Fig. S8), we used the literature value in our estimate of .
We quantified the statistics of noise in kinase activity by measuring FRET in single cells in a constant background of 100 μM MeAsp (Fig. 2F; Supplementary Fig. S6). These fluctuations were well-approximated by an Ornstein-Uhlenbeck process, consistent with previous measurements16,18. Using Bayesian filtering (SI Section 17), we inferred the single-cell parameters of the noise model directly from the time series. These parameters determined the noise autocorrelation function (Fig. 2G; SI Section 9) and power spectrum N(ω) (Supplementary Fig. S7). The population-median parameter values (n = 262 cells) were σn = 0.092 ± 0.002 AU (standard deviation of the noise) and τn = 11.75 ± 0.04 s (noise correlation time) (Supplementary Table S1). These measurements include the effects of all noise sources upstream of CheA, including stochastic ligand arrivals at the cells’ receptors15.
With the signal statistics, response function, and noise, we then computed the information rate from the signal to kinase activity (Fig. 3A). Since the signal power is proportional to g2, the information rate is, as well: . Using our measurements above, we estimated that the E. coli chemotaxis system transfers information to the kinases at a rate β = 0.22 ± 0.03 bits/s/mm−2 (SI Sections 8,9). Thus, in shallow gradients, where concentration varies on millimeter to centimeter length scales, cells only get ~10−2 bits/s. The bound in Eqn. 1 predicts that this is sufficient for a run-and-tumble navigator to climb gradients at a few percent of its swimming speed. However, it is unclear how much of this information is relevant to chemotaxis, communicated to the motors, and used to navigate.
Figure 3. E. coli use information efficiently to navigate.
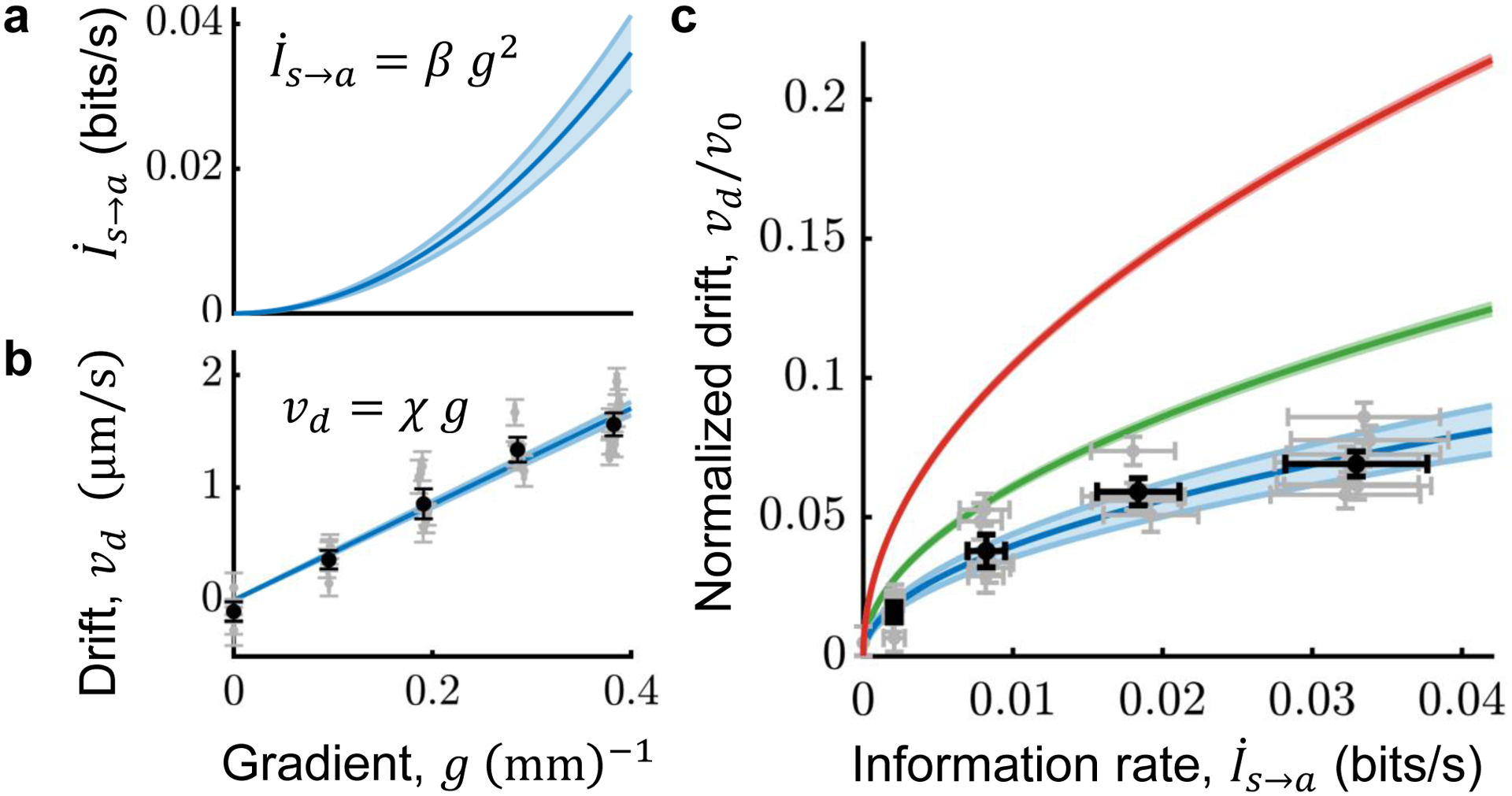
A) The rate of information transfer from signal to kinase activity depends on gradient steepness ; Fig. 2; SI Section 8). Throughout, shading and error bars indicate ± one standard error. B) Chemotactic performance as a function of gradient steepness g, in a background of 100 μM MeAsp. Gray dots: average drift speeds in individual experiments. Black dots: averages over experiments. Error bars on g are smaller than the markers. Population average drift speed increases linearly with gradient steepness, vd = χ g (χ = 4300 ± 150 μm2/s; blue line and shading; SI Section 22). C) From measurements of E. coli cells’ information rates and chemotactic drift speeds, we compared their performance to the theoretical bound (Eqn. 1). Green: predicted maximum performance given information acquisition rate (Eqn. 1), with measured behavioral parameters θ = {λR0, Prun, α} (f(θ) = 0.531 ± 0.005). Blue: measured performance vd/v0 (v0 = 22.61 ± 0.07 μm/s) versus information rate , obtained by eliminating g from the fits of vd(g) = χ g and to plot . Black and gray dots are data points from (B) plotted against . Taking the ratio of the blue and green curves, we find that E. coli achieve drift speeds within a factor η = 0.65 ± 0.05 of the theoretical limit. Red: Theoretical bound (Eqn. 1) if λR0 is optimized, and all other behavioral parameters are held fixed at their measured values (f(θ) = 0.914 ± 0.006, just below the maximum value of 1).
To answer this, we measured E. coli’s average drift speeds by tracking individual cells’ motion in gradients of varying steepness. Static, linear MeAsp gradients were constructed (Methods) in a 100 μM background, with length scales ranging from 10 mm (g = 0.1 mm−1) to 2.5 mm (g = 0.4 mm−1). From >105 seconds of trajectories in each condition, we estimated the drift speed vd as the time-averaged up-gradient velocity over all cells in each experiment (SI Section 22). As expected from theory in shallow gradients, drift speed increased linearly with gradient steepness vd = χ g, with “chemotactic coefficient” χ ~ 4.30 ± 0.15 μm/s/mm−1 (Fig. 3B; Supplementary Fig. S9), consistent with previous measurements48.
With measurements of both their information acquisition rate and performance, we were in a position to quantify how efficiently E. coli use information, η. For each gradient g, we plotted the drift speed vd(g) against the information rate (blue in Fig. 3C). On the same plot, we show the maximum drift speeds, given by the bound in Eqn. 1 (green in Fig. 3C). The ratio of these two curves is the efficiency , which is independent of g in shallow gradients. We find that E. coli achieve an efficiency of η = 0.65 ± 0.05—that is, they climb gradients at ~65% of the maximum possible speed given the rate at which their kinases encode information about environmental signals. Equivalently, comparing information rates (blue and green lines) at the same drift speed in Fig. 3C indicates that ~ 42% of the total information available in kinase activity is relevant, preserved in communication with the motors, and used to navigate.
Many studies of information theory in biology have focused on the maximum amount of information signaling pathways can transmit1–6,49–51. Here, we instead asked how the information an organism transmits limits its performance at functional tasks, using E. coli chemotaxis as a model system. Whereas previous works measured one-shot information transfer by biochemical networks1–6,49–51, in bits, we measured the rate at which E. coli transfer information, in bits/s, between time-varying inputs and outputs, with natural input statistics. Combining this with measurements of E. coli’s chemotactic performance and the theoretical limit (Eqn. 1), we found that about half of the information about concentration changes that flows through CheA is both relevant to and used for navigation.
Achieving high efficiency requires that cells acquire, transmit, and act on information that is relevant to their task10, but which bits are relevant is often not clear. Using rate-distortion theory6,52,53, we found that the relevant bits for bacterial chemotaxis are those that indicate how fast the attractant concentration is currently changing (SI Section 5). Responding to signals experienced multiple signal correlation times, , in the past transmits a significant amount of irrelevant information (SI Section 5). But in typical gradients, which are much longer than their body lengths, E. coli must swim and time integrate to infer signals from stochastic ligand arrivals15,54. This forces E. coli to respond to past signals to learn about the current signal, thus transmitting irrelevant information and preventing them from reaching the bound in Eqn. 1. Between the green and blue lines in Fig. 3C, there is a tighter bound on performance that accounts for the need to time integrate, relative to which E. coli are even more efficient.
The information-performance bound in Eqn. 1 depends on the cell’s swimming behavior. We quantified this bound for a typical cell from a laboratory strain, but behavioral parameters θ vary across individual cells16,36–38,55,39, strains, and growth conditions56. This raises the question of what behavioral phenotype can achieve the highest performance with a given information rate. Maximizing f(θ) with respect to θ (SI Section 6), we find that the optimal agent changes direction by tumbling at the same rate as rotational diffusion33,57,58, (1 − α) λR0 = 2 Dr. Our median phenotype tumbles more frequently than this and misses out two-fold. First, the bound on its performance (green in Fig. 3C) is about half as high as it could be (red in Fig. 3C). Second, a lower average tumble rate λR0 would lengthen the signal correlation time τv, increasing the fraction of relevant information the cell would transfer, and thus increasing η relative to the higher bound. Laboratory strains of E. coli were historically selected for motility in semisolid agar59–61, and our strain’s frequent reorientations may be optimized to navigate that environment62–64. Alternatively, their behavioral parameters may optimize other objectives31,33. While E. coli’s behavioral strategy thus appears sub-optimal for gradient-climbing in liquid, they nevertheless use biochemical information in CheA efficiently to carry it out.
A hallmark of E. coli’s chemotactic response to many ligands—including MeAsp used here—is precise adaptation65: following a stimulus, the kinase activity responds transiently before relaxing back to the pre-stimulus activity (Fig. 2E). However, for some attractants, such as serine, adaptation is imperfect, causing the average tumble rate to decrease with increasing concentration of attractant in the background30,37,57,66–68. By lowering λR0 closer to the optimal value, a background of serine both increases the bound and causes cells to climb gradients of MeAsp faster67 (SI Section 7). However, from an information-usage perspective, if the cell could choose λR0 and its response function independently, our theory shows that it should match its average tumble rate to rotational diffusion, (1 − a) λR0 = 2 Dr, and its response function should adapt perfectly. Imperfect adaptation implies that the cell is responding to concentration changes that occurred far in the past, which transfers irrelevant information.
Because we measured signal statistics and kinase activity in different cells, our estimate of the information rate does not account for the long tail in the run-length distribution that can arise from fluctuations in kinase activity16,69–71. In theory, these fluctuations can improve performance in shallow gradients72,73. However, they only contribute corrections to the information rate that are higher order in g and can be neglected in shallow gradients (SI Section 8).
This work relied on linear theory, which is only valid in shallow gradients. In steep gradients, behavioral feedback can drive large deviations in the tumble rate, leading to ratchet-like gradient climbing25. In this regime, the signal statistics gain multiple time scales due to the very different run durations up and down the gradient. In future studies, it will be interesting to examine how these factors, combined with nonlinearities in the signaling pathway, alter cells’ information-acquisition rates and efficiencies.
Here, we measured information transfer at two very different length scales. The first is the relevant information contained in behavior, which is quantified by the drift speed and behavioral parameters. The second is the total information available in CheA kinase activity. Considering how different these measurements are, there is no reason to expect that they must be similar in magnitude. But their agreement to about a factor of two is a strong indication that information is a limitation on chemotaxis performance. Information transfer is not necessarily the end-goal of biological tasks, but it is needed to perform many of them. Our results suggest organisms may be under selective pressure to efficiently use information from environmental cues to perform tasks necessary for their survival.
Methods
Strains and plasmids
The strain used for the FRET experiments is a derivative of E. coli K-12 strain RP437 (HCB33), a gift of T. Shimizu, and described in detail elsewhere18,42. In brief, the FRET acceptor-donor pair (CheY-mRFP and CheZ-mYFP) is expressed in tandem from plasmid pSJAB10618 under an isopropyl β-D-thiogalactopyranoside (IPTG)-inducible promoter. The glass-adhesive mutant of FliC (FliC*) was expressed from a sodium salicylate (NaSal)-inducible pZR1 plasmid18. The plasmids are transformed in VS115, a cheY cheZ fliC mutant of RP43718 (gift of V. Sourjik). The crosstalk coefficient for spectral bleedthrough was measured using a strain expressing CheZ-YFP from a plasmid, and that for cross-excitation was measured using a strain expressing CheY-mRFP from a plasmid, both of which are gifts from T. Shimizu. RP437, the direct parent of the FRET strain and also a gift from T. Shimizu, was used to measure behavioral parameters and chemotactic drift speeds. A mutant that can’t tumble due to an in-frame deletion of the cheY gene, VS100 (gift of V. Sourjik), was used to measure the rotational diffusion coefficient Dr. All strains are available from the authors upon request.
Cell preparation
Single-cell FRET microscopy and cell culture was carried out essentially as described previously18,42. In brief, cells were picked from a frozen stock at −80°C and inoculated in 2 mL of Tryptone Broth (TB; 1% bacto tryptone, 0.5 % NaCl) and grown overnight to saturation at 30°C and shaken at 250 RPM. Cells from a saturated overnight culture were diluted 100X in 10 mL TB and grown to OD600 0.45–0.47 in the presence of 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, 50 μM IPTG and 3 μM NaSal, at 33.5°C and 250 RPM shaking. Cells were collected by centrifugation (5 min at 5000 rpm, or 4080 RCF) and washed twice with motility buffer (10 mM KPO4, 0.1 mM EDTA, 1 μM methionine, 10 mM lactic acid, pH 7), and then were resuspended in 2 mL motility buffer. Cells were left at 22°C for 90 minutes before loading into the microfluidic device. All experiments, FRET and swimming, were performed at 22–23°C.
For swimming and chemotaxis experiments, cells were prepared identically. Saturated overnight cultures were diluted 100X in 5 mL of TB. After growing to OD600 0.45–0.47, 1 mL of cell suspension was washed twice in motility buffer with 0.05% w/v of polyvinylpyrrolidone (MW 40 kDa) (PVP-40) added. Washes were done by centrifuging the suspension in an Eppendorf tube at 1700 RCF (4000 RPM in this centrifuge) for 3 minutes. After the last wash, cells were resuspended with varying concentrations of MeAsp (see below).
Microfluidic device fabrication and loading for FRET measurements
Microfluidic devices for the FRET experiments42 were constructed from polydimethylsiloxane (PDMS) on 24 × 60 mm cover glasses (#1.5) following standard soft lithography protocols74. Briefly, the master molds for the device were created with a negative SU-8 photoresist on 100-mm silicon wafers. Approximately 16-μm-high master molds were created. To fabricate the device, the master molds were coated with a 5-mm-thick layer of degassed 10:1 PDMS:curing agent mixture (Sylgard 184, Dow Corning). The PDMS layer was cured at 80 °C for 1 hour, and then cut and separated from the wafer, and holes were punched for the inlets and outlet. The punched PDMS layer was further cured at 80 °C for > 2 hours. Then, the PDMS was cleaned with transparent adhesive tape (Magic Tape; Scotch) followed by rinsing with (in order) isopropanol, methanol, and Millipore-filtered water. The glass was rinsed with (in order) acetone, isopropanol, methanol, and Millipore-filtered water. The PDMS device was tape-cleaned an additional time before the surfaces of the device and coverslip were treated in a plasma bonding oven (Harrick Plasma). After 1 min of exposure to plasma under vacuum, the device was laminated to the coverslip and then baked at 80°C hotplate for > 30 min to establish a covalent bond.
Sample preparation in the microfluidic device was conducted as follows. Five inlets of the device (Supplementary Fig. S3) were connected to reservoirs (Liquid chromatography columns, C3669; Sigma Aldrich) filled with motility buffer containing various concentrations of α-methyl-aspartate (MeAsp) through polyethylene tubing (Polythene Tubing, 0.58 mm id, 0.96 mm od; BD Intermedic). The tubing was connected to the PMDS device through stainless steel pins that were directly plugged into the inlets or outlet of the device (New England Tubing). Cells washed and suspended in motility buffer were loaded into the device from the outlet and allowed to attached to the cover glass surface via their sticky flagella by reducing the flow speed inside the chamber. The pressure applied to the inlet solution reservoirs was controlled by computer-controlled solenoid valves (MH1; Festo), which rapidly switched between atmospheric pressure and higher pressure (1.0 kPa) using a source of pressurized air. Only one experiment was conducted per device.
Single-cell FRET imaging system
FRET imaging in the microfluidic device was performed using an inverted microscope (Eclipse Ti-E; Nikon) equipped with an oil-immersion objective lens (CFI Apo TIRF 60X Oil; Nikon). YFP was illuminated by an LED illumination system (SOLA SE, Lumencor) through an excitation bandpass filter (FF01–500/24–25; Semrock) and a dichroic mirror (F01–542/27–25F; Semrock). The fluorescence emission was led into an emission image splitter (OptoSplit II; Cairn) and further split into donor and acceptor channels by a second dichroic mirror (FF580-FDi01–25×36; Semrock). The emission was then collected through emission bandpass filters (FF520-Di02–25×36 and FF593-Di03–25×36; Semrock) by a sCMOS camera (ORCA-Flash4.0 V2; Hamamatsu). RFP was illuminated in the same way as YFP except that an excitation bandpass filter (FF01–575/05–25; Semrock) and a dichroic mirror (FF593-Di03–25×36; Semorock) were used. An additional excitation filter (59026x; Chroma) was used in front of the excitation filters. To synchronize image acquisition and the delivery of stimulus solutions, a custom-made MATLAB program controlled both the imaging system (through the API provided by Micro-Manager75) and the states of the solenoid valves.
Procedure for measuring the linear response functions
All experiments were performed in a background MeAsp concentration of c0 = 100 μM. Measurements were made in single cells. First, the FRET level at minimum kinase activity was measured by delivering a saturating stimulus (1 mM MeAsp plus 100 μM serine76) for 10 seconds. Immediately afterwards, the FRET level at maximum kinase activity was measured by delivering motility buffer with no attractant (0 μM MeAsp, 0 μM serine) for 5 seconds. When cells are adapted to 100 μM MeAsp, removing all attractant is sufficient to elicit a maximal response18,44. Donor excitation interval (i.e., measurements of IDD and IDA; see SI Section 10) was 0.5 seconds and acceptor excitations (i.e., measurements of IAA) were done before and after the set of donor excitations. After this, the concentration of MeAsp was returned to the background c0, and no serine was delivered to the cells for the rest of the experiment. Imaging was then stopped and cells were allowed to adapt to the background for 120 seconds.
After this, a series of stimuli were delivered to the cells in the microfluidic device (see Figure 2E for stimulus protocol). Importantly, the cells were only illuminated and imaged for part of the experiment in order to limit photobleaching. First, cells were imaged for 7.5 seconds in the background concentration c0. Then, the concentration of MeAsp was shifted up to c+ = 110 μM for 30 seconds and imaging continued. Donor excitation interval was 0.75 seconds and acceptor excitations were done before and after the set of donor excitations. After this time, imaging was stopped and the MeAsp concentration returned to c0 for >60 seconds to allow cells to adapt. Then, the same process was repeated, but this time shifting MeAsp concentration down to c− = 90 μM. Alternating up and down stimuli were repeated 10 times each.
The change in concentration experienced by cells in this experiment is significantly larger than the changes experienced by cells swimming in our gradient experiments. Ideally, we could deliver the same magnitude of stimulus to cells in both conditions. However, to detect the responses to stimuli as small as the ones experienced in the gradient experiments above the noise, we would need to deliver and average over responses to many more instances of the stimulus. The photon budget is a major constraint when imaging in single cells, making this impossible. We chose the 10% stimuli to balance the need to measure responses above noise, while also keeping the response in the linear regime44,45 (Supplementary Fig. S5).
FRET levels at minimum and maximum kinase activity were measured again at the end of the experiment. The whole imaging protocol lasted <2200 seconds. In total, cells spent <60 minutes in the device, from loading to the end of imaging. Analyses of these data are described in SI Sections 10–16.
Procedure for measuring the noise statistics
Spontaneous fluctuations in kinase activity were also measured in a background MeAsp concentration of c0 = 100 μM. Measurements were made in single cells. FRET levels at minimum and maximum kinase activity were measured at the beginning and the end of each experiment, as described above. As above, after these measurements, imaging was then stopped and cells were allowed to adapt to the background for 120 seconds. After this, cells were imaged for about 1200 seconds. Throughout, donor excitations (i.e., measurements of IDD and IDA; see SI) were done every 1.0 second, except when it was interrupted by acceptor excitations (i.e., measurements of IAA; see SI), which were conducted every 100 donor excitations. The whole imaging protocol lasted <1400 seconds. In total, cells spent about < 60 minutes in the device, from loading to the end of imaging. Analyses of these data are described in SI Section 10–15, 17.
Procedure to measure swimming and behavioral parameters
After the second wash, cells were centrifuged again and resuspended in motility buffer containing 100 μM MeAsp. Then, the cell suspension was diluted to an OD600 of 0.00025. The cell suspension was then loaded into μ-Slide Chemotaxis devices (ibidi; Martinsried, Germany), the same type of device used to create static gradients, described below. However, instead of tracking cells in the gradient region, we tracked their swimming in one of the large reservoirs, which are roughly 750 μm deep. 1000-s movies of swimming cells were recorded on a Nikon Ti-E Inverted Microscope using a CFI Plan Fluor 4X objective (NA 0.13). This objective’s depth of field is about ±18 μm, much shorter than the depth of the chamber. Adjusting the focal plane to the middle of the chamber made cells that were swimming near the ceiling or floor of the device, which could experience hydrodynamic interactions that affect their behavior77,78, not visible in the movie. At the same time, this lower magnification objective allowed us to collect relatively longer swimming trajectories. Movies were captured around 30 minutes after loading cells into the chamber to mimic the gradient experiments below. Images here and below were captured using a sCMOS camera (ORCA-Flash4.0 V2; Hamamatsu). Analyses of these data are described in SI Sections 18–21. Five biological replicates were done for behavioral parameter measurements, and four biological replicates were done for measuring Dr.
Procedure to measure chemotactic drift speeds
Chemotaxis experiments were performed in μ-Slide Chemotaxis devices (ibidi; Martinsried, Germany). These devices generate a linear gradient between two concentration reservoirs that is stable for a long time. After the second wash, the cell suspension was split into two Eppendorf tubes, 0.5 mL each. After one more centrifugation, one tube of cells was resuspended in 1 mL of motility buffer with 100 μM of MeAsp, to be injected into the “low-concentration reservoir”, and the other was resuspended in 1 mL of motility buffer with 2 μM of fluorescein and varying concentrations of attractant, to be injected into the “high-concentration reservoir”. Cells in both tubes were diluted to OD 0.001 for each experiment. Loading cells in both reservoirs ensured that the concentration of cells throughout the experimental device was approximately uniform. This limited the effects of potential biases that could arise from observing a finite field of view.
Using a background concentration of at least 100 μM MeAsp ensured that the cells were in the log-sensing regime12. The “high” concentrations of MeAsp used were 110.5 μM, 122.1 μM, 135.0 μM, and 149.2 μM. With 1 mm separating the two reservoirs, these concentrations produced linear gradients that approximated shallow exponentials gradients with steepness of roughly: g = {0.1, 0.2, 0.3, 0.4} mm−1. g was calculated from , where Δ log c is the difference in log concentrations between the two reservoirs, and Δx is the distance between them. This is exactly the average steepness of log-concentration across with the width of the channel. To see this, the steady state concentration profile is linear, , where Δc is the difference in concentration between the two reservoirs, x0 is the midpoint between them, and c0 is the concentration at x = x0. From this, the gradient of log concentration depends on position x and can be computed from , where we have defined a reference frame where x0 = 0. Averaging over the channel by integrating over x from −Δx/2 to Δx/2 and dividing by Δx gives, . Close to the low-concentration reservoir, g(x) is larger than g, and vice versa near the high-concentration reservoir, but these errors are small and approximately cancel each other out when we average drift speeds of cells across the channel.
To load the device, first the reservoirs were sealed with the manufacturer’s tabs. Cell suspension with 100 μM MeAsp was injected into the channel where the gradient would form. Excess liquid in the inlets was removed. Then one tab from each reservoir was removed, and the gradient channel was sealed with tabs. The left reservoir was then fully unsealed, and the right reservoir was sealed with tabs. 60–65 μL of cell suspension with 100 μM MeAsp was injected into the left reservoir, and then both inlets of that reservoir were sealed with tape or tabs. Care was taken to make sure there were no bubbles in reservoir at the inlets. Then, the right reservoir was unsealed, and 60–65 μL of cell suspension with the higher concentration of MeAsp was injected. A timer was then immediately started. The right reservoir was then sealed.
Cells were imaged by phase contrast with a CFI Plan Fluor 10X objective (NA 0.30). The depth of the gradient region of the device is 70 μm, and the depth of field of the objective is about ±4 μm. Focusing on the middle of the chamber with this objective filtered out cells that could be interacting with the ceiling or floor surfaces. Images of fluorescein were taken every 5 minutes using a CFI Plan Fluor 4X objective (NA 0.13) through a YFP filter cube (Chroma 49003), illuminated by a LED (SOLA SE, Lumencor) with an exposure time of 100 ms. Since the diffusivity of fluorescein is similar to (slightly lower than) that of MeAsp (MW of fluorescein is 376 kDa; MW of MeAsp is 147 kDa; Sigma Aldrich), we used fluorescein as an indication of when the attractant gradient was stable and linear in the observation region between the two reservoirs, as has been done before55,79. Once the fluorescein profile was stable for several time points (typically around 50–60 minutes after loading), a 1000-second phase contrast movie was recorded at 20 FPS using the 10X phase contrast objective. Before the recording, the transmitted light illumination was adjusted to minimize the number of saturated pixels. After the recording, an additional image of the fluorescein profile was recorded, and the cells were observed to check that they were still swimming normally. Analyses of these data are described below. At least five biological replicates were performed for each gradient steepness.
Supplementary Material
Acknowledgements:
We thank Jeremy Moore and Xiaowei Zhang for help setting up the experimental assays. We also thank Katja Taute and Marianne Grognot for helpful discussions about the gradient experiments. We thank Pieter Rein ten Wolde for providing detailed feedback on an earlier version of this manuscript, Ilya Nemenman and Sosuke Ito for helpful discussions, and Artur Wachtel, Isabella Graf, and Damon Clark for providing comments on the text. We acknowledge Tom Shimizu and Victor Sourjik for bacteria strains and Rafael Gomez-Sjoberg, Microfluidics Lab, for providing information and software to control the solenoid valves in the microfluidic setup. This work was supported by NIH awards R01GM106189 (HM, KK, TE), R01GM138533 (HM, KK, TE), F32GM131583 (HM), and R35GM138341 (BM); by a Yale PEB Seed Grant (TE, BM); and by Simons Investigator Award 624156 (BM).
Footnotes
Competing interests: The authors declare no competing interests.
Code availability: Code to reproduce the main text figures are available with the source data. All algorithms used are described in detail in the Supplementary Information.
Data availability:
Source data for the main text figures are provided online with the manuscript. Source data for the Supplementary Figures are contained in a Supplementary Data file.
References
- 1.Cheong R, Rhee A, Wang CJ, Nemenman I & Levchenko A Information Transduction Capacity of Noisy Biochemical Signaling Networks. Science 334, 354–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uda S et al. Robustness and Compensation of Information Transmission of Signaling Pathways. Science 341, 558–561 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Selimkhanov J et al. Accurate information transmission through dynamic biochemical signaling networks. Science 346, 1370–1373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granados AA et al. Distributed and dynamic intracellular organization of extracellular information. PNAS 115, 6088–6093 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowsher CG & Swain PS Environmental sensing, information transfer, and cellular decision-making. Current Opinion in Biotechnology 28, 149–155 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Tkačik G & Bialek W Information Processing in Living Systems. Annual Review of Condensed Matter Physics 7, 89–117 (2016). [Google Scholar]
- 7.Laughlin S A simple coding procedure enhances a neuron’s information capacity. Z. Naturforsch., C, Biosci 36, 910–912 (1981). [PubMed] [Google Scholar]
- 8.Rieke F, Warland D & Bialek W Coding Efficiency and Information Rates in Sensory Neurons. EPL 22, 151–156 (1993). [Google Scholar]
- 9.Petkova MD, Tkačik G, Bialek W, Wieschaus EF & Gregor T Optimal Decoding of Cellular Identities in a Genetic Network. Cell 176, 844–855.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bialek W, De Ruyter Van Steveninck RR & Tishby N Efficient representation as a design principle for neural coding and computation. in 2006 IEEE International Symposium on Information Theory 659–663 (2006). doi: 10.1109/ISIT.2006.261867. [DOI] [Google Scholar]
- 11.Berg HC E. coli in motion (Springer, 2004). [Google Scholar]
- 12.Lazova MD, Ahmed T, Bellomo D, Stocker R & Shimizu TS Response rescaling in bacterial chemotaxis. PNAS 108, 13870–13875 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu Y Quantitative Modeling of Bacterial Chemotaxis: Signal Amplification and Accurate Adaptation. Annual Review of Biophysics 42, 337–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkinson JS, Hazelbauer GL & Falke JJ Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends in Microbiology 23, 257–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg HC & Purcell EM Physics of chemoreception. Biophysical Journal 20, 193–219 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korobkova E, Emonet T, Vilar JMG, Shimizu TS & Cluzel P From molecular noise to behavioural variability in a single bacterium. Nature 428, 574–578 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Colin R, Rosazza C, Vaknin A & Sourjik V Multiple sources of slow activity fluctuations in a bacterial chemosensory network. eLife 6, e26796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keegstra JM et al. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET. eLife 6, e27455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan G, Sartori P, Neumann S, Sourjik V & Tu Y The energy–speed–accuracy trade-off in sensory adaptation. Nature Physics 8, 422–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clausznitzer D, Micali G, Neumann S, Sourjik V & Endres RG Predicting Chemical Environments of Bacteria from Receptor Signaling. PLOS Computational Biology 10, e1003870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S & Sagawa T Maxwell’s demon in biochemical signal transduction with feedback loop. Nat Commun 6, 7498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Micali G & Endres RG Maximal information transmission is compatible with ultrasensitive biological pathways. Scientific Reports 9, 16898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon CE A Mathematical Theory of Communication. Bell System Technical Journal 27, 379–423 (1948). [Google Scholar]
- 24.Tostevin F & ten Wolde PR Mutual Information between Input and Output Trajectories of Biochemical Networks. Phys. Rev. Lett 102, 218101 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Long J, Zucker SW & Emonet T Feedback between motion and sensation provides nonlinear boost in run- and-tumble navigation. PLOS Computational Biology 13, e1005429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartich D, Barato AC & Seifert U Stochastic thermodynamics of bipartite systems: transfer entropy inequalities and a Maxwell’s demon interpretation. J. Stat. Mech 2014, P02016 (2014). [Google Scholar]
- 27.Meijers M, Ito S & ten Wolde PR Behavior of information flow near criticality. Phys. Rev. E 103, L010102 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Schreiber T Measuring Information Transfer. Phys. Rev. Lett 85, 461–464 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Sigtermans D Towards a Framework for Observational Causality from Time Series: When Shannon Meets Turing. Entropy 22, 426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Gennes P-G Chemotaxis: the role of internal delays. Eur Biophys J 33, 691–693 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Clark DA & Grant LC The bacterial chemotactic response reflects a compromise between transient and steady-state behavior. PNAS 102, 9150–9155 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locsei JT Persistence of direction increases the drift velocity of run and tumble chemotaxis. J. Math. Biol 55, 41–60 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Celani A & Vergassola M Bacterial strategies for chemotaxis response. PNAS 107, 1391–1396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker NB, Mugler A & ten Wolde PR Optimal Prediction by Cellular Signaling Networks. Phys. Rev. Lett 115, 258103 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Nakamura K & Kobayashi TJ Connection between the Bacterial Chemotactic Network and Optimal Filtering. Phys. Rev. Lett 126, 128102 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Spudich JL & Koshland DE Non-genetic individuality: chance in the single cell. Nature 262, 467–471 (1976). [DOI] [PubMed] [Google Scholar]
- 37.Masson J-B, Voisinne G, Wong-Ng J, Celani A & Vergassola M Noninvasive inference of the molecular chemotactic response using bacterial trajectories. PNAS 109, 1802–1807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waite AJ, Frankel NW & Emonet T Behavioral Variability and Phenotypic Diversity in Bacterial Chemotaxis. Annual Review of Biophysics 47, 595–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore JP, Kamino K & Emonet T Non-Genetic Diversity in Chemosensing and Chemotactic Behavior. International Journal of Molecular Sciences 22, 6960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon CE Communication in the Presence of Noise. Proceedings of the IRE 37, 10–21 (1949). [Google Scholar]
- 41.Rieke F, Bodnar DA & Bialek W Naturalistic stimuli increase the rate and efficiency of information transmission by primary auditory afferents. Proceedings of the Royal Society of London. Series B: Biological Sciences 262, 259–265 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Kamino K, Keegstra JM, Long J, Emonet T & Shimizu TS Adaptive tuning of cell sensory diversity without changes in gene expression. Science Advances (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalinin YV, Jiang L, Tu Y & Wu M Logarithmic Sensing in Escherichia coli Bacterial Chemotaxis. Biophysical Journal 96, 2439–2448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sourjik V & Berg HC Receptor sensitivity in bacterial chemotaxis. Proceedings of the National Academy of Sciences 99, 123–127 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu TS, Tu Y & Berg HC A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Molecular Systems Biology 6, 382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francis NR et al. Subunit Organization in a Soluble Complex of Tar, CheW, and CheA by Electron Microscopy. J. Biol. Chem 277, 36755–36759 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Levit MN, Grebe TW & Stock JB Organization of the Receptor-Kinase Signaling Array That Regulates Escherichia coli Chemotaxis. J. Biol. Chem 277, 36748–36754 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Colin R, Zhang R & Wilson LG Fast, high-throughput measurement of collective behaviour in a bacterial population. Journal of The Royal Society Interface 11, 20140486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potter GD, Byrd TA, Mugler A & Sun B Dynamic Sampling and Information Encoding in Biochemical Networks. Biophysical Journal 112, 795–804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders A, Ghosh B, Glatter T & Sourjik V Design of a MAPK signalling cascade balances energetic cost versus accuracy of information transmission. Nature Communications 11, 3494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Razo-Mejia M et al. First-principles prediction of the information processing capacity of a simple genetic circuit. Phys. Rev. E 102, 022404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon CE Coding Theorems for a Discrete Source With a Fidelity Criterion. IRE Nat. Conv. Rec 142–163 (1959). [Google Scholar]
- 53.Andrews BW & Iglesias PA An Information-Theoretic Characterization of the Optimal Gradient Sensing Response of Cells. PLOS Computational Biology 3, e153 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mora T & Wingreen NS Limits of Sensing Temporal Concentration Changes by Single Cells. Phys. Rev. Lett 104, 248101 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Waite AJ et al. Non-genetic diversity modulates population performance. Molecular Systems Biology 12, 895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni B, Colin R, Link H, Endres RG & Sourjik V Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. PNAS 117, 595–601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berg HC & Brown DA Chemotaxis in Escherichia coli analysed by Three-dimensional Tracking. Nature 239, 500–504 (1972). [DOI] [PubMed] [Google Scholar]
- 58.Dufour YS, Fu X, Hernandez-Nunez L & Emonet T Limits of Feedback Control in Bacterial Chemotaxis. PLOS Computational Biology 10, e1003694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armstrong JB, Adler J & Dahl MM Nonchemotactic Mutants of Escherichia coli. Journal of Bacteriology 93, 390–398 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachmann BJ Pedigrees of Some Mutant Strains of Escherichia coli K-12. 36, 33 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barker CS, Prüß BM & Matsumura P Increased Motility of Escherichia coli by Insertion Sequence Element Integration into the Regulatory Region of the flhD Operon. Journal of Bacteriology 186, 7529–7537 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfe AJ & Berg HC Migration of bacteria in semisolid agar. PNAS 86, 6973–6977 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Licata NA, Mohari B, Fuqua C & Setayeshgar S Diffusion of Bacterial Cells in Porous Media. Biophysical Journal 110, 247–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurzthaler C et al. A Geometric Criterion for the Optimal Spreading of Active Polymers in Porous Media. arXiv:2106.05366 [cond-mat] (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alon U, Surette MG, Barkai N & Leibler S Robustness in bacterial chemotaxis. 397, 4 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Neumann S, Vladimirov N, Krembel AK, Wingreen NS & Sourjik V Imprecision of Adaptation in Escherichia coli Chemotaxis. PLOS ONE 9, e84904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong-Ng J, Melbinger A, Celani A & Vergassola M The Role of Adaptation in Bacterial Speed Races. PLOS Computational Biology 12, e1004974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong-Ng J, Celani A & Vergassola M Exploring the function of bacterial chemotaxis. Current Opinion in Microbiology 45, 16–21 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Tu Y & Grinstein G How White Noise Generates Power-Law Switching in Bacterial Flagellar Motors. Phys. Rev. Lett 94, 208101 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Park H, Oikonomou P, Guet CC & Cluzel P Noise Underlies Switching Behavior of the Bacterial Flagellum. Biophysical Journal 101, 2336–2340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huo H, He R, Zhang R & Yuan J Swimming Escherichia coli Cells Explore the Environment by Lévy Walk. Applied and Environmental Microbiology 87, e02429–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flores M, Shimizu TS, ten Wolde PR & Tostevin F Signaling Noise Enhances Chemotactic Drift of E. coli. Phys. Rev. Lett 109, 148101 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Sneddon MW, Pontius W & Emonet T Stochastic coordination of multiple actuators reduces latency and improves chemotactic response in bacteria. PNAS 109, 805–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qin D, Xia Y & Whitesides GM Soft lithography for micro- and nanoscale patterning. Nature Protocols 5, 491–502 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Edelstein A, Amodaj N, Hoover K, Vale R & Stuurman N Computer Control of Microscopes Using μManager. Current Protocols in Molecular Biology 92, 14.20.1–14.20.17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sourjik V & Berg HC Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Frymier PD, Ford RM, Berg HC & Cummings PT Three-dimensional tracking of motile bacteria near a solid planar surface. PNAS 92, 6195–6199 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grognot M & Taute KM A multiscale 3D chemotaxis assay reveals bacterial navigation mechanisms. Commun Biol 4, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colin R, Zhang R & Wilson LG Fast, high-throughput measurement of collective behaviour in a bacterial population. Journal of The Royal Society Interface 11, 20140486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the main text figures are provided online with the manuscript. Source data for the Supplementary Figures are contained in a Supplementary Data file.


