Abstract
The soil bacterium Burkholderia pseudomallei is the causative agent of melioidosis and a significant cause of human morbidity and mortality in many tropical and subtropical countries. The species notoriously survives harsh environmental conditions but the genetic architecture for these adaptations remains unclear. Here we employed a powerful combination of genome-wide epistasis and co-selection studies (2,011 genomes), condition-wide transcriptome analyses (82 diverse conditions), and a gene knockout assay to uncover signals of “co-selection”—that is a combination of genetic markers that have been repeatedly selected together through B. pseudomallei evolution. These enabled us to identify 13,061 mutation pairs under co-selection in distinct genes and noncoding RNA. Genes under co-selection displayed marked expression correlation when B. pseudomallei was subjected to physical stress conditions, highlighting the conditions as one of the major evolutionary driving forces for this bacterium. We identified a putative adhesin (BPSL1661) as a hub of co-selection signals, experimentally confirmed a BPSL1661 role under nutrient deprivation, and explored the functional basis of co-selection gene network surrounding BPSL1661 in facilitating the bacterial survival under nutrient depletion. Our findings suggest that nutrient-limited conditions have been the common selection pressure acting on this species, and allelic variation of BPSL1661 may have promoted B. pseudomallei survival during harsh environmental conditions by facilitating bacterial adherence to different surfaces, cells, or living hosts.
Keywords: Burkholderia pseudomallei, co-selection study, nutrient depletion
Introduction
One of a long-standing research question in evolutionary biology is to understand how natural selection operates across the population. Common selective pressures likely result in consistent patterns across individuals regardless of the background population. An unbiased genome-wide scan for cooccurrence of genetic markers such as single-nucleotide polymorphisms (SNPs), indels, orthologous genes, or pathways can be used to identify functional patterns caused by shared selective pressures. Assuming a perfect linkage equilibrium where genetic markers are inherited independently and combined at random, a nonrandom cooccurrence of these markers likely suggests their interactions that confer a selective advantage and thus are co-selected together. Depending on types of selection, the frequency of co-selected markers may coincrease until they reach fixation (positive selection) or comaintain at low frequency with multiple alleles at selected sites (balancing selection). However, linkage equilibrium is rare with genetic markers often inherited together forming linkage disequilibrium (LD) blocks. These physical blocks can be broken down by recombination events, thereby enabling an opportunity to investigate selective pressures through co-selected markers when LD structure is carefully considered. Although recombination in bacteria does not occur at every generation, some species such as Campylobacter jejuni, Helicobacter pylori, Neisseria gonorrhoeae, Streptococcus pneumoniae, Vibrio parahaemolyticus, and Burkholderia species frequently recombine resulting in disruptions of LD blocks (Nandi et al. 2015; Arnold et al. 2018; Cui et al. 2020; Zhou et al. 2020). Identifications of co-selected genetic markers among these recombinogenic species have shed light on groups of interacting genes required for the species adaptation including antimicrobial resistance genes in host-restricted pathogens or genes essential for cell integrity and mobility in an environmental bacterium (Pensar et al. 2019; Schubert et al. 2019; Cui et al. 2020). These studies motivated us to search for patterns of co-selection in a recombinogenic bacterium, Burkholderia pseudomallei to investigate the selection pressures that have acted on this species, and key genes that allowed its adaptation.
The environmental bacterium B. pseudomallei has been increasingly recognized as an emerging human pathogen and a cause of melioidosis, a rapidly fatal infectious disease that annually affects and kills an estimated number of 165,000 and 89,000 patients, respectively (Limmathurotsakul et al. 2016). The bacterium can be isolated from the soil in many tropical and subtropical regions. It is most abundant at depths ≥ 10 cm from the surface but can move from deeper soil layers to the soil surface during the rainy season and multiply; this has been linked with an increase in disease incidence after heavy rainfalls (Inglis and Sagripanti 2006; Wiersinga et al. 2018). The presence of B. pseudomallei has been associated with nutrient-depleted soil and soil modified by long-term human activity (Coenye and Vandamme 2003; Limmathurotsakul et al. 2013; Baker et al. 2015; Ngamsang et al. 2015; Hantrakun et al. 2016; Manivanh et al. 2017); thereby connecting human manipulation of soil physicochemistry that promotes the abundance of this species. Burkholderiapseudomallei can survive extreme environmental conditions ranging from dry terrain in deserts (Yip et al. 2015) to distilled water (Pumpuang et al. 2011) (no nutrients); the latter ongoing experiment has been run for over 25 years. Such diverse and extreme environmental conditions likely impose unique selection pressures on B. pseudomallei but the key genetic factors that mediate these adaptations are unknown.
We applied a hypothesis-free co-selection analysis, known as genome-wide epistasis and co-selection study (GWES) (Pensar et al. 2019; Schubert et al. 2019; Cui et al. 2020) to detect co-selection signals in a collection of 1,136 B. pseudomallei isolates from 1935 to 2013 and validate our findings on a more recent collection of 875 isolates from 1976 to 2018 (supplementary fig. S1 and table S1, Supplementary Material online), enabling us to determine a set of SNPs under selective pressure. To explore the function of the co-selection signals, SNPs were mapped to genes and noncoding RNA (ncRNA) of B. pseudomallei reference genome K96243 resulting in co-selected gene–gene pairs, gene–nc RNA pairs, and ncRNA–ncRNA pairs. Using condition-wide gene expression data (Ooi et al. 2013; Mohd-Padil et al. 2017), we next searched for conditions at which co-selected gene–gene pairs displayed gene expression correlations, which likely reflect selective conditions B. pseudomallei has been subjected to. Finally, we used gene knockout assays to confirm the function of a gene under co-selection hotspot as well as the conditions that likely shaped B. pseudomallei evolution.
Results and Discussion
Detection of Co-selection Signals
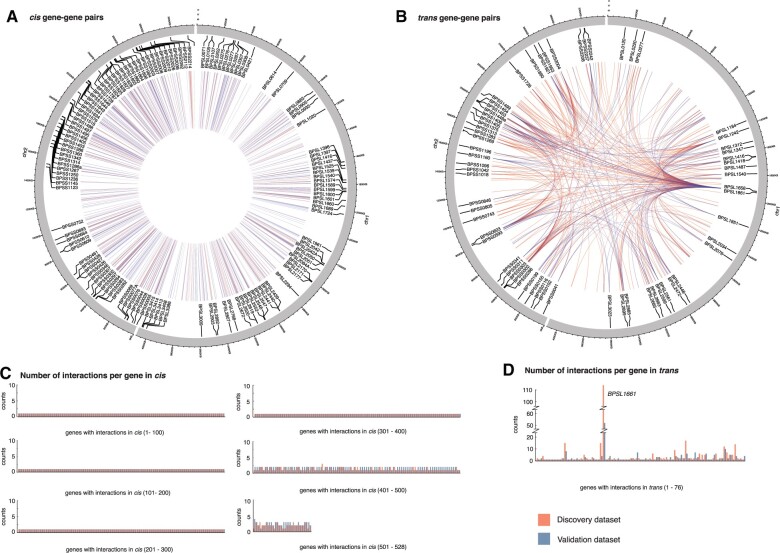
Our search for co-selection signals in the discovery data set resulted in 13,061 SNP–SNP pairs spanning chromosome I (5,550 pairs), chromosome II (7,309 pairs), and between chromosomes I and II (202 pairs); of which 8,035 pairs (61.5%) could be replicated in the validation data set (see Materials and Methods). Although the unique pairs observed in each data set could be due to different sampling timeframe or linkage structure, the congruence of the co-selection signals in the discovery and validation data sets (supplementary table S2, Supplementary Material online) demonstrates that these complex patterns are consistent through time. The co-selected SNPs spanned genes, ncRNA, and intergenic regions, each of which accounted for 69.5%, 6.8%, and 23.7%, respectively in the discovery data set, with similar ratio observed in the validation data set. Most of SNP–SNP pairs were located either within the same gene or in close physical proximity (supplementary fig. S2 and table S2, Supplementary Material online); an observation driven by LD. These pair were separated by small physical distances and are unlikely to have had recombination events between them, resulting in so-called LD-mediated links. Although pairs located in close proximity are most likely explained by LD structure, some of them may represent genuine cis interacting partners. These may include SNPs in regulatory regions upstream of genes, such as DNA binding sites for RNA polymerase and transcription factors, sites in adjacent coding sequences that form protein complex, or ncRNA that predominantly act in cis. As these interacting partners are often cotranscribed and translated in bacteria, we thus grouped co-selected pairs into cis and trans based on the length of transcription fragments reported in B. pseudomallei (Ooi et al. 2013). Here, pairs mapped to different genes or ncRNA but located within 7.68 kb (representing 95th percentile of polycistronic mRNAs) were termed cis pairs, whereas pairs located further than 7.68 kb apart or on separate chromosome were termed trans pairs. This resulted in 334 cis and 252 trans gene–gene pairs, 64 cis and 11 trans gene–ncRNA pairs, and 19 cis ncRNA–ncRNA pairs from the discovery data; of which 383 pairs (56.3%) were replicated in the validation data (fig. 1 and supplementary fig. S3 and table S3, Supplementary Material online). Despite cis pairs being more prevalent, the number of interacting partners per gene was fewer than the number of trans interacting partners (a mean of partners per cis-gene = 1.11, a mean of partners per trans-gene = 2.50) with the highest number observed in BPSL1661 (trans-gene partners = 104 [discovery data], and 52 [validation data]).
Fig. 1.
Co-selected gene–gene pairs in discovery and validation data sets. Circos diagrams showing co-selected gene–gene pairs in cis (A) and trans (B). Any pairs located within 7.68 kb (95th percentile of transcription fragments) were categorized as cis, whereas those located further apart were grouped as trans. Bar charts summarizing the number of interacting partners per gene for cis (C) and trans (D) linkage. Data from discovery and validation data sets were highlighted in red and blue, respectively.
Moderate Functional Conservation of Genes under Co-selection
We assigned functional annotations to genes identified in both cis and trans gene–gene pairs. For both the discovery and validation data set, the majority of the co-selected genes were functionally assigned as cell envelope (21.9%), or transport/binding protein (10.6%), whereas many were left as uncharacterized or hypothetical proteins (26.7%) (supplementary fig. S4A, Supplementary Material online). We observed that a high proportion of the co-selected gene–gene pairs share the same functional annotations (51.3% of cis and 19.9% trans gene–gene pairs). To test whether this observation was driven by chance, we compared the number of pairs under the same or different functional annotations between 100 randomized and real data sets (see Materials and Methods). Excluding pairs with ambiguous annotations, functional conservation of cis gene–gene pairs for both discovery and validation data set were within the range random expectation (supplementary fig. S4B, Supplementary Material online). However, conservation of functional annotation of trans gene–gene pairs was greater than the random expectation for the discovery but not the validation data set (supplementary fig. S4C, Supplementary Material online). The inconsistency in results could be caused by a small sample size after an exclusion of genes with uncharacterized function. Nevertheless, our results indicate that co-selection of genes, at least in trans of the discovery data set, is driven by gene functions.
Expression Patterns of Co-selected Gene–Gene Pairs
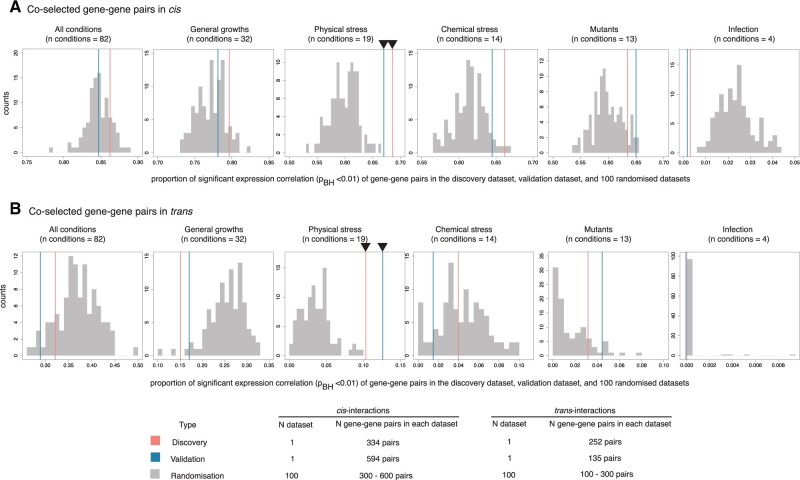
We next sought to identify the conditions that might have driven the selective pressure resulted in observed patterns of co-selected genes. We used B. pseudomallei whole-genome tiling microarray expression data generated by Ooi et al. (2013), which assayed under a broad spectrum of conditions and exposure, including general growth (32 conditions), exposure to physicochemical stress (33 conditions), invasion assays (4 conditions), and defined genetic mutants (13 conditions) (supplementary fig. S1, Supplementary Material online). Here, genes expressed in ≥70 conditions are defined as being constitutively expressed as in Ooi et al. (2013). Approximately 22.4% of the genes that were detected in co-selection pairs in the discovery analysis or validation analysis, or both, were constitutively expressed compared with 39.5% of genes that were not part of any co-selection pairs (two-tailed Fisher’s exact test P = 5.74 × 10−7), indicating that co-selection signals were more strongly associated with condition-specific genes than those constitutively expressed. We next tested if the real data set showed a greater proportion of coexpressed genes compared with 100 randomized data sets (fig. 2). In addition, we examined under which conditions such coexpression patterns were observed. Using normalized gene expression profiles (Ooi et al. 2013), Pearson’s correlation analysis was performed for each gene–gene pair with transcription data from all conditions tested as well as from subsets of those conditions. Except for physical stress conditions, the proportion of coexpressed gene pairs from the real discovery and validation data sets either fell within or lower than the range of 100 randomized controls (significant expression correlation at Benjamini–Hochberg adjusted P value < 0.01). The results were consistent for cis and trans pairs, suggesting that physical stress has largely shaped the co-selection patterns at the time scale considered for this population. Here, physical stress conditions included temperature stress, osmotic stress, UV irradiation, and nutrient deprivation; the environmental conditions B. pseudomallei is regularly or seasonally exposed to (Limmathurotsakul et al. 2013; Baker et al. 2015; Ngamsang et al. 2015; Hantrakun et al. 2016; Manivanh et al. 2017; Bulterys et al. 2018). However, it should be cautioned that the results from trans gene–gene pairs were not completely independent but heavily driven by a co-selection hotspot, BPSL1661, which were linked to many other genes.
Fig. 2.
Expression correlation of co-selected gene–gene pairs. (A and B) Histograms represent the proportion of gene pairs with significant expression correlations for the cis and trans interactions, respectively. The expression conditions tested (from left to right) include all conditions (n = 82), general growth (n = 32), physical stress (n = 19), chemical stress (n = 14), mutants (n = 13), and infection (n = 4). For all plots, horizontal axis denotes the proportion of gene pairs with significant expression correlation (Benjamini–Hochberg adjusted P value <0.01) for the true co-selection signals found in discovery (red), validation (blue), and 100 randomized data sets (gray). Black triangles mark the conditions at which real genes under co-selection display greater proportion of significant expression correlation than 100 randomized controls.
Functional Characterization of Co-selection Hotspot
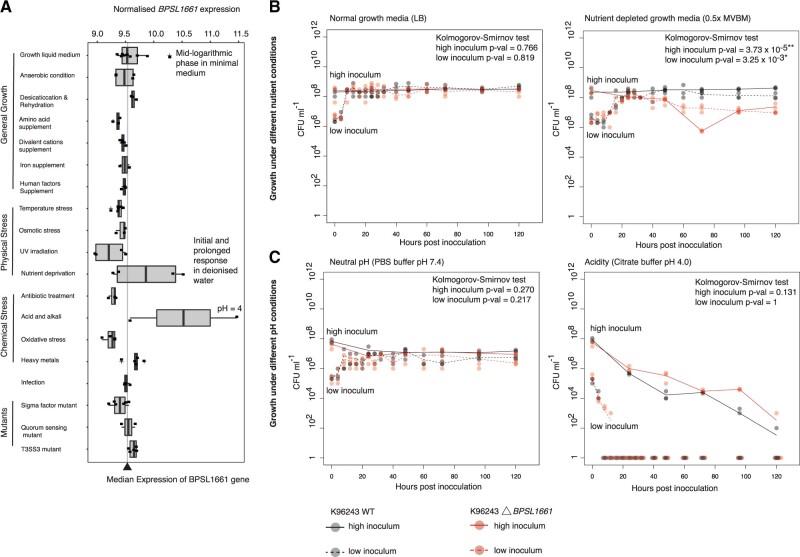
A putative adhesin gene BPSL1661 was the hotspot of the largest co-selected gene network for both discovery and validation data set (fig. 1B and D and supplementary fig. S3, Supplementary Material online). BPSL1661 codes for a secreted protein with a size ranging from 2,594 to 3,230 amino acids (approximately 325 kDa) in different isolates. The gene is located in a large highly variable genomic region termed genomic island 8 (GI8) (Tuanyok et al. 2008), proposed to be acquired through horizontal gene transfer (Holden et al. 2004). This genetic mobility possibly contributes to the presence of multiple BPSL1661 alleles observed in our study. We detected six major alleles of BPSL1661 (n ≥ 5 isolates, see Materials and Methods) consistent with previous reports on multiple protein epitopes (Suwannasaen et al. 2011; Kohler et al. 2016). All BPSL1661 alleles share a conserved hemolysin-type calcium-binding domain which is common in proteins secreted through a type I secretion system, and two copies of the VCBS domain (a repeat domain found in Vibrio, Colwellia, Bradyrhizobium, and Shewanella) known to be involved in cell adhesion. Variations in the presence of bacterial Ig and flagellin domains in BPSL1661 were observed in the study population. Interestingly, previous studies reported heterogeneity in human immune responses to polypeptides generated from different BPSL1661 alleles, ranging from null to strong antibody responses (Felgner et al. 2009; Suwannasaen et al. 2011; Kohler et al. 2016). Such disparity in host recognition of different BPSL1661 alleles potentially suggests that the protein may not principally function in host cell invasion but play other significant roles in B. pseudomallei survival. Transcription assays further revealed that BPSL1661 is downregulated during infection but upregulated in acidic conditions (pH 4), mid-logarithmic phase in minimal media, and nutrient deprivation (fig. 3A); further indicating a role of BPSL1661 in adaptation to environmental stress.
Fig. 3.
Functional characterization of BPSL1661. (A) Expression profile of BPSL1661 across different conditions from Ooi et al. (2013), highlighting the gene upregulation during nutrient limited condition and high acidity. (B) and (C) represent growth- and stationary-phase survival of wild-type and BPSL1661 knockout mutant from K96243 reference strain under changes in nutrition (B) and pH (C). Bacterial survival post inoculation at low (106 CFU ml−1) and high (108 CFU ml−1) concentration was determined as bacterial colony forming unit (CFU) at different time intervals. Three replicates were taken at each timepoint. Black and red dots denote observations from wild-type and BPSL1661 mutant, respectively. Dotted and solid line represent profile of low and high bacterial inoculum, respectively. Difference in growth profiles between wild-type and mutant was measured using two-sided Kolmogorov–Smirnov test.
To better understand the function of BPSL1661, we knocked out the BPSL1661 gene in the K96243 strain and compared the number of live cells of the mutant against the wildtype under the conditions BPSL1661 was upregulated (fig. 3B and C), for 120 h. We observed no significant differences in bacterial survival in nutrient-rich growth media, neutral pH (pH 7.4), or acidic conditions (pH 4). This could be due to functional redundancy that compensates for the loss of a single gene function. However, the BPSL1661 mutant showed a significant reduction in stationary-phase survival compared with the wildtype under nutrient-limited conditions (two-sided Kolmogorov–Smirnov test P value = 3.73 × 10−5 and 3.25 × 10−3 for high and low bacterial inoculum, respectively); confirming an essential role of BPSL1661 under nutrient deprivation. Our observation of BPSL1661 as a hotspot may imply that nutrient depletion is one of the major selective pressures underlying the co-selection patterns. This finding is also consistent with environmental sampling studies in Southeast Asia and Australia which have reported that the bacterium is commonly found in nutrient-depleted soils (Baker et al. 2015; Hantrakun et al. 2016).
The maintenance of different BPSL1661 alleles in the population likely suggests that the gene has been under balancing selection. Although lower nutrient abundance appears to be a common feature across melioidosis endemic areas; the soil physiochemical properties, microbial diversity, temporal disturbances such as monsoon seasons and anthropogenic activities that alter the environmental conditions vary greatly (Kaestli et al. 2015; Musa et al. 2016; Ribolzi et al. 2016; Manivanh et al. 2017; Goodrick et al. 2018). These factors create patterns of spatial and temporal heterogeneity to which B. pseudomallei has adapted and possibly has led to the coexistence of multiple BPSL1661 alleles detected in this study. We also noted geographical differences in BPSL1661 allele frequencies. An allele harboring a flagellin domain (here denoted as allele A, supplementary fig. S5A, Supplementary Material online) was detected at lower frequency in Australia (28.7%), at moderate frequency in the Malay Peninsula (37.6% from Malaysia and Singapore) and higher frequency in the countries bordered by the great Mekong river (59.1% from Thailand, Laos, Cambodia, and Vietnam) (supplementary fig. S5B, Supplementary Material online). Such a difference in allele frequencies could be either driven by different local selection pressures, or by genetic drift (or both). Due to its horizontal mode of inheritance, an ancestral history of each BPSL1661 allele could not be reliably reconstructed. Functional characterization of different BPSL1661 alleles also warrants further future studies.
BPSL1661 Co-selection Network and Putative Bacterial Response under Low Nutrient Abundance
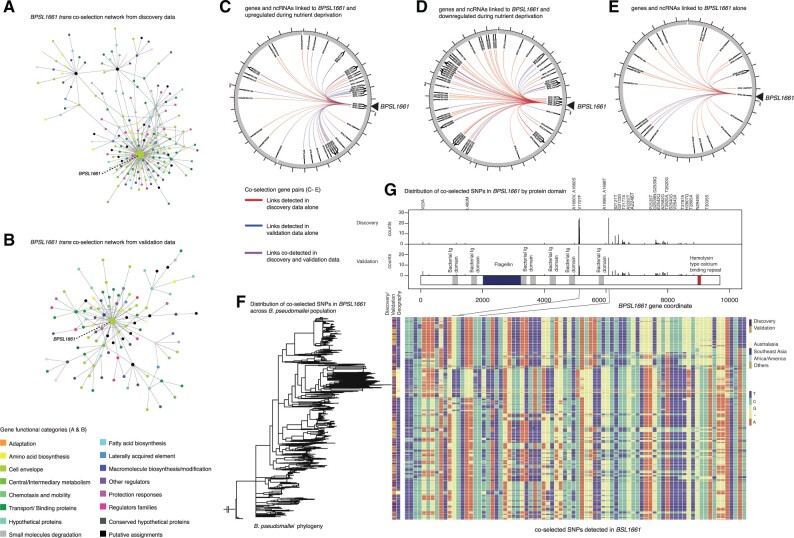
We considered genes and ncRNA co-selected with BPSL1661 in both the discovery (n gene pairs = 105, n ncRNA pairs = 5) and validation data set (n gene pairs = 53, n ncRNA pairs = 2) totaling 136 pairs, of which 29 pairs are shared in both data sets (fig. 4 and supplementary table S4, Supplementary Material online). The majority of these gene and ncRNA pairs were linked to BPSL1661 in trans except for an outer membrane protein BPSL1660 which paired in cis. During nutrient-depleted conditions (Ooi et al. 2013; Mohd-Padil et al. 2017), 43/129 of the trans gene pairs and 1/6 of ncRNA pairs were upregulated with BPSL1661, whereas 64/129 of the trans gene pairs and 2/6 of ncRNA pairs were downregulated, respectively. Many of these genes are predicted to encode proteins that participate in alternative metabolic pathways, energy conservation, uptake of external carbon source, cellular signaling, and transcriptional regulation (supplementary table S4, Supplementary Material online). A pyrophosphohydrolase (spoT or BPSL2561) is upregulated during nutrient starvation and is known to have a dual function to synthesize and hydrolyze guanosine tetra- and pentaphosphates (ppGpp) (Müller et al. 2012). In bacteria, ppGpp serves as a mediator in nutritional surveillance, coordinating a variety of cellular activities in response to changes in nutrient availability (Wang et al. 2007). In B. pseudomallei, deletion of ppGpp synthetase and hydrolase led to reduced survival during stationary phase compared with wildtype (Müller et al. 2012). It is possible that B. pseudomallei switches to alternative carbon sources to maintain cellular energy when complex sources such as glucose are not available. Genes encoding a C4-carboxylate transport transcription regulation protein (BPSL0427), a malate synthase (BPSL2192), and a glycogen branching enzyme (BPSL2076) were co-selected with BPSL1661 but had different expression profiles under nutrient-limited conditions. A transport transcription regulation protein (BPSL0427) was upregulated under nutrient deprivation. Its homolog was shown to facilitate the cellular uptake of four carbon compounds such as aspartate, fumarate, and succinate when common carbon sources such as glucose are scarce (Wösten et al. 2017). Another upregulated gene, malate synthase (BPSL2192) is a key enzyme involved in the glyoxylate cycle and was also shown to be essential to Mycobacterium tuberculosis survival under nutrient starvation (Puckett et al. 2017). Its expression allows cells to utilize two carbon compounds to sustain carbon requirement in the absence of glucose. On the contrary, a glycogen branching enzyme (BPSL2076) was co-selected with BPSL1661 but downregulated during nutrient depletion. A homolog of glycogen branching enzyme is known to facilitate glucose conversion into long-term glycogen storage when there are excessive carbon sources (Wang et al. 2019). The co-selection of spoT, BPSL0427, BPSL2192, and BPSL2076 with BPSL1661 may reflect the energy balance of cells under ranges of nutritional conditions through their evolutionary timeline. We also observed many ncRNA, transcription regulators, and DNA-binding proteins co-selected with BPSL1661. These genes and ncRNA may further regulate downstream responses under changes in nutrient abundance. Together, the BPSL1661 co-selection network seems to suggest that B. pseudomallei has adapted to survive nutrient-limited conditions and/or hostile environments.
Fig. 4.
BPSL1661 co-selected gene and ncRNA network. (A) and (B) represent networks of trans interacting co-selected gene–gene pairs in the discovery and validation data sets, respectively. Each node denotes a gene under co-selection with the node size proportional to numbers of pairs linked to the gene, and color coded by the gene functional category. For both discovery and validation data sets, BPSL1661 consistently acts as hub of the trans co-selected gene network. (C), (D), and (E) summarize genes and ncRNA co-selected with BPSL1661 in both discovery and validation data set: genes and ncRNA co-selected and upregulated under nutrient deprivation (C); genes and ncRNA co-selected but downregulated under nutrient deprivation (D); and genes and ncRNA that are co-selected alone (E). Links identified from the discovery data alone, validation data alone, and both data are colored as red, blue, and purple, respectively. (F) Distribution of BPSL1661 SNPs (rows) across different population (columns). The estimated phylogeny is shown on the left with the first two columns labeled by type of data (discovery or validation data set) and geographical origins of isolates, respectively. The remaining columns demonstrate all nucleotide variants detected in BPSL1661. Variants coincide with peaks of co-selection signals were marked, one of which correspond to V1707F substitution.
Conclusion
This study is the first, in our knowledge, to deploy an integrated approach of GWES, transcriptomic analyses and knockout assays to understand the evolution and unique selective pressures acting on a microorganism. Although GWES detected signals for co-selected loci, transcriptomic data provided condition-dependent information on which selection pressures may have acted upon the detected loci. Nevertheless, our study has some limitations. First, our GWES focused only on nucleotide polymorphisms found by comparison to the B. pseudomallei K96243 reference genome. Co-selected loci on other types of structural variants including indels, genomic inversions, gene duplication, or horizontally acquired genes absent in K96342 will be missed from this analysis. Second, the condition-wide transcriptomic data employed in this study was generated by a microarray platform. A change in the resolution from nucleotide polymorphisms employed in GWES to a gene level used in microarray has led to a loss of signals as only SNPs mapped to genes (69.5%) could be interpreted by gene expression analysis. Because multiple SNPs could be mapped to a single gene, it is possible that each of these SNPs could lead to different product of gene expression. This information is not available with microarray platform. It is also possible that other transcriptional conditions the bacterium is exposed to in its native niche are missing. Together, the incomplete genomic and transcriptomic data warrant further studies to cover more complex genomic variants, and broader transcriptional conditions with higher genetic resolution.
Despite limitations, our integration of data offers stringent predictions on which genes are key to B. pseudomallei survival under specific conditions. In particular, the putative adhesin BPSL1661 was identified as a hotspot of the co-selection map. Our gene knockout experiment confirmed that the gene is essential for survival under nutrient deprivation. This is consistent with the soil conditions in which B. pseudomallei are commonly found and provides an evolutionary evidence that B. pseudomallei has been adapted to nutrient-depleted environments. It is possible that different BPSL1661 alleles may facilitate bacterial adherence to different surfaces, cells, or hosts. This attachment could lead to biofilm formation or the bacterium being internalized by host cells, both of which are known bacterial strategies to withstand nutrient deprivation (Brown et al. 2008; Petrova and Sauer 2012; Chomkatekaew et al. 2020). Our study has a strong implication that the presence of B. pseudomallei in nutrient-depleted soil may define geographical regions where humans are at risk of melioidosis. It is thus necessary to improve environmental health to assist melioidosis prevention.
Materials and Methods
Study Design
We conducted three complementary tests: a co-selection analysis to scan for any SNP–SNP pairs that are mutually detected more frequently than expected as a result of selection pressures; a condition-wide transcriptome analysis to identify conditions under which co-selected gene–gene pairs likely operate; and a gene knockout assay to confirm the function of a hotspot for co-selection signals.
Whole-Genome Sequencing Collections, Mapping, and Annotation
We sought B. pseudomallei whole-genome sequences from the public database (Holden et al. 2004; Hayden et al. 2012; Price et al. 2013; Sahl et al. 2013; Daligault et al. 2014; Bugrysheva et al. 2015; Chen et al. 2015; Hsueh et al. 2015; Johnson, Baker, et al. 2015; Johnson, Bishop-Lilly, et al. 2015; McRobb et al. 2015; Nandi et al. 2015; Sidjabat et al. 2015; Song et al. 2015; Spring-Pearson et al. 2015; Viberg et al. 2015; Chapple et al. 2016; Price et al. 2016; Chewapreecha et al. 2017; Chewapreecha et al. 2019) and combined these with newly sequenced genomes totaling 2,011, and further divided them into a discovery data set (1,136 isolate genomes) and a validation data set (875 isolate genomes) (supplementary fig. S1A and B, Supplementary Material online). Their accession numbers are tabulated in supplementary table S1, Supplementary Material online. The discovery data set came from an older collection (1935–2013), whereas the validation data set represented a recently sequenced collection (1976–2018). The two data sets overlap geographically, spanning two major melioidosis endemic regions of Southeast Asia and Australia. These isolates came from environmental, animal, and human sources with the latter constituted the larger proportion due to availability of microbiology laboratories embedded in the clinical settings. However, over 91% of human cases represent recent acquisition from the environmental sources (Currie et al. 2000; Wiersinga et al. 2012, 2018), thereby reducing the chance of coevolutionary signals being shaped by human infection alone. For newly sequenced data, DNA libraries were sequenced on an Illumina Hiseq2000 with 100-cycle paired-end runs. Short reads were mapped to the reference B.pseudomallei genome K96243 using SMALT 0.7.6 (Ponstingl and Ning 2010). The SNPs were called as in Chewapreecha et al. (2017). For the discovery data set (n = 1,136), the alignment contained 389,476 SNPs, of which 206,019 and 183,457 were located in chromosomes I and II, respectively. For the validation data set (n = 875), the alignment contained 285,543 SNPs, of which 150,499 and 135,044 SNPs were located in chromosomes I and II, respectively. The genome K96243 was reannotated using curated genes, ncRNA, and functional predictions obtained from transcriptional assays described in Ooi et al. (2013) and Mohd-Padil et al. (2017) (supplementary fig. S1C and D, Supplementary Material online).
Co-selection Tests
Co-selection analysis was separately performed on the sequence alignment of discovery and validation data sets using the mutual information-based GWES tool SpydrPick (Pensar et al. 2019) (supplementary table S2, Supplementary Material online). SNPs with minor allele frequency greater than 1% and gap frequency smaller than 15% were included in the analysis. To adjust for the population structure, sequence reweighting was applied using the default similarity threshold of 0.10. Direct links for which the mutual information exceeded the extreme outlier threshold, also after removing the influence of gaps, were selected for further examination. To discriminate signals influenced by the LD structure from the co-selection signals, SNP–SNP pairs were further categorized into cis or trans interaction based on the length of transcription fragments reported in B. pseudomallei (Ooi et al. 2013). Any pairs located on different genes or ncRNA but within 7.68 kb (representing 95th percentile of polycistronic mRNAs) were termed cis pairs, whereas pairs located further than 7.68 kb apart or on separate chromosome were termed trans pairs (fig. 1). These SNPs were mapped to genes and ncRNA from the curated B. pseudomallei reference genome K96243, resulting in cis or trans co-selection of gene–gene, gene–ncRNA, and ncRNA–ncRNA pairs (supplementary table S3 and S4, Supplementary Material online). SNP–SNP pairs located within the same gene, or ncRNA were removed for this analysis.
Generation of Randomized Gene–Gene Cohorts
A hundred randomized data sets were separately generated for cis and trans interactions by randomly pooling any pairs of genes from the reference B.pseudomallei genome K96243. Here, cis randomized data sets were created from pairs located on the same chromosome and <7.68 kb apart, whereas trans randomized data sets were based on genes located >7.68 kb apart or on a different chromosome. The size of each randomized data set matched the size of real discovery and validation data set. For cis gene–gene interactions (n discovery = 334 pairs, n validation = 594 pairs), the sample size of 100 randomized controls ranged between 300 and 600 pairs. For trans gene–gene interactions (n discovery = 252 pairs, n validation = 135 pairs), the number of pairs in the randomized controls ranged between 100 and 300 pairs. This allowed us to generate a distribution of random expectation, which can be used to test the hypotheses for functional conservation and elevated expression correlation of genes under co-selection.
Functional Classification of Genes under Co-selection
Different gene functional categories including a curated Riley’s classification, clusters of orthologous groups, KEGG Pathway (Kanehisa and Sato 2020), and gene ontology (GO terms) (The Gene Ontology Consortium 2019) were assigned to each gene–gene pair. The final analysis was focused on a curated Riley’s classification as it covered more genes detected for co-selection than other classification systems. For both discovery and validation data sets, we searched for enrichment of gene functions among co-selected genes against their distribution in K96243 genome using two-sided Fisher’s exact test (R function fisher.test()) while controlling for false positive from multiple testing using Benjamini–Hochberg method (Benjamini and Hochberg 1995).
To test whether genes under co-selection display functional conservation, we measured the proportion of gene pairs that shared the same functional annotation in the real data sets and compared the observations to the random expectation generated from 100 randomized controls (supplementary fig. S4, Supplementary Material online). Pairs comprising ambiguous gene annotations including uncharacterized or hypothetical proteins were removed from the analysis.
Expression Patterns of Genes and ncRNA under Co-selection
We used B. pseudomallei condition-specific expression comprising 165 array profiles generated from Ooi et al. to elucidate the function of co-selected gene–gene pairs. The data had been log-transformed to fit a Gaussian distribution. Expression correlations between gene–gene pairs were defined by Pearson correlation using R function cor.test(gene1, gene2, method = “pearson”). We applied Benjamini–Hochberg adjusted correction for multiple testing. Expression profiles were categorized into five major categories spanning general growth, physical stresses, chemical stresses, infection, and mutant conditions. For all conditions, and each of five major expression condition; we compared the proportion of pairs with significant expression correlation (Benjamini–Hochberg adjusted P value < 0.01) from the real co-selected gene–gene pairs against the distribution of random expectation generated from 100 randomized controls (fig. 2). Expression profile of BPSL1661 was obtained from Ooi et al. and cross-checked for consistency with (Rodrigues et al. 2006; Chin et al. 2015; Jitprasutwit et al. 2020) where conditions were overlapped.
Additional pattern of ncRNA expression under nutrient limitation was sought from Mohd-Padil et al. (2017). The authors measured and compared ncRNA expressed when B. pseudomallei was subjected to nutrient rich BHIB media and nutrient-depleted M9 condition.
Genetic Variations in BPSL1661 and Their Distribution across the Core Genome Phylogeny
Where complete genomes were available, BLAT v. 36 (Kent 2002) was used to locate the position of BPSL1661 homolog and further confirmed with genome annotations. Illumina-sequenced short reads were assembled as in Chewapreecha et al. (2019) and annotated using Prokka v.1.14.5 (Seemann 2014). Coding sequence of BPSL1661 was identified using BLAT and confirmed with gene annotation. BPSL1661 sequences from all genomes were aligned using MAFFT v.7.407 (Katoh 2002) and assigned into different clusters using CD-HIT-EST v.4.8.1 (Fu et al. 2012) with sequence identity threshold of 0.9. Here, any clusters with ≥5 members were considered representative alleles, resulted in six BPSL1661 alleles across the data sets. Protein domains of each BPSL1661 allele were sought from CDD/SPARCLE v.3.17 conserved domain database (Lu et al. 2020) (supplementary fig. S5, Supplementary Material online).
To investigate the distribution of BPSL1661 variants across B. pseudomallei population, core-genome phylogeny was constructed from core genome SNP alignment. SNPs were called from K96243 mapped genome alignment using SNP sites v.2.5.1 (Page et al. 2016) excluding sites associated with mobile genetic elements (Tuanyok et al. 2008). We next estimated a maximum-likelihood tree using IQ-TREE v.1.6.10 (Minh et al. 2020) using General Time Reversible + Gamma distribution model of nucleotide substitution with default heuristic search options and 1,000 bootstraps (Minh et al. 2013) (supplementary fig. S5, Supplementary Material online).
BPSL1661 Functional Characterization
The hub of co-selection signals, BPSL1661 was next functionally characterized. Bacterial strains, plasmids, and oligonucleotides used in this study are listed in supplementary tables S5 and S6, Supplementary Material online. GF-1 bacterial gDNA extraction kit and deoxynucleotide triphosphates were purchased from Vivantis; Platinum DNA Taq polymerase from Invitrogen; pGEM-T Easy vector systems from Promega; KOD Plus DNA polymerase from Toyobo; restriction enzymes from New England BioLabs; QIAquick Gel Extraction kit, MinElute PCR purification kit from Qiagen; Ampicillin (Ap), Kanamycin (Km), and Gentamycin (Gm) were purchased from Sigma, and Isopropyl β-d-1-thiogalactopyranoside (IPTG), 5-bromo-4-chloro-3-indolyl-β-d-glactopyranoside (X-Gal), and 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) were purchased from Gold Biotechnology.
The culture of B. pseudomallei K96243 wild-type, B. pseudomallei BPSL1661 clean mutant, and the Escherichia coli strains used for construction of the B. pseudomallei mutant were routinely grown in Luria-Bertani (LB) and Luria-Lennox (LB, low salt) medium (Sigma, USA), at 37°C with 200 rpm agitation. When necessary, the medium was supplemented with antibiotics, chemicals, and chromogens at the concentrations of 100 µg ml−1 Ap, 35 µg ml−1 Km, 0.1 mM IPTG, 50 µg ml−1 X-Gal for E. coli; 5 µg ml−1 Gm, 1,000 µg ml−1 Km, 0.1 mM IPTG, 50 µg ml−1 X-Gluc for B. pseudomallei.
Construction of the BPSL1661 Clean Deletion Mutant
The nucleotide sequence encoding (BPS_RS08795 [BPSL1661]) gene (GenBank accession no. WP_045606470.1) of B. pseudomallei K96243 was used to design primers for clean deletion using Primer-BLAST program (Ye et al. 2012). The BPSL1661 clean mutant (ΔBPSL1661) (supplementary fig. S6, Supplementary Material online) was constructed from B. pseudomallei K96243 by double-crossover allelic exchange as described previously (López et al. 2009). Briefly, the upstream and downstream DNA fragments of BPSL1661 were amplified using BPSL1661_PFup/PRup and BPSL1661_PFdown/PRdown primers and then ligated. The ligated amplicons were cloned into TA cloning vector pGEM-T Easy (Promega, USA) and check for its correct insert size before subcloned into an allelic exchange plasmid pEXKM5 (López et al. 2009). The recombinant pEXKM5 plasmids were introduced into B. pseudomallei K96243 by biparental conjugation using conjugal E. coli S17-llpir (Simon et al. 1983). Merodiploid was selected on LB medium supplemented with X-Gluc and Kanamycin that appeared as pale blue colonies. To obtain the clean deletion ΔBPSL1661 mutant, merodiploid colonies were cultured in LB medium to reach stationary phase then subculture into YT broth supplemented with 10% sucrose for overnight and spread on LB medium with 15% sucrose. Suspected B. pseudomallei colonies was verified by PCR and confirmed by DNA sequencing of the region flanking BPSL1661.
BPSL1661 Condition-Specific Assays
Guided by expression profile of BPSL1661 in Ooi et al., the growth of wild-type and ΔBPSL1661 were compared by enumerating the bacterium grown under normal growth (LB broth), nutrient-depleted condition (modified Vogel and Bonner’s medium, MVBM), neutral pH, and acidic pH. The bacterium was grown on Ashdown’s selective agar (Ashdown 1979) before transferring to grow under each condition.
We followed Ooi et al. for normal growth condition using LB broth. We artificially induced nutrient-limited condition in vitro by replacing glucose-limited media (six carbon source) with glycerol (three carbon source) in MVBM. Moreover, MVBM (Lam et al. 1980) further forced cell starvation by inducing biofilm formation. Nutrients will be consumed by cells located on the periphery of the biofilm clusters leading to reduced level of nutrients diffused to the inner cells. Both wild-type and ΔBPSL1661 were grown on 0.05% glucose in 0.5 × MVBM [0.05%G, 0.5×MVBM]) at 37 °C with 200 rpm shaking for overnight. The culture was adjusted and measured an optical density (OD) at OD 600 nm to be 0.2 (OD600 = 0.2) and then inoculated into 10 ml prewarmed 0.05% G, 0.5×MVBM medium at 37 °C with 100 rpm shaking.
Growth in neutral pH was conducted in 10-fold diluted phosphate-buffered saline (PBS) pH 7.4. Growth in a low pH was done as described previously (Inglis and Sagripanti 2006). For both condition, bacteria were cultured for overnight in LB broth at 37 °C with 200 rpm shaking and subcultured in new fresh medium until log phase. Bacterial cells were washed in 1×PBS buffer pH 7.4 or 1×citrate buffer pH 4.0 for three times by centrifugation (8000 × g for 5 min) and resuspended in 10 ml of each buffer. For all conditions, we observed bacterial initial growth and stationary-phase survival for both low (106 CFU ml−1) and high (108 CFU ml−1) inoculum. The colony forming unit (CFU) post inoculation was counted at different time interval. All assays involved three replicates that were independently prepared, cultured, and treated. Difference in growth profile between wild-type and ΔBPSL1661 for each condition was compared by a nonparametric Kolmogorov–Smirnov test (Berger and Zhou 2014) (fig. 3).
Visualization
Visualization of phylogenetic trees was performed in Phandango (Hadfield et al. 2018). Figures were plotted using R (R Core Team 2017) and R applications (Yu et al. 2018).
Statistics and Reproducibility
No sample sizes were predetermined. For categorical data, two-sided Fisher’s exact tests were employed to compare differences between each group. For continuous data, nonparametric Mann–Whitney U tests and a parametric Pearson correlation were used to compare the intergroup distribution and the correlation of the expression levels between each co-selected gene–gene pair, respectively. Where appropriate, we used Benjamini–Hochberg procedure to adjust P value for multiple comparisons.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Dr Alain Pierret, Dr Olivier Ribolzi, and Dr David Dance for insightful discussion on the environmental implication of the study. C.C. was funded by Wellcome International Intermediate Fellowship (216457/Z/19/Z), Sanger International Fellowship, and Thailand National Science and Technology Development Agency. J.C. was funded by the ERC (742158). S.C., C.P., and R.W.S. were funded by Targeted Research Grant, Faculty of Medicine, Khon Kaen University. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust. This project was also funded by a grant awarded to the Wellcome Trust Sanger Institute (098051) and the Wellcome Thailand and African Programme (106698). For the purpose of open access, a CC BY public copyright licence has been applied to this article.
Author Contributions
J.C. conceived the original concept. C.C., R.W.S., and J.C. designed the experiment and oversaw the project and analyses. C.C., S.J.P., N.P.J.D., J.P., N.R.T, R.W.S, and J.C. acquired funding. P.B., A.S., K.S., E.J.F., S.D., N.C., D.L., and N.P.J.D. contributed samples and reagents or assisted in samples preparation and permitting. J.P. and M.P. developed analytical tools. C.C. and J.P. performed genomic analyses. S.C. developed gene knockout mutant. S.C. and C.P. performed experimental validations. C.C., J.P., E.J.F., N.C., D.L., J.P., N.R.T, R.W.S., and J.C. contributed to the interpretation and presentation of results in the main manuscript and supplementary documents. C.C. and J.C. wrote the first draft with input from all the authors.
Resource Availability Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Claire Chewapreecha.
Materials Availability
This study generated new Burkholderia pseudomallei mutant as tabulated in supplementary table S5, Supplementary Material online. The mutant can be requested by contacting Dr Rasana W. Sermswan (rasana@kku.ac.th)
Data Availability
Short reads for isolates underlying this article are archived in ENA or NCBI database. Accession number for each individual isolate in discovery and validation data set are provided in supplementary table S1, Supplementary Material online. Raw results of coevolutionary mutation pairs, co-selected gene–gene pairs, and BPSL1661 co-selection gene and ncRNA network are given in supplementary table S2, S3, and S4, Supplementary Material online, respectively. The code used to detect coevolutionary signals is available in https://github.com/santeripuranen/SpydrPick (last accessed October 24, 2021).
References
- Arnold BJ, Gutmann MU, Grad YH, Sheppard SK, Corander J, Lipsitch M, Hanage WP.. 2018. Weak epistasis may drive adaptation in recombining bacteria. Genetics 208(3):1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashdown LR. 1979. An improved screening technique for isolation of pseudomonas pseudomallei from clinical specimens. Pathology 11(2):293–297. [DOI] [PubMed] [Google Scholar]
- Baker AL, Ezzahir J, Gardiner C, Shipton W, Warner JM.. 2015. Environmental attributes influencing the distribution of Burkholderia pseudomallei in Northern Australia. PLoS One 10(9):e0138953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 57(1):289–300. [Google Scholar]
- Berger VW, Zhou Y.. 2014. Kolmogorov-Smirnov test: overview. In: Balakrishnan N, Colton T, Everitt B, Piegorsch W, Ruggeri F, Teugels JL, editors. Wiley StatsRef: statistics reference online. Chichester (United Kingdom: ): John Wiley & Sons, Ltd. p. stat06558. Available from: http://doi.wiley.com/10.1002/9781118445112.stat06558. Accessed October 24, 2021. [Google Scholar]
- Brown SA, Palmer KL, Whiteley M.. 2008. Revisiting the host as a growth medium. Nat Rev Microbiol. 6(9):657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Sue D, Hakovirta J, Loparev VN, Knipe K, Sammons SA, Ranganathan-Ganakammal S, Changayil S, Srinivasamoorthy G, Weil MR, et al. 2015. Finished annotated genome sequence of Burkholderia pseudomallei strain Bp1651, a multidrug-resistant clinical isolate. Genome Announc. 3(6):e01427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulterys PL, Bulterys MA, Phommasone K, Luangraj M, Mayxay M, Kloprogge S, Miliya T, Vongsouvath M, Newton PN, Phetsouvanh R, et al. 2018. Climatic drivers of melioidosis in Laos and Cambodia: a 16-year case series analysis. Lancet Planet Health. 2(8):e334–e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple SNJ, Sarovich DS, Holden MTG, Peacock SJ, Buller N, Golledge C, Mayo M, Currie BJ, Price EP.. 2016. Whole-genome sequencing of a quarter-century melioidosis outbreak in temperate Australia uncovers a region of low-prevalence endemicity. Microb Genomics. 2:e000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-S, Lin H-H, Hsueh P-T, Liu P-J, Ni W-F, Chung W-C, Lin C-P, Chen Y-L.. 2015. Whole-genome sequence of an epidemic strain of Burkholderia pseudomallei vgh07 in Taiwan. Genome Announc. 3(2):e00345–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C, Holden MTG, Vehkala M, Välimäki N, Yang Z, Harris SR, Mather AE, Tuanyok A, De Smet B, Le Hello S, et al. 2017. Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat Microbiol. 2:16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C, Mather AE, Harris SR, Hunt M, Holden MTG, Chaichana C, Wuthiekanun V, Dougan G, Day NPJ, Limmathurotsakul D, et al. 2019. Genetic variation associated with infection and the environment in the accidental pathogen Burkholderia pseudomallei. Commun Biol. 2:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-Y, Hara Y, Ghazali A-K, Yap S-J, Kong C, Wong Y-C, Rozali N, Koh S-F, Hoh C-C, Puthucheary SD, et al. 2015. Global transcriptional analysis of Burkholderia pseudomallei high and low biofilm producers reveals insights into biofilm production and virulence. BMC Genomics. 16:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomkatekaew C, Boonklang P, Sangphukieo A, Chewapreecha C.. 2020. An evolutionary arms race between Burkholderia pseudomallei and host immune system: what do we know? Front Microbiol. 11:612568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Vandamme P.. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 5(9):719–729. [DOI] [PubMed] [Google Scholar]
- Cui Y, Yang C, Qiu H, Wang H, Yang R, Falush D.. 2020. The landscape of coadaptation in Vibrio parahaemolyticus. eLife 9:e54136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie BJ, Fisher DA, Anstey NM, Jacups SP.. 2000. Melioidosis: acute and chronic disease, relapse and re-activation. Trans R Soc Trop Med Hyg. 94(3):301–304. [DOI] [PubMed] [Google Scholar]
- Daligault HE, Davenport KW, Minogue TD, Bishop-Lilly KA, Broomall SM, Bruce DC, Chain PS, Coyne SR, Frey KG, Gibbons HS, et al. 2014. Whole-genome assemblies of 56 Burkholderia species. Genome Announc. 2:e01106–e01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, Hirst S, Chew JSW, Wang D, et al. 2009. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci U S A. 106(32):13499–13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2019. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrick I, Todd G, Stewart J.. 2018. Soil characteristics influencing the spatial distribution of melioidosis in Far North Queensland. Epidemiol Infect. 146(12):1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR.. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34(2):292–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantrakun V, Rongkard P, Oyuchua M, Amornchai P, Lim C, Wuthiekanun V, Day NPJ, Peacock SJ, Limmathurotsakul D.. 2016. Soil nutrient depletion is associated with the presence of Burkholderia pseudomallei. Appl Environ Microbiol. 82:7086–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, et al. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One. 7(5):e36507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, et al. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 101(39):14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P-T, Chen Y-S, Lin H-H, Liu P-J, Ni W-F, Liu M-C, Chen Y-L.. 2015. Comparison of whole-genome sequences from two colony morphovars of Burkholderia pseudomallei. Genome Announc. 3(5):e01194–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis TJJ, Sagripanti J-L.. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl Environ Microbiol. 72(11):6865–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitprasutwit S, Jitprasutwit N, Hemsley CM, Onlamoon N, Withatanung P, Muangsombut V, Vattanaviboon P, Stevens JM, Ong C, Stevens MP, et al. 2020. Identification of Burkholderia pseudomallei genes induced during infection of macrophages by. Front Microbiol. 11:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Baker AL, Chain PS, Currie BJ, Daligault HE, Davenport KW, Davis CB, Inglis TJJ, Kaestli M, Koren S, et al. 2015a. Whole-genome sequences of 80 environmental and clinical isolates of Burkholderia pseudomallei. Genome Announc. 3(1):e01282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Bishop-Lilly KA, Ladner JT, Daligault HE, Davenport KW, Jaissle J, Frey KG, Koroleva GI, Bruce DC, Coyne SR, et al. 2015b. Complete genome sequences for 59 Burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 3(2):e00159-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestli M, Harrington G, Mayo M, Chatfield MD, Harrington I, Hill A, Munksgaard N, Gibb K, Currie BJ.. 2015. What drives the occurrence of the melioidosis bacterium Burkholderia pseudomallei in domestic gardens? PLoS Negl Trop Dis. 9(3):e0003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y.. 2020. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29(1):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Dunachie SJ, Müller E, Kohler A, Jenjaroen K, Teparrukkul P, Baier V, Ehricht R, Steinmetz I.. 2016. Rapid and sensitive multiplex detection of Burkholderia pseudomallei—specific antibodies in melioidosis patients based on a protein microarray approach. PLoS Negl Trop Dis. 10(7):e0004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J, Chan R, Lam K, Costerton JW.. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 28(2):546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D, Dance DAB, Wuthiekanun V, Kaestli M, Mayo M, Warner J, Wagner DM, Tuanyok A, Wertheim H, Yoke Cheng T, et al. 2013. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis. 7(3):e2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM, Moyes CL, Rolim DB, Bertherat E, Day NPJ, Peacock SJ, et al. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 1:15008. [DOI] [PubMed] [Google Scholar]
- López CM, Rholl DA, Trunck LA, Schweizer HP.. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol. 75(20):6496–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48(D1):D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivanh L, Pierret A, Rattanavong S, Kounnavongsa O, Buisson Y, Elliott I, Maeght J-L, Xayyathip K, Silisouk J, Vongsouvath M, et al. 2017. Burkholderia pseudomallei in a lowland rice paddy: seasonal changes and influence of soil depth and physico-chemical properties. Sci. Rep. 7:3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobb E, Sarovich DS, Price EP, Kaestli M, Mayo M, Keim P, Currie BJ.. 2015. Tracing melioidosis back to the source: using whole-genome sequencing to investigate an outbreak originating from a contaminated domestic water supply. J Clin Microbiol. 53(4):1144–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Padil H, Damiri N, Sulaiman S, Chai S-F, Nathan S, Firdaus-Raih M.. 2017. Identification of sRNA mediated responses to nutrient depletion in Burkholderia pseudomallei. Sci Rep. 7(1):17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CM, Conejero L, Spink N, Wand ME, Bancroft GJ, Titball RW.. 2012. Role of RelA and SpoT in Burkholderia pseudomallei virulence and immunity. Infect Immun. 80(9):3247–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa HI, Hassan L, Shamsuddin ZHj, Panchadcharam C, Zakaria Z, Abdul Aziz S.. 2016. Physicochemical properties influencing presence of Burkholderia pseudomallei in soil from small ruminant farms in Peninsular Malaysia. PLoS One 11:e0162348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi T, Holden MTG, Didelot X, Mehershahi K, Boddey JA, Beacham I, Peak I, Harting J, Baybayan P, Guo Y, et al. 2015. Burkholderia pseudomallei sequencing identifies genomic clades with distinct recombination, accessory, and epigenetic profiles. Genome Res. 25(1):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngamsang R, Potisap C, Boonmee A, Lawongsa P, Chaianunporn T, Wongratanacheewin S, Rodrigues JLM, Sermswan RW.. 2015. The contribution of soil physicochemical properties to the presence and genetic diversity of Burkholderia pseudomallei. Southeast Asian J Trop Med Public Health. 46:38–50. [PubMed] [Google Scholar]
- Ooi WF, Ong C, Nandi T, Kreisberg JF, Chua HH, Sun G, Chen Y, Mueller C, Conejero L, Eshaghi M, et al. 2013. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 9:e1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, Harris SR.. 2016. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics. 2:e000056. Available from: https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000056. Accessed October 24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensar J, Puranen S, Arnold B, MacAlasdair N, Kuronen J, Tonkin-Hill G, Pesonen M, Xu Y, Sipola A, Sánchez-Busó L, et al. 2019. Genome-wide epistasis and co-selection study using mutual information. Nucleic Acids Res. 47(18):e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K.. 2012. Sticky situations: key components that control bacterial surface attachment. J Bacteriol. 194(10):2413–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponstingl H, Ning Z.. 2010. SMALT - a new mapper for DNA sequencing reads. Available from: https://sourceforge.net/projects/smalt/files/smalt_manual.pdf/. Accessed October 24, 2021.
- Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, Kaestli M, Beckstrom-Sternberg SM, Babic-Sternberg JS, Kidd TJ, Bell SC, et al. 2013. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio. 4(4):e00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EP, Sarovich DS, Smith EJ, MacHunter B, Harrington G, Theobald V, Hall CM, Hornstra HM, McRobb E, Podin Y, et al. 2016. Unprecedented melioidosis cases in northern Australia caused by an Asian Burkholderia pseudomallei strain identified by using large-scale comparative genomics. Appl Environ Microbiol. 82(3):954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett S, Trujillo C, Wang Z, Eoh H, Ioerger TR, Krieger I, Sacchettini J, Schnappinger D, Rhee KY, Ehrt S.. 2017. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 114(11):E2225–E2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumpuang A, Chantratita N, Wikraiphat C, Saiprom N, Day NPJ, Peacock SJ, Wuthiekanun V.. 2011. Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans R Soc Trop Med Hyg. 105(10):598–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: https://www.R-project.org/. Accessed October 24, 2021. [Google Scholar]
- Ribolzi O, Rochelle-Newall E, Dittrich S, Auda Y, Newton PN, Rattanavong S, Knappik M, Soulileuth B, Sengtaheuanghoung O, Dance DAB, et al. 2016. Land use and soil type determine the presence of the pathogen Burkholderia pseudomallei in tropical rivers. Environ Sci Pollut Res Int. 23(8):7828–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues F, Sarkar-Tyson M, Harding SV, Sim SH, Chua HH, Lin CH, Han X, Karuturi RKM, Sung K, Yu K, et al. 2006. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J Bacteriol. 188(23):8178–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Stone JK, Gelhaus HC, Warren RL, Cruttwell CJ, Funnell SG, Keim P, Tuanyok A.. 2013. Genome sequence of Burkholderia pseudomallei NCTC 13392. Genome Announc. 1(3):e00183–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert B, Maddamsetti R, Nyman J, Farhat MR, Marks DS.. 2019. Genome-wide discovery of epistatic loci affecting antibiotic resistance in Neisseria gonorrhoeae using evolutionary couplings. Nat Microbiol. 4(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- Sidjabat HE, Cottrell K, Cervin A.. 2015. Draft genome sequences of Burkholderia pseudomallei and Staphylococcus aureus, isolated from a patient with chronic rhinosinusitis. Genome Announc. 3(5):e01075–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A.. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1(9):784–791. [Google Scholar]
- Song L, Yu Y, Feng L, He J, Wang T, Zhu H, Duan Q.. 2015. Draft genome sequence of Burkholderia pseudomallei strain 350105, isolated in Hainan, China, in 1976. Genome Announc. 3:e01162–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring-Pearson SM, Stone JK, Doyle A, Allender CJ, Okinaka RT, Mayo M, Broomall SM, Hill JM, Karavis MA, Hubbard KS, et al. 2015. Pangenome analysis of Burkholderia pseudomallei: genome evolution preserves gene order despite high recombination rates. PLoS One 10(10):e0140274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannasaen D, Mahawantung J, Chaowagul W, Limmathurotsakul D, Felgner PL, Davies H, Bancroft GJ, Titball RW, Lertmemongkolchai G.. 2011. Human immune responses to Burkholderia pseudomallei characterized by protein microarray analysis. J Infect Dis. 203(7):1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, et al. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 9:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS.. 2015. Whole-genome sequences of five Burkholderia pseudomallei isolates from Australian cystic fibrosis patients. Genome Announc. 3(2):e00254–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD.. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128(5):865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Q, Hu J, Asenso J, Wise MJ, Wu X, Ma C, Chen X, Yang J, Tang D.. 2019. Structure and evolution of glycogen branching enzyme N-termini from bacteria. Front Microbiol. 9:3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga WJ, Currie BJ, Peacock SJ.. 2012. Melioidosis. N Engl J Med. 367(11):1035–1044. [DOI] [PubMed] [Google Scholar]
- Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D.. 2018. Melioidosis. Nat Rev Dis Primer. 4:17107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten MMSM, van de Lest CHA, van Dijk L, van Putten JPM.. 2017. Function and regulation of the C4-dicarboxylate transporters in Campylobacter jejuni. Front Microbiol. 8:174. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00174/full. Accessed October 24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL.. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip T-W, Hewagama S, Mayo M, Price EP, Sarovich DS, Bastian I, Baird RW, Spratt BG, Currie BJ.. 2015. Endemic melioidosis in residents of desert region after atypically intense rainfall in central Australia, 2011. Emerg Infect Dis. 21(6):1038–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ouyang Y, Yao W.. 2018. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34(7):1229–1231. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ren H, Hu M, Jing Z, Li B, Kong N, Zhang Q, Jin Y, Liang L, Yue J.. 2020. Characterization of Burkholderia cepacia complex core genome and the underlying recombination and positive selection. Front Genet. 11:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short reads for isolates underlying this article are archived in ENA or NCBI database. Accession number for each individual isolate in discovery and validation data set are provided in supplementary table S1, Supplementary Material online. Raw results of coevolutionary mutation pairs, co-selected gene–gene pairs, and BPSL1661 co-selection gene and ncRNA network are given in supplementary table S2, S3, and S4, Supplementary Material online, respectively. The code used to detect coevolutionary signals is available in https://github.com/santeripuranen/SpydrPick (last accessed October 24, 2021).