Abstract
Context
Familial partial lipodystrophy (FPL), Dunnigan variety is characterized by skeletal muscle hypertrophy and insulin resistance besides fat loss from the extremities. The cause for the muscle hypertrophy and its functional consequences is not known.
Objective
To compare muscle strength and endurance, besides muscle protein synthesis rate between subjects with FPL and matched controls (n = 6 in each group). In addition, we studied skeletal muscle mitochondrial function and gene expression pattern to help understand the mechanisms for the observed differences.
Methods
Body composition by dual-energy X-ray absorptiometry, insulin sensitivity by minimal modelling, assessment of peak muscle strength and fatigue, skeletal muscle biopsy and calculation of muscle protein synthesis rate, mitochondrial respirometry, skeletal muscle transcriptome, proteome, and gene set enrichment analysis.
Results
Despite increased muscularity, FPL subjects did not demonstrate increased muscle strength but had earlier fatigue on chest press exercise. Decreased mitochondrial state 3 respiration in the presence of fatty acid substrate was noted, concurrent to elevated muscle lactate and decreased long-chain acylcarnitine. Based on gene transcriptome, there was significant downregulation of many critical metabolic pathways involved in mitochondrial biogenesis and function. Moreover, the overall pattern of gene expression was indicative of accelerated aging in FPL subjects. A lower muscle protein synthesis and downregulation of gene transcripts involved in muscle protein catabolism was observed.
Conclusion
Increased muscularity in FPL is not due to increased muscle protein synthesis and is likely due to reduced muscle protein degradation. Impaired mitochondrial function and altered gene expression likely explain the metabolic abnormalities and skeletal muscle dysfunction in FPL subjects.
Keywords: lipodystrophy, mitochondria, skeletal muscle hypertrophy, insulin resistance
Familial partial lipodystrophy (FPL), Dunnigan variety is a rare autosomal dominant disorder due to heterozygous missense mutations in LMNA causing selective loss of subcutaneous fat from the extremities and anterior trunk (1, 2). Fat loss typically occurs after puberty and is accompanied by the development of metabolic complications including severe insulin resistance, hyperlipidemia, and hepatic steatosis. Affected individuals also display extreme muscularity. This is not just due to loss of subcutaneous fat, but an actual increase in skeletal muscle mass and volume as revealed by imaging (3), and hypertrophy of the muscle fibers evident on microscopy (4). Muscle hypertrophy represents increased protein accumulation in muscle fibers, either due to increased synthesis or decreased degradation. It remains to be determined whether increased muscle protein synthesis or decreased muscle protein degradation or a combination of both occurs in FPL. The pathophysiology of increased muscularity is likely to impact muscle functions and may determine whether muscle performance is proportional to muscle hypertrophy. Currently it is not known how muscle hypertrophy in individuals with FPL affects their physical strength and endurance. The presence of insulin resistance in FPL individuals (2, 5) does indicate that higher lean mass does not translate to higher insulin sensitivity as noted in nonlipodystrophic subjects (6, 7). It remains to be fully understood whether physical performance is altered proportional to the increased muscle mass in individuals with FPL. We therefore measured muscle strength and endurance in individuals with FPL and compared them with matched controls. In addition, we compared rates of muscle protein synthesis between the 2 subject groups to help understand the mechanism of muscle hypertrophy.
We also sought to determine whether alterations in muscle mitochondrial function, which has been reported in other insulin-resistant states (8-12), also occur in FPL individuals and may impact on their physical performance. Excess mitochondrial emission of H2O2 has been shown to cause high-fat diet–induced insulin resistance (13), and insulin-resistant women are reported to show inefficient mitochondrial coupling, reduced phosphorylation efficiency, and excess oxidative stress that are corrected on improving insulin sensitivity by aerobic exercise (14). Impaired mitochondrial fatty acid oxidation has also been reported to contribute to insulin resistance in healthy, lean, older individuals (15), and in those with type 2 diabetes (10, 16, 17). Altered mitochondrial function has been noted in patients with HIV infection who develop lipodystrophy due to antiretroviral medications (18, 19), but it remains to be determined if patients with genetic lipodystrophies including FPL also have abnormal mitochondrial function in association with insulin resistance. We therefore performed detailed mitochondrial bioenergetics studies in FPL individuals compared with normal controls.
While the occurrence of insulin resistance in FPL concurrent to LMNA mutation is well recognized, the exact mechanism by which LMNA mutations lead to partial fat loss and insulin resistance is not clear, although it is presumed that insulin resistance is secondary to fat loss and the resultant accumulation of ectopic fat in nonadipose tissue (20). This is quite clear in individuals with generalized lipodystrophy, both congenital and acquired, who have near complete fat loss, but not so apparent in patients with partial lipodystrophy who may even have normal amounts of “total” body fat due to excess fat deposition in unaffected areas. Patients with generalized lipodystrophy also have severe hypoleptinemia, and both animal studies (21, 22) and human trials (23) have served to demonstrate the importance of endocrine actions of adipose tissue in preventing metabolic abnormalities. Again, patients with partial lipodystrophy do not uniformly have hypoleptinemia, with many of them noted to have normal leptin and adiponectin levels (24). Thus individuals with partial lipodystrophy are not entirely analogous to those with generalized lipodystrophy, and the cause for their extreme insulin resistance despite the presence of seemingly “adequate” amounts of total body fat needs further investigation. We therefore performed detailed skeletal muscle transcriptome, proteome, and metabolomic studies followed by gene set enrichment analysis to identify metabolic pathways which are affected by lipodystrophy and insulin resistance.
Materials and Methods
The study protocol was approved by the Mayo Clinic Institutional Review Board, and all subjects provided informed, written consent. Figure S1 (25) provides a schematic outline of the study protocol.
Participants
Six individuals with FPL, Dunnigan variety and 6 age, sex, and body mass index (BMI)–matched controls were studied (Table 1 (25)). Lipodystrophy subjects belonged to known FPL pedigrees harboring either the R482W (5 subjects) or the R482Q (1 subject) missense mutation in LMNA. They all showed typical features of FPL including extreme paucity of subcutaneous fat from the extremities and anterior trunk. There were 4 female subjects with age range 26–70 years, and the 2 male subjects were 48 and 24 years of age. All subjects had hyperlipidemia, but 3 of them were not on medications. Three subjects had diabetes, of whom 1 was on diet therapy only, and the other 2 were on medications. Control subjects had no known chronic medical illness and were not on any medications. However, mild hypertriglyceridemia was noted in 2 control subjects and 3 of them had impaired fasting glucose.
Body Composition
Whole-body dual-energy X-ray absorptiometry (DEXA) scan was performed with a multiple detector fan-beam Lunar densitometer (Lunar DPX-L, Lunar Radiation). Data were obtained from the head, trunk, and upper and lower extremities, and the proportion of fat and lean tissue mass in the individual regions as well as the whole body was calculated as percentage of total body mass. Skeletal muscle mass was calculated as the difference between lean mass and bone mineral content.
Magnetic Resonance Imaging and Magnetic Resonance Spectroscopy
High resolution T1 weighted magnetic resonance imaging of the leg and image-guided proton-localized magnetic resonance spectroscopy (1H-MRS) were obtained on a 3.0 T magnetic resonance imaging system using a single voxel PRESS sequence with the following imaging parameters: voxel volume 15 × 15 × 15 mm, repetition time of 2000 ms, echo time of 35 ms, 2048 data points over a 2500 kHz spectral width. Measurements were obtained in the postabsorbtive state (at least 4 hours after the meal), with subjects resting in the supine position. For MRS, voxels were centered over the mid soleus, with care to avoid vascular structures, and to ensure consistent orientation of muscle fibers along the magnetic field. Spectra were processed and resonances quantified using a standard analysis package (LCModel). The intramyocellular lipid concentration was expressed as a percentage of the intensity of the water resonance peak. Leg muscle volume was quantified from the scout images using Analyze Software system (Mayo Clinic Biomedical Imaging Resource).
Muscle Strength and Fatigue
The 1 repetition maximum (1RM) for the recumbent leg press and seated chest press was used to quantify maximal voluntary strength of the lower and upper extremities, respectively, using Keiser A420 pneumatic resistance machine and integrated software (Keiser Sport, Fresno, CA). After a warm-up and familiarization period, participants completed a standardized protocol consisting of progressively increasing resistance. The 1RM was defined as the maximal load lifted through the full range of motion with proper form. Following determination of the 1RM, a fatigue test was performed for the lower and upper extremities. For the leg press, resistance was set at 80% of the 1RM and subjects were instructed to perform as many repetitions as possible through the full range of motion while maintaining proper form. The chest press fatigue measure was similarly performed with the resistance set at 70% of the 1RM.
Mixed Meal Study for Assessment of Muscle Protein Synthesis and Measures of Insulin Sensitivity and Secretion
Study participants were fed a high-fat mixed meal in the form of a protein shake (10 kcal/kg, 45% carbohydrate, 15% protein, and 40% fat) after an overnight 12 hour fast (time 0), immediately after the first muscle biopsy. The protein content of the meal was intrinsically labelled [13C6]phenylalanine (13C6 Phe) casein and whey protein as described previously (26). Muscle protein fractional synthetic rate (FSR) was calculated as described previously (26, 27) after determination of 13C6 Phe enrichment in the 2 muscle biopsy samples obtained before and 5 hours after the study meal, and plasma samples collected throughout the 5 hour period by mass spectrometry as previously described (28). FSR (%/hour) was calculated as [ΔE/(Ep × t)] × 100, where t was hours between biopsies, ΔE was the tissue protein enrichment between biopsies, and Ep was plasma precursor enrichment.
Blood samples for measurement of glucose, insulin, and C-peptide were collected at –30, –20, –10, 0, 5, 10, 15, 20, 30, 40, 50, 60, 75, 90, 120, 150, 160, 210, 240, and 300 minutes, and measures of insulin sensitivity and beta cell function were calculated using the minimal model (29). Serum triglycerides were measured at –30, 0, 30, 60, 90, 120, 150, 180, 240, and 300 minutes, and the area under curve calculated as a measure of fat tolerance.
Mitochondrial Oxidative Capacity
After a 12 hour overnight fast, a percutaneous needle muscle biopsy was obtained from the vastus lateralis muscle under local anesthesia (2% lidocaine). About 300 mg of muscle tissue was obtained using a modified Bergstrom needle as described before (30). Mitochondria were isolated from fresh, nonfrozen muscle tissue using methods described previously (31). Briefly, muscle was transferred into a chilled glass dish containing an ionic homogenizing buffer (100 mM KCl, 50 mM Tris, 5 mM MgCl2, 1.8 mM ATP, and 1 mM EDTA, pH 7.2). Tissue was minced, incubated on ice for 2 minutes with protease, rinsed, and gently homogenized on ice for 10 minutes using a motor-driven Potter-Elvehjem tissue grinder. Mitochondria were separated by differential centrifugation and re-suspended in a nonionic buffer (225 mM sucrose, 44 mM KH2PO4, 12.5 mM Mg acetate, and 6 mM EDTA).
Respiration of isolated mitochondria was studied using the Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria) with continuous stirring and temperature maintained at 37°C as described before (32). Experiments were performed in respiration buffer (MiRO5) containing 110 mM sucrose, 60 mM potassium lactobionate, 0.5 mM EGTA, 1 g/L BSA essentially fat free, 3 mM MgCl2, 20 mM taurine, 10 mM KH2PO4, 20 mM Hepes, pH 7.1. Mitochondria were allowed to equilibrate in MiR05 until stable oxygen flux rates were achieved. After equilibration, a stepwise titration protocol was initiated, starting with the addition of glutamate (10 mM) and malate (2 mM) in the absence of exogenous adenylates to initiate state 2 respiration with electron flow through complex I. State 3 respiration was initiated by the addition of ADP (2.5 mM). The addition of succinate (10 mM) provided convergent electron flow through respiratory chain complexes I and II, followed by rotenone (0.5 μM) to inhibit complex I. State 4 respiration was measured in the presence of oligomycin (2 μg/mL) to inhibit ATP synthase. Subsequently Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added to cause uncoupling of oxidative phosphorylation. Finally, 2.5 µM antimycin A was added to inhibit mitochondrial oxygen consumption to confirm that oxygen consumption was of mitochondrial origin. A parallel experiment was conducted to measure mitochondrial capacity for lipid oxidation. After equilibration with ambient oxygen, palmitoyl-l-carnitine (0.005 mM) and malate (2 mM) were added to assess state 2 respiration in the presence of lipid substrate. This was followed by serial addition of ADP, rotenone, oligomycin, FCCP, and antimycin A to assess state 3 respiration, state 4 respiration, and nonmitochondrial respiration as before. Oxygen flux rates were calculated and corrected for background oxygen kinetics using Datlab software (Oroboros Instruments, Innsbruck, Austria) and expressed per tissue wet weight and protein content. Mitochondrial coupling efficiency was evaluated from the respiratory control ratio as the quotient of state 3 and state 4 respiration rates. Mitochondrial phosphorylation efficiency was evaluated from the ADP:O ratio, determined by quantifying the amount of oxygen consumed in response to a nonsaturating (15 μM) pulse of ADP. The reactive oxygen species emitting potential of isolated mitochondria was also assessed by measuring H2O2 production using a Fluorolog 3 (Horiba Jobin Yvon) spectrofluorometer with temperature control and continuous stirring to monitor Amplex Red (Invitrogen) oxidation as described before (14, 32).
Muscle biopsy and mitochondrial respirometry was repeated on the contralateral leg 5 hours after ingestion of a mixed meal.
Metabolomics
The concentrations of 40 metabolites (Table S2 (25)) in the muscle biopsy specimen were measured by proton nuclear magnetic resonance spectroscopy (1H-NMR). About 30 mg of muscle tissue was pulverized and extracted after treatment with 300 µL of ice-cold 0.6M perchloric acid (HClO4) solution. Sample tubes were vortexed, centrifuged at 10 000g for 10 minutes at 4°C, and supernatants collected. The supernatants obtained from 2 rounds of extraction were combined and neutralized with 140 µl of 2M KHCO3. In a 400 µL aliquot of neutralized extract, 100 μL of 0.1 M phosphate buffer and 50 μL of 1 mM TSP-d4 in D2O were added. Samples were vortexed for 20 seconds and transferred to 5 mm NMR tubes. The NMR spectra were acquired on Bruker AVANCE III 600 MHz instrument (Bruker, Billerica, USA). 1H NMR spectra were recorded using 1D NOESY pulse sequence with presaturation (noesygppr1d) under the following conditions: 90 degree pulse for excitation, acquisition time 3.90 seconds, relaxation delay 5 seconds. All spectra were acquired with 256 scans at room temperature (298 K), with 64k data points and 8417 Hz (14 ppm) spectral width. The spectra were phase and baseline corrected using TopSpin 3.5 software. The spectra were then processed using Chenomx NMR Suite 8.3 software (Chenomx Inc., Edmonton, Canada). The compounds were identified by comparing spectra to database Chenomx 600 MHz Version 10. Quantification was based on internal standard (TSP-d4) peak integral. The metabolites concentrations were exported as µM in NMR sample and recalculated as µmol/g of wet tissue. In addition, long chain acyl carnitines were measured by liquid chromatography–tandem mass spectrometry (LC-MS/MS).
Gene Expression Analysis
We measured mRNA expression by RNA Seq approach as previously described (33). In brief, total RNA was isolated from the muscle biopsies obtained before and after the meal, and sequencing libraries were prepared with TruSeq RNA Sample Prep Kit v2. Libraries were sequenced on a HiSeq 2000 sequencer using TruSeq SBS sequencing kit version 3 and HCS version 2.0.12.0 software. Genes with an FDR-corrected P ≤ .05 and an absolute log2 fold change of ≥0.5 (where 0.0 signifies no change) were considered for further analysis.
Proteomics
Proteins extracted from the muscle biopsy samples were subjected to LC-MS/MS as described previously (33, 34). Label-free LC-MS/MS data were acquired using a high-resolution Lumos MS (Thermo Fisher Scientific). MaxQuant software (v.1.5.1.2) was configured to match MS/MS against RefSeq human sequence database (v.58) and identify peptides and proteins at 1% FDR. Protein groups with a differential expression (corrected) P ≤ .05 and an absolute log2 fold change (FC) of ≥.5 between patients and controls were considered for further analysis.
Pathway and Gene Set Enrichment Analysis
Genes or proteins that were statistically up- or downregulated between patients and controls, before and after the meal were subjected to pathway analysis using WEBGESTALT software. Genes that were upregulated were subjected to Gene Ontology (GO) process enrichment using MetaCore software (Thompson Reuters) and upstream regulator analysis using Ingenuity Pathway Analysis (QIAGEN). Gene set enrichment analysis was performed using Broad’s GSEA software. All enrichment P values were FDR corrected (using Benjamini–Hochberg procedure), and entities with P ≤ .05 were reported. In addition, we also performed a gene set enrichment analysis for each pairwise comparison that does not rely on any fold change or P value thresholds using Preranked method in Broad’s GSEA software. For this, gene-wise ranks were computed for each pairwise comparison using the following formula: –1 × log2(P value) × sign(fold change). Genes and their associated ranks were uploaded into the GSEA software for enrichment analysis using the default parameters. To compare the expression of genes involved in specific metabolic pathways, previously described gene sets were used (35, 36).
Statistical Analysis
Baseline subject data are expressed as mean ± SE. For variables whose distribution are expected to follow a normal distribution, a 2-tailed Student’s t test with unequal variance was utilized to test for differences between the 2 groups. For remaining variables, test for normality was performed and the same 2-tailed Student’s t test with unequal variance was applied for normally distributed variables. For rest of the variables, a nonparametric Wilcoxon rank-sum test was used to compare differences between the 2 groups. Paired t test was used for comparing pre and post meal values within each group. Pearson’s correlation was performed to investigate relationship between mitochondrial respiration and metabolic variables. P < .05 was considered to be statistically significant. Analyses were conducted using JMP Pro 14.1.0 software (SAS Institute Inc.). Additional statistical analysis of genomic and proteomic data has been described in their respective sections.
Results
FPL individuals have characteristic physical and biochemical features including partial fat loss, increased muscularity, and insulin resistance
The genotypic and phenotypic features of study patients were characteristic of FPL, Dunnigan variety, and included missense mutations in LMNA exon 8, and reported fat loss from the limbs, hips, and trunk from early adolescence Table S1 (25). We confirmed that FPL individuals, compared with age, sex, and BMI-matched normal controls, had lower total body fat especially over the limbs, but increased trunk fat as measured by DEXA. The ratio of limb:trunk fat in FPL individuals was <50% of controls (Table 1). Consistent with the fat loss, serum leptin, and adiponectin levels in FPL individuals tended to be lower than controls.
Table 1.
Clinical features and biochemical characteristics of patients with familial partial lipodystrophy (FPL) and controls
| Variable | FPL | Control | P value |
|---|---|---|---|
| Age (y) | 43.2 ± 5.11 | 43.2 ± 4.77 | 1 |
| Sex | 4F:2M | 4F:2M | |
| Weight (kg) | 81.9 ± 2.8 | 84.2 ± 4 | .75 |
| BMI (m/kg2) | 27.9 ± 0.5 | 29.3 ± 0.7 | .3 |
| Body fat (%) | |||
| Total | 22.5 ± 1.5 | 36.9 ± 3 | .02 |
| Trunk | 66.3 ± 0.8 | 52.7 ± 2.1 | .004 |
| Limb | 18.7 ± 0.6 | 33.3 ± 1.8 | .002 |
| Limb:Trunk | 0.41 ± 0.02 | 0.86 ± 0.06 | .004 |
| Lean Mass (%) | |||
| Total | 74.9 ± 1.4 | 60.9 ± 2.8 | .02 |
| Arm | 80 ± 1.2 | 61.8 ± 3.5 | .01 |
| Leg | 81.8 ± 1.1 | 62.5 ± 3.3 | .01 |
| Arm muscle mass (kg) | 7.11 ± 0.4 | 5.58 ± 0.5 | .13 |
| Leg muscle mass (kg) | 18.46 ± 0.9 | 16.94 ± 1 | .45 |
| Calf muscle volume (cc) | 966 ± 29.8 | 776 ± 45 | .04 |
| Serum leptin (ng/mL) | 10.4 ± 1.6 | 28.6 ± 5.9 | .09 |
| Serum adiponectin (ng/mL) | 3474 ± 401 | 9496 ± 1476 | .04 |
| Fasting glucose (mg/dL) | 124.8 ± 10.5 | 93.3 ± 2.5 | .09 |
| Fasting insulin (µIU/mL) | 33.1 ± 5.3 | 9.3 ± 1.4 | .02 |
| HOMA-IR | 10.6 ± 2 | 2.2 ± 0.4 | .03 |
| Serum triglycerides (mg/dL) | 239 ± 32 | 174 ± 28.9 | .3 |
| HDL cholesterol (mg/dL) | 24 ± 1.1 | 47 ± 5.1 | .02 |
| Intramyocellular lipid content (%) | 0.06 ± 0.01 | 0.04 ± 0.003 | .2 |
N = 6 in each group, data are expressed as mean ± SE. Two-tailed Student’s t test with unequal variance was used for comparison between the 2 groups.
Besides the characteristic pattern of fat loss, DEXA studies also showed that in FPL individuals there was a 25% increase in total body percent fat-free mass, an indirect measure of lean tissue mass. Moreover, percent fat-free mass in the arms and legs were also higher by about 22%. While the skeletal muscle mass in both arms and legs (lean mass – bone mineral content) also tended to be higher in FPL individuals by 21.5% and 8.2% respectively, these changes were not statistically significant. However, direct measurement of skeletal muscle volume by magnetic resonance imaging of the calf, showed significantly higher muscle volume in FPL individuals than in controls by nearly 25%.
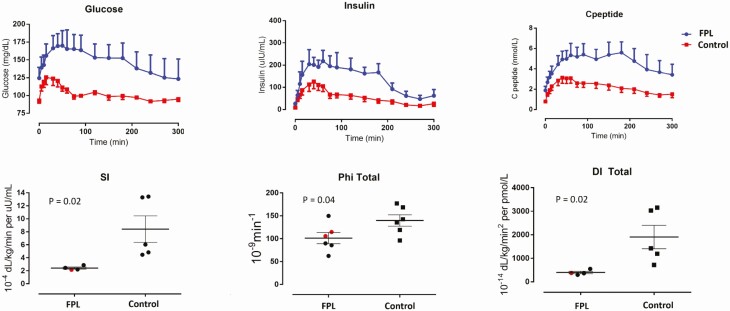
Fasting serum glucose tended to be higher in FPL individuals (of whom 2 were on glucose-lowering therapies including metformin, dapagliflozin, and insulin), and they also had higher insulin resistance as assessed by the Homeostatic Model Assessment of Insulin Resistance (HOMA-IR). Further, minimal modeling of glucose, insulin, and C-peptide excursions during the frequently sampled oral mixed-meal challenge clearly showed insulin resistance in individuals with FPL (Fig. 1). The insulin sensitivity index was lower by nearly 75% (P = .02), and measures of β-cell responsivity to glucose (Φ static and total), and the overall disposition index which expresses β-cell function in relation to insulin sensitivity were also reduced in FPL individuals compared with controls (P = .04 and .02, respectively). Consistent with insulin resistance, fasting, and postmeal serum triglycerides tended to be higher in FPL individuals, and serum HDL cholesterol was significantly lower. Serum free fatty acids tended to be lower in FPL individuals in the fasting state, and did not change after the meal, while in controls it decreased by about 30% (Figure S2 (25)). These differences were however not statistically significant, but it must be noted that half the patients with FPL were on lipid-lowering medications including statins, fibrates, and omega-3 fatty acids.
Figure 1.
Individuals with familial partial lipodystrophy (FPL) show decreased insulin sensitivity and pancreatic response during a mixed meal. Plasma glucose, insulin, and C-peptide excursions following mixed meal in individuals with FPL and controls (top), and corresponding measures of whole body insulin sensitivity (SI), pancreatic sensitivity (phi total), and overall beta-cell function (disposition index, DI) as derived from minimal model. N = 6 in each group but 3 subjects (2 in FPL group and 1 in control group) were excluded from SI calculation as their plasma glucose at 300 minutes did not return to baseline (2 subjects) or insulin profile was abnormal and did not fit the model (1 subject). Data in top panels are shown as mean ± SE, and individual data with mean and SE are shown in the bottom panels. Comparison between the 2 groups was performed using Wilcoxon rank sum test. Red dots in the lower panel indicate patients with diabetes on medications.
Intramyocellular lipid concentration, measured by MRS, tended to be higher in FPL participants, and showed significant correlation with fasting plasma glucose (r = 0.86, P = .0006) and triglyceride levels (r = 0.68, P = .01) as well as postprandial lipemia (triglyceride area under curve after mixed meal ingestion, r = 0.6, P = .04).
Together with the body composition data, these metabolic abnormalities are characteristic of the FPL phenotype.
Skeletal muscle hypertrophy in FPL individuals is not due to increased muscle protein synthesis but likely due to decreased degradation
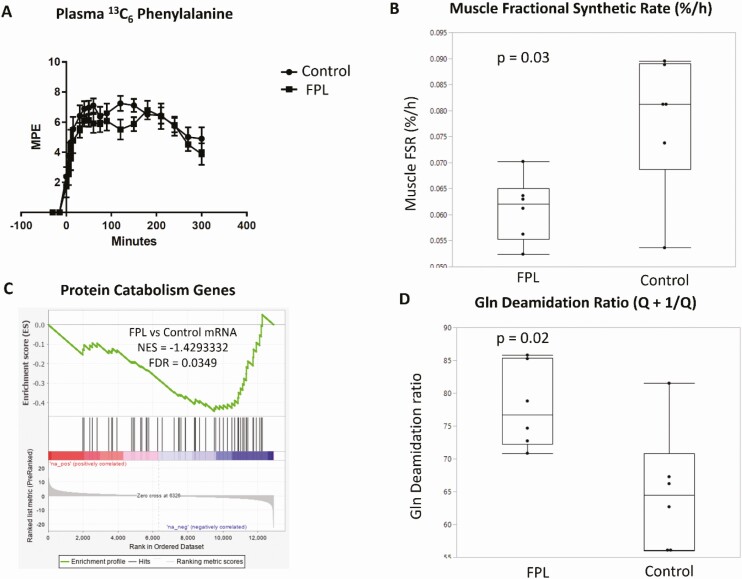
To determine whether the observed increase in skeletal muscle mass is due to increased muscle protein synthesis, we measured the FSR of skeletal muscle mixed protein after oral ingestion of intrinsically labelled (13C6 Phe) casein and whey protein. Surprisingly, the FSR was lower in FPL individuals than in controls and could not account for the higher skeletal muscle mass (Fig. 2B). We then determined if the higher muscle mass was due to decreased protein degradation (catabolism). While this degradation was not directly measured, we found evidence indicating decreased protein catabolism in FPL compared with control individuals based on skeletal muscle gene transcripts specifically showing decreased expression of genes reported to be involved in protein catabolism (http://amigo.geneontology.org/amigo/term/GO:0044257). There was significant downregulation of these genes with a net enrichment score of –1.4 and FDR = 0.04 (Fig. 2C). We also analyzed the proteomics data to determine the extent of glutamine deamidation in the muscle proteome to assess accumulation of proteins increased with age and destined for degradation (37). Normalized intensities of peptides with deamidated and unmodified forms were obtained, and the ratio of deamidation to unmodified form was computed for each peptide. The peptide ratios were summed together to obtain an individual protein deamidation ratio, which is a surrogate measure of protein oxidative damage. We observed a higher ratio of protein deamidation in FPL samples (Fig. 2D) consistent with accumulation of damaged proteins, likely due to reduced catabolism. Together, these observations are consistent with a notion that the increased muscularity in FPL individuals is due to decreased protein breakdown rather than increased synthesis.
Figure 2.
Skeletal muscle hypertrophy in individuals with familial partial lipodystrophy (FPL) is not due to increased protein synthesis but due to reduced protein catabolism. (A) Plasma 13C6 phenylalanine enrichment after labelled mixed meal ingestion in individuals with FPL and controls. (B) Reduced fractional synthetic rate (FSR) of mixed muscle protein in FPL individuals based on plasma and muscle 13C6 enrichment. (C) Downregulation of genes which regulate protein catabolism in FPL individuals compared with controls. (D) Increased ratio of deamidated to unmodified proteins in FPL individuals compared with controls, suggesting higher oxidative damage. N = 6 in each group, each dot represents individual points with the box showing 25th, 50th, and 75th percentile values. Two-tailed Student’s t test with unequal variance was used for comparisons shown in B and D.
FPL individuals do not display increased muscle strength despite increased muscularity, but demonstrate increased fatigability
There was no difference in the peak force generated during both seated chest press and recumbent leg press in FPL individuals compared with controls despite their increased muscularity. When muscle strength was expressed relative to lean mass, it tended to be lower in FPL subjects, though not reaching statistical significance (Table 2). However, the number of chest press repetitions at 80% maximum load was significantly lower by 25% in FPL individuals than in controls, indicating increased fatigability (Figure S3 (25)). Overall, it appears that in FPL individuals, muscle strength is not congruent with muscle mass, and fatigue in a major muscle group sets in earlier.
Table 2.
Measures of muscle strength and fatigue during chest press and leg press exercise in individuals with familial partial lipodystrophy (FPL) and matched controls
| Muscle performance measures | FPL | Controls | P value | |
|---|---|---|---|---|
| Peak strength (N) | Chest press | 686 ± 56.9 | 659 ± 90.6 | .86 |
| Leg press | 2337 ± 138.9 | 2250 ± 191.4 | .8 | |
| Peak strength per lean mass (N/kg) | Chest press | 11 ± 0.5 | 12.7 ± 1.3 | .4 |
| Leg press | 37.9 ± 1.3 | 43.9 ± 1.6 | .07 | |
| Number of repetitions to fatigue | Chest press | 15 ± 0.6 | 20 ± 3.1 | .01 |
| Leg press | 20 ± 0.2 | 20 ± 0.6 | 1 |
Muscle strength was assessed by the maximum load that could be lifted, and fatigue by the maximum number of repetitions at a given load. N = 6 in each group. Data are expressed as mean ± SE, and 2-tailed Student’s t test with unequal variance was used for comparisons.
Impaired mitochondrial function is noted in FPL individuals and is associated with muscle fatigability and metabolic dysfunction
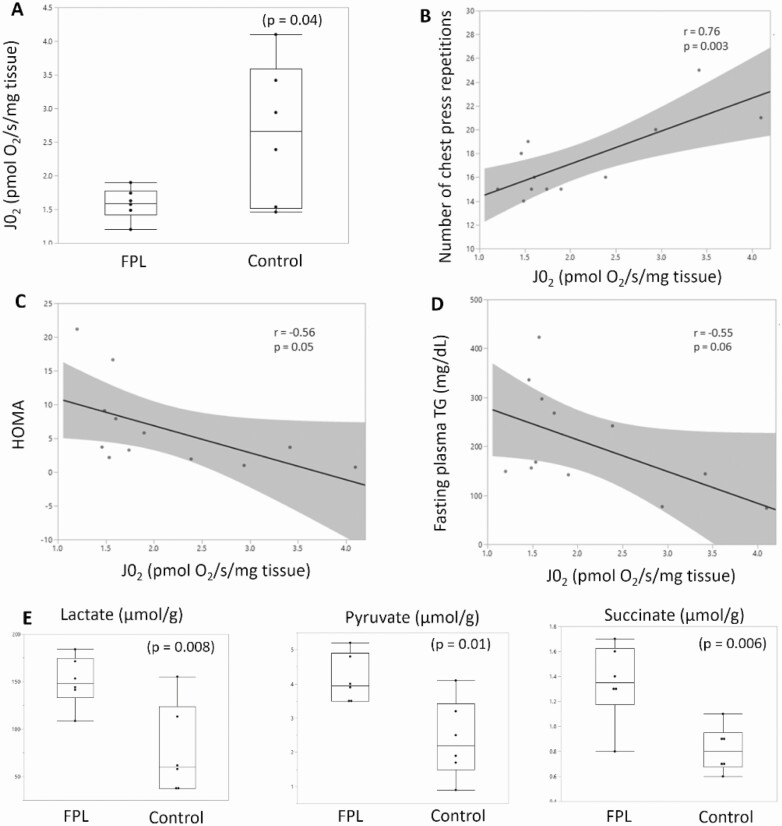
Mitochondrial bioenergetics was studied before and 5 hours after a mixed meal in the presence of both carbohydrate (glutamate, malate and succinate) and lipid (palmitoyl carnitine and malate) substrates. The primary outcome was maximally stimulated state 3 respiration, which was significantly lower in FPL individuals than in controls (Fig. 3A). There was a nearly 40% lower mitochondrial oxygen consumption per tissue mass in the presence of lipid substrate on the addition of ADP in FPL individuals compared with controls (P = .09 premeal, and P = .04 postmeal, respectively). This indicated impaired state 3 respiration in the presence of lipid substrate. It tended to be lower in the presence of carbohydrate substrate, and when expressed per protein content as well, but did not reach statistical significance (Figures S4 and S5 (25)). State 1 respiration was also slightly lower in the presence of both carbohydrate and lipid substrate. The respiratory control ratio tended to be lower in FPL individuals (6.7 ± 1.4 vs 9.7 ± 2.2, P = .07), while there was no difference in the ADP:O ratio, suggesting coupling inefficiency but no effect on phosphorylation efficiency. There was no difference in reactive oxygen species production as assessed by H2O2 emission. Overall, the most striking abnormality noted was a marked reduction in state 3 (maximal) respiration in the presence of lipid substrate.
Figure 3.
Mitochondrial respiration is decreased in individuals with familial partial lipodystrophy (FPL) and correlates with measures of muscle performance and metabolic dysfunction. Oxygen consumption in isolated mitochondria was measured in the presence of palmitoyl carnitine and malate after addition of 2.5 mM of adenosine diphosphate to stimulate maximal state 3 mitochondrial respiration in the presence of fatty acid substrate. Oxygen consumption per tissue mass was significantly reduced in FPL individuals (A), and showed a strong positive correlation with muscle endurance as assessed by number of chest press repetitions (B), and a weak negative correlation with metabolic variables including insulin resistance measured by homeostatic model assessment (HOMA) (C), and fasting plasma triglycerides (D). Muscle lactate, pyruvate, and succinate levels were higher in FPL individuals (E). Each dot represents individual data points, with red dots representing FPL subjects and blue dots control subjects in B-D. Box plots (A,E) show the 25th, 50th, and 75th percentile value along with maximum and minimum values. N = 6 in each group, 2-tailed Student’s t test with unequal variance was used for comparison between the 2 groups, and Pearson correlation coefficient (r) was calculated to measure the strength of linear association between the variables.
Impaired mitochondrial function was also evident from muscle metabolome measurements on the muscle biopsy specimen. Muscle lactate, pyruvate, and succinate levels were nearly doubled in FPL individuals compared with controls (Fig. 3E and Table S3 (25)). No significant difference was seen in other metabolites measured including citrate, creatine, ATP, AMP, and amino acids. Further, long-chain acylcarnitine levels in the muscle, measured by LC-MS were significantly lower in FPL subjects, also reflecting the decreased fatty acid oxidation (Table S4 (25)).
A strong correlation was noted between the number of chest press repetitions and mitochondrial state 3 respiration (r = 0.76, P = .003), suggesting that earlier fatigue could be a result of impaired state 3 respiration which may lead to higher levels of lactate causing muscle fatigue (Fig. 3B). Mitochondrial state 3 respiration also showed a weak negative correlation with fasting serum triglycerides (r = –0.55, P = .06), postprandial lipemia (r = –0.59, P = .04), and insulin resistance (HOMA-IR, r = –0.56, P = .05).
Skeletal muscle gene expression profile provides insight into some of the characteristic metabolic and physical features of FPL
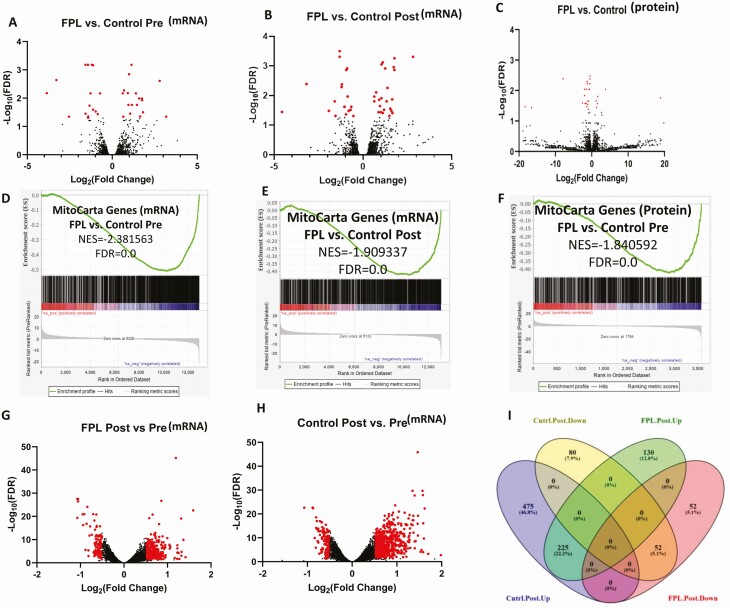
We performed skeletal muscle RNA sequencing to profile genome transcripts and to determine any difference in gene expression pattern between FPL individuals and controls both before and after the meal. There were few differences in the overall skeletal muscle transcriptome and proteome between FPL individuals and controls (Fig. 4A-4C) in the premeal baseline state. However, we noted a marked blunting in the post meal increase in overall gene expression in FPL individuals compared with controls (Fig. 4G-4I). Of the 1014 gene transcripts identified, there was an increase in 700 transcripts in the control group after the meal, while only 355 transcripts increased in FPL individuals. Similarly, 132 gene transcripts decreased in control participants while 52 transcripts decreased in FPL individuals after the meal. We also measured muscle proteome at the baseline state, which showed downregulation in FPL individuals, but did not measure it following the meal as the time frame is not sufficient for any quantitative changes in proteome. Of interest, mitochondrial gene and proteome expression were severely reduced in both the fasting and post meal state with a highly significant negative net enrichment score (Fig. 4D-F).
Figure 4.
Diminished expression of skeletal muscle and mitochondrial genes and proteins in individuals with familial partial lipodystrophy (FPL) compared with controls. N = 6 in each group. (A,B) Volcano plots of skeletal muscle gene expression as revealed by transcriptome analysis before (A) and after (B) ingestion of mixed meal in individuals with FPL and controls. Red dots represent genes whose expression is significantly different (FDR < 0.05 and absolute log2fold change ≥0.5) and black dots represent genes whose expression was not different. (C) Similar comparison of premeal skeletal muscle protein expression in FPL individuals compared with controls. The list of individual proteins differentially expressed is shown in Table S6 (25). (D-F) Downregulation of mitochondrial genes (obtained from MitoCarta, Version 2.0 data base) at both transcript and protein level in FPL individuals compared with controls. (G,H) Comparison of gene expression changes after meal ingestion between FPL individuals (G) and controls (H) showing a blunted increase in gene expression in individuals with FPL compared with controls. (E) Venn diagram showing the actual number of gene transcripts upregulated or downregulated after the meal in the 2 groups, revealing markedly higher number of upregulated gene transcripts following meal ingestion in the control group compared to FPL individuals.
We then rank-ordered gene transcript changes between FPL and control participants and performed Gene Set Enrichment Analysis (GSEA) to identify metabolic pathways upregulated or down regulated. There was significant down regulation of many metabolic pathways including branched chain amino acid degradation, citric acid cycle, mitochondrial respiration, electron transport chain, oxidative phosphorylation, and mitochondrial fatty acid beta oxidation (Table S5 (25)), but no pathways were significantly upregulated.
An intensity-based, label-free proteomics analysis also showed difference in protein abundance between individuals with FPL and controls (Table S6 (25)). The most downregulated protein in FPL individuals was succinate dehydrogenase assembly factor 2, which plays an essential role in the assembly of succinate dehydrogenase enzyme complex linked to both the citric acid cycle and electron transport chain. This further supports decreased mitochondrial function in FPL individuals indicated by mitochondrial functional studies and gene transcriptome analysis.
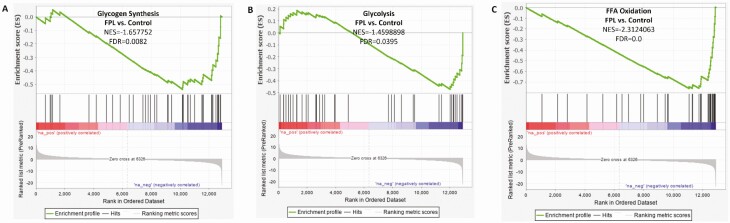
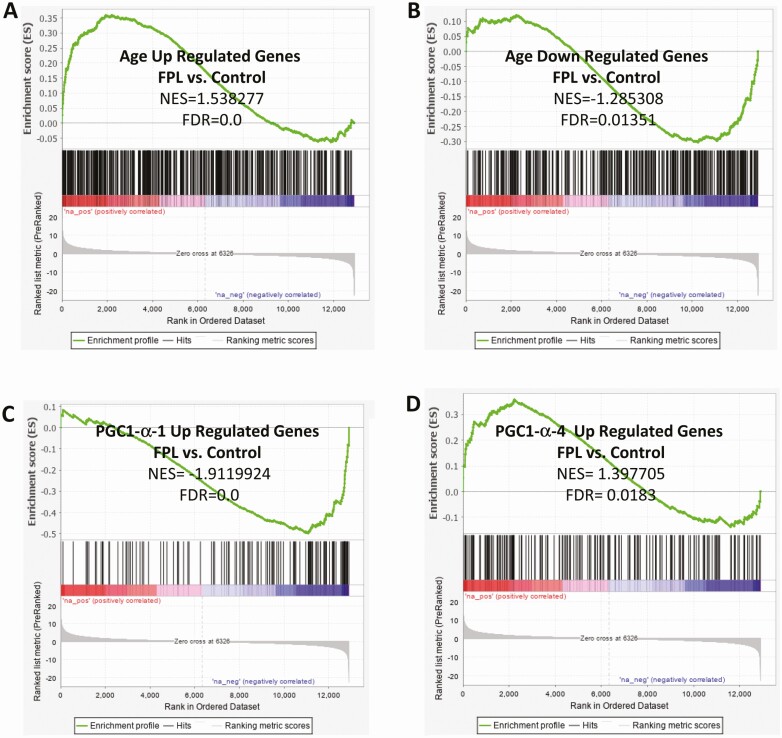
We also examined the gene expression profile in relation to specific metabolic pathways and morphological features which revealed many interesting patterns. The expression of genes regulating key metabolic pathways such as glycolysis, glycogen synthesis and free fatty acid oxidation were compared between FPL individuals and controls. Enrichment analysis showed marked reduction at both the transcript and protein level of key genes involved in these pathways (Fig. 5), and may account for the observed alterations in carbohydrate and fat metabolism. Since LMNA is known to be involved in the aging process, we studied the differential expression of genes which are upregulated or downregulated with normal aging. Gene expression pattern in FPL suggested accelerated aging with a significant positive enhancement of genes normally upregulated with aging, and negative enhancement of genes normally downregulated with aging (Fig. 6A and 6B). In order to gain further insight into the cause for muscular hypertrophy and metabolic dysfunction, we compared the expression of genes which are regulated by the different isoforms of the key transcriptional coactivator PGC-1α. It has been shown previously that PGC-1α1 regulates the expression of genes stimulating mitochondrial biogenesis and fatty acid oxidation, while the isoform PGC-1α4 is involved in regulation of muscle mass and strength (36). We found a significant negative enhancement of genes up-regulated by PGC-1α1 and positive enhancement of genes up-regulated by PGC-1α4 (Fig. 6C and 6D) which may account for the simultaneous decrease in skeletal muscle oxidative capacity despite muscle hypertrophy.
Figure 5.
Differential expression of skeletal muscle genes controlling key metabolic pathways in individuals with familial partial lipodystrophy (FPL) compared with matched controls. There was significant down regulation of genes associated with glycogen synthesis (A), glycolysis (B), and free fatty acid oxidation (C) in FPL individuals compared with controls. N = 6 in each group.
Figure 6.
Differential expression of skeletal muscle genes related to aging and regulation by key transcription factors. Compared with controls, gene expression pattern in individuals with Familial Partial Lipodystrophy (FPL) was reminiscent of aging with increased expression of genes that are normally up regulated with aging (A) and decreased expression of genes that are normally down regulated with aging (B). There was also decreased expression of genes up regulated by the key transcription factor PGC1α1 (C), while there was increased expression of genes up regulated by its isoform PGC1α4. N = 6 in each group.
Discussion
FPL participants in the current study showed characteristic physical features of FPL, Dunnigan variety including fat loss from the extremities and increased muscularity besides metabolic abnormalities related to insulin resistance. Despite the increased muscularity, their muscle protein synthesis was lower, but there was down regulation of protein catabolic pathways indicating decreased muscle protein degradation. They also did not display greater muscle strength proportional to increased muscularity, but had evidence of increased muscle fatigue based on chest press exercise. Increase in muscle lactate and other metabolites such as pyruvate and succinate, and decrease in levels of muscle long chain acyl-carnitines also support decline in mitochondrial function, which is consistent with a reduction in muscle maximal mitochondrial respiration in the presence of lipid substrates. Further, a detailed transcriptome and proteome analysis also revealed down regulation of multiple metabolic pathways including beta oxidation, citric acid cycle, and respiratory electron transport. Together, these observations point to distinct abnormalities in mitochondrial function in individuals with FPL in association with insulin resistance and early muscle fatigue. Reduction in muscle protein turnover, based on reduced synthesis rate and catabolic pathway, indicates a subnormal muscle protein quality control mechanism thus potentially contributing to muscle dysfunction (38, 39). Changes in skeletal muscle gene expression patterns were congruent with the observed alterations in muscle structure and metabolic functions.
While fat loss is the characteristic hallmark of lipodystrophy syndromes, increased muscularity is another well-recognized, but less well–studied feature of this condition. Some of the earliest descriptions of patients with Congenital Generalized Lipodystrophy alluded to the marked “lipodystrophic muscular hypertrophy” and noted that they showed “very good muscle power” (40). This was however not based on objective measure of muscle strength, and subsequent studies by Garg et al. (41) showed that quadriceps muscle strength in 3 female patients with CGL was not increased despite the increased muscularity consistent with the results of the current study in 6 participants. Further, they reported a preponderance of fast twitch (glycolytic) type 2 muscle fibers in comparison to type1 slow twitch (oxidative) fibers which also is consistent with lower oxidative phenotype in FPL in the current study. Our observation of increased muscularity is also consistent with previous reports in FPL, Dunnigan patients of prominent musculature (42), higher muscle mass and volume based on DEXA and magnetic resonance imaging (3), and increased muscle fiber diameter suggestive of muscle hypertrophy (4). The cause for the increased muscularity is however an unresolved issue. It has been speculated that muscle hypertrophy could result from spillover effects of hyperinsulinemia on IGF-1 receptors (3), although it is very rare to see this degree of muscularity in other conditions with hyperinsulinemia or IGF-1 excess. Myostatin, a member of the transforming growth factor-beta (TGF-β) which has a negative influence on skeletal muscle development, has also been implicated in FPL muscle hypertrophy (43, 44). While myostatin gene expression is itself not impaired in individuals with FPL, there was a decrease in nuclear translocation of a small molecule intracellular mediator of myostatin signaling which may account for the phenotypic similarity with myostatin deficiency (4). However unlike myostatin deficiency which is associated with increased strength (45) and insulin sensitivity, FPL patients are severely insulin resistant and do not display increased muscle strength. Further, when the myostatin gene is knocked off in lipodystrophic mice, there is improvement of insulin sensitivity (46), making it unlikely that myostatin is involved in the muscle hypertrophy of FPL. The current study advances the understanding of FPL by clearly showing that muscle hypertrophy is not due to increased protein synthesis but rather due to reduced protein catabolic pathway as suggested by transcriptome data. The results also demonstrate that decreased protein turnover (synthesis and degradation) may represent downregulation of muscle quality control system explaining decreased muscle functions despite increased mass. Higher lean mass is usually associated with greater insulin sensitivity (6, 7), which is not the case in FPL individuals (3) as we have confirmed, suggesting poor protein quality. Maintenance of healthy muscle proteome homeostasis requires removal of damaged proteins by degradation and their replacement by synthesis of new proteins (39). We found muscle deamidated to unmodified peptide ratio was higher in FPL suggesting the accumulation of damaged proteins that occurs when protein turnover is reduced. Reduced muscle protein synthesis concurrent to reduced protein catabolism, based on the gene transcriptome analysis, could potentially contribute to reduced muscle metabolic functions.
The current study also showed a clear increase in muscle fatigability involving the major muscle groups involved in chest press in FPL individuals similar to previous reports in patients with type 2 diabetes (47). Earlier onset of fatigue could result from impaired energy generation especially ATP and phosphocreatine. While we did not detect any difference in ATP, ADP, or phosphocreatine levels, the muscle samples were obtained in the resting state and not after exercise, and the measurement of these highly labile phosphometabolites ex vivo is not very precise. Earlier in vivo investigations using 31P MRS measurements have indeed shown a lower in vivo phosphocreatine recovery after exercise in patients with congenital generalized lipodystrophy indicative of defective mitochondrial oxidative phosphorylation (48), consistent with lower muscle mitochondrial maximal respiration we observed in FPL individuals in the current study. Moreover, we have also noted that even in the resting state, lactate levels were nearly doubled in skeletal muscles of FPL individuals, indicative of altered mitochondrial oxidative metabolism. Lactate serves as the link between glycolytic and oxidative processes (49), and its resting state accumulation in the absence of hypoxia is indicative of defective mitochondrial oxidative metabolism. Traditionally lactate accumulation has itself been recognized as an important factor in the genesis of muscle fatigue, but this view has been challenged by more recent studies (50, 51). It must be noted that these studies were in isolated muscle cells, and lactate accumulation may still play an important role in the in vivo genesis of fatigue (52, 53). We noted earlier onset of fatigue with the chest press maneuver but not during the leg press exercise. The reason for this discrepancy is not clear, but may be due to the involvement of larger muscle groups during the chest maneuver resulting in greater lactate generation. It is also likely that there is greater conditioning of lower extremity musculature compared to trunk musculature due to more frequent use.
Altered mitochondrial oxidative metabolism in the current study was associated with reduced insulin sensitivity and dyslipidemia. These findings are consistent with previous reports of an association between insulin resistance and altered mitochondrial function in other populations, including elderly people (15, 54), obese (55), and offspring of patients with type 2 diabetes (56, 57). Like these studies, our findings also suggest an association between altered mitochondrial function and insulin resistance in FPL patients without proving causation. It is also interesting to consider whether altered mitochondrial function contributes to other morphological changes in lipodystrophy. One of the genetic causes of congenital generalized lipodystrophy is homozygous mutations in the Seipin gene (58), and it has recently been shown that cells with Seipin mutation have impaired mitochondrial TCA cycle leading to defective calcium homeostasis and lipid storage (59). Similarly, mitochondrial dysfunction has also been shown in another mouse model of lipodystrophy due to deficiency of the metalloproteinase ZMPSTE24 involved in LMNA processing (60). Further, selective adipose tissue knockout of mitochondrial transcription factor A (TFAM) in mice has been shown to result in lipodystrophy and insulin resistance (61), suggesting that isolated mitochondrial dysfunction could lead to lipodystrophy syndrome. Overexpression of TFAM in skeletal muscle of mice has also been shown to enhance insulin sensitivity besides mitochondrial biogenesis and beta-oxidation (62). There is paucity of human studies linking lipodystrophy with altered mitochondrial function except in HIV-infected patients receiving antiretroviral medications known to cause mitochondrial toxicity. Nucleoside reverse transcriptase inhibitors, and to a lesser extent non-nucleoside reverse transcriptase inhibitors and protease inhibitors, have been shown to induce mitochondrial respiration defects and oxidative stress leading to fat loss (63), but in our study we did not find any evidence of oxidative stress in the study conditions. It is well known that mitochondria play an important role in adipocyte differentiation and growth (64), and it is possible that altered mitochondrial function may be one of the contributors to fat loss in lipodystrophy. Our current study was however restricted to mitochondrial function in skeletal muscle of adult individuals and does not provide definitive evidence of the role of mitochondrial dysfunction in fat loss occurring during childhood.
While we noticed a significant reduction in state 3 mitochondrial oxygen consumption expressed per tissue mass, this was less evident when oxygen flux data was expressed per tissue protein content. This discrepancy likely reflects the difference in mitochondrial protein content between FPL and controls, and has been noted in some previous studies as well (32), wherein mitochondrial oxygen consumption following high intensity interval aerobic training increased when expressed per tissue weight, but not when expressed per tissue protein content due to simultaneous increase in protein synthesis. Examining mitochondrial oxygen consumption relative to tissue weight may therefore be more instructive of qualitative changes in mitochondrial function, provided there are no technical limitations in sample handling and mitochondrial isolation. Ideally expressing mitochondrial oxygen consumption relative to mitochondrial number would be most accurate, but accurately determining this is very difficult. We did try to assess for differences in mitochondrial density between FPL and controls by examining electron microscopic sections (Figure S6 (25)) and did not notice any significant difference. Overall, these observations point to a qualitative decline in mitochondrial function in subjects with FPL.
In the current study, we demonstrated altered expression of multiple critical genes involved in regulation of metabolic pathways including glycogen synthesis, glycolysis and fatty acid oxidation, muscle structure and function as well as aging. These abnormalities could be potentially attributed to the mutant lamins. LMNA gene encodes intermediate filament proteins called A-type lamins which serve many critical functions including maintenance of cell and nuclear structural integrity, chromatin organization and regulation of gene expression (65). Mutant lamin isoforms in transgenic mice have been reported to affect the expression of key regulators of energy expenditure including PGC1α (66). LMNA is also known to play an important role in normal aging, and our finding of marked increase in the expression of genes normally upregulated during aging raises the possibility that this may play an important role in the phenotypic presentation of FPL. Aging is associated with decline in mitochondrial function, and it is possible that the observed mitochondrial dysfunction in FPL individuals is a consequence of accelerated aging process. Similarly decreased protein turnover can also reflect an accelerated aging process, but it is beyond the scope of the current study to derive definitive conclusions to this effect. Whether LMNA mutation in FPL, Dunnigan syndrome directly affects the expression of genes leading to the genesis of muscle hypertrophy, decreased endurance, and mitochondrial and metabolic dysfunction, or whether the observed changes are secondary to adipocyte loss remains to be established. Further studies which can identify the molecular link between LMNA mutation and the concerted gene expression changes we observed may offer therapeutic clues to the management of these patients. It would also be interesting to examine if the changes in mitochondrial function and gene expression patterns occur in children with LMNA mutation who have not yet experienced fat loss. If these alterations indeed predate fat loss and metabolic defects, it would suggest that LMNA mutation directly affects mitochondrial function which then leads to insulin resistance and other metabolic defects.
The current study has many strengths but also limitations. We studied individuals with partial lipodystrophy as they closely resemble the more common metabolic syndrome characterized by abdominal obesity, relative paucity of peripheral fat, and insulin resistance. To our best knowledge, no previous studies have directly measured skeletal muscle mitochondrial respiration and muscle protein synthesis in FPL individuals in conjunction with measurements of gene transcripts, proteome, and metabolites. Besides measuring muscle mitochondrial function, we also demonstrated similar directional changes in citric acid flux and mitochondrial respiration pathways based on gene transcriptome, proteome and metabolites supporting a notion that mitochondrial functional defect occurs in FPL individuals. A major limitation of our study is the small sample size given the rarity of genetic lipodystrophy syndromes, but significant differences in these relatively small groups is an additional strength. We enrolled a genetically homogenous group of individuals with lipodystrophy, selecting only those with well characterized genetic mutations. FPL, Dunnigan variety due to LMNA mutations has an estimated prevalence of 1: million, making it difficult to enroll large number of individuals, or ensure similarity in severity of metabolic abnormalities and their therapies across all study subjects. Three of the FPL subjects had diabetes and were on glucose- and lipid-lowering therapies, which can potentially affect some of the study variables. To adjust for this, we also compared the individuals who were not on any medications and did not have diabetes, and noted a similar trend and magnitude of difference between FPL individuals and their matched controls (Table S7 (25)). The effect of metformin on mitochondrial respiratory function is unclear (67), with both beneficial (68) and detrimental influences (69) being reported. Similarly the effect of statins on mitochondrial function has been extensively studied with some evidence linking mitochondrial dysfunction with statin-associated muscle disease (70). When we excluded study subjects with diabetes who were on metformin and statin, the difference in state 3 respiration between FPL subjects and matched controls was 44% and 48% in the pre- and postmeal state. The corresponding differences in the whole group were 40% and 38%, respectively which suggests that the observed reduction in mitochondrial function in FPL subjects was not related to their concomitant drug use or diabetes. Another limitation is that we did not measure muscle or plasma lactate levels immediately after exercise, and were unable to quantitate peptide fractions representing in vivo degraded proteins representing protein degradation in the muscle biopsy sample. However multiple lines of evidence including decreased fractional synthetic rate of mixed muscle protein, congruent transcriptome and proteomic data, and increased concentration of modified (deamidated) peptides, all suggest decreased muscle protein turnover, which nonetheless needs to be confirmed by direct measurements of label decay in the muscle proteome in future studies. Further, in order to reduce the complexity, we did exclude the bands (gel electrophoresis) representing the high abundant proteins in mass spectrometry so that the proteomic measurements largely represent the metabolically important proteins rather than structural proteins. Therefore, we did not quantify the many structural proteins contributing to muscle mass.
In summary, we investigated individuals with genetically proven FPL, Dunnigan variety who demonstrated the typical physical and metabolic features of this disorder. Compared with matched controls, they had relatively higher lean mass, which can be attributed to decreased muscle protein catabolism leading to accumulation of muscle proteins. Reduced protein turnover can potentially cause poor muscle quality thus explaining lack of increase in muscle strength proportional to the increased mass, and early muscle fatigue. Easy fatigability is likely related to impaired mitochondrial function based on muscle mitochondrial functional studies and supported by results from RNA sequence and proteome analysis. While our study is limited by a relatively small sample size, the findings support causative interactions between altered mitochondrial function, insulin resistance, and reduced muscle protein turnover on FPL phenotype. The expression of this phenotype may also be influenced by marked changes in gene expression patterns, including accelerated aging due to LMNA mutation.
Acknowledgments
The study was supported by the Mayo Clinic Emslander Career Development Award for VS, and by the Mayo Clinic CTSA through grant number UL1 TR002377 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). We are thankful to Patrick Vanderboom for assistance with mass spectroscopy studies.
Financial Support: The study was supported by the Mayo Clinic Emslander Career Development Award for V.S., and by the Mayo Clinic CTSA through grant number UL1 TR002377 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH).
Glossary
Abbreviations
- 1RM
1 repetition maximum
- BMI
body mass index
- DEXA
dual-energy X-ray absorptiometry
- FPL
familial partial lipodystrophy
- FSR
fractional synthetic rate
- HOMA-IR
Homeostatic Model Assessment of Insulin Resistance
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- MRS
magnetic resonance spectroscopy
- NMR
nuclear magnetic resonance spectroscopy
Additional Information
Disclosures: No conflict of interests for any of the authors
Data Availability
Supplementary data referenced in the manuscript is available at: https://figshare.com/s/4ed149e4e502aa444fe1
References
- 1. Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350(12):1220-1234. [DOI] [PubMed] [Google Scholar]
- 2. Brown RJ, Araujo-Vilar D, Cheung PT, et al. The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J Clin Endocrinol Metab. 2016;101(12):4500-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ji H, Weatherall P, Adams-Huet B, Garg A. Increased skeletal muscle volume in women with familial partial lipodystrophy, Dunnigan variety. J Clin Endocrinol Metab. 2013;98(8): E1410-E1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spuler S, Kalbhenn T, Zabojszcza J, et al. Muscle and nerve pathology in Dunnigan familial partial lipodystrophy. Neurology. 2007;68(9):677-683. [DOI] [PubMed] [Google Scholar]
- 5. Hegele RA. Premature atherosclerosis associated with monogenic insulin resistance. Circulation. 2001;103(18):2225-2229. [DOI] [PubMed] [Google Scholar]
- 6. Atlantis E, Martin SA, Haren MT, Taylor AW, Wittert GA; Members of the Florey Adelaide Male Ageing Study. Inverse associations between muscle mass, strength, and the metabolic syndrome. Metabolism. 2009;58(7):1013-1022. [DOI] [PubMed] [Google Scholar]
- 7. Srikanthan P, Karlamangla AS. Relative muscle mass is inversely associated with insulin resistance and prediabetes. Findings from the third National Health and Nutrition Examination Survey. J Clin Endocrinol Metab. 2011;96(9):2898-2903. [DOI] [PubMed] [Google Scholar]
- 8. Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31(3):364-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stump CS, Henriksen EJ, Wei Y, Sowers JR. The metabolic syndrome: role of skeletal muscle metabolism. Ann Med. 2006;38(6):389-402. [DOI] [PubMed] [Google Scholar]
- 10. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51(10):2944-2950. [DOI] [PubMed] [Google Scholar]
- 11. Ruegsegger GN, Creo AL, Cortes TM, Dasari S, Nair KS. Altered mitochondrial function in insulin-deficient and insulin-resistant states. J Clin Invest. 2018;128(9):3671-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petersen MC, Shulman GI. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98(4):2133-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson EJ, Lustig ME, Boyle KE, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009;119(3):573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Konopka AR, Asante A, Lanza IR, et al. Defects in mitochondrial efficiency and H2O2 emissions in obese women are restored to a lean phenotype with aerobic exercise training. Diabetes. 2015;64(6):2104-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen KF, Befroy D, Dufour S, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300(5622):1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vondra K, Rath R, Bass A, Slabochová Z, Teisinger J, Vitek V. Enzyme activities in quadriceps femoris muscle of obese diabetic male patients. Diabetologia. 1977;13(5):527-529. [DOI] [PubMed] [Google Scholar]
- 17. Bajpeyi S, Pasarica M, Moro C, et al. Skeletal muscle mitochondrial capacity and insulin resistance in type 2 diabetes. J Clin Endocrinol Metab. 2011;96(4):1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feeney ER, Mallon PW. Impact of mitochondrial toxicity of HIV-1 antiretroviral drugs on lipodystrophy and metabolic dysregulation. Curr Pharm Des. 2010;16(30):3339-3351. [DOI] [PubMed] [Google Scholar]
- 19. Pérez-Matute P, Pérez-Martínez L, Blanco JR, Oteo JA. Role of mitochondria in HIV infection and associated metabolic disorders: focus on nonalcoholic fatty liver disease and lipodystrophy syndrome. Oxid Med Cell Longev. 2013;2013:493413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simha V, Garg A. Lipodystrophy: lessons in lipid and energy metabolism. Curr Opin Lipidol. 2006;17(2):162-169. [DOI] [PubMed] [Google Scholar]
- 21. Gavrilova O, Marcus-Samuels B, Graham D, et al. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest. 2000;105(3):271-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401(6748):73-76. [DOI] [PubMed] [Google Scholar]
- 23. Oral EA, Simha V, Ruiz E, et al. Leptin-replacement therapy for lipodystrophy. N Engl J Med. 2002;346(8):570-578. [DOI] [PubMed] [Google Scholar]
- 24. Haque WA, Shimomura I, Matsuzawa Y, Garg A. Serum adiponectin and leptin levels in patients with lipodystrophies. J Clin Endocrinol Metab. 2002;87(5):2395. [DOI] [PubMed] [Google Scholar]
- 25. Simha V, Lanza IR, Dasari S, et al. Data from: Impaired Muscle Mitochondrial Function in Familial Partial Lipodystrophy. Figshare. Deposited August 25, 2021. https://figshare.com/s/4ed149e4e502aa444fe1. [DOI] [PMC free article] [PubMed]
- 26. Soop M, Nehra V, Henderson GC, Boirie Y, Ford GC, Nair KS. Coingestion of whey protein and casein in a mixed meal: demonstration of a more sustained anabolic effect of casein. Am J Physiol Endocrinol Metab. 2012;303(1):E152-E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boirie Y, Dangin M, Gachon P, Vasson MP, Maubois JL, Beaufrère B. Slow and fast dietary proteins differently modulate postprandial protein accretion. Proc Natl Acad Sci U S A. 1997;94(26):14930-14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zabielski P, Ford GC, Persson XM, Jaleel A, Dewey JD, Nair KS. Comparison of different mass spectrometry techniques in the measurement of L-[ring-(13)C6]phenylalanine incorporation into mixed muscle proteins. J Mass Spectrom. 2013;48(2):269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Breda E, Cavaghan MK, Toffolo G, Polonsky KS, Cobelli C. Oral glucose tolerance test minimal model indexes of beta-cell function and insulin sensitivity. Diabetes. 2001;50(1):150-158. [DOI] [PubMed] [Google Scholar]
- 30. Balagopal P, Schimke JC, Ades P, Adey D, Nair KS. Age effect on transcript levels and synthesis rate of muscle MHC and response to resistance exercise. Am J Physiol Endocrinol Metab. 2001;280(2):E203-E208. [DOI] [PubMed] [Google Scholar]
- 31. Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lanza IR, Blachnio-Zabielska A, Johnson ML, et al. Influence of fish oil on skeletal muscle mitochondrial energetics and lipid metabolites during high-fat diet. Am J Physiol Endocrinol Metab. 2013;304(12):E1391-E1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson MM, Dasari S, Konopka AR, et al. Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab. 2017;25(3):581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanza IR, Zabielski P, Klaus KA, et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012;16(6):777-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su J, Ekman C, Oskolkov N, et al. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle. 2015;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruas JL, White JP, Rao RR, et al. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151(6):1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wright HT. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26(1):1-52. [DOI] [PubMed] [Google Scholar]
- 38. Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21(12):1406-1415. [DOI] [PubMed] [Google Scholar]
- 39. James HA, O’Neill BT, Nair KS. Insulin Regulation of Proteostasis and Clinical Implications. Cell Metab. 2017;26(2):310-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. SENIOR B. Lipodystrophic muscular hypertrophy. Arch Dis Child. 1961;36( 188):426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garg A, Stray-Gundersen J, Parsons D, Bertocci LA. Skeletal muscle morphology and exercise response in congenital generalized lipodystrophy. Diabetes Care. 2000;23(10):1545-1550. [DOI] [PubMed] [Google Scholar]
- 42. Wildermuth S, Spranger S, Spranger M, Raue F, Meinck HM. Köbberling-Dunnigan syndrome: a rare cause of generalized muscular hypertrophy. Muscle Nerve. 1996;19(7):843-847. [DOI] [PubMed] [Google Scholar]
- 43. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83-90. [DOI] [PubMed] [Google Scholar]
- 44. McPherron AC, Guo T, Bond ND, Gavrilova O. Increasing muscle mass to improve metabolism. Adipocyte. 2013;2(2):92-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schuelke M, Wagner KR, Stolz LE, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350(26):2682-2688. [DOI] [PubMed] [Google Scholar]
- 46. Guo T, Bond ND, Jou W, Gavrilova O, Portas J, McPherron AC. Myostatin inhibition prevents diabetes and hyperphagia in a mouse model of lipodystrophy. Diabetes. 2012;61(10):2414-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Halvatsiotis P, Short KR, Bigelow M, Nair KS. Synthesis rate of muscle proteins, muscle functions, and amino acid kinetics in type 2 diabetes. Diabetes. 2002;51(8):2395-2404. [DOI] [PubMed] [Google Scholar]
- 48. Sleigh A, Stears A, Thackray K, et al. Mitochondrial oxidative phosphorylation is impaired in patients with congenital lipodystrophy. J Clin Endocrinol Metab. 2012;97(3): E438-E442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brooks GA. Lactate: link between glycolytic and oxidative metabolism. Sports Med. 2007;37(4-5):341-343. [DOI] [PubMed] [Google Scholar]
- 50. Nielsen OB, de Paoli F, Overgaard K. Protective effects of lactic acid on force production in rat skeletal muscle. J Physiol. 2001;536(Pt 1):161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brooks GA. Lactate doesn’t necessarily cause fatigue: why are we surprised? J Physiol. 2001;536(Pt 1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tesch P, Sjödin B, Thorstensson A, Karlsson J. Muscle fatigue and its relation to lactate accumulation and LDH activity in man. Acta Physiol Scand. 1978;103(4):413-420. [DOI] [PubMed] [Google Scholar]
- 53. Ishii H, Nishida Y. Effect of lactate accumulation during exercise-induced muscle fatigue on the sensorimotor cortex. J Phys Ther Sci. 2013;25(12):1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rooyackers OE, Adey DB, Ades PA, Nair KS. Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc Natl Acad Sci U S A. 1996;93(26):15364-15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kelley DE, Goodpaster BH. Skeletal muscle triglyceride. An aspect of regional adiposity and insulin resistance. Diabetes Care. 2001;24(5):933-941. [DOI] [PubMed] [Google Scholar]
- 56. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350(7):664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Short KR, Nair KS, Stump CS. Impaired mitochondrial activity and insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350(23):2419-21; author reply 2419. [DOI] [PubMed] [Google Scholar]
- 58. Magré J, Delépine M, Khallouf E, et al. ; BSCL Working Group. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet. 2001;28(4):365-370. [DOI] [PubMed] [Google Scholar]
- 59. Ding L, Yang X, Tian H, et al. Seipin regulates lipid homeostasis by ensuring calcium-dependent mitochondrial metabolism. EMBO J. 2018;37(17):e97572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peinado JR, Quirós PM, Pulido MR, et al. Proteomic profiling of adipose tissue from Zmpste24-/- mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing. Mol Cell Proteomics. 2011;10(11):M111.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vernochet C, Damilano F, Mourier A, et al. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014;28(10):4408-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koh JH, Johnson ML, Dasari S, et al. TFAM enhances fat oxidation and attenuates high-fat diet-induced insulin resistance in skeletal muscle. Diabetes. 2019;68(8):1552-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caron-Debarle M, Boccara F, Lagathu C, et al. Adipose tissue as a target of HIV-1 antiretroviral drugs. Potential consequences on metabolic regulations. Curr Pharm Des. 2010;16(30):3352-3360. [DOI] [PubMed] [Google Scholar]
- 64. De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175(3):927-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131-164. [DOI] [PubMed] [Google Scholar]
- 66. Lopez-Mejia IC, de Toledo M, Chavey C, et al. Antagonistic functions of LMNA isoforms in energy expenditure and lifespan. EMBO Rep. 2014;15(5):529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vial G, Detaille D, Guigas B. Role of Mitochondria in the Mechanism(s) of Action of Metformin. Front Endocrinol (Lausanne). 2019;10:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Y, An H, Liu T, et al. Metformin improves mitochondrial respiratory activity through activation of AMPK. Cell Rep. 2019;29(6):1511-1523.e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Konopka AR, Laurin JL, Schoenberg HM, et al. Metformin inhibits mitochondrial adaptations to aerobic exercise training in older adults. Aging Cell. 2019;18(1):e12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramachandran R, Wierzbicki AS. Statins, muscle disease and mitochondria. J Clin Med. 2017;6(8):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary data referenced in the manuscript is available at: https://figshare.com/s/4ed149e4e502aa444fe1