Abstract
Childhood posterior fossa-group A ependymomas (PFAs) have limited treatment options and bear dismal prognoses compared to group B ependymomas (PFBs). PFAs overexpress the oncohistone-like protein EZHIP (Enhancer of Zeste Homologs Inhibitory Protein), causing global reduction of repressive histone H3-lysine-trimethylated (H3K27me3), similar to the oncohistone H3K27M. Integrated metabolic analyses in patient-derived cells and tumors, single-cell RNA sequencing of tumors, and non-invasive metabolic imaging in patients demonstrated enhanced glycolysis and tricarboxylic acid (TCA)-cycle metabolism in PFAs. Furthermore, high glycolytic gene expression in PFAs was associated with a poor outcome. PFAs demonstrated high EZHIP expression associated with poor prognosis, and elevated activating mark histone H3-lysine-acetylated (H3K27ac). Genomic H3K27ac was enriched in PFAs at key glycolytic and TCA cycle-related genes including hexokinase-2 and pyruvate dehydrogenase. Similarly, mouse neuronal stem cells (NSCs) expressing wild type EZHIP (EZHIP-WT) versus catalytically-attenuated EZHIP-M406K demonstrated H3K27ac enrichment at hexokinase-2 and pyruvate dehydrogenase, accompanied by enhanced glycolysis and TCA-cycle metabolism. AMPKα-2, a key component of the metabolic regulator AMP-activated protein kinase (AMPK), also showed H3K27ac enrichment in PFAs and EZHIP-WT NSCs. The AMPK activator metformin lowered EZHIP protein concentrations, increased H3K27me3, suppressed TCA-cycle metabolism, and showed therapeutic efficacy in vitro and in vivo in patient-derived PFA xenografts in mice. Our data indicate that PFAs and EZHIP-WT-expressing NSCs are characterized by enhanced glycolysis and TCA cycle metabolism. Repurposing the antidiabetic drug metformin lowered pathogenic EZHIP, increased H3K27me3, and suppressed tumor growth, suggesting that targeting integrated metabolic/epigenetic pathways is a potential therapeutic strategy for treating childhood ependymomas.
One Sentence Summary:
Childhood posterior fossa group A ependymomas demonstrate integrated metabolic and epigenetic pathways that can be disrupted by metformin.
Introduction
Ependymomas can occur throughout the neuraxis including the supratentorial (ST) region, spinal cord and hindbrain [termed posterior fossa (PF)]. Ependymomas are molecularly sub-grouped, and such classification uses both anatomic locations and molecular features. ST ependymomas are classified into ST-RELA (bearing the C11orf95-RELA fusion), ST-YAP1 (bearing YAP1 fusions) and ST-subependymomas (ST-SE) (1). PF ependymomas comprise of PF-group A (PFA), PFB and PF-subependymoma (PF-SE) tumors (1). PFAs are mainly childhood tumors and bear a dismal prognosis based on their poor response to conventional therapies (1-4). In contrast, PFB and PF-SE tumors generally occur in adults and bear an excellent prognosis (1-3).
PFAs are mainly epigenetically driven and are characterized by global reduction of the repressive histone mark H3K27me3, similar to poorly prognostic midline gliomas bearing H3K27M mutations (5-7). H3K27M midline gliomas and PFAs also share similarities in genomic distribution of H3K27me3 (6, 8, 9). Whole exome and genome sequencing of PFAs have revealed that approximately 4% of PFAs show similar H3K27M mutations, while approximately 9% of tumors exhibit mutations in a newly characterized protein termed EZHIP (Enhancer of Zeste Homologs Inhibitory Protein), also referred to as CATACOMB or CXorf67 (5). More than 85% of PFAs do not bear recurrent genetic mutations but rather overexpress wildtype EZHIP (5). Moreover, H3K27M mutations in PFAs are mutually exclusive with EZHIP deregulation (5). EZHIP is also overexpressed in a subset of midline gliomas in a mutually exclusive manner with H3K27M mutations (10). EZHIP mimics H3K27M by binding to the H3K27-methyltransferase EZH2 and inhibiting the function of the PRC2 complex to lower H3K27me3 (8, 9, 11-13). PFAs and H3K27M midline gliomas share similarities including an epigenetic state of low H3K27me3, but the pathogenesis of PFAs remains largely obscure.
Metabolic alterations are a hallmark of cancer. Cancer cells take up and metabolize nutrients in large quantities to generate lipids, proteins, nucleotides and ATP to support their uncontrolled proliferation (14). Moreover, metabolic pathways and epigenetic pathways are intimately related (14). For example, H3K27M mutations can upregulate key metabolic pathways including glycolysis and the tricarboxylic acid (TCA) cycle that in turn modulate H3K27me3 (15). PFA ependymomas exhibit hypoxic niches, and PFA cells grown in hypoxic conditions show an altered and interdependent metabolic/epigenetic state (16, 17). Further, single cell RNA sequencing in PFA ependymomas shows deregulation of metabolic pathways (17, 18).
Because EZHIP mimics H3K27M, and H3K27M mutations have the ability to reprogram metabolism (15), we hypothesized that EZHIP can epigenetically rewire metabolic pathways including glycolysis and TCA cycle metabolism. We also hypothesized that this information could be used to uncover potential therapeutic targets. Our goal was to define metabolic pathways upregulated in PFA ependymomas in relation to EZHIP mediated epigenetic alterations. To address these hypotheses and goals, we: (i) performed comprehensive metabolic analyses of PFA ependymomas using isogenic cell lines, patient-derived cells and tumor tissues, and in vivo, non-invasive metabolic imaging in ependymoma patients, (ii) assessed the relationship between EZHIP-driven epigenetic alterations and metabolism and (iii) defined potential targets to lower EZHIP and inhibit this integrated epigenetic and metabolic pathway in patient-derived cell lines and patient-derived xenografts (PDXs).
Results
PFAs exhibit elevated glycolysis and TCA-cycle metabolism compared to PFB ependymomas
To define metabolic pathways upregulated in PFAs, we used a multi-platform, unbiased approach to assess gene expression and metabolomics in tumor tissues and patient-derived cell lines, and non-invasive in vivo magnetic resonance spectroscopy (MRS) in live patients (patient demographics detailed in table S1). We queried expression of a comprehensive set of 2,754 metabolic genes encoding all known human metabolic enzymes and transporters, defined by Sabatini and colleagues (19), in PFA versus PFB ependymomas. We analyzed gene expression in three independent, non-overlapping published data sets from Bayliss et al. 2016 (PFA n=11, PFB n=4) (6), Witt et. al. 2011 (PFA n=18, PFB n=19) (2) and Pajtler et al. 2015 (PFA n=72 and PFB n=39) (1) (tables S2-S4). Commonly upregulated (n=53, Fig. 1, A and B, and table S5) and downregulated (n=25, fig. S1A and table S6) genes in PFA versus PFB ependymomas were determined from all three data sets. Pathway impact analysis demonstrated that the 53 commonly upregulated genes were mainly related to glycolysis, TCA cycle, glutamate and pentose phosphate pathway (PPP) metabolism (Fig. 1C) including hexokinase-2 (HK2) and PDHB (pyruvate dehydrogenase-B) (Fig. 1D). Because bulk gene expression data are derived from both tumor and non-tumor cells in the microenvironment, we sought to confirm our findings in single cell RNA-seq analyses from Gojo et al. 2020 (18). From these analyses, genes illustrated in Fig. 1D including HK2 and PDHB (Fig. 1E and fig. S1B) were upregulated in PFA versus PFB ependymomas and enriched in many previously defined PF ependymoma cells including metabolic and neuronal stem cell (NSC)-like tumor cells (Fig. 1E and fig. S1, B to D).
Figure 1. PFAs exhibit elevated glycolysis and TCA-cycle metabolism compared to PFB ependymomas.
(A) Schematic indicates epigenetic and clinical differences between PFA and PFBs. (B) To assess metabolic pathways upregulated in PFAs versus PFBs, we queried expression of a comprehensive set of 2,754 metabolic genes encoding all known human metabolic enzymes and transporters (19) in PF ependymomas from three independent, non-overlapping data sets: Bayliss et al. 2016 (PFA n=11, PFB n=4) (6), Witt et. al. 2011 (PFA n=18, PFB n=19) (2) and Pajtler et al. 2015 (PFA n=72 and PFB n=39) (1). Venn diagram illustrates intersection of upregulated genes in all three data sets. (C) Metabo Analyst pathway impact analysis was performed for the 53 commonly upregulated (in all three data sets) metabolic genes in PFA versus PFB ependymomas. (D) Simplified illustration of key metabolites and enzymes related to glycolysis, TCA-cycle and PPP metabolism is illustrated. Enzymes indicated in blue that were upregulated in all three data sets. (E) Single cell RNA-seq expression of HK2 and PDHB in PFA (n=20) versus PFB (n=3) ependymomas from Gojo et al. 2020 is indicated (18). (F) Key metabolites related to glycolysis, TCA-cycle and PPP were measured using liquid chromatography-mass spectroscopy (LC-MS run with technical duplicates) in patient tumor samples from PFA (n=14, red), PFB (n=3, blue), ST (n=3, light blue) and control non-pathologic human pediatric frontal cortex (purple, n=3) and cerebellum (orange, n=3). (G) Steady-state key metabolites related to glycolysis, TCA-cycle and PPP from EPD210 (PFA, red) and EP1NS (ST-RELA, light blue) ependymoma cell lines cultured in neurosphere serum-free conditions are shown (n=8 for all, except n=4 for Pyr, Lac, Cit, Isocit, Fum, Gln and Glu). (H) Metabo Analyst pathway impact analysis was performed using significantly upregulated metabolites from 1G. (I and J) Expression levels of genes in the glycolysis-KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway in 91 PF ependymomas (I) or 55 PFA ependymomas (J) were segregated into high versus low glycolytic gene expression categories using unbiased K-means clustering. Kaplan-Meier analysis was then performed between high versus low glycolytic tumors to determine differences in overall survival. Data were analyzed by the Log rank test. Data represented as mean +/− SD or violin plots (with medians and interquartile ranges and ends of violin plots representing the highest and lowest observations). Statistical significance determined by 2-sided, unpaired, 2-tailed, t-test in 1E and 1G, and by ANOVA in 1F.
(1,3-BPG, 1,3-Bisphosphoglyceric acid; 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglycerate; ACO2, aconitase-2; Ala, alanine; Asp, aspartate; Cit, citrate; CoA, Coenzyme A; DHAP, Dihydroxyacetone phosphate; FBP, fructose-bisphosphate; FUM, fumarate; G-3-P or GAP, Glyceraldehyde 3-phosphate; GLC, glucose; GLN, glutamine; GLUL, glutamine synthetase; GLU, glutamate; GLUD1/2, glutamate dehydrogenase1/2; HK2, hexokinase-2; LAC, lactate; MAL, malate; Met, metabolism; PDH, pyruvate dehydrogenase; PEP, phosphoenolpyruvate; PGAM2, phosphoglycerate mutase 2; PYR, pyruvate; Ribul-5-P, ribulose-5-phosphate; Ribo-5-P, ribose-5-phosphate; Sed-7-P, sedoheptulose 7-phosphate; Suc, succinate; TCA-cycle, tricarboxylic acid cycle; TKT, transketolase; X-5-P, xylulose 5-phosphate)
We assessed metabolites using mass-spectroscopy-based analyses of patient tumor tissues including PFA (n=14), PFB (n=3), and ST-RELA (n=3) ependymomas and non-pathologic pediatric cerebellum (n=3) and cortex (n=3) as controls. Metabolite data mirrored gene expression results and showed elevated concentrations of metabolites related to glycolysis, TCA-cycle and PPP in PFA versus PFB or ST-RELA ependymomas and controls (Fig. 1F). We next assessed steady-state metabolite concentrations in ependymoma cell lines. We used three previously well-characterized PFA cell lines (PFA-EPD210, PFA-MAF811 and PFA-MAF928). Because no PFB cell lines are described in the literature, supratentorial (ST) cell lines (ST-EP1NS and ST-MAF1329 bearing C11orf95-RELA fusions), and immortalized normal human astrocytic cell lines (NHA) were used as controls (20, 21). To rule out the impact of culture conditions on metabolic pathways, we tested both serum-free (PFA-EPD210 and ST-EP1NS) and serum-containing (PFA-MAF811, ST-MAF1329 and NHA) conditions. In both (Fig. 1G) conditions (Fig. 1G and fig. S1E), PFA cells showed higher concentrations of metabolites related to glycolysis, PPP and TCA-cycle metabolism compared to control ST-RELA ependymoma and normal human astrocytic cells. Metabolite pathway impact and enrichment analyses of upregulated metabolites confirmed glycolysis, TCA cycle, and PPP metabolic pathways as top upregulated pathways (Fig. 1H). Moreover, high expression of glycolysis-KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway genes was associated with poor overall prognosis in PF-ependymomas and within PFA ependymomas (Fig. 1, I and J; and fig. S1, F and G). These data together suggest that PFA ependymomas show elevated glycolysis and TCA-cycle metabolism compared to PFB ependymomas.
Non-invasive in vivo MRS imaging shows elevated citrate and glutamate concentrations in PFA ependymomas
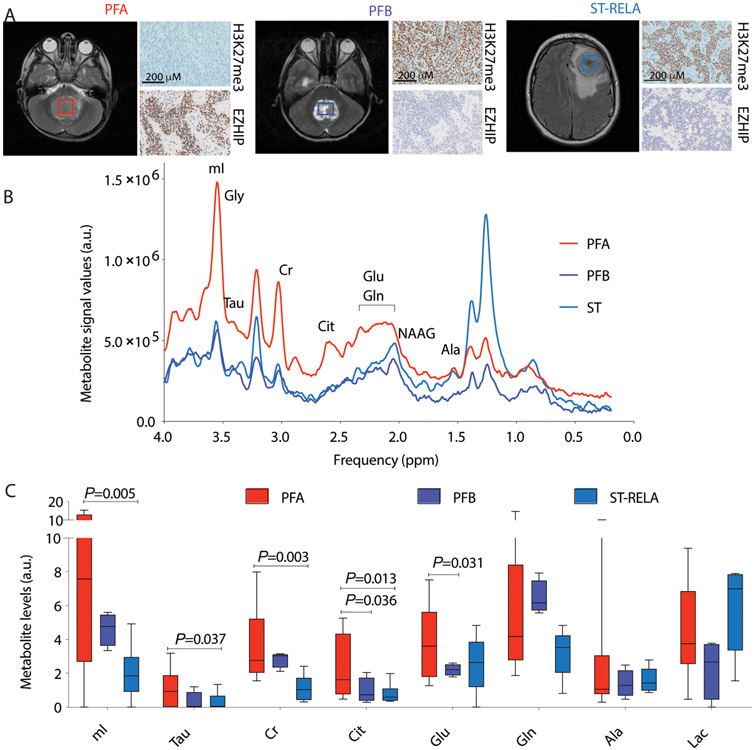
Non-invasive Magnetic Resonance Spectroscopy (MRS) imaging in live patients was used to assay in vivo metabolite concentrations in 15 children with ependymomas in a blinded and retrospective manner. After MRS imaging, corresponding tumor sections from each patient were evaluated for H3K27me3 and EZHIP immunostaining and classified into PFA (n=7, H3K27me3 negative, EZHIP positive), PFB (n=3, H3K27me3 positive, EZHIP negative) and ST-RELA (n=5, H3K27me3 positive, EZHIP negative, with C11orf95-RELA fusions) ependymomas (Fig. 2A). Metabolites were assessed after unblinding and included citrate (Cit), glutamate (Glu), glutamine (Gln), alanine (Ala), creatine (Cr), myoinositol (mI), taurine (Tau) and lactate (Lac). PFA ependymomas contained higher concentrations of citrate and glutamate compared to PFB ependymomas, and myoinositol, taurine and creatine compared to ST ependymomas (Fig. 2, B and C). These data suggest that PFA ependymomas exhibit a unique MRS imaging profile. Whereas glutamate concentrations were high in PFA ependymomas by MRS, we did not observe this in cell lines or tumor samples. However, elevated in vivo citrate concentrations in PFA ependymomas confirmed metabolite data from tumor samples (Fig. 1F) and patient-derived cell lines (Fig. 1G and fig. S1E).
Figure 2. Non-invasive in vivo MRS imaging shows elevated citrate and glutamate concentrations in PFA ependymomas.
(A) Representative axial MRI images from patients with PFA, PFB and ST-RELA are shown. Boxes indicate region of MRS quantification. Representative images from H3K27me3 and EZHIP immunostaining performed on the corresponding tumors are illustrated. Scale bar=200μM. (B) Representative in vivo MRS spectra (TE = 35 ms, TR = 2 s; g) from PFA (red), PFB (blue) and ST (light blue) ependymomas are shown. (C) MRS quantification of myoinositol (mI), taurine (Tau), creatine (Cr), citrate (Cit), glutamate (Glu), glutamine (Gln), alanine (Ala) and lactate (Lac) from PFA (n=7, red), PFB (n=3, blue) and ST (n=5, light blue) patients are plotted. (A.U.- Arbitrary units). Data were analyzed by unpaired t test with Welch's correction and are presented as box and whisker plots.
EZHIP expression is associated with high H3K27ac, location in the fourth ventricle, and prognosis in PF ependymomas
H3K27M mutations enhance glycolysis and TCA-cycle metabolism by epigenetically activating key metabolic genes (15). Because H3K27M and EZHIP show epigenetic similarities, we sought to determine if EZHIP can upregulate these pathways in a similar manner. H3K27M-gliomas demonstrate elevation of the activating mark H3K27ac (22-24). Moreover, EZHIP is associated with elevated H3K27ac in Daoy, 293T and U2OS cells (5, 8, 13), and previous studies from Mack et al. have demonstrated that H3K27ac is deregulated in ependymomas (25). We therefore sought to determine the relationship between EZHIP and H3K27ac in samples from patients with ependymomas in association with various clinical parameters. Western blotting in tumor samples showed an increase in global H3K27ac in PFAs compared to PFBs (Fig. S2A). We used immunohistochemistry to assess EZHIP, H3K27me3 and H3K27ac in PF ependymomas (table S1). EZHIP expression was elevated and corresponded with a reduction in global H3K27me3 and elevated H3K27ac in PFAs versus PFBs (Fig. 3, A and B, fig. S2B). Because PFAs occur more often in young children, we compared expression of these markers in relation to age. EZHIP and H3K27ac were higher in children below 10 years of age compared to those observed in children above 10 years, while H3K27me3 showed the opposite pattern (Fig. 3B, fig. S2B).
Figure 3. EZHIP expression correlates with high H3K27ac, location in the fourth ventricle, and prognosis in PF ependymomas.
(A) Representative images of EZHIP, H3K27me3 and H3K27ac immunostaining in PFA (two samples PFA #1 and PFA#2, illustrating range of EZHIP expression) and PFB ependymomas. Scale bar represents 100 μm. (B) Blinded quantification of EZHIP and H3K27ac in PFA (n=90, red) and PFB (n=65, blue) tumors and association of each marker with age: <10 years (n=78, orange) and >10 years (n=16, purple). (C) Overall and progression free survival in EZHIP low (n=30) versus EZHIP high (n=31, median cutoff) PFAs are indicated. (D) Representative MRI images from PF ependymomas associated with the lateral recess (LR, red arrows), roof (red arrows) and floor (red arrow) of the fourth ventricle (assessed by a radiologist in a blinded manner) are illustrated. (E and F) Representative images and blinded quantification of EZHIP associated with the lateral recess (LR, n=10), roof (n=15) and floor (n=17) of the fourth ventricle; H3K27me3 (LR, n=14), roof (n=25) and floor (n=17); and H3K27ac (LR, n=14), roof (n=25) and floor (n=17) in PF ependymoma are shown. Scale bar=200μm. Data represented as violin plots (with medians and interquartile ranges and ends of violin plots representing the highest and lowest observations). Statistical significance determined by 2-sided, unpaired, 2-tailed, t-test in 3B, Log-rank test in 3C, and by ANOVA in 3F.
We next determined if EZHIP protein abundance was associated with prognosis in 93 PF ependymomas (PFA n=61, PFB n=32) with available clinical information. High EZHIP concentrations (median cutoff) were associated with poor progression-free and overall survival in these tumors (fig. S2C). Moreover, within PFAs (n=61), elevated EZHIP concentrations were associated with poor progression free and overall survival (Fig. 3C). PFAs are thought to arise mainly from the roof and lateral recess of the fourth ventricle, while PFBs are mostly associated with floor of the fourth ventricle (2, 5). We assessed association of EZHIP, H3K27me3, and H3K27ac with tumor-associated regions of the fourth ventricle (radiologically determined in a blinded manner). Consistent with PFAs arising from the roof and lateral recess of the fourth ventricle, EZHIP and H3K27ac were higher in tumors associated with the roof and lateral recess compared to those associated with the floor of the fourth ventricle (Fig. 3, D to F). H3K27me3 showed the opposite pattern (Fig. 3, D to F). These data suggest that high EZHIP protein abundance is associated with worse prognosis in PF ependymomas and within PFAs. Moreover, EZHIP-expressing PF ependymomas with high H3K27ac are associated with tumor locations of the roof and lateral recess of the fourth ventricle.
EZHIP-WT versus EZHIP-M406K NSC demonstrate higher H3K27ac enrichment at Hk2 and Pdh and exhibit enhanced glycolysis and TCA-cycle metabolism
EZHIP bears a methionine (M) residue at position 406 that is essential for PRC2 inhibition similar to the M residue in the H3K27M oncohistone (8, 11, 12). We created an isogenic system by expressing wild-type EZHIP (EZHIP-WT) or by replacing M at position 406 with lysine K (EZHIP-M406K), which reduces EZHIP function (8). We used immortalized mouse NSCs that have been employed to model ependymomas (26). A functionally active EZHIP-M406I (8) was used as control. Both EZHIP-WT and EZHIP-M406I lowered global H3K27me3 compared to EZHIP-M406K NSCs (Fig. 4A). Global H3K27ac was elevated in EZHIP-WT and EZHIP-M406I compared to EZHIP-M406K NSCs (Fig. 4A). EZHIP-WT cells showed increased proliferation compared to EZHIP-M406K NSCs (Fig. 4B) and showed downregulation of differentiation-related cytoskeletal proteins and upregulation of stem cell factors such as Sall3 (Fig. S3A). This isogenic system confirmed data from tumor samples and demonstrated that EZHIP-expressing PFA tumors and NSCs show a global H3K27me3 reduction accompanied by an increase in the activating mark H3K27ac.
Figure 4. EZHIP-WT versus EZHIP-M406K NSCs demonstrate higher H3K27ac enrichment at Hk2 and Pdh and exhibit enhanced glycolysis and TCA-cycle metabolism.
(A) Mouse NSCs were stably transfected with FLAG-tagged EZHIP-WT, EZHIP-M406I or EZHIP-M406K. Representative WB for FLAG, H3K27me3, H3K27ac and total H3 are shown. (B) Bar plot of cell counts (cell numbers, Y-axis) in parental (par, light blue), EZHIP-WT (red) and EZHIP-M406K (blue) NSCs (n=4 with 2-3 technical replicates, each) is shown. (C) Super-enhancer (SE, enhancer rank X-axis) with increased H3K27ac enrichment (EZHIP-WT enhancer strength, Y-axis) in EZHIP-WT, but not EZHIP-M406K NSC is depicted. SE unique to EZHIP-WT that overlap with PFA-specific SE from Mack et al. 2018 (25) are indicated. (D) Representative heatmaps and H3K27ac enrichment (Y-axis) of genes (including both promoters/enhancers) enriched for H3K27ac in EZHIP-WT versus EZHIP-M406K NSC +/−5Kb from peak center (X-axis) are shown. (E) Representative H3K27ac ChIP-seq tracks at Hk2, Prkaa2/Ampkα-2, and Pdhx; gene loci in EZHIP-WT and EZHIP-M406K NSCs are indicated. (F) Representative heatmaps of genes (including both promoters/enhancers) enriched for H3K27ac in PFA versus ST +/−5Kb from peak center are shown. (G) Representative H3K27ac ChIP-seq tracks at HK2, PDHB, and PRKAA2/AMPKα-2 gene loci in PFA and ST ependymoma tumors are indicated. (H-I) Unbiased proteomic analysis (≤1% FDR with adjusted p-value < 0.05) was performed between EZHIP-WT and EZHIP-M406K NSCs. Heatmap illustrates differentially expressed proteins and bar graph shows GSEA pathway analysis of all upregulated proteins in EZHIP-WT compared to EZHIP-M406K NSC (n=3, each, I). (J) Bar graph represents steady state key metabolites related to glycolysis, PPP and TCA-cycle from EZHIP-WT (red) and EZHIP-M406K (blue) NSC (n=3, each). (K) Metabo Analyst pathway impact analysis is illustrated using significantly upregulated metabolites from 4J. Data are represented as violin plots (with medians and interquartile ranges and ends of violin plots representing the highest and lowest observations) or as mean +/− SD. Statistical significance determined by non-parametric, 2-sided, unpaired, 2-tailed, t-test (4J) or one-way ANOVA (4B). Data in C analyzed by the Log rank test.
We determined genome-wide distribution of H3K27ac using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) in EZHIP-WT versus EZHIP-M406K NSCs and PFA and ST samples from patient tumors (tables S7 to S9). Recent studies have suggested that H3K27M tumors show increased global H3K27ac and distinct genomic H3K27ac distribution (22-24) including higher intergenic enrichment (24). Similarly, we found increased H3K27ac enrichment at intergenic regions in EZHIP-WT versus EZHIP-M406K NSCs, and PFA versus control ST tumors (Fig. S3B). H3K27M tumor cells also showed increased H3K27ac enrichment at endogenous retroviral (ERV) elements (24), and we noted increased H3K27ac at the ERV-3 locus in EZHIP-WT versus EZHIP-M406K NSCs and PFA versus ST tumor samples (Fig S3C). H3K27ac enrichment also defines super-enhancers that are specific to each ependymoma molecular subgroup (25). H3K27ac-marked super-enhancers in EZHIP-WT, but not EZHIP-M406K NSCs, overlapped with several PFA-specific super-enhancers described by Mack et al. (25) (Fig. 4C), suggesting that EZHIP-WT-expressing NSCs model several aspects of PFA biology.
We identified genes with increased H3K27ac enrichment (including promoters and enhancers) in EZHIP-WT versus EZHIP-M406K NSC and PFA versus ST ependymomas (. Fig. 4, D to G and fig. S3, D to F). Despite the mouse versus human species difference, GSEA of H3K27ac-enriched genes revealed several common upregulated pathways in EZHIP-WT versus EZHIP-M406K NSCs and PFA versus ST ependymomas, with one of the top common hits being related to glycolysis (Fig. S3E, table S10). HK2 and PDH-related genes identified from gene expression analyses (Fig 1, D and E) showed H3K27ac enrichment in both PFA versus ST ependymomas, EZHIP-WT versus EZHIP-M406K NSCs and PFA versus PFB ependymomas from previously published H3K27ac ChIP-seq data (25) (Figs. 4, E and G, fig. S3F). Moreover, PRKAA2/AMPKα-2, a key component of the central metabolic regulator AMP-activated protein kinase (AMPK) complex, also demonstrated H3K27ac enrichment in EZHIP-WT versus EZHIP-M406K NSCs, PFA versus ST ependymomas, and PFA versus PFB ependymomas (Fig. 4, E and G, fig. S3F). In contrast, pyruvate kinase-M (PKM) was not enriched for H3K27ac in EZHIP-WT versus EZHIP-M406K NSCs, PFA versus ST, and PFA versus PFB ependymomas (fig. S3, F and G) and PKM gene expression was not different in PFAs compared to PFBs (fig. S3H). Genomic H3K27ac in tumors at metabolic gene loci showed a positive correlation with gene expression, HK2, PDHX, PDHB, but not PKM, expression positively correlated with EZHIP and Hk2, Pdhb, and Prkaa-2 were higher in EZHIP-WT versus EZHIP-M406K NSCs (fig. S3, I to K). Because H3K27me3 is a repressive mark and opposes H3K27ac, we compared H3K27ac genomic distribution with previously published H3K27me3 ChIP-seq data obtained from the same PFA and ST ependymomas and EZHIP-WT versus EZHIP M406K NSCs (6). H3K27me3 enrichment was associated with lowered gene expression and showed a weak inverse relationship with H3K27ac genomic distribution (fig. S3, L and M). EZHIP-WT showed variably lowered H3K27me3 at Hk2, Pdhx, and Prkaa2/ Ampkα-2 compared to EZHIP M406K NSCs (Fig. S3N). However, H3K27me3 enrichment was not different between PFA and ST tumors at HK2, PDHX, and PRKAA2/ AMPKα-2 gene loci (Fig. S3O), suggesting that H3K27ac enrichment could be independent of changes in genomic H3K27me3 at these gene loci.
Recent studies have shown that PFA ependymomas exhibit hypoxic niches, and PFA cells grown in hypoxia show altered metabolism (16, 17). To identify the relationship between EZHIP abundance and hypoxia in tumor samples, we combined immunohistochemistry for EZHIP and carbonic anhydrase-9 (CA-9) as a surrogate for hypoxic areas in PFA tumor tissues (n=8). Amongst eight PFAs, four tumors did not exhibit any CA9 staining. The other four PFAs showed focal CA-9 staining distributed around blood vessels and areas of necrosis (fig.. S4, A to C) and constituted overall 5-30% of the entire tissue section (fig. S4C). EZHIP staining was observed in both normoxic and CA-9 positive hypoxic regions (fig.. S4, A to C), with predominance in CA-9 negative, non-hypoxic regions. Furthermore, EZHIP-WT versus EZHIP-M406K NSCs and PFA versus ST cell lines did not show upregulation of master metabolic regulators including activated mTor, Hif-1α, c-Myc or N-Myc (fig.. S4, D and E).
We used an unbiased approach by determining overall changes in the proteome in EZHIP-WT compared to EZHIP-M406K NSCs (table S11). EZHIP-WT showed differential regulation of 2340 (1090 upregulated, 1250 downregulated) proteins compared to EZHIP-M406K cells (Figs. 4G). Pathway analysis of the 1090 upregulated proteins revealed glycolysis and TCA-cycle as the top upregulated pathways (Fig. 4H and table S12). Metabolomic experiments performed in parallel on these cells corroborated these findings and showed upregulation of several key metabolites related to glycolysis, TCA-cycle metabolism and PPP in EZHIP-WT compared to EZHIP-M406K NSCs (Fig. 4, I and J). We used 13C uniformly-labeled isotope tracing to determine if glucose carbons enter the TCA-cycle to serve as an anaplerotic substrate. [U-13C]-glucose labelling studies showed a greater extent of glucose-derived fumarate (m+2), malate (m+2) and TCA-cycle-related metabolite aspartate (m+2) in EZHIP-WT versus EZHIP-M406K NSCs and EPD210-PFA versus EP1NS-ST RELA cells (fig S5).
Overall, these data suggest that EZHIP-WT expression in NSCs elevates global H3K27ac and shares similarities in H3K27ac genomic distribution with PFA ependymomas at intergenic regions, super-enhancers and genic regions. H3K27ac was enriched at HK2 and PDH gene loci in PFA versus ST and PFA versus PFB ependymomas, and EZHIP-WT versus EZHIP-M406K cells and was associated with increased glycolysis and TCA-cycle metabolism.
Metformin suppresses TCA-cycle and PPP metabolism and downregulates EZHIP to increase global H3K27me3 in PFA cells in vitro
From our ChIP-seq analysis, we noted that PRKAA2/AMPKα-2, a critical component of the AMP-activated protein kinase (AMPK) complex, was associated with higher H3K27ac enrichment in EZHIP-WT versus EZHIP-M406K NSCs, PFA versus ST, and PFA versus PFB ependymomas (Fig. 4E, 3M and fig. S3I). PRKAA2/AMPKα-2 was upregulated in PFA versus PFB tumors in the three independent non-overlapping data sets (Fig. 5, A and B, fig. S6A). Single cell RNA-seq data confirmed increased PRKAA2/AMPKα-2 expression in PFA compared to PFB tumor cells and demonstrated increased expression in various previously delineated tumor cells including NSC-like, glial progenitor-like, S-phase and tumor cells (Fig. 5C, fig. S6B). AMPK is a critical metabolic sensor that can be activated by the biguanide metformin. Metformin inhibits mitochondrial complex I to reduce ATP and causes elevation of AMP/ATP ratio resulting in AMPK activation (27). Because metformin can also suppress the TCA-cycle (27), we reasoned that metformin treatment could lower TCA-cycle metabolites in PFA cells. Moreover, metformin is an attractive therapeutic candidate as high-dose metformin shows potent anticancer effects (28), is highly blood-brain barrier penetrable in childhood brain tumor survivors (29), and is currently being tested in more than 250 cancer clinical trials including patients with gliomas (https://clinicaltrials.gov/).
Figure 5. Metformin suppresses TCA-cycle and PPP metabolism and downregulates EZHIP to increase global H3K27me3 in PFA cells.
(A) Bar graph demonstrates PRKAA2/AMPKα-2 expression in PFA (n=11) and PFB (n=4) ependymoma tumor samples from Bayliss et al. 2016 (6). (B) Violin plots indicate PRKAA2/ AMPKα-2 expression in PFA (n=72) and PFB (n=39) from Pajtler et al. (1). (C) Violin plots of single cell RNA-seq expression of PRKAA2/ AMPKα-2 in PFA (n=20) and PFB (n=3) ependymomas from Gojo et al. 2020 are indicated (18). (D) Parental (light blue), EZHIP-M406K (blue), or EZHIP-WT (red) NSCs (n=4, with 2-7 technical replicates each) were plated in a 6-well plate (200,000 cells/well) and treated with vehicle or metformin (indicated doses in mM, X-axis). After 4 days, cells were counted (Y-axis) using Trypan blue exclusion assay. (E, F, and G) PFA cell lines EPD210, MAF928 (E) and MAF811 (F), and control ST01 (supratentorial non-fusion) and NHA (normal human astrocytic cells) (G) were plated in a 6-well plate (100,000 cells/well) and treated with vehicle or metformin (indicated doses, X-axis). After 6 days, cells were counted using Trypan blue exclusion assay (n=4, for all). Cell counts were normalized to untreated controls for the corresponding cell line (Y-axis). Unbiased proteomic analysis was performed in EZHIP-WT NSCs treated with vehicle (veh+, PBS, n=3) or metformin (met+, 4 mM, n=4) for 4 days. (H, I, J and K)Heatmap demonstrates differentially expressed proteins (H) and bar graph shows GSEA pathway analysis of all down-regulated proteins between metformin-treated and vehicle-treated EZHIP-WT NSC (I). Unbiased metabolomics using LC-MS was performed between EPD210 PFA cells treated with vehicle (veh+, PBS, n=4) or metformin (met+, 25 mM, n=3) for 4 days. Heatmap demonstrates differentially expressed metabolites (J). Metabolite enrichment analysis (using Metabo Analyst) was performed on downregulated metabolites between metformin-treated and vehicle-treated EPD210 cells (K). (L) Representative Western blots for phospho-AMPK (pAMPK), total AMPK (tAMPK), VINCULIN, H3K27me3 and Total H3 in EPD210 PFA cells cultured for 5 days with either vehicle (PBS), or metformin (25mM and 50 mM) are shown. (M) Representative Western blots for EZHIP and VINCULIN in EPD210 PFA cells cultured for 4 days with either vehicle (PBS), or AICAR (1mM), or metformin (25 or 50 mM) are illustrated. (N) Representative Western blots for FLAG, Gapdh, H3K27me3 and Total H3 in EZHIP-WT or EZHIP-M406K NSC cultured for 3 days with vehicle (PBS) or 25 mM metformin are shown. (O) Comparison of differential expression of proteins (Log 2 fold change) in EZHIP-WT/ EZHIP-M406K NSC versus Metformin/ Veh EZHIP-WT NSC is shown. Data represented as mean +/− SD or violin plots (with medians and interquartile ranges and ends of violin plots representing the highest and lowest observations). Statistical significance determined by 2-sided, unpaired, 2-tailed, t-test (5A-C), or ANOVA (5D) or Pearson’s correlational analysis (5O).
We assessed sensitivity to metformin in our isogenic cell lines. While all NSCs were sensitive to metformin, cells expressing EZHIP-WT were more sensitive to metformin treatment (at low doses) than EZHIP-M406K or parental NSCs (Fig. 5D). We used well-characterized patient-derived, low passage, PFA ependymoma cells (EPD210, MAF928 and MAF811) that also grow as patient-derived xenografts (PDXs) in vivo (20, 21). All three PDXs clustered with PFA ependymomas by DNA methylation analysis (20, 21), and expressed EZHIP protein accompanied by global H3K27me3 reduction (fig. S6, C and D). High dose metformin treatment in PFA EPD210 cells increased NADH/NAD (consistent with mitochondrial complex I inhibition), AMP/ATP, and lactate/pyruvate ratios (fig. S7, A to C). Two of the PFA cell lines, EPD210 and MAF928 were sensitive to high-dose metformin treatment (Fig. 5E). The metformin IC50 values of MAF928 (7.8 mM) and EPD210 (8.6 mM) were comparable to that observed in metformin-sensitive GBM cells (range 6-13 mM; fig. S7D). EPD210 PFA cells treated with metformin did not show significant changes in percentages of G1 (p=0.06), G2 (p=0.33), or ratio of G2/G1 (p=0.381) cells (fig.. S7E). A small lowering of S phase cells was detected, similar to previous reports in other cancer cells (fig.. S7E) (30). The other PFA cell line, MAF811 (Fig. 5F) and control cell lines ST01 (ST non-RELA) and NHA (Fig. 5G) were relatively resistant to metformin treatment. Metformin is taken up by the OCT/SLC22A family of transporters (fig. S7F) (27). Among these, OCT3/SLC22A3 is highly expressed in the brain, and genetic knockdown of SLC22A3 results in attenuated metformin response (31). Expression (mRNA) of SLC22A3, but not SLC22A1, SLC22A4 or SLC22A5, was higher in metformin-sensitive MAF928 compared to metformin-resistant MAF811 cells (fig. S7G). From single cell RNA-seq data, SLC22A3 expression was higher in PFA versus PFB, and SLC22A3 was expressed in various previously-defined PFA tumor cells including NSC-like and astroependymal-like tumor cells (fig. S7, H and I). This heterogenous expression correlated with overall outcome: high SLC22A3 expression, but not SLC22A4 or SLC22A5, was associated with a poor prognosis in PFAs (fig. S7, J and K). Metformin is known to suppress the TCA-cycle in breast cancer cell models (32). An unbiased proteomic approach in EZHIP-WT NSCs treated with metformin versus vehicle showed downregulation of proteins related to glycolysis, TCA-cycle, pyruvate metabolism and PPP (Fig. 5, H and I and tables S13 and S14). Moreover, metabolomic analysis of metformin-treated versus vehicle-treated EPD210 cells showed downregulation of metabolic pathways related to TCA-cycle and PPP accompanied by a reduction in NADPH/NADP (generated in part by the PPP) (Fig. 5, J and K and fig. S7B).
Treatment with metformin or the AMPK-activating AMP analogue AICAR lowered EZHIP and increased global H3K27me3 in EPD210 PFA cells (Fig. 5, L and M). Similarly, metformin treatment lowered FLAG-tagged EZHIP-WT to increase global H3K27me3 in NSCs (Fig. 5N) and to a lesser extent in EZHIP-M406K cells (Fig. 5N). Moreover, comparison of proteins upregulated in EZHIP-WT/ EZHIP-M406K NSCs with metformin/ veh EZHIP-WT NSCs showed a negative correlation, suggesting that EZHIP-WT-driven differential protein expression is, in part, reversed by metformin (Fig. 5O). We noted heterogeneity in metformin sensitivity, and metformin treatment suppressed TCA-cycle metabolism and the PPP, lowered EZHIP and EZHIP-driven proteins, and increased H3K27me3 in metformin-sensitive PFA cells.
Metformin decreases tumor growth and EZHIP, and increases global H3K27me3, in vivo.
As proof-of-principle, we sought to determine if high-dose metformin would be effective in suppressing PFA tumors in vivo using several animal models. Amongst the more than 250 clinical trials studying metformin for treatment of cancer, 86% (n=217/252) administer a maximal metformin dose ranging from 1000-2000 mg/day (median 1700 mg) including in pediatric brain tumor survivors (maximal dose 2000 mg/day) (29). Similarly, high-dose metformin (250-500mg/kg/day administration for 3-6 weeks) shows efficacy in glioblastoma animal models in vivo (28). To test metformin in vivo, we established 3 PFA PDX (confirmed by DNA methylation, fig. S6, C and D) animal models by engrafting either metformin-sensitive EPD210 (orthotopic), metformin-sensitive MAF928 (subcutaneous), or metformin-resistant MAF811 (subcutaneous) cells in NSG mice (20, 21).
H3K27M midline gliomas and PFA ependymomas share epigenetic similarities including global and genomic H3K27me3 reduction (fig. S8, A and B) (6, 8). We hypothesized that metformin resistance in PFA MAF811 cells could be overcome by therapeutic agents that show efficacy in H3K27M tumors. The pan-HDAC inhibitor panobinostat is currently in clinical trials in H3K27M midline-gliomas (NCT02717455). Panobinostat is toxic to H3K27M cells by increasing global H3K27me3 (hypothesized to be due to acetylation of lysine residues proximal and distal to the H3K27M mutation) (23, 33, 34). Moreover, panobinostat suppressed the PPP in H3K27M cells (35) (fig. S8, C and D). In accordance with these previous observations, panobinostat treatment increased global H3K27me3 (fig. S8E) and suppressed the PPP (fig. S8, F and G) in PFA EPD210 cells. Panobinostat exhibited cytotoxicity in both metformin-sensitive EPD210 (fig. S8H) and metformin-resistant MAF811 (fig. S8I) cells in vitro, but was less specific than metformin as toxicity was also noted in NHA cells (fig. S8J). A recent study by Krug et. al. showed that although panobinostat toxicity may not be specific to H3K27M cells, maximal tumor suppression in vivo was achieved by adding panobinostat as a second agent (24). Because metformin also increased global H3K27me3 (Fig. 5, L to N) and suppressed the PPP in PFA cells (Fig. 5, J and K), we hypothesized that panobinostat alone or combined treatment with metformin could overcome metformin resistance in PFA MAF811 cells.
Animals implanted with all three PFA PDXs were treated (after confirming grafting of tumors) with either metformin [250mg/kg, daily for 4 weeks, based on doses used in glioma models (28)], panobinostat [10mg/kg, three times/week for 4 weeks, based on doses used in H3K27M models (23, 34)] or both (Fig. 6A and S9A-G). PFA-MAF928 PDXs showed slowed tumor growth upon metformin or combined treatment. To a lesser extent, panobinostat also lowered tumor MAF928 growth compared to vehicle-treated animals (Figs. 6B-C and table S15). There was no difference between animals treated with metformin alone or combined metformin/panobinostat (Figs. 6B-C). Similarly, metformin or combined treatment, but not panobinostat by itself, extended overall survival in orthotopic PFA-EPD210 animals (Figs. 6D-F and table S15). There was no difference between animals treated with metformin alone or combined metformin/panobinostat (Figs. 6D-F). Metformin reduced EZHIP, which was accompanied by an increase in global H3K27me3 in vivo in both EPD210 and MAF928 PDXs (Figs. 6G-J and S9A). In both models, panobinostat did not alter EZHIP, but increased H3K27me3 consistent with our in vitro data (Figs. S8E, 6G-J and S9A) and published reports in H3K27M cells (23, 34). The overall weights of mice in both metformin-sensitive models during treatment were not different (S9E-F).
Figure 6. Metformin reduces tumor growth and EZHIP, and increases global H3K27me3, in vivo.
(A) Schematic illustrates treatment schedule with either metformin (250mg/kg, administered daily by oral gavage), or panobinostat (10mg/kg, administered 3 times/week intraperitoneally.), or both agents for four weeks in mice grafted with PFA PDXs (see also fig. S6, C and D, and fig. S9). Treatments were started after confirming tumor engraftment in each model. (SubQ, subcutaneous; BLI, Bioluminescence imaging). (B) Tumor volumes normalized to the initial tumor size (Y-axis) and plotted as a function of time (X-axis, days) in mice grafted subcutaneously with PFA-MAF928 PDX cells and treated with vehicle (DMSO, Veh, blue, n=5), metformin (Met, black, n=8), panobinostat (Pano, n=6, red) or both (Met+Pano, n=6, purple). (C) Kaplan-Meier analysis of percentage of PFA-MAF928 PDXs tumors receiving indicated treatment that grew three-fold in volume (Y-axis) are plotted as a function of time (X-axis, days). (D) NSG Mice were orthotopically implanted with PFA-EPD210 PDXs. Left panel shows representative bioluminescence images in mice before or after treatment treated with either vehicle (Veh, n=7), or metformin (met, n=7), or panobinostat (Pano, n=3) are shown. Right panel shows representative bioluminescence images in mice treated with vehicle or combination of both (Met+Pano, n=8). (E) Bar graph of normalized bioluminescence values in PFA-EPD210 orthotopic mice at 6-weeks post treatment is shown. (F) Kaplan-Meier survival analysis of mice with EPD210 PDXs receiving either vehicle or indicated treatments is indicated. (G and H) Representative images (G) and blinded quantification of EZHIP (H) immunostaining in PFA-EPD210 PDX orthotopic mice treated with vehicle (DMSO, Veh, blue, n=7), panobinostat (Pano, n=3, red), metformin (Met, black, n=5), or both (Met + Pano, n=7, purple) are shown. (I and J) Representative images (I) and blinded quantification of H3K27me3 (J) immunostaining in PFA-EPD210 PDX orthotopic mice treated with vehicle (DMSO, Veh, blue, n=7), panobinostat (Pano, n=3, red), metformin (Met, green, n=7), or both (Met + Pano, n=8, purple) are shown. Data represented as mean +/− SD. Statistical significance was determined by Log-Rank analyses (6C and 6F, P values in comparison with vehicle treated animals are indicated) or ANOVA (6B, 6E, 6H and 6J). Scale bar=50μM.
PFA-MAF811 cells, which were resistant to metformin in vitro (Fig. 5F), did not show tumor suppression with single agent metformin treatment in vivo (fig. S9, B to D). However, both panobinostat alone or combined treatment slowed PFA-MAF811 tumor growth (fig. S9, B to D and table S15). Panobinostat, but not metformin, focally increased global H3K27me3, and EZHIP remained unchanged in MAF811 cells (fig. S9B). There was no difference between animals treated with panobinostat alone or combined metformin/panobinostat (fig. S9, B to D). These results paralleled in vitro studies where PFA-EPD210 and PFA-MAF928, but not PFA-MAF811 cells showed sensitivity to metformin.
Discussion
Children with PFA ependymomas bear a dismal prognosis and are very challenging to treat. [reviewed in (4)]. Radiation therapy and chemotherapy in some cases are the mainstay of treatment (4). Nevertheless, radiation causes substantial long-term toxicity and morbidity. Moreover, a major limitation that has hampered advancement is the dearth of patient-derived cell lines and animal models. Despite decades of research and clinical trials, there have been limited therapeutic advances in the field.
Although PFAs are relatively genomically quiescent, Mack et al. identified epigenetic deregulation as a key pathogenic alteration in PFAs (3). PFAs are defined by a global epigenetic state of H3K27me3 reduction by EZHIP. EZHIP is thought to promote tumorigenesis by lowering H3K27me3 at genes related to neuroglial differentiation pathways, similar to that observed in H3K27M-midline gliomas (5-8, 11, 12). However, low H3K27me3 alone cannot entirely account for the aggressive nature of these tumors. It is not known if EZHIP can activate additional mechanisms to drive PFA ependymoma biology. To identify pathogenic mechanisms in PFA ependymomas, we undertook integrated epigenetic and metabolic analyses to present the following discoveries: (i.) PFAs show high glycolysis and TCA-cycle metabolism. Mechanistic experiments demonstrated that increased expression of metabolic enzymes including HK2 and PDH was associated with H3K27ac enrichment at these gene loci in tumors. Forced expression of EZHIP-WT, but not EZHIP-M406K protein, recapitulated high global H3K27ac and enrichment at metabolic genes, and increased metabolites related to glycolysis and TCA-cycle metabolism. (ii.) PFAs showed H3K27ac enrichment at the AMPKα-2 gene locus accompanied by increased gene expression. AMPK is a central metabolic regulator of both glycolysis and TCA-cycle metabolism (27). We reasoned that targeting AMPK could serve as a therapeutic target by suppressing key metabolic pathways. We choose the antidiabetic biguanide AMPK activator metformin, as it is blood-brain-barrier penetrable, shows efficacy in adult glioma animal models, and is currently being tested in cancer clinical trials. High-dose metformin treatment in vitro showed potent toxicity in two PFA cell lines (EPD210 and MAF928). A third PFA cell line (MAF811) was resistant to metformin treatment and showed low expression of the brain metformin transporter SLC22A3. Metformin lowered EZHIP protein concentrations to increase global H3K27me3 in PFA cells. In PFA EPD210 and MAF928, but not MAF811 PDX models, metformin suppressed tumor growth, decreased EZHIP concentrations and increased H3K27me3 in vivo. Our data suggest that metformin may be toxic to PFA cells by both reducing EZHIP and suppressing the TCA-cycle, however, we were unable to distinguish independent effects of EZHIP reduction versus metabolic suppression by metformin. In light of evidence that AMPK can alter protein function by posttranslational modifications, future studies are geared towards elucidating how metformin downregulates EZHIP.
To develop strategies to overcome metformin resistance, we reasoned that a similar global state of H3K27me3 reduction in PFA and H3K27M-gliomas could create an opportunity for testing the pan-HDAC inhibitor panobinostat that is currently in clinical trials for H3K27M-gliomas. Similar to previous studies in H3K27M glioma models, panobinostat suppressed the PPP and increased global H3K27me3 in PFA cells. In our in vivo models, panobinostat overcame metformin resistance in MAF811 PDXs. Metformin has been used extensively in human subjects with diabetes and in cancer clinical trials, and its safety is well-documented, including a recent study in pediatric brain tumor survivors (29). However, other small molecular activators of AMPK that show greater blood-brain barrier penetrability also need to be considered.
Limitation of our studies include the relatively small number of isogenic and PFA cell lines, in vivo animal models, and patient tumor samples; the number of patients and single voxel spectroscopy in our in vivo MRS analyses is also a limitiation. However, some of these limitations are reflective of the general dearth of cell lines and animal models in the field.
In summary, we demonstrated that PFA ependymomas show enhanced glycolysis and TCA-cycle metabolism in tumor samples and cell lines. Non-invasive MRS imaging in patients showed that PFA ependymomas exhibit a distinct metabolic signature in vivo. MRS imaging could serve as a much-needed, non-invasive clinical tool to metabolically monitor tumor progression/recurrence and assess treatment efficacies in PFA patients, and we are currently expanding our analyses to test potential translation to patients. PFA ependymomas showed enrichment of activating H3K27ac at key genes related to glycolysis and TCA-cycle metabolism. EZHIP, compared to EZHIP-M406K expressing cells, recapitulated these key epigenetic and metabolic phenotypes. The epigenetic mechanisms by which EZHIP drives these metabolic pathways require further elucidation. Importantly, we show as proof-of-principle that EZHIP concentrations can be lowered with metformin treatment, thus targeting integrated epigenetic and metabolic pathways in a subset of PFA ependymomas. Although additional studies will have to be conducted before these findings can be translated to the clinic, our data have the potential to inform future studies to treat lethal childhood PFA ependymomas.
Materials and Methods
Study design
The objective of this study was to define altered metabolic pathways in childhood PFA ependymomas and to use this information to inform therapeutic development. Because more than 95% of PFA ependymomas are driven by EZHIP, we hypothesized that EZHIP would reprogram both epigenetic and metabolic pathways in an integrated manner. We used tumors, patient-derived ependymoma cells, isogenic mouse NSCs expressing EZHIP-WT or EZHIP-M406K, and PFA PDX mouse models. Ependymomas are rare tumors; hence no priori sample size analysis was performed. Sample sizes were determined based on availability of tumor specimens or patients for MRS imaging and a minimum of n=3 was used for all analyses. Sample size for in vivo animal experiments was based on previously published in vivo panobinostat experiments by Grasso et al. in H3K27M xenografted animals which showed a minimum group size of three animals to detect a 50% difference in tumor growth between two groups with an alpha=0.05 and power=0.8. (34) Accordingly, sample size for animal studies ranged from n=3-8/group. No samples were excluded. Individuals blinded to the experimental design assessed MRS data or MRIs in a retrospective manner. Immunostaining analyses were performed by individuals blinded to the experimental design. All studies were conducted in compliance with REMARK and STARD guidelines. All identifiers from cases were removed before analysis. Two independent neuropathologists reviewed H&E sections and H3K27me3/ac immunostaining in a blinded manner. For in vivo experiments, animals (equal males/females) bearing PFA PDXs were randomly assigned to either control or treatment conditions, and in vivo bioluminescence and tumor measurements were assessed in a blinded manner. All in vitro experiments were performed with a minimum of 3 biologic replicates. All studies were performed after Institutional Review Board approval from respective institutions. Animal experiments were performed after approval from the University of Michigan Committee on Use and Care of Animals and were conducted as per NIH guidelines for animal welfare. MRS imaging in patients was conducted at Children’s Hospital Los Angeles and was compliant with the Health Insurance Portability and Accountability Act (the requirement to obtain informed consent was waived).
Statistical analyses
Data are represented as the means ± standard deviation (S.D.). Graphs were plotted and statistical analyses were performed using Prism software (versions 7 and 8, Graphpad,). The sample size (n) along with the statistical test performed and corresponding P-values are indicated in each figure or figure legend. Unpaired, two-tailed, two-sided, Student’s t test or analysis of variance (ANOVA) followed by multiple comparisons analysis were used to analyze data. Survival analyses were performed using Kaplan-Meier analyses with the Log-Rank test. Data were considered significant if P values were below 0.05 (95% confidence intervals). Correlational analyses were performed using 95% confidence intervals. Results from correlational analyses are represented by r, the Spearman’s correlation coefficient.
Supplementary Material
Acknowledgements:
We thank S. Mack for providing access to H3K27ac ChIP-seq files, P. Lewis for providing EZHIP plasmids and V. Basrur and A. Nesvizhskii for proteomics analyses.
Funding:
This work was funded by the Sontag Foundation (S.V.), Clinical Scientist Development Award - Doris Duke Charitable Foundation (S.V.), the Hyundai Hope On Wheels Foundation (S.V.), NINDS R01NS110572 (S.V.), and the Julie Taubman Reys emerging scholar award from the University of Michigan Taubman Institute (S.V.). This work was also supported by a fellowship award from The Robert Connor Dawes Foundation/ CERN Foundation: Ependymoma Cancer Research Network/ National Brain Tumor Society (C.C). The Venneti lab is/was supported by grants from Mathew Larson (S.V.), Sidney Kimmel (S.V.), St Baldrick’s (S.V.), Claire McKenna (S.V.), Chad Tough (S.V.), Alex Lemonade Stand (S.V.), Storm The Heavens (S.V.) and the University of Michigan Chad Carr Pediatric Brain Tumor Initiative (S.V.). M.P.O acknowledges support from the Spectrum Health MD/PhD Fellowship (M.P.O.) and the Aitch Foundation Graduate Fellowship (M.P.O.). C.L. acknowledges support from the Damon Runyon Cancer Research Foundation C.L., Matthew Larson Foundation C.L. and Pew-Stewart Scholars Program for Cancer Research C.L.. The Bluml lab at Children’s Hospital Los Angeles has been supported by the Rudi Schulte Research Institute (S.B.) and the Ian’s Friends Foundation (S.B.). The Lunt lab is supported by the AACR-Incyte Corporation NextGen Grant for Transformative Cancer Research, Grant Number 16-20-46-LUNT (S.Y.L.), and the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program, under Award No. W81XWH-15-1-0453 (S.Y.L.). D.N. was supported by National Institute of Health grants R01-CA222251 (D.N.), R01-CA204969 (D.N.), and R01-CA227622 (D.N.). C.K. acknowledges support from NIH/NINDS K08-NS099427-01 (C.K.). The Foreman lab is supported by the Tanner Seebaum Foundation (N.K.). Resources were provided, in part, by the UCSF Brain Tumor SPORE Biorepository NIH/NCI P50CA097257 (J.J.P.). CC acknowledges support from the DGIST Start-up Fund Program of the Ministry of Science and ICT (2021050001, C.C.)
Footnotes
Competing interests: JH has stock interests in AbbVie. Stephen Yip is a member of advisory boards and has received financial compensations for Amgen, AstraZeneca, Bayer, EMD Serono, Pfizer, and Roche. Stefan Pfister has European Patent Application 16710700.2 Dna-Methylation Based Method For Classifying Tumor Species. All other authors declare no competing interests.
Data and materials availability: H3K27ac ChIP-seq files from this study are deposited in NCBI under GEO: GSE134572. RNA-seq data and H3K27me3 ChIP-seq files from Bayliss et al. 2016 have been deposited in NCBI under GEO: GSE89452. Gene expression microarray data from Pajtler et al. 2015 were obtained from GEO: GSE64415, GSE65362. Gene expression microarray data from Witt et al. 2012 were obtained from GEO: GSE27287. EPD210 cells and PDX were obtained after MTA from J. Olson, Fred Hutchinson Cancer Research Center. MAF811, MAF928 and MAF1329 cells and PDXs were obtained after MTA from N. Foreman, University of Colorado. EP1NS cells were obtained after MTA from S. Pfister, DKFZ. mNSCs were obtained after MTA with St Jude’s Hospital from R.J. Gilbertson. NHA cells were obtained from C. B. Thompson, Memorial Sloan Kettering Cancer Center. Other materials can be obtained after MTA.
References:
- 1.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, Warnatz HJ, Ryzhova M, Mack S, Ramaswamy V, Capper D, Schweizer L, Sieber L, Wittmann A, Huang Z, van Sluis P, Volckmann R, Koster J, Versteeg R, Fults D, Toledano H, Avigad S, Hoffman LM, Donson AM, Foreman N, Hewer E, Zitterbart K, Gilbert M, Armstrong TS, Gupta N, Allen JC, Karajannis MA, Zagzag D, Hasselblatt M, Kulozik AE, Witt O, Collins VP, von Hoff K, Rutkowski S, Pietsch T, Bader G, Yaspo ML, von Deimling A, Lichter P, Taylor MD, Gilbertson R, Ellison DW, Aldape K, Korshunov A, Kool M, Pfister SM, Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27, 728–743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, Jones DT, Northcott PA, Garzia L, Bertrand KC, Wittmann A, Yao Y, Roberts SS, Massimi L, Van Meter T, Weiss WA, Gupta N, Grajkowska W, Lach B, Cho YJ, von Deimling A, Kulozik AE, Witt O, Bader GD, Hawkins CE, Tabori U, Guha A, Rutka JT, Lichter P, Korshunov A, Taylor MD, Pfister SM, Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20, 143–157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, Zhang X, Ramaswamy V, Jager N, Jones DT, Sill M, Pugh TJ, Ryzhova M, Wani KM, Shih DJ, Head R, Remke M, Bailey SD, Zichner T, Faria CC, Barszczyk M, Stark S, Seker-Cin H, Hutter S, Johann P, Bender S, Hovestadt V, Tzaridis T, Dubuc AM, Northcott PA, Peacock J, Bertrand KC, Agnihotri S, Cavalli FM, Clarke I, Nethery-Brokx K, Creasy CL, Verma SK, Koster J, Wu X, Yao Y, Milde T, Sin-Chan P, Zuccaro J, Lau L, Pereira S, Castelo-Branco P, Hirst M, Marra MA, Roberts SS, Fults D, Massimi L, Cho YJ, Van Meter T, Grajkowska W, Lach B, Kulozik AE, von Deimling A, Witt O, Scherer SW, Fan X, Muraszko KM, Kool M, Pomeroy SL, Gupta N, Phillips J, Huang A, Tabori U, Hawkins C, Malkin D, Kongkham PN, Weiss WA, Jabado N, Rutka JT, Bouffet E, Korbel JO, Lupien M, Aldape KD, Bader GD, Eils R, Lichter P, Dirks PB, Pfister SM, Korshunov A, Taylor MD, Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merchant TE, Current Clinical Challenges in Childhood Ependymoma: A Focused Review. J. Clin. Oncol 35, 2364–2369 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B, Hubner JM, Ramaswamy V, Jia S, Dalton JD, Haupfear K, Rogers HA, Punchihewa C, Lee R, Easton J, Wu G, Ritzmann TA, Chapman R, Chavez L, Boop FA, Klimo P, Sabin ND, Ogg R, Mack SC, Freibaum BD, Kim HJ, Witt H, Jones DTW, Vo B, Gajjar A, Pounds S, Onar-Thomas A, Roussel MF, Zhang J, Taylor JP, Merchant TE, Grundy R, Tatevossian RG, Taylor MD, Pfister SM, Korshunov A, Kool M, Ellison DW, Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 136, 211–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, Pekmezci M, McEachin RC, Cieslik M, Tamrazi B, Garcia BA, La Rocca G, Santi M, Lewis PW, Hawkins C, Melnick A, David Allis C, Thompson CB, Chinnaiyan AM, Judkins AR, Venneti S, Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl. Med 8, 366ra161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C, Yip S, Hukin J, Sun Y, Schipper MJ, Chavez L, Margol A, Pekmezci M, Chung C, Banda A, Bayliss JM, Curry SJ, Santi M, Rodriguez FJ, Snuderl M, Karajannis MA, Saratsis AM, Horbinski CM, Carret AS, Wilson B, Johnston D, Lafay-Cousin L, Zelcer S, Eisenstat D, Silva M, Scheinemann K, Jabado N, McNeely PD, Kool M, Pfister SM, Taylor MD, Hawkins C, Korshunov A, Judkins AR, Venneti S, Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol., (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, Bajic A, Juretic N, Deshmukh S, Venneti S, Muir TW, Garcia BA, Jabado N, Lewis PW, PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nature communications 10, 2146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain SU, Rashoff AQ, Krabbenhoft SD, Hoelper D, Do TJ, Gibson TJ, Lundgren SM, Bondra ER, Deshmukh S, Harutyunyan AS, Juretic N, Jabado N, Harrison MM, Lewis PW, H3 K27M and EZHIP Impede H3K27-Methylation Spreading by Inhibiting Allosterically Stimulated PRC2. Mol. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castel D, Kergrohen T, Tauziede-Espariat A, Mackay A, Ghermaoui S, Lechapt E, Pfister SM, Kramm CM, Boddaert N, Blauwblomme T, Puget S, Beccaria K, Jones C, Jones DTW, Varlet P, Grill J, Debily MA, Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3-K27M mutation. Acta Neuropathol. 139, 1109–1113 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Hubner JM, Muller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, Ellison DW, Pfister SM, Pajtler KW, Kool M, EZHIP / CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol., (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piunti A, Smith ER, Morgan MAJ, Ugarenko M, Khaltyan N, Helmin KA, Ryan CA, Murray DC, Rickels RA, Yilmaz BD, Rendleman EJ, Savas JN, Singer BD, Bulun SE, Shilatifard A, CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv 5, eaax2887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragazzini R, Perez-Palacios R, Baymaz IH, Diop S, Ancelin K, Zielinski D, Michaud A, Givelet M, Borsos M, Aflaki S, Legoix P, Jansen P, Servant N, Torres-Padilla ME, Bourc'his D, Fouchet P, Vermeulen M, Margueron R, EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nature communications 10, 3858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venneti S, Thompson CB, Metabolic Reprogramming in Brain Tumors. Annu. Rev. Pathol 12, 515–545 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Chung C, Sweha SR, Pratt D, Tamrazi B, Panwalkar P, Banda A, Bayliss J, Hawes D, Yang F, Lee HJ, Shan M, Cieslik M, Qin T, Werner CK, Wahl DR, Lyssiotis CA, Bian Z, Shotwell JB, Yadav VN, Koschmann C, Chinnaiyan AM, Bluml S, Judkins AR, Venneti S, Integrated Metabolic and Epigenomic Reprograming by H3K27M Mutations in Diffuse Intrinsic Pontine Gliomas. Cancer Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michealraj KA, Kumar SA, Kim LJY, Cavalli FMG, Przelicki D, Wojcik JB, Delaidelli A, Bajic A, Saulnier O, MacLeod G, Vellanki RN, Vladoiu MC, Guilhamon P, Ong W, Lee JJY, Jiang Y, Holgado BL, Rasnitsyn A, Malik AA, Tsai R, Richman CM, Juraschka K, Haapasalo J, Wang EY, De Antonellis P, Suzuki H, Farooq H, Balin P, Kharas K, Van Ommeren R, Sirbu O, Rastan A, Krumholtz SL, Ly M, Ahmadi M, Deblois G, Srikanthan D, Luu B, Loukides J, Wu X, Garzia L, Ramaswamy V, Kanshin E, Sanchez-Osuna M, El-Hamamy I, Coutinho FJ, Prinos P, Singh S, Donovan LK, Daniels C, Schramek D, Tyers M, Weiss S, Stein LD, Lupien M, Wouters BG, Garcia BA, Arrowsmith CH, Sorensen PH, Angers S, Jabado N, Dirks PB, Mack SC, Agnihotri S, Rich JN, Taylor MD, Metabolic Regulation of the Epigenome Drives Lethal Infantile Ependymoma. Cell 181, 1329–1345 e1324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillen AE, Riemondy KA, Amani V, Griesinger AM, Gilani A, Venkataraman S, Madhavan K, Prince E, Sanford B, Hankinson TC, Handler MH, Vibhakar R, Jones KL, Mitra S, Hesselberth JR, Foreman NK, Donson AM, Single-Cell RNA Sequencing of Childhood Ependymoma Reveals Neoplastic Cell Subpopulations That Impact Molecular Classification and Etiology. Cell Rep 32, 108023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gojo J, Englinger B, Jiang L, Hubner JM, Shaw ML, Hack OA, Madlener S, Kirchhofer D, Liu I, Pyrdol J, Hovestadt V, Mazzola E, Mathewson ND, Trissal M, Lotsch D, Dorfer C, Haberler C, Halfmann A, Mayr L, Peyrl A, Geyeregger R, Schwalm B, Mauermann M, Pajtler KW, Milde T, Shore ME, Geduldig JE, Pelton K, Czech T, Ashenberg O, Wucherpfennig KW, Rozenblatt-Rosen O, Alexandrescu S, Ligon KL, Pfister SM, Regev A, Slavc I, Berger W, Suva ML, Kool M, Filbin MG, Single-Cell RNA-Seq Reveals Cellular Hierarchies and Impaired Developmental Trajectories in Pediatric Ependymoma. Cancer Cell 38, 44–59 e49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM, Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brabetz S, Leary SES, Grobner SN, Nakamoto MW, Seker-Cin H, Girard EJ, Cole B, Strand AD, Bloom KL, Hovestadt V, Mack NL, Pakiam F, Schwalm B, Korshunov A, Balasubramanian GP, Northcott PA, Pedro KD, Dey J, Hansen S, Ditzler S, Lichter P, Chavez L, Jones DTW, Koster J, Pfister SM, Kool M, Olson JM, A biobank of patient-derived pediatric brain tumor models. Nat. Med 24, 1752–1761 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Pierce AM, Witt DA, Donson AM, Gilani A, Sanford B, Sill M, Van Court B, Oweida A, Prince EW, Steiner J, Danis E, Dorris K, Hankinson T, Handler MH, Jones KL, Karam SD, Serkova NJ, Vibhakar R, Foreman NK, Griesinger AM, Establishment of patient-derived orthotopic xenograft model of 1q+ posterior fossa group A ependymoma. Neuro Oncol. 21, 1540–1551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi YH, Woodfin AR, Misharin AV, Abshiru NA, Lulla RR, Saratsis AM, Kelleher NL, James CD, Shilatifard A, Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, Li M, Meng W, Ponnuswami A, Sun W, Ma J, Hulleman E, Swigut T, Wysocka J, Tang Y, Monje M, Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell 31, 635–652 e636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, Khazaei S, Koncar RF, Agnihotri S, Faury D, Ellezam B, Weil AG, Ursini-Siegel J, De Carvalho DD, Dirks PB, Lewis PW, Salomoni P, Lupien M, Arrowsmith C, Lasko PF, Garcia BA, Kleinman CL, Jabado N, Mack SC, Pervasive H3K27 Acetylation Leads to ERV Expression and a Therapeutic Vulnerability in H3K27M Gliomas. Cancer Cell 35, 782–797 e788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang X, Erkek S, Federation A, Song A, Lee C, Wang X, McDonald L, Morrow JJ, Saiakhova A, Sin-Chan P, Wu Q, Michaelraj KA, Miller TE, Hubert CG, Ryzhova M, Garzia L, Donovan L, Dombrowski S, Factor DC, Luu B, Valentim CLL, Gimple RC, Morton A, Kim L, Prager BC, Lee JJY, Wu X, Zuccaro J, Thompson Y, Holgado BL, Reimand J, Ke SQ, Tropper A, Lai S, Vijayarajah S, Doan S, Mahadev V, Minan AF, Grobner SN, Lienhard M, Zapatka M, Huang Z, Aldape KD, Carcaboso AM, Houghton PJ, Keir ST, Milde T, Witt H, Li Y, Li CJ, Bian XW, Jones DTW, Scott I, Singh SK, Huang A, Dirks PB, Bouffet E, Bradner JE, Ramaswamy V, Jabado N, Rutka JT, Northcott PA, Lupien M, Lichter P, Korshunov A, Scacheri PC, Pfister SM, Kool M, Taylor MD, Rich JN, Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553, 101–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, White E, Tang B, Orisme W, Gupta K, Rusch M, Chen X, Li Y, Nagahawhatte P, Hedlund E, Finkelstein D, Wu G, Shurtleff S, Easton J, Boggs K, Yergeau D, Vadodaria B, Mulder HL, Becksfort J, Gupta P, Huether R, Ma J, Song G, Gajjar A, Merchant T, Boop F, Smith AA, Ding L, Lu C, Ochoa K, Zhao D, Fulton RS, Fulton LL, Mardis ER, Wilson RK, Downing JR, Green DR, Zhang J, Ellison DW, Gilbertson RJ, C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 506, 451–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shackelford DB, Shaw RJ, The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazurek M, Litak J, Kamieniak P, Kulesza B, Jonak K, Baj J, Grochowski C, Metformin as Potential Therapy for High-Grade Glioma. Cancers (Basel) 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayoub R, Ruddy RM, Cox E, Oyefiade A, Derkach D, Laughlin S, Ades-Aron B, Shirzadi Z, Fieremans E, MacIntosh BJ, de Medeiros CB, Skocic J, Bouffet E, Miller FD, Morshead CM, Mabbott DJ, Assessment of cognitive and neural recovery in survivors of pediatric brain tumors in a pilot clinical trial using metformin. Nat. Med 26, 1285–1294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Kuang Y, Liang S, Wang L, Metformin inhibits cell proliferation in SKM-1 cells via AMPK-mediated cell cycle arrest. J. Pharmacol. Sci 141, 146–152 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Chen EC, Liang X, Yee SW, Geier EG, Stocker SL, Chen L, Giacomini KM, Targeted disruption of organic cation transporter 3 attenuates the pharmacologic response to metformin. Mol. Pharmacol 88, 75–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janzer A, German NJ, Gonzalez-Herrera KN, Asara JM, Haigis MC, Struhl K, Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. U. S. A 111, 10574–10579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown ZZ, Muller MM, Jain SU, Allis CD, Lewis PW, Muir TW, Strategy for "detoxification" of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J. Am. Chem. Soc 136, 13498–13501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, Quist MJ, Davis LE, Huang EC, Woo PJ, Ponnuswami A, Chen S, Johung TB, Sun W, Kogiso M, Du Y, Qi L, Huang Y, Hutt-Cabezas M, Warren KE, Le Dret L, Meltzer PS, Mao H, Quezado M, van Vuurden DG, Abraham J, Fouladi M, Svalina MN, Wang N, Hawkins C, Nazarian J, Alonso MM, Raabe EH, Hulleman E, Spellman PT, Li XN, Keller C, Pal R, Grill J, Monje M, Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med, (2015). [DOI] [PubMed] [Google Scholar]
- 35.Lin GL, Wilson KM, Ceribelli M, Stanton BZ, Woo PJ, Kreimer S, Qin EY, Zhang X, Lennon J, Nagaraja S, Morris PJ, Quezada M, Gillespie SM, Duveau DY, Michalowski AM, Shinn P, Guha R, Ferrer M, Klumpp-Thomas C, Michael S, McKnight C, Minhas P, Itkin Z, Raabe EH, Chen L, Ghanem R, Geraghty AC, Ni L, Andreasson KI, Vitanza NA, Warren KE, Thomas CJ, Monje M, Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci. Transl. Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.