Abstract
Aim
Antisense oligonucleotide (ASO) has the potential to induce off-target effects by inadvertent binding of ASOs to unintended RNAs that have a sequence similar to the target RNA. In the present study, we focused on the association between oligonucleotide length and off-target effects. Oligonucleotide extension is assumed to have bilateral effects on hybridization-dependent changes in gene expression, i.e., one is the decrease of off-target effects based on the reduced number of off-target candidate genes with perfect matches, and the other is the increase of off-target effects based on the increased binding affinity between the ASO and the complementary RNAs that leads to better tolerability for mismatches.
Methods
To determine the effects of oligonucleotide extension of gapmer ASOs on off-target effects, an extensive microarray analysis was performed using human cells treated with a 14-mer gapmer ASO and the extended 18-mer derivatives with the same core 14-mer region.
Results and Discussion
Our data indicated that change in gene expression in the cells treated with 18-mer ASOs was significantly smaller than those with a 14-mer ASO, showing the decrease of off-target effects by oligonucleotide extension.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40291-021-00573-z.
Key Points
| Our extensive microarray analysis indicated that change in gene expression in the cells treated with 18-mer antisense oligonucleotides was significantly smaller than those with a 14-mer antisense oligonucleotide. |
| Our data showed that the extension of antisense oligonucleotides reduced off-target effects in the light of the reduced number of off-target genes and the reduced extent of gene down-regulation. |
| Mismatches, insertions, and deletions between the antisense oligonucleotide and the complementary RNA sequences are better tolerated when longer antisense oligonucleotides cleave complementary RNAs. |
Introduction
Antisense oligonucleotide (ASO) therapeutics have been developed extensively for the treatment of a wide range of diseases, including previously intractable human disorders. ASO therapeutics are single-stranded oligonucleotides that bind to RNA through sequence-specific Watson-Crick base pairings. Most ASO therapeutics that are currently in clinical development are gapmer ASOs. Gapmer ASOs contain sugar-modified nucleotides on both ends and DNA in the central gap region to form the DNA/RNA heteroduplex that is recognized by RNase H [1, 2]. They elicit RNase H-mediated cleavage of the target RNA that leads to subsequent degradation of the cleaved RNAs via nucleases in cells. So far, three gapmer ASOs, mipomersen (Kynamro®), inotersen (Tegsedi®), and volanesorsen (Waylivra®), have been approved for clinical use [3–5]. All of these gapmer ASOs are 20-mer in length, with 5-mer sugar-modified nucleotides, 2′-O-methoxyethyl (2′-MOE) on both ends, and 10-mer DNA in the central gap region [6–8]. Development of sugar-modified nucleotides with high binding affinity to complementary RNA, such as 2′,4′-BNA/LNA and cEt nucleic acids, enables ASOs to be shorter than ever before. Indeed, sugar-modified ASOs currently in clinical development are generally equal to or less than 20-mer in length [9–13].
As with all drugs, ASOs carry the risk of causing unintended toxicity. The mechanisms of ASO-mediated toxicity can be conceptually divided into hybridization-dependent effects and hybridization-independent effects. The hybridization-dependent effects, also called off-target effects, are potentially caused by inadvertent binding of ASOs to unintended RNAs that have a sequence similar to the target RNA. It has been shown that the hepatotoxicity induced by gapmer ASOs is at least partly mediated via the RNase H-dependent reduction of unintended off-target RNAs in mice [14, 15]. Toxicity induced by off-target effects could not be evaluated by conventional animal studies theoretically because of differences in the genome sequence between humans and other animals. Consequently, to predict toxicity induced by off-target effects, assessment of off-target effects with in silico analysis using a human RNA database and in vitro expression analysis using human cells has been proposed [16–18]. The toxicity via hybridization-independent effects is likely to be due to interactions between the ASOs and cellular proteins, which are unrelated to Watson and Crick base-pairing to RNAs. Consistent with this assumption, the toxicity of ASOs is known to be correlated with the binding of ASOs to cellular proteins, and it is assumed that it can be evaluated by animal experiments [19].
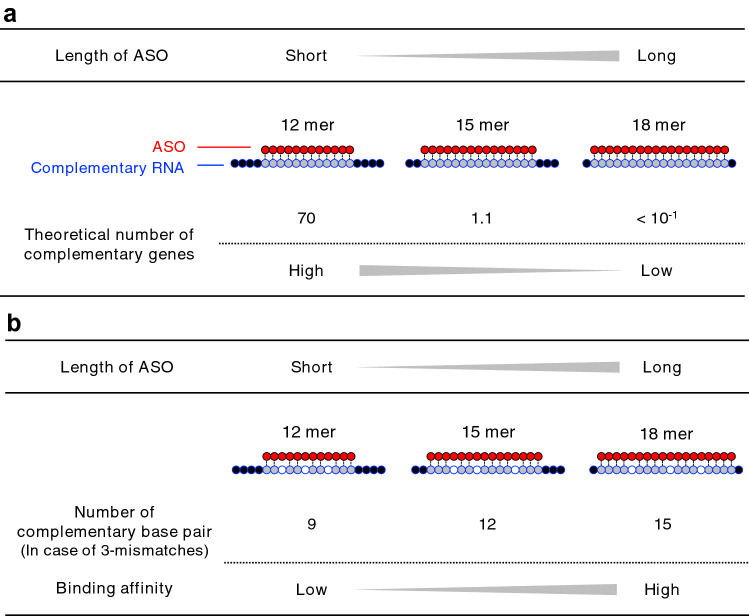
In the present study, we focused on the effects of oligonucleotide extension of gapmer ASOs on off-target effects using human cells with reduction of hybridization-dependent toxicity in mind. The theoretical number of complementary RNA regions, or off-target candidate genes, of the ASOs with perfect matches decreases as the ASOs become longer (Fig. 1a). On the other hand, the binding affinity between the ASO and the complementary RNAs increases as the ASOs become longer (Fig. 1b), and therefore, mismatches between the ASO and the complementary RNA sequences are likely to be better tolerated when longer gapmer ASOs cleave complementary RNAs. This means that, as a result of better tolerability, it is possible that the off-target effects increase as the ASOs become longer. In light of the above considerations, oligonucleotide extension is assumed to have bilateral effects on hybridization-dependent changes in gene expression, i.e., one is the decrease of off-target effects based on the reduced number of off-target candidate genes with perfect matches, and the other is the increase of off-target effects based on the increased binding affinity between the ASO and the complementary RNAs that leads to better tolerability for mismatches. The binding affinity can also be regulated by the introduction of sugar-modified nucleotides into the ASO, and therefore, the number of sugar-modified nucleotides in the ASO is an important factor affecting off-target effects [20–22].
Fig. 1.
The relationship between the length of antisense oligonucleotides (ASOs) and off-target effects. a Theoretical number of complementary genes complementary to ASOs in the case of a perfect match. b The number of complementary base pairs in the case of three mismatches. Light blue: complementary RNA; white: mismatch RNA; red: ASO
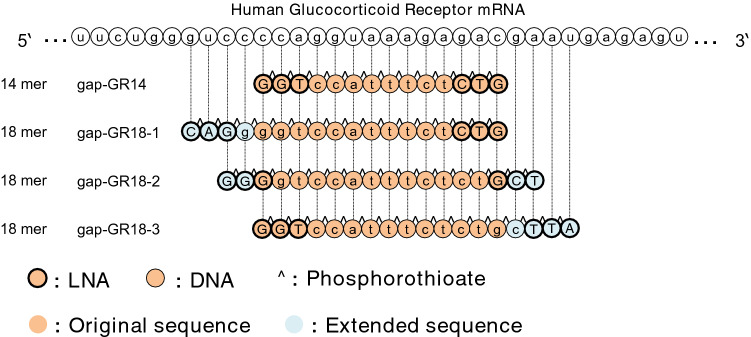
In this study, to reveal the association between oligonucleotide length and off-target effects, we examined the effects of oligonucleotide extension of gapmer ASOs on off-target effects under conditions where the number of sugar-modified nucleotides in the ASOs is constant. Specifically, a 14-mer gapmer ASO (gap-GR14) was first selected as a lead sequence, and extended 18-mer derivatives with the same core 14-mer region (gap-GR18-1, gap-GR18-2 and gap-GR18-3) were designed. All these gapmer ASOs contained three consecutive LNAs on both ends. Exhaustive analyses of changes in gene expression were then performed using microarray profiling for these gapmer ASOs under conditions in which expression of the target gene is down-regulated to similar extents.
Materials and Methods
In silico Analysis
Sequence searches allowing mismatches, insertions, or deletions were performed using GGGenome (https://GGGenome.dbcls.jp/). GGGenome quickly searches short nucleotide sequences using suffix array and FM-index stored on solid state drives, and it was used to query the human pre-mRNA sequences described above that were retrieved from the D3G database (release 20.03; https://d3g.riken.jp/). A complementarity measure called the “distance” (d), which is defined as the total number of mismatches, insertions, or deletions between ASO and the complementary RNA sequences, was used [18, 23]. Pre-mRNA sequences were grouped by d according to the highest complementary site (i.e., with minimal d). For example, if two complementary sites with d = 3 and d = 4 were present in one pre-mRNA sequence, such as one d = 3 site in exon 2 and one d = 4 site in exon 4 in one pre-mRNA sequence, the pre-mRNA was classified into the d = 3 group, because the d = 3 site is more likely to be affected by the ASO than the d = 4 site. The genes that fall into the d ≥ 5 group are all the genes that do not belong to any of the d = 1, 2, 3, or 4 groups. Pre-mRNAs with a complementary RNA sequence to the gapmer ASO are called “Complementary genes (Off-target candidate genes)” in in silico analysis (Table 1).
Table 1.
Statistics from microarray analysis of down-regulated genes (< 50% of control)
| Distance (d) | 0 | 1 | 2 | 3 | ≥ 4 | Total |
|---|---|---|---|---|---|---|
| (a) gap-GR14 | ||||||
| Complementary genes (off-target candidate genes) | 8 | 440 | 5693 | 9583 | 3697 | 19,421 |
| Expressed genes | 6 | 234 | 3530 | 5935 | 1462 | 11,167 |
| Off-target genes | 6 | 168 | 1539 | 668 | 90 | 2471 |
| Percent of off-target genes | 100 | 71.8 | 43.6 | 11.3 | 6.2 | 22.1 |
| Distance (d) | 0 | 1 | 2 | 3 | 4 | ≥ 5 | Total |
|---|---|---|---|---|---|---|---|
| (b) gap-GR18-1 | |||||||
| Complementary genes (off-target candidate genes) | 1 | 0 | 52 | 1432 | 7833 | 10,103 | 19,421 |
| Expressed genes | 1 | 0 | 25 | 850 | 4860 | 5363 | 11,099 |
| Off-target genes | 1 | 0 | 11 | 146 | 259 | 65 | 482 |
| Percent of off-target genes | 100 | NA | 44.0 | 17.2 | 5.3 | 1.2 | 4.3 |
| (c) gap-GR18-2 | |||||||
| Complementary genes (off-target candidate genes) | 1 | 0 | 44 | 1153 | 7567 | 10,656 | 19,421 |
| Expressed genes | 1 | 0 | 22 | 677 | 4677 | 5709 | 11,086 |
| Off-target genes | 1 | 0 | 17 | 329 | 1204 | 430 | 1981 |
| Percent of off-target genes | 100 | NA | 77.3 | 48.6 | 25.7 | 7.5 | 17.9 |
| (d) gap-GR18-3 | |||||||
| Complementary genes (off-target candidate genes) | 1 | 1 | 73 | 1595 | 7805 | 9946 | 19,421 |
| Expressed genes | 1 | 0 | 42 | 947 | 5029 | 5102 | 11,121 |
| Off-target genes | 1 | 0 | 26 | 228 | 311 | 64 | 630 |
| Percent of off-target genes | 100 | NA | 61.9 | 24.1 | 6.2 | 1.3 | 5.7 |
Statistics from microarray analysis of the cells treated with (a) gap-GR14, (b) gap-GR18-1, (c) gap-GR18-2, and (d) gap-GR18-3 ASO. Complementary genes: The number of off-target candidate genes identified by in silico analysis. Expressed genes: The number of “Complementary genes” that are on a microarray and were expressed in Huh-7 cells. Genes with sufficient hybridization signals to be called “present” in at least three of four trials were used in this study. Off-target genes: The number of “Expressed genes” in which gene expression was significantly down-regulated to less than 50% of the level in the control group. Percent of off-target genes: The proportion of genes in “Off-target genes” that were in “Expressed genes.” Note that “Off-target genes” in each d = 0 group included the on-target gene (GR) itself
Antisense Oligonucleotides (ASOs)
A series of four gapmer ASOs targeting human glucocorticoid receptor (GR; also known as Nr3c1) pre-mRNA were synthesized and purified by Gene Design, Inc. (Osaka, Japan). 14- to 18-mer ASOs were phosphorothioated with gapmer designs (3-mer LNA + 8-mer DNA + 3-mer LNA, 3-mer LNA + 12-mer DNA + 3-mer LNA). Note that all the gapmer ASOs contained the same number of LNAs. Detailed sequence information is shown in Fig. 2.
Fig. 2.
The design of antisense oligonucleotides (ASOs) in this study. The 14-mer gapmer ASO targeting human glucocorticoid receptor (GR) RNA, named gap-GR14 in this study, was selected as a lead sequence. Three 18-mer gapmer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3) were designed based on gap-GR14 sequence by extending the oligonucleotides on the 5′-sides, 3′-sides, and both sides. Gap DNA and LNA are indicated in lowercase and uppercase bold, respectively. All oligonucleotides were fully modified with phosphorothioate linkages. Lead sequences are shown in light orange, extended sequences in light blue, and phosphorothioate is indicated by a “^” symbol
Cell Culture and Transfection with ASOs
The human hepatoma cell line Huh-7 was obtained from the Japanese Collection of Research Bioresources (JCRB; Osaka, Japan). The cells were maintained at 37 °C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. Huh-7 cells were seeded in 12-well plates (Corning; Corning, NY, USA) at 1.5 × 104 cells/well (n = 4/group) and transfected with the gapmer ASOs using Lipofectamine® 2000 (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Cells treated with transfection reagent in the absence of ASO were used as a control. After a further 24 h, total RNA was isolated using a Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Cell viability was determined by WST-8 assay according to the manufacturer’s protocol.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed using a One Step TB Green PrimeScript RT-PCR Kit (TaKaRa Bio, Inc., Shiga, Japan) and analyzed with a 7500 Fast Real-Time PCR System with SYBR green (Applied Biosystems, Foster City, CA, USA) and human GR primers (Forward: 5′ CCCTGGTCGAACAGTTTTTTCT 3′ and Reverse: 5′ TGGTCCTGTTGTTGCTGTTGA 3′). The target gene expression level was normalized to that of human GAPDH (Forward: 5′ TCGACAGTCAGCCGCATCTTC 3′ and Reverse: 5′ ACCAAATCCGTTGACTCCGAC 3′).
Microarray Analysis
Genome-wide expression analysis was performed using GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Inc., Santa Clara, CA, USA) according to the manufacturer’s instructions. The scanned data were processed for signal values using Microarray Suite 5.0 software (Affymetrix). Based on the calculated signal value, the ratio of gene expression changes was calculated for all genes in the ASO groups (n = 4) compared with the control group (n = 4). Genes with sufficient hybridization signals to be called “present” in at least three of four trials were used in this study (called “Expressed genes” in Table 1). “Off-target genes” were defined as genes down-regulated to less than 50% of the level in the control group (“Off-target genes” in Table 1). A multiple testing correction was performed using the Benjamini and Hochberg procedure to control the False Discovery Rate (FDR ≤ 0.05), using t-test P value for log2 transformed and normalized ratio data.
Tm Measurements
UV melting experiments were carried out using a SHIMAZU UV-1900i spectrometer (Shimadzu, Inc., Kyoto, Japan) equipped with a Tm analysis accessory. Equimolar amounts of two single-stranded oligonucleotides were dissolved in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl to give a final strand concentration of 4.0 µM. The mixture was annealed by heating at 90 °C followed by slow cooling to room temperature. The melting profile was recorded at 260 nm in the forward and reverse direction from 20 to 90 °C at a scan rate of 0.5 °C/min. All UV melting assays were performed at least three times, and Tm values were calculated by the average method of melting curves [24].
Results
Design of the Gapmer ASOs and Evaluation of On-Target Effects
The purpose of this study was to investigate how oligonucleotide extension of the gapmer ASOs affects the changes in gene expressions of off-target candidate genes. To this end, a previously reported 14-mer gapmer ASO targeting human glucocorticoid receptor (GR) RNA, termed gap-GR14, was selected in this study as a lead sequence [25]. Three 18-mer gapmer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3) were then designed based on the gap-GR14 sequence by extending oligonucleotide on the 5′-sides, 3′-sides, and both sides (Fig. 2). Next, these ASOs were transfected into the human liver-derived Huh-7 cell line (n = 4/group) and their on-target activity was examined by qRT-PCR. All ASOs degraded the target GR RNA efficiently in a concentration-dependent manner (Fig. 3a). Cell viability was not affected substantially at any ASO concentration (Fig. 3b).
Fig. 3.
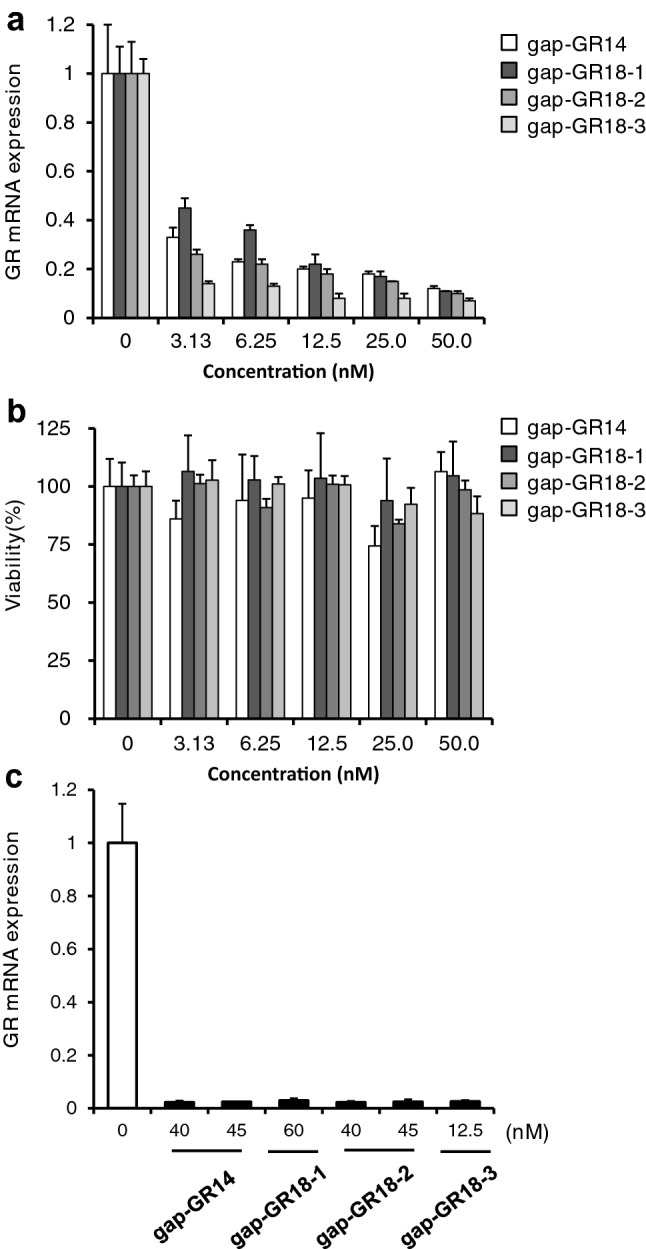
The mRNA expression level of glucocorticoid receptor (GR) and cell viability. a The mRNA expression level of the target gene (GR) in cells treated with each antisense oligonucleotide (ASO) at a concentration in the range of 3.13–50 nM by qRT-PCR. b The cell viability evaluation in cells treated with each ASO at a concentration in the range of 3.13–50 nM by WST-assay. c The mRNA expression level of GR in cells treated with each ASO at concentrations of 40 nM and 45 nM for gap-GR14, 60 nM for gap-GR18-1, 40 nM and 45 nM for gap-GR18-2, and 12.5 nM for gap-GR18-3 by qRT-PCR. The bar graph shows the mean ± standard deviation (n = 4)
To accurately compare the off-target effects between a 14-mer ASO (gap-GR14) and the three 18-mer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3), the ASO concentrations in which the expression of the on-target gene GR is down-regulated to similar extents in all four ASOs were further examined [Electronic Supplemental Material (ESM) Table 1, qRT-PCR]. It was found that GR mRNA was down-regulated in the range of 2.3–3.0% of the control at concentrations of 40–45 nM for gap-GR14, 60 nM for gap-GR18-1, 40 nM and 45 nM for gap-GR18-2, and 12.5 nM for gap-GR18-3 as judged by qRT-PCR (Fig. 3c). Microarray analysis was then performed using the same RNA samples that had been used for qRT-PCR, and it was found that GR mRNA was down-regulated in the range of 2.0–5.1% of the control (ESM Table 1, Microarray). For further analysis, the following microarray data were selected: gap-GR14, 40 nM; gap-GR18-1, 60 nM; gap-GR18-2, 45 nM; and gap-GR18-3, 12.5 nM (ESM Table 1, highlighted by pink background).
Comparative Analysis of Overall Change in Gene Expression
In a previous study, we demonstrated that gapmer ASOs induced off-target effects depending on the degree of complementarity classified by the total number of mismatches, insertions, or deletions on the complementary binding regions, i.e., the distance (d) [18]. To evaluate the overall change in gene expression by gap-GR14, gap-GR18-1, gap-GR18-2, and gap-GR18-3, in silico analysis to identify complementary human pre-mRNAs, or off-target candidate genes, was first performed for each ASO using a search engine, GGGenome, by giving d as the parameter. The numbers of off-target candidate genes with respect to the 14-mer gap-GR14 that were found were: d = 0 (i.e., perfect match), 8; d = 1, 440; d = 2, 5693; d = 3, 9583; and d ≥ 4, 3697 (Table 1a: see the row “Complementary genes”). With 18-mer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3), the only d = 0 gene was the on-target gene (GR) itself. The numbers of off-target candidate genes with respect to gap-GR18-1 that were found were: d = 0, 0; d = 1, 0; d = 2, 52; d = 3, 1432; d = 4, 7833; and d ≥ 5, 10,103 (Table 1b: see the row “Complementary genes”). Equivalent numbers of off-target candidate genes were found for gap-GR18-2 and gap-GR18-3 (Table 1c and d).
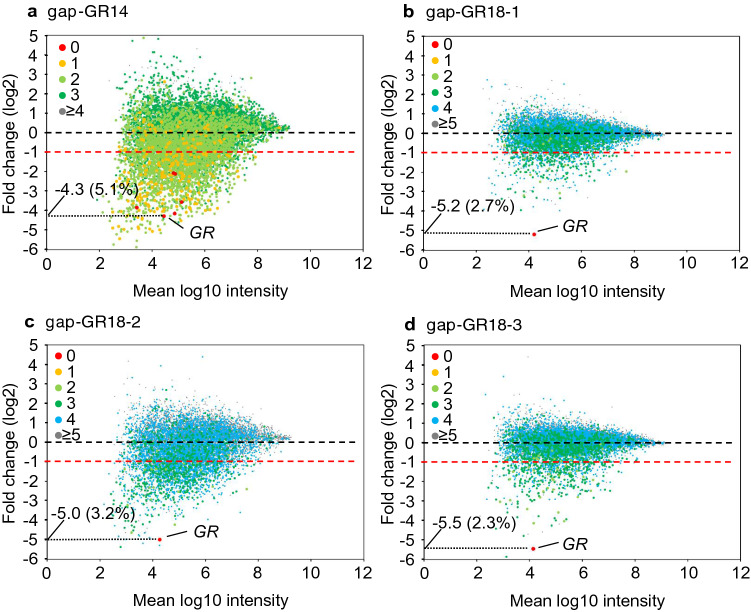
The degree of change in expression is shown as a scatter plot in Fig. 4. As for the core 14-mer gap-GR14, the expression of the highly complementary d = 0 genes indicated by the red dots was particularly strongly suppressed (Fig. 4a). An overall trend toward down-regulation among the d = 1 genes was also observed, shown as orange dots (Fig. 4a). As for the three 18-mer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3), the only d = 0 gene, GR, was down-regulated to less than 4% of the control compared to 5.1% of the control for gap-GR14 (Fig. 4b–d; red dots). It was visually evident that the degree of down-regulation was less in the cells treated with 18-mer ASOs than in those treated with the 14-mer ASO (Fig. 4b–d). In particular, the trend toward down-regulation was dramatically reduced in the gap-GR18-1- and gap-GR18-3-treated groups (Fig. 4b and d). The number of up-regulated genes was also less in the 18-mer ASO-treated groups. To analyze the effects of oligonucleotide extension on overall changes in gene expression, the changes in gene expression were quantified by taking the logarithm of the ratio of gene expression changes and calculating the sum of the absolute value of it (ESM Fig. 1a). While gap-GR14 was down-regulated by 6489, 18-mer gap-GR18-1, gap-GR18-2, and gap-GR18-3 were down-regulated by 2269, 5515, and 2358, respectively (ESM Fig. 1b: Index of overall changes for down-regulated genes). This indicated that oligonucleotide extension reduces the extent of down-regulation. Similar findings were observed for up-regulated genes (ESM Fig. 1: Index of overall changes for up-regulated genes), indicating that oligonucleotide extension suppresses changes in gene expression quantitatively.
Fig. 4.
Scatter plot analysis. Scatter plots from microarray analysis of the cells treated with a gap-GR14, b gap-GR18-1, c gap-GR18-2, and d gap-GR18-3 antisense oligonucleotide (ASO). The amount of gene expression in the control cells is shown on the horizontal axis, and the proportion of change in gene expression as a result of introduction of each ASO (expressed logarithmically) is shown on the vertical axis
Comparative Analysis of Off-Target Effects
In our previous study, we defined “off-target genes” as genes that were down-regulated to less than 50% of the level in the control group [18]. This is because heterozygous knockout animals, in which 50% of gene expression is retained, do not show abnormalities in most cases. Taking the above into account, the number of off-target genes was next analyzed for the 14-mer ASO (gap-GR14) and three 18-mer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3). The statistics from microarray analysis are shown in Table 1. “Off-target genes” in Table 1 shows the number of “Expressed genes” in which gene expression was significantly down-regulated to less than 50% of the level in the control group (see table legend). The numbers of off-target genes in the gap-GR14 group were 6, 168, 1539, 688, and 90 for d = 0, d = 1, d = 2, d = 3, and d ≥ 4, respectively (Table 1a: Off-target genes). The respective numbers of off-target genes in d = 2, d = 3, d = 4, and d ≥ 5 18-mer ASOs were 11, 146, 259, and 65 for gap-GR18-1, 17, 329, 1204, and 430 for gap-GR18-2, and 26, 228, 311, and 64 for gap-GR18-3, respectively (Table 1b–d: Off-target genes). ASO-targeted GR was the only d = 0 gene in 18-mer ASOs. In each ASO, the sum of all genes that were down-regulated to less than 50% of the control was 2471 for gap-GR14, in comparison to 482 for gap-GR18-1, 1981 for gap-GR18-2, and 630 for gap-GR18-3. In other words, extension of ASOs from 14-mer to 18-mer resulted in a reduction in the number of off-target genes. Note that the number of genes with more than a twofold increase in expression was also suppressed by oligonucleotide extension (ESM Table 2).
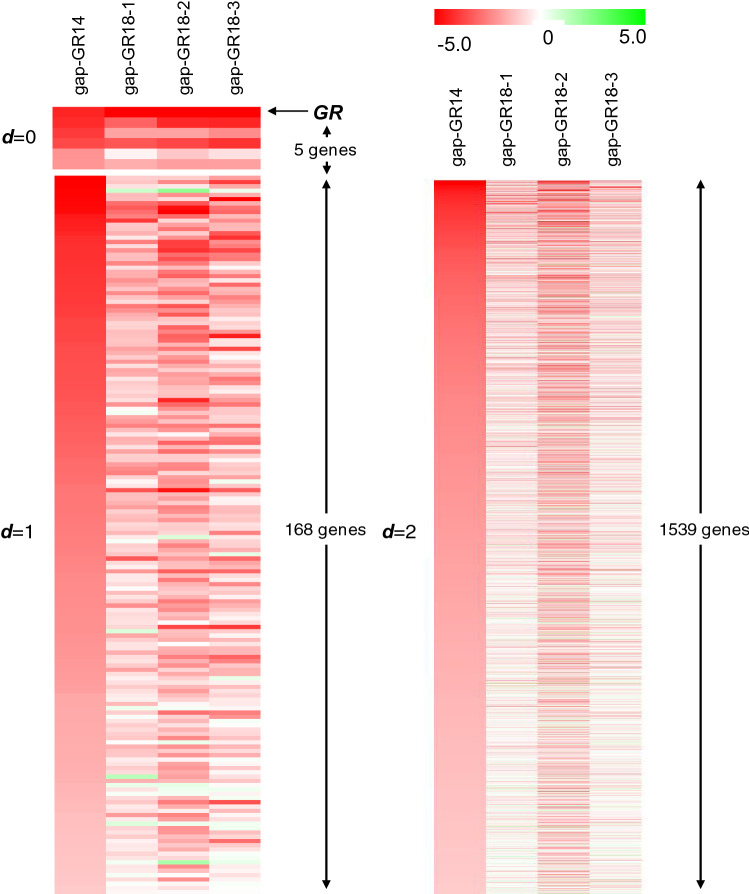
Next, the effects of oligonucleotide extension on each off-target gene in the gap-GR14 group was examined (Fig. 5). As shown in the heatmap in Fig. 5, off-target genes for gap-GR14 were grouped into d = 0 (5 genes excluding GR), d = 1 (168 genes), and d = 2 (1539 genes), and they were ordered by the degree of gene down-regulation from strongest to weakest. Compared with the gap-GR14 column, it is apparent that a large number of genes were not down-regulated in the 18-mer ASOs group. ESM Table 3 shows the number of off-target genes in the gap-GR14 group that were significantly down-regulated to less than 50% of the control in the 18-mer ASOs group. For example, in the gap-GR14-treated cells, 1539 genes in the d = 2 group were down-regulated to less than 50% of the control. Among those 1539 genes, 278, 857, and 307 genes were significantly down-regulated to less than 50% of the control in gap-GR18-1, gap-GR18-2, and gap-GR18-3-treated cells, respectively. That means that on average, 31.2% (481/1539) of off-target genes remained to be down-regulated, while the other 68.8% avoided off-target effects after the extension of oligonucleotides. Off-target genes that were highly complementary with gap-GR14 were more likely to maintain the off-target effects despite oligonucleotide extension (83.3% and 57.1% for d = 0 and d = 1, respectively). In comparison, genes that were less complementary were more likely to avoid the off-target effects (31.2%, 20.5%, and 24.4% for d = 2, d = 3, and d ≥ 4, respectively). Thus, it is likely that the expressions of genes that were less complementary with the gap-GR14 were no longer down-regulated, resulting in the reduction of off-target effects by oligonucleotide extension.
Fig. 5.
Heat map analysis. The expression variation of off-target genes of gap-GR14 in the cells treated with gap-GR18-1, gap-GR18-2, and gap-GR18-3. The off-target genes of gap-GR14 are classified into the d = 0 group [glucocorticoid receptor (GR) and five genes] and d = 1 (168 genes) in the left panel, the d = 2 group (1539 genes) in the right panel. Up-regulated genes are presented in shades of green and down-regulated genes in shades of red
In order to determine what caused the reduction of off-target effects in the 18-mer ASO-treated cells, we focused on 5 d = 0 off-target genes in the gap-GR14-treated cells (PPP1R12B, RAB27B, UCHL5, ZNRF3, ZDHHC18). We examined the change in gene expression of these five genes in the 18-mer ASO-treated cells, as well as the Tm between each ASO and complementary binding site of each gene (ESM Table 4). The findings demonstrated that there was no association between the degree of reduced gene expression and Tm. This may be because reduction of gene expression cannot be attributed solely on the strength of complementary binding between 18-mer ASOs and each gene. This indicates that, in addition to binding affinity, the off-target effect of gapmer ASOs is also related to other factors, such as the structure of RNA around the target region [21, 26, 27] and the length of the gap region of ASOs [28, 29].
Previously, we demonstrated that 13-mer ASOs induced off-target effects on genes with complementarity up to d = 2 [18]. In general, the strength of binding between the ASO and complementary RNA increases as the oligonucleotide becomes longer, so that it is possible that gapmer ASOs longer than 13-mer may induce off-target effects on d = 3 genes in addition to d = 0, 1, and 2 genes. In the present study, changes in gene expression using the 14-mer ASO (gap-GR14) were analyzed, and it was found that it induced off-target effects on genes with complementarity up to d = 2 as judged by cumulative curves, shown in Fig. 6. With 18-mer ASOs (gap-GR18-1, gap-GR18-2, and gap-GR18-3), expressions of d = 3 off-target candidate genes were significantly down-regulated, showing the lines of 18-mer ASOs shifted to the left (Fig. 6b). This means that genes with complementarity up to at least d = 3 were subject to off-target effects in the case of 18-mer ASOs. These data indicated that mismatches, insertions, or deletions between the ASO and the complementary RNA sequences are better tolerated when longer gapmer ASOs cleave complementary RNAs, as described in the Introduction (Sect. 1). However, note that the total number of off-target genes in the 18-mer ASO-treated cells was less than those in 14-mer ASO-treated cells (Table 1).
Fig. 6.
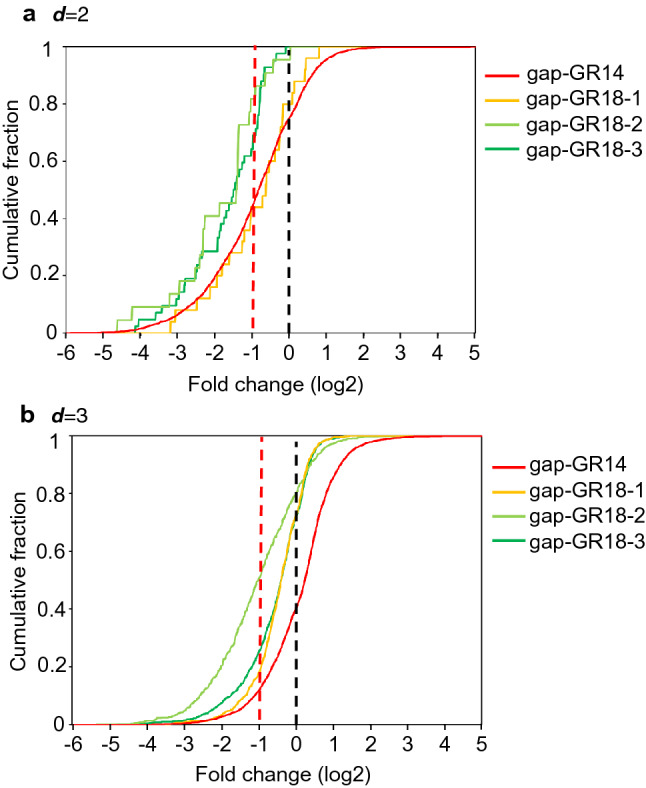
Cumulative plot. Cumulative plot from microarray analysis of the cells treated with gap-GR14, gap-GR18-1, gap-GR18-2, and gap-GR18-3 ASO. The cumulative number of genes is shown on the vertical axis, and the proportion of change in a d = 2 and b d = 3 off-target gene expression due to introduction of the gap-GR14 (red line), gap-GR18-1 (orange line), gap-GR18-2 (green line), and gap-GR18-3 (yellow green line) ASO (expressed logarithmically, respectively) is shown on the horizontal axis
An Attempt to Identify the “Non-Real” Off-Target Genes
As mentioned above, we defined the off-target genes as genes that were down-regulated to less than 50% of the control. These off-target genes include “real” off-target genes that are cleaved by RNase H after binding with ASO, as well as “non-real” off-target genes that are indirectly down-regulated. For example, knockdown of a transcription factor would also affect expression of its downstream genes indirectly. It is generally difficult to distinguish between “real” and “non-real” off-target genes; however, genes with low complementarity, such as those categorized into d ≥ 5 among 18-mer ASOs, were considered more likely to be “non-real” off-target genes, since the ratio of genes that were down-regulated to less than 50% of the control was clearly lower than those categorized into d = 2 and d = 3 (Table 1). To try to identify the “non-real” off-target genes, we further performed the following analysis. Firstly, 184 genes were identified as genes that were commonly down-regulated to less than 50% of the control by treatment of gap-GR14, gap-GR18-1, gap-GR18-2, and gap-GR18-3. Secondly, among 184 genes, 21 genes were identified as genes that were classified into d ≥ 4 groups for all four ASOs. These 21 genes appeared to be down-regulated in a RNase H-independent manner, and thus seem to be “non-real” off-target genes that are indirectly down-regulated. The results of the reanalysis of off-target genes under the conditions excluding the above 21 genes are shown in ESM Table 5.
Discussion
In the present study, an extensive microarray analysis was performed to understand the association between oligonucleotide length and off-target effects using 14-mer and 18-mer ASOs (gap-GR14, gap-GR18-1, gap-GR18-2, gap-GR18-3). So far, Hagedorn et al. examined the melting temperature (Tm) of oligonucleotides (i.e., affinity) and analyzed its association with the LNA modifications and oligonucleotide length [21, 22, 30]. Specifically, they evaluated the off-target effects by changing the number of LNA modifications in the gapmers with equal length. They also analyzed the off-target effects of gapmers with similar binding affinities and potencies by changing both the number of LNA modifications and the oligonucleotide length. In the light of the binding affinity, they showed excellent approaches for optimizing the designs of gapmers to generate versions that are more specific for the intended target. In this study, on the other hand, we examined the effects of oligonucleotide extension of gapmer ASOs on off-target effects under conditions where the number of sugar-modified nucleotides in the ASOs is constant. We selected a 14-mer gapmer ASO (gap-GR14) as a lead sequence, and extended its length to generate 18-mer derivatives with the same core 14-mer region (gap-GR18-1, gap-GR18-2 and gap-GR18-3). In this analysis, we analyzed off-target effects under conditions in which the on-target activities of these ASOs were strictly adjusted to be comparable, and found that the extension of oligonucleotides reduced off-target effects. Given Hagedorn’s findings, we think that adjusting the number of LNAs could also reduce the off-target effects.
In the present study, the on-target activity of each gapmer ASO was strictly adjusted, and the condition in which the on-target activity of 18-mer ASOs was higher than that of 14-mer gap-GR14 was intentionally selected (gap-GR14, 5.1% of control; gap-GR18-1, 2.7% of control; gap-GR18-2, 3.2% of control; gap-GR18-3, 2.3% of control: ESM Table 1). Nevertheless, down-regulation of gene expression was significantly reduced in 18-mer ASO-treated cells (Fig. 4, ESM Fig. 1, and Table 1). Similarly, the extent of up-regulation of genes was clearly suppressed in 18-mer ASO-treated cells. These appeared to be indirect effects on gene expression in a similar manner as described for “non-real” off-target genes (ESM Table 5). “Non-real” off-target genes can be down-regulated indirectly via cleavage of off-target RNAs that encode, for example, transcription factors. If the knockdown of a transcriptional repressor, one of the off-target genes for the 14-mer ASO, are released by oligonucleotide extension, the downstream gene expression is suppressed because the transcriptional repressors restore its function. The combination of indirect effects seems to lead to the overall suppression of gene expression changes by oligonucleotide extension.
In the off-target analysis, we noticed that a relatively strong off-target effect was maintained in gap-GR18-2-treated cells compared to gap-GR18-1- and gap-GR18-3-treated cells. In this regard, we consider the following two possibilities. One is the existence of complementary LNA to off-target genes for gap-GR14. As for gap-GR18-2, complementary LNAs always exist on both ends when gap-GR18-2 binds to the off-target genes for gap-GR14, whereas complementary LNAs were lost with high probability on either end in the case of gap-GR18-1 and gap-GR18-3 (ESM Fig. 2). The other is the complementarity in the central gap region to form the DNA/RNA heteroduplex. Because the gap region of gap-GR18-2 is fully complementary to the off-target genes for gap-GR14, RNase H could be more easily recruited and thus the off-target effect is maintained (ESM Fig. 2).
In the present study, a transfection reagent was used for the delivery of ASOs into the cells. In general, naked delivery without the transfection reagent takes several days to achieve sufficient on-target activity, and, therefore, hybridization-independent secondary changes in gene expression are likely to occur. In addition, in the case of naked delivery, the difference of ASO length affects efficiency of incorporation of ASOs into the cells. In order to avoid the effects of these factors, we selected the transfection reagent-mediated delivery for off-target analysis. As for a negative control, we used the transfection reagent-only group, but not a negative control ASO with the transfection reagent. This is because short LNA ASOs, such as 14-mer and 18-mer ASOs used in this study, always have a significant number of off-target candidate genes with low d values (such as d = 1, d = 2, and d = 3) [17], and these off-target candidate genes are stochastically down-regulated significantly with a certain probability [18]. For example, as shown in Table 1, d = 3 genes are down-regulated to 11.3% (gap-GR14) to 48.6% (gap-GR18-2) of the control. This indicates the difficulty of designing a short LNA gapmer that does not affect gene expression at all. In the present study, we focused on the change in gene expression by off-target effects, thus, we did not use a negative control gapmer. Exhaustive in silico analysis might identify negative control ASO sequences with minimal off-target effects, such as ASOs with no d = 0, d = 1, or d = 2 off-target candidate genes.
The present findings collectively indicate that oligonucleotide extension should be one of the effective strategies to fundamentally reduce the effect of gene expression changes. Since off-target effects induced by gapmer ASOs are known to be one of causes of liver toxicity [14, 15], and such toxicity induced by off-target effects cannot be evaluated in animal models, oligonucleotide extension seems to be effective in lowering the risk of termination of drug development in clinical trials. It is important to note that the present findings were based on in vitro cellular analysis, and that oligonucleotide extension may also affect in vivo pharmacodynamics such as circulation time in blood, as well as tissue and cellular uptake [7, 31]. Future studies are needed to examine the effects of oligonucleotide extension in vivo to determine its effects on a number of factors, such as toxicity and pharmacodynamics.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
Takao Inoue (TI) was supported by the Japan Agency for Medical Research and Development (AMED) under Grant numbers JP20mk0101119, JP20mk0101187, JP20ak0101073, and JP20am0401003.
Conflicts of Interest/Competing interests
The authors declare no conflicts of interest.
Ethics approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of data and material
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession number: GSE188684).
Code availability
Not applicable.
Author Contributions
Conceptualization, Hidenori Yasuhara (HY), Tokuyuki Yoshida (TY), Satoshi Obika (SO) and Takao Inoue (TI); validation, HY and TY; investigation, HY and TY; resources, SO and TI; data curation, HY and TY; writing—original draft preparation, HY, TY and TI; writing—review and editing, HY, TY, Kiyomi Sasaki (KS), SO and TI; supervision, TI; project administration, TY, KS and TI; funding acquisition, TY, SO and TI. All authors have read and agreed to the published version of the manuscript.
Footnotes
Hidenori Yasuhara and Tokuyuki Yoshida contributed equally to this work.
Contributor Information
Satoshi Obika, Email: obika@phs.osaka-u.ac.jp.
Takao Inoue, Email: takao@nihs.go.jp.
References
- 1.Yacyshyn B, Bowen-Yacyshyn MB, Shanahan W. The clinical experience of antisense therapy to ICAM-1 in Crohn’s disease. Curr Opin Mol Ther. 1999;1:332–335. [PubMed] [Google Scholar]
- 2.Crook ST. Antisense drug technology: principles, strategies, and applications. 2. CRC Press; 2008. [Google Scholar]
- 3.Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD, Davidson M. Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: a randomized, double-blind, placebo-controlled trial. J Am Coll Cardiol. 2013;62:2178–2184. doi: 10.1016/j.jacc.2013.07.081. [DOI] [PubMed] [Google Scholar]
- 4.Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PubMed] [Google Scholar]
- 5.Witztum JL, Gaudet D, Freedman SD, Alexander VJ, Digenio A, Williams KR, et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N Engl J Med. 2019;31:531–542. doi: 10.1056/NEJMoa1715944. [DOI] [PubMed] [Google Scholar]
- 6.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geary RS. Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin Drug Metab Toxicol. 2009;5:381–391. doi: 10.1517/17425250902877680. [DOI] [PubMed] [Google Scholar]
- 8.Bennett FC, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 9.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;314:314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury S, Burris HA, Patel M, Infante JR, Jones SF, Voskoboynik M, et al. A phase I dose escalation, safety and pharmacokinetic (PK) study of AZD5312 (IONIS- ARRx), a first-in-class generation 2.5 antisense oligonucleotide targeting the androgen receptor (AR) Eur J Cancer. 2016;69:S145. doi: 10.1186/s40425-018-0436-5. [DOI] [Google Scholar]
- 11.Pfeiffer N, Voykov B, Renieri G, Bell K, Richter P, Weigel M, et al. First-in-human phase I study of ISTH0036, an antisense oligonucleotide selectively targeting transforming growth factor beta 2 (TGF-beta2), in subjects with open-angle glaucoma undergoing glaucoma filtration surgery. PLoS ONE. 2017;12:e0188899. doi: 10.1371/journal.pone.0188899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reilley MJ, McCoon P, Cook C, Lyne P, Kurzrock R, Kim Y, et al. STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. J Immunother Cancer. 2018;6:119. doi: 10.1186/s40425-018-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribrag V, Lee ST, Rizzieri D, Dyer MJS, Fayad L, Kurzrock R, et al. A phase 1b study to evaluate the safety and efficacy of durvalumab in combination with tremelimumab or danvatirsen in patients with relapsed or refractory diffuse large B-cell lymphoma. Clin Lymphoma Myeloma Leuk. 2021;21:309–317. doi: 10.1038/10.1016/j.clml.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Burel SA, Hart CE, Cauntay P, Hsiao J, Machemer T, Katz M, et al. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2016;44:2093–2109. doi: 10.1093/nar/gkv1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasuya T, Hori S, Watanabe A, Nakajima M, Gahara Y, Rokushima M, et al. Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci Rep. 2016;6:30377. doi: 10.1038/srep30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindow M, Vornlocher HP, Riley D, Kornbrust DJ, Burchard J, Whiteley LO, et al. Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat Biotechnol. 2012;30:920–923. doi: 10.1038/nbt.2376. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T, Naito Y, Sasaki K, Uchida E, Sato Y, Naito M, et al. Estimated number of off-target candidate sites for antisense oligonucleotides in human mRNA sequences. Genes Cells. 2018;23:448–455. doi: 10.1111/gtc.12587. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Naito Y, Yasuhara H, Sasaki K, Kawaji H, Kawai J, et al. Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cells. 2019;24:827–835. doi: 10.1111/gtc.12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen W, Hoyos CLD, Migawa MT, Vickers TA, Sun H, Low A, et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat Biotechnol. 2019;37:640–650. doi: 10.1038/s41587-019-0106-2. [DOI] [PubMed] [Google Scholar]
- 20.Kamola PJ, Kitson JD, Turner G, Maratou K, Eriksson S, Panjwani A, et al. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43:8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagedorn PH, Hansen BR, Koch T, Lindow M. Managing the sequence-specificity of antisense oligonucleotides in drug discovery. Nucleic Acids Res. 2017;45:2262–2282. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagedorn PH, Pontoppidan M, Bisgaard TS, Berrera M, Dieckmann A, Ebeling M, et al. Identifying and avoiding off-target effects of RNase H-dependent antisense oligonucleotides in mice. Nucleic Acids Res. 2018;46:5366–5380. doi: 10.1093/nar/gky397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel S, Schirduan K, Shen Y, Klar R, Tost J, Jaschinski F. Using RNA-seq to assess off-target effects of antisense oligonucleotides in human cell lines. Mol Diagn Ther. 2021;25:77–85. doi: 10.1007/s40291-020-00504-4. [DOI] [PubMed] [Google Scholar]
- 24.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 25.Stanton R, Sciabola S, Salatto C, Weng Y, Moshinsky D, Little J, et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22:344–359. doi: 10.1089/nat.2012.0366. [DOI] [PubMed] [Google Scholar]
- 26.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uppuladinne MVN, Sonavane UB, Deka RC, Joshi RR. Structural insight into antisense gapmer-RNA oligomer duplexes through molecular dynamics simulations. J Biomol Struct Dyn. 2019;37:2823–2836. doi: 10.1080/07391102.2018.1498390. [DOI] [PubMed] [Google Scholar]
- 28.Magner D, Biala E, Lisowiec-Wachnicka J, Kierzek R. Influence of mismatched and bulged nucleotides on SNP-preferential RNase H cleavage of RNA-antisense gapmer heteroduplexes. Sci Rep. 2017;7:12532. doi: 10.1038/s41598-017-12844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papargyri N, Pontoppidan M, Andersen MR, Koch T, Hagedorn PH. Chemical diversity of locked nucleic acid-modified antisense oligonucleotides allows optimization of pharmaceutical properties. Mol Ther Nucleic Acids. 2020;19:706–717. doi: 10.1016/j.omtn.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieckmann A, Hagedorn PH, Burki Y, Brugmann C, Berrera M, Ebeling M, et al. A sensitive in vitro approach to assess the hybridization-dependent toxic potential of high affinity gapmer oligonucleotides. Mol Ther Nucleic Acids. 2018;10:45–54. doi: 10.1016/j.omtn.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frazier KS. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol Pathol. 2015;43:78–89. doi: 10.1177/0192623314551840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.