Abstract
Background.
Substance use occurs at a high rate in persons with a psychiatric disorder. Genetically informative studies have the potential to elucidate the etiology of these phenomena. Recent developments in genome-wide association studies (GWAS) allow new avenues of investigation.
Method.
Using results of GWAS meta-analyses, we performed a factor analysis of the genetic correlation structure, a genome-wide search of shared loci, and causally informative tests for six substance use phenotypes (four smoking, one alcohol, and one cannabis use) and five psychiatric disorders (ADHD, anorexia, depression, bipolar disorder, and schizophrenia).
Results.
Two correlated externalizing and internalizing/psychosis factor were found, although model fit was beneath conventional standards. Of 458 loci reported in previous univariate GWAS of substance use and psychiatric disorders, about 50% (230 loci) were pleiotropic with additional 111 pleiotropic loci not reported from past GWAS. Of the 341 pleiotropic loci, 152 were associated with both substance use and psychiatric disorders, implicating neurodevelopment, cell morphogenesis, biological adhesion pathways, and enrichment in 13 different brain tissues. Seventy-five and 114 pleiotropic loci were specific to either psychiatric disorders or substance use phenotypes, implicating neuronal signaling pathway and clathrin-binding functions/structures, respectively. No consistent evidence for phenotypic causation was found across different Mendelian randomization methods.
Conclusions.
Genetic etiology of substance use and psychiatric disorders is highly pleiotropic and involves shared neurodevelopmental path, neurotransmission, and intracellular trafficking. In aggregate, the patterns are not consistent with vertical pleiotropy, more likely reflecting horizontal pleiotropy or more complex forms of phenotypic causation.
Keywords: Addiction, comorbidity, GWAS, Mendelian randomization, pleiotropy, psychopathology
Introduction
Substance use is a leading cause of global mortality (Ezzati, Lopez, Rodgers, Vander Hoorn, & Murray, 2002) and shows a close association with psychiatric disorders. Individuals who have a prior lifetime history of psychiatric disorders (Conway, Swendsen, Husky, He, & Merikangas, 2016) as well as those with a current diagnosis show higher rates of substance use across a variety of substances, including tobacco (Lasser et al., 2000; Weinberger et al., 2017), alcohol (Weitzman, 2004), and cannabis (Blanco et al., 2016). For example, relative to general population, individuals with severe psychotic disorders showed an increased risk of smoking [odds ratio (OR) = 4.6], heavy alcohol use (OR = 4.0), and heavy cannabis use (OR = 3.5) (Hartz et al., 2014). Substance use has been found to predict psychiatric disorders prospectively, including depressive disorders (Brook, Cohen, & Brook, 1998), anxiety disorders (Johnson et al., 2000), and psychotic disorders (van Os et al., 2002). At the same time, early psychiatric problems have also been found to predict later substance use (King, Iacono, & McGue, 2004; Miettunen et al., 2014). The elevated use of substances is associated with poorer outcomes such as medication non-adherence (Margolese, Malchy, Negrete, Tempier, & Gill, 2004), more psychiatric hospitalizations (Dalton, Cate-Carter, Mundo, Parikh, & Kennedy, 2003), and increased suicide rates (Kask et al., 2016), which is partly responsible for the excess mortality due to smoking and alcohol-related diseases (Brown, Inskip, & Barraclough, 2000; Hjorthøj et al., 2015). The high comorbidity and accompanying mortality naturally leads to questions about shared etiology among these behaviors of high public health impact (Barkus & Murray, 2010; Gregg, Barrowclough, & Haddock, 2007).
Numerous studies have demonstrated shared genetic vulnerability to substance use and psychiatric disorder, especially those conceptualized as externalizing disorders such as ADHD and conduct disorder (Hicks, Krueger, Iacono, McGue, & Patrick, 2004; Kendler et al., 2011; Rosenström et al., 2019; Young et al., 2009). Young et al. (2009) reported a moderately heritable factor loading on substance use (alcohol, tobacco, marijuana, and other illicit drugs), conduct disorder, ADHD, and novelty-seeking traits at the start and the end of adolescence, using structural equation ACE models on the family data of 293 twin pairs. Similarly, Hicks et al. (2004) reported a highly heritable, general vulnerability factor that accounts for the correlations among conduct disorder, adult antisocial behavior, and alcohol and drug dependence in a sample of 542 twin families. Of note, most studies reporting such findings are based on known familial relationships (e.g. twins), rather than direct measurement of genetic markers. Genome-wide association studies (GWAS) have now identified hundreds of risk loci for substance use and psychiatric disorders, providing new opportunities to evaluate the pattern of relationships among these phenomena. Moreover, GWAS allows one to test not only the existence of shared genetic risk, but to interrogate individual genomic locations and related genes, with the potential to inform biology associated with the shared liability. The largest substance use GWAS to date identified many sets of genes and relevant tissues contributing to individual differences in alcohol and tobacco use such as nicotinic, dopaminergic, glutamatergic neurotransmission and developmental biology (Liu et al., 2019). Gene-sets relevant to cannabis use also included neurogenesis and dopaminergic neurotransmission (Pasman et al., 2018). Investigating genetic overlap can further elaborate knowledge on how these biological systems relate to psychiatric disorders or more specifically to certain types of substance use measures.
Pleiotropy - that variation within a given locus is associated with variation in two or more phenotypes - is abundant for complex traits (Paaby & Rockman, 2013). Evidence for pleiotropy comes from family studies and population studies of genome-wide genetic covariances among phenotypes, as well as associations between multiple phenotypes and variation within individual genes or genomic loci. These avenues of research are informative about whether pleiotropy exists, but do not readily distinguish different types of pleiotropy. Horizontal pleiotropy arises when genetic variation affects two or more phenotypes independently [or, alternatively, independently affect some intermediate process(es), which then affects the two phenotypes]. Vertical pleiotropy arises when genetic variation affects one phenotype, the experience/expression of which then causes a second phenotype; this is the type of pleiotropy Mendelian randomization (MR) mainly attempts to detect. Genetic covariances can arise under either form of pleiotropy (Solovieff, Cotsapas, Lee, Purcell, & Smoller, 2013). Distinguishing horizontal from vertical pleiotropy would have implications for many existing causal theories of psychopathology and substance use, some of which posit that psychiatric disorders cause substance use (e.g. self-medication; Khantzian, 1997), or that substance use is a causal risk factor of psychiatric disorders (Weiser & Noy, 2005). In contrast, the existence of horizontal pleiotropy would support conceptualizations of psychopathology as a product of more general liabilities to the manifestation of psychopathology and substance use or dependence (Kotov et al., 2017; Krueger & Markon, 2006). Such causal hypotheses can in theory be tested with MR through the use of genetic instruments (Davey Smith & Ebrahim, 2003). Different MR methods provide complementary ways to test causal associations, and here we take advantage of two recently developed methods (O’Connor & Price, 2018; Pickrell et al., 2016).
The aim of this paper is to inform the nature of shared genetic influences on substance use and psychiatric disorders by (1) examining the structure of genetic correlations using genome-wide methods and directly measured genetic variation (i.e. GWAS), (2) characterizing individual loci that are associated with two or more phenotypes, and (3) testing potential causal relations - horizontal or vertical pleiotropy - between substance use and psychiatric disorders. Although several GWAS meta-analyses have been conducted on the genetic overlap between various individual pairs of psychiatric disorders (Lee et al., 2019; van Hulzen et al., 2017), the overlap between multiple forms of substance use and a variety of psychiatric disorders has not been evaluated.
Materials and methods
Quality control
Publicly available European ancestry GWAS summary statistics, the largest to date, were collected for six substance use and five psychiatric disorder phenotypes. Any well-powered meta-analysis GWAS of substance use and psychiatric disorders covering externalizing, internalizing, and psychotic disorders were considered for inclusion. Among these, those reporting at least five significant loci in European ancestry and whose summary statistics were publicly available before mid-2019 were included. Additionally, education and height were included as controls to serve as a reference for interpreting the results. Quality controls, including variants filtering and LD clumping, were applied to ensure that problematic variants were excluded and different sets of summary statistics were as comparable as possible. Study characteristics after QC and details of QC procedure are reported in Table 1 and the Supplementary Note, respectively. Note that LDSC intercept values for the smoking and alcohol use traits are <1.0, consistent with the fact that these summary statistics were originally generated using genomic control (Liu et al., 2019).
Table 1.
Study information
| Phenotype | Type | Number of sentinel GWAS variantsa | Sample sizeb | λ | LDSC interceptc | h 2 | Source |
|---|---|---|---|---|---|---|---|
| Age of initiation of smoking | Substance use | 12 | 341 427 | 1.227 | 0.976 (0.008) | 0.044 (0.003) | Liu et al. (2019) |
| Cigarettes per day | Substance use | 98 | 337 334 | 1.294 | 0.958 (0.010) | 0.074 (0.007) | Liu et al. (2019) |
| Smoking cessation | Substance use | 37 | 547 219 | 1.222 | 0.948 (0.009) | 0.067 (0.03) | Liu et al. (2019) |
| Ever smoker | Substance use | 439 | 1 232 091 | 1.701 | 0.813 (0.011) | 0.085 (0.002) | Liu et al. (2019) |
| Drinks per week | Substance use | 138 | 941 280 | 1.393 | 0.901 (0.009) | 0.036 (0.002) | Liu et al. (2019) |
| Lifetime cannabis use | Substance use | 4 | 162 082 | 1.191 | 1.006 (0.007) | 0.120 (0.008) | Pasman et al. (2018) |
| ADHD | Psychiatric disorder | 12 | 53 293 | 1.247 | 1.029 (0.010 | 0.221 (0.014) | Demontis et al. (2019) |
| Anorexia | Psychiatric disorder | 10 | 72 517 | 1.248 | 1.028 (0.010) | 0.162 (0.011) | Watson et al. (2019) |
| Bipolar disorder | Psychiatric disorder | 16 | 51 710 | 1.326 | 1.022 (0.010) | 0.200 (0.010) | Stahl et al. (2019) |
| Major depressive disorder | Psychiatric disorder | 8 | 173 005 | 1.23 | 0.996 (0.008) | 0.080 (0.005) | Wray et al. (2018) |
| Schizophrenia | Psychiatric disorder | 189 | 105 318 | 1.684 | 1.070 (0.011) | 0.239 (0.008) | Pardiñas et al. (2018) |
| Years of education | Control | 644 | 766 345 | 2.095 | 1.031 (0.014) | 0.107 (0.003) | Lee et al. (2018) |
| Height | Control | 5598 | 709 706 | 3.61 | 1.899 (0.045) | 0.455 (0.019) | Yengo et al. (2018) |
Number of GWAS variants are ascertained by LD clumping (r2 > 0.1) genetic variants with p value <5 × 10−8 using Priority Pruner.
Sample size is based on full summary statistics released in public except GSCAN phenotypes (Liu et al., 2019) which included 23andMe sample.
Smoking and drinking GWAS data from Liu et al. (2019) have LDSC intercept <1 due to genomic control.
Factor analysis
We used genomicSEM (Grotzinger et al., 2019) to perform factor analysis on the multivariate covariance matrix. The matrix was first constructed with SNPs filtered to those polymorphic in HapMap3 Europeans with minor allele frequency >0.01. We fit a hypothesized three-factor model as well as exploratory factor analysis (EFA) with 1–3 factors. Fit for all models were compared using confirmatory factor analysis (CFA). EFA was performed with promax rotation and maximum likelihood estimation using factanal() in the R package stats v.3.6.1. CFA was conducted with Diagonally Weighted Least Square estimation and implemented in genomicSEM, which provides standard fit information including the χ2 fit statistic, Akaike Information Criterion (Akaike, 1974; Vrieze, 2012), comparative fit index (CFI), and standardized root mean square residual (SRMR). While no doubt the confirmatory models based on the EFA were overfitted, the CFA allowed us to evaluate fit indices for best-case (indeed, over-fitted) scenarios.
Bivariate locus-wise association
To identify pleiotropic loci, we used gwas-pw (Pickrell et al., 2016; Ruderfer et al., 2018), a bivariate association method used to identify loci associated with pairs of phenotypes (here, 78 total pairs). The method is a Bayesian hierarchical modeling approach developed to compare the posterior probability of four competing models for a given approximately independent LD block. Models 1 and 2 assume the block harbors a causal variant that is associated with either only the first or only the second phenotype in a pair, respectively. Model 3 assumes the block harbors a variant that is associated with both phenotypes simultaneously, and model 4 assumes the block contains two distinct variants, each associated with only one of the two phenotypes. As in the original publication of the method, genomic regions were defined a priori by splitting chromosomes into approximately independent LD blocks (mean block size: 1.5 M base-pairs) (Berisa & Pickrell, 2016). The method empirically estimates priors for each regional model by using effect size information of variants in a given LD block and applies sample overlap correction (here we used effect size correlations between variants with p value >0.1 in both studies; online Supplementary Table S1) to generate posterior probabilities. A region was deemed pleiotropic if the posterior probability of model 3 was >0.9. SNPs showing the highest posterior probability within the shared locus were chosen as lead SNPs for that region. This method has been shown to detect shared loci at a false discovery rate of 10% (Pickrell et al., 2016).
Functional annotations and tissue/gene-set enrichment analysis
First, we tested whether pleiotropic loci harbor more or less deleterious variants compared to non-pleiotropic loci using Combined Annotation Dependent Depletion (CADD) scores, which predict deleteriousness of variants based on protein function and structure (Kircher et al., 2014). Second, we tested whether variants from pleiotropic loci are more or less likely to harbor variants of different functional classes (e.g. nonsynonymous, intronic, intergenic, etc.). Lead SNPs and their LD partners (r2 > 0.4, ld-window 500 kb) were annotated with CADD and SEQMINER using refGene (retrieved 18 September 2019; Ye et al., 2010). Next, we performed tissue and gene-set enrichment analysis to test whether genes mapped to pleiotropic loci were enriched in certain tissue or gene-sets. A given region was categorized depending on whether it is associated with both substance use and psychiatric or associated with only one or the other. Tissue- and gene-set enrichment analyses were conducted by extracting nearest genes of the lead SNPs in these regions using a hypergeometric association test implemented in FUMA (Watanabe, Taskesen, van Bochoven, & Posthuma, 2017). Gene-sets surviving Bonferroni correction for tissue specificity p < 0.5/54, and FDR of 0.05 for gene-set analysis were considered significantly enriched.
Testing for vertical pleiotropy
To test causal associations, we applied bidirectional MR (Pickrell et al., 2016) and the Latent Causal Variable model (LCV) (O’Connor & Price, 2018), two recently developed methods applicable to GWAS summary statistics. They are designed to be relatively robust to horizontal pleiotropy which is prevalent for complex traits (Verbanck, Chen, Neale, & Do, 2018). Both methods are based on simple intuition: if phenotype A causally influences phenotype B (e.g. smoking causing lung cancer), then any variant associated with A (smoking) will have correlated effects on B (lung cancer). However, variants associated with B (lung cancer) will not necessarily have correlated effects on A (smoking), as there will be many genetic causes of B (lung cancer) that are independent of A (e.g. asbestos exposure). In bidirectional MR, two sets of correlations are calculated on genetic association effect sizes, first using genome-wide significant (GWS) variants of the first phenotype and then repeating the same procedure using GWS variants for the second phenotype. These correlations are used to evaluate evidence for four models: in models 1 and 2, phenotype A or B causes B or A; in model 3, there are no causal relationships; in model 4, two phenotypes are very closely related (e.g. two alternate measurements of a single entity or one could be the major highly penetrant causal factor for the other). A relative likelihood for causal (models 1 and 2) v. non-causal (models 3 and 4) model (rl) <0.01 was interpreted as supporting the causality. In the LCV model, a latent variable mediates the genetic correlation between phenotypes A and B, having causal effects on both traits. A genetic causality proportion (GCP) is estimated (ranging from 0 to 1), which quantifies the degree of genetic causality, using mixed fourth moments of marginal effect sizes of all available SNPs. A high GCP indicates the genetic component of phenotype A is at least partially causal for phenotype B, while low GCP values suggest a lack of causal effects. Phenotype pairs with GCP >0.6 and p values surviving Bonferroni correction (here, p < 0.00064) were identified as potential phenotypes in causal relations (O’Connor & Price, 2018). Finally, the LCV and bidirectional MR results were compared to two-sample MR results obtained using MRbase v.0.5.3 (Hemani et al., 2018) in R 3.6.1 on substance use-psychiatric phenotype pairs, for integration with the broad MR literature (see Supplementary Note for a detailed procedure for each method).
Results
Structure of genetic correlations
The first five eigenvalues of the genetic correlation matrix were 2.9, 1.1, 0.72, 0.13, and 0.07 (online Supplementary Fig. S1). We fit EFAs with 1–4 factors. Only the two-factor model was free of Heywood cases (loadings ⩾1.0), rendering model interpretation difficult. All EFA results are reported in online Supplementary Table S2. The two-factor model consisted of an externalizing factor loading on all three smoking phenotypes and ADHD, and an internalizing-psychosis factor loading on depression, anorexia, schizophrenia, and bipolar disorder (χ2 = 838.92, df = 26, p = 4.31 × 10−160, AIC = 876.92, CFI = 0.80, SRMR = 0.11). This procedure of fitting exploratory models and then similar confirmatory models in the same dataset will lead to overfitting, so these values should be considered an absolute best-case scenario. The four-factor model, roughly having an internalizing, psychosis, externalizing, and substance use factor, achieved the best fit. However, to converge, we had to drop drinks per week and constrain multiple uniquenesses to have >0 variance. Overall, models with 3–4 factors, including our hypothesized three-factor model, resulted in negative variances for several variables. As a sensitivity analysis, we conducted CFA using summary statistics for smoking and drinking to which no genomic controls had been applied, and this did not change the fit. CFA results are presented in online Supplementary Table S3 and Fig. S2.
Shared loci
Complete results on pleiotropic loci are available in online Supplementary Tables S4–8. Of all 30 possible substance use-psychiatric phenotype pairs, ever smoker and ADHD had the highest number of shared loci (N = 40), followed by age of smoking initiation and ADHD (N = 22). Within psychiatric disorder pairs, bipolar disorder and schizophrenia showed the highest number of shared loci (N = 108). Within substance use pairs, age of smoking initiation and ever smoker showed the highest number (N = 58). In contrast, height shared few loci with substance use and psychiatric disorders despite its large number of GWAS signals, but years of education showed substantial overlap with both substance use and psychopathology (Fig. 1). Of note, the number of pleiotropic regions reflects both degree of genetic overlap and statistical power of the original GWAS, rendering between-pair comparisons difficult. The lead SNPs in shared loci mostly agreed with the expected direction based on the genetic correlation (i.e. the more substance use, the higher risk of psychiatric disorder) (Fig. 1).
Fig. 1.
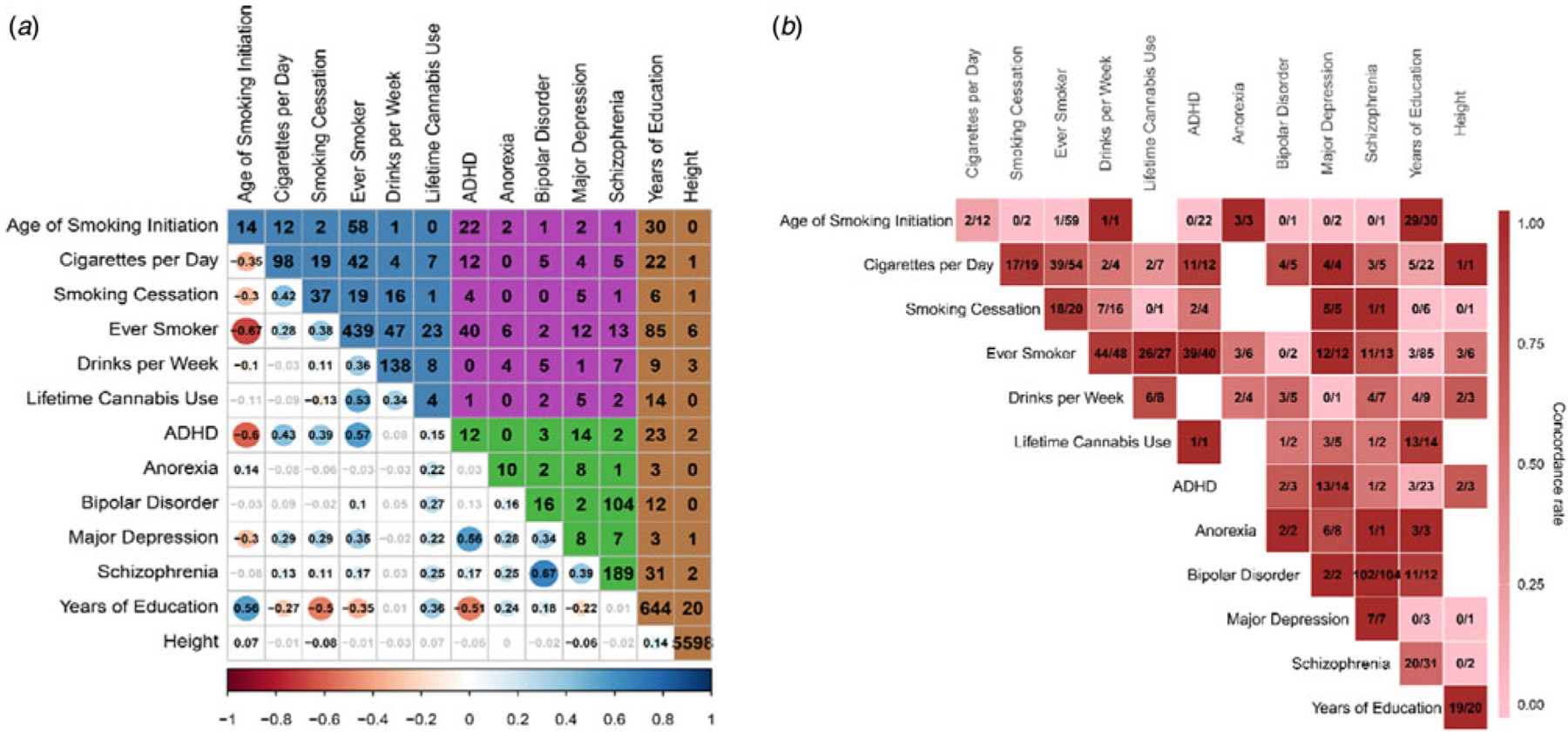
The bottom triangle of panel A shows genetic correlations estimated by bivariate LDSC (online Supplementary Table S14). Non-significant correlations after Bonferroni correction are in gray. The upper triangle shows the number of loci shared between corresponding phenotype pairs. The numbers in the diagonal are the total number of genome-wide significant hits from original, univariate GWAS (ascertained by LD clumping). The background colors of the upper triangle indicate following pairs of domains: blue - between substance uses, purple - between substance use and psychiatric disorders, green - between psychiatric disorders, brown - between control phenotypes and all other phenotypes. Panel B shows the sign concordance of effect for lead SNPs in shared loci for a given phenotype pair. More reddish color indicates higher concordance. In each box, the left-side number indicates the number of lead SNPs having concordant directions of effects while the right-side number indicates the total number of lead SNPs in pleiotropic loci for a given pair. Note that smaller value of Age of Smoking Initiation indicates earlier age of smoking initiation and higher values of Smoking Cessation, Ever Smoker, and Lifetime Cannabis Use indicate current smoker, ever smoker, and ever cannabis user, respectively. Higher values of psychiatric disorders indicate the presence of the disorder.
A total of 341 loci showed evidence for association with two or more substance use or psychiatric disorder phenotypes. This number includes 230 loci reported in past GWAS of respective phenotypes, and 111 additional loci not reported in those studies (online Supplementary Table S8). Of these 341 loci, 152 loci (45%) were associated with both substance use and psychiatric phenotypes; 114 (33%) were associated with multiple substance use phenotypes but no psychiatric disorder, and 75 (22%) were associated with multiple psychiatric disorders but no substance use phenotypes.
When 152 shared loci were classified depending on type of substance, 10 loci were associated with the use of all three types of substances [i.e. smoking, cannabis, and alcohol (Table 2)]. A gene encoding the D2 subtype of the dopamine receptor, DRD2, on chromosome 11 was located in these multi-substance shared loci along with cell-adhesion protein coding genes including NCAM1 and CADM2. Five loci were associated with alcohol and psychiatric disorder, including Utr3 region of FUT2, a gene involved in bifidobacterial diversity in the intestine and plasma levels of B12 vitamins (Mitchell, Conus, & Kaput, 2014). Five loci were associated specifically with cannabis use and psychiatric disorder, including intron region of WSCD2, a gene previously associated with extraversion and risky behaviors (Linnér et al., 2019; Lo et al., 2017). Perhaps notably, nicotinic receptor genes (e.g. CHRNA4 and CHRNA5) did not belong to the 152 shared loci and instead were associated only with smoking (CHRNA4) and smoking and drinking phenotypes (CHRNA5).
Table 2.
Pleiotropic regions associated with all three types of substance use and psychopathology
| Chr | Start | Stop | Associated phenotypes | Genes associated with lead SNPsa | ||
|---|---|---|---|---|---|---|
| Substance use | Psychopathology | Controls | ||||
| 11 | 112459488 | 114256749 | Cigarettes per day, drinks per week, ever smoker, lifetime cannabis use, smoking cessation | Bipolar disorder, major depression, schizophrenia | Years of education | Synonymous:DRD2; Intron: NCAM1 |
| 3b | 84367614 | 87409543 | Age of smoking initiation, cigarettes per day, drinks per week, ever smoker, lifetime cannabis use, smoking cessation | ADHD | Years of education | Intron:CADM2 |
| 4 | 2844097 | 3845571 | Age of smoking initiation, cigarettes per day, drinks per week, ever smoker, lifetime cannabis use, smoking cessation | ADHD | Years of education | Intron:ADD1; Intron:NOP14| NOP14-AS1; Normal_Splice_Site:HTT |
| 1 | 71687454 | 74326484 | Drinks per week, ever smoker, lifetime cannabis use, smoking cessation | ADHD, major depression, schizophrenia | Exon:LINC01360 | |
| 4 | 100678905 | 103220401 | Age of smoking initiation, drinks per week, lifetime cannabis use | ADHD, schizophrenia | Years of education, height | Nonsynonymous:SLC39A8; Intron:DNAJB14 |
| 5 | 87390784 | 88891173 | Age of smoking initiation, cigarettes per day, drink per week, ever smoker, lifetime cannabis use | ADHD | Years of education | Intron:LINC00461; Utr3, Intron:MEF2C |
| 8 | 90638201 | 93554257 | Age of smoking initiation, drinks per week, ever smoker, lifetime cannabis use | Schizophrenia | Intron:RUNX1T1 | |
| 8 | 63349908 | 65232744 | Cigarettes per day, drinks per week, lifetime cannabis use | Schizophrenia | Intergenic | |
| 17 | 1929074 | 3701588 | Drinks per week, ever smoker, lifetime cannabis use | Schizophrenia | Intron:SRR | |
Lead SNP in the locus, their associated phenotype pairs, and the direction of effects can be found in online Supplementary Tables S4 and 5.
Two adjacent regions (3:84367614–85582078, 3:85582231–3:87409543) were combined to form this row.
Genes harboring lead SNPs of the 152 shared loci were overexpressed in all 13 brain tissues in GTEx v8 database and down-regulated in six tissues including substantia nigra, kidney cortex, and liver (Aguet et al., 2019). They are enriched in 90 Gene Ontology (GO) biological processes, 10 molecular function, and 34 cellular components gene-sets. The top 3 most strongly associated biological processes were ‘neurogenesis’, ‘cell projection organization’, ‘regulation of nervous system development’. Enriched molecular functions include protein dimerization, transcription factor binding, and cell adhesion molecule binding, and the enriched cellular components encompass various parts of neuron including glutamatergic synapse. The 75 psychiatric-specific shared loci were overexpressed in four brain tissues (anterior cingulate cortex, frontal cortex, amygdala, and hippocampus) but not downregulated in any tissues. They were enriched in 11 biological processes, five molecular function, and 20 cellular components gene-sets. Most of the enriched gene-sets concerned neuronal signaling-related activity (e.g. voltage-gated ion channel activity) and cell structures (e.g. synaptic membrane). Finally, the 114 substance use-specific shared loci were overexpressed in the cerebellar hemisphere, cortex, and artery tibial, and downregulated in seven tissues including kidney cortex, liver, and pancreas. They are enriched in one molecular function, ‘clathrin binding’ and 12 cellular components most of which are located in neurons, including ‘dopaminergic synapse’ (Fig. 2; see online Supplementary Tables S9 and 10 for full tissue and gene-set enrichment results).
Fig. 2.
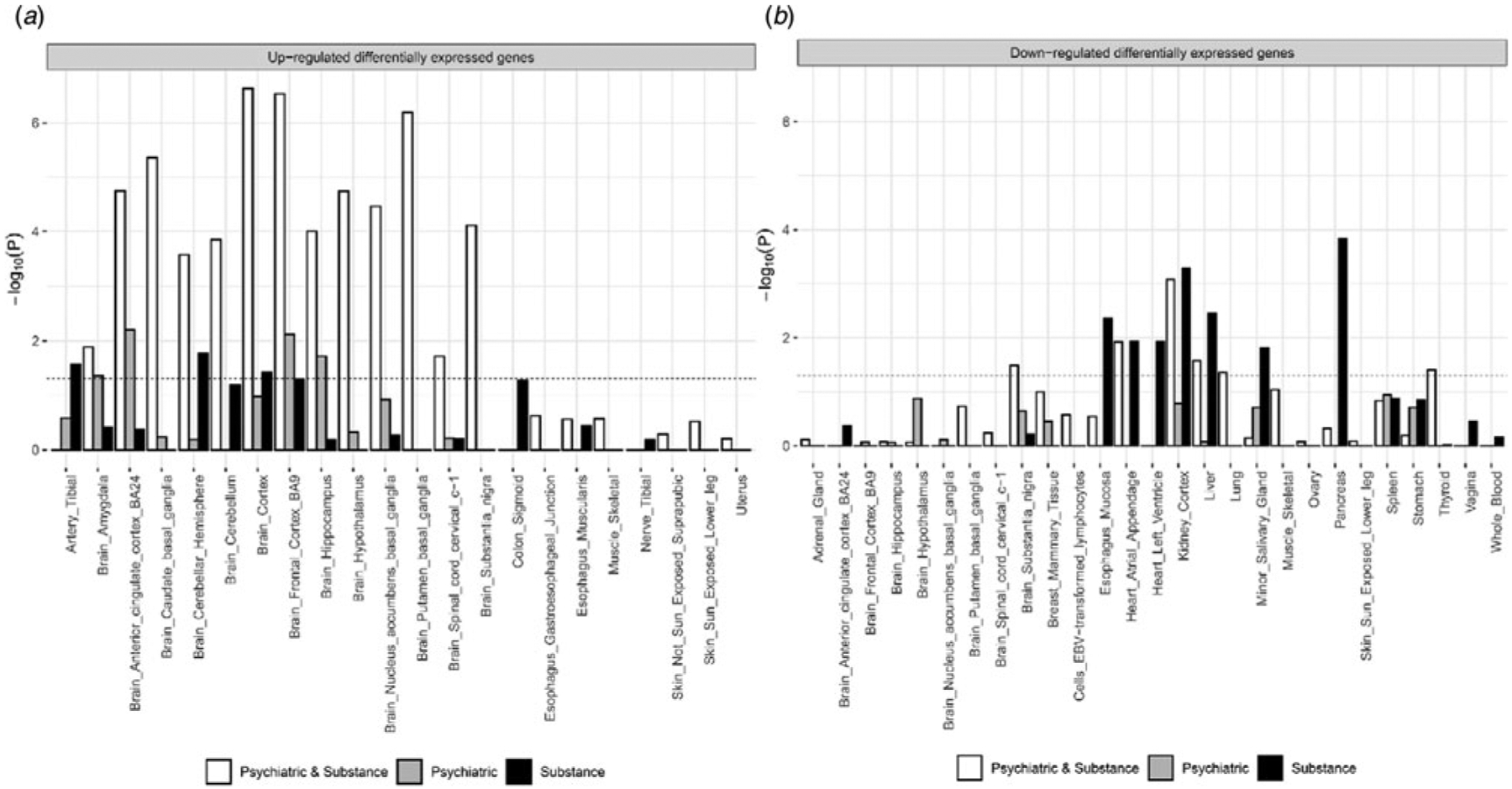
Panel A shows results for up-regulated Differentially Expressed Gene (DEG) set tests while panel B shows results from the down-regulated DEG set test. X-axis and Y-axis each represents tissue types and log10 p value. The bar higher than the dashed line indicates significantly enriched tissue type after Bonferroni correction. The white bar represents genes from loci pleiotropic for both substance use and psychiatric disorders while gray and black bars represent genes from loci pleiotropic within psychiatric disorders or substance use.
The median CADD rank scores were not significantly different across lead SNPs from substance-psychiatric pleiotropic loci, either psychiatric or substance use-specific pleiotropic loci, and non-pleiotropic loci (p = 0.34; online Supplementary Fig. S3). Most of the lead SNPs fell in intergenic or intronic regions (>90%) and their distribution across functional categories did not differ across different groups of pleiotropic and non-pleiotropic loci (p = 0.26; online Supplementary Fig. S4; online Supplementary Table S11).
Causally informative analyses
Of 78 pairs submitted to the analysis, none showed evidence of vertical pleiotropy from bidirectional MR and LCV. As a sensitivity analysis, bidirectional MR was conducted with the GWAS variants originally reported by each study and LCV was repeated with summary statistics for smoking and drinking without genomic control, both yielding the same pattern of results. Eight causal associations were significant in more than half of the four two-sample MR tests performed (Table 3). Full results of bidirectional MR and LCV, and two-sample MR are in online Supplementary Tables S12 and 13, respectively.
Table 3.
Comparison of LCV, bidirectional MR and two-sample MR results
| ADHDa,b Age_Smoke | ADHD Ever smoker | Ever smoker ADHD | MDD Cannabis | Cigarettes per day Schizophrenia | Ever smoker MDD | MDD Ever smoker | Schizophrenia Ever smoker | ||
|---|---|---|---|---|---|---|---|---|---|
| LCV | GCP (s.e.) | −0.08 (0.16) | 0.08 (0.11) | −0.08 (0.11) | 0.25 (0.39) | 0.33 (0.37) | 0.04 (0.06) | −0.04 (0.06) | −0.15 (0.17) |
| p value | 0.61 | 0.50 | 0.50 | 0.56 | 0.28 | 0.54 | 0.54 | 0.12 | |
| Pickrell | Rho1c | −0.63 | 0.8 | 0.66 | 0.64 | 0.33 | 0.5 | 0.5 | 0.28 |
| Rho2 | −0.41 | 0.66 | 0.8 | 0 | 0.19 | 0.5 | 0.5 | 0.38 | |
| Rld | 1.85 | 142.73 | 142.73 | 0.78 | 13.03 | 2.13 | 2.13 | 598.82 | |
| IVW | β (s.e.)e | −0.09 (0.01) | 0.11 (0.02) | 0.67 (0.13) | 0.50 (0.14) | 0.48 (0.16) | 0.46 (0.07) | 0.14 (0.04) | 0.02 (0.01) |
| p valuef | 1.25 × 10−8* | 2.40 × 10−10* | 4.97 × 10−7* | 5.20 × 10−4* | 2.41 × 10−3 | 5.94 ×10−12* | 1.21 × 10−4* | 8.52 × 10−5* | |
| Weighted median | β (s.e.) | −0.09 (0.02) | 0.08 (0.2) | 0.92 (0.19) | 0.58 (0.13) | 0.64 (0.14) | 0.53 (0.10) | 0.11 (0.02) | 0.02 (0.005) |
| p value | 2.38 × 10−5* | 9.72 × 10−6* | 7.53 × 10−7* | 6.64 × 10−6* | 2.56 × 10−6* | 6.70 × 10−8* | 4.38 × 10−6* | 5.67 × 10−5* | |
| Weighted mode | β (s.e.) | −0.09 (0.03) | 0.07 (0.02) | 1.39 (0.45) | 0.58 (0.17) | 0.62 (0.15) | 0.89 (0.26) | 0.10 (0.03) | 0.04 (0.02) |
| p value | 2.75 × 10−2 | 2.13 × 10−2 | 2.48 × 10−3 | 2.80 × 10−2 | 3.67 × 10−4* | 8.24 × 10−4* | 2.17 × 10−2 | 1.08 × 10−2 | |
| MR Egger | β (s.e.) | −0.10 (0.12) | 0.06 (0.14) | 1.01 (0.57) | 0.59 (1.06) | 0.68 (0.30) | 0.23 (0.27) | 0.21 (0.24) | 0.01 (0.03) |
| p value | 4.34 × 10−1 | 7.02 × 10−1 | 7.76 × 10−2 | 6.14 × 10−1 | 2.97 × 10−2 | 4.07 × 10−1 | 4.49 × 10−1 | 6.32 × 10−1 |
The order of phenotypes in the first two rows represents the direction of causality (i.e. top to bottom).
Age_Smoke = age of smoking initiation, Cannabis = lifetime cannabis use, MDD = major depressive disorder.
Rho1, Rho2 = effect size correlations for genome-wide significant variants ascertained with phenotype 1 (Rho1) and phenotype 2 (Rho2).
rl = relative likelihood (rl < 0.01 is considered significant causal association).
Units: log odds ratio and standard deviations for binary and continuous phenotypes, respectively.
Astericks attached to phenotype pairs represent significant causal associations after Bonferroni correction.
Discussion
We coordinated and analyzed GWAS results of six substance use and five psychiatric phenotypes to investigate the genetic correlation structure, multivariate association, and causal links among these phenotypes. The two-factor structure performed better than the single-factor model, but did not achieve a good model fit (e.g. CFI = 0.8, SRMR = 0.11, AIC = 876.92). Modification indices suggested that internalizing factor might not be well captured by covariance between depression and anorexia and residual covariance exists between schizophrenia and bipolar disorder as well as among some substance use phenotypes. In line with this diagnosis, the four-factor model achieved a better fit (e.g. CFI = 0.87, SRMR = 0.08, AIC = 709.62), which additionally fit separate substance use and psychosis factor. This model also aligns with current conceptualizations of the meta-structure of psychopathology (i.e. correlated externalizing, internalizing, and psychosis factors) (Kotov et al., 2017). However, caution is required in interpreting the four-factor model since the solution could be ill-specified (Heywood cases) and over-fitted (same data were used to both construct and test the model). The joint factor structure of substance use and psychiatric disorders should be further tested in independent samples, especially as large-scale GWAS on related phenotypes continue to be published.
We identified a total of 341 loci that showed at least one bivariate pleiotropic association with substance use or psychiatric phenotypes, which included about half of the loci reported in previous GWAS (~50% of the 458 univariate GWAS associations), confirming the presence of extensive pleiotropy. Genes nearest to the loci shared by both substance use and psychiatric disorders simultaneously (152 loci) were over-expressed in broad regions of the brain and enriched in neurodevelopmental pathways, suggesting that general neurodevelopmental processes (e.g. neurogenesis and neuron differentiation) may underlie the risk of both psychiatric disorders and substance use. For example, DCC, a gene involved in axonal growth and white matter projections (Jamuar et al., 2017), was mapped to the most pleiotropic locus in a recent fixed-effect meta-analysis of eight psychiatric disorders (Lee et al., 2019). The same gene was mapped to highly pleiotropic locus in the current analysis, associated with the age of smoking initiation, ever smoker, depression, schizophrenia, and education. Biological adhesion and cellular morphogenesis/organization pathways were prominent among the neurodevelopmental pathways enriched in the 152 shared loci. Cell adhesion molecules have been reported to regulate synapse number, maturation, and plasticity (Sytnyk, Leshchyns’ka, & Schachner, 2017), serving essential functions in neural development. On the other hand, genes nearest to the pleiotropic loci specific to psychiatric disorders were mostly enriched in neuronal signaling pathways such as voltage-gated ion channel activity while those specific to substance use were enriched in clathrin binding-related functions and cell structures. The potential role of clathrin binding is less clear, but it plays a key role in receptor endocytosis and vesicle recycling at th synapse (Jung & Haucke, 2007; Kaksonen & Roux, 2018) and has been implicated in drug-evoked neural plasticity in animal studies of addiction, including amphetamine (Brebner et al., 2005), heroin (Van den Oever et al., 2008), and morphine (Morón et al., 2007). For example, inhibiting clathrin-dependent postsynaptic AMPA receptor endocytosis in ventral mPFC and nucleus accumbens reduced drug-seeking behaviors in rats (Brebner et al., 2005; Van den Oever et al., 2008). Current results provide converging evidence as to the role of clathrin binding as a risk factor relatively specific to drug use and addiction.
Our bidirectional MR and LCV analysis detected no causally associated pairs, consistent with the notion that genetic correlations among these traits arise from horizontal pleiotropy. MR Egger, a classic two-sample method more robust to confounding by horizontal pleiotropy, also detected no pairs in vertical pleiotropy. In contrast, the other two-sample MR analyses more susceptible to such confounding (Verbanck et al., 2018) reported multiple pairs of causal associations, similar to past findings: genetic liability to ADHD associated with increased risk of smoking initiation (Fluharty, Sallis, & Munafò, 2018) and vice versa (Treur et al., 2019), and genetic liability to smoking associated with increased risk of schizophrenia and depression and vice versa (Wootton et al., in press; Yao et al., in press); but see also some null MR findings (Gage et al., 2017; Hodgson et al., 2020; Taylor et al., 2014). Heterogeneity statistics from classic MR were significant for five out of eight associations (p < 0.05), indicating further the potential presence of horizontal pleiotropy for these five pairs (Bowden, Hemani, & Davey Smith, 2018). Three pairs, i.e. ADHD lowering the age of smoking initiation, ever smoker increasing risk for ADHD, and depression, showed non-significant heterogeneity statistics thus may be causally related. However, their genetic correlations were rather high (i.e. rg = −0.6, 0.57, 0.35 for each pair in order), a condition which can produce excess false positives in classic MR (O’Connor & Price, 2018).
On the other hand, LCV and bidirectional MR showed relatively well-calibrated type 1 and 2 errors in such a scenario (O’Connor & Price, 2018). Taken together, vertical pleiotropy, at least a single causal direction, may not explain the well-known high rate of co-occurrence among psychopathology and substance use. Related causal hypotheses such as self-medication or substance use as a causal risk factor for psychopathology are not well supported by current results. For example, GCP for major depression and ever smoker (0.04, S.E.: 0.06) and that for cannabis use and schizophrenia (−0.15, S.E.: 0.20) were close to zero and significantly different from 1.
Our findings should be interpreted in light of several limitations. First, the number of manifest variables may be still insufficient to fit CFA models with more than three factors. We also stress again that CFA fit could be overestimated to some unknown degree since the models were built on EFA results in the same sample. Current results should be tested in replication sample with more manifest variables especially those for internalizing. From a more technical standpoint, some of the constituent GSCAN cohorts used linear mixed models (LMM) and this may have exerted a subtle influence on CFA fit due to model misspecification between linear regression-based LDSC model and LMM (Yang, Zaitlen, Goddard, Visscher, & Price, 2014). The degree and remedy for this potential bias should be further studied. For pleiotropic loci analysis, the size of LD blocks used in this method is relatively large (mean 1.5 Mb) yielding limited resolution, and the posterior probability threshold to detect pleiotropy is almost certainly less stringent than the typical family-wise error rate of 5% in GWAS. We used the more liberal threshold (corresponding to an FDR of ~10%) since the focus of the current study was to assess the scope of pleiotropy in known associated loci, rather than declaring high confidence novel loci. In the simulation performed in the original article, gwas-pw overall gave a slight overestimation for model 3 (pleiotropy). In the presence of high sample overlap, it additionally gave modest overestimation for model 4 (two separate effects in a single locus) over model 1 or 2 (only one effect for either trait in a single locus), requiring caution in interpretation. It is also not possible to distinguish a single variant influencing both traits from two causal variants in strong LD affecting each trait separately. Regarding causal inference, although the results were mainly interpreted in light of shared genetic effects, cyclical feedback loops between phenotypes may exist to some degree, possibly for all pairs of phenotypes. This type of causality can evade LCV and bidirectional MR but can still be detected by two-sample MR methods to some extent. Last, the GWAS were all performed on individuals of European ancestry, which limits the generalizability of current findings. Increasing availability of trans-ancestry GWAS can improve the generalizability of this kind of analyses in the future.
Despite these limitations, the current study provides comprehensive analyses of the genetic overlap between substance use measures and psychiatric disorders based on GWAS results. It offers novel insights into the structure, biology, and causal nature of this overlap and points toward the future directions for methodological and etiology research. Refining our understanding of the biological underpinnings of the externalizing spectrum (e.g. salient developmental processes and periods) will be helpful to tackle adverse outcomes associated with it. Continuing to study multiple layers of biology such as those underlying addiction to various substance types (e.g. clathrin-mediated endocytocis of AMPA receptors) and that pertaining to specific substance-psychiatric disorder association can add rich information to the etiology of the two phenomena. Finally, the current study applied novel causal inference methods developed to address pervasive horizontal pleiotropy in complex traits and adds to the fast-growing causal inference literature.
Supplementary Material
Acknowledgements.
The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper. We would like to thank the research participants and employees of 23andMe for making this work possible. Members of the 23andMe research team are Michelle Agee, Babak Alipanahi, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, David A. Hinds, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Matthew H. McIntyre, Joanna L. Mountain, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, J. Fah Sathirapongsasuti, Olga V. Sazonova, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, and Catherine H. Wilson. We would also like to thank Hoang Nguyen for his advice on factor analysis.
Financial support. This study was supported by funding from US National Institutes of Health awards R01DA037904 and R01DA044283 to S.V.; R01HG008983 and R21DA040177 to D.J.L.
Footnotes
Supplementary material. The supplementary material for this article can be found at https://doi.org/10.1017/S003329172000272X.
Conflict of interest. The authors associated with the 23andMe Research Team are employees of 23andMe Inc. All other authors reported no biomedical financial interests or potential conflicts of interest.
Ethical standards. Ethical review and approval were provided by the University of Minnesota institutional review board.
References
- Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, … Lappalainen T (2019). The GTEx Consortium atlas of genetic regulatory effects across human tissues. BioRxiv. doi:787903. 10.1101/787903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H (1974). A new look at the statistical model identification. IEEE Transactions on Automatic Control, 19(6), 716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Barkus E, & Murray RM (2010). Substance use in adolescence and psychosis: Clarifying the relationship. Annual Review of Clinical Psychology, 6(1), 365–389. doi: 10.1146/annurev.clinpsy.121208.131220. [DOI] [PubMed] [Google Scholar]
- Berisa T, & Pickrell JK (2016). Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics (Oxford, England), 32 (2), 283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco C, Hasin DS, Wall MM, Flórez-Salamanca L, Hoertel N, Wang S, … Olfson M (2016). Cannabis use and risk of psychiatric disorders: Prospective evidence from a US national longitudinal study. JAMA Psychiatry, 73(4), 388–395. doi: 10.1001/jamapsychiatry.2015.3229. [DOI] [PubMed] [Google Scholar]
- Bowden J, Hemani G, & Davey Smith G (2018). Invited commentary: Detecting individual and global horizontal pleiotropy in Mendelian randomization - a job for the humble heterogeneity statistic? American Journal of Epidemiology, 187(12), 2681–2685. doi: 10.1093/aje/kwy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brebner K, Wong TP, Liu L, Liu Y, Campsall P, Gray S, … Wang YT (2005). Nucleus accumbens long-term depression and the expression of behavioral sensitization. Science (New York, N.Y.), 310(5752), 1340–1343. doi: 10.1126/science.1116894. [DOI] [PubMed] [Google Scholar]
- Brook JS, Cohen P, & Brook DW (1998). Longitudinal study of co-occurring psychiatric disorders and substance use. Journal of the American Academy of Child & Adolescent Psychiatry, 37(3), 322–330. doi: 10.1097/00004583-199803000-00018. [DOI] [PubMed] [Google Scholar]
- Brown S, Inskip H, & Barraclough B (2000). Causes of the excess mortality of schizophrenia. The British Journal of Psychiatry, 177(3), 212–217. doi: 10.1192/bjp.177.3.212. [DOI] [PubMed] [Google Scholar]
- Conway KP, Swendsen J, Husky MM, He J-P, & Merikangas KR (2016). Association of lifetime mental disorders and subsequent alcohol and illicit drug use: Results from the national comorbidity survey-adolescent supplement. Journal of the American Academy of Child & Adolescent Psychiatry, 55(4), 280–288. doi: 10.1016/j.jaac.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Dalton EJ, Cate-Carter TD, Mundo E, Parikh SV, & Kennedy JL (2003). Suicide risk in bipolar patients: The role of co-morbid substance use disorders. Bipolar Disorders, 5(1), 58–61. doi: 10.1034/j.1399-5618.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- Davey Smith G, & Ebrahim S (2003). ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology, 32(1), 1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, … Neale BM (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics, 51(1), 63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, & Murray CJ (2002). Selected major risk factors and global and regional burden of disease. The Lancet, 360(9343), 1347–1360. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- Fluharty ME, Sallis H, & Munafò MR (2018). Investigating possible causal effects of externalizing behaviors on tobacco initiation: A Mendelian randomization analysis. Drug and Alcohol Dependence, 191, 338–342. doi: 10.1016/j.drugalcdep.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage SH, Jones HJ, Taylor AE, Burgess S, Zammit S, & Munafò MR (2017). Investigating causality in associations between smoking initiation and schizophrenia using Mendelian randomization. Scientific Reports, 7 (1), 1–8. doi: 10.1038/srep40653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg L, Barrowclough C, & Haddock G (2007). Reasons for increased substance use in psychosis. Clinical Psychology Review, 27(4), 494–510. doi: 10.1016/j.cpr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, … Tucker-Drob EM (2019). Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nature Human Behaviour, 3(5), 513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz SM, Pato CN, Medeiros H, Cavazos-Rehg P, Sobell JL, Knowles JA, … Pato MT (2014). Comorbidity of severe psychotic disorders with measures of substance use. JAMA Psychiatry, 71(3), 248–254. doi: 10.1001/jamapsychiatry.2013.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, … Haycock PC (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife, 7, e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks BM, Krueger RF, Iacono WG, McGue M, & Patrick CJ (2004). Family transmission and heritability of externalizing disorders: A twinfamily study. Archives of General Psychiatry, 61(9), 922–928. doi: 10.1001/archpsyc.61.9.922. [DOI] [PubMed] [Google Scholar]
- Hjorthøj C, Østergaard MLD, Benros ME, Toftdahl NG, Erlangsen A, Andersen JT, & Nordentoft M (2015). Association between alcohol and substance use disorders and all-cause and cause-specific mortality in schizophrenia, bipolar disorder, and unipolar depression: A nationwide, prospective, register-based study. The Lancet Psychiatry, 2(9), 801–808. doi: 10.1016/S2215-0366(15)00207-2. [DOI] [PubMed] [Google Scholar]
- Hodgson K, Coleman JR, Hagenaars SP, Purves KL, Glanville K, Choi SW, … Lewis CM (2020). Cannabis use, depression and self-harm: Phenotypic and genetic relationships. Addiction, 115(3), 482–492. doi: 10.1111/add.14845. [DOI] [PubMed] [Google Scholar]
- Jamuar SS, Schmitz-Abe K, D’Gama AM, Drottar M, Chan WM, Peeva M, … Yu TW (2017). Biallelic mutations in human DCC cause developmental split-brain syndrome. Nature Genetics, 49(4), 606–612. doi: 10.1038/ng.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JG, Cohen P, Pine DS, Klein DF, Kasen S, & Brook JS (2000). Association between cigarette smoking and anxiety disorders during adolescence and early adulthood. JAMA, 284(18), 2348–2351. doi: 10.1001/jama.284.18.2348. [DOI] [PubMed] [Google Scholar]
- Jung N, & Haucke V (2007). Clathrin-mediated endocytosis at synapses. Traffic (Copenhagen, Denmark), 8(9), 1129–1136. doi: 10.1111/j.1600-0854.2007.00595.x. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, & Roux A (2018). Mechanisms of clathrin-mediated endocytosis. Nature Reviews Molecular Cell Biology, 19(5), 313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- Kask J, Ekselius L, Brandt L, Kollia N, Ekbom A, & Papadopoulos FC (2016). Mortality in women with anorexia nervosa: The role of comorbid psychiatric disorders. Psychosomatic Medicine, 78(8), 910–919. doi: 10.1097/PSY.0000000000000342. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Knudsen GP, Røysamb E, Neale MC, & Reichborn-Kjennerud T (2011). The structure of genetic and environmental risk factors for syndromal and subsyndromal common DSM-IV axis I and all axis II disorders. American Journal of Psychiatry, 168(1), 29–39. doi: 10.1176/appi.ajp.2010.10030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khantzian EJ (1997). The self-medication hypothesis of substance use disorders: A reconsideration and recent applications. Harvard Review of Psychiatry, 4(5), 231–244. doi: 10.3109/10673229709030550. [DOI] [PubMed] [Google Scholar]
- King SM, Iacono WG, & McGue M (2004). Childhood externalizing and internalizing psychopathology in the prediction of early substance use. Addiction, 99(12), 1548–1559. doi: 10.1111/j.1360-0443.2004.00893.x. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 46(3), 310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov R, Krueger RF, Watson D, Achenbach TM, Althoff RR, Bagby RM, … Zimmerman M (2017). The Hierarchical Taxonomy of Psychopathology (HiTOP): A dimensional alternative to traditional nosologies. Journal of Abnormal Psychology, 126(4), 454–477. doi: 10.1037/abn0000258. [DOI] [PubMed] [Google Scholar]
- Krueger RF, & Markon KE (2006). Reinterpreting comorbidity: A model-based approach to understanding and classifying psychopathology. Annual Review of Clinical Psychology, 2, 111–133. doi: 10.1146/annurev.clinpsy.2.022305.095213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser K, Boyd JW, Woolhandler S, Himmelstein DU, McCormick D, & Bor DH (2000). Smoking and mental illness: A population-based prevalence study. JAMA, 284(20), 2606–2610. doi: 10.1001/jama.284.20.2606. [DOI] [PubMed] [Google Scholar]
- Lee PH, Anttila V, Won H, Feng YCA, Rosenthal J, Zhu Z, … Smoller JW (2019). Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell, 179(7), 1469–1482. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, … Fontana MA (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics, 50(8), 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnér RK, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, … Nivard MG (2019). Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nature Genetics, 51(2), 245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, … Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51(2), 237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M-T, Hinds DA, Tung JY, Franz C, Fan C-C, Wang Y, … Chen C-H (2017). Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nature Genetics, 49(1), 152–156. doi: 10.1038/ng.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolese HC, Malchy L, Negrete JC, Tempier R, & Gill K (2004). Drug and alcohol use among patients with schizophrenia and related psychoses: Levels and consequences. Schizophrenia Research, 67(2–3), 157–166. doi: 10.1016/S0920-9964(02)00523-6. [DOI] [PubMed] [Google Scholar]
- Miettunen J, Murray GK, Jones PB, Mäki P, Ebeling H, Taanila A, … Moilanen I (2014). Longitudinal associations between childhood and adulthood externalizing and internalizing psychopathology and adolescent substance use. Psychological Medicine, 44(8), 1727–1738. doi: 10.1017/S0033291713002328. [DOI] [PubMed] [Google Scholar]
- Mitchell ES, Conus N, & Kaput J (2014). B vitamin polymorphisms and behavior: Evidence of associations with neurodevelopment, depression, schizophrenia, bipolar disorder and cognitive decline. Neuroscience and Biobehavioral Reviews, 47, 307–320. doi: 10.1016/j.neubiorev.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Morón JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, & Devi LA (2007). Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: A proteomics study focusing on endocytic proteins. Molecular & Cellular Proteomics, 6(1), 29–42. doi: 10.1074/mcp.M600184-MCP200. [DOI] [PubMed] [Google Scholar]
- O’Connor LJ, & Price AL (2018). Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nature Genetics, 50(12), 1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, & Rockman MV (2013). The many faces of pleiotropy. Trends in Genetics, 29(2), 66–73. doi: 10.1016/j.tig.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, … Walters JTR (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nature Genetics, 50(3), 381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, … Vink JM (2018). GWAS Of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nature Neuroscience, 21(9), 1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, & Hinds DA (2016). Detection and interpretation of shared genetic influences on 42 human traits. Nature Genetics, 48(7), 709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenström T, Gjerde LC, Krueger RF, Aggen SH, Czajkowski NO, Gillespie NA, … Ystrom E (2019). Joint factorial structure of psychopathology and personality. Psychological Medicine, 49(13), 2158–2167. doi: 10.1017/S0033291718002982. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, … Kendler KS (2018). Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell, 173(7), 1705–1715.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, & Smoller JW (2013). Pleiotropy in complex traits: Challenges and strategies. Nature Reviews Genetics, 14(7), 483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, & Trubetskoy V, … The Bipolar Disorder Working Group of the Psychiatric Genetics Consortium (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nature Genetics, 51(5), 793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytnyk V, Leshchyns’ka I, & Schachner M (2017). Neural cell adhesion molecules of the immunoglobulin superfamily regulate synapse formation, maintenance, and function. Trends in Neurosciences, 40(5), 295–308. doi: 10.1016/j.tins.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Taylor AE, Fluharty ME, Bjørngaard JH, Gabrielsen ME, Skorpen F, Marioni RE, … Munafò MR (2014). Investigating the possible causal association of smoking with depression and anxiety using Mendelian randomisation meta-analysis: The CARTA consortium. BMJ Open, 4(10), e006141. doi: 10.1136/bmjopen-2014-006141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treur JL, Demontis D, Smith GD, Sallis H, Richardson TG, Wiers RW, … Munafò MR (2019). Investigating causality between liability to ADHD and substance use, and liability to substance use and ADHD risk, using Mendelian randomization. Addiction Biology, e12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Oever MC, Goriounova NA, Wan Li K, Van der Schors RC, Binnekade R, Schoffelmeer ANM, … De Vries TJ (2008). Prefrontal cortex AMPA receptor plasticity is crucial for cue-induced relapse to heroin-seeking. Nature Neuroscience, 11(9), 1053–1058. doi: 10.1038/nn.2165. [DOI] [PubMed] [Google Scholar]
- van Hulzen KJE, Scholz CJ, Franke B, Ripke S, Klein M, McQuillin A, … Reif A (2017). Genetic overlap between attention-deficit/hyperactivity disorder and bipolar disorder: Evidence from genome-wide association study meta-analysis. Biological Psychiatry, 82(9), 634–641. doi: 10.1016/j.biopsych.2016.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os J, Bak M, Hanssen M, Bijl RV, de Graaf R, & Verdoux H (2002). Cannabis use and psychosis: A longitudinal population-based study. American Journal of Epidemiology, 156(4), 319–327. doi: 10.1093/aje/kwf043. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Chen C-Y, Neale B, & Do R (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nature Genetics, 50(5), 693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI (2012). Model selection and psychological theory: A discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychological Methods, 17(2), 228–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, & Posthuma D (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1–11. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JR, Gaspar HA, … Bulik CM (2019). Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nature Genetics, 51(8), 1207–1214. doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AH, Kashan RS, Shpigel DM, Esan H, Taha F, Lee CJ, … Goodwin RD (2017). Depression and cigarette smoking behavior: A critical review of population-based studies. The American Journal of Drug and Alcohol Abuse, 43(4), 416–431. doi: 10.3109/00952990.2016.1171327. [DOI] [PubMed] [Google Scholar]
- Weiser M, & Noy S (2005). Interpreting the association between cannabis use and increased risk for schizophrenia. Dialogues in Clinical Neuroscience, 7(1), 81–85. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3181719/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman ER (2004). Poor mental health, depression, and associations with alcohol consumption, harm, and abuse in a national sample of young adults in college. The Journal of Nervous and Mental Disease, 192(4), 269–277. doi: 10.1097/01.nmd.0000120885.17362.94. [DOI] [PubMed] [Google Scholar]
- Wootton R, Richmond R, Stuijfzand B, Lawn R, Sallis H, Taylor G, … Munafò M (2019). Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: A Mendelian randomisation study. Psychological Medicine, 1–9. doi: 10.1017/S0033291719002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, & Abdellaoui A, … The Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50(5), 668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, & Price AL (2014). Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics, 46(2), 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Xu Y, Cai Z, Liu Q, Ma Y, Li AN, … Li MD (2020). Determination of shared genetic etiology and possible causal relations between tobacco smoking and depression. Psychological Medicine, 1–10. doi: 10.1017/S003329172000063X. [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah M-A, Keime C, Plewniak F, Davidson I, & Tora L (2010). SeqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Research, 39(6), e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, & Weedon MN, … the GIANT Consortium (2018). Meta-analysis of genome-wide association studies for height and body mass index in~ 700000 individuals of European ancestry. Human Molecular Genetics, 27(20), 3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Friedman NP, Miyake A, Willcutt EG, Corley RP, Haberstick BC, & Hewitt JK (2009). Behavioral disinhibition: Liability for externalizing spectrum disorders and its genetic and environmental relation to response inhibition across adolescence. Journal of Abnormal Psychology, 118(1), 117–130. doi: 10.1037/a0014657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


