Abstract
Mutations in breast cancer type 1 susceptibility protein (BRCA1) and its heterodimeric binding partner BARD1 confer a high risk for the development of breast and ovarian cancers. The sole enzymatic function of the BRCA1/BARD1 complex is as a RING-type E3 ubiquitin (Ub) ligase, leading to the deposition of Ub signals onto a variety of substrate proteins. Distinct types of Ub signals deposited by BRCA1/BARD1 (i.e., degradative vs. non-degradative; mono-Ub vs. poly-Ub) on substrate proteins mediate aspects of its function in DNA double-stranded break repair, cell-cycle regulation, and transcriptional regulation. While cancer-predisposing mutations in both subunits lead to the inactivation of BRCA1/BARD1 ligase activity, controversy remains as to whether its loss is directly linked to tumorigenesis. Investigation of BRCA1/BARD1 substrates using rigorous, well-validated mutants and systems will ultimately clarify the role of its ligase activity in cancer and possibly establish prognostic and diagnostic metrics for patients with mutations. In this review, we discuss the Ub ligase function of BRCA1/BARD1, highlighting experimental approaches, mechanistic considerations, and reagents that are useful in the study of substrate ubiquitylation. We also discuss the current understanding of two well-established BRCA1/BARD1 substrates (nucleosomal H2A and estrogen receptor α) and several recently discovered substrates (p50, NF2, Oct1, and LARP7). Lessons from the current body of work should provide a road map to researchers examining novel substrates and biological functions attributed to BRCA1/BARD1 Ub ligase activity.
Introduction
Mutations in the BRCA1 gene were first linked to increased risk for familial breast cancer over 30 years ago1–4.Since then, BRCA1 and its protein product BRCA1 (breast cancer type 1 susceptibility protein) have been investigated by clinical researchers and basic scientists alike. Thousands of BRCA1 variants of unknown significance still require classification as pathogenic or benign, potentially influencing medical decisions of individuals carrying such mutations. More recently, mutations in the BARD1 gene associated with high risk for breast and ovarian cancer have been identified5–7. Together the two gene products form a heterodimeric protein complex, BRCA1/BARD1, that acts as a tumor suppressor by serving as a central regulator and guardian of genomic integrity throughout the cell cycle8. Although most past research has focused mainly on BRCA1, conditional knockout of either BRCA1 or BARD1 in mouse mammary epithelial cells leads to genomic instability and indistinguishable carcinomas, supporting the prevailing view that the proteins’ critical tumor-suppressor functions arise from the heterodimer9.
BRCA1/BARD1 is largely localized to the nucleus and is best characterized in its critical role in double-stranded DNA break repair by promoting homologous recombination (HR). It also functions in transcriptional regulation, cell-cycle control, centrosome regulation, metabolic regulation, and DNA decatenation10–13. In general, these functions are attributed to the molecular scaffolding properties of BRCA1/BARD1, which is a component of numerous extremely large multi-protein complexes14,15. The sole biochemical function defined for BRCA1/BARD1 is as a ubiquitin (Ub) E3 ligase16,17. It is well established that the ligase activity is required for BRCA1/BARD1’s role in the maintenance of genome integrity and transcriptional regulation, but whether the activity is required for other processes in which BRCA1/BARD1 is implicated remains unclear. A direct link between BRCA1/BARD1 E3 ligase activity and its role as a tumor suppressor is not universally accepted, with studies arriving at conflicting conclusions18–20. A major hurdle to a more complete understanding of BRCA1/BARD1 ligase function and its ties to tumor suppression is a lack of clarity regarding how ubiquitylation of each of the numerous cellular protein targets identified to date regulates their function and the processes in which they participate (Table I). Here we focus on aspects of BRCA1/BARD1 related to its E3 ligase function, paying special attention to challenges and strategies that can address remaining gaps in knowledge.
Table I.
BRCA1/BARD1 substrates.
| Substrate | Biological function(s)* | Observed in cells/in vitro? | BRCA1 (BC)/BARD1 (BD) binding site | Substrate Ub-attachment site (linkage-type) | E2-enzyme | Substrate binding site | Ref(s) |
|---|---|---|---|---|---|---|---|
| Aurora Kinase B | CCR | Y/N | n.d. | n.d. | n.d. | n.d. | 116 |
| Cdc25C | CCR | Y/Y | BC: BRCTs | n.d. (poly-Ub, degradative) | Ube2Dα | n.d. | 117 |
| Claspin | DDR | Y/Y | n.d. | K60/K96 (mono-Ub in vitro) | Ube2Dα | 1–331 | 118, 119 |
| CtIP | DDR, TR | Y/Y | BC: BRCTs | n.d. (poly-Ub, non-degradative) | Ube2Dα | 322–333 (pSer327) | 45, 46, 120 |
| Cyclin B1 | CCR | Y/Y | BC: BRCTs | n.d. (poly-Ub, degradative) | Ube2K, Ube2D | n.d. | 117 |
| estrogen receptor ⍺ | TR | Y/Y | BC: IDR BD: IDR |
K302 (mono-Ub) | Ube2D | Ligand binding domain | 29, 44, 86, 90, 91 |
| H2A | DDR, TR | Y/Y | BC: RING BD: RING |
K125/127/129 (mono-Ub) | Ube2D, Ube2E | histone surface, acidic patch | 37, 43, 93, 94 |
| LARP7 | CCR, DDR | Y/Y | BC: BRCTs BD: BRCTs |
n.d. (poly-Ub, K48-linked) | Ube2Dα | around pThr440 | 50 |
| macroH2A1 | CCR, TR | Y/Y | BC: RING BD: RING |
K123 (mono-Ub) | Ube2Dα | likely same as H2A | 85 |
| NF2 | Hippo growth signaling | Y/N | BC: BRCTs | K159/269/274/364/387/396/439/449 (poly-Ub K6-, K27-, K29-, K63-linked) | n.d. | FERM domain | 48 |
| Nuclephosmin | n.d. (colocalizes in mitosis) | Y/Y | BC: 1–222 BD:1–320 |
n.d. (poly-Ub, K6-, K29-linked) | Ube2Dα | n.d. | 121 |
| Oct1 | Metabolic regulation | Y/Y | K9/403 (poly-Ub, K48-linked) | Ube2Dα | n.d. | 107 | |
| P50 | CCR, chromosome stability | Y/Y | BD: BRCTs | K354/356 (mono-Ub) | Ube2Dα | around pSer337 | 49 |
| progesterone receptor | TR | Y/Y | n.d. | n.d. (poly-Ub, degradative) | Ube2D | n.d. | 122 |
| RPB1 | DDR, TR | Y/Y | BC: BRCTs | n.d. (poly-Ub, degradative) | Ube2Dα | C-terminal domain (pSer5) | 47, 123, 124 |
| RPB8 | DDR, TR | Y/Y | n.d. | n.d. (poly-Ub K6-linked) | Ube2Dα, Ube2W | n.d. | 33, 125 |
| TFIIE | TR | N/Y | n.d. | n.d. (likely mono-Ub) | Ube2Dα | n.d. | 124 |
| Topoisomerase IIa | DNA decatenation | Y/N | n.d. | n.d. (poly-Ub, non-degradative) | n.d. | n.d. | 13 |
| γ-tubulin | CCR (centrosome regulation) | Y/Y | BC: BRCTs & IDR | K48/344 (mono-Ub) | Ube2Dα | n.d. | 52, 53, 126 |
DDR, DNA damage repair; TR, transcriptional regulation; CCR, cell-cycle regulation. Reported functions limited to those from references reporting BRCA1/BARD1-mediated Ub of substrate.
(Ube2Dα) other E2s not tested
BRCA1/BARD1 is a RING-type E3 ligase
BRCA1 and BARD1 form a large obligate heterodimer via their N-terminal RING domains, creating a structure that is an archetypal RING-type E3 ligase21,22 (Fig 1A, B). Members of this large E3 class (more than 600 in humans) simultaneously bind a substrate and a Ub-conjugating (E2) enzyme that holds activated Ub (“E2~Ub”) and must therefore contain distinct substrate-and E2-binding regions23. RING domains bind E2s, potentially leaving the remainder of the complex for substrate binding. Intriguingly, BRCA1 and BARD1 are the only two proteins in the human proteome that contain both an N-terminal RING (Really Interesting New Gene) domain and C-terminal tandem BRCT (BRCA1 C-terminal) domains and both contain substantial intrinsically disordered content characteristic of molecular scaffolds (FIG 1a)24. BARD1 also has an ankyrin-repeat domain (Ank) adjacent to its BRCT domain; these are usually associated with protein-protein interactions.
Figure 1. An overview of the features of the BRCA1/BARD1 Ubiquitin ligase.
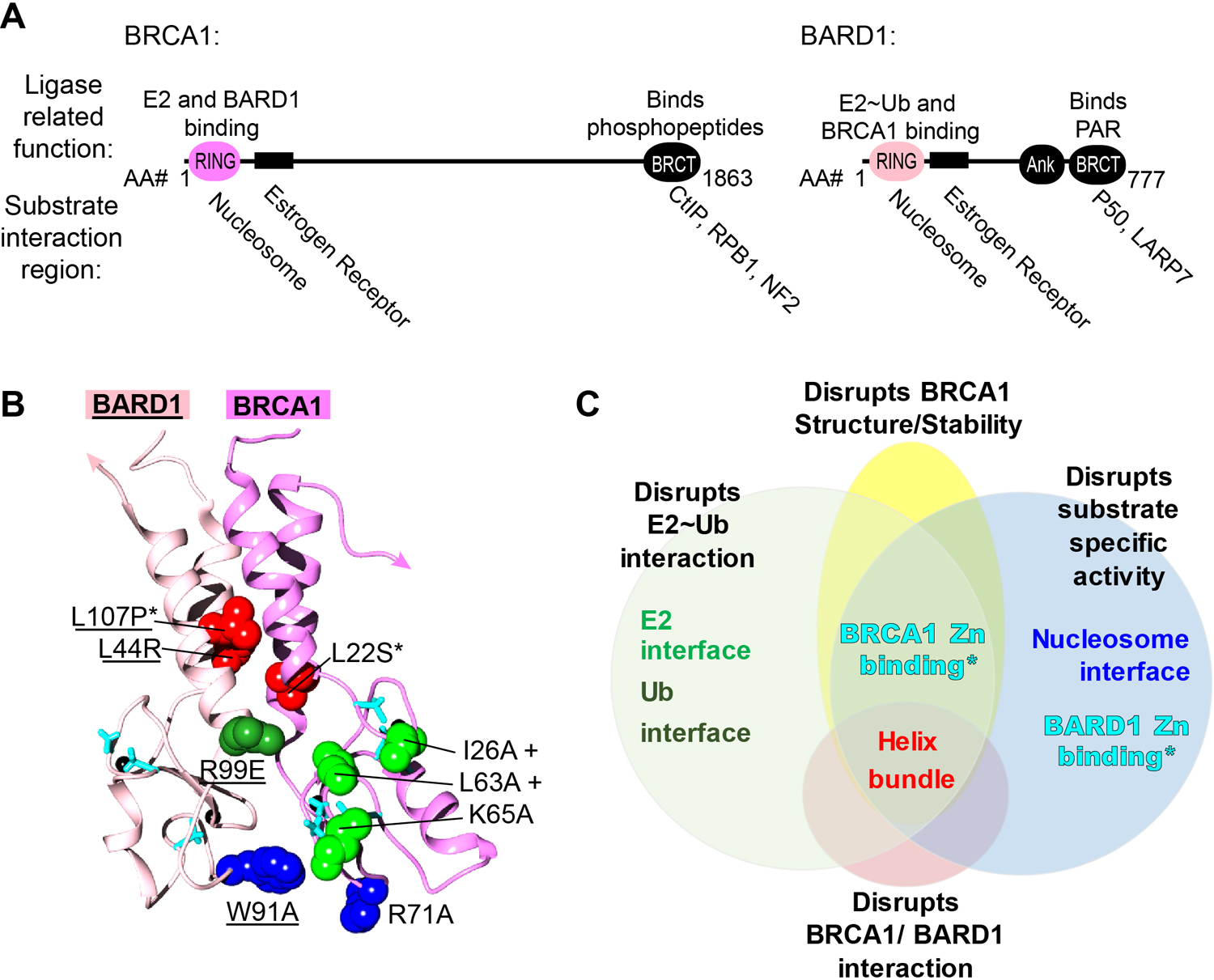
A) The domain structure and function of BRCA1 and BARD1 domains. Folded domains of BRCA1 and BARD1 are depicted with ovals and their corresponding domain names: really interesting new gene (RING), BRCA1 C-terminal (BRCT), and ankyrin repeats (ARD). Substrate binding domains within the intrinsically disordered regions are represented with rectangles. Domain functions related to E3 ligase activity are listed above domains. Substrates are listed below the region of protein with which they interact. B) The solution structure of RING domains from BRCA1 and BARD1 (PDB 1JM7) are shown in magenta and pink respectively. The sidechains at mutation sites used to study the structure/function relationship are shown in spheres or sticks: BARD1 mutations are underlined, cancer-associated mutations are either labeled with an asterisks or in the case of zinc-coordinating mutation sites depicted in cyan sticks (BRCA1 C24R, C39S/R/Y/W, C44S/Y/F, C47S/Y/F, C61G, C64R/Y/W and BARD1 C53W, C71Y, and C83R). The colors of mutation site sidechains correspond with their functions in Panel C. C) Mutations are categorized in a Venn diagram according to the properties and functions they affect.
A challenge faced in the study of RING E3s is that they do not possess a catalytic residue that is directly involved in the Ub transfer reaction. Instead, RINGs facilitate direct transfer of Ub from an E2~Ub to a substrate. While prevalent cancer-associated mutations in BRCA1 RING zinc-coordinating cysteine residues eliminate E3 ligase activity towards all known substrates, they do so by disrupting the structural integrity of the RING and possibly by destabilizing BRCA1 and BARD1 in cells16,25–27. No single mutation has been discovered that can both generate a completely “ligase-dead” BRCA1/BARD1 and leave the structure and other potential functions intact28. Without such a tool, it is difficult to parse out the role of ligase activity from other potential cellular functions. As discussed below, this situation has contributed to the ongoing controversy regarding the role of E3 ligase activity in BRCA1 tumor-suppressor function because a BRCA1 RING mutation presumed to be “ligase-dead” (Ile26Ala) is not truly dead29. There are multiple substrate-binding regions in both subunits of BRCA1/BARD1 (Table I).
Substrates may also bind the ligase indirectly via other binding partners. Numerous BRCA1/BARD1 binding partners have been identified, any of which could be substrates. However, only a subset of reported BRCA1/BARD1-binding proteins have been demonstrated explicitly to be ubiquitylated and an even smaller subset has been rigorously validated as BRCA1/BARD1-dependent substrates. Regions known to interact with established substrates are noted in Figure 1A. The number of possible substrates combined with the large range of possible interaction sites has made characterization of relevant substrates challenging.
Gold standards for understanding Ub E3 ligase function are 1) the ability to reconstitute the activity of interest with high fidelity using specified components and 2) structural information for complexes containing E2, E3, and substrate. Below, we identify challenges posed by features of BRCA1/BARD1 that must be considered when designing in vitro, cellular, or animal studies.
Challenges associated with E2s.
RING E3s must assemble the relevant ubiquitylation machinery to promote substrate modification. Thus, a first step needed to understand RING E3 function requires knowing the relevant E2(s) with which it works. BRCA1/BARD1 can bind to and function with nine of the ~36 human E2s, adding complexity to the study of BRCA1/BARD1 function30. Despite this, the vast majority of studies use only the Ube2D family of E2s. Though general mechanistic principles are likely similar for all the E2s, there are bound to be idiosyncratic differences that can confound interpretation of results. An important case in point is the widely used I26A-BRCA1 variant. Originally reported to lack detectable activity in association with the E2 Ube2D3 (UbcH5c) in vitro, it has subsequently been observed that I26A-BRCA1 retains residual activity with other BRCA1/BARD1-partnering E2s29,31. This raises the possibility that I26A-BRCA1 has sufficient ligase activity to mask potential effects of true loss-of-function in cells or animals.
The general rule is that an E2 specifies the Ub product generated in RING E3-directed reactions: attachment of a single Ub (mono-Ub) or one of seven possible poly-Ub chain types32. Different ubiquitin products lead to distinct cellular outcomes (e.g., degradative and non-degradative) that alter how best to investigate them in cells. Different pairings of E2s and BRCA1/BARD1 produce mono-Ub attached to lysine sidechains (Ube2E1/2/3), mono-Ub attached to the N-terminus of a substrate (Ube2W), and several different poly-Ub chain types (Ube2D1/2/3, Ube2K, Ube2N)30,33. BRCA1/BARD1 preferentially catalyzes atypical, non-degradative K6-linked polyubiquitin chains with the E2 enzymes Ube2D1/2/3 in autoubiquitylation reactions in vitro and in cells34,35. The molecular functions of K6-linked Ub chains synthesized by BRCA1/BARD1 remain a major unanswered question, though they likely play a role in some aspect of DNA DSB repair36. BRCA1/BARD1 catalyzes K48- and K63-linked chain formation with the E2s Ube2K and Ube2N/UBE2V, respectively30. In vitro, BRCA1/BARD1-directed synthesis of these chain linkages requires a different E2 to attach the first Ub to substrate, in a so-called priming step, as these chain-building E2s are inefficient at performing Ub transfer to a lysine that is not on an acceptor Ub. In contrast, Ube2D family members can catalyze both types of reactions, with the specificity determined at least in part by the substrate itself. For example, the E2/E3 pairing of Ube2D1/2/3 and BRCA1/BARD1 generates K6-linked chains in auto-ubiquitylation reactions but mono-ubiquitylates nucleosomal H2A and ERα5,29,34. This indicates that BRCA1/BARD1 and Ube2D assemble into mechanistically distinct structures with the various substrates that allow for either mono- or poly-ubiquitylation. Deciphering the nature of the ubiquitin product deposited on a novel BRCA1/BARD1 substrate is likely to yield insights into the consequence of the modification as well as the E2s that may be involved. This can be accomplished using mass spectrometry, ubiquitin lysine mutations, chain-linkage specific Ub antibodies, or a combination of these tools.
The BRCA1-binding E2s recognize a similar binding surface on the RING domain, making it virtually impossible to find mutations that specifically ablate one E2’s interaction while leaving others intact31. Furthermore, no single-site mutation other than those that disrupt RING structure has been discovered that will render BRCA1/BARD1 “dead” to all its E2s. The E2s that work with BRCA1/BARD1 include several with overlapping and/or redundant activities, especially the three isoforms of Ube2D. This feature may make a strategy of using single E2 knock-out strains uninformative or, at least, confounding. Furthermore, the requirement for both a priming E2 (i.e., an E2 that can place the first Ub onto a substrate) and a chain-building E2, both of which work with BRCA1/BARD1, adds further complications to parsing out BRCA1/BARD1-dependent substrate ubiquitylation. Best practices for in vitro studies are to test the entire panel of E2s, alone and in pairs, with any putative substrate.
Approaches that may overcome challenges.
A triple mutant BRCA1 I26A/L51A/K65A abrogates ligase function with all BRCA1-interacting E2s and does not appear to have defects in BARD1 heterodimerization or protein stability29 (Fig 1B, C). Introduction of the triple-mutant in cells and animals could clarify the controversial role of BRCA1/BARD1 ligase activity in DNA damage, tumor suppression, and other cellular processes. A designed BARD1 mutation (R99E) inactivates E3 activity toward all substrates tested to date for reactions using Ube2D family members as the E237 (dark green, Fig 1B, C). Complementary mutation of ubiquitin suggested that Arg99 helps to stabilize the more active closed state of the E2~Ub conjugate, a general mechanism of action among RING-type E3 ligases37–39. However, it is not known whether the BARD1 Arg99 mutation eliminates activity with all E2 partners of BRCA1/BARD1, so results obtained using this BARD1 mutant in cells should be interpreted with caution.
In addition to approaches that rely on loss of ligase activity to identify substrates and/or to assess the cellular consequences of their BRCA1-directed ubiquitylation, use of a hyperactive E3 ligase may be informative. Such species may yield increased levels of products without the potential pitfalls that over-expression at non-physiological ligase protein levels can entail. A deep mutational screen identified residues in the BRCA1 RING that, when mutated, enhance its ligase activity29,40. Specifically, tandem introduction of L51W and K65R in BRCA1 yields a hyperactive ligase. However, the hyperactive variant has lower specificity for E2 pairings and ubiquitin linkages (mono-Ub vs. chain building) in substrate and autoubiquitylation assays, indicating that wild-type BRCA1 ligase activity is tuned to work with specific E2s to promote different Ub linkages in a context-dependent manner (substrate vs. autoubiquitylation). Notably, combination of hyper-activating mutations with the ligase-dead C61G mutant in cis (L51W/K65R/C61G) restores activity of the Zn2+-coordinating mutant to wild-type levels, implying there could be strategies that stabilize the cancer-associated mutant protein to restore its activity.
Considerations about BRCA1/BARD1 constructs used for in vitro and cellular/in vivo investigations
A minimal BRCA1/BARD1 RING heterodimer consisting of the first ~110 residues of each subunit is sufficient to bind to an E2 and to stimulate the discharge of ubiquitin. These constructs contain the RING domain and flanking α-helices that are necessary and sufficient for formation of the BRCA1/BARD1 heterodimer21. Although both BRCA1 and BARD1 contain RING domains, only the BRCA1 RING binds E2s30,31. The minimal RING constructs of BRCA1/BARD1 have provided important biochemical and structural insights into the underlying mechanism of Ub transfer from an associated E2~Ub. However, additional BRCA1/BARD1-based reagents are required to tackle questions regarding substrate-specific modification.
Robust methodology has been developed to observe RING E3 ligase-mediated ubiquitylation in vitro and all required components to reconstitute a ubiquitylation reaction (E1, E2, Ub) are available commercially or through services such as Addgene41. Biochemically pure recombinant BRCA1/BARD1 heterodimer constructs can be generated by co-expression in E. coli31. There are two widely used versions: 1) a minimal RING construct composed of BRCA1(1–112)/BARD1(26–142) and 2) a longer construct that contains the RING heterodimer and portions of both IDRs BRCA1(1–302)/BARD1(26–327). The minimal construct is sufficient to bind to an E2 and to stimulate the discharge of ubiquitin in so-called E2~Ub discharge reactions that monitor enhancement of Ub release from an E2 by BRCA1/BARD142. This construct is not detectably auto-ubiquitylated and, with the exception of nucleosomal histone H2A, does not contain a substrate-binding region43. The longer construct can serve as a proxy substrate in autoubiquitylation assays, making it useful in direct assessment of the intrinsic ligase activity of point mutations on BRCA1/BARD1 or an E2 of interest. BRCA1(1–302)/BARD1(26–327) is also active in substrate-level ubiquitylation reactions with estrogen receptor-α29,44.
The truncated BRCA1/BARD1 constructs described above likely lack recognition domains for most substrates. For example, the C-terminal BRCTs of BRCA1 or BARD1 are required for binding the substrates CtIP, RPB1, NF2, p50, and LARP745–51 (Fig 1A). Furthermore, substrates may interact with multiple regions of BRCA1/BARD1, necessitating thorough analysis of all possible interaction sites, for example ERα and γ-tubulin44,52,53 (Fig 1A). Production of biochemical quantities of highly purified intact full-length BRCA1/BARD1 has long been a roadblock due to the large size and high amount of intrinsic disorder of the full-length proteins. A robust expression and purification scheme to generate sub-milligram quantities of high-quality full-length BRCA1/BARD1 using a baculovirus-based expression system in insect cells is now available54. This important development provides a new gold standard for production of BRCA1/BARD1 for substrate ubiquitylation assays. The published protocol employs FLAG immunoprecipitation of the complex followed by ion-exchange chromatography, but this can be adapted to incorporate a twin Strep-tag appended to the N-terminus of BARD1, yielding comparable results at a significantly decreased cost55. The ability to generate biochemical-quality full-length BRCA1/BARD1 represents a major advance that should propel studies of native substrates.
Post-translational modifications (PTMs) either on the substrate, on BRCA1/BARD1, or both may affect ligase activity. PTMs including acetylation, phosphorylation, proline isomerization, ubiquitylation, and SUMOylation have been reported on BRCA1/BARD156–60. Several have been observed to modulate ligase function in vitro and possibly in vivo. Phosphorylation and ubiquitylation are often engaged in crosstalk and BRCA1 is modified by DNA damage checkpoint kinases throughout the protein57,61. Phosphorylation of BRCA1/BARD1 by aurora kinase A has been shown to dampen its ligase activity, and the cell-cycle kinase CDK2/cyclin E1 has been observed to suppress the E3 ligase activity of BRCA1/BARD1 indirectly through depletion of its protein levels62,63. The BRCT domains of each subunit bind to phosphorylated substrates and other binding partners, allowing for the possibility that additional BRCA1/BARD1 substrates are recruited via recognition of phosphorylated sites. In the future, methods to generate defined modified protein reagents and/or to manipulate specific sites of modification on BRCA1/BARD1 and/or its substrates will be needed to sort out these important aspects of regulation.
BRCA1/BARD1 itself can serve as a substrate for ubiquitylation, either through its own ligase activity (i.e., autoubiquitylation) or by other cellular ligases64–69. How or if ubiquitylation of BRCA1/BARD1 directly affects its ligase activity remains an open question. The modification could affect intrinsic ligase activity and/or could serve to recruit certain substrates. BRCA1/BARD1 autoubiquitylation is reported to enhance its ligase activity in vitro on free histones as a proxy substrate59. But neither a mechanism by which this effect works nor evidence that it affects ubiquitylation of a bona fide substrate are yet available. Ubiquitylated BRCA1 can be detected in cells after DNA damage. The ubiquitin binding domain of UBXN1 was shown to bind to autoubiquitylated BRCA1/BARD1 and suppress its ligase function in vitro and in cells70. However, the significance of ubiquitylation of BRCA1/BARD1 as a regulatory signal with an outcome other than degradation remains to be clearly defined71.
BRCA1 is SUMOylated at lysine residues proximal to the RING domain60. The responsible SUMO ligases are important for HR following DNA damage, and SUMOylation of BRCA1 was found to enhance its E3 ligase activity in vitro as judged by its ability to form poly-Ub chains. Mutation of a BRCA1 lysine targeted for SUMOylation or of part of the SUMO ligase recognition motif decreased the co-localization of BRCA1 with conjugated ubiquitin, suggesting that this modification may also contribute to its ligase activity in cells. As BRCA1/BARD1 is the target of myriad PTMs that influence its biochemical properties including Ub ligase function, a full understanding of substrate ubiquitylation will require careful and thorough parsing of these effects.
Finally, an important consideration for BRCA1/BARD1 studies is ensuring proper maintenance of the heterodimer. BRCA1/BARD1 heterodimerization is not only required for nuclear localization but also for ensuring the stability of the complex16,72,73. Heterodimerization is thought to mask degron sequences located near the RING domains of each subunit68,74. Attempts to deplete one subunit can result in concomitant depletion of the other75. Subsequent replacement of one subunit by overexpression via transient transfection does not necessarily restore the new BRCA1/BARD1 complex to endogenous levels5,76,77. Therefore, it is important to co-transfect both BRCA1 and BARD1 and to confirm proper heterodimerization of the ectopically-expressed protein by immunoprecipitation when designing rescue experiments to test the effects of mutations. A superior method is to generate stable cell lines expressing siRNA-resistant BRCA1 or BARD1, and subsequently deplete the endogenous protein. This approach has been successfully employed for both BRCA1 and BARD1 to study ligase activity using the Flp-FRT recombination system in multiple cell-types19,37. Additionally, a BARD1 auxin-inducible degron (AID) system in HCT-116 cells has been developed and employed to study its function in DNA damage repair78,79. However, these two methods (Flp-FRT and AID) have yielded conflicting data about the requirements for BRCA1/BARD1 ligase activity in DNA damage repair using the Ube2D-deficient BARD1 Arg99Glu mutant, necessitating further comparison of the two methods and clarification of the effects of this mutant37,78.
Cellular Substrates of BRCA1/BARD1 ligase activity
The biological outcomes of BRCA1/BARD1 ligase activity are ultimately mediated through its cellular substrates. A full understanding of the cellular function of BRCA1/BARD1 requires knowledge of its cellular substrates, the Ub signals generated, and the functional consequences of the substrate modification. In addition to the difficulty of identifying E2s associated with modification of a particular substrate, many substrates may be targeted by more than one E3 ligase, so the effects of depletion of a particular E3 may be difficult to discern. Furthermore, BRCA1/BARD1 can generate non-degradative signals, so the powerful methods that rely on changes in protein levels associated with E3 ablation will not be applicable. Finally, in vitro validation of substrate modification may require use of full-length BRCA1/BARD1 which, until recently, has not been a viable option. Despite these challenges, putative BRCA1/BARD1 substrates have been identified primarily by testing known interaction partners for BRCA1/BARD1-dependent ubiquitylation. Several previous reviews provide in-depth summaries on all but the most recently discovered substrates80–83. The identities of known substrates reveal that BRCA1/BARD1 can target a diverse subset of nuclear proteins and deposit different ubiquitin marks that can serve degradative, stabilizing, or signaling roles (Table I). To illustrate some best practices and highlight the biological functions of several substrates, we elaborate on two exemplary BRCA1/BARD1 substrates – nucleosomal histone H2A and ERα and discuss four recently identified substrates that have not been presented in previous reviews (p50, Oct1, NF2, and LARP7).
Discovery of BRCA1/BARD1 substrates
In addition to candidate-based approaches, putative substrates have been identified by affinity enrichment of ubiquitylated proteins upon overexpression of BRCA1/BARD1 in 293T cells84,85. Over one-hundred proteins with increased ubiquitylation were detected, providing a rich pool of potential substrates to be validated. Among them, the histone variant macroH2A1 was identified and mono-ubiquitylation of a specific lysine residue (K123) was shown to be involved in regulation of cellular senescence85. Two proteins involved in transcriptional regulation and DNA damage repair, GADD45GIP1 and HLTF, were observed to be ubiquitylated in a BRCA1/BARD1-dependent manner in the mass-spectrometry screen and verified by Western blotting, although biological roles for ubiquitylation of these putative substrates have yet to be investigated. However, increased ubiquitylation levels of previously validated BRCA1/BARD1 substrates nucleophosmin (NPM1), ERα, and canonical H2A ubiquitylated at K125/127/129 were not detected. Substrates may be missed due to transient ubiquitylation/deubiquitylation, cell-type specific effects (e.g., mammary vs. non-mammary), or lack of peptide coverage in mass spectrometry analysis.
Considerations in substrate characterization
For substrate ubiquitylation to occur, BRCA1/BARD1 must simultaneously bind an activated E2~Ub conjugate and a substrate protein, bringing the two into proximity. As mentioned previously, substrates may bind throughout BRCA1 or BARD1 or, possibly, may interact indirectly. The possibility of multiple interacting regions should be entertained when characterizing substrate ubiquitylation. Pull-down assays using isolated domains as bait can be an effective method to identify substrate binding domains. Sequential C-terminal truncation constructs of BRCA1 and BARD1 that leave the N-terminal RING heterodimer unit intact are useful reagents for in vitro ubiquitylation assays with substrate candidates. However, relevant substrate-binding interactions and ubiquitylation may be decoupled. For example, the interaction of LARP7, a 7SK RNA binding protein that controls RNAPII pausing, is likely mediated through the BARD1 C-terminal BRCT domain in a phospho-dependent manner in cells50. But a construct that lacked the BRCTs (BRCA1(1–304)/BARD1(26–327)) catalyzed poly-ubiquitylation of LARP7 in vitro, suggesting differential requirements for cellular recruitment compared to in vitro Ub modification. Such considerations are especially important when characterizing the ubiquitylation of putative substrates.
An emerging feature of BRCA1/BARD1-dependent ubiquitylation is that in many cases specific substrate lysine residues are targeted, in contrast to most cases in which poly-Ub chains are attached to a substrate with no apparent lysine discrimination. Specifically-targeted residues may play a critical role in downstream signaling, so their identification using mutagenesis or mass-spectrometry analysis is a worthwhile endeavor. Lysine to arginine mutations that abrogate substrate ubiquitylation may be a viable way to characterize the effects of BRCA1/BARD1 ligase activity. Important considerations for such an approach include the possibility that other substrate lysines may be modified in the absence of its preferred target residues, or that the mutated lysine residues are also targeted for alternative PTMs such as acetylation44,86. An additional strategy in which substrates with Ub genetically fused to their C-terminus has been successfully employed to study substrates that are mono-ubiquitylated by BRCA1/BARD1 at or near their C-terminus. Despite the fact that the N-terminus of Ub is tethered to the substrate in the genetic fusions while bona fide products are attached to the Ub C-terminus, such chimeras have been shown to rescue the effects of BRCA1/BARD1 ligase deficiency for both nucleosomal H2A and p505,37,49,87,88.
Case Studies and recently discovered substrates.
Below we discuss the two best-characterized BRCA1/BARD1 substrates, estrogen receptor (ERα) and histone H2A, and recently discovered substrates. The examples include substrates that are modified with non-degradative Ub signals (mono-ub and K63-linked poly-Ub) and others that are modified with degradative K48-linked chains, highlighting the spectrum of biological outcomes experienced by BRCA1/BARD1 substrates.
Estrogen Receptor-α
Although BRCA1 and BARD1 are expressed in a wide range of cell types, inherited mutations in the genes are associated with breast and ovarian cancers. Therefore, identification of substrates that might provide insight into the tissue-specificity of BRCA1-associated cancers are a key goal. The hormone-responsive estrogen receptor (ERα) was such a candidate, based on early observations that overexpression of wild-type BRCA1, but not a cancer-associated BRCA1 mutant (C61G) results in repressed ERα transcriptional activity89,90. A direct interaction between BRCA1/BARD1 and ERα was inferred from pull-down experiments in which the first 302 residues of BRCA1 co-purified with ERα89.
In reconstituted systems, ERα is mono-ubiquitylated by BRCA1/BARD1 in the linker region between the ERα DNA-binding and ligand-binding domains44. Substitution of the modified residues identified by mass spectrometry (K302/303) with arginine did not completely suppress ERα ubiquitylation, suggesting that BRCA1 can ubiquitylate other lysine residues in the absence of its preferred targets44. Of the E2s that work with BRCA1/BARD1, only Ube2D family members modify ERα in collaboration with BRCA1/BARD1, revealing an E2/substrate specificity that has not been observed with other substrates29,43 (Table I).
BRCA1-dependent ERα monoubiquitylation was subsequently demonstrated in cells overexpressing BRCA1 and by ubiquitin immunoprecipitation with endogenously expressed BRCA1 and ERα86,91. Consistent with the E2 specificity observed in vitro, cells expressing the Ube2D-incompetent BRCA1 mutant (I26A-BRCA1) failed to display repressed ERα transcriptional activity86.
ERα binding regions on BRCA1/BARD1 were mapped using in vitro ubiquitylation assays with purified components. BRCA1/BARD1 constructs that included the minimal RING heterodimer region required for enzymatic activity and incrementally larger regions of the BRCA1/BARD1 IDRs were assayed for their ability to ubiquitylate ERα. The shortest construct that produced detectable ERα ubiquitylation contained BRCA1 residues 1–258, from which it was concluded that an ERα binding site is located between IDR residues 177 and 25844 (Fig 1A). However, more distal regions of either BRCA1 or BARD1 have not yet been tested for ERα binding, so there may be additional sites that contribute to a higher affinity interaction.
Identification of the E3/substrate interaction sites and substrate ubiquitylation sites enabled investigation of BRCA1-dependent ERα ubiquitylation in cells. Mutation of ERα residues Lys 302/303 resulted in lower levels of mono-ubiquitylated ERα and decreased suppression of ERα transcriptional activity by BRCA186. Importantly, acetylation of Lys 302/303 is involved in ERα activation, so mutation of these residues disallows both their ubiquitylation and their acetylation. The balance between acetylation and ubiquitylation may therefore modulate ERα activity in cells. Consistent with this mechanism, overexpression of wild-type BRCA1 in cells, but not I26A-BRCA1, decreases levels of acetylated ERα86. The ERα situation highlights caveats that need to be considered when interpreting effects of substrate lysine mutation, as loss of a ubiquitylation site could have unintended effects that may or may not be relevant to the role of the ubiquitylation process under investigation.
Nucleosomal histone H2A
Early studies identified the core histones (H2A, H2B, H3, H4) and the DNA damage-specific H2A isoform H2Ax as substrates for BRCA1/BARD1 ligase activity in vitro59,76,92. Although BRCA1/BARD1 can attach mono-Ub to these histone proteins when presented individually (i.e., not in the context of histone dimers, octamers, or nucleosomes), these lysine-rich polypeptides likely serve as “proxy-substrates” rather than true cellular targets93. In the context of the nucleosome, BRCA1/BARD1 mono-ubiquitylates histone H2A and this activity is required for maintenance of heterochromatic centers and constitutive transcriptional repression of satellite DNA regions in mice88,94,93,95. Histone H2A ubiquitylation by BRCA1/BARD1 was subsequently shown to play a role in DNA double-strand break repair, promoting cell survival after treatment with a subset of DNA damaging agents37. Specifically, the H2A-Ub promotes end resection of broken DNA ends by recruitment of the SWI/SNF-like chromatin remodeler SMARCAD1 to chromatin, resulting in a repositioning of 53BP1, an opposing DNA repair factor that stimulates non-homologous end joining, at DNA breaks37,87. Thus, BRCA1/BARD1-dependent ubiquitylation of nucleosomal H2A is required for specific steps of homology-directed repair (HDR).
BRCA1/BARD1-dependent H2A-Ub also transcriptionally represses certain estrogen-metabolizing cytochrome P450 (CYP450) genes in breast epithelial cells5. These two enzymes catalyze quinol formation and hydroxylation of estradiol, generating metabolites that can form adducts with DNA and cause double-stranded breaks96. De-repression of CYP450 transcription due to BRCA1/BARD1 deficiency in BARD1+/− CRISPR MCF10a knockout cells was rescued by ectopic expression of an H2A-Ub genetic fusion, but not BARD1 cancer-predisposing RING mutants that are unable to bind H2A in nucleosomes5 (Fig 1B). Notably, these BARD1 mutants do not affect intrinsic ligase function of the heterodimer and are active with other substrates, indicating a direct nucleosome-specific effect (Fig 1C). A comprehensive catalog of genes that are transcriptionally regulated by BRCA1/BARD1-dependent H2A-Ub and how these genes are selected remains an important outstanding question.
Nucleosomal H2A is the only BRCA1/BARD1 substrate for which atomic-level structural information is currently available43. A cryo-EM structure of a BRCA1/BARD1/Ube2D3/nucleosome complex revealed that the non-E2 binding BARD1 RING domain makes unique contacts with the nucleosomal surface (Fig 2). The interaction tilts the BRCA1 E2-binding surface upward and elevates the BRCA1-bound E2 enzyme away from the nucleosomal surface, explaining why lysine residues on the H2A C-terminal tail, rather than the surface, are targeted by the ligase complex. The highly dynamic flexible H2A C-terminal tail retains its flexibility in the E3/E2/nucleosome complex, and the native H2A lysine targets of BRCA1/BARD1 have adequate reach to access the E2 active site for efficient ubiquitin transfer. Importantly, the structure revealed that the non-E2 binding RING domain of BARD1 directs the nucleosomal H2A specificity of BRCA1/BARD1. The study also provided an explanation for known and putative cancer-associated mutations in the BARD1 RING domain, showing that these abrogate nucleosome binding and, therefore, ubiquitylation.
Figure 2: BRCA1 and BARD1 bind the nucleosome substrate and E2 simultaneously.
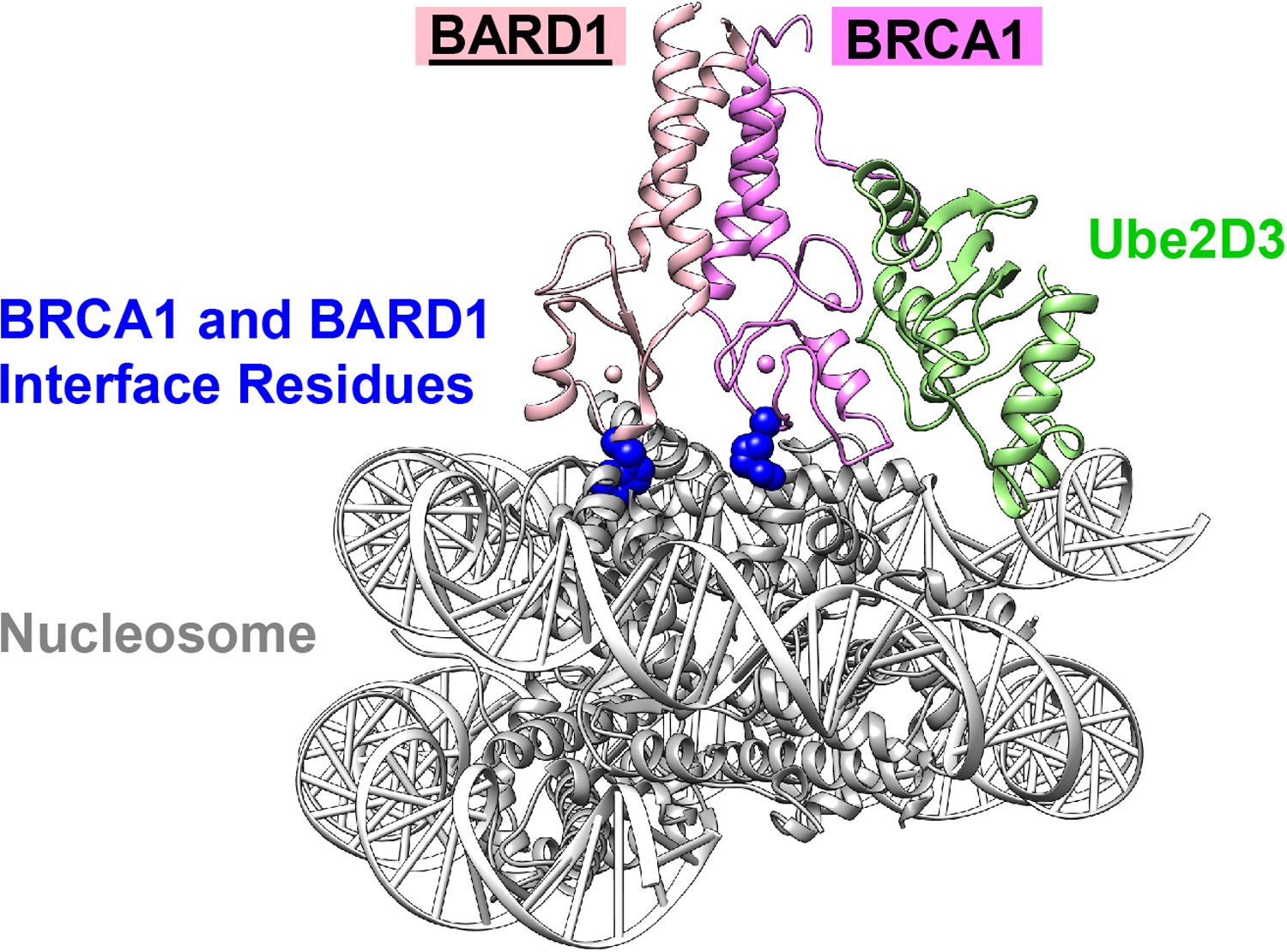
The cryo-EM structure (PDB 7JZV) reveals that both BRCA1 and BARD1 RING domains (magenta and pink, respectively) contain critical residues (BRCA1 Arg 71 and BARD1 Trp 91, blue) that contact the nucleosome surface (gray). The E2, Ube2D3 (green), binds to a distinct interface on BRCA1 allowing the heterodimer to coordinate substrate and E2 simultaneously. The complex allows for transfer of ubiquitin from the E2 onto the dynamic C-terminal end of H2A (unresolved in the structure).
Altogether, the described studies establish histone H2A as an important and specific BRCA1/BARD1 substrate. While greater detail is known about the biological functions of BRCA1/BARD1-mediated H2A ubiquitylation than most other putative cellular substrates, many questions remain. For example, how is BRCA1/BARD1 recruited to nucleosomes for H2A ubiquitylation, and does it involve other histone PTMs? Studies have established that the BARD1 C-terminal domains (Ank-BRCTs) specifically recognize nucleosomes containing H4K20me0 and H2A K15-Ub, binding to a fully overlapping histone surface to that used by the RING heterodimer78,79,51. While these interactions were shown to be essential for DNA DSB repair, a link to BRCA1/BARD1-dependent H2A ubiquitylation has not been established. It is possible that these interactions encode a nucleosome read/write mechanism. Future investigation of this specific H2A-Ub mark and its cellular ramifications are impeded by the lack of a detection system: there are currently no validated antibodies that discern this mark from other H2A-Ub marks. Development of tools and systems to study BRCA1/BARD1-dependent nucleosome ubiquitylation is likely to yield rich insight into its cellular E3 functions and dysregulation in cancer.
p50
The p50 protein is a component of the transcription factor NFkB that is activated by cellular signals that includes cytokines, oxidants, free radicals, ultraviolet irradiation, and bacterial or viral products97. BRCA1 deficiency is associated with dysregulation of NFkB signaling and NFkB signaling is also associated with chemoresistance in BRCA1-proficient tumors98–100. Use of a phosphorylation-resistant p50 mutant in 293T cells revealed that phosphorylated p50 binds to the BARD1 C-terminal BRCT domain49 (Fig 1A, Table I). The interaction facilitates BRCA1/BARD1-dependent mono-ubiquitylation near the p50 C-terminus at K354 or K356, tandem mutation of which abrogated p50 ubiquitylation49. Expression of a p50-Ub genetic fusion as a proxy for p50 that is mono-ubiquitylated at one of its C-terminal lysine residues revealed that mono-ubiquitylation 1) stabilizes p50, 2) decreases its recruitment to chromatin, and 3) promotes its cytoplasmic localization. Furthermore, mice injected with cells expressing a ubiquitylation-resistant mutant of p50 formed larger tumors than wild-type. Ubiquitylation of p50 was also shown to be important for genome maintenance; BARD1 cancer-predisposing mutants in the BRCT domain disrupt the interaction with p50, impair its ubiquitylation, and decrease its stability. Finally, analysis of patient neuroblastoma and breast cancer tumor samples reveal that BARD1 and p50 levels are positively correlated.
The p50 study provides an excellent framework for the investigation of BRCA1/BARD1-dependent ubiquitylation of putative substrates49. The biological findings were contingent on several key insights: (1) identification of the binding sites for both BRCA1/BARD1 and the p50 substrate (including p50 PTM requirements), (2) identification of mutations that selectively disrupt the association, (3) identification of the type of Ub mark and its location on the substrate, and (4) design of Ub-resistant lys to arg mutants and a “constitutively-ubiquitylated” genetic fusion. Together, these findings helped guide design of a robust set of constructs with which to characterize the biological consequences of BRCA1/BARD1-dependent p50 ubiquitylation. While not all substrates may be amenable to all these approaches, it presents a best-case scenario and roadmap for future investigations.
NF2
NF2 is a scaffolding component of the Hippo growth-signaling pathway that controls organ size development by regulating cell proliferation, apoptosis, and stem cell self-renewal101. While components of the Hippo pathway are known to be regulated by ubiquitylation, it has only recently been discovered that BRCA1/BARD1 ubiquitylates NF2 to keep Hippo signaling “off” in the presence of nutrient-rich media conditions48. Ubiquitylated NF2 is unable to interact with the kinase LATS, which in turn prevents the phosphorylation, cytosolic relocation, and proteasomal degradation of the Hippo transcriptional regulator YAP1, in a process mediated by a ligase other than BRCA1/BARD1. Consistent with these findings, luciferase assays revealed that BRCA1 is required for proper YAP1 function. Mass spectrometry analysis of ubiquitylated NF2 from cells revealed mostly non-degradative K63-linked ubiquitin modification at sites throughout the protein. Ubiquitylation of NF2 was decreased upon shRNA knockdown of BRCA1 and BARD1 independently, and a direct interaction between BRCA1 and NF2 was mapped to the C-terminal BRCTs of BRCA1 and the NF2 N-terminal FERM domain (Fig 1A, Table I). However, whether BRCA1/BARD1 collaborates with multiple E2s or other E3s to synthesize the observed K63-linked chains on NF2 is unknown. Although YAP1 stability is contingent on BRCA1-dependent ubiquitylation of NF2, expression of a constitutively active YAP1 mutant resistant to degradation rescued growth defects in BRCA1-deficient breast epithelial cells (MCF10a) and caused invasive structure formation in 3D-culture experiments. Injection of BRCA1-deficient MCF10a cells expressing the constitutively active YAP1 mutant into mice caused tumorigenesis that was not observed upon BRCA1 knockdown alone or in conjunction with knockdown of the known tumor-suppressor p53. These findings indicate that YAP1 reactivation may confer a growth advantage in BRCA1-deficient cells and drive the formation of cancers. Together, these data reveal an important role for BRCA1/BARD1 ligase activity as a regulatory node in Hippo signaling. Remaining questions to be addressed include the relevance and function of the observed K63-linked chains and whether BRCA1/BARD1 are directly and/or solely responsible for them.
Oct1
Oct1 (also called POUF1) is a widely expressed transcription factor that is related to the pluripotency master regulator Oct4102. Oct1 insulates critical genes against oxidative stress, dampens reactive oxygen species, promotes glycolytic metabolism, and promotes normal and cancer stem cell phenotypes. A physical interaction between BRCA1 and Oct1 has been established that regulates the transcriptional activation of multiple genes, notably including ESR1 that encodes ERα103–106. These observations prompted investigation as to whether BRCA1 regulates Oct1 through its ligase activity. An in vitro ubiquitylation assay using Oct1-containing cell lysate as substrate showed that poly-ubiquitylation of Oct1 was dependent on all components of the Ub cascade including BRCA1 and was not observed using a Ub-resistant Oct1 mutant (K9/403R)107. Although exogenous Ube2D was used, it cannot be ruled out that other E2s in the lysate were active in this assay. However, Oct1 levels were stabilized in cells expressing BRCA1-I26A (Ube2D-incompetent form of BRCA1) and cells treated with proteasome inhibitor (MG132) accumulated Oct1 with K48-linked ubiquitin chains. Taken together, the data strongly suggest that BRCA1/BARD1 serves as a degradative ligase for Oct1 by targeting specific lysine residues for Ub attachment. Important gaps in understanding regarding how this occurs remain. Does BRCA1/BARD1 perform both priming and K48-linked chain building reaction with a UBE2D, or does it use another E2 (for example, UBE2K) as the chain extender, or does it perform the critical first step (priming) and another E2/E3 pair builds the chains?
The observed stabilization of Oct1 in BRCA1-I26A-expressing MEFs enabled identification of ~1000 genes whose expression patterns were altered, with “metabolic disease” emerging as a significantly affected pathway107. Consistent with this, the cells had an increased glycolytic phenotype measured by a decrease in oxygen consumption, increase in extracellular acidification rates, and increase in glycolytic metabolites. Notably, Oct1 levels correlate with tumor aggressiveness and inversely correlate with BRCA1 protein levels in patient tumor samples. Overall, the findings point to a link between BRCA1/BARD1 Ub ligase activity and metabolic processes in cancer.
LARP7
La-related protein 7 (LARP7) has been reported to act as a potential tumor suppressor in gastric and breast cancers108,109. LARP7 binds to distinct small nuclear RNAs, stabilizes the 3’ hairpin of non-coding 7SK RNA, and is known to negatively regulate RNAPII pausing release110. LARP7 levels negatively correlate with cell survival upon DNA damage in MEF and HeLa cells50. Following IR treatment, LARP7 is rapidly shuttled from the nucleus and degraded. A potential interaction between LARP7 and BARD1 that appeared in the BioGRID database was confirmed via immunoprecipitation experiments50. While both the BRCA1 and BARD1 BRCTs interact independently with LARP7, only the BARD1 interaction is stimulated by IR treatment (Fig 1A, Table I). BRCA1/BARD1 binding and ubiquitylation of LARP7 was shown to be dependent upon ATM-mediated phosphorylation of LARP7 at T440. BRCA1 and BARD1 co-expression in 293T cells was associated with proteasome-dependent depletion of LARP7. Use of single-lysine Ub mutants revealed the majority of ubiquitylated LARP7 contains K48-linked Ub chains, consistent with a degradation outcome. While full-length BRCA1/BARD1 was needed to induce poly-ubiquitylation of LARP7 in cells, a truncated construct lacking the BRCTs of both E3 subunits was observed to build poly-Ub chains on LARP7 in vitro with Ube2D3 as the E2, raising questions as to why requirements differ in the two contexts. Analysis of LARP7 ubiquitylation using full-length BRCA1/BARD1 and an E2 panel may provide insight into the mechanism by which such K48-linked chains are built. As discussed for other substrates, the possibility that another E2 and/or E3 is involved in LARP7 poly-ubiquitylation remains open.
As LARP7 depletion leads to increased HDR efficiency, loss of BRCA1/BARD1 ligase function towards LARP7 might result in DNA damage defects. Indeed, high LARP7 levels are correlated with increased survival times in patients receiving chemotherapy, and LARP7-overexpressing cells are especially sensitive to ionizing radiation and cisplatin. These data suggest that LARP7 may be a useful drug target or prognostic marker in BRCA1-deficient cancers.
Concluding Remarks
An important initiative in BRCA1/BARD1 research involves the accurate classification of thousands of observed patient variants, of which a majority of missense variants remain unclassified variants of unknown significance in the ClinVar database111. To date, efforts towards this end have relied upon detecting processes such as HDR efficiency, drug resistance, and cell viability112–115. Such phenotypes may be linked to BRCA1/BARD1 function indirectly and, at the very least, the underlying mechanisms are undefined. Given that E3 ligase activity is the sole biochemical function, it is in principle possible that all, or a majority of, known phenotypes have substrate ubiquitylation at their source. Therefore, identification of substrates whose ubiquitylation is associated with processes linked to tumor suppression will provide critical information regarding the potential effects of unclassified variants in either the BRCA1 or BARD1 genes. In principle, quantitative assessment of variant BRCA1/BARD1 species’ ability to ubiquitylate relevant substrates could have the power to predict the impact of a mutation and, ultimately, inform decision-making in prophylactic treatment. Clearly, there will be more than one critical substrate, suggesting an ultimate goal in which multiple substrates are monitored in patient samples. An important intermediate step towards such a goal is the assessment of relevant substrate ubiquitylation by known BRCA1/BARD1 variants and their association with processes such as DNA damage response, transcriptional regulation, etc. With tools and reagents currently available, we may be entering a time when this goal is achievable (though non-trivial) with the existing BRCA1/BARD1 substrates and can be pursued anew for each additional substrate identified in the future.
ACKNOWLEDGEMENTS.
We thank Dr. Peter Brzovic for careful reading and thoughtful comments. This work was supported by the NIH (GM088055). R.E.K. is the Edmond H. Fischer/Washington Research Foundation Endowed Chair in Biochemistry.
References
- 1.Friedman LS et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet 8, 399–404 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Hall J et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684 (1990). [DOI] [PubMed] [Google Scholar]
- 3.King M-C, Marks JH, Mandell JB, & New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Miki Y et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Stewart MD et al. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. U. S. A 115, 1316–1321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W et al. A synergetic effect of BARD1 mutations on tumorigenesis. Nat. Commun 12, 1243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber-Lassalle N et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 21, 55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarsounas M & Sung P The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol 21, 284–299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shakya R et al. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc. Natl. Acad. Sci 105, 7040 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullan PB, Quinn JE & Harkin DP The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25, 5854–5863 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Kais Z & Parvin JD Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol. Ther 7, 1540–1543 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Privat M et al. BRCA1 Induces Major Energetic Metabolism Reprogramming in Breast Cancer Cells. PLOS ONE 9, e102438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou Z, Minter-Dykhouse K & Chen J BRCA1 participates in DNA decatenation. Nat. Struct. Mol. Biol 12, 589–593 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Christou CM & Kyriacou K BRCA1 and Its Network of Interacting Partners. Biology 2, 40–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savage KI & Harkin DP BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J. 282, 630–646 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Hashizume R et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem 276, 14537–14540 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Lorick KL et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci 96, 11364 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drost R et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell 20, 797–809 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Reid LJ et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. U. S. A 105, 20876–20881 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shakya R et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brzovic PS, Rajagopal P, Hoyt DW, King MC & Klevit RE Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol 8, 833–837 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Clark SL, Rodriguez AM, Snyder RR, Hankins GDV & Boehning D Structure-Function of the Tumor Supressor BRCA1. Comput. Struct. Biotechnol. J 1, e201204005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzger MB, Pruneda JN, Klevit RE & Weissman AM RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Ubiquitin-Proteasome Syst. 1843, 47–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mark W-Y et al. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J. Mol. Biol 345, 275–287 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Brzovic PS, Meza JE, King M-C & Klevit RE BRCA1 RING Domain Cancer-predisposing Mutations: Structural Consequences and Effects on Protein-Protein Interactions. J. Biol. Chem 276, 41399–41406 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Ruffner H, Joazeiro CAP, Hemmati D, Hunter T & Verma IM Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci 98, 5134 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson Andrew C. & Holt Jeffrey T.. Impact of RING and BRCT Domain Mutations on BRCA1 Protein Stability, Localization and Recruitment to DNA Damage. Radiat. Res 174, 1–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris JR et al. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet 15, 599–606 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Stewart MD et al. Tuning BRCA1 and BARD1 activity to investigate RING ubiquitin ligase mechanisms. Protein Sci. 26, 475–483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christensen DE, Brzovic PS & Klevit RE E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol 14, 941–948 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Brzovic PS et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. U. S. A 100, 5646–5651 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart MD, Ritterhoff T, Klevit RE & Brzovic PS E2 enzymes: more than just middle men. Cell Res. 26, 423–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vittal V et al. Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat. Chem. Biol 11, 83–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikawa H et al. Mass Spectrometric and Mutational Analyses Reveal Lys-6-linked Polyubiquitin Chains Catalyzed by BRCA1-BARD1 Ubiquitin Ligase*. J. Biol. Chem 279, 3916–3924 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Wu-Baer F, Lagrazon K, Yuan W & Baer R The BRCA1/BARD1 Heterodimer Assembles Polyubiquitin Chains through an Unconventional Linkage Involving Lysine Residue K6 of Ubiquitin*. J. Biol. Chem 278, 34743–34746 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Morris JR & Solomon E BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet 13, 807–817 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Densham RM et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol 23, 647–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruneda JN et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taherbhoy AM, Huang OW & Cochran AG BMI1–RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun 6, 7621 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Starita LM et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics 200, 413–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Q, Liu L & Xie Q In Vitro Protein Ubiquitination Assay. in Plant Signalling Networks: Methods and Protocols (eds. Wang Z-Y & Yang Z) 163–172 (Humana Press, 2012). doi: 10.1007/978-1-61779-809-2_13. [DOI] [PubMed] [Google Scholar]
- 42.Buetow L, Gabrielsen M & Huang DT Single-Turnover RING/U-Box E3-Mediated Lysine Discharge Assays. in The Ubiquitin Proteasome System: Methods and Protocols (eds. Mayor T & Kleiger G) 19–31 (Springer; New York, 2018). doi: 10.1007/978-1-4939-8706-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witus SR et al. BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat. Struct. Mol. Biol 28, 268–277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eakin CM, Maccoss MJ, Finney GL & Klevit RE Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A 104, 5794–5799 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varma AK, Brown RS, Birrane G & Ladias JAA Structural Basis for Cell Cycle Checkpoint Control by the BRCA1−CtIP Complex,. Biochemistry 44, 10941–10946 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Yu X, Fu S, Lai M, Baer R & Chen J BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starita LM et al. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J. Biol. Chem 280, 24498–24505 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Verma S et al. BRCA1/BARD1-dependent ubiquitination of NF2 regulates Hippo-YAP1 signaling. Proc. Natl. Acad. Sci 116, 7363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu L et al. p50 mono-ubiquitination and interaction with BARD1 regulates cell cycle progression and maintains genome stability. Nat. Commun 11, 5007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F et al. L ARP7 Is a BRCA1 Ubiquitinase Substrate and Regulates Genome Stability and Tumorigenesis. Cell Rep. 32, 107974 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Dai L et al. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol. Cell 81, 2765–2777.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Hsu L-C, Doan TP & White RL Identification of a γ-Tubulin-binding Domain in BRCA1. Cancer Res. 61, 7713 (2001). [PubMed] [Google Scholar]
- 53.Starita LM et al. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol 24, 8457–8466 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 550, 360–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan W et al. Preparation and purification of mono-ubiquitinated proteins using Avi-tagged ubiquitin. PLOS ONE 15, e0229000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minten EV et al. SIRT2 promotes BRCA1-BARD1 heterodimerization through deacetylation. Cell Rep. 34, 108921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouchi T BRCA1 phosphorylation: Biological consequences. Cancer Biol. Ther 5, 470–475 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Daza-Martin M et al. Isomerization of BRCA1–BARD1 promotes replication fork protection. Nature 571, 521–527 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Mallery DL, Vandenberg CJ & Hiom K Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21, 6755–6762 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris JR et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Hunter T The Age of Crosstalk: Phosphorylation, Ubiquitination, and Beyond. Mol. Cell 28, 730–738 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Sankaran S, Crone DE, Palazzo RE & Parvin JD Aurora-A Kinase Regulates Breast Cancer–Associated Gene 1 Inhibition of Centrosome-Dependent Microtubule Nucleation. Cancer Res. 67, 11186 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Hayami R et al. Down-regulation of BRCA1-BARD1 Ubiquitin Ligase by CDK2. Cancer Res. 65, 6 (2005). [PubMed] [Google Scholar]
- 64.Wu W et al. HERC2 Is an E3 Ligase That Targets BRCA1 for Degradation. Cancer Res. 70, 6384 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Wang X et al. HUWE1 interacts with BRCA1 and promotes its degradation in the ubiquitin–proteasome pathway. Biochem. Biophys. Res. Commun 444, 549–554 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Lu Y et al. The F-box Protein FBXO44 Mediates BRCA1 Ubiquitination and Degradation*. J. Biol. Chem 287, 41014–41022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu Q, Zhang F-L, Lu D-Y, Shao Z-M & Li D-Q USP9X stabilizes BRCA1 and confers resistance to DNA-damaging agents in human cancer cells. Cancer Med. 8, 6730–6740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song L & Rape M Regulated Degradation of Spindle Assembly Factors by the Anaphase-Promoting Complex. Mol. Cell 38, 369–382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang Y et al. Structural analysis of BRCA1 reveals modification hotspot. Sci. Adv 3, e1701386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Wu-Baer F, Ludwig T & Baer R The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol. Cell. Biol 30, 2787–2798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choudhury AD, Xu H & Baer R Ubiquitination and Proteasomal Degradation of the BRCA1 Tumor Suppressor Is Regulated during Cell Cycle Progression*. J. Biol. Chem 279, 33909–33918 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Fabbro M, Rodriguez JA, Baer R & Henderson BR BARD1 Induces BRCA1 Intranuclear Foci Formation by Increasing RING-dependent BRCA1 Nuclear Import and Inhibiting BRCA1 Nuclear Export*. J. Biol. Chem 277, 21315–21324 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez JA, Schüchner S, Au WWY, Fabbro M & Henderson BR Nuclear–cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene 23, 1809–1820 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Lu Y et al. Ubiquitination and Proteasome-Mediated Degradation of BRCA1 and BARD1 during Steroidogenesis in Human Ovarian Granulosa Cells. Mol. Endocrinol 21, 651–663 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Joukov V, Chen J, Fox EA, Green JBA & Livingston DM Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci 98, 12078 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia Y, Pao GM, Chen H-W, Verma IM & Hunter T Enhancement of BRCA1 E3 Ubiquitin Ligase Activity through Direct Interaction with the BARD1 Protein*. J. Biol. Chem 278, 5255–5263 (2003). [DOI] [PubMed] [Google Scholar]
- 77.McCarthy Ellen E, Celebi Julide T, Baer Richard, & Ludwig Thomas. Loss of Bard1, the Heterodimeric Partner of the Brca1 Tumor Suppressor, Results in Early Embryonic Lethality and Chromosomal Instability. Mol. Cell. Biol 23, 5056–5063 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakamura K et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol 21, 311–318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Becker JR et al. BARD1 links histone H2A Lysine-15 ubiquitination to initiation of BRCA1-dependent homologous recombination. bioRxiv 2020.06.01.127951 (2020) doi: 10.1101/2020.06.01.127951. [DOI] [Google Scholar]
- 80.Starita L & Parvin J Substrates of the BRCA1-Dependent Ubiquitin Ligase. Cancer Biol. Ther 5, 137–41 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Wu W, Koike A, Takeshita T & Ohta T The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Div. 3, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parvin JD The BRCA1-dependent ubiquitin ligase, γ-tubulin, and centrosomes. Environ. Mol. Mutagen 50, 649–653 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Ohta T, Sato K & Wu W The BRCA1 ubiquitin ligase and homologous recombination repair. Ubiquitin Fam. Proteins DNA Damage Response 585, 2836–2844 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Song M, Hakala K, Weintraub ST & Shiio Y Quantitative Proteomic Identification of the BRCA1 Ubiquitination Substrates. J. Proteome Res 10, 5191–5198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim B-J et al. The Histone Variant MacroH2A1 Is a BRCA1 Ubiquitin Ligase Substrate. Cell Rep. 19, 1758–1766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma Y et al. BRCA1 Regulates Acetylation and Ubiquitination of Estrogen Receptor-α. Mol. Endocrinol 24, 76–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Uckelmann M et al. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat. Commun 9, 229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu Q et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fan S et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene 20, 77–87 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Kawai H, Li H, Chun P, Avraham S & Avraham HK Direct interaction between BRCA1 and the estrogen receptor regulates vascular endothelial growth factor (VEGF) transcription and secretion in breast cancer cells. Oncogene 21, 7730–7739 (2002). [DOI] [PubMed] [Google Scholar]
- 91.La Rosa P, Marino M & Acconcia F 17β-estradiol regulates estrogen receptor α monoubiquitination. IUBMB Life 63, 49–53 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Chen A, Kleiman F, Manley J, Ouchi T & Pan Z-Q Autoubiquitination of the BRCA1-BARD1 RING ubiquitin ligase. J. Biol. Chem 277, 22085–92 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Kalb R, Mallery DL, Larkin C, Huang JTJ & Hiom K BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Q et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 70, 842–853.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McGinty RK, Henrici RC & Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Savage KI et al. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 74, 2773–2784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Y, Wan Y & Huang C The biological functions of NF-kappaB1 (p50) and its potential as an anti-cancer target. Curr. Cancer Drug Targets 9, 566–571 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sau A et al. Persistent Activation of NF-κB in BRCA1-Deficient Mammary Progenitors Drives Aberrant Proliferation and Accumulation of DNA Damage. Cell Stem Cell 19, 52–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buckley NE et al. A BRCA1 deficient, NFκB driven immune signal predicts good outcome in triple negative breast cancer. Oncotarget 7, 19884–19896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harte MT et al. NF-κB is a critical mediator of BRCA1-induced chemoresistance. Oncogene 33, 713–723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Misra JR & Irvine KD The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet 52, 65–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vázquez-Arreguín K & Tantin D The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks. Oct Transcr. Factor Fam 1859, 792–804 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan W et al. BRCA1 Regulates GADD45 through Its Interactions with the OCT-1 and CAAT Motifs*. J. Biol. Chem 277, 8061–8067 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Wang R-H, Yu H & Deng C-X A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc. Natl. Acad. Sci. U. S. A 101, 17108 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maekawa T et al. ATF-2 controls transcription of Maspin and GADD45α genes independently from p53 to suppress mammary tumors. Oncogene 27, 1045–1054 (2008). [DOI] [PubMed] [Google Scholar]
- 106.Hosey AM et al. Molecular Basis for Estrogen Receptor α Deficiency in BRCA1-Linked Breast Cancer. JNCI J. Natl. Cancer Inst 99, 1683–1694 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vázquez-Arreguín K et al. BRCA1 through Its E3 Ligase Activity Regulates the Transcription Factor Oct1 and Carbohydrate Metabolism. Mol. Cancer Res. MCR 16, 439–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheng Y et al. LARP7 is a potential tumor suppressor gene in gastric cancer. Lab. Invest 92, 1013–1019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ji X, Lu H, Zhou Q & Luo K LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. eLife 3, e02907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Markert A et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 9, 569–575 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Landrum MJ et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Starita LM et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics 200, 413–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Starita LM et al. A Multiplex Homology-Directed DNA Repair Assay Reveals the Impact of More Than 1,000 BRCA1 Missense Substitution Variants on Protein Function. Am. J. Hum. Genet 103, 498–508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Findlay GM et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adamovich AI et al. Functional analysis of BARD1 missense variants in homology-directed repair and damage sensitivity. PLOS Genet. 15, e1008049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ryser S et al. Distinct Roles of BARD1 Isoforms in Mitosis: Full-Length BARD1 Mediates Aurora B Degradation, Cancer-Associated BARD1β Scaffolds Aurora B and BRCA2. Cancer Res. 69, 1125 (2009). [DOI] [PubMed] [Google Scholar]
- 117.Shabbeer S et al. BRCA1 targets G2/M cell cycle proteins for ubiquitination and proteasomal degradation. Oncogene 32, 5005–5016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin S-Y, Li K, Stewart GS & Elledge SJ Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. U. S. A 101, 6484 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sato K et al. A DNA-damage selective role for BRCA1 E3 ligase in claspin ubiquitylation, CHK1 activation, and DNA repair. Curr. Biol. CB 22, 1659–1666 (2012). [DOI] [PubMed] [Google Scholar]
- 120.Li S et al. Binding of CtIP to the BRCT Repeats of BRCA1 Involved in the Transcription Regulation of p21 Is Disrupted Upon DNA Damage*. J. Biol. Chem 274, 11334–11338 (1999). [DOI] [PubMed] [Google Scholar]
- 121.Sato K et al. Nucleophosmin/B23 Is a Candidate Substrate for the BRCA1-BARD1 Ubiquitin Ligase*. J. Biol. Chem 279, 30919–30922 (2004). [DOI] [PubMed] [Google Scholar]
- 122.Calvo V & Beato M BRCA1 Counteracts Progesterone Action by Ubiquitination Leading to Progesterone Receptor Degradation and Epigenetic Silencing of Target Promoters. Cancer Res. 71, 3422 (2011). [DOI] [PubMed] [Google Scholar]
- 123.Kleiman FE et al. BRCA1/BARD1 inhibition of mRNA 3’ processing involves targeted degradation of RNA polymerase II. Genes Dev. 19, 1227–1237 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Horwitz AA, Affar EB, Heine GF, Shi Y & Parvin JD A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A 104, 6614–6619 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu W et al. BRCA1 Ubiquitinates RPB8 in Response to DNA Damage. Cancer Res. 67, 951 (2007). [DOI] [PubMed] [Google Scholar]
- 126.Satish Sankaran, Starita Lea M., Groen Aaron C., Ko Min Ji, & Parvin Jeffrey D Centrosomal Microtubule Nucleation Activity Is Inhibited by BRCA1-Dependent Ubiquitination. Mol. Cell. Biol 25, 8656–8668 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]


