Abstract
Purpose
The COVID-19 disease with acute respiratory symptoms emerged in 2019. The causal agent of the disease, the SARS-CoV-2 virus, is classified into the Betacoronaviruses family. Coronaviruses (CoVs) are a huge family of viruses. Therefore, homologous recombination studies can help recognize the phylogenetic relationships among these viruses.
Methods
In order to detect possible recombination events in SASRS-CoV-2, the genome sequences of Betacoronaviruses were obtained from the GenBank. The nucleotide sequences with the identity ≥ 60% to SARS-CoV-2 genome sequence were selected and then analyzed using different algorithms.
Results
The results showed two recombination events at the beginning and the end of the genome sequence of SARS-CoV-2. Bat-SL-CoVZC21 (GenBank accession number MG772934) was specified as the minor parent for both events with p-values of 8.66 × 10–87 and 3.29 × 10–48, respectively. Furthermore, two recombination regions were detected at the beginning and the middle of the SARS-CoV-2 spike gene. Pangolin-CoV (PCoV_GX-P4L) and Rattus CoV (ChRCoV-HKU24) were determined as the potential parents with the GenBank accession number MT040333 and KM349742, respectively. Analysis of the spike gene revealed more similarity and less nucleotide diversity between SARS-CoV-2 and pangolin-CoVs.
Conclusion
Detection of the ancestors of SARS-CoV-2 in the coronaviruses family can help identify and define the phylogenetic relationships of the family Coronaviridae. Furthermore, constructing a phylogenetic tree based on the recombination regions made changes in the phylogenetic relationships of Betacoronaviruses.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10529-021-03218-7.
Keywords: Recombination, RDP, SARS-CoV-2, Phylogeny, Pangolin, Rattus
Introduction
Coronaviruses were discovered in the 1960s and classified as the family Coronaviridae (Woo et al. 2010). The family Coronaviridae includes two subfamilies: Orthocoronavirinae and Torovirinae. The subfamily Orthocoronavirinae consists of four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Woo et al. 2010). Alpha and Beta-coronaviruses are related to mammals; for example, bats (Woo et al. 2012) and Gammacoronaviruses are common among bird species (Lin et al. 2016; Zhou et al. 2018; and Mardani et al. 2008). Avian infectious bronchitis virus (IBV), human coronavirus 229E (HCoV-229E), and human coronavirus OC43 (HCoV-OC43) were the first identified coronaviruses. They caused respiratory illnesses in chickens and colds in humans. Since the advent of HCoV-229E and HCoV-OC43 (Van der Hoek 2007), several acute coronaviruses have been discovered, such as severe acute respiratory syndrome (SARS) in 2002 and the Middle East respiratory syndrome (MERS) in 2012.
In December 2019, a report was published about patients with severe viral pneumonia in Wuhan, China (Zhu et al. 2020). In determining the virus sequence obtained from these patients, a new CoV was identified as the causative agent of this respiratory disease (Zhu et al. 2020). Coronavirus 2019 has recently been named by the World Health Organization (WHO) as SARS-CoV-2 that caused COVID-19 disease. Unlike all human COVs that cause mild respiratory symptoms, SARS-CoV, MERS-CoV, and SARS-CoV-2 are associated with severe respiratory illness (Drosten et al. 2003; Zaki et al. 2012). SARS-CoV-2 appeared in Wuhan, Hubei Province, China, with fever, severe respiratory infection, and pneumonia (Chan et al. 2020; Huang et al. 2020a). SARS-CoV-2 is a new member of the Betacoronavirus closely related to bat coronaviruses (Lu et al. 2020; Wu et al. 2020). SARS-CoV appeared in China's Guangdong Province in 2002, infecting 8,098 people and leaving 774 dead. In 2012, MERS-CoV appeared on the Arabian Peninsula, infecting a total of 2,494 people and killing 858 people (Walls et al. 2020). SARS-CoV-2 was transmitted more rapidly from human to human than SARS-CoV and was spread to several continents (Chan et al. 2020; Chen et al. 2020; Li et al. 2020).
CoVs are surrounded by a lipid layer derived from the host cell membrane. CoVs are positive single-stranded RNA viruses characterized by spike proteins in the surface of the virion (Barcena et al. 2009; Neuman et al. 2006). The CoVs genome is the second-largest RNA among viruses, 26 to 32 kbp (Lai 1990). Structural proteins and several non-structural proteins with different functions are encoded by the 3' end of the viral genome (Masters 2006). Two-thirds of the 5' end of RNA strand encodes the non-structural proteins important in viral replication, including RNA-dependent RNA polymerase (RdRP) (Masters, 2006). The proteins encoded by viral RNA include spike proteins (S), membrane proteins (M), surface coat proteins (E), and nucleocapsid proteins (N). However, some beta coronaviruses also include hemagglutinin esterase (HE) (Fehr and Perlman 2015). The homo-trimer structure of the S protein has many N-linked glycans required for proper folding of the protein (Rossen et al. 1998). The S protein consists of two functional subunits. Subunit S1 is responsible for connecting to the host cell receptor, and subunit S2 is responsible for fusing viral and cell membranes (Walls et al. 2016). Combining a viral envelope with a host cell membrane leads to releasing a viral genome into the cytoplasm (He et al. 2006).
Previous studies have shown that bats have CoVs that are the ancestors of SARS-CoV. Also, it has been specified that the Himalayan palm civets had SARS-like CoVs in local Chinese markets (Guan et al. 2003). Therefore, these animals were introduced as mediators of virus transmission between bats and humans (Lau et al. 2005). At the beginning of the outbreak of SARS-CoV-2, researchers hypothesized that the SARS-CoV-2 was attributed to the Huanan Seafood Market in Wuhan, China, where one or more animals traded may have been the direct zoonotic source of the virus (Lam et al. 2020; Wu et al. 2020; Zhou et al. 2020; Zhu et al. 2020). Several reports, however, claimed that the initial occurrence of infection was unrelated to the Huanan Seafood Market (Huang et al. 2020a, 2020b). As a result, initiatives to track down the source of SARS-CoV-2 should not be confined to animals sold in markets but should include a broad spectrum of wild species not sold in markets (Huang et al. 2020b).
As determined in the case of SARS-CoV and MERS-CoV (Li et al. 2005), the bat is considered a likely species of origin for SARS-CoV-2. In 2020, a study on the SARS-CoV-2 genome was performed using Simplot (similarity plotting) software and determined that SARS-CoV-2 was remarkably similar to bat coronavirus (BatCoV-RaTG13) throughout the genome with a 96.2% genome sequence identity. However, there was no evidence of recombination events in the genome of SARS-CoV-2 (Zhou et al. 2020). Several other articles published in 2020 point to the close link between the SARS-CoV-2 genome sequence and the BatCoV-RaTG13, which was isolated from Rhinolophus affinis (Zhang et al. 2020a, 2020b; Xiao et al. 2020). Furthermore, a study described that SARS-CoV-2 was more closely related to two bat-derived coronavirus strains, Bat-SL-CoVZC45 and Bat-SL-CoVZC21 (Lu et al. 2020). The recombinant event in the 1b nucleotide region of the SARS-CoV-2 genome was discovered with the help of Simplot software, and it was suggested that SARS-CoV-2 may have originated in bats (Lu et al. 2020).
Since human-to-human transmission during the SARS-CoV-2 outbreak is attributed to the compatibility of the S protein (especially RBD) to bind to the human ACE2, the possibility of coronavirus transmission through one of these animals is raised. Before infecting humans, SARS-CoV and MERS-CoV typically infected intermediate hosts (Cui et al. 2019). So, SARS-CoV-2 was most likely spread to humans by other animals. Identifying and isolating the intermediate SARS-CoV-2 host is critical to preventing interspecies transmission (Zhang et al. 2020b).
On March 24, 2019, the Guangdong Wildlife Rescue Center received 21 live Malayan pangolins (Manis javanica) from the Anti-Smuggling Customs Bureau. Most of the animals were in poor health, and after rescue operations, 16 of them eventually died (Liu et al. 2019). The majority of the dead pangolins exhibited an enlarged lung filled with a frothy liquid, as well as pulmonary fibrosis symptoms. Analysis of lung samples confirmed the presence of a SARS-like CoV in two out of the 11 cases of dead Malayan pangolins based on a viral metagenomic study (Liu et al. 2019). In another study, during March-August 2019, lung tissues from four Chinese pangolins (Manis pentadactyla) and 25 Malayan pangolins (Manis javanica) were taken from a Wildlife Rescue Center. To identify SARS-related coronaviruses, they used the RT–PCR method with primers targeting a region of Betacoronaviruses. Their results determined pangolin-Cov in 17 cases of Malayan pangolins, while all samples of Chinese pangolins were negative (Xiao et al. 2020).
All publicly available metagenome samples of pangolin-CoV were collected and investigated to learn more about the animal hosts of SARS-CoV-2. Researchers assembled a draught genome of the SARS-CoV-like coronavirus, which showed 73% coverage and 91% sequence identity to the SARS-CoV-2. So, it is suggested that pangolins probably play a role in the evolution of SARS-CoV-2 and its transmission from bats to humans (Zhang et al. 2020a). In another article, researchers re-evaluated previously published data (Liu et al. 2019) about SARS-CoV-like coronaviruses identified in pangolin lung samples to access the genomic and evolutionary evidence of the pangolin-CoV (Zhang et al. 2020b). Their results showed that pangolin-CoV had 91.02% and 90.55% identical to SARS-CoV-2 and BatCoV-RaTG13, respectively, at the whole-genome level. So, it was concluded that pangolin species might serve as a natural reservoir for SARS-CoV-2 (Zhang et al. 2020b).
In this paper, the SARS-CoV-2 gene was investigated by several algorithms to detect recombination events and find its ancestor genes. Then, the spike gene was examined to detect nucleotide sequences with high similarity with SARS-CoV-2.
Materials and methods
Analysis of the SARS-CoV-2 genome sequence to detect recombinant regions
The SARS-CoV-2 genome sequence was obtained from the Genebank (GenBank accession number MT049951). The nucleotide sequences belong to the Betacoronavirus family were selected to compare with SARS-CoV-2. Genome sequences of coronaviruses in Apodemus, Bats, Bos, Camelus, Canis, Equus, Felis, Mustela, Giraffa, Erinaceus, Neovison, Murines, Pangolins, Sus, Rabbits, Rattus, Rusa, Shrew, Hydropotes, Kobus, Odocoileus, and Hippotragus were obtained from the GenBank database of the National Center for Biotechnology Information (NCBI) database. The number of the used nucleotide sequences of each group is given in Supplementary Table 1. The alignment of the whole-genome sequence of the SARS-CoV-2 with other sequences was carried out using MEGA v6.12 software and Clustal Muscle v2.1 server, and percent identity between the SARS-CoV-2 nucleotide sequence and other sequences were determined. The sequences of over 60% identity were used for the rest of the study. RDP v4.5 software was used to detect recombination events (Martin et al. 2015), and the occurrence of all recombination events was examined using RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, and 3Seq programs. For the detection of the recombination regions to be valid, p-value ≤ 0.05 was considered. The beginning and end breakpoints related to each recombination event were determined. Phylogenetic trees were constructed based on the neighbor-joining (NJ) method to examine the possible relations between different viruses. Moreover, the relations between the parents identified in each recombination event were investigated. Once the recombination regions were detected and specified in the SARS-CoV-2 genome, the encoding fragments of the genome were independently studied.
Investigation of the nucleotide sequences of the spike gene
Due to the identification of two different genetic groups as parents in the SARS-CoV-2 spike gene, the nucleotide sequences of this gene were examined and compared. The sequences were first aligned among the members of each group using Align Sequences tool embedded in the server of Virus Pathogen Database and Analysis Resource (Pickett et al. 2012). The Synonymous/Non-synonymous Analysis Program (SNAP v2.1.1) was used to determine the selective pressure which might have happened in the spike gene of the expected groups (group 1: SARS-CoV-2 and pangolin-CoVs; group 2: SARS-CoV-2 and Rattus CoVs). SNAP calculates the non-synonymous (dN) and synonymous (dS) substitution rates of amino acids based on a set of codon-aligned nucleotide sequences (Nei and Gojobori 1986). The rate of nucleotide substitution and the pairwise distances of nucleotide sequences among each group member of the spike gene were estimated using MEGA v6.0 software. Nucleotide and haplotype diversity of the spike gene, Tajima's D-test, and Fu and Li's F* test statistic were examined using DnaSP v6.12 software (Rozas et al. 2017).
Polymorphism identification and motif analysis
For identifying the protein sequence polymorphisms, nucleotide sequences of the spike gene belong to the coronaviruses of groups 1 and 2 (SARS-CoV-2 and pangolin-CoVs; SARS-CoV-2 and Rattus CoVs) were translated into amino acid sequences with the help of a translate tool at the Expasy website. The multiple sequence alignments were made for all sequences using Virus Pathogen Database and Analysis Resource that uses the MUSCLE (Multiple Sequence Comparison by Log-Expectation) algorithms as a preprocessor to enhance the quality and speed of sequence alignment. The outcome obtained from the server was used to find sequence variations (Pickett et al. 2012). Metadata-driven Comparative Analysis Tool (meta-CATS) was also used to ensure that the substitutions occurred between the residues in protein sequences (Pickett et al. 2012). The meta-CATS tool uses a chi-square test to report changes of the amino acids with a p-value ≤ 0.05.
The potential motif discovery and their distribution in the spike protein sequences were investigated using MEME Suite 5.4.1 server (Bailey et al. 2006) for all coronaviruses in groups 1 and 2.
Results and discussion
Detection of recombination events
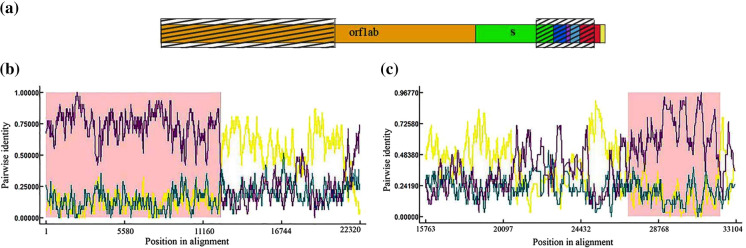
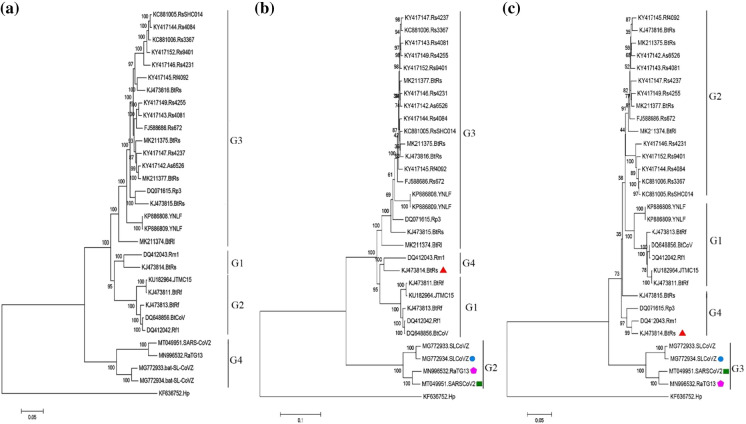
Percentages of identity between the nucleotide sequences of SARS-CoV-2 and other coronaviruses were calculated using the whole-genome alignment (these values are presented in Supplementary Table 1). Based on the results, SARS-CoV-2 had the highest percent identity (96%) to BatCoV-RaTG13 (GenBank accession number MN996532) and the lowest one (51%) to Hydropotes CoV (GenBank accession number MG518518). The nucleotide sequences of over 60% identity to SARS-CoV-2 genome sequence were observed in coronaviruses belonging to shrew, Rusa, Rattus, Rabbits, Erinaceus, Bos, pangolin, Apodemus, and bats. These nucleotide sequences were examined using RDP software to detect putative recombinant events. A recombination event was observed when comparing the SARS-CoV-2 genome sequence to bat, Rattus, and pangolin coronaviruses. The details of these coronaviruses are given in Supplementary Table 2. The genome sequences of SARS-CoV-2 and bat CoVs were analyzed using RDP software, and the results are shown in Fig. 1. The whole genome of coronavirus is illustrated in two Figs. (1b, c) to get a higher resolution. According to the results, two recombination events were detected at the beginning and ending segments of the SARS-CoV-2 genome. The first event was observed at positions 1–12,394 of the aligned nucleotide sequence of SARS-CoV-2 with a p-value = 8.66 × 10–87. As shown in Fig. 1a, this nucleotide region (left hatched area) is associated with the initial segment of the orf1ab gene. The results showed that this segment of the SARS-CoV-2 genome might appear due to the genetic recombination of the bat coronaviruses. Bat-SL-CoVZC21 (GenBank accession number MG772934) and BtRs-BetaCoV-HuB (GenBank accession number KJ473814) were identified as a minor parent (identity 91.6%) and a major parent, respectively. This recombination event was further evaluated using other methods, and the results are shown in Table 1. Comparison of the beginning and ending breakpoints using 3Seq and LARD methods demonstrated that the same positions (1–12,394) were recombined with p-values of 1.34 × 10–96 and 1.72 × 10–145. The position of the second recombination event at the end of the SARS-CoV-2 genome is shown in Fig. 1c. The position 27,088–32,336 of the whole-genome alignment sequence of SARS-CoV-2 was identified as a recombination event in the RDP method. In this case, the p-value was calculated to be 3.29 × 10–48. This nucleotide region was determined by the MaxChi and LARD methods with almost similar beginning and ending breakpoints and p-values of 8.41 × 10–30 and 1.20 × 10–79 (Table 2). The major and minor parents for the event were bat-SL-CoVZC21 (GenBank accession number MG772934) and BtRs-BetaCoV-HuB (GenBank accession number KJ473814), respectively. As illustrated in Fig. 1a, the second recombination region encompassed the ending segments of the spike gene, E gene, M gene, and the beginning segment of N gene (right hatched area). A phylogenetic tree was constructed for the SARS-CoV-2 sequence and 30 genomic sequences of bat CoVs to analyze the recombination occurrence further. The constructed phylogenetic tree without recombination is shown in Fig. 2a. The sequences were categorized into four major clusters (G1-G4) in this mode. The average between the minimum and maximum percent identity of sequences of each cluster was determined. This value for the G1, G2, and G3 cluster members was 99.81%, 98.54%, and 95.82%. SARS-CoV-2 genome sequence was placed in the G4 cluster. The average sequence identity for the members of this cluster was calculated to be 93.11%. Since recombination can affect phylogenetic reconstruction (Sabella et al. 2018), the phylogenetic tree was reconstructed based on the nucleotide sequence of the recombination region, and the relations between the identified parents were investigated for every recombination event after detecting those events in the coronavirus genome sequence. The phylogenetic tree constructed for the first recombinant region (at the beginning of the genome sequence) is shown in Fig. 2b. Categorization was performed based on the average between the minimum and maximum percentage of the sequence identity shared among the members. As it can be seen, the SARS-CoV-2 nucleotide sequence was placed in the G2 cluster. MG772934 coronavirus identified as a minor parent was also grouped in this cluster. The average sequence identity for the members of the cluster was calculated to be 93.74%. KJ473814 (from the species Rhinolophus sinicus (Wu et al. 2016)), serving as a major parent, was in the G4 cluster. MG772934 coronavirus was first found in Rhinolophus sinicus in 2018 (Hu et al. 2018). The phylogenetic tree was constructed based on the recombination region at the end of the SARS-CoV-2 genome sequence, and the resulting phylogram was shown in Fig. 2c. According to the results, SARS-CoV-2 and the minor parent (MG772934) were grouped in the G3 cluster. The average sequence identity for the members of the cluster was calculated to be 93.83%. The major parent of this event was grouped in the G4 cluster, and the nucleotide sequence identity for its members was determined to be 91.33%. The results confirmed that MG772934 was the potential parent of the first and second recombination. After the SAR-CoV outbreak during 2002–2004, researchers were seeking the potential source of the virus among other animals. For this purpose, the species of Rhinolophus was studied more than other live organisms. Forty-seven coronaviruses associated with that species were identified by 2018 (Luk et al. 2019). Extensive research was conducted on this subject due to the pandemic outbreak of SARS-CoV-2 in 2019. Comparing the SAR-Cov-2 genome and other coronavirus revealed some similarities between human and Rhinolophus CoVs (Wassenaar and Zou 2020). Wassenaar and Zou analyzed 253 nucleotides upstream of the start codons of coronaviruses to find the source of the virus. They reported a close genetic relationship among the Sarbecovirus species that contains SARS-CoV-2 and coronaviruses belonging to the species of Rhinolophus (Wassenaar and Zou 2020). Their results indicated a close relationship between the nucleotide sequences of SARS-CoV-2 and MG772933. So, MG772933 might be the source of the SARS-CoV-2 (Wassenaar and Zou 2020). Our results complied with theirs and determined that MG772934, with 97.47% sequence identity to MG772933, might be regarded as the parent of SARS-CoV-2. Furthermore, our results displayed that the recombination events at the beginning and the end of the SARS-CoV-2 genome were observed almost at the same positions in BatCoV-RaTG13 (GenBank accession number MN996532). The analysis of the whole-genome sequence showed 96% identity between BatCoV-RaTG13 and SARS-CoV-2. The BatCoV-RaTG13 was isolated from Rhinolophus affinis in China's Yunnan Province (Zhou et al. 2020). Considering the geographical distance between the location where SARS-CoV-2 emerged and the habitat of the BatCoV-RaTG13 host, it could be hypothesized that another animal acted as a mediator between bats and humans. That hypothesis concerning the virus epidemiology can justify the presence of the coronaviruses isolated from different species or obtained from different geographical regions in a phylogenetic tree (Sabella et al. 2018). The SARS-CoV-2, pangolin-CoVs, and Rattus CoVs genome sequences were analyzed using RDP software, and the recombination regions were detected in the genome of SARS-CoV-2. These events were unique to the SARS-CoV-2 coronavirus and not observed in BatCoV-RaTG13 coronavirus.
Fig. 1.
Examination of possible recombination in full-length segments of SARS-CoV-2. A schematic sequence of SARS-CoV-2 genome with different display colored-parts, mustard, green, blue, purple, and red exhibit orf1ab, spike, E, and N gene regions, respectively. Hatched areas belong to recombination events. B and C plot display graphically illustrating recombination events, areas 15,763 to 22,320 overlap. Pink regions are related to the sequence with a recombinant origin. Yellow line: MG772934-KJ473814. Purple line: MG772934-MT049951. Green line: KJ473814-MT049951
Table 1.
Details of recombination event at the beginning of SARS-CoV-2 genome
| Program | p-value | Beginning breakpoint | End breakpoint | *Parents | |||
|---|---|---|---|---|---|---|---|
| position in alignment | position in genome without gap | position in alignment | position in genome without gap | Major | Minor | ||
| RDP | 8.66 × 10–87 | 1 | 1 | 12,394 | 11,330 | KJ473814 | MG772934 |
| GENECONV | 2.68 × 10–62 | 661 | 620 | 8920 | 7918 | ||
| BootScan | 2.44 × 10–74 | 3690 | 2984 | 12,393 | 11,331 | ||
| MaxChi | 1.10 × 10–45 | 535 | 497 | 12,469 | 11,405 | ||
| Chimaera | 1.09 × 10–48 | 968 | 918 | 12,347 | 11,283 | ||
| SiSscan | 1.34 × 10–96 | 3661 | 2973 | 12,468 | 11,404 | ||
| 3Seq | 6.09 × 10–186 | 1 | 1 | 12,470 | 11,406 | ||
| LARD | 1.72 × 10–145 | 1 | 1 | 12,394 | 11,330 | ||
*Parents belonging to the bat coronavirus
Table 2.
Details of recombination event at the end of SARS-CoV-2 genome
| Program | p-value | Beginning breakpoint | End breakpoint | *Parents | |||
|---|---|---|---|---|---|---|---|
| position in alignment | position in genome without gap | position in alignment | position in genome without gap | Major | Minor | ||
| RDP | 3.29 × 10–48 | 27,088 | 24,212 | 32,336 | 29,194 | KJ473814 | MG772934 |
| GENECONV | 9.06 × 10–44 | 28,834 | 25,921 | 31,394 | 28,284 | ||
| BootScan | 8.77 × 10–44 | 27,897 | 25,022 | 31,546 | 28,419 | ||
| MaxChi | 8.41 × 10–30 | 27,088 | 24,212 | 32,336 | 29,194 | ||
| Chimaera | 5.71 × 10–32 | 26,670 | 23,817 | 32,336 | 29,194 | ||
| SiScan | 4.35 × 10–56 | 28,117 | 25,228 | 32,336 | 29,194 | ||
| Phylpro | 4.05 × 10–13 | 27,088 | 24,212 | 32,336 | 29,194 | ||
| 3Seq | 4.05 × 10–13 | 26,668 | 23,815 | 32,337 | 29,195 | ||
| LARD | 1.20 × 10–79 | 27,088 | 24,212 | 32,336 | 29,194 | ||
*Parents belonging to bat coronavirus
Fig. 2.
Phylogenetic trees of genomic RNA of SARS-CoV-2 and bat coronaviruses (30 items). A full-length RNA, B recombination sequence at the first region of the genome (beginning's part of orf1ab gene), and C recombination sequence at the end region of the genome (The end of the spike gene, E gene, and the beginning of the N gene). (square) putative recombinant, (circle) potential minor parent, (triangle) potential major parent, and (pentagon shape) sequence with evidence of the same recombination event. The phylogenetic trees were constructed by the neighbor-joining (NJ) method with the bootstrap value (1000 replicates)
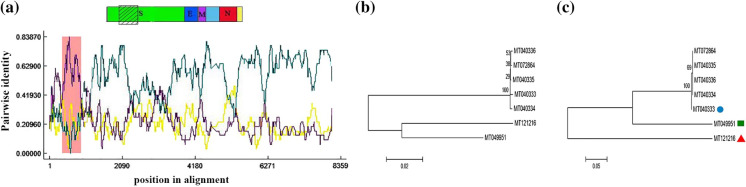
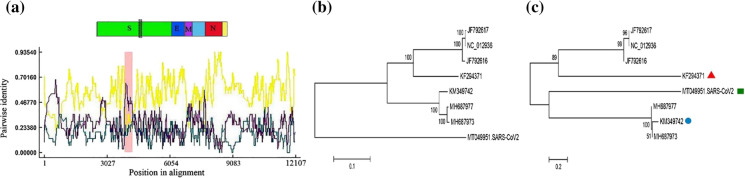
The result of the genome sequence analysis of the SARS-CoV-2 and pangolin-CoVs is shown in Fig. 3a. The recombination event at the spike gene encompassed the position 367–916 in the aligned nucleotide sequence of SARS-CoV-2 with a p-value of 7.74 × 10–18. The recombination occurrence in this region of the genome was also analyzed using other methods. The results given in Table 3 showed that this region of the SARS-CoV-2 genome was identified as the site of recombination occurrence using BootScan and SiScan methods. In this case, the minor and major parents were the coronaviruses with GenBank accession numbers MT040333 (81.1% identity) and MT121216 (91.9% identity), respectively. Both coronaviruses have been isolated from Manis Javanica (Lam et al. 2020). The results obtained by comparing the nucleotide sequence of SARS-CoV-2 with Rattus CoVs are shown in Fig. 4a. The recombination event with a higher rate of occurrence (p-value = 5.79 × 10–5) was detected at 3863–4254 of the aligned SARS-CoV-2 sequence. The minor and major parents identified for this event were the coronaviruses with GenBank accession numbers KM349742 (Rattus norvegicus as host (Lau et al. 2015)) and KF294371 (Rattus losea as host (Wang et al. 2015)), respectively. Since this region was specified as a recombination site using other methods (Table 4), the occurrence of this recombination can be confirmed. Because the recombination events were observed only in SARS-CoV-2 and not in BatCoV-RaTG13, it was likely that the Manis javanica, Rattus norvegicus, and Rattus losea hosting coronaviruses could act as the reservoirs of the SARS-CoV-2 and its transmission route from bats to humans. This hypothesis is not yet proven and needs more investigation to be fully confirmed. The phylogenetic trees were constructed for the nucleotide regions in two modes: without recombination events (Figs. 3b, 4b) and with recombination events (Figs. 3c, 4c). According to a previous study, the researchers believe in the possibility that the virus may be transmitted to humans by infecting another mammal (Wassenaar and Zou 2020). As a result, any animals that may have close contact with humans should be investigated. The studies on this subject introduced pangolins as candidates for transmitting viruses to humans (Liu et al. 2019; Wacharapluesadee et al. 2020; Xiao et al. 2020; Zhang et al. 2020a, 2020b). Previous studies conducted on the primary source of MERS-CoV showed that bats were the primary host of the virus. However, dromedary camels were a reservoir for the virus and transmitted it to humans (Haagmans et al. 2014; Memish et al. 2013). The SARS-CoV and SARS-CoV-2 viruses shared similarities in their genetic sequences and originated from bats (Ge et al. 2013; Yang et al. 2016; Zhou et al. 2020). In SARS-CoV, palm civets are believed to be the reservoir and transmitter of the virus (Kan et al. 2005; Wang et al. 2005), but the intermediate host of SARS-CoV-2 is still unknown.
Fig. 3.
Analysis of the possible homologous recombination in alignment sequences of SARS-CoV-2 and pangolin coronaviruses containing spike, E, and N genes. A plot display graphically illustrating recombination event at 367–916 position in alignment sequences (Pink region). Yellow line: MT121216- MT040333. Purple line: MT040333-MT049951. Green line: MT121216-MT049951. A schematic structure of the SARS-CoV-2 genome has been exhibited in the above plot. B Phylogenetic tree based on the ignore of recombination events, and C Phylogenetic tree based on the recombination event, (square) putative recombinant, (circle) potential minor parent, (triangle) potential major parent. The phylogenetic trees were constructed by the neighbor-joining (NJ) method with the bootstrap value (1000 replicates). (Color figure online)
Table 3.
Details of recombination event at the beginning of the spike gene in SARS-CoV-2
| Program | p-value | Beginning breakpoint | End breakpoint | *Parents | |||
|---|---|---|---|---|---|---|---|
| Position in alignment | Position in genome without gap | Position in alignment | Position in genome without gap | Major | Minor | ||
| RDP | 7.74 × 10–18 | 367 | 352 | 916 | 901 | MT121216 | MT040333 |
| GENECONV | 3.56 × 10–07 | 473 | 458 | 599 | 584 | ||
| BootScan | 2.17 × 10–15 | 367 | 352 | 916 | 901 | ||
| MaxChi | 9.50 × 10–9 | 350 | 335 | 916 | 901 | ||
| Chimaera | 1.05 × 10–02 | 355 | 340 | 916 | 901 | ||
| SiScan | 1.30 × 10–23 | 367 | 352 | 916 | 901 | ||
| Phylpro | |||||||
| 3Seq | 4.65 × 10–03 | 328 | 313 | 798 | 783 | ||
| §LARD | – | – | |||||
*Parents belonging to pangolin coronavirus
§ Unknown
Fig. 4.
Analysis of the possible homologous recombination in the alignment sequences of SARS-CoV-2 and Rattus coronaviruses containing spike, E, and N genes. A plot display graphically illustrating recombination event at 3863–4254 position in alignment sequences (Pink region). Yellow line: KF294371- KM349742. Purple line: KM349742-MT049951. Green line: KF294371-MT049951. A schematic structure of the SARS-CoV-2 genome has been exhibited in the above plot. B Phylogenetic tree based on the ignore of recombination events, and C Phylogenetic tree based on the recombination event, (square) putative recombinant, (circle) potential minor parent, (triangle) potential major parent. The phylogenetic trees were constructed by the neighbor-joining (NJ) method with the bootstrap value (1000 replicates)
Table 4.
Details of the recombination event at the beginning of the spike gene in SARS-CoV-2
| Program | p-value | Beginning breakpoint | End breakpoint | *Parents | |||
|---|---|---|---|---|---|---|---|
| position in alignment | position in genome without gap | position in alignment | position in genome without gap | Major | Minor | ||
| RDP | 5.79 × 10–5 | 3863 | 1472 | 4254 | 1626 | KF294371 | KM349742 |
| §GENECONV | – | – | – | – | – | ||
| §BootScan | – | – | – | – | – | ||
| §MaxChi | – | – | – | – | – | ||
| Chimaera | 6.22 × 10–6 | 3863 | 1472 | 4254 | 1626 | ||
| SiScan | 1.62 × 10–6 | 3863 | 1472 | NF | 1626 | ||
| Phylpro | 6.87 × 10–5 | 3863 | 1472 | 4254 | 1626 | ||
| §3Seq | – | – | – | – | – | ||
| LARD | 6.81 × 10–1 | 3863 | 1472 | 4254 | 1626 | ||
*Parents belonging to Rattus coronavirus
§ Unknown
Analysis of nucleotide sequence of the spike gene
The rate of changes in the nucleotides causing synonymous and non-synonymous amino acid changes are presented in Table 5. In both groups, the nucleotide sequences of SARS-CoV-2 were considered a standard sequence compared to other coronaviruses. At first, the numbers of nucleotide changes resulted in non-synonymous amino acid (dN) and synonymous amino acid (dS) substitutions were calculated. Then, the ratio of dN/dS was measured. The measurement of this parameter is a practical and efficient method for recognizing the natural selection pattern for genes during their evolution (Nei and Kumar 2000). The value of dN/dS > 1 represents positive selection, dN/dS < 1 means purifying selection, and dN/dS = 1 suggests neutral selection (Li 1997). The average value of dN/dS in group 1 (SARS-CoV-2 and pangolin-CoVs) was calculated to be 1.02 ± 0.05, exhibiting the neutral selection pattern during the evolution of the spike gene. Investigation about the selective pressure pattern for the second group (SARS-CoV-2 and Rattus CoVs) revealed that dN/dS value was 0.58 ± 0.01. Hence, the purifying selection pattern was considered for the spike gene in group 2.
Table 5.
Analysis of nucleotide sequence of spike gene
| Groups | Sequence names | §ds | †dn | dn/ds | *Pairwise distance |
|---|---|---|---|---|---|
| αI | MT049951_MT121216 | 0.15 | 0.17 | 1.13 | 0.37 |
| MT049951_MT072864 | 0.18 | 0.18 | 1 | 0.45 | |
| MT049951_MT040336 | 0.18 | 0.18 | 1 | 0.45 | |
| MT049951_MT040335 | 0.18 | 0.18 | 1 | 0.45 | |
| MT049951_MT040334 | 0.18 | 0.18 | 1 | 0.45 | |
| MT049951_MT040333 | 0.18 | 0.18 | 1 | 0.45 | |
| βII | MT049951_MH687977 | 1.19 | 0.69 | 0.58 | 0.72 |
| MT049951_MH687973 | 1.21 | 0.69 | 0.58 | 0.73 | |
| MT049951_JF792616 | 1.14 | 0.65 | 0.57 | 0.73 | |
| MT049951_JF792617 | 1.15 | 0.65 | 0.56 | 0.73 | |
| MT049951_NC-012936 | 1.15 | 0.65 | 0.57 | 0.73 | |
| MT049951_KM349742 | 1.17 | 0.69 | 0.59 | 0.74 | |
| MT049951_KF294371 | 1.10 | 0.67 | 0.61 | 0.75 |
§The rate of nucleotide substitution that caused synonym amino acid changes
†The rate of nucleotide substitution that caused non-synonym amino acid changes
*Pairwise distance refers to the amount of difference between nucleotide sequences
αGroup I including SARS-CoV-2 (MT049951) and pangolin-Covs
βGroup II including SARS-CoV-2 (MT049951) and Rattus Covs
Measuring the pairwise distance for the spike gene among the members of each group (Table 5) showed a shorter distance between the members of group 1 compared to those of group 2. The minimum distance of 0.37 between MT049951- MT121216 and the minimum distance of 0.72 between MT049951- JF792616 were obtained for group 1 and group 2, respectively. This parameter represents the rate of nucleotide substitutions between the sequences under study. The maximum and minimum values of this parameter are 0 and 1.
The results from examining the nucleotide substitutions in the spike gene in both groups are shown in Table 6. These factors (transition and transversion substitutions) are considered as indicators of molecular diversity (Tamura et al. 2004). In group 1, the highest rate of transition substitution belongs to pyrimidine bases; this rate for thymine-cytosine exchange and cytosine-thymine exchange was calculated to be 14% and 23.38%, respectively. Accordingly, the highest rate of change was observed in the C → T exchange due to cytosine methylation. This result is consistent with previous studies that reported the highest substitution rate in pyrimidine bases (Picoult-Newberg et al. 1999; Vignal et al. 2002; Zhang et al. 1994). Analysis of the transition substitutions in group 2 showed higher C → T and G → A exchanges rates. The results showed that transversion substitutions had higher values in group 2 in comparison with group 1. Generally, transversion substitutions exert more effect on nucleotide changes in a gene compared to transition substitutions. Therefore, it seems that members of group 2 have higher nucleotide diversity in the spike gene.
Table 6.
Nucleotide substitution matrix for spike gene
| Groups | A | T/U | C | G | |
|---|---|---|---|---|---|
| αI | A | – | 6.69 | 4.02 | 7.71 |
| T/U | 6.20 | – | 14.37 | 3.80 | |
| C | 6.20 | 23.91 | – | 3.80 | |
| G | 12.59 | 6.69 | 4.02 | – | |
| βII | A | – | 7.57 | 4.03 | 9.65 |
| T/U | 6.16 | – | 11.38 | 4.64 | |
| C | 6.16 | 21.36 | – | 4.64 | |
| G | 12.80 | 7.57 | 4.03 | – |
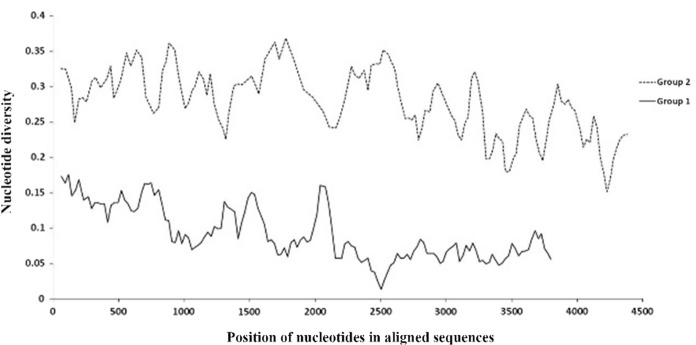
Measurement of nucleotide and haplotype diversity of the spike gene between members of each group was carried out using DnaSP software. In the present study, haplotype diversity was calculated to be 1 ± 0.076 and 1 ± 0.063 for group 1 and group 2, respectively. Haplotype diversity is a suitable marker to determine the rate of genetic diversity among populations. Haplotype diversity can vary from zero (all individuals of a population have similar haplotypes) to one (all individuals of a population have different haplotypes) (Aboim et al. 2005). Nucleotide diversity was determined to be 0.09 ± 0.03 and 0.28 ± 0.05 in group 1 and group 2, respectively. Figure 5 shows that nucleotide diversity is higher at the beginning of the gene (5' end) than at the end region of the gene in both groups. High haplotype diversity and low nucleotide diversity were observed in both groups. In an expanding population, haplotype diversity and the number of polymorphism sites increase rapidly while nucleotide diversity is left behind. As time passes, nucleotide diversity increases when the population expansion ceases.
Fig. 5.
Nucleotide diversity plot of spike gene. Group 1 including SARS-CoV-2 and pangolin-CoVs, and group 2 including of SARS-CoV-2 and Rattus CoVs
Values of Tajima's D-test and Fu-Li's F* test were calculated to be negative numbers for both groups; -0.86 and -0.76 for group 1 and -0.55 and -0.52 for group 2. More negative values of Tajima's D-test and Fu-Li's F* test are present in group 1. Tajima's D is a statistic of population genetics. It is the normalized difference between two estimators, of which one is derived from the average number of pairwise differences and the other from the number of segregating sites (Tajima 1989a). Tajima's D represents the expansion or contraction of population size, the strength of selection, and population structure. Usually, Tajima's D is used to examine whether the population follows three assumptions: (1) constant population size over time, (2) neutral evolution, (3) lack of population structure (for example, subdivision) (Kim et al. 2016). The sign of Tajima's D helps us to interpret natural selection (Biswas and Akey 2006). Natural selection and population dynamics determine the sign of Tajima's D. A positive and negative value suggests decreasing and increasing population size, respectively (Innan and Stephan 2000; Sano and Tachida 2005; Tajima 1989b; Kim et al. 2016). Measurement of the parameter will be highly effective when analyzing pathogens that evolve rapidly, such as RNA viruses, which accumulate random mutations during their epidemic (Duffy et al. 2008). However, Tajima's D is influenced by both population changes and selective pressure. It is not easy to quantify the effectiveness rate of both components on the Tajima's D values (Innan and Stephan 2000; Kim et al. 2016).
Investigation of motifs in the spike protein sequences
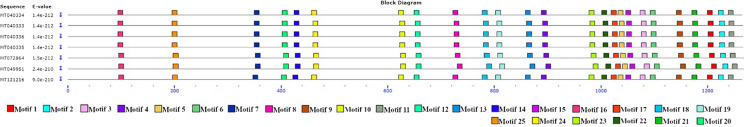
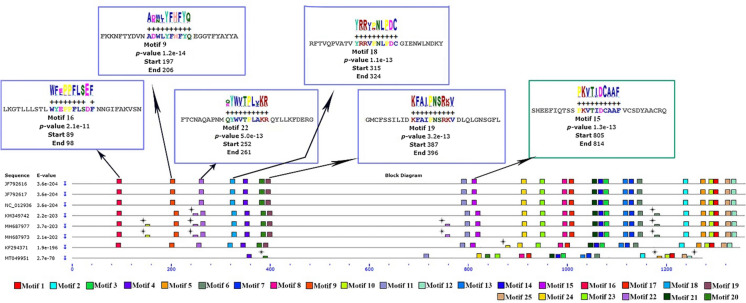
The results obtained from examining the spike protein sequences to find potential motifs between members of group 1 are shown in Fig. 6. Regions of motifs in each sequence are illustrated with colored blocks. All known motifs are shared among members of group 1. Identified motif sequences in the spike protein of SARS-CoV-2 (MT049951) were the same as motif sequences in the spike protein of pangolin-CoVs except for motifs 3, 10, 6, and 12 (Supplementary Fig. 1). The results from the analysis of the spike protein sequences in group 2 showed that motifs 16, 9, 19, 18, 22, and 15 were found only in the spike protein of Rattus CoVs in group 2, and the sequence similar to them was not observed in the MT049951-related protein sequence (Fig. 7). Motif 15 sequence (PKVTIDCAAF) was the same in all studied Rattus CoVs. On the other hand, the lack of this motif in the SARS-CoV-2 sequence makes it a suitable marker for identifying the spike protein of Rattus CoVs. Comparison of the obtained results in Figs. 6, 7 demonstrates the high similarity between the spike protein sequences of SARS-CoV-2 and pangolin-CoVs.
Fig. 6.
The motif analysis in spike protein. The BLOCK diagram shows the sequence of the discovered motifs. Known motifs are common in the protein sequence of SARS-CoV-2 (MT049951) and pangolin-CoVs. Motif sequences were acceptable with a p-value ≤ 1 × 10–5
Fig. 7.
The motif analysis in spike protein. The BLOCK diagram shows the sequence of the discovered motifs. Motifs 16, 9, 22, 18, and 19 were found in the spike protein of Rattus CoVs (blue square). Motif 15 was seen in all Rattus CoVs with the completely same sequence (green square). Motif sequences were acceptable with a p-value ≤ 1 × 10–5. Star markers belong to motifs that are not acceptable
Protein sequence polymorphisms of spike were detected by Analyze Sequence Variation tool. For further analysis, the meta-CATS tool was used to find polymorphisms. In this case, the chi-square test was measured to ensure the accuracy of determining positions, and variations of amino acids with a p-value ≤ 0.05 were reported. The obtained results revealed that 35 amino acids in the spike protein of MT049951 were different from amino acids present in the spike protein of other members of group 1. These altered amino acids resulted from nucleotide changes in their codons. Investigations showed that the altered amino acids were present at positions other than known motifs along the protein sequence. Comparison of the spike protein sequence in MT049951 and other members of group 2 showed 305 altered amino acids, out of which five amino acids were observed in motifs 13, 17, 20, 4, and 25 (Supplementary Fig. 2). Molecular studies have confirmed that protein adaptation is associated with more nucleotide changes in the genome that alter amino acids. (Kryazhimskiy and Plotkin 2008). In general, the results obtained in this paper showed a close relationship between spike proteins in SARS-CoV-2 and pangolin-CoVs.
Conclusion
The epidemic of COVID-19 began in the city of Wuhan, China, in 2019. The outbreak of the disease and its global epidemic status caused many efforts to be done to understand the structure and function of the virus. SARS-CoV-2 is a member of the Betacoronavirus family. Detection of the ancestors of SARS–CoV-2 in the coronaviruses family helps to identify and define the phylogenetic relationships of the family Coronaviridae. In the present paper, the phylogenetic evidence demonstrated that SARS-CoV-2 could develop from bat-SL-CoVZC21. The occurrence of a recombination event in the region of the spike gene specified two recombination regions. PCoV_GX-P4L and ChRCoV-HKU24 were determined as the potential parents for the first and second events, respectively. In this study, it was presumed that pangolins and Rattus could be the parents of SARS-CoV-2. However, this remains a hypothesis and needs further investigations to be proved.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Office of the Vice-chancellor for Research and funded by the Research and Technology Institute of Plant Production (RTIPP), Shahid Bahonar University of Kerman, Kerman, Iran, under grant number [900/41].
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aboim MA, Menezes GM, Schlitt T, Rogers AD. Genetic structure and history of population of the deep-sea fish Helicolenus dactyloptenus (Delaroche 1809 inferred from mtDNA sequence analysis. Mol Ecol. 2005;14:1343–1354. doi: 10.1111/j.1365-294X.2005.02518.x. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, Koster AJ, Bosch BJ. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. PNAS. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Chan JFW, Yuan S, Kok K-H, To KK-H, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang Y, Liu Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change inviruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis Methods. Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–327. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Godeke GJ, Jonges M, Farag E, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Li J, Du L, Yan X, Hu G, Zhou Y, Jiang S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, Yang L, Ding C, Zhu X, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020;6736:30183–30185. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang C, Pearce R, Omenn GS, Zhang Y. Identifying the zoonotic origin of SARS-CoV-2 by modeling the binding affinity between the spike receptor-binding domain and host ACE2. J Proteome Res. 2020;19:4844–4856. doi: 10.1021/acs.jproteome.0c00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Stephan W. The coalescent in an exponentially growingmetapopulation and its application to Arabidopsis thaliana. Genetics. 2000;155:2015–2019. doi: 10.1093/genetics/155.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, Liang W, Zheng H, Wan K, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus- like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Omori R, Ueno K, Iida S, Ito K. Host-specific and segment-specificevolutionary dynamics of avian and human influenza A viruses: a systematicreview. PLoS ONE. 2016 doi: 10.1371/journal.pone.0147021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The population genetics of dN/Ds. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM. Coronavirus: organization replication and expression of genome. Annu Rev Microbio. 1990;44(1):303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lam TT-Y, Jia N, Zhang Y-W, Shum MH-H, Jiang J-F, Zhu H-C, Tong Y-G, Shi Y-X, Ni X-B, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. PNAS. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Li KS, Tsang AK, Fan RY, Luk HK, Cai JP, Chan KH, Zheng BJ, et al. Discovery of a novel coronavirus China Rattus coronavirus HKU24 from Norway rats supports the murine origin of Betacoronavirus. J Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Molecular Evolution. USA: Sinauer Associates Inc; 1997. [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, et al. Early transmission dynamics in Wuhan China of novel coronavirus-infected Pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Chen W, Chen JP. Viral metagenomics revealed sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) Viruses. 2019;11:979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Molecular epidemiology evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardani K, Noormohammadi AH, Hooper P, Ignjatovic J, Browning GF. Infectious bronchitis viruses with a novel genomic organization. J Virol. 2008;82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosa A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, et al. Middle East respiratory syndrome coronavirus in bats Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press, Inc.; 2000. pp. 51–72. [Google Scholar]
- Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80:791. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picoult-Newberg L, Ideker TE, Pohl MG, Taylor SL, Donaldson MA, Nickerson DA, Boyce-Jacino M. Mining SNPs from EST Databases. Genome Res. 1999;9:167–174. doi: 10.1101/gr.9.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen JW, de Beer R, Godeke GJ, Raamsman MJ, Horzinek MC, Vennema H, Rottier PJ. The viral spike protein is not involved in the polarized sorting of coronaviruses in epithelial cells. J Virol. 1998;72:497–503. doi: 10.1128/JVI.72.1.497-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Sabella E, Pierro R, Luvisi A, Panattoni A, D’onofrio C, Scalabrelli G, Nutricati E, Aprile A, De bellis L, phylogenetic analysis of viruses in tuscan vitis vinifera sylvestris (gmeli) hegi. PLoS ONE. 2018;13:e0200875. doi: 10.1371/journal.pone.0200875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano A, Tachida H. Gene genealogy and properties of test statistics of neutrality under population growth. Genetics. 2005;169:1687–1697. doi: 10.1534/genetics.104.032797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoek L. Human coronaviruses: What do they cause? Antivir Ther. 2007;12:651–658. doi: 10.1177/135965350701200S01.1. [DOI] [PubMed] [Google Scholar]
- Vignal A, Milan D, San-Cristobal M, Eggen A. A Review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34:275–305. doi: 10.1186/1297-9686-34-3-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun. 2020;12:972. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Bosch BJ, Frenz B, Rottier PJM, Maio FD, Rey FA, Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici AM, Wall A, McGuire AT, Veesler D. Structure function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B, Chen H, Zheng H, Xu Y, et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin XD, Guo WP, Zhou RH, Wang MR, Wang CQ, Ge S, Mei SH, Li MH, et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, Zou Y. 2019-ncov/sars-cov-2: rapid classification of betacoronaviruses and identification of traditional chinese medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol. 2020;70:342–348. doi: 10.1111/lam.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Ren X, Zhang J, Yang F, Zhang S, Jin Q. ORF8-Related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J Infect Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou J-J, Li N, Guo Y, Li X, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W, Wu LJ, Ge XY, Zhang YZ, et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol. 2016;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human analogue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes Snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wu Q, Zhang Z. Probable Pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with Pneumonia in China 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.