Summary
Large-scale gene sequencing studies for complex traits have the potential to identify causal genes with therapeutic implications. We performed gene-based association testing of blood lipid levels with rare (minor allele frequency < 1%) predicted damaging coding variation by using sequence data from >170,000 individuals from multiple ancestries: 97,493 European, 30,025 South Asian, 16,507 African, 16,440 Hispanic/Latino, 10,420 East Asian, and 1,182 Samoan. We identified 35 genes associated with circulating lipid levels; some of these genes have not been previously associated with lipid levels when using rare coding variation from population-based samples. We prioritize 32 genes in array-based genome-wide association study (GWAS) loci based on aggregations of rare coding variants; three (EVI5, SH2B3, and PLIN1) had no prior association of rare coding variants with lipid levels. Most of our associated genes showed evidence of association among multiple ancestries. Finally, we observed an enrichment of gene-based associations for low-density lipoprotein cholesterol drug target genes and for genes closest to GWAS index single-nucleotide polymorphisms (SNPs). Our results demonstrate that gene-based associations can be beneficial for drug target development and provide evidence that the gene closest to the array-based GWAS index SNP is often the functional gene for blood lipid levels.
Keywords: exome sequencing, association, lipid, cholesterol, gene-based association
Introduction
Blood lipid levels are heritable complex risk factors for atherosclerotic cardiovascular diseases.1 Array-based genome-wide association studies (GWASs) have identified >400 loci as associated with blood lipid levels, explaining 9%–12% of the phenotypic variance of lipid traits.2, 3, 4, 5, 6, 7, 8 These studies have identified mostly common (minor allele frequency [MAF] > 1%) noncoding variants with modest effect sizes and have been instrumental in defining the causal roles of lipid fractions on cardiovascular disease.9, 10, 11, 12, 13 Despite these advances, the mechanisms and causal genes for most of the identified variants and loci can be difficult to determine.
Genetic association studies testing rare coding variants have potential to directly implicate causal genes. Advances in next-generation sequencing over the last decade have facilitated increasingly larger studies with improved power to detect associations of rare variants with complex diseases and traits.14,15 However, most exome sequencing studies to date have been insufficiently powered for rare variant discovery; for example, Flannick et al. estimated that it would require 75,000 to 185,000 sequenced cases of type 2 diabetes (T2D) to detect associations at known drug target genes at exome-wide significance.15
Identifying rare variants with impact on protein function has helped elucidate biological pathways underlying dyslipidemia and atherosclerotic diseases such as coronary artery disease (CAD).14,16, 17, 18, 19, 20, 21, 22, 23, 24, 25 Successes with this approach have led to the development of novel therapeutic targets to modify blood lipid levels and lower risk of atherosclerotic diseases.26,27
The vast majority of participants in previous studies have been of European ancestry, highlighting the need for more diverse study sample. Such diversity can identify associated variants absent or present at very low frequencies in European populations and help implicate new genes with generalizability extending to all populations.
We have assembled exome sequence data from >170,000 individuals across multiple ancestries and systematically tested the association of rare variants in each gene with six circulating lipid phenotypes: low-density lipoprotein cholesterol (LDL-C), high density lipoprotein cholesterol (HDL-C), non-HDL-C, total cholesterol (TC), triglycerides (TG), and the ratio of TG to HDL-C (TG:HDL). We find 35 genes associated with blood lipid levels, show evidence of gene-based signals in array-based GWAS loci, show enrichment of lipid gene-based associations in LDL-C drug targets and genes in close proximity of GWAS index variants, and test lipid genes for association with CAD, T2D, and liver enzymes.
Subjects and methods
Study overview
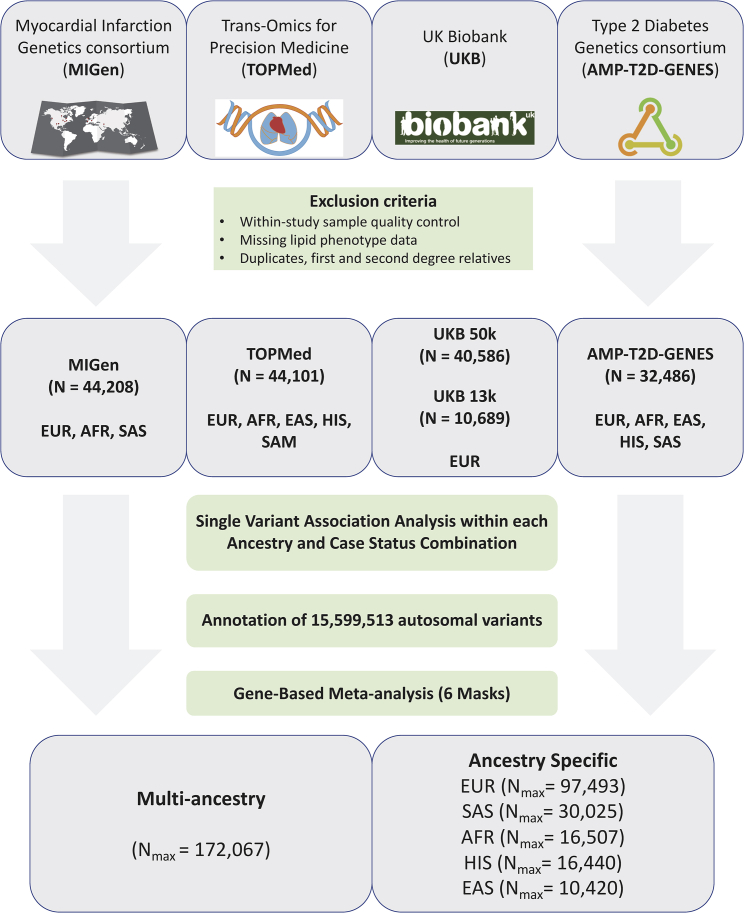
Our study samples were derived from four major data sources with exome or genome sequence data and blood lipid levels: CAD case-control studies from the Myocardial Infarction Genetics Consortium28,29 (MIGen, n = 44,208) and a UK Biobank (UKB) nested case-control study of CAD28 (n = 10,689); T2D cases-control studies from the AMP-T2D-GENES exomes15 (n = 32,486); population-based studies from the TOPMed project30,31 freeze 6a data (n = 44,101) restricted to the exome; and the UKB first tranche of exome sequence data32,33 (n = 40,586) (see supplemental information). Informed consent was obtained from all subjects, and committees approving the studies are available in the supplemental information.
Within each data source, individuals were excluded if they failed study-specific sequencing quality metrics, lacked lipid phenotype data, or were duplicated in other sources. Sequencing and quality control performed in each study is available in the supplemental methods. We additionally removed first- and second-degree relatives across data sources while we kept relatives within each data source because we were able to adjust for relatedness within each data source by using kinship matrices in linear mixed models. If samples from the same study were present in different data sources, we used the samples in the data source that has the largest sample size from the study and removed the overlapping set from the other data source. For instance, samples from the Atherosclerosis Risk in Communities (ARIC) Study were removed from TOPMed and kept in MIGen, which had more sequenced samples from ARIC. Similarly, samples from the Jackson Heart Study were kept in TOPMed and removed from MIGen. To obtain duplicate and kinship information across data sources, we used 14,834 common (MAF > 1%) and no more than weakly dependent (r2 < 0.2) variants by using the make-king flag in PLINK v2.0.
Single-variant association analyses were performed within each data source, case status, and ancestry combination. The data were sequenced and variant calling was performed separately by data source, and this allowed us to look for effects by case status and genetically inferred and/or reported ancestry groups. We performed gene-based meta-analyses by combining single-variant summary statistics and covariance matrices generated from RVTESTS.34 We performed ancestry-specific gene-based meta-analyses by combining single-variant summary data from five major ancestries with >10,000 individuals across all data sources: European, South Asian, African, Hispanic, and East Asian ancestries.
Phenotypes
We studied six lipid phenotypes; total cholesterol (TC), LDL-C, HDL-C, non-HDL-C, triglycerides (TG), and TG:HDL. TC was adjusted by dividing the value by 0.8 in individuals reporting lipid-lowering medication use after 1994 or statin use at any time point. If LDL-C levels were not directly measured, then they were calculated via the Friedewald equation for individuals with TG levels < 400 mg/dL with adjusted TC levels. If LDL-C levels were directly measured then, their values were divided by 0.7 in individuals reporting lipid-lowering medication use after 1994 or statin use at any time point.5 TG and TG:HDL levels were natural logarithm transformed. Non-HDL-C was obtained by subtracting HDL-C from adjusted TC levels. Residuals for each trait in each cohort, ancestry, and case status grouping were created after adjustment for age, age2, sex, principal components, sequencing platform, and fasting status (when available) in a linear regression model. We then inverse-normal transformed the residuals and multiplied them by the standard deviation of the trait to scale the effect sizes to the interpretable units.
Variant annotation
We compiled autosomal variants with call rate > 95% within each case and ancestry-specific analysis dataset with minor allele count (MAC) ≥ 1 (across the combined data). Variants were annotated with the Ensembl Variant Effect Predictor35 and its associated Loss-of-Function Transcript Effect Estimator (LOFTEE)36 and the dbNSFP37 version 3.5a plugins. We limited our annotations to the canonical transcripts. The LOFTEE plugin assesses stop-gained, frameshift, and splice-site-disrupting variants. Loss-of-function variants are classified as either high confidence or low confidence. The dbNSFP is a database that provides functional prediction data and scores for non-synonymous variants by using multiple algorithms.37 We used this database to classify missense variants as damaging by using two different definitions based on bioinformatic prediction algorithms. The first is based on MetaSVM,38 which is derived from ten different component scores (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy, PhyloP). The second is based on five variant prediction algorithms including SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster, and LRT scores. Additionally, we ran a deep neural network analysis (Splice AI) to predict splice-site-altering variants.39 Variant descriptive analysis was performed with a maximal set of variants that were used for analysis of the lipid phenotype with the largest sample size. The counts and proportions of variants—annotated according to the different predicted consequences described above—were obtained out of an overall set of variants.
Single-variant association analysis
Each data source was sub-categorized on the basis of ancestry and CAD or T2D case status in the studies ascertained by disease status. Subgrouping data sources yielded a total of 23 distinct sample sub-categories. As relatives were kept within each sub-group, we performed generalized linear mixed models to analyze the association of single autosomal variants with standard-deviation corrected-inverse-normal transformed traits by using RVTESTS.34 We used RVTESTS to generate summary statistics and covariance matrices using 500 kb sliding windows. To obtain the single-variant associations, we performed a fixed-effects inverse-variance weighted meta-analysis for multi-ancestry and within each of the five major ancestries. We used an exome-wide significance threshold of p < 7.2 × 10−8 (Bonferroni correction for six traits and with previously recommended threshold for coding variants p < 4.3 × 10−7)40 to determine significant coding variants.
Gene-based association analysis
We used summary level score statistics and covariance matrices from autosomal single-variant association results to perform gene-based meta-analyses among all individuals and within each ancestry by using RAREMETALS version 7.2.41 Samoan individuals only contributed to the overall analysis. Gene-based association testing aggregates variants within each gene unit by using burden tests and sequence kernel association tests (SKATs), which allows variable variant effect direction and size.42 The “rareMETALS.range.group” function was used with MAF < 1%, which filters out all variants with combined MAF > 1% in all meta-analytic datasets. All variants with call rates < 95% and not annotated as loss of function (LOF) via LOFTEE, splice-site variants or damaging missense as defined by MetaSVM or by all SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster, and LRT prediction algorithms (damaging 5 out of 5) were excluded in the gene-based meta-analyses.
We used six different variant groupings to determine the set of damaging variants within each gene, (1) high-confidence LOF via LOFTEE, (2) LOF and predicted splice-site-altering variants, (3) LOF and MetaSVM missense variants, (4) LOF, MetaSVM missense, and predicted splice-site-altering variants, (5) LOF and damaging 5 out of 5 missense variants, and (6) LOF, damaging 5 out of 5 missense, and predicted splice-site-altering variants. We used an exome-wide significance threshold of p < 4.3 × 10−7, Bonferroni corrected for the maximum number of annotated genes (n = 19,540) and six lipid traits, to determine significant coding variants. Two gene transcripts, DOCK6 and DOCK7, that overlap with two well-studied lipid genes, ANGPTL8 and ANGPTL3, respectively, met our exome-wide significance threshold. After excluding variation observed in ANGPTL8 and ANGPTL3, DOCK6 and DOCK7, respectively, were no longer significant and have been excluded as associated genes.
We performed a series of sensitivity analyses for our results. We repeated the multi-ancestry gene-based analyses by using an MAF < 0.1% and compared our exome-wide significant gene-based results by using an MAF < 1% to using an MAF < 0.1%. We compared the single variants in our top gene-based associations with respective traits by using GWAS summary data.8 Gene-based tests were repeated excluding variants identified in GWASs with p < 5 × 10−8. Furthermore, all single variants included in each of the top gene-based associations were analyzed in relation to the respective trait. For each exome-wide significant gene-based association, we obtained the association of each single variant within the gene-specific variant groups with the respective phenotype. Then we determined—out of each gene’s overall set of variants—those that had p values at different significance thresholds to identify the percentages of variants contributing to each gene-based signal. To assess whether the most significant variant within each gene was driving the association, we repeated gene-based analyses after removing the respective top single variant from gene-specific variant groups.
To understand whether variants contributing to top gene-based signals were similar or different across different ancestries, we determined the degree of overlap across ancestries for all variants incorporated and then for those with p < 0.05. Finally, we checked for overlap across the most significant (lowest p value) variant from each of the gene-based signals.
Heterogeneity of gene-based estimates in all gene-trait-variant grouping combinations passing exome-wide significant levels was assessed across the five main ancestries (European, South Asian, African, Hispanic, and East Asian) and between T2D and CAD cases and controls via Cochran’s Q.
We performed replication of our top gene-based associations with blood lipid levels in the Penn Medicine BioBank (PMBB) and UK Biobank samples that did not contribute to the discovery analysis (see supplemental methods).
Gene-based analysis of GWAS loci and drug targets
We obtained variants associated with LDL-C, HDL-C, and TG from a recent GWAS in the Million Veterans Program.8 Then we identified genes within ±200 kb of each GWAS index variant and performed gene-based analysis for each of those genes by using the six variant groups. In-silico lookup of gene-based associations for respective lipid traits was then performed for all genes within defined GWAS loci. Drug target genes were obtained from the drug bank database43 with the following search categories: “hypolipidemic agents,” “lipid regulating agents,” “anticholesteremic agents,” “lipid modifying agents,” and/or “hypercholesterolemia.” A liberal definition for drug targets was used—drugs with any number of targets and targets targeted by any number of drugs—and then in-silico lookups were performed for gene-based associations.
Gene-set enrichment analysis
Gene-set enrichment analyses were performed for sets of Mendelian-, protein-altering- and non-protein-altering GWASs, and drug target genes with LDL-C, HDL-C, and TG. 21 genes associated with Mendelian lipid conditions were included on the basis of previous literature:2 LDLR, APOB, PCSK9, LDLRAP1, ABCG5, ABCG8, CETP, LIPC, LIPG, APOC3, ABCA1, APOA1, LCAT, APOA5, APOE, LPL, APOC2, GPIHBP1, LMF1, ANGPTL3, and ANGPTL4. We analyzed GWAS gene sets on the basis of their coding status and their proximity to the most significant signal in the GWAS. Coding variants were defined as missense, frameshift, or stop-gained variants. Gene sets for coding or non-coding variants were then stratified into three categories on the basis of proximity to the most significant variant within each locus—closest, second closest, and greater than second closest gene. For each gene within each set, we obtained the most significant association in the multi-ancestry or ancestry-specific meta-analysis set by using any of the six different variant groups. Then each gene within each gene set was matched to ten other genes on the basis of sample size, total number of variants, cumulative MAC, and variant grouping nearest neighbors via the matchit R function. Then we compared the proportions by using Fisher’s exact test between the main and matched gene sets by applying different p value thresholds.
Association of lipid genes with CAD and T2D data and liver fat/markers
We determined the associations of 40 genes identified in the main and GWAS loci analyses with CAD, T2D, and glycemic and liver enzyme blood measurements. The association with T2D was obtained from the latest gene-based exome association data from the AMP-T2D-GENES consortium.15 The reported associations were obtained from different variant groups on the basis of their previous analyses. We additionally performed gene-based association analyses with CAD by using the MIGen case-control, UKB case-control, and UKB cohort samples with the variant groups described above. Further, six traits including fasting plasma glucose, HbA1c, alanine aminotransferase, aspartate aminotransferase, gamma glutamyl transferase, and albumin were analyzed in the UKB dataset. Single-variant association analyses were performed with RVTESTS. We used linear mixed models incorporating kinship matrices to adjust for relatedness within each study. Covariance matrices were generated with 500 kb sliding windows. We used RAREMETALS to assess associations between aggregated variants (MAF < 1%) in SKATs and burden tests with CAD and each of the six quantitative traits. We used six different variant groupings to determine the set of damaging variants within each gene, (1) high-confidence LOF with LOFTEE, (2) LOF and predicted splice-site-altering variants, (3) LOF and MetaSVM missense variants, (4) LOF, MetaSVM missense, and predicted splice-site-altering variants, (5) LOF and damaging 5 out of 5 missense variants, and (6) LOF, damaging 5 out of 5 missense, and predicted splice-site-altering variants.
Results
Sample and variant characteristics
Individual-level, quality-controlled data were obtained from four sequenced study sources with circulating lipid data for individuals of multiple ancestries (Figure 1). Characteristics of the study samples are detailed in Table S1. We analyzed data on up to 172,000 individuals with LDL-C, non-HDL-C (a calculated measure of TC minus HDL-C), TC, HDL-C, TG, and TG:HDL ratio (a proxy for insulin resistance).44,45 56.7% (n = 97,493) of the sample are of European ancestry, 17.4% (n = 30,025) South Asian, 9.6% (n = 16,507) African American, 9.6% (n = 16,440) Hispanic, 6.1% (n = 10,420) East Asian, and 0.7% (n = 1,182) Samoan, based on genetically estimated and/or self-reported ancestry.
Figure 1.
Study samples and design
Flow chart of the different stages of the study. Exome sequence genotypes were derived from four major data sources: the Myocardial Infarction Genetics consortium (MIGen), the Trans-Omics from Precision Medicine (TOPMed), the UK Biobank, and the Type 2 Diabetes Genetics (AMP-T2D-GENES) consortium. Single-variant association analyses were performed by ancestry and case status in case-control studies and meta-analyzed. Single-variant summary estimates and covariance matrices were used in gene-based analyses with six different variant groups and in multi-ancestry and each of the five main ancestries. AFR, African ancestry; EAS, East Asian ancestry; EUR, European ancestry; HIS, Hispanic ancestry; SAM, Samoan ancestry; SAS, South Asian ancestry.
After sequencing, we observed 15.6 million variants across all studies; we classified 5.0 million (32.6%) as transcript-altering coding variants on the basis of an annotation of frameshift, missense, nonsense, or splice-site acceptor/donor by using the Variant Effect Predictor (VEP).35 A total of 340,214 (6.7%) of the coding variants were annotated as high-confidence LOF via the LOFTEE VEP plugin,36 238,646 (4.7%) as splice-site-altering identified by Splice AI,39 729,098 (14.3%) as damaging missense as predicted by the MetaSVM algorithm,38 and 1,106,309 (21.8%) as damaging missense as predicted by consensus in all five prediction algorithms (SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster, and LRT).37 As expected, we observed a trend of decreasing proportions of putatively deleterious variants with increasing allele count (Figure S2, Table S3).
Single-variant association
We performed inverse-variance weighted fixed-effects meta-analyses of single-variant association results of LDL-C, non-HDL-C, TC, HDL-C, TG, and TG:HDL ratio from each consortium and ancestry group. Meta-analysis results were well controlled with genomic inflation factors ranging between 1.01 and 1.04 (Table S4). Single-variant results were limited to the 425,912 protein-altering coding variants with a total MAC > 20 across all 172,000 individuals. We defined significant associations by a previously established exome-wide significance threshold for coding variants (p < 4.3 × 10−7)40 that was additionally corrected for testing six traits (p = 4.3 × 10−7 divided by 6) within all study samples or within each of the five major ancestries (Tables S5–S10); this yielded in each analysis a significance threshold of p < 7.2 × 10−8. A total of 104 rare coding variants in 57 genes were associated with LDL-C, 95 in 54 genes with non-HDL-C, 109 in 65 genes with TC, 92 in 56 genes with HDL-C, 61 in 36 genes with TG, and 68 in 42 genes with TG:HDL. We identified six missense variants in six genes (TRIM5 p.Val112Phe, ADH1B p.His48Arg, CHUK p.Val268Ile, ERLIN1 p.Ile291Val [rs2862954], TMEM136 p.Gly77Asp, and PPARA p.Val227Ala) >1 Mb away from any index variant previously associated with a lipid phenotype (LDL-C, HDL-C, TC, or TG) in previous genetic discovery efforts (Tables S5–S10).3,7,8 PPARA p.Val227Ala has previously been associated with blood lipids at a nominal significance level in East Asians (p < 0.05), where it is more common than in other ancestries.46 Both TRIM5 and ADH1B LDL-C increasing alleles have been associated with higher risk of CAD in a recent GWAS from CARDIOGRAM (odds ration [OR]: 1.08, p = 2 × 10−9; OR = 1.08, p = 4 × 10−4).47 Single-variant associations were further performed in each of the five main ancestries (Table S11).
Gene-based association
Next, we performed gene-based testing of transcript-altering variants in aggregated SKATs and burden tests42 in all study participants and within each of the five ancestries for six lipid traits: LDL-C, HDL-C, non-HDL-C, TC, TG, and TG:HDL. We excluded the Samoans from the single-ancestry analysis given the small number of individuals. We limited attention to variants with MAF ≤ 1% for each of six variant groups: (1) LOF, (2) LOF and predicted splice-site-altering variants via Splice AI, (3) LOF and MetaSVM missense variants, (4) LOF, MetaSVM missense, and predicted splice-site-altering variants, (5) LOF and damaging 5 out of 5 missense variants, and (6) LOF, damaging 5 out of 5 missense, and predicted splice-site-altering variants. Meta-analyses results were well controlled (Table S12).
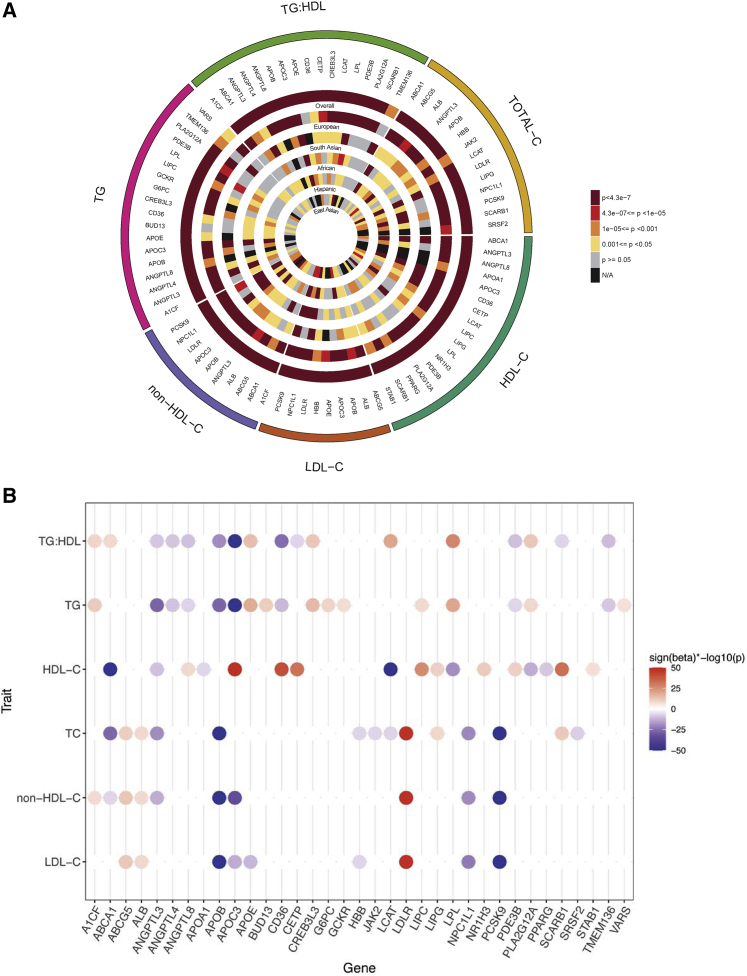
We identified 35 genes reaching exome-wide significance (p = 4.3 × 10−7) for at least one of the six variant groupings (Tables S13–S19). Most of the significant results were from the multi-ancestry analysis where multiple ancestries contributed to the top signals (Figure 2A), and most of the 35 genes were associated with more than one lipid phenotype (Figure 2B). Ten of the 35 genes did not have prior evidence of gene-based links with blood lipid phenotypes (Table 1), and seven genes, including ALB, SRSF2, CREB3L3, NR1H3, PLA2G12A, PPARG, and STAB1, have evidence for a biological connection to circulating lipid levels (Box 1).
Figure 2.
Exome-wide significant associations with blood lipid phenotypes
(A) Circular plot highlighting the evidence of association between the exome-wide significant 35 genes with any of the six different lipid traits (p < 4.3 × 10−7). The most significant associations from any of the six different variant groups are plotted. For almost all of the genes, the most significant associations were obtained from the multi-ancestry meta-analysis.
(B) Strength of association of the 35 exome-wide significant genes based on the most significant variant grouping and ancestry across the six lipid phenotypes studied. Beta (effect size) is obtained from the corresponding burden test for SKAT results. Most of the genes indicated associations with more than one phenotype. Sign(beta)∗−log10(p value) displayed for associations that reached a p < 4.3 × 10−7. When the Sign(beta)∗−log10(p) > 50, they were trimmed to 50.
Table 1.
Genes associated with blood lipids identified in this study
| Gene | Name | Trait | N | cMAC | nVAR | β | SE | p | Mask | Test | Ancestry | UKBB replication | PMBB replication |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALB | albumin | LDL-C | 165,003 | 51 | 18 | 29.51 | 5.11 | 7.76 × 10−9 | LOF | burden | multi-ancestry | <0.005 | N/A |
| ALB | albumin | non-HDL-C | 166,327 | 50 | 17 | 33.91 | 6.07 | 2.27 × 10−8 | LOF | burden | multi-ancestry | N/A | N/A |
| ALB | albumin | TC | 172,103 | 54 | 18 | 33.37 | 5.89 | 1.48 × 10−8 | LOF | burden | multi-ancestry | N/A | N/A |
| SRSF2 | serine and arginine rich splicing factor 2 | TC | 172,103 | 59 | 14 | −30.59 | 5.49 | 2.46 × 10−8 | LOF/DAM5of5/SPLICE AI | burden | multi-ancestry | N/A | <0.005 |
| JAK2 | janus kinase 2 | TC | 975,33 | 441 | 136 | −7.10 | 1.98 | 1.71 × 10−7 | LOF/DAM5of5/SPLICE AI | SKAT | EUR | <0.05 | <0.05 |
| CREB3L3 | camp responsive element binding protein 3 like 3 | TG | 170,239 | 874 | 71 | 0.12 | 0.02 | 2.43 × 10−15 | LOF/DAM5of5/SPLICE AI | burden | multi-ancestry | <0.005 | <0.005 |
| CREB3L3 | camp responsive element binding protein 3 like 3 | TG/HDL-C | 165,380 | 855 | 69 | 0.14 | 0.02 | 5.76 × 10−13 | LOF/DAM5of5/SPLICE AI | burden | multi-ancestry | N/A | N/A |
| TMEM136 | transmembrane protein 136 | TG | 29,571 | 157 | 24 | −0.15 | 0.04 | 3.39 × 10−9 | LOF/DAM5of5/SPLICE AI | SKAT | SAS | N/A | N/A |
| TMEM136 | transmembrane protein 136 | TG/HDL-C | 29,517 | 157 | 24 | −0.20 | 0.05 | 1.76 × 10−11 | LOF/DAM5of5/SPLICE AI | SKAT | SAS | N/A | N/A |
| VARS | valyl-trna synthetase 1 | TG | 56,140 | 67 | 51 | 0.32 | 0.06 | 4.30 × 10−7 | LOF/MetaSVM | burden | EUR | N/A | N/A |
| NR1H3 | nuclear receptor subfamily 1 group h member 3 | HDL-C | 93,044 | 521 | 111 | 3.47 | 0.60 | 1.45 × 10−11 | LOF/MetaSVM/SPLICE AI | SKAT | EUR | <0.005 | <0.05 |
| PLA2G12A | phospholipase a2 group xiia | HDL-C | 166,441 | 1,975 | 47 | −2.28 | 0.31 | 8.12 × 10−14 | LOF/DAM5of5 | burden | multi-ancestry | <0.005 | <0.005 |
| PLA2G12A | phospholipase a2 group xiia | TG | 170,239 | 2,047 | 47 | 0.06 | 0.01 | 1.17 × 10−8 | LOF/DAM5of5 | burden | multi-ancestry | N/A | N/A |
| PLA2G12A | phospholipase a2 group xiia | TG/HDL-C | 165,380 | 1,969 | 46 | 0.11 | 0.01 | 7.56 × 10−13 | LOF/DAM5of5 | burden | multi-ancestry | N/A | N/A |
| PPARG | peroxisome proliferator activated receptor gamma | HDL-C | 166,441 | 147 | 72 | −6.24 | 1.07 | 4.71 × 10−9 | LOF/DAM5of5/SPLICE AI | burden | multi-ancestry | <0.005 | <0.005 |
| STAB1 | stabilin 1 | HDL-C | 166,441 | 6,550 | 804 | 0.83 | 0.16 | 2.58 × 10−7 | LOF/MetaSVM/SPLICE AI | burden | multi-ancestry | <0.005 | N/A |
cMAC, cumulative minor allele count; nVAR, number of variants in test; EUR, European ancestry; SAS, South Asian ancestry; N/A, not applicable.
Box 1. Genes with biological links to lipid metabolism.
ALB
The association between mutations in the albumin gene and elevated cholesterol levels has been previously observed in rare cases of congenital analbuminemia.48 This has been mainly suggested to result from compensatory increases in hepatic production of other non-albumin plasma proteins to maintain colloid osmotic pressure, particularly apolipoprotein B-100, leading to elevations in TC and LDL-C but normal HDL-C levels—which is consistent with our findings—although the exact mechanisms remain uncertain.49 A lipodystrophy-like phenotype has also been linked to analbuminemia, which is consistent with the suggestive tendency for increased risk of T2D with LOF and predicted damaging variants in albumin in the population (OR = 1.85; p = 0.007) (Table S30).
SRSF2
SRSF2 encodes a highly conserved serine/arginine-rich splicing factor and has previously been linked to acute liver failure in liver-specific knockout in mice with accumulation of TC in the mutant liver.50 Thus, this gene could be linked to a non-alcoholic fatty liver phenotype with accumulation of lipids in the liver as observed with other genes as PNPLA3 and TM6SF2.7 Therefore, we looked at association with liver function markers and we found an association between SRSF2 and higher albumin levels (p = 1 × 10−4) and a suggestive tendency for higher gamma glutamyl transferase (GGT) (p = 0.05), consistent with potential liver involvement (Tables S46–S49).
CREB3L3
The association between CREB3L3 and higher TG supports previous evidence from a single family and cohorts with severe hypertriglyceridemia but not sufficient evidence to be classified as a Mendelian lipid gene.51, 52, 53 This has been additionally supported by functional studies where Creb3l3-knockout mice showed hypertriglyceridemia partly due to deficient expression of lipoprotein lipase coactivators (Apoc2, Apoa4, and Apoa5) and increased expression of activator Apoc3.52
NR1H3
The observed association of NR1H3 with higher HDL-C and lower TG is supported by previous evidence of a role in non-alcoholic fatty liver disease in mice.54 This gene encodes a liver X receptor alpha (LXRα), which is a nuclear receptor that acts as a cholesterol sensor and protects from cholesterol overload.55,56 It has previously been shown that disrupting the LXRα phosphorylation at Ser196 in mice prevents non-alcoholic fatty liver disease.54
PLA2G12A
PLA2G12A is in the secretory phospholipase A2 (sPLA2) family, which liberates fatty acids in the −sn2 position of phospholipids. This pattern suggests a previously unreported possible lipolytic role of this phospholipase in a manner similar to another member of the adipose-specific phospholipases, PLA2G16, which has been shown to have a lipolytic role in mice.57,58 Further studies are needed to confirm whether PLA2G12A has a lipolytic role.
PPARG
Rare LOF mutations in PPARG have been previously found to be associated with reduced adipocyte differentiation, lipodystrophy, and increased risk of T2D.59, 60, 61
STAB1
STAB1 is a scavenger receptor that has been shown to mediate uptake of oxidized LDL-C.62,63 There was a suggestive association between LOF variants and higher LDL-C (β = 4.3 mg/dL, p = 2 × 10−3), consistent with its role in LDL-C uptake.
We performed a series of sensitivity analyses on our results. To determine whether low-frequency variants between 0.1%–1% frequency were driving our gene-based association results, we performed the gene-based multi-ancestry meta-analyses by using a maximum MAF threshold of 0.1% instead of 1%. We observed exome-wide significant associations (p < 4.3 × 10−7) for 29 genes with a 0.1% MAF threshold, all observed in our primary analyses with an MAF threshold of 1% (Table S20). We then intersected our 35 lipid-associated genes from 85 gene-based associations observed in the primary analysis with our results with an MAF threshold of 0.1%. All genes remained at least nominally significant (p < 0.05) with an 0.1% MAF threshold, except the A1CF and TMEM136 associations (Table S21). Furthermore, we determined whether those signals were driven by previously reported GWAS hits. We identified a total of seven HDL-C associated variants in six genes, seven LDL-C variants in three genes, three TC variants in one gene, and seven TG variants in six genes that were previously found to be genome-wide significant in the Million Veterans Program (MVP) GWAS results (Table S22).8 Respective gene-based analyses were repeated without those variants. Gene-based signals at A1CF and BUD13 were lost after removal of one variant in each of those genes (Table S23).
The JAK2 signal was further investigated after splitting the 136 contributing variants into those annotated as somatic via the Catalogue of Somatic Mutations in Cancer (COSMIC)64 database and not annotated as a somatic variant. We observed an association only among a set of 26 variants annotated as somatic, while we observed no association when using the remaining 110 variants (Table S24). We also observed that after removal of the most significant variant in JAK2 (p.Val617Phe; rs77375493), a somatic variant, there is no association between JAK2 and total cholesterol (p = 0.10, Table S13).
We also determined which of the 35 genes were outside GWAS regions defined as those within ±200 kb flanking regions of GWAS-indexed single-nucleotide polymorphisms (SNPs) for TC (487 SNPs), LDL-C (531 SNPs), HDL-C, and TG (471 SNPs).8 We identified 1,295 unique genes included in these lipid GWAS regions. Eight out of the 35 associated genes (23%) were not within a GWAS region (Table S13).
To understand whether the gene-based signals were driven by variants that could be identified through single-variant analyses, we looked at the proportion of the 35 genes that were associated with each trait that have at least one single contributing variant that passed the genome-wide significance threshold of 5 × 10−8. Seventeen genes were associated with HDL-C at exome-wide significance (Table S13); eight genes had at least one variant with p < 5 × 10−8 (Table S8). Similarly, we observed 4/9 for LDL-C, 4/10 non-HDL-C, 4/14 TC, 7/18 TG, and 6/17 TG:HDL genes with at least one genome-wide significant variant (Tables S5–S10).
For genes with both gene-based and single-variant signals, we determined the variants that were driving these signals and determined the single-variant associations for all variants contributing to the top 35 genes (Table S25). From a total of 85 gene-based associations, 33 had at least one and 19 had only one single variant with p < 5 × 10−8 (Tables S25 and S26). All of the 19 had at least two variants passing nominal significance (p < 0.05) and 13 had at least ten variants with p < 0.05. Finally, gene-based associations in A1CF, BUD13, JAK2, and TMEM136 were lost after removal of the respective most-significant single variant from the group of variants aggregated in each gene-based association (Table S13).
Comparison of gene-based associations across ancestries
We determined the overlap between single variants included in gene-based signals, which additionally were nominally significant (p < 0.05) in each of the five main ancestries. A large proportion of variants from each ancestry did not overlap with any other ancestry (Figure S3). For example, a total of four genes (CETP, ABCA1, CD36, and LCAT) were observed to have significant gene-based associations with HDL-C in multi-ancestry meta-analyses. A total 68% of variants from European ancestry samples that contributed to HDL-C gene-based associations did not overlap with any other ancestry and nor did 62% in South Asian, 44% in African, 41% in Hispanic, and 59% in East Asian ancestry. When restricted to variants with p < 0.05 in the multi-ancestry meta-analysis, the overlap among ancestries increased (Figure S4). A total of 61% of variants from European ancestry did not overlap with any other ancestry and nor did 46% in South Asian, 27% in African, 27% in Hispanic, and 32% in East Asian ancestry. Finally, we determined the top single variant contributing to each gene-based association (Figure S5). Out of the four HDL-C or the three LDL-C genes, none of the top variants overlapped among any of the ancestries, and at least one out of three variants from the TG genes was shared between two ancestries.
But, the gene-based associations were mostly consistent across the five ancestry groupings: European, South Asian, African, Hispanic, and East Asian. Three of the 17 HDL-C genes showed association in at least two different ancestries at exome-wide significance level (p = 4.3 × 10−7). Similarly, 3/9 LDL-C, 4/10 non-HDL-C, 5/14 TC, 2/18 TG, and 2/17 TG:HDL genes showed association in at least two different ancestries at an exome-wide significance level. Using a less stringent significance level (p < 0.01), across the six lipid traits, 59%–89% of associated genes from the joint analysis were associated in at least two different ancestries.
We tested the top 35 genes for heterogeneity across all 303 gene-trait-variant grouping combinations passing the exome-wide significance threshold (p < 4.3 × 10−7). We observed heterogeneity in effect estimates (pHet < 1.7 × 10−4, accounting for 303 combinations) in 19 (6%) different gene-trait-variant grouping combinations and in six different genes: LIPC, LPL, LCAT, ANGPTL3, APOB, and LDLR (Table S27). Although the LOF gene-based effect sizes were largely consistent across ancestries, there were differences in the cumulative frequencies of LOF variants for several genes, including PCSK9, NPC1L1, HBB, and ABCG5 (Figures S6–S8).
We observed LOF and predicted-damaging variants in TMEM136 associated with TG and TG:HDL only among individuals of South Asian ancestry (pSKAT = 3 × 10−9 and 2 × 10−11, respectively) (Table 1, Figure 2A). With the same variant grouping and ancestry, we observed associations with reduced TG by burden tests (β = −15%, p = 3 × 10−4) and TG:HDL (β = −20%, p = 6 × 10−5) (Tables S18 and S19). Additionally, a single missense variant was associated only among South Asians (rs760568794, 11:120327605-G/A, p.Gly77Asp) with TG (β = −36.9%, p = 2 × 10−8) (Table S9). This variant was present only among individuals with South Asian (MAC = 24) and Hispanic ancestry (MAC = 8) but showed no association among the Hispanic population (p = 0.86). This gene encodes a transmembrane protein of unknown function.
Replication of gene-based associations
We performed replication by using the PMBB and UKB samples that did not contribute to the initial analysis. In PMBB, we observed four out of ten genes without prior evidence of gene-based links with blood lipid phenotypes to have a p < 0.005 (Bonferroni correction for testing ten genes) and in the same direction as the discovery (SRSF2, CREB3L3, PLA2G12A, PPARG) with their respective blood lipids with an additional two genes that met a nominal significance level (p < 0.05; JAK2 and NR1H3). For TMEM136, we found an association of nominal significance for TG and TG:HDL as well but with a beta in the opposite and positive direction. For the other three genes, ALB, VARS, and STAB1, we did not find associations at a nominal significance level for their respective blood lipid traits (Table S28). In UKB, we found six of the ten genes were associated at a p < 0.005 and in the same direction of effect as the discovery analysis (ALB, CREB3L3, NR1H3, PLA2G12A, PPARG, STAB1) (Table S29) with JAK2 reaching a nominal significance threshold (p < 0.05). The only two genes that did not show any evidence of replication in at least one of the replication studies were TMEM136 and VARS. This may indicate these associations are false positives or that we lack power for replication for these associations. Our replication studies did not include individuals of South Asian ancestry, and we observed that our association of TMEM136 with TG and TG:HDL is driven by individuals of South Asian ancestry.
Comparison of gene-based associations by case status
We analyzed heterogeneity by CAD or T2D case status for the top 35 genes. The top 85 signals presented in Table S13 determined in case-status-specific meta-analyses for CAD and T2D. Out of the 85 different gene-based associations, we observed minimal heterogeneity in the results by case status. LDLR, LCAT, and LPL showed significant heterogeneity by CAD case status and LCAT and ANGPTL4 by T2D status (pHet < 6 × 10−4) (Tables S30 and S31).
Gene-based associations in GWAS loci
We determined whether genes near lipid array-based GWAS signals8 were associated with the corresponding lipid measure by using gene-based tests of rare variants with the same traits. We obtained genes from 200 kb flanking regions on both sides of each GWAS signal: 487 annotated to LDL-C GWAS signals, 531 to HDL-C signals, and 471 to TG signals. We analyzed genes within these three sets for gene-based associations with their associated traits. A total of 13, 19, and 13 genes were associated (p < 3.4 × 10−5, corrected for the number of genes tested for the three traits) with LDL-C, HDL-C, or TG, and 32 unique genes were identified in the GWAS loci (Tables S32–S37).
Three of the 32 genes had no prior aggregate rare variant evidence of blood lipid association. Variants annotated as LOF or predicted damaging in EVI5 were associated with LDL-C (pSKAT = 2 × 10−5). The burden test showed association with higher LDL-C levels (β = 1.9 mg/dL, p = 0.008) (Table S32). Variants annotated as LOF or predicted damaging in SH2B3 were associated with lower HDL-C (β = −2.5 mg/dL, p = 1 × 10−6) among Europeans, and variants that were annotated as LOF in PLIN1 were associated with higher HDL-C (β = 3.9 mg/dL, p = 1 × 10−5) (Table S33). Other genes in the regions of EVI5, SH2B3, and PLIN1 did not show an association with the corresponding lipid traits (p > 0.05) in multi-ancestry analyses. A previous report implicated two heterozygous frameshift mutations in PLIN1 in three families with partial lipodystrophy.65 The gene encodes perilipin, the most abundant protein that coats adipocyte lipid droplets and is critical for optimal TG storage.66 We observed a nominal associations of PLIN1 with TG (β = −7.0%, p = 0.02). Our finding is contrary to what would be expected with hypertriglyceridemia in a lipodystrophy phenotype given the association with lower TG. This gene has an additional role where silencing in cow adipocytes has been shown to inhibit TG synthesis and promote lipolysis,67 which may explain those contradictions.
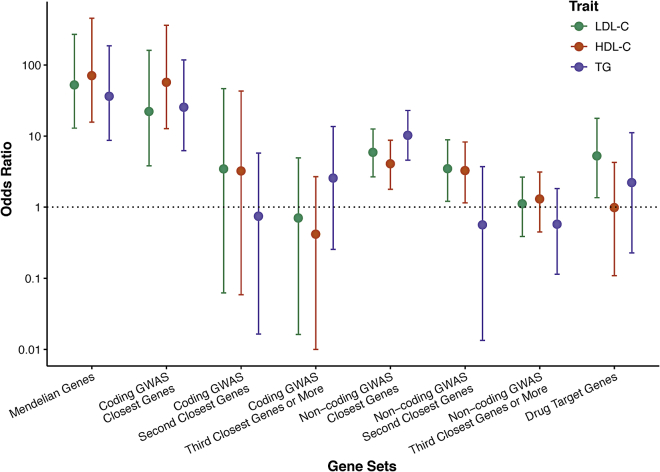
Enrichment of Mendelian, GWAS, and drug targets genes
We next sought to test the utility of genes that showed some evidence for association but did not reach exome-wide significance. Within the genes that reached a sub-threshold level of significant association in this study via SKATs or burden tests (p < 0.005), we determined the enrichment of (1) Mendelian dyslipidemia (N = 21 genes);2 (2) lipid GWAS (N = 487 for LDL-C, N = 531 for HDL-C, and N = 471 for TG);8 and (3) drug target genes (N = 53).43 We stratified genes in GWAS loci according to coding status of the index SNP and proximity to the index SNP (nearest gene, second nearest gene, and genes further away). We tested for enrichment of gene-based signals (p < 0.005) in the gene sets compared to matched genes (Figure 3). For each gene within each gene set, the most significant association in the multi-ancestry or an ancestry-specific analysis was obtained and then matched to ten genes on the basis of sample size, total number of variants, cumulative MAC, and variant grouping. The strongest enrichment was observed for Mendelian dyslipidemia genes within the genes that reached p < 0.005 in our study. For example, 52% of the HDL-C Mendelian genes versus 1.4% of the matched set reached p < 0.005 (OR: 71, 95% CI: 16–455). We also observed that 45.5% of the set of genes closest to an HDL-C protein-altering GWAS variant reached p < 0.005 versus 1.4% in the matched gene set (OR: 57, 95% CI: 13–362). Results were significant but much less striking for genes at non-coding index variants. We observed that 8.9% of the set of genes closest to an HDL-C non-protein-altering GWAS variant reached p < 0.005 versus 2.3% in the matched set (OR: 4.1, 95% CI: 1.8–8.7), while 8% of the set of genes in the second closest to an HDL-C non-protein-altering GWAS variant reached p < 0.005 versus 2.6% in the matched set (OR: 3, 95% CI: 1.1–8.3). There was no significant enrichment in second closest or ≥ third closest genes to protein-altering GWAS signals and in ≥ third closest genes to non-protein-altering GWAS signals. Drug target genes were significantly enriched in LDL-C gene-based associations (OR: 5.3, 95% CI: 1.4–17.8) but not in TG (OR: 2.2, 95% CI: 0.2–11.2) or HDL-C (OR: 1.0, 95% CI: 0.1–4.3) (Figure 3 and Tables S38–S41).
Figure 3.
Enrichment of Mendelian, GWAS, and drug target genes in the gene-based lipid associations
Enrichment of gene sets of Mendelian genes (n = 21), GWAS loci for LDL-C (n = 487), HDL-C (n = 531), and triglycerides (TG) (n = 471) genes, and drug target genes (n = 53). Error bars denote 95% confidence intervals.
Association of lipid genes with CAD, T2D, glycemic traits, and liver enzymes
We tested the genes identified through our discovery (35 genes) and GWAS loci genes (32 genes) for associations with CAD or T2D in our gene-based analyses (40 genes across the two sets). The CAD analyses were restricted to a subset of the overall exome sequence data with information on CAD status, which included the MIGen CAD case-control, UKB CAD nested case-control, and the UKB cohort with a total of 32,981 cases and 79,879 controls. We observed four genes significantly associated with CAD (pCAD < 0.00125, corrected for 40 genes). The four genes associated with lipids and CAD were all primarily associated with LDL-C: LDLR (OR: 2.97, p = 7 × 10−24), APOB (pSKAT = 4 × 10−5), PCSK9 (OR: 0.5, p = 2 × 10−4), and JAK2 (pSKAT = 0.001). Several other known CAD-associated genes (NPC1L1, CETP, APOC3, and LPL) showed nominal significance for association with lipids (p < 0.05). We observed nominal associations with CAD for two of the newly identified lipid genes: PLIN1 (pSKAT = 0.002) and EVI5 (OR: 1.29, p = 0.002; Table S42). None of the 40 lipid genes reached significance for association with T2D in the latest AMP-T2D exome sequence results. We observed nominal associations of T2D with STAB1 (OR: 1.05, pT2D = 0.002) and APOB (OR: 1.08, pT2D = 0.005) (Table S43).15
We additionally tested the 40 genes for association with six glycemic and liver biomarkers in the UKB: blood glucose, HbA1c, alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma glutamyl transferase (GGT), and albumin (Tables S44–S49). Using a significance threshold of p = 0.0012, we found associations between PDE3B and elevated blood glucose, JAK2 and SH2B3 and lower HbA1c, and APOC3 and higher HbA1c. However, JAK2 was no longer associated with Hba1c after removal of the p.Val617Phe missense variant that is known to frequently occur as a somatic mutation (β = 0.22, SE = 0.40, p = 0.47).
We found associations between CREB3L3 and lower ALT and ALB and higher AST and between A1CF and higher GGT. ALB and SRSF2 were associated with lower and higher albumin levels, respectively (Tables S44–S49).
Discussion
We conducted a large multi-ancestry study to identify genes in which protein-altering variants demonstrated association with blood lipid levels. First, we confirm previous associations of genes with blood lipid levels and show that we detect associations across multiple ancestries. Second, we identified gene-based associations that were not observed previously. Third, we show that along with Mendelian lipid genes, the genes closest to both protein-altering and non-protein-altering GWAS signals and LDL-C drug target genes have the highest enrichment of gene-based associations. Fourth, of the new gene-based lipid associations, PLIN1 and EVI5 showed suggestive evidence of an association with CAD.
Our study found evidence of gene-based associations for the same gene in multiple ancestries. The heterogeneity in genetic association with common traits and complex diseases has been discussed extensively. A recent study has shown significant heterogeneity across different ancestries in the effect sizes of multiple GWAS-identified variants.68 However, our study shows that gene-based signals are detected in multiple ancestries with limited heterogeneity in the effect sizes.
Our study highlights enrichment of gene-based associations for Mendelian dyslipidemia genes, genes with protein-altering variants identified by GWASs, and genes that are closest to non-protein-altering GWAS index variants. A previous transcriptome-wide Mendelian randomization study of eQTL variants indicated that most of the genes closest to top GWAS signals (>71%) do not show significant association with the respective phenotype.69 In contrast, our study provides evidence from sequence data that the closest gene to each top non-coding GWAS signal is most likely to be the causal one, indicating an allelic series in associated loci. This has implications for GWAS results, suggesting the prioritization of the closest genes for follow-up studies. We also observed enrichment of drug target genes only among LDL-C gene-based associations and not for HDL-C and TG gene-based associations, consistent with the fact that most approved therapeutics for cardiovascular disease target LDL-C
The gene-based analyses of lipid genes with CAD confirmed previously reported and known associations (LDLR, APOB, and PCSK9). Using a nominal p threshold of 0.05, we also confirmed associations with NPC1L1, CETP, APOC3, and LPL. Of the identified lipid-associated genes, we observed borderline significant signals with EVI5 and higher risk of CAD and between PLIN1 and lower risk of CAD. The putative cardio-protective role of PLIN1 deficiency is supported by previous evidence in mice, which has indicated reduced atherosclerotic lesions with Plin1 deficiency in bone-marrow-derived cells.70 This suggests PLIN1 as a putative target for CAD prevention; however, replication of the CAD association would be needed for confirmation of those signals.
There are limitations to our results. First, we had lower sample sizes for the non-European ancestries, limiting our power to detect ancestry-specific associations and detect replication for TMEM136 that was driven by a variant in South Asians. However, we find consistency of results across ancestries, and when we relax our significance threshold, the majority of associations (59%–89%) are observed in more than one ancestry. Second, it has been reported that there was an issue with the UKB functionally equivalent WES calling.71 This mapping issue may have resulted in under-calling alternative alleles and therefore should not increase false positive findings. Third, we relied on a meta-analysis approach by using summary statistics to perform our gene-based testing because of differences in sequencing platforms and genotyping calling within the multiple consortia contributing to the results. This approach has been shown to be equivalent to a pooled approach for continuous outcomes.41
In summary, we demonstrated association between rare protein-altering variants with circulating lipid levels in >170,000 individuals of diverse ancestries. We identified 35 genes associated with blood lipids, including ten genes not previously shown to have gene-based signals. Our results support the hypothesis that genes closest to a GWAS index SNP are enriched for evidence of association.
Acknowledgements
This work was supported by a grant from the Swedish Research Council (2016-06830) and grants from the National Heart, Lung, and Blood Institute (NHLBI): R01HL142711 and R01HL127564. Please refer to the supplemental information for the full acknowledgements.
Declaration of interests
The authors declare no competing interests for the present work. P.N. reports investigator-initiated grants from Amgen, Apple, and Boston Scientific; is a scientific advisor to Apple, Blackstone Life Sciences, and Novartis; and has spousal employment at Vertex, all unrelated to the present work. A.V.K. has served as a scientific advisor to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color; received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research; and reports a patent related to a genetic risk predictor (20190017119). C.J.W.’s spouse is employed at Regeneron. L.E.S. is currently an employee of Celgene/Bristol Myers Squibb. Celgene/Bristol Myers Squibb had no role in the funding, design, conduct, and interpretation of this study. M.E.M. receives funding from Regeneron unrelated to this work. E.E.K. has received speaker honoraria from Illumina, Inc and Regeneron Pharmaceuticals. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. L.A.C. has consulted with the Dyslipidemia Foundation on lipid projects in the Framingham Heart Study. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular disease. P.T.E. has consulted for Bayer AG, Novartis, MyoKardia, and Quest Diagnostics. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM and has consulted for Bristol Myers Squibb/Pfizer, Bayer AG, and Blackstone Life Sciences. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. M.I.M. has served on advisory panels for Pfizer, NovoNordisk, and Zoe Global and has received honoraria from Merck, Pfizer, Novo Nordisk, and Eli Lilly and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. M.E.J. holds shares in Novo Nordisk A/S. H.M.K. is an employee of Regeneron Pharmaceuticals; he owns stock and stock options for Regeneron Pharmaceuticals. M.E.J. has received research grants form Astra Zeneca, Boehringer Ingelheim, Amgen, and Sanofi. S.K. is founder of Verve Therapeutics.
Published: December 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.11.021.
Data and code availability
Controlled access of the individual-level data is available through dbGAP (please refer to the supplemental information), and the individual-level UK Biobank data are available upon application to the UK Biobank. Summary association results are available on the downloads page of the Cardiovascular Disease Knowledge Portal (broadcvdi.org).
Supplemental information
References
- 1.Di Angelantonio E., Sarwar N., Perry P., Kaptoge S., Ray K.K., Thompson A., Wood A.M., Lewington S., Sattar N., Packard C.J., et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chasman D.I., Paré G., Mora S., Hopewell J.C., Peloso G., Clarke R., Cupples L.A., Hamsten A., Kathiresan S., Mälarstig A., et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M., et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asselbergs F.W., Guo Y., van Iperen E.P., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J., et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C., et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voight B.F., Peloso G.M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M.K., Hindy G., Hólm H., Ding E.L., Johnson T., et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do R., Willer C.J., Schmidt E.M., Sengupta S., Gao C., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hindy G., Engström G., Larsson S.C., Traylor M., Markus H.S., Melander O., Orho-Melander M., Stroke Genetics Network (SiGN) Role of Blood Lipids in the Development of Ischemic Stroke and its Subtypes: A Mendelian Randomization Study. Stroke. 2018;49:820–827. doi: 10.1161/STROKEAHA.117.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J.G., Luk K., Schulz C.A., Engert J.C., Do R., Hindy G., Rukh G., Dufresne L., Almgren P., Owens D.S., et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA. 2014;312:1764–1771. doi: 10.1001/jama.2014.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afshar M., Luk K., Do R., Dufresne L., Owens D.S., Harris T.B., Peloso G.M., Kerr K.F., Wong Q., Smith A.V., et al. Association of Triglyceride-Related Genetic Variants With Mitral Annular Calcification. J. Am. Coll. Cardiol. 2017;69:2941–2948. doi: 10.1016/j.jacc.2017.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C., et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 15.Flannick J., Mercader J.M., Fuchsberger C., Udler M.S., Mahajan A., Wessel J., Teslovich T.M., Caulkins L., Koesterer R., Barajas-Olmos F., et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570:71–76. doi: 10.1038/s41586-019-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Do R., Stitziel N.O., Won H.H., Jørgensen A.B., Duga S., Angelica Merlini P., Kiezun A., Farrall M., Goel A., Zuk O., et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollin T.I., Damcott C.M., Shen H., Ott S.H., Shelton J., Horenstein R.B., Post W., McLenithan J.C., Bielak L.F., Peyser P.A., et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby J., Peloso G.M., Auer P.L., Crosslin D.R., Stitziel N.O., Lange L.A., Lu Y., Tang Z.Z., Zhang H., Hindy G., et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jørgensen A.B., Frikke-Schmidt R., Nordestgaard B.G., Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 20.Musunuru K., Pirruccello J.P., Do R., Peloso G.M., Guiducci C., Sougnez C., Garimella K.V., Fisher S., Abreu J., Barry A.J., et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dewey F.E., Gusarova V., Dunbar R.L., O’Dushlaine C., Schurmann C., Gottesman O., McCarthy S., Van Hout C.V., Bruse S., Dansky H.M., et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N. Engl. J. Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewey F.E., Gusarova V., O’Dushlaine C., Gottesman O., Trejos J., Hunt C., Van Hout C.V., Habegger L., Buckler D., Lai K.M., et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N. Engl. J. Med. 2016;374:1123–1133. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 24.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 25.Kathiresan S., Myocardial Infarction Genetics Consortium A PCSK9 missense variant associated with a reduced risk of early-onset myocardial infarction. N. Engl. J. Med. 2008;358:2299–2300. doi: 10.1056/NEJMc0707445. [DOI] [PubMed] [Google Scholar]
- 26.Sabatine M.S., Giugliano R.P., Keech A.C., Honarpour N., Wiviott S.D., Murphy S.A., Kuder J.F., Wang H., Liu T., Wasserman S.M., et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N. Engl. J. Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 27.Gaudet D., Alexander V.J., Baker B.F., Brisson D., Tremblay K., Singleton W., Geary R.S., Hughes S.G., Viney N.J., Graham M.J., et al. Antisense Inhibition of Apolipoprotein C-III in Patients with Hypertriglyceridemia. N. Engl. J. Med. 2015;373:438–447. doi: 10.1056/NEJMoa1400283. [DOI] [PubMed] [Google Scholar]
- 28.Nomura A., Emdin C.A., Won H.H., Peloso G.M., Natarajan P., Ardissino D., Danesh J., Schunkert H., Correa A., Bown M.J., et al. Heterozygous ABCG5 Gene Deficiency and Risk of Coronary Artery Disease. Circ. Genom. Precis. Med. 2020;13:417–423. doi: 10.1161/CIRCGEN.119.002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peloso G.M., Nomura A., Khera A.V., Chaffin M., Won H.H., Ardissino D., Danesh J., Schunkert H., Wilson J.G., Samani N., et al. Rare Protein-Truncating Variants in APOB, Lower Low-Density Lipoprotein Cholesterol, and Protection Against Coronary Heart Disease. Circ. Genom. Precis. Med. 2019;12:e002376. doi: 10.1161/CIRCGEN.118.002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. bioRxiv. 2019 doi: 10.1101/563866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natarajan P., Peloso G.M., Zekavat S.M., Montasser M., Ganna A., Chaffin M., Khera A.V., Zhou W., Bloom J.M., Engreitz J.M., et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat. Commun. 2018;9:3391. doi: 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szustakowski J.D., Balasubramanian S., Kvikstad E., Khalid S., Bronson P.G., Sasson A., Wong E., Liu D., Wade Davis J., Haefliger C., et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 2021;53:942–948. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
- 33.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.D., Liu D., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N., et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan X., Hu Y., Li B., Abecasis G.R., Liu D.J. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Sveinbjornsson G., Albrechtsen A., Zink F., Gudjonsson S.A., Oddson A., Másson G., Holm H., Kong A., Thorsteinsdottir U., Sulem P., et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet. 2016;48:314–317. doi: 10.1038/ng.3507. [DOI] [PubMed] [Google Scholar]
- 41.Liu D.J., Peloso G.M., Zhan X., Holmen O.L., Zawistowski M., Feng S., Nikpay M., Auer P.L., Goel A., Zhang H., et al. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaughlin T., Abbasi F., Cheal K., Chu J., Lamendola C., Reaven G. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann. Intern. Med. 2003;139:802–809. doi: 10.7326/0003-4819-139-10-200311180-00007. [DOI] [PubMed] [Google Scholar]
- 45.Li C., Ford E.S., Meng Y.X., Mokdad A.H., Reaven G.M. Does the association of the triglyceride to high-density lipoprotein cholesterol ratio with fasting serum insulin differ by race/ethnicity? Cardiovasc. Diabetol. 2008;7:4. doi: 10.1186/1475-2840-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan E., Tan C.S., Deurenberg-Yap M., Chia K.S., Chew S.K., Tai E.S. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis. 2006;187:309–315. doi: 10.1016/j.atherosclerosis.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 47.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minchiotti L., Galliano M., Caridi G., Kragh-Hansen U., Peters T., Jr. Congenital analbuminaemia: molecular defects and biochemical and clinical aspects. Biochim. Biophys. Acta. 2013;1830:5494–5502. doi: 10.1016/j.bbagen.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 49.Koot B.G., Houwen R., Pot D.J., Nauta J. Congenital analbuminaemia: biochemical and clinical implications. A case report and literature review. Eur. J. Pediatr. 2004;163:664–670. doi: 10.1007/s00431-004-1492-z. [DOI] [PubMed] [Google Scholar]
- 50.Cheng Y., Luo C., Wu W., Xie Z., Fu X., Feng Y. Liver-Specific Deletion of SRSF2 Caused Acute Liver Failure and Early Death in Mice. Mol. Cell. Biol. 2016;36:1628–1638. doi: 10.1128/MCB.01071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cefalù A.B., Spina R., Noto D., Valenti V., Ingrassia V., Giammanco A., Panno M.D., Ganci A., Barbagallo C.M., Averna M.R. Novel CREB3L3 Nonsense Mutation in a Family With Dominant Hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 2015;35:2694–2699. doi: 10.1161/ATVBAHA.115.306170. [DOI] [PubMed] [Google Scholar]
- 52.Lee J.H., Giannikopoulos P., Duncan S.A., Wang J., Johansen C.T., Brown J.D., Plutzky J., Hegele R.A., Glimcher L.H., Lee A.H. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med. 2011;17:812–815. doi: 10.1038/nm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dron J.S., Dilliott A.A., Lawson A., McIntyre A.D., Davis B.D., Wang J., Cao H., Movsesyan I., Malloy M.J., Pullinger C.R., et al. Loss-of-Function CREB3L3 Variants in Patients With Severe Hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 2020;40:1935–1941. doi: 10.1161/ATVBAHA.120.314168. [DOI] [PubMed] [Google Scholar]
- 54.Becares N., Gage M.C., Voisin M., Shrestha E., Martin-Gutierrez L., Liang N., Louie R., Pourcet B., Pello O.M., Luong T.V., et al. Impaired LXRalpha Phosphorylation Attenuates Progression of Fatty Liver Disease. Cell Rep. 2019;26:984–995.e6. doi: 10.1016/j.celrep.2018.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao C., Dahlman-Wright K. Liver X receptor in cholesterol metabolism. J. Endocrinol. 2010;204:233–240. doi: 10.1677/JOE-09-0271. [DOI] [PubMed] [Google Scholar]
- 56.Hong C., Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 57.Jaworski K., Ahmadian M., Duncan R.E., Sarkadi-Nagy E., Varady K.A., Hellerstein M.K., Lee H.Y., Samuel V.T., Shulman G.I., Kim K.H., et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat. Med. 2009;15:159–168. doi: 10.1038/nm.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quach N.D., Arnold R.D., Cummings B.S. Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. Biochem. Pharmacol. 2014;90:338–348. doi: 10.1016/j.bcp.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barroso I., Gurnell M., Crowley V.E., Agostini M., Schwabe J.W., Soos M.A., Maslen G.L., Williams T.D., Lewis H., Schafer A.J., et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 60.Agostini M., Schoenmakers E., Mitchell C., Szatmari I., Savage D., Smith A., Rajanayagam O., Semple R., Luan J., Bath L., et al. Non-DNA binding, dominant-negative, human PPARgamma mutations cause lipodystrophic insulin resistance. Cell Metab. 2006;4:303–311. doi: 10.1016/j.cmet.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majithia A.R., Flannick J., Shahinian P., Guo M., Bray M.A., Fontanillas P., Gabriel S.B., Rosen E.D., Altshuler D., GoT2D Consortium. NHGRI JHS/FHS Allelic Spectrum Project. SIGMA T2D Consortium. T2D-GENES Consortium Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc. Natl. Acad. Sci. USA. 2014;111:13127–13132. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goerdt S., Walsh L.J., Murphy G.F., Pober J.S. Identification of a novel high molecular weight protein preferentially expressed by sinusoidal endothelial cells in normal human tissues. J. Cell Biol. 1991;113:1425–1437. doi: 10.1083/jcb.113.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li R., Oteiza A., Sørensen K.K., McCourt P., Olsen R., Smedsrød B., Svistounov D. Role of liver sinusoidal endothelial cells and stabilins in elimination of oxidized low-density lipoproteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G71–G81. doi: 10.1152/ajpgi.00215.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gandotra S., Le Dour C., Bottomley W., Cervera P., Giral P., Reznik Y., Charpentier G., Auclair M., Delépine M., Barroso I., et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N. Engl. J. Med. 2011;364:740–748. doi: 10.1056/NEJMoa1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brasaemle D.L., Subramanian V., Garcia A., Marcinkiewicz A., Rothenberg A. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 2009;326:15–21. doi: 10.1007/s11010-008-9998-8. [DOI] [PubMed] [Google Scholar]
- 67.Zhang S., Liu G., Xu C., Liu L., Zhang Q., Xu Q., Jia H., Li X., Li X. Perilipin 1 Mediates Lipid Metabolism Homeostasis and Inhibits Inflammatory Cytokine Synthesis in Bovine Adipocytes. Front. Immunol. 2018;9:467. doi: 10.3389/fimmu.2018.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L., et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porcu E., Rüeger S., Lepik K., Santoni F.A., Reymond A., Kutalik Z., eQTLGen Consortium. BIOS Consortium Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019;10:3300. doi: 10.1038/s41467-019-10936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X., Gao M., He J., Zou L., Lyu Y., Zhang L., Geng B., Liu G., Xu G. Perilipin1 deficiency in whole body or bone marrow-derived cells attenuates lesions in atherosclerosis-prone mice. PLoS ONE. 2015;10:e0123738. doi: 10.1371/journal.pone.0123738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia T., Munson B., Lango Allen H., Ideker T., Majithia A.R. Thousands of missing variants in the UK Biobank are recoverable by genome realignment. Ann. Hum. Genet. 2020;84:214–220. doi: 10.1111/ahg.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Controlled access of the individual-level data is available through dbGAP (please refer to the supplemental information), and the individual-level UK Biobank data are available upon application to the UK Biobank. Summary association results are available on the downloads page of the Cardiovascular Disease Knowledge Portal (broadcvdi.org).