ABSTRACT
To enhance the utility of the genetically diverse panel of Acinetobacter baumannii isolates reported recently by Galac and coworkers (M. R. Galac, E. Snesrud, F. Lebreton, J. Stam, et al., Antimicrob Agents Chemother 64:e00840-20, 2020, https://doi.org/10.1128/AAC.00840-20) and to identify the novel KL and OCL, all of the gene clusters that direct the biosynthesis of capsular polysaccharide and of the outer core of lipooligosaccharide, respectively, were reexamined. The nine KL and one OCL previously recorded as novel were identified, and nine further novel KL and two OCL were found.
KEYWORDS: Acinetobacter baumannii, K locus, OC locus, diverse panel
INTRODUCTION
Acinetobacter baumannii is a leading cause of infections that are difficult to treat because of the high proportion of isolates that exhibit resistance to all or most of the therapeutically effective antibiotics. Consequently, a variety of potential new treatments are being explored. A recent study (1) compiled a genetically diverse panel of 100 non-redundant, sequenced A. baumannii clinical isolates that can be used to test and develop potential new therapies. The strain collection has been made publicly available and antibiotic resistance profiles, resistance gene families, and sequence types were recorded. As both vaccination and phage therapy approaches are affected by the specific composition and topology of surface polysaccharides (2, 3), information about the capsular polysaccharide (CPS or capsule) and the outer core (OC) of the lipooligosaccharide, the two surface structures known to exhibit considerable variation between isolates (4–7), is also important. Hence, the specific gene clusters responsible for synthesis of the variable portions of these carbohydrate structures, the K locus (KL) and the OC locus (OCL), found in each isolate were also identified using the recently developed Kaptive tool with A. baumannii reference databases (7) and recorded. Genome sequence data were also made available for all isolates in the panel (BioProject PRJNA545079).
KL are assigned a new number when any difference in the gene content between fkpA and lldP is detected (5). The most recent published estimate of the diversity at the K locus reported 128 distinct KL (7), although this number continues to rise. However, currently only 92 reference KL sequences are available in the Kaptive database for A. baumannii. Consequently, a small proportion of KL and OCL in the diverse panel collection were recorded as novel (Fig. 3 in (1)). However, inaccurate assignments can arise in cases where a sequence differs by only a short segment from an available reference sequence and the closest available match is used.
Here, we have re-examined the KL and OCL in the 100 members of the diverse panel and manually re-examined the relevant DNA sequence in all cases where the Kaptive output was not classified as “perfect.” The novel types were also compared to a larger in-house collection of sequences for K and OC loci.
In the diverse panel, 9 isolates were recorded as carrying a novel KL type (1). Only one of these was found among the KL in the larger in-house database of 128 KL types, and 8 were confirmed as novel. Numbers were assigned to all novel KLs as in Table 1. The sequences of the novel KL were annotated according to the standard nomenclature scheme (5, 7), and GenBank accession numbers for the annotated sequences are listed in Table 1. The organization and gene content of these KL is shown in Fig. S1.
TABLE 1.
Isolates with reassigned KL and OCL types
| Isolate name | Assembly accession no. | Previously assigneda | Reassignedb | GenBank accession numberc |
|---|---|---|---|---|
| K locus type | ||||
| MRSN 351524 | GCA_006492685.1 | novel | KL69 | OK052595.1 |
| MRSN 7153 | GCA_006492295.1 | novel | KL151 | OK052589.1 |
| MRSN 15574 | GCA_006494565.1 | novel | KL152 | OK052588.1 |
| MRSN 4943 | GCA_006492305.1 | novel | KL153 | OK052587.1 |
| MRSN 10372 | GCA_006493855.1 | novel | KL154 | OK052586.1 |
| MRSN 21660 | GCA_006494015.1 | novel | KL155 | OK052585.1 |
| MRSN 11816 | GCA_006494285.1 | novel | KL156 | OK052584.1 |
| MRSN 15093 | GCA_006494575.1 | novel | KL157 | OK052583.1 |
| MRSN 32076 | GCA_006493005.1 | novel | KL158 | OK052582.1 |
| MRSN 15075 | GCA_006494055.1 | KL46 | KL121 | OK052594.1 |
| MRSN 32842 | GCA_006492855.1 | KL8 | KL136 | OK052593.1 |
| MRSN 14193 | GCA_006494215.1 | KL19 | KL146 | OK052592.1 |
| MRSN 489678 | GCA_006492055.1 | KL15 | KL147 | OK052591.1 |
| MRSN 351162 | GCA_006492905.1 | KL50 | KL150 | OK052590.1 |
| MRSN 14237 | GCA_006494225.1 | KL32 | KL200-v*d | OK052581.1 |
| MRSN 31196 | GCA_006491855.1 | KL1 | KL1-v* + GI-1 | |
| OC locus type | ||||
| MRSN 423159 | GCA_006492625.1 | novel | OCL3-v* | |
| MRSN 7113 | GCA_006492505.1 | OCL5 | OCL13 | OK052579.1 |
| MRSN 14237 | GCA_006494225.1 | OCL7 | OCL14 | OK052580.1 |
| MRSN 32304 | GCA_006492945.1 | OCL7 | OCL14 |
Previously assigned in Galac et al. (1).
-v* indicates that locus is found across two or more contigs, suggesting a variant type.
Fully annotated sequence of novel KL or OCL gene cluster.
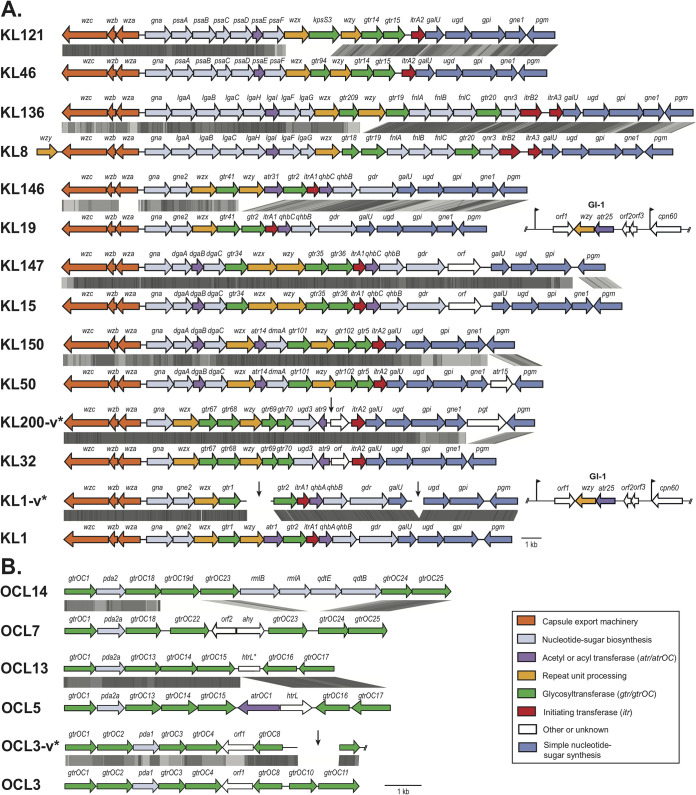
KL200 was found in two contigs that could be directly abutted (see Fig. 1A for position of contig break). A complete sequence with the joined contigs was annotated and submitted to GenBank under accession number OK052581.1.
We found that 75 of the 91 reported loci had been correctly identified by Kaptive and a further 10 were potentially correctly identified but interrupted or potentially interrupted by an insertion sequence as the locus was found in two or more contigs (see Table S1). For three of four KL19 and the two KL24 assignments, which rely on a genomic island (GI) to supply the K unit polymerase Wzy (8, 9), the presence of the appropriate GI (GI-1 for KL19, GenBank accession number KU165787.1; and GI-2 for KL24, GenBank accession number KX756650.1) in the genome was confirmed. The gene cluster in the fourth KL19 assignment was closely related to KL19 but includes a wzy gene (Fig. 1A), and the GI was not present in that isolate. In addition, for one isolate assigned as KL1, a segment normally present in KL1 that includes the wzy and atr1 gene was not found (Fig. 1A) but GI-1 was present in the genome of this isolate.
FIG 1.
Organization of reassigned KL and OCL. (A) The KL gene clusters. (B) The OCL gene clusters. Each reassigned KL or OCL type is compared with the KL or OCL type it was originally assigned to. Figure is drawn to scale with scale bar for KL shown below part A and scale bar for OCL shown below part B. Color scheme (legend bottom right) indicates the functional categories of the gene products. -v* indicates loci that were found across more than one contig, with downward facing black arrows showing the positions of the breaks. Gray shading indicates the level of protein sequence identity 80 to 100%) generated by tblastx comparisons with Easyfig (10).
Six further isolates included KL that were closely related to the assigned KL, but closer matches for one was found in our in-house database and five were novel (Table 1). In all six cases, the new KL differed from the assigned KL only in a short region. A comparison of the gene content and organization of the KL originally assigned and the new KL for each of these pairs of KL is shown in Fig. 1A. KL200 was found in two contigs and may be interrupted by an IS. Two of these KL pairs differ only in the presence/absence of genes that are not known to directly affect the capsule structure (shown in white in Fig. 1), and these KL are predicted to produce the same capsule structure. Two of the remaining include different wzy genes, and the linkage between the oligomeric units that is formed by the encoded Wzy polymerases is likely to differ, changing the topology of the CPS. These differences are likely to be important as a recent study showed that most phage depolymerases examined target the Wzy linkage (2).
Diversity among the OC loci is more limited than for the K loci, with only 16 types identified in the species to date (6, 7), 12 of which are in the current Kaptive OCL reference database (7). One OCL found in isolates in the diverse panel was recorded as novel (1), and our re-analysis found that it was a variant of OCL3 that lacked a 1.2 kb internal segment that includes two glycosyltransferase genes (Table 1, Fig. 1A). Here, we found that the OCL in 89 of the remaining 99 isolates were correctly assigned by Kaptive, and a further 7 were potentially correctly identified but found in two or more contigs, suggesting interruption by an insertion sequence (see Table S1). The remining three OCL cases, one recorded as OCL5 and two assigned as OCL7, were re-assigned to OCL13 or OCL14, respectively (Table 1), which could not have been detected with the current Kaptive database. Fully annotated OCL13 and OCL14 sequences have been deposited in GenBank and their accession numbers are listed in Table 1, and alignments of the two pairs are shown in Fig. 1B. OCL5, OCL7, OCL13, and OCL14 are all members of the OCL B group (6).
The re-analysis of the collection established by Galac et al. (1) has confirmed the utility of the Kaptive tool with current KL and OCL A. baumannii reference databases and highlighted some difficulties encountered when the KL or OCL gene cluster is interrupted by an IS or present in more than one contig. The current diverse panel does not include representatives of all KL types, with only 57 KL types (Fig. S1) and 12 OCL types (Fig. S2) represented. However, most of the KL and OCL found in the globally distributed GC1 and GC2 clones (7) that include most of the multiply, extensively, and pan resistant isolates recovered in hospitals were represented. Nonetheless, compilation of a panel that includes greater KL diversity may be warranted. An update of the Kaptive database that includes these newly identified KL and OCL as well as all KL and OCL found in all genomes of A. baumannii isolates that are publicly available is pending.
Data availability.
Fully annotated sequences of novel KL and OCL were deposited into NCBI GenBank under accession numbers listed in Table 1. Genome sequence data for the diverse panel published by Galac et al. (1) are available in NCBI in BioProject PRJNA545079, and individual accession numbers are listed in Table S1.
ACKNOWLEDGMENTS
We thank Patrick Mc Gann for permission to annotate novel KL and OCL for submission to NCBI GenBank. This work was supported by an Australian Research Council (ARC) DECRA Fellowship DE180101563 to J.J.K., and a National Health and Medical Research Council (NHMRC) Investigator grant GNT1194178 to R.M.H.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Galac MR, Snesrud E, Lebreton F, Stam J, Julius M, Ong AC, Maybank R, Jones AR, Kwak YI, Hinkle K, Waterman PE, Lesho EP, Bennett JW, Mc Gann P. 2020. A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob Agents Chemother 64:e00840-20. 10.1128/AAC.00840-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popova AV, Shneider MM, Arbatsky NP, Kasimova AA, Senchenkova SN, Shashkov AS, Dmitrenok AS, Chizhov AO, Mikhailova YV, Shagin DA, Sokolova OS, Timoshina OY, Kozlov RS, Miroshinikov KA, Knirel YA. 2021. Specific interaction of novel Friunavirus phages encoding tailspike depolymerases with corresponding Acinetobacter baumannii capsular types. J Virol 95:e01714-20. 10.1128/JVI.01714-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang F, Lou T, Kuo S, Wu W, Chern J, Lee Y, Chen S, Zou W, Lin N, Wu S. 2017. A medically relevant capsular polysaccharide in Acinetobacter baumannii is a potential vaccine candidate. Vaccine 35:1440–1447. 10.1016/j.vaccine.2017.01.060. [DOI] [PubMed] [Google Scholar]
- 4.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. 2013. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8:e70329. 10.1371/journal.pone.0070329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. 10.1371/journal.pone.0062160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenyon JJ, Nigro S, Hall RM. 2014. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS One 9:e107833. 10.1371/journal.pone.0107833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. 2020. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb Genom 6:e000339. 10.1099/mgen.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenyon JJ, Shneider MM, Senchenkova SN, Shashkov AS, Siniagina M, Malanin S, Popova AV, Miroshnikov KA, Hall RM, Knirel YA. 2016. K19 capsular polysaccharide of Acinetobacter baumannii is produced via a Wzy polymerase encoded in a small genomic island rather than the KL19 capsule gene cluster. Microbiology (Reading) 162:1479–1489. 10.1099/mic.0.000313. [DOI] [PubMed] [Google Scholar]
- 9.Kenyon JJ, Kasimova AA, Shneider MM, Shashkov AS, Arbatsky NP, Popova AV, Miroshnikov KA, Hall RM, Knirel YA. 2017. The KL24 gene cluster and a genomic island encoding a Wzy polymerase contribute genes needed for synthesis of the K24 capsular polysaccharide by the multiply antibiotic resistant Acinetobacter baumannii isolate RCH51. Microbiology (Reading) 163:355–363. 10.1099/mic.0.000430. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table and figures. Download AAC.01807-21-s0001.pdf, PDF file, 4.3 MB (4.3MB, pdf)
Data Availability Statement
Fully annotated sequences of novel KL and OCL were deposited into NCBI GenBank under accession numbers listed in Table 1. Genome sequence data for the diverse panel published by Galac et al. (1) are available in NCBI in BioProject PRJNA545079, and individual accession numbers are listed in Table S1.