Here, Hada et al. comprehensively analyzed epigenomic features of mouse trophoblast stem cells (TSCs). They used genome-wide, high-throughput analyses to show that the TSC genome contains large-scale (>1-Mb) rigid heterochromatin architectures that have a high degree of histone H3.1/3.2–H3K9me3 accumulation, termed TSC-defined highly heterochromatinized domains (THDs), and are uniquely developed in placental lineage cells that serve to protect them from fate reprogramming to stably maintain placental function.
Keywords: trophoblast stem cell, H3K9me3, H3.1/H3.2, CAF1, somatic cell nuclear transfer
Abstract
The placenta is a highly evolved, specialized organ in mammals. It differs from other organs in that it functions only for fetal maintenance during gestation. Therefore, there must be intrinsic mechanisms that guarantee its unique functions. To address this question, we comprehensively analyzed epigenomic features of mouse trophoblast stem cells (TSCs). Our genome-wide, high-throughput analyses revealed that the TSC genome contains large-scale (>1-Mb) rigid heterochromatin architectures with a high degree of histone H3.1/3.2–H3K9me3 accumulation, which we termed TSC-defined highly heterochromatinized domains (THDs). Importantly, depletion of THDs by knockdown of CAF1, an H3.1/3.2 chaperone, resulted in down-regulation of TSC markers, such as Cdx2 and Elf5, and up-regulation of the pluripotent marker Oct3/4, indicating that THDs maintain the trophoblastic nature of TSCs. Furthermore, our nuclear transfer technique revealed that THDs are highly resistant to genomic reprogramming. However, when H3K9me3 was removed, the TSC genome was fully reprogrammed, giving rise to the first TSC cloned offspring. Interestingly, THD-like domains are also present in mouse and human placental cells in vivo, but not in other cell types. Thus, THDs are genomic architectures uniquely developed in placental lineage cells, which serve to protect them from fate reprogramming to stably maintain placental function.
In eutherian mammals, the separation and formation of the inner cell mass (ICM) and the trophectoderm (TE) represent the first lineage specification event in embryogenesis. After implantation, the ICM and TE develop into embryonic and extraembryonic lineages, respectively. The embryonic lineage can be divided into epiblast and primitive endoderm cells, and the former further differentiate into the three primary germ layers (ectoderm, mesoderm, and endoderm), a process termed pluripotent differentiation ability (Rossant and Tam 2009). The underlying epigenetic mechanisms that ensure the pluripotency of epiblast cells have been studied extensively thanks to the availability of embryonic stem cells (ESCs), especially in mice and humans. In contrast, the extraembryonic lineage differentiates exclusively into trophoblastic cells that go on to constitute the placental tissues. Furthermore, this terminates their ability to support fetal development at parturition. For this reason, extraembryonic cells have received less attention than those from the ICM as a research target, and information on their epigenetic features is much more limited than for the embryonic lineage. However, the extraembryonic cells or trophoblasts represent a cell lineage that has developed specifically in mammals and is likely to harbor special epigenetic features.
Trophoblast stem cells (TSCs), the first extraembryonic stem cells that were established from mouse embryos (Tanaka et al. 1998), have excellent properties; they can proliferate indefinitely while remaining undifferentiated in vitro and can be induced to differentiate into all the trophoblast lineage cells in the placenta in vivo. Therefore, by comparing the epigenetic features of TSCs with those of their embryonic counterparts, ESCs, we expect that the characteristics of extraembryonic cells can be clarified. The major genome-wide epigenetic dynamics in preimplantation mammalian embryos include the accumulation of epigenetic repressive histone marks, including H3K9me3, H3K27me3, and H4K20me3, along with extensive DNA demethylation (Xu et al. 2021). The chromatin assembly factor-1 (CAF1) complex, an H3.1/H3.2–H4 chaperone, is responsible for this accumulation of histone marks into the genome (Hake and Allis 2006; Hatanaka et al. 2015). One of the roles of these repressive histone marks is the suppression of harmful retrotransposon expressions in a low DNA methylation environment (Sharif et al. 2016). Indeed, many types of retrotransposons are expressed ectopically when CAF1 is diminished by knockdown using specific small interfering RNA (siRNA) sequences (Hatanaka et al. 2015). Therefore, it will be interesting to identify the dynamics and functions of these histone variants and histone modifications after implantation. In this study, we focused on modifications to histones H3.1/H3.2 and H3 using TSCs as models to clarify the epigenetic features of extraembryonic cells. By combining genome-wide sequencing and biochemical analyses, we identified that H3.1/H3.2 and H3K9me3 form large-scale (>1-Mb) rigid heterochromatinized domains in TSCs to sustain their phenotype as cells of the placental lineage.
Cloning animals by somatic cell nuclear transfer (SCNT) is a reproductive technology that produces animals from single-donor cells. Until now, cloned mice have been produced using many cell types composing the animal body, but extraembryonic cells including TSCs have never been cloned successfully by SCNT. This is most likely because of the poor development of the resulting reconstructed embryos (Ogawa et al. 2015). As the TSC genome is highly enriched with the H3K9me3 histone, we hypothesized that this excess may inhibit reprogramming of the TSC genome. Here, following demethylation of H3K9me3, we have successfully produced the first mice cloned from TSCs. This result further supports our notion that the TSC genome is protected from fate reprogramming to stably maintain placental function through highly rigid heterochromatinized domains.
Results
The TSC genome contains large H3K9me3 and H3.1/H3.2 domains
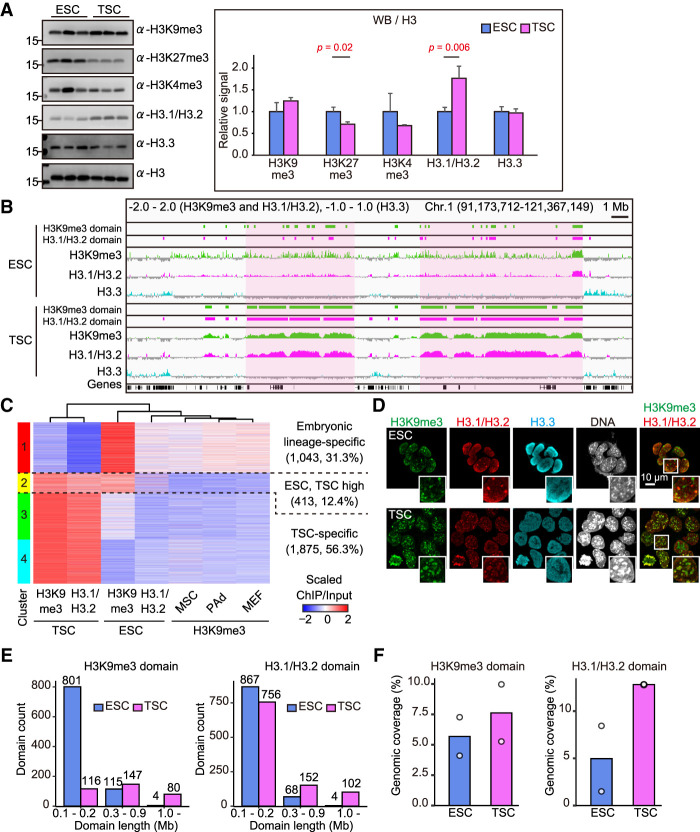
Previous studies have identified that H3 histone variants and their modifications change dynamically in mouse embryos shortly before implantation (Hatanaka et al. 2015; Wang et al. 2018). However, how these epigenetic changes are transmitted to the postimplantation embryonic and extraembryonic lineages is unknown. We performed Western blot analysis for the global abundance of histone H3 variants and their modifications using ESC and TSC lines with the same mouse genetic background ([C57BL/6NCrSlc × DBA/2CrSlc] F1, BDF1). The TSC lines we used were confirmed as being largely composed of normal diploid cells (Supplemental Fig. S1A). The global abundances of H3K4me3, H3K9me3, and H3.3 were indistinguishable between ESCs and TSCs (Fig. 1A). The H3K27me3 level was lower in TSCs than in ESCs, consistent with a previous report (Rugg-Gunn et al. 2010). Importantly, the H3.1/H3.2 level was significantly higher in TSCs, suggesting that these H3 variants might determine the epigenetic feature of TSCs by providing a scaffold for specific histone modifications and interacting proteins. However, because of the high similarity of their amino acid sequences, H3.1 and H3.2 cannot be distinguished using existing antibodies (Hake and Allis 2006; Harada et al. 2012; Hatanaka et al. 2015).
Figure 1.
The TSC genome uniquely contains large H3K9me3- and H3.1/H3.2-enriched domains. (A) Western blot analysis of histone H3 variants and modifications. Representative images (left) and the relative signal levels (right) are shown. Signal intensities were normalized to H3 and are presented as levels relative to ESC (set as 1.0). Data are presented as the mean ± SD (n = 3). P-values from two-tailed, unpaired Student's t-tests are indicated. (B) Integrative genomics viewer (IGV) snapshot of ChIP-seq data shown by log2 transformed enrichment (ChIP/input). Light-magenta areas correspond to large H3K9me3 and H3.1/H3.2 domains specifically formed in TSCs. (C) Heat map showing the enrichment of H3K9me3 and H3.1/H3.2 within H3K9me3 domains in ESCs and TSCs. The numbers and percentages of bins included are indicated for each cluster. (D) Immunostaining for H3K9me3, H3.1/H3.2, and H3.3 in ESCs and TSCs. White boxes show magnified images. There was an extensive overlap of H3K9me3 and H3.1/H3.2 in TSCs, but not in ESCs. (E) The numbers of H3K9me3 and H3.1/H3.2 domains classified by the domain length. (F) Genomic coverage of H3K9me3 and H3.1/H3.2 domains. White dots indicate the genomic coverage of each replicate.
Next, we addressed the genome-wide differences in the H3.1/H3.2 and H3.3 distributions between ESCs and TSCs by chromatin immunoprecipitation sequencing (ChIP-seq) analysis. We also obtained data for H3K9me3, which showed a slightly higher abundance in TSCs than in ESCs (Fig. 1A). We found that H3.1/H3.2 was well colocalized with H3K9me3 in the TSC genome, constructing large domains on the order of megabases in size (Fig. 1B). These large H3.1/H3.2–H3K9me3 domains were rarely observed in ESCs (Fig. 1B) or any other mouse cell lines examined, including mesenchymal stem cells (MSCs), preadipocytes (PAds), and mouse embryonic fibroblast cells (MEFs) (Supplemental Fig. S1B). In contrast, H3.3 was not enriched at these regions, suggesting an exclusive H3.1/H3.2 distribution (Fig. 1B). To gain more precise insight into the H3.1/H3.2 and H3K9me3 localizations, their domains (>0.1 Mb) were defined computationally based on the degree of enrichment (green and magenta rectangles in Fig. 1B; see the Materials and Methods for details). We found that the ESC-defined and TSC-defined H3K9me3 domains were distinct from each other (Supplemental Fig. S1C). H3.1/H3.2 also showed cell type-specific enrichment (Supplemental Fig. S1D). Thus, ESCs and TSCs formed distinct H3.1/H3.2 and H3K9me3 domains in their genomes. This differential distribution of H3K9me3 domains was further confirmed by clustering analysis (Fig. 1C). Cluster 1 displayed higher enrichment of H3K9me3 in ESCs as well as in MSCs, PAds, and MEFs compared with TSCs, indicating that these domains are embryonic lineage-specific (31.3% of all clusters). On the other hand, clusters 3 and 4 (in total, 56.3%) were TSC-specific enriched with both H3K9me3 and H3.1/H3.2. As revealed by an assay for transposase-accessible chromatin and sequencing (ATAC-seq) analysis, the chromatin accessibility of clusters 3 and 4 was significantly lower than that of cluster 1, indicating that TSCs acquired a more closed chromatin status than ESCs in their H3K9me3 domains (Supplemental Fig. S1E). We could further characterize the TSC-specific H3.1/H3.2-H3K9me3 domains by the Jaccard index (a statistical index used to describe the similarity between data sets); in TSCs, the Jaccard index of H3K9me3 domains versus H3.1/H3.2 domains was 0.64, while in ESCs it was 0.32, indicating a more extensive colocalization of H3K9me3 and H3.1/H3.2 in TSCs than in ESCs (Supplemental Fig. S1F). Consistent with these findings, immunostaining for H3K9me3 and H3.1/H3.2 revealed their colocalization in the TSC nuclear foci, but not in the ESCs (Fig. 1D).
We then undertook detailed comparisons of H3.1/H3.2–H3K9me3 domains in ESCs and TSCs based on their sizes and genomic coverage. A size-dependent classification revealed that TSCs contain more of the larger (>1-Mb) H3K9me3 and H3.1/H3.2 domains than ESCs do (80 and 102 vs. four and four, respectively) (Fig. 1E). Notably, most (801 out of 920; 87.1%) of the H3K9me3 domains in ESCs were <0.2 Mb in size. Moreover, the genomic coverages of the H3K9me3 and H3.1/H3.2 domains in TSCs were larger than those in ESCs (Fig. 1F). These data indicate that the H3K9me3 and H3.1/H3.2 domains developed more extensively in TSCs than in ESCs in terms of their size and genomic coverage. However, the H3K9me3 domains, rather than the H3.1/H3.2 domains, might better characterize TSCs because these cells also contained many small (<0.2-Mb) H3.1/H3.2 domains, as did ESCs (Fig. 1E).
To determine whether genes related to H3K9me3, H3.1/H3.2, or heterochromatin formation showed any TSC-specific expression patterns, we performed RNA sequencing (RNA-seq) analysis using TSCs, ESCs, and MEFs. As expected, TSCs displayed higher expressions of the genes encoding H3K9me3 methyltransferases Setdb1 and Suv39h2, and lower expressions for H3K9me3 demethylases Kdm4b/c/d, when compared with ESCs and MEFs (Supplemental Fig. S1G). However, histone H3.1/H3.2 chaperone (CAF1 complex; P150, P60, and P48) and heterochromatin proteins (HP1α, HP1β, and HP1γ) did not show any TSC-specific expression patterns (Supplemental Fig. S1G). Therefore, it is probable that more unidentified factors could be involved in the formation of these TSC-specific heterochromatinized structures in a complicated manner. Thus, H3K9me3 and H3.1/H3.2 were preferentially colocalized in TSCs to form large genomic domains, which appear to endow the TSC genome with highly heterochromatinized structures.
H3K9me3 domains in TSCs are preferentially localized in LINE/L1-rich regions
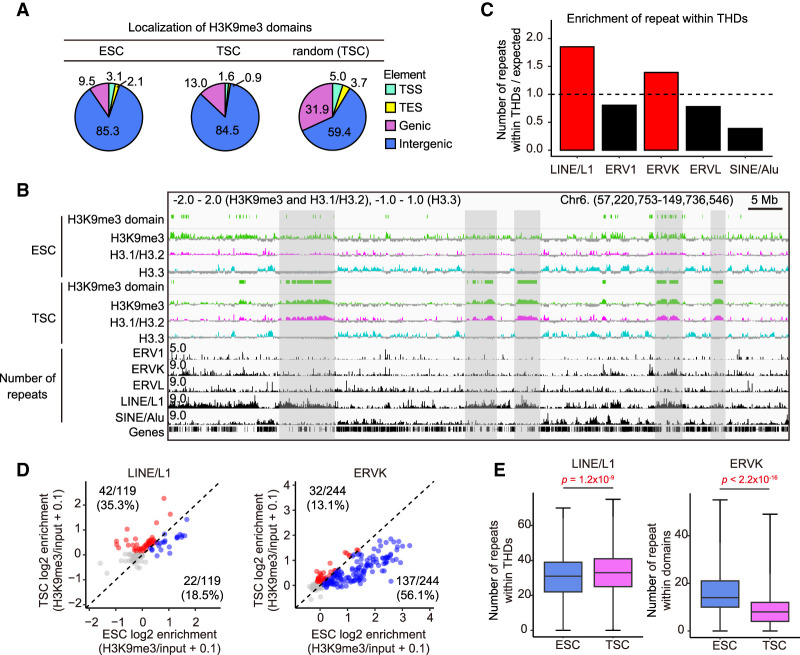
As mentioned above, we found that TSCs showed a high colocalization of H3K9me3 and H3.1/H3.2, but in terms of the length distribution, H3K9me3 rather than H3.1/H3.2 is more characteristic of TSCs. Therefore, we focused on the H3K9me3 domains for subsequent comprehensive analyses using TSCs and ESCs. These TSC-defined domains are referred to here as TSC-defined highly heterochromatinized domains (THDs). We then investigated which types of genomic elements harbored THDs. When the mouse genome was divided into four elements—transcriptional start sites (TSSs), transcriptional end sites (TESs), genic regions, and intergenic regions—the H3K9me3 domains of both TSCs and ESCs were largely localized to the intergenic regions (Fig. 2A). However, as most of the mouse genome is comprised of intergenic regions (Francis and Wörheide 2017), this result might have simply reflected the proportion of intergenic regions in the entire genome. To test this assumption, we randomly mapped THDs against the mouse genome and analyzed their distribution to each element (see the Materials and Methods). As a result, randomly mapped THDs showed a lower localization to intergenic regions (59.4%) compared with the real TSC localization (84.5%), indicating that the THDs are preferentially distributed to the intergenic regions (Fig. 2A).
Figure 2.
The H3K9me3 domains in TSCs (THDs) are preferentially localized in LINE/L1-rich regions. (A) Distributions of H3K9me3 domains in ESCs and TSCs (THDs). The THDs are preferentially distributed in intergenic regions as compared with the distribution by random mapping. (B) IGV snapshot of repeat elements and log2 transformed ChIP-seq data. Gray areas indicate the overlaps of TSC-defined H3K9me3 domains (THDs) and LINE/L1-rich regions. (C) Bar plot showing the enrichment of repetitive elements within the THDs in TSCs. Expected values represent the number of repetitive elements within randomly extracted regions. (D) Scatter plot showing the enrichment of H3K9me3 in ESCs and TSCs to LINE/L1 and ERVK. Each dot represents a subclass of retrotransposons. Red and blue dots indicate the specifically enriched subclasses in TSCs and ESCs, respectively, among subclasses with log2 enrichment >0. (E) Box plots showing the numbers of LINE/L1 and ERVK within H3K9me3 domains in ESCs and TSCs. P-values from the Wilcoxon rank sum test are indicated. There were 1517 bins analyzed for ESCs and 2281 bins analyzed for TSCs.
It is known that intergenic regions are frequently occupied by repetitive sequences including retrotransposons and are the major targets of repressive histone marks including H3K9me3 (Hatanaka et al. 2015; Sharif et al. 2016; Kato et al. 2018). We then compared the numbers of retrotransposons—ERV1, ERVK, ERVL, LINE/L1, and SINE/Alu—distributed within the THDs. There was a relatively higher enrichment of LINE/L1 and ERVK than other retrotransposons (Fig. 2B,C). It has been reported that the ERVK retrotransposons including IAPs are silenced in ESCs by H3K9me3 (Sharif et al. 2016; Kato et al. 2018). Therefore, we next compared ESCs and TSCs for the enrichment of H3K9me3 in these two classes of retrotransposons. As revealed by scatter plots, the TSC-dominant H3K9me3 enrichment in LINE/L1 was 35.3% (42 of 119 subclasses) while that in ERVK was only 13.1% (32 of 244) (Fig. 2D). This was further confirmed by the density of each repeat within the H3K9me3 domains; thus, the number of LINE/L1 in TSC-defined H3K9me3 domains was significantly higher than in ESC-defined domains, but the distribution of ERVK showed the opposite tendency (Fig. 2E). These data indicate that the THDs are distributed predominantly in intergenic regions, especially LINE/L1-rich regions.
THDs mark stable B compartments
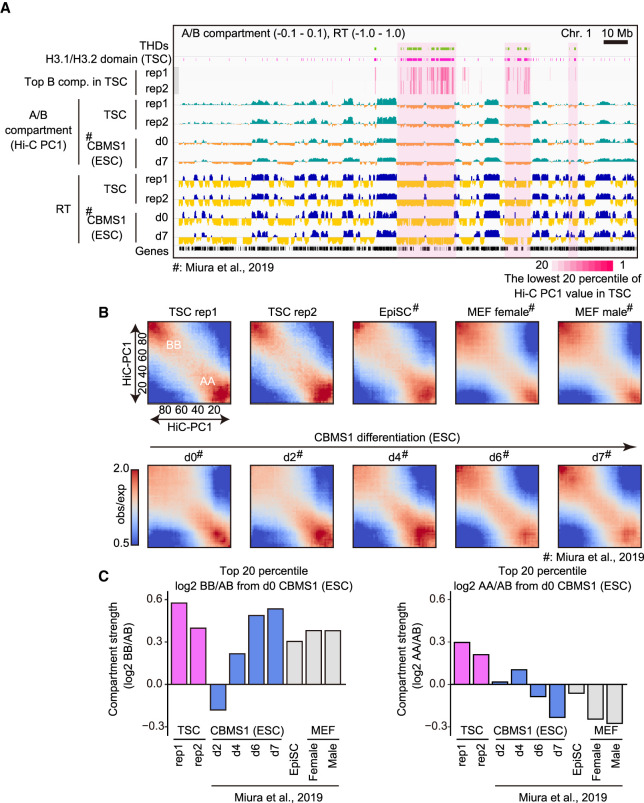
Given that the THD sizes were on the order of several megabases, it is likely that they are involved in the organization of higher-order chromatin structure, rather than in regulating gene repression directly. Mammalian genomic architecture can be divided roughly into two genomic compartments—A (active) and B (inactive)—depending on their transcriptional status (Dixon et al. 2012, 2015). These A and B compartments are well correlated with early and late DNA replication timing, respectively (Gilbert et al. 2010; Ryba et al. 2010). To investigate the relationship between the THDs and higher-order chromatin structure, TSCs were subjected to high-throughput chromosome conformation capture (Hi-C) analysis and DNA replication timing sequencing (Repli-seq) analysis (Marchal et al. 2018). The THDs were well localized with the B compartment and with late replication timing (Fig. 3A). However, these features did not seem to be TSC-specific, because ESCs also displayed B compartments in the same regions. This implies that, although H3K9me3 enrichment was relatively weak in ESCs, these regions were able to construct the B compartment as defined by Hi-C. However, we noted that the THDs overlapped well with the lowest 20 percentile of the Hi-C principal component 1 (Hi-C PC1) value, indicating that these regions were included in an extremely strong B compartment (light-magenta areas in Fig. 3A). We next analyzed the compartment interaction strength to investigate how the H3K9me3 domains can affect chromatin stability. Focusing on the lowest 20 percentile of Hi-C PC1 values, TSCs displayed strong B-to-B compartment interactions, as did differentiated ESCs and MEFs (Fig. 3B,C; Miura et al. 2019). Interestingly, TSCs also showed a high A-to-A interaction, while differentiated ESCs and MEFs did not (Fig. 3B,C). These results suggest that mouse TSCs maintain a specific “bivalent” chromatin architecture that combines the properties of both undifferentiated and differentiated cells.
Figure 3.
The THDs construct stable B compartment domains. (A) IGV snapshot showing the A/B compartments from Hi-C analysis and early/late replication timing from Repli-seq. Light-magenta areas indicate overlapped regions of THDs and the lowest 20 percentile of Hi-C PC1 values in TSCs. d0 and d7 CBMS1 ESCs represent the statuses of undifferentiated ESCs and differentiated ESCs for neurectoderm (7 d after differentiation), respectively. Hi-C and Repli-seq data for ESCs were obtained from Miura et al. (2019). (B) Heat map showing average contact enrichment between pairs of 200-kb bins sorted by their Hi-C PC1 values, from the lowest (the most extreme B) to the highest (the most extreme A). Names of cells, time points of ESC differentiation, and the scale bar of observed/expected ratio are as indicated. (C) Bar plots showing the differential ratio of B–B/A–B (left) and A–A/A–B (right) interactions between each cell and d0 ESCs. Note that TSCs show strong B–B compartment interaction as well as A–A compartment interaction.
H3.1/H3.2 are responsible for maintenance of H3K9me3 in THDs
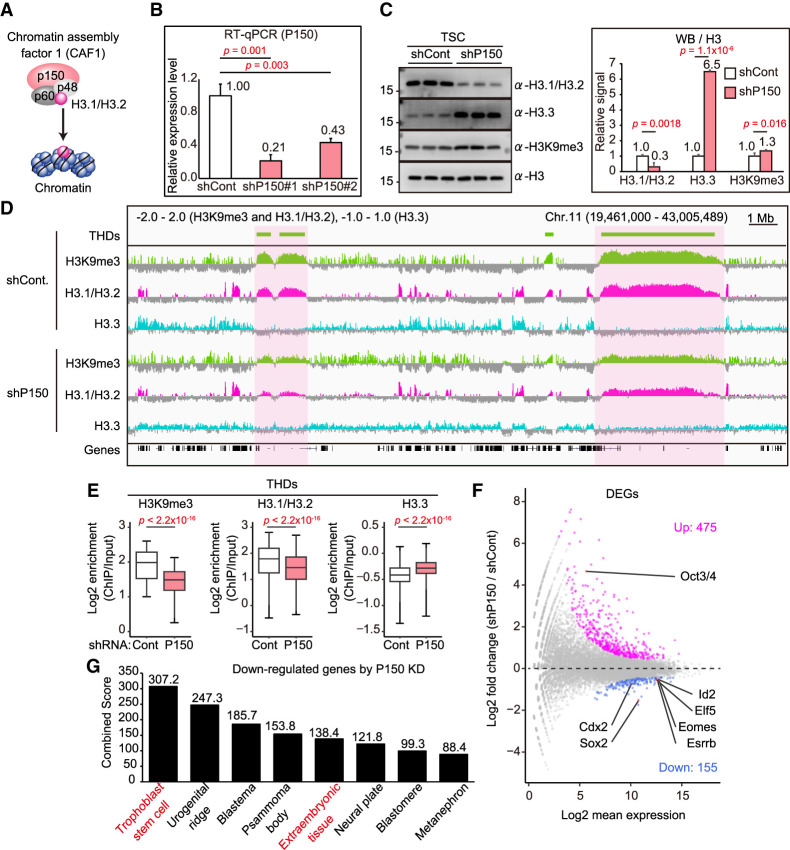
That H3K9me3 and H3.1/H3.2 are colocalized in the large domains of the TSC genome prompted us to investigate their functional relationships at the molecular level. To this end, we attempted to diminish H3.1/H3.2 from TSC chromatin by knocking down P150, the large subunit of the CAF1 complex responsible for H3.1/H3.2 deposition (Fig. 4A; Hake and Allis 2006). We introduced two different short hairpin (sh) RNA sequences targeting P150 (shP150#1 and shP150##2) into TSCs using a lentivirus vector. Based on the knockdown efficiencies of these two shRNAs, we used shP150#1 for further experiments (Fig. 4B). Depletion of P150 successfully decreased the amounts of H3.1/H3.2 proteins (Fig. 4C). The P150 knockdown did not induce any apparent changes in the morphology of TSC colonies (Supplemental Fig. S3A). Consistent with previous studies, depletion of H3.1/H3.2 triggered the accumulation of H3.3, probably to compensate for the loss of H3.1/H3.2 (Fig. 4C; Hatanaka et al. 2015; Gomes et al. 2019). Our ChIP-seq analysis further confirmed the reciprocal behaviors of H3.1/H3.2 and H3.3 (Fig. 4D,E). Importantly, it was apparent that the enrichment of H3.1/H3.2 and H3K9me3 in the THDs was attenuated by P150 knockdown (Fig. 4D,E). These data indicate that H3.1/H3.2 play an important role in maintaining the H3K9me3 domains. Unexpectedly, the global amount of H3K9me3 was slightly increased by P150 knockdown (Fig. 4B), perhaps reflecting the abundant H3K9me3 modification on the H3.3 region, as reported for mouse ESCs (Elsässer et al. 2015). To investigate the possibility of regulating H3K9me3 levels by H3.1/H3.2, we examined the expression levels of H3K9me3-related genes in P150 knockdown TSCs. We found that the expression levels of genes encoding H3K9me3 methyltransferases (Setdb1 and Suv39h1/2) and demethylases (Kdem4a/b/c/d) were largely unaffected by P150 knockdown (Supplemental Fig. S2A). These results are consistent with our finding that the global H3K9me3 level was not decreased following P150 knockdown treatment (Fig. 4C).
Figure 4.
H3.1/H3.2 regulates the THDs, as revealed by P150 knockdown. (A) Illustration of the CAF1 complex. P150 is a large subunit of the CAF1 complex and is responsible for H3.1/H3.2 assembly. (B) The effectiveness of knockdown of P150 in TSCs by RT-qPCR analysis. Two different shRNAs (shP150#1 and shP150#2) were used for the knockdown. (C) Western blot analysis of P150 knockdown TSCs. Representative images (left) and the relative signal levels (right) are shown. Signal intensities were normalized to H3 and are presented as levels relative to ESCs (set as 1.0). Data are presented as the mean ± SD. P-values were calculated from two-tailed, unpaired Student's t-tests. (D) IGV snapshots of large H3K9me3 (THDs) and H3.1/H3.2 domains in TSCs following P150 knockdown. The THDs, as well as the H3.1/H3.2 domains, were diminished by P150 knockdown. (E) Box plots showing enrichment of H3K9me3, H3.1/H3.2, and H3.3 within the THDs. P-values from the Wilcoxon rank sum test are indicated. There were 3434 bins analyzed. (F) Differential expression analysis of P150 knockdown TSCs. Significantly up-regulated (475) and down-regulated (155) genes in P150 knockdown TSCs are highlighted by magenta and blue, respectively. Up-regulated ESC marker gene (Oct3/4) and down-regulated TSC marker genes (Cdx2, Sox2, Elf5, Id2, Eomes, and Esrrb) are indicated. Genes with a Padj-value < 0.01 were extracted as differentially expressed genes (DEGs). (G) Gene set enrichment analysis using down-regulated genes in P150 knockdown TSCs. Down-regulated genes were subjected to Jensen Tissues through the Enrichr website. The combined score was calculated from the P-value and the z-score.
Based on our findings above, we hypothesized that THDs might be responsible for silencing repetitive elements, especially LINE/L1. Therefore, we next addressed whether P150 knockdown could derepress LINE/L1 expression. Knockdown of P150 in TSCs resulted in significant up-regulation of 15 repeats (Supplemental Fig. S2B), but they did not contain any LINE/L1 family repeats (Supplemental Fig. S2C). One possible explanation for this might be the involvement of DNA methylation in the repression of repetitive elements in TSCs, as reported for ESCs and germ cells (Hatanaka et al. 2015; Sharif et al. 2016; Yang and Wang 2016). To test this, we treated TSCs with 5-aza-2′-deoxycytidine (5-AZA-CdR), a DNA methylation inhibitor. Treatment with 5-AZA-CdR induced significantly higher expression of not only LINE but also SINE, IAP, and MERVL (Supplemental Fig. S2D). As the toxic effect of 5-AZA-CdR might have damaged TSCs, we also attempted to down-regulate DNA methylation by Dnmt1 knockdown. This significantly increased the expression of all the repetitive elements examined (LINE, SINE, IAP, and MERVL) (Supplemental Fig. S2E). In addition, we also found that, when combined with P150 knockdown, DNMT1 knockdown further increased the expression levels of LINE and SINE, indicating that H3.1/H3.2–H3K9me3 and DNA methylation cooperate in silencing these retrotransposons (Supplemental Fig. S2E). These data indicate that DNA methylation plays a major role in silencing repetitive elements in TSCs, and that H3.1/H3.2–H3K9me3 may play a minor role.
We also noted that P150 knockdown TSCs displayed decreased expression levels of TSC marker genes (Cdx2, Sox2, Elf5, Id2, Esrrb, and Eomes) (Fig. 4F; Supplemental Fig. S3B). Consistent with this, gene ontology (GO) analysis using 155 down-regulated genes in P150 knockdown TSCs revealed that the down-regulated differentially expressed genes (DEGs) were enriched for the GO terms “trophoblast stem cell” and “extraembryonic tissue,” indicating that TSCs’ character was compromised by the depletion of P150 (Fig. 4G). However, this was not caused by TSC differentiation because differentiation marker genes including Hand1, ASCl2, Tpbpa, and Gcm1 were not up-regulated by P150 knockdown (Supplemental Fig. S3B). We confirmed that the TSC line we used had normal differentiation ability from changes in colony morphology and increased expressions of marker genes for differentiated TSCs such as Hand1 and Mash2 (Supplemental Fig. S3A–C). Therefore, we speculate that TSCs might have lost their identity as the trophoblast lineage by P150 knockdown. Indeed, we also found that P150 knockdown TSCs showed increased expression of Oct3/4, one of the major undifferentiated markers of ESCs, although the levels of other pluripotency markers such as Nanog, Gdf3, and Eras were unchanged (Fig. 4F; Supplemental Fig. S3B,D). We also confirmed the protein expression of Oct3/4 in P150 knockdown TSCs by immunostaining (Supplemental Fig. S3F). To determine whether Oct3/4 expression was the direct consequence of H3.1/H3.2 depletion, we examined the enrichment of H3.1/H3.2 around the Oct3/4 locus in TSCs. However, there was no such accumulation of H3.1/H3.2, indicating that the up-regulation of Oct3/4 by P150 knockdown was not the direct consequence of H3.1/H3.2 depletion, but rather ectopic activation following the decay of the TSC-specific gene network (Supplemental Fig. S3G). Taken together, these data suggest that TSCs require the H3.1/H3.2 groundwork to maintain their identity as the trophoblast lineage through the formation of THDs. However, it will be important to identify the mechanisms by which THDs maintain the expressions of TSC marker genes because THDs represent repressive chromatin structures that do not play active roles in gene expression.
Extraembryonic cells share the H3K9me3 domains in vivo
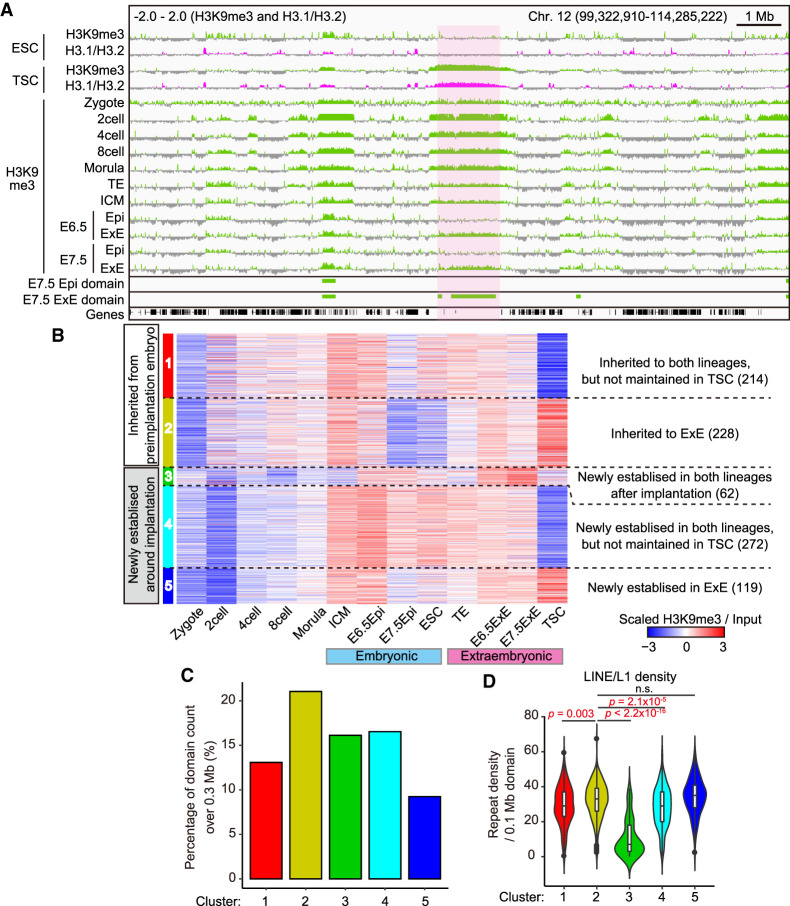
The existence of THDs in the TSC genome raises two important questions: Are they also observed in extraembryonic cells in vivo? If so, when are they established during embryonic development? To address these questions, we analyzed published H3K9me3 ChIP-seq data from zygotes to embryonic day 7.5 (E7.5) postimplantation mouse embryos consisting of the epiblast (Epi) and extraembryonic ectoderm (ExE) (Wang et al. 2018). Of note, embryos from the two-cell stage to blastocysts (the ICM and TE) displayed H3K9me3-enriched regions that appeared to be slightly broader than those of TSCs (light-magenta area in Fig. 5A). Importantly, we found that some H3K9me3 domains were maintained in both ExE and Epi cell lines while others were found only in the ExE after implantation. This prompted us to investigate the lineage-specific dynamics of H3K9me3 domains during development. The H3K9me3 domains in E7.5 Epi and ExE were defined in the same way for ESCs and TSCs (above) and subjected to k-means clustering. About half of the domains were inherited from preimplantation embryos (clusters 1 and 2 [442 of 895]) while the rest were newly established around the time of implantation (clusters 3–5 [453 of 895]) (Fig. 5B). Clusters 2 and 5 were specifically maintained in extraembryonic cells, while others were maintained in both lineages. Cluster 2 contained a higher percentage of large H3K9me3 domains (>0.3 Mb) than the other clusters did, suggesting that the large H3K9me3 domains for the most part are constructed during preimplantation development and tend to be inherited by the extraembryonic lineage (Fig. 5C). Finally, we computed the LINE/L1 density within each cluster and found that the ExE-specific clusters (clusters 2 and 5) showed significantly higher LINE/L1 densities than other clusters did (Fig. 5D). These data indicate that in vivo extraembryonic cells possess the H3K9me3 domains preferentially covering the LINE/L1-rich regions, as also seen in TSCs.
Figure 5.
TSCs and the extraembryonic linage share the H3K9me3 domains, which originate from preimplantation embryos. (A) IGV snapshots showing enrichment of H3K9me3 in ESCs, TSCs, preimplantation embryos, and postimplantation embryos. H3K9me3 ChIP-seq data against preimplantation and postimplantation embryos were obtained from Wang et al. (2018). The light-magenta area indicates a THD that seems to be inherited from preimplantation embryos by the extraembryonic tissues (ExE). (TE) Trophectoderm, (ICM) inner cell mass, (Epi) epiblast, (ExE) extraembryonic ectoderm. (B) Heat map showing H3K9me3 dynamics of the H3K9me3 domains defined by either E7.5 Epi or ExE. The domains are classified into five clusters by their pattern before and after implantation. The number of domains in each cluster is indicated in parenthesis. (C) Bar plot showing the distribution of the domains over 0.3 Mb among the clusters. (D) Violin plot showing the LINE/L1 density in each cluster. P-values from the Wilcoxon rank-sum test are indicated.
Human placental cells also develop large H3K9me3 domains in their intergenic regions
We next sought to evaluate whether the extraembryonic lineage-specific H3K9me3 domains are also conserved in human cells. The human placenta is composed of three major types of trophoblast cells: cytotrophoblast (CT) cells, extravillous cytotrophoblast (EVT) cells, and syncytiotrophoblast (ST) cells. CT cells are undifferentiated cells that can differentiate into EVT and ST cells (Knöfler and Pollheimer 2013). EVT cells are invasive cells that serve to anchor the chorioallantoic placenta to the uterine wall (Chang et al. 2018). Multinucleated ST cells provide the main site for placental functions such as gas exchange (Knöfler and Pollheimer 2013).The ChIP-seq data for H3K9me3 in these trophoblast cells and two human TSC lines established from blastocyst cells and CT cells, respectively, are available at the International Human Epigenome Consortium site (http://ihec-epigenomes.org/about), so we compared the enrichment of H3K9me3 in these human placental cells with that of human ESCs, IMR90 cells (human fetal lung fibroblasts) (Soufi et al. 2012), and stroma cells isolated from the uterine endometrium. As shown in Supplemental Figure S4A, human placental cells had distinct H3K9me3 domains regardless of cell type and developmental stage. IMR90 and stroma cells also had H3K9me3-enriched regions, but they were characterized by broader, less dense distributions compared with those of placental trophoblast cells (Supplemental Fig. S4A). Unexpectedly, human TSCs—whether derived from blastocyst cells or CT cells—showed an H3K9me3 distribution pattern similar to that of human ESCs, indicating that human TSCs most likely lose trophoblast-specific H3K9me3 enrichment during their establishment, unlike mouse TSCs (see Fig. 1B). The H3K9me3 domains in human placental cells were predominantly distributed in intergenic regions, as found in mouse TSCs (Fig. 2A; Supplemental Fig. S4B). To determine the specificity of H3K9me3 distribution among the cell types tested, we performed hierarchical clustering analysis using the Jaccard index based on the H3K9me3 domains. Importantly, human placental trophoblast cells were categorized into a close cluster, whereas IMR90 cells, stroma cells, ESCs, and TSCs were outside this cluster, indicating that human placental trophoblast cells also construct specific H3K9me3 domains in their genome (Supplemental Fig. S4C). Two human TSC lines were classified into the same cluster as ESCs (Supplemental Fig. S4C).
H3K9me3 in the TSC genome is the major barrier to genomic reprogramming by SCNT
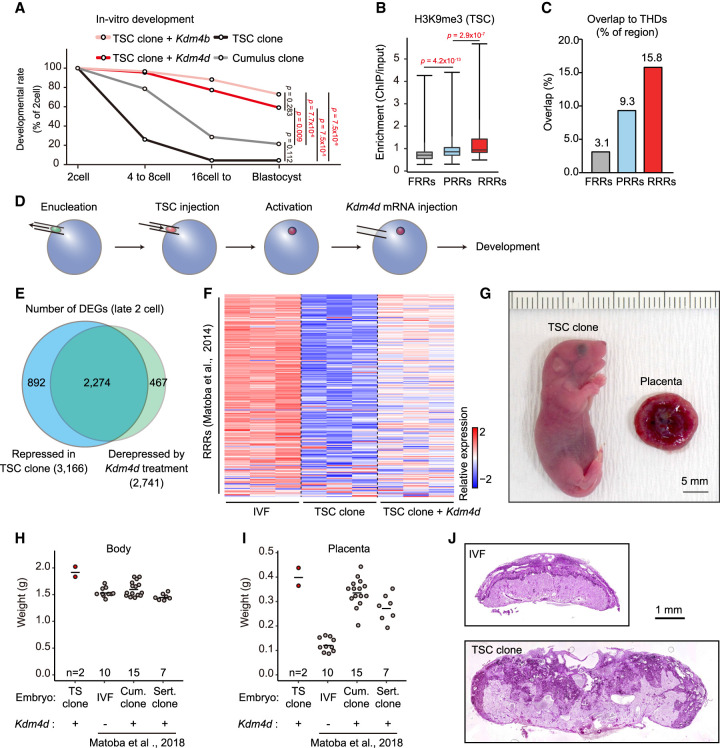
Genomic reprogramming by SCNT is a powerful technology used to produce cloned animals from somatic cells and for assessment of the genomic plasticity of cells of interest. As far as we know, there is no report on the production of offspring by nuclear transfer using TSCs or extraembryonic cells, while many other cell types have been cloned successfully by SCNT (Loi et al. 2016; Matoba and Zhang 2018; Wakayama et al. 2019; Ogura et al. 2021). Indeed, when we examined the in vitro development of embryos cloned from mouse TSCs, most of them showed arrested development before the morula/blastocyst stages (Fig. 6A; Supplemental Table S1). Previous studies have shown that H3K9me3-enriched regions in donor somatic cells are prone to persist after genomic reprogramming by SCNT (reprogramming-resistant regions [RRRs]) and strongly interfere with the development of cloned embryos (Matoba et al. 2014). Indeed, the removal of H3K9me3 from RRRs in the donor genome by the overexpression of H3K9me3 demethylase Kdm4d significantly improved the cloning efficiency. Therefore, we hypothesized that the high H3K9me3 contents of the TSC genome might have hampered efficient reprogramming. We confirmed that, as with other somatic cells, more H3K9me3 was enriched in the RRRs than in fully reprogrammed regions (FRRs) or partially reprogrammed regions (PRRs) (Matoba et al. 2014) in TSCs (Fig. 6B). Furthermore, RRRs were preferentially overlapped with THDs (Fig. 6C). To investigate the effect of H3K9me3 removal on the development of TSC cloned embryos, we introduced Kdm4d mRNA into reconstructed oocytes after activation (Fig. 6D). In the nuclei of the reconstructed oocytes treated with Kdm4d, H3K9me3 and H3K9me2 were diminished because Kdm4d has a demethylase activity for both H3K9me3 and H3K9me2 (Supplemental Fig. S5A; Whetstine et al. 2006). The developmental ability of TSC cloned embryos was greatly improved by the Kdm4d treatment: Approximately 60% of these embryos reached the blastocyst stage (Fig. 6A; Supplemental Table S1).
Figure 6.
H3K9me3 strongly impaired the developmental efficiency of TSC cloned embryos. (A) In vitro development of SCNT embryos derived from TSCs and cumulus cells. Injection of Kdm4d or Kdm4b mRNA greatly improved the blastocyst formation rate of TSC cloned embryos. P-values were calculated using Fisher's exact test. (B) Box plot showing H3K9me3 enrichment within FRRs, PRRs, and RRRs in TSCs. These regions are referred from Matoba et al. (2014). P-values were calculated using the Wilcoxon rank sum test. (C) Bar plot showing the percentage of overlapped regions between THDs and FRRs, PRRs, or RRRs. (D) Schematic illustration of SCNT procedure. Kdm4d mRNA was injected to erase H3K9me3 from the genome of TSC cloned embryos. (E) Venn diagram showing the overlap between the genes that failed to be activated in TSC cloned embryos and those derepressed in Kdm4d-treated TSC cloned embryos. Genes with Padj < 0.01 and fold change > 1.5 were classed as DEGs. (F) Heat map showing relative gene expression levels of the RRRs at the late two-cell stage. (G) A pup and a placenta derived by cloning from TSCs with Kdm4d mRNA injection. (H,I) Scatter plots showing the weight of the body (H) or the placenta (I) at birth (E19.5). Each dot represents a sample. Data for IVF-derived embryos, cumulus cell cloned embryos, and Sertoli cell cloned embryos are from Matoba et al. (2018). (J) Representative images of histological sections of term placentas stained with periodic acid Schiff (PAS). The TSC cloned placenta shows an expansion of the PAS-positive spongiotrophoblast layer and an irregular boundary with the PAS-negative labyrinthine layer.
To confirm the improved developmental efficiency of these Kdm4d-treated cloned embryos at the transcriptomic level, we analyzed them at the late two-cell stage by RNA-seq. It is known that major zygotic gene activation (ZGA) occurs at the late two-cell stage in mice and that SCNT embryos frequently fail to induce major ZGA (Inoue et al. 2015; Yang et al. 2021). Differential expression analysis revealed that 3166 genes were significantly suppressed in TSC cloned embryos compared with in vitro fertilization (IVF)-derived embryos, whereas 2274 genes (71.8% of them) were recovered by Kdm4d treatment (Fig. 6E). We next tested how much major ZGA was improved in TSC cloned embryos following the Kdm4d treatment. Hierarchical clustering analysis revealed that 2993 reported major ZGA genes could be divided into two groups based on their differential expression levels in TSC cloned embryos compared with IVF-derived embryos; group 1 (G1) genes were repressed in TSC cloned embryos while group 2 (G2) genes were hyperactivated in these embryos (Abe et al. 2018). Following Kdm4d treatment, the expression levels of G1 genes recovered to the IVF-derived embryo level, whereas G2 genes largely remained hyperactivated (Supplemental Fig. S5B). We also analyzed the reprogramming efficiency of RRRs and found that the gene expression levels within RRRs were at least partially restored by Kdm4d treatment (Fig. 6F). Furthermore, genes within THDs, which were suppressed in TSC cloned embryos, were activated by Kdm4d treatment (Supplemental Fig. S5C). Collectively, our transcriptome analysis confirmed that the reprogramming efficiency of the TSC genome by nuclear transfer was strongly impeded by H3K9me3 enrichment but could be largely ameliorated by Kdm4d treatment leading to activation of developmentally important genes.
However, as mentioned above, it is true that Kdm4d can also demethylate H3K9me2, another strong repressive histone mark (Supplemental Fig. S5A). To further evaluate the major involvement of H3K9me3 in the genomic stability of TSCs, we next examined the developmental ability of TSC cloned embryos treated with Kdm4b, a more specific demethylase for H3K9me3. As expected, H3K9me3 was diminished in the nuclei of Kdm4b-treated TSC cloned embryos, whereas H3K9me2 remained positive to various extents in these nuclei (Supplemental Fig. S5D). Importantly, the Kdm4b-treated TSC cloned embryos developed into blastocysts at a rate similar to, or even higher than, those treated with Kdm4d (73% vs. 59%, P > 0.05) (Fig. 6A; Supplemental Table S1). These findings further support the notion that H3K9me3 plays the major role in the genomic stability of TSCs.
Because we found that P150 knockdown in TSCs led to a decrease of H3K9me3 within THDs, we next sought to investigate the effect of P150 knockdown on SCNT cloning efficiency. However, the blastocyst formation rates of embryos cloned from P150 knockdown and from control TSCs were indistinguishable (Supplemental Fig. S5E; Supplemental Table S1). Consistent with this result, our RNA-seq analysis revealed that among the 3166 genes repressed in TSC cloned embryos, only two (0.01%) had their expression restored by P150 knockdown in the donor TSCs (Supplemental Fig. S5F). Taken together, P150 knockdown in the donor TSCs did not improve reprogramming efficiency by SCNT, unlike the pronounced effects of Kdm4d or Kdm4b treatment (see Fig. 6A,E). The inability of P150 knockdown to improve reprogramming efficiency might have been caused by the slight increase in genome-wide H3K9me3 in TSCs following P150 knockdown (Fig. 4C). In contrast, Kdm4d and Kdm4b target H3K9me3 directly and remove it from the genome (Supplemental Fig. S5A).
Finally, we transferred Kdm4d-treated TSC cloned embryos into recipient females to determine their developmental ability in vivo. Following transfer of 37 embryos, two live cloned pups were born (5%) (Fig. 6G). Their body and placental weights were slightly greater than those of other cloned pups (Fig. 6H,I). This result indicates that TSC clones also develop SCNT-specific large placentas (Matoba et al. 2018; Matoba and Zhang 2018; Wakayama et al. 2019). Their placentas also showed SCNT-specific histology with an enlarged PAS-positive spongiotrophoblast layer (Fig. 6J). Unfortunately, two TSC-derived pups did not survive until weaning for unknown causes, although such neonatal deaths are common in SCNT-derived newborn cloned mice. Thus, our embryo transfer experiments confirmed that the TSC genome can be fully reprogrammed to the totipotent state by the removal of high H3K9me3 contents.
Discussion
Here, we show that THDs are broadly deposited in the TSC genome, predominantly within intergenic regions. One of the important features of THDs is the coexistence of H3K9me3 with H3.1/H3.2. This clearly distinguishes TSCs from ESCs, which showed only weak association of the large H3K9me3 domains with H3.1/H3.2, as revealed by ChIP-seq analysis and immunostaining (Fig. 1). Clustering analysis based on the distribution of the H3K9me3 domains also showed that TSCs are clearly distinct from ESCs and other somatic cells (Fig. 1C). In addition, the ATAC-seq data showed that the TSC-specific clusters (clusters 3 and 4) have lower chromatin accessibility than the embryonic lineage-specific cluster (cluster 1). These findings indicate that the THDs in TSCs consist of a distinctively closed chromatin state that is not found in other cell types.
We then tested why the THDs are formed specifically in TSCs. They largely overlapped with the LINE/L1 regions, suggesting that they might be responsible for silencing endogenous retrotransposons. Indeed, we and others have found previously that knockdown of P150, the large subunit of the CAF1 complex, led to ectopic derepression of different classes of retrotransposons in preimplantation embryos, in association with reduced repressive histone marks including H3K9me3 (Hatanaka et al. 2015; Wang et al. 2018). This adverse phenotype occurs because the genomes of late preimplantation embryos are largely devoid of DNA methylation, and thus the silencing of the endogenous retrotransposons is dependent on repressive histone marks deposited by CAF1. In contrast, in the present study, depletion of P150 from the TSC genome led to very limited up-regulation of retrotransposons. The most probable cause of this discrepancy is the presence of DNA methylation because inhibition of DNA methylation by 5-AZA-CdR and Dnmt1 knockdown resulted in ectopic expression of endogenous retrotransposons in TSCs (Supplemental Fig. S2D,E). Importantly, Dnmt1 knockdown combined with P150 knockdown further derepressed some retrotransposons (LINE, SINE, and MERVL) (Supplemental Fig. S2E). Thus, repression of retrotransposons in trophoblast cells can be achieved by two layers of repressive systems, DNA methylation, and H3.1/H3.2–H3K9me3, which act synergistically depending on the type of retrotransposon.
Knockdown of P150 also resulted in down-regulation of major TSC marker genes including Cdx2, Sox2, and Elf5. As their down-regulation was not associated with the up-regulation of differentiated trophoblast genes such as Hand1 and Ascl2, this phenomenon might reflect deviation from the extraembryonic lineage, rather than differentiation from the stem cell state. Consistent with this, Oct3/4, a marker of pluripotent stem cells such as ESCs, was expressed ectopically, probably from dysregulation of the TSC gene network. Thus, CAF1 contributes to the establishment of the epigenetic basis of the extraembryonic lineage by providing specific histone molecules including H3K9me3. In contrast, in ESCs, P150 knockdown resulted in the increase of “two-cell embryo-like” cells while maintaining the ESC characteristics (Ishiuchi et al. 2015). Thus, it is interesting to note that H3.1/H3.2 in ESCs and TSCs play such diverse roles in relation to their stem cell characteristics.
We found that large H3K9me3 domains were also established in the intergenic regions of different trophoblast types from human tissues, indicating that the acquisition of this large-scale intergenic H3K9me3 enrichment might have been coincident with the evolution of the placenta in mammals. Therefore, we assume that this epigenetic feature of extraembryonic cells is related to the special features of the placenta—specifically, to its restricted ability to differentiate only into placental cells (trophoblasts) and its gestation-limited function. H3K9me3 is generally considered to be responsible for the formation of closed chromatin regions and may act as a barrier to cell fate changes by occluding the DNA from transcription factor binding (Burton and Torres-Padilla 2014; Becker et al. 2016). Indeed, THDs involve a distinctively closed chromatin state as revealed by our Hi-C and ATAC-seq analyses (Figs. 1C, 3A,B). Thus, we postulate that the rigid THDs in extraembryonic cells may serve to protect their genomic architecture from unnecessary rearrangements during placental development and steer differentiation in the correct direction. Importantly, Hi-C analysis used to identify A/B compartments revealed that the genomic architecture of TSCs possesses characteristics not only of undifferentiated ESCs (high A-to-A interaction) but also of differentiated ESCs (high B-to-B interaction) (Fig. 3B,C). It is known that in mouse ESCs, high A-to-A interactions involve large chromatin loops bound by Polycomb-related proteins and pluripotency transcription factors such as Nanog. During neuronal differentiation, B-to-B interactions appear gradually in association with disruption of the A-to-A-specific networks and newly formed loop contacts within neural genes (Bonev et al. 2017). Thus, for better understanding of the bivalent property of TSCs (both A-to-A and B-to-B interactions), it will be necessary to know what transcription factors are involved in the A-to-A interactions and what genes occupy the B-to-B-interacting regions. This should be the subject of future studies. Taken together, in TSCs, and perhaps among mammalian placental cell lineages in general, such lineage-specific differentiation ability may rely on THDs in their genome. However, the THDs in TSCs and the H3K9me3 regions in extraembryonic tissues are not always matched exactly. Therefore, to make this idea more plausible, we should identify how H3K9me3, a repressive histone mark, ensures the epigenetic status of trophoblast cells.
In this context, the loss of large H3K9me3 domains in the human TSCs was unexpected. Human TSCs should have lost these domains during their establishment as cell lines. Our hierarchical clustering analysis revealed that human TSCs are very similar to human ESCs. Consistent with this, it was reported that human TSCs can be established easily from human ESCs or iPSCs (Zhou et al. 2021). It is possible that the barrier between embryonic and extraembryonic lineages might be lower in humans than that in mice.
Finally, we aimed to determine the genomic plasticity of TSCs by transferring their nuclei into enucleated oocytes using SCNT technology. At present, >15 cell types can be used for SCNT cloning in >20 mammalian species (Loi et al. 2016; Matoba and Zhang 2018; Wakayama et al. 2019; Ogura et al. 2021). However, there has been no report on the birth of clones from extraembryonic cells including TSCs (Ogawa et al. 2015). We also confirmed that the developmental ability of TSC cloned embryos in vitro was worse than we had ever experienced using >10 cell types (Fig. 6A; Ogura et al. 2013; Mizutani et al. 2015). However, when we removed H3K9me3 from the genome of TSC cloned embryos by Kdm4d mRNA injection, their developmental ability was greatly improved, and two live pups were born following embryo transfer. As far as we know, these are the first cloned animals derived from extraembryonic cells. We identified that the THDs largely overlapped with the RRRs reported previously (Matoba et al. 2014). These findings further support our hypothesis that H3K9me3 in the TSC genome acts as a safeguard against undesirable cell fate transitions discussed above.
The SCNT-specific placental enlargement (placentomegaly) is known to be caused by the loss of H3K27me3-dependent imprinting (noncanonical imprinting) (Inoue et al. 2017, 2020; Wang et al. 2020). This noncanonical imprinting is erased in the embryonic lineage but remains in the extraembryonic lineage (Inoue et al. 2017). Therefore, the finding that cloned pups derived from TSCs had enlarged placentas was unexpected. One possibility is that the epigenetic status of a subset of DNA methylation-dependent canonical imprinted genes had been compromised in TSCs during culture and led to placental enlargement. This might also explain the overweight of TSC-derived pups. Similar adverse effects of imprinting disorders in donor ESCs on the phenotypes of the ESC-derived cloned pups and placentas have been documented (Eggan et al. 2001; Humpherys et al. 2001).
In conclusion, the TSC genome is characterized by abundant H3.1/H3.2–H3K9me3 sequences, forming large domains in the intergenic regions. These genomic domains are responsible for the formation of a specific genomic architecture that ensures the restricted differentiation of TSCs into placental cells, rather than the repression of the endogenous retrotransposons. This epigenetic feature and its role might be shared with extraembryonic cells across mammalian species. The abundance of H3K9me3 in the TSC genome is a strong barrier to reprogramming by SCNT, but following removal of this barrier, the first TSC-derived cloned mice were born, indicating that the TSC genome can indeed be fully reprogrammed.
Materials and methods
Animal experimentation
Experiments using mice were approved by the institutional animal care and use committee of RIKEN Tsukuba Branch and were performed in accordance with the committee's guiding principles. Mice were housed under controlled lighting conditions (daily light, 07:00–21:00) and were maintained under specific pathogen-free conditions.
Establishment of ESC and TSC lines
ESC and TSC lines were established from blastocysts generated by IVF using oocytes from C57BL/6NCrSlc female mice and spermatozoa from DBA/2 or JF1/Ms male mice. For establishment of ESCs, developing IVF-derived blastocysts were treated with acidic Tyrode's solution (Merk Millipore MR-004-D) to remove the zona pellucida, and then transferred to ESC medium containing knockout DMEM (Gibco 10829018), 15% knockout serum replacement (KSR; Thermo Fisher 10828028), MEM nonessential amino acid solution (Gibco 1140050), GlutaMAX supplement (Gibco 35050061), 100 μM 2-mercaptoethanol (2-ME; Merk Millipore M-7522), ESGRO recombinant mouse LIF protein (Merk Millipore ESG1107), 1 μM PD0325901 (Stemgent 04-0006), and 1 μM CHIR99021 (Stemgent 04-0004). Blastocysts were placed onto layers of primary mouse embryonic fibroblasts treated with mitomycin C (MMC MEFs). After several passages, cell lines with typical ESC colonies were selected and used for further analysis. For TSCs, developing blastocysts were transferred directly to TSC medium containing the same amount of neurobasal medium (Gibco 21103049) and Dulbecco's modified Eagle's medium/F12 (Ham's; 1:1; Gibco 11320033), N2 supplement (Gibco 17502048), B27 supplement (Gibco 17504044), GlutaMAX supplement, 150 μM 2-ME, 1 μg/mL heparin (Merk Millipore H3149), 0.05% bovine serum albumin (Merk Millipore 12657), 1% KSR, 50 ng/mL recombinant mouse basic FGF (Wako 062-05181), 20 ng/mL recombinant human activin A (R&D Systems 338-AC), 10 μM XAV939 (Calbiochem 575545), and 5 μM Y27632 (Wako 036-24023) (Ohinata and Tsukiyama 2014; Hirose et al. 2018). After several passages, cell lines with typical TSC colony morphology and normal karyotypes were selected for further experiments (Supplemental Fig. S1A).
Maintenance of ESCs and TSCs
Established ESC and TSC lines were adapted for feeder-free conditions. Briefly, ESCs were placed onto gelatin-coated dishes with ESC medium lacking PD0325901 and CHIR99021, and 1 μM GSK-3 inhibitor IX (Merk Millipore 361550) was added. TSCs were placed onto dishes coated with 15 mg/mL human plasma fibronectin (Merk Millipore FC010) (Ohinata and Tsukiyama 2014; Hirose et al. 2018). Medium was replaced every day and cells were passaged every 2 d.
Western blot analyses
Collected cell pellets were suspended in lysis buffer (20 mM Tris-HCl at pH 7.4, 500 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, plus a protease inhibitor cocktail [Roche 11873580001]) and then incubated for 5 min on ice. Chromatin pellets were collected by centrifugation at 20,400g for 5 min at 4°C and then suspended in SDS sampling buffer (50 mM Tris-HCl at pH 6.8, 2% SDS, 0.01% bromophenol blue, 10% glycerol, 5% 2-mercaptoethanol) for boiling at 95°C. Protein samples were resolved with 12% TGX FastCast acrylamide kits (Bio-Rad 1610175) and then transferred to PVDF membranes (Amersham 10600057). Membranes were blocked using 5% skim milk (BD 232100) in PBST (0.1% Tween 20 in phosphate-buffered saline [PBS]) for 30 min and then incubated with primary antibodies (Supplemental Table S3) diluted in 5% skim milk in PBST. Membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Supplemental Table S3), and then chemiluminometric signals were detected using ECL (Amersham RPN2232) and C-Digit (Li-cor 3600). Acquired images were processed using Image Studio (Li-cor) and ImageJ software (https://imagej.net/Welcome).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
RT-qPCR was performed as described (Hirose et al. 2018). Briefly, RNAs extracted using RNeasy mini kits were reverse-transcribed using the SuperScript III first strand synthesis system for RT-qPCR with oligo(dT) 20 primers (Invitrogen 18080051). Expression levels were quantified using a QuantStudio 7 Flex real-time PCR system with a PowerUp SYBR Green master mix (Applied Biosystems A25742) and normalized against the level of mouse Gapdh protein (NM_001289726).
Immunostaining of ESCs and TSCs
Cells were seeded into eight-well coverglass chambers (Iwaki 5232-008) and then cultured for 2 or 3 d. For fixation, 4% paraformaldehyde (PFA) in PBS was added and then incubated for 10 min at room temperature. Fixed cells were washed using PBST and then permeabilized using 1% Triton X-100 in PBS for 15 min. The cells were blocked using 1% bovine serum albumin (BSA) in PBST for 30 min and then incubated with primary antibodies (Supplemental Table S3) overnight at 4°C. After washing, secondary antibodies conjugated with Alexa fluor dyes (1:500) were added and incubated for 1 h at room temperature. DNA was stained by Hoechst 33342 (Dojindo 346-07951). Images were acquired using a BZ9000 (Keyence) or C-2 confocal laser scanning microscope (Nikon). Data were processed using ImageJ software.
Immunostaining of SCNT-derived embryos
Immunostaining of IVF and TSC cloned embryos was performed as described previously (Matoba et al. 2014). Briefly, one-cell-stage embryos were fixed with 4% PFA/0.1% Triton X-100 in PBS for 20 min at room temperature. Washed embryos were incubated in 1% BSA/PBS for 1 h for blocking and then stained with the primary antibodies (Supplemental Table S3) overnight at 4°C. After washing, the embryos were incubated for 1 h in 1% BSA/PBS containing secondary antibodies conjugated with Alexa fluor dyes (1:500). DNA was stained by mounting medium with DAPI (VectaShield H-1200). Images were acquired using a Nikon C-2 confocal laser scanning microscope. The obtained fluorescent intensity data were quantified using ImageJ software.
Lentiviral vector construction
The lentiviral vectors (CS-RfA-EVBsd), the packaging plasmids (pCAG-HIVgp and pCMV-VSV-G-RSV-Rev), and a Gateway entry vector pENTR4-H1tetOx1 were purchased from RIKEN Bioresource Research Center (BRC). The shRNA target sequences for genes encoding LacZ as well as and mouse P150 (NM_013733) are summarized in Supplemental Table S3. The annealed DNA oligos targeting each gene were inserted into pENTR4-H1tetOx1 at BglII/XbaI sites. The pENTR4-H1tetOx1 plasmids and CS-RfA-EVBsd were then recombined with LR clonase (Invitrogen 11791-020). In the case of shDnmt1, the blasticidin S resistance gene was replaced with the hygromycin resistance gene using a seamless ligation cloning extract (SLiCE) reaction (Motohashi 2015, 2017; Okegawa and Motohashi 2015a,b).
Lentivirus infection
Lentiviral vectors were cotransfected with the packaging plasmids into HEK293T cells purchased from RIKEN BRC (RCB2202). After incubation, the supernatants were collected twice at days 2 and 3, passed through a 0.45-μm filter, and centrifuged at 8000g for at least 14 h at 4°C. The virus pellets were suspended in TSC medium and mixed with the single-cell suspensions of TSCs derived by crossing C57BL/6NCrSlc female mice with JF1 male mice. After incubation for 4 h, the cells were washed and seeded onto fibronectin-coated dishes.
Doxycycline (DOX)-inducible knockdown
To establish stable TSC lines that express shRNA for LacZ and P150 in a DOX-dependent manner, 10 μg/mL blasticidin S (Fujifilm 026-18711) was added the next day after lentiviral infection, and cells were then cultured for at least 1 wk. In the Dnmt1 knockdown experiment, 150 μg/mL hygromycin B (Nakalai Tesque 02987-84) was added for 1 wk and then reduced to 50 μg/mL. To start knockdown, 2 μg/mL DOX was added to the cultures.
ChIP-seq library preparation
ChIP experiments were performed as described (Harada et al. 2012). Briefly, 1 × 107 ESCs or TSCs were collected by trypsinization and fixed using truChIP chromatin shearing kits (Covaris 520154). Aliquots of 2 × 107 cells were suspended in 2 mL of ChIP buffer (10 mM Tris-HCL at pH 8.0, 200 mM KCL, 1 mM CaCl2, 0.5% NP40, protease inhibitor cocktail) and incubated for 5 min on ice. Cells were sonicated using Handy Sonic (Tomy Seiko UR-20P) with 5-sec sonication (intensity 7) and 15-sec intervals three times and then incubated with micrococcal nuclease (NEB M0247) for 25 min at 37°C. Reaction was stopped by EDTA, and then lysates were centrifuged at 20,400g for 15 min at 4°C. Aliquots of the supernatants were kept for input. Antibodies were prebound to Dynabeads Protein G or Protein A (Invitrogen 10001D and 10004D) for at least 2 h at 4°C with rotation. Supernatants were incubated with bead complexes at 4°C with rotation, and then the immunocomplexes were washed twice using ChIP buffer, twice using wash buffer (10 mM Tris-HCL at pH 8.0, 500 mM KCL, 1 mM CaCl2, 0.5% NP40, protease inhibitor cocktail), and once using TE buffer. These complexes were eluted with elution buffer (10 mM Tris-HCl, 1% SDS, 5 mM EDTA, 300 mM NaCl) for 30 min at 65°C. Cross-linking was reversed by incubation for 6 h at 65°C, after which RNAs and proteins were degraded by adding RNase (Ambion AM2286) for 30 min at 37°C and Proteinase K (Zymo Research D3001-2) for 2 h at 55°C, respectively. chromatin-immunoprecipitated DNAs were purified using QIAquick PCR purification kits (Qiagen 28106). ChIP-seq libraries were prepared using NEBNext Ultra II DNA library preparation kits for Illumina (NEB E7645) according to the manufacturer's instructions. Single-end 65-bp sequencing was performed on an Illumina HiSeq 2500 platform.
RNA-seq library preparation for cultured cells
RNeasy mini kits (Qiagen 74134) were used for RNA extraction according to the manufacturer's instructions. RNA-seq library preparation and sequencing were performed at Macrogen Japan Corporation. Briefly, libraries were prepared using TruSeq stranded mRNA library (Illumina 20020594) and then sequenced by NovaSeq6000 for paired-end 100 bp.
RNA-seq library preparation for embryos
SMART-seq v4 Plus kits (Takara R400752) were used to construct RNA-seq libraries from late two-cell embryos. We used five embryos per reaction and subjected them to construction of libraries according to the manufacturer's instructions. Next-seq was used for sequencing in a 75-bp single-end mode.
Hi-C library preparation
Libraries for Hi-C were generated as described (Kadota et al. 2020). The detailed protocol for Hi-C library preparation is available at Protocols.io (https://www.protocols.io/view/iconhi-c-protocol-ver-1-0-4mjgu4n). Briefly, collected cells were washed with 5% FBS/PBS and then fixed with 1% formaldehyde (Polyscience 18814-10) in 5% FBS/PBS for 10 min. To stop fixation, 2.5 M glycine was added and then incubated for 5 min. Fixed cells were washed and stored at –80°C. In total, 2 × 106 fixed cells were lysed, permeabilized, and then digested with DpnII enzyme (4-bp cutter). Digested fragment ends were next filled with biotinylated nucleotides, and the resulting blunt-end fragments were ligated. After reverse cross-linking, ligation products were purified to obtain Hi-C DNA. One microgram of Hi-C DNA was fragmented by sonication, and the biotinylated ligation junctions were pulled down with streptavidin beads. Then, library preparation was performed on beads, and Hi-C libraries were sequenced by HiSeq X Ten for 150-bp paired-end reads.
Repli-seq library preparation
We followed the BrdU immunoprecipitation (IP)-based protocol, as described (Ryba et al. 2011). Cells were labeled with BrdU (Merk Millipore B5002) for 2 h and then collected by trypsinization. Cell pellets were resuspended in 5% FBS/PBS, and EtOH was added to a final concentration of 75% for fixation. The fixed cells were sorted into early and late S-phase fractions (20,000 cells per fraction) by FACS using a Sony SH800 cell sorter as described (Takahashi et al. 2019). Genomic DNA was extracted from each fraction and fragmented by sonication. Then, immunoprecipitation with anti-BrdU mouse monoclonal antibody (BD Biosciences 555627) was performed to enrich BrdU-labeled replicating DNA. After BrdU IP, immunoprecipitated DNA samples were subjected to whole-genome amplification (WGA) followed by library preparation (Miura et al. 2020). Constructed libraries from early- and late-replicating DNA after WGA were sequenced by HiSeq X Ten for 150-bp paired-end reads.
ATAC-seq
Libraries for ATAC-seq were prepared as described (Corces et al. 2017). Collected cells were washed and subjected to fragmentation by Tn5 transposase using Illumina Nextera DNA library preparation kits (Illumina 15032354) for 30 min at 37°C. Transposed DNA was purified using Qiagen MinElute PCR purification kits (Qiagen 28204) and amplified by PCR using published primers (Buenrostro et al. 2015). Purified libraries were sequenced by HiSeq X Ten for 150-bp paired-end reads.
SCNT
SCNT was performed as described (Matoba et al. 2018; Inoue et al. 2020). Briefly, MII oocytes were collected from adult B6D2F1 female mice via superovulation and enucleated in HEPES-buffered KSOM medium containing 7.5 μg/mL cytochalasin B (Calbiochem 250233) using a Piezo-driven micromanipulator (Primetech PMM-150FU). The donor nuclei of TSCs or cumulus cells were injected into the enucleated oocytes using a Piezo-driven micromanipulator. After 1-h incubation in KSOM, reconstructed SCNT oocytes were activated by incubation in Ca-free KSOM containing 3 mM strontium chloride, 5 μg/mL cytochalasin B, and 50 nM Trichostatin A (TSA) for 1 h, and further cultured in KSOM with 5 µg/mL cytochalasin B and 50 nM TSA for 4 h. Some SCNT embryos were injected with 10 pL of 1500 ng/μL mouse Kdm4d or Kdm4b mRNA at 5–6 h postactivation (hpa) using a Piezo-driven micromanipulator. The embryos were cultured with KSOM containing 50 nM TSA for an additional 3–5 h (i.e., 8–10 hpa), followed by further culture in KSOM. To evaluate the developmental capacity, some SCNT embryos were cultured in KSOM until 96 hpa, while others were transferred to the oviducts of pseudopregnant (E0.5) ICR female mice at the two-cell stage. The transferred embryos were recovered by cesarean section on the day of delivery (E19.5).
Chromosomal number analysis
Cells were seeded into six-well tissue culture dishes and cultured overnight. KaryoMax Colcemid (Thermo Fisher Scientific) was added to the medium and incubated for 5 h. A pellet of single cells dispersed by trypsin treatment was mixed with 75 mM KCl and incubated for 20 min at 37°C. For fixation, a double volume of Carnoy's solution was added and stored overnight at 4°C. The cells were centrifuged at 2250 g for 5 min and the supernatant was discarded. The fixed cells were dispersed in Carnoy's solution and washed again by centrifugation. The cells dispersed in Carnoy's solution were placed onto a slide glass. After drying, sample slides were stained with Giemsa solution for 10 min and washed with water.
ChIP-seq and RNA-seq data processing
Adaptor sequences and low-quality reads were excluded using Trimmomatic (v. 0.36) (Bolger et al. 2014). ChIP-seq reads were aligned to the mouse genome (mm10) or human genome (hg38) using “bowtie command” software (v. 1.2.2) with “-n 2 -M 1 –best –strata” option (Supplemental Table S2; Langmead et al. 2009). PCR duplicates were removed by the “samtools rmdup” command. Signal tracks for each sample were generated using “deeptools” (v. 3.4.3) “bamCompare” or “bamCoverage” commands with default parameters and were normalized to reads per kilobase of transcript per million mapped reads (RPKM) values. Signals were visualized by the integrative genomics viewer (v. 2.5.3) (Robinson et al. 2011). Reproducibility of each biological replicate was checked, and each read count was adjusted using the RSeQC “divide_bam.py” module before merging them with the “samtools merge” command.
RNA-seq reads were aligned to the mouse genome (mm10) using the HISAT2 command (v. 2.1.0) with default parameters (Supplemental Table S2; Kim et al. 2015). For repeat element analysis, STAR (v. 2.7.5c) was used for mapping with the “–twopassMode Basic –outFilterType BySJout –outFilterMultimapNmax 1” options, and a best-matched transposable elements file was used as a gtf file (Dobin et al. 2013; Sakashita et al. 2020). The read counts for all RefSeq genes were calculated using the “featureCounts” command (v. 1.6.2) with default parameters, whereas expression levels were quantified to transcripts per million (TPM) using the “StringTie” command (v. 2.1.1) (Liao et al. 2014; Pertea et al. 2015).
Hi-C data processing
Hi-C data processing was done using the Docker for 4DN Hi-C pipeline (v. 43). The pipeline includes alignment (using mm10) and filtering steps. After filtering valid Hi-C alignments with MAPQ > 10 (.pair format file), .cool format Hi-C matrix files were generated by cooler (v. 0.8.7) (Supplemental Table S2; Abdennur and Mirny 2020). To correct the bias of Hi-C matrices, the “cooler balance” command with the “—ignore-diags 1” option was applied to each .cool file. The A/B compartment (Hi-C PC1) profiles (in 200-kb bins) in each chromosome were calculated from each .cool file by the “cooltools call-compartments” command of cooltools (v. 0.3.0) with slight modifications to perform the original A/B compartment analysis (Lieberman-Aiden et al. 2009), including the computation of Pearson correlation and covariance matrix followed by eigen vector decomposition. The sign of Hi-C PC1 in each chromosome was corrected using gene density tracks (the sign of PC1 with higher gene density assigned as A compartment showing positive PC1 values, and the other assigned as a B compartment showing negative PC1 values). To visualize average contact enrichments (average observed/expected interactions) between pairs of 200-kb bins according to the percentile rank of the A/B compartment profile, a saddle plot (heat map of 50 × 50 bins) for each .cool file was generated by the “cooltools compute-saddle” command with default parameters using the A/B compartment profile and an expected interaction profile (the average distance decay) for each chromosome generated by the “cooltools compute-expected” command. A–A or B–B compartment strength was defined as the ratio of A–A/A–B or B–B/A–B interactions. The values of these ratios were determined by calculating the mean value of the 10 bins (top 20 percentile) in each corner of the saddle plot.
Repli-seq data processing
For Repli-seq data processing, the scRepli-seq pipeline (v. 1.3) was used (Miura et al. 2020). This version is applicable to population-based BrdU-IP Repli-seq data processing, which includes adapter trimming, alignment (using mm10), normalization, and computing early/late replication timing scores as described (Supplemental Table S2; Takahashi et al. 2019). Briefly, the mapped reads of early and late S-phase BrdU IP samples were counted in sliding windows of 200 kb at 40-kb interval followed by reads per million (rpm) normalization. Then, the ratio of early S phase to total read counts [(early S reads)/(early S reads + late S reads)] was calculated for each bin, and total read counts within the bottom 5% of all bins were filtered out. The value of the ratio was converted to fit within a ±1 scale, and this scaled value was defined as the early/late replication timing score of each bin.
Identification of broad H3K9me3 and H3.1/2 domains
To identify broad domains, we first divided the genome into 100-kb consecutive bins and calculated the normalized signal to 10 million reads using the “parse2wig” command (v. 3.7.0) with “-n GR -f BAM -of 3 -np 10000000 –showzero” options (Nakato et al. 2013). Sex chromosomes were removed for further analysis. Bins with no value in the input sample or <300 in the ChIP sample were removed, and then the ChIP signals were divided by the input signals using corresponding bines. Regions in which the ChIP/input ratio was >2.0 (H3K9me3) or 1.5 (H3.1/H3.2) were classified as domains.
Localization of H3K9me3 domains for TSS, TES, genic, and intergenic regions
The procedure for finding the percentage of H3K9me3 domains for TSS, genic, TES, and intergenic regions was as described previously, and was performed using an in-house pipeline (Hada et al. 2017; Yamaguchi et al. 2018). The TSS and TES regions were defined as ±2.5 kb from the transcription start site and the transcription end site of each gene. The gene regions were defined as those obtained from the UCSC Genome Browser (https://genome.ucsc.edu) without TSS and TES regions. The intergenic regions were defined as genome regions without TSS, TES, and gene regions. The percentages of H3K9me3 domains at these regions were computed using the “bedtools” intersect command and R, and then visualized using the “ggplot2” library by R. H3K9me3 domains in TSCs as well as early CT cells were mapped randomly using the “bedtools shuffle” command, and the percentage for each region was computed as a control (Quinlan and Hall 2010).
Differential expression analysis
The read counts of each RNA-seq sample were subjected to DESeq2 (v. 1.26.0) to perform differential expression analysis (Love et al. 2014). The threshold P-values (Padj) and fold changes are described in the corresponding figure legends. Enrichr was used for GO analysis (Kuleshov et al. 2016).
Heat map showing the Jaccard index
The Jaccard index was computed by using the “bedtools jaccard” command. An R library component named “pheatmap” was used to draw the heat map with hierarchical clustering.
Heat map showing domain dynamics
H3K9me3 domains in which the input value was 0 were removed, and ChIP/input enrichment was computed. Rows were then divided by k-means clustering into the indicated number. Heat maps were drawn using the “pheatmap” library in R.
Enrichment of repeat clusters within H3K9me3 domains
A best-matched transposable element file was used to calculate the repeat number within H3K9me3 domains. We also counted the repeat number within randomly mapped TSC-defined H3K9me3 domains using the “bedtools shuffle” command. Then, the number of repeats within H3K9me3 domains was divided by those of randomly mapped ones and visualized using the “ggplot2” library in R.
Statistics and reproducibility
Error bars in the bar plot graphs represent the standard deviation (SD), and two-tailed paired Student's t-tests were used for statistics. For box plot graphs in this study, the central line represents the median value, and lower and upper lines represent the first and third quartiles, respectively. Outliers are included in line. The numbers for the box plots are described in the corresponding figure legends. The Wilcoxon rank sum test was used for statistics in the box plot data. All the ChIP-seq, RNA-seq, ATAC-seq, Hi-C, and Repli-seq analyses in this study were performed twice, thus confirming their reproducibility.
Data availability
All the ChIP-seq and RNA-seq data generated in this study are summarized in Supplemental Table S3 and have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE175646 (ChIP-seq: GSE175638, RNA-seq: GSE175644, Hi-C: GSE175934, Repli-seq: GSE175935, and ATAC-seq: GSE175632). Public data analysis for ChIP-seq, RNA-seq, Hi-C, and Repli-seq data sets were downloaded from NCBI GEO. The relevant accession numbers are summarized in Supplemental Table S3. All analyses were made based on the Python3 and R codes and are available on request.
Supplementary Material
Acknowledgments
We thank Dr. A. Harada and Dr. Y. Ohkawa (Kyushu University) for sharing the ChIP-seq protocol using anti-H3.1/H3.2 and anti-H3.3 antibodies, and Dr. Y. Okada (The University of Tokyo) for helpful discussions. We also thank members of the Bioresource Engineering Division, Bioresource Research Center, RIKEN, for helpful assistance. The JF1/Ms mouse strain (RBRC00639) used in this study was provided by the RIKEN Bioresource Research Center with the support of the National Bioresource Project of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan. This work was supported by Japan Society of the Promotion of Science (JSPS) Grant-in-Aid for JSPS Fellows (JP18J00122 to M. Hada), Grant-in-Aid for Early-Career Scientists (JP20K15705 to M. Hada), MEXT Grants-in-Aid for Scientific Research in Innovative Areas (JP19H05758 to A.O., JP19H05757 to H.O., and JP18H05530 to I.H.), Japan Science and Technology (JST) CREST (JPMJCR20S5 to I.H), and RIKEN Genome Building Project of the All-RIKEN projects (to A.O.).
Author contributions: M. Hada and A.O. conceived this project. H.M., A.T., and I.H. performed Hi-C and Repli-seq analysis. S.M., K.I., N.O., N.W., and K.M. produced SCNT embryos, and S.M. analyzed TSC cloned embryo and placental tissue sections. M. Hirose and N.W. helped with maintenance of ESCs and TSCs. M. Hada performed bioinformatic analysis with the help from R.N. K.F. performed sequencing of ChIP-seq data sets. A.H. produced B6/JF1 embryos. N.W. performed karyotype analysis of TSCs. D.S. converted the mm9 RRR list to mm10. A.S. provided best-matched TE files and helped with bioinformatic analysis concerning repeat elements. H.O. helped with the analysis of H3K9me3 ChIP-seq data obtained from human placental cells and human TSCs. M. Hada and A.O. wrote the manuscript with the help of T.A. and K.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348782.121.
Competing interest statement
The authors declare no competing interests.
References
- Abdennur N, Mirny LA. 2020. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36: 311–316. 10.1093/bioinformatics/btz540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Funaya S, Tsukioka D, Kawamura M, Suzuki Y, Suzuki MG, Schultz RM, Aoki F. 2018. Minor zygotic gene activation is essential for mouse preimplantation development. Proc Natl Acad Sci 115: E6780–E6788. 10.1073/pnas.1804309115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, Zaret KS. 2016. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet 32: 29–41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21 29 21–21 29 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. 2014. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15: 723–735. 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- Chang CW, Wakeland AK, Parast MM. 2018. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J Endocrinol 236: R43–R56. 10.1530/JOE-17-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM III, Yanagimachi R, Jaenisch R. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci 98: 6209–6214. 10.1073/pnas.101118898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. 10.1038/nature14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis WR, Wörheide G. 2017. Similar ratios of introns to intergenic sequence across animal genomes. Genome Biol Evol 9: 1582–1598. 10.1093/gbe/evx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. 2010. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153. 10.1101/sqb.2010.75.011 [DOI] [PubMed] [Google Scholar]
- Gomes AP, Ilter D, Low V, Rosenzweig A, Shen ZJ, Schild T, Rivas MA, Er EE, McNally DR, Mutvei AP, et al. 2019. Dynamic incorporation of histone H3 variants into chromatin Is essential for acquisition of aggressive traits and metastatic colonization. Cancer Cell 36: 402–417 e413. 10.1016/j.ccell.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada M, Masuda K, Yamaguchi K, Shirahige K, Okada Y. 2017. Identification of a variant-specific phosphorylation of TH2A during spermiogenesis. Sci Rep 7: 46228. 10.1038/srep46228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: the ‘H3 barcode hypothesis’. Proc Natl Acad Sci 103: 6428–6435. 10.1073/pnas.0600803103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, Kurumizaka H, et al. 2012. Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J 31: 2994–3007. 10.1038/emboj.2012.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, Ohkawa Y, Tsukada Y, Ogura A. 2015. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci 112: 14641–14646. 10.1073/pnas.1512775112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose M, Hada M, Kamimura S, Matoba S, Honda A, Motomura K, Ogonuki N, Shawki HH, Inoue K, Takahashi S, et al. 2018. Aberrant imprinting in mouse trophoblast stem cells established from somatic cell nuclear transfer-derived embryos. Epigenetics 13: 693–703. 10.1080/15592294.2018.1507199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM III, Biniszkiewicz D, Yanagimachi R, Jaenisch R. 2001. Epigenetic instability in ES cells and cloned mice. Science 293: 95–97. 10.1126/science.1061402 [DOI] [PubMed] [Google Scholar]
- Inoue K, Oikawa M, Kamimura S, Ogonuki N, Nakamura T, Nakano T, Abe K, Ogura A. 2015. Trichostatin A specifically improves the aberrant expression of transcription factor genes in embryos produced by somatic cell nuclear transfer. Sci Rep 5: 10127. 10.1038/srep10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. 2017. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547: 419–424. 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Kamimura S, Inoue H, Matoba S, Hirose M, Honda A, Miura K, Hada M, Hasegawa A, et al. 2020. Loss of H3K27me3 imprinting in the Sfmbt2 miRNA cluster causes enlargement of cloned mouse placentas. Nat Commun 11: 2150. 10.1038/s41467-020-16044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Kadota M, Nishimura O, Miura H, Tanaka K, Hiratani I, Kuraku S. 2020. Multifaceted Hi-C benchmarking: what makes a difference in chromosome-scale genome scaffolding? Gigascience 9: giz158. 10.1093/gigascience/giz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Takemoto K, Shinkai Y. 2018. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat Commun 9: 1683. 10.1038/s41467-018-04132-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöfler M, Pollheimer J. 2013. Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet 4: 190. 10.3389/fgene.2013.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi P, Iuso D, Czernik M, Ogura A. 2016. A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol 34: 791–797. 10.1016/j.tibtech.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Sasaki T, Vera D, Wilson K, Sima J, Rivera-Mulia JC, Trevilla-García C, Nogues C, Nafie E, Gilbert DM. 2018. Genome-wide analysis of replication timing by next-generation sequencing with E/L Repli-seq. Nat Protoc 13: 819–839. 10.1038/nprot.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Zhang Y. 2018. Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 23: 471–485. 10.1016/j.stem.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. 2014. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159: 884–895. 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. 2018. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23: 343–354.e5. 10.1016/j.stem.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura H, Takahashi S, Poonperm R, Tanigawa A, Takebayashi SI, Hiratani I. 2019. Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization. Nat Genet 51: 1356–1368. 10.1038/s41588-019-0474-z [DOI] [PubMed] [Google Scholar]
- Miura H, Takahashi S, Shibata T, Nagao K, Obuse C, Okumura K, Ogata M, Hiratani I, Takebayashi SI. 2020. Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing. Nat Protoc 15: 4058–4100. 10.1038/s41596-020-0378-5 [DOI] [PubMed] [Google Scholar]
- Mizutani E, Oikawa M, Kassai H, Inoue K, Shiura H, Hirasawa R, Kamimura S, Matoba S, Ogonuki N, Nagatomo H, et al. 2015. Generation of cloned mice from adult neurons by direct nuclear transfer. Biol Reprod 92: 81. 10.1095/biolreprod.114.123455 [DOI] [PubMed] [Google Scholar]
- Motohashi K. 2015. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol 15: 47. 10.1186/s12896-015-0162-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K. 2017. Seamless ligation cloning extract (SLiCE) method using cell lysates from laboratory Escherichia coli strains and its application to SLiP site-directed mutagenesis. Methods Mol Biol 1498: 349–357. 10.1007/978-1-4939-6472-7_23 [DOI] [PubMed] [Google Scholar]
- Nakato R, Itoh T, Shirahige K. 2013. DROMPA: easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells 18: 589–601. 10.1111/gtc.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Watanabe H, Fukuda A, Kono T. 2015. Deficiency of genomic reprogramming in trophoblast stem cells following nuclear transfer. Cell Reprogram 17: 115–123. 10.1089/cell.2014.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A, Inoue K, Wakayama T. 2013. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 368: 20110329. 10.1098/rstb.2011.0329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A, Matoba S, Inoue K. 2021. 25th anniversary of cloning by somatic-cell nuclear transfer: epigenetic abnormalities associated with somatic cell nuclear transfer. Reproduction 162: F45–F58. 10.1530/REP-21-0013 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Tsukiyama T. 2014. Establishment of trophoblast stem cells under defined culture conditions in mice. PLoS One 9: e107308. 10.1371/journal.pone.0107308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y, Motohashi K. 2015a. Evaluation of seamless ligation cloning extract preparation methods from an Escherichia coli laboratory strain. Anal Biochem 486: 51–53. 10.1016/j.ab.2015.06.031 [DOI] [PubMed] [Google Scholar]
- Okegawa Y, Motohashi K. 2015b. A simple and ultra-low cost homemade seamless ligation cloning extract (SLiCE) as an alternative to a commercially available seamless DNA cloning kit. Biochem Biophys Rep 4: 148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Tam PP. 2009. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136: 701–713. 10.1242/dev.017178 [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. 2010. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci 107: 10783–10790. 10.1073/pnas.0914507107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM. 2011. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc 6: 870–895. 10.1038/nprot.2011.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita A, Maezawa S, Takahashi K, Alavattam KG, Yukawa M, Hu YC, Kojima S, Parrish NF, Barski A, Pavlicev M, et al. 2020. Endogenous retroviruses drive species-specific germline transcriptomes in mammals. Nat Struct Mol Biol 27: 967–977. 10.1038/s41594-020-0487-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind'Amour J, Sun MA, Sun Z, et al. 2016. Activation of endogenous retroviruses in Dnmt1−/− ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell 19: 81–94. 10.1016/j.stem.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Miura H, Shibata T, Nagao K, Okumura K, Ogata M, Obuse C, Takebayashi SI, Hiratani I. 2019. Genome-wide stability of the DNA replication program in single mammalian cells. Nat Genet 51: 529–540. 10.1038/s41588-019-0347-5 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282: 2072–2075. 10.1126/science.282.5396.2072 [DOI] [PubMed] [Google Scholar]
- Wakayama S, Kishigami S, Wakayama T. 2019. Improvement of mouse cloning from any type of cell by nuclear injection. Methods Mol Biol 1874: 211–228. 10.1007/978-1-4939-8831-0_12 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, et al. 2018. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat Cell Biol 20: 620–631. 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- Wang LY, Li ZK, Wang LB, Liu C, Sun XH, Feng GH, Wang JQ, Li YF, Qiao LY, Nie H, et al. 2020. Overcoming intrinsic H3K27me3 imprinting barriers improves post-implantation development after somatic cell nuclear transfer. Cell Stem Cell 27: 315–325.e5. 10.1016/j.stem.2020.05.014 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481. 10.1016/j.cell.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Xu R, Li C, Liu X, Gao S. 2021. Insights into epigenetic patterns in mammalian early embryos. Protein Cell 12: 7–28. 10.1007/s13238-020-00757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Hada M, Fukuda Y, Inoue E, Makino Y, Katou Y, Shirahige K, Okada Y. 2018. Re-evaluating the localization of sperm-retained histones revealed the modification-dependent accumulation in specific genome regions. Cell Rep 23: 3920–3932. 10.1016/j.celrep.2018.05.094 [DOI] [PubMed] [Google Scholar]
- Yang F, Wang PJ. 2016. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin Cell Dev Biol 59: 118–125. 10.1016/j.semcdb.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhang L, Liu W, Qiao Z, Shen S, Zhu Q, Gao R, Wang M, Wang M, Li C, et al. 2021. Dux-mediated corrections of aberrant H3K9ac during 2-cell genome activation optimize efficiency of somatic cell nuclear transfer. Cell Stem Cell 28: 150–163.e5. 10.1016/j.stem.2020.09.006 [DOI] [PubMed] [Google Scholar]
- Zhou J, West RC, Ehlers EL, Ezashi T, Schulz LC, Roberts RM, Yuan Y, Schust DJ. 2021. Modeling human peri-implantation placental development and functiondagger. Biol Reprod 105: 40–51. 10.1093/biolre/ioab080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the ChIP-seq and RNA-seq data generated in this study are summarized in Supplemental Table S3 and have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under accession number GSE175646 (ChIP-seq: GSE175638, RNA-seq: GSE175644, Hi-C: GSE175934, Repli-seq: GSE175935, and ATAC-seq: GSE175632). Public data analysis for ChIP-seq, RNA-seq, Hi-C, and Repli-seq data sets were downloaded from NCBI GEO. The relevant accession numbers are summarized in Supplemental Table S3. All analyses were made based on the Python3 and R codes and are available on request.