Abstract
Metabolic reprogramming has been shown to occur in uveal melanoma (UM), the most common intraocular tumor in adults. Mechanisms driving metabolic reprogramming in UM are poorly understood. Elucidation of these mechanisms could inform development of new therapeutic strategies for metastatic UM, which has poor prognosis because existing therapies are ineffective. Here, we determined whether metabolic reprogramming is driven by constitutively active mutant α-subunits of the heterotrimeric G proteins Gq or G11 (Gq/11), the oncogenic drivers in ∼90% of UM patients. Using PET–computed tomography imaging, microphysiometry, and GC/MS, we found that inhibition of oncogenic Gq/11 with the small molecule FR900359 (FR) attenuated glucose uptake by UM cells in vivo and in vitro, blunted glycolysis and mitochondrial respiration in UM cell lines and tumor cells isolated from patients, and reduced levels of several glycolytic and tricarboxylic acid cycle intermediates. FR acutely inhibited glycolysis and respiration and chronically attenuated expression of genes in both metabolic processes. UM therefore differs from other melanomas that exhibit a classic Warburg effect. Metabolic reprogramming in UM cell lines and patient samples involved protein kinase C and extracellular signal–regulated protein kinase 1/2 signaling downstream of oncogenic Gq/11. Chronic administration of FR upregulated expression of genes involved in metabolite scavenging and redox homeostasis, potentially as an adaptive mechanism explaining why FR does not efficiently kill UM tumor cells or regress UM tumor xenografts. These results establish that oncogenic Gq/11 signaling is a crucial driver of metabolic reprogramming in UM and lay a foundation for studies aimed at targeting metabolic reprogramming for therapeutic development.
Keywords: Gq/11, uveal melanoma, metabolism, oncogenes, signaling
Abbreviations: 2-DG, 2-deoxyglucose; 2-DG6P, 2-deoxy-d-glucose-6-phosphate; 18F-FDG, 18F-fluorodeoxyglucose; AMPK, AMP-activated protein kinase; ATCC, American Type Culture Collection; BAP1, BRCA1-associated protein 1; BRAF, B-Raf proto-oncogene serine/threonine-protein kinase; CT, computed tomography; DMSO, dimethyl sulfoxide; ECAR, extracellular acidification rate; Erk, extracellular signal–regulated protein kinase; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; FR, FR900359; GST, Glycolytic Stress Test; Gq/11, Gq or G11; I.I., integrated fluorescence intensities; MEF, mouse embryonic fibroblast; MEK, mitogen-activated protein kinase/extracellular signal–regulated kinase kinase; MEKi, MEK inhibitor; MST, Mito Stress Test; mTORC1, mechanistic target of rapamycin complex 1; NSG, NOD scid gamma; OCR, oxygen consumption rate; OxPhos, oxidative phosphorylation; PDX, patient-derived xenograft; PKCi, PKC inhibitor; ROI, region of interest; RRID, Research Resource Identifier; TBST, Tris-buffered saline with Tween-20; TCA, tricarboxylic acid; UM, uveal melanoma
Metabolic reprogramming is a hallmark of cancer that enables tumor cells to meet biosynthetic demands for proliferation and survival (1). In many tumors, oncogenic signaling mechanisms reprogram metabolism transcriptionally and post-transcriptionally (2, 3). Signaling by oncogenic HRAS, KRAS, or B-Raf proto-oncogene serine/threonine-protein kinase (BRAF), for example, increases expression of glucose transporters and glycolytic genes (3, 4, 5), decreases flux through the tricarboxylic acid (TCA) cycle, and represses mitochondrial oxidative phosphorylation (OxPhos) (6, 7), inducing a Warburg effect (7, 8, 9). Such knowledge provides avenues for developing new therapeutic options and combating resistance to targeted therapies (10).
Extensive evidence indicates that metabolic reprogramming occurs in uveal melanoma (UM) (11), the most common intraocular tumor in adults (12). Relative to normal melanocytes, for example, UM cells highly express genes involved in glycolysis, the TCA cycle, mitochondrial biogenesis and respiration, amino acid metabolism, and lipid metabolism (13, 14, 15). Metastatic UM tumors exhibit high levels of glucose uptake and glycolytic activity compared with surrounding tissues, which correlate inversely with overall survival as shown by PET/computed tomography (CT) with 18F-fluorodeoxyglucose (18F-FDG) (16, 17, 18, 19, 20, 21).
Such evidence and the poor prognosis of metastatic UM because of ineffective therapy (22, 23) have motivated studies to elucidate molecular mechanisms of metabolic reprogramming that potentially could open new avenues for therapeutic discovery (11). Studies thus far have shown that metastatic risk associated with monosomy of chromosome 3 in UM correlates with greater mitochondrial activity and resistance to OxPhos inhibitors (24), and that UM cell lines lacking the BRCA1-associated protein 1 (BAP1) metastasis suppressor can be stratified by metabolic phenotypes and differential sensitivity to metabolic inhibitors (25).
Nevertheless, the physiological characteristics and genetic drivers of metabolic reprogramming in UM are poorly understood, unlike cutaneous melanomas driven by oncogenic mutant BRAF or NRAS. In ∼90% of UM patients, constitutively active mutant α-subunits of the heterotrimeric G proteins Gq or G11 (Gq/11) initiate tumorigenesis (26, 27). Gq and G11 function similarly in UM because they cause equivalent clinical outcomes (28, 29) and largely are functionally interchangeable (30). Several well-characterized secondary genetic events subsequently drive tumor progression (22, 23). Whether oncogenic Gq/11 or other genetic events during UM tumorigenesis and progression drive or sustain metabolic reprogramming is unknown.
Here, we have addressed these questions by using FR900359 (FR), a bioavailable cyclic depsipeptide that is a potent and highly selective inhibitor of the Gq/11 subfamily of heterotrimeric G protein α-subunits (31, 32). We have used FR to characterize in detail the metabolic consequences of inhibiting oncogenic Gq/11 in mouse xenografted UM tumors, UM cell lines, and UM tumor cells freshly isolated from patients, and to identify signaling mechanisms downstream of oncogenic Gq/11 that drive these metabolic effects. Our studies establish that oncogenic Gq/11 signaling is a principal driver of metabolic reprogramming in UM, reveal atypical features of metabolic reprogramming characteristic of UM, and suggest new avenues for therapeutic development in UM.
Results
Oncogenic Gq/11 activity drives glucose uptake in UM tumor cells in vivo and in vitro
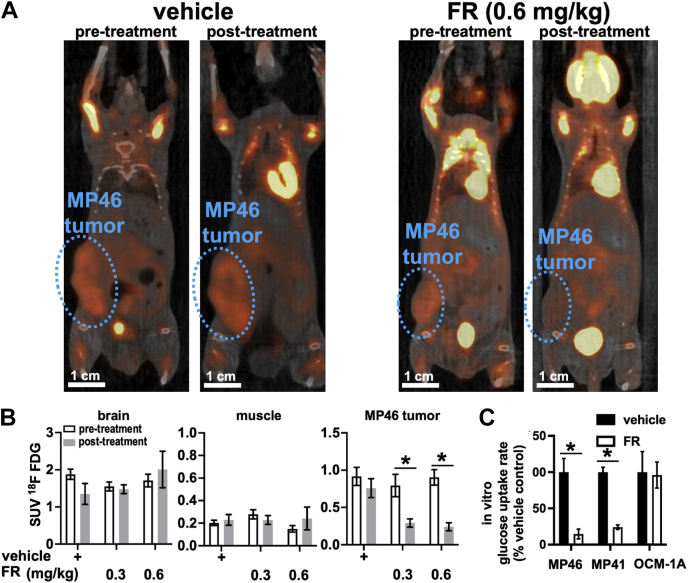
As an initial means of exploring whether oncogenic Gq/11 signaling drives metabolic activity in UM tumors, we determined whether FR administered systemically in mice affected uptake of 18F-FDG measured by PET/CT imaging. In these experiments, MP46 UM tumor cells (Gq-Q209L; patient-derived xenograft [PDX]-derived model of BAP1-deficient class 2 tumors with high metastatic potential (33)) were implanted subcutaneously in the flanks of immunodeficient NOD scid gamma (NSG) mice and allowed to form tumors. Tumor-bearing mice were injected with 18F-FDG, and tissue uptake was analyzed by PET/CT (Fig. 1, A and B). Results indicated that MP46 tumor xenografts displayed robust uptake of 18F-FDG, consistent with studies of UM patients (16, 17, 18, 19, 20, 21). Tumor-bearing mice subsequently were injected every other day for 1 week with vehicle or FR at levels (0.3 or 0.6 mg/kg s.c.) that arrest tumor growth but preserve host viability (34), and then injected with 18F-FDG for analysis by PET/CT (Fig. 1, A and B). Relative to pretreatment controls, FR administered at either level significantly reduced 18F-FDG uptake in MP46 UM tumors ∼2.5-fold (p < 0.01) but had insignificant effect on 18F-FDG uptake in brain or skeletal muscle.
Figure 1.
Oncogenic Gq/11 drives glucose uptake in UM tumors.A, MP46 (Gq Q209L) UM cells were injected into the flanks of NSG mice and allowed to form tumors. Tumor-bearing mice were injected with 18F-FDG and imaged by PET/CT to measure the metabolic activities of the tumors and reference tissues (brain and muscle). MP46 UM tumors (circled) showed higher 18F-FDG uptake than surrounding tissues. Systemic treatment with FR reduced 18F-FDG uptake compared with control. B, standardized uptake values (SUVs) were calculated for 18F-FDG from PET/CT imaging of MP46 tumor-bearing mice treated with vehicle (n = 3), 0.3 mg/kg FR (n = 3), and 0.6 mg/kg FR (n = 3). MP46 UM tumors in FR-treated mice showed significant reduction of 18F-FDG uptake compared with nontumor tissues (∗p < 0.01). C, representative graph from three independent experiments of glucose uptake assays performed on UM cell lines in culture. MP46 (Gq Q209L) and MP41 (G11 Q209L) cells showed significant reduction of glucose uptake with FR treatment (∗p < 0.01), whereas OCM-1A (BRAF V600E) cells showed no significant response to FR. 18F-FDG, NOD scid gamma; BRAF, B-Raf proto-oncogene serine/threonine-protein kinase; CT, computed tomography; FR, FR900359; Gq/11, Gq or G11; NSG, NOD scid gamma; UM, uveal melanoma.
Subsequent analysis in vitro indicated that FR inhibited glucose uptake by Gq/11-driven UM cell lines (Fig. 1C) irrespective of BAP1 status. Glucose transport was measured in cultured UM cell lines treated 18 h with vehicle or FR at a maximally effective dose (100 nM (32, 34)). We used two PDX-derived cell lines: MP46 cells derived from class 2 UM cells with high metastatic potential and MP41 cells derived from class 1 UM cells with low metastatic potential. Results indicated that FR reduced glucose uptake threefold to fivefold (p < 0.01) in MP46 and MP41 UM cell lines. In contrast, FR had insignificant effect on glucose uptake in a UM cell line (OCM-1A) driven by BRAF-V600E (Fig. 1C), which is not targeted by FR (32, 34). These results are the first to indicate that oncogenic Gq/11 signaling is required to drive or sustain glucose uptake in UM tumor cells in vivo and in vitro.
Oncogenic Gq/11 signaling acutely drives glycolysis and respiration in UM cell lines
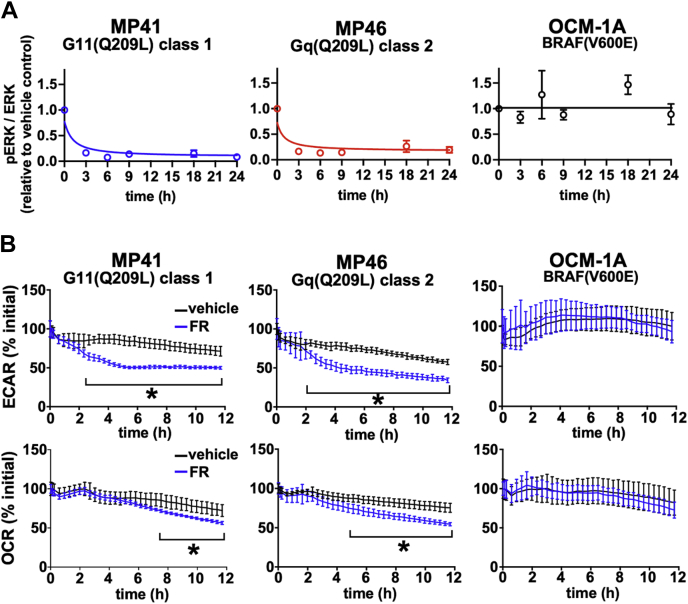
Next, we investigated whether oncogenic Gq/11 signaling drives metabolic activity acutely or chronically in UM cell lines in vitro. An acutely acting mechanism would be suggested if FR inhibits oncogenic Gq/11 signaling and metabolism on similar and shorter time scales (minutes to hours). A chronically acting mechanism would be suggested if FR requires long time scales (many hours to days) to inhibit UM tumor cell metabolism, as is the case for imposition of UM tumor cell cycle arrest and redifferentiation by FR (32, 34). We addressed these possibilities by comparing the effects of FR over time on oncogenic signaling downstream of Gq/11 (extracellular signal–regulated protein kinase [Erk] phosphorylation), glycolysis (extracellular acidification rate [ECAR]), and mitochondrial respiration (oxygen consumption rate [OCR]) in vitro (Fig. 2). In Gq/11-driven MP41 (UM class 1; low metastatic potential) and MP46 cells (UM class 2; high metastatic potential), we found that FR inhibited oncogenic Gq/11 signaling in less than 3 h as indicated by reduction of Erk phosphorylation (Figs. 2A and S1). Similarly, FR blunted ECAR within 2 h and attenuated OCR within 5 to 7 h (Fig. 2B). These effects on UM cell lines were specific for oncogenic Gq/11 because FR did not affect Erk phosphorylation, ECAR, or OCR in a UM cell line (OCM-1A) driven by BRAF-V600E (Fig. 2, A and B). These results supported the hypothesis that oncogenic Gq/11 signaling in UM cell lines acutely drives both glycolysis and mitochondrial respiration.
Figure 2.
Oncogenic Gq/11 acutely drives metabolism in c cells. The effects of FR over time on oncogenic Gq/11 signaling (Erk phosphorylation), glycolysis (extracellular acidification rate [ECAR]), and mitochondrial respiration (oxygen consumption rate [OCR]) were measured in vitro. Each graph shows representative data from at least three independent experiments for each cell line. A, MP41, MP46, and OCM-1A cells were treated with 100 nM FR for the indicated time and then lysed to collect protein. Lysates were subjected to SDS-PAGE and immunoblotted for pERK1/2 or total ERK1/2. Fluorescence intensity measurements were performed on a LI-COR Odyssey system. For each experiment, the ratio of pERK to total ERK for each lane was normalized to the ratio in untreated cells. FR (100 nM) reduced ERK1/2 phosphorylation acutely in Gq/11-driven UM cells within 3 h of treatment. B, ECAR and OCR were measured over 12 h time courses, and the data were normalized to the mean starting values for each experiment. Three 5 min baseline measurements were taken before injection of inhibitors into each well (final concentration of FR, 100 nM), after which measurements were taken at 20 min intervals. FR-treated MP41 and MP46 cells showed significant reductions in ECAR compared with untreated cells 2 h after injection (∗p < 0.01). In contrast, significant reductions in OCR were not seen until 5 h (MP46) and 7 h (MP41) after FR injection (∗p < 0.01). OCM-1A cells showed no significant metabolic response to FR treatment compared with untreated cells. Erk, extracellular signal–regulated protein kinase; FR, FR900359; Gq/11, Gq or G11; UM, uveal melanoma.
Oncogenic Gq/11 signaling drives several aspects of glycolytic and respiratory activity in UM tumor cell lines
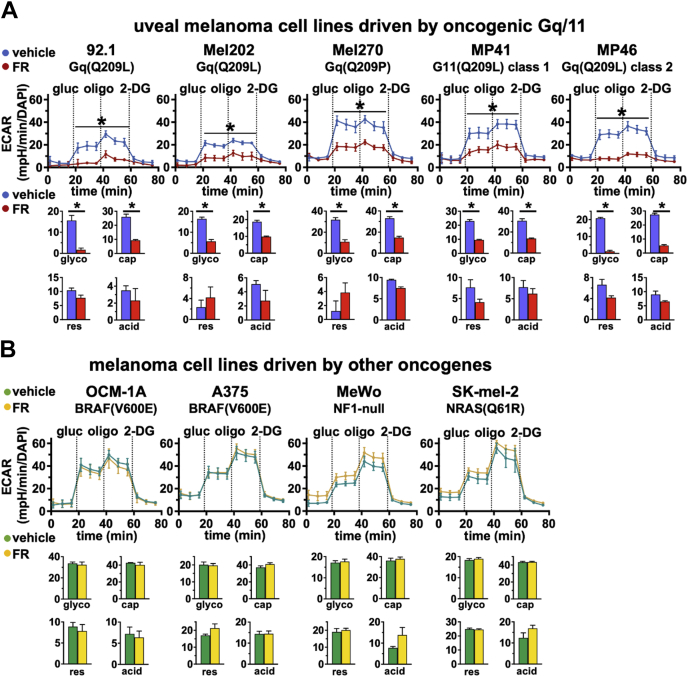
To determine which aspects of glycolysis and respiration are driven by oncogenic Gq/11 signaling, we performed glycolytic and mitochondrial stress tests. We determined whether FR has similar metabolic effects in UM cells driven specifically by oncogenic Gq/11 by comparing a panel of Gq/11-driven UM cell lines (92.1, Mel202, Mel270, and PDX-derived MP41 and MP46 cells) with several melanoma cell lines (OCM-1A, A375, MeWo, and SK-mel-2) driven by oncogenes other than Gq/11. Glycolytic stress tests were used to measure (i) basal activity (ECAR in glucose-free media); (ii) glycolysis (ECAR after addition of glucose minus ECAR after addition of 2-deoxyglucose [2-DG]); (iii) glycolytic capacity (ECAR following addition of glucose to stimulate glycolysis and oligomycin [oligo] to block oxidative phosphorylation minus ECAR after addition of 2-DG); (iv) glycolytic reserve (ECAR upon addition of glucose and oligomycin minus ECAR after addition of glucose); and (v) nonglycolytic acidification (ECAR upon addition of 2-DG). Results indicated that FR significantly reduced glycolysis and glycolytic capacity in all Gq/11-driven UM cell lines (Fig. 3A) with potencies in the nanomolar range (Fig. S2), similar to the potency of FR as an inhibitor of UM cell proliferation (32, 34). In contrast, FR had insignificant effect on glycolysis in melanoma cell lines driven by oncogenes other than Gq/11 (Fig. 3B).
Figure 3.
Oncogenic Gq/11 drives glycolysis in UM cells. Cellular metabolic physiology was measured with an Agilent Seahorse XF analyzer using the Glycolysis Stress Test. First, cells are acclimated to glucose-free medium to measure nonglycolytic acidification (first three measurements). Second, glucose is added to measure basal glycolysis. Third, oligomycin is added to block oxidative phosphorylation and measure maximal glycolytic capacity. Fourth, 2-deoxyglucose (2-DG) is added to inhibit glycolysis and return to baseline nonglycolytic acidification. Each graph shows representative data from at least three independent experiments for each cell line. Top panels show raw data traces of ECAR normalized to number of cells per well (by DAPI straining); bar graphs below each panel show glycolysis (glyco), glycolytic capacity (cap), glycolytic reserve (res), and nonglycolytic acidification (acid) rates derived from the aforementioned data. A, older Gq/11-driven UM cell lines (92.1, Mel202, and Mel270) and newer PDX-derived Gq/11-driven UM cell lines (MP41 and MP46) show significant reductions in glycolysis (glyco) and glycolytic capacity (cap) in response to FR (∗p < 0.01). B, melanoma cell lines driven by other oncogenes (OCM-1A, A375, MeWo, and SK-mel-2) show no response to FR in glycolysis or glycolytic capacity. DAPI, 4′,6-diamidino-2-phenylindole; ECAR, extracellular acidification rate; FR, FR900359; Gq/11, Gq or G11; PDX, patient-derived xenograft; UM, uveal melanoma.
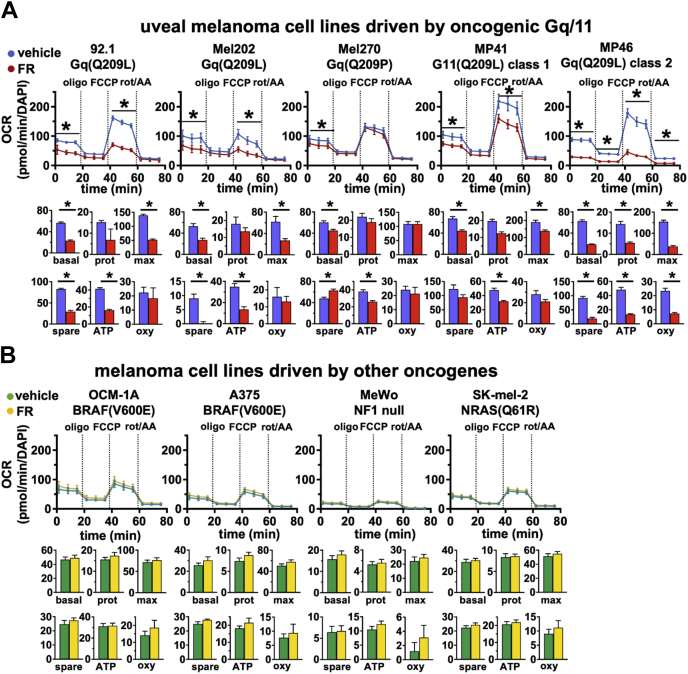
Similar studies indicated that FR affected several aspects of mitochondrial respiration only in Gq/11-driven UM cell lines (Fig. 4). Here, we used mitochondrial stress tests to measure the effects of FR on (i) basal respiration (OCR in media containing glucose and pyruvate); (ii) proton leak (OCR after blocking respiratory complex V with oligomycin [oligo]); (iii) respiratory ATP production (basal OCR minus proton leak); (iv) maximal respiratory capacity (OCR after collapsing the mitochondrial proton gradient with carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP); (v) spare respiratory capacity (maximal respiration minus basal respiration); and (vi) nonmitochondrial oxygen consumption (OCR after inhibiting complexes I and III with rotenone and antimycin A). In every Gq/11-driven UM cell line, we found that FR reduced basal respiration (Fig. 4A) with potencies in the nanomolar range (Fig. S2). Other modes of mitochondrial function and respiratory activity were affected variably by FR among these Gq/11-driven UM cell lines (Fig. 4A). In melanoma cell lines driven by other oncogenes, all aspects of mitochondrial respiration and function were insensitive to FR (Fig. 4B). These results demonstrated that oncogenic Gq/11 signaling in UM cell lines drives several aspects of both glycolytic activity and mitochondrial respiration, rather than eliciting a classical Warburg effect in which glycolysis is driven and respiration repressed.
Figure 4.
Oncogenic Gq/11 drive oxidative phosphorylation in UM cells. Oxidative metabolic physiology was measured with an Agilent Seahorse XF analyzer using the Mitochondrial Stress Test. First, cells are grown in medium containing glucose and pyruvate to measure basal respiration (first three measurements). Second, oligomycin is added to block respiratory complex V (ATPase) and measure proton leak. Third, FCCP is added to uncouple the proton gradient across the mitochondrial inner membrane and measure maximal respiratory capacity. Fourth, rotenone and antimycin A are added to inhibit complexes I and III to measure baseline nonmitochondrial oxygen consumption. Each graph shows representative data from at least three independent experiments for each cell line. Top panels show raw data traces of OCR normalized to number of cells per well (by DAPI straining); bar graphs below each panel show basal respiration (basal), proton leak (prot), maximal respiration (max), spare respiratory capacity (spare), ATP production (ATP), and nonmitochondrial oxygen consumption (oxy) rates derived from the aforementioned data. A, older Gq/11-driven UM cell lines (92.1, Mel202, and Mel270) and newer PDX-derived Gq/11-driven UM cell lines (MP41 and MP46) show significant reductions in basal respiration (basal) in response to FR. Four of the Gq/11-driven lines (92.1, Mel202, MP41, and MP46) also show significant reductions in maximal respiration (max); this was not significant in Mel270 cells. B, melanoma cell lines driven by other oncogenes (OCM-1A, A375, MeWo, and SK-mel-2) show no respiratory responses to FR. DAPI, 4′,6-diamidino-2-phenylindole; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; FR, FR900359; Gq/11, Gq or G11; OCR, oxygen consumption rate; PDX, patient-derived xenograft; UM, uveal melanoma.
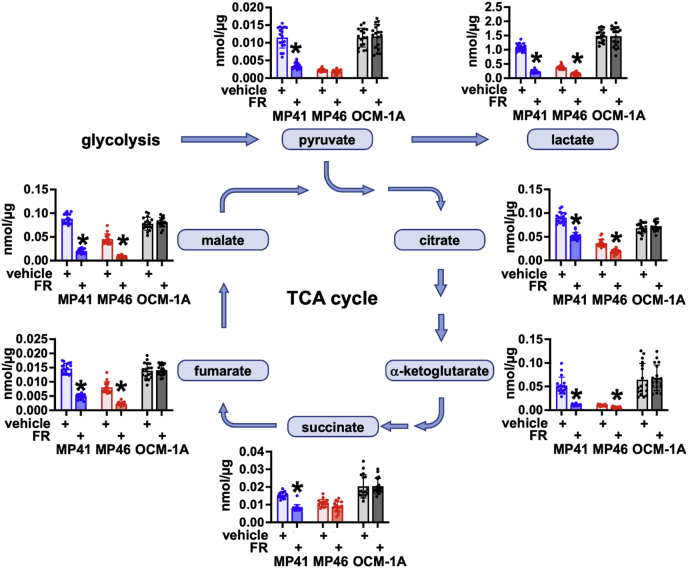
Oncogenic Gq/11 signaling drives production of glycolytic and TCA intermediates in UM cell lines
To determine whether biochemical events associated with glycolysis and mitochondrial respiration are regulated by oncogenic Gq/11 signaling, we measured the effects of FR on the abundance of pyruvate, lactate, and TCA cycle intermediates. MP41, MP46, and OCM-1A cells were treated with vehicle or FR (100 nM) for 24 h and analyzed by GC–MS. In G11-driven MP41 cells, FR significantly decreased the levels of each metabolite studied (Fig. 5), several of which have complex roles as “oncometabolites” in various types of cancer (35, 36). Similar results were obtained with Gq-driven MP46 cells, except that reduction of pyruvate and succinate levels did not quite achieve statistical significance (Fig. 5). In BRAF-driven OCM-1A cells, FR had insignificant effect on levels of all metabolites studied (Fig. 5). These results demonstrated that oncogenic Gq/11 signaling drives production of glycolytic and TCA cycle intermediates in UM cell lines.
Figure 5.
Glycolytic and TCA intermediates regulated by oncogenic Gq/11 in UM cells. MP41, MP46, and OCM-1A cells were grown in 6-well plates for 18 h in separate plates for vehicle (six wells) or FR (six wells) and then extracted in ice-cold methanol. Each well was processed separately as a technical replicate, and the experiment was performed on three independent sets of samples. Extracts were spiked with labeled metabolite standards and then subjected to GC–MS analysis. Graphs show results for pyruvate, lactate, and TCA cycle intermediates. In MP41 cells, FR induced significant decreases in all metabolites examined (∗p < 0.01). In MP46 cells, FR induced significant decreases in lactate, citrate, α-ketoglutarate, fumarate, and malate (∗p < 0.01) but did not affect pyruvate or succinate levels significantly. FR had no significant effect on metabolite levels in OCM-1A cells. FR, FR900359; Gq/11, Gq or G11; TCA, tricarboxylic acid; UM, uveal melanoma.
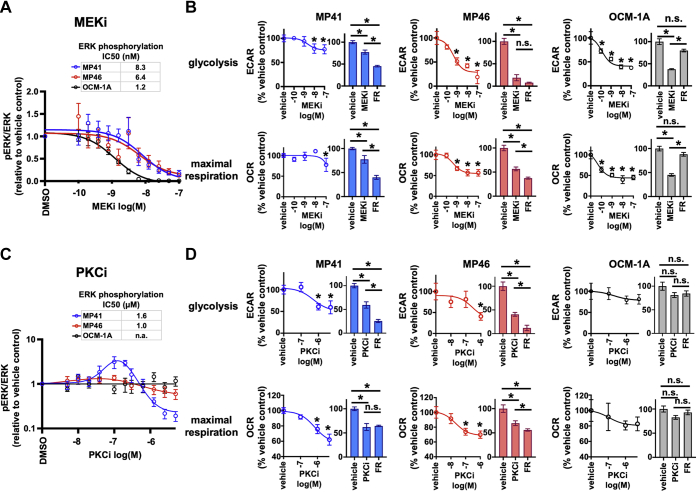
Oncogenic Gq/11 signaling drives metabolic activity in UM cell lines through protein kinase C and Erk
To identify signaling mechanisms downstream of oncogenic Gq/11 that drive metabolic activity in UM cell lines, we employed inhibitors used in UM clinical trials to target protein kinase C and the Erk pathway (37, 38). As a prerequisite for metabolic studies, we first determined the potencies and efficacies of the mitogen-activated protein kinase/extracellular signal–regulated kinase kinase (MEK) inhibitor (MEKi) trametinib (GSK1120212; MEKi) and the PKC subtype-nonselective inhibitor sotrastaurin (AEB071; PKC inhibitor [PKCi]) by measuring their effects on Erk1/2 phosphorylation in UM cell lines (Fig. 6, A and C). We subsequently compared the potency and efficacy of MEKi, PKCi, and FR as inhibitors of glycolysis and mitochondrial respiration.
Figure 6.
PKC and Erk signaling downstream of oncogenic Gq/11 regulate metabolic activity in UM cells. Gq/11 signals through PLCβ and PKC to activate the Ras/MAPK pathway resulting in phosphorylation of ERK1/2 by MEK1/2. For each experiment in A and C, cells were treated with MEKi or PKCi at the indicated doses for 18 h and then lysed to collect protein. Lysates were subjected to SDS-PAGE and immunoblotted for pERK1/2 or total ERK1/2. Fluorescence intensity measurements were performed on a LI-COR Odyssey system, and the ratio of pERK to total ERK for each lane was normalized to the ratio in untreated cells; data points show the mean ratio from four experiments. Each graph in B and D shows representative data from at least three independent experiments for each cell line. A, the MEKi trametinib reduces ERK1/2 phosphorylation in UM cells at nanomolar (nM) concentrations regardless of driver mutation. B, graphs showing the effects of MEKi on glycolysis and maximal respiration in MP41 (blue), MP46 (red), and OCM-1A (black) cell lines. Each pair of graphs shows the dose response of the metabolic activity to MEKi (left) and a comparison of MEKi to FR treatment at maximum doses (right) for each cell line. C, the PKCi sotrastaurin shows a bell-shaped dose–response curve, resulting from a stimulatory response at low concentrations and an inhibitory response at high concentrations. At micromolar (μM) concentrations, PKCi reduces ERK1/2 phosphorylation in Gq/11-driven UM cells but does not affect BRAF-driven cells. For PKCi, IC50 values were calculated based on the untreated levels of phosphorylation and final levels of phosphorylation at maximal inhibitor concentration, ignoring the stimulated levels at noninhibitory concentrations. D, graphs showing the effects of PKCi on glycolysis and maximal respiration in MP41 (blue), MP46 (red), and OCM-1A (black) cell lines. Each pair of graphs shows the dose response of the metabolic activity to PKCi (left) and a comparison of PKC inhibition to FR treatment at maximum doses (right) for each cell line. Both MEK and PKC inhibition significantly reduced glycolysis and oxidative phosphorylation (∗p < 0.01) and at similar concentrations to inhibition of ERK phosphorylation. BRAF, B-Raf proto-oncogene serine/threonine-protein kinase; ERK1/2, extracellular signal–regulated protein kinase1/2; FR, FR900359; Gq/11, Gq or G11; MAPK, mitogen-activated protein kinase; MEK1/2, mitogen-activated protein kinase/extracellular signal–regulated kinase kinase; MEKi, MEK inhibitor; PKCi, PKC inhibitor; PLCβ, phospholipase β; UM, uveal melanoma.
MEKi inhibited Erk1/2 phosphorylation both in Gq/11-driven (MP41 and MP46) and BRAF-driven (OCM-1A) cells with nanomolar potency and full efficacy (Figs. 6A and S3). In glycolytic and mitochondrial stress tests, MEKi in the nanomolar range significantly reduced glycolysis and maximal respiration in both Gq/11- and BRAF-driven UM cell lines (Fig. 6B). However, MEKi efficacy as an inhibitor of glycolysis and respiration in both Gq/11-driven UM cell lines was markedly less than that of FR (Fig. 6B). The Erk1/2 pathway therefore may be only one of several effectors downstream of oncogenic Gq/11 that drive glycolysis and respiration in these UM cell lines.
PKCi inhibited Erk1/2 phosphorylation in G11-driven MP41 cells with micromolar potency and nearly full efficacy, whereas in Gq-driven MP46 UM cells, it inhibited Erk1/2 phosphorylation with similar potency but limited efficacy (Figs. 6C and S3). As expected, PKCi had insignificant effect on Erk1/2 phosphorylation in BRAF-driven OCM-1A cells (Figs. 6C and S3) because oncogenic BRAF activates Erk1/2 independently of PKC. Metabolically, PKCi in the micromolar range significantly inhibited glycolysis and maximal respiration in Gq/11, but not BRAF-driven UM cell lines (Fig. 6D). PKCi efficacy was significantly less than that of FR as an inhibitor of glycolysis, suggesting that PKC isoforms provide one group within a set of effectors downstream of oncogenic Gq/11 that drive metabolic activity in UM cell lines.
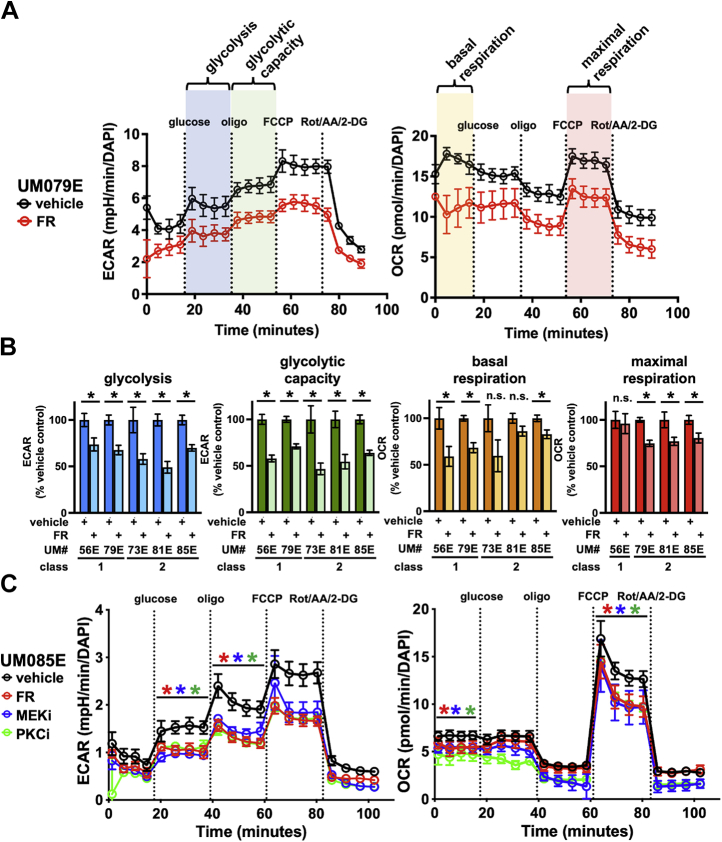
Oncogenic Gq/11 signaling drives glycolysis and respiration in freshly isolated UM tumor cells
Response to therapeutic agents can differ significantly between tumor cell lines and tumor cells freshly isolated from patients (39). This understanding underscored the importance of determining the metabolic effects of FR, MEKi, and PKCi in UM tumor cells freshly isolated from a group of patients whose primary tumors warranted surgical enucleation. The modest yields and limited proliferative capacities of primary UM tumor cells obtained from enucleated patients required us to develop novel protocols in which the effects of FR, MEKi, and PKCi could be determined in triplicate with a single modified glycolytic and mitochondrial stress test (Fig. 7A). These methods were used to study UM tumor cells isolated within 2 years from five patients, two of whom had class 1 choroidal tumors (low metastatic potential) and three had class 2 choroidal tumors (high metastatic potential). UM tumor cells freshly isolated from all five patients expressed oncogenic forms of Gq or G11 because in every case FR significantly reduced glycolysis and glycolytic capacity relative to vehicle controls (Fig. 7B). These metabolic effects occurred mainly or exclusively in UM tumor cells because stromal cells are minor component of these samples (34) (e.g., UM085E contained 77% melanoma cells and 23% stromal cells as indicated by tyrosinase immunofluorescence staining), and stromal cells otherwise do not respond to FR (34). Glycolytic reprogramming therefore appeared to be a hallmark of Gq/11-driven primary UM tumors irrespective of tumor class.
Figure 7.
Oncogenic Gq/11 drives metabolic activity in primary human UM samples. Seahorse metabolic analyses were performed on samples of human UM tumor tissue collected from enucleated eyes at the time of surgery. A, to address limitations of sample quantities, a novel injection protocol was devised to measure both glycolytic and mitochondrial stress in a single Seahorse experiment: basal ECAR and basal OCR (orange) were measured, and then, glucose was injected to measure glycolysis (blue); next, oligomycin was injected to measure glycolytic capacity (green); then, FCCP was injected to measure maximal respiration (red); finally, a cocktail of rotenone, antimycin A, and 2-DG was injected to measure metabolic baselines. Five wells were measured per condition for each sample as technical replicates. B, five enucleation samples were available with sufficient material for Seahorse assays. All primary cells were in culture for <72 h from sample collection to analysis. Significant reductions in glycolytic rates and glycolytic capacities were seen in all five samples in FR-treated samples compared with vehicle controls (∗p < 0.01). Significant reductions in basal respiration and maximal respiration in response to FR treatment were seen in three of the five samples; maximal respiration data were not available for UM073E, treated or untreated. GEP class (Castle Biosciences) information is indicated for each sample. C, one of the enucleation samples, UM085E, had sufficient material to measure the effects of FR (red), MEK inhibitor (blue), and PKC inhibitor (green) on metabolism. All three inhibitors caused significant reductions in all metabolic activities measured. 2-DG, 2-deoxyglucose; ECAR, extracellular acidification rate; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; FR, FR900359; Gq/11, Gq or G11; MEK, mitogen-activated protein kinase/extracellular signal–regulated kinase kinase; OCR, oxygen consumption rate; UM, uveal melanoma.
In contrast, the effects of FR on mitochondrial respiration differed somewhat among these five UM tumor samples (Fig. 7B). FR significantly reduced basal respiration in three of the five UM tumor samples and attenuated maximal respiration in all but one. Gq/11-driven primary UM tumors therefore appeared to be somewhat heterogeneous at the level of respiratory reprogramming.
To identify signaling pathways downstream of oncogenic Gq/11 that drive metabolic reprogramming in UM tumor cells from patients, we compared the metabolic effects of FR, MEKi, and PKCi in tumor cells from the sole UM patient sample (UM085E) that yielded tumor cells in quantity sufficient for such studies. We found that FR, MEKi, and PKCi inhibited glycolysis, glycolytic capacity, basal respiration, and maximal respiration with similar efficacy (Fig. 7C). These results suggested that this patient's tumor cells used PKC-dependent Erk activation as the principal pathway downstream of oncogenic Gq/11 to drive glycolysis and respiration. Downstream signaling mechanisms that drive metabolic reprogramming in UM cells from this enucleated patient therefore appeared to be simpler than the pathways used in UM cell lines.
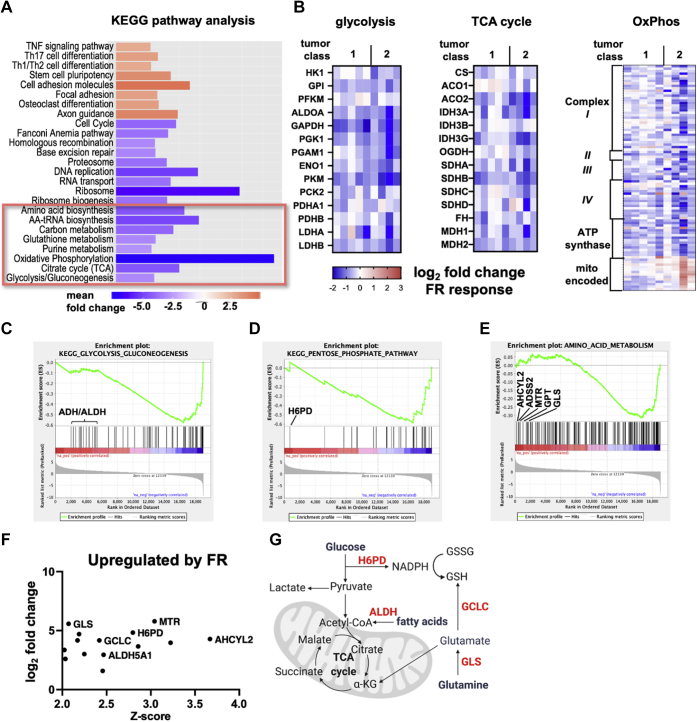
Oncogenic Gq/11 signaling sustains metabolic reprogramming by driving expression of TCA cycle and oxidative phosphorylation genes
In many tumors, oncogenic signaling chronically reprograms metabolism in part by upregulating metabolic gene expression (2). We therefore determined whether oncogenic Gq/11 signaling drives expression of metabolic genes by examining our published RNA-Seq data (GSE165552) derived from human UM tumor samples treated chronically (7 days) ex vivo with FR (34). Analysis showed that genes encoding components of several metabolic pathways were among the top 25 Kyoto Encyclopedia of Genes and Genomes processes downregulated significantly by chronic treatment with FR (Fig. 8A and Data File S7). Gene-by-gene inspection of data from class 1 (low metastatic potential) and class 2 (high metastatic potential) UM tumor cells indicated that genes encoding nearly every enzyme involved in glycolysis, the TCA cycle, and oxidative phosphorylation were downregulated significantly (Fig. 8B and Data File S8). These results suggested that oncogenic Gq/11 signaling is required chronically to sustain metabolic reprogramming in UM.
Figure 8.
Metabolic genes regulated by oncogenic Gq/11 in primary human UM tumor samples. KEGG pathway analysis of published RNA-Seq data from human UM tumor samples (GSE165552) treated with FR for 7 days was performed. A, the top 25 KEGG signaling pathways showing differential gene expression in FR-treated UM tumor samples include several metabolic pathways that are significantly downregulated (boxed). B, further analysis of the dataset showed that chronic treatment with FR caused downregulation of central genes required for glycolysis, the TCA cycle, and oxidative phosphorylation. Gene names are indicated for glycolysis and TCA cycle; electron transport chain complexes are indicated for oxidative phosphorylation (OxPhos). Colors correspond to log2 fold change in RNA expression (counts per million) in FR-treated versus untreated for each tumor sample. C–E, genes were reranked by z-score, and metabolic pathways were analyzed by gene set enrichment analysis to identify upregulated genes. C, the glycolysis/gluconeogenesis gene set is not only significantly enriched in downregulated genes (NES = −2.39; FWER p < 0.001) but also identifies several alcohol and aldehyde dehydrogenases upregulated with chronic FR treatment (indicated). D, the pentose phosphate pathway gene set is significantly enriched in downregulated genes (NES = −2.05; FWER p < 0.001) but identifies significant upregulation of GDP-dependent hexose-6-phosphate dehydrogenase (H6PD). E, the amino acid metabolism gene set is significantly enriched in downregulated genes (NES = −1.57; FWER p < 0.01) but identifies upregulation of genes required for metabolism of glutamine and cysteine (indicated). F, plot of significance (z-score of FR response; x-axis) versus expression (log2 fold change of FR response; y-axis) of metabolic genes significantly upregulated in response to FR (z-score > 2; log2 fold change > 1). G, schematic showing the relationships among the FR responsive genes identified in C–F as part of the metabolite scavenging pathways. Oxidation of glucose-6-phosphate by H6PD generates NADPH, which maintains cytosolic reducing potential and is a cofactor for glutathione reductase. Glutathione is synthesized by glutamate-cysteine ligase (GCLC). Glutamate levels are maintained by glutaminase (GLS). FWER, family-wise error rate; NES, normalized enrichment score.
Chronic inhibition of oncogenic Gq/11 signaling upregulates expression of metabolite scavenging and redox homeostasis genes
Finally, we explored whether UM tumor cells potentially develop adaptive responses to long-term treatment with FR, potentially enabling them to survive chronic inhibition of oncogenic Gq/11 signaling. We considered this hypothesis because FR arrests UM tumor cell proliferation and markedly attenuates glycolysis and oxidative phosphorylation, but it does not regress UM tumor xenografts or efficiently kill UM tumor cell lines or tumor cells freshly isolated from primary or metastatic tumors (32, 34). Accordingly, we analyzed our published RNA-Seq data (GSE165552) derived from human UM tumor samples treated chronically (7 days) ex vivo with vehicle or FR (34). We ranked FR-regulated genes based on z-score, identified genes encoding central metabolic pathways by gene set enrichment analysis, and performed gene-by-gene analysis to determine which metabolic genes were upregulated significantly. This analysis indicated that FR significantly upregulated genes encoding several alcohol and aldehyde dehydrogenases (Fig. 8, C and F), GDP-dependent hexose-6-phosphate dehydrogenase (Fig. 8, D and F), and enzymes involved in glutamine and cysteine metabolism (Fig. 8, E and F). All these genes play important roles in scavenging metabolites and maintaining pools of reduced glutathione (Fig. 8G and Data File S8) (40). Therefore, when FR chronically inhibits oncogenic Gq/11 signaling, proliferation, glycolysis, and mitochondrial respiration, these upregulated genes potentially could promote UM survival by facilitating nutrient scavenging and maintenance of redox homeostasis.
Discussion
Here, we have shown by using the highly selective inhibitor FR (31) that oncogenic Gq/11 signaling is a principal driver of metabolic reprogramming in a mouse model of UM, UM cell lines, and UM tumor cells isolated from patients. Our results illustrate the advantage of using FR to discriminate between acute and chronic effects of interrupting Gq/11 signaling, which would be difficult to determine by using knockdown or knockout approaches. The implications of our findings on UM biology and therapy are discussed later.
Oncogenic Gq/11 is the principal driver of UM metabolism
We found that inhibition of oncogenic Gq/11 by FR dramatically reduced glycolysis and mitochondrial respiration in Gq/11-driven UM cell lines and class 1 (low metastatic potential) and class 2 (high metastatic potential) primary UM tumor samples, indicating that metabolic reprogramming in UM is driven by oncogenic Gq/11 irrespective of tumor class. Prior studies have indicated that glycolysis and glycolytic capacity are somewhat greater in BAP1+ (class 1) than in BAP1-deficient (class 2) UM cell lines (24, 25). However, we found that glycolysis, glycolytic capacity, and basal and maximal respiration were all affected more by FR than by tumor cell class (BAP1 status) (Figs. 3A, 4A, and 6B). Moreover, metabolic activity, levels of glycolytic and TCA intermediates, and expression of metabolic genes were all reduced by FR irrespective of BAP1 status (Figs. 5 and 8A). Thus, BAP1 status may modify some aspects of metabolism (24, 25), whereas BAP1 is dispensable for oncogenic Gq/11 signaling to drive glycolytic and respiratory reprogramming in UM.
Oncogenic Gq/11 signaling drives glucose uptake and glycolysis
We found that FR inhibited glucose uptake and glycolysis on short and long time scales, suggesting that oncogenic Gq/11 signaling drives these metabolic processes acutely and chronically by post-transcriptional and transcriptional mechanisms, respectively. FR also reduced levels of pyruvate, lactate, and several TCA intermediates. FR might attenuate glucose transport in UM cells as an indirect consequence of reducing glucose utilization via glycolysis, thereby increasing cytosolic glucose levels that in turn impair facilitative glucose transport (41). However, FR potentially inhibits glucose transport as a direct consequence of attenuating signaling mechanisms that regulate the GLUT4 glucose transporter (SLC2A4). We suggest this mechanism because forced expression in adipocytes of the same constitutively active Gq mutant found in UM can drive plasma membrane targeting of GLUT4 through a post-translational mechanism involving Arf6 (42, 43, 44). Furthermore, we found that FR reduced GLUT4 mRNA expression threefold (z = −2.0) in freshly isolated primary UM tumor cells (Data File S8) but did not affect mRNA expression of GLUT1 (SLC2A1) or GLUT3 (SLC2A3) (Data File S8), which are induced by oncogenic BRAF in cutaneous melanoma (45, 46, 47).
Atypical metabolic reprogramming in UM: Lack of a classical Warburg effect
We have discovered that oncogenic Gq/11 signaling in UM tumor cells drives both glycolysis and respiration, in contrast to a classic Warburg effect characteristic of many tumors including BRAF-driven cutaneous melanoma (5, 6, 8) in which oncogenic signaling augments glycolysis and attenuates respiration. A non-Warburg effect was indicated by our finding that FR attenuated rather than increased mitochondrial respiration (Fig. 4A), in contrast to the induction of mitochondrial respiration by BRAF inhibitors in cutaneous melanoma cells (5, 6). Similar to what we found with Gq/11-driven UM cells treated with FR, mouse embryonic fibroblasts (MEFs) lacking Gq and G11 exhibit reduced mitochondrial membrane potential, respiratory capacity, ATP levels, and OxPhos-dependent growth (48), indicating that Gq/11-driven respiration is not unique to UM cells. In principle, the lack of a classical Warburg effect in UM compared with cutaneous melanoma might be due to the distinct identities of their respective oncogenic drivers. By extension, differences in developmental histories or environmental niches of melanoma cells arising in the uveal tract versus skin may select for oncogenic signaling that drives or attenuates mitochondrial respiration. This possibility is supported by a recent report from Urtatiz et al. (49) showing that tissue microenvironment determines whether forced expression of constitutively active Gq promotes or inhibits melanocyte growth, and that growth inhibition may be caused by mitochondrial stress. Accordingly, high oxygen tension in the highly vascularized uveal tract (50) might select for maintenance of oncogenic signaling–driven mitochondrial respiration (51, 52) to drive ATP production and consume oxygen as protection against oxidative stress, thereby supporting growth and/or survival of uveal melanocytic lesions driven by Gq/11.
Implications for UM therapy
Combinatorial therapy is likely to be required for treating UM because single agents targeting PKC or MEK downstream of oncogenic Gq/11 have shown little therapeutic benefit in clinical trials (37, 38), and molecules that target oncogenic Gq/11 (FR or its close relative YM-254890) arrest but do not regress xenografted UM tumors in mouse models (34, 53, 54). Harmonizing our results to those reported elsewhere suggests that blocking autophagy might improve the therapeutic efficacy of Gq/11 inhibitors in UM. We have shown that inhibition of Gq/11 attenuates metabolic reprogramming and induces metabolite scavenging in UM cells from patients (Fig. 8). These metabolic effects may explain why knockdown of oncogenic Gq in UM cells (55) or deletion of Gq and G11 in MEFs (56) induces autophagy, potentially as a parallel adaptive survival response to degrade and recycle cytoplasmic constituents as precursors for macromolecular synthesis (57). Mechanisms that induce protective autophagy in FR-treated UM cells potentially function through AMP-activated protein kinase (AMPK) and mechanistic target of rapamycin complex 1 (mTORC1), opposing regulators of autophagy (57), because knockdown of oncogenic Gq in UM cells activates AMPK (55) and deletion of Gq and G11 in MEFs reduces mTORC1 activity (56). FR is likely to exert similar effects in UM cells because FR reduces mitochondrial ATP production, which would activate AMPK (57), and attenuates Erk1/2 activation, which would reduce Erk1/2-dependent activation of mTORC1 (57). Induction of autophagy through these pathways may explain why combining FR and the lysosomal inhibitor chloroquine synergistically kills UM cell lines (58) and suggests that combining FR with inhibitors of autophagy should be explored for synergistic effect in UM.
Experimental procedures
Reagents
FR was purified from Ardisia crenata according to published methods (59). The MEKi trametinib (GSK1120212) and the PKC subtype-nonselective inhibitor sotrastaurin (AEB071; PKCi) were purchased from Selleck Chemicals.
18F-FDG PET/CT
All animal experiments were performed using protocols approved by the Animal Studies Committee of Washington University in St Louis School of Medicine. Animal experiments were performed using the NOD scid gamma (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ: NSG) mice purchased from The Jackson Laboratory (catalog no.: 005557). MP46 cells were inoculated subcutaneously (2 × 10⁶ cells in100 μl in ice-cold PBS/Matrigel Matrix [50/50 v/v] [Fisher Scientific; catalog no.: CB40234A]) into the flanks of 5-week-old male NSG mice (n = 10). MP46 tumors were allowed to grow to ∼30 mm2 basal area measured with calipers and using the formula (L × 2W)/2, where L and W are the longest and shortest basal diameters of the tumor, respectively. For micro-PET imaging studies, animals were anesthetized in a Plexiglas induction chamber flowing oxygen and 1.5 to 2.0% isoflurane. Approximately 250 μCi of 18F-FDG in 100 μl was counted in a dose calibrator (Capintec, Inc) and injected into the tail vein of each anesthetized mouse using a 0.300 cc, 29 gauge × ½” insulin syringe. The syringe was counted again to quantify uninjected radioactivity. After injection, each mouse was weighed and returned to the cage to restore normal and awake state. At 1 h post radionuclide injection, each mouse was anesthetized in the isoflurane induction chamber and placed on a mouse imaging hotel with four nose-coned bed positions for imaging. Micro-CT scanning was performed using the microPET image system (Siemens Medical Solutions USA, Inc). Anatomical placement was determined, and a 10 min emission scan was performed at 1 h for PET. Each region of interest (ROI) was selected in microPET encompassing the desired tissue, and 18F positron emission counts were calculated. Mean counts per volume (becquerel/milliliter), and standard deviations were calculated for each ROI. The 18F decay from injection to start of scan was calculated, and standard uptake values were calculated for each tissue as the mean of the ROI (becquerel/milliliter) times the total mass of the animal (g) divided by the decay-adjusted injected dose (converted to becquerel). After pretreatment FDG–PET, animals were randomly grouped (n = 3/group) for vehicle (1% dimethyl sulfoxide [DMSO]) or FR treatments. FR was prepared from a 15 mM stock solution in DMSO and injected at doses of 0.3 and 0.6 mg/kg and dextrose at a final volume of 200 μl. FR or vehicle was administered subcutaneously every other day for a total of 7 days, and FDG–PET was repeated as described previously. Mice were euthanized at the end of the experiment before tumor burden became harmful.
UM cell lines
Cell lines are summarized in Table S1. Human UM cell lines 92.1 (Research Resource Identifier [RRID]: CVCL_8607), Mel202 (RRID: CVCL_C301), Mel270 (RRID: CVCL_C302), and OCM-1A (RRID: CVCL_6934) were derived and the generous gifts of Drs Martine Jager (Laboratory of Ophthalmology, Leiden University), Bruce Ksander (Schepens Eye Institute, Massachusetts Eye and Ear Infirmary), and June Kan-Mitchell (Biological Sciences, University of Texas at El Paso). The human UM PDX-derived cell lines MP41 (American Type Culture Collection [ATCC]; catalog no.: CRL-3297; RRID: CVCL_4D12) and MP46 (ATCC; catalog no.: CRL-3298, RRID: CVCL_4D13) (33) were purchased from ATCC. The human cutaneous cell lines A375 (RRID: CVCL_0132), MeWo (RRID: CVCL_0445), and SK-mel-2 (RRID: CVCL_0069) were the generous gifts of Dr Lynn Cornelius (Division of Dermatology, Washington University). All cell lines were grown at 37 °C in 5% CO2 in RPMI1640 medium (Life Technologies) supplemented with antibiotics and 25% fetal bovine serum (MP41 and MP46) or 10% fetal bovine serum (all other lines). Cells were not used above passage 35.
Glucose uptake assays
MP41, MP46, and OCM-1A cells were seeded in white and clear bottom 96-well plates at 1.5 × 104 cells/well and allowed to attach overnight. The next day, cells were treated with vehicle (DMSO) or FR (100 nM in DMSO) in growth medium; six wells for each condition per cell line per experiment. Five independent experiments were performed with these cell lines. The next day, glucose uptake assays were performed using the Promega Glucose Uptake-Glo Assay (Promega) following the manufacturer's protocol. Briefly, growth medium was removed, and cells were washed once with PBS, and then, for each treatment condition, 1 mM 2-DG was added to three wells, and PBS was added to the other three wells as a negative control for accumulation of 2-deoxy-d-glucose-6-phosphate (2-DG6P). The plates were shaken briefly to mix and then incubated for 10 min at room temperature. Stop buffer was added immediately to stop the reaction, followed immediately by neutralization buffer. The wells were aspirated, and 2-DG6P detection reagent (luciferase reagent, NADP+, G6PDH, reductase, and reductase substrate) was added and allowed to incubate at room temperature for 30 min. After incubation, sample luminescence was measured, and rates of glucose uptake (fmol/min/cell) were interpolated in GraphPad Prism (GraphPad Software, Inc) using a 2-DG6P standard curve with concentrations from 0.5 to 30 μM in PBS.
UM patients and tumor collection
Human primary UM enucleation samples were obtained with patient written informed consent and with approval of the institutional review board of Washington University in St Louis. Removal of tumor-bearing eyes by enucleation was performed as part of standard of care, which also included collecting samples for molecular classification (Castle Biosciences). After excision from the patient, eyes were transilluminated to localize the tumor mass and then opened opposite the tumor. Vitreous was carefully removed, and tumor samples were collected through the retina from the tumor apex. All samples were collected directly into 0.05% trypsin–EDTA (Gibco) on ice in the operating room. Samples were transported to the laboratory and incubated on ice for a total of 1 h post excision. Samples were then changed to medium containing 5 mg/ml collagenase I (Sigma–Aldrich) and incubated at 37 °C for 45 min, followed by centrifugation at 3000g for 5 min. The supernatants were removed gently, leaving enough medium behind to cover the pellet, and then resuspended by trituration with fresh collagenase solution. This cycle was repeated four times until the samples were fully dissociated to single cells and then resuspended once in phosphate-buffered saline before being resuspended finally in melanoblast defined medium plus fibroblast growth factor growth medium: HAM's F12 (Lonza) supplemented with 1 mg/ml bovine serum albumin (Sigma–Aldrich), 2 mM l-glutamine (Lonza), 1× SITE (Sigma–Aldrich), 1× B27 (Gibco), 20 ng/ml basic fibroblast growth factor (PeproTech, Inc), and 50 μg/ml gentamicin (Sigma–Aldrich) (60). Enucleated samples yielded less than a total of 106 intact dissociated cells, most of which were used for Seahorse assays at 2 × 104 cells/well. Cells were maintained in suspension at 37 °C for 24 h and then plated as described later for Seahorse experiments.
Microphysiometry experiments
Cells were plated on Seahorse XFe96 Cell Culture Microplates (Agilent Technologies) coated with Cell-Tak (Corning Life Sciences) in 50 μl of appropriate medium for each cell line or sample. Cell density was set near 80% confluency. Cells were plated 2 days prior to assay and treated with FR, MEKi, PKCi, or vehicle (0.1% DMSO) 18 h prior to assay. For dilution curves, FR, MEKi, or PKCi was diluted in the appropriate RPMI medium for the treated cell line at the indicated concentrations with 0.1% DMSO as a vehicle control for each drug. Each treatment group had a minimum of three replicate wells, and each plate had a minimum of four background wells. Seahorse XF DMEM pH 7.4 media were used to run all experiments, with a final well volume of 180 μl. Basal respiration was measured using the Mito Stress Test (MST) Assay Kit, and glycolysis was measured using the Glycolytic Stress Test (GST) Assay Kit, both from Agilent. Samples were run according to Agilent assay protocol on Seahorse XF96 and XFe96 analyzers. For enucleation samples, one GST kit and one MST kit were combined and loaded into the injection ports of a Seahorse Sensor Cartridge as follows: port A was loaded with 10 mM glucose (from the GST kit); port B was loaded with 1.5 μM oligomycin (from the GST kit); port C was loaded with 1 μM FCCP (from the MST kit); port D was loaded with 0.5 μM rotenone/antimycin A (from the MST kit) dissolved into 50 mM 2-DG (from the GST kit). Samples were then run on a Seahorse XF96 or XFe96 analyzer using a four-port injection protocol using standard cycles (3 min mix, 4 min wait, and 3 min measure). FR time-course experiments were run using custom assays. Cells were plated as described previously the day prior to the assay. The day of the assay, 20 μl of FR (prepared in Seahorse XF DMEM, pH 7.4 media to 1 μM [100 nM final well concentration]) and vehicle control (prepared in Seahorse XF DMEM, pH 7.4 media adding equal volume of DMSO as used for FR) were loaded into port A. Each treatment group had a minimum of six replicate wells. Three baseline cycles (3 min mix, 30 s wait, and 3 min measure) were run prior to the injection of the drugs in port A, and 33 cycles (3 min mix, 14 min wait, and 3 min measure) were run post injection of port A for a total of 12 h of run time (including calibration). Normalization of all data to cell number/well was accomplished through measurement of 4′,6-diamidino-2-phenylindole fluorescence. After the Seahorse assay was completed, cells were fixed with cold methanol and stained with 4′,6-diamidino-2-phenylindole. Plates were read at 358/20 nm excitation and 461/20 nm emission on a Cytation 5 imaging reader using Gen5 (version 3.08) software from BioTek. Rates for glycolysis and basal respiration were gathered from GST and MST report generators created by Wave Desktop Software (version 2.6.1), available from Agilent, and utilized in Microsoft Excel 2016.
Measurement of glycolytic and TCA cycle intermediates by GC–MS
MP41, MP46, and OCM-1A cells were cultured to 70% confluency in 6-well plates and treated for 24 h with 100 nM FR or vehicle control. Each well corresponded to one technical replicate, and three independent experiments were performed for each cell line. Plates were placed on wet ice, growth media were aspirated, and the plates were rinsed twice with ice-cold PBS (gently) without disturbing the cells, and then transferred to a bed of dry ice. About 200 μl of 80% methanol was added to the well along with 10 μl of a cocktail of labeled amino acid and TCA cycle standards (Tables S2 and S3). Each well was scraped in the aqueous methanol to suspend and extract the adherent cell material and pipetted into an Eppendorf tube. An additional 200 μl of 80% methanol was added to each well and scraped to recover more cell extract and combined with the material from the first scraping. The cell suspension extract was centrifuged at 15,000 rpm to pellet the cell debris. Cell extract supernatants were added to a GC insert vial (National C4011-631) and placed in an Eppendorf tube with a needle puncture in the lid and evaporated to dryness in a Labconco CentriVap Concentrator. Pellets were saved for protein quantification using the Pierce bicinchoninic acid assay.
The GC vial inserts containing the cell extract dried residues were placed in a glass vial (National C4000-1), derivatized with 10 μl of 20 mg/ml methoxyamine HCl in pyridine and incubated on a heating block set to 38 °C for 90 min. Immediately following the incubation, 40 μl of N-methyl-N-tert-butyldimethylsilyltrifluoroacetamide (with 1% t-butyldimethylchlorosilane) (catalog number: M-108-5x1mL) were added to each vial and incubated at 38 °C for an additional 30 min (61). Derivatized samples were then run on an Agilent 7890A GC coupled to an Agilent 5975C MS, and data were acquired and analyzed in MSD ChemStation E.02.02.1431. All data were collected in SIM mode with an initial 80 °C hold for 2 min, followed by a temperature rate change of 10 °C/min to 300 °C, and a hold for 6 min.
Cell pellets were suspended in 200 μl of lysis buffer cocktail (Santa Cruz Biotechnology) containing protease inhibitor cocktail, 2 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate provided in the Santa Cruz Biotechnology radioimmunoprecipitation assay lysis buffer kit. Metabolite peak areas in each sample were quantified relative to nanomoles of internal standards and normalized to mass of protein in the cell pellet.
Statistical analyses
All statistical analyses were performed in GraphPad Prism (version 8.2.1 [441]). Summary data values on all graphs represent means. Error bars on graphs represent standard error of the mean for immunoblot and Seahorse experiments or standard deviation for GC–MS metabolomic data. Stars indicate significance as determined by statistical analysis (∗<0.01). All datasets were checked for normality prior to analysis via Shapiro–Wilk and Kolmogorov–Smirnov (with Lilliefors's significance correction) tests. If groups in the dataset differed in normality, all were first run through nonparametric statistics, with normal groups being run through the parametric version of the test after to detect additional significant relationships. If all groups followed a Gaussian distribution, parametric statistics were performed. Data for baseline measurements of glycolysis and basal respiration were compared using one-way ANOVAs with Bonferroni post hoc tests or Kruskal–Wallis with Dunn post hoc tests. Data for treated and untreated samples were analyzed via unpaired t tests or Mann–Whitney tests. Nonlinear regression log(inhibitor) versus parameter model tests were used for dose–response curves and used to determine IC50 values. Drug monitoring experiments also utilized both nonlinear and linear regressions in order to detect significance between treated and untreated samples. Nonlinear regressions used a one phase decay model with plateau comparisons.
Gene expression analyses
All gene expression analyses were performed using published transcriptional data (GSE165552) on human UM tumor samples treated for 7 days with FR (34). Kyoto Encyclopedia of Genes and Genomes pathway analysis was performed using gene set enrichment analysis (62). Reranking of gene in the published dataset by z-score, based on responses to FR, used the equation: Z-score = (meanFR − meanvehicle)/SQRT[(StDevFR2/nFR) + (StDevvehicle2/nvehicle)], where “mean” is the average gene expression across the set of samples, “StDev” is the standard deviation of the gene expression across the set of samples, and “n” is the number of samples in the corresponding set. Z-scores were calculated for each expressed gene in the dataset and used to generate preranked gene lists, which were used for metabolic pathway analysis by gene set enrichment analysis (62).
Immunoblot assays
Cells were grown in 10 cm dishes and treated for 18 h with FR, PKCi, MEKi, or vehicle alone (DMSO) at the indicated concentrations for the indicated durations. Cells were lysed in 1× cell lysis buffer. Lysates were sonicated on ice for 2 min (30 s on, 30 s off, 60% A), rotated end over end for 30 min, and cleared by centrifugation at 16,000g for 15 min. Protein concentration was determined using Bio-Rad Protein Assay Dye Reagent (Bio-Rad; catalog no.: 5000006). About 15 μg of tumor protein was resolved on 12% SDS-PAGE gels and transferred to Immobilon-FL poly(vinylidene fluoride) membrane (Millipore; catalog no.: IPFL00010). Membranes were blocked with 5% (w/v) bovine serum albumin in TBST (20 mM Tris, pH 7.6, 137 mM NaCl, 0.1% v/v Tween-20) and incubated with primary antibodies (Phospho-p44/42 MAPK [Erk1/2], Cell Signaling Technology; catalog no.: 4370S; lot no.: 24 and p44/42 MAPK [Erk1/2], Cell Signaling Technology; catalog no.: 9107S; lot no.: 10). Membranes were washed with TBST at least three times and incubated with IRDye 680–coupled goat anti-rabbit (LI-COR; catalog no.: 926-6807; lot no.: C90618-09) and IRDye 800 goat antimouse (LI-COR; catalog no.: 926-32210: lot no.: C91210-09 antibodies [LI-COR Biosciences]). After incubation, membranes were washed at least three times with TBST, and fluorescence intensity signals were detected using Odyssey model 9120 imaging system (LI-COR Biosciences). Integrated fluorescence intensities (I.I.) were calculated, and background fluorescence was subtracted for the selected bands during imaging. The sum of the I.I. for the pERK bands was divided by the sum of the I.I. for the total ERK bands, and these ratios were normalized to the ratios of control cells. Data were collected from four independent experiments.
Data availability
All RNA-Seq data used in the present study were published previously and deposited on the National Center for Biotechnology Information Gene Expression Omnibus server with the accession #GSE165552. All other data associated with this study are presented in the main text or supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
K. J. B. and M. D. O. are listed as coinventors on a provisional patent application on targeted pharmacological therapeutics in uveal melanoma that is owned by Washington University in St Louis. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St Louis, MO, for the use of the Small Animal Cancer Imaging shared resource, which provided FDG–PET services. The Siteman Cancer Center is supported in part by the National Cancer Institute Cancer Center Support grant (P30 CA091842). We also thank the Department of Genetics Tissue Culture Support Center and the Washington University Diabetes Research Center (National Institutes of Health grant [P30 DK020579]) for providing Agilent Seahorse Metabolic Assay services.
Author contributions
K. J. B. conceptualization; M. D. O., S. E. N., K. M. K., C. F., C. M. M., N. F., J. E. I., and K. J. B. formal analysis; M. D. O., S. E. N., K. M. K., C. F., C. M. M., and N. F. investigation; K. D. P. and P. L. C. resources; M. D. O., J. E. I., and K. J. B. writing–original draft; M. D. O. and K. J. B. writing–review & editing; K. J. B. funding acquisition.
Funding and additional information
This work was supported by the National Institutes of Health grants GM124093 and CA234533 (awarded to K. J. B.); CA218869 and CA242221 (awarded to J. E. I.); and GM118171 (awarded to M. D. O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Henrik Dohlman
Supporting information
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ward P.S., Thompson C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng W., Tayyari F., Gowda G.A.N., Raftery D., Mclamore E.S., Porterfield D.M., Donkin S.S., Bequette B., Teegarden D. Altered glucose metabolism in Harvey-ras transformed MCF10A cells. Mol. Carcinog. 2015;54:111–120. doi: 10.1002/mc.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchetti P., Trinh A., Khamari R., Kluza J. Melanoma metabolism contributes to the cellular responses to MAPK/ERK pathway inhibitors. Biochim. Biophys. Acta Gen. Subj. 2018;1862:999–1005. doi: 10.1016/j.bbagen.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Haq R., Shoag J., Andreu-Perez P., Yokoyama S., Edelman H., Rowe G.C., Frederick D.T., Hurley A.D., Nellore A., Kung A.L., Wargo J.A., Song J.S., Fisher D.E., Arany Z., Widlund H.R. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haq R., Fisher D.E., Widlund H.R. Molecular pathways: BRAF induces bioenergetic adaptation by attenuating oxidative phosphorylation. Clin. Cancer Res. 2014;20:2257–2263. doi: 10.1158/1078-0432.CCR-13-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liberti M.V., Locasale J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida G.J. Metabolic reprogramming: The emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han A., Schug Z.T., Aplin A.E. Metabolic alterations and therapeutic opportunities in rare forms of melanoma. Trends Cancer. 2021;7:671–681. doi: 10.1016/j.trecan.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh A.D., Topham A. Incidence of uveal melanoma in the United States: 1973-1997. Ophthalmology. 2003;110:956–961. doi: 10.1016/S0161-6420(03)00078-2. [DOI] [PubMed] [Google Scholar]
- 13.An J., Wan H., Zhou X., Hu D.N., Wang L., Hao L., Yan D., Shi F., Zhou Z., Wang J., Hu S., Yu J., Qu J. A comparative transcriptomic analysis of uveal melanoma and normal uveal melanocyte. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng X., Chen Z., Farshidfar F., Xu X., Lorenzi P.L., Wang Y., Cheng F., Tan L., Mojumdar K., Du D., Ge Z., Li J., Thomas G.V., Birsoy K., Liu L., et al. Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep. 2018;23:255–269.e4. doi: 10.1016/j.celrep.2018.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B., Ma R., Ren H., Qian J. Genome-wide analysis of uveal melanoma metastasis-associated LncRNAs and their functional network. DNA Cell Biol. 2018;37:99–108. doi: 10.1089/dna.2017.4015. [DOI] [PubMed] [Google Scholar]
- 16.Modorati G., Lucignani G., Landoni C., Freschi M., Trabucchi G., Fazio F., Brancato R. Glucose metabolism and pathological findings in uveal melanoma: Preliminary results. Nucl. Med. Commun. 1996;17:1052–1056. doi: 10.1097/00006231-199612000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Klingenstein A., Haug A.R., Nentwich M.M., Tiling R., Schaller U.C. Whole-body F-18-fluoro-2-deoxyglucose positron emission tomography/computed tomography imaging in the follow-up of metastatic uveal melanoma. Melanoma Res. 2010;20:511–516. doi: 10.1097/CMR.0b013e3283403d6c. [DOI] [PubMed] [Google Scholar]
- 18.Finger P.T., Kurli M., Reddy S., Tena L.B., Pavlick A.C. Whole body PET/CT for initial staging of choroidal melanoma. Br. J. Ophthalmol. 2005;89:1270–1274. doi: 10.1136/bjo.2005.069823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orcurto V., Denys A., Voelter V., Schalenbourg A., Schnyder P., Zografos L., Leyvraz S., Delaloye A.B., Prior J.O. 18F-fluorodeoxyglucose positron emission tomography/computed tomography and magnetic resonance imaging in patients with liver metastases from uveal melanoma: Results from a pilot study. Melanoma Res. 2012;22:63–69. doi: 10.1097/CMR.0b013e32834d3dcb. [DOI] [PubMed] [Google Scholar]
- 20.Cohen V., Pavlidou E., Costa J., Arora A., Szyszko T., Sagoo M., Szlosarek P. Staging uveal melanoma with whole-body positron-emission tomography/computed tomography and abdominal ultrasound: Low incidence of metastatic disease, high incidence of second primary cancers. Middle East Afr. J. Ophthalmol. 2018;25:91–95. doi: 10.4103/meajo.MEAJO_96_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eldredge-Hindy H., Ohri N., Anne P.R., Eschelman D., Gonsalves C., Intenzo C., Bar-Ad V., Dicker A., Doyle L., Li J., Sato T. Yttrium-90 microsphere brachytherapy for liver metastases from uveal melanoma clinical outcomes and the predictive value of fluorodeoxyglucose positron emission tomography. Am. J. Clin. Oncol. 2016;39:189–195. doi: 10.1097/COC.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krantz B.A., Dave N., Komatsubara K.M., Marr B.P., Carvajal R.D. Uveal melanoma: Epidemiology, etiology, and treatment of primary disease. Clin. Ophthalmol. 2017;11:279–289. doi: 10.2147/OPTH.S89591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A.D., Zabor E.C., Radivoyevitch T. Estimating cured fractions of uveal melanoma. JAMA Ophthalmol. 2021;139:174–181. doi: 10.1001/jamaophthalmol.2020.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay C., Oba J., Roszik J., Marszalek J.R., Chen K., Qi Y., Eterovic K., Gordon Robertson A., Burks J.K., McCannel T.A., Grimm E.A., Woodman S.E. Elevated endogenous SDHA drives pathological metabolism in highly metastatic uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2019;60:4187–4195. doi: 10.1167/iovs.19-28082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han A., Purwin T.J., Bechtel N., Liao C., Chua V., Seifert E., Sato T., Schug Z.T., Speicher D.W., William Harbour J., Aplin A.E. BAP1 mutant uveal melanoma is stratified by metabolic phenotypes with distinct vulnerability to metabolic inhibitors. Oncogene. 2021;40:618–632. doi: 10.1038/s41388-020-01554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O'Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N., Sozen M.M., Baimukanova G., Roy R., Heguy A., Dolgalev I., et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onken M.D., Worley L.A., Long M.D., Duan S., Council M.L., Bowcock A.M., Harbour J.W. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 2008;49:5230–5234. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koopmans A.E., Vaarwater J., Paridaens D., Naus N.C., Kilic E., De Klein A. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br. J. Cancer. 2013;109:493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offermanns S., Zhao L.P., Gohla A., Sarosi I., Simon M.I., Wilkie T.M. Embryonic cardiomyocyte hypoplasia and craniofacial defects in Gα(q)/Gα11-mutant mice. EMBO J. 1998;17:4304–4312. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patt J., Alenfelder J., Pfeil E.M., Voss J.H., Merten N., Eryilmaz F., Heycke N., Rick U., Inoue A., Kehraus S., Deupi X., Müller C.E., König G.M., Crüsemann M., Kostenis E. An experimental strategy to probe Gq contribution to signal transduction in living cells. J. Biol. Chem. 2021;296:100472. doi: 10.1016/j.jbc.2021.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onken M.D., Makepeace C.M., Kaltenbronn K.M., Kanai S.M., Todd T.D., Wang S., Broekelmann T.J., Rao P.K., Cooper J.A., Blumer K.J. Targeting nucleotide exchange to inhibit constitutively active G protein a subunits in cancer cells. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aao6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amirouchene-Angelozzi N., Nemati F., Gentien D., Nicolas A., Dumont A., Carita G., Camonis J., Desjardins L., Cassoux N., Piperno-Neumann S., Mariani P., Sastre X., Decaudin D., Roman-Roman S. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol. Oncol. 2014;8:1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onken M.D., Makepeace C.M., Kaltenbronn K.M., Choi J., Hernandez-Aya L., Weilbaecher K.N., Piggott K.D., Kumar Rao P., Yuede C.M., Dixon6 A.J., Osei-Owusu P., Cooper J.A., Blumer K.J. Targeting primary and metastatic uveal melanoma with a G protein inhibitor. J. Biol. Chem. 2021;296:100403. doi: 10.1016/j.jbc.2021.100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang M., Soga T., Pollard P.J. Oncometabolites: Linking altered metabolism with cancer. J. Clin. Invest. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowicki S., Gottlieb E. Oncometabolites: Tailoring our genes. FEBS J. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luke J.J., Triozzi P.L., McKenna K.C., Van Meir E.G., Gershenwald J.E., Bastian B.C., Gutkind J.S., Bowcock A.M., Streicher H.Z., Patel P.M., Sato T., Sossman J.A., Sznol M., Welch J., Thurin M., et al. Biology of advanced uveal melanoma and next steps for clinical therapeutics. Pigment Cell Melanoma Res. 2015;28:135–147. doi: 10.1111/pcmr.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chattopadhyay C., Kim D.W., Gombos D.S., Oba J., Qin Y., Williams M.D., Esmaeli B., Grimm E.A., Wargo J.A., Woodman S.E., Patel S.P. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer. 2016;122:2299–2312. doi: 10.1002/cncr.29727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cree I.A., Glaysher S., Harvey A.L. Efficacy of anti-cancer agents in cell lines versus human primary tumour tissue. Curr. Opin. Pharmacol. 2010;10:375–379. doi: 10.1016/j.coph.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Sponagel J., Jones J.K., Frankfater C., Zhang S., Tung O., Cho K., Tinkum K.L., Gass H., Nunez E., Spitz D.R., Chinnaiyan P., Schaefer J., Patti G.J., Graham M.S., Mauguen A., et al. Sex differences in brain tumor glutamine metabolism reveal sex-specific vulnerabilities to treatment. bioRxiv. 2021 doi: 10.1101/2021.09.29.461531. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barron C.C., Bilan P.J., Tsakiridis T., Tsiani E. Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism. 2016;65:124–139. doi: 10.1016/j.metabol.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Imamura T., Vollenweider P., Egawa K., Clodi M., Ishibashi K., Nakashima N., Ugi S., Adams J.W., Brown J.H., Olefsky J.M. G alpha-q/11 protein plays a key role in insulin-induced glucose transport in 3T3-L1 adipocytes. Mol. Cell. Biol. 1999;19:6765–6774. doi: 10.1128/mcb.19.10.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishibashi K.I., Imamura T., Sharma P.M., Huang J., Ugi S., Olefsky J.M. Chronic endothelin-1 treatment leads to heterologous desensitization of insulin signaling in 3T3-L1 adipocytes. J. Clin. Invest. 2001;107:1193–1202. doi: 10.1172/JCI11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bose A., Cherniack A.D., Langille S.E., Nicoloro S.M.C., Buxton J.M., Park J.G., Chawla A., Czech M.P. G α 11 signaling through ARF6 regulates F-actin mobilization and GLUT4 glucose transporter translocation to the plasma membrane. Mol. Cell. Biol. 2001;21:5262–5275. doi: 10.1128/MCB.21.15.5262-5275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parmenter T.J., Kleinschmidt M., Kinross K.M., Bond S.T., Li J., Kaadige M.R., Rao A., Sheppard K.E., Hugo W., Pupo G.M., Pearson R.B., McGee S.L., Long G.V., Scolyer R.A., Rizos H., et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4:423–433. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theodosakis N., Held M.A., Marzuka-Alcala A., Meeth K.M., Micevic G., Long G.V., Scolyer R.A., Stern D.F., Bosenberg M.W. BRAF inhibition decreases cellular glucose uptake in melanoma in association with reduction in cell volume. Mol. Cancer Ther. 2015;14:1680–1692. doi: 10.1158/1535-7163.MCT-15-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardeman K.N., Peng C., Paudel B.B., Meyer C.T., Luong T., Tyson D.R., Young J.D., Quaranta V., Fessel J.P. Dependence on glycolysis sensitizes BRAF-mutated melanomas for increased response to targeted BRAF inhibition. Sci. Rep. 2017;7:42604. doi: 10.1038/srep42604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benincá C., Planagumà J., de Freitas Shuck A., Acín-Perez R., Muñoz J.P., de Almeida M.M., Brown J.H., Murphy A.N., Zorzano A., Enríquez J.A., Aragay A.M. A new non-canonical pathway of Gαq protein regulating mitochondrial dynamics and bioenergetics. Cell. Signal. 2014;26:1135–1146. doi: 10.1016/j.cellsig.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urtatiz O., Haage A., Tanentzapf G., Van Raamsdonk C.D. Crosstalk with keratinocytes causes GNAQ oncogene specificity in melanoma. bioRxiv. 2021 doi: 10.1101/2021.07.26.453858. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bill A., Sperber G., Ujiie K. Physiology of the choroidal vascular bed. Int. Ophthalmol. 1983;6:101–107. doi: 10.1007/BF00127638. [DOI] [PubMed] [Google Scholar]
- 51.McKeown S.R. Defining normoxia, physoxia and hypoxia in tumours - implications for treatment response. Br. J. Radiol. 2014;87:20130676. doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhandari V., Li C.H., Bristow R.G., Boutros P.C. Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat. Commun. 2020;11:737. doi: 10.1038/s41467-019-14052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Annala S., Feng X., Shridhar N., Eryilmaz F., Patt J., Yang J.H., Pfeil E.M., Cervantes-Villagrana R.D., Inoue A., Häberlein F., Slodczyk T., Reher R., Kehraus S., Monteleone S., Schrage R., et al. Direct targeting of Gα q and Gα 11 oncoproteins in cancer cells. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aau5948. [DOI] [PubMed] [Google Scholar]
- 54.Hitchman T.D., Bayshtok G., Ceraudo E., Moore A.R., Lee C., Jia R., Wang N., Pachai M.R., Shoushtari A.N., Francis J.H., Guan Y., Chen J., Chang M.T., Taylor B.S., Sakmar T.P., et al. Combined inhibition of Gαq and MEK enhances therapeutic efficacy in uveal melanoma. Clin. Cancer Res. 2020;27:1476–1490. doi: 10.1158/1078-0432.CCR-20-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ambrosini G., Musi E., Ho A.L., de Stanchina E., Schwartz G.K. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol. Cancer Ther. 2013;12:768–776. doi: 10.1158/1535-7163.MCT-12-1020. [DOI] [PubMed] [Google Scholar]
- 56.Cabezudo S., Sanz-Flores M., Caballero A., Tasset I., Rebollo E., Diaz A., Aragay A.M., Cuervo A.M., Mayor F., Ribas C. Gαq activation modulates autophagy by promoting mTORC1 signaling. Nat. Commun. 2021;12:4540. doi: 10.1038/s41467-021-24811-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu Y., Chang Y., Lu W., Sheng X., Wang S., Xu H., Ma J. Regulation of autophagy by glycolysis in cancer. Cancer Manag. Res. 2020;12:13259–13271. doi: 10.2147/CMAR.S279672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Truong A., Yoo J.H., Scherzer M.T., Sanchez J.M.S., Dale K.J., Kinsey C.G., Richards J.R., Shin D., Ghazi P.C., Onken M.D., Blumer K.J., Odelberg S.J., McMahon M. Chloroquine sensitizes GNAQ/11-mutated melanoma to MEK1/2 inhibition. Clin. Cancer Res. 2020;26:6374–6386. doi: 10.1158/1078-0432.CCR-20-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrage R., Schmitz A.L., Gaffal E., Annala S., Kehraus S., Wenzel D., Büllesbach K.M., Bald T., Inoue A., Shinjo Y., Galandrin S., Shridhar N., Hesse M., Grundmann M., Merten N., et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 2015;6:10156. doi: 10.1038/ncomms10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landreville S., Agapova O.A., Kneass Z.T., Salesse C., William Harbour J. ABCB1 identifies a subpopulation of uveal melanoma cells with high metastatic propensity. Pigment Cell Melanoma Res. 2011;24:430–437. doi: 10.1111/j.1755-148X.2011.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016;114:30.4.1–30.4.32. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq data used in the present study were published previously and deposited on the National Center for Biotechnology Information Gene Expression Omnibus server with the accession #GSE165552. All other data associated with this study are presented in the main text or supporting information.