Visual Abstract
Keywords: kidney disease, proteomics, kidney biopsy, histopathology, Boston, cohort studies, biomarkers, nephrectomy, causality, plasma
Abstract
Background and objectives
Biomarkers for noninvasive assessment of histopathology and prognosis are needed in patients with kidney disease.
Design, setting, participants, & measurements
Using a proteomics assay, we measured a multimarker panel of 225 circulating plasma proteins in a prospective cohort study of 549 individuals with biopsy-confirmed kidney diseases and semiquantitative assessment of histopathology. We tested the associations of each biomarker with histopathologic lesions and the risks of kidney disease progression (defined as ≥40% decline in eGFR or initiation of KRT) and death.
Results
After multivariable adjustment and correction for multiple testing, 46 different proteins were associated with histopathologic lesions. The top-performing markers positively associated with acute tubular injury and interstitial fibrosis/tubular atrophy were kidney injury molecule-1 (KIM-1) and V-set and Ig domain-containing protein 2 (VSIG2), respectively. Thirty proteins were significantly associated with kidney disease progression, and 35 were significantly associated with death. The top-performing markers for kidney disease progression were placental growth factor (hazard ratio per doubling, 5.4; 95% confidence interval, 3.4 to 8.7) and BMP and activin membrane-bound inhibitor (hazard ratio, 3.0; 95% confidence interval, 2.1 to 4.2); the top-performing markers for death were TNF-related apoptosis-inducing ligand receptor-2 (hazard ratio, 2.9; 95% confidence interval, 2.0 to 4.0) and CUB domain-containing protein-1 (hazard ratio, 2.4; 95% confidence interval, 1.8 to 3.3).
Conclusion
We identified several plasma protein biomarkers associated with kidney disease histopathology and adverse clinical outcomes in individuals with a diverse set of kidney diseases.
Podcast
This article contains a podcast at https://www.asn-online.org/media/podcast/CJASN/2021_12_28_CJN09380721.mp3
Introduction
The two measures used to diagnose and stage kidney diseases, albuminuria and eGFR, do not provide specificity about kidney histopathologic lesions and are imperfect prognostic markers. Novel biomarkers with greater specificity for histopathologic lesions will enhance clinical phenotyping of kidney diseases and may provide an important milestone toward precision medicine.
Several biomarkers of kidney inflammation and tubular injury have been shown to prognosticate the risk of adverse clinical outcomes in patients with kidney diseases (1–4), but these biomarkers only represent a small segment of the human blood proteome. Proteomics-based approaches add to this by allowing for an unsupervised protein discovery that does not entirely depend on preexisting pathophysiologic knowledge. The history of biomarker studies in other clinical disciplines illustrates how large-scale proteomics approaches can identify important protein markers for improved risk prediction (5) and the development of new therapeutic agents (6). Fewer studies, however, have made use of these techniques for investigation of human kidney disease (7–9).
In this study, we used a novel proteomics assay to measure a multimarker panel of 225 plasma proteins in a prospective cohort study of individuals with biopsy-confirmed kidney diseases and adjudicated semiquantitative assessment of histopathology. We tested the associations of each biomarker with kidney histopathologic lesions and the risks of subsequent kidney disease progression and death and explored pathways with potential biologic relevance for patients at risk of kidney disease progression and death using the Reactome pathway database.
Materials and Methods
Study Population
The Boston Kidney Biopsy Cohort (BKBC) is a prospective, observational cohort study of adult patients undergoing native kidney biopsy between September 2006 and October 2018 at three tertiary care hospitals in Boston, Massachusetts, including Brigham and Women’s Hospital, Massachusetts General Hospital, and Beth Israel Deaconess Medical Center. Details of the study design have been previously described (10). Patients provided blood and urine samples on the day of kidney biopsy. For this study, we evaluated 549 participants with available plasma samples that included both common forms (kidney disease attributed to diabetes or hypertension) and more rare forms of kidney diseases (all other etiologies, such as other glomerular, tubulointerstitial, or vascular diseases). The Partners Human Research Committee (the Brigham and Women’s Hospital Institutional Review Board) approved the study protocol, which is in accordance with the principles of the Declaration of Helsinki.
Sample Collection, Proteomics Assays, and Exposures
Blood samples were collected from study participants on the day of biopsy, aliquoted, and immediately stored at −80°C. Aliquots were analyzed at Olink using high-throughout, multiplex immunoassays (11) on three commercially available panels named Inflammation, Organ Damage, and Cardiovascular II. Each panel consists of 92 biomarker proteins that were chosen on the basis of their potential relevance in various pathologic processes. All protein values are expressed as normalized protein expression values on a log2 scale. We included 5% blind split replicates in addition to BKBC samples. Of the 276 proteins included in the three panels, we used 225 biomarkers as the primary exposures for statistical analyses that were nonoverlapping across the panels and passed our quality control metrics (Supplemental Material).
Histopathologic Outcomes
Methods to evaluate and score histopathologic lesions were previously described in detail (10). Kidney biopsy specimens were adjudicated under light microscopy by two experienced kidney pathologists who provided semiquantitative scores of kidney inflammation, fibrosis, vascular sclerosis, and acute tubular injury (ATI). Global and segmental glomerulosclerosis, endocapillary and extracapillary glomerular inflammation, focal glomerular necrosis, fibrocellular crescents, interstitial fibrosis/tubular atrophy (IFTA), and inflammation in the fibrosed and nonfibrosed interstitium were scored from zero to three, reflecting the percentage of affected total cortical volume of ≤10%, 11%–25%, 26%–50%, and >50%, respectively. Mesangial expansion, ATI, and arterial and arteriolar sclerosis were scored from zero to three, reflecting none, mild, moderate, and severe lesions, respectively (Supplemental Table 1). Additional details on evaluation and scoring of histopathologic lesions are provided in Supplemental Material. All participants’ charts were reviewed alongside histopathologic evaluations to provide the final primary clinicopathologic diagnosis.
Clinical Outcomes
The primary outcome was kidney disease progression, defined as ≥40% decline in eGFR or kidney failure (defined as the initiation of dialysis or kidney transplantation). The secondary outcome was death. To ascertain information on vital status, change in creatinine, or need for dialysis, we reviewed the electronic medical record (EMR) of the respective hospital as well as other linked EMR systems. This review consisted of reviewing progress notes, discharge summary, laboratory tests, and other documentation in EMR. Data on eGFR during follow-up were obtained from the EMR, and KRT status was confirmed by reviewing the EMR and linkage with the United States Renal Data System database. Mortality status was confirmed with the Social Security Death Index. Participants were followed until the occurrence of death, voluntary study withdrawal, loss to follow-up (determined by the absence of clinical information during follow-up), or February 1, 2020.
Covariates
Detailed patient information was collected at the biopsy visit, including demographics, medical history, medication lists, and pertinent laboratory data. We obtained serum creatinine (SCr) from the EMR on the day of biopsy. In participants for whom this was unavailable, we measured SCr in available blood samples collected on the day of biopsy. We obtained spot urine protein-creatinine ratio (UPCR) or urine albumin-creatinine ratio (UACR) from the date of kidney biopsy to 3 months before biopsy from the EMR. If both were available, UACR was used. If a participant did not have any of these values, we measured UACR from urine collected on the day of the kidney biopsy. SCr and urine creatinine were measured using a Jaffe-based method, and urine albumin was measured by an immunoturbidometric method. The creatinine-based Chronic Kidney Disease Epidemiology Collaboration equation was used to calculate the eGFR (12).
Pathway Analyses
To obtain basic functional information on our proteins of interest and to investigate potentially relevant biologic pathways, we used Reactome, an open-source database for pathway analyses (13) for biomarkers that were significantly associated with kidney disease progression and death after correction for multiple testing. Reactome uses the Benjamini–Hochberg approach to provide a false discovery rate that accounts for the number of tests performed and ranks pathways on the basis of P values calculated using a permutation approach that permutes group labels using 1000 permutations as well as the false discovery rate.
Statistical Analyses
We summarized descriptive statistics as count with percentage for categorical variables and mean±SD or median with interquartile range for continuous variables. For skewed data distributions, we performed natural logarithmic transformation as appropriate. Unadjusted and adjusted multivariable linear regression models were used to assess associations of each plasma biomarker protein with histopathologic lesions. Histopathologic lesions were dichotomized as described in Supplemental Table 1. Percentage differences of plasma biomarkers by histopathologic lesion were calculated by raising two to the power of the β-coefficient, subtracting one, and multiplying by 100 [(2β−1)×100)] for each respective histopathologic lesion. We limited statistical analyses on histopathologic lesions to participants with adjudicated histopathology by both kidney pathologists (n=411; 75%), except for analyses of global or segmental glomerulosclerosis because they were taken from the biopsy report (n=549).
We performed time-to-event analyses to examine the associations of biomarkers with kidney disease progression and death. Cox proportional hazards models using kidney disease progression or death as the dependent variable were stratified by site and adjusted for age, race, sex, log(proteinuria), eGFR (modeled continuously), and primary clinicopathologic diagnostic category of kidney disease. For the outcome of kidney disease progression, we treated the data as interval censored because the exact date of the event may not be known. We evaluated the association between plasma biomarkers and subsequent kidney disease progression using a nonparametric survival function for interval-censored data (14). We confirmed no violations of the proportional hazards assumption through assessment of Schoenfeld residuals and used complete case analysis for the analyses as there were <5% missing data. To counteract the problem of multiple testing, we used Bonferroni correction for all reported P values (adjusted P value = 225 × Puncorrected). Statistical analyses were performed using R Version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Baseline Characteristics
Baseline characteristics of the study cohort are shown in Table 1. The mean age was 52±17 years, and 52% were women. The mean eGFR was 58±36 ml/min per 1.73 m2, and the median proteinuria (interquartile range) was 1.7 (0.4–3.8) g/g creatinine. The most common primary clinicopathologic diagnoses were glomerulopathies (47%), diabetic nephropathy (12%), advanced glomerulosclerosis (11%), vascular disease (9%), and tubulointerstitial disease (9%).
Table 1.
Baseline characteristics of participants in the Boston Kidney Biopsy Cohort
| Baseline Characteristics | Boston Kidney Biopsy Cohort Participants, n=549 |
|---|---|
| Clinical characteristics | |
| Age, yr | 52±17 |
| Women | 288 (52) |
| Race | |
| Black | 105 (19) |
| White | 347 (63) |
| Other | 97 (18) |
| eGFR, ml/min per 1.73 m2 | 58±36 |
| eGFR categories | |
| Category 1 (eGFR>90 ml/min per 1.73 m2) | 132 (24) |
| Category 2 (eGFR=60–89 ml/min per 1.73 m2) | 92 (17) |
| Category 3A (eGFR=45–59 ml/min per 1.73 m2) | 81 (15) |
| Category 3B (eGFR=30–44 ml/min per 1.73 m2) | 91 (17) |
| Category 4 (eGFR=15–29 ml/min per 1.73 m2) | 101 (18) |
| Category 5 (eGFR<15 ml/min per 1.73 m2) | 52 (9) |
| Proteinuria, g/g creatinine | 1.7 [0.4–3.8] |
| Proteinuria categories, g/g creatinine | |
| Category 1, <0.15 | 73 (13) |
| Category 2, 0.16–1 | 139 (25) |
| Category 3, 1.1–3.5 | 175 (32) |
| Category 4, >3.5 | 162 (30) |
| Reason for biopsy | |
| Proteinuria | 312 (57) |
| Hematuria | 134 (24) |
| Nephrotic syndrome | 69 (13) |
| Nephritic syndrome | 14 (3) |
| Abnormal eGFR/other | 336 (61) |
| Primary clinicopathologic diagnosis | |
| Proliferative glomerulonephritis | 162 (30) |
| Nonproliferative glomerulopathy | 96 (17) |
| Diabetic nephropathy | 64 (12) |
| Advanced glomerulosclerosis | 62 (11) |
| Vascular disease | 52 (9) |
| Tubulointerstitial disease | 47 (9) |
| Paraprotein-related disease | 35 (6) |
| Other | 31 (6) |
| Comorbid conditions | |
| Diabetes mellitus | 122 (22) |
| Hypertension | 287 (52) |
| SLE | 90 (16) |
| Hepatitis C | 11 (2) |
| Hepatitis B | 4 (1) |
| HIV | 6 (1) |
| Malignancy | 80 (15) |
| Medications | |
| ACEi/ARB | 257 (47) |
| MRA | 14 (3) |
| Calcium channel blockers | 137 (25) |
| β-blockers | 166 (30) |
| Immunosuppression | 102 (19) |
| Corticosteroids | 123 (22) |
Data are presented as mean±SD, median [interquartile range], or count with frequency (percentage) for binary and categorical variables. Data on proteinuria were missing for seven individuals. Percentages do not add to 100 as there may have been more than one reason for a kidney biopsy. ACEi, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blocker; MRA, mineralocorticoid receptor blocker.
Plasma Protein Biomarkers Associated with Histopathologic Lesions
Associations between plasma biomarkers and histopathologic lesions are shown in Figure 1 and Table 2. After multivariable adjustment and correction for multiple testing, 46 different plasma proteins (57 associations total) were independently associated with different histopathologic lesions. By level of statistical significance, the top-performing biomarkers positively associated with more severe ATI and IFTA were kidney injury molecule-1 (KIM-1) and V-set and Ig domain-containing protein 2 (VSIG2), respectively (Table 2). The top-performing biomarkers positively associated with other lesions included protein S100-A12 (EN-RAGE) with greater severity of glomerular inflammation, C-X-C motif chemokine 9 with inflammation in the nonfibrosed interstitium, tissue factor with glomerular sclerosis, and TNF-related apoptosis-inducing ligand (TRAIL) with mesangial expansion. Matrix metalloproteinase-7 and fatty acid–binding protein 2 (FABP2) were the top-performing markers associated with more severe inflammation in the fibrosed interstitium and arteriolar sclerosis, respectively. Seven different biomarkers (nine associations total) were inversely associated with histopathologic lesions (Table 2). Among those, stem cell factor (SCF) associated inversely with both ATI and inflammation in the nonfibrosed interstitium. There was no significant association between any of the biomarkers and segmental glomerulosclerosis or arterial sclerosis after multivariable adjustment.
Figure 1.
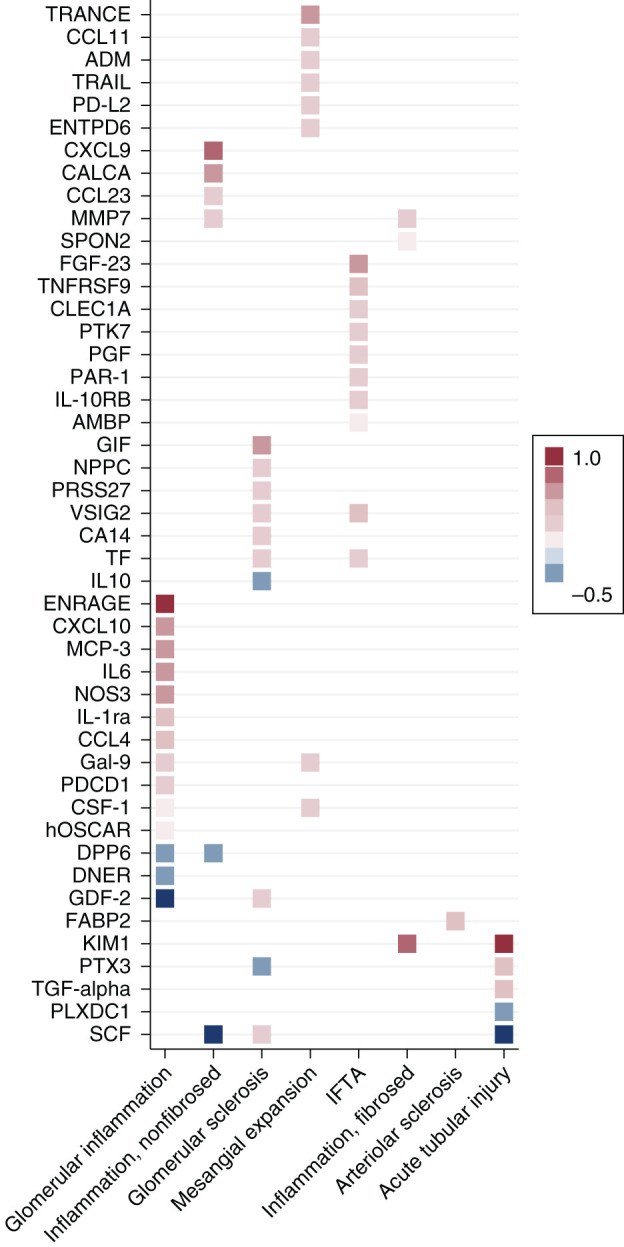
Circulating plasma proteins associated with histopathologic lesions. The heat map shows associations between histopathologic lesions and biomarkers after Bonferroni correction. β-coefficients are derived from multivariable linear regression models adjusted for age, sex, race, and eGFR (modeled continuously), and they are displayed as colors ranging from blue to red. Reference is none/mild lesion severity for acute tubular injury and arteriolar sclerosis. Reference is absence of lesion for mesangial expansion. Reference is 0%–10% of cortical volume or glomeruli affected for inflammation in the fibrosed and nonfibrosed interstitium and glomerular inflammation. Reference is 0%–25% of cortical volume or glomeruli affected for glomerular sclerosis and interstitial fibrosis/tubular atrophy (IFTA). Endocapillary glomerular inflammation, extracapillary cellular crescents, focal glomerular necrosis, and fibrocellular crescents were combined into a single dichotomous variable named “glomerular inflammation.”
Table 2.
Associations of histopathologic lesions with plasma biomarkers
| Plasma Protein | Percentage Difference in Plasma Protein Associated with Histopathologic Lesion (95% Confidence Interval)a | P Value |
|---|---|---|
| Glomerular inflammation | ||
| EN-RAGE | 98 (61 to 144) | <0.001 |
| DNER | −15 (–20 to –10) | <0.001 |
| MCP-3 | 51 (30 to 77) | <0.001 |
| NOS3 | 46 (24 to 71) | 0.002 |
| Gal-9 | 19 (10 to 28) | 0.003 |
| IL-1ra | 35 (18 to 55) | 0.005 |
| CSF-1 | 11 (5 to 16) | 0.02 |
| IL-6 | 48 (21 to 80) | 0.03 |
| GDF-2 | −23 (–32 to –12) | 0.03 |
| CCL4 | 32 (15 to 52) | 0.03 |
| hOSCAR | 9 (4 to 13) | 0.03 |
| DPP6 | −14 (–20 to –7) | 0.04 |
| CXCL10 | 53 (23 to 90) | 0.04 |
| PDCD1 | 16 (7 to 25) | 0.05 |
| Inflammation, nonfibrosed interstitium | ||
| SCF | −22 (–31 to –13) | 0.003 |
| CXCL9 | 72 (34 to 120) | 0.01 |
| MMP-7 | 22 (11 to 33) | 0.01 |
| DPP6 | −17 (–24 to –9) | 0.02 |
| CCL23 | 24 (11 to 38) | 0.04 |
| CALCA | 55 (23 to 95) | 0.04 |
| Glomerular sclerosis | ||
| TF | 17 (11 to 24) | <0.001 |
| PRSS27 | 24 (14 to 34) | <0.001 |
| SCF | 23 (14 to 32) | <0.001 |
| GIF | 43 (25 to 65) | <0.001 |
| CA14 | 18 (9 to 28) | 0.01 |
| PTX3 | −17 (–24 to –9) | 0.01 |
| GDF-2 | 23 (11 to 35) | 0.02 |
| VSIG2 | 21 (10 to 32) | 0.02 |
| IL-10 | −20 (–28 to –10) | 0.04 |
| NPPC | 25 (11 to 41) | 0.04 |
| Mesangial expansion | ||
| TRAIL | 20 (10 to 30) | 0.01 |
| Gal-9 | 22 (11 to 34) | 0.01 |
| CSF-1 | 13 (7 to 20) | 0.01 |
| ENTPD6 | 14 (7 to 21) | 0.02 |
| PD-L2 | 19 (9 to 29) | 0.02 |
| TRANCE | 42 (19 to 70) | 0.03 |
| ADM | 24 (11 to 39) | 0.03 |
| CCL11 | 24 (11 to 39) | 0.04 |
| Interstitial fibrosis/tubular atrophy | ||
| VSIG2 | 30 (16 to 46) | 0.003 |
| TF | 17 (9 to 26) | 0.01 |
| AMBP | 10 (5 to 15) | 0.01 |
| IL-10RB | 13 (7 to 20) | 0.01 |
| PGF | 19 (9 to 29) | 0.02 |
| FGF-23 | 47 (22 to 79) | 0.02 |
| CLEC1A | 23 (11 to 36) | 0.02 |
| PTK7 | 22 (10 to 35) | 0.03 |
| PAR-1 | 17 (8 to 27) | 0.03 |
| TNFRSF-9 | 33 (15 to 54) | 0.04 |
| Inflammation, fibrosed interstitium | ||
| MMP-7 | 21 (10 to 33) | 0.02 |
| SPON2 | 8 (4 to 13) | 0.03 |
| KIM-1 | 65 (28 to 114) | 0.04 |
| Arteriolar sclerosis | ||
| FABP2 | 35 (16 to 56) | 0.02 |
| Acute tubular injury | ||
| SCF | −30 (–38 to –20) | <0.001 |
| KIM-1 | 101 (52 to 166) | <0.001 |
| TGF-α | 27 (14 to 42) | 0.004 |
| PLXDC1 | −16 (–22 to –9) | 0.01 |
| PTX3 | 33 (15 to 54) | 0.04 |
Reference is none/mild lesion severity for acute tubular injury and arteriolar sclerosis. Reference is absence of lesion for mesangial expansion. Reference is 0%–10% of cortical volume or glomeruli affected for inflammation in the fibrosed and nonfibrosed interstitium and glomerular inflammation. Reference is 0%–25% of cortical volume or glomeruli affected for glomerular sclerosis and interstitial fibrosis/tubular atrophy. Endocapillary glomerular inflammation, extracapillary cellular crescents, focal glomerular necrosis, and fibrocellular crescents were combined into a single dichotomous variable named “glomerular inflammation.” Shown are significant associations after applying Bonferroni correction. Abbreviations and definitions of all proteins are provided in Supplemental Table 5.
Percentage differences were derived from linear regression models using plasma protein biomarkers as the dependent variable and each histopathologic lesion as the independent variable. Models were further adjusted for age, sex, race, and eGFR (modeled continuously).
Plasma Protein Biomarkers Associated with Kidney Disease Progression
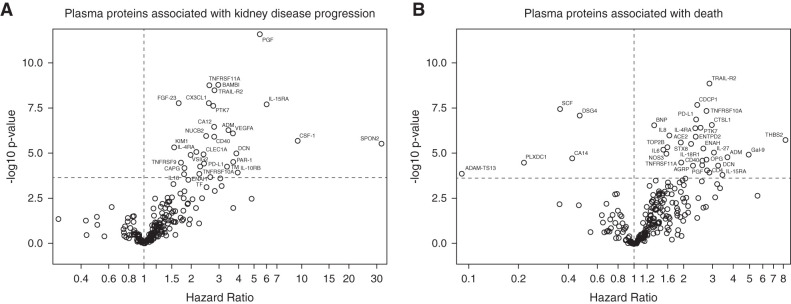
In total, 170 individuals suffered kidney disease progression during a median follow-up of 52 months. Higher levels of 30 plasma proteins were independently associated with greater risk of kidney disease progression (Figure 2A, Supplemental Table 2). In multivariable-adjusted models, higher levels of the following biomarkers were associated with greater risks of kidney disease progression (top five findings in the order of level of significance): placental growth factor (PGF; hazard ratio [HR] per doubling, 5.4; 95% confidence interval [95% CI], 3.4 to 8.7), BMP and activin membrane-bound inhibitor (BAMBI; HR, 3.0; 95% CI, 2.1 to 4.2), TNF-receptor superfamily-11A (TNFRSF-11A), TNF-related apoptosis-inducing ligand-R2 (TRAIL-R2/TNFRSF-10B), and C-X3-C motif chemokine ligand 1. Among all 30 proteins, several were ILs and IL receptors (IL-4RA, IL-10RB, IL-15RA, and IL-16), hemostatic markers (thrombomodulin, protease-activated receptor-1, and tissue factor), and members of the TNF superfamily (TNFRSF-9, TNFRSF-10A, TRAIL-R2/TNFRSF-10B, and TNFRSF-11A). Cytokine signaling and activation of the clotting cascade were among the top-ranked pathways associated with kidney disease progression (Supplemental Table 3).
Figure 2.
Circulating plasma proteins associated with kidney disease progression and death. (A) Kidney disease progression, (B) death. Results are derived from proportional hazards models adjusted for eGFR, race, age, sex, log(proteinuria), and primary clinicopathologic diagnosis. Horizontal dotted lines show Bonferroni-adjusted significance thresholds, and vertical dotted lines mark the hazard ratio of one.
Plasma Protein Biomarkers Associated with Death
In total, 82 participants died during a median follow-up of 59 months. Higher levels of 30 proteins and lower levels of five proteins were independently associated with greater risk of death (Figure 2B, Supplemental Table 4). The top five proteins positively associated with greater risk of death according to level of significance were TRAIL-R2/TNFRSF-10B (HR per doubling, 2.9; 95% CI, 2.0 to 4.0), CUB domain-containing protein 1 (HR, 2.4; 95% CI, 1.8 to 3.3), TNFRSF-10A, programmed cell death-ligand 1 (CD274), and brain natriuretic peptide. The five plasma proteins inversely associated with greater risk of death were SCF, desmoglein-4, carbonic anhydrase 14, plexin domain-containing protein, and disintegrin and metalloproteinase with thrombospondin motifs-13 (ADAMTS13). Among all proteins associated with death, there were several members of the TNF superfamily (TNFRSF-10A, TNFRSF-10B/TRAIL-R2, and TNFRSF-11A) as well as cytokines and cytokine receptors involved in Th2 inflammation (IL-4RA and IL-6), NK cell regulation (IL-18R1, IL-15RA, and IL-27), and neutrophil recruitment (C-X-C motif chemokine 8/IL-8). Cytokine and IL signaling pathways were among the top-ranked pathways associated with death (Supplemental Table 3).
Discussion
This study provides an assessment of a subset of the plasma proteome in a cohort of individuals with biopsy-confirmed kidney diseases. We evaluated associations of 225 plasma biomarkers with histopathologic findings in native kidney biopsies and subsequent risks of kidney disease progression and death and identified several new promising biomarker candidates. Pathways enriched in the plasma proteome of patients at risk of disease progression may point toward key mechanisms involved in disease pathogenesis, including inflammation, extracellular matrix remodeling, disturbances of hemostasis, cell growth, and apoptosis.
Previous large-scale proteomics studies have primarily focused on evaluation of the urine CKD proteome (15–19). Fewer studies have utilized proteomics to interrogate plasma in patients with CKD (9,20–22). Some of the individual proteins that we investigated have previously been shown to be associated with histopathologic lesions and adverse clinical outcomes in individuals with kidney disease. Among those are plasma KIM-1 (1,23) and fibroblast growth factor-23 (4). Many others are new findings that represent areas for future investigation.
In line with results from previous studies, KIM-1 was the biomarker most strongly associated with more severe ATI, and it was also associated with a higher risk of kidney disease progression, which may provide additional evidence for the importance of kidney tubular injury in CKD progression (1,2,23,24). VSIG2, a protein of unknown function, was the top-performing marker for IFTA, and higher VSIG2 levels associated with a higher risk of kidney disease progression. VSIG2 has been associated with incident heart failure (25) and prevalent diabetes in the general population (26), but there are no studies, to our knowledge, that have linked VSIG2 to kidney diseases. Although we are unable to demonstrate a causal association, these results could point toward a potential role of VSIG2 in the progression of kidney fibrosis. Several inflammatory markers were associated with histopathologic lesions. EN-RAGE, a proinflammatory marker that leads to IL-1β and TNF-α release, had the strongest association with glomerular inflammation. In rodent models of CKD, EN-RAGE was reported to play a key role in the pathogenesis of vascular calcification through modulation of Pi cotransporter expression (27). This finding is supported by previous studies in humans that described associations between elevated plasma EN-RAGE levels and atherosclerosis as well as cardiovascular and all-cause mortality in patients on dialysis (28–30). Our study adds that EN-RAGE not only may be crucial for sustaining chronic systemic inflammation in individuals with kidney disease but also may be an important marker specific for glomerular inflammation. The single marker that remained significantly associated with vascular pathology after multivariable adjustment was FABP2, a protein expressed in epithelial cells of the small intestines and involved in the reabsorption and distribution of long-chain fatty acids (31). Polymorphisms in the FABP2 gene have been linked to insulin resistance, dyslipidemia, and cardiovascular diseases (31,32). We are not aware of any prior studies that have evaluated plasma FABP2 in kidney disease. Both EN-RAGE and FABP2 were not associated with adverse clinical events in this study. If the distinct signal for histopathologic lesions we observed can be confirmed in additional studies, these markers may be able to serve as specific tools to assess for vascular or glomerular inflammatory lesions.
A previous plasma proteomics study by Carlsson et al. (20) of individuals from the general population identified 20 plasma proteins that were associated with incident CKD. Although the associations in the study by Carlsson et al. (20) lost statistical significance after adjustment for baseline eGFR, seven of these proteins (TRAIL-R2, PGF, CD40, protease-activated receptor-1, fibroblast growth factor-23, CSF-1, and thrombomodulin) were also associated with kidney disease progression in our study and remained statistically significant after multivariable adjustment. We observed a particularly strong association for PGF, a member of the vascular endothelial growth factor family that has previously been studied in the setting of preeclampsia, where lower circulating maternal PGF levels were found to be associated with the disease (33,34). A prior study in kidney disease demonstrated that higher PGF levels associated with lower risks of AKI and mortality in patients undergoing cardiac surgery (35), but another study identified that higher PGF levels associated with a higher risk of incident CKD (20). We observed a higher risk of kidney disease progression and more severe IFTA with higher PGF levels. It is possible that in patients with established CKD, PGF levels may not have a pathogenic role but rather, reflect a compensatory response to sustained angiogenic imbalance that is present in the setting of chronic injury and more severe kidney fibrosis. We also observed a strong association between higher levels of BAMBI and greater risk of kidney disease progression. BAMBI serves as a regulator of angiogenesis and endothelial homeostasis through modulation of alternative TGF-β signaling pathways, such as extracellular signal–related kinase 1/2 and Smad1/5 (36,37). Diabetic BAMBI−/− mice develop more severe albuminuria with increased activation of alternative TGF-β pathways compared with wild-type animals (36). In this study, we observed an association between higher, not lower, BAMBI levels and greater risks of kidney disease progression. Because our cohort primarily included patients with established CKD, these findings may point toward an important role of BAMBI in regulating antifibrotic responses, whereas its elevation could be due to the active and ongoing fibrotic processes in CKD. This, however, remains to be tested in additional cohort studies of CKD.
The top-performing biomarker associated with higher risks of death was TRAIL-R2, a key mediator of apoptosis, which was also associated with kidney disease progression. TRAIL-R2 is a death domain-containing receptor that is capable of transmitting apoptotic signals in response to TRAIL binding (38). TRAIL, a member of the TNF gene superfamily, has been shown to associate with albuminuria in diabetes and plays an important role in the progression of diabetic nephropathy by inducing kidney cell death (20,38).
We found lower levels of five plasma proteins to be associated with a higher risk of death. Among those was SCF, a regulator of bone marrow–derived stem cell migration and survival, known to act on early hematopoietic stem cells and primitive hematopoietic progenitor cells. Findings from rodent models suggest that SCF is crucial to mediate tubular epithelial cell survival after hypoxic injury (39). Similar findings were reported in mice in which local injection of SCF improved cardiac function after myocardial infarction (40). In the Malmö Diet and Cancer study, lower circulating levels of SCF associated with significantly higher risks of stroke and heart failure as well as cardiovascular and all-cause mortality (41). We observed similar findings in this study and found lower SCF levels to be associated with a higher risk of death in patients with kidney diseases. Evaluation of inflammatory parameters in the Malmö Diet and Cancer study also revealed that C-reactive protein levels and white blood cell counts correlated inversely with SCF (41). In line with this, we showed that SCF was inversely associated with both ATI and interstitial inflammation, which could suggest that SCF may attenuate inflammation in patients with kidney disease or that, in turn, inflammation may function as a negative regulator of SCF. We also found that lower levels of ADAMTS13 were associated with a greater risk of death. ADAMTS13 is a vWf-cleaving protease, and deficiency of plasma ADAMTS13 activity causes thrombotic thrombocytopenic purpura. In a case-control study of patients with malignant hypertension, ADAMTS13 levels correlated inversely with creatinine and were significantly lower in patients with more severe thrombotic microangiopathy and hypertension (42). Although the functional relevance and putative prognostic value of these proteins need to be further explored, several biomarkers shared common biologic pathways with pathophysiologic relevance for kidney diseases, including inflammation and extracellular matrix remodeling, apoptosis, angiogenesis, and endothelial dysfunction.
Significant strengths of our study include the relatively large number of protein biomarkers included in the analyses. Adjudicated histopathologic scores on lesion severity and the prospective study design with long-term follow-up data allowed us to test associations not only with histopathology, but also with major adverse clinical events, including kidney disease progression and death. Our study has several limitations that warrant consideration as well. First, we were not able to take historical SCr values prior to biopsy into account and did not account for BP and therapy at baseline, which could alter levels of biomarkers or an individual’s risk of disease progression or death. Although almost all participants had proteinuria data, we did not convert UPCR to UACR for participants who only had measurements of UPCR, which should still provide similar risk estimation (43). Furthermore, although the majority of BKBC participants had two or more follow-up eGFR values, we cannot rule out that kidney disease progression was more likely to be detected in individuals with more eGFR values ordered by their clinicians. We were also not able to compare plasma protein measurements with protein measurements in the urine. Limitations of the proteomics assay include that protein values are obtained in relative quantification units (normalized protein expression values) rather than as absolute values, which may limit comparability between studies. As we selected three Olink proteomics panels on the basis of their potential relevance for kidney disease, the protein biomarkers included in the study necessarily represent processes related to cardiovascular disease, inflammation, and organ injury. Thus, our pathway analyses are limited by the narrow range and nature of the protein markers studied. Lastly, we were not able to replicate our findings in a validation cohort, which needs to be addressed in future studies.
In summary, we identified a number of protein biomarkers associated with histopathologic lesions on native kidney biopsy specimens and subsequent kidney disease progression and death. Several of our biomarker findings merit development of quantitative assays for further replication in other prospective cohort studies to develop markers for noninvasive diagnosis and optimal prognosis. Studies using techniques such as Mendelian randomization may identify which of our biomarker findings represent therapeutic targets for patients with specific kidney diseases. Our study demonstrates how findings from a cohort of individuals with available kidney pathology and prospective clinical follow-up can uncover relevant pathways and biomarker candidates for understanding, diagnosing, and providing prognosis of kidney diseases.
Disclosures
B.D. Humphreys reports consultancy agreements with Chinook Therapeutics, Janssen, and Pfizer; ownership interest in Chinook Therapeutics; receiving research funding from Chinook Therapeutics and Janssen; receiving honoraria from the American Society of Nephrology; patents and inventions with Evotec, AG; serving on editorial boards for American Journal of Physiology Renal Physiology, JCI Insight, Kidney International, and Seminars in Nephrology; serving as Vice President of the American Society of Clinical Investigation (ASCI) and as an associate editor for JASN; and serving as a scientific advisor or member of Chinook Therapeutics SAB, the National Institute of Diabetes and Digestive and Kidney Diseases Board of Scientific Advisors, and RegMed XB Regenerative Medicine Crossing Borders SAB. H.G. Rennke reports consultancy agreements with Ionis Pharmaceuticals Inc. and receiving honoraria from Wolters Kluwer for book royalties for Renal Pathophysiology: The Essentials, 5th Ed. I.M. Schmidt is supported by the American Philosophical Society Daland Fellowship in Clinical Investigation. A. Srivastava reports consultancy agreements with CVS Caremark and Tate & Latham (medicolegal consulting); receiving honoraria from AstraZeneca, Bayer, and Horizon Therapeutics PLC; and speakers bureau for AstraZeneca. A. Srivastava is also supported by National Institutes of Health grant K23DK120811, National Institute of Diabetes and Digestive and Kidney Diseases Kidney Precision Medicine Project Opportunity Pool grant U2CDK114886, and core resources from George M. O’Brien Kidney Research Center at Northwestern University (Northwestern University-GoKIDNEY) grant P30DK114857. I.E. Stillman reports serving as a board member of the Organization for Renal Care in Haiti, Torch Inc. (no compensation), and legal consulting. S.S. Waikar reports consultancy agreements with Allena, BioMarin, CVS, GSK, JNJ, Mallinckrodt, Mass Medical International, Metro Biotechnology, Oxidien, Pfizer, Regeneron, Roth Capital Partners, Sironax, Strataca/3ive, Venbio, and Wolters Kluewer; receiving research funding from Vertex; serving as a scientific advisor or member of Kantum (scientific advisory board); receiving personal fees from Barron and Budd (versus Fresenius), Bunch and James, Cerus, CVS, GE Health Care, GSK, the Harvard Clinical Research Institute (also known as Baim), JNJ, Kantum Pharma, Mallinckrodt, Mass Medical International, Pfizer, the Public Health Advocacy Institute, Roth Capital Partners, Strataca, Takeda, Venbio, and Wolters Kluewer; receiving grants and personal fees from Allena Pharmaceuticals; and serving as an expert witness for litigation related to GE product Omniscan, an expert witness for litigation related to the Fresenius product Granuflo, an expert witness for litigation involving cisplatin toxicity, an expert witness for litigation related to the Gilead product Tenofovir, and an expert witness for litigation related to DaVita laboratory testing. S.S. Waikar is also supported by National Institutes of Health grants UH3DK114915, U01DK085660, U01DK104308, R01DK103784, and R21DK119751. P.C. Wilson reports receiving research funding from NovoNordisk. M. Zhuo is supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases T32 award DK007199. All remaining authors have nothing to disclose.
Funding
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK093574 (to S.S. Waikar). This work was conducted with support from Harvard Catalyst, the Harvard Clinical and Translational Science Center (National Institutes of Health, National Center for Advancing Translational Sciences award UL1TR001102), and financial contributions from Harvard University and its affiliated academic health care centers.
Supplementary Material
Acknowledgments
We thank the members of the laboratory of S.S. Waikar for their invaluable assistance in the BKBC.
Part of this work was presented as an oral presentation at the 2020 American Society of Nephrology Scientific Session on October 22.
The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health.
B.D. Humphreys, R. Palsson, S. Sarvode Mothi, I.M. Schmidt, A. Srivastava, S.S. Waikar, and P.C. Wilson were responsible for the concept and design of the study; Z.A. Kibbelaar, R. Palsson, A. Srivastava, S.S. Waikar, and M. Zhuo adjudicated clinical outcomes; H.G. Rennke and I.E. Stillman were responsible for the adjudication of histopathology; S. Sarvode Mothi, I.M. Schmidt, and S.S. Waikar designed the computational framework; I.F. Onul, S. Sarvode Mothi, I.M. Schmidt, and S.S. Waikar were responsible for statistical analyses; all authors interpreted the data; I.M. Schmidt and S.S. Waikar drafted the manuscript; and all authors contributed to critical revisions of the manuscript for important intellectual content.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
Supplemental Material
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.09380721/-/DCSupplemental.
Supplemental Material. Olink assay and adjudication of histopathologic outcomes.
Supplemental Table 1. Histopathologic scoring system for light microscopy.
Supplemental Table 2. Associations of plasma biomarkers with kidney disease progression.
Supplemental Table 3. Top-ranked pathways for plasma biomarkers associated with adverse clinical outcomes.
Supplemental Table 4. Associations of plasma biomarkers with death.
Supplemental Table 5. Abbreviations and list of all plasma biomarkers included in the analyses.
References
- 1.Sabbisetti VS, Waikar SS, Antoine DJ, Smiles A, Wang C, Ravisankar A, Ito K, Sharma S, Ramadesikan S, Lee M, Briskin R, De Jager PL, Ngo TT, Radlinski M, Dear JW, Park KB, Betensky R, Krolewski AS, Bonventre JV: Blood kidney injury molecule-1 is a biomarker of acute and chronic kidney injury and predicts progression to ESRD in type I diabetes. J Am Soc Nephrol 25: 2177–2186, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waikar SS, Sabbisetti V, Ärnlöv J, Carlsson AC, Coresh J, Feldman HI, Foster MC, Fufaa GD, Helmersson-Karlqvist J, Hsu CY, Kimmel PL, Larsson A, Liu Y, Lind L, Liu KD, Mifflin TE, Nelson RG, Risérus U, Vasan RS, Xie D, Zhang X, Bonventre JV; Chronic Kidney Disease Biomarkers Consortium Investigators : Relationship of proximal tubular injury to chronic kidney disease as assessed by urinary kidney injury molecule-1 in five cohort studies. Nephrol Dial Transplant 31: 1460–1470, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, Park J, Nair V, Schlafly A, Saulnier PJ, Satake E, Simeone CA, Shah H, Qiu C, Looker HC, Fiorina P, Ware CF, Sun JK, Doria A, Kretzler M, Susztak K, Duffin KL, Nelson RG, Krolewski AS: A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25: 805–813, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isakova T: Fibroblast growth factor 23 and adverse clinical outcomes in chronic kidney disease. Curr Opin Nephrol Hypertens 21: 334–340, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA: Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 315: 2532–2541, 2016 [DOI] [PubMed] [Google Scholar]
- 6.Loffredo FS, Steinhauser ML, Jay SM, Gannon J, Pancoast JR, Yalamanchi P, Sinha M, Dall’Osso C, Khong D, Shadrach JL, Miller CM, Singer BS, Stewart A, Psychogios N, Gerszten RE, Hartigan AJ, Kim MJ, Serwold T, Wagers AJ, Lee RT: Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153: 828–839, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonomini M, Sirolli V, Pieroni L, Felaco P, Amoroso L, Urbani A: Proteomic investigations into hemodialysis therapy. Int J Mol Sci 16: 29508–29521, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsalik EL, Willig LK, Rice BJ, van Velkinburgh JC, Mohney RP, McDunn JE, Dinwiddie DL, Miller NA, Mayer ES, Glickman SW, Jaehne AK, Glew RH, Sopori ML, Otero RM, Harrod KS, Cairns CB, Fowler VG, Rivers EP, Woods CW, Kingsmore SF, Langley RJ: Renal systems biology of patients with systemic inflammatory response syndrome. Kidney Int 88: 804–814, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldreich T, Nowak C, Fall T, Carlsson AC, Carrero JJ, Ripsweden J, Qureshi AR, Heimbürger O, Barany P, Stenvinkel P, Vuilleumier N, Kalra PA, Green D, Ärnlöv J: Circulating proteins as predictors of cardiovascular mortality in end-stage renal disease. J Nephrol 32: 111–119, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava A, Palsson R, Kaze AD, Chen ME, Palacios P, Sabbisetti V, Betensky RA, Steinman TI, Thadhani RI, McMahon GM, Stillman IE, Rennke HG, Waikar SS: The prognostic value of histopathologic lesions in native kidney biopsy specimens: Results from the Boston Kidney Biopsy Cohort Study. J Am Soc Nephrol 29: 2213–2224, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, Andersson AC, Lindstedt P, Stenvang J, Gullberg M, Fredriksson S: Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 9: e95192, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May B, Milacic M, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P: The reactome pathway knowledgebase. Nucleic Acids Res 48[D1]: D498–D503, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Sun J: Interval censoring. Stat Methods Med Res 19: 53–70, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindhardt M, Persson F, Zürbig P, Stalmach A, Mischak H, de Zeeuw D, Lambers Heerspink H, Klein R, Orchard T, Porta M, Fuller J, Bilous R, Chaturvedi N, Parving HH, Rossing P: Urinary proteomics predict onset of microalbuminuria in normoalbuminuric type 2 diabetic patients, a sub-study of the DIRECT-Protect 2 study. Nephrol Dial Transplant 32: 1866–1873, 2017 [DOI] [PubMed] [Google Scholar]
- 16.Good DM, Zürbig P, Argilés A, Bauer HW, Behrens G, Coon JJ, Dakna M, Decramer S, Delles C, Dominiczak AF, Ehrich JH, Eitner F, Fliser D, Frommberger M, Ganser A, Girolami MA, Golovko I, Gwinner W, Haubitz M, Herget-Rosenthal S, Jankowski J, Jahn H, Jerums G, Julian BA, Kellmann M, Kliem V, Kolch W, Krolewski AS, Luppi M, Massy Z, Melter M, Neusüss C, Novak J, Peter K, Rossing K, Rupprecht H, Schanstra JP, Schiffer E, Stolzenburg JU, Tarnow L, Theodorescu D, Thongboonkerd V, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P: Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics 9: 2424–2437, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schanstra JP, Zürbig P, Alkhalaf A, Argiles A, Bakker SJ, Beige J, Bilo HJ, Chatzikyrkou C, Dakna M, Dawson J, Delles C, Haller H, Haubitz M, Husi H, Jankowski J, Jerums G, Kleefstra N, Kuznetsova T, Maahs DM, Menne J, Mullen W, Ortiz A, Persson F, Rossing P, Ruggenenti P, Rychlik I, Serra AL, Siwy J, Snell-Bergeon J, Spasovski G, Staessen JA, Vlahou A, Mischak H, Vanholder R: Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol 26: 1999–2010, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tofte N, Lindhardt M, Adamova K, Bakker SJL, Beige J, Beulens JWJ, Birkenfeld AL, Currie G, Delles C, Dimos I, Francová L, Frimodt-Møller M, Girman P, Göke R, Havrdova T, Heerspink HJL, Kooy A, Laverman GD, Mischak H, Navis G, Nijpels G, Noutsou M, Ortiz A, Parvanova A, Persson F, Petrie JR, Ruggenenti PL, Rutters F, Rychlík I, Siwy J, Spasovski G, Speeckaert M, Trillini M, Zürbig P, von der Leyen H, Rossing P; PRIORITY investigators : Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): A prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol 8: 301–312, 2020 [DOI] [PubMed] [Google Scholar]
- 19.Critselis E, Lambers Heerspink H: Utility of the CKD273 peptide classifier in predicting chronic kidney disease progression. Nephrol Dial Transplant 31: 249–254, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Carlsson AC, Ingelsson E, Sundström J, Carrero JJ, Gustafsson S, Feldreich T, Stenemo M, Larsson A, Lind L, Ärnlöv J: Use of proteomics to investigate kidney function decline over 5 years. Clin J Am Soc Nephrol 12: 1226–1235, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glorieux G, Mullen W, Duranton F, Filip S, Gayrard N, Husi H, Schepers E, Neirynck N, Schanstra JP, Jankowski J, Mischak H, Argilés À, Vanholder R, Vlahou A, Klein J: New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol Dial Transplant 30: 1842–1852, 2015 [DOI] [PubMed] [Google Scholar]
- 22.Kammer M, Heinzel A, Willency JA, Duffin KL, Mayer G, Simons K, Gerl MJ, Klose C, Heinze G, Reindl-Schwaighofer R, Hu K, Perco P, Eder S, Rosivall L, Mark PB, Ju W, Kretzler M, McCarthy MI, Heerspink HL, Wiecek A, Gomez MF, Oberbauer R; BEAt-DKD Consortium : Integrative analysis of prognostic biomarkers derived from multiomics panels helps discrimination of chronic kidney disease trajectories in people with type 2 diabetes. Kidney Int 96: 1381–1388, 2019 [DOI] [PubMed] [Google Scholar]
- 23.Nowak N, Skupien J, Niewczas MA, Yamanouchi M, Major M, Croall S, Smiles A, Warram JH, Bonventre JV, Krolewski AS: Increased plasma kidney injury molecule-1 suggests early progressive renal decline in non-proteinuric patients with type 1 diabetes. Kidney Int 89: 459–467, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt IM, Srivastava A, Sabbisetti V, McMahon GM, He J, Chen J, Kusek J, Taliercio J, Ricardo AC, Hsu CY, Kimmel PL, Liu KD, Mifflin TE, Nelson RG, Ramachandran VS, Xie D, Zhang X, Palsson R, Stillman IE, Rennke HG, Feldman HI, Bonventre JV, Waikar SS: Plasma kidney injury molecule 1 in CKD: Findings from the Boston Kidney Biopsy Cohort and CRIC studies [published online ahead of print June 25, 2021]. Am J Kidney Dis 10.1053/j.ajkd.2021.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira JP, Verdonschot J, Collier T, Wang P, Pizard A, Bär C, Björkman J, Boccanelli A, Butler J, Clark A, Cleland JG, Delles C, Diez J, Girerd N, González A, Hazebroek M, Huby AC, Jukema W, Latini R, Leenders J, Levy D, Mebazaa A, Mischak H, Pinet F, Rossignol P, Sattar N, Sever P, Staessen JA, Thum T, Vodovar N, Zhang ZY, Heymans S, Zannad F: Proteomic bioprofiles and mechanistic pathways of progression to heart failure. Circ Heart Fail 12: e005897, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beijer K, Nowak C, Sundström J, Ärnlöv J, Fall T, Lind L: In search of causal pathways in diabetes: A study using proteomics and genotyping data from a cross-sectional study. Diabetologia 62: 1998–2006, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belmokhtar K, Ortillon J, Jaisson S, Massy ZA, Boulagnon Rombi C, Doué M, Maurice P, Fritz G, Gillery P, Schmidt AM, Rieu P, Touré F: Receptor for advanced glycation end products: A key molecule in the genesis of chronic kidney disease vascular calcification and a potential modulator of sodium phosphate co-transporter PIT-1 expression. Nephrol Dial Transplant 34: 2018–2030, 2019 [DOI] [PubMed] [Google Scholar]
- 28.Kim JK, Park S, Lee MJ, Song YR, Han SH, Kim SG, Kang SW, Choi KH, Kim HJ, Yoo TH: Plasma levels of soluble receptor for advanced glycation end products (sRAGE) and proinflammatory ligand for RAGE (EN-RAGE) are associated with carotid atherosclerosis in patients with peritoneal dialysis. Atherosclerosis 220: 208–214, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Nakashima A, Carrero JJ, Qureshi AR, Miyamoto T, Anderstam B, Bárány P, Heimbürger O, Stenvinkel P, Lindholm B: Effect of circulating soluble receptor for advanced glycation end products (sRAGE) and the proinflammatory RAGE ligand (EN-RAGE, S100A12) on mortality in hemodialysis patients. Clin J Am Soc Nephrol 5: 2213–2219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiotsu Y, Mori Y, Nishimura M, Sakoda C, Tokoro T, Hatta T, Maki N, Iida K, Iwamoto N, Ono T, Matsuoka E, Kishimoto N, Tamagaki K, Matsubara H, Kosaki A: Plasma S100A12 level is associated with cardiovascular disease in hemodialysis patients. Clin J Am Soc Nephrol 6: 718–723, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratley RE, Baier L, Pan DA, Salbe AD, Storlien L, Ravussin E, Bogardus C: Effects of an Ala54Thr polymorphism in the intestinal fatty acid-binding protein on responses to dietary fat in humans. J Lipid Res 41: 2002–2008, 2000 [PubMed] [Google Scholar]
- 32.Abbas S, Raza ST, Chandra A, Rizvi S, Ahmed F, Eba A, Mahdi F: Association of ACE, FABP2 and GST genes polymorphism with essential hypertension risk among a North Indian population. Ann Hum Biol 42: 461–469, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Almaani SJ: Placental growth factor in pre-eclampsia: Friend or foe? Kidney Int 95: 730–732, 2019 [DOI] [PubMed] [Google Scholar]
- 34.Bramham K, Seed PT, Lightstone L, Nelson-Piercy C, Gill C, Webster P, Poston L, Chappell LC: Diagnostic and predictive biomarkers for pre-eclampsia in patients with established hypertension and chronic kidney disease. Kidney Int 89: 874–885, 2016 [DOI] [PubMed] [Google Scholar]
- 35.Mansour SG, Zhang WR, Moledina DG, Coca SG, Jia Y, Thiessen-Philbrook H, McArthur E, Inoue K, Koyner JL, Shlipak MG, Wilson FP, Garg AX, Ishibe S, Parikh CR; TRIBE-AKI Consortium : The association of angiogenesis markers with acute kidney injury and mortality after cardiac surgery. Am J Kidney Dis 74: 36–46, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Li X, Xiao W, Fu J, Harris RC, Lindenmeyer M, Cohen CD, Guillot N, Baron MH, Wang N, Lee K, He JC, Schlondorff D, Chuang PY: BAMBI elimination enhances alternative TGF-β signaling and glomerular dysfunction in diabetic mice. Diabetes 64: 2220–2233, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillot N, Kollins D, Gilbert V, Xavier S, Chen J, Gentle M, Reddy A, Bottinger E, Jiang R, Rastaldi MP, Corbelli A, Schlondorff D: BAMBI regulates angiogenesis and endothelial homeostasis through modulation of alternative TGFβ signaling. PLoS One 7: e39406, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorz C, Benito-Martín A, Boucherot A, Ucero AC, Rastaldi MP, Henger A, Armelloni S, Santamaría B, Berthier CC, Kretzler M, Egido J, Ortiz A: The death ligand TRAIL in diabetic nephropathy. J Am Soc Nephrol 19: 904–914, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokman G, Stroo I, Claessen N, Teske GJ, Weening JJ, Leemans JC, Florquin S: Stem cell factor expression after renal ischemia promotes tubular epithelial survival. PLoS One 5: e14386, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutz M, Rosenberg M, Kiessling F, Eckstein V, Heger T, Krebs J, Ho AD, Katus HA, Frey N: Local injection of stem cell factor (SCF) improves myocardial homing of systemically delivered c-kit + bone marrow-derived stem cells. Cardiovasc Res 77: 143–150, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Björkbacka H, Yao Mattisson I, Wigren M, Melander O, Fredrikson GN, Bengtsson E, Gonçalves I, Almgren P, Lagerstedt JO, Orho-Melander M, Engström G, Nilsson J: Plasma stem cell factor levels are associated with risk of cardiovascular disease and death. J Intern Med 282: 508–521, 2017 [DOI] [PubMed] [Google Scholar]
- 42.van den Born BJ, van der Hoeven NV, Groot E, Lenting PJ, Meijers JC, Levi M, van Montfrans GA: Association between thrombotic microangiopathy and reduced ADAMTS13 activity in malignant hypertension. Hypertension 51: 862–866, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Fisher H, Hsu CY, Vittinghoff E, Lin F, Bansal N: Comparison of associations of urine protein-creatinine ratio versus albumin-creatinine ratio with complications of CKD: A cross-sectional analysis. Am J Kidney Dis 62: 1102–1108, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.