Abstract
Chromatin remodeling, specifically the tissue-specific regulation in mineralized tissues, is an understudied avenue of gene regulation. Here we show that Baf45a and Baf45d, two Baf45 homologs belong to ATPase-dependent SWI/SNF chromatin remodeling complex, preferentially expressed in osteoblasts and odontoblasts compared to Baf45b and Baf45c. Recently, biochemical studies revealed that BAF45A associates with Polybromo-associated BAF (PBAF) complex. However, the BAF45D subunit belongs to the polymorphic canonical BRG1-associated factor (cBAF) complex. Protein profiles of osteoblast and odontoblast differentiation uncovered a significant increase of BAF45A and PBAF subunits during early osteoblast and odontoblast maturation. Chromatin immunoprecipitation sequencing (ChIP-seq) during the bone marrow stromal cells (BMSCs) differentiation showed higher histone H3K9 and H3K27 acetylation modifications in the promoter of Baf45a and Baf45d and increased binding of bone and tooth specific transcription factor RUNX2. Overexpression of Baf45a in osteoblasts activates genes essential for the progression of osteoblast maturation and mineralization. Furthermore, shRNA-mediated knockdown of Baf45a in odontoblasts leads to markedly altered genes responsible for the proliferation, apoptosis, DNA repair, and modest decrease in dentinogenic marker gene expression. Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) assay in Baf45a knockout osteoblasts revealed a noticeable reduction in chromatin accessibility of osteoblast and odontoblast specific genes, along with transcription factor Atf4 and Klf4. Craniofacial mesenchyme-specific loss of Baf45a modestly reduced the mineralization of the tooth and mandibular bone. These findings indicated that BAF45A-dependent mineralized tissue-specific chromatin remodeling through PBAF-RUNX2 crosstalk results in transcriptional activation is critical for early differentiation and matrix maturation of mineralized tissues.
Keywords: Baf45a, Brg1/Smarca4, osteoblastogenesis, chromatin remodeling, PBAF complex, dentinogenesis
Introduction
During cellular differentiation from stem cells to progenitors to differentiated cells, must orchestrate a definite gene expression program to ensure accurate patterning and tissue specificity. This gene expression program is fundamentally regulated largely by tissue-specific epigenetic regulation that results in spatiotemporal gene expression (1, 2).
Chromatin regulation occurs through a series of crosstalk between cell-type-specific regulators and chromatin remodelers/modifiers. The BRG-/BRM-associated factors (BAF) chromatin remodeling complex is an ATPase-dependent epigenetic machine with affinity to bind and slide or eject nucleosomes (3). This process allows accessibility of lineage-specific transcriptional machinery enabling timely and appropriate control of gene expression.
In mammals, BAF factors (4, 5) are highly polymorphic and form complexes composed of a single central ATPase, either BRG1 (SMARCA4) or BRM1 (SMARCA2) (6). Similar to the BAF complex, a family of Polybromo-associated BAF (PBAF) complex has been identified as major tumor suppressors in several malignancies (7–9). Gatchalian et al. recently defined a smaller, non-canonical GBAF complex composed of bromodomain containing protein 9 (BRD9) and glioma tumor suppressor candidate region gene 1 (GLTSCR1) or its paralog GLTSCR1-like (GLTSCR1L) in mouse ESCs that is distinct from the canonical ESC BAF complex (esBAF). Further mechanistic insights demonstrated that BRD9 interacts with BRD4, which guides the recruitment of GBAF complexes to chromatin to maintain the transcriptional network of pluripotency (10).
Among four families of chromatin remodeling complex (SWI/SNF, CHD ISWI, and INO80), the SWI/SNF (switching defective/sucrose nonfermenting) family remodelers are composed of 8 to 15 BRG1 associated non-catalytic subunits. The catalytic ATPase includes an HSA (helicase-SANT), a post-HSA, and a C-terminal bromodomain. The non-catalytic subunits with BRG1 or BRM generate conserved, modular, or defining BAF complexes for cell and tissue-specific function. These complexes include BAF, PBAF, embryonic stem cell BAF (esBAF), neural progenitor BAF (npBAF), and neuron BAF (nBAF). This family has many activities that include sliding and ejecting nucleosomes at many loci. However, they lack roles in chromatin assembly (11, 12). Collectively the mammalian BAF (mSWI/SNF) and PBAF complexes are comprised eight bromodomains (six on PBRM1, one on either BRG or BRM, and one on BRD7), two PHD finger proteins (BAF45 subunits), two chromodomains (BAF155 and BAF170), and between seven and nine DNA binding domain. A ligand-activated in vitro transcription assay was used to biochemically identify the PBAF complex, which was included with specific subunits such as BAF180 (polybromo PBRM1), BAF200 (ARID2), BAF45A (PHF10), and BRD7. The name polybromo BAF (PBAF) came from the discovery that this complex contained polybromo (PBRM1 or BAF180) and BAF200 (ARID2), as well as BRG1 (but not BRM) and several other subunits of the BAF complex (9, 13). At a given time, the BAF complex consists of about 15 BAF subunits. Tissue-specific regulation by the BAF complex arises from the combinatorial assembly of the 29 genes that make up this 2-MDa protein complex (9). The BAF complex promotes gene expression around active promoters, enhancers, and tissue-specific enhancers. To date, many studies have been performed to characterize the roles of ubiquitous and tissue-specific BAF subunits/complexes at the biochemical and physiological levels. Specific examples include during neurogenesis (14), hematopoiesis (15), immune responses (16), in development and disease (17) and embryonic stem cell functions (18).
In multicellular organisms, plant homology factor 10/BRG1 associated factor 45A (PHF10/BAF45A) is a distinct subunit of the PBAF subfamily, which is among the three classes of mSWI/SNF complexes (BAF, PBAF, and ncBAF) (19–21). Cross-linking mass spectrometry on BRD7-and PHF10/BAF45A-bound complexes with stepwise modular assembly authenticated that PBAF complexes contain the same common BAF core module, ARID2, BRD7, and PHF10/BAF45A for tissue-specific combinatorial diversity (21). BAF45A possesses the double tandem PHD finger domain at its C-terminus. Besides BAF45A, similar domains have been identified in other BAF45 homologs; the tandem domains can function through lysine acetylation (H3K14ac) and methylation (H3K4me3) to control the activation of gene transcription in a cooperative and inter-dependent manner (22). The PHD domain can facilitate lysine acetylation at 14, K9, and K27 in histone 3 (H3K14, H3K9, and H3K27) to regulate chromatin for gene activation (23). Originally PHF10 was discovered in Drosophila melanogaster, and homozygous deletion resulted in lethality. Weak mutation led to decreased viability, reduced yellow gene transcription, and sterility (24–26). Conditional BAF45A homozygote null mice survived until embryonic day 19.5. Survived animals have impairment in the frequency of repopulating long-term hematopoietic stem cells and committed myeloid progenitors (27). BAF45A plays a crucial role in transitioning from proliferating neural stem/progenitor cells to post-mitotic neurons (28), nonredundant and specialized functions within the developing hemopoietic tissue (27), and transcriptional activation in myelogenesis (29). However, the precise mechanism of chromatin remodeling is not fully understood, and very little is known about the BAF complex function during the differentiation of mineralized tissue.
Bone formation occurs through a series of transcriptional and signaling events aligned with epigenetic rulings that take mesenchymal stem cells into mature osteoblasts, osteocytes, and chondrocytes. Mature osteoblasts are differentiated from committed osteoprogenitors and pre-osteoblasts to form the bone matrix. This matrix further mineralizes ultimately forming mineralized bone. During this time, a unique gene expression program is needed to grow and differentiate osteoblast cells, which must be tightly regulated in a tissue-specific manner. Much of what we know about the regulation of this gene program comes from our understanding of the bone gene transcriptional program and bone-tissue-specific signaling (30, 31). Furthermore, growing studies about the epigenetic regulation involved in osteogenesis primarily focus on histone post-translational modifications, miRNA, and non-coding RNAs (32–39). However, the tissue-specific mechanisms involved in the openness of the chromatin around active genes are understudied in osteogenesis.
Dentinogenesis occurs, similar to osteogenesis, during the maturation of neural crest cells, dental pulp cells, or pre-odontoblasts differentiated into mature, mineral depositing odontoblasts. These cells form pre-dentin before calcification and maturation. After maturation and calcification, pre-dentin actively is transformed into dentin, the central mineral portion of the tooth (40–42). Like osteoblasts, a highly regulated gene expression program is needed to grow and differentiate odontoblast cells in a tissue-specific manner (43). Little is known about the epigenetic regulation of the odontoblasts, with less being known about chromatin remodeling in either tissue type (44, 45).
Our lab’s recent preliminary findings have demonstrated that the PBAF subunit BAF45A is preserving epigenetic balance for osteogenesis (37). There are four BAF45 homologs, Baf45(a-d). Baf45d is thought to be expressed ubiquitously and is part of the canonical BAF complex. Baf45b and Baf45c are known to regulate neurogenesis and belong to npBAF (19).
Baf45a is a member of the polybromo-BAF (PBAF) complex and is unique from the other BAF45 subunits because of its N-terminal SAY domain, which could potentially enable its transcriptional activity (27).
Here, we investigated the chromatin remodeling mechanism by which the Baf45a promotes tissue-specific gene expression for the maturation and mineralization of osteoblasts and odontoblasts. Deletion of Baf45a in osteoblasts altered the chromatin state around gene promoters and enhancers that are important for osteoblast and odontoblast maturation. Furthermore, we begin to dissect further the temporal expression of Baf45a with other members of the PBAF complex that specifically regulate the chromatin remodeling in the osteoblasts and odontoblasts. Overall, our findings indicate that remodeling of tissue-specific chromatin by Baf45a is required to understand the chromatin accessibility and gene activation of bone and tooth formation.
Materials and Methods
Generation of Baf45a Early Craniofacial Mesenchyme Specific Conditional Knockout Mice
We purchased Baf45a‐floxed (Phf10/Baf45af/f ) mice from The Jackson Laboratory (Stock Phf10tm1.1Grc/J, Stock No: 019480) containing loxP sites flanking exons 3 and 4 as previously described (27). To excise Baf45a exclusively in early craniofacial mesenchyme, Baf45af/f mice were bred with paired related homeobox 1 promoter Cre mice (Prrx1-Cre transgenic mice, Stock No: 005584, from The Jackson laboratory) (33). The subsequent Baf45awt/f; Prrx1-Cre mice were bred to obtain WT (Baf45awt/wt, Prrx1-cre), Baf45aKO (Baf45a f/f , Prrx1-cre) male (n=3) and female(n-3) mice. Mice were genotyped by polymerase chain reaction (PCR) on tail genomic DNA with Accuris™ Mammalian Genotyping Kit (Accuris, Edison, NJ, USA). The Baf45a wild‐type and floxed alleles and the Cre transgene were detected by PCR using the specific primers listed in the Supplementary Table S1 . Eight-week-old mice were used to study tooth phenotype.
Animal Care and Use Statement
Animals were used for this research in accordance with guidelines implemented by the Institutional Animal Care and Use Committee and Animal Resource Program at UAB. Mice were housed in a facility with 12/12 lighting in standard rodent cages with free access to rodent chow and water. Pups were euthanized by decapitation prior to calvarial isolation at post-natal day 2-5.
Cell Culture
The culture and maintenance of osteo and odontoblast cells were carried out as described previously (44, 46–49). HEK-293T cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle Medium (Corning Cellgro, Manassas, VA, USA); MC3T3-E1 cells and primary rat calvarial osteoblasts were cultured in Minimum Essential Media – Alpha (α-MEM; Hyclone, Cytiva, Marlborough, MA, USA); OD-21 cells (ATCC, Manassas, VA) were cultured in DMEM. All media was supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin. Osteogenic and odontogenic differentiation was induced with the addition of 50 µg/mL ascorbic acid and 3 – 5 mM β-glycerophosphate (βGP; Sigma Aldrich, St. Louis, MO, USA). Cells were refreshed with fresh differentiation media at every 48 hrs.
Rationale for using various in vitro cellular models: This study utilized multiple in vitro cellular models of osteoblast and odontoblast differentiation: murine bone marrow stromal cells (BMSCs); murine preosteoblast MC3T3-E1 cells; rat OD-21 odontoblast differentiation, and primary calvarial preosteoblast differentiation. The primary goal of our study is to present evidence of the role of BAF45 homologs in cell-type-specific differences in their expression and chromatin remodeling potentials across the mineralizing cells of different tissue origins. We assume that having different cell lines will help to complement experimental data and add additional layers of experimental evidence. The nuanced differences between cell lines also yield new insights in addition to strengthening our overall findings. BMSCs and OD-21 cells (dental pulp stem cells) have shown remarkable similarities in their transcriptomic profiles. However, differential gene expression might affect their capacities of osteogenesis. Studying both OD-21 and BMSCs allow for complimentary analysis while still recognizing critical differences in the odontogenic vs. osteogenic pathways. MC3T3-E1 cells represent preosteoblast cells – a cell line more committed to osteogenesis than BMSCs. Primary calvarial cells likewise represent preosteoblasts but as a primary cell source and have been used as an ideal cellular model for many years. This adds additional confidence to study Baf45a’s contribution to osteoblastic differentiation and functional activity. Interpreting the difference of cell differentiation stages and kinetic among different cell lines, we validate and authenticate the differentiation stages for MC3T3-E1, mouse calvarial osteoblasts, and BMSCs and OD-21 cells by analyzing gene markers for proliferation, differentiation, and osteoblastic activity, mineralization potential, and osteoclast activity using ALP staining. The stages are indicated as proliferation (day 0-3), matrix maturation (day 10-14), and mineralization (day 15-20).
RNA Isolation & RT-qPCR
TRIzol reagent extraction (Invitrogen, NY, USA) followed by DNAse1-treatment was used to isolate total RNA according to the manufacturer protocol from MC3T3-E1 cells, mouse calvarial osteoblasts, and OD-21 cells during osteogenic differentiation. RNA from short hairpin RNA transfected (Control shRNA and Baf45a-shRNA) odontoblasts was extracted by Direct-zol RNA kits (Zymo Research, CA, USA). RNA quantity and quality were measured using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Boston, MA, USA) and samples diluted to the same RNA concentration prior to cDNA synthesis using the Primescript RT Reagent Kit (TaKaRa, Mountain View, CA, USA). Luna Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA) was utilized for all qPCR reactions according to the manufacturer’s protocol. Gene expression levels were normalized to U6 small RNA or Gapdh mRNA expression. Results were calculated using the ΔΔCT method and expressed as Log2 Fold Change ± SEM. Student’s t-tests where α = 0.05 were performed to compare experimental versus control groups.
Baf45a Overexpression
The full-length Baf45a cDNA (1663 bp cDNA, 497 aa, transcription ID # ENSMUST00000024657.12) with 12 exons was synthesized from Life Technologies Corporation (Carlsbad, CA) and cloned in pCDH-EF1-MCS-T2A-GFP vector (https://www.systembio.com/pcdh-ef1-mcs-t2a-gfp-cloning-and-expression-lentivector). Control empty vector and Baf45a overexpression constructs at concentrations of 1-2 ug/well of a 6-well plate were transfected into MC3T3-E1 cells at ∼50% confluence using FuGENE 6 transfection reagent (Promega Corp.), following the manufacturer’s instructions, and harvested after 72 h for protein and RNA analysis.
Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
The ChIP-seq data was obtained with the gracious permission of authors of “Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis” (50) from GEO (Series GSE76074). Data was uploaded to IGV and qualitatively analyzed using area under curve measurement with Adobe Photoshop. Data was normalized based on gene length and scale and plotted on a three-dimensional axis with Origin Pro 2019.
According to the protocol (50) ChIP-seq was performed on BMSCs undergoing osteogenic differentiation for 0, 7, 14, and 21 days. Formaldehyde was used to crosslink DNA to histone proteins and bound transcription factors before BMSC (5 × 107 MSCs/timepoint) lysis and sonication, which produced protein-bound DNA fragments ranging from 200 to 600 bp. Chromatins were prepared and sheared as previously described (50). Sheared chromatin was then immunoprecipitated with Runx2 antibody (M-70, Santa Cruz), H3K4me (ab8895, Abcam, Cambridge, MA, USA), H3K9me3 (Ab8898, Abcam), H3K9ac (39137, Active Motif), H3K27me3 (07-449, Millipore), H3K27ac (07-360, Millipore), or immunoglobulin G (IgG) (12-370, Millipore). Protein-DNA complexes were purified using Protein-G Dynabeads (Invitrogen). DNA libraries were sequenced on an Illumina Genome Analyzer II or Illumina HiSeq-1500. Base calls and sequence reads were generated by Illumina CASAVA or bcl2fastq software (version 1.8 and 1.8.4, respectively, Illumina). Two independent biological repeats of ChIP-Seq libraries were prepared for each time point. Two input libraries were prepared with sonicated DNA for each time point to compensate for a possible variation of chromatin structure during MSC differentiation. For ChIP qPCR, primer sequences corresponding to genomic regions were generated and evaluated by RT-qPCR on an Applied Biosystems Viia 7 thermocycler (Thermo-Fisher).
Assay for Transposase Accessible Chromatin (ATAC) Sequencing
Baf45a wild type and knockout calvarial cells were assayed for chromatin accessibility using the Assay for Transposase Accessible Chromatin sequencing (ATAC-seq) technique as described by Buenrostro et al. (51). For inducible deletion of Baf45a, the tamoxifen-inducible ubiquitously expressing CAGGCre-ER™ (JAX stock #004682) (36) mouse was bread into the Baf45af/f mouse line. Primary calvarial cells were subsequently isolated and either treated with 1µM 4-OH tamoxifen (72 hours, refreshed media daily) or with an equal volume of ethanol as a mock treatment and differentiated in osteogenic media for 3 and 10 days.
Cells were lysed in ice-cold lysis buffer (10mM Tris-HCl pH 7.4, 10mM NaCl, 3mM MgCl2, and 0.1% Igepal CA-630). Lysates were incubated with Tn5 transposase (Illumina Tagment DNA Enzyme and Buffer Kit, Illumina, San Diego, CA, USA) for 30 min at 37°C, followed by DNA purification using the MinElute PCR Purification kit (Qiagen, Germantown, MD, USA). Transposed DNA was amplified with barcoded Nextera primers (Illumina, San Diego, CA, USA) using the NEBNext High-Fidelity 2X PCR Master Mix (New England Biolabs, Ipswich, MA, USA). The barcoded DNA libraries were sequenced using the Illumina sequencing platform and analyzed by the Genomics Core Lab at the University of Alabama at Birmingham Heflin Center for Genomic Sciences (Birmingham, AL).
MicroCT Analysis
Baf45a was deleted at embryonic day E9.5 using Baf45af/f mice and Prrx1-Cre mice. For micro-CT analysis, 2-month-old male and female Baf45af/f and WT mice were dissected, and the whole head was fixed in 70% ethanol. The bone volume of the whole molar, the outer enamel layer, the inner dentin layer, and the whole mandible was measured using a Scanco 40 µCT. Samples were placed in a 12 mm diameter sample holder and scanned at 12 µm resolution, 55kVp, 145 µA with 200ms integration time. Data were analyzed using the µCT Evaluation Program (v.6.5-2, Scanco Medical). MicroCT was performed at the Small Animal Imaging Shared Facility at the University of Alabama, Birmingham.
Stable Transduction of shRNA in OD-21 Cells
pLKO.1-puro lentiviral backbone (Addgene, MA) was used to clone shRNA targeting exons 3 and 4 of rnoBaf45a mRNA (sequence: rno shRNA Baf45a F: 5’-CCG GAA GCG GAA ATA TCC AGA TTT ACT CGA GTA AAT CTG GAT ATT TCC GCT TTT TTT G-3’ and rno shRNA Baf45a R: 5’-AAT TCA AAA AAA GCG GAA ATA TCC AGA TTT ACT CGA GTA AAT CTG GAT ATT TCC GCT T-3’) to generate Baf45a-specific shRNA. The shRNA sequences are designed from the N-terminal SAY (Supporter of Activation of Yellow) domain encompassing exons 3 and 4. Structurally, this particular domain is not present in the other three BAF homologs. pLKO.1 empty vector was used as control. Viral particles were generated in HEK-293T cells by co-transfection of pLKO.1 shRNA plasmid [(52), Addgene plasmid #10878] with packaging vectors pCMVΔR-8.91 [(53), Addgene plasmid # 12263] and pMD2.G [(54), Addgene plasmid 12259] using polyethylenimine (PEI) (Polysciences). Forty-eight hours post-transfection, viral supernatants were collected, filtered through a 0.45 µm syringe filter, and added to OD-21 cells for spin infection followed by 24 hours incubation before adding fresh medium. The cells were grown on puromycin (2ug/ml) selected media for 72 hours. The gene expression changes due to Baf45a knockdown were studied by RNA-seq analysis.
Library Generation and RNA Sequencing (RNA-Seq) Analysis
To examine the potential gene regulatory function of Baf45a during dentinogenesis, we performed high-throughput RNA sequencing. Briefly, total RNA was isolated from control, and Baf45a shRNA transfected OD21 cells using Direct-zol RNA MiniPrep kit protocol (Cat # R2052, ZYMO Research) and treated with DNase I (1u/ug of RNA) to remove the genomic DNA. Preparation of cDNA library generation and transcriptome sequencing was conducted by Novogene Co., LTD (Beijing, China). A total of 0.5-1 μg of RNA per sample was used as input material. Ribosomal RNA was removed using rRNA Removal Kit or poly(A) capture, and the rRNA-free total RNA was recovered via ethanol precipitation. Total RNA was fragmented to a specific size range for whole transcriptome or mRNA sequencing using RNAse III. Afterwards, sequencing libraries were generated using the rRNA-depleted fragmented RNA using a cDNA Library Prep Kit for Illumina following Novogene mRNA-seq services and recommendations. First-strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H minus). Subsequently, second-strand cDNA synthesis was performed using DNA polymerase I and RNase H. The size-selected, adaptor-ligated cDNA was amplified with High-Fidelity DNA polymerase, Universal PCR primers, and multiplex oligo adapters. The library quality was assessed on the Bioanalyzer system to perform sequencing. The correct adapters to capture the directionality of RNA in the resulting cDNA is important to maintain the RNA strand information and reduce the gene overlaps. RNA-seq Pair-end reads were aligned to the mouse genome (mm10). A reference genome index was built using Bowtie v.2.0.6 (55). Raw count data per gene was calculated using HTSeq (56). The raw count matrix was then used by DESeq2 (57) to quantify gene expression levels as normalized counts. Cuffdiff57 was used to detect differentially expressed genes between control shRNA and Baf45a shRNA (58). Transcripts with an adjusted P < 0.05 were considered differentially expressed. The differential expression analysis was performed by edgeR software and shown as a volcano plot. Gene Ontology (GO) functional categories were annotated and top significantly differentially expressed genes (DEG) enriched in apoptosis, DNA replication, and cell cycle were extracted and presented in a heatmap using Heatmap2 R package (59).
Results
Increased Histone H3K9 and H3K27 Acetylation and RUNX2 Occupancy Indicate Baf45a and Baf45d Promoters Are Highly Accessible During Induction of BMSCs to Osteoblast Differentiation
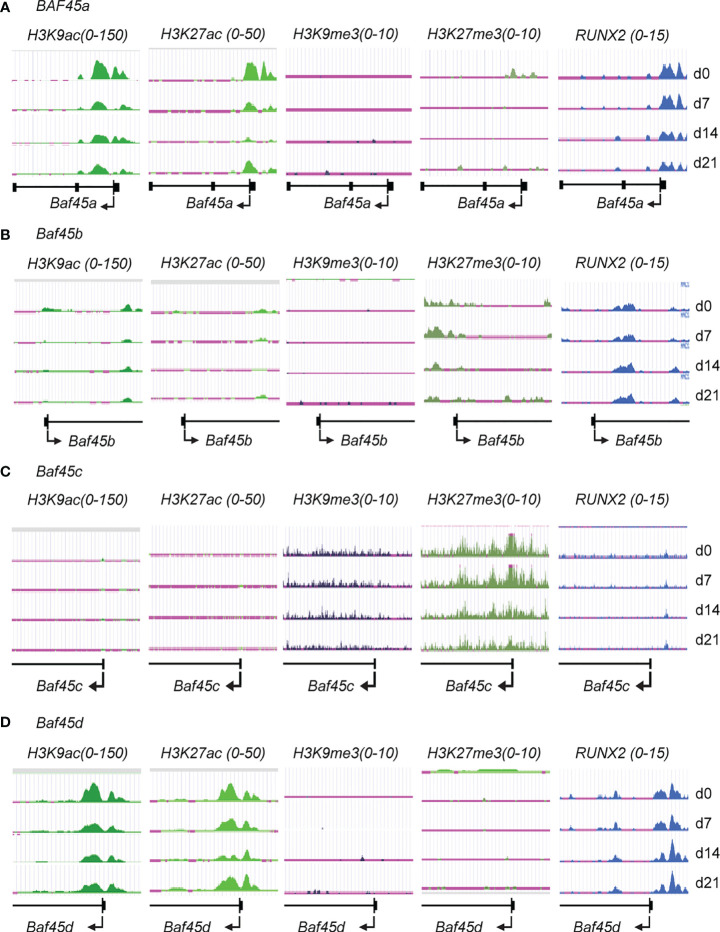
Studies previously suggested that Baf45a associated chromatin remodeling facilitates neurogenic differentiation (28). We currently identified that Baf45a remodeled chromatin implicate the transcriptional activation of osteoblast-specific genes (37). Lately, we found that osteoblast-specific deletion of Baf45a reduced bone formation in vivo. Originally, Baf45a was discovered as a target of miR-23a, and the knockdown of the miR-23a cluster has a high bone mass phenotype (manuscript in preparation/unpublished). Furthermore, we found that the bone tissue-specific RUNX2-BAF45A-EZH2 molecular epigenetic axis regulates osteogenesis in vivo (37, 38). These are the scientific premises behind choosing Baf45a to study further. However, the process of chromatin remodeling events and transcriptional activation mediated by Baf45a in osteoblast or odontoblast maturation and differentiation are not well-defined. We examined ChIP-seq data from primary bone marrow stromal cells (BMSCs) over a differentiation time course (50). BMSCs ChIP-seq analysis demonstrated that the genomic region around Baf45a and Baf45d gene promoters is active during osteogenic differentiation. High levels of the active chromatin marks H3K9ac and H3K27ac localized to the promoter regions of the Baf45a and Baf45d genes when compared with Baf45b and Baf45c ( Figures 1A, D vs. 1B, C ). Because of the presence of the active chromatin modification marks, it is apparent that there would be a low or undetectable level of the repressive chromatin marks, including H3K27me3 and H3K9me3 around these genes. Furthermore, BMSCs ChIP-seq analysis with anti H3K9me3 and H3K27me3 showed very low occupancy levels at the promoter regions of Baf45a and Baf45d compared to Baf45b and Baf45c ( Figures 1A, D vs. 1B, C ). Of note, Baf45b displayed moderate levels of both active (H3K9ac) and repressive (H3K27me3) chromatin marks in the gene body ( Figure 1B ), typically characteristic of a poised transcriptional state. The Baf45c promoter region displayed the highest and most pronounced marks of H3K27me3 modification and lacked any activating chromatin marks ( Figure 1C ). Evaluation of the RUNX2 binding on Baf45 homolog promoters revealed a significant binding of RUNX2 to the Baf45a and the Baf45d promoters ( Figures 1A, D ). Conversely, little to no RUNX2 binding was observed on Baf45b and Baf45c promoters ( Figures 1B, C ).
Figure 1.
Histone H3 modifications and RUNX2 binding on the promoter regions indicate the accessibility of Baf45 homolog promoters during Bone Marrow Stromal Cells (BMSCs) differentiation. Comparison of ChIP-Seq data acquired by Illumina sequencing. BMSCs were isolated from the bone marrow of mice, grown, and differentiated under osteogenic differentiation conditions, harvested at days 0, 7, 14, and 21, and ChIP-sequencing was performed. H3K27ac, H3K27me3, H3K9ac, H3K9me3, and RUNX2 antibodies were used to immunoprecipitate protein-DNA interactome (A) Genomic tracks display ChIP-seq data showing enrichment profiles for H3K27ac, H3K27me3, H3K9ac, H3K9me3, and RUNX2 at (A) Baf45a, (B) Baf45b, (C) Baf45c and (D) Baf45d gene promoters. Quantitative peaks representing active chromatin marks H3K9ac and H3K27ac and peaks representing repressive chromatin marks H3K9me3 and H3K27me3.
When taken together, our results showed that increased Baf45a and Baf45d chromatin accessibility are implicated for BMSC’s commitment to the osteoblast lineage and reinforce osteogenic differentiation.
Baf45a Promotes Osteogenic Differentiation
Baf45a is a subunit of the PBAF complex, while Baf45d is functionally complexed to the canonical BAF (cBAF) complexes (21, 60). Our current study will specifically focus on Baf45a to identify its function linked to osteo and dentinogenesis.
To study the function of Baf45a in osteogenic differentiation, we first analyzed the expression of Baf45a during MC3T3-E1 osteoblast differentiation. Additionally, we overexpressed Baf45a in MC3T3-E1 pre-osteoblasts to ascertain its effect on the osteoblast marker genes.
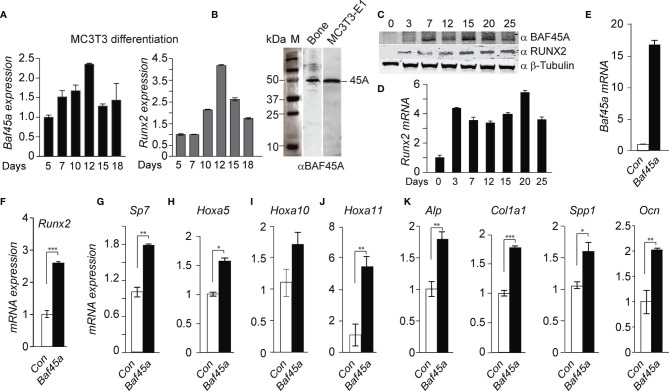
The Baf45a mRNA analysis by RT-qPCR demonstrated a temporal pattern of Baf45a expression during osteoblast differentiation, with a sharp increase in early osteoblast’s growth (day 5-7), followed by peak expression levels in mature osteoblasts (day 10-12) and decreased expression during the mineralization stage (day 15-18) ( Figure 2A ). Runx2 is a well-characterized bone-tissue-specific transcription factor. As a marker, the temporal increase of Runx2 during osteoblast matrix maturation (day 10-12) and mineralization (day 15-20) supports the osteoblast differentiation program ( Figure 2A , right panel). This temporal pattern of increased expression indicated that Baf45a remodeling is required to activate the osteoblast differentiation program.
Figure 2.
Baf45a expression levels induce gene expression profiles for osteoblast differentiation. (A) Relative expression levels of Baf45a (left panel) and Runx2 (right panel) were obtained by real-time RT-qPCR using DNase I treated total RNA isolated from murine preosteoblast MC3T3-E1 cells induced to differentiate for 18 days. Gapdh mRNA profile was used as an experimental control. (B) Representative Western blot analysis with anti-BAF45A antibody of lysates from the femur and MC3T3-E1 cells. Molecular weight markers were indicated in kDa. (C) Relative protein levels of BAF45A (upper panel) and RUNX2 (middle panel) were obtained by Western blot analysis using total cell lysate isolated from murine calvarial osteoblast cells induced to differentiate for 25 days. Beta Tubulin protein profile (lower panel) was used as loading control. (D) Relative expression levels of Runx2 were obtained by real-time RT-qPCR using DNase I treated total RNA isolated from murine calvarial cells induced to differentiate for 25 days. (E) Overexpression of Baf45a in MC3T3-E1 osteoblast cells normalized to Gapdh after 72 hours by RT-qPCR. (F–J) Expressions of Runx2, Sp7 (osterix) osterix, Hoxa5, Hoxa10, and Hoxa11 were assessed by RT-qPCR and normalized to Gapdh. (J) Early markers Alp and Col1A1 and (K) late markers Spp1(Opn), and Ocn were assayed. Statistical significance was determined by Student’s t-test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 versus matched control). Gapdh expression was used as the control.
Analysis of protein levels of BAF45A in long bone and osteoblast cells, using anti-BAF45A antibody ( Supplementary Table S3 , NovusBio: Cat# NBP2-19795; Lot# 42837) indicated a specific 45 kDa BAF45A protein in both mouse bone and MC3T3-E1 osteoblast cellular lysate ( Figure 2B ). Further western verified a similar temporal pattern of BAF45A protein during calvaria-derived osteoblast differentiation, with an increase in protein levels in mature osteoblasts (day 7-15) and maintained the level during the mineralization stage (day 20-25) ( Figure 2C ). We also immunodetected additional BAF45A species in our western analysis on osteoblast cells. In 2017, Tatarskiy et al. reported that PHF10/BAF45A, expressed as four ubiquitous isoforms, has different domain structures, and two of them (PHF10-S isoforms) lack C-terminal PHD domains, which enables their phosphorylation by CK-1, and is degraded by β-TrCP (61). Further studies are needed to uncover the role of these BAF45A isoforms in chromatin remodeling and tissue-specific gene expression. The protein ( Figure 2C , middle panel) and mRNA ( Figure 2D ) profiles of the Runx2 gene were used as a marker of mouse calvarial osteoblast differentiation time course. With the observed increase in expression of Baf45a during osteoblast differentiation, we hypothesized that Baf45a overexpression would promote osteoblast maturation by activating bone marker genes through their chromatin remodeling. Cells were transfected with control (empty vector) and Baf45a overexpression construct and then allowed to differentiate for 72hrs. Compared with the control, Baf45a overexpression substantially increased the expression of Baf45a-encoded mRNA ( Figure 2E ). Analysis of osteogenic transcription factors revealed that Baf45a noticeably induced Runx2, Sp7, Hoxa5, Hoxa10, and Hoxa11 expression from 1.5 to 6-fold to promote osteoblast differentiation ( Figures 2F–J ). Among bone-specific markers, Baf45a increased early markers, including alkaline phosphatase (Alp) and Col1a1 (1.5-2.0-fold) ( Figure 2K , left panel), as well as Spp1 (osteopontin) and Ocn (Bglap), the late markers of osteoblast mineralization ( Figure 2K , right panel). When taken together, these results revealed that Baf45a remodels osteoblast chromatin and promotes osteoblast differentiation through activating genes essential for osteoblast maturation and mineralization.
Baf45a Subunit Is Necessary for Odontoblast Differentiation
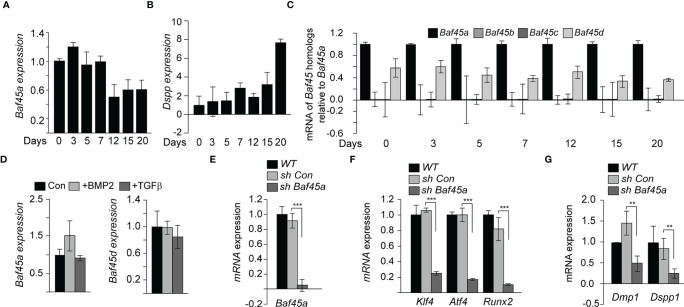
Odontoblasts, the primary cell type in the tooth pulp, similarly deposit minerals to form detin as osteoblasts synthesize cortical and trabecular bone. We hypothesized that Baf45a mediated chromatin accessibility is critical for tooth-specific gene expression during odontoblast differentiation. To identify the role of Baf45a for odontoblast function, we studied rat odontoblast OD-21 cells to investigate differentiation. Baf45a mRNA was expressed at high levels during early differentiation and reduced by approximately 50% at day 12, corresponding to the mineral maturation period ( Figure 3A ). Dentin Sialophosphoprotein (Dspp) is a well-characterized dentin extracellular matrix phosphoprotein. The temporal increase of Dspp during OD-21 maturation and mineralization was used as a marker for odontoblast differentiation ( Figure 3B ). Relative to Baf45a expression, Baf45b and Baf45c were negligible ( Figure 3C ). Baf45d was also expressed at high levels during early odontoblast differentiation similar to Baf45a and consistently maintained later during mineralization stage ( Figure 3B ). These results indicate that like osteoblasts, Baf45a and Baf45d are preferred subunits of the PBAF and BAF complex in odontoblast cells. BMP2 treatment of OD-21 cells modestly (non-significantly) increased Baf45a expression ( Figure 3C , left panel); however, no effect with BMP2 treatment in the expression of Baf45d was observed ( Figure 3D , left and right panel). Transforming growth factor-β (TGF-β) is a secreted factor that modulates osteoprogenitors differentiation (62, 63). In odontoblasts, TGF-β1 signaling has a pivotal role in the transcriptional regulation of dentin sialophosphoprotein (DSPP) and dentin matrix protein 1 (DMP1) (64). It is unclear whether TGF-β directly modulates chromatin remodeler BAF45A and OD-21 induction for differentiation; therefore, we treated the cells with TGFβ to examine the effect of TGFβ on Baf45a expression. Treatment of OD-21 cells with TGF-β did not reduce expression of Baf45a nor Baf45d ( Figure 3D , left and right panels).
Figure 3.
Baf45a functions promote odontoblast differentiation. (A) Relative expression levels of Baf45a were obtained by real-time RT-qPCR using total RNA isolated from murine odontoblast OD-21 cells induced to differentiate for 20 days. β-Actin cDNA was used as a loading control. (B) Real-time RT-qPCR analysis for the relative expression levels of Dspp using total RNA isolated from murine odontoblast OD-21 cells differentiated for 20 days. (C) Representative of mRNA analysis in triplicate (n=3) of Baf45 homologs in OD-21 cells induced to differentiate for 20 days. (D) Baf45a and Baf45d responsiveness to BMP2 and TGFβ in OD-21 odontoblast cells. RT-qPCR analysis from OD-21 cells treated with BMP2 (100 ng/ml) and TGFβ (10 nM) at 80-90% confluence of culture and harvested 24 h post-treatment. (E) OD-21 cells were treated without shRNA (WT), control shRNA (con sh), and shRNA specific to Baf45a (sh Baf45a). After 72 hrs. post-treatment, RT-qPCR was performed on the RNA isolated from the harvested cells to assay the expression of Baf45a. Gapdh expression was used to normalize the relative gene expression. (F) Effect of Baf45a shRNA was assayed on the expression of Atf4, Klf4, and Runx2. (G) odontoblast differentiation markers Dmp1 and Dspp1 were assayed. Gapdh expression was assessed to normalize the relative expression. Statistical significance was determined by Student’s t-test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 versus matched control).
To further investigate the role of Baf45a in odontoblasts, a short hairpin RNA (shRNA) was used to reduce the expression of Baf45a by nearly 90% ( Figure 3E ). We then hypothesized that Baf45a remodeling facilitates the expression of transcription factors Klf4, Atf4, and Runx2, affecting the expression of downstream odontoblast markers. With the shRNA mediated knockdown of Baf45a, Klf4, Atf4, and Runx2’s expression decreased significantly ( Figure 3F ). Analysis of odontogenic marker genes required for proper dentinogenesis showed decreased levels of Dmp1 and Dspp1 with loss of Baf45a ( Figure 3G ). This data suggests Baf45a is an essential subunit linked to odontogenic transcriptional induction and differentiation. Hence, when taken together, these findings suggest that similar to osteoblasts, Baf45a is important for odontoblast cells.
BAF45A Led PBAF Complex Orchestrates Osteogenic Differentiation
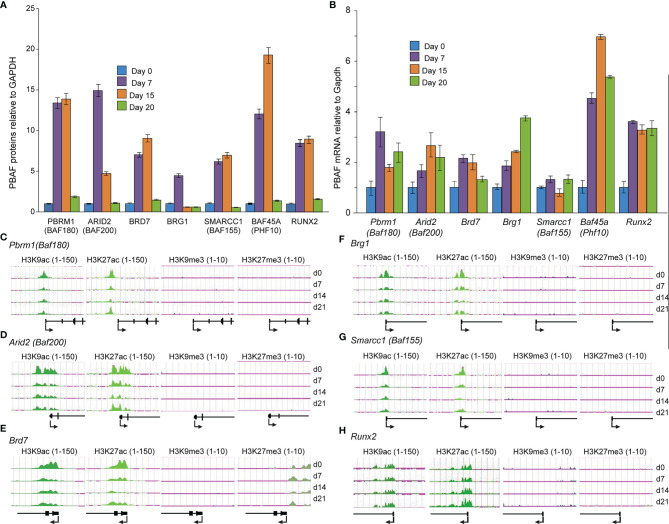
The functions and roles of PBAF in osteoblast and odontoblast differentiation remain largely undefined. In light of uncovering that BAF45A was a key factor of the PBAF complex in osteoblasts, we hypothesized that the BAF45A, including the members of the PBAF complex, are vital chromatin regulatory complex for determining chromatin accessibility for mineralized cells commitment and differentiation. To address our hypothesis, we first profiled the protein levels of the PBAF subunits during primary calvarial preosteoblast differentiation to address our hypothesis. We found a temporal protein profile during osteoblast differentiation, with moderate protein expression for all members of PBAF subunits between proliferation and early differentiation (day 0-5). This pattern was followed by significantly higher proteins starting at matrix maturation to early mineralization (day 7-15) and then decreased expression at the mineralization stage (day 20) ( Figure 4A ). Interestingly, the increase of BAF45A, PBRM1, and BAF200 proteins were noticeably higher, ranging from 5 to 17-fold and RUNX2 with 5-7-fold. When taken together, the expression of PBAF subunits during differentiation indicates a requirement of PBAF mediated remodeling for determined osteoblast-specific chromatin accessibility to facilitate the osteogenesis program. A temporal pattern of PBAF subunits and Runx2 mRNA expression was observed during murine calvarial osteoblast differentiation, with significantly higher levels for all PBAF subunits and Runx2 between early differentiation and matrix maturation stages (day 7-15), followed by lower (Pbrm1, Baf200, Brd7 and Baf45a), steady (Baf155 and Runx2) and higher (Brg1) mRNA levels in the mineralization (day 15-20) stage ( Figure 4B ). Taken together, these findings indicate that the chromatin remodeling event associated with PBAF functions may induce and maintain osteoblast/odontoblast differentiation. Next, we compared the osteoblast’s PBAF protein ( Figure 4A ) and mRNA profiles ( Figure 4B ) during differentiation to the chromatin modifications on PBAF subunit promoters ( Figures 4C–H ) in BMSCs to understand the epigenetic clues for the differentiation of two mineralized cells. BMSCs ChIP-seq analysis demonstrated that the chromatin modification landscape around PBAF subunits promoters consists of Pbrm1 (Baf180), Arid2 (Baf200), Brd7, Smarca4 (Brg1), and Smarcc1 (Baf155) displayed high levels of active H3K9ac and H3K27ac chromatin modifications ( Figures 4C–H left two panels). Contrary to that, ChIP-seq analysis with anti H3K9me3 and H3K27me3 ChIP on the PBAF proteins mentioned above promoters showed low and undetectable levels ( Figures 4C–G , right two panels). During differentiation, as expected, we found a reliable increase in H3K9 and H3K27 acetylation on the promoter region of the Runx2 ( Figure 4G ). Hence, we posit that higher Runx2 expression and RUNX2 binding to Baf45a promoter ( Figure 1D ) may promote a regulatory loop with PBAF subunits set the foundation for accessibility of chromatin and may activate transcription.
Figure 4.
PBAF complex is critical for early differentiation and matrix maturation. (A) Quantitative Western blot analysis of PBAF factors from primary rat calvarial osteoblast cells induced to differentiate for 20 days. (B) Relative mRNA levels of PBAF complex, obtained by real-time RT-qPCR using DNase I treated total RNA isolated from murine calvarial osteoblast cells induced to differentiate for 20 days. (C-H) Histone H3K9 and H3K27 modifications on the promoter regions of PBAF complex members and Runx2 during Bone Marrow Stromal Cells (BMSCs) differentiation. Illumina ChIP-seq genomic tracks at (C) Pbrm1 (Baf180), (D) Arid2 (Baf200), (E) Brd7, (F) Brg1, (G) Smarcc1 (Baf155) and (H) Runx2 promoter regions. Quantitative peaks representing active chromatin marks H3K9ac and H3K27ac, as well as peaks representing repressive chromatin marks H3K9me3 and H3K27me3.
BAF45A-PBAF Dependent Chromatin Accessibility Is Critical for the Expression of Mineralized Tissue Marker Genes
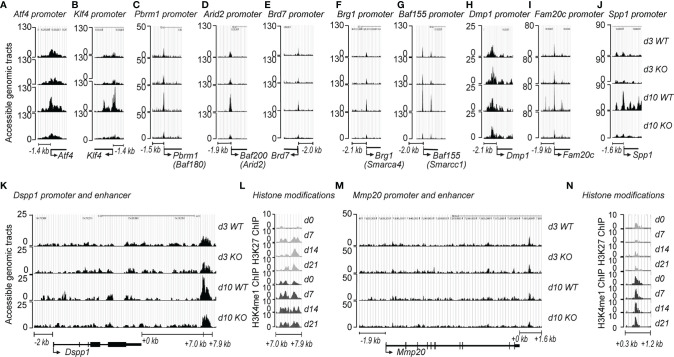
The binding of transcription factors (TFs) and RNA polymerase machinery to the promoter and enhancer regions of DNA is critical for regulating gene expression. BAF45A constitutes an essential subunit of the PBAF complex that promotes determined genomic accessibility for TFs and RNA Pol II for tissue specificity. To study how BAF45A mediated chromatin remodeling changes contribute to accessibility, we assessed chromatin accessibility in mouse calvarial osteoblasts (MOBs) derived from wild-type (WT) and Baf45a deleted mice during differentiation using ATAC-seq assays. We found that chromatin accessibility (promoter region surrounding -1.4 kb upstream) of the three crucial osteogenic transcription factors, Runx2 (data not shown), Klf4, and Atf4, were significantly reduced in Baf45a knockout cells when compared to WT cells at day 10 ( Figures 5A, B ) (65–67). Little or modest change was observed on day 3. We assume that at day 3, the promoter accessibilities for differentiation are limited. Still, some cells would reflect accessibility for transcription initiation for the start of differentiation. Therefore, the BAF45A remodeling effect is modest. Therefore, this trend indicates that Baf45a deletion decreased promoter accessibility of these vital transcription factors required for induction and maturation of osteoblasts. The chromatin remodeling contributed by the BAF/PBAF complex in promoting osteoblast differentiation to activate bone tissue-specific gene expression is not clearly defined (68–70). With the idea that BAF45A has a strong association with the rest of the PBAF subunits for increased accessible chromatin, we next sought to test whether Baf45a deletion would cause subsequent changes in chromatin accessibility of PBAF complex members. We observed that chromatin accessibility of signature PBAF subunits, namely Pbrm1, Arid2, Brd7, Brg1, and Baf155 ( Figures 5C–G ), showed non-significant changes at proliferation (day 3) compared to a noticeable high degree of diminished chromatin accessibilities at differentiation (day 10) at the promoter regions (encompassing - 1.4 to -2.1 kb upstream) in Baf45a deleted cells when compared to WT cells. We next assessed the promoter accessibility on osteoblast/odontoblast-specific promoters due to Baf45a deletion. We identified promoter regions for Dmp1 (-2.1 kb upstream), Fam20c (-1.9 kb upstream), and Spp1 (-1.6 kb upstream) are showing significantly diminished accessibility in Baf45a deleted cells compared to WT cells ( Figures 5H–J ). Interestingly, we did not observe any changes in the accessibility of the two essential marker genes, namely Dspp1 and Mmp20, at their promoter regions. Therefore, we hypothesized that the cell-type-specificity of osteoblast/odontoblast cells might be controlled additionally by enhancer-like activity. We localized three distinct enhancer regions with accessibility changes when analyzed using ATAC sequencing of WT and Baf45a knockout cells. The regions located +7.0 to +7.9 kb downstream of Dspp1 and +0.3 to +1.2 kb downstream of Mmp20 genes ( Figures 5K, M ). These enhancers were further characterized using ChIP-sequencing with anti-H3K27ac and anti- H3K4me1 modifications during 21 days of BMSCs differentiation to support our ATAC results. These two enhancer-associated histone modifications were typical to define specific regions as enhancers contributing to gene expression changes. We observed high H3K27ac and H3K4me1 modifications surrounding the regions of Dspp1 (+7-7.9 kb distal) and Mmp20 (+0-1.6Kb distal) ( Figures 5L, N ) during differentiation. Taken together, our findings indicate that the PBAF complex, including BAF45A remodeling, is critical for osteoblast/odontoblast cell-type-specific transcriptional regulatory networks.
Figure 5.
BAF45A-PBAF complex regulates chromatin accessibility and bone-specific gene transcriptional network. Baf45a floxed allele was deleted in primary calvarial osteoblasts via 4-hydroxytamoxifen inducible CAG-Cre genetic background. The cells were then subjected to osteogenic differentiation. Osteoblasts were harvested at day 3 and day 10 of differentiation. ATAC-seq was performed on WT and Baf45a knockout osteoblasts. (A, B) Chromatin accessibility at the gene loci of transcriptional factors Atf4 and Klf4, (C–G) Genomic accessibility profiles of PBAF subunits, and tooth and bone-related genes (H) Dmp1, (I) Fam20c, (J) Spp1, (K) Dspp1, and Mmp20 (M). ChIP-sequencing analysis was performed in BMSCs using the anti-H3K27ac and anti-H3K4me1 antibodies to identify downstream enhancer regions of bone and tooth-related genes (L) Dspp1, (N) Mmp20. “Y-axis” indicates accessible genomic tracts, and the “X” axis denotes the genomic regions.
Deletion of Baf45a in a Subset of Craniofacial Mesenchyme Modestly Reduced the Mineralization of the Tooth and Mandibular Bone
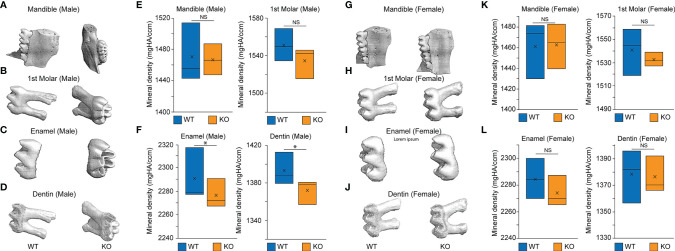
We hypothesized that Baf45a deletion would reduce tooth mineral density. Thus, we used Prx1-Cre (Prrx1Cre, Jackson Cat # 005584) transgenic mice to delete BAf45a in a subset of craniofacial mesenchyme (71). Molars from two-month-old male Baf45a deleted mice (n=3) had a modest reduction of tooth mineralization in the enamel and dentin layers ( Figures 6C, D, F ). However, a significant difference in the mandibular bone density of these same mice was not apparent ( Figures 6A, B, E ). In 2-month-old Baf45a deleted female mice, no difference in the mineral density in either the mandible or the molars ( Figures 6G–L ) were observed. Furthermore, a compensatory regulation from other BAF45 homologs might play a role in this regulation. Evidence of homolog switching comes from previous studies showing that the expression of BAF45A and BAF45D switch to BAF45B and BAF45C during development in early neurogenesis (28, 72). Thus, further mechanistic study of BAF45 homolog gene expression from dental pulp cells of Prrx1-Cre; Baf45af/f mice would help us understand the compensatory mechanism.
Figure 6.
MicroCT of the mandible, first molar, enamel, and dentin in 2-month-old Baf45a conditional knockout mice compared to wild type. Baf45af/f was deleted at embryonic day E9.5 mating with Prrx1-Cre mice. (A–F) Homozygous Baf45af/f (n = 3) and age-matched mice of 2 months were analyzed. Mice were euthanized, and teeth were extracted and stored in ethanol. The bone volume of the whole molar, the outer enamel layer, the inner dentin layer, and the whole mandible from WT and Baf45a KO male mice, (G–L) Bone volume of the whole molar, the outer enamel layer, the inner dentin layer, and the whole mandible from WT and Baf45a KO female mice. Males (n=3) and females (n=3) at 2-months littermates. Significance was determined using Student’s one-tailed t-test: *p < 0.05; NS (non significant). Mineral density (mgHA/ccm).
shRNA Knockdown of Baf45a In Vitro Regulates Odontoblast Growth, DNA Repair, and Apoptosis
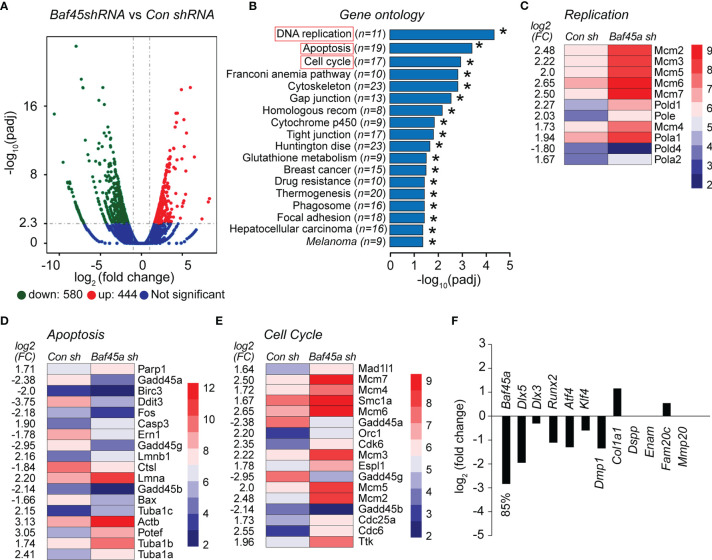
To evaluate the effect of Baf45a in odontoblast cells, we transduced OD-21 cells using lentiviral Baf45a shRNA and performed RNA-seq after 72hrs. Volcan Plot shows changes in mRNA expression between control (non-specific shRNA) and Baf45a shRNA ( Figure 7A ). Next, we compared the relative gene expression changes between control and Baf45a-specific shRNA transduced OD-21 cells using the RNA sequencing platform. Based on log2 fold change (FC), 444 upregulated (red dots) and 580 (green dots) downregulated mRNAs. Gene ontology (GO) functional annotations of significantly changed genes, using Database for Annotation Visualization and Integrated Discovery (DAVID) bioinformatics demonstrated the enrichment of pathways associated mainly with DNA replication, apoptosis, and cell cycle ( Figure 7B ). Amongst these genes, heatmap analysis found the highly conserved mini-chromosome maintenance proteins (MCM), component of the pre-replication complex (Mcm2-7), and DNA Polymerase subunits α1 and α2, δ1, and ϵ were induced, but Polδ4 was reduced in Baf45a depleted OD-21 cells ( Figure 7C ).
Figure 7.
The shRNA mediated knockdown of Baf45a in odontoblast OD-21 cells affect growth, DNA repair, and apoptosis. (A) Volcano plot comparison of differential gene expression between control shRNA (non-specific) and Baf45a-specific shRNA. Red dots highlight upregulated, and green dots highlight downregulated differentially expressed genes with threshold FDR < 0.01 and Log2 Fold Change [≥ 1]. The blue zone indicates the number of transcripts that do not show significant differential expression. (B) Functional annotation based on Gene Ontology (GO) categorization. The horizontal bar plot represents differentially enriched genes linked to biological processes (FDR < 0.05) identified in control shRNA vs. Baf45a specific shRNA. *P < 0.05. (C–E). Heatmap of differentially expressed genes belongs to DNA replication, apoptosis, and cell cycle. (F) The relative changes in the expression of osteoblast and odontoblast-specific transcription factors and markers genes due to Baf45a knockdown.
Further heatmap analysis of differentially expressed genes identified 18 factors associated with apoptosis and 17 factors linked to the cell cycle that were significantly altered in Baf45a deleted RNA compared with control shRNA ( Figures 7D, E ). Interestingly, we found that Gadd45 homologs including 45a, 45b, and 45g were significantly downregulated. These factors are associated with vital cellular processes, including growth control, maintenance of genomic stability, DNA repair, cell cycle control, and apoptosis (73, 74). We next hypothesized that Baf45a knockdown would affect early odontoblast differentiation. Therefore, we analyzed RNA-seq data for changes in the expression of odontoblast-specific transcription factors that induce differentiation and odontoblast differentiation marker genes. A significant knockdown of Baf45a mRNA decreased transcription factors Dlx5, Dlx3, Runx2, Atf4, and Klf4. Tooth matrix gene Dmp1 has been reduced; however, we did not observe any significant changes in the expression of Col1a1, Dspp1, Enam, Fam20c, and Mmp20 resulting from Baf45a depletion ( Figure 7F ). Taken together, our data support that BAF45A promotes transcriptional activation in odontoblasts during differentiation, as we observed in osteoblasts.
Discussion
Mineralized tissue phenotypes result from osteoblast or odontoblast cellular functions, controlled by bone and tooth-specific gene expression. Persistent upstream signals followed by regulatory interactions of chromatin remodelers, mediators, and insulators with aligned histone modifications mark in the promoter and enhancer DNA lead to the entry of transcriptional machinery. Most genetic and biochemical evidence indicates that BAF chromatin remodeling orchestrates the predetermined genome accessibility to control transcriptional gene activity in a tissue-specific manner (5–7, 9, 14–16). It is also evident that BAF remodelers’ function disruption resulted in inappropriate gene expression (17). The SWI/SNF chromatin remodeling complexes in mammals exist in three different forms: canonical BAF (cBAF), Polybromo BAF (PBAF), and non-canonical BAF (ncBAF).
Additionally, the PBAF complex is similar in function to the canonical BAF (cBAF) complexes with a few exceptions, one of which is incorporating BAF45A as a subunit in place of BAF45D. We focused our attention on PBAF from the perspective of BAF45A activity in mineralized tissue for specificity. Understanding the functions of chromatin remodeling complexes and the mechanisms underpinning their roles in bone and tooth formation are not defined. Here we studied the role of BAF45A, an essential subunit of the PBAF remodeling complex, in osteoblast and odontoblast growth, maturation, and differentiation.
BAF45A and Osteoblast Chromatin Regulation
This study provides evidence that the function of BAF45A guided remodeling is vital to increase the chromatin potential for bone-tissue-specific gene expression. BAF45A, a member of the PBAF remodeling complex, was highly transcriptionally active, as shown by the deposition of H3K9 and H3K27 acetylation on the promoter. This deposition is indicating that BAF45A chromatin remodeling activity was necessary for BMSCs induction and osteogenic differentiation. Furthermore, increased RUNX2 occupancy with higher histone H3K9 and H3K27 acetylation in Baf45a promoter at days 0 to 7 demonstrated that osteoblast-specific RUNX2 promotes Baf45a transcription and hence mediates the bone-tissue-specific chromatin remodeling to induce and promote BMSCs for osteogenic differentiation. Overall, these findings indicate a collaborative crosstalk between BAF45A, PBAF complex members, and the transcription factor RUNX2 may exist to program the chromatin accessibility and transcriptional activation to initiate transcription of bone-specific genes necessary for bone formation. The relatively high expression of Baf45a up to osteoblast maturation (day 12) compared with the lower expression at mineralization (day 15-18) indicates that BAF45A mediated chromatin accessibility is essential to induce and accelerate osteoblast differentiation. BAF45A protein profile indicated a substantial amount of protein present in osteoblasts and bone tissue. Overexpression of Baf45a promotes the differentiation program of osteoblasts. Forced expression of Baf45a activates homeobox and tissue-specific transcription factors, early and late bone marker genes, and secreted phosphoprotein 1 necessary for bone formation. Thus, our research indicates that the BAF45A remodeled chromatins of transcription factors and marker genes in osteoblast cells may promote osteoblast transcriptional program at stages of differentiation. PBAF is a multi-subunit SWI/SNF chromatin remodeling complex, essential for cell proliferation and differentiation by directing eukaryotic gene transcription. In primary calvarial osteoblasts, we found that the increase of BAF45A, PBRM1, BAF200, BRD7, BRG1, and BAF155 proteins was noticeably higher with RUNX2 protein. This expression of PBAF proteins during differentiation indicates a requirement of PBAF mediated remodeling for determined osteoblast-specific chromatin accessibility. Further chromatin modification profiles indicate that BAF45A may act as a high-affinity H3 acetylation reader for histone H3 acetylation and, together with RUNX2, generates an epigenetic code for gene expression in osteoblast cell function and activity (75). Chromatin accessibility is vital for transcriptional machinery to the promoter and enhancer regions. BAF45A constitutes an essential subunit of the PBAF complex that promotes genomic accessibility for tissue-specificity, including bone and tooth. In Baf45a deleted mouse calvarial osteoblasts, where chromatin accessibility within 2kb upstream of osteoblast-specific transcription factors, marker genes, and PBAF factors were significantly diminished. Interestingly, we found that BAF45A deficiency is also reduced the accessibility of possible osteoblast-specific enhancers that may contribute to gene expression changes. These findings indicate that the BAF45A led PBAF remodeling forms accessible chromatins that are important to establish and maintain osteoblast cell-type and tissue-specificity.
BAF45A and Odontoblast Chromatin Regulation
Our results in another mineralized cell, OD-21, showed BAF45A remodeled chromatin accessibility is critical for tooth-specific gene expression during odontoblast differentiation. We evidence that BAF45A and BAF45D are preferred subunits of the PBAF and BAF complex in odontoblast cells. BMP2 induction modestly increased Baf45a, but no effect was seen on Baf45d expression; interestingly, TGFβ signaling neither inhibits Baf45a expression nor Baf45d.
Additionally, knockdown of Baf45a significantly decreased mRNA of Klf4, Atf4, and Runx2, which are essentially required for activating tooth-specific genes transcription. Baf45a knockdown noticeably decreased Dmp1 and Dspp1 expression, critical for proper mineralization of dentin matrix and dentin. Hence, our findings suggest that, like osteoblasts, BAF45A mediates remodeling of odontoblast signature gene promoters directly or might be through the reduced transcriptional potential of RUNX2, ATF4, and KLF4 for odontoblast cell function. Hence, we believe that the regulation of tissue-specific mineralization via PBAF across these mineralized cells is identical. Baf45a deletion by Prrx1-Cre, at 2-months, showed subtle tooth abnormalities in males; however, in female mice, we did not observe any significant difference in the mineral density in either the mandible or the molars. Even though we noticed significant changes in gene expression and chromatin accessibility in multiple molecular biological approaches, the outcomes of such changes did not reflect tooth development and formation, particularly in females. One possibility might be due to limited Cre expression observed in female germline (oocytes). Additionally, the Prrx1-Cre effect might be affecting a wide variety of cells, including cells in cranial bone, chondrocranium, coronal suture, and lower jaw skeleton, which may not potentially commit to tooth formation. Similar minor changes were observed in molar dentin when Foster et al. deleted ALPL with Prrx1-Cre driver (76). Furthermore, as previously shown during neurogenesis, a compensatory regulation from other Baf45 homologs might play a role in this regulation (28, 72). Gene ontology bioinformatics demonstrated the enrichment of pathways associated mainly with DNA replication, apoptosis, and the cell cycle. Analysis of differentially expressed genes identified 11, linked to DNA replication, 18 factors associated with apoptosis, and 17 factors related to cell cycle due to Baf45a knockdown. Furthermore, Baf45a shRNA decreased transcription factors Dlx5, Dlx3, Runx2, Atf4, and Klf4. Tooth matrix gene Dmp1 has been reduced; however, we did not observe any significant changes in the expression of Col1a1, Dspp, Enam, Fam20c, and Mmp20 resulting from Baf45a depletion. Hence, we concluded that BAF45A promotes transcriptional activation primarily by remodeling the promoters of genes that transcriptionally induce odontoblasts differentiation.
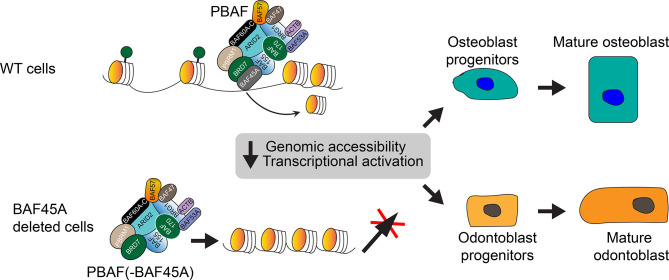
Our mechanistic model ( Figure 8 ) shows that BAF45A evicts nucleosomes, so chromatin can be accessible to initiate transcription by PBAF remodeling. Depletion or ablation of Baf45a in osteoblasts and odontoblasts destabilizes or switches composition, resulting in defective PBAF complex formation, which leads to the inaccessibility of chromatin. Recruitment of such altered PBAF complex inhibits relevant bone or tooth tissue-specific transcriptional programs for gene expression. Thus, inappropriate remodeling preventing the replication, growth, and differentiation of osteo or odonto-progenitor cells. Further compositional assessment and rescue experiments for the members of the PBAF complex will better understand chromatin remodeling in mineralized tissues.
Figure 8.
Working model: BAF45A led PBAF chromatin remodeling activity regulates transcriptional activation in mineralized cells. The model shows that the BAF45A is an integral subunit of the mammalian PBAF chromatin remodeling complex. BAF45a, with the help of PBRM1, ARID2, BRD7, and BAF155, evict nucleosome and promote chromatin accessibility to initiates transcription. Depletion, mutation, or complete knockout of Baf45a in osteoblasts and odontoblasts destabilizes or switches the PBAF complex composition. Recruitment of such defective PBAF complex inhibits relevant bone or tooth tissue-specific transcriptional events and gene expression. Finally, this inappropriate remodeling preventing the maturation of osteo or odonto-progenitor cells. Further biochemical assessment of BAF45A function with other PBAF subunits in vivo will help to understand chromatin remodeling in mineralized tissues better.
Data Availability Statement
The BMSCs ChIP-sequencing data is deposited to the NCBI GEO database with the Accession number GSE76074 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76074).
Ethics Statement
The animal study was reviewed and approved by University of Alabama, IACUC.
Author Contributions
TB designed the experiment, collected data, contributed to data analysis, and drafted the first version of the manuscript. YC performed and designed the experiment, troubleshoot the experiment, and contributed to data analysis. TG performed and analyzed ATAC-seq data and edited the manuscript. MR designed experiments, collected, and analyzed data. CS and BW performed experiments, analyzed data, and helped in manuscript editing. QH supervised the experimental design, reviewed data, supervised reproducibility of analysis, designed, and made figures, and supervised the manuscript writing for submission. All authors contributed to the article and approved the submitted version.
Author Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the School of Dentistry Research Faculty members for assistance with experimentation, critical comments, technical advice, and sharing of reagents and general support, as well as Delores Stacks for editing the manuscript. We gratefully acknowledge the receipt of OD-21 cells from Jacques E. Nör (University of Michigan School of Density, MI). We thankfully acknowledge Gary Stein, Jane Lian and Jonathan Gordon for sharing the BMSCs ChIP-sequencing data. We also thank UAB Heflin Centre for Genomic Sciences and UAB Small Animal Phenotyping and Histomorphometry Cores for RNA sequencing and microCT and histomorphometric analyses. Research reported in this publication was supported by the National Institute of Arthritis And Musculoskeletal And Skin Diseases of the National Institutes of Health under Award Number R01AR069578, National Institute of Dental and Craniofacial Research under award number T90DE022736 and 1F30 DE027280.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.763392/full#supplementary-material
References
- 1. Wang GG, Allis CD, Chi P. Chromatin Remodeling and Cancer, Part II: ATP-Dependent Chromatin Remodeling. Trends Mol Med (2007) 13(9):373–80. doi: 10.1016/j.molmed.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yadav T, Quivy JP. Chromatin Plasticity: A Versatile Landscape That Underlies Cell Fate and Identity. Science (2018) 361(6409):1332–6. doi: 10.1126/science.aat8950 [DOI] [PubMed] [Google Scholar]
- 3. Clapier C, Iwasa J, Cairns B, Peteron CL. Mechanisms of Action and Regulation of ATP-Dependent Chromatin-Remodeling Complexes. Nat Rev Mol Cell Biol (2017) 18:407–22. doi: 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, et al. Purification and Biochemical Heterogeneity of the Mammalian Swi-Snf Complex. EMBO J (1996) 15:5370–82. doi: 10.1002/j.1460-2075.1996.tb00921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and Specialization of Mammalian Swi/Snf Complexes. Genes Dev (1996) 10:2117–30. doi: 10.1101/gad.10.17.2117 [DOI] [PubMed] [Google Scholar]
- 6. Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 Contains a Conserved Domain of the SWI2/SNF2 Family Necessary for Normal Mitotic Growth and Transcription. Nature (1993) 366:170–4. doi: 10.1038/366170a0 [DOI] [PubMed] [Google Scholar]
- 7. Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, et al. Cumulative Haploinsufficiency and Triplosensitivity Drive Aneuploidy Patterns and Shape the Cancer Genome. Cell (2013) 155:948–62. doi: 10.1016/j.cell.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and Bioinformatic Analysis of Mammalian SWI/SNF Complexes Identifies Extensive Roles in Human Malignancy. Nat Genet (2013) 45:592–601. doi: 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kadoch C, Crabtree GR. Mammalian SWI/SNF Chromatin Remodeling Complexes and Cancer: Mechanistic Insights Gained From Human Genomics. Sci Adv (2015) 1(5):e1500447. doi: 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gatchalian J, Malik S, Ho J, Lee D, Kelso T, Shokhirev M, et al. A Non-Canonical BRD9-Containing BAF Chromatin Remodeling Complex Regulates Naive Pluripotency in Mouse Embryonic Stem Cells. Nat Commun (2018) 9:5139. doi: 10.1038/s41467-018-07528-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varga-Weisz P. Chromatin Remodeling: A Collaborative Effort. Nat Struct Mol Biol (2014) 21:14–6. doi: 10.1038/nsmb.2748 [DOI] [PubMed] [Google Scholar]
- 12. Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annu Rev Biochem (2009) 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 13. Lemon B, Inouye C, King D, Tjian R. Selectivity of Chromatin-Remodelling Cofactors for Ligand-Activated Transcription. Nature (2001) 414:924–8. doi: 10.1038/414924a [DOI] [PubMed] [Google Scholar]
- 14. Bachmann C, Nguyen H, Rosenbusch J, Pham L, Rabe T, Patwa M, et al. Mswi/SNF (BAF) Complexes are Indispensable for the Neurogenesis and Development of Embryonic Olfactory Epithelium. PloS Genet (2016) 12(9):e1006274. doi: 10.1371/journal.pgen.1006274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buscarlet M, Krasteva V, Ho L, Simon C, Hébert J, Wilhelm B, et al. Essential Role of BRG, the Atpase Subunit of BAF Chromatin Remodeling Complexes, in Leukemia Maintenance. Blood (2014) 123(11):1720–8. doi: 10.1182/blood-2013-02-483495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chi T. A BAF-Centred View of the Immune System. Nat Rev Immunol (2004) 4(12):965–77. doi: 10.1038/nri1501 [DOI] [PubMed] [Google Scholar]
- 17. Alfert A, Moreno N, Kerl K. The BAF Complex in Development and Disease. Epigenet Chromatin (2019) 12(1):19. doi: 10.1186/s13072-019-0264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, et al. An Embryonic Stem Cell Chromatin Remodeling Complex, Esbaf, is Essential for Embryonic Stem Cell Self-Renewal and Pluripotency. Proc Natl Acad Sci USA (2009) 106(13):5181–6. doi: 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Middeljans E, Wan X, Jansen PW, Sharma V, Stunnenberg HG, Logie C. SS18 Together With Animal-Specific Factors Defines Human BAF-Type SWI/SNF Complexes. PloS One (2012) 7(3):e33834. doi: 10.1371/journal.pone.0033834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brechalov AV, Georgieva SG, Soshnikova NV. Mammalian Cells Contain Two Functionally Distinct PBAF Complexes Incorporating Different Isoforms of PHF10 Signature Subunit. Cell Cycle (2014) 13(12):1970–9. doi: 10.4161/cc.28922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mashtalir N, D'Avino AR, Michel BC, Luo J, Pan J, Otto JE, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell (2018) 175(5):1272–88.e20. doi: 10.1016/j.cell.2018.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and Regulation of Acetylated Histone Binding by the Tandem PHD Finger of DPF3b. Nature (2010) 466(7303):258–62. doi: 10.1038/nature09139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soshnikova NV, Sheynov AA, Tatarskiy EV, Georgieva SG. The DPF Domain as a Unique Structural Unit Participating in Transcriptional Activation, Cell Differentiation, and Malignant Transformation. Acta Naturae (2020) 12(4):57–65. doi: 10.32607/actanaturae.11092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, Ermolaeva MA, et al. A Novel Multidomain Transcription Coactivator SAYP can Also Repress Transcription in Heterochromatin. EMBO J (2005) 24(1):97–107. doi: 10.1038/sj.emboj.7600508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, et al. The Transcriptional Coactivator SAYP is a Trithorax Group Signature Subunit of the PBAP Chromatin Remodeling Complex. Mol Cell Biol (2008) 28(9):2920–9. doi: 10.1128/MCB.02217-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, et al. Transcription Coactivator SAYP Combines Chromatin Remodeler Brahma and Transcription Initiation Factor TFIID Into a Single Supercomplex. Proc Natl Acad Sci USA (2009) 106(27):11049–54. doi: 10.1073/pnas.0901801106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krasteva V, Crabtree GR, Lessard JA. The BAF45a/PHF10 Subunit of SWI/SNF-Like Chromatin Remodeling Complexes is Essential for Hematopoietic Stem Cell Maintenance. Exp Hematol (2017) 48:58–71.e15. doi: 10.1016/j.exphem.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 28. Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex During Neural Development. Neuron (2007) 55(2):201–15. doi: 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Viryasova GM, Tatarskiy VV, Jr, Sheynov AA, Tatarskiy EV, Sud'ina GF, Georgieva SG, et al. PBAF Lacking PHD Domains Maintains Transcription in Human Neutrophils. Biochim Biophys Acta Mol Cell Res (2019) 1866(12):118525. doi: 10.1016/j.bbamcr.2019.118525 [DOI] [PubMed] [Google Scholar]
- 30. Rauch A, Haakonsson AK, Madsen JGS, Larsen M, Forss I, Madsen M, et al. Osteogenesis Depends on Commissioning of a Network of Stem Cell Transcription Factors That Act as Repressors of Adipogenesis. Nat Genet (2019) 51:716–27. doi: 10.1038/s41588-019-0359-1 [DOI] [PubMed] [Google Scholar]
- 31. Salhotra A, Shah HN, Levi B, Longaker MT. Mechanisms of Bone Development and Repair. Nat Rev Mol Cell Biol (2020) 21:696–711. doi: 10.1038/s41580-020-00279-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montecino M, Stein G, Stein J, Zaidi K, Aguilar R. Multiple Levels of Epigenetic Control for Bone Biology and Pathology. Bone (2015) 81:733–8. doi: 10.1016/j.bone.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon JA, Stein JL, Westendorf JJ, van Wijnen AJ. Chromatin Modifiers and Histone Modifications in Bone Formation, Regeneration, and Therapeutic Intervention for Bone-Related Disease. Bone (2015) 81:739–45. doi: 10.1016/j.bone.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tye CE, Hassan MQ, Stein GS, Lian JB. Non-Coding Rnas: Epigenetic Regulators of Bone Development and Homeostasis. Bone (2015) 81:746–56. doi: 10.1016/j.bone.2015.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pike JW, Meyer MB, St John HC, Benkusky NA. Epigenetic Histone Modifications and Master Regulators as Determinants of Context Dependent Nuclear Receptor Activity in Bone Cells. Bone (2015) 81:757–64. doi: 10.1016/j.bone.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wildman BJ, Godfrey TC, Rehan M, Chen Y, Afreen LH, Hassan Q. Micromanagement of Runx2 Function in Skeletal Cells. Curr Mol Biol Rep (2019) 5(1):55–64. doi: 10.1007/s40610-019-0115-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Godfrey TC, Wildman BJ, Javed A, Lengner CJ, Hassan MQ. Epigenetic Remodeling and Modification to Preserve Skeletogenesis In Vivo . Connect Tissue Res (2018) 59(sup1):52–4. doi: 10.1080/03008207.2017.1408599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Godfrey TC, Wildman BJ, Beloti MM, Kemper AG, Ferraz EP, Roy B, et al. The Microrna-23a Cluster Regulates the Developmental Hoxa Cluster Function During Osteoblast Differentiation. J Biol Chem (2018) 293(45):17646–60. doi: 10.1074/jbc.RA118.003052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. Microrna Control of Bone Formation and Homeostasis. Nat Rev Endocrinol (2012) 8(4):212–27. doi: 10.1038/nrendo.2011.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Chen Z, Song Y, Liu C, Chen YP. Making a Tooth: Growth Factors, Transcription Factors, and Stem Cells. Cell Res (2005) 15:301–16. doi: 10.1038/sj.cr.7290299 [DOI] [PubMed] [Google Scholar]
- 41. Landin MA, Shabestari M, Babaie E, Reseland JE, Osmundsen H. Gene Expression Profiling During Murine Tooth Development. Front Genet (2012) 3:139. doi: 10.3389/fgene.2012.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lisi S, Peterková R, Peterka M, Vonesch JL, Ruch JV, Lesot H. Tooth Morphogenesis and Pattern of Odontoblast Differentiation. Connect Tissue Res (2003) 44 Suppl 1:167–70. doi: 10.1080/03008200390152278 [DOI] [PubMed] [Google Scholar]
- 43. Olley RC, Xavier GM, Seppala M, Volponi AA, Geoghegan F, Sharpe PT, et al. Expression Analysis of Candidate Genes Regulating Successional Tooth Formation in the Human Embryo. Front Physiol (2014) 5:445. doi: 10.3389/fphys.2014.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heair HM, Kemper AG, Roy B, Lopes HB, Rashid H, Clarke JC, et al. Microrna 665 Regulates Dentinogenesis Through Microrna-Mediated Silencing and Epigenetic Mechanisms. Mol Cell Biol (2015) 35(18):3116–30. doi: 10.1128/MCB.00093-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li C, Cui Y, Zhou C, Sun J, Zhou X. Epigenetics in Odontogenesis and its Influences. Curr Stem Cell Res Ther (2018) 13(2):110–7. doi: 10.2174/1574888X12666170530100524 [DOI] [PubMed] [Google Scholar]
- 46. Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, et al. Dlx3 Transcriptional Regulation of Osteoblast Differentiation: Temporal Recruitment of Msx2, Dlx3, and Dlx5 Homeodomain Proteins to Chromatin of the Osteocalcin Gene. Mol Cell Biol (2004) 24(20):9248–61. doi: 10.1128/MCB.24.20.9248-9261.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, et al. A Network Connecting Runx2, SATB2, and the Mir-23a~27a~24-2 Cluster Regulates the Osteoblast Differentiation Program. Proc Natl Acad Sci USA (2010) 107(46):19879–84. doi: 10.1073/pnas.1007698107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, et al. HOXA10 Controls Osteoblastogenesis by Directly Activating Bone Regulatory and Phenotypic Genes. Mol Cell Biol (2007) 27(9):3337–52. doi: 10.1128/MCB.01544-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taylor SE, Shah M, Orriss IR. Generation of Rodent and Human Osteoblasts. Bonekey Rep (2014) 3:585. doi: 10.1038/bonekey.2014.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu H, Gordon JAR, Whitfield TW, Tai PWL, van Wijnen AJ, Stein JL, et al. Chromatin Dynamics Regulate Mesenchymal Stem Cell Lineage Specification and Differentiation to Osteogenesis. Biochim Biophys Acta (BBA) - Gene Regul Mech (2017) 1860(4):438–49. doi: 10.1016/j.bbagrm.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-Seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol (2001) 109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. And Mouse Genes Applied to an Arrayed Viral High-Content Screen. Cell (2006) 124(6):1283–98. doi: 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- 53. Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply Attenuated Lentiviral Vector Achieves Efficient Gene Delivery In Vivo . Nat Biotechnol (1997) 15:871–5. doi: 10.1038/nbt0997-871 [DOI] [PubMed] [Google Scholar]
- 54. Gordon JA, Hassan MQ, Koss M, Montecino M, Selleri L, van Wijnen AJ, et al. Epigenetic Regulation of Early Osteogenesis and Mineralized Tissue Formation by a HOXA10-PBX1-Associated Complex. Cells Tissues Organs (2011) 194(2-4):146–50. doi: 10.1159/000324790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Langmead B, Salzberg SL. Fast Gapped-Read Alignment With Bowtie 2. Nat Methods (2012) 9(4):357–9. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders S, Pyl PT, Huber W. Htseq—a Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trapnell C, Hendrickson D, Sauvageau M, Goff L, Rinn JL, Patcher L. Differential Analysis of Gene Regulation at Transcript Resolution With RNA-Seq. Nat Biotechnol (2013) 31:46–53. doi: 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments With Tophat and Cufflinks. Nat Protoc (2012) 7:562–78. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang L, Feng Z, Wang X, Wang X, Zhang X. Degseq: An R Package for Identifying Differentially Expressed Genes From RNA-Seq Data. Bioinformatics (2010) 26:136–8. doi: 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 60. Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (Mswi/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med (2016) 6(8). doi: 10.1101/cshperspect.a026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tatarskiy VV, Simonov YP, Shcherbinin DS, Brechalov AV, Georgieva SG, Soshnikova NV. Stability of the PHF10 Subunit of PBAF Signature Module is Regulated by Phosphorylation: Role of β-Trcp. Sci Rep (2017) 7:5645. doi: 10.1038/s41598-017-05944-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bonewald LF, Dallas SL. Role of Active and Latent Transforming Growth Factor β in Bone Formation. J Cell Biochem (1994) 55:350–7. doi: 10.1002/jcb.240550312 [DOI] [PubMed] [Google Scholar]
- 63. Centrella M, Horowitz MC, Wozney JM, McCarthy TL. Transforming Growth Factor-β Gene Family Members and Bone. Endocr Rev (1994) 15:27–39. doi: 10.1210/edrv-15-1-27 [DOI] [PubMed] [Google Scholar]
- 64. Unterbrink A, O'Sullivan M, Chen S, MacDougall M. TGF Beta-1 Downregulates DMP-1 and DSPP in Odontoblasts. Connect Tissue Res (2002) 43(2-3):354–8. doi: 10.1080/03008200290000565 [DOI] [PubMed] [Google Scholar]
- 65. Kawane T, Qin X, Jiang Q, Miyazaki T, Komori H, Yoshida CA, et al. Runx2 Is Required for the Proliferation of Osteoblast Progenitors and Induces Proliferation by Regulating Fgfr2 and Fgfr3. Sci Rep (2018) 8:13551. doi: 10.1038/s41598-018-31853-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, et al. ATF4 is a Substrate of RSK2 and an Essential Regulator of Osteoblast Biology; Implication for Coffin-Lowry Syndrome. Cell (2004) 117(3):387–98. doi: 10.1016/s0092-8674(04)00344-7 [DOI] [PubMed] [Google Scholar]
- 67. Kim JH, Kim K, Youn BU, Lee J, Kim I, Shin HI, et al. Kruppel-Like Factor 4 Attenuates Osteoblast Formation, Function, and Cross Talk With Osteoclasts. J Cell Biol (2014) 204(6):1063–74. doi: 10.1083/jcb.201308102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xu F, Flowers S, Moran E. Essential Role of ARID2 Protein-Containing SWI/SNF Complex in Tissue-Specific Gene Expression. J Biol Chem (2012) 287(7):5033–41. doi: 10.1074/jbc.M111.279968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sinha S, Biswas M, Chatterjee SS, Kumar S, Sengupta A. Pbrm1 Steers Mesenchymal Stromal Cell Osteolineage Differentiation by Integrating PBAF-Dependent Chromatin Remodeling and BMP/TGF-β Signaling. Cell Rep (2020) 31(4):107570. doi: 10.1016/j.celrep.2020.107570 [DOI] [PubMed] [Google Scholar]
- 70. Pico MJ, Hashemi S, Xu F, Nguyen KH, Donnelly R, Moran E, et al. Glucocorticoid Receptor-Mediated Cis-Repression of Osteogenic Genes Requires BRM-SWI/SNF. Bone Rep (2016) 5:222–7. doi: 10.1016/j.bonr.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Elefteriou F, Yang X. Genetic Mouse Models for Bone Studies–Strengths and Limitations. Bone (2011) 49(6):1242–54. doi: 10.1016/j.bone.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin Remodeling BAF. (SWI/SNF) Complexes in Neural Development and Disorders. Front Mol Neurosci (2017) 10:243. doi: 10.3389/fnmol.2017.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, et al. Genomic Instability in Gadd45a-Deficient Mice. Nat Genet (1999) 23:176–84. doi: 10.1038/13802 [DOI] [PubMed] [Google Scholar]
- 74. Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, et al. P53-Mediated DNA Repair Responses to UV Radiation: Studies of Mouse Cells Lacking P53, P21, and/or Gadd45 Genes. Mol Cell Biol (2000) 20:3705–14. doi: 10.1128/MCB.20.10.3705-3714.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Makowski MM, Gräwe C, Foster BM, Nguyen NV, Bartke T, Vermeulen M. Global Profiling of Protein–DNA and Protein–Nucleosome Binding Affinities Using Quantitative Mass Spectrometry. Nat Commun (2018) 9:1653. doi: 10.1038/s41467-018-04084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Foster BL, Kuss P, Yadav MC, Kolli TN, Narisawa S, Lukashova L, et al. Conditional Alpl Ablation Phenocopies Dental Defects of Hypophosphatasia. J Dent Res (2017) 96(1):81–91. doi: 10.1177/0022034516663633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BMSCs ChIP-sequencing data is deposited to the NCBI GEO database with the Accession number GSE76074 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76074).