Abstract
Although hybridization plays a large role in speciation, some unknown fraction of hybrid individuals never reproduces, instead remaining as genetic dead-ends. We investigated a morphologically distinct and culturally important Chinese walnut, Juglans hopeiensis, suspected to have arisen from hybridization of Persian walnut (J. regia) with Asian butternuts (J. cathayensis, J. mandshurica, and hybrids between J. cathayensis and J. mandshurica). Based on 151 whole-genome sequences of the relevant taxa, we discovered that all J. hopeiensis individuals are first-generation hybrids, with the time for the onset of gene flow estimated as 370,000 years, implying both strong postzygotic barriers and the presence of J. regia in China by that time. Six inversion regions enriched for genes associated with pollen germination and pollen tube growth may be involved in the postzygotic barriers that prevent sexual reproduction in the hybrids. Despite its long-recurrent origination and distinct traits, J. hopeiensis does not appear on the way to speciation.
Keywords: chromosomal rearrangements, gene flow, hybridization, postzygotic reproductive barriers, speciation, walnuts
Reproductive isolation is a key process in speciation and plays a major role in population divergence (Coyne and Orr 1989; Baack et al. 2015). In plants, reproductive isolation usually involves extrinsic and intrinsic pre- and postzygotic barriers that evolve over time (Coyne and Orr 2004; Rieseberg and Willis 2007). Generally, prezygotic barriers are considered to contribute more to total reproductive isolation than postzygotic barriers since the latter have higher reproductive costs, including wasted gametes and energy invested in unfit hybrid progeny (Lowry et al. 2008; Baack et al. 2015; Ramirez-Aguirre et al. 2019). Nevertheless, postzygotic barriers, such as hybrid nonviability and sterility, are hallmarks of most “good” species (Coughlan and Matute 2020). Hybrid nonviability refers to hybrid seeds being less likely to germinate and survive than parental seeds, whereas hybrid sterility refers to hybrid individuals having reduced pollen or ovule fertility. Hybrid individuals can often be recognized by their intermediate morphology, and suspected first-generation (F1) hybrids have been reported from several tree genera, including Eucalyptus (Robins et al. 2021), Quercus (Burgarella et al. 2009), and mangrove genera (Qiu et al. 2008; Zhou et al. 2008; Lo 2010). In studies that have used molecular markers, F1 hybrids are always heterozygous for parental alleles (Lo 2010; Robins et al. 2021). It is easy for hybrids to be viewed as independent evolutionary lineages, that is, species, because of their persistence as a morphologically distinct cohort, perhaps with high heterozygosity, and at least in Europe, there is a long tradition of formally naming arborescent hybrids as taxa at the species rank (Robertson et al. 2010; Feulner et al. 2013).
Whether morphologically distinct “hybrid species” are reproductively isolated from their parents, however, can only be decided with molecular data from numerous individuals. With such data, Worth et al. (2016) revealed that Athrotaxis laxifolia, described by J. Hooker in 1843 and long suspected to be a homoploid hybrid species descending from A. cupressoides and A. selaginoides, actually consists of rare F1 hybrids and advanced generation backcrosses within the range of the pure species, suggesting that the long-lived A. laxifolia hybrids will eventually be “reabsorbed” by the parental species via backcrossing, a process that might take millennia. This case highlights that conclusions about the large role of hybridization in speciation (Taylor and Larson 2019), at least as regards homoploid tree hybrids, as well as the time required for their reproductive isolation, need more empirical data (Schumer et al. 2014, 2018).
The time required for successful sexual reproduction of hybrid individuals will depend on the genetic architecture underlying their postzygotic reproductive barriers. In the past decades, besides Bateson–Dobzhansky–Muller (BDM) incompatibilities (Bateson 1909; Dobzhansky 1937; Muller 1942), which involve interactions between nuclear genes or between nuclear and organellar genes (Coyne and Orr 2004), chromosomal rearrangements have been recognized as another kind of important postzygotic isolating barrier in plants (Levin 2002; Fishman et al. 2013; Baack et al. 2015). They can directly affect hybrid fitness or also may increase the strength of genic barriers by selection against recombinant gametes (Rieseberg 2001).
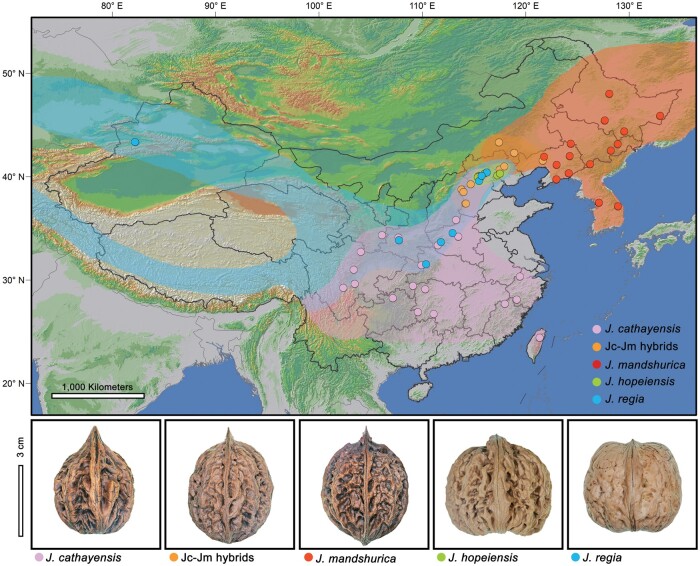
Juglans hopeiensis Hu, the Ma walnut, in Chinese “Mahetao,” can reach 20 m in height and is morphologically intermediate between Persian walnut (J. regia) and J. mandshurica, one of the two Chinese butternut species. Its leaves have 7–15 almost glabrous leaflets, which is intermediate between J. regia (5–11 glabrescent leaflets) and J. mandshurica (7–19 glandular-pubescent leaflets), and its fruits mostly have two ridges on the outer surface and four internal nut chambers similar to J. regia, but the thick shell and lacunate septa resemble J. mandshurica (Manning 1978) (fig. 1). Like J. regia and both Chinese butternuts (J. cathayensis and J. mandshurica), J. hopeiensis has a diploid chromosome number of 2n=32 (Mu et al. 1990), but its pollen viability is low (8–30%) (Mu et al. 1990; Ma et al. 2014; Chen et al. 2015) and so is its fruit set (2–23%) (Dai et al. 2014; Zhu et al. 2020). In former times, J. hopeiensis timber was used for ladders and rifle stocks and its fruits for small carvings (Liu 2014). The species was described by H. H. Hu (1894–1968), a genetics pioneer in China, who first considered it highly distinct (Hu 1932), but who in a follow-up paper (Hu 1934) mentioned its possible hybrid origin.
Fig. 1.
The geographic locations and representative nuts of Juglans hopeiensis and its parents. The Asian butternuts, J. cathayensis, Jc–Jm hybrids, and J. mandshurica all have similar nuts (ovoid with a distinct apical, shell rough ridged, and deeply pitted). The species geographic distributions were mainly referred from the Global Biodiversity Information Facility (GBIF: http://www.gbif.org/).
Molecular studies so far have not resolved the status of J. hopeiensis and phylogenetic relationship within the genus (Cheng and Yang 1987; Aradhya et al. 2007; Hu et al. 2016, 2017; Dong et al. 2017; Zhao et al. 2018), with some suggesting that it is the sister species to J. mandshurica (Hu et al. 2017), others that it is a hybrid species between J. regia and J. mandshurica, that is, an evolutionary lineage (Cheng and Yang 1987; Mu et al. 1990, 2017; Wu et al. 1999; Zhao et al. 2018).
The low pollen viability and fruit set, and the taxon’s natural distribution confined to regions where both putative parents are present (fig. 1) led us to suspect that J. hopeiensis might consist of spontaneous hybrids instead of being a species (a reproducing gene pool), a hypothesis that bears on the time since when Persian walnuts have been present in China, because hybrids could only have begun forming once J. regia overlapped with the ranges of butternuts. Juglans mandshurica itself is close to, and sometimes forms hybrids with, a more tropical butternut species, J. cathayensis (Bai et al. 2016) (fig. 1); together with J. ailantifolia, a Japanese variety of J. mandshurica, these three entities form the Asian butternuts. It was long thought that Persian walnuts were introduced to China from Central Asia only during the Han Dynasty, about 2,000–2,500 years ago, but this view has been challenged (Feng et al. 2018; Zhang, Xu, et al. 2019). To test our hypothesis, we applied four population genetic methods to a sample of over 150 morphologically identified tree individuals from the relevant geographic range. We also used comparative and population-genomic approaches to investigate the architecture of postzygotic isolation between Asian butternuts and Persian walnut.
Results
We carried out whole-genome resequencing for 151 individuals of J. cathayensis, J. mandshurica, hybrids between J. cathayensis and J. mandshurica (Jc–Jm hybrids), J. hopeiensis, and J. regia across northern China (fig. 1 and supplementary table 1, Supplementary Material online; see Materials and Methods). The average effective depth for our data set was 23-fold, with an average mapping rate of 63.70% coverage of the Pterocarya stenoptera reference genome (supplementary notes 1 and 2 and supplementary tables 1 and 2, Supplementary Material online). The allele frequency distribution in J. hopeiensis is bimodal whereas allele frequency spectra in the other species are unimodal (supplementary fig. 1, Supplementary Material online), implying that there are more alleles with medium frequencies in J. hopeiensis than in the other groups, as is typical for F1 hybrids.
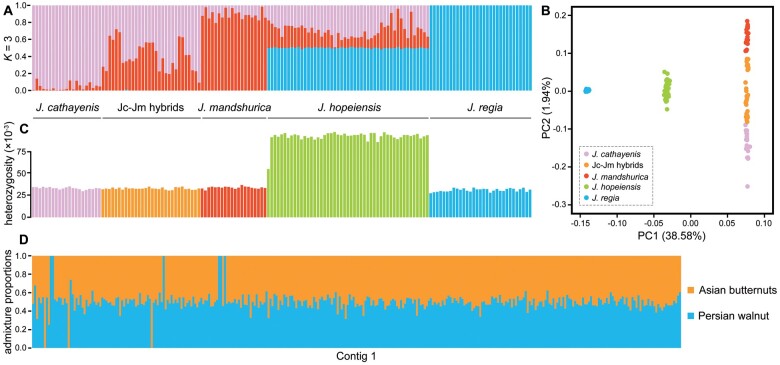
Analysis of the genetic structure of the 151 individuals (using the program STRUCTURE) showed that K = 3 was the optimal number of populations when using the parsimony method of Wang (2019) or K = 2 when using the deltaK method of Evanno et al. (2005). At K = 2, the Asian butternuts formed one group, whereas J. regia formed another, and J. hopeiensis was assigned approximately 50% ancestry to each group (supplementary fig. 2, Supplementary Material online). At K = 3, there are three distinct groups for J. cathayensis, J. mandshurica, and J. regia, respectively, but J. hopeiensis was still assigned approximately 50% ancestry to J. regia, with the other 50% assigned to Asian butternuts (fig. 2A). Principal component analysis (PCA) is consistent with the STRUCTURE result, showing an intermediate position for J. hopeiensis between Asian butternuts and J. regia along PC1 whereas the separation along PC2 is not distinct (fig. 2B and supplementary fig. 3, Supplementary Material online). Admixture analysis across the whole genome revealed that J. hopeiensis has a high heterozygosity (fig. 2C), with windows assigned equally to Asian butternuts and J. regia (fig. 2D and supplementary fig. 4, Supplementary Material online).
Fig. 2.
Genomic variation in Juglans hopeiensis and its parents. (A) Results of a STRUCTURE analysis with the optimal K showing individual ancestry proportions. (B) PCA showing the first two principal components (excluding four outlying individuals of J. regia). (C) Individual heterozygosity for polymorphic sites across the genome. (D) NgsAdmix analysis assignments for 50-kb stepping windows across contig 1.
Analysis with NewHybrids identified all individuals of J. hopeiensis as F1 hybrids and none as F2 hybrids, backcrosses, Asian butternuts, or J. regia, whereas Asian butternuts and J. regia were assigned as pure parents. The initial gene flow most likely happened in Southwest China, where the range of J. regia abuts that of J. cathayensis (fig. 1).
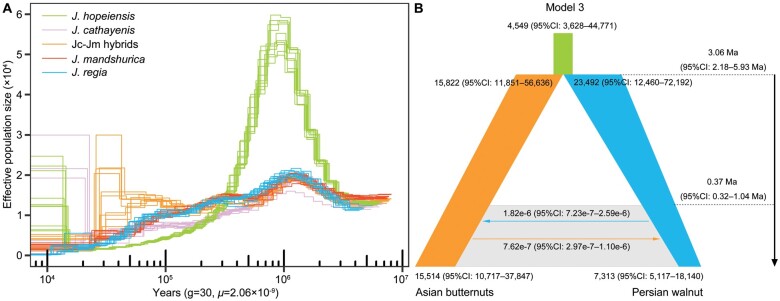
Pairwise sequentially Markovian coalescent (PSMC) plots of F1 hybrids (hPSMC) can be used to infer the divergence time between the two parents because coalescence between the two alleles of an F1 hybrid can only occur in the ancestral population and cannot coalesce more recently than the speciation of the two parents (Cahill et al. 2016). These plots show a transition from an infinite population size during the time of lineage divergence to population sizes that reflect the shared ancestry period prior to divergence. However, hPSMC is highly sensitive to postdivergence gene flow (Cahill et al. 2016). If there is gene flow between two parents after their divergence, or if the time of gene flow cessation is too recent, the PSMC plots of F1 hybrids will not increase to infinity (Mather et al. 2020). Thus, we used hPSMC to infer the timing of the end of gene flow between the two parents, Asian butternuts and Persian walnut. This approach suggests that the plot of J. hopeiensis began to deviate significantly from Asian butternuts and Persian walnut some 1–2 Ma (fig. 3A and supplementary fig. 5, Supplementary Material online) and became much larger than the Ne of either parental lineage, after which it declined consistently to the present (fig. 3A and supplementary fig. 5, Supplementary Material online). Since the inferred plot of J. hopeiensis never goes to infinity, gene flow must have existed recently, consistent with divergence with secondary contact between Asian butternuts and Persian walnut (fig. 3B and supplementary fig. 6 and supplementary table 3, Supplementary Material online).
Fig. 3.
Inferring population demographic history. (A) Changes in effective population size (Ne) over the past 4.5 My in Asian butternuts and Juglans regia as inferred with PSMC. Within J. hopeiensis and Jc–Jm hybrids, results were obtained by using J. mandshurica as the reference genome. Each color and line represent one group and individual, respectively, and g, generation time (year), μ, mutation rate (per site per year). (B) Simplified graphical summary of the best-fitting demographic model inferred by fastsimcoal2 based on SNPs from Asian butternuts and Persian walnut. The joint past population is shown as the light green bar. Gray-shaded area and arrows indicate the bidirectional gene flow. The orange and blue bars represent Asian butternuts and Persian walnut. Populations of Asian butternuts and Persian walnut were set to decline from past to present based on the PSMC results.
Using our genome data of Asian butternuts and Persian walnut, we also inferred divergence time and gene flow by means of fastsimcoal simulation analysis. The best-fit model suggested their divergence at ∼3.06 Ma (95% CI: 2.18–5.93 Ma) and gene flow initiation at ∼0.37 Ma (95% CI: 0.32–1.04 Ma). Although the best-fit model suggests secondary contact and gene flow between Asian butternuts and J. regia, the migration rates (m) are extremely low, only 7.62e-7 (95% CI: 2.97e-7–1.10e-6) from Asian butternuts to J. regia, and 1.82e-6 (95% CI: 7.23e-7–2.59e-6) in the opposite direction. We also evaluated contemporary gene flow using a Bayesian framework as implemented in the software BA3-SNPs (Mussmann et al. 2019), which suggested no significant contemporary gene flow between Asian butternuts and Persian walnut (bidirectional migration rates [4 Nem]: 0.0046 ± 0.0088; 0.0101 ± 0.0191).
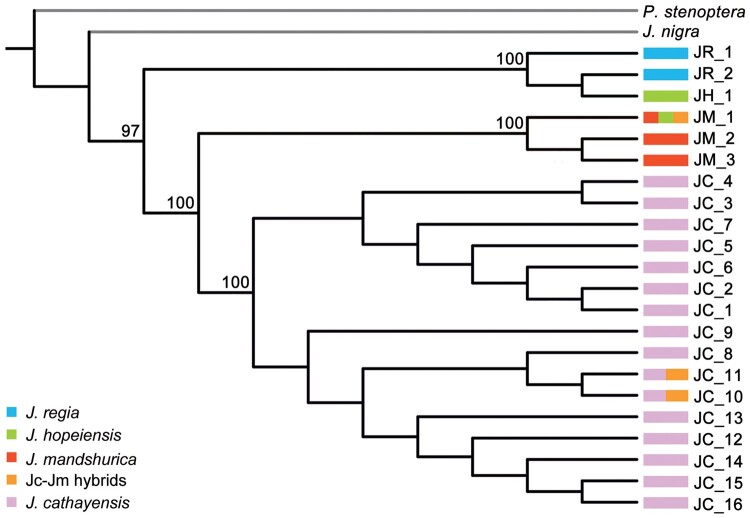
To infer the direction of gene flow, we used chloroplast haplotypes, which are maternally inherited. A total of 22 haplotypes in 80 chloroplast protein-coding genes were found in the 151 individuals, namely 16 in J. cathayensis, three in J. mandshurica, two in J. regia, and two in J. hopeiensis, one of them shared with J. mandshurica (supplementary table 1, Supplementary Material online). Of the 49 individuals of J. hopeiensis, 48 had a haplotype from J. mandshurica (JM_1), whereas one individual had a distinct haplotype (JH_1) that clustered with two haplotypes (JR_1 and JR_2) of J. regia. In a maximum likelihood (ML) phylogeny, 19 haplotypes from Asian butternuts formed a monophyletic group, whereas two haplotypes of J. regia and one haplotype (JH_1) of J. hopeiensis formed another (fig. 4).
Fig. 4.
The maternal origin of hybrids. ML phylogeny obtained from 22 chloroplast haplotypes. ML bootstrap values (MLBS) ≥95% are labeled on each node.
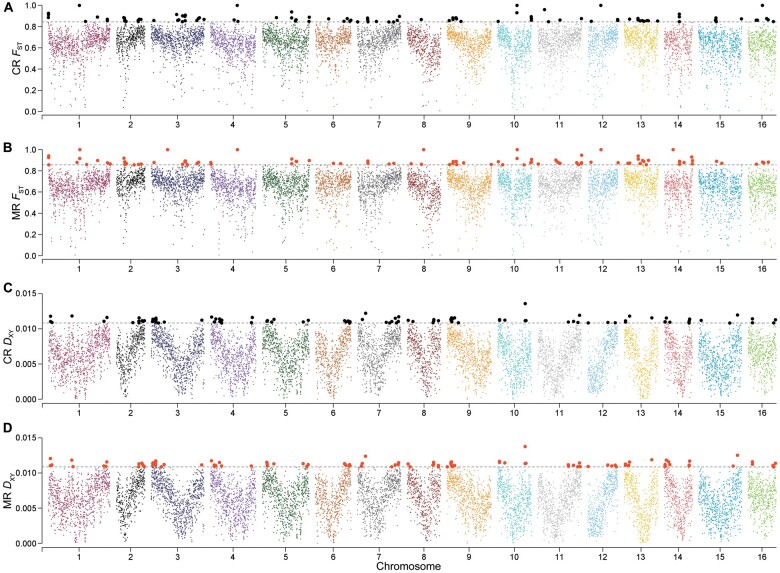
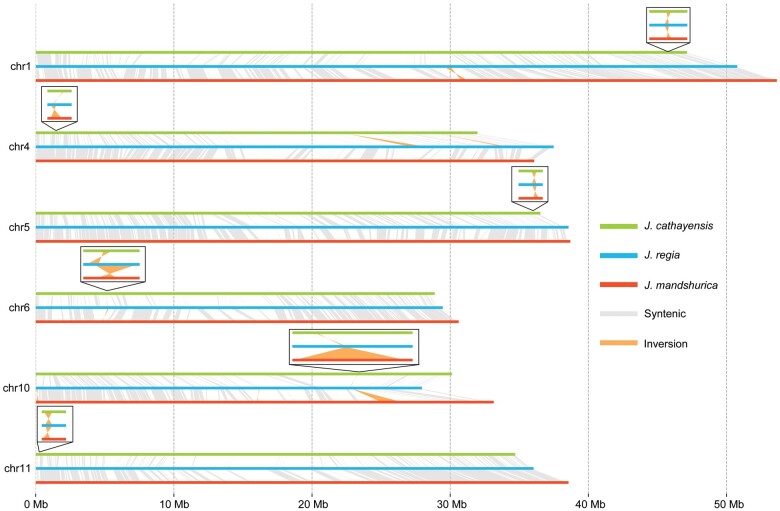
To understand what might cause the reproductive barrier between the parental species that has prevented the hybrids from becoming a distinct species despite the long available time, we assessed the genome-wide divergence between Asian butternuts and J. regia to identify outlier regions potentially associated with reproductive isolation (Materials and Methods). The mean FST across the genomes was 0.641 and 0.649, with 101 regions above the 99th percentile between J. cathayensis and J. regia (FST > 0.844) and 103 between J. mandshurica and J. regia (FST > 0.856) identified as outlier regions (fig. 5A and B). The mean DXY was 0.0063 and 0.0063, with 105 regions above the 99th percentile between J. cathayensis and J. regia (DXY > 0.011) and 106 between J. mandshurica and J. regia (DXY > 0.011) identified as outliers (fig. 5C and D). In addition, we identified 51 inversions between J. cathayensis and J. regia and 35 inversions between J. mandshurica and J. regia (supplementary table 4, Supplementary Material online). The outlier regions and inversions shared by J. mandshurica and J. regia, and by J. cathayensis and J. regia could have been involved in these species’ reproductive isolation. We therefore checked the gene content of 49 FST outlier regions, 86 DXY outlier regions, and six inversion regions (longer than 10 kb) (figs. 5 and 6). The FST and DXY outlier regions contained 133 and 473 genes spread across the 16 chromosomes, whereas the six inversion regions contained 14 genes (supplementary table 5, Supplementary Material online). To infer the function of these genes, we used Gene Ontology (GO) enrichment analysis. Some of the 133 genes in FST outlier regions are implicated in fruit development, such as the abscisic acid-activated signaling pathway, cellular response to abscisic acid stimulus, and fruit morphogenesis, as well as with pollen development, for example, regulation of pollen tube growth (supplementary fig. 7, Supplementary Material online). The 14 genes in the six long inversion regions are implicated in pollen germination and pollen tube growth (fig. 6 and supplementary fig. 8, Supplementary Material online). The remaining 473 genes are not associated with particular biological processes.
Fig. 5.
High genetic differentiation regions with 50-kb stepping windows across the 16 chromosomes. (A) FST analysis between Juglans cathayensis and J. regia. (B) FST analysis between J. mandshurica and J. regia. (C) DXY analysis between J. cathayensis and J. regia. (D) DXY analysis between J. mandshurica and J. regia. Windows that exceeded the gray-dotted line were identified as FST and DXY outliers, represented by black or red dots. C, M, and R are the abbreviations of J. cathayensis, J. mandshurica, and J. regia.
Fig. 6.
Six inversions longer than 10 kb in syntenic chromosome regions between Asian butternuts and Juglans regia.
These outlier regions and inversions might be due to selection, local reduction in recombination, or to genetic drift resulting from the small effective population sizes of Asian butternuts and J. regia (above). To assess the possibility of selection, we performed McDonald–Kreitman (MK) tests (McDonald and Kreitman 1991) and Tajima’s D tests (Tajima 1989) for the 133 FST outlier genes, 473 DXY outlier genes, and 14 genes in inversion regions between these parental genomes (see Positive Selection Analysis section in Materials and Methods; supplementary table 5, Supplementary Material online). Among all genes, 12 in J. cathayensis, five in J. mandshurica, and six in J. regia were significant by both Tajima’s D and the MK test. To assess the possibility of low recombination rates causing high levels of genetic divergence, we checked correlations between FST and the population-level recombination rate (ρ = 4 Ner) in J. cathayensis and J. mandshurica as well as J. regia. Negative correlations were found between FST and recombination rate in J. regia (n = 10,020, r = −0.0377, P < 0.0001 and n = 10,037, r = −0.0547, P < 0.0001) and positive correlations between FST and recombination rate in J. cathayensis (n = 10,020, r = 0.0328, P = 0.0006), but nonsignificant negative correlations between FST and recombination rate in J. mandshurica (n = 10,037, r = 0.0098, P = 0.1614) (supplementary note 3 and fig. 9, Supplementary Material online).
Discussion
Four lines of evidence support the F1 status of today’s entire cohort of J. hopeiensis trees. First, both nuclear diversity and individual heterozygosity of J. hopeiensis are twice that in each of its parents (fig. 2C). Second, J. hopeiensis is an admixed group (fig. 2D and supplementary fig. 2, Supplementary Material online) and has no unique genetic composition. Third, the Bayesian-assignment analysis of NewHybrids categorized all J. hopeiensis individuals as F1 hybrids. Fourth, hPSMC analysis matched theoretical expectations for first-generation hybrids (fig. 3A), and J. hopeiensis fits a scenario of divergence with secondary contact between its parents (fig. 3B).
The chloroplast phylogeny implies that J. mandshurica is usually the maternal parent of J. hopeiensis (fig. 4). Walnuts are monoecious and strongly dichogamous, with male and female catkins being produced about a week apart from each other. The flowering period of J. regia in China is mid to late April (Zhao et al. 2014), whereas that of J. mandshurica is late April to early May (Bai et al. 2006). Juglans mandshurica female catkins are therefore usually still available when J. regia sheds pollen, but not the other way around.
In former times, Ma walnuts (J. hopeiensis) had cultural importance. The nuts were used for walnut-shell carvings that were traditional gifts for aristocrats and noblemen as early as the Han Dynasty (206 BC–220 AD) (Liu 2014), and these sculptures, along with the valuable timber, created a market for Ma walnuts for at least 2,000 years. Although Asian butternuts have occurred in China since the Tertiary (Bai et al. 2016), J. regia is much younger, with phylogenomic analyses revealing that it arose as a hybrid between American and Asian lineages in the late Pliocene, about 3.45 Ma (Zhang, Xu, et al. 2019). Our inference that J. regia hybridized with Asian butternuts by the mid-Pleistocene (∼0.37 Ma; fig. 3B) implies an overlap in geographic ranges of the parents and rejects the view that Persian walnuts were introduced from Central Asia only during the Han Dynasty (Xi 1990; Deng and Xie 2006; Jiang and He 2019), some 2,000–2,500 years ago, instead supporting the view that J. regia was present in China much earlier (Feng et al. 2018; Zhang, Xu, et al. 2019).
That J. hopeiensis consist entirely of F1 individuals highlights strong postzygotic isolation barriers between Asian butternuts and Persian walnuts, as also suggested with three nuclear loci and 17 EST-SSRs (Dang et al. 2021). These barriers might be due to the six long inversion regions that we found to be enriched for genes associated with pollen germination and pollen tube growth (fig. 6 and supplementary fig. 8, Supplementary Material online). Juglans hopeiensis has low pollen viability (8–30%) (Mu et al. 1990; Ma et al. 2014; Chen et al. 2015) and fruit set (2–23%) (Ma et al. 2014; Zhu et al. 2020), and its pollen mother cells show abnormal meiosis and irregular chromosome arrangement, whereas its embryo sacs are often atrophic (Mu et al. 1990; Dai et al. 2014). Chromosomal rearrangements, especially inversions, can facilitate the evolution of postzygotic isolation between hybridizing species (Noor et al. 2001; Rieseberg 2001; Hoffmann and Rieseberg 2008; Baack et al. 2015), even in the presence of gene flow (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). In addition, 12 genes in J. cathayensis, five in J. mandshurica, and six in J. regia that were found to be subject to positive selection may contribute to genome divergence and formation of postzygotic isolation (supplementary table 5, Supplementary Material online). In J. regia, local reductions of the recombination rate seem to play an additional role in the build-up of reproductive isolation since negative correlations were found between FST and recombination rate.
In other tree species of inferred homoploid hybrid origin, successful reproductive isolation appears to have required at least a few million years, for example, 6 Ma for Picea purpurea (Ru et al. 2018), 1.8 Ma for Ostryopsis intermedia (Wang et al. 2021), 6.64 Ma for Pinus densata (Gao et al. 2012), and 3.45 Ma for Juglans regia (Zhang, Xu, et al. 2019). Compared with these species, Asian butternuts and J. regia have been hybridizing since 0.37 Ma, which may be too short to form a hybrid species. To better understand the frequency of homoploid speciation, more genomic studies with comprehensive population genomic analysis will need to distinguish between nonreproducing transient first-generation hybrids and successfully reproducing hybrids in clades for which the genomic resources now exist, such as Juglans. In particular, our work shows that persistence of hybrids over time does not imply a stabilized independent hybrid lineage or species.
Materials and Methods
Reference Genomes and Genome Assembly
We used five reference genomes. For P. stenoptera, which is equally related to all species of Juglans species (Zhang, Xu, et al. 2019), we sequenced an individual for de novo assembly. DNA was extracted from fresh young leaves of an adult P. stenoptera tree collected from Beijing, China (39.983°N/116.209°E). For P. stenoptera, a total of 117 Gb (∼208×) PacBio single-molecule long reads and 75 Gb (∼134×) Illumina short reads were used in the initial assembly and subsequent correction, which produced a total assembly length of 555.14 Mb with an N50 of 3.76 Mb (see details in supplementary note 1 and supplementary table 2, Supplementary Material online). The new P. stenoptera genome assembly is better than a previous one (v1.0) (Zhang, Xu, et al. 2019), judging by its smaller total genome size and larger N50. PacBio long reads combined with previous Illumina paired-end and mate-pair reads were also used to update the genomes of J. regia, J. mandshurica, and J. nigra (v2.0; see details in supplementary note 1, Supplementary Material online), which were used to identify chromosomal rearrangements. The J. regia genome was 525.44 Mb with N50 of 35.86 Mb, consistent with previous published genome (Marrano et al. 2020). The J. mandshurica genome had a total length of 537.15 Mb with an N50 of 35.99 Mb (supplementary table 2, Supplementary Material online), and the J. nigra genome comprised 531.15 Mb with an N50 of 35.28 Mb. A chromosome-level genome of J. cathayensis has been made available by Zhang et al. (2020). Herbarium vouchers for these species are listed in supplementary table 1, Supplementary Material online, and a voucher for J. hopeiensis has been deposited in the BNU herbarium as W. N. Bai DLG1 (BNU).
Gene Prediction and Functional Annotation of Protein-Coding Genes
To annotate the genomes of P. stenoptera, J. regia, J. mandshurica, and J. nigra, a combination of homology-based inference, ab initio prediction, and transcripts from RNA sequencing (RNA-Seq) was used (see details in supplementary note 2, Supplementary Material online). The final gene sets were functionally annotated using BlastP (minimum mapping length of 50 bp, minimum identity of 50%, minimum coverage of 50%, and minimum e-value of 1e-5) against the NCBI NR, UniProt-TrEMBL, and KEGG databases (supplementary table 6, Supplementary Material online). GO annotation was performed using Blast2GO (Conesa et al. 2005), and pathway annotation were performed using KAAS (Moriya et al. 2007).
Sampling Design and Resequencing
In 2018 and 2019, we visited all known locations of J. hopeiensis in the vicinity of Beijing, Tianjin, and Hebei provinces, and sampled 49 wild individuals. The range of J. hopeiensis overlaps with the ranges of J. mandshurica and J. cathayensis, which are sister species (Bai et al. 2016), and hybrids between J. mandshurica and J. cathayensis (Jc–Jm hybrids) may also have contributed to its genome. Ten individuals of Jc–Jm hybrids and 19 individuals of J. regia in sympatric distribution with J. hopeiensis were also sampled. In addition, ten trees of J. regia were sampled from the provinces Henan, Hubei, Shaanxi, and Xinjiang. All investigated adult trees were located in remote mountainous areas away from cities. Morphological characteristics of the leaves, fruits, and trunks were used for identification in the field. For each individual, six to eight flesh leaflets were dried and stored in silica gel. DNA was isolated according to the manufacturer’s protocol using the HP Plant DNA Kit D2485-02 (Omega Bio-Tek). Whole-genome sequencing was performed on Illumina NovaSeq 6000 instruments by NovoGene (Beijing, China). All individuals were sequenced to an expected average depth of 30×, with paired-end libraries of 350 bp insert size and read length of 150 bp. The genome resequencing data of 61 individuals of Asian butternuts (21 J. cathayensis, 20 Jc–Jm hybrids, and 20 J. mandshurica) (Xu et al. 2021) and two individuals of J. regia (Zhang, Xu, et al. 2019) were also included in our study (fig. 1 and supplementary table 1, Supplementary Material online). In total, 49 wild individuals of J. hopeiensis, 21 individuals of J. cathayensis, 30 individuals of Jc–Jm hybrids, 20 individuals of J. mandshurica, and 31 individuals of J. regia were included in this study.
Mapping and Variant Calling
The raw reads were trimmed for adapters and low-quality reads using Trimmomatic v0.32 (Bolger et al. 2014). Because P. stenoptera is equally related to Juglans species (Zhang, Xu, et al. 2019), all clean reads were mapped to a P. stenoptera reference genome (supplementary table 2, Supplementary Material online) using BWA-MEM algorithm of BWA v0.7.12 with default settings (Li 2013). Only uniquely mapped and properly paired reads were used in the analyses. The SAMtools v.0.1.19 (Li 2011) were used to convert the Sequence Alignment Map to a Binary Alignment Map format file and to remove polymerase chain reaction duplicates. Subsequently, the SENTIEON DNAseq software package v. 201808.08 (Weber et al. 2016) was used to realign indels, call SNPs from each individual, and to joint SNPs from all individuals. To control the quality of genome-wide SNPs, sites with a mapping depth of less than a third or more than double of an individual’s average depth, nonbiallelic sites, and sites with missing data were removed. Next, the Q20 filter was applied, and heterozygous genotypes called if the proportion of the nonreference allele was between 20% and 80% for a sequencing depth >20× (Nielsen et al. 2011), or if the proportion of the nonreference allele was between 10% and 90% for a sequencing depth >10×; otherwise, a homozygous genotype would be called. To obtain neutral and independent SNPs, those located in a coding sequence or its 10-kb extension region were discarded. Besides, singletons were excluded to reduce false positive effects caused by sequencing error. Linkage disequilibrium (LD) for each group was calculated using PopLDdecay v3.40 (Zhang, Dong, et al. 2019). Finally, these SNPs were thinned using a distance filter of interval >10 kb based on LD results.
The individuals mapping to P. stenoptera were prepared for two SNP data sets: 1) A five-group SNP data set including all sampled individuals, which was used to conduct analysis of STRUCTURE, PCA, and NEWHYBRID, and 2) a four-group SNP data set including two putative parents, Asian butternuts and Persian walnut, which was used to conduct analysis of fastsimcoal2 and BA3-SNPs. For the five-group and four-group SNP data sets, a total of 1,353 SNPs and 3,076 SNPs were obtained after the series of filtering methods described above.
Genetic Diversity and Structure
The nucleotide diversity (π) of the five groups was calculated in stepping windows 50 kb in size by VCFtools v0.1.13 (Danecek et al. 2011). A 50-kb window size was chosen for stepping window analyses because LD decays within this distance (the same below). Individual heterozygosity (H) was calculated as the number of polymorphic sites divided by the total length of the P. stenoptera reference genome. The folded site frequency spectrum (SFS) of each group was calculated by using a custom perl script.
Stepping window analysis with a size of 50 kb for each of the five groups suggested that J. hopeiensis had the highest nucleotide diversity (π = 0.0089 ± 0.0042), followed by J. cathayensis (π = 0.0040 ± 0.0023), Jc–Jm hybrids (π = 0.0040 ± 0.0023), J. mandshurica (π = 0.0040 ± 0.0022), and J. regia (π = 0.0037 ± 0.0022). The individual heterozygosity of J. hopeiensis (0.0088 ± 0.0002) is two times higher than that of J. cathayensis (0.0031 ± 0.0001), Jc–Jm hybrids (0.0031 ± 0.0001), J. mandshurica (0.0032 ± 0.0001), and J. regia (0.0030 ± 0.0002) (fig. 2C). The folded SFS showed that allele frequency distribution in J. hopeiensis is bimodal (supplementary fig. 1, Supplementary Material online) whereas allele frequency spectra in the other groups are unimodal, implying that there are more alleles with medium frequencies in J. hopeiensis than in the other groups, as is typical for F1s (supplementary fig. 1, Supplementary Material online).
To investigate the population structure of J. hopeinesis and its closest relatives, a PCA was performed using the R package SNPRelate v. 1.6.2 (Zheng et al. 2012) with default settings. STRUCTURE v. 2.3.4 (Pritchard et al. 2000) was used to cluster individuals based on K = 1–8, using the admixture model with correlated allele frequencies. To control unequal sample sizes among species, we set POPALPHAS = 1 with an initial value of ALPHA = 0.25 as suggested by Meirmans (2019). The optimal value of K was determined using both STRUCTURE HARVESTER v.0.6.94 (Earl and Vonholdt 2012) according to the delta K method of Evanno et al. (2005) and KFinder v1.0 according to the parsimony method of Wang (2019).
Genomic Admixture Source
The NgsAdmix v. 32 (Skotte et al. 2013) was used to estimate admixture source of J. hopeiensis individuals from its two parents, Asian butternuts and J. regia, across the whole genomes. Genotype likelihoods were calculated from bam files in ANGSD v 0.921 (Korneliussen et al. 2013) with the parameters “-doGlf 2, -doMajorMinor 1, -SNP_pval 1e-6, -doMaf 1,” and the result file was then input into NgsAdmix with 50-kb stepping windows and K = 2 ancestral populations. The longest five contigs (>10 Mb) were chosen to visualize the proportions of the parents’ ancestry for each stepping window.
Hybrid Identification
The program EasyParallel (Zhao et al. 2020), utilizing a multithread parallel algorithm to process multiple iterations of NewHybrids (Anderson and Thompson 2002), was used to assign the 49 individuals of J. hopeiensis to six genotype classes: Asian butternuts as pure parent A (Pure A), Persian walnut as pure parent B (Pure B), F1 progeny (F1), F2 progeny (F2), backcrosses with Asian butternuts (F1 × A), and backcrosses with Persian walnut (F1 × B). Three independent runs were performed on the same SNPs data with population structure analysis. Runs were performed with 50,000 Markov Chain Monte Carlo sweeps following 50,000 burn-in sweeps, and the Jeffries-like priors were used for both the allele frequency (θ) and mixing proportion (π) parameters. Juglans hopeiensis was set as the unknown, and other groups as pure A and pure B.
Population Demographic Analysis
We used the PSMC model (Li and Durbin 2011) to infer the timing of the end of gene flow between Persian walnut and Asian butternuts. The recommendations of using sequencing data with a mean genome coverage of ≥18×, a per-site filter of ≥10 reads, and no more than 25% of missing data were followed (Nadachowska-Brzyska et al. 2016). Therefore, ten individuals each of J. cathayensis, J. mandshurica, and J. regia were mapped to their own reference genome (supplementary table 2, Supplementary Material online), and ten individuals of Jc–Jm hybrids were mapped to both J. cathayensis and J. mandshurica reference genomes. The reads of J. hopeiensis were mapped to all three J. cathayensis, J. mandshurica, and J. regia reference genomes to assess if the reference genome used had any effect on the PSMC results. Regardless which genome was used, PSMC results were similar. The parameters in PSMC were set with quality adjusted to 50, the minimum mapping quality to 20, the minimum depth to one-third of average depth genome coverage, and maximum depth to 2-fold average depth genome coverage. For five of the groups, we used the default bin of 100-bp regions. A generation time of 30 years and a mutation rate of 2.06 × 10−9 site/year were used (Bai et al. 2018).
Chloroplast Genome Analysis
Reads of each individual of the five groups were mapped to the P. stenoptera chloroplast genome (NC_046428.1) using BWA-MEM algorithm of BWA v. 0.7.12 (Li 2013). The shared 80 protein-coding genes for the five groups and two outgroup species, J. nigra and P. stenoptera, were chosen from their chloroplast genome annotations. All 80 protein-coding genes were aligned with MAFFT v. 7.017 (Katoh and Standley 2013) and then converted to a coding sequence alignment with PAL2NAL v. 14 (Suyama et al. 2006). The haplotypes of all individuals were identified from DnaSP v6 (Rozas et al. 2017). The first, second, and third codon positions from each gene were treated as different subsets. The best partitioning scheme was determined using PartitionFinder v. 2.1 (Lanfear et al. 2017) with the GTRGAMMA model of substitution. We carried out phylogenetic reconstructions under the ML criterion in RAxML v 8.0.26 (Stamatakis 2014), with 1,000 rapid bootstraps and using P. stenoptera and J. nigra as the outgroup.
Testing Gene Flow and Divergence Time between the Putative Parents, Asian Butternuts, and J. regia
To test whether there is gene flow between the parent lineages, we used the coalescence-based method implemented in fastsimcoal2 (Excoffier et al. 2013). The 2D joint site frequency spectra were converted by easySFS.py (https://github.com/isaacovercast/easySFS) for a variant call format file for Asian butternuts and J. regia. Five evolutionary models were compared (supplementary fig. 6, Supplementary Material online), all of which represented dichotomous topologies with or without bidirectional gene flow after divergence. For each model, 100,000 coalescent simulations were performed to compute log-likelihoods, and global ML estimates were obtained from 100 independent runs, with 50 expectation conditional maximization algorithm cycles. The model with the smallest Akaike information criterion value was determined as the best. A parametric bootstrapping approach was used to construct 95% CI based on the best-fit model with 100 independent runs.
For two putative parents, we evaluated contemporary gene flow using a Bayesian framework as implemented by the software BA3-SNPs v1.1 (Wilson and Rannala 2003; Mussmann et al. 2019). Migration rates, allele frequencies, and inbreeding coefficients were adjusted to achieve acceptance rates between 0.2 and 0.6 as recommended by Wilson and Rannala (2003). After finding optimal mixing parameters for each run, Markov Chain Monte Carlo was applied for 20 million iterations, discarding the first two million iterations and sampling every 100th iteration. Five exploratory analyses were conducted with different random seeds to be sure of concordance. The 95% credible sets were constructed using the mean migration rate ± 1.96 mean standard deviation. Migrations rates were considered statistically significant if the credible set did not include zero.
Identifying Barrier Genomic Regions
To test for population differentiation between species of Asian butternuts and J. regia, two 2-taxon SNPs data sets (21 J. cathayensis and 31 J. regia and 20 J. mandshurica and 31 J. regia) were prepared (supplementary table 1, Supplementary Material online). After keeping only biallelic sites, removing missing data, and correcting quality in any genome of the both species, a total of 5,014,130 SNPs and 4,932,665 SNPs were retained between J. cathayensis and J. regia and J. mandshurica and J. regia, respectively. Estimates for FST and DXY were computed in 50-kb stepping windows with VCFtools v0.1.13 (Danecek et al. 2011) and python3 scripts (Sun et al. 2020), respectively. For each interspecific comparison between the either Asian butternuts species and J. regia, regions with relative and absolute genetic divergence falling above the 99th percentile were designated as outliers. Lastly, FST and DXY outliers were taken from shared regions by J. cathayensis and J. regia and J. mandshurica and J. regia.
To test for the presence of chromosome rearrangements that might contribute to postzygotic isolation, synteny and homology between J. cathayensis and J. regia as well as J. mandshurica and J. regia were identified. First, reference genomes of J. cathayensis and J. mandshurica were aligned to the J. regia reference genome using the nucmer program in the MUMmer4 package (Marcais et al. 2018) with the parameters set to “-c 500 -b 500 -l 100.” Repetitive sequences were removed in the reference genome because it was extremely time demanding for nucmer to deal with repeats. Alignments of a length <100 and identity <90% were filtered out. Chromosomal rearrangements including inversions, translocations, and duplications were then identified by using SyRI (Goel et al. 2019). To infer which inversions might be original to J. cathayensis, J. mandshurica, or J. regia, we made J. nigra an outgroup (supplementary table 2, Supplementary Material online). We also examined synteny between J. nigra and J. regia. A plotsr tool provided by SyRI was used to visualize pairwise alignments between species.
GO Enrichment Analysis
The R package clusterProfiler (Yu et al. 2012) was used to perform GO enrichment analysis for genes in the regions of FST and DXY outlier regions and inversions with length >10 kb. The Benjamini–Hochberg method was used for multitest correction, and overrepresented GO biological process terms were selected for those with a false discovery rate less than 0.05.
Positive Selection Analysis
To detect positive selection for genes in the regions of inversions and FST and DXY outliers, we carried out MK tests (McDonald and Kreitman 1991) and Tajima’s D tests (Tajima 1989) for each gene. MK tests were run using a custom python3 script that used Fisher’s exact tests to assess the statistical significance. Test statistics for Tajima’s D were calculated for each gene using DnaSP v6 (Rozas et al. 2017) and compared with 5,000 simulated samples to test for significance. For J. cathayensis, Tajima’s D was significantly negative (P < 0.05) for 23 genes in FST outlier regions, 63 genes in DXY outlier regions, and two genes in inversion regions, and it was significantly positive (P < 0.05) for two genes in DXY outlier regions; MK tests were significant (P < 0.05) for six and 32 genes in FST and DXY outlier regions, respectively. For J. mandshurica, Tajima’s D was significantly negative (P < 0.05) for nine genes in FST outlier regions, 21 genes in DXY outlier regions, and no gene in inversion regions; it was significantly positive (P < 0.05) for one gene in DXY outlier regions; MK tests were significant (P < 0.05) for three and 27 genes in FST and DXY outlier regions, respectively. For J. regia, Tajima’s D was significantly negative (P < 0.05) for nine genes in FST outlier regions, 19 genes in DXY outlier regions, and no gene in inversion regions, and it was significantly positive (P < 0.05) for one gene in FST outlier regions and 44 genes in DXY outlier regions; MK tests were significant (P < 0.05) for six and 24 genes in FST and DXY outlier regions, respectively.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2017YFA0605104), the National Natural Science Foundation of China (41671040 and 31421063), the “111” Program of Introducing Talents of Discipline to Universities (B13008), and a key project of State Key Laboratory of Earth Surface Processes and Resource Ecology.
Author Contributions
W.N.B. and D.Y.Z. conceived of the study. W.N.B., D.Y.Z., and S.S.R. conceptualized and wrote the manuscript. W.P.Z. and L.C. assembled the genomes. W.P.Z., W.N.B., L.C., X.R.L., Y.M.D, Y.L., and E.L.P. performed the analyses. E.L.P. contributed ideas, and assisted in editing the manuscript.
Data Availability
The newly resequenced raw reads from 88 individuals and four assembled genomes have been deposited at GenBank under the accession PRJNA356989 and the later are also available at the website (http://cmb.bnu.edu.cn/juglans).
References
- Anderson EC, Thompson EA.. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160(3):1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aradhya MK, Potter D, Gao F, Simon CJ.. 2007. Molecular phylogeny of Juglans (Juglandaceae): a biogeographic perspective. Tree Genet Genomes. 3(4):363–378. [Google Scholar]
- Baack E, Melo MC, Rieseberg LH, Ortiz-Barrientos D.. 2015. The origins of reproductive isolation in plants. New Phytol. 207(4):968–984. [DOI] [PubMed] [Google Scholar]
- Bai WN, Wang WT, Zhang DY.. 2016. Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. New Phytol. 209(4):1757–1772. [DOI] [PubMed] [Google Scholar]
- Bai WN, Yan PC, Zhang BW, Woeste KE, Lin K, Zhang DY.. 2018. Demographically idiosyncratic responses to climate change and rapid Pleistocene diversification of the walnut genus Juglans (Juglandaceae) revealed by whole-genome sequences. New Phytol. 217(4):1726–1736. [DOI] [PubMed] [Google Scholar]
- Bai WN, Zeng YF, Liao WJ, Zhang DY.. 2006. Flowering phenology and wind-pollination efficacy of heterodichogamous Juglans mandshurica (Juglandaceae). Ann Bot. 98(2):397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. 1909. Heredity and variation in modern lights. In: Seward AC editor. Darwin and modern science. Cambridge: Cambridge University Press. p. 85–101. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit RJ, Soto A, Gil L.. 2009. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity 102(5):442–452. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Soares AE, Green RE, Shapiro B.. 2016. Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic data. Philos Trans R Soc B. 371(1699):20150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Jin L, Zhao D, Li B, Zhang X.. 2015. Study on pollen physiological characteristics in Juglans hopeiensis Hu and Juglans regia (In Chinese). North Hortic. 23:42–44. [Google Scholar]
- Cheng S, Yang W.. 1987. Taxonomic studies of ten species of the genus Juglans based on isozymic zymograms (In Chinese). Acta Hortic Sin. 12:90–96. [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Coughlan JM, Matute DR.. 2020. The importance of intrinsic postzygotic barriers throughout the speciation process. Philos Trans R Soc Lond B Biol Sci. 375(1806):20190533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1989. Patterns of speciation in Drosophila. Evolution 43(2):362–381. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Dai SJ, Qi JX, Duan CR, Wang YP, Chen P, Li Q, Hao YB, Leng P.. 2014. Abnormal development of pollen and embryo sacs contributes to poor fruit set in walnut (Juglans hopeiensis). J Hortic Sci BioTech. 89(3):273–278. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang M, Zhou HJ, Woeste KE, Yue M, Zhang Y, Zhao GF, Zhang SX, Zhao P.. 2021. Comparative phylogeography of Juglans regia and J. mandshurica combining organellar and nuclear DNA markers to assess genetic diversity and introgression in regions of sympatry. Trees. 10.1007/s00468-021-02167-y. [DOI] [Google Scholar]
- Deng Y, Xie BX.. 2006. Origin and distribution of tree species of Juglandaceae (In Chinese). Nonwood for Res. 24:35–37. [Google Scholar]
- Dobzhansky TG. 1937. Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- Dong WP, Xu C, Li WQ, Xie XM, Lu YZ, Liu YL, Jin XB, Suo ZL.. 2017. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, Vonholdt BM.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4(2):359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14(8):2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M.. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9(10):e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XJ, Zhou HJ, Zulfiqar S, Luo X, Hu YH, Feng L, Malvolti ME, Woeste K, Zhao P.. 2018. The phytogeographic history of common walnut in China. Front Plant Sci. 9:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feulner M, Liede-Schumann S, Meve U, Weig A, Aas G.. 2013. Genetic structure of three Sorbus latifolia (Lam.) Pers. taxa endemic to northern Bavaria. Plant Syst Evol. 299(6):1065–1074. [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, Hill JP.. 2013. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkey flowers). Evolution 67(9):2547–2560. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang B, Mao JF, Ingvarsson P, Zeng QY, Wang XR.. 2012. Demography and speciation history of the homoploid hybrid pine Pinus densata on the Tibetan Plateau. Mol Ecol. 21(19):4811–4827. [DOI] [PubMed] [Google Scholar]
- Goel M, Sun H, Jiao WB, Schneeberger K.. 2019. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20(1):277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH.. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH. 1932. Identification of Chinese Hardwoods. Bull Fan Mem Inst Bio. 3:173–174. [Google Scholar]
- Hu HH. 1934. Notulae systematicae ad Florem Sinensium V. Bull Fan Mem Inst Biol. 5:305–306. [Google Scholar]
- Hu Y, Dang M, Feng XJ, Woeste K, Zhao P.. 2017. Genetic diversity and population structure in the narrow endemic Chinese walnut Juglans hopeiensis Hu: implications for conservation. Tree Genet Genomes. 13:91. [Google Scholar]
- Hu Y, Woeste KE, Zhao P.. 2016. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front Plant Sci. 7:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, He Z.. 2019. Textual research on origin and dissemination of Juglans regia L. in China (In Chinese). Agric Archaeol. 6:148–154. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Moltke I, Albrechtsen A, Nielsen R.. 2013. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics 14:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution. New York: Oxford University Press. [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint:arXiv:1303.3997v1302.
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. 2014. Wenwan walnut. World Chin 4:74. [Google Scholar]
- Lo EY. 2010. Testing hybridization hypotheses and evaluating the evolutionary potential of hybrids in mangrove plant species. J Evol Biol. 23(10):2249–2261. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Modliszewski JL, Wright KM, Wu CA, Willis JH.. 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos Trans R Soc Lond B Biol Sci. 363(1506):3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Jin L, Zhang X, Li B, Chen M.. 2014. Study on phenology observations and pollen characteristics of different Juglans hopeiensis Hu cultivars (In Chinese). North Hortic. 15:17–21. [Google Scholar]
- Manning WE. 1978. The classification within the Juglandaceae. Ann Mo Bot Gard. 65(4):1058–1087. [Google Scholar]
- Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A.. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14(1):e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrano A, Britton M, Zaini PA, Zimin AV, Workman RE, Puiu D, Bianco L, Pierro EAD, Allen BJ, Chakraborty S, et al. 2020. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 9(5):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather N, Traves SM, Ho SYW.. 2020. A practical introduction to sequentially Markovian coalescent methods for estimating demographic history from genomic data. Ecol Evol. 10(1):579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the adh locus in Drosophila. Nature 351(6328):652–654. [DOI] [PubMed] [Google Scholar]
- Meirmans PG. 2019. Subsampling reveals that unbalanced sampling affects STRUCTURE results in a multi-species dataset. Heredity 122(3):276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server Issue):W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu XY, Sun M, Yang PF, Lin QW.. 2017. Unveiling the identity of wenwan walnuts and phylogenetic relationships of Asian Juglans species using restriction site-associated DNA-sequencing. Front Plant Sci. 8:1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Xi R, Lv Z.. 1990. Microsporogenesis observation and karyotype analysis of some species in genus Juglans L. (In Chinese). J Wuhan Bot Res. 8:301–310. [Google Scholar]
- Muller HJ. 1942. Isolating mechanisms, evolution, and temperature. Biol Symp. 6:71–125. [Google Scholar]
- Mussmann SM, Douglas MR, Chafin TK, Douglas ME, Jarman S.. 2019. BA3‐SNPs: contemporary migration reconfigured in BayesAss for next‐generation sequence data. Methods Ecol Evol. 10(10):1808–1813. [Google Scholar]
- Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H.. 2016. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol Ecol. 25(5):1058–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH.. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57(3):447–459. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS.. 2011. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 12(6):443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J.. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 98(21):12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Zhou RC, Li YQ, Havanond S, Jaengjai C, Shi SH.. 2008. Molecular evidence for natural hybridization between Sonneratia alba and S. griffithii. J Syst Evol. 46:391–395. [Google Scholar]
- Ramirez-Aguirre E, Marten-Rodriguez S, Quesada-Avila G, Quesada M, Martinez-Diaz Y, Oyama K, Espinosa-Garcia FJ.. 2019. Reproductive isolation among three sympatric Achimenes species: pre- and post-pollination components. Am J Bot. 106(7):1021–1031. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16(7):351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH.. 2007. Plant speciation. Science 317(5840):910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Rich TCG, Allen AM, Houston L, Roberts C, Bridle JR, Harris SA, Hiscock SJ.. 2010. Hybridization and polyploidy as drivers of continuing evolution and speciation in Sorbus. Mol Ecol. 19(8):1675–1690. [DOI] [PubMed] [Google Scholar]
- Robins TP, Binks RM, Byrne M, Hopper SD.. 2021. Landscape and taxon age are associated with differing patterns of hybridization in two Eucalyptus (Myrtaceae) subgenera. Ann Bot. 127(1):49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A.. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302. [DOI] [PubMed] [Google Scholar]
- Ru D, Sun Y, Wang D, Chen Y, Wang T, Hu Q, Abbott RJ, Liu J.. 2018. Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Mol Ecol. 27(23):4875–4887. [DOI] [PubMed] [Google Scholar]
- Schumer M, Rosenthal GG, Andolfatto P.. 2014. How common is homoploid hybrid speciation? Evolution 68(6):1553–1560. [DOI] [PubMed] [Google Scholar]
- Schumer M, Rosenthal GG, Andolfatto P.. 2018. What do we mean when we talk about hybrid speciation? Heredity 120(4):379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A.. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195(3):693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lu Z, Zhu X, Ma H.. 2020. Genomic basis of homoploid hybrid speciation within chestnut trees. Nat Commun. 11(1):3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server Issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL.. 2019. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat Ecol Evol. 3(2):170–177. [DOI] [PubMed] [Google Scholar]
- Wang J. 2019. A parsimony estimator of the number of populations from a STRUCTURE-like analysis. Mol Ecol Resour. 19(4):970–981. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang Y, Bi H, Lu Z, Ma Y, Yang X, Chen N, Tian B, Liu B, Mao X, et al. 2021. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol Plant. 14(2):208–222. [DOI] [PubMed] [Google Scholar]
- Weber JA, Aldana R, Gallagher BD, Edwards JS.. 2016. Sentieon DNA pipeline for variant detection – software-only solution, over 20× faster than GATK 3.3 with identical results. PeerJ PrePrints. 4:e1672v1672. [Google Scholar]
- Wilson GA, Rannala B.. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163(3):1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth JR, Larcombe MJ, Sakaguchi S, Marthick JR, Bowman DM, Ito M, Jordan GJ.. 2016. Transient hybridization, not homoploid hybrid speciation, between ancient and deeply divergent conifers. Am J Bot. 103(2):246–259. [DOI] [PubMed] [Google Scholar]
- Wu Y, Pei D, Xi S, LI J.. 1999. Analysis of the origin and the taxonmic position of Juglans hopeiensis using RAPD markers (In Chinese). Sci Silva Sin. 35:25–30. [Google Scholar]
- Xi R. 1990. Textural criticism of walnut (Juglans regia L.) origin in China (In Chinese). J Agric Univ Hebei. 13:89–94. [Google Scholar]
- Xu LL, Yu RM, Lin XR, Zhang BW, Li N, Lin K, Zhang DY, Bai WN.. 2021. Different rates of pollen and seed gene flow cause branch-length and geographic cytonuclear discordance within Asian butternuts. New Phytol. 232(1):388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GC, Wang LG, Han YY, He QY.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BW, Xu LL, Li N, Yan PC, Jiang XH, Woeste KE, Lin K, Renner SS, Zhang DY, Bai WN.. 2019. Phylogenomics reveals an ancient hybrid origin of the Persian walnut. Mol Biol Evol. 36(11):2451–2461. [DOI] [PubMed] [Google Scholar]
- Zhang C, Dong SS, Xu JY, He WM, Yang TL.. 2019. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35(10):1786–1788. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Zhang WT, Ji FY, Qiu J, Song XB, Bu DC, Pan G, Ma QG, Chen JX, Huang RM, et al. 2020. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol J. 18(9):1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Beck B, Fuller A, Peatman E.. 2020. EasyParallel: a GUI platform for parallelization of STRUCTURE and NEWHYBRIDS analyses. PLoS One 15(4):e0232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Li MM, Zhao FD, Ma HB, Li BG, Qi GH.. 2014. Research on flowering and pollinating characteristics of precocious walnut (In Chinese). Hebei J Orch Res. 29:148–154. [Google Scholar]
- Zhao P, Zhou HJ, Potter D, Hu YH, Feng XJ, Dang M, Feng L, Zulfiqar S, Liu WZ, Zhao GF, et al. 2018. Population genetics, phylogenomics and hybrid speciation of Juglans in China determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS). Mol Phylogenet Evol. 126:250–265. [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Gong X, Boufford D, Wu CI, Shi S.. 2008. Testing a hypothesis of unidirectional hybridization in plants: observations on Sonneratia, Bruguiera and Ligularia. BMC Evol Biol. 8:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang H, liu K, Zhang Z.. 2020. Regularity and influence factors of fruits drop in Juglans hopeiensis Hu (in Chinese). North Hortic. 9:46–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly resequenced raw reads from 88 individuals and four assembled genomes have been deposited at GenBank under the accession PRJNA356989 and the later are also available at the website (http://cmb.bnu.edu.cn/juglans).