Abstract
Background
Numerous serology assays are available for detection of SARS-CoV-2 antibodies but are limited in that only 1 or 2 target antigen(s) can be tested at a time. Here, we describe a novel multiplex assay that simultaneously detects and quantifies IgG antibodies to SARS-CoV-2 antigens, spike (S), nucleocapsid (N), receptor-binding domain (RBD), and N-terminal domain (NTD) in a single well.
Methods
Sensitivity was determined using samples (n = 124) from confirmed SARS-CoV-2 RT-PCR positive individuals. Prepandemic (n = 100) and non-COVID respiratory infection positive samples (n = 100) were used to evaluate specificity. Samples were analyzed using COVID-19 IgG multiplex serology assay from Meso Scale Discovery (MSD) and using commercial platforms from Abbott, EUROIMMUN, and Siemens.
Results
At >14 days post-PCR, MSD assay displayed >98.0% sensitivity [S 100% (95% CI 98.0%–100.0%); N 98.0% (95% CI 97.2%–98.9%); RBD 94.1% (95% CI 92.6%–95.6%); NTD 98.0% (95% CI, 97.2%–98.9%)] and 99% specificity (95% CI 99.3%–99.7%) for antibodies to all 4 antigens. Parallel assessment of antibodies to more than 1 antigen improved the sensitivity to 100% (95% CI 98.0%–100.0%) while maintaining 98% (95% CI 97.6%–98.4%) specificity regardless of the combinations used. When AU/mL concentrations of IgG antibodies from the MSD assay were compared against the corresponding IgG signals acquired from the single target commercial assays, the following correlations were observed: Abbott (vs MSD N, R2 = 0.73), Siemens (vs MSD RBD, R2 = 0.92), and EUROIMMUN (vs MSD S, R2 = 0.82).
Conclusion
MSD assay offers an accurate and a comprehensive assessment of SARS-CoV-2 antibodies with higher sensitivity and equivalent specificity compared to the commercial IgG serology assays.
Keywords: multiplex immunoassays, immunology, infectious disease, microbiology
Introduction
IMPACT STATEMENT
Herein, we evaluated the performance characteristics of a novel multiplex assay by Meso Scale Discoveries, with abilities to quantitatively measure IgG antibodies to 4 major SAR-CoV-2 antigens in a very small sample volume. Antibody response to SARS-CoV-2 is multifaceted, but most commercial assays are directed only against a single target. Using a cohort of 324 samples, our data showed higher sensitivity and equivalent specificity across all 4 IgG antibodies in comparison to commercial platforms. We believe that this multiplex assay will enable comprehensive understanding of an individual’s antibody signature in the context of immunity to vaccines and emerging SAR-CoV-2 variants.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing COVID-19 was first identified in Wuhan, China, in December 2019 (1). By June 2021, the outbreak had led to >170 000 000 total confirmed cases and >3 600 000 deaths globally (2). The SARS-CoV-2 infection is diagnosed through quantitative RT-PCR analysis using nasopharyngeal, nasal, oral swabs, or saliva. While not suitable for diagnosis, serology tests can be particularly useful in mild or asymptomatic cases where the RT-PCR results may be negative due to low viral load (3). It is also important for seroprevalence studies, in the estimation of vaccine-acquired humoral immunity, in the identification of convalescent plasma donors and for epidemiological studies (4).
The SARS-CoV-2 virus has a trimeric spike (S) protein on the surface and can be cleaved by the host proteases into S1 and S2 subunits. S2 subunit is responsible for membrane fusion and S1 subunit has a unique region called the receptor-binding domain (RBD), which the virus uses to achieve entry into the host cell through recognition of the angiotensin-converting enzyme 2 receptor. The N-terminal domain (NTD) does not bind to the receptor but contains elements for recognition of specific sugar moieties for initial attachment and fusion. The nucleocapsid (N) protein helps in the viral assembly (5).
The antibody response to SARS-CoV-2 infection is nonhomogenous targeting different epitopes of several proteins. The efficiency of serological testing is dependent on the dynamics and kinetics of the diverse SARS-CoV-2 antibody responses (6). A wide range of serological immunoassays have been developed against different SARS-CoV-2 antigens; however, most assays are directed against single targets or utmost 2 targets (7–9). Identification of seropositive individuals using a single cutoff antibody levels is not representative of the entire serological signature of infection. Additionally, all IgG assays that are currently available are qualitative or semiquantitative in nature (10). Quantitative tests are traceable to a specific international standard that ensures the result produced correlates to a numeric quantity. Semiquantitative assays provide an approximate estimation in relative amounts and sometimes traceable to a specific standard to ensure consistent results. Different assays detecting different antigenic targets and current lack of a standard reference material for detecting antibodies against individual targets pose challenges while comparing results from 2 or more SARS-CoV-2 IgG assays.
In this study, we evaluated the performance characteristics of a novel multiplex assay. The assay has the ability to quantitatively measure antibodies to multiple SARS-CoV-2 proteins simultaneously. We conducted an in-depth evaluation of the diagnostic characteristics of the 4 major IgG antibodies to the SAR-CoV-2 antigens individually and combined. To understand whether the multiplex assay demonstrated characteristics in par with the commercially available IgG serology assays, we compared the performance of this assay to 3 commercial assays, Abbott (N), EUROIMMUN (S), and Siemens (RBD), which are all directed against a single SARS-CoV-2 target antigen.
Methods and Materials
Samples
A total of 124 samples collected from 81 individuals confirmed positive for SARS-CoV-2 infection by an Food and Drug Administration–authorized RT-PCR assay at ARUP Laboratories were used to evaluate clinical sensitivity. Samples were collected 0 to 36 days post-PCR testing. Specificity was evaluated using samples collected from 100 healthy donors before August 2019 (prepandemic). Of these, 80 were from adults (age range 20–68 years), and 20 were from pediatric patients (age range 2–18 years). To assess cross-reactivity (analytical specificity), a cohort of 100 samples, which included samples collected from individuals with respiratory illnesses other than COVID-19 (n = 82) as well as individuals positive for rheumatoid factor (n = 12) and heterophile antibody (n = 6), were used (analytical specificity). All samples were collected, handled, and deidentified in accordance with University of Utah Institutional Review Board (protocol 00007275).
IgG Antibody Testing
A Meso Scale Discovery (MSD) COVID-19 serology kit (Meso Scale Discovery) was used to simultaneously detect and quantify anti-IgG binding antibodies to the full-length S, N, RBD, and NTD of the SARS-CoV-2 virus. The assay uses an electrochemiluminescent detection system that reads signals on a solid-phase antigen-printed 96-well plate. Assay was performed as described in the package insert (11). Briefly, diluted sample serum, reference standards, and controls were incubated in the wells. After washing, kit antihuman IgG antibody was added. Following another incubation and washing steps, kit buffer was added, and plates immediately read using the MESO® QuickPlex SQ 120 reader. The assay includes a reference standard that is calibrated against the WHO International Standard (NIBSC code: 20/136) for a quantitative detection all 4 SARS-CoV-2 antibodies.
The MSD IgG assay was compared to 3 other commercial IgG serology platforms: Abbott SARS-CoV-2 IgG assay, performed on the Abbott Architect i2000 (Abbott Laboratories Inc.); the EUROIMMUN Anti-SARS-CoV-2 IgG ELISA Assay (EUROIMMUN US), performed both manually and automated on the Dynex Agility (Dynex Technologies); and the Siemens SARS-CoV-2 IgG assay, performed on the ADVIA Centaur XPT (Siemens Medical Solutions). All assays were performed following manufacturer’s directions.
Statistical Analysis
Statistical analysis was performed using Excel (AnalyzeIT, version 5.66) and GraphPad Prism (version 8.4.3). ROC analysis and Youden J statistics was used to establish specificity (≥99%) optimized cut points for the MSD assay. The area under the ROC curve (AUC), predictive values and the likelihood ratios were determined via ROC analysis using the Excel AnalyzeIT software. Predictive values were determined using a presumed prevalence of 5% COVID-19 to account for the low prevalence settings and high false positivity in certain areas of the United States. Positive thresholds for the commercial assays, EUROIMMUN, Abbott, and Siemens, were adopted from the manufacturer’s package inserts. Sensitivity was calculated using the SARS-CoV-2 RT-PCR as the reference method and specificity was assessed using results from the healthy and other infection groups. Overall agreement was determined from the proportion of total positive and negative concordant results. Simple linear regression analysis was used to determine the correlation coefficients.
Results
ROC Curve Analysis and Cutoff Estimation of IgG to Each SARS-CoV-2 Antigen
To evaluate the ability of the assay to discriminate between the COVID-19 patients and healthy controls, ROC curves were plotted (Fig. 1, A and B). All 4 IgG to the SARS-CoV-2 antigens displayed high area under the curve values indicating high levels of accuracy in the delineation of positive and negative results. Antibodies to S [AUC 0.962 (95% CI 0.938–0.986)] exhibited the highest accuracy followed by N [AUC 0.934 (95% CI, 0.902–0.967)], NTD [AUC 0.931 (95% CI 0.895–0.967)], and RBD [AUC: 0.912 (95% CI 0.873–0.952)] antigens (Fig. 1, A). The trade-off between sensitivity and specificity was assessed by Youden J statistics and cut points (S > 869, N > 7225, RBD > 592, NTD > 19) that resulted in a specificity of at least 99.0% was chosen (Fig. 1, B). These specificity optimized cut points were selected keeping in mind of the potential false positivity in the low prevalence settings.
Fig. 1.
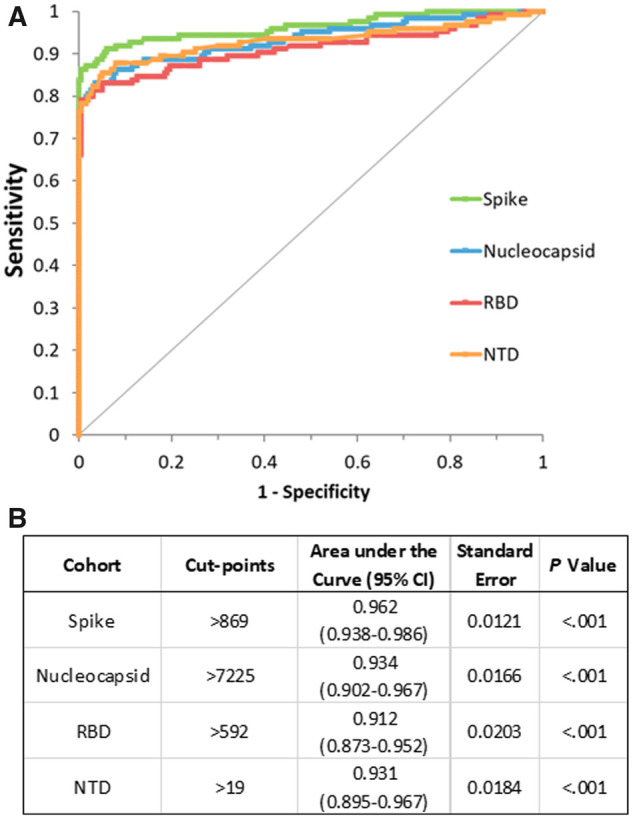
ROC analysis for the IgG antibodies to 4 SARS-CoV-2 antigens detected by MSD assay. (A) ROC plots for the IgG antibodies to SARS-CoV-2 antigens. (B) Cut points that resulted in a specificity of at least 99.0% was chosen. AUC values for each individual antibodies, corresponding standard errors and P values are listed. P denotes statistical significance of difference from AUC = 0.5 or line of no discrimination based on z test, calculated using the Excel AnalyzeIT software.
Evaluation of Sensitivity at Different Time Points after the Initial COVID-19 Diagnosis
At the established cut points, sensitivity was assessed on a total of 124 specimens collected at different time points after a positive COVID-19 diagnosis confirmed by RT-PCR. At >14 days, which is within the CDC-recommended time frame for serology testing, the S component of the MSD assay displayed a 100% sensitivity, N antigen showed a sensitivity of 98.0% (95% CI 97.2%–98.9%), RBD exhibited a sensitivity of 94.1% (95% CI, 92.6%–95.6%) and NTD displayed a 98.0% (95% CI 97.2%–98.9%) sensitivity. The sensitivities observed at earlier time points are listed in the Table 1. In samples collected between 7 and 14 days after a positive COVID-19 diagnosis by RT-PCR, S, N, and RBD components of the assay displayed sensitivities >90% except NTD, which displayed a sensitivity of 88.9% (95% CI, 86.5%–91.3%) (Table 1). Sixteen out of 37 samples collected less than 7 days post-PCR confirmation tested negative to antibodies against the S component of the MSD assay resulting in a sensitivity of 56.8% (95% CI 52.0%–61.5%) (Table 1). Components N, RBD, and NTD of the MSD assay displayed a < 41% sensitivity at these early time-points (Table 1).
Table 1.
Diagnostic accuracy of MSD’s SARS-CoV-2 IgG multiplex assay in COVID-19 patients and healthy controls.
| Cohort | Positive MSD | Negative MSD | Sensitivity % (95% CI) | Specificity % (95% CI) | Positive likelihood Ratio | Negative likelihood Ratio | PPVa % at 5% prevalence | NPVb % at 5% prevalence | |
|---|---|---|---|---|---|---|---|---|---|
| MSD S (cutoff: >869) | |||||||||
| Total (n = 324) | 107/124 | 199/200 | 86.3 (84.8–87.7) | 99.5 (99.3–99.7) | 172.6 | 0.14 | 99 | 92 | |
| PCR Positive (n = 124) | <7 days post PCR + (n = 37) | 21/37 | 16/37 | 56.8 (52.0–61.5) | |||||
| 7–14 days post PCR + (n = 36) | 35/36 | 1/36 | 97.2 (96.0–98.4) | ||||||
| 14 days post PCR + (n = 51) | 51/51 | 0/51 | 100 (98.0–100) | ||||||
| Healthy (n = 100) | 0/100 | 100/100 | 100 (98.0–100) | ||||||
| Other Infections (n = 100) | 1/100 | 99/100 | 99.0 (98.6–99.4) | ||||||
| MSD RBD (cutoff: >592) | |||||||||
| Total (n = 324) | 98/124 | 199/200 | 79.0 (77.2–80.8) | 99.5 (99.3–99.7) | 158.1 | 0.21 | 99 | 88 | |
| PCR Positive (n = 124) | <7 days post PCR + (n = 37) | 15/37 | 22/37 | 40.5 (35.0–46.1) | |||||
| 7–14 days post PCR + (n = 36) | 33/36 | 3/36 | 91.7 (89.6–93.8) | ||||||
| 14 days post PCR + (n = 51) | 50/51 | 1/51 | 98.0 (97.2–98.9) | ||||||
| Healthy (n = 100) | 1/100 | 99/100 | 99.0 (98.6–99.4) | ||||||
| Other Infections (n = 100) | 0/100 | 100/100 | 100 (98.0–100) | ||||||
| MSD N (cutoff: >7225) | |||||||||
| Total (n = 324) | 97/124 | 199/200 | 78.2 (76.4–80.1) | 99.5 (99.3–99.7) | 156.5 | 0.22 | 99 | 88 | |
| PCR Positive (n = 124) | <7 days post PCR + (n = 37) | 14/37 | 23/37 | 37.8 (32.2–43.5) | |||||
| 7–14 days post PCR + (n = 36) | 35/36 | 1/36 | 97.2 (96.0–98.4) | ||||||
| 14 days post PCR + (n = 51) | 48/51 | 3/51 | 94.1 (92.6–95.6) | ||||||
| Healthy (n = 100) | 1/100 | 99/100 | 99.0 (98.6–99.4) | ||||||
| Other Infections (n = 100) | 0/100 | 100/100 | 100 (98.0–100) | ||||||
| MSD NTD (cutoff: >19) | |||||||||
| Total (n = 324) | 97/124 | 199/200 | 78.2 (76.4–80.1) | 99.5 (99.3–99.7) | 156.5 | 0.22 | 99 | 88 | |
| PCR Positive (n = 124) | <7 days post PCR + (n = 37) | 15/37 | 22/37 | 40.5 (35.0–46.1) | |||||
| 7–14 days post PCR + (n = 36) | 32/36 | 4/36 | 88.9 (86.5–91.3) | ||||||
| 14 days post PCR + (n = 51) | 50/51 | 1/51 | 98.0 (97.2–98.9) | ||||||
| Healthy (n = 100) | 0/100 | 100/100 | 100 (98.0–100) | ||||||
| Other Infections (n = 100) | 1/100 | 99/100 | 99.0 (98.6–99.4) | ||||||
Positive predictive value, calculated in a presumed 5% prevalence setting.
Negative predictive value, calculated in a presumed 5% prevalence setting.
Evaluation of Specificity and Other Assay Performance Characteristics
The clinical specificity was evaluated using 100 samples from healthy individuals whose samples were collected prior to the pandemic. At the established cut points, S and NTD antigens of the assay showed 100% specificity and N and RBD antigens each showed 99% (95% CI 98.6–99.4) specificity (Table 1). To evaluate the analytical specificity, samples positive for either other respiratory infections or that are positive for some of the common analytical interferences such as heterophile antibodies and rheumatoid factors were tested. MSD N and RBD antigens showed 100% specificity, and S and NTD antigens each showed 99% (95% CI 98.6–99.4) specificity at the cut points established (Table 1). The positive predictive values, negative predictive values, and likelihood ratios for the detection of antibodies against all 4 SARS-CoV-2 target antigens are summarized in Table 1. Consistent with the high specificity, all 4 antibodies displayed an estimated positive predictive value of 99% at a set prevalence of 5% (Table 1). The S component of the assay displayed a high positive likelihood ratio of 172.6 and a negative likelihood ratio of 0.14, RBD showed a positive likelihood ratio of 158.1 and negative likelihood ratio of 0.21, N and NTD showed a positive likelihood ratio of 156.5 and negative likelihood ration of 0.22 (Table 1).
Association between the Concentrations of the IgG Antibodies against the Antigen Components of the MSD Assay and Their Combined Performance Characteristics
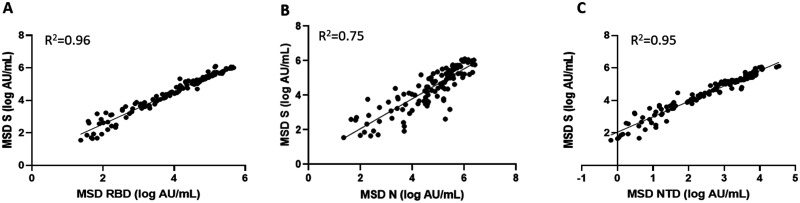
The concentrations of the antibodies against the different SARS-CoV-2 antigens of the MSD assay were compared against each other. All 4 antibodies to S, N, RBD, and NTD correlated significantly (P < 0.0001) (Fig. 2, A–C). The logarithmic concentrations of anti-S and anti-RBD (R2 = 0.96) as well as anti-S and anti-NTD (R2 = 0.95) showed a very strong association, and anti-S with anti-N (R2 = 0.75) showed a slightly lower correlation (Fig. 2, A–C). The combined performance characteristics of parallel assessment of antibodies to 2 antigens at a time was evaluated. Anti-S and anti-N results or anti-RBD and anti-N results combined increased the overall sensitivity of predicting the disease status to 87.1% and 83.9% in comparison to using the concentration of the antibodies alone (see Supplemental Table 1 in the online Data Supplement). This improvement in sensitivity was accompanied without much loss of specificity (98.0%) (Supplemental Table 1). However, combining anti-S and anti-RBD results did not show any improvement in sensitivity or specificity. Finally, parallel assessment of antibodies to more than 1 antigen at greater than 14 days improved the sensitivity to 100% regardless of which combinations were used.
Fig. 2.
Correlation between antibodies to individual antigens on the MSD multiplex platform. The log ratio of the arbitrary units (AU/mL) of the IgG concentrations to each antigen was plotted against each other and the correlation coefficients were calculated for each combination. (A) MSD S (log AU/mL) vs MSD RBD (log AU/mL). (B) MSD S (log AU/mL) vs MSD N (log AU/mL). (C) MSD S (log AU/mL) vs MSD NTD (log AU/mL).
Comparison of the IgG Antibody Concentrations against the Antigen Components of the MSD Assay with the Commercial SARS-CoV-2 Serology Assays
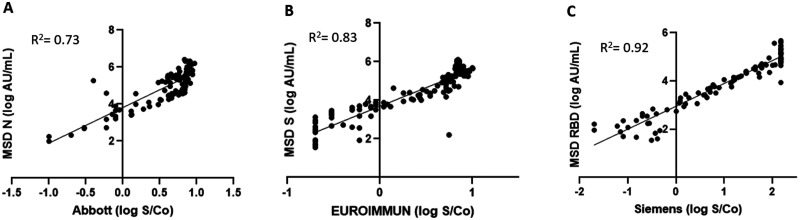
Each antigen component of the MSD assay was compared against the commercial serology assays, Abbott, EUROIMMUN, and Siemens, that are directed against the individual antigens, N, S, and RBD, respectively. MSD assay is designed for a quantitative measurement of antibodies; hence, we compared the logarithmic concentrations (AU/mL) of the IgG antibodies on the MSD platform with the logarithmic concentrations of the signal to noise ratios measured using the commercial assays. As shown in Fig. 3, A–C, linear regression analysis showed very good correlation between assays with an R2 = 0.73 for MSD N vs Abbott, R2 = 0.82 for MSD S vs EUROIMMUN, and R2 = 0.92 for MSD RBD vs Siemens.
Fig. 3.
Correlation between antibodies on the MSD multiplex platform to the commercial assay. The log ratio of the arbitrary units (AU/mL) of the IgG concentrations to each antigen was plotted against each other and the correlation coefficients were calculated for each combination. (A) MSD N (log AU/mL) vs Abbott (log S/Co). (B) MSD S (log AU/mL) vs EUROIMMUN (log S/Co). (C) MSD RBD (log AU/mL) vs Siemens (log S/Co).
When compared against each other qualitatively, the overall agreement for MSD N and Abbott was 97.5% with a positive percent agreement of 95.7% and a negative percent agreement of 98.2% (Table 3). The overall agreement for MSD S and MSD RBD in comparison to EUROIMMUN was 92.6% and 95.2% and the negative percent agreement was 93.5% and 97.2%, respectively (Table 3). The positive percent agreement was 90.4% for both comparisons with EUROIMMUN. For MSD S and MSD RBD, in comparison to Siemens, the overall agreement was 94.0% and 96.4%, respectively. For each comparison of MSD S and MSD RBD with Siemens, the positive percent agreement was 91.8% and 90.6%, respectively, and the negative percent agreement was 94.9% and 98.6%, respectively (Table 3).
Table 3.
Overall agreement between MSD and commerical serology assays.
| Assays a | Overall agreement % (95% CI) | Overall agreement % (95% CI) | Overall agreement % (95% CI) |
|---|---|---|---|
| MSD N vs Abbott (N) (n = 316) | 97.5 (97.1–97.9) | 95.7 (94.8–96.6) | 98.2 (97.8–98.6) |
| MSD S vs EUROIMMUN (S) (n = 310) | 92.6 (91.9–93.3) | 90.4 (89.0–91.8) | 93.5 (92.8–94.3) |
| MSD RBD vs EUROIMMUN (S) (n = 310) | 95.2 (94.6–95.7) | 90.4 (89.0–91.8) | 97.2 (96.7–97.7) |
| MSD S vs Siemens (RBD) (n = 301) | 94.0 (93.4–94.6) | 91.8 (90.4–93.1) | 94.9 (94.2–95.6) |
| MSD RBD vs Siemens (RBD) (n = 301) | 96.4 (95.9–96.8) | 90.6 (89.1–92.0) | 98.6 (98.3–99.0) |
IgG antibodies detected in the MSD multiplex assay were compared with the results obtained from the commercial serology assays directed against individual antigens.
The discrepant results largely represented samples that were collected <14 days after a positive PCR confirmation for COVID-19 diagnosis. Of note, several PCR-confirmed COVID-19 samples that tested negative for antibodies on the commercial IgG assays were captured by 1 or more antigens of the MSD assay in samples collected at >7 days post-PCR. Antibodies to at least 1 antigen of the MSD assay were detected in the PCR-confirmed COVID-19 samples, and none missed detection on the MSD platform in samples collected as early as 8 days or later after a positive PCR confirmation (Table 2). To dissect this further, sensitivities and specificities of the commercial serology assays were calculated and compared with the MSD platform in the same set of samples (Supplemental Table 2). Due to sample volume limitations, few samples were not tested on all 3 commercial assays. The overall sensitivity of Abbott [76.9% (95% CI 75.0%–78.9%)], EUROIMMUN [71.1% (95% CI 68.9%–73.2%)], and Siemens [75.0% (95% CI, 72.9%–77.1%)] was less than the overall sensitivity of antibodies to MSD antigens individually (≥78.2%) or combined (≥83.9%) (Supplemental Tables 1 and 2). At >14 days post-PCR confirmation, except the antibodies to the N antigen [94.1% (95% CI, 92.6–95.6)], all other components of the MSD platform displayed higher sensitivities (≥98.0%) than Abbott, EUROIMMUN, or Siemens assay (Table 1; also see Supplemental Table 2). The antibodies to the individual antigens of the MSD platform also displayed higher specificity [99.5% (95% CI, 99.3%–99.7%)] than all 3 commercial assays at the manufacturer-established cut points, and the combined specificity of antibodies to 2 antigens on the MSD platform was still higher [98.0% (95% CI 99.3%–99.7%)] than the specificity of EUROIMMUN [94.0% (95% CI 93.2–94.7)] and Siemens [96.5% (95% CI 95.9%–97.0%)] IgG assays (Table 1; also see Supplemental Table 2).
Table 2.
Broader coverage of antibody responses to SARS-CoV-2 antigens in individual patients using MSD multiplex platform.a
| Patient ID | Days post-PCR | Abbott (≥1.4 S/Co b ) | EUROIMMUN (≥1.1 S/Co) | Siemens (≥1.0 S/Co) | MSD S (>869 AU/mL) | MSD N (>7225 AU/mL) | MSD RBD (>592 AU/mL) |
|---|---|---|---|---|---|---|---|
| 7–14 days post-PCR | |||||||
| 632 | 7 | POSc | NEGd | NEG | POS | POS | POS |
| 336 | 8 | NEG | NEG | NEG | POS | POS | NEG |
| 322 | 10 | POS | NEG | POS | POS | POS | POS |
| 619 | 11 | POS | NEG | NEG | POS | POS | NEG |
| 929 | 14 | POS | NEG | NTe | POS | POS | POS |
| >14 days post-PCR | |||||||
| 941 | 19 | POS | NEG | POS | POS | POS | NEG |
| 945 | 22 | NEG | INDf | POS | POS | NEG | POS |
| 622 | 26 | POS | IND | POS | POS | POS | POS |
| 929 | 30 | NEG | POS | POS | POS | NEG | POS |
Patients with negative results from each commercial IgG assays were compared qualitatively to antibodies detected on MSD platform individually and combined.
Signal/calibrator.
Positive.
Negative.
Not tested.
Indeterminate.
Discussion
Herein, we evaluated a novel multiplex assay, which offers simultaneous and quantitative assessment of patient sera for antibodies to multiple SARS-CoV-2 antigens. We focused on the evaluation of IgG antibodies to the 4 major SARS-CoV-2 antigen targets, full-length S, RBD, N, and NTD based on their individual immunogenicity potential (12). A previous study had also evaluated the MSD multiplex platform for the IgG detection across these 4 SARS-CoV-2 antigens (13). In that study, controls and calibrators were established by the investigators and their work predated the actual production and supply of reagents by the manufacturer. Thus, the assigned values for the arbitrary units (AU/mL) of the calibrators and controls were largely different from the values outlined in the current package insert of the assay (11). Their data showed good sensitivity for IgG detection over 14 days since onset of symptoms for 3 SARS-CoV-2 antigens (S, RBD, and N). Here, we evaluated for the first time the performance characteristics of the currently available multiplex MSD IgG assay. Our data demonstrated a higher sensitivity, specificity, and performance characteristics across all IgG antibodies (13). In addition, we also compared the diagnostic characteristics of the IgG antibodies to all SARS-CoV-2 antigens individually and combined with 3 commercially available IgG serology assays (Abbott, EUROIMMUN, and Siemens) that are directed against single antigen targets. Our in-depth analysis demonstrated excellent performance characteristics in par with the commercial serology assays. The benefits of the MSD multiplex assay include smaller sample volume (10 μL), a relatively shorter assay time (4 h), high-throughput capabilities, and, most important, the ability to evaluate multiple viral targets, which thereby could serve as a quality control for the SARS-CoV-2 immunity.
Although serology assays are not recommended for diagnostic purposes, there is a continuous demand for accurate and robust serology tests in both the pre- and postvaccination phases for number of reasons (14–17). It is currently used to monitor natural- and vaccine-acquired humoral immunity in desired situations at the clinician’s discretion (18–20). The field is rapidly evolving to develop models to correlate the convalescent and vaccination antibody titers for the prediction of an individual’s immunity course against the SARS-CoV-2 virus (21–24). In that light, there is an anticipation for increased utility for identifying individuals susceptible to infection or reinfection. The S protein is the focus for all the 3 COVID-19 vaccines (from Johnson and Johnson, Pfizer/BioNTech, and Moderna) currently in use in the Unites States (25). However, some studies suggest the inclusion of N protein antibodies in addition to S antibodies as COVID-19 vaccine targets due to its higher homology between coronaviruses and its lower mutation rates (26, 27). While the existing commercial serology assays that are directed against 1 antigen and that are qualitative or semiquantitative in nature can be beneficial, a quantitative multiplex assay can be of a much greater value in all the previously outlined premises. Depending on the vaccine targets employed, the simultaneous detection of antibodies can help in the accurate identification of an individual’s antibody signature as well as aid in distinguishing natural from vaccine-mediated acquired immunity. The quantitative nature of the multiplex assay can be an additional plus in aiding with the clinical management in the context of resistance to infection and in the accurate identification of convalescent plasma donors by accounting for the polyclonal antibody responses in the individual donors.
Our data showed all 4 antigenic targets included in the MSD assay exhibit a high-level accuracy for the detection of the IgG responses against the antigens. The sensitivity and specificity of currently available serology assays that has been authorized by the Food and Drug Administration ranges from 90% to 100% (28). These performance characteristics while reasonable can still result in a number false-positive and false-negative results. The CDC suggests the use of tests with a specificity ≥99.5% to minimize the potential for false-positive results (29). Even with a specificity (≥99.5%) optimized cutoff, MSD assay still displayed a high sensitivity of 94% to 100% in samples collected >14 days post positive PCR confirmation across all 4 major antigens. Recently, MSD had released a set of recommended cut points for antibodies to different antigens detected in this assay, and the sensitivity and specificity using MSD-recommended and our derived cut points were compared. Notably, the comparisons using both sets of cut points resulted in no major differences in the performance characteristics of the assay (Supplemental Table 3).
Parallel assessment of antibodies to 2 antigens at timepoints >14 days after a positive PCR confirmation increased the sensitivity of the assay to 100% while still retaining a 98% specificity. MSD S assay individually or combined with N antigen displayed sensitivity that was higher than all 3 commercial assays in samples collected >14 days post positive PCR confirmation. Similarly, MSD N individually displayed comparable sensitivity to Abbott but in combination with RBD displayed higher sensitivity than all 3 commercial assays. Noticeably, none of the COVID-19 positive samples that were collected >7 days after a PCR confirmation, including the ones that tested negative on the commercial platforms, missed detection on the MSD platform. At least 1 or more antibodies were positive by MSD assay in these early PCR-positive samples that were classified seronegative by 1 or more commercial IgG assays. Additionally, several studies have reported the nonhomogenous nature of the antibody responses (30, 31). Assays directed against individual targets can fail to capture the full spectrum of an individual’s immunoreactivity. Several patients included in our analysis demonstrated this pattern; for example, patient 619 (Table 2) was negative for anti-RBD IgG by both MSD and Siemens assay but was positive for anti-N IgG by both MSD and Abbott assays. Similarly, patient 945 was negative for anti-N IgG by both MSD and Abbott but was positive for anti-RBD IgG by MSD and Siemens assays. Patient 629 was negative for anti-N IgG by both MSD and Abbott but was positive for anti-S IgG by MSD and EUROIMMUN assays and positive for anti-RBD IgG by MSD and Siemens assays. Besides the increased sensitivity of the MSD platform, the ability to simultaneously detect multiple antigens at a given time provides broader coverage of a patient’s antibody signature, which could aid in accurate clinical management.
Our study is limited due to the lack clinical data, including symptoms and severity. Future studies on well-characterized patient cohorts using the MSD multiplex technology will be extremely beneficial to evaluate the different antibody signatures to SARS-CoV-2 antigens in correlation to disease severity, vaccine responses, viral neutralization titers, and responses to therapies. Our study cohort included samples that were collected before detection of the SARS-CoV-2 variants of concern (32–34). The propensity for the SARS-CoV-2 proteins, such as SARS-CoV-2 S and RBD, that are more susceptible to mutations may continue to pose increasing challenges for its use as targets in serological assays (16). Developing new serology assays to keep up with the emerging variants is less ideal, and a more practical approach will be to develop assays that provides a broader coverage encompassing viral regions that are less prone to mutations. SARS-CoV-2 N gene has been described to be conserved and stable with fewer mutations over time (35). A recent study had confirmed that Abbott COVID-19 molecular antigen and serological assays effectively detect SARS-CoV-2 B.1.1.7, B.1.351, and P.1 variant infections and B.1.1.7 antibodies (36). Further studies will be required to evaluate the ability of the MSD platform to detect IgG antibodies targeted to the variant-specific antigenic regions. Nevertheless, the simultaneous detection of 4 different antigens may provide a broader coverage of the mutated and native epitopes, increasing the frequency of detection of the SARS-CoV-2 variant strains.
In conclusion, the MSD multiplex assay demonstrated excellent performance characteristics for the simultaneous detection of IgG antibodies against all 4 SARS-CoV-2 antigens with a higher sensitivity and a comparable specificity to the existing commercial IgG serology assays. In addition, the simultaneous detection of all 4 major IgG antibody responses to SARS-CoV-2 antigens provides a comprehensive signature of an individual’s antibody responses, which may aid in the accurate management of clinical decisions.
Supplemental Material
Supplemental material is available at The Journal of Applied Laboratory Medicine online.
Supplementary Material
Nonstandard Abbreviations: SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; S, spike; N, nucleocapsid; RBD, receptor-binding domain; NTD, N-terminal domain; AUC, area under the ROC curve; MSD, Meso Scale Discovery; FDA, Food and Drug Administration.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest: Employment or Leadership: None declared. Consultant or Advisory Role: None declared. Stock Ownership: None declared. Honoraria: P. Slev, Grand Rounds-MD Anderson Cancer Center: SARS-CoV-2 serology testing. Research Funding: Meso Scale Discovery (Piscataway, NJ) for the donation of the COVID-19 Coronavirus 2 Panel 2 kits. Expert Testimony: None declared. Patents: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments: The study team would like to thank Meso Scale Discovery (Piscataway, NJ) for the donation of the COVID-19 Coronavirus 2 Panel 2 kits that allowed us to complete this work. We would also like to thank Hailey Wells at the ARUP Laboratories (SLC, Utah) for her help with collecting and aliquoting samples.
REFERENCES
- 1.Lu H, Stratton CW, Tang YW.. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol 2020;92:401–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . WHO coronavirus (COVID-19) dashboard with vaccination data. https://covid19.who.int/ (Accessed June 2021).
- 3.Peeling RW, Wedderburn CJ, Garcia PJ, Boeras D, Fongwen N, Nkengasong J, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis 2020;20:e245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hossain A, Nasrullah SM, Tasnim Z, Hasan MK, Hasan MM.. Seroprevalence of SARS-CoV-2 IgG antibodies among health care workers prior to vaccine administration in Europe, the USA and East Asia: a systematic review and meta-analysis. EClinicalMedicine 2021;33:100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Yang C, Xu XF, Xu W, Liu SW.. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 2020;41:1141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assis R, Jain A, Nakajima R, Jasinskas A, Kahn S, Palma A, et al. Distinct SARS-CoV-2 antibody reactivity patterns elicited by natural infection and mRNA vaccination. NPJ Vaccines 2021;4;6(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vashist SK. 2020. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics 2020;10:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jääskeläinen AJ, Kuivanen S, Kekäläinen E, Ahava MJ, Loginov R, Kallio-Kokko H, et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Elslande J, Decru B, Jonckheere S, Van Wijngaerden E, Houben E, Vandecandelaere P, et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect 2020;26:1557.e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gundlapalli AV, Salerno RM, Brooks JT, Averhoff F, Petersen LR, McDonald LC, Iademarco MF.. SARS-CoV-2 serologic assay needs for the next phase of the US COVID-19 pandemic response. Open Forum Infect Dis 2021;8:ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meso Scale Discovery. MSD MULTI-SPOT Assay System: V-PLEX COVID-19 Coronavirus Panel 2 (IgG) Kit. https://www.mesoscale.com/products/covid-19-coronavirus-panel-2-igg-k15369u/ (Accessed September 2020).
- 12.Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J 2020;39:198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson M, Wagstaffe HR, Gilmour KC, Mai AL, Lewis J, Hunt A, et al. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J Clin Virol 2020;130:104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney N, Merrick B, Pedro Galão R, Pickering S, Botgros A, Wilson HD, et al. Clinical utility of targeted SARS-CoV-2 serology testing to aid the diagnosis and management of suspected missed, late or post-COVID-19 infection syndromes: results from a pilot service implemented during the first pandemic wave. PLoS One 2021;16:e0249791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter AK, Hegde ST.. The important role of serology for COVID-19 control. Lancet Infect Dis 2020;20:758–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galipeau Y, Greig M, Liu G, Driedger M, Langlois MA.. Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol 2020;11:610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danese E, Montagnana M, Salvagno GL, Peserico D, Pighi L, De Nitto S, et al. Comprehensive assessment of humoral response after Pfizer BNT162b2 mRNA Covid-19 vaccination: a three-case series. Clin Chem Lab Med 2021;59:1585–91. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat Commun 2021;12:1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damluji AA, Christenson RH, deFilippi C.. Clinical application of serologic testing for coronavirus disease 2019 in contemporary cardiovascular practice. J Am Heart Assoc 2021;10:e019506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sewell HF, Agius RM, Kendrick D, Stewart M.. Covid-19 vaccines: delivering protective immunity. BMJ 2020;371:m4838. [DOI] [PubMed] [Google Scholar]
- 21.Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ 2020;369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly protective of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021;27:1205–11. [DOI] [PubMed] [Google Scholar]
- 23. Bubar KM, Reinholt K, Kissler SM, Lipsitch M, Cobey S, Grad YH, Larremore DB.. Model-informed COVID-19 vaccine prioritization strategies by age and serostatus. Science 2021;371:916–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Wu B, Topcu U.. Control strategies for COVID-19 epidemic with vaccination, shield immunity and quarantine: a metric temporal logic approach. PLoS One 2021;16:e0247660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai L, Gao GF.. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021;21:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahlén G, Frelin L, Nikouyan N, Weber F, Höglund U, Larsson O, et al. The SARS-CoV-2 N protein is a good component in a vaccine. J Virol 2020;94:e01279–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batra M, Tian R, Zhang C, Clarence E, Sacher CS, Miranda JN, et al. Role of IgG against N-protein of SARS-CoV2 in COVID19 clinical outcomes. Sci Rep 2021;11:3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Food and Drug Administration. EUA authorized serology test performance. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance (Accessed September 2020).
- 29.CDC. Interim guidance for antigen testing for SARS-CoV-2. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (Accessed September 2020).
- 30.Weisberg SP, Connors TJ, Zhu Y, Baldwin MR, Lin WH, Wontakal S, et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol 2021;22:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y, et al. ; MGH COVID-19 Collection & Processing Team. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020;370:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Food and Drug Administration. SARS-CoV-2 viral mutations: impact on COVID-19 tests. 2021. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (Accessed April 2021).
- 33.Diamond M, Chen R, Xie X, Case J, Zhang X, VanBlargan L, et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Preprint at https://pubmed.ncbi.nlm.nih.gov/33594356/ (2021). [DOI] [PMC free article] [PubMed]
- 34.Jungnick S, Hobmaier B, Mautner L, Hoyos M, Haase M, Baiker A, et al. Detection of the new SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in five SARS-CoV-2 rapid antigen tests (RATs), Germany, March 2021. Euro Surveill 2021;26:2100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutta NK, Mazumdar K, Gordy JT.. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol 2020;94:e00647–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodgers MA, Batra R, Snell LB, Daghfal D, Roth R, Huang S, et al. Detection of SARS-CoV-2 variants by Abbott molecular, antigen, and serological tests. Preprint at https://medrxiv.org/content/10.1101/2021.04.24.21256045v1 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.