Abstract
Background
Protection offered by coronavirus disease 2019 (COVID-19) vaccines wanes over time, requiring an evaluation of different boosting strategies to revert such a trend and enhance the quantity and quality of Spike-specific humoral and cellular immune responses. These immunological parameters in homologous or heterologous vaccination boosts have thus far been studied for mRNA and ChAdOx1 nCoV-19 vaccines, but knowledge on individuals who received a single dose of Ad26.COV2.S is lacking.
Methods
We studied Spike-specific humoral and cellular immunity in Ad26.COV2.S-vaccinated individuals (n = 55) who were either primed with Ad26.COV2.S only (n = 13) or were boosted with a homologous (Ad26.COV2.S, n = 28) or heterologous (BNT162b2, n = 14) second dose. We compared our findings with the results found in individuals vaccinated with a single (n = 16) or double (n = 44) dose of BNT162b2.
Findings
We observed that a strategy of heterologous vaccination enhanced the quantity and breadth of both Spike-specific humoral and cellular immunity in Ad26.COV2.S-vaccinated individuals. In contrast, the impact of the homologous boost was quantitatively minimal in Ad26.COV2.S-vaccinated individuals, and Spike-specific antibodies and T cells were narrowly focused to the S1 region.
Conclusions
Despite the small sample size of the study and the lack of well-defined correlates of protection against COVID-19, the immunological features detected support the utilization of a heterologous vaccine boost in individuals who received Ad26.COV2.S vaccination.
Funding
This study is partially supported by the Singapore Ministry of Health’s National Medical Research Council under its COVID-19 Research Fund (COVID19RF3-0060, COVID19RF-001, and COVID19RF-008), The Medical College St. Bartholomew’s Hospital Trustees – Pump Priming Fund for SMD COVID-19 Research.
Keywords: COVID-19, vaccines, antiviral immunity, Adenovirus vector, heterologous immunity
Graphical abstract
Context and significance
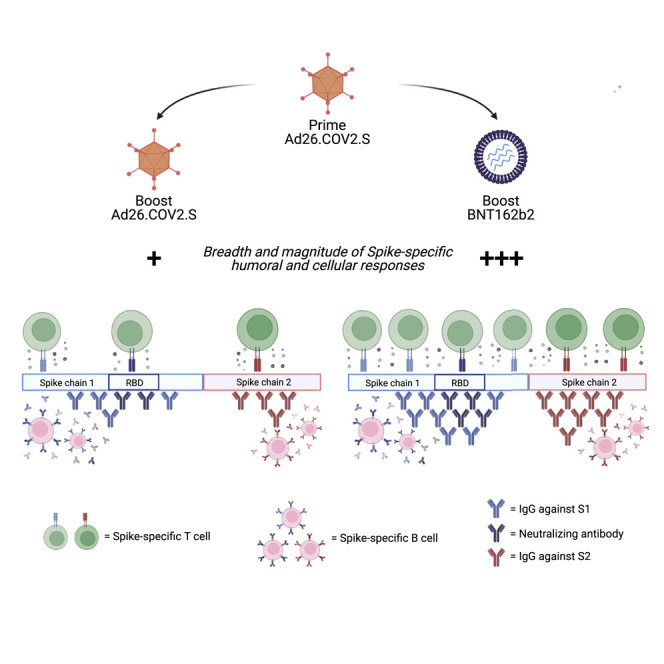
Protection offered by COVID-19 vaccines wanes over time. We evaluated whether individuals vaccinated with a single dose of Johnson & Johnson’s vaccine Ad26.COV2.S would benefit from a second dose of Ad26.COV2.S (homologous boost) or of the Pfizer-BioNTech mRNA vaccine BNT162b2 (heterologous boost). The study of different vaccination regimens is critical to optimize future vaccine strategies. We analyzed the quantity and quality of antibodies and T cells specific for Spike, demonstrating that Spike-specific antibodies and T cells were augmented preferentially following a boost with BNT162b2. A heterologous boost also expanded the ability of antibodies and T cells to recognize different sections of Spike. These data support the use of a heterologous mix-and-match strategy in individuals vaccinated with a single dose of Ad26.COV2.S.
Homologous versus heterologous boosting strategies were evaluated in single-dose Ad26.COV2.S vaccines. Magnitude of Spike-specific humoral and cellular immunity was augmented following a boost with BNT162b2. Heterologous vaccination expanded the ability of both humoral and cellular immunity to recognize multiple regions of Spike. Convalescent individuals responded equally to both boosting strategies.
Introduction
Vaccination has been the key strategy to reduce the incidence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and to protect from severe coronavirus disease 2019 (COVID-19) worldwide. Accelerated vaccine development efforts led to the approval of SARS-CoV-2 vaccines utilizing several different technological platforms that displayed varying clinical efficacy, with the highest being associated with adenoviral vector- and mRNA-based vaccines.1, 2, 3, 4, 5 Vaccine-induced protective efficacy is associated with their ability to induce neutralizing anti-Spike antibodies and Spike-specific T cells.3 , 6 , 7 Unfortunately, the appearance of the Delta variant and the progressive waning of antibody titers8 observed over time has reduced the protective efficacy of COVID-19 vaccines,5 particularly in individuals vaccinated with a single dose of Ad26.COV2.9 These findings have ignited a debate about the need for possible booster vaccinations.
Ad26.COV2 (Johnson & Johnson) is a single-dose vaccine10 with protective efficacy against severe disease.4 , 11 A single immunization with Ad26.COV2.S induced rapid cellular immune responses as well as binding and neutralizing antibodies, including an induction of receptor binding domain (RBD)-specific binding antibodies in 90% of vaccine recipients4 , 11 , 12 that persists over time.13 A recent report also indicated that vaccination with Ad26.COV2.S leads to a persistence of protective efficacy.14 On the flip side, there are other evidences that a single dose of Ad26.COV2.S may not be sufficient and may lead to a higher incidence of breakthrough infections.9 Additionally, a reduced ability of Ad26.COV2.S to induce antibody responses was reported in immunocompromised individuals.15 As a result, it has been proposed that individuals vaccinated with Ad26.COV2.S should receive a second dose, similar to the two-dose regimen recommended for mRNA-based vaccines (BNT162b2 and mRNA1273) and the adenoviral-vector-based vaccine (ChAdOx1 nCov-19).
Vaccine-induced protective efficacy is associated with the ability to induce neutralizing anti-Spike antibodies and Spike-specific T cells.16 Data in animal models and in healthy individuals vaccinated with the other adenoviral-vector-based vaccine, ChAdOx1 nCov19, and BNT162b2 have shown that a heterologous vaccine boost enhances both cellular and humoral immunity17, 18, 19, 20, 21, 22 and might be even more protective than homologous BNT162b2 vaccination.23 Data recently reported showed an ability of both homologous and heterologous boosts after Ad26.COV2.S to increase Spike-specific antibodies; however, a parallel analysis for cellular immunity was not performed.24
Therefore, to gain more comprehensive information on the best boosting strategy in individuals vaccinated with a single dose of Ad26.COV2.S, we studied here the Spike-specific T and B cell immunogenicity after a homologous or heterologous second vaccination dose. We compared the results with those obtained from individuals vaccinated with a single or double dose of BNT162b2. The quantity and breadth of Spike-specific antibodies and T cells were studied. Collectively, our data show the enhanced immunogenicity of the heterologous boosting strategy in individuals primed with Ad26.COV2.S.
Results
Cohorts of vaccinated individuals
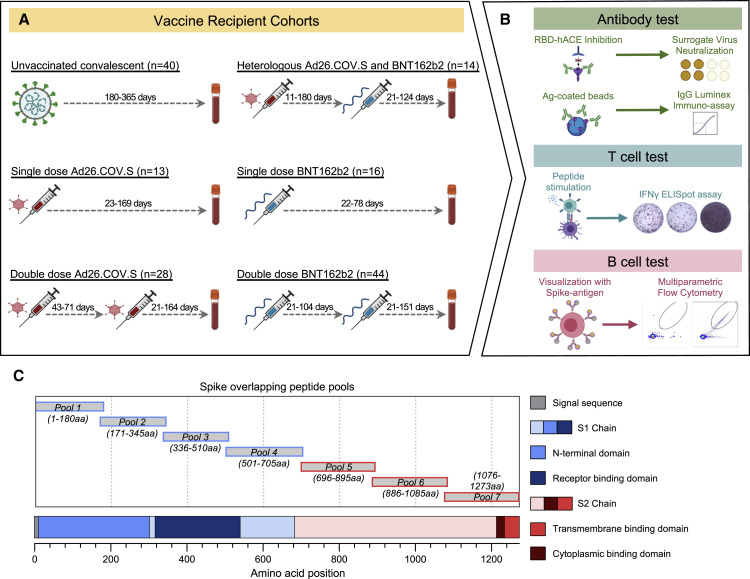
We studied humoral and cellular immunity in a total of 115 individuals who received different vaccination schedules (Figure 1 A): single-dose Ad26.COV2.S (indicated as J for Johnson & Johnson; n = 13), homologous double-dose Ad26.COV2.S (J+J; n = 28), single-dose BNT162b2 (indicated as P for Pfizer/Biontech; n = 16), homologous double-dose BNT162b2 (P+P; n = 44), and heterologous switch-dose Ad26.COV2.S followed by BNT162b2 (J+P; n = 14). Epidemiological characteristics of the studied population are summarized in Table 1 . There were no significant differences in sex and age among the different vaccinated groups except between J (39.2 ± 12.0) and J+J (51.2 ± 14.2) and J+J (51.2 ± 14.2) and P+P (42 ± 9.0). Importantly. no statistically significant differences were present between homologous (J+J: 51.2 ± 14.2) and heterologous (J+P: 45.5 ± 15.5) vaccinated boost cohorts.
Figure 1.
Vaccinated individuals and immune characterization
(A) Immune characterization was done in six cohorts: single-dose Ad26.COV2.S (J), double-dose Ad26.COV2.S (J+J), heterologous Ad26.COV2.S + BNT162b2 (J+P), single-dose BNT162b2 (P), and double-dose BNT162b2 (P+P). Unvaccinated cohort was used as a control (Unvac). The number of individuals for each cohort, the interval between first and second dose of vaccine, and the range of days for blood collection after last dose of vaccine are indicated. Sampling for immune analysis (humoral and cellular) was done only after the last vaccination dose both in homologous and heterologous vaccinated groups.
(B) Schematic of humoral and cellular analysis.
(C) Schematic representation of the location of the seven Spike-specific peptide pools containing 15-mer overlapping peptides spanning the entire Spike protein. Pools 1 to 4 contain peptides from the signal peptide and the S1 chain. Pools 5 to 7 encompass the S2 chain together with the transmembrane and cytoplasmic domains.
See also Figure S1.
Table 1.
Demographics of vaccinated individuals
| J |
J+J |
P |
P+P |
J+P |
|||
|---|---|---|---|---|---|---|---|
| Characteristics | Ad26.COV2.S, single dose | Ad26.COV2.S, double dose | BNT162b2, single dose | BNT162b2, double dose | Ad26.COV2.S and BNT162b2 | Total | Statistical analysisa |
| No. of participants | 13 | 28 | 16 | 44 | 14 | 115 | N/A |
| Sexb | |||||||
| Male (%) | 6 (46.2) | 18 (64.2) | 7 (43.7) | 15 (34.1) | 9 (64.3) | 55 (47.8) | no statistical difference |
| Female (%) | 7 (53.8) | 10 (35.8) | 9 (56.3) | 29 (65.9) | 5 (35.7) | 60 (52.1) | no statistical difference |
| Infection statusb | |||||||
| Naïve (%) | 12 (92.3) | 23 (82.1) | 10 (62.5) | 37 (84.1) | 9 (64.3) | 91 (79.1) | no statistical difference |
| Infected (%) | 1 (7.7) | 5 (17.9) | 6 (37.5) | 7 (15.) | 5 (35.7) | 24 (20.9) | no statistical difference |
| Agec | |||||||
| Mean, years | 39.2 | 51.2 | 42.8 | 42 | 45.5 | 44.2 | J and J+J∗ |
| Range, years | 25–69 | 25–75 | 32–53 | 23–62 | 25–70 | 23–75 | J+J and P+P∗ |
| Days between prime and boost | |||||||
| Median | – | 57 | – | 21 | 42 | – | N/A |
| Range | – | 43–71 | – | 21–104 | 11–180 | – | N/A |
| Days post-last dose | |||||||
| Median | 80 | 49.5 | 60 | 94 | 32 | 77.5 | N/A |
| Range | 23–169 | 21–164 | 22–78 | 21–151 | 21–124 | 7–169 | N/A |
| Race or ethnic group | |||||||
| White (other/British/Irish) (%) | 11 (84.6) | 21 (75) | 8 (50) | 5 (11.4) | 11 (75.6) | 56 (48.7) | N/A |
| Black (%) | 1 (7.6) | – | – | – | – | 1 (0.9) | N/A |
| Asian or South Asian (Indian/Chinese/other) (%) | 1 (7.6) | 4 (14.3) | 6 (37.5) | 36 (81.8) | 2 (14.3) | 49 (42.6) | N/A |
| Caucasian/mixed/others (%) | – | 2 (7.2) | 1 (6.2) | 3 (6.8) | 1 (7.1) | 7 (6.1) | N/A |
| Unknown/undisclosed (%) | – | 1 (3.5) | 1 (6.2) | – | – | 2 (1.7) | N/A |
Statistical analysis was carried out where applicable. ∗p ≤ 0.05. NA, not applicable.
Only results shown for significant multiple comparisons.
Chi-square test was used.
One-way ANOVA was used.
For controls, we studied 10 to 22 unvaccinated healthy individuals and 40 unvaccinated SARS-CoV-2 convalescents. In the homologous double-dose Ad26.COV2.S cohort, the median time between the first and second dose was 56 days (43–71 days). In the heterologous switch-dose Ad26.COV2.S and BNT162b2 cohort, the median number of days between first and second dose was 31 days but with a wider range (11–180 days). The time between the first and second dose of BNT162b2 was 21 days (21–104 days). Note also that the analysis of humoral and cellular immune parameters was performed at variable intervals after the second dose with a median number of days indicated in Table 1. Humoral responses were characterized by measuring immunoglobulin G (IgG) and IgA against the whole Spike, S1 and S2 domains, and neutralizing antibodies (using the surrogate virus neutralization test [sVNT]) and by quantification of Spike-specific memory B cells (Figure 1B). T cell response was analyzed by quantification of interferon (IFN)-γ-secreting cells in reaction to peptides covering the whole Spike protein in an ELISpot assay. Seven pools of 15-mer peptides were used to detect Spike-specific T cells, Spike pools 1 to 4 were derived from the S1 chain, including the signal sequence. Spike pools 5 to 7 were derived from the S2 chain (STAR Methods; Figure 1C).
Furthermore, to ensure that we could differentiate individuals who had prior exposure to SARS-CoV-2 infection, the presence of SARS-CoV-2 membrane- and nucleoprotein-specific T cells were determined.[25] We detected 24 individuals who tested positive for at least one of the three peptide pools (membrane, NP1, and NP2). They were classified as vaccination in SARS-CoV-2 convalescents in further analyses (Figure S1).
Quantification of humoral and cellular Spike-specific immune responses
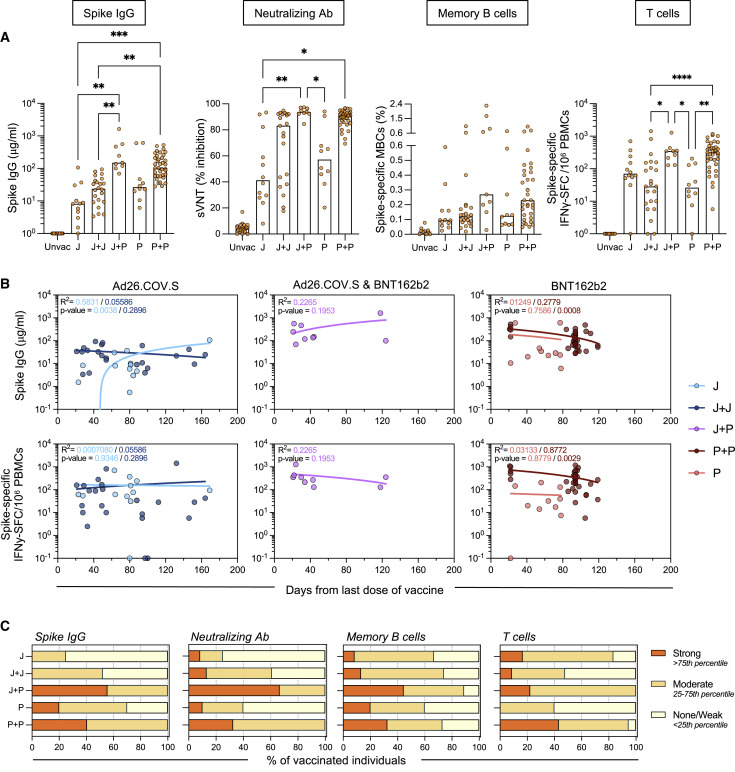
We first quantified Spike-specific humoral and cellular immune responses in the five groups of vaccinated naive individuals (Figure 2 A). Vaccine boosts increased the overall profile of humoral immune responses in all individuals irrespective of their first vaccination (J or P). In individuals vaccinated with Ad26.COV2.S, both homologous (J+J) and heterologous (J+P) vaccination increased the quantity of total Spike IgG and neutralizing antibodies (sVNT). However, heterologous (J+P) vaccination induced a higher quantity of anti-Spike IgG and IgA antibodies than did homologous (J+J) vaccination (Figures 2A and S2A). The level of neutralizing antibodies was also higher in heterologous versus homologous vaccinated individuals even though it did not reach statistical significance. Remarkably, all heterologous vaccinated individuals (9/9) had neutralizing antibodies that achieved more than 80% of inhibition in the sVNT. In contrast, 7/21 of homologous vaccinated (J+J) had sVNT levels below 50%. Analysis of the frequency of circulating Spike-specific memory B cells was instead poorly indicative of the level of antibodies detected. Many individuals showed no or minimal increase of Spike-specific memory B cell frequency after the booster, yet individuals with a higher frequency were among the heterologous (J+P) vaccination cohort (Figure 2A).
Figure 2.
Quantification of humoral and cellular Spike-specific immune responses
(A) Spike immunoglobulin G (IgG) antibody, sVNT, Spike-specific memory B cells (MBCs), and T cell responses were tested in five cohorts of naive vaccinated individuals: J (n = 12), J+J (n = 23), J+P (n = 9), P (n = 10), and P+P (n = 37). A naive unvaccinated cohort was used as a control (Unvac; n = 10–22). Bars denote the median value of each group. Each dot represents an individual. Significant differences in each group were analyzed by one-way ANOVA, and the adjusted p values (adjusted for multiple comparison) are shown. No significance is not shown, ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.
(B) Linear regression analysis between neutralizing antibody activity (top panels) or Spike-specific T cell frequency (bottom panels) and time of testing after last vaccine dose (days). Goodness of fit and p values are shown in plots.
(C) Bar graphs show the proportion of vaccinees with varying levels of Spike-IgG antibodies, neutralizing antibodies, and Spike-specific B and T cell frequencies. The type of responders (strong/moderate/none-weak) was expressed as a fraction of the number of vaccine recipients in each cohort. Type of responders were determined by percentile score calculated from all the vaccinees (n = 87–91).
The analysis of Spike-specific T cells showed a higher frequency in heterologous (J+P) versus homologous (J+J) vaccine recipients (347.5 versus 152 SFC/106 PBMC). This was similar to the level observed in double-dose BNT161b2-vaccinated individuals. Notably, while Spike-specific T cells were clearly detected in 11/12 individuals vaccinated with a single dose of Ad26.COV2.S, 10/23 individuals who received a homologous (J+J) booster displayed a weak level of Spike-specific T cells (<30 SFC/106 PBMC) with a frequency similar to that observed in single-dose BNT162b2-vaccinated individuals (Figure 2A). These J+J individuals with weak Spike-specific T cells were of different ages (Table S1), ruling out that age was the cause of the observed low response to the homologous boost.
To ensure that differences in time of sampling after vaccination did not interfere with the observed trends, we investigated the effect of time after vaccination on immunogenicity by plotting the Spike-IgG quantity and the frequency of Spike-specific T cells against the day of testing after the last dose of vaccine (Figure 2B). Overall, as has been reported in previous studies, the Spike-specific T cell frequency and neutralizing antibodies in vaccinated individuals were not significantly reduced over time at least in the first 3–4 months post-vaccination.26 , 27 Importantly, even though we tested the majority of individuals boosted with the heterologous (J+P) vaccine within 30–40 days, Spike-IgG quantity and T cell frequency remained high in the individuals tested at day 120 post-second dose (Figure 2B). Moreover, the individuals vaccinated with a single dose of Ad26.COV2.S or with the homologous boost (J+J) displayed a pattern of Spike-IgG and T cells that was not influenced by the time of testing (Figure 2B). We also analyzed Spike-specific T cell frequency in relation to the age and sex of the vaccine recipients (Figure S3). Undetectable/low frequency of Spike-specific T cells was observed in the homologous vaccinated individuals who were above the age of 50 years. However, individuals of similar age (above 50 years old) in the heterologous (J+P) vaccination cohort all displayed a high frequency of Spike-specific T cells (>100 SFC/106 PBMC). Finally, the level of antibodies and T cell responses to the different vaccine regimens were categorized into none/weak, moderate, or strong based on their percentile ranking (Figure 2C). The heterologous (J+P) vaccination cohort had the highest proportion of strong responders both in antibodies and T cell responses. Instead, the homologous (J+J) group had the highest proportion of weak responders for T cells, lower than in single-dose Ad26.COV2.S-vaccinated and equivalent to single-dose BNT162b2-vaccinated individuals.
Qualitative analysis of humoral and cellular Spike-specific immune responses
The ability to produce a polyclonal antibody response targeting different regions of the Spike protein is essential to maintain the protective efficacy of humoral immunity against SARS-CoV-2 variants.16 , 28 , 29 Similarly, T cell responses targeting multiple sites of Spike will reduce the chances of viral variants escaping T cell recognition.16 , 30 Therefore, we analyzed qualitative aspects of both humoral and cellular immunity induced by the different vaccination strategies.
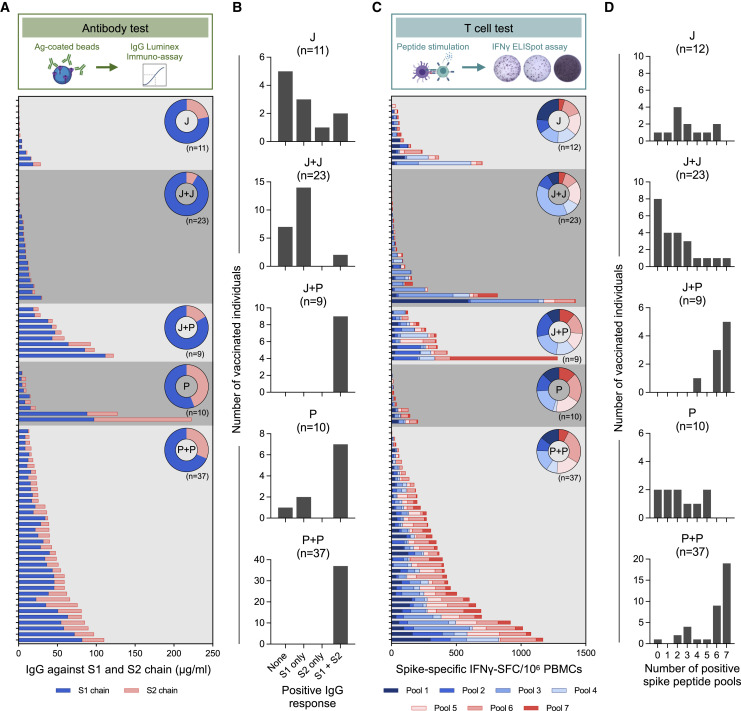
First, we analyzed the breadth of antibody responses. Antibodies (IgG and IgA) against the S1 (containing RBD) and the S2 regions of Spike were quantified. Homologous or single-dose BNT162b2 vaccination elicited antibodies targeting both chains of Spike, while Ad26.COV2.S vaccination mounted an antibody response targeting preferentially the S1 chain (Figures 3 A and S2B). However, the heterologous (J+P) vaccination appears to broaden the antibody repertoire against Spike since all of the heterologous vaccinated individuals (9/9) had antibodies against both domains, while only 2/23 of homologous (J+J) vaccinated individuals displayed such antibody diversity (Figures 3B and S2B).
Figure 3.
Qualitative profile of Spike-specific humoral and cellular immune responses
(A) Stacked bars represent IgG antibody titers against the S1 (blue) and S2 (pink) chains of SARS-CoV-2 Spike antigen. Each column represents an individual. Donut plots represent the mean of percentage of IgG antibodies against S1 or S2.
(B) Frequency of Spike-specific IgG antibodies recognizing none, S1, S2, or both S1 and S2 regions of Spike. IgG titers >1.35 μg/mL were considered positive.
(C) Stacked bars represent frequency of IFN-γ-spot-forming cells (SFCs) reactive to the individual Spike peptide pools (1 to 7) in each vaccinee. Donut charts represent the percentage mean of IFN-γ-SFCs reactive to the individual Spike peptide pools.
(D) Bar graphs show the frequency of vaccinees with varying breadths of Spike-specific T cell responses determined by the number of positive Spike peptide pools. Responses >7.5 SFC/106 PBMCs were considered positive.
The analysis of the breadth of the Spike-specific T cells also confirmed the ability of the heterologous vaccination to broaden the immune response against Spike. Figure 3C shows the frequencies of Spike-specific T cells reactive to the distinct peptide pools covering the different regions of Spike (Figure 1C). While heterologous (J+P) vaccination expanded a population of T cells able to recognize both S1 and S2 regions of Spike (a similar pattern was observed in the homologous BNT162b2 vaccination group), homologous (J+J) and single-dose (J) vaccination induced T cells primarily targeting the S1 chain.
The number of Spike-peptide pools recognized by T cells was also different (Figure 3D). Individuals on the J+P vaccine regimen had T cells recognizing at least four Spike-specific peptide pools. Furthermore, 8/9 had highly multi-specific T cells recognizing six or seven different peptide pools. In contrast, a homologous Ad26.COV2.S booster did not expand the ability of Spike-specific T cells to recognize different regions (Figure 3D). Only 4/23 individuals on homologous J+J had T cells recognizing four or more distinct regions of Spike. A similar pattern was observed in single-dose Ad26.COV2.S-vaccinated individuals. Of concern, 8/23 homologous (J+J) vaccinated individuals did not develop a T cell response against any region of Spike (<7.5 SFC/106 PBMC). In contrast, this was observed in only 1/12 single-dose Ad26.COV2.S vaccine recipients.
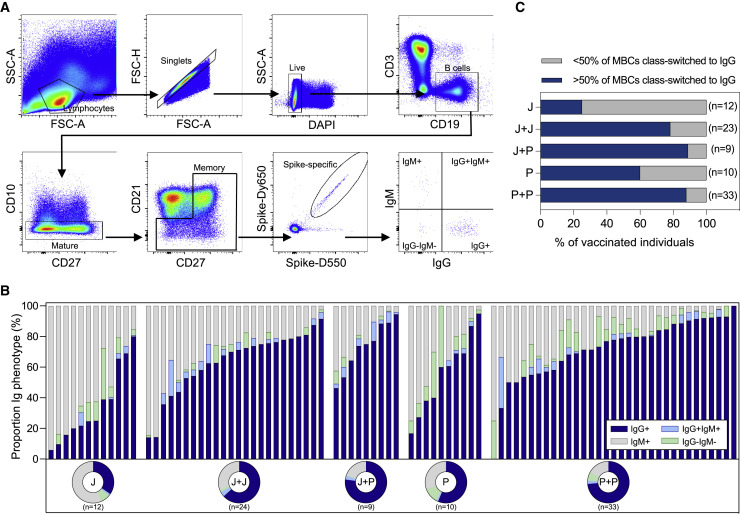
Class switching of Spike-specific memory B cells
High affinity antibodies are produced by memory B cells (MBCs) upon re-encounter with viral antigen.31 , 32 Particularly, class-switched IgG+ MBCs are responsible for durable humoral responses.33 , 34 Having observed that a subset of homologous J+J vaccine recipients had none/weak antibody and T cell responses (Figure 2C), we were prompted to analyze the profile of Spike-specific B cell maturation.
The proportion of class-switched IgG+ Spike-specific MBCs is shown in Figure 4 . In all of the cohorts, we observed a vast heterogeneity in the proportion of class-switched MBCs among the individuals. Both homologous (J+J; P+P) and heterologous (J+P) booster vaccination increased the proportion of class-switched Spike-specific MBCs compared with a single dose of Ad26.COV2.S (J) or BNT162b2 (P) (Figure 4B). However, there was no significant difference observed between homologous and heterologous boosting (Figure 4C).
Figure 4.
Phenotypic analysis of immunoglobulin isotypes of Spike-specific memory B cells
(A) Representative gating strategy of Spike-specific MBCs expressing different Ig types.
(B) Stacked bars represent the frequency of indicated Ig isotypes expressed on Spike-specific MBCs. Corresponding donut plots represent the mean proportion of the four Ig categories (IgG+, IgG–IgM–, IgG+IgM+, and IgM+).
(C) Frequency of individuals with >50% of MBCs class-switched to IgG (blue).
Characterization of Spike-specific humoral and cellular immune responses in vaccinated SARS-CoV-2 convalescents
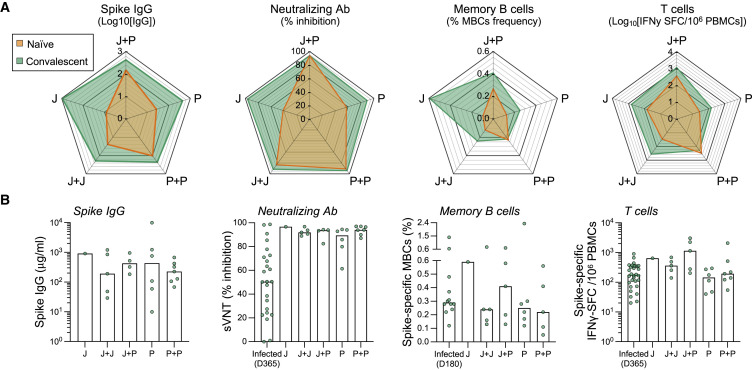
The presence of SARS-CoV-2 membrane- and nucleoprotein-specific T cells allowed us to identify a total of 23 individuals who were vaccinated with different regimens but were likely infected by SARS-CoV-2 before or during the vaccination regimens.25 The limited number of these individuals categorized within the different vaccination strategies did not allow us to perform statistically significant comparisons. Note that in the cohort of single-dose Ad26.COV2.S (J) individuals, there was only one convalescent individual, hence any measurement of “possible boosting effect” by the second dose could not be performed. Nevertheless, we compared the quantity of antibodies (total anti-Spike IgG, neutralizing antibodies [sVNT]) and Spike-specific T cells induced by the different vaccination regime in naive versus convalescent individuals (Figure 5 A). We observed that all convalescent individuals elicited a stronger humoral and cellular immunity than naive individuals irrespective of the vaccination regime (Figure 5A), in line with recent data.35, 36, 37
Figure 5.
Spike-specific humoral and cellular immune response in convalescent vaccinated individuals
Spike IgG antibody, sVNT, Spike-specific MBCs, and T cell responses were tested in five cohorts of naive vaccinated individuals: J (n = 12), J+J (n = 23), J+P (n = 9), P (n = 10), and P+P (n = 37). Same analysis was carried out in SARS-CoV-2 convalescent vaccinated individuals: J (n = 1), J+J (n = 5), P (n = 6), P+P (n = 7), and J+P (n = 5). A convalescent unvaccinated cohort was used as a control (Infected; n = 10–30).
(A) The four radar plots show, respectively, the median levels of Spike IgG antibody titers, neutralizing antibodies, frequency of Spike-specific memory B cells and T cells in convalescent (green; n = 24), and naive (orange; n = 87–91) vaccinated individuals. Vaccination strategies (J, J+J, J+P, P, and P+P) are indicated in the vertex of each pentagon.
(B) Quantities of Spike IgG antibody, neutralizing antibodies, and spike-specific memory B cells and T cells from convalescent vaccinated individuals. Bars denote the median value of each group. Each dot represents an individual. Significant differences in each group were analyzed by one-way ANOVA, and the adjusted p values (adjusted for multiple comparison) are shown. No significance is not shown; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.
In addition, the different vaccination strategies in convalescent individuals boosted neutralizing antibodies above 90% inhibition compared with the 50% inhibition present in non-vaccinated convalescents 1 year post-infection. In contrast, their efficacy in boosting cellular immunity was minimal. The frequency of Spike-specific T cells was marginally increased (J+J) or even lower (P and P+P) than their level detected in unvaccinated convalescent individuals (Figure 5B). However, a high frequency of Spike-specific T cells was detected in heterologous vaccinated individuals. 3 out of 5 individuals in this group displayed a quantity of Spike-specific T cells exceeding 1,000 SFC/106 PBMC.
Discussion
Our study provides information on how to boost Spike-specific humoral and cellular immunity in individuals vaccinated with a single dose of Ad26.COV2.S.
Heterologous booster vaccination with a dose of BNT162b2 resulted in elevated titers of anti-Spike IgG and neutralizing antibodies and a high frequency of Spike-specific T cells in all of the individuals tested.
In addition, heterologous vaccination expanded the breadth of both humoral and T cell immunity. The results of Spike-specific T cells were particularly robust since we observed that 8 out of 9 of the heterologous vaccinated individuals possessed T cells widely scattered along the whole length of the Spike protein. The immunological correlates of protection induced by the vaccines are still only hypothesized.38 Thus, we cannot conclude that the enhanced immunogenicity of heterologous vaccination will translate in a superior protective efficacy. However, it is likely that future prospective trials of vaccine efficacy will find that the quantity and quality of humoral and cellular immunity directly translate into protective efficacy against SARS-CoV-2 infection and COVID-19 disease.
Our observation that a heterologous vaccination strategy in Ad26.COV2.S-vaccinated individuals is more immunogenic than a homologous boost was also confirmed in convalescents and is in line with the results obtained in individuals vaccinated with ChadOx1 nCov-19, another adenoviral-based vaccine. Heterologous vaccination after a single dose of ChadOx1 nCov-19 was recommended in several countries due to the problem of intermittent supplies and of rare thrombotic events associated with ChadOx1 nCov-19.39 Analysis of immunogenicity in individuals boosted with mRNA-based vaccines after a single dose of ChadOx1 nCov-19 revealed an enhanced quantitative profile of antibody and T cell responses compared with those who received a homologous booster vaccination.18 , 19 The superior immunogenicity of heterologous vaccination with a combination of vaccines utilizing different expression vectors 40 has also been seen in other vaccination strategies against different viruses (Ebola, human immunodeficiency virus, hepatitis B virus 41, 42, 43) and other pathogens (malaria, tubercolosis 44 , 45). This further suggests that heterologous prime-boost vaccination strategies involving different types of vaccines should be considered. For instance, using an mRNA-based followed by an adenoviral-vector-based vaccine might be able to delay the reduction of anti-Spike humoral immunity observed after homologous mRNA vaccination5 since such a strategy has been reported to induce improvements in cellular immunity compared with a homologous mRNA boost.22 , 46
Importantly, although we observed that homologous vaccination with Ad26.COV2.S enhances the quantity of antibody production, only heterologous boosting expanded antibodies and T cells able to recognize the S2 region of Spike, in addition to the S1 region. The ability of BNT162b2 to broaden the humoral and cellular Spike-specific repertoire is a likely reflection of the structural differences of the Spike proteins synthesized by the two different vaccines. Ad26.COV2.S encodes a prefusion-stabilized Spike with a mutation in both the furin cleavage site and two consecutive proline substitutions.10 This results in a more stable Spike protein that preferentially triggers the production of antibodies targeting mainly the S1 chain. In contrast, BNT162b2 encodes for a Spike protein that has stabilizing mutations in two prolines but still contains its furin cleavage site that allows for the production of separate S1 and S2 chains.47
We also noted that a second dose of Ad26.COV2.S did not enhance the quantity and breadth of Spike-specific T cells. More than one-third (8/23) of individuals boosted with the homologous Ad26.COV2.S vaccine did not display detectable Spike-specific T cell responses. This proportion of individuals with a very weak T cell response was higher than in individuals who received a single dose of Ad26.COV2.S (1/12).
The inability of a homologous second dose Ad26.COV2.S to boost cellular immunity was also observed in non-human primates48 as well as in individuals receiving homologous ChadOx1 nCov-19 vaccination,19 , 20 and it is likely primarily caused by the induction of a robust immunity against the adenoviral vector, which may potentially reduce or perhaps even abort the expression of the Spike protein.49 , 50 However, the low level of Spike-specific T cells detected here in the individuals with the Ad26.COV2.S homologous boost was unexpected and calls for a careful evaluation of such a strategy in individuals who already received a single dose of Ad26.COV2.S.
Note that these individuals were studied at a median of 80 days post-vaccination, while the ones receiving the Ad26.COV2.S homologous boost were analyzed at a median of 50 days post-second dose. As such, the dynamic process of waning of T cell responses might not explain the low Spike-specific T cell frequency detected in the Ad26.COV2.S homologous boost. Furthermore, while the individuals who received the Ad26.COV2.S homologous boost were on average older (age 51.2 ± 14.2) than the ones receiving the single Ad26.COV2.S dose (age 39.2 ± 12), low Spike-specific T cells were found in individuals ranging from 31 to 75 years of age.
In animals vaccinated with two doses of Ad26.COV2.S, the Spike-specific T cell response was found to be more stable.48 Thus, a prospective parallel analysis of both humoral and cellular immune parameters in larger group of Ad26.COV2.S-vaccinated individuals is needed to confirm these data and possibly understand the real causes.
The discrepancy between levels of Spike-specific humoral and cellular immune responses has been also frequently observed in SARS-CoV-2 convalescents and vaccine recipients (reviewed in Bertoletti et al.51 and Le Bert et al. 52). If we exclude the early phases of convalescence and vaccination, the level of Spike-specific antibodies and T cells appears independently regulated in numerous studies.26 , 27 , 53 As such, the data gathered here further support the concept that the immunogenicity of vaccines should be comprehensively evaluated in its cellular and humoral branches. Humoral analysis alone cannot be used to evaluate overall vaccine responses.
In conclusion, while the Ad26.COV2.S vaccine has been initially proposed as a single-dose vaccine, the progressive reduction of its protective efficacy against SARS-CoV-2 infection over time have warranted a better definition of the best boosting strategy.54 Here, we provide data that demonstrate the enhanced immunogenicity of heterologous versus homologous boost vaccination in Ad26.COV2.S vaccine recipients. Despite representing a minority within the vaccinated individuals worldwide, the Ad26.COV2.S vaccine recipients deserve information that can guide their future vaccination choices.
Limitations of study
There are some important limitations in our study. There is heterogeneity in the timing between first and second vaccine dosing in the homologous and heterologous vaccinations. Furthermore, the cross-sectional nature of the study did not allow for the precise evaluation of the modifications of Spike-specific T cell frequency induced by the second dose, and the immunological characterization in the different vaccinated cohorts was performed at different time points after vaccination. Even though we showed that the level of Spike-specific T cells was minimally reduced within the first 6 months after vaccination and did not appear to influence the different pattern of Spike-specific T cells, an analysis at identical time points after boosting is indicated to demonstrate the enhanced immunogenicity of heterologous vaccination and its durability over time. Finally, the limited quantity of PBMCs collected only allowed us to perform an ELISpot analysis of the T cell response, a method that cannot discriminate whether the Spike-specific T cells induced by different vaccination regimens are CD4 or CD8 T cells. Such information would be needed to better characterize the possible further qualitative differences in T cell responses induced by the different vaccination regimens.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 605 anti-human CD3 Antibody | Biolegend | Cat# 317322; RRID: AB_2561911 |
| PE-CF594 Mouse Anti-Human CD10 | BD | Cat# 562396; RRID: AB_11154416 |
| BV510 Mouse Anti-Human CD19 | BD | Cat# 562947; RRID: AB_2737914 |
| BV421 Mouse Anti-Human CD21 | BD | Cat# 562966; RRID: AB_2737921 |
| BV650 Mouse Anti-Human CD27 | BD | Cat# 563228; RRID: AB_2744352 |
| PE/Cyanine7 anti-human CD38 Antibody | Biolegend | Cat# 356608; RRID: AB_2561904 |
| PerCP/Cyanine5.5 anti-human CD40 Antibody | Biolegend | Cat# 334316; RRID: AB_1186044 |
| FITC anti-human CD71 Antibody | Biolegend | Cat# 334104; RRID: AB_2201482 |
| APC-Cy7 Mouse Anti-Human CD69 | BD | Cat# 557756; RRID: AB_396862 |
| BUV737 Mouse Anti-Human CD95 | BD | Cat# 564710; RRID: AB_2738907 |
| Alexa Fluor® 700 anti-human IgD Antibody | Biolegend | Cat# 348230; RRID: AB_2563335 |
| BV786 Mouse Anti-Human IgG | BD | Cat# 564230; RRID: AB_2738684 |
| Brilliant Violet 711 anti-human IgM Antibody | Biolegend | Cat# 314540; RRID: AB_2687215 |
| Anti-human IFN-g coating antibody | Mabtech | Cat# 3420-3-1000; RRID: AB_907282 |
| Anti-human IFN-g biotin | Mabtech | Cat# 3420-6-1000; RRID: AB_907272 |
| Streptavidin Protein, DyLight 550 | ThermoFisher | Cat# 84542 |
| Streptavidin Protein, DyLight 650 | ThermoFisher | Cat# 84547 |
| Biological samples | ||
| Blood from individuals who received SARS-CoV2 vaccine (Pfizer/BNT162b2 and/or Johnson&Johnson/Ad26.COV2.S) | Singapore General Hospital; Queen Mary University of London | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant SARS-CoV-2 Spike His-tag Biotin Protein, CF | R&D Systems | Cat# BT10549 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | ThermoFisher | Cat# L23105 |
| Streptavidin-ALP | Mabtech | Cat# 3310-10-1000 |
| KPL BCIP/NBT Phosphatase substrate | SeraCare | Cat# 5420-0038 |
| 15-mer SARS-COV2 overlapping Spike, Nucleoprotein and Membrane peptides | Genscript | N/A |
| Luminex MagPlex-C Microspheres, Region 033 | Luminex | MC10033-01 |
| Luminex MAGPIX Performance Verification Kit (IVD) | Luminex | MPXIVD-PVER-K25 |
| Luminex MAGPIX Calibration Kit (IVD) | Luminex | MPXIVD-CAL-K25 |
| Luminex MAGPIX Drive Fluid, 4 Pack | Luminex | MPXDF-4PK-1 |
| xMAP Antibody Coupling Kit | Luminex | 40-50016 |
| Critical commercial assays | ||
| cPASS™ SARS-CoV-2 Neutralization Antibody Detection Kit | GenScript | L00847-B |
| Deposited data | ||
| No unique dataset was generated | N/A | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Immunospot software | Cellular Technology Limited | https://immunospot.com/immunospot-software.html |
| FlowJo Software | BD | https://www.flowjo.com/solutions/flowjo/downloads |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antonio Bertoletti (antonio@duke-nus.edu.sg)
Material availability
This study did not generate new unique reagents.
Experimental model and subject details
This study was approved by the NUS Institutional Review Board (NUS-IRB-2021-292), the SingHealth Centralised Institutional Review Board (CIRB ref.: 2018/2387; 2018/3045; 2021/2014) and the Queen Mary University of London Review Board (REC Ref: 20/EE/0154). We recruited 115 study participants who received different vaccination regimens against COVID-19 and blood and serum was taken at various time points. Details are presented in Table 1.
Method details
Peripheral blood mononuclear cell isolation
Peripheral blood of all individuals was collected and peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Paque density gradient centrifugation.
T cell analysis
Peptides
15-mer peptides that are overlapping by 10 amino acids (AA) spanning the entire SARS-CoV-2 Spike protein, Nucleoprotein and Membrane protein were synthesized (Genscript) and pooled into 7, 2 and 1 pools of approximately 40 peptides in each pool, respectively.25 , 26
SARS-CoV-2 Spike-specific T cell quantification
The frequency of SARS-CoV-2-specific T cells was quantified as described previously.55 Briefly, cryopreserved PBMCs were stimulated with peptide pools in an IFN-γ ELISpot assay. ELISpot plates (Millipore) were coated with human IFN-γ antibody overnight at 4°C. 400,000 PBMCs were seeded per well and stimulated for 18h with the distinct peptide pools at 2 μg/ml. The plates were then incubated with human biotinylated IFN-γ detection antibody, followed by Streptavidin-AP and developed using the KPL BCIP/NBT Phosphatase Substrate. To quantify positive peptide-specific responses, 2x mean spots of the unstimulated wells were subtracted from the peptide-stimulated wells, and the results expressed as spot forming cells (SFC)/106 PBMC. Results were excluded if negative control wells had > 30 SFC/106 PBMC or if positive control wells (PMA/Ionomycin) were negative.
Serological analysis
Surrogate Virus Neutralization Test (sVNT)
The sVNT assay is a proxy measurement of antibodies inhibiting SARS-CoV-2 virus binding to the host cell receptor, human angiotensin-converting enzyme 2 (hACE2), and has been shown to correlate closely with antibody neutralization of SARS-CoV-2.56 sVNT was measured using a commercial RBD-hACE2 binding inhibition assay called cPASS (GenScript) as per manufacturer guidelines. Briefly, serum was diluted 1:10 in the kit sample buffer, was mixed 1:1 with HRP-conjugated RBD and incubated for 30 mins at 37°C. RBD-antibody mixtures were then transferred and incubated for 15 mins at 37°C in enzyme-linked immunosorbent assay (ELISA) plates coated with recombinant hACE2 receptor. Following incubation, plates were washed with wash solution, incubated with TMB substrate for 12-15 mins and reaction stopped with stop solution. Absorbance was measured at OD450nm. Percent inhibition of RBD-hACE2 binding was computed using the following equation:
SARS-CoV-2-specific Luminex Antibody assay
Antigen-specific IgG and IgA responses in serum samples were measured using a previously described bead-based immune-assay with some adjustments.55 Briefly, SARS-CoV-2 recombinant proteins Spike, S1 or S2 (AcroBiosystems) were covalently conjugated to Magpix Luminex beads. Antigen-conjugated beads were then blocked with 1% BSA (bovine serum albumin, before being probed with either diluted human serum or antibody standards for 1 hr at 37C. Beads were then washed and probed with either anti-human IgG-PE (Invitrogen) or anti-human IgA-Biotin (Southern Biotech) followed by Streptavidin-PE (Southern Biotech) for measuring human IgG and IgA, respectively. IgG and IgA binding to antigen were measured as Median Fluorescence Intensity (MFI) using a Magpix instrument (Luminex). MFI values of serum samples were converted to antibody quantity (i.e., g/ml) using anti-Spike IgG and IgA antibody standards (AcroBiosystems). Serum samples were first tested at dilutions 1:100 and 1:2000, if MFI values were above the range of the antibody standards, then serum samples were further diluted to 1:10,000 and tested.
B cell analysis
Detection of SARS-CoV2-specific Memory B cells
To detect SARS-CoV-2-specific memory B cells, biotinylated protein antigens were individually multimerized with streptavidin (SA) fluorophore conjugates, as described here.57 Briefly, full length Spike protein (RnD Systems) was multimerized with SA-Dylight 550 or SA-Dylight 650 (Thermo Fisher Scientific) in buffer containing 50/50 mixture of 2% FBS and Brilliant Buffer (BD Bioscience) for 1 hour at 4°C. Spike protein (RnD Systems) and SA-Dy550/SA-Dy650 were mixed at a 10:1 mass ratio (∼4:1 molar ratio) freshly before every staining. Cells were first stained for 10 minutes at RT with Live/Dead Fixable Blue Stain Reagent (Life Technologies). Subsequently cells were stained with 50μl of antigen probe cocktail containing 100ng of Spike per probe (i.e., 100ng of Spike-Biotin/SA-Dylight 550 and 100ng of Spike-Biotin/SA-Dylight 650) for 1 hour at 4°C. In parallel, SA-Dylight 550 and SA-Dylight 650 probes (100ng each) not conjugated to protein were used as decoy probes to gate out non-specific streptavidin-binding B cells. Next, cells were stained with an antibody cocktail (against CD3, CD10, CD19, CD21, CD27, CD38, CD40, CD69, CD71, CD95, IgD, IgG, IgM, see Table S2) for 30 mins at 4°C. Finally, cells were washed and fixed with 1% formaldehyde before acquisition on a LSR-Fortessa flow cytometer (BD). Analysis of flow cytometry data was performed using FlowJo software, version 10 (BD).
Quantification and statistical analysis
All statistical analyses were performed in Prism (GraphPad Software). Where applicable, the statistical tests used, the definition of center and statistical significance were indicated in the figure legends. In all instances, “n” refers to the number of individuals analyzed.
Acknowledgments
We would like to thank all of the clinical and nursing staff who provided care to all vaccinated individuals and aided in coordinating collection of the samples. We are also grateful to all clinical trial coordinators and staff at Barts Vaccine Center for their assistance in the recruitment organization of vaccine recipients. This study is partially supported by the Singapore Ministry of Health’s National Medical Research Council under its COVID-19 Research Fund (COVID19RF3-0060, COVID19RF-001, and COVID19RF-008) and The Medical College St. Bartholomew’s Hospital Trustees – Pump Priming Fund for SMD COVID-19 Research. In addition, U.S.G. is supported by grant funding from an Academy of Medical Sciences Starter Grant (SGL021/1030), Seedcorn funding from Rosetrees/Stoneygate Trust (A2903), and an Early Career Research Award from The Medical Research Foundation (MRF-044-0004-F-GILL-C0823). P.T.F.K. is supported by funding from Barts Charity Project Grants (723/1795 and MGU/0406) and an NIHR Research for Patient Benefit award (PB-PG-0614-34087).
Author contributions
Method used to assign authorship order among co-first authors was based on experimental contribution. U.S.G., N.L.B., P.T.F.K., and A.B. designed the experiments. U.S.G., J.E.A., C.U., and J.G.H.L. aided in the recruitment of the vaccinated individuals. N.K.H.K., J.M.E.L., and N.T. performed the B and T cell experiments. E.E.O., R.d.A., and J.Z.N.T. performed the antibody experiments. N.K.H.K., J.M.E.L., U.S.G., E.E.O., J.G.H.L., N.L.B., P.T.F.K., and A.B. analyzed and interpreted all the data. N.K.H.K. and J.M.E.L. prepared the figures. N.K.H.K., J.M.E.L., U.S.G., N.L.B., P.T.F.K., and A.B. prepared the first draft and reviewed and edited it. A.B., N.L.B., N.K.H.K., J.M.E.L., and U.S.G. had unrestricted access to all the data. All of the authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility for its content, including the accuracy of the data.
Declaration of interests
N.L.B. and A.B. reported a patent for a method to monitor SARS-CoV-2-specific T cells in biological samples pending. The other authors declare no competing interests.
Published: January 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2021.12.004.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This study did not generate any new codes.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- 1.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Oxford COVID Vaccine Trial Group Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. ENSEMBLE Study Group Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemaitelly H., Tang P., Hasan M.R., AlMukdad S., Yassine H.M., Benslimane F.M., Al Khatib H.A., Coyle P., Ayoub H.H., Al Kanaani Z., et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N. Engl. J. Med. 2021;385:e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 7.Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altmann D.M., Boyton R.J. Comment Waning immunity to SARS-CoV-2 : implications for vaccine booster strategies. Lancet Respir. 2021;2600:10–11. doi: 10.1016/S2213-2600(21)00458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohn B.A., Cirillo P.M., Murphy C.C., Krigbaum N.Y., Arthur W. Breakthrough SARS-CoV-2 infections in 620, 000 U. S. Veterans, February 1, 2021 to August 13, 2021. medRxiv. 2021 doi: 10.1101/2021.10.13.21264966. [DOI] [Google Scholar]
- 10.Bos R., Rutten L., van der Lubbe J.E.M., Bakkers M.J.G., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A.H., Koornneef A., Verwilligen A., et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. Vaccines (Basel) 2020;5:1–11. doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephenson K.E., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., et al. Immunogenicity of the Ad26.COV2.S Vaccine for COVID-19. JAMA. 2021;325:1535–1544. doi: 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barouch D.H., Stephenson K.E., Sadoff J., Yu J., Chang A., Gebre M., McMahan K., Liu J., Chandrashekar A., Patel S., et al. Durable Humoral and Cellular Immune Responses 8 Months after Ad26.COV2.S Vaccination. N. Engl. J. Med. N. Engl. J. Med. 2021;385:951–953. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polinski J.M., Weckstein A.R., Batech M., Kabelac C., Kamath T., Harvey R., Jain S., Rassen J.A., Khan N., Schneeweiss S. Effectiveness of the Single-Dose Ad26.COV2.S COVID Vaccine. medRxiv. 2021 doi: 10.1101/2021.09.10.21263385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia P., Anand S., Han J., Montez-Rath M., Sun S., Shang T., Parsonnet J., Chertow G., Schiller B., Abra G. COVID19 Vaccine Type and Humoral Immune Response in Patients Receiving Dialysis. J. Am. Soc. Nephrol. 2021 doi: 10.1101/2021.08.02/21261516. Published online August 4, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cevik M., Grubaugh N.D., Iwasaki A., Openshaw P. COVID-19 vaccines: Keeping pace with SARS-CoV-2 variants. Cell. 2021;184:5077–5081. doi: 10.1016/j.cell.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer A.J., McKay P.F., Belij-Rammerstorfer S., Ulaszewska M., Bissett C.D., Hu K., Samnuan K., Blakney A.K., Wright D., Sharpe H.R., et al. Heterologous vaccination regimens with self-amplifying RNA and adenoviral COVID vaccines induce robust immune responses in mice. Nat. Commun. 2021;12:2893. doi: 10.1038/s41467-021-23173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt T., Klemis V., Schub D., Mihm J., Hielscher F., Marx S., Abu-Omar A., Ziegler L., Guckelmus C., Urschel R., et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021;27:1530–1535. doi: 10.1038/s41591-021-01464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros-Martins J., Hammerschmidt S.I., Cossmann A., Odak I., Stankov M.V., Morillas Ramos G., Dopfer-Jablonka A., Heidemann A., Ritter C., Friedrichsen M., et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat. Med. 2021;27:1525–1529. doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillus D., Schwarz T., Tober-Lau P., Vanshylla K., Hastor H., Thibeault C., Jentzsch S., Helbig E.T., Lippert L.J., Tscheak P., et al. EICOV/COVIM Study Group Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: a prospective cohort study. Lancet Respir. Med. 2021;9:1255–1265. doi: 10.1016/S2213-2600(21)00357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borobia A.M., Carcas A.J., Pérez-Olmeda M., Castaño L., Bertran M.J., García-Pérez J., Campins M., Portolés A., González-Pérez M., García Morales M.T., et al. CombiVacS Study Group Immunogenicity and reactogenicity of BNT162b2 booster in ChAdOx1-S-primed participants (CombiVacS): a multicentre, open-label, randomised, controlled, phase 2 trial. Lancet. 2021;398:121–130. doi: 10.1016/S0140-6736(21)01420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Shaw R.H., Stuart A.S.V., Greenland M., Aley P.K., Andrews N.J., Cameron J.C., Charlton S., Clutterbuck E.A., Collins A.M., et al. Com-COV Study Group Safety and immunogenicity of heterologous versus homologous prime-boost schedules with an adenoviral vectored and mRNA COVID-19 vaccine (Com-COV): a single-blind, randomised, non-inferiority trial. Lancet. 2021;398:856–869. doi: 10.1016/S0140-6736(21)01694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pozzetto B., Legros V., Djebali S., Barateau V., Guibert N., Villard M., Peyrot L., Allatif O., Fassier J.-B., Massardier-Pilonchéry A., et al. Covid-Ser study group Immunogenicity and efficacy of heterologous ChAdOx1-BNT162b2 vaccination. Nature. 2021 doi: 10.1038/s41586-021-04120-y. Published online October 21, 2021. [DOI] [PubMed] [Google Scholar]
- 24.Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., et al. Heterologous SARS-CoV-2 Booster Vaccinations: Preliminary Report. medRxiv. 2021 doi: 10.1101/2021.10.10.21264827. [DOI] [Google Scholar]
- 25.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan A.T., Lim J.M.E., Le Bert N., Kunasegaran K., Chia A., Qui M.D.C., Tan N., Chia W.N., de Alwis R., Ying D., et al. Rapid measurement of SARS-CoV-2 spike T cells in whole blood from vaccinated and naturally infected individuals. J. Clin. Invest. 2021;131 doi: 10.1172/JCI152379. 2021.06.29.450293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurosaki T., Kometani K., Ise W. Memory B cells. Nat. Rev. Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- 32.Palm A.E., Henry C. Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Front. Immunol. 2019;10:1787. doi: 10.3389/fimmu.2019.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amanna I.J., Carlson N.E., Slifka M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 34.Ogega C.O., Skinner N.E., Blair P.W., Park H.S., Littlefield K., Ganesan A., Dhakal S., Ladiwala P., Antar A.A.R., Ray S.C., et al. Durable SARS-CoV-2 B cell immunity after mild or severe disease. J. Clin. Invest. 2021;131:e145516. doi: 10.1172/JCI145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds C.J., Pade C., Gibbons J.M., Butler D.K., Otter A.D., Menacho K., Fontana M., Smit A., Sackville-West J.E., Cutino-Moguel T., et al. UK COVIDsortium Immune Correlates Network. UK COVIDsortium Investigators Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372:1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prendecki M., Clarke C., Brown J., Cox A., Gleeson S., Guckian M., Randell P., Pria A.D., Lightstone L., Xu X.N., et al. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. Lancet. 2021;397:1178–1181. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozano-Ojalvo D., Camara C., Lopez-Granados E., Nozal P., Del Pino-Molina L., Bravo-Gallego L.Y., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep. 2021;36:109570. doi: 10.1016/j.celrep.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 39.ECDC . 2021. Overview of EU/EEA country recommendations on COVID-19 vaccination with Vaxzevria, and a scoping review of evidence to guide decision-making.https://www.ecdc.europa.eu/en/publications-data/overview-eueea-country-recommendations-covid-19-vaccination-vaxzevria-and-scoping [Google Scholar]
- 40.Woodland D.L. Jump-starting the immune system: prime-boosting comes of age. Trends Immunol. 2004;25:98–104. doi: 10.1016/j.it.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Backes S., Jäger C., Dembek C.J., Kosinska A.D., Bauer T., Stephan A.S., Dišlers A., Mutwiri G., Busch D.H., Babiuk L.A., et al. Protein-prime/modified vaccinia virus Ankara vector-boost vaccination overcomes tolerance in high-antigenemic HBV-transgenic mice. Vaccine. 2016;34:923–932. doi: 10.1016/j.vaccine.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 42.Anywaine Z., Whitworth H., Kaleebu P., Praygod G., Shukarev G., Manno D., Kapiga S., Grosskurth H., Kalluvya S., Bockstal V., et al. Safety and Immunogenicity of a 2-Dose Heterologous Vaccination Regimen With Ad26.ZEBOV and MVA-BN-Filo Ebola Vaccines: 12-Month Data From a Phase 1 Randomized Clinical Trial in Uganda and Tanzania. J. Infect. Dis. 2019;220:46–56. doi: 10.1093/infdis/jiz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baden L.R., Stieh D.J., Sarnecki M., Walsh S.R., Tomaras G.D., Kublin J.G., McElrath M.J., Alter G., Ferrari G., Montefiori D., et al. Traverse/HVTN 117/HPX2004 Study Team Safety and immunogenicity of two heterologous HIV vaccine regimens in healthy, HIV-uninfected adults (TRAVERSE): a randomised, parallel-group, placebo-controlled, double-blind, phase 1/2a study. Lancet HIV. 2020;7:e688–e698. doi: 10.1016/S2352-3018(20)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider J., Gilbert S.C., Blanchard T.J., Hanke T., Robson K.J., Hannan C.M., Becker M., Sinden R., Smith G.L., Hill A.V.S. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat. Med. 1998;4:397–402. doi: 10.1038/nm0498-397. [DOI] [PubMed] [Google Scholar]
- 45.McShane H., Hill A. Prime-boost immunisation strategies for tuberculosis. Microbes Infect. 2005;7:962–967. doi: 10.1016/j.micinf.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Tan C.S., Collier A.Y., Liu J., Yu J., Wan H., McMahan K., He X., Jacob-Dolan C., Chandrashekar A., Sellers D., et al. Ad26.COV2.S or BNT162b2 Boosting of BNT162b2 Vaccinated Individuals. medRxiv. 2021 doi: 10.1101/2021.12.02.21267198. [DOI] [Google Scholar]
- 47.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solforosi L., Kuipers H., Jongeneelen M., Rosendahl Huber S.K., van der Lubbe J.E.M., Dekking L., Czapska-Casey D.N., Izquierdo Gil A., Baert M.R.M., Drijver J., et al. Immunogenicity and efficacy of one and two doses of Ad26.COV2.S COVID vaccine in adult and aged NHP. J. Exp. Med. 2021;218:e20202756. doi: 10.1084/jem.20202756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein N., Bockstal V., Bart S., Luhn K., Robinson C., Gaddah A., Callendret B., Douoguih M. Safety and Immunogenicity of Heterologous and Homologous 2-Dose Regimens of Adenovirus Serotype 26– and Modified Vaccinia Ankara–Vectored Ebola Vaccines: A Randomized, Controlled Phase 1 Study. J. Infect. Dis. 2020:jiaa586. doi: 10.1093/infdis/jiaa586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J.X., Hou L.H., Meng F.Y., Wu S.P., Hu Y.M., Liang Q., Chu K., Zhang Z., Xu J.J., Tang R., et al. Immunity duration of a recombinant adenovirus type-5 vector-based Ebola vaccine and a homologous prime-boost immunisation in healthy adults in China: final report of a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Glob. Health. 2017;5:e324–e334. doi: 10.1016/S2214-109X(16)30367-9. [DOI] [PubMed] [Google Scholar]
- 51.Bertoletti A., Le Bert N., Qui M., Tan A.T. SARS-CoV-2-specific T cells in infection and vaccination. Cell. Mol. Immunol. 2021;18:2307–2312. doi: 10.1038/s41423-021-00743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Bert N., Chia W.N., Wan W.Y., Teo A.K.J., Chong S.Z.-R., Tan N., Tan D.S.C., Chia A., Tan I.B., Kunasegaran K., et al. Widely heterogeneous humoral and cellular immunity after mild SARS-CoV-2 infection in a homogeneous population of healthy young men. Emerg. Microbes Infect. 2021;10:2141–2150. doi: 10.1080/22221751.2021.1999777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chia W.N., Zhu F., Ong S.W.X., Young B.E., Fong S.W., Le Bert N., Tan C.W., Tiu C., Zhang J., Tan S.Y., et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021;2:e240–e249. doi: 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown C.M., Vostok J., Johnson H., Burns M., Gharpure R., Sami S., Sabo R.T., Hall N., Foreman A., Schubert P.L., et al. Outbreak of SARS-CoV-2 Infections, Including COVID-19 Vaccine Breakthrough Infections, Associated with Large Public Gatherings - Barnstable County, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.-C., Syenina A., Zhang S.L., Le Bert N., et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med (N Y) 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.W., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 57.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:1–19. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This study did not generate any new codes.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.